Introduction
MicroRNAs (miRNAs/miRs) are 21–24 nucleotide
single-stranded non-coding small RNAs that play a crucial role in
the regulation of gene expression (1). miRNAs have been shown to play a key
role in the cellular response to environmental stresses, such as
starvation, hypoxia, oxidative stress and DNA damage. The
biological importance of miRNAs has been highlighted in recent
decades by the deregulation of miRNA expression in a variety of
human diseases, including cancer (2,3).
Several studies have found that the dysregulation of miRNA
expression is closely linked to tumor initiation, growth and
metastasis (4–6). Numerous miRNAs can function as
oncogenes or tumor suppressors, causing cancer or suppressing tumor
growth (7,8). Several miRNAs have been found to be
involved in epigenetic, transcriptional and post-transcriptional
processes (9,10). miRNA dysfunction disrupts the
expression of oncogenic or tumor-suppressing target genes, which is
implicated in cancer pathogenesis. The majority of mature miRNAs
are known to interact with the 3′UTR of target mRNAs via complete
or incomplete pairing to induce mRNA degradation or translational
inhibition (11,12). miRNAs are loaded onto Argonaute
(AGO) proteins in the cytoplasm to direct the RNA-induced silencing
complex (RISC), which is composed of AGO, DICER, TAR-RNA-binding
protein (TRBP) and trinucleotide repeat containing 6 (TNRC6), to
bind to the 3′UTR (13).
Post-transcriptional gene silencing in the cytoplasm is a
well-known function mediated by miRNAs through the miRNA-induced
silencing complex (miRISC). Furthermore, certain studies have
suggested that it occurs in the 5′-UTR (14) and even in protein-coding sequences
(15). However, interactions of
miRNAs with other regions, such as gene promoters, have been
reported (16,17). miRNAs are active in the nuclei,
according to research, and several miRNAs that are prominently
localized in the nucleus are associated with transcriptional
regulation. To support this, the majority of miRNAs have recently
been reported to be imported back into the nucleus following
maturation, with abundant putative miRNA target sites in gene
promoters in both the sense and antisense orientations, which
regulate transcriptions in the nucleus by binding to complementary
sequences in the promoters (18–20)
of target genes. miRNAs have been found to have two functions:
Transcriptional gene activation (TGA) and transcriptional gene
silencing (TGS) (21). It is now
clear that the transcriptional regulation of nuclear miRNAs can
promote the occurrence and progression of a variety of malignant
tumors, including gastric, prostate cancer (Pca) and lung (22–24).
In recent years, there has been a growing body of evidence
indicating an epigenetic interaction between DNA methylation
modification and nuclear miRNA expression in cancer (24–26).
Several mature miRNAs have been presently identified to be enriched
in the nuclei of various tissues and cell types using evolving
technologies such as high-throughput sequencing (27). These findings shed new light onto
the mechanisms and treatment strategies of miRNAs in the control of
tumor gene expression (23).
However, the types and numbers of miRNAs found in the nuclei of
different cells, as well as their potential biological regulatory
functions, remain unknown (28).
The understanding of the time frame and associated mechanisms
through which nuclear miRNAs regulate transcription in tumor cells
is particularly limited. As a result, it is of utmost importance to
investigate the regulatory networks of gene expression that not
only cover nuclear miRNA expression, but also its immediate effects
on different tumor cell types for cancer treatment by targeting or
manipulating the expression profile of nuclear miRNAs.
The present review article discusses a number of
processes involved in the biogenesis of miRNAs and their transport
into the nucleus. The structure, function and roles of miRNA in the
regulatory effects on AGO proteins are also discussed. In addition,
some software tools for predicting miRNA binding sites in order to
identify and predict the corresponding miRNAs are noted and
summarized. It is demonstrated that nuclear miRNAs, through
epigenetic modifications, mediate the regulatory function of TGA or
TGS, as well as their importance in tumorigenesis and development.
The aim of the present review was to discuss the endogenous
function of nuclear miRNAs in cancer cell growth and to propose
potential applications in disease treatment, such as the
development of drug targets via miRNAs (29).
The process of miRNAs entering the
nuclei
The majority of miRNAs have their own promoters that
exist in the genome on their own (30). The promoter, in conjunction with
RNA polymerase II (RNAP II), transcribes the genes encoding miRNAs
into primary miRNA transcripts (pri-miRNAs) (31). A microprocessor complex (32) converts these pri-miRNAs into short
70-nucleotide stem-loop structures known as precursor miRNAs
(pre-miRNAs) in the nucleus. The DROSHA RNase III enzyme and the Di
George syndrome critical region 8 double-stranded-RNA binding
protein comprise the microprocessor complex (DGCR8). DROSHA cleaves
the primary miRNAs in order to remove a portion of the unpaired RNA
sequences (33).
The nuclear export machinery, which consists of
Exportin-5 complexed with GTPase (Ran GTP), transports the
pre-miRNAs to the cytoplasm, where they are converted into mature
miRNAs, a 22-nt miRNA duplex, by cleavage of the hairpins by the
enzyme DICER (34). The asymmetry
rule is a strand-selection mechanism in which only one strand is
preferentially retained to form the functional miRISC, while the
other strand is degraded. In most cases, the miRISC induces mRNA
decay and translational suppression by interacting with
complementary sequences in the 3′UTR of target gene mRNAs (Fig. 1). Several previous studies have
found that miRISC is imported into the nucleus of mammalian cells,
and that nuclear RISC (an ~158 kDa complex) is smaller than its
cytoplasmic counterpart (a 20-fold larger complex of nearly 3 MDa).
Moreover, several RISC components have been identified to function
in the nucleus (21). Western blot
analysis has revealed the presence of AGO protein, DICER, TRBP and
GW182/TNRC6 in the nucleus, where they combine to form
multi-protein complexes (35).
Furthermore, miRISC plays a role in the transcriptional regulation
of a variety of life activities, including cell proliferation,
differentiation, development and apoptosis (11).
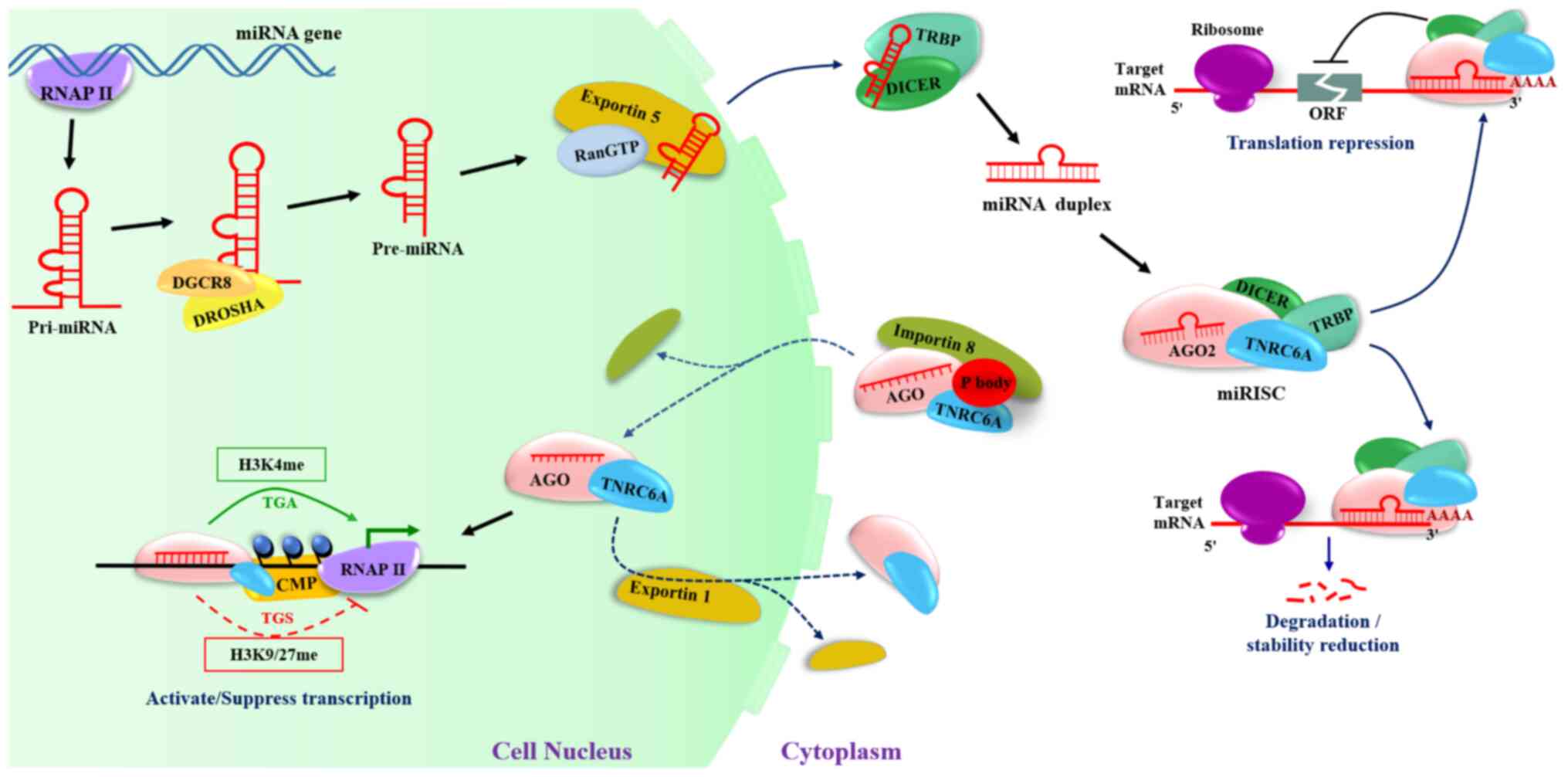 | Figure 1.Biogenesis of miRNAs and their return
to the nuclei. miRNAs, microRNAs; RNAP II, RNA polymerase II;
pri-mRNA, primary miRNA transcript; pre-miRNAs, precursor miRNAs;
H3K, histone H3; CMP, chromatin-modifying protein; AGO, Argonaute;
TNRC6A, trinucleotide repeat containing 6; TRBP, TAR-RNA-binding
protein; ORF, open reading frame; miRISC, miRNA-induced silencing
complex. |
A large number of nuclear transport receptor
proteins are directly involved in the process of miRNA import to
the nucleus (36). Importin 8 and
exportin 1, importin family members that can recognize nuclear
localization sequences in protein cargoes and assist their active
transport through the nuclear pore complex, have been found to be
involved in the nuclear and cytoplasmic shuttling of miRNAs and RNA
interference/RNA activation (RNAa) components (37,38).
