Introduction
Numerous targeted therapeutic agents have been
developed for cancer treatment in the past two decades. For
example, the anti-Erb-B2 receptor tyrosine kinase 2 (ERBB2/HER2)
monoclonal antibody Trastuzumab (Herceptin), is commonly used
nowadays for treating patients with ERBB2+
(HER2+) breast tumor. On the other hand, the epidermal
growth factor receptor (EGFR) inhibitor Gefitinib (Iressa), is used
to manage patients with advanced non-small cell lung cancer, which
is often EGFR+. Despite the early success of various
targeted therapeutics, drug resistance remains a major problem in
clinical practice. As cancer cells can acquire resistance to
targeted therapeutic agents when the designated-targets or their
downstream signaling molecules develop protein conformational or
activity changes due to gene mutations or amplifications, agents
specificity targeting a single molecule or pathway often fails
particularly in patients after prolonged treatment (1). This leads to the recent interest in
poly-pharmacology, which refers to the design or use of
pharmacological agents that act on multiple targets or disease
pathways (2,3).
The microRNA (miR)-125a-5p is a recently discovered
tumor suppressor. In clinical situations, low expression levels of
miR-125a-5p were associated with enhanced malignant potential and
poor prognosis in patients with head and neck, gastric, and breast
cancer (4-6). At the cellular level, ectopic
overexpression of miR-125a-5p inhibits the proliferation (or
promotes apoptosis), migration, invasion, and epithelialmesenchymal
transition (EMT) of breast, colorectal and lung cancer cells
(7-12). Ectopic overexpression of
miR-125a-5p also restores the sensitivity to cisplatin in the
cisplatin-resistant cervical cancer cells and counteracts
EGF-induced cell proliferation and invasion in cervical cancer
cells (13,14). At the molecular level, miR-125a-5p
negatively regulates the expression of ERBB2, baculoviral IAP
repeat containing 5 (BIRC5/SURVIVIN), Sp1 transcription factor
(SP1), LIM kinase 1 (LIMK1), and polypeptide
N-acetylgalactosaminyltransferase 7 (GALNT7) in cells, in which
upregulation of these molecules is known to promote tumorigenesis,
tumor metastasis and drug resistance (4,6,14-18).
BIRC5 is a member of the inhibitor of apoptosis
proteins (IAPs) family known for its inhibitory effects on caspase
activity. Physiologically, BIRC5 plays an important role in brain
development during embryogenesis (19). Unlike other IAPs, BIRC5 is highly
expressed in different tumor types (Fig. S1A) but its expression remains
low/undetectable in the differentiated normal and undamaged tissues
(20-24). BIRC5 is also highly expressed in
cancer stem cells (25,26). Clinically, high expression levels
of BIRC5 is associated with poor prognosis in patients with cancer
(Fig. S1B) (26,27). As upregulation of BIRC5 promotes
tumorigenesis and tumor drug resistance, various efforts have been
made in the development of the BIRC5-targeting anticancer
therapies, including targeted therapy and vaccination (28-35). However, none of these
BIRC5-targeting therapies has yet been approved by FDA for clinical
application, mainly due to the lack of efficacy during clinical
trials.
Polymeric gene delivery systems offer increased
amounts of plasmid DNA uptake and the possibility of controlling
the rate and conditions of release of plasmid DNA after
administration. Plasmid DNA complexed with poly-L-lysine can be
protected against digestion by nucleases present in the
physiological environment (36).
It was previously revealed that liposomal transfection of a
BIRC5 promoter-driven antisense BIRC5-expressing
plasmid DNA (pSur/AS-Sur) induces apoptosis in
BIRC5-expressing (BIRC5+) cancer cells but not in
the BIRC5 non-expressing (BIRC5−) human umbilical
vein endothelial cells (HUVEC) in vitro (37). In the present study, the
multi-molecules/pathways-targeting BIRC5 promoter-driven
miR-125a-5p expressing plasmid DNA (pSur-125a) loaded
nanoparticles was created, in which the biodegradable and
biocompatible poly-L-lysine polymer was used to encapsulate
pSur-125a. The feasibility of using these nanoparticles to suppress
the expression of various known miR-125a-5p-targeting
cancer-related molecules such as ERBB2, SP1 and BIRC5 in
BIRC5+ cancer cells was demonstrated. It was found that
overexpression of miR-125a-5p downregulates the cellular expression
of histone deacetylase 5 (HDAC5) and tryptophan 2,3-dioxygenase
(TDO2), which is an enzyme that facilitates the production of
kynurenine and the related induction of immunosuppression in
tumors. Overexpression of ATP Binding Cassette Subfamily B Member 1
(ABCB1/MDR1/P-gp) induces multidrug resistance in cancer cells. In
the present study, it was also demonstrated that the anticancer
efficacy and the molecular effects of these nanoparticles are not
affected by the expression of ABCB1 in cancer cells in vitro
and in vivo.
Materials and methods
Cell lines and cell culture
conditions
Human KB (cervical carcinoma), MCF7 (breast
adenocarcinoma), and MDA-MB-231 (breast adenocarcinoma) cells were
originally obtained from the American Type Culture Collection.
Human KB cells were cultured in RPMI-1640 medium (cat. no.
31800-022; Gibco; Thermo Fisher Scientific, Inc.) containing 5%
fetal bovine serum (FBS) (cat. no. 04-001-1A; Biological
Industries) and penicillin/streptomycin/glutamine (PSG). The human
NTUB1 and the NTUB1-derived ABCB1-expressing NTU0.017 bladder
carcinoma cells were kindly provided by Dr Jang-Yang Chang of
Institute of Biotechnology and Pharmaceutical Research, National
Health Research Institutes, Miaoli, Taiwan (38). Human NTUB1 and MDA-MB-231 cells
were cultured in RPMI-1640 medium containing 10% FBS and PSG. The
KB- and NTUB1-derived ABCB1-expressing, multidrug resistant
KB-TAX50 and NTU0.017 cells were generated by the paclitaxel-driven
selection and cultured in medium containing 50 and 17 nM
paclitaxel, respectively, as previously described (37,39-41). Human MCF7 cells were cultured in
α-MEM containing 5% FBS, PSG, and insulin-transferring-selenium
supplement (ITS) (cat. no. 11074547001; Diagnostics). MCF7-TamC3
cells were created by prolonged culture of MCF7 cells under
estrogen-depleted conditions. The cellular and molecular phenotypes
of the MCF7-dervided estrogen-independent and tamoxifen-resistant
MCF7-TamC3 breast cancer cells have already been characterized in
previous studies (15,42). MCF7-TamC3 cells were cultured in
phenol-red-free RPMI containing 5% charcoal-stripped FBS, PSG and
ITS. The human HMEC-1 dermal microvascular endothelial cells were
kindly provided by Dr Ben-Kuen Chen of the Department of
Pharmacology of National Cheng Kung University, Taiwan. All cells
were cultured at 37°C in a humidified incubator containing 5%
CO2 and were revealed to be mycoplasma free. The use of
the aforementioned human cell lines in the present study was
approved by the review board of Ministry of Science and Technology
(Taiwan) and the biosafety committee of National Cheng Kung
University (Taiwan).
Construction of the BIRC5 promoter driven
miR-125a-5p expressing plasmid DNA
PCR was used to amplify the miR-125a-5p fragment and
to insert the BspHI and EcoRI endonuclease restriction site on the
5′ and 3′ end of the PCR products (i.e. the newly synthesized
miR-125a-5p fragments), respectively, with the use of the plasmid
DNA pLV-[hsa-mir-125a] (cat. no. p087; BioSettia, Inc.) as
template. The PCR cycle was carried out as follows: 98°C for 30
sec, followed up by 30 cycles of 98°C for 10 sec, 64.5°C for 30
sec, 72°C for 30 sec and then 72°C for 10 min using the following
set of primers: forward, [designed to bind on the hsa-miR-125a
precursor sequence (miRNA accession no. MI0000469) carried by the
plasmid DNA pLV-(hsa-mir-125a)] 5′-AAT CAT GAT CGA GGA TCC TCG
TTT-3′ and reverse, 5′-AAG
AAT TCG GTC AGG TTT CAG TTG-3′. The BspHI and EcoRI
enzymatic sites are underlined. The PCR product was incubated with
the restriction endonuclease BspHI and EcoRI to generate sticky
ends. The destination vector pSur was created by incubating the
plasmid DNA pDRIVE-hSurvivin (a vector that harbors a
BIRC5/survivin gene promoter and a luciferase gene)
(cat. no. pdrive-hsurvivin; InvivoGen) with BspHI and EcoRI for the
removal of the luciferase gene. Then, the digested PCR product was
ligated onto the linearized pSur in a molar ratio of 1:3.
Successful ligation of the PCR product onto pSur (i.e. the creation
of pSur-125a) was validated by DNA sequencing. The plasmid DNA
pSur-125a was transformed into the DH5α E. coli cells for
long term storage.
In vitro cell viability analysis
Cells (5,040 cells/well) were seeded onto each well
of 96-well plates overnight before being transfected with the
plasmid DNA for 48 h or treated with the nanoparticles for 96 h.
After treatment, 200 µl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
solution (cat. no. 0793; diluted in phenol-red free RPMI in a ratio
of 1:10; Amresco, LLC) was added to each well and incubated at 37°C
for 4 h. Then, 100 µl MTT lysis buffer containing 500 ml/l
dimethylformamide and 100 g/l sodium dodecyl sulfate, was added to
each well and incubated for 16 h. Cell viability was quantified by
measuring the absorbance of the solution at 570 nm using a
SpectraMax® M5 microplate reader (Molecular Devices,
LLC). The percentage of viable cells for each treatment group was
calculated by adjusting the control group to 100%. Samples were
assayed in duplicate and the experiments were repeated at least
three times.
Wound healing (cell migration) assay
Cells (2.2×104) were seeded onto each
well of the culture inserts (Ibidi GmbH) for 24 h. Cell-free gaps
(500 mm) were created after removing the culture inserts. Cells
were treated with either pL-MNP-pSur-Emp or 0.5 × IC50
pL-MNP-pSur-125a. Images of the wound areas were captured by using
an invert light microscope (CKX53; Olympus Corporation) after 9 h
(NTU0.017 cells) or 12 h (NTUB1 cells). The average width of the
wound was measured and analyzed using ImageJ 1.52a software
(National Institutes of Health) to calculate the cell
migration.
Transwell invasion (cell invasion)
assay
The upper chambers of the Transwell plates with
8-µm pore size were coated with 20% Matrigel at 37°C for 1 h
(BD Medical Technology). Cells (5×105) were seeded onto
the upper chamber of the Transwell (cat. no. 353182; Falcon;
Corning Life Sciences) in serum-free culture medium containing
indicated concentration of pL-MNP-pSur-Emp and pL-MNP-pSur-125a.
Cell culture medium was added to the lower chamber containing the
same concentration of pL-MNP-pSur-Emp and pL-MNP-pSur-125a as upper
chamber. At 20 h post-treatment, cells attached on the revere side
of the PET membrane were fixed with 4% paraformaldehyde solution at
room temperature for 15 min and subsequently stained with 0.2%
crystal violet solution at room temperature for 10 min. Images were
captured by using an invert microscope (OLYMPUS CKX53). The crystal
violet was dissolved with 33% acetic acid, and the absorbance was
measured (570 nm). The related invasion ability was calculated by
comparing the absorbance intensity.
MicroRNA (miR-125a-5p) and BIRC5 mRNA
expression analysis
Total RNAs were extracted using TRIzol®
reagent (cat. no. 15596-026; Thermo Fisher Scientific, Inc.). To
detect the expression level of miR-12a-5p in cells, complementary
DNA (cDNA) was synthesized from the extracted RNA using TaqMan™
microRNA-specific primers, following protocol as described in the
TaqMan® MicroRNA Reverse transcription kit (cat. no.
