Introduction
Heart failure (HF) is becoming an increasingly
serious public health concern (1). Despite recent advances in
treatment, HF remains a fatal clinical syndrome. In the mouse, rat
and human adult heart, sarco/endoplasmic reticulum
Ca2+-ATPase (SERCA2a) is the major cardiac isoform,
which pumps Ca2+ from the cytosol to the sarcoplasmic
reticulum (SR) lumen utilizing the energy obtained by hydrolyzing
ATP. HF is associated with the decreased expression and activity of
SERCA2a (2-4). For this reason, SERCA2a has become
an attractive target for the gene targeted therapy of HF. The
abnormal calcium flux, and contraction and relaxation of
cardiomyocytes in a failing heart may be improved by the transfer
of SERCA2a (5). The
improvement in cardiac contractility following SERCA2a
transfer has been confirmed in a number of small and large animal
models of HF induced by pressure overload, volume overload,
ischemia, rapid ventricular pacing, or long-term isoproterenol
stimulation. In a porcine volume-overload HF model (6), rAAV1-mediated intracoronary gene
transfer in vivo has been reported to maintain the
contractile function and improve cardiac remodeling. In both
transgenic mice and rats, the overexpression of SERCA2 has
been shown to enhance calcium transients, myocardial contractility
and the relaxation in the absence or presence of pressure overload
(7-12). In addition to its beneficial
effects on myocardial contractility, the transfer of SERCA2a
revives energy metabolism in the heart (13-15), decreases the Ca2+ leak
from the SR (16), restores
electrical stability (17),
reduces arrhythmic aftercontractions (18), decreases ventricular arrhythmias
(16,19,20), suppresses cellular alternans
(21) and increases coronary
flow by activating endothelial nitric oxide synthase in endothelial
cells (22). Moreover, the
Calcium Upregulation by Percutaneous Administration of Gene Therapy
in Cardiac Disease (CUPID) study demonstrated the safety of
SERCA2a therapy in patients with advanced HF and unraveled
the benefits of this therapy (23-25). However, in the CUPID 2 study
(26), AAV1-SERCA2a did not
improve the clinical course of HF.
Misfolded proteins in the endoplasmic reticulum (ER)
can induce the unfolded protein response (UPR). The UPR is composed
of at least three branches (27). In resting cells, the three
ER-located stress sensors, namely double-stranded RNA-dependent
protein kinase (PKR)-like ER kinase (PERK), inositol-requiring 1α
(IRE1α), and activating transcription factor (ATF)6, are associated
with immunoglobulin heavy chain-binding protein (BiP) and are
maintained in an inactive state. In response to ER stress (ERS),
PERK phosphorylates the α subunit of the eukaryotic protein
synthesis initiation factor 2 (eIF2α), resulting in the inhibition
of translation of the majority of mRNAs, but allowing for the
translation of ATF4 mRNA. Under ERS conditions, IRE1α
autophosphorylates and activates its RNase activity, resulting in
the splicing of X-box binding protein-1 (XBP1) mRNA and the
production of an active spliced XBP1 isoform. In parallel,
following its release from BiP, ATF6 migrates to the Golgi
apparatus, where it is cleaved by site-1 protease (S1P) and site-2
protease (S2P). The functional cleaved fragment of ATF6 is then
released and migrates to the nucleus. The UPR leads to apoptosis
when cells fail to address the protein folding defects and cannot
re-establish homeostasis in the ER.
It has been shown that the UPR and nuclear factor
κ-light-chain-enhancer of activated B cells (NF-κB) interact at
multiple levels and are interconnected through the production of
reactive oxygen species (ROS), the release of calcium ion from the
ER, activation of NF-κB and c-Jun N-terminal kinases (JNK) and the
induction of the acute-phase response (27). Under ERS conditions, the
PERK-induced phosphorylation of eIF2α inhibits the translation of
nuclear factor of κ light polypeptide gene enhancer in B-cells
inhibitor-α (IκBα), decreasing the export of nuclear NF-κB to the
cytoplasm. In response to ERS, the phosphorylated cytoplasmic
domain of IRE1α recruits tumor necrosis
factor-α-receptor-associated factor 2 (TRAF2). The IRE1α-TRAF2
complex interacts with IκB kinase (IKK) and/or JNK to activate
these kinases. Activated IKK activates NF-κB through
phosphorylation of IκB, initiating the degradation of IκB.
Activated JNK activates the transcription factor activator
protein-1 (AP-1) through phosphorylation. Activated NF-κB and AP-1
translocate to the nucleus and induce the transcription of
inflammation-related genes. ATF6 can activate NF-κB through the
protein kinase B (Akt) pathway. In addition, the ERS-triggered
release of calcium from the ER and ROS can activate NF-κB (28).
Liu et al (29) revealed that the
cardiomyocyte-specific tamoxifen-inducible disruption of
SERCA2 induced ER/SR structural changes, UPR and apoptosis.
As also previously demonstrated, in a porcine myocardial ischemia
model, the overexpression of SERCA2a significantly attenuated the
activation of UPR and decreased ERS-associated apoptosis (30).
In the above context, it was hypothesized that the
overexpression of SERCA2a could attenuate ERS by maintaining
calcium homeostasis, thereby attenuating ERS-associated
inflammation. The present study was thus conducted to explore this
premise by overexpressing SERCA2a in neonatal rat cardiomyocytes
(NRCMs).
Materials and methods
Cell culture and experimental
protocol
All animal experiments were carried out in
accordance with the Guide for the Care and Use of Laboratory
Animals (8th Edition, 2011) of National Research Council (US)
(31) and approved by the
Institutional Animal Care and Use Committee (IACUC) of PLA General
Hospital (approval no. 2013-x7-28). The NRCMs were isolated from
1-day-old Sprague-Dawley rats. Pups were anesthetized with 5%
isoflurane and sacrificed by cervical dislocation. Hearts were
removed and immediately placed in cold phosphate-buffered saline
(NaCl 136.75 mmol/l, KCl 2.68 mmol/l, Na2HPO4
9.75 mmol/l, KH2PO4 1.47 mmol/l, glucose 5.50
mmol/l, pH 7.4). The ventricles were minced and digested with 0.15%
trypsin for 6-10 min at 37°C, and the supernatant was then
transferred to a centrifuge tube containing Dulbecco's modified
Eagle's medium (cat. no. 31600-034; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Shandong Yin Xiang Wei Ye
Group Co., Ltd.), 100 IU/ml penicillin, and 100 µg/ml
streptomycin (cat. no. 15140-122; Thermo Fisher Scientific, Inc.).
The digestion was repeated ~10 times. Following centrifugation for
10 min, the supernatant was aspirated off and the cell pellet was
resuspended in complete culture medium. The suspended cells were
plated and incubated in a 5% CO2, 37°C incubator for 1
h. Thereafter, the culture medium containing non-adherent cells was
collected, and these enriched cardiomyocytes were seeded in cell
culture flasks with 0.1 mmol/l 5-bromo-2-deoxyuridine added to the
medium to inhibit fibroblast proliferation. After two days, the
cardiomyocytes were trypsinized and counted; the aliquots of
cardiomyocyte suspension were then seeded. Following another day of
culture, the cells were kept in serum-free medium overnight for
cell cycle synchronization. On the following day, the NRCMs were
infected with adenoviral vectors carrying human SERCA2a or enhanced
green fluorescent protein (EGFP) gene at an MOI of 2 pfu/cell
(unless otherwise stated), the latter used as a control. At 48 h
following infection, the cells were subjected to the corresponding
treatments. Both rAd-SERCA2a and rAd-EGFP were purchased from
Beijing FivePlus Molecular Medicine Institute Co., Ltd.
In the ERS model induced by tunicamycin (TM), the
NRCMs were assigned to one of the four groups: i) The vehicle
control, dimethyl sulfoxide was added to the complete culture
medium; ii) the TM group, the culture medium was changed to fresh
complete culture medium with 10 µg/ml TM; iii) the TM +
rAd-EGFP group, at 48 h following rAd-EGFP infection, the culture
medium was changed to fresh complete culture medium with 10
µg/ml TM; and iv) the TM + rAd-SERCA2a group, at 48 h
following rAd-SERCA2a infection, the culture medium was changed to
fresh complete culture medium with 10 µg/ml TM.
In another ERS model induced by
hypoxia/reoxygenation (H/R), following infection with adenoviral
vectors for 48 h, for the control group, the culture medium was
replaced with fresh complete medium and the culture flask was
maintained in a normal cell culture incubator with 95% air and 5%
CO2; by contrast, for the H/R, H/R + rAd-EGFP, and H/R +
rAd-SERCA2a groups, the culture medium was replaced with fresh
low-glucose DMEM medium (cat. no. 31600-034; Thermo Fisher
Scientific, Inc.) without calf serum, and the flasks were
transferred to a tri-gas incubator (5% O2, 5%
CO2, 90% N2) (Thermo Fisher Scientific, Inc.)
for 8 h, and then returned to a normal cell culture incubator for
16 h, without changing the serum-free low-glucose DMEM medium.
Cell viability assessment
Cell viability was assessed using the Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to
the manufacturer's instructions. Briefly, to each well seeded with
the cells, 10 µl of CCK-8 solution was added, avoiding the
formation of bubbles during the process. The plate was then
incubated at 37°C for 1 h. The absorbance of solution in each well
was detected at 450 nm using a microplate reader (SN: 1106007713;
Tecan Group Ltd.).
Lactate dehydrogenase (LDH) activity
assay
The extent of cellular injury was assessed by
measuring the release of LDH in the culture medium using a
commercial kit (JianCheng Bioengineering Institute). The LDH
activity was quantified by measuring the level of pyruvic acid at
450 nm using a Tecan microplate reader.
Determination of the optimal multiplicity
of infection (MOI)
O'Donnell et al (32) found out that the exogenous
expression of SERCA in the NRCMs reduced the viability of the
cells, with cell floaters occurring even if the MOI was as low as 5
pfu/cell. The apoptotic index in myocytes infected with adenoviral
vectors carrying the wild-type SERCA1 gene was 7% at 2 pfu/cell and
31% at 10 pfu/cell. The expression of exogenous SERCA and
acceleration of Ca2+ transients could be achieved with
minimal cell damage in rat myocytes when the MOI was in the range
of 1 to 4 pfu/cell. O'Donnell et al (32) also performed in situ
immunofluorescence staining with specific antibodies against the
exogenous SERCA1. It was found out that SERCA1 was densely packed
within sarco/endoplasmic reticulum even in apparently normal cells.
Severe structural changes occurred in cytopathic cells. It should
be highlighted that both wild-type SERCA and inactive SERCA mutant
produced cytotoxic effects. Thus, the investigators proposed that
the dense accumulation of SERCA within a very limited
sarco/endoplasmic reticulum space will disturb membrane structure
and function and perturb calcium homeostasis (32). Therefore, the present study
decided to perform a titration test on the MOIs in the NRCMs
transfected with rAd-SERCA2a, with the expression level of SERCA2a,
cell viability and LDH in the cell culture supernatant evaluated.
The present study hoped to determine a certain MOI value, at which
the high expression of SERCA2a could be achieved, while the
cytotoxicity would be minimized to prevent the impact on cell
inflammation.
