Introduction
Vitiligo is the most common pigmentary skin disorder
affecting 0.5-1% of the population worldwide. It is characterized
by selective loss of epidermal melanocytes resulting in the
occurrence of patchy depigmentation (1). Though not entirely clear, the
pathogenesis of vitiligo is widely believed to reflect a complex
interplay between genetic, immunologic, and environmental factors
interacting to promote a model of melanocyte-directed autoimmunity
(1-3). The current thought is that vitiligo
is a multifactorial, polygenic disorder, suggesting that different
genes may be involved in the onset and evolution of the disease
(4).
Vitamin D has been shown to be involved in multiple
biologic processes and pathways. Its role in human carcinogenesis,
as well as in osteoporosis is well known (5,6).
Given the effects of vitamin D against autoimmunity (7), a potential link between vitamin D
deficiency and immune-mediated dermatoses, such as psoriasis and
atopic dermatitis, has already been suggested (8,9).
In this context, due to the stimulatory and protective action of
1,25(OH)2D3 on melanocytes, as well as its
antioxidant and immunomodulatory properties, topical vitamin D and
its analogs have been used as repigmentation agents in vitiligo,
either alone or combined with other modalities, but reported
outcomes have been conflicting (10,11).
Currently, great attention has also been focused on
gene-disease associations, opening new perspectives for precision
medicine (12,13). As the nuclear vitamin D receptor
(VDR) mediates most of the genomic effects of
1,25(OH)2D3, VDR might represent a
susceptibility gene for vitiligo (14).
The VDR gene, located on chromosome 12q13.11,
has been found to contain more than 200 single-nucleotide
poly-morphisms (SNPs) (15).
Among these, the rs2228570 (Fokl), rs1544410 (Bsml),
and rs731236 (Taql) have been widely studied. The
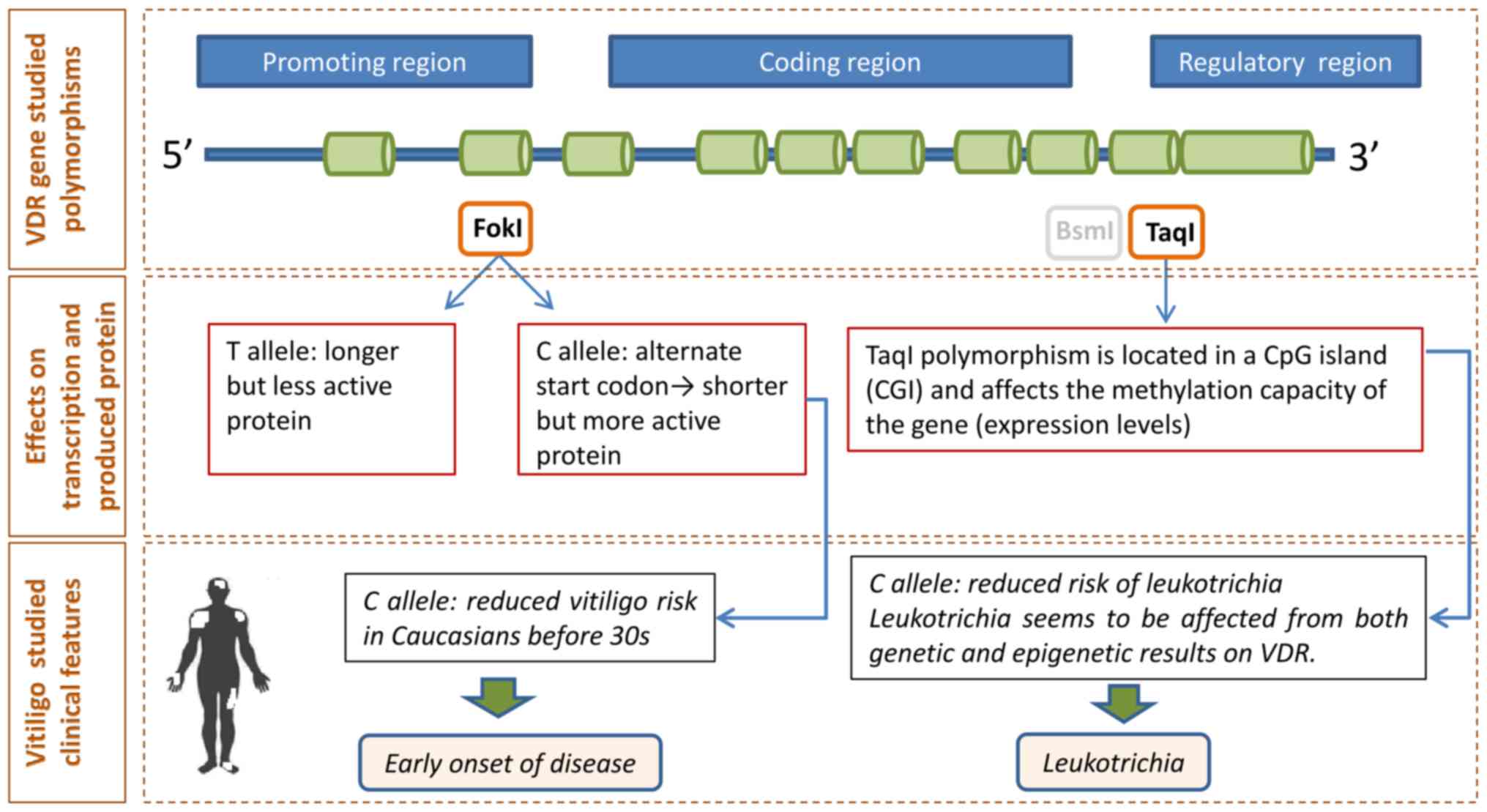
FokI polymorphism, located at exon 2 initiation codon, might
result in two forms of the VDR protein, i.e., a long (allele f) and
a short (allele F) version, with diverse transcriptional capacity.