4427975; Thermo Fisher Scientific, Inc.). A TaqMan reverse
transcription-quantitative PCR (RT-qPCR)-based microRNA assay (ID
002198-hsa-miR-125a-5p; ID 001093-RNU6B) was used to determine the
expression of miR-125a-5p in cells. The target fragment was
amplified according to the following protocol: preheating at 95°C
for 10 min, 40 cycles at 95°C for 15 sec and 60°C for 1 min. The
miRNA expression level was normalized with RNU6B, which was broadly
used as the endogenous reference microRNA in different miRNA
quantification studies. To detect the expression level of BIRC5
mRNA, total RNA was extracted using TRIzol® reagent and
complementary DNA was synthesized from RNA using the RevertAid H
Minus first strand cDNA synthesis kit (cat. no. K1631; Thermo
Fisher Scientific, Inc.). The relative expression levels of BIRC5
and ACTA1 mRNA were determined by qPCR using primers as previously
described (15). The specific
primers with the following sequences were used in the present
study: human BIRC5 forward, 5′-CTG CCT GGC AGC CCT TT-3′ and
reverse, 5′-CCT CCA AGA AGG GCC AGT TC-3′; human ACTA1 forward,
5′-GGC GGC ACC ACC ATG TAC CCT-3′ and reverse, 5′-AGG GGC CGG ACT
CGT CAT ACT-3′. The target genes were quantified using the
comparative threshold cycle (Ct) values 2−ΔΔCq method
(43) (ΔCq=Cq Target
gene-CtRNU6B, ΔΔCq=ΔCq Treatment-ΔCq Control). Experiments were
repeated thrice.
Western blot analysis
Cells were lysed using CelLytic™ M cell lysis
reagent (cat. no. C2978; Sigma-Aldrich; Merck KGaA) containing 1 mM
phenylmethylsulfonyl fluoride, 1 mM sodium fluoride and cocktail
protease inhibitor cocktail (cat. no. 05892791001; Roche
Diagnostics). The protein concentration was measured by the
bicinchoninic acid (BCA) protein assay kit. Equal amounts of
protein (30 µg) were subjected to sodium dodecyl-sulfate
polyacrylamide gel electrophoresis (SDS-PAGE) on a 6, 10 or 12%
acrylamide gel. The resolved proteins were transferred onto a PVDF
membrane (cat. no. IPVH00010; Merck KGaA) and incubated with a
blocking buffer TBST (5% non-fat dried milk in Tris-buffered saline
with Tween-20 (cat. no. 9480; Calbiochem; Merck KGaA) for 1 h at
room temperature before an overnight incubation at 4°C with the
following primary antibodies: anti-cleaved CASP3 antibody (1:1,000;
cat. no. 9664; Cell Signaling Technology, Inc.), anti-ERBB2
antibody (1:1,000; cat. no. UM570036; UltraMAB), anti-BIRC5
(Survivin) antibody (1:700; cat. no. AF886; R&D Systems, Inc.),
anti-CDH1 (1:1,000; cat. no. 24E10; Cell Signaling Technology,
Inc.), anti-HDAC5 antibody (1:1,000; cat. no. 161661-AP;
ProteinTech Group, Inc.), anti-TDO2 (1:500; cat. no. GTX114831;
GeneTex, Inc.) anti-PARP antibody (1:1,000; cat. no. 9532; Cell
Signaling Technology, Inc.), and anti-ACTA1 antibody (1:20,000;
cat. no. MAB1501; Merck Millipore). The PVDF membrane was then
washed thrice with TBS containing 0.1% Tween-20 before incubation
for 1 h at room temperature with horseradish peroxidase-conjugated
goat anti-rabbit antibodies (1:10,000; cat. no. AP132P;
MilliporeSigma), mouse (1:10,000; cat. no. AP124P) or goat
(1:10,000; cat. no. AP106P; both from MilliporeSigma)
immunoglobulin G. Immunoreactive proteins were visualized using
western blot enhanced chemiluminescence reagents (cat. no.
WBKLS05000; Merck Millipore) and protein signals were detected by
luminescence readers (FUJI LAS-100). The intensity of protein bands
was determined by using the ImageJ 1.52a software (National
Institutes of Health). Experiments were repeated at least three
times.
Preparation of
NH2-Fe3O4 magnetic nanoparticles
(MNPs)
First, 1 M ferric chloride hexahydrate
(FeCl3·6H2O) and 2 M ferrous chloride
tetrahydrate (FeCl2·4H2O) were prepared by
dissolving iron salts in 2 M hydrochloric acid (HCl) solution.
Next, 4 ml of 1 M FeCl3 solution and 1 ml 2 M
FeCl2 solution were mixed, and 1 ml of organic acid
aqueous solution (0.5 g glycine dissolved in 1 ml deionized water)
was then added to prepare the mixture solution. After vigorous
stirring of the mixture solution, 5 M sodium hydroxide (NaOH)
solution was then added drop by drop until the solution turned
black. The solution was vigorously stirred again for 15 min at room
temperature. Using a permanent magnet, the precipitated magnetic
powder was fractionated, and the solution was subsequently
discarded. Deionized water was then added to wash the precipitates
thrice to remove excess salt. Subsequently, 3 g glycine, which was
dissolved in 50 ml HCl, was added to the washed precipitates. The
mixture solution was stirred for 5 min and then sonicated for 30
min. After adding deionized water and acetone in a volume ratio of
Mixture solution: Deionized water: Acetone=5:2:3, the solution was
centrifuged at 6,200 g for 10 min to discard the supernatant. Next,
the following steps were repeated twice to remove excess organic
acid in the suspension: 7 ml deionized water was added to dissolve
and wash the precipitates, 3 ml acetone was added, and the mixture
solution was centrifuged at 6,200 g for 10 min. In the end,
NH2-Fe3O4 MNPs were produced after
the precipitates were dispersed in deionized water. An inductively
coupled plasma analysis of the Fe ion concentration for the
NH2-Fe3O4 MNPs was measured by a
spectro analyzer (Jobin-Yvon JY138).
Preparation of the plasmid DNA loaded
poly-L-lysine-conjugated MNPs (pL-MNPs)
The poly-L-lysine-conjugated MNPs (pL-MNPs) were
prepared by mixing 5 ml of 0.1% (w/v) low molecular weight
poly-L-lysine (MW 1,000 to 5,000; cat. no. P0879; Sigma-Aldrich;
Merck KGaA) with 0.1 ml of 0.1 mM MNPs. After the mixture solution
was stirred for 30 min at room temperature and centrifuged at
17,000 g for 10 min, the precipitates were isolated by removing the
supernatant. The precipitates were then washed twice with deionized
water. Finally, the precipitates (pL-MNPs) were dispersed and
stored in deionized water. A total of 500 ng/µl of plasmid
DNA (in aqueous solution) were incubated with 0.22 µM
pL-MNPs. Next, deionized water and rhodamine 6G (R6G; final
concentration, 10 µM) were added to the plasmid DNA solution
to achieve the final volume depending on the concentration of
nanoparticles in the experiments. The volume ratio of 500
ng/µl of plasmid DNA:0.22 µM pL-MNP: deionized
water=2:1:7. All samples were gently stirred at room temperature
for at least 30 min before stirring at 4°C for 16 h.
Characterizations of the plasmid
DNA-loaded pL-MNPs
Dynamic light scattering (DLS) was used to measure
the size of plasmid DNA-loaded pL-MNPs, by Zetasizer Nano ZS90
(Malvern Instruments, Inc.). The zeta potential of pL-MNPs were
determined by ELSZ-2000 (Otsuka Electronics Co., Ltd.). The surface
morphology, shape and size of nanoparticles were measured by the
scanning electron microscopy (SEM) at an accelerating voltage of 10
kV at a working distance of 9 mm. Samples prepared for SEM were
firstly dropped on the cooper coin and collected after the solvent
was evaporated at room temperature. Subsequently, samples were
coated with gold for the sputter coating. Liquid transmission
electron microscopy (liquid-TEM) was used for the in situ
TEM image inspection for analyzing the structural and chemical
properties, including size, structure and elements of plasmid
DNA-loaded pL-MNPs. In the present study, the liquid-TEM was
operated under the acceleration voltage of 200 kV (A JEOL JEM 2100
TEM).
DNase I protection assay
pSur-125a and PL-MNP-pSur-125a were incubated with
DNase I at 37°C for 15, 30 min, 1 and 2 h in a final volume of 10
µl. The digestions were halted by incubating at 75°C for 10
min. The integrity of plasmid DNA was assessed by gel
electrophoresis (0.8% agarose gel, 100 V), which was stained with
the Healthview™ nucleic acid stain (cat. no. GN-NAS-100;
Genomic).
In vitro plasmid DNA release assay
In vitro release of plasmid DNA from the
plasmid DNA-loaded nanoparticle was performed in phosphate buffer
saline (PBS) with different pH values (pH=7.4, 6, 5 and 4). 15
µg pL-MNP-pSur-Emp were incubated in 1 ml PBS with different
pH value for various durations (1, 3, 6, 9, 12 and 24 h). After
incubation, pL-MNP-pSur-Emp was stratified from the solution by
using a magnet and the supernatant was collected to a new tube. The
concentration of the plasmid DNA pSur-Emp in the supernatant was
determined using spectrophotometry (MaestroGen, Inc.), and the
total amount of the plasmid DNA presence was calculated using the
following equation: Amount of plasmid DNA releasing
(µg)=concentration of plasmid DNA (µg/µl) x
1,000 µl. The release percentage of plasmid DNA was
determined using the following equation:
In vivo drug potency evaluation
The animal protocol was approved (approval no.
109273) by the Institutional Animal Care and Use Committee (IACUC)
of National Cheng Kung University (Tainan, Taiwan). Wild type
zebrafish (Danio rerio, strain: AB) embryos were purchased
from the Laboratory Animal Center, College of Medicine, National
Cheng Kung University. Human KB, KB-TAX50 and MDA-MB-231 cancer
cells were labeled with PKH67 to track tumor growth in vivo.
Zebrafish embryos were anesthetized with 0.01% tricaine 48 h
post-fertilization (hpf) and subsequently transplanted with cancer
cells. Total of 500 cancer cells were transplanted into the yolk
sac of zebrafish embryos. A total of 1 h after cells
transplantation, saline (negative control, N=24), pL-MNP-pSur-Emp
(negative control, N=24), or pL-MNP-pSur-125a (N=24) were
microinjected into yolk sac of zebrafish embryos. The saline or
nanoparticles-treated zebrafish embryos were kept at 35°C for 48 h.
Tumor size was measured from images captured using fluorescence
microscopy every 12 h post-treatment.
In vivo hepatotoxicity analysis
Zebrafish hepatotoxicity analysis was performed by
the Taiwan zebrafish core facility of National Health Research
Institute (NHRI). Briefly, 48 hpf transgenic zebrafish
(fabp10a:mCherry) embryos were treated (i.e. microinjected) with
saline (N=24), or the indicated concentrations of pL-MNP-pSur-Emp
(N=24) and pL-MNP-pSur-125a (N=24) for 48 h. The size of liver was
measured from images captured using fluorescence microscopy.
Statistical analysis
Each experiment was performed at least three times.
