Introduction
Cerebral ischemia/reperfusion (I/R)-related
disability, which is characterized by inflammation, necrosis and
apoptosis of cells, represents a significant healthcare burden
worldwide, causing 6.5 million deaths every year (1-3).
Autophagy plays important roles in the pathology of cerebral I/R
(4). Numerous neuroprotective
agents have been shown to be effective in animal experiments;
however, they were reported to be ineffective in clinical trials
(5-7). Thus, a deeper exploration of the
molecular mechanisms underlying cerebral I/R injury and the
identification of novel therapeutic targets to control cerebral
I/R-associated brain injury is urgently required.
During the last decade, substantial efforts have
been undertaken to isolate and identify microRNAs (miRNAs/miRs),
and investigate their functions in vitro and in vivo
(8-10). As functional RNA molecules that
can inhibit protein expression by targeting the 3′-untranslated
region (3′-UTR) of target mRNAs, miRNAs are involved in numerous
physiological and pathological processes (11). Cerebral miRNA profiles are
reportedly significantly altered and affect disease outcomes in
central nervous system (CNS) diseases, which has led to an
explosion of potential biomarkers for these diseases (12,13). miRNAs associated with the
regulation of apoptosis and autophagy during ischemic stroke have
been identified (14,15). miRNA binding sites in the coding
sequences (CDSs) of mRNAs have been predicted by combining
computational approaches and human mRNA expression data (16). Numerous studies have provided
experimental evidence of the existence of functional miRNA binding
sites in mammalian CDSs (17-20). For example, miR-148 inhibits DNA
methyltransferase 3β expression by targeting a conserved site in
its CDS (17).
Seipin is an endoplasmic reticulum membrane protein
that is encoded by the Berardinelli-Seip congenital lipodystrophy
type 2 (BSCL2/Bscl2) gene (21-23). Seipin is a widely expressed
protein but with highest levels found in the CNS of humans
(24). Using
immunohistochemistry, Seipin expression has been detected in
neurons of the frontal lobe cortex of the brain, including the
motor and somatosensory cortex (25). Recently, the transcript encoding
the long Seipin isoform (BSCL2-203, 462 amino acids) was reported
to be expressed primarily in the brain, and its expression was
inversely correlated with age in neuronal cells (24). Seipin deficiency has been reported
to aggravate I/R-induced cerebral damage (26). Seipin mutations at glycosylation
sites result in the activation of autophagy (27,28). Our recent study reported that
neuronal Seipin deletion reduces autophagosome formation to cause
hyperphosphorylation and aggregation of tau protein through reduced
inhibition of peroxisome proliferator-activated receptor γ in
Akt/mTOR signaling (29).
Furthermore, miRNAs have been associated with autophagy and
apoptosis in brains following ischemic stroke (30,31). Therefore, if cerebral ischemia
alters miRNA and Seipin levels, it is of great interest to
investigate whether alterations in miRNA expression affect the
expression of Seipin, and autophagy and apoptosis.
The present study evaluated the involvement of
miR-187-3p in Seipin expression, autophagic flux and apoptosis
after ischemic stroke, or in PC12 cells treated with oxygen-glucose
deprivation and reoxygenation (OGD/R), and explored the underlying
molecular mechanisms. The results of the present study indicated
that the OGD/R-induced increase in the levels of miR-187-3p
suppressed Seipin expression, leading to a deficit in autophagic
flux and neuronal apoptosis.
Materials and methods
Cells, culture and OGD/R model
establishment
PC12 cells and 293T cells were purchased from the
Cell Bank of Shanghai Institute of Cell Biology. PC12 cells and
293T cells were incubated in Dulbecco's modified Eagle's medium
(DMEM; Invitrogen; Thermo Fisher Scientific, Inc.) containing 10%
fetal bovine serum, 100 µg/ml streptomycin and 100 U/ml
penicillin (all from Gibco; Thermo Fisher Scientific, Inc.) at 37°C
with 5% CO2. For OGD/R treatment, PC12 cells were
incubated in glucose-free DMEM (Invitrogen; Thermo Fisher
Scientific, Inc.) and incubated at 37°C in the presence of 5%
CO2 and 95% N2 for 4 h, followed by 18 h of
reoxygenation.
Identification of miRNAs targeting
Seipin
Total RNA, including miRNA, was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) from control and 4:18-h OGD/R-exposed PC12 cells.
High-throughput next-generation sequencing was used for miRNA
sequencing to achieve optimal PC12 cell miRNA profiles. miRNA
sequencing was carried out using a high-throughput Illumina HiSeq
2000 sequencing platform (BGI Group). miRNAs with a Q-value ≤0.01
and |log (fold-change)|>2 were considered differentially
expressed. miRNAs targeting the CDS of Seipin were predicted using
the miRanda database (http://www.microrna.org/microrna/home.do).
Seipin short hairpin (sh)RNA design and
lentivirus construction
Seipin expression was knocked down in PC12 cells
using a targeted shRNA. Short hairpin RNA (shRNA) specific for
Seipin (5′-ACG CTC GGT GAT GCT TC ATT A-3′) and a non-targeting
scramble shRNA sequence (5′-TTC TCC GAA CGT GTC ACG T-3′) construct
were designed and cloned into a
hU6-MCS-Ubiquitin-EGFP-IRES-puromycin vector, and packaged into a
lentivirus (Shanghai GeneChem Co., Ltd.). The lentiviral constructs
(MOI=70) were infected into PC12 cells (5×105
cells/cm2) for the generation of stable clones according
to the manufacturer's instructions. At 48 h after transduction,
PC12 cells were treated with 4:18-h OGD/R. For experiments
involving rapamycin (Sigma-Aldrich; Merck KGaA) treatment,
rapamycin (20 nM) was added to the culture medium 2 h before PC12
cells were subjected to OGD/R.
Cell transfection
miR-187-3p mimics (sense, 5′-UCG UGU CUU GUG UUG CAG
CCG G-3′; antisense, 5′-CCG GCU GCA ACA CAA GAC ACG A-3′),
miR-187-3p inhibitor (5′-CCG GCU GCA ACA CAA GAC ACG A-3′), mimics
negative control (NC; sense, 5′-UUU GUA CUA CAC AAA AGU ACU G-3′;
antisense, 5′-CAG UAC UUU UGU GUA GUA CAA A-3′) and inhibitor NC
(5′-CAG UAC UUU UGU GUA GUA CAA A-3′) were purchased from Guangzhou
RiboBio Co., Ltd. and transfected into PC12 cells (5×105
cells/cm2) using riboFECT™ CP transfection reagent
(Guangzhou RiboBio Co., Ltd.) according to the manufacturer's
instructions (50 nM). Cells were collected for further experiments
after 48 h of transfection.
Adenovirus (Ad)-mRFP-GFP-LC3 transfection
and confocal microscopy
PC12 cells were cultured in 24-well plates at a
density of 1×104 cells/well and transfected with
miR-187-3p mimics, inhibitor or mimics NC. After 12 h, cells were
infected with an Ad-mRFP-GFP-LC3 vector (Hanbio Biotechnology Co.,
Ltd.) at MOI=30 for a further 24 h. Then, PC12 cells were incubated
in glucose-free DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
and incubated at 37°C in the presence of 5% CO2 and 95%
N2 for 4 h, followed by 18 h of reoxygenation.
Fluorescent puncta were detected in PC12 cells infected with
Ad-mRFP-GFP-LC3 via Olympus laser scanning confocal microscopy
(magnification, ×100; Olympus Corporation). Yellow and red
represent autophagolysosomes and autophagosomes, respectively; 20
cells/group were analyzed.
Reverse transcription-quantitative
(RT-q)PCR
Total RNAs (including miRNAs) were extracted from
PC12 cells or brain tissues using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. miRNAs were transcribed into cDNA (25°C for 5 min, 50°C
for 15 min and 85°C for 5 min) using an miRNA 1st-Strand cDNA
Synthesis kit (Vazyme Biotech Co., Ltd.). miR-187-3p expression was
quantified by RT-qPCR using an miRNA Universal SYBR qPCR Master Mix
kit (Vazyme Biotech Co., Ltd.), and U6 small nuclear RNA served as
an internal reference for normalization. For mRNA expression
analysis, cDNA was synthesized (42°C for 15 min and 85°C for 5 min)
using StarScript II First-strand cDNA Synthesis Mix With gDNA
Remover kit (GenStar). qPCR was performed using SYBR-Green SuperMix
(Vazyme Biotech Co., Ltd.) as follows: At 95°C for 3 min, followed
by 39 cycles of 60°C for 30 sec and 70°C for 15 sec. The following
primers were used (Sangon Biotech Co., Ltd.): U6, forward, 5′-CTC
GCT TCG GCA GC ACA-3′ and reverse, AAC GCT TCA CGA ATT TGC GT;
miR-187-3p, forward 5′-AGC GTC GTG TCT TGT GTT GC-3′ and reverse,
5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACC CGG
CT-3′; Seipin, forward, 5′-AGG TGG CCG AAT CAT CTC CA-3′ and
reverse, 5′-GAG AAC ACC AGC GTG TCG AG-3′; GAPDH, forward, 5′-AGG
TCG GTG TGA ACG GAT TTG-3′ and reverse, 5′-GGG GTC GTT GAT GGC AAC
A-3′. The relative expression levels of miRNAs and Seipin mRNA were
calculated using the 2−ΔΔCq method (32).
