1. Introduction
Severe acute respiratory syndrome coronavirus 2, or
SARS-CoV-2, is the name of the virus responsible for the pandemic
disease Coronavirus Disease 2019 (COVID-19) discovered in Wuhan
(China) in December, 2019, from where it spread to the whole
province of Hubei and and subsequently worldwide (1,2).
The Coronaviridae family of the order Nidovirales is
divided into 2 subfamilies Orthocoronavirinae and
Torovirinae. The family Orthocoronavirinae has 4
described genera: Alphacoronavirus, betacoronavirus,
gammacoronavirus and deltacoronavirus (3). In total, 7 species are known to
infect humans, four of which (HCoV 229E, HCoV NL63, HCoV HKU1 and
HCoV OC43) have been linked to mild cold-like symptoms in
immunocompetent individuals, while two of them, SARS-CoV
originating in China and Middle East respiratory syndrome
coronavirus (MERS-CoV) in the Middle East, were responsible for a
large number of deaths due to severe respiratory infections in 2003
and 2012, respectively (4,5).
SARS-CoV-2, similar to SARS-CoV and MERS-CoV, belongs to the genus
betacoronavirus, whose species cause zoonotic infections that may
affect humans (6). Nevertheless,
it is SARS-CoV that shares most characteristics with SARS-CoV-2,
having approximately 80% homology with its genome (7).
Bats seem to be a natural reservoir for coronavirus.
However, the mechanisms whereby these viruses can affect other
species, including human beings, are not yet well understood
(8). Some scientists have
suggested that, as has occurred with SARS-CoV and MERS-CoV, there
is an intermediate as yet unidentified host that acts as the real
reservoir for SARS-CoV2. Identifying this host is essential to
prevent the transmission of this virus. Thus far, the pangolin was
considered to be the most likely candidate, although other animals
such as minks, snakes or even some turtle species have not been
ruled out (9-11). However, despite its origin as a
zoonotic disease, the most frequent mechanism through which
SARS-CoV-2 causes infection is via human-to-human transmission. The
main route of this spread is through droplets containing viral
particles produced when a person sneezes or coughs. These droplets
are able to settle on mucous membranes in the mouth, nose and eyes
of individuals in close proximity, or the virus is acquired through
contact with contaminated objects (fomites) or surfaces (12). Other proposed routes are the
fecal-oral route (13) or even
vertical transmission (14).
The incubation period for the virus has been
estimated to be approximately 5-7 days, although in certain cases
it can be as long as 14 days. The reproduction rate of SARS-CoV-2
(reflecting its contagiousness) is approximately 2.2-2.6, and its
average serial interval (time elapsed between when symptoms appear
in a transmitting person until they appear in an infected
individual) is 7.5 days (15-17). Viral load is another important
factor for the transmission of this virus. It is known that the
greater viral load is found in the upper respiratory tract until 3
days after symptoms begin. However, transmission has been also
described in some individuals at 1-3 days prior to the onset of
symptoms, indicating the viral load may be sufficient for its
transmission even before symptoms arise (18). Furthermore, it is also considered
that asymptomatic individuals can also transmit this virus to other
individuals (19). These data
need to be kept in mind to prevent the spread of SARS-CoV-2.
2. Molecular features of SARS-CoV-2
infection
Coronaviruses are large (80-220 nm), enveloped,
positive-sense single-stranded RNA (+ssRNA) viruses. Their genome
is the largest of all RNA viruses and can be up to 33.5 kb.
SARS-CoV-2 is a Baltimore group IV virus, meaning its genome is a
+ssRNA whose mRNA is identical to the viral genome (20). The term coronavirus refers to the
crown shape that its proteins make up when observed by electron
microscopy (21). Structurally,
coronaviruses are composed of nucleoproteins that envelope the
+ssRNA, forming a nucleocapsid. This nucleocapsid is bounded by a
lipid envelope with 2 or 3 structural proteins anchored to the
membrane. A major characteristic of betacoronaviruses is that they
usually have haemagglutininesterases (22). The genome of coronaviruses is a
highly conserved structure characterized by the presence of a
replicase gene preceding structural and accessory genes. This last
group of genes will express non-structural proteins by ribosomal
frameshifting (23), while 4
structural proteins are encoded by coronaviruses: E, N, M and S
protein. The sequence of the genome is: 5′-replicase- S-E-M-N-3′
(22). Of note, Phan (24) reported the presence of a wide
range of mutations and deletions in coding and non-coding regions
in 86 genomes of SARS-CoV-2, confirming its mutagenic capacity and
the rapid evolution of this coronavirus.
S protein, or spike surface protein, plays a
prominent role in the entry of SARS-CoV-2 in the host cell by
inter-acting via the viral receptor binding domain (RBD) with the
human angiotensin converting enzyme 2 (ACE-2) receptor. The
structure of this domain is strongly conserved and exhibits a high
degree of similarity with that of SARS-CoV. Nonetheless, some
differences have been described at the C-terminal residue
conferring SARS-CoV-2 a greater affinity for ACE-2 (25). Importantly, this homology means
that some therapies targeting the spike protein used against
SARS-CoV may also prevent the entry of SARS-CoV-2 into the cell
(26). Recently, neuropilin-1
(NRP-1), a protein strongly expressed in the olfactory tract, has
been identified as a novel molecular target of SARS-CoV-2 entry
(27,28). Walls et al (29) demonstrated that S glycoprotein has
a furin cleavage site between subunits S1 and S2, which is
important in the processing of the protein. Similarly, these
authors also described multiple conformations of S protein through
cryo-electron microscopy, highlighting the importance of structural
studies for the development of future vaccines and specific
inhibitors.
Once inside the cell, the virus replicates its
genome starting with the replicase gene. Coronaviruses contains 2
or 3 proteases, which will model the initial polyprotein forming
this transcript. Non-structural proteins then bind to form a
replicase-transcriptase complex, promoting RNA replication and
transcription. During this process, genomic and subgenomic RNA are
produced serving as templates for structural and accessory genes
(18).
Coronaviruses are distinguished through their strong
recombination capacity favoring the appearance of novel
coronaviruses with unpredictable consequences for humans (30). This ability is due to the
non-structural protein, nsp12, also known as RNA-dependent RNA
polymerase (RdRp) and has been proposed as a promising target for
COVID-19 therapy (31).
Finally, novel molecular targets are emerging as
therapies for SARS-CoV-2 infection. Examples are Mpro protease, a
key enzyme for coronavirus replication and transcription (32), or M protein, which plays a main
role in coronavirus morphology and assembly (33) through its interaction with other
components, such as E and N protein. M and E protein are
responsible for formation of the lipid envelope and N protein is
needed for nucleocapsid stabilization, giving rise to N protein-RNA
and an inner core of virions, thus concluding this process
(34,35).
As has been reported, when infecting a cell,
SARS-CoV-2 gains entry by attaching to the host ACE-2 receptor.
ACE-2 expression is widely distributed in different human tissues,
although not all cells that express this enzyme are susceptible or
may be involved in the pathogenesis of COVID-19. There are other
factors, such as host age, sex, ethnicity, or the presence of
comorbidities that may modulate ACE-2 expression and its role in
COVID-19 (36). Pulmonary
epithelial cells, particularly alveolar epithelial cells or type 2
pneumocytes, exhibit a high ACE-2 expression and are considered to
act as a reservoir for viral replication and invasion (37). Type 2 pneumocytes constitute 60%
of alveolar epithelial cells. These pneumocytes are essential for
surfactant synthesis and secretion, xenobiotic metabolism, and
water flow on the lung surface and alveolar epithelial regeneration
following lung damage (38).
Thus, damage to these type 2 pneumocytes can lead to
irreversible lung damage. Moreover, some histological features
related to SARS-CoV-2 have been found in common with SARS-CoV, such
as diffuse bilateral alveolar damage, the formation of hyaline
membranes and pulmonary edema in response to pneumocyte type 1
damage (39). This means that
SARS-CoV-2 may infect any alveolar epithelial cell. Apart from
ACE-2, transmembrane protease serine 2 (TMPRSS2) seems equally
important for the entry of SARS-CoV-2 into the cell. The virus
enters the cytosol via the proteolytic cleavage of viral spike
surface protein through human proteases, such as cathepsin or
TMPRRS2 followed by viral particle-to-membrane fusion (18). Notably, the expression of both
human receptors, ACE-2 and TMPRSS2, has been detected in bronchial
transitory secretory cells, and a higher activity of RHO GTPase has
been found to promote susceptibility to COVID infection (40). In an ex vivo model of
pulmonary SARS-CoV-2 infection, the quantity of viral antigen (N)
is markedly higher than in SARS-CoV models, producing up to
3.2-fold more viral particles, thus explaining its greater
infectivity (41).
The emergence and rapid spread of SARS-CoV-2 has led
to devastating consequences on global health and the economy. Its
clinical features indicate that it is able to evade human immune
surveillance much more effectively than SARS-CoV. Its high
infectivity, mutagenic capacity and ability to evade the immune
system are contributing to its worldwide spread. Understanding the
molecular mechanisms responsible for these characteristics of the
virus are key to gaining control of the virus through novel
treatments, vaccines, diagnostic and screening techniques and
preventive measures. Only through a coordinated multidisciplinary
approach on the part of researchers, healthcare and non-healthcare
professionals will we be able to curb the spread of SARS-CoV-2
(42,43). The present review article
summarizes the relevant data on the epidemiology and social and
clinical management of COVID-19, and discusses the more notable
discoveries of basic and translational research proposing measures
to effectively combat this global concern.
3. Epidemiology and economic burden of
COVID-19: Examples of Italy and Spain
While SARS-CoV was responsible for approximately
8,000 infections and 800 deaths, MERS-CoV affected fewer
individuals and was restricted to Saudi Arabia and a small area of
South Korea (44). To date
(November 17, 2020), by contrast, SARS-CoV-2 has infected
54,771,888 individuals and has been responsible for a staggering
1,324,249 deaths (45). According
to these latest figures, the mortality ratio is 2.46%, much lower
than the 9.5% reported for SARS-CoV, or the 30% attributed to
MERS-CoV (46). Nevertheless,
this percentage notably varies, depending on the country or the
moment; presently, this ratio has been estimated at 15.23% in
several countries, such as France and at <% in Israel or Russia
(47). In addition, it is proving
difficult to accurately identify individuals affected by SARS-CoV-2
and hence, its real associated mortality. For instance, individuals
with no access to diagnostic tests or with false negative test
results are not taken into consideration, as are asymptomatic
individuals or those not seen at a health-care center. Furthermore,
variations in the criteria selected to report probable and
confirmed deaths due to COVID-19 may notably complicate estimates
(48). The vast majority of
affected individuals fall within the age group of 30-79 years, and
in these individuals, COVID-19 usually presents with mild symptoms
or as non-severe pneumonia (49).
By contrast, elderly individuals or individuals with a chronic
disease or cardiopulmonary insufficiency, often require monitoring
primarily 1-2 weeks following the onset of symptoms (50). In children, SARS-CoV-2 infection
is less usual, and symptoms are less severe. This may be due to
lower exposure to sources of contagion or maybe they become
infected in a manner similar to adults; however, cases are more
often subclinical due to biological differences in the functioning
of their immune system. Despite all these factors, children do not
seem to be an important reservoir of SARS-CoV-2 (51,52). Other observations indicate that
males are more severely affected than females. The different
explanations put forward for this have been, for instance, a
greater exposure of males to certain work environment or social
factors, such as smoking, the better efficacy of the innate immune
response in females or even biological differences in ACE-2
activity (53). Moreover,
pregnant women with COVID-19 seem to exhibit the same clinical
manifestations as adults, although SARS-CoV-2 infection may
increase the risk of complications during pregnancy or labor
(54,55).
According to the latest data from the World Health
Organization (WHO), >43% of all cases of COVID-19 have occurred
in the USA (22,438,205 cases) followed by Europe (14,487,598 cases)
and South-East Asia (9,908,674 cases) (45). In Europe, Italy and Spain were the
most affected countries during the first wave along with France,
where the first case of COVID-19 was reported in individuals
arriving from China in mid-January, 2020 (56). Initially, Italy, and particularly,
its northern region, was the hardest-hit country in Europe, and
rapidly became the second country with the majority of cases and
deaths associated with COVID-19 worldwide. Containment measures
began on March 9-11, and March 21st was the day with the highest
number of infections recorded in Italy (57,58). According to the Italian Ministry
of Health, the average age of COVID-19 patients was 62 years, while
81 year-old-patients exhibited the highest mortality rate, those
with 3 or more comorbidities being particularly susceptible
(59). Spain went into full
lockdown on March 14th, 13 days after the start of the exponential
rise in infections (60).
Patients 50 to 59 years of age were the most commonly affected and
the disease was described as potentially lethal in individuals ≥80
years of age (61). In Italy,
lockdown was continued until May 3rd, and measures to curb local or
regional outbreaks were introduced, such as social distancing
(maintaining a 1.5 m safety distance), the use of masks, avoiding
crowds and limiting the number of persons in social events
(62). These measures were also
implemented in Spain, where lockdown easing was conducted in 4
stages (0 to 3) until reaching a state of 'new normality' (63). According to the last data
collected by the WHO from January-February to November, 2020, Italy
had 1,178,529 confirmed cases and suffered 45,229 deaths; however,
of note, from May to October of this year, the number of new
infections and mortality have plateaued (64). During the same time period, Spain
has similarly registered 40,769 deaths and a total of 1,458,591
cases although many of these cases have arisen in the last 3 months
(65). Italy and Spain are
examples among several other countries of a lack of effective
measures taken, identifying a need to redirect measures for better
management of the situation. Apart from causing human suffering,
this pandemic has also affected the economy of each country.
According to the International Monetary Fund (IMF), a 12.8%
reduction in the GDP growth rate is predicted for Spain and Italy
(66,67). To minimize economic impacts on
businesses, the Spanish government has pledged a 200 billion Euros
rescue package whereas the package proposed for Italy is 25 billion
Euros (68). The economic burden
of COVID-19 is a serious concern worldwide and a concerted approach
is required to mitigate its impact on all sectors. In the face of
such turmoil, the hopes of country leaders, researchers and
healthcare systems are turning to the introduction of an effective
vaccine against SARS-CoV-2.
4. Diagnostic and preventive methods
A diagnosis of SARS-CoV-2 can be made according to
its clinical manifestations, such as fever (88.7%), unproductive
cough (67.7%), dyspnea (45.6%), fatigue (29.4%) and other less
frequent symptoms, such as myalgia, sore throat, productive cough,
headache, diarrhea, sickness and/or vomiting and dizziness, in that
order (69). Even so, as
described below, there is also another set of clinical
manifestations that may be associated with COVID-19; however, the
presence of these symptoms may not be sufficient to make an
accurate differential diagnosis; thus, it is necessary to be able
to rely on efficient molecular tests. There are 2 most commonly
used molecular tests to diagnose the virus: Cell cultures and PCR.
In the case of SARS-CoV-2, the performance of cell cultures has
exhibited a very low efficiency due to a delay of at least 3 days
to detect the presence of this virus (70). Thus, RT-qPCR is the most feasible
molecular test owing to the early characterization of the viral
genome. The first RT-qPCR protocol was published on January 23rd,
and targeted the genes RdRp, E and N, whose relevance will be
detailed below (71). Since then,
multiple kits have been developed, allowing for a more rapid and
more sensitive detection of COVID-19. An example is a slight
modification of the initial protocol proposed by Chan et al
(72) that targets RdRp/helicase
(hel), S and N genes to avoid cross-reactivity with other
coronaviruses. Its efficacy, however, varies according to the
laboratory conducting the test. These techniques pursue a high
sensitivity and quick diagnosis (73). They have effectively exhibited a
high efficiency to detect viral particles in different clinical
samples like bronchoalveolar lavage, sputum, saliva, throat and
nasopharyngeal samples (25).
Other diagnostic methods may be especially useful, as is the case
of imaging techniques used to examine lung structure, e.g.,
computed tomography, which can be faster, more sensitive and hence
more reliable, particularly for individuals with fever as a clear
symptom (74). Imaging tests are
less useful, however, for population screening as they require
visits to a hospital.
