Epitranscriptomics is an emerging research field of
biology, in which a recently discovered novel mechanism of
post-transcriptional regulation of RNA has been suggested to play a
vital role in the regulation of RNA function and to be involved in
various biological processes, including disease progression
(1). RNA methylation modification
is the most common type of RNA modification in eukaryotes,
accounting for 60% of total RNA modification. The identified RNA
modifications have been found to contribute to the structural
complexity of RNA and participate in multiple biological functions
(2).
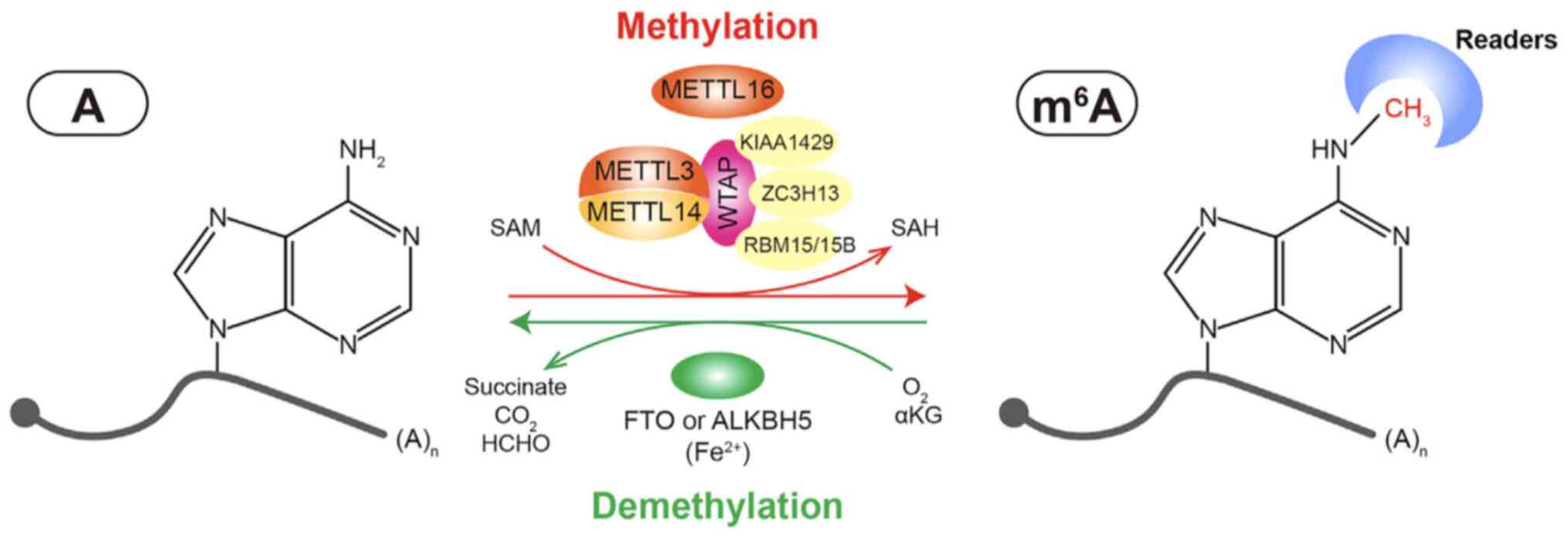
Writers [i.e., methyltransferase-like 3 (METTL3),
METTL14 and Wilms tumor 1-associated protein (WTAP)] and erasers
[i.e., fat mass and obesity-associated protein (FTO) and
a-ketoglutarate-dependent dioxygenase (ALKB) homolog 5 (ALKBH5)]
are responsible for catalyzing and removing m6A,
respectively (34-37). The regulatory mechanisms of
m6A are complex. Moreover, an increasing number of
m6A modulators have been discovered, particularly
methyltransferases and m6A binding proteins.
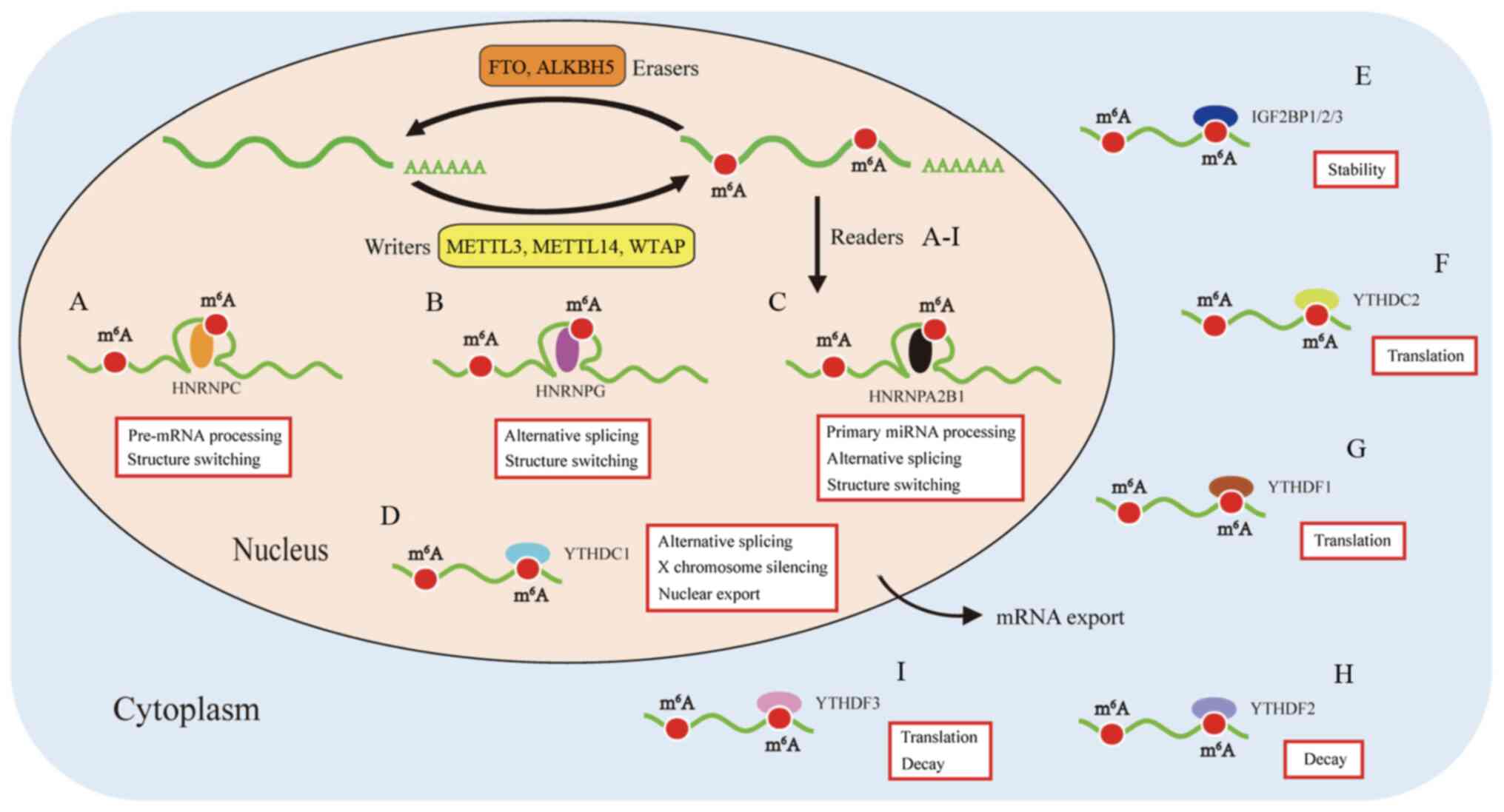
m6A readers have both stimulatory and inhibitory effects
on translation dynamics. YTHDF2 can accelerate the degradation of
m6A-modified maternal transcripts, while YTHDF1
increases the translation efficiency of m6A-marked
dynamic transcript (38-40). The site-specific m6A
maps of transcripts and the role of site-specific methylation in
translation warrant further investigation.