Electrophoretic mobility shift assay
(EMSA)
Nuclear extracts were prepared using the NProtein
Extraction kit (Exprogen Biotechnologies, Inc.). The sequences of
the probes used for the assay were as follows: Ds-Bio-NF-κB probe,
Bio-5′-AGT TGA GGG GAC TTT CCC AGG C-3′-Bio; Ds-Bio-AP1 probe,
Bio-5′-CGC TTG ATG AGT CAG CCG GAA-3′-Bio; Ds-Bio-OCT1 probe,
Bio-5′-TGT CGA ATG CAA ATC ACT AGAA-3′-Bio. EMSA was carried out
using the BiotinLight™ Chemiluminescent EMSA kit (Exprogen
Biotechnologies, Inc.) according to the instruction manual.
Competition experiments with 100-fold excess of unlabeled probe
used as a specific competitor were performed to confirm the
specificity of protein-DNA binding. Antibodies against NF-κB p50
(cat. no. sc-1190), NF-κB p65 (cat. no. sc-372), c-Jun (cat. no.
sc-1694), and c-Fos (cat. no. sc-52) were purchased from Santa Cruz
Biotechnology, Inc. A total of 4 µl of undiluted antibodies
were added to 15-µl binding reactions. The samples were
incubated for 20 min at room temperature. The sample was
electrophoresed on a 1% agarose gel in 0.5X Tris-borate-EDTA buffer
at 120 V for 1.5 h, and then electrophoretically blotted onto a
nylon membrane at 380 mA for 1 h. The membrane was cross-linked in
a UV-light cross-linker (Analytik Jena AG) for 10 min, and the
biotin-labeled DNA was detected by chemiluminescence.
Western blot analysis
Primary antibodies against BiP (cat. no. 3183;
1:1,000), phospho-PERK (cat. no. 3179; 1:1,000), PERK (cat. no.
3192; 1:1,000), phospho-eIF2α (cat. no. 3398; 1:1,000), eIF2α (cat.
no. 9722; 1:1,000), phospho-NF-κB p65 (Ser536) (cat. no. 3033;
1:1,000), NF-κB p65 (cat. no. 8242; 1:1,000), SERCA2 (cat. no.
9580; 1:1,000) and histone H3 (cat. no. 4499; 1:2,000) were
purchased from Cell Signaling Technology, Inc., those against
phospho-IRE1 (cat. no. ab48187; 1:1,000) and caspase-12 (cat. no.
ab62484; 1:500) were from Abcam, that against IRE1 (cat. no.
NB100-2324; 1:1,000) was from Novus Biologicals, LLC, that against
CHOP (cat. no. sc-7351; 1:200) was from Santa Cruz Biotechnology,
Inc. and that against GAPDH (cat. no. 60004-1-Ig; 1:2,000) was from
Proteintech Group, Inc. HRP-conjugated secondary antibodies of goat
anti-mouse IgG (cat. no. sc-2005; 1:3,000) and goat anti-rabbit IgG
(cat. no. sc-2004; 1:3,000) were purchased from Santa Cruz
Biotechnology, Inc. Whole-cell extracts were prepared using
radioimmunoprecipitation assay lysis buffer (cat. no. CW2333;
Beijing Cowinbioscience Co., Ltd.) containing a protease inhibitor
cocktail (cat. no. CW2200; Beijing Cowinbioscience Co., Ltd.) and
phosphatase inhibitors (cat. no. CW2383; Beijing Cowinbioscience
Co., Ltd.). Cytoplasmic and nuclear extracts were prepared using
the NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (cat. no.
78833; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The protein concentration in each
sample was determined using the BCA Protein Assay kit (cat. no.
CW0014; Beijing Cowinbioscience Co., Ltd.) with bovine serum
albumin as a standard. Equal amounts of protein (100 µg)
lysate per sample were denatured in 5X Sodium Dodecyl
Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) loading
buffer (cat. no. CW0027; Beijing Cowinbioscience Co., Ltd.).
Denatured proteins were separated on an 8-12% resolving gel and
transferred onto nitrocellulose membranes (Pall Life Sciences)
using a semidry transfer apparatus (Beijing Liuyi Biotechnology
Co., Ltd.). After being blocked with 5% bovine serum albumin (cat.
no. 0332-100G; Amresco Inc.) in Tris-buffered saline with 0.1%
Tween-20 (TBST) for 1 h at room temperature, the membranes were
probed with primary antibodies with gentle agitation overnight at
4°C. After washing with TBST buffer, the membranes were incubated
with appropriate HRP-conjugated secondary antibodies at room
temperature for 1 h. After washing three times with TBST buffer,
immunolabeled bands were detected by enhanced chemiluminescence.
The integrated optical density (IOD) of the analyzed bands on the
film was quantified using ImageJ software (National Institutes of
Health; version 1.46). GAPDH and histone H3 served as cytoplasmic
and nuclear internal controls, respectively. The levels of analyzed
proteins were normalized to those of the internal control.
Semi-quantitative RT-PCR
Total RNA was isolated using the TRNzol method (cat.
no. DP421; Tiangen Biotech Co., Ltd.). RNA (2 µg) was
reverse transcribed with TransScript® First-Strand cDNA