The Bsml (in intron 8) and Taql (in exon 9) alleles
occur in the 3′-untranslated region (3′-UTR) of the gene. Although
allelic variations within or near the VDR locus could modify
the VDR gene expression and protein function, their clinical
relevance remains largely unknown (16,17).
So far, implication of VDR in vitiligo has
not been thoroughly explored among Caucasians (16,18), although an association between
VDR SNPs and immune-mediated skin diseases, i.e., psoriasis
and atopic dermatitis, has been supported (19,20). Moreover, the majority of studies
evaluating the influence of VDR polymorphisms on vitiligo
susceptibility in certain population settings have reported
inconsistent results (16),
indicating that ethnicity might be a potential source of
heterogeneity.
The current study investigated whether three common
SNPs in the VDR gene (FokI, BsmI, and
TaqI) may confer susceptibility to vitiligo and influence
its main clinical features in a Southeastern European Caucasian
(SEC) population.
Patients and methods
Patients and setting
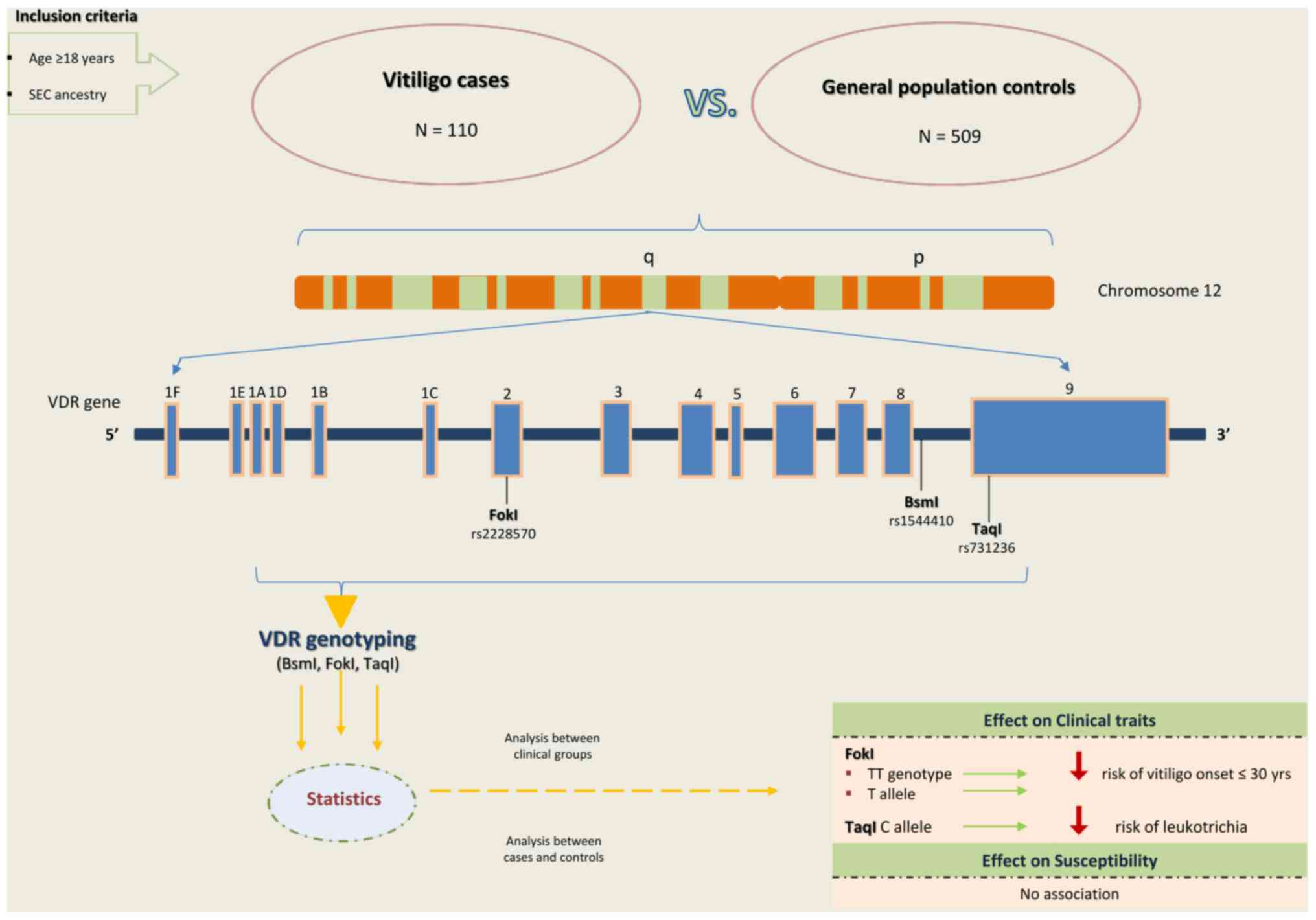
In this single-center, case-control study, a total
of 110 unrelated vitiligo patients (42 males, 68 females) with mean
± standard deviation (SD) age of 45.1±13.4 years (age range 18-70
years) were recruited as a case group at the Vitiligo Outpatient
Unit of 'A. Sygros' Hospital, Athens, Greece, from October 2018 to
November 2019. Inclusion criteria were: i) age ≥18 years; ii)
clinically diagnosed vitiligo; and iii) SEC ancestry. First- to
third-degree blood relatives were considered ineligible. A group of
509 SEC individuals from the general population (220 males, 289
females) with mean ± SD age of 40.5±11.3 years (age range 18-70
years) served as controls.