Luciferase reporter assay
Fragments of the wild-type Seipin CDS containing two
miR-187-3p target sites, and mutants of these respective sites were
amplified and cloned into a psiCHECK™-2 vector (Promega
Corporation). 293T cells were seeded in 96-well plates 1 day before
transfection. When the cell density reached ~50%, the cells were
co-transfected with the psiCHECK-2 vector (0.16 µg)
containing the CDS of Seipin [with wild-type (Seipin-wt), mutant
miR-187-3p binding site 1 (Seipin-mu1) or mutant miR-187-3p binding
site 2 (Seipin-mu2)], and miR-187-3p mimics (5 pmol) or mimics NC
(5 pmol) with riboFECT CP. After transfection for 48 h, the cells
were analyzed using a Dual-Luciferase Reporter Assay System
(Promega Corporation) according to the manufacturer's protocols.
Data were calculated as the ratio of Renilla to firefly
luciferase activity.
Western blotting
Cells were seeded in a 6-well plate and washed three
times with pre-cooled PBS. Then, 60 µl RIPA buffer (Thermo
Fisher Scientific, Inc.) containing protease inhibitor cocktail
(Sigma-Aldrich; Merck KGaA) was added to each well, and the plate
was placed on ice for ~2 h; Total proteins were extracted from the
rat cortical peri-infarct region using RIPA buffer (Thermo Fisher
Scientific, Inc.) containing protease inhibitor cocktail
(Sigma-Aldrich; Merck KGaA). Proteins were quantified using a
bicinchoninic acid assay kit (cat. no. P0009; Beyotime Institute of
Biotechnology). Proteins (25 µg) were separated by 12%
SDS-PAGE and transferred onto nitrocellulose membranes. The
membrane was blocked with 5% nonfat milk at room temperature for 2
h and then incubated with primary antibodies against the
autophagy-related proteins phosphorylated (p)-mTOR (1:1,000; cat.
no. 5536S; Cell Signaling Technology, Inc.), mTOR (1:1,000; cat.
no. 2972S; Cell Signaling Technology, Inc.), p62 (1:1,000; cat. no.
23214; Cell Signaling Technology, Inc.) and light chain (LC)3II/I
(1:1,000; cat. no. 4108S; Cell Signaling Technology, Inc.), the
apoptosis-related proteins cleaved caspase-3 (1:1,000; cat. no.
9661S; Cell Signaling Technology, Inc.), Bax (1:1,000; cat. no.
ab32503; Abcam), and Seipin (1:1,000; cat. no. ab106793; Abcam) and
β-actin (1:1,000; cat. no. 3700S; Cell Signaling Technology, Inc.)
at 4°C overnight. Membranes were then incubated at room temperature
for 2 h with the horseradish peroxidase-conjugated secondary
antibody (1:1,000; cat. no. 7074S; Cell Signaling Technology,
Inc.), and an enhanced chemiluminescence reagent kit (Amersham;
Cytiva) was used for visualization. Gray values of autophagy- and
apoptosis-related proteins were measured using ImageJ software
(version 1.42q, National Institutes of Health) and normalized to
that of β-actin as an internal control.
Detection of apoptosis by flow
cytometry
Seipen shRNA-expressing PC12 cells were seeded in a
6-well plate at 1×106 cells/well and exposed to
miR-187-3p inhibitor or inhibitor NC (50 nM) for 48 h. Then,
normoxic control and OGD/R PC12 cells were prepared as described
above. After three washes with pre-cooled PBS, 100 µl of the
cell suspension was collected. The cells were stained with 5
µl of Annexin V-allophycocyanin and 10 µl of
7-aminoactinomyosin D (MultiSciences Biotech Co., Ltd.) at 37°C for
10 min in the dark. Binding buffer (400 µl) was subsequently
added to each sample, and apoptosis was detected by flow cytometry
(BD Accuri C6 Flow Cytometer; BD Biosciences). Flow cytometric data
were analyzed using FlowJo version 8.6 (FlowJo LLC). The apoptosis
rate was calculated as the percentage of early + late apoptotic
cells.
Rat model of focal cerebral I/R
The present study was approved by the Animal Care
and Use Committee of Guizhou Medical University (permit no.
1900813). A total of 54 male Sprague-Dawley rats (2 months old;
weight, 250-300 g) were obtained from the Experimental Animals
Center at Guizhou Medical University. Rats were housed under
comfortable conditions (12:12-h light/dark cycle, humidity 55-60%,
20-25°C, and access to food and water ad libitum). 18 rats
were used in each group (6 rats for infarct size measurement, 6
rats for expression analysis, 6 rats for immunofluorescence).
Reperfusion was established 1 h after middle cerebral artery
occlusion (MCAO) as previously reported (33). The animals were anesthetized with
chloral hydrate 10% (300 mg/kg) and the rectal temperature was
maintained at 37.0±0.5°C throughout the operation. No rats
exhibited signs of peritonitis after administration of 10% chloral
hydrate. At 24 h after reperfusion, the rats were sacrificed, and
their brains were collected. Sham rats underwent the same surgical
procedure, except without MCAO. Rat euthanasia was conducted via
CO2 inhalation; rats were placed into enclosed flow
cages for 5 min, and 100% CO2 was then added to the
cages at a displacement rate of 30% volume/min (34). After euthanasia, death was
confirmed via decapitation and brains were collected.
Intracerebroventricular injection
miR-187-3p antagomir and antagomir NC (1.5 nmol/200
g), synthesized by Guangzhou RiboBio Co., Ltd. (same sequences as
miR-187-3p inhibitor and inhibitor NC, respectively), were
dissolved in 0.9% saline (1.5 nmol/6 µl saline) and
administered to rats 2 h before MCAO though intraventricular
injection as previously described (35). Rats were anesthetized and placed
on a stereotactic head frame (RWD Life Science). miR-187-3p
antagomir and antagomir NC were infused slowly into the right
lateral ventricle using a micro-infusion pump (KDS 310; KD
Scientific) at a rate of 0.2 µl/min. To prevent possible
leakage, the burr hole was sealed with bone wax. The incision was
sutured and rats were placed into cages for recovery, then rats
were given free access to food and water.
Determination of infarct size
According to a previously described method (33), after 24 h reperfusion, the brains
were collected and stored at -20°C for 20 min before sectioning
sectioned coronally at a thickness of 1 mm. Briefly, the sections
were stained with tetrazolium chloride (0.2%) at 37°C for 30 min
and imaged with a digital camera. The infarct volume was corrected
for possible interference from brain edema and presented as a ratio
of the infarct volume compared with the contralateral hemisphere
volume.
Immunofluorescence assay
Rats were perfused transcardially with 4%
paraformaldehyde. The frozen brains were mounted on a freezing
(-80°C) microtome and cut into 30-µm sections (Leica
Microsystems GmbH). Frozen sections (n=4/rat) were blocked and
permeabilized with 5% normal goat serum (Sigma-Aldrich; Merck KGaA)
and 0.3% Triton X-100 for 1 h at room temperature. The slices were
then incubated overnight at 4°C with a primary antibody against LC3
(1:200; cat. no. 4108S; Cell Signaling Technology, Inc.) followed
by incubation with Alexa Fluor® 568-conjugated goat
anti-rabbit IgG (H + L) cross-adsorbed secondary antibody (1:100;
cat. no. A-11011; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. After washing, nuclei were counterstained with DAPI
(Beyotime Institute of Biotechnology) for 5 min at room
temperature. Images were collected with a Laser confocal microscope
(magnification, ×20; Olympus FV 1000; Olympus Corporation); 20
fields/section were randomly analyzed.
