Introduction
Mesenchymal stem cells (MSCs) are mesoderm-derived
adult SCs that possess multidirectional differentiation potential
(1). Although bone marrow (BM)
is the primary site for MSC isolation, MSCs are also located in the
perivascular space of various types of tissue expressing CD146,
including dental pulp, adipose tissue, neonatal placenta, amniotic
membrane and umbilical cord (2).
Our previous study isolated tonsil-derived MSCs (T-MSCs) (3) and showed their various lineage
differentiation potentials and immune modulatory effects (4). Compared with MSCs of other origins,
T-MSCs possess distinctive features, such as CD106 (vascular cell
adhesion protein 1), CD166 (5)
and CD274 [programmed death ligand-1 (PD-L1)] (6) expression.
MSCs migrate to injured tissue and secrete a range
of paracrine factors that induce regeneration in damaged tissue.
MSCs contribute to tissue repair primarily via paracrine factors
and stimulation of target cells, but not by replacement of injured
tissue (7). MSCs mediate
fibrosis [keratinocyte growth factor, hepatocyte growth factor
(HGF), vascular endothelial (VE GF, Angiopoietin-1, stromal
cell-derived factor-1, insulin-like (I GF-1, epidermal (E)GF, nerve
GF and transforming (T)GF-α], angiogenesis [angiogenin, VEGF,
tissue inhibitor of metalloproteinases 1 (TIMP-1), TIMP-2 and
matrix metalloproteinase (MMP)-1], immune modulation (IL-10, IL-13,
IFN-γ, IL-12, prostaglandin E2, indoleamine 2,3-dioxygenase, heme
oxygenase-1 and galectins), chemotaxis [C-C motif chemokine ligand
(CCL)5, C-X-C motif chemokine ligand (CXCL)12 and CCL8], apoptosis
[HGF, IGF-1, osteopontin, growth hormone) and proliferation (EGF,
TGF-α, HGF, bFGF, IGF binding protein and macrophage
colony-stimulating factor) through various factors (8). Consequently, culture medium
conditioned by MSCs produces therapeutic effects similar to those
observed in cell delivery studies using mouse models of acute
myocardial infarction (9) and
lung injury (10). The active
fraction of conditioned medium (CM) contains particles released
from the cells, collectively called extracellular vesicles (EVs)
(11). Cells produce EVs,
including both microvesicles (>200 nm) and exosomes (50-200 nm),
via intracellular vesicle sorting processes. EVs do not contain
functional nuclei and are only surrounded by lipid bilayer. EV are
secreted by endothelial, immune and smooth muscle cells and
platelets (11). Common MSC EV
markers include CD9, CD63, CD81, tumor susceptibility gene 101
(TSG101) protein and heat shock protein 70 (12,13). MSC-derived EVs act on target
cells by transferring mRNAs, microRNAs (miRNAs or miRs), lipids and
proteins, which alter the activity of target cells (14).
Cross-talk between the tumor and tumor
microenvironment may be key for tumor growth and development
(15). MSCs may contribute to
tumor development by migrating to inflammatory or cancer sites and
evolve into tumor-associated MSCs and fibroblasts, thereby
activating cell proliferation, invasion, angiogenesis and
metastasis (16). This
interaction between tumor cells and MSCs is primarily mediated by
EVs (17). MSCs also produce EVs
that mediate information transfer in the tumor microenvironment;
for example, BM-MSC-derived exosomes induce apoptosis and cell
cycle arrest in HepG2 cells (18). Although the effect of MSC-derived
EVs on tumor development and progression is still unknown, the use
of MSC-derived EVs as cancer modulators is preferable to using the
cellular form of MSCs due to the small size and homogeneity of EVs
(19). The present study aimed
to identify the potential effect of T-MSC-derived EVs on tumor
development using the human liver cancer cell line HepG2.
Materials and methods
Cell culture and CM collection
HepG2 cells were purchased from American Type
Culture Collection and additional STR profiling was performed
(Fig. S1). T-MSCs previously
obtained from patients (Ewha University Medical Center
Institutional Review Board; approval no. EUMC 2018-01-011-002) at
Ewha Womans University Seoul Hospital (Seoul, Korea) were
maintained as previously described (20). Patients provided informed written
consent for the use of their tissue.
Phenotype analysis
To analyze cell phenotype, T-MSCs were washed with
FACS buffer (0.5 FBS and 0.1% NaN3 in PBS), blocked with
0.5 µg/ml purified rat anti-mouse CD16/CD32 (BD Pharmingen)
at 4°C for 5 min and stained with FITC anti-CD11b (cat. no. ICRF44,
mouse IgG1, κ), FITC anti-CD45 (2D1, mouse
IgG1, κ), FITC anti-CD73 (AD2, mouse IgG1,
κ), FITC anti-CD90 (5E10, mouse IgG1, κ), FITC
anti-CD105 (43A3, mouse IgG1, κ) and FITC mouse
IgG1, κ isotype (MOPC-21) antibodies (all BioLegend,
Inc.) at 0.5 µg/ml for 20 min at room temperature. After
staining, the cells were fixed with 4% paraformaldehyde (PFA,
Sigma-Aldrich; Merck KGaA) in phosphate-buffered saline (PBS) to
final 0.5% PFA. Stained cells were acquired using a Novocyte flow
cytometer (ACEA Bioscience, Inc.). Acquired cells were analyzed by
FlowJo software (v10, TreeStar, Inc.).
Adipogenic differentiation
For adipogenic differentiation, T-MSCs were seeded
at a density of 1×104 cells/well in a 12-well plate and
cultured with adipogenic medium (Invitrogen; Thermo Fisher
Scientific, Inc.) in a humid atmosphere with 5% CO2 at
37°C for 3 weeks. Medium was replaced every 3-4 days. After 3
weeks, adipogenic cultures of T-MSCs were rinsed with PBS and fixed
in 4% PFA for 5 min at room temperature. The wells were dried
completely and stained with Oil red O (Sigma-Aldrich; Merck KGaA)
for 10 min at room temperature. The Oil red O solution was removed
and wells were immediately washed with distilled water four times.
Wharton's jelly-derived (WJ-)MSCs were purchased from PromoCell
GmbH. Ethics approval was received for the use of primary cells
(Ewha Institutional Biosafety Committee; approval no.
IBC-past-096). HepG2 cells were cultured in Minimum Essential
Medium Eagle (MEM; Welgene, Inc.) with 10 FBS (Welgene, Inc.) and
1% penicillin/streptomycin solution (P/S; Capricorn Scientific
GmbH) in a humid atmosphere with 5% CO2 at 37°C. T-MSCs
and WJ-MSCs were cultured in low-glucose Dulbecco's modified Eagle
medium (DMEM; Welgene, Inc.) with 10 FBS (Welgene, Inc.) and 1% P/S
in 100-mm cell culture plates. For preparation of T-MSC CM (T-CM),
T-MSCs at 80% confluence were washed four times with PBS (Welgene,
Inc.) and medium was replaced with serum-free DMEM. The medium was
collected after 48 h, centrifuged at 190 x g for 5 min at room
temperature, passed through a 0.2-µm filter (MilliporeSigma)
and concentrated 20-fold using a 3-kDa Amicon Ultra centrifugal
filter unit (EMD Millipore) with high-speed centrifugation (Sorvall
LYNX4000; Thermo Fisher Scientific, Inc.) at 5,000 x g for 1 h at
4°C. T-CM for animal experiments was frozen, whereas T-CM and CM
from WJ-MSCs for exosome isolation and reverse
transcription-quantitative (RT-q)PCR were used immediately.
Isolation of T-MSC EVs
For EV isolation, Minimal Information for Studies of
EVs (MISEV) 2018 guidelines proposed by the International Society
for EVs (ISEV) were referred to for separation and characterization
(11).