Measures implemented to prevent and stop the spread
of SARS-CoV-2 have focused on avoiding infected individuals
spreading the virus and also preventing non-affected individuals to
come into contact with the virus (75). One of the measures that is having
more impact is as simple as hand washing; 15 sec can be sufficient
to kill up to 90% of pathogens on a hand, and 30 sec to eliminate
99.9%. The conclusion is that appropriate hand hygiene can prevent
16 to 21% of respiratory viral infections in the general population
(76). However, this precaution
may not be sufficient to protect healthcare personnel from
SARS-CoV-2, as this virus seem, similar to other coronaviruses,
exhibits a high resistance on a wide variety of surfaces such as
metal, glass or plastic. The accurate inhibition and elimination of
the virus is necessary, as it has been shown that contact with
contaminated surfaces can lead to the inadvertent self-inoculation
of mucous membranes in the nose, eyes or mouth (77,78). A review article written by Kampf
et al (79) examines the
most appropriate measures for the correct decontamination of
fomites. One example is the use of ethanol 62-71%, hydrogen
peroxide 0.5% and sodium hypochlorite 0.1% over a 1-min time
period. The efficiency of this method was described as greater than
the use of benzalkonium chloride 0.05-0.2%, chlorhexidine
digluconate 0.02%. Other preventive measures recommended by the WHO
for the general population consist of avoiding transmission to
other individuals by restraining drops from sneezing or coughing
using masks, apart from maintaining a safety distance of 3 m
(80). The protective equipment
recommended include masks, particularly surgical masks or N95
respirators that prevent contact with contaminated droplets from
the mouth and nose, which supposedly are the 2 main entry routes of
SARS-CoV-2 (75).
At the community level, other types of measures have
been implemented, as is the case of quarantine, one of the most
effective measures used to contain the spread of various infectious
diseases throughout history, with the objective of attenuating the
contagion curve. In infected individuals, quarantine should be at
least 2 weeks to prevent infection spreading to other individuals
(81). In healthy individuals,
different countries have imposed different lengths of quarantine
based on factors, such as population screening. Models, such as
that of South Korea have been particularly successful. One of its
secrets has been the rapid development of massive diagnosis
centers, where in merely 10 min, a person can be tested. This
screening is organized as a registration center and a brief
questionnaire, whereby individuals are referred to a test center
for sample collection and are provided with instructions and
detailed information (82).
Measures such as this serve to control not only the infected
population, but also the healthy one, facilitating counts and
avoiding dissemination. Notwithstanding, this strategy and
epidemiological surveillance has met with some difficulties. For
example, patients who have had the disease will continue to return
a positive RT-PCR test, which is not the case with other infectious
diseases (83). Consequently,
continuous communication and coordination between all countries is
warranted, apart from governments, institutions and professionals
from different fields, to establish accurate measures and reliable
information for the population, also highlighting a sense of social
responsibility.
5. Immune system and SARS-CoV-2
The connection between SARS-CoV-2 and the immune
system is one of the main points of study to fully understand the
pathogenesis and impact that COVID-19 may have on patients, as well
as to develop novel effective therapies. When SARS-CoV-2 binds to a
cell through the host receptors ACE-2 and TMPRSS2, it replicates
inside the cell and following the release of virus particles, the
virus promotes host cell entry into pyroptosis. Infected cells
signal various pattern recognition receptors (PRRs), such as
Toll-like receptor 7 (TLR-7), retinoic acid-inducible gene I
(RIG-1), melanoma differentiation-associated protein 5 (MDA5) and
the cyclic GMP-AMP synthase (cGAS)/stimulator of interferon genes
(STING) pathway, thus recognizing viral components such as RNA or
even cytosolic DNA of the affected cell, promoting the production
of type I interferon (IFN-I) and other inflammatory cytokines
(84). Simultaneously,
damage-associated molecular patterns (DAMPs) are also produced,
such as adenosine triphosphate (ATP) or genetic content. The
presence of DAMPs in the tissue will activate the innate immune
response in the lungs, mainly mediated by granulocytes, antigen
presenting cells (APCs) and pro-inflammatory macrophages (85,86). In a proper immune response, this
system is coordinated and supported by an adaptive immune response,
represented by CD4+ T cells, CD8+ T cells,
Tregs and B cells. The activation and proliferation of
CD4+ T cells fosters CD8+ T cells, crucial
for antiviral responses through the induction of cell death.
Likewise, B cells secrete antibodies to neutralize the virus or
induce the lysis of infected cells by complement activation or
antibody-dependent cellular cytotoxicity (mediated by natural
killer (NK) cells) (85). The
effectiveness and coordination of this response will finally
determine the recovery of the patient. Notwithstanding, severe
cases of COVID-19 exhibit a low efficacy of the immune response,
causing cell and tissue damage with systemic implications
characteristic of this disease when compared to other respiratory
infections, such as severe lymphopenia and eosinopenia, pneumonia
with lung damage, acute respiratory distress syndrome (ARDS), along
with other systemic alterations (87,88).
The course of COVID-19 presents in 3 main stages:
Stage I, characterized by primary production of IgM antibodies, and
stage II characterized by exacerbated lung inflammation accompanied
by cytokine storm (89). Stage I
with mild manifestations is the incubation time and the time of
preparation for the adaptive immune response to try to eliminate
the virus. This response is mobilized if the virus outpaces the
innate immune response in situations of a poor health state or if
there are other comorbidities or in the elderly. Firstly, the
innate immune response begins with the activation of TLRs 3,7 and 8
signaling transduction and boosted IFN production. The humoral, or
adaptive, response commences with the production of antibodies
against viral N and S proteins, IgG and IgM, which may appear 4-8
days later. However, if this initial protective phase fails, the
virus disseminates causing multi-organ involvement through the
ACE-2 receptor. In stage II, or the so-called macrophage activation
syndrome, an intense inflammatory response is established featuring
the overexpression of cytokines and chemokines (87,90). Eventually, a third stage may be
distinguished if therapy is ineffective against stage II, including
multiorgan failure and hyperinflammation with typically more severe
symptoms. At this point, anti-viral treatment will not be effective
unless combined with anti-inflammatory and anti-rheumatic drugs
(91). These are the 3 stages of
COVID-19 proposed by Siddiqi and Mehra (92) as summarized in Fig. 1. An awareness of these events is
crucial to design strategies for the prevention of severe disease,
the better management of patients (89) and to ensure the safety of
physicians (90). With the
purpose of containing the virus and preventing the disease
progressing to a severe physiopathology, the most appropriate time
for the administration of each drug is under debate, as it
discussed below.
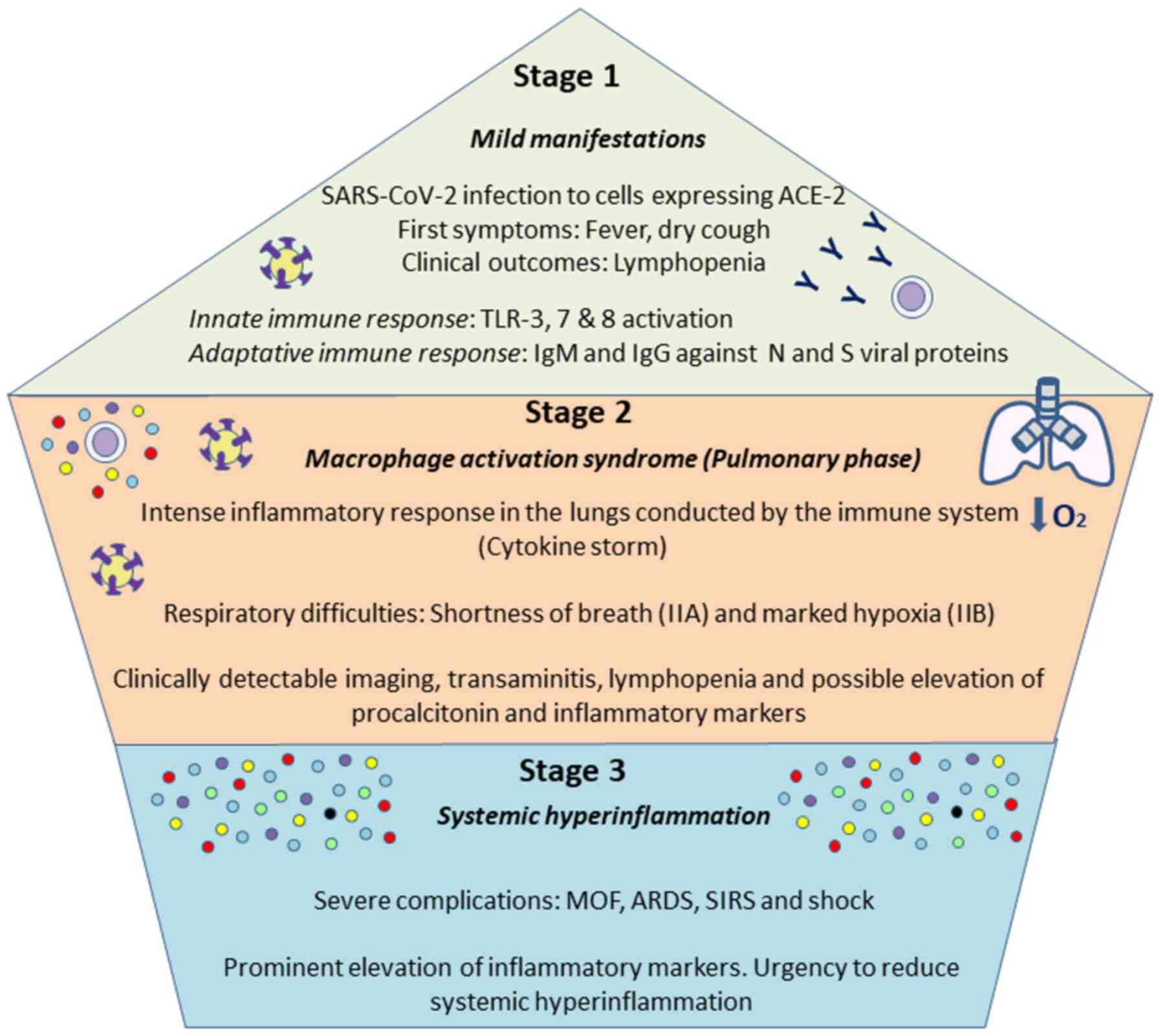 | Figure 1Stages of SARS-CoV-2 course. As
proposed by Siddiqi and Mehra (92), 3 main phases can be distinguished
in COVID-19. The first one is associated with mild symptoms, and
individuals infected will develop and innate and adaptatively
responses, being able to eliminate the virus. If not, a more severe
pulmonary stage will take place, causing a pneumonia without (IIA)
or with hypoxia (IIB), with an aberrant release of cytokines. The
third stage is an hyper-inflammation status, defined by a cytokine
storm and possibly leading to the most severe symptoms, including
ARDS, SIRS or MOF. Addressing the different complications during
the various phases will be vital for a proper management of the
disease. ARDS, acute respiratory distress syndrome; SIRS, systemic
inflammatory response syndrome; MOF, multi-organ failure. |
The main event responsible for an aberrant response
is the phenomenon known as the cytokine storm. Beta corona-viruses
are capable of interacting with the immune system via the membrane
and non-structural proteins forming the virion, hence disrupting
IFN-I signaling (93). This
failure is a trigger for the massive uncontrolled secretion of
cytokines, which will cause the cytokine storm. In addition, it is
known that the higher the viral load, the more likely this
defective response, which is equally related to severity (94). Epithelial respiratory cells,
dendritic cells and macrophages produce and secrete greater levels
of cytokines, among which the following have been highlighted:
Interleukin (IL)-1β, IL-1RA, IL-2, IL-6, IL-7, IL-10, IFN-γ (type
II) tumor necrosis factor-α (TNF-α), IL-8, monocyte chemoattractant
peptide (MCP)-1, macrophage inflammatory protein (MIP)-1A, MIP-1B
and granulocyte-colony stimulating factor (G-CSF) (95).
The effect of cytokines in COVID-19 will depend on
the stage of the disease in which they are produced as well as host
features. For instance, it is known that IL-6 is one of the most
important molecules in the immune system for combatting SARS-CoV-2,
although the role of this cytokine in COVID-19 has not yet been
completely elucidated. Firstly, IL-6 seems to promote a proper
adaptive immunity response by boosting Tregs, CD8+ T
cells and B lymphocytes, amongst other effects. By contrast, IL-6
can promote the polarization of Th lymphocytes to Th2 or Th17 at
the expense of Th1 lymphocytes, which may have aberrant effects on
tissue integrity, extracellular matrix (ECM) structure and the
maintenance of pro-inflammatory neutrophils and macrophages
(96). Consequently, in patients
with severe ARDS, the overactivation of T cells seems to occur,
with a considerable increase in Th17 and CD8+ T cell
cytotoxicity (97). This effect
may be associated with IL-6 hypersecretion. Consistent with
previous research, Zhang et al (98) reported an IL-6 concentration
>10 pg/ml in all samples collected from 82 patients that
succumbed to COVID-19, thus linking higher levels of this cytokine
with its detrimental effects during the cytokine storm. Notably,
these authors also reported findings, such as a
neutro-phil/lymphocyte ratio >5 or elevated levels of
inflammatory markers, such as C reactive protein (CRP). Similarly,
a decrease in CD4+ T cells and CD8+
lymphocytes has been described in severe cases of COVID-19
(99).
This cytokine storm, along with disrupted IFN-I
signaling, will further induce the apoptosis of lung epithelial and
endothelial cells, thus leading to ARDS and multi-organ failure
(MOF) (100,101). ARDS leads to major respiratory
damage, and is responsible for up to 70% of the total demise
produced by COVID-19 (98).
During ARDS, epithelial-endothelial barrier integrity is disrupted,
promoting the entry of macrophages, and aggravating lung damage
which in turn will facilitate virus dissemination (102). ARDS caused by COVID-19 has some
peculiarities compared to other infections, including greater
damage to epithelial than endothelial alveolar cells and clinical
manifestations such as a lower exudation or a delayed onset of the
symptoms from 8 to 12 days (103).
The susceptibility of the main risk groups to severe
COVID-19 may also have its origin in the immune system response.
Non-communicable diseases (NCDs) such as obesity, type 2 diabetes
mellitus, renal insufficiency, liver disease, etc. are importantly
related to low-grade chronic inflammation, characterized by immune
system malfunction with systemic implications (104). This state favors the
exacerbation of the immune response, the knowledge of which may be
essential for proper clinical management of these patients
(105,106). Excessive adipose tissue, typical
of NCDs, is considered to act as a reservoir for SARS-CoV-2, and
this may have a synergic effect with chronic inflammation (107). Elderly individuals present
further significant alterations in their immune function. Aging
causes immunosenescence and disruptions in innate and adaptive
immune responses involving Th involution, which reduces efficiency
to recognize antigens and coordinate defense mechanisms, such as
the defective production of anti-bodies, thus providing a reason
for the vulnerability of these patients to SARS-CoV-2 infection
(84,108).
On the contrary, the immune system may be reinforced
and prepared to minimize its negative effect against SARS-CoV-2.
Immunity is the result of complex interactions of genetic and
acquired factors, such as hormone, metabolic, psychological and
lifestyle factors or an individual's microbiota. Diet has been
identified as a relevant factor, in that the western pattern of
consuming 'fast food' and limited vegetable consumption promotes
the activation of the innate immune response and simultaneously
impairs the performance of the adaptive system (109). A healthy dietary pattern, such
as the Mediterranean diet which is abundant in plant-based foods
and healthy fats could contribute to an appropriate immune
response, due to the intake of components, such as omega-3 fatty
acids. Omega-3 has been attributed a role in immune system
resolution during sepsis (110).
Immune resolution is one of the most intriguing mechanisms proposed
to slow down the cytokine storm responsible for COVID-19
progression (111). Thereby, a
healthy diet may play a key role in the prevention of SARS-CoV-2
infection or reduce its severity (109). Likewise, adherence to physical
activity recommendations and a positive psychology are equally
critical for maintaining a healthy immune system (112).
On the whole, the immune system response in patients
with COVID-19 remains an essential topic of study. The
overactivation of the immune system produces the cytokine storm,
which is one of the most important alterations in severe cases of
COVID-19 (Fig. 2). An early
diagnosis, pharmacotherapy and a healthy lifestyle can reverse or
control this aberrant response, thus supporting the clinical
management of SARS-CoV-2 infection.
 | Figure 2Immune response during SARS-CoV-2
infection. When there is a proper defense mechanism, the innate
immunity of an individual will give rise to cytokines, while
carrying out antigenic presentation, promoting and enhancing a
coordinated adaptive immune response to combat the virus.
