Introduction
Parkinson's disease (PD) is the second most common
neuro-degenerative disease worldwide affecting 1% of the population
that is over 60 years of age. The main pathological feature is the
progressive loss of dopaminergic neurons of the substantia nigra in
the mid-brain region. PD is characterized clinically by several
motor signs, including bradykinesia, rigidity, tremor, and postural
instability, but also by non-motor symptoms such as dementia and
sleep and mood disorders. The intraneuronal accumulation and
misfolding of α-synuclein is a well-characterized mechanism that
leads to dopaminergic neurotoxicity (1-3).
It is known that PD onset is strongly associated with age and its
clinical diagnosis is difficult, in particular during the earlier
stages of the disease, when the risk of misdiagnosis between PD and
other neurodegenerative diseases is high. For this reason, the
identification of relevant early molecular biomarkers is of
paramount importance to allow an accurate diagnosis of the
disease.
Extracellular vesicles (EVs) are nanoparticles that
are released in systemic circulation by most cell types. Their role
in cell-to-cell communication has been widely documented and they
have been implicated in several pathological conditions, including
neurodegenerative diseases (4,5).
Three main EV types have been characterized according to their
biogenesis: Apoptotic bodies, microvesicles and exosomes. However,
the most recent guidelines highlight that these traditional
definitions do not take into account the overlapping size range,
similar morphology and variable composition of such EV
subpopulations. Thus, a more appropriate nomenclature is emerging
in literature, based on the classification of EVs by multiple
parameters, such as size, density, protein composition, lipid
content, RNA and DNA cargo, morphology, description of conditions
and cell origin (6). EVs are
stable and easily detectable in different body fluids and their
content has become of interest for the development of EV-based
molecular biomarkers for neurodegenerative diseases, including PD
(7). Indeed, EV cargo is
characterized by a repertoire of molecules that can be precisely
isolated and their alteration may be associated with disease
development and progression.
MicroRNAs (miRNAs) are members of EV cargo molecules
and are small RNA molecules (18-22 nt) that mainly act as negative
regulators of gene expression. They generally target the 3′UTR of
mRNA by inducing the block of translation or its degradation.
miRNAs are present in high amounts in EVs and different miRNA
profiles in exosomes have been associated with pathological
conditions. Gui et al developed a miRNA profiling strategy
for exosomal miRNAs isolated from cerebrospinal fluid (CSF) of
patients with PD (8). Exosomes
were isolated from CSF using an ultracentrifugation method and,
among the miRNA examined, miR-1 and miR-19b-3p were downregulated;
while miR-153, miR-409-3p, miR-10a-5p and let-7g-3p were
overexpressed in PD CSF exosomes. In recent studies, Cao et
al and Yao et al reported a differential expression of
given miRs in exosomes isolated by a commercial kit from the serum
and plasma, respectively, of PD patients versus healthy
individuals. Cao et al reported the down-regulation of
miR-19b and the overexpression of miR-195 and miR-24 (9), while Yao et al showed the
differential expression of miR-331-5p and miR-505 (10).
Generally, many studies highlighted the different
levels of selected miRNAs present in EVs indicating their role as
possible molecular biomarkers. However, technical issues, including
different protocols for EV isolation and optimization of
methodologies to characterize profiles with low quantities of RNA,
often make the characterization of the EV-RNA content inconsistent.
According to the ISEV position article on the necessity of dealing
with these issues (11), the
current study was conducted to test the expression level of
miR-34a-5p, not only in the whole plasma, but also in all the EV
subpopulations of PD patients and control subjects to verify the
contention that this strategy may be used to examine possible
expression variations between PD and control subjects. To carry out
this study, the strategy to isolate and characterize the different
EV subpopulations using a standardized methodology (serial
ultracentrifugations followed by density gradient) was employed.
miR-34a-5p was selected because of its involvement in
neurophysiology and neuropathology (12). miR-34a-5p is abundantly expressed
in the adult mammalian brain and there is emerging evidence that
miR-34a-5p plays key roles in mammalian neurogenesis,
synaptogenesis and neural differentiation. Furthermore, miR-34a-5p
has been recently described as a common dysregulated miRNA in
different disorders of the central nervous system, including
Alzheimer's disease, schizophrenia and major depression disorder
(13).
In the current study, to the best of our knowledge,
for the first time the expression levels of miR-34a-5p in whole
plasma and in its different EV subpopulations (Large, LEVs; Medium,
MEVs; Small EVs, SEVs; pure SEVs) isolated from PD patients and
control subjects were examined.
Materials and methods
Clinicopathological features of subjects
enrolled in this study
Peripheral blood was obtained from PD patients
(n=15; Neurology Unit, Civil Hospitals of Brescia, Italy) and
control subjects (n=14; Geriatrics Unit, Civil Hospitals of
Brescia, Italy). The study was approved by the ethics committee of
Civil Hospitals of Brescia on 8th June 2016. Informed consent was
obtained from all the subjects enrolled in the study.
The PD patients were male, with an age range of
70-92 years. Control subjects were age- and sex-matched to PD
patients and they had no evidence of PD, parkinsonism or dementia.
Clinical characteristics are reported in Table I for controls and PD patients.
Unified Parkinson's Disease Rating Scale (UPDRS) and modified Hoehn
and Yahr scale were used to evaluate PD symptoms (14,15). Beck Depression Inventory (BDI) was
used for the evaluation of depression of PD patients. Conventional
cut-off values used were: 0-9 for normal range, 10-18 for mild to
moderate depression, 19-29 for moderate to severe depression, and
30-63 for severe depression (16).
 | Table IClinical features of subjects
enrolled in this study. |
Table I
Clinical features of subjects
enrolled in this study.
| Parameters | Controls | PD patients |
|---|
| No. of subjects
enrolled | 14 | 15 |
| Sex | | |
| Male | 14 | 15 |
| Female | 0 | 0 |
| Age, years | 78.5±7.3 | 75.7±3.0 |
| Age at onset,
years | - | 69.5±4.4 |
| Duration of
disease, years | - | 5.1±2.9 |
| H and Y stage | - | 2.2±0.6 |
| UPDRS I | - | 10.5±3.2 |
| UPDRS II | - | 7.3±6.3 |
| UPDRS III | - | 21.7±6.8 |
| BDI score | - | 10.4±7.6 |
| Levodopa dose,
mg/day | - | 312.5±236.6 |
Plasma pre-analytical processing
Plasma is the most suitable medium to extract blood
physiological EVs because their total amount is not influenced by
platelet-derived EVs released upon clot formation. We used samples
containing EDTA, avoiding heparin based anticoagulants that may
interfere further analyses (17,18). Blood samples were processed within
2 h from the withdrawal and kept at room temperature. Careful tube
transportation was ensured to avoid unnecessary agitation. For this
purpose, a box maintaining blood tubes in a steady vertical
position was used. Plasma-EDTA (plasma) was centrifuged at 800 × g
for 10 min (5804R Eppendorf Centrifuge, A-4-44 rotor, 15 ml tubes),
2,500 × g for 15 min and then centrifuged a second time at 2,500 ×
g for 15 min. All centrifugations steps were made at room
temperature with low acceleration and avoiding application of the
centrifuge brake. After each centrifugation plasma was collected in
a fresh plastic tube, leaving 1 cm of plasma above the buffy layer
so as not to disturb it (19).
Plasma was finally transferred into crio-vials in 1 ml aliquots and
stored at -80°C.
EV preparations from plasma of controls
and PD subjects
All relevant data of the experiments have been
submitted to the EV-TRACK knowledge base (EV-TRACK ID: EV200031)
(20). Serial ultracentrifugation
steps were: For each subject 4 vials of 1 ml each of plasma were
thawed and pooled. Pooled plasma (3 ml) was then aliquoted in 3
Eppendorf tubes, 1 ml each. One tube of 1 ml aliquot was left at
4°C until further processing. Two tubes of 1 ml aliquot were
processed, in parallel, with serial centrifugation steps as
previously described (21).
Briefly, 1 ml plasma was centrifuged at 800 × g for 30 min (5417C
Eppendorf Centrifuge, 45-30-11 rotor, 1.5 ml Eppendorf tubes, 1 ml
each tube). Supernatant (1 ml) was transferred to a new tube and
centrifuged at 16,000 × g for 45 min (5417C Eppendorf Centrifuge,
45-30-11 rotor, 1.5 ml Eppendorf tubes, 1 ml each tube). Finally,
supernatant (1 ml) was transferred to an appropriate tube and
centrifuged at 100,000 × g for 2 h (Optima MAX, TLA-55 rotor, 1.5
ml polypropylene microfuge tube, Beckman). The 800 × g
centrifugation step allows sedi-mentation of LEVs, the 16,000 × g
step allows us to pellet MEVs, while the 100,000 × g
ultracentrifugation enriches SEVs. Pellets were washed with 1 ml
sterile H2O (Milli-Q, Merck Millipore) as described in
Paolini et al (6).
Discontinuous sucrose gradient was carried out as
follows: SEV pellet, obtained as described above, was further
processed adapting the protocols developed in previous studies
(22,23) to obtain pure SEVs. Briefly, SEVs
were re-suspended in 1,000 µl buffer A (10 mM Tris-HCl 250
mM sucrose, pH 7.4), loaded on top of a discontinuous sucrose
gradient 15% (600 µl), 20, 25, 30, 40, 60% (400 µl),
70% (800 µl) sucrose in 10 mM Tris-HCl, pH 7.4) and
centrifuged at 100,000 × g for 16 h at 4°C (rotor MLS 50; Beckman
Optima MAX). Twelve fractions with equal volumes (400 µl)
were collected from the top of the gradient and ultracentrifuged
(100,000 × g for 2 h at 4°C). Western blot analysis fractions were
precipitated by incubation with 10% trichloracetic acid (TCA)
(Sigma) overnight as described in Paolini et al (24).
For RNaseA treatment (6.25 µg/ml; Qiagen),
pure SEVs were resuspended in PBS. The RNase-treated samples were
incubated at room temperature for 30 min, followed by an
inactivation step in ice for 1 min; the non-treated samples were
kept in ice. Total RNA was immediately isolated from the two
samples and miR-34a-5p levels were analyzed as described below. No
difference in miR-34a-5p levels was detected between the treated
and non-treated pure SEVs indicating that miR-34a-5p is contained
in vesicles (data not shown).
