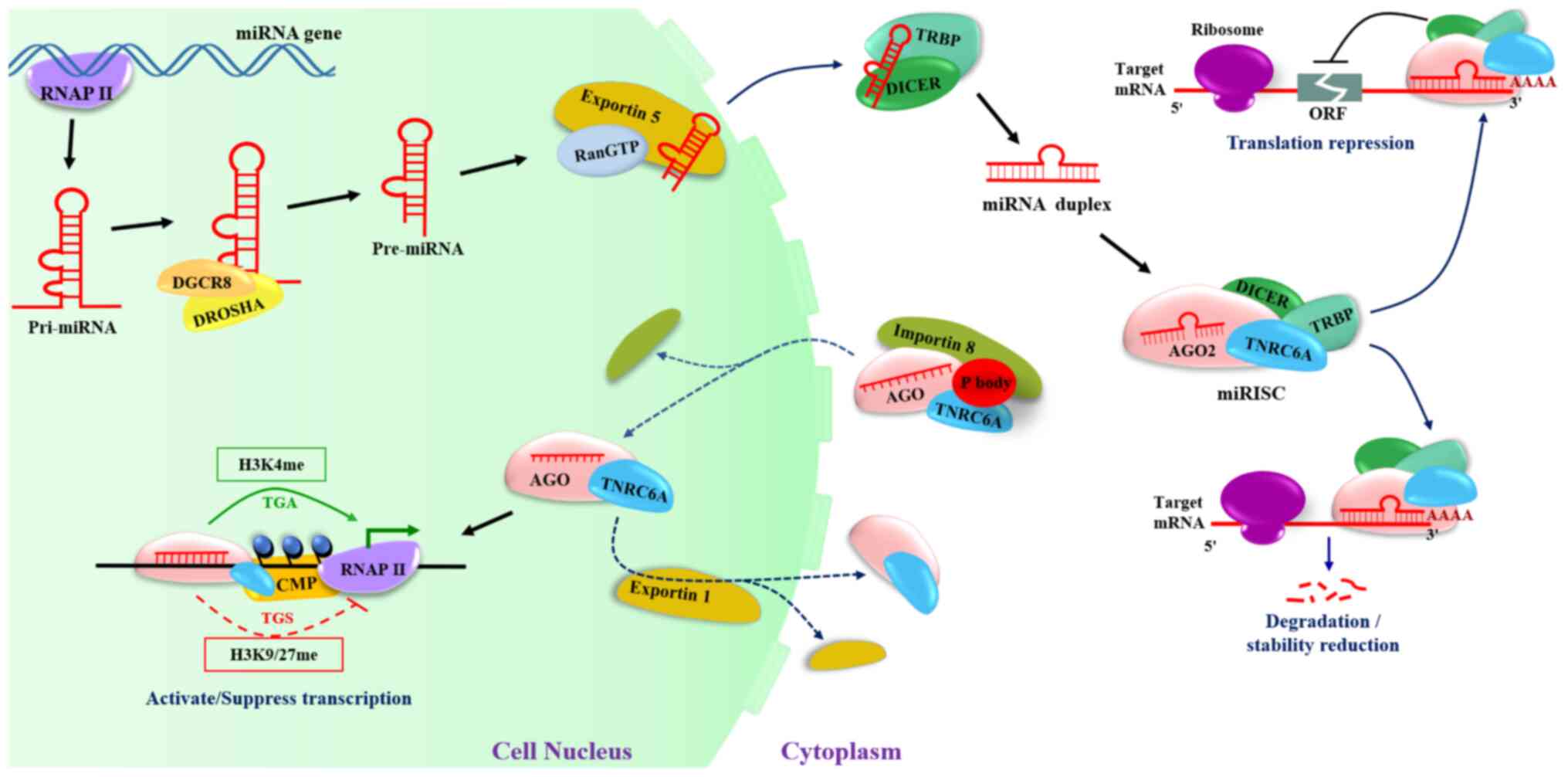
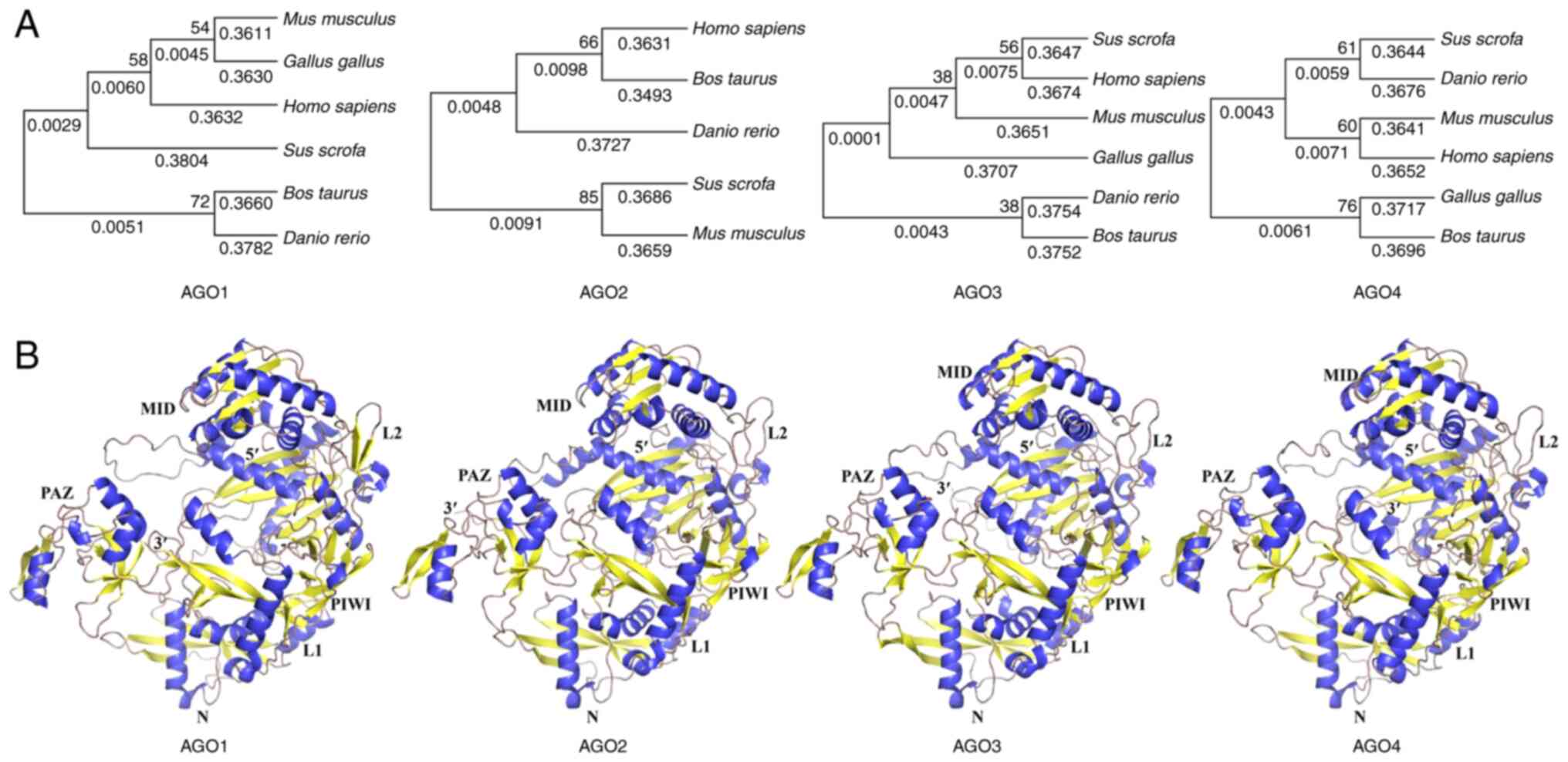
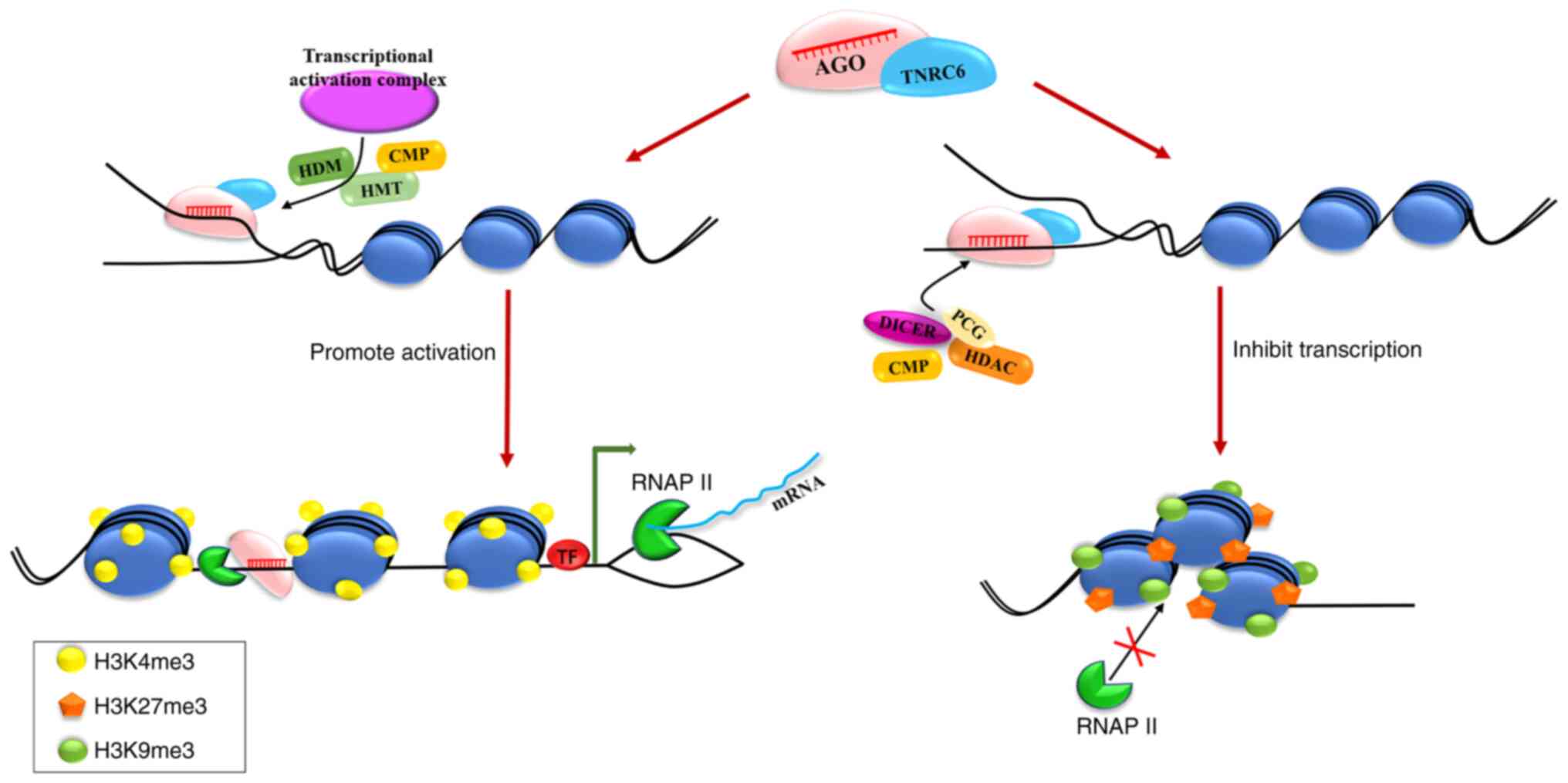
|
1
|
Huang V and Li LC: miRNA goes nuclear. RNA
Biol. 9:269–273. 2012. View Article : Google Scholar
|
|
2
|
Syeda ZA, Langden SS, Munkhzul C, Lee M
and Song SJ: Regulatory mechanism of microRNA expression in cancer.
Int J Mol Sci. 21:17232020. View Article : Google Scholar
|
|
3
|
Bhat IP, Rather TB, Bhat GA, Maqbool I,
Akhtar K, Rashid G, Parray FQ, Besina S and Mudassar S: TEAD4
nuclear localization and regulation by miR-4269 and miR-1343-3p in
colorectal carcinoma. Pathol Res Pract. 231:1537912022. View Article : Google Scholar
|
|
4
|
Zheng T, Zhou Y, Xu X, Qi X, Liu J, Pu Y,
Zhang S, Gao X, Luo X, Li M, et al: MiR-30c-5p loss-induced PELI1
accumulation regulates cell proliferation and migration via
activating PI3K/AKT pathway in papillary thyroid carcinoma. J
Transl Med. 20:202022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li L, Wei D, Zhang J, Deng R, Tang J and
Su D: MiR-641 inhibited cell proliferation and induced apoptosis by
