1. Introduction
Tumor immunotherapy has shown great application
prospects by stimulating the body autoimmune system to balance the
immunosuppressive microenvironment, thereby improving the antitumor
effect (1). Immunotherapy
includes adoptive cell immunotherapy, immune checkpoint inhibitors,
cancer vaccines, costimulatory receptor agonists, monoclonal
anti-bodies (mAbs), and oncolytic virus therapy (2). In recent years, research on immune
checkpoint blockade has become a hotspot study in tumor
immunotherapy (3). Immune
checkpoint molecules are receptors on the surface of immune cells
that, after binding with their ligand, transduce inhibitory signals
or stimulatory signals (4). Drugs
that target immune checkpoint molecules, which can transduce the
inhibitory signals, are called checkpoint inhibitors. Among these
receptors, the most studied ones are cytotoxic
T-lymphocyte-associated protein 4 (CTLA-4), programmed death
receptor 1 (PD-1), programmed death-ligand 1 (PD-L1), T-cell
immunoglobulin and mucin domain-containing-3 (TIM-3), and
lymphocyte-activation gene 3 (LAG-3) (4,5).
Blocking tumor immune escape and tolerance mechanisms through
immune checkpoints is an effective way to enhance antitumor effect.
An immune checkpoint inhibitor, especially the mAb-based one, as an
immunoregulatory factor, can specifically bind to T cells or tumor
cells, thus enhancing the antitumor ability of T cells (6-8).
The traditional mAb-based immune checkpoint blockade
is the main method of tumor treatment and detection. Results of
preclinical studies and follow-up of clinical trials led to the
approval of various checkpoint inhibitors by the Food and Drug
Administration (FDA) for the treatment of renal cell carcinoma,
melanoma, lymphoma, classic Hodgkin lymphoma, pancreatic ductal
adenocarcinoma, cervical cancer, non-small cell lung cancer,
squamous cell carcinoma of the head and neck, and breast ca, ncer.
Some immune checkpoint inhibitors approved, include mAbs targeting
PD-1 such as nivolumab (Opdivo®), pembrolizumab
(Keytruda®), cemiplimab (Sanofi, Regeneron); mAbs
targeting PD-L1 such as atezolizumab (Tecentriq®),
avelumab (Bavencio®) and durvalumab
(Imfinzi®); and mAb targeting CTLA-4 is represented by
ipilimumab (4,9-14).
Most tumor patients have benefitted from these antibodies, however
some have not responded to them (15). In addition, the characteristics of
mAbs such as poor stability, high production costs, poor tissue
penetration, and immune-related adverse events has limited their
utility (16-18).
To improve the therapeutic effect of antibodies for
immune checkpoint blockade, immune checkpoint expression should be
analyzed in patients before and during treatment. Due to the
heterogeneous and highly dynamic expression of the immune
checkpoint molecules in primary or metastatic tumors, traditional
immunohistochemical methods are limited because they cannot detect
the dynamic information of immune checkpoint molecules in the tumor
environment (19-30). Therefore, a real-time, dynamic,
and accurate detection method with high sensitivity resolution is
urgently required.
Methods involving imaging of labeled antibody
molecules have been reported, but the poor tissue penetration of
these antibodies, the long circulation time, and the high-contrast
imaging are obstacles that prevent them from becoming ideal imaging
agents (16,19,31). Thus, the development of tracers
with faster kinetics is of utmost importance.
To increase the effectiveness of immunotherapy,
including immune checkpoint blockade therapy, miniaturization of
antibodies, has been introduced. Nanobodies have a small molecular
weight conferring them a strong tissue penetration where they bind
their antigens quickly and specifically, while unbound nanobodies
can be quickly cleared through renal excretion. Therefore, compared
to mAbs, nanobodies produce higher target-to-background signals
soon after their administration (18).
Similar to mAb-based immune checkpoint inhibitors,
nanobodies that target immune checkpoint molecules have been
developed as effective tools for studying tumor immunotherapy and
immunoimaging (32). In this
review, some of the recent advances in the development of
nanobodies and nanobody-based immune checkpoint inhibitors for
immunotherapy and immunoimaging (Table I) were examined, as well as the
challenges faced to achieve successful use.
 | Table ISummary of nanobodies targeting
immune checkpoints for immunotherapy and immunoimaging. |
Table I
Summary of nanobodies targeting
immune checkpoints for immunotherapy and immunoimaging.
| Target | Nanobody name | Target species | Application | Referred
studies |
|---|
| CTLA-4 | Nb16 | Human | Immunotherapy | (112) |
| CTLA-4 | Nb36 | Human | Immunoimaging | (135,136) |
| PD-L1 | B3 | Murine | Immunotherapy | (116) |
| PD-L1 | KN035 | Human | Immunotherapy | (117) |
| PD-L1 | Nb97 | Human | - | (118) |
| PD-L1 | - | Human | - | (119) |
| PD-L1 | Nb109 | Human | Immunoimaging | (132) |
| PD-L1 C | 3/E2 | Murine | Immunoimaging | (133) |
| PD-L1 | sdAb K2 | Human |
Immunotherapy/immunoimaging | (115,134) |
| TIM3 | - | Human | - | (124) |
| TIM3 | - | Human | - | (125) |
| LAG3 | 3131/3206 | Murine | Immunoimaging | (131) |
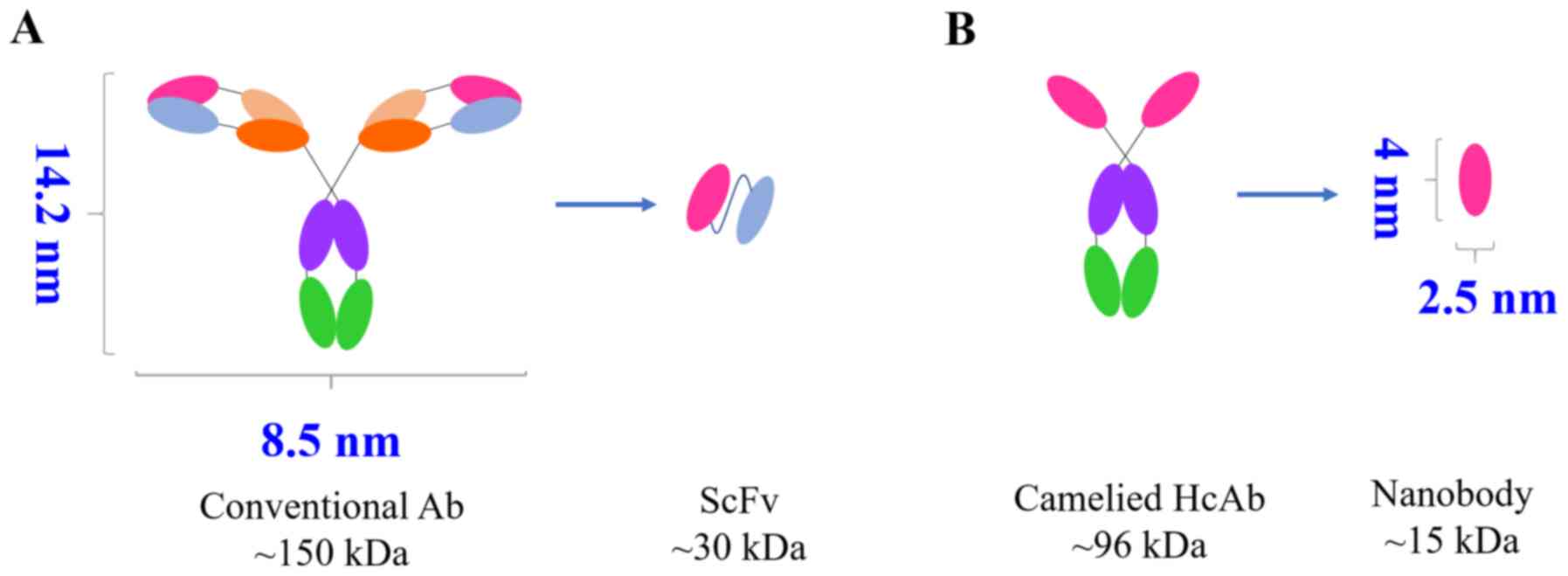
2. Biophysical properties of nanobodies
In 1993, a special antibody was revealed in the
blood of camelids (camel, alpaca, llama) and sharks (33). This antibody is different from the
traditional one with a tetrapeptide chain structure because it
lacks a light chain, thus, it is called a heavy chain antibody. Due
to the absence of the CH1 domain and the light chain, the
antigen-binding region of a heavy chain antibody consists only of
the heavy chain variable region of the heavy chain antibody
(34). For this reason, it is
called single domain antibody (sdAb), or VHH antibody or nanobody
(Fig. 1), and can be obtained
through cloning and expression. The special structure and unique
biological properties of nanobodies have attracted the attention of
numerous scholars, and several research institutions are screening
new nanobodies as new drugs for cancer treatment (35-37) and diagnosis (38).
Small molecular weight and low
immunogenicity
The crystal structure of nanobodies is similar to a
rugby ball with a diameter of approximately 2.5 nm and a length of
approximately 4.2 nm. The relative molecular mass is approximately
15 kDa, which is one-tenth of the size of conventional antibodies.
In fact, it is the smallest antibody with fully functional
properties that currently exists (39-41). Some methods using nanobodies have
better results than conventional antibodies, such as imaging tracer
agents (42-46), microscopic imaging (47), enzyme inhibitors (48,49), and electrochemical biosensors
(50-57). Due to their small size, the
binding region between the nanobody and the epitope forms a
high-density binding, providing a significant advantage in
increasing the sensitivity of the binding signal.