SARS-CoV-2 could inhibit type I interferon-mediated signaling,
affecting the innate immune system by additionally acquiring more
DAMPs, thus leading to a cytokine storm. This aberrant response
could promote tissue damage as well as viral sepsis, which may be
detected in blood tests. A high viral load, advanced age or
underlying disease are usually associated with severe cases of
COVID-19, whereas a healthy lifestyle or diet, successful
pharmacotherapy and an early diag-nosis could prevent the
complications of COVID-19 promoting a rapid recovery by controlling
the immune response. DAMPs, damage-associated molecular patterns;
IL, interleukin; IFN-γ, interferon γ; MCP-1, monocyte
chemoattractant protein-1; MIP, macrophage inflammatory protein;
G-CSF, granulocyte colony-stimulating factor; TNF-α, tumor necrosis
factor α. |
6. Endothelial cell involvement and effects
of SARS-CoV-2
The endothelium is one of the most affected
structures associated with COVID-19, having important consequences
when there is an underlying disease, such as thrombosis, renal
disease or cardiovascular compromise, among others (113,114). The vascular endothelium is one
of the organs that more significantly expresses the ACE-2 receptor
(115). Consequently, the
endothelium is proposed as a SARS-COV-2 target. In the study by
Varga et al (116), the
presence of viral particles was detected in endothelial cells,
along with an accumulation of inflammatory cells.
Both the inflammatory response and the infection can
activate the endothelium, leading finally to its dysfunction
associated with greater platelet and leucocyte activation, along
with altered anticoagulant and fibrinolytic mechanisms detected
through some serum markers such as increased levels of D-dimer,
C-reactive protein or TNF-α (117). In fact, elevated serum levels of
some of these compounds have been linked to a poorer prognosis and
a greater severity commencing early on (118). Accordingly, therapy against
endothelial damage and its complications is an interesting target
for severe cases of COVID-19 (119).
Neutrophils can be recruited by activated
endothelial cells and they can also activate the coagulation
cascade through the production of neutrophil extracellular traps
(NETs) in a coordinated manner with monocytes and platelets
(120). These NETs are comprised
of a DNA blend with different toxic compounds present in the
cytosolic granules of these neutrophils that are released in an
explosive way in a process known as NETosis (121).
Investigators are beginning to propose that fibrin
formation occurs in the bosom of NETS after coagulation begins in
its intrinsic and extrinsic pathways (122). The activation of endothelial
cells renders them and monocytes, with their microvesicles, to
express the tissue factor and activate the extrinsic pathway of
coagulation that leads to fibrin deposition and clotting (123). NETs are regulated by their
associated proteins, such as NE, cathepsin G and tissue factor
pathway inhibitor (TFPI), the negative regulator per excellence of
the extrinsic pathway (124). In
the case of the intrinsic pathway, the histones present in NETs
interact with platelets, both with phospholipids and TLRs (125,126). According to research, vascular
occlusions are caused by the inability of the organism to eliminate
these NETS, particularly during sepsis (127). In fact, using different
techniques these NETs have been used as reliable measures of sepsis
and thrombosis (128). Likewise,
NETs are also involved in the cytokine storm, contributing to the
secretion of IL1β, which in turn increases NETs and the production
of other important cytokines like IL-6 (129). Thus, NETs are yet other
interesting target for managing the complications of COVID-19.
NETs are not the only mechanisms that promote these
alterations. Zhang et al (130) reported that increased serum
levels of antiphospholipid antibodies may be involved in thrombus
formation. In parallel, Escher et al (131) described increased levels and
activity of the von Willebrand factor (VWF), arising from
activation and damage to the endothelium promoting its release from
Weibel-Palade bodies. The levels of coagulation factor VIII are
also increased in these patients. Furthermore, fibrin deposits have
been found in pulmonary parenchyma due to the expression of tissue
factor in this tissue with a decrease in plasminogen activator
inhibitor 1 (PAI-1) creating a hypofibrinolytic status (132).
Therefore, on the whole, endothelial dysfunction and
its impacts give rise to some of the major complications of
COVID-19 (Fig. 3). Improving
knowledge of the alterations produced in the endothelium during
infection and trying to control its response could benefit
patients.
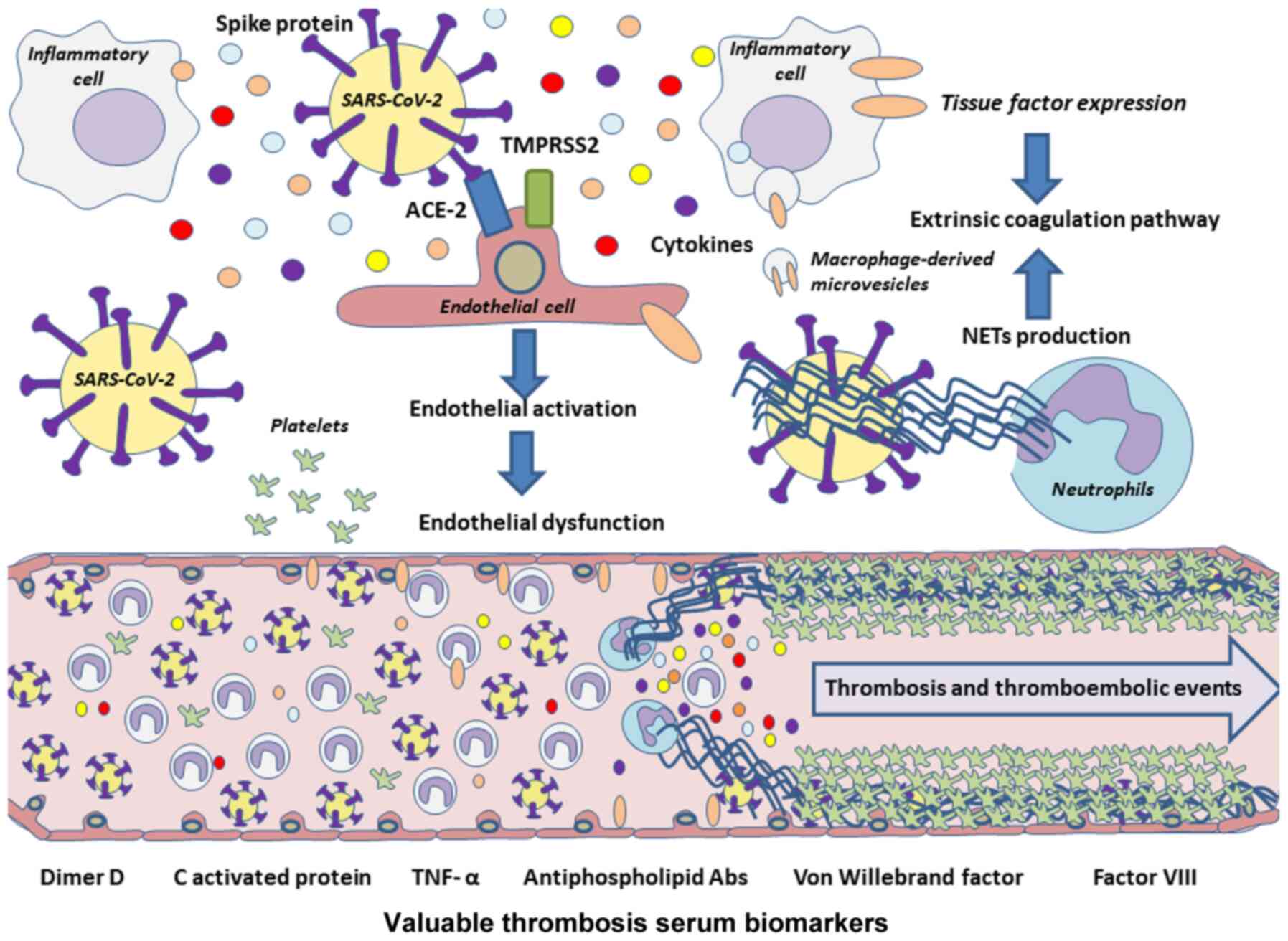 | Figure 3Pathophysiology of thrombotic and
thromboembolic events associated with COVID-19. Owing to the high
expression of the ACE-2 receptor by endothelial cells, they could
be a potential targets of SARS-CoV-2, which may lead to endothelial
cell activation and finally to endothelial dysfunction. Cytokines
released during a cytokine storm also promote endothelial damage,
and along with inflammatory cells and their microvesicles, the
extrinsic coagulation pathway may be activated through further
tissue factor expression. In addition, NETs could lead to
activation of coagulation pathways, acting as inhibitors of the
negative extrinsic pathway regulator TFPI, besides boosting the
production of proinflammatory cytokines, such as IL-6 or IL-1β,
both leading to platelet coagulation and therefore to thrombosis or
thromboembolisms in these patients. NETs, neutrophil extracellular
traps; IL, interleukin; TFPI, tissue factor pathway inhibitor;
TNF-α, tumor necrosis factor α. |
7. Systemic manifestations of
SARS-CoV-2
In December, 2019, scientists began to suspect that
a novel coronavirus was emerging due to a series of patients in the
Chinese city of Wuhan who presented with pneumonia of unknown
etiology as the main clinical manifestation, along with dyspnea,
fever and cough (133,134). These cases resembled the
symptoms produced by SARS-CoV and MERS-CoV in previous years
(135). Following the worldwide
spread of the pandemic, further clinical manifestations have been
added to define a complex syndrome. Several of these manifestations
are related to expression of the ACE-2 receptor in the different
tissues of the human body (136-139) and include a notably array of
respiratory symptoms along with cardiovascular, neurological,
gastrointestinal, hematological, renal and dermatological
alterations (Fig. 4).
However, it should be highlighted that diagnostic
tests have confirmed the existence of individuals with COVID-19
that remain asymptomatic, whose role as carriers of this infection
has become a controversial issue (140,141).
Respiratory manifestations
Respiratory manifestations were first used as
criteria for suspected cases of infection. These manifestations are
related to the presence of the ACE-2 receptor in the respiratory
tract and the special tropism of the virus for this site (137). Fever and cough are the most
common symptoms. Other symptoms are fatigue, anorexia, myalgia; as
well as expectoration, dyspnea and chest tightness (118,142,143). The majority of patients develop
symptoms of pneumonia, characterized by being uni- or bilateral
(more frequently), typically peripheral, with ground glass
opacities and >3 lobules affected. In severe cases, the
infection can lead to ARDS associated with severe hypoxemia and
respiratory failure that requires mechanical ventilation (142). Two different clinical phenotypes
have been described: Type 'L' ('low elastance') and type 'H' ('high
elastance'). Phenotype 'L', apart from a low elastance (i.e., high
compliance) is characterized by a low ventilation-to-perfusion
ratio, and low lung recruitability; thus, perfusion is the main
factor limiting pulmonary function. Phenotype 'H' is characterized
by a high elastance, a high right-to-left shunt and high lung
recruitability. In this phenotype, ventilation is the main limiting
factor of pulmonary function (144).
Cardiovascular manifestations
During the course of infection, biomarkers, such as
troponin, BNP and NT-proBNP may rise. However, there are no
associated electrocardiographic changes or symptoms (145). Moreover, the increase produced
in these biomarkers is related to clinical severity (146). The presence of cardiovascular
risk factors, such as hypertension, diabetes or obesity has been
linked to a poor outcome (139,147). Some authors claim that obesity
is an essential prognostic factor, as obese patients exhibit higher
mortality rates and greater intubation and oxygen requirements
(148). Endothelial cells also
feature the ACE-2 receptor. The virus has been held responsible for
endothelial dysfunction and endotheliitis, leading to a variety of
symptoms according to effects on the different organs and systems
of the human body (116), as
described below. A higher prevalence of distal vein thrombosis has
been described in patients with COVID-19 (149), and this has also been associated
with higher levels of D-dimer (150).
Gastrointestinal manifestations
Since the beginning of the pandemic,
gastrointestinal manifestations, such as nausea, vomiting,
abdominal pain and diarrhea have been associated with COVID-19
(151) due to viral tropism and
the presence of the ACE-2 receptor in the gastrointestinal tract
(138,139). These symptoms may be attributed
to gastritis and enteritis caused by virus infection. In the
majority of patients with gastrointestinal manifestations, diarrhea
seems to be the most common and is usually limited to a median
duration of 4 days. All these manifestations are usually associated
with other typically described symptoms, such as fever or cough
(133,143,151). Gastrointestinal symptoms have
been associated with a fever up to 38.5°C and family clustering
(151). A higher ratio of
chronic liver disease has also been described, as well as increased
bilirubin, ALT and aspartate aminotransferase (AST) levels
(133,152,153). Through PCR, the presence of the
virus has been detected in saliva (154) and feces (155), indicating the important effects
SARS-CoV-2 may have on the gastrointestinal tract.
Hematological manifestations
During the course of infection, normal leucocyte
counts are present in 60% of patients, and a number of patients
exhbit lymphopenia (47%) and high PCR levels (65.9%) (133,156). Patients also feature elevated
levels of transaminases (AST, ALT and lactate dehydrogenase (LDH))
and cardiac enzymes, D-dimer, VSG and procalcitonin. Leukocytosis
and increased levels of procalcitonin indicate the viral origin of
the infection, as is already known. Increases in these parameters
can also be related to disease severity (145). Low platelet counts have been
described in patients with severe COVID-19 and this may be
considered a poor prognostic factor (157).
Neurological manifestations
The presence of the ACE-2 receptor in the vascular
endothelium explains the specific neurotropism of SARS-CoV-2
(158). Neurological symptoms
are present in 36.4% of patients with COVID-19 (159). Headaches are the most common
symptom, while other neurological symptoms, such as dizziness, loss
of consciousness, stroke and convulsions have been described.
Notably, Liguori et al (159) described subjective neurological
symptoms, particularly sleep disturbances in 90% of a cohort of 103
patients hospitalized due to COVID-19. Manifestations affecting the
peripheral nervous system include hypogeusia, hyposmia, vision loss
and neuralgia in the absence of other typical symptoms, such as
fever (160). Other severe
neurological manifestations have been described, such as
encephalitis and meningoencephalitis (161-163). The presence of these
neurological manifestations is a poor prognostic factor in patients
with severe COVID-19 infection.
Renal manifestations
Kidney injury has been observed, with the elevation
of serum creatinine levels in up to 10.9% of cases (133,156). According to Pei et al
(164), patients can develop
renal complications during infection, such as proteinuria,
hematuria or acute renal failure. Furthermore, while almost 50% of
patients recover normal renal function, mortality has been
estimated to be 11.2% higher in patients with renal symptoms than
without these symptoms.
Dermatological manifestations
Cutaneous manifestations of COVID-19 infection
include maculopapular eruptions as the most frequent, followed by
acral erythema with vesicles and pustules (resembling chilblain),
urticarial lesions and vesicular eruptions (165). The more common skin
manifestations seem to be exanthematous eruptions which are
typically heterogeneous and show high variability on presentation
(166).
Other clinical manifestations
Recent investigations have associated ocular
manifestations with COVID-19 infection. In a study conducted by
Chen et al (167),
conjunctival congestion was recorded in up to 5% of patients. Other
ocular symptoms described in that study were ocular pain,
photophobia, dry eye and tearing. Their multivariate regression
analysis revealed an association between ocular symptoms and hand
contact with ocular tissues, which is a risk factor for developing
the infection. In the meta-analysis conducted by Loffredo et
al (168), incidence rates
differed and the incidence rate of conjunctivitis was 1.1%.
However, this latter study demonstrated that hospitalized patients
with severe COVID-19 infection exhibited an increased incidence of
conjunctivitis. Accordingly, conjunctivitis is considered a
possible sign of COVID-19 infection.
Clinical manifestations in pediatric
patients
In children, COVID-19 infection exhibits different
typical clinical characteristics to those described in adults. This
may be due to the immaturity of their tissues and immune system or
to possible cross-immunity produced by the infection with other
respiratory viruses (169). Its
presentation in children is often asymptomatic or with milder
symptoms than those in adults.
