Introduction
As the largest organ of the human body, the skin
provides protection against environmental injury and microbial
infection, and has sensory and metabolic functions (1). Injury caused to the skin can cause
both physical and mental damage to patients. An impaired skin wound
healing process may lead to infection, prolonged wound healing and
scarring (2). The classic stages
of wound repair include inflammation, new tissue formation and
remodeling (3). During the
proliferation phase, neoangiogenesis plays a crucial role (4).
Exosomes (Exos) from mesenchymal stem cells (MSCs)
originating from bone marrow, adipose tissue and the umbilical cord
have been demonstrated to accelerate cutaneous wound healing by
enhancing angiogenesis, re-epithelialization and collagen
deposition (5-7). Exos are nanovesicles of endocytic
origin secreted by the majority of cells in culture, with a
diameter of 30-150 nm; they are known as messengers that transmit
bioactive molecules, such as nucleic acids and proteins to the
recipient cells for cell-cell communications (8). They contain abundant cargoes which
participate in regeneration. From a material science perspective,
these nanocarriers with a particular composition of functional
molecules are natural carriers, featuring an extraordinarily
improved biocompatibility and bioavailability than any conventional
manufactured materials (9).
Exosomal proteins, such as Jagged 1, Ang-2 and DMBT1 actively
participate in therapeutic angiogenesis for tissue repair (10-12). However, the use of stem cells is
often limited as the source and harvesting of stem cells is highly
invasive. Thus, it may prove useful to search for a new parent cell
source from which it may be easier to obtain abundant Exos for
tissue repair.
Dental pulp stem cells (DPSCs) exhibit MSC-like
behavior, including immense potential for angiogenic effects
(13). These cells can be
harvested from extracted human third molars non-invasively, and are
an extremely accessible cell resource for the development of
therapeutic approaches. Although DPSCs are less abundant in tissue,
they are easy to culture and expand in vitro along with
their stemness (14,15). Indeed, accumulating evidence
suggests that DPSCs exhibit immense potential in central nervous
system repair, stroke recovery, diabetes treatment, muscle
regeneration and wound healing (16-18). Exos derived from DPSCs (DPSC-Exos)
have been reported to exhibit regenerative therapeutic potential
for applications in dental pulp tissue regeneration,
neuroprotection and craniofacial bone healing (19-21). Considering that Exos are critical
mediators of cell activity, it would be meaningful to explore
whether DPSC-Exos have the ability to accelerate the healing
process of cutaneous wounds.
The present study thus examined the effects of
DPSC-Exos on the angiogenesis of human umbilical vein endothelial
cells (HUVECs), which are related to cutaneous wound healing.
Tandem mass tag (TMT)-labeled quantitative proteomics analysis was
employed to identify the protein expression profiles in DPSC-Exos
compared with their parent cells. Gene Ontology (GO) annotation and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were used to evaluate biological functions and pathways
for the differentially expressed proteins in DPSC-Exos. The aims of
the present study were to clarify the following: i) The role of
DPSC-Exos in the cutaneous wound healing in vivo; ii) the
effects of DPSC-Exos on the angiogenic activities of HUVECs in
vitro; and iii) the potential signaling pathways involved in
the DPSC-Exo-induced angiogenesis of HUVECs.
Materials and methods
Cells and cell culture
The present study was approved by the Ethics
Committee of Sun Yat-sen University [No. ERC-(2017)-34]. Human
DPSCs were obtained from healthy pulp tissues harvested from wisdom
teeth without caries, which were extracted from 10 donors (both
male and female, 22-36 years old) between September, 2018 and
March, 2019 at the Hospital of Stomatology, Sun Yat-sen University,
Guangzhou, China. Dental pulp tissues were minced and digested for
the isolation of DPSCs as previously described (22). DPSCs were cultured in α-MEM with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin (MilliporeSigma). HUVECs were
purchased from AllCells, LLC and expanded in complete endothelial
cell growth medium (EGM; AllCells Biotech Shanghai Co., Ltd.). For
the exosome co-culture experiment, 10 µg/ml exosomes were
added to the culture medium of HUVECs. All cells were maintained at
37°C in an incubator contained 5% CO2.
Exosome isolation and identification
The DPSCs were washed with phosphate-buffered saline
(PBS; Biosharp) and incubated in fresh serum-free α-MEM 48 h prior
to the isolation of exosomes. The cell culture supernatant was then
harvested and subjected to two centrifugation steps (500 × g for 10
min at 25°C; 2,000 × g for 30 min at 25°C) to remove dead cells and
cell debris. Exosome pellets were purified from the cleared
supernatant by ultracentrifugation at 100,000 × g for 1 h at 4°C.
An exosome pellet from about 12×106 cells was
resuspended in 100 µl cold PBS. Exosomes were either used
immediately or stored at −80°C until use. The exosome protein
concentration was quantified using a BCA Protein Assay kit (Jinan
Bocai Chemical Technology Co., Ltd.). The expression levels of the
exosomal markers, CD9 and CD63 (Affinity Biosciences) were measured
using western blot analysis, as described below. Nanoparticle
tracking analysis (NTA) was performed to determine the size
distribution of exosomes.
Transmission electron microscopy
(TEM)
The morphology of the exosomes was visualized using
TEM. Exosomes suspended in PBS were placed onto
formvar/carbon-coated nickel grids and incubated for 30 min at room
temperature. Images of the grids were acquired using an H-7650
transmission electron microscope (Hitachi corporation).
Mouse skin wound model and treatment
All animal experiments were performed in accordance
with institutionally approved protocols for animal research (Animal
Care and Use Committee of Sun Yat-sen University) (No.
SYSU-IACUC-2022-000248). A total of 6 female C57BL/6 mice (8 weeks
old; weighing 18-20 g) were purchased from the Guangdong Medical
Laboratory Animal Center. Age-matched 8-week-old female mice from
the same background were used in the present study. During the
experiment, the mice were kept in a specific pathogen-free
environment (temperature, 22±2°C; humidity, 55±10%), with free
access to food and water. All mice were anesthetized with an
intraperitoneal injection of sodium pentobarbital (75 mg/kg) and
euthanized by cervical dislocation, while being deeply anesthetized
with sodium pentobarbital. Death verification included the absence
of heartbeat, breathing or respiration. In the animal experiments,
the humane endpoints was reached when the animal lost 20% of its
original weight according to predefined criteria (23,24). In the present study, the maximum
percentage of body weight loss observed was not >10%. After
shaving, a 1×1 cm full-thickness excision skin wound was created
with a pair of surgical scissors, on the backs of the mice. After
the surgery on day 1, the 6 mice were randomly divided into two
groups and were subcutaneously injected with either DPSC-Exos (120
µg per mouse) as the treatment group or an equal volume of
PBS (control group) around the wounds at four equally spaced
injection sites.
Wound healing evaluation
To observe the wound healing process, images of the
wounds were captured using a digital camera (iPhone 12, Apple) on
days 0, 3, 5, 7, 9, 12 and 14, and the wound healing rates were
calculated according to the following formula: Wound healing rate
(%)=(A0-An)/A0 ×100, where 'A0' represents the area of the initial
wound (t=day 0) and 'An' represents the residual area of the wound
on a the certain day (t=day n). The rates of wound closure were
quantified using ImageJ software (v.1.52a, National Institutes of
Health).
Hematoxylin and eosin (H&E) and
immunohistochemical staining
To examine the formation of new blood vessels, the
mice were sacrificed on day 14 post-wounding. The skin specimens
were harvested and photographed using a stereo-microscope (MZ10F,
Lecia Microsystems GmbH), and then analyzed using H&E and
immunohistochemical staining. The use of H&E was preferred for
viewing cellular and tissue structure details in the wound area.
Hematoxylin precisely stains nuclear components which helps to
identify inflammation, while eosin stains cytoplasmic components,
including collagen and red blood cells (25-27). Immunostaining for CD31 was
utilized to identify the endothelial cells of blood vessels in
order to quantify the newly formed blood vessels in the wound.
CD31, a 130-kDa glycoprotein expressed on endothelial cells, is
commonly used as a biomarker for detecting vascular endothelium
(28). The tissues were fixed in
4% paraformaldehyde solution, dehydrated with a series of graded
ethanol and embedded in paraffin. The sections (10-µm-thick)
were stained with H&E (Wuhan Servicebio Technology Co., Ltd.)
or angiogenesis-related protein CD31 (1:600, cat. no. GB11063-2,
Wuhan Servicebio Technology Co., Ltd.) to evaluate vascularization.
For H&E staining, the sections were stained with hematoxylin
for 3 min and eosin for 1 min at room temperature. For the
immunohistochemical staining of CD31, the sections were incubated
with first antibody at 4°C overnight, then followed by incubation
with the horseradish peroxidase-conjugated anti-rabbit IgG antibody
(1:200, cat. no. G1215-3, Wuhan Servicebio Technology Co., Ltd.)
for 1 h at room temperature. After rinsing with PBS, the
3,3'-diaminobenzidine (DAB) Staining kit (Wuhan Servicebio
Technology Co., Ltd.) was employed with hematoxylin counterstain.