SDS-PAGE and western blot analysis
SDS-PAGE and western blotting (WB) were performed by
standard procedures on the plasma EDTA samples (30 µg,
protein content determined by Bradford assay) (25), the LEV, MEV and SEV pellets and
the gradient fractions positive to EV markers (from 5 to 9,
collected together, nominated Pure SEV). LEV, MEV, SEV pellets and
gradient fractions were resuspended in 50 µl sterile
H2O (Milli-Q, Merck Millipore). Ten µl of
reducing sample buffer (480 mM Tris pH 6.8, 12% SDS, 45% glycerin,
0,06% bromophenol blue and 12% of β-mercaptoethanol) were added to
samples and they were boiled for 5 min at 95°C. Then, 20 µl
of each sample were electrophoresed, transferred to membrane and
developed with antibodies. The following antibodies (1:500
dilution) were used to characterize EVs (26,27): mouse anti-Flotillin 1 (Santa Cruz
Biotechnology, clone C-2, sc-74566), mouse anti-Annexin V
(Santa-Cruz Biotechnology, clone H-3, sc-74438), rabbit anti-ADAM10
(Origene, AP05830PU-N), mouse anti-CD81 (Santa Cruz Biotechnology,
clone B11, sc-166029), mouse anti-CD63 (Millipore, clone RFAC4,
CBL553), mouse anti-Alix (Santa Cruz Biotechnology, clone 2H12,
sc-53539), mouse anti TSG101 (Santa Cruz Biotechnology, clone C-2,
sc-7964), rabbit anti-Actinin-4 (Genetex, clone N2C1, GTX113115).
Mouse anti-GM130 Cis-Golgi protein diluted 1:250 (BD Transduction,
clone 35/130, 610822) and rabbit anti-APOA1 diluted at 1:700
(Thermo Scientific Fisher, 701239) were used as negative
controls.
Atomic force microscopy (AFM) imaging and
size distribution
AFM imaging and image analysis were performed
adapting protocols developed in previous studies (28-30). Briefly, the LEV, MEV and SEV
pellets and the gradient fractions positive to EV markers (from 5
to 9, collected together and nominated Pure SEVs) were resuspended
in 50 µl sterile H2O (Milli-Q; Merck Millipore)
and diluted 1:10 in H2O. Five µl of each sample
were then spotted onto freshly cleaved mica sheets (Grade V-1,
thickness 0.15 mm, size 15×15 mm2) and the samples were
allowed to dry at room temperature. They were imaged with a NaioAFM
(Nanosurf AG) equipped with Multi75-AI-G tip (Budget Sensors).
Images were acquired in tapping mode, with a scan size ranging from
1.5 µm to 25 µm and a scan speed of 1 sec per
scanning line. EV size distributions were obtained by image
analysis of at least five representative AFM images with a scan
size of 8.7×8.7 µm. Image analysis was performed using WSxM
version 5.0 and ImageJ (31).
EV subpopulations purity assessment
EV subpopulations were checked for purity from
protein contaminants applying the COlorimetric NANoplasmonic
(CONAN) assay, which exploits the nanoplasmonic properties of
colloidal gold nanoparticles (AuNPs) and their peculiar interaction
with proteins and lipid bilayers (32). LEV, MEV and SEV pellets and
gradient fractions positive to EV markers (from 5 to 9, collected
together; Pure SEV), were resuspended in 50 µl of sterile
H2O (Milli-Q; Merck Millipore). Then, 2 µl of
each sample were used for the assay. Three independent measurements
of at least two representative subjects both for controls and PD
patients were performed. The assay consisted of an aqueous solution
of bare gold nanoparticles (AuNPs) at 6 nM concentration. When
mixed with pure EV formulations, the AuNPs clustered on the EV
membrane, whereas in EV formulations which contained exogenous
protein contaminants (EPCs) the AuNPs were preferentially cloaked
by such EPCs (an AuNP-EPC corona forms), which prevents AuNPs from
clustering to the EV membrane. When AuNPs cluster (are in tight
proximity), their localized surface plasmon resonance (LSPR) red
shifts and broadens, resulting in a color change of the AuNP
solution from red to blue, which can be accurately monitored
through UV-Vis spectroscopy. The assay red shift is therefore
directly related to the purity grade of the added EV formulation
and can be conveniently quantified by describing the AuNP
UV/Vis/NIR absorption spectra with the nanoparticle Aggregation
Index (AI), defined as the ratio between the absorbance intensity
at the LSPR peak and the intensity at 650 nm plus the intensity at
850 nm (Abs LSPR/(Abs 650+Abs 850) (33).
Total RNA isolation
Total RNA was isolated from 200 µl of plasma
and each EV fraction using the miRNeasy mini kit (Qiagen).
According to the manufacturers' protocol, plasma and EV pellets
were lysed in 1 ml of QIAzol. Then, 2.5 µl of 5 nM synthetic
cel-miR-39 were added to each sample and 200 µl of
chloroform were used to separate the aqueous phase (34) including intellectual disability,
congenital heart disease, childhood leukemia and immune defects.
Specific microRNAs (miRNAs/miR. After centrifugation 12,000 × g for
15 min at 4°C, the upper aqueous phase was transferred into a new
tube and the miRNeasy kit protocol was followed. RNA was eluted in
35 µl of RNase-free H2O. The total RNA
concentration was measured using a NanoDrop spectrophotometer
(Thermo Fisher Scientific) and RNA quality was assessed using the
260/280 ratio. For RNA samples isolated from plasma and each EV
type of a subgroup of random subjects (including controls and PD
subjects), Agilent 2100 Bioanalyser using small RNA assay (Agilent)
was used to analyze the concentration of small RNA and miRNA, miRNA
percentage, and quality. This assay arbitrarily defined two
distinct regions: Small RNA region from 0 to 150 nt and miRNA
region from 10 to 40 nt.
Bioinformatics analysis
The miRNA target genes were downloaded from miRWALK
3.0 (35) and the targets
reported in the miRDB platform (781 genes) were selected for
subsequent analysis. For functional enrichment analysis of gene
ontology terms, Database for Annotation Visualization and
Integrated Discovery (DAVID v6.8) was used (36,37). Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway and Gene Ontology categories (Biological
Process, Molecular Function and Cellular Component) were
considered.
Quantitative PCR (qPCR)
Reverse transcription (RT) and qPCR were performed
as previously described (38,39). cDNA was synthesized from 5
µl of RNA isolated from plasma or EV subpopulations using
the TaqMan microRNA Reverse Transcription kit (Thermo Fisher
Scientific) in a 15-µl reaction containing 0.15 µl of
dNTPs, 1.00 µl of 50 U/µl MultiScribe™ Reverse
Transcriptase, 1.50 µl of 10X Reverse Transcription Buffer,
0.19 µl of RNase Inhibitor, 4.16 µl of nuclease-free
water and 3 µl of RT-specific stem-loop primers. The RT
reaction was performed at 16°C for 30 min, followed by incubation
at 42°C for 30 min and 85°C for 5 min. The qPCR reaction (20
µl/tube) contained 1.3 µl of reverse transcription
product, 10 µl of Taq-Man 2X Universal PCR Master Mix and 1
µl of the appropriate TaqMan MicroRNA Assay (20X) for
miR-34a-5p (Thermo Fisher Scientific, assay ID 000426), miR-23b-3p
(Thermo Fisher Scientific, assay ID 000400) and the spike-in
cel-miR-39 (Thermo Fisher Scientific, assay ID 000200). The PCR
reactions were incubated at 95°C for 10 min, followed by 40 cycles
at 95°C for 15 sec and 60°C for 60 sec. PCRs were performed in
triplicate using the 7500 real time PCR system (Applied
Biosystems). Due to the lack of a known miRNA reference that would
allow the normalization of circulating miRNAs, qPCR data were
reported as the average raw cycle thresholds (Ct) in both plasma
and in the different EVs types.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 6.01. Data are presented as average ± SEM. The unpaired
Student's t-test was used to determine whether the differences of
miRNA expression between PD patients and controls were significant.
Receiving operating characteristic (ROC) curve was conducted to
assess the ability of pure SEVs miR-34a-5p to distinguish PD
patients from controls. The differences in miR-34a-5p levels among
PD patients groups with different disease severity were determined
using one-way ANOVA followed by Bonferroni's test. The correlations
between the miR-34a-5p levels in pure SEVs and clinical
characteristics were assessed by Pearson's correlation test.
Differences between groups were considered significant with P-value
≤0.05.
Results
Different EV separation and
characterization
We separated different subtypes of EVs from either
control subjects or patients with Parkinson's disease via the
serial ultracentrifugation protocol (800 × g for 30 min at 4°C,
16,000 × g for 45 min at 4°C and 100,000 × g for 2 h at 4°C)
(Fig. 1A). This protocol allows
the EV separation to be dependent on their different density and
diameter. The 800 × g centrifugation step allows sedimentation of
cell debris and Large EVs (labelled LEVs), the 16,000 × g step
allows to pellet Medium EVs (labelled MEVs), while the final
100,000 × g ultracentrifugation (labelled SEVs) enriches Small EVs,
exogenous protein contaminants (EPCs) and other nanoparticles that
can co-precipitate after the last ultracentrifugation step. To
further enrich small EVs, SEV pellet was loaded on top of a
discontinuous sucrose gradient. Twelve fractions of equal volumes
(400 µl) were collected from the top after 16 h of
ultracentrifugation at 100,000 × g. For further analyses we
selected fraction from 5 to 9, designated as pure SEVs, as
described below.
