Introduction
Ovarian cancer, a lethal gynaecological tumour, is
the fifth most common cause of tumour-associated mortalities among
females (1). Ovarian cancer
ranks first among gynaecological malignancies in terms of
mortality, with a 5-year survival rate of <35% (2). Every year, approximately 150,000
patients succumb to ovarian cancer, and this number is increasing
annually (3). Epithelial ovarian
cancer (EOC) is the major type of ovarian cancer, accounting for
approximately 85-90% of all ovarian cancer cases and is associated
with poor clinical outcomes (4).
Currently, EOC is primarily treated with surgical resection along
with chemotherapy, radiotherapy and immunological therapy; however,
although numerous patients with EOC experience favourable
therapeutic effects, EOC patients diagnosed at an advanced stage
have poor prognosis after treatment with first-line therapies
(5,6). Furthermore, due to its subtle
symptoms and signs, EOC is usually diagnosed in later stages, with
pelvic dissemination or distant metastasis already present; thus,
patients miss the optimal timing for surgical intervention and have
reduced survival (7). Therefore,
understanding the mechanisms of EOC pathogenesis is urgently
required to identify promising targets for tumour diagnosis and
management.
Long noncoding RNAs (lncRNAs) are a heterogeneous
class of RNA transcripts consisting of over 200 nucleotides
(8). Although lncRNAs lack the
ability to code proteins, they participate in controlling gene
expression (9). Functionally,
lncRNAs have been confirmed to be crucial modulators of diverse
physiological and pathological processes, especially during
tumorigenesis and cancer progression (10). Increasing evidence has indicated
that an imbalance in lncRNA expression is closely associated with
EOC progression (11-13). The dysregulation of lncRNAs has
been clearly demonstrated to exert cancer-inhibiting or
cancer-facilitating effects and to contribute to the aggressive
properties of EOC (14-16).
MicroRNAs (miRNAs) belong to a group of small,
nonprotein-coding RNA transcripts that are known to negatively
regulate gene expression by base pairing with target genes and
consequently causing mRNA degradation or translation suppression
(17). Convincing evidence has
revealed the important regulatory activities of miRNAs in EOC
oncogenesis through their pro- or anti-tumourigenic effects
(18-20). Recently, the competing endogenous
RNA (ceRNA) theory was proposed to explain the mechanisms of lncRNA
action; lncRNAs directly sequester miRNAs and thereby modulate gene
expression by weakening the miRNA-mediated suppression of gene
expression (21,22). Thus, exploring cancer-related
lncRNAs and miRNAs, as well as the ceRNA theory, could be a
feasible approach for understanding the mechanisms underlying EOC
progression.
Long intergenic non-protein-coding RNA 1132
(LINC01132) expression in EOC and the underlying mechanisms have
not yet been explored. Thus, the expression level of LINC01132 in
EOC was detected and its correlation with patient prognosis was
examined. Next, functional experiments were performed to assess the
effects of LINC01132 on the aggressive behaviours of EOC cells.
Furthermore, the mechanisms that occurred downstream of LINC01132
in EOC cells were elucidated.
Materials and methods
Tissue specimens and cell lines
The collection and use of human tissues was approved
(approval no. EC-WFPH.20150106) by the Ethics Committee of Weifang
People's Hospital (Weifang, China). After obtaining the written
informed consent, 51 pairs of EOC tissues and adjacent normal
tissues were obtained from patients (age range, 46-69 years) at
Weifang People's Hospital from February 2015 to January 2016. The
inclusion criteria included: i) Diagnosed with EOC and agreed to
take part in this research; and ii) had not received chemotherapy
or radiotherapy prior to surgery. Patients who had other types of
human cancer or had been treated with chemotherapy or radiotherapy
were excluded. ES-2, an EOC cell line, was obtained from American
Type Culture Collection (ATCC) and was cultured in McCoy's 5A
Medium that was supplemented with 10% foetal bovine serum (FBS)
(Gibco; Thermo Fisher Scientific, Inc.). The 3 EOC cell lines,
namely, OVCAR3, CAOV-3 and SK-OV-3 were obtained from the National
Collection of Authenticated Cell Cultures (Shanghai, China). The
culture conditions for the SK-OV-3 cell line were the same as those
for the ES-2 cell line. CAOV-3 and OVCAR3 cells were maintained in
10% FBS-supplemented DMEM and 20% FBS-supplemented RPMI-1640 medium
(both from Gibco; Thermo Fisher Scientific, Inc.), respectively.
Bovine insulin (0.01 mg/ml; Gibco; Thermo Fisher Scientific, Inc.)
was also used for OVCAR3 cells. The human ovarian surface
epithelial cell line (OSE) was grown in ovarian epithelial cell
medium (both from ScienCell Research Laboratories, Inc.). All cell
lines aforementioned were cultured at 37°C an incubator containing
a humidified (100%) atmosphere with 5% CO2.
Transfection experiment
To overexpress miR-431-5p and SOX9, transfection of
the miR-431-5p mimic (100 pmol; Shanghai GenePharma Co., Ltd.) and
pcDNA3.1-SOX9 (4 μg; Sangon Biotech Co., Ltd.) was conducted
with Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), and the miRNA mimic control (100 pmol;
miR-NC; Shanghai GenePharma Co., Ltd.) and pcDNA3.1 (4 μg;)
were used as controls. All transfections were performed at room
temperature. The transfection duration was 8 h, after which the
culture medium was replaced with fresh culture medium.
Small interfering RNAs (siRNAs) targeting LINC01132
(si-LINC01132; 100 pmol) and the miR-431-5p inhibitor were prepared
by Shanghai GenePharma Co., Ltd. and used to knock down LINC01132
and miR-431-5p. The negative control (NC) siRNA (si-NC; 100 pmol)
and NC inhibitor functioned as the controls for si-LINC01132 and
the miR-431-5p inhibitor, respectively. The miR-431-5p mimic
sequence was 5′-ACGUACUGCCGGACGUUCUGU-3′ and the miR-NC sequence
was 5′-UUGUACUACACAAAAGUACUG-3′. The miR-431-5p inhibitor sequence
was 5′-UGCAUGACGGCCUGCAAGACA-3′ and the NC inhibitor sequence was
5′-ACUACUGAGUGACAGUAGA-3′. The si-LINC01132#1 sequence was
5′-AGGAGATAAAAATTTTAAATTAC-3′; the si-LINC01132#2 sequence was
5′-TTCTGTTTTTTGTTTTTTTAAGA-3′; and the si-NC sequence was
5′-CACGATAAGACAATGTAT TT-3′.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was employed for total RNA isolation from
tissues, cells or tumor xenografts. The RNA was quantified with a
NanoDrop™ 2000 spectrophotometer (Invitrogen; Thermo Fisher
Scientific, Inc.) at a 260-nm wavelength. For the determination of
miRNA expression, first-strand cDNA was synthesized by applying
Mir-X miRNA First-Strand Synthesis kit (Takara Biotechnology Co.,
Ltd.), according to the manufacturer's instructions. Next, PCR
quantification was implemented utilizing Mir-X miRNA RT-qPCR TB
Green® kit (Takara Biotechnology Co., Ltd.) in
accordance with the manufacturer's instructions. U6 small nuclear
RNA acted as the internal reference for miRNAs.
For the determination of LINC01132 and SOX9 levels,
PrimeScript™ RT reagent kit with gDNA Eraser and TB
Green® Premix Ex Taq™ II (Takara Biotechnology Co.,
Ltd.) were, respectively, adopted for reverse transcription and PCR
amplification. Both kits were used according to the manufacturer's
instructions. The thermocycling conditions for PCR amplification
were as follows: Initial hold at 95°C for 30 sec; 40 cycles of
amplification at 95°C for 3 sec, and annealing for 30 sec at 60°C
and extension at 72°C for 30 sec. GAPDH acted as the internal
reference for LINC01132 and SOX9 in this assay. The
2-ΔΔCq (23)
quantification method was used to calculate the relative mRNA
expression levels. The primers were designed as follows: LINC01132
forward, 5′-CGGAAGCAGGGACTGCTATT-3′ and reverse, 5′-TCCTGGTGGCTCTGT
CCTCTC-3′; SOX9 forward, 5′-GACGTCATCTCCAACATCGAGACCT-3′ and
reverse, 5′-GCTGCCCGT GTAGGTGACCT-3′; GAPDH forward,
5′-CGGAGTCAACGG ATTTGGTCGTAT-3′ and reverse, 5′-AGCCTTCTC CATGGTGGT
GAA GAC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3; miR-125b-5p forward,
5′-TCGGCAGGUCCCUGAGACC-3′ and reverse, 5′-CACTCAACTGGTGTCGTGGA-3′;
miR-134-3p forward, 5′-TCGGCAGGAACCACUGAUC-3′ and reverse,
5′-CACTCAACTGGTGTCGTGGA-3′; miR-199a-3p forward,
5′-TCGGCAGGAUUGGUUACACG-3′ and reverse, 5′-CACTCAACT
GGTGTCGTGGA-3′; miR-199b-3p forward, 5′-TCGGCAGGAUUG GUUACACG-3′
and reverse, 5′-CACTCAACTGGT GTCGTGGA-3′; miR-431-5p forward,
5′-TCGGCAGGUGUCUUGCAGG-3′ and reverse,
5′-CACTCAACTGGTGTCGTGGA-3′.
