Introduction
Ovarian cancer (OC) was the fifth most prevalent
malignancy in females, and had the highest mortality rate among all
types of gynecological cancer in the United States in 2015
(1). Conventional treatment
strategies for OC include cytoreduc-tive surgery accompanied by
intermittent administration of chemotherapeutic agents, such as
platinum or taxane (TAX), at the maximum tolerated doses (2). Docetaxel (DTX) is a TAX which is a
type of FDA-approved drug for use in OC, and is frequently used in
a clinical setting (3). Despite
significant research investments and efforts, the 5-year overall
survival rate for patients with OC has only increased slightly
since 1995 (4). Chemoresistance
in OC is potentially caused by several mechanisms in a
heterogeneous tumor cell population (5). Mesenchymal stem cells (MSCs) are a
group of multipotent stromal cells which reside in several areas,
such as the bone marrow, fat or the dental pulp (6). MSCs have been demonstrated to
function in the tumor microenvironment, and are involved in tumor
progression, metastasis and chemotherapeutic resistance (7). The human umbilical cord is a
suitable source of MSCs, and unlike bone marrow-derived MSCs, human
umbilical cord MSCs (hUCMSCs) are collected in a painless manner
and exhibit faster self-renewal properties (8). MSCs potentially promote drug
resistance either through direct contact with the tumor cells or
via systemic mechanisms with the involvement of secreted factors
(9). Exosomes, a subgroup of
extracellular vesicles, may be released by virtually all mammalian
cell types, and transfer a variety of molecules, including mRNAs
and microRNAs (miRNAs/miRs), as well as long non-coding RNAs, any
of which may confer resistance to drug-sensitive cancer cells
(10).
miR-146a is an established anti-inflammatory miRNA
and has been implicated as a therapeutic target for several
inflammation-associated disorders, including peripartum
cardiomyopathy (11) and obesity
(12). Furthermore,
hUCMSCs-derived exosomes have been reported to exert
anti-inflammatory effects on human trophoblast cells by
transferring miR-146a-5p (13).
miR-146a has been demonstrated to serve a significant role in
exosomes secreted by colorectal cancer stem cells, where exosomal
miR-146a promotes stem-like properties and tumorigenicity (14). In addition, the oncogenic role of
laminin γ2 (LAMC2) in OC has been reported previously, and it
increased p38 expression via miR-125a-5p (15). Therefore, it was hypothesized that
exosomal miR-146a derived from hUCMSCs may affect chemoresistance
to TAX and DTX by regulating LAMC2 in OC. The aim of the present
study was to determine the association between hUCMSCs-derived
exosomal miR-146a and LAMC2 in OC cells, and to examine the
relevance of exosomal miR-146a/LAMC2 in TAX- and DTX-resistant
OC.
Materials and methods
Reagents, antibodies and lentiviral
vectors
The following antibodies were used to identify
hUCMSC surface markers: FITC-conjugated anti-CD90 (cat. no. 13801),
anti-CD14 (cat. no. 29943), anti-CD73 (cat. no. 13160) and
anti-CD105 (cat. no. 14606), and phycoerythrin (PE)-conjugated
anti-CD34 (cat. no. 79253), anti-CD29 (cat. no. 34971), anti-CD44
(cat. no. 38200), anti-CD45-PC7 (cat. no. 28418) and anti-human
leukocyte antigen DR-1 (HLA-DR; cat. no. 17634) (all Cell Signaling
Technology, Inc.). The hUCMSC osteogenic differentiation kit
OriCell™ (OriCell™; cat. no. HUXUC-90021) and adipogenic
differentiation kit (cat. no. GUXMX-90031) were obtained from
Cyagen Biosciences, Inc. ELISA kits for the detection of PI3K (cat.
no. ab207485) and Akt (cat. no. ab176657) were purchased from
Abcam. miR-146a inhibitor and inhibitor control were purchased from
Shanghai GenePharma Co., Ltd. LAMC2 lentiviral overexpression
vector (cat. no. RC222076L4) and empty vector control were
purchased from OriGene Technologies, Inc. The specific sequences
were: miR-146a inhibitor, 5'-CCC AUG GAA UUC AGU UCU CA-3'; and
inhibitor control, 5'-CAG AGG UCA GGC CUU GCA AA-3'.
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis
(http://gepia.cancer-pku.cn/) was used to
analyze the expression levels of LAMC2 in tumor tissues in The
Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) database (https://cancergenome.nih.gov) and normal ovarian
tissues in the GTEX (https://www.gtexportal.org/) database. Significant
differences were determined by Limma analysis (version 3.11;
https://bioconductor.org/packages/limma/).
Cell culture and identification of
hUCMSCs
hUCMSCs (SC2020011402) were purchased from
FenghuiShengwu and cultured at 37°C with 5% CO2 in DMEM
(cat. no. 11965-084; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (cat. no. 10099141; Gibco; Thermo Fisher
Scientific, Inc.), 100 g/l streptomycin and 100 U/ml penicillin
(Sangon Biotech Co., Ltd.). Subsequently, cells were blocked with
5% bovine serum albumin (cat. no. E661003; Sangon Biotech Co.,
Ltd.) at 37°C for 40 min and incubated with a series of mouse
anti-human FITC-conjugated anti-CD90, anti-CD14, anti-CD73,
anti-CD105 and anti-CD29, and PE-conjugated anti-CD34, anti-CD44
and anti-HLA-DR antibodies (all dilutions, 1:50) at 37°C for 2 h. A
flow cytometer (BD FACSCanto II; BD Biosciences) was utilized for
analysis using FlowJo v10.0 software (BD Biosciences). Following
culturing with culture media from the hUCMSC osteogenic and
adipo-genic differentiation kits, hUCMSCs were stained with 0.1%
alizarin red stain and 0.5% oil red O stain at 37°C for 2 h.
Extraction and identification of
exosomes
The extraction and identification of hUCMSCs-derived
exosomes was performed as described in Data S1. The isolated exosomes were
labeled using the PKH26 red fluorescent cell labelling kit
(Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocol. Subsequently, unlabeled or PKH26 red fluorescence-labeled
exosomes were added to A2780 and SKOV3 cell lines and incubated at
37°C with 5% CO2 for 24 h. The cells were then fixed at
room temperature with 4% paraformaldehyde for 30 min and stained
with 4',6-diamidino-2-phenylindole at room temperature for 5 min.
The phagocytose of exosomes from SKOV3 and A2780 cell lines was
observed using a confocal microscope (Zeiss AG).
Western blotting
The exosome marker proteins CD9 (1:1,000; cat. no.
ab92726), CD81 (1:1,000; cat. no. ab79559), CD63 (1:1,000; cat. no.
ab217345), heat shock protein 70 (HSP70; 1:1,000; cat. no. ab2787)
and GAPDH (1:100; cat. no. ab8245) were detected by western
blotting as previously described (16). All antibodies were from purchased
from Abcam. Briefly, radioimmunoprecipitation assay lysis buffer
containing phenylmethanesulfonyl fluoride (Sangon Biotech Co.,
Ltd.) was used to extract exosomes. Pierce™ 660 nm protein assay
reagent (Thermo Fisher Scientific, Inc.) was applied to determine
the protein concentration. The same amount of protein was isolated
using 10% Gradi-Gel™ II gradient gel electrophoresis (ElpisBiotech,
Inc.) from exosomes (20 µg) and cell lysate (60 µg)
and transferred to a polyvinylidene fluoride membrane (EMD
Millipore). The membrane was subsequently blocked at room
temperature for 1 h in tris buffer saline-0.1% tween 20 containing
5% bovine serum albumin and incubated overnight at 4°C with the
primary antibodies (1:2,000). The goat anti-mouse secondary
antibody against IgG (cat. no. sc-2005; 1:5,000; Santa Cruz
Biotechnology, Inc.) conjugated to horseradish peroxidase was added
for a 1-h incubation at room temperature. Protein blots were
visual-ized using an enhanced chemiluminescence kit (Santa Cruz
Biotechnology, Inc.) and analyzed using Glyko BandScan 5.0 software
(Agilent Technologies, Inc.).
Construction of OC cell lines resistant
to chemotherapeutics
The establishment of OC resistant cell lines
SKOV3/DTX and A2780/TAX is described in Data S1.
Cell treatment
Parental and resistant A2780 and SKOV3 cells in the
logarithmic growth phase were selected for transfection using 100
ng miR-146a inhibitor or corresponding inhibitor control with
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Reverse
transcription-quantitative (RT-q)PCR was used to verify the success
of transfection at 48 h post-trans-fection. Subsequently, cells
were treated using 20 µg/ml hUCMSC-derived exosomes. Cells
were collected after 24 h for subsequent experiments.