Nanobody backbone regions have more than 80%
sequence homology with human VH regions, and their
three-dimensional structures can overlap. The camel VHH germline
gene sequence is highly homologous to the human VH3 family
sequence. Thus, it has the advantages of weak immunogenicity and
good biocompatibility, and humanizing VHH is relatively simple
(58-61).
High stability
Nanobodies are markedly smaller than traditional
antibodies and they are characterized by the presence of a
disulfide bond, rendering their structure more stable and making
them more resistant to heat and an acid environment (62,63). Under extreme environmental
conditions, such as high temperatures or extreme acid and alkaline
environments, the structure of the traditional polyclonal antibody
changes, exposing its hydrophobic surface; the exposed hydrophobic
molecules aggregate with each other to form large molecules that
precipitate, losing their original function (64). Unlike the traditional antibody,
the nanobody forms different conformational patterns to protect the
stability of amino acids. After chemical and thermal denaturation,
the nanobody refolds and forms a disulfide bond between the
complementarity determination region-1 (CDR1) and CDR3 to improve
the stability of its structure and ensure the stability of its
functional activity (65-68). The stability of nanobodies
establishes them as a potential drug in the treatment of
gastrointestinal diseases, and their excellent characteristics
render them an effictive probe molecule for biosensor applications
(38).
Improved solubility
There are some important differences between the VHH
and the VH of the traditional antibodies. The VH structure of the
conventional antibodies easily form inclusion bodies when expressed
alone, or when the exposed hydrophobic regions adhere to each
other, making the anti-body markedly poor in water solubility. The
four hydrophilic amino acids in FR2 of the nanobodies replace the
hydrophobic amino acids of conventional antibody FR2, such as the
42nd amino acid in the VH of traditional antibodies is often Val,
while in VHH it is often Phe or Tyr. The 49th amino acid in the VH
of the traditional antibodies is often Gly, while in VHH it is
often Glu. The 50th amino acid in the VH of the traditional
antibodies is often Leu, while in VHH it is often Arg or Cys. The
52nd amino acid in the VH of the traditional antibodies is often
Trp, while in VHH it is often Gly. These 4 amino acids in VHH are
hydrophilic, thus rendering the surface of the nanobody more
hydrophilic and increasing its water solubility (40,65,69-72).
High affinity and cavity binding
Similar to traditional VH, VHH includes 4 FRs and 3
CDRs. CDR1 and CDR3 of the VHH are longer than the ones of the VH,
which makes up for the lack of antigen-binding ability caused by
the deletion of the light chains, at least to a certain extent
(40). The cysteine in the VHH
CDR3 also forms disulfide bonds with the cysteine in CDR1 or FR2.
These increased sequences and loop structures expand the area of
antibody-antigen binding and the diversity of antibodies, and
concurrently lead to a markedly stable structure that tolerates
high temperatures and harsh extreme environments (67,70-73). In addition, the nanobody does not
have a traditional Fc segment, thereby avoiding complement
reactions caused by this segment (74).
Conventional Fab fragments and typical ScFv have
concave or planar antigen-binding sites, thus, only surface
antigens can be identified. The nanobody has CDR3 loops that are
generally longer than conventional VH, allowing it to bind to
unconventional epitopes, such as protein clefts and some hidden
epitopes, which are not recognized by traditional anti-bodies
(46,75). Therefore, the nanobody is more
suitable than the ScFv antibody binding site to bind to the
recessed portion of the antigen surface, such as the catalytic
reaction site of the enzyme, thereby blocking its catalytic
activity (70,71,76,77).
Strong tissue penetrability
Nanobodies are small and highly soluble, thus, they
have strong and fast tissue penetration capabilities, and can enter
dense tissues such as solid tumors to play their role (72,78). In addition, nanobodies can
penetrate the blood-brain barrier (71,79,80) and become potential new treatments
for brain diseases such as dementia. Studies have revealed that
camel-derived nanobodies immunize cerebro-vascular endothelial
cells and they can be released on the outer side of vascular
endothelial cells through transcytosis. Their small size allows
better penetration through the tissue- and immune-like synaptic
cell interface. Furthermore, nanobodies are easily filtered by the
glomerulus, and the blood clearing rate is fast so that the excess
of free nanobodies is quickly removed without adversely affecting
the body due to long-term retention. Compared with the shortcoming
of monoclonal anti-bodies, which have poor penetrating power and
are not easily removed, such characteristics are more useful in the
diagnosis of diseases (31,39,46). Currently, several nanobody-based
imaging technologies, such as radionuclides, optics, and
ultra-sound, have been used to visualize target protein expression
levels in multiple types of disease models (31,39).
High expression yields
Nanobodies are markedly simpler in chemical
composition and shape than traditional anti-bodies, which render
nanobodies easily cloned, chemically or genetically modifiable, and
recombinantly produced in various cells (18,40,71,72). It is easier to obtain them from
prokaryotic cells and a soluble expression (81). Recombinant nanobodies are usually
highly expressed in E. coli, reaching 10 mg/l-200 mg/l
(82-84). A biopharmaceutical company
(Ablynx) reported that they greatly increased the production of
nanobodies produced by the yeast reactor to one gram per liter
(85). Nanobodies are easily
genetically manipulated to form monovalent, bivalent, bispecific,
and multivalent anti-bodies, and they can also form fusion proteins
for targeted therapy (31).
Easy modification and functional
modification
Nanobodies are VHH genes cloned from the camel or
alpaca blood by genetic engineering and then expressed by
prokaryotic or eukaryotic cells. Therefore, nanobodies are easily
modified or genetically modified (86). Wang et al (87) added right-handed coiled-coil,
cartilage oligomeric matrix protein, and C4-binding protein to the
C-terminus of the three nanobodies, respectively, and generated
tetramers, pentamers, and heptamers with these added peptides.
Toxins, biotin molecules, and reporter molecules can
also be added to the tail of the nanobody for functional
modification (46,88). In addition, genetic modification
of VHH can transform monovalent nanobodies into various forms, such
as bivalent nanobodies, bispecific nanobodies, and multivalent
nanobodies (89,90).
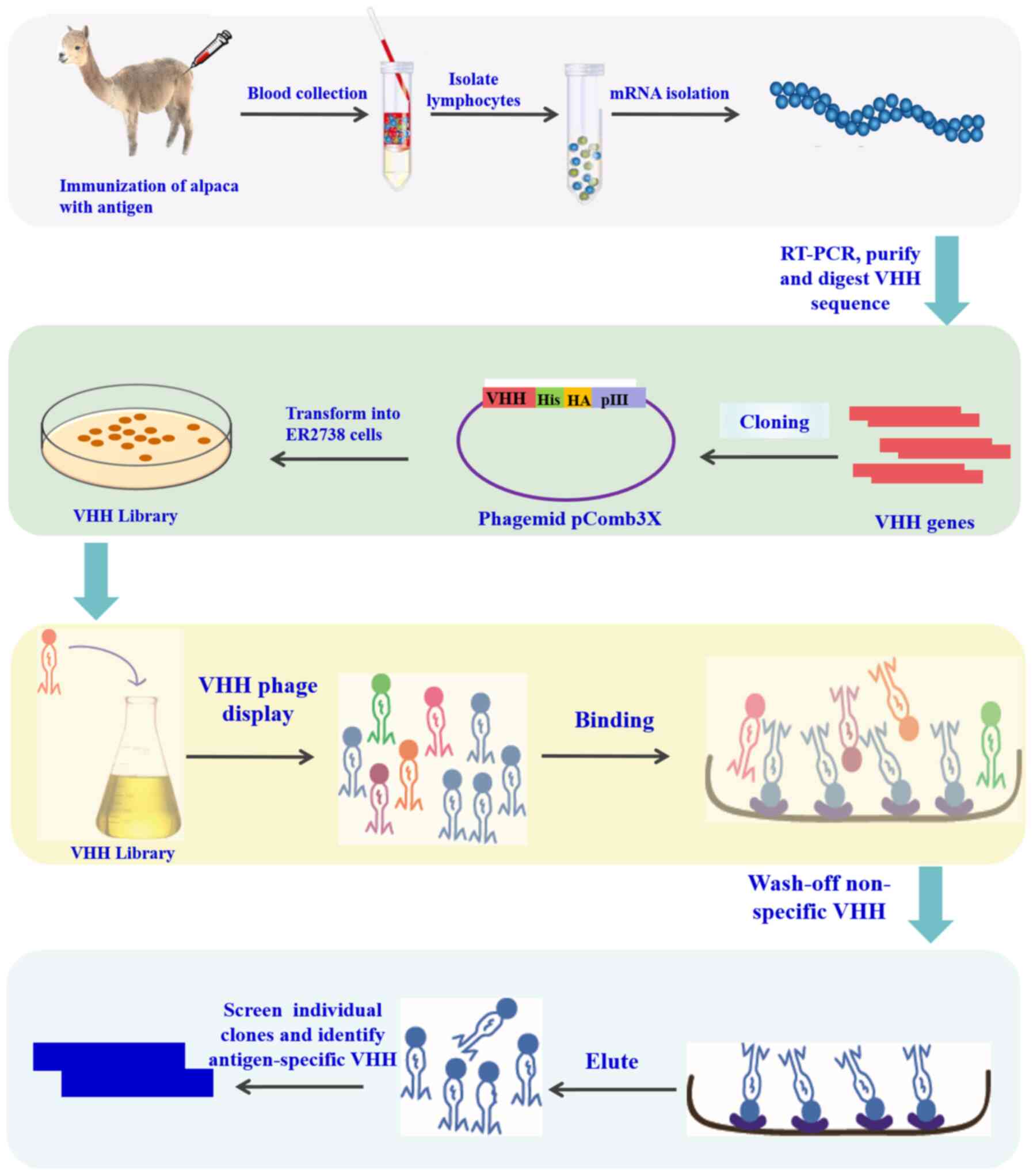
3. Construction of library and panning of
nanobodies
The merits of molecular properties such as affinity
and specificity of nanobodies depend on two factors: The capacity
and diversity of the library. The preparation and screening of
nanobodies are commonly used in a phage display library (40), which contains three library types:
Natural, immune, and synthetic (91,92). The natural library is formed by
amplifying the variable region genes of heavy chain antibodies from
camel peripheral blood mononuclear lymphocytes that have not yet
been immunized, then these variable region genes are recombined
into phagemid vectors, and transformed into host bacteria to form
antibody libraries (93,94). The immune library is an antibody
library obtained by immunizing an alpaca or camel with an antigen
protein, using peripheral blood mononuclear cells of the camel to
amplify the variable region gene of the heavy chain antibody, and
then recombining this variable region gene into a phagemid vector
(94). The capacity of the immune
library is lower than that of natural libraries, but numerous
functional antibodies in the library recognize specific antigens
for immunity, making screening of high-affinity antibodies a
reality. For the construction of synthetic libraries (95), a certain VHH framework (such as
cAbBCIll0) is generally selected as the backbone structure.
