In recent years, great technological leaps in
sequencing technologies have enabled the in-depth investigation of
the genetic basis of a multitude of human disorders, and they have
paved the way for a new era of personalized medicine (1,2).
Such breakthrough developments in sequencing technologies have
substantiated the deep complexity of associations between genotype
and phenotype and revealed unexpected cases, such as those of
identical twins carrying the same disease mutations, but exhibiting
different clinical features, such as balance problems and the
development of blindness (3).
Such discrepancies can be attributed to epigenetic and/or
epitranscriptomic differences. The term epigenetics, first
introduced by C.H. Waddington in 1942, refers to the study of the
mechanisms and molecules that can perpetuate variable gene activity
states in the context of the same DNA sequence (4). Epigenetic mechanisms include DNA
methylation, chromatin remodeling, histone modifications, gene
activity regulation by non-coding RNA (ncRNA) molecules and
protein-protein interactions (5). These mechanisms, which include a
vast array of different molecules and pathways, regulate genomic
structure and transcriptional activity in response to the
ever-changing profiles of cell-intrinsic, cell-cell and
cell-extrinsic signals (6).
Epigenetic regulation has been increasingly gaining
interest due to its strong relationship with environmental
adaptation. As new insights are gained, novel distinctions are also
formed, leading to the emerging field of epitranscriptomics.
Instead of encompassing all epigenetic regulation,
epitranscriptomics focuses on modifications at the RNA level
(7). Due to the vast array of
effects that coding and ncRNAs exert in regulating the differential
response of organisms to environmental stimuli, as well as
homeostasis maintenance, epitranscriptomics has turned into an
explosive field of research. Several different RNA modification
databases have been established throughout the years in an effort
to catalogue the plethora of RNA modifications that are
continuously being detected. These include databases such as
Modomics (https://iimcb.genesilico.pl/modomics/) (8-11), RMBase v2.0 (http://rna.sysu.edu.cn/rmbase/) (12), DARNED (https://darned.ucc.ie/) (13), the RNA Modification Database
(https://mods.rna.albany.edu/) (14) and REDIportal v2.0 (http://srv00.recas.ba.infn.it/atlas/)
(15), encompassing >172 RNA
modifications to date.
Epitranscriptomic changes induced by such mechanisms
have been implicated in various diseases and most of them display
reversible chemistry, making epitranscriptomics a promising
candidate for providing novel therapeutics (16). As such, a number of reviews have
already been published discussing the ever-expanding field of
epitranscriptomic modifications, with a limited number focusing on
their effects under the prism of cardiovascular disease (CVD). Most
notable reviews have focused on N6-methyladenosine (m6A)
modifications, as the most prevalent epitranscriptomic modification
and its role in CVD (17,18).
Although, Kumari et al (17) featured a section about
m6A readers, Chen et al (18) also discussed the potential for
m6A modification to influence CVD risk factors. Focusing
more on clinical trials investigating epigenetic-sensitive drugs
for heart failure (HF), Napoli et al (19) also outlined the discovery of
epigenetic biomarkers and signatures of cardiac remodeling. On the
other hand, Fischer and Vondriska (20) focused their discussion on
epigenetic changes occurring in CVD, but did not expand into RNA
modifications, as was also the case for Schiano et al
(21), who discussed epigenetic
mechanisms underlying the various pathologies encompassed by the
CVD umbrella-term. Although the authors mentioned CVD
epitranscriptomics as an emerging layer of epigenetic regulation in
CVD, they also highlighted the need for further research that
covers this subject matter.
In the present review, the most prevalent
epitranscriptomic modifications that have been shown to be involved
in the field of CVD have been outlined (22), in an effort to extensively cover
the area of RNA modifications, without focusing on a single one.
This study also briefly discussed the mode of action of each
modification and then explored their respective effect on both
coding and ncRNAs, including microRNAs (miRNAs/miRs) and long
ncRNAs (lncRNAs), in the context of CVD. Furthermore, the current
methods of RNA modification detection that have been on the
forefront of epitranscriptomic research were also explored in
brief. Finally, available data on genetic associations of RNA
modifications, as well as therapeutic implications of
epitranscriptomic approaches, in the heart were discussed.
CVD is currently the leading cause of death
worldwide, accounting for almost half the total number of deaths
(23). CVD encompasses a wide
array of heart and vessel-related pathologies, including, but not
limited to HF, coronary heart disease, hypertension, hypertrophic
and dilated cardiomyopathy, as well as congenital heart disease
(24). Accumulating data have
shown that cardiovascular risk factors may alter epigenomic
patterns and that several cardiovascular biomarkers are associated
with epigenetic modifications (25). DNA methylation appears to
contribute to processes underlying CVDs, such as atherosclerosis,
hypertension and inflammation (26-28). Moreover, epidemiological studies
imply that methylation of repetitive sequences such as
long-interspersed nucleotide repetitive elements-1 (LINE-1) and Alu
elements are associated with CVD (26). Specifically, patients with
prevalent ischemic heart disease (IHD) and stroke displayed lower
blood LINE-1 methylation, while elevated methylation of Alu
elements was associated with CVD and obesity in Chinese individuals
(26). Histone modifications
have also been implicated in processes, such as hypertension and
atherosclerosis, while histone deacetylase 4 overexpression
following myocardial infarction (MI) has been shown to increase
myocardial fibrosis and cardiac hypertrophy, eventually leading to
cardiac dysfunction (29).
Although epigenetic regulation has been the focus of attention, RNA
modifications have only recently started becoming the focus of CVD
researchers.
Epitranscriptomic regulation manifests through the
action of different enzymes. Enzymes that modify the RNA itself are
called 'writers', while the ones that recognize and remove
modifications are termed 'erasers'. Finally, 'readers' are the
group of enzymes that bind the modifications themselves (30,31). These different modifications are
classified into groups based on their different characteristics.
These groups include classification into reversible and
non-reversible (where erasers are lacking), substitutional and
non-substitutional (32), cap
(where the modifications happen to the 5′-end of the RNAs) or
internal modifications [where the modifications occur within the
5′- or 3′-untranslated regions (UTRs) or within transcript introns]
(33), and finally,
modifications on coding or ncRNAs (34). NcRNAs have now been studied
extensively and have been proven to have important regulatory
effects in both physiological and pathological conditions. The term
ncRNAs encompasses a large array of RNA molecules, including, but
not limited to the major classes, such as miRNAs, lncRNAs and
circular RNAs (circRNAs), as well as transfer RNAs (tRNAs),
ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), small
nuclear RNAs (snRNAs) and others (35). ncRNA regulatory roles extend from
interacting with RNA/DNA-binding proteins, being part of complex
structures, interacting with messenger RNA (mRNA) molecules to
participating in translation and guiding chemical modifications
(35). Through
post-transcriptional modifications, ncRNAs display discrete
temporal and spatial expression patterns, reflecting a precise
regulation of their expression (36).
Epitranscriptomic editing of ncRNAs is quite
prevalent during physiological, but also pathological conditions
(37). miRNA editing is capable
of creating alternative miRNAs, known as isomiRs (38). isomiR Bank, a database
integrating >300,000 detected isomiRs (https://mcg.ustc.edu.cn/bsc/isomir/) (39), gives an estimate of the extent of
additional layers of regulation that this editing process can
generate. Although, isomiRs were first dismissed as artifacts,
follow-up research has shown that almost half of miRNA transcripts
are edited, and these edited transcripts can be loaded into the
RNA-induced silencing complex and exert their regulatory activity
(40). miRNA modifications
happen either in the 3′-end or in the 5′-end sequences. Although,
3′-end editing is more prevalent (41), it mostly influences miRNA
stability and activity. 5′-end editing, on the other hand,
introduces modifications in the seed sequence, altering the target
set of the miRNA and regulating new pathways (42-45). Moreover, circRNA efficiency and
translation have been shown to be subject to regulation by distinct
RNA modifications, such as m6A, 5-methylcytosine
(m5C) and pseudouridylation (Ψ) modifications (46), as was also the case for numerous
lncRNAs, which have been found to have roles in various CVD-related
pathways, such as atherosclerosis and pulmonary hypertension
(47).
In the present study, the epitranscriptomic
modifications are classified into three major categories. The most
prevalent form of epitranscriptomic modification, as in
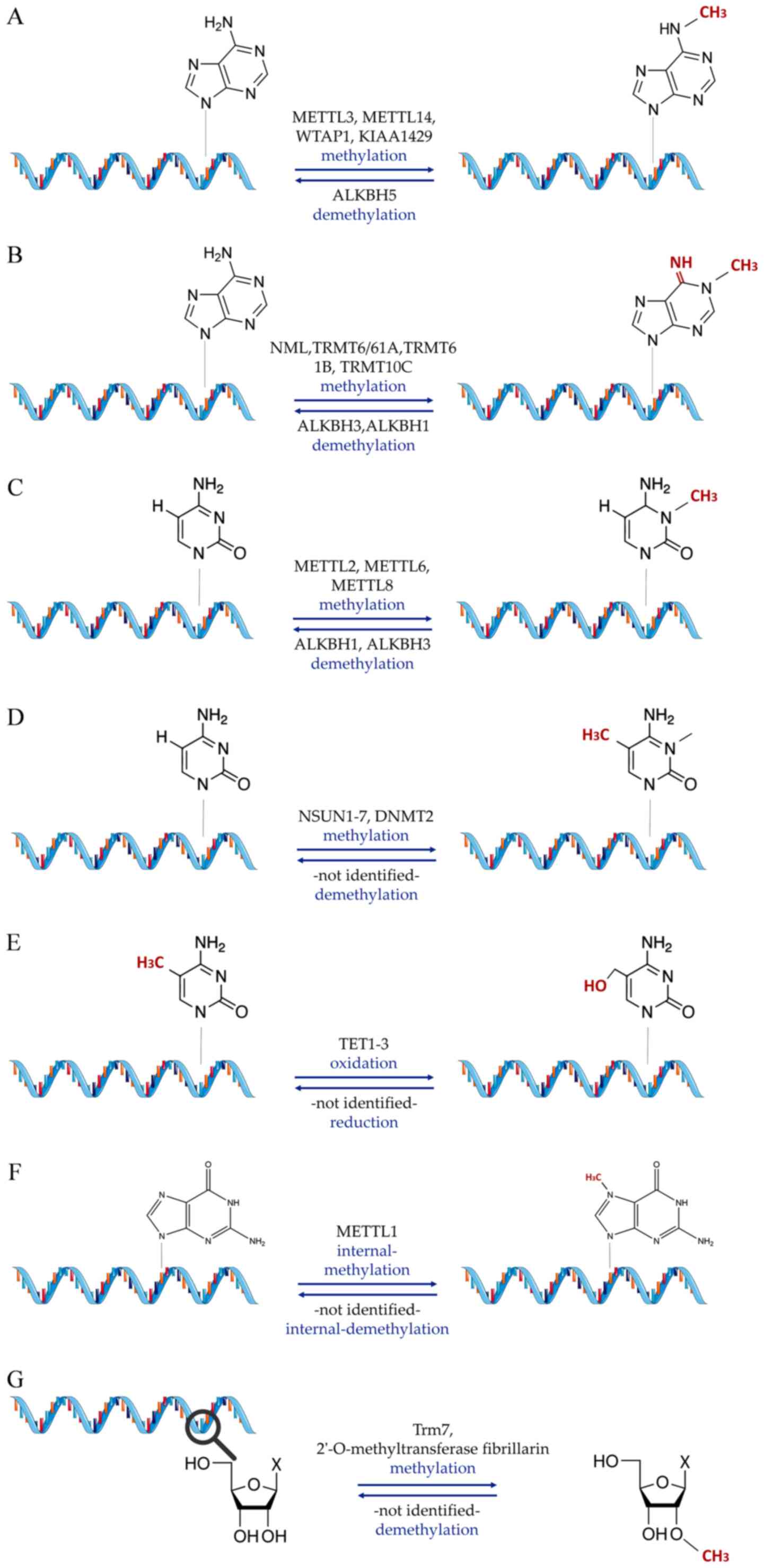
epigenetics, is RNA methylation (Fig. 1), which can affect adenosines in
different positions [N1-methyladenosine
(m1A), m6A, 2′-O-methylation (Nm)], cytosines
[m5C, 5-hydroxymethylcytosine (hm5C)] or
guanosines [7-methylguanosine (m7G)] (48). The second group encompasses
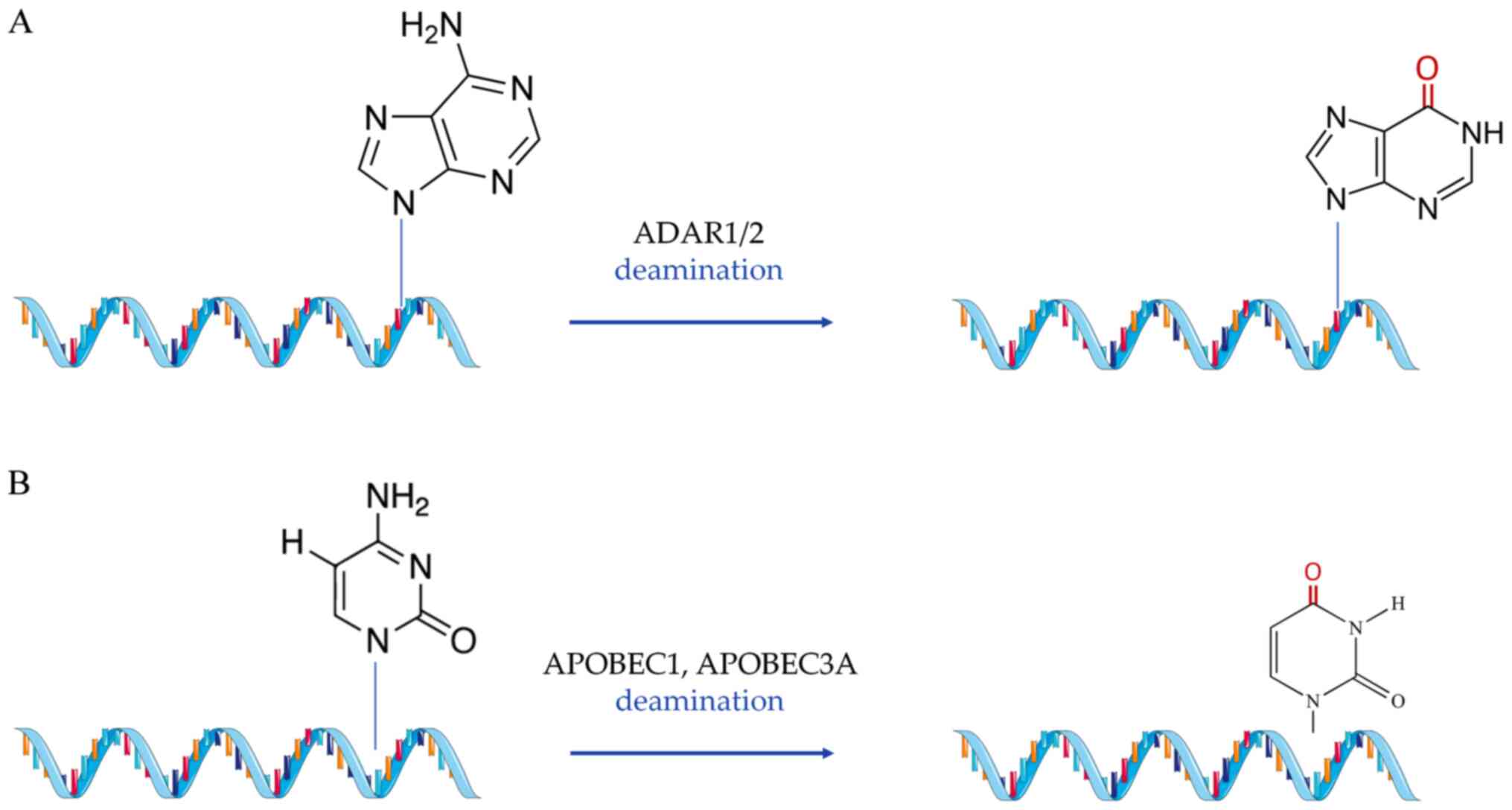
substitutional modifications (Fig.
