Introduction
Ovarian cancer is the 7th most common form of cancer
encountered in women worldwide, with an estimated 295,000 new cases
and 185,000-related deaths in 2018 (1). Epithelial ovarian cancer is the
most prevalent type of ovarian cancer, which can be further
classified into endometrioid, clear-cell, mucinous, high-grade and
low-grade serous carcinomas (2).
Despite advancements being made in the understanding of ovarian
cancer pathology and the application of novel early diagnostic and
treatment strategies for patients with ovarian cancer, the majority
of the patients are diagnosed at an advanced stage of the disease
and the 5-year overall survival rate has been estimated to <50%
(3). Therefore, the
identification of novel targets for the diagnosis and treatment of
patients with ovarian cancer is of utmost importance.
Long non-coding RNAs (lncRNAs) are single-stranded
RNA molecules that are ≥200 nucleotides in length (4). According to the competing
endogenous RNA hypothesis, certain lncRNAs contain miRNA response
elements and can compete for binding to miRNAs with mRNAs,
resulting in the regulation of gene expression (5). In doing so, lncRNAs are implicated
in almost all physiological processes (6). Recent studies have indicated that
the aberrant expression of lncRNAs is involved in cancer initiation
and development, and notably in cancer cell proliferation and
apoptosis (7). For example,
lncRNA DNM3OS has been shown to be involved in the
epithelial-to-mesenchymal transition process and to promote ovarian
cancer cell migration and invasion (8). lncRNA ABHD11-AS1 has been
shown to facilitate ovarian cancer cell proliferation, migration
and invasion, and inhibit cell apoptosis via the upregulation of
RhoC expression (9). lncRNA
ASAP1-IT1 is an intronic transcript of the ASAP1 gene
(10). A previous study
indicated that ASAP1-IT1 expression was decreased in
high-grade ovarian tumors compared with the corresponding
expression noted in low-grade tumors (11). A high expression of
ASAP1-IT1 has been shown to be associated with the optimal
prognosis of patients with ovarian cancer (11). However, the mechanisms through
which ASAP1-IT1 contributes to ovarian cancer progression
remain unknown.
Cell number and cell size are tightly controlled by
the Hippo pathway (12). The
dysregulation of the Hippo pathway leads to the overgrowth of cells
and resistance to cell apoptosis (13). Accumulating evidence has
suggested that the overexpression of Yes associated protein 1
(YAP1), which is the downstream effector of the Hippo pathway,
contributes to cancer development and initiation in different
organs, including the colon, bladder, breast and ovaries (14-17). The higher expression of YAP1 has
been observed in ovarian cancer tissues and cells, and is
considered critical for cancer cell proliferation and survival
(18). Among several tumor
suppressive kinases of the Hippo pathway, the loss of large tumor
suppressor 2 (LATS2) expression has been considered a major cause
of YAP1 overexpression in cancer cells (19,20). Decreased expression of LATS2 was
also reported in ovarian cancer (21). However, the regulation of
LATS2/YAP1 signaling by lncRNAs in ovarian cancer remains
elusive.
In the present study, the overexpression of
ASAP1-IT1 was found to inhibit cell proliferation and induce
apoptosis by sponging miR-2278. Concomitantly, it was able to
increase LATS2 expression in ovarian cancer. The findings presented
herein indicate that the ASAP1-IT1/miR-2278/LATS2 axis may
play a key role in ovarian cancer.
Materials and methods
Clinical samples
Consecutive patients with ovarian cancer (n=58) who
were treated at the China-Japan Union Hospital of Jilin University
between January, 2015 and January, 2017, were used in the present
study. The healthy tissues (n=58) were those 2 cm away from the
tumors. The clinical stage and metastatic status of the cancer was
determined by surgical evaluation, whereas histopathological
analysis was conducted by 2 gynecological pathologists to assess
cancer type and grade, independently. The extracted tissue samples
were stored at -80°C. All the participants in the present study
provided written informed consent prior to the surgery and the
study conduct. Ethical approval was also provided for their
participation by the Ethical Committee of The China-Japan Union
Hospital of Jilin University.
Cell lines and cell culture
The human ovarian surface epithelial cell line
HOSEpiC was purchased from ScienCell. The human ovarian serous
cystadenocarcinoma cell line, SKOV3, and the human ovarian serous
adenocarcinoma cell line, OVCAR3, were obtained from the American
Type Culture Collection (ATCC). These cells were cultured in DMEM
(Invitrogen; Thermo Fisher Scientific, Inc.) and supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
The cells were maintained in an incubator at 37°C with 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the HOSEpiC, OVCAR3,
SKOV3 cells and tissues using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol. RNA
was reverse transcribed into first-stranded cDNA using the
RevertAid RT Reverse Transcription kit (cat. no. K1691, Thermo
Fisher Scientific, Inc.). RT-qPCR was carried out with TB
Green® Premix Ex Taq™. The reaction conditions included
the following steps: Step 1: 95°C for 30 sec; step 2: 35 cycles of
95°C for 10 sec and 60°C for 30 sec. GAPDH was used as the internal
control for lncRNA and mRNA, while U6 was used as the control for
miRNA expression. The relative expression of each gene was
calculated with the 2−ΔΔCq method (22). The primer sequences are listed in
Table I.
 | Table ISequences of primer used for
RT-qPCR. |
Table I
Sequences of primer used for
RT-qPCR.
| Primer | Sequence |
|---|
| LATS2-forward |
5′-ACTTTTCCTGCCACGACTTATTC-3′ |
| LATS2-reverse |
5′-GATGGCTGTTTTAACCCCTCA-3′ |
| YAP1-forward |
5′-TAGCCCTGCGTAGCCAGTTA-3′ |
| YAP1-reverse |
5′-TCATGCTTAGTCCACTGTCTGT-3′ |
| SGK1-forward |
5′-CATATTATGTCGGAGCGGAATGT-3′ |
| SGK1-reverse |
5′-TGTCAGCAGTCTGGAAAGAGA-3′ |
| FGF2-forward |
5′-AGAAGAGCGACCCTCACATCA-3′ |
| FGF2-reverse |
5′-CGGTTAGCACACACTCCTTTG-3′ |
| ETV5-forward |
5′-TCAGCAAGTCCCTTTTATGGTC-3′ |
| ETV5-reverse |
5′-GCTCTTCAGAATCGTGAGCCA-3′ |
|
ASAP1-IT1-forward |
5′-TCTGGTCCAAAAAGATTTCTGA-3′ |
|
ASAP1-IT1-reverse |
5′-CTTTGCAGAAAGCTTTTACCATA-3′ |
| GAPDH-forward |
5′-ACAACTTTGGTATCGTGGAAGG-3′ |
| GAPDH-reverse |
5′-GCCATCACGCCACAGTTTC-3′ |
| Stem-loop
primer |
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCAGGA-3′ |
|
miR-2278-forward |
5′-GCCGAGGAGAGCAGTGTGTGTT-3′ |
|
miR-2278-reverse |
5′-CTCAACTGGTGTCGTGGA-3′ |
| U6-forward |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6-reverse |
5′-AACGCTTCACGAATTTGCGT-3′ |
Overexpression of ASAP1-IT1
The full length of ASAP1-IT1 was amplified
from HOSEpiC cDNA, digested by restriction endonucleases
EcoRI (NEB) and XhoI (NEB) and ligated into the
pcDNA3.1 plasmid (Invitrogen; Thermo Fisher Scientific). The primer
sequences were as follows: ASAP1-IT1 forward, 5′-GGG TAC CCA
AAT GGG AAA AAA A-3′ and reverse, 5′-GGA ATT CCA AAG ACT ATC
ACA-3′. For overexpression experiments, 2 µg
pcDNA3.1-ASAP1-IT1 were transfected into the OVCAR3 and
SKOV3 cells using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. The cells
were allowed to grow for 48 h and RT-qPCR was performed to confirm
the overexpression of ASAP1-IT1 as described above.