WTAP is a mammalian splicing factor responsible for
the interaction with the METTL3-METTL14 complex. WTAP is critical
for initiating and guiding the localization of nuclear speckles,
which is required for m6A methylation activation. WTAP
also regulates their recruitment to mRNA targets (46). Numerous regulatory enzymes have
been found to interact with the heterodimer, such as Vir-like
METTL16, KIAA1429 (47),
RNA-binding motif 15 (RBM15)/RBM15B (48), Vir-like m6A methyltransferase
associated (49), E3
ubiquitin-protein ligase Hakai and zinc finger CCCH domain
containing protein 13 (50). The
above-mentioned enzymes bind to mRNA and recruit the METTL3-METTL14
complex, guiding the heterodimer to target regions (Fig. 1).
A number of AlkB protein family members serve as RNA
methylation erasers through the oxidative demethylation of
m6A marks; however, only ALKBH5 has been identified to
exhibit efficient demethylation activity towards m6A in
animals (54). ALKBH5 has a tight
interaction with mRNA and other RNA substrates. The alanine-rich
sequence and potential coiled-coil structure in the N-terminus of
ALKBH5 are important for its localization. ALKBH5 can affect both
the synthesis and splicing rate of mRNAs (36) (Fig.
1).
The YTH domain was identified in the human splicing
factor YT521-B and is typically found in 174 different proteins
expressed in eukaryotes, with a residue size of 100-150. It is
featured by 14 invariant residues with an α-helix/β-fold to bind to
RNA (57,58). The YTH domain-containing family
can be divided into two subfamilies, YTHDF (YTHDF1-3) and YTHDC
(YTHDC1-2) (59). YTHDF1 is known
as an important m6A reader promoting the cap-independent
translation regulation of m6A-modified RNA transcripts
in the 5′UTR region. YTHDF1 relocates from the cytoplasm to the
nucleus and initiates and augments translation in an eIF3
initiation factor-dependent manned (39). Coversely, YTHDF2 trans-ports the
mRNA targets to the cytoplasmic processing body and promotes its
degradation (18). Specifically,
Du et al (60) reported
the specific mechanism of deadenylation mediated by YTHDF2 in
mammalian cells. YTHDF2 distinguishes m6A-bearing RNAs,
and then interacts with CNOT1 to recruit the CCR4-NOT complex,
which is critical for the demethylation of m6A-marks by
CAF1 and CCR4. YTHDF3 plays a cooperative role in RNA stability and
translation among proteins from the YTHDF family to influence the
metabolism of m6A-methylated mRNA (24).
Nuclear binding protein YTHDC1 regulates alternative
nuclear mRNA splicing by recruiting SRSF3 splicing factor and
inhibiting SRSF10 binding to mRNA (61). YTHDC1 has also been found to be
involved in nuclear transport (62) and gene translation silencing
(48). YTHDC2 has several defined
domains, including the YTH domain, R3H domain and ankyrin repeats
(63). Phillip reported that
YTHDC2 is a critical m6A reader involved in
spermatogenesis. YTHDC2 selectively binds to m6A
residues, decreases their mRNA abundance and enhances the
translation efficiency of mRNAs (64).
Several proteins from the HNRNP family have been
found to have the potential of recognizing m6A-modified
mRNA, including HNRNPA2B1, HNRNPC and HNRNPG. HNRNPA2B1 is a
nuclear m6A-reader that directly binds to nuclear
transcripts, elicits regulatory effects on RNA splicing (55) and promotes primary microRNA
processing (13). HNRNPC plays a
regulatory role in the acceleration of pre-miRNA processing
(65). HNRNPG interacts with RNA
polymerase II and m6A-modified pre-mRNAs to modulate the
alternative splicing and expression of target mRNAs (66).
The rapid emergence of next-generation sequencing
technology has increased our understanding of epigenetic research
and helped us assess the in vivo methylation state of
m6A sites. The two novel m6A modification
site detection technologies, m6A sequencing
(m6A-seq) and m6A-specific methylated RNA
immunoprecipitation (MeRIP-seq) were created. m6A-seq,
presented by Dominissini et al (8), is a novel approach based on
m6A antibody-mediated capture and high-throughput
sequencing. m6A-Modified are highly conserved between
human and mice (8). Meyer et
al (68) presented the
MeRIP-seq method for transcriptome-wide m6A
localization, and m6A methylated RNA was
immunoprecipitated, followed by next-generation sequencing.
m6A-seq and MeRIP-seq identified thousands of mRNAs with
m6A modification and revealed that m6A is a
pervasive and dynamically reversible internal chemical modification
of mRNA. However, there exist some limitations in the detection
technology. This method has a relatively low accuracy in the
identification of m6A modification sites in the whole
transcriptome for the following reasons: First, only RNA fragments
100-200 nt long could be captured through these mapping approaches.
Besides, two similar m6A sites could not be
identified.
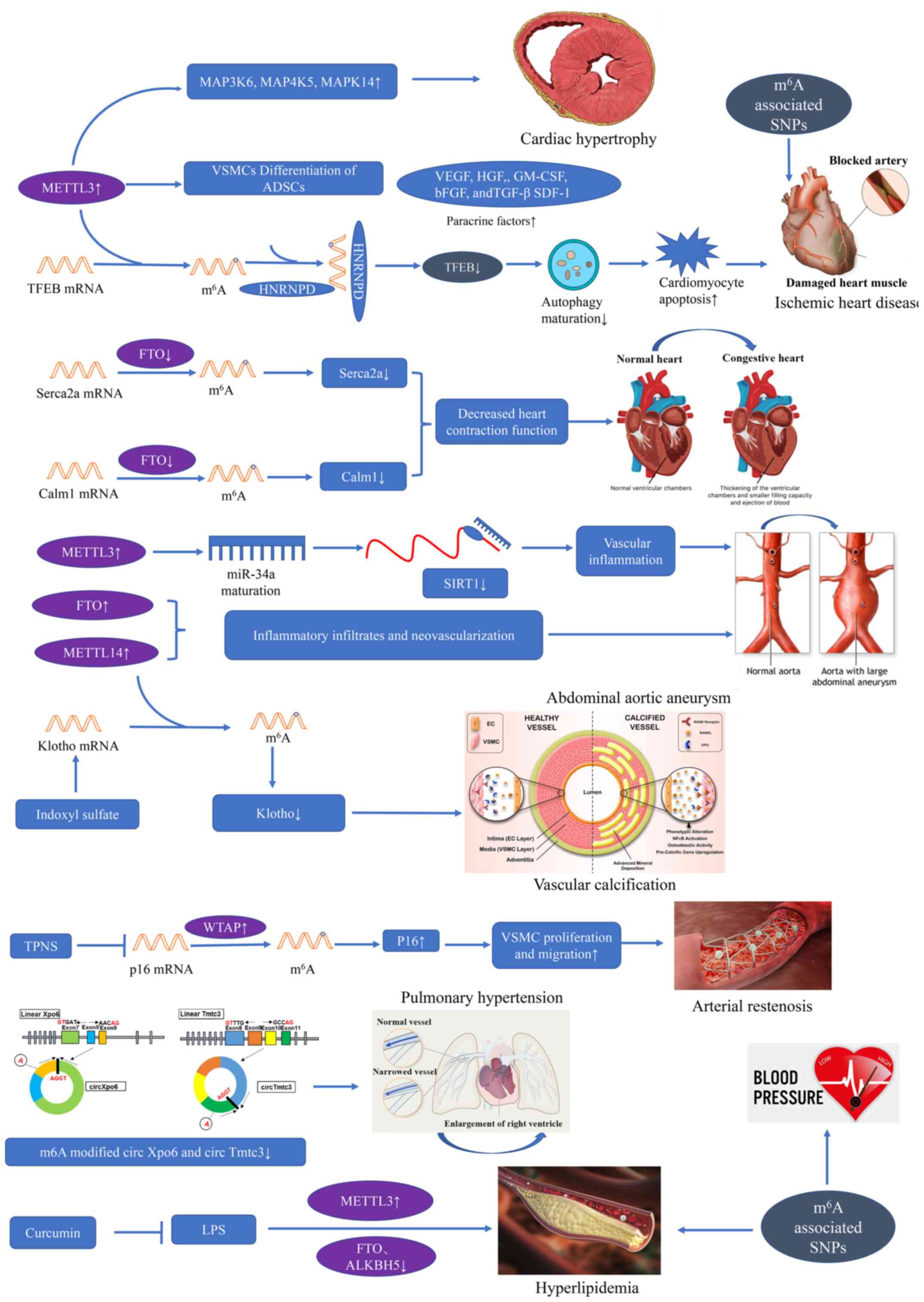
Adipose-derived stem cells (ADSCs) are a source of
mesenchymal stem cells that can be used to develop biological
treatment strategies for tissue regeneration. ADSCs can
differentiate into vascular smooth muscle cells (VSMCs) under
certain conditions. METTL3 mRNA and protein expression are
increased in the VSMC differentiation of ADSCs. The downregulation
of METTL3 contributes to a decrease in the m6A
modification of VSMC-specific markers, thus leading to a decline in
their mRNA and protein expression. The silencing of METTL3 inhibits
ADSC differentiation into VSMCs. In addition, an increased
expression of paracrine factors has been observed in ADSC
differentiation into VSMCs, including vascular endothelial growth
factor, hepatocyte growth factor, granulocyte-macrophage
colony-stimulating factor, basic fibroblast growth factor and
stromal cell-derived factor-1 (77). An investigation of the role of
METTL3 in the regulation of VSMC differentiation has suggested that
METTL3 is involved in ADSC differentiation to VSMCs, providing a
promising perspective for novel therapeutic strategies for vascular
network regeneration (77).