Cell Counting Kit-8 (CCK-8) assay
EOC cell proliferation was assessed using a CCK-8
assay (Dojindo Molecular Technologies, Inc.). In detail, every well
of 96-well plates was seeded with 2,000 transfected cells that were
resuspended in 100 μl of complete culture medium. At 0, 1,
2, and 3 days after adherence to the wells, 10 μl of CCK-8
solution was added, and the cells were cultured for an additional 2
h under the conditions aforementioned. The absorbance was monitored
at a wavelength of 450 nm via a microplate reader.
Flow cytometric analysis
Transfected cells (1×106) were digested
with trypsin (Gibco; Thermo Fisher Scientific, Inc.), rinsed with
ice-cooled phosphate-buffered saline and centrifuged at room
temperature at 1,000 × g for 5 min. The apoptosis of these cells
was analysed by employing an Annexin V-fluorescein isothiocyanate
(FITC) apoptosis detection kit (Beyotime Institute of
Biotechnology) in accordance with the manufacturer's instructions.
Briefly, the cells were resuspended in 195 μl of Annexin
V-FITC binding buffer and cultured with 5 μl of Annexin
V-FITC and 10 μl of PI at 25°C. The incubation was performed
in the dark and continued for 20 min. All samples were analysed
using a flow cytometer (FACSCalibur; BD Biosciences). CellQuest
software (version 2.9; BD Biosciences) was applied for data
analysis.
Cell migration and invasion assays
The EOC cell migration and invasion abilities were
evaluated using the Transwell method (8-μm pores; BD
Biosciences). In the migration assay, the upper chambers were
filled with 200 μl of serum-free RPMI-1640 medium (for
OVCAR3) or DMEM medium (for CAOV-3) containing 5×104
cells. By serving as a chemoattractant, a volume of 20%
FBS-supplemented 600 μl of culture medium was added to the
lower compartments. After 1 day of culture, the non-migrated cells
were wiped off by applying a cotton swab, and the migrated cells
that crossed the pores were fixed using 100% methanol at room
temperature for 30 min and stained using 0.1% crystal violet. In
the invasion assay, Matrigel-coated Transwell chambers (BD
Biosciences) were utilized, and the experimental procedures were
the same as those of the migration assay. The precoating with
Matrigel was conducted at 37°C for 2 h. The number of stained cells
in 5 random fields of view was counted with a light microscope
(magnification, x200).
Tumor xenograft model
A second-generation lentiviral system was applied
for the production of lentiviruses. LINC01132 short hairpin RNA
(shRNA; sh-LINC01132) and NC shRNA (sh-NC) were prepared by
Shanghai GenePharma Co., Ltd. After these molecules were inserted
into a pLKO.1 lentiviral vector (Addgene Inc.), the vectors were
transfected into 293T cells (National Collection of Authenticated
Cell Cultures) with psPAX2 and pMD2.G. A total of 30 μg of
plasmids were used for lentivirus packaging, and the ratio of
lentiviral plasmid: pLKO.1: psPAX2: pMD2.G was 2:1:1. Approximately
48 h after transfection, the lentiviruses were collected and used
to infect CAOV-3 cells (MOI=5). After 3 days, fresh complete
culture medium containing puromycin was used to further incubate
the CAOV-3 cells, yielding cells stably overexpressing sh-LINC01132
or sh-NC. The concentrations of puromycin used for selection and
maintenance were 2 and 0.3 μg/ml, respectively.
All experimental steps involving animals were
conducted with the approval from the Institutional Animal Care and
Use Committee of Weifang People's Hospital (Weifang, China).
Four-week-old female BALB/c nude mice (n=6; 20 g) were acquired
from Shanghai SLAC Laboratory Animal Co. Ltd. (Shanghai, China).
The mice were housed under specific pathogen-free conditions at
25°C and 50% humidity, with a 10:14 light/dark cycle and ad
libitum access to food and water. Stably transfected cells
(2×106) were harvested and subcutaneously injected into
the flanks of nude mice. One week later, the sizes of the
subcutaneous tumours were recorded every 4 days, and their volumes
were calculated with the formula: Volume (mm3)=1/2 ×
length × width2. The mice were euthanized by cervical
dislocation on day 31, and the subcutaneous tumours were harvested
for subsequent use.
Bioinformatics prediction
The online software miRDB (http://mirdb.org/) was adopted to identify the
downstream target of LINC01132. The direct target of miR-431-5p was
predicted utilizing TargetScan (version 7.2; http://www.targetscan.org) and miRDB.
TCGA and GTEx databases
TCGA (https://portal.gdc.cancer.gov/) and GTEx (https://www.genome.gov/Funded-Programs-Projects/Genotype-Tissue-Expression-Project)
were utilized to analyse LINC01132 expression in ovarian
cancer.
Subcellular fractionation assay
The lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) was
utilized to predict subcellular distribution of LINC01132 in human
cells. The prediction was then verified by a Cytoplasmic and
Nuclear RNA Purification kit (Norgen Biotek Corp.), which was used
according to the manufacturer's instructions. EOC cells were
processed with 200 μl ice-cold cell fractionation buffer for
5 min to separate the cytoplasm and nucleus. The relative
expression of LINC01132 in both fractions was measured by
RT-qPCR.
Luciferase reporter assay
LINC01132 and the SOX9 3′-UTR fragments harbouring
wild-type (wt) miR-431-5p-binding sequences were amplified and
cloned into the downstream region of the psiCHECK™-2 vector
(Promega Corporation). The luciferase reporter vectors were
labelled LINC01132-wt and SOX9-wt. The corresponding mutant (mut)
luciferase reporter vectors, namely, psiCHECK™-2-LINC01132-mut and
psiCHECK™-2-SOX9-mut, were generated in the same manner. For the
reporter assay, EOC cells were inoculated into 24-well plates with
a density of 1.5×105 cells/well, and incubated at 37°C
overnight. Cells were transfected with miR-431-5p mimic or miR-NC
alongside wt or mut reporter vectors utilizing
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The transfected cells were lysed at 48 h
post-transfection with a Dual-Luciferase Reporter Assay System
(Promega Corporation) for luciferase activity determination.
Renilla luciferase activity was used to normalize the
firefly luciferase activity.
RNA immunoprecipitation (RIP)
The EZ-Magna RIP™ RNA-Binding Protein
Immunoprecipitation kit (cat. no. 17-701; EMD Millipore) was used
in the assay, according to the manufacturer's instructions. After
lysing the cells in complete RIP lysis buffer (EMD Millipore), the
whole-cell lysates were collected and treated with magnetic beads
coupled to an anti-Ago2 antibody or normal mouse IgG (cat. no.
17-701; dilution, 1:5,000; both from EMD Millipore) at 4°C
overnight. In addition, 10 μl of whole-cell lysates was
aliquoted for use as the input and served as the positive control.
IgG acted as the negative control. Proteinase K (EMD Millipore)
treatment (30 min at 55°C) was applied to detach the proteins, and
the immunoprecipitated RNAs were extracted. Finally, the relative
enrichment of LINC01132, miR-431-5p and SOX9 in the
immunoprecipitated RNAs was examined via RT-qPCR.
Protein extraction and western
blotting
Total protein was extracted using RIPA lysis buffer
(Beyotime Institute of Biotechnology). After total protein
quantification using a bicinchoninic acid kit (Beyotime Institute
of Biotechnology), equal amounts of proteins (30 μg) were
separated by 10% SDS-PAGE electrophoresis and transferred onto PVDF
membranes. The membranes were blocked at room temperature in 5%
skimmed milk for 2 h and then incubated overnight at 4°C with
primary antibodies against SOX9 (product code ab185966; 1:1,000
dilution; Abcam) or GAPDH (product code ab181602; 1:1,000 dilution;
Abcam), followed by 1 h incubation at room temperature with an
HRP-conjugated secondary antibody (product code ab205718; 1:5,000
dilution; Abcam). Ultimately, a BeyoECL plus detection kit
(Beyotime Institute of Biotechnology) was used to visualize the
protein bands. Quantity One software (version 4.62; Bio-Rad
Laboratories, Inc.) was employed for densitometric analysis.
