Introduction
Cervical cancer is one of the most frequent
malignancies in gynecology, and it is the second leading cause of
cancer-related mortality in women, greatly endangering their health
(1). Approximately 569,847
individuals are diagnosed with cervical cancer annually, and
311,365 individuals succumb to this malignancy (2). The currently available therapies,
including surgical treatment, chemotherapy and radiotherapy,
effectively treat cervical cancer in situ. However, these
treatment modalities have a poor therapeutic efficacy in patients
with cervical cancer that is diagnosed at an advanced stage,
particularly in patients with metastatic tumors (3). Notably, only approximately 40% of
patients with cervical cancer survive for >5 years partially due
to the highly invasive, uncontrolled growth and metastatic capacity
of cervical cancer (4). The
initiation and progression of cervical cancer involve a wide range
of complex changes (5). However,
the exact molecular events are largely undefined, which severely
limits the exploration of novel treatment methods. Therefore, an
in-depth elucidation of the mechanisms underlying the pathogenesis
of cervical cancer is imperative for the development of effective
therapies and improving clinical outcomes.
Long non-coding RNAs (lncRNAs) have gained
increasing attention in recent years (6). These molecules are comprised of
>200 nucleotides and are non-protein coding in nature (7). lncRNAs positively or negatively
affect gene expression at the transcriptional or
post-transcriptional level (8).
It has been demonstrated that lncRNAs function as modulators to
regulate physiological and pathological activities (9). Differentially expressed lncRNAs are
observed in the majority of human diseases, including cancer
(10). An increasing number of
dysregulated lncRNAs have been identified in cervical cancer and
exhibit a close association with malignant phenotypes (11,12). lncRNAs play critical roles in the
oncogenesis and progression of cervical cancer, and play oncogenic
or antioncogenic roles in this type of cancer (13). These properties suggest that
lncRNAs function as potential diagnostic biomarkers and therapeutic
targets.
MicroRNAs (miRNAs or miRs) are endogenous and short
RNA transcripts of approximately 22 nucleotides in length (14). These molecules play a role in
post-transcriptional gene regulation by base pairing with the
3′-untranslated regions of their target genes to ultimately trigger
translational suppression and/or mRNA degradation (15). The abnormal expression of miRNAs
is a hallmark of cancer, including cervical cancer (16). miRNAs are crucial regulators
during the genesis and development of cervical cancer (17,18). The competing endogenous RNA
(ceRNA) theory was introduced and it states that lncRNAs function
as miRNA sponges to lower the inhibition of gene expression induced
by miRNAs (19).
Previous studies have confirmed the abnormal
expression of the long intergenic non-protein-coding RNA 1006
(LINC01006) in prostate (20),
pancreatic (21) and gastric
(22) cancers. However, whether
LINC01006 plays important roles in cervical cancer remains unclear.
Therefore, the present study investigated the expression status and
detailed roles of LINC01006 in cervical cancer, and aimed to
elucidate the mechanisms underlying the functions of LINC01006 in
cervical cancer.
Materials and methods
Patient samples
The Ethics Committee of Renmin Hospital of Wuhan
University approved the present study. All experiments involving
human samples were performed in accordance with the principles of
the Declaration of Helsinki. All participators provided written
informed consent. A total of 67 pairs of cervical cancer tissues
and matched adjacent normal tissues were acquired from the patients
at Renmin Hospital of Wuhan University. No patient had undergone
chemotherapy, radiotherapy, or other anticancer treatments prior to
surgical resection. Immediately after tissue excision, all tissues
were stored in liquid nitrogen until further analysis.
Cell lines
The normal human cervical epithelial cell line,
Ect1/E6E7 (ATCC® CRL-2614™), was purchased from the
American Type Culture Collection (ATCC). Keratinocyte serum-free
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 0.1 ng/ml
human recombinant epidermal growth factor, 0.05 mg/ml bovine
pituitary extract and 0.4 mM calcium chloride was used to culture
the Ect1/E6E7 cells. In addition, 4 cervical cancer cell lines,
SiHa (TCHu113), CaSki (TCHu137), C33A (TCHu176) and HeLa (TCHu187),
were acquired from the Cell Bank of the Chinese Academy of
Sciences. The SiHa, CaSki and HeLa cells were grown in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific,
Inc.). The C33A cells were cultured in minimum essential medium
(Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS. All cells
were routinely grown at 37°C in a humidified atmosphere supplied
with 5% CO2.
Cell transfection
To silence LINC01006, specific small interfering
RNAs (siRNAs) targeting LINC01006 (si-LINC01006) were devised and
synthesized by Shanghai GenePharma Co., Ltd. The si-LINC01006#1
sequence was 5′-CGCAAAGTTTTCCTATTAACTCT-3′; the si-LINC01006#2
sequence was 5′-TTCAAATTTTGACTTATTTTACA-3′; and the si-NC sequence
was 5′-CACGATAAGACAATGTATTT-3′. A nonsense sequence (si-NC) was
used as the negative control (NC). The miR-28-5p mimic, NC mimic,
miR-28-5p inhibitor (anti-miR-28-5p) and NC inhibitor (anti-NC)
were obtained from RiboBio Co., Ltd. To overexpress P21-activated
kinase 2 (PAK2), the sequences of PAK2 were inserted into the
pcDNA3.1 plasmid to obtain the PAK2 overexpression plasmid
pcDNA3.1-PAK2 (pc-PAK2; Shanghai GenePharma Co., Ltd.). Cervical
cancer cells were cultivated in 6-well plates. As per the
manufacturer's instructions, cells were transfected with siRNAs
(100 pmol), mimics (100 pmol), miRNA inhibitors (100 pmol) or
plasmids (4 μg) using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol reagent (Beyotime Institute of Biotechnology)
was used to extract total RNA, followed by the assessment of RNA
concentration and purity using a NanoDrop 2000 spectrometer (Thermo
Fisher Scientific, Inc.). To determine miRNA expression, reverse
transcription was performed using the Mir-X miRNA First-Strand
Synthesis kit (cat. no. 638315; Takara Biotechnology Co., Ltd.).
Using complementary DNA (cDNA) as a template, the Mir-X miRNA
qRT-PCR TB Green® kit (cat. no. 638314; Takara
Biotechnology Co., Ltd.) was used to perform qPCR. U6 small nuclear
RNA was used as the internal control. To quantify the mRNA
expression levels of LINC01006 and PAK2, cDNA was obtained by
performing reverse transcription using the PrimeScript™ RT Reagent
klit with gDNA Eraser (cat. no. RR047A; Takara Biotechnology Co.).
Thereafter, PCR amplification was performed using TB Green Premix
Ex Taq (cat. no. RR420A; Takara Biotechnology Co.). The mRNA
expression levels of LINC01006 and PAK2 were normalized to
glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Gene expression
was calculated using the 2−ΔΔCq method (23). The sequences of all primers are
presented in Table I.
 | Table IPrimer sequences used for
RT-qPCR. |
Table I
Primer sequences used for
RT-qPCR.
| Gene | Sequences
(5′→3′) |
|---|
| LINC01006 | Forward:
GTGTGACACATCGGAGTGAATTGAG |
| Reverse:
AACCCTGCACTATTTGTGGCGT |
| PAK2 | Forward:
ATAACGGAGAACTGGAAGATAAGC |
| Reverse:
AGATGATTTTATGCCTGGGCTT |
| GAPDH | Forward:
ACCTGACCTGCCGTCTAGAAAA |
| Reverse:
TTGAAGTCAGAGGAGACCACCTG |
| U6 | Forward:
CTCGCTTCGGCAGCACA |
| Reverse:
AACGCTTCACGAATTTGCGT |
| hsa-miR-28-5p | Forward:
TCGGCAGGAAGGAGCUCACAGUC |
| Reverse:
CACTCAACTGGTGTCGTGGA |
| hsa-miR-154-3p | Forward:
TCGGCAGGGCUUCCGUUGUGCC |
| Reverse:
CACTCAACTGGTGTCGTGGA |
Cell proliferation assay
Transfected cells were collected after 24 h, and the
cell number was counted using a blood cell counting chamber. Cells
were resuspended in 10% FBS-supplemented culture medium. A
100-μl cell suspension containing 2×103 cells was
seeded into 96-well plates, followed by culturing in the
aforementioned conditions for different time periods of 0, 24, 48
and 72 h. For the cell proliferation assay, 10 μl of cell
counting kit-8 (CCK-8) reagent were added to each well, followed by
incubation for a further 2 h at 37°C. The absorbance was measured
at a wavelength of 450 nm using a Multiskan Spectrum Microplate
spectrophotometer (Thermo Fisher Scientific, Inc.).
