Introduction
Oral tongue squamous cell carcinoma, the sixth most
frequent cancer worldwide, is one of the most aggressive
pathological types of head and neck squamous cell carcinoma
(1,2). OTSCC is a highly malignant cancer
that presents with rapid local invasion and distal metastasis and
is frequently diagnosed in young individuals, particularly in males
(3,4). Although the therapeutic regimens
for OTSCC have been greatly improved over the years, clinicians
have not observed evident improvement of the 5-year survival rate
in patients with OTSCC due to the high recurrence rates (5,6).
Generally, OTSCC is a complex disease with extensive
genetic/epigenetic alterations (7). Therefore, it is urgent to
investigate its potential carcinogens with the hope of identifying
new therapeutic targets.
Long non-coding RNAs refers to a group of non-coding
RNAs with a length of >200 nucleotides (8). When first discovered, lncRNAs were
regarded as transcriptional 'noise' generated during the
transcription process and held no biological functions (9,10). However, in recent years, more
studies have reported that lncRNAs play an important role in a wide
range of cellular processes, including cell differentiation, cell
apoptosis, cell proliferation, gene transcription, epigenetic
modification as well as others (11-13). Furthermore, lncRNAs have been
demonstrated to act as crucial regulators in various malignancies
(14-16). HOTTIP, one lncRNA in OTSCC, has
been reported to suppress the proliferation, migration and invasion
of OTSCC by regulating the HMGA2-mediated Wnt/b-Catenin pathway
(17,18). Therefore, it is essential to
identify the key lncRNAs involved in OTSCC progression in order to
help understand the mechanisms underlying this disease.
The present study investigated the expression levels
of MIR4713HG in OSTCC tissues and cell lines, and evaluated its
influence on cell proliferation, invasion, migration and
tumorigenesis of OTSCC. Furthermore, the interaction between
MIR4713HG and let-7c-5p, and their downstream target genes were
investigated. The findings may help elucidate the molecular
mechanisms underlying the carcinogenic process of OTSCC and provide
potential therapeutic targets.
Materials and methods
RNA sequence data analysis
MIR4713HG and let-7c-5p RNASeq data from OTSCC
samples were downloaded from the Cancer Genome Atlas (TCGA)
database (https://cancergenome.nih.gov/). All the data are
publicly available. MIR4713HG and let-7c-5p were identified
according to the Ensembl data-base (http://www.ensembl.org/index.html, version 89) The
edgeR package (19) was used to
normalize gene expression of MIR4713HG and let-7c-5p in TSCC and
normal tissues.
Cell culture and patient samples
The human OTSCC cell lines (CAL-27, SCC-9, SCC-4,
SCC-15 and SCC-25) were purchased from Shanghai Cell Bank of the
Chinese Academy of Sciences and the human periodontal ligament
fibroblast (HPLF) cell line was purchased from ScienCell Research
Laboratories, Inc. (cat. no. 2630). Cells were cultured at 37°C in
an atmosphere containing 5% CO2 in RPMI-1640 medium
supplemented with 10% FBS and 1% penicillin-streptomycin solution
(all from Gibco; Thermo Fisher Scientific, Inc.).
With approval from the institutional review board of
the Joint Ethics Committee of the Southern Medical University
Health Authority, freshly resected OTSCC tissues (20 pairs) and
paraffin-embedded OTSCC sections were obtained from the archives of
the Department of Oral Surgery, Stomatological Hospital, Southern
Medical University (Guangzhou, China). Each participant provided
written informed consent.
RNA interference assay
The short hairpin (sh) RNA targeting MIR4713HG and
small interfering (si)RNA targeting transmembrane channel like 7
(TMC7) were synthesized by Shanghai GenePharma Co., Ltd. Sequences
of shRNA or siRNAs against specific targets are listed in Table SI. The sequence with the best
interfering effect was selected and used in subsequent
experiments.
Cell transfection
CAL-27 and SCC-9 cells were transfected with 1
µg pcDNA3.1-MIR4713HG and 1 µg control pcDNA3.1
plasmids (control-plasmids; both from Shanghai GenePharma Co.,
Ltd.); 1 µg shRNA (sh-MIR4713HG), 1 µg negative
control shRNA (sh-NC), 100 nM let-7c-5p inhibitors
(miR20000064-1-5), and 100 nM let-7c-5p inhibitor controls
(miR2N0000001-1-5); 100 nM let-7c-5p-mimics (miR10000064-1-5) and
100 nM let-7c-5p mimic negative controls (miR1N0000001-1-5; all
from Guangzhou RiboBio Co., Ltd.) for 48 h at 37°C using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Transfection efficiency was determined 48 h post-transfection via
reverse transcription-quantitative (RT-q) PCR. Sequences of
let-7c-5p inhibitor or let-7c-5p mimics against specific targets
are listed in Table SI.
Cell viability assay
To assess cell viability, CAL-27 and SCC-9 cells
were seeded in triplicate in 96-well plates at a density of
5×103 cells/well in 100 µl of culture medium. The
cell proliferation index was measured using an MTT assay, which was
performed at 0, 24, 48 and 72 h after transfection, respectively. A
total of 10 µl of MTT reagent (5 mg/ml; Sigma-Aldrich; Merck
KGaA) was added for 3 h. MTT was solubilized using dimethyl
sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA) and plates were read at
490 nm using Tecan Sunrise microplate reader (Tecan F50; Tecan
Group, Ltd.).
EdU proliferation assay
To assess cell proliferation, CAL-27 and SCC-9 cells
(5×103 cells/well) were seeded in 96-well plates. The
cells were incubated under standard conditions in complete media.
Transfection of the cells was performed the following day. A total
of 48 h after transfection, cell proliferation was detected using
the incorporation of EdU with an EdU Cell Proliferation Assay kit
(Guangzhou RiboBio Co., Ltd.). Briefly, the cells were incubated
with 50 µM EdU for 6 h before fixation, permeabilization and
EdU staining, which were performed according to the manufacturer's
protocol. The cell nuclei were stained with DAPI (Sigma-Aldrich;
Merck KGaA) at a concentration of 1 µg/ml for 20 min. The
proportion of the EdU-incorporated cells was determined with
fluorescence microscopy (×200).
Clone formation assay
Single cells at low density (52.63 cells per
cm2) were seeded over a coating of 0.6% agarose solution
containing supplemented media in normal conditions. The cells were
allowed to sit for 10 min at room temperature and were then covered
with a 0.3% agarose solution. Plates were incubated at 37°C in a 5%
CO2 humidified atmosphere and media were replaced every
3-4 days. After 15 days, The cells were then immobilized with 4%
paraformaldehyde for 15 min, and soaked in Giemsa stain (G4640,
Beijing Solarbio Science & Technology Co., Ltd.) for 30 min at
room temperature and the cells were then washed twice with
ultra-pure water. Images were acquired using a camera [DSC-HX90;
SONY (China), Co., Ltd.].
Lung metastasis model
A total of 12 male BALB/C nude mice (5 weeks old),
with a weight of 17-19 g were obtained from the Guangdong Medical
Laboratory Animal Center, were used in this study. All mice were
placed in an environment with 50-60% humidity and a temperature of
22-24°C. The mice were given free access to water and food, and a
12-h light/dark cycle was applied. Following 7 days of
environmental adaptation, the mice were randomly assigned into two
groups; a sh-NC group and a sh-MIR4713HG group, with 6 mice in each
group. For the in vivo metastasis assays, MIR4713-knockdown
OTSCC cells and OTSCC cells were injected into the caudal veins of
BALB/C nude mice. Animal health and behavior was monitored every
two days. Preliminary experiments verified that OTSCC cells
metastasized to the lungs of mice after four weeks, which was in
line with the one-month processing time of lung metastasis model
reported in most literature (20,21). Therefore, in the present study,
the mice were sacrificed after four weeks. The mice were
anesthetized by intraperitoneal injection of 1% sodium
pentobarbital (50 mg/kg) and then subjected to cervical dislocation
for euthanasia. The lungs of the mice were separated and fixed with
4% paraformaldehyde for 7 days at 4°C, then embedded in paraffin
and sectioned at 4-µm. The sections were subjected to
hematoxylin and eosin (H&E) staining. Paraffin sections were
baked at 59°C for 30 min, dewaxed in xylene and hydrated in a
series of gradient alcohol, followed by immersion in hematoxylin
dye solution at room temperature for 5 min to stain the nuclei, in
hydrochloric acid for 5 sec for differentiation, and in lithium
carbonate to stain the nuclei blue. They were then immersed in
eosin at room temperature for 5 min to stain the cytoplasm.