To function in the nucleus, miRNAs must first form an AGO-miRNA
complex with the AGO protein, then bind to importin 8 and TNRC6A
(also known as GW182) in the cytoplasmic-processing body (P body)
for introduction into the nucleus (35,36).
Unlike other members of the cytoplasmic RISC, AGO2 lacks a nuclear
localization signal. TNRC6A also contains a nuclear localization
signal (NLS) and a nuclear export signal (NES), which can be
transported between the cytoplasm and the nucleus by importin 8 or
exportin 1 (39,40). As a result, by combining with
TNRC6A, which possesses NLS, AGO2 can function as a
nuclear-cytoplasmic shuttling protein to re-localize the miRNA to
the nucleus. TNRC6A functions as a scaffold in the nucleus,
allowing other effector proteins to be recruited (41). TNRC6A directs AGO-miRNA to promoter
region of target the gene, eventually cleaving the target RNA or
retaining it in the nucleus. As a result, TNRC6A is yet another
core protein involved in miRNA-mediated gene transcription
(42,43). Furthermore, exportin 1 can
transport AGO-TNRC6 complexes back to the cytoplasm. Consequently,
inhibiting exportin 1 leads to an accumulation of TNRC6A and AGO2
in the nucleus (44). DICER and
TRBP, on the other hand, are not attached to the RISC during their
import into the nucleus. Kalantari et al (45) used semi-quantitative mass
spectrometry to confirm that the association of AGO2 with
GW182/TNRC6 and AGO3 was most well preserved and more stable in
both the nuclei and cytoplasm. The interactions of AGO2 with DICER
and TRBP, on the other hand, are restricted to the cytoplasm.
Although cytoplasm-processed miRNAs can be introduced into the
nucleus to regulate gene expression, the time frame and mechanisms
through which miRNAs perform their functions in the nucleus remain
to be fully elucidated.
Argonaute protein family and its structural
characteristics
AGO proteins are members of the Argonaute family,
which is highly specialized and conserved (46). In several organisms, they are the
primary participants in the small non-coding RNA-mediated gene
silencing pathway, and they are primarily responsible for
regulating gene expression (47).
The Argonaute family is divided into two groups: The ubiquitously
expressed AGO subfamily proteins, also known as the AGO clade, and
the gonad-expressed PIWI subfamily proteins, also known as the PIWI
clade (48). AGO subfamily
proteins are typically associated with miRNAs and short-interfering
RNAs (siRNAs) and collaborate in the process of
post-transcriptional gene silencing (49). The PIWI clade proteins are only
found in another type of small RNA known as PIWI-interacting RNA
(piRNA). Unlike the AGO clade, PIWI proteins are primarily
expressed in germ cells and help to silence male germline
transposons by interacting with piRNAs (50).
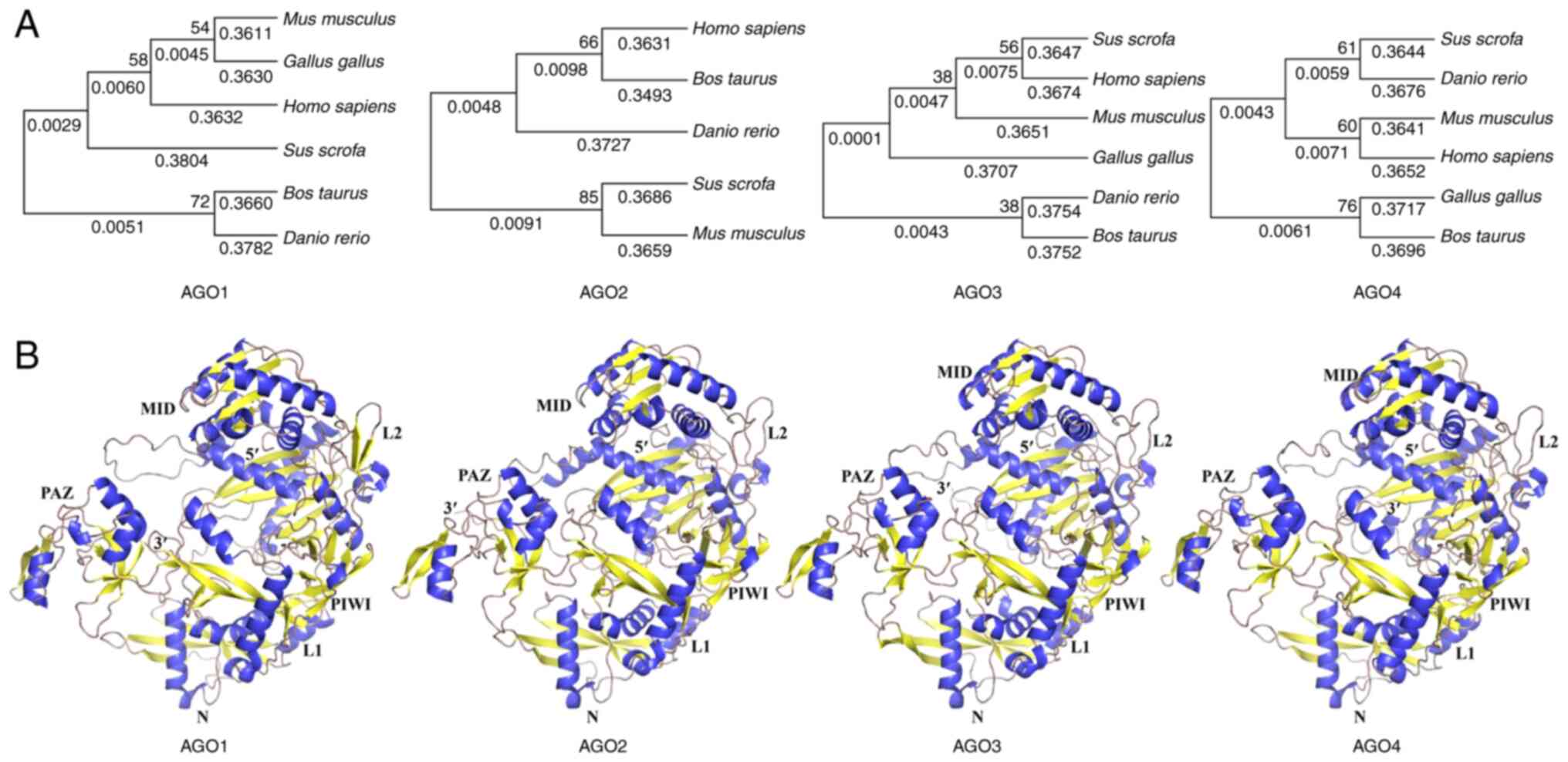
The majority of AGO proteins have 700-1,000 amino
acid residues and have molecular weights of ~100 kDa, with 80%
amino acid sequence identity (51). AGO protein amino acid sequences are
highly conserved across species (Fig.
2A). The AGO proteins have an amino-terminal (N), a
PIWI/Argonaute/Zwille (PAZ) domain, a middle (MID) domain and a
PIWI domain (52–54). It should be noted, however, that
the N-terminal domain is required for small RNA loading and
double-stranded small RNA unwinding (55). Mammalian cells contain four AGO
proteins (AGO1-4), all of which are loaded with siRNA and miRNA
duplexes (56). They have similar
biochemical preferences for binding to duplex RNA (45,57),
but only AGO2 cleaves completely complementary mRNA targets
(58).
AGO proteins have been previously shown to play
critical roles in germ cell maintenance and division, as well as in
transcriptional and translational regulation (59). These proteins are also involved in
the repair of double-strand breaks in DNA through homologous
recombination, chromatin modifications and alternative splicing.
The AGO proteins, on the other hand, are incapable of catalyzing
any reaction on their own. They are the core component of the RISC,
interacting directly or indirectly with various partners, such as
DICER, TRBP and GW182 family proteins, which are responsible for
binding to short RNA and the cleavage or inhibition of target mRNA
translation (60). A growing body
of evidence suggests that AGO not only functions as a critical
regulator of gene expression in the cytoplasm, but also mediates
the repression of miRNA transcription in the nucleus, with importin
8 acting as an essential co-factor and assisting in the
localization of AGO2 in the nucleus. miRNAs have also been found to
colocalize in the nucleus with the AGO-TNRC6A complex and to
exhibit gene-silencing activity (39). AGO1 and AGO2 proteins, in
particular, promote histone H3 lysine 9 (H3K9) methylation and play
critical roles in miRNA-mediated gene regulation (61). Notably, while previous research
provided knowledge of the nuclear functions of AGO clade proteins,
the mechanisms underlying the transport of cytoplasmic AGO proteins
into the nucleus remain largely unknown.
Furthermore, the association of AGO-bound miRISC
with various cellular organelles or structures remains unknown.
Data on the cellular localization of the numerous steps involved in
the assembly, function and recycling of these complexes are
limited. It is well understood that AGO-mediated epigenetic
modification is a common phenomenon that plays a role in the
transcriptional regulation of nuclear miRNAs. However, further and
more in-depth knowledge of the types and quantities of modified
proteins recruited by the AGO complex, such as epigenetic enzymes
and transcription factors (TFs) is required.