Trinucleotide cassettes are used as a unit of raw materials to
generate CDRs sequences. Then, the DNA of each region of VHH is
assembled by PCR to form a complete VHH gene. The gene is cloned
into a phagemid vector and transformed into an E. coli TG1
cell to form a library. The synthetic library, as an artificial
library constructed by genetic engineering technology, has a great
complementary role in the immune libraries and natural libraries
and is an important source for screening high-affinity antibodies,
with significant importance for the development of antibody drugs
(91,95).
The phage display technology is used to screen
binders for various targets from diverse and large libraries and is
widely employed by various research teams. Since the difference
between VHH and VH is mainly due to the absence of the CH1 region,
the work of building an immune and natural library focuses on
isolating antibodies lacking CH1 from IgG, which can be achieved by
one-step (96) or two-step nested
PCR amplification (97). The key
of PCR is to design PCR primers using the conserved nucleotide
sequence of the antibody back-bone region. With regard to the
one-step PCR, the sequence can be amplified from the FR1 to the
hinge region, and restriction sites are introduced on the primers.
For two-step nested PCR, two bands are obtained through the first
step of PCR amplification, and the sizes are 700 and 900 bp,
respectively. Among them, the 700-bp band encodes the VHH-H-CH2
fragment of the heavy chain antibody. This fragment is recovered
using a Gel Extraction kit and used as a template for the second
PCR. The VHH fragment is amplified by the second PCR (98). The amplified antibody sequence is
ligated to the phagemid vector, such as pMECS (95,97), pHEN (99), pAX50 (100), and pComb3 (96,101). The remaining steps are the same
as the ordinary process of constructing an antibody library
(98). The specific protocol is
presented in Fig. 2. In general,
the nanobody library capacity can easily reach 1x108 or
1x109, and diversity is greater than 95%. Furthermore,
because VHH has only one variable region, specific antibodies can
be screened at 1x106. However, in ordinary antibody Fab
or ScFv libraries, the library needs to be large enough to more
easily screen for antibodies (93,96,97,102,103).
In addition to phage display, some other techniques
are applied to screen nanobodies, such as mRNA (104), ribosome (105-107), yeast (108,109) and bacterial surface displays
(110,111). Salema et al (112) reported an E. coli display
system, which combines the advantages of both a phage display and
yeast display system. Flow cytometric analysis of nanobodies on the
surface of E. coli during the screening process can monitor
the selection process in real-time and identify antigen-binding
characteristics.
Nanobodies are generally screened from libraries
using specific immobilized antigens. A specific antigen is coated
in a microtiter plate to specifically bind VHH-displayed phages in
the library, then unbound phages are removed, and the bound phages
are eluted, and are used for further amplification. After several
rounds of panning and enrichment, VHH with high affinity is
obtained, and a positive VHH is identified by indirect ELISA
(98). Or, as an alternative, it
can also be panned in the liquid phase (96). After blocking, binding, elution
and amplification, specific binding between streptavidin-coated
magnetic beads and biotin-labeled antigen can also be used to
obtain antigen-specific antibodies. Cells expressing specific
antigens are similar to protein molecules and can also be used for
nanobody panning (113). The
panning step is the same as the panning step for protein molecules.
To improve the affinity of the antibodies obtained by panning,
during the panning process, the concentration of immobilized
antigen or the number of cells in each panning can be continuously
reduced, and the number of washings can be gradually increased.
4. Nanobodies targeting immune checkpoints
for immunotherapy
The large size of mAbs limits their penetration and
distribution in tumor tissues in certain clinical situations.
Compared to mAbs, the unique structure and biological activity of
the small nanobody molecule make it an effective tool for
successful immunotherapy (60).
CTLA-4
Ipilimumab (10,114-117), a fully human IgG1κ anti-CTLA-4
mAb, was the first immune checkpoint inhibitor against CTLA4
approved for non-small cell lung carcinoma, renal cell carcinoma,
prostate cancer, and metastatic melanoma by the FDA in 2011.
Tremelimumab is another fully human IgG2 anti-CTLA-4 mAb, which is
used in clinical trials (10).
Considering that mAbs have some drawbacks, such as a
markedly poor tissue penetration, production cost and unstable
behavior, Tang et al (118) immunized the camel using the
recombinant human CTLA4 protein, constructed a VHH library and
screened nanobodies using phage display technology. Four
CTLA-4-specific nanobodies were obtained from the repertoire of an
immunized dromedary camel using phage display technology. These
nanobodies recognized unique epitopes of CTLA-4 and exhibited high
binding ability. The Nb16 treatment for melanoma-bearing mice could
reduce tumor growth and prolong their survival time. However, the
study revealed that both the Nb16 and mAb groups have anti-tumor
effects, with no difference between these two groups. Thus, the
antitumor mechanism of Nb16 should be analyzed to facilitate the
clinical application of antibodies.
PD1/PD-L1
The world's first PD-1 inhibitor Opdivo®
was approved for use in 2014. By the end of 2018, the FDA had
approved 6 PD-1/PD-L1 mAbs such as nivolumab (Opdivo®;
Bristol-Myers Squibb), pembrolizumab (Keytruda®; Merck),
and cemiplimab (Libtayo®; Sanofi and Regeneron), and
three PD-L1 inhibitors, such as atezolizumab
(Tecentriq®; Roche), avelumab (Bavencio®;
Pfizer and Merck) and durvalumab (Imfinzi®; AstraZeneca)
(9,10,119). To increase the effectiveness of
immunotherapy, including the immune checkpoint blockade therapy,
miniaturization of antibodies has been introduced. Some
nanobody-based PD1/PD-L1 inhibitors were developed (19).
Broos et al developed a PD-L1 specific sdAb
called K2, which blocks the interaction between PD1 and PD-L1
(120). This property enhances
the ability of dendritic cells to stimulate T-cell activation and
cytokine production. sdAb K2 combined with dendritic cell vaccine
treatment may be more beneficial than PD-L1 mAb against cancer
diseases. The reason that PD-L1 mAbs fail to enhance T-cell
activation may be that they have low efficacy in binding to PD-L1
on DCs, while sdAb K2 has a high ability to bind PD-L1 on both
immune and non-immune cells.
Nanobodies can also be used as a carrier for
cytokines to remodel the tumor microenvironment. In one context,
Fang et al (121)
developed a functional chemokine-VHH fusion protein using a PD-L1
specific nanobody B3 and a model chemo-kine CCL21 to deliver CCL21
to a PD-L1-positive environment and recruit the relevant leukocyte
for improving immunotherapy.
KN035 (122), an
anti-PD-L1 nanobody, was screened using a camel immunological
library. It is the first nanobody research project used in the
field of immunotherapy in the world. Its binding surface of PD-L1
is smaller than other PD-L1 anti-bodies, and its affinity is
similar to other antibodies (3 nm). KN035 can bind the PD-L1
molecule with a high affinity and effectively block the action
between PD-L1 and PD1. Similar to other PD-L1 antibodies, it can
effectively compete for the five hotspot sites where PD-L1 binds to
PD-1. In addition, KN035 can effectively activate PBMCs in
vitro and induce interferon secretion. As an antitumor drug,
its preliminary results regarding its efficacy are favorable.
Xian et al (123) screened anti-PD1 nanobody Nb97 by
phage display, and then Nb97 was used to develop the
Nb97-Nb97-Human serum albumin fusion protein (MY2935), which
exhibited a more efficient blocking effect to that of a humanized
Nb97-Fc (MY2626), and Human serum albumin fusion extended the serum
half-life of nanobody Nb97. Li et al (124) obtained three anti-PDL1
nanobodies from a high quality dromedary camel immune library by
phage display, and analyzed the binding activity and affinity of
the three nanobodies, but did not research the PD1/PDL1 pathway
blocking effect.
TIM3 and LAG3
Some cancer patients do not respond to PD1/PDL1 and
CTLA4 inhibitors. To obtain a greater number of patients benefiting
from immune checkpoint blockade immunotherapy, some other immune
checkpoint molecules have been developed such as TIM3 (125,126) and LAG3 (127,128).