Overexpression and downregulation of
miR-2278
miR-NC mimic, miR-2278 mimic, miR-NC inhibitor and
miR-2278 inhibitor were purchased from Suzhou GenePharma Co., Ltd.
For transfection, 50 nM miR-NC mimic (5′-GUG GAU UUU CCU CUA UGA
UUU-3′), miR-2278 mimic (5′-GAG AGC AGU GUG UGU UGC CUG G-3′),
miR-NC inhibitor (5′-UUC UCC GAA CGU GUC ACG UUU-3′) or miR-2278
inhibitor (5′-CCA GGC AAC ACA CAC UGC UCU C-3′) were transfected
into 1×106 OVCAR3 or SKOV3 cells in 6-well plates using
Lipofectamine 300 (Invitrogen; Thermo Fisher Scientific, Inc.).
Following 48 h of cell growth, the cells were harvested and the
transfection efficiency was determined by RT-qPCR as described
above.
Small interference RNA mediated silencing
of LATS2
Control siRNA (5′-UUC UCC GAA CGU GUC ACG UTT-3′)
and LATS2 siRNA (5′-UAC CAUA AAU ACA AUC UUC TT-3′) were
synthesized and purchased from Suzhou GenePharma Co., Ltd. A total
of 50 nM control siRNA or LATS2 siRNA were transfected into
1×106 OVCAR3 or SKOV3 cells in 6-well plates using
Lipofectamine 300 (Invitrogen; Thermo Fisher Scientific, Inc.).
After 48 h, cells were harvested and the transfection efficiency
was determined by western blot analysis as described below.
Cell proliferation assay
The proliferative ability of the OVCAR3 and SKOV3
cells was detected using the CCK-8 kit (Dojindo Molecular
Technologies, Inc.). Briefly, 10 µl CCK-8 solution were
added into the culture medium and sustained for 1 h at 37°C. The
absorbance at 450 nM of the medium was measured using a microplate
reader (iMark™, Bio-Rad Laboratories, Inc.).
Cell apoptosis assay
The percentage of apoptotic cells was detected by
flow cytometry. OVCAR3 and SKOV3 cells were incubated for 48 h at
37°C following transfection, harvested and stained with propidium
iodide (PI) and Annexin-V provided by the Annexin-V Apoptosis
Detection kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The stained cells were
examined by flow cytometry on a BD FACSVerse flow cytometer (BD
Biosciences). PI+/Annexin-V+ and
PI-/Annexin-V+ cells were considered as apoptotic
cells.
Bioinformatics analysis
The sequence of ASAP1-IT1 was entered into
the online miRDB software (http://mirdb.org/) (23) to predict potential binding
miRNAs. The potential target genes of miR-2278 were predicted with
the TargetScan software (http://www.targetscan.org/vert_72/) (24).
Western blot analysis
Tissues and cells were subjected to protein
extraction using RIPA lysis buffer (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. The
lysates were quantified with a BCA protein assay kit (Thermo Fisher
Scientific, Inc.). A total of 20 µg proteins were loaded
into each lane of the 8% SDS-PAGE gel and electrophoresis was
performed. The proteins were transferred to a PVDF membrane.
Subsequently, the membrane was blocked in 5% non-fat milk for 1 h
at room temperature and incubated with primary and secondary
antibodies at room temperature for 1 h at 37°C. The blots were
developed using the ECL Western blot substrate (Pierce; Thermo
Fisher Scientific, Inc.). The results were quantified using ImageJ
software (version 1.8, National Institutes of Health). The
antibodies used were as follows: YAP1 (cat. no. 14074, 1:1,000),
p-YAP1 (Ser127l; cat. no. 13008, 1:1,000), LATS2 (cat. no. 5888,
1:1,000) and GAPDH (cat. no. 97166, 1:10,000) antibodies were
purchased from Cell Signaling Technology, Inc; p-LATS2 (T1079 +
T1041; cat. no. ab111344, 1:1,000) antibody was bought from Abcam.
HRP-conjugated secondary antibodies against mouse (cat. no.
L3032-2, 1:5,000) and rabbit (cat. no. L3012-2, 1:5,000) were
products of Signalway Antibody LLC.
Dual luciferase reporter assay
The full length of ASAP1-IT1 was sub-cloned
from pcDNA3.1 into the pmirGLO plasmid (Promega Corporation). The
3′UTR of LATS2 was amplified from HOSEpiC cDNA, digested with
restriction endonucleases NheI (NEB) and SalI (NEB)
and then ligated into the pmirGLO plasmid. Point mutations were
introduced into the putative binding site for miR-2278 on
ASAP1-IT1 and LATS2 3′UTR with a QuickChange Lightning
Site-Directed Mutagenesis kit (Agilent Technologies, Inc.)
following the manufacturer's protocol. OVCAR3 and SKOV3 cells were
seeded at a density of 5×105 cells in 24-well plates and
transfected with 1 µg pmirGLO-ASAP1-IT1-WT,
pmirGLO-ASAP1-IT1-Mut, pmirGLO-LATS2 3′UTR-WT or
pmirGLO-LATS2 3′UTR-Mut. The cells were incubated for 48 h
following transfection and subsequently collected for the
determination of the relative luciferase activity, which was
detected with a Dual Luciferase Reporter System (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity.
Statistical analysis
Graphpad Prism 6 was used to analyze the data and
the data are presented as the means ± SD. Differences between 2
groups were compared with the Student's t-test and those between
multiple groups were compared with one-way ANOVA followed by a
Tukey's post hoc test. The log rank test and Kaplan-Meier curve
were used for survival analysis and the graphs were prepared using
Graphpad Prism 6. The association between ASAP1-IT1, miR-2278 and
LATS2 expression with the clinicopathological characteristics of 58
ovarian tumors was examined using the Chi-squared test. The
correlation between the differences in the mRNA expression levels
of ASAP1-IT1, miR-2278 and LATS2 were analyzed by the
Pearson's correlation analysis. A P-value <0.05 (P<0.05) was
considered to indicate a statistically significant difference.