To isolate EVs from T-MSCs, 1/5 volume of
ExoQuick-TC reagent (System Biosciences) was added to 10 ml T-CM
and mixed by vigorous inverting. Following incubation at 4°C
overnight, the mixture was centrifuged at 1,500 x g for 30 min at
4°C. The supernatant was removed, and final centrifugation at 1,500
x g was performed for 5 min at room temperature. The visible
EV-containing pellet was resuspended in 100-500 µl PBS for
Nanosight particle tracking analysis (Nanosight NS300; Malvern
Instruments, Ltd.) and for protein concentration analysis via
bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific,
Inc.). Following BCA assay, 6X SDS-PAGE loading buffer was added
for sample preparation for immunoblotting and stored at −80°C until
use.
Transmission electron microscopy
(TEM)
Purified EVs were diluted to 1:1,000 in PBS. A total
of 5 µl diluted EVs was dropped on Formvar-carbon-coated EM
grids. The grids were stained with 2% uranyl acetate for 2 min at
room temperature and removed using filter paper. Finally, the grids
were viewed using a H-7650 TEM (Hitachi, Ltd.) at 80 kV. Digital
images were captured at a magnification of 70,000-200,000.
Scanning EM (SEM)
Diluted EVs were dropped on Poly L-lysine cover
glass and prefixed with 0.25% glutaraldehyde for 30 min at room
temperature. After washing in PBS, samples were maintained in 1%
osmium tetroxide for 30 min for final fixation at room temperature.
Then, samples were washed and dehydrated by serial dilution with
ethanol and critical point drying. Finally, samples were mounted
onto stubs, sputter-coated with gold by Quorum Technologies and
examined with a Sigma-300 microscope (Zeiss GmbH) at a
magnification of ×70,000.
Animal experiments
Male BALB/c nude mice (n=6, age, 5 weeks; weight,
18±2 g) were purchased from OrientBio. All animals were housed at
21-23°C and 51-54% humidity in a pathogen-free environment on a
12/12-h light/dark cycle and allowed free access to food and water.
All animals were monitored every day for health and behavior. The
method of euthanasia was carbon dioxide inhalation followed by
cervical dislocation (20% vol/min for a cage size of 8×13×5 inches)
and animal death was confirmed by cardiac and respiratory arrest.
The experimental procedures were approved by the Animal Care and
Use Committee of the College of Medicine, Ewha Womans University
(Seoul, Korea; approval no. EUM18-0408). To construct a hepatoma
xenograft model, 3×106 HepG2 cells were suspended in 100
µl low-glucose DMEM and injected subcutaneously into the
right back of each animal (n=3) (21). To assess the effect of T-CM,
HepG2 cells were suspended in 100 µl CM from
5×105 T-MSCs and injected at the same position (n=3).
Five days after the injection, mice were euthanized by carbon
dioxide and cervical dislocation and the tumor was isolated,
chopped and seeded onto a 100-mm cell culture plate. Images were
captured following 2 and 9 days of culturing in a humid atmosphere
with 5% CO2 at 37°C using an inverted light microscope
(Olympus Corporation) at ×100 magnification, and cell clusters were
calculated manually by counting cluster of >50 cells.
Immunoblotting
Equal concentrations of EV (5 µg/lane) from
T-MSCs were loaded onto 5% stacking/10% separating sodium dodecyl
sulfate (SDS)-PAGE, separated by electrophoresis, transferred to
polyvinylidene difluoride (PVDF) membranes, blocked with 5% skimmed
milk in TBST (50 mM Tris HCl, pH 7.6, 150 mM NaCl, 0.1% Tween-20)
for 1 h at room temperature and incubated with primary antibodies
overnight at 4°C. Gels were stained with Coomassie blue solution
(0.1 Coomassie brilliant blue R-250, 40.0 methanol and 10.0% acetic
acid in water) overnight at room temperature followed by incubation
with de-staining solution (40 methanol and 10% acetic acid in
water) for 2 h at room temperature. All primary antibodies were
prepared by diluting in 3.00 BSA (Bovogen Biologicals Pty, Ltd.)
and 0.02% sodium azide (Sigma-Aldrich; Merck KGaA) in TBST.
Anti-CD63 mouse monoclonal antibody (cat. no. ab193349; 1:1,000;
IgG1κ) was purchased from Abcam; anti-CD81 (cat. no.
sc-166029; 1:200; IgG2bκ) and anti-β-actin mouse
monoclonal antibody (cat. no. sc-47778; 1:3,000; IgG1κ)
were purchased from Santa Cruz Biotechnology, Inc. PVDF membranes
were washed three times for 10 min each in TBST and incubated with
horseradish peroxidase-conjugated goat anti-mouse IgG (H+L)
antibody (cat. no. #1706516; BioRad Laboratories, Inc.), diluted in
TBST (1:4,000), for 1 h at room temperature. Following incubation,
the membranes were washed three times for 10 min each in TBST and
developed using an EZ-Western Lumi Femto Western blot kit (Doo Gene
Bio Co., Ltd.). Images were obtained using ImageQuant LAS 3000
(FUJIFILM Wako Pure Chemical Corporation). The pixel densities of
protein bands were analyzed using UN-SCAN-IT-gel 6.1 software (Silk
Scientific, Inc.).
Cell transfection
METAFECTENE PRO (Biontex Laboratories GmbH) was used
according to manufacturer's protocol to transfect cells. Briefly,
mixed solutions containing 1 µg AccuTarget™
fluorescein-labeled miRNA negative control, inhibitor #1 (cat. no.
SMC-4101, Bioneer, bioneer.co.kr/20-smc-4101.html) or
has-miR-199a-3p inhibitor (Bioneer) were added to 50 µl
serum-free medium. Then, 3 µl METAFECTENE PRO was added to
50 µl serum-free medium at room temperature for 20 min. The
sequence of has-miR-199a-3p inhibitor was 5′-ACA GUA GUC UGC ACA
UUG GUU A-3. ′ Following incubation, the mixture was carefully
added to cells. After 5 h, cells were collected for RT-qPCR and
wound healing assay was performed.
miRNA sequencing and target gene
prediction
CM from five different donor originated T-MSCs and
DMEM (negative control) were sent to Macrogen, Inc. for miRNA
sequencing by SMARTer smRNA-Seq method (22) using TruSeq Small RNA Library Prep
kit (RS-200-0012, Illumina, Inc.).Ribosomal RNA removed reads were
aligned to reference genome (miRBase v22.1) and non-coding RNA
database (RNAcentral 14.0) to classify known miRNA and other type
of RNA. Novel miRNA prediction is performed by miRDeep2 (v2.0.0.8).
To reduce systemic bias, size factors from the count data were
estimated and Trimmed Mean of M-values normalization with edgeR R
library was applied. Statistical analysis was performed using Fold
Change, exact-Test using edgeR per comparison pair. Hierarchical
clustering analysis was also performed using complete linkage and
Euclidean distance as a measure of similarity to display the
expression patterns of differentially expressed miRNAs that
satisfied the |fold-change|≥2 and P<0.05 criteria. All data
analysis and visualization of differentially expressed genes was
performed using R 3.3.1 (r-project.org). miRNA target gene prediction was
performed by TargetScanHuman 7.2 (targetscan.org/vert_72/). In order to analyze
signaling pathways associated with miRNA from exosomes,
miRNA-target genes were analyzed by Kyoto Encylcopedia of Genes and
Genomes (KEGG) analysis. Genes derived from mirDIP were further
analyzed by Database for Annotation, Visualization and Integrated
Discovery v6.8 to identify enriched biological pathways.
