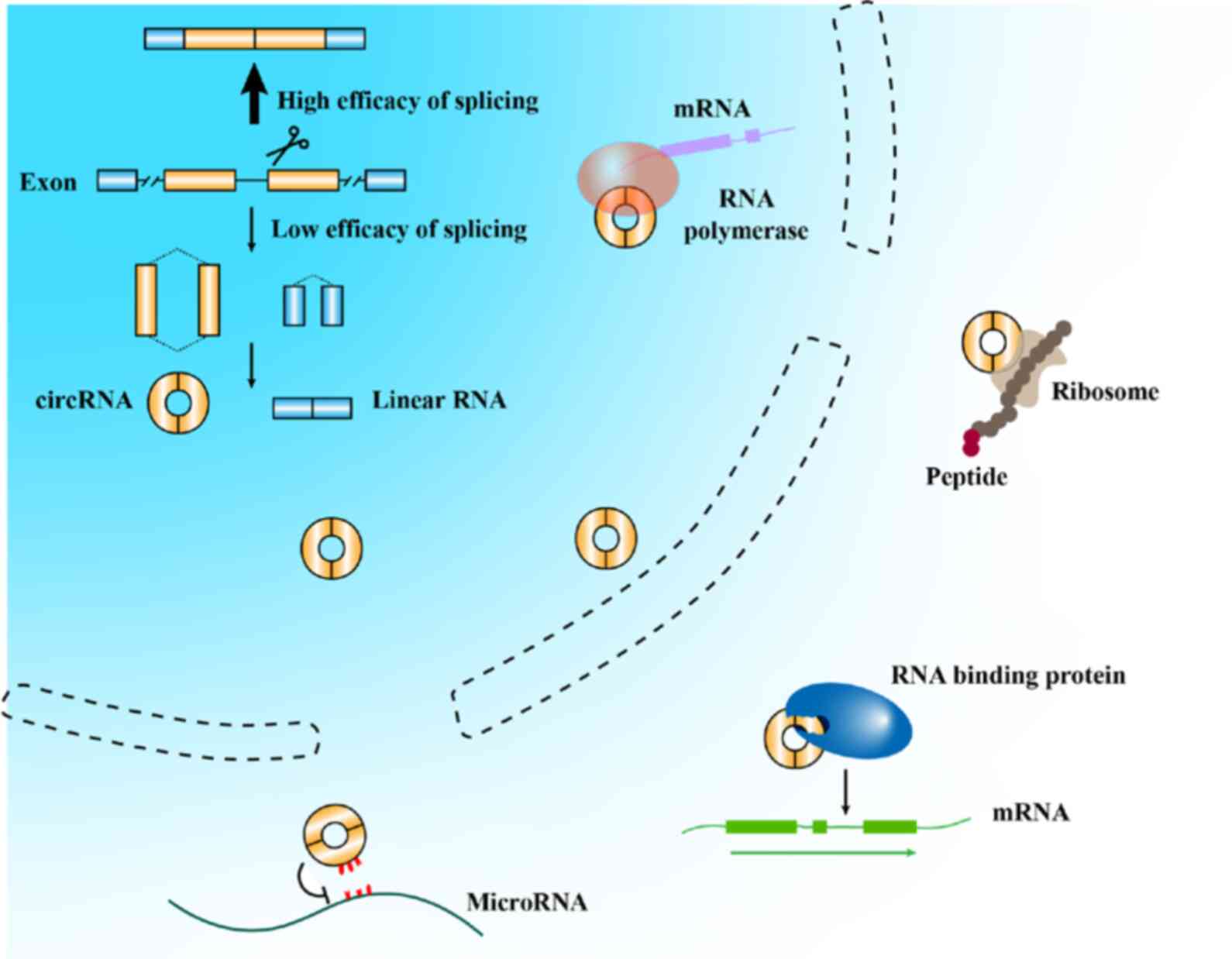
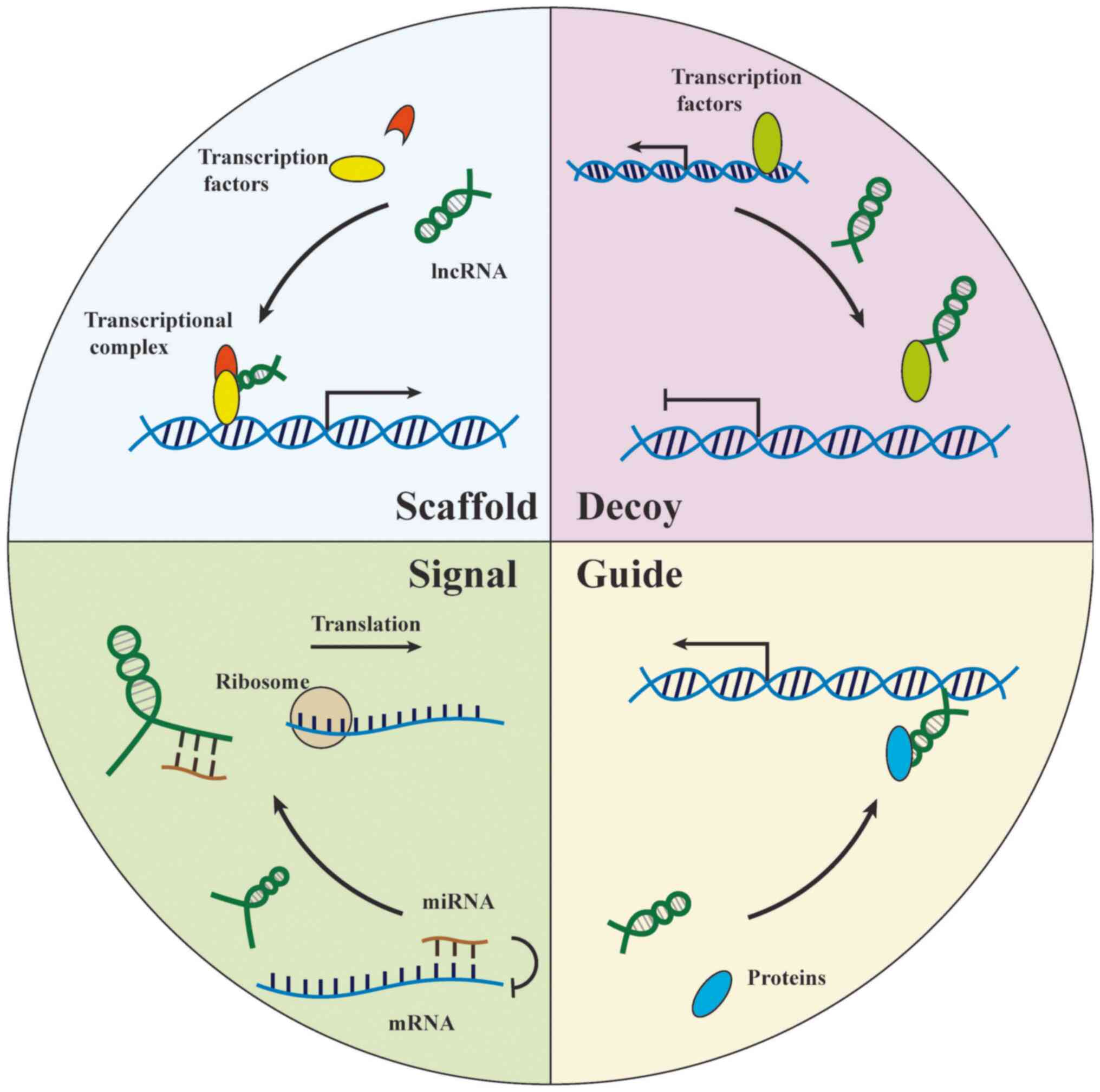
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ilson DH: Advances in the treatment of
gastric cancer. Curr Opin Gastroenterol. 33:473–476. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zurleni T, Gjoni E, Altomare M and Rausei
S: Conversion surgery for gastric cancer patients: A review. World
J Gastrointest Oncol. 10:398–409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pernot S, Voron T, Perkins G,
Lagorce-Pages C, Berger A and Taieb J: Signet-ring cell carcinoma
of the stomach: Impact on prognosis and specific therapeutic
challenge. World J Gastroenterol. 21:11428–11438. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kolakofsky D: Isolation and
characterization of Sendai virus DI-RNAs. Cell. 8:547–555. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cocquerelle C, Mascrez B, Hetuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kristensen LS, Hansen TB, Veno MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
10
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
14
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pan J, Meng X, Jiang N, Jin X, Zhou C, Xu
D and Gong Z: Insights into the noncoding RNA-encoded peptides.
Protein Pept Lett. 25:720–727. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin X, Feng CY, Xiang Z, Chen YP and Li
YM: CircRNA expression pattern and circRNA-miRNA-mRNA network in
the pathogenesis of nonalcoholic steatohepatitis. Oncotarget.
7:66455–66467. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Piwecka M, Glazar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao M, Gao F, Zhang D, Wang S, Zhang Y,
Wang R and Zhao J: Altered expression of circular RNAs in Moyamoya
disease. J Neurol Sci. 381:25–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R,
Yang SY, Yang DC and Wang XL: Circular RNA hsa_circ_0001982
promotes breast cancer cell carcinogenesis through decreasing
miR-143. DNA Cell Biol. 36:901–908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX,
Liao ZJ and Nan KJ: Over-expression of CircRNA_100876 in non-small
cell lung cancer and its prognostic value. Pathol Res Pract.