Finally, paraffin sections were observed under an optical
microscope at a magnification (×200; Nikon Corporation).
Subcutaneous tumor xenograft assay
A total of 10 male BALB/C nude mice (5 weeks old),
with a weight of 17-19 g were obtained from the Guangdong Medical
Laboratory Animal Center, were used in this study. All mice were
placed in an environment with 50-60% humidity and a temperature of
22-24°C. The mice were given free access to water and food, and a
12-h light/dark cycle was applied. Following 7 days of
environmental adaptation, the mice were randomly assigned into two
groups; a sh-NC group and a sh-MIR4713HG group, with 5 mice in each
group. The mice were inoculated subcutaneously in the right flank
with 2×106 cells. Animal health and behavior was
monitored every two days. Tumor growth was observed and measured
weekly, and tumors were finally excised according to the schedule.
Tumors were harvested for the measurement of volume and weight
after 30 days. The mice were anesthetized by intraperitoneal
injection of 1% sodium pentobarbital (50 mg/kg) and then subjected
to cervical dislocation for euthanasia.
Luciferase reporter assay
The fragment of MIR4713HG containing the target
sequence of let-7c-5p was amplified via RT-qPCR and then inserted
into a pmirGLO vector (Promega Corporation) to form the wild-type
MIR4713HG reporter vector (MIR4713HG-WT). An additional expression
vector was also constructed by inserting a mutated binding site and
was termed MIR4713HG-mutated-type (MIR4713HG-MUT). CAL-27 and SCC-9
cells were seeded into 24-well plates at a density of
5×104 cells/well. When the confluency reached ~80%,
cells were co-transfected with MIR4713HG-WT or MIR4713HG-MUT and
let-7c-5p mimics or the negative control at 37°C for 48 h using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), in accordance with the manufacturer's protocol.
The relative luciferase activity was then determined using a
Dual-Luciferase Reporter assay system (Promega Corporation)
according to the manufacturer's protocol. Luciferase activity was
normalized to the activity of Renilla.
To detect the direct binding of let-7c-5p to the
target gene TMC7, the entire 3′-untranslated region (3′-UTR) of
human TMC7 was amplified via PCR using human genomic DNA as a
template. The PCR products were inserted into the p-MIR-reporter
plasmid (Ambion; Thermo Fisher Scientific, Inc.). The insertion was
confirmed as correct by sequencing. To test the binding
specificity, the sequences that interact with the let-7c-5p seed
sequence were mutated, and the mutant TMC7 3′-UTR was inserted into
an equivalent luciferase reporter. For luciferase reporter assays,
2×105 cells were cultured in 24-well plates, and each
well was transfected with 1 µg firefly luciferase reporter
plasmid, 1 µg β-galactosidase (β-gal) expression plasmid
(Ambion; Thermo Fisher Scientific, Inc.), and equal amounts (100
pmol) of pre-let-7c-5p, anti-let-7c-5p or the scrambled negative
control RNA using Lipofectamine® 2000 at 37°C. The β-gal
plasmid was used as a transfection control. After 24 h
post-transfection, the cells were assayed using a Dual-Luciferase
Reporter assay system (Promega Corporation).
Immunohistochemistry
Samples were fixed in 10% formalin, embedded in
paraffin, and cut into 4-µm-thick sections. The specimens
were deparaffinized in xylene and dehydrated using a graded series
of ethanol. The specimens were then heated in Tris-EDTA buffer (pH
9.0) for 15 min (except for PCNA-stained samples). After antigen
retrieval, the specimens were treated with 3%
H2O2 in methanol for 30 min at room
temperature to inhibit the activity of endogenous peroxidase, and
incubated with 10% goat serum for 20 min at room temperature to
block non-specific binding of the immunoreagents. Samples were
incubated at 4°C overnight with a primary antibody against
E-cadherin (1:300; cat. no. 20874-1-AP; Proteintech Group, Inc.),
Ki67 (1:200; ab16667; Abcam) and vimentin (1:300; ab137321; Abcam),
followed by incubation with the secondary antibody (code number
K4001; Dako EnVision+System-horseradish peroxidase-labelled
polymer, anti-mouse; Agilent Technologies, Inc.). The sections were
subjected to the hyper-sensitive polymer method, and 4′,
6-diamidino-2-phenylindole (DAPI) was used as the chromogen. Images
were captured using an optical microscope (×200; Nikon
Corporation).
RT-qPCR
Total RNA was extracted from CAL-27 and SCC-9 cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RNA quality was assessed using the 260/280 nm
absorbance ratio, and concentration was quantified using a
microplate reader. Subsequently, isolated RNA was reverse
transcribed into cDNA using a PrimeScript RT reagent kit (Takara
Bio, Inc.). The conditions of RT were as follows: 38°C for 15 min
and 85°C for 5 sec. RT-qPCR analysis was performed using Maxima
SYBR-Green/ROX qPCR Master Mix (Invitrogen; Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were used
for the RT-qPCR: 95°C for 10 min, 95°C for 15 sec, 62°C for 30 sec,
and 72°C for 30 sec. The relative mRNA expression was calculated
using the 2−∆∆Cq method (22), GAPDH and U6 were set as internal
controls. Primer sequences are listed in Table SII.
Protein extraction and western blot
analysis
OTSCC tissues were homogenized using a Beads
crusher. Homogenized tissues and adherent cells were lysed using
RIPA buffer (product no. 08714-04; Nacalai Tesque, Inc.). Protein
quantification was carried out using a BCA protein assay kit.
Proteins (30 µg) were separated via 10% SDS-PAGE and then
transferred to polyvinylidene difluoride membranes. The membranes
were washed, then blocked with 5% skimmed milk for 1 h at room
temperature and incubated overnight at 4°C with a primary antibody
specific for E-cadherin (1:1,000; cat. no. 20874-1-AP; ProteinTech
Group, Inc.), N-cadherin (1:1,000; product code ab76011), vimentin
(1:1,000; product code ab137321) and TMC7 (1:1,000; product code
ab191521; all from Abcam) and GAPDH (1:2,000; product no. 5174S;
Cell Signaling Technology, Inc.), followed by incubation with
horseradish peroxidase-conjugated secondary antibodies for 60 min
at 37°C (both 1:500; product codes ab6789 or ab6721; both from
Abcam). Immunocomplexes were visualized using an enhanced
chemiluminescence assay kit (ECL Plus Western Blotting Detection
Reagents; GE Healthcare Life Sciences).
Wound healing assay
Cells (1×103 cells/well) were seeded into
6-well plates and grown until 100% confluent. A scratch was
produced with a sterile 200-µl pipette tip in the cell
monolayer followed by culture in DMEM for an additional 24 h at
37°C. Images were captured using an optical microscope (×100; Nikon
Corporation), and the migration distance was measured at 0 and 24 h
after scratching using ImageJ software (version 1.48; National
Institutes of Health).
Transwell migration assay
For the detection of migration capacity, CAL-27 and
SCC-9 cells (1×104; 150 µl) were collected in
serum-free medium and spread onto the upper chamber of a Transwell
plate. The lower chamber was filled with 700 µl of medium
containing 10% FBS. The plates were incubated at 37°C for 24 h. The
membranes were then fixed with 4% methanol at 4°C, stained with
0.1% crystal violet for 30 min at room temperature, and the
staining was observed under an optical microscope (×200). For the
invasion assay, 80 µl Matrigel solution (BD Biosciences) was
used to precoat the Transwell membrane for 30 min at 37°C.
Statistical analysis
Student's t-test, One-way ANOVA and Spearman's rank
correlation coefficient were performed using Graph Pad Prism 8
(GraphPad Software, Inc.). The post hoc test used was Tukey's test.
Paired t-test was used for the comparison between the tumor and
adjacent non-tumor tissues of the same patients. Experiments were
performed in triplicate, with three independent experiments. Data
are presented as the mean ± SEM. P<0.05 was considered to
indicate a statistically significant difference.
Results
lncRNA MIR4713HG is upregulated in OTSCC
tissue and cell lines
In order to investigate the potential role of
MIR4713HG in OTSCC, the present study first examined the expression
pattern of MIR4713HG in OTSCC using the information acquired from
online databases (23).