Statistical analysis
All experiments were repeated thrice and conducted
in triplicate, and the experimental data are expressed as the mean
± standard deviation. Comparison of the significance between two
groups was conducted with both paired and unpaired Student's
t-tests. The statistical significance among multiple groups was
examined utilizing one-way analysis of variance (ANOVA) with
Tukey's post hoc test. The overall survival curves were calculated
with the Kaplan-Meier method, after which they were analysed with
the log-rank test. Pearson's correlation analysis was used to
examine the expression correlations. All tests were two-sided, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Silencing of LINC01132 suppresses EOC
progression
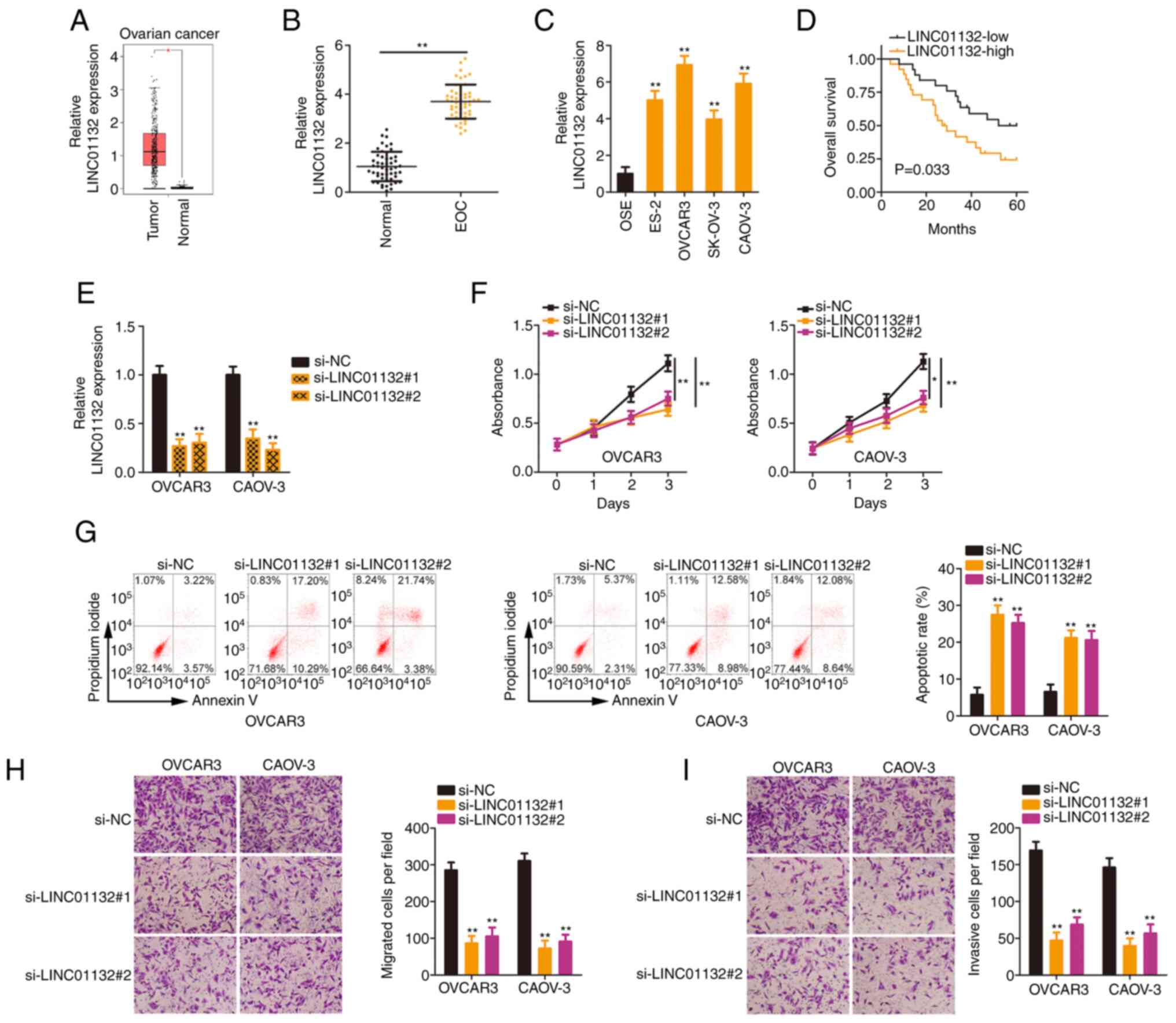
The present study firstly analysed LINC01132
expression in ovarian cancer tissues through The Cancer Genome
Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) data-bases.
LINC01132 was significantly overexpressed in ovarian cancer tissues
(Fig. 1A). Next, as compared
with adjacent normal tissues, a higher LINC01132 expression level
in EOC tissues was confirmed (Fig.
1B). Similarly, LINC01132 was highly expressed in the EOC cell
lines (Fig. 1C) compared with
OSE cells. Next, all patients in these datasets were classified
into either the LINC01132-high expression group or the
LINC01132-low expression group on the basis of the median value of
LINC01132 in EOC tissues. Patients with EOC characterized by a high
LINC01132 expression level exhibited poorer overall survival than
those with EOC characterized by a low LINC01132 expression level
(Fig. 1D).
Given the upregulation of LINC01132 in EOC, it was
questioned whether the dysregulation of LINC01132 was related to
the aggressiveness of EOC cells. To this end, the OVCAR3 and CAOV-3
cell lines, which exhibited higher LINC01132 expression than the
other EOC cell lines, were selected for subsequent experiments. To
avoid off-target effects, si-LINC01132#1 and si-LINC01132#2, were
separately transfected into OVCAR3 and CAOV-3 cells to effectively
induce LINC01132 silencing. RT-qPCR revealed that the 2 siRNAs both
exerted satisfactory silencing effects (Fig. 1E). Silencing of LINC01132
significantly inhibited the proliferative ability of EOC cells
(Fig. 1F). Additionally, the
transfection of si-LINC01132 led to a significant increase in the
apoptosis of EOC cells (Fig.
1G). Conversely, cell migratory (Fig. 1H) and invasive (Fig. 1I) capacities were significantly
suppressed due to LINC01132 downregulation. Hence, LINC01132 was
considered to exert a tumorigenic role in EOC.
LINC01132 sequesters miR-431-5p in
EOC
To reveal the mechanisms exerted by LINC01132, the
location of LINC01132 was predicted by applying lncLocator. The
outcomes revealed that most LINC01132 was distributed in cytoplasm
(Fig. 2A), which was
subsequently confirmed by applying subcellular fractionation
(Fig. 2B). These results
indicated that LINC01132 functioned via a ceRNA mechanism. The
miRDB tool was utilized to identify the potential target miRNAs of
LINC01132. In total, 75 miRNAs contained binding sites for
LINC01132. Through TCGA database, a total of 5 miRNAs, including
miR-125b-5p, miR-134-3p (Fig.
2C), miR-199a-3p, miR-199b-3p, and miR-431-5p (Fig. 2D), were found to be downregulated
in ovarian cancer tissues and thus were selected for experimental
verification. Furthermore, the expression of these candidates in
EOC cells upon LINC01132 silencing was assessed by RT-qPCR. Only
miR-431-5p was overexpressed by si-LINC01132 transfection (Fig. 2E). The influence of LINC01132 on
the expression levels of miR-34c-5p was also evaluated and it was
demonstrated that LINC01132 failed to affect miR-34c-5p (Fig. 2E). Conversely, miR-431-5p was
significantly weakly expressed in EOC tissues (Fig. 2F), and its expression was
negatively correlated with LINC01132 (Fig. 2G).
