Hepatocellular carcinoma (HCC) is one of the most
common, aggressive malignancies and a frequent cause of
cancer-related mortality worldwide, especially in many Asian and
African countries (1,2). The high recurrence rate of HCC is
mainly due to the spread of intrahepatic metastasis (3,4).
Metastasis is a complex and multistep process. In this process,
tumour metastasis-related gene abnormalities,
epithelial-mesenchymal transition (EMT), and tumour
microenvironment alterations, such as angiogenesis, are believed to
be important for the metastasis process (5). However, the underlying molecular
mechanisms that mediate the metastatic cascade remain unclear.
Recently, accumulated evidence suggests that long noncoding RNAs
(lncRNAs) may play a crucial role in the metastasis process of
HCC.
lncRNAs are defined as RNA molecules with a length
of 200-100,000 bp that lack protein-coding potential (6,7).
lncRNAs have been reported to be involved in gene regulation,
including transcriptional and post-transcriptional regulation,
epigenetic regulation, and siRNA-directed gene regulation (8). Hundreds of lncRNAs have been
identified (http://rfam.xfam.org) by computational
predictions, and some of them have been experimentally verified and
show tissue-specific expression (9-11). Moreover, these novel lncRNAs were
elucidated to be involved in some pivotal biological processes,
including cell growth, proliferation, apoptosis and metastasis and
angiogenesis (12). In addition,
recent evidence suggests that some HCC-related lncRNAs can act
either as oncogenes or tumour suppressor genes, and expression
profiling has revealed characteristic lncRNA signatures in HCC
(13,14). Similarly, some HCC-related
lncRNAs have been suggested to be useful as novel potential markers
for HCC diagnosis and prognosis (15). This review focused on the
relationship between lncRNAs and EMT, HCC metastasis-related genes
and tumour angiogenesis and highlights many pathways of lncRNAs
involved in these processes.
Gene expression profiling analyses have shown that
numerous lncRNAs are dysregulated in HCC cell lines or cancer
tissues compared with liver normal cell lines or matched normal
liver tissue (16,17). Importantly, some of these
aberrantly expressed lncRNAs have been confirmed to be associated
with hepatocarcinogenesis (18,19). In addition, lncRNA expression
signatures were correlated with clinicopathological characteristics
of HCC, such as metastasis and prognosis (20,21). In the present review, the
aberrant expression of lncRNAs and their biological roles in HCC
were summarized (Table I).
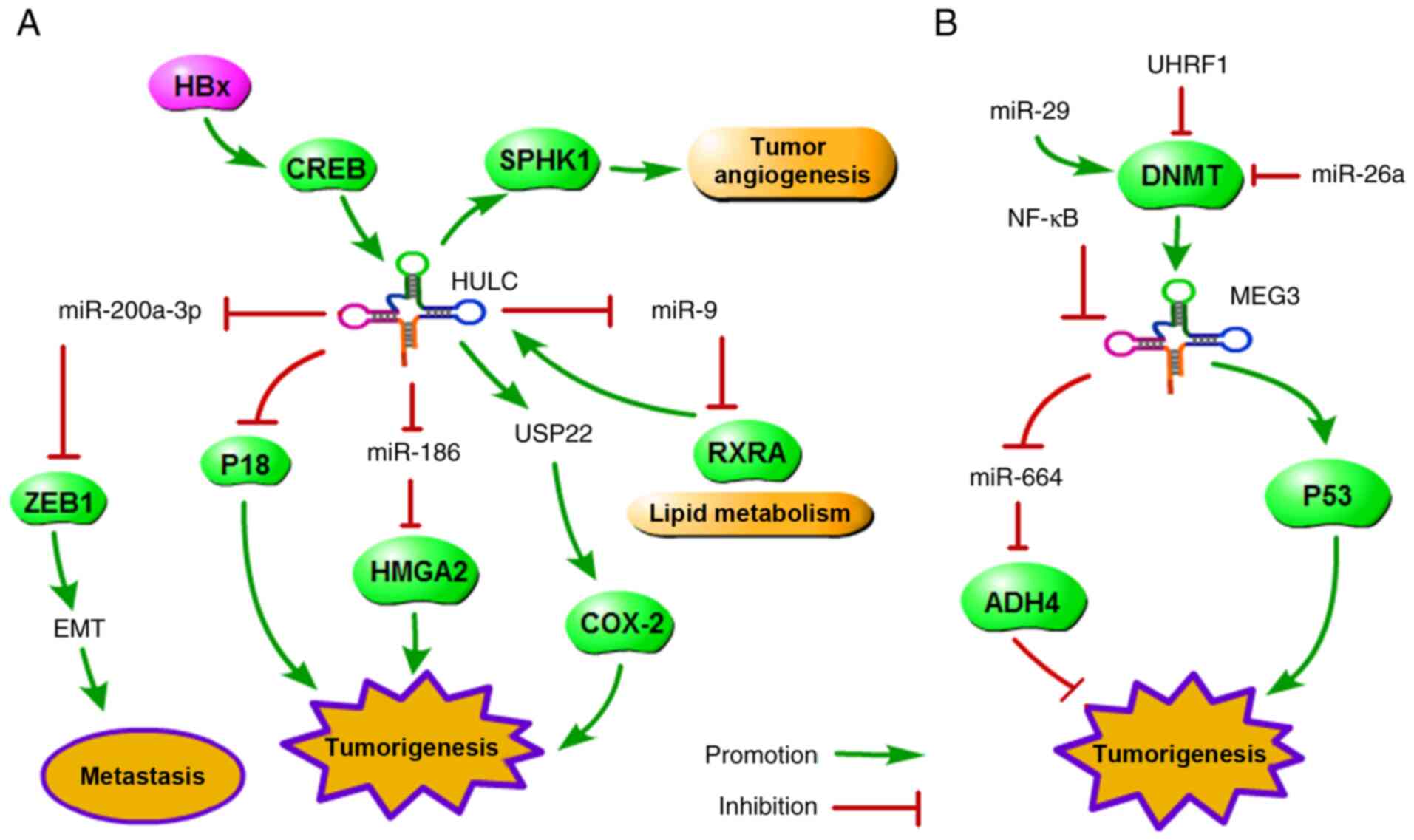
Among the upregulated lncRNAs, highly upregulated in
liver cancer (HULC) was identified and reported by more than one
study. As Fig. 1A shows, HULC,
as an oncogene, promotes tumorigenesis by controlling multiple
pathways, such as upregulation of HMGA2 expression (13), promotion of EMT via miR-200a-3p
(22), promotion of abnormal
lipid metabolism via RXRA (23),
and upregulation of sphingosine kinase 1 (SPHK1) expression to
promote tumour angiogenesis (24). The reports indicated that these
signalling pathways broadly interact with each other and highlight
the complexity of gene regulation by lncRNAs. Interestingly, HULC
was also upregulated in other cancer types, including gastric
cancer, osteosarcoma, pancreatic cancer and breast cancer (25). The aforementioned studies
suggested that HULC plays an important pathogenic role and clinical
value in human cancer. Among the downregulated lncRNAs, as shown in
Fig. 1B, maternally expressed
gene 3 (MEG3), as a tumour suppressor, inhibits tumorigenesis and
progression by regulating multiple pathways, such as the
MEG3/miR-664/ADH4 (14) and
DNMT1/MEG3/P53 axes (26,27).
Taken together, these data indicate that dysregulation of lncRNAs
results in abnormal expression of target genes and activity of
signalling pathways, eventually leading to tumorigenesis and
metastasis.
Epithelial-mesenchymal transition (EMT) is a highly
conserved molecular reprogramming process that causes polarized
immotile epithelial cells to change to motile mesenchymal cells
(28). This process was
initially recognized during embryonic development and was
implicated in the early events of tumour cell metastasis by
endowing cells with a more motile, invasive potential (29,30). EMT is mediated by key
transcription factors, including zinc-finger E-box-binding (ZEB),
SNAIL, and basic helix-loop-helix (bHLH) transcription factors
(31). These key transcription
factors are regulated at the transcriptional and translational
levels. In addition to EMT transcription factors, different
signalling pathways have leading roles in the initiation and
progression of EMT (32). Thus,
lncRNAs may play a crucial role in EMT progression.