RT-qPCR
To isolate miRNA, T-CM and CM from WJ-MSCs was added
to an appropriate volume of Exo2D™ for RNA (ExosomePlus) according
to the manufacturer's protocol, and inverted several times. The
mixture was incubated at 4°C for 30 min and centrifuged at 3,000 x
g for 30 min at 4°C. The supernatant was removed and resuspended in
100 µl PBS and the RNA concentration was measured by
BioPhotometer D30 (Eppendorf) and adjusted to 1
µg/µl. miRNA was converted into complementary DNA
(cDNA) using MystiCq™ microRNA cDNA Synthesis Mix (Sigma-Aldrich;
Merck KGaA) according to the manufacturer's protocol. MystiCq
Universal PCR Primer (Sigma-Aldrich; Merck KGaA) was used as a
reverse primer for RT-qPCR. MystiCq microRNA qPCR Control Primer
RNU6-1 (cat. no. MIRCP00001) and MystiCq microRNA qPCR Assay Primer
hsa-miR-199a-3p (cat. no. MIRAP00244; both Sigma-Aldrich; Merck
KGaA) were used as forward primers. Total RNA was extracted using
an RNeasy Plus Mini kit (Qiagen GmbH) to isolate RNA of transfected
HepG2 cells. Total RNA (1 µg) was transcribed into cDNA
using RT reagent (ElpisBiotech, Inc.), and RT-qPCR was performed.
Primer sequences were as follows: CD151 forward, 5′-ATG GGT GAG TTC
AAC GAG AAG A-3′ and reverse, 5′-GCA GGC TGA TGT AGT CAC TCT -3′;
integrin α3 (ITGA3) forward, 5′-TGT GGC TTG GAG TGA CTG TG-3′ and
reverse, 5′-TCA TTG CCT CGC ACG TAG C-3′; ITGA6 forward, 5′-ATG CAC
GCG GAT CGA GTT T-3′ and reverse, 5′-TTC CTG CTT CGT ATT AAC ATG
CT-3′ and human GAPDH (192 bp) forward, 5′-GGT AAA GTG GAT ATT GTT
GCC ATC AAT G-3′and reverse, 5′-GGA GGG ATC TCG CTC CTG GAA GAT GGT
G-3′. The mixture was prepared in each well of a Fast Optical
96-well reaction plate (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using 0.5 reverse/forward primer each, 10.0 1X
SensiFAST SYBR Hi-ROX mix (Bioline; Meridian Bioscience, Inc.), 8.0
deionized water and 1.0 µl cDNA. Amplification was performed
in triplicate with 40 cycles of 15 sec denaturation at 95°C, 1 min
annealing at 62°C and 30 sec extension at 72°C using StepOnePlus
Real-Time RCR System (Applied Biosystems; Thermo Fischer
Scientific, Inc.). The relative fold expression and changes were
calculated using the 2−∆∆Cq method (23).
Wound healing assay
A scratch wound healing assay was performed to
compare the effect of T-CM or exosomes on the migration capability
of HepG2. HepG2 cells (2×105) were seeded onto a 12-well
cell culture plate with MEM containing 10% FBS and 1% P/S and
incubated to 90-100% confluence at 37°C in a 5% CO2
incubator. When the confluence of cells was reached, a scratch
wound was made in the cell monolayers using a cut cell scraper. The
transfected cells were allowed to grow for an additional 24 and 48
h in the presence of CM or EVs (10 µg/ml) with serum-free
MEM. Images were taken at 0, 24 and 48 h using an inverted light
microscope (Olympus Corporation; ×40 magnification). The migration
distance was quantified using ImageJ software v1.53 g (imagej.nih.gov). The percentage of area closure was
calculated as follows: Final wound width/initial wound width
×100.
Statistical analysis
Data are presented as the mean ± SEM (n>3).
Statistical significance was determined by one-way ANOVA with
multiple comparison by Sidak test applied to the wound healing
assay with T-CM or miRNA inhibitors. Paired t-test was used for
wound area closure after T-CM treatment. Student t-test (unpaired)
was used for the comparison of miR expression between WJ-MSCs and
T-MSCs. All analyses were performed using Prism 9.2 (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
T-CM suppresses tumor formation and
migration of HepG2 cells
Investigation of the effect of subcutaneous
injection of CM of T-MSCs at the right back of mice showed that
T-CM with HepG2 cells formed a smaller tumor mass than injection of
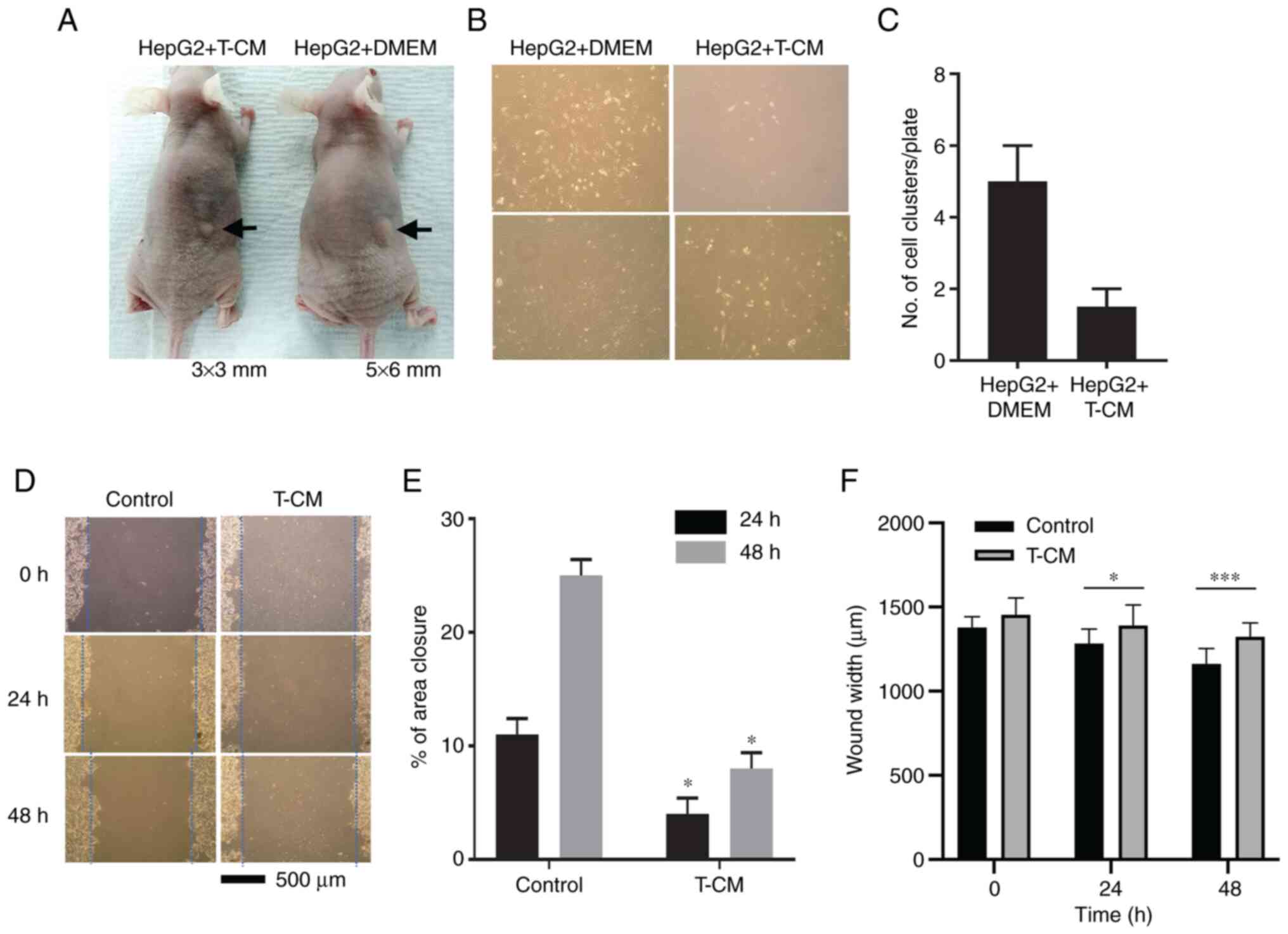
DMEM with HepG2 cells after 9 days (Fig. 1A). At 5 days after the injection,
no mice had died and the plated cells from tumor masses were
separated, chopped and seeded for culture. Formation of cell
clusters was assessed at 2 and 9 days. The cell clusters in the
HepG2 + T-CM group tumor mass were less prolific than those from
the control group (Fig. 1B).