2), which include A-to-I and C-to-U RNA editing (49,50). Finally, the third group of
modifications includes all epitranscriptomic changes that do not
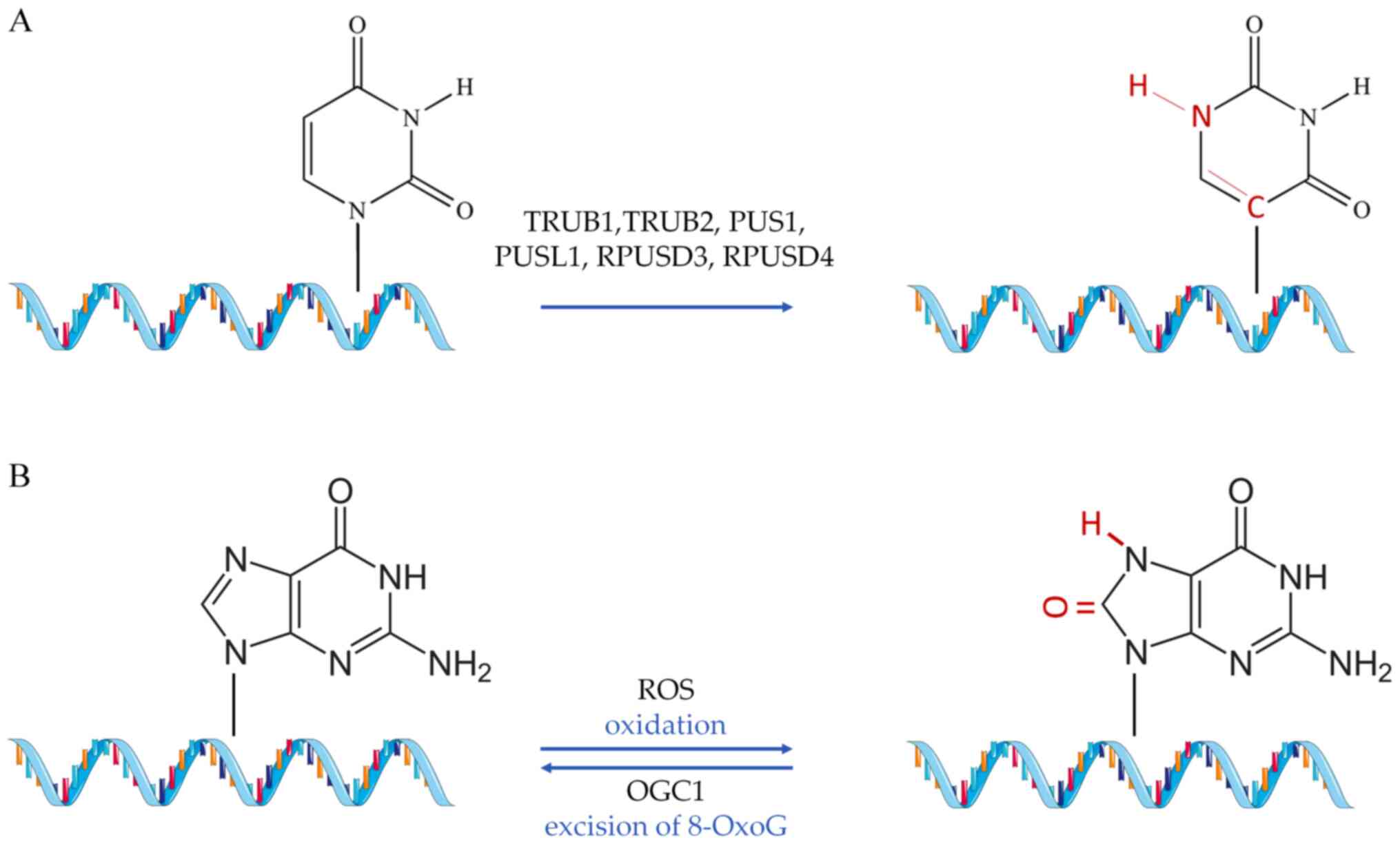
fall into any of the previous two categories [such as Ψ (51) and 8-oxoguanine (8-OxoG)], but
nevertheless, have a proven or implied role in CVD (52) (Fig. 3).
Nm (N meaning any nucleotide in this case) is a
modification of RNA occurring co-transcriptionally or
post-transcriptionally, where a methyl group is added to the
2′-hydroxyl of the ribose moiety (154). This type of modification is
recurrent and observed in numerous RNA classes, such as sncRNAs,
mRNA and tRNA (154). This
modification can be carried out by stand-alone methyltransferases
(155), such as tRNA
[cytidine(32)/guanosine(34)-2′-O]-methyltransferase
or by the fibrillarin enzyme, which requires guiding by box C/D
snoRNAs (156). Loss of
snoRNA-guided Nm modifications on snRNAs reportedly leads to
significant defects in the splicing of cardiac mRNA and the
development of the heart (157,158). In cardiometabolic disease,
small nucleolar RNA C/D Box 32A, a subtype of ncRNA from the Rpl13a
locus, was found to target the mRNA of peroxidasin for Nm,
indicating a role in the functional altering of peroxidase activity
in the heart (159). As in the
case of m7G modifications, research in this modification
area is still limited, but cardiovascular effect implications
exist, capable of driving future research avenues.
A-to-I RNA editing is the most common form of
substitutional RNA editing in mammals (160). During this process, two
conserved mammalian enzymes, adenosine deaminase acting on RNA
(ADAR)1 and 2, hydrolyze the adenosine residues into
double-stranded RNA regions (mRNAs and ncRNAs) in order to convert
them into inosines (161). Due
to the similar chemical content inosines share with guanines, they
are misread by the endogenous translational complex during reverse
transcription and thus pair with cytosines (162). A-to-I editing has been shown to
be indispensable both for physiological development and the
emergence of pathological conditions in the heart, while an average
of ~80,000 A-to-I editing sites have been identified in human
cardiac tissue (163).
ADAR1 has been shown to edit cathepsin S (CTSS), a
cysteine protease associated with atherosclerosis and angiogenesis
(164-166). The editing event occurred in
the 3′-UTR region of CTSS mRNA, enabling the recruitment of ELAV
like RNA binding protein 1, which in turn regulated the mRNA
expression and stability of CTSS. Of note, both ADAR1-mediated
editing and CTSS mRNA expression were elevated in blood samples
from patients with coronary artery disease (167). In mice, ADAR1 was increased
during oxidative stress in neonatal cardiomyocytes (168), while a knockout study in the
developing heart showed that ADAR1 cardiac deletion is associated
with embryonic lethality, establishing the importance of A-to-I RNA
editing during cardiac embryonic development for both proliferation
and survival (169).
Additionally, El Azzouzi et al (170) were able to bypass embryonic
lethality and knock out ADAR1 in adult cardiomyocytes by using an
inducible knockout method under the control of the α-myosin heavy
chain promoter, which is specifically expressed in cardiomyocytes.
Their results showed increased lethality in Adar1-null mice,
accompanied by a decrease in global miRNA expression, worsening of
cardiac function and severe ventricular remodeling, via a pathway
involving miR-199a-5p and the unfolded protein response (170). In a study by van der Kwast
et al (44), an edited
version of miR-478b-3p, a miRNA present in smooth muscle cells,
fibroblasts and vascular endothelial cells, was responsible for
neovascularization in response to ischemia. The A-to-I modification
of miR-478b-3p was located in the seed sequence and modified its
target set by enriching for proangiogenic pathways (44). Moreover, Filamin A (FLNA) mRNA
has been previously shown to be one of the substrates for ADAR2
editing (171). In a study by
Jain et al (172), mice
with impaired FLNA editing developed left ventricular hypertrophy
and cardiac remodeling, accompanied by elevated blood pressure.
Additionally, FLNA mRNA editing in patients with CVD was found to
be decreased by up to 50%, making ADAR2-mediated FLNA mRNA editing
one of the first studies to highlight an editing event associated
with cardiac disease in humans (172). In terms of occurrence, there is
limited information about A-to-I modifications in lncRNAs.
Nevertheless, ANRIL, a lncRNA acting as a regulator of coronary
heart disease, was shown to undergo A-to-I editing at the site of
its Alu motifs, potentially affecting its interaction with
chromatin and its downstream effects (47).
Although the activity of APOBEC1 is responsible for
editing apolipoprotein B, it has been previously reported that
another member of the subfamily, APOBEC2, is exclusively expressed
in the heart and skeletal muscle, and maintains low, but definite
deaminase activity (182).
Meanwhile, APOBEC3A is capable of C-to-U editing under hypoxic
conditions (183), while its
overexpression induced editing, among others, of primary pulmonary
hypertension genes in an in vitro experiment (184). All of the above imply a yet
undiscovered potential for RNA editing of cardiac-specific
transcripts in a C-to-U editing manner, similar to the one observed
for apolipoprotein modifications in the liver.
As in the previous modifications, the mutations in
PUS are related to various diseases, such as cancer and
mitochondrial myopathy (195,196). Of note, the absence of eraser
proteins for the Ψ modifications, coupled with the inactivity of
the C-C bond between the base and the sugar (Ψ), suggest that this
is a potentially irreversible modification (188). Analysis of TRUB1 levels in
human tissue revealed its high expression in the heart and skeletal
muscle, with still unexplored modification potential mainly in
tRNAs (197). Moreover, during
both Ψ and Nm methylation modifications, snoRNAs have been found to
act as guides for the modification process (159,198). A special class of guide RNAs
concentrated in the Cajal body are responsible for guiding
spliceosomal U modifications, these snRNAs are termed scaRNAs
(199). In this regard, scaRNAs
are responsible for regulated alternative splicing, with extensive
implications for response to variable environmental conditions
(158). Notably, in a study by
Nagasawa et al (200),
infants born with a common congenital cardiac defect termed
Tetralogy of Fallot, were shown to have decreased spliceosomal
pseudouridylation levels in their right ventricle, which in turn
depended on scaRNA1 levels, as exhibited in an in vitro
experiment in primary cardiomyocytes (200). These findings imply that
spliceosomal pseudouridylation depends on scaRNA levels in human
tissue, revealing a novel potential regulatory mechanism for the
alternative splicing of genes important in embryogenesis and
cardiogenesis. KCNQ1 overlapping transcript 1, a lncRNA and a
biomarker for MI, has also been shown to be able to be modified by
Ψ (201). Establishing studies
with a larger number of samples and the examination of additional
RNA modifications and epigenetic factors is necessary for deeper
investigation into the cardiovascular effects of
pseudouridylation.
Finally, 8-OxoG is conventionally formed through
the interaction of the guanine base in DNA molecules with reactive
oxygen species, under conditions of oxidative stress (202). Repair of this type of base
lesion is executed by the enzyme 8-OxoG glycosylase (OGG1), which
excises 8-OxoG (203). A study
by Shah et al (204)
documented the detrimental effects of 8-OxoG on the function of
vascular smooth cells, reporting a reduction of human
atherosclerotic plaque development when the activity of 8-OxoG
glycosylase was restored. By sequencing oxidized miRNAs in rat
models, 8-OxoG modifications at specific positions in miR-1 were
found to promote cardiac hypertrophy (205). Additionally, 8-OxoG DNA
glycosylase 1 overexpression was found to lower cardiac
mitochondrial levels of DNA 7,8-dihydro-8-OxoG (8-oxo-dG) in mouse
models (206). The same study
evidenced the decrease in transverse aortic constriction-induced
cardiac fibrosis in a state of OGG1 overexpression, suggesting that
increased repair of 8-oxo-dG in mtDNA leads to decreased cardiac
pathology (206). In a study by
Noren Hooten et al (207), 8-oxo-dG levels were found to be
associated with clinical cardiovascular risk factors, such as high
sensitivity C-reactive protein, systolic blood pressure, IL-23
levels and body/mass index. Moreover, strong association between
8-oxo-dG and the levels of systolic blood pressure have been
documented (207). Although
there are implications for important regulatory effects mediated by
8-oxo-dG modification in the cardiac tissue, this field of research
remains in its infancy.
In the past decade, dramatic advances in the
development of powerful sequencing technologies have facilitated
transcriptomic investigation in a faster, more efficient and more
in-depth manner than ever before. Such advances have also assisted
greatly in the study of epigenetics and epitranscriptomics. The use
of RNases constitutes one of the earliest methods of mapping mRNA
modifications and still exhibits the highest sensitivity for
m6A mapping (208).
In the same manner, the more recent MAZTER-seq (209) and m6A-REF-seq
(210) technologies exploit the
discovery of methylation-blocked endoRNases. Another method, termed
site-specific cleavage and radioactive-labeling followed by
ligation-assisted extraction and thin-layer chromatography, also
known as SCARLET, utilizes site-specific cleavage and splint
ligation and has also been extensively used to detect
m6A modifications in both coding RNA and lncRNAs
(211). Furthermore, antibody
incorporating techniques have been established for the detection of
RNA modifications. These include the m6A-LAIC-seq or
m6A-level and isoform-characterization sequencing
method, which uses immunoprecipitation in total RNA samples
(56), as well as the widely
used MeRIP-Seq technology, which maps m6A-methylated RNA
through the use of m6A-specific antibodies (212). Combining the aforementioned
technique with Ab cross-linking, allowed the enhancement of the
resolution of the technique, giving rise to methylation
individual-nucleotide resolution UV cross-linking and
immunoprecipitation (124).
Applications of the same principle of cross-linking include
PA-m6A-seq (213),
but also m1A-MAP (110) in the case of m1A
modification mapping.