According to the RNA-sequencing data, the expression
of MIR4713HG was significantly upregulated in OTSCC tissues
compared with normal tissues and its expression was stage-dependent
in OTSCC, indicating its potential role in tumor progression
(Fig. 1A and B). To investigate
the prognostic value of MIR4713HG, the present study extracted the
expression data of MIR4713HG from 113 patients with OTSCC.The
expression level of MIR4713HG was significantly higher in patients
with OTSCC (Fig. 1C).
Furthermore, the high expression of MIR4713HG indicated an
unfavorable prognosis in OTSCC (Fig.
1D). To confirm the online data, the present study performed
RT-PCR in order to determine the expression level of MIR4713HG in
20 paired OTSCC and adjacent non-tumor tissues (Fig. 1E). Additionally, the present
study also examined the expression levels of MIR4713HG in multiple
OTSCC cell lines (CAL-27, SCC-9, SCC-4, SCC-15 and SCC-25) and HPLF
cell line. It was revealed that the expression of MIR4713HG was
significantly increased in OTSCC cell lines as well (Fig. 1F). The differential expression of
MIR4713HG in OTSCC and adjacent non-tumor tissues suggests that
MIR4713HG may have an important role in the progression of
OTSCC.
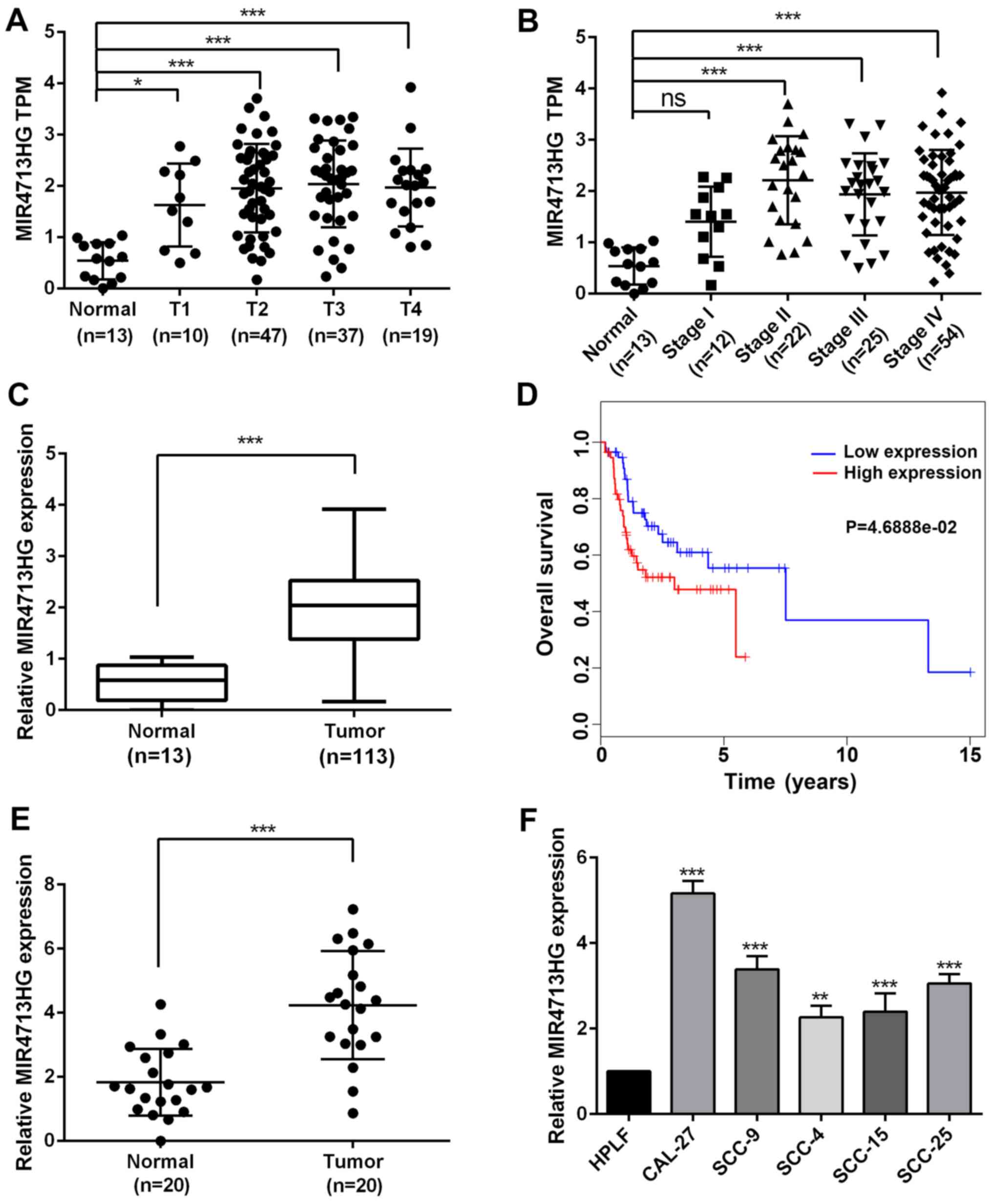 | Figure 1lncRNA MIR4713HG is specifically
upregulated in OTSCC tissues and cell lines. (A) The expression of
MIR4713HG (TCGA expression data, TPM) in normal oral epithelium
tissues compared with OTSCC tissues (T1-T4). (B) The expression of
MIR4713HG (TPM data) in normal oral epithelium tissues compared
with OTSCC tissues (Stage I-IV). (C) The relative expression level
of MIR4713HG in OTSCC and adjacent non-tumor tissues. (D) Survival
analysis of patients with OTSCC with high/low expression of
MIR4713HG. (E) The relative expression level of MIR4713HG in 20
paired OTSCC and adjacent non-tumor tissues. (F) The relative
expression level of MIR4713HG in human OTSCC cell lines (CAL-27,
SCC-9, SCC-4, SCC-15 and SCC-25, and the HPLF cell line. Data are
presented as the mean ± SEM. *P<0.05,
**P<0.01 and ***P<0.001. OTSCC, οral
tongue squamous cell carcinoma; TCGA, The Cancer Genome Atlas;
HPLF, human periodontal ligament fibroblast; ns, not significant;
TPM, Transcripts Per Kilobase of exon model per Million mapped
reads. |
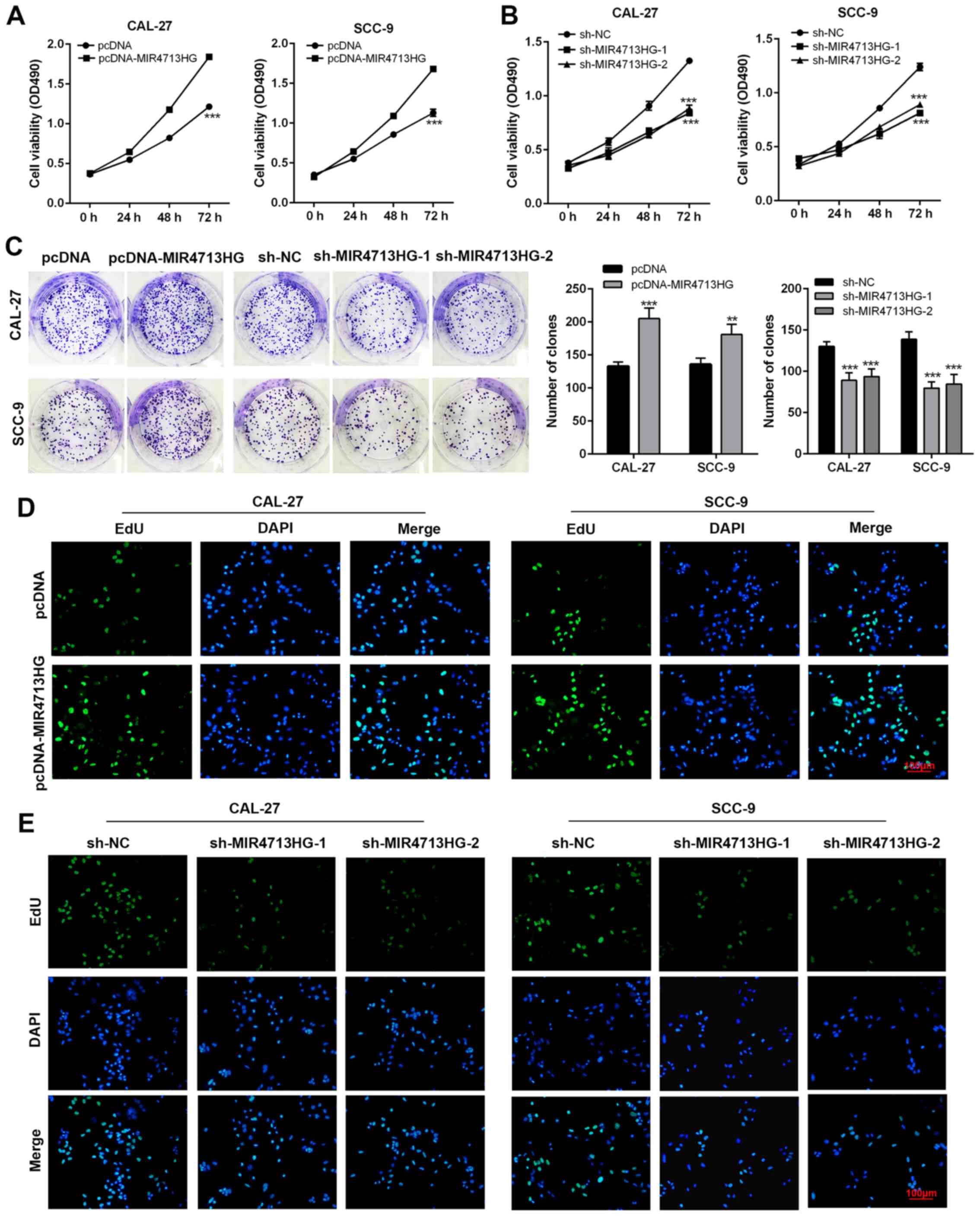
lncRNA MIR4713HG regulates the
proliferation and migration of OTSCC cells in vitro
To further investigate the regulating role of
MIR4713HG in OTSCC, the present study performed a series of
malignancy-associated experiments on CAL-27 and SCC-9 cell lines.