The slides were observed and photographed under an epifluorescence
microscope (Zeiss AG). To compare the number of newly formed blood
vessels from the different groups, five random fields per section
near wound edges were counted using Image-Pro Plus 6 software
(Media Cybernetics, Inc.).
Exosome uptake assay
To visualize the endocytosis of exosomes by HUVECs,
exosomes were stained with PKH26 (MilliporeSigma) according to the
manufacturer's instructions. Isolated exosomes were resuspended in
250 µl diluent C, and 1 µl PKH26 was added and
immediately mixed by gentle pipetting. The staining was terminated
by the addition of an equal volume of exosome-free FBS following
incubation for 5 min at room temperature. The mixture was washed
twice with PBS by ultracentrifugation (100,000 × g for 1 h at 4°C)
and resuspended in 100 µl exosome-free culture medium. The
PKH26-labeled exosomes were then added to the HUVECs and incubated
for 24 h at 37°C. Following incubation with PKH26-labeled exosomes,
the cells were washed with PBS for three times and fixed with 4%
paraformaldehyde for 10 min at room temperature, the cell nuclei
were stained with 4,6-diamidino-2-phenylindole (DAPI) for 5 min at
room temperature. The images of fluorescence-labeled exosome
internalization were obtained using a confocal laser scanning
microscope (Zeiss AG).
Transwell migration assay
For the Transwell migration assay, Transwell inserts
with 8-µm pore filters (Corning, Inc.) were used. The HUVECs
(1×104 cells/well) were seeded onto upper chambers
containing 200 µl medium supplemented with DPSC-Exos,
SB203580 (10 µM; Selleck Chemicals), SB203580 (10 µM)
+ DPSC-Exos, or PBS. Concurrently, 500 µl cultured medium
were added to the bottom chamber and incubated at 37°C for 24 h.
The non-migrated cells in the upper chamber were gently removed
using a cotton swab and the lower chamber was fixed with 4%
paraformaldehyde for 10 min at room temperature, stained with 0.1%
crystal violet (MYM Technologies, Ltd.) for 30 min at room
temperature, washed with PBS twice, and imaged using an inversion
microscope (Zeiss AG). The quantity of migrated cells was counted
and analyzed using ImageJ software (v.1.52a, National Institutes of
Health).
Scratch wound assay
The HUVECs (2×105 cells/well) were plated
in 12-well plates and incubated at 37°C until reaching 90%
confluency. The assay was conducted in the absence of serum. A
micro-injury was scratched on the monolayer using a 200 µl
pipette tip and washed with PBS to remove the debris. The cells
were treated with DPSC-Exos, SB203580 (10 µM), SB203580 (10
µM) + DPSC-Exos, or PBS prior to obtaining microscopic
images at 0 h. The level of migration was measured by the ratio of
closure area to initial wound area (t=0 h) as follows: Migration
area (%)=(A0-A6)/A0 ×100, where 'A0' represents the area of initial
wound area and 'A6' represents the residual area of wound at the
metering point after 6 h (t=6 h). The wound area was quantified
using ImageJ software (v.1.52a, National Institutes of Health) to
calculate the percentage wound closure and the migration rate.
Cell proliferation assay
A Cell Counting Kit-8 (Dojindo Laboratories, Inc.)
was used to assess cell proliferation. Concisely, the HUVECs were
seeded in 96-well plates (1×103 cells/well) in EGM
supplemented with DPSC-Exos and SB203580 (10 µM). On days 1,
2, 3 and 4, CCK-8 solution was added to HUVECs (10 µl per
well) and the cells were incubated at 37°C for 1 h. The absorbance
was measured at 450 nm using a microplate reader (BioTek
Instruments, Inc.) and the optical density values represented the
proliferation level of the HUVECs.
Tube formation assay
The HUVECs (1×104 cells/well) were seeded
in a 96-well plate coated with 50 µl Matrigel (cat. No.
354277, Corning, Inc.) and treated with DPSC-Exos, SB203580 (10
µM), SB203580 (10 µM) + DPSC-Exos, or PBS,
respectively. After 6 h of culture incubation at 37°C, gels were
observed to examine the image of tube formation. The total tube
length and total loops were measured as the mean sum length and
loops of capillary-like structures per well using ImageJ software
(v.1.52a, National Institutes of Health).
Protein extraction for proteomics
analysis
The DPSCs seeded in 75 cm2 cell culture
flasks were incubated in serum-free α-MEM for 48 h at 37°C and then
washed with PBS prior to use. DPSC-Exos were isolated from the
culture medium of DPSCs, respectively. Both the DPSCs and DPSC-Exo
samples contained three biological replicates. TMT-based
quantitative proteomics analysis was conducted by Jingjie PTM
BioLab (Hangzhou) Co., Inc. All samples were sonicated on ice in
lysis buffer (8 M urea, 1% Protease Inhibitor Cocktail) three times
prior to centrifugation (12,000 × g for 10 min) at 4°C to remove
the remaining debris. Subsequently, the supernatant was collected.
A BCA protein assay (Beyotime Institute of Biotechnology) was
performed to determine the protein concentration.
Trypsin digestion and TMT labeling
The protein solution was reduced with 5 mM
dithiothreitol at 56°C for 30 min, alkylated with 11 mM
iodoacetamide (Millipore Sigma) in the dark at room temperature for
15 min, and then diluted with 100 mM tetraethyl ammonium bromide
(TEAB; Millipore Sigma) until the urea concentration was <2 M.
TEAB is a dissolution buffer for isobaric mass tag labeling
experiments (29,30). Trypsin was added to the solution
at 1:50 (w/w) trypsin-to-protein mass ratio and incubated at 37°C
overnight, and the trypsin-to-protein mass ratio was then changed
to 1:100 (w/w). Following trypsin digestion, the peptide was
desalted, vacuum-dried, reconstituted in 0.5 M TEAB, and processed
using a TMT kit (Thermo Fisher Scientific, Inc.).
High-performance liquid chromatography
(HPLC) fractionation and liquid chromatography-mass spectrometry
(LC-MS)/MS analysis
A Thermo Betasil C18 column (5 µm particles,
10 mm ID, 250 mm length; Thermo Fisher Scientific, Inc.) was
utilized to fractionate tryptic peptides into fractions using high
pH reverse-phase HPLC. In brief, peptides were initially separated
with a gradient of 8 to 32% acetonitrile (pH 9.0) over a period of
60 min into 60 fractions, and then combined into six fractions
before being dried by vacuum centrifugation (300 × g for 2.5 h at
25°C).
For LC-MS/MS analysis, the tryptic peptides were
dissolved in solvent A (0.1% formic acid) and separated using a
homemade reversed-phase analytical column (15-cm length, 75
µm i.d.). The gradient comprised an increase from 6 to 23%
solvent B (0.1% formic acid in 98% acetonitrile) over a period of
26 min, 23 to 35% in 8 min and climbing to 80% in 3 min then
holding at 80% for the last 3 min. All processes were performed on
an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific, Inc.) at a
constant flow rate of 400 nl/min. The peptides were subjected to
NSI source and analyzed using tandem mass spectrometry (MS/MS)
using the UPLC system coupled online to Q ExactiveTM Plus (Thermo
Fisher Scientific, Inc.). Intact peptides were detected in the
Orbitrap at 70,000 resolution and selected for MS/MS with the NCE
setting as 28, the fragments were then detected at a resolution of
17,500. MS and MS/MS spectra were acquired in a data-dependent
manner. The automatic gain control (AGC) was set at
5×104. An electrospray voltage of 2.0 kV was applied.
The scan range was set from 350 to 1,800 m/z for full scan and the
fixed first mass was set as 100 m/z.
Database search and bioinformatics
analysis
The acquired MS/MS data were processed using
Maxquant search engine (v.1.5.2.8, Max Planck Institute of
Biochemistry, Germany). Tandem mass spectra were searched against
the human UniProt database concatenated with reverse decoy
database. Trypsin/P was designated as the cleavage enzyme; up to
four missing cleavages were allowed. The mass tolerance for
precursor ions was set to 20 ppm (first search) and 5 ppm (main
search), and the mass tolerance for fragment ions to 0.02 Da,
respectively. The false discovery rate was adjusted to <1% and a
minimum score of 40 was set for the identification of modified
peptides.
GO annotation proteome was carried out using the
UniProt-GOA database (http://www.ebi.ac.uk/GOA/). Proteins were classified
by GO annotation into three categories: Biological Process,
Cellular Compartment and Molecular Function. InterProScan was used
for the functional annotation of identified protein domains based
on protein sequence alignment and the InterPro (http://www.ebi.ac.uk/interpro/) domain database.
For functional enrichment analysis, the KEGG, http://www.genome.jp/kaas-bin/kaas_main)
and GO (http://geneontology.org/) were
applied.