Cardiac remodeling occurs in the heart following
stress stimulation or injury, and it involves molecular, cellular
and interstitial alterations, clinically manifested as changes in
size, shape and function (78).
Stress stimulation initially induces adaptive hypertrophic response
to produce sufficient force to match increased wall tension or
increased pressure/volume overload in cardiomyocytes (79). Currently, post-transcriptional
regulation is recognized as similarly critical to transcriptional
regulation in cardiac hypertrophy (80).
The lysosomal-mediated degradation pathway
(autophagy) is an important evolution-arily conserved degradation
mechanism in eukaryotic cells that removes unnecessary and harmful
parts to maintain homeostasis (82). Autophagy is associated with
numerous human diseases, including CVD (83). Song et al (83) first researched the role of
m6A in autophagy and discovered that m6A
modification is significantly upregulated in hypoxia/reoxygenation
(H/R)-treated cardiomyocytes and ischemia/reperfusion (I/R)-treated
mouse hearts. The key member of the methyltransferase complex,
METTL3, was abnormally upregulated in infraction heart tissue, as
compared with healthy heart tissue. Consistently, an increased
expression of METTL3 was identified in cardiomyocytes treated with
H/R. The silencing of METTL3 increased the I/R-impaired autophagic
flux and inhibited apoptosis (83).
It has been well documented that transcription
factor EB (TFEB) is an important gene in lysosomal biogenesis,
which also drives the expression of autophagy and lysosomal genes
(84,85). Decreased TFEB mRNA stability and
TFEB protein expression were previously discovered in H/R-induced
cardiomyocytes. The silencing of METTL3 significantly promoted the
expression and transcriptional activity of TFEB and translocation
to the nucleus. TFEB knockdown inhibited the enhanced autophagy
induced by the downregulation of METTL3, indicating that the
mediation of autophagic flux by METTL3 is TFEB-dependent (86). The RNA binding protein
heterogeneous nuclear ribonucleoprotein D (HNRNPD) was reported to
bind to m6A-modified RNA (25), and an increased HNRNPD was found
in patients with heart failure (87). RIP-PCR confirmed the interaction
between HNRNPD and TFEB pre-mRNA. H/R further enhanced the binding
(86). In accordance with
previous research (88), the
H/R-induced HNRNPD elevation has been shown to promote TFEB mRNA
degradation, and reduce the protein expression and transcriptional
activity of TFEB. Furthermore, TFEB can suppress METTL3 expression
by downregulating the stability of METTL3 mRNA, thereby forming
negative feedback between METTL3 and TFEB (86). On the whole, research has revealed
that the METTL3-TFEB feedback loop plays an essential role in
autophagy, and has laid a foundation for dynamic m6A
modification in ischemic heart disease.
There is currently no effective approach available
for adverse cardiac remodeling post-ischemia. mRNA and protein
expression levels in the right ventricles during heart failure are
inconsistent, suggesting a role for post-transcriptional regulation
in failing hearts (89).
Mathiyalagan et al (90)
discovered that FTO played a vital role in cardiac contractile
function during homeostasis and remodeling. An increased
m6A modification and significantly decreased FTO
expression were found in infarct and peri-infarct regions of
failing hearts, as compared with healthy heart tissues. FTO
overexpression attenuated the ischemia-induced elevation in
m6A modification. FTO-knockdown exhibited aberrantly
increased arrhythmic events in cardiomyocytes. In vivo,
FTO-overexpression significantly improved cardiac function at the
chronic stage of post-myocardial infraction. Enhanced angiogenesis
and alleviated fibrosis were also observed. A more rapid
progression of heart failure was witnessed with a lower ejection
fraction and more severe dilatation in FTO knockout mice,
indicating the indispensable role of FTO in heart failure (90). Mechanistically, the
Ca2+-handling transcript Serca2a has been shown to be
hypermethylated in failing hearts, and FTO demethylated Serca2a and
enhanced the stability of Serca2a mRNA (91) or exerted co-transcriptional
regulation (38), leading to an
increase in SERCA2a expression, ultimately improving cardiac
contractile function. Collectively, these results provide
compelling evidence that FTO-mediated m6A methylation
plays an important role in cardiac function during heart failure
and recommend FTO as a novel therapeutic approach (90).
AAA is a common vascular condition among the
elderly with a high mortality rate (94). AAA is characterized by chronic
inflammation in the tunica media and adventitia, with the
upregulation of numerous cytokines activating a large amount of
proteolytic enzymes, ultimately leading to the rapid expansion and
rupture of AAA (95). The exact
mechanisms contributing to AAA have not yet been elucidated. He
et al first investigated the role of m6A
modification in AAA and provided a potential epigenetic mechanism
in AAA (96). They discovered
that m6A modification significantly increased in AAA, as
compared to healthy aortic tissues. METTL14 was shown to be
associated with inflammatory infiltrates and neovascularization in
AAA, and increased FTO was also involved in aberrant m6A
modification. A higher m6A level was also found to be
associated with a higher risk of rupture. They subsequently
discovered significant correlations between the m6A
modification level and the mRNA expression level of the writers,
erasers and readers, indicating a tight crosstalk among these
modulators. In clinical data, a higher m6A level was
found to be positively associated with the AAA diameter and
hematological parameters (96).
Hence, abnormal m6A modification is crucial to the
occurrence and progression of AAA.
Chronic kidney disease (CKD) is a worldwide public
health concern. Patients with CKD have a 2-fold higher risk of
suffering from CVD and also exhibit a higher mortality rate, as
compared to the healthy population (98). Patients with CKD are more likely
to suffer from vascular calcification, and vascular calcification
is recognized as the main cause of increased mortality from CVD
(99). It has been confirmed that
indoxyl sulfate (IS), a vital protein-bound uremic toxin, is
associated with the development of renal and vascular progression
(100). Chen et al
investigated the epigenetic translation underlying IS-induced
vascular calcification and noted marked in vitro, in
vivo and translational evidence, indicating the essential role
of METTL14 in IS-induced vascular calcification (101). A significantly elevated
m6A level of total RNA was discovered in the radical
arteries of patients with end-stage renal disease and preclinical
calcified mouse arteries. An increased METTL14 mRNA and protein
expression was detected in calcified arteries, indicating an
IS-induced increase in the levels of METTL14 and METTL14, which
mediated the increase in global m6A modification, which
may be a hallmark of vascular calcification. In vitro, the
overexpression of METTL14 caused the loss of repair function.