First, the present study constructed
MIR4713HG-overexpressing/knockdown cell lines via pcDNA-MIR4713HG
and sh-MIR4713HG, respectively. The overexpression and knockdown
efficiency were evaluated using RT-PCR. The expression of MIR4713HG
was significantly upregulated/downregulated in constructed cell
lines (Fig. S1A and B). MTT
assays were performed to assess the effect of MIR4713HG expression
level on OTSCC cell proliferation. After 72 h of observation, the
present study clearly identified that the overexpression of
MIR4713HG improved the OTSCC cell proliferation rate, and the
administration of sh-MIR4713HG achieved the exact opposite effect
(Fig. 2A and B). In the colony
formation assay, MIR4713HG-overexpressing OTSCC cells formed more
clones on the culture dish after 14 days compared with the control
group, while the clonogenicity of OTSCC cells was compromised after
MIR4713HG-knockdown (Fig. 2C).
In order to ensure the reliability of tumor cell proliferation
experiments, EdU assays were used to detect the cell proliferation
rate. In the EdU cell proliferation assay, the staining intensity
was significantly stronger in MIR4713HG-overexpressing OTSCC cells
and weaker in MIR4713HG-knockdown OTSCC cells, confirming the
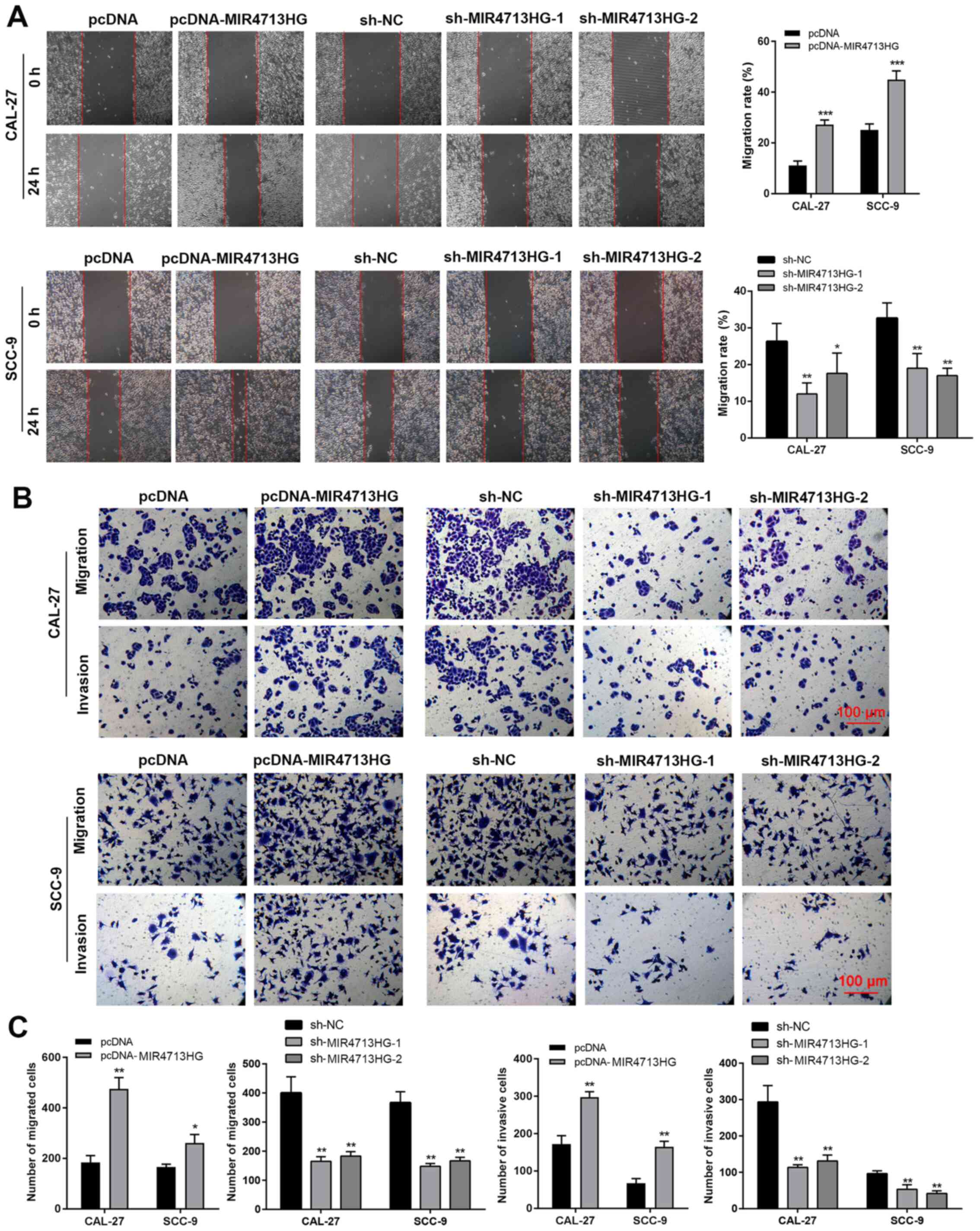
positive effect of MIR4713HG on OTSCC growth (Fig. 2D and E). OTSCC is a highly
metastatic cancer. To evaluate the regulatory role of MIR4713HG in
cellular migration and metastasis in OTSCC, the present study
performed a wound healing assay and Transwell assay. The wound
healing results revealed that the overexpression of MIR4713HG
accelerated the migration rate of OTSCC cells while the
downregulation of MIR4713HG slowed down the migration rate
(Fig. 3A). The Transwell assay
results indicated that MIR4713HG was a contributing factor in OTSCC
metastasis, as overexpression of MIR4713HG improved the invasion
rate of OTSCC cells, while the downregulation of MIR4713HG
decreased the invasion rate (Fig. 3B
and C).