Flow cytometric analysis
After 48 h, the transfected cells were digested with
0.25% trypsin and collected following centrifugation at 1,000 × g
for 5 min at room temperature. As per the instructions of the
Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of
Biotechnology), the harvested cells were resuspended in 195
μl Annexin V-FITC buffer, followed by probing with 5
μl of Annexin V-FITC and 10 μl of propidium iodide.
Following 20 min of incubation at 20-25°C in the dark, apoptosis
was detected using a FACSCalibur flow cytometer (BD
Biosciences).
Transwell migration and invasion
assays
For the migration assay, transfected cells were
treated with 0.25% trypsin, rinsed with phosphate-buffered saline,
and centrifuged at 1,000 × g for 5 min at room temperature. The
collected cells were resuspended in culture medium without FBS. The
upper chambers (8 μm pores; Corning Inc.) were covered with
100 μl of the cell suspension containing 5×104
cells. The lower chamber included 20% FBS-supplemented culture
medium, followed by incubation at 37°C for 24 h. The cells in the
inner membrane were cleaned, and the cells that passed through the
pores were fixed with 100% methanol and dyed with 0.1% crystal
violet (Beyotime Institute of Biotechnology) at room temperature
for 30 min. The number of migrated cells was counted under a light
microscope (Olympus Corporation). A total of 5 visuals were
randomly selected for microscopic observation. Cell invasion was
examined using the same experimental procedures, with the exception
that the chambers were precoated with Matrigel (BD
Biosciences).
Tumor xenograft model
To inhibit LINC01006 expression, short hairpin RNAs
(shRNAs) against LINC01006 (sh-LINC01006) and NC shRNA (sh-NC) were
constructed and synthesized by Shanghai GenePharma Co., Ltd. The
sh-LINC01006 sequence was
5′-CCGGTTCAAATTTTGACTTATTTTACACTCGAGTGTAAAATAAGTCAAAATTTGAATTTTTG-3′;
and the sh-NC sequence was
5′-CCGGCACGATAAGACAATGTATTTCTCGAGAAATACATTGTCTTATCGTGTTTTTG-3′. The
synthesized shRNAs were inserted into the pLKO.1 vector (Biosettia,
Inc.) and transfected into 293T cells (Cell Bank of Chinese Academy
of Sciences) in parallel with psPAX2 and pMD2.G. Following 2 days
of incubation at 37°C, HeLa cells were transfected with
lentiviruses expressing sh-LINC01006 or sh-NC. The infected HeLa
cells were incubated with puromycin (5 μg/ml; Sigma-Aldrich;
Merck KGaA) to select stable cell lines.
The Institutional Animal Care and Use Committee of
Renmin Hospital of Wuhan University approved the experimental steps
involving animals. A total of 6 BALB/c female nude mice (4-5 weeks
old; weighing 20 g; Shanghai Laboratory Animal Center of Chinese
Academy of Sciences) were subcutaneously injected with HeLa cells
that were stably transfected with sh-LINC01006 or sh-NC. Each group
contained 3 nude mice. The animals were kept under specific
pathogen-free conditions at 25°C and 50% humidity, with a 10:14
light/dark cycle and ad libitum access to food and water.
The volume of the tumor xenografts was monitored weekly and
recorded. Tumor volume was determined using the following formula:
Volume=0.5× (length × width2). The mice were euthanized
by means of cervical dislocation at week 4, and tumor xenografts
were obtained, imaged and weighed.
Subcellular fractionation assay
The Cytoplasmic and Nuclear RNA Purification kit
(Norgen Biotek Corp.) was used to prepare the cytoplasmic and
nuclear fractions of the cervical cancer cells. The RNA of both
fractions was used to determine the LINC01006 distribution by
performing RT-qPCR. GAPDH and U6 were considered the cytoplasmic
and nuclear internal references, respectively.
Bioinformatics analysis and luciferase
reporter assay
The binding sequences between miR-28-5p and
LINC01006 were predicted using StarBase 3.0 (http://starbase.sysu.edu.cn/). TargetScan (http://www.targetscan.org/vert_60/) and miRDB
(http://mirdb.org/miRDB/index.html)
were used to search for the putative targets of miR-28-5p. The
Cancer Genome Atlas (TCGA) database was applied to analyze
LINC01006 expression in cervical squamous cell carcinoma and
endocervical adenocarcinoma (CESC).
LINC01006 fragments containing the wild-type (WT) or
mutant (MUT) miR-28-5p binding sites were amplified by Shanghai
GenePharma Co., Ltd. and fused into the psiCHECK™-2 luciferase
reporter vector (Promega Corporation). The obtained luciferase
reporter vectors were labeled WT-LINC01006 and MUT-LINC01006. The
WT-PAK2 and MUT-PAK2 reporter vectors were produced in a similar
manner. For the reporter assay, the synthesized reporter vectors
(0.2 μg) were co-transfected with miR-28-5p mimic (20 pmol)
or NC mimic (20 pmol) into cervical cancer cells. Luciferase
activity was detected at 48 h following transfection using the
Dual-Luciferase Reporter Analysis system (Promega Corporation).
Renilla luciferase activity was used for the Firefly
luciferase activity normalization.
RNA immunoprecipitation (RIP) assay
RIP assay was performed using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (cat. no. 03-110;
Merck-Millipore). The lysate of cervical cancer cells was obtained
by cultivating the cells with RIP lysis buffer, followed by the
addition of magnetic beads conjugated with human anti-argonaute 2
(Ago2) or control anti-IgG antibodies (1:5,000 dilution; both from
cat. no. 03-110; Merck Millipore). Following overnight incubation
at 4°C, the magnetic beads were rinsed with washing buffer. After
probing with proteinase K, the relative enrichment of LINC01006,
miR-28-5p, and PAK2 in the immunoprecipitate was assessed by
RT-qPCR.
Western blot analysis
RIPA lysis buffer (Beyotime Institute of
Biotechnology) was used to extract total proteins from the cells.
Total protein concentration was quantified using the Pierce™
bicinchoninic acid Protein Assay kit (cat. no. 23225; Thermo Fisher
Scientific, Inc.). The equivalent protein (30 μg) was
separated using 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis. The resolved proteins were electrotransferred onto
polyvinylidene fluoride membranes. Membrane blocking was performed
using 5% defatted milk powder at room temperature for 2 h, followed
by incubation with primary antibodies overnight at 4°C. Primary
antibodies specific for PAK2 (cat. no. ab76293; 1:1,000 dilution)
or GAPDH (cat. no. ab181603; 1:1,000 dilution) were purchased from
Abcam. After the membranes were incubated with the horseradish
peroxidase-conjugated anti-rabbit secondary antibody (cat. no.
ab205718; 1:5,000 dilution; Abcam) at room temperature for 2 h,
imprinting was performed using Pierce™ ECL Western blotting
Substrate (Pierce; Thermo Fisher Scientific, Inc.). Densitometry of
the protein signals was implemented utilizing Quantity One software
version 4.62 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Experimental data are presented as the means ±
standard deviation. A Student's t-test was used to determine
differences between 2 groups, and multiple group comparisons were
performed using one-way analysis of variance with Tukey's post hoc
test. Gene expression correlations were examined using Pearson's
correlation coefficient. Kaplan-Meier survival curves and the log
rank test were used for survival analyses. P<0.05 indicated a
statistically significant difference.