lncRNAs that selectively bind mRNAs, miRNAs, and
proteins, thus inhibiting their transcription, promoting their
degradation or suppressing their translation, also regulate EMT
progression (6,33). A decrease in E-cadherin
expression is considered a crucial step and fundamental event in
the progression of EMT. We know that SNAIL and ZEB transcription
factors, as EMT drivers, can bind to and repress the activity of
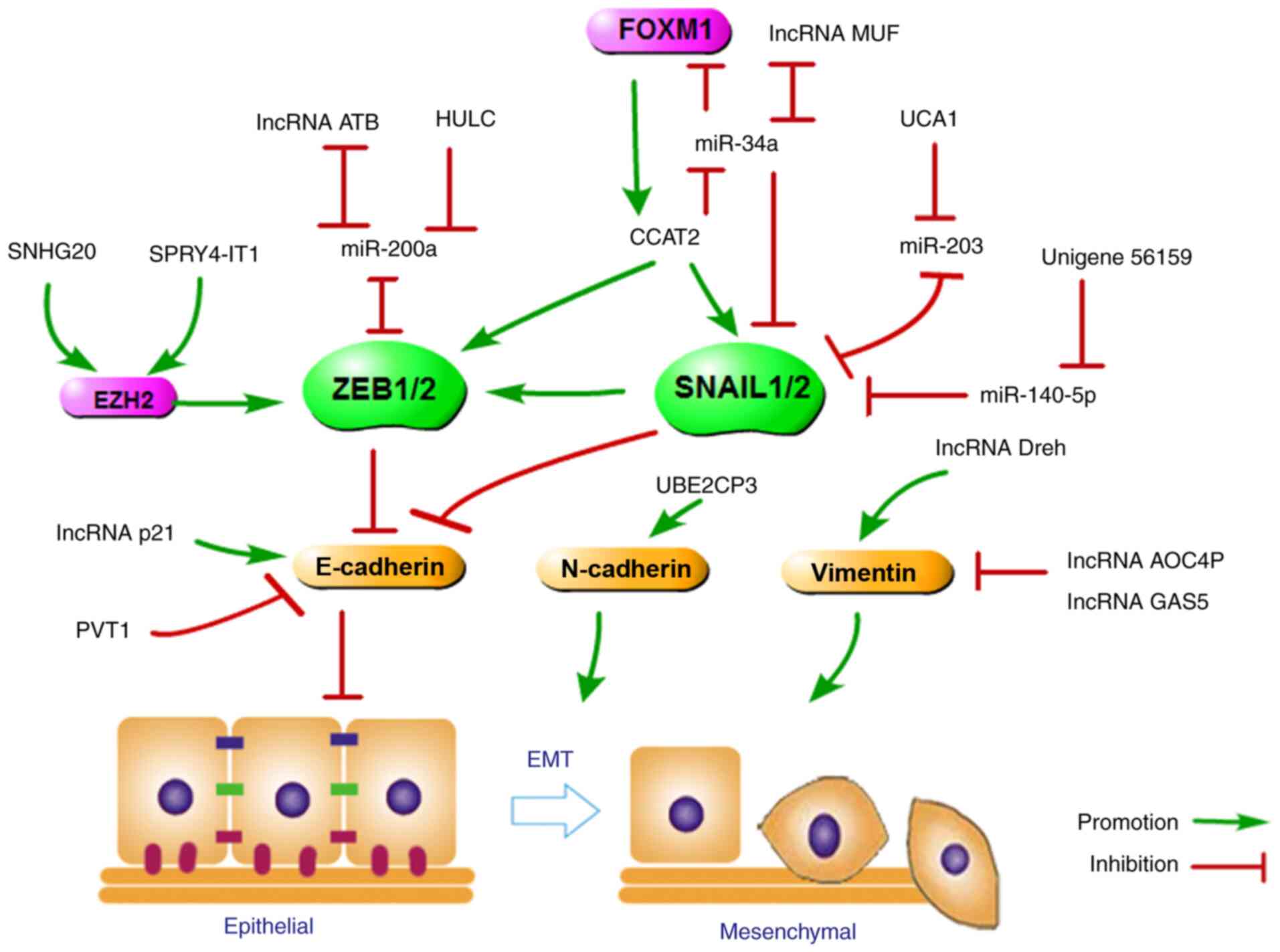
the E-cadherin promoter, leading to EMT (31,34). Accumulated evidence has shown
that some lncRNAs control the expression of EMT master
transcription factors. A schematic model and regulatory network of
lncRNA functions during the process of EMT were summarized and
generated (Fig. 2). For example,
lncRNA-ATB and lncRNA HULC upregulated ZEB1 and ZEB2 expression by
competitively binding miR-200a; hence, lncRNA-ATB and lncRNA HULC
depletion or the loss of ZEB by siRNA can reverse EMT and inhibit
HCC invasion and metastasis (22,35). Additionally, lncRNA CCAT2, as an
oncogene, promotes SNAIL2 and ZEB1 expression, and decreased
expression of E-cadherin leads to EMT (36,37). Interestingly, lncRNA CCAT2 can
promote HCC progression by competitively binding to miR-34a and
upregulating FOXM1 expression while FOXM1 binds to the CCAT2
promoter to activate its transcription, resulting in a
double-positive feedback loop between CCAT2 and FOXM1 (38). SNHG20 promoted ZEB1/2 and
N-cadherin expression and downregulated E-cadherin expression in
HCC by binding to enhancer of zeste homolog 2 (39). A similar regulatory mechanism
also exists between SPRY4-IT1 and EZH2 and E-cadherin (40).
UCA1 upregulation promotes the translation of SNAIL2
mRNA and induces EMT progression in HCC by effectively sponging
miR-203. MiR-203 and SNAIL2 regulation is a double-negative
feedback loop, with miR-203 suppressing SNAIL2 expression and
SNAIL2 protein repressing the expression of miR-203 (41). During EMT, increased HULC or
UBE2CP3 expression results in upregulation in SNAIL1 levels and EMT
progression (42,43). Additionally, lncRNA-MUF
overexpression accelerated EMT progression by binding Annexin A2
and activating the Wnt/β-catenin pathway. lncRNA-MUF functions as a
competing endogenous RNA for miR-34a, leading to SNAIL1
upregulation and EMT activation (44). Finally, SNAIL2, as a target of
miR-140-5p (which induces EMT in HCC cells), is upregulated by
Unigene56159 in HCC (45).
Apart from regulating EMT transcription factor
expression, lncRNAs target genes that encode adhesion junction and
polarity complex proteins and signalling mediators. For example,
high levels of lncRNA UBE2CP3, which are related to poor prognosis
in HCC patients, inhibit E-cadherin expression but enhance SNAIL1
and N-cadherin expression and promote EMT progression by increasing
HCC cell invasion and migration (43). Similarly, high levels of PVT1
promote HCC cell EMT progression by repressing E-cadherin
expression (46). Conversely,
lncRNA-p21 inhibits HCC invasion and metastasis through the
miR-9/E-cadherin cascade signalling pathway (47). N-cadherin expression is repressed
by SNHG20, the expression of which is positively associated with
larger tumour size and is negatively correlated with OS in HCC
patients. Additionally, decreased SNHG20 expression in HCC cells
results in increased N-cadherin expression levels and induces EMT
progression (39). During EMT,
high expression of ZEB2 AS1, which was correlated with tumour
metastasis in HCC, enhanced HCC metastasis by regulating ZEB2,
E-cadherin and N-cadherin expression (48).
Several lncRNAs could repress the expression of the
intermediate filament protein vimentin, which changes the normal
cytoskeleton structure, to inhibit HCC cell EMT progression and
metastasis. Among these, lncRNA-Dreh, which is downregulated by
HBx, targets vimentin mRNA, resulting in decreased vimentin
expression and reduced dissolution of tight junctions, to block EMT
(49). Vimentin is also
controlled by lncRNA AOC4P and lncRNA GAS5, both of which are
tumour suppressors that bind to vimentin and promote its
degradation and therefore inhibit HCC invasion and metastasis
(50,51). Additionally, CASC2 could inhibit
HCC cell invasion and EMT progression by targeting the
miR-367/FBXW7 axis, and lncRNA-p21 decreased EMT and metastasis by
targeting Notch (52). Clearly,
abnormal expression and activities of lncRNAs represent an
extensive regulatory network by changing the gene expression
programme to control EMT progression.