Mechanistically, MeRIP analysis revealed that the
vascular-protecting transcript Klotho mRNA was hyper-methylated in
calcified arteries, resulting in the degradation and decreased mRNA
expression of Klotho. Therefore, this investigation demonstrated
the functional significance of METTL14 in IS-induced vascular
calcification and provided proof of concept for anti-vascular
calcification therapy through the modulation of METTL14 (101).
Pulmonary hypertension (PH) is a complex and
multidisciplinary pathophysi-ological disorder defined as a resting
mean pulmonary artery pressure(PAP) of >25 mmHg, as assessed by
right heart catheterization (102,103). Hypoxic PH (HPH) is a progressive
disease due to lung diseases and categorized as group III PH.
Chronic obstructive pulmonary disease and interstitial lung
diseases are common causes (2).
Currently there is no specific therapy for the increased PAP and
structural abnormalities of HPH (104).
circRNAs mostly play the role of a sponge for
miRNAs, and are involved in the regulation of target miRNAs
(111). A circRNA-miRNA-mRNA
network was previously constructed to explore the regulation of
gene expression in HPH. Key mRNAs associated with the Wnt (112) and FoxO signaling pathways
(113), and miRNAs reported to
be involved in HPH were filtered, and the most enriched novel
circRNA Xpo6 and Tmtc3 were then identified (110). RIP-PCR confirmed that the
expression of circXpo6 and circTmtc3 were significantly decreased
in both hypoxia-induced pulmo-nary artery smooth muscle cells and
endothelial cells (110).
Collectively, the study (110)
first revealed that m6A affected the stability of
circRNAs, subsequently influencing the circRNA-miRNA-mRNA network,
leading to the activation of the Wnt and FoxO signaling pathways
and ultimately promoting HPH. The results of that study (110) suggested that
m6A-modified circRNAs were associated with pulmonary
hypertension. However, the research was just an expression file of
circRNAs in lung tissues from HPH and control mice. The exact
molecular mechanisms underlying the role of m6A
methylation in HPH remain to be elucidated.
High-throughput sequencing technology has
identified millions of SNPs across multiple genomes. Distinguishing
the disease-associated functional variants, which can regulate
amino acids at the protein level (114), RNA secondary structure (115), RNA-protein interactions and RNA
splicing (116) or editing
(117) at the transcriptional or
post-transcriptional level, is a major challenge. SNPs can
interfere with m6A methylation by altering the RNA
sequences of target sites or key flanking nucleotides (118). Putative m6A-related
SNPs (m6A-SNPs), which are close to or at the exact
methylation site, can disrupt m6A modification and
corresponding biological processes through multiple mechanisms, due
to the location of m6A-SNPs, and can subsequently affect
disease progression (119).
m6A-SNPs have been recognized as a fundamental class of
crucial multifunctional genetic variants associated with CVDs,
which is concerning. Numerous m6A-SNPs associated with
CVDs have been identified using genome-wide association studies
(GWAS) and the genetic functional mechanisms have been explored.
These m6A-SNPs and genes may be candidates and promote
novel therapeutic approaches (120,121). The present study reviewed the
relevant research on m6A-SNPs and CVD.
Genetic factors are significant, and heritability
contributes to 40-60% of BP cases (122). A large number of SNPs for BP
have been identified by a GWAS (123). Zheng et al (118) reported a comprehensive database
(m6Avar), which is a useful tool for the investigation
of the association between m6A-related variants and
disease. A previous study first made efforts to investigate the
association between m6A-SNPs and BP by excavating data
from a large-scale GWAS, and demonstrated that m6A-SNP
may play a pivotal role in BP regulation (124). Some identified BP-related SNPs
in genes have been found to be associated with coronary artery
disease (CAD) (125). The
majority of m6A-SNPs were closely associated with gene
expression. For example, rs9847953 and rs197922 were identified as
potential functional variants and strongly associated with gene
expression, ultimately contributing to the regulation of blood
pressure.
CAD is the leading cause of mortality and
disability worldwide, with both genetic and environmental factors
playing an essential role in its pathogenesis (131). The effect of m6A-SNPs
on CAD was previously explored and 304 m6A-SNPs were
found to be associated with CAD (120). Some m6A-SNPs not only
alter m6A methylation and local gene expression, but
also regulate protein binding, indicating that m6A-SNPs
may be functional variants of CAD. Among these, rs12286 was
markedly associated with CAD. Mechanistically, rs12286 may
influence the m6A methylation level and regulate the
expression of downstream gene ADAMTS7 to exert its function
(120). On the whole, these
findings elucidated the role of m6A-SNPs in CAD;
however, the specific regulatory mechanisms remain to be
explored.
Blood cholesterol is one of the most important
heritable risk factors for CAD, and is positively associated with
mortality due to CAD (132). The
role of m6A modification in lipid metabolism has been
reported (133). Mo et al
(121) investigated the effect
of m6A-SNPs on lipid levels using GWAS summary datasets
of 188,578 individuals. m6A-SNP rs6859 at the 3′UTR of
poliovirus receptor-related 2 was found to be significantly
expressed at the genome-wide level and associated with
triglycerides, total cholesterol and high/low-density lipoprotein
cholesterol, suggesting that rs6859 may be a lipid
metabolism-related multifunctional SNP and an important candidate
for further functional studies. That study found a large amount of
lipid-related m6A-SNPs (121). Further studies are required to
elucidate the mechanisms.
Characteristic biomarkers play a crucial role in
the prevention, diagnosis and treatment of disease. Previous
studies have demonstrated that methylation serves as a potential
biomarker and therapeutic target in cancer (134). Fustin et al demonstrated
that m6A editing through specific METTL3 inhibition
contributed to eliciting circadian period elongation and RNA
processing delay (10).
Total Panax notoginseng saponin (TPNS) has been
reported to inhibit balloon injury-induced intimal hyperplasia and
VSMC proliferation (135). Zhu
et al (136) observed a
reduced WTAP expression and decreased m6A modification
in balloon catheter-injured rat carotid artery. Furthermore, TPNS
alleviated the proliferation and migration of VSMCs through the
upregulation of WTAP and downstream target p16 m6A
modification. Therefore, WTAP and m6A regulatory p16
expression may serve as a novel molecular target for the assessment
of arterial stenosis risk and a novel therapeutic target of
arterial stenosis.
It is widely acknowledged that lipopolysaccharide
(LPS) is a trigger for inflammation and metabolic diseases
(137). Rao et al
(138) first found that curcumin
reduced the blood cholesterol level in normal animals. Lu et
al discovered that the lipid metabolism disorder in the liver
and increase in total cholesterol induced by LPS injection were all
attenuated after dietary supplementation with curcumin (139). Mechanistically, the LPS
injection increased the m6A methylation level,
accompanied by an increased METTL3 level, and decreased the mRNA of
ALKBH5 and FTO in the liver. Of note, curcumin affected the
expression of some crucial m6A-related modulators,
including METTL3, METTL14, ALKBH5, FTO and YTHDF2. Further
upregulated m6A methylation in the liver was discovered
in piglets that had received dietary curcumin, as compared with the
LPS injection group, suggesting that the protective effect of
curcumin in LPS-induced hepatic lipid metabolism may result from
increased m6A RNA modification (139). The precise mechanisms of how
curcumin affects gene-specific m6A modification require
further investigation. Collectively, the findings indicate that
m6A could be a promising therapeutic target for
hyperlipidemia.