Cell survival assay
SKOV3 and A2780 cells were plated in 96-well plates
at a density of 8x103 cells/well. After 4 h, the cells
were treated with either DTX (serial dilution, 0-5 µM;
Sigma-Aldrich; Merck KGaA) or TAX (serial dilution, 0-10 µM;
Sigma-Aldrich; Merck KGaA) for 72 h at 37°C. A Cell Counting Kit-8
(CCK-8; Roche Diagnostics) assay was used to measure cell survival
according to the manufacturer's protocol.
CCK-8 assay
Cells (5x103 cells/well) were treated
with 10 µl CCK-8 (Roche Diagnostics) for 2 h at 37°C
followed by cell proliferation assays. The proliferation of cells
was evaluated after 0, 24, 48 or 72 h of incubation at 37°C
according to the manufacturer's protocol.
5-Ethynyl-2'-deoxyuridine (EdU)
labeling
EdU analysis was performed according to the
instructions of the Cell-Light™ EdU apollo643 in vitro kit
(cat. no. C10310-1; Guangzhou RiboBio Co., Ltd.). After fixation
with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) for 30 min at
37°C, the cells were treated with Apollo reaction mixture for 30
min at 37°C and stained with 4',6-diamidino-2-phenylindole at 37°C
for 2 h for DNA staining. A2780 and SKOV3 cell proliferation was
analyzed using randomly selected images obtained under a
fluorescence microscope and was expressed as the ratio of
EdU+ cells to all cells.
Apoptosis analysis
Flow cytometry was used to detect apoptosis using a
FITC-Annexin V Apoptosis Detection kit (BD Biosciences) according
to the manufacturer's protocol. Apoptotic cells were loaded onto a
BD FACSCanto II flow cytometer (BD Biosciences) and evaluated using
FlowJo v10.0 software (BD Biosciences). In addition, SKOV3,
SKOV3/DTX, A2780 and A2780/TAX cells (1x106 cells/ml)
were stained with 1X Hoechst 33258 staining solution at room
temperature for 3-5 min. Apoptotic cells were observed using a
Hoechst 33258 staining kit (Thermo Fisher Scientific, Inc.) after a
48-h exosome treatment at 37°C.
Oligonucleotide microarray
Three groups of PBS-treated and MSC-derived
exosome-treated SKOV3/DTX cells were collected and total RNA was
extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.).
Subsequently, 0.5 µg RNA was used for hybridization using
the Human miRNA Expression Array V4.0 (Arraystar, Inc.). The
extracted and isolated miRNAs were subsequently washed and scanned
using the GeneChip™ Scanner3000 7G system (Thermo Fisher
Scientific, Inc.).
RT-qPCR
Total RNA was extracted from cells using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was
synthesized using the Transcriptor First Strand cDNA Synthesis kit
(Toyobo Life Science) at 42°C for 50 min and at 72°C for 10 min. A
total of 2 µg cDNA was used as a template for qPCR to detect
the target genes using SYBR Green Real-Time PCR master mix (Thermo
Fisher Scientific, Inc.). The thermocycling conditions were: 95°C
for 30 sec, followed by 40 (two-step) cycles of 95°C for 5 sec and
60°C for 35 sec, and a final extension at 72°C for 15 min. mRNA
expression levels were measured using the 2-ΔΔCq method
(17) after normalization to the
expression levels of U6 or GAPDH. The primer sequences used were:
miR-146a forward, 5'-TGA GAA CTG AAT TCC ATG GGT-3' and reverse,
5'-TAT GGC ACT GGT AGA ATT CAC T-3'; U6 forward, 5'-GAC CTC TAT GCC
AAC ACA GT-3' and reverse, 5'-AGT ACT TGC GCT CAG GA G GA-3'; LAMC2
forward, 5'-TAC CAG AGC CAA GAA CG C TG-3' and reverse, 5'-CGC AGT
TGG CTG TTG ATC TG-3'; and GAPDH forward, 5'-CCA CTA GGC GCT CAC
TGT TCT C-3' and reverse, 5'-CAT GGT GGT GAA GAC GCC AG-3'.
Luciferase gene reporter assay
Initially, the targeting mRNAs of miR-146a were
screened using StarBase (http://starbase.sysu.edu.cn/) and miRsearch
(https://www.exiqon.com/ls/Pages/). The
complementary sequence for miR-146a was synthesized and inserted
into a psiCHECK2 vector (Promega Corporation) downstream of the
Renilla reporter gene to generate psiCHECK2-miR-146a. The
LAMC2 3' untranslated region (3'UTR) containing the putative
miR-146a binding sites was cloned into the same vector to create
psiCHECK2-LAMC2. The psiCHECK2 vectors contained a second reporter
gene (firefly luciferase) designed for end point lysis assays. The
reporter plasmid (100 ng) was transfected into cells using
Lipofectamine LTX (Thermo Fisher Scientific, Inc.). Luciferase
activity was measured after 48 h using the dual-luciferase reporter
assay (Promega Corporation). Values were normalized to firefly
luciferase activity.
ELISA
Specific ELISA kits were used to determine the
protein expression levels of PI3K and Akt in cell lysates according
to the manufacturer's protocol.
Statistical analysis
SPSS version 21.0 (IBM Corp.) software was used for
statistical analysis. Data are presented as the mean ± standard
deviation. Each assay was repeated at least three times, and the
comparisons between two groups were performed using an unpaired
t-test. When comparing one factor among multiple groups, a one-way
ANOVA was utilized, and when comparing two factors among multiple
groups, two-way ANOVA was applied, both were followed by Tukey's
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Isolation and characterization of hUCMSCs
and extracted exosomes
Flow cytometry was used to detect the expression
levels of surface markers of hUCMSCs. The presence of CD29, CD44,
CD73, CD90 and CD105 was confirmed; while the cells were negative
for CD14, CD34, HLA-DR and CD45 (Fig.
1A). The cell surface marker proteins expressed by the purified
hUCMSCs met the current criteria for the definition of MSCs
according to the Minimal Criteria for Defining Multipotent MSCs
(18). In addition, the
adipogenic and osteo-genic differentiation abilities of hUCMSCs
were assessed by Oil-red O staining and alizarin-red staining,
respectively (Fig. 1B-D).
