Introduction
Lung cancer is a malignant and fatal human cancer
and is the leading cause of cancer deaths worldwide (1). Lung cancer is divided into 2 main
types: Non-small cell lung cancer (NSCLC) and small cell lung
cancer (SCLC). NSCLC is the most common type of lung cancer,
accounting for approximately 85% of lung cancers, while the more
malignant SCLC accounts for approximately 15% of cases (2). The majority of lung cancers are
diagnosed at an advanced stage, which reduces the survival rate of
patients (3,4). Despite progress being made in the
treatment of lung cancer, the long-term survival rates have not
been improved significantly (5).
A small fraction of patients with lung cancer undergo surgery at
the initial stage; however, the majority of patients require
chemotherapy as diagnosis when the disease is at an advanced stage.
Survival rates are better when lung cancer is identified earlier
during its progression.
Ectodermal-neural cortex 1 (ENC1) is an
actin-binding protein expressed primarily in nerve cells (6). ENC1 is essential for adipocyte and
neural crest cell differentiation (6,7).
Of note, ENC1 reduces the aggregation and neurotoxicity of mutant
Huntingtin proteins through the downregulation of p62 expression
(8). Recently, ENC1 was shown to
play a key role in malignancies. Studies have confirmed that ENC1
is upregulated in various types of brain tumors, as well as in
colon, ovarian and breast cancers (9-12). However, ENC1 has been shown to
significantly increase the level of reactive oxygen species in
ovarian cancer cells, inhibiting the proliferation, invasion and
migration of these cells (11).
The mechanisms of action of ENC1 in the development
of lung cancer remain unclear. In the present study, it was found
that the ENC1 levels were increased in lung cancer. The
downregulation of ENC1 significantly inhibited the proliferation
and invasion of lung cancer cells, and altered the expression
levels of a variety of other proteins. The silencing of ENC1
expression in A549 cells inhibited the growth of tumors when
introduced into a mouse lung tumor model. These results are
consistent with published evidence that ENC1 plays an important
role in the regulation of the growth, proliferation and metastasis
of cancers (9-12). ENC1 may be used as an effective
marker for the presence of certain cancers and may be key to the
discovery of effective treatments for cancer.
Materials and methods
Patients and sample collection
A total of 28 lung tumor tissues were obtained from
patients with NSCLC at Yijishan Hospital (affiliated with Wannan
Medical College) from April, 2018 to April, 2020 upon initial
diagnosis of the cancer in each patient. None of the patients
received any other specific treatment. Samples from tissues
adjacent to the tumors were collected for use as controls. The
present study was approved by the Ethics Committee of Yijishan
Hospital. Informed consent was obtained from all patients.
Cells, cell culture and transfection
Lung cancer cell lines A549 (SCSP-503), H1299
(SCSP-589) and human bronchial epithelial cells 16HBE (FS-0400)
were obtained from the cell bank of the Chinese Academy of Sciences
(Shanghai, China). All cells were cultured in RPMI-1640 (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere with 5% CO2.
For transfection, cells at approximately 60% confluency were
transfected with 8 nmol/l siRNAs targeting ENC1 or ENC1
overexpression vector using the GenMute Transfection kit according
to the manufacturer's instructions (SignaGen Laboratories) for 48
h. Following 12-24 h of transfection, the culture medium was
replaced with fresh medium for an additional 48 h of incubation at
37°C in a humidified atmosphere with 5% CO2. siRNA
against ENC1 (si-ENC1), non-specific siRNA (si-NC), ENC1
overexpression vector and vector were designed and synthesized by
Guangzhou RiboBio Co., Ltd. The sequence of the siRNA (si-ENC1) was
5′-CTG CTA CGA TCC AAC ATT A-3′.
Histological analysis and
immunohistochemistry (IHC)
For hematoxylin and eosin (H&E) staining (G1005,
Servicebio), tissues were fixed by 10% neutral formalin for 1 day,
dehydrated and embedded in paraffin, and then cut into
5-µm-thick sections. After being dewaxed and treated with
gradient alcohol, the sections were stained with hematoxylin for 5
min at room temperature and were washed with running water.
Subsequently, alcohol solution with 1% hydrochloric acid and
ammonia was employed. Subsequently, 1% eosin was utilized for
further staining for 3 min at room temperature, gradient ethanol
for section dehydration, and neutral balsam for section mounting,
followed by microscope visualization (Nikon Eclipse E100; Nikon
Corporation).
For IHC, the specimens were cut into sections
(5-µm-thick). All tissue samples were fixed with 10% neutral
formalin and embedded in paraffin. Paraffin sections were dewaxed
and the endogenous peroxides quenched with 0.3% hydrogen peroxide.
They were then incubated with anti-ENC1 antibody (1:200, ab124902;
Abcam) overnight (at 4°C) and secondary antibody (1:200, 7074S;
Cell Signaling Technology, Inc.) for 30 min (at 37°C). The sections
were stained with diaminobenzidine (G1212; Servicebio). Images were
collected under a microscope (Nikon Eclipse E100; Nikon
Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA (1 µg) was extracted from the cells
and tissues using TRIzol reagent (Ambion; Thermo Fisher Scientific,
Inc.) was reverse-transcribed into cDNA following the instructions
provided with the cDNA synthesis kit (K1622; Fermentas; Thermo
Fisher Scientific, Inc.). The concentration and quality of the RNA
were measured usign a NanoDrop 2000 spectrophotometer (Thermo
Fisher Scientific, Inc.), paired samples were adjusted to the
similar concentration for use. The mRNA level of GAPDH was used as
an internal control. Relative mRNA expression levels calculated
normalized to GAPDH. All experiments were performed in triplicate.
qPCR was performed at 95°C for 10 min followed by 40 cycles at 95°C
for 15 sec and at 60°C for approximately 1 min by using the
QuantiNova™ SYBR®-Green PCR kit according to the
manufacturer's instructions (Qiagen GmbH). The ENC1 primers were as
follows: Forward, 5′-TGG TTG GAG GAT ACT TTG GCA TTC AG-3′ and
reverse, 5′-TAG GAA TCA GCG AGT ACG GGA CAG-3′. The GAPDH primers
were as follows: Forward, 5′-CTG GGC TAC ACT GAG CAC C-3′ and
reverse, 5′-AAG TGG TCG TTG AGG GCA ATG-3′. Relative mRNA
expression was calculated using the 2−ΔΔCq method, as
previously described (13).
GAPDH served as an internal control.