Alternative methods for the detection of RNA
modifications take advantage of chemical reactions that are limited
to a certain type of RNA modification, combining them with
short-read sequencing. RNA-BisSeq, aimed toward the mapping of
m5C, involves the chemical deamination of cytidines
except for m5C (118). Modern library preparation
protocols, yield RNA fragments with nucleotide modifications at the
5′- or 3′-end, which can be used for the enrichment of the RNA
fragments in RNA seq libraries (214,215). The Nm-Seq and RibOxi-Seq
techniques used to map internal Nm modifications entail the
treatment of RNA fragments with NaIO4 oxidation, which
along with additional steps, leads to the enrichment of
3′-Nm-containing fragments and improvement of the final
transcriptome-wide RNA analysis (166,216). Using the same principle,
RiboMethSeq is based on the protection of the phosphodiester bond
in RNA when Nm occurs at the 5′-neighboring ribose (217). Following alkaline hydrolysis,
library preparation and 5′- and 3′-extremity counting, the
aforementioned protection is translated into a signal.
Mass spectrometry (MS) has also been an invaluable
tool for RNA modification analysis. State-of-the-art MS methods are
being employed for the detection and quantification of chemical
modifications in RNA, yielding different types of information based
on the type of MS analysis (218). Top-down MS analysis can
identify and localize mass-altering RNA modifications in undigested
RNA, while also allowing de novo sequencing to be performed.
Nevertheless, non-altering mass modifications, such as
m1A, m6A and mass-silent modifications, such
as pseudouridine, remain a major challenge (219,220). Bottom-up MS is conducted for
the mass mapping of partially hydrolyzed RNA, and MS approaches can
generate oligonucleotides and sequencing ladders that can be
subsequently interpreted into RNA sequences and localization of the
modifications (221). Another
MS-based method is nucleoside MS, which is performed on complete
RNA hydrolysates, followed by liquid chromatography separation of
the nucleoside mixtures. While highly accurate for the detection of
chemical modifications, it cannot provide sequence information or
localization of the modification (222). Still, each method's advantages
can be combined to overcome limitations and drawbacks on
high-throughput RNA modification mapping, while appropriate
software for MS data processing should always be incorporated
(223).
A-to-I modifications are either investigated via
the traditional method of screening for A-to-G mismatches in
reverse transcribed RNAs (224), by the use of the more recently
developed inosine chemical erasing (ICE) methods, or by the use of
transgenic mice where ADAR knockdown is followed by
deep-sequencing. In the case of ICE, reverse transcription is
blocked by the formation of N1-cyanoethylinosine after
acrylonitrile processing. This method combined with deep-sequencing
gave rise to ICE-seq, for high-throughput investigation of A-to-I
modifications (225). In the
case of Ψ modification profiling, several high-throughput
sequencing techniques are utilized, wherein treatment with
N-cyclohe
xyl-N′-(2-morpholinoethyl)-carbodiimide-metho-p-toluenesulfonate
specifically modifies Ψ, G and U residues on RNA. Although the G
and U modifications are later removed, the chemically induced
modification on Ψ is stable and blocks reverse transcription
(226). Such methods include
Ψ-seq (227), PSI-seq (228), Pseudo-seq (229) and CeU-seq (230).
Novel sequencing approaches enable direct RNA
sequencing without amplification or cDNA conversion. The rapidly
developing technology of nanopore sequencers, such as the one
created by Oxford Nanopore Technologies (ONT), includes the use of
a synthetic membrane with embedded nanopores in an ionic solution
(231). As an ionic current
passes through the nanopore, an individual read is recorded by a
sensor and the corresponding data is acquired by the sequencer's
implemented software. Characteristic changes in the current reads
during the movement of a nucleic acid strand, as it traverses the
nanopore from one chamber to the other, enable the identification
of the strand's nucleic acid sequence, in a process known as
'base-calling' (232).
Nucleotide modifications in ONT reads can be determined with the
use of specialized software, such as Nanopolish and the ONT
integrated CpG-methylation calling software (233).
High-throughput sequencing techniques have not only
promoted the field of epitranscriptomic profiling, but have,
through Genome-wide Association Studies (GWAS), facilitated the
identification of single nucleotide polymorphisms (SNPs) in a
variety of diseases, including CVD (234). These studies have led to the
identification of >5,000 associations with CVD (https://www.ebi.ac.uk/gwas/) (235), exhibiting the importance of
SNPs in CVD emergence. Several databases have also been developed
in an effort to catalogue disease-associated polymorphisms that
affect epitranscriptomic modifications. These databases include
m6Avar (236) and
m6ASNP (237), both
of which catalogue m6A-related polymorphisms,
m7GHub (238)
focusing on m7G-related SNPs, RMDisease encompassing
>200,000 human SNPs that affect m6A, m1A,
m6Am, m5U, m7G, Ψ and Nm
modifications (239) and the
RNA Framework, which is a rounded toolkit for the analysis of
post-transcriptional modifications (240).
In terms of epitranscriptomic genetic variation,
research remains at an early stage. As expected due to the greater
emphasis given so far on m6A-related modifications, in
the context of CVD, a number of m6A-related SNPs have
been recognized as genetic variants associated with CVD. Multiple
GWAS studies by Mo et al (241) have paved the way in this field
and associated m6A-SNPs with a variety of CVD factors.
More specifically, m6A-SNPs were shown to be associated
with coronary artery disease (241) and have a potential role in the
regulation of blood pressure (242), as well as in the regulation of
lipid metabolism (243).
Furthermore, several m6A-related SNPs were found to
affect the expression of multiple disease-causing genes, with
potential adverse effects for ischemic stroke in humans (244). In this context, the genetic
variant rs12286, which is strongly associated with coronary artery
disease, was shown to be able to affect ADAMTS7 expression, by
regulating the upstream m6A methylation (241). Ali et al (113) analyzed the levels of
m1A/G methylation in mitochondrial-encoded RNA across
multiple tissue types, followed by the identification of overlaps
between peak associated nuclear variants and disease-associated
variants with significance on a genome-wide level. Nuclear genetic
variants (rs13874, rs1084535), which are associated with inferred
methylation levels at mt-RNR2 and several mt-tRNA P9 sites, were in
linkage disequilibrium (LD) with rs34080181, which has been linked
to atrial fibrillation (245).
Furthermore, the intronic variant in polyribonucleotide
nucleotidyltransferase 1 mitochondrial (rs2627773) that is
associated with inferred methylation levels of mt-RNR2, is in LD
with rs1975487, which is associated with diastolic pressure
(246). Franzén et al
(247) mapped A-to-I RNA
editing quantitative trait loci (edQTLs) in order to identify
clinical features associated with RNA editing. Subsequently, they
evaluated the disease relevance of RNA editing by intersecting the
edQTLs with GWAS data (247).
More specifically, the authors intersected edSNPs with lead SNPs
from published GWAS data. Of note, the rs10847434 SNP, which is
associated with coronary artery disease (248) had an edQTL with an editing site
in the 3′ exon of apolipoprotein C1 pseudogene 1, a locus that has
been linked to coronary artery disease (249). Additionally, the SNP rs4739066,
a polymorphism associated with MI (250), was also found to have two
edQTLs with editing sites in the 3′-UTR of the α-tocopherol
transfer protein gene, a gene associated with the level of severity
of atherosclerotic lesions in the area of the proximal aorta
(251). Taken together, the
aforementioned studies suggest that there is still a large
unexplored area of genetic variation related to CVD pathogenesis,
especially in regards to epitranscriptomic modifications.
Although the field of epitranscriptomics is still
in its infancy, there are already efforts being made to utilize
such new regulatory knowledge for the development of novel
therapeutic approaches, both for epigenetic and epitranscriptomic
modifications in the context of CVD (19). As previously discussed, most
epitranscriptomic research in CVD has so far been focused on
m6A modifications and, as such, methods have focused on
identifying ways to manipulate m6A methylation levels in
the context of various therapeutic approaches. In a seminal study
by Lu et al (252), it
was established that curcumin was able to attenuate the effects of
lipid metabolism disorder and increase total cholesterol in the
liver, via the increase of m6A methylation, suggesting a
protective role for this modification against hyperlipidemia.
Recently, a large scale epitranscriptomic study has been
established to identify IHD biomarkers in circulation, termed the
IHD-EPITRAN study. This study is expected to include 200 patients,
split into two cohorts of IHD and non-IHD patients, focusing on the
identification of m6A and A-to-I modification biomarkers
(253). Additionally, limited
approaches have also been taken in an effort to modulate
m6A demethylation. Inhibition of demethylation was
selectively blocked by the use of meclofenamic acid (MA) in
vitro (254), while using
the recently developed CRISPR-Cas13b technology, Li et al
(255) attempted to manipulate
m6A modified transcripts and specifically demethylate
m6A marks. In a similar manner, Cox et al
(256) developed the RNA
Editing for Programmable A to I Replacement system, also utilizing
CRISPR-Cas13b technology, to address disease-causing mutations.
Although such tools are still outside the reach of clinical
practice, as multiple technical and ethical concerns remain
unaddressed, they pave the way for the development of future
personalized CVD therapeutics.
The field of epitranscriptomics has been rapidly
emerging, as the focus regarding disease development, environmental
adaptation and homeostasis maintenance, shifts from the rigid
genomic structure to the much more dynamic transcriptomic
landscape. Although there have been major advances in
transcriptomic profiling, understanding the mechanisms in which the
transcriptome itself is differentially regulated through
modifications, will allow for the development of novel and precise
pharmacological interventions. The additional level of regulatory
sensitivity that epitranscriptomic modifications are shown to
offer, corresponds to the increased level of specificity required
for any successful therapeutic intervention. To date,
epitranscriptomic modifications are nearing 200, but not all of
them have been thoroughly evaluated, nor do they all appear with
equal frequency. Although epitranscriptomic research progresses
rapidly in the fields of cancer and neurodegenerative disorders
(257,258), in the context of CVD the number
of modifications that have a significant impact are just beginning
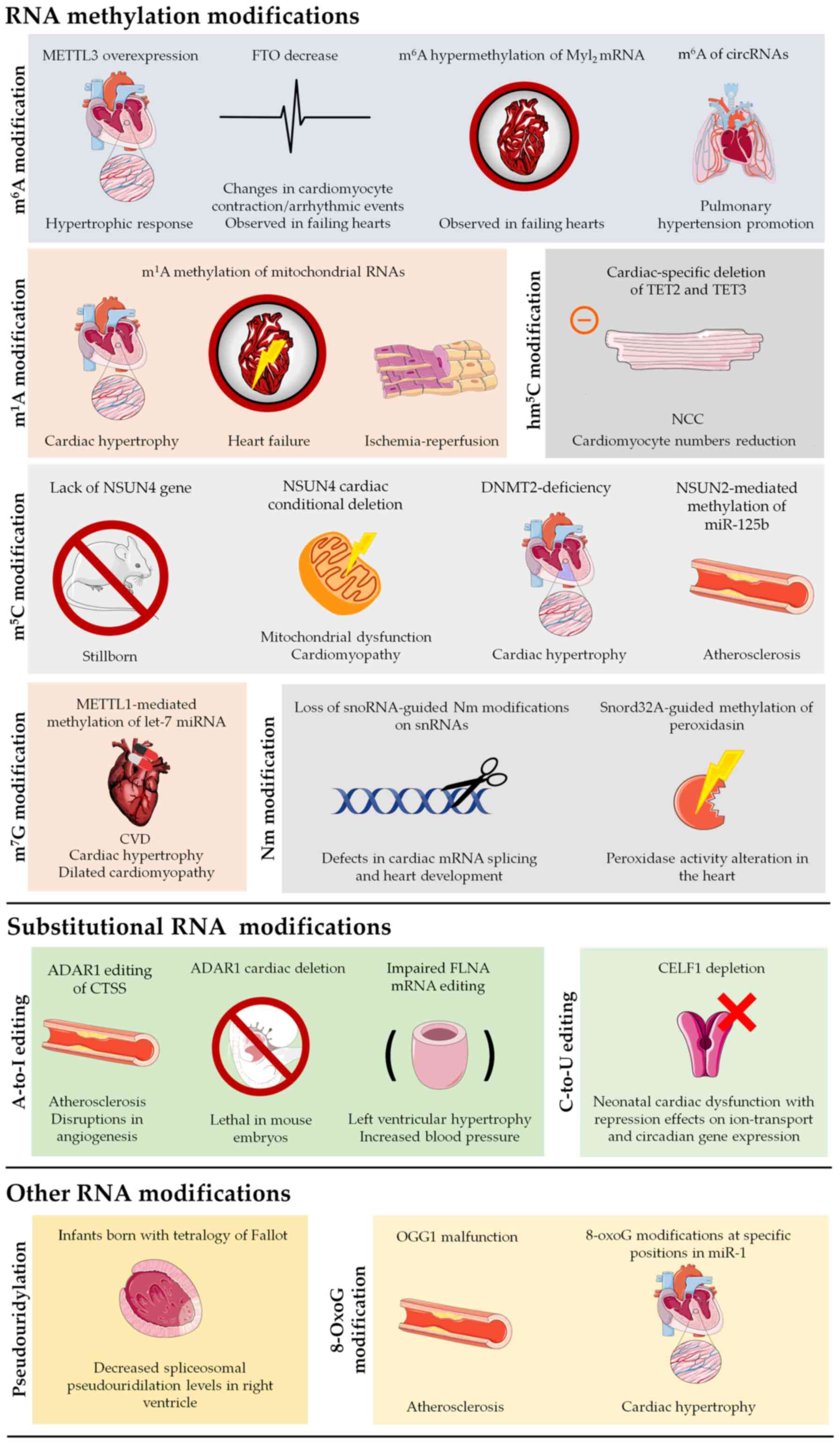
to be elucidated (Table I).
However, their biological and clinical significance cannot be
denied, as shown by the plethora of studies published in the past
couple of years, showing the effect of RNA modifications in CVDs
(Fig. 4).
In this context, cardiac aging has recently emerged
as an exciting new field, exploring among others, the possible
connection between RNA modifications and the various morphological
and biomolecular changes that take place during the cardiac aging
process. Increased cardiac fibrosis, left ventricular hypertrophy
and valvular degeneration are just some of the main physiological
changes that occur during human cardiac aging (261). Of note, cardiac fibrosis
emergence has been linked to changes in RNA modifications, such as
m6A and inosine (262), while cardiomyocyte aging has
been found to be affected by RNA methylation (263). Cardiomyocyte hypertrophy,
responsible for the thickening of the left ventricle walls during
cardiac aging, has also been linked to m6A methylation.
More specifically, in vitro and in vivo experiments
have shown that m6A RNA methylase METTL3 could promote
cardiomyocyte hypertrophy, whereas METTL3 inhibition inhibited the
hypertrophic potential of cardiomyocytes (78). All of the above provide just a
glimpse of the still unexplored effects that epitranscriptomic
regulation may potentially have during cardiac aging.
