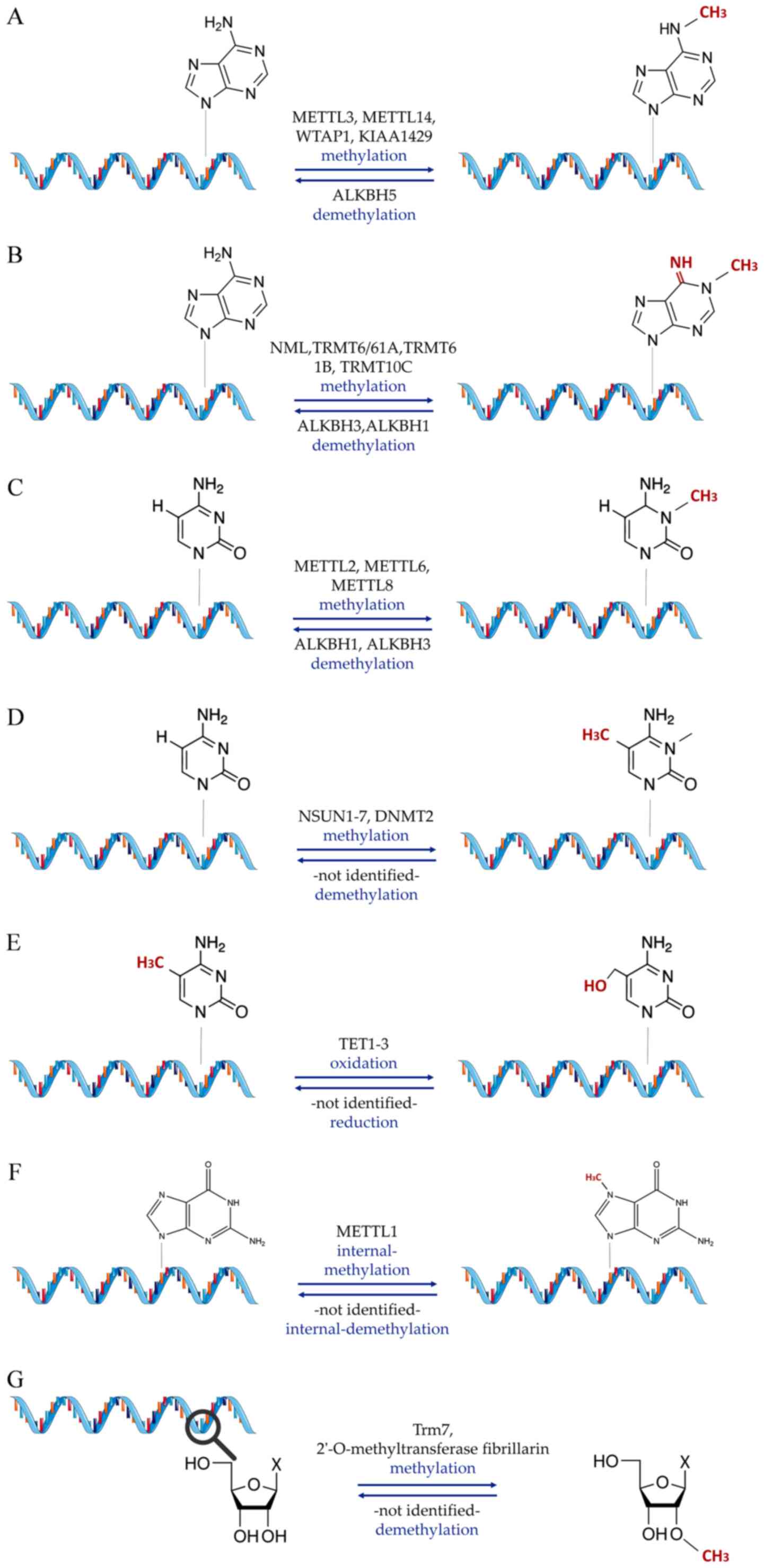
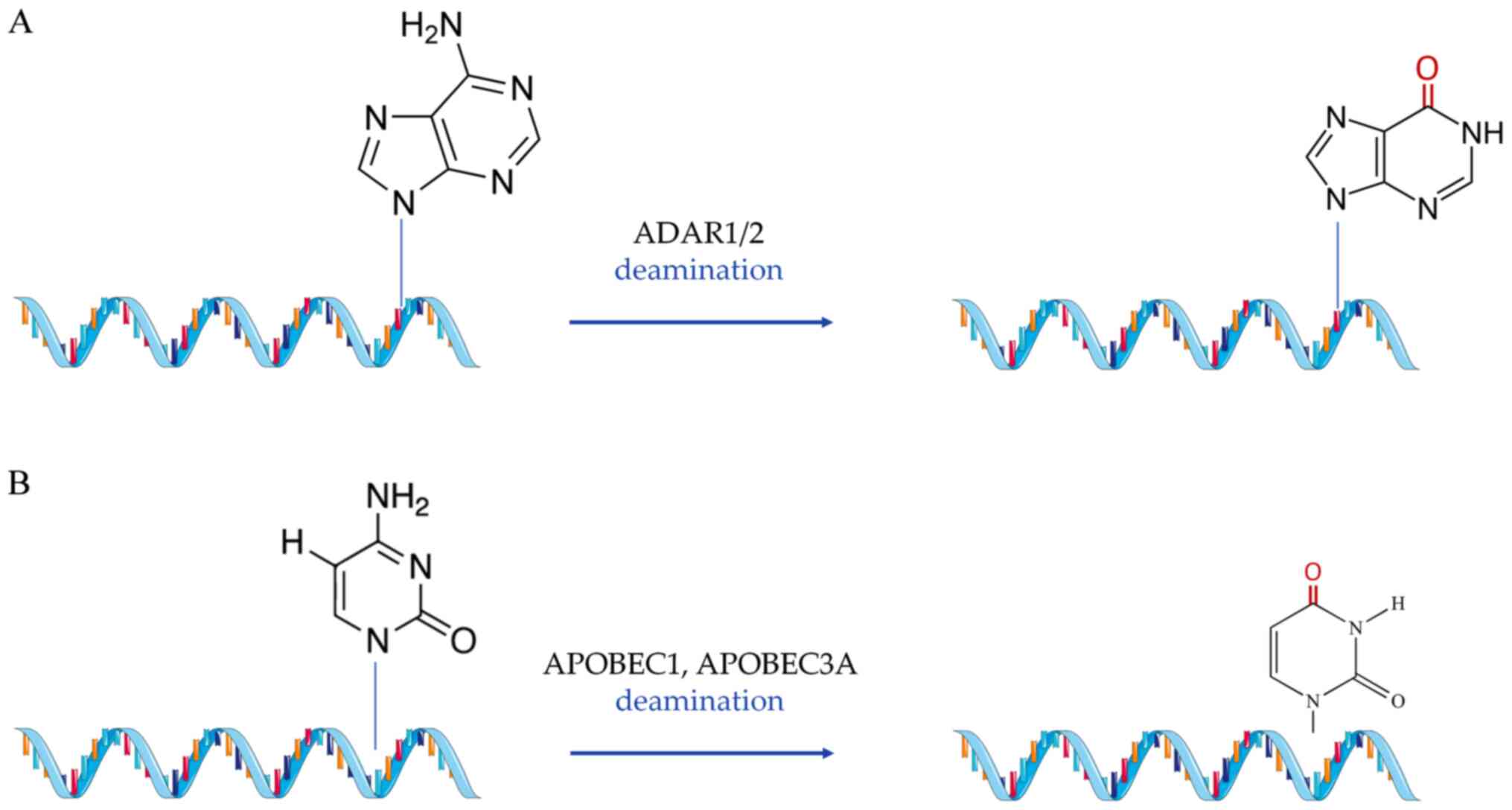
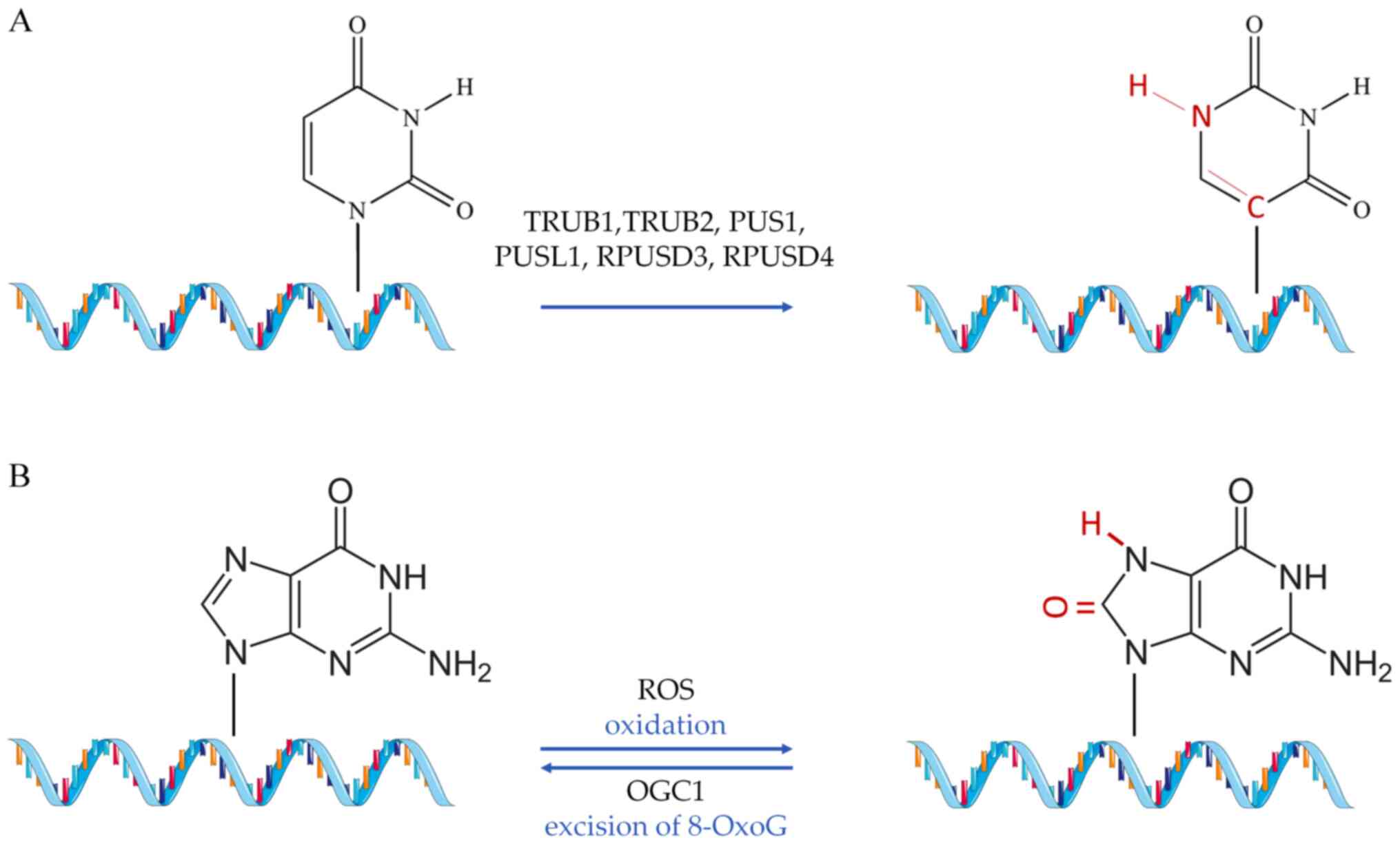
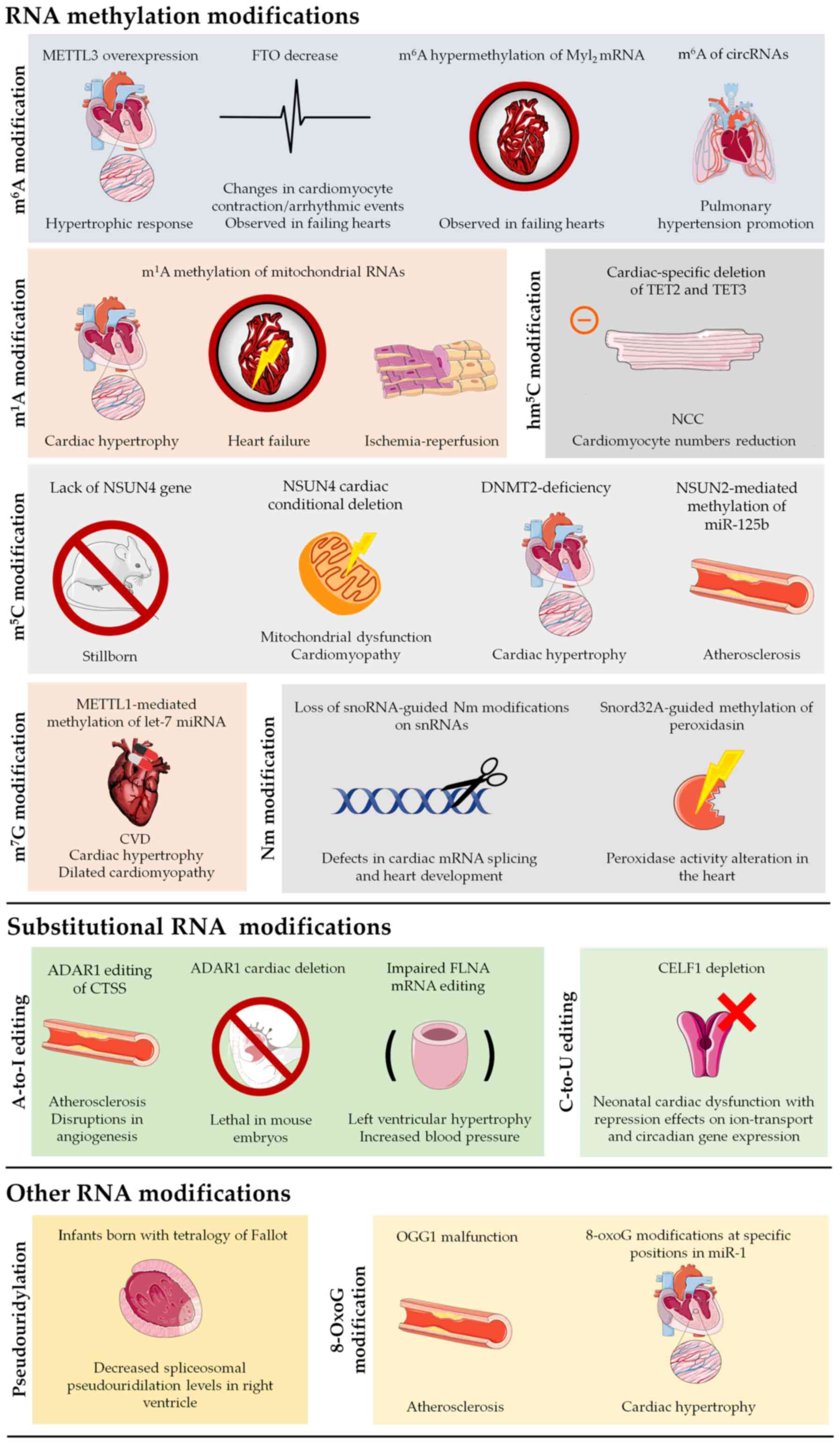
|
1
|
Manzoni C, Kia DA, Vandrovcova J, Hardy J,
Wood NW, Lewis PA and Ferrari R: Genome, transcriptome and
proteome: The rise of omics data and their integration in
biomedical sciences. Brief Bioinform. 19:286–302. 2018. View Article : Google Scholar :
|
|
2
|
Buriani A, Fortinguerra S, Sorrenti V,
Gabbia D and Carrara M: Single-cell omics in personalized medicine.
Single-Cell Omics. Barh D and Azevedo V: Academic Press; Cambridge,
MA: pp. 221–236. 2019, View Article : Google Scholar
|
|
3
|
Korenke GC, Fuchs S, Krasemann E, Doerr
HG, Wilichowski E, Hunneman DH and Hanefeld F: Cerebral
adrenoleukodystrophy (ALD) in only one of monozygotic twins with an
identical ALD genotype. Ann Neurol. 40:254–257. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cavalli G and Heard E: Advances in
epigenetics link genetics to the environment and disease. Nature.
571:489–499. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sarkies P: Molecular mechanisms of
epigenetic inheritance: Possible evolutionary implications. Semin
Cell Dev Biol. 97:106–115. 2020. View Article : Google Scholar :
|
|
6
|
Qureshi IA and Mehler MF: Epigenetic
mechanisms underlying nervous system diseases. Handb Clin Neurol.
147:43–58. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schwartz S: Cracking the epitranscriptome.
RNA. 22:169–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boccaletto P, Machnicka MA, Purta E,
Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R,
Limbach PA, Kotter A, et al: MODOMICS: A database of RNA
modification pathways. 2017 update. Nucleic Acids Res.
46:D303–D307. 2018.
|
|
9
|
Czerwoniec A, Dunin-Horkawicz S, Purta E,
Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H and Rother K:
MODOMICS: A database of RNA modification pathways. 2008 update.
Nucleic Acids Res. 37:D118–D121. 2009. View Article : Google Scholar :
|
|
10
|
Dunin-Horkawicz S, Czerwoniec A, Gajda MJ,
Feder M, Grosjean H and Bujnicki JM: MODOMICS: A database of RNA
modification pathways. Nucleic Acids Res. 34:D145–D149. 2006.
View Article : Google Scholar :
|
|
11
|
Machnicka MA, Milanowska K, Osman Oglou O,
Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S,
Dunin-Horkawicz S, Rother KM, et al: MODOMICS: A database of RNA
modification pathways–2013 update. Nucleic Acids Res. 41:D262–D267.
2013. View Article : Google Scholar
|
|
12
|
Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S,
Zheng LL, Qu LH and Yang JH: RMBase v2.0: Deciphering the map of