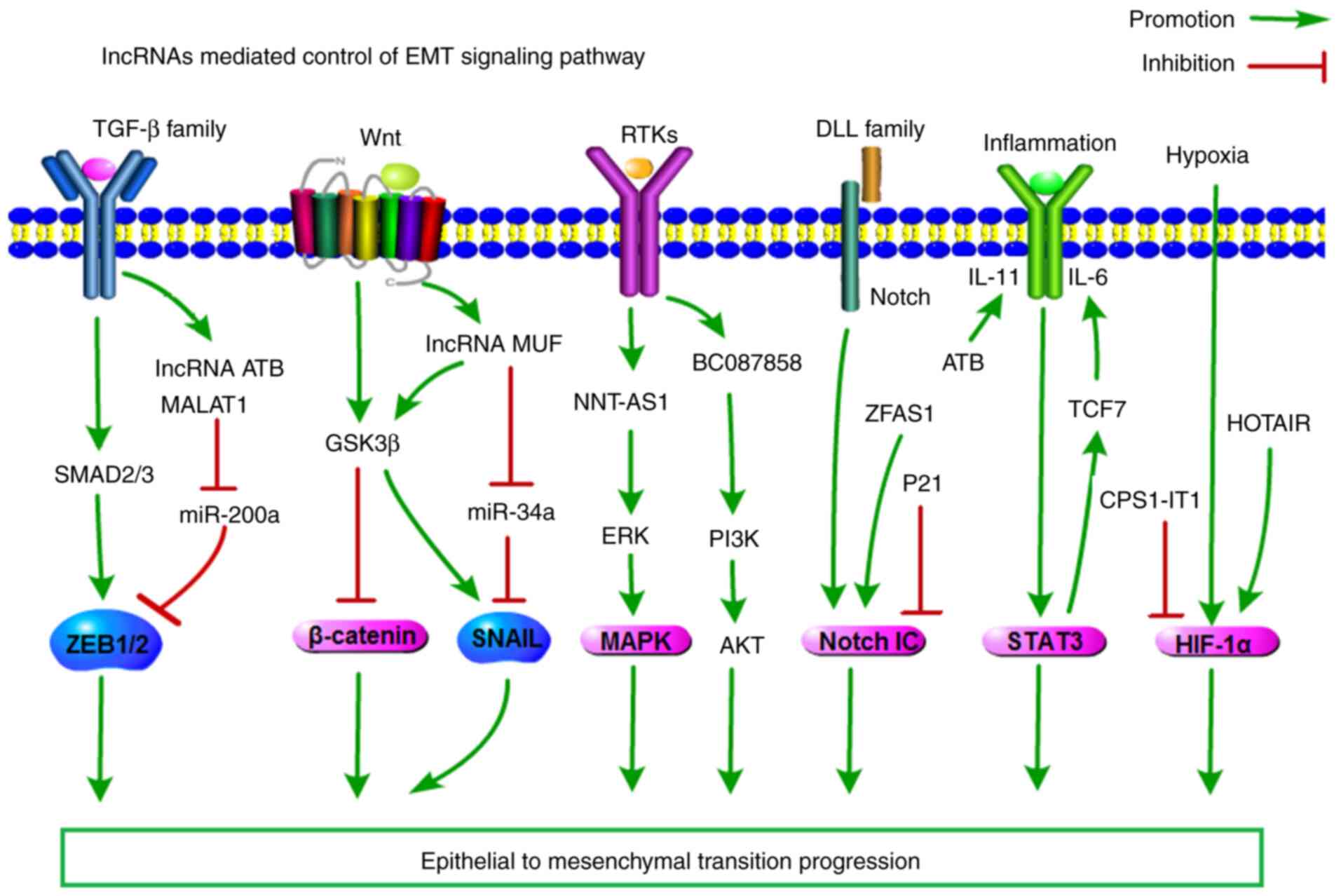
Long noncoding RNAs are commonly known as potential
gene expression modifiers that alter several phases of expression,
including transcription, post-transcriptional processing,
translation, and epigenetic regulation (6). Recently, the number of known
lncRNAs that have been directly or indirectly associated with EMT
signalling pathways has become extensive (Fig. 3). Multiple signalling pathways
trigger EMT in the initiation and progression of tumour metastasis,
including the transforming growth factor-β (TGF-β) family,
Wnt/β-catenin, Notch, EGF, HGF, FGF, and HIF. Among these pathways,
the TGF-β signalling pathway has a predominant role, but the others
are also required. Most importantly, the convergence of these
signalling pathways is essential for EMT (53).
As the role of TGF-β-induced EMT in cancer cell
dissemination is well established, Yuan et a. reported that
lncRNA-ATB promotes HCC cell metastasis by competitively binding
miR-200a, upregulating ZEB1 and ZEB2, and then inducing EMT
(35). Notably, several similar
reports in other tumour types (lung cancer, breast cancer and
endometrial carcinoma) have been reported. For example, Li et
a. showed that MALAT1 expression was increased by TGF-β
induction, showed that, lnc00673 activated by TGF-β induced EMT,
promoted lung cancer cell invasion by inhibiting miR-150-5p
expression, and then upregulated ZEB1 to finally induce EMT
progression (55). Additionally,
studies with similar regulatory mechanisms were also reported
(56,57).
The WNT signalling pathway promotes EMT, and
β-catenin-mediated gene expression is increased in tumour cells and
tissues. lncRNA-MUF is highly expressed in HCC. Decreased
lncRNA-MUF expression repressed EMT; conversely, lncRNA-MUF
overexpression accelerated EMT progression. Mechanistic
investigations suggested that lncRNA-MUF combined with Annexin A2
to activate Wnt/β-catenin and EMT. In addition, lncRNA-MUF can bind
to miR-34a, leading to Snail1 upregulation and EMT activation
(44).
The activation of EMT by receptor tyrosine kinases
(RTKs) highlights the roles of the PI3K-AKT and MAPK pathways in
this transitional process. MAPK pathway activation by not only
growth factors but also mutations in the genes encoding RAS or RAF,
also contributes to EMT. lncRNA NNT-AS1 promoted CRC cell migration
and invasion by activating the ERK/MAPK pathway and EMT (58). lncRNA BC087858 could promote cell
invasion by activating the PI3K/AKT and MEK/ERK pathways and EMT by
upregulating ZEB1 and Snail expression in NSCLC (59).
The Notch signalling pathway also triggers EMT.
Overexpression of lincRNA-p21 inhibited Notch signalling and EMT,
while its downregulation led to the reverse result (60). Upregulation of ZFAS1 expression
in glioma tissues was significantly associated with poor overall
survival. EMT and the Notch signalling pathway were inhibited in
glioma cells after ZFAS1 knockdown. Therefore, ZFAS1 could exhibit
an oncogenic role by regulating EMT and the Notch signalling
pathway (61).
Activation of the HIF-α/STAT3 signalling pathway
also switches on the EMT process. lncRNA-ATB promoted the
colonization of disseminated tumour cells to organs by binding
IL-11 and triggering STAT3 signalling (35). IL-6 could induce lncTCF7
expression in HCC cells by activating STAT3. Importantly, knocking
down STAT3 and repressing STAT3 activation reduced lncTCF7
expression; as a result, decreased lncTCF7 prevented IL-6-induced
EMT (62). Decreased expression
of CPS1-IT1 was significantly correlated with poor prognosis.
CPS1-IT1 acts as a tumour suppressor in HCC by decreasing HIF-1α
activation and inhibiting the EMT process (63). Another report describes HOTAIR
and RCC metastasis. The results showed that HOTAIR promotes RCC
tumorigenesis and metastasis via miR-217/HIF-1α/AXL signalling both
in vitr. and in viv. (64).
Clearly, these intricate networks are closely
connected, and changes in any of these networks have a profound
effect on other networks. Once the network of balance is disrupted,
one or several signalling pathways may be switched on and
activated, finally leading to EMT or MET.
The role of lncRNAs either as oncogenes or tumour
suppressors in human cancers, including HCC, has been previously
established. Genome-wide transcriptomic analyses have identified a
series of metastasis-associated genes, and their aberrant
expression in primary HCC correlates with metastasis and poor
prognosis. Recently, an increasing number of lncRNAs associated
with metastasis based on lncRNA profiling of early-stage versus
advanced HCC tissues have been identified and verified as upstream
regulators of metastasis-associated genes. For the present review,
the available evidence on these lncRNAs in HCC was summarized.