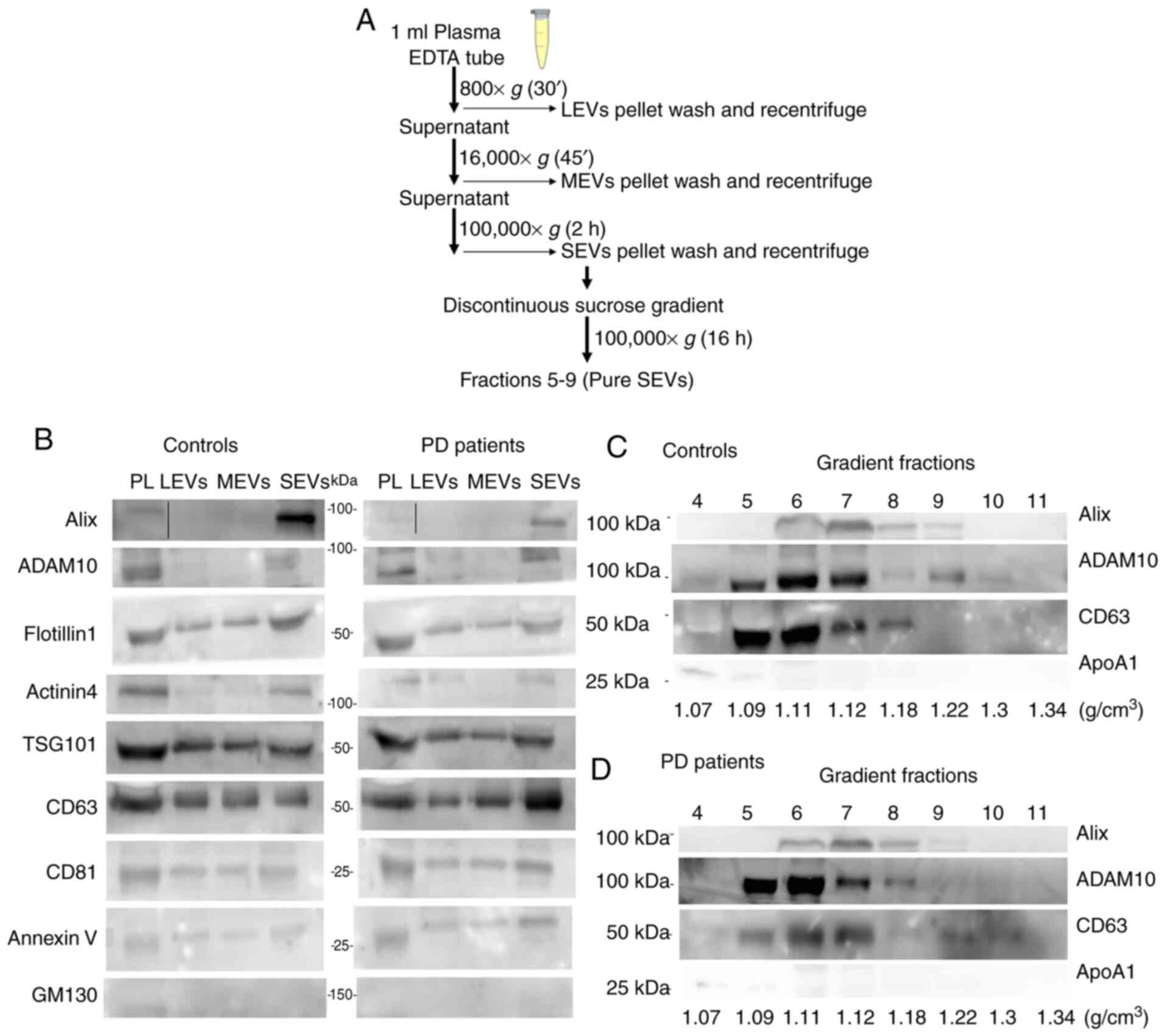 | Figure 1Biochemical characterization of EVs
by western blot analysis. (A) Scheme of the serial
ultracentrifugation protocol used to isolate the different EV
subtypes. (B) Plasma (PL, 30 µg), EVs pelleted at 800 × g
for 30 min (LEVs), EVs pelleted at 16,000 × g for 45 min (MEVs),
EVs pelleted at 100,000 × g for 2 h (SEVs) preparations (20
µl) both from controls and PD patients were loaded. Gradient
fractions from either controls (C) or PD patients (D) obtained with
discontinuous sucrose gradient. Top numbers refer to the
corresponding gradient fraction. Bottom numbers refer to sucrose
density (g/cm3) for each fraction. Samples were
electrophoresed on SDS-PAGE gel and analyzed using the antibodies
described in the figures. PL, plasma; LEVs, large extracellular
vesicles; MEVs, medium extracellular vesicles; SEVs, small
extracellular vesicles. |
Western blot (WB) analysis for different EV markers
was performed on plasma and different preparations from either
control subjects or PD patients in order to biochemically
char-acterize them (Fig. 1B-D).
Markers were chosen according to MISEV 2018 guidelines (40) and recent studies that compared the
protein composition of different EV subtypes from different cell
lines (26,27,40). In our experimental conditions and
using plasma as biological fluid we found an enrichment of Alix
(intracytoplasmatic protein, involved in the regulation of the
endosomal trafficking), ADAM10 (membrane protein belonging to the
family of matrix metalloproteinases involved in several biological
events, from the release of specific proteins or their fragments to
shedding through EVs) and Actinin-4 (major vault protein) only in
SEV preparation. Flotillin-1 (protein involved in the vesicular
trafficking), tumor susceptibility gene 101 protein (TSG101)
(component of the ESCRT-I complex, a regulator of the vesicular
trafficking process and exosome biogenesis), Annexin V (cytosolic
protein with membrane binding ability) and the tetraspanins CD63,
CD81, were detectable in all preparations, even if with different
degrees of enrichment (Fig. 1B).
The cis-Golgi marker GM130 was somewhat visible in total plasma
only, indicating an undetectable presence of intracytoplasmic
membranous components in all preparations (Fig. 1B). In addition, small EV markers
(Alix, ADAM10 and CD63) were visualized by WB in gradient fractions
from 5 to 9 (pure SEVs) (1.09-1.22 g/cm3) either in
controls (Fig. 1C) or PD patients
(Fig. 1D). The lipoprotein marker
APOA1 was detectable in fraction 4 and 5 confirming that, using our
separation protocol, most of the HDLs do not co-purify with pure
SEVs.
EV preparations were analyzed by dry atomic force
microscopy (AFM) to check the morphology and differences in size
among LEVs, MEVs, SEVs and pure SEVs. All the preparations analyzed
were composed of round-shaped objects and the size distribution for
each preparation was estimated (Fig.
2A). AFM analysis confirmed that our separation protocol
enriched EVs with different diameter distributions in both control
and PD subjects. Enriched-LEVs sample exhibited the largest
diameter distribution from 200 to 600 nm; MEVs from 50 to 400 nm;
SEVs from 30 to 200 nm, pure SEVs from 50 to 100 nm (Fig. 2B). Purity from protein
contaminants was checked with the colorimetric nanoplasmonic
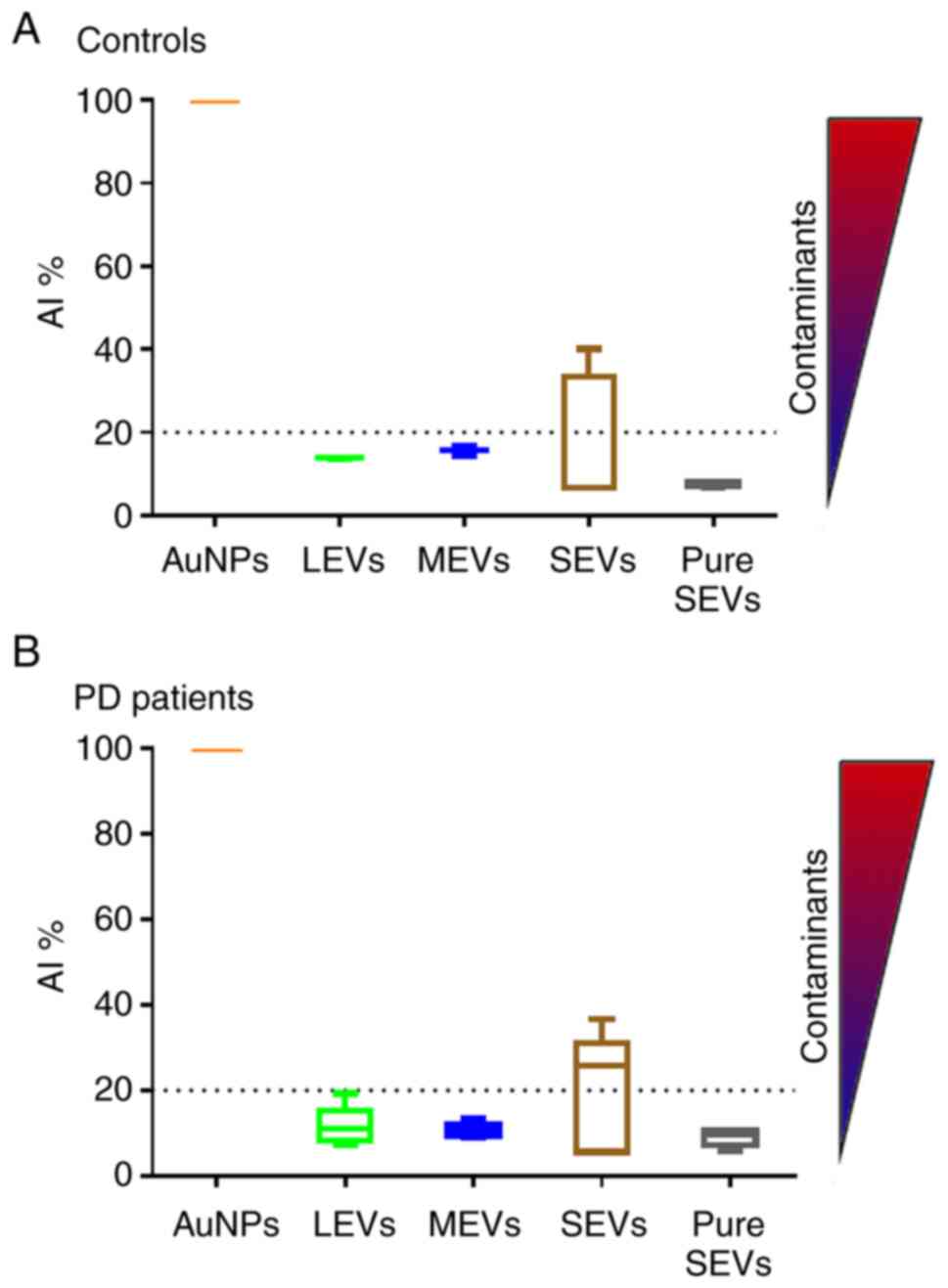
(CONAN) assay (Fig. 3). For LEVs,
MEVs and pure SEVs the mean AI values for both controls and PD
patients, resulted in lower than 20% of the reference AI (i.e., the
dispersed AuNPs solution, AI normalized at 100). This proves that
the EV formulations contained negligible amounts of EPCs (33). However, SEV fraction returned a
mean AI higher than 20%, with high standard deviation, indicating
that most of the SEV samples contain EPCs, even if in different
amounts.
