The pathogenesis of thyroid cancer (TC), the most
frequent type of endocrine cancer, is related to a variety of gene
mutations, such as those in v-raf murine sarcoma viral oncogene
homolog B1 (BRAF), human telomerase reverse transcriptase promoter,
TP53 and NRAS. However, almost no gene mutation has been proven to
be able to guide treatment or influence clinical outcomes (1). In addition to age, sex, ethnicity
and residential region of patients and subjects with a family
history of TC, radiation exposure, obesity, smoking and even body
height may be risk predictors of TC (2). In the last decade (2007-2016), the
incidence of TC rose at an annual rate of ~3% among 20- to
39-year-olds and 4% among 15- to 19-year-olds. The 5-year survival
rate of TC >99% (3) is
related to numerous factors, including the unique type of TC
(4), therapeutic treatment and
overdiagnosis (2,5). According to the origin and
histological characteristics of cancer cells, TC may be divided
into the following types: Differentiated TC, originating from
follicular cells, which accounts for >90% of all cases of TC
(6) and includes two major
subtypes, papillary thyroid carcinoma (PTC) and follicular thyroid
carcinoma; cancer originating from parafollicular cells is known as
medullary thyroid carcinoma. However, anaplastic TC (ATC), the most
heterogeneous tumor type among all TC subtypes, accounting for 1%
of TC cases, is the cause of the majority of all TC-associated
deaths and the chance of a cure is slim (7). In South Korea, the prevalence rate
has increased 15-fold over the past decade, which may be related to
overdiagnosis due to thyroid ultrasound examination, while the
TC-related mortality rate has been stable (8). Although fine-needle aspiration
biopsy is the gold standard for TC diagnosis, its invasiveness
limits its application; therefore, there is an urgent requirement
for a novel, less invasive method to balance the specificity and
sensitivity to improve the accuracy of TC diagnosis.
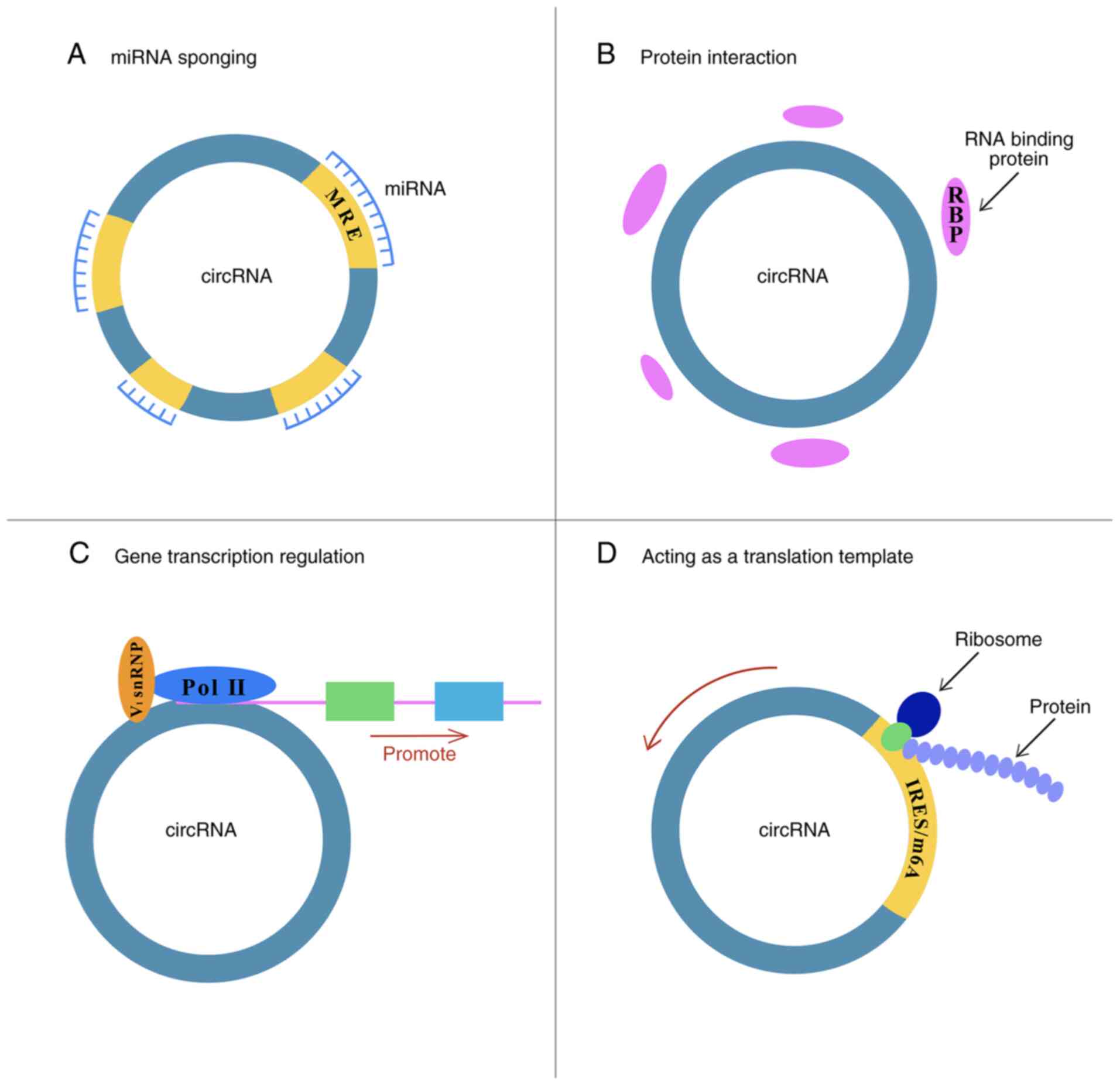
Due to their capacity to broadly regulate cell
conditions, circRNAs may be associated with tumor progression.
Abnormal expression of circRNAs has been detected in numerous
cancer types, such as esophageal cancer (16), gastric cancer (17), colorectal cancer (18), hepatocarcinoma (19), glioma (20), bladder cancer (21) and TC (22). CircRNAs function as cell activity
regulators, mainly through the following four aspects discussed
below.
MicroRNAs (miRNAs) are single-stranded noncoding
RNAs ~22 nucleotides in length and mediate post-transcriptional
gene silencing in the cytoplasm by interacting with the 6 nt seed
sequence on the 3′-untranslated region (3′UTR) of their target
mRNAs (23,24). miRNAs have extensively vital
roles in the post-transcriptional regulation of gene expression,
including cell proliferation, migration, differentiation and
apoptosis (25-31). Serving as the upstream molecules
of miRNAs, circRNAs, which contain miRNA response elements, are
able to bind various miRNAs and suppress their activity via a
mechanism known as miRNA sponging (30,31). For instance, the first discovered
miRNA sponge, antisense transcript of cerebellar
degeneration-related protein 1 (CDR1), also termed CIRS-7, derived
from the antisense transcript of the CDR1 gene, is able to
negatively regulate miR-7 (30,31), which may be detected in gastric
cancer and hepatocellular carcinoma (32,33). This effect is also commonly
observed in TC: CircFAT1 (e2) has a function similar to that of
CIRS-7 and has a binding site for miR-873. CircFAT1 is highly
expressed in PTC tissues and cells and serves as an miRNA sponge to
vastly downregulate miR-873 expression, thus upregulating the
activity of zinc finger E-box binding homeobox 1 and ultimately
promoting the growth, migration and invasion of PTC cells (34).
In addition to miRNA sponging, protein interaction
is another important function of circRNAs. Among other proteins,
RNA binding proteins (RBPs) are the most famous proteins involved
in the regulation of RNA metabolism in different fields, such as
formation, transportation, localization and translation (35). Errichelli et al (36) reported that fused in sarcoma was
able to modify the formation of numerous circRNAs in mouse motor
neurons derived in vitro by binding the introns flanking the
back-splicing junctions. Sharing consensus sequences with other
RBPs, the c-MYC protein is able to interact with circ-Amotl1,
easing nuclear translocation of c-MYC, which appears to promote its
stability and upregulate c-MYC targets (37). Similarly, nuclear translocation
of signal transducer and activator of transcription 3 is also
associated with circ-Amotl1 in a parallel manner (38). In addition, numerous circRNAs
have been reported to interact with proteins in TC. CircRNA_102171
is highly expressed in PTC tissues, directly interacts with
β-catenin interacting protein 1 (CTNNBIP1) and obstructs its
association with the β-catenin/T cell factor (TCF)3/TCF4/lymphoid
enhancer factor 1 complex to facilitate PTC progression (39). Existing evidence affirms that
specific circRNAs are able to interact with different proteins
(9,40), while certain proteins may also
dynamically bind to different circRNAs (41,42). To date, research on circRNAs
interacting with proteins in TC remains limited, probably due to it
being more challenging to analyze the binding sequences of circRNAs
in RBPs than those of mRNAs.