RNA modifications from epitranscriptome sequencing data. Nucleic
Acids Res. 46:D327–D334. 2018. View Article : Google Scholar
|
|
13
|
Kiran AM, O'Mahony JJ, Sanjeev K and
Baranov PV: Darned in 2013: Inclusion of model organisms and
linking with Wikipedia. Nucleic Acids Res. 41:D258–D261. 2013.
View Article : Google Scholar :
|
|
14
|
Cantara WA, Crain PF, Rozenski J,
McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D and Agris
PF: The RNA Modification Database, RNAMDB: 2011 update. Nucleic
Acids Res. 39:D195–D201. 2011. View Article : Google Scholar
|
|
15
|
Picardi E, D'Erchia AM, Lo Giudice C and
Pesole G: REDIportal: A comprehensive database of A-to-I RNA
editing events in humans. Nucleic Acids Res. 45:D750–D757. 2017.
View Article : Google Scholar
|
|
16
|
Schuebel K, Gitik M, Domschke K and
Goldman D: Making sense of epigenetics. Int J Neuropsychopharmacol.
19:pyw0582016. View Article : Google Scholar
|
|
17
|
Kumari R, Ranjan P, Suleiman ZG, Goswami
SK, Li J, Prasad R and Verma SK: mRNA modifications in
cardiovascular biology and disease: With a focus on m6A
modification. Cardiovasc Res. May 6–2021.Epub ahead of print.
View Article : Google Scholar
|
|
18
|
Chen YS, Ouyang XP, Yu XH, Novák P, Zhou
L, He P and Yin K: N6-adenosine methylation (m(6)A) RNA
modification: An emerging role in cardiovascular diseases. J
Cardiovasc Transl Res. Feb 25–2021.Epub ahead of print. View Article : Google Scholar
|
|
19
|
Napoli C, Benincasa G, Donatelli F and
Ambrosio G: Precision medicine in distinct heart failure
phenotypes: Focus on clinical epigenetics. Am Heart J. 224:113–128.
2020. View Article : Google Scholar
|
|
20
|
Fischer MA and Vondriska TM: Clinical
epigenomics for cardiovascular disease: Diagnostics and therapies.
J Mol Cell Cardiol. 154:97–105. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schiano C, Benincasa G, Franzese M, Della
Mura N, Pane K, Salvatore M and Napoli C: Epigenetic-sensitive
pathways in personalized therapy of major cardiovascular diseases.
Pharmacol Ther. 210:1075142020. View Article : Google Scholar
|
|
22
|
Vlachakis C, Dragoumani K, Raftopoulou S,
Mantaiou M, Papageorgiou L, Champeris Tsaniras S, Megalooikonomou V
and Vlachakis D: Human emotions on the onset of cardiovascular and
small vessel related diseases. In Vivo. 32:859–870. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Timmis A, Townsend N, Gale CP, Torbica A,
Lettino M, Petersen SE, Mossialos EA, Maggioni AP, Kazakiewicz D,
May HT, et al: European Society of Cardiology: European Society of
Cardiology: Cardiovascular Disease Statistics 2019. Eur Heart J.
