1. Introduction
Genitourinary and gynecological cancers are a wide
group of cancers with differences in etiology, rapidity of
progression and treatment strategies (1-7).
Among these, prostate and cervical cancers (CCs) are associated
with high incidence and mortality rates worldwide (8). The common feature of the majority
of defined tumors is a lack of characteristic symptoms in the early
stages, which often leads to a diagnosis of invasive or metastatic
disease and treatment difficulties (1,3,9,10). Therefore, novel prognostic and
predictive clinical and molecular targets for modern drugs are
required to improve the therapeutic process.
The Sonic Hedgehog (SHH) signaling pathway is an
evolutionary conserved molecular cascade discovered by
Nusslein-Volhard and Wieschaus during their studies on D.
melanogaster body segmentation (11). Further research has revealed that
this signaling plays an important role in human embryonic
development, as well as in maintaining the homeostasis of organisms
in postnatal life (12-14). The canonical signaling pathway
includes several proteins involved in signal transmission from the
cell membrane to the nucleus (Fig.
1) (15). The activity of
the pathway is regulated by the SHH signaling ligand, which can
bind to patched 1 (PTCH1) receptor (16). This interaction results in the
translocation of smoothened, frizzled class receptor (SMO)
(17) from the cytoplasm to the
cell membrane in the region of the primary cilium (18). The single non-motile cell
protrusion can be found in almost all cell types. The core of the
primary cilium is composed of nine microtubule doublets, without
central microtubule pairs and dynein arms, which are found in the
motile cilia (19). The ciliary
localization of SMO promotes intracellular signal transmission to
the cytoplasm, protein complex composed of SUFU negative regulator
of hedgehog signaling (SUFU) protein and GLI family zinc finger 2
and 3 (GLI2/3) transcription factors (20). Consequently, SUFU undergoes
proteolytic degradation and GLIs (the SHH pathway effectors)
translocate to the cell nucleus and act as transcription factors
for various target genes involved in cell survival (i.e.,
BCL2), proliferation [cyclin D (CCND1) and MYC
proto-oncogene, bHLH transcription factor (MYC)] (15), epithelial-mesenchymal transition
[snail family transcriptional repressor 1 (Snail)] and
angiogenesis (vascular endothelial growth factor A), or genes that
regulate SHH signaling, such as GLI1 (positive feedback
loop) and PTCH1 (negative feedback loop) (21). The upregulation of SHH pathway
components and, particularly GLI transcription factors, is
frequently associated with the progression of various types of
cancer, including retinoblastoma, breast, colorectal and non-small
cell lung cancer (22,27), acute myeloid leukemia (AML), as
well as basal cell carcinoma (BCC) (28,29). Drugs that inhibit SMO have been
introduced for BCC and AML and tested in other malignancies;
however, since GLI activation may occur in an SMO-independent
manner, drug resistance occurs frequently during treatment
(17,30). To date, no SHH pathway-targeted
drugs have been introduced for the treatment of gynecological or
genitourinary tract cancers, at least to the best of our knowledge.
The present review includes a comprehensive description of SHH
signaling components and their role as potential molecular targets,
which may prove useful for the treatment of genitourinary and
gynecological cancers. The present review also aimed to discuss the
upstream regulation of the SHH pathway, as well as its
correspondence with other cellular pathways, which may support the
introduction of a combination of drugs targeting different
tumor-related pathways.
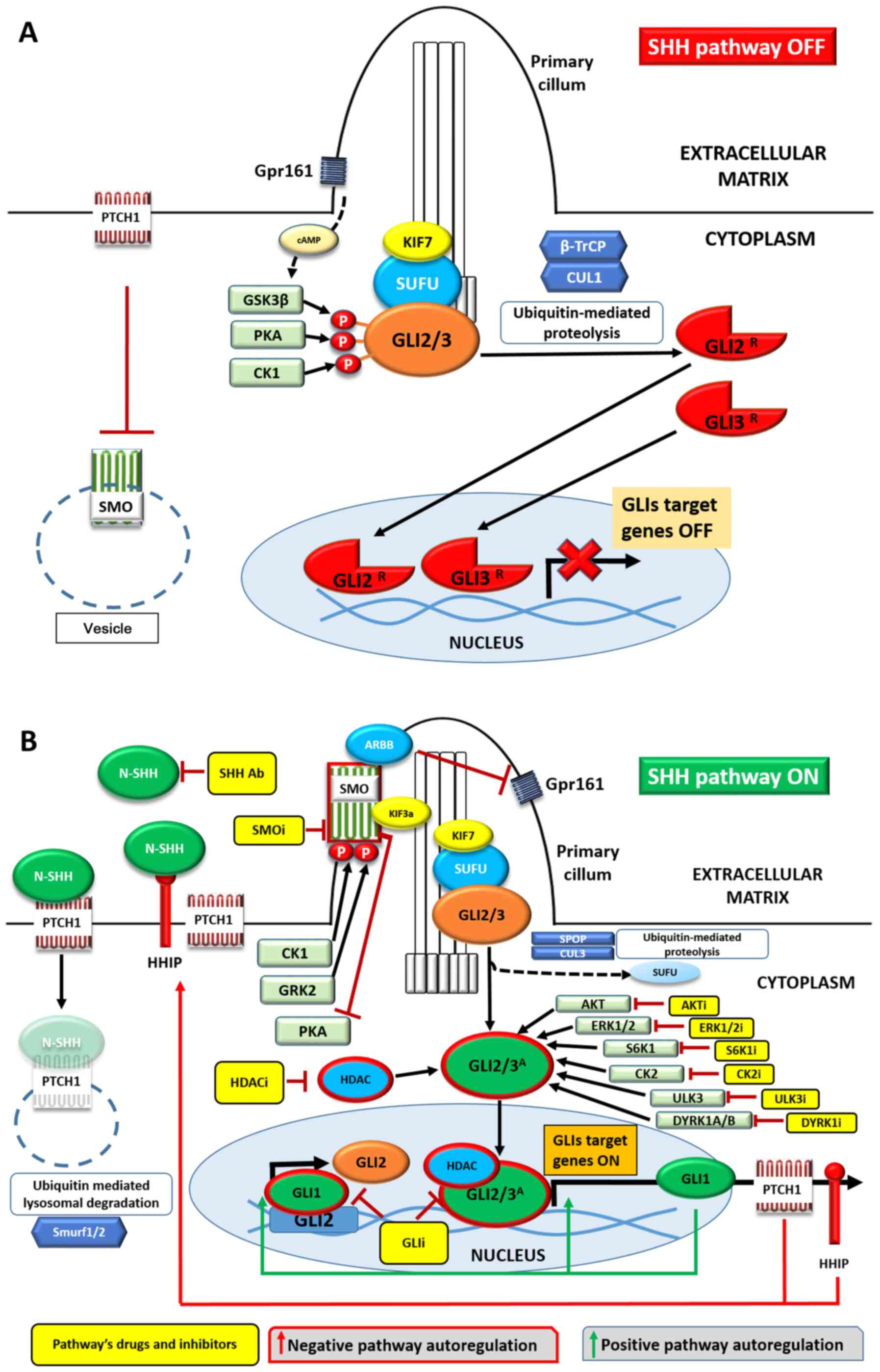 | Figure 1Overview of the SHH pathway in the
(A) absence or (B) presence of the SHH ligand. Negative signaling
regulators are presented in red and positive regulators in green.
Transmembrane proteins are shown as rods or trails, SHH pathway
elements and proteins forming complexes with them as ovals, kinases
as rectangles and proteolytic proteins as hexagons. Yellow
rectangles represent drugs inhibiting/blocking the specific
cellular components. Activated proteins are surrounded by red
borders. See main text for details. Ab, antibody; i, inhibitor;
SHH, Sonic Hedgehog; PITCH1, pitched 1; SMO, smoothened, frizzled
class receptor; Gpr161, G protein-coupled receptor 161; GSK-3β,
glycogen synthase kinase 3β; PKA, protein kinase A; KIF7, kinesin
family member 7 motor protein; CUL1, β-TrCP, β transducin
repeat-containing protein; cullin 1; GRK2, G protein-coupled
receptor kinase 2; HDAC, histone deacetylase. |
2. Mammalian Sonic Hedgehog canonical
pathway
Sonic hedgehog signaling molecule
SHH signaling transfers signals from the
extracellular environment and activates the expression of genes
involved in cell survival and proliferation (28). A schematic presentation of the
pathway is shown in Fig. 1A and
B, and the core elements of the pathway are briefly presented
in Table I.
 | Table IMain components of the Sonic Hedgehog
pathway in mammals. |
Table I
Main components of the Sonic Hedgehog
pathway in mammals.
| Mammalian gene | Protein, full name
(aliases) | Post-translational
protein modifications (Refs.) | Protein function
(Refs.) |
|---|
| SHH | SHH, Sonic Hedgehog
signaling molecule | Autocatalytic
cleavage into C-SHH and N-SHH Addition of cholesterol and palmitic
acid moiety to N-SHH (39-42) | Upstream, positive
regulator of SHH signaling; ligand for PTCH1 receptor (16,20,46) |
| PTCH1 | PTCH, patched 1
(PTC, BCNS, PTC1) | Conformational
changes of protein to enable binding of N-palmitoyled residue of
SHH ligand (48) | Receptor for SHH
protein; negative SHH signaling regulator; suppress the activity of
SMO protein (20,45) |
| SMO | SMO, smoothened,
frizzled class receptor (Gx, CRJS, SMOH) | Phosphorylation by
PKA, GSK3β and CK1 Translocation into primary cilia with ARBB
(18,59) | Atypical G-coupled
receptor; positive, SHH pathway signal carrier (17,55) |
| GLI1 | GLI1, GLI family
zinc finger 1 (GLI, PPD1) | Translocation into
primary cilia (21) Dissociation
from SUFU (21) GLI2 and GLI3
proteolytic truncation | Downstream effector
of SHH signaling; zinc-finger transcriptional activator (20,21,70) |
| GLI2 | GLI2, GLI family
zinc finger 2 (CJS; HPE9) | suppression
(70) Phosphorylation,
ubiquitination, sumoylation, acetylation, deacetylation (70) | Downstream effector
of SHH signaling; zinc-finger transcriptional activator/repressor
(20,21,70) |
| GLI3 | GLI3, GLI family
zinc finger 3 | | Downstream effector
of SHH signaling; zinc-finger transcriptional activator/repressor
(20,21,70) |
SHH signaling is triggered by the cell membrane
binding of the functional SHH glycoprotein. It acts as a classic
morphogen during embryonic development, where it is involved in the
crucial phases, such as patterning of the ventral neural tube, the
anterior-posterior limb axis and ventral somites (20). Germinal mutations of the
SHH gene, located at 7q36.3, lead to congenital defects,
such as holoprosencephaly (31-33). Recent research on genomic DNA of
patients affected by holoprosencephaly has revealed that eight
synonymous single-nucleotide variants in the SHH gene are
associated with a reduced level of SHH protein (34). A recent in vivo study on
Cre-modified mice demonstrated that SHH expression was also
crucial for proper fetal development of the tongue and mandible
(35). SHH is the most
well-known among other hedgehog family proteins, comprising Desert
Hedgehog (DHH) and Indian hedgehog (IHH) molecules (36). Although all hedgehog family
members can bind to the PTCH1 receptor, their tissue distribution
and roles are different (20,37). It has been proven that SHH
protein plays a significant role in central nervous system
development (38). The activity
of IHH in skeletal tissue formation has been reported, whereas DHH
is present only in granulosa cells of ovaries and Sertoli cells of
the testis (20,32). Post-translational modifications
of all three hedgehog protein family members are required for their
attachment to the PTCH1 receptor (15). During this molecular process, the
full-length SHH protein (~45 kDa) undergoes autoproteolysis and
cleavage into the C- (C-SHH; ~25 kDa) and N- (N-SHH; ~19 kDa)
terminal domains (39,40). C-SHH is an auto-processing
molecule that participates in the attachment of cholesterol to the
C-terminal end of N-SHH. Furthermore, the N-terminal end of N-SHH
binds to palmitic acid moiety through the reaction induced by
hedgehog acyltransferase (HHAT), which is necessary for its full
biological activity (41,42).
The activity of HHAT may be blocked by the use of RU-SKI inhibitors
(RU-SKI 41, 43, 101 and 201; not shown in the figures) (43); however, overall cytotoxicity was
observed for RU-SKI 41, 43 and 101 in an in vitro study
(44). Currently, there are no
data available regarding the use of RU-SKI inhibitors in clinical
studies, at least to the best of our knowledge. Finally, through
its interaction with dispatched resistance-nodulation-division
(RND) transporter family member 1, modified N-SHH is secreted to
the extracellular matrix (ECM) and may act as a biologically active
upstream regulator of the SHH pathway (Fig. 1B) (15,45,46). Therefore, the binding of N-SHH
may present another target in cancer drug studies. For that reason,
5E1 antibody against N-SHH (Fig.
1B) was analyzed in a mouse model of pancreatic cancer, and was
found to have a promising effect in the reduction of tumor size and
angiogenesis (47). The final
interaction between N-SHH ligand and PTCH1 may occur in either an
auto- or paracrine way (40),
which will be discussed below in the present review.
PTCH1 protein
The PTCH1 receptor, encoded by the PTCH1 gene
at 9q22.32, is composed of 1,447 amino acids, arranged in 12
transmembrane helices, two extracellular domains (1 and 2) that can
attach extracellular ligand N-SHH and one cytoplasmic
carboxyl-terminal domain (48).
Mutations in the PTCH1 gene lead to an autosomal dominant,
multisystem disorder known as Nevoid basal cell carcinoma syndrome,
also known as the Gorlin-Goltz syndrome (49). The patched protein family also
includes PTCH2 receptor (50).
Although both PTCH1 and PTCH2 can bind to Hedgehog ligands with the
same affinity, PTCH2 appears to have a lower ability to inhibit the
SMO protein (20,45,51). PTCH1 acts as a negative regulator
of the SHH pathway by inhibiting SMO protein from translocating to
the plasma membrane (Fig. 1A).
The mechanism of this inhibition is not yet fully understood;
however, recent studies have suggested the involvement of
cholesterol or another sterol lipid in this regulation (52,53). Following SHH pathway activation,
the blockade of PTCH1 is abolished and the receptor undergoes
internalization, while SMO protein is exposed on the cell surface
in the primary cilium (54).
PTCH1 subsequently undergoes endocytosis, followed by
ubiquitination and lysosomal degradation through E3 ubiquitin
ligase SMAD specific E3 ubiquitin protein ligase 1/2 (Smurf1-2;
Fig. 1B).
Smoothened protein
Smoothened protein belongs to the F-class of the
G-protein-coupled receptor superfamily and is a key intracellular
positive SHH pathway regulator (55,56). Research on SHH signaling in D.
melanogaster has indicated that biochemical processes, such as
phosphorylation or sumoylation, are required to obtain full SMO
activity (57). In mammalian
cells, ciliary localization of this molecule appears to be crucial
for SMO activation, as well as post-translational SMO
modifications, which are analogous to those in Drosophila
cells (58). The ciliary
translocation of SMO occurs following phosphorylation by a
β-adrenergic-receptor kinase (G protein-coupled receptor kinase 2)
and is followed by an interaction between cytosolic β-arrestin
(ARBB) and clathrins (59).
Following ciliary translocation, SMO is further phosphorylated by
casein kinase 1α, and then SMO-β-arrestin complex recruits motor
protein kinesin family member 3A (Kif3A), which consequently
interacts with the kinesin family member 7 motor protein
(KIF7)-SUFU-GLI2/3 ciliary located complex (Fig. 1B) (60). The SMO protein is the main
anticancer treatment target among all SHH pathway proteins
(61). Several SMO inhibitors
are in clinical trials, and three of them (vismodegib, sonidegib
and, in November 2018, glasdegib) have been approved for selected
cancer treatment by the US Food and Drug Administration (FDA)
(61,62). However, this molecular treatment
has certain limitations, including the development of drug
resistance due to frequent SMO mutations, as well as the presence
of alternative SMO-independent mechanisms of GLI transcription
factor activation (63,64), which are discussed in the
following paragraphs.
GLI proteins
Cubitus interruptus has been identified as a
transcription factor of the Hedgehog pathway in D.
melanogaster (41,65). In mammals, this protein has three
analogs (the GLI1, GLI2 and GLI3 molecules), which belong to the
Kruppel zinc-finger transcription factor family (66). The lack of a repressor domain in
GLI1 protein structure suggests that this molecule may act only as
a transcription activator, while GLI2 and GLI3 possess both
repressive (GLI2/3R; Fig.
