Gastric cancer (GC) is one of the most common
malignancies worldwide, and is ranked as the fourth most common
type of cancer and the third highest cause of cancer-related
mortality. Despite a slight decline in incidence, ~1,000,000
individuals are diagnosed with GC and >700,000 patients succumb
to the disease each year (1).
Surgery remains the main treatment strategy for GC. In recent
years, the wide application of neoadjuvant chemoradiotherapy has
improved the survival rates and quality of life of patients, and
has reduced the chances of a necessary gastrectomy (2,3).
If patients are diagnosed with GC at early stages and receive
treatment promptly, then their chances of recovery are optimal.
However, the majority of patients are initially diagnosed with GC
at an advanced stage due to atypical symptoms and the lack of
sensitive examinations (4). Poor
prognosis calls for more effective diagnostic methods with improved
sensitivity and specificity. There is also a need for developing
novel target medicine to enhance the efficacy of cancer treatment
and minimize the side-effects of conventional perioperative
regimens.
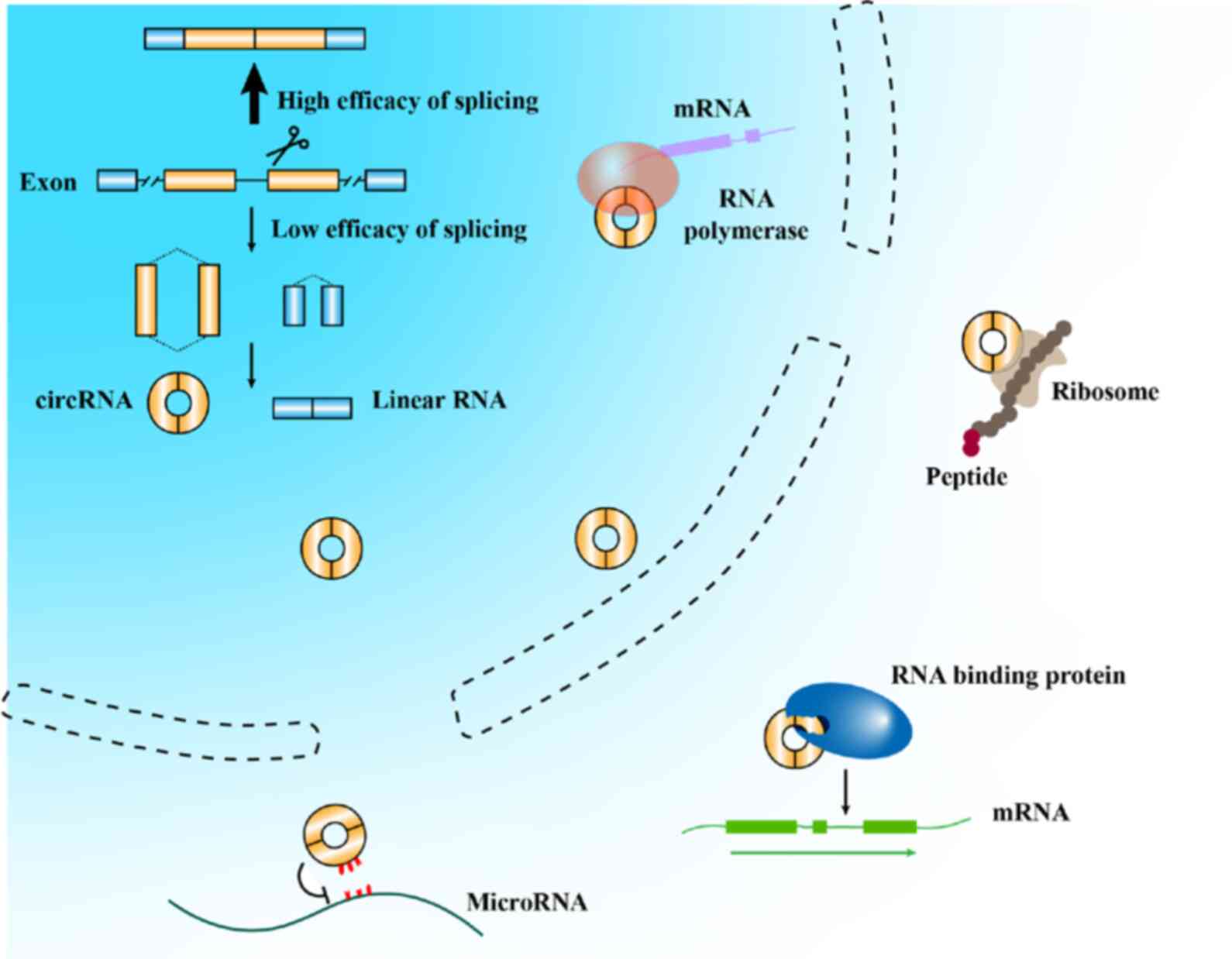
circRNAs have been demonstrated to play an
indispensable role in physiological and pathological changes
(20-22). In recent years, attention has been
paid to the association between circRNAs and cancer. Some circRNAs
may participate in the carcinogenesis, progression and metastasis
of numerous types of cancer (9,23,24), which suggests that they have
potential diagnostic and prognostic value for patients with cancer
(25,26). Despite the fact that a relatively
small number of studies have been conducted on this topic, circRNAs
have already demonstrated their optimal function in the clinical
translation of tumors.
A number of circRNAs were identified serendipitously
and were largely disregarded as non-specific byproducts when they
were found to be expressed at low levels. Nonetheless, circRNAs
appear to be abundant in both normal and cancer cells. Some types
of circRNAs are present at levels comparable to those of their
canonical linear counterparts. Furthermore, Salzman et al
(27) even reported that the
abundance of circular molecules exceeded that of associated linear
mRNAs by >10-fold in some cases. These results suggest that
circRNA levels are absolutely detectable. Carcinogenesis and
progression may lead to the alteration of circRNA profiles, which
can serve as a novel indicator of GC.
The first step in exploring the association of
circRNAs with GC is to screen the circRNA profiles of patients with
GC and normal individuals. The identification of circRNAs at
differential expression levels may contribute to a better
understanding of the mechanisms of GC, which also suggests their
use as potential biomarkers for diagnosis and prognosis. The
existing types of specimens that have been studied include tumor
tissues, blood and gastric juices. Previous studies have reported
GC expression profiles, and some of them have constructed
circRNA-miRNA or circRNA-miRNA-mRNA regulatory networks by
bioinformatics analysis (28-31). These results provide fundamental
evidence for searching potential targets of circRNAs and potential
downstream of RNAs. For instance, Gu et al (29) found that circ_101504 played a
central role in the regulatory network. circ_101504 was predicted
to affect some mRNAs by inhibiting miR-454-3p and miR-301a-3p.
Future studies should be conducted to verify the signaling
pathways. Huang et al (30) screened three sets of tissues and
found that only circ_0026 was significantly downregulated.
Bioinformatics analysis indicated a potential role of circ_0026 in
gastric carcinogenesis and its potential use as a diagnostic
biomarker. Despite the limited number of GC samples, that study
provided a direction of research points (30).
Numerous studies have indicated the potential
diagnostic values of circRNAs in GC. The majority of these have
focused on altered circRNA expression levels in GC and adjacent
tissues. The area under the curve (AUC) of a ROC curve is a
comprehensive indicator of diagnostic values. Previous studies have
revealed that circ_0001017 has the highest diagnostic accuracy in
tissues, with an AUC of 0.871. The sensitivity and specificity
could reach, respectively, 79.4 and 81.1% (32). Li et al (33) demonstrated that the
down-regulation of circ_002059 had a potential diagnostic value in
GC, with its AUC being 0.73. Its sensitivity and specificity were,
respectively, 0.81 and 0.62. Samples stored at various temperatures
for different periods of time exhibited the same expression levels
of circ_002059, which demon-strated its optimal stability as a
clinical biomarker (33).
circ_0000190 was previously demonstrated to be downregulated in GC
tissues, and had a higher AUC (0.75), than that of circ_002059
(34). Tian et al
(35) found that circ_0003159
could also become a potential GC biomarker. The AUC, sensitivity
and specificity were 0.75, 0.852 and 0.565, respectively. As far as
circ_0074362 is concerned, despite its low sensitivity and
specificity, the levels of circ_0074362 have been shown to be
closely associated with lymphatic metastasis, suggesting it may be
an auxiliary biomarker for predicting the advancement of GC
(36).
Some researchers have noted the clinical value of
circRNAs in plasma. Previously, the circ_0000745 expression level
in plasma was assessed, and its sensitivity reached 0.855. However,
the current diagnostic use of circ_0000745 cannot meet clinical
standards due to its relatively low specificity (37). The ubiquitous expression of
circRNAs can be regarded as the main reason behind this low
specificity. The accuracy of circRNAs in tissues is commonly higher
than that in plasma partly due to spatial position. Notably, Sun
et al (38) reported that
the diagnostic accuracy of circ_0000520 was much higher than the
one in tumor tissues. The AUC, sensitivity and specificity were
0.8967, 0.8235 and 0.8444, respectively. According to the
afore-mentioned studies, the diagnosis of a single circRNA does not
meet clinical translation. Li et al (32) combined the four biomarkers
(circ_0001017 and circ_0061276 both in tissues and in plasma) as a
panel. The sensitivity and specificity were 95.5 and 95.7%,
respectively. Thus, the circRNA panel is regarded as the most
promising for possible and effective translation into clinical
application.