Synthesis SuperMix (cat. no. AT301; TransGen Biotech Co., Ltd.).
The forward and reverse primers used for PCR were as follows: Tumor
necrosis factor (TNF)-α forward, 5′-CGT AGC CCA CGT CGT AGC AAA
CCA-3′ and reverse, 5′-CGC CAG TCG CCT CAC AGA GCA AT-3′; XBP-1(u)
forward, 5′-CTG GAG CAG CAA GTG GTG GAT TT-3′ and reverse, 5′-GTC
CTT CTG GGT AGA CCT CTG GGAG-3′; XBP1(s) forward, 5′-CTG AGT CCG
CAG CAG GTGC-3′ and reverse, 5′-CAG GGT CCA ACT TGT CCA GAA TG-3′;
GAPDH forward, 5′-TGC TGA GTA TGT CGT GGAG-3′ and reverse, 5′-GTC
TTC TGA GTG GCA GTGAT-3′. The primers were purchased from Sangon
Biotech Co., Ltd. The PCR reaction conditions were as follows:
94°C, 3 min; (4°C, 30 sec; 55°C, 30 sec; 72°C, 1 min) ×30 cycles;
72°C, 5 min. The amplified products were separated on 1.5% agarose
gels mixed with GoodView™ nucleic acid dye (cat. no. GV-2; Beijing
SBS Genetech Co., Ltd.). Following electrophoresis, the agarose gel
was visualized on a UV transilluminator and photographed. The IODs
of the bands observed on the image were quantified using ImageJ
software (National Institutes of Health; version 1.46). The
transcription levels of the analyzed genes were normalized to those
of GAPDH.
Statistical analysis
Data are expressed as the mean ± SD. Statistical
analyses of the data were carried out by one-way ANOVA, followed by
post hoc Tukey's tests. A value of P<0.05 was considered to
indicate a statistically significant difference. All analyses were
performed using SPSS 19.0 software (IBM, Inc.).
Results
Overexpression of SERCA2a attenuates the
upregulation of nuclear NF-κB and AP-1 DNA-binding activities
following treatment of NRCMs with TM
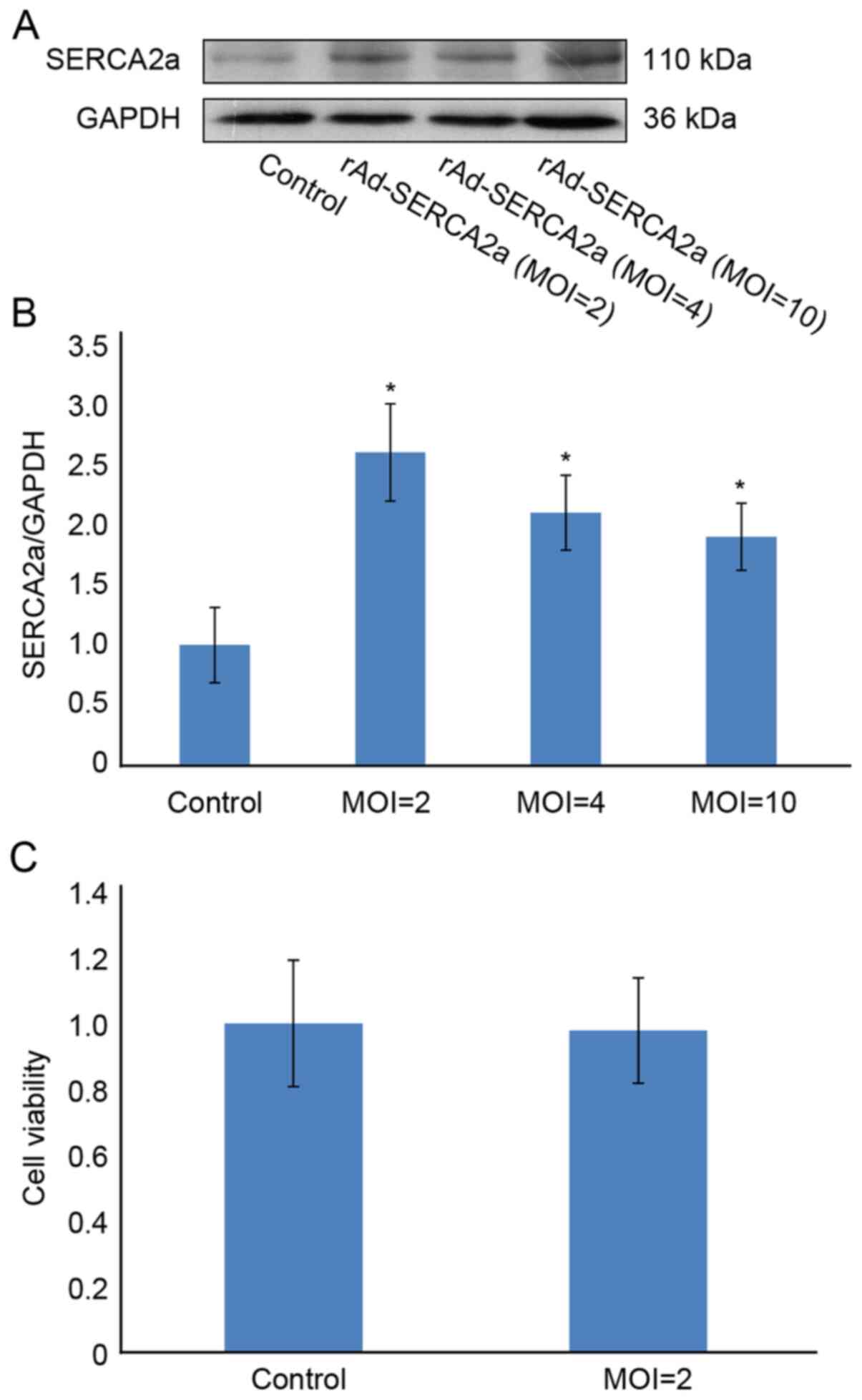
At MOIs of 2, 4 and 10 pfu/cell, the expression
level of SERCA2a was increased by 160, 110 and 90%, respectively,
compared with that of the control group (Fig. 1). At an MOI of 2 pfu/cell, the
viability of the NRCMs was 97.8%, similar to that of the control
group. Unless otherwise stated, 2 pfu/cell was used as the
preferred MOI in the subsequent experiments. When the MOI was >2
pfu/cell, the expression of SERCA2a was decreased, suggesting that
the overexpression of SERCA2a may be cytotoxic. Following treatment
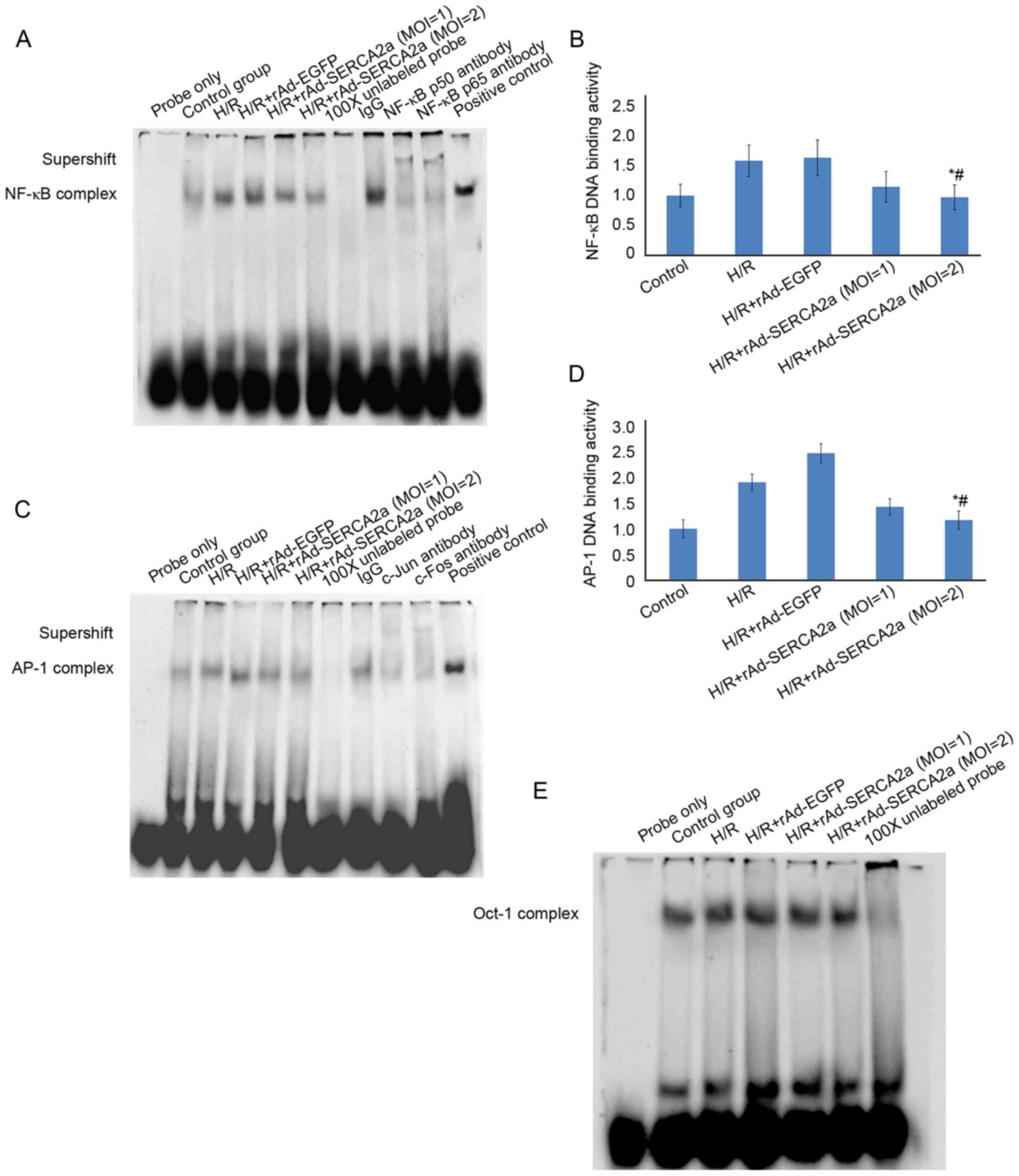
with TM for 24 h, the DNA-binding activity of NF-κB in the TM group
was increased by 4.1-fold (P<0.01; Fig. 2). Compared with the TM + rAd-EGFP
and TM groups, the DNA-binding activity of NF-κB in the TM +
rAd-SERCA2a group was decreased by 43.6% (P<0.01) and 66.0%
(P<0.01), respectively. Following treatment with TM for 24 h,
the DNA-binding activity of AP-1 in the TM group was increased by
26.9-fold (P<0.01). Compared with the TM + rAd-EGFP and TM
groups, the DNA-binding activity of AP-1 in the TM + rAd-SERCA2a
group was decreased by 60.2% (P<0.01) and 26.3% (P<0.01),
respectively.
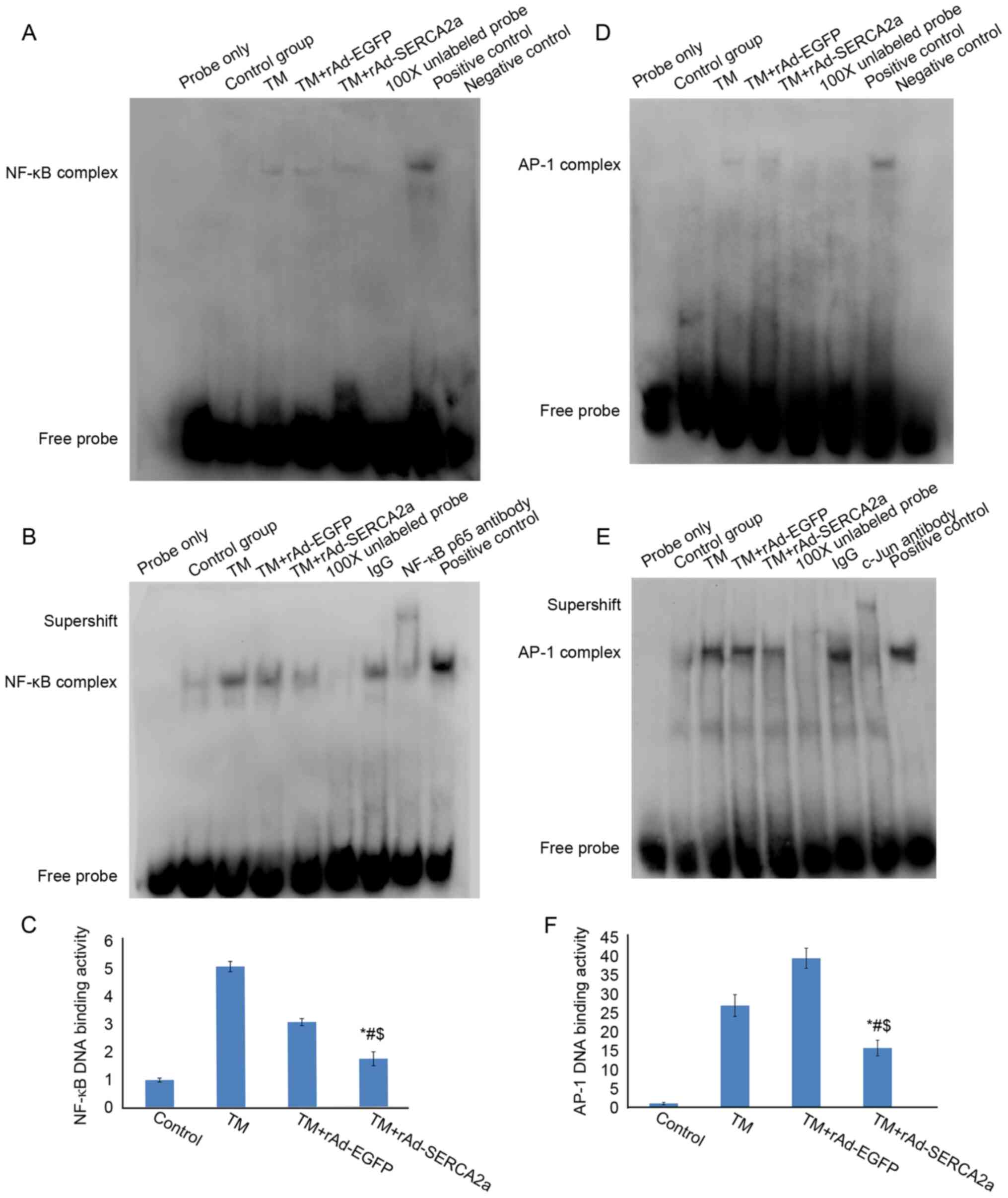 | Figure 2Effects of the overexpression of
SERCA2a on the nuclear NF-κB and AP-1 DNA-binding activities in
NRCMs following treatment with TM for 8 and 24 h, as assessed by
EMSA. (A) EMSA gel showing the NF-κB DNA-binding activity following
treatment with TM for 8 h. The amount of nuclear extract loaded was
3.5 µg. The eight lanes from left to right represent the
following: Blank control (probe only), vehicle control group
(Control), tunicamycin group (TM), tunicamycin + rAd-EGFP group (TM
+ rAd-EGFP), tunicamycin + rAd-SERCA2a group (TM + rAd-SERCA2a),
100X cold probe, positive control and negative control,
respectively. (B) EMSA gel showing the NF-κB DNA-binding activity
following treatment with TM for 24 h. The amount of nuclear extract
loaded was 10 µg. The nine lanes from left to right
represent the blank control (probe only), vehicle control group
(Control), TM, TM + rAd-EGFP, TM + rAd-SERCA2a, 100X cold probe,
non-specific IgG antibody, NF-κB p65 antibody and positive control,
respectively. (C) The NF-κB complex bands from panel B were
analyzed by densitometry using ImageJ software. (D) EMSA gel
showing the AP-1 DNA-binding activity following treatment with TM
for 8 h. The amount of nuclear extract loaded was 3.5 µg.
(E) EMSA gel showing the AP-1 DNA-binding activity following
treatment with TM for 24 h. The amount of nuclear extract loaded
was 10 µg. (F) The AP-1 complex bands from panel E were
analyzed by densitometry using ImageJ software. Data are
representative of three independent experiments (mean ± SD).
*P<0.01 vs. TM + rAd-EGFP; #P<0.01 vs.