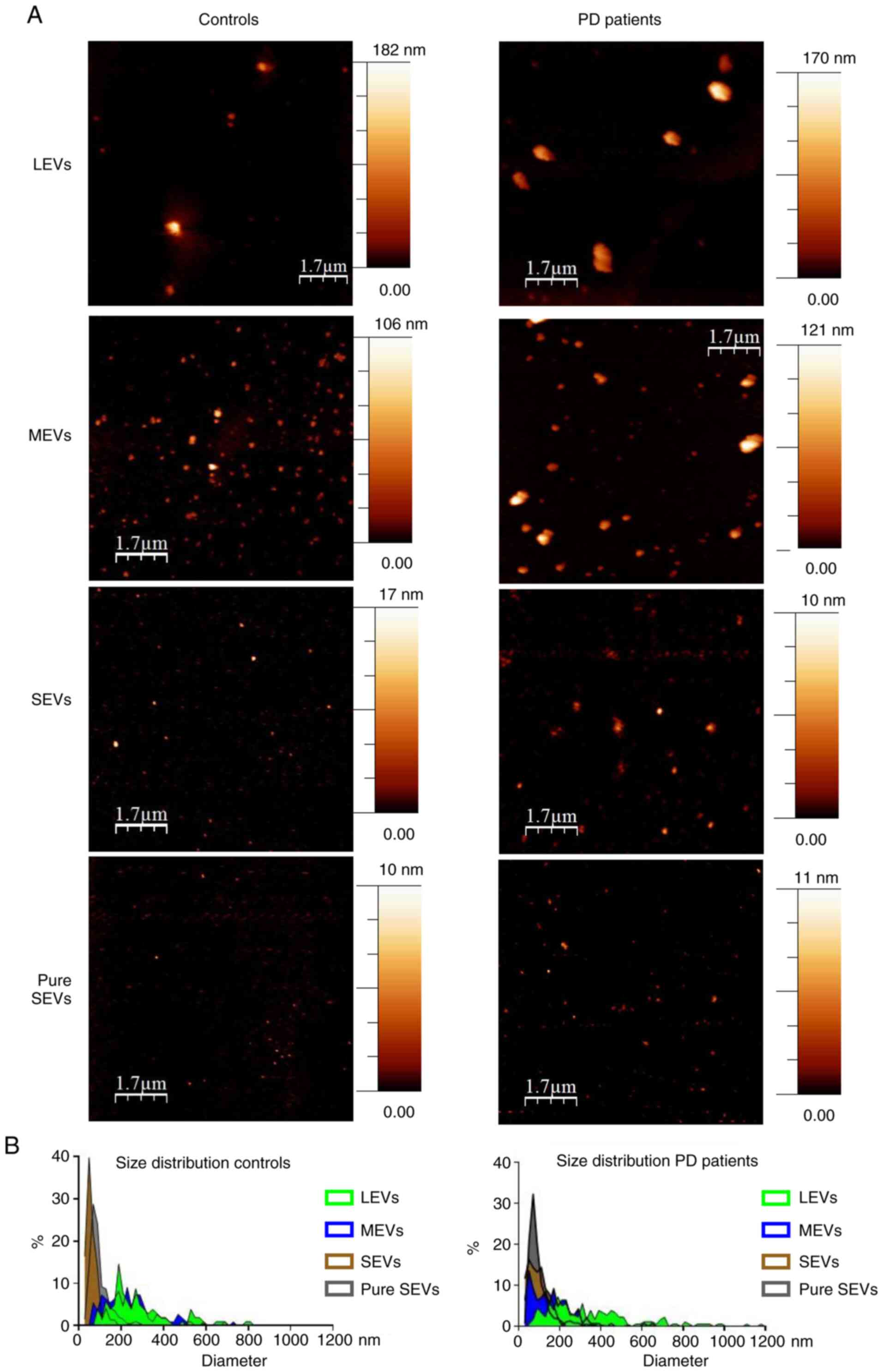 | Figure 2Imaging of different EV preparations.
(A) Atomic force microscopy (AFM) topography image of the LEVs,
MEVs, SEVs and gradient fractions positive to EV markers (from 5 to
9, pure SEVs), preparations from controls and PD patients. Samples
were adsorbed onto mica sheets, as mentioned in Materials and
methods section (scale bar is indicated in each image; colorimetric
scale, on the right of AFM images, indicates the maximum height
detected in each image). (B) Size distribution obtained from
analysis of AFM images such as in (A) A total of >500 objects
(both for controls and PD patients) were analyzed for LEVs (green),
MEVs (blue), SEVs (brown) and pure SEVs (grey) fractions. Numbers
on graphs indicate the diameter (in nm), of each preparation from
controls and PD patients respectively. LEVs, large extracellular
vesicles; MEVs, medium extracellular vesicles; SEVs, small
extracellular vesicles. |
Small RNA profiles in plasma and EV
fraction
To explore the profile of small RNA content in
plasma and in different EV subpopulations, total RNA was isolated
from 200 µl of whole plasma and from the various EV
fractions purified from 1 ml of plasma. Small RNA and miRNA yield
and miRNA percentage were detected in plasma and each fraction
using Agilent 2100 Bioanalyser with small RNA assay. As shown in
Table II, a different yield of
small RNA and miRNA was obtained for the different samples
indicating plasma as the sample with a minor content of small RNA,
including miRNA, both in controls and in PD patients. In addition,
30% or more miRNA percentage was detected in plasma samples and all
fractions from the two groups studied. Finally, the comparison
among the different EV subpopulations showed that the miRNA yield
(in ng) obtained from pure SEVs was greater than the one obtained
from LEVs, MEVs or SEVs in the two groups. These data confirm that
the small RNA cargo is different when different EV types are
compared and highlight our protocol as a good standardized method
to isolate miRNAs from the EV fractions.
 | Table IIYield of small RNA and miRNA
extracted from plasma and the different EV subpopulations derived
from plasma of either controls or PD patients. |
Table II
Yield of small RNA and miRNA
extracted from plasma and the different EV subpopulations derived
from plasma of either controls or PD patients.
| Controls
|
|---|
| Plasma | LEVs | MEVs | SEVs | Pure SEVs |
|---|
| Small RNA (ng) | 2.26±1.27 | 4.32±1.04 | 4.90±0.95 | 10.34±3.51 | 14.51±8.03 |
| miRNA (ng) | 0.82±0.49 | 1.70±0.45 | 2.12±0.26 | 4.16±1.17 | 9.87±6.72 |
| miRNA/small RNA
(%) | 29.67±5.90 | 37.07±3.45 | 39.27±1.83 | 39.43±1.82 | 51.37±12.64 |
|
| PD patients
|
| Plasma | LEVs | MEVs | SEVs | Pure SEVs |
|
| Small RNA (ng) | 8.37±4.32 | 13.15±2.15 | 16.18±3.94 | 11.76±3.94 | 19.51±4.10 |
| miRNA (ng) | 4.03±2.14 | 5.38±0.96 | 7.00±1.48 | 4.74±1.48 | 8.92±2.40 |
| miRNA/small RNA
(%) | 32.80±2.40 | 35.15±0.25 | 36.50±0.60 | 35.30±0.60 | 35.15±0.35 |
Biological rationale for the selection of
miR-34a-5p in the present study
In order to identify possible and relevant
alterations of miRNA content in plasma and plasmatic EV
subpopulations in PD, we selected miR-34a-5p due to its
well-documented involvement in neurobiology. In Table III we summarized the biological
processes modulated by miR-34a-5p in the nervous system. miR-34a-5p
plays an important role in neurodevelopment by regulating different
aspects of neurogenesis and synaptogenesis (41-44). In addition, miR-34a-5p is involved
in neuronal differentiation and brain aging by targeting key genes,
including cyclin D1, SIRT1, and Bcl-2 (45-50). Accordingly, the functional
annotation analysis performed in the current study on the target
genes of the miR-34a-5p using Database for Annotation Visualization
and Integrated Discovery (DAVID v6.8) strongly supported the
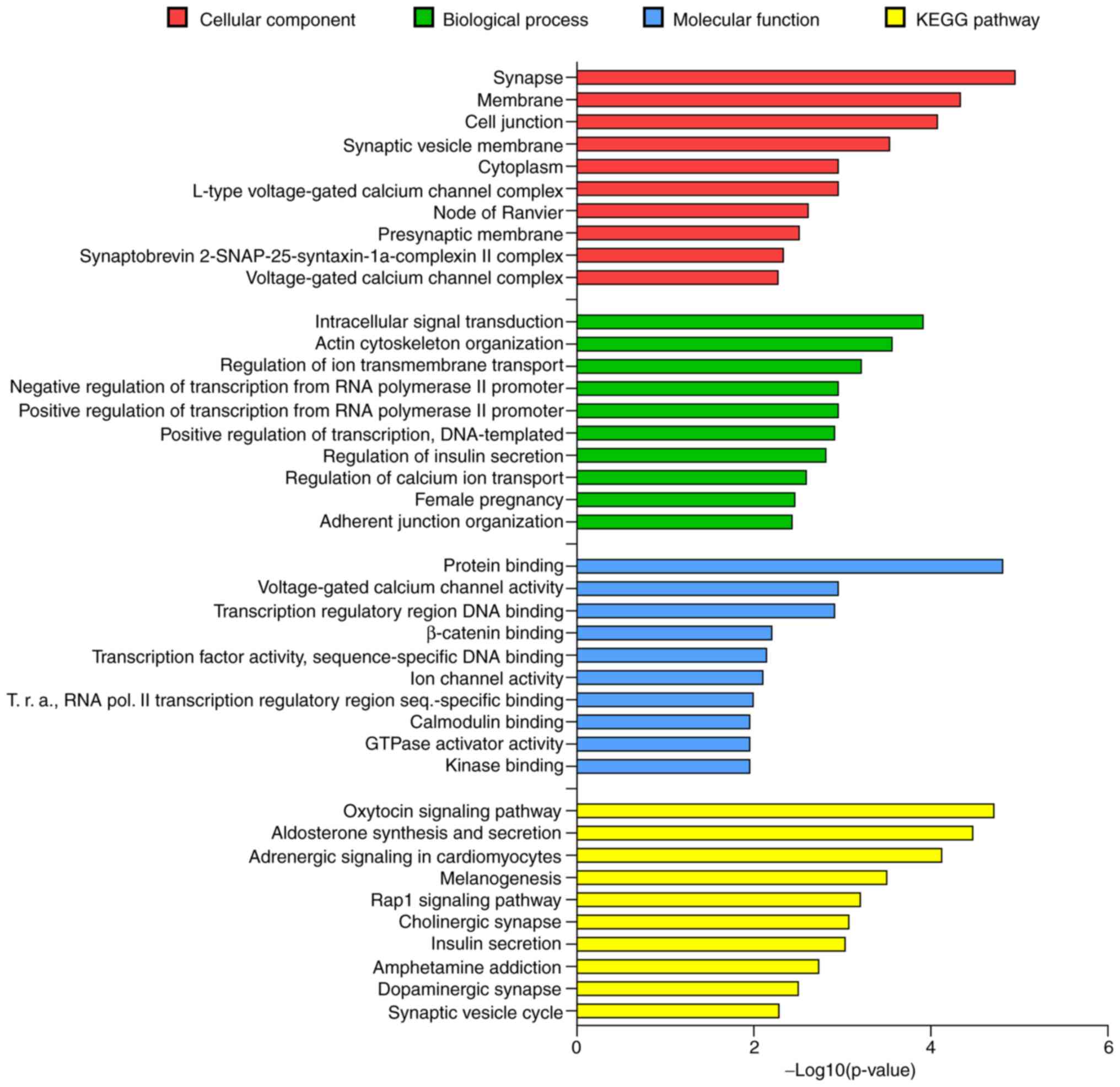
relevant role of miR-34a-5p in the nervous system (Fig. 4). In the cellular component
category, 'Synapse' was the first term with 22 targets (P<0.05)
and 'synaptic vesicle membrane', 'node of Ranvier', 'presynaptic
membrane' and 'synaptobrevin 2-SN AP-25-syntaxin-1a-complexin II
complex' were among the first 10 significant terms. For the
biological process category, 'regulation of ion transmembrane
transport' and 'regulation of calcium ion transport' were the third
and eighth biological term, respectively. In the molecular function
category, 414 predicted target genes were included in 'protein
binding' term and the other enriched terms concerned to ion
channels activity and transcription regulation. The KEGG pathway
enrichment analysis outlined 51 biological terms (P<0.05). Among
these, 'Cholinergic synapse', 'Dopaminergic synapse' and 'Synaptic
vesicle cycle' represented the sixth, ninth and tenth biological
term, respectively. The 'Cholinergic synapse' and 'Dopaminergic
synapse' terms were both significantly enriched with 14 predicted
genes and 'Synaptic vesicle cycle' with 9 predicted genes. Taken
together, these data sustained the biological evidence to select
this miRNA and to evaluate its levels in the whole plasma and its
EV fractions in PD patients.