Results
High ASAP1-IT1 levels predict the optimal
prognosis of patients with ovarian cancer
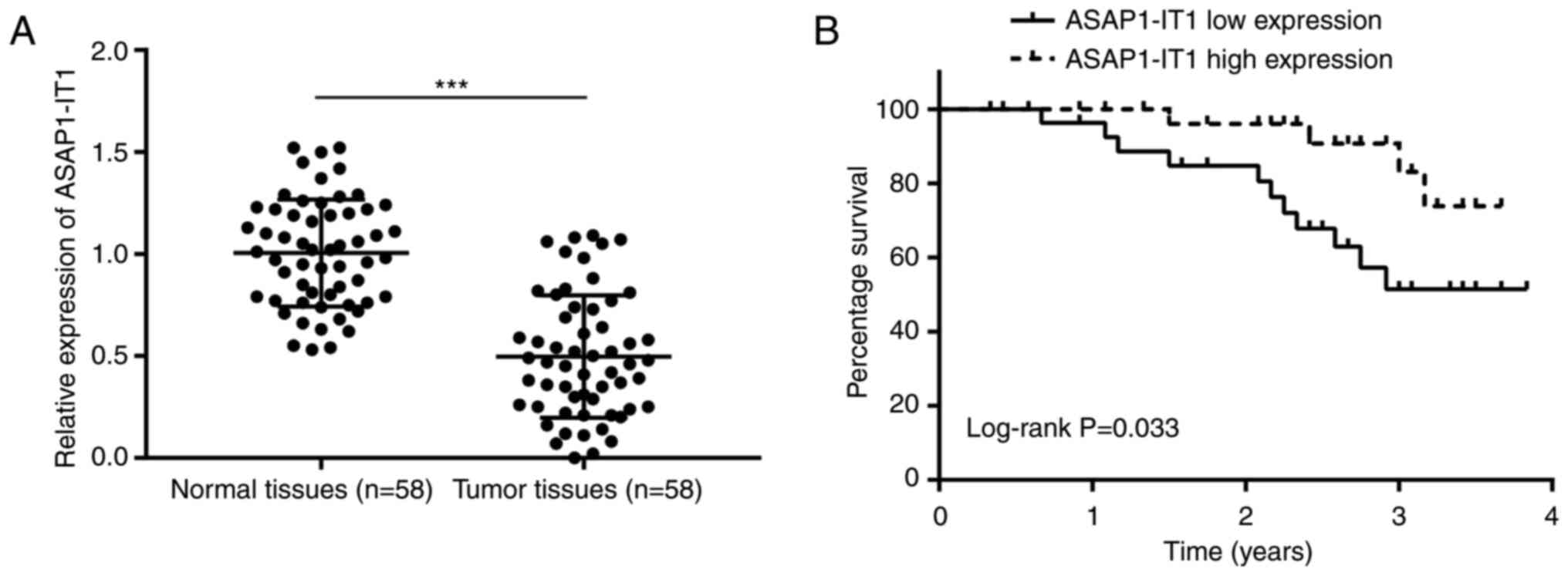
To examine the role of ASAP1-IT1 in ovarian
cancer, 58 pairs of ovarian tumors were collected and matched with
healthy tissues from the patients. The expression levels of
ASAP1-IT1 were downregulated in ovarian tumor tissues
compared with the non-cancer specimens (Fig. 1A). No significant associations
were observed between ASAP1-IT1 expression and the clinical
parameters of pathological type, patient age and lymph node
metastasis (Table II). However,
disease stage and differentiation status were associated with
ASAP1-IT1 expression (Table
II). Subsequently, the patients were divided into the
ASAP1-IT1 high and low expression groups according to the
median expression of ASAP1-IT1. The results of Kaplan-Meier
analysis indicated that a high expression of ASAP1-IT1 was
associated with the optimal prognosis of patients with ovarian
cancer, whereas the 3-year survival rate was estimated at 85 and
45% in the high and low expression groups, respectively (Fig. 1B). These results highlighted the
potential involvement of ASAP1-1T1 in the progression of
ovarian cancer.
 | Table IIAssociation of ASAP1-IT1, miR-2278
and LATS2 expression with the clinicopathological characteristics
of 58 patients with ovarian tumors. |
Table II
Association of ASAP1-IT1, miR-2278
and LATS2 expression with the clinicopathological characteristics
of 58 patients with ovarian tumors.
| Clinicopathological
features | Total number | ASAP1-IT1
expression
| P-value | miR-2278 expression
| P-value | LATS2 expression
| P-value |
|---|
| Low | High | Low | High | Low | High |
|---|
| Pathological
type | | | | 0.395 | | | 0.155 | | | 0.777 |
| Serous
carcinoma | 40 | 22 | 18 | | 17 | 23 | | 19 | 21 | |
| Other pathology
types | 18 | 7 | 11 | | 12 | 6 | | 10 | 8 | |
| Age, years | | | | 0.999 | | | 0.292 | | | 0.599 |
| >50 | 31 | 16 | 15 | | 18 | 13 | | 14 | 17 | |
| ≤50 | 27 | 13 | 14 | | 11 | 16 | | 15 | 12 | |
| FIGO stage | | | | 0.019 | | | 0.003 | | | 0.248 |
| I-II | 17 | 4 | 13 | | 14 | 3 | | 11 | 6 | |
| III-IV | 41 | 25 | 16 | | 15 | 26 | | 18 | 23 | |
| Differentiation
status | | | | 0.021 | | | 0.747 | | | 0.331 |
| Well | 12 | 2 | 10 | | 5 | 7 | | 8 | 4 | |
| Moderate and
poor | 46 | 27 | 19 | | 24 | 22 | | 21 | 25 | |
| Lymph node
metastasis | | | | 0.229 | | | 0.549 | | | 0.229 |
| Negative | 15 | 5 | 10 | | 6 | 9 | | 10 | 5 | |
| Positive | 43 | 24 | 19 | | 23 | 20 | | 19 | 24 | |
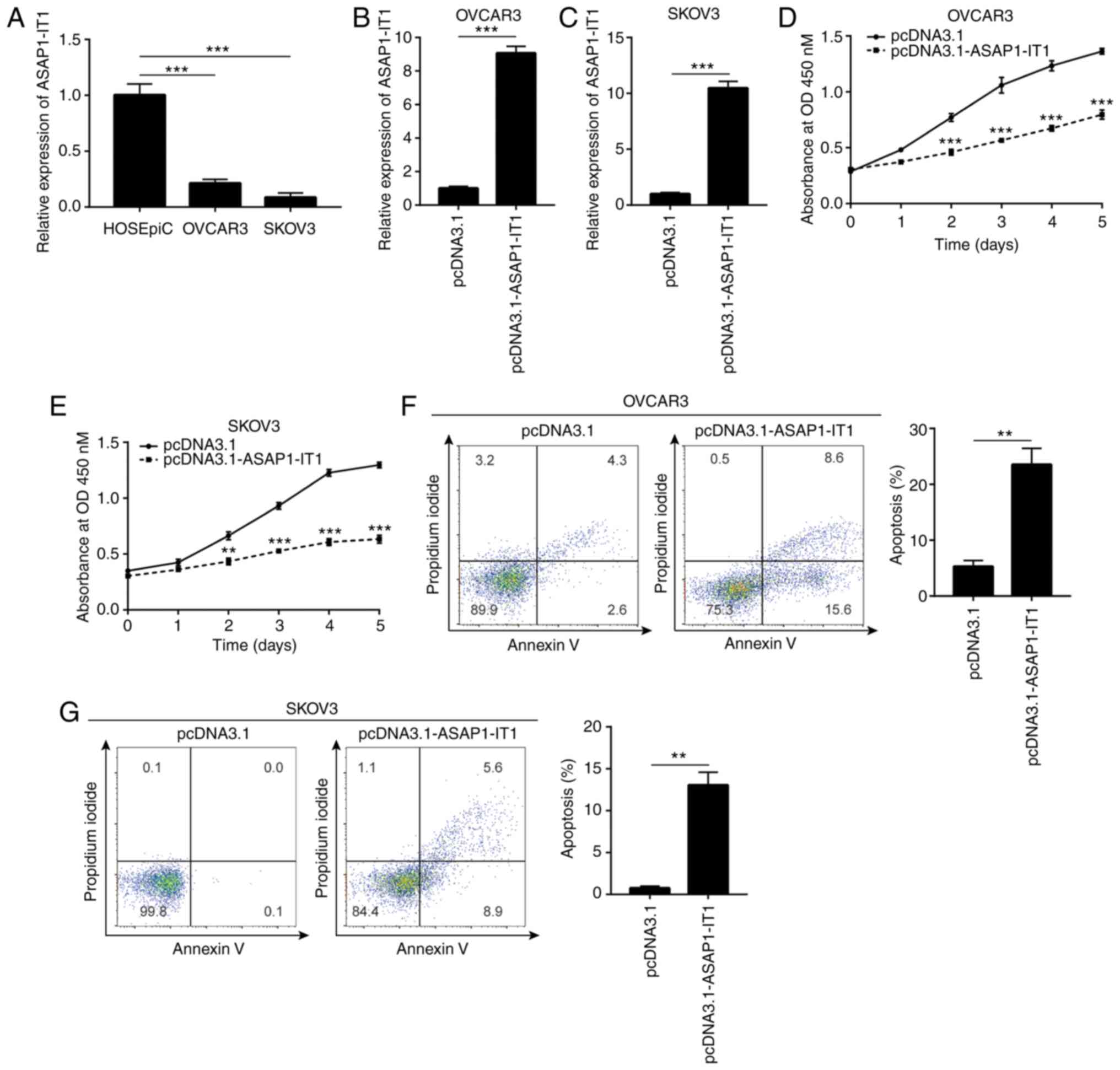
Overexpression of ASAP1-IT1 inhibits
ovarian cancer cell proliferation and induces apoptosis
To investigate the biological function of
ASAP1-IT1 in ovarian cancer cells, RT-qPCR was conducted to
detect ASAP1-IT1 expression in normal ovarian cells and
ovarian cancer cells. It was found that ASAP1-IT1 expression
was decreased in OVCAR3 and SKOV3 cells, which are 2 ovarian cancer
cell lines, compared with the corresponding expression noted in the
HOSEpiC cell line (Fig. 2A). The
vector containing the full length of ASAP1-IT1 was
transfected into the OVCAR3 and SKOV3 cells in order to increase
ASAP1-IT1 expression (Fig. 2B
and C). The proliferative ability of the OVCAR3 and SKOV3 cells
was detected by CCK-8 assay and the data indicated that it was
significantly inhibited following ASAP1-IT1 overexpression
(Fig. 2D and E). Moreover, flow
cytometric analysis indicated an increase in the number of
apoptotic cells observed in the OVCAR3 and SKOV3
pcDNA3.1-ASAP1-IT1-transfected cell lines (Fig. 2F and G). These data suggested
that ASAP1-IT1 was involved in ovarian cancer cell
proliferation and survival.