The study was conducted according to the principles
outlined in the Declaration of Helsinki after obtaining ethics
approval from the Institutional Review Board (protocol no.
3044/6-9-2018). All participants provided written informed consent
for using their genetic data after de-identification and
anonymization of the DNA samples, following the European Medicines
Agency guidelines (EMEA/CPMP/3070/01).
Study assessments
Vitiligo was clinically diagnosed by experienced
dermatologists after physical examination of the affected skin,
often under the Wood's lamp. Clinical types of the disease were
categorized as focal (one or few lesions in a non-dermatomal
pattern), segmental (unilateral segmental distribution), acrofacial
(limited to the face and/or distal extremities), generalized or
vulgaris (scattered across the body), and universal (over 90%
depigmentation), based on the latest criteria of classification
(21). Stable vitiligo was
defined as no appearance of new and/or progression of existing
lesions for at least 1 year before inclusion in the study. Patients
were considered to have i) early-onset vitiligo if the age at
disease onset was prior or equal to 30 years, and ii) family
history if they reported one or more first- to third-degree
relatives affected by the condition.
Demographic and clinical disease-specific
characteristics (i.e., age, sex, nationality, clinical type, age of
onset, family history, duration of vitiligo, disease activity,
presence of leukotrichia, Koebner phenomenon, and Sutton nevi, body
surface area (BSA), and associated diseases) were retrieved for
each patient via a predesigned questionnaire. Peripheral blood
samples (5 ml) of all patients were collected in tubes containing
ethylenediaminetetraacetic acid (EDTA).
DNA isolation, sample storage and
genotyping
Whole blood samples were collected in 5 ml EDTA
collection tubes followed by DNA isolation, using
PureLink® Genomic DNA Mini kit (Thermo Fisher
Scientific, Inc), according to the manufacturer's instructions. All
DNA samples were stored in −20°C until genotyping.
Three polymorphisms of VDR [FokI
(rs2228570), BsmI (rs1544410), and TaqI (rs731236)]
were analyzed both in patients and controls. Genotyping was
performed by qPCR (Light Cycler 480; Roche) using simple probes for
each SNP (LightSnip Assays; TIBMOLBIOL) and melting curve
analysis.
Statistical analysis
Descriptive statistics are presented as mean ± SD,
or frequencies (numbers) with percentages (%), as appropriate.
Distributions in genotype and allele frequencies in vitiligo cases
versus controls were evaluated using the Chi-squared and Fisher's
exact tests. The Hardy-Weinberg equilibrium (HWE) for each SNP was
calculated among controls. ORs and 95% CIs, were used to
investigate the association of the selected polymorphisms with
clinical aspects and risk of vitiligo under five genetic models
(allele, dominant, recessive, homozygous and heterozygous). All
statistical tests were carried out using IBM SPSS Statistics
version 22.0 (IBM Corp.). The significance level was set to
P<0.05.
Results
Demographic and clinical data
The demographic and clinical disease-specific data
of the case group are presented in Table I. Vitiligo vulgaris was the most
common clinical form (n=84; 76.4%), followed by acrofacial (n=16;
14.5%), focal (n=7; 6.4%) and universal (n=3; 2.7%) patterns. The
mean ± SD age at disease onset was 33.5±15.1 (age range 5-66
years), while vitiligo onset before 30 years of age was reported in
52 (47.3%) cases. Thirty-eight (34.5%) patients with family history
of vitiligo were recorded. Koebner phenomenon, leukotrichia, and
Sutton nevi were present in 45 (40.9%), 36 (32.7%), and 12 (10.9%)
cases, respectively. Stable vitiligo was reported by 69 (62.7%)
patients. Regarding comorbidities, thyroid disease was the most
common associated disorder (n=50; 45.5%).
 | Table IDemographic and clinical data of
vitiligo group (n=110). |
Table I
Demographic and clinical data of
vitiligo group (n=110).