Comparison of the number of clusters on culture plates (Fig. 1C) indicated that T-CM exerted an
inhibitory effect on HepG2 cell tumor growth and expansion in
vitro. The in vitro scratch assay compared the effect of
T-CM on the migration of HepG2 cells were obtained after 24 and 48
h (Fig. 1D); the distance
between the scratch of the control group (DMEM) was closer and the
area became smaller as time passed. By contrast, in the T-CM group,
wound area remained almost same as that at 0 h. These results
suggested that T-CM had an inhibitory effect on the migration of
HepG2 cells (Fig. 1E and F).
Isolation and characterization of T-MSC
exosomes
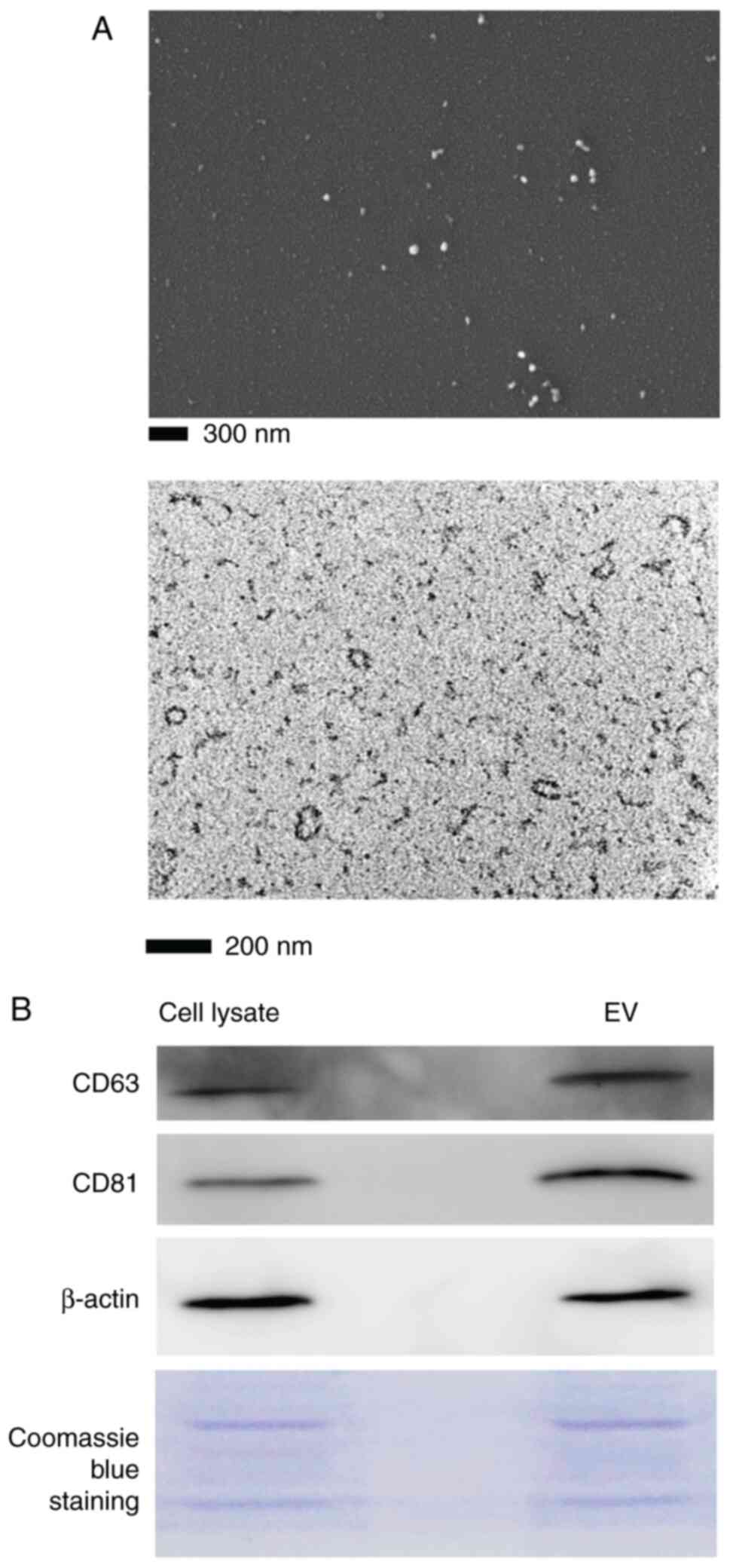
The morphology of T-MSC small EVs isolated from CM
and observed by TEM and SEM showed a round or oval shape with
diameter <200 nm (Fig. 2A).
Immunoblot analysis indicated positive expression of CD63 and CD81
(Fig. 2B). The particle size
distribution and concentration of isolated exosomes were analyzed
by Nanosight particle tracking analysis (Fig. S2). The mean diameter of
particles was 192.3±21.6 nm and the concentration was
1.28×108±2.38×107 particles/ml.
Exosomal miRNA hsa-miR-199a-3p suppresses
migration of HepG2 cells
To search the highly expressed miRNAs in T-MSC EVs,
five different origins of T-MSCs established in the lab were
selected. These five T-MSCs showed typical surface markers of MSCs
(Fig. S3A). For the comparison
of differentiation potential, adipogenic differentiation of these
five different T-MSCs was induced (Fig. S3B). After confirming these cells
possessed chracteristics of MSCs [plastic adherence, phenotype
marker expression (CD73+, CD90+,
CD105+, CD45− and CD11b−) and
adipocyte differentiation], miRNA sequening was performed.
Mycoplasma contamination was not detected in small RNA
composition report from Macrogen. The highly expressed miRNAs are
listed in Table I. A heatmap
(Fig. S4A) was constructed to
demonstrate the results of hierarchical clustering analysis
(Euclidean method, complete Linkage), which clusters the similarity
of mature miRNAs and samples by expression level (normalized value)
from a significant list. Fig.
S4B shows the number of up- and downregulated mature miRNAs
based on fold change compared with those of negative control. The
upregulated miRNAs with similar expression levels were grouped
together using the normalized value of each sample (Fig. S4C). Enriched signaling pathways
among top 20 miRNA-target genes were analyzed by KEGG analysis;
genes belonging to cancer pathway were highly enriched (Fig. S5). Exosomal RNA was extracted
from T-MSC and WJ-MSC CM. To validate miRNA sequencing,
hsa-miR-199a-3p, hsa-miR-214-3p, hsa-miR-199a-5p and
hsa-miR-199b-5p expression levels were compared with expression of
exosomal RNA from T-MSCs and WJ-MSCs by RT-qPCR. Expression of
miR-199a-3p, mir-214-3p, and miR-199-5p was higher in T-MSCs than
in WJ-MSCs (Fig. 3A). The
exosomes of T-MSCs had higher levels of hsa-miR-199a-3p than those
of WJ-MSCs, as indicated by RT-qPCR and miRNA sequencing (Fig. 3B; Table I). To identify the effect of
miR-199a-3p on HepG2 cell migration in vitro, transfected
cells were treated with negative control or miR-199a-3p inhibitor.