41:12–85. 2020. View Article : Google Scholar
|
|
24
|
Flora GD and Nayak MK: A brief review of
cardiovascular diseases, associated risk factors and current
treatment regimes. Curr Pharm Des. 25:4063–4084. 2019. View Article : Google Scholar
|
|
25
|
Baccarelli A, Rienstra M and Benjamin EJ:
Cardiovascular epigenetics: Basic concepts and results from animal
and human studies. Circ Cardiovasc Genet. 3:567–573. 2010.
View Article : Google Scholar
|
|
26
|
Zhong J, Agha G and Baccarelli AA: The
role of DNA methylation in cardiovascular risk and disease:
Methodological aspects, study design, and data analysis for
epidemiological studies. Circ Res. 118:119–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vlachakis D, Zacharaki EI, Tsiamaki E,
Koulouri M, Raftopoulou S, Papageorgiou L, Chrousos GP, Ellul J and
Megalooikonomou V: Insights into the molecular mechanisms of stress
and inflammation in ageing and frailty of the elderly. J Mol
Biochem. 6:41–44. 2017.
|
|
28
|
Guerra J, Valadao AL, Vlachakis D, Polak
K, Vila IK, Taffoni C, Prabakaran T, Marriott AS, Kaczmarek R,
Houel A, et al: Lysyl-tRNA synthetase produces diadenosine
tetraphosphate to curb STING-dependent inflammation. Sci Adv.
6:eaax33332020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soler-Botija C, Gálvez-Montón C and
Bayés-Genís A: Epigenetic biomarkers in cardiovascular diseases.
Front Genet. 10:950. 2019. View Article : Google Scholar
|
|
30
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and eras. Mol Cell. 74:640–650. 2019. View Article : Google Scholar
|
|
31
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gatsiou A and Stellos K: Dawn of
epitranscriptomic medicine. Circ Genom Precis Med. 11:e0019272018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anreiter I, Mir Q, Simpson JT, Janga SC
and Soller M: New twists in detecting mRNA modification dynamics.
Trends Biotechnol. 39:72–89. 2021. View Article : Google Scholar
|
|
34
|
Mongelli A, Atlante S, Bachetti T,
Martelli F, Farsetti A and Gaetano C: Epigenetic signaling and RNA
regulation in cardiovascular diseases. Int J Mol Sci. 21:5092020.
View Article : Google Scholar :
|
|
35
|
Zhang P, Wu W, Chen Q and Chen M:
Non-coding RNAs and their integrated networks. J Integr Bioinform.
16:162019. View Article : Google Scholar
|
|
36
|
Gomes CPC, Schroen B, Kuster GM, Robinson
EL, Ford K, Squire IB, Heymans S, Martelli F and Emanueli C:
Regulatory RNAs in heart failure. Circulation. 141:313–328. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Esteller M and Pandolfi PP: The
epitranscriptome of noncoding RNAs in cancer. Cancer Discov.
7:359–368. 2017. View Article : Google Scholar
|
|
38
|
Blow MJ, Grocock RJ, van Dongen S, Enright
AJ, Dicks E, Futreal PA, Wooster R and Stratton MR: RNA editing of
human microRNAs. Genome Biol. 7:R272006. View Article : Google Scholar :
|
|
39
|
Zhang Y, Zang Q, Xu B, Zheng W, Ban R,
Zhang H, Yang Y, Hao Q, Iqbal F, Li A, et al: IsomiR Bank: A
research resource for tracking IsomiRs. Bioinformatics.
32:2069–2071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Neilsen CT, Goodall GJ and Bracken CP:
IsomiRs - the overlooked repertoire in the dynamic microRNAome.
Trends Genet. 28:544–549. 2012. View Article : Google Scholar
|
|
41
|
Newman MA, Mani V and Hammond SM: Deep
sequencing of microRNA precursors reveals extensive 3′ end
modification. RNA. 17:1795–1803. 2011. View Article : Google Scholar :
|
|
42
|
Cloonan N, Wani S, Xu Q, Gu J, Lea K,
Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al:
MicroRNAs and their isomiRs function cooperatively to target common
biological pathways. Genome Biol. 12:R1262011. View Article : Google Scholar
|
|
43
|
Manzano M, Forte E, Raja AN, Schipma MJ
and Gottwein E: Divergent target recognition by coexpressed
5′-isomiRs of miR-142-3p and selective viral mimicry. RNA.
21:1606–1620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
van der Kwast RV, van Ingen E, Parma L,
Peters HA, Quax PH and Nossent AY: Adenosine-to-inosine editing of
MicroRNA-487b alters target gene selection after ischemia and
promotes Neovascularization. Circ Res. 122:444–456. 2018.
View Article : Google Scholar
|
|
45
|
van der Kwast RV, Woudenberg T, Quax PHA
and Nossent AY: MicroRNA-411 and its 5′-IsomiR have distinct
targets and functions and are differentially regulated in the
vasculature under ischemia. Mol Ther. 28:157–170. 2020. View Article : Google Scholar
|
|
46
|
Gilbert WV, Bell TA and Schaening C:
Messenger RNA modifications: Form, distribution, and function.
Science. 352:1408–1412. 2016. View Article : Google Scholar :
|
|
47
|
Yin L, Zhu X, Novák P, Zhou L, Gao L, Yang
M, Zhao G and Yin K: The epitranscriptome of long noncoding RNAs in
metabolic diseases. Clin Chim Acta. 515:80–89. 2021. View Article : Google Scholar
|
|
48
|
Zhou Y, Kong Y, Fan W, Tao T, Xiao Q, Li N
and Zhu X: Principles of RNA methylation and their implications for
biology and medicine. Biomed Pharmacother. 131:1107312020.
View Article : Google Scholar
|
|
49
|
Wulff BE and Nishikura K: Substitutional
A-to-I RNA editing. Wiley Interdiscip Rev RNA. 1:90–101. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Blanc V and Davidson NO: C-to-U RNA
editing: Mechanisms leading to genetic diversity. J Biol Chem.
278:1395–1398. 2003. View Article : Google Scholar
|
|
51
|
Adachi H, De Zoysa MD and Yu YT:
Post-transcriptional pseudouridylation in mRNA as well as in some
major types of noncoding RNAs. Biochim Biophys Acta Gene Regul
Mech. 1862:230–239. 2019. View Article : Google Scholar
|
|
52
|
Martinet W, De Meyer GR, Herman AG and
Kockx MM: RNA damage in human atherosclerosis: Pathophysiological
significance and implications for gene expression studies. RNA
Biol. 2:4–7. 2005. View Article : Google Scholar
|
|
53
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar
|
|
54
|
Perry RPK and Kelley DE: Existence of
methylated messenger RNA in mouse L cells. Cell. 1:37–42. 1974.
View Article : Google Scholar
|
|
55
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar
|
|
56
|
Molinie B, Wang J, Lim KS, Hillebrand R,
Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon
P, et al: m(6)A-LAIC-seq reveals the census and complexity of the
m(6)A epitranscriptome. Nat Methods. 13:692–698. 2016. View Article : Google Scholar
|
|
57
|
Sun H, Li K, Zhang X, Liu J, Zhang M, Meng
H and Yi C: m6Am-seq reveals the dynamic m6Am methylation in the
human transcriptome. Nat Commun. 12:47782021. View Article : Google Scholar
|
|
58
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
60
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)a promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
Modulates Messenger RNA Translation Efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar
|
|
62
|
Kierzek E and Kierzek R: The thermodynamic
stability of RNA duplexes and hairpins containing
N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic
Acids Res. 31:4472–4480. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N(6)-methyladenosine-dependent RNA structural switches
regulate RNA-protein interactions. Nature. 518:560–564. 2015.
View Article : Google Scholar :
|
|
64
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Balatsos NA, Vlachakis D, Maragozidis P,
Manta S, Anastasakis D, Kyritsis A, Vlassi M, Komiotis D and
Stathopoulos C: Competitive inhibition of human poly(A)-specific
ribonuclease (PARN) by synthetic fluoro-pyranosyl nucleosides.
Biochemistry. 48:6044–6051. 2009. View Article : Google Scholar
|
|
66
|
Balatsos N, Vlachakis D, Chatzigeorgiou V,
Manta S, Komiotis D, Vlassi M and Stathopoulos C: Kinetic and in
silico analysis of the slow-binding inhibition of human
poly(A)-specific ribonuclease (PARN) by novel nucleoside analogues.
Biochimie. 94:214–221. 2012. View Article : Google Scholar
|
|
67
|
Fustin JM, Doi M, Yamaguchi Y, Hida H,
Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I,
et al: RNA-methylation-dependent RNA processing controls the speed
of the circadian clock. Cell. 155:793–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xiang Y, Laurent B, Hsu CH, Nachtergaele
S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al: RNA m6A
methylation regulates the ultraviolet-induced DNA damage response.
Nature. 543:573–576. 2017. View Article : Google Scholar :
|
|
69
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
70
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification destabilizes
developmental regulators in embryonic stem cells. Nat Cell Biol.
16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar :
|
|
72
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar :
|
|
73
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Meyer KD and Jaffrey SR: Rethinking m6A
readers, writers, and eras. Annu Rev Cell Dev Biol. 33:319–342.
2017. View Article : Google Scholar
|
|
75
|
Longenecker JZ, Gilbert CJ, Golubeva VA,
Martens CR and Accornero F: Epitranscriptomics in the heart: A
focus on m6A. Curr Heart Fail Rep. 17:205–212. 2020. View Article : Google Scholar
|
|
76
|
Qin Y, Li L, Luo E, Hou J, Yan G, Wang D,
Qiao Y and Tang C: Role of m6A RNA methylation in cardiovascular
disease (Review). Int J Mol Med. 46:1958–1972. 2020. View Article : Google Scholar
|
|
77
|
Zheng N, Su J, Hu H, Wang J and Chen X:
Research progress of N6-methyladenosine in the cardiovascular
system. Med Sci Monit. 26. pp. e9217422020, View Article : Google Scholar
|
|
78
|
Dorn LE, Lasman L, Chen J, Xu X, Hund TJ,
Medvedovic M, Hanna JH, van Berlo JH and Accornero F: The
N6-methyladenosine mRNA methylase METTL3 controls cardiac
homeostasis and hypertrophy. Circulation. 139:533–545. 2019.