 | Table IIIBiological processes modulated by
miR-34a-5p in the nervous system. |
Table III
Biological processes modulated by
miR-34a-5p in the nervous system.
| Neurological
process | Molecular mechanism
mediated by miR-34a-5p | Experimental
model | (Refs.) |
|---|
| Neurogenesis | Regulation of
proliferation in neural progenitors.
Modulation of morphology and electrophysiological properties in
differentiating neurons. | Rat CN,
rAAV-miR-34a-infused rat | (41) |
| Synaptogenesis | Negative regulation
of the synaptic targets SYT-1 and STX1A.
Modulation of the expression of the presynaptic and postsynaptic
neuronal markers, including SYP and SYN1.
Alteration of neurite outgrowth and spinal morphology. | Mouse CN Mouse
NSC
Human iPSC-derived neurons | (42)
(43)
(44) |
| Neuronal
differentiation | Suppression of cell
cycle entry by targeting cyclin D1 in mature neurons.
Influence of neuronal survival by the direct regulation of the
anti-apoptotic gene Bcl-2 and the protein deacetylase
SIRT-1 which has a neuroprotective activity. | Rat CN SH-SY5Y
cells, APPswe/PSE9 mouse
Mouse NSC | (45)
(46)
(47) |
| Cognitive
functions | Impairment of cued
fear memory consolidation through the modulation of Notch pathway
in the basolateral amygdala.
Alteration of learning abilities and emotionality in rats. | miR-34a
sponge-infused mouse
rAAV-miR-34a-infused rat | (48)
(41) |
| Brain aging | Modulation of
neural deterioration with age and alteration of aging-associated
transcriptional profile. Support of healthy brain aging through the
inhibition of PRC2 and the decrease of H3K27me3 that determines the
reduction of small heat shock proteins. |
miR-34-/-Drosophila | (49)
(50) |
miR-34a-5p levels are higher in pure SEVs
from PD patients compared to controls
As already mentioned, in this study we evaluated the
level of miR-34a-5p in plasma and its EV fractions of PD patients.
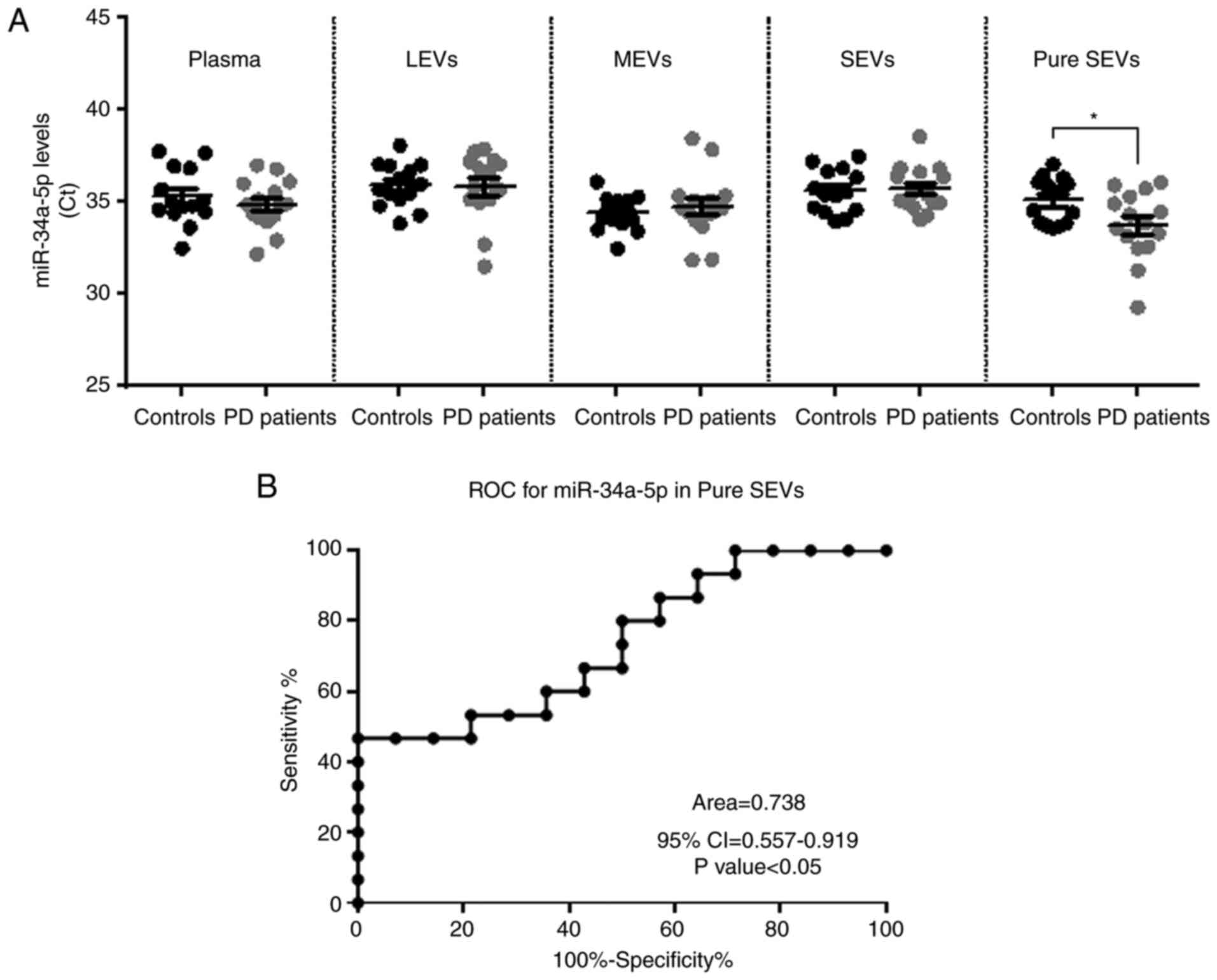
As shown in Fig. 5A, our findings
revealed that miR-34a-5p was detectable in plasma, LEVs, MEVs,
SEVs, but no differences in its levels were found between the two
groups of subjects in these biological samples. Interestingly, the
miR-34a-5p levels resulted in a significantly higher in the pure
exosomes fraction from PD patients compared to controls (Fig. 5A, CtPD average=33.74
versus CtCONTROLS average=35.07; P<0.05). As
mentioned in Materials and methods, the results of these
experiments were reported as Ct average since no endogenous miRNAs
were established as normalizers for plasma and the different EV
fractions. Thus, in order to verify the good technical performance
of RNA isolation, RT reaction and the specificity of observed
changes in miR-34a-5p, we measured in the same samples the levels
of the spike-in cel-miR-39 and has-miR-23b-3p, for which no roles
in NCS have been reported yet. The Ct mean of each miRNA was nearly
the same between PD patients and controls in plasma and in the EVs
fractions (Fig. S1). The
diagnostic significance of miR-34a-5p in pure SEVs was further
tested by ROC curve analysis. The calculated Area under ROC curve
(AUC) of 0.738 (95% CI=0.557-0.919; P<0.05) may indicate that
the levels of this miRNA in pure SEVs may discriminate controls
from PD patients with good accuracy in the examined cohort
(Fig. 5B).
Correlations between pure SEVs miR-34a-5p
levels and clinical parameters of PD patients
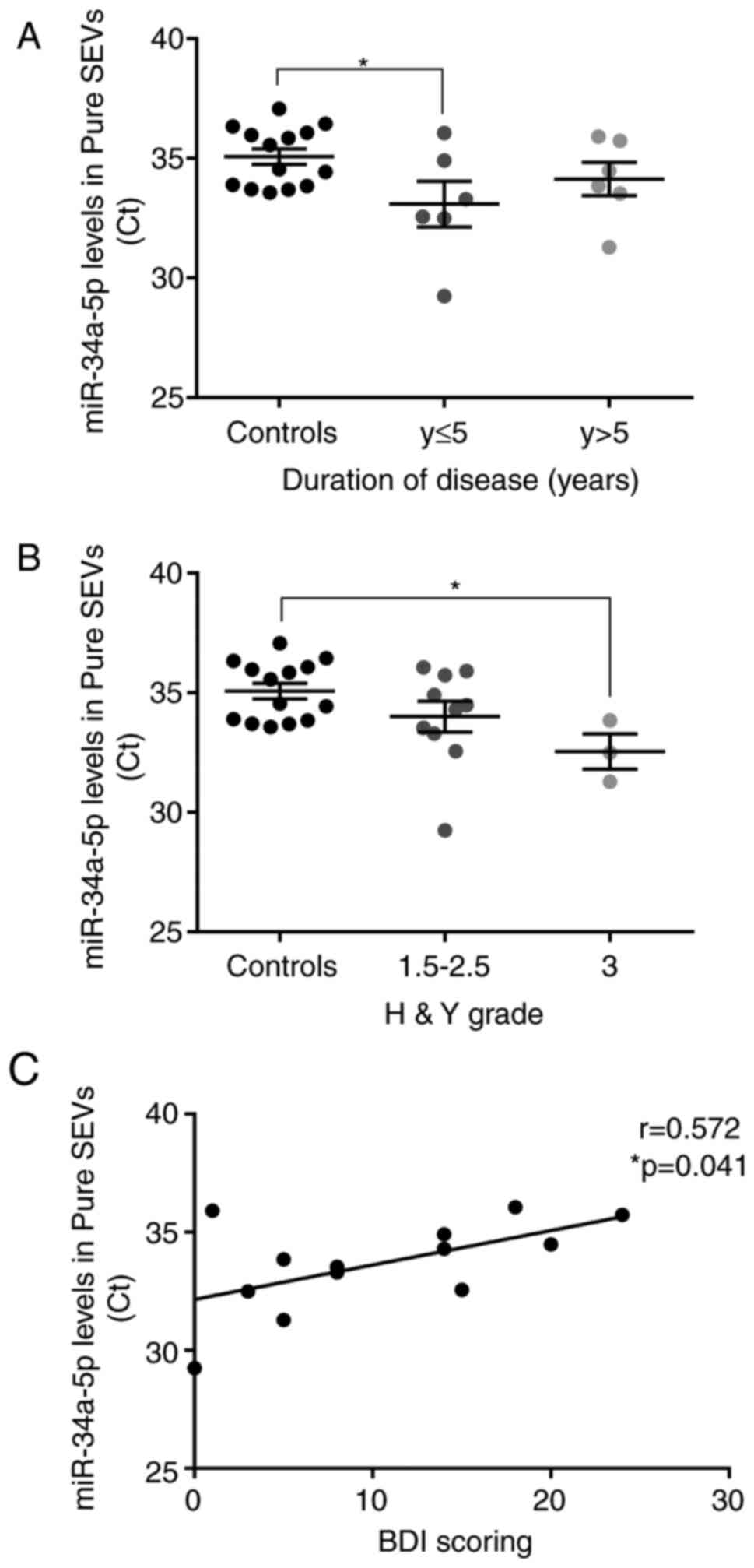
We considered and explored whether miR-34a-5p levels
detected in pure SEVs could be related to clinical parameters,
including disease duration, the scores of clinical disease severity
and depression, and levodopa dose. To carry out this analysis, the
PD patients were divided into sub-groups according to disease
duration and Hoehn&Yahr grade. The miR-34a-5p mean Ct value was
significantly lower in PD patients with shorter disease duration
(≤5 years; P<0.05), with respect to control subjects. By
considering the group of PD patients with disease duration >5
years, a trend of decrease of miR-34a-5p Ct values was found in
respect to controls (not significant) (Fig. 6A). Interestingly, PD patients with
higher Hoehn&Yahr score (H&Y score=3) showed a significant
decrease of the miR-34a-5p mean Ct value compared to control
subjects (P<0.05; Fig. 6B).