Identification and prediction of nuclear
miRNAs
A number of mature miRNAs have been found to be
enriched in the nucleus using high-throughput profiling
technologies, such as microarray and deep sequencing. The number
and types of nuclear miRNAs found in different species, tissues and
cells are also distinct, emphasizing the specificity and importance
of nuclear miRNA regulation (Table
I).
 | Table I.Identification of nuclear miRNAs. |
Table I.
Identification of nuclear miRNAs.
| Tissue/cell | Method | Number of nuclear
miRNAs | Highly expressed
miRNAs | (Refs.) |
|---|
| Neural stem | ChIP/RT-qPCR | 21 | miR-16 | (67) |
|
|
|
| miR-30c |
|
| Mouse
hemopoietic | ChIP/RT-qPCR | 4 | miR-709/706 | (68) |
|
|
|
| miR-690/467a* |
|
| HeLa | MicroRNA
sequencing | 346 | miR-30a-5p | (35) |
|
|
|
| miR-191-5p |
|
|
|
|
| miR-148a-3p |
|
| HeLa | RT-qPCR
profiling | 750 | miR-484 | (69) |
|
|
|
| miR-191 |
|
| Human
megakaryocyte | Perl script | 21 | miR-19a-3p | (70) |
|
|
|
| miR-197-5p |
|
|
|
|
| miR-30c |
|
| Human
nasopharyngeal carcinoma | Deep
sequencing | 339 | miR-29b | (71) |
|
|
|
| miR-32 |
|
|
|
|
| miR-148a |
|
|
|
|
| miR-148b |
|
| Rat myoblasts | Microarray
assay/RT-qPCR | 5 | miR-340-5p | (72) |
|
|
|
| miR-351 |
|
|
|
|
| miR-494 |
|
| Mouse livers | miRNA microarray
assay | 44 | miR-30e | (73) |
|
|
|
| miR-709 |
|
|
|
|
| miR-690 |
|
| Pig granulosa | Digital droplet
RT-PCR | 12 | miR-122 | (74) |
|
|
|
| miR-142-5p |
|
|
|
|
| miR-192 |
|
|
|
|
| miR-378 |
|
The database RNALocate (http://www.rna-society.org/rnalocate/) is the first
database which comprehensively focuses on RNA subcellular
localization. miRNA symbols from the miRBase database are also
provided in RNALocate. This database also includes some miRNAs with
nuclear localization (62). Other
databases, such as microPIR2 (https://tools4mirs.org/software/mirna_databases/micropir2/)
(63), Tools4miRs (https://tools4mirs.org/) (64) and miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/mirwalk2) (65) provide predicted and validated
information of miRNA-target interaction. Furthermore, the program
mirSTP (miRNA transcription Start sites Tracking Program) was
created to predict the transcription start sites (TSSs) of miRNAs.
mirSTP can be found at http://bioinfo.vanderbilt.edu/mirSTP/ (66). The knowledge of nuclear-localized
miRNAs is critical for locating the core promoters of miRNAs and
integrating the control of miRNA transcription into complex
regulatory networks.
In Table I, a list
of various nuclear miRNAs identified in various studies (35,67–74)
is presented.
Transcriptional regulation by nuclear
miRNA-mediated epigenetic modification
The mechanism of nuclear miRNA-mediated
transcription regulation is similar to that of piRNA/PIWI-mediated
transposon silencing (75). piRNAs
assemble with PIWI members of AGO proteins to form piRNA-induced
RNA silencing complexes (piRISCs) and modulate key signaling
pathways at the transcriptional or post-transcriptional level in
the piRNA/PIWI pathway (76).
Studies have demonstrated that the nuclear PIWI-piRNA complexes at
the cytoplasmic PIWI proteins silence their targets
post-transcriptionally via piRNA-directed cleavage and the
transcriptional ping-pong cycle. PIWI is translocated to the
nucleus after piRISC assembly and represses transposons
co-transcriptionally by inducing the formation of local
heterochromatin at target transposon loci (77). CG9754 is a critical downstream
pathway protein of PIWI/piRNA that works with PIWI to repress
transposon transcription (78).
Its effect on PIWI/piRNA-guided transcriptional silencing is
realized after histone H3K9 is methylated as a result of its
binding to the target DNA or RNA and the inhibition of genetic
transposition by PIWI (79). The
overexpression of the PIWI/piRNA pathway in tumor cells can lead to
dysregulated methylation at H3K9, and some tumor suppressor genes
may be inhibited (80). Similarly,
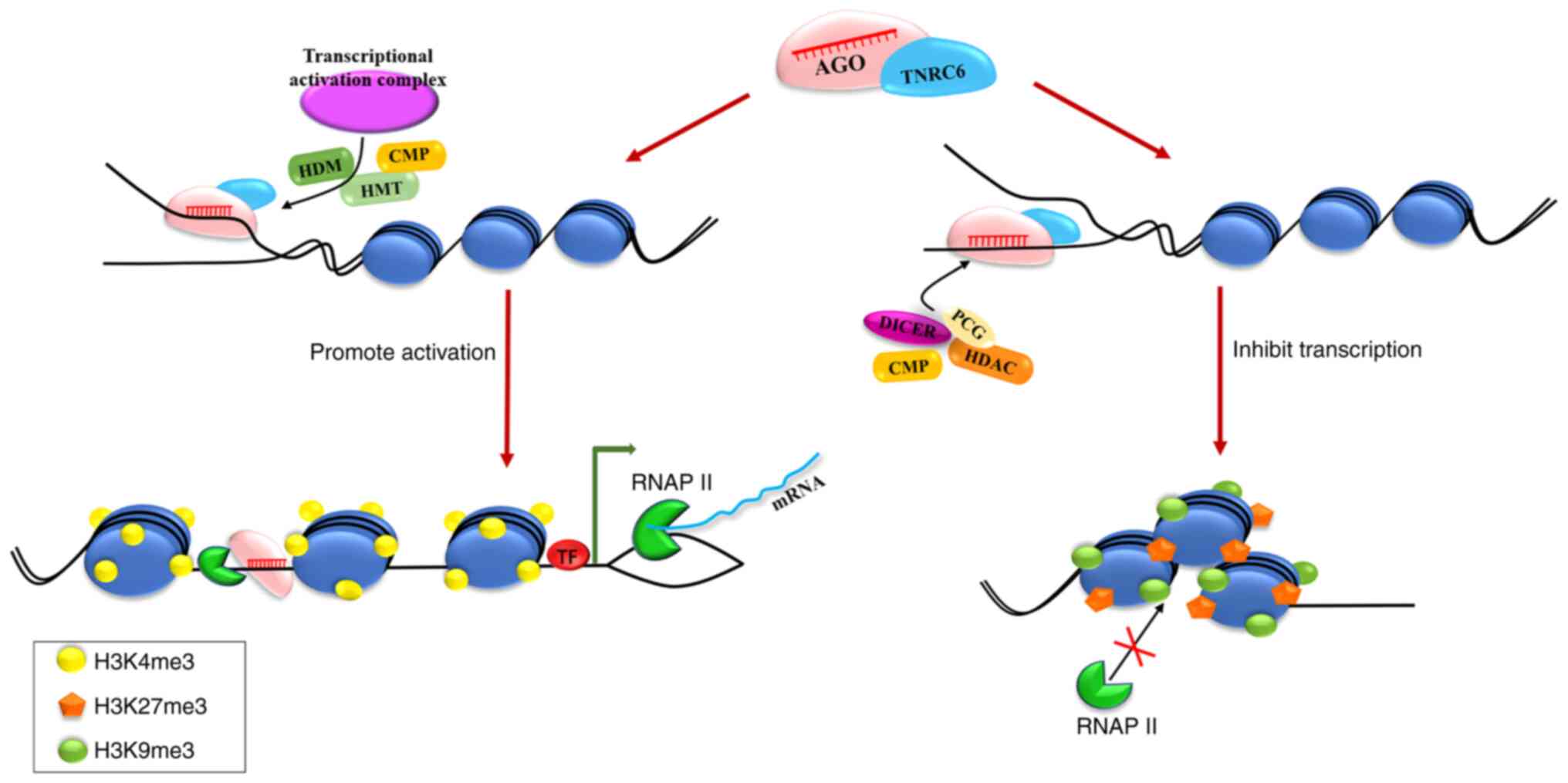
the transcriptional activation or inhibition of genes by nuclear
miRNAs is primarily accomplished through the mediation of
epigenetic modifications such as H3K9 methylation and H3 lysine 4
(H3K4) acetylation. AGO proteins, in particular, as well as
proteins from the epigenetic machinery, alter the structure of
chromatin (Fig. 3). This process
is critical for tumor cell growth, proliferation, invasion and
migration, which has become a hot topic in tumor regulation
research.
Following maturation, the majority of miRNAs are
imported back into the nucleus, according to sequencing and
bioinformatics analyses (81).
miRNAs guide AGO proteins to promoter targets (82), such as the TATA-box motif or
regions associated with transcription factors, owing to an
abundance of putative miRNA target sites in gene promoters
(83,84). The recruitment of AGO proteins
(primarily AGO2) and the enrichment of RNAP II at regulated gene
promoters then alter histone epigenetic modification and
transcription factor binding. During the initiation of
transcription, the DNA double helix unwinds. miRNAs typically
mediate acetylation at H3K9 and histone H3 lysine 27 (H3K27), as
well as methylation at H3K4, which stabilizes the promoter region
and increases the recognition and binding efficiency of RNAP II,
thereby activating gene transcription (85–87).
Some miRNAs, such as miR-26a-1, miRNA-558, miR-195-5p and miR-24-1,
have been shown to induce target gene expression. Recent research,
for example, has demonstrated that nucleus-enriched miR-195-5p may
regulate Foxo3 expression in porcine ovarian granulosa cells,
thereby regulating follicular development and atresia. The
mechanism of action is considered to involve miR-195-5p binding to
the Foxo3 promoter TATA box, transcription activation, histone
acetylation and hypomethylation and transcriptional regulation. Of
note, it was found that AGO2, rather than other AGO proteins,
regulates the effect of miR-195-5p on Foxo3 transcription (88). However, PcG protein recruitment
increases H3K27 trimethylation (H3K27 me3) and decreases H3K4
trimethylation (H3K4 me3), and DNA methylation is frequently
observed at some miRNA-targeted promoter regions. The chromatin
structure would then be altered, and the promoter region would be
tightened to establish a non-permissive transcriptional status
(89,90). Several studies have found that
nuclear miRNAs exert an inhibitory effect on transcriptional
regulation. The lysosome is the critical organelle of autophagy,
according to a study on autophagy and lysosome biogenesis, and the
TF EB (TFEB) is the primary regulator of lysosome biogenesis.
