Introduction
Matrix metalloproteinases (MMPs) are zinc-dependent
endopeptidases that have proteolytic activity and play vital roles
in a number of physiopathological processes, such as experimental
autoimmune encephalomyelitis and breast cancer (1-3).
Tissue remodeling processes involve the thinning of the basement
membrane, glandular changes and accumulation of the extracellular
matrix (4). MMPs can degrade
specific extracellular matrix (ECM) components, suggesting that
MMPs play a vital role in tissue remodeling (5,6).
Alterations to the ECM can induce a range of cell behaviors, such
as cell proliferation, migration and apoptosis (7,8).
In addition, MMPs are regulated by a variety of biological
processes, including transcription and posttranscription processes,
and their main regulators are tissue inhibitors of matrix
metalloproteinases (TIMPs) (9).
Numerous studies have found that MMPs and TIMPs are specifically
expressed in chronic rhinosinusitis with nasal polyps (CRSwNPs);
for example, the levels of MMP-2 and MMP-9 are significantly
increased in CRSwNPs, and the levels of TIMP-1 and TIMP-4 are
significantly decreased (10-13). In addition, corticosteroids and
budesonide contribute to ameliorating inflammation by
downregulating the expression of MMP-2 and MMP-9 (14) and upregulating the levels of
TIMP-1, TIMP-2 and TIMP-4 (15).
Thus, the balance between MMPs and TIMPs is important in tissue
homeostasis within CRSwNPs (5).
Tubulin and microtubules (MTs) are the largest
cytoskeletal components (16),
and posttranslational modifications of tubulin are found in all
cells with MTs (17,18). For example, acetyl-α-tubulin
regulates MT stabilization and cell morphology (19,20). In addition, the loss of
acetyl-α-tubulin is associated with TGF-β-induced
epithelial-mesenchymal transition and sulfur mustard-induced
chronic airway remodeling (21,22). A recent study showed that MMP-9
and integrins both act as regulators of α-tubulin acetylation and
detyrosination (23), and Smith
(24) found that MMP-9 can
directly bind integrins to activate signaling pathways that involve
cell adhesion molecules and pro-forms of growth factors (24). These studies suggested that MMP-9
binds to integrin proteins to regulate α-tubulin acetylation and
deacetylation, and is involved in regulating polyp formation. Of
note, a recent study showed that integrin β1 and α-tubulin proteins
were potential MMP-9-interacting proteins (25). Thus, the present study aimed to
pursue these ideas further.
It has been reported that microRNAs (miRNAs/miRs)
can regulate the synthesis and degradation of ECM via the
regulation of MMPs and TIMPs (26,27). For example, miR-29b-3p can
directly and indirectly regulate MMP-2 and MMP-9 expression
(28,29). A recent study showed that
ciliogenesis and cilia function are significantly impaired in the
CRSwNPs epithelium, presumably due to the altered expression of
miRNAs (30). Zhang et al
(31) found that overexpression
of miR-30a-5p can attenuate the epithelial-mesenchymal transition
by repressing CDK6 expression in nasal polyps (31). However, the relationships between
miR-29b-3p and MMP-2/MMP-9 in regulating the progression of CRSwNPs
are unclear.
Lee et al (23) found that MMP-9 and integrin β1
activity can increase α-tubulin acetylation. Of note, Smith
(24) and Yin et al
(25) found that integrin β1 is
a potential MMP-9-interacting protein. Thus, we hypothesized that
downregulation of miR-29b-3p promotes α-tubulin acetylation by
increasing MMP-9 binding to integrin β1, and the present study
aimed to provide novel insight into the etiology and pathogenesis
of CRSwNPs.
Materials and methods
Patient tissue samples
The study group consisted of 100 patients (35 female
and 65 male, median age of 42.7 years, age range of 18.2-83.6) who
underwent functional endoscopic sinus surgery or septoplasty by a
single surgeon at the Department of Otolaryngology, The First
People's Hospital of Qujing (Qujing, China) between July 2018 and
June 2019. Patients younger than 18 years old, with unilateral
nasal polyps or with associated diseases, such as cystic fibrosis,
inverted papilloma and ciliary dyskinesia were excluded from the
present study. Each tissue was divided into four parts: One part
was reserved for cell culture, one part was fixed for
immunofluorescence evaluation using formalin for paraffin
sectioning section or frozen sectioning, and the last two parts
were stored at −80°C for protein and mRNA extraction. The study was
approved by the medical ethics committee of The First People's
Hospital of Qujing, and written informed consent was obtained from
each patient before participation in the study.
Isolation of primary human nasal
epithelial cells (PHNECs) and cell lines
A human nasal epithelial cell line (NP69; cat. no.
BNCC338439) and Escherichia coli BL21 competent cells (BL21;
cat. no. BNCC353591) was purchased from BeNa Culture Collection;
Beijing Beina Chuanglian Biotechnology Research Institute, human
embryonic kidney 293T cells were purchased from The Cell Bank of
Type Culture Collection of The Chinese Academy of Sciences. The
MMP-2 and MMP-9 protein expression of 100 CRSwNPs tissues was
determined via western blotting, and the CRSwNPs tissues with the
lowest (Fig. S1; green box) and
highest (Fig. S1; red box)
expression of MMP-2 and MMP-9 were used to isolate PHNECs with the
lowest and highest expression of MMP-2 and MMP-9 (L-PHNECs and
H-PHNECs, respectively) as previously described (32). In brief, CRSwNPs tissue samples
of ~1 ml volume were rinsed with normal saline, transferred into 10
ml DMEM/F12 medium (Thermo Fisher Scientific, Inc.) containing 1%
penicillin/streptomycin (Sangon Biotech Co., Ltd.), digested with
0.1% protease from Streptomyces griseus (Thermo Fisher Scientific,
Inc.) and 0.1 mg/ml deoxyribonuclease I (Sangon Biotech Co., Ltd.),
and incubated at 4°C overnight. Epithelial cells were removed by
gentle scraping and dispersed into a single cell suspension. The
medium was then transferred into a 15-ml conical tube and
centrifuged at 300 × g for 5 min at room temperature. The
supernatant was decanted, and the pellet was resuspended in
DMEM/F12 medium.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from CRSwNPs tissues or
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. First-strand cDNA
was synthesized from the total RNA according to the instructions of
the PrimeScript™ RT Reagent Kit (Takara Biotechnology Co., Ltd.),
and RT-qPCR was subsequently performed using the SYBR-Green qPCR
kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols. The following primer sequences were used
for RT-qPCR: miR-29b-3p forward, 5′-ACA CTC CAG CTG GGT AGC ACC ATT
TGA AAT C-3′ and reverse, 5′-TGG TGT CGT GGA GTC G-3′; and U6
forward, 5′-CTC GCT TCG GCA GCA CAT A-3′ and reverse, 5′-AAC GAT
TCA CGA ATT TGC GT-3′. The RT-qPCR experiments were performed on an
Applied Biosystems 7900HT Fast Real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used for RT-qPCR: Initial
denaturation at 95°C for 7 min; followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec and
elongation at 72°C for 20 sec; and a final extension at 72°C for 10
min. The relative expression levels were calculated using the
2−∆∆Cq method (33)
and normalized to those of internal reference gene U6.
Western blot assay
Proteins were extracted from CRSwNPs tissues or
cells using radioimmunoprecipitation assay (Beyotime Institute of
Biotechnology), and the concentrations were determined according to
the standard protocols of BCA protein assay kit (Beyotime Institute
of Biotechnology), respectively. The total protein (30
µg/well) in the supernatant was separated via SDS-PAGE on
10% gel, and then transferred to PVDF membranes. After blocking
with 5% skimmed milk for 1 h at room temperature, the membranes
were incubated overnight at 4°C with rabbit anti-MMP-2 (1:1,000;
no. ab92536; Abcam), rabbit anti-MMP-9 (1:1,000; no. ab76003;
Abcam), rabbit anti-TIMP-1 (1:1,000; no. ab211926; Abcam), rabbit
anti-integrin β1 (1:1,000; no. ab52971; Abcam), rabbit
anti-α-tubulin (1:5,000; no. ab18207; Abcam), rabbit
anti-acetyl-α-tubulin (1:1,000; no. ab179484; Abcam) and rabbit
anti-β-actin (1:5,000; no. ab8227; Abcam) antibodies. After three
washes with TBS with 0.1% Tween-20, the immunoblots were incubated
for 1 h at room temperature with goat alkaline phosphatase-labeled
anti-rabbit antibody (1:1,000; cat. no. 14708; Cell Signaling
Technology, Inc.). The immunoreactive bands were visualized using
an enhanced chemiluminescence reagent (Beyotime Institute of
Biotechnology). The blots were semi-quantified using ImageJ
software (version 1.47; National Institutes of Health).