The detection of circRNAs in gastric juice is
another potential non-invasive type of clinical examination. Shao
et al (39) recruited 38
healthy volunteers, 30 patients with gastric ulcer, 15 patients
with chronic atrophic gastritis and 39 patients with GC in order to
evaluate the diagnostic values of circ_0014717. Although no
significant differences in circ_0014717 levels were observed
amongst the healthy, gastric ulcer and GC groups, a marked
reduction was observed in the chronic atrophic gastritis group.
Chronic atrophic gastritis is a possible precancerous lesion of GC
(40); thus, the aforementioned
study suggested that circ_0014717 may be a predictive indicator of
GC. This result broadens the horizons that circRNAs can exist in an
extreme environment for a long period of time. Their considerable
stability suggests that circRNAs are likely to become an excellent
non-invasive biomarker for GC.
Accumulating evidence has demonstrated that circRNAs
may play a promising role in predicting the prognosis of GC. A
common method used for the prognosis of GC is defining the cut-off
value of circRNAs. Recruited patients can be divided into positive
(high expression) and negative (low expression) groups.
Kaplan-Meier survival analysis is then employed to evaluate the
association between circRNAs and prognosis. Univariate analysis,
further multivariate analysis and the Cox proportional hazard model
are also used to assess the prognostic role of circRNAs.
circ_100269, circ_0001017 and circ_0061276 have been demonstrated
to serve as potential biomarkers for predicting prognosis in GC by
these methods (32,41). For instance, Zhang et al
(42) indicated that circLARP4
(circ_101057) was closely associated with the prognosis of patients
with GC at the early stage rather than at the late stage of the
disease. Patients at the early stage of the disease with a high
expression of circLARP4 had a better overall survival (OS) and a
better response to adjuvant chemotherapy with oxaliplatin and
5-fluorouracil. The result revealed that circRNA may have the
potential to direct clinical medication to a certain extent. The
ability of circRNAs alone to determine the prognosis of GC may not
meet clinical needs. Chen et al (43) shared a novel approach for this
field in 2017. They simultaneously evaluated the predictive values
of circPVT1 and PVT1 expression. The combination yielded more
accurate results than those obtained by either of the two alone.
Future studies are required however, to further concentrate on
panels of combined biomarkers to maintain an ideal balance between
efficiency and economy.
Considerable progress has been made in the
identification of the mechanisms through which circRNAs participate
in gastric carcino-genesis. Numerous studies have focused on
identifying circRNAs that may play potential roles in the treatment
of GC (Table I). Multiple
circRNAs were selected, and the majority of these were demonstrated
to be involved in GC by acting as miRNA sponges. Binding to target
miRNAs can downregulate miRNA expression and subsequently affect
the functions of downstream molecules. A number of circRNAs have
been reported to be upregulated in GC, such as circPVT1,
circ_0047905, circ_0138960, circ_7690-15, circHIPK3 (circ_0000284),
circ_0023642 and circ_001569. They may participate in cancer
proliferation, migration or invasion. Their downregulation can
inhibit malignant behaviors (43-47). However, other circRNAs, such as
circLARP4 and circ_100269 have been demonstrated to be expressed at
lower levels in GC cancer tissues. They have both been demonstrated
to be involved in cancer growth, and may function as suppressors of
GC through sponging miR-424-5p and miR-630. The upregulation of
these two circRNAs significantly reverses carcinogenesis and
proliferation (41,42).
The aforementioned circRNAs naturally exist in
cancer or normal cells. Their regulation can interrupt or reverse
the development of tumors. However, previous studies on GC have
focused on basic research, which is far from clinical translation
due to complex mechanisms in vivo. Liu et al
(54) provided a novel approach
of the clinical translation of circRNAs. Since miRNA sponges rely
on the antisense sequences, synthetic molecules with these
sequences are likely to exhibit similar effects on miRNA
inhibition. The researcher can decide the circRNA levels and
quantity of binding sites in each circRNA. Controllable modulation
ability should provide an optimal balance between efficiency and
safety. Liu et al designed a synthetic circRNA functioning
as a miR-21 sponge. The administration of this circRNA led to the
inhibition of proliferation and induction of apoptosis (54). Downstream proteomic screening
revealed that proteins which should have been downregulated by
miR-21 were effectively restored (54). Moreover, the reduction of the
cancer burden may not be the sole direction of research. The
inhibition of ciRS-133 has been shown to alleviate GC-associated
cachexia by repressing miR-133 (59). The elucidation of the mechanisms
responsible for complications associated with GC is also expected
to be meaningful, helping to prolong the lifetime and comfort of
patients with late-stage GC. In addition to miRNA sponges, circRNAs
in the nucleus can initiate the expression of transcriptional
factors to promote cancer growth (60). Future studies are required to pay
greater attention to this field and explore additional approaches
with which to improve the prognosis and quality of life of
patients.
lncRNAs belong to a characterized group of RNAs
without the capacity of transcription, which are >200 nt in
length. In the 1990s, lncRNAs were identified without optimal
origins and functions (65).
lncRNAs were initially regarded as transcriptional noises (66). However, accumulating evidence
suggested that lncRNAs may play a crucial role in physiological
activities and various diseases (67-69). Compared with their small-length
counterparts, miRNAs, the main functional mechanism of which is
binding to 3′ untranslated regions and activating the degradation
of target mRNAs (70), lncRNAs
have richer regulatory methods, including levels of transcription,
post-transcription, alternative splicing and translation (71). For instance, lncRNA h5S-OT
modulates retrotransposons jump into alternative-splicing by virtue
of the Alu element. Alternative splicing produces variants of
proteins responsible for distinct physiological or pathological
processes (72,73). lncRNA RoR has been reported to
interact with c-Myc mRNA and increase its stability, leading to
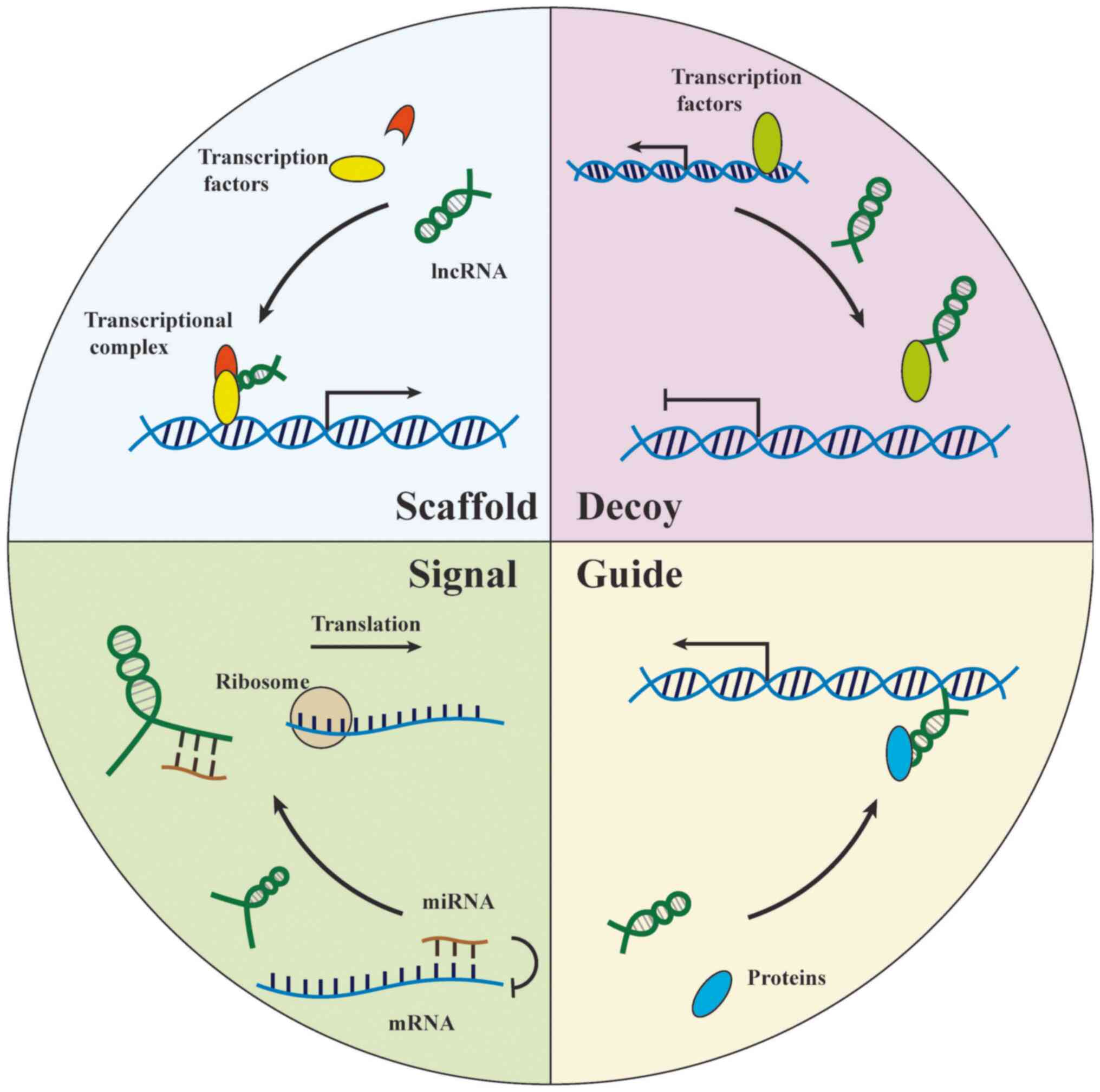
cell proliferation and tumorigenesis (74). lncRNA-protein regulatory
mechanisms can be generally summarized by four aspects: i) Signal,
lncRNAs reflect on the spatiotemporal expression level of genes via
alterations of transcription factors or signaling path-ways; ii)
decoy, lncRNAs titrate specific proteins in the cell nucleus and
perform concrete functions; iii) guide, lncRNAs direct
ribonucleoprotein complex to locate on specific regions; and iv)
scaffold, the lncRNA scaffold is structural and stabilizes nuclear
structures or signaling complexes (75) (Fig.