Data are presented as the mean ± standard error of the mean. A
two-tailed unpaired Student's t-test was used for comparisons
between two groups. One-way ANOVA with Tukey's post hoc test were
used for multi-group comparisons. All statistical analyses were
performed using GraphPad Prism version 8 (GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
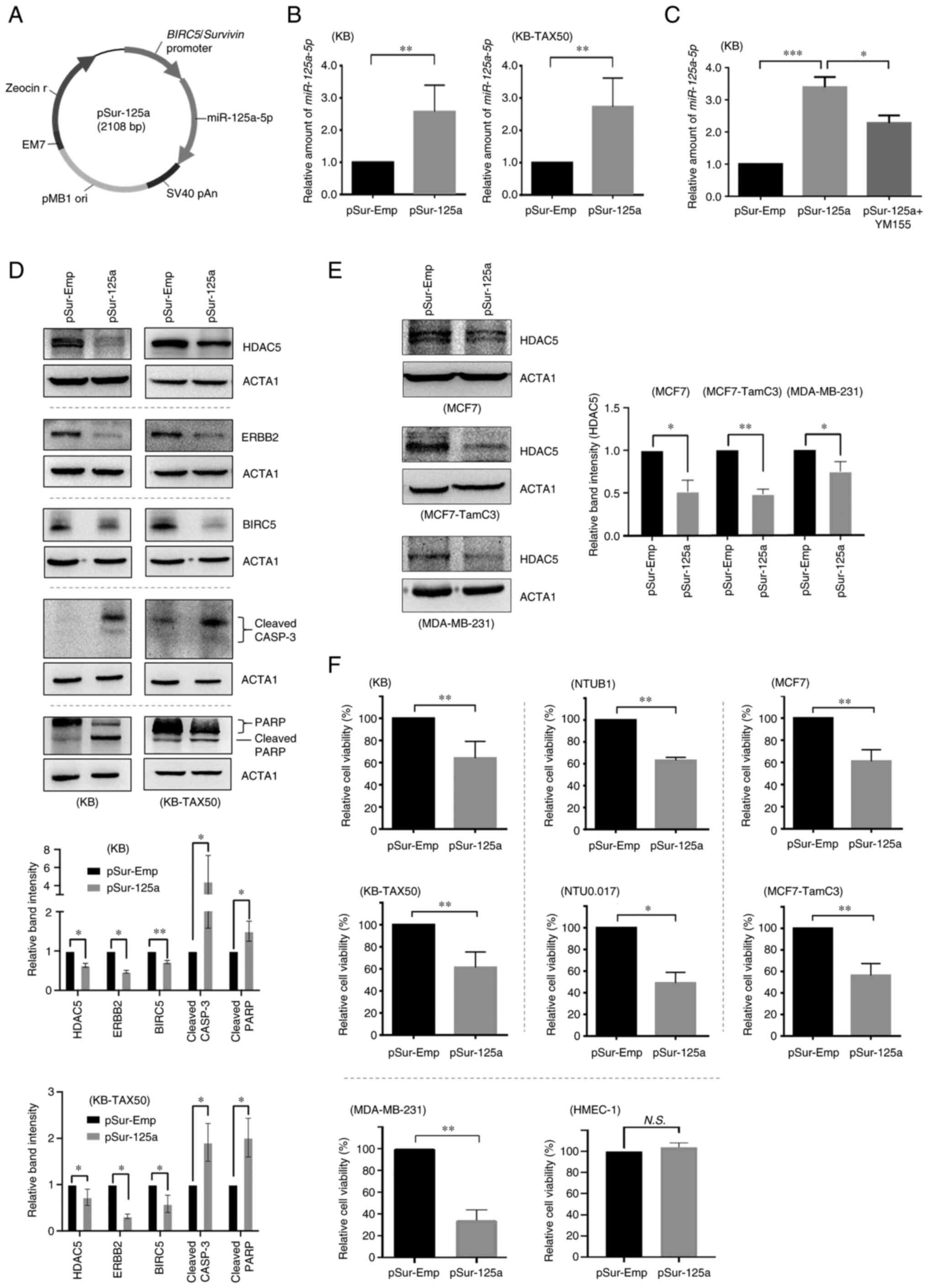
Transfection of pSur-125a decreases the
viability of various BIRC5-expressing cancer cells
A BIRC5 gene promoter driven
miR-125a-5p expressing, multiple oncoproteins
downregulating, plasmid DNA (i.e. pSur-125a) was constructed. To
confirm if pSur-125a functions as designed (Fig. 1A), the expression level of
miR-125a-5p was examined in BIRC5+ cancer cells with or
without liposomal delivery (i.e. transfection) of pSur-125a in
vitro. The human KB and the KB-derived ABCB1-expressing,
multidrug-resistant, KB-TAX50 cervical cancer cells (Fig. S2A) are known to express BIRC5
protein (37,39,44). Results of the qPCR analysis showed
that transfection of pSur-125a significantly increased the amount
of miR-125a-5p presence in both KB and KB-TAX50 cells compared with
those transfected with pSur-Emp (i.e. the control plasmid
DNA-BIRC5 promoter containing, but without the miR-125a-5p
insert) (Fig. 1B). YM155 is a
small molecule BIRC5 inhibitor that suppresses BIRC5 protein
expression at the transcriptional level through direct interactions
with the BIRC5 promoter region (45). In the present study, co-treatment
with YM155 at a sub-lethal concentration (0.25 × IC50 of
cell viability) partially attenuated the expression effects of
pSur-125a on miR-125a-5p in KB cells, confirming that the increased
expression of miR-125a-5p was at least in part driven by the
BIRC5 promoter region located on pSur-125a (Fig. 1C). As revealed in Fig. 1D, transfection of pSur-125a
decreased the expression of various known miR-125a-5p-targeting
oncoproteins including ERBB2 and BIRC5 in KB and KB-TAX50 cancer
cells. Transfection of pSur-125a also induced the protein cleavage
of CASP3/caspase-3 and Poly (ADP-ribose) polymerase (PARP), which
are markers for apoptosis, confirming the pro-apoptotic property of
pSur-125a in BIRC5+ cancer cells (Fig. 1D). It was previously demonstrated
that HDAC5 downregulation increases miR-125a-5p expression in the
human estrogen receptor-positive (ER+) MCF7 and the
ER+ MCF7-dervied estrogen-independent, tamoxifen
resistant, MCF7-TamC3 breast cancer cells (15). Results of the western blot
analysis showed that ectopic overexpression of miR-125a-5p
decreased the expression of HDAC5 not only in KB and KB-TAX50, but
also in MCF7, MCF7-TamC3 and MDA-MB-231 (triple-negative breast)
cancer cells examined in the present study (Fig. 1E). These finding suggested that
HDAC5 is a possible downstream affecting molecule of miR-125a-5p
and a negative feedback loop possibly exists between HDAC5 and
miR-125a-5p.
The viability of a panel of BIRC5+ cancer
cells transfected with or without pSur-125a was examined (37,39,46). Results of the cell viability
analysis revealed that transfection of pSur-125a significantly
decreased the viability of KB, KB-TAX50, MCF7, MCF7-TamC3,
MDA-MB-231, NTUB1 (bladder), and NTU0.017 (NTUB1-derived
ABCB1-expressing) (Fig. S2A and
B) cancer cells by 40-60% (Fig.
1F). In addition, the levels of effect of pSur-125a
transfection on miR-125a-5p expression appear to be associated with
the endogenous BIRC5 gene transcription levels (by using
amounts of BIRC5 mRNA transcripts presence as an indicator)
in the examined NTUB1, NTU0.017, and MDA-MB-231 cells (Fig. S2B and C). By contrast,
transfection of pSur-125a did not affect the viability of the
BIRC5-non-expressing (or only expressing at a relatively low level)
human dermal microvascular endothelial HMEC-1 cells (Figs. S2D and 1F) and this result was
unlikely caused by the limited transfection efficiency as bright
green fluorescence signal could still be observed in HMEC-1 cells
transfected with pCMV6-AC-GFP using the same plasmid DNA-liposome
formulation (Fig. S2E).
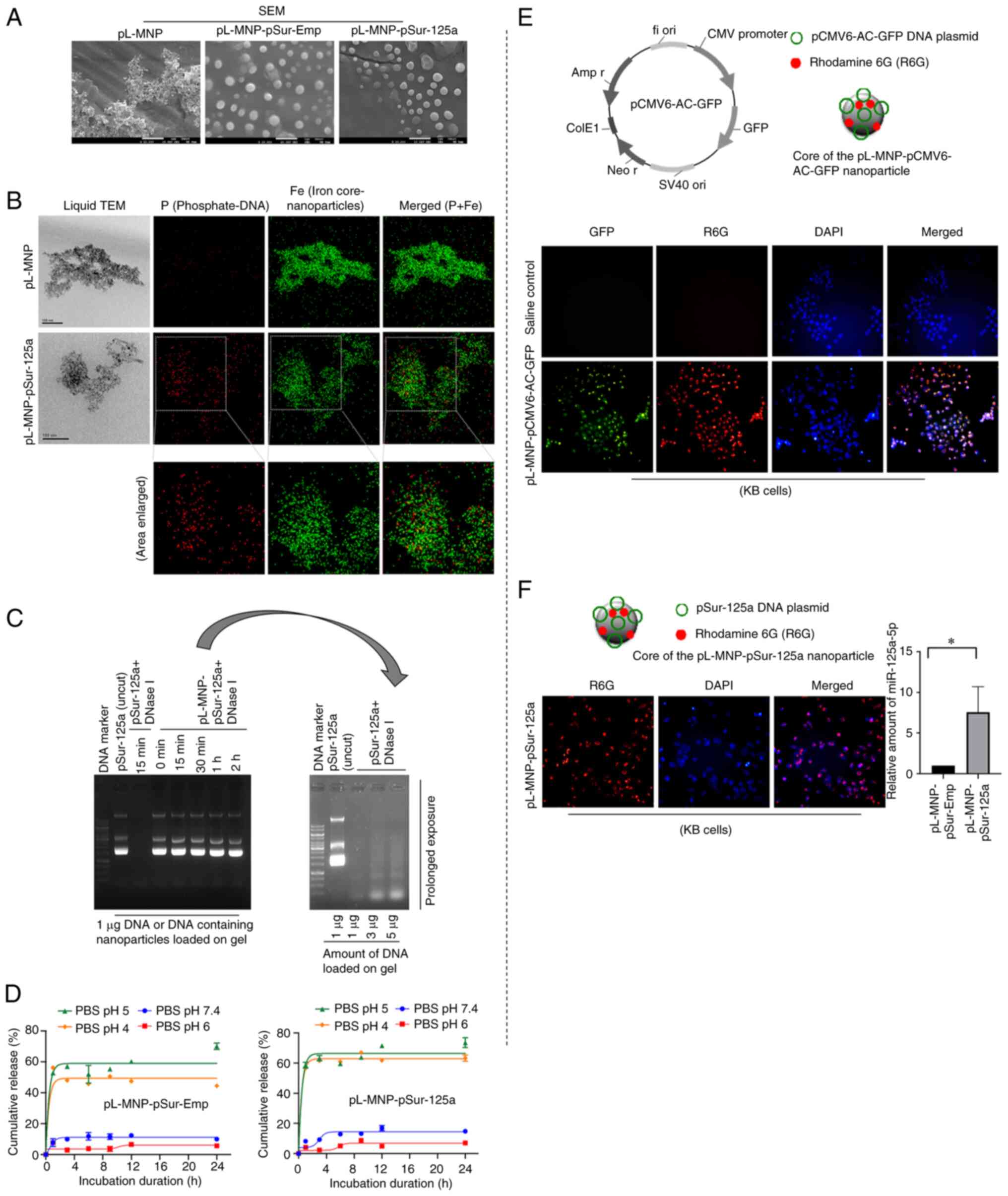
Physicochemical characterizations of the
pSur-125a-loaded poly-L-lysine-modified magnetic iron oxide
nanoparticles (pL-MNP-pSur-125a)
To increase the feasibility of utilizing pSur-125a
as a therapeutic agent, the pSur-125a-loaded poly-L-lysine-modified
magnetic iron oxide nanoparticles (pL-MNPs) (i.e. pL-MNP-pSur-125a)
was developed. SEM images demonstrated that the pSur-Emp- and the
pSur-125a-loaded nanoparticles (i.e. pL-MNP-pSur-Emp and
pL-MNP-pSur-125a) were mostly in round shape under dehydrated
conditions (Fig. 2A). The mean
particle size of the nanoparticles pL-MNP (plasmid DNA-free),
pL-MNP-pSur-Emp and pL-MNP-pSur-125a was ~289.1±9.5, 332.9±25.7 and
327.1±27.0 nm, respectively, as determined by the DLS analysis
(Table I). The mean zeta (ζ)
potential of pL-MNP, pL-MNP-pSur-Emp, and pLMNP-pSur-125a was
+23.4±3.0, -45.0±4.3, and -39.8±3.4 mV, respectively (Table I). The chemical composition of
pL-MNP-pSur-125a was determined by liquid TEM together with the
energy dispersive X-ray spectroscopy (EDS) analysis. Images
obtained by liquid TEM and results of the EDS analysis mapping
together showed overlapping between the P element (representing the
phosphate group of the DNA backbone) and the Fe element
(representing the iron core of the nanoparticles), indicating
successful loading of pSur-125a into the nanoparticles under the
optimized conditions (Fig. 2B).