Determination and inhibition of the p38
mitogen-activated protein kinase (MAPK) signaling pathway
A pharmacological p38 MAPK inhibitor (SB203580;
Selleck Chemicals), was used to assess the participation of the
MAPK signaling pathway in the DPSC-Exo-induced effects on HUVECs.
The compound was resuspended to 10 mM in dimethyl sulfoxide (DMSO;
MP Biomedicals France) and used at 10 µM. For control
treatments (0 µM) 0.1% DMSO was used. For experiments
assessing the expression of target proteins using western blot
analysis, the HUVECs were plated in six-well plates and pre-treated
with 10 µM SB203580 for 6 h. Since SB203580 inhibited p38
MAPK catalytic activity instead of the phosphorylation of p38, the
effects of SB203580 on the p38 MAPK pathway were assessed by the
expression of hsp27 as a substrate (31).
To determine the participation of the MAPK signaling
pathway in the therapeutic effect of DPSC-Exos on cutaneous wound
healing in vivo, a total of 12 age-matched 8-week-old female
C57BL/6 mice were randomly divided into four groups as follows: PBS
+ vehicle, PBS + SB203580, DPSC-Exos + vehicle and DPSC-Exos +
SB203580 group. After the cutaneous wound had been formed for 6 h,
mice in the PBS/DPSC-Exos + SB203580 group were administered an
intraperitoneal injection of SB203580 (5 mg/kg). The SB203580 was
dissolved in the vehicle (4% DMSO + 30% PEG300 + 5% Tween-80 + 61%
ddH2O) according to the manufacturer's instructions. The
dose of SB203580 was selected from a previous study (32).
Western blot analysis
Total protein was extracted from either DPSCs or
HUVECs using RIPA buffer (Jinan Bocai Chemical Technology Co.,
Ltd.) and quantified using a BCA Protein Assay kit (Jinan Bocai
Chemical Technology Co., Ltd.). The protein samples (5 µg
per lane) were electrophoresed with 4-20% Bis-Tris sodium dodecyl
sulfate polyacrylamide gel and transferred onto a 0.2 µm
PVDF membrane (MilliporeSigma). After blocking with 5% (w/v)
non-fat milk for 1 h at room temperature, the membranes were
incubated with diluted primary antibodies overnight at 4°C. The
primary antibodies and concentrations used in the present study
were as follows: Phosphorylated (p-)p38 (1:1,000; cat. no. 4511,
Cell Signaling Technology, Inc.), p38 (1:1,000; cat. no. 8690, Cell
Signaling Technology, Inc.), p-hsp27 (1:1,000; cat. no. AF3080),
hsp27 (1:1,000; cat. no. AF6082), cell division control protein 42
(Cdc42; 1:1,000; cat. no. DF6322), β-tubulin (1:8,000; cat. no.
AF7011), FYVE, RhoGEF and PH domain containing 5 (FGD5; 1:500; cat.
no. DF13013), CD63 (1:1,000; cat. no. AF5117), CD9 (1:1,000; cat.
no. AF5139) (all from Affinity Biosciences) and β-actin (1:8,000;
cat. no. MA5-11869, Invitrogen; Thermo Fisher Scientific, Inc.).
Subsequently, the membranes were washed with Tris-buffered saline
with Tween-20 (TBST) and incubated with goat anti-rabbit IgG-HRP
(1:2,000; cat. no. As006, Asbio Technology, Inc.) as the secondary
antibody for 1 h at room temperature. Bands on the blots were
visualized using GeneGnome XRQ (Syngene). ImageJ software (v.1.52a,
National Institutes of Health) was utilized to quantify the density
of the bands.
Statistical analysis
All experiments were performed with at least three
replicates per group and each in vitro experiment was
repeated three times. All values are expressed as the mean ±
standard deviation (SD). Differences between two groups were
evaluated using an unpaired Student's t-test and those between more
than two groups were compared using one-way ANOVA with a Bonferroni
post hoc test using SPSS 25.0 software (IBM Corp.). P<0.05 was
considered to indicate a statistically significant difference.
Results
Identification of DPSC-Exos
DPSC-Exos were characterized using TEM, western blot
analysis and NTA. TEM images revealed that the vesicles isolated
from the culture supernatant of the DPSCs exhibited a spherical
morphology (Fig. S1A). Western
blot analysis demonstrated that these vesicles were positive for
the exosomal surface markers, CD9 and CD63 (Fig. S1B). The results of NTA revealed
that the diameters of these nanoparticles ranged from 50-100 nm
(Fig. S1C). Taken together,
these results confirmed their identity as exosomes.
DPSC-Exos promote cutaneous wound healing
in mice by promoting angiogenesis
The present study investigated the effects of
DPSC-Exos on cutaneous wound healing using a full-thickness wound
model on mice. A significantly more rapid wound closure rate was
observed when DPSC-Exos were injected (Fig. 1A and B). Skin images from the
underside of the wounds, which were collected 14 days post-wounding
demonstrated that the mice treated with the DPSC-Exos exhibited a
greater amount of newly formed blood vessels compared to the
control group (Fig. 1C).
Furthermore, H&E and immunohistochemical staining was performed
at the wound site. A considerable amount of capillary-structure
blood vessels (Fig. 1D and E) and
a higher expression of CD31 in the DPSC-Exo-treated group (Fig. 1F and G) confirmed that the
DPSC-Exos enhanced local angiogenesis. These findings suggested
that the DPSC-Exos accelerated wound healing in vivo by
promoting angiogenesis.
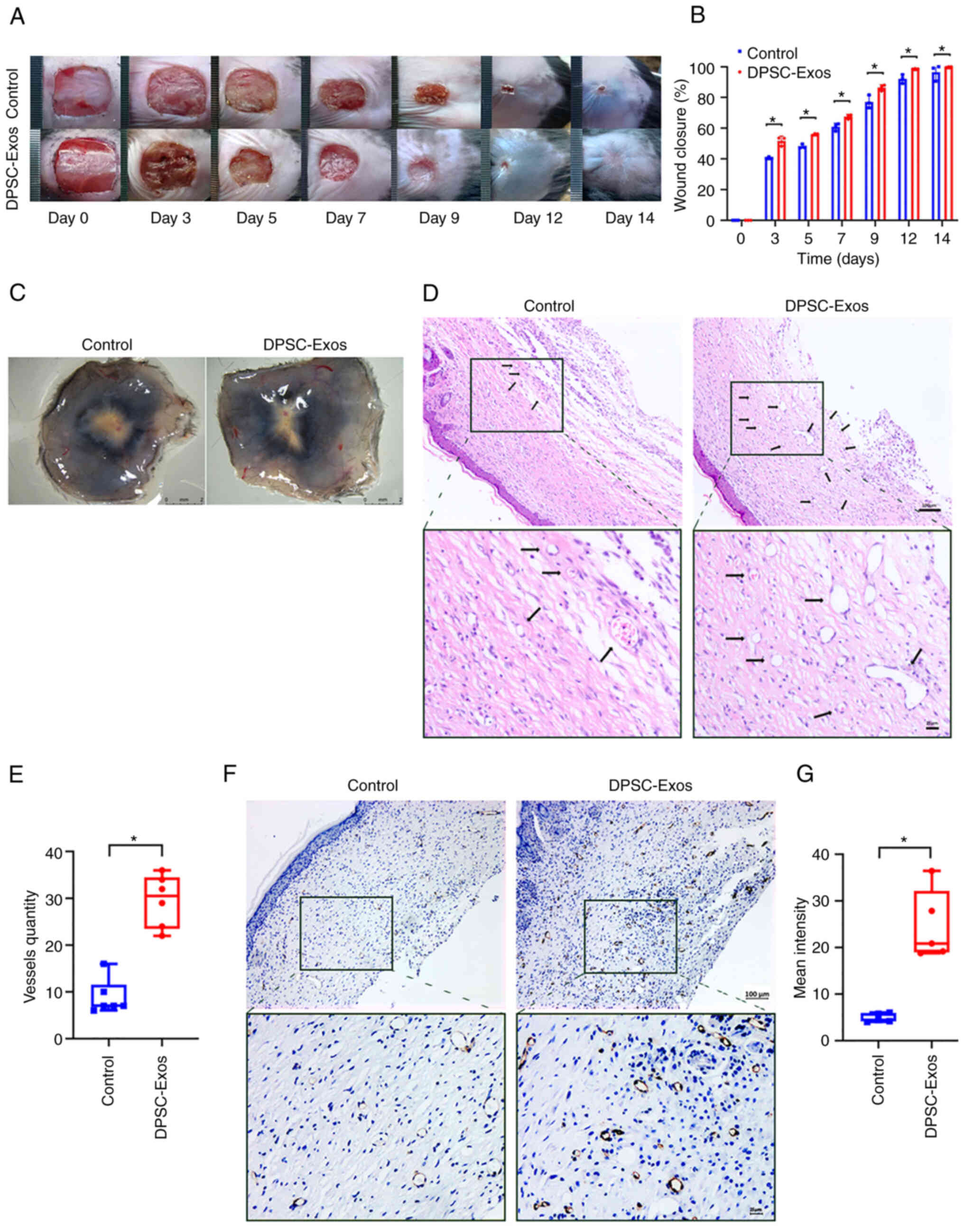 | Figure 1DPSC-Exos accelerate cutaneous wound
healing in mice by promoting angiogenesis. (A) Gross view and
quantification of wound area of mice treated with PBS and DPSC-Exos
on days 3, 5, 7, 9, 12 and 14 post-wounding. Scale bar, 1 cm. (B)
The rate of wound closure in wounds receiving DPSC-Exos treatments
was significantly higher at the indicated time points (n=3). (C)
Gross view of wounds of mice treated with PBS and DPSC-Exos on day
14 post-wounding from the undersurface. More newly formed blood
vessels were detected in the wound sites of the DPSC-Exo-treated
group. Scale bar, 2 mm. (D and E) Hematoxylin & eosin staining
of the wound sections treated with PBS and DPSC-Exos on day 14
after the operation. The black arrows indicate newly formed blood
vessels. The vessel quantity in the DPSC-Exo-treated group was
larger than that of the control group (n=3). Scale bar, 100
µm. (F and G) Immunohistochemical staining for CD31 in wound
sections of mice treated with PBS and DPSC-Exos at 14 days after
the operation. A higher expression of CD31 was found in the
DPSC-Exo-treated group (n=3). Scale bar, 100 µm.
