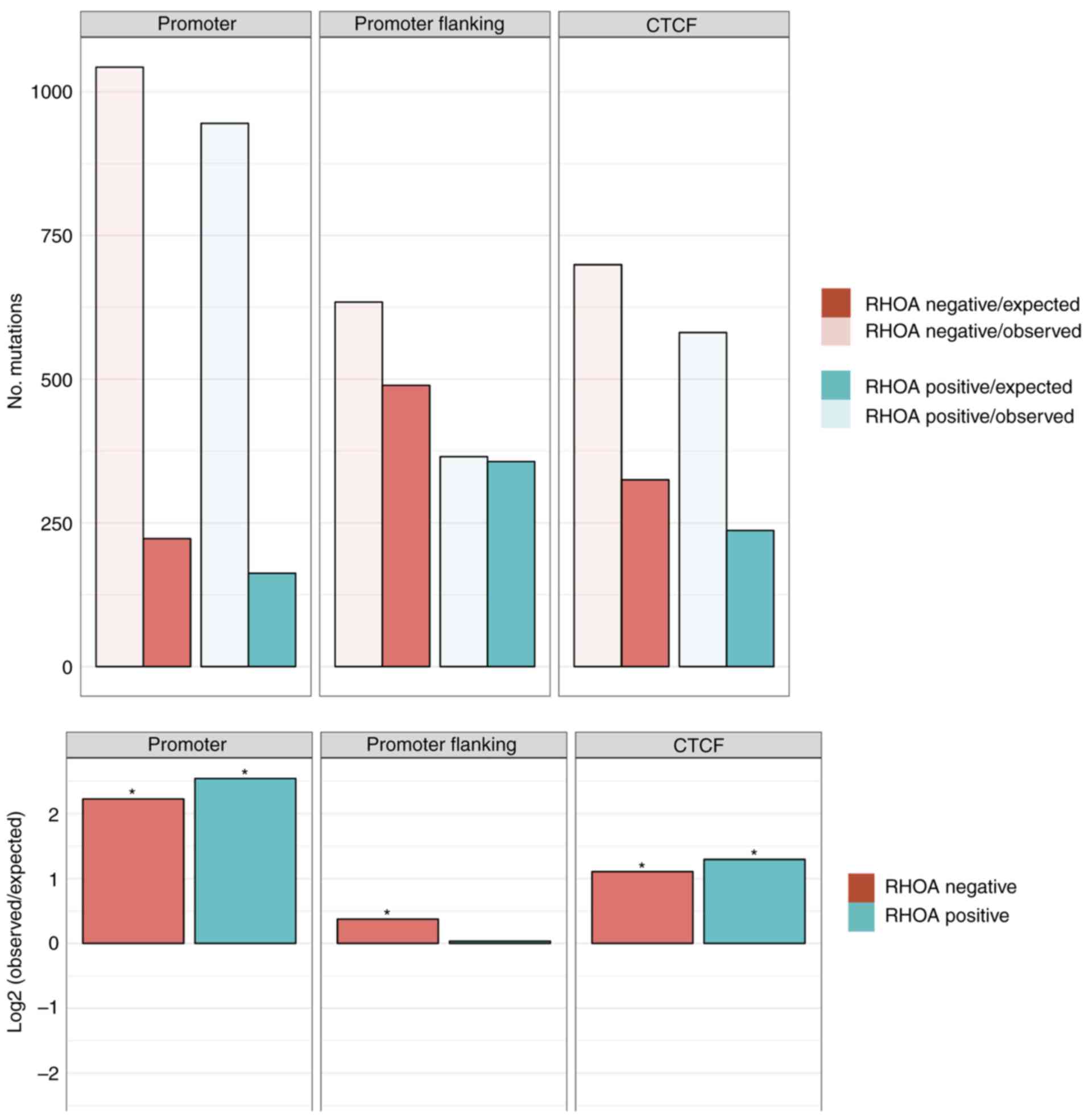
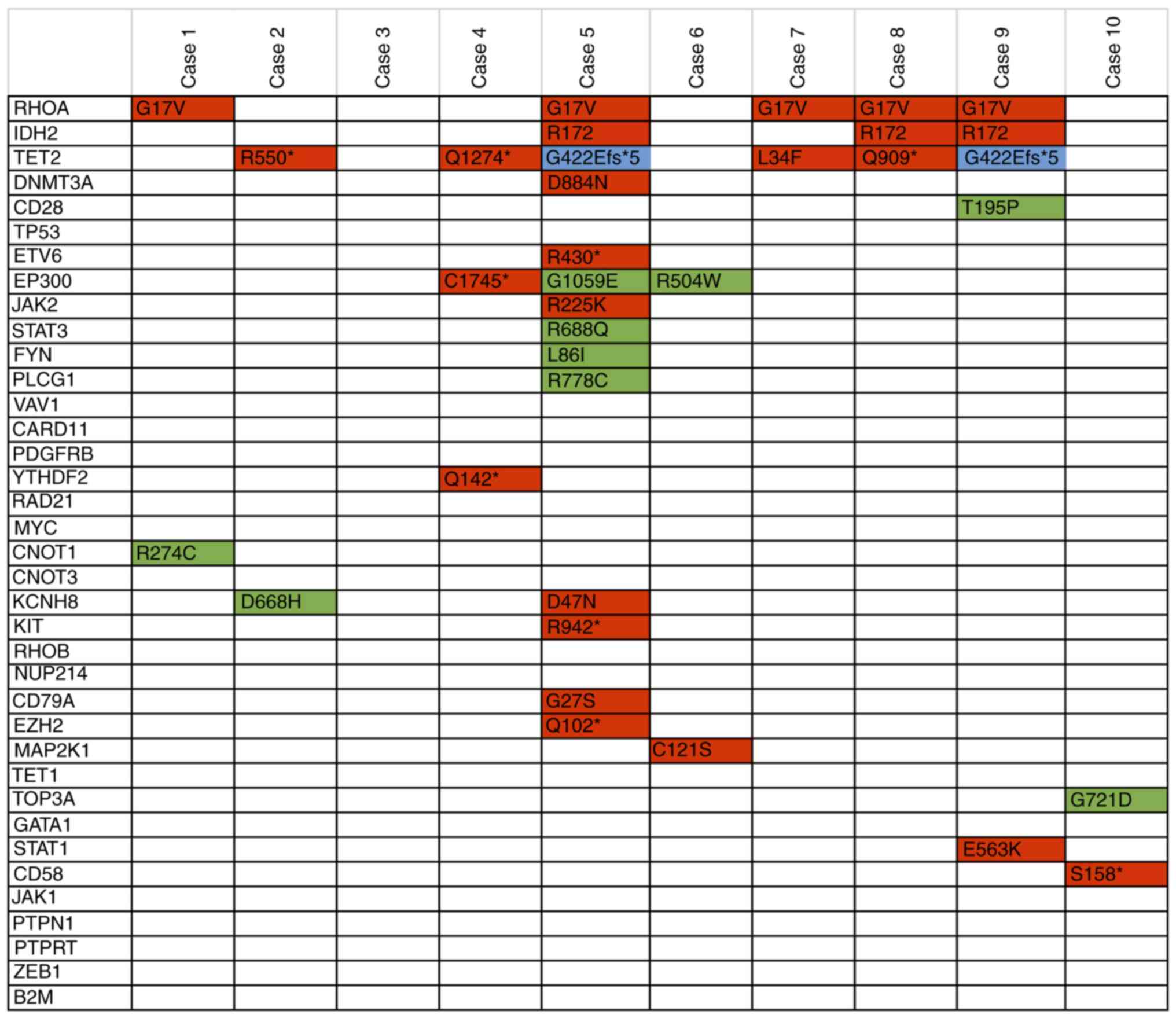
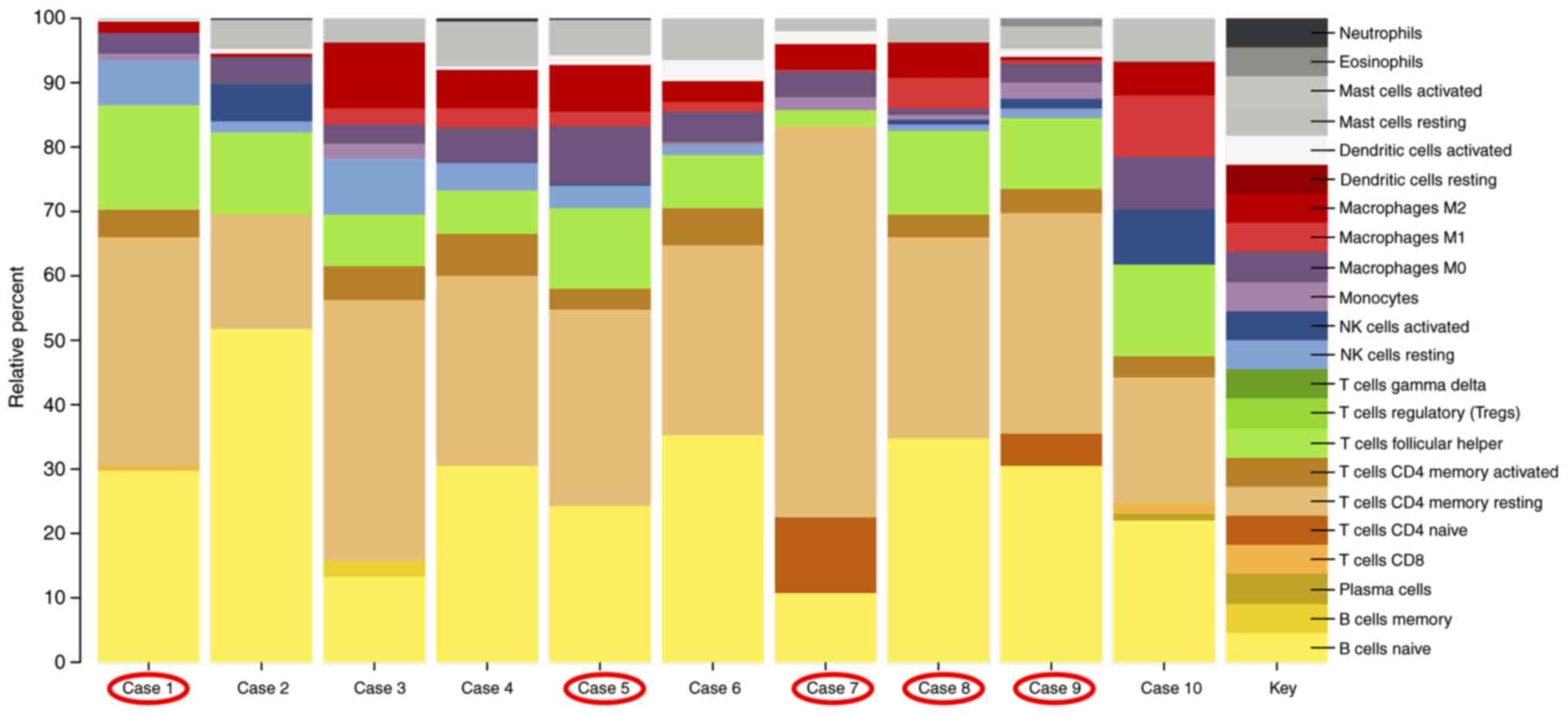
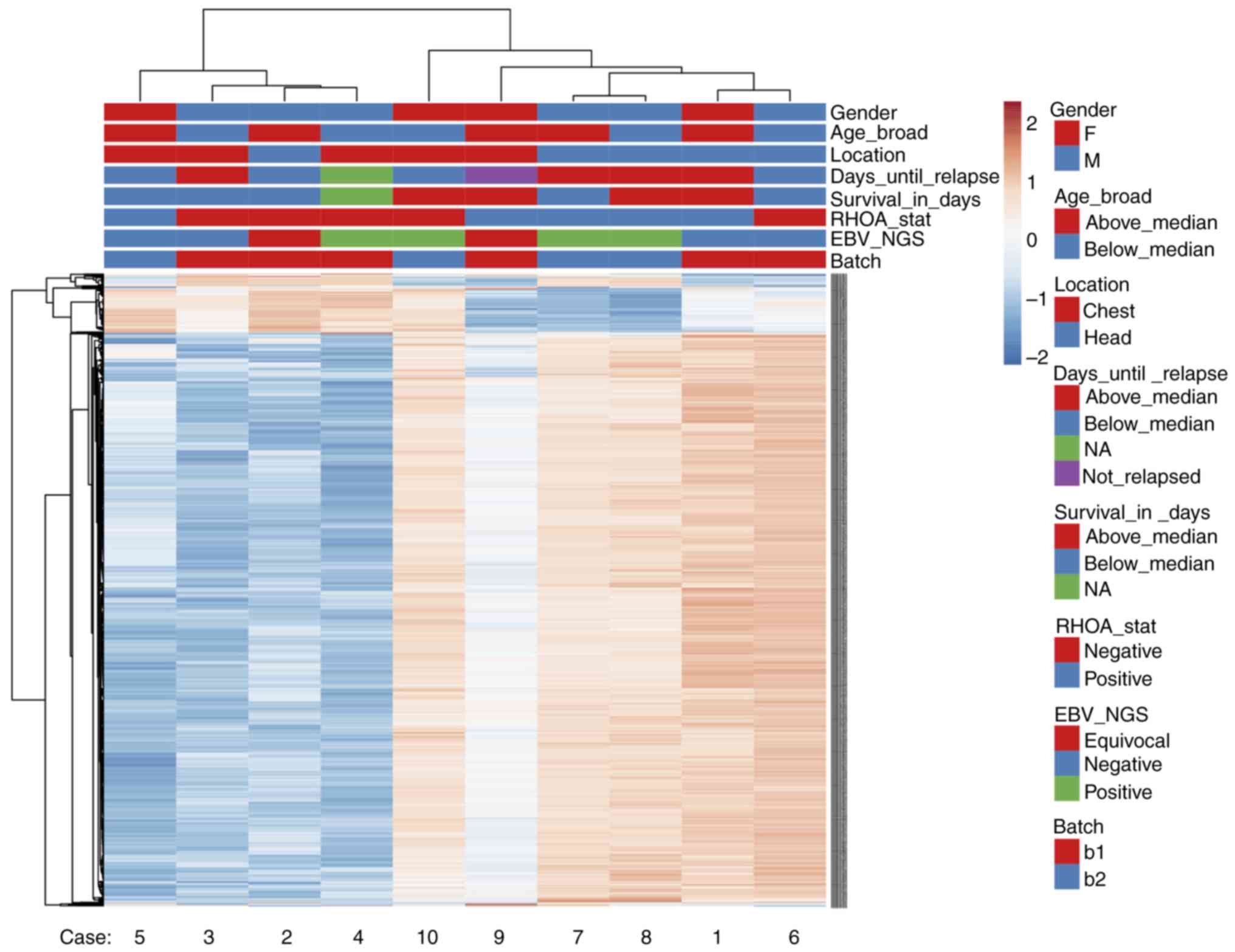
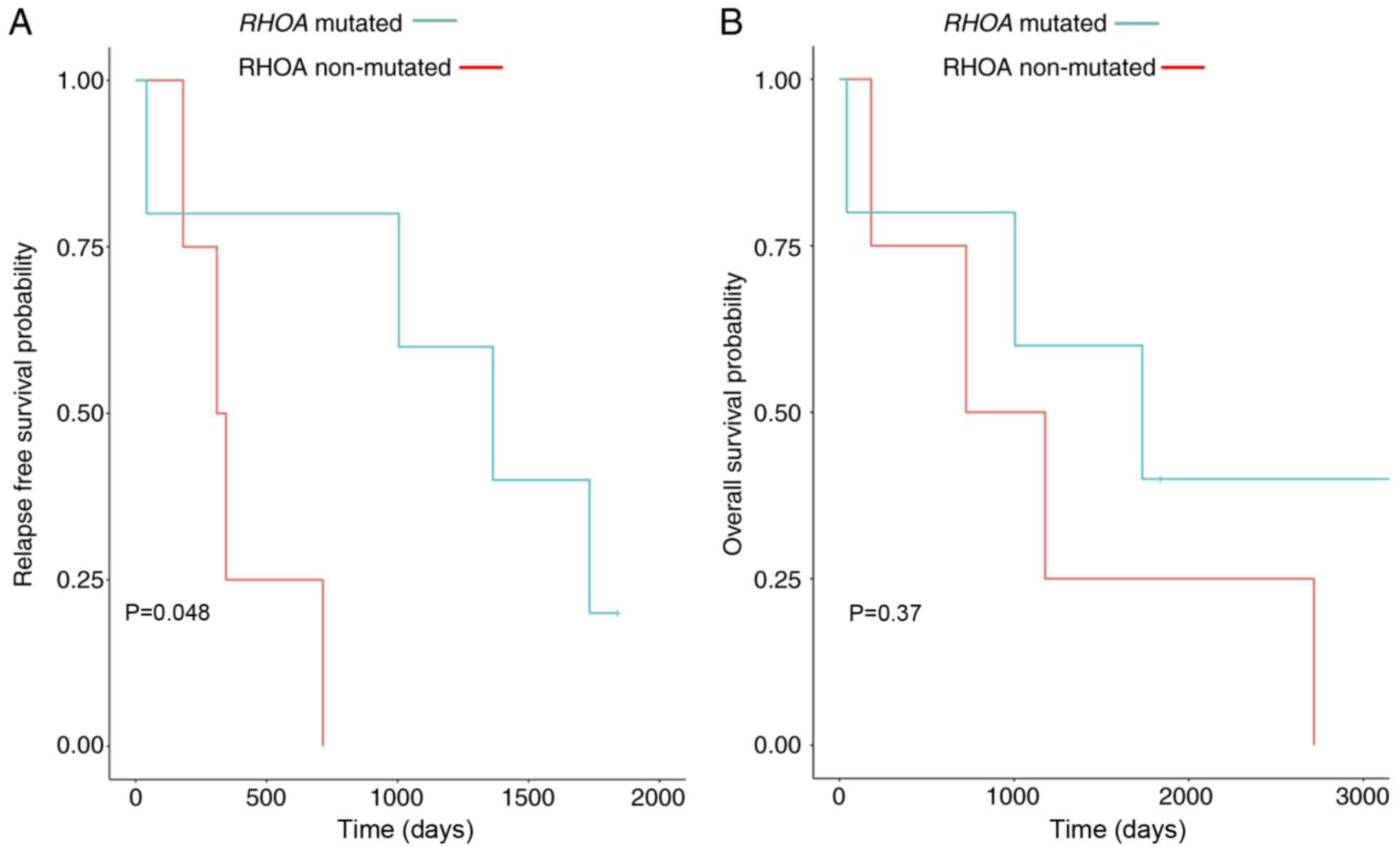
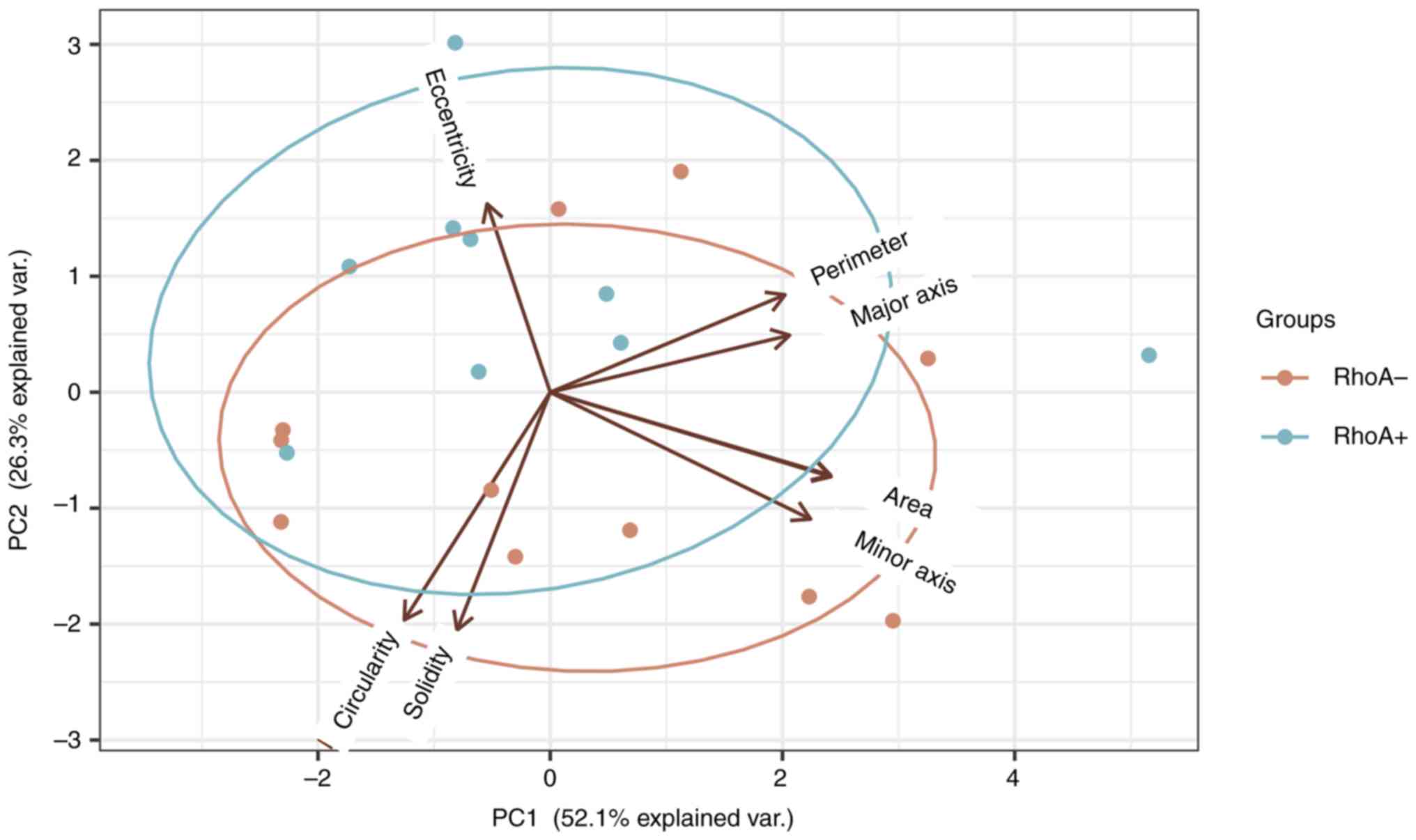
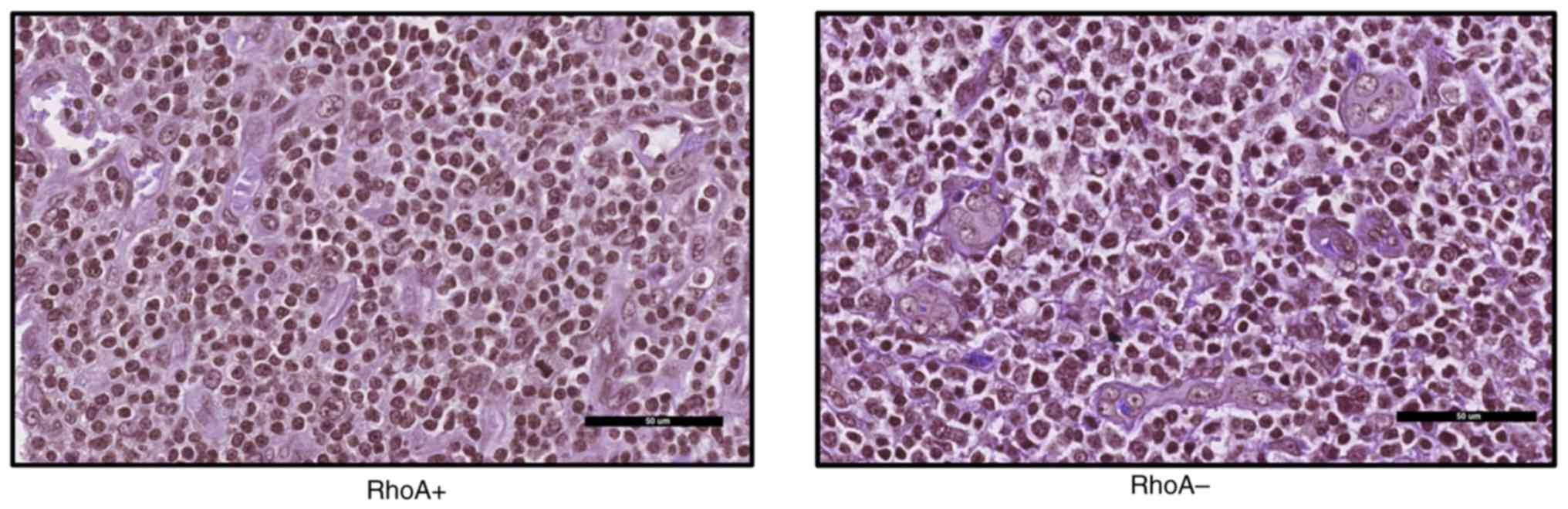
|
1
|
Attygalle AD, Kyriakou C, Dupuis J, Grogg
KL, Diss TC, Wotherspoon AC, Chuang SS, Cabeçadas J, Isaacson PG,
Du MQ, et al: Histologic evolution of angioimmunoblastic T-cell
lymphoma in consecutive biopsies: Clinical correlation. Am J Surg
Pathol. 31:1077–1088. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dogan A, Atygalle AD and Kyriakou C:
Angioimmunoblastic T-cell lymphoma. Br J Haematol. 121:681–691.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Attygalle A, Al-Jehani R, Diss TC, Munson