Homayouni et al (129) immunized a six-month Camelus
dromedarius with a human TIM3 protein and developed a novel
anti-human TIM-3 (CD366) nanobody from an immune library. This
nanobody exhibited a high binding capacity to TIM-3, and a high
antiproliferative effect on the acute myeloid leukemia cell line
HL-60 by blocking the galectin/TIM-3 signal, with an inhibitory
effect comparable to or better than that of anti-TIM-3 antibodies.
However, the researchers did not detect the difference in tissue
penetration between nanobodies and mAbs. Ma et al (130) immunized a camel, constructed the
phage display library, and then screened ten anti-TIM3 nanobodies
with high specificity and high affinity using flow cytometry.
However, the researchers did not detect the function of the
anti-TIM3 nanobody, thus, it is not known whether blocking the TIM3
inhibitory signal by anti-TIM3 nanobodies can activate the
antitumor function of T cells. Nevertheless, the aforementioned
studies provide the basis for the development of specific nanobody
drugs blocking TIM-3.
There are seven anti-LAG3 mAbs and two bispecific
anti-bodies targeting LAG3 (131-133), which are at different stages of
clinical development. However, studies on the application of
anti-LAG3 nanobodies in antitumor research have yet to be
published.
5. Nanobodies targeting immune checkpoints
for immunoimaging
Molecular imaging can intuitively detect changes at
the molecular level during and after the treatment of diseases,
therefore it is one of the important methods for evaluating the
effect of tumor therapies. With the continuous development of
targeted therapies, it is becoming increasingly important to
visualize the expression of tumor antigens and the level of immune
infiltrations to predict the course of the treatment. Molecular
imaging based on mAbs has been extensively studied, however the
poor tissue penetration and long half-life severely hinder the
development of successful molecular imaging (60). Nanobody molecular probes bind to
the target with high specificity, and the unbound part is quickly
excreted by the kidney. In addition, nanobody molecular probes have
a deep tumor penetration and high tumor to background ratio soon
after their administration, which clearly reveals the dynamic
changes of target molecules (60,134,135). Therefore, nanobodies have
recently become a powerful tool for in vivo and in
vitro imaging diagnostics.
Imaging of LAG-3
In 2019, Lecocq et al reported an anti-LAG3
nanobody used for noninvasive imaging (136). They immunized alpaca with mouse
LAG3 protein, bio-panned the phage-displayed library, and obtained
nine nanobodies 3132, 3134, 3141, 3204, 3206, 3208, 3209, 3210, and
3366, which exhibited high specificity and high affinity validated
by ELISA, flow cytometry, and surface plasmon resonance methods.
These nanobodies were labeled with Technetium-99m
(99mTc) at the His tail, and then, these nanobodies were
injected into naive C57BL/6 mice intravenously. The results
revealed the nanobody 3132 exhibited specific uptake by LAG3-low
expression immune peripheral organs, such as the spleen and lymph
nodes. To detect whether the nine 99mTc-labeled
nanobodies bind to the tumor overexpressing LAG-3, SPECT/CT imaging
was used to detect the biodistribution of 99mTc-labeled
nanobodies in mice harboring a subcutaneous tumor modified to
overexpress mouse LAG-3. The result revealed that it was possible
to visualize the nanobody uptake using SPECT/CT imaging with high
contrast levels immediately, even 1 h after injection, and this
result was confirmed by flow cytometry and immunohistochemistry.
The results demonstrated that the nanobodies 3132 and 3206 are both
effective diagnostic tools for noninvasively evaluating LAG-3
expression within the tumor environment before and during
immunotherapy.
Imaging of PD1/PDL1
Lv et al developed a nanobody tracer using
the PD-L1 targeted nanobody (Nb109) and the radionuclide 68Ga
through the chelator 1,4,7-triazacyclononane-1,4,7-triac-etic acid
(NOTA), and 68Ga-NOTA-Nb109 exhibited a high affinity for PD-L1
with a KD of 2.9x10-9 M (137). The competitive binding assay
demonstrated a different binding epitope between Nb109 and PD-L1 or
PD1 mAb, suggesting no impact on the tumor uptake of
68Ga-NOTA-Nb109 before and after the treatment with KN035. The
68Ga-NOTA-Nb109 tracer specifically accumulated in mice model
harboring the A375-human PD-L1 tumor, with a maximum uptake of
5.0±0.35% ID/g at 1 h as determined through the PET imaging,
biodistribution, immunohistochemical staining, and autoradiography
assay, indicating that 68Ga-NOTA-Nb109 is a promising nanobody
tracer for noninvasive PET imaging of PD-L1 in the tumor
microenvironment and promising in evaluating the effectiveness of
immune checkpoint blockade immunotherapy in real-time.
In 2017, Broos et al developed an anti-PD-L1
nanobody for noninvasive imaging of the murine PDL1 (138). They immunized 10 million PD-L1
high-expressing mouse macro-phage RAW264.7 cells, and 37 mouse
PD-L1 nanobodies were identified by biopanning of the Nb-phage
display library. Among those Technetium-99m
(99mTc)-labeled nanobodies, four nanobodies were
selected to evaluate their biodistribution in PD-L1-knockout and
wild-type mice using SPECT/CT. According to the results,
Technetium-99m (99mTc)-labeled nanobodies C3 and E2 were
used to image PD-L1 in a synge-neic mouse model because C3 and E2
have high specific antigen binding and beneficial biodistribution.
Their work demonstrated that 99mTc-labeled nanobody
tracers identified PD-L1-expressing tumors, but not in the
PD-L1-knockout tumors, suggesting that these
99mTc-labeled nanobodies can be used in SPECT/CT imaging
to assess the PD-L1 expression markedly soon even one hour after
injection. Owing to the fast tumor-penetrating properties of the
nanobodies, the results confirmed that a 99mTc-labeled
nanobody tracer is a good method to image PD-L1 inhibitory signals
during the treatment of the tumor environment.
In 2019, Broos et al (139) developed another tracer for
noninvasive imaging of the human PDL1. They generated a new panel
of sdAbs by alpaca immunizations, biopanning, and screenings on
recombinant human PD-L1 protein, and they obtained a nanobody
called sdAb K2, which binds to the same epitope on the PD-L1
molecule as the mAb avelumab. Thus, this nanobody is able to block
the PD1/PDL1 inhibitory signal resulting in activated T cells and
enhanced antitumor activity. The nanobody tracer also labeled with
Technetium-99m (99mTc) to develop
99mTc-labeled sdAb K2 tracer, was intravenously injected
into mice bearing melanoma and breast tumors to detect the PD-L1 by
SPECT/CT imaging. This assay revealed a 99mTc-labeled
sdAb K2 tracer with a high signal-to-noise ratio and a strong
ability to image PD-L1. Collectively, sdAb K2 has a dual function
as a diagnostic and therapeutic agent, offering broad prospects in
a variety of tumor immunotherapy and immunoimaging techniques.
Imaging of CTLA4
In 2018, Wan et al reported four
CTLA-4-specific nanobodies from a camel immune library by phage
display technology (140). One
of these nanobodies called Nb36 was conjugated to the carbon
quantum to synthesize a CTLA-4-specific nanobody-fluorescent carbon
quantum dot complex (QDs-Nb36) (141) in 2019 by the same group. Because
anti-CTLA-4 nanobodies specifically bind to CTLA-4+ T
cells and QDs provide a sensitive fluorescent signal for accurate
detection, the QDs-Nb36 complex revealed a high sensitivity
detection of CTLA-4+ T cells by flow cytometry and
immunofluorescence staining. Owing to the small size of the
nanobody, the QDs-Nb36 complex was superior to mAbs in detecting
the positive cells. Thus, nanobody-QDs is a promising method for
the detection of some other biological targets, although this
method cannot monitor the dynamic changes of the target molecule in
real-time.
6. Conclusion
In view of the past few decades, monoclonal
antibodies have shown considerable success in cancer treatment and
diagnosis (44). However, the
large and complex structure of the monoclonal anti-bodies limits
their clinical utility (44). As
revealed in this review, nanobodies have a small molecular weight
conferring them a strong tissue penetration where they bind their
antigens quickly and specifically, while unbound nanobodies can be
quickly cleared through renal excretion, resulting in high
target-to-background signals soon after their administration
(134,135). Therefore, the introduction of
nanobodies has demonstrated that they can overcome certain
shortcomings of monoclonal antibody-based immunotherapy and
immunoimaging.
Currently, nanobodies targeting immune checkpoints
are mainly concentrated in PD1/PDL1. In the future, it is necessary
to develop more research on nanobodies targeting other immune
checkpoints, including TIM3, LAG3, OX40 and VISTA. These immune
checkpoints and their corresponding nanobodies should be
characterized for clinical application. In addition, in order to
prolong the half-life of nanobodies in vivo, nanobody dimers
and multimeric nanobodies have been produced. The moderate relative
molecular mass can better meet the requirements of deep tissue
penetration, targeted aggregation, and blood clearance. With
in-depth research on nanobodies, the application of nanobodies in
tumor immunotherapy and immunodiagnosis is promising.
Funding
The present work was supported by grants from the
National Natural Scientific Foundation of China (grant nos.
81760551 and 82003250), the Guangxi Natural Science Foundation
Project (grant no. 2018GXNSFAA294117), and the Guangxi Medical and
Health Appropriate Technology Development and Application Project
(grant no. S2020005).
Availability of data and materials
Not applicable.