213:453–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gu W, Sun Y, Zheng X, Ma J, Hu XY, Gao T
and Hu MJ: Identification of gastric cancer-related circular RNA
through microarray analysis and bioinformatics analysis. Biomed Res
Int. 2018:23816802018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang YS, Jie N, Zou KJ and Weng Y:
Expression profile of circular RNAs in human gastric cancer
tissues. Mol Med Rep. 16:2469–2476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J,
Lin H, Liu F and Dai Y: Circular RNA and gene expression profiles
in gastric cancer based on microarray chip technology. Oncol Rep.
37:1804–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W,
Yu R, Xiao B and Guo J: Plasma circular RNA profiling of patients
with gastric cancer and their droplet digital RT-PCR detection. J
Mol Med (Berl). 96:85–96. 2018. View Article : Google Scholar
|
|
33
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. 32:e222812018. View Article : Google Scholar
|
|
36
|
Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao
B and Guo J: Downregulated expression of hsa_circ_0074362 in
gastric cancer and its potential diagnostic values. Biomark Med.
12:11–20. 2018. View Article : Google Scholar
|
|
37
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun H, Tang W, Rong D, Jin H, Fu K, Zhang
W, Liu Z, Cao H and Cao X: Hsa_circ_0000520, a potential new
circular RNA biomarker, is involved in gastric carcinoma. Cancer
Biomark. 21:299–306. 2018. View Article : Google Scholar
|
|
39
|
Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B
and Guo J: Global circular RNA expression profile of human gastric
cancer and its clinical significance. Cancer Med. 6:1173–1180.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamamichi N, Hirano C, Ichinose M,
Takahashi Y, Minatsuki C, Matsuda R, Nakayama C, Shimamoto T,
Kodashima S, Ono S, et al: Atrophic gastritis and enlarged gastric
folds diagnosed by double-contrast upper gastrointestinal barium
X-ray radiography are useful to predict future gastric cancer
development based on the 3-year prospective observation. Gastric
Cancer. 19:1016–1022. 2016. View Article : Google Scholar
|
|
41
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: CircRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017. View Article : Google Scholar
|
|
42
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar
|
|
44
|
Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng
J, Hou J, Lin L and Cai J: Regulatory network of circRNA-miRNA-mRNA
contributes to the histological classification and disease
progression in gastric cancer. J Transl Med. 16:2162018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lai Z, Yang Y, Yan Y, Li T, Li Y, Wang Z,
Shen Z, Ye Y, Jiang K and Wang S: Analysis of co-expression
networks for circular RNAs and mRNAs reveals that circular RNAs
hsa_ circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are
candidate oncogenes in gastric cancer. Cell Cycle. 16:2301–2311.
2017. View Article : Google Scholar
|
|
46
|
Shen F, Liu P, Xu Z, Li N, Yi Z, Tie X,
Zhang Y and Gao L: CircRNA_001569 promotes cell proliferation
through absorbing miR-145 in gastric cancer. J Biochem. 165:27–36.
2019. View Article : Google Scholar
|
|
47
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
48
|
Lu J, Zhang PY, Li P, Xie JW, Wang JB, Lin
JX, Chen QY, Cao LL, Huang CM and Zheng CH: Circular RNA
hsa_circ_0001368 suppresses the progression of gastric cancer by
regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun.
512:29–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang
W and Cao H: CircPSMC3 suppresses the proliferation and metastasis
of gastric cancer by acting as a competitive endogenous RNA through
sponging miR-296-5p. Mol Cancer. 18:252019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang L, Shen J and Jiang Y:
Circ_0027599/PHDLA1 suppresses gastric cancer progression by
sponging miR-101-3p.1. Cell Biosci. 8:582018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li
H, Li D, Song H, Wang J, Hong M, et al: Circular RNA circAGO2
drives cancer progression through facilitating HuR-repressed
functions of AGO2-miRNA complexes. Cell Death Differ. 26:1346–1364.
2019. View Article : Google Scholar :
|
|
53
|
Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin
M, Liang H and Gao L: A novel circular RNA, circFAT1(e2), inhibits
gastric cancer progression by targeting miR-548g in the cytoplasm
and interacting with YBX1 in the nucleus. Cancer Lett. 442:222–232.
2019. View Article : Google Scholar
|
|
54
|
Liu X, Abraham JM, Cheng Y, Wang Z, Wang
Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al:
Synthetic Circular RNA Functions as a miR-21 Sponge to suppress
gastric carcinoma cell proliferation. Mol Ther Nucleic Acids.