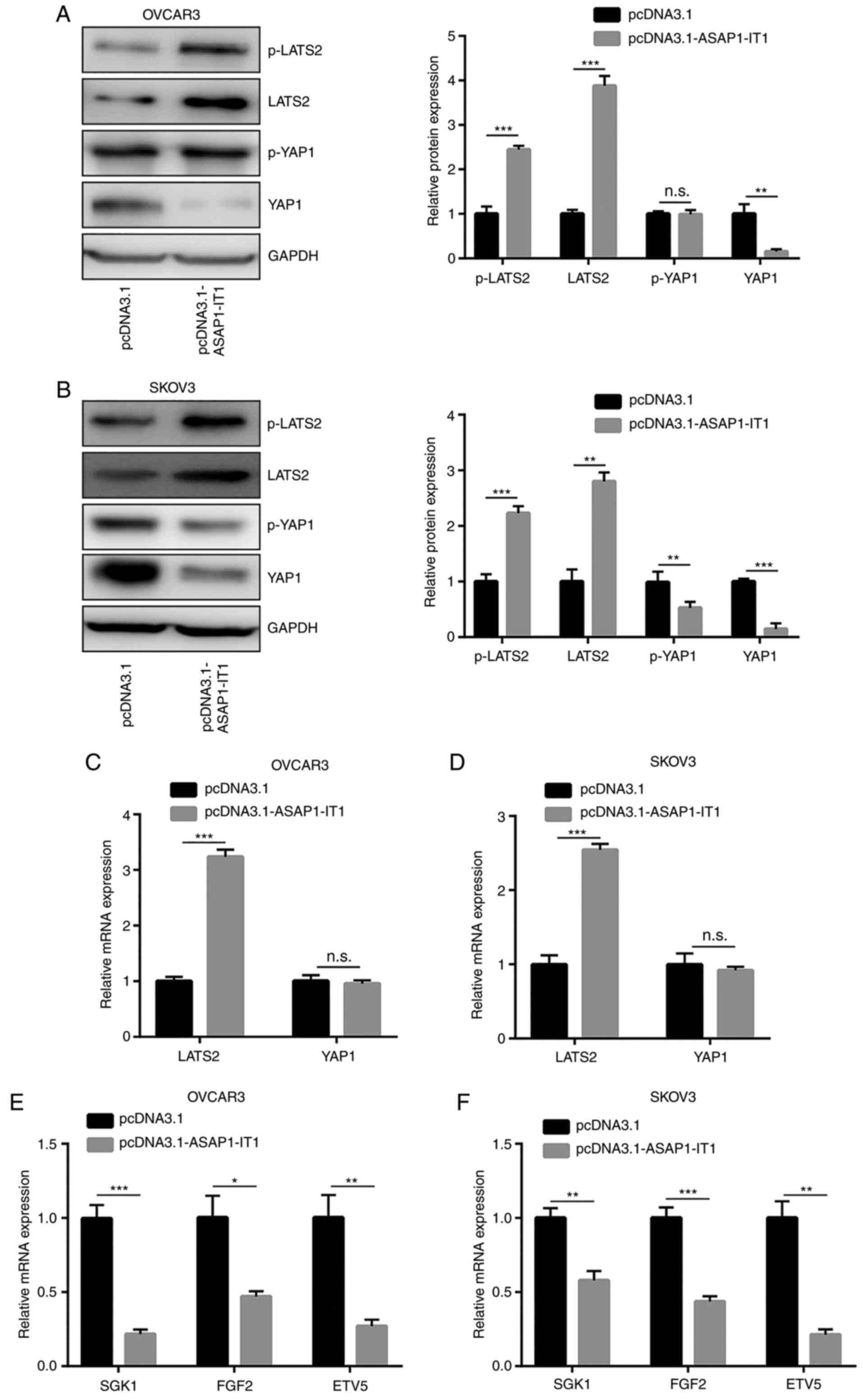
ASAP1-IT1 inactivates YAP1 signaling in
ovarian cancer cells
Hippo/YAP1 signaling is critical for ovarian cancer
cell proliferation and survival (17). In the present study, western blot
analysis was used to detect the expression levels of LATS2, which
is the major kinase component of the Hippo pathway (19). Moreover, the expression of YAP1
was detected in ovarian cancer cells following ASAP1-IT1
overexpression. ASAP1-IT1 overexpression increased LATS2 and
p-LATS2 expression, and decreased YAP1 protein expression in the
OVCAR3 and SKOV3 cells (Fig. 3A and
B). An increase in the LATS2 mRNA levels was also observed
(Fig. 3C and D). In addition,
the mRNA expression levels of SGK1, FGF2 and
ETV5, which are 3 conserved YAP1 downstream genes involved
in ovarian cancer cell apoptosis (25-28), were decreased in the OVCAR3 and
SKOV3 cells (Fig. 3E and F).
These data indicated that ASAP1-IT1 upregulated LATS2
expression and inactivated YAP1 signaling in ovarian cancer
cells.