Through an intricate mechanism, circRNAs are able to
modify gene transcription. With the assistance of U1 small nuclear
ribonucleoproteins, exon-intron circular RNAs (EIciRNAs) influence
the activity of RNA polymerase II (RNA pol II) via RNA-RNA
interactions and then regulate the transcription of their parental
gene, ultimately affecting protein translation (9,43). The nuclear circRNAs EIciPAIP2 and
EIciEIF3J promote parental gene transcription in a similar manner
(43). In addition to long
non-coding RNAs, ciRNAs may also act as activators of RNA pol II
and upregulate gene transcription. For instance, ci-ankrd52
enhances the expression of ankyrin repeat domain 52 protein
(44). Certain circRNAs have the
capacity to regulate gene expression at the translation level, in
addition to the transcription level. Recently, Wu et al
(45) reported that circYap
impaired the interaction of PABP on the poly(A) tail with eIF4G on
the 5′-cap of Yap mRNA. As a consequence, circYap suppressed gene
translation at the initiation stage. It was also demonstrated that
circRNAs may be associated with DNA modification. DNA
methyltransferase 1 is a methyltransferase that controls DNA
methylation and its promoter may be downregulated by circFECR1.
Furthermore, circFECR1 is also able to recruit the demethylase TET1
to the Friend leukemia virus integration 1 promoter, resulting in
the demethylation of DNA (46).
As a consequence, circRNAs may regulate gene expression in
different ways, while the effect of circRNAs on DNA replication
remains unexplored.
The two ends of a circRNA are connected by one
covalent bond, which impairs its translation function. Furthermore,
due to their loop structure lacking a 3′ and 5′UTR, circRNAs have
been classified as noncoding RNAs. However, most circRNAs are
ecircRNAs with an open reading frame, which implies their
translational potential (47-49). To date, their translational
activity has been proven in diverse organisms (50). With regard to certain groups of
circRNAs, specific elements are indispensable for cap-independent
translation mechanisms for translation initiation, such as internal
ribosome entry sites (IRESs) or N6-methyladenosine (m6A) (49). For instance, with the existence
of an IRES and necessary splicing elements, circZNF609 may be
translated into zinc finger protein 609 in a cap-independent way,
but the translation activity is much lower than that of its linear
counterpart (47). Another
cap-independent translation mechanism for circRNA is m6A in the
circRNA 5′UTR, which interacts with eIF4G2 and eIF3A and recruits
the 43S preinitiation complex to trigger translation. m6A
modification may be found in more than one-tenth of circRNAs, the
level of which is associated with circRNA translation efficiency
(51,52). It has been inferred that rolling
circle amplification may be a mechanism of enhancing the
translation productivity of circRNAs, but this mechanism only
produces long, repetitive peptides (53). Increasing evidence suggests the
direct translation function of circRNAs (54,55). However, the proteins encoded by
circRNAs remained to be analyzed. Perhaps these proteins are
persistently produced at low levels due to the resistance to
degradation and low translation activity characteristics of
circRNAs.
In conclusion, circRNAs may regulate downstream gene
and protein expression through various mechanisms and have numerous
unique advantages over other noncoding RNAs. Although circRNAs have
better stability and longer half-lives than their linear
transcripts, the possibility of circRNA degradation in their in
vivo transport when they are used as a therapeutic intervention
remains to be investigated. The effects of circRNA degradation on
disease and the biological effects on surrounding cells and tissues
require further study. Previous studies have revealed that circRNAs
are more conserved among mammals than other RNAs (12), but of note, in-depth study and
continuous discovery of novel circRNAs have indicated that numerous
circRNAs expressed are species-specific and do not have any
sequence homology (56).
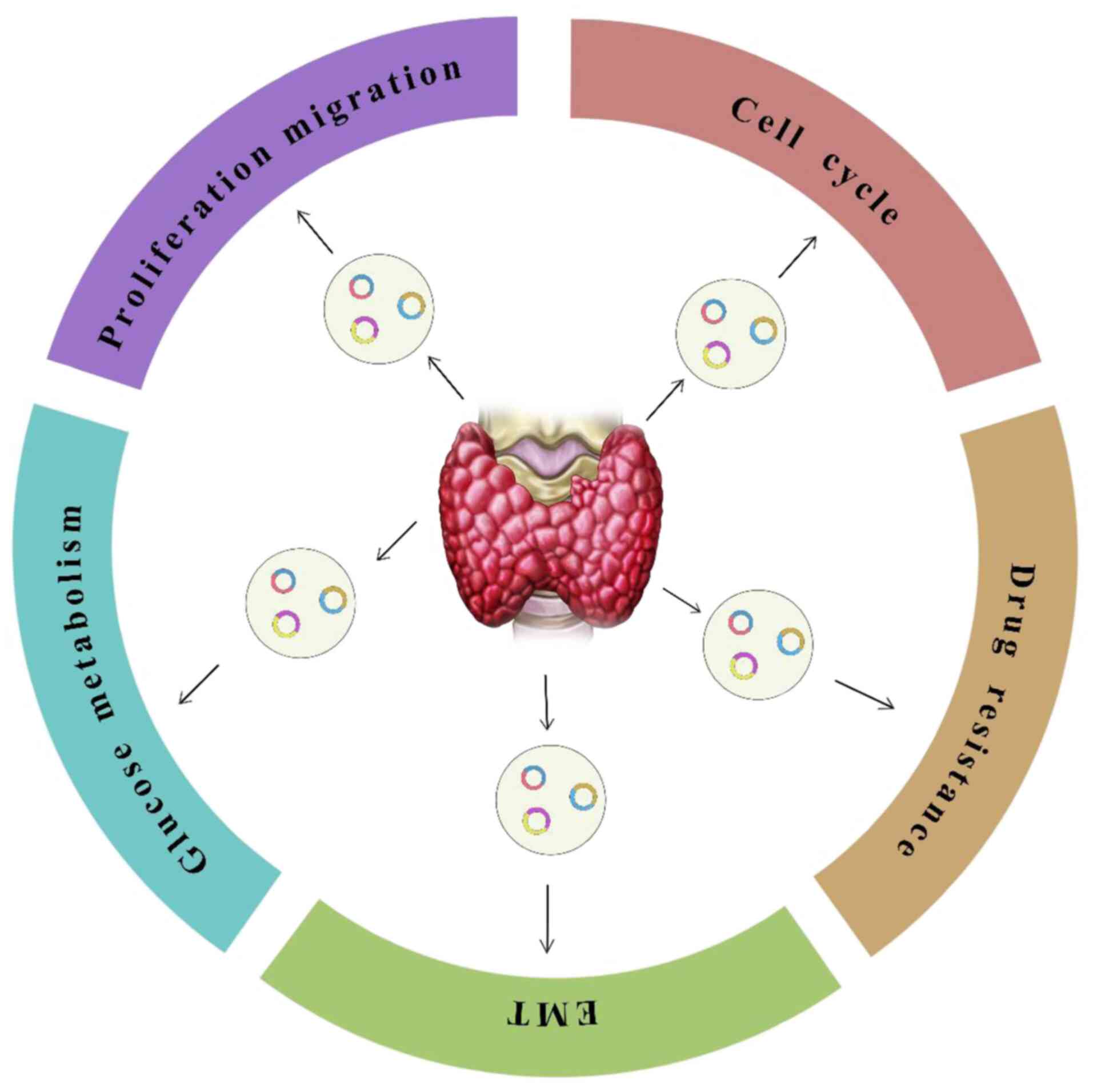
Increasing evidence suggests that noncoding RNAs,
including circRNAs, contribute to the formation of the tumor
microenvironment (TME), communication between cancer cells and
surrounding stromal cells (fibroblasts, immune cells, endothelial
cells) and the impact of physicochemical parameters (57). Numerous studies have revealed
that multiple functions of the TME may be influenced by circRNAs,
not only in tumor invasiveness but also in tumor angiogenesis,
epithelial to mesenchymal transition (EMT) and drug resistance
(57,58). Thus, circRNAs may regulate the
progression of TC invasion and evolution through different
approaches (Fig. 2).
Regulating cell invasiveness is of great importance
in tumor progression. In recent years, certain oncogenic circRNAs
have been indicated to regulate the invasiveness of TC. The
upregulation of circ-ABCB10 in TC is negatively correlated with the
expression of Krüppel-like zinc finger transcription factor 6
(KLF6) (59), which encodes a
series of proteins that engage in the regulation of cancer
development through alternative splicing, suggesting that
circ-ABCB10 may promote the proliferation and invasion of TC by
targeting KLF6 (60). As
mentioned previously, circRNA_102171 is able to directly bind
CTNNBIP1 and activate the Wnt/β-catenin pathway. Ultimately,
upregulated circRNA_102171 enhances the invasiveness of PTC
(39). In addition to directly
interacting with proteins, circRNAs also promote the progression of
TC by sponging miRNAs. For instance, overexpressed circBACH2 in PTC
sponges miR-139-5p, which is able to interact with the LIM-only
protein 4 (LMO4) 3′UTR, resulting in the inhibition of LMO4.
Therefore, the invasiveness of PTC is enhanced (61,62). Similarly, circRASSF2 is enriched
in PTC. By downregulating miR-1178, circRASSF2 facilitates tumor
growth and promotes invasiveness (63). To date, circ_0008274 (64), circ_0103552 (65), circ_FOXM1 (66), hsa_circ_0058124 (67), circ_FNDC3B (68) and circ_EIF3I (69) have been reported to participate
in the regulation of different signaling pathways associated with
the proliferation and apoptosis of TC cells.