P, Liu H, Du MQ, Isaacson PG and Dogan A: Neoplastic T cells in
angioimmunoblastic T-cell lymphoma express CD10. Blood. 99:627–633.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Manso R, Sánchez-Beato M, Monsalvo S,
Gómez S, Cereceda L, Llamas P, Rojo F, Mollejo M, Menárguez J,
Alves J, et al: The RHOA G17V gene mutation occurs frequently in
peripheral T-cell lymphoma and is associated with a characteristic
molecular signature. Blood. 123:2893–2894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cairns RA, Iqbal J, Lemonnier F, Kucuk C,
de Leval L, Jais JP, Parrens M, Martin A, Xerri L, Brousset P, et
al: IDH2 mutations are frequent in angioimmunoblastic T-cell
lymphoma. Blood. 119:1901–1903. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Enami T, Yoshida K, Shiraishi Y, Ishii R,
Miyake Y, Muto H, Tsuyama N, Okuno Y, Sakata S, Kamada Y, et al:
Somatic G17V Rhoa mutation specifies angioimmunoblastic T-Cell
lymphoma. Blood. 122:8152013. View Article : Google Scholar
|
|
7
|
Sakata-Yanagimoto M, Enami T, Yoshida K,
Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A,
Okuno Y, et al: Somatic RHOA mutation in angioimmunoblastic T cell
lymphoma. Nat Genet. 46:171–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fujisawa M, Sakata-Yanagimoto M, Nishizawa
S, Komori D, Gershon P, Kiryu M, Tanzima S, Fukumoto K, Enami T,
Muratani M, et al: Activation of RHOA-VAV1 signaling in
angioimmunoblastic T-cell lymphoma. Leukemia. 32:694–702. 2018.
View Article : Google Scholar
|
|
9
|
Cortes JR, Ambesi-Impiombato A, Couronné
L, Quinn SA, Kim CS, da Silva Almeida AC, West Z, Belver L, Martin