According to the meta-analysis conducted by Mustafa
and Selim (170), the most
common symptoms in the pediatric patients analyzed were similar to
those observed in adults: Cough, fever and odynophagia; and in
almost 60% of patients, the infection developed into pneumonia. The
majority of patients presented with mild clinical infection and
only 5% required admission to intensive care. Similar findings have
been reported by Panahi et al (169) including cough, fever and other
respiratory tract symptoms, as well as gastrointestinal symptoms
such as abdominal pain, nausea, vomiting and diarrhea. Laboratory
findings were leucopenia (19%), lymphopenia (21%), increased levels
of C-reactive protein (28%) and procalcitonin (28%); and only 4 %
of patients presented thrombocytopenia (170). In another study by Liguoro et
al (171) these findings
were also reported along with increased levels of CPK and
transaminases. That study, with a total sample size of 7,480
children from Italy, USA and China, also revealed that in 49.1% of
pediatric patients, abnormalities were detected in a radiological
examination even if they were asymptomatic. That study also
estimates a mortality rate of 0.08%.
In pediatric patients with severe infection, the
disease duration is longer than that in adults, and they suffer
septic shock and multiple organ failure as main complications
(172).
Clinical manifestations during
pregnancy
The most common symptoms of COVID-19 infection
during pregnancy are fever, cough, dyspnea, myalgia and fatigue.
The majority of patients exhibit abnormalities in radiological
studies and also feature the same hematological manifestations
described for non-pregnant patients (173). This latter study also suggested
that COVID-10 does not increase the risk of adverse pregnancy
outcomes. There was no evidence of vertical transmission detected
as the virus was not found in amniotic fluid, cord blood,
oropharyngeal samples in the newborn or breast milk.
There has been a description, however, of a neonate
that tested positive for SARS-CoV-2 according to a nasopharyngeal
swab. The mother had a severe case of COVID-19 infection; thus,
possible vertical transmission cannot be ruled out (174). This possibility may be supported
by the presence of the ACE-2 receptor in placental tissue (175).
8. Therapies in use and under
development
Since the beginning of the pandemic, the race
against time has begun in order to identify an appropriate
treatment that is safe and effective against the infection caused
by SARS-CoV-2. Over these months, numerous clinical trials have
included a huge variety of drugs, spanning from already known
antivirals used for other infections to monoclonal antibodies.
According to the results obtained and the current short experience
of outcomes in clinical practice, these therapeutic strategies have
been adapted; however, there are currently no specific treatments
available for infection with SARS-CoV-2 and a number of suggested
drugs are controversial. In Spain, the majority of hospitals have
internal protocols of clinical management that follow the
recommendations for potential treatments of the Spanish Ministry of
Health and Agencia Española de Medicamentos y Productos Sanitarios
(AEMPS; Spanish Agency Of Drugs and Medical Devices) (176).
The WHO has elaborated an interim guidance for the
clinical management of COVID-19. For patients with mild COVID-19,
the updated guidance (177)
recommends symptomatic treatment with antipyretics and adequate
nutrition and rehydration. Patients with risk factors or
complications should be monitored closely and should prompt urgent
care. This guidance argues against antibiotic therapy or
prophylaxis for patients with mild COVID-19. In addition,
antibiotics should not be prescribed for patients with confirmed
moderate COVID-19 unless there is bacterial infection. Supplemental
oxygen therapy must be immediately administered when
SpO2 <90% or when emergency signs are present.
Furthermore, fluid management must be carried out with caution, as
it may worsen oxygenation.
While the clinical management of SARS-CoV-2
infection is based on general measures (close monitoring, oxygen
therapy, analgesia, etc.) and support treatment in the Intensive
Care Unit, numerous drugs are being investigated as possible
therapeutics against this emerging disease (178,179).
In stage I, intravenous immunoglobulin (IVIG) has
been proposed for patients with a deficient humoral response
(180) to avoid viral entry into
new cells, along with chloroquine and hydroxychloroquine, two
agents that reduce glycosylation of ACE-2 receptors (181). Another option is to attenuate
cytokine production by disrupting the kinase signaling pathway
activated following endocytosis. This treatment includes inhibitors
of Janus kinase (JAK) and numb-associated kinase (NAK), such as
baricitinib (182). In stage II,
proposed options are anti-IL1, anti-IL-6 and anti-TNFα agents to
reduce acute inflammation. Colchicine as an anti-IL-1 is being
tested in several clinical trials (NCT04322682, NCT04527562 and
NCT04360980). The WHO does not recommend corticosteroids as a prime
option, although some physicians made use of these drugs when
episodes of ARDS were alarming (183,184).
Remdesivir has been considered a potential drug. It
is a nucleotide analogue prodrug that inhibits RNA-dependent RNA
polymerase, whose effects against Ebola virus were investigated by
Mulangu et al (185). Its
efficacy against other viruses, such as MERS has been demonstrated
in vivo in rhesus macaques (186) or against Nipah virus in African
green monkeys (187). Its use
against SARS-CoV-2, security profile and dosing are still under
investigation; however, the preliminary results in a cohort study
by Grein et al in patients with severe COVID-19 indicated
clinical improvement in 36 out of 53 patients (68%) (188). In a large proportion of
patients, remdesivir has been associated with adverse outcomes,
such as hypotension, elevated liver enzymes, diarrhea and kidney
failure, among others (189).
Its use in Spain has been limited to pregnant patients and
pediatric patients with severe disease (176).
Virus protease activity has been examined as a
possible therapeutic target. Lopinavir and ritonavir are protease
inhibitors used against HIV combined with other retroviral agents
(190). Once their efficacy
against SARS-Co-1 virus was established in vitro by
Groneberg et al(191),
these protease inhibitors were a priori used for the treatment of
SARS-CoV-2 infection. The results of a clinical trial by Cao et
al indicated no therapeutic benefits of these agents in
patients with severe COVID-19 infection (192). A number of authors (193) have responded to the article in
question, arguing that further information is required on its
efficacy prior to abandoning its use. This controversy is being
addressed in the RECOVERY clinical trial that includes 4,972
patients (1,596 treated with lopinavir/ritonavir vs. 3,376 control
patients). The preliminary results again suggest no clinical
benefits and advise against their widespread use (194).
Chloroquine is another drug that has been
extensively used for the management of SARS-CoV-2 infection. Due to
its immunomodulatory effects, this drug has been successfully used
for the treatment of malaria and rheumatic diseases (195). Its action against SARS
coronavirus virus was investigated in vitro in 2005 by
Vincent et al (196) and
it has also exhibited activity against SARS-CoV-2 and in regulating
the immunological response against this virus in vitro
(197). In addition, when
combined with a macrolide, hydroxychloroquine has exhibited
immunomodulatory synergy in in vivo and in vitro
studies (198,199).
However, other authors have come to different
conclusions, reporting that hydroxychloroquine, associated or not
with a macrolide, as a treatment for SARS-CoV-2 infection has no
clinical benefits (200,201). These latter results are
consistent with preliminary findings of the RECOVERY clinical trial
in which 1,542 patients were treated with
hydroxychloroquine-macrolide versus 3,132 control patients; as no
clinical benefits were observed, the widespread use of this
approach is discouraged (202).
Immunotherapy involving convalescent plasma
therapy, mesenchymal stem cell therapy and intravenous
immunoglobulin has also been used for the treatment of or for the
prevention of viral infection. Intravenous immunoglobulin (IVIG)
has offered good outcomes in some patients with COVID-19 by
improving passive immunity and the anti-inflammatory response.
Mesenchymal stem cell therapy could exert an immunomodulatory
effect via the secretion of anti-inflamma-tory factors (203).
Other agents that should be highlighted as possible
candidates for COVID-19 therapy are biological drugs against IL-6,
such as tocilizumab and sarilumab, or IL-1 receptor antagonists,
such as anakinra (204,205). Both classes of agents block the
inflammatory cascade and have been successfully used in refractory
rheumatoid arthritis and other autoimmune diseases (206,207). Tocilizumab has exhibited
potential in reducing the risk of invasive mechanical ventilation
and mortality in patients with severe COVID-19 with pneumonia
(208), and recent results have
indicate that it may palliate symptoms rapidly (209). Anakinra has been tested in
patients with ARDS, suggesting its potential use prior to stage III
disease to curb the worsening of the cytokine storm; however, its
benefits/drawbacks are still under discussion (210).
The association between ILs and the cytokine
release syndrome in severe infection due to SARS CoV-2 has been
described by McGonagle et al, setting up the theoretical
framework for the potential use of this therapy (211). At the clinical level, the
preliminary results of Xu et al in 20 patients with severe
COVID-19 infection indicated a good response to tocilizumab in all
patients in the absence of severe adverse effects (212). Furthermore, in a systematic
review, Alzghari and Acuña recommend the compassionate use of
anti-IL6 in patients with SARS-CoV-2 infection, as its safety and
efficacy remain to be clarified (213). Likewise, numerous therapies have
targeted blocking cell pathways of the inflammatory cascade, such
as the JAK 1 and JAK 2 pathways (with ruxolitinib or baricitinib),
and other ILs (e.g., anti-CD-23) and TNF inhibitors are still being
assessed as possible therapeutic options (214,215).
The systematic review of Mansourabadi et al
(203) concluded that
immunotherapy may be effective in improving the clinical outcomes
of patients with COVID-19. The WHO guidelines recommend that these
drugs should not be administered as prophylaxis outside the context
of clinical trials due to their high rates of adverse effects
(177).
Finally, some authors have advocated the use of
certain drugs in paucisymptomatic and non-hospitalized patients. In
this context, lactoferrin is one of the most promising molecules
for the prevention or aid in the treatment of COVID-19. Chang et
al (216) described the
potential mechanisms whereby this glycoprotein may be used against
SARS-CoV-2, including direct binding of lactoferrin to the virus,
through binding to host cell receptors, such as heparan sulphate
proteoglycans to reduce SARS-CoV-2 surfing and entry and through
the inhibition of viral replication via activation of IFN-α/β.
Pidotimod may be equally effective for the prevention of clinical
worsening by rebalancing the immune status and reducing systemic
effects of disease in ambulatory adult patients without pneumonia
(217). Additional strategies,
such as vitamin C and quercetin co-administration have exhibited
potent overlapping immunomodulatory and antiviral effects, and
these may be useful prophylactic agents in high-risk populations or
may be used as co-adjuvants with other drugs, such as remdesivir
(218).
9. Nanotechnology-based strategies for the
treatment, prevention and diagnosis of COVID-19
The advent of the electron microscopy in the late
1930s was a breakthrough in the history of virology (219). Its high resolving power enabled
the detailed study of viruses and confirmed their nanometric size
of between 20 and 300 nm in diameter (220). Antiviral therapies working at
this scale, such as those provided by nanotechnology, enable
efficient interaction with viral particles and thus maximize the
efficacy of the therapy. The size of nanoparticles (NPs), which
influences bioavailability and circulation time, and their large
surface area-to-volume ratio where large drug payloads can be
attached are promising properties that may overcome the challenges
of traditional antiviral strategies.
Nanotechnology has long been explored for the
treatment of viral infections (221). In particular, strategies
targeting respiratory viruses such as influenza (H1N1) or
respiratory syncytial virus (RSV) are currently in the spotlight
due to similarities to SARS-CoV-2 (222). Among the multiple strategies
available, three main categories of NPs are taking the lead
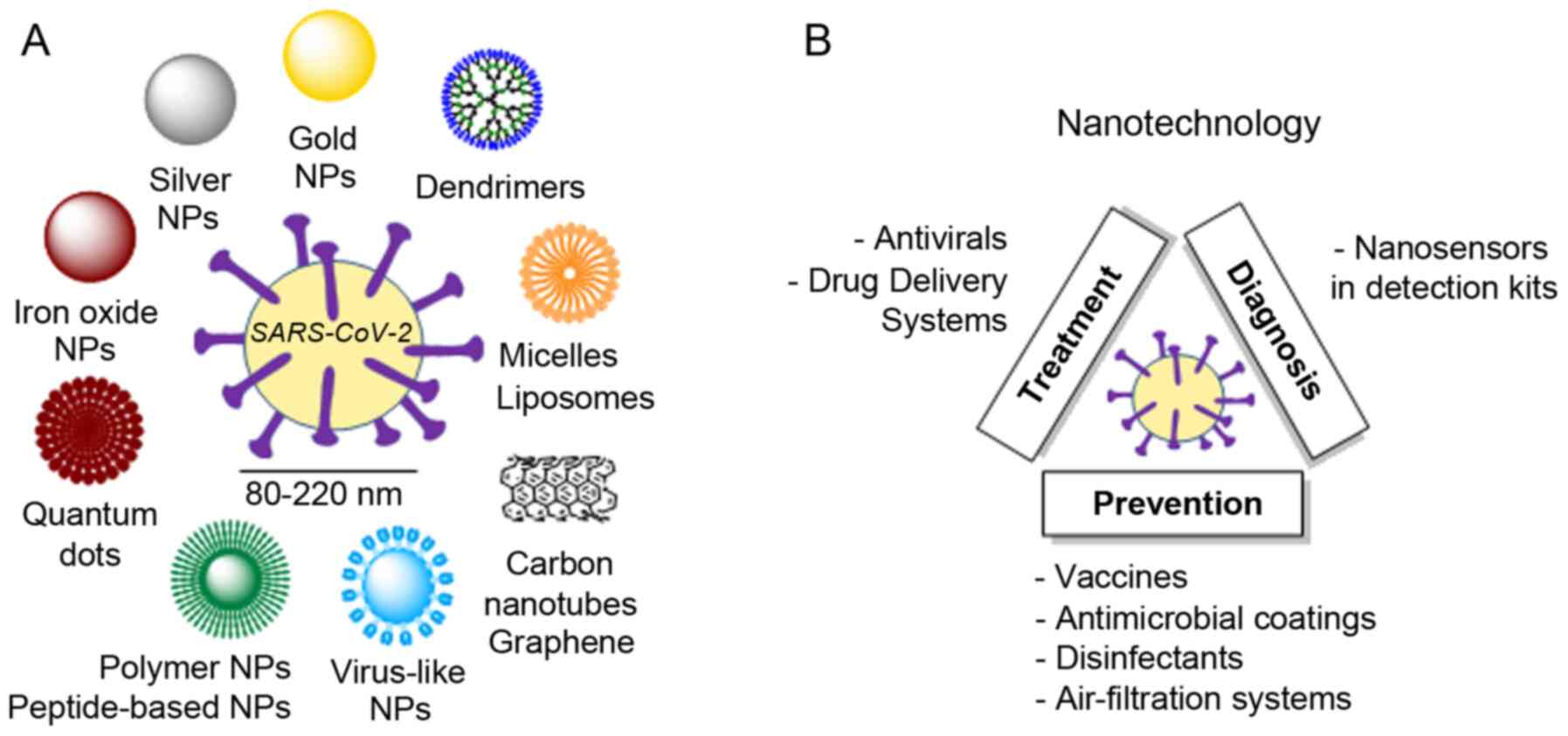
(Fig. 5A): Polymeric NPs,
inorganic NPs and peptide-based NPs (223). The battle against COVID-19 must
consider 3 key principles: Early detection, monitoring and
targeting (224). Relevant
examples of the use of nanotechnology for the treatment, prevention
and diagnosis of SARS-CoV-2 are presented herein (Fig. 5B).
Nanotechnology-based treatment
NPs play an unquestionable role as drug delivery
systems (DDS) (225). By either
encapsulating or binding drugs, NPs improve delivery to the desired
tissue and offer controlled drug release, thus decreasing the
required dose and side-effects. Typical antiviral mechanisms for
NPs range from direct inactivation of the virus (e.g., graphene
oxide), interference in virus attachment to the cell (e.g., silver
or gold NPs, graphene oxide), blockage of viral RNA synthesis
(e.g., Ag2S nanoclusters) or oxidation of viral proteins
(e.g., copper oxide NPs), among others (226).
An example of nano-sized DDS for the treatment of
SARS-CoV-2 relies on chitosan-based polymeric NPs (Novochizol™).
These NPs strongly adhere to lung epithelial tissues, as one of the
main targets of this virus. The potential to provide safe, local
sustained intra-pulmonary drug release is currently being tested
for the delivery of losartan aimed at reducing inflammation and
viral infection. The strategy based on self-assembled peptide NPs
developed by Cui et al (226) is currently being explored using
a peptide that specifically and strongly binds to the spike protein
of coronavirus. These NPs not only protect the peptide from
degradation, but also have water-filled channels to carry antiviral
drugs. COVID-19 infections frequently lead to a hyperinflammatory
state characterized by a fulminant cytokine storm. To combat this
situation, stable squalene-based multidrug NPs are a new approach
for the targeted treatment of acute inflammation with reduced-side
effects (227). Nanobodies, or
single-domain antibodies, are peptide-based NPs with sizes in the
range 2-4 nm that exhibit high stability, low immunogenicity and
excellent affinity and specificity towards the epitope (228). In this context, synthetic
nanobodies against the receptor-binding domain of SARS-CoV-2 spike
protein are being developed as potential inhalable prophylactic
formulations (229).
