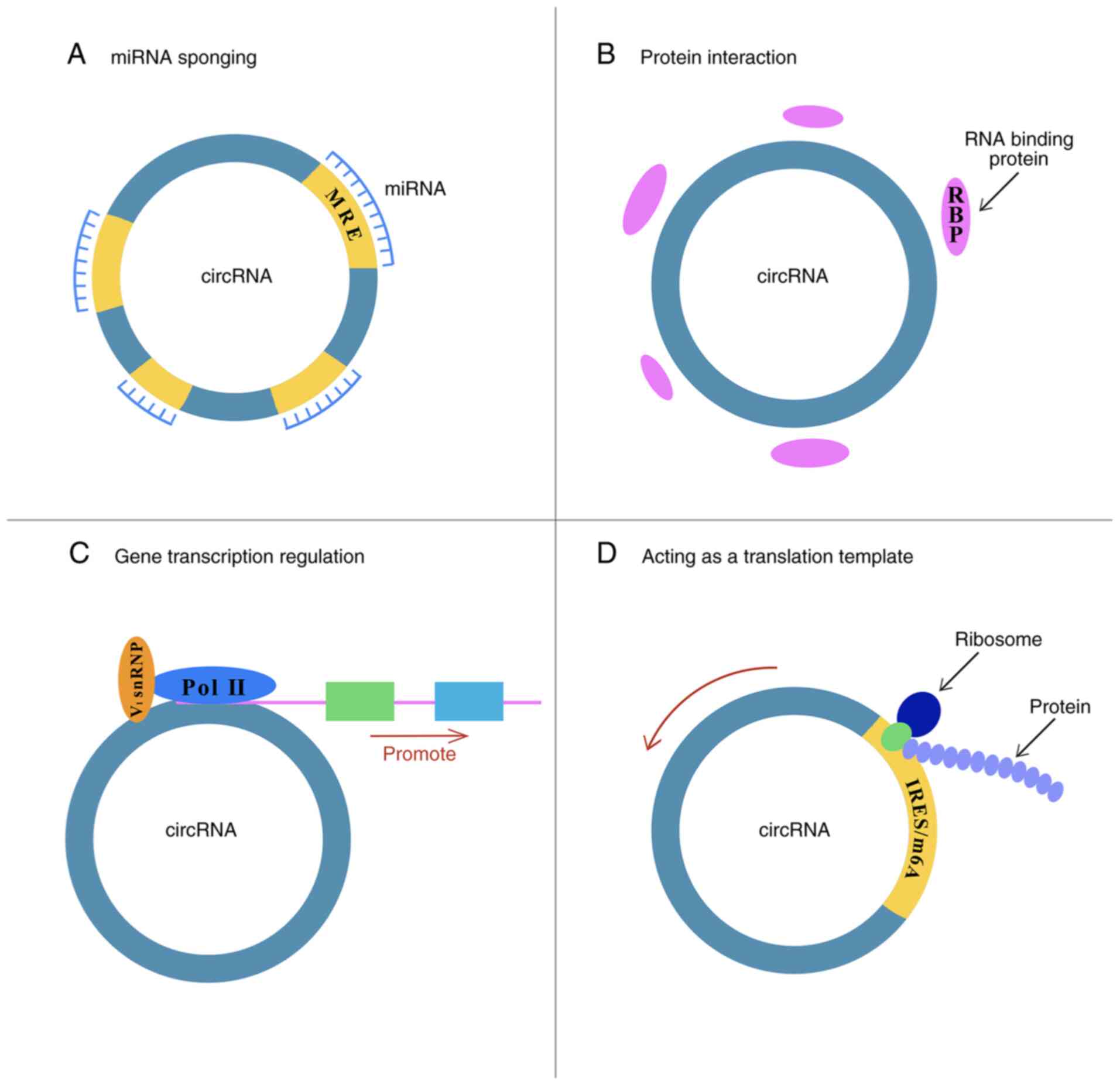
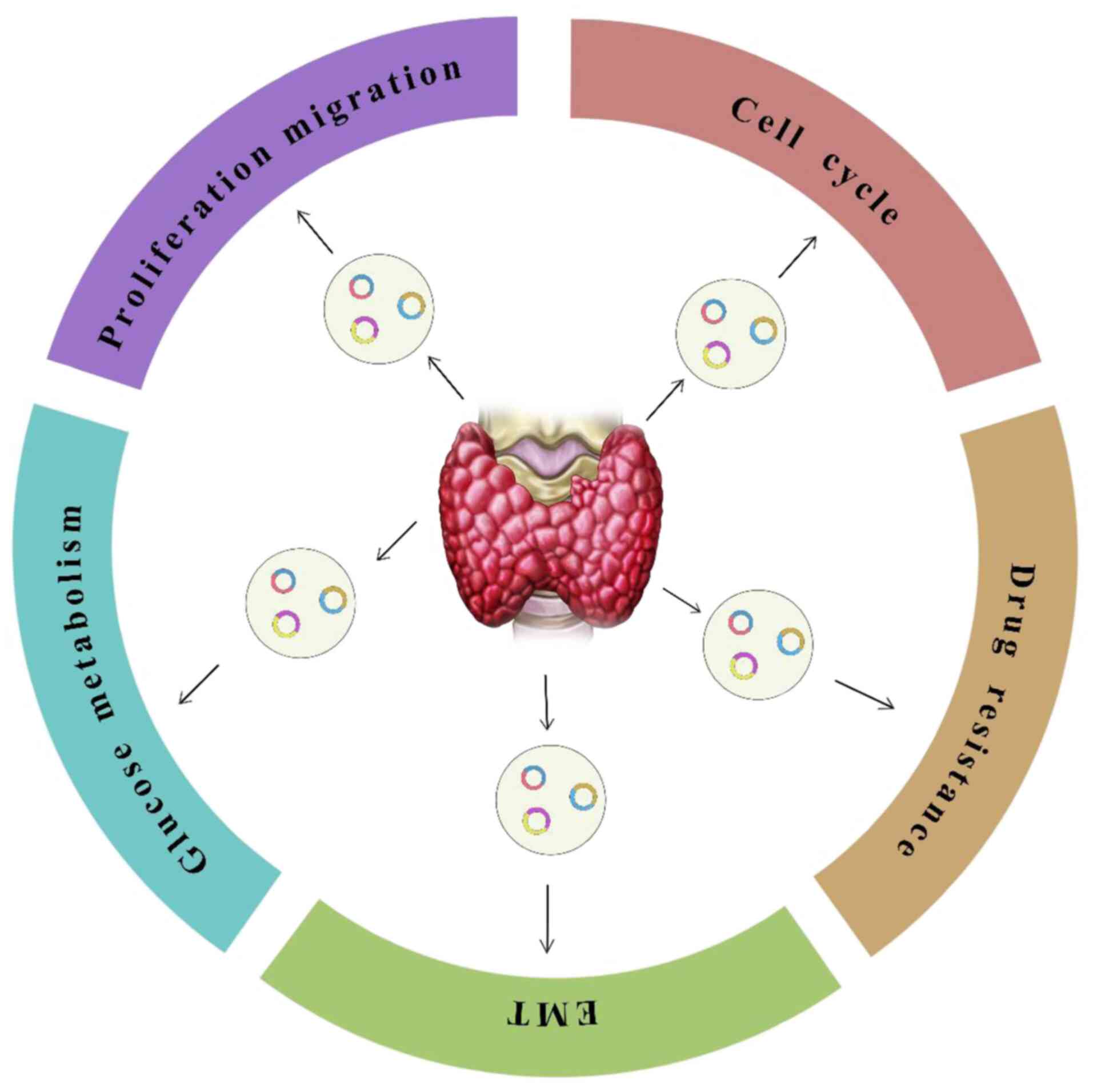
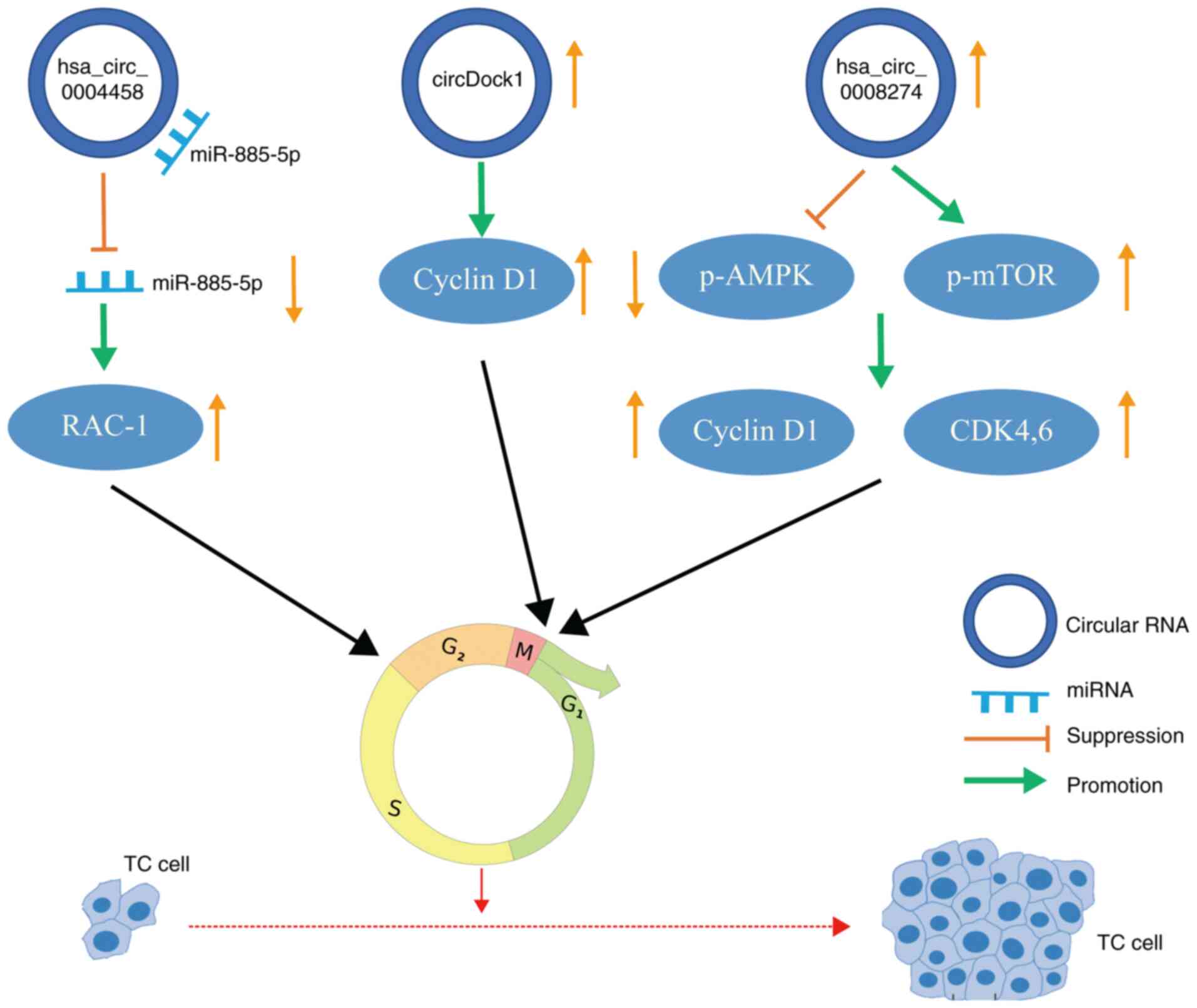
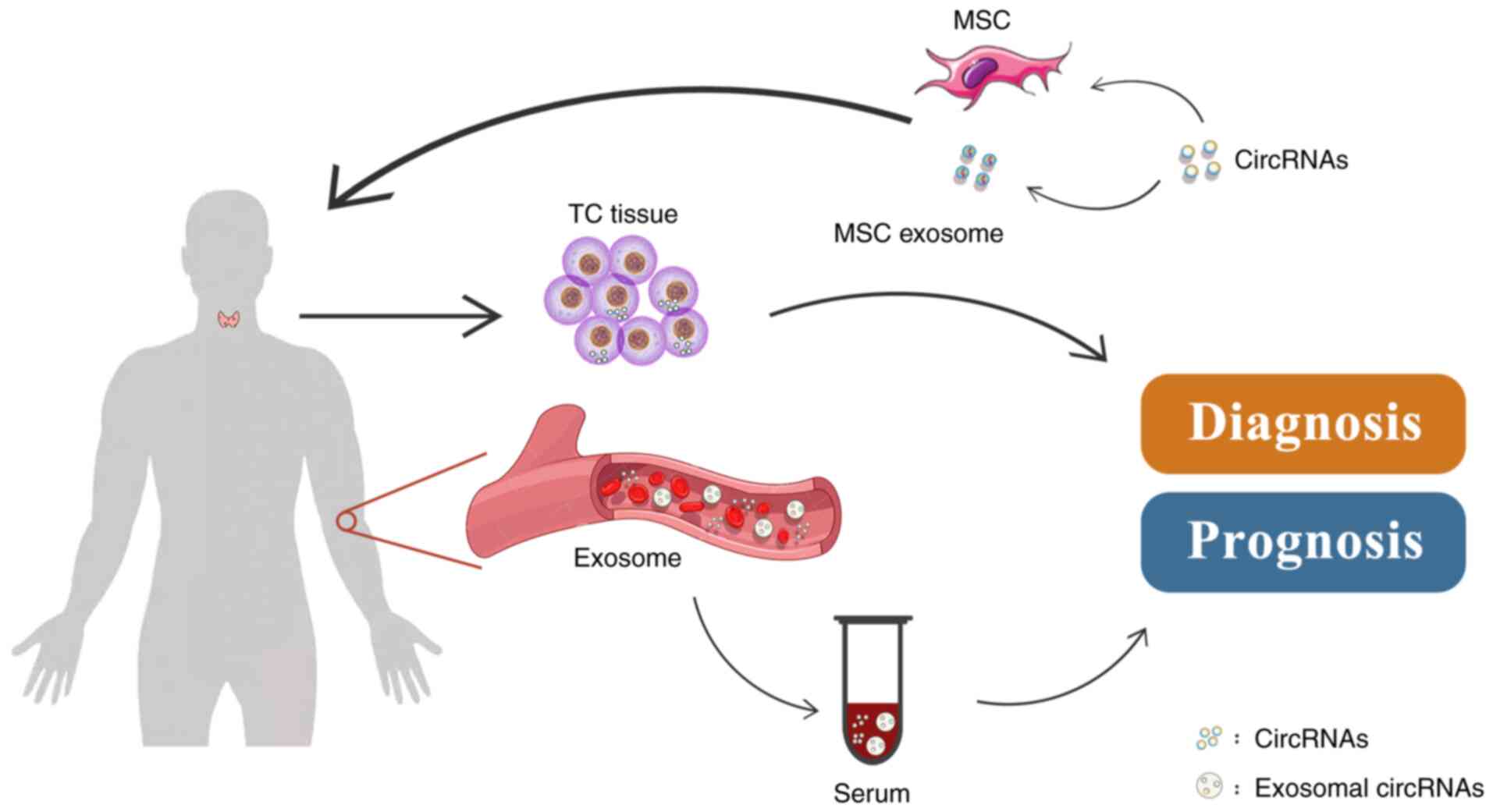
|
1
|
Prete A, Borges de Souza P, Censi S, Muzza
M, Nucci N and Sponziello M: Update on fundamental mechanisms of
thyroid cancer. Front Endocrinol (Lausanne). 11:1022020. View Article : Google Scholar
|
|
2
|
Kitahara CM and Sosa JA: The changing
incidence of thyroid cancer. Nat Rev Endocrinol. 12:646–653. 2016.
View Article : Google Scholar
|
|
3
|
Miller KD, Fidler-Benaoudia M, Keegan TH,
Hipp HS, Jemal A and Siegel RL: Cancer statistics for adolescents
and young adults, 2020. CA Cancer J Clin. 70:443–459. 2020.
View Article : Google Scholar
|
|
4
|
ASCO Thyroid cancer: Statistics.
https://www.cancer.net/cancer-types/thyroid-cancer/statistics.
