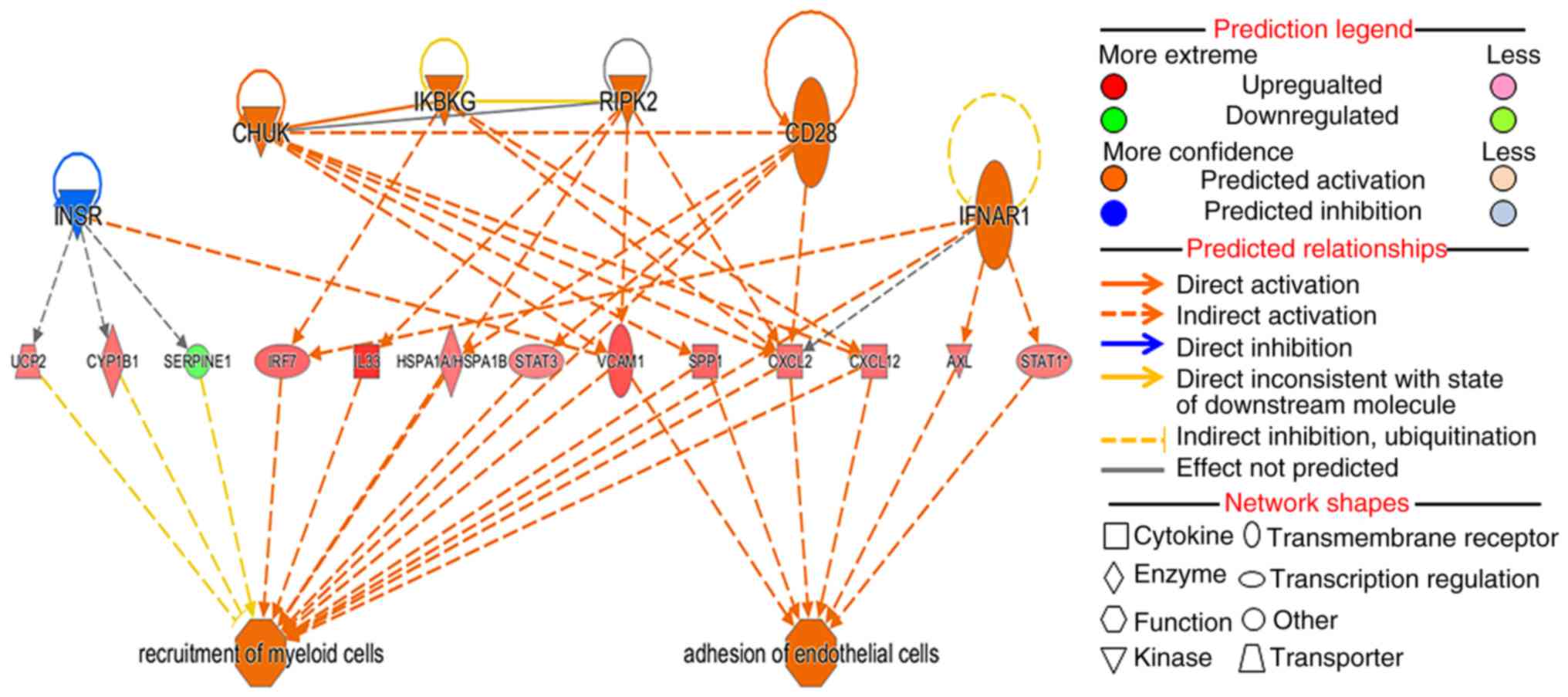
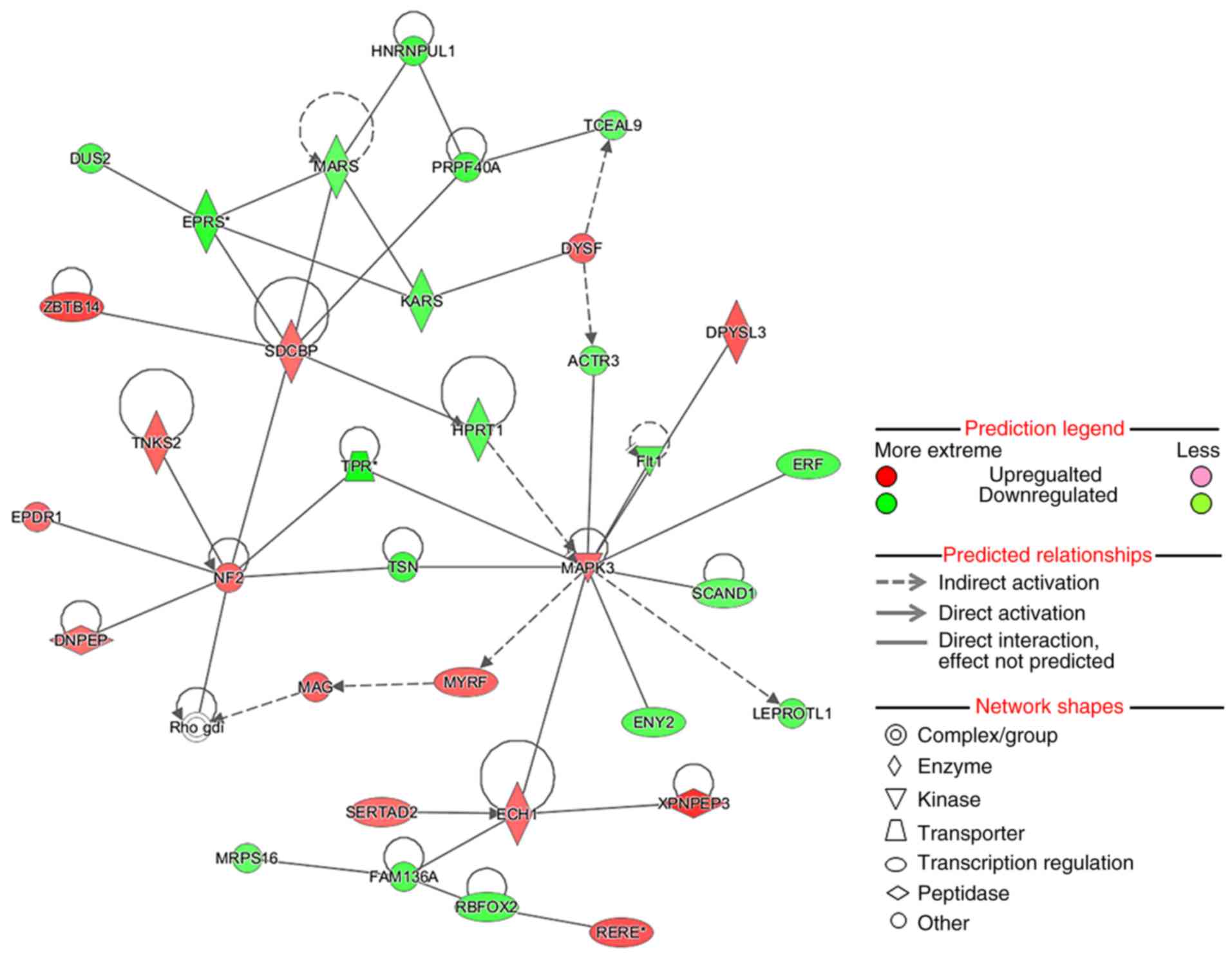
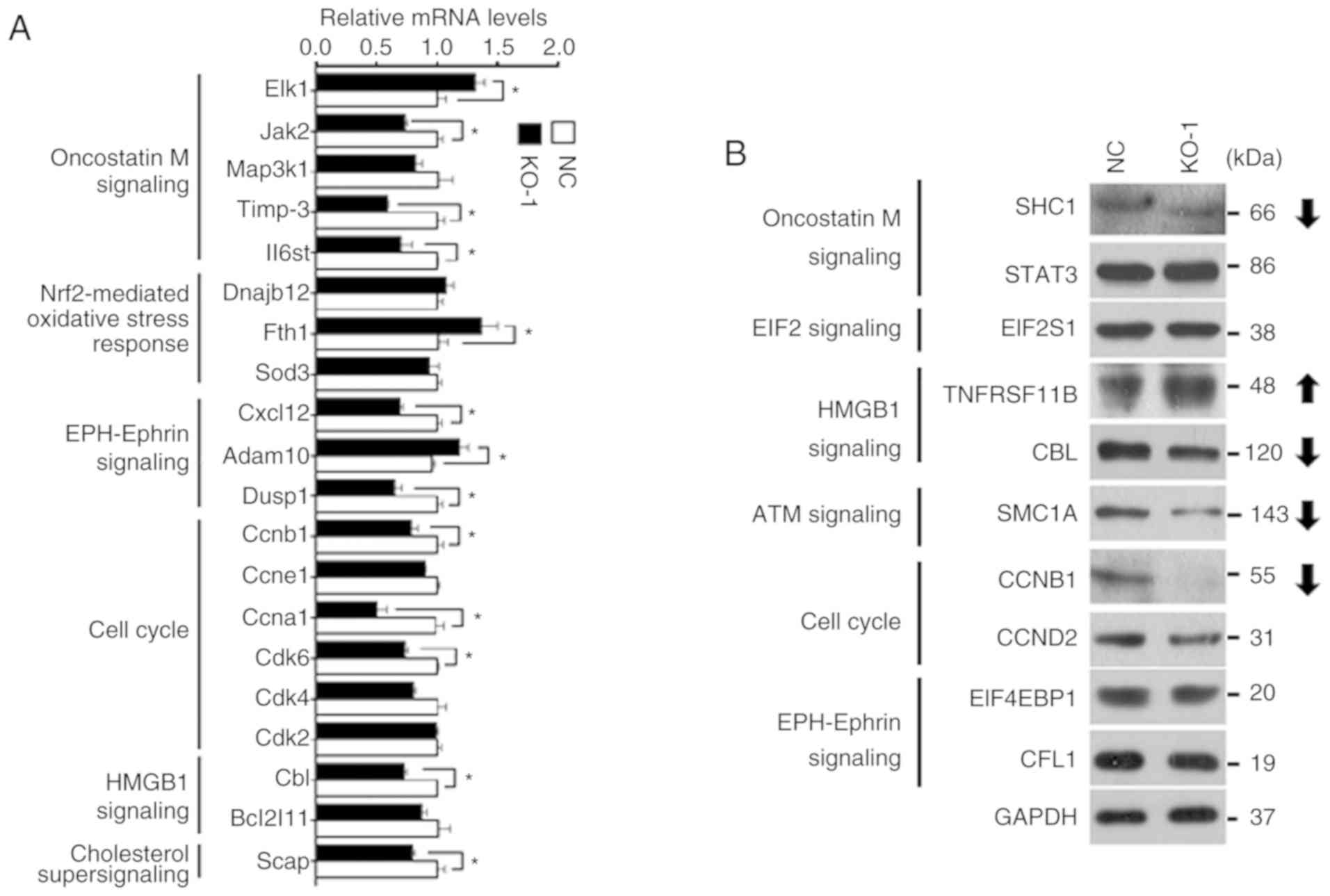
|
1
|
Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J
and Settleman J: Identification of a novel human rho protein with
unusual properties: GTPase deficiency and in vivo farnesylation.