Cell transfection
miR-29b-3p mimic (50 nM; 5′-UAG CAC CAU UUG AAA UCA
GUG UU-3′), NC mimics (50 nM; 5′-UUG UAC UAC ACA AAA GUA CUG-3′),
miR-29b-3p inhibitor (100 nM; 5′-UUC UCC GAA CGU GUC ACG UTT-3′),
NC inhibitor (100 nM; 5′-CAG UAC UUU UGU GUA GUA-3′), specific
small interfering RNAs (siRNAs) targeting MMP-9 (si-MMP-9; 1.0
µg; 5′-ACC ACA ACA TCA CCT ATT GGA TC-3′), siRNA-negative
control (si-NC; 1.0 µg; 5′-UUC UCC GAA CGU GUC ACG UTT-3′)
were purchased from Shanghai GenePharma Co., Ltd. To overexpress
MMP-9, the sequences of MMP-9 were inserted into a pcDNA3.1 plasmid
to obtain the MMP-9 overexpression plasmid pcDNA3.1-MMP-9
(OE-MMP-9; 1.0 µg; Shanghai GenePharma Co., Ltd.), and an
empty pcDNA3.1 plasmid was used as the negative control (OE-NC; 1.0
µg). Plasmid DNA, siRNA, miR-mimic or miR-inhibitor was
transfected into L-PHNECs, H-PHNECs or NP69 cells
(1×105), which were subcultured at a density of 80%,
with Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C. After transfection for 48 h, the
transfection efficiency was detected via RT-qPCR and western
blotting, and then subsequent experiments were performed.
Lipopolysaccharide (LPS) stimulation
NP69 cells (1×105 cells/well) were seeded
in 12-well plates and transfected as described above. When the NP69
cells reached 80-90% confluence, the cells were washed with
phosphate-buffered saline (PBS; 37°C, pH 7.4), and fresh culture
medium was added, along with LPS (Sangon Biotech Co., Ltd.) at a
concentration of 1 µg/ml and incubated at 37°C for 48 h.
Bioinformatics and luciferase reporter
assays
StarBase (Version 2.0; http://starbase.sysu.edu.cn/) online software was used
to predict the binding sites of miRNAs to target mRNAs.
pmirGLO-MMP-9-wild-type (WT)/mutant (Mut) and pmirGLO-MMP-2-WT/Mut
reporter plasmids were provided by Shanghai GenePharma Co., Ltd.
293T cells (2×105/well) were co-transfected with
pmirGLO-MMP-9-Wt/Mut or pmirGLO-MMP-2-Wt/Mut plasmid (1.0
µg) and NC mimic or miR-29b-3p mimic (50 nM) using
Lipofectamine® 2000 reagent at 37°C. At 48 h
post-transfection, luciferase activity was determined using the
dual-luciferase reporter assay system (Promega Corporation).
Firefly luciferase activities were normalized to Renilla
luciferase activities.
Immunofluorescence staining
For immunofluorescence staining, 10-µm-thick
tissue sections of CRSwNPs samples were fixed with 4%
paraformaldehyde for 2 h at 4°C. CRSwNPs were incubated with
blocking solution [5% bovine serum albumin (Thermo Fisher
Scientific, Inc.) with 0.2% Triton X-100] for 1 h at room
temperature, and incubated overnight at 4°C with antibodies against
MMP-9 (1:500; cat. no. ab76003; Abcam) and integrin β1 (1:100; cat.
no. ab52971; Abcam). After washing, the sections were incubated
with Alexa Fluor 555-conjugated anti-rabbit IgG (1:500; no.
ab150062; Abcam) at room temperature for 1 h, and the nuclei were
counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 30 min
at room temperature. Fluorescence images were collected with a
Nikon Eclipse 80i microscope (Nikon Corporation).
Co-immunoprecipitation (Co-IP)
assays
Myc-integrin β1, Myc-α-tubulin and/or MMP-9-WT-HA
plasmids were purchased from Transomic Technologies, Inc., and were
transiently transfected into 293T cells with
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. At 24 h posttransfection, the cells were
harvested and lysed with 500 µl IP lysis buffer containing
protease inhibitor cocktail (Thermo Fisher Scientific, Inc.). After
incubating on ice for 5 min, the cell lysates were centrifuged
(4°C) at 13,000 × g for 10 min. Then, ~25% of the supernatant was
subjected to input assays, and the remaining supernatant was used
for the Co-IP assay with an anti-HA agarose affinity gel (Shanghai
Yeasen Biotechnology Co., Ltd.) according to the manufacturer's
instructions. Rabbit anti-HA (1 µg; cat. no. ab9110; Abcam)
and rabbit anti-Myc (1 µg; cat. no. ab9106; Abcam) were used
for IP. Beads alone were used as the negative control. Briefly, 500
µl agarose affinity gel was centrifuged at 13,000 × g for 30
sec at 4°C to remove the glycerol and washed with cold TBS. Then,
500 µl cell lysate was added to the equilibrated resin and
rocked gently on a rotating platform for 2 h at 4°C. The resin was
washed with cold TBS, and the protein samples were evaluated by
western blotting.
GST-pull down assays
GST pull-down assays were carried out as previously
described (34). Briefly, the
pGEX-GST-MMP-9 plasmid was transformed into BL21 cells to GST-MMP-9
proteins. The pcDNA-Myc-integrin β1 or pcDNA-Myc-α-tubulin plasmid
was transfected into 293T cells to express Myc-integrin β1 and
Myc-α-tubulin proteins. The Pierce™ GST Protein Interaction
Pull-Down Kit (cat. no. 21516; Pierce; Thermo Fisher Scientific,
Inc.) was used according to the manufacturer's instructions. BL21
cells expressing GST-MMP-9 proteins were treated with pull-down
lysis buffer and immobilized on equilibrated glutathione agarose
resin at 4°C for 2 h. The resin was washed with wash solution (TBS
with Pull-down lysis buffer), and 293T lysates containing
Myc-integrin β1 or Myc-α-tubulin protein were added, followed by
incubation at 4°C for 12 h. After washing with a wash solution (TBS
with Pull-down lysis buffer), the resin was eluted with glutathione
elution buffer. The protein samples were evaluated via western
blotting.
Cell viability assays
After 24 h of treatment with 1 µg/ml LPS,
cell viability was assessed using Cell Counting Kit-8 (CCK-8;
Beyotime Institute of Biotechnology). Briefly, cells were seeded in
96-well plates at a density of 1×105 cells/well; 10
µl CCK-8 solution was added to each well and incubated for 4
h. The OD value at 450 nm was measured utilizing a microplate
reader (BioTek Instruments, Inc.).
Detection of cell apoptosis
Apoptosis was detected using an Annexin V combined
fluorescein isothiocyanate (FITC)/propidine iodide (PI) cell
apoptosis detection kit (Beijing Solarbio Science & Technology
Co., Ltd.). In brief, cells were collected using cold PBS buffer
and then cultured with 5 µl Annexin V-FITC reagent and 5
µl PI in the dark for 15 min at room temperature.
Subsequently, 400 µl 1X binding buffer was added, and the
cells were analyzed using a BD FACSCanto II flow cytometer (BD
Biosciences) with FlowJo software (version 10; FlowJo LLC). Early
and late apoptosis were both analyzed.
ELISA
The cell culture media was centrifuged at 2,000 × g
for 10 min to remove debris, and the cell-free culture supernatants
were collected after treatment. The secretory levels of IL-6 (cat.
no. 900-T16), TNF-α (cat. no. 900-M25) and IL-1β (cat. no. 900-M95)
were analyzed using their corresponding ELISA kits (PeproTech
China.) according to the manufacturer's protocol. The
concentrations of inflammatory cytokines were measured using a
microplate spectrophotometer (BioTek Instruments, Inc.) at a
wavelength of 450 nm.
miR-29b-3p differential analysis by Gene
Expression Omnibus (GEO)
Differential expression of miR-29b-3p between three
nasal cell samples and four nasal polyps cell samples from airway
epithelia samples was analyzed using the NCBI GEO DataSets portal:
http://www.ncbi.nlm.nih.gov/geo/. The
miRNA expression data (GEO accession no. GSE159708) from
high-throughput sequencing were submitted by Pommier et al
(35). The web-accessible
analysis tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was utilized
on its default settings to screen the miR-29b-3p expression of each
dataset. The cut-off value for the filtration criteria was set at
FDR<0.05 and log2 fold-change>1.
Bivariate correlation analysis
In the 100 CRSwNPs tissue samples, the levels of
MMP-2, MMP-9 and TIMP-1 protein were detected via western blotting,
the blots were semi-quantified by ImageJ software and normalized to
those of the internal reference gene β-actin. The levels of
miR-29b-3p were determined via RT-qPCR, and the relative expression
levels were calculated using the 2−ΔΔCq method and
normalized to those of internal reference gene U6. Bivariate
correlation analyses of the correlations between MMP-2 and MMP-9
expression and miR-29b-3p, or TIMP-1 expression were performed.
Statistical analysis
All the experiments were repeated three times.