MS, Scourzic L, et al: RHOA G17V induces T follicular helper cell
specification and promotes lymphomagenesis. Cancer Cell.
33:259–273, e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ondrejka SL, Grzywacz B, Bodo J, Makishima
H, Polprasert C, Said JW, Przychodzen B, Maciejewski JP and His ED:
Angioimmunoblastic T-cell lymphomas with the RHOA p. Gly17Val
mutation have classic clinical and pathologic features. Am J Surg
Pathol. 40:335–341. 2016. View Article : Google Scholar
|
|
11
|
Swerdlow SH, Campo E, Pileri SA, Harris
NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz
AD and Jaffe ES: The 2016 revision of the World Health Organization
classification of lymphoid neoplasms. Blood. 127:2375–2391. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H and Thiele J: WHO Classification of Tumours of
Haematopoietic and Lymphoid Tissues. 4th edition. International
Agency for Research in Cancer (IARC); Lyon: 2017
|
|
13
|
Kumar J, Butzmann A, Wu S, Easly S,
Zehnder JL, Warnke RA, Bangs CD, Jangam D, Cherry A, Lau J, et al:
Indolent in situ B-cell neoplasms with MYC rearrangements show
somatic mutations in MYC and TNFRSF14 by next-generation
sequencing. Am J Surg Pathol. 43:1720–1725. 2019.PubMed/NCBI
|
|
14
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The Genome Analysis Toolkit: A
MapReduceframework for analyzing next-generation DNA sequencing
data. Genome Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan2: Somatic mutation and copy number alteration discovery in
cancerby exome sequencing. Genome Res. 22:568–576. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karczewski KJ, Francioli LC, Tiao G,
Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A,
Birnbaum DP, et al: The mutational constraint spectrum quantified
from variation in 141,456 humans. Nature. 581:434–443. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rehm HL, Berg JS, Brooks LD, Bustamante
CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL,
Nussbaum RL, et al: ClinGen-the clinical genome resource. N Engl J
Med. 372:2235–2242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
1000 Genomes Project Consortium; Auton A,
Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL,
McCarthy S, McVean GA and Abecasis GR: A global reference for human
genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Exome Variant Server. http:////evs.gs.washington.edu/EVS/urisimple//evs.gs.washington.edu/EVS/Accessed
February 16 2020.