|
Characteristics | Total, n=110 |
|---|
| Sex, n (%) | |
| Male | 42 (38.2) |
| Female | 68 (61.8) |
| Age (years), mean ±
SD (range) | 45.1±13.4
(18-70) |
| Family History
(yes), n (%) | 38 (34.5) |
| Age at vitiligo
onset (years), | 33.5±15.1
(5-66) |
| mean ± SD
(range) | |
| Early onset
vitiligo, n (%) | 52 (47.3) |
| Clinical type, n
(%) | |
| Vulgaris | 84 (76.4) |
| Acrofacial | 16 (14.5) |
| Focal | 7 (6.4) |
| Universal | 3 (2.7) |
| BSA (% of body), n
(%) | |
| ≤5 | 74 (67.3) |
| 5-20 | 25 (22.7) |
| >20 | 11 (10.0) |
| Koebner phenomenon,
n (%) | 45 (40.9) |
| Leukotrichia, n
(%) | 36 (32.7) |
| Sutton nevi, n
(%) | 12 (10.9) |
| Stable disease, n
(%) | 69 (62.7) |
| Comorbidities
(yes), n (%) | 77 (70.0) |
| Most common | |
| Thyroid disease, n
(%) | 50 (45.5) |
| Skin
diseasea, n (%) | 13 (11.8) |
Associations between VDR SNPs and
vitiligo phenotypes
Subgroup analyses of subsets of patients based on
clinical features indicated a significant correlation between the
VDR FokI and age at vitiligo onset (Table II). Both the CC geno-type and C
allele of FokI SNP were overpresented in cases with vitiligo
onset after the age of 30 compared to those with earlier disease
onset (48.28 vs. 30.77%, P=0.041, respectively; 69.83 vs. 55.77%,
P=0.031, respectively), conferring protection against early-onset
vitiligo. Patients carrying the FokI CC genotype or C allele
may thus be at lower risk for development of vitiligo up to 30
years of age compared to carriers of the FokI TT genotype or
T allele, respectively (TT vs. CC: OR=0.286, 95% CI: 0.083-0.984,
P=0.041; T vs. C: OR=0.545, 95% CI: 0.313-0.948, P=0.031).
 | Table IIGenotype and allele frequencies of
VDR FokI polymorphism in vitiligo cases according to age at
disease onset. |
Table II
Genotype and allele frequencies of
VDR FokI polymorphism in vitiligo cases according to age at
disease onset.
| Onset >30 years
(n=58), n (%) | Onset ≤30 years
(n=52), n (%) | OR (95% CI) | P-value |
|---|
| FokI
(rs2228570) | | | | |
| TT | 5 (8.62) | 10 (19.23) | 1.0
(reference) | |
| CT | 25 (43.10) | 26 (50.00) | 0.520
(0.156-1.736) | 0.283 |
| CC | 28 (48.28) | 16 (30.77) | 0.286
(0.083-0.984) | 0.041 |
| CT + CC | 53 (91.38) | 42 (80.77) | 0.396
(0.126-1.248) | 0.105 |
| CC | 28 (48.28) | 16 (30.77) | 1.0
(reference) | |
| CT + TT | 30 (51.72) | 36 (69.23) | 2.100
(0.960-4.592) | 0.061 |
| CT | 25 (43.10) | 26 (50.00) | 1.0
(reference) | |
| TT + CC | 33 (56.90) | 26 (50.00) | 0.758
(0.357-1.607) | 0.469 |
| T allele
frequency | 30.17 | 44.23 | 1.0
(reference) | |
| C allele
frequency | 69.83 | 55.77 | 0.545
(0.313-0.948) | 0.031 |
For TaqI polymorphism, the variant C allele
frequency was significantly higher in vitiligo cases devoid of
leukotrichia compared to cases with concurrent leukotrichia (41.89
vs. 27.78%, P=0.042). Using the T allele as reference, the C allele
was found to adversely affect the risk for occurrence of
leukotrichia; TaqI C allele carriers were ~1.9 times less
prone to develop leukotrichia compared to T allele carriers (T vs.
C: OR=1.874, 95% CI: 1.018-3.451, P=0.042) (Table III). Either FokI,
BsmI, or TaqI loci had no obvious correlation with
other vitiligo-related clinical variables in our sample (data not
shown).
 | Table IIIGenotype and allele frequencies of
VDR TaqI polymorphism in vitiligo cases according to
presence of leukotrichia. |
Table III
Genotype and allele frequencies of
VDR TaqI polymorphism in vitiligo cases according to
presence of leukotrichia.
| With leukotrichia
(n=36), n (%) | Without
leukotrichia (n=74), n (%) | OR (95% CI) | P-value |
|---|
| TaqI
(rs731236) | | | | |
| TT | 19 (52.78) | 26 (35.14) | 1.0
(reference) | |
| CT | 14 (38.89) | 34 (45.95) | 1.775
(0.752-4.188) | 0.189 |
| CC | 3 (8.33) | 14 (18.92) | 3.410
(0.858-13.557) | 0.071 |
| CT + CC | 17 (47.22) | 48 (64.86) | 2.063
(0.918-4.638) | 0.077 |
| CC | 3 (8.33) | 14 (18.92) | 1.0
(reference) | |
| CT + TT | 33 (91.67) | 60 (81.08) | 0.390
(0.104-1.455) | 0.150 |
| CT | 14 (38.89) | 34 (45.95) | 1.0
(reference) | |
| TT + CC | 22 (61.11) | 40 (54.05) | 0.749
(0.333-1.685) | 0.484 |
| T allele
frequency | 72.22 | 58.11 | 1.0
(reference) | |
| C allele
frequency | 27.78 | 41.89 | 1.874
(1.018-3.451) | 0.042 |
Genotypic and allelic distributions of VDR
polymorphisms.The genotype and allele frequencies of the
selected VDR SNPs among cases and general population
controls, as well as their associations with vitiligo
susceptibility are summarized in Table IV. The relevant genotype
frequencies were in accor-dance with the HWE equilibrium among the
controls (P=0.909 for FokI; P=0.966 for BsmI; and
P=0.970 for TaqI).