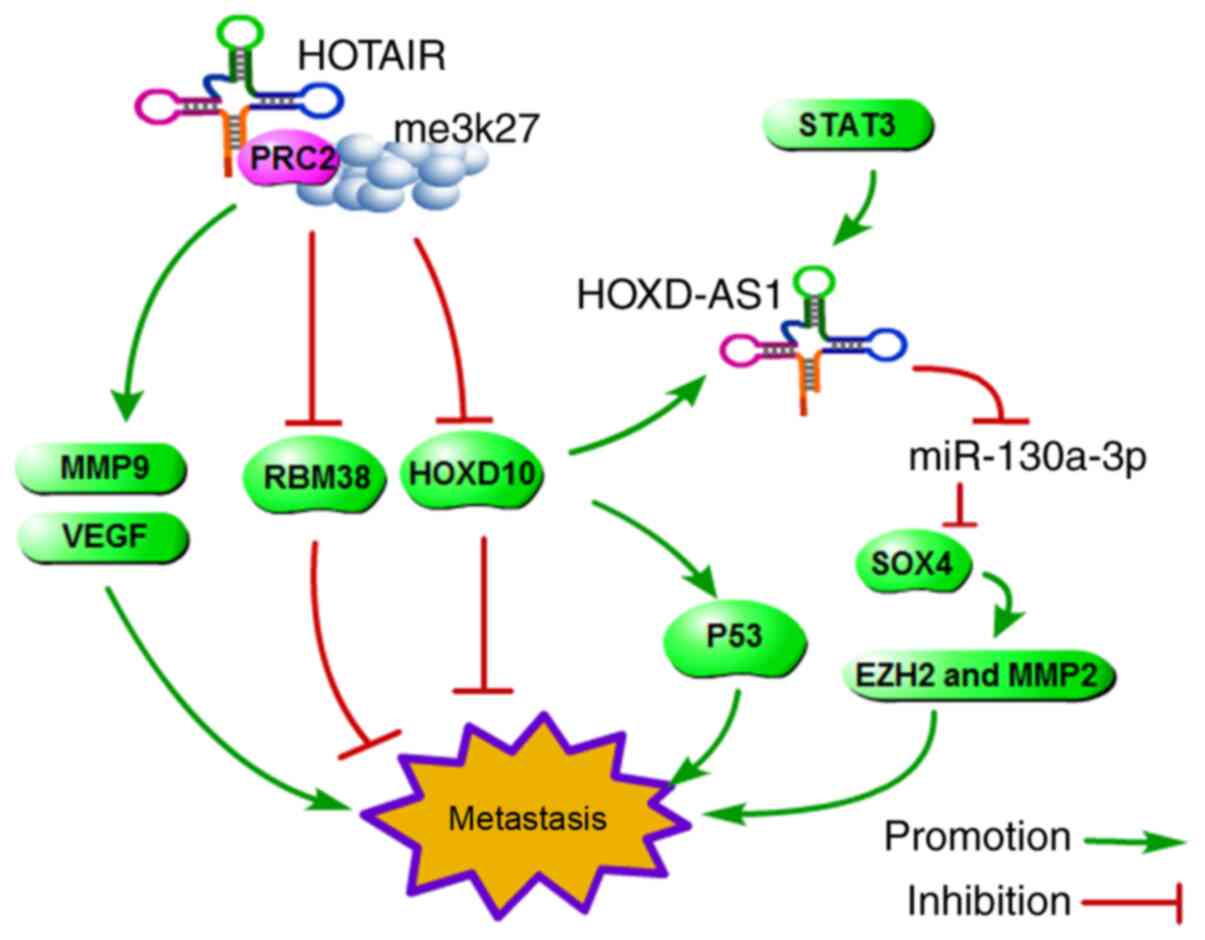
High expression of HOX transcript antisense RNA
(HOTAIR) in human breast cancer was first reported by Gupta et
a. (65). Overexpression of
HOTAIR promoted cancer cell metastasis in vitr. and in
viv.. Following investigation of the mechanism, they reported
that HOTAIR is selectively required to target polycomb repressive
complex 2 (PRC2) and induce methylation of histone H3 on lysine 27
(H3K27me3), resulting in the downregulation of multiple metastasis
suppressor genes, including HOXD1. and many others.
Moreover, HOTAIR is highly expressed in breast cancer tissues, and
upregulation of HOTAIR expression in primary breast cancer is a
powerful predictor of prognosis (65). Similarly, HOTAIR is correlated
with tumour size and lymph node metastasis in HCC. Knockdown of
HOTAIR decreases matrix metallopeptidase-9 (MMP-9) and vascular
endothelial growth factor (VEGF) expression, which play an
important role in metastasis (66). The HOTAIR/PRC2/H3K27me3/HOXD axis
is a classic example of metastasis-related interplay between
lncRNAs and cancer metastasis-associated genes.
As described previously, HOXD may be a key lncRNA in
the cancer metastatic pathway. lncRNA HOXD-AS1 is transcribed in
the antisense orientation of the protein-coding gene HOXD..
lncRNA HOXD-AS1 functions as an oncogene and can promote cell
invasion and metastasis by affecting signalling pathways in various
cancer cell lines. Global gene expression analysis revealed that
HOXD-AS1 upregulation was associated with poor prognosis in HCC
patients and may represent an independent prognostic biomarker in
HCC. Mechanistically, the transcription factor STAT3 could activate
the transcription of HOXD-AS1, which protects SOX4 against
miRNA-mediated degradation and thus activates the expression of
EZH2 and MMP2 to facilitate HCC metastasis (67). Lu et a. reported that
HOXD-AS1 promotes HCC metastasis and that its pro-metastatic
phenotype can partially be attributed to the
HOXD-AS1/miR-19a/ARHGAP11A signalling axis (68). Accordingly, the
HOTAIR/PRC2/H3K27me3/HOXD, HOXD-AS1/miR-130a-3p/SOX4/EZH2 and MMP2
and HOXD-AS1/miR-19a/ARHGAP11A signalling pathways form a cascade
effect for HCC metastasis (Fig.
4).
The long noncoding RNA metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1), the expression of which is
upregulated in lung cancer tissues and cells, is a critical
regulator of the metastasis phenotype of lung cancer (9). Similarly, findings have shown that
MALAT1 is a pro-metastatic lncRNA in HCC invasion and metastasis.
First, the level of MALAT1 expression correlated significantly with
advanced clinical stage, metastasis and poor prognosis in HCC.
Moreover, MALAT1 was found to promote HCC cell invasion and
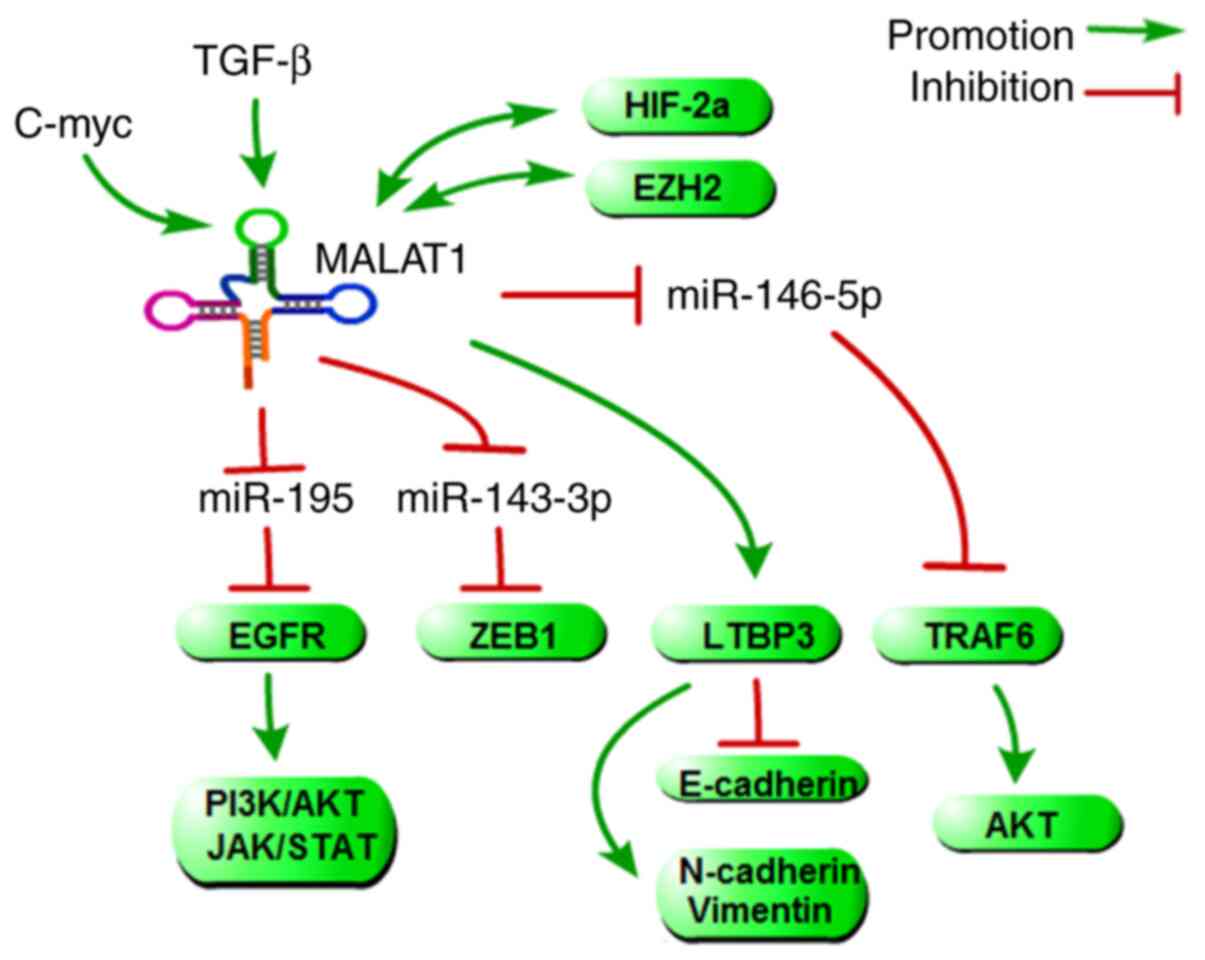
metastasis both in vitr. and in viv. (69). In addition, many targets of
MALAT1, which include some metastasis-associated genes that
directly or indirectly regulate HCC metastasis, have been
identified by computational predictions, and some of them have been
experimentally verified (Fig.
5). Transforming growth factor β-binding protein 3 (LTBP3),
which has been identified as a target gene of MALAT1, could promote
HCC cell migration and invasion, which could also be upregulated by
HBx. Mechanistically, one group found that HBx could upregulate
MALAT1 and that MALAT1 could further activate the expression of
LTBP3, resulting in the promotion of HCC metastasis (70). EGFR is also an important target
of MALAT1, not only because EGFR is a validated driver gene and an
important protein in tumour metastasis, but also because EGFR can
promote the PI3K/AKT and JAK/STAT signalling pathway activity.