2).
The association between lncRNAs and GC has been
gradually revealed. Numerous lncRNAs have been found to participate
in carcinogenesis and progression. Diverse biological functions and
wide distribution determines the complexity of lncRNA networks in
GC. One lncRNA can regulate multiple cancer behaviors through
different pathways (76,77). One cancer behavior can be under
the control of several lncRNAs and their respective downstream
molecules (78,79). Additionally, intersectional lncRNA
profiles and regulatory mechanisms vary in different cancer types
(80,81). Their theoretical links with GC
provide substantial biomarkers and targets with potential clinical
values. Therefore, lncRNAs are one of the most promising approaches
for GC diagnosis and treatment.
The biological characteristics of lncRNAs facilitate
the speed of translation into the clinical diagnosis of GC. Optimal
diagnostic biomarkers require remarkable stability of molecular
structures and expression levels. Given the existing number of
studies, lncRNAs are suitable candidates for this purpose. lncRNAs
have been found to stably exist in extreme environments, such as
gastric juice, urine and hair follicles (82-84). lncRNAs have distinct half-life
periods, which range from <2 to >16 h (85). A long half-life period would lead
to the accumulation and would impair the accuracy of the biomarker.
On the other hand, an excessively short detectable time exerts more
pressure on the limitation of technology to form a reliable
clinical diagnosis. In a previous study, the median half-life
period of lncRNAs was ~3.5 h shorter than that of protein-coding
RNAs (85). The appropriate
half-life period indicates their potential to function as
diagnostic biomarkers due to the prompt reactions of lncRNAs
consistent with the degrees of primary focus. The characteristics
and severity of diseases can be reflected by these reliable
indicators.
The majority of studies concerning lncRNA diagnosis
have focused on the detection of cancerous tissues. Alterations in
lncRNA expression are significantly associated with numerous
clinicopathological features. For instance, Sun et al
(86) found that lncRNA
AC096655.1-002 expression was associated with TNM stage,
differentiation, lymph node metastasis and depth of invasion.
lncRNA ABHD11-AS1 has also been proven to be associated with
differentiation, Lauren histological classification and
carbohydrate antigen 19-9 (CA19-9) (87). Testing the expression levels of
some lncRNAs may provide a reference for the evaluation of disease
severities and the selection of treatment regimens. However, the
association between lncRNAs and clinicopathological features is not
stable. Baratieh et al (88) analyzed data from patients with GC
in The Cancer Genome Atlas (TCGA) database. Their results revealed
limited features associated with FAM83H-AS1 expression; however,
FAM83H-AS1 mean and median gene expression data in the TCGA cohort
exhibited a significant association with M-classification, tumor
stage, grade and different Lauren's classes. Thus, further
investigations into more reliable associations of lncRNAs with
clinical characteristics are required using larger sample numbers
and standardizing experimental procedures.
Non-invasive and pain-free detection methods of
lncRNAs are also pursued, such as in the case of circRNAs. lncRNA
performance in GC diagnosis is commonly better than that of
classical biomarkers, as highlighted from current methods, such as
CEA and CA19-9 (89). A
meta-analysis (90) indicated
that the clinical values of lncRNAs are limited to screening tools
rather than diagnosis with high accuracy. This conclusion may not
be convincing enough, partly due to the inclusion of relatively old
studies and the ignorance of the respective discussions of each
lncRNA. Some lncRNAs have exhibited great potential for clinical
translation. lncRNA H19 and HOTAIR were demonstrated to be
effective biomarkers with high AUC. Their combination with CEA
significantly enhanced their diagnostic capacity (91-93). Furthermore, a multi-lncRNA
diagnostic panel in plasma is a feasible approach to compensate for
the comparably low sensitivity of a single lncRNA. Dong et
al (89) created a panel with
three plasma lncRNAs (CUDR, LSINCT-5 and PTENP1) in 2015, which was
sensitive and specific to the discrimination of healthy controls
from patients with GC, patients with peptic ulcers from patients
with GC, and patients with stage I and II-IV disease from healthy
controls. Zhang et al (94) employed the genome-wide profiling
identifies TINCR, CCAT2, AOC4P, BANCR and LINC00857 in plasma. The
diagnostic panel was estimated to be an excellent method for the
discrimination of patients with GC from both precancerous
individuals and gastrointestinal stromal tumors. Another advantage
of this research is that these data were translated into Fagan's
nomogram, a tool used to calculate the probability that an
individual has GC based on this panel. A convenient evaluation
method is a developing direction of GC diagnosis. In addition, Li
et al (95) found that the
levels of LINC00152 in plasma and exosomes were consistent, which
suggested that lncRNAs detected in exosomes may also have optimal
clinical values as plasma biomarkers.
Current treatment methods cannot reach curative
goals for patients at advanced stages of the disease. A basic
method with which to resolve GC-associated mortality and the poor
prognosis of affected patient is to search for efficient diagnostic
biomarkers for the early stages of the disease. H. pylori is
an important carcinogenic factor, estimated to be prevalent in
developing countries (102,103). NR_026827 has been demonstrated
to be downregulated in gastric epithelial cells infected with H.
pylori (104). Despite the
wide application of C13 or C14 examinations,
NR_026827 may be a predictive role of GC risks. H19 in plasma
enabled the discrimination of early-stage GC from controls with an
AUC of 0.877 (93). Lu et
al (105) also developed a
panel of five lncRNAs in tumor tissues with high values of early
diagnosis, which helps to identify indistinguishable focus in
endoscopy. The single nucleotide polymorphism (SNP) rs4759314,
which contributes to a genotype-specific effect on the expression
of the host gene HOTAIR, has been shown to be closely associated
with the risk of developing GC in Chinese populations (106).
The number of lncRNAs for the prediction of
prognosis remains smaller than that for GC diagnosis. A substantial
number of lncRNAs have been screened out, which have remarkable
predicting performance, theoretically. The majority of previous
studies suggest the potential values of tissue lncRNAs in
evaluating the survival of patients with GC. Liu and Shangguan
(107) employed the Kaplan-Meier
curve to prove that high level of lncRNA CARLo-5 was associated
with overall survival (OS) and recurrence-free survival (RFS).
Further univariate and multi-variate analyses indicated that
CARLo-5 could serve as an independent predictor of OS and RFS. Feng
et al (108) reported
that patients with a high expression of lncRNA AFAP1-AS1 had a
significantly poorer OS. Some other biomarkers for the prediction
of prognosis have been identified in recent years, such as CASC15,
UPF1, ZEB1-AS1 and PANDAR (109-112). They were all found to be
independent prognostic factors of survival time.
The degree of metastasis is an important factor for
patient prognosis, and there is a close association between lncRNAs
and GC metastasis. Xia et al (113) found that MALAT1 both in tissue
and plasma could serve as a prognostic biomarker of distant
metastasis. CARLo-5 has also been reported to be associated with
lymph node involvement and distant metastasis (107).
Nevertheless, the selected predictors are
considerable, while the pace of clinical translation has stagnated
for a long time. None of these lncRNAs can reach the criterion of
wide application. A previous study even demonstrated that a
combi-nation of 24 lncRNAs had the clinical value of predicting
prognosis (114). Superfluous
lncRNAs in a panel may decrease the prognostic values and increase
the economic burden of patients. Overall, the value of lncRNAs is
not regarded as a preferable indicator for the prediction of
prognosis based on current studies.