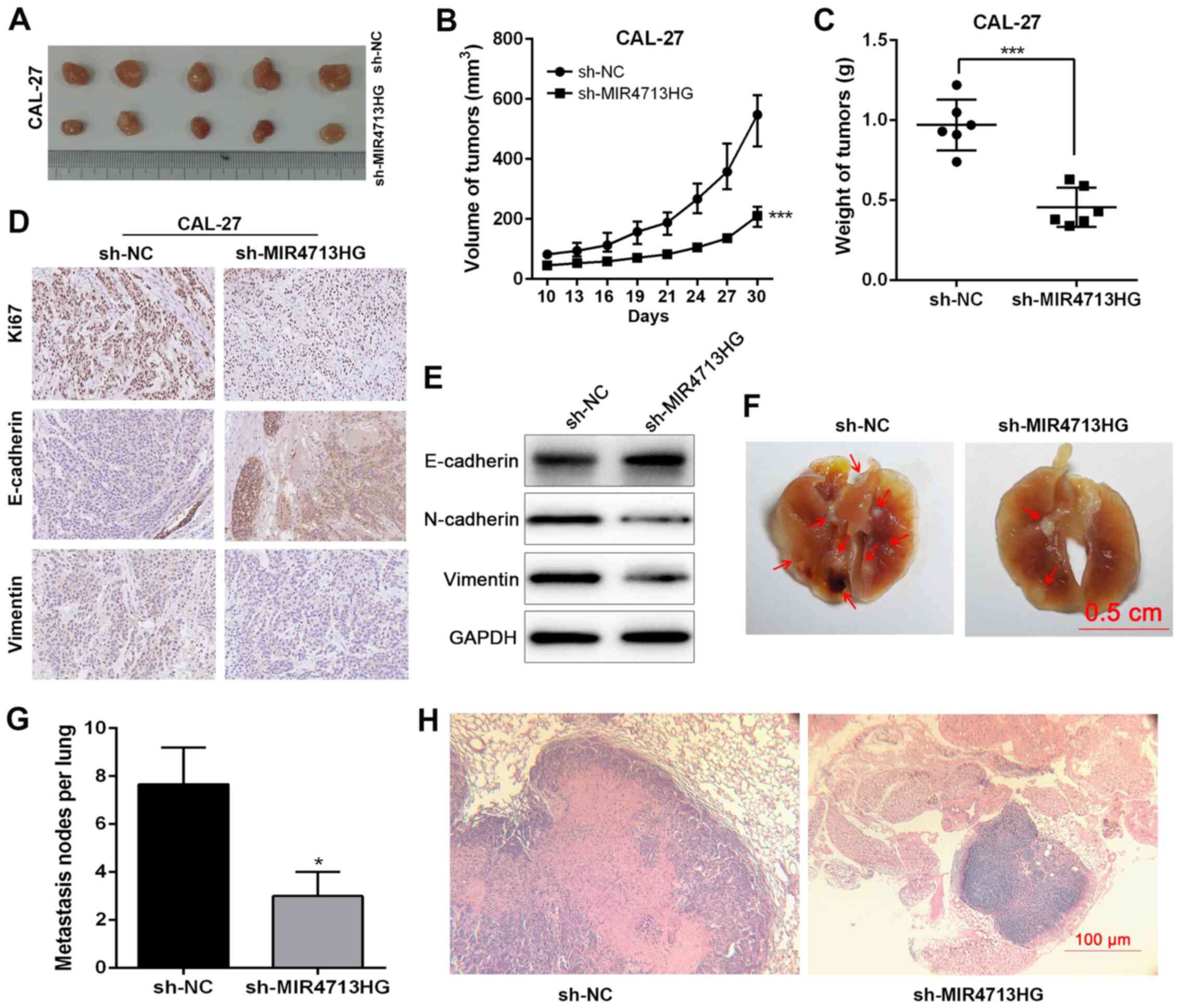
Knockdown of lncRNA MIR4713HG inhibits
tumor growth in vivo
To further investigate the impact of MIR4713HG on
the tumorigenicity of OTSCC in vivo, 12 nude mice were
injected with transfected OTSCC cells (6 sh-MIR4713HG mice and 6
sh-NC mice). At 30 days post-inoculation, tumors were harvested for
the measurement of volume and weight (Fig. 4A). The largest tumor was 612.4
mm3 (10.5×10.8 mm3). The volume of tumors in
the sh-MIR4713HG group was smaller than that of the sh-NC group
(Fig. 4B). The tumor weight was
decreased in the sh-MIR4713HG group compared with the sh-NC group
(Fig. 4C). The
epithelial-mesenchymal transition (EMT) is a crucial process in
cancer metastasis (24,25). Therefore, the present study
examined the expression levels of EMT-associated proteins,
E-cadherin/vimentin in the harvested tumors via
immunohistochemistry and western blotting. The knockdown of
MIR4713HG increased the expression levels of E-cadherin and
suppressed the expression of vimentin (Fig. 4D and E), suggesting that
MIR4713HG affects the metastatic process of OTSCC via accelerating
EMT. Furthermore, the present study also constructed lung
metastasis models via injecting OTSCC cells into the caudal veins
of nude mice. Macroscopically, the mice injected with
MIR4713HG-knockdown OTSCC cells exhibited statistically less lung
metastasis than the control mice (Fig. 4F-H). In addition, the expression
of MIR4713HG was downregulated in the subcutaneous tumor tissue
transfected with sh-MIR4173HG (Fig.
S1C).
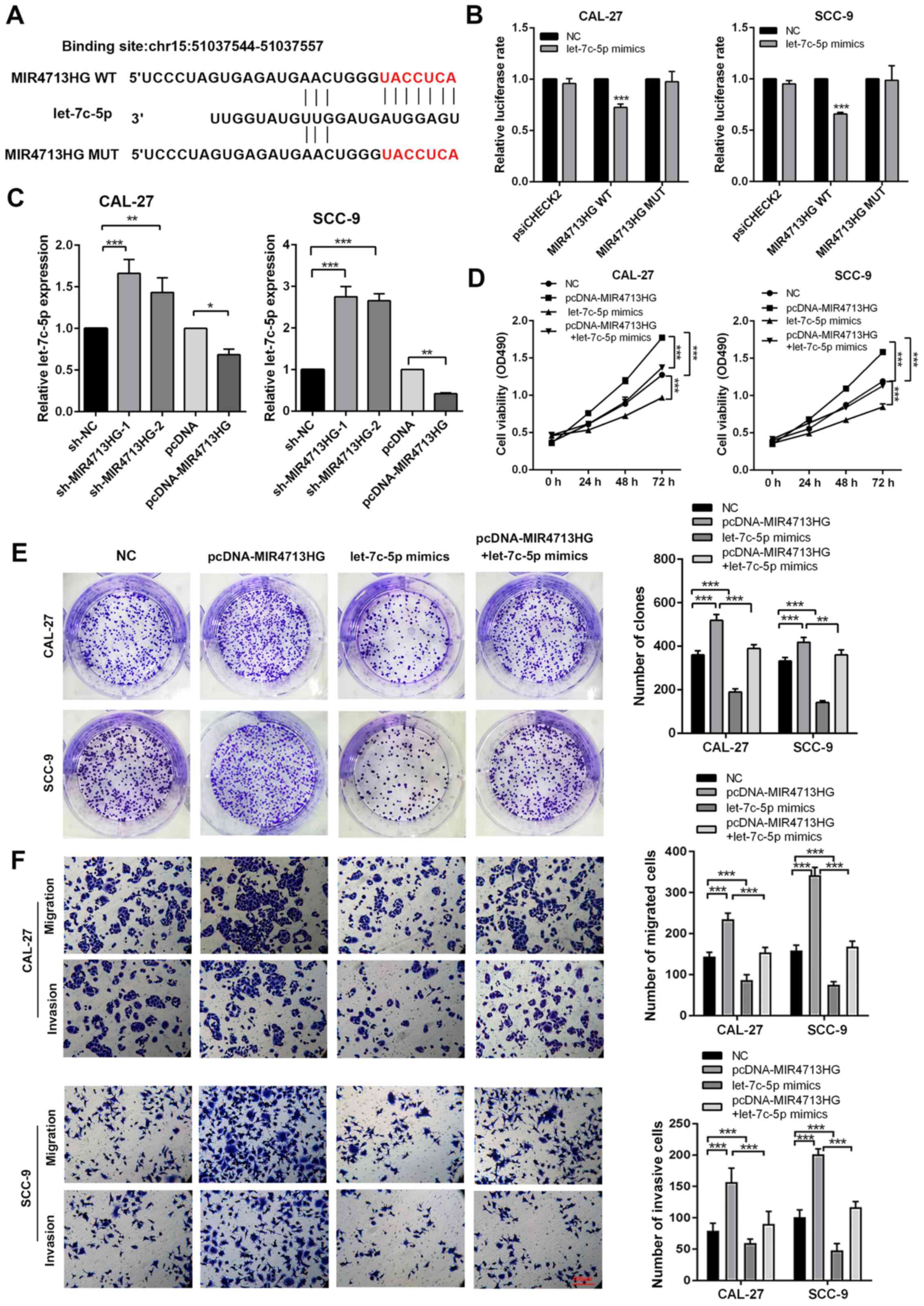
lncRNA MIR4713HG regulates malignant
behaviors of OTSCC via binding with miRNA let-7c-5p
lncRNAs commonly act as competing endogenous RNAs to
regulate cellular functions. Using publicly available DIANA Tools
(26), the present study
successfully observed let-7c-5p directly binding with MIR4713HG
(Fig. 5A). To test the direct
binding of let-7c-5p to MIR4713HG, a luciferase reporter assay was
performed as previously described. The administration of let-7c-5p
mimics resulted in a 0-20% reduction of luciferase reporter
activity compared with the control group (Fig. 5B). Furthermore, the present study
introduced point mutations into the corresponding sites in
MIR4713HG to eliminate the predicted let-7c-5p binding sites. This
mutated luciferase reporter was unaffected by the overexpression of
let-7c-5p (Fig. 5B). According
to the RT-PCR results, the expression of let-7c-5p increased after
MIR4713HG knockdown and decreased after MIR4713HG overexpression
(Fig. 5C). Based on The Cancer
Genome Atlas (TCGA) database (23), the expression of let-7c-5p was
downregulated in OTSCC tumors and its expression pattern was
confirmed in 20 paired OTSCC tumor and adjacent nontumor tissues
(Fig. S2A and B). The
expression of let-7c-5p was negatively correlated with MIR4713HG
(Fig. S2C and D). CAL-27/SCC-9
cells were transfected with let-7c-5p mimics and inhibitor, and the
expression of let-7c-5p was significantly upregulated/downregulated
in constructed cell lines (Fig. S2E
and F). To assess the effect of let-7c-5p expression levels on
OTSCC cell proliferation, the present study performed rescue
experiments via MTT and clone formation assays. MIR4713HG
overexpression significantly improved cell proliferation and
let-7c-5p mimics suppressed the cell viability and clonogenicity
compared with the NC group. The let-7c-5p mimics partially rescued
the stimulative effects of MIR4713HG overexpression on OTSCC cell
proliferation (Fig. 5D and E).