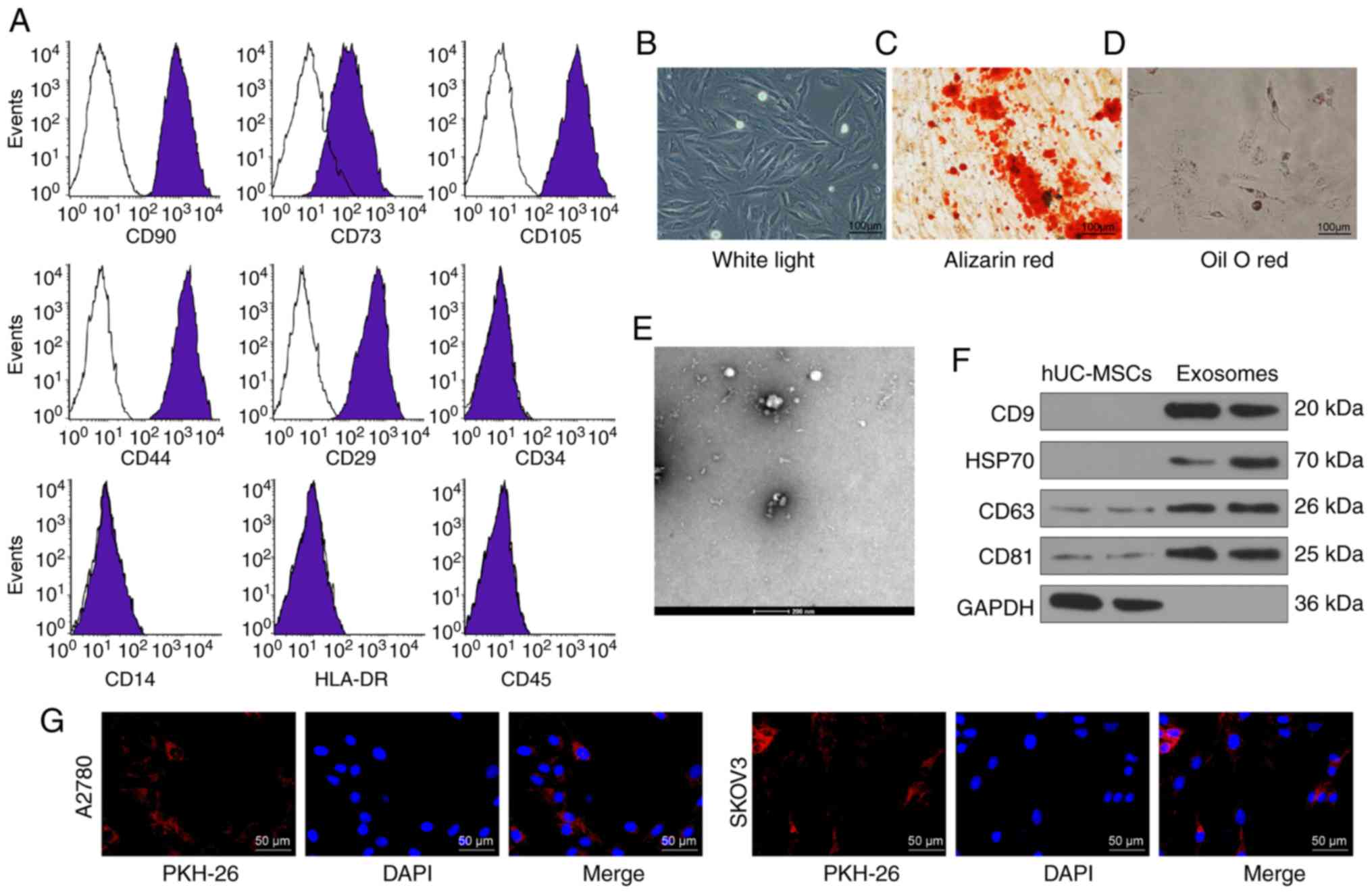 | Figure 1Identification of hUCMSCs and their
derived exosomes. (A) Expression of MSCs surface markers, including
CD29, CD44, CD73, CD90, CD105, CD14, CD34, CD45 and HLA-DR,
analyzed by flow cytometry. (B) hUCMSC morphology at passage 3
observed under a light microscope. Scale bar, 100 µm. (C)
Representative images of osteogenic differentiation of hUCMSCs
using Alizarin Red staining. Scale bar, 100 µm. (D)
Representative images of adipogenic differentiation of hUCMSCs
using Oil red O staining. Scale bar, 100 µm. (E)
Morphological analysis of hUCMSC by transmission electronic
microscopy. Scale bar, 200 nm. (F) Detection of exosomal marker
expression in hUCMSC-released exosomes by western blotting. (G)
Internalization of PKH26-labeled exosomes by SKOV3 and A2780 cells
was observed under a fluorescence microscope. Scale bar, 50
µm. hUCMSCs, human umbilical cord mesenchymal stem
cells. |
The exosomes exhibited an elliptical or cup-shaped
morphology under an electron transmission microscope, with a
particle size of ~100 nm (Fig.
1E). Western blotting illustrated that the expression levels of
CD9, CD63, CD81 and HSP70 were upregulated in exosomes compared
with cell lysates, and only a small quantity of GAPDH was expressed
(Fig. 1F). Furthermore, the
protein concentration of the extracted exosomes was 332.81
µg/ml. According to the definition of Exo by Minimal
Information for Studies of Extracellular Vesicles 2018 (19), the extracted exosomes were
considered to be extracellular vesicles. Subsequently, the exosomes
were diluted to 20 µg/ml for subsequent use. After
co-culturing SKOV3 and A2780 cells for 24 h with labeled exosomes,
a fluorescence microscope was used to observe the cells and it was
shown that the cells had a large quantity of red fluorescence,
which was primarily distributed in the cytoplasm. The results
revealed that the exosomes were internalized by SKOV3 and A2780
cells (Fig. 1G).
hUCMSC-derived exosomes increase drug
sensitivity in OC cells
First, the successful induction of the SKOV3/DTX and
A2780/TAX cell lines was assessed using a CCK-8 assay (P<0.05;
Fig. 2A). Subsequently, the
extracted hUCMSC-derived exosomes were co-cultured with the
parental SKOV3 or A2780 cells and the corresponding resistant cell
lines at a dose of 20 µg/ml for 24 h, and then cultured with
2 µM DTX or 5 µM TAX for 2 h, followed by CCK-8, EdU
staining and apoptosis detection experiments. The treatment of
hUCMSC-derived exosomes reduced the resistance of OC parental cells
to chemotherapy and the resistance of drug-resistant cells
(P<0.05; Fig. 2B-E). In
general, hUCMSC-derived exosomes suppressed resistance to
chemotherapy.
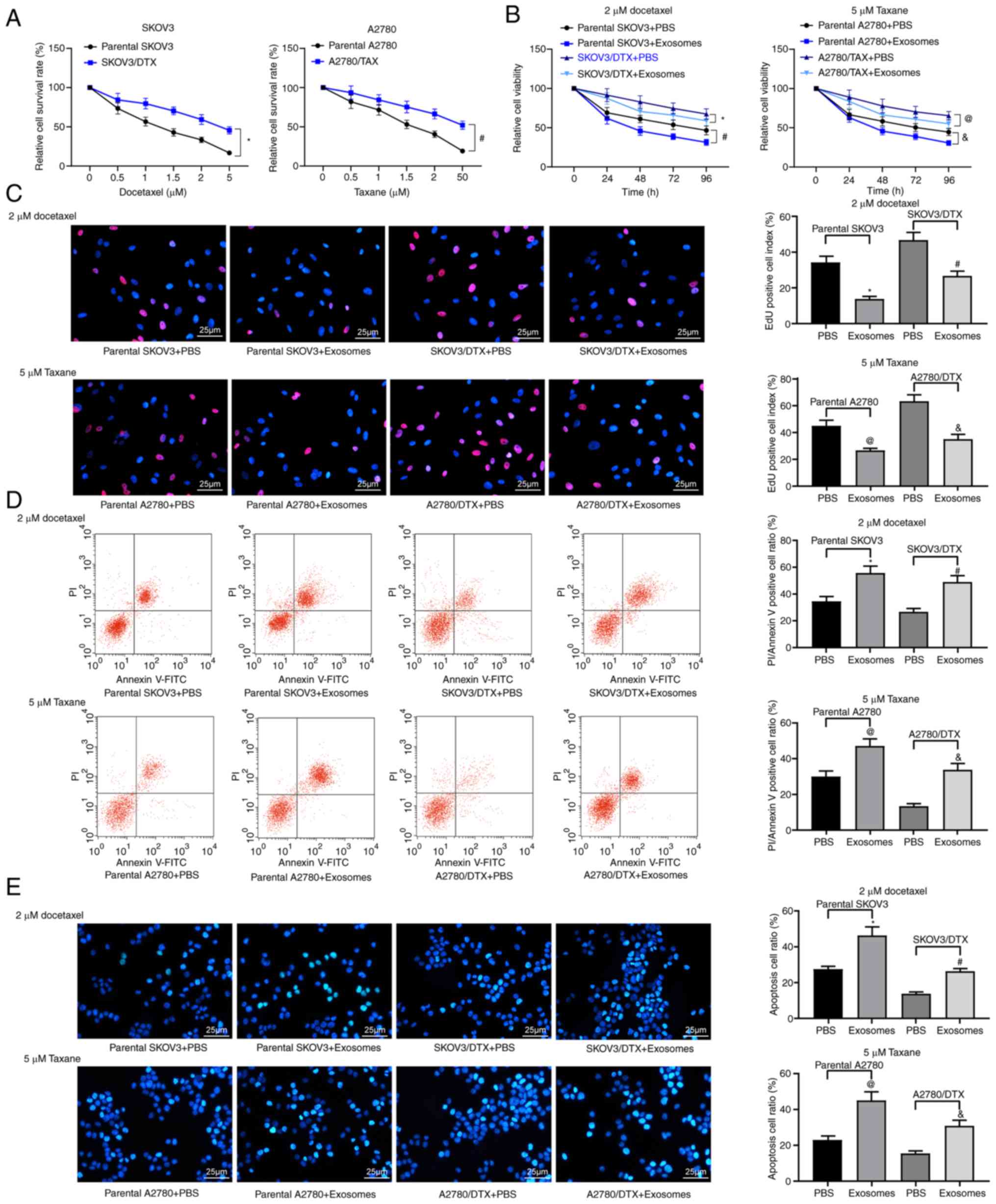 | Figure 2hUCMSC-derived exosomes inhibit OC
cell drug resistance. SKOV3/DTX and A2780/TAX cells were induced by
the exposure to gradient concentrations of drugs. (A) Cell survival
rate examined by CCK-8 cell survival assays. Subsequently, the
parental or drug-resistant OC cells were treated with 20
µg/ml MSCs-derived exosomes or PBS as a negative control.