*P<0.05. DPSC, dental pulp stem cell; Exo, exosome;
PBS, phosphate-buffered saline. |
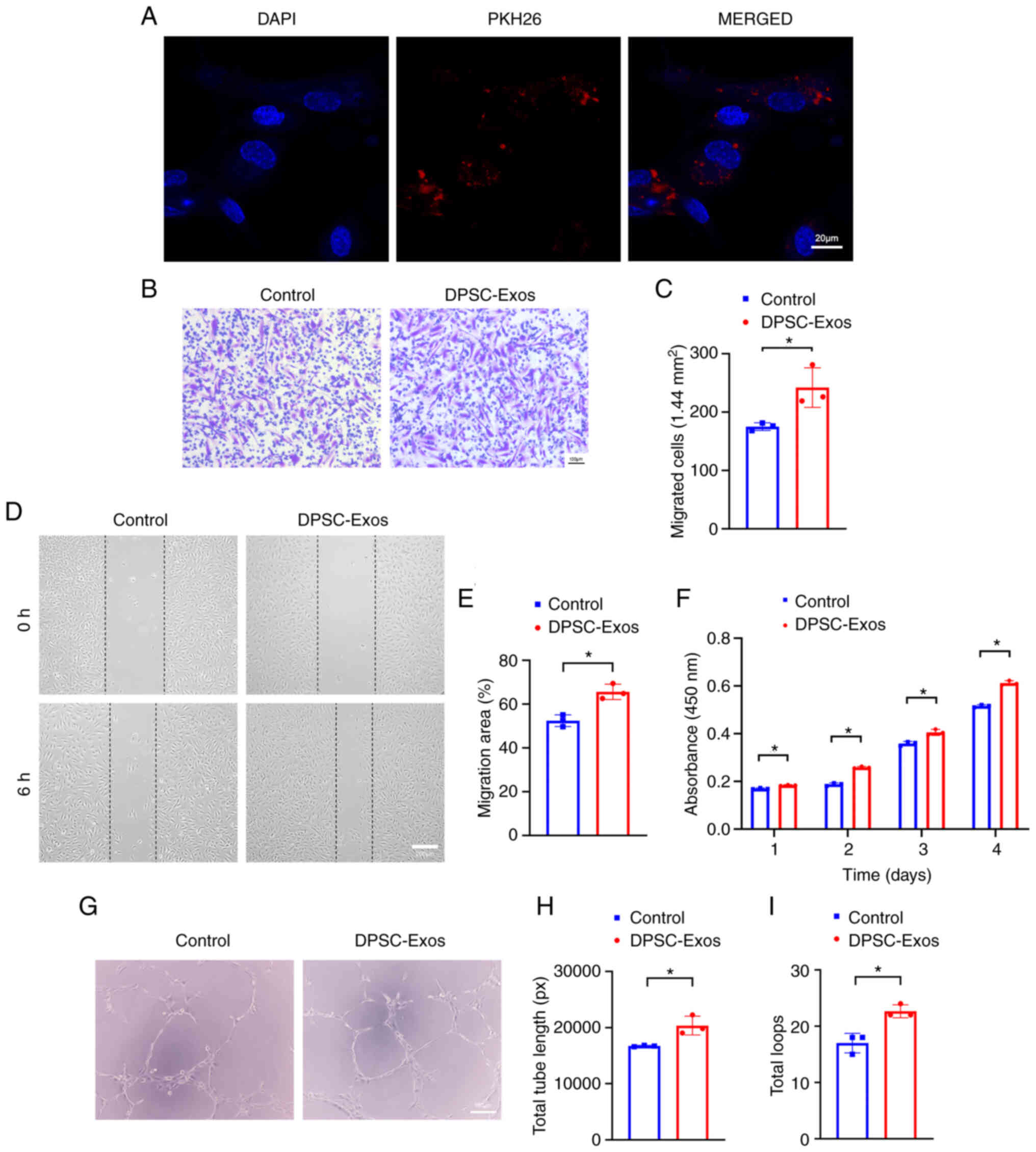
DPSC-Exos enhance the angiogenic
activities of HUVECs
To examine the effects of DPSC-Exos on HUVECs
behavior in vitro, the internalization of exosomes into
these cells was first monitored. Following 24 h of incubation with
PKH26-labeled DPSC-Exos, fluorescence microscopy analysis revealed
red fluorescent spots in the cytoplasm around the nucleus of
HUVECs, which indicated that the HUVECs were able to take up
DPSC-Exos (Fig. 2A). For
functional analysis, the present study then assessed how DPSC-Exos
modulated HUVEC angiogenesis, including migration, proliferation
and tube formation. Transwell migration assay (Fig. 2B and C) and scratch wound assay
revealed that the DPSC-Exos significantly increased the motility of
HUVECs (Fig. 2D and E). CCK-8
assay was utilized to examine the proliferation of the cells within
4 days of culture time, which revealed that the DPSC-Exos increased
the proliferative capacity of the HUVECs (Fig. 2F). DPSC-Exo treatment also
improved tube formation in the HUVECs as the total tube length and
total loops increased in the DPSC-Exo-treated group (Fig. 2G-I). On the whole, these results
indicated that the DPSC-Exos enhanced the migration, proliferation
and tube formation ability of HUVECs.
Proteomics analysis of DPSCs and
DPSC-Exos
Since the mechanisms involved in the promoting
effects of DPSC-Exos on angiogenesis remained unknown, the present
study employed TMT-labeled quantitative proteomics analysis to
identify the protein expression profiles in DPSC-Exos compared with
their parent cells. All proteins and peptides identified and
quantified using LC-MS/MS in DPSC-Exos and DPSCs are presented in
Table SI. A total of 4,592
proteins were quantified, including 1,159 upregulated and 460
downregulated proteins (Fig. 3A).
In addition, a volcano plot illustrating the statistically sizable
difference of expressed proteins between DPSC-Exos and DPSCs was
constructed (Fig. 3B). A change
in differential expression levels exceeding 1.5 was regarded as the
change threshold for significance.