View Article : Google Scholar :
|
|
79
|
Song H, Pu J, Wang L, Wu L, Xiao J, Liu Q,
Chen J, Zhang M, Liu Y, Ni M, et al: ATG16L1 phosphorylation is
oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein
phosphatase 1 which determines the fate of cardiomyocytes during
hypoxia/reoxygenation. Autophagy. 11:1308–1325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Pastore N, Brady OA, Diab HI, Martina JA,
Sun L, Huynh T, Lim JA, Zare H, Raben N, Ballabio A, et al: TFEB
and TFE3 cooperate in the regulation of the innate immune response
in activated macrophages. Autophagy. 12:1240–1258. 2016. View Article : Google Scholar
|
|
81
|
Zhao E and Czaja MJ: Transcription factor
EB: A central regulator of both the autophagosome and lysosome.
Hepatology. 55:1632–1634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Song H, Feng X, Zhang H, Luo Y, Huang J,
Lin M, Jin J, Ding X, Wu S, Huang H, et al: METTL3 and ALKBH5
oppositely regulate m6A modification of TFEB mRNA, which dictates
the fate of hypoxia/reoxygenation-treated cardiomyocytes.
Autophagy. 15:1419–1437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kmietczyk V, Riechert E, Kalinski L,
Boileau E, Malovrh E, Malone B, Gorska A, Hofmann C, Varma E,
Jürgensen L, et al: m6A-mRNA methylation regulates cardiac gene
expression and cellular growth. Life Sci Alliance.
2:e2018002332019. View Article : Google Scholar :
|
|
84
|
Berulava T, Buchholz E, Elerdashvili V,
Pena T, Islam MR, Lbik D, Mohamed BA, Renner A, von Lewinski D,
Sacherer M, et al: Changes in m6A RNA methylation contribute to
heart failure progression by modulating translation. Eur J Heart
Fail. 22:54–66. 2020. View Article : Google Scholar
|
|
85
|
Fischer J, Koch L, Emmerling C, Vierkotten
J, Peters T, Brüning JC and Rüther U: Inactivation of the Fto gene
protects from obesity. Nature. 458:894–898. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-dependent N6-methyladenosine regulates
cardiac function during remodeling and repair. Circulation.
139:518–532. 2019. View Article : Google Scholar
|
|
87
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Shihong Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-dependent m6A regulates cardiac function
during remodeling and repair. Circulation. 139:518–532. 2019.
View Article : Google Scholar
|
|
88
|
Michlewski G and Cáceres JF:
Post-transcriptional control of miRNA biogenesis. RNA. 25:1–16.
2019. View Article : Google Scholar :
|
|
89
|
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH,
Wang F, Wang TT, Xu QG, Zhou WP and Sun SH: METTL14 suppresses the
metastatic potential of hepatocellular carcinoma by modulating
N6-methyladenosine-dependent primary MicroRNA processing.
Hepatology. 65:529–543. 2017. View Article : Google Scholar
|
|
90
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu
HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor
proliferation of bladder cancer by accelerating pri-miR221/222
maturation in m6A-dependent manner. Mol Cancer. 18:1102019.
View Article : Google Scholar :
|
|
91
|
Baarsma H and Königshoff M: 'WNT-er is
coming': WNT signalling in chronic lung diseases. Thorax.
72:746–759. 2017. View Article : Google Scholar
|
|
92
|
Savai R, Al-Tamari HM, Sedding D,
Kojonazarov B, Muecke C, Teske R, Capecchi MR, Weissmann N,
Grimminger F, Seeger W, et al: Pro-proliferative and inflammatory
signaling converge on FoxO1 transcription factor in pulmonary
hypertension. Nat Med. 20:1289–1300. 2014. View Article : Google Scholar
|
|
93
|
Lin B, Xu J, Wang F, Wang J, Zhao H and
Feng D: LncRNA XIST promotes myocardial infarction by regulating
FOS through targeting miR-101a-3p. Aging (Albany NY). 12:7232–7247.
2020. View Article : Google Scholar
|
|
94
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar
|
|
95
|
Sun R and Zhang L: Long non-coding RNA
MALAT1 regulates cardiomyocytes apoptosis after hypoxia/reperfusion
injury via modulating miR-200a-3p/PDCD4 axis. Biomed Pharmacother.
111:1036–1045. 2019. View Article : Google Scholar
|
|
96
|
Liu N, Parisien M, Dai Q, Zheng G, He C
and Pan T: Probing N6-methyladenosine RNA modification status at
single nucleotide resolution in mRNA and long noncoding RNA. RNA.
19:1848–1856. 2013. View Article : Google Scholar
|
|
97
|
Spitale RC, Flynn RA, Zhang QC, Crisalli
P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET,
et al: Erratum: Structural imprints in vivo decode RNA regulatory
mechanisms. Nature. 527:2642015. View Article : Google Scholar
|
|
98
|
Coker H, Wei G and Brockdorff N: m6A
modification of non-coding RNA and the control of mammalian gene
expression. Biochim Biophys Acta Gene Regul Mech. 1862:310–318.
2019. View Article : Google Scholar
|
|
99
|
Mendel M, Chen KM, Homolka D, Gos P,
Pandey RR, McCarthy AA and Pillai RS: Methylation of structured RNA
by the m6A writer METTL16 is essential for mouse embryonic
development. Mol Cell. 71:986–1000.e11. 2018. View Article : Google Scholar :
|
|
100
|
Brown JA, Kinzig CG, DeGregorio SJ and
Steitz JA: Methyltransferase-like protein 16 binds the 3′-terminal
triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA.
113:14013–14018. 2016. View Article : Google Scholar
|
|
101
|
Dunn DB: The occurrence of 1-methyladenine
in ribonucleic acid. Biochim Biophys Acta. 46:198–200. 1961.
View Article : Google Scholar
|
|
102
|
Agris PF: The importance of being
modified: Roles of modified nucleosides and Mg2+ in RNA
structure and function. Prog Nucleic Acid Res Mol Biol. 53:79–129.
1996. View Article : Google Scholar
|
|
103
|
Xiong X, Li X and Yi C: N1-methyladenosine
methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol.
45:179–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Waku T, Nakajima Y, Yokoyama W, Nomura N,
Kako K, Kobayashi A, Shimizu T and Fukamizu A: NML-mediated rRNA
base methylation links ribosomal subunit formation to cell
proliferation in a p53-dependent manner. J Cell Sci. 129:2382–2393.
2016.PubMed/NCBI
|
|
105
|
Chujo T and Suzuki T: Trmt61B is a
methyltransferase responsible for 1-methyladenosine at position 58
of human mitochondrial tRNAs. RNA. 18:2269–2276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Vilardo E, Nachbagauer C, Buzet A,
Taschner A, Holzmann J and Rossmanith W: A subcomplex of human
mitochondrial RNase P is a bifunctional methyltransferase -
extensive moon-lighting in mitochondrial tRNA biogenesis. Nucleic
Acids Res. 40:11583–11593. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ozanick S, Krecic A, Andersland J and
Anderson JT: The bipartite structure of the tRNA m1A58
methyltransferase from S. cerevisiae is conserved in humans. RNA.
11:1281–1290. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhang C and Jia G: Reversible RNA
modification N1-methyladenosine (m1A) in mRNA and tRNA. Genomics
Proteomics Bioinformatics. 16:155–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Shi Q, Xue C, Yuan X, He Y and Yu Z: Gene
signatures and prognostic values of m1A-related regulatory genes in
hepatocellular carcinoma. Sci Rep. 10:150832020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Li X, Xiong X, Zhang M, Wang K, Chen Y,
Zhou J, Mao Y, Lv J, Yi D, Chen XW, et al: Base-resolution mapping
reveals distinct m1A methylome in nuclear- and
mitochondrial-encoded transcripts. Mol Cell. 68:993–1005.e9. 2017.
View Article : Google Scholar
|
|
111
|
Siasos G, Tsigkou V, Kosmopoulos M,
Theodosiadis D, Simantiris S, Tagkou NM, Tsimpiktsioglou A,
Stampouloglou PK, Oikonomou E, Mourouzis K, et al: Mitochondria and
cardiovascular diseases-from pathophysiology to treatment. Ann
Transl Med. 6:2562018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Poznyak AV, Ivanova EA, Sobenin IA, Yet SF
and Orekhov AN: The role of mitochondria in cardiovascular
diseases. Biology (Basel). 9:92020.
|
|
113
|
Ali AT, Idaghdour Y and Hodgkinson A:
Analysis of mitochondrial m1A/G RNA modification reveals links to
nuclear genetic variants and associated disease processes. Commun
Biol. 3:1472020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Xu L, Liu X, Sheng N, Oo KS, Liang J,
Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, et al: Three distinct
3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in
mice and humans. J Biol Chem. 292:14695–14703. 2017. View Article : Google Scholar
|
|
115
|
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J,
Lu Z, Zheng Z, Dai Q and Wang H: Transfer RNA demethylase ALKBH3
promotes cancer progression via induction of tRNA-derived small
RNAs. Nucleic Acids Res. 47:2533–2545. 2019. View Article : Google Scholar
|
|
116
|
Ma CJ, Ding JH, Ye TT, Yuan BF, Feng YQ
and Alk B: AlkB homologue 1 demethylates N3-methylcytidine in mRNA
of mammals. ACS Chem Biol. 14:1418–1425. 2019. View Article : Google Scholar
|
|
117
|
Lentini JM, Alsaif HS, Faqeih E, Alkuraya
FS and Fu D: DALRD3 encodes a protein mutated in epileptic
encephalopathy that targets arginine tRNAs for 3-methylcytosine
modification. Nat Commun. 11:25102020. View Article : Google Scholar :
|
|
118
|
Yang X, Yang Y, Sun BF, Chen YS, Xu JW,
Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al: 5-methylcytosine
promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as
an m5C reader. Cell Res. 27:606–625. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Gambaryan AS, Venkstern TV and Baev AA:
Use of the method of mixed substrates to study the specificity of
tRNA methylases. Mol Biol (Mosk). 10:697–705. 1976.
|
|
120
|
Amort T, Rieder D, Wille A,
Khokhlova-Cubberley D, Riml C, Trixl L, Jia XY, Micura R and Lusser
A: Distinct 5-methylcytosine profiles in poly(A) RNA from mouse
embryonic stem cells and brain. Genome Biol. 18:12017. View Article : Google Scholar
|
|
121
|
Squires JE, Patel HR, Nousch M, Sibbritt
T, Humphreys DT, Parker BJ, Suter CM and Preiss T: Widespread
occurrence of 5-methylcytosine in human coding and non-coding RNA.