Statistical analysis
Statistical analysis was conducted using GraphPad
Prism software 7.0 (GraphPad Software, Inc.). Data are presented as
the mean ± SD. Comparisons among multiple groups were performed
using one-way ANOVA followed by Tukey's post hoc test; unpaired
t-tests were used for comparisons between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-187-3p is upregulated and Seipin
protein is downregulated in OGD/R PC12 cells
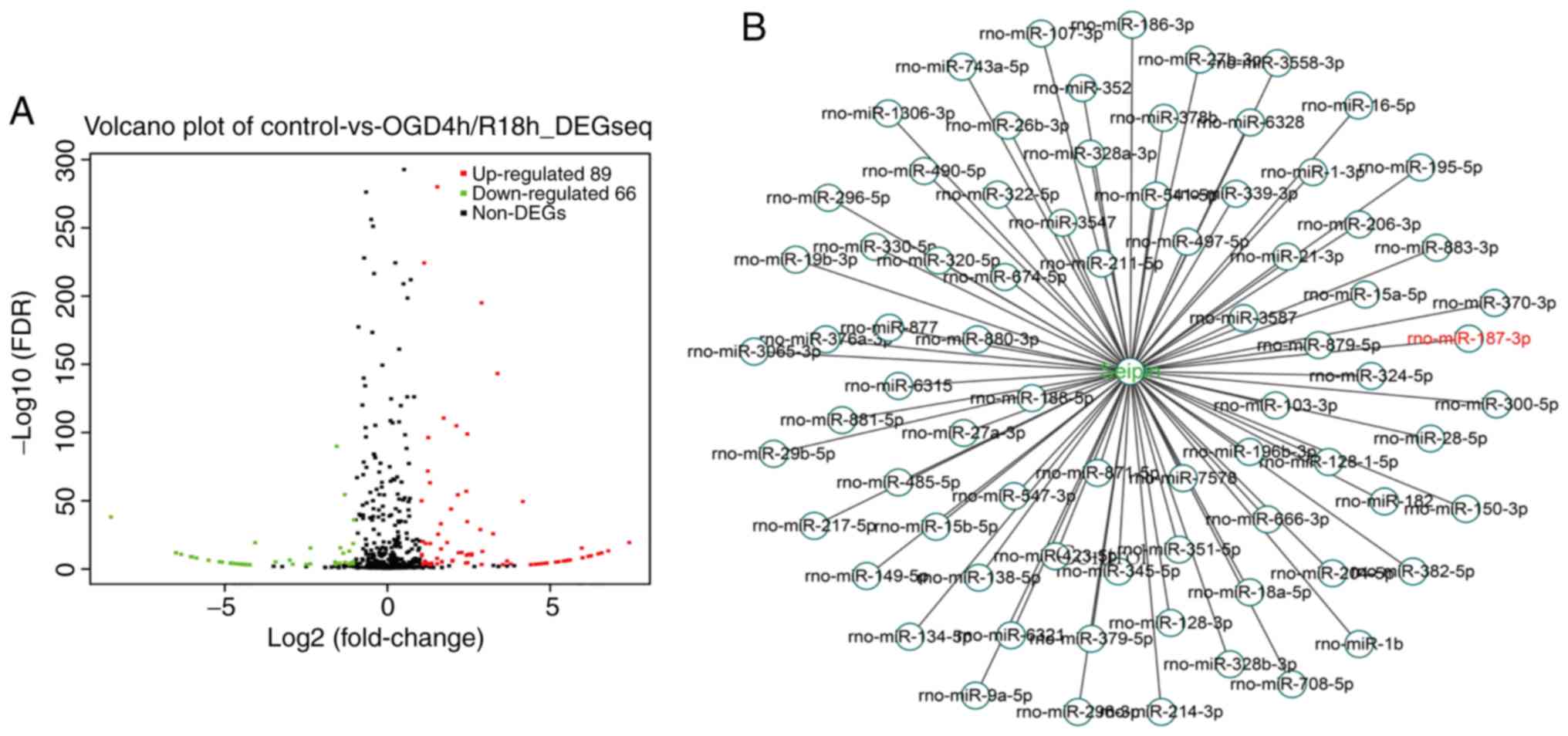
RNA-seq analysis in 4:18-h OGD/R-treated PC12 cells
revealed that 89 miRNAs were upregulated and 66 miRNAs were
downregulated (Fig. 1A). Using
bioinformatics analysis, 76 miRNAs targeting the CDS of Seipin were
identified (Fig. 1B). A heatmap
revealed that 18 miRNAs were significantly differentially expressed
with a fold change >1, including miR-187-3p (Fig. 1C). RT-qPCR analysis showed that
miR-187-3p levels were increased in OGD/R PC12 cells compared with
control cells (n=3; P<0.05; Fig.
1D), whereas the level of Seipin mRNA was unchanged (n=3;
P>0.05; Fig. 1E). However, the
results from western blotting found that the levels of Seipin
protein were significantly decreased in OGD/R PC12 cells (n=3;
P<0.05; Fig. 1F). The results
indicated that the increased miR-187-3p may directly alter the
Seipin expression.
Seipin is a downstream target of
miR-187-3p
To test whether miR-187-3p reduced Seipin expression
in OGD/R PC12 cells, two miR-187-3p binding sites were predicted in
the CDS of Seipin using miRanda (Fig.
2A). As shown in Fig. 2B, a
luciferase assay validated Seipin as a target of miR-187-3p,
indicating that miR-187-3p can regulate Seipin expression and
function. Co-transfection of a dual-luciferase reporter plasmid
containing the Seipin-wt or Seipin-mu1 CDS of Seipin and miR-187-3p
mimics caused a decrease in the reporter activity (P<0.01;
Fig. 2B); only the Seipin-mu2
mutant effectively produced reporter activity. Fig. S1A demonstrated that miR-187-3p
mimic increased the expression of miR-187-3p compared with mimic NC
in the absence of any other treatment, whereas miR-187-3p inhibitor
decreased the expression of miR-187-3p compared with inhibitor NC
group in the absence of any other treatment (Fig. S1B). Neither overexpression of
miR-187-3p by mimics nor inhibition of miR-187-3p by inhibitor in
PC12 cells altered the levels of Seipin mRNA (n=3; P>0.05;
Fig. 2C). However, the
administration of miR-187-3p mimics in PC12 cells reduced the
levels of Seipin protein (n=3; P<0.05; Fig. 2D), whereas miR-187-3p inhibitor
partially reversed the reduction in Seipin protein expression in
OGD/R PC12 cells (n=3; P<0.01; Fig. 2E). The results indicated that the
increase in miR-187-3p suppressed Seipin expression in OGD/R PC12
cells.
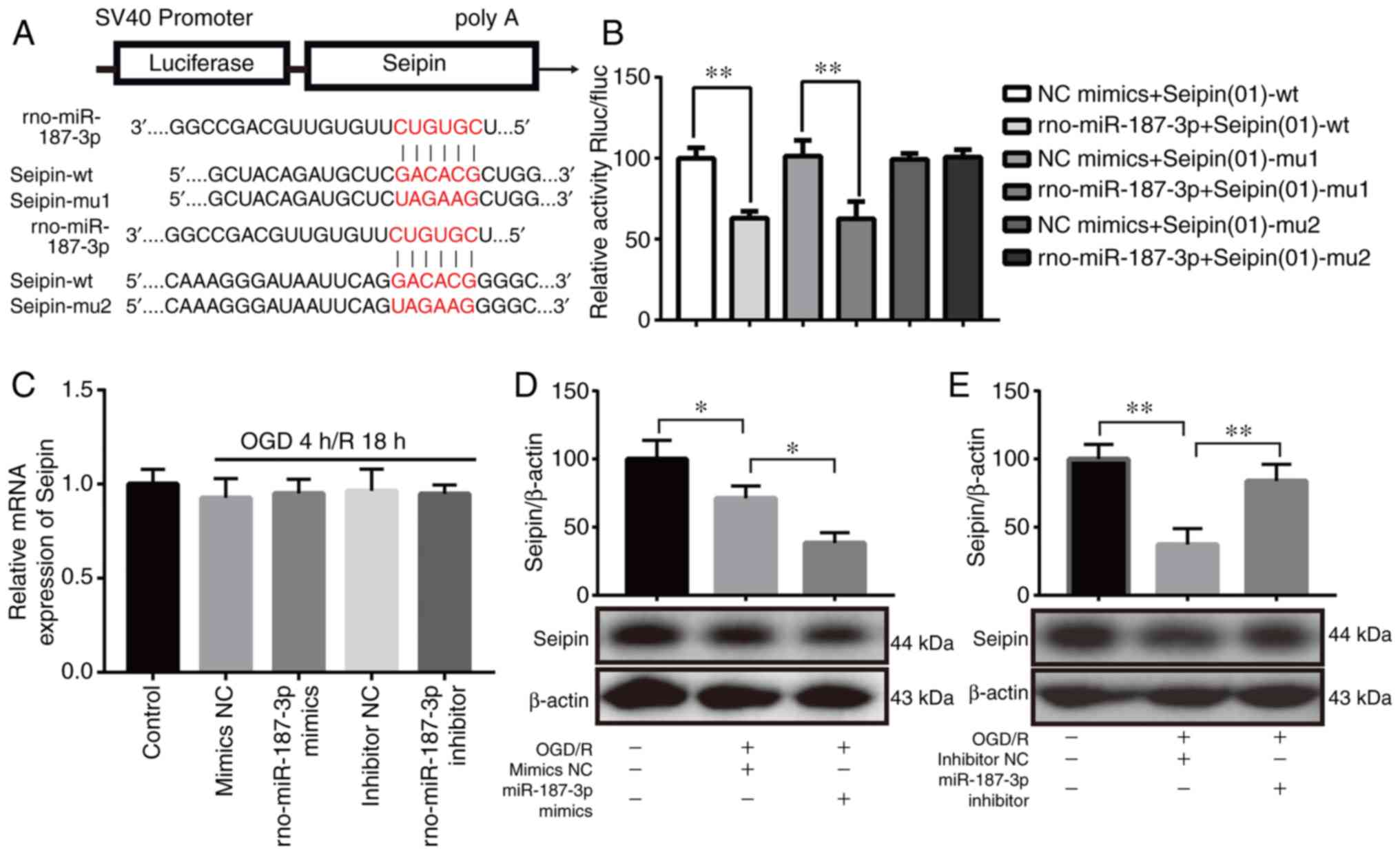 | Figure 2miR-187-3p regulates Seipin
expression by directly targeting its CDS. (A) Schematic diagram
showing the potential miR-187-3p binding sites in the seipin CDS.