Results
LINC01006 interference induces cell
apoptosis and restrains the proliferation, migration, and invasion
of cervical cancer cells
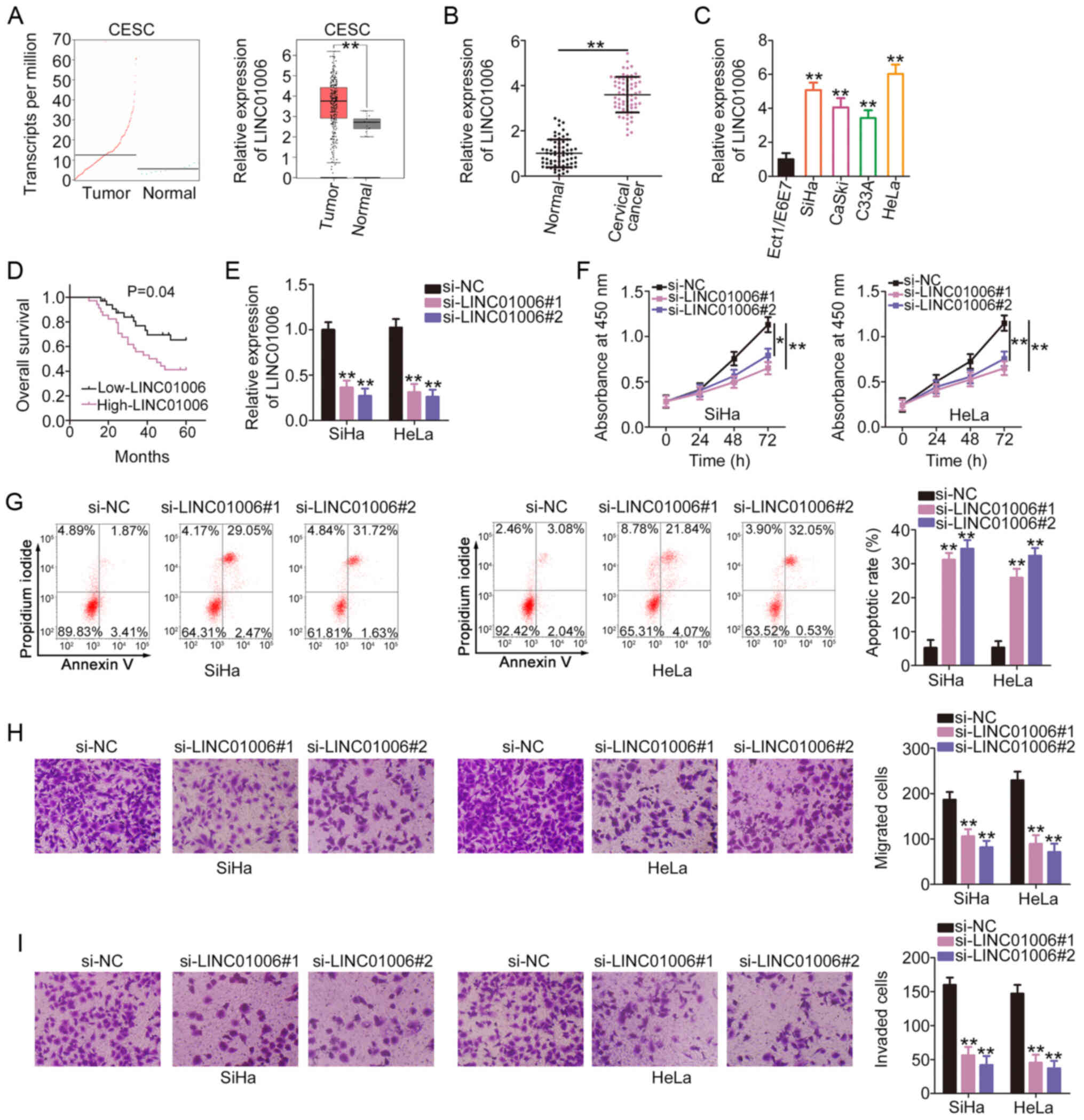
To determine the expression pattern of LINC01006 in
cervical cancer, its expression levels in CESC were examined using
The Cancer Genome Atlas (TCGA) database. The expression level of
LINC01006 was higher in CESC tissues than in normal tissues
(Fig. 1A). Consistently, the
expression level of LINC01006 was higher in cervical cancer tissues
than in adjacent normal tissues (Fig. 1B). RT-qPCR was then performed to
determine the expression level of LINC01006 in cervical cancer cell
lines. Compared to the Ect1/E6E7 cells, a relatively higher
LINC01006 expression level was confirmed in the 4 tested cervical
cancer cell lines (Fig. 1C). The
association between LINC01006 expression and the overall survival
of patients with cervical cancer was also addressed. Patients with
cervical cancer who exhibited a high LINC01006 expression had a
shorter overall survival than patients who exhibited a low
LINC01006 expression (Fig.
1D).
Of the 4 cervical cancer cell lines, the SiHa and
HeLa cells exhibited a relatively higher LINC01006 expression.
Therefore, these cells were selected to determine the specific
functions of LINC01006. The SiHa and HeLa cells were transfected
with si-LINC01006. To avoid off-target effects, 2 siRNAs were used,
and the efficiency of RNA interference was assessed by RT-qPCR
(Fig. 1E). Cell proliferation
assay then demonstrated that the proliferation of the SiHa and HeLa
cells was suppressed following the knockdown of LINC01006 (Fig. 1F). Flow cytometric analysis
further demonstrated that the knockdown of LINC01006 promoted the
apoptosis of the SiHa and HeLa cells (Fig. 1G). In addition, si-LINC01006
evidently decreased the migration (Fig. 1H) and invasion (Fig. 1I) of the SiHa and HeLa cells.
Taken together, these results suggest that LINC01006 is upregulated
in cervical cancer and functions as a promoter of cancer
progression.
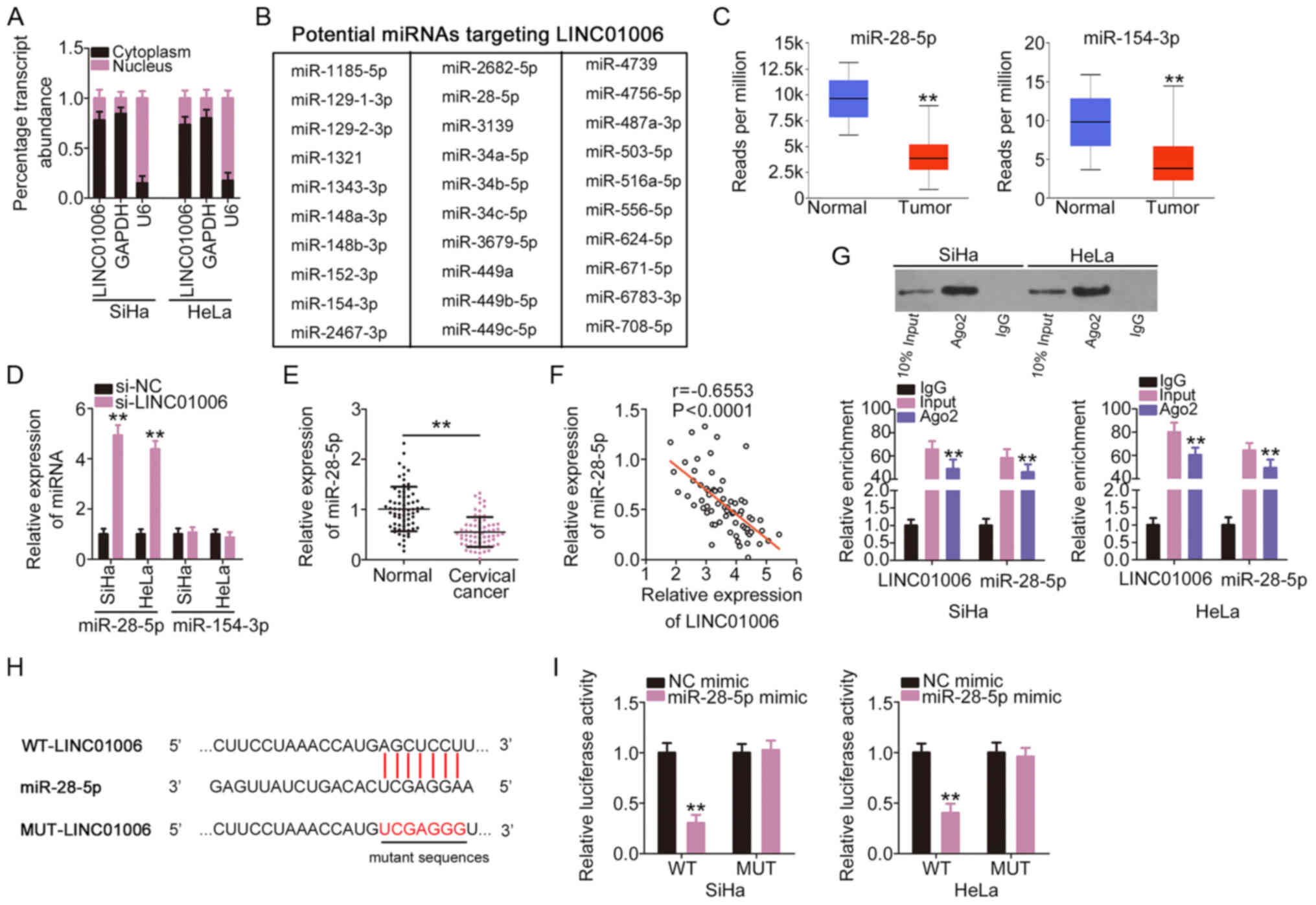
LINC01006 directly interacts with
miR-28-5p and functions as a miR-28-5p sponge
To determine the downstream mechanism of LINC01006,
the cellular localization of LINC01006 in cervical cancer cells was
analyzed using a subcellular fractionation assay. The data
confirmed that LINC01006 was a cytoplasmic lncRNA in cervical