Nucleic Acids Res. 40:5023–5033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Sharma S, Yang J, Watzinger P, Kötter P
and Entian KD: Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the
25S rRNA, respectively. Nucleic Acids Res. 41:9062–9076. 2013.
View Article : Google Scholar :
|
|
123
|
Brzezicha B, Schmidt M, Makalowska I,
Jarmolowski A, Pienkowska J and Szweykowska-Kulinska Z:
Identification of human tRNA:m5C methyltransferase catalysing
intron-dependent m5C formation in the first position of the
anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res.
34:6034–6043. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Hussain S, Sajini AA, Blanco S, Dietmann
S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et
al: NSun2-mediated cytosine-5 methylation of vault noncoding RNA
determines its processing into regulatory small RNAs. Cell Rep.
4:255–261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Haag S, Sloan KE, Ranjan N, Warda AS,
Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S,
Rehling P, et al: NSUN3 and ABH1 modify the wobble position of
mt-tRNAMet to expand codon recognition in mitochondrial
translation. EMBO J. 35:2104–2119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Metodiev MD, Spåhr H, Loguercio Polosa P,
Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG and
Ruzzenente B: NSUN4 is a dual function mitochondrial protein
required for both methylation of 12S rRNA and coordination of
mitoribosomal assembly. PLoS Genet. 10:e10041102014. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Schosserer M, Minois N, Angerer TB, Amring
M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP,
Pfeifenberger S, et al: Methylation of ribosomal RNA by NSUN5 is a
conserved mechanism modulating organismal lifespan. Nat Commun.
6:61582015. View Article : Google Scholar
|
|
128
|
Haag S, Warda AS, Kretschmer J, Günnigmann
MA, Höbartner C and Bohnsack MT: NSUN6 is a human RNA
methyltransferase that catalyzes formation of m5C72 in specific
tRNAs. RNA. 21:1532–1543. 2015. View Article : Google Scholar :
|
|
129
|
Kaiser S, Jurkowski TP, Kellner S,
Schneider D, Jeltsch A and Helm M: The RNA methyltransferase Dnmt2
methylates DNA in the structural context of a tRNA. RNA Biol.
14:1241–1251. 2017. View Article : Google Scholar
|
|
130
|
Goll MG, Kirpekar F, Maggert KA, Yoder JA,
Hsieh CL, Zhang X, Golic KG, Jacobsen SE and Bestor TH: Methylation
of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science.
311:395–398. 2006. View Article : Google Scholar
|
|
131
|
Abbasi-Moheb L, Mertel S, Gonsior M,
Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM,
Esmaeeli-Nieh S, Cremer K, et al: Mutations in NSUN2 cause
autosomal-recessive intellectual disability. Am J Hum Genet.
90:847–855. 2012. View Article : Google Scholar
|
|
132
|
Frye M and Watt FM: The RNA
methyltransferase Misu (NSun2) mediates Myc-induced proliferation
and is upregulated in tumors. Curr Biol. 16:971–981. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Yi J, Gao R, Chen Y, Yang Z, Han P, Zhang
H, Dou Y, Liu W, Wang W, Du G, et al: Overexpression of NSUN2 by
DNA hypomethylation is associated with metastatic progression in
human breast cancer. Oncotarget. 8:20751–20765. 2017. View Article : Google Scholar
|
|
134
|
Forbes SA, Beare D, Boutselakis H, Bamford
S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al:
COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids
Res. 45:D777–D783. 2017. View Article : Google Scholar
|
|
135
|
Elhardt W, Shanmugam R, Jurkowski TP and
Jeltsch A: Somatic cancer mutations in the DNMT2 tRNA
methyltransferase alter its catalytic properties. Biochimie.
112:66–72. 2015. View Article : Google Scholar
|
|
136
|
Ghanbarian H, Wagner N, Polo B, Baudouy D,
Kiani J, Michiels JF, Cuzin F, Rassoulzadegan M and Wagner KD:
Dnmt2/Trdmt1 as mediator of RNA polymerase II transcriptional
activity in cardiac growth. PLoS One. 11:e01569532016. View Article : Google Scholar :
|
|
137
|
Luo Y, Feng J, Xu Q, Wang W and Wang X:
NSun2 deficiency protects endothelium from inflammation via mRNA
methylation of ICAM-1. Circ Res. 118:944–956. 2016. View Article : Google Scholar
|
|
138
|
Wang N, Tang H, Wang X, Wang W and Feng J:
Homocysteine upregulates interleukin-17A expression via
NSun2-mediated RNA methylation in T lymphocytes. Biochem Biophys
Res Commun. 493:94–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Yuan S, Tang H, Xing J, Fan X, Cai X, Li
Q, Han P, Luo Y, Zhang Z, Jiang B, et al: Methylation by NSun2
represses the levels and function of microRNA 125b. Mol Cell Biol.
34:3630–3641. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wen P, Cao H, Fang L, Ye H, Zhou Y, Jiang
L, Su W, Xu H, He W, Dai C, et al: miR-125b/Ets1 axis regulates
transdifferentiation and calcification of vascular smooth muscle
cells in a high-phosphate environment. Exp Cell Res. 322:302–312.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Goettsch C, Rauner M, Pacyna N, Hempel U,
Bornstein SR and Hofbauer LC: miR-125b regulates calcification of
vascular smooth muscle cells. Am J Pathol. 179:1594–1600. 2011.
View Article : Google Scholar :
|
|
142
|
Trixl L and Lusser A: The dynamic RNA
modification 5-methylcytosine and its emerging role as an
epitranscriptomic mark. Wiley Interdiscip Rev RNA. 10:e15102019.
View Article : Google Scholar
|
|
143
|
Jacob R, Zander S and Gutschner T: The
dark side of the epitranscriptome: Chemical modifications in long
non-coding RNAs. Int J Mol Sci. 18:182017. View Article : Google Scholar
|
|
144
|
Fu L, Guerrero CR, Zhong N, Amato NJ, Liu
Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, et al: Tet-mediated
formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc.
136:11582–11585. 2014. View Article : Google Scholar :
|
|
145
|
Fang S, Li J, Xiao Y, Lee M, Guo L, Han W,
Li T, Hill MC, Hong T, Mo W, et al: Tet inactivation disrupts YY1
binding and long-range chromatin interactions during embryonic
heart development. Nat Commun. 10:42972019. View Article : Google Scholar :
|
|
146
|
Ramanathan A, Robb GB and Chan S-H: mRNA
capping: Biological functions and applications. Nucleic Acids Res.
44:7511–7526. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Furuichi Y: Discovery of m(7)G-cap in
eukaryotic mRNAs. Proc Jpn Acad, Ser B, Phys Biol Sci. 91:394–409.
2015. View Article : Google Scholar
|
|
148
|
Pandolfini L, Barbieri I, Bannister AJ,
Hendrick A, Andrews B, Webster N, Murat P, Mach P, Brandi R, Robson
SC, et al: METTL1 promotes let-7 MicroRNA processing via m7G
methylation. Mol Cell. 74:1278–1290.e9. 2019. View Article : Google Scholar :
|
|
149
|
Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y
and Zhou HH: Let-7 in cardiovascular diseases, heart development
and cardiovascular differentiation from stem cells. Int J Mol Sci.
14:23086–23102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Bernstein DL, Jiang X and Rom S: let-7
microRNAs: Their role in cerebral and cardiovascular diseases,
inflammation, cancer, and their regulation. Biomedicines. 9:92021.
View Article : Google Scholar
|
|
151
|
Tolonen A-M, Magga J, Szabó Z, Viitala P,
Gao E, Moilanen AM, Ohukainen P, Vainio L, Koch WJ, Kerkelä R, et
al: Inhibition of Let-7 microRNA attenuates myocardial remodeling
and improves cardiac function postinfarction in mice. Pharmacol Res
Perspect. 2:e00056. 2014. View Article : Google Scholar
|
|
152
|
Yang Y, Ago T, Zhai P, Abdellatif M and
Sadoshima J: Thioredoxin 1 negatively regulates angiotensin
II-induced cardiac hypertrophy through upregulation of
miR-98/let-7. Circ Res. 108:305–313. 2011. View Article : Google Scholar
|
|
153
|
Satoh M, Minami Y, Takahashi Y, Tabuchi T
and Nakamura M: A cellular microRNA, let-7i, is a novel biomarker
for clinical outcome in patients with dilated cardiomyopathy. J
Card Fail. 17:923–929. 2011. View Article : Google Scholar
|
|
154
|
Dimitrova DG, Teysset L and Carré C: RNA
2′-O-Methylation (Nm) modification in human diseases. Genes
(Basel). 10:102019. View Article : Google Scholar
|
|
155
|
Somme J, Van Laer B, Roovers M, Steyaert
J, Versées W and Droogmans L: Characterization of two homologous
2′-O-methyltransferases showing different specificities for their
tRNA substrates. RNA. 20:1257–1271. 2014. View Article : Google Scholar
|
|
156
|
Cavaillé J, Nicoloso M and Bachellerie
J-P: Targeted ribose methylation of RNA in vivo directed by
tailored antisense RNA guides. Nature. 383:732–735. 1996.
View Article : Google Scholar
|
|
157
|
O'Brien JE Jr, Kibiryeva N, Zhou X-G,
Marshall JA, Lofland GK, Artman M, Chen J and Bittel DC: Noncoding
RNA expression in myocardium from infants with tetralogy of Fallot.
Circ Cardiovasc Genet. 5:279–286. 2012. View Article : Google Scholar
|
|
158
|
Nagasawa C, Ogren A, Kibiryeva N, Marshall
J, O'Brien JE Jr, Kenmochi N and Bittel DC: The role of scaRNAs in
adjusting alternative mRNA splicing in heart development. J
Cardiovasc Dev Dis. 5:262018. View Article : Google Scholar
|
|
159
|
Elliott BA, Ho H-T, Ranganathan SV,
Vangaveti S, Ilkayeva O, Abou Assi H, Choi AK, Agris PF and Holley
CL: Modification of messenger RNA by 2′-O-methylation regulates
gene expression in vivo. Nat Commun. 10:34012019. View Article : Google Scholar
|
|
160
|
Gagnidze K, Rayon-Estrada V, Harroch S,
Bulloch K and Papavasiliou FN: A new chapter in genetic medicine:
RNA Editing and its role in disease pathogenesis. Trends Mol Med.