Treated cells were scratched and observed for 24 and 48 h following
the addition of 10 µg/ml exosomes (Fig. 3C). The migrated distance in the
control group (without exosomes) was greater and the wound area
became smaller as time passed. By contrast, the wound area in the
exosome-exposed group remained similar throughout the experiment.
miR-199a-3p inhibitor reversed the effect of exosomes on migration
(Fig. 3D). miR-199a-3p inhibitor
reversed the effect of exosomes on migration after 48 h (Fig. 3E).
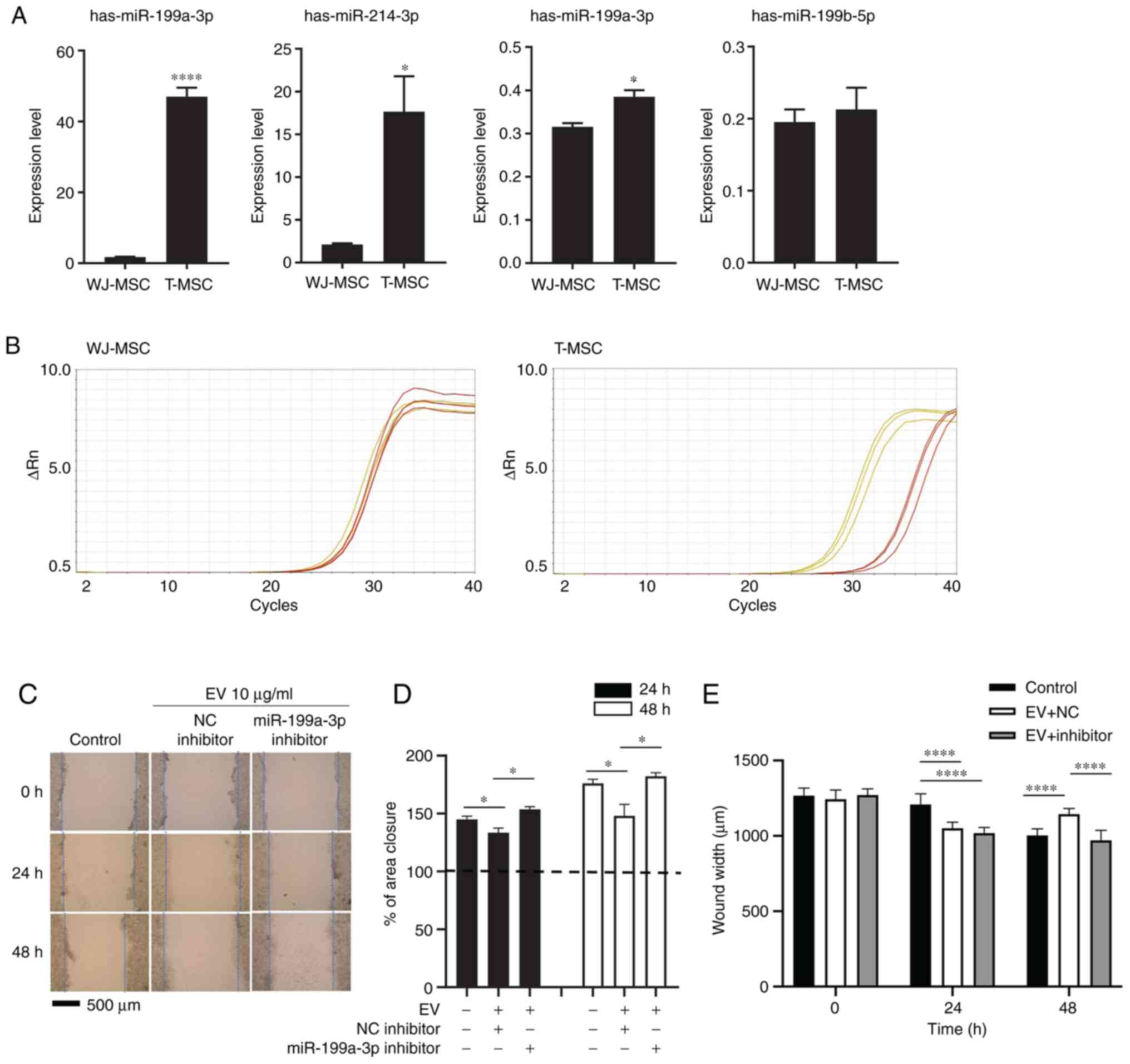 | Figure 3miR-199a-3p inhibitor attenuates the
effect of T-MSC EVs on scratch wound healing assay. (A) EV RNA from
WJ-MSCs and T-MSCs was extracted at 80% confluence and analyzed by
reverse transcription-qquantitative PCR. Data are presented as the
mean ± SEM and analyzed by Student t-test *P<0.05,
****P<0.0001. (B) Amplification plot. Yellow,
hsa-miR-199a-3p; red, control (RNU6-1). (C) Migration of HepG2
cells following transfection and addition of EVs with NC or
miR-199a-3p inhibitor. Magnification, ×40. (D) Closure area was
calculated after 24 or 48 h. A total of >10 fields of view were
analyzed and each experiment was repeated three times. Statistical
significance was determined by t test. (E) Wound width was measured
and distance was calculated (n=10 per field). Data are presented as
the mean ± SEM and analyzed by one-way ANOVA with multiple
comparison by Sidak test *P<0.05,
****P<0.0001 vs. WJ-MSC. Untreated, miRNA NC- and
miR-199a-3p inhibitor-treated HepG2 cells were used as the control,
NC and inhibitor, respectively. miR, microRNA; T-MSC,
tonsil-derived mesenchymal stem cell; EV, extracellular vesicle;
WJ, Wharton's jelly; NC, negative control. |
 | Table ITop 30 miRs highly expressed in
tonsil-derived mesenchymal stem cell extracellular vesicles. |
Table I
Top 30 miRs highly expressed in
tonsil-derived mesenchymal stem cell extracellular vesicles.