cancer (Fig. 2A), which suggests
that it functions as a molecular sponge for miRNA and plays its
pro-oncogenic roles at the post-transcription level. A search of
StarBase 3.0 revealed a total of 30 miRNAs (Fig. 2B) that included binding sites for
LINC01006. The analysis of the TCGA database identified 2 miRNAs
(miR-28-5p and miR-154-3p) that were weakly expressed (Fig. 2C), and 7 miRNAs that were
overexpressed in CESC tissues (data not shown). miR-28-5p and
miR-154-3p were selected for further experiments. The expression of
miR-28-5p and miR-154-3p was detected in cervical cancer cells in
which LINC01006 was silenced. miR-28-5p expression increased
prominently in the SiHa and HeLa cells following the silencing of
LINC01006; however, there was no difference in the levels of
miR-154-3p (Fig. 2D). The
expression of miR-28-5p was downregulated in cervical cancer
tissues (Fig. 2E) and Pearson's
correlation coefficient revealed that the expression of miR-28-5p
in cervical cancer tissues inversely correlated with that of
LINC01006 (Fig. 2F). Notably,
miR-28-5p and LINC01006 were enriched in Ago2-containing
immunoprecipitate complexes compared to IgG control
immunoprecipitate complexes (Fig.
2G). The direct binding between LINC01006 and miR-28-5p
(Fig. 2H) was confirmed by
luciferase reporter assay. The outcomes revealed that miR-28-5p
overexpression led to a notable decrease in the luciferase activity
of WT-LINC01006 (Fig. 2I).
However, no evident change was identified in the cells transfected
with MUT-LINC01006. Taken together, these results suggest that
LINC01006 directly interacts with miR-28-5p and functions as an
miR-28-5p sponge in cervical cancer.
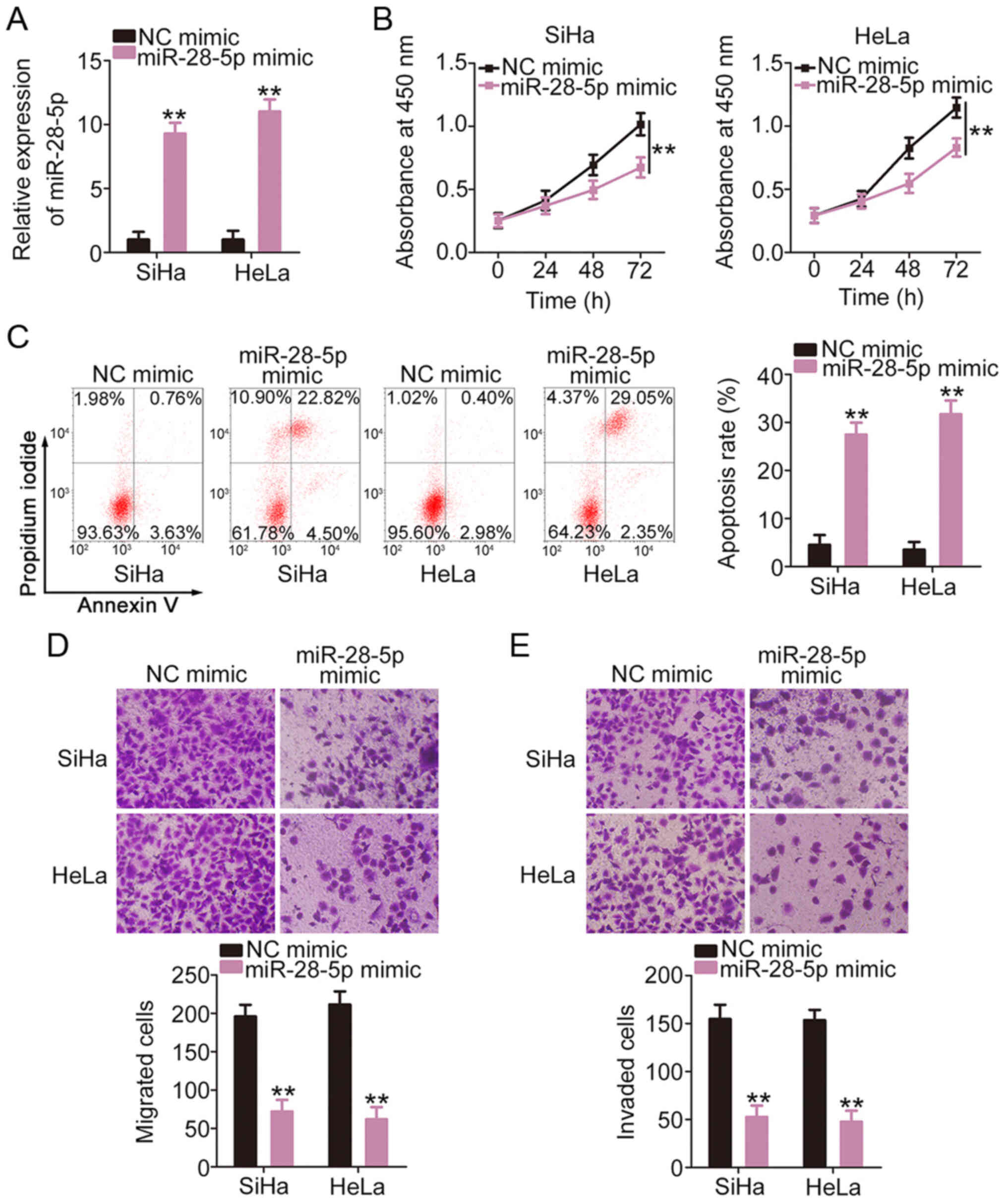
miR-28-5p is a tumor-suppressor in
cervical cancer
To determine the contribution of miR-28-5p,
functional changes in cervical cancer cells were examined following
the overex-pression of miR-28-5p (Fig. 3A). The increased expression of
miR-28-5p clearly hindered the proliferation (Fig. 3B) and promoted the apoptosis
(Fig. 3C) of the SiHa and HeLa
cells, as demonstrated in by cell proliferation assay and flow
cytometric analysis. In addition, the migratory (Fig. 3D) and invasive (Fig. 3E) abilities of the SiHa and HeLa
cells were inhibited with the enforced expression of miR-28-5p.
Taken together, these results suggest that miR-28-5p exerts
antioncogenic effects in cervical cancer cells.