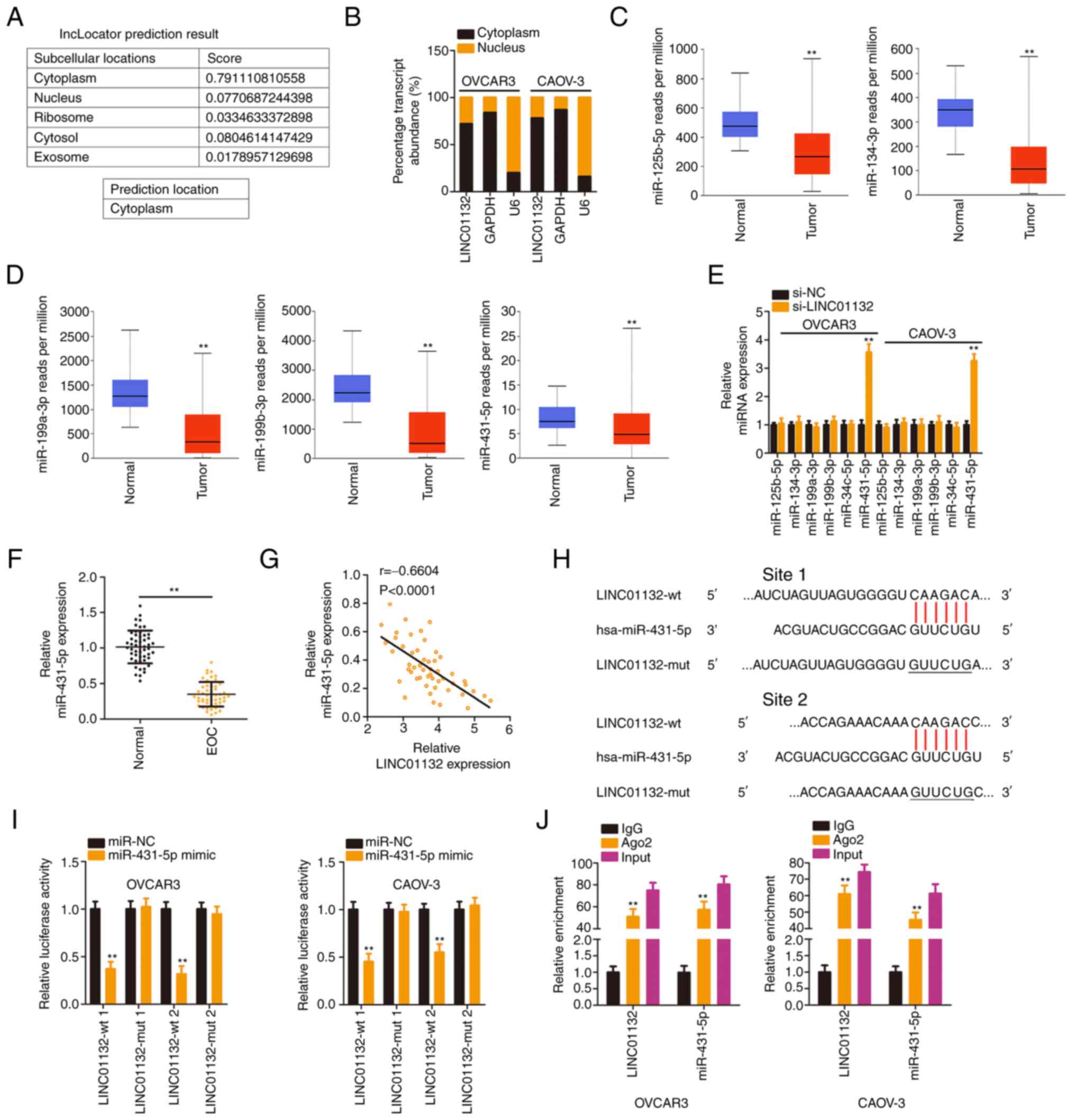 | Figure 2LINC01132 is capable of sponging
miR-431-5p in EOC cells. (A) The distribution of LINC01132
predicted by lncLocator. (B) Localization of LINC01132 in EOC
cells. (C and D) Expression of miR-125b-5p, miR-134-3p,
miR-199a-3p, miR-199b-3p, and miR-431-5p in ovarian cancer was
analysed via The Cancer Genome Atlas database. (E) Expression of
the 5 candidates in EOC cells following LINC01132 silencing was
measured by RT-qPCR. (F) miR-431-5p level in EOC tissues was
examined by RT-qPCR. (G) The expression relationship between
LINC01132 and miR-431-5p in EOC tissues. (H) The predicted binding
sequences between LINC01132 and miR-431-5p were demonstrated. The
sequences containing underscores were the site that was mutated in
LINC01132. (I) Luciferase activity of wt or mut LINC01132 reporter
plasmids was quantified in OVCAR3 and CAOV-3 cells in the presence
of miR-431-5p mimic or miR-NC. (J) RNA immunoprecipitation
demonstrated the co-existence of LINC01132 and miR-431-5p in
immunoprecipitated RNA. **P<0.01. LINC01132, long
intergenic non-protein coding RNA 1132; miR, microRNA; EOC,
epithelial ovarian cancer; RT-qPCR, reverse
transcription-quantitative PCR; wt, wild-type; mut, mutant; si-,
small interfering; NC, negative control. |
Luciferase reporter assays served to certify the
direct binding between miR-431-5p and LINC01132 (Fig. 2H). The upregulation of miR-431-5p
significantly decreased the luciferase activity of
psiCHECK™-2-LINC01132-wt (1 and 2), but these regulatory effects on
luciferase activity were offset when the binding sites were mutated
(1 and 2; Fig. 2I). Furthermore,
as demonstrated by the RIP assay, LINC01132 and miR-431-5p were
significantly enriched in the Ago2 group (Fig. 2J), indicating the coexistence of
two RNAs in the RNA-induced silencing complexes. In summary,
LINC01132 sponged miR-431-5p in EOC.
LINC01132 regulates SOX9 expression via
sequestering miR-431-5p
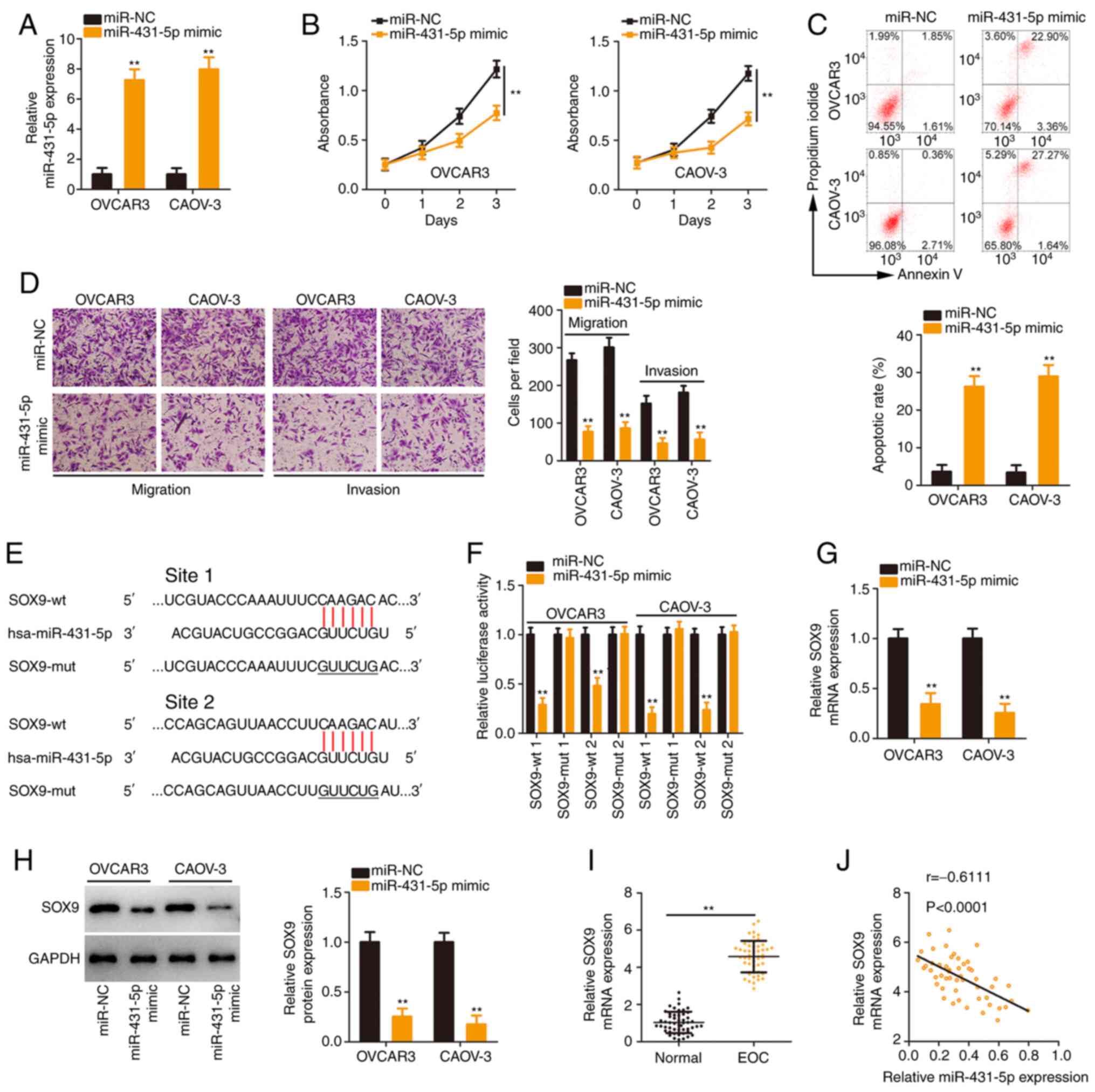
The detailed roles of miR-431-5p in EOC were further
examined. The overexpression of miR-431-5p in EOC cells by
transfection with the miR-431-5p mimic was verified (Fig. 3A). The proliferation of EOC cells
was reduced following miR-431-5p overexpression (Fig. 3B). Flow cytometric analysis
confirmed that apoptosis was significantly promoted in
miR-431-5p-overexpressed EOC cells (Fig. 3C). Additionally, exogenous
miR-431-5p expression caused a notable decrease in EOC cell
migration and invasion (Fig.