The present study also performed rescue experiments via Transwell
assays. The addition of let-7c-5p mimics partially rescued the
improved metastatic potential of OTSCC cells induced by MIR4713HG
overexpression compared with pcDNA-MIR4713HG group (Fig. 5F).
TMC7 is the downstream target gene of
miRNA let-7c-5p
To identify the downstream target of let-7c-5p, the
present study used online bioinformatics platforms, DIANA tools and
TargetScan to predict the potential target genes of let-7c-5p
(26,27). By overlapping the aforementioned
results with OTSCC-overexpressing genes, a list of 79 genes was
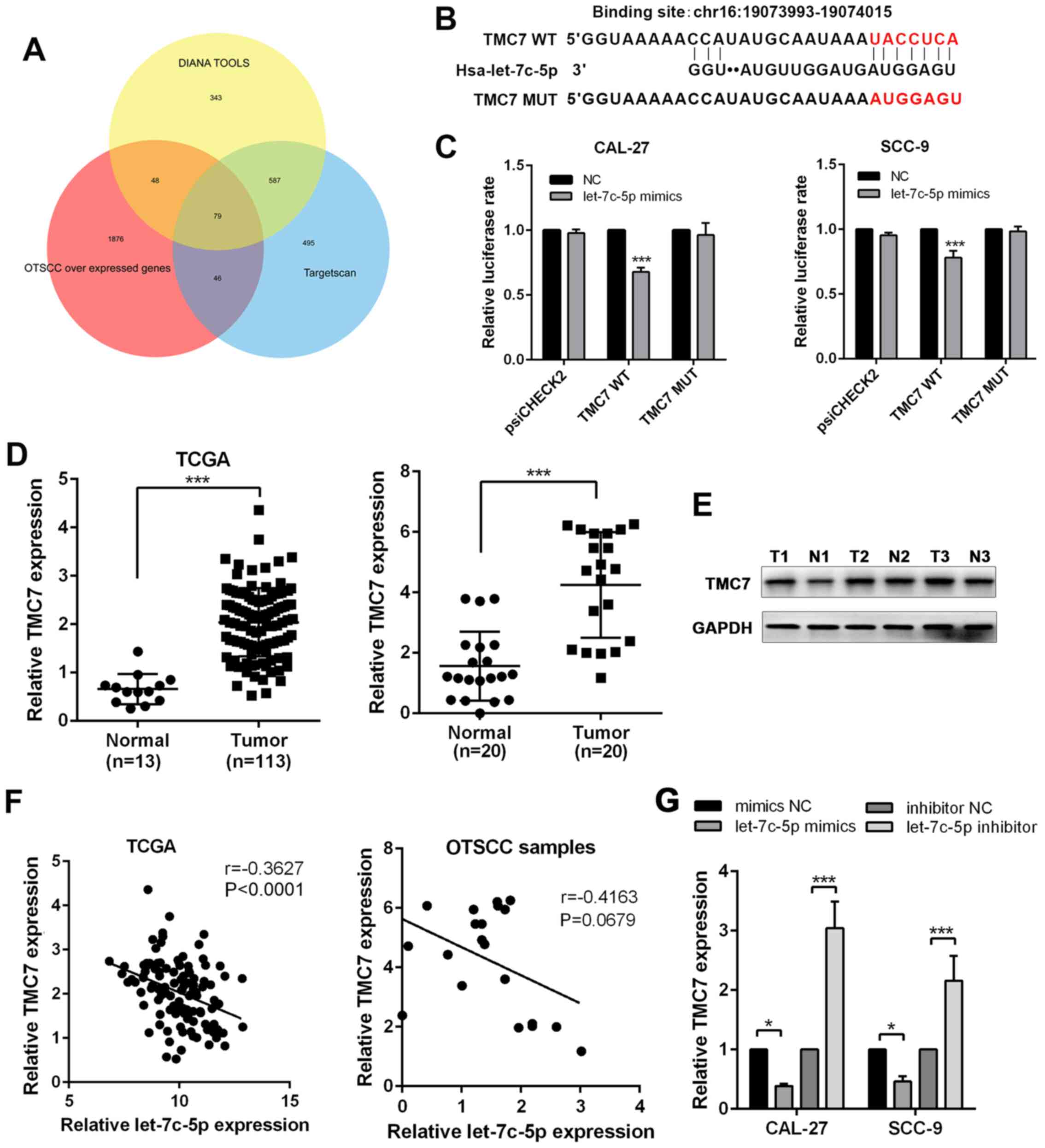
obtained (Fig. 6A). Among the 79
genes, TMC7 was found to directly bind with let-7c-5p (Fig. 6B). According to the results of
the luciferase reporter assay, the administration of let-7c-5p
mimics resulted in a reduction of luciferase reporter activity
compared with the control group, and the introduction of point
mutations into TMC7 eliminated the predicted let-7c-5p binding
sites (Fig. 6C), confirming the
direct binding between let-7c-5p and TMC7. Based on TCGA database,
the expression of TMC7 was upregulated in OTSCC tumors and its
expression pattern was confirmed in 20 paired OTSCC tumor and
adjacent non-tumor tissues (Fig.
6D). The same expression pattern of TMC7 was also confirmed at
the protein level (Fig. 6E). The
expression of TMC7 was negatively associated with let-7c-5p
(Fig. 6F). Furthermore, the
addition of let-7c-5p mimics significantly suppressed the
expression of TMC7 while the addition of let-7c-5p inhibitor
greatly boosted the expression of TMC7 (Fig. 6G).