Mechanistically, MALAT promotes HCC metastasis by targeting
miR-195-mediated EGFR phosphorylation (71). Similar to EGFR, ZEB1 and TRAF6
(as targets of MALAT1 mediated by miR-143-3p and miR-146-5p)
promote HCC metastasis (72,73). Aside from its role in regulating
downstream genes, MALAT1 is regulated by several tumour
metastasis-associated genes. TGF-β, c-MYC, and HIF-2α could induce
or activate the expression of MALAT1 by different molecular
mechanisms (74-76). With the reciprocal positive
feedback loop between MALAT1 and EZH2 or HIF-2α, these upstream
regulators and downstream targets, such as EGFR, ZEB1, LEBP3 and
TRAF6, compose a complex network to regulate HCC metastasis.
Recently, several other lncRNAs were proposed to
promote HCC growth and metastasis. Yang et a. reported that
a high lncRNA expression in HCC was significantly associated with
recurrence and is an independent prognostic factor for survival.
lncRNA-HEIH promotes HCC growth by targeting enhancer of zeste
homolog 2 (EZH2) (77).
Moreover, lncRNA-MVIH (lncRNA associated with microvascular
invasion in HCC) can also regulate HCC growth and intrahepatic
metastasis by inhibiting the secretion of phosphoglycerate kinase 1
(PGK1). CCAT1 promotes HCC progression by functioning as a let-7
sponge and leads to release of the repression of its endogenous
targets HMGA2 and c-Myc (78).
Another pro-metastatic lncRNA, ZEB1-AS1, is frequently upregulated
in HCC, especially in metastatic HCC tissues. ZEB1-AS1 acts as an
oncogene in HCC and promotes tumour growth and metastasis by
positively regulating ZEB1 expression (79). PCAT-1 can promote HCC cell
proliferation and viability by targeting the kinase CRK-like
proto-oncogene adaptor protein (CRKL), and, in turn, PCAT-1 is
regulated by miR-215, a P53-inducible miRNA. Therefore, the
TP53-PCAT-1-CRKL axis may be an important regulatory pathway in HCC
(80). Taken together, the data
regarding these pro-metastatic lncRNAs and their targets suggest
the existence of a complex network controlling HCC growth and
metastasis.
Another metastasis suppressor, lncRNA GAS5, was
found to be downregulated in HCC tissues compared to adjacent
normal tissues. Moreover, a decreased expression of GAS5 was
significantly correlated with differentiation and portal vein
tumour metastasis and was an independent predictor for overall
survival. In addition, it was demonstrated that GAS5 suppressed
proliferation and invasion in HCC by negatively regulating vimentin
expression (51). GAS5 was
downregulated in other cancers, and restoring its expression
significantly inhibited cancer progression. EZH2, miR-21 and
miR-222 were further verified as GAS5 target genes, and knocking
down these genes can reduce the in vitr. invasive ability
and in viv. metastatic potential (82-84). The downregulation of long
noncoding RNA FTX (lnc-FTX) was also found in HCC. Lnc-FTX can
inhibit HCC cell epithelial-mesenchymal transition and invasion by
physically binding miR-374a and MCM2 (85). Of note, Furlan et a.
reported that lnc-FTX is required in cis to promote XIST
transcriptional activation and establishment of X chromosome
inactivation (XCI) (86).
Tumour angiogenesis is considered a crucial step and
one of the cornerstones for helping to sustain expanding neoplastic
growth (5). In this regard, HCC
is one of the most vascular solid tumours with a high tendency for
vascular invasion (93). A
compelling body of evidence indicates that the angiogenesis switch
is triggered by cellular stress factors such as hypoxia, the
activation of oncogenes such as RAS and C-MYC or the inactivation
of tumour suppressors, such as P53. One of the well-known
regulatory pathways of angiogenesis is via signalling proteins that
bind to cell-surface receptors demonstrated by vascular endothelial
cells, such as VEGF and FGF (94). VEGF gene expression is induced
and upregulated by hypoxia through hypoxia inducible factor-1
(HIF-1), which in turn activates VEGFR2 to stimulate cell migration
and to initiate angiogenesis. The role of VEGF in the formation and
development of neovascularization in HCC has been extensively
reported and established in the process between the early and
advanced stage of HCC. Furthermore, VEGF correlates with HCC
progression, metastasis, a tendency towards portal invasion and a
higher recurrence rate (95).
Moreover, current clinical practice shows extensively established
and accepted VEGFR-targeted therapies for patients with advanced
HCC, such as the multi-kinase inhibitor sorafenib, which is well
known to inhibit the kinase activities of VEGFR and PDGFR (96).
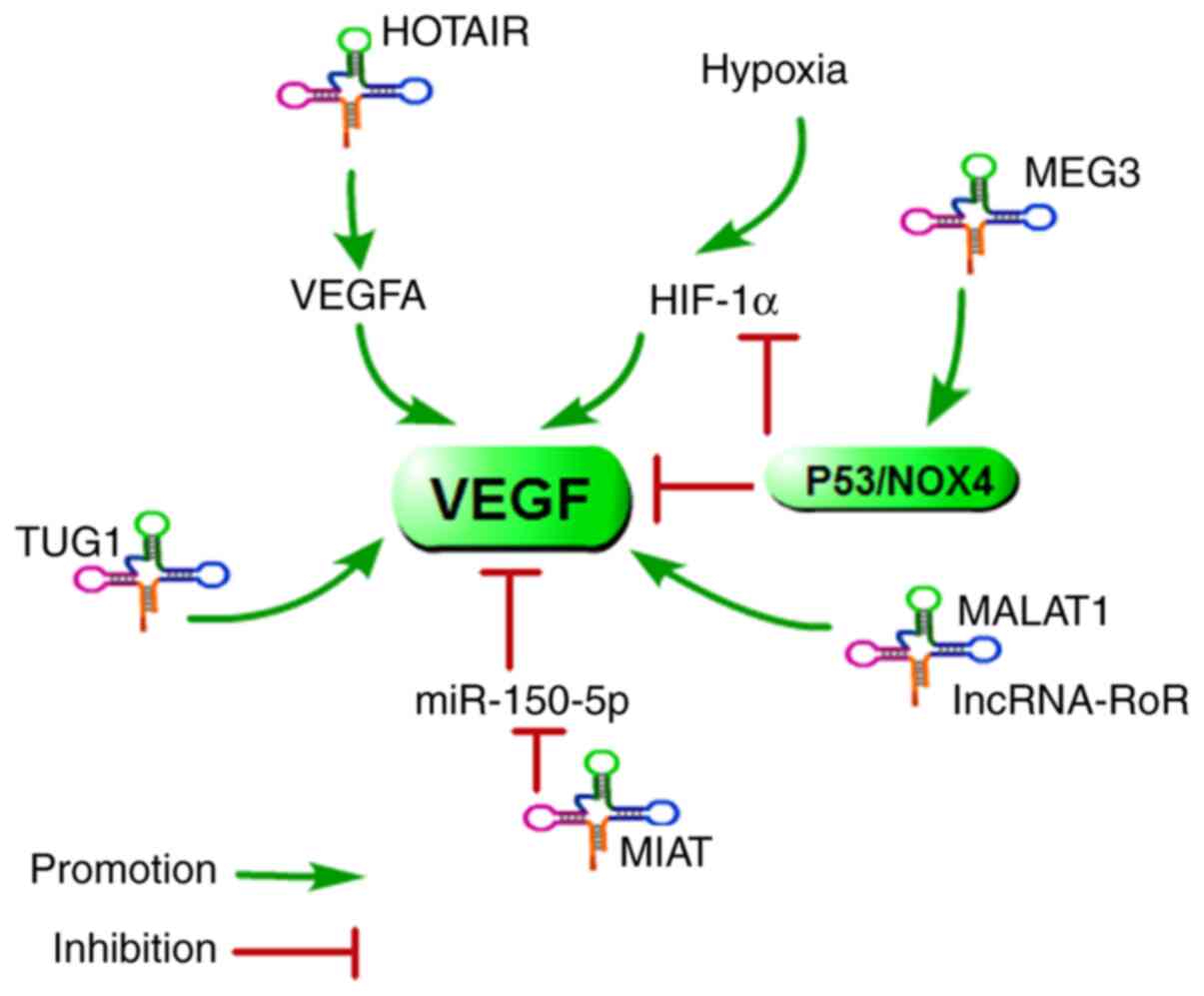
Recently, accumulating evidence has shown that
aberrantly expressed lncRNAs could be linked to cancer-associated
angiogenesis (Fig. 6). Fu et
a. reported that the upregulation of HOTAIR promoted tumour
cell growth and angiogenesis by directly activating VEGFA and Ang2
expression (97). A similar
result was also reported: HOTAIR could enhance angiogenesis by
inducing VEGFA expression in glioma cells (98). Su et a. found that MEG3
was downregulated and inversely associated with VEGF expression
levels in osteoarthritis (99).