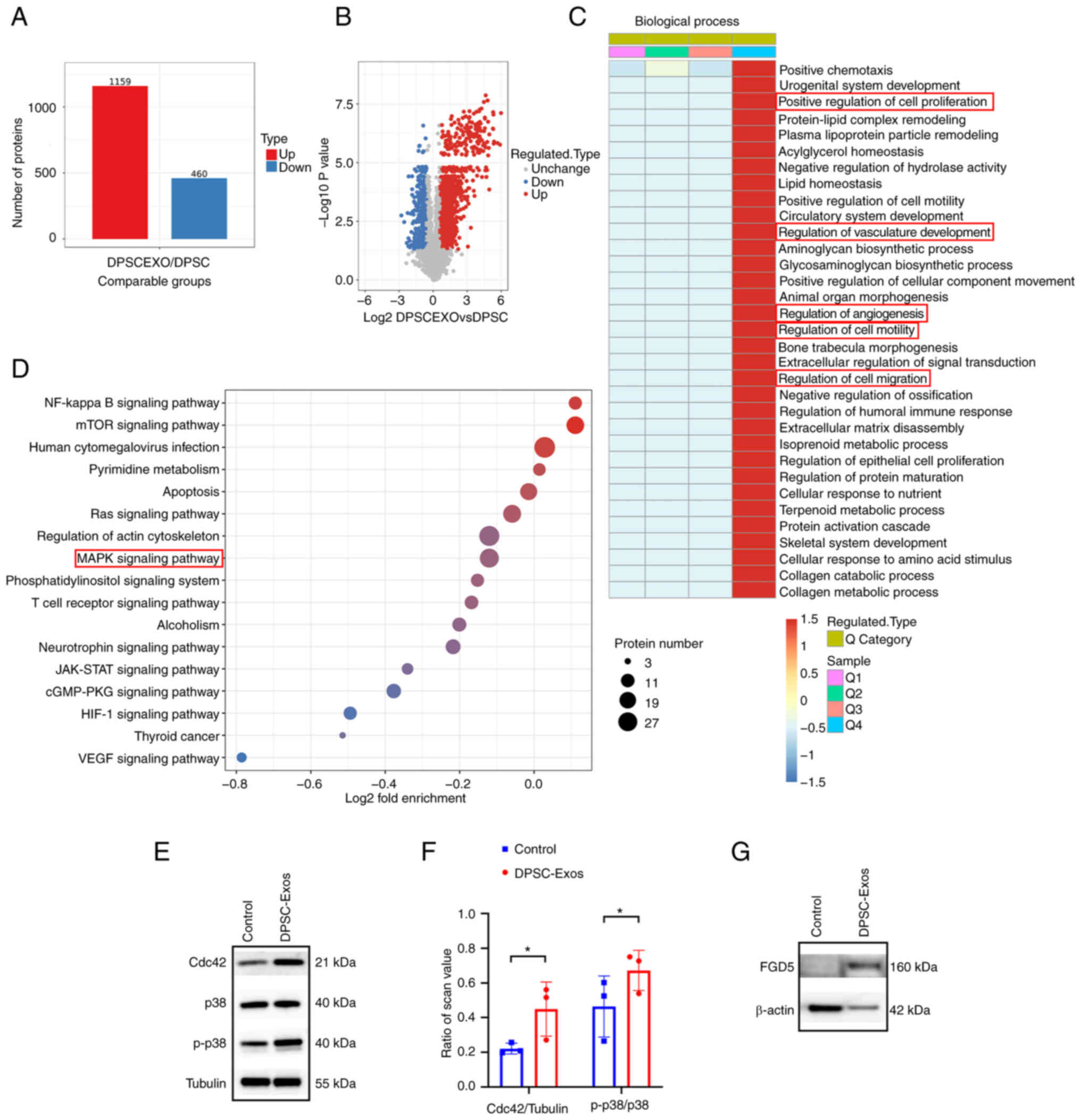 | Figure 3Expression and function of the
proteins identified by TMT-labeled quantitative proteomic in
DPSC-Exos. (A) The number of differentially expressed proteins in
DPSC-Exos compared with DPSCs was identified using TMT-labeled
quantitative proteomics analysis. (B) Volcano map of differentially
expressed proteins. (C) Differentially expressed proteins were
categorized according to the biological process (Gene Ontology
term) they were involved in; several of these were related to wound
healing. (D) Kyoto Encyclopedia of Genes and Genomes pathway
analysis of DPSC-Exos revealed that the enriched proteins involved
in multiple signal transductions, including the MAPK signaling
pathway. (E and F) The expression of Cdc42, p38 and p-p38 in HUVECs
was higher following treatment with DPSC-Exos (n=3).
*P<0.05. (G) Western blot analysis of pro-angiogenic
protein (FGD5) and cytosolic marker (β-actin) expression from DPSC
and DPSC-Exos. TMT, tandem mass tags; DPSC, dental pulp stem cell;
Exo, exosome; p-, phosphorylated; MAPK, mitogen-activated protein
kinase; HUVEC, human umbilical vein endothelial cell; Cdc42, cell
division control protein 42; FGD5, FYVE, RhoGEF and PH domain
containing 5. |
To thoroughly determine the proteins obtained in
these data, GO analysis and KEGG pathway analysis were performed to
classify the biological functions and their characteristics, as
well as their molecular interaction networks. The results of GO
classification analysis revealed that proteins involved in various
biological processes in relation to wound healing, such as positive
regulation of cell motility, migration, proliferation, vasculature
development and angiogenesis, were markedly increased in the
DPSC-Exos (Fig. 3C). KEGG pathway
analysis revealed that proteins contained in DPSC-Exos were
involved in several pathways, including the MAPK signaling pathway
(Fig. 3D and Table SII). As shown in Fig. 3E, tubulin was used as an internal
control to normalize the western blot analysis data. The ratios of
Cdc42, p38 and p-p38 protein levels to tubulin expression were used
for comparisons. HUVECs were treated with DPSC-Exos and a
significant increase was found in the expression of Cdc42 and p-p38
(Fig. 3E and F). Cdc42 is
required for the activation of p38 and plays a key role in cell
migration and proliferation (33,34). Taken together, these findings
prompted the hypothesis that the Cdc42/p38 MAPK signaling pathway
may play an essential role in DPSC-Exo-induced angiogenesis.
Of note, the proteomics data indicated that the
expression of FGD5, a pro-angiogenesis protein, was 2.299-fold
higher in the DPSC-Exos than in the DPSCs (Table SIII). FGD5 is known to mediate
pro-angiogenic action of vascular endothelial growth factor (VEGF)
in human vascular endothelial cells by inducing the activation of
Cdc42 (35). In the present
study, as shown in Fig. 3G,
β-actin was used as an internal control to normalize the western
blot analysis data. The ratios of the FGD5 protein level to β-actin
expression were used for comparisons. Western blot analysis also
verified that FGD5 was enriched in the DPSC-Exos (Fig. 3G), which may be a potential
candidate mediating pro-angiogenic function of DPSC-Exos.
Inhibition of the p38 MAPK signaling
pathway decreases DPSC-Exo-induced angiogenesis
To examine whether the p38 MAPK signaling pathway is
involved in DPSC-Exo-induced angiogenesis, HUVECs were treated with
SB203580 to inhibit the p38 MAPK pathway prior to stimulation with
the DPSC-Exos. hsp27 is a terminal substrate of the p38 MAPK
pathway (36,37), which is related to the positive
regulation of angiogenesis, as well as the positive regulation of
blood vessel endothelial cell migration (38). In the present study, as shown in
Fig. 4A, tubulin was used as an
internal control to normalize the western blot analysis data. The
ratios of p38, p-p38, hsp27, p-hsp27 protein levels to tubulin
expression were used for comparisons. Once the p38 MAPK signaling
pathway was inhibited by SB203580, the protein levels of hsp27 and
p-hsp27 were significantly decreased in the HUVECs (Fig. 4A). The ratio of p-p38/p-38 and
p-hsp27/hsp27 was also shown (Fig.
4B). Transwell migration assay (Fig. 4C and D) and scratch wound assay
(Fig. 4E and F) revealed a
decreased capacity of the DPSC-Exos to promote cell migration when
the HUVECs were pre-treated with SB203580. As shown by CCK-8 assay,
the proliferation of the HUVECs which was promoted by DPSC-Exos was
also suppressed by the inhibition of p38 by SB203580 (Fig. 4G). In addition, as shown by the
results of tube formation assay, the enhanced tube formation
ability of the HUVECs stimulated with the DPSC-Exos was reduced by
SB203580 (Fig. 4H-J). These
outcomes all confirmed that the p38 MAPK signaling pathway was
involved in DPSC-Exo-induced angiogenesis.