13:312–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ouyang Y, Li Y, Huang Y, Li X, Zhu Y, Long
Y, Wang Y, Guo X and Gong K: CircRNA circPDSS1 promotes the gastric
cancer progression by sponging miR-1865p and modulating NEK2. J
Cell Physiol. 234:10458–10469. 2019. View Article : Google Scholar
|
|
56
|
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou
J, Jin H, Zhao A, Tang WW and Cao XF: Down-regulation of circPVRL3
promotes the proliferation and migration of gastric cancer cells.
Sci Rep. 8:101112018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J,
Jin H, Zheng W, Tang W, Cao H and Cao X: Circ-SFMBT2 promotes the
proliferation of gastric cancer cells through sponging miR-182-5p
to enhance CREB1 expression. Cancer Manag Res. 10:5725–5734. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA hsa_circ_0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 496:455–461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar
|
|
60
|
Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu
X, Li Z, Wei J, Liu M and Li G: Circular RNA circ-DONSON
facilitates gastric cancer growth and invasion via NURF complex
dependent activation of transcription factor SOX4. Mol Cancer.
18:452019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang J, Hou L, Liang R, Chen X, Zhang R,
Chen W and Zhu J: CircDLST promotes the tumorigenesis and
metastasis of gastric cancer by sponging miR-502-5p and activating
the NRAS/MEK1/ERK1/2 signaling. Molecular Cancer. 18:802019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhu Z, Rong Z, Luo Z, Yu Z, Zhang J, Qiu Z
and Huang C: Circular RNA circNHSL1 promotes gastric cancer
progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer.
18:1262019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang L, Song X, Chen X, Wang Q, Zheng X,
Wu C and Jiang J: Circular RNA CircCACTIN promotes gastric cancer
progression by Sponging MiR-331-3p and regulating TGFBR1
expression. Int J Biol Sci. 15:1091–1103. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang S, Zhang X, Li Z, Wang W, Li B, Huang
X, Sun G, Xu J, Li Q, Xu Z, et al: Circular RNA profile identifies
circOSBPL10 as an oncogenic factor and prognostic marker in gastric
cancer. Oncogene. 38:6985–7001. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Huang X, Luo YL, Mao YS and Ji JL: The
link between long noncoding RNAs and depression. Prog
Neuropsychopharmacol Biol Psychiatry. 73:73–78. 2017. View Article : Google Scholar
|
|
68
|
Marin-Bejar O, Mas AM, Gonzalez J,
Martinez D, Athie A, Morales X, Galduroz M, Raimondi I, Grossi E,
Guo S, et al: The human lncRNA LINC-PINT inhibits tumor cell
invasion through a highly conserved sequence element. Genome Biol.
18:2022017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Neppl RL, Wu CL and Walsh K: lncRNA
Chronos is an aging-induced inhibitor of muscle hypertrophy. J Cell
Biol. 216:3497–3507. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu Y, Gonzalez-Porta M, Santos S, Brazma
A, Marioni JC, Aebersold R, Venkitaraman AR and Wickramasinghe VO:
Impact of alternative splicing on the human proteome. Cell Rep.
20:1229–1241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Luco RF: Retrotransposons jump into
alternative-splicing regulation via a long noncoding RNA. Nat
Struct Mol Biol. 23:952–954. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Huang J, Zhang A, Ho TT, Zhang Z, Zhou N,
Ding X, Zhang X, Xu M and Mo YY: Linc-RoR promotes c-Myc expression
through hnRNP I and AUF1. Nucleic Acids Res. 44:3059–3069. 2016.
View Article : Google Scholar :
|
|
75
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mao Z, Li H, Du B, Cui K, Xing Y, Zhao X
and Zai S: LncRNA DANCR promotes migration and invasion through
suppression of lncRNA-LET in gastric cancer cells. Biosci Rep.
37:BSR201710702017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Pan L, Liang W, Gu J, Zang X, Huang Z, Shi
H, Chen J, Fu M, Zhang P, Xiao X, et al: Long noncoding RNA DANCR
is activated by SALL4 and promotes the proliferation and invasion
of gastric cancer cells. Oncotarget. 9:1915–1930. 2017. View Article : Google Scholar
|
|
78
|
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu
Y, Shan B, Zheng D and Shi J: The lncRNA SNHG5/miR-32 axis
regulates gastric cancer cell proliferation and migration by
targeting KLF4. FASEB J. 31:893–903. 2017. View Article : Google Scholar
|
|
79
|
Zhu H, Zhao H, Zhang L, Xu J, Zhu C, Zhao
H and Lv G: Dandelion root extract suppressed gastric cancer cells
proliferation and migration through targeting lncRNA-CCAT1. Biomed
Pharmacother. 93:1010–1017. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li Y, Zhu G, Ma Y and Qu H: LncRNA CCAT1
contributes to the growth and invasion of gastric cancer via
targeting miR-219. J Cell Biochem. Sep 3–2019.Epub ahead of print.