As a newly identified type of post-transcriptional
regulation, the dynamic and reversible m6A modification
is the most prevalent type of internal modification of RNA
methylation. m6A modification plays a significant role
not only in various cellular biological processes but also in the
pathogenesis of CVDs. The dysregulated m6A methylation
has enhanced our understanding of epigenetic regulation in the
pathogenesis in cardiovascular disorders. However, the
investigation of m6A modification in CVDs remains in its
infancy and a more comprehensive understanding of the biological
function of m6A is needed. m6A modification
may further contribute to the recognition of molecular mechanisms
underlying cardio-vascular pathogenesis, and might provide novel
insight into the potential biomarkers and therapeutic approaches
for CVD in the future.
Not applicable.
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81970237 and
81600227).
Not applicable.
YQin, LL, EL, JH, GY, DW, YQiao and CT contributed
to the conception and design of the study. YQin, LL, EL and JH
searched the relevant literature. YuQ wrote the manuscript. GY,
YQiao and DW provided advice and were responsible for revising the
manuscript. All authors have read and approved the final version of
the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Stellos K: The rise of epitranscriptomic
era: Implications for cardiovascular disease. Cardiovasc Res.
113:e2–e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang H, Weng H and Chen J: m6A
modification in coding and non-coding RNAs: Roles and therapeutic
implications in cancer. Cancer Cell. 37:270–288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei CM, Gershowitz A and Moss B:
Methylated nucleotides block 5′ terminus of HeLa cell messenger
RNA. Cell. 4:379–386. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee M, Kim B and Kim VN: Emerging roles of
RNA modification: m(6)A and U-tail. Cell. 158:980–987. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhat SS, Bielewicz D, Jarmolowski A and
Szweykowska- Kulinska Z: N6-methyladenosine
(m6A): Revisiting the old with focus on new, an
arabidopsis thaliana centered review. Genes (Basel). 9:5962018.
View Article : Google Scholar
|
|
7
|
Zhao W, Qi X, Liu L, Ma S, Liu J and Wu J:
Epigenetic regulation of m6A modifications in human
cancer. Mol Ther. 19:405–412. 2020.
|
|
8
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Molinie B, Wang J, Lim KS, Hillebrand R,
Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon
P, et al: m(6)A-LAIC-seq reveals the census and complexity of the
m(6)A epitranscriptome. Nat Methods. 13:692–698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fustin JM, Doi M, Yamaguchi Y, Hida H,
Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I
and Okamura H: RNA-methylation-dependent RNA processing controls
the speed of the circadian clock. Cell. 155:793–806. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
12
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6) A promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen XY, Zhang J and Zhu JS: The role of
m6A RNA methylation in human cancer. Mol Cancer.
18:1032019. View Article : Google Scholar
|
|
15
|
Edupuganti RR, Geiger S, Lindeboom RGH,
Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen PWTC, Rossa M,
et al: N6-methyladenosine (m6A) recruits and
repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol.
24:870–878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Visvanathan A and Somasundaram K: mRNA
traffic control reviewed: N6-Methyladenosine (m6A) takes
the driver's seat. Bioessays. Dec 4–2017.Epub ahead of print.
View Article : Google Scholar
|
|
17
|
Meyer KD and Jaffrey SR: The dynamic
epitranscriptome: N6-methyladenosine and gene expression control.
Nat Rev Mol Cell Biol. 15:313–326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tong J, Flavell RA and Li HB: RNA
m6A modification and its function in diseases. Front
Med. 12:481–489. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N,
Han D, Dominissini D, Dai Q, Pan T and He C: High-resolution N(6)-
methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6)
A sequencing. Angew Chem Int Ed Engl. 54:1587–1590. 2015.
View Article : Google Scholar
|
|
21
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m(6)A mRNA methylation directs translational
control of heat shock response. Nature. 526:591–594. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barbieri I, Tzelepis K, Pandolfini L, Shi
J, Millán-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister
AJ, Han N, et al: Promoter-bound METTL3 maintains myeloid leukaemia
by m6A-dependent translation control. Nature.
552:126–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X and He C: Dynamic RNA modifications
in posttranscriptional regulation. Mol Cell. 56:5–12. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N-methyladenosine-modified RNA. Cell Res. 27:315–328. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang S, Zhao BS, Zhou A, Lin K, Zheng S,
Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, et al: m6A
demethylase ALKBH5 maintains tumorigenicity of glioblastoma
stem-like cells by sustaining FOXM1 expression and cell
proliferation program. Cancer Cell. 31:591–606.e6. 2017. View Article : Google Scholar
|
|
26
|
Zhu S, Wang JZ, Chen D, He YT, Meng N,
Chen M, Lu RX, Chen XH, Zhang XL and Yan GR: An oncopeptide
regulates m6A recognition by the m6A reader
IGF2BP1 and tumorigenesis. Nat Commun. 11:16852020. View Article : Google Scholar
|
|
27
|
Wu Y, Yang X, Chen Z, Tian L, Jiang G,
Chen F, Li J, An P, Lu L, Luo N, et al: m6A-induced
lncRNA RP11 triggers the dissemi-nation of colorectal cancer cells
via upregulation of Zeb1. Mol Cancer. 18:872019. View Article : Google Scholar
|
|
28
|
Hess ME, Hess S, Meyer KD, Verhagen LA,
Koch L, Brönneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento
O, et al: The fat mass and obesity associated gene (Fto) regulates
activity of the dopaminergic midbrain circuitry. Nat Neurosci.
16:1042–1048. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du K, Zhang L, Lee T and Sun T: m(6)A RNA
methylation controls neural development and is involved in human
diseases. Mol Neurobiol. 56:1596–1606. 2019. View Article : Google Scholar
|
|
30
|
Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang
Y, Li J, Sheng R, Deng P, Wang Y, et al: Mettl3-mediated
m6A RNA methylation regulates the fate of bone marrow
mesenchymal stem cells and osteoporosis. Nat Commun. 9:47722018.
View Article : Google Scholar
|
|
31
|
Kane SE and Beemon K: Precise localization
of m6A in Rous sarcoma virus RNA reveals clustering of methylation
sites: Implications for RNA processing. Mol Cell Biol. 5:2298–2306.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Roignant JY and Soller M: m6A
in mRNA: An ancient mechanism for fine-tuning gene expression.
Trends Genet. 33:380–390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao Y, Shi Y, Shen H and Xie W:
m6A-binding proteins: The emerging crucial performers in
epigenetics. J Hematol Oncol. 13:352020. View Article : Google Scholar
|
|
34
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yao QJ, Sang L, Lin M, Yin X, Dong W, Gong
Y and Zhou BO: Mettl3-Mettl14 methyltransferase complex regulates
the quiescence of adult hematopoietic stem cells. Cell Res.
28:952–954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar :
|
|
37
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltrans-ferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar :
|
|
38
|
Slobodin B, Han R, Calderone V, Vrielink
JAFO, Loayza-Puch F, Elkon R and Agami R: Transcription impacts the
efficiency of mRNA translation via co-transcriptional N6-adenosine
methylation. Cell. 169:326–337.e12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Choi J, Ieong KW, Demirci H, Chen J,
Petrov A, Prabhakar A, O'Leary SE, Dominissini D, Rechavi G, Soltis
SM, et al: N(6)-methyladenosine in mRNA disrupts tRNA selection and
translation-elongation dynamics. Nat Struct Mol Biol. 23:110–115.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bujnicki JM, Feder M, Radlinska M and
Blumenthal RM: Structure prediction and phylogenetic analysis of a
functionally diverse family of proteins homologous to the MT-A70
subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol.
55:431–444. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Śledź P and Jinek M: Structural insights
into the molecular mechanism of the m(6)A writer complex. Elife.