1A) and activating (GLI2/3A; Fig. 1B) properties (20). Furthermore, several isoforms of
GLI1 and GLI2 (called GLIΔN or tGLI), which are the products of
alternative splicing, have been identified in human tissues
(65). tGLI1 has been detected
only in cancer samples and been associated with aggressive behavior
of the disease (67-69). In the absence of SHH, GLI2/3 are
attached to the SUFU molecule in the ciliary location by KIF7
(Fig. 1A). GLI2/3 underwent
phosphorylation by glycogen synthase kinase (GSK)-3β, protein
kinase A (PKA) and CK1, which is triggered by cyclic AMP produced
by G protein-coupled receptor 161 (Gpr161). Such action causes
proteolytic cleavage of GLIs' C terminus by cullin 1 (CUL1) and β
transducin repeat-containing protein (β-TrCP), leading to the
removal of their transcriptional activation domain. Cleaved GLI2/3,
in the form of GLI2R and GLI3R, translocates
to the nucleus and act as inhibitors/repressors after binding to
regulatory regions of SHH target genes (21,70).
Following the activation of the canonical SHH
pathway, the SMO-β-arrestin complex inhibits Gpr161 and cyclic
adenosine monophosphate-dependent PKA (Fig. 1B) (71), which blocks the phosphorylation
and proteolytic cleavage of GLI2/3. Subsequently, the GLI2/3
KIF7/SUFU/GLI2/3 complex dissociates, and full-length GLIs undergo
several posttranslational modifications, including phosphorylation,
ubiquitination and sumoylation (70), and may simultaneously undergo
proteolysis mediated by CUL3 and speckle-type POZ protein (72). The activity of GLI2A
and GLI3A transcription factors may then be upregulated
by various cytoplasmic factors on their way to the nucleus. There
are several protein kinases [casein kinase II (CK2), protein kinase
B (AKT), extracellular signal-regulated kinase 1/2 (ERK1/2),
ribosomal protein S6 kinase 1 (S6K1), dual specificity tyrosine
phosphorylation regulated kinase 1B (DYRK1B) or unc-51 like kinase
3 (ULK3)] (73), which
phosphorylate GLI2/3A (Fig. 1B), thus promoting GLI
translocation into the nucleus. Acetylation/deacetylation of GLIs
is another important factor that regulates their transcriptional
activity (70). Acetylation of
GLI1/2 by p300/CBP complex prevents GLIs from attaching to DNA and
provides nuclear export through exportin 1 and LAP2 proteins
(70,74). On the contrary, GLI deacetylation
by histone deacetylase HDAC1 enables them to interact with genomic
DNA (75). Of note, HDAC1
is upregulated by GLIs; therefore, the HDAC1-GLIs interaction forms
a positive feedback loop with the SHH pathway (70,75). A significant role of primary
cilium in the functioning of the GLI proteins has been recently
reported. In the absence of the SHH ligand, GLI2/3-SUFU complexes
are transported to the tip of the cilium by kinesin through the
microtubule cytoskeleton, while GLI2 translocation to the cell
nucleus following ligand stimulation occurs through dynein-2
(76). The in vivo
studies by Wong et al (77) and Han et al (78) revealed that the removal of the
Kif3a allele, which is essential for cilia formation, leads
to the inhibition of both BCC and medulloblastoma, respectively.
However, this effect was observed only in lesions overexpressing
the SMO gene, but not the constitutively active GLI2
gene. Therefore, the primary cilium components could be a molecular
target for SMO-dependent neoplasms (77,78).
Due to frequent mutations in the SMO receptor, which
lead to cancer resistance to previously mentioned SMO-targeted
drugs (64), blocking
cytoplasmic/nuclear GLI-activator proteins is one of the recently
identified targets (30,79). It has been found that CK2, DYRK1B
and S6K1 protein kinases, as well as HDAC1, do not require
SMO-dependent activation of the SHH pathway to activate GLIs
(64). This observation provides
reasoning for examining the activity of several potential drugs,
including CIGB-300 and CX-4945 targeting CK2 (80), CCI-779 and RAD001 targeting S6K1
(81), BVD-523 targeting ERK1/2
(82), MK2206 targeting AKT
(83), AZ191 inhibiting DYRK1B
(84) and SU6668 targeting ULK3
(85). It has also been reported
that HDAC1 deacetylation activity is successfully blocked by
4SC-202 (17), with
GLI-DNA-interaction inhibitors (glabrescione B and GANT61) as well
as GLI2 destabilizers (arsenic trioxide and pirfenidone) (Fig. 1B) (17). It is worth noting that the GLI
proteins are also involved in other cancer-related pathways, which
are discussed below in the present review.
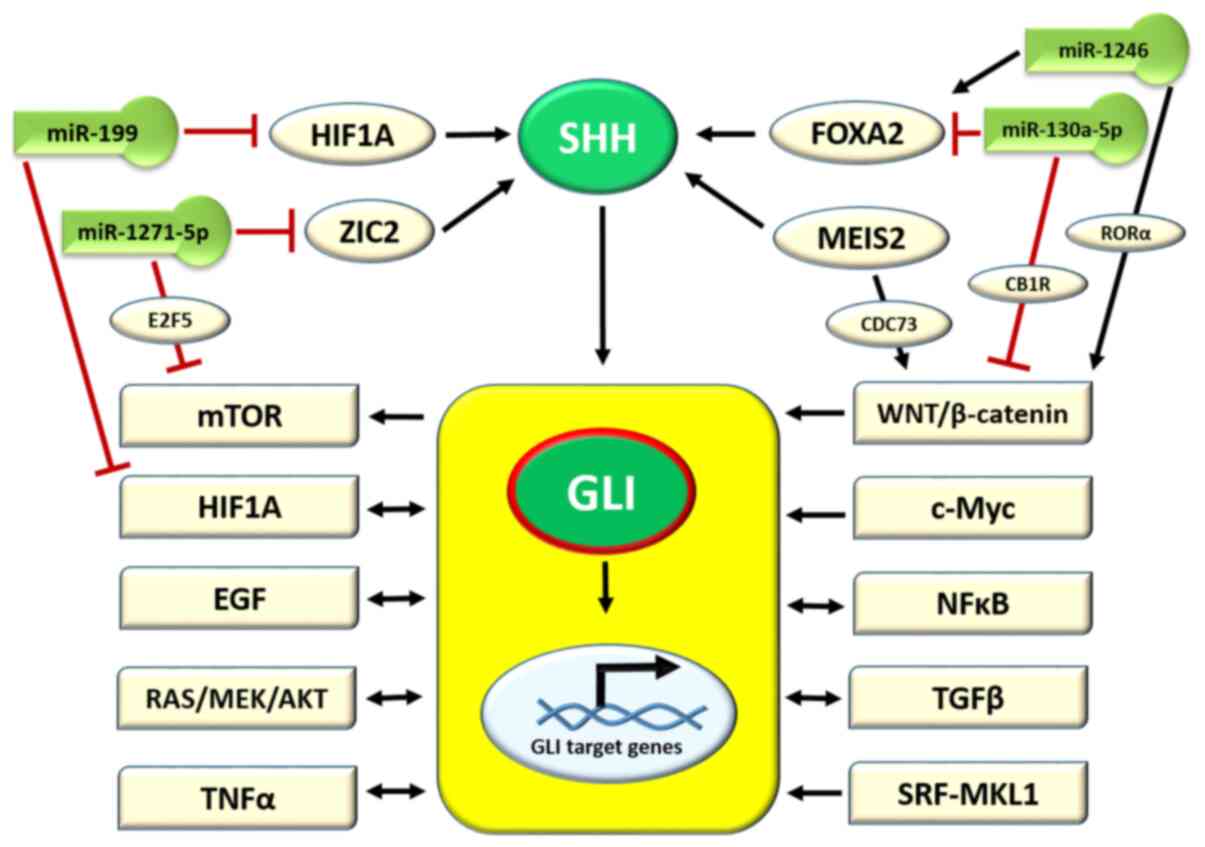
3. Role of microRNAs (miRNAs/miRs) in
upstream SHH gene regulation
The SHH ligand is a major molecule that activates
SHH signaling. It is, therefore, of no surprise, that studies
regarding the upstream regulation of the SHH gene have been
performed to complete the understanding of the role of the SHH
pathway in carcinogenesis. The available data are schematically
presented in Fig. 2. Due to the
important role of SHH signaling in the CNS formation during fetal
life, the majority of studies are based on CNS diseases associated
with SHH pathway alterations. In this regard, Schachter and Krauss
(86) observed, in a mouse model
of holoprosencephaly, that the activation of the SHH gene
was regulated by zinc finger protein 2 (ZIC2). ZIC2 protein belongs
to the zinc-finger transcription factor family, and its deficiency
results in holoprosencephaly 5, as observed by Barratt and Arkell
(87), also in a murine model.
In addition, ZIC2 activity is regulated by the miRNA/miR molecule,
miR-1271-5p, as reported by Chen et al (88) in an in vitro study on AML.
In turn, miR-1271-5p inhibits carcinogenesis in ovarian cancer (OC)
and negatively regulates the mechanistic target of rapamycin kinase
(mTOR) pathway through the E2F5 transcription factor protein
(89). Another study on
SHH gene activation in a CNS murine model revealed the
positive role of forkhead box protein A2 (FOXA2) transcription
factor in this process (Fig. 2),
while the lack of FOXA2 was found to result in a lethal birth
defect known as congenital diaphragmatic hernia (CDH) (90). The FOXA2/SHH axis is also
negatively regulated by the miR-130a-5p molecule, which has been
found to be overexpressed in CDH (90). The progression of gastric cancer
is associated with a decreased expression of miR-130a-5p; in turn,
this deficiency causes an upregulation of Wnt/β-catenin signaling
by targeting cannabinoid receptor 1 (Fig. 2) (91). Other studies on melanoma
progression and hepatocellular carcinoma demonstrated that FOXA2
was activated by the miR-1246 molecule and, in turn, triggered the
Wnt/β-catenin pathway by retinoid-related orphan receptor α nuclear
receptor (Fig. 2) (92,93). A myeloid ecotropic insertion site
2 (MEIS2) transcription factor is another molecule that activates
the expression of the SHH gene for patterning the mandibular
arch during fetal development, as observed by Fabik et al
(35) in a mouse model. The
upregulation of MEIS2 was observed in castration-resistant prostate
cancer (PC) (94) and
hepatocellular carcinoma, where its isoform MEIS2C activates the
Wnt/β-catenin pathway by interacting with the CDC73 molecule
(95). The hypoxia-inducible
factor 1-α (HIF1A) transcription factor is an important molecule
triggered by hypoxia in the cells and tissues of fetal and mature
organisms. It has been observed that HIF1A activates SHH secretion
in the frontonasal ectodermal zone during upper jaw development
(96). Furthermore, the
upregulation of HIF1A and, indirectly, the HIF1A pathway is halted
by the miR-199b molecule (Fig.
2) (96). In conclusion, SHH
secretion is regulated by cellular transcription factors, which in
turn are mostly regulated by miRNA molecules involved in the
regulation of various cellular pathways (Fig. 2).
4. Activation of the target genes of the SHH
pathway via GLI factors and crosstalk with other cellular
pathways
Several dozen target genes of GLI1-3 have been
identified, which are summarized in Table II. Of note, GLI2/3A
stimulates the expression of GLI1, which in turn recognizes
the same DNA motive in target genes (5′-GACCACCCA-3′) as
GLI3A, with GLI2A recognizing an almost
identical sequence (5′-GAACCACCCA-3′) (15). Therefore, the expression of
GLI1 acts as a positive feedback loop for SHH signaling
(Fig. 1B) (41). On the contrary, two genes of the
SHH pathway negative loop are simultaneously activated by GLIs:
PTCH1 (97) and hedgehog
interacting protein (HHIP); once their protein products
reach the plasma membrane, PTCH and HHIP may decrease the rate of
SHH signaling, due to their binding to the extracellular N-SHH
ligand (41) (Fig. 1B). Other genes activated by
GLI1-3 encode proteins that are involved in the processes of cell
proliferation (MYCN proto-oncogene, bHLH transcription factor),
cell cycle regulation (CCND1), angiogenesis (VEGF)
and cell survival (BCL2) (37,98). They are also responsible for the
stimulation of mechanisms strongly associated with tumorigenesis,
such as activating invasion and metastasis [genes encoding matrix
metallopeptidases and transforming growth factor (TGF)-β], cell
immortality maintenance (gene encoding telomerase reverse
transcriptase) or avoiding immune destruction [genes encoding
interleukin (IL)-4 and suppressor of cytokine signaling 1].
Therefore, since the SHH pathway interacts with the molecular
events important for cancer development and progression, it may be
a promising target for anti-tumor therapy (37).
 | Table IISonic Hedgehog signaling target genes
and their impact on cells or the SHH pathway. |
Table II
Sonic Hedgehog signaling target genes
and their impact on cells or the SHH pathway.
| Gene | Protein, full
name | function | (Refs.) |
|---|
| ABCG2 | ABCG2, ATP binding
cassette subfamily G member 2 (Junior blood group) | ABC transporters,
cellular defense mechanism of xenobiotics removal | (197) |
| ALDH1A1 | ALDH1A1, aldehyde
dehydrogenase 1 family member A1 | Metabolism of
alcohol and retinol, stemness of cancer cells | (177,198) |
| BCL2 | BCL2, BCL2
apoptosis regulator | Inhibition of
apoptosis | (199) |
| BIRC5 | baculoviral IAP
repeat containing 5, survivin | Inhibition of
apoptosis | (173) |
| BMP4 | BMP4, bone
morphogenetic protein 4 | Ligand of the TGF-β
superfamily of proteins, regulation of heart and teeth development
and adipogenesis | (200) |
| CCND2 | Cyclin D2 | Cell cycle
inhibition | (37) |
| CD24 | CD24 | Modulation of
growth and differentiation of B cells, neutrophils and neuroblasts;
association with stemness state of cancer stem cells | (201) |
| CDH2 | CDH2,
N-cadherin | Cell adhesion
molecule; development of nervous system and formation of bone and
cartilage; EMT in cancer development | (190) |
| CDK1 | CDK1,
cyclin-dependent kinase 1 | Essential kinase
for G1/S and G2/M phase transitions; cell cycle control | (202) |
| FGF3/4 | FGF3/4, fibroblast
growth factor 3/4 | Mitogenic and cell
survival activities | (200) |
| FOXM1 | FOXM1, Forkhead box
M1 | Transcription
factor; cell proliferation | (181,203) |
| GLI1 | GLI1, GLI family
zinc finger 1 | Positive feedback
of SHH signaling | (28) |
| HDAC1 | HDAC1, histone
deacetylase 1 | Key role in
regulation of gene expression, modulates p53, activates GLIs
forming positive loop | (75) |
| HHIP | HHIP, hedgehog
interacting protein | Decoy for N-SHH
ligand; negative regulator of SHH | (51) |
| JAG1 | JAG1, jagged
canonical Notch ligand 1 | Notch ligand and
Wnt signaling pathway; hematopoiesis | (204) |
| MMP7 | MMP7, matrix
metalloproteinase 7 | Cancer invasion and
angiogenesis by the proteolytic cleavage of ECM and basement
membrane proteins; activated by GLI2 | (175) |
| MYCN | MYCN
proto-oncogene, bHLH transcription factor | Cell proliferation,
neoplastic transformation | (205) |
| NANOG | NANOG, Nanog
homeobox | Transcription
factor involved in embryonic stem (ES) cell proliferation, renewal,
and pluripotency | (17) |
|
PAX6/7/9 | PAX6/7/9, paired
box 6, 7, 9 | Fetal development
of organs: Eye (PAX6), skeletal muscle (PAX7), tooth (PAX9) | (206,207) |
| PTCH1 | PTCH1, patched
1 | Negative regulator
of SHH pathway | (41,97) |
| SNAI1 | SNAI1, snail family
transcriptional repressor 1 | Transcriptional
repressor which downregulates the expression of ectodermal genes
within the mesoderm; EMT in cancer development | (205) |
| SOX2 | SOX2, SRY-box
transcription factor 2 | Transcription
factors involved in the regulation of embryonic development and in
the determination of cell fate | (208) |
| VEGFA | VEGFA, vascular
endothelial growth factor A | Angiogenesis;
induction of proliferation and migration of vascular endothelial
cells | (209) |
GLI-activated genes (Table II) are associated with various
pathways in the cell, which determine the cell's fate and play an
important role in tumorigenesis. As previously mentioned, certain
cellular pathways are regulated by miRNA molecules, which
indirectly act on the SHH pathway through the regulation of the
SHH gene expression. Furthermore, GLIs activate the
expression of genes involved in cellular signaling. However, it has
also been observed that different pathways may upregulate
components of the SHH pathway, and these interactions are
schematically presented in Fig.
2. The hypoxia-induced HIF1A pathway triggers SHH,
SMO and GLI expression, thus influencing cell
stemness and epithelial-to-mesenchymal transition (EMT) in
cholangiocarcinoma (99). On the
contrary, GLI1 is necessary for hypoxia-modulated EMT and
invasiveness of MDA-MB-231 breast cancer cells (100). It was observed in a previous
study that the KRAS proto-oncogene of the MAPK/ERK pathway
increases GLI1 transcriptional activity and the expression of SHH
pathway target genes in gastric cancer (101). The epidermal growth factor
(EGF) pathway is associated with SHH in a complex way: The
simultaneous activation of the SHH/GLI and EGF pathway
synergistically induced oncogenic transformation of human
keratinocytes, an effect that was dependent on the activation of
MAPK/ERK signaling (21). The
influence of AKT protein on PI3K/AKT/mTOR signaling leads to
nuclear translocation, and elevated activity and stability of GLI1
(Fig. 1B) in melanoma (102) and OC cells (103). Moreover, certain studies have
revealed that the main tumor suppressor protein, p53, plays a role
in the inhibition of transcriptional activity, nuclear
translocation, protein stability and the disruption of the DNA
binding ability of GLI1 (63).