Dephosphorylated TFEB enters the nucleus and binds to the
palindromic sequence (GTCACGTGAC) at the promoter of autophagy and
lysosomal biogenesis-related genes, i.e., the coordinated lysosomal
expression and regulation (CLEAR) element, to activate
transcription. The miR-30b-5 represses the transcription of
TFEB-dependent downstream genes by targeting CLEAR, the TFEB
binding motif, further inhibiting the flux of autophagy and
lysosome biogenesis (91).
miRNAs are now known to be potent gene regulators.
However, there is a limited understanding of the time frame and
mechanisms through which miRNAs regulate transcription (92). Several in vitro studies have
been conducted over the past few decades, which have involved the
transfection of pre-miRNAs or mature mRNA mimics into immortalized
and cancer cell lines. However, further research is required to
determine how well these findings reflect endogenous miRNA
functions in vivo. Emerging evidence regarding miRNAs has
demonstrated the significance of the regulatory mechanisms of
nuclear miRNA expression in cancer (93,94).
The critical role of nuclear miRNAs in tumor initiation, growth and
metastasis is discussed below.
Role of nuclear miRNAs in the
transcriptional regulation of tumor cells
Previous research has suggested that changes in
miRNA expression and function may contribute to cancer formation
and progression, as well as to cellular responses to various types
of stress (95,96). The Let-7 family, for example,
influences tumor differentiation, migration, invasion and
proliferation, and its members are dysregulated in various types of
cancer. They can be activated or deactivated, and they can function
as tumor suppressors or oncogenes (97). miR-124 has the potential to be a
potent tumor suppressor in cancers with a high myc expression. A
previous study discovered that miR-124 had an RNA-activating
function, which resulted in the direct induction of p27 protein
levels by binding to and inducing transcription on the p27 promoter
region, causing G1 arrest. In breast and ovarian cancers, miR-124
can not only reverse the myc/p27/phospho-Rb protein signature to
target cell proliferation, but it can also block integrin 1 and
increase sensitivity to etoposide (98). Another family of onco-suppressor
miRNAs, miR-34, may target cell cycle-related genes (including
BCL2, MET, MYC, AXL and CDK4) and has tumor-suppressive properties
that mediate apoptosis, cell cycle arrest and senescence. miR-34a,
in particular, has been shown to be a direct transcriptional target
of p53, which is frequently mutated in epithelial ovarian
carcinomas (99,100). Furthermore, the miR-21 and
miR-17-92 clusters are potent carcinogenic factors. miR-21 is a
robust modulator of tumor behavior and malignant transformation.
The expression of miR-21 in non-small cell lung cancer (NSCLC)
tissues was regulated by the transcriptional factor nuclear factor
(NF)-B binding to its promoter element. As a result, inhibiting
NF-B via RNA silencing protects cells from cisplatin by decreasing
miR-21 expression (101).
Clusters of miR-17-92, on the other hand, are linked to cell
proliferation, migration and angiogenesis. According to the
findings of previous studies, miR-17-5p may inhibit the
proliferation and induce the apoptosis of NSCLC H460 cells by
targeting TGFR2 (102,103).
A number of factors, including matrix
metalloproteinase 14 (MMP-14), circular RNAs (circRNAs_ and
Cockayne syndrome protein B (CSB), play a role in the
miRNA-mediated regulation of tumorigenesis (26,104,105). MMP-14, a membrane-anchored MMP
that promotes tumorigenesis and aggressiveness, has been found to
be highly expressed in neuroblastoma (NB) and gastric cancer. Xiang
et al (26) identified one
binding site of miRNA-584-5p (miR-584-5p) within the MMP-14
promoter through mining computational algorithm program and
Argonaute-chromosome interaction dataset. Their results indicate
that promoter-targeting miR-584-5p exerts tumor suppressive
functions in NB through repressing the transcription of MMP-14.
Zheng et al (106)
identified adjacent binding sites of the myeloid zinc finger 1
(MZF1) and miRNA-337-3p (miR-337-3p) in the MMP-14 promoter by
mining computational algorithm programs and chromatin
immunoprecipitation datasets. According to the findings, miR-337-3p
binds directly to the MMP-14 promoter to repress MZF1-mediated
MMP-14 expression, thereby suppressing GC progression (106). circ-PLEKHM3 was identified as one
of the most significantly downregulated circRNAs in ovarian cancer
tissues by Zhang et al (107). circPLEKHM3 overexpression was
found to inhibit cell growth, migration and epithelial-mesenchymal
transition (EMT). Mechanistically, when circPLEKHM3 was
downregulated in ovarian cancer, the competitive binding of miR-9
was attenuated, resulting in a decrease in miR-9-targeted gene
expression, which promoted tumor cell proliferation (107). It has been have demonstrated that
the binding sites of Let-7 and miR-29 in the proximal 3′UTR of CSB
are highly conserved. Let-7 and miR-29 suppress CSB expression by
directly targeting the 3′UTR of CSB in lung cancer cells, resulting
in NSCLC cell apoptosis (104).
The importance of the regulatory mechanisms of nuclear miRNA
expression in cancer has been demonstrated by emerging evidence
exhibiting genetic and epigenetic dysregulations of the machinery
of miRNA biogenesis and the regulatory mechanisms of miRNAs.
Nuclear miRNAs activate
transcriptional regulation in tumor cells
Under certain conditions, nuclear miRNAs have been
shown to activate gene transcription and increase the expression of
target genes. The human miR-373, which induces gene transcription
by targeting the E-cadherin (CDH1) and cold-shock domain-containing
protein 2 (CSDC2) promoters, was the first to be identified as an
activator of gene transcription (25).
Nuclear miRNAs typically act at the promoter site to
achieve molecular regulation. It has been reported, have found that
RNAP II is enriched at the E-cadherin and CSDC2 promoters following
miR-373 transfection (25).
Furthermore, there is direct evidence that RISC interacts with
miRNA targets in the nucleus, allowing for miRNA enrichment.
Christofides et al (108)
found that miR-548c-5p is highly enriched in human podocyte nuclei
and can promote podocyte differentiation. The RISC complex, which
is guided by miR-548c-5p, interacts with the promoter region of the
gene to increase FOXC2 expression. There is evidence to suggest
that miRNAs may function as nucleoplasmic gene regulators via a
mechanism other than classic post-transcriptional repression. The
nuclear miRNA-mediated regulation of gene transcriptional
activation contradicts previously discovered miRNA functions
involving only the degradation of genes and the inhibition of gene
expression (25,109,110). Deep sequencing analysis has
revealed that AGO2 is responsible for the majority of miRNAs
entering the nucleus (Table II).
AGO can recruit TFs and epigenetic enzymes to the promoter region
of a gene, such as chromatin-modifying protein (CMP), histone
demethylase and histone methyltransferase (111,112), to initiate histone modification
and activate gene transcription (Fig.
3). For example, miR-589 has been shown to bind to the
cyclooxygenase-2 (COX-2) promoter RNA and activate transcription.
Specifically, the recognition of the promoter RNA by the
miR-589-AGO2-GW182 complex may result in the recruitment of other
factors, such as WDR5 and the activation of COX-2 and phospholipase
A2 group IVA (PLA2G4A) gene expression (112). Turner et al (113) also provided the first in
vivo evidence of RNAa by an endogenous miRNA in
Caenorhabditis elegans. It has also been demonstrated that
miRNA lin-4 can attract RNAP II to its promoter region to initiate
transcription. This lin-4 miRNA overexpression is sufficient for
autoactivation (113,114).
 | Table II.Advances in the transcriptional
activation of miRNAs. |
Table II.
Advances in the transcriptional
activation of miRNAs.
| miRNA | Type of cell | Type of AGO | Target
location | Mechanism | (Refs.) |
|---|
| miR-373 | Prostate
cancer | AGO2 | E-cadherin
promoter/CSDC2 promoter | / | (25) |
| miR-24-1 | 293T | AGO2 | FBP1 and
FANCC enhancer | Increase in
H3K27ac/H3K4me1 modification and p300/CBP combination; decrease in
H3K9me3 modification | (86) |
| miR-589 | Lung
adenocarcinoma | AGO2 | COX-2
promoter | TFs are
recruited | (112) |
| miRNA-558 | Neuroblastoma | AGO1 | HPSE
promoter | Increase in H3K4me3
modification and decrease in H3K9me2/H3K27me3 modification | (115) |
| miR-6734 | Colorectal
cancer | / | p21
promoter | Increase in H2B and
H3 acetylation modification and decrease in H3K9me2
modification | (126) |
| miR-H3 | CD4+
T-lymphocytes | / | HIV-15′
LTR | Pol II
pre-initiation complexes (PICs) are assembled at TATA box | (127) |
| miR-877-3p | Bladder cancer | / | p16
promoter | / | (128) |
| miR-195-5p | Porcine
granulosa | AGO2 | Foxo3
promoter | Activation of RNA
polymerase II initiates transcription or epigenetic
modification | (88) |
Nuclear miRNAs are currently being studied in cancer
therapeutics and tumorigenesis research, in addition to regulating
gene transcription activation as previously described (110–112). As previously demonstrated, in NB,
AGO1 interacts with RNAP II at active promoters throughout the
genome. Through the binding site in the promoter, miR-558 induces
the transcriptional activation of heparanase (HPSE), facilitating
tumorigenesis and NB aggressiveness. miR-558, in an AGO1-dependent
manner, induces the enrichment of the active epigenetic marker and
RNA pol II on the HPSE promoter in NB cells, which is inhibited by
repressing the miR-558-promoter interaction (115). Huang et al (111) discovered that several miRNAs
(e.g., miR-370, miR-1180 and miR-1236) with predicted target sites
in the Ccnb1 promoter activated the expression of cyclin B1 (CCNB1)
in mouse cells and influenced tumor development or growth in
vivo. AGO1 transports miR-744 to the CCNB1 promoter region,
where it promotes RNAP II and H3K4 me3 trimethylation at the CCNB1
TSS. Ccnb1 overexpression has been shown to promote tumorigenesis
and to function as a putative oncogene in a variety of cancers
(111,116). Wang et al (116) discovered that miR-370, miR-1180
and miR-1236 bound to the P21 promoter and upregulated p21
transcription in human bladder cancer cells. p21 expression
attenuated the cell cycle of bladder cancer cells, preventing
invasion and metastasis (116).