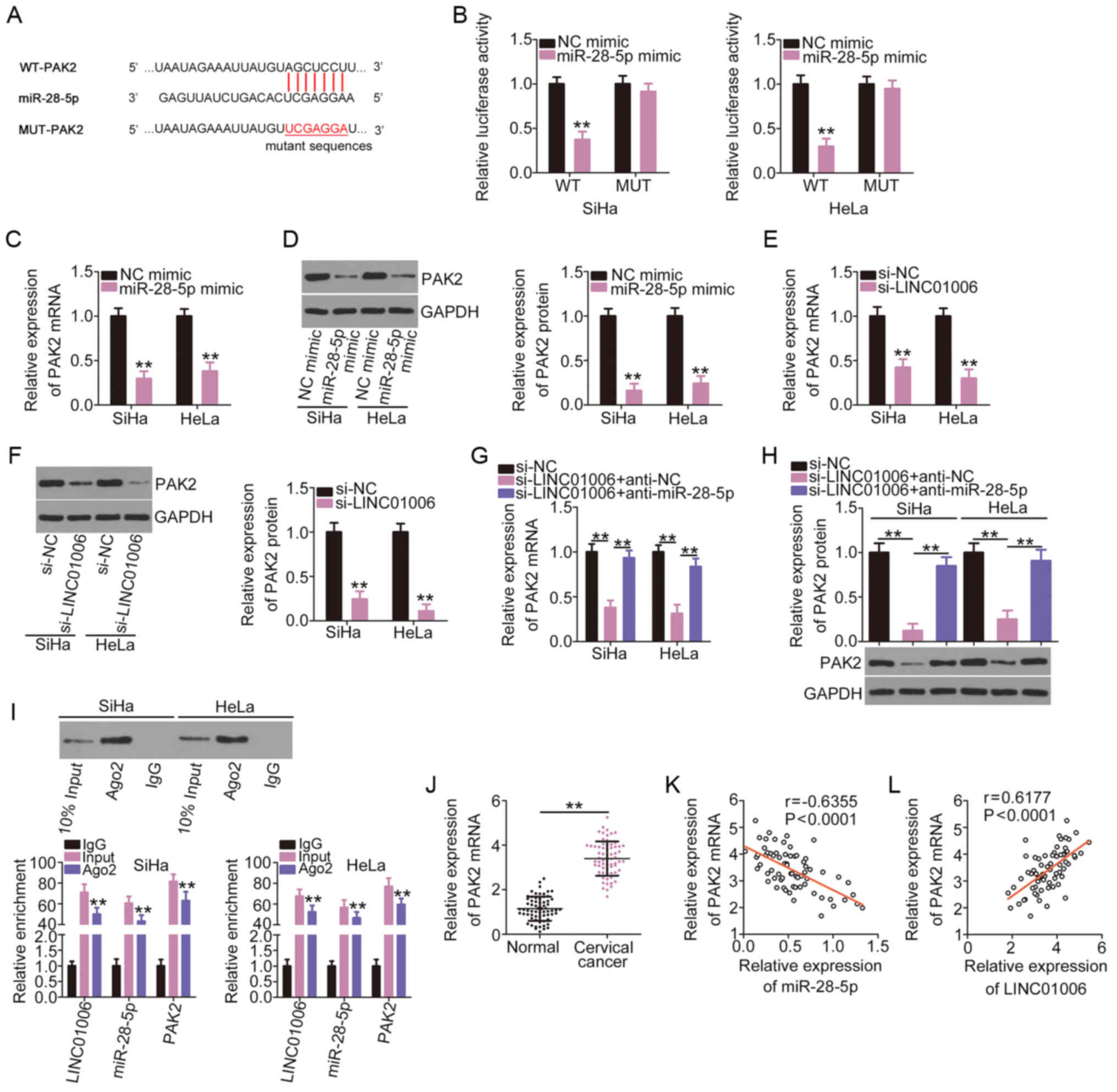
LINC01006 positively regulates PAK2 in
cervical cancer by acting as a decoy to miR-28-5p
Online bioinformatics prediction databases were used
to determine the potential target of miR-28-5p and PAK2 (Fig. 4A) was selected for subsequent
confirmation due to its critical roles in carcinogenesis and cancer
progression. A luciferase reporter assay was performed to
demonstrate the direct binding of miR-28-5p and PAK2. miR-28-5p did
not bind MUT-PAK2 or influence its luciferase activity. By
contrast, the luciferase activity of WT-PAK2 was evidently
decreased in the SiHa and HeLa cells co-transfected the miR-28-5p
mimic (Fig. 4B). miR-28-5p
overexpression evidently diminished the mRNA (Fig. 4C) and protein (Fig. 4D) expression levels of PAK2 in
the SiHa and HeLa cells.
Subsequent analyses were performed to determine the
association between LINC01006 and miR-28-5p in PAK2 regulation. The
regulatory effects of LINC01006 on PAK2 expression in cervical
cancer cells were determined by RT-qPCR and western blot analysis.
Notably, the knockdown of LINC01006 clearly decreased PAK2
expression in the SiHa and HeLa cells at the mRNA (Fig. 4E) and protein (Fig. 4F) levels, and co-transfection
with anti-miR-28-5p counteracted these inhibitory effects (Fig. 4G and H). LINC01006, miR-28-5p and
PAK2 were abundant in the RNA immunoprecipitated with anti-Ago2
antibody (Fig. 4I), which
suggests that these three molecules co-exist in the same
RNA-induced silencing complex. Compared to the adjacent normal
tissues, the cervical cancer tissues had a higher PAK2 expression
(Fig. 4J). PAK2 expression
negatively correlated with miR-28-5p expression (Fig. 4K) and positively correlated with
LINC01006 expression (Fig. 4L)
in the cervical cancer tissues. Taken together, these results
suggest that LINC01006 functions as a ceRNA for miR-28-5p and
positively regulates PAK2 expression in cervical cancer.
LINC01006 drives the oncogenicity of
cervical cancer via the miR-28-5p/PAK2 axis
Rescue experiments were performed to determine
whether LINC01006 achieved its tumor-promoting roles in cervical
cancer cells by affecting the miR-28-5p/PAK2 axis. RT-qPCR was
performed to examined the transfection efficiency of
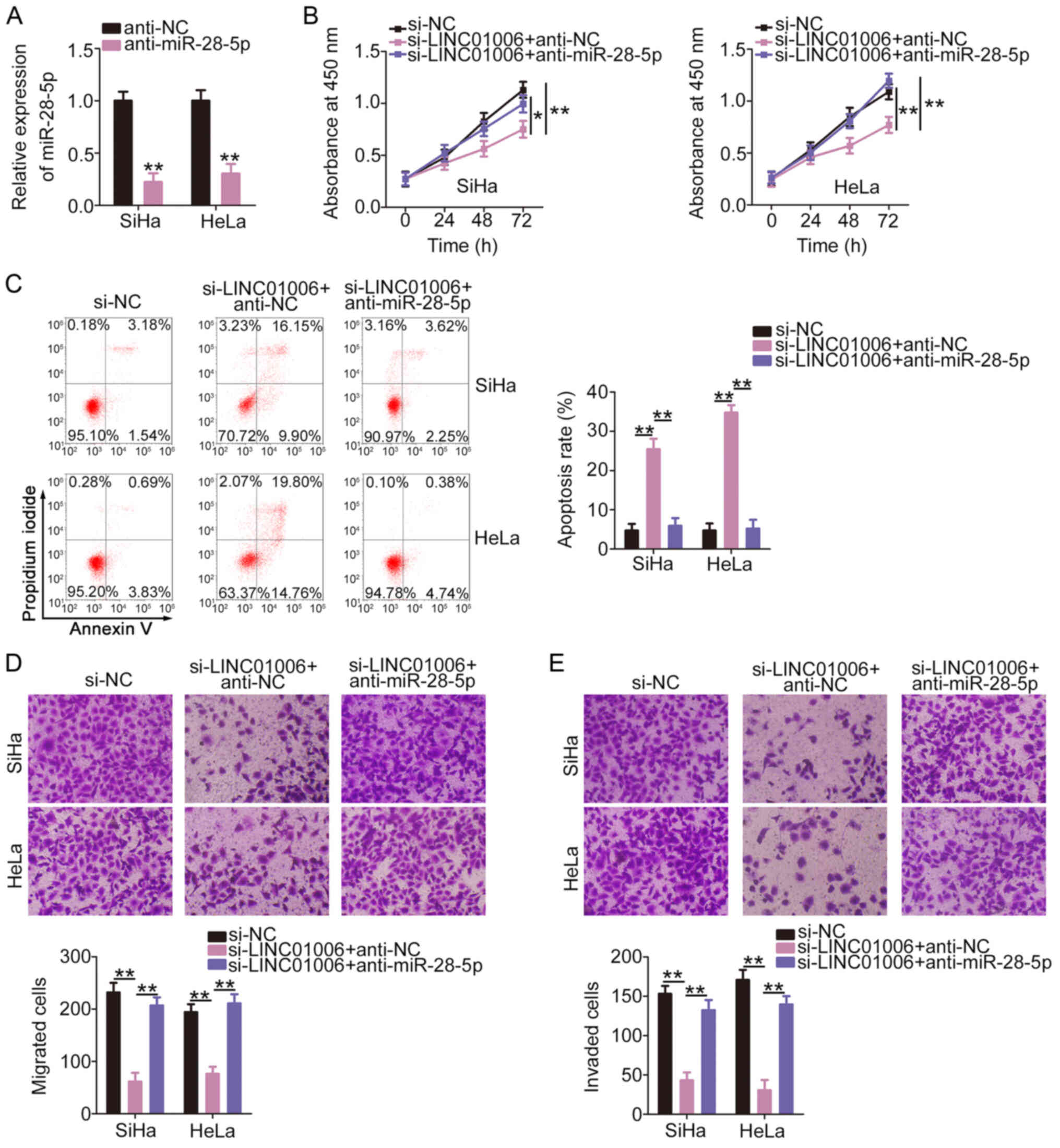
anti-miR-28-5p. Transfection with anti-miR-28-5p resulted in a
marked decrease in miR-28-5p expression in the SiHa and HeLa cells
(Fig. 5A). Anti-miR-28-5p or
anti-NC and si-LINC01006 were transfected into the SiHa and HeLa
cells. The loss of LINC01006 evidently restricted the proliferation
(Fig. 5B) and promoted the
apoptosis (Fig. 5C) of the SiHa
and HeLa cells. Anti-miR-28-5p co-transfection counteracted the
regulatory effects. LINC01006 interference induced a significant
decrease in cell migration (Fig.