The induction of the expression of SNAIL,
proto-oncogene Int-1 homolog and secreted frizzled-related protein
1 by GLIs indicates the impact of SHH on the Wnt/β-catenin pathway
(104). Different analyses of
hair follicle morphogenesis and development have revealed a key
regulation of the NF-κB pathway upon Wnt and SHH signaling
(105). Research on
gastrointestinal stromal tumors has indicated an association
between SHH and PI3K and mitogen-activated protein kinase pathways
(106). The activation of the
c-MYC pathway induces the upregulation of GLI1, while both
10058-F4 and GANT61, c-MYC and GLI1 inhibitors respectively, have
been found to increase apoptosis and reduce the viability of the
Burkitt lymphoma cells (107).
Research on drug-resistant BCC cells has revealed a novel
activation of GLI1 expression triggered by transcription
factor serum response factor together with its co-activator,
megakaryoblastic leukemia 1 (108).
The differential activation of the SHH pathway has
been observed in systemic sclerosis. The enzyme HHAT, which
catalyzes the attachment of palmitate onto the SHH molecule, is
regulated in a TGF-β-dependent manner and, in turn, stimulates
TGF-β-induced long-range hedgehog signaling to promote fibroblast
activation and tissue fibrosis (109). Last but not least, research on
PC3 and DU145 PC cell lines has demonstrated that the tumor
necrosis factor α-triggered mammalian target of rapamycin
(TNFα/mTOR) pathway is connected with GLI activation by S6K1
(Fig. 1B) (110). The list of the complex
associations between SHH and other pathways involved in
tumorigenesis is still growing, suggesting the pivotal role of GLI
modulation in cancer development (21).
5. Non-canonical, GLI-independent activation
of SHH signaling
Previous studies have revealed that the SHH
canonical SHH/PTCH1/SMO/GLI pathway may trigger different cellular
mechanisms without activating GLI transcription factors (20,111). This activity was divided into
two modules: Module 1 included those not demanding SMO protein, and
module 2 those activated by SMO but not requiring GLIs (20,111). However, it should be noted that
other studies merged 'non-canonical SHH activation' with 'GLI
activation' via other (not SHH/PTCH1/SMO) cellular pathways
(63), interactions that were
discussed in the previous section. Both modules are presented in
Fig. 3. According to module 1,
in the absence of the SHH ligand (Fig. 3A), phosphorylated cyclin B1
[active mitosis promoting factor (MPF)] is bound to PTCH1 during
G2/M cell cycle transition, thus decreasing the cellular
proliferation rate, as observed in 293T cells (112). On the contrary, PTCH1-mutant or
SHH-stimulated BCC cells (with wild-type p PTCH1) were
characterized by MPF nuclear translocation and an increased
proliferation rate (Fig. 3A,
right panel) (113). The impact
of PTCH1 activation on apoptosis relies on caspase-3 activity
(Fig. 3A, left panel). In the
absence of SHH, it cleaves C-terminal PTCH1 domain
(Asp1392), thus releasing caspase recruitment domain
family member 8 (CARD8) protein, and the adaptor protein four and a
half LIM domains 2/DRAL (111).
This action activates caspase-9, which in turn speeds up the
formation of this complex by promoting the activation of caspase-3,
leading to caspase-9-dependent apoptosis (114,115). When PTCH1 is inactivated by
SHH-binding, CARD dissociates to protein components without
caspase-9 activation. This leads to a decreased apoptotic ratio
(Fig. 3A, right panel), as
observed in 293T cells and in a chicken embryo model (115).
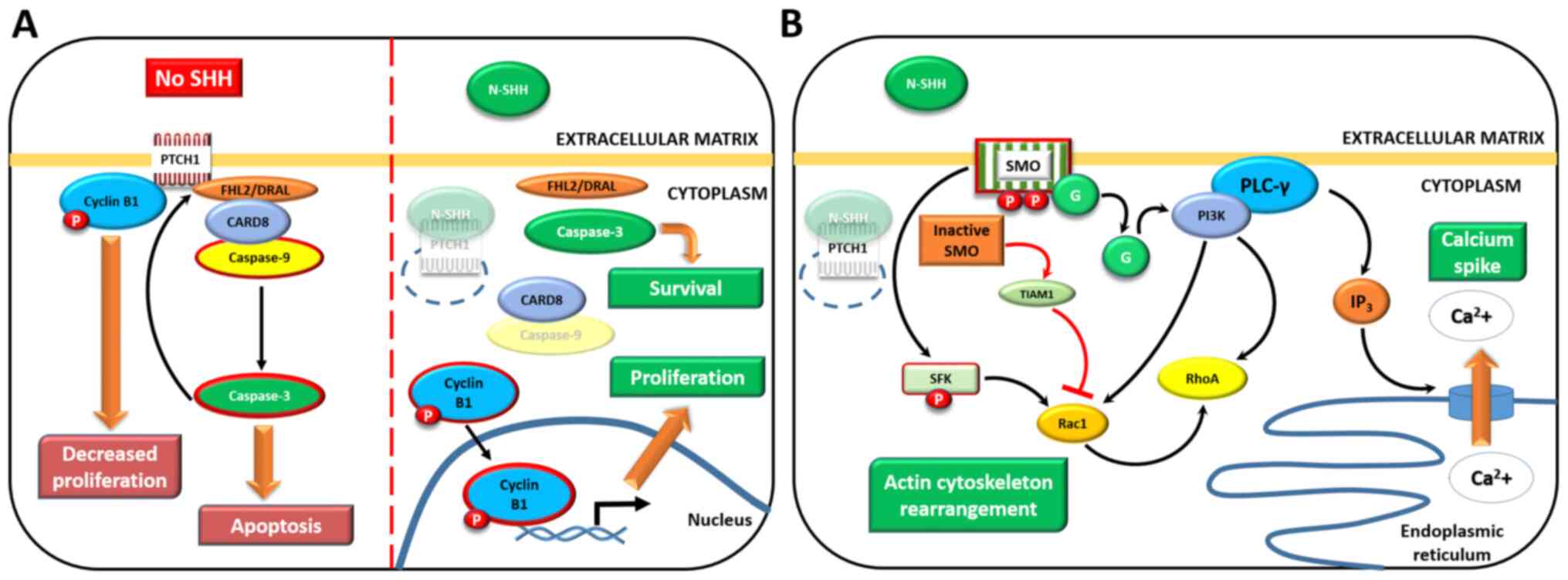 | Figure 3GLI-independent, non-canonical
activation of the SHH pathway. (A) Type 1: SMO-independent
mechanism (left panel, in the absence of the SHH ligand; right
panel, SHH is bound to PTCH1). (B) Type 2: SMO downstream effectors
that do not require GLIs; in the presence of SHH only (with the
exception of SMO-TIAM1 activity, red arrows). Activated proteins
are surrounded by red borders, degraded proteins are partially
transparent and thick brown arrows point at activated mechanisms
(in cropped rectangles). See main text for details. Adapted from a
previous study (112). SHH,
Sonic Hedgehog; GLI, GLI family zinc finger; SMO, smoothened,
frizzled class receptor; PTCH1, patched 1; TIAM1, TIAM Rac1
associated GEF 1. |
Another model of non-canonical SHH activation
involves the SMO protein and its downstream effectors, except GLIs
(Fig. 3B). Phosphorylated SMO
(please see Fig. 1B) uses
Gi proteins to activate PI3K kinase, followed by
Ras-related C3 botulinum toxin substrate 1 (Rac1) and Ras
homologous (Rho) protein activation. Furthermore, Rac1 may be
triggered by SMO by phosphorylated SFK kinase. As part of the
feedback SMO-Rho pathway, inactive, dephosphorylated SMO inhibits
Rac1 through the TIAM Rac1 associated GEF 1 protein (Fig. 3B, red arrows) (111,116). Such pathways give a
considerably faster cellular response than GLI activation, and
result in the rebuilding of the Rho-dependent actin cytoskeleton;
stress fiber formation and tubulogenesis, as observed in
endotheliocytes, result in tumor-dependent angiogenesis (117). SHH-SMO-regulated Rho-dependent
actin cytoskeleton rear-rangement resulting in fibroblast migration
(118) has been found to be
critical to dendrite spine formation in hippocampal and cerebellar
neurons (116). The regulation
of calcium ions significantly affects the proliferation,
differentiation, apoptosis and migration of neuronal and neuronal
precursor cells (111).
SHH-SMO-G protein activation of phospholipase C-γ has been shown to
result in the production of PI3K secondary messenger in Rohon-Beard
embryonic neurons, which opened calcium channels in SER membrane,
thus leading to concentration-dependent Ca2+ transport
from SER to cytosol ('calcium spike'; Fig. 3B) (119). Of note, the latter actions of
the SHH-SMO non-canonical pathway on nervous tissue play a
similarly important role to that of GLI canonical activation during
CNS formation (20,111,116,119).
6. SHH signaling in cancer cells and its
implications for the tumor microenvironment
The different modes of SHH pathway activity in
various neoplasms can be divided into three types, which are shown
in Fig. 4. Type I (Fig. 4A) is caused by activating
mutations in the SMO gene and inactivating mutations in the
PTCH1 or SUFU genes in tumor cells. This leads to the
uncontrolled stimulation of GLI transcription factors and,
ultimately, SHH pathway target genes. Consequently, the cells
acquire the ability to increase the rate of proliferation,
intensify angiogenesis and suppress apoptosis (120). Type I SHH signaling activation
has mainly been observed in BCCs, either in sporadic cases or
hereditary disorders, such as Gorlin-Goltz syndrome (15). A study that included 42 BCC tumor
samples, revealed PTCH1 gene inactivation in 67% cases,
increased SMO gene expression in 10% cases and a SUFU
gene mutation in 5% cases (121). Furthermore, non-epithelial
tumors, such as medulloblastoma and rhabdomyosarcoma, are another
type of neoplasm that may be associated with type I SHH pathway
dysregulation (15). Since this
type of regulation is ligand-independent, targeted SHH therapy
should affect downstream pathway effectors such as GLI
transcription factors (120).
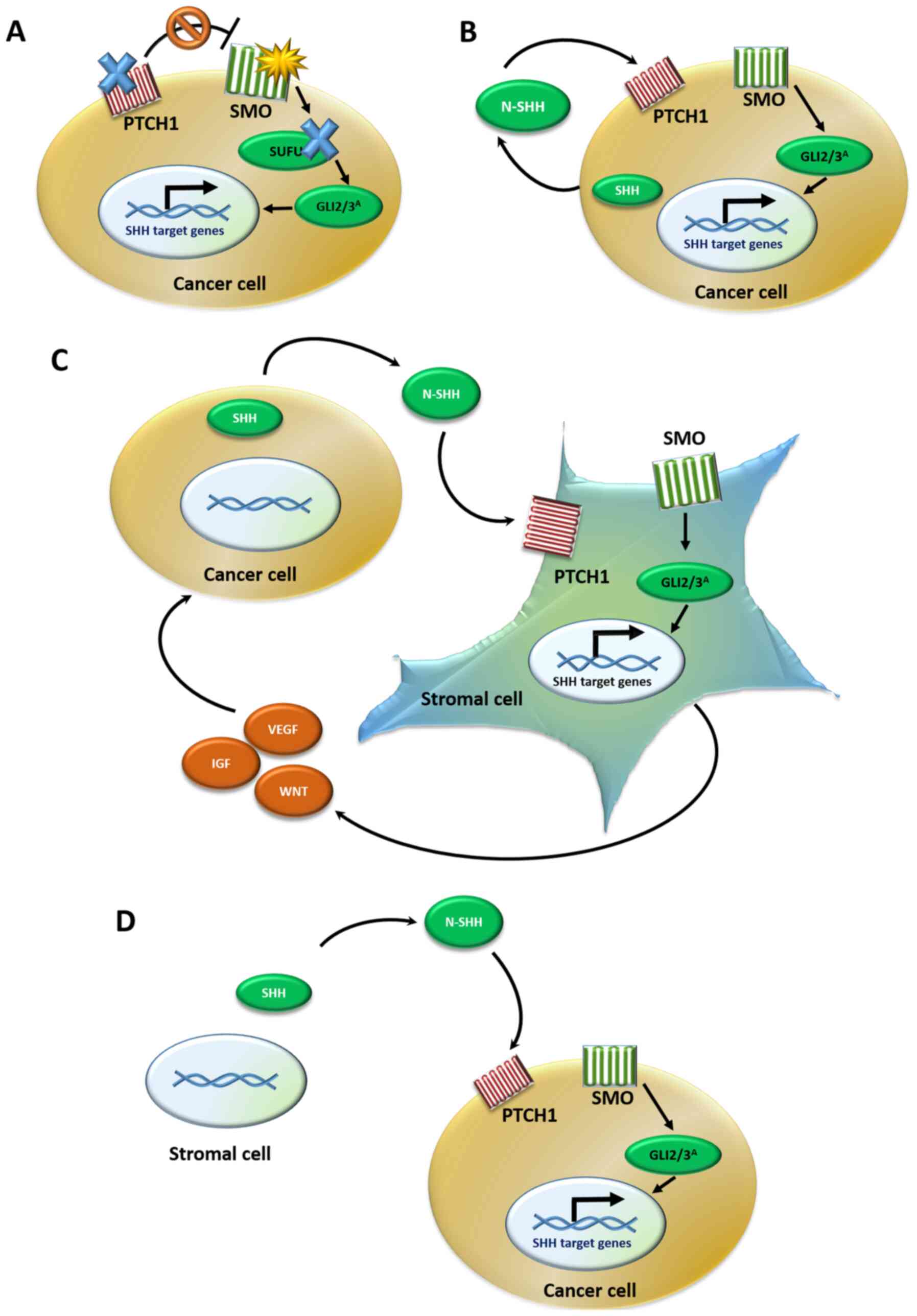 | Figure 4Models of the SHH signaling pathway
in cancer. (A) Type I: Ligand-independent activation occurs due to
either PTCH1 or SUFU inactivating mutations (blue X)
or SMO activating mutations (yellow star), which lead to the
constitutive activation of GLI effectors, even in the absence of
the N-SHH ligand. (B) Type II: Ligand-dependent autocrine
activation. Cancer cells both synthesize and bind to the SHH
ligand, resulting in a positive auto-loop activation of the SHH
pathway. (C) Type III: Ligand-dependent paracrine activation.
Cancer cells secrete the SHH ligand, which is bound by the stromal
cells leading to SHH pathway activation in the stroma. The stroma
reacts by secreting back various cancer-stimulating signals, such
as growth factors to the tumor tissue. (D) Type IIIb: Reverse
paracrine activation. Cancer cells receive SHH ligand secreted from
the stroma, leading to SHH signaling activation in the tumor cells
and upregulation of survival signals. SHH, Sonic hedgehog; PTCH1,
patched 1; SUFU, SUFU negative regulator of hedgehog signaling;
SMO, smoothened, frizzled class receptor; GLI, GLI family zinc
finger. |
In type II SHH signaling activation (Fig. 4B), the SHH (or IHH) ligand is
exposed on the cancer cell surface and may act on the adjacent
cancer cells in either an autocrine or juxtacrine manner.
Consequently, the SHH pathway becomes reactivated in target tumor
cells, and the final effects are the same as those in type I, since
they result in cancer development and progression (120). Type II SHH signaling activation
in cancer is characterized by the overexpression of SHH components
at the mRNA level in cancer cells (but not in stromal cells), as
found in four hepatoma cell lines, using the reverse transcription
PCR method. Moreover, the immunoreactivity of SHH, PTCH1 and GLI2
proteins was significantly elevated in human hepatocellular
carcinoma samples derived from 57 patients, compared to
non-cancerous liver tissues (122).
Paracrine, ligand-dependent signaling between tumor
and surrounding stromal cells is involved in type III
cancer-related SHH alterations (Fig.
4C). The SHH protein can be secreted in excess by cancer cells
into the tumor stroma, which leads to the activation of SHH
signaling in stromal cells. In response, stromal cells release
various SHH signaling target proteins to their microenvironment,
which stimulate tumor growth and progression. Furthermore, a
reverse paracrine type III mechanism has been observed, in which
the PTCH1 receptor on cancer cells binds to the SHH ligand, that is
synthesized by stromal cells, which also increases cancer cell
viability (15,120). This type of regulation
(Fig. 4D) was observed in a
pancreatic ductal adenocarcinoma mouse model (123) and human pancreatic and
metastatic cancer specimens (123). The expression of SHH and
IHH was elevated in tumor cells; however, stromal
GLI1 mRNA levels were found to be 13-150-fold higher than
those in cancer cells, suggesting a paracrine SHH signaling
activation in stromal cells (123). In addition, the association
between SHH and GLI1 mRNA levels has been found in
stromal cells, but not in tumor cells derived from 22 samples of
primary human tumor colorectal adenocarcinoma xenografts (124).