Western blot analysis
Whole cells (A549 and H1299) and tissues (lung
cancer and para-cancerous tissues) were lysed using cell lysis
buffer (KeyGen Biotech. Co. Ltd.). The protein concentration was
measured using a BCA protein assay kit (Beyotime Institute of
Biotechnology). Protein samples (30 µg) were separated by
10% SDS-PAGE and proteins transferred to PVDF membranes (Bio-Rad
Laboratories, Inc.). Membranes were blocked with 5% non-fat milk
(Bio-Rad Laboratories, Inc.) for 1 h at room temperature, and
subsequently incubated with diluted primary antibody at 4°C
overnight. The membranes were incubated with HRP-conjugated
secondary antibodies (1:2,000, 7074S, Cell Signaling Technology,
Inc.) for 1 h at room temperature. Primary antibodies used in the
present study included the following: ENC1 was obtained from Abcam
(1:1,000, ab124902); N-Cadherin (1:1,000, 13116S), E-Cadherin
(1:1,000, 3195S), matrix metalloproteinase (MMP)2 (1:1,000,
40994S), MMP9 (1:1,000, 13667S), p-extracellular signal-regulated
kinase (ERK) (1:1,000, AP0485), ERK (1:1,000, 4695T), p-c-Jun
N-terminal kinase (JNK) (1:1,000, 4668T), JNK (1:1,000, 9252T),
p-p38 (1:1,000, 4511T), p-38 (1:1,000, 8690T) and β-actin (1:1,000,
3700T) were obtained from Cell Signaling Technology, Inc. The blots
were visualized using an ECL kit (Bio-Rad Laboratories, Inc.).
Bands were visualized by an enhanced chemiluminescence detection
system (Bio-Rad Laboratories, Inc.) and quantified using ImageJ
software (NIH) and normalized to the internal control, β-actin.
Cell viability and proliferation
assay
The viability of lung cancer cell lines (A549 and
H1299) was determined by CCK-8 assay (Bioss). Following
transfection with the siRNAs for 24, 48 and 72 h, 10 µl
CCK-8 were added to each well followed by incubation for 3 h at
37°C according to manufacturer's instructions. The absorbance at
450 nm was determined using a multi-detection microplate reader
(Bio-Tek Instruments, Inc.). The proliferation of lung cancer cell
lines (A549 and H1299) was evaluated using the EdU kit
(C10310-1/-2/-3; Guangzhou RiboBio Co., Ltd.). The transfected lung
cancer cell lines (A549 and H1299) were seeded in 96-well plates
and incubated for 24 h at 37°C in a humidified atmosphere with 5%
CO2. The cell culture medium was then supplemented with
100 µl 50 µM EdU and cells were incubated for a
further 2 h at 37°C in a humidified atmosphere with 5%
CO2. The cells were then fixed with 4% paraformaldehyde
at room temperature for 30 min, followed by 0.5% Triton X-100 to
permeabilize cell. Nucleic acids were stained with DAPI dye at room
temperature for 5 min. Signals were detected using an inverted
fluorescence microscope (Olympus IX83, Olympus Corporation).
Following transfection with the siRNAs for 48 h, the proportion of
proliferating cells in each group was determined. The intensity was
determined using ImageJ software (NIH).
Cell invasion and migration assay
Transwell membrane filters (8 µm pore size,
EMD Millipore) were used for the invasion and migration assays. For
cell invasion assay, 50 µl Matrigel (BD Biosciences) were
added to the upper chambers and incubated at 37°C for 1 h, while
Matrigel was not added for cell migration assay. Approximately
5×104 si-ENC1-transfected A549 and H1299 cells in a
volume of 100 µl were seeded in the upper chamber without
serum. The Transwell chambers were then placed in 24-well plates
and 600 µl RPIM-1640 medium with 10% FBS were added to the
lower chambers. Following incubation for 24 h at 37°C in a
humidified atmosphere with 5% CO2, the non-migrated
cells were removed using a cotton swab. Migrated cells were fixed
with 4% PFA for 30 min, then stained with crystal violet solution
for 20 min. After washing with phosphate-buffered saline, 5 random
fields of cells were imaged and quantified under a CNikon Eclipse
E100; Nikon Corporation). The average of the cells from 5 fields
was used for statistical analysis.
Tumor xenotransplantation experiment
Subcutaneous xenografts were created in the ventral
region of female BALB/C nude mice (weighting approximately 20-22 g
each) at 4 weeks of age. The mice were allowed to adapt in the SPF
environment (temperature: 23-24°C, humidity: 30-50%), and food and
water were freely available throughout the study. BALB/C nude mice
were divided randomly into 2 groups (n=5 per group). A total of
5×106 A549 cells with stable expression of sh-NC or
sh-ENC1 were implanted. Tumor nodules were monitored once a week
and tumor volumes were estimated using the following formula:
Volume=longest diameter x (smallest diameter)2/2.
The mice were sacrificed after 6 weeks and the
tumors were collected. All animals were euthanized by an
intravenous injection of 100 mg/kg pentobarbital sodium. The weight
of the tumors was recorded and the tumors fixed in 4% PFA
overnight, dehydrated and embedded in paraffin. The samples were
sectioned and stained with H&E as described above. The animal
research was approved by the Ethics Review Committee of Wannan
Medical College.
TCGA validation of ENC1
The expression level of ENC1 was verified using the
UALCAN lung cancer database (http://ualcan.path.uab.edu/index.html). The TCGA
survival data and Kaplan plot for ENC1 were verified using the
OncoLnc lung cancer database (http://www.oncolnc.org/).
Microarray data
The microarray data have been deposited on the GEO
database (GEO-GSE165972).
RNA sequencing
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The RNA concentration was determined
using a Bioanalyzer 2100 and RNA 1000 Nano LabChip kit (Agilent
Technologies, Inc.) with RIN number >7.0. RNA sequencing and
analysis were provided by LC-bio. Bioinformatics analyses (GO and
KEGG analyses) were performed using the OmicStudio tools at
https://www.omicstudio.cn/tool.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 5.0. All values represent at least 3 independent experiments
and are presented as the means ± SD. Differences between means were
analyzed using a Student's t-test. P-values <0.05 were
considered to indicate statistically significantly differences.