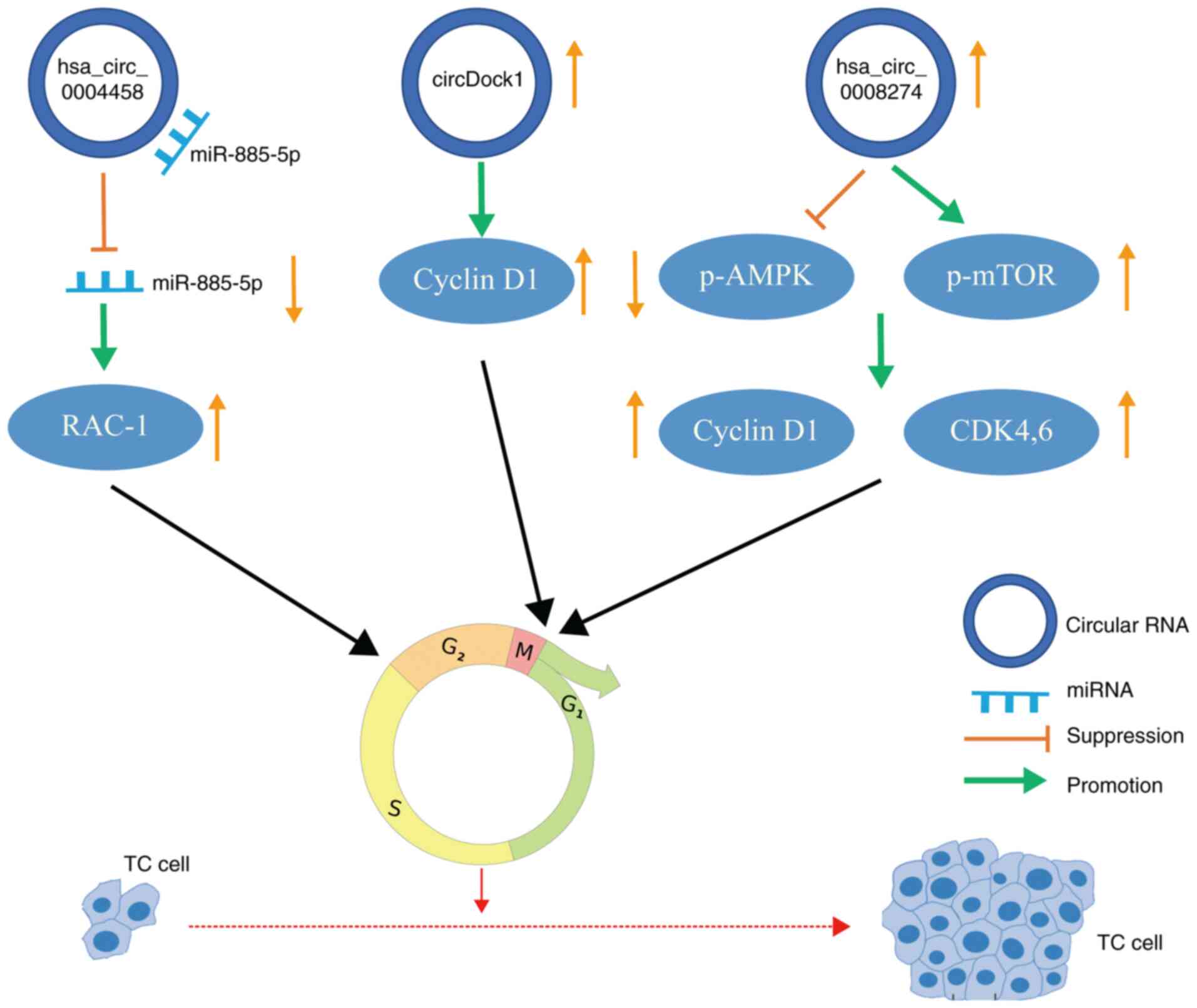
The cell cycle refers to the time interval, as
determined by experiments, during which cells prepare and then
equally replicate their genomes to form two daughter cells
(70). Cancer is characterized
by the abnormal activity of various cyclins that lead to
uncontrolled proliferation of tumor cells (Fig. 3). The upregulation of circDOCK1
in TC is accompanied by increased expression of cyclin D1 and
downregulation of p53, which leads to an imbalance of
cyclin-dependent kinase (CDK) activity and rapid growth of tumor
cells (71). Zhou et al
(64) indicated that the
expression of hsa_circ_0008274 in PTC was upregulated following
activation of the adenosine monophosphate-activated protein kinase
(AMPK)/mammalian target of rapamycin signaling pathway. AMPK
upregulates the p53-p21 axis and leads to cell cycle stagnation in
G1/S phase (72). In PTC,
circ_0004458 is upregulated. After circ_0004458 silencing, the cell
cycle distribution was substantially decreased compared with that
of the control group. Furthermore, hsa_circ_0004458 is able to
regulate the expression of Rac1 through the specific sponging of
miR-885-5p (73). Rac1 protein
is able to activate extracellular regulated protein kinase 1/2
signaling induced by γ-irradiation and the subsequent G2/M
checkpoint response (74).
Circ_0001666 is highly expressed in PTC cells. After circ_0001666
downregulation, the cell cycle was blocked in G1 phase, decreasing
the expression of cyclin D1 and cyclin E (75). When the overexpression of
circ_PSD3 in PTC cells was abolished, the expression of cyclin D1
and CDK4 also decreased significantly, which inhibited the cell
cycle progression of PTC cells (76). CircRNAs have a significant role
in cell cycle regulation, bypassing cell cycle checkpoints in
different ways to accelerate the progression of the cell cycle.
Most current findings are from studies that performed knockout of
cancer-promoting circRNAs and these types of experiments may reveal
the effects of circRNAs on TC. The direct interactions between
circRNA/miRNA or circRNA/protein and cell cycle proteins remain to
be determined.
EMT is a process whereby epithelial cells lose their
characteristics, such as cell-to-cell adhesion and cell polarity,
and instead acquire interstitial characteristics during the process
of cell culture; EMT is universally involved in physiological and
pathological processes, particularly tumor metastasis (77-79). Therefore, analyzing the
contribution of circRNAs to EMT and tumor metastasis may reveal
them to be potential targets to inhibit the malignant progression
of TC. CircRNA of low-density lipoprotein receptor (circ_LDLR) in
PTC tissues was significantly upregulated compared with that in
normal tissues and served as an miR-195-5p sponge to upregulate the
expression of LDLR mRNA, which led to a decrease in the level of
E-cadherin and an increase in the level of Twist1. Twist is able to
downregulate epithelial markers such as E-cad and upregulate
interstitial cell markers such as vimentin by altering the
transcription of EMT-related genes (80,81), which promotes EMT transformation
in PTC. In PTC cells with high expression of circ_102002, the
expression of E-cadherin was downregulated and the expression of
N-cadherin and mesenchymal phenotypic markers was upregulated,
while PTC cells with low expression of circ_102002 produced the
opposite result (82). CIRS-7
had a similar effect: E-cadherin was downregulated in PTC cells
with high expression of CIRS-7, while vimentin levels were
significantly increased (83).
Similarly, another study suggested that circ_0067934 may promote TC
by regulating EMT and PI3K/AKT signaling pathways (84). By sponging downstream targets,
circRNAs may promote the invasion and metastasis of TC through
their regulatory role in the EMT pathway, but the specific process
and downstream pathway are largely elusive and require to be
further explored.
The glucose metabolism in tumor cells differs from
that in normal cells. Only when oxygen is scarce do normal cells
rely on glycolysis rather than oxygen-consuming mitochondrial
metabolism to create energy. However, tumor cells prefer to perform
glycolysis in the cytosol regardless of whether oxygen is
sufficient (85-87). This phenomenon was termed the
Warburg effect and describes the ability of tumor cells to generate
energy at a rapid rate, accompanied by low efficiency in adenosine
triphosphate (ATP) production per molecule of glucose (88). Cancer cells undergo rapid growth
and proliferation, and their energy demand may be satisfied via
glycolysis. The Warburg effect has been suggested to be associated
with the regulation of certain oncogenes and tumor suppressors,
such as Akt, PI3K and Ras (89).
However, the relationship between circRNAs and glucose metabolism
in cancer remains to be fully proven. After circCCDC66 gene
knockout, the glucose metabolism of PTC cells was significantly
inhibited. Further analysis determined that circCCDC66 is able to
act as a sponge for miR-211-5p to promote the expression of
pyruvate dehydrogenase kinase 4, thereby increasing the level of
glucose metabolism in PTC cells (90). However, circPUM1 was highly
expressed in PTC. After downregulation of circPUM1, the expression
of hexokinase 2 was downregulated and glycolysis in PTC was blocked
(91). Similar research
suggested that hsa_circ_0011290 was significantly upregulated in
PTC (92). After specific
silencing of hsa_circ_0011290 in cells, the glucose metabolism
spectrum indicated that glucose uptake was inhibited, lactic acid
production decreased, the ATP content increased, and cell
proliferation and cell viability were significantly inhibited.