Therefore, HOTAIR and MEG3 may play a major role in the control of
angiogenesis. Next, a report demonstrated that MEG3 could regulate
angiogenesis through activation of the p53/NOX4 axis, which would
downregulate HIF-1α and VEGF expression (100). Furthermore, upregulation of
lncRNA TUG1 in hepatoblastoma promotes tumour growth and
angiogenesis by enhancing the expression of VEGFA, which is
regulated by miR-34a-5p (101).
lncRNA-MIAT was able to promote endothelial cell proliferation,
migration, and tube formation by increasing the expression of VEGF
via direct binding to miR-150-5p (102). Additionally, a previous report
indicated that MALAT1 could promote angiogenesis and
immunosuppressive properties of mesenchymal stem cells by inducing
VEGF and IDO expression (103).
Zhu et a. demonstrated that HULC could promote angiogenesis
in human gliomas by regulating ESM-1 via the PI3K/Akt/mTOR
signalling pathway (104).
Furthermore, lncRNA associated with microvascular invasion in
hepatocellular carcinoma (lncRNA MVIH) is well known to play an
important role in HCC by inducing angiogenesis. lncRNA MVIH could
activate tumour-inducing angiogenesis by repressing the secretion
of PKG1 and increasing microvessel density in hepatocellular
carcinoma (78).
Another well-known switch of angiogenesis is
hypoxia. Under hypoxic conditions, angiogenesis can be activated by
a family of transcription factors known as hypoxia inducible
factors (HIFs), such as HIF-1. Dysregulation of HIF-1 expression is
associated with tumour metastasis and poor clinical outcome in HCC
(105). In recent years, a
series of hypoxia-induced lncRNAs, such as HOTAIR, MALAT1, and
PVT1, have been identified using RNA expression profiling and
high-throughput approaches. Based on RNA expression profiling data,
Takahashi et a. identified 89 differentially expressed
lncRNAs in HepG2 HCC cells under hypoxic conditions; 20 lncRNAs
were upregulated by at least a 2-fold change, while 11 were
downregulated under hypoxic conditions. Those authors further
confirmed that lncRNA-RoR, as an oncogene, could drive tumour
growth via the lncRNA-RoR/miR-145/HIF-1α axis. These
hypoxia-induced lncRNAs have important functions in controlling
tumour phenotypes, such as angiogenesis (106).
Accumulating evidence has shown that several
hypoxia-induced lncRNAs regulate tumour angiogenesis and metastasis
by modulating the HIF-1α signalling pathway. For example, Yang
et a. reported that lncRNA low expression in tumour
(lncRNA-LET) was downregulated in hepatocellular carcinomas.
Mechanistically, lncRNA-LET is suppressed under hypoxic conditions
due to the activation of histone deacetylase 3 (HDAC3). lncRNA-LET
was able to interact with nuclear factor 90 (NF90), which led to
the degradation of NF90, therefore increasing HIF-1A under hypoxic
conditions and leading to hypoxia-induced cancer cell invasion.
Notably, lncRNA-LET downregulation was significantly correlated
with tumour micrometastases in HCC (81). A similar result for lncRNA-LET as
a prognostic marker for metastasis was also reported in primary
gallbladder cancer (107).
Another report showed that lncRNA-RERT downregulation was
significantly correlated with HCC occurrence. lncRNA-RERT decreased
the expression of HIF-1α by upregulating EGLN2 mRNA levels
(108). In addition, two other
lncRNAs, HIF1A-AS1 and HIF1A-AS2, which are antisense transcripts
transcribed from the 3′-UTR of the sense HIF-1A mRNA negatively
regulated HIF1A mRNA expression (109,110). Therefore, the association
between HIF-1A mRNA levels and HIF1A-AS exhibits a negative
feedback loop.
It has been well established that lncRNAs involved
in the biological processes of HCC angiogenesis interact with the
key angiogenesis regulators VEGF and HIF-1A and appear to be
promising targets for anti-angiogenesis therapy. More novel
lncRNA-associated angiogenesis and its mechanisms need to be
identified and elucidated. Understanding the networks of
interactions between lncRNAs and target genes may pave the way for
new therapeutic strategies.
In this review, we have shown that, lncRNAs, acting
as tumour suppressor genes or oncogenes, play an important role in
HCC tumorigenesis and metastasis. Based on the corresponding
relationships between lncRNAs, miRNAs and mRNAs, the regulatory
relationships between lncRNAs and EMT, lncRNAs and HCC
metastasis-related genes, lncRNAs and tumour angiogenesis were
summarized. In these regulatory networks, once one of the important
lncRNAs is out of balance, a chain reaction ultimately affects the
relationship between multiple lncRNAs and several target genes of
different pathways. Accordingly, with the identification of new
lncRNAs associated with HCC metastasis, this regulatory network
becomes increasingly complex. However, the molecular mechanism of
HCC metastasis remains to be revealed more thoroughly. Based on
current studies and additional lncRNAs that will be identified and
verified in the future, lncRNAs may serve as novel diagnostic
markers, prognostic markers and therapeutic targets.
Not applicable.
LL and HP conceived the review. XZ and LL were
involved in collecting the references, writing and reviewing the
manuscript. XZ and LL were responsible for confirming the
authenticity of all the raw data. The authors contributed to the
final version.
Not applicable.
All authors consent to publication.
The authors declare that they have no competing
interests.
Not applicable.
This review was supported by the Science and Technology Project
of Zhejiang Province (2016KYA023, 2013RCA006, 2016ZA025,
2018KY466), the Natural Science Foundation of Zhejiang Province
(LQ18H160025) and the National Natural Science Foundation of China
(81572592).
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J and Sherman M; American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
9
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar
|
|
10
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nissan A, Stojadinovic A,
Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A,
Dayanc BE, Ritter G, Gomceli I, et al: Colon cancer associated
transcript-1: A novel RNA expressed in malignant and pre-malignant
human tissues. Int J Cancer. 130:1598–1606. 2012. View Article : Google Scholar
|
|
12
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Chen F, Zhao M, Yang Z, Li J,
Zhang S, Zhang W, Ye L and Zhang X: The long noncoding RNA HULC
promotes liver cancer by increasing the expression of the HMGA2
oncogene via sequestration of the microRNA-186. J Biol Chem.
292:15395–15407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv
YB, Li YG and Cao MR: Overexpression of long non-coding RNA MEG3
inhibits proliferation of hepatocellular carcinoma Huh7 cells via
negative modulation of miRNA-664. J Cell Biochem. 118:3713–3721.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv
XW and Li J: Long noncoding RNAs: Novel insights into hepatocelluar
carcinoma. Cancer Letters. 344:20–27. 2014. View Article : Google Scholar
|
|
16
|
Gong X, Wei W, Chen L, Xia Z and Yu C:
Comprehensive analysis of long non-coding RNA expression profiles
in hepatitis B virus-related hepatocellular carcinoma. Oncotarget.
7:42422–42430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y, Chen L, Gu J, Zhang H, Yuan J,
Lian Q, Lv G, Wang S, Wu Y, Yang YT, et al: Recurrently deregulated
lncRNAs in hepatocellular carcinoma. Nat Commun. 8:144212017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J,
Qiao Y and Zhan Q: Long noncoding RNA HULC modulates the
phosphorylation of YB-1 through serving as a scaffold of
extracellular signal-regulated kinase and YB-1 to enhance
hepatocarcinogenesis. Hepatology. 65:1612–1627. 2017. View Article : Google Scholar
|
|
19
|
He T, Zhang L, Kong Y, Huang Y, Zhang Y,
Zhou D, Zhou X, Yan Y, Zhang L, Lu S, et al: Long non-coding RNA
CASC15 is upregulated in hepatocellular carcinoma and facilitates
hepatocarcinogenesis. Int J Oncol. 51:1722–1730. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar :
|
|
22
|
Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ,
Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al: lncRNA HULC enhances
epithelial-mesenchymal transition to promote tumorigenesis and
metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1
signaling pathway. Oncotarget. 7:42431–42446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z,
Li J, Ye L and Zhang X: Long non-coding RNA HULC promotes tumor
angiogenesis in liver cancer by up-regulating sphingosine kinase 1
(SPHK1). Oncotarget. 7:241–254. 2016. View Article : Google Scholar :
|
|
25
|
Yu X, Zheng H, Chan MT and Wu WK: HULC: An
oncogenic long non-coding RNA in human cancer. J Cell Mol Med.
21:410–417. 2017. View Article : Google Scholar
|
|
26
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Wei L, Jin Y, Fu H, Wu Y and Zheng X: Long noncoding RNA MEG3
interacts with p53 protein and regulates partial p53 target genes
in hepatoma cells. PLoS One. 10:e01397902015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moreno-Bueno G, Portillo F and Cano A:
Transcriptional regulation of cell polarity in EMT and cancer.