24:294–303. 2018. View Article : Google Scholar
|
|
161
|
Nishikura K: A-to-I editing of coding and
non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 17:83–96. 2016.
View Article : Google Scholar :
|
|
162
|
Uchida S and Jones SP: RNA editing:
Unexplored opportunities in the cardiovascular system. Circ Res.
122:399–401. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Picardi E, Manzari C, Mastropasqua F,
Aiello I, D'Erchia AM and Pesole G: Profiling RNA editing in human
tissues: Towards the inosinome Atlas. Sci Rep. 5:149412015.
View Article : Google Scholar :
|
|
164
|
Sukhova GK, Zhang Y, Pan JH, Wada Y,
Yamamoto T, Naito M, Kodama T, Tsimikas S, Witztum JL, Lu ML, et
al: Deficiency of cathepsin S reduces atherosclerosis in LDL
receptor-deficient mice. J Clin Invest. 111:897–906. 2003.
View Article : Google Scholar
|
|
165
|
Wang B, Sun J, Kitamoto S, Yang M, Grubb
A, Chapman HA, Kalluri R and Shi GP: Cathepsin S controls
angiogenesis and tumor growth via matrix-derived angiogenic
factors. J Biol Chem. 281:6020–6029. 2006. View Article : Google Scholar
|
|
166
|
Zhu Y, Pirnie SP and Carmichael GG:
High-throughput and site-specific identification of
2′-O-methylation sites using ribose oxidation sequencing
(RibOxi-seq). RNA. 23:1303–1314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Stellos K, Gatsiou A, Stamatelopoulos K,
Perisic Matic L, John D, Lunella FF, Jaé N, Rossbach O, Amrhein C,
Sigala F, et al: Adenosine-to-inosine RNA editing controls
cathepsin S expression in atherosclerosis by enabling HuR-mediated
post-transcriptional regulation. Nat Med. 22:1140–1150. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Wang Y, Men M, Xie B, Shan J, Wang C, Liu
J, Zheng H, Yang W, Xue S and Guo C: Inhibition of PKR protects
against H2O2-induced injury on neonatal
cardiac myocytes by attenuating apoptosis and inflammation. Sci
Rep. 6:387532016. View Article : Google Scholar
|
|
169
|
Moore JB IV, Sadri G, Fischer AG, Weirick
T, Militello G, Wysoczynski M, Gumpert AM, Braun T and Uchida S:
The A-to-I RNA editing enzyme Adar1 is essential for normal
embryonic cardiac growth and development. Circ Res. 127:550–552.
2020. View Article : Google Scholar :
|
|
170
|
El Azzouzi H, Vilaça AP, Feyen DA, Gommans
WM, de Weger RA, Doevendans PA and Sluijter JP: Cardiomyocyte
specific deletion of ADAR1 causes severe cardiac dysfunction and
increased lethality. Front Cardiovasc Med. 7:302020. View Article : Google Scholar
|
|
171
|
Stulić M and Jantsch MF: Spatio-temporal
profiling of Filamin A RNA-editing reveals ADAR preferences and
high editing levels outside neuronal tissues. RNA Biol.
10:1611–1617. 2013. View Article : Google Scholar
|
|
172
|
Jain M, Mann TD, Stulić M, Rao SP, Kirsch
A, Pullirsch D, Strobl X, Rath C, Reissig L, Moreth K, et al: RNA
editing of Filamin A pre-mRNA regulates vascular contraction and
diastolic blood pressure. EMBO J. 37:372018. View Article : Google Scholar
|
|
173
|
Vu LT and Tsukahara T: C-to-U editing and
site-directed RNA editing for the correction of genetic mutations.
Biosci Trends. 11:243–253. 2017. View Article : Google Scholar
|
|
174
|
Gerber AP and Keller W: RNA editing by
base deamination: More enzymes, more targets, new mysteries. Trends
Biochem Sci. 26:376–384. 2001. View Article : Google Scholar
|
|
175
|
Rosenberg BR, Dewell S and Papavasiliou
FN: Identifying mRNA editing deaminase targets by RNA-Seq. Methods
Mol Biol. 718:103–119. 2011. View Article : Google Scholar
|
|
176
|
Powell LM, Wallis SC, Pease RJ, Edwards
YH, Knott TJ and Scott J: A novel form of tissue-specific RNA
processing produces apolipoprotein-B48 in intestine. Cell.
50:831–840. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Teng B, Burant CF and Davidson NO:
Molecular cloning of an apolipoprotein B messenger RNA editing
protein. Science. 260:1816–1819. 1993. View Article : Google Scholar
|
|
178
|
Blanc V and Davidson NO: APOBEC-1-mediated
RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2:594–602. 2010.
View Article : Google Scholar
|
|
179
|
Anant S, Henderson JO, Mukhopadhyay D,
Navaratnam N, Kennedy S, Min J and Davidson NO: Novel role for
RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2
modulates C to U editing of apolipoprotein B mRNA by interacting
with apobec-1 and ACF, the apobec-1 complementation factor. J Biol
Chem. 276:47338–47351. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Blech-Hermoni Y, Dasgupta T, Coram RJ and
Ladd AN: Identification of targets of CUG-BP, Elav-like family
member 1 (CELF1) regulation in embryonic heart muscle. PLoS One.
11:e01490612016. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Giudice J, Xia Z, Li W and Cooper TA:
Neonatal cardiac dysfunction and transcriptome changes caused by
the absence of Celf1. Sci Rep. 6:355502016. View Article : Google Scholar
|
|
182
|
Liao W, Hong SH, Chan BH, Rudolph FB,
Clark SC and Chan L: APOBEC-2, a cardiac - and skeletal
muscle-specific member of the cytidine deaminase supergene family.
Biochem Biophys Res Commun. 260:398–404. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Sharma S, Patnaik SK, Taggart RT, Kannisto
ED, Enriquez SM, Gollnick P and Baysal BE: APOBEC3A cytidine
deaminase induces RNA editing in monocytes and macrophages. Nat
Commun. 6:68812015. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Sharma S, Patnaik SK, Kemer Z and Baysal
BE: Transient overexpression of exogenous APOBEC3A causes C-to-U
RNA editing of thousands of genes. RNA Biol. 14:603–610. 2017.
View Article : Google Scholar :
|
|
185
|
Li X, Ma S and Yi C: Pseudouridine: The
fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol.
33:108–116. 2016. View Article : Google Scholar
|
|
186
|
Cohn WE and Volkin E:
Nucleoside-5′-phosphates from ribonucleic acid. Nature.
167:483–484. 1951. View Article : Google Scholar
|
|
187
|
Sengupta R, Vainauskas S, Yarian C,
Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM and Agris PF:
Modified constructs of the tRNA TPsiC domain to probe substrate
conformational requirements of m(1)A(58) and m(5)U(54) tRNA
methyltransferases. Nucleic Acids Res. 28:1374–1380. 2000.
View Article : Google Scholar
|
|
188
|
Charette M and Gray MW: Pseudouridine in
RNA: What, where, how, and why. IUBMB Life. 49:341–351. 2000.
View Article : Google Scholar
|
|
189
|
Davis DR: Stabilization of RNA stacking by
pseudouridine. Nucleic Acids Res. 23:5020–5026. 1995. View Article : Google Scholar
|
|
190
|
Karijolich J and Yu YT: Converting
nonsense codons into sense codons by targeted pseudouridylation.
Nature. 474:395–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Spenkuch F, Motorin Y and Helm M:
Pseudouridine: Still mysterious, but never a fake (uridine)! RNA
Biol. 11:1540–1554. 2014. View Article : Google Scholar
|
|
192
|
Safra M, Nir R, Farouq D, Vainberg
Slutskin I and Schwartz S: TRUB1 is the predominant pseudouridine
synthase acting on mammalian mRNA via a predictable and conserved
code. Genome Res. 27:393–406. 2017. View Article : Google Scholar
|
|
193
|
Antonicka H, Choquet K, Lin ZY, Gingras
AC, Kleinman CL and Shoubridge EA: A pseudouridine synthase module
is essential for mitochondrial protein synthesis and cell
viability. EMBO Rep. 18:28–38. 2017. View Article : Google Scholar
|
|
194
|
Zaganelli S, Rebelo-Guiomar P, Maundrell
K, Rozanska A, Pierredon S, Powell CA, Jourdain AA, Hulo N,
Lightowlers RN, Chrzanowska-Lightowlers ZM, et al: The
pseudouridine synthase RPUSD4 is an essential component of
mitochondrial RNA granules. J Biol Chem. 292:4519–4532. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu
L, Li RY, Ji L, Dorsey SG, Jiang ZR, et al: Small nucleolar RNA 42
acts as an oncogene in lung tumorigenesis. Oncogene. 31:2794–2804.
2012. View Article : Google Scholar
|
|
196
|
Bykhovskaya Y, Casas K, Mengesha E, Inbal
A and Fischel-Ghodsian N: Missense mutation in pseudouridine
synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic
anemia (MLASA). Am J Hum Genet. 74:1303–1308. 2004. View Article : Google Scholar
|
|
197
|
Zucchini C, Strippoli P, Biolchi A, Solmi
R, Lenzi L, D'Addabbo P, Carinci P and Valvassori L: The human TruB
family of pseudouridine synthase genes, including the Dyskeratosis
Congenita 1 gene and the novel member TRUB1. Int J Mol Med.
11:697–704. 2003.PubMed/NCBI
|
|
198
|
Jády BE and Kiss T: A small nucleolar
guide RNA functions both in 2′-O-ribose methylation and
pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541–551.
2001. View Article : Google Scholar
|
|
199
|
Darzacq X, Jády BE, Verheggen C, Kiss AM,
Bertrand E and Kiss T: Cajal body-specific small nuclear RNAs: A
novel class of 2′-O-methylation and pseudouridylation guide RNAs.
EMBO J. 21:2746–2756. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Nagasawa CK, Kibiryeva N, Marshall J,
O'Brien JE Jr and Bittel DC: scaRNA1 levels alter pseudouridylation
in spliceosomal RNA U2 affecting alternative mRNA splicing and
embryonic development. Pediatr Cardiol. 41:341–349. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Wilusz JE: Long noncoding RNAs: Re-writing
dogmas of RNA processing and stability. Biochim Biophys Acta.