CircRNAs have a certain effect on glycolysis in TC and may directly
or indirectly regulate the activities of key enzymes in glucose
metabolism, but their regulatory mode of action remains to be
proven.
Drug resistance refers to the tolerance of
microorganisms, parasites and tumor cells to the effect of
chemotherapy drugs. Drug resistance may develop prior to treatment
or be acquired during treatment by tumors, which is a significant
obstacle to overcome in tumor treatment (93). Platinum drugs such as cisplatin
are extensively applied in treating human cancers and are
considered successful standard therapies (94), but their effect is limited in ATC
due to drug resistance (95-98). Liu et al (99) proved that circEIF6 acted as a
regulator of drug resistance in ATC. CircEIF6 was upregulated in
ATC tissues and cells and controlled transforming growth factor
(TGF)-α by sponging miR-144-3p in cells treated with cisplatin.
This signaling pathway (circEIF6/miR-144-3p/TGF-α axis) was
confirmed to be associated with lowering the sensitivity of ATCs to
cisplatin resistance. However, the study did not identify the TGF-α
downstream signaling molecules, which requires further
research.
In summary, numerous studies suggest that various
circRNAs may promote or inhibit the occurrence of TC through
different mechanisms (Table I).
Most circRNAs function as miRNA sponges and by interacting with
RBPs and a single circRNA may participate in different mechanisms
of cancer development through sponging a variety of miRNAs. Thus,
understanding the regulatory mechanism of circRNAs involved in the
TC process may reveal novel therapeutic targets. However, there are
limited studies on the subcellular localization and degradation
mechanism of circRNAs in TC cells, which may become a novel
starting point for research. The current research mainly focused on
the regulatory network of circRNAs in PTC, but studies on circRNAs
involved in other types and subtypes of TC are scarce, suggesting
that other types of TC may become the focus of future research. At
the same time, the lack of diversity in experimental models to
study the correlation between circRNAs and TC and other
uncontrollable factors may also have a negative impact on the
repeatability and reliability of the experimental results; thus,
unifying the relevant experimental models may lead to more
convincing results.
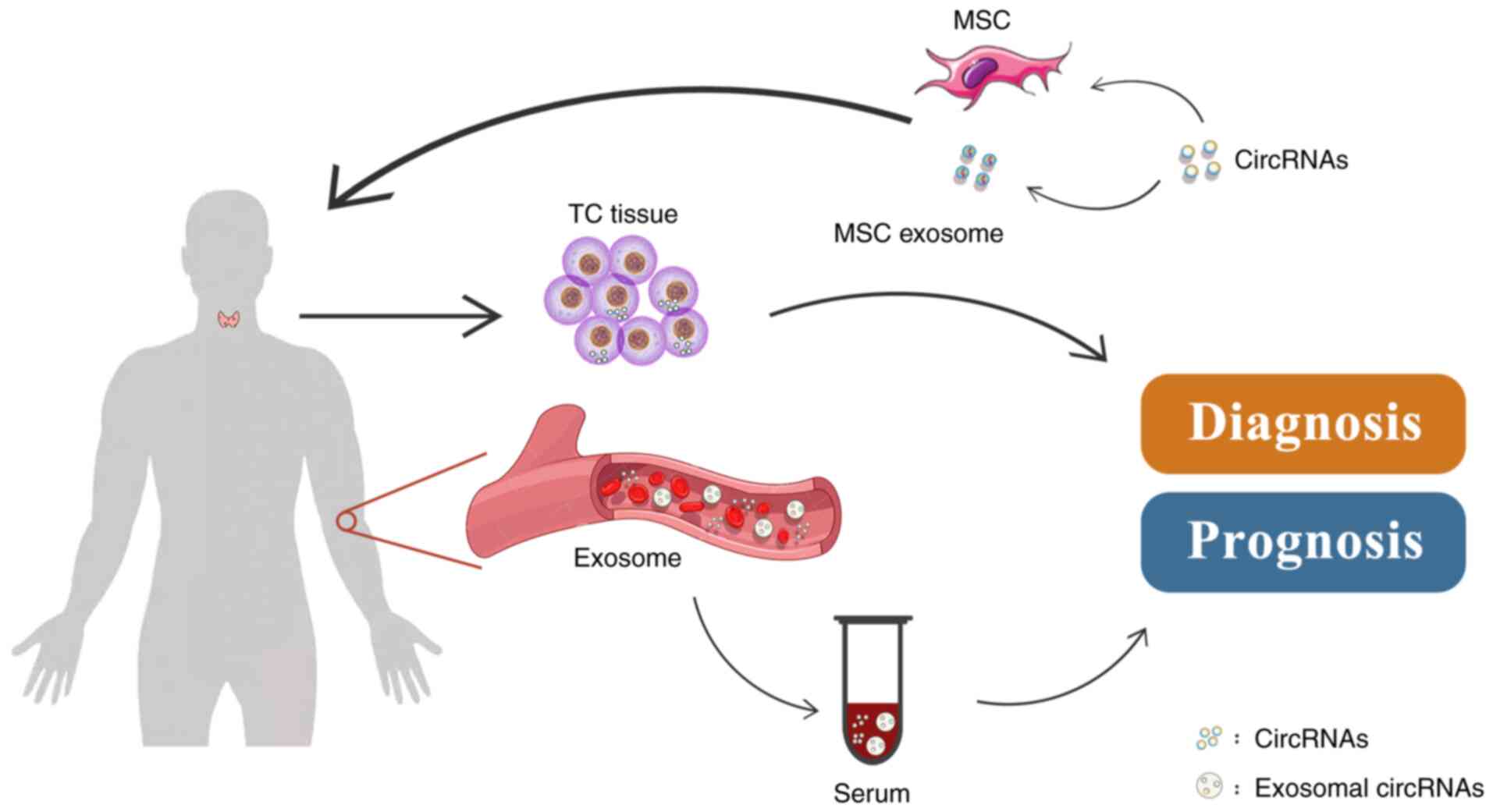
It is universally acknowledged that most cancer
types may be cured if diagnosed at an early stage. Due to the high
incidence of TC, the demand for early diagnosis is increasing. The
features of circRNAs, such as their stability and diverse nature,
enable them to accumulate in body fluids and tissues (12,100,101), making it possible for circRNAs
to serve as cancer biomarkers. The biomarker value of circRNAs has
been proven in different cancer types, such as breast cancer
(102), lung cancer (103) and gastric cancer (104). Several circRNAs have been
investigated as potential biomarkers through broad clinical sample
testing, including tissues, serum and exosomes from patients with
TC and healthy controls. These circRNAs have the potential to
enable the early diagnosis of TC and predict recurrence and
metastasis (Fig. 4).
Numerous studies have reported that circRNAs are
able to stably accumulate in peripheral blood. Hence, serum
circRNAs may be suitable and less invasive biomarkers. A study
suggested that two upregulated circRNAs (circRAPGEF5 and
hsa_circ_0058124) were stably enriched in the peripheral blood of
patients with PTC (107). The
results of a further expression analysis of circRAPGEF5 and
hsa_circ_0058124 in serum samples during the treatment of patients
with PTC indicated that the two circRNAs were markedly decreased
along with systematic treatment, indicating that circRNAs may allow
researchers to monitor the PTC process dynamically. The AUC for the
ability to discriminate PTC from healthy controls was 0.711 for
circRAPGEF5 and 0.790 for hsa_circ_0058124. Furthermore, the
combination of the two circRNAs (circRAPGEF5 and hsa_circ_0058124)
demonstrated a better diagnostic ability than a single circRNA in
PTC identification, with an AUC value of 0.860. Therefore, the
appropriate selection of a group of several circRNAs may be a novel
diagnostic approach that may increase the sensitivity, specificity
and accuracy of TC diagnosis and prognosis.
Exosomes are small vesicles with a single membrane,
exhibiting the same topology as cells, and their diameter varies
from 30 to 200 nm. Select proteins, lipids, nucleic acids and
glycoconjugates may be packaged in exosomes (108). Several studies have proven that
exosomes may act as carriers of circRNAs and transfer them between
cancer cells (109), suggesting
that circRNAs in exosomes have the potential to act as biomarkers.
Wu et al (63) isolated
serum exosomes from 60 serum samples collected from patients with
PTC and healthy subjects and indicated that circRASSF2 was
overexpressed in serum exosomes from patients with PTC. Logistic
regression analysis demonstrated that upregulation of circRASSF2
was markedly associated with tumor stage and LNM. In addition,
another study using high-throughput sequencing determined that
circRNAs were differentially expressed in serum-derived exosomes
collected from patients with PTC and patients with benign thyroid
goiter. A total of three upregulated circRNAs and 19 downregulated
circRNAs were detected in the former (110). However, no clinicopathologic
factor association analysis was performed in that study.