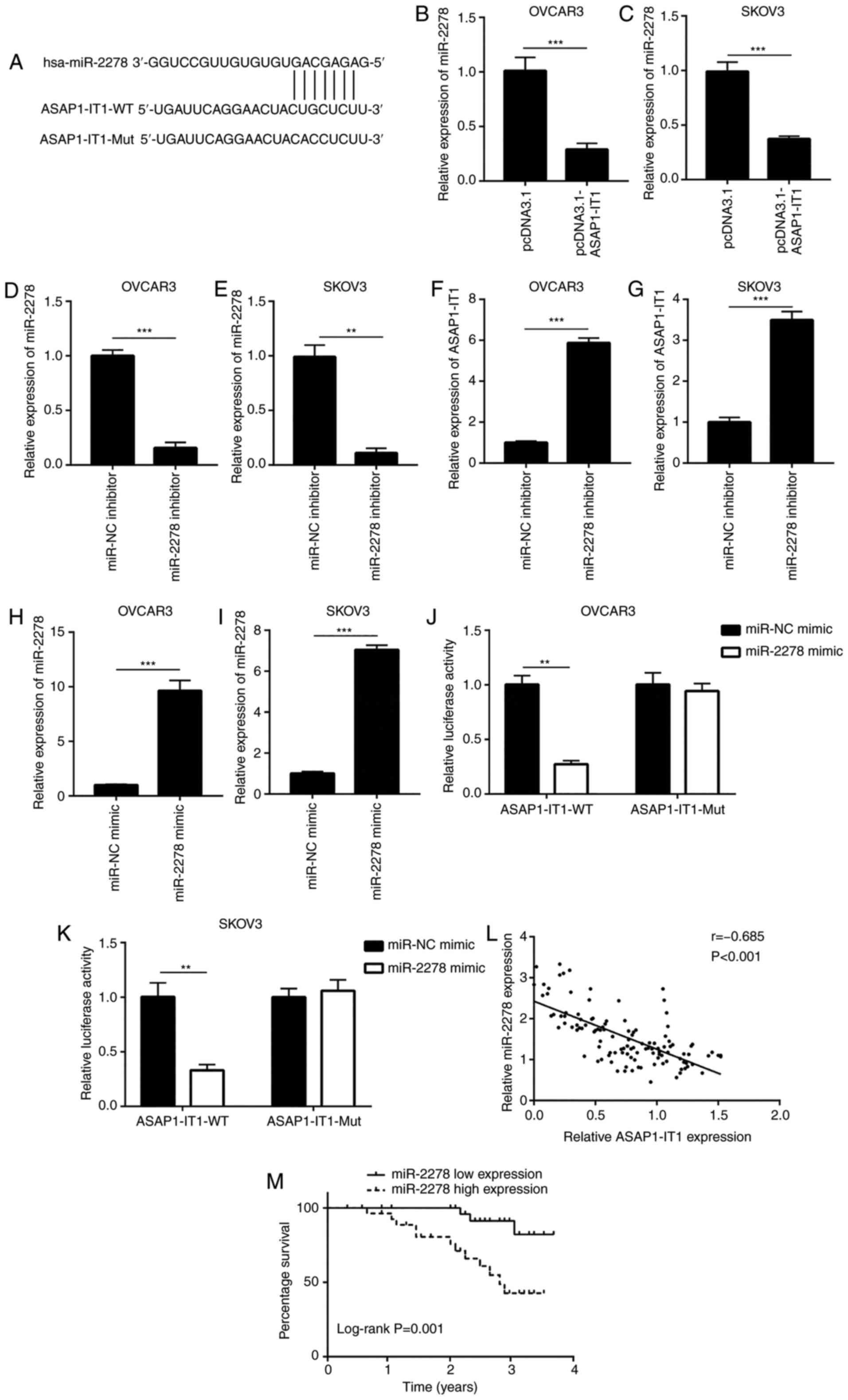
ASAP1-IT1 sponges miR-2278 in ovarian
cancer cells
Bioinformatic analysis was used to predict potential
miRNAs that may interact with ASAP1-IT1. Among the candidate
miRNAs, the ASAP1-IT1 sequence harbored the miRNA responsive
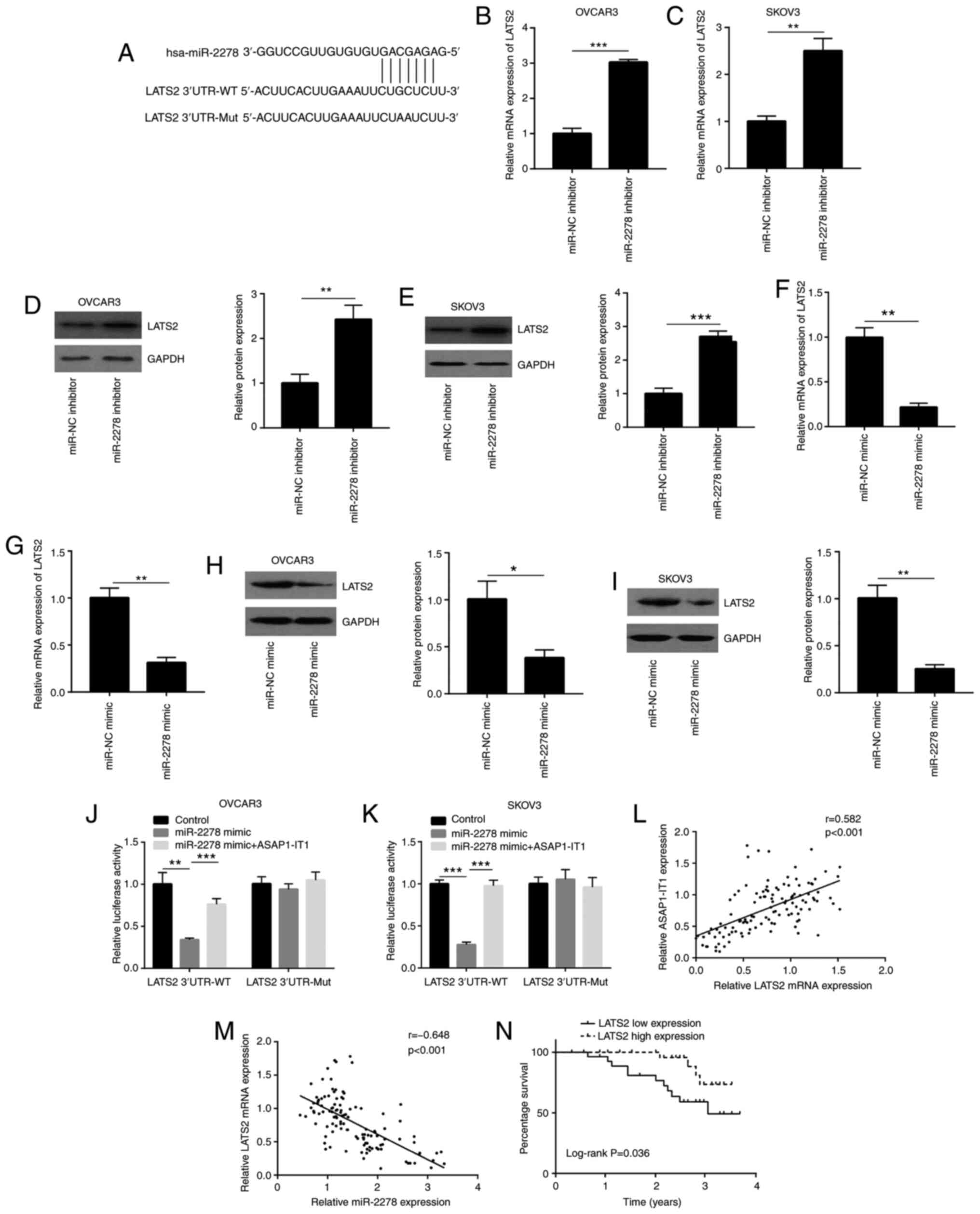
element for miR-2278 (Fig. 4A).
The expression of the latter has been shown to be upregulated in
the serum from patients with ovarian cancer (29). It is interesting to note that in
the present study, high expression levels of miR-2278 were
associated with an advanced FIGO tumor stage (Table II). The overexpression of
ASAP1-IT1 decreased miR-2278 expression in OVCAR3 and SKOV3
cells (Fig. 4B and C).
Subsequently, miR-2278 inhibitor was used to decrease miR-2278
expression in OVCAR3 and SKOV3 cells (Fig. 4D and E). The downregulation of
miR-2278 increased ASAP1-IT1 expression in these cells
(Fig. 4F and G). In addition,
transfection with miR-2278 mimic increased miR-2278 expression in
the OVCAR3 and SKOV3 cells (Fig. 4H
and I). In the OVCAR3 cells, the overexpression of miR-2278
decreased the relative luciferase activity of the
ASAP1-IT1-WT cells, whereas this effect was not noted in the
ASAP1-IT1-Mut cells that harbored mutations in the putative
binding site (Fig. 4J). Similar
results were observed in SKOV3 cells (Fig. 4K), suggesting a direct
interaction between ASAP1-IT1 and miR-2278. Furthermore, a
significant negative correlation between ASAP1-IT1 and
miR-2278 expression was noted in the clinical samples (Fig. 4L). In contrast to the
ASAP1-IT1 cells, a high expression of miR-2278 was
associated with a poor prognosis of patients with ovarian cancer
(Fig. 4M).