Results
High expression of ENC1 in lung cancer
tissues and cells
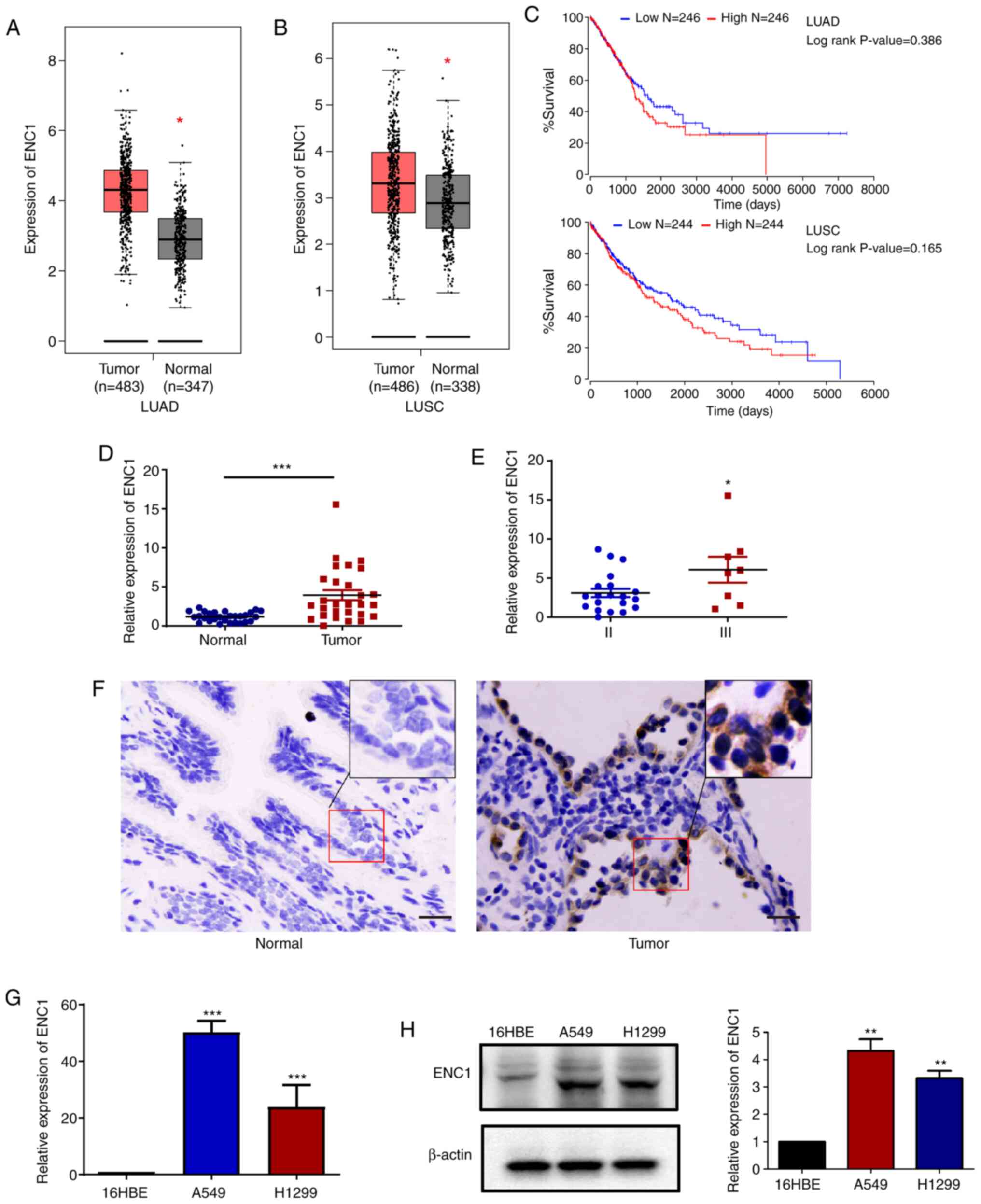
The mRNA expression levels of ENC1 between the lung
cancer and para-cancerous tissues were compared using the TCGA
database. As shown in Fig. 1A and
B, the mRNA levels of ENC1 in the cancer tissues were increased
compared to those in the para-cancerous tissues. Analysis using the
TCGA data-base also revealed that the lung cancer samples with a
higher ENC1 expression (red bar) exhibited a worse overall
survival, but the difference was not significant (Fig. 1C). Subsequently, the mRNA
expression of ENC1 was compared between cancer and para-cancerous
tissues obtained from lung cancer patients. The expression of ENC1
was determined by RT-qPCR and IHC (Fig. 1D-F). As shown in Fig. 1D and E, the expression of ENC1
was significantly increased in lung tumor tissues compared to
para-cancerous tissues, and the expression of ENC1 was
significantly increased in stage III cancer tissues compared to
stage II cancer tissues. The association between the
clinicopathological features of patients with NSCLC and the
expression levels of ENC1 was also investigated (Table I), and the expression of ENC1 was
only related to the M phase, not to sex, age, tumor size or N
stage. In addition, the expression of ENC1 in a normal human
bronchial epithelium cell line (16HBE), and lung cancer cell lines
(A549 and H1299) was examined by RT-qPCR and western blot analysis
(Fig. 1G and H). The results
revealed that the expression of ENC1 in the A549 and H1299 cells
was markedly higher than that in the 16HBE cells. These results
suggest that ENC1 is highly expressed in lung cancer cells and
tissues.
 | Table IAssociations between ENC1 expression
and the clinicopathological parameters of patients with lung