Authors' contributions
SD and XB conceived and designed this review. GX
and SZ researched and wrote sections 1 and 2 of the present review.
SY researched and wrote section 3 of the present review. YT and HT
researched and wrote section 4 of the present review. KW, HL and KL
researched and wrote sections 5 and 6 of the present review. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
We would like to express our gratitude to all those
who helped during the writing of this review.
References
|
1
|
Frankel T, Lanfranca MP and Zou W: The
role of tumor micro-environment in cancer immunotherapy. Adv Exp
Med Biol. 1036:51–64. 2017. View Article : Google Scholar
|
|
2
|
Fan CA, Reader J and Roque DM: Review of
immune therapies targeting ovarian cancer. Curr Treat Options
Oncol. 19:742018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marin-Acevedo JA, Soyano AE, Dholaria B,
Knutson KL and Lou Y: Cancer immunotherapy beyond immune checkpoint
inhibitors. J Hematol Oncol. 11:82018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marin-Acevedo JA, Dholaria B, Soyano AE,
Knutson KL, Chumsri S and Lou Y: Next generation of immune
checkpoint therapy in cancer: New developments and challenges. J
Hematol Oncol. 11:392018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ok CY and Young KH: Checkpoint inhibitors
in hematological malignancies. J Hematol Oncol. 10:1032017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baghdadi M, Takeuchi S, Wada H and Seino
K: Blocking mono-clonal antibodies of TIM proteins as orchestrators
of anti-tumor immune response. MAbs. 6:1124–1132. 2014. View Article : Google Scholar
|
|
7
|
Rodallec A, Sicard G, Fanciullino R,
Benzekry S, Lacarelle B, Milano G and Ciccolini J: Turning cold
tumors into hot tumors: Harnessing the potential of tumor immunity
using nanoparticles. Expert Opin Drug Metab Toxicol. 14:1139–1147.
2018.PubMed/NCBI
|
|
8
|
Nishino M, Ramaiya NH, Hatabu H and Hodi
FS: Monitoring immune-checkpoint blockade: Response evaluation and
biomarker development. Nat Rev Clin Oncol. 14:655–668. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darvin P, Toor SM, Sasidharan Nair V and
Elkord E: Immune checkpoint inhibitors: Recent progress and
potential biomarkers. Exp Mol Med. 50:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hargadon KM, Johnson CE and Williams CJ:
Immune checkpoint blockade therapy for cancer: An overview of
FDA-approved immune checkpoint inhibitors. Int Immunopharmacol.
62:29–39. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen H, Yang ES, Conry M, Fiveash J,
Contreras C, Bonner JA and Shi LZ: Predictive biomarkers for immune
checkpoint blockade and opportunities for combination therapies.
Genes Dis. 6:232–246. 2019. View Article : Google Scholar
|
|
12
|
Kamath SD, Kalyan A and Benson AB III:
Pembrolizumab for the treatment of gastric cancer. Expert Rev
Anticancer Ther. 18:1177–1187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hsu FS, Su CH and Huang KH: A
comprehensive review of US FDA-approved immune checkpoint
inhibitors in urothelial carcinoma. J Immunol Res.
2017:69405462017. View Article : Google Scholar
|
|
14
|
Song MK, Park BB and Uhm J: Understanding
immune evasion and therapeutic targeting associated with PD-1/PD-L1
pathway in diffuse large B-cell lymphoma. Int J Mol Sci.
20:13262019. View Article : Google Scholar :
|
|
15
|
Michot JM, Bigenwald C, Champiat S,
Collins M, Carbonnel F, Postel-Vinay S, Berdelou A, Varga A,
Bahleda R, Hollebecque A, et al: Immune-related adverse events with
immune checkpoint blockade: A comprehensive review. Eur J Cancer.
54:139–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bannas P, Hambach J and Koch-Nolte F:
Nanobodies and nanobody-based human heavy chain antibodies as
antitumor therapeutics. Front Immunol. 8:16032017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ubah OC, Buschhaus MJ, Ferguson L,
Kovaleva M, Steven J, Porter AJ and Barelle CJ: Next-generation
flexible formats of VNAR domains expand the drug platform's utility
and developability. Biochem Soc Trans. 46:1559–1565. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang H, Meng AM, Li SH and Zhou XL: A
nanobody targeting carcinoembryonic antigen as a promising
molecular probe for non-small cell lung cancer. Mol Med Rep.
16:625–630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Broos K, Lecocq Q, Raes G, Devoogdt N,
Keyaerts M and Breckpot K: Noninvasive imaging of the PD-1:PD-L1
immune checkpoint: Embracing nuclear medicine for the benefit of
personalized immunotherapy. Theranostics. 8:3559–3570. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mayer AT, Natarajan A, Gordon SR, Maute
RL, McCracken MN, Ring AM, Weissman IL and Gambhir SS: Practical
immuno-PET radiotracer design considerations for human immune
checkpoint imaging. J Nucl Med. 58:538–546. 2017. View Article : Google Scholar :
|
|
21
|
Natarajan A, Mayer AT, Reeves RE, Nagamine
CM and Gambhir SS: Development of novel ImmunoPET tracers to image
human PD-1 checkpoint expression on tumor-infiltrating lymphocytes
in a humanized mouse model. Mol Imaging Biol. 19:903–914. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li M, Ehlerding EB, Jiang D, Barnhart TE,
Chen W, Cao T, Engle JW and Cai W: In vivo characterization of
PD-L1 expression in breast cancer by immuno-PET with
89Zr-labeled avelumab. Am J Transl Res. 12:1862–1872.
2020.
|
|
23
|
Kikuchi M, Clump DA, Srivastava RM, Sun L,
Zeng D, Diaz-Perez JA, Anderson CJ, Edwards WB and Ferris RL:
Preclinical immunoPET/CT imaging using Zr-89-labeled anti-PD-L1
monoclonal antibody for assessing radiation-induced PD-L1
upregulation in head and neck cancer and melanoma. Oncoimmunology.
6:e13290712017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li D, Zou S, Cheng S, Song S, Wang P and
Zhu X: Monitoring the response of PD-L1 expression to epidermal
growth factor receptor tyrosine kinase inhibitors in nonsmall-cell
lung cancer xenografts by immuno-PET imaging. Mol Pharm.
16:3469–3476. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
González Trotter DE, Meng X, McQuade P,
Rubins D, Klimas M, Zeng Z, Connolly BM, Miller PJ, O'Malley SS,
Lin SA, et al: In vivo imaging of the programmed death ligand 1 by
18F PET. J Nucl Med. 58:1852–1857. 2017. View Article : Google Scholar
|
|
26
|
Hettich M, Braun F, Bartholomä MD,
Schirmbeck R and Niedermann G: High-resolution PET imaging with
therapeutic antibody-based PD-1/PD-L1 checkpoint tracers.
Theranostics. 6:1629–1640. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heskamp S, Hobo W, Molkenboer-Kuenen JD,
Olive D, Oyen WJ, Dolstra H and Boerman OC: Noninvasive imaging of
tumor PD-L1 expression using radiolabeled anti-PD-L1 antibodies.
Cancer Res. 75:2928–2936. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li D, Cheng S, Zou S, Zhu D, Zhu T, Wang P
and Zhu X: Immuno-PET imaging of 89Zr labeled anti-PD-L1
domain anti-body. Mol Pharm. 15:1674–1681. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Josefsson A, Nedrow JR, Park S, Banerjee
SR, Rittenbach A, Jammes F, Tsui B and Sgouros G: Imaging,
biodistribution, and dosimetry of radionuclide-labeled PD-L1
antibody in an immunocompetent mouse model of breast cancer. Cancer
Res. 76:472–479. 2016. View Article : Google Scholar :
|
|
30
|
Natarajan A, Patel CB, Habte F and Gambhir
SS: Dosimetry prediction for clinical translation of
64Cu-pembrolizumab ImmunoPET targeting human PD-1
expression. Sci Rep. 8:6332018. View Article : Google Scholar
|
|
31
|
Van Audenhove I and Gettemans J:
Nanobodies as versatile tools to understand, diagnose, visualize
and treat cancer. EBioMedicine. 8:40–48. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lecocq Q, De Vlaeminck Y, Hanssens H,
D'Huyvetter M, Raes G, Goyvaerts C, Keyaerts M, Devoogdt N and
Breckpot K: Theranostics in immunooncology using nanobody
derivatives. Theranostics. 9:7772–7791. 2019. View Article : Google Scholar :
|
|
33
|
Hamers-Casterman C, Atarhouch T,
Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N and
Hamers R: Naturally occurring antibodies devoid of light chains.
Nature. 363:446–448. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Muyldermans S: Nanobodies: Natural
single-domain antibodies. Annu Rev Biochem. 82:775–797. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Könning D, Zielonka S, Grzeschik J,
Empting M, Valldorf B, Krah S, Schröter C, Sellmann C, Hock B and
Kolmar H: Camelid and shark single domain antibodies: Structural
features and therapeutic potential. Curr Opin Struct Biol.
45:10–16. 2017. View Article : Google Scholar
|
|
36
|
Krah S, Schröter C, Zielonka S, Empting M,
Valldorf B and Kolmar H: Single-domain antibodies for biomedical
applications. Immunopharmacol Immunotoxicol. 38:21–28. 2016.
View Article : Google Scholar
|
|
37
|
Steeland S, Vandenbroucke RE and Libert C:
Nanobodies as therapeutics: Big opportunities for small antibodies.