|
|
20
|
Kopanos C and Tsiolkas V: VarSome: The
human genomic variant search engine. Bioinformatics. 35:1978–1980.
2019. View Article : Google Scholar :
|
|
21
|
Blokzijl F, Janssen R, Van Boxtel R and
Cuppen E: MutationalPatterns: Comprehensive genome-wide analysis of
mutational processes. Genome Med. 10:332018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z,
Meirelles GV, Clark NR and Ma'ayan A: Enrichr: Interactive and
collaborative HTML5 gene list enrichment analysis tool. BMC
Bioinformatics. 14:1282013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kamburov A, Stelzl U, Lehrach H and Herwig
R: The ConsensusPathDB interaction database: 2013 update. Nucleic
Acids Res. 41(Database Issue): D793–D800. 2013. View Article : Google Scholar :
|
|
26
|
Gyarmati P, Kjellander C, Aust C, Song Y,
Öhrmalm L and Giske CG: Metagenomic analysis of bloodstream
infections in patients with acute leukemia and therapy-induced
neutropenia. Sci Rep. 21:235322016. View Article : Google Scholar
|
|
27
|
Baheti S, Tang X, O'Brien DR, Chia N,
Roberts LR, Nelson H, Boughey JC, Wang L, Goetz MP, Kocher JA and
Kalari KR: HGT-ID: An efficient and sensitive workflow to detect
human-viral insertion sites using next-generation sequencing data.
BMC Bioinformatics. 19:2712018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim D, Paggi JM, Park C, Bennett C and
Salzberg SL: Graph-based genome alignment and genotyping with
HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anders S, Pyl PT and Huber W: HTSeq-a
Python frame-work to work with high-throughoutput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar
|
|
30
|
Integrated Development for R. Version
125001. Mode: desktop. RStudio, PBC; Boston, MA: 2020, http://www.rstudio.com/.
Accessed February 16 2020.
|
|
31
|
Metsalu T and Vilo J: ClustVis: A web tool
visualizing clustering of multivariate data using Prinicpal
Component Analysis and heatmap. Nucleic Acids Res. 43(W1):
W566–W570. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jangam D, Sridhar K, Butzmann A,
Sambhagati P, Plowey ED and Ohgami RS: TBL1XR1 Mutations in primary
marginal zone lymphomas of occular adnexa are associated with
unique morphometric phenotypes. Curr Eye Res. Jun 19–2020.Epub
ahead of print. View Article : Google Scholar
|
|
34
|
Wong MA and Hartigan JA: Algorithm AS 136:
A K-means clustering algorithm. J R Stat Soc Ser C. 28:100–108.
1979.
|
|
35
|
Fouad S, Randell D, Galton A, Mehanna H
and Landini G: Unsupervised morphological segmentation of tissue
compartments in histopathological images. PLoS One.
12:e01887172017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hart M, Lauer J, Selig M, Hanak M, Walters
B and Rolauffs B: Shaping the cell and the future: Recent
advancements in biophysical aspects relevant to regenerative
medicine. J Funct Morphol Kinesiol. 3:22017. View Article : Google Scholar
|
|
37
|
Lyons SM, Alizadeh E, Mannheimer J,
Schuamberg K, Castle J, Schroder B, Turk P, Thamm D and Prasad A:
Changes in cell shape are correlated with metastatic potential in
murine and human osteosarcomas. Biol Open. 5:289–299. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chiba S, Enami T, Ogawa S and
Sakata-Yanagimoto M: G17V RHOA: Genetic evidence of GTP-unbound
RHOA playing a role in tumorigenesis in T cells. Small GTPases.