targeting NUCKS1/PI3K/AKT signaling pathway in breast cancer.
Comput Math Methods Med. 2022:52038392022.PubMed/NCBI
|
|
6
|
Mirzaei S, Zarrabi A, Asnaf SE, Hashemi F,
Zabolian A, Hushmandi K, Raei M, Goharrizi MASB, Makvandi P,
Samarghandian S, et al: The role of microRNA-338-3p in cancer:
Growth, invasion, chemoresistance, and mediators. Life Sci.
268:1190052021. View Article : Google Scholar
|
|
7
|
El Fatimy R, Zhang Y, Deforzh E, Ramadas
M, Saravanan H, Wei Z, Rabinovsky R, Teplyuk NM, Uhlmann EJ and
Krichevsky AM: A nuclear function for an oncogenic microRNA as a
modulator of snRNA and splicing. Mol Cancer. 21:172022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo X, Dong J, He X, Shen L, Long C, Liu
F, Liu X, Lin T, He D and Wei G: MiR-155-5p exerts
tumor-suppressing functions in Wilms tumor by targeting IGF2 via
the PI3K signaling pathway. Biomed Pharmacother. 125:1098802020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gong R and Jiang Y: Non-coding RNAs in
pancreatic ductal adenocarcinoma. Front Oncol. 10:3092020.
View Article : Google Scholar
|
|
10
|
Gregorova J, Vychytilova-Faltejskova P and
Sevcikova S: Epigenetic regulation of MicroRNA clusters and
families during tumor development. Cancers (Basel). 13:13332021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of microRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar
|
|
12
|
Yang YL, Chang YH, Li CJ, Huang YH, Tsai
MC, Chu PY and Lin HY: New insights into the role of miR-29a in
hepatocellular carcinoma: Implications in mechanisms and
theragnostics. J Pers Med. 11:2192021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kobayashi H and Tomari Y: RISC assembly:
Coordination between small RNAs and Argonaute proteins. Biochim
Biophys Acta. 1859:71–81. 2016. View Article : Google Scholar
|
|
14
|
Zhang J, Zhou W, Liu Y, Liu T, Li C and
Wang L: Oncogenic role of microRNA-532-5p in human colorectal
cancer via targeting of the 5′UTR of RUNX3. Oncol Lett.
15:7215–7220. 2018.
|
|
15
|
Liu M, Roth A, Yu M, Morris R, Bersani F,
Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, et al: The
IGF2 intronic miR-483 selectively enhances transcription from IGF2
fetal promoters and enhances tumorigenesis. Genes Dev.