Oncogene. 27:6958–6969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nieto MA, Huang RY, Jackson RA and Thiery
JP: Emt: 2016. Cell. 166:21–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-beta promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou N, Si Z, Li T, Chen G, Zhang Z and Qi
H: Long non-coding RNA CCAT2 functions as an oncogene in
hepatocellular carcinoma, regulating cellular proliferation,
migration and apoptosis. Oncol Lett. 12:132–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu Y, Wang B, Zhang F, Wang A, Du X, Hu P,
Zhu Y and Fang Z: Long non-coding RNA CCAT2 is associated with poor
prognosis in hepatocellular carcinoma and promotes tumor metastasis
by regulating Snail2-mediated epithelial-mesenchymal transition.
Onco Targets Ther. 10:1191–1198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen F, Bai G, Li Y, Feng Y and Wang L: A
positive feedback loop of long noncoding RNA CCAT2 and FOXM1
promotes hepatocellular carcinoma growth. Am J Cancer Res.
7:1423–1434. 2017.PubMed/NCBI
|
|
39
|
Liu J, Lu C, Xiao M, Jiang F, Qu L and Ni
R: Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and
promotes cell invasion by regulating the epithelial-to-mesenchymal
transition. Biomed Pharmacother. 89:857–863. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou M, Zhang XY and Yu X: Overexpression
of the long non-coding RNA SPRY4-IT1 promotes tumor cell
proliferation and invasion by activating EZH2 in hepatocellular
carcinoma. Biomed Pharmacother. 85:348–354. 2017. View Article : Google Scholar
|
|
41
|
Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu
SW, Que HX, Liu ZP and Jiang JH: Long non-coding RNA UCA1 regulates
the expression of Snail2 by miR-203 to promote hepatocellular
carcinoma progression. J Cancer Res Clin Oncol. 143:981–990. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q
and Zhang L: Molecular mechanism of HEIH and HULC in the
proliferation and invasion of hepatoma cells. Int J Clin Exp Med.
8:12956–12962. 2015.PubMed/NCBI
|
|
43
|
Cao SW, Huang JL, Chen J, Hu YW, Hu XM,
Ren TY, Zheng SH, Lin JD, Tang J, Zheng L and Wang Q: Long
non-coding RNA UBE2CP3 promotes tumor metastasis by inducing
epithelial-mesenchymal transition in hepatocellular carcinoma.
Oncotarget. 8:65370–65385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y,
Yan F, Zhu P, Wu J, Huang G, et al: Mesenchymal stem cells promote
hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and
miR-34a. Cancer Res. 77:6704–6716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang
Y, Liu M and Tang H: Long non-coding RNA Unigene 56159 promotes
epithelial-mesenchymal transition by acting as a ceRNA of
miR-140-5p in hepatocellular carcinoma cells. Cancer Lett.
382:166–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gou X, Zhao X and Wang Z: Long noncoding
RNA PVT1 promotes hepatocellular carcinoma progression through
regulating miR-214. Cancer Biomark. 20:511–519. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ding G, Peng Z, Shang J, Kang Y, Ning H
and Mao C: LincRNA-p21 inhibits invasion and metastasis of
hepatocellular carcinoma through miR-9/E-cadherin cascade signaling
pathway molecular mechanism. Onco Targets Ther. 10:3241–3247. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lan T, Chang L, Wu L and Yuan Y:
Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in
hepatocellular carcinoma. Mol Med Rep. 14:4606–4612. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar
|
|
50
|
Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL,
Hsieh TH, Shieh TM, Hsueh C and Chen TC: Long non-coding RNA AOC4P
suppresses hepatocellular carcinoma metastasis by enhancing
vimentin degradation and inhibiting epithelial-mesenchymal
transition. Oncotarget. 6:23342–23357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q
and Liu Z: Decreased expression of long non-coding RNA GAS5
indicates a poor prognosis and promotes cell proliferation and
invasion in hepatocellular carcinoma by regulating vimentin. Mol
Med Rep. 13:1541–1550. 2016. View Article : Google Scholar :
|
|
52
|
Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang
C, Dou C, Xu M, Liu Q and Tu K: Long non-coding RNA CASC2
suppresses epithelial-mesenchymal transition of hepatocellular
carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer.
16:1232017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Q, Zhang C, Chen R, Xiong H, Qiu F, Liu
S, Zhang M, Wang F, Wang Y, Zhou X, et al: Disrupting
MALAT1/miR-200c sponge decreases invasion and migration in
endometrioid endometrial carcinoma. Cancer Lett. 383:28–40. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA linc00673
regulated non-small cell lung cancer proliferation, migration,
invasion and epithelial mesenchymal transition by sponging
miR-150-5p. Mol Cancer. 16:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rodriguez-Mateo C, Torres B, Gutierrez G
and Pintor-Toro JA: Downregulation of Lnc-Spry1 mediates
TGF-beta-induced epithelial-mesenchymal transition by
transcriptional and post-transcriptional regulatory mechanisms.
Cell Death Differ. 24:785–797. 2017. View Article : Google Scholar
|
|
57
|
Li Z, Dong M, Fan D, Hou P, Li H, Liu L,
Lin C, Liu J, Su L, Wu L, et al: lncRNA ANCR down-regulation
promotes TGF-beta-induced EMT and metastasis in breast cancer.
Oncotarget. 8:67329–67343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Q, Yang L, Hu X, Jiang Y, Hu Y, Liu
Z, Liu J, Wen T, Ma Y, An G and Feng G: Upregulated NNT-AS1, a long
noncoding RNA, contributes to proliferation and migration of
colorectal cancer cells in vitro and in vivo. Oncotarget.
8:3441–3453. 2017. View Article : Google Scholar :
|
|
59
|
Pan H, Jiang T, Cheng N, Wang Q, Ren S, Li
X, Zhao C, Zhang L, Cai W and Zhou C: Long non-coding RNA BC087858
induces non-T790M mutation acquired resistance to EGFR-TKIs by
activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell
lung cancer. Oncotarget. 7:49948–49960. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jia M, Jiang L, Wang YD, Huang JZ, Yu M
and Xue HZ: lincRNA-p21 inhibits invasion and metastasis of
hepatocellular carcinoma through Notch signaling-induced
epithelial-mesenchymal transition. Hepatol Res. 46:1137–1144. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gao K, Ji Z, She K, Yang Q and Shao L:
Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and
promotes glioma cell progression by activation of the Notch
signaling pathway. Biomed Pharmacother. 87:555–560. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu J, Zhang J, Shen B, Yin K, Xu J, Gao W
and Zhang L: Long noncoding RNA lncTCF7, induced by IL-6/STAT3
transactivation, promotes hepatocellular carcinoma aggressiveness
through epithelial-mesenchymal transition. J Exp Clin Cancer Res.
34:1162015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT,
Liang KH, Shieh TM, Chen CY and Hsueh C: Long noncoding RNA
CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by
regulating HIF-1alpha activity and inhibiting
epithelial-mesenchymal transition. Oncotarget. 7:43588–43603. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hong Q, Li O, Zheng W, Xiao WZ, Zhang L,
Wu D, Cai GY, He JC and Chen XM: lncRNA HOTAIR regulates
HIF-1alpha/AXL signaling through inhibition of miR-217 in renal
cell carcinoma. Cell Death Dis. 8:e27722017. View Article : Google Scholar
|
|
65
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar
|
|
67
|
Wang H, Huo XS, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Molecular Cancer. 16:1362017.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lu S, Zhou J, Sun Y, Li N, Miao M, Jiao B
and Chen H: The noncoding RNA HOXD-AS1 is a critical regulator of
the metastasis and apoptosis phenotype in human hepatocellular
carcinoma. Molecular Cancer. 16:1252017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Malakar P, Shilo A, Mogilevsky A, Stein I,
Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV
and Karni R: Long noncoding RNA MALAT1 promotes hepatocellular
carcinoma development by SRSF1 upregulation and mTOR activation.
Cancer Research. 77:1155–1167. 2017. View Article : Google Scholar
|
|
70
|
Hou Z, Xu X, Fu X, Tao S, Zhou J, Liu S
and Tan D: HBx-related long non-coding RNA MALAT1 promotes cell
metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am
J Cancer Res. 7:845–856. 2017.PubMed/NCBI
|
|
71
|
Liu DL, Zhu Y, Pang JK, Weng X, Feng XR
and Guo YB: Knockdown of long non-coding RNA MALAT1 inhibits growth
and motility of human hepatoma cells via modulation of miR-195. J
Cell Biochem. 119:1368–1380. 2018. View Article : Google Scholar
|
|
72
|
Chen LS, Yao HB, Wang K and Liu XF: Long
non-coding RNA MALAT1 regulates ZEB1 expression by sponging
miR-143-3p and promotes hepatocellular carcinoma progression. J
Cell Biochem. 118:4836–4843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sun HB, Lin DC, Cao Q, Pang B, Gae DD, Lee
VKM, Lim HJ, Doan N, Said JW, Gery S, et al: Identification of a
novel SYK/c-MYC/MALAT1 signaling pathway and its potential
therapeutic value in Ewing sarcoma. Clin Cancer Res. 23:4376–4387.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yuan P, Cao WB, Zang QL, Li GX, Guo XF and
Fan JH: The HIF-2 alpha-MALAT1-miR-216b axis regulates multi-drug
resistance of hepatocellular carcinoma cells via modulating
autophagy. Biochem Bioph Res Commun. 478:1067–1073. 2016.