| Mature ID | Fold-change |
|---|
|
hsa-miR-199a-3p | 3609 |
| hsa-miR-145-5p | 3433 |
| hsa-miR-24-3p | 2612 |
| hsa-miR-214-3p | 2603 |
| hsa-let-7b-3p | 2448 |
|
hsa-miR-125a-5p | 2196 |
|
hsa-miR-125b-5p | 2068 |
| hsa-miR-29b-3p | 2020 |
| hsa-miR-19b-3p | 1902 |
| hsa-miR-424-5p | 1779 |
| hsa-let-7a-3p | 1648 |
| hsa-miR-29a-3p | 1575 |
|
hsa-miR-151a-3p | 1517 |
| hsa-let-7i-5p | 1449 |
| hsa-miR-126-5p | 1390 |
|
hsa-miR-199a-5p | 1220 |
|
hsa-miR-376c-3p | 1213 |
| hsa-miR-30a-3p | 1060 |
| hsa-miR-19a-3p | 1039 |
| hsa-miR-143-3p | 915 |
|
hsa-let-7f-1-3p | 913 |
|
hsa-miR-130a-3p | 887 |
| hsa-miR-30e-3p | 794 |
|
hsa-miR-199b-5p | 705 |
| hsa-miR-409-3p | 669 |
| hsa-miR-92b-3p | 618 |
| hsa-miR-654-3p | 607 |
| hsa-miR-6126 | 603 |
| hsa-miR-98-5p | 584 |
| hsa-miR-483-3p | 567 |
Exosomal microRNA hsa-miR-199a-3p
suppresses expression of target genes in HepG2 cells
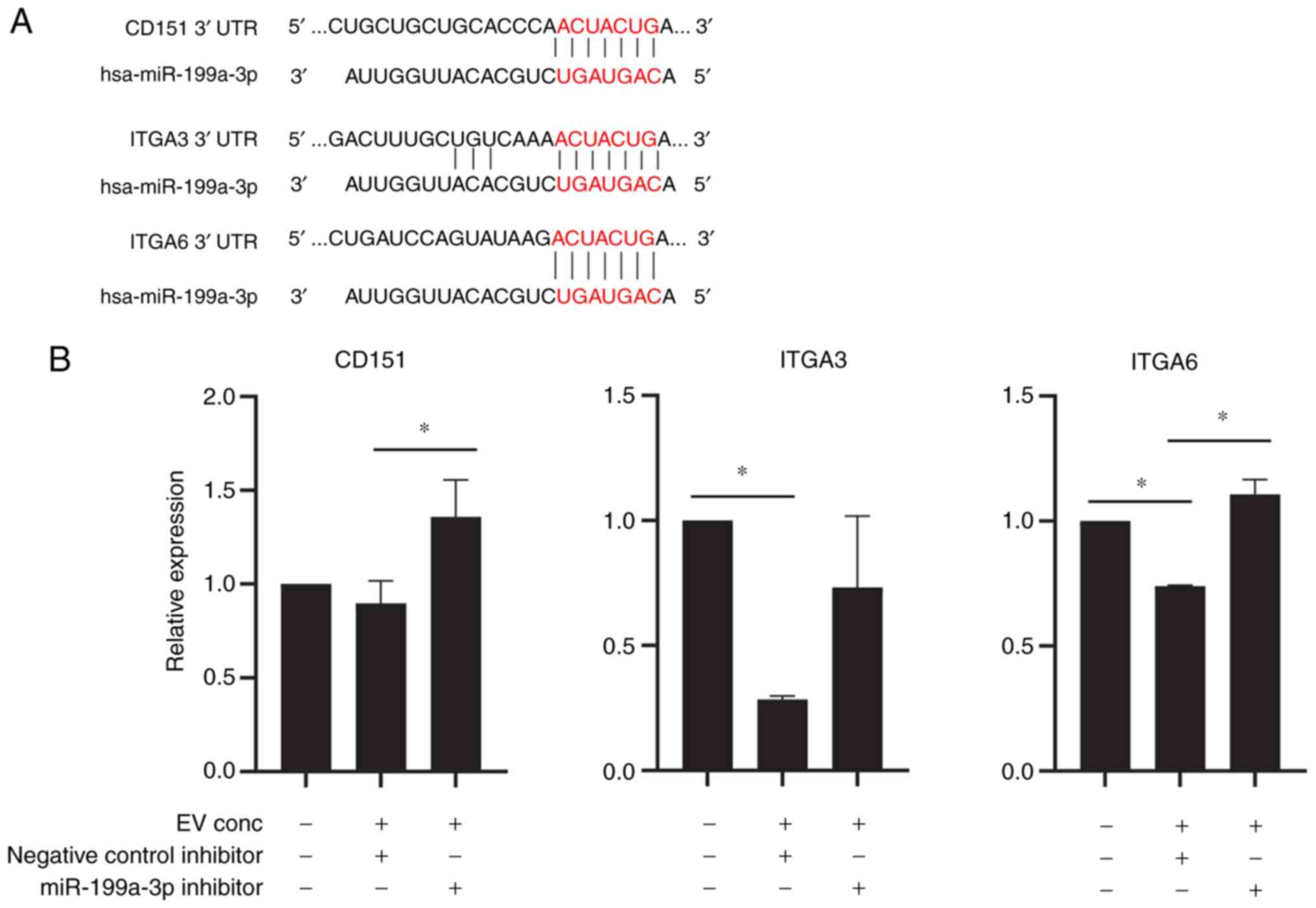
To investigate the molecular mechanism of how
miR-199a-3p affects HepG2 cells, miR-199a-3p targets were predicted
using the TargetScanHuman 7.2 database (targetscan.org/vert_72/). Hsa-miR-199a-3p was
predicted to target Kelch-like family member 3, serpin family E
member 2, pro-apoptotic WT1 regulator, vesicle-associated membrane
protein 3, G protein subunit α12, BCAR3, CDK7, CD2-associated
protein, CD151, FGF7, CXCL11, ITGB8, G3BP stress granule assembly
factor 2, CDK17, mesenteric estrogen-dependent adipogenesis,
collagen (COL) type IV α5 chain, RAB GTPase-activating protein 1,
IL13RA1, ITGA3, SP1, TAO kinase 1, ITGA6, COL12A1 and histamine
N-methyltransferase. The seed sequence of hsa-miR-199a-3p matched
the sequence of the 3′ untranslated regions of CD151, ITGA3 and
ITGA6, suggesting that CD151, ITGA3 and ITGA6 were potential
targets of miR-199a-3p (Fig.
4A). To confirm the effect of miR-199a-3p on T-MSCs, HepG2
cells were transfected with miR-199a-3p inhibitor or negative
control inhibitor. Transfection efficiency of miR-199a-3p inhibitor
was confirmed by the 87% decrease of the miR-199a-3p expression
(Fig. S6). Expression of CD151,
ITGA3 and ITGA6 in HepG2 cells was downregulated by EV treatment
(Fig. 4B).
Discussion
Our previous studies reported several novel immune
modulatory effects of T-MSCs and T-CM. For examples, tumor necrosis
factor α inducible gene 6 protein expression in T-CM attenuates the
acute graft-versus-host reaction (24); EBV-induced gene 3 stimulates
regulatory B cells (25); and
PD-L1 inhibits Th17-mediated autoimmune or skin inflammatory
responses (6). Because all of
these disease models for the immune regulatory effect of T-MSCs or
T-CM are inflammatory conditions, the effect of T-CM on tumor
growth was assessed in an animal model; T-MSCs inhibited tumor
growth and HepG2 cell migration, potentially via miRNA containing
EVs, in particular, miR-199a-3p.
EVs are nanometer-sized particles and mostly below
the detection limit of conventional analysis methods, such as
ultracentrifugation, filtration or precipitation. Therefore, to
meet minimal experimental requirement, ISEV proposed MISEV
guidelines for EV studies. EV are <100 or 200 nm (small EVs) or
>200 nm (medium/large). EVs can be derived from tissue culture
CM, biofluid or tissue. EV separation and concentration methods
includes centrifugation, density gradient, chromatography,
precipitation, filtration and antibody-based. For EV
characterization, quantification (protein amount, particle number,
lipid amount), global characterization (transmembrane or
glycosylphosphatidylinositol-anchored protein or expected
contaminants) and single EV characterization can be image-(electron
microscopy) or non-image-based (nanoparticle tracking analysis,
flow cytometry or Raman spectroscopy) (11). Functional studies can be
quantitative comparison of activity of total fluid, EV-depleted
fluid and EVs (11,26).
MSC regeneration is a secretome-based paracrine
effect, and the use of MSC CM has become more common as a strategy
to discover novel therapeutic targets. However, the heterogeneity
of the MSC population promotes the use of its refined form, EVs,
instead of CM or MSCs. Protein- and miRNA-enriched MSC-derived EV
serve a role in maintaining homeostasis as stromal cells as well as
in response to stimuli, such as injury or disease state.
MSC-derived EVs serve a role in immune regulation, angiogenesis,
proliferation and other processes (27). For immune regulation, MSC-derived
EVs deliver anti-inflammatory cytokine IL-10 and anti-inflammatory
miRNAs (miRNA-21, miRNA-146a, miRNA-181c, miRNA-124a and
miRNA-125b) (28).