5:e184342016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schöller E, Weichmann F, Treiber T, Ringle
S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G:
Interactions, localization, and phosphorylation of the
m6A generating METTL3-METTL14-WTAP complex. RNA.
24:499–512. 2018. View Article : Google Scholar
|
|
45
|
Yue Y, Liu J and He C: RNA
N6-methyladenosine methylation in post-transcriptional gene
expression regulation. Genes Dev. 29:1343–1355. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
47
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang
Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential
m6A mRNA methylation in 3′UTR and near stop codon and
associates with alternative polyadenylation. Cell Discov. 4:102018.
View Article : Google Scholar
|
|
50
|
Knuckles P, Lence T, Haussmann IU, Jacob
D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, et
al: Zc3h13/Flacc is required for adenosine methylation by bridging
the mRNA-binding factor Rbm15/Spenito to the m6A
machinery component Wtap/Fl(2)d. Genes Dev. 32:415–429. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tang C, Klukovich R, Peng H, Wang Z, Yu T,
Zhang Y, Zheng H, Klungland A and Yan W: ALKBH5-dependent m6A
demethylation controls splicing and stability of long 3′-UTR mRNAs
in male germ cells. Proc Natl Acad Sci USA. 115:E325–E333. 2018.
View Article : Google Scholar
|
|
52
|
Boissel S, Reish O, Proulx K,
Kawagoe-Takaki H, Sedgwick B, Yeo GS, Meyre D, Golzio C, Molinari
F, Kadhom N, et al: Loss-of-function mutation in the
dioxygenase-encoding FTO gene causes severe growth retardation and
multiple malformations. Am J Hum Genet. 85:106–111. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Su R, Dong L, Li C, Nachtergaele S,
Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al: R-2HG
exhibits anti-tumor activity by targeting
FTO/m6A/MYC/CEBPA signaling. Cell. 172:90–105.e23. 2018.
View Article : Google Scholar
|
|
54
|
A Alemu E, He C and Klungland A:
ALKBHs-facilitated RNA modifications and de-modifications. DNA
Repair (Amst). 44:87–91. 2016. View Article : Google Scholar
|
|
55
|
Alarcón CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of
m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang Z, Theler D, Kaminska KH, Hiller M,
de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM,
Allain FH and Stamm S: The YTH domain is a novel RNA binding
domain. J Biol Chem. 285:14701–14710. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Stoilov P, Rafalska I and Stamm S: YTH: A
new domain in nuclear proteins. Trends Biochem Sci. 27:495–497.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liao S, Sun H and Xu C: YTH domain: A
family of N6-methyladenosine (m6A) readers.
Genomics Proteomics Bioinformatics. 16:99–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M,
Ma J and Wu L: YTHDF2 destabilizes m(6)A-containing RNA through
direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun.
7:126262016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Roundtree IA, Luo GZ, Zhang Z, Wang X,
Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al: YTHDC1
mediates nuclear export of N6-methyladenosine methylated
mRNAs. Elife. 6:e313112017. View Article : Google Scholar
|
|
63
|
Kretschmer J, Rao H, Hackert P, Sloan KE,
Höbartner C and Bohnsack MT: The m6A reader protein
YTHDC2 interacts with the small ribosomal subunit and the 5′-3′
exoribonuclease XRN1. RNA. 24:1339–1350. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y,
Qi M, Lu Z, Shi H, Wang J, et al: Ythdc2 is an
N6-methyladenosine binding protein that regulates
mammalian spermatogenesis. Cell Res. 27:1115–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zarnack K, König J, Tajnik M, Martincorena
I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM and Ule
J: Direct competition between hnRNP C and U2AF65 protects the
transcriptome from the exonization of Alu elements. Cell.
152:453–466. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu N, Zhou KI, Parisien M, Dai Q,
Diatchenko L and Pan T: N6-methyladenosine alters RNA structure to
regulate binding of a low-complexity protein. Nucleic Acids Res.
45:6051–6063. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bell JL, Wächter K, Mühleck B, Pazaitis N,
Köhn M, Lederer M and Hüttelmaier S: Insulin-like growth factor 2
mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of
cancer progression? Cell Mol Life Sci. 70:2657–2675. 2013.
View Article : Google Scholar :
|
|
68
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Linder B, Grozhik AV, Olarerin-George AO,
Meydan C, Mason CE and Jaffrey SR: Single-nucleotide-resolution
mapping of m6A and m6Am throughout the transcriptome. Nat Methods.
12:767–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ke S, Alemu EA, Mertens C, Gantman EC, Fak
JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al:
A majority of m6A residues are in the last exons, allowing the
potential for 3′ UTR regulation. Genes Dev. 29:2037–2053. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu N, Parisien M, Dai Q, Zheng G, He C
and Pan T: Probing N6-methyladenosine RNA modification status at
single nucleo-tide resolution in mRNA and long noncoding RNA. RNA.
19:1848–1856. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Garcia-Campos MA, Edelheit S, Toth U,
Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis
A, et al: Deciphering the 'm6A Code' via
antibody-independent quantitative profiling. Cell. 178:731–747.e16.
2019. View Article : Google Scholar
|
|
74
|
Liu Q and Gregory RI: RNAmod: An
integrated system for the annotation of mRNA modifications. Nucleic
Acids Res. 47:W548–W555. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang SY, Zhang SW, Fan XN, Zhang T, Meng
J and Huang Y: FunDMDeep-m6A: Identification and prioritization of
functional differential m6A methylation genes. Bioinformatics.
35:i90–i98. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang Z, Chen LQ, Zhao YL, Yang CG,
Roundtree IA, Zhang Z, Ren J, Xie W, He C and Luo GZ: Single-base
mapping of mA by an antibody-independent method. Sci Adv.
5:eaax02502019. View Article : Google Scholar
|
|
77
|
Lin J, Zhu Q, Huang J, Cai R and Kuang Y:
Hypoxia promotes vascular smooth muscle cell (VSMC) differentiation
of adipose-derived stem cell (ADSC) by regulating Mettl3 and
paracrine factors. Stem Cells Int. 2020:28305652020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Cohn JN, Ferrari R and Sharpe N: Cardiac
remodeling-concepts and clinical implications: A consensus paper
from an international forum on cardiac remodeling. Behalf of an
International Forum on Cardiac Remodeling. J Am Coll Cardiol.
35:569–582. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kehat I and Molkentin JD: Molecular
pathways underlying cardiac remodeling during pathophysiological
stimulation. Circulation. 122:2727–2735. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Maier T, Guell M and Serrano L:
Correlation of mRNA and protein in complex biological samples. FEBS
Lett. 583:3966–3973. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Dorn LE, Lasman L, Chen J, Xu X, Hund TJ,
Medvedovic M, Hanna JH, van Berlo JH and Accornero F: The
N6-Methyladenosine mRNA methylase METTL3 controls
cardiac homeostasis and hypertrophy. Circulation. 139:533–545.
2019. View Article : Google Scholar
|
|
82
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Song H, Pu J, Wang L, Wu L, Xiao J, Liu Q,
Chen J, Zhang M, Liu Y, Ni M, et al: ATG16L1 phosphorylation is
oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein
phos-phatase 1 which determines the fate of cardiomyocytes during
hypoxia/reoxygenation. Autophagy. 11:1308–1325. 2015. View Article : Google Scholar
|
|
84
|
Pastore N, Brady OA, Diab HI, Martina JA,
Sun L, Huynh T, Lim JA, Zare H, Raben N, Ballabio A and Puertollano
R: TFEB and TFE3 cooperate in the regulation of the innate immune
response in activated macrophages. Autophagy. 12:1240–1258. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhao E and Czaja MJ: Transcription factor
EB: A central regulator of both the autophagosome and lysosome.