ASAP1-IT1 upregulates LATS2 expression
via sponging miR-2278
Bioinformatics analysis indicated a putative binding
site of miR-2278 in the 3′UTR of LATS2 mRNA (Fig. 5A), suggesting that
ASAP1-IT1 may regulate LATS2 via miR-2278. The
downregulation of miR-2278 increased LATS2 mRNA expression
in OVCAR3 and SKOV3 cells (Fig. 5B
and C). In addition, western blot analysis confirmed an
elevation of LATS2 protein expression in OVCAR3 and SKOV3 cells
transfected with the miR-2278 inhibitor (Fig. 5D and E). In contrast to these
observations, the overexpression of miR-2278 decreased LATS2
mRNA (Fig. 5F and G) and protein
expression levels (Fig. 5H and
I). Dual luciferase reporter assay indicated that miR-2278
reduced the relative luciferase activity of the plasmid containing
the LATS2 3′UTR sequence, which was reversed when the
ASAP1-IT1 overexpression plasmid was transfected into OVCAR3
and SKOV3 cells (Fig. 5J and K).
Furthermore, a positive correlation was noted between LATS2
mRNA and ASAP1-IT1 expression in the clinical samples
(Fig. 5L). In contrast to these
observations, the miR-2278 expression negatively correlated with
the LATS2 mRNA levels in these samples (Fig. 5M). LATS2 expression was
also negatively associated with the overall survival status of the
patients (Fig. 5N). However,
LATS2 mRNA expression was not associated with the parameters
pathological type, age, FIGO stage, differentiation status and
lymph node metastasis (Table
II). The data indicated that miR-2278 directly suppressed LATS2
expression and suggested that ASAP1-IT1 may inactivate YAP1
signaling by binding to miR-2278.
ASAP1-IT1 inactivates YAP1 signaling via
the upregulation of LATS2 expression
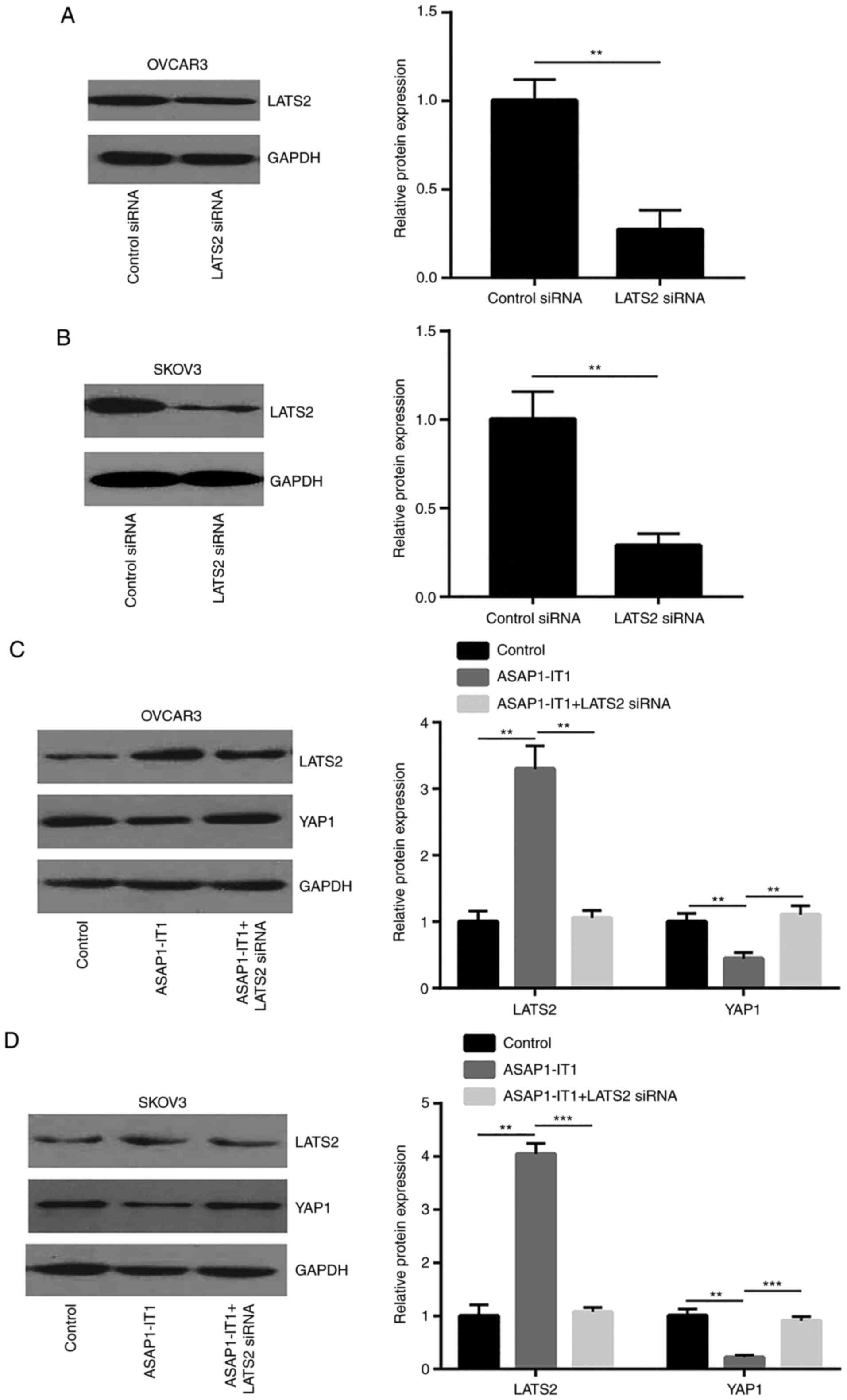
To explore whether ASAP1-IT1 can downregulate
YAP1 via LATS2, LATS2 siRNA was used to silence LATS2 expression in
OVCAR3 and SKOV3 cells (Fig. 6A and
B). The silencing of LATS2 attenuated the downregulation of
YAP1 protein expression induced by ASAP1-IT1 overexpression
in OVCAR3 and SKOV3 cells (Fig. 6C
and D). The Hippo pathway suppressed YAP1 activity via the
phosphorylation of YAP1 to induce its translocation from the
nucleus to the cytoplasm. Taken together, these data indicated that
ASAP1-IT1 inactivated YAP1 signaling via the upregulation of
LATS2 expression.