Accessed May 5, 2021.
|
|
5
|
Massimino M, Evans DB, Podda M, Spinelli
C, Collini P, Pizzi N and Bleyer A: Thyroid cancer in adolescents
and young adults. Pediatr Blood Cancer. 65:e270252018. View Article : Google Scholar
|
|
6
|
Fleeman N, Houten R, Bagust A, Richardson
M, Beale S, Boland A, Dundar Y, Greenhalgh J, Hounsome J, Duarte R
and Shenoy A: Lenvatinib and sorafenib for differentiated thyroid
cancer after radioactive iodine: A systematic review and economic
evaluation. Health Technol Assess. 24:1–180. 2020. View Article : Google Scholar
|
|
7
|
Molinaro E, Romei C, Biagini A, Sabini E,
Agate L, Mazzeo S, Materazzi G, Sellari-Franceschini S, Ribechini
A, Torregrossa L, et al: Anaplastic thyroid carcinoma: From
clinicopathology to genetics and advanced therapies. Nat Rev
Endocrinol. 13:644–660. 2017. View Article : Google Scholar
|
|
8
|
Ahn HS, Kim HJ and Welch HG: Korea's
thyroid-cancer 'epidemic'-screening and overdiagnosis. N Engl J
Med. 371:1765–1767. 2014. View Article : Google Scholar
|
|
9
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar
|
|
10
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar
|
|
11
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar
|
|
12
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar
|
|
13
|
Suzuki H, Zuo Y, Wang J, Zhang MQ,
Malhotra A and Mayeda A: Characterization of RNase R-digested
cellular RNA source that consists of lariat and circular RNAs from
pre-mRNA splicing. Nucleic Acids Res. 34:e632006. View Article : Google Scholar
|
|
14
|
Enuka Y, Lauriola M, Feldman ME, Sas-Chen
A, Ulitsky I and Yarden Y: Circular RNAs are long-lived and display
only minimal early alterations in response to a growth factor.
Nucleic Acids Res. 44:1370–1383. 2016. View Article : Google Scholar
|
|
15
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar
|
|
16
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar
|
|
17
|
Zhang M and Du X: Noncoding RNAs in
gastric cancer: Research progress and prospects. World J
Gastroenterol. 22:6610–6618. 2016. View Article : Google Scholar
|
|
18
|
Xiong W, Ai YQ and Li YF, Ye Q, Chen ZT,
Qin JY, Liu QY, Wang H, Ju YH, Li WH and Li YF: Microarray analysis
of circular RNA expression profile associated with
5-fluorouracil-based chemoradiation resistance in colorectal cancer
cells. Biomed Res Int. 2017:84216142017. View Article : Google Scholar
|
|
19
|
Yao T, Chen Q, Fu L and Guo J: Circular
RNAs: Biogenesis, properties, roles, and their relationships with
liver diseases. Hepatol Res. 47:497–504. 2017. View Article : Google Scholar
|
|
20
|
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z,
Que Z and Liu Y: TTBK2 circular RNA promotes glioma malignancy by
regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol.
10:522017. View Article : Google Scholar
|
|
21
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar
|
|
22
|
Guo D, Li F, Zhao X, Long B, Zhang S, Wang
A, Cao D, Sun J and Li B: Circular RNA expression and association
with the clinicopathological characteristics in papillary thyroid
carcinoma. Oncol Rep. 44:519–532. 2020. View Article : Google Scholar
|
|
23
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
24
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar
|
|
25
|
Zhou X and Yang PC: MicroRNA: A small
molecule with a big biological impact. Microrna. 1:12012.
View Article : Google Scholar
|
|
26
|
Lee KP, Shin YJ, Panda AC, Abdelmohsen K,
Kim JY, Lee SM, Bahn YJ, Choi JY, Kwon ES, Baek SJ, et al: miR-431
promotes differentiation and regeneration of old skeletal muscle by
targeting Smad4. Genes Dev. 29:1605–1617. 2015. View Article : Google Scholar
|
|
27
|
Panda AC, Abdelmohsen K and Gorospe M:
SASP regulation by noncoding RNA. Mech Ageing Dev. 168:37–43. 2017.
View Article : Google Scholar
|
|
28
|
Panda AC, Sahu I, Kulkarni SD, Martindale
JL, Abdelmohsen K, Vindu A, Joseph J, Gorospe M and Seshadri V:
miR-196b-mediated translation regulation of mouse insulin2 via the
5′UTR. PLoS One. 9:e1010842014. View Article : Google Scholar
|
|
29
|
Munk R, Panda AC, Grammatikakis I, Gorospe
M and Abdelmohsen K: Senescence-associated MicroRNAs. Int Rev Cell
Mol Biol. 334:177–205. 2017. View Article : Google Scholar
|
|
30
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar
|
|
31
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar
|
|
32
|
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang
Z, Yu H and Kong D: Overexpression of sircular RNA ciRS-7 abrogates
the tumor suppressive effect of miR-7 on gastric cancer via
PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 119:440–446. 2018.
View Article : Google Scholar
|
|
33
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar
|
|
34
|
Liu J, Li H, Wei C, Ding J, Lu J, Pan G
and Mao A: circFAT1(e2) promotes papillary thyroid cancer
proliferation, migration, and invasion via the miRNA-873/ZEB1 axis.
Comput Math Methods Med. 2020:14593682020. View Article : Google Scholar
|
|
35
|
Conlon EG and Manley JL: RNA-binding
proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev.
31:1509–1528. 2017. View Article : Google Scholar
|
|
36
|
Errichelli L, Dini Modigliani S, Laneve P,
Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R,
Peruzzi G, et al: FUS affects circular RNA expression in murine
embryonic stem cell-derived motor neurons. Nat Commun. 8:147412017.