27:2543–2548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu W, San LA, Wang Z and Liu Y:
Identifying microRNA targets in different gene regions. BMC
Bioinformatics. 15 (Suppl 7):S42014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Z, Lan X, Han R, Wang J, Huang Y, Sun
J, Guo W and Chen H: MiR-2478 inhibits TGFβ1 expression by
targeting the transcriptional activation region downstream of the
TGFβ1 promoter in dairy goats. Sci Rep. 7:426272017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo D, Barry L, Lin SSH, Huang V and Li
LC: RNAa in action: From the exception to the norm. RNA Biol.
11:1221–1225. 2014. View Article : Google Scholar
|
|
19
|
Stavast CJ and Erkeland SJ: The
non-canonical aspects of microRNAs: Many roads to gene regulation.
Cells Basel. 8:14652019. View Article : Google Scholar
|
|
20
|
Fan L, Lai R, Ma N, Dong Y, Li Y, Wu Q,
Qiao J, Lu H, Gong L, Tao Z, et al: MiR-552-3p modulates
transcriptional activities of FXR and LXR to ameliorate hepatic
glycolipid metabolism disorder. J Hepatol. 74:8–19. 2021.
View Article : Google Scholar
|
|
21
|
Liu H, Lei C, He Q, Pan Z, Xiao D and Tao
Y: Nuclear functions of mammalian microRNAs in gene regulation,
immunity and cancer. Mol Cancer. 17:642018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu L, Tian YC, Mao G, Zhang YG and Han L:
MiR-675 is frequently overexpressed in gastric cancer and enhances
cell proliferation and invasion via targeting a potent anti-tumor
gene PITX1. Cell Signal. 62:1093522019. View Article : Google Scholar
|
|
23
|
Majid S, Dar AA, Saini S, Yamamura S,
Hirata H, Tanaka Y, Deng G and Dahiya R: MicroRNA-205-directed
transcriptional activation of tumor suppressor genes in prostate
cancer. Cancer. 116:5637–5649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kumar R and Xi Y: MicroRNA, epigenetic
machinery and lung cancer. Thorac Cancer. 2:35–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Place RF, Li LC, Pookot D, Noonan EJ and
Dahiya R: MicroRNA-373 induces expression of genes with
complementary promoter sequences. Proc Natl Acad Sci USA.
105:1608–1613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiang X, Mei H, Qu H, Zhao X, Li D, Song
H, Jiao W, Pu J, Huang K, Zheng L and Tong Q: MiRNA-584-5p exerts
tumor suppressive functions in human neuroblastoma through
repressing transcription of matrix metalloproteinase 14. Biochim
Biophys Acta. 1852:1743–1754. 2015. View Article : Google Scholar
|
|
27
|
Bai B, Liu H and Laiho M: Small RNA
expression and deep sequencing analyses of the nucleolus reveal the
presence of nucleolus-associated microRNAs. FEBS Open Bio.
4:441–449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Catalanotto C, Cogoni C and Zardo G:
MicroRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17:17122016. View Article : Google Scholar
|
|
29
|
Saito Y, Liang G, Egger G, Friedman JM,
Chuang JC, Coetzee GA and Jones PA: Specific activation of
microRNA-127 with downregulation of the proto-oncogene BCL6 by
chromatin-modifying drugs in human cancer cells. Cancer Cell.
9:435–443. 2006. View Article : Google Scholar
|
|
30
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
31
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar
|
|
32
|
Lai .Eric C: Micro RNAs are complementary
to 3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar
|
|
33
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Gene Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gagnon KT, Li L, Chu Y, Janowski BA and
Corey DR: RNAi factors are present and active in human cell nuclei.
Cell Rep. 6:211–221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weinmann L, Höck J, Ivacevic T, Ohrt T,
Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H and Meister G:
Importin 8 is a gene silencing factor that targets argonaute
proteins to distinct mRNAs. Cell. 136:496–507. 2009. View Article : Google Scholar
|
|
37
|
Wei Y, Li L, Wang D, Zhang CY and Zen K:
Importin 8 regulates the transport of mature microRNAs into the
cell nucleus. J Biol Chem. 289:10270–10275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Azmi AS, Uddin MH and Mohammad RM: The
nuclear export protein XPO1-from biology to targeted therapy. Nat
Rev Clin Oncol. 18:152–169. 2021. View Article : Google Scholar
|
|
39
|
Nishi K, Nishi A, Nagasawa T and Ui-Tei K:
Human TNRC6A is an argonaute-navigator protein for
microRNA-mediated gene silencing in the nucleus. RNA. 19:17–35.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Daniel S, Schindler SG, Johannes D,
Elisabeth K, Janina P, Stefan H, Reinhard D and Gunter M:
Importin-β facilitates nuclear import of human GW proteins and
balances cytoplasmic gene silencing protein levels. Nucleic Acids
Res. 43:7447–7461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Behm-Ansmant I, Rehwinkel J, Doerks T,
Stark A, Bork P and Izaurralde E: MRNA degradation by miRNAs and
GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping
complexes. Genes Dev. 20:1885–1898. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nishi K, Takahashi T, Suzawa M, Miyakawa
T, Nagasawa T, Ming Y, Tanokura M and Ui-Tei K: Control of the
localization and function of a miRNA silencing component TNRC6A by
argonaute protein. Nucleic Acids Res. 43:9856–9873. 2015.PubMed/NCBI
|
|
43
|
Hicks JA, Li L, Matsui M, Chu Y, Volkov O,
Johnson KC and Corey DR: Human GW182 paralogs are the central
organizers for RNA-Mediated control of transcription. Cell Rep.