TM; $P<0.05 vs. Control. SERCA2a, sarco/endoplasmic
reticulum Ca2+-ATPase; AP-1, activator protein-1; NRCMs,
neonatal rat cardiomyocytes; TM, tunicamycin; EMSA, electrophoretic
mobility shift assay; EGFP, enhanced green fluorescent protein. |
Overexpression of SERCA2a at an MOI of 1
pfu/cell still attenuates the upregulation of nuclear NF-κB and
AP-1 DNA-binding activities following treatment of NRCMs with
TM
As previously demonstrated, under limited exposure
to calf serum, compared with the non-infected control group, the
size, protein content and protein synthesis rate in the infected
rat myocytes exhibited a more rapid increase (32). Tauroursodeoxycholic acid (TUDCA),
a recognized ERS inhibitor, was used as a control in this
experiment. The synchronization time was delayed to that prior to
the addition of TM (final concentration, 10 µg/ml), instead
of that prior to infection. Considering that NRCMs are prone to the
ER overload response (EOR) induced by rAd-SERCA2a infection, the
MOI was reduced to 1.0 pfu/cell. Compared with the TM + rAd-EGFP
and TM groups, the overexpression of SERCA2a significantly
attenuated the upregulation of NF-κB (both P<0.05) and the AP-1
DNA-binding activities (both P<0.05), respectively. The results
were similar to those observed at 2 pfu/cell (Fig. 3). The results of EMSA revealed
that TUDCA significantly attenuated the upregulation of NF-κB and
AP-1 DNA-binding activities induced by TM, corroborating the
successful construction of the cellular model of ERS. The
supershift assays revealed that activated NF-κB in the nucleus
contained p50 and p65 subunits, and activated AP-1 in the nucleus
contained c-Jun and c-Fos subunits (Fig. 3).
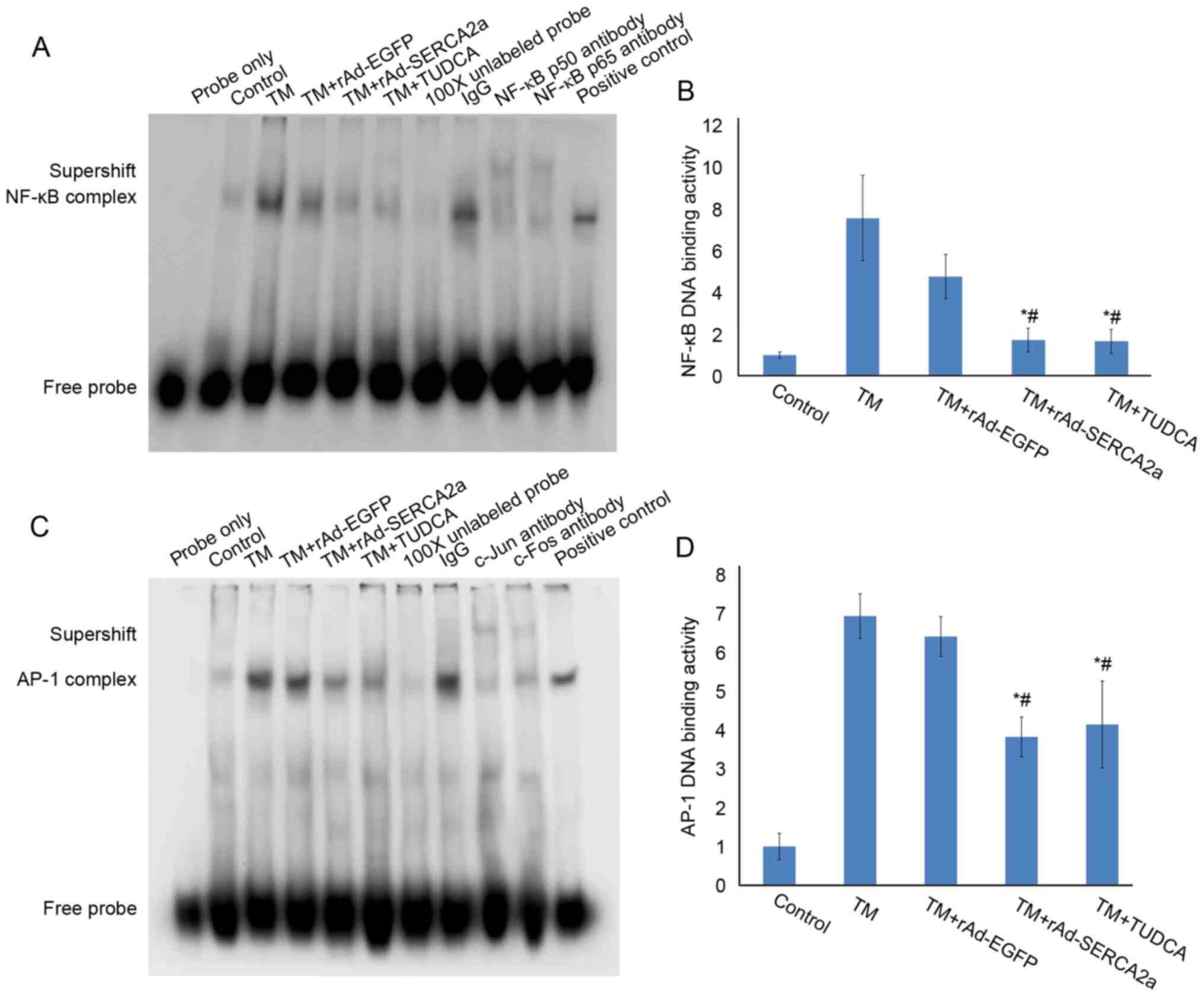 | Figure 3Effects of overexpression of SERCA2a
at 1 pfu/cell on the NF-κB and AP-1 DNA-binding activities in NRCMs
following treatment with TM for 24 h when the synchronization time
was delayed to that prior to treatment with TM. (A) EMSA gel
showing the NF-κB DNA-binding activity. The lanes from left to
right represent the following: Blank control (probe only), vehicle
control group (Control), TM, TM + rAd-EGFP, TM + rAd-SERCA2a, TM +
TUDCA (500 µmol/l), 100X cold probe, IgG, NF-κB p50
antibody, NF-κB p65 antibody and positive control, respectively.
(B) NF-κB complex bands from (A) were analyzed by densitometry
using ImageJ software. (C) EMSA gel showing the AP-1 DNA-binding
activity. The lanes from left to right represent blank control
(probe only), vehicle control (Control), TM, TM + rAd-EGFP, TM +
rAd-SERCA2a, TM + TUDCA, 100X cold probe, IgG, c-Jun antibody,
c-Fos antibody and positive control, respectively. (D) The AP-1
complex bands from panel C were analyzed by densitometry using
ImageJ software. Data are representative of three independent
experiments (mean ± SD). *P<0.05 vs. TM + rAd-EGFP;
#P<0.05 vs. TM. SERCA2a, sarco/endoplasmic reticulum