Hepatology. 55:1632–1634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Song H, Feng X, Zhang H, Luo Y, Huang J,
Lin M, Jin J, Ding X, Wu S, Huang H, et al: METTL3 and ALKBH5
oppositely regu-late m6A modification of TFEB mRNA,
which dictates the fate of hypoxia/reoxygenation-treated
cardiomyocytes. Autophagy. 15:1419–1437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Misquitta CM, Iyer VR, Werstiuk ES and
Grover AK: The role of 3′-untranslated region (3′-UTR) mediated
mRNA stability in cardiovascular pathophysiology. Mol Cell Biochem.
224:53–67. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gratacós FM and Brewer G: The role of AUF1
in regulated mRNA decay. Wiley Interdiscip Rev RNA. 1:457–473.
2010. View Article : Google Scholar
|
|
89
|
Su YR, Chiusa M, Brittain E, Hemnes AR,
Absi TS, Lim CC and Di Salvo TG: Right ventricular protein
expression profile in end-stage heart failure. Pulm Circ.
5:481–497. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-Dependent N6-Methyladenosine
regulates cardiac function during remodeling and repair.
Circulation. 139:518–532. 2019. View Article : Google Scholar :
|
|
91
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification destabilizes
developmental regulators in embryonic stem cells. Nat Cell Biol.
16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Berulava T, Buchholz E, Elerdashvili V,
Pena T, Islam MR, Lbik D, Mohamed BA, Renner A, von Lewinski D,
Sacherer M, et al: Changes in m6A RNA methylation contribute to
heart failure progression by modulating translation. Eur J Heart
Fail. 22:54–66. 2020. View Article : Google Scholar
|
|
93
|
Kmietczyk V, Riechert E, Kalinski L,
Boileau E, Malovrh E, Malone B, Gorska A, Hofmann C, Varma E,
Jürgensen L, et al: m6A-mRNA methylation regulates
cardiac gene expression and cellular growth. Life Sci Alliance.
2:e2018002332019. View Article : Google Scholar
|
|
94
|
Hirsch AT, Haskal ZJ, Hertzer NR, Bakal
CW, Creager MA, Halperin JL, Hiratzka LF, Murphy WR, Olin JW,
Puschett JB, et al: ACC/AHA 2005 Practice Guidelines for the
management of patients with peripheral arterial disease (lower
extremity, renal, mesenteric, and abdominal aortic): A
collaborative report from the American Association for Vascular
Surgery/Society for Vascular Surgery, Society for Cardiovascular
Angiography and Interventions, Society for Vascular Medicine and
Biology, Society of Interventional Radiology, and the ACC/AHA Task
Force on Practice Guidelines (Writing Committee to Develop
Guidelines for the Management of Patients With Peripheral Arterial
Disease): Endorsed by the American Association of Cardiovascular
and Pulmonary Rehabilitation; National Heart, Lung, and Blood
Institute; Society for Vascular Nursing; TransAtlantic
Inter-Society Consensus; and Vascular Disease Foundation.
Circulation. 113:e463–e654. 2006.PubMed/NCBI
|
|
95
|
Reeps C, Pelisek J, Seidl S, Schuster T,
Zimmermann A, Kuehnl A and Eckstein HH: Inflammatory infiltrates
and neovessels are relevant sources of MMPs in abdominal aortic
aneurysm wall. Pathobiology. 76:243–252. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
He Y, Xing J, Wang S, Xin S, Han Y and
Zhang J: Increased m6A methylation level is associated with the
progression of human abdominal aortic aneurysm. Ann Transl Med.
7:7972019. View Article : Google Scholar
|
|
97
|
Zhong L, He X, Song H, Sun Y, Chen G, Si
X, Sun J, Chen X, Liao W, Liao Y and Bin J: METTL3 induces AAA
development and progression by modulating
N6-methyladenosine-dependent primary miR34a processing. Mol Ther
Nucleic Acids. 21:394–411. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Thomas B, Matsushita K, Abate KH, Al-Aly
Z, Ärnlöv J, Asayama K, Atkins R, Badawi A, Ballew SH, Banerjee A,
et al: Global Cardiovascular and Renal Outcomes of Reduced GFR. J
Am Soc Nephrol. 28:2167–2179. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Fang Y, Ginsberg C, Sugatani T,
Monier-Faugere MC, Malluche H and Hruska KA: Early chronic kidney
disease-mineral bone disorder stimulates vascular calcification.
Kidney Int. 85:142–150. 2014. View Article : Google Scholar
|
|
100
|
Cao XS, Chen J, Zou JZ, Zhong YH, Teng J,
Ji J, Chen ZW, Liu ZH, Shen B, Nie YX, et al: Association of
indoxyl sulfate with heart failure among patients on hemodialysis.
Clin J Am Soc Nephrol. 10:111–119. 2015. View Article : Google Scholar :
|
|
101
|
Chen J, Ning Y, Zhang H, Song N, Gu Y, Shi
Y, Cai J, Ding X and Zhang X: METTL14-dependent m6A regulates
vascular calcification induced by indoxyl sulfate. Life Sci.
239:1170342019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
McLaughlin VV, Archer SL, Badesch DB,
Barst RJ, Farber HW, Lindner JR, Mathier MA, McGoon MD, Park MH,
Rosenson RS, et al: ACCF/AHA 2009 expert consensus document on
pulmonary hypertension a report of the American College of
Cardiology Foundation Task Force on Expert Consensus Documents and
the American Heart Association developed in collaboration with the
American College of Chest Physicians; American Thoracic Society,
Inc.; and the Pulmonary Hypertension Association. J Am Coll
Cardiol. 53:1573–1619. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Galiè N, Humbert M, Vachiery JL, Gibbs S,
Lang I, Torbicki A, Simonneau G, Peacock A, Vonk Noordegraaf A,
Beghetti M, et al: 2015 ESC/ERS Guidelines for the diagnosis and
treatment of pulmonary hypertension: The Joint Task Force for the
Diagnosis and Treatment of Pulmonary Hypertension of the European
Society of Cardiology (ESC) and the European Respiratory Society
(ERS): Endorsed by: Association for European Paediatric and
Congenital Cardiology (AEPC), International Society for Heart and
Lung Transplantation (ISHLT). Eur Heart J. 37:67–119. 2016.
View Article : Google Scholar
|
|
104
|
Weitzenblum E, Sautegeau A, Ehrhart M,
Mammosser M and Pelletier A: Long-term oxygen therapy can reverse
the progression of pulmonary hypertension in patients with chronic
obstructive pulmonary disease. Am Rev Respir Dis. 131:493–498.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L,
Shen Q, Xu P, Zeng L, Zhou Y, et al: YTHDF1 links hypoxia
adaptation and non-small cell lung cancer progression. Nat commun.
10:48922019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang C, Samanta D, Lu H, Bullen JW, Zhang
H, Chen I, He X and Semenza GL: Hypoxia induces the breast cancer
stem cell phenotype by HIF-dependent and ALKBH5-mediated
m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA.
113:E2047–E2056. 2016. View Article : Google Scholar
|
|
107
|
Fry NJ, Law BA, Ilkayeva OR, Holley CL and
Mansfield KD: N6-methyladenosine is required for the
hypoxic stabilization of specific mRNAs. RNA. 23:1444–1455. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang J, Zhu MC, Kalionis B, Wu JZ, Wang
LL, Ge HY, Chen CC, Tang XD, Song YL, He H and Xia SJ:
Characteristics of circular RNA expression in lung tissues from
mice with hypoxiainduced pulmonary hypertension. Int J Mol Med.
42:1353–1366. 2018.PubMed/NCBI
|
|
110
|
Su H, Wang G, Wu L, Ma X, Ying K and Zhang
R: Transcriptome-wide map of m6A circRNAs identified in
a rat model of hypoxiamediated pulmonary hypertension. BMC
Genomics. 21:392020. View Article : Google Scholar
|
|
111
|
Chan JJ and Tay Y: Noncoding RNA: RNA
regulatory networks in cancer. Int J Mol Sci. 19:2018. View Article : Google Scholar
|
|
112
|
Baarsma HA and Königshoff M: 'WNT-er is
coming': WNT signalling in chronic lung diseases. Thorax.