Exosome-treated parental or drug-resistant OC cells were exposed to
2 µM DTX or 5 µM TAX for 2 h, respectively.
*P<0.05 vs. parental SKOV3 cells,
#P<0.05 vs. parental A2780 cells. (B) Cell viability
evaluated by CCK-8 cell viability assays. (C) EdU staining for cell
proliferation. Scale bar, 25 µm. (D) Flow cytometry analysis
for cells labelled with PI/Annexin V. (E) Hoechst 33258 staining
for cell apoptosis. Scale bar, 25 µm. Data are presented as
the mean ± SD. One-way (panels C, D and E) or two-way (panel A)
ANOVA followed by Tukey's multiple comparisons test was used to
determine statistical significance. Each assessment was performed
in triplicate with three repetitions to ensure minimum deviation.
In panels B, C, D and E: *P<0.05 vs. parental SKOV3
cells treated with PBS; #P<0.05 vs. SKOV3/DTX cells
treated with PBS; @P<0.05 vs. parental A2780 cells
treated with PBS, &P<0.05 vs. A2780/TAX cells
treated with PBS. CCK-8, Cell Counting Kit-8; DTX, docetaxel;
hUCMSCs, human umbilical cord mesenchymal stem cells; OC, ovarian
cancer; PI, propidium iodide; TAX, taxane. |
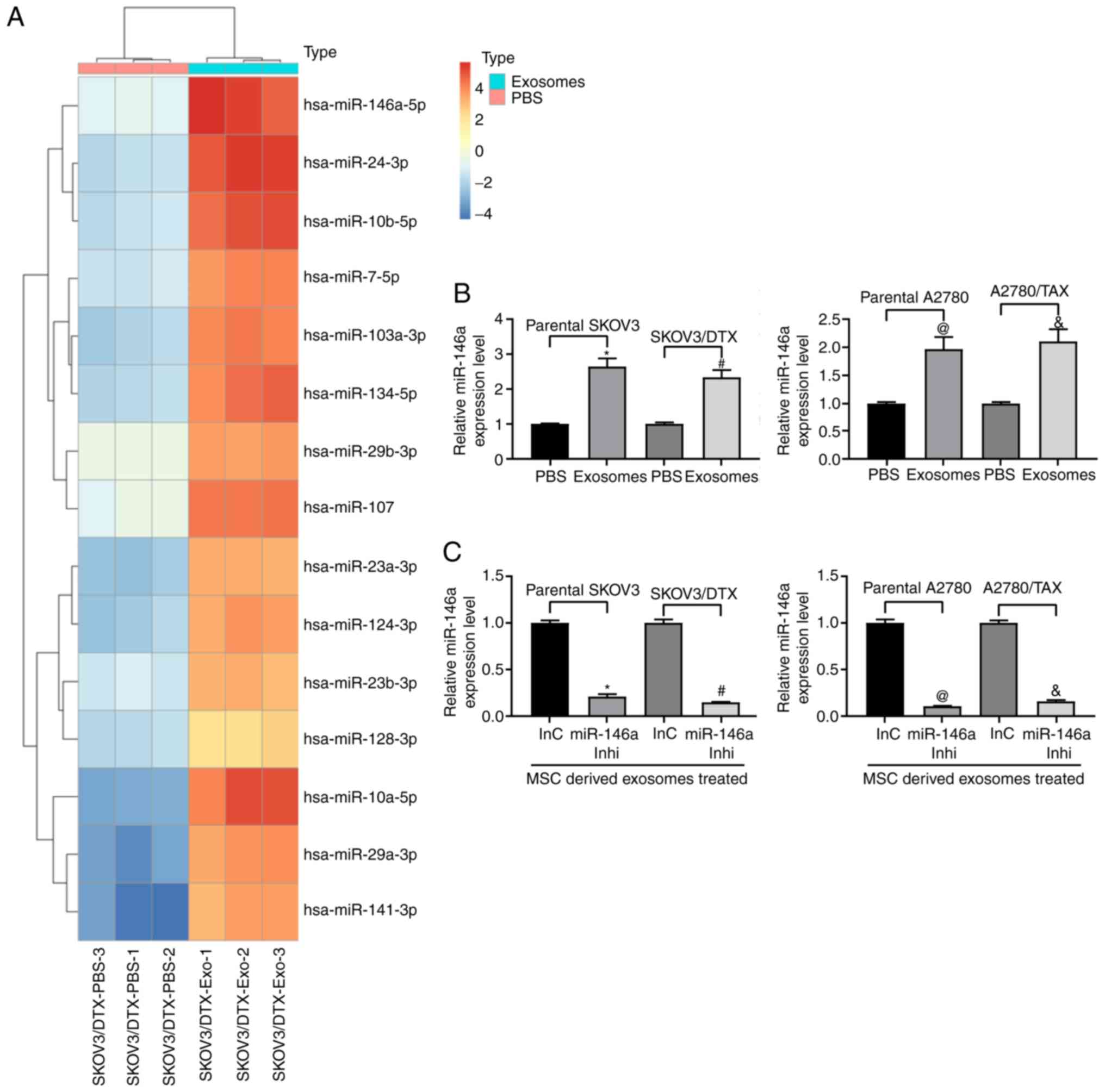
Expression of miR-146a is increased
following treatment with exosomes
Microarray analysis was performed to analyze the
differentially expressed miRNAs in SKOV3/DTX cells before and after
exosome treatment. miR-146a expression was demonstrated to be
increased following exosome treatment (Fig. 3A). Furthermore, RT-qPCR was used
to detect the expression levels of miR-146a in parental SKOV3 and
A2780 cells and the corresponding drug-resistant cell lines. A
similar trend in miR-146a expression was observed in both cell
lines relative to their parental SKOV3 and A2780 cells following
treatment with exosomes (Fig.
3B). Therefore, to further clarify the role of miR-146a in OC
cell chemoresistance, miR-146a inhibitor was transfected into
exosome-treated parental SKOV3 and A2780 cells and the
corresponding resistant cells (Fig.
3C).
Inhibition of miR-146a expression in OC
cells reduces the effects of hUCMSC-derived exosomes
Exosome-treated parental SKOV3 and A2780 cells and
the corresponding resistant cells were treated with 2 µM DTX
or 5 µM TAX, respectively, and a CCK-8 assay, EdU staining
and apoptosis analysis were performed. The results revealed that
miR-146a inhibitor suppressed the sensitizing role of
hUCMSC-derived exosomes in OC cells. Following knockdown of
miR-146a in hUCMSCs, the drug resistance of SKOV3/DTX cells to DTX
and the drug resistance of A2780/TAX to TAX were increased, the
numbers of EdU positive cells were increased and the proportions of
apoptotic cells were decreased in the parental and drug-resistant
cells following treatment with 2 µM DTX and 5 µM TAX
(P<0.05; Fig. 4A-D).
Therefore, transferring miR-146a may be one of the primary
mechanisms by which hUCMSC-derived exosomes reduce
chemoresistance.