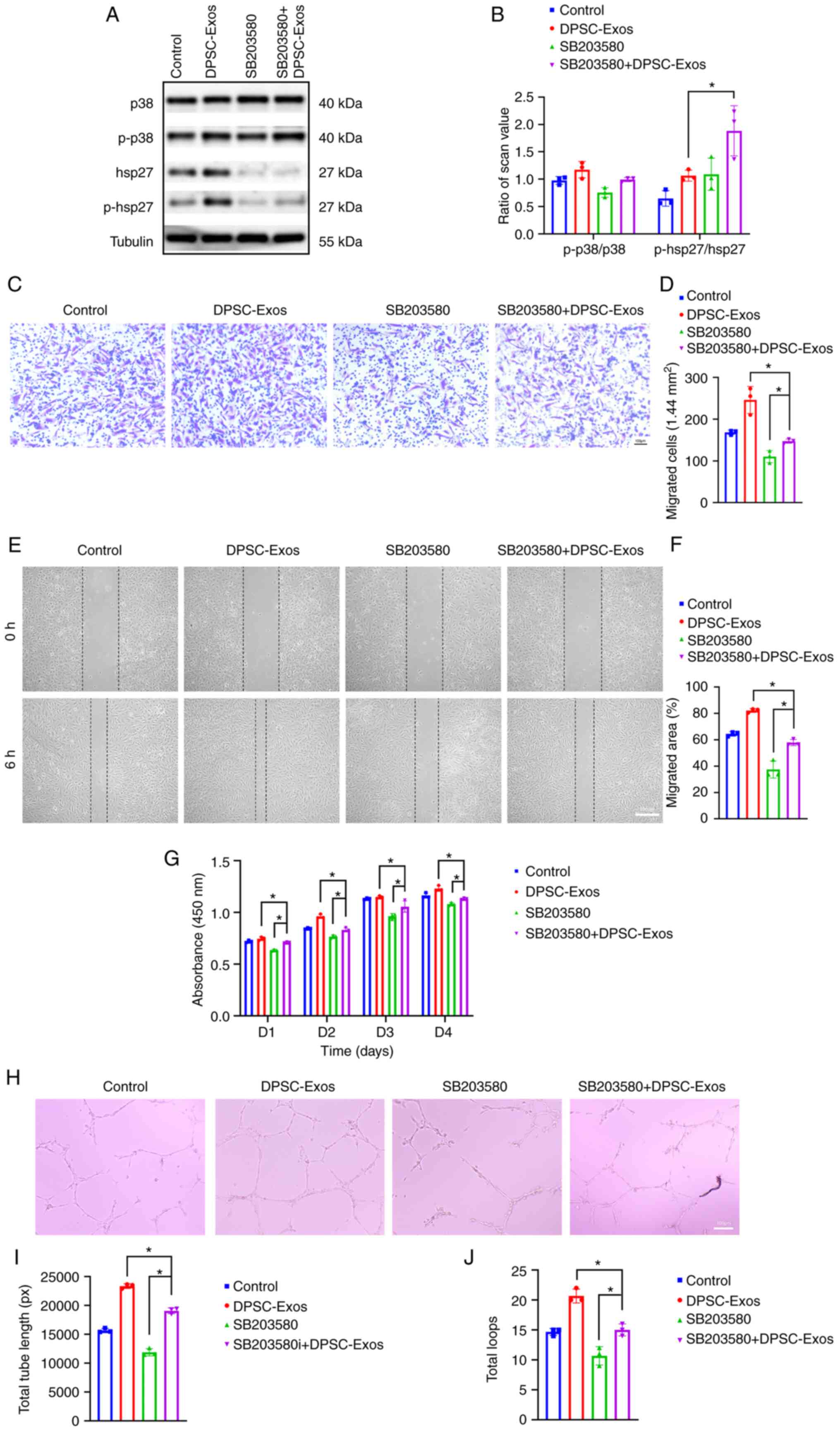 | Figure 4DPSC-Exos promote angiogenesis via
p38 MAPK to promote wound healing. (A and B) HUVECs were treated
with DPSC-Exos in the presence or absence of SB203580. The
expression of p38, p-p38, hsp27 and p-hsp27 was detected using
western blot analysis (n=3). (C and D) The migration of HUVECs
stimulated by DPSC-Exos or an equal volume of PBS in the presence
or absence of SB203580 (n=3). Scale bar, 100 µm. (E and F)
Representative images of scratch wound assay in HUVECs treated with
DPSC-Exos or PBS in the presence or absence of SB203580 (n=3).
Scale bar, 100 µm. (G) The proliferation of HUVECs receiving
different treatments were examined using CCK-8 assay (n=3). (H-J)
Representative images of the tube formation assay on Matrigel in
HUVECs treated with DPSC-Exos or PBS (n=3). Scale bar, 100
µm. *P<0.05. DPSC, dental pulp stem cell; Exo,
exosome; p-, phosphorylated; MAPK, mitogen-activated protein
kinase; HUVEC, human umbilical vein endothelial cell; hsp27, heat
shock protein 27; PBS, phosphate-buffered saline. |
Inhibition of the p38 MAPK signaling
pathway attenuates the therapeutic and pro-angiogenic effects of
DPSC-Exos
The present study also examined the effects of p38
inhibition on the healing process of murine cutaneous wounds. The
subcutaneous injection of DPSC-Exos accelerated the cutaneous wound
closure rate, while this therapeutic effect was attenuated after
the p38 MAPK signaling pathway was blocked by SB203580 (Fig. 5A and B). the wound closure rate in
all groups reached almost 100% on day 14; thus, a statistically
significant difference was not observed at this time point. On the
other hand, wounds in the DPSC-Exos + vehicle group were almost
completely closed on day 12, which exhibited a much more rapid
healing rate than other three groups. Skin images from the
underside of the wounds were collected 14 days post-wounding.
Compared with the DPSC-Exos + vehicle group, treatment with
SB203580 attenuated the development of newly formed blood vessels,
as shown in the DPSC-Exos + SB203580 group (Fig. 5C). The quantification of newly
formed blood vessels was performed using H&E and
immunohistochemical staining. With the presence of SB203580, the
amount of capillary-structure blood vessels (Fig. 5D and E) and the expression of CD31
(Fig. 5F and G) were suppressed
in the mice treated with either PBS or DPSC-Exos. The formed
vessels and positive rate of CD31 were decreased in the DPSC-Exos +
SB203580 group as compared to the DPSC-Exos + vehicle group.
Accordingly, these results substantiated that DPSC-Exo-induced
angiogenesis was attenuated by the inhibition of the p38 MAPK
signaling pathway in vivo.
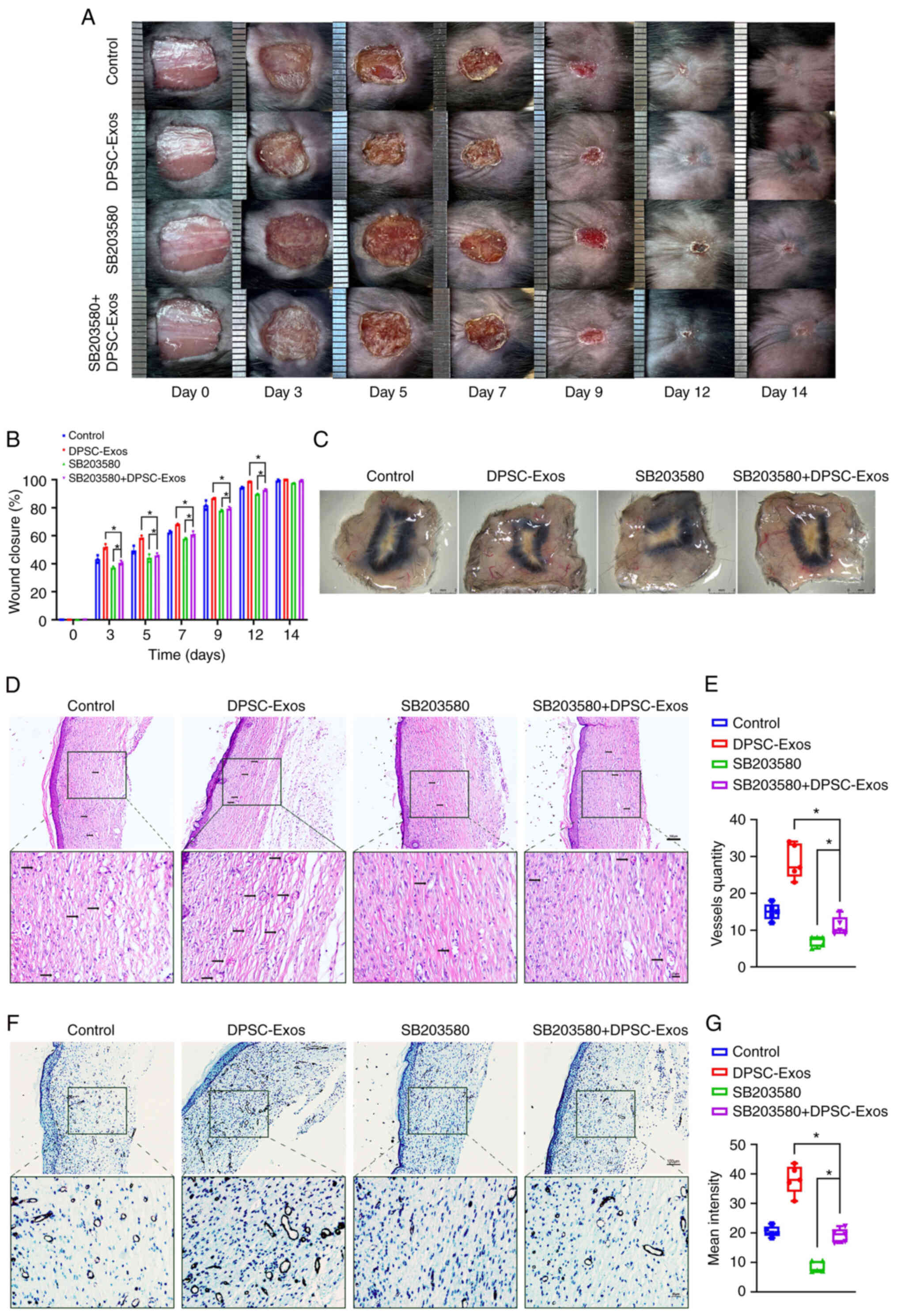 | Figure 5DPSC-Exos accelerate cutaneous wound
healing in mice via p38 MAPK. (A) Gross view and quantification of
the wound area of mice treated with DPSC-Exos in the presence or
absence of SB203580 on days 3, 5, 7, 9, 12 and 14 post-wounding.