1859:128–138. 2016. View Article : Google Scholar
|
|
202
|
Kanvah S, Joseph J, Schuster GB, Barnett
RN, Cleveland CL and Landman U: Oxidation of DNA: Damage to
nucleobases. Acc Chem Res. 43:280–287. 2010. View Article : Google Scholar
|
|
203
|
Faucher F, Doublié S and Jia Z:
8-oxoguanine DNA glycosylases: One lesion, three subfamilies. Int J
Mol Sci. 13:6711–6729. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Shah A, Gray K, Figg N, Finigan A, Starks
L and Bennett M: Defective base excision repair of oxidative DNA
damage in vascular smooth muscle cells promotes atherosclerosis.
Circulation. 138:1446–1462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Seok H, Lee H, Lee S, Ahn SH, Lee HS, Kim
GD, Peak J, Park J, Cho YK, Jeong Y, et al: Position-specific
oxidation of miR-1 encodes cardiac hypertrophy. Nature.
584:279–285. 2020. View Article : Google Scholar
|
|
206
|
Wang J, Wang Q, Watson LJ, Jones SP and
Epstein PN: Cardiac overexpression of 8-oxoguanine DNA glycosylase
1 protects mitochondrial DNA and reduces cardiac fibrosis following
transaortic constriction. Am J Physiol Heart Circ Physiol.
301:H2073–H2080. 2011. View Article : Google Scholar :
|
|
207
|
Noren Hooten N, Ejiogu N, Zonderman AB and
Evans MK: Association of oxidative DNA damage and C-reactive
protein in women at risk for cardiovascular disease. Arterioscler
Thromb Vasc Biol. 32:2776–2784. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Motorin Y and Helm M: RNA nucleotide
methylation. Wiley Interdiscip Rev RNA. 2:611–631. 2011. View Article : Google Scholar
|
|
209
|
Garcia-Campos MA, Edelheit S, Toth U,
Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis
A, et al: Deciphering the 'm6A Code' via antibody-independent
quantitative profiling. Cell. 178:731–747.e16. 2019. View Article : Google Scholar
|
|
210
|
Zhang Z, Chen L-Q, Zhao Y-L, Yang CG,
Roundtree IA, Zhang Z, Ren J, Xie W, He C and Luo GZ: Single-base
mapping of m6A by an antibody-independent method. Sci Adv.
5:eaax02502019. View Article : Google Scholar :
|
|
211
|
Liu N and Pan T: Probing
N6-methyladenosine (m6A) RNA modification in total RNA with
SCARLET. Methods Mol Biol. 1358:285–292. 2016. View Article : Google Scholar
|
|
212
|
Dominissini D, Moshitch-Moshkovitz S,
Salmon-Divon M, Amariglio N and Rechavi G: Transcriptome-wide
mapping of N(6)-methyladenosine by m(6)A-seq based on
immunocapturing and massively parallel sequencing. Nat Protoc.
8:176–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N,
Han D, Dominissini D, Dai Q, Pan T, et al: High-resolution
N(6)-methyladenosine (m(6) A) map using photo-crosslinking-assisted
m(6) A sequencing. Angew Chem Int Ed Engl. 54:1587–1590. 2015.
View Article : Google Scholar
|
|
214
|
Marchand V, Bourguignon-Igel V, Helm M and
Motorin Y: Analysis of pseudouridines and other RNA modifications
using HydraPsiSeq protocol. Methods. Sep 1–2021.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Motorin Y and Helm M: Methods for RNA
modification mapping using deep sequencing: Established and new
emerging technologies. Genes (Basel). 10:352019. View Article : Google Scholar
|
|
216
|
Dai Q, Moshitch-Moshkovitz S, Han D, Kol
N, Amariglio N, Rechavi G, Dominissini D and He C: Nm-seq maps
2′-O-methylation sites in human mRNA with base precision. Nat
Methods. 14:695–698. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Birkedal U, Christensen-Dalsgaard M, Krogh
N, Sabarinathan R, Gorodkin J and Nielsen H: Profiling of ribose
methylations in RNA by high-throughput sequencing. Angew Chem Int
Ed Engl. 54:451–455. 2015.
|
|
218
|
Yoluç Y, Ammann G, Barraud P, Jora M,
Limbach PA, Motorin Y, Marchand V, Tisné C, Borland K and Kellner
S: Instrumental analysis of RNA modifications. Crit Rev Biochem Mol
Biol. 56:178–204. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Huang TY, Liu J and McLuckey SA: Top-down
tandem mass spectrometry of tRNA via ion trap collision-induced
dissociation. J Am Soc Mass Spectrom. 21:890–898. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Durairaj A and Limbach PA: Improving
CMC-derivatization of pseudouridine in RNA for mass spectrometric
detection. Anal Chim Acta. 612:173–181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Kowalak JA, Pomerantz SC, Crain PF and
McCloskey JA: A novel method for the determination of
post-transcriptional modification in RNA by mass spectrometry.
Nucleic Acids Res. 21:4577–4585. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Heiss M, Borland K, Yoluç Y and Kellner S:
Quantification of Modified nucleosides in the context of NAIL-MS.
Methods Mol Biol. 2298:279–306. 2021. View Article : Google Scholar
|
|
223
|
Wein S, Andrews B, Sachsenberg T,
Santos-Rosa H, Kohlbacher O, Kouzarides T, Garcia BA and Weisser H:
A computational platform for high-throughput analysis of RNA
sequences and modifications by mass spectrometry. Nat Commun.
11:9262020. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Marceca GP, Tomasello L, Distefano R,
Acunzo M, Croce CM and Nigita G: Detecting and characterizing
A-To-I microRNA editing in cancer. Cancers (Basel). 13:16992021.
View Article : Google Scholar
|
|
225
|
Okada S, Ueda H, Noda Y and Suzuki T:
Transcriptome-wide identification of A-to-I RNA editing sites using
ICE-seq. Methods. 156:66–78. 2019. View Article : Google Scholar
|
|
226
|
Adachi H, DeZoysa MD and Yu YT: Detection
and quantification of pseudouridine in RNA. Epitranscriptomics:
Methods and Protocols. Wajapeyee N and Gupta R: Springer; New York,
NY: pp. 219–235. 2019, View Article : Google Scholar
|
|
227
|
Schwartz S, Bernstein DA, Mumbach MR,
Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M,
Satija R, Lander ES, et al: Transcriptome-wide mapping reveals
widespread dynamic-regulated pseudouridylation of ncRNA and mRNA.
Cell. 159:148–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Lovejoy AF, Riordan DP and Brown PO:
Transcriptome-wide mapping of pseudouridines: Pseudouridine
synthases modify specific mRNAs in S. cerevisiae. PLoS One.
9:e1107992014. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Carlile TM, Rojas-Duran MF, Zinshteyn B,
Shin H, Bartoli KM and Gilbert WV: Pseudouridine profiling reveals
regulated mRNA pseudouridylation in yeast and human cells. Nature.
515:143–146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Li X, Zhu P, Ma S, Song J, Bai J, Sun F
and Yi C: Chemical pulldown reveals dynamic pseudouridylation of
the mammalian transcriptome. Nat Chem Biol. 11:592–597. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Banerjee A, Mikhailova E, Cheley S, Gu LQ,
Montoya M, Nagaoka Y, Gouaux E and Bayley H: Molecular bases of
cyclodextrin adapter interactions with engineered protein
nanopores. Proc Natl Acad Sci USA. 107:8165–8170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Wang Y, Yang Q and Wang Z: The evolution
of nanopore sequencing. Front Genet. 5:449. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Simpson JT, Workman RE, Zuzarte PC, David
M, Dursi LJ and Timp W: Detecting DNA cytosine methylation using
nanopore sequencing. Nat Methods. 14:407–410. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Vlachakis D, Papakonstantinou E, Sagar R,
Bacopoulou F, Exarchos T, Kourouthanassis P, Karyotis V, Vlamos P,
Lyketsos C, Avramopoulos D, et al: Improving the utility of
polygenic risk scores as a biomarker for Alzheimer's disease.
Cells. 10:16272021. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Buniello A, MacArthur JAL, Cerezo M,
Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy
E, Sollis E, et al: The NHGRI-EBI GWAS Catalog of published
genome-wide association studies, targeted arrays and summary
statistics 2019. Nucleic Acids Res. 47:D1005–D1012. 2019.
View Article : Google Scholar
|
|
236
|
Zheng Y, Nie P, Peng D, He Z, Liu M, Xie
Y, Miao Y, Zuo Z and Ren J: m6AVar: A database of functional
variants involved in m6A modification. Nucleic Acids Res.
46:D139–D145. 2018. View Article : Google Scholar :
|
|
237
|
Jiang S, Xie Y, He Z, Zhang Y, Zhao Y,
Chen L, Zheng Y, Miao Y, Zuo Z and Ren J: m6ASNP: A tool for
annotating genetic variants by m6A function. Gigascience. 7:72018.
View Article : Google Scholar
|
|
238
|
Song B, Tang Y, Chen K, Wei Z, Rong R, Lu
Z, Su J, de Magalhães JP, Rigden DJ and Meng J: m7GHub: Deciphering
the location, regulation and pathogenesis of internal mRNA
N7-methylguanosine (m7G) sites in human. Bioinformatics.
36:3528–3536. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Chen K, Song B, Tang Y, Wei Z, Xu Q, Su J,
de Magalhães JP, Rigden DJ and Meng J: RMDisease: A database of
genetic variants that affect RNA modifications, with implications
for epitranscriptome pathogenesis. Nucleic Acids Res.
49:D1396–D1404. 2021. View Article : Google Scholar
|
|
240
|
Incarnato D, Morandi E, Simon LM and
Oliviero S: RNA Framework: An all-in-one toolkit for the analysis
of RNA structures and post-transcriptional modifications. Nucleic
Acids Res. 46:e972018. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Mo XB, Lei SF, Zhang YH and Zhang H:
Detection of m6A-associated SNPs as potential functional variants
for coronary artery disease. Epigenomics. 10:1279–1287. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Mo XB, Lei SF, Zhang YH and Zhang H:
Examination of the associations between m6A-associated
single-nucleotide polymorphisms and blood pressure. Hypertens Res.
42:1582–1589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Mo X, Lei S, Zhang Y and Zhang H:
Genome-wide enrichment of m6A-associated single-nucleotide
polymorphisms in the lipid loci. Pharmacogenomics J. 19:347–357.