Moreover, the miR-34a-5p mean Ct value in plasmatic pure SEVs from
PD patients resulted correlated positively with Beck Depression
Inventory (BDI) score (P<0.05; Fig. 6C). However, no significant
correlation was found with age at onset, Levodopa dose, and unified
Parkinson's disease rating scale (UPDRS) in PD patients (Fig. S2).
Discussion
Circulating Extracellular Vesicles (EVs) containing
disease-specific molecular signatures (including miRNAs) have
recently emerged as promising biomarkers in neurode-generative
diseases. Circulating microRNAs encapsulated in EVs remain stable
in body fluids and are easily detected using standard molecular
techniques, such as RT-qPCR, allowing the identification of
specific circulating miRNAs that are differentially expressed
between patients and control subjects (51).
For Parkinson's disease very few data have been
reported concerning the detection of miRNAs within EVs and their
potential role as biomarkers (52,53). Moreover, no study has taken into
consideration the miRNAs content in all the different vesicle types
isolated from plasma/serum or cerebrospinal fluid of PD patients.
To the best of our knowledge, this is the first study that used
differential centrifugation followed by density gradient to isolate
all the EVs fractions from the plasma of PD patients, including
LEVs, MEVs, SEVs and pure SEVs. Regarding the RNA content of EVs,
it has been known that different RNA profiles are detectable in the
different subpopulations of EVs. The collective data from the few
published studies on this topic indicate that the exosomes released
by different cell lines carry a significant amount of small RNAs
respect to apoptotic bodies and microvesicles. On the other hand,
these fractions contain ribosomal RNA (displayed as two peaks
corresponding to subunits 18S and 28S in RNA profiles) that is
totally absent in exosomes (54,55). In agreement with this, in the
present study, the small RNA profiles obtained from plasmatic EVs
of controls and PD subjects showed that the pure SEVs contained
relatively more miRNAs than the other fractions tested in this
study. In addition, the miRNAs yield in pure SEVs was similar
between controls and patients groups and it could be considered a
very appreciable quantity because it was higher than the range
value of 1-10 ng/ml plasma of exosomal RNA yield obtained using
spin column method (56), but
also the mean yield reported by Lunavat et al for miRNAs
from plasmatic exosomes isolated using ultracen-trifugation or
commercial kits (55). The
results shown in the present study proved that the experimental
workflow used is a reliable method to isolate EV-associated miRNAs,
i.e., the SEV miRNAs, from plasma also supporting its potential use
for the preparation of the small non-coding RNA samples for the
discovery of biomarkers of a given disease.
In the current study, the levels of circulating
miR-34a-5p were evaluated to identify possible alterations between
plasma and plasmatic EV subpopulations in PD patients and between
patients and control subjects. In this context, various strategies
have been used to select the miRNAs to be studied as circulating
molecules in PD patients (57).
In the current study, the selection of miR-34a-5p was based on its
fundamental role in the regulation of neuronal gene expression and
its important functions in the central nervous system, as
summarized in Table III. In
addition, our data derived from the functional annotation analysis
of miR-34a-5p predicting target genes strongly supported the
important functions of this miRNA in pathways and biological
structures clearly associated to the nervous system. It is known
that miR-34a-5p is abundantly expressed in the adult brain as
indicated by miRNAs expression profiling studies in rodents
(58,59). and its expression level in neurons
needs to be finely regulated to allow proper neuronal development
and aging.
Regarding the role of miR-34a-5p in neuropathology,
several in vitro and in vivo studies have linked
miR-34a-5p to neurodegenerative diseases. Expression of miR-34a-5p
was elevated in the hippocampus and temporal cortex of 3×Tg AD
mice, a model of familial Alzheimer's disease (AD) (60). Moreover in the human brain,
miR-34a-5p was upregulated in the hippocampal region of AD patients
compared to controls (61).
Regarding PD, no evidence has revealed any dysregulation of
miR-34a-5p in the human brain thus far (62). However, some studies have reported
the over-expression of miR-34a-5p both in in vitro and in
vivo models of PD, such as differentiated PC12 cells treated
with the dopaminergic neurotoxin MPP+ and rats
chronically exposed to rotenone (63-65), respectively. In addition, the
downregulation of miR-34a-5p induced by lithium or the use of
anti-miR molecules improved cell survival and showed
neuroprotective effects in neuronal cells treated with
PD-associated neurotoxins (paraquat and rotenone) (66,67).
In the current study, we showed that miR-34a-5p can
be easily detected both in plasma, such as miRNA vesicle-free
primarily, and in plasmatic EVs from control subjects and PD
patients suggesting that this miRNA is secreted by cells and
remains stable in circulation. In light of this, Cosín-Tomás et
al reported that miR-34a-5p was downregulated in plasma of AD
patients, but not in that of PD patients (68). Furthermore, the miR-34a-5p ectopic
overexpression in primary cortical neurons revealed the secretion
of miR-34a-5p containing exosomes that were taken up by neighboring
cells as demonstrated using co-culture experiments (60). Mao et al showed that EVs
from astrocytes contain increased miR-34a-5p after the stimulation
of the stress condition induced by Lipopolysaccharide (LPS)
(69). Of note, the secreted
miR-34a-5p in astrocytic EVs enhanced the vulnerability of SH-SY5Y
cells to neurotoxins exposure by inhibiting Bcl-2 expression.
Blocking the upregulation of miR-34a-5p in LPS-stimulated astrocyte
EVs alleviated the loss of dopaminergic neurons in vitro and
in vivo (69). All these
data seem to suggest that cell stress conditions or neurotoxic
exposures increase the intracellular miR-34a-5p in specific brain
regions. Moreover, the altered miR-34a-5p level can be delivered in
EVs and then modulate some significant biological functions in the
receiving cell in disease.
In the present study, for the first time, we found
that miR-34a-5p levels were significantly overexpressed in pure
SEVs from plasma of PD patients compared to controls and that its
expression in pure SEVs revealed a good ability of this miRNA to
distinguish PD patients from control subjects suggesting its
potential consideration as a marker of diagnosis at molecular
level. Moreover, pure SEVs miR-34a-5p levels were higher in PD
patients even at the beginning stage of PD when the disease
duration is less than 5 years. Finally, high levels of pure SEV
miR-34a-5p were detected in PD patients with mild/progressive
symptoms of disease and were associated with minimal/absent
depression. In our opinion, the dysregulation of circulating
miR-34a-5p depending on neuro-pathological conditions, such as
depression, is not unexpected. Indeed, elevated levels of
miR-34a-5p were found in serum of patients with major depressive
disorders and in post-mortem cerebellar tissues and iPSC-derived
neurons of bipolar disorder patients (44,70). However, further studies are needed
to clarify whether the analysis of circulating miR-34a-5p in the
different biological samples may be useful for the identification
of depressive states.
In conclusion, our data showed the differential
expression between PD patients and control subjects of miR-34a-5p
in plasmatic pure SEVs, but not in the other EV fractions,
suggesting the necessity to consider not only the whole plasma, but
each EV subpopulation in order to improve the possibility to
identify relevant differences of specific miRNAs levels. For this
purpose, the SEVs purification protocol is crucial since the
presence of protein contaminants in the sample seems to conceal
those differences (21). We have
clearly defined this study as an exploratory one and further
investigations are necessary to verify the potential diagnostic
value of circulating miR-34a-5p in PD. Since our findings derive
from an exploratory study, they trace the starting point for future
research. Indeed, they address the direct use of pure SEVs: i) To
detect the miR-34a-5p levels in larger cohorts that also include
female PD patients allowing the reduction of the number of samples
to be analysed; ii) To perform RNA-seq experiments in order to
identify novel potential biomarkers among the entire repertoire of
circulating miRNAs in PD patients. Finally, we think that the
exploration on the origin of pure SEVs [i.e., from brain: neuron-,
astrocyte- or oligodendrocyte-derived exosomes (71)], could be useful for the
comprehension of the role of secreted miR-34a-5p in PD.