Two mutations were introduced in the Seipin CDS by replacing the wt
binding sequence (GACACG) with a mutant sequence (UAGAAG). (B)
Inhibition of the relative luciferase activity of the Seipin CDS
reporter in 293T cells by miR-187-3p. (C) Seipin mRNA levels are
unchanged after the addition of miR-187-3p mimics or inhibitor. (D)
Western blot assay showing decreased Seipin protein in OGD/R +
miR-187-3p mimic-treated cells when compared with OGD/R + mimic
NC-treated cells. (E) Western blot assay showing increased Seipin
protein in OGD/R + miR-187-3p inhibitor-treated cells when compared
with OGD/R + inhibitor NC-treated cells. Data are the mean ± SD;
n=3/group. *P<0.05, **P<0.01. miR,
microRNA; OGD/R, oxygen-glucose deprivation and reoxygenation; NC,
negative control; rno, Rattus norvegicus; CDS, coding
sequence; wt, wild-type; mu, mutant; Rluc, Renilla
luciferase; fluc, firefly luciferase. |
Increased miR-187-3p disturbs autophagic
flux and aggravates apoptosis in OGD/R PC12 cells
To test whether miR-187-3p is involved in
alterations in autophagic flux and apoptosis, OGD/R-treated PC12
cells were transfected with miR-187-3p mimics or inhibitor. It was
determined by qPCR that miR-187-3p mimics caused a significant
increase in the levels of miR-187-3p in OGD/R PC12 cells compared
with mimics NC-transfected OGD/R-treated PC12 cells (n=3;
P<0.05), whereas miR-187-3p inhibitor reduced miR-187-3p levels
in OGD/R PC12 cells (n=3; P<0.05; Fig. 3A). In comparison with mimics
NC-transfected OGD/R PC12 cells, exogenously overexpressing
miR-187-3p in OGD/R-treated PC12 cells aggravated the impairment of
autophagic flux and increased apoptosis; the levels of p62 were
higher (n=3; P<0.05; Fig. 3C and
D) and the LC3II/I ratio was lower (n=3; P<0.05; Fig. 3C and E), but also the levels of
cleaved caspase-3 (n=3; P<0.05; Fig. 3C and F) and Bax (n=3; P<0.05;
Fig. 3C and G) were higher. By
contrast, the miR-187-3p inhibitor in OGD/R PC12 cells decreased
the levels of p62 (n=3; P<0.05), increased the LC3II/I ratio
(n=3; P<0.05), and decreased the levels of cleaved caspase-3
(n=3; P<0.05) and Bax (n=3; P<0.05), indicating that
autophagic flux was recovered and apoptosis was decreased. The
results indicated that increased miR-187-3p causes impairment of
autophagic flux and apoptosis in OGD/R-treated PC12 cells.
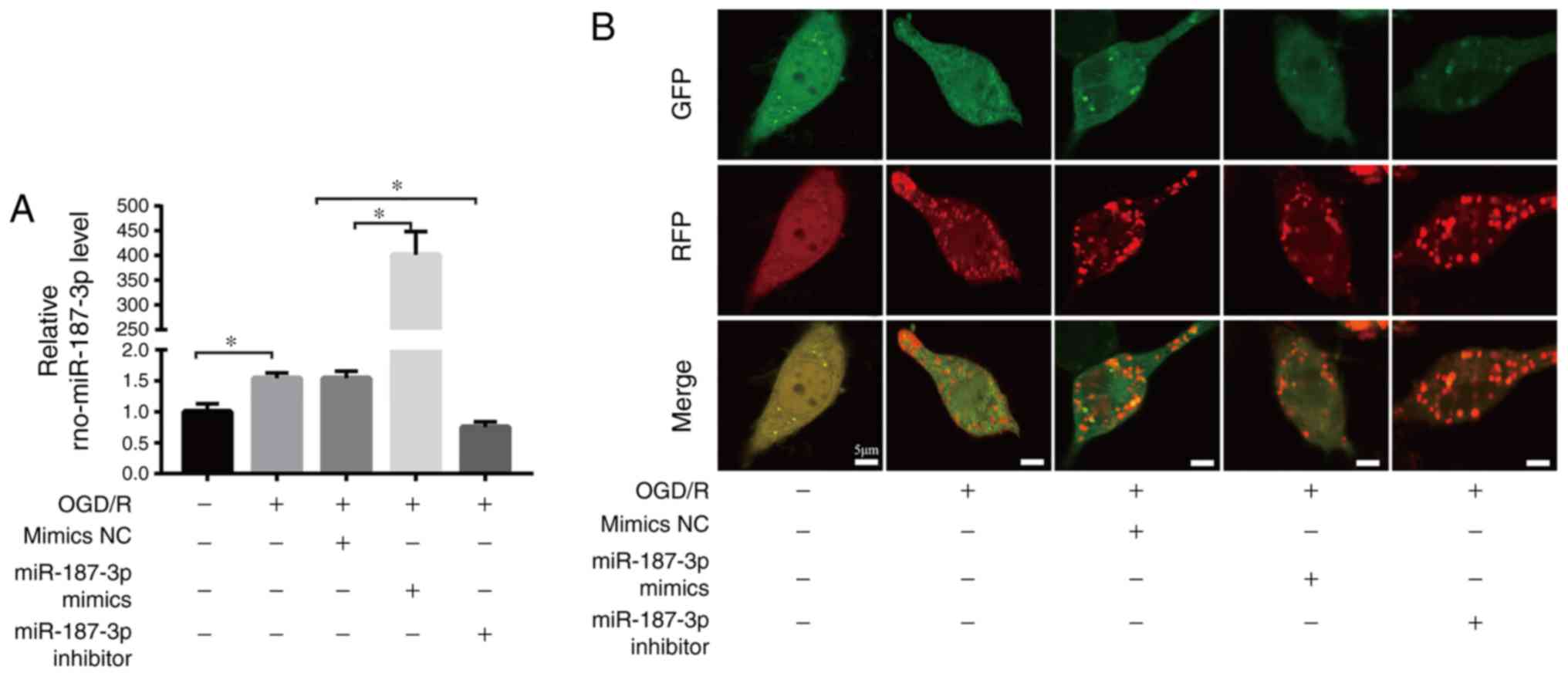 | Figure 3miR-187-3p regulates autophagic flux
and apoptosis in OGD/R. (A) Transfection efficiency was determined
by quantitative PCR. (B) Representative confocal images of GFP and
RFP fluorescent puncta were observed by laser scanning confocal
microscopy in PC12 cells treated with Ad-mRFP-GFP-LC3 plasmid.
Scale bar=5 µm. (C) Representative western blots of
autophagy- and apoptosis-related proteins in the different groups.
Quantification (D) p62, (E) LC3II/LC3I, (F) c-caspase-3 and (G) Bax
levels in OGD/R PC12 cells. Data are the mean ± SD; n=3/group.