View Article : Google Scholar
|
|
37
|
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang
L, Ma J, Li X, Zeng Y, Yang Z, et al: A circular RNA promotes
tumorigenesis by inducing c-myc nuclear translocation. Cell Death
Differ. 24:1609–1620. 2017. View Article : Google Scholar
|
|
38
|
Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu
D, Gupta S, Yang W and Yang BB: The circular RNA interacts with
STAT3, increasing its nuclear translocation and wound repair by
modulating Dnmt3a and miR-17 function. Mol Ther. 25:2062–2074.
2017. View Article : Google Scholar
|
|
39
|
Bi W, Huang J, Nie C, Liu B, He G, Han J,
Pang R, Ding Z, Xu J and Zhang J: CircRNA circRNA_102171 promotes
papillary thyroid cancer progression through modulating
CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin
Cancer Res. 37:2752018. View Article : Google Scholar
|
|
40
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
41
|
Feng Y, Yang Y, Zhao X, Fan Y, Zhou L,
Rong J and Yu Y: Circular RNA circ0005276 promotes the
proliferation and migration of prostate cancer cells by interacting
with FUS to transcriptionally activate XIAP. Cell Death Dis.
10:7922019. View Article : Google Scholar
|
|
42
|
Garikipati VNS, Verma SK, Cheng Z, Liang
D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et
al: Circular RNA CircFndc3b modulates cardiac repair after
myocardial infarction via FUS/VEGF-A axis. Nat Commun. 10:43172019.
View Article : Google Scholar
|
|
43
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar
|
|
44
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar
|
|
45
|
Wu N, Yuan Z, Du KY, Fang L, Lyu J, Zhang
C, He A, Eshaghi E, Zeng K, Ma J, et al: Translation of
yes-associated protein (YAP) was antagonized by its circular RNA
via suppressing the assembly of the translation initiation
machinery. Cell Death Differ. 26:2758–2773. 2019. View Article : Google Scholar
|
|
46
|
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang
S, Song W, Li X, Li L, Du Z, et al: A novel FLI1 exonic circular
RNA promotes metastasis in breast cancer by coordinately regulating
TET1 and DNMT1. Genome Biol. 19:2182018. View Article : Google Scholar
|
|
47
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e29. 2017. View Article : Google Scholar
|
|
48
|
Stagsted LV, Nielsen KM, Daugaard I and
Hansen TB: Noncoding AUG circRNAs constitute an abundant and
conserved subclass of circles. Life Sci Alliance. 2:e2019003982019.
View Article : Google Scholar
|
|
49
|
Wawrzyniak O, Zarębska Ż, Kuczyński K,
Gotz-Więckowska A and Rolle K: Protein-related circular RNAs in
human pathologies. Cells. 9:18412020. View Article : Google Scholar
|
|
50
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of circRNAs. Mol
Cell. 66:9–21.e27. 2017. View Article : Google Scholar
|
|
51
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)a promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar
|
|
52
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N(6)-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar
|
|
53
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar
|
|
54
|
Liang ZX, Liu HS, Xiong L, Yang X, Wang
FW, Zeng ZW, He XW, Wu XR and Lan P: A novel NF-κB regulator
encoded by circ-PLCE1 inhibits colorectal carcinoma progression by
promoting RPS3 ubiquitin-dependent degradation. Mol Cancer.
20:1032021. View Article : Google Scholar
|
|
55
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar
|
|
56
|
Huang C, Liang D, Tatomer DC and Wilusz
JE: A length-dependent evolutionarily conserved pathway controls
nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018.
View Article : Google Scholar
|
|
57
|
Natua S, Dhamdhere SG, Mutnuru SA and
Shukla S: Interplay within tumor microenvironment orchestrates
neoplastic RNA metabolism and transcriptome diversity. Wiley
Interdiscip Rev RNA. 9:e16762021.
|
|
58
|
Viralippurath Ashraf J, Sasidharan Nair V,
Saleh R and Elkord E: Role of circular RNAs in colorectal tumor
microenvironment. Biomed Pharmacother. 137:1113512021. View Article : Google Scholar
|
|
59
|
Han XT, Jiang JQ, Li MZ and Cong QM:
Circular RNA circ-ABCB10 promotes the proliferation and invasion of
thyroid cancer by targeting KLF6. Eur Rev Med Pharmacol Sci.
24:97742020.
|
|
60
|
DiFeo A, Martignetti JA and Narla G: The
role of KLF6 and its splice variants in cancer therapy. Drug Resist
Updat. 12:1–7. 2009. View Article : Google Scholar
|
|
61
|
Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J,
Shen Y, Kang J and Li J: Circular RNA circBACH2 plays a role in
papillary thyroid carcinoma by sponging miR-139-5p and regulating
LMO4 expression. Cell Death Dis. 10:1842019. View Article : Google Scholar
|
|
62
|
Racevskis J, Dill A, Sparano JA and Ruan
H: Molecular cloning of LMO41, a new human LIM domain gene. Biochim
Biophys Acta. 1445:148–153. 1999. View Article : Google Scholar
|
|
63
|
Wu G, Zhou W, Lin X, Sun Y, Li J, Xu H,
Shi P, Gao L and Tian X: circRASSF2 acts as ceRNA and promotes
papillary thyroid carcinoma progression through miR-1178/TLR4
signaling pathway. Mol Ther Nucleic Acids. 19:1153–1163. 2020.