3D).
Bioinformatics analysis was implemented to identify
the potential downstream target of miR-431-5p. It was demonstrated
that SOX9 contained two binding sites for miR-431-5p (Fig. 3E) and was further studied due to
its critical tumour-promoting activities during EOC oncogenesis
(24-26). Transfection with miR-431-5p mimic
significantly reduced the luciferase activity of
psiCHECK™-2-SOX9-wt (1 and 2) but produced no regulatory effect on
the activity of psiCHECK™-2-SOX9-mut (1 and 2; Fig. 3F). Subsequently, it was revealed
that the SOX9 expression (Fig. 3G
and H) was downregulated after miR-431-5p mimic transfection.
Furthermore, high SOX9 expression in EOC tissues (Fig. 3I) exhibited an inverse
relationship with miR-431-5p levels (Fig. 3J). Collectively, these results
validated SOX9 as a direct target of miR-431-5p.
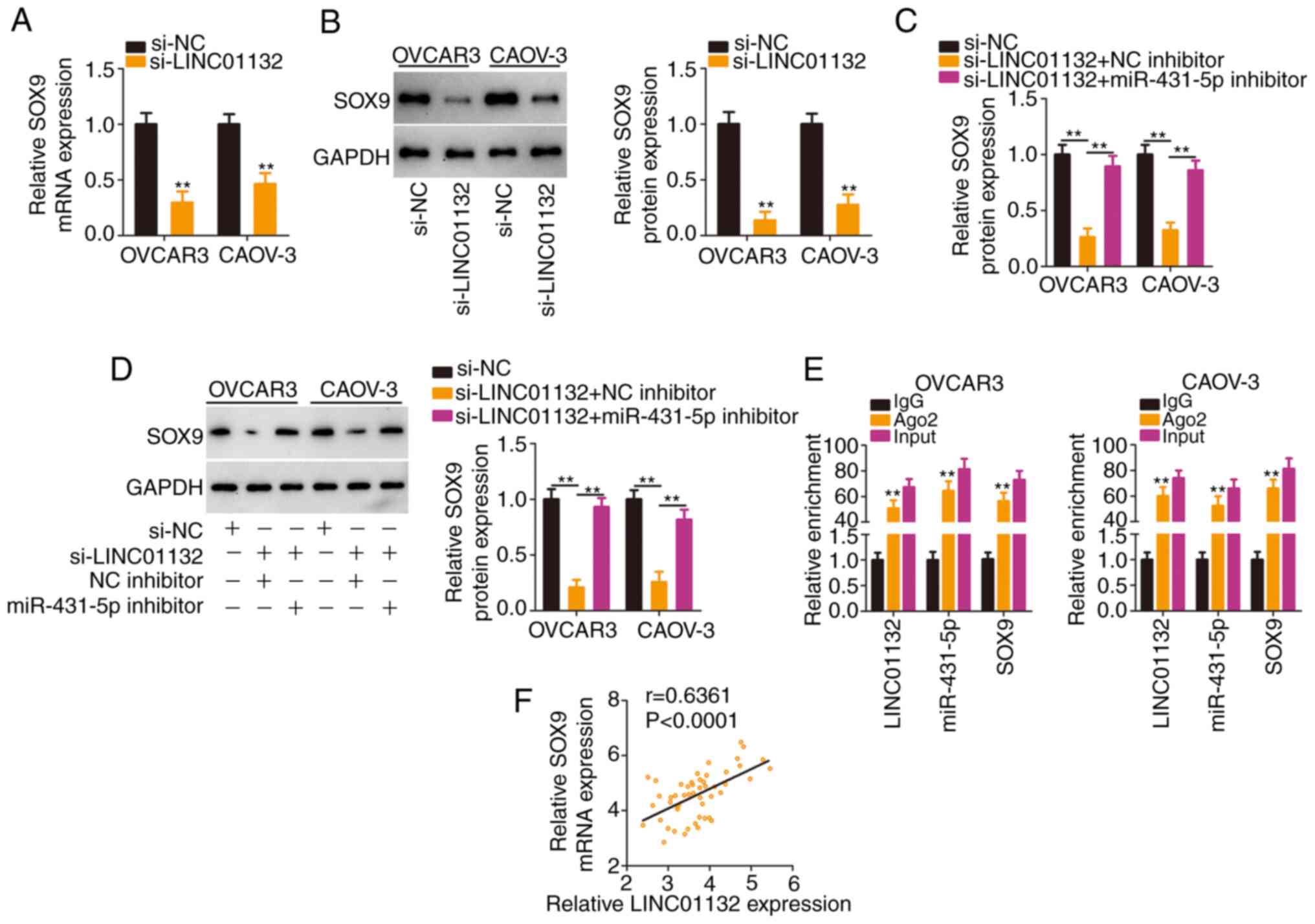
Then, it was evaluated whether LINC01132 functioned
as a miR-431-5p molecular sponge to regulate SOX9 expression. The
absence of LINC01132 significantly reduced SOX9 expression
(Fig. 4A and B), whereas this
regulation was largely counteracted after miR-431-5p inhibitor
treatment (Fig. 4C and D). More
importantly, the RIP assay confirmed that LINC01132, miR-431-5p and
SOX9 coexisted in RNA-induced silencing complexes (Fig. 4E). In addition, a positive
expression relationship between LINC01132 and SOX9 was verified in
EOC tissues (Fig. 4F).
Collectively, LINC01132 acted as a ceRNA by sequestering miR-431-5p
and consequently increasing SOX9 expression.