Nanotechnology-based prevention
The fierce race towards the development of an
efficient vaccine for the novel corona-virus can undoubtedly
benefit from the use of nanocarriers that protect their sensitive
cargo, namely RNA, DNA or protein subunits. Viral vectors, a common
approach in vaccine development (230), are also being explored against
SARS-CoV-2 (e.g., Janssen's AdVac®; TNX-1800, Tonix
Pharmaceuticals; Ad5-nCoV, CanSino Biologics, currently in Phase 2;
or GV-MVA-VLPTM, GeoVax). As an alternative to viral vectors, NPs
have extensively been explored as gene carriers (231). In particular, different
approaches are currently under pre-clinical or clinical assessment
for SARS-CoV-2. In the design of RNA-based vaccines, lipid NPs are
promising nanocarriers for mRNA (232), as currently explored with
mRNA-1273 (Moderna Inc., Phase 1), which codes for a prefusion form
of the spike protein; or with self-amplifying RNA, which is
responsible for sustained protein expression inside the body and
thus a vaccine response at much lower doses than traditional mRNA
vaccines (233)
(LUNAR®, in the preclinical stage). DNA-based vaccines
produce a potent immune response after the direct introduction of a
plasmid encoding the desired antigens into the cells, which later
will produce this antigen. For example, proteo-liposomes (Fusogenix
Platform, in the preclinical stage) are being explored for the
delivery of plasmids encoding multiple protein epitopes from key
immunogenic SARS-CoV-2 proteins. Vaccines based on protein subunits
also employ nanocarriers. NVX-CoV2373 (Novovax Inc., Phase 1/2) is
a recombinant protein NP vaccine (234) able to generate antigens derived
from the spike protein. Virus-like particles (VLPs) induce potent
immune responses and are effective vaccines (235). The VLP strategy is used in
1c-SApNP (Ufovax LLC), comprising a one-component
self-assembling protein NP with SARS-CoV-2 protein spikes
protruding from the scaffold. The molecular clamp platform strategy
consists of the self-assembly of a polypeptide to form an
artificial enveloped virus fusion protein complex which locks the
spike protein into a shape to enable the immune system to recognize
it (University of Queensland). This approach has been successfully
tested for viruses, such as influenza, HIV and Ebola.
While vaccines are a medium-term solution, several
nanotechnology products are currently available to halt the spread
of COVID-19. Nanotechnology enables or enhances the properties of
different medical supplies. A broad variety of these products
employ nanosilver technology, with antimicrobial, antibacterial and
antiviral properties. Examples include cloths (NanoTech
Micro®); adhesive bandages; hand sanitizers (Human
Glove™); textiles (Xstatic®, CERTAINTYTM®);
medical fabrics such as labcoats, curtains, bed sheets (Silver Care
Plus® with X-Static®); toilet paper and paper
towels (Nanotex®). Nanosilver-containing disinfectants
are also commercialized as sanitizers, soaps and detergents. Other
antimicrobial technologies include cationic nanosword coatings
(NANO4-HYGIENE LIFE®), which attract and destroy the
negatively charged membranes of microorganisms; photocatalytic
coatings that destroy any microbe upon activation with light, such
as the mineral nano-crystals (NanoSeptic® self-cleaning
surfaces) and nano-TiO2 (FN-NANO®), used in
hospitals, acute care facilities, airports, etc.; or the dual
TiO2/silver zeolite coating (Invisi Smart™), which has a
duration of 5 years.
High-efficiency nano-based filters have been
implemented in air filtration systems to prevent airborne
contamination in hospitals by trapping or destroying most
infectious particles (bacteria, viruses). A dense network of
nanofibres is the basis behind HyperHEPA® technology
(IQAir) and nanoparticle-covered catalytic filters for photo
electrochemical oxidation technology (Molekule), which destroy air
pollutants through the production of free radicals upon irradiation
with UV-A light. Nanofiber technology, in combination with copper
dioxide NPs, is also implemented in face masks
(ReSpimask®) that trap and destroy the virus.
Self-sterilizing face masks (Guardian G-Volt) are based on
laser-induced graphene filters which eliminate trapped
microorganisms through an electrical charge.
Nanotechnology-based diagnosis
The control of the COVID-19 pandemic requires the
rapid, yet precise detection of novel coronavirus cases.
Traditional detection methods that identify nucleic acids have
several drawbacks including low sensitivity, lengthy protocols, a
high rate of false negatives and a lack of specificity towards a
particular virus. The nanometric size and multivalence of NPs can
overcome these drawbacks by greatly amplifying the signal, thus
decreasing the amount of samples needed and increasing the
effectiveness of detection kits (236). Examples of NPs used for the
diagnosis of SARS-CoV-2 include polymer-coated iron oxide NPs
(237), which potently bind to
the viral RNA and can be later extracted from the solution using a
magnet, simplifying the purification process before RT-PCR
reaction; and gold nanorods or nanoparticles (Sona Nanotech Inc.
and SureScreen Diagnostics Ltd.) implemented in disposable
quick-response lateral-flow tests, which detect COVID-19
biomarkers, IgG and IgM, within 5 to 15 min. A different
nanotechnological strategy is the use of nanopore sequencing, an
electrophoresis-based tool for the sequencing of a single molecule
of DNA or RNA without the need for PCR amplification. Portable DNA
and RNA sequencers (MinION®) employ this strategy for
the rapid and simultaneous detection of SARS-CoV-2 and other
respiratory viruses within 6 to 10 h (238).
10. Proposals for control
SARS-CoV-2 is an emerging infectious agent that has
had a marked impact on society in recent months. Although its
exponential increase has diminished, the implementation of an
appropriate surveillance and action strategy is required, which may
aid in the control of disease outbreaks which, as had occurred with
the Spanish flu, can be fatal (239). This strategy will involve a
gradual return to normality while continuing to follow WHO hygiene
and prevention recommendations along with individual and population
screening to assess the situation at all times. Citizen awareness
and appropriate coordination of governments, countries and
institutions will play a key role in this ongoing process.
Funding
The present review article was funded by grants
from the B2017/BMD-3804 MITIC-CM (Community of Madrid, Spain) and
University of Alcalá (COVID-19 UAH 2019/00003/016/001/023),
co-financed by the European Development Regional Fund 'A way to
achieve Europe' (ERDF).
Availability of data and materials
Not applicable.
Authors' contributions
MAO, OFM, CGM, SGG, MAAM, LST, LP, AA and BDLT were
involved in the writing of the original draft and in the literature
search. MAO, OFM, DTC, NGH, MAM, JB and AA were involved in the
writing, reviewing and editing of the manuscript. MAM, AA and BDLT
provided supervision. MAO, OFM, LP and DTC created the figures.
MAM, AA and BDLT provided resources. MAO, OFM, SGG, DTC and AA were
involved in the conceptualization of the study.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, Lu R, et al: A Novel coronavirus from
patients with pneumonia in China, 2019. N Engl J Med. 382:727–733.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Q, Guan X, Wu P, Wang X, Zhou L, Tong
Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al: Early transmission
dynamics in Wuhan, China, of novel coronavirus-infected pneu-monia.
N Engl J Med. 382:1199–1207. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Woo PC, Huang Y, Lau SK and Yuen KY:
Coronavirus genomics and bioinformatics analysis. Viruses.
2:1804–1820. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Wit E, van Doremalen N, Falzarano D and
Munster VJ: SARS and MERS: Recent insights into emerging
coronaviruses. Nat Rev Microbiol. 14:523–534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hasöksüz M, Kiliç S and Saraç F:
Coronaviruses and SARS-COV-2. Turk J Med Sci. 50(SI-1): 549–556.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ashour HM, Elkhatib WF, Rahman MM and
Elshabrawy HA: Insights into the recent 2019 novel coronavirus
(SARS-CoV-2) in light of past human coronavirus outbreaks.
Pathogens. 9:1862020. View Article : Google Scholar :
|
|
7
|
Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan
S and Yuen KY: Genomic characterization of the 2019 novel
human-pathogenic coronavirus isolated from a patient with atypical
pneumonia after visiting Wuhan. Emerg Microbes Infect. 9:221–236.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Banerjee A, Kulcsar K, Misra V, Frieman M
and Mossman K: Bats and coronaviruses. Viruses. 11:412019.
View Article : Google Scholar :
|
|
9
|
Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H,
Zhu J, Zhang Q, Wu J and Liu L: Composition and divergence of
coronavirus spike proteins and host ACE2 receptors predict
potential intermediate hosts of SARS-CoV-2. J Med Virol.
92:595–601. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu J, Zhao S, Teng T, Abdalla AE, Zhu W,
Xie L, Wang Y and Guo X: Systematic comparison of two
animal-to-human trans-mitted human coronaviruses: SARS-CoV-2 and
SARS-CoV. Viruses. 12:2442020. View Article : Google Scholar
|
|
11
|
Zheng J: SARS-CoV-2: An emerging
coronavirus that causes a global threat. Int J Biol Sci.
16:1678–1685. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang Y, Peng F, Wang R, Yange M, Guan K,
Jiang T, Xu G, Sun J and Chang C: The deadly coronaviruses: The
2003 SARS pandemic and the 2020 novel coronavirus epidemic in
China. J Autoimmun. 109:1024342020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuen KS, Ye ZW, Fung SY, Chan CP and Jin
DY: SARS-CoV-2 and COVID-19: The most important research questions.
Cell Biosci. 10:402020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peyronnet V, Sibiude J, Deruelle P,
Huissoud C, Lescure X, Lucet JC, Mandelbrot L, Nisand I, Vayssière
C, Yazpandanah Y, et al: SARS-CoV-2 infection during pregnancy.
Information and proposal of management care. CNGOF Gynecol Obstet
Fertil Senol. 48:436–443. 2020.In French.
|
|
15
|
Jiang X, Rayner S and Luo MH: Does
SARS-CoV-2 has a longer incubation period than SARS and MERS? J Med
Virol. 92:476–478. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lai A, Bergna A, Acciarri C, Galli M and
Zehender G: Early phylogenetic estimate of the effective
reproduction number of SARS-CoV-2. J Med Virol. 92:675–679. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai CC, Shih TP, Ko WC, Tang HJ and Hsueh
PR: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
and coronavirus disease-2019 (COVID-19): The epidemic and the
challenges. Int J Antimicrob Agents. 55:1059242020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Malik YA: Properties of coronavirus and
SARS-CoV-2. Malays J Pathol. 42:3–11. 2020.PubMed/NCBI
|
|
19
|
Yu X and Yang R: COVID-19 transmission
through asymptomatic carriers is a challenge to containment.
Influenza Other Respir Viruses. 14:474–475. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baltimore D: Expression of animal virus
genomes. Bacteriol Rev. 35:235–241. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Englund JA, Kim YJ and McIntosh K: Human
coronaviruses, including Middle East respiratory syndrome
coronavirus. Feigin and Cherry's textbook of pediatric infectious
disease. Cherry J, Demmler Harrison GJ, Kaplan SL, Steinbach WJ and
Hotez PJ: 8th edition. Elsevier Inc; Philadelphia, PA, pp: pp.
1846–1854. 2019
|
|
22
|
Park SE: Epidemiology, virology, and
clinical features of severe acute respiratory
syndrome-coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19). Clin
Exp Pediatr. 63:119–124. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fehr AR and Perlman S: Coronaviruses: An
overview of their replication and pathogenesis. Methods Mol Biol.
1282:1–23. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Phan T: Genetic diversity and evolution of
SARS-CoV-2. Infect Genet Evol. 81:1042602020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Q, Zhang Y, Wu L, Niu S, Song C,
Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, et al: Structural and
functional basis of SARS-CoV-2 entry by using human ACE2. Cell.
181:894–904.e9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S,
Zhang Q, Shi X, Wang Q, Zhang L and Wang X: Structure of the
SARS-CoV-2 spike receptor-binding domain bound to the ACE2
receptor. Nature. 581:215–220. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cantuti-Castelvetri L, Ojha R, Pedro LD,
Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya
T, Anastasina M, et al: Neuropilin-1 facilitates SARS-CoV-2 cell
entry and infectivity. Science. 370:856–860. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Daly JL, Simonetti B, Klein K, Chen KE,
Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer
M, Hollandi R, et al: Neuropilin-1 is a host factor for SARS-CoV-2
infection. Science. 370:861–865. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Walls AC, Park YJ, Tortorici MA, Wall A,
McGuire AT and Veesler D: Structure, function, and antigenicity of
the SARS-CoV-2 spike glycoprotein. Cell. 181:281–292.e6. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou
J, Liu W, Bi Y and Gao GF: Epidemiology, genetic recombination, and
pathogenesis of coronaviruses. Trends Microbiol. 24:490–502. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Elfiky AA: SARS-CoV-2 RNA dependent RNA
polymerase (RdRp) targeting: An in silico perspective. J Biomol
Struct Dyn. 1–9. 2020.Epub ahead of print.
|
|
32
|
Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y,
Zhang B, Li X, Zhang L, Peng C, et al: Structure of Mpro
from SARS-CoV-2 and discovery of its inhibitors. Nature.
82:289–293. 2020. View Article : Google Scholar
|
|
33
|
Neuman BW, Kiss G, Kunding AH, Bhella D,
Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, et
al: A structural analysis of M protein in coronavirus assembly and
morphology. J Struct Biol. 174:11–22. 2011. View Article : Google Scholar
|
|
34
|
Vennema H, Godeke GJ, Rossen JW, Voorhout
WF, Horzinek MC, Opstelten DJ and Rottier PJ:
Nucleocapsid-independent assembly of coronavirus-like particles by
co-expression of viral envelope protein genes. EMBO J.
15:2020–2028. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Escors D, Ortego J, Laude H and Enjuanes
L: The membrane M protein carboxy terminus binds to transmissible
gastroenteritis coronavirus core and contributes to core stability.
J Virol. 75:1312–1324. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bourgonje AR, Abdulle AE, Timens W,
Hillebrands JL, Navis GJ, Gordijn SJ, Bolling MC, Dijkstra G, Voors
AA, Osterhaus AD, et al: Angiotensin-converting enzyme-2 (ACE2),
SARS-CoV-2 and the pathophysiology of coronavirus disease 2019
(COVID-19). J Pathol. 251:228–248. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang H, Penninger JM, Li Y, Zhong N and
Slutsky AS: Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2
receptor: Molecular mechanisms and potential therapeutic target.
Intensive Care Med. 46:586–590. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Castranova V, Rabovsky J, Tucker JH and
Miles PR: The alveolar type II epithelial cell: A multifunctional
pneumocyte. Toxicol Appl Pharmacol. 93:472–483. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rockx B, Kuiken T, Herfst S, Bestebroer T,
Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den
Brand J, Okba NMA, et al: Comparative pathogenesis of COVID-19,
MERS, and SARS in a nonhuman primate model. Science. 368:1012–1015.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lukassen S, Chua RL, Trefzer T, Kahn NC,
Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, et
al: SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in
bronchial transient secretory cells. EMBO J. 39:e1051142020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chu H, Chan JF, Wang Y, Yuen TT, Chai Y,
Hou Y, Shuai H, Yang D, Hu B, Huang X, et al: Comparative
replication and immune activation profiles of SARS-CoV-2 and
SARS-CoV in human lungs: An ex vivo study with implications for the
patho-genesis of COVID-19. Clin Infect Dis. 71:1400–1409. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pachetti M, Marini B, Benedetti F, Giudici
F, Mauro E, Storici P, Masciovecchio C, Angeletti S, Ciccozzi M,
Gallo RC, et al: Emerging SARS-CoV-2 mutation hot spots include a
novel RNA-dependent-RNA polymerase variant. J Transl Med.
18:1792020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Holmes EA, O'Connor RC, Perry VH, Tracey
I, Wessely S, Arseneault L, Ballard C, Christensen H, Cohen Silver
R, Everall I, et al: Multidisciplinary research priorities for the
COVID-19 pandemic: A call for action for mental health science.
Lancet Psychiatry. 7:547–560. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Roussel Y, Giraud-Gatineau A, Jimeno MT,
Rolain JM, Zandotti C, Colson P and Raoult D: SARS-CoV-2: Fear
versus data. Int J Antimicrob Agents. 55:1059472020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
World Health Organization (WHO): WHO
Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/urisimplehttps://covid19.who.int/.