20:1543–1552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Castanotto D, Lingeman R, Riggs AD and
Rossi JJ: CRM1 mediates nuclear-cytoplasmic shuttling of mature
microRNAs. Proc Natl Acad Sci USA. 106:21655–21659. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kalantari R, Hicks JA, Li L, Gagnon KT,
Sridhara V, Lemoff A, Mirzaei H and Corey DR: Stable association of
RNAi machinery is conserved between the cytoplasm and nucleus of
human cells. RNA. 22:1085–1098. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kuhn CD and Joshua-Tor L: Eukaryotic
argonautes come into focus. Trends Biochem Sci. 38:263–271. 2013.
View Article : Google Scholar
|
|
47
|
Ryazansky S, Kulbachinskiy A and Aravin
AA: The expanded universe of prokaryotic argonaute proteins. mBio.
9:e01935–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Peters L and Meister G: Argonaute
proteins: Mediators of RNA silencing. Mol Cell. 26:611–623. 2007.
View Article : Google Scholar
|
|
49
|
Hutvagner G and Simard MJ: Argonaute
proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol.
9:22–32. 2008. View Article : Google Scholar
|
|
50
|
Siomi MC, Sato K, Pezic D and Aravin AA:
PIWI-interacting small RNAs: The vanguard of genome defence. Nat
Rev Mol Cell Biol. 12:246–258. 2011. View Article : Google Scholar
|
|
51
|
Sasaki T, Shiohama A, Minoshima S and
Shimizu N: Identification of eight members of the argonaute family
in the human genome. Genomics. 82:323–330. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Faehnle CR, Elkayam E, Haase AD, Hannon GJ
and Joshua-Tor L: The making of a slicer: Activation of human
argonaute-1. Cell Rep. 3:1901–1909. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schirle NT, Sheu-Gruttadauria J,
Chandradoss SD, Joo C and MacRae IJ: Water-mediated recognition of
t1-adenosine anchors argonaute2 to microRNA targets. Elife.
4:e076462015. View Article : Google Scholar
|
|
54
|
Park MS, Phan HD, Busch F, Hinckley SH,
Brackbill JA, Wysocki VH and Nakanishi K: Human argonaute3 has
slicer activity. Nucleic Acids Res. 45:11867–11877. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kwak PB and Tomari Y: The N domain of
argonaute drives duplex unwinding during RISC assembly. Nat Struct
Mol Biol. 19:145–151. 2012. View Article : Google Scholar
|
|
56
|
Czech B and Hannon GJ: Small RNA sorting:
Matchmaking for argonautes. Nat Rev Genet. 12:19–31. 2011.
View Article : Google Scholar
|
|
57
|
Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki
S, Liu Q and Tomari Y: ATP-dependent human RISC assembly pathways.
Nat Struct Mol Biol. 17:17–23. 2010. View Article : Google Scholar
|
|
58
|
Liu J, Carmell MA, Rivas FV, Marsden CG,
Thomson JM, Song JJ, Hammond SM, Joshua-Tor L and Hannon GJ:
Argonaute2 is the catalytic engine of mammalian RNAi. Science.
305:1437–1441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang V and Li LC: Demystifying the
nuclear function of argonaute proteins. RNA Biol. 11:18–24. 2014.
View Article : Google Scholar
|
|
60
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar
|
|
61
|
Younger ST and Corey DR: Transcriptional
gene silencing in mammalian cells by miRNA mimics that target gene
promoters. Nucleic Acids Res. 39:5682–5691. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang T, Tan P, Wang L, Jin N, Li Y, Zhang
L, Yang H, Hu Z, Zhang L, Hu C, et al: RNALocate: A resource for
RNA subcellular localizations. Nucleic Acids Res. 45:D135–D138.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Piriyapongsa J, Bootchai C, Ngamphiw C and
Tongsima S: MicroPIR2: A comprehensive database for human-mouse
comparative study of microRNA-promoter interactions. Database
(Oxford). 2014:bau1152014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lukasik A, Wójcikowski M and Zielenkiewicz
P: Tools4miRs-one place to gather all the tools for miRNA analysis.
Bioinformatics. 32:2722–2724. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Parveen A, Gretz N and Dweep H: Obtaining
miRNA-target interaction information from miRWalk2.0. Curr Protoc
Bioinformatics. 55:12.15.1–12.15.27. 2016. View Article : Google Scholar
|
|
66
|
Liu Q, Wang J, Zhao Y, Li CI, Stengel KR,
Acharya P, Johnston G, Hiebert SW and Shyr Y: Identification of
active miRNA promoters from nuclear run-on RNA sequencing. Nucleic
Acids Res. 45:e1212017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jeffries CD, Fried HM and Perkins DO:
Nuclear and cytoplasmic localization of neural stem cell microRNAs.