Prognosis is a prediction of the course and outcome
of a disease. Prognostic evaluation in the early stage of a disease
helps mitigate the effects of negative predictive factors. In
addition, cancer patients heavily rely on prognostic evaluation to
determine their life expectancy. The prognostic value of circRNAs
has been proven in diverse cancer types, such as colon cancer
(111), lung cancer (112) and triple-negative breast cancer
(113). Sun et al
(114) indicated that
hsa_circ_0124055 and hsa_circ_0101622 were overexpressed in the
plasma of patients with TC, which was significantly associated with
larger tumor size, poorer TNM stage or histological grade and LNM.
Compared with the levels in plasma of patients prior to
tumorectomy, hsa_circ_0124055 and hsa_circ_0101622 were markedly
decreased after surgery. The patients with high hsa_circ_0124055 or
hsa_circ_0101622 expression exhibited shorter overall survival. The
AUCs were 0.836 for hsa_circ_0124055 and 0.805 for
hsa_circ_0101622, which indicated their TC diagnostic significance.
Furthermore, the AUC rose to 0.911 after combining hsa_circ_0124055
with hsa_circ_0101622. In addition, in patients with PTC,
circFNDC3B was demonstrated to be upregulated in serum exosomes and
tissues. The AUC for circFNDC3B was 0.891. Patients with PTC with
low expression of circFNDC3B exhibited longer overall survival than
those with high expression of circFNDC3B according to Kaplan-Meier
survival curve analysis (68).
Guo et al (22)
discovered 8 circRNAs expressed abnormally in PTC tissues, 5 of
which were associated with BRAFV600E, capsular invasion, advanced
pT stage and LNM. These circRNAs, whose expression levels were
associated with the overall survival of patients, may be potential
prognostic biomarkers. However, to date, compared to the value
reported for circRNA as diagnostic biomarkers, the value of
prognostic circRNA biomarkers has not been widely demonstrated.
CircRNAs have attracted increasing attention in the
field of tumor research and have potential for interventional,
expression or regulatory therapy. There are certain problems
associated with circRNAs due to their low targeting, potential
off-target effects and biodegradation in vivo. Therefore,
novel strategies to improve their regulatory effect and targeting
of circRNAs require to be developed. The application of mesenchymal
stem cell (MSC) therapy in the field of cancer has attracted
increasing attention due to its unique chemotaxis, which brings
hope to improve the accuracy of targeted therapy. In addition,
exosomes secreted by MSCs are considered promising by researchers
due to their special biological characteristics and superiority
over MSCs. MSCs are multipotent stem cells with significant
potential for regenerative medicine. The therapeutic potential of
MSCs may be attributed to the key mechanism of homing, i.e. they
are able to migrate to the injured site and differentiate into
local components there (115-117).
Exosomes are small, lipid-membrane extracellular
vesicles (EVs) that are formed by endocytosis, integration and
efflux; they have a diameter of 30-150 nm, are stable in a variety
of biological fluids, such as urine, plasma and serum (121), and may be used as drug or gene
carriers. Therefore, to a certain extent, exosomes derived from
MSCs may be a good and promising cargo-loading carrier for the
treatment of tumors. Various studies have emphasized the function
of miRNAs in TC. However, research concerning the influence of
exosomal miRNAs on TC remains limited. Tang et al (122) aimed to uncover the regulatory
effects of exosomal miR152 on TC and the underlying mechanisms.
They isolated exosomal miR-152 from bone marrow mesenchymal stem
cells (BMSCs) and cocultured it with TC cells to explore its
potential for therapy. The results indicated that BMSC-derived
exosomal miR-152 inhibited the proliferation, invasion and
migration of TC cells and promoted cell apoptosis. The therapeutic
potential of MSC-derived exosomes has been proven in non-small cell
lung carcinoma (123), ischemic
muscle injury (124) and liver
fibrosis (125). However, the
use of exosome-loaded circRNAs in TC has remained unexplored, to
the best of our knowledge, suggesting that future studies may
further explore related treatments for TC using similar
strategies.
Taken together, the application of exosomes for the
treatment of tumors is currently a major research hotspot and the
utility of exosomes as carriers has also been widely and deeply
discussed in the field of cancer. At present, diverse technologies
may be used to isolate and analyze EVs, such as density gradient
zone centrifugation (119),
immunocapture by magnetic beads (126), exosome precipitation or
chromatography (127). However,
emptying native contents and loading the desired cargos may
represent limitations in EV applications. Whether the biological
efficacy of exosomes is only related to the cargo loaded into them
or whether the effects of exosomes themselves on the delivery of
goods have a role, as well as their molecular mechanisms, requires
to be further studied. Exosomes are vesicles that mediate cellular
communication via paracrine signaling (128); thus, whether the genes loaded
into exosomes, such as circRNAs and miRNAs, are able to mediate
intercellular communication and the underlying mechanisms of their
mode of action require to be investigated.
CircRNAs are closed circular noncoding RNAs formed
by reverse splicing of precursor RNAs. In recent years, circRNAs
have received increasing attention in cancer research. The present
study reviewed the role of circRNAs in TC, with an emphasis on
their ability to promote and inhibit proliferation, invasion and
metastasis of TC cells. CircRNAs affect tumor cells in different
ways, such as through regulating the cell cycle, EMT and drug
resistance. Of note, the mechanism by which circRNAs regulate
glucose metabolism in TC has also been indicated to have an
important role. At present, the clinical application of circRNAs is
mainly focused on diagnostic and prognostic biomarkers. CircRNAs
have been widely studied as diagnostic and prognostic biomarkers.
For the prognosis of patients with TC, regular detection of the
expression level of specific circRNAs may bring new improvements to
prognosis. In the present review, the possibility of using exosomes
from MSCs and MSCs themselves as carriers to load circRNAs for
treatment was described. At present, research on the biomarker
function of circRNAs in TC is mainly focused on invasive TC tissue
biopsy. Attention should be paid to noninvasive body fluids to
improve their practicability as diagnostic and prognostic markers.
In addition, the immunomodulatory effects of circRNAs in the TC
tumor microenvironment, their effects on angiogenesis of TC cells
and radiotherapy resistance have not been previously reported,
which may become potential research directions in the future. In
addition, the downstream signals of certain differentially
expressed circRNAs in thyroid carcinoma have not been fully studied
and the regulatory circRNA-miRNA-mRNA network mechanism and its
effects on the tumor microenvironment, extracellular matrix and
cellular communication require to be further studied. CircRNAs may
become a novel research direction and may provide therapeutic
targets. To date, studies have focused on the discovery signaling
pathways of circRNAs acting as miRNA sponges in PTC. However,
studies on other aspects, such as circRNAs affecting gene
expression and directly affecting proteins, their role as a
transcriptional template and the function of circRNAs in other
types of TC are currently scarce. In general, circRNAs have great
application prospects in the clinical treatment of tumors and the
molecular mechanisms of their effect on tumor cells require further
experimental research.
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
GZ and XC contributed to the literature search and
wrote the original draft of the manuscript. YK and XZ contributed
to the revised version of the manuscript. XT and CM generated all
the figures. SF contributed by performing the conceptualization.
All authors read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The study was supported by the Project of Local Science and
Technology Development guided by the Central Government (Innovative
Platform for Improving the Ability of Prevention and Treatment of
Multiple Diseases in Gansu), the Construction Plan of Gansu
Endocrine Disease Clinical Medical Research Center (grant no.
20JR10FA667), the Project of Gansu Natural Science Foundation
(grant no. 20JR10RA681), Lanzhou Science and Technology Development
Guiding Plan Project (grant no. 2019-ZD-38), College Students'
Innovation, Entrepreneurship and Excellence Program of Lanzhou
University in 2020 (grant no. 20200060103) and the College
Students' Innovation and Entrepreneurship in Lanzhou University in
2021 (grant no. 20210060155).
|
1
|
Prete A, Borges de Souza P, Censi S, Muzza
M, Nucci N and Sponziello M: Update on fundamental mechanisms of
thyroid cancer. Front Endocrinol (Lausanne). 11:1022020. View Article : Google Scholar
|
|
2
|
Kitahara CM and Sosa JA: The changing
incidence of thyroid cancer. Nat Rev Endocrinol. 12:646–653. 2016.
View Article : Google Scholar
|
|
3
|
Miller KD, Fidler-Benaoudia M, Keegan TH,
Hipp HS, Jemal A and Siegel RL: Cancer statistics for adolescents
and young adults, 2020. CA Cancer J Clin. 70:443–459. 2020.
View Article : Google Scholar
|
|
4
|
ASCO Thyroid cancer: Statistics.
https://www.cancer.net/cancer-types/thyroid-cancer/statistics.