The identification of the elusive mechanisms of
carcinogenesis and progression remains at a preliminary stage. Both
explored and unexplored pathological processes contain multiple
molecules with clinical values. Unlike diagnostic and prognostic
biomarkers, a therapeutic candidate requires the marked involvement
of important mechanisms; thus, there is a great difficulty.
Numerous studies have paid attention to lncRNAs in the view of the
established regulatory network of GC. Large quantities of targets
have been successfully selected to act as potential targets of
clinical treatment. For instance, Chen et al (115) measured the expression of
lncRNA-ATB in a pathological specimen in GC cell lines by RT-qPCR.
lncRNA-ATB was found to be significantly upregulated in cancer
tissues and cell lines. Its knockdown led to the alteration of
clinico-pathological features, including proliferation, invasion
and migration (115).
Long intergenic non-coding RNAs (lincRNAs) are one
of the four defined categories of lncRNAs (116). lincRNAs and lncRNAs share
similar features. However, the difference between the two types of
molecules should be emphasized, owing to frequent errors made by
numerous studies, including gene expression analyses, evolutionary
conservation patterns and targeted gene disruptions that did not
alter adjacent protein-coding genes or genic RNAs (117). Previous studies have suggested
that lincRNAs may be potential targets of treatment. HOTAIR is
recognized as a key lincRNA in GC, which can modulate cancer
development and fate determination through multiple molecules and
pathways. HOTAIR, as a type of non-coding RNA sponge, can
competitively inhibit several miRNAs and affect downstream
functioning molecules, such as miR-217, miR-152, miR-454-3p and
miR-17-5p (118-121). The interaction of HOTAIR with
crucial proteins also plays an indispensable role in various
biological functions of GC. Runx3 endows gastric cells with the
capacity of excessive proliferation and invasion (122). The combination of HOTAIR and
Mex3b, a type of E3 ligase possessing RNA binding domains, can
attenuate the degradation of Runx3, thus regulating cancer
migration and invasion (123).
Zeng et al (124) found
that LINC00675 could enhance the phosphorylation of vimentin on
Ser8 and the p53 signaling pathway. The downregulation of LINC00675
facilitated cancer proliferation, migration and invasion in
vitro and in vivo (124). Previous studies have validated
that LINC00052, Linc00152, Linc00483 and H19 are attributable to
the genesis and development of GC (125-128). The intervention of their
expression may lead to the reversion of cancer progression and an
improvement in patient prognosis.
Drug resistance can limit the efficacy and
effectiveness of GC treatments. The prognosis and quality of life
of numerous patients deteriorate at the late stages of the disease
due to the failure of existing chemotherapeutics or targeted
medicine (129). Previous
studies have indicated that lncRNAs may be capable of preventing
and reversing drug resistance. A high expression of GHET1 and ANRIL
was detected in GC tissues. Further experiments demonstrated that
these two lncRNAs were associated with multi-drug resistance
(MDR)-related genes. The attenuation of these sensitized the
reactions of GC cells (130,131). CASC2 overexpression has also
been shown to overcome cisplatin resistance by binding to miR-19a,
whereas MALAT1 potentiates cisplatin resistance through sponging
miR-30b (132,133).
An increase in the apoptotic protein, cleaved
caspase-3, has been shown to restore the sensitivity to multiple
drugs, including doxorubicin, cisplatin and 5-fluorouracil
(134,135). Autophagy is a complex and highly
regulated process that delivers cellular material to lysosomes for
degrading, recycling, and generating molecules that fuel cellular
metabolism (136). Recent
research has revealed that there is a close association between
autophagy and MDR (137). Two
studies separately explained the mechanisms of MALAT1-induced
autophagy concerning the formation of GC resistance. MALAT1 served
as a miRNA sponge to target miR-23-3p and miR-30b, and related
proteins downstream of autophagy functioned subsequently (132,138). The inhibition of MALAT1 was
considered as a promising approach to alleviating MDR at late
stages.
The regulatory mechanisms of lncRNAs re not simply
considered as a 'one-to-many' mode. Previous evidence suggests that
lncRNAs share the same regulated targets. These common targets
cannot only help delineate sophisticated networks of non-coding
RNAs and GC, but also serve as intervention sites with great
values. YB-1 is a multifunctional protein that regulates apoptosis,
cell proliferation, differentiation and stress response (139). lncRNAs GAS5 and HOXC-AS3 can
directly bind to YBX1 proteins, promoting the conversion of YBX1
configuration. The inhibition of these two lncRNAs in GC cells has
been shown to abolish G1 phase cell cycle arrest, and the cell
proliferative capacity has been shown to be considerably enhanced
(140,141). Moreover, EZH2, which is
associated with genetic abnormalities, has been found to
participate in the regulation of epigenetics and transcription.
HOTAIR, MALAT1, UCA1 and LINC00673 can interact with EZH2 and
suppress downstream E-cadherin, PCDH10, AKT and KLF4, respectively
(142-145). The inhibition of the
inter-section of the mechanism means partially refraining functions
of upstream lncRNAs. Therapeutic efficacy may be enlarged
manifold.
Despite progress being made in determining the role
of lncRNAs in the treatment of GC, none of these lncRNAs have
reached the standard of clinical translation to date. A few key
lncRNAs, including HOTAIR, UCA1 and MALAT1, have been reported to
regulate various downstream molecules and suppress tumors in
vitro and in vivo. Relevant agents for clinical use were
kept at a slow pace due to the distinction between human
physiological processes and simulation environment of cells and
animals. Furthermore, the existing mechanisms were scattered and
unable to constitute an integral network. The inability to
recognize the overall perspective of may set obstacles for finding
regimens of curing GC thoroughly. Additional in-depth
investigations are warranted to mine more effective therapeutic
targets.
Exosomes are nm-sized vesicles in the extracellular
fluid, which span 40-150 nm and contain numerous functional
molecules such as proteins, miRNAs, lncRNAs and circRNAs. The
exploration of exosome-relevant surface markers and transmission
electron microscope contribute to the development of detection and
further research. The processes of formation, content selection,
loading, trafficking and release of exosomes are strictly under
physical control (146-148).
On the other hand, alteration in the levels and
modified states of lncRNAs and circRNAs have potential for use in
the diagnosis and prognosis of GC. Exosomes encapsulate biomarkers
and protect them from RNase degradation, which endow them with
extraordinary stability and capability of representativeness. For
example, Lin et al found that lncUEGC1 was early
GC-specific, and could discriminate patients with early GC from
healthy individuals and those with premalignant chronic atrophic
gastritis (150). Moreover,
LINC00152, lncRNA HOTTIP, circ-KIAA1244 and circ_0065149 were
identified as biomarkers for GC diagnosis and prognosis (95,151-153).
circRNAs and lncRNAs both belong to the family of
non-coding RNAs. Similar constructions determine consistent
characteristics: i) Relative incapacity of expressing proteins; ii)
some highly conserved sequences; iii) stable existence in extreme
environments for a relatively long period of time; iv) wide
varieties and distribution; v) diversity and complexity of the
involved regulatory mechanisms; and vi) association with multiple
diseases. The isoforms derived from the same genes have been
demonstrated to play important roles in cancer. For instance,
lncRNA PVT1 and circPVT1 both play oncogenic roles in GC
progression, and have great diagnostic and therapeutic potential
(43,154). The biological functions also
overlap. circRNAs and lncRNAs can bind to miRNAs through
complementation, which serves as a negative regulatory method for
target miRNAs. During the past decade, numerous studies concerning
miRNAs have been published (20,21,44,52). The network of miRNAs in
tumorigenesis has begun to take shape. The association of miRNAs
with lncRNAs and circRNAs suggests a vast number of uncovered
regulatory pathways. The classical view indicates that circRNAs and
lncRNAs lack the capacity of coding proteins. However, recent
evidence supports coding competence for these (155), which universally overturns
traditional impression. Additionally, although limited studies have
been published regarding the crosstalk of circRNAs and lncRNAs, the
shared features indicated an abundance of underlying interactions
in carcinogenesis and development. Further basic and translational
studies are required on this topic.
Researchers strive to identify a novel method of
clinical translation. As aforementioned, circRNAs and lncRNAs have
great potential for use in the diagnosis and treatment of GC.
Extensive focus has been placed on lncRNAs over the past decade,
with multiple studies suggesting effective clinical performance for
GC. Although circRNAs have been discov-ered for a long time, they
have only increased in popularity in recent years. However, unique
translational methods of circRNAs provide a new direction for
clinical use and research. Both ncRNAs have their own strengths and
shortcomings.
The location association of lncRNAs and circRNAs is
important for the exploration of their potential clinical
translation. Distinct location endows these molecules with
different biological functions. lncRNAs were estimated to be mostly
located in the nucleus, while circRNAs are mostly located in the
cytoplasm, acting as the miRNA sponges to regulate downstream
signaling pathways. This location association determines their
potential for use in clinical research or practice. Researchers
should not only verify their experimental capability of interfering
GC progression, but also ascertain the natural location of these
molecules in cancer cells.