Mol Cell Biol. 16:2689–2699. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jie W, Andrade KC, Lin X, Yang X, Yue X
and Chang J: Pathophysiological functions of Rnd3/Rhoe. Compr
Physiol. 6:169–186. 2016.PubMed/NCBI
|
|
3
|
Riento K, Villalonga P, Garg R and Ridley
A: Function and regulation of RhoE. Biochem Soc Trans. 33:649–651.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chardin P: Function and regulation of rnd
proteins. Nat Rev Mol Cell Biol. 7:54–62. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Paysan L, Piquet L, Saltel F and Moreau V:
Rnd3 in Cancer: A review of the evidence for tumor promoter or
suppressor. Mol Cancer Res. 14:1033–1044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guasch RM, Scambler P, Jones GE and Ridley
AJ: RhoE regulates actin cytoskeleton organization and cell
migration. Mol Cell Biol. 18:4761–4771. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Klein RM and Aplin AE: Rnd3 regulation of
the actin cytoskeleton promotes melanoma migration and invasive
outgrowth in three dimensions. Cancer Res. 69:2224–2233. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yue X, Lin X, Yang T, Yang X, Yi X, Jiang
X, Li X, Li T, Guo J, Dai Y, et al: Rnd3/RhoE modulates
hypoxia-inducible factor 1alpha/vascular endothelial growth factor
signaling by stabilizing hypoxia-inducible factor 1alpha and
regulates responsive cardiac angiogenesis. Hypertension.
67:597–605. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu B, Lin X, Yang X, Dong H, Yue X,
Andrade KC, Guo Z, Yang J, Wu L, Zhu X, et al: Downregulation of
RND3/RhoE in glioblastoma patients promotes tumorigenesis through
augmentation of notch transcriptional complex activity. Cancer Med.
4:1404–1416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang X, Wang T, Lin X, Yue X, Wang Q, Wang
G, Fu Q, Ai X, Chiang DY, Miyake CY, et al: Genetic deletion of
Rnd3/RhoE results in mouse heart calcium leakage through
upregulation of protein kinase a signaling. Circ Res. 116:e1–e10.
2015. View Article : Google Scholar :
|
|
11
|
Lin X, Liu B, Yang X, Yue X, Diao L, Wang
J and Chang J: Genetic deletion of Rnd3 results in aqueductal
stenosis leading to hydrocephalus through up-regulation of notch
signaling. Proc Natl Acad Sci USA. 110:8236–8241. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai Y, Song J, Li W, Yang T, Yue X, Lin X,
Yang X, Luo W, Guo J, Wang X, et al: RhoE fine-tunes inflammatory
response in myocardial infarction. Circulation. 139:1185–1198.
2019. View Article : Google Scholar :
|
|
13
|
Breslin JW, Daines DA, Doggett TM, Kurtz
KH, Souza-Smith FM, Zhang XE, Wu MH and Yuan SY: Rnd3 as a novel
target to ameliorate microvascular leakage. J Am Heart Assoc.
5:e0033362016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu S, Zheng F, Cai Y, Zhang W and Dun Y:
Effect of long-term exercise training on lncrnas expression in the
vascular injury of insulin resistance. J Cardiovasc Transl Res.
11:459–469. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yue X, Yang X, Lin X, Yang T, Yi X, Dai Y,
Guo J, Li T, Shi J, Wei L, et al: Rnd3 haploinsufficient mice are
predisposed to hemodynamic stress and develop apoptotic
cardiomyopathy with heart failure. Cell Death Dis. 5:e12842014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao Y, Wang Z, Hao Q, Li W, Xu Y, Zhang J,
Zhang W, Wang S, Liu S, Li M, et al: Loss of ERalpha induces
amoeboid-like migration of breast cancer cells by downregulating
vinculin. Nat Commun. 8:144832017. View Article : Google Scholar
|
|
17
|
Park J, Bae S and Kim JS: Cas-Designer: A
web-based tool for choice of CRISPR-Cas9 target sites.
Bioinformatics. 31:4014–4016. 2015.PubMed/NCBI
|
|
18
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of affymetrix genechip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thomas S and Bonchev D: A survey of
current software for network analysis in molecular biology. Hum
Genomics. 4:353–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Calvano SE, Xiao W, Richards DR, Felciano
RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK,
et al: A network-based analysis of systemic inflammation in humans.