ASAP1-IT1 suppresses ovarian cancer
progression via the regulation of LATS2 expression
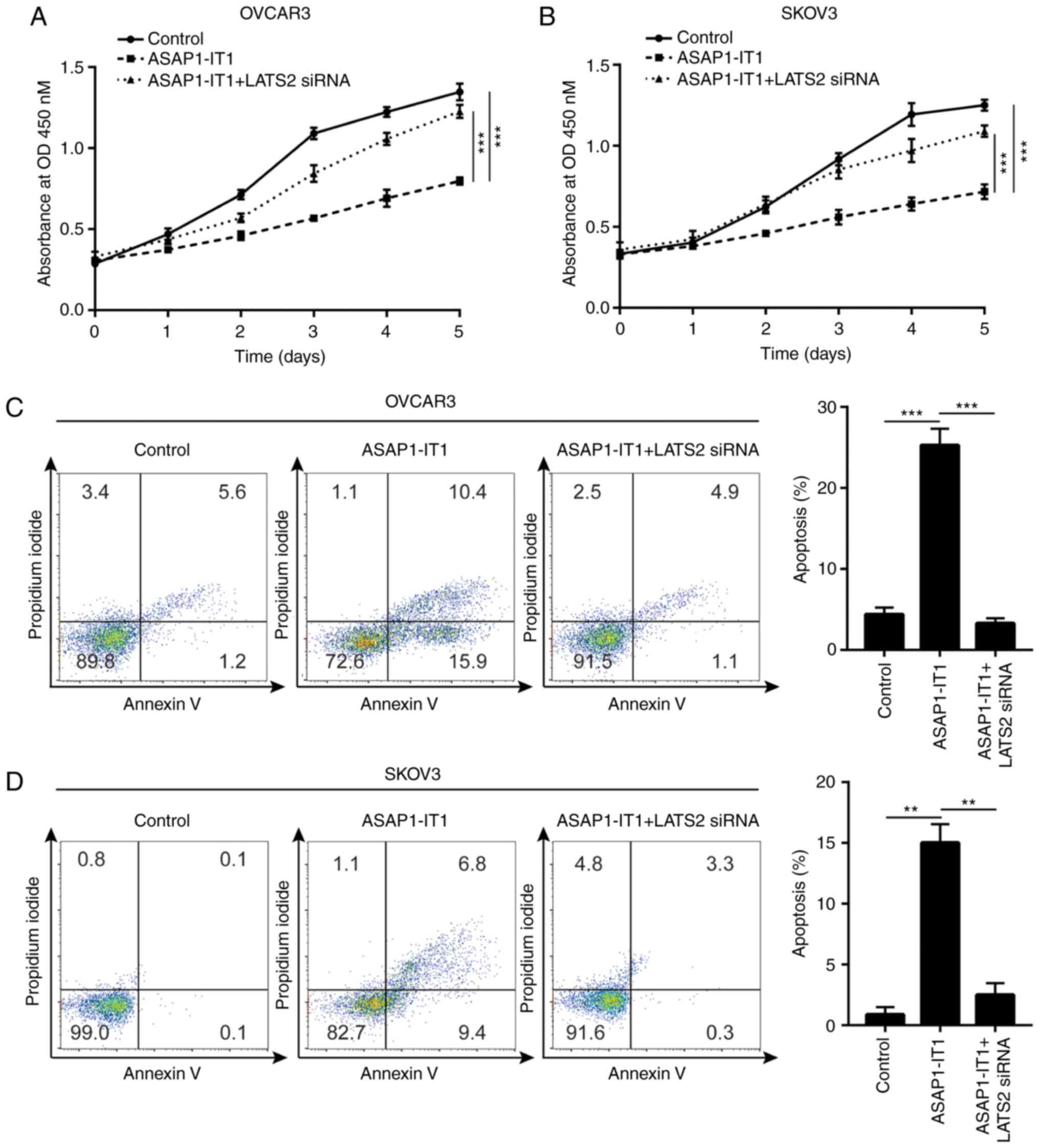
The results of cell proliferation assay indicated
that the silencing of LATS2 attenuated the growth inhibitory
effects of ASAP1-IT1 overexpression on the OVCAR3 and SKOV3
cells (Fig. 7A and B).
Furthermore, LATS2 silencing inhibited the apoptosis of OVCAR3 and
SKOV3 cells induced by ASAP1-IT1 overexpression (Fig. 7C and D), suggesting that LATS2 is
essential for the tumor suppressive role of ASAP1-IT1 in
ovarian cancer.
Discussion
Accumulating evidence has indicated that lncRNAs are
mainly involved in the initiation and development of ovarian cancer
by controlling signaling networks and promoting sustained cancer
cell proliferation and resistance to cell apoptosis (18,30,31). The inactivation of uncontrolled
cell growth signaling and the induction of cancer cell death are
critical for improving the prognosis of patients with ovarian
cancer (32). Therefore,
investigations into the biological roles and molecular mechanisms
of lncRNAs is crucial for providing insight into the treatment of
ovarian cancer.
Due to the oncogenic potential of ASAP1 (33), ASAP1-IT1, the intronic
transcript of ASAP1 (11), has
gained considerable attention in cancer research over the past
years. The aberrant upregulation of ASAP1-IT1 expression has
been observed in several cancer types, including bladder cancer
(10), non-small cell lung
cancer (34) and
cholangiocarcinoma (35). In
non-small cell lung cancer cells, ASAP1-IT1 facilitates cell
proliferation, migration and invasion via the activation of the
PI3K/AKT pathway (34).
ASAP1-IT1 positively regulates Hedgehog signaling in
cholangiocarcinoma cells, leading to an enhanced cell proliferation
and migration (35). In contrast
to these observations, ASAP1-IT1 expression is downregulated
in ovarian cancer and is associated with the optimal prognosis of
patients with this disease (11). Moreover, the results demonstrated
that the upregulation of ASAP1-IT1 expression in ovarian
tumors was associated with optimal prognosis. In addition, high
expression levels of ASAP1-IT were associated with early-stage
tumors and an optimal differentiation status. The overexpression of
ASAP1-IT1 significantly inhibited ovarian cancer cell
proliferation and induced cell apoptosis. The data suggested the
involvement of ASAP1-IT1 in ovarian cancer progression.
The molecular mechanisms of ASAP1-IT1 in
ovarian cancer cells have not yet been previously examined, at
least to the best of our knowledge. The transcription co-activator,
YAP1, plays a pivotal role in ovarian cancer initiation,
proliferation and survival (36). The loss of LATS2 is essential for
the malignant transformation of ovarian cells via the regulation of
YAP1 expression (37). The
present study indicated that ASAP1-IT1 upregulated LATS2
mRNA and protein expression, while it suppressed YAP1 protein
expression. This suggested that ASAP1-IT1 may promote LATS2
mRNA stability, activate the Hippo pathway, destabilize YAP1
protein and inactivate YAP1 signaling. By using bioinformatic
analysis, several miRNAs were predicted to interact with
ASAP1-IT1. Among the list of miRNAs, miR-2278 was a highly
ranked potential target of ASAP1-IT1. According to the
microarray data derived from the sera of 168 high-grade serous
ovarian carcinoma patients and 65 healthy controls, miR-2278 has
been shown to be one of the top upregulated miRNAs identified
(29). Currently, the
association between miR-2278 and LATS2 has not yet been reported in
any cell type, at least to the best of our knowledge.
Bioinformatics analysis indicated that miR-2278 may target
LATS2. By using RT-qPCR, western blot analysis and dual
luciferase reporter assays, the data of the present study confirmed
that LATS2 was a target of miR-2278 and that
ASAP1-IT1 upregulated LATS2 expression. The Hippo pathway
phosphorylates YAP1, leading to translocation of YAP1 from the
nucleus to the cytoplasm, which is required for subsequent
degradation (19). In the
present study, ASAP1-IT1 overexpression induced the
translocation of YAP1, which was reversed following LATS2 the
silencing in OVCAR3 cells. Furthermore, silencing of LATS2
attenuated the biological function of ASAP1-IT1 in ovarian
cancer cells. Previous studies have indicated that YAP1 is directly
regulated by miRNA/lncRNA in ovarian cancer (17,18). The findings of the present study
suggested that ASAP1-IT1 inhibited ovarian cancer
progression by sponging miR-2278 and inducing the upregulation of
LATS2 expression, the activation of the Hippo pathway and the
inactivation of YAP1.
Taken together, the present study revealed that the
ASAP1-IT1/miR-2278/LATS2 axis was involved in the regulation
of ovarian cancer progression. Due to the complexity of signaling
transduction in cancer cells, the current findings will be further
extended in future studies in order to examine the molecular
mechanisms of ASAP1-IT1 in ovarian cancer.
Funding
No funding was received.
Availability of data and materials
They are available at special request.
Authors' contributions
KW and YZ contributed to the experiment design and
the performance of the experiments. KW and YBH collected the
specimens and clinical information of the patients. CY supervised
the study, analyzed the data and wrote the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in the present study
involving human participants were supervised and approved by the
Ethical Committee of The China-Japan Union Hospital of Jilin
University. Written informed consent for the publication of all
data was obtained from all patients prior to participate the
current study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Prat J: New insights into ovarian cancer
pathology. Ann Oncol. 23(Suppl 10): x111–x117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lheureux S, Gourley C, Vergote I and Oza
AM: Epithelial ovarian cancer. Lancet. 393:1240–1253. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Castro-Oropeza R, Melendez-Zajgla J,
Maldonado V and Vazquez-Santillan K: The emerging role of lncRNAs
in the regulation of cancer stem cells. Cell Oncol (Dordr).