A novel therapeutic method for GC relies on the
elucidation of the mechanisms responsible for the development of
GC. Previous studies have screened out numerous circRNAs and
lncRNAs with therapeutic potential (41-43,47-64,115,118-121,123-128,132,133,138,140-145), mainly by small interference
in vitro and by inhibition of tumor burden in vivo.
Various new signaling pathways were identified, which were
connected with established cancer promoters or suppressors. The
reported efficacy of regulating circRNAs and lncRNAs was
demonstrated to be effective. It is worth mentioning that the
application of synthetic circRNAs offers a brand-new perspective
for researchers, which is considered as a more direct and
controllable method for GC treatment. The limitations of these
studies are evident. Basic studies cannot replace clinical trials.
Unknown efficacy, administration method and dosage of human agents
hinder the further use of these RNAs. The therapeutic regimens
should also refer to the cost-benefit principle. No studies
available to date have reported the side-effects associated with
the interference, at least to the best of our knowledge. In short,
detailed information on therapeutic applications requires more
in-depth investigations.
The following are some suggestions for the
development of circRNAs and lncRNAs. Firstly, sample sizes should
be enlarged. The majority of studies collected tissue and plasma
samples from <200 GC patients with GC. The limited number of
sources definitely increases the contingency of false results.
Despite the lack of evidence, circRNA and lncRNA profiles may vary
in different groups of age, ethnicity, and living conditions.
Therefore, additional large-scale, multi-center studies conducted
under strict supervision are required, which will draw more
convincible and compelling conclusions. More reliable information
is also required for individual diagnosis.
Secondly, clinical trials concerning optimal
biomarkers should be implemented as early as possible. Despite
findings of therapeutic targets at the preliminary stages, the
diag-nostic and prognostic biomarkers have been demonstrated to be
more convenient and efficient than compared to classical use.
Plasma detection is a promising future due to non-inva-sive
examination, a high acceptance by patients and better sensitivity.
With the development of detection technologies, circRNAs and
lncRNAs can be measured in gastric juice, which has exhibited great
efficacy in the diagnosis of GC. Their high stability endows these
biomarkers with the ability to endure extreme environments.
Furthermore, despite the limited number of studies available
concerning lncRNAs and circRNAs in exosomes of GC, exosomes can
serve as shields to protect non-coding RNAs from RNases and extreme
environments. The potential diagnostic values of lncRNAs and
circRNAs in exosomes need to be more deeply investigated. Exosomes
may also be effective transporters of RNA interference drugs.
Additional detection media for GC diagnosis and prognosis, such as
feces, warrant further exploration in the future. These
gastrointestinal-specific, alteration-immediate biomarkers will be
more promising for future applications.
Thirdly, the distinction of cancer locations in GC
should be recognized. Previous research has suggested that
carcino-genesis in different sites of GC, such as the cardia and
antral stomach, involves different mechanisms (156). However, to the best of our
knowledge, to date, there are no studies available differentiating
the location discrepancy. Studies have explored circRNA and lncRNA
profiles, and the regulatory pathways in the whole stomach, which
may be an important factor leading to partially contradictory
results, attenuating the development of clinical translation, and
promoting the unbalance of clinical accuracy and
practicability.
Fourthly, the disturbance of non-cancer factors
should be emphasized. circRNAs and lncRNAs exist in both normal and
pathological tissues, and are secreted into the extracellular
environment. Previous studies showed that the expression of these
two kinds of RNAs can be influenced by numerous non-cancer factors,
including inflammation, neurodegen-erative diseases, drugs and
circadian rhythms (157-161). The fluctuation of circRNA and
lncRNA levels leads to inaccurate results. It is advisable to
remove background disturbances by mathematical modulation or
molecular biology techniques. On the other hand, diagnostic and
prognostic translation of circRNA and lncRNA levels requires
normative criterion of sampling time, methods and criteria of
patients' inclusion and exclusion.
Fifthly, the specificity of circRNAs and lncRNAs can
be poor, which decreases down the accuracy and safety of non-coding
RNA interference in patients. This may be an important reason for
the slow pace of relevant therapeutic drugs. An efficient approach
is to administer drugs in situ, since this can reduce the
effects on non-targets and optimize the sufficient dose of drugs
for tumor focus. The efficacy and accompanying side-effects can be
easily observed in animal models. However, administration in
situ definitely would lead to more difficulties in clinical
practice and patient compliance. Biomaterial serves as a robust
star for modern biomedicine. A large number of novel transporter
systems have been established, and have been demonstrated to be
effective for maximizing the specificity and minimizing side
effects of interference drugs, such as nanoparticles and nanotube
sponges. More efficient and reliable mediators should be
constructed to improve the specificity and reduce the side-effects
of lncRNA and circRNA treatment.
The advantages and disadvantages of these two types
of RNA have been compared in the present review based on their
similar biological features and research achievements. Accumulating
evidence has revealed their essential role in the diagnosis,
prognosis and treatment of GC. Certain circRNAs and lncRNAs have
been reported to exhibit optimal effects and act as convenient
detection methods. Panels of combined biomarkers serve as a novel
trend in early diagnosis and prognostic prediction. Furthermore,
the identification of the mechanisms responsible for GC can promote
the development of GC therapeutic targets. Previous studies have
identified numerous circRNA and lncRNA molecules (28-31,82,85,94,114), as well as relevant signaling
pathways. The interference of target expression significantly
affects GC cell behavior and tumor burden.
However, there are still several limitations
concerning circRNAs and lncRNAs. The number of previous studies on
these molecules is relatively small, and numerous unknown molecules
require further exploration. Publication bias may partly cover the
potential targets. The small number of collected samples and the
different GC locations impair the reliability of the results. A
number of translational questions remain to be answered, such as:
i) the clinical position compared with that of classical biomarkers
and therapeutic regimens; ii) the development of strategies with
which to combine biomarkers to reach the cost-benefit balance; and
iii) the development of strategies with which to administer circRNA
and lncRNA disruptors into the human body with minimal side-effects
and optimal efficacy. It is too early to assert definite values in
clinical application.
In conclusion, the present review summarized the
current achievements of circRNAs and lncRNAs in GC. Numerous
biomarkers and targets were selected with theoretically optimal
performance. However, several issues remain to be resolved. The
translational procedures will encounter setbacks and difficulties.
Nevertheless, it is expected that circRNAs and lncRNAs will play
crucial roles in the diagnosis and treatment of GC in the
future.
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81773135)
and Beijing Science and Technology Commission Bio-Medicine and
Bio-Science Innovation Research Major Subject (grant no.
Z171100000417023).
Not applicable.
BC and GL were mainly responsible for collecting
relevant information and completing this review. WZ and YS were
mainly responsible for consulting literature materials and revising
the manuscript. BW was responsible for the conception of this
review and the assignment of tasks. There was no additional
assistance with manuscript preparation. All authors read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ilson DH: Advances in the treatment of
gastric cancer. Curr Opin Gastroenterol. 33:473–476. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zurleni T, Gjoni E, Altomare M and Rausei
S: Conversion surgery for gastric cancer patients: A review. World
J Gastrointest Oncol. 10:398–409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pernot S, Voron T, Perkins G,
Lagorce-Pages C, Berger A and Taieb J: Signet-ring cell carcinoma
of the stomach: Impact on prognosis and specific therapeutic
challenge. World J Gastroenterol. 21:11428–11438. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kolakofsky D: Isolation and
characterization of Sendai virus DI-RNAs. Cell. 8:547–555. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cocquerelle C, Mascrez B, Hetuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kristensen LS, Hansen TB, Veno MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
10
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
14
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pan J, Meng X, Jiang N, Jin X, Zhou C, Xu
D and Gong Z: Insights into the noncoding RNA-encoded peptides.
Protein Pept Lett. 25:720–727. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin X, Feng CY, Xiang Z, Chen YP and Li
YM: CircRNA expression pattern and circRNA-miRNA-mRNA network in
the pathogenesis of nonalcoholic steatohepatitis. Oncotarget.
7:66455–66467. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Piwecka M, Glazar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao M, Gao F, Zhang D, Wang S, Zhang Y,
Wang R and Zhao J: Altered expression of circular RNAs in Moyamoya
disease. J Neurol Sci. 381:25–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R,
Yang SY, Yang DC and Wang XL: Circular RNA hsa_circ_0001982
promotes breast cancer cell carcinogenesis through decreasing
miR-143. DNA Cell Biol. 36:901–908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX,
Liao ZJ and Nan KJ: Over-expression of CircRNA_100876 in non-small
cell lung cancer and its prognostic value. Pathol Res Pract.
213:453–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gu W, Sun Y, Zheng X, Ma J, Hu XY, Gao T
and Hu MJ: Identification of gastric cancer-related circular RNA
through microarray analysis and bioinformatics analysis. Biomed Res
Int. 2018:23816802018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang YS, Jie N, Zou KJ and Weng Y:
Expression profile of circular RNAs in human gastric cancer
tissues. Mol Med Rep. 16:2469–2476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J,
Lin H, Liu F and Dai Y: Circular RNA and gene expression profiles
in gastric cancer based on microarray chip technology. Oncol Rep.