miR-431-5p/SOX9 axis is required for the
carcinogenic actions of LINC01132 in EOC cells
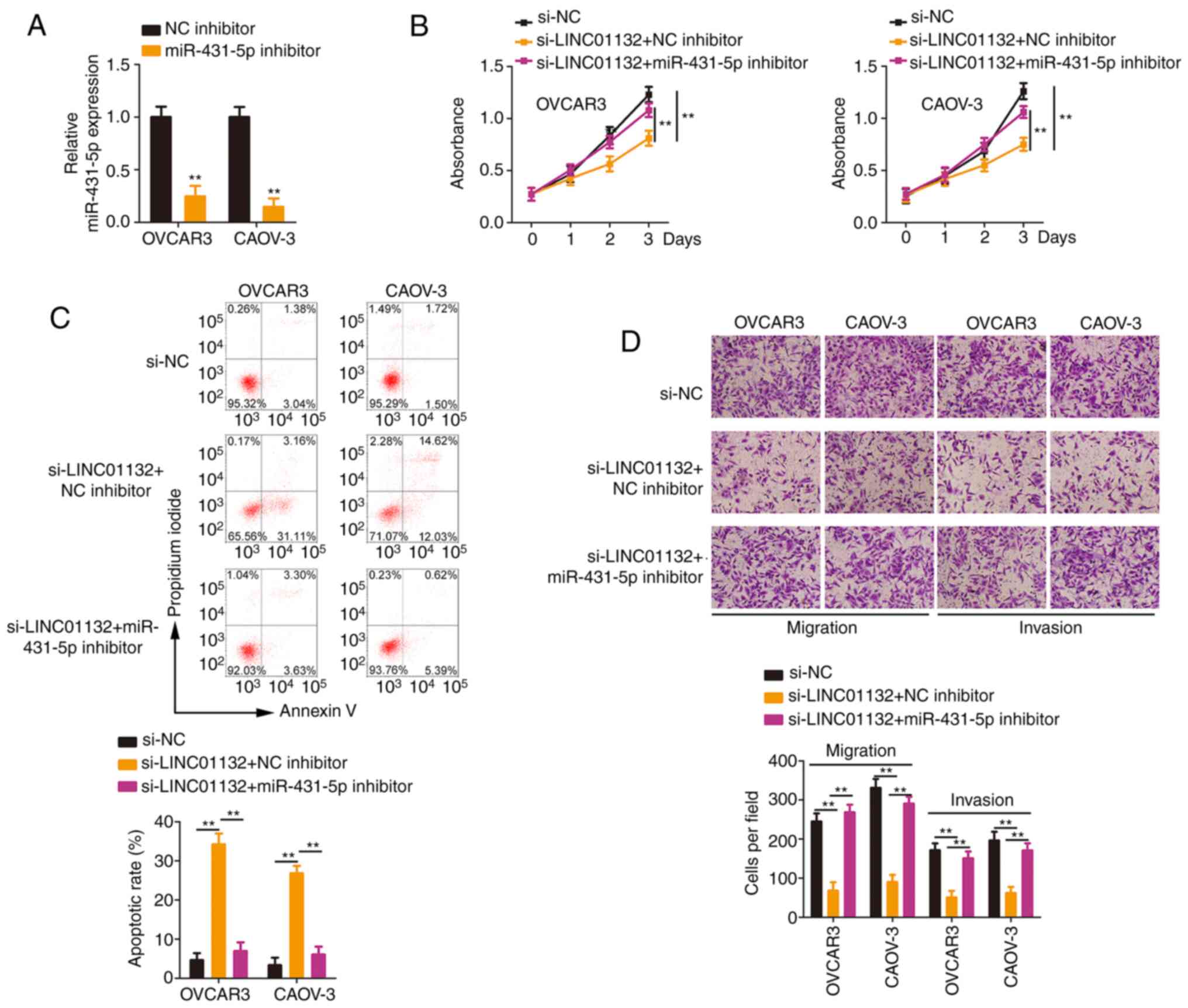
A series of rescue experiments were designed to
determine whether modulatory actions of LINC01132 are attributed to
the miR-431-5p/SOX9 axis. Transfection of the miR-431-5p inhibitor
decreased the expression level of miR-431-5p, suggesting that
miR-431-5p was successfully silenced in EOC cells (Fig. 5A). The loss of LINC01132 reduced
cell proliferation, which was largely restored after miR-431-5p
inhibitor treatment (Fig. 5B).
Co-transfection with miR-431-5p inhibitor abolished the increase in
apoptosis in LINC01132-silenced EOC cells (Fig. 5C). Furthermore, treatment with
the miR-431-5p inhibitor recovered the migratory and invasive
(Fig. 5D) capacities that were
impaired by LINC01132 knockdown.
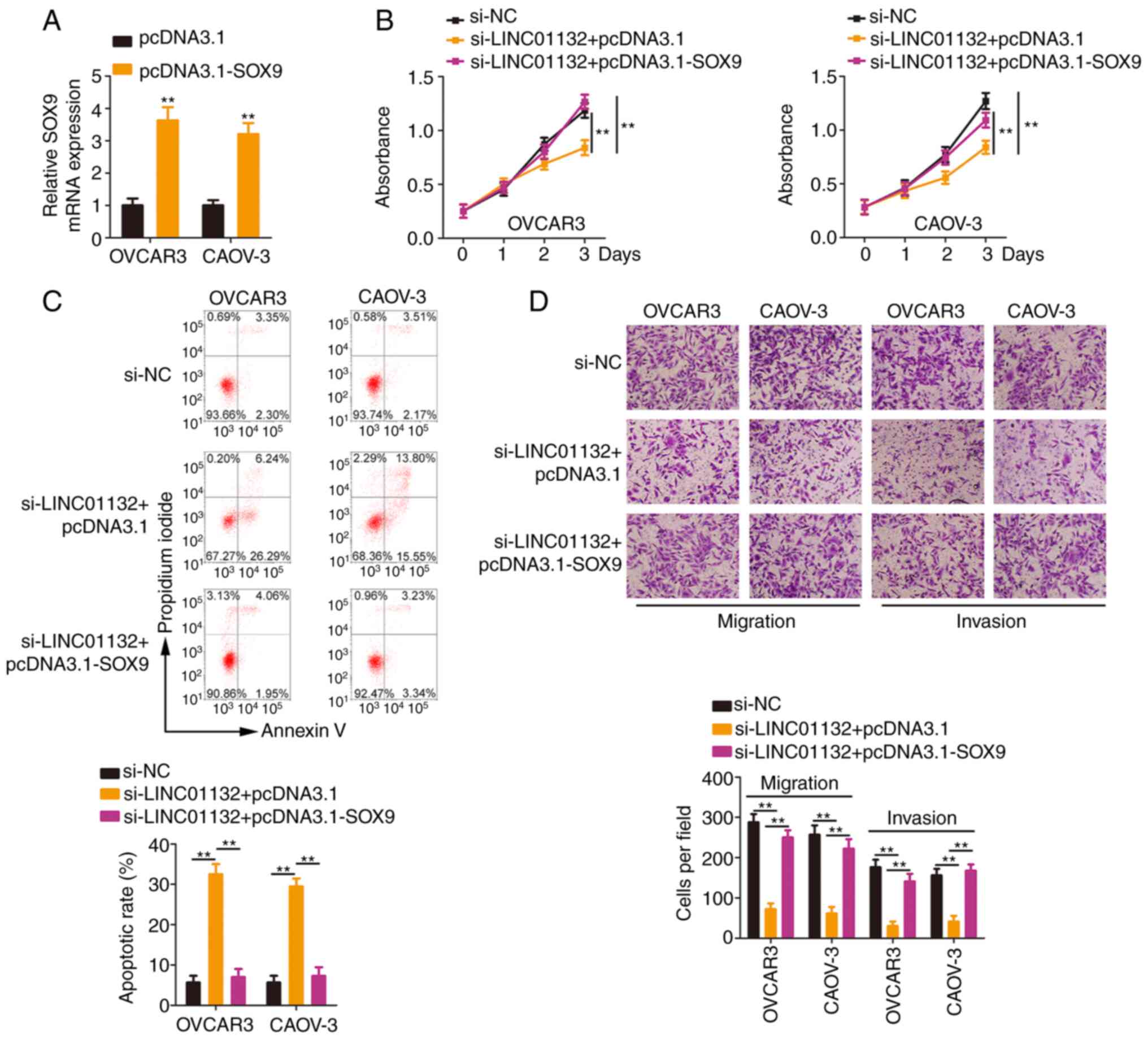
In addition, the SOX9 overexpression plasmid
pcDNA3.1-SOX9 (Fig. 6A) was
introduced into LINC01132-deficient EOC cells. Re-expression of
SOX9 antagonized the cell proliferation suppression and cell
apoptosis promotion induced by si-LINC01132 (Fig. 6B and C). Moreover, reduced cell
migration and invasion caused by si-LINC01132 was neutralized by
overexpressing SOX9 (Fig. 6D).
Collectively, the aforementioned results indicated that LINC01132
played tumour-promoting roles in EOC cells via controlling the
miR-431-5p/SOX9 axis.