With regards to types II and III SHH pathway
activity in tumor tissues, therapy including both anti-SHH ligand
molecules, such as an anti-SHH antibody, and SMO and GLI protein
inhibitors, may be effective (120). As described above with regards
to the activation of GLIs, the existence of both canonical and
non-canonical SHH pathways should always be considered in studies
on potential SHH pathway-targeted treatments. For certain types of
neoplasms, combination therapy, such as treatment with an SHH
signaling inhibitor and an inhibitor of another signaling pathway,
may be effective. For example, an ongoing clinical phase II trial
is evaluating the combination of sonidegib (SMO inhibitor) with
buparlisib (PI3K inhibitor) in patients with locally advanced or
metastatic BCC (18).
Among the stromal cells of the tumor
microenvironment involved in the type III mechanism of SHH
signaling in cancer tissue, cancer-associated fibroblasts (CAFs)
appear to play an important role (125). CAFs resemble myofibroblasts in
terms of morphology and molecular features. They can originate from
different cell types, such as resident fibroblasts, mesenchymal
stem cells or epithelial cells, resulting in a significant CAF
heterogeneity. The signals for CAFs activation may be derived both
from factors secreted by cancer cells, such as TGF-β1 and IL-6, as
well as physical properties of the tumor micro-environment,
including hypoxia and ECM stiffness (126). There have been reports on SHH
pathway paracrine stimulation in CAFs, either by tumor cells
(17) or cancer stem cells
(CSCs) (127). Subsequently,
CAFs are stimulated to secrete molecules that promote
VEGF-dependent tumor angiogenesis and self-renewal in CSCs
(17,127). The association between SHH
signaling and CAFs was observed in pancreatic ductal adenocarcinoma
(128) and mammary gland tumors
(127).
Other cells of the tumor microenvironment that can
be indirectly affected by the SHH pathway are tumor-associated
macrophages (TAMs) (125).
Although the role of TAMs in tumor development is still not
well-described, certain studies have suggested that the cellular
PTCH1/SMO/SUFU/GLI1-3 cascade not only elevates TAM infiltration
within the tumor stroma, but also promotes the acquisition of the
anti-inflammatory M2 phenotype responsible for tumor tissue
avoidance of immune destruction (129). The proposed mechanism
responsible for recruiting TAMs to the neoplastic niche includes
SHH-ligand-driven CAFs, which secrete molecules, such as
granulocyte-macrophage colony-stimulating factor, C-C motif
chemokine ligand (CCL)2, CCL5 and C-X-C motif chemokine ligand 12.
Consequently, the number of cells with immunosuppressive
properties, including M2 phenotype-TAMs, myeloid-derived suppressor
cells and regulatory T cells, increase, which leads to a reduction
in immune effector cell infiltration (17). The significant role of TAMs in
the tumorigenesis of BCC (130)
and the subgroup of medulloblastomas with upregulated SHH signaling
has been reported (131). The
association between TAMs and SHH pathways, as well as their impact
on cancer-related immunosuppression, may lead to the discovery of
novel cancer immunotherapeutic strategies (131).
7. Sonic Hedgehog signaling in cancers of
the urinary tract
Kidney cancer
Kidney cancers, otherwise known as renal cell
cancers (RCCs), are a group of histologically different tumors
(132), which rank 14th in
incidence among other neoplasms worldwide (8). Clear cell RCC (ccRCC) is the most
common subtype (6) and is
associated with unfavorable outcomes (133). ccRCC development is strongly
associated with the inactivation of the von Hippel-Lindau tumor
suppressor (VHL) gene, which can be hereditary (VHL
syndrome) or occurs spontaneously during life (10,134-136). Other alterations in genes such
as PBRM1 or mTOR have been identified; however, no
specific prognostic or predictive molecular markers of RCC can be
recommended for clinical use (6,10). RCC therapy includes surgical and
pharmacological treatment in the advanced stages of the disease,
including tyrosine kinase inhibitors (TKIs) with sunitinib, the
first such drugs to be introduced, and mTOR kinase inhibitors
(everolimus) and several others, introduced into clinical treatment
over the past decade (6,137).
The first report regarding the expression of SHH
pathway components in ccRCC was published in 2009 (138). Dormoy et al (138) found that the SHH signaling
genes were expressed at the mRNA level in various RCC cell lines,
independently of VHL gene status. In that study the
overexpression of SMO and GLI1 mRNAs was also
revealed by RT-qPCR in RCC tumor tissues, compared with
corresponding normal kidney samples in the group of 8 patients.
Furthermore, incubation with cyclopamine (SMO inhibitor) decreased
ccRCC cell proliferation and increased apoptosis, as well as
induced the regression of ccRCC tumors in nude mice (138).
Further studies conducted on human RCC samples have
demonstrated the association between SHH signaling and cancer
progression. A study on 140 ccRCC specimens derived from patients
with non-metastatic disease revealed a significantly elevated DHH,
SHH, PTCH1 and GLI3 protein immunoreactivity in samples assessed as
G3 or G4 in Fuhrman's grading system [grades 3 and 4 in
International Society of Urologic Pathologists (ISUP) grading
(139)] than in those with
grade G1 or G2 (ISUP grades 1 and 2) (140). An elevated immunoreactivity of
the GLI2 transcription factor was found to be associated with a
poor prognosis in a group of 39 patients with metastatic ccRCC
treated with sunitinib (141).
In addition, in vitro experiments revealed a decrease in the
GLI2 protein level by western blot analysis in ACHN cells treated
with sunitinib, but not in sunitinib-resistant ACHN cells.
Therefore, these results suggested that GLI2 protein may be
involved in the mechanism of drug resistance associated with TKI
inhibitors in RCC (141).
Behnsawy et al (142)
demonstrated an association between the activity of SHH signaling
and EMT, an important step of cancer progression, in RCC cell
lines. The recombinant SHH ligand (r-SHH) not only significantly
increased proliferation in RenCa and ACHN cells, but also reduced
the mRNA level of E-cadherin, the epithelial marker of EMT,
suggesting a stimulating role of the SHH pathway in EMT (142).
Since several studies have reported a SHH
gene upregulation in RCC (140,142,143), the present review focused on
the occurrence of the upstream protein and miRNA regulators of SHH
expression in RCC. Shang et al (144) analyzed the mRNA expression
rates of the ZIC2 gene in 533 ccRCC and 72 normal kidney
samples (TCGA database), and found that the overexpression of
ZIC2 mRNA was associated with age, TNM, histological grade
and a shorter overall survival; thus, this gene can therefore be
used as an independent prognostic factor in ccRCC. Jia et al
(145) also analyzed TCGA data
from 525 patients with ccRCC focusing on SHH-associated FOX family
genes, and it was found that FOXA2 mRNA overexpression was
associated with poor outcomes.
As previously mentioned, RCC initiation is strongly
associated with the VHL gene status, which is inactivated in
a broad range of ccRCC cases. The gene encoding VHL protein, which
acts as an E3 ubiquitin ligase, is the enzyme responsible for
hypoxia-inducible factor (HIF1α and HIF2α) degradation under
normoxic conditions (136).
Therefore, Zhou et al (146) investigated the expression of
SHH pathway genes in normoxia and hypoxia, as well as the
association between the SHH signaling components and HIF2α. The
mRNA expression of all SHH signaling genes was significantly
elevated in RCC cell lines that were cultured under hypoxia,
compared with normoxic control RCC cells. Of note, the
re-activation of the SHH pathway under hypoxic conditions was
independent of VHL expression, with the dual inhibition of
HIF2α and GLI1 activity. Furthermore, the treatment with sh-HIF2a
and GLI1 inhibitor GANT61 significantly sensitized RCC cells to
ionic radiation. These results demonstrated that the SHH pathway
together with HIF2α protein may be involved in the molecular
mechanisms of RCC radioresistance. In addition, the SMO inhibitor,
cyclopamine, was not found to reduce the observed overexpression of
GLI1 under hypoxic conditions, which suggested that
GLI1 expression in RCC cells does not depend on upstream SHH
signaling components, but could be induced by different molecular
signaling (non-canonical activation) (146). Further evidence provided by
Zhou et al (147)
confirmed this conclusion and demonstrated an involvement of the
PI3K/AKT cascade on the main effectors of the SHH pathway in RCC
cells. PI3K/AKT signaling stimulation or inhibition induced or
decreased the expression of GLI1 and GLI2,
respectively. It was also demonstrated in vitro and in
vivo that the combination of GANT61 with the AKT specific
inhibitor, perifosine, was associated with a significantly enhanced
therapeutic potential, compared with that of the use of each
substance alone (147).
The efficacy of several other SHH inhibitors on
kidney cancer treatment has been under investigation over the past
few years. Erismodegib, a SMO antagonist, was previously shown to
inhibit the survival of the human 786-O RCC line, either alone or,
more effectively, in combination with sunitinib and everolimus
(148). This antitumor effect
was also observed in sunitinib-resistant RCC cells (786-O SuR
cells), revealing a novel research direction for RCC therapy. It
was also observed that erismodegib combined with sunitinib or
everolimus decreased the tumor volume and increased the survival of
nude mice with 786-O SuR cell-derived tumor xenografts, confirming
previously described results. However, unlike erismodegib, GANT61
had no inhibitory effect on RCC cells (148), indicating that SMO is a more
promising selective RCC therapy target than GLI transcription
factors.
In a previous study by the authors (143), the expression of SHH pathway
components in 37 ccRCC tissue samples, a significant correlation
was identified among the expression of almost all SHH signaling
genes at the mRNA level. Although the mRNA level of SHH,
SMO and GLI1 was increased in ccRCC samples, compared
to the morphologically unaltered kidney tissues, no association was
observed between the expression rates of genes and the pathological
features of patients. However, at the protein level, western blot
analysis of SHH revealed a significant increase of full-length SHH
and a decrease of the C-SHH domain in ccRCC tissues (143). This novel observation may
suggest an involvement of the SHH ligand in ccRCC development, and
indicate changes in the post-translational modification of this
protein during tumor progression.
Bladder cancer
According to the GLOBOCAN database, there were
549,393 new cases of bladder cancer in 2018, which renders this
type of cancer as the 11th most common type of cancer worldwide
(149). Approximately 90% of
bladder carcinomas are derived from the transitional epithelium
(1). Several studies have proven
the significance of nicotine and industrial gases in the
pathogenesis of this type of cancer (150,151). The disease risk assessment is
performed using clinical patient examination, medical imaging and
microscopic examination of the resected tumor tissues. The bladder
cancer guidelines recommend the tumor-node-metastasis (TNM) system
as an appropriate classification system for tumor staging. The
treatment of bladder cancer includes transurethral resection of the
bladder tumor for initial bladder neoplasms; however, for more
advanced tumors, radical cystectomy with lymphadenectomy and
additional radio- or chemotherapy are required (1). Genetic and epigenetic alterations
of bladder cancer cells, which may be useful prognostic factors or
targets for personalized therapy, are under investigation. The
molecular profile of non-muscle invasive bladder cancer (NMIBC)
differs significantly from muscle invasive bladder cancer (MIBC).
In addition, genetic alterations characteristic of low-grade NMIBC,
such as fibroblast growth factor receptor 3 (FGFR3) or
RAS mutations, can be distinguished among NMIBCs.
FGFR1, FGFR3, PNEN, CCND1 or MDM2
proto-oncogene genes have been identified as potential therapeutic
targets, whereas TSC complex subunit 1 or
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
alpha (PIK3CA) mutations may be predictive targets for mTOR
or PIK3CA/mTOR inhibitors, respectively (1,152).
In 2012, He et al (153) performed immunohistochemistry
(IHC) on 118 human bladder cancer samples. The expression of
proteins encoded by the SHH, PTCH1 and GLI1
genes was significantly elevated in tumor tissues, compared to 30
adjacent normal bladder tissues. The increased immunore-activity of
SHH pathway proteins was observed in samples derived from patients
with a high pathological stage, the presence of venous invasion and
lymph node metastasis. Patients with a positive SHH,
PTCH1 and GLI1 expression also exhibited poorer
disease-free survival rates, according to Kaplan-Meier analysis
(153). Further studies
suggested the prognostic value of the SHH pathway protein level in
bladder cancer. Nedjadi et al (154) revealed that a high SHH protein
immunoreactivity in urothelial bladder cancer tissues was
associated with the presence of lymph node metastasis; however, no
association was identified between SHH expression and other
clinicopathological parameters or patient survival. SHH
overexpression can be associated with the upregulation of MEIS2 (an
upstream SHH gene regulator) in bladder cancer lymph node
metastasis, as observed by Xie et al (155) in a clinical study on 104
patients with bladder cancer.
The significant role of SHH pathway proteins was
observed in the EMT of bladder cancer cells. An HTB-9 transitional
bladder cancer cell line, with acquired mesenchymal features due to
TGFβ1 stimulation (T-HTB-9), exhibited an overexpression of the
SHH and GLI2 genes at the mRNA and protein levels.
Furthermore, following incubation with cyclopamine, and GDC-0449,
SMO and GLI1-3 inhibitors, a decrease in the migration, invasion
and clonogenicity of T-HTB-9 cells was observed (156). This evidence suggested that
inhibitors of the SHH pathway may effectively decrease bladder
cancer invasive potential and may thus prove to be useful to
bladder cancer treatment. Islam et al (156) examined 22 specimens derived
from human bladder cancer. An elevated immunoreactivity of SHH,
GLI2, Ki-67 proliferation marker and N-cadherin (mesenchymal cell
marker), and a decrease in E-cadherin (epithelial cell marker),
were observed in high-grade tumors compared with low-grade tumors,
further confirming the participation of SHH signaling proteins in
the EMT of human bladder cancer cells (156). Another analysis concerning the
association between EMT and the SHH pathway was performed on
muscle-invasive T24 and 5637 bladder cancer and non-muscle-invasive
KK47 cell lines. The incubation of the cells with recombinant SHH
protein decreased the expression of E-cadherin and enhanced that of
N-cadherin and vimentin in all three cell lines. Cyclopamine was
found to inhibit cell proliferation and invasiveness; however, the
effect was more pronounced in T24 and 5637 cell lines. In
vivo studies on nude mice with induced bladder cancer revealed
a significant inhibition of muscle-invasive-derived tumor
development, which indicated the potential benefits of using SHH
pathway-targeted therapy in advanced stages of bladder cancers
(157). Of note, Kim et
al (158) found that the
CpG hypermethylation-induced decrease in SHH gene expression
in bladder cancer cells led to an increase in tumor invasiveness.
The lack of SHH ligand decreased the activity of SHH signaling in
stromal cells, inhibiting the expression of bone morphogenetic
proteins and ultimately stimulating bladder cancer progression
(158). Furthermore, the
pharmacological inhibition of DNA methylation inhibited the
initiation of invasive urothelial carcinoma at the premalignant
stage of progression, through the increase in SHH expression in
cancer cells (158). These
findings were not consistent with previously presented results;
thus, further research on the cell-to-cell interactions between
bladder cancer and stromal cells in bladder tumors would improve
the understanding of the molecular basis of the role of the SHH
pathway in bladder cancer (158).
8. SHH pathway in gynecological cancers
Cancers of the female reproductive tract include
OC, CC and fallopian tube, uterine, vaginal and vulvar cancers, as
well as gestational trophoblastic neoplasms, according to the AJCC
Cancer Staging Manual, 8th Edition (159). The involvement of the SHH
pathway in the latter has barely been studied since its discovery
(160). Ho et al
(160) focused on the
expression of Kif7 motor protein and GLI1-3 transcription factors,
and reported a strong downregulation of the GLI1-3 genes at
the mRNA level in 4 choriocarcinomas, as well as 50 hydatidiform
moles, compared with 19 normal placentas. Although it was proven in
that study that the overexpression of Kif7 in the
choriocarcinoma cell lines, JAR and JEG-3, suppressed cell
migration, the role of SHH in the development of gestational
trophoblastic neoplasms remains unclear (160). Furthermore, only one study
focused on SHH expression in vulvar squamous cell carcinoma (VSCC);
Yap et al (161)
performed semi-quantitative IHC of tissue specimens from 91 VSCC
cases for SHH, PTCH1 and GLI1 proteins. Although an increased
immunoreactivity of one or more of the assessed proteins was
reported, only the decreased expression of PTCH1 was
associated with an increased risk of developing a local disease
recurrence (161).
OC
OC ranks 8th in incidence and mortality among
cancers affecting women (18th and 14th in both sexes, respectively)
worldwide, with almost 300,000 cases and 185,000 deaths in 2018,
according to the GLOBOCAN data (8). Epithelial OC accounts for >90%
of all ovarian malignancies and is classified into five
histological subtypes: Serous, mucinous, endometrioid,
undifferentiated and clear cell subtypes (162), while OC advancement is based on
the International Federation of Gynecology and Obstetrics (FIGO)
staging. Molecular patterns of SHH upstream regulators in OC have
only been analyzed by a few studies.