RNA. 17:675–86. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wong JJ, Ritchie W, Gao D, Lau KA,
Gonzalez M, Choudhary A, Taft RJ, Rasko JE and Holst J:
Identification of nuclear-enriched miRNAs during mouse
granulopoiesis. J Hematol Oncol. 7:422014. View Article : Google Scholar
|
|
69
|
Li ZF, Liang YM, Lau PN, Shen W, Wang DK,
Cheung WT, Xue CJ, Poon LM and Lam YW: Dynamic localisation of
mature microRNAs in human nucleoli is influenced by exogenous
genetic materials. PLoS One. 8:e708692013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sahu I, Hebalkar R, Kar S, Sreevidya TS,
Gutti U and Gutti RK: Systems biology approach to study the role of
miRNA in promoter targeting during megakaryopoiesis. Exp Cell Res.
366:192–198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H,
Shao P, Chen YQ and Qu LH: Deep sequencing of human nuclear and
cytoplasmic small RNAs reveals an unexpectedly complex subcellular
distribution of miRNAs and tRNA 3′trailers. PLoS One. 5:e105632010.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Politz JCR, Hogan EM and Pederson T:
MicroRNAs with a nucleolar location. RNA. 15:1705–1715. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Tang R, Li L, Zhu D, Hou D, Cao T, Gu H,
Zhang J, Chen J, Zhang CY and Zen K: Mouse miRNA-709 directly
regulates miRNA-15a/16-1 biogenesis at the posttranscriptional
level in the nucleus: Evidence for a microRNA hierarchy system.
Cell Res. 22:504–515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Toms D, Pan B, Bai Y and Li J: Small RNA
sequencing reveals distinct nuclear microRNAs in pig granulosa
cells during ovarian follicle growth. J Ovarian Res. 14:542021.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sato K and Siomi MC: The piRNA pathway in
Drosophila ovarian germ and somatic cells. Proc Jpn Acad Ser B Phys
Biol Sci. 96:32–42. 2020. View Article : Google Scholar
|
|
76
|
Gunawardane LS, Saito K, Nishida KM,
Miyoshi K, Kawamura Y, Nagami T, Siomi H and Siomi MC: A
slicer-mediated mechanism for repeat-associated siRNA 59 end
formation in drosophila. Science. 315:1587–1590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Brennecke J, Aravin AA, Stark A, Dus M,
Kellis M, Sachidanandam R and Hannon GJ: Discrete small
RNA-generating loci as master regulators of transposon activity in
drosophila. Cell. 128:1089–1103. 2007. View Article : Google Scholar
|
|
78
|
Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J,
Czech B and Hannon GJ: Panoramix enforces piRNA-dependent
cotranscriptional silencing. Science. 350:339–342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sienski G, Donertas D and Brennecke J:
Transcriptional silencing of transposons by Piwi and maelstrom and
its impact on chromatin state and gene expression. Cell.
151:964–980. 2012. View Article : Google Scholar
|
|
80
|
Watanabe T, Tomizawa S, Mitsuya K, Totoki
Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ,
Toyoda A, et al: Role for piRNAs and noncoding RNA in de novo DNA
methylation of the imprinted mouse Rasgrf1 locus. Science.
332:848–852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang N, Hu G, Myers TG and Williamson PR:
Protocols for the analysis of microRNA expression, biogenesis, and
function in immune cells. Curr Protoc Immunol. 126:e782019.
View Article : Google Scholar
|
|
82
|
Fu Y, Zhang L, Zhang R, Xu S, Wang H, Jin
Y and Wu Z: Enterovirus 71 suppresses miR-17-92 cluster through
up-regulating methylation of the miRNA promoter. Front Microbiol.
10:6252019. View Article : Google Scholar
|
|
83
|
Younger ST, Pertsemlidis A and Corey DR:
Predicting potential miRNA target sites within gene promoters.
Bioorg Med Chem Lett. 19:3791–3794. 2009. View Article : Google Scholar
|
|
84
|
Chellini L, Frezza V and Paronetto MP:
Dissecting the transcriptional regulatory networks of
promoter-associated noncoding RNAs in development and cancer. J Exp
Clin Cancer Res. 39:512020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang Y and Zhang H: RNAa induced by TATA
box-targeting microRNAs. Adv Exp Med Biol. 983:91–111. 2017.
View Article : Google Scholar
|
|
86
|
Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y,
Zhang L, Ding C, Luo H, Li Y, et al: MicroRNAs activate gene
transcription epigenetically as an enhancer trigger. RNA Biol.
14:1326–1334. 2017. View Article : Google Scholar
|
|
87
|
Zhang Y, Liu W, Chen Y, Liu J, Wu K, Su L,
Zhang W, Jiang Y, Zhang X, Zhang Y, et al: A cellular microRNA
facilitates regulatory t lymphocyte development by targeting the
FOXP3 promoter TATA-box motif. J Immunol. 200:1053–1063. 2017.
View Article : Google Scholar
|
|
88
|
Bai Y, Pan B, Zhan X, Silver H and Li J:
MicroRNA 195-5p targets foxo3 promoter region to regulate its
expression in granulosa cells. Int J Mol Sci. 22:67212021.