Pro-angiogenic miRNAs delivered by MSC-derived EVs include
miRNA-126, miRNA-130a, miRNA-125a (inhibits angiogenic inhibitor
Δ-like 4) and miRNA-31 [suppresses hypoxia-inducible factor
(HIF)-1] (29). MSC-derived EVs
also deliver mRNA from the PI3K/AKT/endothelial (eNOS pathway and
trophic factors FGF1, VEGFA, VEGFR2, IL8, angiopoietin 1,
E-selectin, CXCL16 and eNOS (30). MSC-derived EVs induce β-catenin
activation but decrease MMP-9 mRNA in target cells (31). The targets of MSC-derived EV
miRNAs are associated with cell death and growth and fibrosis via
Wnt signaling, platelet-derived GF and TGF-β (32).
Previous reports indicated MSC-derived EVs both
promote and inhibit tumor progression (33,34). The miRNAs or non-coding RNAs that
are involved in cancer promotion include miR-1587
(glioma-associated MSCs), miRNA-21 and miRNA34a (blood and breast
cancer MSCs), miR-221 (gastric cancer tissue-derived MSCs) and
LINC00461 (multiple myeloma BM-MSCs) (35). Anti-tumor miRNAs from MSC-derived
EVs include: miRNA-145 (from adipose tissue-derived MSCs in
prostate cancer), miR-124 and miR-145 (from glioma cells), miR-100
(suppresses in vitro angiogenesis via mTOR/HIF-1α/VEGF in
breast cancer), miR-23b and miR-222/223 (promote dormancy in breast
cancer) (36,37). Therefore, T-MSC EVs may regulate
cancer progression as indicated by highly enriched cancer pathway
in KEGG analysis of miRNA target genes (Fig. S5).
miRNA-199a-3p was the most highly expressed miRNA in
EVs isolated from T-MSCs and was more highly expressed in EVs from
T-MSCs compared with those from WJ-MSCs. miRNA-199a-3p was also
highly expressed in BM-MSC-derived EVs and has been found to
inhibit cardiomyocyte apoptosis (32). Another report found the potential
target of miR-199a-3p is ITGB8, which enhances cisplatin
sensitivity in ovarian cancer (38). Because ITGs may affect cell
growth and motility, CD151, ITGA3, and ITGA6 were selected from
miRNA-199a-3p target genes for further investigation.
Tetraspanins do not have enzymatic activity or a
canonical signaling pathway, but organize cytokine receptors,
adhesion receptors and proteases (39). Tetraspanin CD151 is normally
expressed in endothelial cells and platelets and overexpressed in
cancer cells (40) and involved
in membrane fusion, trafficking, cell motility and tumor
development. CD151 interacts with MMP-14, cadherins, immunoglobulin
proteins, other tetraspanins and ITGA3 and α6 (39). CD151-targeted monoclonal antibody
1A5 inhibits tumor cell motility and metastasis (41). CD151 association with ITGA6ß1
activates angiogenesis signaling (PI3K, Akt and NOS) and the
invasion pathway (JNK, JUN and MMP-9). In addition, CD151 binds to
α3ß1 and causes invasion and cytoskeletal reorganization (PKC, cell
division cycle 42 and actin) (40). Although loss of CD151 decreases
migration on laminin (42),
ITG-free tetraspanin CD151 clustering is a strong regulator of
motility in the absence of α3 expression but requires PKCα
(43). Disruption of CD151-ITGA3
complex inhibits migration and invasion of lung adenocarcinoma
cells in vitro via the FAK/p130Cas signaling pathway
(44).
The present results suggested that
miR-199a-3p-containing EVs from T-MSCs may exert a suppressive
effect on HepG2 cell motility. Because EVs possess membrane
structures that fuse cell membranes of target cells, EVs or
modified EVs may enhance the therapeutic effect and cancer
targeting (45) of isolated
T-MSC-derived or miR-199a-3p-containing EVs, resulting in a tumor
suppressive effect.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DWC and KAC performed experiments and wrote the
manuscript. JK performed the experiments. HJL, YHK and JWP analyzed
the data and wrote the manuscript. SYW designed the experiments and
wrote the manuscript. KAC and SYW confirm the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
The experimental procedures were approved by the
Animal Care and Use Committee of the College of Medicine, Ewha
Womans University (approval no. EUM19-446) and by the Institutional
Review Board of the Ewha University Medical Center (approval no.
EUMC 2018-01-011-002).
Patent consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Caplan AI: Mesenchymal stem cells. J
Orthop Res. 9:641–650. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crisan M, Yap S, Casteilla L, Chen CW,
Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, et al: A
perivascular origin for mesenchymal stem cells in multiple human
organs. Cell Stem Cell. 3:301–313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ryu KH, Cho KA, Park HS, Kim JY, Woo SY,
Jo I, Choi YH, Park YM, Jung SC, Chung SM, et al: Tonsil-derived
mesenchymal stromal cells: Evaluation of biologic, immunologic and
genetic factors for successful banking. Cytotherapy. 14:1193–1202.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oh SY, Choi YM, Kim HY, Park YS, Jung SC,
Park JW, Woo SY, Ryu KH, Kim HS and Jo I: Application of
tonsil-derived mesenchymal stem cells in tissue regeneration:
Concise review. Stem Cells. 37:1252–1260. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho KA, Park M, Kim YH, Woo SY and Ryu KH:
RNA sequencing reveals a transcriptomic portrait of human
mesenchymal stem cells from bone marrow, adipose tissue, and
palatine tonsils. Sci Rep. 7:171142017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim JY, Park M, Kim YH, Ryu KH, Lee KH,
Cho KA and Woo SY: Tonsil-derived mesenchymal stem cells (T-MSCs)
prevent Th17-mediated autoimmune response via regulation of the
programmed death-1/programmed death ligand-1 (PD-1/PD-L1) pathway.
J Tissue Eng Regen Med. 12:e1022–e1033. 2018. View Article : Google Scholar
|
|
7
|
Caplan AI and Correa D: The MSC: An injury
drugstore. Cell Stem Cell. 9:11–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Andrzejewska A, Lukomska B and Janowski M:
Concise review: Mesenchymal stem cells: From roots to boost. Stem
Cells. 37:855–864. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gnecchi M, He H, Liang OD, Melo LG,
Morello F, Mu H, Noiseux N, Zhang L, Pratt RE, Ingwall JS and Dzau
VJ: Paracrine action accounts for marked protection of ischemic
heart by Akt-modified mesenchymal stem cells. Nat Med. 11:367–368.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goolaerts A, Pellan-Randrianarison N,
Larghero J, Vanneaux V, Uzunhan Y, Gille T, Dard N, Planès C,
Matthay MA and Clerici C: Conditioned media from mesenchymal
stromal cells restore sodium transport and preserve epithelial
permeability in an in vitro model of acute alveolar injury. Am J
Physiol Lung Cell Mol Physiol. 306:L975–L985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Théry C, Witwer KW, Aikawa E, Alcaraz MJ,
Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F,
Atkin-Smith GK, et al: Minimal information for studies of
extra-cellular vesicles 2018 (MISEV2018): A position statement of
the international society for extracellular vesicles and update of
the MISEV2014 guidelines. J Extracell Vesicles. 7:15357502018.
View Article : Google Scholar
|
|
12
|
Nakamura Y, Miyaki S, Ishitobi H,
Matsuyama S, Nakasa T, Kamei N, Akimoto T, Higashi Y and Ochi M:
Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle
regeneration. FEBS Lett. 589:1257–1265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Asgarpour K, Shojaei Z, Amiri F, Ai J,
Mahjoubin-Tehran M, Ghasemi F, ArefNezhad R, Hamblin MR and Mirzaei
H: Exosomal microRNAs derived from mesenchymal stem cells:
Cell-to-cell messages. Cell Commun Signal. 18:1492020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mathivanan S, Fahner CJ, Reid GE and
Simpson RJ: ExoCarta 2012: Database of exosomal proteins, RNA and
lipids. Nucleic Acids Res. 40(Database Issue): D1241–D1244. 2012.