One of the first studies on SHH pathway components
in OC was conducted by Levanat et al in 2004 (163). Although an upregulation of
GL1 mRNA expression was not observed in a group of 11
ovarian fibromas and 15 ovarian dermoids, higher mRNA levels of
SMO and SHH were observed. A frequent mutation of the
PTCH1 gene was also identified in the majority of ovarian
fibromas, but it was not found to be associated with the expression
level of this gene (163).
Marchini et al (164)
observed the overexpression of ZIC2 in the malignant form of
epithelial OC (n=193), compared to low-malignant potential OCs
(n=39). In OC cell lines, ZIC2 overexpression was found to
increase the growth rate and foci formation of NIH3T3 cells and
stimulate anchorage-independent colony formation (164). The data on FOXA2
expression in OC are inconclusive: Salem et al (165) found that its lower mRNA levels
promoted OC tumorigenesis, while Peng et al (166) reported high FOXA2 levels in
OCs. Loss of heterozygosity of the PTCH1 gene was a frequent
observation in OC (167,168),
suggesting that the mechanism of SHH pathway activation in OC is
type I. Moreover, the studies regarding somatic mutations in SHH
signaling components counted 14% frequency in a MyPathway study
(169).
Further studies identified the association between
SHH signaling and OC progression; Liao et al (170) observed the overexpression of
SHH and patched proteins (assessed by IHC in 80 patients with OC)
and GLI1 mRNA (quantified by qPCR in 37 OCs) in tumor
specimens, whereas no changes were observed in ovarian tissue. In
addition, the observed molecular alterations were associated with
the poorer outcome of OC patients. Liao et al (170) also performed a GLI1 ectopic
expression experiment on SKOV3 and OVCAR3 OC cell lines and
reported the upregulation of tumorigenesis-related genes (i.e.,
BCL2, VEGF and genes encoding vimentin and
E-cadherin). The incubation of SKOV3, OVCAR3 and OVCA433 cells with
KAAD-cyclopamine, an inhibitor of SMO protein, suppressed cancer
cell viability, induced apoptosis, and decreased the expression of
the aforementioned cancer-related genes (170). However, contrasting results
were obtained by Yang et al (171), who did not report higher levels
of SHH pathway components nor the target genes in 34 OC tumor
samples. Based on the SHH, PTCH1, GLI1,
HHIP, SMO and SUFU mRNA semi-quantification
results (assessed by PCR and qPCR for GLI1), as well as
patched 1, GLI1 and HHIP proteins (assessed by IHC), the results of
that study suggested infrequent involvement of the SHH pathway in
OC development (171). In a
study by Schmid et al (172), inconclusive results of the
expression of SHH signaling and target genes in OC were obtained.
In a group of 16 FIGO stage III serous tumors, various expression
levels of SHH genes (GLI1/2, PTCH1, SHH and
SMO; assessed by qPCR) were observed, while IHH and
PTCH2 genes were upregulated in the majority of cases
(172).
More recent data have confirmed, however, the
impact of the SHH pathway in the progression of OC; Ozretić et
al (97) analyzed SHH
pathway genes in 23 OCs, including 16 carcinomas (CA) and 7
atypical proliferative (borderline) tumors. However, higher mRNA
levels of GLI1 and SUFU were observed in OCs, and
SUFU levels were found to decrease with increasing FIGO
stages. Moreover, a strong positive correlation was observed
between the SMO and GLI1 mRNA levels. In the primary
culture of tumor cells obtained from a high-grade ovarian tumor
sample (FIGO IIIC), cyclopamine exerted an inhibitory effect on
cell proliferation, but only in the first 24 h, whereas GANT61
decreased the proliferation rates of both primary and SKOV-3 cell
lines after 72 h (97).
Furthermore, GANT61, unlike cyclopamine, led to the downregulation
of GLI2 transcription factor in the cells at the molecular level,
rendering it a more effective SHH signaling inhibitor in OC
treatment (97).
Recent studies have highlighted the importance of
GLI-regulated anti-apoptotic protein survivin (BIRC5) (97,173,174) and matrix metalloproteinase
(MMP)-7 (175) as putative
markers for OC progression. Zhang et al (175) reported a high immunoreactivity
of MMP-7 and GLI2 in tumor tissues from 95 OC patients, and the
high expression of MMP-7 protein was found to be associated with
poor patient outcomes. The association between the SHH pathway and
MMP-7 expression was proven by demonstrating that ectopic
stimulation of SHH in an SK-OV-3 OC cell line increased
MMP-7 expression (175).
BIRC5 is an anti-apoptotic protein that acts as a
negative regulatory protein that prevents apoptotic cell death; the
gene is highly expressed during fetal development and in cancer
tissues (176). Trnski et
al (173) and Vlčková et
al (174) analyzed the
association between BIRC5 gene activation and the SHH
pathway. The first team worked on A549 and the other experimented
on SKOV-3 OC cell lines. Based on BIRC5 promoter inactivation by
GANT61 rather than cyclopamine, Vlčková et al (174) proved that BIRC5 was regulated
by the GLI2 transcription factor. Trnski et al (173) further revealed, by the addition
of the GLI1 activator, that GLI3 was not associated with survivin
expression.
Recently, the associations between the SHH pathway
and CSC have been studied in high-grade serous OC (HGSOC) (177). Sneha et al (177) analyzed the effects of SHH
pathway inhibitors on cell viability and spheroid formation through
primary cultures of tumor cells from HGSOC and in nine OC cell
lines. The treatment of cells with SHH inhibitors reduced the
formation of spheroids with the higher efficacy of GANT61, compared
with LDE225 (sonidegib) and salinomycin. In a xenograft model, the
formation of tumors with an OVCAR3 origin was inhibited by GANT61
treatment. It was also found that the stemness marker, ALDH1A1, was
at least partially dependent on the SHH pathway (177). The association between ALDH1A1
and the SHH pathway through the inhibition of GLIs was also
observed in bladder (178) and
breast (179) cancer. In
conclusion, data have demonstrated that the SHH pathway plays an
important role in OC development with GLI1/2 downstream effectors
as the key points.
CC
The worldwide incidence and mortality numbers of CC
in 2018 were approximately 590,000 and 311,000, respectively, with
CC ranking fourth in both categories among other malignancies
(8). Although it is known that
the pathogenesis and progression of CC are associated with human
papillomavirus (HPV) infection, the involvement of the SHH pathway
has also been described. The study by Rojo-León et al
focused on the impact of HPV E6/E7 oncogenes on the SHH pathway in
transgenic mice that carry eight GLI1-binding sites bound to the
firefly luciferase gene (180).
An increased GLI1 expression was observed in the cervix and
skin either after exogenous estradiol or E6/E7 oncogene activation
(180). Chen et al
(181), using a microarray
assay, found an increased expression of GLI1, SMO,
SHH, PTCH1 and FOXM1 (GLI target gene) in 70
tumor CCs, compared to 10 normal cervical tissues; the expression
patterns of those genes were associated with either the clinical or
pathological progression of CC.
The majority of studies describing the role of the
SHH pathway in CC have been performed using CC cell lines. Vishnoi
et al (182) reported a
connection between E6/E7 oncoproteins and SHH activation by
analyzing HPV-16 positive SiHa CC cells. In SiHa cells, the SHH
components, GLI, SMO and PTCH1, were found to
be overexpressed, while their reduced expression was observed
following either the addition of cyclopamine or siRNA-mediated E6
gene silencing (182). Wang
et al (183)
demonstrated that, in a hypoxic environment, the GLI1 mRNA
level in HeLa cells was increased and was accompanied by an
enhanced invasion ability, whereas GLI1 silencing reversed
these effects, compromising the invasiveness of HeLa cells.
Furthermore, Wang et al (183) observed that the ectopic
increase of mir-129-5p resulted in the lower mRNA and protein
levels of ZIC2, SHH, GLI1 and GLI2, together with SHH target genes
CXCL1, VEGF and ANG2, as well as the inhibition of tumor formation
in a mouse xenograft model. These results indicated that mir-129-5p
may be a promising target for CC treatment (184). In combination, the available
evidence suggested that the SHH pathway is involved in CC
progression.
9. SHH pathway in cancers of the male
reproductive system
The testis, penis and prostate may be affected by
neoplastic transformation, leading to cancers of the male
reproductive system, according to AJCC Cancer Staging Manual, 8th
Edition (159). Although DHH is
involved in the differentiation of peritubular myoid cells and
consequent formation of the testis cord (185), while the SHH is involved in
penile development (186),
there are no data available on the SHH pathway during testicular or
penile tumorigenesis, at least to the best of our knowledge.
PC
Prostate gland tumors rank 2nd in the worldwide
cancer incidence among males (4th among all cancers in both sexes)
with almost 1.3 million new cases, and 5th in worldwide cancer
mortality in males (8th among all cancers in both sexes) with
~359,000 deaths in 2018 (8). The
majority of PCs are associated with defective DNA damage repair
molecules, while androgen receptor (AR) signaling also plays an
important role in PC pathogenesis, particularly in metastasized
cases (187). During fetal
life, the AR and SHH pathways play a crucial role in the
development of the prostate gland (188,189). Le et al (188) reported that, during prostate
development, growth and regeneration, both pathways are
indispensable; the AR signaling pathway is superior since, in the
murine in vivo model, the expression of AR was essential for
urogenital mesenchymal and epithelial cell differentiation, even if
the cells overexpressed GLI1.
Yamamichi et al (190) reported that in PC epithelial
cells (LNCap) and prostate fibroblast cell lines, normal (NPF) and
PC-associated (CPF), dihydrotestosterone (DHT) enhanced cell
proliferation in all cell types while the inhibition of SHH
signaling by cyclopamine decreased this rate in CPF cells only. The
activation of both androgen and SHH signaling enhanced EMT,
accelerating PC development, while cyclopamine blocked cancer
progression. In addition, DHT (but not SHH) induced the expression
of osteonectin, and a high GLI1 expression and stromal
osteonectin expression (as found by IHC) in tumor tissues from 25
patients with PC, were associated with PSA recurrence (190).
A recent study by Zhang et al (191) analyzed the AR and SHH pathways
in PC clinical cases. In a large group of 443 patients with primary
PC and 96 with benign prostatic hyperplasia, the increased
immunoreactivity of SHH protein was observed in more aggressive
tumors (Gleason score of >7), which was much higher in
AR-positive than in AR-negative cancer. Furthermore, SHH was
overexpressed in high-grade PC and positively correlated with the
expression of both GRP78 (the molecule involved in endoplasmic
reticulum stress response) and AR; this suggested that the
assessment of SHH protein could be beneficial as a prognostic
factor in PC, since SHH overexpression in all patients with PC with
AR+ tumors was associated with a shorter
disease-specific survival (191). Describing the expression
pattern of SHH pathway components in PC, Tzelepi et al
(192) analyzed SHH, SMO, PTCH,
GLI1, VEGF, CD31 and ki67 protein levels using western blot
analysis, IHC and tissue microarrays in large groups consisting of
141 hormone-naive primary PC and 53 castrate-resistant bone marrow
metastases, compared to 119 prostate non-neoplastic peripheral
zone. First, they observed the crosstalk between prostate cells in
healthy tissues; SHH and PTCH1 were primarily expressed in
epithelial and stromal cells, respectively, while SMO and GLI1 were
expressed in both epithelial and stromal cells. This observation
suggested paracrine signaling between epithelial (donor) and
stromal (acceptor) cells, followed by SHH pathway activation in all
cells (192). The expression
pattern was continued in primary PCs with higher SHH and SMO
protein levels in PC epithelial cells than those in the
non-neoplastic peripheral prostate zone. Of note, in PC metastases,
a higher PTCH1 expression was observed in epithelial cells compared
with that in stromal cells, while the expression of SHH and
GLI1 did not differ between the two (192). These results suggested an
alteration in the mechanisms of SHH signaling in PC and its
metastases, as well as its involvement in PC development.
In combination, the available data demonstrate that
the SHH pathway plays an important role in PC development,
indicating that SHH pathway-targeting drugs should be introduced
into PC treatment. Indeed, two phase I and one phase II clinical
trials that used LDE225, vismodegib or itraconazole (SMO
inhibitors) have been performed (193-195). Although decreased levels of
GLI1 were recorded in tumor tissues from patients treated with
vismodegib or LDE255, there was no apparent effect on clinical
activity. In addition, vismodegib caused side-effects, such as
fatigue or nausea, and LDE255 increased the prostate-specific
antigen (PSA) serum level (193,194). Treatment with itraconazole, an
FDA-approved antifungal drug, demonstrated that a high dose (600
mg) may be beneficial for progress-free survival. However, such a
dose has been found to cause hypokalemia (195,196). In summary, drugs targeting the
SHH pathway should be further evaluated as an additional modality
of PC treatment, given that more studies associated with the
interactions between stromal and PC cells in relation to the AR and
SHH signaling pathways are being carried out.
10. Conclusions and future perspectives
The SHH signaling pathway was identified 40 years
ago, and since then, the understanding of the functions of and
cellular associations between its components has been considerably
increased. Although the SHH-PTCH-SMO-GLI cellular cascade has been
widely discussed in several studies, the aim of the present review
was to also describe the upstream genetic regulation of the SHH
ligand expression. Of note, the activation of SHH biosynthesis
relies on proteins with transcription factor properties that are
involved in fetal development, tissue renewal and remodeling in the
adult body. Indirectly, SHH is regulated by miRNAs, which also
interact with other cellular pathways. GLIs are the main downstream
effectors of SHH signaling and their transcriptional activity
depends mainly on their release from the SUFU-KIF7 complex
triggered by the SMO receptor. Since the upregulation of the SHH
pathway, particularly GLIs, is associated with the progression of
several types of cancer, specific drugs inhibiting this signaling
have been developed. Most of them target the SMO receptor; however,
due to frequent SMO/PTCH1 mutations that may lead to drug
resistance, GLIs can be also activated through other cellular
pathways.
In the present review, the focus was placed on
analyzing the SHH pathway components in the kidney, urinary
bladder, OC, CC and PC. In all these cancers, including sex
hormone-dependent ovarian and prostate tumors, deregulations of SHH
pathway components were observed by several authors. Furthermore,
the interaction between viral proteins and SHH signaling molecules
has been noted in cervical types of cancer, mostly originating from
HPV infection. The alterations of the SHH pathway components in
these cancers have often been found to be associated with either
the clinical or pathological status of patients. Despite these
findings, the SHH components have not yet been considered as
prognostic or therapeutic molecular parameters in gynecological and
urogenital cancers. This may have been caused by the unsatisfactory
results of older clinical trials with SMO or GLI inhibitors.
However, since the knowledge of SHH pathway interactions with other
cellular signaling pathways in these malignancies is accumulating
and new molecules targeting the SHH pathway are being developed, it
can be expected that new clinical trials will soon be performed. It
is also worth noting that limited data are available on the
involvement of the SHH pathway in the pathogenesis of penile,
fallopian tube, vaginal and vulvar cancer.
Availability of data and materials
Not applicable.
Authors' contributions
AKC, ZK and PMW performed the literature search,
wrote the manuscript and prepared the figures. All the authors
confirm the authenticity of all the raw data. All the authors have
read and approved the final version of this manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
The ORCID numbers of the authors of the present
study are as follows: AKC, 0000-0002-2942-6270; ZK,
0000-0002-9801-8166; and PMW, 0000-0002-4310-1616.
Acknowledgments
Not applicable.
Funding
The present study was funded by the ST-12 internal funds of the
Medical University of Gdańsk, Poland.
References
|
1
|
Bellmunt J, Orsola A, Leow JJ, Wiegel T,
De Santis M and Horwich A: Bladder cancer: ESMO Practice Guidelines
for diagnosis, treatment and follow-up. Ann Oncol. 25:iii40–iii48.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parker C, Gillessen S, Heidenreich A and
Horwich A: Cancer of the prostate: ESMO Clinical Practice
Guidelines for diagnosis, treatment and follow-up. Ann Oncol.
26:v69–v77. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marth C, Landoni F, Mahner S, McCormack M,
Gonzalez-Martin A and Colombo N: Cervical cancer: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 28(suppl-4): iv72–iv83. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Colombo N, Preti E, Landoni F, Carinelli
S, Colombo A and Marini C; ESMO Guidelines Working Group:
Endometrial cancer: ESMO Clinical Practice Guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 24(Suppl 6):
vi33–vi38. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ray-Coquard I, Morice P, Lorusso D, Prat
J, Oaknin A, Pautier P and Colombo N; ESMO Guidelines Committee:
Non-epithelial ovarian cancer: ESMO Clinical Practice Guidelines
for diagnosis, treatment and follow-up. Ann Oncol. 29(Suppl 4):
iv1–iv18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Escudier B, Porta C, Schmidinger M,
Rioux-Leclercq N, Bex A, Khoo V, Grünwald V, Gillessen S, Horwich
A, et al ESMO Guidelines Committee: Renal cell carcinoma: ESMO
Clinical Practice Guidelines for diagnosis, treatment and
follow-up. Ann Oncol. 30:706–720. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Colombo N, Sessa C, du Bois A, Ledermann
J, McCluggage WG, McNeish I, Morice P, Pignata S, Ray-Coquard I,
Vergote I, et al: ESMO-ESGO consensus conference recommendations on
ovarian cancer: Pathology and molecular biology, early and advanced
stages, borderline tumours and recurrent disease. Ann Oncol.