View Article : Google Scholar
|
|
89
|
Cao R and Zhang Y: The functions of
E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr
Opin Genet Dev. 14:155–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mellor J, Dudek P and Clynes D: A glimpse
into the epigenetic landscape of gene regulation. Curr Opin Genet
Dev. 18:116–122. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Guo H, Pu M, Tai Y, Chen Y and Ren J:
Nuclear miR-30b-5p suppresses TFEB-mediated lysosomal biogenesis
and autophagy. Cell Death Differ. 28:320–336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li LC: Chromatin remodeling by the small
RNA machinery in mammalian cells. Epigenetics. 9:45–52. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Barlak N, Capik O, Kilic A, Sanli F,
Aytatli A, Yazici A, Karatas EA, Ortucu S and Karatas OF:
MicroRNA-145 transcriptionally regulates semaphorin 3A expression
in prostate cancer cells. Cell Biol Int. 45:1082–1090. 2021.
View Article : Google Scholar
|
|
94
|
Song M, Wang Y, Zhou P, Wang J, Xu H and
Zheng J: MicroRNA-361-5p aggravates acute pancreatitis by promoting
interleukin-17A secretion via impairment of nuclear factor
IA-dependent hes1 downregulation. J Med Chem. 64:16541–16552. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang K, Wang YY, Xu Y, Zhang L, Zhu J, Si
PC, Wang YW and Ma R: A two-miRNA signature of upregulated
miR-185-5p and miR-362-5p as a blood biomarker for breast cancer.
Pathol Res Pract. 222:1534582021. View Article : Google Scholar
|
|
96
|
Van Roosbroeck K and Calin GA: Cancer
hallmarks and MicroRNAs: The therapeutic connection. Adv Cancer
Res. 135:119–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kolenda T, Przybyla W, Teresiak A,
Mackiewicz A and Lamperska KM: The mystery of let-7d-a small RNA
with great power. Contemp Oncol (Pozn). 18:293–301. 2014.PubMed/NCBI
|
|
98
|
Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T,
Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, et al: Functional
proteomics identifies miRNAs to target a p27/Myc/phospho-Rb
signature in breast and ovarian cancer. Oncogene. 35:691–701. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Schmid G, Notaro S, Reimer D, Abdel-Azim
S, Duggan-Peer M, Holly J, Fiegl H, Rossler J, Wiedemair A, Concin
N, et al: Expression and promotor hypermethylation of miR-34a in
the various histological subtypes of ovarian cancer. BMC Cancer.
16:1022016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wong KY, Yu L and Chim CS: DNA methylation
of tumor suppressor miRNA genes: A lesson from the miR-34 family.
Epigenomics. 3:83–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yang Z, Fang S, Di Y, Ying W, Tan Y and Gu
W: Modulation of NF-κB/miR-21/PTEN pathway sensitizes non-small
cell lung cancer to cisplatin. PLoS One. 10:e01215472015.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Xu X, Zhu S, Tao Z and Ye S: High
circulating miR-18a, miR-20a, and miR-92a expression correlates
with poor prognosis in patients with non-small cell lung cancer.
Cancer Med. 7:21–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li H, Zhou H, Luo J and Huang J:
MicroRNA-17-5p inhibits proliferation and triggers apoptosis in
non-small cell lung cancer by targeting transforming growth factor
β receptor 2. Exp Ther Med. 13:2715–2722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Yang Z, Liu C, Wu H, Xie Y and Zhang X:
CSB affected on the sensitivity of lung cancer cells to
platinum-based drugs through the global decrease of let-7 and
miR-29. BMC Cancer. 19:9482019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Kristensen LS, Andersen MS, Stagsted L,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar
|
|
106
|
Zheng L, Jiao W, Mei H, Song H, Li D,
Xiang X, Chen Y, Yang F, Li H, Huang K and Tong Q: MiRNA-337-3p
inhibits gastric cancer progression through repressing myeloid zinc
finger 1-facilitated expression of matrix metalloproteinase 14.
Oncotarget. 7:40314–40328. 2016. View Article : Google Scholar
|
|
107
|
Zhang L, Zhou Q, Qiu Q, Hou L and Lu Y:
CircPLEKHM3 acts as a tumor suppressor through regulation of the
miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer.
18:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Christofides A, Papagregoriou G, Dweep H,
Makrides N, Gretz N, Felekkis K and Deltas C: Evidence for
miR-548c-5p regulation of FOXC2 transcription through a distal
genomic target site in human podocytes. Cell Mol Life Sci.
77:2441–2459. 2020. View Article : Google Scholar
|
|
109
|
Dharap A, Pokrzywa C, Murali S, Pandi G
and Vemuganti R: MicroRNA miR-324-3p induces promoter-mediated
expression of RelA gene. PLoS One. 8:e794672013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Huang V: Endogenous miRNAa: MiRNA-mediated
gene upregulation. Adv Exp Med Biol. 983:65–79. 2017. View Article : Google Scholar
|
|
111
|
Huang V, Place RF, Portnoy V, Wang J, Qi
Z, Jia Z, Yu A, Shuman M, Yu J and Li LC: Upregulation of Cyclin B1
by miRNA and its implications in cancer. Nucleic Acids Res.