Ca2+-ATPase; AP-1, activator protein-1; NRCMs, neonatal
rat cardiomyocytes; TM, tunicamycin; EMSA, electrophoretic mobility
shift assay; EGFP, enhanced green fluorescent protein; TUDCA,
tauroursodeoxycholic acid. |
Overexpression of SERCA2a attenuates the
upregulation of nuclear NF-κB and AP-1 DNA-binding activities
induced by H/R
In the H/R model, the overexpression of SERCA2a
significantly attenuated the upregulation of NF-κB and AP-1
DNA-binding activities (Fig. 4),
similar to the findings observed with the TM model.
Overexpression of SERCA2a attenuates the
activation of the IRE1α signaling pathway induced by TM in the
NRCMs
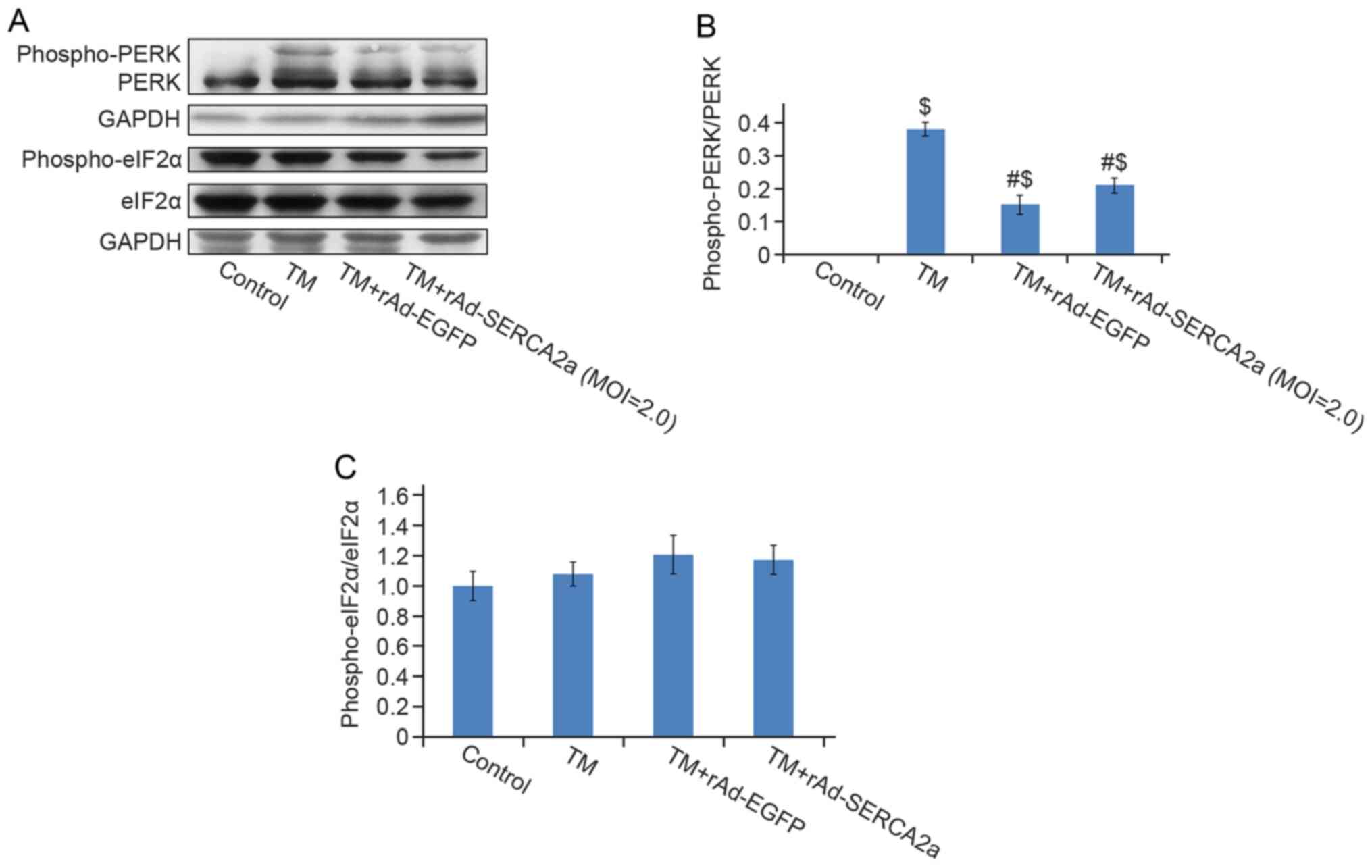
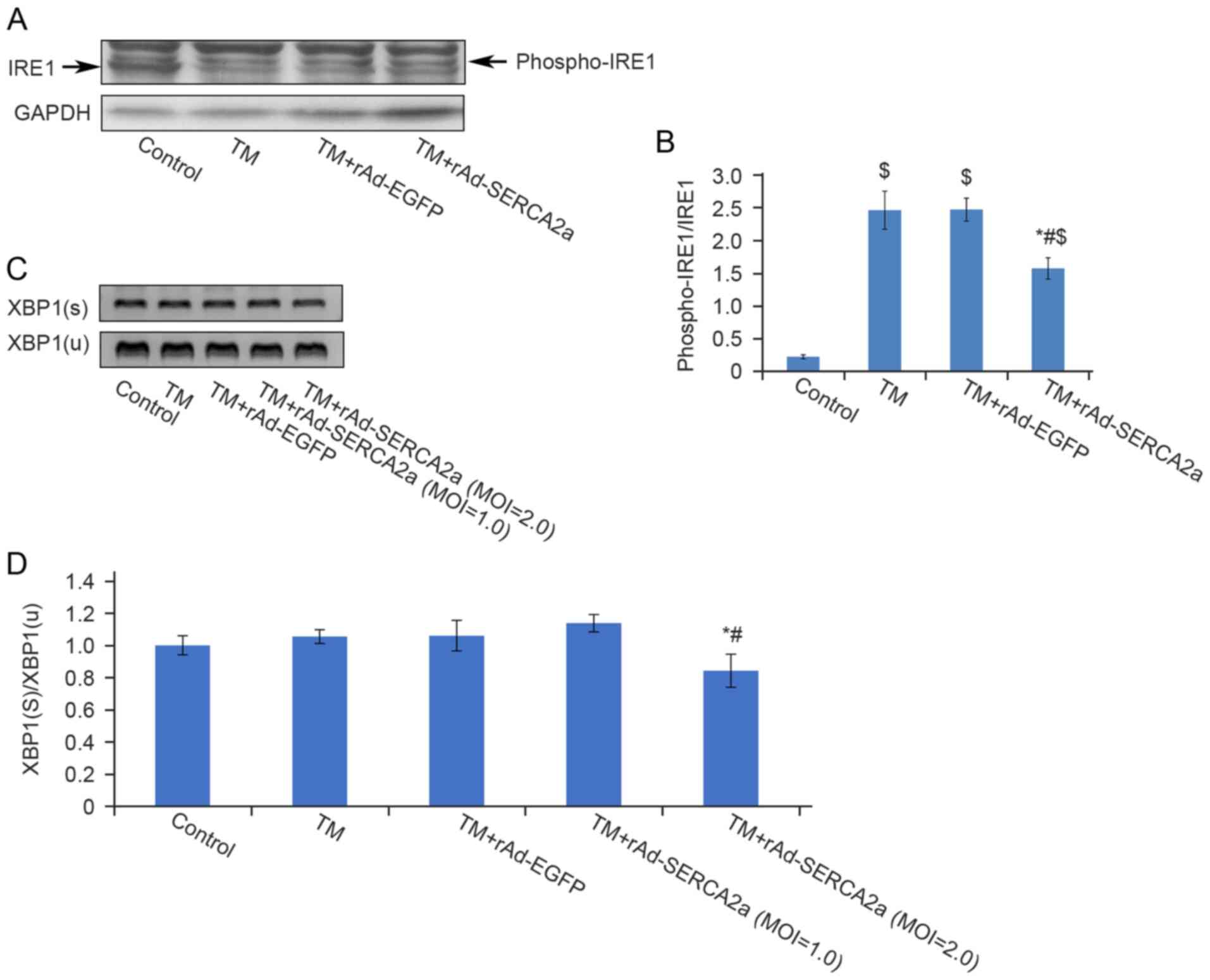
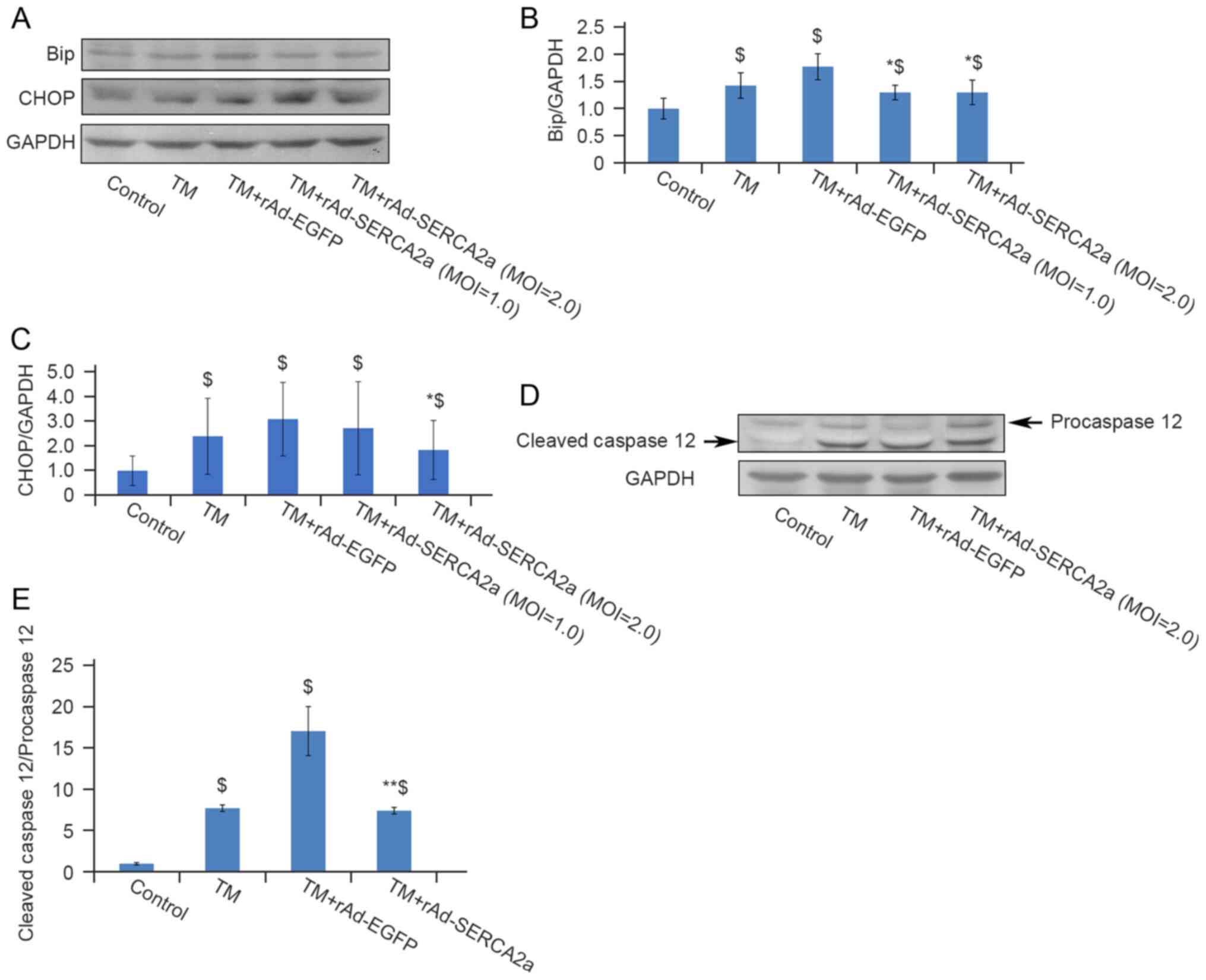
Compared with the vehicle control group, the protein
levels of phospho-PERK (Thr980) (Fig. 5), phospho-IRE1 (Ser724) (Fig. 6), BiP, CHOP and cleaved
caspase-12 (Fig. 7) in the TM
group were significantly increased. No significant decreases were
observed in the phospho-PERK (Thr980) and phospho-eIF2α (Ser51)
levels in the TM + rAd-SERCA2a group compared with the TM +
rAd-EGFP group (Fig. 5).
Compared with the TM + rAd-EGFP and TM groups, the ratio of
phospho-IRE1 to unphosphorylated IRE1 in the TM + rAd-SERCA2a group
was decreased by 25% (P<0.01) and 31.8% (P<0.01),
respectively (Fig. 6). The
results of semi-quantitative RT-PCR revealed that compared with the
TM + rAd-EGFP and TM groups, the ratio of spliced active XBP1 to
unspliced inactive XBP1 in the TM + rAd-SERCA2a (MOI=2.0) group was
reduced by 20.5% (P<0.01) and 20% (P<0.05), respectively.
Overexpression of SERCA2a attenuates
ERS-associated apoptosis
BiP, also known as Grp78, is one of the molecular
markers of ERS. The overexpression of SERCA2a decreased the
expression of BiP, compared with that in the TM + rAd-EGFP group.
CHOP (also known as GADD153) and caspase-12 are relevant to
ERS-associated apoptosis. Compared with the TM + rAd-EGFP group,
the expression of CHOP and the ratio of cleaved caspase-12 to
pro-caspase-12 in the TM + rAd-SERCA2a group were decreased by 40%
(P<0.05) and 56% (P<0.01), respectively (Fig. 7). Compared with the TM group, the
expression of CHOP and the ratio of cleaved caspase-12 to
pro-caspase-12 were decreased by 23% (P>0.05) and 3.9%
(P>0.05), respectively. These findings indicated that the
overexpression of SERCA2a attenuated ERS-associated apoptosis.
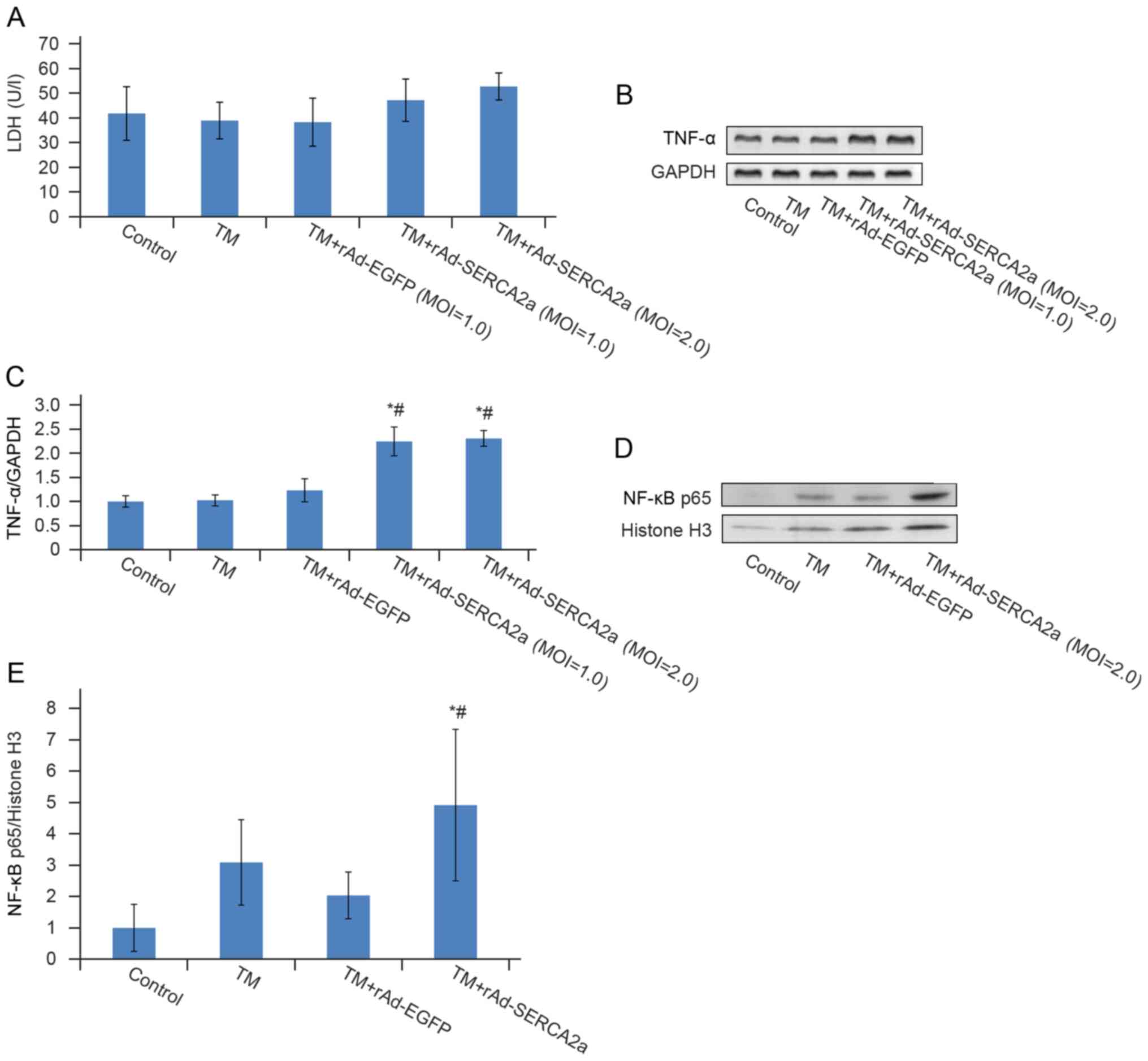
Overexpression of SERCA2a induces
EOR
The overexpression of molecules resident in the ER
can lead to EOR. EOR is characterized by NF-κB activation. In the
TM + rAd-SERCA2a group, the nuclear translocation of NF-κB was
significantly increased by 1.40-fold (P<0.05), the transcription
level of TNF-α increased by 87.4% (P<0.05) and LDH leakage
exhibited an increasing trend (P>0.05) compared with the TM +
rAd-EGFP group, which suggested that the overexpression of SERCA2a
induced EOR (Fig. 8).