The encapsulation efficiency of pL-MNP-pSur-Emp and
pL-MNP-pSur-125a was 42.6±3.2 and 45.8±2.8%, respectively. The
loading capacity of pL-MNP-pSur-Emp and pL-MNP-pSur-125a was
59.6±4.5 and 64.1±3.9% (Table
I).
 | Table IPhysicochemical properties of
different nanoparticles. |
Table I
Physicochemical properties of
different nanoparticles.
| Nanoparticles | pL-MNP |
pL-MNP-pSur-Emp |
pL-MNP-pSur-125a |
|---|
| Carried plasmid
DNA | None | pSur-Emp | pSur-125a |
| Zeta potential
(mV) | +23.4±3.0 | -45.0±4.3 | -39.8±3.4 |
| Size (nm) | 289.1±9.5 | 332.9±25.7 | 327.1±27.0 |
| Encapsulation
efficiency (%) | N/A | 42.6±3.2 | 45.8±2.8 |
| Loading capacity
(%) | N/A | 59.6±4.5 | 64.1±3.9 |
Circulating DNA and RNA are susceptible to
degradation by nucleases in the body. As revealed in Fig. 2C, the DNase I protection assay
results demonstrated that the plasmid DNA pSur-125a, which is
encapsulated in our formulated pL-MNPs, remained intact in the
presence of DNase I for up to 2 h incubation. By contrast, the
naked plasmid DNA pSur-125a was completely digested by the same
amount of DNase I within 15 min of incubation, suggesting that
pL-MNPs could protect the encapsulated pSur-125a from nuclease
digestion (Fig. 2C). The plasmid
DNA pSur-125a needs to be released from the nanoparticles
pL-MNP-pSur-125a within the targeted cancer cells before turning on
its miR-125a-5p-expressing function. The in vitro plasmid
DNA release assay showed that ~50-65% of the loaded plasmid DNAs
(i.e. pSur-Emp and pSur-125a) were released in the medium at pH 4-5
(Fig. 2D). By contrast, only
~5-10% of the loaded plasmid DNAs were released in the medium at pH
6-7.4, suggesting the release of the plasmid DNAs is pH sensitive,
and the loaded plasmid DNAs are likely to be released in the
endo/lysosomal compartments of cells (Fig. 2D) (47). The plasmid DNA pCMV6-AC-GFP (a
GFP-expressing construct) and R6G-containing nanoparticles (i.e.
pL-MNP-pCMV6-AC-GFP) were created to confirm if the nanoparticles
could penetrate and release the loaded plasmid DNAs in cells. These
nanoparticles were synthesized using the same formulation and under
the same conditions as for the production of pL-MNP-pSur-125a.
Incorporating R6G in pL-MNP-pCMV6-AC-GFP enabled the tracing of
these nanoparticles during the transfection process. To prevent the
leaching of R6G from nanoparticles, R6G-isocyanate was covalently
bonded to the amino groups of pL-MNP. Microscopic images
demonstrated that most KB cells treated with pL-MNP-pCMV6-AC-GFP
emitted both the red (i.e. R6G) and green (i.e. GFP) fluorescent
signals, indicating that pL-MNP-pCMV6-AC-GFP nanoparticles are
capable of penetrating the cells and releasing the loaded
pCMV6-AC-GFP for ectopic expression of GFP (Fig. 2E). Similarly, fluorescence
microscopy results revealed successful binding/penetration of
pL-MNP-pSur-125a on/into KB cells (Fig. 2F). Of note, results of the qPCR
analysis showed that the amount of the miR-125a-5p transcripts
present in the pL-MNP-pSur-125a treated cells was significantly
increased as compared with cells treated with the control
nanoparticles pL-MNP-pSur-Emp, confirming that the plasmid DNA
pSur-125a was successfully released by the nanoparticles and was
also activated to express miR-125a-5p in the treated cells
(Fig. 2F).
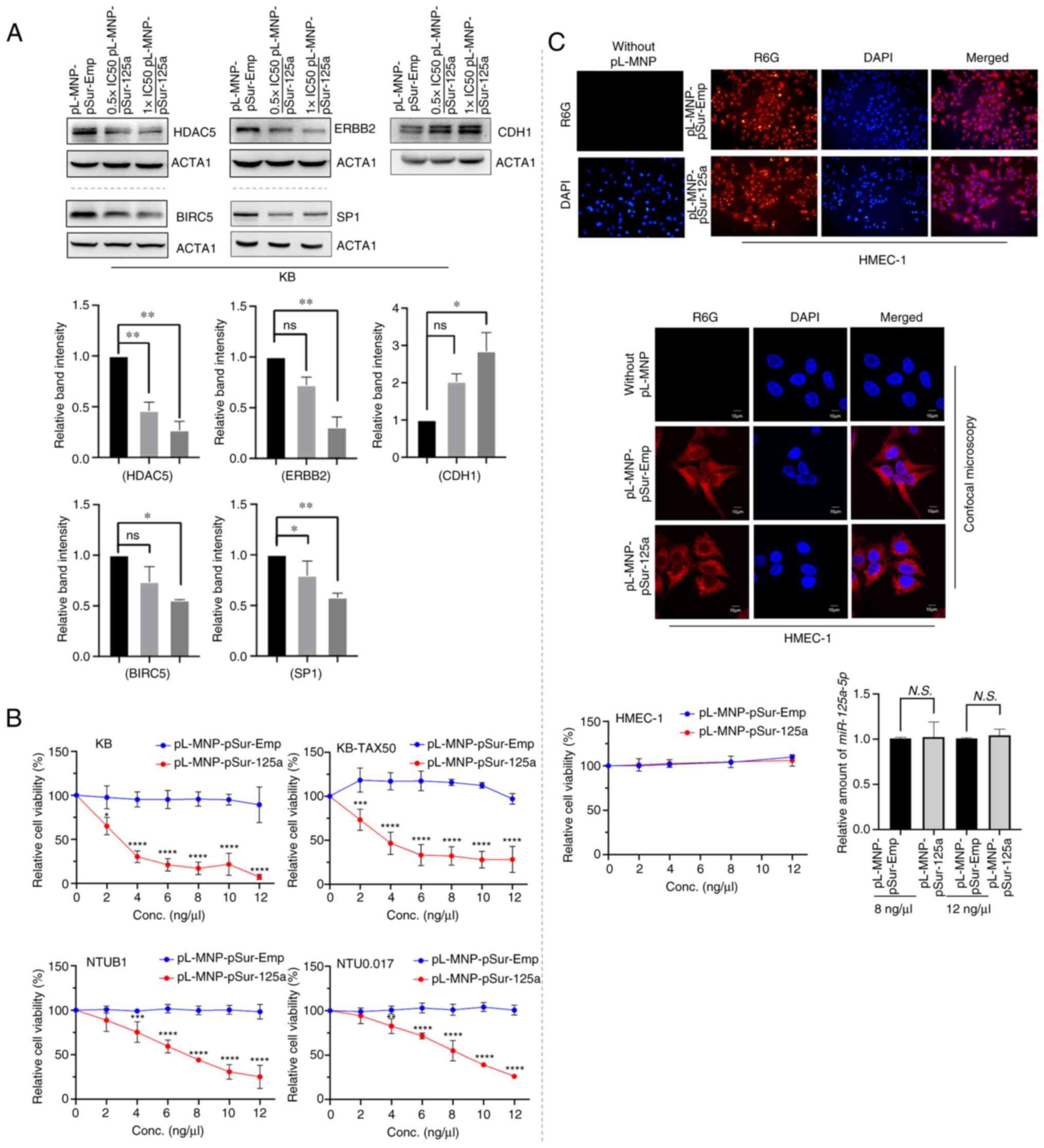
pL-MNP-pSur-125a exhibits the designated
molecular and cellular functions in BIRC5-expressing cancer
cells
At the molecular level and similar to the results of
cells transfected with the plasmid DNA pSur-125a, KB cells treated
with pL-MNP-pSur-125a also showed decreased protein expression
levels of BIRC5, ERBB2, and HDAC5 (Fig. 3A). PL-MNP-pSur-125a also decreased
the protein expression of HDAC5 in KB-TAX50 cells, confirming that
HDAC5 is a downstream target of miR-125a-5p (Fig. S3A). By contrast, pL-MNP-pSur-125a
increased the expression of CDH1/E-cadherin in KB cells, suggesting
that the reduced expression of BIRC5, ERBB2 and HDAC5 protein was
unlikely caused by the general reduction in the rate of protein
synthesis in cells undergoing apoptosis and cell death (Fig. 3A). It was previously demonstrated
that ectopic overexpression of miR-125a-5p decreases the expression
of SP1, a transcription factor that is known to promote
tumorigenesis upon upregulation in human SK-BR-3, MCF7, and
MCF7-TamC3 breast cancer cells (15). In the present study,
pL-MNP-pSur-125a decreased the expression of SP1 in KB cells,
further confirming the poly-pharmacological effects (on various
known miR-125a-5p-affecting molecules) of the nanodrug (Fig. 3A). Upregulation of tryptophan
2,3-dioxygenase (TDO2) promotes cancer immune evasion, and
recently, TDO2 has been a hot therapeutic target for cancer
(48,49). Intriguingly, it was found that
pL-MNP-pSur-125a decreased the expression of TDO2 in KB but not in
other examined cancer cells (Fig.
S3B). To confirm if the downregulation of TDO2 was caused by
miR-125a-5p overexpression, and not by the chemicals used for the
nanoparticle production, cells were transfected with or without the
plasmid DNA pSur-125a using liposomal reagents and the expression
of TDO2 was examined. Similar to cells treated with
pL-MNP-pSur-125a, transfection of pSur-125a also decreased the
expression of TDO2 in KB cells (Fig.
S3B). Collectively, these results suggested that miR-125a-5p
differentially regulates TDO2 expression in different cancer
cells.
The cellular effects of pL-MNP-pSur-125a were
examined in both the BIRC5-expressing KB, KB-TAX50, NTUB1, NTU0.017
cancer cells and the BIRC5-non-expressing (or expressing at a
relatively low level) HMEC-1 cells (50). In the present study, the cell
viability assay results showed that pL-MNP-pSur-125a, but not
pL-MNP-pSur-Emp, decreased the viability of KB, KB-TAX50, NTUB1 and
NTU0.017 cells in a concentration-dependent manner (Fig. 3B). Importantly, pL-MNP-125a-5p is
equally potent in inhibiting the growth of the multidrug resistance
protein ABCB1 expressing (i.e. KB-TAX50 and NTU0.017) and their
parental ABCB1 non-expressing (i.e. KB and NTUB1) cancer cells
(Table II). Besides KB, NTUB1,
and their derived ABCB1-expressing sublines, pL-MNP-pSur-125a (12
ng/µl, i.e. the maximum concentration used in the
aforementioned cell viability assay) also decreased the viability
(>50%) of various BIRC5+ cancer cell lines including
MCF7, MCF7-TamC3, MDA-MB-231 and MIA-PaCa 2, as compared with those
treated with pL-MNP-pSur-Emp (Fig.