Finally, a large number of modifications have also
been detected in mitochondria. Due to the importance of
mitochondria for physiological cardiac function, these editing
events can have severe implications for both homeostasis and
disease emergence. HF, despite its various complications, has
historically been studied as a left ventricular disease. As such,
m1A modifications in mitochondrial 16s rRNA, as well as
tRNAs, but also components of the mitochondrial complex I (such as
mt-ND5) in the left ventricle, can have severe implications for
both HF and various other CVDs. Additionally, NSUN4-mediated
mitochondrial m5C methylation is required for
physiological function, as shown by the emergence of
cardiomyopathy, after NSUN4 cardiac-specific deletion. While
examining mitochondrial RNA methylation, Van Haute et al
(264) demonstrated NSUN3 as a
novel human m5C RNA methyltransferase, specializing in
mitochondrial tRNAMet. Mutations of NSUN3 caused reduced
methylation and absence of formulation of cytosine residues at
position 34 of the mitochondrial tRNAMet, leading to
reduced mitochondrial translation and the development of
mitochondrial disease. Thus, mitochondrial RNA methylation seems to
affect mitochondrial function as well as the translation of
mitochondrial proteins, leading to the emergence of pathology
(264). Even though other
mitochondrial RNA modifications have not been implicated in CVD, it
is safe to assume that we are only starting to scratch the surface,
as various more mitochondrial modifications have been described,
such as Ψ modifications. As already reviewed by Bohnsack and Sloan
(265), the mitochondrial
epitranscriptome is rapidly gaining interest as a key regulator of
dynamic, efficient and accurate responses to metabolic needs.
Mutations in mitochondrial RNase P protein 2, a mitochondrial RNase
P subcomplex cofactor, participating in m1A and m1G
mt-tRNA modifications, were shown to cause cardiomyopathy (106,266). Last, but not least, a few RNA
modifications, such as C-to-U editing by APOBEC3A or m6A
modification of the FTO protein were shown to be manifesting during
hypoxic conditions. Taking into account the extensive role of
hypoxia in metabolic regulation, mitochondrial biogenesis and
cardiac remodeling, such modifications further cement the role of
epitranscriptomic regulation in the adaptation to ever-changing
environmental stimuli both in physiological, but also in
pathological conditions.
This new knowledge is now paving the way towards a
new chapter in personalized medicine (267), where an in-depth understanding
of epitranscriptomic modifications could not only enable more
accurate patient classification based on epitranscriptomic
'profiles' or specific epitranscriptomic biomarkers, but, more
importantly, allow for early predictions of response to treatment.
Early evidence in this direction stems from the area of oncology,
where specific RNA modifications (e.g. m6A) appear to be
associated with therapeutic response and/or resistance (268). Significant promise also lies in
the development of novel targeted epitranscriptomic therapies
against CVD. The fine mapping of the role of epitranscriptomic
changes in different aspects of CVD development, is likely to
unveil a multitude of promising therapeutic targets, that could
subsequently be modulated by targeted approaches, such as small
molecule inhibitors. A number of such approaches are currently
being pursued against FTO in the milieu of cancer (269). For example, the US Food and
Drug Administration-approved nonsteroidal anti-inflammatory drug
ethyl ester form of MA, MA2, was found to be an FTO inhibitor,
which led to elevated levels of m6A modification in
mRNAs in glioblastoma cells, suppressing tumor progression and
prolonging the lifespan of glioblastoma stem cell-grafted mice
(270). MO-I-500 was developed
to selectively inhibit the m6A demethylase activity of
FTO and was found to successfully inhibit the survival and/or
colony formation of a triple-negative inflammatory breast cancer
cells (271,272). R-2HG was found to bind directly
to FTO and inhibit m6A demethylase activity leading to
the inhibition of leukemic cell growth/survival and leukemia
progression (273). FB23-2 was
also effective in inhibiting the progression of human AML in
xenotransplantation mice, by achieving the potent inhibition of FTO
(274). In an effort to
expedite the discovery of such inhibitors different predictive
in silico approaches are also employed (275-279). The majority of the
aforementioned approaches are based on conventional pipelines on
databases' data management (280,281). However, at present in the
post-genomic era, state-of-the-art approaches based on artificial
intelligence are being employed, thus providing novel and radical
solutions for the management and analysis of high amounts of data,
where algorithms and convolutional networks not only decipher
information by removing noise and reducing dimensionality, but also
produce new knowledge and associations (282). The tremendous clinical
potential of these advancements is supported by the early
establishment of multiple companies focusing on epitranscriptomics,
such as Accent, Gotham and Storm Therapeutics. It is only a matter
of time before similar avenues are explored in the field of CVD, as
evidenced by recent studies on epitranscriptomic modification-based
therapy solutions.
All of the above serve to show we currently stand
at the shore of cardiac epitranscriptomic research, where a vast
ocean of information still remains unexplored. We still have to
understand the relationship between the number of modifications
that each coding or ncRNA carries, to their respective effect, or
the methods of action for tissue-specific RNA modifications in
response to physiological or pathological stimuli. As we delve
deeper into CVD epitranscriptomics, we are sure to come closer to
the 'holy grail' of personalized medicine and targeted
therapeutics.
Not applicable.
SL, EP, KID, KP, TM, KD, FB, DS, GPC and DV
contributed to conceptualization, designing, writing, drafting,
revising, editing and reviewing of the manuscript. Data
authentication is not applicable. All authors have read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The authors would like to acknowledge funding from the following
organizations: i) CURE-PLaN grant from the Leducq Foundation for
Cardiovascular Research (grant no. 18CVD01); ii) AdjustEBOVGP-Dx
(grant no. RIA2018EF-2081): Biochemical Adjustments of native EBOV
Glycoprotein in Patient Sample to Unmask target Epitopes for Rapid
Diagnostic Testing. A European and Developing Countries Clinical
Trials Partnership (EDCTP2) under the Horizon 2020 'Research and
Innovation Actions' DESCA; and iii) 'MilkSafe: A novel pipeline to
enrich formula milk using omics technologies', a research
co-financed by the European Regional Development Fund of the
European Union and Greek national funds through the Operational
Program Competitiveness, Entrepreneurship and Innovation, under the
call RESEARCH-CREATE-INNOVATE (project no. T2EDK-02222).
|
1
|
Manzoni C, Kia DA, Vandrovcova J, Hardy J,
Wood NW, Lewis PA and Ferrari R: Genome, transcriptome and
proteome: The rise of omics data and their integration in
biomedical sciences. Brief Bioinform. 19:286–302. 2018. View Article : Google Scholar :
|
|
2
|
Buriani A, Fortinguerra S, Sorrenti V,
Gabbia D and Carrara M: Single-cell omics in personalized medicine.
Single-Cell Omics. Barh D and Azevedo V: Academic Press; Cambridge,
MA: pp. 221–236. 2019, View Article : Google Scholar
|
|
3
|
Korenke GC, Fuchs S, Krasemann E, Doerr
HG, Wilichowski E, Hunneman DH and Hanefeld F: Cerebral
adrenoleukodystrophy (ALD) in only one of monozygotic twins with an
identical ALD genotype. Ann Neurol. 40:254–257. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cavalli G and Heard E: Advances in
epigenetics link genetics to the environment and disease. Nature.
571:489–499. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sarkies P: Molecular mechanisms of
epigenetic inheritance: Possible evolutionary implications. Semin
Cell Dev Biol. 97:106–115. 2020. View Article : Google Scholar :
|
|
6
|
Qureshi IA and Mehler MF: Epigenetic
mechanisms underlying nervous system diseases. Handb Clin Neurol.
147:43–58. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schwartz S: Cracking the epitranscriptome.
RNA. 22:169–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boccaletto P, Machnicka MA, Purta E,
Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R,
Limbach PA, Kotter A, et al: MODOMICS: A database of RNA
modification pathways. 2017 update. Nucleic Acids Res.
46:D303–D307. 2018.
|
|
9
|
Czerwoniec A, Dunin-Horkawicz S, Purta E,
Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H and Rother K:
MODOMICS: A database of RNA modification pathways. 2008 update.
Nucleic Acids Res. 37:D118–D121. 2009. View Article : Google Scholar :
|
|
10
|
Dunin-Horkawicz S, Czerwoniec A, Gajda MJ,
Feder M, Grosjean H and Bujnicki JM: MODOMICS: A database of RNA
modification pathways. Nucleic Acids Res. 34:D145–D149. 2006.
View Article : Google Scholar :
|
|
11
|
Machnicka MA, Milanowska K, Osman Oglou O,
Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S,
Dunin-Horkawicz S, Rother KM, et al: MODOMICS: A database of RNA
modification pathways–2013 update. Nucleic Acids Res. 41:D262–D267.
2013. View Article : Google Scholar
|
|
12
|
Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S,
Zheng LL, Qu LH and Yang JH: RMBase v2.0: Deciphering the map of
RNA modifications from epitranscriptome sequencing data. Nucleic
Acids Res. 46:D327–D334. 2018. View Article : Google Scholar
|
|
13
|
Kiran AM, O'Mahony JJ, Sanjeev K and
Baranov PV: Darned in 2013: Inclusion of model organisms and
linking with Wikipedia. Nucleic Acids Res. 41:D258–D261. 2013.
View Article : Google Scholar :
|
|
14
|
Cantara WA, Crain PF, Rozenski J,
McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D and Agris
PF: The RNA Modification Database, RNAMDB: 2011 update. Nucleic
Acids Res. 39:D195–D201. 2011. View Article : Google Scholar
|
|
15
|
Picardi E, D'Erchia AM, Lo Giudice C and
Pesole G: REDIportal: A comprehensive database of A-to-I RNA
editing events in humans. Nucleic Acids Res. 45:D750–D757. 2017.
View Article : Google Scholar
|
|
16
|
Schuebel K, Gitik M, Domschke K and
Goldman D: Making sense of epigenetics. Int J Neuropsychopharmacol.
19:pyw0582016. View Article : Google Scholar
|
|
17
|
Kumari R, Ranjan P, Suleiman ZG, Goswami
SK, Li J, Prasad R and Verma SK: mRNA modifications in
cardiovascular biology and disease: With a focus on m6A
modification. Cardiovasc Res. May 6–2021.Epub ahead of print.
View Article : Google Scholar
|
|
18
|
Chen YS, Ouyang XP, Yu XH, Novák P, Zhou
L, He P and Yin K: N6-adenosine methylation (m(6)A) RNA
modification: An emerging role in cardiovascular diseases. J
Cardiovasc Transl Res. Feb 25–2021.Epub ahead of print. View Article : Google Scholar
|
|
19
|
Napoli C, Benincasa G, Donatelli F and
Ambrosio G: Precision medicine in distinct heart failure
phenotypes: Focus on clinical epigenetics. Am Heart J. 224:113–128.
2020. View Article : Google Scholar
|
|
20
|
Fischer MA and Vondriska TM: Clinical
epigenomics for cardiovascular disease: Diagnostics and therapies.
J Mol Cell Cardiol. 154:97–105. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schiano C, Benincasa G, Franzese M, Della
Mura N, Pane K, Salvatore M and Napoli C: Epigenetic-sensitive
pathways in personalized therapy of major cardiovascular diseases.
Pharmacol Ther. 210:1075142020. View Article : Google Scholar
|
|
22
|
Vlachakis C, Dragoumani K, Raftopoulou S,
Mantaiou M, Papageorgiou L, Champeris Tsaniras S, Megalooikonomou V
and Vlachakis D: Human emotions on the onset of cardiovascular and
small vessel related diseases. In Vivo. 32:859–870. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Timmis A, Townsend N, Gale CP, Torbica A,
Lettino M, Petersen SE, Mossialos EA, Maggioni AP, Kazakiewicz D,
May HT, et al: European Society of Cardiology: European Society of
Cardiology: Cardiovascular Disease Statistics 2019. Eur Heart J.
41:12–85. 2020. View Article : Google Scholar
|
|
24
|
Flora GD and Nayak MK: A brief review of
cardiovascular diseases, associated risk factors and current
treatment regimes. Curr Pharm Des. 25:4063–4084. 2019. View Article : Google Scholar
|
|
25
|
Baccarelli A, Rienstra M and Benjamin EJ:
Cardiovascular epigenetics: Basic concepts and results from animal
and human studies. Circ Cardiovasc Genet. 3:567–573. 2010.
View Article : Google Scholar
|
|
26
|
Zhong J, Agha G and Baccarelli AA: The
role of DNA methylation in cardiovascular risk and disease:
Methodological aspects, study design, and data analysis for
epidemiological studies. Circ Res. 118:119–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vlachakis D, Zacharaki EI, Tsiamaki E,
Koulouri M, Raftopoulou S, Papageorgiou L, Chrousos GP, Ellul J and
Megalooikonomou V: Insights into the molecular mechanisms of stress
and inflammation in ageing and frailty of the elderly. J Mol
Biochem. 6:41–44. 2017.
|
|
28
|
Guerra J, Valadao AL, Vlachakis D, Polak
K, Vila IK, Taffoni C, Prabakaran T, Marriott AS, Kaczmarek R,
Houel A, et al: Lysyl-tRNA synthetase produces diadenosine
tetraphosphate to curb STING-dependent inflammation. Sci Adv.
6:eaax33332020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Soler-Botija C, Gálvez-Montón C and
Bayés-Genís A: Epigenetic biomarkers in cardiovascular diseases.
Front Genet. 10:950. 2019. View Article : Google Scholar
|
|
30
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and eras. Mol Cell. 74:640–650. 2019. View Article : Google Scholar
|
|
31
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gatsiou A and Stellos K: Dawn of
epitranscriptomic medicine. Circ Genom Precis Med. 11:e0019272018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anreiter I, Mir Q, Simpson JT, Janga SC
and Soller M: New twists in detecting mRNA modification dynamics.
Trends Biotechnol. 39:72–89. 2021. View Article : Google Scholar
|
|
34
|
Mongelli A, Atlante S, Bachetti T,
Martelli F, Farsetti A and Gaetano C: Epigenetic signaling and RNA
regulation in cardiovascular diseases. Int J Mol Sci. 21:5092020.
View Article : Google Scholar :
|
|
35
|
Zhang P, Wu W, Chen Q and Chen M:
Non-coding RNAs and their integrated networks. J Integr Bioinform.
16:162019. View Article : Google Scholar
|
|
36
|
Gomes CPC, Schroen B, Kuster GM, Robinson
EL, Ford K, Squire IB, Heymans S, Martelli F and Emanueli C:
Regulatory RNAs in heart failure. Circulation. 141:313–328. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Esteller M and Pandolfi PP: The
epitranscriptome of noncoding RNAs in cancer. Cancer Discov.