Supplementary Data
Abbreviations:
|
PD
|
Parkinson's disease
|
|
miRNAs
|
microRNAs
|
|
EVs
|
extracellular vesicles
|
|
CSF
|
cerebrospinal fluid
|
|
ISEV
|
International Society for
Extracellular Vesicles
|
|
LEVs
|
large extracellular vesicles
|
|
MEVs
|
medium extracellular vesicles
|
|
SEVs
|
small extracellular vesicles
|
|
EPCs
|
exogenous protein contaminants
|
|
MISEV
|
minimal information for studies of
extracellular vesicles
|
|
WB
|
western blotting
|
|
ESCRT
|
Endosomal Sorting Complexes Required
for Transport
|
|
AFM
|
atomic force microscopy
|
|
CONAN
|
COlorimetric NANoplasmotic assay
|
|
NCS
|
nervous central system
|
|
AUC
|
area under curve
|
|
BDI
|
Beck depression inventory
|
|
UPDRS
|
unified Parkinson disease rating
scale
|
Funding
This work was supported by a BIOMANE grant
(University of Brescia, Brescia, Italy) to MP, GDP, AP and AM; by
FFRB grants (Italian Ministry of University and Research) to AS and
AR; by research local funds (University of Brescia) to GDP and AS;
by a CIB grant (Biotechnology Inter-university Consortium) to
GDP.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GDP conceived this study. GDP, AS, AR, IG, LP
designed the experimental workflow. APad, APil; and AM provided the
clinical samples and the clinical characteristics of the enrolled
subjects. VP performed the plasma-pre-analytical processing. LP and
AR performed the EV characterization experiments of WB, AFM imaging
and CONAN assay. IG performed the RNA isolation from plasma and
each EV subpopulation, the qPCR experiments and the statistical
analyses. AS performed the Small RNA assay using the Agilent 2100
Bioanalyser. IG and LP wrote the manuscript. All co-authors, IG,
AR, LP, VP, APad, APil, AM, ABel, ABar, MP, AS and GDP,
collaborated in the discussion of the data and in the critical
revision of the manuscript. IG, AR, AS and GDP drafted the final
version of the manuscript. All co-authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The study was approved by the ethics committee of
Spedali Civili di Brescia on 8th June 2016. Informed consent was
obtained from all the subjects enrolled in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing of interest.
Acknowledgments
We would like to thank Dr M. Crosatti (University of
Leicester; UK) for the linguistic revision of the manuscript.
References
|
1
|
Tysnes OB and Storstein A: Epidemiology of
Parkinson's disease. J Neural Transm (Vienna). 12:901–905. 2017.
View Article : Google Scholar
|
|
2
|
Spillantini MG and Goedert M:
Neurodegeneration and the ordered assembly of α-synuclein. Cell
Tissue Res. 373:137–148. 2018. View Article : Google Scholar
|
|
3
|
Bellucci A, Mercuri NB, Venneri A,
Faustini G, Longhena F, Pizzi M, Missale C and Spano P: parkinson's
disease: From synaptic loss to connectome dysfunction. Neuropathol
Appl Neurobiol. 42:77–94. 2016. View Article : Google Scholar
|
|
4
|
Thompson AG, Gray E, Heman-Ackah SM, Mäger
I, Talbot K, El Andaloussi S, Wood MJ and Turner MR: Extracellular
vesicles in neurodegenerative disease-pathogenesis to biomarkers.
Nat Rev Neurol. 12:346–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tofaris GK: A critical assessment of
exosomes in the pathogenesis and stratification of parkinson's
disease. J Parkinsons Dis. 7:569–579. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paolini L, Federici S, Consoli G, Arceri
D, Radeghieri A, Alessandri I and Bergese P: Fourier-Transform
infrared (FT-IR) spectroscopy fingerprints subpopulations of
extracellular vesicles of different sizes and cellular origin. J
Extracell Vesicles. 9:17411742020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomlinson PR, Zheng Y, Fischer R, Heidasch
R, Gardiner C, Evetts S, Hu M, Wade-Martins R, Turner MR, Morris J,
et al: Identification of distinct circulating exosomes in
parkinson's disease. Ann Clin Transl Neurol. 2:353–361. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gui YX, Liu H, Zhang LS, Lv W and Hu XY:
Altered microRNA profiles in cerebrospinal fluid exosome in
parkinson disease and alzheimer disease. Oncotarget.
10:37043–37053. 2015. View Article : Google Scholar
|
|
9
|
Cao XY, Lu JM, Zhao ZQ, Li MC, Lu T, An XS
and Xue LJ: MicroRNA biomarkers of Parkinson's disease in serum
exosome-like microvesicles. Neurosci Lett. 644:94–99. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao YF, Qu MW, Li GC, Zhang FB and Rui HC:
Circulating exosomal miRNAs as diagnostic biomarkers in parkinson's
disease. Eur Rev Med Pharmacol Sci. 22:5278–5283. 2018.PubMed/NCBI
|
|
11
|
Mateescu B, Kowal EJ, van Balkom BW,
Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow
FW, Das S, et al: Obstacles and opportunities in the functional
analysis of extracellular vesicle RNA-an ISEV position paper. J
Extracell vesicles. 6:12860952017. View Article : Google Scholar
|
|
12
|
Chua CE and Tan BL: MiR-34a in
neurophysiology and neuropathology. J Mol Neurosci. 67:235–246.
2019. View Article : Google Scholar
|
|
13
|
van den Berg MM, Krauskopf J, Ramaekers
JG, Kleinjans JC, Prickaerts J and Briedé JJ: Circulating microRNAs
as potential biomarkers for psychiatric and neurodegenerative
disorders. Prog Neurobiol. 185:1017322020. View Article : Google Scholar
|
|
14
|
Goetz CG, Fahn S, Martinez-Martin P, Poewe
W, Sampaio C, Stebbins GT, Stern MB, Tilley BC, Dodel R, Dubois B,
et al: Movement disorder society-sponsored revision of the unified
parkinson's disease rating scale (MDS-UPDRS): Process, format, and
clinimetric testing plan. Mov Disord. 22:41–47. 2007. View Article : Google Scholar
|
|
15
|
Goetz CG, Poewe W, Rascol O, Sampaio C,
Stebbins GT, Counsell C, Giladi N, Holloway RG, Moore CG, Wenning
GK, et al: Movement disorder society task force report on the hoehn
and yahr staging scale: Status and recommendations. Mov Disord.
19:1020–1028. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Beck AT, Ward CH, Mendelson M, Mock J and
Erbaugh J: An inventory for measuring depression. Arch Gen
Psychiatry. 4:561–571. 1961. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Witwer KW, Buzás EI, Bemis LT, Bora A,
Lässer C, Lötvall J, Nolte-'t Hoen EN, Piper MG, Sivaraman S, Skog
J, et al: Standardization of sample collection, isolation and
analysis methods in extracellular vesicle research. J Extracell
Vesicles. 2:272013. View Article : Google Scholar
|
|
18
|
Coumans FA, Brisson AR, Buzas EI,
Dignat-George F, Drees EE, El-Andaloussi S, Emanueli C, Gasecka A,
Hendrix A, Hill AF, et al: Methodological guidelines to study
extracellular vesicles. Circ Res. 120:1632–1648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lacroix R, Judicone C, Mooberry M,
Boucekine M, Key NS and Dignat-George F; The ISTH SSC Workshop:
Standardization of pre-analytical variables in plasma microparticle
determination: Results of the international society on thrombosis
and haemostasis SSC collaborative workshop. J Thromb Haemost.
11:11902013. View Article : Google Scholar
|
|
20
|
Van Deun J, Mestdagh P, Agostinis P, Akay
Ö, Anand S, Anckaert J, Martinez ZA, Baetens T, Beghein E, Bertier
L, et al: EV-TRACK: Transparent reporting and centralizing
knowledge in extracellular vesicle research. Nat Methods.
28:228–232. 2017. View Article : Google Scholar
|
|
21
|
Paolini L, Zendrini A, Di Noto G, Busatto
S, Lottini E, Radeghieri A, Dossi A, Caneschi A, Ricotta D and
Bergese P: Residual matrix from different separation techniques
impacts exosome biological activity. Sci Rep. 6:235502016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Di Noto G, Chiarini M, Paolini LL,
Mazzoldi EL, Giustini V, Radeghieri A, Caimi L and Ricotta D:
Immunoglobulin free light chains and GAGs mediate multiple myeloma
extracellular vesicles uptake and secondary NfκB nuclear
translocation. Front Immunol. 5:5172014. View Article : Google Scholar
|
|
23
|
Di Noto G, Paolini L, Zendrini A,
Radeghieri A, Caimi L and Ricotta D: C-Src enriched serum
microvesicles are generated in malignant plasma cell dyscrasia.
PLoS One. 8:e708112013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paolini L, Radeghieri A, Civini S, Caimi L
and Ricotta D: The epsilon hinge-ear region regulates membrane
localization of the AP-4 complex. Traffic. 12:1604–1619. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alvisi G, Paolini L, Contarini A, Zambarda
C, Di Antonio V, Colosini A, Mercandelli N, Timmoneri M, Palù G,
Caimi L, et al: Intersectin goes nuclear: Secret life of an
endocytic protein. Biochem J. 475:1455–1472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jeppesen DK, Fenix AM, Franklin JL,
Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q,
Evans R, et al: Reassessment of exosome composition. Cell.
177:428–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kowal J, Arras G, Colombo M, Jouve M,
Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M and Théry
C: Proteomic comparison defines novel markers to characterize
heterogeneous populations of extracellular vesicle subtypes. Proc
Natl Acad Sci USA. 113:E968–E977. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Radeghieri A, Savio G, Zendrini A, Di Noto
G, Salvi A, Bergese P and Piovani G: Cultured human amniocytes
express hTERT, which is distributed between nucleus and cytoplasm
and is secreted in extracellular vesicles. Biochem Biophys Res
Commun. 483:706–711. 2017. View Article : Google Scholar
|
|
29
|
Berardocco M, Radeghieri A, Busatto S,
Gallorini M, Raggi C, Gissi C, D'Agnano I, Bergese P, Felsani A and
Berardi AC: RNA-Seq reveals distinctive RNA profiles of small
extracellular vesicles from different human liver cancer cell
lines. Oncotarget. 8:82920–82939. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vescovi R, Monti M, Moratto D, Paolini L,
Consoli F, Benerini L, Melocchi L, Calza S, Chiudinelli M, Rossi G,
et al: Collapse of the plasmacytoid dendritic cell compartment in
advanced cuta-neous melanomas by components of the tumor cell
secretome. Cancer Immunol Res. 7:12–28. 2019. View Article : Google Scholar
|
|
31
|
Horcas I, Fernández R, Gómez-Rodríguez JM,
Colchero J, Gómez-Herrero J and Baro AM: WSXM: A software for
scanning probe microscopy and a tool for nanotechnology. Rev Sci
Instrum. 78:0137052007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maiolo D, Paolini L, Di Noto G, Zendrini
A, Berti D, Bergese P and Ricotta D: Colorimetric nanoplasmonic
assay to determine purity and titrate extracellular vesicles. Anal
Chem. 87:4168–4176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zendrini A, Paolini L, Busatto S,
Radeghieri A, Romano M, Wauben MH, van Herwijnen MJ, Nejsum P,
Borup A, Ridolfi A, et al: Augmented COlorimetric NANoplasmonic
(CONAN) method for grading purity and determine concentration of EV
microliter volume solutions. Front Bioeng Biotechnol. 7:4522019.