Retrieved November 17, 2020.
|
|
46
|
Sohrabi C, Alsafi Z, O'Neill N, Khan M,
Kerwan A, Al-Jabir A, Iosifidis C and Agha R: World Health
Organization declares global emergency: A review of the 2019 novel
coronavirus (COVID-19). Int J Surg. 76:71–76. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bulut C and Kato Y: Epidemiology of
COVID-19. Turk J Med Sci. 50:563–570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
New York City Department of Health and
Mental Hygiene (DOHMH) COVID-19 Response Team: Preliminary estimate
of excess mortality during the COVID-19 outbreak-New York City,
March 11-May 2, 2020. MMWR Morb Mortal Wkly Rep. 69:603–605. 2020.
View Article : Google Scholar
|
|
49
|
The epidemiological characteristics of an
outbreak of 2019 novel coronavirus diseases (COVID-19) in China.
Zhonghua Liu Xing Bing Xue Za Zhi. 41:145–151. 2020.In Chinese.
|
|
50
|
Goh KJ, Choong MC, Cheong EH, Kalimuddin
S, Duu Wen S, Phua GC, Chan KS and Haja Mohideen S: Rapid
progression to acute respiratory distress syndrome: Review of
current under-standing of critical illness from COVID-19 infection.
Ann Acad Med Singap. 49:108–118. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Balasubramanian S, Rao NM, Goenka A,
Roderick M and Ramanan AV: Coronavirus Disease 2019 (COVID-19) in
children-what we know so far and what we do not? Indian Pediatr.
57:435–442. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zimmermann P and Curtis N: Coronavirus
infections in children including COVID-19: An overview of the
epidemiology, clinical features, diagnosis, treatment and
prevention options in children. Pediatr Infect Dis J. 39:355–368.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sharma G, Volgman AS and Michos ED: Sex
differences in mortality from COVID-19 pandemic: Are men vulnerable
and women protected? JACC Case Rep. 2:1407–1410. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu Y, Chen H, Tang K and Guo Y: Clinical
manifestations and outcome of SARS-CoV-2 infection during
pregnancy. J Infect. Mar 4–2020.Epub ahead of print.
|
|
55
|
Rasmussen SA, Smulian JC, Lednicky JA, Wen
TS and Jamieson DJ: Coronavirus Disease 2019 (COVID-19) and
pregnancy: What obstetricians need to know. Am J Obstet Gynecol.
222:415–426. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lescure FX, Bouadma L, Nguyen D, Parisey
M, Wicky PH, Behillil S, Gaymard A, Bouscambert-Duchamp M, Donati
F, Le Hingrat Q, et al: Clinical and virological data of the first
cases of COVID-19 in Europe: A case series. Lancet Infect Dis.
20:697–706. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Remuzzi A and Remuzzi G: COVID-19 and
Italy: What next? Lancet. 395:1225–1228. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Saglietto A, D'Ascenzo F, Zoccai GB and De
Ferrari GM: COVID-19 in Europe: The Italian lesson. Lancet.
395:1110–1111. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
COVID-19 Situazione in Italia. http://www.salute.gov.it/portale/nuovocoronavirus/dettaglioContenutiNuovoCorona-virus.jsp?lingua=italiano&id=5351&area=nuovoCoronavirus&menu=vuotourisimplehttp://www.salute.gov.it/portale/nuovocoronavirus/dettaglioContenutiNuovoCorona-virus.jsp?lingua=italiano&id=5351&area=nuovoCoronavirus&menu=vuoto.
Retrieved May 29, 2020.
|
|
60
|
Saez M, Tobias A, Varga D and Barceló MA:
Effectiveness of the measures to flatten the epidemic curve of
COVID-19. The case of Spain. Sci Total Environ. 727:1387612020.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Update no. 120. Coronavirus disease
(COVID-19). 29–05. 2020, https://www.mscbs.gob.es/en/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Actualizacion_120_COVID-19.pdfurisimplehttps://www.mscbs.gob.es/en/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Actualizacion_120_COVID-19.pdf.
Retrieved May 29, 2020.
|
|
62
|
Castaldi S, Romano L, Pariani E, Garbelli
C and Biganzoli E: COVID-19: The end of lockdown what next? Acta
Biomed. 91:236–238. 2020.PubMed/NCBI
|
|
63
|
Mateos R, Fernández M, Franco M and
Sánchez M: COVID-19 in Spain. Coming back to the 'new normality'
after 2 months of confinement. Int Psychogeriatr. 32:1169–1172.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
World Health Organization (WHO): Dashboard
Italy From January 29 to 17 November 2020. https://covid19.who.int/region/euro/country/iturisimplehttps://covid19.who.int/region/euro/country/it.
|
|
65
|
World Health Organization (WHO): Dashboard
Spain From January 29 to 17 November 2020. https://covid19.who.int/region/euro/country/esurisimplehttps://covid19.who.int/region/euro/country/es.
|
|
66
|
International Monetary Fund Italy.
https://www.imf.org/en/Countries/ITAurisimplehttps://www.imf.org/en/Countries/ITA.
Retrieved 12 August 12, 2020.
|
|
67
|
International Monetary Fund Spain.
https://www.imf.org/en/Countries/ESP#countrydataurisimplehttps://www.imf.org/en/Countries/ESP#countrydata.
Retrieved August 12, 2020.
|
|
68
|
Nicola M, Alsafi Z, Sohrabi C, Kerwan A,
Al-Jabir A, Iosifidis C, Agha M and Agha R: The socio-economic
implications of the coronavirus pandemic (COVID-19): A review. Int
J Surg. 78:185–193. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rabaan AA, Al-Ahmed SH, Haque S, Sah R,
Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK and
Rodriguez-Morales AJ: SARS-CoV-2, SARS-CoV, and MERS-COV: A
comparative overview. Infez Med. 28:174–184. 2020.PubMed/NCBI
|
|
70
|
Zhou P, Yang XL, Wang XG, Hu B, Zhang L,
Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al: A pneumonia outbreak
associ-ated with a new coronavirus of probable bat origin. Nature.
579:270–273. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Corman VM, Landt O, Kaiser M, Molenkamp R,
Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML,
et al: Detection of 2019 novel coronavirus (2019-nCoV) by real-time
RT-PCR. Euro Surveill. 25:20000452020. View Article : Google Scholar :
|
|
72
|
Chan JF, Yip CC, To KK, Tang TH, Wong SC,
Leung KH, Fung AY, Ng AC, Zou Z, Tsoi HW, et al: Improved molecular
diagnosis of COVID-19 by the novel, highly sensitive and specific
COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR assay
validated in vitro and with clinical specimens. J Clin Microbiol.
58:e00310–20. 2020. View Article : Google Scholar :
|
|
73
|
Artika IM, Wiyatno A and Ma'roef CN:
Pathogenic viruses: Molecular detection and characterization.
Infect Genet Evol. 81:1042152020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dai WC, Zhang HW, Yu J, Xu HJ, Chen H, Luo
SP, Zhang H, Liang LH, Wu XL, Lei Y and Lin F: CT Imaging and
differential diagnosis of COVID-19. Can Assoc Radiol J. 71:195–200.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX,
Wang QZ, Sun C, Sylvia S, Rozelle S, Raat H and Zhou H:
Epidemiology, causes, clinical manifestation and diagnosis,
prevention and control of coronavirus disease (COVID-19) during the
early outbreak period: A scoping review. Infect Dis Poverty.
9:292020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yan Y, Shin WI, Pang YX, Meng Y, Lai J,
You C, Zhao H, Lester E, Wu T and Pang CH: The First 75 Days of
Novel Coronavirus (SARS-CoV-2) Outbreak: Recent advances,
prevention, and treatment. Int J Environ Res Public Health.
17:23232020. View Article : Google Scholar :
|
|
77
|
Otter JA, Donskey C, Yezli S, Douthwaite
S, Goldenberg SD and Weber DJ: Transmission of SARS and MERS
coronaviruses and influenza virus in healthcare settings: The
possible role of dry surface contamination. J Hosp Infect.
92:235–250. 2016. View Article : Google Scholar
|
|
78
|
Dowell SF, Simmerman JM, Erdman DD, Wu JS,
Chaovavanich A, Javadi M, Yang JY, Anderson LJ, Tong S and Ho MS:
Severe acute respiratory syndrome coronavirus on hospital surfaces.
Clin Infect Dis. 39:652–657. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kampf G, Todt D, Pfaender S and Steinmann
E: Persistence of coronaviruses on inanimate surfaces and their
inactivation with biocidal agents. J Hosp Infect. 104:246–251.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
World Health Organization (WHO): Novel
Coronavirus (2019-nCoV) Advice for the public. WHO; Geneva:
2020
|
|
81
|
Güner R, Hasanoğlu I and Aktaş F:
COVID-19: Prevention and control measures in community. Turk J Med
Sci. 50:571–577. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kwon KT, Ko JH, Shin H, Sung M and Kim JY:
Drive-Through screening center for COVID-19: A safe and efficient
screening system against massive community outbreak. J Korean Med
Sci. 35:e1232020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lan L, Xu D, Ye G, Xia C, Wang S, Li Y and
Xu H: Positive RT-PCR test results in patients recovered from
COVID-19. JAMA. 323:1502–1503. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Nikolich-Zugich J, Knox KS, Rios CT, Natt
B, Bhattacharya D and Fain MJ: SARS-CoV-2 and COVID-19 in older
adults: What we may expect regarding pathogenesis, immune
responses, and outcomes. Geroscience. 42:505–514. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Tay MZ, Poh CM, Rénia L, MacAry PA and Ng
LFP: The trinity of COVID-19: Immunity, inflammation and
intervention. Nat Rev Immunol. 20:363–374. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Shi Y, Wang Y, Shao C, Huang J, Gan J,
Huang X, Bucci E, Piacentini M, Ippolito G and Melino G: COVID-19
infection: The perspectives on immune responses. Cell Death Differ.
27:1451–1454. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ahmadpoor P and Rostaing L: Why the immune
system fails to mount an adaptive immune response to a COVID-19
infection. Transpl Int. 33:824–825. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Azkur AK, Akdis M, Azkur D, Sokolowska M,
van de Veen W, Brüggen MC, O'Mahony L, Gao Y, Nadeau K and Akdis
CA: Immune response to SARS-CoV-2 and mechanisms of
immuno-pathological changes in COVID-19. Allergy. 75:1564–1581.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
de Simone G and Mancusi C: Finding the
right time for anti-inflammatory therapy in COVID-19. Int J Infect
Dis. 101:247–248. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Shah VK, Firmal P, Alam A, Ganguly D and
Chattopadhyay S: Overview of immune response during SARS-CoV-2
infection: Lessons from the past. Front Immunol. 11:19492020.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Soy M, Keser G, Atagündüz P, Tabak F,
Atagündüz I and Kayhan S: Cytokine storm in COVID-19: Pathogenesis
and overview of anti-inflammatory agents used in treatment. Clin
Rheumatol. 39:2085–2094. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Siddiqi HK and Mehra MR: COVID-19 illness
in native and immunosuppressed states: A clinical-therapeutic
staging proposal. J Heart Lung Transplant. 39:405–407. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Prompetchara E, Ketloy C and Palaga T:
Immune responses in COVID-19 and potential vaccines: Lessons
learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol.
38:1–9. 2020.PubMed/NCBI
|
|
94
|
Hadjadj J, Yatim N, Barnabei L, Corneau A,
Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux
C, et al: Impaired type I interferon activity and exacerbated
inflammatory responses in severe COVID-19 patients. Science.
369:718–724. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tufan A, Avanoğlu Güler A and
Matucci-Cerinic M: COVID-19, immune system response,
hyperinflammation and repurposing antirheumatic drugs. Turk J Med
Sci. 50(SI-1): 620–632. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gubernatorova EO, Gorshkova EA, Polinova
AI and Drutskaya MD: IL-6: Relevance for immunopathology of
SARS-CoV-2. Cytokine Growth Factor Rev. 53:13–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Xu Z, Shi L, Wang Y, Zhang J, Huang L,
Zhang C, Liu S, Zhao P, Liu H, Zhu L, et al: Pathological findings
of COVID-19 associ-ated with acute respiratory distress syndrome.
Lancet Respir Med. 8:420–422. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang B, Zhou X, Qiu Y, Song Y, Feng F,
Feng J, Song Q, Jia Q and Wang J: Clinical characteristics of 82
cases of death with COVID-19. PLoS One. 15:e02354582020. View Article : Google Scholar
|
|
99
|
He R, Lu Z, Zhang L, Fan T, Xiong R, Shen
X, Feng H, Meng H, Lin W, Jiang W and Geng Q: The clinical course
and its correlated immune status in COVID-19 pneumonia. J Clin
Virol. 127:1043612020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Coperchini F, Chiovato L, Croce L, Magri F
and Rotondi M: The cytokine storm in COVID-19: An overview of the
involvement of the chemokine/chemokine-receptor system. Cytokine
Growth Factor Rev. 53:25–32. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ye Q, Wang B and Mao J: The pathogenesis
and treatment of the ʻCytokine Storm' in COVID-19. J Infect.
80:607–613. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li H, Liu L, Zhang D, Xu J, Dai H, Tang N,
Su X and Cao B: SARS-CoV-2 and viral sepsis: Observations and
hypotheses. Lancet. 395:1517–1520. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li X and Ma X: Acute respiratory failure
in COVID-19: Is it 'typical' ARDS? Crit Care. 24:1982020.
View Article : Google Scholar
|
|
104
|
Hand TW, Vujkovic-Cvijin I, Ridaura VK and
Belkaid Y: Linking the microbiota, chronic disease, and the immune
system. Trends Endocrinol Metab. 27:831–843. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Stoian AP, Banerjee Y, Rizvi AA and Rizzo
M: Diabetes and the COVID-19 pandemic: How insights from recent
experience might guide future management. Metab Syndr Relat Disord.
18:173–175. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Dietz W and Santos-Burgoa C: Obesity and
its implications for COVID-19 mortality. Obesity (Silver Spring).
28:10052020. View Article : Google Scholar
|
|
107
|
Ryan PM and Caplice NM: Is adipose tissue
a reservoir for viral spread, immune activation and cytokine
amplification in Coronavirus Disease 2019? Obesity (Silver Spring).
8:1191–1194. 2020. View Article : Google Scholar
|
|
108
|
Nikolich-Žugich J: The twilight of
immunity: Emerging concepts in aging of the immune system. Nat
Immunol. 19:10–19. 2018. View Article : Google Scholar
|
|
109
|
Butler MJ and Barrientos RM: The impact of
nutrition on COVID-19 susceptibility and long-term consequences.
Brain Behav Immun. 87:53–54. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Körner A, Schlegel M, Theurer J,
Frohnmeyer H, Adolph M, Heijink M, Giera M, Rosenberger P and
Mirakaj V: Resolution of inflammation and sepsis survival are
improved by dietary Ω-3 fatty acids. Cell Death Differ. 25:421–431.
2018. View Article : Google Scholar
|
|
111
|
Panigrahy D, Gilligan MM, Huang S, Gartung
A, Cortés-Puch I, Sime PJ, Phipps RP, Serhan CN and Hammock BD:
Inflammation resolution: A dual-pronged approach to averting
cytokine storms in COVID-19? Cancer Metastasis Rev. 39:337–340.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Chen P, Mao L, Nassis GP, Harmer P,
Ainsworth BE and Li F: Coronavirus disease (COVID-19): The need to
maintain regular physical activity while taking precautions. J
Sport Health Sci. 9:103–104. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Sardu C, Gambardella J, Morelli MB, Wang
X, Marfella R and Santulli G: Hypertension, thrombosis, kidney
failure, and diabetes: Is COVID-19 an endothelial disease? A
comprehensive evaluation of clinical and basic evidence. J Clin
Med. 9:E14172020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Guzik TJ, Mohiddin SA, Dimarco A, Patel V,
Savvatis K, Marelli-Berg FM, Madhur MS, Tomaszewski M, Maffia P,
D'Acquisto F, et al: COVID-19 and the cardiovascular system:
Implications for risk assessment, diagnosis, and treatment options.