LINC01132 downregulation inhibits
xenograft tumour growth in vivo
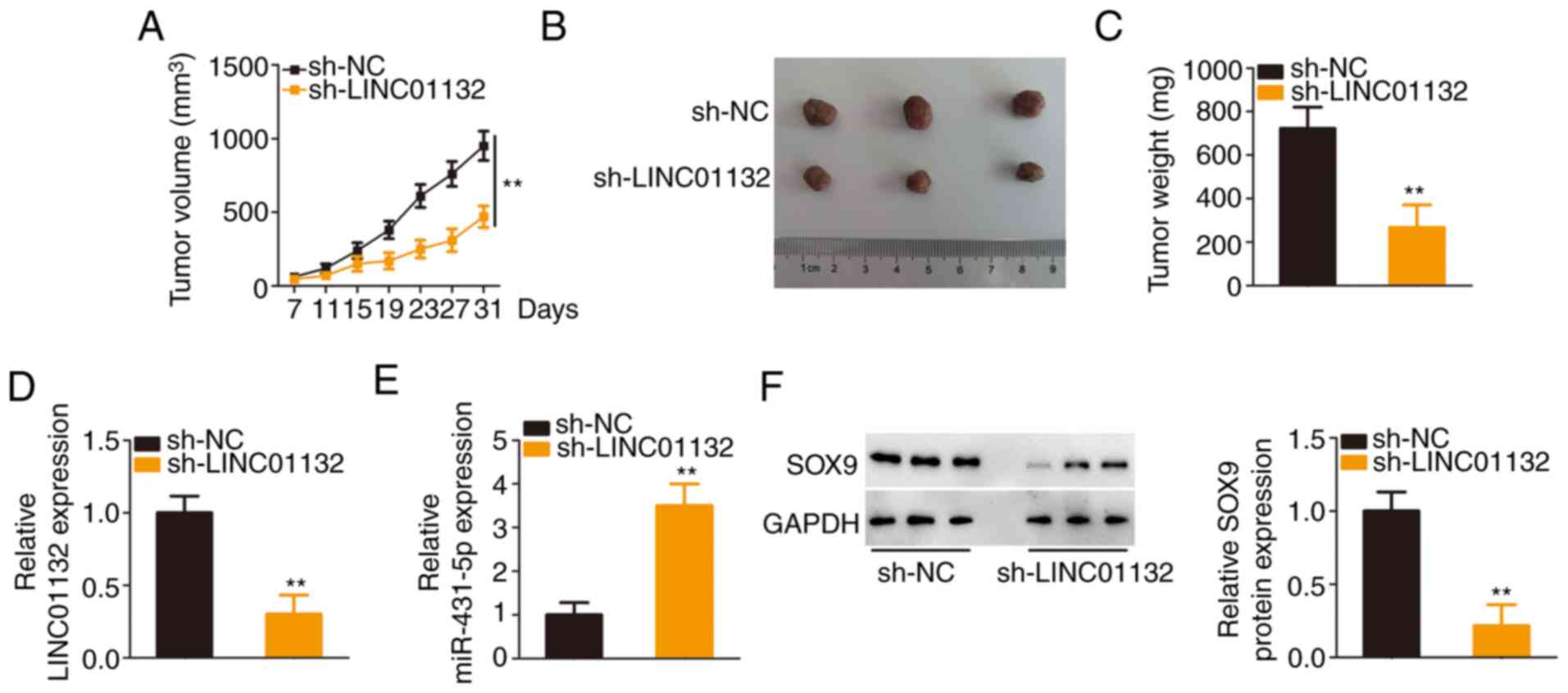
A tumour xenograft model was implemented to confirm
the function of LINC01132 in EOC tumour growth in vivo.
Downregulation of LINC01132 significantly slowed tumour growth
(Fig. 7A) and reduced tumour
size (Fig. 7B). The subcutaneous
tumours were harvested at day 31 after injection. It was revealed
that the cells transfected with sh-LINC01132 formed tumours with
significantly decreased weights (Fig. 7C). Further detection of LINC01132
and miR-431-5p demonstrated that the subcutaneous tumours in the
sh-LINC01132 group exhibited a low expression of LINC01132 and a
high expression of miR-431-5p (Fig.
7D and E). Furthermore, the LINC01132-downregulated
cell-derived tumours exhibited decreased SOX9 expression compared
with sh-NC cell-derived tumours (Fig. 7F). Collectively, these results
indicated that the depletion of LINC01132 impaired the tumour
growth of EOC cells in vivo.
Discussion
At present, the clinical relationship between
lncRNAs and human cancer is a popular research topic in tumour
molecular biology given their importance in tumorigenesis and
tumour development (27-29). Therefore, lncRNAs are attractive
potential therapeutic targets for intervention. As high-throughput
sequencing technologies progress, the number of lncRNAs known to be
dysregulated in EOC is rapidly increasing (30); however, the detailed
contributions of the majority of these lncRNAs remain poorly
understood. In the present study, the cellular function and
underlying mechanisms of LINC01132 in EOC were revealed in order to
provide novel directions and insights for managing EOC.
Multiple studies performed by different scientists
have revealed the critical roles of lncRNAs in the oncogenicity of
EOC. For instance, NORAD (31),
LINC00673 (32) and TC0101441
(33) were upregulated in EOC
and have been identified as tumour promoters. Conversely, the low
expression of WDFY3-AS2 (34),
MAGI2-AS3 (35) and AOC4P
(36) in EOC resulted in
anti-tumourigenic effects. However, it is unknown whether LINC01132
is involved in EOC malignancy. In the present study, high levels of
LINC01132 in EOC were observed in the TCGA database and our own
cohort. The overall survival of EOC patients with high LINC01132
levels was significantly lower than patients with low LINC01132
levels. Cell experiments revealed that the knockdown of LINC01132
was capable of inhibiting the proliferation, migration and invasion
of EOC cells and promoting apoptosis. The results of the
tumour-forming experiment in nude mice were consistent with the
in vitro results.
Next, an attempt was made to understand the detailed
mechanism by which LINC01132 participates in EOC, which remains
mostly uncharacterized. The important roles of lncRNAs in
regulating physiological and pathological behaviours are achieved
through various mechanisms. The lncRNA-mediated ceRNA theory is a
classic mechanism that explains the mechanism of lncRNA action in
EOC (12). Cytoplasmic lncRNAs
function as endogenous sponges to sequester certain miRNAs and
indirectly modulate gene expression by protecting mRNAs from
miRNA-induced mRNA dysregulation or translation inhibition
(37). Accordingly, lncLocator
and subcellular fractionation assays were used to demonstrate that
LINC01132 is primary located in the EOC cell cytoplasm. Then,
bioinformatics analysis indicated that miR-431-5p may be a target
of LINC01132. A luciferase reporter assay verified that LINC01132
possessed a miR-431-5p-binding site.
To explore the ceRNA network mediated by LINC01132
in EOC, the downstream target of miR-431-5p was explored, and the
mechanistic analysis identified SOX9 as a target of miR-431-5p.
Notably, the data of the present study further revealed that SOX9
was positively controlled by LINC01132 in EOC cells. In rescue
experiments, the effects of LINC01132 knockdown on SOX9 expression
were abrogated after the inhibition of miR-431-5p. More
importantly, LINC01132, miR-431-5p and SOX9 were confirmed to
coexist in RNA-induced silencing complexes. Collectively, a new
ceRNA network comprising LINC01132, miR-431-5p and SOX9 was
identified in EOC.
Differentially expressed miR-431-5p has been
validated in several types of human cancer, including EOC (38). Consistent with these studies, the
present research confirmed the downregulation of miR-431-5p in EOC
and the cancer-inhibiting roles of miR-431-5p in EOC progression.
SOX9 was identified as a downstream effector of miR-431-5p in EOC
cells. Although the effects of SOX9 in driving EOC initiation and
progression have been well elucidated (24-26), the upstream regulatory mechanisms
that cause the overexpression of SOX9 in EOC remain largely
unknown. Herein, the experimental results revealed a novel
LINC01132/miR-431-5p axis that controlled SOX9 expression in EOC
cells. Furthermore, miR-431-5p inhibition or SOX9 re-expression
eliminated the tumour-suppressive effects of LINC01132
downregulation on the pathological behaviours of EOC cells,
demonstrating that SOX9 played critical roles in mediating the
LINC01132/miR-431-5p axis-triggered biological functions in
EOC.
In the present study, the cell lines CAOV3 and
OVCAR3 were used, both of which are highly sensitive to platinum
therapy, such as cisplatin or carboplatin (26). The regulation of SOX9 has also
been demonstrated to be crucial to cisplatin resistance (26). Additionally, OVCAR8 is a
cisplatin-resistant EOC cell line. Thus, the
LINC01132/microRNA-431-5p/SOX9 pathway may perform crucial roles in
the regulation of cisplatin sensitivity in EOC cells. The present
study did not evaluate this effect, which is a limitation that will
be addressed in the near future.
In summary, the present study was the first to
report, to the best of our knowledge, that LINC01132 was
upregulated in EOC and exhibited a significant relationship with
poor clinical prognosis. LINC01132 was revealed to exert oncogenic
effects in EOC cells by controlling the outcome of the
miR-431-5p/SOX9 axis. LINC01132/miR-431-5p/SOX9 is anticipated to
be an attractive target for the treatment and prognostic evaluation
of EOC.