GraphPad Prism 8 (GraphPad Software, Inc.) was used for statistical
analysis, and the data are presented as the mean ± standard
deviation. Data between two groups were analyzed using an unpaired
Student's t-test, and data among multiple groups were analyzed by
one-way analysis of variance (ANOVA) followed by a Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Correlations between miR-29b-3p, MMP-2,
MMP-9 and TIMP-1 expression in CRSwNPs
Previous studies have shown that miR-29b-3p
regulates the expression of MMP-2 and MMP-9 (28,29), and the balance between MMPs and
TIMPs is important in tissue homeostasis within CRSwNPs (5). To examine the relationships between
miR-29b-3p, MMP-2/MMP-9 and TIMP-1 in CRSwNPs, the mRNA expression
of miR-29b-3p was detected using RT-qPCR, and the protein
expression levels of MMP-2, MMP-9 and TIMP-1 were determined via
western blotting in 100 CRSwNPs tissue samples (Fig. S1). The results showed that
miR-29b-3p expression was moderately positively correlated with the
expression of MMP-2 (r=0.4420; P<0.001; Fig. 1A) and MMP-9 (r=0.4799;
P<0.001; Fig. 1B), and TIMP-1
expression was moderately negatively correlated with the expression
of MMP-2 (r=-0.4672; P<0.001; Fig. 1C) and MMP-9 (r=−0.4484;
P<0.001; Fig. 1D).
Immunofluorescent images showed MMP-2, MMP-9 and TIMP-1 protein
expression in CRSwNPs tissue samples (Fig. 1E). These observations suggested
that miR-29b-3p plays a protective role in the regulation of the
balance in MMP and TIMP expression.
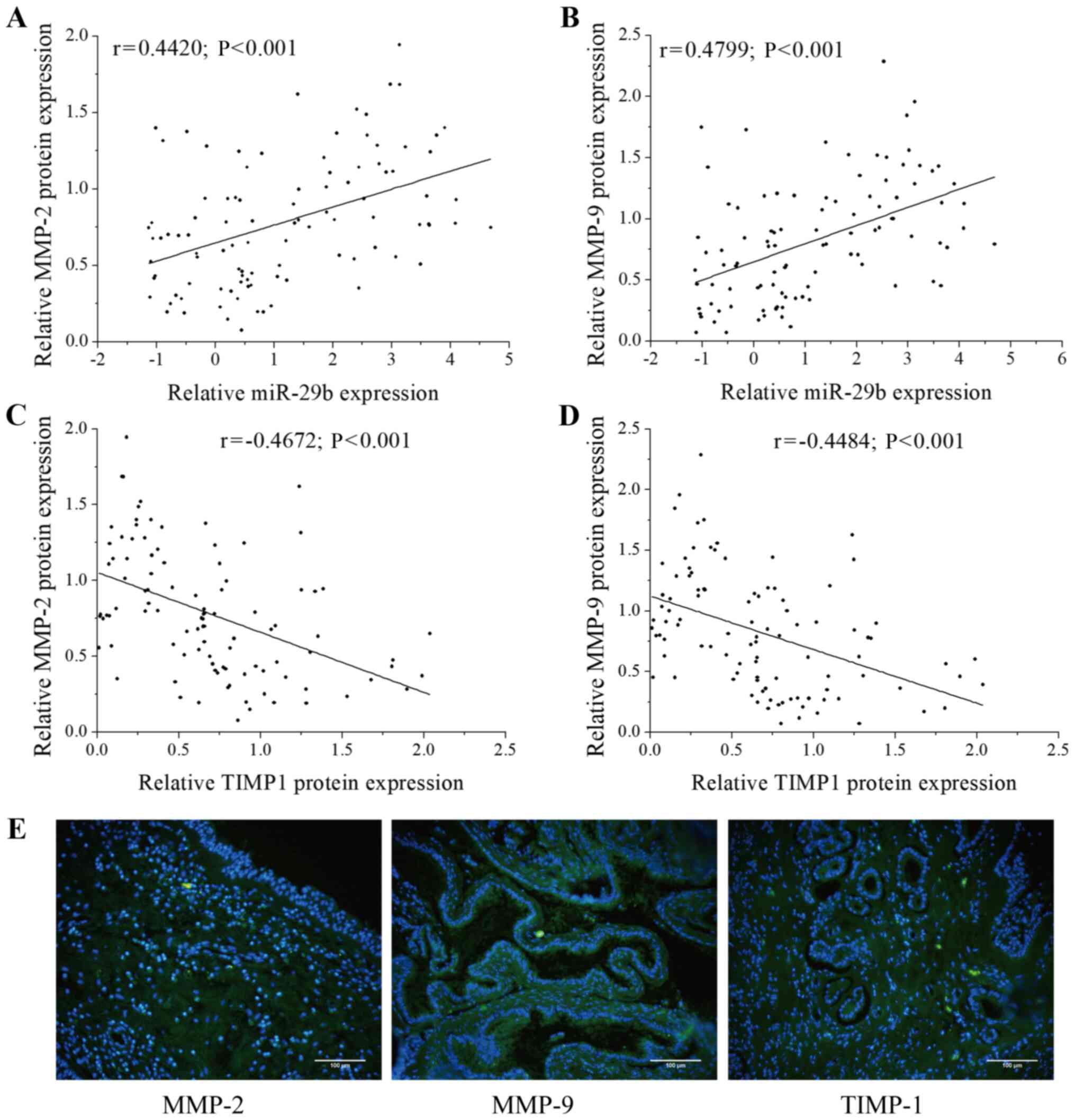 | Figure 1Correlation between miR-29b-3p,
MMP-2, MMP-9 and TIMP-1 expression in CRSwNPs. Correlation between
miR-29b-3p expression and (A) MMP-2 and (B) MMP-9 expression in
CRSwNPs samples (n=100). The X-axis represents the expression of
miR-29b-3p, the Y-axis represents the expression of MMP-2/MMP-9,
each point in the figure represents a sample, and the P-value and
the correlation coefficient (r value) are stated. The data were
normalized to U6 expression and are shown as the Cq value.
Correlation between TIMP-1 expression and (C) MMP-2 and (D) MMP-9
expression in CRSwNPs samples (n=100). The X-axis represents the
expression of TIMP-1, the Y-axis represents the expression of the
MMP-2/MMP-9, each point in the figure represents a sample, and the
P-value and the correlation coefficient (r value) are stated. (E)
The expression of MMP-2, MMP-9 and TIMP-1 based on
immunofluorescence staining. Green staining shows positive
expression of MMP-2, MMP-9 and TIMP-1, and blue (DAPI) indicates
nuclear staining. Scale bar, 100 µm. miR, microRNA; MMP,
matrix metalloproteinase; TIMP-1, tissue inhibitor of
metalloproteinase 1; CRSwNPs, chronic rhinosinusitis with nasal
polyps. |
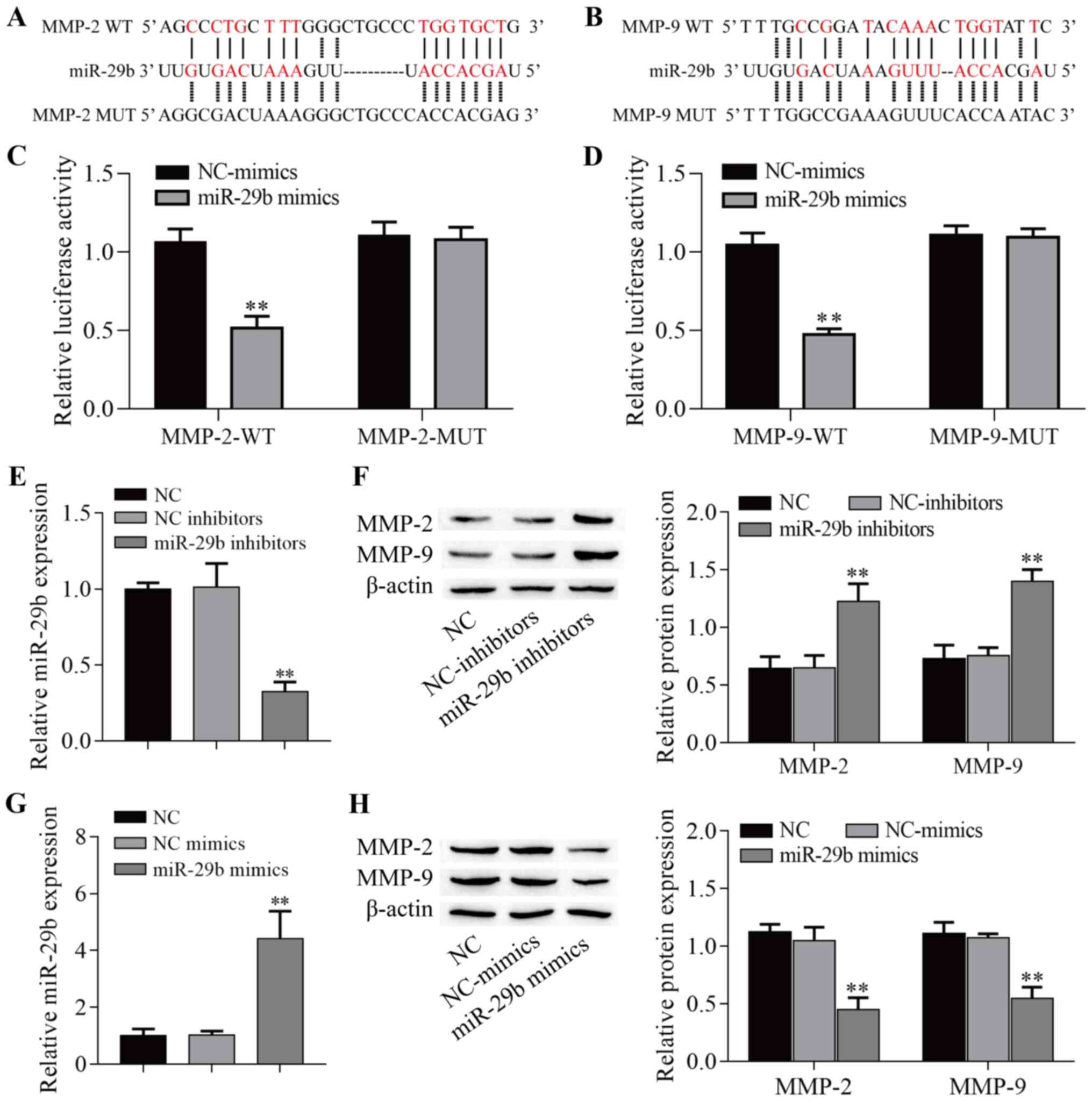
miR-29b-3p targets MMP-2 and MMP-9 in
PHNECs
Previous studies have shown that miR-29b-3p targets
the 3′-untranslated region of MMP-2 and MMP-9 (28,29). Using the bioinformatics database
StarBase to search for potential targets, a putative interaction
between miR-29b-3p and MMP-2/MMP-9 was found, and the target
binding sequence is shown in Fig. 2A
and B. A dual-luciferase reporter assay demonstrated that