 | Table IVGenotype and allele frequencies of
selected VDR polymorphisms among cases and controls and
their association with vitiligo risk. |
Table IV
Genotype and allele frequencies of
selected VDR polymorphisms among cases and controls and
their association with vitiligo risk.
| Cases (n=110), n
(%) | Controls (n=509), n
(%) | OR (95% CI) | P-value |
|---|
| FokI
(rs2228570) | | | | |
| TT | 15 (13.64) | 51 (10.02) | 1.0
(reference) | |
| CT | 51 (46.36) | 222 (43.61) | 1.280
(0.668-2.455) | 0.456 |
| CC | 44 (40.00) | 236 (46.37) | 1.589
(0.816-3.051) | 0.173 |
| CT + CC | 95 (86.36) | 458 (89.98) | 1.418
(0.765-2.627) | 0.265 |
| CC | 44 (40.00) | 236 (46.37) | 1.0
(reference) | |
| CT + TT | 66 (60.00) | 273 (53.63) | 0.771
(0.507-1.173) | 0.224 |
| CT | 51 (46.36) | 222 (43.61) | 1.0
(reference) | |
| TT + CC | 59 (53.64) | 287 (56.39) | 1.117
(0.739-1.690) | 0.599 |
| T allele
frequency | 36.82 | 31.83 | 1.0
(reference) | |
| C allele
frequency | 63.18 | 68.17 | 1.248
(0.921-1.692) | 0.152 |
| BsmI
(rs1544410) | | | | |
| GG | 39 (35.45) | 186 (36.54) | 1.0
(reference) | |
| AG | 49 (44.55) | 243 (47.74) | 1.040
(0.655-1.650) | 0.868 |
| AA | 22 (20.00) | 80 (15.72) | 0.762
(0.425-1.368) | 0.362 |
| AG + AA | 71 (64.55) | 323 (63.46) | 0.954
(0.620-1.467) | 0.830 |
| AA | 22 (20.00) | 80 (15.72) | 1.0
(reference) | |
| AG + GG | 88 (80.00) | 429 (84.28) | 1.341
(0.793-2.265) | 0.272 |
| AG | 49 (44.55) | 243 (47.74) | 1.0
(reference) | |
| GG + AA | 61 (55.45) | 266 (52.26) | 0.879
(0.581-1.331) | 0.543 |
| G allele
frequency | 57.73 | 60.41 | 1.0
(reference) | |
| A allele
frequency | 42.27 | 39.59 | 0.895
(0.666-1.203) | 0.461 |
| TaqI
(rs731236) | | | | |
| TT | 45 (40.91) | 197 (38.70) | 1.0
(reference) | |
| CT | 48 (43.64) | 239 (46.95) |
1.1137(0.726-1.781) | 0.573 |
| CC | 17 (15.45) | 73 (14.34) | 0.981
(0.528-1.822) | 0.951 |
| CT + CC | 65 (59.09) | 312 (61.30) | 1.096
(0.721-1.669) | 0.667 |
| CC | 17 (15.45) | 73 (14.34) | 1.0
(reference) | |
| CT + TT | 93 (84.55) | 436 (85.66) | 1.092
(0.615-1.937) | 0.764 |
| CT | 48 (43.64) | 239 (46.95) | 1.0
(reference) | |
| TT + CC | 62 (53.36) | 270 (53.05) | 0.875
(0.577-1.325) | 0.527 |
| T allele
frequency | 62.73 | 62.18 | 1.0
(reference) | |
| C allele
frequency | 37.27 | 37.82 | 1.024
(0.757-1.383) | 0.879 |
In overall analysis, no statistically significant
differences between vitiligo cases and general population controls
were observed for the VDR FokI, BsmI, and TaqI
polymorphisms in any of the genetic models used, indicating a lack
of association between the studied SNPs and susceptibility to
vitiligo in this cohort.
A schematic diagram of study design and summary of
results is provided in Fig.
1.
Discussion
This study investigated associations of the
FokI, BsmI, and TaqI SNPs in the VDR
gene with the risk of vitiligo and its clinical features. The
results showed that the VDR FokI SNPs (CC genotype and C
allele) seemed to reduce the risk of vitiligo onset until the age
of 30, while the TaqI C allele may confer a lower risk for
leukotrichia. Our data also showed that either FokI,
BsmI, or TaqI loci are not involved in the occurrence
and development of vitiligo.
To date, genetic studies have identified over 50
vitiligo susceptibility loci, emphasizing the importance of genetic
factors in the onset and evolution of the depigmentation process
(2,4). In this regard, a limited number of
studies covering mostly Latin American, African, and Asian
populations have previously examined the role of VDR gene
polymorphisms in vitiligo (16,18,22-27), but the reported findings have been
controversial and inconclusive (Table
V). In addition, studies investigating the BsmI,
TaqI, and FokI SNPs in vitiligo patients are fewer
compared to those conducted on ApaI (18), making it more difficult to yield
meaningful results, especially among Caucasian subjects.
 | Table VCharacteristics of main studies on
the effect of VDR gene polymorphisms (BsmI,
FokI, and TaqI) on vitiligo risk. |
Table V
Characteristics of main studies on
the effect of VDR gene polymorphisms (BsmI,
FokI, and TaqI) on vitiligo risk.