41:585–603. 2018. View Article : Google Scholar
|
|
5
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang X, Hamblin MH and Yin KJ: The long
noncoding RNA malat1: Its physiological and pathophysiological
functions. RNA Biol. 14:1705–1714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitra R, Chen X, Greenawalt EJ, Maulik U,
Jiang W, Zhao Z and Eischen CM: Decoding critical long non-coding
RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat
Commun. 8:16042017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu DD, Chen X, Sun KX, Wang LL, Chen S and
Zhao Y: Role of the lncRNA ABHD11-AS1 in the tumorigenesis and
progression of epithelial ovarian cancer through targeted
regulation of RhoC. Mol Cancer. 16:1382017. View Article : Google Scholar :
|
|
10
|
Yang L, Xue Y, Liu J, Zhuang J, Shen L,
Shen B, Yan J and Guo H: Long noncoding RNA ASAP1-IT1 promotes
cancer stemness and predicts a poor prognosis in patients with
bladder cancer. Neoplasma. 64:847–855. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fu Y, Biglia N, Wang Z, Shen Y, Risch HA,
Lu L, Canuto EM, Jia W, Katsaros D and Yu H: Long non-coding RNAs,
ASAP1-IT1, FAM215A, and LINC00472, in epithelial ovarian cancer.
Gynecol Oncol. 143:642–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu FX, Zhao B, Panupinthu N, Jewell JL,
Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al:
Regulation of the Hippo-YAP pathway by G-protein-coupled receptor
signaling. Cell. 150:780–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Overholtzer M, Zhang J, Smolen GA, Muir B,
Li W, Sgroi DC, Deng CX, Brugge JS and Haber DA: Transforming
properties of YAP, a candidate oncogene on the chromosome 11q22
amplicon. Proc Natl Acad Sci USA. 103:12405–12410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pan Y, Tong JHM, Lung RWM, Kang W, Kwan
JSH, Chak WP, Tin KY, Chung LY, Wu F, Ng SSM, et al: RASAL2
promotes tumor progression through LATS2/YAP1 axis of hippo
signaling pathway in colorectal cancer. Mol Cancer. 17:1022018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dong L, Lin F, Wu W, Liu Y and Huang W:
Verteporfin inhibits YAP-induced bladder cancer cell growth and
invasion via Hippo signaling pathway. Int J Med Sci. 15:645–652.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu S, Cai X, Wu C, Wu L, Wang Y, Liu Y, Yu
Z, Qin S, Ma F, Thiery JP and Chen L: Adhesion glycoprotein CD44
functions as an upstream regulator of a network connecting ERK, AKT
and Hippo-YAP pathways in cancer progression. Oncotarget.
6:2951–2965. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan H, Li H, Silva MA, Guan Y, Yang L, Zhu
L, Zhang Z, Li G and Ren C: LncRNA FLVCR1-AS1 mediates miR-513/YAP1
signaling to promote cell progression, migration, invasion and EMT
process in ovarian cancer. J Exp Clin Cancer Res. 38:3562019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yan H, Li H, Li P, Li X, Lin J, Zhu L,
Silva MA, Wang X, Wang P and Zhang Z: Long noncoding RNA MLK7-AS1
promotes ovarian cancer cells progression by modulating
miR-375/YAP1 axis. J Exp Clin Cancer Res. 37:2372018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo C, Wang X and Liang L: LATS2-mediated
YAP1 phosphorylation is involved in HCC tumorigenesis. Int J Clin
Exp Pathol. 8:1690–1697. 2015.PubMed/NCBI
|
|
20
|
Guo Y, Cui J, Ji Z, Cheng C, Zhang K,
Zhang C, Chu M, Zhao Q, Yu Z, Zhang Y, et al: miR-302/367/LATS2/YAP
pathway is essential for prostate tumor-propagating cells and
promotes the development of castration resistance. Oncogene.
36:6336–6347. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia Y and Gao Y: MicroRNA-181b promotes
ovarian cancer cell growth and invasion by targeting LATS2. Biochem
Biophys Res Commun. 447:446–451. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48(D1): D127–D131. 2020. View Article : Google Scholar :
|
|
24
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar :
|
|
25
|
Cordenonsi M, Zanconato F, Azzolin L,
Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR,
Poletti A, et al: The Hippo transducer TAZ confers cancer stem
cell-related traits on breast cancer cells. Cell. 147:759–772.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Melhem A, Yamada SD, Fleming GF, Delgado
B, Brickley DR, Wu W, Kocherginsky M and Conzen SD: Administration
of glucocorticoids to ovarian cancer patients is associated with
expression of the anti-apoptotic genes SGK1 and MKP1/DUSP1 in
ovarian tissues. Clin Cancer Res. 15:3196–3204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li R, Wang Y, Xu Y, He X and Li Y:
Silencing the long noncoding RNA, TINCR, a molecular sponge of
miR335, inhibits the malignant phenotype of epithelial ovarian
cancer via FGF2 suppression. Int J Oncol. 55:1110–1124.
2019.PubMed/NCBI
|
|
28
|
Llaurado M, Abal M, Castellvi J, Cabrera
S, Gil-Moreno A, Pérez-Benavente A, Colás E, Doll A, Dolcet X,
Matias-Guiu X, et al: ETV5 transcription factor is overexpressed in
ovarian cancer and regulates cell adhesion in ovarian cancer cells.
Int J Cancer. 130:1532–1543. 2012. View Article : Google Scholar
|
|
29
|
Todeschini P, Salviato E, Paracchini L,
Ferracin M, Petrillo M, Zanotti L, Tognon G, Gambino A, Calura E,
Caratti G, et al: Circulating miRNA landscape identifies miR-1246
as promising diagnostic biomarker in high-grade serous ovarian
carcinoma: A validation across two independent cohorts. Cancer
Lett. 388:320–327. 2017. View Article : Google Scholar
|
|
30
|
Li J, Yang C, Li Y, Chen A, Li L and You
Z: LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome
formation. Biosci Rep. 38:BSR201711502018. View Article : Google Scholar :
|
|
31
|
An J, Lv W and Zhang Y: LncRNA NEAT1
contributes to paclitaxel resistance of ovarian cancer cells by
regulating ZEB1 expression via miR-194. Onco Targets Ther.
10:5377–5390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Muller T, Stein U, Poletti A, Garzia L,
Rothley M, Plaumann D, Thiele W, Bauer M, Galasso A, Schlag P, et
al: ASAP1 promotes tumor cell motility and invasiveness, stimulates
metastasis formation in vivo, and correlates with poor survival in
colorectal cancer patients. Oncogene. 29:2393–2403. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang L, Shi SB, Zhu Y, Qian TT and Wang
HL: Long non-coding RNA ASAP1-IT1 promotes cell proliferation,
invasion and metastasis through the PTEN/AKT signaling axis in
non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 22:142–149.
2018.PubMed/NCBI
|
|
35
|
Guo L, Zhou Y, Chen Y, Sun H, Wang Y and
Qu Y: LncRNA ASAP1-IT1 positively modulates the development of
cholangiocarcinoma via hedgehog signaling pathway. Biomed
Pharmacother. 103:167–173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Justilien V, Brennan KI, Jamieson
L, Murray NR and Fields AP: PKCiota regulates nuclear YAP1
localization and ovarian cancer tumorigenesis. Oncogene.
36:534–545. 2017. View Article : Google Scholar
|
|
37
|
He C, Lv X, Huang C, Hua G, Ma B, Chen X,
Angeletti PC, Dong J, Zhou J, Wang Z, et al: YAP1-LATS2 feedback
loop dictates senescent or malignant cell fate to maintain tissue
homeostasis. EMBO Rep. 20:e449482019. View Article : Google Scholar : PubMed/NCBI
|