7:359–368. 2017. View Article : Google Scholar
|
|
38
|
Blow MJ, Grocock RJ, van Dongen S, Enright
AJ, Dicks E, Futreal PA, Wooster R and Stratton MR: RNA editing of
human microRNAs. Genome Biol. 7:R272006. View Article : Google Scholar :
|
|
39
|
Zhang Y, Zang Q, Xu B, Zheng W, Ban R,
Zhang H, Yang Y, Hao Q, Iqbal F, Li A, et al: IsomiR Bank: A
research resource for tracking IsomiRs. Bioinformatics.
32:2069–2071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Neilsen CT, Goodall GJ and Bracken CP:
IsomiRs - the overlooked repertoire in the dynamic microRNAome.
Trends Genet. 28:544–549. 2012. View Article : Google Scholar
|
|
41
|
Newman MA, Mani V and Hammond SM: Deep
sequencing of microRNA precursors reveals extensive 3′ end
modification. RNA. 17:1795–1803. 2011. View Article : Google Scholar :
|
|
42
|
Cloonan N, Wani S, Xu Q, Gu J, Lea K,
Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al:
MicroRNAs and their isomiRs function cooperatively to target common
biological pathways. Genome Biol. 12:R1262011. View Article : Google Scholar
|
|
43
|
Manzano M, Forte E, Raja AN, Schipma MJ
and Gottwein E: Divergent target recognition by coexpressed
5′-isomiRs of miR-142-3p and selective viral mimicry. RNA.
21:1606–1620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
van der Kwast RV, van Ingen E, Parma L,
Peters HA, Quax PH and Nossent AY: Adenosine-to-inosine editing of
MicroRNA-487b alters target gene selection after ischemia and
promotes Neovascularization. Circ Res. 122:444–456. 2018.
View Article : Google Scholar
|
|
45
|
van der Kwast RV, Woudenberg T, Quax PHA
and Nossent AY: MicroRNA-411 and its 5′-IsomiR have distinct
targets and functions and are differentially regulated in the
vasculature under ischemia. Mol Ther. 28:157–170. 2020. View Article : Google Scholar
|
|
46
|
Gilbert WV, Bell TA and Schaening C:
Messenger RNA modifications: Form, distribution, and function.
Science. 352:1408–1412. 2016. View Article : Google Scholar :
|
|
47
|
Yin L, Zhu X, Novák P, Zhou L, Gao L, Yang
M, Zhao G and Yin K: The epitranscriptome of long noncoding RNAs in
metabolic diseases. Clin Chim Acta. 515:80–89. 2021. View Article : Google Scholar
|
|
48
|
Zhou Y, Kong Y, Fan W, Tao T, Xiao Q, Li N
and Zhu X: Principles of RNA methylation and their implications for
biology and medicine. Biomed Pharmacother. 131:1107312020.
View Article : Google Scholar
|
|
49
|
Wulff BE and Nishikura K: Substitutional
A-to-I RNA editing. Wiley Interdiscip Rev RNA. 1:90–101. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Blanc V and Davidson NO: C-to-U RNA
editing: Mechanisms leading to genetic diversity. J Biol Chem.
278:1395–1398. 2003. View Article : Google Scholar
|
|
51
|
Adachi H, De Zoysa MD and Yu YT:
Post-transcriptional pseudouridylation in mRNA as well as in some
major types of noncoding RNAs. Biochim Biophys Acta Gene Regul
Mech. 1862:230–239. 2019. View Article : Google Scholar
|
|
52
|
Martinet W, De Meyer GR, Herman AG and
Kockx MM: RNA damage in human atherosclerosis: Pathophysiological
significance and implications for gene expression studies. RNA
Biol. 2:4–7. 2005. View Article : Google Scholar
|
|
53
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar
|
|
54
|
Perry RPK and Kelley DE: Existence of
methylated messenger RNA in mouse L cells. Cell. 1:37–42. 1974.
View Article : Google Scholar
|
|
55
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar
|
|
56
|
Molinie B, Wang J, Lim KS, Hillebrand R,
Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon
P, et al: m(6)A-LAIC-seq reveals the census and complexity of the
m(6)A epitranscriptome. Nat Methods. 13:692–698. 2016. View Article : Google Scholar
|
|
57
|
Sun H, Li K, Zhang X, Liu J, Zhang M, Meng
H and Yi C: m6Am-seq reveals the dynamic m6Am methylation in the
human transcriptome. Nat Commun. 12:47782021. View Article : Google Scholar
|
|
58
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
60
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)a promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
Modulates Messenger RNA Translation Efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar
|
|
62
|
Kierzek E and Kierzek R: The thermodynamic
stability of RNA duplexes and hairpins containing
N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic
Acids Res. 31:4472–4480. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N(6)-methyladenosine-dependent RNA structural switches
regulate RNA-protein interactions. Nature. 518:560–564. 2015.
View Article : Google Scholar :
|
|
64
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao
YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A
reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Balatsos NA, Vlachakis D, Maragozidis P,
Manta S, Anastasakis D, Kyritsis A, Vlassi M, Komiotis D and
Stathopoulos C: Competitive inhibition of human poly(A)-specific
ribonuclease (PARN) by synthetic fluoro-pyranosyl nucleosides.
Biochemistry. 48:6044–6051. 2009. View Article : Google Scholar
|
|
66
|
Balatsos N, Vlachakis D, Chatzigeorgiou V,
Manta S, Komiotis D, Vlassi M and Stathopoulos C: Kinetic and in
silico analysis of the slow-binding inhibition of human
poly(A)-specific ribonuclease (PARN) by novel nucleoside analogues.
Biochimie. 94:214–221. 2012. View Article : Google Scholar
|
|
67
|
Fustin JM, Doi M, Yamaguchi Y, Hida H,
Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I,
et al: RNA-methylation-dependent RNA processing controls the speed
of the circadian clock. Cell. 155:793–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xiang Y, Laurent B, Hsu CH, Nachtergaele
S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al: RNA m6A
methylation regulates the ultraviolet-induced DNA damage response.
Nature. 543:573–576. 2017. View Article : Google Scholar :
|
|
69
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
70
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification destabilizes
developmental regulators in embryonic stem cells. Nat Cell Biol.
16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar :
|
|
72
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar :
|
|
73
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Meyer KD and Jaffrey SR: Rethinking m6A
readers, writers, and eras. Annu Rev Cell Dev Biol. 33:319–342.
2017. View Article : Google Scholar
|
|
75
|
Longenecker JZ, Gilbert CJ, Golubeva VA,
Martens CR and Accornero F: Epitranscriptomics in the heart: A
focus on m6A. Curr Heart Fail Rep. 17:205–212. 2020. View Article : Google Scholar
|
|
76
|
Qin Y, Li L, Luo E, Hou J, Yan G, Wang D,
Qiao Y and Tang C: Role of m6A RNA methylation in cardiovascular
disease (Review). Int J Mol Med. 46:1958–1972. 2020. View Article : Google Scholar
|
|
77
|
Zheng N, Su J, Hu H, Wang J and Chen X:
Research progress of N6-methyladenosine in the cardiovascular
system. Med Sci Monit. 26. pp. e9217422020, View Article : Google Scholar
|
|
78
|
Dorn LE, Lasman L, Chen J, Xu X, Hund TJ,
Medvedovic M, Hanna JH, van Berlo JH and Accornero F: The
N6-methyladenosine mRNA methylase METTL3 controls cardiac
homeostasis and hypertrophy. Circulation. 139:533–545. 2019.
View Article : Google Scholar :
|
|
79
|
Song H, Pu J, Wang L, Wu L, Xiao J, Liu Q,
Chen J, Zhang M, Liu Y, Ni M, et al: ATG16L1 phosphorylation is
oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein
phosphatase 1 which determines the fate of cardiomyocytes during
hypoxia/reoxygenation. Autophagy. 11:1308–1325. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Pastore N, Brady OA, Diab HI, Martina JA,
Sun L, Huynh T, Lim JA, Zare H, Raben N, Ballabio A, et al: TFEB
and TFE3 cooperate in the regulation of the innate immune response
in activated macrophages. Autophagy. 12:1240–1258. 2016. View Article : Google Scholar
|
|
81
|
Zhao E and Czaja MJ: Transcription factor
EB: A central regulator of both the autophagosome and lysosome.
Hepatology. 55:1632–1634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Song H, Feng X, Zhang H, Luo Y, Huang J,
Lin M, Jin J, Ding X, Wu S, Huang H, et al: METTL3 and ALKBH5
oppositely regulate m6A modification of TFEB mRNA, which dictates
the fate of hypoxia/reoxygenation-treated cardiomyocytes.
Autophagy. 15:1419–1437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kmietczyk V, Riechert E, Kalinski L,
Boileau E, Malovrh E, Malone B, Gorska A, Hofmann C, Varma E,
Jürgensen L, et al: m6A-mRNA methylation regulates cardiac gene
expression and cellular growth. Life Sci Alliance.
2:e2018002332019. View Article : Google Scholar :
|
|
84
|
Berulava T, Buchholz E, Elerdashvili V,
Pena T, Islam MR, Lbik D, Mohamed BA, Renner A, von Lewinski D,
Sacherer M, et al: Changes in m6A RNA methylation contribute to
heart failure progression by modulating translation. Eur J Heart
Fail. 22:54–66. 2020. View Article : Google Scholar
|
|
85
|
Fischer J, Koch L, Emmerling C, Vierkotten
J, Peters T, Brüning JC and Rüther U: Inactivation of the Fto gene
protects from obesity. Nature. 458:894–898. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-dependent N6-methyladenosine regulates
cardiac function during remodeling and repair. Circulation.
139:518–532. 2019. View Article : Google Scholar
|
|
87
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Shihong Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-dependent m6A regulates cardiac function
during remodeling and repair. Circulation. 139:518–532. 2019.
View Article : Google Scholar
|
|
88
|
Michlewski G and Cáceres JF:
Post-transcriptional control of miRNA biogenesis. RNA. 25:1–16.
2019. View Article : Google Scholar :
|
|
89
|
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH,
Wang F, Wang TT, Xu QG, Zhou WP and Sun SH: METTL14 suppresses the
metastatic potential of hepatocellular carcinoma by modulating
N6-methyladenosine-dependent primary MicroRNA processing.
Hepatology. 65:529–543. 2017. View Article : Google Scholar
|
|
90
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu
HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor
proliferation of bladder cancer by accelerating pri-miR221/222
maturation in m6A-dependent manner. Mol Cancer. 18:1102019.
View Article : Google Scholar :
|
|
91
|
Baarsma H and Königshoff M: 'WNT-er is
coming': WNT signalling in chronic lung diseases. Thorax.
72:746–759. 2017. View Article : Google Scholar
|
|
92
|
Savai R, Al-Tamari HM, Sedding D,
Kojonazarov B, Muecke C, Teske R, Capecchi MR, Weissmann N,
Grimminger F, Seeger W, et al: Pro-proliferative and inflammatory
signaling converge on FoxO1 transcription factor in pulmonary
hypertension. Nat Med. 20:1289–1300. 2014. View Article : Google Scholar
|
|
93
|
Lin B, Xu J, Wang F, Wang J, Zhao H and
Feng D: LncRNA XIST promotes myocardial infarction by regulating
FOS through targeting miR-101a-3p. Aging (Albany NY). 12:7232–7247.
2020. View Article : Google Scholar
|
|
94
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar
|
|
95
|
Sun R and Zhang L: Long non-coding RNA
MALAT1 regulates cardiomyocytes apoptosis after hypoxia/reperfusion
injury via modulating miR-200a-3p/PDCD4 axis. Biomed Pharmacother.
111:1036–1045. 2019. View Article : Google Scholar
|
|
96
|
Liu N, Parisien M, Dai Q, Zheng G, He C
and Pan T: Probing N6-methyladenosine RNA modification status at
single nucleotide resolution in mRNA and long noncoding RNA. RNA.
19:1848–1856. 2013. View Article : Google Scholar
|
|
97
|
Spitale RC, Flynn RA, Zhang QC, Crisalli
P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET,
et al: Erratum: Structural imprints in vivo decode RNA regulatory
mechanisms. Nature. 527:2642015. View Article : Google Scholar
|
|
98
|
Coker H, Wei G and Brockdorff N: m6A
modification of non-coding RNA and the control of mammalian gene
expression. Biochim Biophys Acta Gene Regul Mech. 1862:310–318.
2019. View Article : Google Scholar
|
|
99
|
Mendel M, Chen KM, Homolka D, Gos P,
Pandey RR, McCarthy AA and Pillai RS: Methylation of structured RNA
by the m6A writer METTL16 is essential for mouse embryonic
development. Mol Cell. 71:986–1000.e11. 2018. View Article : Google Scholar :
|
|
100
|
Brown JA, Kinzig CG, DeGregorio SJ and
Steitz JA: Methyltransferase-like protein 16 binds the 3′-terminal
triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA.
113:14013–14018. 2016. View Article : Google Scholar
|
|
101
|
Dunn DB: The occurrence of 1-methyladenine
in ribonucleic acid. Biochim Biophys Acta. 46:198–200. 1961.
View Article : Google Scholar
|
|
102
|
Agris PF: The importance of being
modified: Roles of modified nucleosides and Mg2+ in RNA
structure and function. Prog Nucleic Acid Res Mol Biol. 53:79–129.
1996. View Article : Google Scholar
|
|
103
|
Xiong X, Li X and Yi C: N1-methyladenosine
methylome in messenger RNA and non-coding RNA. Curr Opin Chem Biol.
45:179–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Waku T, Nakajima Y, Yokoyama W, Nomura N,
Kako K, Kobayashi A, Shimizu T and Fukamizu A: NML-mediated rRNA
base methylation links ribosomal subunit formation to cell
proliferation in a p53-dependent manner. J Cell Sci. 129:2382–2393.
2016.PubMed/NCBI
|
|
105
|
Chujo T and Suzuki T: Trmt61B is a
methyltransferase responsible for 1-methyladenosine at position 58
of human mitochondrial tRNAs. RNA. 18:2269–2276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Vilardo E, Nachbagauer C, Buzet A,
Taschner A, Holzmann J and Rossmanith W: A subcomplex of human
mitochondrial RNase P is a bifunctional methyltransferase -
extensive moon-lighting in mitochondrial tRNA biogenesis. Nucleic
Acids Res. 40:11583–11593. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ozanick S, Krecic A, Andersland J and
Anderson JT: The bipartite structure of the tRNA m1A58
methyltransferase from S. cerevisiae is conserved in humans. RNA.