Scale bar, 1 cm. (B) The rate of wound closure in wounds receiving
DPSC-Exos or PBS treatments in the presence or absence of SB203580
at the indicated time points (n=3). (C) Gross view of wounds
treated with PBS and DPSC-Exos in the presence or absence of
SB203580 on day 14 post-wounding from the undersurface. Scale bar,
2 mm. (D and E) Hematoxylin and eosin staining of wound sections
treated with PBS and DPSC-Exos in the presence or absence of
SB203580 at 14 days after the operation (n=3). The black arrows
indicate newly formed blood vessels. Scale bar, 100 µm. (F
and G) Immunohistochemical staining for CD31 in wound sections
treated with PBS and DPSC-Exos in the presence or absence of
SB203580 at 14 days after the operation (n=3). Scale bar, 100
µm. *P<0.05. DPSC, dental pulp stem cell; Exo,
exosome; MAPK, mitogen-activated protein kinase; PBS,
phosphate-buffered saline. |
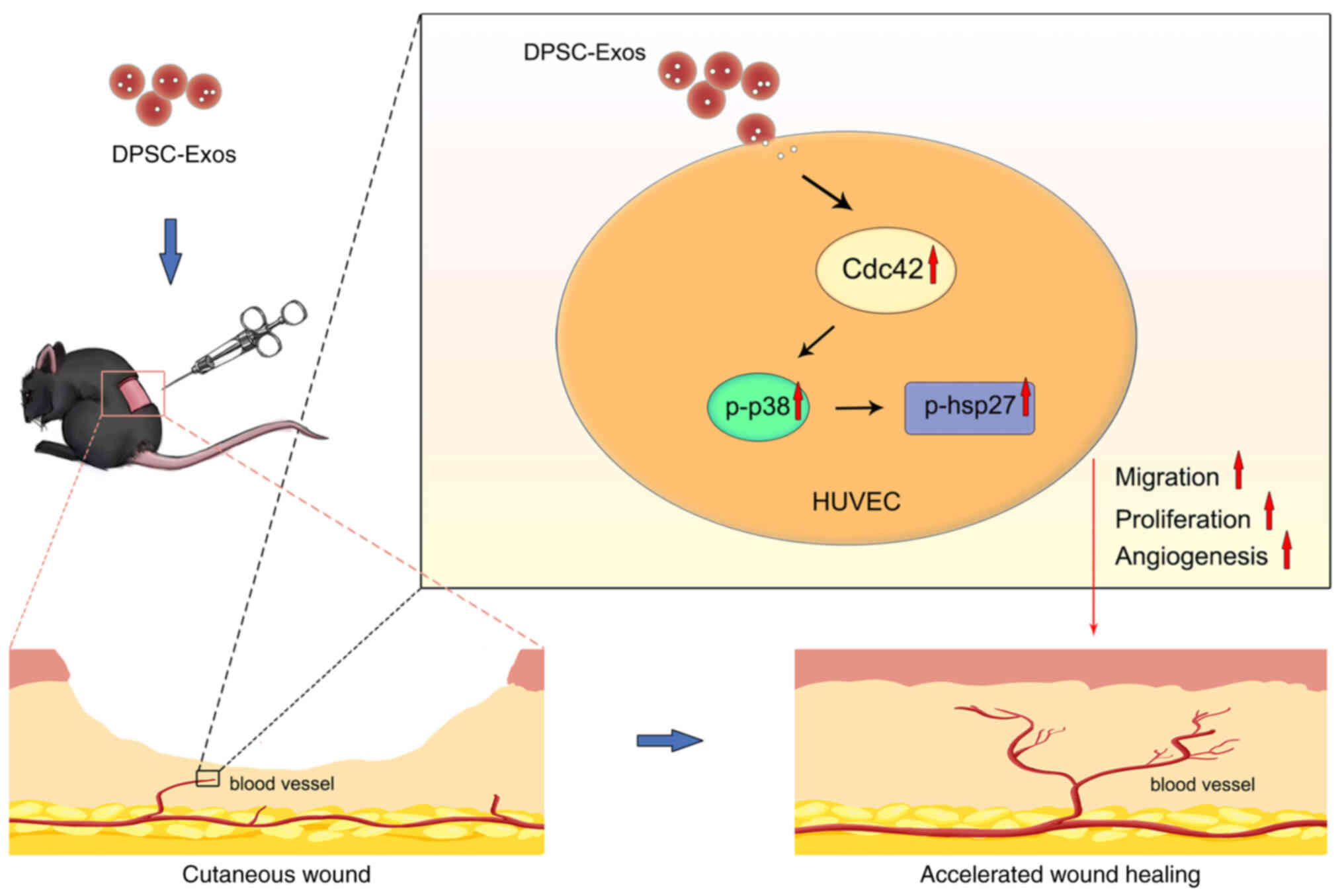
Discussion
The present study, to the best of our knowledge, is
the first to demonstrate that DPSC-Exos could effectively enhanced
the functional properties of HUVECs and accelerated cutaneous wound
healing in mice. In the process of angiogenesis induced by
DPSC-Exos, the Cdc42/p38 MAPK pathway played a crucial role, as the
pro-angiogenic effects of DPSC-Exos were attenuated by the
inhibition of the p38 MAPK pathway (Fig. 6).
In the present study, it was found that DPSC-Exos
accelerated cutaneous wound healing by enhancing the development of
new blood vessels in the wound site. As all mice used were female,
and estrogen levels can affect the rate of wound healing, more care
should be taken in the selection of female animals for future
studies, as the levels of estrogens and progesterone cab affect
rate of wound healing (39). The
results of the present study are unlikely to be affected, since the
animal groups were created by chance and the likelihood of the
effects of the estrous cycle on outcomes would have averaged out.
As has been reported, DPSC-Exos exhibit great potential in bone
regeneration, neuroprotection and anti-inflammation (40-42). In a previous study by the authors,
DPSC-Exos were found to promote odontogenic differentiation via the
TGFβ1/smad signaling pathway by downregulating latent TGF-β-binding
protein 1 (LTBP1) (22). The
present study demonstrated that the injection of DPSC-Exos
triggered angiogenesis by evaluating the vascular marker, CD31, in
the wound site. Angiogenesis at the wound site is a critical
determinant of wound healing processes, since neovascularization
ensures oxygen and nutrition delivery, thus establishing a
favorable environment for wound healing. The stimulation of new
blood vessel development is a helpful therapeutic target for tissue
regeneration.
Recently, exosomes from hypoxia-preconditioned human
adipose MSCs have been found to carry >30 types of
angiogenesis-related proteins and improve neovascularization around
the graft tissue (43). Exosomes
from MSCs derived from the human umbilical cord, expressing α2M,
TLN1, ANK1 and other proteins, are involved in damage repairing
(44). In the present study,
proteomics analysis revealed that 1,619 differentially expressed
proteins were detected in DPSC-Exos. This suggested that there may
be potential functional proteins among these, which are involved in
different processes of wound healing, and are thus worthy of
further exploration.
GO term enrichment analysis revealed that the
DPSC-Exos were enriched in the proteins that are involved in the
regulation of wound healing-related biological processes, such as
the positive regulation of cell motility, migration, proliferation,
vasculature development and angiogenesis. Additionally, several
in vitro experiments were conducted to demonstrate that
DPSC-Exos effectively enhanced the migration, proliferation and
tube formation ability of HUVECs. New blood vessel formation, which
involves endothelial cell proliferation, migration and branching to
form capillaries, requires a dynamic regulated interaction between
endothelial cells, angiogenesis factors and the surrounding
extracellular matrix (45). It is
known that some exosomes can regulate the angiogenic function of
recipient endothelial cells by transferring exosomal proteins, RNAs
and miRNAs into the cytoplasm of these cells (46). Such exosomes represent a highly
attractive delivery vehicle for any proteins or RNAs through which
they can exert their therapeutic effects.
Previous studies have demonstrated that MSC-Exos can
facilitate wound repair; however, some of the underlying mechanisms
remain unclear (44,47,48). The present study focused on
identifying the underlying mechanisms through which the infusion of
DPSC-Exos initiates endothelial cell-mediated wound healing. KEGG
pathway analysis revealed that exosomal proteins were enriched in
several pathways, including the MAPK pathway. Western blot analysis
confirmed that the DPSC-Exos induced significant increases in the
protein levels of Cdc42 and the phosphorylation of p38 in HUVECs.