S3C). Of note, despite high levels of localization (and
internalization) of the nanoparticles on cells, neither
pL-MNP-pSur-Emp nor pL-MNP-pSur-125a exhibited any inhibitory
effects on the viability of HMEC-1 cells in vitro (Fig. 3C). In addition, pL-MNP-pSur-125a,
even at high concentrations (i.e. at 8 and 12 ng/µl), did
not increase the expression of miR-125a-5p in HMEC-1 cells, further
confirming that pL-MNP-pSur-125a is inactivated in BIRC5
non-expressing cells as designated (Fig. 3C).
 | Table IIpL-MNP-pSur-125a exhibits
anti-proliferative activity against various types of cancer cells
regardless of the expression of ABCB1. |
Table II
pL-MNP-pSur-125a exhibits
anti-proliferative activity against various types of cancer cells
regardless of the expression of ABCB1.
| Cell line | HMEC-1 | KB | KB-TAX50 | NTUB1 | NTU0.017 |
|---|
| Tissue type | Microvascular
endothelial | Cervical
cancer | Cervical
cancer | Bladder cancer | Bladder cancer |
| Drug
resistance | N/A | - | Paclitaxel | - | Paclitaxel |
| Vincristine | | Vincristine |
| YM155 | | YM155 |
| ABCB1 status | Negative | Negative | Positive | Negative | Positive |
| pL-MNP-pSur-125a
(IC50, ng/µl) | >12 (unable to
determine) | 2.8±0.5 | 3.6±1.3 | 7.2±0.6 | 8.6±0.4 |
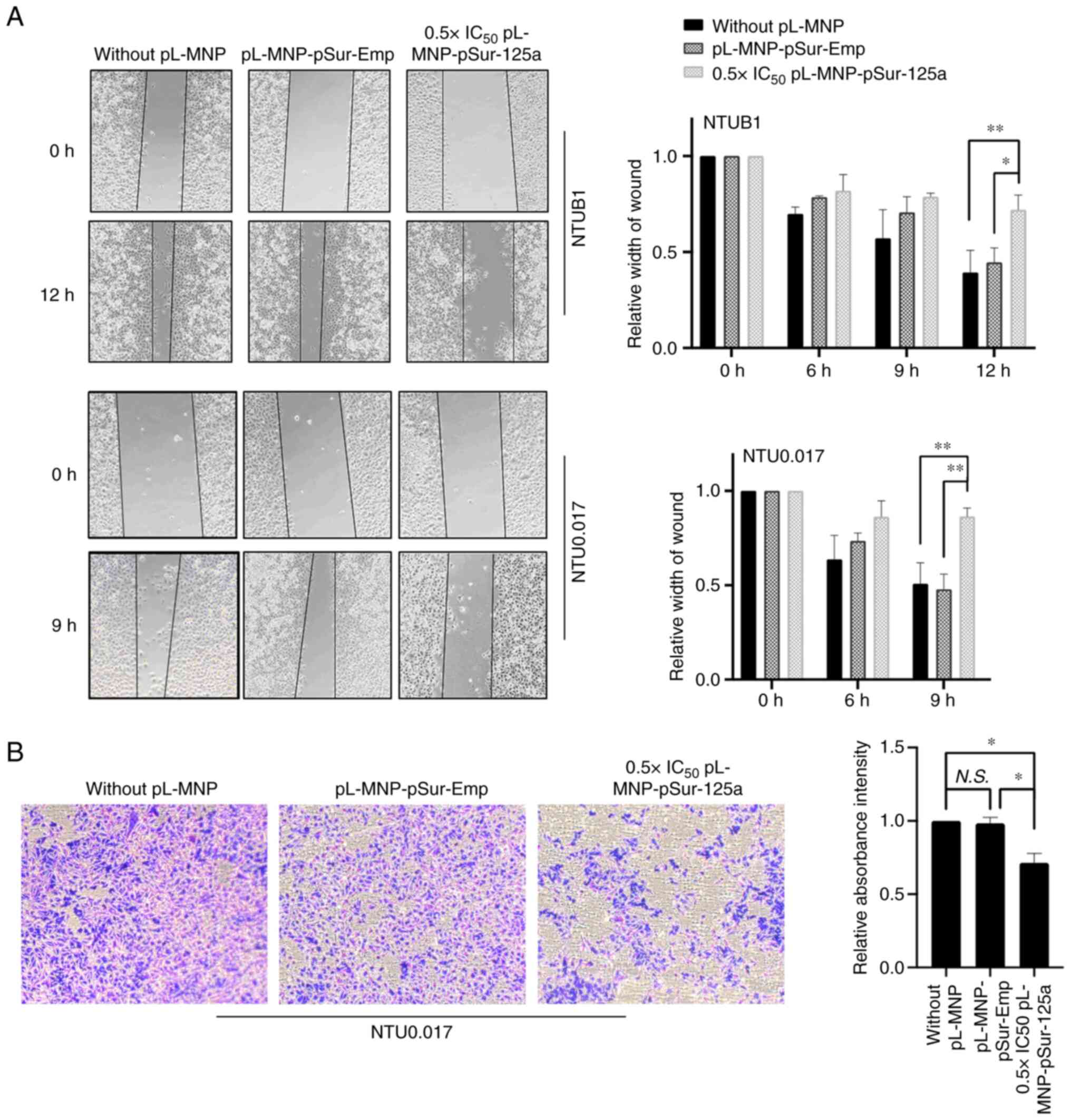
Upregulation of BIRC5 and SP1 promotes cancer cells
migration, invasion, and tumor metastasis (51). Notably, CDH1 downregulation is
frequently observed in cancer cells undergoing EMT and
pL-MNP-pSur-125a increased the expression of CDH1 in KB cells
(Fig. 3A). In the present study,
despite the migration rate of NTUB1 and NTU0.017 cells under
culturing conditions being slightly different, results of the
wound-healing assay showed that pL-MNP-pSur-125a at a low cytotoxic
concentration (i.e. 0.5 × IC50) reduced the migration of
both NTUB1and NTU0.017 cells (Fig.
4A). As NTUB1 is not suitable for use in the cell invasion
assay, the potential effects of pL-MNP-pSur-125a on cell invasion
were examined in NTU0.017 cells. In the present study,
0.5×IC50 pL-MNP-pSur-125a also decreased the
invasiveness of NTU0.017 cancer cells in vitro (Fig. 4B).
pL-MNP-pSur-125a exhibits antitumor
formation effects in vivo
The anticancer efficacy of pL-MNP-pSur-125a was
assessed in zebrafish, which is a xenograft model widely used for
studies of tumor development and preclinical testing of anticancer
drugs (52-54). The PKH67-stained (green
fluorescent) KB and KB-TAX50 cancer cells were microinjected into
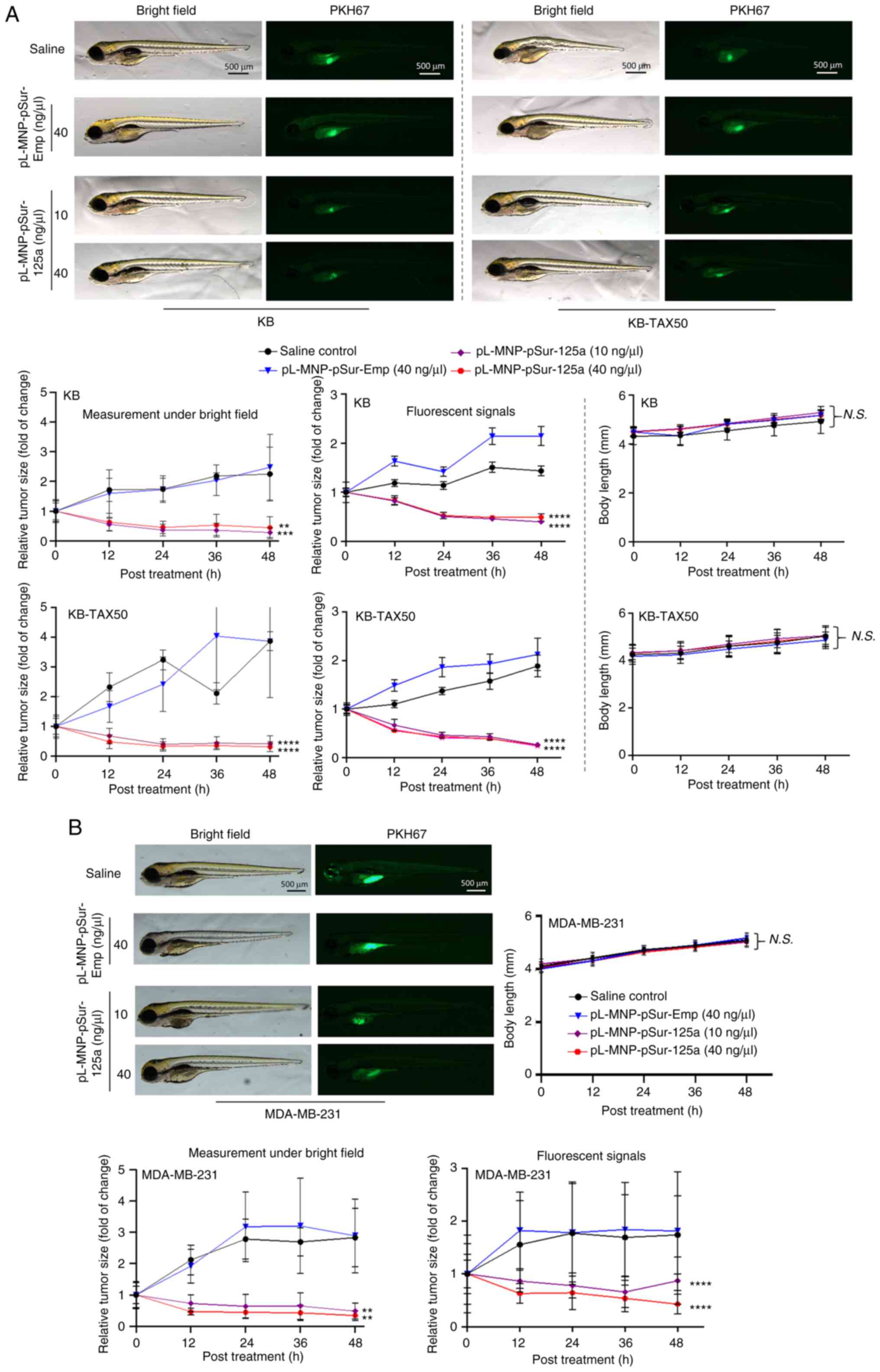
the yolk sac of zebrafish embryos. As revealed in Fig. 5, pL-MNP-pSur-125a reduced the size
of the KB, KB-TAX50 and MDA-MB-231 xenograft tumors in zebrafish.
By contrast, neither saline nor pL-MNP-pSur-Emp showed any
inhibitory effects on the growth of tumors (Fig. 5A and B). Notably, pL-MNP-pSur-125a
was well-tolerated at the examined concentrations with no signs of
severe toxicity in the KB, KB-TAX50 and MDA-MB-231 xenograft tumor
models as the overall body length of zebrafish was similar (i.e.
changes were insignificant) between the treatment and the control
groups (i.e. saline and pL-MNP-pSur-Emp) (Fig. 5A and B).
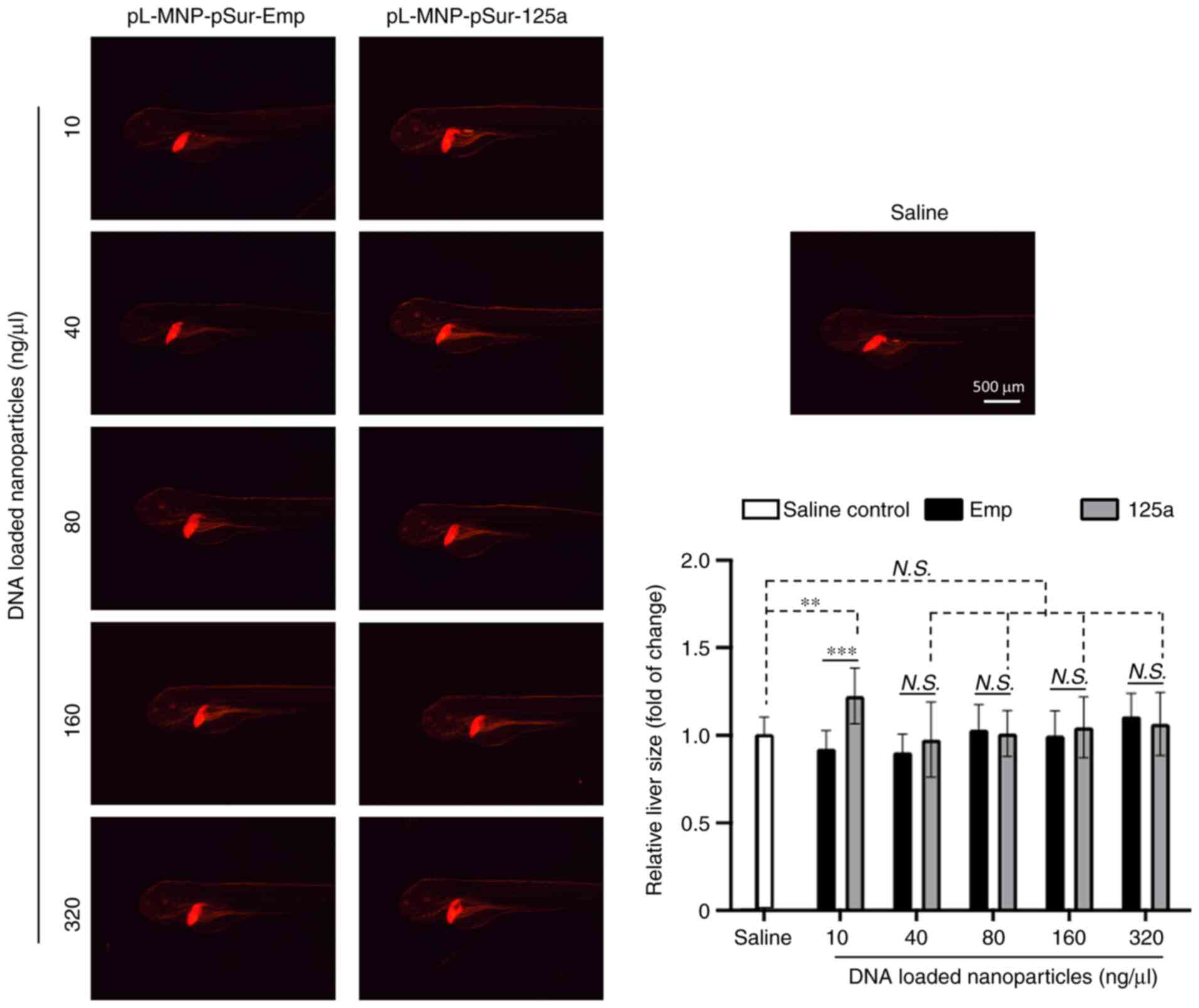
The possible hepatotoxicity effect of
pL-MNP-pSur-125a was also examined in zebrafish, as it is an in
vivo model commonly used to study drug-induced liver injury
(55,56). Decreased liver size is a sign of
liver damage. In the present study, pL-MNP-pSur-125a did not show
any negative-effects on the size of the liver at most examined
concentrations (except for 10 and 640 ng/µl, with slightly
increased liver size), suggesting that the use of pL-MNP-pSur-125a
may not induce liver damage during the treatment (Fig. 6).