View Article : Google Scholar
|
|
64
|
Zhou GK, Zhang GY, Yuan ZN, Pei R and Liu
DM: Has_ circ_0008274 promotes cell proliferation and invasion
involving AMPK/mTOR signaling pathway in papillary thyroid
carcinoma. Eur Rev Med Pharmacol Sci. 22:8772–8780. 2018.
|
|
65
|
Zheng FB, Chen D, Ding YY, Wang SR, Shi DD
and Zhu ZP: Circular RNA circ_0103552 promotes the invasion and
migration of thyroid carcinoma cells by sponging miR-127. Eur Rev
Med Pharmacol Sci. 24:2572–2578. 2020.
|
|
66
|
Ye M, Hou H, Shen M, Dong S and Zhang T:
Circular RNA circFOXM1 plays a role in papillary thyroid carcinoma
by sponging miR-1179 and regulating HMGB1 expression. Mol Ther
Nucleic Acids. 19:741–750. 2020. View Article : Google Scholar
|
|
67
|
Yao Y, Chen X, Yang H, Chen W, Qian Y, Yan
Z, Liao T, Yao W, Wu W, Yu T, et al: Hsa_circ_0058124 promotes
papillary thyroid cancer tumorigenesis and invasiveness through the
NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 38:3182019. View Article : Google Scholar
|
|
68
|
Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H,
Shi P, Li J, Gao L and Tian X: Circular RNA profiling reveals
exosomal circ_0006156 as a novel biomarker in papillary thyroid
cancer. Mol Ther Nucleic Acids. 19:1134–1144. 2020. View Article : Google Scholar
|
|
69
|
Wang YF, Li MY, Tang YF, Jia M, Liu Z and
Li HQ: Circular RNA circEIF3I promotes papillary thyroid carcinoma
progression through competitively binding to miR-149 and
upregulating KIF2A expression. Am J Cancer Res. 10:1130–1139.
2020.
|
|
70
|
Qie S and Diehl JA: Cyclin D1, cancer
progression, and opportunities in cancer treatment. J Mol Med
(Berl). 94:1313–1326. 2016. View Article : Google Scholar
|
|
71
|
Cui W and Xue J: Circular RNA DOCK1
downregulates microRNA-124 to induce the growth of human thyroid
cancer cell lines. Biofactors. 46:591–599. 2020. View Article : Google Scholar
|
|
72
|
Motoshima H, Goldstein BJ, Igata M and
Araki E: AMPK and cell proliferation-AMPK as a therapeutic target
for atherosclerosis and cancer. J Physiol. 574:63–71. 2006.
View Article : Google Scholar
|
|
73
|
Jin X, Wang Z, Pang W, Zhou J, Liang Y,
Yang J, Yang L and Zhang Q: Upregulated hsa_circ_0004458
contributes to progression of papillary thyroid carcinoma by
inhibition of miR-885-5p and activation of RAC1. Med Sci Monit.
24:5488–5500. 2018. View Article : Google Scholar
|
|
74
|
Yan Y, Greer PM, Cao PT, Kolb RH and Cowan
KH: RAC1 GTPase plays an important role in γ-irradiation induced
G2/M checkpoint activation. Breast Cancer Res. 14:R602012.
View Article : Google Scholar
|
|
75
|
Qi Y, He J, Zhang Y, Wang L, Yu Y, Yao B
and Tian Z: Circular RNA hsa_circ_0001666 sponges
miR_x001E_330_x001E_5p, miR_x001E_193a_x001E_5p and miR_x001E_326,
and promotes papillary thyroid carcinoma progression via
upregulation of ETV4. Oncol Rep. 45:502021. View Article : Google Scholar
|
|
76
|
Li Z, Huang X, Liu A, Xu J, Lai J, Guan H
and Ma J: Circ_PSD3 promotes the progression of papillary thyroid
carcinoma via the miR-637/HEMGN axis. Life Sci. 264:1186222021.
View Article : Google Scholar
|
|
77
|
Feldkoren B, Hutchinson R, Rapoport Y,
Mahajan A and Margulis V: Integrin signaling potentiates
transforming growth factor-beta 1 (TGF-β1) dependent
down-regulation of E-Cadherin expression-important implications for
epithelial to mesenchymal transition (EMT) in renal cell carcinoma.
Exp Cell Res. 355:57–66. 2017. View Article : Google Scholar
|
|
78
|
Nishiyama M, Tsunedomi R, Yoshimura K,
Hashimoto N, Matsukuma S, Ogihara H, Kanekiyo S, Iida M, Sakamoto
K, Suzuki N, et al: Metastatic ability and the
epithelial-mesenchymal transition in induced cancer stem-like
hepatoma cells. Cancer Sci. 109:1101–1109. 2018. View Article : Google Scholar
|
|
79
|
Derynck R and Weinberg RA: EMT and cancer:
More than meets the eye. Dev Cell. 49:313–316. 2019. View Article : Google Scholar
|
|
80
|
Gui X, Li Y, Zhang X, Su K and Cao W:
Circ_LDLR promoted the development of papillary thyroid carcinoma
via regulating miR-195-5p/LIPH axis. Cancer Cell Int. 20:2412020.
View Article : Google Scholar
|
|
81
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar
|
|
82
|
Zhang W, Liu T, Li T and Zhao X:
Hsa_circRNA_102002 facilitates metastasis of papillary thyroid
cancer through regulating miR-488-3p/HAS2 axis. Cancer Gene Ther.