adenocarcinoma. |
Table I
Associations between ENC1 expression
and the clinicopathological parameters of patients with lung
adenocarcinoma.
| Characteristic | No. of patients | ENC1 expression
(2−ΔΔCq), means ± SD | P-value |
|---|
| Sex | | | 0.42 |
| Male | 17 | 4.828±3.672 | |
| Female | 11 | 3.530±4.688 | |
| Age, years | | | 0.262 |
| ≤65 | 18 | 3.641±4.080 | |
| >65 | 10 | 5.462±4.035 | |
| Tumor size (cm) | | | 0.728 |
| ≤3 | 7 | 3.809±2.644 | |
| >3 | 21 | 4.461±4.507 | |
| N stage | | | 0.794 |
| N0 | 13 | 4.374±2.920 | |
| N1-N3 | 15 | 4.841±5.409 | |
| M stage | | | 0.037 |
| II | 20 | 3.094±2.405 | |
| III | 8 | 6.067±4.399 | |
Gene expression profiling of
si-ENC1-transfected A549 cells
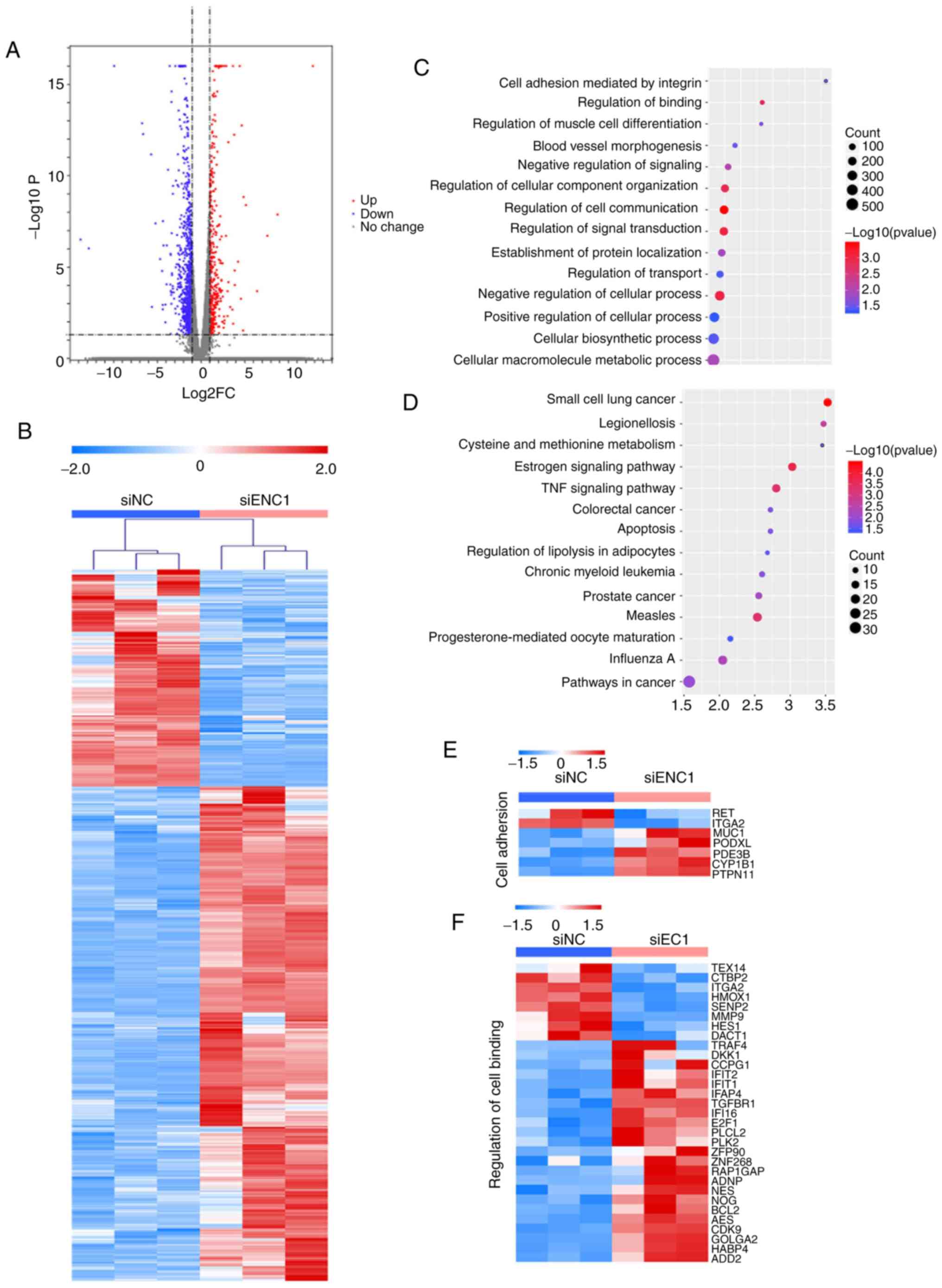
Overall gene expression was evaluated in
siRNA-transfected A549 cells. The transcriptome of the A549 cells
transfected with si-NC and si-ENC1 was compared. In total, there
were 1,404 differentially expressed genes (DEGs) between the cells
transfected with si-NC and si-ENC1 (Fig. 2A). Hierarchical clustering of the
DEGs separated the 2 groups (Fig.
2B). GO and KEGG analyses revealed changes in gene expression
related to cancer development, including cell adhesion, cell
communication, cell transduction, apoptosis and small cell lung
cancer (Fig. 2C-F). These
results suggested that downregulated ENC1 could affect the
progression of lung cancer.
Downregulation of ENC1 inhibits the
migration and invasion of NSCLC cells
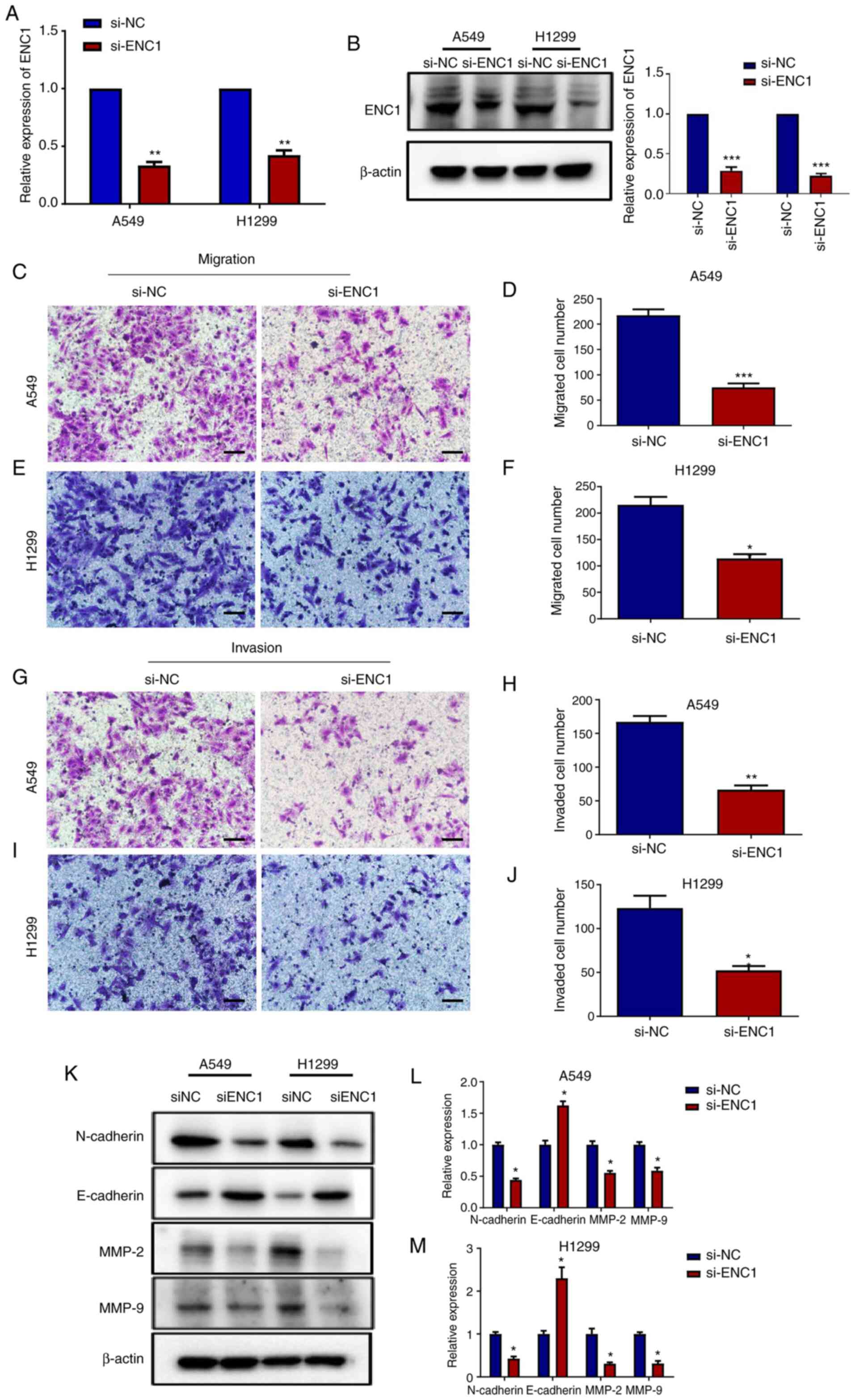
To examine the effects of si-ENC1 on the invasion
and migration of A549 and H1299 cells, Transwell assay was
performed. Following transfection with si-ENC1, the mRNA and
protein levels of ENC1 were determined in the A549 and H1299 cells.