Discussion
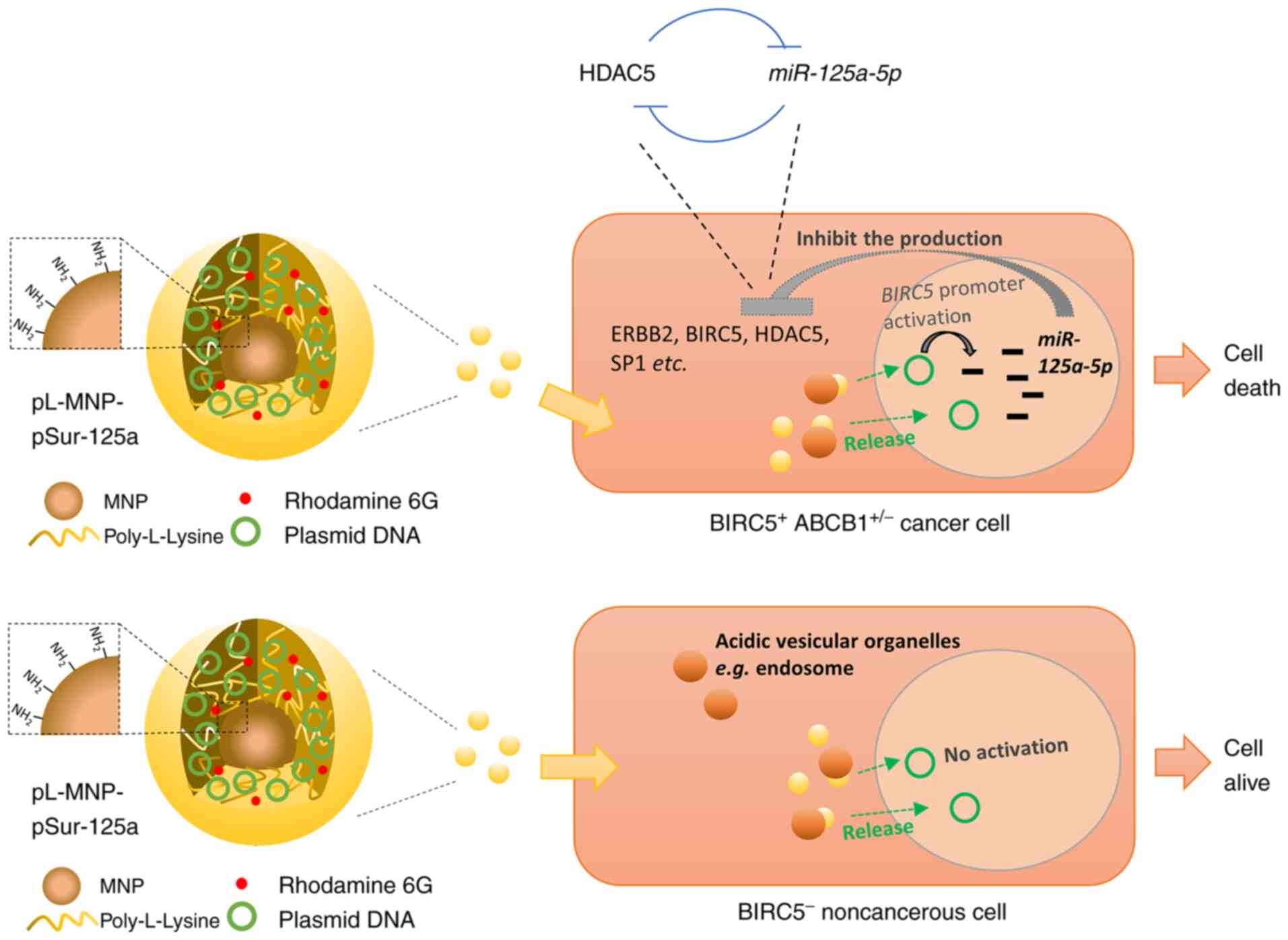
In the present study, pL-MNP was successfully
utilized as a transfection agent and the high gene expression
efficiency of DNA-loaded pL-MNP (pL-MNP-pSur-125a) in
BIRC5-expressing cancer cells was demonstrated. In this DNA-carried
system, DNA was adsorbed on pL-MNP by strong electrostatic
interactions and it could be released from pL-MNP by protonation of
the phosphate group in DNA under acidic conditions (Fig. 7). During the production of
pL-MNP-pSur-125a from pL-MNP, the increased hydrodynamic diameter
(from ~289 to ~327 nm) and the charge transformation (from positive
to negative charge) of zeta potential indicated the successful
adsorption of pSur-125a on pL-MNP. Moreover, a high degree of
overlap between the two chemical elements, phosphorus and iron, in
pL-MNP-pSur-125a as shown by the elemental distribution maps also
suggests that pSur-125a was successfully adsorbed onto pL-MNP.
Functionally, pL-MNP-pSur-125a exhibits
poly-pharmacological properties as it downregulates the expression
of various cancer-related molecules (HDAC5, ERBB2, BIRC5 and SP1)
and induces cell death in BIRC5+ cancer cells (Fig. 7). It was previously identified
that HDAC5 negatively regulates the expression of miR-125a-5p in
cancer cells (15). Notably, in
the present study, it was found that miR-125a-5p negatively
regulates the expression of HDAC5. Co-incubation with the BIRC5
expression-suppressant (through the binding on the BIRC5
gene promotor region), YM155, only partially suppressed the
increased-expression of miR-125a-5p in cells transfected with
pSur-125a. The use of YM155 at low concentration (i.e. sublethal
concentration) in the experiment may be one of the reasons behind
the incomplete suppression of the increased-expression of
miR-125a-5p in the pSur-125a-transfected cells. It is also possible
that the increased amount of miR-125a-5p transcripts present in the
pSur-125a-transfected cells was not solely caused by the
BIRC5 promotor-driven miR-125a-5p expressing function of
pSur-125a but also partly caused by the decreased expression of
HDAC5 (induced by the pSur-125a-expressing miR-125a-5p), leading to
the increased expression of the endogenous miR-125a-5p in cells
(i.e. activating the endogenous feedback loop between HDAC5 and
miR-125a-5p). Although the expression of the well-known
anti-apoptotic molecule, B-cell lymphoma 2 (BCL2), was not examined
in the pL-MNP-pSur-125a-treated cancer cells in the present study,
it was demonstrated in a previous study that ectopic overexpression
of miR-125a-5p decreases the expression of BCL2 in MCF7, MCF7-TamC3
and SK-BR-3 breast cancer cells (15). As BCL2 mRNA contains a
putative miR-125a-5p binding site in the 3′ untranslated region
(position 2419-2426 of BCL2 3′UTR), it is considered that
pL-MNP-pSur-125a should also downregulate the expression of BCL2 in
various BIRC5+ cancer cells.
Two of the most important pharmacological features
of pL-MNP-pSur-125a are the following: i) this nanodrug is designed
to be activated primarily in tumors, but not in the differentiated
normal tissues, as BIRC5 is highly expressed in cancer cells, but
not in the differentiated normal cells, and ii) the potency of this
nanodrug is not affected by the expression of the multidrug
resistant protein ABCB1 in cancer cells. Drug resistance is known
to be a major clinical problem in cancer treatment. Upregulation of
a drug efflux transporter ABCB1 causes cancer cells to become
refractory to various chemotherapeutic, targeted therapeutic and
hormonal drugs such as paclitaxel (mitotic inhibitor), doxorubicin
(topoisomerase inhibitor), olaparib [poly ADP-ribose polymerase
inhibitor (PARPi)], and tamoxifen [selective estrogen receptor
modulator (SERM)] (57-60). Besides ABCB1, upregulation of
BIRC5 is also known to promote multidrug resistance in cancer
cells. For example, Park et al demonstrated that
overexpression of BIRC5 promotes vincristine (mitotic inhibitor)
resistance, whereas downregulation of BIRC5 increases the
sensitivity to vincristine in acute lymphoblastic leukemia cells
(61). It was previously
demonstrated that dysregulation of the HDAC5-BIRC5-signaling
pathway contributes to the development of both estrogen
independence and hormone therapy resistance in estrogen
receptor-positive (ER+) breast cancer cells (15). Overexpression of HDAC5 has also
been shown to promote SOX9 deacetylation and nuclear translocation,
contributing to tamoxifen resistance in ER+ breast
cancer (62). In the case of
ERBB2, overexpression or hyperactivation of this EGFR family member
has been demonstrated to regulate the NRF2-dependent
transcriptional activation, leading to the induction of antioxidant
response and drug resistance in cancer cells (63). The expression of ERBB2 and the
activation of its downstream signaling pathway is also known to
play an important role in the survival of ERBB2+ breast
cancer (i.e. the HER2-enriched subtype). On the other hand, TDO2
plays a pivotal role in regulating the immune microenvironment in
tumors and overexpression of TDO2 promotes tumor immune evasion
(64,65). Thus, various efforts have been
made to the development of HDAC5 (e.g., LMK-235), BIRC5 (e.g.,
YM155, SPC3042), ERBB2 (e.g., Herceptin, Irbinitinib) and TDO2
(e.g., 680C91 and LM10) modulators/inhibitors for cancer treatment
and several of them have reached clinical trials (30,31,35,45,48,66). Not to mention that the anti-ERBB2
monoclonal antibody, Herceptin, is already being used clinically in
treating patients with ERBB2+ (i.e. HER2+)
breast cancer. Given the roles of BIRC5, ERBB2, HDAC5 and TDO2 in
survival of tumor cells, pL-MNP-pSur-125a is a promising nanodrug
that has potential for the management of various malignancies,
particularly for patients with ABCB1-related multidrug resistance
after prolonged chemotherapeutic treatments. In addition, since
miR-125a-5p modulates the expression of various proteins, it will
be interesting to investigate in the future if pL-MNP-pSur-125a can
also modulate the expression of different major histocompatibility
class I molecules, in which dysregulation of these molecules is
known to affect the effectiveness of anticancer immunotherapy
(67).