Of note, some enhancer markers, such as H3K27ac, are
found in miRNA genes. miRNAs, according to Zou et al
(117), interact at genomic loci,
where enhancer-derived RNA (eRNA) is transcribed, and increase the
mRNA levels of adjacent genes by promoting a transcriptionally
active chromatin state. The overexpression of miR-24-1, for
example, in 293T cells can increase the expression of its
neighboring genes, FBP1 and FANC. Increased levels of miR-24-1 will
result in the enrichment of RNA polymerase II, p300/CBP and
enhancer RNAs, as well as an increase in histone H3K27ac/H3K4 me1
modification and a decrease in H3K9 me3 modification (86). Notably, ChIP-qPCR has revealed that
AGO2 is present at the enhancer locus (86). Furthermore, the protein-coding
genes, ITGA9, CTDSPL, VILL and PLCD1, surround the miR26a-1 gene.
However, when the seed region of the miRNAs is deleted or mutated,
or when the enhancer locus is deleted, the activation is disrupted,
suggesting that this function is dependent on the base-pairing of
miRNA-enhancers.
Inhibitory effect of nuclear miRNAs on
the transcriptional regulation of tumor cells
It is now clear that miRNAs regulate gene expression
in mammalian somatic cells at both the transcriptional and
post-transcriptional levels. Apart from post-transcriptional gene
silencing via the RISC pathway in the cytoplasm, it is well
established that the regulatory function of miRNAs in the nucleus
is primarily achieved through transcriptional activation or
inhibition of genes via epigenetic modifications such as H3K9
methylation and H3K4 acetylation (118). Following miRNA transport into the
nuclei, AGO proteins recruit inhibitory complexes to the
miRNA-targeted promoter region (Fig.
3), which are primarily composed of RISC (AGO and DICER1
proteins), PcG elements (YY1, EZH2 and SUZ12), CMPs and histone
deacetylase. This interaction allows the protein inhibitor complex
to move closer to the targeted promoter region, causing an increase
in H3K27 me3 modifications and a decrease in H3K4 me3
modifications, altering the chromatin structure and establishing a
non-permissive transcriptional status (119). Some examples of the regulatory
mechanisms of nuclear miRNA-mediated transcriptional repression are
presented in Table III.
 | Table III.Advances in the transcriptional
repression of miRNAs. |
Table III.
Advances in the transcriptional
repression of miRNAs.
| miRNA | Type of cell | Type of AGO | Target
location | Mechanism | (Refs.) |
|---|
| miR-10a | Breast cancer | AGO1/AGO3 | HOXD4
promoter | Increase in DNA
methylation and H3K27me3 modification | (125) |
| miR-320 | Cervical
carcinoma | AGO1 | POLR3D
promoter | EZH2 binding and
increase in H3K27me3 modification | (129) |
| miR-423-5p | Breast cancer | AGO2 | PR
promoter | Increase in H3K9me2
modification | (61) |
| miR-939 | Neuroblastoma | / | NF-κB | / | (130) |
| miR-584-5p | Neuroblastoma | AGO2 | MMP-14
promoter | Promote enhancer
enrichment of zeste homolog 2 and the modification of
H3K9me2/H3K27me3 | (26) |
| miRNA-337-3p | Gastric cancer | AGO2 | MMP-14
promoter | Increase in
H3K9me2/H3K27me3 modification and decrease in H3K4me3
modification | (106) |
| miR-223 | Myeloid
progenitors | AGO1/AGO2 | NFI-A
promoter | Increased levels of
H3K4me3 activation marks and decreased H3K27me3 repressive
marks | (131) |
| miR-130a | Astrocytes | / | AQP4 M1
promoter | / | (132) |
| miR-552 | Hepatoma | / | CYP2E1
promoter | Inhibited Pol II
binding to the CYP2E1 | (133) |
Studies revealing the epigenetic dysregulations of
nuclear miRNAs, biogenesis-based machinery and nuclear miRNA
regulatory mechanisms have demonstrated the importance of miRNA
expression regulation in cancer (120). This may lead to the discovery of
novel predictive markers for the prognosis of cancer patients. For
example, in gastric cancer, Guo et al (121) demonstrated for the first time,
using a dual-luciferase report system, that YWHAZ was a target gene
of miR-375 and played a role in miR-375 regulation in the malignant
phenotype of gastric cancer. However, the precise mechanism through
which YWHAZ regulates gastric cancer cell migration and invasion
remains unknown. The overexpression of miR-375 was shown to inhibit
gastric cancer cell migration invasion, EMT, and the activation of
the Wnt/β-catenin signaling pathway, indicating that the
miR-375/YWHAZ axis may be a novel therapeutic target for gastric
cancer (121). Similarly, Liu
et al (22) found that
miR-675 aided gastric cancer cell proliferation and invasion by
targeting the paired-like homeodomain transcription factor 1
(PITX1) and promoting EMT and the activation of the Wnt/β-catenin
signaling pathway. Furthermore, the role of miR-652 as an oncogene
in gastric cancer in targeting the RORA pathway can promote gastric
cancer cell proliferation, migration and invasion and may be a
novel predictive marker for the poor prognosis of patients with
gastric cancer (122).
Furthermore, the role of miR-652 in targeting the VEGF pathway can
impede the formation of vascular networks and may be used to
develop anti-angiogenetic agents. The molecular mechanisms of
nuclear miRNAs regulating the occurrence of cancer have been
explained using newly developed techniques and more detailed
exploration.
Majid et al (23) used miRNA-205 to transfect three
human PCa cell lines (LNCaP, PC3 and Du145), as well as a
non-malignant epithelial PCa cell line (RWPE-1). Their findings
revealed that miR-205 functioned as a tumor suppressor miRNA in Pca
by directly targeting the tumor suppressor genes, IL24 and IL32,
via the apoptotic and cell-survival pathways, and that its
overexpression easily induced the expression of both genes. Further
investigations revealed that miR-205 directly targeted tumor
suppressor genes (IL-24 and IL-32) to suppress tumorigenesis via
active histone modifications, such as 2H3K4, 3H3K4, the acetylation
of Lys 9/14 of histone 3 and pol II (23). miR-584-5p has been shown to inhibit
MMP-14 expression in NB cells. AGO2 has been shown to transport
miR-584-5p to bind to the MMP-14 promoter, promoting EZH2
enrichment and the methylation of H3K27 and H3K9. It has also been
shown to prevent MMP-14 transcription from suppressing NB cell
growth, migration, invasion and angiogenesis (26). ABCG2 repression has been linked to
aberrant promoter methylation in a certain type of cancer,
including renal carcinoma and multiple myeloma. In colorectal
cancer (CRC), the de novo DNA methyltransferase (DNMT3B) is
a direct target of miR-203. Notably, miR-203 expression in CRC can
be downregulated by reducing DNMT3B repression, resulting in a
lower miR-203 expression in CRC and causing methylation of the
ABCG2 promoter, which significantly inhibits ABCG2 expression and
promotes cancer development (123). Notably, miR-215-5p has been found
to target both the PCDH9 gene promoter and the mRNA 3′UTR,
inhibiting PCDH9 expression, while promoting glioma cell growth and
proliferation (124).
Furthermore, while several studies have demonstrated the oncogenic
or tumor-suppressing roles of nuclear miRNAs, there are some
unresolved issues regarding the possible regulatory mechanisms
modulating nuclear miRNA expression in cancers.
Conclusions and future perspectives
miRNAs have been shown to be extensively deregulated
in human cancers over the past few decades, highlighting their
critical role in tumor initiation, growth and metastasis.
Similarly, the regulatory mechanisms that control miRNA expression
are strongly linked to cancer diagnosis, prognosis and treatment,
as well as to cancer pathogenesis. miRNAs typically suppress target
gene expression by partially binding to complementary sequences in
the 3′-UTR of target mRNAs, inhibiting translation and degrading
the target mRNAs. However, additional evidence suggests that
miRNA-mediated gene expression regulation also has other functions,
and the 5′-UTR coding sequence or promoter of the genes could be
the target region. Several small RNAs within miRNA response
elements, such as circRNAs and long non-coding RNAs, have been
shown to interact with miRNAs to regulate target gene expression.
Current research on the regulation of miRNAs, however, is still
focused on the traditional cytoplasmic pathways and how miRNAs
specifically control specific types of cancer. The present review
article summarized the series of processes involved in the entry of
miRNAs into the nucleus, particularly new mechanisms of nuclear
miRNA expression regulation in tumors. Mature miRNAs that are
carried into the nucleus and the target gene promoter region by AGO
proteins to activate epigenetic modification and regulate
transcription perform the majority of the regulatory functions of
miRNAs in the nucleus. A better understanding of nuclear miRNA
regulation and the potential underlying mechanisms may aid in the
understanding of the association between miRNAs and tumorigenesis.
These new signs of progress will lead to the development of more
precise therapeutic targets and the identification of novel
biological markers for the diagnosis of tumors and cancers, as well
as new avenues for future research on a variety of malignant
diseases.
Acknowledgements
Not applicable.