28:279–293. 2021. View Article : Google Scholar
|
|
83
|
Han JY, Guo S, Wei N, Xue R, Li W, Dong G,
Li J, Tian X, Chen C, Qiu S, et al: ciRS-7 promotes the
proliferation and migration of papillary thyroid cancer by
negatively regulating the miR-7/Epidermal growth factor receptor
axis. Biomed Res Int. 2020:98756362020. View Article : Google Scholar
|
|
84
|
Wang H, Yan X, Zhang H and Zhan X: CircRNA
circ_0067934 overexpression correlates with poor prognosis and
promotes thyroid carcinoma progression. Med Sci Monit.
25:1342–1349. 2019. View Article : Google Scholar
|
|
85
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar
|
|
86
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar
|
|
87
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.
|
|
88
|
Lunt SY and Vander Heiden MG: Aerobic
glycolysis: Meeting the metabolic requirements of cell
proliferation. Annu Rev Cell Dev Biol. 27:441–464. 2011. View Article : Google Scholar
|
|
89
|
Dang CV, Hamaker M, Sun P, Le A and Gao P:
Therapeutic targeting of cancer cell metabolism. J Mol Med (Berl).
89:205–212. 2011. View Article : Google Scholar
|
|
90
|
Ren H, Song Z, Chao C and Mao W:
circCCDC66 promotes thyroid cancer cell proliferation, migratory
and invasive abilities and glycolysis through the miR-211-5p/PDK4
axis. Oncol Lett. 21:4162021. View Article : Google Scholar
|
|
91
|
Li Y, Qin J, He Z, Cui G, Zhang K and Wu
B: Knockdown of circPUM1 impedes cell growth, metastasis and
glycolysis of papillary thyroid cancer via enhancing MAPK1
expression by serving as the sponge of miR-21-5p. Genes Genomics.
43:141–150. 2021. View Article : Google Scholar
|
|
92
|
Hu Z, Zhao P, Zhang K, Zang L, Liao H and
Ma W: Hsa_ circ_0011290 regulates proliferation, apoptosis and
glycolytic phenotype in papillary thyroid cancer via miR-1252/FSTL1
signal pathway. Arch Biochem Biophys. 685:1083532020. View Article : Google Scholar
|
|
93
|
Longley DB and Johnston PG: Molecular
mechanisms of drug resistance. J Pathol. 205:275–292. 2005.
View Article : Google Scholar
|
|
94
|
Shaili E: Platinum anticancer drugs and
photochemotherapeutic agents: Recent advances and future
developments. Sci Prog. 97:20–40. 2014. View Article : Google Scholar
|
|
95
|
Hundahl SA, Fleming ID, Fremgen AM and
Menck HR: A national cancer data base report on 53-856 cases of
thyroid carcinoma treated in the U.S., 1985-1995[see commetns].
Cancer. 83:2638–2648. 1998. View Article : Google Scholar
|
|
96
|
Kitamura Y, Shimizu K, Nagahama M, Sugino
K, Ozaki O, Mimura T, Ito K, Ito K and Tanaka S: Immediate causes
of death in thyroid carcinoma: Clinicopathological analysis of 161
fatal cases. J Clin Endocrinol Metab. 84:4043–4049. 1999.
View Article : Google Scholar
|
|
97
|
Antonelli A, Miccoli P, Derzhitski VE,
Panasiuk G, Solovieva N and Baschieri L: Epidemiologic and clinical
evaluation of thyroid cancer in children from the Gomel region
(Belarus). World J Surg. 20:867–871. 1996. View Article : Google Scholar
|
|
98
|
Zheng X, Cui D, Xu S, Brabant G and
Derwahl M: Doxorubicin fails to eradicate cancer stem cells derived
from anaplastic thyroid carcinoma cells: Characterization of
resistant cells. Int J Oncol. 37:307–315. 2010.
|
|
99
|
Liu F, Zhang J, Qin L, Yang Z, Xiong J,
Zhang Y, Li R, Li S, Wang H, Yu B, et al: Circular RNA EIF6
(Hsa_circ_0060060) sponges miR-144-3p to promote the
cisplatin-resistance of human thyroid carcinoma cells by autophagy
regulation. Aging (Albany NY). 10:3806–3820. 2018. View Article : Google Scholar
|
|
100
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar
|
|
101
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar
|
|
102
|
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W
and Zhang RP: Circulating circular RNA hsa_circ_0001785 acts as a
diagnostic biomarker for breast cancer detection. Clin Chim Acta.
487:363–368. 2018. View Article : Google Scholar
|
|
103
|
Chen Y, Wei S, Wang X, Zhu X and Han S:
Progress in research on the role of circular RNAs in lung cancer.
World J Surg Oncol. 16:2152018. View Article : Google Scholar
|
|
104
|
Wei J, Wei W, Xu H, Wang Z, Gao W, Wang T,
Zheng Q, Shu Y and De W: Circular RNA hsa_circRNA_102958 may serve
as a diagnostic marker for gastric cancer. Cancer Biomark.
27:139–145. 2020. View Article : Google Scholar
|
|
105
|
Lan X, Cao J, Xu J, Chen C, Zheng C, Wang
J, Zhu X, Zhu X and Ge M: Decreased expression of hsa_circ_0137287
predicts aggressive clinicopathologic characteristics in papillary
thyroid carcinoma. J Clin Lab Anal. 32:e225732018. View Article : Google Scholar
|
|
106
|
Ren H, Liu Z, Liu S, Zhou X, Wang H, Xu J,
Wang D and Yuan G: Profile and clinical implication of circular
RNAs in human papillary thyroid carcinoma. PeerJ. 6:e53632018.