View Article : Google Scholar
|
|
81
|
Zhang J and Gao Y: CCAT-1 promotes
proliferation and inhibits apoptosis of cervical cancer cells via
the Wnt signaling pathway. Oncotarget. 8:68059–68070. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Du L, Duan W, Jiang X, Zhao L, Li J, Wang
R, Yan S, Xie Y, Yan K, Wang Q, et al: Cell-free lncRNA expression
signatures in urine serve as novel non-invasive biomarkers for
diagnosis and recurrence prediction of bladder cancer. J Cell Mol
Med. 22:2838–2845. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Si Y, Bai J, Wu J, Li Q, Mo Y, Fang R and
Lai W: LncRNA PlncRNA1 regulates proliferation and differentiation
of hair follicle stem cells through TGFβ1-mediated Wnt/β-catenin
signal pathway. Mol Med Rep. 17:1191–1197. 2018.
|
|
84
|
Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X,
Xu C, Xiao B, Sun Y and Guo J: Using gastric juice
lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of
gastric cancer. Tumour Biol. 37:1183–1188. 2016. View Article : Google Scholar
|
|
85
|
Clark MB, Johnston RL, Inostroza-Ponta M,
Fox AH, Fortini E, Moscato P, Dinger ME and Mattick JS: Genome-wide
analysis of long noncoding RNA stability. Genome Res. 22:885–898.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T,
Xiao B and Guo J: Decreased expression of long noncoding RNA
AC096655.1-002 in gastric cancer and its clinical significance.
Tumour Biol. 34:2697–2701. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lin X, Yang M, Xia T and Guo J: Increased
expression of long noncoding RNA ABHD11-AS1 in gastric cancer and
its clinical significance. Med Oncol. 31:422014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Baratieh Z, Khalaj Z, Honardoost MA,
Emadi-Baygi M, Khanahmad H, Salehi M and Nikpour P: Aberrant
expression of PlncRNA-1 and TUG1: Potential biomarkers for gastric
cancer diagnosis and clinically monitoring cancer progression.
Biomark Med. 11:1077–1090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu
QH, Weng WW, Tan C, Sheng WQ, Zhou XY and Du X: Circulating CUDR,
LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish
patients with gastric cancer from healthy controls. Int J Cancer.
137:1128–1135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hu QY, Zhao ZY, Li SQ, Li L and Li GK: A
meta-analysis: The diagnostic values of long non-coding RNA as a
biomarker for gastric cancer. Mol Clin Oncol. 6:846–852. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Hashad D, Elbanna A, Ibrahim A and Khedr
G: Evaluation of the role of circulating long non-coding RNA H19 as
a promising novel biomarker in plasma of patients with gastric
cancer. J Clin Lab Anal. 30:1100–1105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Elsayed ET, Salem PE, Darwish AM and Fayed
HM: Plasma long non-coding RNA HOTAIR as a potential biomarker for
gastric cancer. Int J Biol Markers. Apr 1–2018.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y,
Liang W, Hu C, Liu Y, Li J, et al: Genome-Wide lncRNA microarray
profiling identifies novel circulating lncRNAs for detection of
gastric cancer. Theranostics. 7:213–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar
|
|
96
|
Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C,
Ye H, Zhou B, Chen JJ and Chen P: Aberrant expression of UCA1 in
gastric cancer and its clinical significance. Clin Transl Oncol.
17:640–646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu
Z, Ye G, Zhang X, Xiao B and Guo J: Gastric juice long noncoding
RNA used as a tumor marker for screening gastric cancer. Cancer.
120:3320–3328. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Pang Q, Ge J, Shao Y, Sun W, Song H, Xia
T, Xiao B and Guo J: Increased expression of long intergenic
non-coding RNA LINC00152 in gastric cancer and its clinical
significance. Tumour Biol. 35:5441–5447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fei ZH, Yu XJ, Zhou M, Su HF, Zheng Z and
Xie CY: Upregulated expression of long non-coding RNA LINC00982
regulates cell proliferation and its clinical relevance in patients
with gastric cancer. Tumour Biol. 37:1983–1993. 2016. View Article : Google Scholar
|
|
101
|
Chen JS, Wang YF, Zhang XQ, Lv JM, Li Y,
Liu XX and Xu TP: H19 serves as a diagnostic biomarker and
up-regulation of H19 expression contributes to poor prognosis in
patients with gastric cancer. Neoplasma. 63:223–230. 2016.
|
|
102
|
Hunt RH, Xiao SD, Megraud F, Leon-Barua R,
Bazzoli F, van der Merwe S, Vaz Coelho LG, Fock M, Fedail S, Cohen
H, et al: Helicobacter pylori in developing countries. World
gastroenterology organisation global guideline. J Gastrointestin
Liver Dis. 20:299–304. 2011.PubMed/NCBI
|
|
103
|
Amieva M and Peek RM Jr: Pathobiology of
Helicobacter pylori-induced gastric cancer. Gastroenterology.