MMP-2-WT/MMP-9-WT and miR-29b-3p mimic co-transfection
significantly decreased the luciferase activities in 293T cells
(P<0.01; Fig. 2C and D),
while MMP-2-MUT/MMP-9-MUT and miR-29b-3p mimic co-transfection
failed to affect the luciferase activity in 293T cells (P>0.05;
Fig. 2C and D). Next,
transfection of miR-29b-3p inhibitor and miR-29b-3p mimic into
PHNECs significantly decreased and increased miR-29b-3p expression,
respectively (P<0.01; Fig. 2E and
G). Notably, transfection of miR-29b-3p inhibitor into PHNECs
significantly increased MMP-2 and MMP-9 protein expression in
L-PHNECs (P<0.01; Fig. 2F),
and transfection with miR-29b-3p mimic significantly decreased
MMP-2 and MMP-9 protein expression in H-PHNECs (P<0.01; Fig. 2H). Altogether, the aforementioned
results indicated that miR-29b-3p directly targeted both the MMP-2
and MMP-9 genes.
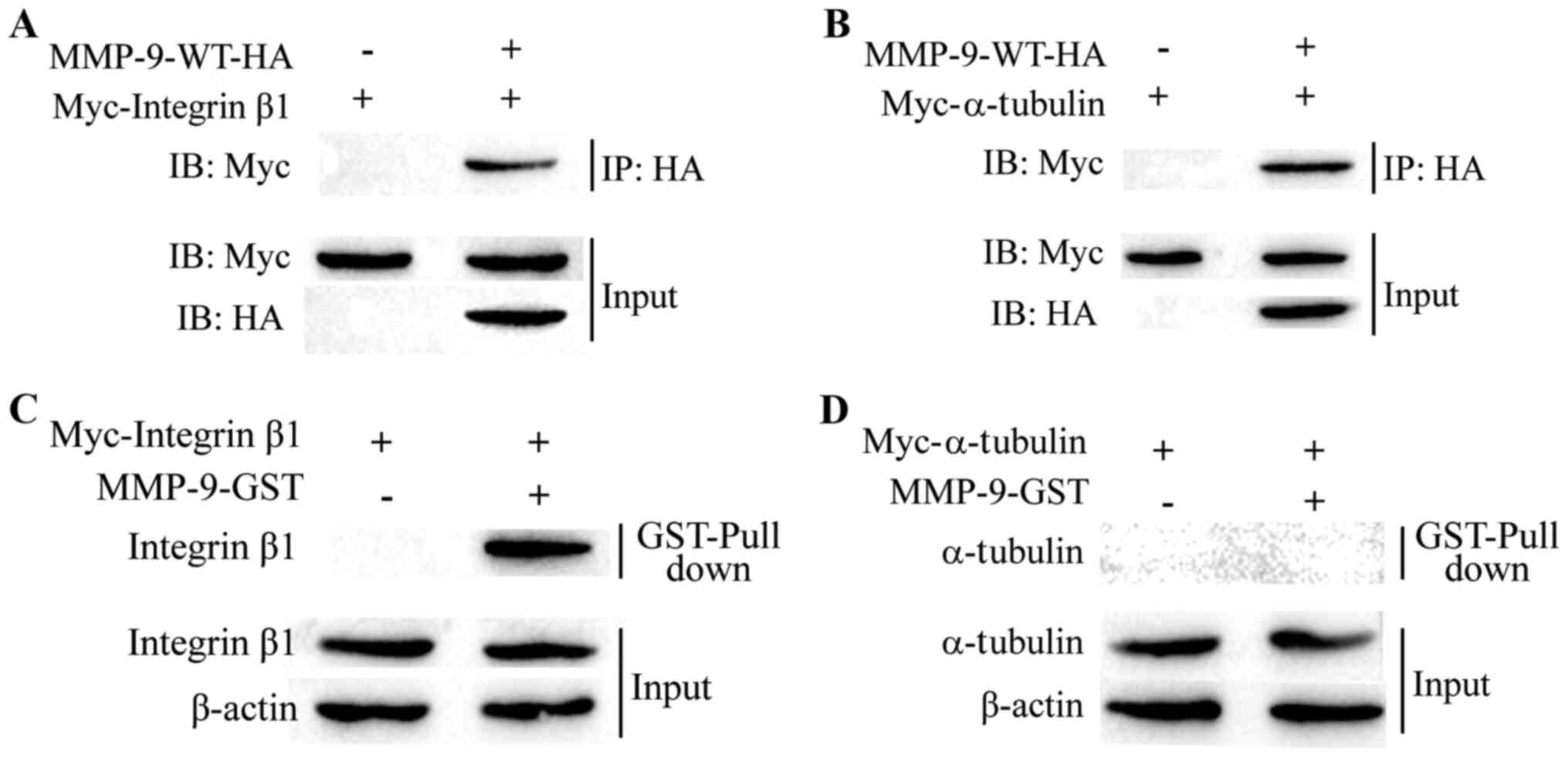
MMP-9 binds to integrin b1
Compared with MMP-2, the r value (Fig. 1A and B) and the targeting
capability (Fig. 2F and H) were
higher for MMP-9. Thus, the miR-29b-3p/MMP-9 axis was investigated
in subsequent assays. In a recent study, integrin β1 and α-tubulin
were identified as potential MMP-9-interacting proteins (25). To examine the interaction between
MMP-9 and integrin β1/α-tubulin, Myc-integrin β1/Myc-α-tubulin and
HA-MMP-9 were transiently co-expressed in 293T cells and Co-IP
assays were performed using a HA antibody. Indeed, MMP-9 readily
immunoprecipitated Myc-tagged integrin β1 and α-tubulin (Fig. 3A and B). To further determine
whether the interaction between integrin β1/α-tubulin and MMP-9 was
direct, GST pull-down assays were performed using the GST-MMP-9
protein expressed in and purified from bacteria. As shown in
Fig. 3C and D, GST-fused MMP-9
pulled down integrin β1, but not α-tubulin. These observations
suggested that integrin β1 could directly interact with MMP-9, but
that α-tubulin indirectly interacts with MMP-9. These results
suggested that MMP-9 binds with integrin β1.
miR-29b-3p affects the acetyl-α-tubulin
levels by increasing MMP-9 and integrin b1 binding
MMP-9 and integrin β1 activation can increase
α-tubulin acetylation (23), and
Smith (24) and the present
study found that MMP-9 directly bound to integrin β1 (Fig. 3C). We hypothesized that MMP-9
binds to integrin β1 to promote α-tubulin acetylation by
downregulating miR-29b-3p in CRSwNPs. MMP-9 expression was knocked
down in H-PHNECs by transfection with si-MMP-9 (P=0.024; Fig. 4A), whereas expression of MMP-9
was upregulated in L-PHNECs by transfection with OE-MMP-9
(P<0.01; Fig. 4B). To test
this hypothesis, miR-29b-3p was overexpressed in H-PHNECs. The
western blotting results showed that MMP-9 expression was reduced
after overexpression of miR-29b-3p (P<0.01), but this effect was
reversed after overexpression of MMP-9 (P<0.01; Fig. 4C). By contrast, the expression of
acetyl-α-tubulin was elevated after overexpression of miR-29b-3p
(P<0.01), which was repressed by overexpression of MMP-9
(P<0.01). In addition, miR-29b-3p expression was inhibited in
L-PHNECs. These results showed the expression of MMP-9 was elevated
by miR-29b-3p inhibitors (P<0.01), but this effect was reversed
by MMP-9 knockdown (P=0.002; Fig.