| First author,
year | Population | Participants (n)
| Sex F/M (%)
| Age (years), mean
(SD)
| Effect on vitiligo
risk
| Refs. |
|---|
| Cases | Controls | Cases | Controls | Cases | Controls | FokI | BsmI | TaqI |
|---|
| Hassan, 2019 | South Asian | 100 | 100
(age/sex-matched) | 61/39 | 60/40 | 28.7 (11.98) | - | No relation | No relation | No relation | (24) |
| Ochoa-Ramírez,
2019 | Latin American | 173 | 184
(age/sex-matched) | 53.2/46.8 | - | - | - | - | No relation | No relation | (26) |
| Sobeih, 2016 | African | 75 | 75
(age/sex-matched) | - | - | 31.5 (13.5) | - | No relation | - | CC genotype
↑(br1)CT genotype ↓ | (27) |
| Aydıngöz, 2012 | Asian | 98 | 216
(age/sex-matched) | 46.9/53.1 | 56.5/48.2 | 39 (12.05) | 37.1 (9.8) | No relation | No relation | C allele ↑(br1)CC
genotype ↑ | (22) |
| Li, 2012 | East Asian | 749 | 763
(age/sex-matched) | 44.7/55.3 | 45.9/54.1 | 24.7 (13.6) | 26.4 (13.3) | No relation | A allele ↓(br1)GA
genotype ↓ | C allele ↓(br1)CT
genotype ↓ | (25) |
| Birlea, 2006 | Caucasian | 31 | 33 | 67.7/32.3 | | 53 (17.1) | - | No relation | - | No relation | (23) |
| Zhang, 2018a | | 7b | | - | - | - | - | No relation | No relation | No relation | (18) |
| Li, 2015a | | 4b | | - | - - - - | | | | GG genotype ↑ - (in
East Asians) | | (16) |
| This study | Caucasian | 110 | 509 (general
population) | 68/42 | 289/220 | 45.1 (13.4) | 40.5 | No relation | No relation | No relation | |
With respect to VDR SNPs effect on vitiligo
phenotypes, two studies investigating the relationship of the
VDR FokI, BsmI, and TaqI with clinical
characteristics of vitiligo failed to demonstrate a link between
the studied VDR SNPs and age at disease onset (22,26). In contrast, our intra-patient
analysis showed that the VDR FokI SNPs could have a
protective effect against early-onset vitiligo, as carriers of the
CC genotype or C allele of FokI exhibited a 71.4% and a
45.5% decreased risk of developing vitiligo before the age of 30
compared to patients carrying the FokI TT genotype or T
allele, respectively. Although not related to vitiligo per
se, VDR FokI appears to delay the onset of vitiligo
until the age of 30 and may thus be a potential biomarker providing
prognostic clues for early detection of this condition.
In addition, using the T allele as reference, we
observed a protective effect of the VDR TaqI C allele
against leukotrichia. This finding contrasts with previous data
demonstrating no association between the VDR
FokI/BsmI/TaqI poly-morphisms and leukotrichia
(22). Given that the active
melanocytes located into the black hair follicles can serve as a
main source of perifollicular repigmentation, the presence of
leukotrichia is known to predict a poor response to traditional
treatments, pointing towards a surgical solution (28-30). In this sense, it seems possible
that the VDR TaqI might indirectly be implicated in
treatment decision making despite its lack of association with
vitiligo per se. Moreover, since the TaqI
polymorphism is located in the gene's regulatory region (CpG site),
leukotrichia also appears as a clinical feature that can be
influenced not only by genetic regulation but also by epigenetic
factors that may affect VDR expression levels (Fig. 2) (31).
Contrary to Ochoa-Ramírez et al (26), no correlation between the VDR
BsmI and Koebner phenomenon was observed in our sample.
Although, similar to prior studies (22,26), we could not reveal further
associations between the studied VDR polymorphisms and other
clinical manifestations of vitiligo, our findings underline the
need for analyzing VDR variants according to disease-related
clinical features in order to identify genetically-based subsets of
patients that may benefit from personalized approaches to vitiligo
diagnosis or treatment.
This study presents some limitations to consider.
Apart from the relatively small sample size (110 cases), our
analyses included data only from SEC subjects, thus making the
results applicable only to this ethnotic group, especially
considering that genetic variability differs within European
populations (32-34). Moreover, investigations on other
common VDR SNPs, such as the ApaI, as well as
haplotype analysis was not conducted, thus limiting the acquisition
of further genetic information. Regarding other clinical aspects,
i.e., Sutton nevi, the limited data available did not allow us to
analyze each feature separately, which might have influenced the
results.
In conclusion, the current study provided evidence
to support a potential implication of the studied VDR SNPs
in the clinical course and treatment decision making of vitiligo.
The FokI CC genotype and C allele seemed to play a
protective role in early onset of vitiligo (≤30 years), while the
TaqI C allele may reduce the risk for future development of
leukotrichia. Moreover, no evidence was found to support an
association between the VDR BsmI, TaqI, and
FokI loci and vitiligo susceptibility in our cohort.