30:672–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar
|
|
9
|
Bankhead CR, Kehoe ST and Austoker J:
Symptoms associated with diagnosis of ovarian cancer: A systematic
review. BJOG. 112:857–865. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nüsslein-Volhard C and Wieschaus E:
Mutations affecting segment number and polarity in Drosophila.
Nature. 287:795–801. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rivell A, Petralia RS, Wang YX, Clawson E,
Moehl K, Mattson MP and Yao PJ: Sonic hedgehog expression in the
postnatal brain. Biology Open. 8:bio0405922019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fattahi S, Pilehchian Langroudi M and
Akhavan-Niaki H: Hedgehog signaling pathway: Epigenetic regulation
and role in disease and cancer development. J Cell Physiol.
233:5726–5735. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang C, Cassandras M and Peng T: The Role
of hedgehog signaling in adult lung regeneration and maintenance. J
Dev Biol. 7:142019. View Article : Google Scholar :
|
|
15
|
Skoda AM, Simovic D, Karin V, Kardum V,
Vranic S and Serman L: The role of the Hedgehog signaling pathway
in cancer: A comprehensive review. Bosn J of Basic Med Sci.
18:8–20. 2018. View Article : Google Scholar
|
|
16
|
Cohen M, Kicheva A, Ribeiro A, Blassberg
R, Page KM, Barnes CP and Briscoe J: Ptch1 and Gli regulate Shh
signalling dynamics via multiple mechanisms. Nat Commun.
6:67092015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Katoh M: Genomic testing, tumor
microenvironment and targeted therapy of Hedgehog-related human
cancers. Clin Sci. 133:953–970. 2019. View Article : Google Scholar
|
|
18
|
Girardi D, Barrichello A, Fernandes G and
Pereira A: Targeting the hedgehog pathway in cancer: Current
evidence and future perspectives. Cells. 8:1532019. View Article : Google Scholar :
|
|
19
|
Hua K and Ferland RJ: Primary cilia
proteins: Ciliary and extraciliary sites and functions. Cell Mol
Life Sci. 75:1521–1540. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carballo GB, Honorato JR, de Lopes GPF and
Spohr TCL DESE: A highlight on Sonic hedgehog pathway. Cell Commun
Signal. 16:112018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Didiasova M, Schaefer L and Wygrecka M:
Targeting GLI transcription factors in cancer. Molecules.
23:10032018. View Article : Google Scholar :
|
|
22
|
ten Haaf A, Bektas N, von Serenyi S, Losen
I, Arweiler EC, Hartmann A, Knüchel R and Dahl E: Expression of the
glioma-associated oncogene homolog (GLI) 1in human breast cancer is
associated with unfavourable overall survival. BMC Cancer.
9:2982009. View Article : Google Scholar
|
|
23
|
Im S, Choi HJ, Yoo C, Jung JH, Jeon YW,
Suh YJ and Kang CS: Hedgehog related protein expression in breast
cancer: Gli-2 is associated with poor overall survival. Korean J
Pathol. 47:116–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Choe JY, Yun JY, Jeon YK, Kim SH, Choung
HK, Oh S, Park M and Kim JE: Sonic hedgehog signalling proteins are
frequently expressed in retinoblastoma and are associated with
aggressive clinicopathological features. J Clin Pathol. 68:6–11.
2015. View Article : Google Scholar
|
|
25
|
Al Ghamdi D, Gomaa W, Abulaban A, Al-Ahwal
M, Buhmeida A, Al-Qahtani M and Al-Maghrabi J: The significance of
sonic hedgehog immunohistochemical expression in colorectal
carcinoma. J Microsc Ultrastruct. 3:169–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bai XY, Lin JY, Zhang XC, Xie Z, Yan HH,
Chen ZH, Xu CR, An SJ, Sheng GM and Wu YL: High expression of
truncated GLI3 is associated with poor overall survival in patients
with non-small cell lung cancer. Cancer Biomark. 13:37–47. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ding YL, Wang QS, Zhao WM and Xiang L:
Expression of smoothened protein in colon cancer and its prognostic
value for postoperative liver metastasis. Asian Pac J Cancer Prev.
13:4001–4005. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aberger F and Frischauf AM: GLI Genes and
Their Targets in Epidermal Development and Disease. Landes
Bioscience. 2013.
|
|
29
|
Lesiak A, Sobolewska-Sztychny D,
Danilewicz M, Rogowski-Tylman M, Sysa-Jedrzejowska A, Sobjanek M,
Olejniczak-Staruch I and Narbutt J: Sonic hedgehog pathway
dysregulation in skin basal-cell carcinoma of a Polish population.
Folia Histochem Cytobiol. 51:219–224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu F, Jiang W, Sui Y, Meng W, Hou L, Li
T, Li M, Zhang L, Mo J, Wang J, et al: CDK7 inhibition suppresses
aberrant hedgehog pathway and overcomes resistance to smoothened
antagonists. Proc Natl Acad Sci USA. 116:12986–12995. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fernandes-Silva H, Correia-Pinto J and
Moura RS: Canonical sonic hedgehog signaling in early lung
development. J Dev Biol. 5:32017. View Article : Google Scholar
|
|
32
|
Memi F, Zecevic N and Radonjić N: Multiple
roles of Sonic Hedgehog in the developing human cortex are
suggested by its widespread distribution. Brain Struct Funct.
223:2361–2375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Odent S, Attie-Bitach T, Blayau M, Mathieu
M, Aug J, Delezo de AL, Gall JY, Le Marec B, Munnich A, David V and
Vekemans M: Expression of the Sonic hedgehog (SHH) gene during
early human development and phenotypic expression of new mutations
causing holoprosencephaly. Hum Mol Genet. 8:1683–1689. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim A, Le Douce J, Diab F, Ferovova M,
Dubourg C, Odent S, Dupé V, David V, Diambra L, Watrin E and de
Tayrac M: Synonymous variants in holoprosencephaly alter codon
usage and impact the Sonic Hedgehog protein. Brain. 143:2027–2038.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fabik J, Kovacova K, Kozmik Z and Machon
O: Neural crest cells require Meis2 for patterning the mandibular
arch via the Sonic hedgehog pathway. Biol Open. 9:bio0520432020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bürglin TR: The Hedgehog protein family.
Genome Biol. 9:2412008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hanna A and Shevde LA: Hedgehog signaling:
Modulation of cancer properies and tumor mircroenvironment. Mol
Cancer. 15:242016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nikolopoulou E, Galea GL, Rolo A, Greene
NDE and Copp AJ: Neural tube closure: Cellular, molecular and
biomechanical mechanisms. Development. 144:552–566. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roessler E, Belloni E, Gaudenz K, Vargas
F, Scherer SW, Tsui LC and Muenke M: Mutations in the C-terminal
domain of sonic hedgehog cause holoprosencephaly. Hum Mol Genet.
6:1847–1853. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mimeault M and Batra SK: Frequent
deregulations in the hedgehog signaling network and cross-talks
with the epidermal growth factor receptor pathway involved in
cancer progression and targeted therapies. Pharmacol Rev.
62:497–524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Varjosalo M and Taipale J: Hedgehog:
Functions and mechanisms. Genes Dev. 22:2454–2472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chamoun Z, Mann RK, Nellen D, von Kessler
DP, Bellotto M, Beachy PA and Basler K: Skinny hedgehog, an
acyltransferase required for palmitoylation and activity of the
hedgehog signal. Science. 293:2080–2084. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lanyon-Hogg T, Masumoto N, Bodakh G,
Konitsiotis AD, Thinon E, Rodgers UR, Owens RJ, Magee AI and Tate
EW: Synthesis and characterisation of
5-acyl-6,7-dihydrothieno[3,2-c] pyridine inhibitors of Hedgehog
acyltransferase. Data Brief. 7:257–281. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rodgers UR, Lanyon-Hogg T, Masumoto N,
Ritzefeld M, Burke R, Blagg J, Magee AI and Tate EW:
Characterization of hedgehog acyltransferase inhibitors identifies
a small molecule probe for hedgehog signaling by cancer cells. ACS
Chem Biol. 11:3256–3262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei
J and Yan N: Structural basis for the recognition of Sonic Hedgehog
by human Patched1. Science. 361:eaas89352018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Choudhry Z, Rikani AA, Choudhry AM, Tariq
S, Zakaria F, Asghar MW, Sarfraz MK, Haider K, Shafiq AA and
Mobassarah NJ: Sonic hedgehog signalling pathway: A complex
network. Ann Neurosci. 21:28–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bausch D, Fritz S, Bolm L, Wellner UF,
Fernandez-Del-Castillo C, Warshaw AL, Thayer SP and Liss AS:
Hedgehog signaling promotes angiogenesis directly and indirectly in
pancreatic cancer. Angiogenesis. 23:479–492. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Qi X, Schmiege P, Coutavas E, Wang J and
Li X: Structures of human Patched and its complex with native
palmitoylated sonic hedgehog. Nature. 560:128–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Martinez MF, Romano MV, Martinez AP,
González A, Muchnik C, Stengel FM, Mazzuoccolo LD and Azurmendi PJ:
Nevoid Basal Cell Carcinoma Syndrome: PTCH1 mutation profile and
expression of genes involved in the hedgehog pathway in argentinian
patients. Cells. 8:1442019. View Article : Google Scholar :
|
|
50
|
Fleet AJ and Hamel PA: The
protein-specific activities of the transmembrane modules of Ptch1
and Ptch2 are determined by their adjacent protein domains. J Biol
Chem. 293:16583–16595. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Holtz AM, Peterson KA, Nishi Y, Morin S,
Song JY, Charron F, McMahon AP and Allen BL: Essential role for
ligand-dependent feedback antagonism of vertebrate hedgehog
signaling by PTCH1, PTCH2 and HHIP1 during neural patterning.
Development. 140:3423–3434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kowatsch C, Woolley RE, Kinnebrew M,
Rohatgi R and Siebold C: Structures of vertebrate Patched and
Smoothened reveal intimate links between cholesterol and Hedgehog
signalling. Curr Opin Struct Biol. 57:204–214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xiao X, Tang JJ, Peng C, Wang Y, Fu L, Qiu
ZP, Xiong Y, Yang LF, Cui HW, He XL, et al: Cholesterol
modification of smoothened is required for hedgehog signaling. Mol
Cell. 66:154–162.e10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Denef N, Neubüser D, Perez L and Cohen SM:
Hedgehog induces opposite changes in turnover and subcellular
localization of patched and smoothened. Cell. 102:521–531. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jiang K and Jia J: Smoothened regulation
in response to Hedgehog stimulation. Front Biol. 10:475–486. 2015.
View Article : Google Scholar
|
|
56
|
Huang P, Zheng S, Wierbowski BM, Kim Y,
Nedelcu D, Aravena L, Liu J, Kruse AC and Salic A: Structural basis
of smoothened activation in hedgehog signaling. Cell.
174:312–324.e16. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Qi Y, Liu H and Lin X: Sumoylation
stabilizes smoothened to promote hedgehog signaling. Dev Cell.
39:385–387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang B, Zhuang T, Lin Q, Yang B, Xu X,
Xin G, Zhu S, Wang G, Yu B, Zhang T, et al:
Patched1-ArhGAP36-PKA-Inversin axis determines the ciliary
translocation of Smoothened for Sonic Hedgehog pathway activation.
Proc Natl Acad Sci USA. 116:874–879. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rohatgi R and Scott MP: Cell biology.
Arrestin' movement in cilia. Science. 320:1726–1727. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cochrane CR, Szczepny A, Watkins DN and
Cain JE: Hedgehog signaling in the maintenance of cancer stem
cells. Cancers (Basel). 7:1554–1585. 2015. View Article : Google Scholar
|
|
61
|
Carpenter RL and Ray H: Safety and
tolerability of sonic hedgehog pathway inhibitors in cancer. Drug
Saf. 42:263–279. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hoy SM: Glasdegib: First Global Approval.
Drugs. 79:207–213. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Pietrobono S, Gagliardi S and Stecca B:
Non-canonical hedgehog signaling pathway in cancer: Activation of
GLI transcription factors beyond smoothened. Front Genet.
10:5562019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Peer E, Tesanovic S and Aberger F:
Next-generation hedgehog/GLI pathway inhibitors for cancer therapy.
Cancers. 11:5382019. View Article : Google Scholar :
|
|
65
|
Sabol M, Trnski D, Musani V, Ozretić P and
Levanat S: Role of GLI Transcription factors in pathogenesis and
their potential as new therapeutic targets. Int J Mol Sci.
19:25622018. View Article : Google Scholar :
|
|
66
|
Ruppert JM, Kinzler KW, Wong AJ, Bigner
SH, Kao FT, Law ML, Seuanez HN, O'Brien SJ and Vogelstein B: The
GLI-Kruppel family of human genes. Mol Cell Biol. 8:3104–3113.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Carpenter RL and Lo HW: Hedgehog Pathway
and GLI1 Isoforms in Human Cancer. Discov Med. 13:105–113.
2013.
|
|
68
|
Cao X, Geradts J, Dewhirst MW and Lo HW:
Upregulation of VEGF-A and CD24 gene expression by the tGLI1
transcription factor contributes to the aggressive behavior of
breast cancer cells. Oncogene. 31:104–115. 2012. View Article : Google Scholar
|
|
69
|
Carpenter RL and Lo HW: Identification,
Functional Characterization, and Pathobiological Significance of
GLI1 Isoforms in Human Cancers. Vitamins, Hormones. 88. Elsevier;
pp. 115–140. 2012, View Article : Google Scholar
|
|
70
|
Niewiadomski P, Niedziółka SM, Markiewicz
Ł, Uśpieński T, Baran B and Chojnowska K: Gli Proteins: Regulation
in development and cancer. Cells. 8:1472019. View Article : Google Scholar :
|
|
71
|
Pal K, Hwang SH, Somatilaka B, Badgandi H,
Jackson PK, DeFea K and Mukhopadhyay S: Smoothened determines
β-arrestin-mediated removal of the G protein-coupled receptor
Gpr161 from the primary cilium. J Cell Biol. 212:861–875. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Briscoe J and Thérond PP: The mechanisms
of Hedgehog signalling and its roles in development and disease.
Nat Rev Mol Cell Biol. 14:416–429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Montagnani V and Stecca B: Role of protein
kinases in hedgehog pathway control and implications for cancer
therapy. Cancers. 11:4492019. View Article : Google Scholar :
|
|
74
|
Mirza AN, McKellar SA, Urman NM, Brown AS,
Hollmig T, Aasi SZ and Oro AE: LAP2 proteins chaperone GLI1
movement between the lamina and chromatin to regulate
transcription. Cell. 176:198–212.e15. 2019. View Article : Google Scholar
|
|
75
|
Canettieri G, Di Marcotullio L, Greco A,
Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E,
Ferretti E, Miele E, et al: Histone deacetylase and
Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog
signalling through Gli acetylation. Nat Cell Biol. 12:132–142.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bangs F and Anderson KV: Primary cilia and
mammalian hedgehog signaling. Cold Spring Harb Perspect Biol.
9:a0281752017. View Article : Google Scholar
|
|
77
|
Wong SY, Seol AD, So PL, Ermilov AN,
Bichakjian CK, Epstein EH Jr, Dlugosz AA and Reiter JF: Primary
cilia can both mediate and suppress Hedgehog pathway-dependent
tumorigenesis. Nat Med. 15:1055–1061. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Han YG, Kim HJ, Dlugosz AA, Ellison DW and
Alvarez-Buylla A: Dual and opposing roles of primary cilia in
medulloblastoma development. Nat Med. 15:1062–1065. 2010.
View Article : Google Scholar
|
|
79
|
Quaglio D, Infante P, Di Marcotullio L,
Botta B and Mori M: Hedgehog signaling pathway inhibitors: An
updated patent review (2015-present). Expert Opin Ther Pat.
30:235–250. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Perea SE, Baladrón I, Valenzuela C and
Perera Y: CIGB-300: A peptide-based drug that impairs the Protein
Kinase CK2-mediated phosphorylation. Semin Oncol. 45:58–67. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Huang S and Houghton PJ: Targeting mTOR
signaling for cancer therapy. Curr Opin Pharmacol. 3:371–377. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sullivan RJ, Infante JR, Janku F, Wong
DJL, Sosman JA, Keedy V, Patel MR, Shapiro GI, Mier JW, Tolcher AW,
et al: First-in-Class ERK1/2 Inhibitor Ulixertinib (BVD-523) in
Patients with MAPK mutant advanced solid tumors: Results of a phase
i dose-escalation and expansion study. Cancer Discov. 8:184–195.