150:64–78. 2016. View Article : Google Scholar
|
|
104
|
Zhong F, Zhu M, Gao K, Xu P, Yang H, Hu D,
Cui D, Wang M, Xie X, Wei Y, et al: Low expression of the long
non-coding RNA NR_026827 in gastric cancer. Am J Transl Res.
10:2706–2711. 2018.PubMed/NCBI
|
|
105
|
Lu Q, Yu T, Ou X, Cao D, Xie T and Chen X:
Potential lncRNA diagnostic biomarkers for early gastric cancer.
Mol Med Rep. 16:9545–9552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J,
Ma G, Chu H, Tong N, Zhu H, et al: The association analysis of
lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese
population. Oncotarget. 6:31255–31262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Liu JN and Shangguan YM: Long non-coding
RNA CARLo-5 upregulation associates with poor prognosis in patients
suffering gastric cancer. Eur Rev Med Pharmacol Sci. 21:530–534.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Feng Y, Zhang Q, Wang J and Liu P:
Increased lncRNA AFAP1-AS1 expression predicts poor prognosis and
promotes malignant phenotypes in gastric cancer. Eur Rev Med
Pharmacol Sci. 21:3842–3849. 2017.PubMed/NCBI
|
|
109
|
Li L, Geng Y, Feng R, Zhu Q, Miao B, Cao J
and Fei S: The human RNA surveillance factor UPF1 modulates gastric
cancer progression by targeting long non-coding RNA MALAT1. Cell
Physiol Biochem. 42:2194–2206. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Li Y, Wen X, Wang L, Sun X, Ma H, Fu Z and
Li L: LncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric
cancer. Surg Oncol. 26:527–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ma P, Xu T, Huang M and Shu Y: Increased
expression of LncRNA PANDAR predicts a poor prognosis in gastric
cancer. Biomed Pharmacother. 78:172–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yao XM, Tang JH, Zhu H and Jing Y: High
expression of LncRNA CASC15 is a risk factor for gastric cancer
prognosis and promote the proliferation of gastric cancer. Eur Rev
Med Pharmacol Sci. 21:5661–5667. 2017.PubMed/NCBI
|
|
113
|
Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang
Q, Ren M, Chen L, Yuan D, Zhang Y, et al: The lncRNA MALAT1 is a
novel biomarker for gastric cancer metastasis. Oncotarget.
7:56209–56218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zhu X, Tian X, Yu C, Shen C, Yan T, Hong
J, Wang Z, Fang JY and Chen H: A long non-coding RNA signature to
improve prognosis prediction of gastric cancer. Mol Cancer.
15:602016. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chen Y, Wei G, Xia H, Tang Q and Bi F:
Long noncoding RNA-ATB promotes cell proliferation, migration and
invasion in gastric cancer. Mol Med Rep. 17:1940–1946. 2018.
|
|
116
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar :
|
|
118
|
Dong X, He X, Guan A, Huang W, Jia H,
Huang Y, Chen S, Zhang Z, Gao J and Wang H: Long non-coding RNA
Hotair promotes gastric cancer progression via miR-217-GPC5 axis.
Life Sci. 217:271–282. 2019. View Article : Google Scholar
|
|
119
|
Song B, Guan Z, Liu F, Sun D, Wang K and
Qu H: Long non-coding RNA HOTAIR promotes HLA-G expression via
inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res
Commun. 464:807–813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Jiang D, Li H, Xiang H, Gao M, Yin C, Wang
H, Sun Y and Xiong M: Long chain non-coding RNA (lncRNA) HOTAIR
knockdown increases miR-454-3p to suppress gastric cancer growth by
targeting STAT3/Cyclin D1. Med Sci Monit. 25:1537–1548. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Jia J, Zhan D, Li J, Li Z, Li H and Qian
J: The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and
Shenqifuzheng injection on chemosensitivity of gastric cancer
cells. J Cell Mol Med. 23:656–669. 2019. View Article : Google Scholar
|
|
122
|
Lotem J, Levanon D, Negreanu V, Bauer O,
Hantisteanu S, Dicken J and Groner Y: Runx3 in immunity,
inflammation and cancer. Adv Exp Med Biol. 962:369–393. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Xue M, Chen LY, Wang WJ, Su TT, Shi LH,
Wang L, Zhang W, Si JM, Wang LJ and Chen SJ: HOTAIR induces the
ubiquitination of Runx3 by interacting with Mex3b and enhances the
invasion of gastric cancer cells. Gastric Cancer. 21:756–764. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zeng S, Xie X, Xiao YF, Tang B, Hu CJ,
Wang SM, Wu YY, Dong H, Li BS and Yang SM: Long noncoding RNA
LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress
gastric cancer progression. Cancer Lett. 412:179–187. 2018.