View Article : Google Scholar
|
|
77
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of Zeste Homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z,
Deng X, Chen H, Shen B, Peng C, et al: Upregulation of long
noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor
prognosis in hepatocellular carcinoma. Oncogene. 35:1575–1584.
2016. View Article : Google Scholar
|
|
80
|
Ren YL, Shang JH, Li JL, Liu W, Zhang Z,
Yuan J and Yang M: The long noncoding RNA PCAT-1 links the microRNA
miR-215 to oncogene CRKL-mediated signaling in hepatocellular
carcinoma. J Biol Chem. 292:17939–17949. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP,
Wang F and Sun SH: Repression of the long noncoding RNA-LET by
histone deacetylase 3 contributes to hypoxia-mediated metastasis.
Mol Cell. 49:1083–1096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang M, Guo C, Wang L, Luo G, Huang C, Li
Y, Liu D, Zeng F, Jiang G and Xiao X: Long noncoding RNA GAS5
promotes bladder cancer cells apoptosis through inhibiting EZH2
transcription. Cell Death Dis. 9:2382018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C,
Xu M, Wu F and Mo YY: Negative regulation of lncRNA GAS5 by miR-21.
Cell Death Differ. 20:1558–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhao XH, Wang P, Liu J, Zheng J, Liu Y,
Chen J and Xue Y: Gas5 exerts tumor-suppressive functions in human
glioma cells by targeting miR-222. Mol Ther. 23:1899–1911. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Liu F, Yuan JH, Huang JF, Yang F, Wang TT,
Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, et al: Long noncoding RNA
FTX inhibits hepatocellular carcinoma proliferation and metastasis
by binding MCM2 and miR-374a. Oncogene. 35:5422–5434. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Furlan G, Gutierrez Hernandez N, Huret C,
Galupa R, van Bemmel JG, Romito A, Heard E, Morey C and Rougeulle
C: The Ftx noncoding locus controls X chromosome inactivation
independently of its RNA products. Mol Cell. 70:462–472.e8. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhuang LK, Yang YT, Ma X, Han B, Wang ZS,
Zhao QY, Wu LQ and Qu ZQ: MicroRNA-92b promotes hepatocellular
carcinoma progression by targeting Smad7 and is mediated by long
non-coding RNA XIST. Cell Death Dis. 7:e22032016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chang SZ, Chen BH, Wang XY, Wu KQ and Sun
YQ: Long non-coding RNA XIST regulates PTEN expression by sponging
miR-181a and promotes hepatocellular carcinoma progression. BMC
Cancer. 17:2482017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Kong Q, Zhang S, Liang C, Zhang Y, Kong Q,
Chen S, Qin J and Jin Y: lncRNA XIST functions as a molecular
sponge of miR-194-5p to regulate MAPK1 expression in hepatocellular
carcinoma cell. J Cell Biochem. 119:4458–4468. 2018. View Article : Google Scholar
|
|
90
|
Ma L, Zhou YJ, Luo XJ, Gao H, Deng XB and
Jiang YJ: Long non-coding RNA XIST promotes cell growth and
invasion through regulating miR-497/MACC1 axis in gastric cancer.
Oncotarget. 8:4125–4135. 2017. View Article : Google Scholar :
|
|
91
|
Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng
ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ and Xu RH: Long
noncoding RNA XIST expedites metastasis and modulates
epithelial-mesenchymal transition in colorectal cancer. Cell Death
Dis. 8:e30112017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wu XL, Dinglin XX, Wang X, Luo W, Shen Q,
Li Y, Gu L, Zhou Q, Zhu H, Li Y, et al: Long noncoding RNA XIST
promotes malignancies of esophageal squamous cell carcinoma via
regulation of miR-101/EZH2. Oncotarget. 8:76015–76028. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar
|
|
94
|
Schmitt M, Horbach A, Kubitz R, Frilling A
and Häussinger D: Disruption of hepatocellular tight junctions by
vascular endothelial growth factor (VEGF): A novel mechanism for
tumor invasion. J Hepatol. 41:274–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Choi SB, Han HJ, Kim WB, Song TJ and Choi
SY: VEGF overexpression predicts poor survival in hepatocellular
carcinoma. Open Med (Wars). 12:430–439. 2017. View Article : Google Scholar
|
|
96
|
Cao G, Li X, Qin C and Li J: Prognostic
value of VEGF in hepatocellular carcinoma patients treated with
Sorafenib: A meta-analysis. Med Sci Monit. 21:3144–3151. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang
HD, Li G and Zhang JF: Long noncoding RNA Hotair mediated
angiogenesis in nasopharyngeal carcinoma by direct and indirect
signaling pathways. Oncotarget. 7:4712–4723. 2016. View Article : Google Scholar :
|
|
98
|
Ma X, Li Z, Li T, Zhu L, Li Z and Tian N:
Long non-coding RNA HOTAIR enhances angiogenesis by induction of
VEGFA expression in glioma cells and transmission to endothelial
cells via glioma cell derived-extracellular vesicles. Am J Transl
Res. 9:5012–5021. 2017.PubMed/NCBI
|
|
99
|
Su W, Xie W, Shang Q and Su B: The long
noncoding RNA MEG3 is downregulated and inversely associated with
vegf levels in osteoarthritis. Biomed Res Int. 2015:3568932015.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhan R, Xu K, Pan J, Xu Q, Xu S and Shen
J: Long noncoding RNA MEG3 mediated angiogenesis after cerebral
infarction through regulating p53/NOX4 axis. Biochem Biophys Res
Commun. 490:700–706. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu L, Chen X, Zhang Y, Hu Y, Shen X and
Zhu W: Long non-coding RNA TUG1 promotes endometrial cancer
development via inhibiting miR-299 and miR-34a-5p. Oncotarget.
8:31386–31394. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Jiang Q, Shan K, Qun-Wang X, Zhou RM, Yang
H, Liu C, Li YJ, Yao J, Li XM, Shen Y, et al: Long non-coding
RNA-MIAT promotes neurovascular remodeling in the eye and brain.
Oncotarget. 7:49688–49698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li X, Song Y, Liu F, Liu D, Miao H, Ren J,
Xu J, Ding L, Hu Y, Wang Z, et al: Long non-coding RNA MALAT1
promotes proliferation, angiogenesis, and immunosuppressive
properties of mesenchymal stem cells by inducing VEGF and IDO. J
Cell Biochem. 118:2780–2791. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhu Y, Zhang X, Qi L, Cai Y, Yang P, Xuan
G and Jiang Y: HULC long noncoding RNA silencing suppresses
angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling
pathway in human gliomas. Oncotarget. 7:14429–14440. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Xiao H, Tong R, Cheng S, Lv Z, Ding C, Du
C, Xie H, Zhou L, Wu J and Zheng S: BAG3 and HIF-1 alpha
coexpression detected by immunohistochemistry correlated with
prognosis in hepatocellular carcinoma after liver transplantation.
Biomed Res Int. 2014:5165182014. View Article : Google Scholar
|
|
106
|
Takahashi K, Yan IK, Haga H and Patel T:
Modulation of hypoxia-signaling pathways by extracellular linc-RoR.
J Cell Sci. 127:1585–1594. 2014.PubMed/NCBI
|
|
107
|
Zhuang J, Shen L, Yang L, Huang X, Lu Q,
Cui Y, Zheng X, Zhao X, Zhang D, Huang R, et al: TGFβ1 promotes
gemcitabine resistance through regulating the
lncRNA-LET/NF90/miR-145 signaling axis in bladder cancer.
Theranostcs. 7:3053–3067. 2017. View Article : Google Scholar
|
|
108
|
Zhu Z, Gao X, He Y, Zhao H, Yu Q, Jiang D,
Zhang P, Ma X, Huang H, Dong D, et al: An insertion/deletion
polymorphism within RERT-lncRNA modulates hepatocellular carcinoma
risk. Cancer Res. 72:6163–6172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Mineo M, Ricklefs F, Rooj AK, Lyons SM,
Ivanov P, Ansari KI, Nakano I, Chiocca EA, Godlewski J and Bronisz
A: The long non-coding RNA HIF1A-AS2 facilitates the maintenance of
mesenchymal glioblastoma stem-like cells in hypoxic Niches. Cell
Rep. 15:2500–2509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang J, Chen L, Li H, Yang J, Gong Z, Wang
B and Zhao X: Clopidogrel reduces apoptosis and promotes
proliferation of human vascular endothelial cells induced by
palmitic acid via suppression of the long non-coding RNA HIF1A-AS1
in vitro. Mol Cell Biochem. 404:203–210. 2015. View Article : Google Scholar : PubMed/NCBI
|