Cardiovasc Res. 116:1666–1687. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Hamming I, Timens W, Bulthuis ML, Lely AT,
Navis G and van Goor H: Tissue distribution of ACE2 protein, the
functional receptor for SARS coronavirus. A first step in
understanding SARS pathogenesis. J Pathol. 203:631–637. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Varga Z, Flammer AJ, Steiger P, Haberecker
M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka
F and Moch H: Endothelial cell infection and endotheliitis in
COVID-19. Lancet. 395:1417–1418. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Alvarado-Moreno JA and Majluf-Cruz A:
COVID-19 and dysfunctional endothelium: The Mexican Scenario. Arch
Med Res. 51:587–588. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhou X, Zhu J and Xu T: Clinical
characteristics of corona-virus disease 2019 (COVID-19) patients
with hypertension on renin-angiotensin system inhibitors. Clin Exp
Hypertens. 42:656–660. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Bikdeli B, Madhavan MV, Jimenez D, Chuich
T, Dreyfus I, Driggin E, Nigoghossian C, Ageno W, Madjid M, Guo Y,
et al: COVID-19 and thrombotic or thromboembolic disease:
Implications for prevention, antithrombotic therapy, and follow-up:
JACC State-of-the-Art Review. J Am Coll Cardiol. 75:2950–2973.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
von Brühl ML, Stark K, Steinhart A,
Chandraratne S, Konrad I, Lorenz M, Khandoga A, Tirniceriu A,
Coletti R, Köllnberger M, et al: Monocytes, neutrophils, and
platelets cooperate to initiate and propagate venous thrombosis in
mice in vivo. J Exp Med. 209:819–835. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Gunzer M: Escaping the traps of your own
hunters. Science. 358:1126–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
de Bont CM, Boelens WC and Pruijn GJM:
NETosis, complement, and coagulation: A triangular relationship.
Cell Mol Immunol. 16:19–27. 2019. View Article : Google Scholar :
|
|
123
|
Merad M and Martin JC: Pathological
inflammation in patients with COVID-19: A key role for monocytes
and macrophages. Nat Rev Immunol. 20:355–362. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Massberg S, Grahl L, von Bruehl ML,
Manukyan D, Pfeiler S, Goosmann C, Brinkmann V, Lorenz M, Bidzhekov
K, Khandagale AB, et al: Reciprocal coupling of coagulation and
innate immunity via neutrophil serine proteases. Nat Med.
16:887–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Oehmcke S, Mörgelin M and Herwald H:
Activation of the human contact system on neutrophil extracellular
traps. J Innate Immun. 1:225–230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Semeraro F, Ammollo CT, Morrissey JH, Dale
GL, Friese P, Esmon NL and Esmon CT: Extracellular histones promote
thrombin generation through platelet-dependent mechanisms:
Involvement of platelet TLR2 and TLR4. Blood. 118:1952–1961. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Jiménez-Alcázar M, Rangaswamy C, Panda R,
Bitterling J, Simsek YJ, Long AT, Bilyy R, Krenn V, Renné C, Renné
T, et al: Host DNases prevent vascular occlusion by neutrophil
extracellular traps. Science. 358:1202–1206. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Lee KH, Cavanaugh L, Leung H, Yan F,
Ahmadi Z, Chong BH and Passam F: Quantification of NETs-associated
markers by flow cytometry and serum assays in patients with
thrombosis and sepsis. Int J Lab Hematol. 40:392–399. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Barnes BJ, Adrover JM, Baxter-Stoltzfus A,
Borczuk A, Cools-Lartigue J, Crawford JM, Daßler-Plenker J, Guerci
P, Huynh C, Knight JS, et al: Targeting potential drivers of
COVID-19: Neutrophil extracellular traps. J Exp Med.
217:e202006522020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zhang Y, Xiao M, Zhang S, Xia P, Cao W,
Jiang W, Chen H, Ding X, Zhao H, Zhang H, et al: Coagulopathy and
antiphos-pholipid antibodies in patients with covid-19. N Engl J
Med. 382:e382020. View Article : Google Scholar
|
|
131
|
Escher R, Breakey N and Lämmle B: Severe
COVID-19 infection associated with endothelial activation. Thromb
Res. 190:622020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Whyte CS, Morrow GB, Mitchell JL, Chowdary
P and Mutch NJ: Fibrinolytic abnormalities in acute respiratory
distress syndrome (ARDS) and versatility of thrombolytic drugs to
treat COVID-19. J Thromb Haemost. 18:1548–1555. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Zhu J, Zhong Z, Ji P, Li H, Li B, Pang J,
Zhang J and Zhao C: Clinicopathological characteristics of 8697
patients with COVID-19 in China: A meta-analysis. Fam Med Community
Health. 8:e0004062020. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Novel Coronavirus-China. http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/urisimplewww.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/.
Retrieved May 30, 2020.
|
|
135
|
Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y,
Zhu H, Zhao W, Han Y and Qin C: From SARS to MERS, thrusting
coronavi-ruses into the spotlight. Viruses. 11:592019. View Article : Google Scholar
|
|
136
|
Fu J, Zhou B, Zhang L, Balaji KS, Wei C,
Liu X, Chen H, Peng J and Fu J: Expressions and significances of
the angiotensin-converting enzyme 2 gene, the receptor of
SARS-CoV-2 for COVID-19. Mol Biol Rep. 47:4383–4392. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Hui KPY, Cheung MC, Perera RAPM, Ng KC,
Bui CHT, Ho JCW, Ng MMT, Kuok DIT, Shih KC, Tsao SW, et al:
Tropism, replication competence, and innate immune responses of the
coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva:
An analysis in ex-vivo and in-vitro cultures. Lancet Respir Med.
8:687–695. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng
X, Li T and Chen Q: High expression of ACE2 receptor of 2019-nCoV
on the epithelial cells of oral mucosa. Int J Oral Sci. 12:82020.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zhang H, Kang Z, Gong H, Xu D, Wang J, Li
Z, Li Z, Cui X, Xiao J, Zhan J, et al: Digestive system is a
potential route of COVID-19: An analysis of single-cell
coexpression pattern of key proteins in viral entry process. Gut.
69:1010–1018. 2020. View Article : Google Scholar :
|
|
140
|
Gao Y, Li T, Han M, Li X, Wu D, Xu Y, Zhu
Y, Liu Y, Wang X and Wang L: Diagnostic utility of clinical
laboratory data determinations for patients with the severe
COVID-19. J Med Virol. 92:791–796. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Wang Y, Kang H, Liu X and Tong Z:
Asymptomatic cases with SARS-CoV-2 infection. J Med Virol.
92:1401–1403. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Fu L, Wang B, Yuan T, Chen X, Ao Y,
Fitzpatrick T, Li P, Zhou Y, Lin YF, Duan Q, et al: Clinical
characteristics of coronavirus disease 2019 (COVID-19) in China: A
systematic review and meta-analysis. J Infect. 80:656–665. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Lian J, Jin X, Hao S, Jia H, Cai H, Zhang
X, Hu J, Zheng L, Wang X, Zhang S, et al: Epidemiological,
clinical, and virological characteristics of 465 hospitalized cases
of coronavirus disease 2019 (COVID-19) from Zhejiang province in
China. Influenza Other Respir Viruses. 14:564–574. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Gattinoni L, Chiumello D, Caironi P,
Busana M, Romitti F, Brazzi L and Camporota L: COVID-19 pneumonia:
Different respiratory treatments for different phenotypes?
Intensive Care Med. 46:1099–1102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
The European Society for Cardiology (ESC):
ESC Guidance for the Diagnosis and Management of CV Disease during
the COVID-19 Pandemic. https://www.escardio.org/Education/COVID-19-and-Cardiology/ESCCOVID-19-Guidanceurisimplehttps://www.escardio.org/Education/COVID-19-and-Cardiology/ESCCOVID-19-Guidance.
Last updated May 28, 2020.
|
|
146
|
Shang W, Dong J, Ren Y, Tian M, Li W, Hu J
and Li Y: The value of clinical parameters in predicting the
severity of COVID-19. J Med Virol. 92:2188–2192. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Cai Q, Chen F, Wang T, Luo F, Liu X, Wu Q,
He Q, Wang Z, Liu Y, Liu L, et al: Obesity and COVID-19 Severity in
a Designated Hospital in Shenzhen, China. Diabetes Care.
43:1392–1398. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Palaiodimos L, Kokkinidis DG, Li W,
Karamanis D, Ognibene J, Arora S, Southern WN and Mantzoros CS:
Severe obesity, increasing age and male sex are independently
associated with worse in-hospital outcomes, and higher in-hospital
mortality, in a cohort of patients with COVID-19 in the Bronx, New
York. Metabolism. 108:1542622020. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhang L, Feng X, Zhang D, Jiang C, Mei H,
Wang J, Zhang C, Li H, Xia X, Kong S, et al: Deep vein thrombosis
in hospitalized patients with (COVID-19) in Wuhan, China:
Prevalence, risk factors, and outcome. Circulation. 142:114–128.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Stoneham SM, Milne KM, Nuttall E, Frew GH,
Sturrock BR, Sivaloganathan H, Ladikou EE, Drage S, Phillips B,
Chevassut TJ and Eziefula AC: Thrombotic risk in COVID-19: A case
series and case-control study. Clin Med (Lond). 20:e76–e81. 2020.
View Article : Google Scholar
|
|
151
|
Jin X, Lian JS, Hu JH, Gao J, Zheng L,
Zhang YM, Hao SR, Jia HY, Cai H, Zhang XL, et al: Epidemiological,
clinical and virological characteristics of 74 cases of
coronavirus-infected disease 2019 (COVID-19) with gastrointestinal
symptoms. Gut. 69:1002–1009. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Lin L, Jiang X, Zhang Z, Huang S, Zhang Z,
Fang Z, Gu Z, Gao L, Shi H, Mai L, et al: Gastrointestinal symptoms
of 95 cases with SARS-CoV-2 infection. Gut. 69:997–1001. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Mao R, Qiu Y, He JS, Tan JY, Li XH, Liang
J, Shen J, Zhu LR, Chen Y, Iacucci M, et al: Manifestations and
prognosis of gastro-intestinal and liver involvement in patients
with COVID-19: A systematic review and meta-analysis. Lancet
Gastroenterol Hepatol. 5:667–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
To KK, Tsang OT, Yip CC, Chan KH, Wu TC,
Chan JM, Leung WS, Chik TS, Choi CY, Kandamby DH, et al: Consistent
detection of 2019 novel coronavirus in saliva. Clin Infect Dis.
71:841–843. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Holshue ML, DeBolt C, Lindquist S, Lofy
KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural
A, et al: First Case of 2019 Novel Coronavirus in the United
States. N Engl J Med. 382:929–936. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z,
Xiang J, Wang Y, Song B, Gu X, et al: Clinical course and risk
factors for mortality of adult inpatients with COVID-19 in Wuhan,
China: A retrospective cohort study. Lancet. 395:1054–1062. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Jiang SQ, Huang QF, Xie WM, Lv C and Quan
XQ: The association between severe COVID-19 and low platelet count:
Evidence from 31 observational studies involving 7613 participants.
Br J Haematol. 190. pp. e29–e33. 2020, View Article : Google Scholar
|
|
158
|
Doobay MF, Talman LS, Obr TD, Tian X,
Davisson RL and Lazartigues E: Differential expression of neuronal
ACE2 in transgenic mice with overexpression of the brain
renin-angiotensin system. Am J Physiol Regul Integr Comp Physiol.
292:R373–R381. 2007. View Article : Google Scholar :
|
|
159
|
Liguori C, Pierantozzi M, Spanetta M,
Sarmati L, Cesta N, Iannetta M, Ora J, Mina GG, Puxeddu E, Balbi O,
et al: Subjective neurological symptoms frequently occur in
patients with SARS-CoV2 infection. Brain Behav Immun. 88:11–16.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Kim GU, Kim MJ, Ra SH, Lee J, Bae S, Jung
J and Kim SH: Clinical characteristics of asymptomatic and
symptomatic patients with mild COVID-19. Clin Microbiol Infect.
26:948.e1–948.e3. 2020. View Article : Google Scholar
|
|
161
|
Ye M, Ren Y and Lv T: Encephalitis as a
clinical manifestation of COVID-19. Brain Behav Immun. 88:945–946.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Bernard-Valnet R, Pizzarotti B, Anichini
A, Demars Y, Russo E, Schmidhauser M, Cerutti-Sola J, Rossetti AO
and Du Pasquier R: Two patients with acute meningoencephalitis
concomitant to SARS-CoV-2 infection. Eur J Neurol. 27:e43–e44.
2020. View Article : Google Scholar
|
|
163
|
Moriguchi T, Harii N, Goto J, Harada D,
Sugawara H, Takamino J, Ueno M, Sakata H, Kondo K, Myose N, et al:
A first case of meningitis/encephalitis associated with
SARS-Coronavirus-2. Int J Infect Dis. 94:55–58. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Pei G, Zhang Z, Peng J, Liu L, Zhang C, Yu
C, Ma Z, Huang Y, Liu W, Yao Y, et al: Renal involvement and early
prognosis in patients with COVID-19 pneumonia. J Am Soc Nephrol.
31:1157–1165. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Galván Casas C, Català A, Carretero
Hernández G, Rodríguez-Jiménez P, Fernández Nieto D,
Rodríguez-Villa Lario A, Navarro Fernández I, Ruiz-Villaverde R,
Falkenhain-López D, Llamas Velasco M, et al: Classification of the
cutaneous manifestations of COVID-19: A rapid prospective
nationwide consensus study in Spain with 375 cases. Br J Dermatol.
183:71–77. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Tang K, Wang Y, Zhang H, Zheng Q, Fang R
and Sun Q: Cutaneous manifestations of the Coronavirus Disease 2019
(COVID-19): A brief review. Dermatol Ther. 33:e135282020.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Chen L, Deng C, Chen X, Zhang X, Chen B,
Yu H, Qin Y, Xiao K, Zhang H and Sun X: Ocular manifestations and
clinical characteristics of 535 cases of COVID-19 in Wuhan, China:
a cross-sectional study. Acta Ophthalmol. 98:e951–e959. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Loffredo L, Pacella F, Pacella E, Tiscione
G, Oliva A and Violi F: Conjunctivitis and COVID-19: A
meta-analysis. J Med Virol. 92:1413–1414. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Panahi L, Amiri M and Pouy S: Clinical
characteristics of COVID-19 infection in newborns and pediatrics: A
systematic review. Arch Acad Emerg Med. 8:e502020.PubMed/NCBI
|
|
170
|
Mustafa NM and A Selim L: Characterisation
of COVID-19 Pandemic in Paediatric Age Group: A systematic review
and meta-analysis. J Clin Virol. 128:1043952020. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Liguoro I, Pilotto C, Bonanni M, Ferrari
ME, Pusiol A, Nocerino A, Vidal E and Cogo P: SARS-COV-2 infection
in children and newborns: A systematic review. Eur J Pediatr.
179:1029–1046. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Sun D, Li H, Lu XX, Xiao H, Ren J, Zhang
FR and Liu ZS: Clinical features of severe pediatric patients with
coronavirus disease 2019 in Wuhan: A single center's observational
study. World J Pediatr. 16:251–259. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Juan J, Gil MM, Rong Z, Zhang Y, Yang H
and Poon LC: Effects of coronavirus disease 2019 (COVID-19) on
maternal, perinatal and neonatal outcomes: A systematic review.
Ultrasound Obstet Gynecol. 56:15–27. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Alzamora MC, Paredes T, Caceres D, Webb
CM, Valdez LM and La Rosa M: Severe COVID-19 during pregnancy and
possible vertical transmission. Am J Perinatol. 37:861–865. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Valdés G, Neves LA, Anton L, Corthorn J,
Chacón C, Germain AM, Merrill DC, Ferrario CM, Sarao R, Penninger J
and Brosnihan KB: Distribution of angiotensin-(1-7) and ACE2 in
human placentas of normal and pathological pregnancies. Placenta.
27:200–207. 2006. View Article : Google Scholar
|
|
176
|
Spanish Ministry: Available treatments
subjected to special access conditions for the managing of the
respiratory infection by SARS-CoV-2. Spanish Agency for Medicines
and Health Products; 2020, https://www.aemps.gob.es/la-aemps/ultima-informacion-de-la-aemps-acerca-del-covid%E2%80%9119/tratamientos-disponibles-para-el-manejo-de-la-infeccion-respiratoria-por-sars-cov-2/?lang=enurisimplehttps://www.aemps.gob.es/la-aemps/ultima-informacion-de-la-aemps-acerca-del-covid%E2%80%9119/tratamientos-disponibles-para-el-manejo-de-la-infeccion-respiratoria-por-sars-cov-2/?lang=en.