11:1281–1290. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhang C and Jia G: Reversible RNA
modification N1-methyladenosine (m1A) in mRNA and tRNA. Genomics
Proteomics Bioinformatics. 16:155–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Shi Q, Xue C, Yuan X, He Y and Yu Z: Gene
signatures and prognostic values of m1A-related regulatory genes in
hepatocellular carcinoma. Sci Rep. 10:150832020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Li X, Xiong X, Zhang M, Wang K, Chen Y,
Zhou J, Mao Y, Lv J, Yi D, Chen XW, et al: Base-resolution mapping
reveals distinct m1A methylome in nuclear- and
mitochondrial-encoded transcripts. Mol Cell. 68:993–1005.e9. 2017.
View Article : Google Scholar
|
|
111
|
Siasos G, Tsigkou V, Kosmopoulos M,
Theodosiadis D, Simantiris S, Tagkou NM, Tsimpiktsioglou A,
Stampouloglou PK, Oikonomou E, Mourouzis K, et al: Mitochondria and
cardiovascular diseases-from pathophysiology to treatment. Ann
Transl Med. 6:2562018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Poznyak AV, Ivanova EA, Sobenin IA, Yet SF
and Orekhov AN: The role of mitochondria in cardiovascular
diseases. Biology (Basel). 9:92020.
|
|
113
|
Ali AT, Idaghdour Y and Hodgkinson A:
Analysis of mitochondrial m1A/G RNA modification reveals links to
nuclear genetic variants and associated disease processes. Commun
Biol. 3:1472020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Xu L, Liu X, Sheng N, Oo KS, Liang J,
Chionh YH, Xu J, Ye F, Gao YG, Dedon PC, et al: Three distinct
3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in
mice and humans. J Biol Chem. 292:14695–14703. 2017. View Article : Google Scholar
|
|
115
|
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J,
Lu Z, Zheng Z, Dai Q and Wang H: Transfer RNA demethylase ALKBH3
promotes cancer progression via induction of tRNA-derived small
RNAs. Nucleic Acids Res. 47:2533–2545. 2019. View Article : Google Scholar
|
|
116
|
Ma CJ, Ding JH, Ye TT, Yuan BF, Feng YQ
and Alk B: AlkB homologue 1 demethylates N3-methylcytidine in mRNA
of mammals. ACS Chem Biol. 14:1418–1425. 2019. View Article : Google Scholar
|
|
117
|
Lentini JM, Alsaif HS, Faqeih E, Alkuraya
FS and Fu D: DALRD3 encodes a protein mutated in epileptic
encephalopathy that targets arginine tRNAs for 3-methylcytosine
modification. Nat Commun. 11:25102020. View Article : Google Scholar :
|
|
118
|
Yang X, Yang Y, Sun BF, Chen YS, Xu JW,
Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al: 5-methylcytosine
promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as
an m5C reader. Cell Res. 27:606–625. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Gambaryan AS, Venkstern TV and Baev AA:
Use of the method of mixed substrates to study the specificity of
tRNA methylases. Mol Biol (Mosk). 10:697–705. 1976.
|
|
120
|
Amort T, Rieder D, Wille A,
Khokhlova-Cubberley D, Riml C, Trixl L, Jia XY, Micura R and Lusser
A: Distinct 5-methylcytosine profiles in poly(A) RNA from mouse
embryonic stem cells and brain. Genome Biol. 18:12017. View Article : Google Scholar
|
|
121
|
Squires JE, Patel HR, Nousch M, Sibbritt
T, Humphreys DT, Parker BJ, Suter CM and Preiss T: Widespread
occurrence of 5-methylcytosine in human coding and non-coding RNA.
Nucleic Acids Res. 40:5023–5033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Sharma S, Yang J, Watzinger P, Kötter P
and Entian KD: Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the
25S rRNA, respectively. Nucleic Acids Res. 41:9062–9076. 2013.
View Article : Google Scholar :
|
|
123
|
Brzezicha B, Schmidt M, Makalowska I,
Jarmolowski A, Pienkowska J and Szweykowska-Kulinska Z:
Identification of human tRNA:m5C methyltransferase catalysing
intron-dependent m5C formation in the first position of the
anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res.
34:6034–6043. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Hussain S, Sajini AA, Blanco S, Dietmann
S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et
al: NSun2-mediated cytosine-5 methylation of vault noncoding RNA
determines its processing into regulatory small RNAs. Cell Rep.
4:255–261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Haag S, Sloan KE, Ranjan N, Warda AS,
Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S,
Rehling P, et al: NSUN3 and ABH1 modify the wobble position of
mt-tRNAMet to expand codon recognition in mitochondrial
translation. EMBO J. 35:2104–2119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Metodiev MD, Spåhr H, Loguercio Polosa P,
Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG and
Ruzzenente B: NSUN4 is a dual function mitochondrial protein
required for both methylation of 12S rRNA and coordination of
mitoribosomal assembly. PLoS Genet. 10:e10041102014. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Schosserer M, Minois N, Angerer TB, Amring
M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP,
Pfeifenberger S, et al: Methylation of ribosomal RNA by NSUN5 is a
conserved mechanism modulating organismal lifespan. Nat Commun.
6:61582015. View Article : Google Scholar
|
|
128
|
Haag S, Warda AS, Kretschmer J, Günnigmann
MA, Höbartner C and Bohnsack MT: NSUN6 is a human RNA
methyltransferase that catalyzes formation of m5C72 in specific
tRNAs. RNA. 21:1532–1543. 2015. View Article : Google Scholar :
|
|
129
|
Kaiser S, Jurkowski TP, Kellner S,
Schneider D, Jeltsch A and Helm M: The RNA methyltransferase Dnmt2
methylates DNA in the structural context of a tRNA. RNA Biol.
14:1241–1251. 2017. View Article : Google Scholar
|
|
130
|
Goll MG, Kirpekar F, Maggert KA, Yoder JA,
Hsieh CL, Zhang X, Golic KG, Jacobsen SE and Bestor TH: Methylation
of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science.
311:395–398. 2006. View Article : Google Scholar
|
|
131
|
Abbasi-Moheb L, Mertel S, Gonsior M,
Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM,
Esmaeeli-Nieh S, Cremer K, et al: Mutations in NSUN2 cause
autosomal-recessive intellectual disability. Am J Hum Genet.
90:847–855. 2012. View Article : Google Scholar
|
|
132
|
Frye M and Watt FM: The RNA
methyltransferase Misu (NSun2) mediates Myc-induced proliferation
and is upregulated in tumors. Curr Biol. 16:971–981. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Yi J, Gao R, Chen Y, Yang Z, Han P, Zhang
H, Dou Y, Liu W, Wang W, Du G, et al: Overexpression of NSUN2 by
DNA hypomethylation is associated with metastatic progression in
human breast cancer. Oncotarget. 8:20751–20765. 2017. View Article : Google Scholar
|
|
134
|
Forbes SA, Beare D, Boutselakis H, Bamford
S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al:
COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids
Res. 45:D777–D783. 2017. View Article : Google Scholar
|
|
135
|
Elhardt W, Shanmugam R, Jurkowski TP and
Jeltsch A: Somatic cancer mutations in the DNMT2 tRNA
methyltransferase alter its catalytic properties. Biochimie.
112:66–72. 2015. View Article : Google Scholar
|
|
136
|
Ghanbarian H, Wagner N, Polo B, Baudouy D,
Kiani J, Michiels JF, Cuzin F, Rassoulzadegan M and Wagner KD:
Dnmt2/Trdmt1 as mediator of RNA polymerase II transcriptional
activity in cardiac growth. PLoS One. 11:e01569532016. View Article : Google Scholar :
|
|
137
|
Luo Y, Feng J, Xu Q, Wang W and Wang X:
NSun2 deficiency protects endothelium from inflammation via mRNA
methylation of ICAM-1. Circ Res. 118:944–956. 2016. View Article : Google Scholar
|
|
138
|
Wang N, Tang H, Wang X, Wang W and Feng J:
Homocysteine upregulates interleukin-17A expression via
NSun2-mediated RNA methylation in T lymphocytes. Biochem Biophys
Res Commun. 493:94–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Yuan S, Tang H, Xing J, Fan X, Cai X, Li
Q, Han P, Luo Y, Zhang Z, Jiang B, et al: Methylation by NSun2
represses the levels and function of microRNA 125b. Mol Cell Biol.
34:3630–3641. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wen P, Cao H, Fang L, Ye H, Zhou Y, Jiang
L, Su W, Xu H, He W, Dai C, et al: miR-125b/Ets1 axis regulates
transdifferentiation and calcification of vascular smooth muscle
cells in a high-phosphate environment. Exp Cell Res. 322:302–312.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Goettsch C, Rauner M, Pacyna N, Hempel U,
Bornstein SR and Hofbauer LC: miR-125b regulates calcification of
vascular smooth muscle cells. Am J Pathol. 179:1594–1600. 2011.
View Article : Google Scholar :
|
|
142
|
Trixl L and Lusser A: The dynamic RNA
modification 5-methylcytosine and its emerging role as an
epitranscriptomic mark. Wiley Interdiscip Rev RNA. 10:e15102019.
View Article : Google Scholar
|
|
143
|
Jacob R, Zander S and Gutschner T: The
dark side of the epitranscriptome: Chemical modifications in long
non-coding RNAs. Int J Mol Sci. 18:182017. View Article : Google Scholar
|
|
144
|
Fu L, Guerrero CR, Zhong N, Amato NJ, Liu
Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, et al: Tet-mediated
formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc.
136:11582–11585. 2014. View Article : Google Scholar :
|
|
145
|
Fang S, Li J, Xiao Y, Lee M, Guo L, Han W,
Li T, Hill MC, Hong T, Mo W, et al: Tet inactivation disrupts YY1
binding and long-range chromatin interactions during embryonic
heart development. Nat Commun. 10:42972019. View Article : Google Scholar :
|
|
146
|
Ramanathan A, Robb GB and Chan S-H: mRNA
capping: Biological functions and applications. Nucleic Acids Res.
44:7511–7526. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Furuichi Y: Discovery of m(7)G-cap in
eukaryotic mRNAs. Proc Jpn Acad, Ser B, Phys Biol Sci. 91:394–409.
2015. View Article : Google Scholar
|
|
148
|
Pandolfini L, Barbieri I, Bannister AJ,
Hendrick A, Andrews B, Webster N, Murat P, Mach P, Brandi R, Robson
SC, et al: METTL1 promotes let-7 MicroRNA processing via m7G
methylation. Mol Cell. 74:1278–1290.e9. 2019. View Article : Google Scholar :
|
|
149
|
Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y
and Zhou HH: Let-7 in cardiovascular diseases, heart development
and cardiovascular differentiation from stem cells. Int J Mol Sci.
14:23086–23102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Bernstein DL, Jiang X and Rom S: let-7
microRNAs: Their role in cerebral and cardiovascular diseases,
inflammation, cancer, and their regulation. Biomedicines. 9:92021.
View Article : Google Scholar
|
|
151
|
Tolonen A-M, Magga J, Szabó Z, Viitala P,
Gao E, Moilanen AM, Ohukainen P, Vainio L, Koch WJ, Kerkelä R, et
al: Inhibition of Let-7 microRNA attenuates myocardial remodeling
and improves cardiac function postinfarction in mice. Pharmacol Res
Perspect. 2:e00056. 2014. View Article : Google Scholar
|
|
152
|
Yang Y, Ago T, Zhai P, Abdellatif M and
Sadoshima J: Thioredoxin 1 negatively regulates angiotensin
II-induced cardiac hypertrophy through upregulation of
miR-98/let-7. Circ Res. 108:305–313. 2011. View Article : Google Scholar
|
|
153
|
Satoh M, Minami Y, Takahashi Y, Tabuchi T
and Nakamura M: A cellular microRNA, let-7i, is a novel biomarker
for clinical outcome in patients with dilated cardiomyopathy. J
Card Fail. 17:923–929. 2011. View Article : Google Scholar
|
|
154
|
Dimitrova DG, Teysset L and Carré C: RNA
2′-O-Methylation (Nm) modification in human diseases. Genes
(Basel). 10:102019. View Article : Google Scholar
|
|
155
|
Somme J, Van Laer B, Roovers M, Steyaert
J, Versées W and Droogmans L: Characterization of two homologous
2′-O-methyltransferases showing different specificities for their
tRNA substrates. RNA. 20:1257–1271. 2014. View Article : Google Scholar
|
|
156
|
Cavaillé J, Nicoloso M and Bachellerie
J-P: Targeted ribose methylation of RNA in vivo directed by
tailored antisense RNA guides. Nature. 383:732–735. 1996.
View Article : Google Scholar
|
|
157
|
O'Brien JE Jr, Kibiryeva N, Zhou X-G,
Marshall JA, Lofland GK, Artman M, Chen J and Bittel DC: Noncoding
RNA expression in myocardium from infants with tetralogy of Fallot.
Circ Cardiovasc Genet. 5:279–286. 2012. View Article : Google Scholar
|
|
158
|
Nagasawa C, Ogren A, Kibiryeva N, Marshall
J, O'Brien JE Jr, Kenmochi N and Bittel DC: The role of scaRNAs in
adjusting alternative mRNA splicing in heart development. J
Cardiovasc Dev Dis. 5:262018. View Article : Google Scholar
|
|
159
|
Elliott BA, Ho H-T, Ranganathan SV,
Vangaveti S, Ilkayeva O, Abou Assi H, Choi AK, Agris PF and Holley
CL: Modification of messenger RNA by 2′-O-methylation regulates
gene expression in vivo. Nat Commun. 10:34012019. View Article : Google Scholar
|
|
160
|
Gagnidze K, Rayon-Estrada V, Harroch S,
Bulloch K and Papavasiliou FN: A new chapter in genetic medicine:
RNA Editing and its role in disease pathogenesis. Trends Mol Med.
24:294–303. 2018. View Article : Google Scholar
|
|
161
|
Nishikura K: A-to-I editing of coding and
non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 17:83–96. 2016.
View Article : Google Scholar :
|
|
162
|
Uchida S and Jones SP: RNA editing:
Unexplored opportunities in the cardiovascular system. Circ Res.
122:399–401. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Picardi E, Manzari C, Mastropasqua F,
Aiello I, D'Erchia AM and Pesole G: Profiling RNA editing in human
tissues: Towards the inosinome Atlas. Sci Rep. 5:149412015.
View Article : Google Scholar :
|
|
164
|
Sukhova GK, Zhang Y, Pan JH, Wada Y,
Yamamoto T, Naito M, Kodama T, Tsimikas S, Witztum JL, Lu ML, et
al: Deficiency of cathepsin S reduces atherosclerosis in LDL
receptor-deficient mice. J Clin Invest. 111:897–906. 2003.