4D). By contrast, the expression of acetyl-α-tubulin was
decreased by the inhibition of miR-29b-3p (P<0.01), but it was
enhanced by MMP-9 knockdown. Of note, the protein expression of
integrin β1 was not affected (P>0.05). Immunofluorescence was
performed to examine the protein expression of MMP-9 and integrin
β1 in tissues with lower and higher MMP-9 expression. As shown in
Fig. 4E, there was no notable
difference in the expression of integrin β1 in low and high MMP
tissues. Next, the correlation between MMP-9 and acetyl-α-tubulin
expression was further analyzed in 100 CRSwNPs tissue samples, and
a moderate negative correlation was observed between MMP-9 and
acetyl-α-tubulin (r=-0.3435; P=0.004; Fig. 4F). Taken together, we propose a
novel model in which MMP-9 binds to integrin β1 to promote
α-tubulin deacetylation-mediated tissue remodeling by
downregulating miR-29b-3p in CRSwNPs (Fig. 4G).
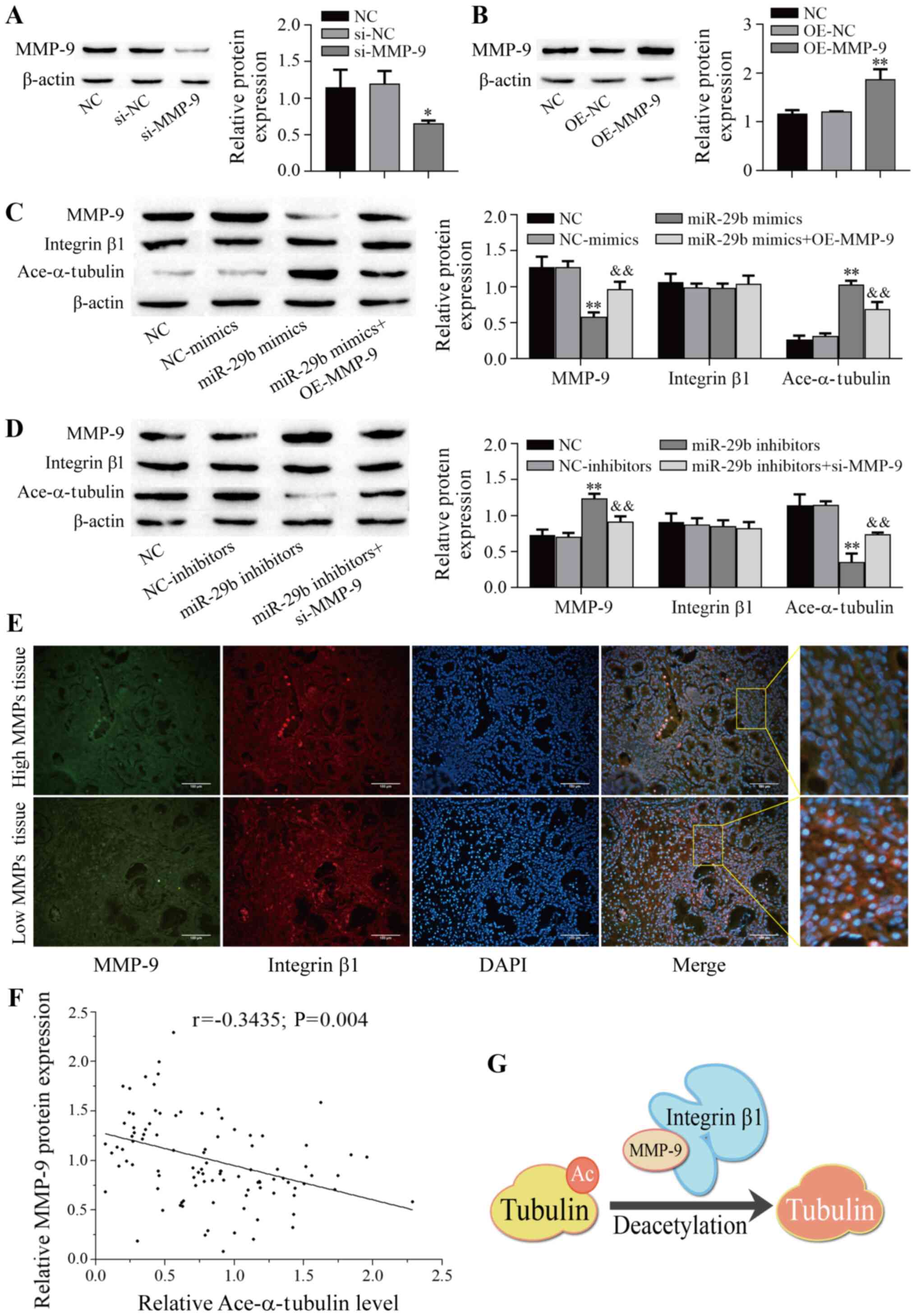 | Figure 4miR-29b-3p affects the
acetyl-α-tubulin levels by increasing the MMP-9 and integrin β1
binding in CRSwNPs. (A-D) Protein expression was detected via
western blotting (n=3). (E) The expression of MMP-9 and integrin β1
was determined by immunofluorescence staining. Green indicates
positive expression of MMP-9, red indicates positive expression of
integrin β1, and blue (DAPI) indicates nuclear staining. Scale bar,
100 µm. (F) Correlation between MMP-9 expression with
acetyl-α-tubulin expression in CRSwNPs samples (n=100). The X-axis
represents the expression of acetyl-α-tubulin, the Y-axis
represents the expression of MMP-9, each point in the figure
represents a sample, and the P-value and the correlation
coefficient (r value) are stated. (G) Working model showing that
MMP-9 binds to integrin β1 to promote α-tubulin deacetylation.
*P<0.05, **P<0.01 vs. NC;
&&P<0.01 vs. miR-29b inhibitors or miR-29b
mimics. miR, microRNA; MMP, matrix metalloproteinase; CRSwNPs,
chronic rhinosinusitis with nasal polyps; NC, negative control;
si-, small interfering RNA; OE, overexpression vector. |
miR-29b-3p regulates LPS-induced
viability, apoptosis and inflammation of NP69 cells by targeting
MMP-9
MMP-9 expression was downregulated in NP69 cells by
transfection with si-MMP-9 (P=0.043; Fig. 5A), whereas expression was
upregulated by transfection with OE-MMP-9 (P=0.006; Fig. 5B). To explore the potential role
of miR-29b-3p in CRSwNPs, miR-29b-3p expression in nasal polyps was
investigated using the GEO database. This analysis showed that
miR-29b-3p was expressed at lower levels in nasal polyps (P=0.008;
Fig. 5C). Next, miR-29b-3p
expression in PHNECs and NP69 cells was measured via RT-qPCR. As
shown in Fig. 5D, the expression
of miR-29b-3p was downregulated in both L-PHNECs and H-PHNECs
compared with NP69 cells (P<0.001), and it was expressed at
lower levels in H-PHNECs than in L-PHNECs (P=0.005). These results
suggested that miR-29b-3p was downregulated in CRSwNPs. Next, a
cell model of LPS-induced CRSwNPs was investigated. The results
showed that miR-29b-3p expression was significantly lower in
LPS-induced NP69 cells than in the control cells (P<0.01;
Fig. 5E) and was significantly
increased by the upregulation of miR-29b-3p compared with the LPS
group (P<0.01). Notably, overexpression of MMP-9 reduced
miR-29b-3p expression compared with the LPS + miR-29 mimics group
(P<0.01) and knockdown of MMP-9 elevated miR-29b-3p expression
compared with the LPS group (P<0.01). In addition, LPS also
significantly increased MMP-9 expression in NP69 cells (P<0.01;
Fig. 5F), which were effectively
reversed by upregulation of miR-29b-3p and knockdown of MMP-9
(P<0.01), whereas overexpression of MMP-9 reversed the
inhibitory effect of miR-29b-3p upregulation (P<0.01).
Conversely, LPS significantly decreased acetyl-α-tubulin level
(P<0.01; Fig. 5F), which was
effectively elevated by upregulation of miR-29b-3p and knockdown of
MMP-9 (P<0.05), but overexpression of MMP-9 reversed the effects
of miR-29b-3p overexpression (P<0.01). Of note, the protein
expression of integrin β1 was not altered (P>0.05). The results
suggested that miR-29b-3p regulated acetyl-α-tubulin levels by
targeting MMP-9. Next, cell viability, apoptosis and inflammatory
cytokines (IL-1β, IL-6 and TNF-α) of LPS-induced NP69 cells were
determined. As presented in Fig.