72:746–759. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Savai R, Al-Tamari HM, Sedding D,
Kojonazarov B, Muecke C, Teske R, Capecchi MR, Weissmann N,
Grimminger F, Seeger W, et al: Pro-proliferative and inflammatory
signaling converge on FoxO1 transcription factor in pulmonary
hypertension. Nat Med. 20:1289–1300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Haraksingh RR and Snyder MP: Impacts of
variation in the human genome on gene regulation. J Mol Biol.
425:3970–3977. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Mao F, Xiao L, Li X, Liang J, Teng H, Cai
W and Sun ZS: RBP-Var: A database of functional variants involved
in regulation mediated by RNA-binding proteins. Nucleic Acids Res.
44:D154–D163. 2016. View Article : Google Scholar :
|
|
116
|
Wu X and Hurst LD: Determinants of the
usage of splice-associated cis-Motifs predict the distribution of
human pathogenic SNPs. Mol Biol Evol. 33:518–529. 2016. View Article : Google Scholar :
|
|
117
|
Ramaswami G, Deng P, Zhang R, Anna Carbone
M, Mackay TFC and Billy Li J: Genetic mapping uncovers
cis-regulatory land-scape of RNA editing. Nat Commun. 6:81942015.
View Article : Google Scholar
|
|
118
|
Zheng Y, Nie P, Peng D, He Z, Liu M, Xie
Y, Miao Y, Zuo Z and Ren J: m6AVar: A database of functional
variants involved in m6A modification. Nucleic Acids Res.
46:D139–D145. 2018. View Article : Google Scholar :
|
|
119
|
Yang N, Ying P, Tian J, Wang X, Mei S, Zou
D, Peng X, Gong Y, Yang Y, Zhu Y, et al: Genetic variants in m6A
modification genes are associated with esophageal squamous-cell
carcinoma in the Chinese population. Carcinogenesis. 41:761–768.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Mo XB, Lei SF, Zhang YH and Zhang H:
Detection of m6A-asso-ciated SNPs as potential
functional variants for coronary artery disease. Epigenomics.
10:1279–1287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Mo X, Lei S, Zhang Y and Zhang H:
Genome-wide enrichment of m6A-associated
single-nucleotide polymorphisms in the lipid loci. Pharmacogenomics
J. 19:347–357. 2019. View Article : Google Scholar
|
|
122
|
Kupper N, Willemsen G, Riese H, Posthuma
D, Boomsma DI and de Geus EJC: Heritability of daytime ambulatory
blood pressure in an extended twin design. Hypertension. 45:80–85.
2005. View Article : Google Scholar
|
|
123
|
Evangelou E, Warren HR, Mosen-Ansorena D,
Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman
I, et al: Genetic analysis of over 1 million people identifies 535
new loci associated with blood pressure traits. Nat Genet.
50:1412–1425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Mo XB, Lei SF, Zhang YH and Zhang H:
Examination of the associations between m6A-associated
single-nucleotide polymorphisms and blood pressure. Hypertens Res.
42:1582–1589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Lee JY, Lee BS, Shin DJ, Woo Park K, Shin
YA, Joong Kim K, Heo L, Young Lee J, Kyoung Kim Y, Jin Kim Y, et
al: A genome-wide association study of a coronary artery disease
risk variant. J Hum Genet. 58:120–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Gerken T, Girard CA, Tung YC, Webby CJ,
Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill
LA, et al: The obesity-associated FTO gene encodes a
2-oxoglutarate-dependent nucleic acid demethylase. Science.
318:1469–1472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Guyenet PG: The sympathetic control of
blood pressure. Nat Rev Neurosci. 7:335–346. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Pausova Z, Syme C, Abrahamowicz M, Xiao Y,
Leonard GT, Perron M, Richer L, Veillette S, Smith GD, Seda O, et
al: A common variant of the FTO gene is associated with not only
increased adiposity but also elevated blood pressure in French
Canadians. Circ Cardiovasc Genet. 2:260–269. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Frayling TM, Timpson NJ, Weedon MN,
Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H,
Rayner NW, et al: A common variant in the FTO gene is associated
with body mass index and predisposes to childhood and adult
obesity. Science. 316:889–894. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Marcadenti A, Fuchs FD, Matte U, Sperb F,
Moreira LB and Fuchs SC: Effects of FTO RS9939906 and MC4R
RS17782313 on obesity, type 2 diabetes mellitus and blood pressure
in patients with hypertension. Cardiovasc Diabetol. 12:1032013.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
O'Donnell CJ and Nabel EG: Genomics of
cardiovascular disease. N Engl J Med. 365:2098–2109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Prospective Studies Collaboration;
Lewington S, Whitlock G, Clarke R, Sherliker P, Emberson J, Halsey
J, Qizilbash N, Peto R and Collins R: Blood cholesterol and
vascular mortality by age, sex, and blood pressure: A meta-analysis
of individual data from 61 prospective studies with 55,000 vascular
deaths. Lancet. 370:1829–1839. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Yadav PK, Rajvanshi PK and Rajasekharan R:
The role of yeast m6A methyltransferase in peroxisomal
fatty acid oxidation. Curr Genet. 64:417–422. 2018. View Article : Google Scholar
|
|
134
|
Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G,
Yuan W, Kan Q and Sun Z: The interplay between m6A RNA methylation
and noncoding RNA in cancer. J Hematol Oncol. 12:1212019.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhang W, Chen G and Deng CQ: Effects and
mechanisms of total Panax notoginseng saponins on proliferation of
vascular smooth muscle cells with plasma pharmacology method. J
Pharm Pharmracol. 64:139–145. 2012. View Article : Google Scholar
|
|
136
|
Zhu B, Gong Y, Shen L, Li J, Han J, Song
B, Hu L, Wang Q and Wang Z: Total Panax notoginseng saponin
inhibits vascular smooth muscle cell proliferation and migration
and intimal hyperplasia by regulating WTAP/p16 signals via
m6A modulation. Biomed Pharmacother. 124:1099352020.
View Article : Google Scholar
|
|
137
|
Nakarai H, Yamashita A, Nagayasu S,
Iwashita M, Kumamoto S, Ohyama H, Hata M, Soga Y, Kushiyama A,
Asano T, et al: Adipocyte-macrophage interaction may mediate
LPS-induced low-grade inflammation: Potential link with metabolic
complications. Innate Immun. 18:164–170. 2012. View Article : Google Scholar
|
|
138
|
Rao DS, Sekhara NC, Satyanarayana MN and
Srinivasan M: Effect of curcumin on serum and liver cholesterol
levels in the rat. J Nutri. 100:1307–1315. 1970. View Article : Google Scholar
|
|
139
|
Lu N, Li X, Yu J, Li Y, Wang C, Zhang L,
Wang T and Zhong X: Curcumin attenuates lipopolysaccharide-induced
hepatic lipid metabolism disorder by modification of m6
A RNA methylation in piglets. Lipids. 53:53–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou
H, Gan J, Jiang H, Jia GF, Luo C and Yang CG: Meclofenamic acid
selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic
Acids Res. 43:373–384. 2015. View Article : Google Scholar :
|
|
141
|
Li J, Chen Z, Chen F, Xie G, Ling Y, Peng
Y, Lin Y, Luo N, Chiang CM and Wang H: Targeted mRNA demethylation
using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids
Res. 48:5684–5694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Lawson DA, Kessenbrock K, Davis RT,
Pervolarakis N and Werb Z: Tumour heterogeneity and metastasis at
single-cell resolution. Nat Cell Biol. 20:1349–1360. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Paramasivam A, Vijayashree Priyadharsini J
and Raghunandhakumar S: N6-adenosine methylation (m6A): A promising
new molecular target in hypertension and cardiovascular diseases.
Hypertens Res. 43:153–154. 2020. View Article : Google Scholar
|