Nature. 437:1032–1037. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Krämer A, Green J, Pollard J Jr and
Tugendreich S: Causal analysis approaches in ingenuity pathway
analysis. Bioinformatics. 30:523–530. 2014. View Article : Google Scholar :
|
|
22
|
Wang K, Ding R, Ha Y, Jia Y, Liao X, Wang
S, Li R, Shen Z, Xiong H, Guo J and Jie W: Hypoxia-Stressed
cardiomyocytes promote early cardiac differentiation of cardiac
stem cells through HIF-1alpha/Jagged1/Notch1 signaling. Acta Pharm
Sin B. 8:795–804. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen Z, Liao X, Shao Z, Feng M, Yuan J,
Wang S, Gan S, Ha Y, He Z and Jie W: Short-Term stimulation with
histone deacetylase inhibitor trichostatin a induces
epithelial-mesenchymal transition in nasopharyngeal carcinoma cells
without increasing cell invasion ability. BMC Cancer. 19:2622019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Wu Y, Shen Z, Wang K, Ha Y, Lei H, Jia Y,
Ding R, Wu D, Gan S, Li R, et al: High FMNL3 expression promotes
nasopha-ryngeal carcinoma cell metastasis: Role in
TGF-beta1-induced epithelia-to-mesenchymal transition. Sci Rep.
7:425072017. View Article : Google Scholar
|
|
26
|
Yasuda M, Tanaka Y, Nishiguchi KM, Ryu M,
Tsuda S, Maruyama K and Nakazawa T: Retinal transcriptome profiling
at transcription start sites: A cap analysis of gene expression
early after axonal injury. BMC Genomics. 15:9822014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu C, Chen M, Zhang Q, Yu L, Zhu J and Gao
X: Genomic and genechip expression profiling reveals the inhibitory
effects of amorphophalli rhizoma in TNBC cells. J Ethnopharmacol.
235:206–218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang F, Wen Y and Guo X: CRISPR/Cas9 for
genome editing: Progress implications and challenges. Hum Mol
Genet. 23:R40–R46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doudna JA and Charpentier E: Genome
editing. The new frontier of genome engineering with CRISPR-Cas9.
Science. 346:12580962014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Park S, Mathis KW and Lee IK: The
physiological roles of apolipoprotein J/clusterin in metabolic and
cardiovascular diseases. Rev Endocr Metab Disord. 15:45–53. 2014.
View Article : Google Scholar
|
|
31
|
Willems S, Hoefer I and Pasterkamp G: The
role of the interleukin 1 receptor-like 1 (ST2) and interleukin-33
pathway in cardiovascular disease and cardiovascular risk
assessment. Minerva Med. 103:513–524. 2012.PubMed/NCBI
|
|
32
|
Sudhahar V, Okur MN, Bagi Z, O'Bryan JP,
Hay N, Makino A, Patel VS, Phillips SA, Stepp D, Ushio-Fukai M and
Fukai T: Akt2 (Protein Kinase B Beta) stabilizes ATP7A, a copper
transporter for extracellular superoxide dismutase, in vascular
smooth muscle: Novel mechanism to limit endothelial dysfunction in
type 2 diabetes mellitus. Arterioscler Thromb Vasc Bio. 38:529–541.
2018. View Article : Google Scholar
|
|
33
|
Fang CY, Chen MC, Chang TH, Wu CC, Chang
JP, Huang HD, Ho WC, Wang YZ, Pan KL, Lin YS, et al: Idi1 and
hmgcs2 are affected by stretch in HL-1 atrial myocytes. Int J Mol
Sci. 19:40942018. View Article : Google Scholar
|
|
34
|
Tabaczar S, Wolosiewicz M, Filip A,
Olichwier A and Dobrzyń P: The role of stearoyl-CoA desaturase in
the regulation of cardiac metabolism. Postepy Biochem. 64:183–189.