View Article : Google Scholar
|
|
107
|
Shi E, Ye J, Zhang R, Ye S, Zhang S, Wang
Y, Cao Y and Dai W: A combination of circRNAs as a diagnostic tool
for discrimination of papillary thyroid cancer. Onco Targets Ther.
13:4365–4372. 2020. View Article : Google Scholar
|
|
108
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019. View Article : Google Scholar
|
|
109
|
van der Pol E, Böing AN, Harrison P, Sturk
A and Nieuwland R: Classification, functions, and clinical
relevance of extracellular vesicles. Pharmacol Rev. 64:676–705.
2012. View Article : Google Scholar
|
|
110
|
Yang C, Wei Y, Yu L and Xiao Y:
Identification of altered circular RNA expression in serum exosomes
from patients with papillary thyroid carcinoma by high-throughput
sequencing. Med Sci Monit. 25:2785–2791. 2019. View Article : Google Scholar
|
|
111
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar
|
|
112
|
Chen D, Ma W, Ke Z and Xie F: CircRNA
hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung
cancer progression. Cell Cycle. 17:2080–2090. 2018. View Article : Google Scholar
|
|
113
|
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M,
Liu X, Sun H, Pan Y and Wang S: The pro-metastasis effect of
circANKS1B in breast cancer. Mol Cancer. 17:1602018. View Article : Google Scholar
|
|
114
|
Sun JW, Qiu S, Yang JY, Chen X and Li HX:
Hsa_circ_0124055 and hsa_circ_0101622 regulate proliferation and
apoptosis in thyroid cancer and serve as prognostic and diagnostic
indicators. Eur Rev Med Pharmacol Sci. 24:4348–4360. 2020.
|
|
115
|
Rüster B, Göttig S, Ludwig RJ, Bistrian R,
Müller S, Seifried E, Gille J and Henschler R: Mesenchymal stem
cells display coordinated rolling and adhesion behavior on
endothelial cells. Blood. 108:3938–3944. 2006. View Article : Google Scholar
|
|
116
|
De Becker A and Riet IV: Homing and
migration of mesenchymal stromal cells: How to improve the efficacy
of cell therapy? World J Stem Cells. 8:73–87. 2016. View Article : Google Scholar
|
|
117
|
Nitzsche F, Müller C, Lukomska B,
Jolkkonen J, Deten A and Boltze J: Concise review: MSC adhesion
cascade-insights into homing and transendothelial migration. Stem
Cells. 35:1446–1460. 2017. View Article : Google Scholar
|
|
118
|
Toh WS, Lai RC, Zhang B and Lim SK: MSC
exosome works through a protein-based mechanism of action. Biochem
Soc Trans. 46:843–853. 2018. View Article : Google Scholar
|
|
119
|
Keshtkar S, Azarpira N and Ghahremani MH:
Mesenchymal stem cell-derived extracellular vesicles: Novel
frontiers in regenerative medicine. Stem Cell Res Ther. 9:632018.
View Article : Google Scholar
|
|
120
|
Kalimuthu S, Oh JM, Gangadaran P, Zhu L,
Lee HW, Jeon YH, Jeong SY, Lee SW, Lee J and Ahn BC: Genetically
engineered suicide gene in mesenchymal stem cells using a Tet-On
system for anaplastic thyroid cancer. PLoS One. 12:e01813182017.
View Article : Google Scholar
|
|
121
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar
|
|
122
|
Tang M, Wang Q, Wang K and Wang F:
Mesenchymal stem cells-originated exosomal microRNA-152 impairs
proliferation, invasion and migration of thyroid carcinoma cells by
interacting with DPP4. J Endocrinol Invest. 43:1787–1796. 2020.
View Article : Google Scholar
|
|
123
|
Zhang C, Cao J, Lv W and Mou H:
CircRNA_100395 carried by exosomes from adipose-derived mesenchymal
stem cells inhibits the malignant transformation of non-small cell
lung carcinoma through the miR-141-3p-LATS2 axis. Front Cell Dev
Biol. 9:6631472021. View Article : Google Scholar
|
|
124
|
Yan B, Zhang Y, Liang C, Liu B, Ding F,
Wang Y, Zhu B, Zhao R, Yu XY and Li Y: Stem cell-derived exosomes
prevent pyroptosis and repair ischemic muscle injury through a
novel exosome/circHIPK3/FOXO3a pathway. Theranostics. 10:6728–6742.
2020. View Article : Google Scholar
|
|
125
|
Zhu M, Liu X, Li W and Wang L: Exosomes
derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis
via activating autophagy. Hum Exp Toxicol. 39:1619–1627. 2020.
View Article : Google Scholar
|
|
126
|
Kamerkar S, LeBleu VS, Sugimoto H, Yang S,
Ruivo CF, Melo SA, Lee JJ and Kalluri R: Exosomes facilitate
therapeutic targeting of oncogenic KRAS in pancreatic cancer.
Nature. 546:498–503. 2017. View Article : Google Scholar
|
|
127
|
Zarovni N, Corrado A, Guazzi P, Zocco D,
Lari E, Radano G, Muhhina J, Fondelli C, Gavrilova J and Chiesi A:
Integrated isolation and quantitative analysis of exosome shuttled
proteins and nucleic acids using immunocapture approaches. Methods.
87:46–58. 2015. View Article : Google Scholar
|
|
128
|
Yu LL, Zhu J, Liu JX, Jiang F, Ni WK, Qu
LS, Ni RZ, Lu CH and Xiao MB: A comparison of traditional and novel
methods for the separation of exosomes from human samples. Biomed
Res Int. 2018:36345632018. View Article : Google Scholar
|