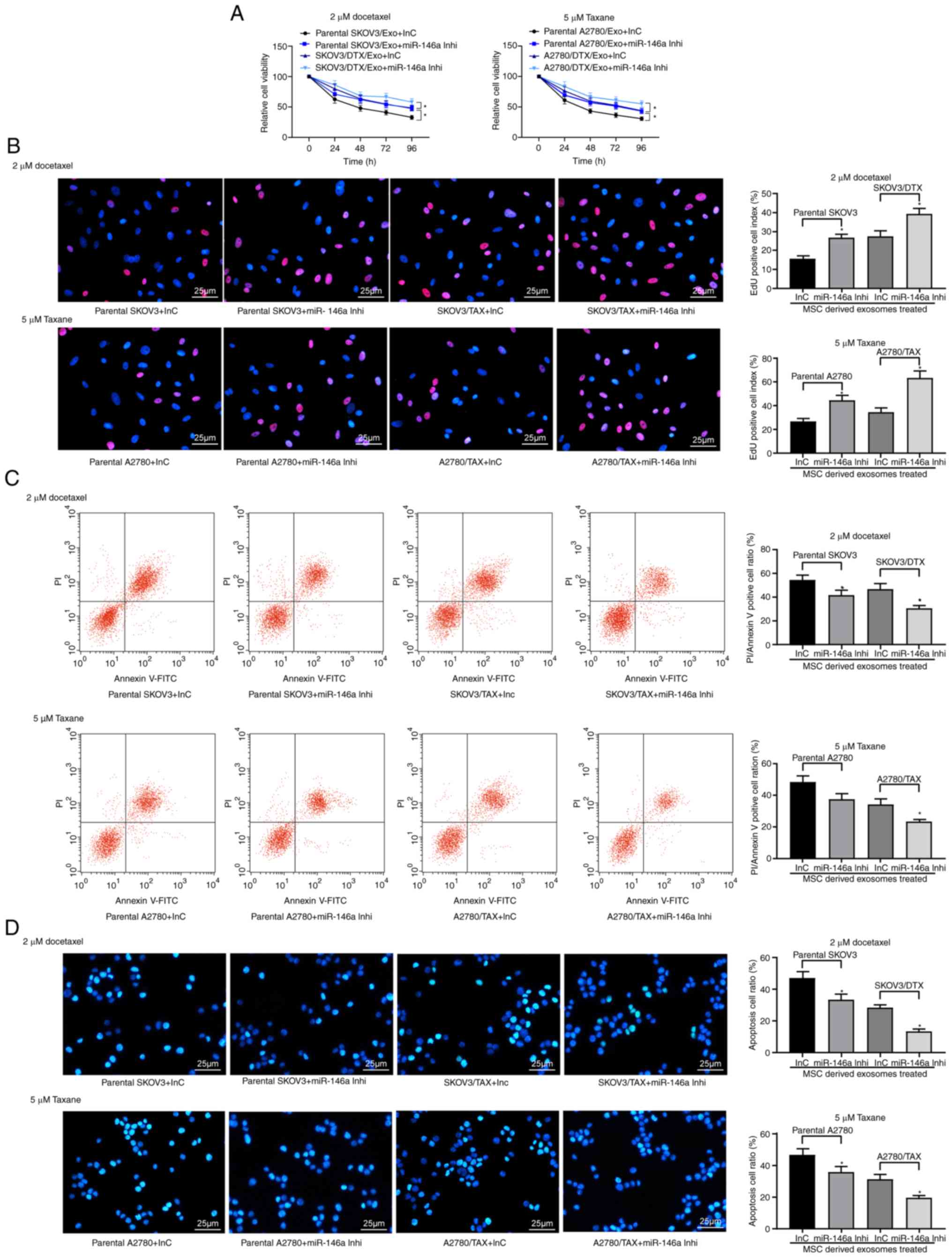 | Figure 4Exosomes from hUCMSCs promote OC cell
chemosensitivity partly by transferring miR-146a. hUCMSC-derived
exosome-treated parental or drug resistant OC cells were exposed to
2 µM DTX or 5 µM TAX for 2 h, respectively. (A) Cell
viability assessed by Cell Counting Kit-8 cell viability assays.
(B) EdU staining for cell proliferation. Scale bar, 25 µm.
(C) Flow cytometry analysis for cells labelled with PI/Annexin V.
(D) Hoechst 33258 staining for cell apoptosis. Scale bar, 25
µm. Data are presented as the mean ± SD. One-way (panels C,
D and E) or two-way (panel A) ANOVA followed by Tukey's post hoc
test was used to determine statistical significance. Each
assessment was performed in triplicate with three repetitions to
ensure minimum deviation. *P<0.05 vs. parental cells
co-cultured with exosomes derived from InC-transfected MSCs. DTX,
docetaxel; hUCMSCs, human umbilical cord mesenchymal stem cells;
inhi, inhibitor; InC, inhibitor control; miR, microRNA; OC, ovarian
cancer; PI, propidium iodide; TAX, taxane. |
miR-146a downregulates the PI3K signaling
pathway by targeting LAMC2
To identify the targets of miR-146a that may have
contributed to the increased chemosensitivity in OC, the putative
targets of miR-146a were predicted in silico using StarBase
and miRSearch. Among the potential candidates, LAMC2 was selected
for further analysis due to the complementary structures with
miR-146a (Fig. 5A). To further
identify whether miR-146a directly binds to the 3'UTR of LAMC2,
chimeric constructs harboring LAMC2-wild type (LAMC2-WT) or
LAMC2-mutant (LAMC2-MT) were constructed. As shown in Fig. 5B, overexpression of miR-146a
inhibited the luciferase activity of the reporter gene in the WT
construct but not the LAMC2-MT construct. Furthermore, gene
expression profiling interactive analysis illustrated that LAMC2
expression was significantly increased in the TCGA-OV dataset
compared with in normal ovarian tissues in the GTEX database
(Fig. 5C). LAMC2 expression was
upregulated in resistant OC cell lines, whereas, following the
addition of exosomes, LAMC2 expression was significantly decreased.
After further inhibition of miR-146a expression, LAMC2 expression
was restored (Fig. 5D). ELISA was
used to detect PI3K expression and Akt phosphorylation, and it was
demonstrated that exosome treatment reduced PI3K expression and Akt
phosphorylation, and miR-146a inhibitor treatment increased these
(Fig. 5E). To determine the role
of LAMC2 in OC cell resistance mediated by hUCMSC-released
exosomes, LAMC2 was overexpressed in SKOV3/DTX and A2780/TAX cells.
Transfection efficiency was determined by RT-qPCR (Fig. 5F).
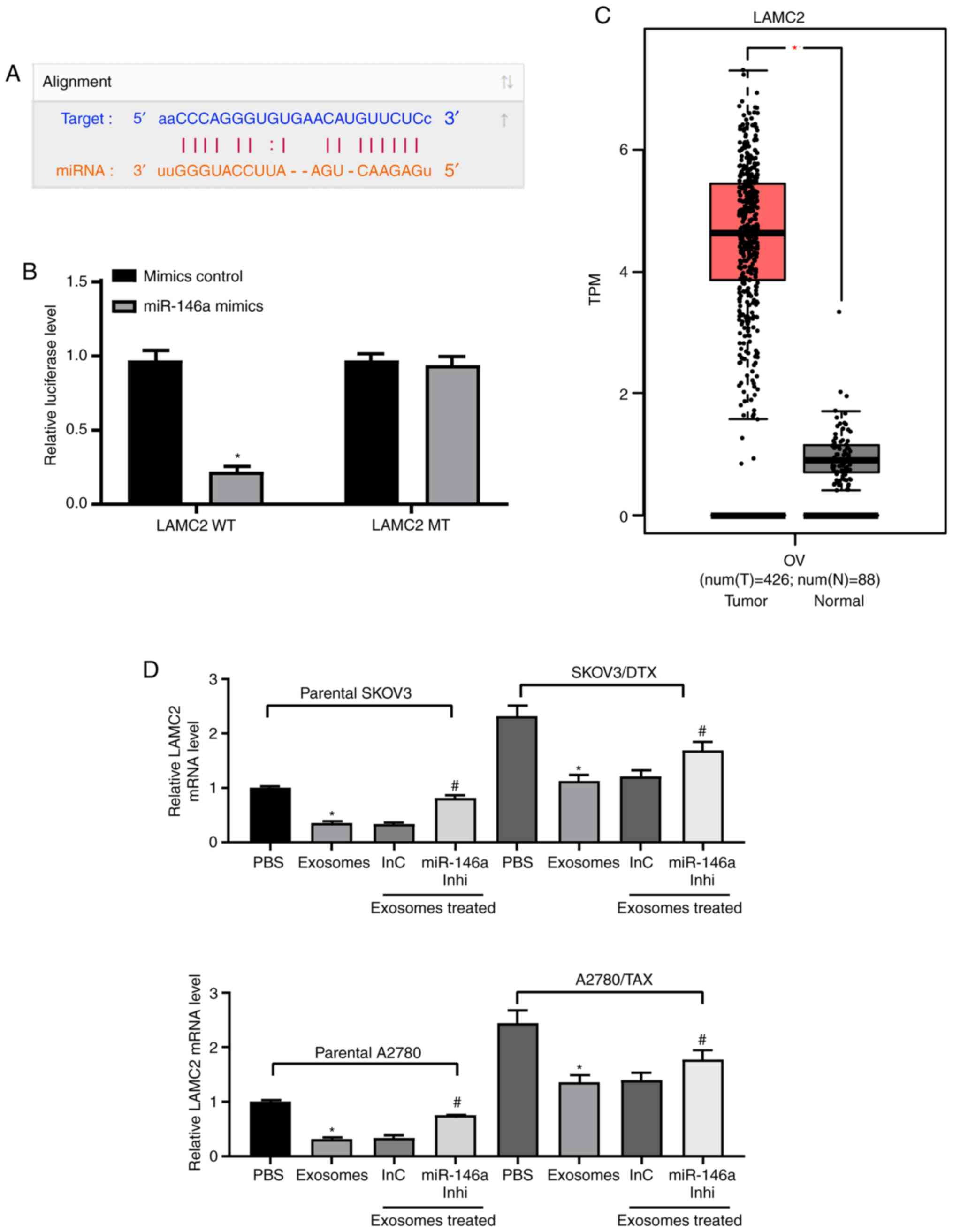 | Figure 5miR-146a blocks the PI3K signaling
pathway by inhibiting LAMC2. (A) Possible miR-146 binding sites in
the LAMC2 3' untranslated region. (B) Impact of miR-146a on the
luciferase activity of LAMC2-WT and LAMC2-MT evaluated by dual
luciferase assays. *P<0.05 vs. LAMC2 MT. (C) LAMC2
expression in The Cancer Genome Atlas-ovarian cancer dataset (n=426
for tumor samples and n=88 for normal tissues) predicted by gene
expression profiling interactive analysis. *P<0.05.