Large-sized, DNA-loaded, nanoparticles
(hydrodynamic diameter of 300-600 nm) have been revealed to mediate
a higher transfection and gene expression efficiency in cells as
compared with the DNA-loaded nanoparticles of smaller size (<100
nm) (68,69). However, nanoparticles with a
hydrodynamic diameter of 100-400 nm, particularly for those with a
diameter of less than 300 nm, have widely been considered optimal
for passive tumor targeting due to the enhanced permeability and
retention effect (70). In
addition, it has been demonstrated that nanoparticles of 100-200 nm
in size can escape from recognition by the reticuloendothelial
system, which prolongs the half-life of nanoparticles in blood
circulation (71). As revealed by
the DLS results, the average hydrodynamic diameter of
pL-MNP-pSur-125a is ~360 nm. Despite the fact that the size of
pL-MNP-pSur-125a is slightly larger than 300 nm, which may hamper
the efficiency of 'tumor site-penetration', several studies showed
that nanoparticles with size larger than 300 nm still exhibit
potent anticancer effects in vivo. For example, Talekar
et al (72) demonstrated
that the wild-type TP53/p53 and miR-125b co-expressing plasmid
DNA-loaded hyaluronic acid-based nanoparticles, in a size range of
200-400 nm, were capable of inhibiting tumor growth and inducing
apoptosis in a mouse model of lung cancer. In addition, a study
showed that co-delivery of doxorubicin and the BIRC5
shRNA-expressing plasmid DNA by using mesoporous silica
nanoparticles of around 350 nm in size were potent in targeting
cancer cells in vitro and in vivo (73). It has also been demonstrated that
chemotherapeutic drugs loaded to iron oxide mesoporous magnetic
microparticles, with an average hydrodynamic size of 765 nm, were
capable of penetrating the deep tumor cell layers of the dissected
breast tumor tissues in culturing conditions of oxygen, nutrient
and energy gradients similar to those found in vivo
(74). Notably, fewer large-sized
nanoparticles (i.e. 200-400 nm in size) were shown to be uptaken by
macrophages than those of smaller size (75). As BIRC5 was expressed in
atherosclerotic macrophages, the currently developed
pL-MNP-pSur-125a nanoparticles in a size range of 300-400 nm may
escape from being uptaken by macrophages, thereby limiting the
induction of the unwanted immune responses (76). Further in vivo studies
(e.g., by using genetically engineered mice) are needed to
determine if pL-MNP-pSur-125a, at the current size, remains
functional in targeting BIRC5+ tumors in a more complex
tumor environment.
In conclusion, a poly-pharmacological nanoparticle
pL-MNP-pSur-125a has been successfully produced and demonstrated to
exert biological activity in eliminating BIRC5-expressing cancer
cells, regardless of the tissue origins and the expression of the
multidrug efflux pump ABCB1. PL-MNP-pSur-125a is a promising
anticancer nanodrug that has the potential for the management of
various malignancies, particularly for patients with ABCB1-related
drug resistance after prolonged chemotherapeutic treatments.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
Y-CC, M-CS, Y-HC, W-LH, W-CS, F-YC and CHAC
conceived and designed the experiments. Y-CC, Y-HC and F-YC
performed the experiments. Y-CC, Y-HC and F-YC analyzed the data.
Y-CC and Y-HC confirm the authenticity of all the raw data. F-YC
and CHAC wrote and proofread the paper. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The animal protocol was approved (approval no.
109273) by the Institutional Animal Care and Use Committee (IACUC)
of National Cheng Kung University (Tainan, Taiwan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank the technical
services provided by the 'Bio-image Core Facility of the National
Core Facility Program for Biotechnology, Ministry of Science and
Technology, Taiwan'.
Funding
The present study was supported by the Ministry of Science and
Technology of Taiwan (grant nos. 109-2320-B-006-031 and
110-2320-B-006-047-MY3) and the Ditmanson Medical Foundation
Chia-Yi Christian Hospital of Taiwan (grant no. CYC109006).
References
|
1
|
Groenendijk FH and Bernards R: Drug
resistance to targeted therapies: Déjà vu all over again. Mol
Oncol. 8:1067–1083. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xie L and Bourne PE: Developing
multi-target therapeutics to fine-tune the evolutionary dynamics of
the cancer ecosystem. Front Pharmacol. 6:2092015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Antolin AA, Workman P, Mestres J and
Al-Lazikani B: Polypharmacology in precision oncology: Current
applications and future prospects. Curr Pharm Design. 22:6935–6945.
2016. View Article : Google Scholar
|
|
4
|
Nishida N, Mimori K, Fabbri M, Yokobori T,
Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y and Mori M:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai
CY, Hou MF, Lee JN, Wu DC, Wang SC and Tsai EM: miR-125a-5p is a
prognostic biomarker that targets HDAC4 to suppress breast
tumorigenesis. Oncotarget. 6:4942014. View Article : Google Scholar
|
|
6
|
Vo DT, Karanam NK, Ding L, Saha D, Yordy
JS, Giri U, Heymach JV and Story MD: miR-125a-5p functions as tumor
suppressor microRNA and is a marker of locoregional recurrence and
poor prognosis in head and neck cancer. Neoplasia. 21:849–862.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang Z, Pan Q, Zhang Z, Huang C, Yan Z,
Zhang Y and Li J: MicroRNA-125a-5p controls the proliferation,
apoptosis, migration and PTEN/MEK1/2/ERK1/2 signaling pathway in
MCF-7 breast cancer cells. Mol Med Rep. 20:4507–4514. 2019.
|
|
8
|
Yan L, Yu MC, Gao GL, Liang HW, Zhou XY,
Zhu ZT, Zhang CY, Wang YB and Chen X: MiR-125a-5p functions as a
tumour suppressor in breast cancer by downregulating BAP1. J Cell
Biochem. 119:8773–8783. 2018. View Article : Google Scholar
|
|
9
|
Tang L, Zhou L, Wu S, Shi X, Jiang G, Niu
S and Ding D: miR-125a-5p inhibits colorectal cancer cell
epithelial-mesenchymal transition, invasion and migration by
targeting TAZ. Onco Targets Ther. 12:3481–3489. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tong Z, Liu N, Lin L, Guo X, Yang D and
Zhang Q: miR-125a-5p inhibits cell proliferation and induces
apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1.
Biomed Pharmacother. 75:129–136. 2015. View Article : Google Scholar
|
|
11
|
Zhong L, Sun S, Shi J, Cao F, Han X and
Chen Z: MicroRNA-125a-5p plays a role as a tumor suppressor in lung
carcinoma cells by directly targeting STAT3. Tumor Biol.
39:10104283176975792017. View Article : Google Scholar
|
|
12
|
Hsieh TH, Hsu CY, Tsai CF, Long CY, Wu CH,
Wu DC, Lee JN, Chang WC and Tsai EM: HDAC inhibitors target HDAC5,
upregulate microRNA-125a-5p and induce apoptosis in breast cancer
cells. Mol Ther. 23:656–666. 2015. View Article : Google Scholar :
|
|
13
|
Xu Y, Zheng Y, Duan Y, Ma L and Nan P:
MicroRNA-125a-5p targets LIM kinase 1 to inhibit cisplatin
resistance of cervical cancer cells. Oncol Lett. 21:3922021.
View Article : Google Scholar :
|
|
14
|
Cao Q, Wang N, Ren L, Tian J, Yang S and
Cheng H: miR-125a-5p post-transcriptionally suppresses GALNT7 to
inhibit proliferation and invasion in cervical cancer cells via the
EGFR/PI3K/AKT pathway. Cancer Cell Int. 20:1172020. View Article : Google Scholar :
|
|
15
|
Huang WT, Tsai YH, Chen SH, Kuo CW, Kuo
YL, Lee KT, Chen WC, Wu PC, Chuang CY, Cheng SM, et al: HDAC2 and
HDAC5 up-regulations modulate survivin and miR-125a-5p expressions
and promote hormone therapy resistance in estrogen receptor
positive breast cancer cells. Front Pharmacol. 8:9022017.
View Article : Google Scholar
|
|
16
|
Zhang Y, Li A, Shi J, Fang Y, Gu C, Cai J,
Lin C, Zhao L and Liu S: Imbalanced LIMK1 and LIMK2 expression
leads to human colorectal cancer progression and metastasis via
promoting β-catenin nuclear translocation. Cell Death Dis.
9:7492018. View Article : Google Scholar
|
|
17
|
Cheung CH, Chen HH, Kuo CC, Chang CY,
Coumar MS, Hsieh HP and Chang JY: Survivin counteracts the
therapeutic effect of microtubule de-stabilizers by stabilizing
tubulin polymers. Mol Cancer. 8:432009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mahalaxmi I and Santhy KS: Role and
hallmarks of Sp1 in promoting ovarian cancer. J Oncol Sci.
4:102–105. 2018. View Article : Google Scholar
|
|
19
|
Jiang Y, de Bruin A, Caldas H, Fangusaro
J, Hayes J, Conway EM, Robinson ML and Altura RA: Essential role
for survivin in early brain development. J Neurosci. 25:6962–6970.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vischioni B, van der Valk P, Span SW,
Kruyt FAE, Rodriguez JA and Giaccone G: Nuclear localization of
survivin is a positive prognostic factor for survival in advanced
non-small-cell lung cancer. Ann Oncol. 15:1654–1660. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang L, Yan R, Zhang Q, Wang H, Kang X,
Li J, Yang S, Zhang J, Liu Z and Yang X: Survivin, a key component
of the Wnt/β-catenin signaling pathway, contributes to traumatic
brain injury-induced adult neurogenesis in the mouse dentate gyrus.
Int J Mol Med. 32:867–875. 2013. View Article : Google Scholar
|
|
23
|
Bao R, Connolly DC, Murphy M, Green J,
Weinstein JK, Pisarcik DA and Hamilton TC: Activation of
cancer-specific gene expression by the survivin promoter. J Natl
Cancer Inst. 94:522–528. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang L, Cao Z, Li F, Post DE, Van Meir EG,
Zhong H and Wood WC: Tumor-specific gene expression using the
survivin promoter is further increased by hypoxia. Gene Ther.
11:1215–1223. 2004. View Article : Google Scholar
|
|
25
|
Siddharth S, Das S, Nayak A and Kundu CN:
Survivin as a marker for quiescent-breast cancer stem cells-An
intermediate, adherent, pre-requisite phase of breast cancer
metastasis. Clin Exp Metastasis. 33:661–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carter BZ, Qiu Y, Huang X, Diao L, Zhang
N, Coombes KR, Mak DH, Konopleva M, Cortes J, Kantarjian HM, et al:
Survivin is highly expressed in CD34+38-leukemic stem/progenitor
cells and predicts poor clinical outcomes in AML. Blood.
120:173–180. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Yan H, Li R, Guo Y and Zheng R:
High expression of survivin predicts poor prognosis in cervical
squamous cell carcinoma treated with paclitaxel and carboplatin.
Medicine (Baltimore). 98:e156072019. View Article : Google Scholar
|
|
28
|
Onodi F, Maherzi-Mechalikh C, Mougel A,
Hamouda NB, Taboas C, Gueugnon F, Tran T, Nozach H, Marcon E, Gey
A, et al: High therapeutic efficacy of a new survivin LSP-cancer
vaccine containing CD4+ and CD8+ T-cell epitopes. Front Oncol.
8:5172018. View Article : Google Scholar :
|
|
29
|
Voges Y, Michaelis M, Rothweiler F,
Schaller T, Schneider C, Politt K, Mernberger M, Nist A, Stiewe T,
Wass MN, et al: Effects of YM155 on survivin levels and viability
in neuroblastoma cells with acquired drug resistance. Cell Death
Dis. 7:e24102016. View Article : Google Scholar :
|
|
30
|
Nakahara T, Kita A, Yamanaka K, Mori M,
Amino N, Takeuchi M, Tominaga F, Hatakeyama S, Kinoyama I,
Matsuhisa A, et al: YM155, a novel small-molecule survivin
suppressant, induces regression of established human
hormone-refractory prostate tumor xenografts. Cancer Res.
67:8014–8021. 2007. View Article : Google Scholar
|
|
31
|
Hansen JB, Fisker N, Westergaard M,
Kjaerulff LS, Hansen HF, Thrue CA, Rosenbohm C, Wissenbach M, Orum
H and Koch T: SPC3042: A proapoptotic survivin inhibitor. Mol
Cancer Ther. 7:2736–2745. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tolcher AW, Mita A, Lewis LD, Garrett CR,
Till E, Daud AI, Patnaik A, Papadopoulos K, Takimoto C, Bartels P,
et al: Phase I and pharmacokinetic study of YM155, a small-molecule
inhibitor of survivin. J Clin Oncol. 26:5198–5203. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheung CHA, Sun X, Kanwar JR, Bai JZ,
Cheng L and Krissansen GW: A cell-permeable dominant-negative
survivin protein induces apoptosis and sensitizes prostate cancer
cells to TNF-α therapy. Cancer Cell Int. 10:362010. View Article : Google Scholar
|
|
34
|
Tsai SL, Chang YC, Sarvagalla S, Wang S,
Coumar MS and Cheung CHA: Cloning, expression, and purification of
the recombinant pro-apoptotic dominant-negative survivin T34A-C84A
protein in Escherichia coli. Protein Expr Purif. 160:73–83. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Quispe PA, Lavecchia MJ and León IE: On
the discovery of a potential survivin inhibitor combining
computational tools and cytotoxicity studies. Heliyon.