As shown in Fig. 3A and B, the
mRNA levels of ENC1, as well as the protein levels were
significantly reduced by transfection with si-ENC1. The results of
Transwell assay revealed that the downregulation of ENC1
significantly inhibited the migratory and invasive ability of the
A549 and H1299 cells compared to the control group (Fig. 3C-J). MMP2 and MMP9 are the
primary enzymes that degrade type IV collagen, playing an important
role in the vascularization, invasion, and metastases of tumor
cells (14,15). They have been shown to be closely
related to the development of lung cancer (16). In the present study, following
transfection with si-ENC1, the MMP2 and MMP9 protein levels were
determined in the A549 and H1299 cells. As shown in Fig. 3K-M, the protein levels of MMP2
and MMP9 were decreased following transfection with si-ENC1.
Epithelial-mesenchymal transition (EMT) plays an important role in
the process of tumor cell metastasis, which has been widely
recognized. The decrease in E-cadherin expression and the increase
in N-cadherin expression are considered as key factors of EMT. In
the present study, as shown in Fig.
3K-M, following transfection with si-ENC1, the expression of
N-cadherin was decreased, while that of E-cadherin was
increased.
Downregulation of ENC1 inhibits the
proliferation of NSCLC cells
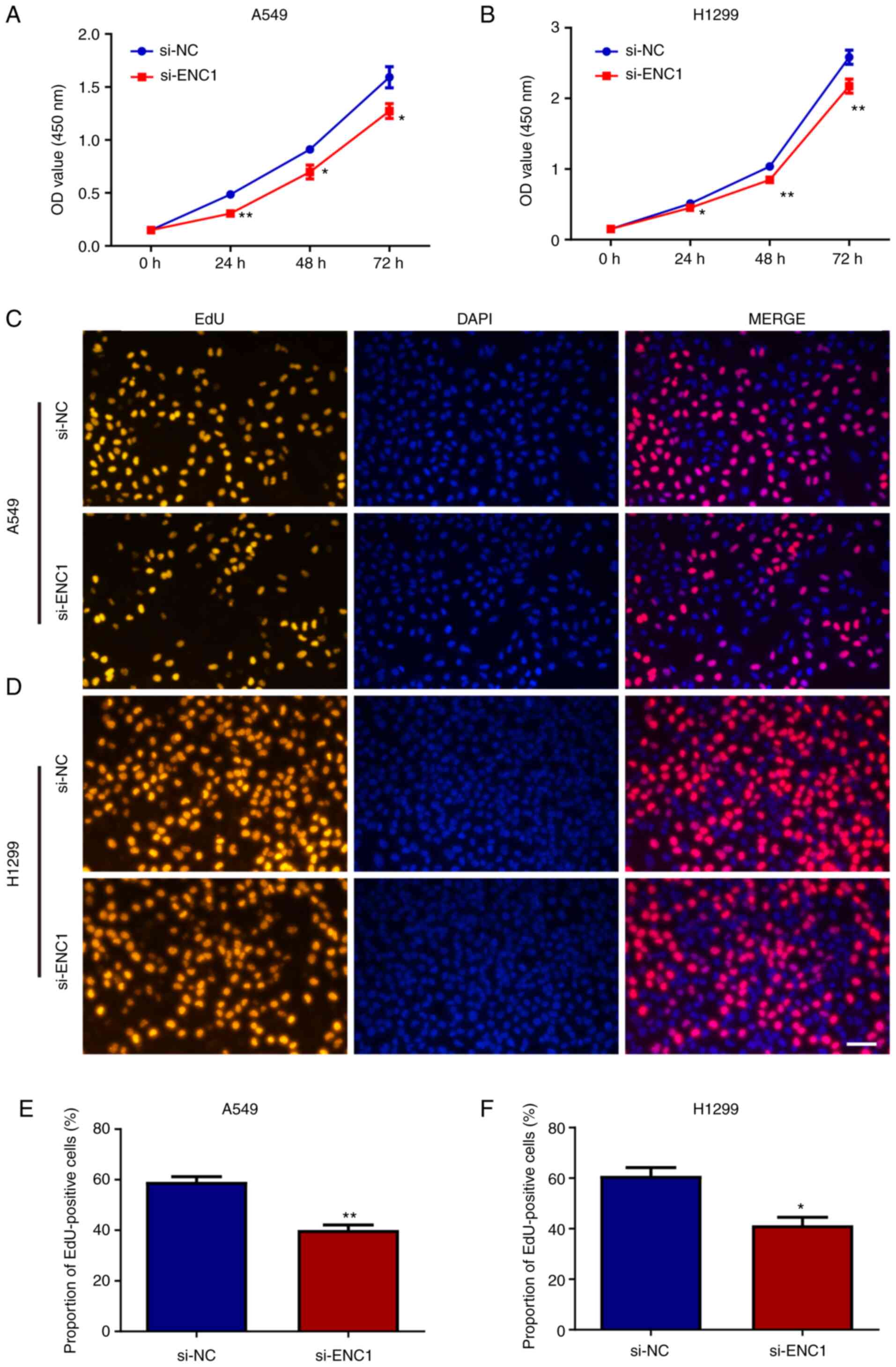
The proliferation of the A549 and H1299 cells
following transfection with si-ENC1 was assessed. The results of
CCK-8 assay revealed that the downregulated expression of ENC1
inhibited the proliferation of the A549 and H1299 cells (Fig. 4A and B). Similar results were
also obtained using EdU detection (Fig. 4C-F). These results suggest that
ENC1 plays a significant role in the proliferation of NSCLC
cells.