*P<0.05, **P<0.01. miR, microRNA;
OGD/R, oxygen-glucose deprivation and reoxygenation; NC, negative
control; rno, Rattus norvegicus; Ad, adenovirus; LC3, light
chain 3; c-, cleaved. |
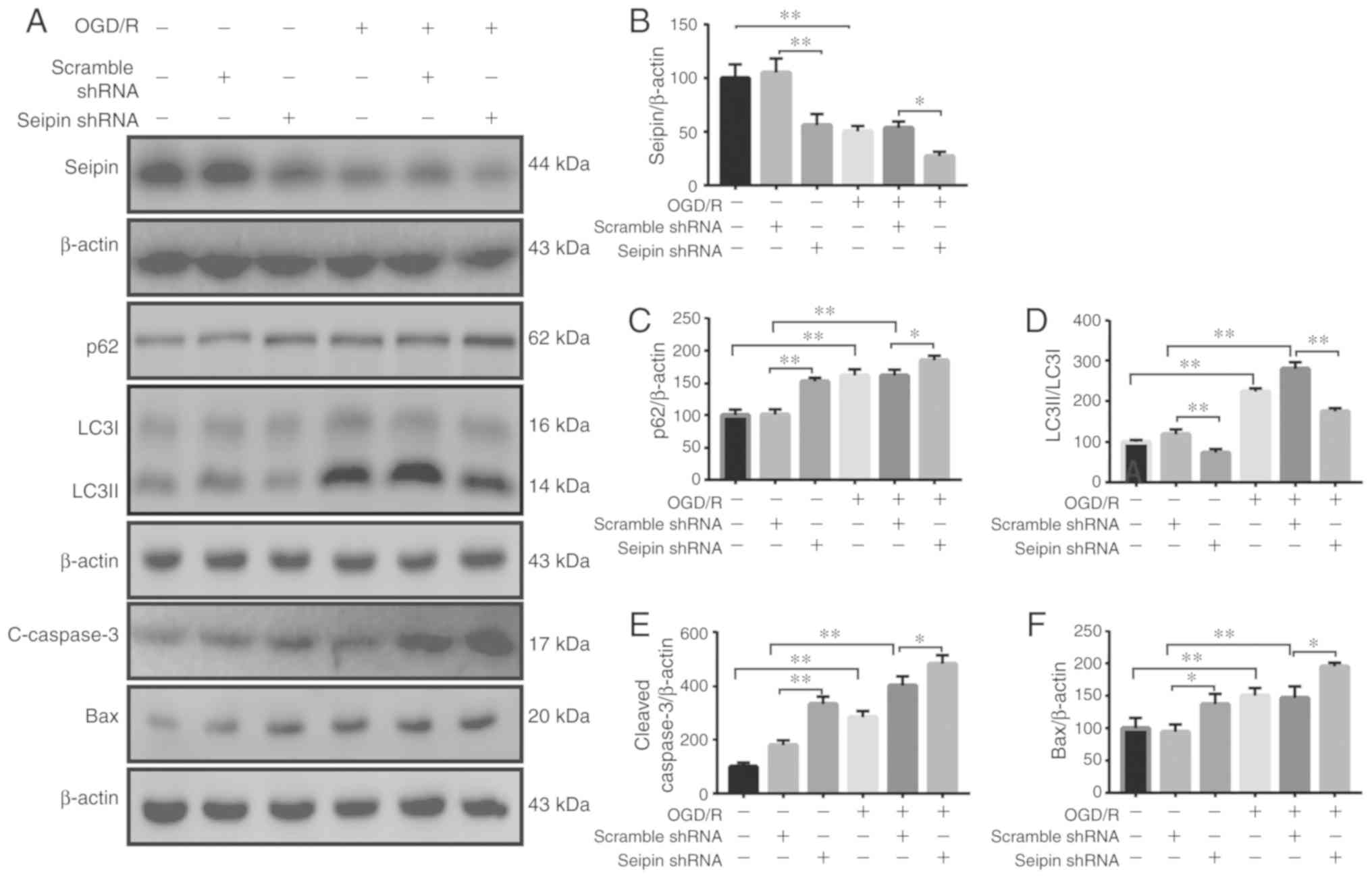
Reduced Seipin disturbs autophagic flux
and aggravates apoptosis in OGD/R PC12 cells
These results indicated that increased miR-187-3p
suppresses Seipin expression in OGD/R PC12 cells. Therefore,
experiments were performed to examine whether reduced Seipin
expression is associated with impaired autophagic flux and
increased apoptosis in OGD/R PC12 cells. Seipin expression was
decreased in normoxic control (n=3; P<0.05) and OGD/R PC12 cells
(n=3; P<0.05; Fig. 4A and B)
following transfection of Seipin shRNA. In comparison with
vehicle-treated PC12 cells, the levels of p62 (n=3; P<0.05;
Fig. 4A and C) were elevated, the
LC3II/I ratio was decreased (n=3; P<0.05; Fig. 4A and D), and the levels of cleaved
caspase-3 (n=3; P<0.05; Fig. 4A
and E) and Bax (n=3; P<0.05; Fig. 4A and F) were increased in normoxic
control and OGD/R-treated PC12 cells following Seipin shRNA
transfection. These results indicated that reduced Seipin
expression disturbed autophagic flux and aggravated apoptosis in
OGD/R PC12 cells.
Rapamycin does not rescue autophagy flux
and apoptosis in Seipin knockdown PC12 cell after OGD/R
Seipin knockdown has been reported to reduce
hippocampal autophagosome formation by enhancing Akt/mTOR activity
(29). To investigate whether
Seipin in PC12 cells regulated mTOR activity to regulate autophagic
flux and apoptosis, the levels of phosphorylated (p-)mTOR were
examined in PC12 cells. The levels of p-mTOR were lower in
OGD/R-treated PC12 cells than normoxic controls; however, Seipin
knockdown partially upregulated p-mTOR (n=3; P<0.05; Fig. 5A and B). Indeed, the
administration of mTOR inhibitor rapamycin in Seipin
shRNA-transfected PC12 cells failed to correct the increase in the
level of p62 (n=3; P<0.05; Fig. 5A
and C) and decrease in LC3II/I (n=3; P<0.05; Fig. 5A and D). Furthermore, rapamycin
failed to correct the increase in cleaved caspase-3 (n=3;
P<0.05; Fig. 5A and E) and Bax
(n=3; P<0.05; Fig. 5A and F).
The results indicated that reductions in Seipin expression
disturbed autophagic flux and aggravated apoptosis in PC12
cells.
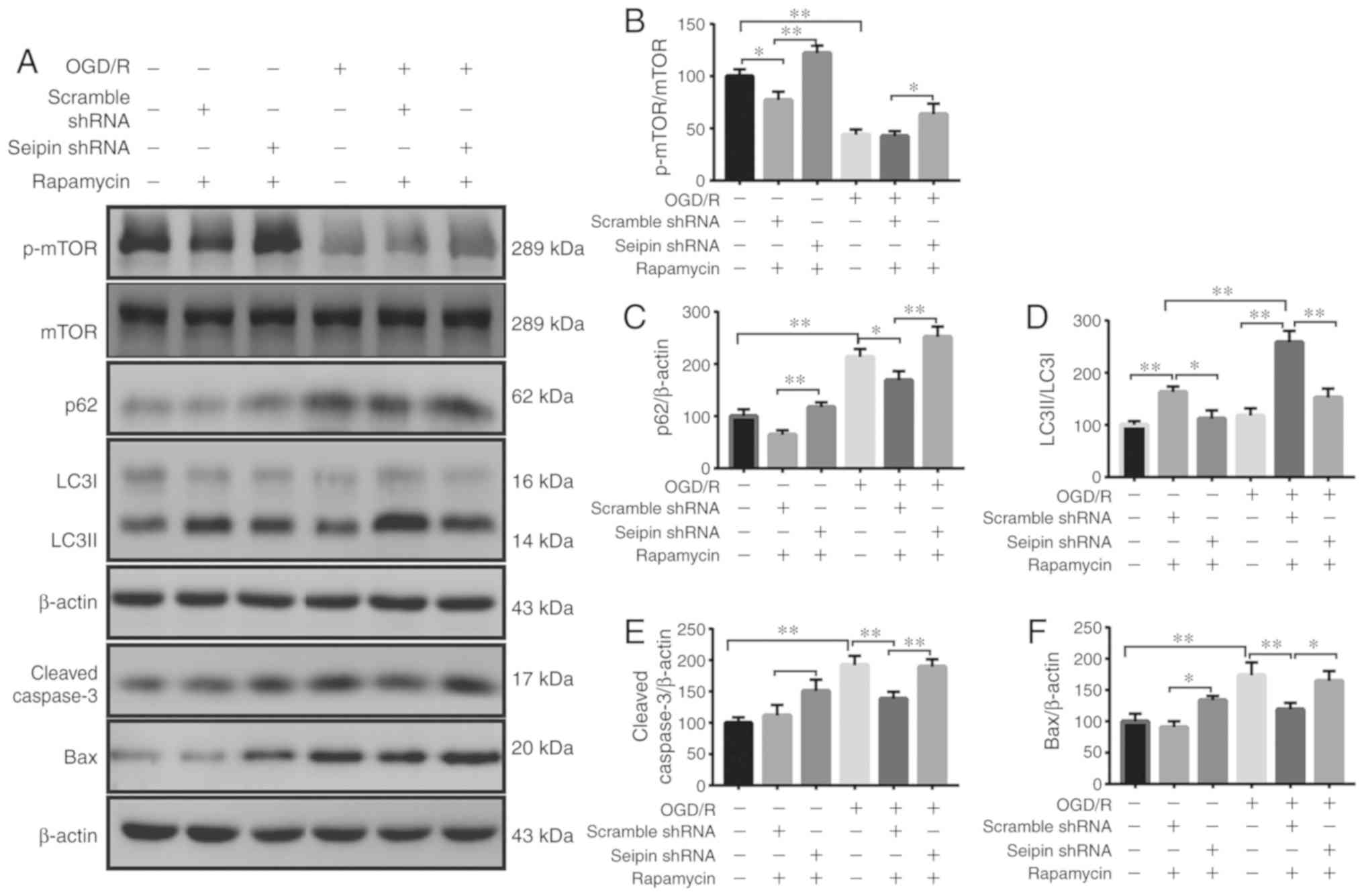 | Figure 5Effects of rapamycin on the
expression of autophagy- and apoptosis-related proteins in
Seipin-silenced PC12 cells. (A) Representative western blot images
of the autophagy-related proteins p-mTOR, p62 and LC3II/I, and the
apoptosis-related proteins c-caspase-3 and Bax in the different
groups. Quantitative analysis of (B) p-mTOR/mTOR, (C) p62, (D)
LC3II/LC3I, (E) c-caspase 3 and (F) Bax levels. Data are the mean ±
SD; n=3/group. *P<0.05, **P<0.01. miR,
microRNA; OGD/R, oxygen-glucose deprivation and reoxygenation;
shRNA, short hairpin RNA; LC3, light chain 3; c-, cleaved; p-,
phosphorylated. |
Seipin knockdown blocks miR-187-3p
inhibitor-mediated neuroprotection in vitro
Next, it was examined whether Seipin knockdown would
attenuate the effects on autophagic flux and apoptosis mediated by
miR-187-3p inhibitor. The levels of p62 (n=3; P<0.05; Fig. 6A and B), cleaved caspase-3 (n=3;
P<0.05; Fig. 6A and D) and Bax
(n=3; P<0.05; Fig. 6A and E)
were higher in OGD/R-treated PC12 cells than normoxic controls,
whereas the LC3II/I ratio (n=3; P<0.05; Fig. 6A and C) was lower. Furthermore,
flow cytometry revealed increased apoptosis in OGD/R-treated PC12
cells (Fig. 6F and G).