37:1804–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W,
Yu R, Xiao B and Guo J: Plasma circular RNA profiling of patients
with gastric cancer and their droplet digital RT-PCR detection. J
Mol Med (Berl). 96:85–96. 2018. View Article : Google Scholar
|
|
33
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. 32:e222812018. View Article : Google Scholar
|
|
36
|
Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao
B and Guo J: Downregulated expression of hsa_circ_0074362 in
gastric cancer and its potential diagnostic values. Biomark Med.
12:11–20. 2018. View Article : Google Scholar
|
|
37
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang
W, Liu Z, Cao H and Cao X: Hsa_circ_0000520, a potential new
circular RNA biomarker, is involved in gastric carcinoma. Cancer
Biomark. 21:299–306. 2018. View Article : Google Scholar
|
|
39
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamamichi N, Hirano C, Ichinose M,
Takahashi Y, Minatsuki C, Matsuda R, Nakayama C, Shimamoto T,
Kodashima S, Ono S, et al: Atrophic gastritis and enlarged gastric
folds diagnosed by double-contrast upper gastrointestinal barium
X-ray radiography are useful to predict future gastric cancer
development based on the 3-year prospective observation. Gastric
Cancer. 19:1016–1022. 2016. View Article : Google Scholar
|
|
41
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: CircRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017. View Article : Google Scholar
|
|
42
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar
|
|
44
|
Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng
J, Hou J, Lin L and Cai J: Regulatory network of circRNA-miRNA-mRNA
contributes to the histological classification and disease
progression in gastric cancer. J Transl Med. 16:2162018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lai Z, Yang Y, Yan Y, Li T, Li Y, Wang Z,
Shen Z, Ye Y, Jiang K and Wang S: Analysis of co-expression
networks for circular RNAs and mRNAs reveals that circular RNAs
hsa_ circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are
candidate oncogenes in gastric cancer. Cell Cycle. 16:2301–2311.
2017. View Article : Google Scholar
|
|
46
|
Shen F, Liu P, Xu Z, Li N, Yi Z, Tie X,
Zhang Y and Gao L: CircRNA_001569 promotes cell proliferation
through absorbing miR-145 in gastric cancer. J Biochem. 165:27–36.
2019. View Article : Google Scholar
|
|
47
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
48
|
Lu J, Zhang PY, Li P, Xie JW, Wang JB, Lin
JX, Chen QY, Cao LL, Huang CM and Zheng CH: Circular RNA
hsa_circ_0001368 suppresses the progression of gastric cancer by
regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun.
512:29–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang
W and Cao H: CircPSMC3 suppresses the proliferation and metastasis
of gastric cancer by acting as a competitive endogenous RNA through
sponging miR-296-5p. Mol Cancer. 18:252019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang L, Shen J and Jiang Y:
Circ_0027599/PHDLA1 suppresses gastric cancer progression by
sponging miR-101-3p.1. Cell Biosci. 8:582018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li
H, Li D, Song H, Wang J, Hong M, et al: Circular RNA circAGO2
drives cancer progression through facilitating HuR-repressed
functions of AGO2-miRNA complexes. Cell Death Differ. 26:1346–1364.
2019. View Article : Google Scholar :
|
|
53
|
Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin
M, Liang H and Gao L: A novel circular RNA, circFAT1(e2), inhibits
gastric cancer progression by targeting miR-548g in the cytoplasm
and interacting with YBX1 in the nucleus. Cancer Lett. 442:222–232.
2019. View Article : Google Scholar
|
|
54
|
Liu X, Abraham JM, Cheng Y, Wang Z, Wang
Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al:
Synthetic Circular RNA Functions as a miR-21 Sponge to suppress
gastric carcinoma cell proliferation. Mol Ther Nucleic Acids.
13:312–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ouyang Y, Li Y, Huang Y, Li X, Zhu Y, Long
Y, Wang Y, Guo X and Gong K: CircRNA circPDSS1 promotes the gastric
cancer progression by sponging miR-1865p and modulating NEK2. J
Cell Physiol. 234:10458–10469. 2019. View Article : Google Scholar
|
|
56
|
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou
J, Jin H, Zhao A, Tang WW and Cao XF: Down-regulation of circPVRL3
promotes the proliferation and migration of gastric cancer cells.
Sci Rep. 8:101112018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J,
Jin H, Zheng W, Tang W, Cao H and Cao X: Circ-SFMBT2 promotes the
proliferation of gastric cancer cells through sponging miR-182-5p
to enhance CREB1 expression. Cancer Manag Res. 10:5725–5734. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA hsa_circ_0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 496:455–461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar
|
|
60
|
Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu
X, Li Z, Wei J, Liu M and Li G: Circular RNA circ-DONSON
facilitates gastric cancer growth and invasion via NURF complex
dependent activation of transcription factor SOX4. Mol Cancer.
18:452019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang J, Hou L, Liang R, Chen X, Zhang R,
Chen W and Zhu J: CircDLST promotes the tumorigenesis and
metastasis of gastric cancer by sponging miR-502-5p and activating
the NRAS/MEK1/ERK1/2 signaling. Molecular Cancer. 18:802019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhu Z, Rong Z, Luo Z, Yu Z, Zhang J, Qiu Z
and Huang C: Circular RNA circNHSL1 promotes gastric cancer
progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer.
18:1262019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang L, Song X, Chen X, Wang Q, Zheng X,
Wu C and Jiang J: Circular RNA CircCACTIN promotes gastric cancer
progression by Sponging MiR-331-3p and regulating TGFBR1
expression. Int J Biol Sci. 15:1091–1103. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang S, Zhang X, Li Z, Wang W, Li B, Huang
X, Sun G, Xu J, Li Q, Xu Z, et al: Circular RNA profile identifies
circOSBPL10 as an oncogenic factor and prognostic marker in gastric
cancer. Oncogene. 38:6985–7001. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Huang X, Luo YL, Mao YS and Ji JL: The
link between long noncoding RNAs and depression. Prog
Neuropsychopharmacol Biol Psychiatry. 73:73–78. 2017. View Article : Google Scholar
|
|
68
|
Marin-Bejar O, Mas AM, Gonzalez J,
Martinez D, Athie A, Morales X, Galduroz M, Raimondi I, Grossi E,
Guo S, et al: The human lncRNA LINC-PINT inhibits tumor cell
invasion through a highly conserved sequence element. Genome Biol.
18:2022017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Neppl RL, Wu CL and Walsh K: lncRNA
Chronos is an aging-induced inhibitor of muscle hypertrophy. J Cell
Biol. 216:3497–3507. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu Y, Gonzalez-Porta M, Santos S, Brazma
A, Marioni JC, Aebersold R, Venkitaraman AR and Wickramasinghe VO:
Impact of alternative splicing on the human proteome. Cell Rep.
20:1229–1241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Luco RF: Retrotransposons jump into
alternative-splicing regulation via a long noncoding RNA. Nat
Struct Mol Biol. 23:952–954. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Huang J, Zhang A, Ho TT, Zhang Z, Zhou N,
Ding X, Zhang X, Xu M and Mo YY: Linc-RoR promotes c-Myc expression
through hnRNP I and AUF1. Nucleic Acids Res. 44:3059–3069. 2016.
View Article : Google Scholar :
|
|
75
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X
and Zai S: LncRNA DANCR promotes migration and invasion through
suppression of lncRNA-LET in gastric cancer cells. Biosci Rep.
37:BSR201710702017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Pan L, Liang W, Gu J, Zang X, Huang Z, Shi
H, Chen J, Fu M, Zhang P, Xiao X, et al: Long noncoding RNA DANCR
is activated by SALL4 and promotes the proliferation and invasion
of gastric cancer cells. Oncotarget. 9:1915–1930. 2017. View Article : Google Scholar
|
|
78
|
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu
Y, Shan B, Zheng D and Shi J: The lncRNA SNHG5/miR-32 axis
regulates gastric cancer cell proliferation and migration by
targeting KLF4. FASEB J. 31:893–903. 2017. View Article : Google Scholar
|
|
79
|
Zhu H, Zhao H, Zhang L, Xu J, Zhu C, Zhao
H and Lv G: Dandelion root extract suppressed gastric cancer cells
proliferation and migration through targeting lncRNA-CCAT1. Biomed
Pharmacother. 93:1010–1017. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li Y, Zhu G, Ma Y and Qu H: LncRNA CCAT1
contributes to the growth and invasion of gastric cancer via
targeting miR-219. J Cell Biochem. Sep 3–2019.Epub ahead of print.