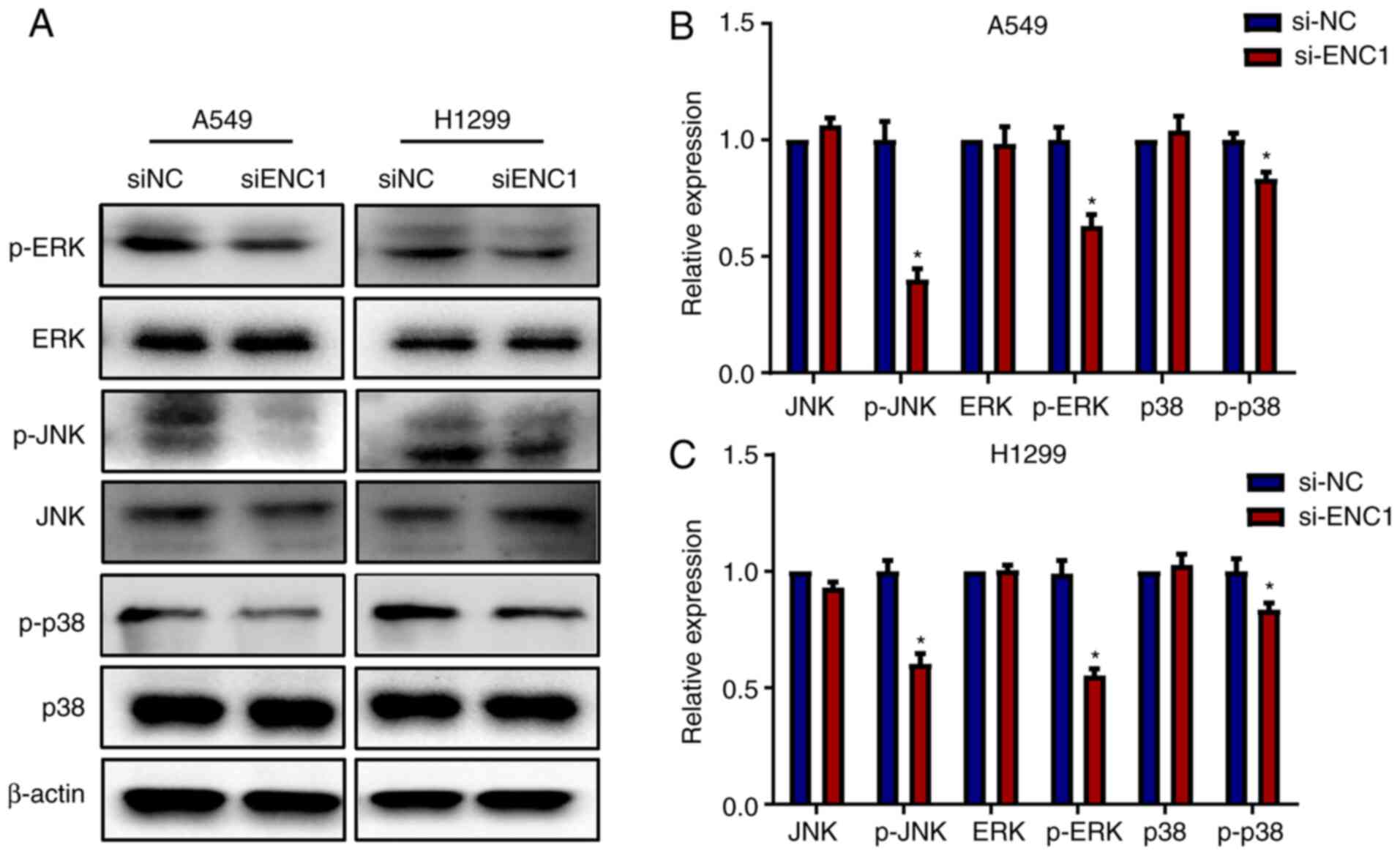
Downregulation of ENC1 affects the MAPK
pathway
Subsequently, the present study investigated whether
si-ENC1 can affect the regulation of the MAPK pathway. As shown in
Fig. 5, transfection of the A459
and H1299 cells with si-ENC1 significantly inhibited the
phosphorylation of JNK, ERK and p38. However, there was no
significant change in the total protein levels of JNK, ERK and p38.
These results suggest that ENC1 inhibits the migration, invasion
and proliferation of NSCLC cells by affecting the pathway
downstream of MAPK.
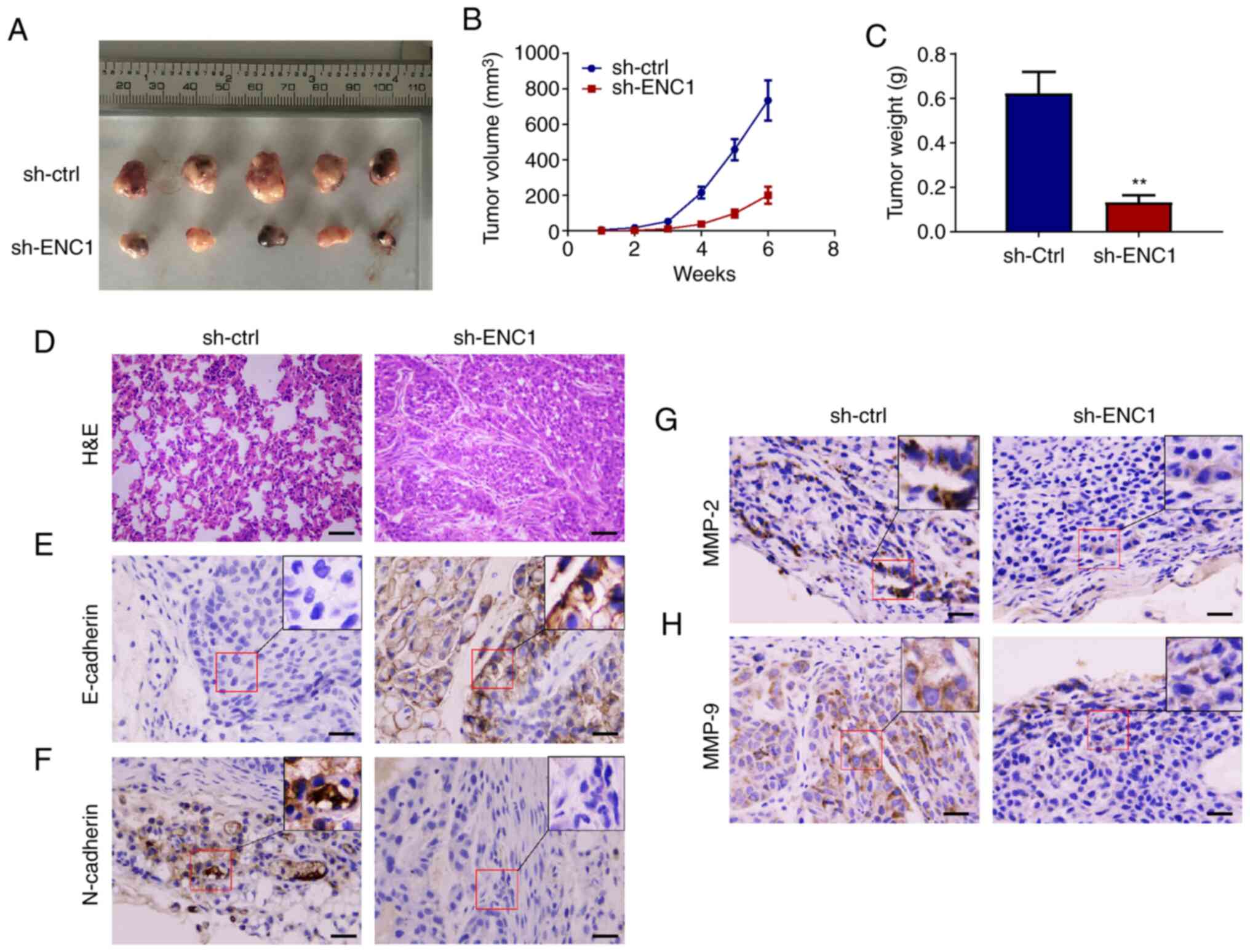
Downregulation of ENC1 inhibits the
growth of NSCLC in a mouse model
To determine the effect of ENC1 on the growth of
tumors in vivo, a subcutaneous xenograft tumor model was
established. The A549 cells, which were transfected with sh-ENC1 or
sh-ctrl, were introduced into nude mice. As shown in Fig. 6A and B, the growth of the tumors
in the sh-ENC1 group was significantly inhibited and the weight of
the tumors was significantly lower than that in the sh-ctrl group
(Fig. 6C). In addition, tumor
tissue structures are shown in Fig.