Accessed May 5, 2021.
|
|
5
|
Massimino M, Evans DB, Podda M, Spinelli
C, Collini P, Pizzi N and Bleyer A: Thyroid cancer in adolescents
and young adults. Pediatr Blood Cancer. 65:e270252018. View Article : Google Scholar
|
|
6
|
Fleeman N, Houten R, Bagust A, Richardson
M, Beale S, Boland A, Dundar Y, Greenhalgh J, Hounsome J, Duarte R
and Shenoy A: Lenvatinib and sorafenib for differentiated thyroid
cancer after radioactive iodine: A systematic review and economic
evaluation. Health Technol Assess. 24:1–180. 2020. View Article : Google Scholar
|
|
7
|
Molinaro E, Romei C, Biagini A, Sabini E,
Agate L, Mazzeo S, Materazzi G, Sellari-Franceschini S, Ribechini
A, Torregrossa L, et al: Anaplastic thyroid carcinoma: From
clinicopathology to genetics and advanced therapies. Nat Rev
Endocrinol. 13:644–660. 2017. View Article : Google Scholar
|
|
8
|
Ahn HS, Kim HJ and Welch HG: Korea's
thyroid-cancer 'epidemic'-screening and overdiagnosis. N Engl J
Med. 371:1765–1767. 2014. View Article : Google Scholar
|
|
9
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar
|
|
10
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar
|
|
11
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar
|
|
12
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar
|
|
13
|
Suzuki H, Zuo Y, Wang J, Zhang MQ,
Malhotra A and Mayeda A: Characterization of RNase R-digested
cellular RNA source that consists of lariat and circular RNAs from
pre-mRNA splicing. Nucleic Acids Res. 34:e632006. View Article : Google Scholar
|
|
14
|
Enuka Y, Lauriola M, Feldman ME, Sas-Chen
A, Ulitsky I and Yarden Y: Circular RNAs are long-lived and display
only minimal early alterations in response to a growth factor.
Nucleic Acids Res. 44:1370–1383. 2016. View Article : Google Scholar
|
|
15
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar
|
|
16
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar
|
|
17
|
Zhang M and Du X: Noncoding RNAs in
gastric cancer: Research progress and prospects. World J
Gastroenterol. 22:6610–6618. 2016. View Article : Google Scholar
|
|
18
|
Xiong W, Ai YQ and Li YF, Ye Q, Chen ZT,
Qin JY, Liu QY, Wang H, Ju YH, Li WH and Li YF: Microarray analysis
of circular RNA expression profile associated with
5-fluorouracil-based chemoradiation resistance in colorectal cancer
cells. Biomed Res Int. 2017:84216142017. View Article : Google Scholar
|
|
19
|
Yao T, Chen Q, Fu L and Guo J: Circular
RNAs: Biogenesis, properties, roles, and their relationships with
liver diseases. Hepatol Res. 47:497–504. 2017. View Article : Google Scholar
|
|
20
|
Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z,
Que Z and Liu Y: TTBK2 circular RNA promotes glioma malignancy by
regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol.
10:522017. View Article : Google Scholar
|
|
21
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar
|
|
22
|
Guo D, Li F, Zhao X, Long B, Zhang S, Wang
A, Cao D, Sun J and Li B: Circular RNA expression and association
with the clinicopathological characteristics in papillary thyroid
carcinoma. Oncol Rep. 44:519–532. 2020. View Article : Google Scholar
|
|
23
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
24
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar
|
|
25
|
Zhou X and Yang PC: MicroRNA: A small
molecule with a big biological impact. Microrna. 1:12012.
View Article : Google Scholar
|
|
26
|
Lee KP, Shin YJ, Panda AC, Abdelmohsen K,
Kim JY, Lee SM, Bahn YJ, Choi JY, Kwon ES, Baek SJ, et al: miR-431
promotes differentiation and regeneration of old skeletal muscle by
targeting Smad4. Genes Dev. 29:1605–1617. 2015. View Article : Google Scholar
|
|
27
|
Panda AC, Abdelmohsen K and Gorospe M:
SASP regulation by noncoding RNA. Mech Ageing Dev. 168:37–43. 2017.
View Article : Google Scholar
|
|
28
|
Panda AC, Sahu I, Kulkarni SD, Martindale
JL, Abdelmohsen K, Vindu A, Joseph J, Gorospe M and Seshadri V:
miR-196b-mediated translation regulation of mouse insulin2 via the
5′UTR. PLoS One. 9:e1010842014. View Article : Google Scholar
|
|
29
|
Munk R, Panda AC, Grammatikakis I, Gorospe
M and Abdelmohsen K: Senescence-associated MicroRNAs. Int Rev Cell
Mol Biol. 334:177–205. 2017. View Article : Google Scholar
|
|
30
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar
|
|
31
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar
|
|
32
|
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang
Z, Yu H and Kong D: Overexpression of sircular RNA ciRS-7 abrogates
the tumor suppressive effect of miR-7 on gastric cancer via
PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 119:440–446. 2018.
View Article : Google Scholar
|
|
33
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar
|
|
34
|
Liu J, Li H, Wei C, Ding J, Lu J, Pan G
and Mao A: circFAT1(e2) promotes papillary thyroid cancer
proliferation, migration, and invasion via the miRNA-873/ZEB1 axis.
Comput Math Methods Med. 2020:14593682020. View Article : Google Scholar
|
|
35
|
Conlon EG and Manley JL: RNA-binding
proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev.
31:1509–1528. 2017. View Article : Google Scholar
|
|
36
|
Errichelli L, Dini Modigliani S, Laneve P,
Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfò R,
Peruzzi G, et al: FUS affects circular RNA expression in murine
embryonic stem cell-derived motor neurons. Nat Commun. 8:147412017.
View Article : Google Scholar
|
|
37
|
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang
L, Ma J, Li X, Zeng Y, Yang Z, et al: A circular RNA promotes
tumorigenesis by inducing c-myc nuclear translocation. Cell Death
Differ. 24:1609–1620. 2017. View Article : Google Scholar
|
|
38
|
Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu
D, Gupta S, Yang W and Yang BB: The circular RNA interacts with
STAT3, increasing its nuclear translocation and wound repair by
modulating Dnmt3a and miR-17 function. Mol Ther. 25:2062–2074.
2017. View Article : Google Scholar
|
|
39
|
Bi W, Huang J, Nie C, Liu B, He G, Han J,
Pang R, Ding Z, Xu J and Zhang J: CircRNA circRNA_102171 promotes
papillary thyroid cancer progression through modulating
CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin
Cancer Res. 37:2752018. View Article : Google Scholar
|
|
40
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
41
|
Feng Y, Yang Y, Zhao X, Fan Y, Zhou L,
Rong J and Yu Y: Circular RNA circ0005276 promotes the
proliferation and migration of prostate cancer cells by interacting
with FUS to transcriptionally activate XIAP. Cell Death Dis.
10:7922019. View Article : Google Scholar
|
|
42
|
Garikipati VNS, Verma SK, Cheng Z, Liang
D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et
al: Circular RNA CircFndc3b modulates cardiac repair after
myocardial infarction via FUS/VEGF-A axis. Nat Commun. 10:43172019.
View Article : Google Scholar
|
|
43
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar
|
|
44
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar
|
|
45
|
Wu N, Yuan Z, Du KY, Fang L, Lyu J, Zhang
C, He A, Eshaghi E, Zeng K, Ma J, et al: Translation of
yes-associated protein (YAP) was antagonized by its circular RNA
via suppressing the assembly of the translation initiation
machinery. Cell Death Differ. 26:2758–2773. 2019. View Article : Google Scholar
|
|
46
|
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang
S, Song W, Li X, Li L, Du Z, et al: A novel FLI1 exonic circular
RNA promotes metastasis in breast cancer by coordinately regulating
TET1 and DNMT1. Genome Biol. 19:2182018. View Article : Google Scholar
|
|
47
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e29. 2017. View Article : Google Scholar
|
|
48
|
Stagsted LV, Nielsen KM, Daugaard I and
Hansen TB: Noncoding AUG circRNAs constitute an abundant and
conserved subclass of circles. Life Sci Alliance. 2:e2019003982019.
View Article : Google Scholar
|
|
49
|
Wawrzyniak O, Zarębska Ż, Kuczyński K,
Gotz-Więckowska A and Rolle K: Protein-related circular RNAs in
human pathologies. Cells. 9:18412020. View Article : Google Scholar
|
|
50
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of circRNAs. Mol
Cell. 66:9–21.e27. 2017. View Article : Google Scholar
|
|
51
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)a promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar
|
|
52
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N(6)-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar
|
|
53
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar
|
|
54
|
Liang ZX, Liu HS, Xiong L, Yang X, Wang
FW, Zeng ZW, He XW, Wu XR and Lan P: A novel NF-κB regulator
encoded by circ-PLCE1 inhibits colorectal carcinoma progression by
promoting RPS3 ubiquitin-dependent degradation. Mol Cancer.
20:1032021. View Article : Google Scholar
|
|
55
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar
|
|
56
|
Huang C, Liang D, Tatomer DC and Wilusz
JE: A length-dependent evolutionarily conserved pathway controls
nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018.
View Article : Google Scholar
|
|
57
|
Natua S, Dhamdhere SG, Mutnuru SA and
Shukla S: Interplay within tumor microenvironment orchestrates
neoplastic RNA metabolism and transcriptome diversity. Wiley
Interdiscip Rev RNA. 9:e16762021.
|
|
58
|
Viralippurath Ashraf J, Sasidharan Nair V,
Saleh R and Elkord E: Role of circular RNAs in colorectal tumor
microenvironment. Biomed Pharmacother. 137:1113512021. View Article : Google Scholar
|
|
59
|
Han XT, Jiang JQ, Li MZ and Cong QM:
Circular RNA circ-ABCB10 promotes the proliferation and invasion of
thyroid cancer by targeting KLF6. Eur Rev Med Pharmacol Sci.