View Article : Google Scholar
|
|
81
|
Zhang J and Gao Y: CCAT-1 promotes
proliferation and inhibits apoptosis of cervical cancer cells via
the Wnt signaling pathway. Oncotarget. 8:68059–68070. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Du L, Duan W, Jiang X, Zhao L, Li J, Wang
R, Yan S, Xie Y, Yan K, Wang Q, et al: Cell-free lncRNA expression
signatures in urine serve as novel non-invasive biomarkers for
diagnosis and recurrence prediction of bladder cancer. J Cell Mol
Med. 22:2838–2845. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Si Y, Bai J, Wu J, Li Q, Mo Y, Fang R and
Lai W: LncRNA PlncRNA1 regulates proliferation and differentiation
of hair follicle stem cells through TGFβ1-mediated Wnt/β-catenin
signal pathway. Mol Med Rep. 17:1191–1197. 2018.
|
|
84
|
Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X,
Xu C, Xiao B, Sun Y and Guo J: Using gastric juice
lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of
gastric cancer. Tumour Biol. 37:1183–1188. 2016. View Article : Google Scholar
|
|
85
|
Clark MB, Johnston RL, Inostroza-Ponta M,
Fox AH, Fortini E, Moscato P, Dinger ME and Mattick JS: Genome-wide
analysis of long noncoding RNA stability. Genome Res. 22:885–898.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T,
Xiao B and Guo J: Decreased expression of long noncoding RNA
AC096655.1-002 in gastric cancer and its clinical significance.
Tumour Biol. 34:2697–2701. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lin X, Yang M, Xia T and Guo J: Increased
expression of long noncoding RNA ABHD11-AS1 in gastric cancer and
its clinical significance. Med Oncol. 31:422014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Baratieh Z, Khalaj Z, Honardoost MA,
Emadi-Baygi M, Khanahmad H, Salehi M and Nikpour P: Aberrant
expression of PlncRNA-1 and TUG1: Potential biomarkers for gastric
cancer diagnosis and clinically monitoring cancer progression.
Biomark Med. 11:1077–1090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu
QH, Weng WW, Tan C, Sheng WQ, Zhou XY and Du X: Circulating CUDR,
LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish
patients with gastric cancer from healthy controls. Int J Cancer.
137:1128–1135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hu QY, Zhao ZY, Li SQ, Li L and Li GK: A
meta-analysis: The diagnostic values of long non-coding RNA as a
biomarker for gastric cancer. Mol Clin Oncol. 6:846–852. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Hashad D, Elbanna A, Ibrahim A and Khedr
G: Evaluation of the role of circulating long non-coding RNA H19 as
a promising novel biomarker in plasma of patients with gastric
cancer. J Clin Lab Anal. 30:1100–1105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Elsayed ET, Salem PE, Darwish AM and Fayed
HM: Plasma long non-coding RNA HOTAIR as a potential biomarker for
gastric cancer. Int J Biol Markers. Apr 1–2018.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y,
Liang W, Hu C, Liu Y, Li J, et al: Genome-Wide lncRNA microarray
profiling identifies novel circulating lncRNAs for detection of
gastric cancer. Theranostics. 7:213–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar
|
|
96
|
Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C,
Ye H, Zhou B, Chen JJ and Chen P: Aberrant expression of UCA1 in
gastric cancer and its clinical significance. Clin Transl Oncol.
17:640–646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu
Z, Ye G, Zhang X, Xiao B and Guo J: Gastric juice long noncoding
RNA used as a tumor marker for screening gastric cancer. Cancer.
120:3320–3328. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Pang Q, Ge J, Shao Y, Sun W, Song H, Xia
T, Xiao B and Guo J: Increased expression of long intergenic
non-coding RNA LINC00152 in gastric cancer and its clinical
significance. Tumour Biol. 35:5441–5447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fei ZH, Yu XJ, Zhou M, Su HF, Zheng Z and
Xie CY: Upregulated expression of long non-coding RNA LINC00982
regulates cell proliferation and its clinical relevance in patients
with gastric cancer. Tumour Biol. 37:1983–1993. 2016. View Article : Google Scholar
|
|
101
|
Chen JS, Wang YF, Zhang XQ, Lv JM, Li Y,
Liu XX and Xu TP: H19 serves as a diagnostic biomarker and
up-regulation of H19 expression contributes to poor prognosis in
patients with gastric cancer. Neoplasma. 63:223–230. 2016.
|
|
102
|
Hunt RH, Xiao SD, Megraud F, Leon-Barua R,
Bazzoli F, van der Merwe S, Vaz Coelho LG, Fock M, Fedail S, Cohen
H, et al: Helicobacter pylori in developing countries. World
gastroenterology organisation global guideline. J Gastrointestin
Liver Dis. 20:299–304. 2011.PubMed/NCBI
|
|
103
|
Amieva M and Peek RM Jr: Pathobiology of
Helicobacter pylori-induced gastric cancer. Gastroenterology.
150:64–78. 2016. View Article : Google Scholar
|
|
104
|
Zhong F, Zhu M, Gao K, Xu P, Yang H, Hu D,
Cui D, Wang M, Xie X, Wei Y, et al: Low expression of the long
non-coding RNA NR_026827 in gastric cancer. Am J Transl Res.
10:2706–2711. 2018.PubMed/NCBI
|
|
105
|
Lu Q, Yu T, Ou X, Cao D, Xie T and Chen X:
Potential lncRNA diagnostic biomarkers for early gastric cancer.
Mol Med Rep. 16:9545–9552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J,
Ma G, Chu H, Tong N, Zhu H, et al: The association analysis of
lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese
population. Oncotarget. 6:31255–31262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Liu JN and Shangguan YM: Long non-coding
RNA CARLo-5 upregulation associates with poor prognosis in patients
suffering gastric cancer. Eur Rev Med Pharmacol Sci. 21:530–534.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Feng Y, Zhang Q, Wang J and Liu P:
Increased lncRNA AFAP1-AS1 expression predicts poor prognosis and
promotes malignant phenotypes in gastric cancer. Eur Rev Med
Pharmacol Sci. 21:3842–3849. 2017.PubMed/NCBI
|
|
109
|
Li L, Geng Y, Feng R, Zhu Q, Miao B, Cao J
and Fei S: The human RNA surveillance factor UPF1 modulates gastric
cancer progression by targeting long non-coding RNA MALAT1. Cell
Physiol Biochem. 42:2194–2206. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Li Y, Wen X, Wang L, Sun X, Ma H, Fu Z and
Li L: LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric
cancer. Surg Oncol. 26:527–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ma P, Xu T, Huang M and Shu Y: Increased
expression of LncRNA PANDAR predicts a poor prognosis in gastric
cancer. Biomed Pharmacother. 78:172–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yao XM, Tang JH, Zhu H and Jing Y: High
expression of LncRNA CASC15 is a risk factor for gastric cancer
prognosis and promote the proliferation of gastric cancer. Eur Rev
Med Pharmacol Sci. 21:5661–5667. 2017.PubMed/NCBI
|
|
113
|
Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang
Q, Ren M, Chen L, Yuan D, Zhang Y, et al: The lncRNA MALAT1 is a
novel biomarker for gastric cancer metastasis. Oncotarget.
7:56209–56218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zhu X, Tian X, Yu C, Shen C, Yan T, Hong
J, Wang Z, Fang JY and Chen H: A long non-coding RNA signature to
improve prognosis prediction of gastric cancer. Mol Cancer.
15:602016. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chen Y, Wei G, Xia H, Tang Q and Bi F:
Long noncoding RNA-ATB promotes cell proliferation, migration and
invasion in gastric cancer. Mol Med Rep. 17:1940–1946. 2018.
|
|
116
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar :
|
|
118
|
Dong X, He X, Guan A, Huang W, Jia H,
Huang Y, Chen S, Zhang Z, Gao J and Wang H: Long non-coding RNA
Hotair promotes gastric cancer progression via miR-217-GPC5 axis.
Life Sci. 217:271–282. 2019. View Article : Google Scholar
|
|
119
|
Song B, Guan Z, Liu F, Sun D, Wang K and
Qu H: Long non-coding RNA HOTAIR promotes HLA-G expression via
inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res
Commun. 464:807–813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Jiang D, Li H, Xiang H, Gao M, Yin C, Wang
H, Sun Y and Xiong M: Long chain non-coding RNA (lncRNA) HOTAIR
knockdown increases miR-454-3p to suppress gastric cancer growth by
targeting STAT3/Cyclin D1. Med Sci Monit. 25:1537–1548. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Jia J, Zhan D, Li J, Li Z, Li H and Qian
J: The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and
Shenqifuzheng injection on chemosensitivity of gastric cancer
cells. J Cell Mol Med. 23:656–669. 2019. View Article : Google Scholar
|
|
122
|
Lotem J, Levanon D, Negreanu V, Bauer O,
Hantisteanu S, Dicken J and Groner Y: Runx3 in immunity,
inflammation and cancer. Adv Exp Med Biol. 962:369–393. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Xue M, Chen LY, Wang WJ, Su TT, Shi LH,
Wang L, Zhang W, Si JM, Wang LJ and Chen SJ: HOTAIR induces the
ubiquitination of Runx3 by interacting with Mex3b and enhances the
invasion of gastric cancer cells. Gastric Cancer. 21:756–764. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zeng S, Xie X, Xiao YF, Tang B, Hu CJ,
Wang SM, Wu YY, Dong H, Li BS and Yang SM: Long noncoding RNA
LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress
gastric cancer progression. Cancer Lett. 412:179–187. 2018.