lncRNA MIR4713HG performs its regulatory
roles by affecting the let-7c-5p/TMC7 axis
In order to investigate the regulation of MIR4713HG
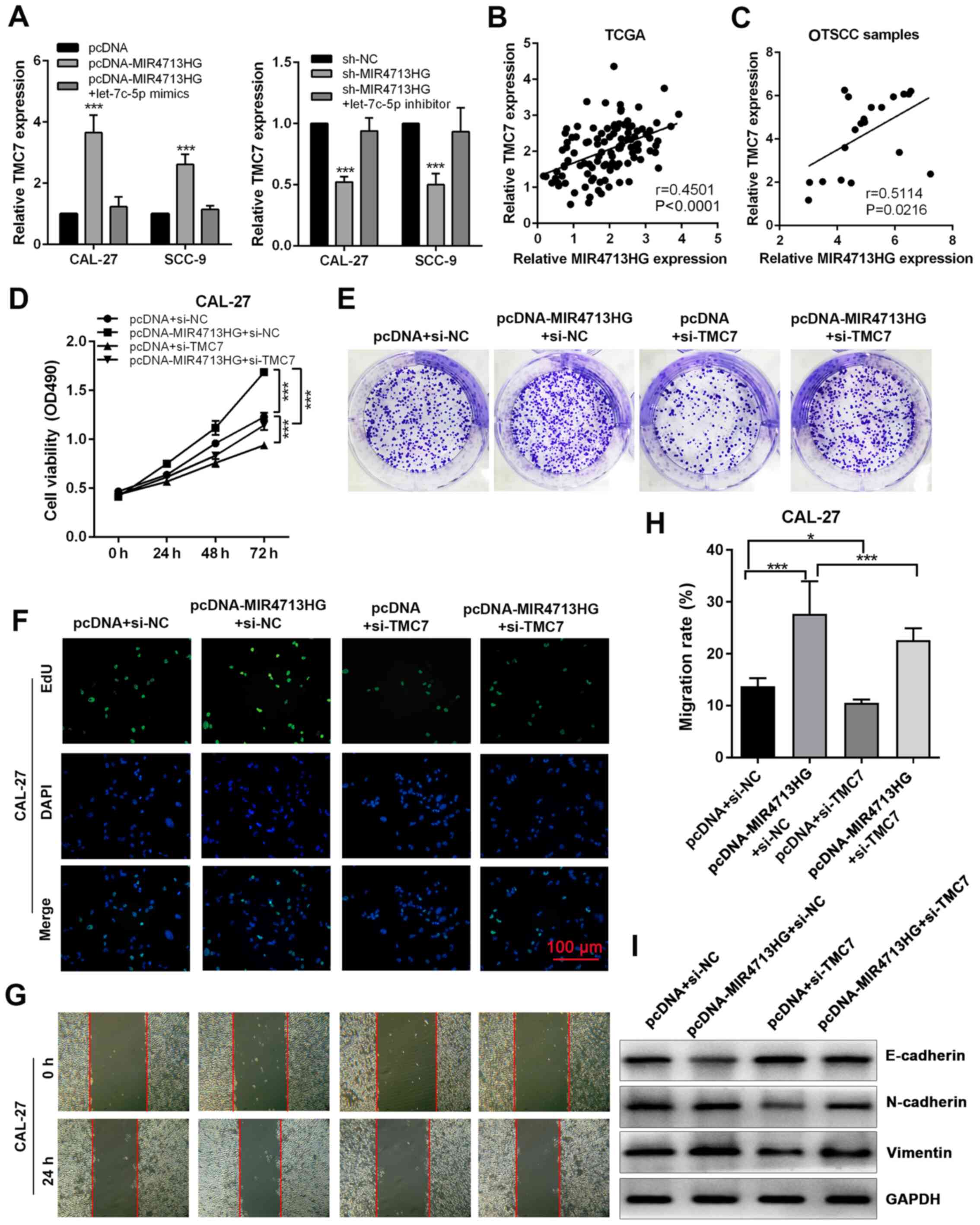
on TMC7 expression, the present study performed RT-qPCR. The
overexpression of MIR4713HG significantly increased the expression
of TMC7, while the knockdown of MIR4713HG suppressed the expression
of TMC7 and both effects were counteracted by let-7c-5p mimics and
inhibitor, respectively (Fig.
7A). Based on TCGA database, the expression of TMC7 was
positively correlated with MIR4713HG in OTSCC tumors and its
expression pattern was confirmed in 20 paired OTSCC tumor and
adjacent non-tumor tissues (Fig. 7B
and C). The knockdown efficiency was evaluated using RT-qPCR,
and the expression of TMC7 was significantly downregulated in
constructed cell lines (Fig.
S2G). In order to assess the effect of TMC7 expression level on
OTSCC cell proliferation, the present study performed rescue
experiments via MTT, clone formation and EdU cell proliferation
assays. It was revealed that the knockdown of TMC7 could reverse
the improved cell proliferation and clonogenicity in OTSCC induced
by MIR4713HG overexpression (Fig.
7D-F). In the wound healing assay, the improved cell migration
ability induced by MIR4713HG overexpression was counteracted by
TMC7 knockdown (Fig. 7G and H).
In addition, according to the western blotting results, the
overexpression of MIR4713HG accelerated the EMT process in OTSCC,
while TMC7 knockdown reversed this effect (Fig. 7I).
Discussion
Globally, head and neck squamous cell carcinoma
(HNSCC) is one of the most common types of cancer and the number of
cases of this disease increases by 500,000 annually (28). The therapeutic strategies of oral
tongue squamous cell carcinoma include surgical resection,
chemotherapy, radiotherapy and targeted therapy. Although great
improvements have been made in all of these fields, the prognosis
of patients with OTSCC remains unsatisfactory (29,30). Local invasion and metastasis
account for the poor prognosis of patients with OTSCC (31). Thus, OTSCC remains a challenging
field in cancer and the underlying molecular mechanisms require
further investigation.
Long non-coding RNAs and microRNAs are two subtypes
of non-coding RNAs with different lengths. It has been well
documented that lncRNAs can act as endogenous competing RNAs that
interact with miRNAs and participate in human diseases, such as
cancer (32,33). In OTSCC, the potential regulating
role of lncRNAs remains unclear, except for one lncRNA, HOTTIP,
which has been reported to act as an oncogenic factor in OTSCC
(17). The present study first
revealed that MIR4713HG was upregulated in OTSCC tissues and
exhibited itself as an unfavorable prognostic factor in OTSCC
patients. To further investigate the function of MIR4713HG, the
present study performed gain- or loss-of-function experiments by
transfecting OTSCC cell lines with MIR4713HG-overexpressing plasmid
or sh-MIR4713HG. The results revealed that MIR4713HG positively
regulated the proliferation and clonogenicity of OTSCC cells.
Furthermore, in the wound healing and Transwell assays, it was
revealed that the upregulation of MIR4713HG was correlated with
improved migration and metastasis of OTSCC cells.
To test the tumor regulating role of MIR4713HG in
vivo, the present study constructed xenograft tumor mouse
models and lung metastasis mouse models. Notably, the knockdown of
MIR4713HG significantly slowed down the process of tumor growth and
suppressed the progression of tumor metastasis in OTSCC in
vivo. OTSCC cells have strong invasion and migration
capabilities, and they experience enhanced invasion and migration
capabilities during EMT, which is an important step in tumor
metastasis and spread (29,30). EMT is a multi-step process in
which epithelial cells acquire a mesenchymal phenotype, which is
characterized by increased exercise capacity, downregulation of
epithelial proteins including E-cadherin and α-catenin, and
overexpression of mesenchymal phenotype proteins, such as vimentin
and N-cadherin (34). The
present study revealed that knockdown of MIR4713HG upregulated
E-cadherin expression and downregulated the expression of
N-cadherin and vimentin, which indicated that knockdown of
MIR4713HG significantly inhibited EMT. With further investigation,
the present study successfully identified that let-7c-5p physically
bonds with MIR4713HG. To the best of our knowledge, the function of
miRNA in OTSCC remains unclear. To examine the impact of the
expression level of let-7c-5p on OTSCC cells, the present study
performed a series of rescue experiments. The results revealed that
the addition of let-7c-5p counteracted the improved cell
proliferation and clonogenicity induced by MIR4713HG, and
suppressed the increased metastasis potential in OTSCC cells caused
by MIR4713HG overexpression. Furthermore, it was revealed that
expression of let-7c-5p was downregulated in OTSCC and negatively
correlated with MIR4713HG. According to the results of the
bioinformatic prediction, it was revealed that TMC7 directly bound
with let-7c-5p. The expression pattern of TMC7 exhibited the
opposite to that of let-7c-5p. The administration of let-7c-5p
mimics or inhibitor decreased or increased the expression level of
TMC7, respectively. Furthermore, the overexpression of MIR4713HG
could increase the expression of TMC7, and in rescue experiments,
the knockdown of TMC7 could counteract the effect induced by
MIR4713HG overexpression on OTSCC cells.