View Article : Google Scholar
|
|
125
|
Li D, Yang M, Liao A, Zeng B, Liu D, Yao
Y, Hu G, Chen X, Feng Z, Du Y, et al: Linc00483 as ceRNA regulates
proliferation and apoptosis through activating MAPKs in gastric
cancer. J Cell Mol Med. 22:3875–3886. 2018. View Article : Google Scholar :
|
|
126
|
Shan Y, Ying R, Jia Z, Kong W, Wu Y, Zheng
S and Jin H: LINC00052 promotes gastric cancer cell proliferation
and metastasis via activating the Wnt/β-catenin signaling pathway.
Oncol Res. 25:1589–1599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang
J, Wang J, Zhang Q and Xu Z: Linc00152 promotes proliferation in
gastric cancer through the EGFR-dependent pathway. J Exp Clin
Cancer Res. 34:1352015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Yan J, Zhang Y, She Q, Li X, Peng L, Wang
X, Liu S, Shen X, Zhang W, Dong Y, et al: Long noncoding RNA
H19/miR-675 axis promotes gastric cancer via FADD/Caspase 8/Caspase
3 signaling pathway. Cell Physiol Biochem. 42:2364–2376. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Marin JJ, Al-Abdulla R, Lozano E, Briz O,
Bujanda L, Banales JM and Macias RI: Mechanisms of resistance to
chemo-therapy in gastric cancer. Anticancer Agents Med Chem.
16:318–334. 2016. View Article : Google Scholar
|
|
130
|
Zhang X, Bo P, Liu L, Zhang X and Li J:
Overexpression of long non-coding RNA GHET1 promotes the
development of multi-drug resistance in gastric cancer cells.
Biomed Pharmacother. 92:580–585. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lan WG, Xu DH, Xu C, Ding CL, Ning FL,
Zhou YL, Ma LB, Liu CM and Han X: Silencing of long non-coding RNA
ANRIL inhibits the development of multidrug resistance in gastric
cancer cells. Oncol Rep. 36:263–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Xi Z, Si J and Nan J: LncRNA MALAT1
potentiates autophagyassociated cisplatin resistance by regulating
the microRNA30b/autophagyrelated gene 5 axis in gastric cancer. Int
J Oncol. 54:239–248. 2019.
|
|
133
|
Li Y, Lv S, Ning H, Li K, Zhou X, Xv H and
Wen H: Down-regulation of CASC2 contributes to cisplatin resistance
in gastric cancer by sponging miR-19a. Biomed Pharmacother.
108:1775–1782. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Fang Q, Chen X and Zhi X: Long non-coding
RNA (LncRNA) urothelial carcinoma associated 1 (UCA1) increases
multi-drug resistance of gastric cancer via downregulating miR-27b.
Med Sci Monit. 22:3506–3513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Shang C, Guo Y, Zhang J and Huang B:
Silence of long noncoding RNA UCA1 inhibits malignant proliferation
and chemotherapy resistance to adriamycin in gastric cancer. Cancer
Chemother Pharmacol. 77:1061–1067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Thorburn A: Autophagy and disease. J Biol
Chem. 293:5425–5430. 2018. View Article : Google Scholar :
|
|
137
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye
WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in
cancer. Chin J Cancer. 36:522017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
YiRen H, YingCong Y, Sunwu Y, Keqin L,
Xiaochun T, Senrui C, Ende C, XiZhou L and Yanfan C: Long noncoding
RNA MALAT1 regulates autophagy associated chemoresistance via
miR-23b-3p sequestration in gastric cancer. Mol Cancer. 16:1742017.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Suresh PS, Tsutsumi R and Venkatesh T:
YBX1 at the cross-roads of non-coding transcriptome, exosomal, and
cytoplasmic granular signaling. Eur J Cell Biol. 97:163–167. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhang E, He X, Zhang C, Su J, Lu X, Si X,
Chen J, Yin D, Han L and De W: A novel long noncoding RNA HOXC-AS3
mediates tumorigenesis of gastric cancer by binding to YBX1. Genome
Biol. 19:1542018. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang
G and Zhang Y: lncRNA GAS5 enhances G1 cell cycle arrest via
binding to YBX1 to regulate p21 expression in stomach cancer. Sci
Rep. 5:101592015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Chen WM, Chen WD, Jiang XM, Jia XF, Wang
HM, Zhang QJ, Shu YQ and Zhao HB: HOX transcript antisense
intergenic RNA represses E-cadherin expression by binding to EZH2
in gastric cancer. World J Gastroenterol. 23:6100–6110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan
S, Yu Q, Li YY, Kang Y, Li H, et al: MALAT1 long ncRNA promotes
gastric cancer metastasis by suppressing PCDH10. Oncotarget.