View Article : Google Scholar
|
|
165
|
Wang B, Sun J, Kitamoto S, Yang M, Grubb
A, Chapman HA, Kalluri R and Shi GP: Cathepsin S controls
angiogenesis and tumor growth via matrix-derived angiogenic
factors. J Biol Chem. 281:6020–6029. 2006. View Article : Google Scholar
|
|
166
|
Zhu Y, Pirnie SP and Carmichael GG:
High-throughput and site-specific identification of
2′-O-methylation sites using ribose oxidation sequencing
(RibOxi-seq). RNA. 23:1303–1314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Stellos K, Gatsiou A, Stamatelopoulos K,
Perisic Matic L, John D, Lunella FF, Jaé N, Rossbach O, Amrhein C,
Sigala F, et al: Adenosine-to-inosine RNA editing controls
cathepsin S expression in atherosclerosis by enabling HuR-mediated
post-transcriptional regulation. Nat Med. 22:1140–1150. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Wang Y, Men M, Xie B, Shan J, Wang C, Liu
J, Zheng H, Yang W, Xue S and Guo C: Inhibition of PKR protects
against H2O2-induced injury on neonatal
cardiac myocytes by attenuating apoptosis and inflammation. Sci
Rep. 6:387532016. View Article : Google Scholar
|
|
169
|
Moore JB IV, Sadri G, Fischer AG, Weirick
T, Militello G, Wysoczynski M, Gumpert AM, Braun T and Uchida S:
The A-to-I RNA editing enzyme Adar1 is essential for normal
embryonic cardiac growth and development. Circ Res. 127:550–552.
2020. View Article : Google Scholar :
|
|
170
|
El Azzouzi H, Vilaça AP, Feyen DA, Gommans
WM, de Weger RA, Doevendans PA and Sluijter JP: Cardiomyocyte
specific deletion of ADAR1 causes severe cardiac dysfunction and
increased lethality. Front Cardiovasc Med. 7:302020. View Article : Google Scholar
|
|
171
|
Stulić M and Jantsch MF: Spatio-temporal
profiling of Filamin A RNA-editing reveals ADAR preferences and
high editing levels outside neuronal tissues. RNA Biol.
10:1611–1617. 2013. View Article : Google Scholar
|
|
172
|
Jain M, Mann TD, Stulić M, Rao SP, Kirsch
A, Pullirsch D, Strobl X, Rath C, Reissig L, Moreth K, et al: RNA
editing of Filamin A pre-mRNA regulates vascular contraction and
diastolic blood pressure. EMBO J. 37:372018. View Article : Google Scholar
|
|
173
|
Vu LT and Tsukahara T: C-to-U editing and
site-directed RNA editing for the correction of genetic mutations.
Biosci Trends. 11:243–253. 2017. View Article : Google Scholar
|
|
174
|
Gerber AP and Keller W: RNA editing by
base deamination: More enzymes, more targets, new mysteries. Trends
Biochem Sci. 26:376–384. 2001. View Article : Google Scholar
|
|
175
|
Rosenberg BR, Dewell S and Papavasiliou
FN: Identifying mRNA editing deaminase targets by RNA-Seq. Methods
Mol Biol. 718:103–119. 2011. View Article : Google Scholar
|
|
176
|
Powell LM, Wallis SC, Pease RJ, Edwards
YH, Knott TJ and Scott J: A novel form of tissue-specific RNA
processing produces apolipoprotein-B48 in intestine. Cell.
50:831–840. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Teng B, Burant CF and Davidson NO:
Molecular cloning of an apolipoprotein B messenger RNA editing
protein. Science. 260:1816–1819. 1993. View Article : Google Scholar
|
|
178
|
Blanc V and Davidson NO: APOBEC-1-mediated
RNA editing. Wiley Interdiscip Rev Syst Biol Med. 2:594–602. 2010.
View Article : Google Scholar
|
|
179
|
Anant S, Henderson JO, Mukhopadhyay D,
Navaratnam N, Kennedy S, Min J and Davidson NO: Novel role for
RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2
modulates C to U editing of apolipoprotein B mRNA by interacting
with apobec-1 and ACF, the apobec-1 complementation factor. J Biol
Chem. 276:47338–47351. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Blech-Hermoni Y, Dasgupta T, Coram RJ and
Ladd AN: Identification of targets of CUG-BP, Elav-like family
member 1 (CELF1) regulation in embryonic heart muscle. PLoS One.
11:e01490612016. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Giudice J, Xia Z, Li W and Cooper TA:
Neonatal cardiac dysfunction and transcriptome changes caused by
the absence of Celf1. Sci Rep. 6:355502016. View Article : Google Scholar
|
|
182
|
Liao W, Hong SH, Chan BH, Rudolph FB,
Clark SC and Chan L: APOBEC-2, a cardiac - and skeletal
muscle-specific member of the cytidine deaminase supergene family.
Biochem Biophys Res Commun. 260:398–404. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Sharma S, Patnaik SK, Taggart RT, Kannisto
ED, Enriquez SM, Gollnick P and Baysal BE: APOBEC3A cytidine
deaminase induces RNA editing in monocytes and macrophages. Nat
Commun. 6:68812015. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Sharma S, Patnaik SK, Kemer Z and Baysal
BE: Transient overexpression of exogenous APOBEC3A causes C-to-U
RNA editing of thousands of genes. RNA Biol. 14:603–610. 2017.
View Article : Google Scholar :
|
|
185
|
Li X, Ma S and Yi C: Pseudouridine: The
fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol.
33:108–116. 2016. View Article : Google Scholar
|
|
186
|
Cohn WE and Volkin E:
Nucleoside-5′-phosphates from ribonucleic acid. Nature.
167:483–484. 1951. View Article : Google Scholar
|
|
187
|
Sengupta R, Vainauskas S, Yarian C,
Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM and Agris PF:
Modified constructs of the tRNA TPsiC domain to probe substrate
conformational requirements of m(1)A(58) and m(5)U(54) tRNA
methyltransferases. Nucleic Acids Res. 28:1374–1380. 2000.
View Article : Google Scholar
|
|
188
|
Charette M and Gray MW: Pseudouridine in
RNA: What, where, how, and why. IUBMB Life. 49:341–351. 2000.
View Article : Google Scholar
|
|
189
|
Davis DR: Stabilization of RNA stacking by
pseudouridine. Nucleic Acids Res. 23:5020–5026. 1995. View Article : Google Scholar
|
|
190
|
Karijolich J and Yu YT: Converting
nonsense codons into sense codons by targeted pseudouridylation.
Nature. 474:395–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Spenkuch F, Motorin Y and Helm M:
Pseudouridine: Still mysterious, but never a fake (uridine)! RNA
Biol. 11:1540–1554. 2014. View Article : Google Scholar
|
|
192
|
Safra M, Nir R, Farouq D, Vainberg
Slutskin I and Schwartz S: TRUB1 is the predominant pseudouridine
synthase acting on mammalian mRNA via a predictable and conserved
code. Genome Res. 27:393–406. 2017. View Article : Google Scholar
|
|
193
|
Antonicka H, Choquet K, Lin ZY, Gingras
AC, Kleinman CL and Shoubridge EA: A pseudouridine synthase module
is essential for mitochondrial protein synthesis and cell
viability. EMBO Rep. 18:28–38. 2017. View Article : Google Scholar
|
|
194
|
Zaganelli S, Rebelo-Guiomar P, Maundrell
K, Rozanska A, Pierredon S, Powell CA, Jourdain AA, Hulo N,
Lightowlers RN, Chrzanowska-Lightowlers ZM, et al: The
pseudouridine synthase RPUSD4 is an essential component of
mitochondrial RNA granules. J Biol Chem. 292:4519–4532. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu
L, Li RY, Ji L, Dorsey SG, Jiang ZR, et al: Small nucleolar RNA 42
acts as an oncogene in lung tumorigenesis. Oncogene. 31:2794–2804.
2012. View Article : Google Scholar
|
|
196
|
Bykhovskaya Y, Casas K, Mengesha E, Inbal
A and Fischel-Ghodsian N: Missense mutation in pseudouridine
synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic
anemia (MLASA). Am J Hum Genet. 74:1303–1308. 2004. View Article : Google Scholar
|
|
197
|
Zucchini C, Strippoli P, Biolchi A, Solmi
R, Lenzi L, D'Addabbo P, Carinci P and Valvassori L: The human TruB
family of pseudouridine synthase genes, including the Dyskeratosis
Congenita 1 gene and the novel member TRUB1. Int J Mol Med.
11:697–704. 2003.PubMed/NCBI
|
|
198
|
Jády BE and Kiss T: A small nucleolar
guide RNA functions both in 2′-O-ribose methylation and
pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541–551.
2001. View Article : Google Scholar
|
|
199
|
Darzacq X, Jády BE, Verheggen C, Kiss AM,
Bertrand E and Kiss T: Cajal body-specific small nuclear RNAs: A
novel class of 2′-O-methylation and pseudouridylation guide RNAs.
EMBO J. 21:2746–2756. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Nagasawa CK, Kibiryeva N, Marshall J,
O'Brien JE Jr and Bittel DC: scaRNA1 levels alter pseudouridylation
in spliceosomal RNA U2 affecting alternative mRNA splicing and
embryonic development. Pediatr Cardiol. 41:341–349. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Wilusz JE: Long noncoding RNAs: Re-writing
dogmas of RNA processing and stability. Biochim Biophys Acta.
1859:128–138. 2016. View Article : Google Scholar
|
|
202
|
Kanvah S, Joseph J, Schuster GB, Barnett
RN, Cleveland CL and Landman U: Oxidation of DNA: Damage to
nucleobases. Acc Chem Res. 43:280–287. 2010. View Article : Google Scholar
|
|
203
|
Faucher F, Doublié S and Jia Z:
8-oxoguanine DNA glycosylases: One lesion, three subfamilies. Int J
Mol Sci. 13:6711–6729. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Shah A, Gray K, Figg N, Finigan A, Starks
L and Bennett M: Defective base excision repair of oxidative DNA
damage in vascular smooth muscle cells promotes atherosclerosis.
Circulation. 138:1446–1462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Seok H, Lee H, Lee S, Ahn SH, Lee HS, Kim
GD, Peak J, Park J, Cho YK, Jeong Y, et al: Position-specific
oxidation of miR-1 encodes cardiac hypertrophy. Nature.
584:279–285. 2020. View Article : Google Scholar
|
|
206
|
Wang J, Wang Q, Watson LJ, Jones SP and
Epstein PN: Cardiac overexpression of 8-oxoguanine DNA glycosylase
1 protects mitochondrial DNA and reduces cardiac fibrosis following
transaortic constriction. Am J Physiol Heart Circ Physiol.
301:H2073–H2080. 2011. View Article : Google Scholar :
|
|
207
|
Noren Hooten N, Ejiogu N, Zonderman AB and
Evans MK: Association of oxidative DNA damage and C-reactive
protein in women at risk for cardiovascular disease. Arterioscler
Thromb Vasc Biol. 32:2776–2784. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Motorin Y and Helm M: RNA nucleotide
methylation. Wiley Interdiscip Rev RNA. 2:611–631. 2011. View Article : Google Scholar
|
|
209
|
Garcia-Campos MA, Edelheit S, Toth U,
Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis
A, et al: Deciphering the 'm6A Code' via antibody-independent
quantitative profiling. Cell. 178:731–747.e16. 2019. View Article : Google Scholar
|
|
210
|
Zhang Z, Chen L-Q, Zhao Y-L, Yang CG,
Roundtree IA, Zhang Z, Ren J, Xie W, He C and Luo GZ: Single-base
mapping of m6A by an antibody-independent method. Sci Adv.
5:eaax02502019. View Article : Google Scholar :
|
|
211
|
Liu N and Pan T: Probing
N6-methyladenosine (m6A) RNA modification in total RNA with
SCARLET. Methods Mol Biol. 1358:285–292. 2016. View Article : Google Scholar
|
|
212
|
Dominissini D, Moshitch-Moshkovitz S,
Salmon-Divon M, Amariglio N and Rechavi G: Transcriptome-wide
mapping of N(6)-methyladenosine by m(6)A-seq based on
immunocapturing and massively parallel sequencing. Nat Protoc.
8:176–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N,
Han D, Dominissini D, Dai Q, Pan T, et al: High-resolution
N(6)-methyladenosine (m(6) A) map using photo-crosslinking-assisted
m(6) A sequencing. Angew Chem Int Ed Engl. 54:1587–1590. 2015.
View Article : Google Scholar
|
|
214
|
Marchand V, Bourguignon-Igel V, Helm M and
Motorin Y: Analysis of pseudouridines and other RNA modifications
using HydraPsiSeq protocol. Methods. Sep 1–2021.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Motorin Y and Helm M: Methods for RNA
modification mapping using deep sequencing: Established and new
emerging technologies. Genes (Basel). 10:352019. View Article : Google Scholar
|
|
216
|
Dai Q, Moshitch-Moshkovitz S, Han D, Kol
N, Amariglio N, Rechavi G, Dominissini D and He C: Nm-seq maps
2′-O-methylation sites in human mRNA with base precision. Nat
Methods. 14:695–698. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Birkedal U, Christensen-Dalsgaard M, Krogh
N, Sabarinathan R, Gorodkin J and Nielsen H: Profiling of ribose
methylations in RNA by high-throughput sequencing. Angew Chem Int
Ed Engl. 54:451–455. 2015.
|
|
218
|
Yoluç Y, Ammann G, Barraud P, Jora M,
Limbach PA, Motorin Y, Marchand V, Tisné C, Borland K and Kellner
S: Instrumental analysis of RNA modifications. Crit Rev Biochem Mol
Biol. 56:178–204. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Huang TY, Liu J and McLuckey SA: Top-down
tandem mass spectrometry of tRNA via ion trap collision-induced
dissociation. J Am Soc Mass Spectrom. 21:890–898. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Durairaj A and Limbach PA: Improving
CMC-derivatization of pseudouridine in RNA for mass spectrometric
detection. Anal Chim Acta. 612:173–181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Kowalak JA, Pomerantz SC, Crain PF and
McCloskey JA: A novel method for the determination of
post-transcriptional modification in RNA by mass spectrometry.
Nucleic Acids Res. 21:4577–4585. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Heiss M, Borland K, Yoluç Y and Kellner S:
Quantification of Modified nucleosides in the context of NAIL-MS.
Methods Mol Biol. 2298:279–306. 2021. View Article : Google Scholar
|
|
223
|
Wein S, Andrews B, Sachsenberg T,
Santos-Rosa H, Kohlbacher O, Kouzarides T, Garcia BA and Weisser H:
A computational platform for high-throughput analysis of RNA
sequences and modifications by mass spectrometry. Nat Commun.
11:9262020. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Marceca GP, Tomasello L, Distefano R,
Acunzo M, Croce CM and Nigita G: Detecting and characterizing
A-To-I microRNA editing in cancer. Cancers (Basel). 13:16992021.