24:97742020.
|
|
60
|
DiFeo A, Martignetti JA and Narla G: The
role of KLF6 and its splice variants in cancer therapy. Drug Resist
Updat. 12:1–7. 2009. View Article : Google Scholar
|
|
61
|
Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J,
Shen Y, Kang J and Li J: Circular RNA circBACH2 plays a role in
papillary thyroid carcinoma by sponging miR-139-5p and regulating
LMO4 expression. Cell Death Dis. 10:1842019. View Article : Google Scholar
|
|
62
|
Racevskis J, Dill A, Sparano JA and Ruan
H: Molecular cloning of LMO41, a new human LIM domain gene. Biochim
Biophys Acta. 1445:148–153. 1999. View Article : Google Scholar
|
|
63
|
Wu G, Zhou W, Lin X, Sun Y, Li J, Xu H,
Shi P, Gao L and Tian X: circRASSF2 acts as ceRNA and promotes
papillary thyroid carcinoma progression through miR-1178/TLR4
signaling pathway. Mol Ther Nucleic Acids. 19:1153–1163. 2020.
View Article : Google Scholar
|
|
64
|
Zhou GK, Zhang GY, Yuan ZN, Pei R and Liu
DM: Has_ circ_0008274 promotes cell proliferation and invasion
involving AMPK/mTOR signaling pathway in papillary thyroid
carcinoma. Eur Rev Med Pharmacol Sci. 22:8772–8780. 2018.
|
|
65
|
Zheng FB, Chen D, Ding YY, Wang SR, Shi DD
and Zhu ZP: Circular RNA circ_0103552 promotes the invasion and
migration of thyroid carcinoma cells by sponging miR-127. Eur Rev
Med Pharmacol Sci. 24:2572–2578. 2020.
|
|
66
|
Ye M, Hou H, Shen M, Dong S and Zhang T:
Circular RNA circFOXM1 plays a role in papillary thyroid carcinoma
by sponging miR-1179 and regulating HMGB1 expression. Mol Ther
Nucleic Acids. 19:741–750. 2020. View Article : Google Scholar
|
|
67
|
Yao Y, Chen X, Yang H, Chen W, Qian Y, Yan
Z, Liao T, Yao W, Wu W, Yu T, et al: Hsa_circ_0058124 promotes
papillary thyroid cancer tumorigenesis and invasiveness through the
NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 38:3182019. View Article : Google Scholar
|
|
68
|
Wu G, Zhou W, Pan X, Sun Z, Sun Y, Xu H,
Shi P, Li J, Gao L and Tian X: Circular RNA profiling reveals
exosomal circ_0006156 as a novel biomarker in papillary thyroid
cancer. Mol Ther Nucleic Acids. 19:1134–1144. 2020. View Article : Google Scholar
|
|
69
|
Wang YF, Li MY, Tang YF, Jia M, Liu Z and
Li HQ: Circular RNA circEIF3I promotes papillary thyroid carcinoma
progression through competitively binding to miR-149 and
upregulating KIF2A expression. Am J Cancer Res. 10:1130–1139.
2020.
|
|
70
|
Qie S and Diehl JA: Cyclin D1, cancer
progression, and opportunities in cancer treatment. J Mol Med
(Berl). 94:1313–1326. 2016. View Article : Google Scholar
|
|
71
|
Cui W and Xue J: Circular RNA DOCK1
downregulates microRNA-124 to induce the growth of human thyroid
cancer cell lines. Biofactors. 46:591–599. 2020. View Article : Google Scholar
|
|
72
|
Motoshima H, Goldstein BJ, Igata M and
Araki E: AMPK and cell proliferation-AMPK as a therapeutic target
for atherosclerosis and cancer. J Physiol. 574:63–71. 2006.
View Article : Google Scholar
|
|
73
|
Jin X, Wang Z, Pang W, Zhou J, Liang Y,
Yang J, Yang L and Zhang Q: Upregulated hsa_circ_0004458
contributes to progression of papillary thyroid carcinoma by
inhibition of miR-885-5p and activation of RAC1. Med Sci Monit.
24:5488–5500. 2018. View Article : Google Scholar
|
|
74
|
Yan Y, Greer PM, Cao PT, Kolb RH and Cowan
KH: RAC1 GTPase plays an important role in γ-irradiation induced
G2/M checkpoint activation. Breast Cancer Res. 14:R602012.
View Article : Google Scholar
|
|
75
|
Qi Y, He J, Zhang Y, Wang L, Yu Y, Yao B
and Tian Z: Circular RNA hsa_circ_0001666 sponges
miR_x001E_330_x001E_5p, miR_x001E_193a_x001E_5p and miR_x001E_326,
and promotes papillary thyroid carcinoma progression via
upregulation of ETV4. Oncol Rep. 45:502021. View Article : Google Scholar
|
|
76
|
Li Z, Huang X, Liu A, Xu J, Lai J, Guan H
and Ma J: Circ_PSD3 promotes the progression of papillary thyroid
carcinoma via the miR-637/HEMGN axis. Life Sci. 264:1186222021.
View Article : Google Scholar
|
|
77
|
Feldkoren B, Hutchinson R, Rapoport Y,
Mahajan A and Margulis V: Integrin signaling potentiates
transforming growth factor-beta 1 (TGF-β1) dependent
down-regulation of E-Cadherin expression-important implications for
epithelial to mesenchymal transition (EMT) in renal cell carcinoma.
Exp Cell Res. 355:57–66. 2017. View Article : Google Scholar
|
|
78
|
Nishiyama M, Tsunedomi R, Yoshimura K,
Hashimoto N, Matsukuma S, Ogihara H, Kanekiyo S, Iida M, Sakamoto
K, Suzuki N, et al: Metastatic ability and the
epithelial-mesenchymal transition in induced cancer stem-like
hepatoma cells. Cancer Sci. 109:1101–1109. 2018. View Article : Google Scholar
|
|
79
|
Derynck R and Weinberg RA: EMT and cancer:
More than meets the eye. Dev Cell. 49:313–316. 2019. View Article : Google Scholar
|
|
80
|
Gui X, Li Y, Zhang X, Su K and Cao W:
Circ_LDLR promoted the development of papillary thyroid carcinoma
via regulating miR-195-5p/LIPH axis. Cancer Cell Int. 20:2412020.
View Article : Google Scholar
|
|
81
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar
|
|
82
|
Zhang W, Liu T, Li T and Zhao X:
Hsa_circRNA_102002 facilitates metastasis of papillary thyroid
cancer through regulating miR-488-3p/HAS2 axis. Cancer Gene Ther.
28:279–293. 2021. View Article : Google Scholar
|
|
83
|
Han JY, Guo S, Wei N, Xue R, Li W, Dong G,
Li J, Tian X, Chen C, Qiu S, et al: ciRS-7 promotes the
proliferation and migration of papillary thyroid cancer by
negatively regulating the miR-7/Epidermal growth factor receptor
axis. Biomed Res Int. 2020:98756362020. View Article : Google Scholar
|
|
84
|
Wang H, Yan X, Zhang H and Zhan X: CircRNA
circ_0067934 overexpression correlates with poor prognosis and
promotes thyroid carcinoma progression. Med Sci Monit.
25:1342–1349. 2019. View Article : Google Scholar
|
|
85
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar
|
|
86
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar
|
|
87
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.
|
|
88
|
Lunt SY and Vander Heiden MG: Aerobic
glycolysis: Meeting the metabolic requirements of cell
proliferation. Annu Rev Cell Dev Biol. 27:441–464. 2011. View Article : Google Scholar
|
|
89
|
Dang CV, Hamaker M, Sun P, Le A and Gao P:
Therapeutic targeting of cancer cell metabolism. J Mol Med (Berl).
89:205–212. 2011. View Article : Google Scholar
|
|
90
|
Ren H, Song Z, Chao C and Mao W:
circCCDC66 promotes thyroid cancer cell proliferation, migratory
and invasive abilities and glycolysis through the miR-211-5p/PDK4
axis. Oncol Lett. 21:4162021. View Article : Google Scholar
|
|
91
|
Li Y, Qin J, He Z, Cui G, Zhang K and Wu
B: Knockdown of circPUM1 impedes cell growth, metastasis and
glycolysis of papillary thyroid cancer via enhancing MAPK1
expression by serving as the sponge of miR-21-5p. Genes Genomics.
43:141–150. 2021. View Article : Google Scholar
|
|
92
|
Hu Z, Zhao P, Zhang K, Zang L, Liao H and
Ma W: Hsa_ circ_0011290 regulates proliferation, apoptosis and
glycolytic phenotype in papillary thyroid cancer via miR-1252/FSTL1
signal pathway. Arch Biochem Biophys. 685:1083532020. View Article : Google Scholar
|
|
93
|
Longley DB and Johnston PG: Molecular
mechanisms of drug resistance. J Pathol. 205:275–292. 2005.