Even with limitations, this study will enrich the
evolving field of vitiligo genetics, especially among SEC subjects,
providing a reference for subsequent investigations in order to
translate all genetically derived data into clinical applications
for early diagnosis and individualized, precision treatment of
vitiligo. Further research with larger sample sizes is needed to
validate our results and elucidate whether the same associations
are also eligible in vitiligo patients among other Caucasian
populations or diverse ethnotic groups.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
MSK, ND, EN conceived the presented idea, while DI
and PS developed the whole theory. MSK designed the experiments and
DI, ML, PS prepared the samples and performed the experiments. PS,
EN, SM, MS, SG, AT and DR contributed to clinical data and sample
collection. KX made the data entry and CD performed the statistical
analyses. DI, MSK, PS, ND interpreted the results and designed the
figures. PS, DI, AN and LK wrote the manuscript under the
supervision of ND and EN, and DAS contributed to the final editing.
All the authors discussed the results and contributed to the final
manuscript.
Ethics approval and consent to
participate
Written informed consent for participation in the
study and use of their genetic data was obtained from all
participants.
Patient consent for publication
Written informed consent for publication of any
associated data was obtained from all participants.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
Acknowledgments
We would like to thank GENOMED for providing
anonymized and de-identified genotyping data of SEC
individuals.
Abbreviations:
|
BSA
|
body surface area
|
|
CI
|
confidence interval
|
|
EDTA
|
Ethylenediaminetetraacetic acid
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
PCR
|
polymerase chain reaction
|
|
SD
|
standard deviation
|
|
SEC
|
Southeastern European Caucasian
|
|
SNPs
|
single-nucleotide polymorphisms
|
|
VDR
|
vitamin D receptor
|
References
|
1
|
Ezzedine K, Eleftheriadou V, Whitton M and
van Geel N: Vitiligo. Lancet. 386:74–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roberts GHL, Santorico SA and Spritz RA:
The genetic architecture of vitiligo. Pigment Cell Melanoma Res.
33:8–15. 2020. View Article : Google Scholar
|
|
3
|
Wu J, Zhou M, Wan Y and Xu A:
CD8+ T cells from vitiligo perilesional margins induce
autologous melanocyte apoptosis. Mol Med Rep. 7:237–241. 2013.
View Article : Google Scholar
|
|
4
|
Spritz RA and Andersen GH: Genetics of
Vitiligo. Dermatol Clin. 35:245–255. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Krasanakis T, Nikolouzakis TK, Sgantzos M,
Mariolis-Sapsakos T, Souglakos J, Spandidos DA, Tsitsimpikou C,
Tsatsakis A and Tsiaoussis J: Role of anabolic agents in colorectal
carcinogenesis: Myths and realities (Review). Oncol Rep.
42:2228–2244. 2019.PubMed/NCBI
|
|
6
|
Barbu CG, Arsene AL, Florea S, Albu A,
Sirbu A, Martin S, Nicolae AC, Burcea-Dragomiroiu GTA, Popa DE,
Velescu BS, et al: Cardiovascular risk assessment in osteo-porotic
patients using osteoprotegerin as a reliable predictive biochemical
marker. Mol Med Rep. 16:6059–6067. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Murdaca G, Tonacci A, Negrini S, Greco M,
Borro M, Puppo F and Gangemi S: Emerging role of vitamin D in
autoimmune diseases: An update on evidence and therapeutic
implications. Autoimmun Rev. 18:1023502019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kechichian E and Ezzedine K: Vitamin D and
the skin: an update for dermatologists. Am J Clin Dermatol.
19:223–235. 2018. View Article : Google Scholar
|
|
9
|
Upala S and Sanguankeo A: Low
25-hydroxyvitamin D levels are associated with vitiligo: A
systematic review and meta-analysis. Photodermatol Photoimmunol
Photomed. 32:181–190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Birlea SA, Costin GE and Norris DA: New
insights on therapy with vitamin D analogs targeting the
intracellular pathways that control repigmentation in human
vitiligo. Med Res Rev. 29:514–546. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Parsad D and Kanwar AJ: Topical vitamin D
analogues in the treatment of vitiligo. Pigment Cell Melanoma Res.
22:487–488. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Siokas V, Aslanidou P, Aloizou AM,
Peristeri E, Stamati P, Liampas I, Arseniou S, Drakoulis N, Aschner
M, Tsatsakis A, et al: Does the CD33 rs3865444 polymorphism confer
susceptibility to Alzheimer's disease? J Mol Neurosci. 70:851–860.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dardiotis E, Aloizou AM, Siokas V, Tsouris
Z, Rikos D, Marogianni C, Aschner M, Kovatsi L, Bogdanos DP and
Tsatsakis A: Paraoxonase-1 genetic polymorphisms in
organo-phosphate metabolism. Toxicology. 411:24–31. 2019.
View Article : Google Scholar
|
|
14
|
Anbar TS, Hegazy RA, Picardo M and Taieb
A: Beyond vitiligo guidelines: Combined stratified/personalized
approaches for the vitiligo patient. Exp Dermatol. 23:219–223.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nejentsev S, Godfrey L, Snook H, Rance HS,
Walker NM, Lam AC, Guja C, Ionescu-Tirgoviste C, Undlien DE, et al:
Comparative high-resolution analysis of linkage disequilibrium and
tag single nucleotide polymorphisms between populations in the
vitamin D receptor gene. Hum Mol Genet. 13:1633–1639. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Wu Y, Li L, Cai YF, Geng L, Gao XH
and Chen HD: Association of ApaI and BsmI polymorphisms with
vitiligo risk: A meta-analysis. Clin Exp Dermatol. 40:794–803.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uitterlinden AG, Fang Y, Van Meurs JB,
Pols HA and Van Leeuwen JP: Genetics and biology of vitamin D
receptor polymorphisms. Gene. 338:143–156. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang JZ, Wang M, Ding Y, Gao F, Feng YY,
Yakeya B, Wang P, Wu XJ, Hu FX, Xian J, et al: Vitamin D receptor
gene polymorphism, serum 25-hydroxyvitamin D levels, and risk of
vitiligo: A meta-analysis. Medicine (Baltimore). 97:e115062018.