5G, cell viability was inhibited after LPS induction
(P<0.01), and upregulation of miR-29b-3p and knockdown of MMP-9
alleviated the inhibitory effect of LPS (P<0.01). Overexpression
of MMP-9 reduced the alleviatory effect of miR-29b-3p upregulation
on cell viability (P<0.01). Moreover, cell apoptosis and
inflammatory cytokine levels were increased after LPS induction
(P<0.01; Fig. 5H and I), and
these effects were reduced by the upregulation of miR-29b-3p and
knockdown of MMP-9 (P<0.01); however, the overexpression of
MMP-9 repressed the inhibitory effect of miR-29b-3p upregulation
(P<0.01). These data indicated that overexpression of miR-29b-3p
alleviated LPS-induced inflammation in NP69 cells by targeting
MMP-9.
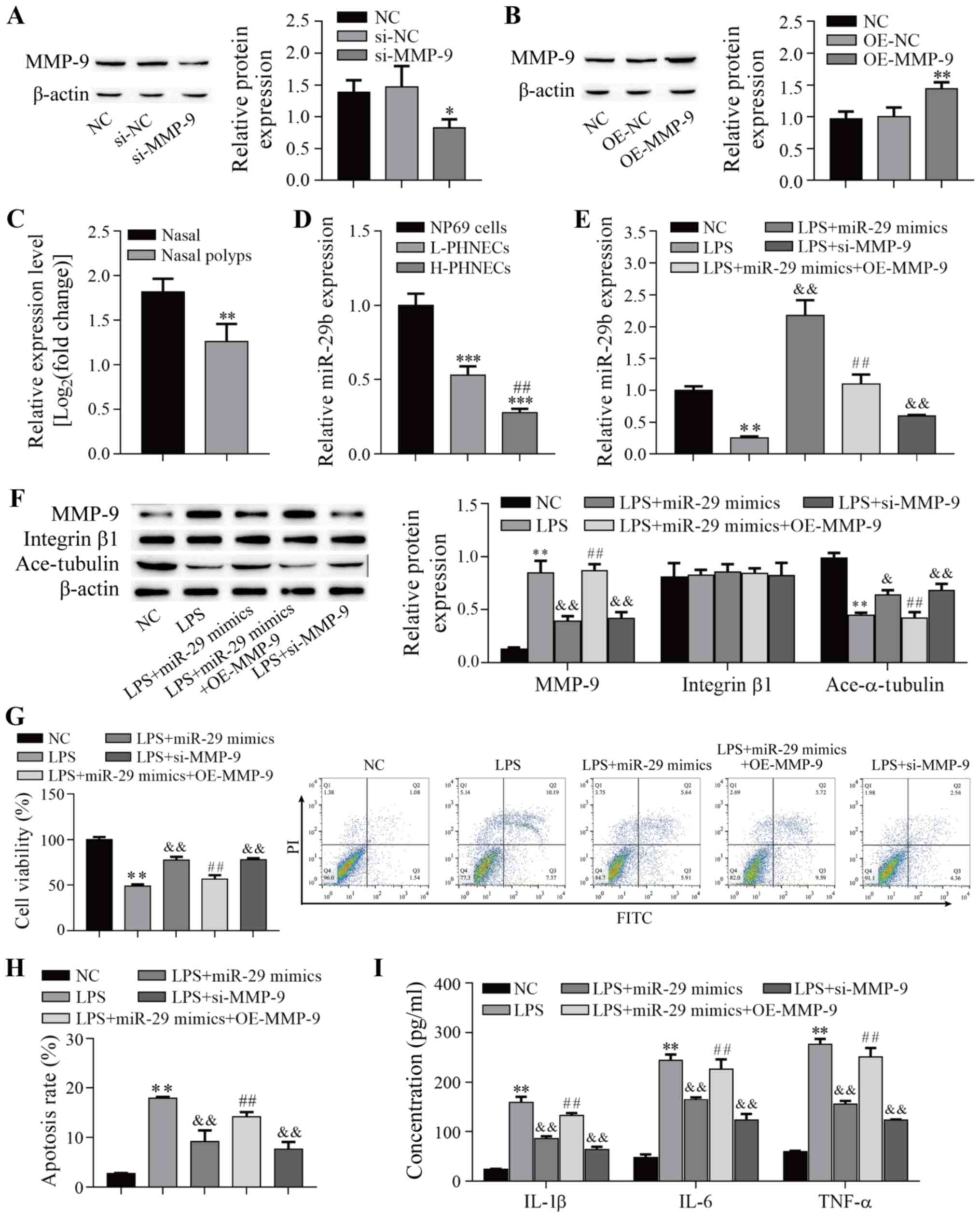 | Figure 5miR-29b-3p alleviates LPS-induced
inflammation in NP69 cells by targeting MMP-9. (A and B) Protein
expression was detected via western blotting (n=3). (C) The
expression of miR-29b-3p was downregulated in the nasal polyps
cells (n=4) compared with nasal cells (n=3) from airway epithelia
samples. The GEO2R analysis tool was used to analyze the miRNA
expression dataset (GEO accession no. GSE159708) to screen the
miR-29b-3p expression of each dataset. The cut-off value for the
filtration criteria was set at FDR <0.05 and log2 fold-change
>1. **P<0.01 vs. Nasal. (D) The expression of
miR-29b-3p was downregulated in both L-PHNECs and H-PHNECs compared
with NP69 cells (n=3). ***P<0.001 vs. NP69 cells;
##P<0.01 vs. L-PHNECs. (E) LPS treatment decreased
miR-29b-3p expression (n=3). (F) Protein expression was determined
via western botting (n=3). (G) Cell viability was determined via
Cell Counting Kit-8 assay (n=3). (H) Cell apoptosis was determined
by flow cytometry (n=3). (I) The inflammatory cytokine levels were
determined by ELISA (n=3). *P<0.05,
**P<0.01 vs. NC; &P<0.05,
&&P<0.01 vs. LPS; ##P<0.01 vs.
LPS + miR-29b mimics. L-PHNECs, PHNECs with the lowest MMP-2 and
MMP-9 expression; H-PHNECs, PHNECs with highest MMP-2 and MMP-9
expression; miR/miRNA, microRNA; LPS, lipopolysaccharide; MMP,
matrix metalloproteinase; NC, negative control; GEO, Gene
Expression Omnibus; si-, small interfering RNA; OE, overexpression
vector. |
Discussion
Ciliogenesis and cilia function are significantly
impaired in the CRSwNPs epithelium, presumably due to the altered
expression of miRNAs (30). The
function of miRNAs in CRSwNPs remains unclear. In previous reports,
miR-29b-3p was shown to directly and indirectly regulate MMP-2 and
MMP-9 expression (28,29). It is commonly known that MMPs are
likely to be associated with airway remodeling in CRSwNPs based on
the increased expression of MMP-2 and MMP-9 in patients with
CRSwNPs compared with control subjects (36-38). Numerous studies have found that
the levels of MMPs are significantly increased in CRSwNPs compared
with healthy control tissues and that the levels of TIMPs are
significantly decreased (10-13). In the present study, no healthy
samples were analyzed as a control group, so it cannot be concluded
that the expression levels of MMP-2, MMP-9, TIMP-1 and miR-29b-3p
differ in CRSwNPs compared with healthy individuals. However, it
was found that TIMP-1 expression was negatively correlated with the
expression of MMP-2 and MMP-9 (Fig.
1C and D), and the results suggested that the imbalance in MMPs
and TIMPs is closely associated with the development of CRSwNPs. In
addition, miR-29b-3p expression was positively correlated with the
expression of MMP-2 and MMP-9 in CRSwNPs (Fig. 1A and B), and miR-29b-3p directly
targeted both the MMP-2 and MMP-9 genes (Fig. 2). Chen et al (28) found that oxidized low-density
lipoprotein upregulates miR-29b-3p and increases MMP-2 and MMP-9
expression by reducing DNA methylation in cardiovascular diseases.
Of note, a recent study showed that MMP-2 and MMP-9 were target
genes of miR-29b-3p, and metformin alleviates polycystic ovary
syndrome by decreasing the expression of MMP-2 and MMP-9 via
upregulation of miR-29b-3p expression (29). Thus, bioinformatics analysis and
luciferase activity assays identified MMP-2 and MMP-9 as functional
targets of miR-29b-3p in CRSwNPs.