5D) and invasion (Fig. 5E),
which was abolished by miR-28-5p inhibition.
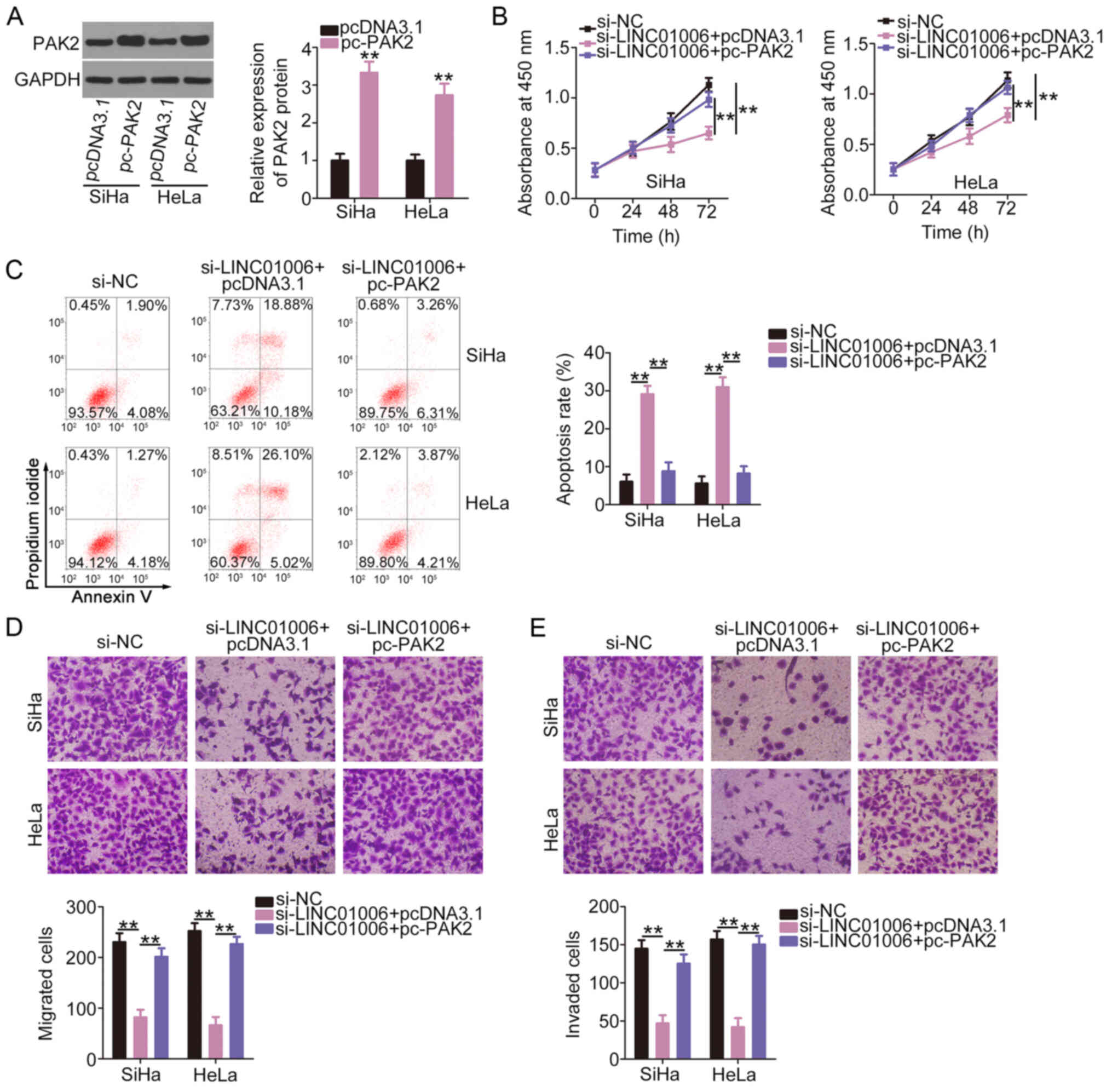
The protein levels of PAK2 in the
pcDNA3.1-transfected and pc-PAK2-transfected SiHa and HeLa cells
were determined by western blot analysis. The data confirmed that
PAK2 protein was markedly overexpressed in the SiHa and HeLa cells
following pc-PAK2 transfection (Fig.
6A). PAK2 upregulation recovered cell proliferation which was
impaired due to the silencing of LINC01006 (Fig. 6B). PAK2 overexpression attenuated
the promoting effect of si-LINC01006 on cell apoptosis (Fig. 6C). The re-introduction of PAK2
also restored the cell migration (Fig. 6D) and invasive (Fig. 6E) abilities that were impaired by
si-LINC01006. Therefore, the miR-28-5p/PAK2 axis may act as a
downstream effector of LINC01006 in cervical cancer.