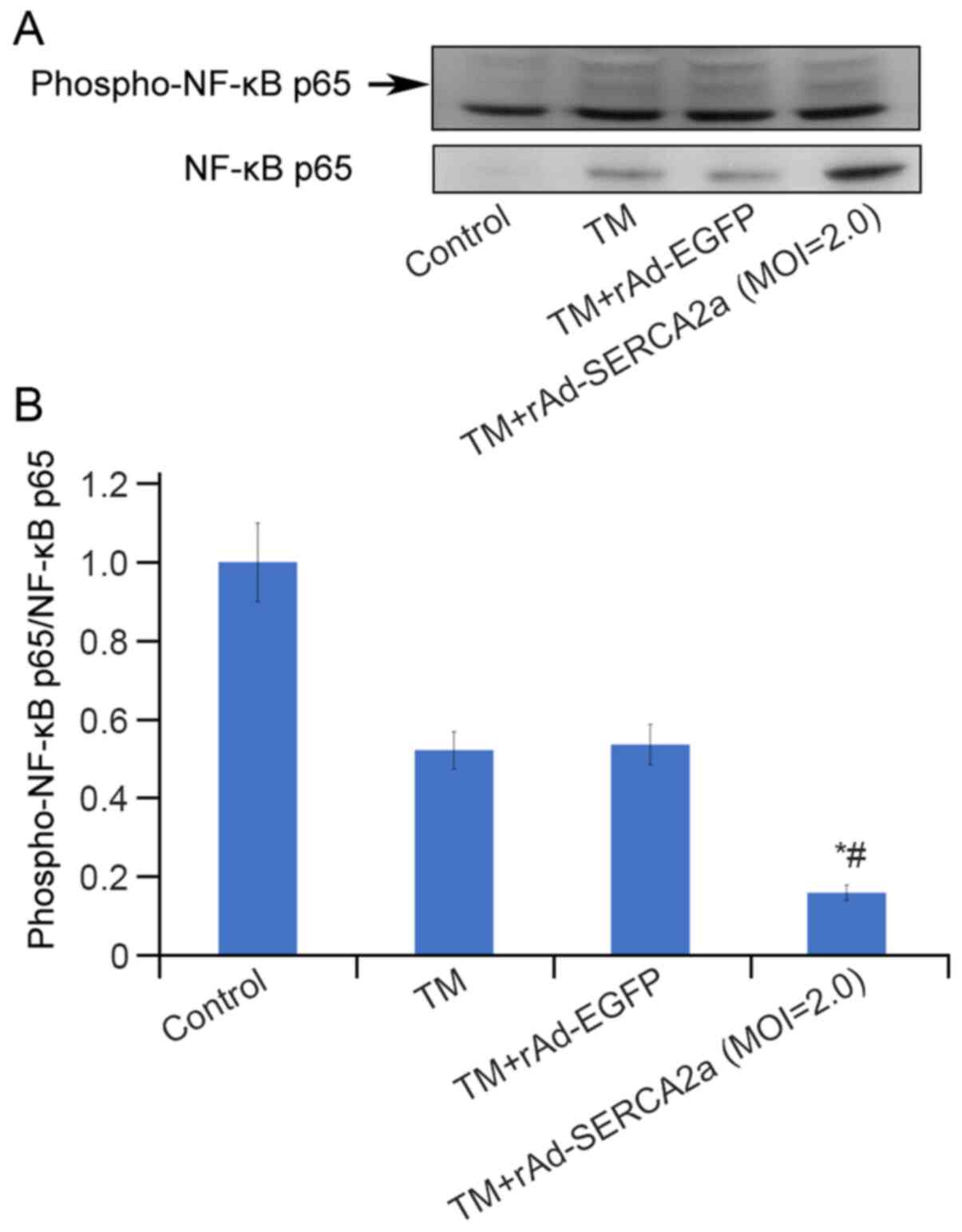
Overexpression of SERCA2a decreases the
level of nuclear phospho-p65 (Ser536)
The increase in the NF-κB p65 nuclear translocation
and the attenuation of the upregulation of NF-κB p65 DNA-binding
activity due to the overexpression of SERCA2a appeared paradoxical.
To address this issue, the effects of overexpression of SERCA2a on
post-translational modifications of NF-κB p65 were further
explored. Compared with that in the TM + rAd-EGFP group, the ratio
of nuclear phospho-NF-κB p65 (ser536) to NF-κB p65 in the TM +
rAd-SERCA2a group was significantly decreased by 59.6% (P<0.05;
Fig. 9).
Discussion
H9c2 cells lack NF-κB p50 expression (33); therefore, this cell line was not
selected as the study object. TM blocks N-linked glycosylation and
is widely used to induce UPR. In this cell-based study,
ERS-associated inflammation was induced by TM, thus preventing
interference from tissue and circulating immune cells.
The present study demonstrated that TM induced a
significant increase in the NF-κB DNA-binding activity and in the
nuclear translocation of NF-κB. The addition of the ERS protectant,
TUDCA, prior to treatment with TM significantly attenuated the
upregulation of DNA-binding activity of NF-κB and AP-1. These
findings indicate that the cellular TM-induced ERS model was
successfully constructed.
Hamid et al (33) revealed that in HF, persistent
activation of NF-κB p65 in myocytes aggravates ventricular
remodeling by conferring pro-inflammatory, profibrotic and
pro-apoptotic effects. It appears important to control the
activation of NF-κB in HF. The UPR and NF-κB are interconnected
through various mechanisms. In the present study, it was found that
the overexpression of SERCA2a attenuated ERS and the activation of
the IRE1α signaling pathway in the NRCMs induced by TM, resulting
in the attenuation of the upregulation of NF-κB and AP-1
DNA-binding activities.
The accumulation of wild-type or misfolded proteins
in the ER results in the release of Ca2+ from the ER.
This leads to the generation of ROS, activating NF-κB. This process
is called the EOR (34). Some
viral proteins, such as the virion surface hemagglutinin (35), C-terminal truncation of the
middle surface antigen from hepatitis B virus (36), adenovirus E3/19K protein
(37) and human hepatitis C
virus NS5A protein (38), can
cause the EOR. The overexpression of SERCA2a in COS cells increases
the calcium uptake rate; however, the overexpression of SERCA2a
also induces cellular calcium overload and death (39). O'Donnell et al (32) proposed that in neonatal
cardiomyocytes, the SR system was not well developed, and the SR
volume was limited. A several-fold increase in SERCA within 2- to
3-day period can induce the dense accumulation of SERCA molecules
in the limited SR space and leads to the disorder of membrane
structure and function, resulting in perturbation of calcium
homeostasis (32). These earlier
findings indicate that exogenous expression of SERCA can cause EOR,
although NF-κB activation and TNF-α transcription have not been
investigated. The window of MOIs between exogenous gene expression
and production of cellular toxicity is narrower for the
overexpression of SERCA than for EGFP. Wu et al found that
at an MOI of 4 pfu/cell, the overexpression of SERCA1 induced the
loss of NRCMs and DNA fragmentation (40). O'Donnell et al (32) suggested that the optimal MOI of
adenoviral vector carrying wild-type SERCA1 is in the range of 2 to
4 pfu/cell in NRCMs. This titer increased SERCA activity by
>2-fold and enhanced the kinetics of Ca2+
transients.
The present study identified 2 pfu/cell as the
preferred MOI based on the expression level of exogenous SERCA2a,
cell viability and LDH leakage, thus minimizing the cytopathic
effects. However, at this MOI, the detachment of cells can still be
observed under an inverted microscope. It was found that the
nuclear translocation of NF-κB p65 in the TM + rAd-SERCA2a group
was significantly increased following treatment with TM compared
with that in the TM + rAd-EGFP group, which was consistent with the
occurrence of the EOR.
However, the mechanisms through which the
accumulation of proteins in the ER membrane increase
Ca2+ permeability remain unclear. Pahl (34) proposed that the accumulation of
membrane proteins may impair SERCA function, or the Ca2+
permeability of the ER membrane may be aggravated due to an
increase in the protein-to-lipid ratio. As earlier cell-based
studies have demonstrated that the overexpression of SERCA2a can
enhance its pump function, the latter possibility is more
reasonable in the case of overexpression of SERCA2a-induced
EOR.
Hu et al (41) confirmed that the production of
TNF-α induced by ER stress was dependent on IRE1α and NF-κB. The
inhibition of the TNF receptor 1 signaling pathway significantly
decreased ER stress-associated cell death (41). Hamid et al (42) demonstrated that TNFR1 augmented
the activation of NF-κB in H9c2 cells, and the pro-apoptotic
effects of NF-κB overexpression required TNF elaboration and
concomitant TNFR1 signaling. The present study demonstrated an
increase in TNF-α transcription in the group overexpressing SERCA2a
(Fig. 8), and it was thus
hypothesized that EOR induced NF-κB p65 activation, which in turn
induced an increase in TNF-α transcription. The transcription level
of TNF-α was not significantly altered following TM treatment in
the TM group, which may be due to IRE1-dependent decay of mRNA
(RIDD) induced by ERS. It would thus be ideal to perform real-time
fluorescent quantitative PCR at different time points to further
verify this finding.