In the present study, it was found that LPS
treatment increased MMP-9 expression (Fig. 5F) and that overexpression of
miR-29b-3p alleviated LPS-induced inflammation in NP69 cells by
targeting MMP-9 (Fig. 5I). This
study further demonstrated that the miR-29b-3p/MMP-9 axis may
regulate remodeling in CRSwNPs. A recent study showed that MMP-9
and integrin activity both regulate fibroblast migration by
increasing α-tubulin acetylation (23), which can directly bind to
integrins to activate signaling, which involves cell adhesion
molecules and pro-forms of growth factors (24). Previous studies have shown that
the hemopexin domain of MMP-9 can interact with different integrin
subunits to promote enhanced cancer cell migration, invasion and
proliferation in various cancer cell types, such as colon cancer
cells, B cell chronic lymphocytic leukemia and breast cancer cells
(39-42). Notably, the integrin β1, Src,
elongation factor 1-α 2, α-tubulin, actin and histone H2B proteins
are potential MMP-9-interacting proteins (25). In the present study, it was shown
that integrin β1 and MMP-9 bind directly (Fig. 3A and C), but α-tubulin and MMP-9
indirectly interact (Fig. 3B and
D). Furthermore, overexpression and knockdown of MMP-9 did not
just alter MMP-9 expression, but, as a novel finding, it also
altered α-tubulin acetylation levels (Fig. 4C and D). However, overexpression
and knockdown of MMP-9 failed to regulate integrin β1 expression
(Fig. 4C and D). In addition, a
moderate negative correlation between MMP-9 and acetyl-α-tubulin
expression levels was also found (Fig. 4F). Our observations demonstrated
that MMP-9-integrin β1 complexes promoted α-tubulin deacetylation
(Fig. 4G). Of note, this result
contrasts with Lee et al (23), who found that MMP-9 and integrin
activation can increase α-tubulin acetylation. Previous research
suggests that integrins inhibit the activation of histone
deacetylase 6 by activating AKT signaling, which then suppresses
acetyl-α-tubulin deacetylation (43,44). We hypothesize that MMP-9 binds to
integrin β1 to promote acetyl-α-tubulin deacetylation by inhibiting
the activation of integrin signaling. Each integrin is a
heterodimer composed of an α- and a β-subunit and must be assembled
as a heterodimer within the endoplasmic reticulum in order to be
expressed on the cell surface (45). Thus, we speculate that MMP-9
binds to integrin αβ1 homodimers to promote acetyl-α-tubulin
deacetylation. These are all areas of interest that we will
continue to investigate in further studies.
Although the cellular mechanisms underlying the
development of CRSwNPs remain uncertain, polyp formation occurs due
to the protrusion of connective tissue through an initial
epithelial defect and remodeling (46). The present study further
demonstrated that MMP-9-integrin β1 complexes promote α-tubulin
deacetylation involved in the airway remodeling of CRSwNPs. Few
studies have investigated whether the acetylation of α-tubulin
plays an important role in MT stabilization and cell morphology,
and loss of α-tubulin acetylation is associated with
epithelial-mesenchymal transition and chronic airway remodeling
(21,22). In addition, inhibition of HDACs
improves endothelial barrier function and attenuates the
progression of osteoarthritis by suppressing α-tubulin
deacetylation (47,48). In the present study, it was shown
that the miR-29b-3p/MMP-9 axis decreased acetyl-α-tubulin levels
and that overexpression of miR-29b-3p significantly decreased MMP-9
expression and increased the acetyl-α-tubulin levels in PHNECs.
LPS-induced inhibition of acetyl-α-tubulin levels and
overexpression of miR-29b-3p not only increased the
acetyl-α-tubulin levels (Fig.
5F), but also alleviated LPS-induced inflammation in NP69 cells
(Fig. 5I). Thus, targeting
miR-29b-3p/MMP-9 is a novel strategy for the clinical treatment of
CRSwNPs.
In conclusion, miR-29b-3p expression was positively
correlated with the expression of MMP-2 and MMP-9 in CRSwNPs, and
TIMP-1 expression was negatively correlated with the expression of
MMP-2 and MMP-9. miR-29b-3p affects the acetyl-α-tubulin levels by
increasing MMP-9 and integrin β1 interaction. The miR-29b-3p/MMP-9
pathway was identified as a novel protective axis in CRSwNPs and
provided a significant theoretical foundation for developing novel
therapies for CRSwNPs.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the study, performed the experiments,
and prepared and revised the final manuscript. HL designed the
study, collected the clinical samples, analyzed and interpreted the
data and revised the final manuscript. DY performed the
experiments, commented on the data analysis and revised the
manuscript. JG performed the experiments, analyzed and interpreted
the data, and finalized the manuscript. BR designed the study, and
prepared and revised the final manuscript. RL designed the study,
and prepared and revised the final manuscript. BR and RL confirm
the authenticity all the raw data. All the authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All the experimental protocols were checked and
approved by the Medical Ethics Committee of The First People's
Hospital of Qujing (Qujing, China), and written informed consent
was obtained from each patient before participation in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This study was funded by the National Natural Science Foundation
of China (NSFC; grant no. 81960187), Applied basic research of
Yunnan Province [grant no. 2017FE468(−195)] and Scientific Research
Fund of Yunnan Education Department (grant no. 2019J1262).
References
|
1
|
Sarig-Nadir O and Seliktar D: The role of
matrix metalloproteinases in regulating neuronal and nonneuronal
cell invasion into PEGylated fibrinogen hydrogels. Biomaterials.
31:6411–6416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fang CY, Zhang J, Yang HL, Peng LL, Wang
K, Wang YJ, Zhao X, Liu HJ, Dou CH, Shi LH, et al: Leucine
aminopeptidase 3promotes migration and invasion of breast cancer
cells through upregulation of fascin and matrix
metalloproteinases-2/9 expression. J Cell Biochem. 120:3611–3620.
2019. View Article : Google Scholar
|
|
3
|
Gerwien H, Hermann S, Zhang X, Korpos E,
Song J, Kopka K, Faust A, Wenning C, Gross CC, Honold L, et al:
Imaging matrix metalloproteinase activity in multiple sclerosis as
a specific marker of leukocyte penetration of the blood-brain
barrier. Sci Transl Med. 8:364ra1522016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eifan AO, Orban NT, Jacobson MR and Durham
SR: Severe persistent allergic rhinitis. Inflammation but no
histologic features of structural upper airway remodeling. Am J
Respir Crit Care Med. 192:1431–1439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kahveci OK, Derekoy FS, Yilmaz M, Serteser
M and Altuntas A: The role of MMP-9 and TIMP-1 in nasal polyp
formation. Swiss Med Wkly. 138:684–688. 2008.PubMed/NCBI
|
|
6
|
Kelly EA, Busse WW and Jarjour NN:
Increased matrix metalloproteinase-9 in the airway after allergen
challenge. Am J Respir Crit Care Med. 162:1157–1161. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen Y, Peng W, Raffetto JD and Khalil RA:
Matrix metalloproteinases in remodeling of lower extremity veins
and chronic venous disease. Prog Mol Biol Transl Sci. 147:267–299.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pinet K and McLaughlin K: Mechanisms of
physiological tissue remodeling in animals: Manipulating tissue,
organ, and organism morphology. Dev Biol. 451:134–145. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alaseem A, Alhazzani K, Dondapati P,
Alobid S, Bishayee A and Rathinavelu A: Matrix metalloproteinases:
A challenging paradigm of cancer management. Semin Cancer Biol.
56:100–115. 2019. View Article : Google Scholar
|
|
10
|
Can IH, Ceylan K, Caydere M, Samim EE,
Ustun H and Karasoy DS: The expression of MMP-2, MMP-7, MMP-9, and
TIMP-1 in chronic rhinosinusitis and nasal polyposis. Otolaryngol
Head Neck Surg. 139:211–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang JH, Kwon HJ and Jang YJ:
Staphylococcus aureus increases cytokine and matrix
metalloproteinase expression in nasal mucosae of patients with
chronic rhinosinusitis and nasal polyps. Am J Rhinol Allergy.
24:422–427. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Meng J, Qiao X, Liu Y, Liu F, Zhang
N, Zhang J, Holtappels G, Luo B, Zhou P, et al: Expression of TGF,
matrix metalloproteinases, and tissue inhibitors in Chinese chronic
rhinosinusitis. J Allergy Clin Immunol. 125:1061–1068. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guerra G, Testa D, Salzano F, Tafuri D,
Hay E, Schettino BA, Iovine R, Marcuccio G and Motta G: Expression
of matrix metalloproteinases and their tissue inhibitors in chronic
rhinosinusitis with nasal polyps: Etiopathogenesis and recurrence.
Ear Nose Yhroat J. 145561319896635:2020.
|
|
14
|
Yigit O, Acioğlu E, Gelişgen R, Server EA,
Azizli E and Uzun H: The effect of corticosteroid on
metalloproteinase levels of nasal polyposis. Laryngoscope.
121:667–673. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang C, Lou H, Wang X, Wang Y, Fan E, Li
Y, Wang H, Bachert C and Zhang L: Effect of budesonide transnasal
nebulization in patients with eosinophilic chronic rhinosinusitis
with nasal polyps. J Allergy Clin Immunol. 135:922–929.e926. 2015.