(D) LAMC2 expression in SKOV3 and A2780 cells determined by
RT-qPCR. *P<0.05 vs. cells treated with PBS;
#P<0.05 vs. cells co-cultured with exosomes extracted
from InC-treated MSCs. (E) Protein expression levels of PI3K
measured by ELISA. *P<0.05 vs. cells treated with
PBS; #P<0.05 vs. cells co-cultured with exosomes
extracted from InC-treated MSCs. (F) LAMC2 expression in SKOV3/DTX
and A2780/TAX cells determined by RT-qPCR. Data are presented as
the mean ± SD. *P<0.05 vs. cells transfected with EV.
One-way (panels D, E and F) or two-way (panel B) ANOVA and Tukey's
multiple comparisons test were used to determine statistical
significance. Each assessment was performed in triplicate with
three repetitions to ensure minimum deviation. DTX, docetaxel; InC,
inhibitor control; inhi, inhibitor; LAMC2, laminin γ2; miR,
microRNA; MSCs, mesenchymal stem cells; MT, mutant; RT-qPCR,
reverse transcription-quantitative PCR; TAX, taxane; WT, wild
type. |
Overexpression of LAMC2 reduces drug
sensitivity in OC cells
LAMC2 lentivirus-packed overexpression vector was
added to exosome-treated parental SKOV3 and A2780 cells, and
RT-qPCR was used to verify successful infection (Fig. 6A). Subsequently, the drug
sensitivity of the cells to DTX and TAX was observed. Following
overexpression of LAMC2, the drug resistance of OC cells treated
with hUCMSC-derived exosomes was reduced, which was demonstrated by
increased cell viability after DTX or TAX treatment (Fig. 6B) and increased numbers of EdU
positive cells (Fig. 6C), but
decreased numbers of apoptotic cells (Fig. 6D and E). Overall, these results
revealed that hUCMSC-released exosomes reduced LAMC2 expression by
transferring miR-146a, which is involved in the modulation of OC
cell chemosensitivity.
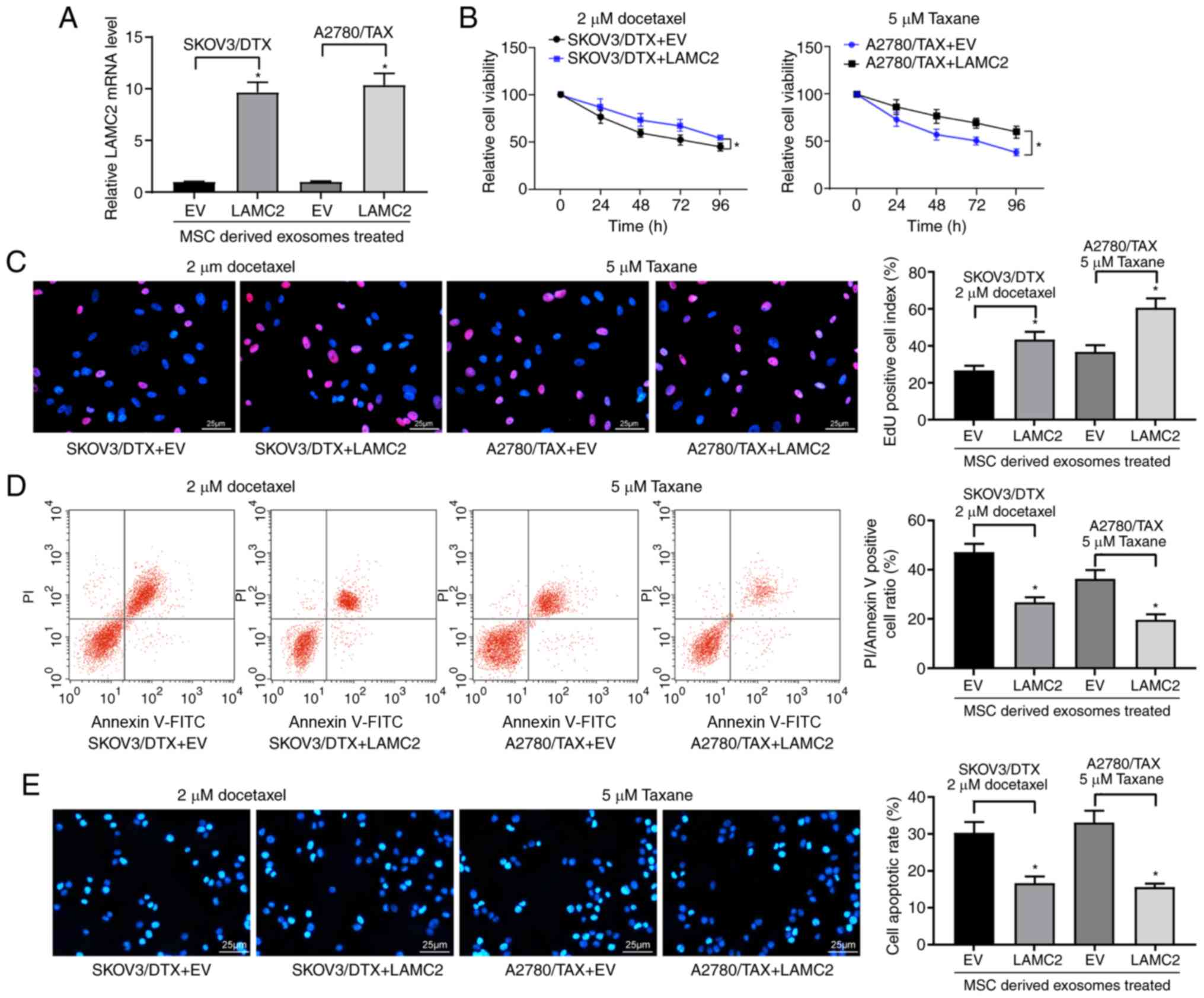 | Figure 6LAMC2 is involved in
miR-146a-mediated OC cell sensitivity. hUCMSCs-derived
exosome-treated parental or drug resistant OC cells were exposed to
2 µM DTX or 5 µM TAX for 2 h, respectively. (A) LAMC2
mRNA expression measured by reverse transcription-quantitative PCR.
(B) Cell viability examined by Cell Counting Kit-8 cell viability
assays. (C) EdU staining of OC cells. Scale bar, 25 µm. (D)
Flow cytometry analysis for cells labelled with propidium
iodide/Annexin V. Scale bar, 25 µm. (E) Hoechst 33258
staining for cell apoptosis. Data are presented as the mean ± SD.
One-way (panels A, C, D and E) or two-way (panel B) ANOVA followed
by Tukey's multiple comparisons test was used.
*P<0.05 vs. cells transfected with EV. Each
assessment was performed in triplicate with three repetitions to
ensure minimum deviation. DTX, docetaxel; EV, empty vector;
hUCMSCs, human umbilical cord mesenchymal stem cells; LAMC2,
laminin γ2; miR, microRNA; OC, ovarian cancer; TAX, taxane. |
Discussion
Exosomes, endogenous nanoparticles with a diameter
of 40-100 nm, are produced by most cell types and released into the
extracellular space. Exosomes have been demonstrated to deliver
mRNAs, miRNAs and other non-coding RNAs to neighboring cells and/or
distant cells when in circulation (20). The use of MSC-secreted exosomes as
biological vehicles to carry tumor suppressor miRNAs or
chemotherapeutic agents is a potential approach for cancer
treatment (21). In the present
study, hUCMSCs effectively transferred miR-146a into secreted
exosomes, which increased the sensitivity to the chemotherapeutic
agents DTX and TAX by reducing LAMC2 expression via the PI3K/Akt
signaling pathway in OC cells. The primary results suggested a
feasible therapeutic regimen for use of hUCMSCs-derived exosomes as
a potential vehicle for delivery of miR-146a to OC cells.