Retrieved May 28, 2020.
|
|
177
|
World Health Organization (WHO): Clinical
management of COVID-19: interim guidance. https://apps.who.int/iris/handle/10665/332196urisimplehttps://apps.who.int/iris/handle/10665/332196.
License: CC BY-NC-SA 3.0 IGO Accessed May 27, 2020.
|
|
178
|
Wu R, Wang L, Kuo HD, Shannar A, Peter R,
Chou PJ, Li S, Hudlikar R, Liu X, Liu Z, et al: An update on
current therapeutic drugs treating COVID-19. Curr Pharmacol Rep.
May 11–2020.Epub ahead for print. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Zhou M, Zhang X and Qu J: Coronavirus
disease 2019 (COVID-19): A clinical update. Front Med. 14:126–135.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Dima A, Balaban DV, Jurcut C, Berza I,
Jurcut R and Jinga M: Physicians' Perspectives on COVID-19: An
International Survey. Healthcare (Basel). 8:2502020. View Article : Google Scholar
|
|
181
|
Jawhara S: Could intravenous
immunoglobulin collected from recovered coronavirus patients
protect against COVID-19 and strengthen the immune system of new
patients? Int J Mol Sci. 21:22722020. View Article : Google Scholar :
|
|
182
|
Qiu T, Liang S, Dabbous M, Wang Y, Han R
and Toumi M: Chinese guidelines related to novel coronavirus
pneumonia. J Mark Access Health Policy. 8:18184462020. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Gutiérrez-Lorenzo M and Cuadros-Martínez
CM: Baricitinib in treatment of SARS-CoV-2 infection. Rev Esp
Quimioter. 33:294–295. 2020.In Spanish. View Article : Google Scholar
|
|
184
|
Singh AK, Majumdar S, Singh R and Misra A:
Role of corticosteroid in the management of COVID-19: A systemic
review and a Clinician's perspective. Diabetes Metab Syndr.
14:971–978. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Mulangu S, Dodd LE, Davey RT Jr, Tshiani
Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D,
Tshomba Oloma A, Ibanda A, et al: A randomized, controlled trial of
Ebola virus disease therapeutics. N Engl J Med. 381:2293–2303.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
de Wit E, Feldmann F, Cronin J, Jordan R,
Okumura A, Thomas T, Scott D, Cihlar T and Feldmann H: Prophylactic
and therapeutic remdesivir (GS-5734) treatment in the rhesus
macaque model of MERS-CoV infection. Proc Natl Acad Sci USA.
117:6771–6776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Lo MK, Feldmann F, Gary JM, Jordan R,
Bannister R, Cronin J, Patel NR, Klena JD, Nichol ST, Cihlar T, et
al: Remdesivir (GS-5734) protects African green monkeys from Nipah
virus challenge. Sci Transl Med. 11:eaau92422019. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Grein J, Ohmagari N, Shin D, Diaz G,
Asperges E, Castagna A, Feldt T, Green G, Green ML, Lescure FX, et
al: Compassionate use of remdesivir for patients with severe
Covid-19. N Engl J Med. 382:2327–2336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Ferner RE and Aronson JK: Remdesivir in
covid-19. BMJ. m1610:3692020.
|
|
190
|
Jean SS, Lee PI and Hsueh PR: Treatment
options for COVID-19: The reality and challenges. J Microbiol
Immunol Infect. 53:436–443. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Groneberg DA, Poutanen SM, Low DE, Lode H,
Welte T and Zabel P: Treatment and vaccines for severe acute
respiratory syndrome. Lancet Infect Dis. 5:147–155. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Cao B, Wang Y, Wen D, Liu W, Wang J, Fan
G, Ruan L, Song B, Cai Y, Wei M, et al: A Trial of
Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N
Engl J Med. 382:1787–1799. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Kunz KM: A Trial of Lopinavir-Ritonavir in
Covid-19. N Engl J Med. 382:e682020. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Statement from the Chief Investigators of
the Randomised Evaluation of COVid-19 thERapY (RECOVERY) Trial on
lopinavir-ritonavir. https://www.recoverytrial.net/files/lopi-navir-ritonavir-recovery-statement-29062020_final.pdfurisimplehttps://www.recoverytrial.net/files/lopi-navir-ritonavir-recovery-statement-29062020_final.pdf
Accessed June 29, 2020.
|
|
195
|
Al-Bari MA: Chloroquine analogues in drug
discovery: New directions of uses, mechanisms of actions and toxic
manifestations from malaria to multifarious diseases. J Antimicrob
Chemother. 70:1608–1621. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Vincent MJ, Bergeron E, Benjannet S,
Erickson BR, Rollin PE, Ksiazek TG, Seidah NG and Nichol ST:
Chloroquine is a potent inhibitor of SARS coronavirus infection and
spread. Virol J. 2:692005. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Wang M, Cao R, Zhang L, Yang X, Liu J, Xu
M, Shi Z, Hu Z, Zhong W and Xiao G: Remdesivir and chloroquine
effectively inhibit the recently emerged novel coronavirus
(2019-nCoV) in vitro. Cell Res. 30:269–271. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Andreani J, Le Bideau M, Duflot I, Jardot
P, Rolland C, Boxberger M, Wurtz N, Rolain JM, Colson P, La Scola B
and Raoult D: In vitro testing of combined hydroxychloroquine and
azithromycin on SARS-CoV-2 shows synergistic effect. Microb Pathog.
145:1042282020. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Gautret P, Lagier JC, Parola P, Hoang V,
Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira V,
et al: Hydroxychloroquine and azithromycin as a treatment of
COVID-19: Results of an open- label non-randomized clinical trial.
Int J Antimicrob Agents. 56:1059492020. View Article : Google Scholar
|
|
200
|
Molina JM, Delaugerre C, Goff JL,
Mela-Lima B, Ponscarme D, Goldwirt L and de Castro N: No evidence
of rapid antiviral clearance or clinical benefit with the
combination of hydroxy-chloroquine and azithromycin in patients
with severe COVID-19 infection. Med Mal Infect. 50:3842020.
View Article : Google Scholar
|
|
201
|
Rosenberg ES, Dufort EM, Udo T,
Wilberschied LA, Kumar J, Tesoriero J, Weinberg P, Kirkwood J, Muse
A, DeHovitz J, et al: Association of treatment with
hydroxychloroquine or azithro-mycin with in-hospital mortality in
patients with COVID-19 in New York State. JAMA. 323:2493–2502.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Hornby P and Landray M: Statement from the
Chief Investigators of the Randomised Evaluation of COVid-19
thERapY (RECOVERY) Trial on hydroxychloroquine. https://www.recov-erytrial.net/files/hcq-recovery-statement-050620-final-002.pdfurisimplehttps://www.recov-erytrial.net/files/hcq-recovery-statement-050620-final-002.pdf.
Accessed June 5, 2020.
|
|
203
|
Mansourabadi AH, Sadeghalvad M,
Mohammadi-Motlagh HR and Rezaei N: The immune system as a target
for therapy of SARS-CoV-2: A systematic review of the current
immunotherapies for COVID-19. Life Sci. 258:1181852020. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Aouba A, Baldolli A, Geffray L, Verdon R,
Bergot E, Martin-Silva N and Justet A: Targeting the inflammatory
cascade with anakinra in moderate to severe COVID-19 pneu-monia:
Case series. Ann Rheum Dis. 79:1381–1382. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Zhang C, Wu Z, Li JW, Zhao H and Wang GQ:
Cytokine release syndrome in severe COVID-19: Interleukin-6
receptor antago-nist Tocilizumab may be the key to reduce the
mortality. Int J Antimicrob Agents. 55:1059542020. View Article : Google Scholar
|
|
206
|
Sheppard M, Laskou F, Stapleton PP, Hadavi
S and Dasgupta B: Tocilizumab (Actemra). Hum Vaccin Immunother.
13:1972–1988. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Vastert SJ, Jamilloux Y, Quartier P,
Ohlman S, Osterling Koskinen L, Kullenberg T, Franck-Larsson K,
Fautrel B and de Benedetti F: Anakinra in children and adults with
Still's disease. Rheumatology (Oxford). 58(Suppl 6): vi9–vi22.
2018. View Article : Google Scholar
|
|
208
|
Guaraldi G, Meschiari M, Cozzi-Lepri A,
Milic J, Tonelli R, Menozzi M, Franceschini E, Cuomo G, Orlando G,
Borghi V, et al: Tocilizumab in patients with severe COVID-19: A
retrospective cohort study. Lancet Rheumatol. 2:e474–e484. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Tomasiewicz K, Piekarska A,
Stempkowska-Rejek J, Serafińska S, Gawkowska A, Parczewski M,
Niścigorska-Olsen J, Lstrok;apiński TW, Zarębska-Michaluk D,
Kowalska JD, et al: Tocilizumab for patients with severe COVID-19:
A retrospective, multi-center study. Expert Rev Anti Infect Ther.
Aug 1–2020.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Cauchois R, Koubi M, Delarbre D, Manet C,
Carvelli J, Blasco VB, Jean R, Fouche L, Bornet C, Pauly V, et al:
Early IL-1 receptor blockade in severe inflammatory respiratory
failure complicating COVID-19. Proc Natl Acad Sci USA.
117:18951–18953. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
McGonagle D, Sharif K, O'Regan A and
Bridgewood C: The role of cytokines including interleukin-6 in
COVID-19 induced pneumonia and macrophage activation syndrome-like
disease. Autoimmun Rev. 19:1025372020. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Xu X, Han M, Li T, Sun W, Wang D, Fu B,
Zhou Y, Zheng X, Yang Y, Li X, et al: Effective treatment of severe
COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA.
117:10970–10975. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Alzghari SK and Acuña VS: Supportive
treatment with tocilizumab for COVID-19: A systematic review. J
Clin Virol. 127:1043802020. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Galimberti S, Baldini C, Baratè C, Ricci
F, Balducci S, Grassi S, Ferro F, Buda G, Benedetti E, Fazzi R, et
al: The CoV-2 outbreak: How hematologists could help to fight
Covid-19. Pharmacol Res. 157:1048662020. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Peterson D, Damsky W and King B: The use
of Janus kinase inhibitors in the time of severe acute respiratory
syndrome coronavirus 2 (SARS-CoV-2). J Am Acad Dermatol.
82:e223–e226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Chang R, Ng TB and Sun WZ: Lactoferrin as
potential preventative and adjunct treatment for COVID-19. Int J
Antimicrob Agents. 56:1061182020. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Ucciferri C, Barone M, Vecchiet J and
Falasca K: Pidotimod in paucisymptomatic SARS-CoV2 infected
patients. Mediterr J Hematol Infect Dis. 12:e20200482020.
View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Colunga Biancatelli RML, Berrill M,
Catravas JD and Marik PE: Quercetin and vitamin C: An experimental,
synergistic therapy for the prevention and treatment of SARS-CoV-2
related disease (COVID-19). Front Immunol. 11:14512020. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Mettenleiter TC: Chapter One-The First
'Virus Hunters'. Advances in Virus Research. Beer M and Höper D:
Academic Press; pp. 1–16. 2017
|
|
220
|
Goldsmith CS and Miller SE: Modern uses of
electron microscopy for detection of viruses. Clin Microbiol Rev.
22:552–563. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Singh L, Kruger HG, Maguire GEM, Govender
T and Parboosing R: The role of nanotechnology in the treatment of
viral infections. Ther Adv Infect Dis. 4:105–131. 2017.PubMed/NCBI
|
|
222
|
Sivasankarapillai VS, Pillai AM, Rahdar A,
Sobha AP, Das SS, Mitropoulos AC, Mokarrar MH and Kyzas GZ: On
facing the SARS-CoV-2 (COVID-19) with combination of nanomaterials
and medicine: Possible strategies and first challenges.
Nanomaterials (Basel). 10:8522020. View Article : Google Scholar
|
|
223
|
Kostarelos K: Nanoscale nights of
COVID-19. Nat Nanotechnol. 15:343–344. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Patra JK, Das G, Fraceto LF, Campos EV R,
Rodriguez-Torres MDP, Acosta-Torres LS, Diaz-Torres LA, Grillo R,
Swamy MK, Sharma S, et al: Nano based drug delivery systems: Recent
developments and future prospects. J Nanobiotechnology. 16:712018.
View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Chen L and Liang J: An overview of
functional nanoparticles as novel emerging antiviral therapeutic
agents. Mater Sci Eng C Mater Biol Appl. 112:1109242020. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Cui H, Webber MJ and Stupp SI:
Self-assembly of peptide amphiphiles: From molecules to
nanostructures to biomaterials. Biopolymers. 94:1–18. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Dormont F, Brusini R, Cailleau C, Reynaud
F, Peramo A, Gendron A, Mougin J, Gaudin F, Varna M and Couvreur P:
Squalene-based multidrug nanoparticles for improved mitigation of
uncontrolled inflammation. Sci Adv. 6:eaaz54662020. View Article : Google Scholar
|
|
228
|
Hu Y, Liu C and Muyldermans S:
Nanobody-based delivery systems for diagnosis and targeted tumor
therapy. Front Immunol. 8:14422017. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Walter JD, Hutter CAJ, Zimmermann I, Earp
J, Egloff P, Sorgenfrei M, Hürlimann LM, Gonda I, Meier G, Remm S,
et al: Synthetic nanobodies targeting the SARS-CoV-2
receptor-binding domain. BioRxiv. 1–18. 2020.
|
|
230
|
Ura T, Okuda K and Shimada M: Developments
in viral vector-based vaccines. Vaccines (Basel). 2:624–641. 2014.
View Article : Google Scholar
|
|
231
|
Xu H, Li Z and Si J: Nanocarriers in gene
therapy: A review. J Biomed Nanotechnol. 10:3483–3507. 2014.
View Article : Google Scholar
|
|
232
|
Feldman RA, Fuhr R, Smolenov I, Ribeiro A,
Panther L, Watson M, Senn JJ, Smith M, Almarsson Ӧ, Pujar HS, et
al: mRNA vaccines against H10N8 and H7N9 influenza viruses of
pandemic potential are immunogenic and well tolerated in healthy
adults in phase 1 randomized clinical trials. Vaccine.
37:3326–3334. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
McKay PF, Hu K, Blakney AK, Samnuan K,
Bouton CR, Rogers P, Polra K, Lin PJC, Barbosa C, Tam Y and
Shattock RJ: Self-amplifying RNA SARS-CoV-2 lipid nanoparticle
vaccine induces equivalent preclinical antibody titers and viral
neutralization to recovered COVID-19 patients. https://doi.org/10.1101/2020.04.22.055608urisimplehttps://doi.org/10.1101/2020.04.22.055608.
|
|
234
|
Raghunandan R, Lu H, Zhou B, Xabier MG,
Massare MJ, Flyer DC, Fries LF, Smith GE and Glenn GM: An insect
cell derived respiratory syncytial virus (RSV) F nanoparticle
vaccine induces antigenic site II antibodies and protects against
RSV challenge in cotton rats by active and passive immunization.
Vaccine. 32:6485–6492. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
He L, de Val N, Morris CD, Vora N, Thinnes
TC, Kong L, Azadnia P, Sok D, Zhou B, Burton DR, et al: Presenting
native-like trimeric HIV-1 antigens with self-assembling
nanoparticles. Nat Commun. 7:120412016. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Foundation TWN: COVID-19 testing report
& analysis, 2020. http://www.worldnanofoundation.com/covid-19-report-analysisurisimplewww.worldnanofoundation.com/covid-19-report-analysis
Accessed July 24, 2020.
|
|
237
|
Zhao Z, Cui H, Song W, Ru X, Zhou W and Yu
X: A simple magnetic nanoparticles-based viral RNA extraction
method for efficient detection of SARS-CoV-2. bioRxiv. https://doi.org/10.1101/2020.02.22.961268urisimplehttps://doi.org/10.1101/2020.02.22.961268.
|
|
238
|
Wang M, Fu A, Hu B, Tong Y, Liu R, Liu Z,
Gu J, Xiang B, Liu J, Jiang W, et al: Nanopore target sequencing
for accurate and comprehensive detection of SARS-CoV-2 and other
respiratory viruses. Small. 16:e20021692020. View Article : Google Scholar
|
|
239
|
Lüthy IA, Ritacco V and Kantor IN: One
hundred years after the 'Spanish' flu. Medicina (B Aires).
78:113–118. 2018.In Spanish.
|