6:100–103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nagao R, Kikuti YY, Carreras J, Kikuchi T,
Miyaoka M, Matsushita H, Kojima M, Ando K, Sakata-Yanagimoto M,
Chiba S and Nakamura N: Clinicopathologic analysis of
angioimmunoblastic T-cell lymphoma with or without RHOA G17V
mutation using formalin-fixed paraffin-embedded sections. Am J Surg
Pathol. 40:1041–1050. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Willemsen M, Hamid MA, Winkens B and Zur
Hausen A: Mutational heterogeneity of angioimmunoblastic T-cell
lymphoma indicates distinct lymphomagenic pathways. Blood Cancer J.
8:62018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoo HY, Sung MK, Lee SH, Kim S, Lee H,
Park S, Kim SC, Lee B, Rho K, Lee JE, et al: A recurrent
inactivating mutation in RHOA GTPase in angioimmunoblastic T cell
lymphoma. Nat Genet. 46:371–375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang M, Zhang S, Chuang SS, Ashton-Key M,
Ochoa E, Bolli N, Vassiliou G, Gao Z and Du MQ: Angioimmunoblastic
T cell lymphoma: Novel molecular insights by mutation profiling.
Oncotarget. 8:17763–17770. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ng SY, Brown L, Stevenson K, deSouza T,
Aster JC, Louissaint A Jr and Weinstock DM: RhoA G17V is sufficient
to induce autoimmunity and promotes T-cell lymphomagenesis in mice.
Blood. 132:935–947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Abate F, Da Silva-Almeida AC, Zairis S,
Zairis S, Robles-Valero J, Couronne L, Khiabanian H, Quinn SA, Kim
MY, Laginestra MA, et al: Activating mutations and trans-locations
in the guanine exchange factor VAV1 in peripheral T-cell lymphomas.
Proc Natl Acad Sci USA. 114:764–769. 2017. View Article : Google Scholar
|
|
45
|
Cornish AJ, Hoang PH, Dobbins SE, Law PJ,
Chubb D, Orlando G and Houlston RS: Identification of recurrent
non-coding mutations in B-cell lymphoma using Hi-C. Blood Adv.
3:21–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang X, Zhang Y, Ba Z, Kyritsis N,
Casellas R and Alt FW: Fundamental roles of chromatin loop
extrusion in antibody class switching. Nature. 575:385–389. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Weiss LM, Jaffe ES, Liu XF, Chen YY,
Shibata D and Medeiros LJ: Detection and localization of
Epstein-Barr viral genomes in angioimmunoblastic lymphadenopathy
and angioimmunoblastic lymphadenopathy-like lymphoma. Blood.
79:1789–1795. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Khan G, Norton AJ and Slavin G:
Epstein-Barr virus in angioimmunoblastic T-cell lymphomas.
Histopathology. 22:145–149. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou Y, Attygalle AD, Chuang SS, Diss T,
Ye H, Liu H, Hamoudi RA, Munson P, Bacon CM, Dogan A and Du MQ:
Angioimmunoblastic T-cell lymphoma: Histological progression
associates with EBV and HHV6B viral load. Br J Haematol. 138:44–53.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Eladl AE, Shimada K, Suzuki Y, Takahara T,
Kato S, Kohno K, Elsayed AA, Wu CC, Tokunaga T, Kinoshita T, et al:
EBV status has prognostic implication among young patients with
angioimmunoblastic T-cell lymphoma. Cancer Med. 9:678–688. 2020.
View Article : Google Scholar
|
|
51
|
Hoffmann JC, Chisholm KM, Cherry A, Chen
J, Arber DA, Natkunam Y, Warnke RA and Ohgami RS: An analysis of
MYC and EBV in diffuse large B-cell lymphomas associated with
angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma
not otherwise specified. Hum Pathol. 48:9–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bal M, Gujral S, Gandhi J, Shet T, Epari S
and Subramanian PG: Angioimmunoblastic T-cell lymphoma: A critical
analysis of clinical, morphologic and immunophenotypic features.
Indian J Pathol Microbiol. 53:640–645. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
De Leval L, Rickman DS, Thielen C, Reynies
Ad, Huang YL, Delsol G, Lamant L, Leroy K, Brière J, Molina T, et
al: The gene expression profile of nodal peripheral T-cell lymphoma
demonstrates a molecular link between angioimmunoblastic T-cell
lymphoma (AITL) and follicular helper T (TFH) cells. Blood.
109:4952–4963. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Piccaluga PP, Agostinelli C, Califano A,
Carbone A, Fantoni L, Ferrari S, Gazzola A, Gloghini A, Righi S,
Rossi M, et al: Gene expression analysis of angioimmunoblastic
lymphoma indicates derivation from T follicular helper cells and
vascular endothelial growth factor deregulation. Cancer Res.
67:10703–10713. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nguyen TB, Sakata-Yanagimoto M, Asabe Y,
Matsubara D, Kano J, Yoshida K, Shiraishi Y, Chiba K, Tanaka H,
Miyano S, et al: Identification of cell-type-specific mutations in
nodal T-cell lymphomas. Blood Cancer J. 7:e5162017. View Article : Google Scholar : PubMed/NCBI
|