View Article : Google Scholar
|
|
19
|
Lee YH: Vitamin D receptor ApaI, TaqI,
BsmI, and FokI polymorphisms and psoriasis susceptibility: An
updated meta-analysis. Clin Exp Dermatol. 44:498–505. 2019.
View Article : Google Scholar
|
|
20
|
Heine G, Hoefer N, Franke A, Nöthling U,
Schumann RR, Hamann L and Worm M: Association of vitamin D receptor
gene polymorphisms with severe atopic dermatitis in adults. Br J
Dermatol. 168:855–858. 2013. View Article : Google Scholar
|
|
21
|
Ezzedine K, Lim HW, Suzuki T, Katayama I,
Hamzavi I, Lan CCE, Goh BK, Anbar T, Silva de Castro C, Lee AY, et
al: Vitiligo Global Issue Consensus Conference Panelists: Revised
classification/nomenclature of vitiligo and related issues: The
Vitiligo Global Issues Consensus Conference. Pigment Cell Melanoma
Res. 25:E1–E13. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aydıngöz IE, Bingül I, Doğru-Abbasoğlu S,
Vural P and Uysal M: Analysis of vitamin D receptor gene
polymorphisms in vitiligo. Dermatology. 224:361–368. 2012.
View Article : Google Scholar
|
|
23
|
Birlea S, Birlea M, Cimponeriu D, Apostol
P, Cosgarea R, Gavrila L, Tigan S, Costin G and Das P: Autoimmune
diseases and vitamin D receptor Apa-I polymorphism are associated
with vitiligo in a small inbred Romanian community. Acta Derm
Venereol. 86:209–214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hassan I, Bhat YJ, Majid S, Sajad P,
Rasool F, Malik RA and Ul Haq I: Association of vitamin D receptor
gene polymorphisms and serum 25-hydroxyvitamin D levels in vitiligo
- a case-control study. Indian Dermatol Online J. 10:131–138.
2019.PubMed/NCBI
|
|
25
|
Li K, Shi Q, Yang L, Li X, Liu L, Wang L,
Li Q, Wang G, Li CY and Gao TW: The association of vitamin D
receptor gene polymorphisms and serum 25-hydroxyvitamin D levels
with generalized vitiligo. Br J Dermatol. 167:815–821. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ochoa-Ramírez LA, Díaz-Camacho SP,
Becerra-Loaiza DS, Verdugo-Nieto L, Muñoz-Estrada VF,
Servín-Vázquez LA, Osuna-Ramírez I, Rodríguez-Millán J and
Velarde-Félix JS: Catalase but not vitamin D receptor gene
polymorphisms are associated with nonsegmental vitiligo in
Northwestern Mexicans. Int J Dermatol. 58:1264–1269. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sobeih S, Mashaly HM, Gawdat H, Amr K,
Hamid MF and Shaalan E: Evaluation of the correlation between serum
levels of vitamin D and vitamin D receptor gene polymorphisms in an
Egyptian population. Int J Dermatol. 55:1329–1335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee DY, Kim CR, Park JH and Lee JH: The
incidence of leukotrichia in segmental vitiligo: Implication of
poor response to medical treatment. Int J Dermatol. 50:925–927.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
van Geel N, Grine L, De Wispelaere P,
Mertens D, Prinsen CAC and Speeckaert R: Clinical visible signs of
disease activity in vitiligo: A systematic review and
meta-analysis. J Eur Acad Dermatol Venereol. 33:1667–1675. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saccone D, Asani F and Bornman L:
Regulation of the vitamin D receptor gene by environment, genetics
and epigenetics. Gene. 561:171–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang D, Xu X, Ma H, Yue X, Li C and Zhu W:
Optimization of the method for the culture of melanocyte precursors
from hair follicles and their activation by 1,25-dihydroxyvitamin
D3. Exp Ther Med. 6:967–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Katsarou MS, Karathanasopoulou A,
Andrianopoulou A, Desiniotis V, Tzinis E, Dimitrakis E, Lagiou M,
Charmandari E, Aschner M, Tsatsakis AM, et al: Beta 1, Beta 2 and
Beta 3 adrenergic receptor gene polymorphisms in a Southeastern
European population. Front Genet. 9:5602018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Katsarou MS, Papasavva M, Latsi R, Toliza
I, Gkaros AP, Papakonstantinou S, Gatzonis S, Mitsikostas DD,
Kovatsi L, Izotov BN, et al: Population-based analysis of cluster
headache-associated genetic polymorphisms. J Mol Neurosci.
65:367–376. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Katsarou MS, Latsi R, Papasavva M,
Demertzis N, Kalogridis T, Tsatsakis AM, Spandidos DA and Drakoulis
N: Population-based analysis of the frequency of HFE gene
polymorphisms: Correlation with the susceptibility to develop
hereditary hemochromatosis. Mol Med Rep. 14:630–636. 2016.
View Article : Google Scholar : PubMed/NCBI
|