Administration of miR-187-3p inhibitor in Seipin shRNA-transfected
PC12 cells failed to rescue the increased levels of p62, apoptosis,
cleaved caspase-3 and Bax, and the decreased LC3II/I ratio. The
results suggested that the regulation of autophagic flux and
apoptosis by miR-187-3p was dependent upon Seipin.
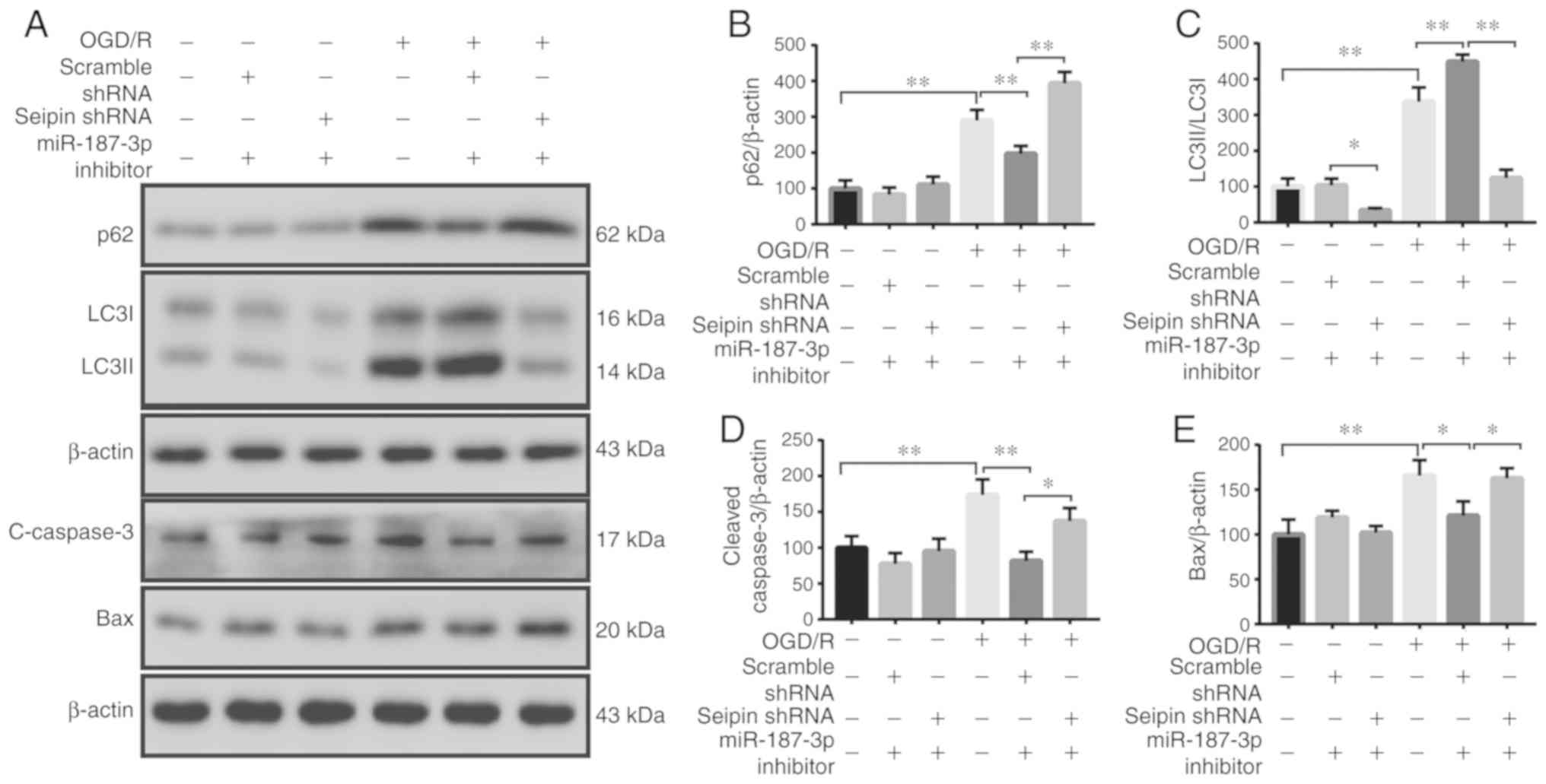 | Figure 6Effects of Seipin knockdown on
autophagy and apoptosis mediated by miR-187-3p inhibitor in OGD/R
PC12 cells. (A) Representative western blot images of the
autophagy-related proteins p62 and LC3II/I, and the
apoptosis-related proteins c-caspase-3 and Bax in the different
groups. Quantitative analysis of (B) p62, (C) LC3II/LC3I, (D)
c-caspase 3 and (E) Bax levels. (F) Flow cytometry assay and (G)
quantification showing that Seipin knockdown abolishes miR-187-3p
inhibitor-mediated neuroprotection in OGD/R-treated PC12 cells.
Data are the mean ± SD; n=3/group. *P<0.05,
**P<0.01. miR, microRNA; OGD/R, oxygen-glucose
deprivation and reoxygenation; shRNA, short hairpin RNA; LC3, light
chain 3; c-, cleaved; APC, allophycocyanin; 7-AAD,
7-aminoactinomyosin D. |
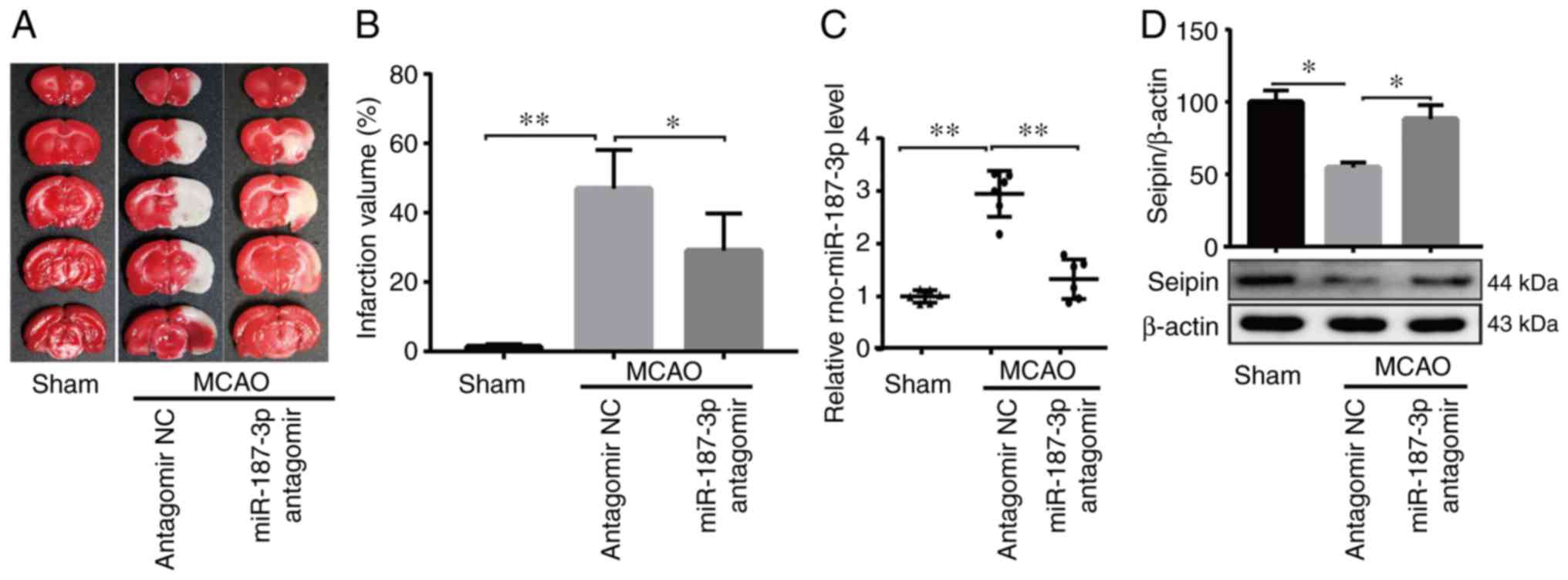
miR-187-3p antagomir reduces
ischemia-induced infarction and upregulates Seipin-mediated
autophagy in rats
A previous study reported that Seipin knockout in
mice exacerbated cerebral damage induced by I/R (26). The in vitro results from
the present study indicated that the OGD/R-induced increase in
miR-187-3p expression and suppression of Seipin expression promoted
neuronal apoptosis. Therefore, in vitro experiments examined
the effect of miR-187-3p antagomir on ischemia-induced brain injury
24 h after 60 min of MCAO. The infarct volume was observed in MCAO
rats, which was reduced by treatment with miR-187-3p antagomir
(n=6; P<0.05; Fig. 7A and B).
miR-187-3p antagomir was used to inhibit miR-187-3p expression in
rats (n=6; P<0.05; Fig. 7C),
which resulted in upregulation of Seipin (n=6; P<0.05; Fig. 7D) and LC3 (n=6; Fig. 7E).