Depletion of LINC01006 inhibits tumor
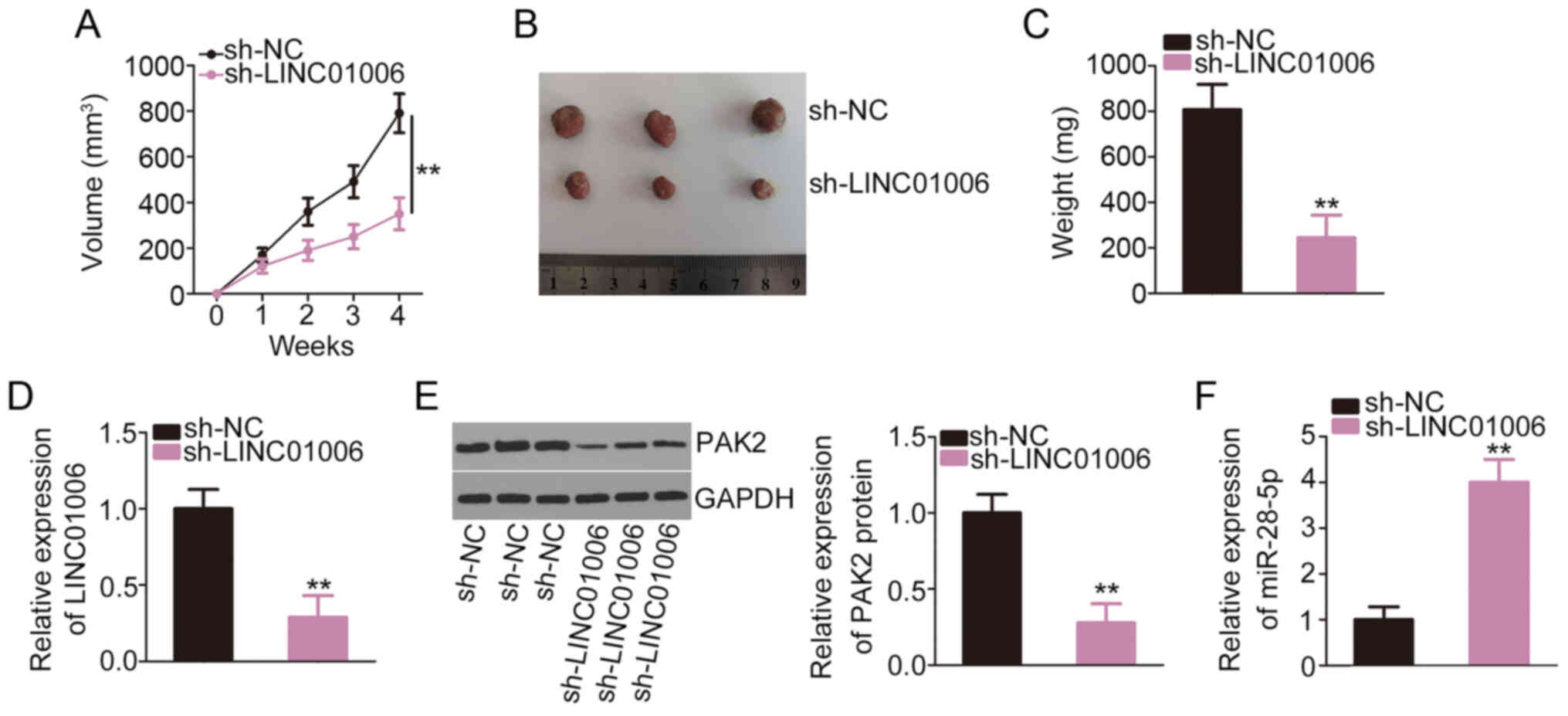
growth in vivo
HeLa cells that were stably transfected with
sh-LINC01006 or sh-NC were subcutaneously injected into nude mice
to establish a tumor xenograft model. Tumor growth was evidently
lower in mice in the sh-LINC01006 group than the sh-NC group
(Fig. 7A). The volume (Fig. 7B) and weight (Fig. 7C) of the tumor xenografts were
evidently decreased in the LINC01006-silenced group compared to the
sh-NC group. The levels of LINC01006, miR-28-5p and PAK2 were
analyzed in the tumor tissues. The expression of LINC01006
(Fig. 7D) and PAK2 (Fig. 7E) was downregulated, and
miR-28-5p (Fig. 7F) was
overexpressed in the tumor xenografts of the LINC01006 deficiency
group. Overall, these results suggest that LINC01006 depletion
decreases tumor growth in vivo.
Discussion
Dysfunctional lncRNAs are frequently observed in
cervical cancer. These molecules have a close association with
cervical carcinogenesis and cancer progression (24-26). Considering the importance of
lncRNAs, it is essential to examine the roles of cancer-related
lncRNAs in the malignancy of cervical cancer and elucidate the
underlying mechanisms, which are of the utmost importance for the
development of attractive targets for cancer diagnosis, prognosis,
and management. Over 50,000 lncRNAs are present in the human genome
(27); however, the majority of
these have not been studied in cervical cancer and thus require
clarification. Therefore, the present study examined the precise
roles of LINC01006 in cervical cancer and determined its regulatory
mechanism.
LINC01006 is upregulated in prostate (20) and pancreatic (21) cancers and plays pro-oncogenic
roles. By contrast, this lncRNA is weakly expressed in gastric
cancer and exhibits a notable association with age, tumor location,
tumor size and venous invasion (22). These observations indicate tissue
specificity in the expression profile and function of LINC01006 in
human cancers. However, the expression pattern and detailed roles
of LINC01006 in cervical cancer remain largely ambiguous. The
present study demonstrated that the expression level of LINC01006
was visibly higher in cervical cancer tissues and cell lines.
Survival analysis confirmed that a high LINC01006 expression was
significantly associated with a worse overall survival of patients
with cervical cancer. The silencing of LINC01006 suppressed the
proliferation, migration and invasion of cervical cancer cells, but
induced cell apoptosis in vitro. The absence of LINC01006
impeded tumor growth in vivo. Taken together, these results
highlight LINC01006 as a potential target for cervical cancer
diagnosis, prognosis and therapy.
The ceRNA theory was recently established, and it
describes ceRNAs as a novel group of post-transcriptional
regulators that participate in tumorigenesis and tumor development
(28,29). The ceRNA network involving
lncRNAs, miRNAs and mRNAs is a widely accepted mechanism of
post-transcriptional regulation of lncRNAs (30). Notably, the subcellular
distribution of lncRNAs determines their roles; i.e., whether an
lncRNA functions as a ceRNA is largely based on its localization
(31). lncRNAs that are
primarily located in the nucleus generally affect gene expression
at the pre-transcriptional or transcriptional levels (32). By contrast, cytoplasmic lncRNAs
sequester certain miRNAs via the same miRNA response elements to
indirectly modulate mRNA expression at the post-transcriptional
level (33). The majority of
LINC01006 expression was observed in the cytoplasm of cervical
cancer cells in the present study. Therefore, it was hypothesized
that LINC01006 exerts its tumor-promoting actions in cervical
cancer in a ceRNA manner.
Data from bioinformatics analysis suggested a
possible binding interaction between LINC01006 and miR-28-5p. To
test this hypothesis, luciferase reporter and RIP assays were
performed. The results confirmed that LINC01006 physically
interacted with miR-28-5p and sponged miR-28-5p in cervical cancer.
Emerging studies have reported that miR-28-5p is differentially
expressed in multiple human cancer types and plays an important
role in tumorigenesis (34-36). Consistently, the data of the
present study revealed the antioncogenic roles of miR-28-5p in
cervical cancer. Additional in-depth mechanistic analyses revealed
that PAK2 was a direct target of miR-28-5p. LINC01006 deficiency
reduced the expression of PAK2 in cervical cancer cells, and this
regulatory effect was achieved by sponging miR-28-5p. Notably, 3
molecules, namely, LINC01006, miR-28-5p and PAK2, co-existed in the
same RNA-induced silencing complex. Correlation analysis revealed
that PAK2 negatively correlated with miR-28-5p, but positively
correlated with LINC01006 expression in cervical cancer tissues.
The present study demonstrated a novel ceRNA pathway comprised of
LINC01006, miR-28-5p and PAK2 in cervical cancer.
PAKs are a family of serine/threonine kinases that
constitute 2 distinct subgroups: Subgroup 1 contains PAK 1-3 and
subgroup 2 contains PAKs (37).
PAK2 has an autoinhibitory domain and may be activated by the small
GTP-binding proteins Cdc42 and Rac (38). Several studies have confirmed the
implication of PAK2 in the course of cancer oncogenesis and
progression (39-41). The downregulation of PAK2 by
miRNAs abates the aggressiveness of human cancer cells. The present
study observed a similar trend in cervical cancer. The final rescue
experiments demonstrated that suppression of miR-28-5p or PAK2
overexpression abrogated the impacts of LINC01006 downregulation on
malignant cellular functions in cervical cancer. Jointly, these
results confirm that the miR-28-5p/PAK2 axis is a crucial mediator
in which LINC01006 functions as a novel carcinogenic lncRNA in
cervical cancer.
The present study revealed that LINC01006 increased
PAK2 expression by acting as a ceRNA for miR-28-5p, which
aggravated the oncogenicity of cervical cancer cells. These
findings offer a basis for the identification of LINC01006-targeted
clinical therapy.
Funding
The present study was supported by the Renmin
Hospital of Wuhan University.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors (LT, FH, JY, XM and LC) made significant
contributions to the findings, analysis and methodology of the
study. All authors read and approved the final draft.
Ethics approval and consent to
participate
The Ethics Committee of Renmin Hospital of Wuhan
University approved the present study. All experiments involving
human samples were performed in accordance with the principles of
the Declaration of Helsinki. All participants provided written
informed consent. The Institutional Animal Care and Use Committee
of Renmin Hospital of Wuhan University approved the experimental
steps involving animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fang J, Zhang H and Jin S: Epigenetics and
cervical cancer: From pathogenesis to therapy. Tumour Biol.