View Article : Google Scholar
|
|
225
|
Okada S, Ueda H, Noda Y and Suzuki T:
Transcriptome-wide identification of A-to-I RNA editing sites using
ICE-seq. Methods. 156:66–78. 2019. View Article : Google Scholar
|
|
226
|
Adachi H, DeZoysa MD and Yu YT: Detection
and quantification of pseudouridine in RNA. Epitranscriptomics:
Methods and Protocols. Wajapeyee N and Gupta R: Springer; New York,
NY: pp. 219–235. 2019, View Article : Google Scholar
|
|
227
|
Schwartz S, Bernstein DA, Mumbach MR,
Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M,
Satija R, Lander ES, et al: Transcriptome-wide mapping reveals
widespread dynamic-regulated pseudouridylation of ncRNA and mRNA.
Cell. 159:148–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Lovejoy AF, Riordan DP and Brown PO:
Transcriptome-wide mapping of pseudouridines: Pseudouridine
synthases modify specific mRNAs in S. cerevisiae. PLoS One.
9:e1107992014. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Carlile TM, Rojas-Duran MF, Zinshteyn B,
Shin H, Bartoli KM and Gilbert WV: Pseudouridine profiling reveals
regulated mRNA pseudouridylation in yeast and human cells. Nature.
515:143–146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Li X, Zhu P, Ma S, Song J, Bai J, Sun F
and Yi C: Chemical pulldown reveals dynamic pseudouridylation of
the mammalian transcriptome. Nat Chem Biol. 11:592–597. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Banerjee A, Mikhailova E, Cheley S, Gu LQ,
Montoya M, Nagaoka Y, Gouaux E and Bayley H: Molecular bases of
cyclodextrin adapter interactions with engineered protein
nanopores. Proc Natl Acad Sci USA. 107:8165–8170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Wang Y, Yang Q and Wang Z: The evolution
of nanopore sequencing. Front Genet. 5:449. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Simpson JT, Workman RE, Zuzarte PC, David
M, Dursi LJ and Timp W: Detecting DNA cytosine methylation using
nanopore sequencing. Nat Methods. 14:407–410. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Vlachakis D, Papakonstantinou E, Sagar R,
Bacopoulou F, Exarchos T, Kourouthanassis P, Karyotis V, Vlamos P,
Lyketsos C, Avramopoulos D, et al: Improving the utility of
polygenic risk scores as a biomarker for Alzheimer's disease.
Cells. 10:16272021. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Buniello A, MacArthur JAL, Cerezo M,
Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy
E, Sollis E, et al: The NHGRI-EBI GWAS Catalog of published
genome-wide association studies, targeted arrays and summary
statistics 2019. Nucleic Acids Res. 47:D1005–D1012. 2019.
View Article : Google Scholar
|
|
236
|
Zheng Y, Nie P, Peng D, He Z, Liu M, Xie
Y, Miao Y, Zuo Z and Ren J: m6AVar: A database of functional
variants involved in m6A modification. Nucleic Acids Res.
46:D139–D145. 2018. View Article : Google Scholar :
|
|
237
|
Jiang S, Xie Y, He Z, Zhang Y, Zhao Y,
Chen L, Zheng Y, Miao Y, Zuo Z and Ren J: m6ASNP: A tool for
annotating genetic variants by m6A function. Gigascience. 7:72018.
View Article : Google Scholar
|
|
238
|
Song B, Tang Y, Chen K, Wei Z, Rong R, Lu
Z, Su J, de Magalhães JP, Rigden DJ and Meng J: m7GHub: Deciphering
the location, regulation and pathogenesis of internal mRNA
N7-methylguanosine (m7G) sites in human. Bioinformatics.
36:3528–3536. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Chen K, Song B, Tang Y, Wei Z, Xu Q, Su J,
de Magalhães JP, Rigden DJ and Meng J: RMDisease: A database of
genetic variants that affect RNA modifications, with implications
for epitranscriptome pathogenesis. Nucleic Acids Res.
49:D1396–D1404. 2021. View Article : Google Scholar
|
|
240
|
Incarnato D, Morandi E, Simon LM and
Oliviero S: RNA Framework: An all-in-one toolkit for the analysis
of RNA structures and post-transcriptional modifications. Nucleic
Acids Res. 46:e972018. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Mo XB, Lei SF, Zhang YH and Zhang H:
Detection of m6A-associated SNPs as potential functional variants
for coronary artery disease. Epigenomics. 10:1279–1287. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Mo XB, Lei SF, Zhang YH and Zhang H:
Examination of the associations between m6A-associated
single-nucleotide polymorphisms and blood pressure. Hypertens Res.
42:1582–1589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Mo X, Lei S, Zhang Y and Zhang H:
Genome-wide enrichment of m6A-associated single-nucleotide
polymorphisms in the lipid loci. Pharmacogenomics J. 19:347–357.
2019. View Article : Google Scholar
|
|
244
|
Mo X-B, Lei S-F, Zhang Y-H and Zhang H:
Integrative Analysis identified IRF6 and NDST1 as potential causal
genes for ischemic stroke. Front Neurol. 10:5172019. View Article : Google Scholar :
|
|
245
|
Nielsen JB, Thorolfsdottir RB, Fritsche
LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM,
Sveinbjornsson G, et al: Biobank-driven genomic discovery yields
new insight into atrial fibrillation biology. Nat Genet.
50:1234–1239. 2018. View Article : Google Scholar
|
|
246
|
Ehret GB, Ferreira T, Chasman DI, Jackson
AU, Schmidt EM, Johnson T, Thorleifsson G, Luan J, Donnelly LA,
Kanoni S, et al CHARGE-EchoGen consortium; CHARGE-HF consortium:
Wellcome Trust Case Control Consortium: The genetics of blood
pressure regulation and its target organs from association studies
in 342,415 individuals. Nat Genet. 48:1171–1184. 2016. View Article : Google Scholar :
|
|
247
|
Franzén O, Ermel R, Sukhavasi K, Jain R,
Jain A, Betsholtz C, Giannarelli C, Kovacic JC, Ruusalepp A,
Skogsberg J, et al: Global analysis of A-to-I RNA editing reveals
association with common disease variants. PeerJ. 6:e4466. 2018.
View Article : Google Scholar :
|
|
248
|
Lee JY, Lee BS, Shin DJ, Woo Park K, Shin
YA, Joong Kim K, Heo L, Young Lee J, Kyoung Kim Y, Jin Kim Y, et
al: A genome-wide association study of a coronary artery disease
risk variant. J Hum Genet. 58:120–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Jeemon P, Pettigrew K, Sainsbury C,
Prabhakaran D and Padmanabhan S: Implications of discoveries from
genome-wide association studies in current cardiovascular practice.
World J Cardiol. 3:230–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Wakil SM, Ram R, Muiya NP, Mehta M, Andres
E, Mazhar N, Baz B, Hagos S, Alshahid M, Meyer BF, et al: A
genome-wide association study reveals susceptibility loci for
myocardial infarction/coronary artery disease in Saudi Arabs.
Atherosclerosis. 245:62–70. 2016. View Article : Google Scholar
|
|
251
|
Terasawa Y, Ladha Z, Leonard SW, Morrow
JD, Newland D, Sanan D, Packer L, Traber MG and Farese RV Jr:
Increased atherosclerosis in hyperlipidemic mice deficient in alpha
-tocopherol transfer protein and vitamin E. Proc Natl Acad Sci USA.
97:13830–13834. 2000. View Article : Google Scholar
|
|
252
|
Lu N, Li X, Yu J, Li Y, Wang C, Zhang L,
Wang T and Zhong X: Curcumin attenuates lipopolysaccharide-induced
hepatic lipid metabolism disorder by modification of m6A RNA
methylation in piglets. Lipids. 53:53–63. 2018. View Article : Google Scholar
|
|
253
|
Sikorski V, Karjalainen P, Blokhina D,
Oksaharju K, Khan J, Katayama S, Rajala H, Suihko S, Tuohinen S,
Teittinen K, et al: Epitranscriptomics of ischemic heart
disease-the IHD-EPITRAN study design and objectives. Int J Mol Sci.
22:222021. View Article : Google Scholar
|
|
254
|
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou
H, Gan J, Jiang H, Jia GF, Luo C, et al: Meclofenamic acid
selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic
Acids Res. 43:373–384. 2015. View Article : Google Scholar
|
|
255
|
Li J, Chen Z, Chen F, Xie G, Ling Y, Peng
Y, Lin Y, Luo N, Chiang CM and Wang H: Targeted mRNA demethylation
using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids
Res. 48:5684–5694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Cox DBT, Gootenberg JS, Abudayyeh OO,
Franklin B, Kellner MJ, Joung J and Zhang F: RNA editing with
CRISPR-Cas13. Science. 358:1019–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Papageorgiou L, Cuong NT and Vlachakis D:
Antibodies as stratagems against cancer. Mol Biosyst. 12:2047–2055.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Vlachakis D: Antibody clustering and 3D
modelling for neurodegenerative diseases. Computational
Neurodegeneration. Vlamos P, Kotsireas I and Tarnanas I: Springer;
New York, NY: pp. 1–13. 2020
|
|
259
|
Altaf F, Vesely C, Sheikh AM, Munir R,
Shah ST and Tariq A: Modulation of ADAR mRNA expression in patients
with congenital heart defects. PLoS One. 14:e02009682019.
View Article : Google Scholar : PubMed/NCBI
|
|
260
|
van der Kwast RV, Quax PH and Nossent AY:
An emerging role for isomiRs and the microRNA epitranscriptome in
neovascularization. Cells. 9:92019. View Article : Google Scholar
|
|
261
|
Dai DF, Chen T, Johnson SC, Szeto H and
Rabinovitch PS: Cardiac aging: From molecular mechanisms to
significance in human health and disease. Antioxid Redox Signal.
16:1492–1526. 2012. View Article : Google Scholar
|
|
262
|
McMahon M, Forester C and Buffenstein R:
Aging through an epitranscriptomic lens. Nat Aging. 1:335–346.
2021. View Article : Google Scholar
|
|
263
|
Komal S, Zhang LR and Han SN: Potential
regulatory role of epigenetic RNA methylation in cardiovascular
diseases. Biomed Pharmacother. 137:1113762021. View Article : Google Scholar
|
|
264
|
Van Haute L, Dietmann S, Kremer L, Hussain
S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S,
et al: Deficient methylation and formylation of mt-tRNA(Met) wobble
cytosine in a patient carrying mutations in NSUN3. Nat Commun.
7:120392016. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Bohnsack MT and Sloan KE: The
mitochondrial epitranscriptome: The roles of RNA modifications in
mitochondrial translation and human disease. Cell Mol Life Sci.
75:241–260. 2018. View Article : Google Scholar
|
|
266
|
Vilardo E and Rossmanith W: Molecular
insights into HSD10 disease: Impact of SDR5C1 mutations on the
human mitochondrial RNase P complex. Nucleic Acids Res.
43:66492015. View Article : Google Scholar
|
|
267
|
Vlachakis D, Tsagrasoulis D,
Megalooikonomou V and Kossida S: Introducing Drugster: A
comprehensive and fully integrated drug design, lead and structure
optimization toolkit. Bioinformatics. 29:126–128. 2013. View Article : Google Scholar
|
|
268
|
Xu Z, Peng B, Cai Y, Wu G, Huang J, Gao M,
Guo G, Zeng S, Gong Z and Yan Y: N6-methyladenosine RNA
modification in cancer therapeutic resistance: Current status and
perspectives. Biochem Pharmacol. 182:1142582020. View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Tsaniras S and Vlachakis D: Diet, obesity
and cancer. J Mol Biochem. 4:202016.
|
|
270
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun
G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation regulates the
self-renewal and tumorigenesis of glioblastoma stem cells. Cell
Rep. 18:2622–2634. 2017. View Article : Google Scholar :
|
|
271
|
Singh B, Kinne HE, Milligan RD, Washburn
LJ, Olsen M and Lucci A: Important role of FTO in the survival of
rare panresistant triple-negative inflammatory breast cancer cells
facing a severe metabolic challenge. PLoS One. 11:e01590722016.
View Article : Google Scholar : PubMed/NCBI
|
|
272
|
Charalampopoulou M, Syrigos K, Filopoulos
E, Megalooikonomou V, Vlachakis D, Chrousos G and Darviri C:
Reliability and validity of the instrument newly diagnosed breast
cancer stress scale in the Greek population. J Mol Biochem. 9:5–12.
2020.
|
|
273
|
Su R, Dong L, Li C, Nachtergaele S,
Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al: R-2HG
exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA
signaling. Cell. 172:90–105.e23. 2018. View Article : Google Scholar
|
|
274
|
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu
H, Ni T, Zhang ZS, Zhang T, Li C, et al: Small-molecule targeting
of oncogenic FTO demethylase in acute myeloid leukemia. Cancer
Cell. 35:677–691.e10. 2019. View Article : Google Scholar :
|
|
275
|
Floresta G, Pistarà V, Amata E, Dichiara
M, Damigella A, Marrazzo A, Prezzavento O, Punzo F and Rescifina A:
Molecular modeling studies of pseudouridine isoxazolidinyl
nucleoside analogues as potential inhibitors of the pseudouridine
5′-mono-phosphate glycosidase. Chem Biol Drug Des. 91:519–525.
2018. View Article : Google Scholar
|
|
276
|
Vlachakis D, Fakourelis P, Megalooikonomou
V, Makris C and Kossida S: DrugOn: A fully integrated pharmacophore
modeling and structure optimization toolkit. PeerJ. 3:e7252015.
View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Vlachakis D and Kossida S: Molecular
modeling and pharmacophore elucidation study of the Classical Swine
Fever virus helicase as a promising pharmacological target. PeerJ.
1:e852013. View Article : Google Scholar : PubMed/NCBI
|
|
278
|
Papageorgiou L, Loukatou S, Sofia K,
Maroulis D and Vlachakis D: An updated evolutionary study of
Flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase
reveals novel invariable motifs as potential pharmacological
targets. Mol Biosyst. 12:2080–2093. 2016. View Article : Google Scholar
|
|
279
|
Papageorgiou L, Loukatou S, Koumandou VL,
Makałowski W, Megalooikonomou V, Vlachakis D and Kossida S:
Structural models for the design of novel antiviral agents against
Greek Goat Encephalitis. PeerJ. 2:e6642014. View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Amidi S, Amidi A, Vlachakis D, Paragios N
and Zacharaki EI: Automatic single- and multi-label enzymatic
function prediction by machine learning. PeerJ. 5:e30952017.
View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Marinou M, Platis D, Ataya FS,
Chronopoulou E, Vlachakis D and Labrou NE: Structure-based design
and application of a nucleotide coenzyme mimetic ligand:
Application to the affinity purification of nucleotide dependent
enzymes. J Chromatogr A. 1535:88–100. 2018. View Article : Google Scholar
|
|
282
|
Vlachakis D and Vlamos P: Mathematical
multidimensional modelling and structural artificial intelligence
pipelines provide insights for the designing of highly specific
antiSARS-CoV2 agents. Math Comput Sci. 15:877–888. 2021. View Article : Google Scholar
|