Discussion
The present study present for the first time, to our
knowledge, in vivo evidence that the ischemia-induced
increase in miR-187-3p and subsequent suppression of Seipin
expression led to deficits in autophagic flux and increased
neuronal apoptosis. This conclusion is based on the following
experimental data: i) OGD/R caused an increase in miR-187-3p
expression and a decrease in Seipin protein levels; ii)
bioinformatics analysis found miR-187-3p binding sites in the CDS
of Seipin, and the reduction of Seipin protein in OGD/R-treated
PC12 cells could be reversed by the inhibition of miR-187-3p; iii)
autophagic flux was reduced in OGD/R-treated PC12 cells along with
an increase in apoptosis-related protein expression, which was
sensitive to the inhibition of miR-187-3p expression; iv) the
impairment of autophagic flux and increase in neuronal apoptosis
were aggravated in OGD/R-treated PC12 cells following Seipin
knockdown, which was insensitive to inhibition of miR-187-3p; and
v) the inhibition of miR-187-3p in a mouse model of cerebral I/R
decreased the infarct volume.
In the present study, the level of Seipin mRNA was
unchanged, but the level of Seipin protein was significantly
decreased in OGD/R PC12 cells, which could be partially reversed by
miR-187-3p inhibitor. The overexpression of miR-187-3p in PC12
cells reduced the level of Seipin protein. The results indicated
that the increased miR-187-3p levels suppressed Seipin expression
in OGD/R PC12 cells. miR-187-3p has been identified as a
cancer-related miRNA that plays promotive or suppressive roles in
different malignancies (36-40). miR-187-3p inhibits the migration
and invasion of hepatocellular carcinoma by suppressing S100
calcium-binding protein A4 (41).
In mice, miR-187-3p mimic relieved I/R-induced pain
hypersensitivity by inhibiting P2X7 receptor expression and
subsequent mature IL-1β release (42).
Co-expression of human Seipin and ubiquitin showed
that Seipin is polyubiquitinated, and its ubiquitination is
enhanced by mutation (43);
treatment of cells with a proteasome inhibitor increased the
quantity of mutant Seipin in the cells, suggesting that it is
degraded through the ER-associated degradation pathway. Of note,
the N88S and S90L mutations are in the N-glycosylation motif, and
these mutations enhance ubiquitination and degradation of Seipin
via the ubiquitin-proteasome system (44). Conversely, the transcript encoding
the long Seipin isoform (BSCL2-203, 462 aa) is expressed primarily
in the brain, and its expression is inversely correlated with age
in neuronal cells (24).
Autophagy plays a role in the pathophysiological
process of ischemia-induced brain injury (45). The monitoring of autophagic flux
is essential for estimating autophagic activity. Autophagic flux is
a dynamic process that involves the formation of autophagosomes,
delivery of autophagic substrates to the autophagosomes and the
degradation of these substrates (46). LC3I is post-translationally
modified during autophagy induction to form LC3II; thus, LC3II/I is
an index of autophagosome formation (47). LC3II/I, as an index of
autophagosome formation, was increased in OGD/R-treated PC12 cells.
The mammalian protein p62 is a selective autophagy substrate,
continuously degraded by autophagy (48). It has been demonstrated that p62
binds directly to LC3, and this motif is required for autophagic
degradation of p62 (49).
Consistent with previous studies (50,51), the level of p62 was increased in
OGD/R PC12 cells, indicating impaired autophagic flux. Of note, the
decreased Seipin expression and impairment of autophagic flux in
OGD/R PC12 cells could be ameliorated by the inhibition of
miR-187-3p. In addition, knockdown of Seipin in OGD/R PC12 cells
further aggravated the impairment of autophagic flux, which was
insensitive to inhibition of miR-187-3p. The results supported the
hypothesis that Seipin deficiency leads to an impairment in
autophagic flux. mTOR signaling is a major negative regulator of
autophagy (52). Neuronal Seipin
deletion has been found to reduce autophagosome formation by
enhancing mTOR signaling (29).
The present findings indicated that a reduction in Seipin
expression disturbed autophagic flux and aggravated apoptosis in in
PC12 cells. Long-lasting autophagy or impaired autophagic flux
induces type II programmed cell death (53-55). It was previously observed that the
knockdown of Seipin exacerbated cerebral I/R-induced damage in mice
(26). The increased expression
of apoptosis-related proteins in OGD/R PC12 cells was corrected by
the inhibition of miR-187-3p. Additionally, the knockdown of Seipin
in OGD/R-treated PC12 cells further increased apoptosis, which was
insensitive to the inhibition of miR-187-3p.
In conclusion, it was demonstrated that miR-187-3p
inhibitor protected against OGD/R-induced apoptosis by upregulating
Seipin-mediated autophagic flux in PC12 cells. It is hypothesized
that enhanced activation of autophagic flux supports functional
recovery, whereas impaired autophagic flux is associated with
increased apoptosis. The present findings suggested that the
miR-187-3p/Seipin axis has potential for the treatment of cerebral
I/R injury in the future.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81360199), Special
Grant for Central Government Supporting Local Science and
Technology Development, Science and Technology Department of
Guizhou Province [grant no. (2019) 4008], Science and Technology
Plan Project of Guizhou Province (Basic Science and Technology
Cooperation) [grant no. (2020)1Z060], the Science and Technology
Fund Project of Guizhou Health and Health Commission (grant no.
gzwjkj2019-1-039), and the Science and Technology Fund Project of
Southwest Guizhou Autonomous Prefecture (grant no. 2019-1-10).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WY and ZR designed the study. PX, JL and YH
performed the experiments. ZR drafted the manuscript. ZG and LC
contributed to the analysis of data and designed the animal
experiment. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Use Committee of Guizhou Medical University (approval no.
1900813).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the Global
Burden of Disease Study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Deng YH, He HY, Yang LQ and Zhang PY:
Dynamic changes in neuronal autophagy and apoptosis in the ischemic
penumbra following permanent ischemic stroke. Neural Regen Res.
11:1108–1114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheon SY, Kim EJ, Kim SY, Kim JM, Kam EH,
Park JK and Koo BN: Apoptosis signal-regulating kinase 1 silencing
on astroglial inflammasomes in an experimental model of ischemic
stroke. Neuroscience. 390:218–230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang S, An Q, Wang T, Gao S and Zhou G:
Autophagy- and MMP-2/9-mediated reduction and redistribution of
ZO-1 contribute to Hyperglycemia-increased blood-brain barrier
permeability during early reperfusion in stroke. Neuroscience.
377:126–137. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cassidy JM and Cramer SC: Spontaneous and
therapeutic-induced mechanisms of functional recovery after stroke.
Transl Stroke Res. 8:33–46. 2017. View Article : Google Scholar
|
|
6
|
Chan SJ, Love C, Spector M, Cool SM,
Nurcombe V and Lo EH: Endogenous regeneration: Engineering growth
factors for stroke. Neurochem Int. 107:57–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spector WD, Limcangco R, Furukawa MF and
Encinosa WE: The marginal costs of adverse drug events associated
with exposures to anticoagulants and hypoglycemic agents during
hospitalization. Med Care. 55:856–863. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo WT and Wang Y: Dgcr8 knockout
approaches to understand microRNA functions in vitro and in vivo.
Cell Mol Life Sci. 76:1697–1711. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu H, Zhang Y, Tang R, Qu H, Duan X and
Jiang Y: Banana sRNAome and degradome identify microRNAs
functioning in differential responses to temperature stress. BMC
Genomics. 20:332019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Blenkiron C, Askelund KJ, Shanbhag ST,
Chakraborty M, Petrov MS, Delahunt B, Windsor JA and Phillips A:
MicroRNAs in mesenteric lymph and plasma during acute pancreatitis.
Ann Surg. 260:341–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fatimy El R, Li S, Chen Z, Mushannen T,
Gongala S, Wei Z, Balu DT, Rabinovsky R, Cantlon A, Elkhal A, et
al: MicroRNA-132 provides neuroprotection for tauopathies via
multiple signaling pathways. Acta Neuropathol. 136:537–555. 2018.
View Article : Google Scholar
|
|
13
|
Huang W: MicroRNAs: Biomarkers,
diagnostics, and therapeutics. Methods Mol Biol. 1617:57–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han B, Zhang Y, Zhang Y, Bai Y, Chen X,
Huang R, Wu F, Leng S, Chao J, Zhang JH, et al: Novel insight into
circular RNA HECTD1 in astrocyte activation via autophagy by
targeting MIR142-TIPARP: Implications for cerebral ischemic stroke.
Autophagy. 14:1164–1184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du K, Zhao C, Wang L, Wang Y, Zhang KZ,
Shen XY, Sun HX, Gao W and Lu X: MiR-191 inhibit angiogenesis after
acute ischemic stroke targeting VEZF1. Aging (Albany NY).
11:2762–2786. 2019. View Article : Google Scholar
|
|
16
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Duursma AM, Kedde M, Schrier M, le-Sage C
and Agami R: MiR-148 targets human DNMT3b protein coding region.