40:1695–1707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Matsui M, Chu Y, Zhang H, Gagnon KT,
Shaikh S, Kuchimanchi S, Manoharan M, Corey DR and Janowski BA:
Promoter RNA links transcriptional regulation of inflammatory
pathway genes. Nucleic Acids Res. 41:10086–10109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Turner M, Jiao A and Slack FJ:
Autoregulation of lin-4 microRNA transcription by RNA activation
(RNAa) in C. Elegans. Cell Cycle. 13:772–781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Vera H, Yi Q, Ji W, Xiaoling W, Place RF,
Guiting L, Lue TF, Long-Cheng L and Dong-Yan J: RNAa is conserved
in mammalian cells. PLoS One. 5:e88482010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao
X, Li D, Li S, Mao L, Huang K and Tong Q: MiRNA-558 promotes
tumorigenesis and aggressiveness of neuroblastoma cells through
activating the transcription of heparanase. Hum Mol Genet.
24:2539–2551. 2015. View Article : Google Scholar
|
|
116
|
Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, Xu
H, Ye Z and Li LC: Up-regulation of p21(WAF1/CIP1) by miRNAs and
its implications in bladder cancer cells. FEBS Lett. 588:4654–4664.
2014. View Article : Google Scholar
|
|
117
|
Zou Q, Liang Y, Luo H and Yu W:
MiRNA-mediated RNAa by targeting enhancers. Adv Exp Med Biol.
983:113–125. 2017. View Article : Google Scholar
|
|
118
|
Huang YP, Qiu LZ and Zhou GP: MicroRNA-939
down-regulates CD2-associated protein by targeting promoter in
HEK-293T cells. Renal Failure. 38:508–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Mao H, Zhu C, Zong D, Weng C, Yang X,
Huang H, Liu D, Feng X and Guang S: The nrde pathway mediates
small-RNA-directed histone H3 lysine 27 Trimethylation in
Caenorhabditis elegans. Curr Biol. 25:2398–2403. 2015. View Article : Google Scholar
|
|
120
|
Liu X, Fan Z, Li Y, Li Z, Zhou Z, Yu X,
Wan J, Min Z, Yang L and Li D: MicroRNA-196a-5p inhibits testicular
germ cell tumor progression via NR6A1/E-cadherin axis. Cancer Med.
9:9107–9122. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Guo F, Gao Y, Sui G, Jiao D, Sun L, Fu Q
and Jin C: MiR-375-3p/YWHAZ/β-catenin axis regulates migration,
invasion, EMT in gastric cancer cells. Clin Exp Pharmacol Physiol.
46:144–152. 2019. View Article : Google Scholar
|
|
122
|
Li J and Zou X: MiR-652 serves as a
prognostic biomarker in gastric cancer and promotes tumor
proliferation, migration, and invasion via targeting RORA. Cancer
Biomark. 26:323–331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
To KK, Leung WW and Ng SS: A novel
miR-203-DNMT3b-ABCG2 regulatory pathway predisposing colorectal
cancer development. Mol Carcinog. 56:4642016. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Wang C, Chen Q, Li S, Li S and Zhao Z:
Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in
gliomas. Oncotarget. 8:10287–10297. 2016. View Article : Google Scholar
|
|
125
|
Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X,
Guo X, He S and Chen R: Transcriptional inhibiton of Hoxd4
expression by miRNA-10a in human breast cancer cells. BMC Mol Biol.
10:122009. View Article : Google Scholar
|
|
126
|
Kang MR, Park KH, Yang JO, Lee CW and Kang
JS: MiR-6734 up-regulates p21 gene expression and induces cell
cycle arrest and apoptosis in colon cancer cells. PLoS One.
11:e1609612016. View Article : Google Scholar
|
|
127
|
Zhang Y, Fan M, Geng G, Liu B, Huang Z,
Luo H, Zhou J, Guo X, Cai W and Zhang H: A novel HIV-1-encoded
microRNA enhances its viral replication by targeting the TATA box
region. Retrovirology. 11:232014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Li S, Zhu Y, Liang Z, Wang X and Xie L:
Up-regulation of p16 by miR-877-3p inhibits proliferation of
bladder cancer. Oncotarget. 7:51773–51783. 2016. View Article : Google Scholar
|
|
129
|
Kim DH, Saetrom P, Snove O Jr and Rossi
JJ: MicroRNA-directed transcriptional gene silencing in mammalian
cells. Proc Natl Acad Sci USA. 105:16230–16235. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Cui C, Yu J, Huang S, Zhu H and Huang Z:
Transcriptional regulation of gene expression by microRNAs as
endogenous decoys of transcription factors. Cell Physiol Biochem.
33:1698–1714. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zardo G, Ciolfi A, Vian L, Starnes LM,
Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N,
et al: Polycombs and microRNA-223 regulate human granulopoiesis by
transcriptional control of target gene expression. Blood.
119:4034–4046. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Sepramaniam S, Ying LK, Armugam A, Wintour
EM and Jeyaseelan K: MicroRNA-130a represses transcriptional
activity of aquaporin 4 M1 promoter. J Biol Chem. 287:12006–12015.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Miao L, Yao H, Li C, Pu M, Yao X, Yang H,
Qi X, Ren J and Wang Y: A dual inhibition: MicroRNA-552 suppresses
both transcription and translation of cytochrome P450 2E1. Biochim
Biophys Acta. 1859:650–662. 2016. View Article : Google Scholar
|