Drug Discov Today. 21:1076–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stijlemans B, De Baetselier P, Caljon G,
Van Den Abbeele J, Van Ginderachter JA and Magez S: Nanobodies as
tools to understand, diagnose, and treat African trypanosomiasis.
Front Immunol. 8:7242017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hassanzadeh-Ghassabeh G, Devoogdt N, De
Pauw P, Vincke C and Muyldermans S: Nanobodies and their potential
applications. Nanomedicine (Lond). 8:1013–1026. 2013. View Article : Google Scholar
|
|
40
|
Arezumand R, Alibakhshi A, Ranjbari J,
Ramazani A and Muyldermans S: Nanobodies as novel agents for
targeting angio-genesis in solid cancers. Front Immunol.
8:17462017. View Article : Google Scholar
|
|
41
|
Van Heeke G, Allosery K, De Brabandere V,
De Smedt T, Detalle L and de Fougerolles A: Nanobodies®
as inhaled biotherapeutics for lung diseases. Pharmacol Ther.
169:47–56. 2017. View Article : Google Scholar
|
|
42
|
Massa S, Xavier C, Muyldermans S and
Devoogdt N: Emerging site-specific bioconjugation strategies for
radioimmunotracer development. Expert Opin Drug Deliv.
13:1149–1163. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Massa S, Vikani N, Betti C, Ballet S,
Vanderhaegen S, Steyaert J, Descamps B, Vanhove C, Bunschoten A,
van Leeuwen FW, et al: Sortase A-mediated site-specific labeling of
camelid single-domain antibody-fragments: A versatile strategy for
multiple molecular imaging modalities. Contrast Media Mol Imaging.
11:328–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Oliveira S, Heukers R, Sornkom J, Kok RJ,
van Bergen EN and Henegouwen PM: Targeting tumors with nanobodies
for cancer imaging and therapy. J Control Release. 172:607–617.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Iezzi ME, Policastro L, Werbajh S,
Podhajcer O and Canziani GA: Single-domain antibodies and the
promise of modular targeting in cancer imaging and treatment. Front
Immunol. 9:2732018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hu Y, Liu C and Muyldermans S:
Nanobody-based delivery systems for diagnosis and targeted tumor
therapy. Front Immunol. 8:14422017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Beghein E and Gettemans J: Nanobody
technology: A versatile toolkit for microscopic imaging,
protein-protein interaction analysis, and protein function
exploration. Front Immunol. 8:7712017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Menzel S, Rissiek B, Haag F, Goldbaum FA
and Koch-Nolte F: The art of blocking ADP-ribosyltransferases
(ARTs): Nanobodies as experimental and therapeutic tools to block
mammalian and toxin ARTs. FEBS J. 280:3543–3550. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Unger M, Eichhoff AM, Schumacher L,
Strysio M, Menzel S, Schwan C, Alzogaray V, Zylberman V, Seman M,
Brandner J, et al: Selection of nanobodies that block the
enzy-matic and cytotoxic activities of the binary clostridium
difficile toxin CDT. Sci Rep. 5:78502015. View Article : Google Scholar
|
|
50
|
Mars A, Bouhaouala-Zahar B and Raouafi N:
Ultrasensitive sensing of androctonus australis hector scorpion
venom toxins in biological fluids using an electrochemical graphene
quantum dots/nanobody-based platform. Talanta. 190:182–187. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Singh A, Pasha SK, Manickam P and Bhansali
S: Single-domain antibody based thermally stable electrochemical
immunosensor. Biosens Bioelectron. 83:162–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhu Z, Shi L, Feng H and Zhou HS: Single
domain antibody coated gold nanoparticles as enhancer for
Clostridium difficile toxin detection by electrochemical impedance
immunosensors. Bioelectrochemistry. 101:153–158. 2015. View Article : Google Scholar
|
|
53
|
Li G, Zhu M, Ma L, Yan J, Lu X, Shen Y and
Wan Y: Generation of small single domain nanobody binders for
sensitive detection of testosterone by electrochemical impedance
spectroscopy. ACS Appl Mater Interfaces. 8:13830–13839. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li H, Sun Y, Elseviers J, Muyldermans S,
Liu S and Wan Y: A nanobody-based electrochemiluminescent
immunosensor for sensitive detection of human procalcitonin.
Analyst. 139:3718–3721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu X, Wen Y, Wang W, Zhao Z, Han Y, Tang
K and Wang D: Nanobody-based electrochemical competitive
immunosensor for the detection of AFB1 through
AFB1-HCR as signal amplifier. Mikrochim Acta.
187:3522020. View Article : Google Scholar
|
|
56
|
Liu A, Yin K, Mi L, Ma M, Liu Y, Li Y, Wei
W, Zhang Y and Liu S: A novel photoelectrochemical immunosensor by
integration of nanobody and ZnO nanorods for sensitive detection of
nucleoside diphosphatase kinase-A. Anal Chim Acta. 973:82–90. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhou Q, Li G, Zhang Y, Zhu M, Wan Y and
Shen Y: Highly selective and sensitive electrochemical immunoassay
of Cry1C using nanobody and π-π stacked graphene oxide/thionine
assembly. Anal Chem. 88:9830–9836. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Steeland S, Puimège L, Vandenbroucke RE,
Van Hauwermeiren F, Haustraete J, Devoogdt N, Hulpiau P,
Leroux-Roels G, Laukens D, Meuleman P, et al: Generation and
characterization of small single domain antibodies inhibiting human
tumor necrosis factor receptor 1. J Biol Chem. 290:4022–4037. 2015.
View Article : Google Scholar :
|
|
59
|
Kazemi-Lomedasht F, Pooshang-Bagheri K,
Habibi-Anbouhi M, Hajizadeh-Safar E, Shahbazzadeh D, Mirzahosseini
H and Behdani M: In vivo immunotherapy of lung cancer using
cross-species reactive vascular endothelial growth factor
nano-bodies. Iran J Basic Med Sci. 20:489–496. 2017.PubMed/NCBI
|
|
60
|
Salvador JP, Vilaplana L and Marco MP:
Nanobody: Outstanding features for diagnostic and therapeutic
applications. Anal Bioanal Chem. 411:1703–1713. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
De Munter S, Van Parys A, Bral L, Ingels
J, Goetgeluk G, Bonte S, Pille M, Billiet L, Weening K, Verhee A,
et al: Rapid and effective generation of nanobody based CARs using
PCR and gibson assembly. Int J Mol Sci. 21:8832020. View Article : Google Scholar :
|
|
62
|
Ren W, Li Z, Xu Y, Wan D, Barnych B, Li Y,
Tu Z, He Q, Fu J and Hammock BD: One-step ultrasensitive
bioluminescent enzyme immunoassay based on nanobody/nanoluciferase
fusion for detection of aflatoxin B1 in cereal. J Agric
Food Chem. 67:5221–5229. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Allegra A, Innao V, Gerace D, Vaddinelli
D, Allegra AG and Musolino C: Nanobodies and cancer: Current status
and new perspectives. Cancer Invest. 36:221–237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
De Genst E, Chan PH, Pardon E, Hsu SD,
Kumita JR, Christodoulou J, Menzer L, Chirgadze DY, Robinson CV,
Muyldermans S, et al: A nanobody binding to non-amyloido-genic
regions of the protein human lysozyme enhances partial unfolding
but inhibits amyloid fibril formation. J Phys Chem B.
117:13245–13258. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Gonzalez-Sapienza G, Rossotti MA and
Tabares-da Rosa S: Single-domain antibodies as versatile affinity
reagents for analytical and diagnostic applications. Front Immunol.
8:9772017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Akazawa-Ogawa Y, Uegaki K and Hagihara Y:
The role of intra-domain disulfide bonds in heat-induced
irreversible denaturation of camelid single domain VHH antibodies.
J Biochem. 159:111–121. 2016. View Article : Google Scholar :
|
|
67
|
Goldman ER, Liu JL, Zabetakis D and
Anderson GP: Enhancing stability of camelid and shark single domain
antibodies: An overview. Front Immunol. 8:8652017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kunz P, Zinner K, Mücke N, Bartoschik T,
Muyldermans S and Hoheisel JD: The structural basis of nanobody
unfolding reversibility and thermoresistance. Sci Rep. 8:79342018.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Schumacher D, Helma J, Schneider AFL,
Leonhardt H and Hackenberger CPR: Nanobodies: Chemical
functionalization strategies and intracellular applications. Angew
Chem Int Ed Engl. 57:2314–2333. 2018. View Article : Google Scholar :
|
|
70
|
Wang Y, Fan Z, Shao L, Kong X, Hou X, Tian
D, Sun Y, Xiao Y and Yu L: Nanobody-derived nanobiotechnology tool
kits for diverse biomedical and biotechnology applications. Int J
Nanomedicine. 11:3287–3303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Jovčevska I and Muyldermans S: The
therapeutic potential of nanobodies. BioDrugs. 34:11–26. 2020.
View Article : Google Scholar
|
|
72
|
Zottel A, Jovčevska I, Šamec N, Mlakar J,
Šribar J, Križaj I, Skoblar Vidmar M and Komel R: Anti-vimentin,
anti-TUFM, anti-NAP1L1 and anti-DPYSL2 nanobodies display cytotoxic
effect and reduce glioblastoma cell migration. Ther Adv Med Oncol.