View Article : Google Scholar :
|
|
15
|
Whiteside TL: The tumor microenvironment
and its role in promoting tumor growth. Oncogene. 27:5904–5912.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Balkwill FR, Capasso M and Hagemann T: The
tumor microenvironment at a glance. J Cell Sci. 125:5591–5596.
2012. View Article : Google Scholar
|
|
17
|
Keshtkar S, Azarpira N and Ghahremani MH:
Mesenchymal stem cell-derived extracellular vesicles: Novel
frontiers in regenerative medicine. Stem Cell Res Ther. 9:632018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bruno S, Collino F, Deregibus MC, Grange
C, Tetta C and Camussi G: Microvesicles derived from human bone
marrow mesenchymal stem cells inhibit tumor growth. Stem Cells Dev.
22:758–771. 2013. View Article : Google Scholar
|
|
19
|
Alzahrani FA, El-Magd MA,
Abdelfattah-Hassan A, Saleh AA, Saadeldin IM, El-Shetry ES, Badawy
AA and Alkarim S: Potential effect of exosomes derived from cancer
stem cells and MSCs on progression of DEN-induced HCC in rats. Stem
Cells Int. 2018:80589792018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim YH, Cho KA, Lee HJ, Park M, Shin SJ,
Park JW, Woo SY and Ryu KH: Conditioned medium from human
tonsil-derived mesenchymal stem cells enhances bone marrow
engraftment via endothelial cell restoration by pleiotrophin.
Cells. 9:2212020. View Article : Google Scholar :
|
|
21
|
He XX, Chang Y, Meng FY, Wang MY, Xie QH,
Tang F, Li PY, Song YH and Lin JS: MicroRNA-375 targets AEG-1 in
hepatocellular carcinoma and suppresses liver cancer cell growth in
vitro and in vivo. Oncogene. 31:3357–3369. 2012. View Article : Google Scholar
|
|
22
|
Ramsköld D, Luo S, Wang YC, Li R, Deng Q,
Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et
al: Full-length mRNA-Seq from single-cell levels of RNA and
individual circulating tumor cells. Nat Biotechnol. 30:777–782.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Cho KA, Kim YH, Park M, Kim HJ, Woo SY,
Park JW and Ryu KH: Conditioned medium from human palatine tonsil
mesenchymal stem cells attenuates acute graft-vs-host disease in
mice. Mol Med Rep. 19:609–616. 2019.
|
|
25
|
Cho KA, Lee JK, Kim YH, Park M, Woo SY and
Ryu KH: Mesenchymal stem cells ameliorate B-cell-mediated immune
responses and increase IL-10-expressing regulatory B cells in an
EBI3-dependent manner. Cell Mol Immunol. 14:895–908. 2017.
View Article : Google Scholar :
|
|
26
|
Poupardin R, Wolf M and Strunk D:
Adherence to minimal experimental requirements for defining
extracellular vesicles and their functions. Adv Drug Deliv Rev.
176:1138722021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ferreira JR, Teixeira GQ, Santos SG,
Barbosa MA, Almeida-Porada G and Gonçalves RM: Mesenchymal stromal
cell secretome: Influencing therapeutic potential by cellular
pre-conditioning. Front Immunol. 9:28372018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tahamtan A, Teymoori-Rad M, Nakstad B and
Salimi V: Anti-inflammatory MicroRNAs and their potential for
inflammatory diseases treatment. Front Immunol. 9:13772018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suárez Y and Sessa WC: MicroRNAs as novel
regulators of angiogenesis. Circ Res. 104:442–454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gu H, Ji R, Zhang X, Wang M, Zhu W, Qian
H, Chen Y, Jiang P and Xu W: Exosomes derived from human
mesenchymal stem cells promote gastric cancer cell growth and
migration via the activation of the Akt pathway. Mol Med Rep.
14:3452–3458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu P, Zhang B, Shi H, Qian H and Xu W:
MSC-exosome: A novel cell-free therapy for cutaneous regeneration.
Cytotherapy. 20:291–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ferguson SW, Wang J, Lee CJ, Liu M,
Neelamegham S, Canty JM and Nguyen J: The microRNA regulatory
landscape of MSC-derived exosomes: A systems view. Sci Rep.
8:14192018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao R, Chen X, Song H, Bie Q and Zhang B:
Dual role of MSC-derived exosomes in tumor development. Stem Cells
Int. 2020:88447302020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Christodoulou I, Goulielmaki M, Devetzi M,
Panagiotidis M, Koliakos G and Zoumpourlis V: Mesenchymal stem
cells in preclinical cancer cytotherapy: A systematic review. Stem
Cell Res Ther. 9:3362018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vallabhaneni KC, Penfornis P, Dhule S,
Guillonneau F, Adams KV, Mo YY, Xu R, Liu Y, Watabe K, Vemuri MC
and Pochampally R: Extracellular vesicles from bone marrow
mesenchymal stem/stromal cells transport tumor regulatory microRNA,
proteins, and metabolites. Oncotarget. 6:4953–4967. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Takahara K, Ii M, Inamoto T, Nakagawa T,
Ibuki N, Yoshikawa Y, Tsujino T, Uchimoto T, Saito K, Takai T, et
al: microRNA-145 mediates the inhibitory effect of adipose
tissue-derived stromal cells on prostate cancer. Stem Cells Dev.
25:1290–1298. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cai QQ, Dong YW, Wang R, Qi B, Guo JX, Pan
J, Liu YY, Zhang CY and Wu XZ: MiR-124 inhibits the migration and
invasion of human hepatocellular carcinoma cells by suppressing
integrin αV expression. Sci Rep. 7:407332017. View Article : Google Scholar
|
|
38
|
Cui Y, Wu F, Tian D, Wang T, Lu T, Huang
X, Zhang P and Qin L: miR-199a-3p enhances cisplatin sensitivity of
ovarian cancer cells by targeting ITGB8. Oncol Rep. 39:1649–1657.
2018.PubMed/NCBI
|
|
39
|
Hemler ME: Tetraspanin functions and
associated microdomains. Nat Rev Mol Cell Biol. 6:801–811. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kumari S, Devi GV, Badana A, Dasari VR and
Malla RR: CD151-A striking marker for cancer therapy. Biomark
Cancer. 7:7–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zijlstra A, Lewis J, Degryse B, Stuhlmann
H and Quigley JP: The inhibition of tumor cell intravasation and
subsequent metastasis via regulation of in vivo tumor cell motility
by the tetraspanin CD151. Cancer Cell. 13:221–234. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Winterwood NE, Varzavand A, Meland MN,
Ashman LK and Stipp CS: A critical role for tetraspanin CD151 in
alpha3beta1 and alpha6beta4 integrin-dependent tumor cell functions
on laminin-5. Mol Biol Cell. 17:2707–2721. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zevian SC, Johnson JL, Winterwood NE,
Walters KS, Herndon ME, Henry MD and Stipp CS: CD151 promotes α3β1
integrin-dependent organization of carcinoma cell junctions and
restrains collective cell invasion. Cancer Biol Ther. 16:1626–1640.
2015. View Article : Google Scholar
|
|
44
|
Peng D, Li PC, Liu T, Zeng HS, Fei YJ, Liu
ZX and Zuo HJ: Key role of CD151-integrin complex in lung cancer
metastasis and mechanisms involved. Curr Med Sci. 40:1148–1155.
2020. View Article : Google Scholar
|
|
45
|
You B, Xu W and Zhang B: Engineering
exosomes: A new direction for anticancer treatment. Am J Cancer
Res. 8:1332–1342. 2018.PubMed/NCBI
|