Availability of data and materials
All datasets generated and analysed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
JC and WZ devised the current research. All the
experiments were performed by WZ, XX and JC. JC and WZ analysed the
obtained data. The manuscript was drafted by JC and WZ. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The collection and use of human tissues was approved
by the Ethics Committee of Weifang People's Hospital (Weifang,
China). All experimental steps involving animals were conducted
with approval from the Institutional Animal Care and Use Committee
of Weifang People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37,513,025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Di Lorenzo G, Ricci G, Severini GM, Romano
F and Biffi S: Imaging and therapy of ovarian cancer: Clinical
application of nanoparticles and future perspectives. Theranostics.
8:4279–4294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kuroki L and Guntupalli SR: Treatment of
epithelial ovarian cancer. BMJ. 371:m37732020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gourley C and Bookman MA: Evolving
concepts in the management of newly diagnosed epithelial ovarian
cancer. J Clin Oncol. 37:2386–2397. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rojas V, Hirshfield KM, Ganesan S and
Rodriguez-Rodriguez L: Molecular characterization of epithelial
ovarian cancer: Implications for diagnosis and treatment. Int J Mol
Sci. 17:21132016. View Article : Google Scholar
|
|
7
|
Sundar S, Neal RD and Kehoe S: Diagnosis
of ovarian cancer. BMJ. 351:h44432015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qiu L, Tang Q, Li G and Chen K: Long
non-coding RNAs as biomarkers and therapeutic targets: Recent
insights into hepato-cellular carcinoma. Life Sci. 191:273–282.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Zhang M and Zhou F: Biological
functions and clinical applications of exosomal long non-coding
RNAs in cancer. J Cell Mol Med. 24:11656–11666. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pei C, Gong X and Zhang Y: LncRNA MALAT-1
promotes growth and metastasis of epithelial ovarian cancer via
sponging microrna-22. Am J Transl Res. 12:6977–6987.
2020.PubMed/NCBI
|
|
12
|
Braga EA, Fridman MV, Moscovtsev AA,
Filippova EA, Dmitriev AA and Kushlinskii NE: LncRNAs in ovarian
cancer progression, metastasis, and main pathways: ceRNA and
alternative mechanisms. Int J Mol Sci. 21:88552020. View Article : Google Scholar :
|
|
13
|
Wambecke A, Ahmad M, Lambert B, Joly F,
Poulain L, Denoyelle C and Meryet-Figuiere M: The influence of long
non-coding RNAs on the response to chemotherapy in ovarian cancer.
Gynecol Oncol. 156:726–733. 2020. View Article : Google Scholar
|
|
14
|
Zeng S, Liu S, Feng J, Gao J and Xue F:
Upregulation of lncRNA AB073614 functions as a predictor of
epithelial ovarian cancer prognosis and promotes tumor growth in
vitro and in vivo. Cancer Biomark. 24:421–428. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu W, Gao H, Li X, Zhu Y, Peng S, Yu J,
Zhan G, Wang J, Liu N and Guo X: LncRNA TPT1-AS1 promotes
tumorigenesis and metastasis in epithelial ovarian cancer by
inducing TPT1 expression. Cancer Sci. 110:1587–1598. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao J and Liu HR: Down-regulation of long
noncoding RNA DLX6-AS1 defines good prognosis and inhibits
proliferation and metastasis in human epithelial ovarian cancer
cells via Notch signaling pathway. Eur Rev Med Pharmacol Sci.
23:3243–3252. 2019.PubMed/NCBI
|
|
17
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar :
|
|
18
|
Nguyen VHL, Yue C, Du KY, Salem M, O'Brien
J and Peng C: The role of microRNAs in epithelial ovarian cancer
metastasis. Int J Mol Sci. 21:70932020. View Article : Google Scholar :
|
|
19
|
Staicu CE, Predescu DV, Rusu CM, Radu BM,
Cretoiu D, Suciu N, Crețoiu SM and Voinea SC: Role of microRNAs as
clinical cancer biomarkers for ovarian cancer: A short overview.
Cells. 9:1692020. View Article : Google Scholar :
|
|
20
|
Khan S, Ayub H, Khan T and Wahid F:
MicroRNA biogenesis, gene silencing mechanisms and role in breast,
ovarian and prostate cancer. Biochimie. 167:12–24. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Niu ZS, Wang WH, Dong XN and Tian LM: Role
of long noncoding RNA-mediated competing endogenous RNA regulatory
network in hepatocellular carcinoma. World J Gastroenterol.
26:4240–4260. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weng W, Zhang Z, Huang W, Xu X, Wu B, Ye
T, Shan Y, Shi K and Lin Z: Identification of a competing
endogenous RNA network associated with prognosis of pancreatic
adenocarcinoma. Cancer Cell Int. 20:2312020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Raspaglio G, Petrillo M, Martinelli E, Li
Puma DD, Mariani M, De Donato M, Filippetti F, Mozzetti S, Prislei
S, Zannoni GF, et al: Sox9 and Hif-2α regulate TUBB3 gene
expression and affect ovarian cancer aggressiveness. Gene.
542:173–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malki S, Bibeau F, Notarnicola C, Roques
S, Berta P, Poulat F and Boizet-Bonhoure B: Expression and
biological role of the prostaglandin D synthase/SOX9 pathway in
human ovarian cancer cells. Cancer Lett. 255:182–193. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiao S, Li Y, Pan Q, Ye M, He S, Tian Q
and Xue M: MiR-34c/SOX9 axis regulates the chemoresistance of
ovarian cancer cell to cisplatin-based chemotherapy. J Cell
Biochem. 120:2940–2953. 2019. View Article : Google Scholar
|
|
27
|
McCabe EM and Rasmussen TP: lncRNA
involvement in cancer stem cell function and epithelial-mesenchymal
transitions. Semin Cancer Biol. Dec 17–2020.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qian Y, Shi L and Luo Z: Long non-coding
RNAs in cancer: Implications for diagnosis, prognosis, and therapy.
Front Med ( Lausanne). 7:6123932020. View Article : Google Scholar
|
|
29
|
Chen S and Shen X: Long noncoding RNAs:
Functions and mechanisms in colon cancer. Mol Cancer. 19:1672020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nikpayam E, Tasharrofi B, Sarrafzadeh S
and Ghafouri-Fard S: The role of long non-coding RNAs in ovarian
cancer. Iran Biomed J. 21:3–15. 2017. View Article : Google Scholar :
|
|
31
|
Tong L, Ao Y, Zhang H, Wang K, Wang Y and
Ma Q: Long noncoding RNA NORAD is upregulated in epithelial ovarian
cancer and its downregulation suppressed cancer cell functions by
competing with miR-155-5p. Cancer Med. 8:4782–4791. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng T, Qiu J, Li C, Lin X, Tang X and
Hua K: Long noncoding RNA LINC00673 promotes the proliferation and
metastasis of epithelial ovarian cancer by associating with opioid
growth factor receptor. Onco Targets Ther. 12:6145–6156. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiu JJ, Lin XJ, Tang XY, Zheng TT, Zhang
XY and Hua KQ: Long noncoding RNA TC0101441 induces
epithelial-mesenchymal transition in epithelial ovarian cancer
metastasis by downregulating KiSS1. Int J Cancer. 146:2588–2598.
2020. View Article : Google Scholar
|
|
34
|
Li W, Ma S, Bai X, Pan W, Ai L and Tan W:
Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting
as a competing endogenous RNA of microRNA-18a in ovarian cancer. J
Cell Physiol. 235:1141–1154. 2020. View Article : Google Scholar
|
|
35
|
Gokulnath P, de Cristofaro T, Manipur I,
Di Palma T, Soriano AA, Guarracino MR and Zannini M: Long
non-coding RNA MAGI2-AS3 is a new player with a tumor suppressive
role in high grade serous ovarian carcinoma. Cancers (Basel). 11.
pp. 20082019, View Article : Google Scholar
|
|
36
|
Lin X, Tang X, Zheng T, Qiu J and Hua K:
Long non-coding RNA AOC4P suppresses epithelial ovarian cancer
metastasis by regulating epithelial-mesenchymal transition. J
Ovarian Res. 13:452020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qu J, Li M, Zhong W and Hu C: Competing
endogenous RNA in cancer: A new pattern of gene expression
regulation. Int J Clin Exp Med. 8:17110–17116. 2015.
|
|
38
|
Yang L, Lv Q, Liu J, Qi S and Fu D:
miR-431 regulates granulosa cell function through the IRS2/PI3K/AKT
signaling pathway. J Reprod Dev. 66:231–239. 2020. View Article : Google Scholar : PubMed/NCBI
|