2019. View Article : Google Scholar
|
|
244
|
Mo X-B, Lei S-F, Zhang Y-H and Zhang H:
Integrative Analysis identified IRF6 and NDST1 as potential causal
genes for ischemic stroke. Front Neurol. 10:5172019. View Article : Google Scholar :
|
|
245
|
Nielsen JB, Thorolfsdottir RB, Fritsche
LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM,
Sveinbjornsson G, et al: Biobank-driven genomic discovery yields
new insight into atrial fibrillation biology. Nat Genet.
50:1234–1239. 2018. View Article : Google Scholar
|
|
246
|
Ehret GB, Ferreira T, Chasman DI, Jackson
AU, Schmidt EM, Johnson T, Thorleifsson G, Luan J, Donnelly LA,
Kanoni S, et al CHARGE-EchoGen consortium; CHARGE-HF consortium:
Wellcome Trust Case Control Consortium: The genetics of blood
pressure regulation and its target organs from association studies
in 342,415 individuals. Nat Genet. 48:1171–1184. 2016. View Article : Google Scholar :
|
|
247
|
Franzén O, Ermel R, Sukhavasi K, Jain R,
Jain A, Betsholtz C, Giannarelli C, Kovacic JC, Ruusalepp A,
Skogsberg J, et al: Global analysis of A-to-I RNA editing reveals
association with common disease variants. PeerJ. 6:e4466. 2018.
View Article : Google Scholar :
|
|
248
|
Lee JY, Lee BS, Shin DJ, Woo Park K, Shin
YA, Joong Kim K, Heo L, Young Lee J, Kyoung Kim Y, Jin Kim Y, et
al: A genome-wide association study of a coronary artery disease
risk variant. J Hum Genet. 58:120–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Jeemon P, Pettigrew K, Sainsbury C,
Prabhakaran D and Padmanabhan S: Implications of discoveries from
genome-wide association studies in current cardiovascular practice.
World J Cardiol. 3:230–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Wakil SM, Ram R, Muiya NP, Mehta M, Andres
E, Mazhar N, Baz B, Hagos S, Alshahid M, Meyer BF, et al: A
genome-wide association study reveals susceptibility loci for
myocardial infarction/coronary artery disease in Saudi Arabs.
Atherosclerosis. 245:62–70. 2016. View Article : Google Scholar
|
|
251
|
Terasawa Y, Ladha Z, Leonard SW, Morrow
JD, Newland D, Sanan D, Packer L, Traber MG and Farese RV Jr:
Increased atherosclerosis in hyperlipidemic mice deficient in alpha
-tocopherol transfer protein and vitamin E. Proc Natl Acad Sci USA.
97:13830–13834. 2000. View Article : Google Scholar
|
|
252
|
Lu N, Li X, Yu J, Li Y, Wang C, Zhang L,
Wang T and Zhong X: Curcumin attenuates lipopolysaccharide-induced
hepatic lipid metabolism disorder by modification of m6A RNA
methylation in piglets. Lipids. 53:53–63. 2018. View Article : Google Scholar
|
|
253
|
Sikorski V, Karjalainen P, Blokhina D,
Oksaharju K, Khan J, Katayama S, Rajala H, Suihko S, Tuohinen S,
Teittinen K, et al: Epitranscriptomics of ischemic heart
disease-the IHD-EPITRAN study design and objectives. Int J Mol Sci.
22:222021. View Article : Google Scholar
|
|
254
|
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou
H, Gan J, Jiang H, Jia GF, Luo C, et al: Meclofenamic acid
selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic
Acids Res. 43:373–384. 2015. View Article : Google Scholar
|
|
255
|
Li J, Chen Z, Chen F, Xie G, Ling Y, Peng
Y, Lin Y, Luo N, Chiang CM and Wang H: Targeted mRNA demethylation
using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids
Res. 48:5684–5694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Cox DBT, Gootenberg JS, Abudayyeh OO,
Franklin B, Kellner MJ, Joung J and Zhang F: RNA editing with
CRISPR-Cas13. Science. 358:1019–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Papageorgiou L, Cuong NT and Vlachakis D:
Antibodies as stratagems against cancer. Mol Biosyst. 12:2047–2055.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Vlachakis D: Antibody clustering and 3D
modelling for neurodegenerative diseases. Computational
Neurodegeneration. Vlamos P, Kotsireas I and Tarnanas I: Springer;
New York, NY: pp. 1–13. 2020
|
|
259
|
Altaf F, Vesely C, Sheikh AM, Munir R,
Shah ST and Tariq A: Modulation of ADAR mRNA expression in patients
with congenital heart defects. PLoS One. 14:e02009682019.
View Article : Google Scholar : PubMed/NCBI
|
|
260
|
van der Kwast RV, Quax PH and Nossent AY:
An emerging role for isomiRs and the microRNA epitranscriptome in
neovascularization. Cells. 9:92019. View Article : Google Scholar
|
|
261
|
Dai DF, Chen T, Johnson SC, Szeto H and
Rabinovitch PS: Cardiac aging: From molecular mechanisms to
significance in human health and disease. Antioxid Redox Signal.
16:1492–1526. 2012. View Article : Google Scholar
|
|
262
|
McMahon M, Forester C and Buffenstein R:
Aging through an epitranscriptomic lens. Nat Aging. 1:335–346.
2021. View Article : Google Scholar
|
|
263
|
Komal S, Zhang LR and Han SN: Potential
regulatory role of epigenetic RNA methylation in cardiovascular
diseases. Biomed Pharmacother. 137:1113762021. View Article : Google Scholar
|
|
264
|
Van Haute L, Dietmann S, Kremer L, Hussain
S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S,
et al: Deficient methylation and formylation of mt-tRNA(Met) wobble
cytosine in a patient carrying mutations in NSUN3. Nat Commun.
7:120392016. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Bohnsack MT and Sloan KE: The
mitochondrial epitranscriptome: The roles of RNA modifications in
mitochondrial translation and human disease. Cell Mol Life Sci.
75:241–260. 2018. View Article : Google Scholar
|
|
266
|
Vilardo E and Rossmanith W: Molecular
insights into HSD10 disease: Impact of SDR5C1 mutations on the
human mitochondrial RNase P complex. Nucleic Acids Res.
43:66492015. View Article : Google Scholar
|
|
267
|
Vlachakis D, Tsagrasoulis D,
Megalooikonomou V and Kossida S: Introducing Drugster: A
comprehensive and fully integrated drug design, lead and structure
optimization toolkit. Bioinformatics. 29:126–128. 2013. View Article : Google Scholar
|
|
268
|
Xu Z, Peng B, Cai Y, Wu G, Huang J, Gao M,
Guo G, Zeng S, Gong Z and Yan Y: N6-methyladenosine RNA
modification in cancer therapeutic resistance: Current status and
perspectives. Biochem Pharmacol. 182:1142582020. View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Tsaniras S and Vlachakis D: Diet, obesity
and cancer. J Mol Biochem. 4:202016.
|
|
270
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation regulates the
self-renewal and tumorigenesis of glioblastoma stem cells. Cell
Rep. 18:2622–2634. 2017. View Article : Google Scholar :
|
|
271
|
Singh B, Kinne HE, Milligan RD, Washburn
LJ, Olsen M and Lucci A: Important role of FTO in the survival of
rare panresistant triple-negative inflammatory breast cancer cells
facing a severe metabolic challenge. PLoS One. 11:e01590722016.
View Article : Google Scholar : PubMed/NCBI
|
|
272
|
Charalampopoulou M, Syrigos K, Filopoulos
E, Megalooikonomou V, Vlachakis D, Chrousos G and Darviri C:
Reliability and validity of the instrument newly diagnosed breast
cancer stress scale in the Greek population. J Mol Biochem. 9:5–12.
2020.
|
|
273
|
Su R, Dong L, Li C, Nachtergaele S,
Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al: R-2HG
exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA
signaling. Cell. 172:90–105.e23. 2018. View Article : Google Scholar
|
|
274
|
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu
H, Ni T, Zhang ZS, Zhang T, Li C, et al: Small-molecule targeting
of oncogenic FTO demethylase in acute myeloid leukemia. Cancer
Cell. 35:677–691.e10. 2019. View Article : Google Scholar :
|
|
275
|
Floresta G, Pistarà V, Amata E, Dichiara
M, Damigella A, Marrazzo A, Prezzavento O, Punzo F and Rescifina A:
Molecular modeling studies of pseudouridine isoxazolidinyl
nucleoside analogues as potential inhibitors of the pseudouridine
5′-mono-phosphate glycosidase. Chem Biol Drug Des. 91:519–525.
2018. View Article : Google Scholar
|
|
276
|
Vlachakis D, Fakourelis P, Megalooikonomou
V, Makris C and Kossida S: DrugOn: A fully integrated pharmacophore
modeling and structure optimization toolkit. PeerJ. 3:e7252015.
View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Vlachakis D and Kossida S: Molecular
modeling and pharmacophore elucidation study of the Classical Swine
Fever virus helicase as a promising pharmacological target. PeerJ.
1:e852013. View Article : Google Scholar : PubMed/NCBI
|
|
278
|
Papageorgiou L, Loukatou S, Sofia K,
Maroulis D and Vlachakis D: An updated evolutionary study of
Flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase
reveals novel invariable motifs as potential pharmacological
targets. Mol Biosyst. 12:2080–2093. 2016. View Article : Google Scholar
|
|
279
|
Papageorgiou L, Loukatou S, Koumandou VL,
Makałowski W, Megalooikonomou V, Vlachakis D and Kossida S:
Structural models for the design of novel antiviral agents against
Greek Goat Encephalitis. PeerJ. 2:e6642014. View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Amidi S, Amidi A, Vlachakis D, Paragios N
and Zacharaki EI: Automatic single- and multi-label enzymatic
function prediction by machine learning. PeerJ. 5:e30952017.
View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Marinou M, Platis D, Ataya FS,
Chronopoulou E, Vlachakis D and Labrou NE: Structure-based design
and application of a nucleotide coenzyme mimetic ligand:
Application to the affinity purification of nucleotide dependent
enzymes. J Chromatogr A. 1535:88–100. 2018. View Article : Google Scholar
|
|
282
|
Vlachakis D and Vlamos P: Mathematical
multidimensional modelling and structural artificial intelligence
pipelines provide insights for the designing of highly specific
antiSARS-CoV2 agents. Math Comput Sci. 15:877–888. 2021. View Article : Google Scholar
|