View Article : Google Scholar
|
|
125
|
Li D, Yang M, Liao A, Zeng B, Liu D, Yao
Y, Hu G, Chen X, Feng Z, Du Y, et al: Linc00483 as ceRNA regulates
proliferation and apoptosis through activating MAPKs in gastric
cancer. J Cell Mol Med. 22:3875–3886. 2018. View Article : Google Scholar :
|
|
126
|
Shan Y, Ying R, Jia Z, Kong W, Wu Y, Zheng
S and Jin H: LINC00052 promotes gastric cancer cell proliferation
and metastasis via activating the Wnt/β-catenin signaling pathway.
Oncol Res. 25:1589–1599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang
J, Wang J, Zhang Q and Xu Z: Linc00152 promotes proliferation in
gastric cancer through the EGFR-dependent pathway. J Exp Clin
Cancer Res. 34:1352015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Yan J, Zhang Y, She Q, Li X, Peng L, Wang
X, Liu S, Shen X, Zhang W, Dong Y, et al: Long noncoding RNA
H19/miR-675 axis promotes gastric cancer via FADD/Caspase 8/Caspase
3 signaling pathway. Cell Physiol Biochem. 42:2364–2376. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Marin JJ, Al-Abdulla R, Lozano E, Briz O,
Bujanda L, Banales JM and Macias RI: Mechanisms of resistance to
chemo-therapy in gastric cancer. Anticancer Agents Med Chem.
16:318–334. 2016. View Article : Google Scholar
|
|
130
|
Zhang X, Bo P, Liu L, Zhang X and Li J:
Overexpression of long non-coding RNA GHET1 promotes the
development of multi-drug resistance in gastric cancer cells.
Biomed Pharmacother. 92:580–585. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lan WG, Xu DH, Xu C, Ding CL, Ning FL,
Zhou YL, Ma LB, Liu CM and Han X: Silencing of long non-coding RNA
ANRIL inhibits the development of multidrug resistance in gastric
cancer cells. Oncol Rep. 36:263–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Xi Z, Si J and Nan J: LncRNA MALAT1
potentiates autophagyassociated cisplatin resistance by regulating
the microRNA30b/autophagyrelated gene 5 axis in gastric cancer. Int
J Oncol. 54:239–248. 2019.
|
|
133
|
Li Y, Lv S, Ning H, Li K, Zhou X, Xv H and
Wen H: Down-regulation of CASC2 contributes to cisplatin resistance
in gastric cancer by sponging miR-19a. Biomed Pharmacother.
108:1775–1782. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Fang Q, Chen X and Zhi X: Long non-coding
RNA (LncRNA) urothelial carcinoma associated 1 (UCA1) increases
multi-drug resistance of gastric cancer via downregulating miR-27b.
Med Sci Monit. 22:3506–3513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Shang C, Guo Y, Zhang J and Huang B:
Silence of long noncoding RNA UCA1 inhibits malignant proliferation
and chemotherapy resistance to adriamycin in gastric cancer. Cancer
Chemother Pharmacol. 77:1061–1067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Thorburn A: Autophagy and disease. J Biol
Chem. 293:5425–5430. 2018. View Article : Google Scholar :
|
|
137
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye
WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in
cancer. Chin J Cancer. 36:522017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
YiRen H, YingCong Y, Sunwu Y, Keqin L,
Xiaochun T, Senrui C, Ende C, XiZhou L and Yanfan C: Long noncoding
RNA MALAT1 regulates autophagy associated chemoresistance via
miR-23b-3p sequestration in gastric cancer. Mol Cancer. 16:1742017.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Suresh PS, Tsutsumi R and Venkatesh T:
YBX1 at the cross-roads of non-coding transcriptome, exosomal, and
cytoplasmic granular signaling. Eur J Cell Biol. 97:163–167. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhang E, He X, Zhang C, Su J, Lu X, Si X,
Chen J, Yin D, Han L and De W: A novel long noncoding RNA HOXC-AS3
mediates tumorigenesis of gastric cancer by binding to YBX1. Genome
Biol. 19:1542018. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang
G and Zhang Y: lncRNA GAS5 enhances G1 cell cycle arrest via
binding to YBX1 to regulate p21 expression in stomach cancer. Sci
Rep. 5:101592015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Chen WM, Chen WD, Jiang XM, Jia XF, Wang
HM, Zhang QJ, Shu YQ and Zhao HB: HOX transcript antisense
intergenic RNA represses E-cadherin expression by binding to EZH2
in gastric cancer. World J Gastroenterol. 23:6100–6110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan
S, Yu Q, Li YY, Kang Y, Li H, et al: MALAT1 long ncRNA promotes
gastric cancer metastasis by suppressing PCDH10. Oncotarget.
7:12693–12703. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wang ZQ, Cai Q, Hu L, He CY, Li JF, Quan
ZW, Liu BY, Li C and Zhu ZG: Long noncoding RNA UCA1 induced by SP1
promotes cell proliferation via recruiting EZH2 and activating AKT
pathway in gastric cancer. Cell Death Dis. 8:e28392017. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu
YB and Tu YN: Long noncoding RNA LINC00673 epigenetically
suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric
cancer. Oncotarget. 8:95542–95553. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Vlassov AV, Magdaleno S, Setterquist R and
Conrad R: Exosomes: Current knowledge of their composition,
biological functions, and diagnostic and therapeutic potentials.
Biochim Biophys Acta. 1820:940–948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Rajagopal C and Harikumar KB: The origin
and functions of exosomes in cancer. Front Oncol. 8:662018.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Pan L, Liang W, Fu M, Huang ZH, Li X,
Zhang W, Zhang P, Qian H, Jiang PC, Xu WR and Zhang X:
Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes
gastric cancer progression. J Cancer Res Clin Oncol. 143:991–1004.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Lin LY, Yang L, Zeng Q, Wang L, Chen ML,
Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, et al: Tumor-originated
exosomal lncUEGC1 as a circulating biomarker for early-stage
gastric cancer. Mol Cancer. 17:842018. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Zhao R and Zhang Y, Zhang X, Yang Y, Zheng
X, Li X, Liu Y and Zhang Y: Exosomal long noncoding RNA HOTTIP as
potential novel diagnostic and prognostic biomarker test for
gastric cancer. Mol Cancer. 17:682018. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Tang W, Fu K, Sun H, Rong D, Wang H and
Cao H: CircRNA microarray profiling identifies a novel circulating
biomarker for detection of gastric cancer. Mol Cancer. 17:1372018.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao
B, Ye G and Guo J: Hsa_circ_0065149 is an indicator for early
gastric cancer screening and prognosis prediction. Pathol Oncol
Res. 26:1475–1482. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ,
Liu M and Wang B: The long noncoding RNA PVT1 functions as a
competing endogenous RNA by sponging miR-186 in gastric cancer.
Biomed Pharmacother. 88:302–308. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Li LJ, Leng RX, Fan YG, Pan HF and Ye DQ:
Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and
circRNAs. Exp Cell Res. 361:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Gulmann C, Hegarty H, Grace A, Leader M,
Patchett S and Kay E: Differences in proximal (cardia) versus
distal (antral) gastric carcinogenesis via the retinoblastoma
pathway. World J Gastroenterol. 10:17–21. 2004. View Article : Google Scholar
|
|
157
|
Pan Z, Li GF, Sun ML, Xie L, Liu D, Zhang
Q, Yang XX, Xia S, Liu X, Zhou H, et al: MicroRNA-1224 splicing
circularRNA-Filip1l in an Ago2-Dependent manner regulates chronic
inflammatory pain via targeting Ubr5. J Neurosci. 39:2125–2143.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Du M, Yuan L, Tan X, Huang D, Wang X,
Zheng Z, Mao X, Li X, Yang L, Huang K, et al: The LPS-inducible
lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun.
8:20492017. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Coon SL, Munson PJ, Cherukuri PF, Sugden
D, Rath MF, Møller M, Clokie SJ, Fu C, Olanich ME, Rangel Z, et al:
Circadian changes in long noncoding RNAs in the pineal gland. Proc
Natl Acad Sci USA. 109:13319–13324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Zhao Y, Alexandrov PN, Jaber V and Lukiw
WJ: Deficiency in the ubiquitin conjugating enzyme UBE2A in
Alzheimer's disease (AD) is linked to deficits in a natural
circular miRNA-7 Sponge (circRNA; ciRS-7). Genes (Basel).
7:1162016. View Article : Google Scholar
|
|
161
|
Guo H, Liu J, Ben Q, Qu Y, Li M, Wang Y,
Chen W and Zhang J: The aspirin-induced long non-coding RNA OLA1P2
blocks phosphorylated STAT3 homodimer formation. Genome Biol.
17:242016. View Article : Google Scholar : PubMed/NCBI
|