2018. View Article : Google Scholar
|
|
83
|
Xing Y, Lin NU, Maurer MA, Chen H, Mahvash
A, Sahin A, Akcakanat A, Li Y, Abramson V, Litton J, et al: Phase
II trial of AKT inhibitor MK-2206 in patients with advanced breast
cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN
loss/PTEN mutation. Breast Cancer Res. 21:782019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chen H, Shen J, Choy E, Hornicek FJ, Shan
A and Duan Z: Targeting DYRK1B suppresses the proliferation and
migration of liposarcoma cells. Oncotarget. 9:13154–13166. 2017.
View Article : Google Scholar
|
|
85
|
Piirsoo A, Kasak L, Kauts ML, Loog M,
Tints K, Uusen P, Neuman T and Piirsoo M: Protein kinase inhibitor
SU6668 attenuates positive regulation of Gli proteins in cancer and
multipotent progenitor cells. Biochim Biophys Acta. 1843:703–714.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Schachter KA and Krauss RS: Murine models
of holoprosencephaly. Curr Top Dev Biol. 84:139–170. 2008.
View Article : Google Scholar
|
|
87
|
Barratt KS and Arkell RM: ZIC2 in
Holoprosencephaly. Zic family: Evolution, Development and Disease.
Aruga J: Springer; Singapore: pp. 269–299. 2018, View Article : Google Scholar
|
|
88
|
Chen X, Yang S, Zeng J and Chen M:
miR-1271-5p inhibits cell proliferation and induces apoptosis in
acute myeloid leukemia by targeting ZIC2. Mol Med Rep. 19:508–514.
2019.
|
|
89
|
Li Q, Shi J and Xu X: MicroRNA-1271-5p
inhibits the tumorigenesis of ovarian cancer through targeting E2F5
and negatively regulates the mTOR signaling pathway. Panminerva
Med. May 14–2020.Online ahead of print.
|
|
90
|
Li X, Liu H, Lv Y, Yu W, Liu X and Liu C:
MiR-130a-5p/Foxa2 axis modulates fetal lung development in
congenital diaphragmatic hernia by activating the Shh/Gli1
signaling pathway. Life Sci. 241:1171662020. View Article : Google Scholar
|
|
91
|
Xian X, Tang L, Wu C and Huang L:
miR-23b-3p and miR-130a-5p affect cell growth, migration and
invasion by targeting CB1R via the Wnt/β-catenin signaling pathway
in gastric carcinoma. Onco Targets Ther. 11:7503–7512. 2018.
View Article : Google Scholar :
|
|
92
|
Huang JL, Fu YP, Gan W, Liu G, Zhou PY,
Zhou C, Sun BY, Guan RY, Zhou J, Fan J, et al: Hepatic stellate
cells promote the progression of hepatocellular carcinoma through
microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett. 476:140–151.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Yu Y, Yu F and Sun P: MicroRNA-1246
promotes melanoma progression through targeting FOXA2. Onco Targets
Ther. 13:1245–1253. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jeong JH, Park SJ, Dickinson SI and Luo
JL: A constitutive intrinsic inflammatory signaling circuit
composed of miR-196b, Meis2, PPP3CC, and p65 drives prostate cancer
castration resistance. Mol Cell. 65:154–167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Guan L, Li T, Ai N, Wang W, He B, Bai Y,
Yu Z, Li M, Dong S, Zhu Q, et al: MEIS2C and MEIS2D promote tumor
progression via Wnt/β-catenin and hippo/YAP signaling in
hepatocellular carcinoma. J Exp Clin Cancer Res. 38:4172019.
View Article : Google Scholar
|
|
96
|
Richbourg HA, Hu DP, Xu Y, Barczak AJ and
Marcucio RS: miR-199 family contributes to regulation of sonic
hedgehog expression during craniofacial development. Dev Dyn.
249:1062–1076. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Ozretić P, Trnski D, Musani V, Maurac I,
Kalafatić D, Orešković S, Levanat S and Sabol M: Non-canonical
Hedgehog signaling activation in ovarian borderline tumors and
ovarian carcinomas. Int J Oncol. 51:1869–1877. 2017. View Article : Google Scholar
|
|
98
|
Mishra P, Panda A, Bandyopadhyay A, Kumar
H and Mohiddin G: Sonic hedgehog signalling pathway and
ameloblastoma-a review. J Clin Diagn Res. 9:ZE10–ZE13.
2015.PubMed/NCBI
|
|
99
|
Bhuria V, Xing J, Scholta T, Bui KC,
Nguyen MLT, Malek NP, Bozko P and Plentz RR: Hypoxia induced Sonic
Hedgehog signaling regulates cancer stemness,
epithelial-to-mesenchymal transition and invasion in
cholangiocarcinoma. Exp Cell Res. 385:1116712019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lei J, Fan L, Wei G, Chen X, Duan W, Xu Q,
Sheng W, Wang K and Li X: Gli-1 is crucial for hypoxia-induced
epithelial- mesenchymal transition and invasion of breast cancer.
Tumour Biol. 36:3119–3126. 2015. View Article : Google Scholar
|
|
101
|
Seto M, Ohta M, Asaoka Y, Ikenoue T, Tada
M, Miyabayashi K, Mohri D, Tanaka Y, Ijichi H, Tateishi K, et al:
Regulation of the hedgehog signaling by the mitogen-activated
protein kinase cascade in gastric cancer. Mol Carcinog. 48:703–712.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Stecca B, Mas C, Clement V, Zbinden M,
Correa R, Piguet V, Beermann F, Ruiz I and Altaba A: Melanomas
require HEDGEHOG-GLI signaling regulated by interactions between
GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA.
104:5895–5900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Singh R, Dhanyamraju PK and Lauth M:
DYRK1B blocks canonical and promotes non-canonical Hedgehog
signaling through activation of the mTOR/AKT pathway. Oncotarget.
8:833–845. 2017. View Article : Google Scholar :
|
|
104
|
Stemmer V, de Craene B, Berx G and Behrens
J: Snail promotes Wnt target gene expression and interacts with
beta-catenin. Oncogene. 27:5075–5080. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Rishikaysh P, Dev K, Diaz D, Qureshi WMS,
Filip S and Mokry J: Signaling involved in hair follicle
morphogenesis and development. Int J Mol Sci. 15:1647–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Qi Y, Zhao W, Wang Z, Xie Q, Cao J and
Meng X: Cross regulation of signaling pathways in gastrointestinal
stromal tumor. Oncol Lett. 16:6770–6776. 2018.PubMed/NCBI
|
|
107
|
Yoon JW, Gallant M, Lamm ML, Iannaccone S,
Vieux KF, Proytcheva M, Hyjek E, Iannaccone P and Walterhouse D:
Noncanonical regulation of the hedgehog mediator GLI1 by c-MYC in
burkitt lymphoma. Mol Cancer Res. 11:604–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Whitson RJ, Lee A, Urman NM, Mirza A, Yao
CY, Brown AS, Li JR, Shankar G, Fry MA, Atwood SX, et al:
Noncanonical hedgehog pathway activation through SRF-MKL1 promotes
drug resistance in basal cell carcinomas. Nat Med. 24:271–281.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Liang R, Kagwiria R, Zehender A, Dees C,
Bergmann C, Ramming A, Krasowska D, Michalska-Jakubus M, Kreuter A,
Kraner ME, et al: Acyltransferase skinny hedgehog regulates
TGFβ-dependent fibroblast activation in SSc. Ann Rheum Dis.
78:1269–1273. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yang H, Hu L, Liu Z, Qin Y, Li R, Zhang G,
Zhao B, Bi C, Lei Y and Bai Y: Inhibition of Gli1-mediated prostate
cancer cell proliferation by inhibiting the mTOR/S6K1 signaling
pathway. Oncol Lett. 14:7970–7976. 2017.PubMed/NCBI
|
|
111
|
Robbins DJ, Fei DL and Riobo NA: The
hedgehog signal transduction network. Sci Signal. 5:re62012.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Barnes EA, Kong M, Ollendorff V and
Donoghue D: Patched1 interacts with cyclin B1 to regulate cell
cycle progression. EMBO J. 20:2214–2223. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Barnes EA, Heidtman KJ and Donoghue DJ:
Constitutive activation of the shh-ptc1 pathway by a patched1
mutation identified in BCC. Oncogene. 24:902–915. 2005. View Article : Google Scholar
|
|
114
|
Thibert C: Inhibition of neuroepithelial
patched-induced apoptosis by sonic hedgehog. Science. 301:843–846.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Mille F, Thibert C, Fombonne J, Rama N,
Guix C, Hayashi H, Corset V, Reed JC and Mehlen P: The Patched
dependence receptor triggers apoptosis through a DRAL-caspase-9
complex. Nat Cell Biol. 11:739–746. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sasaki N, Kurisu J and Kengaku M: Sonic
hedgehog signaling regulates actin cytoskeleton via Tiam1-Rac1
cascade during spine formation. Mol Cell Neurosci. 45:335–344.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Chinchilla P, Xiao L, Kazanietz MG and
Riobo NA: Hedgehog proteins activate pro-angiogenic responses in
endothelial cells through non-canonical signaling pathways. Cell
Cycle. 9:570–579. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Polizio AH, Chinchilla P, Chen X, Kim S,
Manning DR and Riobo NA: Heterotrimeric G i proteins
link hedgehog signaling to activation of rho small GTPases to
promote fibroblast migration. J Biol Chem. 286:19589–19596. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Belgacem YH and Borodinsky LN: Sonic
hedgehog signaling is decoded by calcium spike activity in the
developing spinal cord. Proc Natl Acad Sci USA. 108:4482–4487.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Gupta S, Takebe N and LoRusso P: Targeting
the Hedgehog pathway in cancer. Ther Adv Med Oncol. 2:237–250.
2010. View Article : Google Scholar
|
|
121
|
Reifenberger J, Wolter M, Knobbe CB,
Köhler B, Schönicke A, Scharwächter C, Kumar K, Blaschke B, Ruzicka
T and Reifenberger G: Somatic mutations in the PTCH, SMOH, SUFUH
and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol.
152:43–51. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Cheng WT, Xu K, Tian DY, Zhang ZG, Liu LJ
and Chen Y: Role of Hedgehog signaling pathway in proliferation and
invasiveness of hepatocellular carcinoma cells. Int J Oncol.
34:829–836. 2009.PubMed/NCBI
|
|
123
|
Tian H, Callahan CA, DuPree KJ, Darbonne
WC, Ahn CP, Scales SJ and de Sauvage FJ: Hedgehog signaling is
restricted to the stromal compartment during pancreatic
carcinogenesis. Proc Natl Acad Sci USA. 106:4254–4259. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Yauch RL, Gould SE, Scales SJ, Tang T,
Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, et al: A
paracrine requirement for hedgehog signalling in cancer. Nature.
455:406–410. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Jeng KS, Chang CF and Lin SS: Sonic
hedgehog signaling in organogenesis, tumors, and tumor
microenvironments. Int J Mol Sci. 21:7582020. View Article : Google Scholar :
|
|
126
|
Santi A, Kugeratski FG and Zanivan S:
Cancer Associated Fibroblasts: The Architects of Stroma Remodeling.
Proteomics. 18:e17001672018. View Article : Google Scholar
|
|
127
|
Valenti G, Quinn HM, Heynen GJJE, Lan L,
Holland JD, Vogel R, Wulf-Goldenberg A and Birchmeier W: Cancer
stem cells regulate cancer-associated fibroblasts via activation of
hedgehog signaling in mammary gland tumors. Cancer Res.
77:2134–2147. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
von Ahrens D, Bhagat TD, Nagrath D, Maitra
A and Verma A: The role of stromal cancer-associated fibroblasts in
pancreatic cancer. J Hematol Oncol. 10:762017. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Petty AJ, Li A, Wang X, Dai R, Heyman B,
Hsu D, Huang X and Yang Y: Hedgehog signaling promotes
tumor-associated macrophage polarization to suppress intratumoral
CD8+ T cell recruitment. J Clin Invest. 129:5151–5162. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Fan Q, He M, Sheng T, Zhang X, Sinha M,
Luxon B, Zhao X and Xie J: Requirement of TGFbeta Signaling for
SMO-mediated carcinogenesis. J Biol Chem. 285:36570–36576. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Maximov V, Chen Z, Wei Y, Robinson MH,
Herting CJ, Shanmugam NS, Rudneva VA, Goldsmith KC, MacDonald TJ,
Northcott PA, et al: Tumour-associated macrophages exhibit
anti-tumoural properties in Sonic Hedgehog medulloblastoma. Nat
Commun. 10:24102019. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part a: Renal, penile, and
testicular tumours. Eur Urol. 70:93–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Cheville JC, Lohse CM, Zincke H, Weaver AL
and Blute ML: Comparisons of outcome and prognostic features among
histologic subtypes of renal cell carcinoma. Am J Surg Pathol.
27:612–624. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Sanchez DJ and Simon MC: Genetic and
metabolic hallmarks of clear cell renal cell carcinoma. Biochim
Biophys Acta Rev Cancer. 1870:23–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Brugarolas J: Molecular genetics of
clear-cell renal cell carcinoma. J Clin Oncol. 32:1968–1976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Gossage L, Eisen T and Maher ER: VHL, the
story of a tumour suppressor gene. Nat Rev Cancer. 15:55–64. 2015.
View Article : Google Scholar
|
|
137
|
Le Tourneau C, Raymond E and Faivre S:
Sunitinib: A novel tyrosine kinase inhibitor. A brief review of its
therapeutic potential in the treatment of renal carcinoma and
gastrointestinal stromal tumors (GIST). Ther Clin Risk Manag.
3:341–348. 2007. View Article : Google Scholar
|
|
138
|
Dormoy V, Danilin S, Lindner V, Thomas L,
Rothhut S, Coquard C, Helwig JJ, Jacqmin D, Lang H and Massfelder
T: The sonic hedgehog signaling pathway is reactivated in human
renal cell carcinoma and plays orchestral role in tumor growth. Mol
Cancer. 8:1232009. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Samaratunga H, Gianduzzo T and Delahunt B:
The ISUP system of staging, grading and classification of renal
cell neoplasia. J Kidney Cancer VHL. 1:26–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Jäger W, Thomas C, Fazli L, Hurtado-Coll
A, Li E, Janssen C, Gust KM, So AI, Hainz M, Schmidtmann I, et al:
DHH is an independent prognosticator of oncologic outcome of clear
cell renal cell carcinoma. J Urol. 192:1842–1848. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Furukawa J, Miyake H and Fujisawa M: GLI2
expression levels in radical nephrectomy specimens as a predictor
of disease progression in patients with metastatic clear cell renal
cell carcinoma following treatment with sunitinib. Mol Clin Oncol.
5:186–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Behnsawy HM, Shigemura K, Meligy FY,
Yamamichi F, Yamashita M, Haung WC, Li X, Miyake H, Tanaka K,
Kawabata M, et al: Possible role of sonic hedgehog and
epithelial-mesenchymal transition in renal cell cancer progression.
Korean J Urol. 54:547–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Kotulak-Chrzaszcz A, Klacz J, Matuszewski
M, Kmiec Z and Wierzbicki P: Expression of the Sonic Hedgehog
pathway components in clear cell renal cell carcinoma. Oncol Lett.
18:5801–5810. 2019.PubMed/NCBI
|
|
144
|
Shang Z, Zhao T, Ou T, Yan H, Cui B, Wang
Q, Wu J, Jia C, Cui X and Li J: The level of zinc finger of the
cerebellum 2 is predictive of overall survival in clear cell renal
cell carcinoma. Transl Androl Urol. 9:614–620. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Jia Z, Wan F, Zhu Y, Shi G, Zhang H, Dai B
and Ye D: Forkhead-box series expression network is associated with
outcome of clear-cell renal cell carcinoma. Oncol Lett.
15:8669–8680. 2018.PubMed/NCBI
|
|
146
|
Zhou J, Wu K, Gao D, Zhu G, Wu D, Wang X,
Chen Y, Du Y, Song W, Ma Z, et al: Reciprocal regulation of
hypoxia-inducible factor 2α and GLI1 expression associated with the
radioresistance of renal cell carcinoma. Int J Radiat Oncol Biol
Phys. 90:942–951. 2014. View Article : Google Scholar
|
|
147
|
Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y,
Wu D, Wang X, Hsieh JT, He D and Wu K: Non-canonical GLI1/2
activation by PI3K/AKT signaling in renal cell carcinoma: A novel
potential therapeutic target. Cancer Lett. 370:313–323. 2016.
View Article : Google Scholar
|
|
148
|
D'Amato C, Rosa R, Marciano R, D'Amato V,
Formisano L, Nappi L, Raimondo L, Di Mauro C, Servetto A, Fulciniti
F, et al: Inhibition of hedgehog signalling by NVP-LDE225
(Erismodegib) interferes with growth and invasion of human renal
cell carcinoma cells. Br J Cancer. 111:1168–1179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zhang X and Zhang Y: Bladder cancer and
genetic mutations. Cell Biochem Biophys. 73:65–69. 2015. View Article : Google Scholar
|
|
151
|
Mobley D and Baum N: Smoking: Its impact
on urologic health. Rev Urol. 17:220–225. 2015.
|
|
152
|
Jalanko T, de Jong JJ, Gibb EA, Seiler R
and Black PC: Genomic subtyping in bladder cancer. Curr Urol Rep.