12:17588359209153022020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Peyron I, Kizlik-Masson C, Dubois MD,
Atsou S, Ferrière S, Denis CV, Lenting PJ, Casari C and Christophe
OD: Camelid-derived single-chain antibodies in hemostasis:
Mechanistic, diagnostic, and therapeutic applications. Res Pract
Thromb Haemost. 4:1087–1110. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kijanka M, Dorresteijn B, Oliveira S and
van Bergen en Henegouwen PM: Nanobody-based cancer therapy of solid
tumors. Nanomedicine (Lond). 10:161–174. 2015. View Article : Google Scholar
|
|
75
|
Huen J, Yan Z, Iwashkiw J, Dubey S,
Gimenez MC, Ortiz ME, Patel SV, Jones MD, Riazi A, Terebiznik M, et
al: A novel single domain antibody targeting FliC flagellin of
salmonella enterica for effective inhibition of host cell invasion.
Front Microbiol. 10:26652019. View Article : Google Scholar :
|
|
76
|
Zavrtanik U, Lukan J, Loris R, Lah J and
Hadži S: Structural basis of epitope recognition by heavy-chain
camelid antibodies. J Mol Biol. 430:4369–4368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lauwereys M, Arbabi Ghahroudi M, Desmyter
A, Kinne J, Hölzer W, De Genst E, Wyns L and Muyldermans S: Potent
enzyme inhibitors derived from dromedary heavy-chain anti-bodies.
EMBO J. 17:3512–3520. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Arbabi-Ghahroudi M: Camelid single-domain
antibodies: Historical perspective and future outlook. Front
Immunol. 8:15892017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Muruganandam A, Tanha J, Narang S and
Stanimirovic D: Selection of phage-displayed llama single-domain
antibodies that transmigrate across human blood-brain barrier
endothelium. FASEB J. 16:240–242. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Abulrob A, Sprong H, Van Bergen en
Henegouwen P and Stanimirovic D: The blood-brain barrier
transmigrating single domain antibody: Mechanisms of transport and
antigenic epitopes in human brain endothelial cells. J Neurochem.
95:1201–1214. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Menzel S, Schwarz N, Haag F and Koch-Nolte
F: Nanobody-based biologics for modulating purinergic signaling in
inflammation and immunity. Front Pharmacol. 9:2662018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhu M, Hu Y, Li G, Ou W, Mao P, Xin S and
Wan Y: Combining magnetic nanoparticle with biotinylated nanobodies
for rapid and sensitive detection of influenza H3N2. Nanoscale Res
Lett. 9:5282014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zarschler K, Witecy S, Kapplusch F,
Foerster C and Stephan H: High-yield production of functional
soluble single-domain antibodies in the cytoplasm of Escherichia
coli. Microb Cell Fact. 12:972013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
He T, Zhu J, Nie Y, Hu R, Wang T, Li P,
Zhang Q and Yang Y: Nanobody technology for mycotoxin detection in
the field of food safety: Current status and prospects. Toxins
(Basel). 10:1802018. View Article : Google Scholar
|
|
85
|
Detalle L, Stohr T, Palomo C, Piedra PA,
Gilbert BE, Mas V, Millar A, Power UF, Stortelers C, Allosery K, et
al: Generation and characterization of ALX-0171, a potent novel
therapeutic nanobody for the treatment of respiratory syncytial
virus infection. Antimicrob Agents Chemother. 60:6–13. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sheng Y, Wang K, Lu Q, Ji P, Liu B, Zhu J,
Liu Q, Sun Y, Zhang J, Zhou EM and Zhao Q: Nanobody-horseradish
peroxidase fusion protein as an ultrasensitive probe to detect
antibodies against newcastle disease virus in the immunoassay. J
Nanobiotechnology. 17:352019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang L, Liu X, Zhu X, Wang L, Wang W, Liu
C, Cui H, Sun M and Gao B: Generation of single-domain antibody
multimers with three different self-associating peptides. Protein
Eng Des Sel. 26:417–423. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Behdani M, Zeinali S, Karimipour M,
Khanahmad H, Schoonooghe S, Aslemarz A, Seyed N, Moazami-Godarzi R,
Baniahmad F, Habibi-Anbouhi M, et al: Development of
VEGFR2-specific nanobody pseudomonas exotoxin A conjugated to
provide efficient inhibition of tumor cell growth. N Biotechnol.
30:205–209. 2013. View Article : Google Scholar
|
|
89
|
Sadeghnezhad G, Romão E, Bernedo-Navarro
R, Massa S, Khajeh K, Muyldermans S and Hassania S: Identification
of new DR5 agonistic nanobodies and generation of multivalent
nanobody constructs for cancer treatment. Int J Mol Sci.
20:48182019. View Article : Google Scholar :
|
|
90
|
Huet HA, Growney JD, Johnson JA, Li J,
Bilic S, Ostrom L, Zafari M, Kowal C, Yang G, Royo A, et al:
Multivalent nano-bodies targeting death receptor 5 elicit superior
tumor cell killing through efficient caspase induction. Mabs.
6:1560–1570. 2014. View Article : Google Scholar
|
|
91
|
Liu W, Song H, Chen Q, Yu J, Xian M, Nian
R and Feng D: Recent advances in the selection and identification
of antigen-specific nanobodies. Mol Immunol. 96:37–47. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wagner HJ, Wehrle S, Weiss E, Cavallari M
and Weber W: A two-step approach for the design and generation of
nanobodies. Int J Mol Sci. 19:34442018. View Article : Google Scholar :
|
|
93
|
Yan J, Wang P, Zhu M, Li G, Romão E, Xiong
S and Wan Y: Characterization and applications of nanobodies
against human procalcitonin selected from a novel naïve Nanobody
phage display library. J Nanobiotechnology. 13:332015. View Article : Google Scholar
|
|
94
|
Itoh K, Reis AH, Hayhurst A and Sokol SY:
Isolation of nanobodies against xenopus embryonic antigens using
immune and nonimmune phage display libraries. PLoS One.
14:e02160832019. View Article : Google Scholar
|
|
95
|
Yan J, Li G, Hu Y, Ou W and Wan Y:
Construction of a synthetic phage-displayed Nanobody library with
CDR3 regions randomized by trinucleotide cassettes for diagnostic
applications. J Transl Med. 12:3432014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Cui Y, Li D, Morisseau C, Dong JX, Yang J,
Wan D, Rossotti MA, Gee SJ, González-Sapienza GG and Hammock BD:
Heavy chain single-domain antibodies to detect native human soluble
epoxide hydrolase. Anal Bioanal Chem. 407:7275–7283. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Gong X, Zhu M, Li G, Lu X and Wan Y:
Specific determination of influenza H7N2 virus based on
biotinylated single-domain antibody from a phage-displayed library.
Anal Biochem. 500:66–72. 2016. View Article : Google Scholar
|
|
98
|
Vincke C, Gutiérrez C, Wernery U, Devoogdt
N, Hassanzadeh-Ghassabeh G and Muyldermans S: Generation of single
domain antibody fragments derived from camelids and generation of
manifold constructs. Methods Mol Biol. 907:145–176. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Behar G, Sibéril S, Groulet A, Chames P,
Pugnière M, Boix C, Sautès-Fridman C, Teillaud JL and Baty D:
Isolation and characterization of anti-FcgammaRIII (CD16) llama
single-domain antibodies that activate natural killer cells.
Protein Eng Des Sel. 21:1–10. 2018. View Article : Google Scholar
|
|
100
|
Maussang D, Mujić-Delić A, Descamps FJ,
Stortelers C, Vanlandschoot P, Stigter-van Walsum M, Vischer HF,
van Roy M, Vosjan M, Gonzalez-Pajuelo M, et al: Llama-derived
single variable domains (nanobodies) directed against chemokine
receptor CXCR7 reduce head and neck cancer cell growth in vivo. J
Biol Chem. 288:29562–29572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Farajpour Z, Rahbarizadeh F, Kazemi B and
Ahmadvand D: A nanobody directed to a functional epitope on VEGF,
as a novel strategy for cancer treatment. Biochem Biophys Res
Commun. 446:132–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kim HJ, McCoy MR, Majkova Z, Dechant JE,
Gee SJ, Tabares-da Rosa S, González-Sapienza GG and Hammock BD:
Isolation of alpaca anti-hapten heavy chain single domain
antibodies for development of sensitive immunoassay. Anal Chem.
84:1165–1171. 2012. View Article : Google Scholar :
|
|
103
|
Li K, Zettlitz KA, Lipianskaya J, Zhou Y,
Marks JD, Mallick P, Reiter RE and Wu AM: A fully human scFv phage
display library for rapid antibody fragment reformatting. Protein
Eng Des Sel. 28:307–316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Doshi R, Chen BR, Vibat CR, Huang N, Lee
CW and Chang G: In vitro nanobody discovery for integral membrane
protein targets. Sci Rep. 4:67602014. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ferrari D, Garrapa V, Locatelli M and
Bolchi A: A novel nanobody scaffold optimized for bacterial
expression and suitable for the construction of ribosome display
libraries. Mol Biotechnol. 62:43–55. 2020. View Article : Google Scholar
|
|
106
|
Yau KY, Groves MA, Li S, Sheedy C, Lee H,
Tanha J, MacKenzie CR, Jermutus L and Hall JC: Selection of
hapten-specific single-domain antibodies from a non-immunized llama
ribosome display library. J Immunol Methods. 281:161–175. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Bencurova E, Pulzova L, Flachbartova Z and
Bhide M: A rapid and simple pipeline for synthesis of
mRNA-ribosome-V(H)H complexes used in single-domain antibody
ribosome display. Mol Biosyst. 11:1515–1524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
McMahon C, Baier AS, Pascolutti R,
Wegrecki M, Zheng S, Ong JX, Erlandson SC, Hilger D, Rasmussen SGF,
Ring AM, et al: Yeast surface display platform for rapid discovery
of conformationally selective nanobodies. Nat Struct Mol Biol.