View Article : Google Scholar
|
|
34
|
Salvi A, Vezzoli M, Busatto S, Paolini L,
Faranda T, Abeni E, Caracausi M, Antonaros F, Piovesan A, Locatelli
C, et al: Analysis of a nanoparticle-enriched fraction of plasma
reveals miRNA candidates for down syndrome pathogenesis. Int J Mol
Med. 43:2303–2318. 2019.PubMed/NCBI
|
|
35
|
Sticht C, De La Torre C, Parveen A and
Gretz N: Mirwalk: An online resource for prediction of microrna
binding sites. PLoS One. 18:e02062392018. View Article : Google Scholar
|
|
36
|
Dennis G, Sherman BT, Hosack DA, Yang J,
Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Grossi I, Salvi A, Abeni E, Marchina E and
De Petro G: Biological function of MicroRNA193a-3p in health and
disease. Int J Genomics. 2017:59131952017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Faranda T, Grossi I, Manganelli M,
Marchina E, Baiocchi G, Portolani N, Crosatti M, De Petro G and
Salvi A: Differential expression profiling of long non-coding RNA
GAS5 and miR-126-3p in human cancer cells in response to sorafenib.
Sci Rep. 9:91182019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Grossi I, Arici B, Portolani N, De Petro G
and Salvi A: Clinical and biological significance of miR-23b and
miR-193a in human hepatocellular carcinoma. Oncotarget.
8:6955–6969. 2017. View Article : Google Scholar :
|
|
40
|
Théry C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extra-cellular vesicles 2018 (MISEV2018): A position statement of
the international society for extracellular vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar
|
|
41
|
Mollinari C, Racaniello M, Berry A, Pieri
M, De Stefano MC, Cardinale A, Zona C, Cirulli F, Garaci E and
Merlo D: MiR-34a regulates cell proliferation, morphology and
function of newborn neurons resulting in improved behavioural
outcomes. Cell Death Dis. 6:e16222015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Agostini M, Tucci P, Steinert JR,
Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW,
Ventura A, Concepcion CP, et al: microRNA-34a regulates neurite
outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA.
108:21099–21104. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Morgado AL, Xavier JM, Dionísio PA,
Ribeiro MF, Dias RB, Sebastião AM, Solá S and Rodrigues CM:
MicroRNA-34a modulates neural stem cell differentiation by
regulating expression of synaptic and autophagic proteins. Mol
Neurobiol. 51:1168–11183. 2015. View Article : Google Scholar
|
|
44
|
Bavamian S, Mellios N, Lalonde J, Fass DM,
Wang J, Sheridan SD, Madison JM, Zhou F, Rueckert EH, Barker D, et
al: Dysregulation of miR-34a links neuronal development to genetic
risk factors for bipolar disorder. Mol Psychiatry. 20:573–584.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Modi PK, Jaiswal S and Sharma P:
Regulation of neuronal cell cycle and apoptosis by MicroRNA 34a.
Mol Cell Biol. 36:84–94. 2016.
|
|
46
|
Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X,
Huang L, Liu Y, Zhang L and Qin C: MiR-34a, a microRNA up-regulated
in a double transgenic mouse model of alzheimer's disease, inhibits
bcl2 translation. Brain Res Bull. 28:268–273. 2009. View Article : Google Scholar
|
|
47
|
Aranha MM, Santos DM, Solá S, Steer CJ and
Rodrigues MP: MiR-34a regulates mouse neural stem cell
differentiation. PLoS One. 6:e213962011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dias BG, Goodman JV, Ahluwalia R, Easton
AE, Andero R and Ressler KJ: Amygdala-Dependent Fear Memory
Consolidation via miR-34a and Notch Signaling. Neuron. 20:906–918.
2014. View Article : Google Scholar
|
|
49
|
Liu N, Landreh M, Cao K, Abe M, Hendriks
GJ, Kennerdell JR, Zhu Y, Wang LS and Bonini NM: The microRNA
miR-34 modulates ageing and neurodegeneration in drosophila.
Nature. 15:519–523. 2012. View Article : Google Scholar
|
|
50
|
Kennerdell JR, Liu N and Bonini NM: MiR-34
inhibits poly-comb repressive complex 2 to modulate chaperone
expression and promote healthy brain aging. Nat Commun.
10:41882018. View Article : Google Scholar
|
|
51
|
Cheng L, Sharples RA, Scicluna BJ and Hill
AF: Exosomes provide a protective and enriched source of miRNA for
biomarker profiling compared to intracellular and cell-free blood.
J Extracell Vesicles. 3:262014. View Article : Google Scholar
|
|
52
|
Leggio L, Vivarelli S, L'Episcopo F,
Tirolo C, Caniglia S, Testa N, Marchetti B and Iraci N: MicroRNAs
in parkinson's disease: From pathogenesis to novel diagnostic and
therapeutic approaches. Int J Mol Sci. 18:26982017. View Article : Google Scholar
|
|
53
|
Wang L and Zhang L: Circulating exosomal
miRNA as diag-nostic biomarkers of neurodegenerative diseases.
Front Mol Neurosci. 13:532020. View Article : Google Scholar
|
|
54
|
Crescitelli R, Lässer C, Szabó TG, Kittel
A, Eldh M, Dianzani I, Buzás EI and Lötvall J: Distinct RNA
profiles in subpopulations of extracellular vesicles: Apoptotic
bodies, microvesicles and exosomes. J Extracell vesicles. 2:122013.
View Article : Google Scholar
|
|
55
|
Lunavat TR, Cheng L, Kim DK, Bhadury J,
Jang SC, Lässer C, Sharples RA, López MD, Nilsson J, Gho YS, et al:
Small RNA deep sequencing discriminates subsets of extracellular
vesicles released by melanoma cells-Evidence of unique microRNA
cargos. RNA Biol. 12:810–823. 2015. View Article : Google Scholar :
|
|
56
|
Enderle D, Spiel A, Coticchia CM, Berghoff
E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM,
Lader E, Skog J and Noerholm M: Characterization of RNA from
exosomes and other extracellular vesicles isolated by a novel spin
column-based method. PLoS One. 10:e01361332015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ravanidis S, Bougea A, Papagiannakis N,
Maniati M, Koros C, Simitsi A, Bozi M, Pachi I, Stamelou M,
Paraskevas GP, et al: Circulating brain-enriched MicroRNAs for
detection and discrimination of idiopathic and genetic parkinson's
disease. Mov Disord. 35:457–467. 2020. View Article : Google Scholar
|
|
58
|
Bak M, Silahtaroglu A, Møller M,
Christensen M, Rath MF, Skryabin B, Tommerup N and Kauppinen S:
MicroRNA expression in the adult mouse central nervous system. RNA.
14:432–444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jauhari A, Singh T, Singh P, Parmar D and
Yadav S: Regulation of miR-34 family in neuronal development. Mol
Neurobiol. 55:936–945. 2018. View Article : Google Scholar
|
|
60
|
Sarkar S, Jun S, Rellick S, Quintana DD,
Cavendish JZ and Simpkins JW: Expression of microRNA-34a in
alzheimer's disease brain targets genes linked to synaptic
plasticity, energy metabolism, and resting state network activity.
Brain Res. 1646:139–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Agostini M, Tucci P, Killick R, Candi E,
Sayan BS, di Val Cervo PR, Nicoterad P, McKeon F, Knight RA, Mak TW
and Melino G: Neuronal differentiation by TAp73 is mediated by
microRNA-34a regulation of synaptic protein targets. Proc Natl Acad
Sci USA. 108:21093–21098. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Briggs CE, Wang Y, Kong B, Woo TU, Iyer LK
and Sonntag KC: Midbrain dopamine neurons in parkinson's disease
exhibit a dysregulated miRNA and target-gene network. Brain Res.
1618:111–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Delavar RM, Baghi M, Safaeinejad Z,
Kiani-Esfahani A, Ghaedi K and Nasr-Esfahani MH: Differential
expression of miR-34a, miR-141, and miR-9 in
MPP+-treated differentiated PC12 cells as a model of
parkinson's disease. Gene. 662:54–65. 2018. View Article : Google Scholar
|
|
64
|
Horst CH, Schlemmer F, de Aguiar
Montenegro N, Domingues AC, Ferreira GG, da Silva Ribeiro CY, de
Andrade RR, Del Bel Guimarães E, Titze-de-Almeida SS and
Titze-de-Almeida R: Signature of aberrantly expressed microRNAs in
the striatum of rotenone-induced parkinsonian rats. Neurochem Res.
43:2132–2140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ba Q, Cui C, Wen L, Feng S, Zhou J and
Yang K: Schisandrin B shows neuroprotective effect in
6-OHDA-induced parkinson's disease via inhibiting the negative
modulation of miR-34a on Nrf2 pathway. Biomed Pharmacother.
75:165–172. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Alural B, Ozerdem A, Allmer J, Genc K and
Genc S: Lithium protects against paraquat neurotoxicity by NRF2
activation and miR-34a inhibition in SH-SY5Y cells. Front Cell
Neurosci. 9:2092015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Horst CH, Titze-De-Almeida R and
Titze-De-Almeida SS: The involvement of eag1 potassium channels and
miR-34a in rotenone-induced death of dopaminergic SH-SY5Y cells.
Mol Med Rep. 15:1479–1488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cosín-Tomás M, Antonell A, Lladó A,
Alcolea D, Fortea J, Ezquerra M, Lleó A, Martí MJ, Pallàs M,
Sanchez-Valle R, et al: Plasma miR-34a-5p and miR-545-3p as early
biomarkers of alzheimer's disease: Potential and limitations. Mol
Neurobiol. 54:5550–5562. 2017. View Article : Google Scholar
|
|
69
|
Mao S, Sun Q, Xiao H, Zhang C and Li L:
Secreted miR-34a in astrocytic shedding vesicles enhanced the
vulnerability of dopaminergic neurons to neurotoxins by targeting
Bcl-2. Protein Cell. 6:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wan Y, Liu Y, Wang X, Wu J, Liu K, Zhou J,
Liu L and Zhang C: Identification of differential microRNAs in
cerebrospinal fluid and serum of patients with major depressive
disorder. PLoS One. 10:e01219752015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ohmichi T, Mitsuhashi M, Tatebe H, Kasai
T, El-Agnaf OA and Tokuda T: Quantification of brain-derived
extracellular vesicles in plasma as a biomarker to diagnose
parkinson's and related diseases. Park Relat Disord. 61:82–87.
2019. View Article : Google Scholar
|