In view of the paradox between the increase in NF-κB
nuclear translocation and the attenuation of the upregulation of
NF-κB DNA-binding activity, it was hypothesized that different
post-translational modifications may account for this issue. RelA
is phosphorylated at Ser536 by IKKβ, IKKα, IKKε, NF-κB activating
kinase and RSK1. The stimulatory modifications of RelA enhance the
transcriptional activity and capability of interaction with
coactivators, such as histone acetyltransferase p300 (p300) and
CREB-binding protein (CBP) (43). p300 and CBP acetylate RelA at
several sites. Acetylation of K310 is necessary for complete
transcriptional activity of NF-κB. The acetylation of K221
increases the DNA-binding affinity of RelA for κB sites. The
present exploratory study revealed a reduction in the level of
phosphorylated P65 (Ser536) in the group overexpressing SERCA2a;
however, the details of further post-translational modifications
warrant further investigations. NF-κB luciferase reporter assays
should be helpful in clarifying the effects of the overexpression
of SERCA2a on the transcriptional activity of NF-κB.
Sensitivity to subsequent TNF stimulation is
lessened with pre-exposure to TNF, which is known as the 'TNF
tolerance phenomenon'. Zwergal et al (44) demonstrated that
CCAAT-enhancer-binding proteins (C/EBP) is necessary for the
inhibition of NF-κB induced transcription in TNF-tolerant cells,
which is mediated by the inhibition of p65 phosphorylation. Hu
et al (41) revealed that
ERS induced the downregulation of TRAF2 expression, leading to the
attenuation of the TNF-induced activation of NF-κB and JNK.
The studies by Kitamura (45), and Nakajima and Kitamura
(46) reported that preceding
ERS may attenuate the subsequent activation of NF-κB by
inflammatory cytokines and reviewed several possibilities. ERS can
induce the selective degradation of TRAF2 (a key component involved
in the TNF signaling), thereby inhibiting NF-κB activation by
TNF-α. ERS can also induce the expression of C/EBPβ, which
interacts with the NF-κB p65 subunit. The C/EBPβ-p65 complexes
contribute to the inhibition of activation of NF-κB by cytokines.
In addition, ERS can induce the production of alpha induced protein
3 (A20), IκBα, GRP78, and NO and dephosphorylation of Akt, which
are involved in the suppression of NF-κB through various
mechanisms.
In a preliminary experiment, it was found out that
the expression level of TRAF2 was significantly reduced in the
group overexpressing SERCA2a (data not shown); however, further
repeated experiments are required to confirm this conclusion. It
was hypothesized that the preceding EOR induced by accumulation of
exogenous SERCA2a in sarcoplasmic reticulum might precondition the
cells against subsequent TM-induced upregulation of NF-κB and AP-1
DNA-binding activities. Further studies to investigate C/EBP-p65
complexes and TRAF2 are required to substantiate this view.
It remains unclear as to whether EOR induced by
SERCA2a overexpression was involved in alleviating ERS-related
apoptosis in the present study. As it is well known that the
increased SERCA2a expression can maintain calcium homeostasis and
attenuate ERS, it could not be determined whether EOR can
precondition the NRCMs against subsequent ERS-induced apoptosis. It
is best to include another group to block NF-κB and/or TNFα
receptor signaling pathway to test this hypothesis.
Wu et al (40) revealed that the effects of
adenoviral vector carrying SERCA1 on NRCMs and adult rat
cardiomyocytes (ARCMs) were differed significantly. The infection
of NRCMs at an MOI of 4 pfu/cell led to apoptosis. At an optimal
MOI, the protein level of SERCA1 in NRCMs was 4-fold higher than
that in the ARCMs, and the activity of Ca2+-ATPase
increased by 4-fold in the NRCMs, but only by 1.5-fold in the
ARCMs. It should be pointed out that since adenoviral vector
carrying SERCA1 has no apoptotic effect on ARCMs (40), the findings of the present study
using NRCMs cannot be extrapolated to explain the results of
AAV1-SERCA2a gene therapy in the CUPID 2 study. In a previous rat
pressure overload HF model, the intracoronary delivery of
adenoviral vector carrying SERCA2a induced reductions in the
serum levels of interleukin (IL)-1, IL-6 and TNF-α; however, local
inflammation of the heart was not investigated (47). To prevent the interference from
EOR, it is better to undertake similar experiments in ARCMs.
There are some limitations associated with the
present study. At the beginning of the experiment, it was not
expected that the EOR would have such a profound impact on the
experimental results. After obtaining the results, it was
determined that the overexpression of SERCA2a leads to EOR, which
would greatly interfere with the study of ERS-related inflammation.
The authors thus aim to perform further research on ARCMs in the
future. As shown in Fig. 9B,
compared with the other three groups, the total p65 content in the
nuclear compartment of untreated cardiomyocytes was minimal. When
calculating the ratio of phosphorylated p65 to p65 in the control
group, the ratio may become unreliable. IL-1β, IL-6 and MCP-1 were
detected in the culture medium supernatant in the present study;
however, since these experiments were not repeated a sufficient
number of times, the data were not presented. It is preferable to
use more sensitive methods, such as reporter gene plasmid
transfection to confirm the conclusions. In addition to caspase-12,
it is preferable to evaluate more indicators related to apoptosis,
such as caspase-3, poly(ADP-ribose) polymerase and Annexin V, in
order to strengthen these conclusions.
In conclusion, in the cellular TM-induced
ERS-associated inflammation model, the overexpression of SERCA2a in
the NRCMs induced EOR, approximately two days prior to TM-induced
UPR. The results suggested that the overexpression of SERCA2a had a
'double-edged sword' effect on ERS-associated inflammation. On the
one hand, the overexpression of SERCA2a attenuated ERS and the
activation of IRE1α signaling pathway induced by TM, resulting in
the attenuation of the upregulation of NF-κB and AP-1 DNA-binding
activities. However, on the other hand, the overexpression of
SERCA2a induced EOR, leading to the further nuclear translocation
of NF-κB and the transcription of TNF-α. The preceding EOR may
precondition the NRCMs against subsequent ERS-associated
inflammation induced by TM. The findings of the present study may
enhance the current understanding of the pros and cons of the
overexpression of SERCA2a in the NRCMs and inspire the further
exploration of the underlying mechanisms of the preconditioning
effects induced by the EOR. Elucidating the aforementioned
mechanisms may help to identify novel treatments for heart diseases
in the future. Further studies performed using ARCMs are required
to prevent the interference of the EOR, in which SERCA2a
overexpression can be achieved through AAV1-SERCA2a transfection or
constructing transgenic animal models.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XLu and XLi were involved in the conception of the
study, applying for funds and revising the manuscript. ZQ, YQ and
TT performed the experiments. ZQ prepared the draft of the
manuscript. XLiu was involved in designing part of the study and
revising the manuscript. ZQ and XLu confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal experiments were performed in accordance
with the Guide for the Care and Use of Laboratory Animals (8th
Edition, 2011) and the animal experimentation guidelines of the
Chinese PLA General Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Nature Science
Foundation of China (grant no. 81170228).
Abbreviations:
|
AP-1
|
activator protein-1
|
|
ARCMs
|
adult rat cardiomyocytes
|
|
ATF6
|
activating transcription factor 6
|
|
BiP
|
immunoglobulin heavy chain-binding
protein
|
|
CCK-8
|
Cell Counting Kit-8
|
|
C/EBP
|
CCAAT-enhancer-binding proteins
|
|
EGFP
|
enhanced green fluorescent protein
|
|
eIF2α
|
eukaryotic protein synthesis
initiation factor 2
|
|
EMSA
|
electrophoretic mobility shift
assay
|
|
EOR
|
endoplasmic reticulum overload
response
|
|
ER
|
endoplasmic reticulum
|
|
ERS
|
endoplasmic reticulum stress
|
|
HF
|
heart failure
|
|
H/R
|
hypoxia/reoxygenation
|
|
IOD
|
integrated optical density
|
|
IRE1α
|
inositol-requiring 1α
|
|
MOI
|
multiplicity of infection
|
|
NRCMs
|
neonatal rat cardiomyocytes
|
|
PERK
|
double-stranded RNA-dependent protein
kinase (PKR)-like ER kinase
|
|
SERCA2a
|
sarco/endoplasmic reticulum
Ca2+-ATPase
|
|
SR
|
sarcoplasmic reticulum
|
|
TM
|
tunicamycin
|
|
TNF-α
|
tumor necrosis factor-α
|
|
TUDCA
|
tauroursodeoxycholic acid
|
|
UPR
|
unfolded protein response
|
|
XBP1
|
X-box binding protein-1
|
References
|
1
|
WRITING GROUP MEMBERS; Lloyd-Jones D,
Adams RJ, Brown TM, Carnethon M, Dai S, De Simone G, Ferguson TB,
Ford E, Furie K, et al: Heart disease and stroke statistics-2010
Update: A report from the American Heart Association. Circulation.
121:e46–e215. 2010.
|
|
2
|
Gwathmey JK, Copelas L, MacKinnon R,
Schoen FJ, Feldman MD, Grossman W and Morgan JP: Abnormal
intracellular calcium handling in myocardium from patients with
end-stage heart failure. Circ Res. 61:70–76. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hasenfuss G, Reinecke H, Studer R, Meyer
M, Pieske B, Holtz J, Holubarsch C, Posival H, Just H and Drexler
H: Relation between myocardial function and expression of
sarcoplasmic reticulum Ca(2+)-ATPase in failing and nonfailing
human myocardium. Circ Res. 75:434–442. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meyer M, Schillinger W, Pieske B,
Holubarsch C, Heilmann C, Posival H, Kuwajima G, Mikoshiba K, Just
H, Hasenfuss G, et al: Alterations of sarcoplasmic reticulum
proteins in failing human dilated cardiomyopathy. Circulation.
92:778–784. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hajjar RJ, Kang JX, Gwathmey JK and
Rosenzweig A: Physiological effects of adenoviral gene transfer of
sarcoplasmic reticulum calcium ATPase in isolated rat myocytes.
Circulation. 95:423–429. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kawase Y, Ly HQ, Prunier F, Lebeche D, Shi
Y, Jin H, Hadri L, Yoneyama R, Hoshino K, Takewa Y, et al: Reversal
of cardiac dysfunction after long-term expression of SERCA2a by
gene transfer in a pre-clinical model of heart failure. J Am Coll
Cardiol. 51:1112–1119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He H, Giordano FJ, Hilal-Dandan R, Choi
DJ, Rockman HA, McDonough PM, Bluhm WF, Meyer M, Sayen MR, Swanson
E, et al: Overexpression of the rat sarcoplasmic reticulum
Ca2+ ATPase gene in the heart of transgenic mice
accelerates calcium transients and cardiac relaxation. J Clin
Invest. 100:380–389. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Baker DL, Hashimoto K, Grupp IL, Ji Y,
Reed T, Loukianov E, Grupp G, Bhagwhat A, Hoit B, Walsh R, et al:
Targeted overexpression of the sarcoplasmic reticulum
Ca2+-ATPase increases cardiac contractility in
transgenic mouse hearts. Circ Res. 83:1205–1214. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dillmann WH: Influences of increased
expression of the Ca2+ ATPase of the sarcoplasmic
reticulum by a transgenic approach on cardiac contractility. Ann N
Y Acad Sci. 853:43–48. 1998. View Article : Google Scholar
|
|
10
|
Maier LS, Wahl-Schott C, Horn W, Weichert
S, Pagel C, Wagner S, Dybkova N, Müller OJ, Näbauer M, Franz WM and
Pieske B: Increased SR Ca2+ cycling contributes to
improved contractile performance in SERCA2a-overexpres sing
transgenic rats. Cardiovasc Res. 67:636–646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Müller OJ, Lange M, Rattunde H, Lorenzen
HP, Müller M, Frey N, Bittner C, Simonides W, Katus HA and Franz
WM: Transgenic rat hearts overexpressing SERCA2a show improved
contractility under baseline conditions and pressure overload.