7:12693–12703. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wang ZQ, Cai Q, Hu L, He CY, Li JF, Quan
ZW, Liu BY, Li C and Zhu ZG: Long noncoding RNA UCA1 induced by SP1
promotes cell proliferation via recruiting EZH2 and activating AKT
pathway in gastric cancer. Cell Death Dis. 8:e28392017. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu
YB and Tu YN: Long noncoding RNA LINC00673 epigenetically
suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric
cancer. Oncotarget. 8:95542–95553. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Vlassov AV, Magdaleno S, Setterquist R and
Conrad R: Exosomes: Current knowledge of their composition,
biological functions, and diagnostic and therapeutic potentials.
Biochim Biophys Acta. 1820:940–948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Rajagopal C and Harikumar KB: The origin
and functions of exosomes in cancer. Front Oncol. 8:662018.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Pan L, Liang W, Fu M, Huang ZH, Li X,
Zhang W, Zhang P, Qian H, Jiang PC, Xu WR and Zhang X:
Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes
gastric cancer progression. J Cancer Res Clin Oncol. 143:991–1004.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Lin LY, Yang L, Zeng Q, Wang L, Chen ML,
Zhao ZH, Ye GD, Luo QC, Lv PY, Guo QW, et al: Tumor-originated
exosomal lncUEGC1 as a circulating biomarker for early-stage
gastric cancer. Mol Cancer. 17:842018. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Zhao R and Zhang Y, Zhang X, Yang Y, Zheng
X, Li X, Liu Y and Zhang Y: Exosomal long noncoding RNA HOTTIP as
potential novel diagnostic and prognostic biomarker test for
gastric cancer. Mol Cancer. 17:682018. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Tang W, Fu K, Sun H, Rong D, Wang H and
Cao H: CircRNA microarray profiling identifies a novel circulating
biomarker for detection of gastric cancer. Mol Cancer. 17:1372018.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao
B, Ye G and Guo J: Hsa_circ_0065149 is an indicator for early
gastric cancer screening and prognosis prediction. Pathol Oncol
Res. 26:1475–1482. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Huang T, Liu HW, Chen JQ, Wang SH, Hao LQ,
Liu M and Wang B: The long noncoding RNA PVT1 functions as a
competing endogenous RNA by sponging miR-186 in gastric cancer.
Biomed Pharmacother. 88:302–308. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Li LJ, Leng RX, Fan YG, Pan HF and Ye DQ:
Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and
circRNAs. Exp Cell Res. 361:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Gulmann C, Hegarty H, Grace A, Leader M,
Patchett S and Kay E: Differences in proximal (cardia) versus
distal (antral) gastric carcinogenesis via the retinoblastoma
pathway. World J Gastroenterol. 10:17–21. 2004. View Article : Google Scholar
|
|
157
|
Pan Z, Li GF, Sun ML, Xie L, Liu D, Zhang
Q, Yang XX, Xia S, Liu X, Zhou H, et al: MicroRNA-1224 splicing
circularRNA-Filip1l in an Ago2-Dependent manner regulates chronic
inflammatory pain via targeting Ubr5. J Neurosci. 39:2125–2143.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Du M, Yuan L, Tan X, Huang D, Wang X,
Zheng Z, Mao X, Li X, Yang L, Huang K, et al: The LPS-inducible
lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun.
8:20492017. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Coon SL, Munson PJ, Cherukuri PF, Sugden
D, Rath MF, Møller M, Clokie SJ, Fu C, Olanich ME, Rangel Z, et al:
Circadian changes in long noncoding RNAs in the pineal gland. Proc
Natl Acad Sci USA. 109:13319–13324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Zhao Y, Alexandrov PN, Jaber V and Lukiw
WJ: Deficiency in the ubiquitin conjugating enzyme UBE2A in
Alzheimer's disease (AD) is linked to deficits in a natural
circular miRNA-7 Sponge (circRNA; ciRS-7). Genes (Basel).
7:1162016. View Article : Google Scholar
|
|
161
|
Guo H, Liu J, Ben Q, Qu Y, Li M, Wang Y,
Chen W and Zhang J: The aspirin-induced long non-coding RNA OLA1P2
blocks phosphorylated STAT3 homodimer formation. Genome Biol.
17:242016. View Article : Google Scholar : PubMed/NCBI
|