6D. Subcutaneous transplantation of tumor in nude mice was
successful. The cancer cells were diffusely distributed and formed
adenoid structures, with obvious atypia, pathological mitosis and
focal necrosis. The expression of E-cadherin increased and that of
N-cadherin decreased in the sh-ENC1 group compared with the sh-ctrl
group (Fig. 6E and F). In
addition, the expression of both MMP2 and MMP9 was decreased in the
sh-ENC1 group (Fig. 6G and H).
These experiments suggest that ENC1 is a key gene mediating the
growth and metastasis of lung cancer.
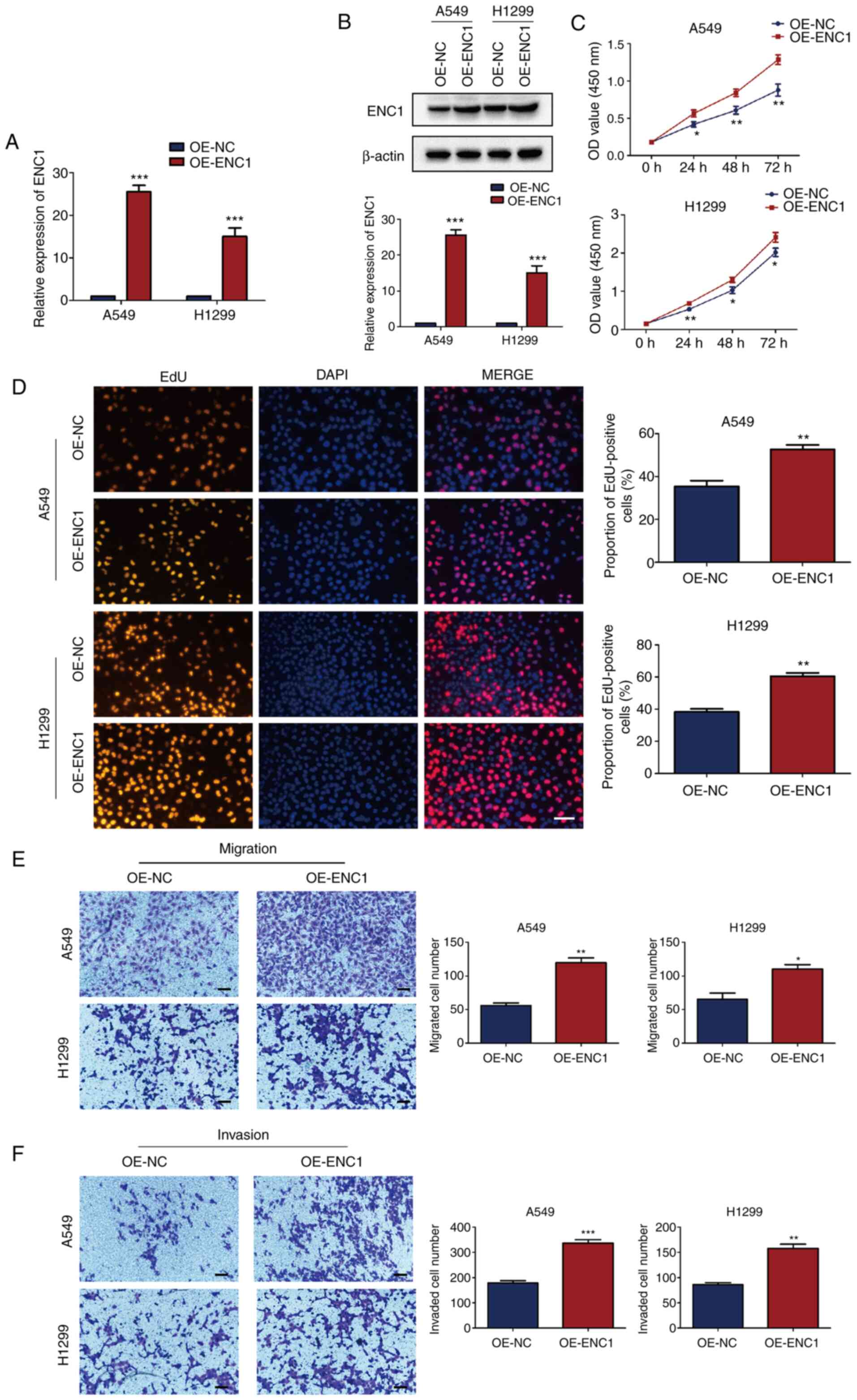
Upregulation of ENC1 enhances the
proliferation, migration and invasion of NSCLC cells
The effects of a high expression of ENC1 on NSCLC
cells were then investigated. An ENC1-overexpression plasmid
(OE-ENC1 group) and an NC plasmid (OE-NC group) were transfected
into the A549 and H1299 cells. The ENC1 mRNA and protein levels
were significantly increased in both cell lines following
transfection with OE-ENC1 (Fig. 7A
and B). The increased expression of ENC1 induced the
proliferation of A549 and H1299 cells, as shown by CCK-8 assay
(Fig. 7C). Similar results were
obtained using the EdU detection kit (Fig. 7D). As shown in Fig. 7E and F, the upregulation of ENC1
significantly enhanced the migration and invasion of A549 and H1299
cells compared with the control group. These results suggest that
ENC1 plays a significant role in proliferation, migration and
invasion of NSCLC cells.
Discussion
With over a million deaths annually worldwide, lung
cancer is the leading cause of cancer-related mortality (17,18). NSCLC accounts for approximately
85% of all lung cancers (19).
Despite tremendous advances being made in the treatment using a
combination of surgical techniques, chemotherapy and radiotherapy,
NSCLC is still associated with a dismal prognosis due to its
resistance to therapy and its local recurrence. Consequently,
patients with NSCLC have a median survival of <1 year and a
2-year survival rate of <20%. The lack of effective means for
the early diagnosis of NSCLC is the cause for the high mortality
rate associated with this disease (20,21). It is thus critical to identify
accurate and sensitive biomarkers for NSCLC in order to increase
the survival rate of patients and to lower the costs of
treatment.
ENC1 encodes an actin-binding protein and plays an
important role in early gastrulation and in the formation of the
nervous system. ENC1 is emerging as a novel biomarker due to its
conservation, abundance and roles in cancer progression. It is
overexpressed in various types of cancer, including ovarian cancer
and breast cancer, increasing tumor metastases in patients
(11,12). Previous studies have demonstrated
controversial associations between ENC1 expression and different
types of cancers. ENC1 expression is downregulated in null cell
adenomas and oncocytomas (22),
but is overexpressed in breast cancer tissue compared with normal
breast tissues (12). A high
ENC1 expression is associated with a high metastatic ability of
breast cancer cells, which was consistent with the finding that
ENC1 was associated with the invasiveness of both pituitary null
cell adenoma and oncocytoma (12). However, there is a lack of
studies demonstrating the association between ENC1 and lung cancer.