The present study demonstrated that hUCMSC-derived
exosomes, successfully engineered to package miR-146a into the
exosomes, has the potential to deliver miR-146 to OC cells in
vitro. Exosomal miR-146a effectively reduced the
chemo-resistance of OC cells to DTX and TAX. In agreement with the
results of the present study, Asare-Werehene et al (22) have demonstrated that
exosome-transported plasma gelsolin results in increased
chemoresistance in OC cells in an autocrine manner, and confers
cisplatin resistance to chemosensitive OC cells (22). Furthermore, cancer-associated
fibroblasts-secreted exosomes transferred miR-98-5p to poten-tiate
OC cell resistance to cisplatin, which was associated with
upregulated expression of the anticancer drug target CDKN1A in
cisplatin-sensitive OC cells (23). Therefore, it was hypoth-esized
that the exosomal content conferred chemoresistance in OC cells.
Macrophage-secreted exosomes containing elevated levels of
miR-146a-5p have been delivered to endothelial cells, and resulted
in reduced migration of monocytes by targeting junctional adhesion
molecule C (24). In vivo,
circulating endo-thelial cell-derived extracellular vesicles from
tumor-bearing mice have been demonstrated to contain upregulated
levels of miR-146a compared with the control mice (25). Additionally, overexpression of
miR-146a results in increased cell proliferation, and increased
apoptosis and sensitivity to DTX in epithelial OC cells (26). Furthermore, hUCMSC-delivered
exogenous miR-145-5p reduces pancreatic ductal adenocarci-noma cell
proliferation and invasion, and increases apoptosis and cell cycle
arrest (27). In vitro and
in vivo assays have confirmed the tumor suppressor role of
hUCMSCs-released exosomes carrying miR-148b-3p in breast cancer
(28). The present study revealed
that hUCMSC-delivered exosomes with increased expression levels of
miR-146a sensitized the OC cells to DTX and TAX by targeting
LAMC2.
LAMC2, a component of Ln-332, is a potent tumor
biomarker which is upregulated in urothelial carcinoma (29), hepatocel-lular carcinoma (30) and pancreatic adenocarcinoma
(31). In addition, Ln-332
increases chemoresistance and quiescence of cancer stem cells in
hepatocellular carcinoma (32).
The bioin-formatics analysis performed in the present study
suggested that LAMC2 expression was increased in TCGA-OV, and this
upregulation was reversed by hUCMSCs-delivered exosomes. In
addition, miR-146a inhibitor reduced the expression levels of LAMC2
in SKOV3 and A2780 parental and resistant cells. Furthermore,
upregulation of LAMC2 significantly reduced the inhibitory role of
hUCMSCs-delivered exosomes in OC cell chemoresistance, as suggested
by the reduced apoptosis and increased proliferation, further
demonstrating that LAMC2 was involved in the mediation of exosomal
miR-146a in OC cell chemoresistance. Similarly, LAMC2 expression is
upregulated and facilitates metastasis in esophageal squamous cell
carcinoma, whereas LAMC2 restoration partially reduces the decrease
in cell migration and invasion induced by cancer susceptibility 9
knockdown (33). The ELISA
experiments performed in the present study revealed that exosome
treatment reduced the activity of the PI3K/Akt signaling pathway in
both resistant and sensitive OC cells, as demonstrated by the
decreased PI3K expression and Akt phosphorylation levels; whereas
miR-146a inhibitor reversed these trends. PI3K is the control hub
of growth signal transmission, and activated PI3K is a hallmark of
several types of cancer, including OC (34). Consistent with the results of the
present study, PI3K gene expression is significantly increased in
cervical cancer tissues and the derived exosomes relative to the
corresponding adjacent tissues (35). Furthermore, PI3K has been reported
to be a putative target of miR-146a in chondrocytes, whereas PI3K
expression at both the protein and mRNA levels is decreased
following transfection with a miR-146a mimic (36).
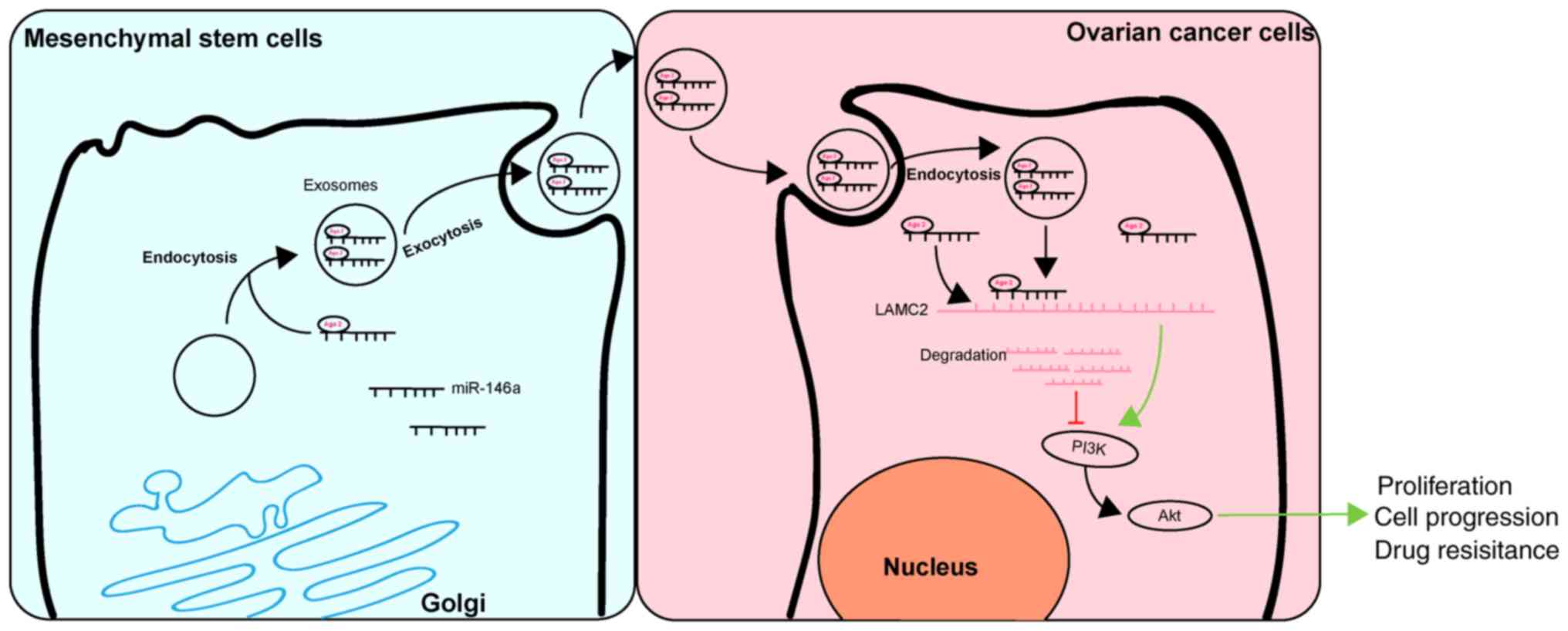
In conclusion, the results of the present study
demonstrated that exosomal miR-146a released by hUCMSCs contributed
to hUCMSC-derived exosome-mediated chemosensitivity, and this
effect may be mediated by interactions between miR-146a and LAMC2
via the PI3K/Akt signaling pathway (Fig. 7). These results may improve the
understanding of the intercel-lular communications between cancer
cells and MSCs through an MSC-exosome-miRNA axis. The present study
suggests that management of hUCMSCs and their released exosomes may
be a therapeutic strategy with potential clinical applications.