Funding
The present study was supported by a Special Project on Key-Area
R&D Program of scientific research institutions in Guangdong,
China (grant no. 2021B0707010007).
Availability of data and materials
Not applicable.
Authors' contributions
JL was mainly responsible for collecting all
relevant information and completing the review article. TY was were
mainly responsible for performing the literature search. ZH and HC
were mainly responsible for revising the manuscript. YB was
responsible for the conception of the review and the assignment of
tasks. There was no additional assistance with manuscript
preparation. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang V and Li LC: miRNA goes nuclear. RNA
Biol. 9:269–273. 2012. View Article : Google Scholar
|
|
2
|
Syeda ZA, Langden SS, Munkhzul C, Lee M
and Song SJ: Regulatory mechanism of microRNA expression in cancer.
Int J Mol Sci. 21:17232020. View Article : Google Scholar
|
|
3
|
Bhat IP, Rather TB, Bhat GA, Maqbool I,
Akhtar K, Rashid G, Parray FQ, Besina S and Mudassar S: TEAD4
nuclear localization and regulation by miR-4269 and miR-1343-3p in
colorectal carcinoma. Pathol Res Pract. 231:1537912022. View Article : Google Scholar
|
|
4
|
Zheng T, Zhou Y, Xu X, Qi X, Liu J, Pu Y,
Zhang S, Gao X, Luo X, Li M, et al: MiR-30c-5p loss-induced PELI1
accumulation regulates cell proliferation and migration via
activating PI3K/AKT pathway in papillary thyroid carcinoma. J
Transl Med. 20:202022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li L, Wei D, Zhang J, Deng R, Tang J and
Su D: MiR-641 inhibited cell proliferation and induced apoptosis by
targeting NUCKS1/PI3K/AKT signaling pathway in breast cancer.
Comput Math Methods Med. 2022:52038392022.PubMed/NCBI
|
|
6
|
Mirzaei S, Zarrabi A, Asnaf SE, Hashemi F,
Zabolian A, Hushmandi K, Raei M, Goharrizi MASB, Makvandi P,
Samarghandian S, et al: The role of microRNA-338-3p in cancer:
Growth, invasion, chemoresistance, and mediators. Life Sci.
268:1190052021. View Article : Google Scholar
|
|
7
|
El Fatimy R, Zhang Y, Deforzh E, Ramadas
M, Saravanan H, Wei Z, Rabinovsky R, Teplyuk NM, Uhlmann EJ and
Krichevsky AM: A nuclear function for an oncogenic microRNA as a
modulator of snRNA and splicing. Mol Cancer. 21:172022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo X, Dong J, He X, Shen L, Long C, Liu
F, Liu X, Lin T, He D and Wei G: MiR-155-5p exerts
tumor-suppressing functions in Wilms tumor by targeting IGF2 via
the PI3K signaling pathway. Biomed Pharmacother. 125:1098802020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gong R and Jiang Y: Non-coding RNAs in
pancreatic ductal adenocarcinoma. Front Oncol. 10:3092020.
View Article : Google Scholar
|
|
10
|
Gregorova J, Vychytilova-Faltejskova P and
Sevcikova S: Epigenetic regulation of MicroRNA clusters and
families during tumor development. Cancers (Basel). 13:13332021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of microRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar
|
|
12
|
Yang YL, Chang YH, Li CJ, Huang YH, Tsai
MC, Chu PY and Lin HY: New insights into the role of miR-29a in
hepatocellular carcinoma: Implications in mechanisms and
theragnostics. J Pers Med. 11:2192021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kobayashi H and Tomari Y: RISC assembly:
Coordination between small RNAs and Argonaute proteins. Biochim
Biophys Acta. 1859:71–81. 2016. View Article : Google Scholar
|
|
14
|
Zhang J, Zhou W, Liu Y, Liu T, Li C and
Wang L: Oncogenic role of microRNA-532-5p in human colorectal
cancer via targeting of the 5′UTR of RUNX3. Oncol Lett.
15:7215–7220. 2018.
|
|
15
|
Liu M, Roth A, Yu M, Morris R, Bersani F,
Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, et al: The
IGF2 intronic miR-483 selectively enhances transcription from IGF2
fetal promoters and enhances tumorigenesis. Genes Dev.
27:2543–2548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu W, San LA, Wang Z and Liu Y:
Identifying microRNA targets in different gene regions. BMC
Bioinformatics. 15 (Suppl 7):S42014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Z, Lan X, Han R, Wang J, Huang Y, Sun
J, Guo W and Chen H: MiR-2478 inhibits TGFβ1 expression by
targeting the transcriptional activation region downstream of the
TGFβ1 promoter in dairy goats. Sci Rep. 7:426272017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo D, Barry L, Lin SSH, Huang V and Li
LC: RNAa in action: From the exception to the norm. RNA Biol.
11:1221–1225. 2014. View Article : Google Scholar
|
|
19
|
Stavast CJ and Erkeland SJ: The
non-canonical aspects of microRNAs: Many roads to gene regulation.
Cells Basel. 8:14652019. View Article : Google Scholar
|
|
20
|
Fan L, Lai R, Ma N, Dong Y, Li Y, Wu Q,
Qiao J, Lu H, Gong L, Tao Z, et al: MiR-552-3p modulates
transcriptional activities of FXR and LXR to ameliorate hepatic
glycolipid metabolism disorder. J Hepatol. 74:8–19. 2021.
View Article : Google Scholar
|
|
21
|
Liu H, Lei C, He Q, Pan Z, Xiao D and Tao
Y: Nuclear functions of mammalian microRNAs in gene regulation,
immunity and cancer. Mol Cancer. 17:642018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu L, Tian YC, Mao G, Zhang YG and Han L:
MiR-675 is frequently overexpressed in gastric cancer and enhances
cell proliferation and invasion via targeting a potent anti-tumor
gene PITX1. Cell Signal. 62:1093522019. View Article : Google Scholar
|
|
23
|
Majid S, Dar AA, Saini S, Yamamura S,
Hirata H, Tanaka Y, Deng G and Dahiya R: MicroRNA-205-directed
transcriptional activation of tumor suppressor genes in prostate
cancer. Cancer. 116:5637–5649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kumar R and Xi Y: MicroRNA, epigenetic
machinery and lung cancer. Thorac Cancer. 2:35–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Place RF, Li LC, Pookot D, Noonan EJ and
Dahiya R: MicroRNA-373 induces expression of genes with
complementary promoter sequences. Proc Natl Acad Sci USA.
105:1608–1613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiang X, Mei H, Qu H, Zhao X, Li D, Song
H, Jiao W, Pu J, Huang K, Zheng L and Tong Q: MiRNA-584-5p exerts
tumor suppressive functions in human neuroblastoma through
repressing transcription of matrix metalloproteinase 14. Biochim
Biophys Acta. 1852:1743–1754. 2015. View Article : Google Scholar
|
|
27
|
Bai B, Liu H and Laiho M: Small RNA
expression and deep sequencing analyses of the nucleolus reveal the
presence of nucleolus-associated microRNAs. FEBS Open Bio.
4:441–449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Catalanotto C, Cogoni C and Zardo G:
MicroRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17:17122016. View Article : Google Scholar
|
|
29
|
Saito Y, Liang G, Egger G, Friedman JM,
Chuang JC, Coetzee GA and Jones PA: Specific activation of
microRNA-127 with downregulation of the proto-oncogene BCL6 by
chromatin-modifying drugs in human cancer cells. Cancer Cell.
9:435–443. 2006. View Article : Google Scholar
|
|
30
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
31
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar
|
|
32
|
Lai .Eric C: Micro RNAs are complementary
to 3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar
|
|
33
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Gene Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gagnon KT, Li L, Chu Y, Janowski BA and
Corey DR: RNAi factors are present and active in human cell nuclei.
Cell Rep. 6:211–221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weinmann L, Höck J, Ivacevic T, Ohrt T,
Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H and Meister G:
Importin 8 is a gene silencing factor that targets argonaute
proteins to distinct mRNAs. Cell. 136:496–507. 2009. View Article : Google Scholar
|
|
37
|
Wei Y, Li L, Wang D, Zhang CY and Zen K:
Importin 8 regulates the transport of mature microRNAs into the
cell nucleus. J Biol Chem. 289:10270–10275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Azmi AS, Uddin MH and Mohammad RM: The
nuclear export protein XPO1-from biology to targeted therapy. Nat
Rev Clin Oncol. 18:152–169. 2021. View Article : Google Scholar
|
|
39
|
Nishi K, Nishi A, Nagasawa T and Ui-Tei K:
Human TNRC6A is an argonaute-navigator protein for
microRNA-mediated gene silencing in the nucleus. RNA. 19:17–35.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Daniel S, Schindler SG, Johannes D,
Elisabeth K, Janina P, Stefan H, Reinhard D and Gunter M:
Importin-β facilitates nuclear import of human GW proteins and
balances cytoplasmic gene silencing protein levels. Nucleic Acids
Res. 43:7447–7461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Behm-Ansmant I, Rehwinkel J, Doerks T,
Stark A, Bork P and Izaurralde E: MRNA degradation by miRNAs and
GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping
complexes. Genes Dev. 20:1885–1898. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nishi K, Takahashi T, Suzawa M, Miyakawa
T, Nagasawa T, Ming Y, Tanokura M and Ui-Tei K: Control of the
localization and function of a miRNA silencing component TNRC6A by
argonaute protein. Nucleic Acids Res. 43:9856–9873. 2015.PubMed/NCBI
|
|
43
|
Hicks JA, Li L, Matsui M, Chu Y, Volkov O,
Johnson KC and Corey DR: Human GW182 paralogs are the central
organizers for RNA-Mediated control of transcription. Cell Rep.
20:1543–1552. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Castanotto D, Lingeman R, Riggs AD and
Rossi JJ: CRM1 mediates nuclear-cytoplasmic shuttling of mature
microRNAs. Proc Natl Acad Sci USA. 106:21655–21659. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kalantari R, Hicks JA, Li L, Gagnon KT,
Sridhara V, Lemoff A, Mirzaei H and Corey DR: Stable association of
RNAi machinery is conserved between the cytoplasm and nucleus of
human cells. RNA. 22:1085–1098. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kuhn CD and Joshua-Tor L: Eukaryotic
argonautes come into focus. Trends Biochem Sci. 38:263–271. 2013.