In the present study, it was demonstrated that the inhibition of
ENC1 may be a possible therapeutic target for the treatment of lung
cancer. Furthermore, it was found that the downregulation of ENC1
in A549 and H1299 cells by siRNA significantly inhibited their
proliferative, migratory and invasive ability, and also affected
the expression of proteins MPP2, MMP9, N-cadherin and E-cadherin,
which are closely related to the invasion and migration of cancer
cells. These results suggested that ENC1 may become another novel
diagnostic, metastatic and prognostic biomarker, and even a target
for lung cancer in the future.
The MAPK pathway regulates important cellular
processes, including gene expression, cell proliferation, cell
movement and apoptosis, which play important roles in the
progression of NSCLC (23). In
the present study, the knockdown of ENC1 expression significantly
reduced the phosphorylation levels of 3 MAPK family members, JNK,
ERK and p38 in A549 and H1299 cells. Thus, si-ENC1 reduces the
proliferation, migration and invasion of lung cancer cells through
the MAPK pathway. However, further investigations into the details
of ENC1 function are warranted to support these findings. Mouse
models will be a valuable resource for identifying therapeutic
targets, preclinical screening and the evaluation of different
therapies for various types of cancer. It was confirmed that ENC1
regulated the growth and invasion of tumors in nude mice. The
results obtained in vivo demonstrated that tumor sizes in
nude mice injected with cells transfected with sh-ENC1 were smaller
than those of the control group.
In conclusion, the present study demonstrates that
ENC1 is significantly overexpressed in lung cancer and has
diagnostic and prognostic value. ENC1 may thus be a target for the
detection and treatment of NSCLC, as well as other types of
cancer.
Availability of data and materials
The microarray data has been deposited at the GEO
database (GEO-GSE165972). Other raw data supporting the conclusions
of the present study will be made available by the authors, without
undue reservation, to any qualified researcher.
Authors' contributions
CW and XC conceived and designed the study. CW, XWa
and XWu performed the experiments. CW and XWa analyzed the data. CW
wrote the manuscript. XC edited the manuscript. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Yijishan Hospital. Informed consent was obtained from
all patients. The animal research was approved by the Ethics Review
Committee of Wannan Medical College.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pfannschmidt J, Muley T, Bülzebruck H,
Hoffmann H and Dienemann H: Prognostic assessment after surgical
resection for non-small cell lung cancer: Experiences in 2083
patients. Lung Cancer. 55:371–377. 2007. View Article : Google Scholar
|
|
4
|
Reungwetwattana T, Weroha SJ and Molina
JR: Oncogenic pathways, molecularly targeted therapies, and
highlighted clinical trials in non-small-cell lung cancer (NSCLC).
Clin Lung Cancer. 13:252–266. 2012. View Article : Google Scholar
|
|
5
|
Karuppasamy R, Veerappapillai S, Maiti S,
Shin WH and Kihara D: Current progress and future perspectives of
polypharmacology: From the view of non-small cell lung cancer.
Semin Cancer Biol. 68:84–91. 2021. View Article : Google Scholar
|
|
6
|
Hernandez MC, Andres-Barquin PJ, Holt I
and Israel MA: Cloning of human ENC-1 and evaluation of its
expression and regulation in nervous system tumors. Exp Cell Res.
242:470–477. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao L, Gregoire F and Sul HS: Transient
induction of ENC-1, a Kelch-related actin-binding protein, is
required for adipocyte differentiation. J Biol Chem.
275:16845–16850. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee H, Ahn HH, Lee W, Oh Y, Choi H, Shim
SM, Shin J and Jung YK: ENC1 modulates the aggregation and
neurotoxicity of mutant huntingtin through p62 under ER stress. Mol
Neurobiol. 53:6620–6634. 2016. View Article : Google Scholar
|
|
9
|
Kim TA, Ota S, Jiang S, Pasztor LM, White
RA and Avraham S: Genomic organization, chromosomal localization
and regulation of expression of the neuronal nuclear matrix protein
NRP/B in human brain tumors. Gene. 255:105–116. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fujita M, Furukawa Y, Tsunoda T, Tanaka T,
Ogawa M and Nakamura Y: Up-regulation of the ectodermal-neural
cortex 1 (ENC1) gene, a downstream target of the
beta-catenin/T-cell factor complex, in colorectal carcinomas.
Cancer Res. 61:7722–7726. 2001.PubMed/NCBI
|
|
11
|
Fan S, Wang Y, Sheng N, Xie Y, Lu J, Zhang
Z, Shan Q, Wu D, Sun C, Li M, et al: Low expression of ENC1
predicts a favorable prognosis in patients with ovarian cancer. J
Cell Biochem. 120:861–871. 2019. View Article : Google Scholar
|
|
12
|
Zhou Y, Tang X, Niu L, Liu Y, Wang B and
He J: Ectodermal-neural cortex 1 as a novel biomarker predicts poor
prognosis and induces metastasis in breast cancer by promoting
Wnt/β-catenin pathway. J Cell Mol Med. 24:8826–8835. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
14
|
Jodele S, Chantrain CF, Blavier L, Lutzko
C, Crooks GM, Shimada H, Coussens LM and Declerck YA: The
contribution of bone marrow-derived cells to the tumor vasculature
in neuroblastoma is matrix metalloproteinase-9 dependent. Cancer
Res. 65:3200–3208. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mott JD and Werb Z: Regulation of matrix
biology by matrix metalloproteinases. Curr Opin Cell Biol.
16:558–564. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leinonen T, Pirinen R, Böhm J, Johansson
R, Ropponen K and Kosma VM: Expression of matrix metalloproteinases
7 and 9 in non-small cell lung cancer. Relation to
clinicopathological factors, beta-catenin and prognosis. Lung
Cancer. 51:313–321. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li MY, Liu LZ and Dong M: Progress on
pivotal role and application of exosome in lung cancer
carcinogenesis, diagnosis, therapy and prognosis. Mol Cancer.
20:222021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han J, Liu Y, Yang S, Wu X, Li H and Wang
Q: MEK inhibitors for the treatment of non-small cell lung cancer.
J Hematol Oncol. 14:12021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baade PD, Youlden DR and Krnjacki LJ:
International epidemiology of prostate cancer: Geographical
distribution and secular trends. Mol Nutr Food Res. 53:171–184.
2009. View Article : Google Scholar
|
|
20
|
Reck M, Heigener DF, Mok T, Soria JC and
Rabe KF: Management of non-small-cell lung cancer: Recent
developments. Lancet. 382:709–719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gottesman MM, Lavi O, Hall MD and Gillet
JP: Toward a better understanding of the complexity of cancer drug
resistance. Annu Rev Pharmacol Toxicol. 56:85–102. 2016. View Article : Google Scholar
|
|
22
|
Feng J, Hong L, Wu Y, Li C, Wan H, Li G,
Sun Y, Yu S, Chittiboina P, Montgomery B, et al: Identification of
a subtype-specific ENC1 gene related to invasiveness in human
pituitary null cell adenoma and oncocytomas. J Neurooncol.
119:307–315. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang L and Karin M: Mammalian MAP kinase
signalling cascades. Nature. 410:37–40. 2001. View Article : Google Scholar : PubMed/NCBI
|