Cardiovasc Res. 59:380–389. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suarez J, Gloss B, Belke DD, Hu Y, Scott
B, Dieterle T, Kim YK, Valencik ML, McDonald JA and Dillmann WH:
Doxycycline inducible expression of SERCA2a improves calcium
handling and reverts cardiac dysfunction in pressure
overload-induced cardiac hypertrophy. Am J Physiol Heart Circ
Physiol. 287:H2164–H2172. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
del Monte F, Williams E, Lebeche D,
Schmidt U, Rosenzweig A, Gwathmey JK, Lewandowski ED and Hajjar RJ:
Improvement in survival and cardiac metabolism after gene transfer
of sarcoplasmic reticulum Ca(2+)-ATPase in a rat model of heart
failure. Circulation. 104:1424–1429. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sakata S, Lebeche D, Sakata N, Sakata Y,
Chemaly ER, Liang LF, Tsuji T, Takewa Y, del Monte F, Peluso R, et
al: Restoration of mechanical and energetic function in failing
aortic-banded rat hearts by gene transfer of calcium cycling
proteins. J Mol Cell Cardiol. 42:852–861. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitsuyama S, Takeshita D, Obata K, Zhang
GX and Takaki M: Left ventricular mechanical and energetic changes
in long-term isoproterenol-induced hypertrophied hearts of SERCA2a
transgenic rats. J Mol Cell Cardiol. 59:95–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Davia K, Bernobich E, Ranu HK, del Monte
F, Terracciano CM, MacLeod KT, Adamson DL, Chaudhri B, Hajjar RJ
and Harding SE: SERCA2A overexpression decreases the incidence of
aftercontractions in adult rabbit ventricular myocytes. J Mol Cell
Cardiol. 33:1005–1015. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cutler MJ, Wan X, Plummer BN, Liu H,
Deschenes I, Laurita KR, Hajjar RJ and Rosenbaum DS: Targeted
sarcoplasmic reticulum Ca2+ ATPase 2a gene delivery to restore
electrical stability in the failing heart. Circulation.
126:2095–2104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lyon AR, Bannister ML, Collins T, Pearce
E, Sepehripour AH, Dubb SS, Garcia E, O'Gara P, Liang L,
Kohlbrenner E, et al: SERCA2a gene transfer decreases sarcoplasmic
reticulum calcium leak and reduces ventricular arrhythmias in a
model of chronic heart failure. Circ Arrhythm Electrophysiol.
4:362–372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Prunier F, Kawase Y, Gianni D, Scapin C,
Danik SB, Ellinor PT, Hajjar RJ and Del Monte F: Prevention of
ventricular arrhythmias with sarcoplasmic reticulum Ca2+
ATPase pump overexpression in a porcine model of ischemia
reperfusion. Circulation. 118:614–624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
del Monte F, Lebeche D, Guerrero JL, Tsuji
T, Doye AA, Gwathmey JK and Hajjar RJ: Abrogation of ventricular
arrhythmias in a model of ischemia and reperfusion by targeting
myocardial calcium cycling. Proc Natl Acad Sci USA. 101:5622–5627.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cutler MJ, Wan X, Laurita KR, Hajjar RJ
and Rosenbaum DS: Targeted SERCA2a gene expression identifies
molecular mechanism and therapeutic target for arrhythmogenic
cardiac alternans. Circ Arrhythm Electrophysiol. 2:686–694. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hadri L, Bobe R, Kawase Y, Ladage D,
Ishikawa K, Atassi F, Lebeche D, Kranias EG, Leopold JA, Lompré AM,
et al: SERCA2a gene transfer enhances eNOS expression and activity
in endothelial cells. Mol Ther. 18:1284–1292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jaski BE, Jessup ML, Mancini DM, Cappola
TP, Pauly DF, Greenberg B, Borow K, Dittrich H, Zsebo KM and Hajjar
RJ: Calcium upregulation by percutaneous administration of gene
therapy in cardiac disease (CUPID Trial), a first-in-human phase
1/2 clinical trial. J Card Fail. 15:171–181. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jessup M, Greenberg B, Mancini D, Cappola
T, Pauly DF, Jaski B, Yaroshinsky A, Zsebo KM, Dittrich H and
Hajjar RJ; Calcium Upregulation by Percutaneous Administration of
Gene Therapy in Cardiac Disease (CUPID) Investigators: Calcium
upregulation by percutaneous administration of gene therapy in
cardiac disease (CUPID) A phase 2 trial of intracoronary gene
therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with
advanced heart failure. Circulation. 124:304–313. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zsebo K, Yaroshinsky A, Rudy JJ, Wagner K,
Greenberg B, Jessup M and Hajjar RJ: Long-term effects of
AAV1/SERCA2a gene transfer in patients with severe heart failure
analysis of recurrent cardiovascular events and mortality. Circ
Res. 114:101–108. 2014. View Article : Google Scholar
|
|
26
|
Greenberg B, Butler J, Felker GM,
Ponikowski P, Voors AA, Desai AS, Barnard D, Bouchard A, Jaski B,
Lyon AR, et al: Calcium upregulation by percutaneous administration
of gene therapy in patients with cardiac disease (CUPID 2): A
randomised, multinational, double-blind, placebo-controlled, phase
2b trial. Lancet. 387:1178–1186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang K and Kaufman RJ: From
endoplasmic-reticulum stress to the inflammatory response. Nature.
454:455–462. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schmitz ML, Shaban MS, Albert BV, Goekcen
A and Kracht M: The crosstalk of endoplasmic reticulum (ER) stress
pathways with NF-κB: Complex mechanisms relevant for cancer,
inflammation and infection. Biomedicines. 6:582018. View Article : Google Scholar
|
|
29
|
Liu XH, Zhang ZY, Andersson KB, Husberg C,
Enger UH, Ræder MG, Christensen G and Louch WE:
Cardiomyocyte-specific disruption of Serca2 in adult mice causes
sarco(endo) plasmic reticulum stress and apoptosis. Cell Calcium.
49:201–207. 2011. View Article : Google Scholar
|
|
30
|
Xin W, Lu X, Li X, Niu K and Cai J:
Attenuation of endoplasmic reticulum stress-related myocardial
apoptosis by SERCA2a gene delivery in ischemic heart disease. Mol
Med. 17:201–210. 2011. View Article : Google Scholar
|
|
31
|
National Research Council (U.S.):
Committee for the Update of the Guide for the Care and Use of
Laboratory Animals. Guide for the Care and Use of Laboratory
Animals. 8th edition. Washington DC: National Academies Press;
2011
|
|
32
|
O'Donnell JM, Sumbilla CM, Ma H, Farrance
IK, Cavagna M, Klein MG and Inesi G: Tight control of exogenous
SERCA expression is required to obtain acceleration of calcium
transients with minimal cytotoxic effects in cardiac myocytes. Circ
Res. 88:415–421. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hamid T, Guo SZ, Kingery JR, Xiang X, Dawn
B and Prabhu SD: Cardiomyocyte NF-κB p65 promotes adverse
remodelling, apoptosis, and endoplasmic reticulum stress in heart
failure. Cardiovasc Res. 89:129–138. 2011. View Article : Google Scholar
|
|
34
|
Pahl HL: Signal transduction from the
endoplasmic reticulum to the cell nucleus. Physiol Rev. 79:683–701.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pahl HL and Baeuerle PA: Expression of
influenza virus hemagglutinin activates transcription factor
NF-kappa B. J Virol. 69:1480–1484. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Meyer M, Caselmann WH, Schluter V, Schreck
R, Hofschneider PH and Baeuerle PA: Hepatitis B virus
transactivator MHBst: Activation of NF-kappa B, selective
inhibition by antioxidants and integral membrane localization. EMBO
J. 11:2991–3001. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pahl HL, Sester M, Burgert HG and Baeuerle
PA: Activation of transcription factor NF-kappaB by the adenovirus
E3/19K protein requires its ER retention. J Cell Biol. 132:511–522.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gong G, Waris G, Tanveer R and Siddiqui A:
Human hepatitis C virus NS5A protein alters intracellular calcium
levels, induces oxidative stress, and activates STAT-3 and NF-kappa
B. Proc Natl Acad Sci USA. 98:9599–9604. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma TS: Sarcoplasmic reticulum calcium
ATPase overexpression induces cellular calcium overload and cell
death. Ann NY Acad Sci. 853:325–328. 1998. View Article : Google Scholar
|
|
40
|
Wu GM, Long XL and Marin-Garcia J:
Adenoviral SERCA1 overexpression triggers an apoptotic response in
cultured neonatal but not in adult rat cardiomyocytes. Mol Cell
Biochem. 267:123–132. 2004. View Article : Google Scholar
|
|
41
|
Hu P, Han Z, Couvillon AD, Kaufman RJ and
Exton JH: Autocrine tumor necrosis factor alpha links endoplasmic
reticulum stress to the membrane death receptor pathway through
IRE1 alpha-mediated NF-kappa B activation and down-regulation of
TRAF2 expression. Mol Cell Biol. 26:3071–3084. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hamid T, Gu Y, Ortines RV, Bhattacharya C,
Wang G, Xuan YT and Prabhu SD: Divergent tumor necrosis factor
receptor-related remodeling responses in heart failure role of
nuclear factor-kappa B and inflammatory activation. Circulation.
119:1386–1397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Perkins ND: Post-translational
modifications regulating the activity and function of the nuclear
factor kappa B pathway. Oncogene. 25:6717–6730. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zwergal A, Quirling M, Saugel B, Huth KC,
Sydlik C, Poli V, Neumeier D, Ziegler-Heitbrock HW and Brand K:
C/EBP beta blocks p65 phosphorylation and thereby NF-kappa
B-mediated transcription in TNF-tolerant cells. J Immunol.
177:665–672. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kitamura M: Biphasic, bidirectional
regulation of NF-kappaB by endoplasmic reticulum stress. Antioxid
Redox Signal. 11:2353–2364. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nakajima S and Kitamura M: Bidirectional
regulation of NF-κB by reactive oxygen species: A role of unfolded
protein response. Free Radic Biol Med. 65:162–174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gupta D, Palma J, Molina E, Gaughan JP,
Long W, Houser S and Macha M: Improved exercise capacity and
reduced systemic inflammation after adenoviral-mediated SERCA-2a
gene transfer. J Surg Res. 145:257–265. 2008. View Article : Google Scholar : PubMed/NCBI
|