View Article : Google Scholar
|
|
47
|
Ryazansky S, Kulbachinskiy A and Aravin
AA: The expanded universe of prokaryotic argonaute proteins. mBio.
9:e01935–18. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Peters L and Meister G: Argonaute
proteins: Mediators of RNA silencing. Mol Cell. 26:611–623. 2007.
View Article : Google Scholar
|
|
49
|
Hutvagner G and Simard MJ: Argonaute
proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol.
9:22–32. 2008. View Article : Google Scholar
|
|
50
|
Siomi MC, Sato K, Pezic D and Aravin AA:
PIWI-interacting small RNAs: The vanguard of genome defence. Nat
Rev Mol Cell Biol. 12:246–258. 2011. View Article : Google Scholar
|
|
51
|
Sasaki T, Shiohama A, Minoshima S and
Shimizu N: Identification of eight members of the argonaute family
in the human genome. Genomics. 82:323–330. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Faehnle CR, Elkayam E, Haase AD, Hannon GJ
and Joshua-Tor L: The making of a slicer: Activation of human
argonaute-1. Cell Rep. 3:1901–1909. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schirle NT, Sheu-Gruttadauria J,
Chandradoss SD, Joo C and MacRae IJ: Water-mediated recognition of
t1-adenosine anchors argonaute2 to microRNA targets. Elife.
4:e076462015. View Article : Google Scholar
|
|
54
|
Park MS, Phan HD, Busch F, Hinckley SH,
Brackbill JA, Wysocki VH and Nakanishi K: Human argonaute3 has
slicer activity. Nucleic Acids Res. 45:11867–11877. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kwak PB and Tomari Y: The N domain of
argonaute drives duplex unwinding during RISC assembly. Nat Struct
Mol Biol. 19:145–151. 2012. View Article : Google Scholar
|
|
56
|
Czech B and Hannon GJ: Small RNA sorting:
Matchmaking for argonautes. Nat Rev Genet. 12:19–31. 2011.
View Article : Google Scholar
|
|
57
|
Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki
S, Liu Q and Tomari Y: ATP-dependent human RISC assembly pathways.
Nat Struct Mol Biol. 17:17–23. 2010. View Article : Google Scholar
|
|
58
|
Liu J, Carmell MA, Rivas FV, Marsden CG,
Thomson JM, Song JJ, Hammond SM, Joshua-Tor L and Hannon GJ:
Argonaute2 is the catalytic engine of mammalian RNAi. Science.
305:1437–1441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Huang V and Li LC: Demystifying the
nuclear function of argonaute proteins. RNA Biol. 11:18–24. 2014.
View Article : Google Scholar
|
|
60
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar
|
|
61
|
Younger ST and Corey DR: Transcriptional
gene silencing in mammalian cells by miRNA mimics that target gene
promoters. Nucleic Acids Res. 39:5682–5691. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang T, Tan P, Wang L, Jin N, Li Y, Zhang
L, Yang H, Hu Z, Zhang L, Hu C, et al: RNALocate: A resource for
RNA subcellular localizations. Nucleic Acids Res. 45:D135–D138.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Piriyapongsa J, Bootchai C, Ngamphiw C and
Tongsima S: MicroPIR2: A comprehensive database for human-mouse
comparative study of microRNA-promoter interactions. Database
(Oxford). 2014:bau1152014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lukasik A, Wójcikowski M and Zielenkiewicz
P: Tools4miRs-one place to gather all the tools for miRNA analysis.
Bioinformatics. 32:2722–2724. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Parveen A, Gretz N and Dweep H: Obtaining
miRNA-target interaction information from miRWalk2.0. Curr Protoc
Bioinformatics. 55:12.15.1–12.15.27. 2016. View Article : Google Scholar
|
|
66
|
Liu Q, Wang J, Zhao Y, Li CI, Stengel KR,
Acharya P, Johnston G, Hiebert SW and Shyr Y: Identification of
active miRNA promoters from nuclear run-on RNA sequencing. Nucleic
Acids Res. 45:e1212017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jeffries CD, Fried HM and Perkins DO:
Nuclear and cytoplasmic localization of neural stem cell microRNAs.
RNA. 17:675–86. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wong JJ, Ritchie W, Gao D, Lau KA,
Gonzalez M, Choudhary A, Taft RJ, Rasko JE and Holst J:
Identification of nuclear-enriched miRNAs during mouse
granulopoiesis. J Hematol Oncol. 7:422014. View Article : Google Scholar
|
|
69
|
Li ZF, Liang YM, Lau PN, Shen W, Wang DK,
Cheung WT, Xue CJ, Poon LM and Lam YW: Dynamic localisation of
mature microRNAs in human nucleoli is influenced by exogenous
genetic materials. PLoS One. 8:e708692013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sahu I, Hebalkar R, Kar S, Sreevidya TS,
Gutti U and Gutti RK: Systems biology approach to study the role of
miRNA in promoter targeting during megakaryopoiesis. Exp Cell Res.
366:192–198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H,
Shao P, Chen YQ and Qu LH: Deep sequencing of human nuclear and
cytoplasmic small RNAs reveals an unexpectedly complex subcellular
distribution of miRNAs and tRNA 3′trailers. PLoS One. 5:e105632010.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Politz JCR, Hogan EM and Pederson T:
MicroRNAs with a nucleolar location. RNA. 15:1705–1715. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Tang R, Li L, Zhu D, Hou D, Cao T, Gu H,
Zhang J, Chen J, Zhang CY and Zen K: Mouse miRNA-709 directly
regulates miRNA-15a/16-1 biogenesis at the posttranscriptional
level in the nucleus: Evidence for a microRNA hierarchy system.
Cell Res. 22:504–515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Toms D, Pan B, Bai Y and Li J: Small RNA
sequencing reveals distinct nuclear microRNAs in pig granulosa
cells during ovarian follicle growth. J Ovarian Res. 14:542021.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sato K and Siomi MC: The piRNA pathway in
Drosophila ovarian germ and somatic cells. Proc Jpn Acad Ser B Phys
Biol Sci. 96:32–42. 2020. View Article : Google Scholar
|
|
76
|
Gunawardane LS, Saito K, Nishida KM,
Miyoshi K, Kawamura Y, Nagami T, Siomi H and Siomi MC: A
slicer-mediated mechanism for repeat-associated siRNA 59 end
formation in drosophila. Science. 315:1587–1590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Brennecke J, Aravin AA, Stark A, Dus M,
Kellis M, Sachidanandam R and Hannon GJ: Discrete small
RNA-generating loci as master regulators of transposon activity in
drosophila. Cell. 128:1089–1103. 2007. View Article : Google Scholar
|
|
78
|
Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J,
Czech B and Hannon GJ: Panoramix enforces piRNA-dependent
cotranscriptional silencing. Science. 350:339–342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sienski G, Donertas D and Brennecke J:
Transcriptional silencing of transposons by Piwi and maelstrom and
its impact on chromatin state and gene expression. Cell.
151:964–980. 2012. View Article : Google Scholar
|
|
80
|
Watanabe T, Tomizawa S, Mitsuya K, Totoki
Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ,
Toyoda A, et al: Role for piRNAs and noncoding RNA in de novo DNA
methylation of the imprinted mouse Rasgrf1 locus. Science.
332:848–852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang N, Hu G, Myers TG and Williamson PR:
Protocols for the analysis of microRNA expression, biogenesis, and
function in immune cells. Curr Protoc Immunol. 126:e782019.
View Article : Google Scholar
|
|
82
|
Fu Y, Zhang L, Zhang R, Xu S, Wang H, Jin
Y and Wu Z: Enterovirus 71 suppresses miR-17-92 cluster through
up-regulating methylation of the miRNA promoter. Front Microbiol.
10:6252019. View Article : Google Scholar
|
|
83
|
Younger ST, Pertsemlidis A and Corey DR:
Predicting potential miRNA target sites within gene promoters.
Bioorg Med Chem Lett. 19:3791–3794. 2009. View Article : Google Scholar
|
|
84
|
Chellini L, Frezza V and Paronetto MP:
Dissecting the transcriptional regulatory networks of
promoter-associated noncoding RNAs in development and cancer. J Exp
Clin Cancer Res. 39:512020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang Y and Zhang H: RNAa induced by TATA
box-targeting microRNAs. Adv Exp Med Biol. 983:91–111. 2017.
View Article : Google Scholar
|
|
86
|
Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y,
Zhang L, Ding C, Luo H, Li Y, et al: MicroRNAs activate gene
transcription epigenetically as an enhancer trigger. RNA Biol.
14:1326–1334. 2017. View Article : Google Scholar
|
|
87
|
Zhang Y, Liu W, Chen Y, Liu J, Wu K, Su L,
Zhang W, Jiang Y, Zhang X, Zhang Y, et al: A cellular microRNA
facilitates regulatory t lymphocyte development by targeting the
FOXP3 promoter TATA-box motif. J Immunol. 200:1053–1063. 2017.
View Article : Google Scholar
|
|
88
|
Bai Y, Pan B, Zhan X, Silver H and Li J:
MicroRNA 195-5p targets foxo3 promoter region to regulate its
expression in granulosa cells. Int J Mol Sci. 22:67212021.
View Article : Google Scholar
|
|
89
|
Cao R and Zhang Y: The functions of
E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr
Opin Genet Dev. 14:155–164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mellor J, Dudek P and Clynes D: A glimpse
into the epigenetic landscape of gene regulation. Curr Opin Genet
Dev. 18:116–122. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Guo H, Pu M, Tai Y, Chen Y and Ren J:
Nuclear miR-30b-5p suppresses TFEB-mediated lysosomal biogenesis
and autophagy. Cell Death Differ. 28:320–336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li LC: Chromatin remodeling by the small
RNA machinery in mammalian cells. Epigenetics. 9:45–52. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Barlak N, Capik O, Kilic A, Sanli F,
Aytatli A, Yazici A, Karatas EA, Ortucu S and Karatas OF:
MicroRNA-145 transcriptionally regulates semaphorin 3A expression
in prostate cancer cells. Cell Biol Int. 45:1082–1090. 2021.