35:5083–5093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsikouras P, Zervoudis S, Manav B, Tomara
E, Iatrakis G, Romanidis C, Bothou A and Galazios G: Cervical
cancer: Screening, diagnosis and staging. J BUON. 21:320–325.
2016.PubMed/NCBI
|
|
5
|
Lin M, Ye M, Zhou J, Wang ZP and Zhu X:
Recent advances on the molecular mechanism of cervical
carcinogenesis based on systems biology technologies. Comput Struct
Biotechnol J. 17:241–250. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sarfi M, Abbastabar M and Khalili E: Long
noncoding RNAs biomarker-based cancer assessment. J Cell Physiol.
234:16971–16986. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ransohoff JD, Wei Y and Khavari PA: The
functions and unique features of long intergenic non-coding RNA.
Nat Rev Mol Cell Biol. 19:143–157. 2018. View Article : Google Scholar :
|
|
8
|
Shi X, Sun M, Wu Y, Yao Y, Liu H, Wu G,
Yuan D and Song Y: Post-transcriptional regulation of long
noncoding RNAs in cancer. Tumour Biol. 36:503–513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ramakrishnaiah Y, Kuhlmann L and Tyagi S:
Towards a comprehensive pipeline to identify and functionally
annotate long noncoding RNA (lncRNA). Comput Biol Med.
127:1040282020. View Article : Google Scholar
|
|
11
|
Luo F, Wen Y, Zhou H and Li Z: Roles of
long non-coding RNAs in cervical cancer. Life Sci. 256:1179812020.
View Article : Google Scholar
|
|
12
|
He J, Huang B, Zhang K, Liu M and Xu T:
Long non-coding RNA in cervical cancer: From biology to therapeutic
opportunity. Biomed Pharmacother. 127:1102092020. View Article : Google Scholar
|
|
13
|
Galvão M and Coimbra EC: Long noncoding
RNAs (lncRNAs) in cervical carcinogenesis: New molecular targets,
current prospects. Crit Rev Oncol Hematol. 156:1031112020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer-a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar
|
|
15
|
Adams BD, Kasinski AL and Slack FJ:
Aberrant regulation and function of microRNAs in cancer. Curr Biol.
24:R762–R776. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miao J, Regenstein JM, Xu D, Zhou D, Li H,
Zhang H, Li C, Qiu J and Chen X: The roles of microRNA in human
cervical cancer. Arch Biochem Biophys. 690:1084802020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen S, Zhang S, Liu P, Wang J and Du H:
Potential role of microRNAs in the treatment and diagnosis of
cervical cancer. Cancer Genet. 248-249:25–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tornesello ML, Faraonio R, Buonaguro L,
Annunziata C, Starita N, Cerasuolo A, Pezzuto F, Tornesello AL and
Buonaguro FM: The role of microRNAs, long non-coding RNAs, and
circular RNAs in cervical cancer. Front Oncol. 10:1502020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ye Y, Shen A and Liu A: Long non-coding
RNA H19 and cancer: A competing endogenous RNA. Bull Cancer.
106:1152–1159. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma E, Wang Q, Li J, Zhang X, Guo Z and
Yang X: LINC01006 facilitates cell proliferation, migration and
invasion in prostate cancer through targeting miR-34a-5p to
up-regulate DAAM1. Cancer Cell Int. 20:5152020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Wang Y, Zhang L, You G, Li C,
Meng B, Zhou M and Zhang M: LINC01006 promotes cell proliferation
and metastasis in pancreatic cancer via miR-2682-5p/HOXB8 axis.
Cancer Cell Int. 19:3202019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu X, Chen F, Shao Y, Xu D and Guo J:
Long intergenic non-protein coding RNA 1006 used as a potential
novel biomarker of gastric cancer. Cancer Biomark. 21:73–80. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Duan DM, Zhang L and Hua F: LncRNA UCA1
inhibits proliferation and promotes apoptosis of cervical cancer
cells by regulating beta-catenin/TCF-4. Eur Rev Med Pharmacol Sci.
24:5963–5969. 2020.
|
|
25
|
Yan A, Chen G and Nie J: DGUOK-AS1
promotes the proliferation cervical cancer through regulating
miR-653-5p/EMSY. Cancer Biol Ther. 1–9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin L, Xin B, Jiang T, Wang XL, Yang H and
Shi TM: Long non-coding RNA LINC00460 promotes proliferation and
inhibits apoptosis of cervical cancer cells by targeting
microRNA-503-5p. Mol Cell Biochem. 475:1–13. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu J, Bai J, Zhang X, Lv Y, Gong Y, Liu L,
Zhao H, Yu F, Ping Y, Zhang G, et al: A comprehensive overview of
lncRNA annotation resources. Brief Bioinform. 18:236–249. 2017.
|
|
28
|
Niu ZS, Wang WH, Dong XN and Tian LM: Role
of long noncoding RNA-mediated competing endogenous RNA regulatory
network in hepatocellular carcinoma. World J Gastroenterol.
26:4240–4260. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Raziq K, Cai M, Dong K, Wang P, Afrifa J
and Fu S: Competitive endogenous network of lncRNA, miRNA, and mRNA
in the chemoresistance of gastrointestinal tract adenocarcinomas.
Biomed Pharmacother. 130:1105702020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang XZ, Liu H and Chen SR: Mechanisms of
long non-coding RNAs in cancers and their dynamic regulations.
Cancers (Basel). 12:12452020. View Article : Google Scholar
|
|
31
|
Du H and Chen Y: Competing endogenous RNA
networks in cervical cancer: Function, mechanism and perspective. J
Drug Target. 27:709–723. 2019. View Article : Google Scholar
|
|
32
|
Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX
and Hong W: The ways of action of long non-coding RNAs in cytoplasm
and nucleus. Gene. 547:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rashid F, Shah A and Shan G: Long
non-coding RNAs in the cytoplasm. Genomics Proteomics
Bioinformatics. 14:73–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma L, Zhang Y and Hu F: miR285p inhibits
the migration of breast cancer by regulating WSB2. Int J Mol Med.
46:1562–1570. 2020.PubMed/NCBI
|
|
35
|
Zhang L, Wang X, Liu X, Lv M, Shen E, Zhu
G and Sun Z: miR-28-5p targets MTSS1 to regulate cell proliferation
and apoptosis in esophageal cancer. Acta Biochim Biophys Sin
(Shanghai). 52:842–852. 2020. View Article : Google Scholar
|
|
36
|
Fazio S, Berti G, Russo F, Evangelista M,
D'Aurizio R, Mercatanti A, Pellegrini M and Rizzo M: The miR-28-5p
targetome discovery identified SREBF2 as one of the mediators of
the miR-28-5p tumor suppressor activity in prostate cancer cells.
Cells. 9:3542020. View Article : Google Scholar :
|
|
37
|
Nuche-Berenguer B, Ramos-Alvarez I and
Jensen RT: The p21-activated kinase, PAK2, is important in the
activation of numerous pancreatic acinar cell signaling cascades
and in the onset of early pancreatitis events. Biochim Biophys
Acta. 1862:1122–1136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Knaus UG, Wang Y, Reilly AM, Warnock D and
Jackson JH: Structural requirements for PAK activation by Rac
GTPases. J Biol Chem. 273:21512–21518. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu H, Shin SH, Chen H, Liu T, Li Z, Hu Y,
Liu F, Zhang C, Kim DJ, Liu K and Dong Z: CDK12 and PAK2 as novel
therapeutic targets for human gastric cancer. Theranostics.
10:6201–6215. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee JS, Mo Y, Gan H, Burgess RJ, Baker DJ,
van Deursen JM and Zhang Z: Pak2 kinase promotes cellular
senescence and organismal aging. Proc Natl Acad Sci USA.
116:13311–13319. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gupta A, Ajith A, Singh S, Panday RK,
Samaiya A and Shukla S: PAK2-c-Myc-PKM2 axis plays an essential
role in head and neck oncogenesis via regulating Warburg effect.
Cell Death Dis. 9:8252018. View Article : Google Scholar : PubMed/NCBI
|