5:e022382019. View Article : Google Scholar :
|
|
36
|
Arigita C, Zuidam NJ, Crommelin DJ and
Hennink WE: Association and dissociation characteristics of
polymer/DNA complexes used for gene delivery. Pharm Res.
16:1534–1541. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin KY, Cheng SM, Tsai SL, Tsai JY, Lin CH
and Cheung CHA: Delivery of a survivin promoter-driven antisense
survivin-expressing plasmid DNA as a cancer therapeutic: A
proof-of-concept study. Onco Targets Ther. 9:2601–2613.
2016.PubMed/NCBI
|
|
38
|
Cheung CH, Lin WH, Hsu JTA, Hour TC, Yeh
TK, Ko S, Lien TW, Coumar MS, Liu JF, Lai WY, et al: BPR1K653, a
novel Aurora kinase inhibitor, exhibits potent anti-proliferative
activity in MDR1 (P-gp170)-mediated multidrug-resistant cancer
cells. PLoS One. 6:e234852011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chang YC, Kondapuram SK, Yang TH, Syed SB,
Cheng SM, Lin TY, Lin YC, Coumar MS, Chang JY, Leung E and Cheung
CHA: The SMAC mimetic LCL161 is a direct ABCB1/MDR1-ATPase activity
modulator and BIRC5/Survivin expression down-regulator in cancer
cells. Toxicol Appl Pharmacol. 401:1150802020. View Article : Google Scholar
|
|
40
|
Lee PC, Lee HJ, Kakadiya R, Sanjiv K, Su
TL and Lee TC: Multidrug-resistant cells overexpressing
P-glycoprotein are susceptible to DNA crosslinking agents due to
attenuated Src/nuclear EGFR cascade-activated DNA repair activity.
Oncogene. 32:1144–1154. 2013. View Article : Google Scholar
|
|
41
|
Yu HJ, Tsai TC, Hsieh TS and Chiu TY:
Characterization of a newly established human bladder carcinoma
cell line, NTUB1. J Formos Med Assoc. 91:608–613. 1992.PubMed/NCBI
|
|
42
|
Leung E, Kannan N, Krissansen GW, Findlay
MP and Baguley BC: MCF-7 breast cancer cells selected for tamoxifen
resistance acquire new phenotypes differing in DNA content,
phospho-HER2 and PAX2 expression, and rapamycin sensitivity. Cancer
Biol Ther. 9:717–724. 2010. View Article : Google Scholar
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
44
|
Jiang G, Ren B, Xu L, Song S, Zhu C and Ye
F: Survivin may enhance DNA double-strand break repair capability
by up-regulating Ku70 in human KB cells. Anticancer Res.
29:223–228. 2009.PubMed/NCBI
|
|
45
|
Cheng Q, Ling X, Haller A, Nakahara T,
Yamanaka K, Kita A, Koutoku H, Takeuchi M, Brattain MG and Li F:
Suppression of survivin promoter activity by YM155 involves
disruption of Sp1-DNA interaction in the survivin core promoter.
Int J Biochem Mol Biol. 3:179–197. 2012.PubMed/NCBI
|
|
46
|
Al-Sharif I, Remmal A and Aboussekhra A:
Eugenol triggers apoptosis in breast cancer cells through
E2F1/survivin down-regulation. BMC Cancer. 13:6002013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Meng F, Cheng R, Deng C and Zhong Z:
Intracellular drug release nanosystems. Materials Today.
15:436–442. 2012. View Article : Google Scholar
|
|
48
|
de Iudicibus RC, Tomek P, Palmer BD,
Tijono SM, Flanagan JU and Ching LM: Parallel discovery of
selective and dual inhibitors of tryptophan dioxygenases IDO1 and
TDO2 with a newly-modified enzymatic assay. Bioorg Med Chem.
39:1161602021. View Article : Google Scholar
|
|
49
|
Sari S, Tomek P, Leung E and Reynisson J:
Discovery and characterisation of dual inhibitors of tryptophan
2,3-Dioxygenase (TDO2) and indoleamine 2,3-dioxygenase 1 (IDO1)
using virtual screening. Molecules. 24:43462019. View Article : Google Scholar :
|
|
50
|
Gong Y, Li Y, Abdolmaleky HM, Li L and
Zhou JR: Tanshinones inhibit the growth of breast cancer cells
through epigenetic modification of aurora a expression and
function. PLoS One. 7:e336562012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tai CJ, Chin-Sheng H, Kuo LJ, Wei PL, Lu
HH, Chen HA, Liu TZ, Liu JJ, Liu DZ, Ho YS, et al:
Survivin-mediated cancer cell migration through GRP78 and
epithelial-mesenchymal transition (EMT) marker expression in
mahlavu cells. Ann Surg Oncol. 19:336–343. 2012. View Article : Google Scholar
|
|
52
|
Al-Thani HF, Shurbaji S and Yalcin HC:
Zebrafish as a model for anticancer nanomedicine studies.
Pharmaceuticals (Basel). 14. pp. 6252021, View Article : Google Scholar
|
|
53
|
Letrado P, de Miguel I, Lamberto I,
Díez-Martínez R and Oyarzabal J: Zebrafish: Speeding up the cancer
drug discovery process. Cancer Res. 78:6048–6058. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hason M and Bartůněk P: Zebrafish models
of cancer-new insights on modeling human cancer in a non-mammalian
vertebrate. Genes (Basel). 10. pp. 9352019, View Article : Google Scholar
|
|
55
|
He JH, Guo SY, Zhu F, Zhu JJ, Chen YX,
Huang CJ, Gao JM, Dong QX, Xuan YX and Li CQ: A zebrafish
phenotypic assay for assessing drug-induced hepatotoxicity. J
Pharmacol Toxicol Methods. 67:25–32. 2013. View Article : Google Scholar
|
|
56
|
Vliegenthart ADB, Tucker CS, Pozo JD and
Dear JW: Zebrafish as model organisms for studying drug-induced
liver injury. Br J Clin Pharmacol. 78:1217–1227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mechetner E, Kyshtoobayeva A, Zonis S, Kim
H, Stroup R, Garcia R, Parker RJ and Fruehauf JP: Levels of
multidrug resistance (MDR1) P-glycoprotein expression by human
breast cancer correlate with in vitro resistance to taxol and
doxorubicin. Clin Cancer Res. 4:389–398. 1998.PubMed/NCBI
|
|
58
|
Duan Z, Brakora KA and Seiden MV:
Inhibition of ABCB1 (MDR1) and ABCB4 (MDR3) expression by small
interfering RNA and reversal of paclitaxel resistance in human
ovarian cancer cells. Mol Cancer Ther. 3:833–838. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Krisnamurti DGB, Louisa M, Anggraeni E and
Wanandi SI: Drug efflux transporters are overexpressed in
short-term tamoxifen-induced MCF7 breast cancer cells. Adv
Pharmacol Sci. 2016:67024242016.PubMed/NCBI
|
|
60
|
Vaidyanathan A, Sawers L, Gannon AL,
Chakravarty P, Scott AL, Bray SE, Ferguson MJ and Smith G: ABCB1
(MDR1) induction defines a common resistance mechanism in
paclitaxel- and olaparib-resistant ovarian cancer cells. Br J
Cancer. 115:431–441. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Park E, Gang EJ, Hsieh YT, Schaefer P,
Chae S, Klemm L, Huantes S, Loh M, Conway EM, Kang ES, et al:
Targeting survivin overcomes drug resistance in acute lymphoblastic
leukemia. Blood. 118:2191–2199. 2011. View Article : Google Scholar :
|
|
62
|
Xue Y, Lian W, Zhi J, Yang W, Li Q, Guo X,
Gao J, Qu H, Lin W, Li Z, et al: HDAC5-mediated deacetylation and
nuclear localisation of SOX9 is critical for tamoxifen resistance
in breast cancer. Br J Cancer. 121:1039–1049. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kang HJ, Yi YW, Hong YB, Kim HJ, Jang YJ,
Seong YS and Bae I: HER2 confers drug resistance of human breast
cancer cells through activation of NRF2 by direct interaction. Sci
Rep. 4:72012014. View Article : Google Scholar :
|
|
64
|
Liu Q, Zhai J, Kong X, Wang X, Wang Z,
Fang Y and Wang J: Comprehensive analysis of the expressionand
prognosis for TDO2 in breast cancer. Mol Ther Oncolytics.
17:153–168. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Miyazaki T, Chung S, Sakai H, Ohata H,
Obata Y, Shiokawa D, Mizoguchi Y, Kubo T, Ichikawa H, Taniguchi H,
et al: Stemness and immune evasion conferred by the TDO2-AHR
pathway are associated with liver metastasis of colon cancer.
Cancer Sci. 113:170–181. 2022. View Article : Google Scholar
|
|
66
|
Wanek J, Gaisberger M, Beyreis M, Mayr C,
Helm K, Primavesi F, Jäger T, Fazio PD, Jakab M, Wagner A, et al:
Pharmacological inhibition of class IIA HDACs by LMK-235 in
pancreatic neuroendocrine tumor cells. Int J Mol Sci. 19:31282018.
View Article : Google Scholar :
|
|
67
|
Dhatchinamoorthy K, Colbert JD and Rock
KL: Cancer immune evasion through loss of MHC class I antigen
presentation. Front Immunol. 12:6365682021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ogris M, Steinlein P, Kursa M, Mechtler K,
Kircheis R and Wagner E: The size of DNA/transferrin-PEI complexes
is an important factor for gene expression in cultured cells. Gene
Ther. 5:1425–1433. 1998. View Article : Google Scholar
|
|
69
|
Ogris M, Steinlein P, Carotta S, Brunner S
and Wagner E: DNA/polyethylenimine transfection particles:
Influence of ligands, polymer size, and PEGylation on
internalization and gene expression. AAPS PharmSci. 3:E212001.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kalyane D, Raval N, Maheshwari R, Tambe V,
Kalia K and Tekade RK: Employment of enhanced permeability and
retention effect (EPR): Nanoparticle-based precision tools for
targeting of therapeutic and diagnostic agent in cancer. Mater Sci
Eng C Mater Biol Appl. 98:1252–1276. 2019. View Article : Google Scholar
|
|
71
|
Kulkarni SA and Feng SS: Effects of
particle size and surface modification on cellular uptake and
biodistribution of polymeric nanoparticles for drug delivery. Pharm
Res. 30:2512–2522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Talekar M, Trivedi M, Shah P, Ouyang Q,
Oka A, Gandham S and Amiji MM: Combination wt-p53 and MicroRNA-125b
transfection in a genetically engineered lung cancer model using
dual CD44/EGFR-targeting nanoparticles. Mol Ther. 24:759–769. 2016.
View Article : Google Scholar :
|
|
73
|
Li Z, Zhang L, Tang C and Yin C:
Co-delivery of doxorubicin and survivin shRNA-expressing plasmid
via microenvironment-responsive dendritic mesoporous silica
nanoparticles for synergistic cancer therapy. Pharm Res.
34:2829–2841. 2017. View Article : Google Scholar
|
|
74
|
El-Boubbou K, Ali R, Al-Zahrani H,
Trivilegio T, Alanazi AH, Khan AL, Boudjelal M and AlKushi A:
Preparation of iron oxide mesoporous magnetic microparticles as
novel multidrug carriers for synergistic anticancer therapy and
deep tumor penetration. Sci Rep. 9:94812019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Behzadi S, Serpooshan V, Tao W, Hamaly MA,
Alkawareek MY, Dreaden EC, Brown D, Alkilany AM, Farokhzad OC and
Mahmoudi M: Cellular uptake of nanoparticles: Journey inside the
cell. Chem Soc Rev. 46:4218–4244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Blanc-Brude OP, Teissier E, Castier Y,
Lesèche G, Bijnens AP, Daemen M, Staels B, Mallat Z and Tedgui A:
IAP survivin regulates atherosclerotic macrophage survival.
Arterioscler Thromb Vasc Biol. 27:901–907. 2007. View Article : Google Scholar
|