2018. View Article : Google Scholar
|
|
35
|
Ma W, Sze KM, Chan LK, Lee JM, Wei LL,
Wong CM, Lee TK, Wong CC and Ng IO: RhoE/ROCK2 regulates
chemoresistance through NF-kappaB/IL-6/ STAT3 signaling in
hepatocellular carcinoma. Oncotarget. 7:41445–41459. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guasch RM, Blanco AM, Perez-Arago A,
Miñambres R, Talens-Visconti R, Peris B and Guerri C: RhoE
participates in the stimulation of the inflammatory response
induced by ethanol in astrocytes. Exp Cell Res. 313:3779–3788.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gordon JW, Shaw JA and Kirshenbaum LA:
Multiple facets of NF-kappaB in the heart: To be or not to
NF-kappaB. Circ Res. 108:1122–1132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Panday A, Inda ME, Bagam P, Sahoo MK,
Osorio D and Batra S: Transcription factor NF-kappaB: An update on
intervention strategies. Arch Immunol Ther Exp (Warsz). 64:463–483.
2016. View Article : Google Scholar
|
|
39
|
Chou R, Dana T, Blazina I, Daeges M and
Jeanne TL: Statins for prevention of cardiovascular disease in
adults: Evidence report and systematic review for the us preventive
services task force. JAMA. 316:2008–2024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu Y, Ge Z, Zhang E, Zuo Q, Huang S, Yang
N, Wu D, Zhang Y, Chen Y, Xu H, et al: The lncRNA TUG1 modulates
proliferation in trophoblast cells via epigenetic suppression of
RND3. Cell Death Dis. 8:e31042017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang H, Wang Y, Liang B, He F, Li Y, Che
J, Li X, Zhao H and Shi G: The Rho GTPase RhoE exerts
tumor-suppressing effects in human esophageal squamous cell
carcinoma via negatively regulating epidermal growth factor
receptor. J Cancer Res Ther. 12:60–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Grise F, Sena S, Bidaud-Meynard A, Baud J,
Hiriart JB, Makki K, Dugot-Senant N, Staedel C, Bioulac-Sage P,
Zucman-Rossi J, et al: Rnd3/RhoE is down-regulated in
hepatocellular carcinoma and controls cellular invasion.
Hepatology. 55:1766–1775. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao H, Yang J, Fan T, Li S and Ren X:
RhoE functions as a tumor suppressor in esophageal squamous cell
carcinoma and modulates the PTEN/PI3K/Akt signaling pathway. Tumour
Biol. 33:1363–1374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu Y, Zhou J, Xia H, Chen X, Qiu M, Huang
J, Liu S, Tang Q, Lang N, Liu Z, et al: The rho GTPase rhoe is a
p53-regulated candidate tumor suppressor in cancer cells. Int J
Oncol. 44:896–904. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bektic J, Pfeil K, Berger AP, Ramoner R,
Pelzer A, Schäfer G, Kofler K, Bartsch G and Klocker H: Small
G-protein rhoe is underexpressed in prostate cancer and induces
cell cycle arrest and apoptosis. Prostate. 64:332–340. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu B, Dong H, Lin X, Yang X, Yue X, Yang
J, Li Y, Wu L, Zhu X, Zhang S, et al: RND3 promotes Snail 1 protein
degradation and inhibits glioblastoma cell migration and invasion.
Oncotarget. 7:82411–82423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou J, Li K, Gu Y, Feng B, Ren G, Zhang
L, Wang Y, Nie Y and Fan D: Transcriptional up-regulation of RhoE
by hypoxia-inducible factor (HIF)-1 promotes epithelial to
mesenchymal transition of gastric cancer cells during hypoxia.
Biochem Biophys Res Commun. 415:348–354. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang Q, Li M, Chen L, Bi F and Xia H:
MiR-200b/c targets the expression of RhoE and inhibits the
proliferation and invasion of non-small cell lung cancer cells. Int
J Oncol. 53:1732–1742. 2018.PubMed/NCBI
|