Supplementary Data
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LQ conceived and designed the present study. JW
contributed to the design and definition of intellectual content of
this study. MC, FC and WT contributed to the experimental studies,
data acquisition and statistical analysis. LQ and WT contributed to
the manuscript preparation. All authors read and approved the final
manuscript and guarantee the integrity of the study.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
References
|
1
|
Tawde SA, Chablani L, Akalkotkar A and
D'Souza MJ: Evaluation of microparticulate ovarian cancer vaccine
via transdermal route of delivery. J Control Release. 235:147–154.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zahedi P, De Souza R, Huynh L,
Piquette-Miller M and Allen C: Combination drug delivery strategy
for the treatment of multi-drug resistant ovarian cancer. Mol
Pharm. 8:260–269. 2011. View Article : Google Scholar
|
|
3
|
Lohse I, Azzam DJ, Al-Ali H, Volmar CH,
Brothers SP, Ince TA and Wahlestedt C: Ovarian cancer treatment
stratification using ex vivo drug sensitivity testing. Anticancer
Res. 39:4023–4030. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Secord AA: Ovarian cancer: Time to move
beyond one size fits all. Lancet Oncol. 20:754–755. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Borley J and Brown R: Epigenetic
mechanisms and therapeutic targets of chemotherapy resistance in
epithelial ovarian cancer. Ann Med. 47:359–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ridge SM, Sullivan FJ and Glynn SA:
Mesenchymal stem cells: Key players in cancer progression. Mol
Cancer. 16:312017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li P, Gong Z, Shultz LD and Ren G:
Mesenchymal stem cells: From regeneration to cancer. Pharmacol
Ther. 200:42–54. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding DC, Shyu WC, Lin SZ, Liu HW, Chiou SH
and Chu TY: Human umbilical cord mesenchymal stem cells support
nontu-morigenic expansion of human embryonic stem cells. Cell
Transplant. 21:1515–1527. 2012. View Article : Google Scholar
|
|
9
|
Houthuijzen JM, Daenen LG, Roodhart JM and
Voest EE: The role of mesenchymal stem cells in anti-cancer drug
resistance and tumour progression. Br J Cancer. 106:1901–1906.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sundararajan V, Sarkar FH and Ramasamy TS:
Correction to: The versatile role of exosomes in cancer
progression: Diagnostic and therapeutic implications. Cell Oncol
(Dordr). 41:4632018. View Article : Google Scholar
|
|
11
|
Halkein J, Tabruyn SP, Ricke-Hoch M,
Haghikia A, Nguyen NQ, Scherr M, Castermans K, Malvaux L, Lambert
V, Thiry M, et al: MicroRNA-146a is a therapeutic target and
biomarker for peri-partum cardiomyopathy. J Clin Invest.
123:2143–2154. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roos J, Enlund E, Funcke JB, Tews D,
Holzmann K, Debatin KM, Wabitsch M and Fischer-Posovszky P:
miR-146a-mediated suppression of the inflammatory response in human
adipocytes. Sci Rep. 6:383392016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang C, Lim W, Park J, Park S, You S and
Song G: Anti-inflammatory effects of mesenchymal stem cell-derived
exosomal microRNA-146a-5p and microRNA-548e-5p on human trophoblast
cells. Mol Hum Reprod. 25:755–771. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng WC, Liao TT, Lin CC, Yuan LE, Lan
HY, Lin HH, Teng HW, Chang HC, Lin CH, Yang CY, et al:
RAB27B-activated secretion of stem-like tumor exosomes delivers the
biomarker microRNA-146a-5p which promotes tumorigenesis and
associates with an immunosuppressive tumor microenvironment in
colorectal cancer. Int J Cancer. 145:2209–2224. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang D, Guo H, Feng W and Qiu H: LAMC2
regulated by microRNA-125a-5p accelerates the progression of
ovarian cancer via activating p38 MAPK signalling. Life Sci.
232:1166482019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu Z, Cai M, Zhang Y, Tao L and Guo R:
miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian
cancer by regulating FOXP1/ATG14 pathway. Cell Cycle. 19:193–206.
2020. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop DJ and Horwitz E: Minimal criteria for defining multipotent
mesen-chymal stromal cells. The international society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar
|
|
19
|
Thery C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extracellular vesicles 2018 (MISEV2018): A position statement of
the international society for extracellular vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar
|
|
20
|
Zhang J, Li S, Li L, Li M, Guo C, Yao J
and Mi S: Exosome and exosomal microRNA: Trafficking, sorting, and
function. Genomics Proteomics Bioinformatics. 13:17–24. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lai RC, Yeo RW, Tan KH and Lim SK:
Exosomes for drug delivery-a novel application for the mesenchymal
stem cell. Biotechnol Adv. 31:543–551. 2013. View Article : Google Scholar
|
|
22
|
Asare-Werehene M, Nakka K, Reunov A, Chiu
CT, Lee WT, Abedini MR, Wang PW, Shieh DB, Dilworth FJ, Carmona E,
et al: The exosome-mediated autocrine and paracrine actions of
plasma gelsolin in ovarian cancer chemoresistance. Oncogene.
39:1600–1616. 2020. View Article : Google Scholar :
|
|
23
|
Guo H, Ha C, Dong H, Yang Z, Ma Y and Ding
Y: Cancer-associated fibroblast-derived exosomal microRNA-98-5p
promotes cisplatin resistance in ovarian cancer by targeting
CDKN1A. Cancer Cell Int. 19:3472019. View Article : Google Scholar :
|
|
24
|
Hu W, Xu B, Zhang J, Kou C, Liu J, Wang Q
and Zhang R: Exosomal miR-146a-5p from treponema
pallidum-stimulated macrophages reduces endothelial cells
permeability and mono-cyte transendothelial migration by targeting
JAM-C. Exp Cell Res. 388:1118232020. View Article : Google Scholar
|
|
25
|
McCann JV, Liu A, Musante L, Erdbrugger U,
Lannigan J and Dudley AC: A miRNA signature in endothelial
cell-derived extracellular vesicles in tumor-bearing mice. Sci Rep.
9:167432019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cui Y, She K, Tian D, Zhang P and Xin X:
miR-146a inhibits proliferation and enhances chemosensitivity in
epithelial ovarian cancer via reduction of SOD2. Oncol Res.
23:275–282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ding Y, Cao F, Sun H, Wang Y, Liu S, Wu Y,
Cui Q, Mei W and Li F: Exosomes derived from human umbilical cord
mesen-chymal stromal cells deliver exogenous miR-145-5p to inhibit
pancreatic ductal adenocarcinoma progression. Cancer Lett.
442:351–361. 2019. View Article : Google Scholar
|
|
28
|
Yuan L, Liu Y, Qu Y, Liu L and Li H:
Exosomes derived from MicroRNA-148b-3p-overexpressing human
umbilical cord mesenchymal stem cells restrain breast cancer
progression. Front Oncol. 9:10762019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kamada M, Koshikawa N, Minegishi T, Kawada
C, Karashima T, Shuin T and Seiki M: Urinary laminin-g2 is a novel
biomarker of non-muscle invasive urothelial carcinoma. Cancer Sci.
106:1730–1737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Korbakis D, Dimitromanolakis A, Prassas I,
Davis GJ, Barber E, Reckamp KL, Blasutig I and Diamandis EP: Serum
LAMC2 enhances the prognostic value of a multi-parametric panel in
non-small cell lung cancer. Br J Cancer. 113:484–491. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kosanam H, Prassas I, Chrystoja CC, Soleas
I, Chan A, Dimitromanolakis A, Blasutig IM, Ruckert F, Gruetzmann
R, Pilarsky C, et al: Laminin, gamma 2 (LAMC2): A promising new
putative pancreatic cancer biomarker identified by proteomic
analysis of pancreatic adenocarcinoma tissues. Mol Cell Proteomics.
12:2820–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Govaere O, Wouters J, Petz M, Vandewynckel
YP, Van den Eynde K, Van den Broeck A, Verhulst S, Dolle L,
Gremeaux L, Ceulemans A, et al: Laminin-332 sustains
chemo-resistance and quiescence as part of the human hepatic cancer
stem cell niche. J Hepatol. 64:609–617. 2016. View Article : Google Scholar
|
|
33
|
Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang
K, Guan X, Yang K and Bai Y: LncRNA CASC9 promotes esophageal
squamous cell carcinoma metastasis through upregulating LAMC2
expression by interacting with the CREB-binding protein. Cell Death
Differ. 25:1980–1995. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Binju M, Amaya-Padilla MA, Wan G,
Gunosewoyo H, Suryo Rahmanto Y and Yu Y: Therapeutic inducers of
apoptosis in ovarian cancer. Cancers (Basel). 11. pp. 17862019,
View Article : Google Scholar
|
|
35
|
Zhang W, Zhou Q, Wei Y, Da M, Zhang C,
Zhong J, Liu J and Shen J: The exosome-mediated PI3k/Akt/mTOR
signaling pathway in cervical cancer. Int J Clin Exp Pathol.
12:2474–2484. 2019.
|
|
36
|
Li H, Xie S, Li H, Zhang R and Zhang H:
LncRNA MALAT1 mediates proliferation of LPS treated-articular
chondrocytes by targeting the miR-146a-PI3K/Akt/mTOR axis. Life
Sci. Aug 28, 2019 (Epub ahead of print).
|