View Article : Google Scholar
|
|
94
|
Song M, Wang Y, Zhou P, Wang J, Xu H and
Zheng J: MicroRNA-361-5p aggravates acute pancreatitis by promoting
interleukin-17A secretion via impairment of nuclear factor
IA-dependent hes1 downregulation. J Med Chem. 64:16541–16552. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang K, Wang YY, Xu Y, Zhang L, Zhu J, Si
PC, Wang YW and Ma R: A two-miRNA signature of upregulated
miR-185-5p and miR-362-5p as a blood biomarker for breast cancer.
Pathol Res Pract. 222:1534582021. View Article : Google Scholar
|
|
96
|
Van Roosbroeck K and Calin GA: Cancer
hallmarks and MicroRNAs: The therapeutic connection. Adv Cancer
Res. 135:119–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kolenda T, Przybyla W, Teresiak A,
Mackiewicz A and Lamperska KM: The mystery of let-7d-a small RNA
with great power. Contemp Oncol (Pozn). 18:293–301. 2014.PubMed/NCBI
|
|
98
|
Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T,
Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, et al: Functional
proteomics identifies miRNAs to target a p27/Myc/phospho-Rb
signature in breast and ovarian cancer. Oncogene. 35:691–701. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Schmid G, Notaro S, Reimer D, Abdel-Azim
S, Duggan-Peer M, Holly J, Fiegl H, Rossler J, Wiedemair A, Concin
N, et al: Expression and promotor hypermethylation of miR-34a in
the various histological subtypes of ovarian cancer. BMC Cancer.
16:1022016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wong KY, Yu L and Chim CS: DNA methylation
of tumor suppressor miRNA genes: A lesson from the miR-34 family.
Epigenomics. 3:83–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yang Z, Fang S, Di Y, Ying W, Tan Y and Gu
W: Modulation of NF-κB/miR-21/PTEN pathway sensitizes non-small
cell lung cancer to cisplatin. PLoS One. 10:e01215472015.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Xu X, Zhu S, Tao Z and Ye S: High
circulating miR-18a, miR-20a, and miR-92a expression correlates
with poor prognosis in patients with non-small cell lung cancer.
Cancer Med. 7:21–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li H, Zhou H, Luo J and Huang J:
MicroRNA-17-5p inhibits proliferation and triggers apoptosis in
non-small cell lung cancer by targeting transforming growth factor
β receptor 2. Exp Ther Med. 13:2715–2722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Yang Z, Liu C, Wu H, Xie Y and Zhang X:
CSB affected on the sensitivity of lung cancer cells to
platinum-based drugs through the global decrease of let-7 and
miR-29. BMC Cancer. 19:9482019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Kristensen LS, Andersen MS, Stagsted L,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar
|
|
106
|
Zheng L, Jiao W, Mei H, Song H, Li D,
Xiang X, Chen Y, Yang F, Li H, Huang K and Tong Q: MiRNA-337-3p
inhibits gastric cancer progression through repressing myeloid zinc
finger 1-facilitated expression of matrix metalloproteinase 14.
Oncotarget. 7:40314–40328. 2016. View Article : Google Scholar
|
|
107
|
Zhang L, Zhou Q, Qiu Q, Hou L and Lu Y:
CircPLEKHM3 acts as a tumor suppressor through regulation of the
miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer.
18:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Christofides A, Papagregoriou G, Dweep H,
Makrides N, Gretz N, Felekkis K and Deltas C: Evidence for
miR-548c-5p regulation of FOXC2 transcription through a distal
genomic target site in human podocytes. Cell Mol Life Sci.
77:2441–2459. 2020. View Article : Google Scholar
|
|
109
|
Dharap A, Pokrzywa C, Murali S, Pandi G
and Vemuganti R: MicroRNA miR-324-3p induces promoter-mediated
expression of RelA gene. PLoS One. 8:e794672013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Huang V: Endogenous miRNAa: MiRNA-mediated
gene upregulation. Adv Exp Med Biol. 983:65–79. 2017. View Article : Google Scholar
|
|
111
|
Huang V, Place RF, Portnoy V, Wang J, Qi
Z, Jia Z, Yu A, Shuman M, Yu J and Li LC: Upregulation of Cyclin B1
by miRNA and its implications in cancer. Nucleic Acids Res.
40:1695–1707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Matsui M, Chu Y, Zhang H, Gagnon KT,
Shaikh S, Kuchimanchi S, Manoharan M, Corey DR and Janowski BA:
Promoter RNA links transcriptional regulation of inflammatory
pathway genes. Nucleic Acids Res. 41:10086–10109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Turner M, Jiao A and Slack FJ:
Autoregulation of lin-4 microRNA transcription by RNA activation
(RNAa) in C. Elegans. Cell Cycle. 13:772–781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Vera H, Yi Q, Ji W, Xiaoling W, Place RF,
Guiting L, Lue TF, Long-Cheng L and Dong-Yan J: RNAa is conserved
in mammalian cells. PLoS One. 5:e88482010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao
X, Li D, Li S, Mao L, Huang K and Tong Q: MiRNA-558 promotes
tumorigenesis and aggressiveness of neuroblastoma cells through
activating the transcription of heparanase. Hum Mol Genet.
24:2539–2551. 2015. View Article : Google Scholar
|
|
116
|
Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, Xu
H, Ye Z and Li LC: Up-regulation of p21(WAF1/CIP1) by miRNAs and
its implications in bladder cancer cells. FEBS Lett. 588:4654–4664.
2014. View Article : Google Scholar
|
|
117
|
Zou Q, Liang Y, Luo H and Yu W:
MiRNA-mediated RNAa by targeting enhancers. Adv Exp Med Biol.
983:113–125. 2017. View Article : Google Scholar
|
|
118
|
Huang YP, Qiu LZ and Zhou GP: MicroRNA-939
down-regulates CD2-associated protein by targeting promoter in
HEK-293T cells. Renal Failure. 38:508–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Mao H, Zhu C, Zong D, Weng C, Yang X,
Huang H, Liu D, Feng X and Guang S: The nrde pathway mediates
small-RNA-directed histone H3 lysine 27 Trimethylation in
Caenorhabditis elegans. Curr Biol. 25:2398–2403. 2015. View Article : Google Scholar
|
|
120
|
Liu X, Fan Z, Li Y, Li Z, Zhou Z, Yu X,
Wan J, Min Z, Yang L and Li D: MicroRNA-196a-5p inhibits testicular
germ cell tumor progression via NR6A1/E-cadherin axis. Cancer Med.
9:9107–9122. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Guo F, Gao Y, Sui G, Jiao D, Sun L, Fu Q
and Jin C: MiR-375-3p/YWHAZ/β-catenin axis regulates migration,
invasion, EMT in gastric cancer cells. Clin Exp Pharmacol Physiol.
46:144–152. 2019. View Article : Google Scholar
|
|
122
|
Li J and Zou X: MiR-652 serves as a
prognostic biomarker in gastric cancer and promotes tumor
proliferation, migration, and invasion via targeting RORA. Cancer
Biomark. 26:323–331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
To KK, Leung WW and Ng SS: A novel
miR-203-DNMT3b-ABCG2 regulatory pathway predisposing colorectal
cancer development. Mol Carcinog. 56:4642016. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Wang C, Chen Q, Li S, Li S and Zhao Z:
Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in
gliomas. Oncotarget. 8:10287–10297. 2016. View Article : Google Scholar
|
|
125
|
Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X,
Guo X, He S and Chen R: Transcriptional inhibiton of Hoxd4
expression by miRNA-10a in human breast cancer cells. BMC Mol Biol.
10:122009. View Article : Google Scholar
|
|
126
|
Kang MR, Park KH, Yang JO, Lee CW and Kang
JS: MiR-6734 up-regulates p21 gene expression and induces cell
cycle arrest and apoptosis in colon cancer cells. PLoS One.
11:e1609612016. View Article : Google Scholar
|
|
127
|
Zhang Y, Fan M, Geng G, Liu B, Huang Z,
Luo H, Zhou J, Guo X, Cai W and Zhang H: A novel HIV-1-encoded
microRNA enhances its viral replication by targeting the TATA box
region. Retrovirology. 11:232014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Li S, Zhu Y, Liang Z, Wang X and Xie L:
Up-regulation of p16 by miR-877-3p inhibits proliferation of
bladder cancer. Oncotarget. 7:51773–51783. 2016. View Article : Google Scholar
|
|
129
|
Kim DH, Saetrom P, Snove O Jr and Rossi
JJ: MicroRNA-directed transcriptional gene silencing in mammalian
cells. Proc Natl Acad Sci USA. 105:16230–16235. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Cui C, Yu J, Huang S, Zhu H and Huang Z:
Transcriptional regulation of gene expression by microRNAs as
endogenous decoys of transcription factors. Cell Physiol Biochem.
33:1698–1714. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zardo G, Ciolfi A, Vian L, Starnes LM,
Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N,
et al: Polycombs and microRNA-223 regulate human granulopoiesis by
transcriptional control of target gene expression. Blood.
119:4034–4046. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Sepramaniam S, Ying LK, Armugam A, Wintour
EM and Jeyaseelan K: MicroRNA-130a represses transcriptional
activity of aquaporin 4 M1 promoter. J Biol Chem. 287:12006–12015.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Miao L, Yao H, Li C, Pu M, Yao X, Yang H,
Qi X, Ren J and Wang Y: A dual inhibition: MicroRNA-552 suppresses
both transcription and translation of cytochrome P450 2E1. Biochim
Biophys Acta. 1859:650–662. 2016. View Article : Google Scholar
|