View Article : Google Scholar
|
|
94
|
Shaili E: Platinum anticancer drugs and
photochemotherapeutic agents: Recent advances and future
developments. Sci Prog. 97:20–40. 2014. View Article : Google Scholar
|
|
95
|
Hundahl SA, Fleming ID, Fremgen AM and
Menck HR: A national cancer data base report on 53-856 cases of
thyroid carcinoma treated in the U.S., 1985-1995[see commetns].
Cancer. 83:2638–2648. 1998. View Article : Google Scholar
|
|
96
|
Kitamura Y, Shimizu K, Nagahama M, Sugino
K, Ozaki O, Mimura T, Ito K, Ito K and Tanaka S: Immediate causes
of death in thyroid carcinoma: Clinicopathological analysis of 161
fatal cases. J Clin Endocrinol Metab. 84:4043–4049. 1999.
View Article : Google Scholar
|
|
97
|
Antonelli A, Miccoli P, Derzhitski VE,
Panasiuk G, Solovieva N and Baschieri L: Epidemiologic and clinical
evaluation of thyroid cancer in children from the Gomel region
(Belarus). World J Surg. 20:867–871. 1996. View Article : Google Scholar
|
|
98
|
Zheng X, Cui D, Xu S, Brabant G and
Derwahl M: Doxorubicin fails to eradicate cancer stem cells derived
from anaplastic thyroid carcinoma cells: Characterization of
resistant cells. Int J Oncol. 37:307–315. 2010.
|
|
99
|
Liu F, Zhang J, Qin L, Yang Z, Xiong J,
Zhang Y, Li R, Li S, Wang H, Yu B, et al: Circular RNA EIF6
(Hsa_circ_0060060) sponges miR-144-3p to promote the
cisplatin-resistance of human thyroid carcinoma cells by autophagy
regulation. Aging (Albany NY). 10:3806–3820. 2018. View Article : Google Scholar
|
|
100
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar
|
|
101
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar
|
|
102
|
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W
and Zhang RP: Circulating circular RNA hsa_circ_0001785 acts as a
diagnostic biomarker for breast cancer detection. Clin Chim Acta.
487:363–368. 2018. View Article : Google Scholar
|
|
103
|
Chen Y, Wei S, Wang X, Zhu X and Han S:
Progress in research on the role of circular RNAs in lung cancer.
World J Surg Oncol. 16:2152018. View Article : Google Scholar
|
|
104
|
Wei J, Wei W, Xu H, Wang Z, Gao W, Wang T,
Zheng Q, Shu Y and De W: Circular RNA hsa_circRNA_102958 may serve
as a diagnostic marker for gastric cancer. Cancer Biomark.
27:139–145. 2020. View Article : Google Scholar
|
|
105
|
Lan X, Cao J, Xu J, Chen C, Zheng C, Wang
J, Zhu X, Zhu X and Ge M: Decreased expression of hsa_circ_0137287
predicts aggressive clinicopathologic characteristics in papillary
thyroid carcinoma. J Clin Lab Anal. 32:e225732018. View Article : Google Scholar
|
|
106
|
Ren H, Liu Z, Liu S, Zhou X, Wang H, Xu J,
Wang D and Yuan G: Profile and clinical implication of circular
RNAs in human papillary thyroid carcinoma. PeerJ. 6:e53632018.
View Article : Google Scholar
|
|
107
|
Shi E, Ye J, Zhang R, Ye S, Zhang S, Wang
Y, Cao Y and Dai W: A combination of circRNAs as a diagnostic tool
for discrimination of papillary thyroid cancer. Onco Targets Ther.
13:4365–4372. 2020. View Article : Google Scholar
|
|
108
|
Pegtel DM and Gould SJ: Exosomes. Annu Rev
Biochem. 88:487–514. 2019. View Article : Google Scholar
|
|
109
|
van der Pol E, Böing AN, Harrison P, Sturk
A and Nieuwland R: Classification, functions, and clinical
relevance of extracellular vesicles. Pharmacol Rev. 64:676–705.
2012. View Article : Google Scholar
|
|
110
|
Yang C, Wei Y, Yu L and Xiao Y:
Identification of altered circular RNA expression in serum exosomes
from patients with papillary thyroid carcinoma by high-throughput
sequencing. Med Sci Monit. 25:2785–2791. 2019. View Article : Google Scholar
|
|
111
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar
|
|
112
|
Chen D, Ma W, Ke Z and Xie F: CircRNA
hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung
cancer progression. Cell Cycle. 17:2080–2090. 2018. View Article : Google Scholar
|
|
113
|
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M,
Liu X, Sun H, Pan Y and Wang S: The pro-metastasis effect of
circANKS1B in breast cancer. Mol Cancer. 17:1602018. View Article : Google Scholar
|
|
114
|
Sun JW, Qiu S, Yang JY, Chen X and Li HX:
Hsa_circ_0124055 and hsa_circ_0101622 regulate proliferation and
apoptosis in thyroid cancer and serve as prognostic and diagnostic
indicators. Eur Rev Med Pharmacol Sci. 24:4348–4360. 2020.
|
|
115
|
Rüster B, Göttig S, Ludwig RJ, Bistrian R,
Müller S, Seifried E, Gille J and Henschler R: Mesenchymal stem
cells display coordinated rolling and adhesion behavior on
endothelial cells. Blood. 108:3938–3944. 2006. View Article : Google Scholar
|
|
116
|
De Becker A and Riet IV: Homing and
migration of mesenchymal stromal cells: How to improve the efficacy
of cell therapy? World J Stem Cells. 8:73–87. 2016. View Article : Google Scholar
|
|
117
|
Nitzsche F, Müller C, Lukomska B,
Jolkkonen J, Deten A and Boltze J: Concise review: MSC adhesion
cascade-insights into homing and transendothelial migration. Stem
Cells. 35:1446–1460. 2017. View Article : Google Scholar
|
|
118
|
Toh WS, Lai RC, Zhang B and Lim SK: MSC
exosome works through a protein-based mechanism of action. Biochem
Soc Trans. 46:843–853. 2018. View Article : Google Scholar
|
|
119
|
Keshtkar S, Azarpira N and Ghahremani MH:
Mesenchymal stem cell-derived extracellular vesicles: Novel
frontiers in regenerative medicine. Stem Cell Res Ther. 9:632018.
View Article : Google Scholar
|
|
120
|
Kalimuthu S, Oh JM, Gangadaran P, Zhu L,
Lee HW, Jeon YH, Jeong SY, Lee SW, Lee J and Ahn BC: Genetically
engineered suicide gene in mesenchymal stem cells using a Tet-On
system for anaplastic thyroid cancer. PLoS One. 12:e01813182017.
View Article : Google Scholar
|
|
121
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar
|
|
122
|
Tang M, Wang Q, Wang K and Wang F:
Mesenchymal stem cells-originated exosomal microRNA-152 impairs
proliferation, invasion and migration of thyroid carcinoma cells by
interacting with DPP4. J Endocrinol Invest. 43:1787–1796. 2020.
View Article : Google Scholar
|
|
123
|
Zhang C, Cao J, Lv W and Mou H:
CircRNA_100395 carried by exosomes from adipose-derived mesenchymal
stem cells inhibits the malignant transformation of non-small cell
lung carcinoma through the miR-141-3p-LATS2 axis. Front Cell Dev
Biol. 9:6631472021. View Article : Google Scholar
|
|
124
|
Yan B, Zhang Y, Liang C, Liu B, Ding F,
Wang Y, Zhu B, Zhao R, Yu XY and Li Y: Stem cell-derived exosomes
prevent pyroptosis and repair ischemic muscle injury through a
novel exosome/circHIPK3/FOXO3a pathway. Theranostics. 10:6728–6742.
2020. View Article : Google Scholar
|
|
125
|
Zhu M, Liu X, Li W and Wang L: Exosomes
derived from mmu_circ_0000623-modified ADSCs prevent liver fibrosis
via activating autophagy. Hum Exp Toxicol. 39:1619–1627. 2020.
View Article : Google Scholar
|
|
126
|
Kamerkar S, LeBleu VS, Sugimoto H, Yang S,
Ruivo CF, Melo SA, Lee JJ and Kalluri R: Exosomes facilitate
therapeutic targeting of oncogenic KRAS in pancreatic cancer.
Nature. 546:498–503. 2017. View Article : Google Scholar
|
|
127
|
Zarovni N, Corrado A, Guazzi P, Zocco D,
Lari E, Radano G, Muhhina J, Fondelli C, Gavrilova J and Chiesi A:
Integrated isolation and quantitative analysis of exosome shuttled
proteins and nucleic acids using immunocapture approaches. Methods.
87:46–58. 2015. View Article : Google Scholar
|
|
128
|
Yu LL, Zhu J, Liu JX, Jiang F, Ni WK, Qu
LS, Ni RZ, Lu CH and Xiao MB: A comparison of traditional and novel
methods for the separation of exosomes from human samples. Biomed
Res Int. 2018:36345632018. View Article : Google Scholar
|