21:92020. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
He HC, Chen JH, Chen XB, Qin GQ, Cai C,
Liang YX, Han ZD, Dai QS, Chen YR, Zeng GH, et al: Expression of
hedgehog pathway components is associated with bladder cancer
progression and clinical outcome. Pathol Oncol Res. 18:349–355.
2012. View Article : Google Scholar
|
|
154
|
Nedjadi T, Salem N, Khayyat D, Al-Sayyad
A, Al-Ammari A and Al-Maghrabi J: Sonic hedgehog expression is
associated with lymph node invasion in urothelial bladder cancer.
Pathol Oncol Res. 25:1067–1073. 2019. View Article : Google Scholar :
|
|
155
|
Xie R, Chen X, Chen Z, Huang M, Dong W, Gu
P, Zhang J, Zhou Q, Dong W, Han J, et al: Polypyrimidine tract
binding protein 1 promotes lymphatic metastasis and proliferation
of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer
Lett. 449:31–44. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Islam SS, Mokhtari RB, Noman AS, Uddin M,
Rahman MZ, Azadi MA, Zlotta A, van der Kwast T, Yeger H and Farhat
WA: Sonic hedgehog (Shh) signaling promotes tumorigenicity and
stemness via activation of epithelial-to-mesenchymal transition
(EMT) in bladder cancer. Mol Carcinog. 55:537–551. 2016. View Article : Google Scholar
|
|
157
|
Kitagawa K, Shigemura K, Sung SY, Chen KC,
Huang CC, Chiang YT, Liu MC, Huang TW, Yamamichi F, Shirakawa T and
Fujisawa M: Possible correlation of sonic hedgehog signaling with
epithelial-mesenchymal transition in muscle-invasive bladder cancer
progression. J Cancer Res Clin Oncol. 145:2261–2271. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Kim S, Kim Y, Kong J, Kim E, Choi JH, Yuk
HD, Lee H, Kim HR, Lee KH, Kang M, et al: Epigenetic regulation of
mammalian Hedgehog signaling to the stroma determines the molecular
subtype of bladder cancer. Elife. 8:e430242019. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC cancer staging manual. 8th edition.
Springer International Publishing; Chicago, IL: 2017, View Article : Google Scholar
|
|
160
|
Ho J, Du Y, Wong OG, Siu MK, Chan KK and
Cheung AN: Downregulation of the gli transcription factors
regulator Kif7 facilitates cell survival and migration of
choriocarcinoma cells. PLoS One. 9:e1082482014. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Yap J, Fox R, Narsia N, Pinheiro-Maia S,
Pounds R, Woodman C, Luesley D, Ganesan R, Kehoe S and Dawson C:
Under expression of the Sonic Hedgehog receptor, Patched1 (PTCH1),
is associated with an increased risk of local recurrence in
squamous cell carcinoma of the vulva arising on a background of
Lichen Sclerosus. PLoS One. 13:e02065532018. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Rosen DG, Yang G, Liu G, Mercado-Uribe I,
Chang B, Xiao XS, Zheng J, Xue FX and Liu J: Ovarian cancer:
Pathology, biology, and disease models. Front Biosci (Landmark Ed).
14:2089–2102. 2009. View
Article : Google Scholar
|
|
163
|
Levanat S, Musani V, Komar A and Orešković
S: Role of the Hedgehog/Patched Signaling Pathway in Oncogenesis: A
new polymorphism in the PTCH gene in ovarian fibroma. Ann N Y Acad
Sci. 1030:134–143. 2004. View Article : Google Scholar
|
|
164
|
Marchini S, Poynor E, Barakat RR, Clivio
L, Cinquini M, Fruscio R, Porcu L, Bussani C, D'Incalci M, Erba E,
et al: The zinc finger gene ZIC2 has features of an oncogene and
its overexpression correlates strongly with the clinical course of
epithelial ovarian cancer. Clin Cancer Res. 18:4313–4324. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Salem M, O'Brien JA, Bernaudo S, Shawer H,
Ye G, Brkić J, Amleh A, Vanderhyden BC, Refky B, Yang BB, et al:
miR-590-3p Promotes ovarian cancer growth and metastasis via a
Novel FOXA2-versican pathway. Cancer Res. 78:4175–4190. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Peng Q, Qin J, Zhang Y, Cheng X, Wang X,
Lu W, Xie X and Zhang S: Autophagy maintains the stemness of
ovarian cancer stem cells by FOXA2. J Exp Clin Cancer Res.
36:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Musani V, Sabol M, Car D, Ozretić P,
Kalafatić D, Maurac I, Orešković S and Levanat S: PTCH1 gene
polymorphisms in ovarian tumors: Potential protective role of
c.3944T allele. Gene. 517:55–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Cretnik M, Musani V, Oreskovic S, Leovic D
and Levanat S: The Patched gene is epigenetically regulated in
ovarian dermoids and fibromas, but not in basocellular carcinomas.
Int J Mol Med. 19:875–883. 2007.PubMed/NCBI
|
|
169
|
Hainsworth JD, Meric-Bernstam F, Swanton
C, Hurwitz H, Spigel DR, Sweeney C, Burris H, Bose R, Yoo B, Stein
A, et al: Targeted therapy for advanced solid tumors on the basis
of molecular profiles: Results from mypathway, an open-label, phase
IIa multiple basket study. J Clin Oncol. 36:536–542. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Liao X, Siu MK, Au CW, Wong ES, Chan HY,
Ip PP, Ngan HY and Cheung AN: Aberrant activation of hedgehog
signaling pathway in ovarian cancers: Effect on prognosis, cell
invasion and differentiation. Carcinogenesis. 30:131–140. 2009.
View Article : Google Scholar
|
|
171
|
Yang L, He J, Huang S, Zhang X, Bian Y, He
N, Zhang H and Xie J: Activation of hedgehog signaling is not a
frequent event in ovarian cancers. Mol Cancer. 8:1122009.
View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Schmid S, Bieber M, Zhang F, Zhang M, He
B, Jablons D and Teng NN: Wnt and hedgehog gene pathway expression
in serous ovarian cancer. Int J Gynecol Cancer. 21:975–980. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Trnski D, Gregorić M, Levanat S, Ozretić
P, Rinčić N, Vidaković TM, Kalafatić D, Maurac I, Orešković S,
Sabol M and Musani V: Regulation of survivin isoform expression by
GLI proteins in ovarian cancer. Cells. 8:1282019. View Article : Google Scholar :
|
|
174
|
Vlčková K, Ondrušová L, Vachtenheim J,
Réda J, Dundr P, Zadinová M, Žáková P and Poučková P: Survivin, a
novel target of the Hedgehog/GLI signaling pathway in human tumor
cells. Cell Death Dis. 7:e20482016. View Article : Google Scholar
|
|
175
|
Zhang H, Wang Y, Chen T, Zhang Y, Xu R,
Wang W, Cheng M and Chen Q: Aberrant activation of hedgehog
signalling promotes cell migration and invasion via matrix
metalloproteinase-7 in ovarian cancer cells. J Cancer. 10:990–1003.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Xu M, Hu X, Zhang M and Ge Y: What is the
impact of BIRC5 gene polymorphisms on urinary cancer
susceptibility? Evidence from 9348 subjects. Gene. 733:1442682020.
View Article : Google Scholar
|
|
177
|
Sneha S, Nagare RP, Sidhanth C,
Krishnapriya S, Garg M, Ramachandran B, Murhekar K, Sundersingh S
and Ganesan TS: The hedgehog pathway regulates cancer stem cells in
serous adenocarcinoma of the ovary. Cell Oncol (Dordr). 43:601–616.
2020. View Article : Google Scholar
|
|
178
|
Sun X, Song J, Li E, Geng H, Li Y, Yu D
and Zhong C: (-)-Epigallocatechin-3-gallate inhibits bladder cancer
stem cells via suppression of sonic hedgehog pathway. Oncol Rep.
42:425–435. 2019.PubMed/NCBI
|
|
179
|
Li X, Wang X, Xie C, Zhu J, Meng Y, Chen
Y, Li Y, Jiang Y, Yang X, Wang S, et al: Sonic hedgehog and
Wnt/β-catenin pathways mediate curcumin inhibition of breast cancer
stem cells. Anticancer Drugs. 29:208–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Rojo-León V, García C, Valencia C, Méndez
MA, Wood C and Covarrubias L: The E6/E7 oncogenes of human
papilloma virus and estradiol regulate hedgehog signaling activity
in a murine model of cervical cancer. Exp Cell Res. 381:311–322.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Chen H, Wang J, Yang H, Chen D and Li P:
Association between FOXM1 and hedgehog signaling pathway in human
cervical carcinoma by tissue microarray analysis. Oncol Lett.
12:2664–2673. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Vishnoi K, Mahata S, Tyagi A, Pandey A,
Verma G, Jadli M, Singh T, Singh SM and Bharti AC: Cross-talk
between human papillomavirus oncoproteins and hedgehog signaling
synergistically promotes stemness in cervical cancer cells. Sci
Rep. 6:343772016. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Wang XH, He X, Jin HY, Liang JX and Li N:
Effect of hypoxia on the Twist1 in EMT of cervical cancer cells.
Eur Rev Med Pharmacol Sci. 22:6633–6639. 2018.PubMed/NCBI
|
|
184
|
Wang YF, Yang HY, Shi XQ and Wang Y:
Upregulation of microRNA-129-5p inhibits cell invasion, migration
and tumor angiogenesis by inhibiting ZIC2 via downregulation of the
Hedgehog signaling pathway in cervical cancer. Cancer Biol Ther.
19:1162–1173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Pereira J, Johnson WE, O'Brien SJ, Jarvis
ED, Zhang G, Gilbert MT, Vasconcelos V and Antunes A: Evolutionary
Genomics and Adaptive Evolution of the Hedgehog Gene Family (Shh,
Ihh and Dhh) in Vertebrates. PLoS One. 9:e741322014. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Joodi M, Amerizadeh F, Hassanian SM,
Erfani M, Ghayour-Mobarhan M, Ferns GA, Khazaei M and Avan A: The
genetic factors contributing to hypospadias and their clinical
utility in its diagnosis. J Cell Physiol. 234:5519–5523. 2019.
View Article : Google Scholar
|
|
187
|
Aurilio G, Cimadamore A, Santoni M, Nolè
F, Scarpelli M, Massari F, Lopez-Beltran A, Cheng L and Montironi
R: New frontiers in prostate cancer treatment: Are we ready for
drug combinations with novel agents? Cells. 9:15222020. View Article : Google Scholar :
|
|
188
|
Le V, He Y, Aldahl J, Hooker E, Yu EJ,
Olson A, Kim WK, Lee DH, Wong M, Sheng R, et al: Loss of androgen
signaling in mesenchymal sonic hedgehog responsive cells diminishes
prostate development, growth, and regeneration. PLoS Genet.
16:e10085882020. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Datta S and Datta MW: Sonic Hedgehog
signaling in advanced prostate cancer. Cell Mol Life Sci.
63:435–448. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Yamamichi F, Shigemura K, Behnsawy HM,
Meligy FY, Huang WC, Li X, Yamanaka K, Hanioka K, Miyake H, Tanaka
K, et al: Sonic hedgehog and androgen signaling in tumor and
stromal compartments drives epithelial-mesenchymal transition in
prostate cancer. Scand J Urol. 48:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Zhang X, Zhang Y, Lin F, Shi X, Xiang L
and Li L: Shh overexpression is correlated with GRP78 and AR
expression in primary prostate cancer: Clinicopathological features
and outcomes in a chinese cohort. Cancer Manag Res. 12:1569–1578.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Tzelepi V, Karlou M, Wen S, Hoang A,
Logothetis C, Troncoso P and Efstathiou E: Expression of hedgehog
pathway components in prostate carcinoma microenvironment: Shifting
the balance towards autocrine signalling: Hedgehog pathway in
prostate cancer. Histopathology. 58:1037–1047. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
U.S. Natinal Library of Medicine: A
Pre-surgical Study of LDE225 in Men With High-risk Localized
Prostate Cancer (LDE225). ClinicalTrials.gov Identifier: NCT02111187.
https://clinicaltrials.gov/ct2/show/NCT02111187.
Last updated March 7, 2019.
|
|
194
|
U.S. Natinal Library of Medicine: A Study
of Vismodegib in Men With Metastatic CRPC With Accessible
Metastatic Lesions for Tumor Biopsy. ClinicalTrials.gov Identifier: NCT02115828.
https://clinicaltrials.gov/ct2/show/results/NCT02115828.
Last updated July 20, 2018.
|
|
195
|
Antonarakis ES, Heath EI, Smith DC,
Rathkopf D, Blackford AL, Danila DC, King S, Frost A, Ajiboye AS,
Zhao M, et al: Repurposing itraconazole as a treatment for advanced
prostate cancer: A noncomparative randomized phase II trial in men
with metastatic castration-resistant prostate cancer. Oncologist.
18:163–173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Xie H, Paradise BD, Ma WW and
Fernandez-Zapico ME: Recent advances in the clinical targeting of
hedgehog/GLI signaling in cancer. Cells. 8:3942019. View Article : Google Scholar :
|
|
197
|
Kim JE, Singh RR, Cho-Vega JH, Drakos E,
Davuluri Y, Khokhar FA, Fayad L, Medeiros LJ and Vega F: Sonic
hedgehog signaling proteins and ATP-binding cassette G2 are
aberrantly expressed in diffuse large B-cell lymphoma. Mod Pathol.
22:1312–1320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Ciccone V, Morbidelli L, Ziche M and
Donnini S: How to conjugate the stemness marker ALDH1A1 with tumor
angiogenesis, progression, and drug resistance. Cancer Drug Resist.
3:26–37. 2020.
|
|
199
|
Bigelow RLH, Chari NS, Unden AB, Spurgers
KB, Lee S, Roop DR, Toftgard R and McDonnell TJ: Transcriptional
regulation of bcl-2 mediated by the sonic hedgehog signaling
pathway through gli-1. J Biol Chem. 279:1197–1205. 2004. View Article : Google Scholar
|
|
200
|
Li J, Xu J, Cui Y, Wang L, Wang B, Wang Q,
Zhang X, Qiu M and Zhang Z: Mesenchymal sufu regulates development
of mandibular molars via shh signaling. J Dent Res. 98:1348–1356.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Wang D, Nagle PW, Wang HH, Smit JK, Faber
H, Baanstra M, Karrenbeld A, Chiu RK, Plukker JTM and Coppes RP:
Hedgehog pathway as a potential intervention target in esophageal
cancer. Cancers (Basel). 11:8212019. View Article : Google Scholar
|
|
202
|
Cai H, Li H, Li J, Li X, Li Y, Shi Y and
Wang D: Sonic hedgehog signaling pathway mediates development of
hepatocellular carcinoma. Tumour Biol. Oct 15–2016.Epub ahead of
print. View Article : Google Scholar
|
|
203
|
Park AK, Lee JY, Cheong H, Ramaswamy V,
Park SH, Kool M, Phi JH, Choi SA, Cavalli F, Taylor MD and Kim SK:
Subgroup-specific prognostic signaling and metabolic pathways in
pediatric medulloblastoma. BMC Cancer. 19:5712019. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Polychronidou G, Kotoula V, Manousou K,
Kostopoulos I, Karayannopoulou G, Vrettou E, Bobos M, Raptou G,
Efstratiou I, Dionysopoulos D, et al: Mismatch repair deficiency
and aberrations in the Notch and Hedgehog pathways are of
prognostic value in patients with endometrial cancer. PLoS One.
13:e02082212018. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Katoh Y and Katoh M: Hedgehog target
genes: Mechanisms of carcinogenesis induced by aberrant hedgehog
signaling activation. Curr Mol Med. 9:873–886. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Jin W, Peng J and Jiang S: The epigenetic
regulation of embryonic myogenesis and adult muscle regeneration by
histone methylation modification. Biochem Biophys Rep. 6:209–219.
2016.PubMed/NCBI
|
|
207
|
Nasrallah I and Golden JA: Brain, eye, and
face defects as a result of ectopic localization of Sonic hedgehog
protein in the developing rostral neural tube. Teratology.
64:107–113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Castillo-Azofeifa D, Seidel K, Gross L,
Golden EJ, Jacquez B, Klein OD and Barlow LA: SOX2 regulation by
hedgehog signaling controls adult lingual epithelium homeostasis.
Development. 145:dev1648892018. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Henno P, Grassin-Delyle S, Belle E, Brollo
M, Naline E, Sage E, Devillier P and Israël-Biet D: In smokers,
Sonic hedgehog modulates pulmonary endothelial function through
vascular endothelial growth factor. Respir Res. 18:1022017.
View Article : Google Scholar : PubMed/NCBI
|