In conclusion, the present study demonstrated that
MIR4713HG was significantly upregulated in both OTSCC tissues and
cell lines. MIR4713HG acted as a pro-tumor factor, facilitating
cell proliferation and metastasis. Furthermore, the present study
identified that MIR4713HG directly bound with let-7c-5p, and that
let-7c-5p regulated malignant behaviors of OTSCC via affecting the
expression of TMC7. Collectively, MIR4713HG aggravated the
malignant behaviors in OTSCC via affecting the let-7c-5p/TMC7
signaling pathway, and could be a promising therapeutic target in
OTSCC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BJ and JZ conceived the research idea and designed
the whole experimental plan. BJ, XQ and SX performed the
experiments. BJ and XZ analyzed the in vitro and in
vivo results and discussed the findings with GC, who wrote the
initial draft of the manuscript. XJ, JL and ZH collected and
analyzed the data, revised and finalized the manuscript. GC and SX
provided constructive suggestions for the work and proofread the
language of the manuscript. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The research protocols associated with the
experimental mice were approved by the Ethics Committee of the
Southern Medical University. With approval from the institutional
review board of the Joint Ethics Committee of the Southern Medical
University Health Authority, freshly resected OTSCC tissues (20
pairs) and paraffin-embedded OTSCC sections were obtained from the
archives of the Department of Oral Surgery, Stomatological
Hospital, Southern Medical University (Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was financially supported by the Guangzhou
Science and Technology Program Key Projects (grant no.
201802020018), the China Postdoctoral Science Foundation (grant no.
2019M652979), the Guangdong Science and Technology Program (grant
no. 2019A1515010408), the Program of Stomatologic Hospital,
Southern Medical University, China (grant no. PY2019003) and the
Medical Scientific Research Foundation of Guangdong Province of
China (grant no. B2017103).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Franceschi D, Gupta R, Spiro RH and Shah
JP: Improved survival in the treatment of squamous carcinoma of the
oral tongue. Am J Surg. 166:360–365. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wong WM, Parvathaneni U, Jewell PD,
Martins RG, Futran ND, Laramore GE and Liao JJ: Squamous cell
carcinoma of the oral tongue in a patient with Fanconi anemia
treated with radiotherapy and concurrent cetuximab: A case report
and review of the literature. Head Neck. 35:E292–E298. 2013.
|
|
4
|
Amichetti M: Squamous cell carcinoma of
the oral tongue in patients less than fifteen years of age. Report
of a case and review of the literature. J Craniomaxillofac Surg.
17:75–77. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fakih AR, Rao RS, Borges AM and Patel AR:
Elective versus therapeutic neck dissection in early carcinoma of
the oral tongue. Am J Surg. 158:309–313. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He Q, Chen Z, Dong Q, Zhang L, Chen D,
Patel A, Koya A, Luan X, Cabay RJ, Dai Y, et al: MicroRNA-21
regulates prostaglandin E2 signaling pathway by targeting
15-hydroxyprostaglandin dehydrogenase in tongue squamous cell
carcinoma. BMC Cancer. 16:6852016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
St Laurent G, Wahlestedt C and Kapranov P:
The landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Nie FQ, Wang ZX and De W:
Involvement of lncRNA dysregulation in gastric cancer. Histol
Histopathol. 31:33–39. 2016.
|
|
12
|
Xia P, Gu R, Zhang W and Sun YF: lncRNA
CEBPA-AS1 overexpression inhibits proliferation and migration and
stimulates apoptosis of OS cells via notch signaling. Mol Ther
Nucleic Acids. 19:1470–1481. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao Y, Xiong JB, Zhang GY, Liu Y, Jie ZG
and Li ZR: Long noncoding RNA UCA1 regulates PRL-3 expression by
sponging MicroRNA-495 to promote the progression of gastric cancer.
Mol Ther Nucleic Acids. 19:853–864. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu SJ, Malatesta M, Lien BV, Saha P,
Thombare SS, Hong SJ, Pedraza L, Koontz M, Seo K, Horlbeck MA, et
al: CRISPRi-based radiation modifier screen identifies long
non-coding RNA therapeutic targets in glioma. Genome Biol.
21:832020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren X, Chen C, Luo Y, Liu M, Li Y, Zheng
S, Ye H, Fu Z, Li M, Li Z and Chen R: lncRNA-PLACT1 sustains
activation of NF-κB pathway through a positive feedback loop with
IκBα/E2F1 axis in pancreatic cancer. Mol Cancer. 19:352020.
View Article : Google Scholar
|
|
16
|
Pan J, Fang S, Tian H, Zhou C, Zhao X,
Tian H, He J, Shen W, Meng X, Jin X and Gong Z: lncRNA
JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis
of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer.
19:92020. View Article : Google Scholar
|
|
17
|
Xiong L, Tang Y, Tang J, Liu Z and Wang X:
Downregulation of lncRNA HOTTIP suppresses the proliferation,
migration, and invasion of oral tongue squamous cell carcinoma by
regulation of HMGA2-Mediated Wnt/β-catenin pathway. Cancer Biother
Radiopharm. 35:720–730. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mu M, Li Y, Zhan Y, Li X and Zhang B:
Knockdown of HOXA transcript at the distal tip suppresses the
growth and invasion and induces apoptosis of oral tongue squamous
carcinoma cells. Onco Targets Ther. 11:8033–8044. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar
|
|
20
|
Tang X, Shi L, Xie N, Liu Z, Qian M, Meng
F, Xu Q, Zhou M, Cao X, Zhu WG and Liu B: SIRT7 antagonizes TGF-β
signaling and inhibits breast cancer metastasis. Nat Commun.
8:3182017. View Article : Google Scholar
|
|
21
|
Wei C, Yang C, Wang S, Shi D, Zhang C, Lin
X, Liu Q, Dou R and Xiong B: Crosstalk between cancer cells and
tumor associated macrophages is required for mesenchymal
circulating tumor cell-mediated colorectal cancer metastasis. Mol
Cancer. 18:642019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Wang S, Ke H, Zhang H, Ma Y, Ao L, Zou L,
Yang Q, Zhu H, Nie J, Wu C and Jiao B: lncRNA MIR100HG promotes
cell proliferation in triple-negative breast cancer through triplex
formation with p27 loci. Cell Death Dis. 9:8052018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cui Y, Song Y, Yan S, Cao M, Huang J, Jia
D, Liu Y, Zhang S, Fan W, Cai L, et al: CUEDC1 inhibits
epithelial-mesenchymal transition via the TβRI/Smad signaling
pathway and suppresses tumor progression in non-small cell lung
cancer. Aging (Albany NY). 12:20047–20068. 2020. View Article : Google Scholar :
|
|
25
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Paraskevopoulou MD, Vlachos IS and
Hatzigeorgiou AG: DIANA-TarBase and DIANA suite tools: Studying
experimentally supported microRNA targets. Curr Protoc
Bioinformatics. 55:12.14.1–12.14.18. 2016. View Article : Google Scholar
|
|
27
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Camisasca DR, Silami MA, Honorato J, Dias
FL, de Faria PA and Lourenco Sde Q: Oral squamous cell carcinoma:
Clinicopathological features in patients with and without
recurrence. ORL J Otorhinolaryngol Relat Spec. 73:170–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lindenblatt Rde C, Martinez GL, Silva LE,
Faria PS, Camisasca DR and Lourenco Sde Q: Oral squamous cell
carcinoma grading systems-analysis of the best survival predictor.
J Oral Pathol Med. 41:34–39. 2012. View Article : Google Scholar
|
|
31
|
Chen YH, Chien CY, Fang FM, Huang TL, Su
YY, Luo SD, Huang CC, Lin WC and Li SH: Nox4 overexpression as a
poor prognostic factor in patients with oral tongue squamous cell
carcinoma receiving surgical resection. J Clin Med. 7:4972018.
View Article : Google Scholar
|
|
32
|
Wang W, Li J, Zhang Z, Ma H, Li Q, Yang H,
Li M and Liu L: Genome-wide analysis of acute traumatic spinal cord
injury-related RNA expression profiles and uncovering of a
regulatory axis in spinal fibrotic scars. Cell Prolif.
54:e129512021. View Article : Google Scholar
|
|
33
|
Zhang M, Han Y, Zheng Y, Zhang Y, Zhao X,
Gao Z and Liu X: ZEB1-activated LINC01123 accelerates the
malignancy in lung adenocarcinoma through NOTCH signaling pathway.
Cell Death Dis. 11:9812020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hong W, Ying H, Lin F, Ding R, Wang W and
Zhang M: lncRNA LINC00460 silencing represses EMT in colon cancer
through downregulation of ANXA2 via upregulating miR-433-3p. Mol
Ther Nucleic Acids. 19:1209–1218. 2020. View Article : Google Scholar : PubMed/NCBI
|