RNA. 14:872–877. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang K, Li W, Bai Y, Yang W, Ling Y and
Fang M: ssc-miR-7134-3p regulates fat accumulation in castrated
male pigs by targetingMARK4gene. Int J Biol Sci. 13:189–197. 2017.
View Article : Google Scholar :
|
|
20
|
Ma L, Chen X, Li C, Cheng R, Gao Z, Meng
X, Sun C, Liang C and Liu Y: miR-129-5p and -3p co-target WWP1 to
suppress gastric cancer proliferation and migration. J Cell
Biochem. Nov 11–2018.Epub ahead of print.
|
|
21
|
Su WC, Lin YH, Pagac M and Wang CW: Seipin
negatively regulates sphingolipid production at the ER-LD contact
site. J Cell Biol. 218:3663–3680. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Castro IG, Eisenberg-Bord M, Persiani E,
Rochford JJ, Schuldiner M and Bohnert M: Promethin is a conserved
seipin partner protein. Cells. 8:2682019. View Article : Google Scholar :
|
|
23
|
Ding L, Yang X, Tian H, Liang J, Zhang F,
Wang G, Wang Y, Ding M, Shui G and Huang X: Seipin regulates lipid
homeostasis by ensuring calcium-dependent mitochondrial metabolism.
EMBO J. 37:e975722018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sánchez-Iglesias S, Fernández-Liste A,
Guillín-Amarelle C, Rábano A, Rodriguez-Cañete L, González-Méndez
B, Fernández-Pombo A, Senra A and Araújo-Vilar D: Does seipin play
a role in oxidative stress protection and peroxisome biogenesis?
New insights from human brain autopsies. Neuroscience. 396:119–137.
2019. View Article : Google Scholar
|
|
25
|
Ito D, Fujisawa T, Iida H and Suzuki N:
Characterization of seipin/BSCL2, a protein associated with spastic
paraplegia 17. Neurobiol Dis. 31:266–277. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Y, Wei L, Tian J, Wang YH, Liu G and
Wang C: Seinpin knockout exacerbates cerebral ischemia/reperfusion
damage in mice. Biochem Biophys Res Commun. 474:377–383. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zowalaty AEE and Ye X: Seipin deficiency
leads to defective parturition in mice. Biol Reprod. 97:378–386.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan HD, Chen SP, Sun YX, Xu SH and Wu LJ:
Seipin mutation at glycosylation sites activates autophagy in
transfected cells via abnormal large lipid droplets generation.
Acta Pharmacol Sin. 36:497–506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang H, Di T, Wang Y, Zeng X, Li G, Wan
Q, Yu W and Chen L: Seipin deletion in mice enhances
phosphorylation and aggregation of tau protein through reduced
neuronal PPARγ and insulin resistance. Neurobiol Dis. 127:350–361.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tao J, Liu W, Shang G, Zheng Y, Huang J,
Lin R and Chen L: miR-207/352 regulate lysosomal-associated
membrane proteins and enzymes following ischemic stroke.
Neuroscience. 305:1–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen F, Zhang L, Wang E, Zhang C and Li X:
LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA
for miR-137 to regulate the Notch1 signaling pathway. Biochem
Biophys Res Commun. 496:184–190. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
33
|
Yan H, Yuan J, Gao L, Rao J and Hu J: Long
noncoding RNA MEG3 activation of p53 mediates ischemic neuronal
death in stroke. Neuroscience. 337:191–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Slott VL, Linder RE and Dyer CJ: Method of
euthanasia does not affect sperm motility in the laboratory rat.
Reprod Toxicol. 8:371–374. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu Q, Manaenko A, Bian H, Guo Z, Huang JL,
Guo ZN, Yang P, Tang J and Zhang JH: Hyperbaric oxygen reduces
infarction volume and hemorrhagic transformation through
ATP/NAD+/Sirt1 pathway in hyperglycemic middle cerebral
artery occlusion rats. Stroke. 48:1655–1664. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun C, Li S, Yang C, Xi Y, Wang L, Zhang F
and Li D: MicroRNA-187-3p mitigates non-small cell lung cancer
(NSCLC) development through down-regulation of BCL6. Biochem
Biophys Res Commun. 471:82–88. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y,
Fan S and Liu Y: MicroRNA-187, down-regulated in clear cell renal
cell carcinoma and associated with lower survival, inhibits cell
growth and migration though targeting B7-H3. Biochem Biophys Res
Commun. 438:439–444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Casanova-Salas I, Masiá E, Armiñán A,
Calatrava A, Mancarella C, Rubio-Briones J, Scotlandi K, Vicent MJ
and López-Guerrero JA: MiR-187 targets the androgen-regulated gene
ALDH1A3 in prostate cancer. PLoS One. 10:e01255762015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu
Y, Shi J, Sun X, Liu Y, Ding Y and Zhao L: MicroRNA-187, a
downstream effector of TGFβ pathway, suppresses Smad-mediated
epithelial-mesenchymal transition in colorectal cancer. Cancer
Lett. 373:203–213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mulrane L, Madden SF, Brennan DJ, Gremel
G, McGee SF, McNally S, Martin F, Crown JP, Jirström K, Higgins DG,
et al: miR-187 is an independent prognostic factor in breast cancer
and confers increased invasive potential in vitro. Clin Cancer Res.
18:6702–6713. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q,
Yang W, Zheng X, Tu K and Liu Q: MiR-187-3p inhibits the metastasis
and epithelial-mesenchymal transition of hepatocellular carcinoma
by targeting S100A4. Cancer Lett. 381:380–390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li XQ, Yu Q, Zhang ZL, Sun XJ and Ma H:
MiR-187-3p mimic alleviates ischemia-reperfusion-induced pain
hypersensitivity through inhibiting spinal P2X7R and subsequent
mature IL-1β release in mice. Brain Behav Immun. 79:91–101. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ito D and Suzuki N: Seipin/BSCL2-related
motor neuron disease: Seipinopathy is a novel conformational
disease associated with endoplasmic reticulum stress. Rinsho
Shinkeigaku. 47:329–335. 2007.In Japanese. PubMed/NCBI
|
|
44
|
Ito D, Yagi T, Ikawa M and Suzuki N:
Characterization of inclusion bodies with cytoprotective properties
formed by seipinopathy-linked mutant seipin. Hum Mol Genet.
21:635–646. 2012. View Article : Google Scholar
|
|
45
|
Wang P, Xu TY, Wei K, Guan YF, Wang X, Xu
H, Su DF, Pei G and Miao CY: ARRB1/β-arrestin-1 mediates
neuroprotection through coordination of BECN1-dependent autophagy
in cerebral ischemia. Autophagy. 10:1535–1548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Iwai-Kanai E, Yuan H, Huang C, Sayen MR,
Perry-Garza CN, Kim L and Gottlieb RA: A method to measure cardiac
autophagic flux in vivo. Autophagy. 4:322–329. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Holt SV, Wyspianska B, Randall KJ, James
D, Foster JR and Wilkinson RW: The development of an
immunohistochemical method to detect the autophagy-associated
protein LC3-II in human tumor xenografts. Toxicol Pathol.
39:516–523. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pankiv S, Clausen TH, Lamark T, Brech A,
Bruun JA, Outzen H, Øvervatn A, Bjørkøy G and Johansen T:
p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of
ubiquitinated protein aggregates by autophagy. J Biol Chem.
282:24131–24145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Johansen T and Lamark T: Selective
autophagy mediated by autophagic adapter proteins. Autophagy.
7:279–296. 2011. View Article : Google Scholar :
|
|
50
|
Liu X, Tian F, Wang S, Wang F and Xiong L:
Astrocyte autophagy flux protects neurons against oxygen-glucose
deprivation and ischemic/reperfusion injury. Rejuvenation Res.
21:405–415. 2018. View Article : Google Scholar
|
|
51
|
Jiang H, Ma Y, Yan J, Liu J and Li L:
Geniposide promotes autophagy to inhibit insulin resistance in
HepG2 cells via P62/NF-κB/GLUT-4. Mol Med Rep. 16:7237–7244. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao Z, Han F, Yang S, Wu J and Zhan W:
Oxamate-mediated inhibition of lactate dehydrogenase induces
protective autophagy in gastric cancer cells: Involvement of the
Akt-mTOR signaling pathway. Cancer Lett. 358:17–26. 2015.
View Article : Google Scholar
|
|
53
|
Adhami F, Liao G, Morozov YM, Schloemer A,
Schmithorst VJ, Lorenz JN, Dunn RS, Vorhees CV, Wills-Karp M, Degen
JL, et al: Cerebral ischemia-hypoxia induces intravascular
coagulation and autophagy. Am J Pathol. 169:566–583. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT,
Liu B and Bao JK: Programmed cell death pathways in cancer: A
review of apoptosis, autophagy and programmed necrosis. Cell
Prolif. 90:487–498. 2012. View Article : Google Scholar
|
|
55
|
McCrary MR, Jiang MQ, Giddens MM, Zhang
JY, Owino S, Wei ZZ, Zhong W, Gu X, Xin H, Hall RA, et al:
Protective effects of GPR37 via regulation of inflammation and
multiple cell death pathways after ischemic stroke in mice. FASEB
J. 33:10680–10691. 2019. View Article : Google Scholar : PubMed/NCBI
|