View Article : Google Scholar
|
|
16
|
Song Y and Brady S: Post-translational
modifications of tubulin: Pathways to functional diversity of
microtubules. Trends Cell Biol. 25:125–136. 2015. View Article : Google Scholar
|
|
17
|
Janke C and Bulinski JC:
Post-translational regulation of the microtubule cytoskeleton:
Mechanisms and functions. Nat Rev Mol Cell Biol. 12:773–786. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wloga D and Gaertig J: Post-translational
modifications of microtubules. J Cell Sci. 123:3447–3455. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vo NT and Bols NC: Demonstration of
primary cilia and acetylated α-tubulin in fish endothelial,
epithelial and fibroblast cell lines. Fish Physiol Biochem.
42:29–38. 2016. View Article : Google Scholar
|
|
20
|
Lee CC, Cheng YC, Chang CY, Lin CM and
Chang JY: Alpha-tubulin acetyltransferase/MEC-17 regulates cancer
cell migration and invasion through epithelial-mesenchymal
transition suppression and cell polarity disruption. Sci Rep.
8:174772018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu S, Liu Y, Zhu B, Ding K, Yao TP, Chen
F, Zhan L, Xu P, Ehrlich M, Liang T, et al: Loss of α-Tubulin
acetylation is associated with TGF-β-induced epithelial-mesenchymal
transition. J Biol Chem. 291:5396–5405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McGraw M, Rioux J, Garlick R, Rancourt R,
White C and Veress L: From the cover: Impaired proliferation and
differentiation of the conducting airway epithelium associated with
bronchiolitis obliterans after sulfur mustard inhalation injury in
rats. Toxicol Sci. 157:399–409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee HN, Bosompra OA and Coller HA: RECK
isoforms differentially regulate fibroblast migration by modulating
tubulin post-translational modifications. Biochem Biophys Res
Commun. 510:211–218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Smith AC: A glitch in the matrix: Aberrant
extracellular matrix proteolysis contributes to alcohol seeking.
Biol Psychiatry. 81:900–902. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin L, Li FQ, Li J, Yang XR, Xie XY, Xue
LY, Li YL and Zhang C: Chronic intermittent ethanol exposure
induces upregulation of matrix metalloproteinase-9 in the rat
medial prefrontal cortex and hippocampus. Neurochem Res.
44:1593–1601. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang W, Yang C, Wang X, Zhou LY, Lao GJ,
Liu D, Wang C, Hu MD, Zeng TT, Yan L and Ren M: MicroRNA-129 and
-335 promote diabetic wound healing by inhibiting sp1-Mediated
MMP-9 expression. Diabetes. 67:1627–1638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
del Campo SEM, Latchana N, Levine KM,
Grignol VP, Fairchild ET, Jaime-Ramirez AC, Dao TV, Karpa VI,
Carson M, Ganju A, et al: MiR-21 enhances melanoma invasiveness via
inhibition of tissue inhibitor of metalloproteinases 3 expression:
In vivo effects of MiR-21 inhibitor. PLoS One. 10:e01159192015.
View Article : Google Scholar
|
|
28
|
Chen KC, Wang YS, Hu CY, Chang WC, Liao
YC, Dai CY and Juo SH: OxLDL up-regulates microRNA-29b, leading to
epigenetic modifications of MMP-2/MMP-9 genes: A novel mechanism
for cardiovascular diseases. FASEB J. 25:1718–1728. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen Z, Wei H, Zhao X, Xin X, Peng L, Ning
Y, Wang YP, Lan YL and Zhang QH: Metformin treatment alleviates
polycystic ovary syndrome by decreasing the expression of MMPand
MMPvia H19/miRb and AKT/mTOR/autophagy signaling pathways. J Cell
Physiol. 234:19964–19976. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Callejas-Díaz B, Fernandez G, Fuentes M,
Martínez-Antón A, Alobid I, Roca-Ferrer J, Picado C, Tubita V and
Mullol J: Integrated mRNA and microRNA transcriptome profiling
during differentiation of human nasal polyp epithelium reveals an
altered ciliogenesis. Allergy. 75:2548–2561. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang T, Zhou Y, You B, You YW, Yan YB,
Zhang J, Pei Y, Zhang W and Chen J: miR-30a-5p inhibits
epithelial-to-mesenchymal transition by targeting CDK6 in nasal
polyps. Am J Rhinol Allergy. 35:152–163. 2021. View Article : Google Scholar
|
|
32
|
Yang P, Chen S, Zhong G, Kong W and Wang
Y: Agonist of PPAR-γ reduced epithelial-mesenchymal transition in
eosinophilic chronic rhinosinusitis with nasal polyps via
inhibition of high mobility group box 1. Int J Med Sci.
16:1631–1641. 2019. View Article : Google Scholar :
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Zhang L, Zhao D, Jin M, Song MZ, Liu SC,
Guo KK and Zhang YM: Rab18 binds to classical swine fever virus
NS5A and mediates viral replication and assembly in swine umbilical
vein endothelial cells. Virulence. 11:489–501. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pommier A, Varilh J, Bleuse S, Delétang K,
Bonini J, Bergougnoux A, Brochiero E, Koenig M, Claustres M and
Taulan-Cadars M: miRNA repertoires of cystic fibrosis ex vivo
models highlight miR-181a and miR-101 that regulate WISP1
expression. J Pathol. 253:186–197. 2021. View Article : Google Scholar
|
|
36
|
Mudd P, Katial R, Alam R, Hohensee S,
Ramakrishnan V and Kingdom T: Variations in expression of matrix
metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 in
nasal mucosa of aspirin-sensitive versus aspirin-tolerant patients
with nasal polyposis. Ann Allergy Asthma Immunol. 107:353–359.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Y, Langhammer T, Westhofen M and
Lorenzen J: Relationship between matrix metalloproteinases MMP-2,
MMP-9, tissue inhibitor of matrix metalloproteinases-1 and IL-5,
IL-8 in nasal polyps. Allergy. 62:66–72. 2007. View Article : Google Scholar
|
|
38
|
Yeo N, Eom D, Oh M, Lim H and Song Y:
Expression of matrix metalloproteinase 2 and 9 and tissue inhibitor
of metalloproteinase 1 in nonrecurrent vs recurrent nasal polyps.
Ann Allergy Asthma Immunol. 111:205–210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Karadag A, Fedarko N and Fisher L: Dentin
matrix protein 1 enhances invasion potential of colon cancer cells
by bridging matrix metalloproteinase-9 to integrins and CD44.
Cancer Res. 65:11545–11552. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Redondo-Muñoz J and Ugarte-Berzal E:
Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia b
cell survival through its hemopexin domain. Cancer Cell.
17:160–172. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ugarte-Berzal E, Bailón E, Amigo-Jiménez
I, Vituri CL, del Cerro MH, Terol MJ, Albar JP, Rivas G,
García-Marco JA and García-Pardo A: A 17-residue sequence from the
matrix metalloproteinase-9 (MMP-9) hemopexin domain binds α4β1
integrin and inhibits MMP-9-induced functions in chronic
lymphocytic leukemia B cells. J Biol Chem. 287:27601–27613. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alford V, Kamath A, Ren X, Kumar K, Gan Q,
Awwa M, Tong M, Seeliger MA, Cao J, Ojima I and Sampson NS:
Targeting the hemopexin-like domain of latent matrix
metalloproteinase-9 (proMMP-9) with a small molecule inhibitor
prevents the formation of focal adhesion junctions. ACS Chem Biol.
12:2788–2803. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen S, Owens GC, Makarenkova H and
Edelman DB: HDAC6 regulates mitochondrial transport in hippocampal
neurons. PLoS One. 5:e108482010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tan HF and Tan SM: The focal adhesion
protein kindlin-2 controls mitotic spindle assembly by inhibiting
histone deacetylase 6 and maintaining α-tubulin acetylation. J Biol
Chem. 295:5928–5943. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bouvard D, Pouwels J, De Franceschi N and
Ivaska J: Integrin inactivators: Balancing cellular functions in
vitro and in vivo. Nat Rev Mol Cell Biol. 14:430–442. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mudd PA, Katial RK, Alam R, Hohensee S,
Ramakrishnan V and Kingdom TT: Variations in expression of matrix
metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 in
nasal mucosa of aspirin-sensitive versus aspirin-tolerant patients
with nasal polyposis. Ann Allergy Asthma Immunol. 107:353–359.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kovacs-Kasa A, Kovacs L, Cherian-Shaw M,
Patel V, Meadows M, Fulton D, Su YC and Verin AD: Inhibition of
class IIa HDACs improves endothelial barrier function in
endotoxin-induced acute lung injury. J Cell Physiol. 236:2893–2905.
2021. View Article : Google Scholar
|
|
48
|
Zheng Y, Chen Y, Lu X, Weng QH, Dai GL, Yu
Y, Yu KH and Gao WY: Inhibition of histone deacetylase 6 by
Tubastatin a attenuates the progress of osteoarthritis via
improving mitochondrial function. Am J Pathol. 190:2376–2386. 2020.
View Article : Google Scholar : PubMed/NCBI
|