25:289–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Uchański T, Zögg T, Yin J, Yuan D,
Wohlkönig A, Fischer B, Rosenbaum DM, Kobilka BK, Pardon E and
Steyaert J: An improved yeast surface display platform for the
screening of nanobody immune libraries. Sci Rep. 9:3822019.
View Article : Google Scholar
|
|
110
|
Salema V and Fernández LÁ: Escherichia
coli surface display for the selection of nanobodies. Microb
Biotechnol. 10:1468–1484. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Salema V, Marín E, Martínez-Arteaga R,
Ruano-Gallego D, Fraile S, Margolles Y, Teira X, Gutierrez C,
Bodelón G and Fernández LÁ: Selection of single domain antibodies
from immune libraries displayed on the surface of E. coli cells
with two β-domains of opposite topologies. PLoS One. 8:e751262013.
View Article : Google Scholar
|
|
112
|
Salema V, Mañas C, Cerdán L, Piñero-Lambea
C, Marín E, Roovers RC, Van Bergen En Henegouwen PM and Fernández
LÁ: High affinity nanobodies against human epidermal growth factor
receptor selected on cells by E. coli display. MAbs. 8:1286–1301.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Tang J, Li J, Zhu X, Yu Y, Chen D, Yuan L,
Gu Z, Zhang X, Qi L, Gong Z, et al: Novel CD7-specific
nanobody-based immunotoxins potently enhanced apoptosis of
CD7-positive malignant cells. Oncotarget. 7:34070–34083. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Hassel JC, Heinzerling L, Aberle J, Bähr
O, Eigentler TK, Grimm MO, Grünwald V, Leipe J, Reinmuth N, Tietze
JK, et al: Combined immune checkpoint blockade
(anti-PD-1/anti-CTLA-4): Evaluation and management of adverse drug
reactions. Cancer Treat Rev. 57:36–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Gupta A, De Felice KM, Loftus EV Jr and
Khanna S: Systematic review: Colitis associated with anti-CTLA-4
therapy. Aliment Pharmacol Ther. 42:406–417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Savoia P, Astrua C and Fava P: Ipilimumab
(Anti-Ctla-4 Mab) in the treatment of metastatic melanoma:
Effectiveness and toxicity management. Hum Vaccin Immunother.
12:1092–1101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Gao X and McDermott DF: Ipilimumab in
combination with nivolumab for the treatment of renal cell
carcinoma. Expert Opin Biol Ther. 18:947–957. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Tang Z, Mo F, Liu A, Duan S, Yang X, Liang
L, Hou X, Yin S, Jiang X, Vasylieva N, et al: A nanobody against
cytotoxic T-lymphocyte associated antigen-4 increases the
anti-tumor effects of specific CD8+ T cells. J Biomed
Nanotechnol. 15:2229–2239. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Mahoney KM, Freeman GJ and McDermott DF:
The next immune-checkpoint inhibitors: PD-1/PD-L1 blockade in
melanoma. Clin Ther. 37:764–782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Broos K, Lecocq Q, Keersmaecker B, Raes G,
Corthals J, Lion E, Thielemans K, Devoogdt N, Keyaerts M and
Breckpot K: Single domain antibody-mediated blockade of programmed
death-ligand 1 on dendritic cells enhances CD8 T-cell activation
and cytokine production. Vaccines (Basel). 7:852019. View Article : Google Scholar
|
|
121
|
Fang T, Li R, Li Z, Cho J, Guzman JS, Kamm
RD and Ploegh HL: Remodeling of the tumor microenvironment by a
chemokine/Anti-PD-L1 nanobody fusion protein. Mol Pharm.
16:2838–2844. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhang F, Wei H, Wang X, Bai Y, Wang P, Wu
J, Jiang X, Wang Y, Cai H, Xu T and Zhou A: Structural basis of a
novel PD-L1 nanobody for immune checkpoint blockade. Cell Discov.
3:170042017. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Xian Z, Ma L, Zhu M, Li G, Gai J, Chang Q,
Huang Y, Ju D and Wan Y: Blocking the PD-1-PD-L1 axis by a novel
PD-1 specific nanobody expressed in yeast as a potential
therapeutic for immunotherapy. Biochem Biophys Res Commun.
519:267–273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Li S, Jiang K, Wang T, Zhang W, Shi M,
Chen B and Hua Z: Nanobody against PDL1. Biotechnol Lett.
42:727–736. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Liu J, Zhang S, Hu Y, Yang Z, Li J, Liu X,
Deng L, Wang Y, Zhang X, Jiang T and Lu X: Targeting PD-1 and Tim-3
path-ways to reverse CD8 T-cell exhaustion and enhance ex vivo
T-cell responses to autologous dendritic/tumor vaccines. J
Immunother. 39:171–180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Liu JF, Ma SR, Mao L, Bu LL, Yu GT, Li YC,
Huang CF, Deng WW, Kulkarni AB, Zhang WF and Sun ZJ: T-cell
immu-noglobulin mucin 3 blockade drives an antitumor immune
response in head and neck cancer. Mol Oncol. 11:235–247. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Chang X, Lu X, Guo J and Teng GJ:
Interventional therapy combined with immune checkpoint inhibitors:
Emerging opportunities for cancer treatment in the era of
immunotherapy. Cancer Treat Rev. 74:49–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Ascione A, Arenaccio C, Mallano A, Flego
M, Gellini M, Andreotti M, Fenwick C, Pantaleo G, Vella S and
Federico M: Development of a novel human phage display-derived
anti-LAG3 scFv antibody targeting CD8+ T lymphocyte
exhaustion. BMC Biotechnol. 19:672019. View Article : Google Scholar
|
|
129
|
Homayouni V, Ganjalikhani-Hakemi M, Rezaei
A, Khanahmad H, Behdani M and Lomedasht FK: Preparation and
characterization of a novel nanobody against T-cell immunoglobulin
and mucin-3 (TIM-3). Iran J Basic Med Sci. 19:1201–1208.
2016.PubMed/NCBI
|
|
130
|
Ma LL, Zhu M, Li GH, Li YF, Gai JW and Wan
YK: Construction and screening of phage display library for TIM-3
nanobody. Acta Pharmaceutica Sinica. 53:388–395. 2018.
|
|
131
|
Long L, Zhang X, Chen F, Pan Q,
Phiphatwatchara P, Zeng Y and Chen H: The promising immune
checkpoint LAG-3: From tumor microenvironment to cancer
immunotherapy. Genes Cancer. 9:176–189. 2018. View Article : Google Scholar
|
|
132
|
Everett KL, Kraman M, Wollerton FPG,
Zimarino C, Kmiecik K, Gaspar M, Pechouckova S, Allen NL, Doody JF
and Tuna M: Generation of Fcabs targeting human and murine LAG-3 as
building blocks for novel bispecific antibody therapeutics.
Methods. 154:60–69. 2019. View Article : Google Scholar
|
|
133
|
Puhr HC and Ilhan-Mutlu A: New emerging
targets in cancer immunotherapy: The role of LAG3. ESMO Open.
4:e0004822019. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Dmitriev OY, Lutsenko S and Muyldermans S:
Nanobodies as probes for protein dynamics in vitro and in cells. J
Biol Chem. 291:3767–3775. 2016. View Article : Google Scholar :
|
|
135
|
Chanier T and Chames P: Nanobody
engineering: Toward next generation immunotherapies and
immunoimaging of cancer. Antibodies (Basel). 8:132019. View Article : Google Scholar
|
|
136
|
Lecocq Q, Zeven K, De Vlaeminck Y, Martens
S, Massa S, Goyvaerts C, Raes G, Keyaerts M, Breckpot K and
Devoogdt N: Noninvasive imaging of the immune checkpoint LAG-3
using nanobodies, from development to pre-clinical use.
Biomolecules. 9:5482019. View Article : Google Scholar :
|
|
137
|
Lv G, Sun X, Qiu L, Sun Y, Li K, Liu Q,
Zhao Q, Qin S and Lin J: PET imaging of tumor PD-L1 expression with
a highly specific nonblocking single-domain antibody. J Nucl Med.
61:117–122. 2020. View Article : Google Scholar :
|
|
138
|
Broos K, Keyaerts M, Lecocq Q, Renmans D,
Nguyen T, Escors D, Liston A, Raes G, Breckpot K and Devoogdt N:
Non-invasive assessment of murine PD-L1 levels in syngeneic tumor
models by nuclear imaging with nanobody tracers. Oncotarget.
8:41932–41946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Broos K, Lecocq Q, Xavier C, Bridoux J,
Nguyen TT, Corthals J, Schoonooghe S, Lion E, Raes G, Keyaerts M,
et al: Evaluating a single domain antibody targeting human PD-L1 as
a nuclear imaging and therapeutic agent. Cancers (Basel).
11:8722019. View Article : Google Scholar
|
|
140
|
Wan R, Liu A, Hou X, Lai Z, Li J, Yang N,
Tan J, Mo F, Hu Z, Yang X, et al: Screening and antitumor effect of
an anti-CTLA-4 nanobody. Oncol Rep. 39:511–518. 2018.
|
|
141
|
Wang W, Hou X, Yang X, Liu A, Tang Z, Mo
F, Yin S and Lu X: Highly sensitive detection of CTLA-4-positive
T-cell subgroups based on nanobody and fluorescent carbon quantum
dots. Oncol Lett. 18:109–116. 2019.PubMed/NCBI
|