Cdc42 is a Rho-family GTPase regulating actin dynamics and cell
proliferation (49). Moreover, it
is an important activator of the p38 MAPK pathway (50). The sequential activation of the
Cdc42/p38 MAPK signaling pathway is essential for VEGF-induced
actin reorganization in HUVECs (51,52). Previous studies have indicated
that p38 MAPK activation mediates the angiogenesis of endothelial
cells by modulating cell migration and proliferation (36,53). Thus, it was hypothesized that
DPSC-Exos increased the angiogenic activities of HUVECs through the
p38 MAPK pathway. It was found that the stimulation of DPSC-Exos
increased the angiogenic activities of HUVECs, whereas these
effects were attenuated with the inhibition of the p38 MAPK
signaling pathway. Consistent with the phenomenon observed in
vitro, the in vivo experiment with the SB203580
inhibitor illustrated that blocking the p38 MAPK signaling pathway
diminished DPSC-Exo-induced angiogenesis in the wound site.
SB203580 was used at 10 µM in in vitro experiments,
and for the in vivo experiments, SB203580 at 5 mg/kg
dissolved in the vehicle was used according to the manufacturer's
instructions, which has been proven to be non-toxic (32,54,55). Taken together, the findings
demonstrated that DPSC-Exos increased the migration, proliferation
and capillary formation capacity of endothelial cells via the
Cdc42/p38 MAPK signaling pathway, thereby enhancing cutaneous wound
healing in mice.
It has been reported that exosomal proteins can
activate the p38 MAPK pathway and promote angiogenesis (56). Moreover, the AKT/mTOR, JAK2/STAT3
and PKA signaling pathways are associated with MSC-exo-induced
angiogenesis (57-59). The data of the present study
confirmed that the positive effects of DPSC-Exos on angiogenesis
were not entirely abolished by the inhibition of the p38 MAPK
signaling pathway both in vitro and in vivo, which
suggested that other pathways are involved in the regulation of
neovascularization.
Furthermore, the present study sought to determine
the key component that participates in the modulation of
DPSC-Exo-stimulated angiogenic activities of HUVECs. It was found
that the expression of FGD5, a pro-angiogenic protein, was markedly
higher in DPSC-Exos. FGD5, known to be conducive to
pro-angiogenesis processes in endothelial cells, is a Rho
guanine-nucleotide exchange factor (Rho GEF) which catalyzes the
exchange of GDP for GTP and leads to the activation of its target
Rho protein Cdc42 (60). A
previous study demonstrated that FGD5 regulates Cdc42 activity and
plays a key role in the mediation of the proangiogenic action of
VEGF (35). The FGD5-mediated
activation of Cdc42 in endothelial cells can protect VEGFR2 from
degradation and regulates cytoskeletal dynamics (61). Thus, based on such mechanisms
provided by previous research, it was hypothesized that exosomal
FGD5 from DPSC-Exos may accelerate cutaneous wound healing by
enhancing angiogenesis through the Cdc42/p38 MAPK signaling
pathway. To investigate the molecular mechanisms of FGD5 from
DPSC-Exos on the angiogenesis of HUVECs, future studies are
required.
In conclusion, the present demonstrated that
DPSC-Exos promoted the angiogenic properties of endothelial cells
via theCdc42/p38 MAPK signaling pathway, thereby enhancing
cutaneous wound healing in mice. DPSC-Exo based therapy may
possibly represent a useful tool in the field of soft tissue
regeneration. However, further studies are required before DPSC-Exo
can be used in clinical practice.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XH and JZ designed the study. ZZ and JZ performed
the experiments and collected the data. DL, RX and YC assisted with
the experiments. ZZ analyzed the data and prepared the manuscript.
XH and JZ revised the manuscript. All authors have read and
approved the manuscript. XH and JZ confirm the authenticity of all
the raw data.
Ethic approval and consent to
participate
The present study was approved by the Ethics
Committee of Sun Yat-sen University [No. ERC-(2017)-34]. Animal
experiments followed the Ethical Guidelines for Laboratory Animal
Welfare determined by the Institutional Animal Care and Use
Committee, Sun Yat-Sen University (no. SYSU-IACUC-2022-000248).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 11772361 and 81700950), and the
Guangdong Basic and Applied Basic Research Foundation (grant nos.
2022A1515011266 and 2022A1515012531).
Abbreviations:
|
DPSCs
|
dental pulp stem cells
|
|
HUVECs
|
human umbilical vein endothelial
cells
|
|
MSCs
|
mesenchymal stem cells
|
|
Exos
|
exosomes
|
|
TMT
|
tandem mass tags
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
FBS
|
fetal bovine serum
|
|
PBS
|
phosphate-buffered saline
|
|
H&E
|
hematoxylin and eosin
|
|
Cdc42
|
cell division control protein 42
|
|
MAPK
|
mitogen-activated protein kinase
|
|
FGD5
|
FYVE, RhoGEF and PH domain containing
5
|
|
VEGF
|
vascular endothelial growth factor
|
|
NTA
|
nanoparticle tracking analysis
|
|
TEM
|
transmission electron microscopy
|
References
|
1
|
Strecker-McGraw MK, Jones TR and Baer DG:
Soft tissue wounds and principles of healing. Emerg Med Clin North
Am. 25:1–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rodrigues M, Kosaric N, Bonham CA and
Gurtner GC: Wound healing: A cellular perspective. Physiol Rev.
99:665–706. 2019. View Article : Google Scholar :
|
|
3
|
Gurtner GC, Werner S, Barrandon Y and
Longaker MT: Wound repair and regeneration. Nature. 453:314–321.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li J, Zhang YP and Kirsner RS:
Angiogenesis in wound repair: Angiogenic growth factors and the
extracellular matrix. Microsc Res Tech. 60:107–114. 2003.
View Article : Google Scholar
|
|
5
|
Ding J, Wang X, Chen B, Zhang J and Xu J:
Exosomes derived from human bone marrow mesenchymal stem cells
stimulated by deferoxamine accelerate cutaneous wound healing by
promoting angiogenesis. Biomed Res Int. 2019:97427652019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li X, Xie X, Lian W, Shi R, Han S, Zhang
H, Lu L and Li M: Exosomes from adipose-derived stem cells
overexpressing Nrf2 accelerate cutaneous wound healing by promoting
vascularization in a diabetic foot ulcer rat model. Exp Mol Med.
50:1–14. 2018.
|
|
7
|
Hu Y, Rao SS, Wang ZX, Cao J, Tan YJ, Luo
J, Li HM, Zhang WS, Chen CY and Xie H: Exosomes from human
umbilical cord blood accelerate cutaneous wound healing through
miR-21-3p-mediated promotion of angiogenesis and fibroblast
function. Theranostics. 8:169–184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang B, Chen Y and Shi J: Exosome
biochemistry and advanced nanotechnology for next-generation
theranostic platforms. Adv Mater. 31:e18028962019. View Article : Google Scholar
|
|
10
|
Gonzalez-King H, García NA, Ontoria-Oviedo
I, Ciria M, Montero JA and Sepúlveda P: Hypoxia inducible factor-1α
potentiates jagged 1-mediated angiogenesis by mesenchymal stem
cell-derived exosomes. Stem Cells. 35:1747–1759. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu J, Yan Z, Yang F, Huang Y, Yu Y, Zhou
L, Sun Z, Cui D and Yan Y: Exosomes derived from human umbilical
cord mesenchymal stem cells accelerate cutaneous wound healing by
enhancing angiogenesis through delivering angiopoietin-2. Stem Cell
Rev Rep. 17:305–317. 2021. View Article : Google Scholar
|
|
12
|
Chen CY, Rao SS, Ren L, Hu XK, Tan YJ, Hu
Y, Luo J, Liu YW, Yin H, Huang J, et al: Exosomal DMBT1 from human
urine-derived stem cells facilitates diabetic wound repair by
promoting angiogenesis. Theranostics. 8:1607–1623. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsutsui TW: Dental pulp stem cells:
Advances to applications. Stem Cells Cloning. 13:33–42.
2020.PubMed/NCBI
|
|
14
|
Yoon JK, Kang ML, Park JH, Lee KM, Shin
YM, Lee JW, Kim HO and Sung HJ: Direct control of stem cell
behavior using biomaterials and genetic factors. Stem Cells Int.
2018:86429892018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim BC, Bae H, Kwon IK, Lee EJ, Park JH,
Khademhosseini A and Hwang YS: Osteoblastic/cementoblastic and
neural differentiation of dental stem cells and their applications
to tissue engineering and regenerative medicine. Tissue Eng Part B
Rev. 18:235–244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Botelho J, Cavacas MA, Machado V and
Mendes JJ: Dental stem cells: Recent progresses in tissue
engineering and regenerative medicine. Ann Med. 49:644–651. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martínez-Sarrà E, Montori S, Gil-Recio C,
Núñez-Toldrà R, Costamagna D, Rotini A, Atari M, Luttun A and
Sampaolesi M: Human dental pulp pluripotent-like stem cells promote
wound healing and muscle regeneration. Stem Cell Res Ther.
8:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mead B, Logan A, Berry M, Leadbeater W and
Scheven BA: Concise review: Dental pulp stem cells: A novel cell
therapy for retinal and central nervous system repair. Stem Cells.
35:61–67. 2017. View Article : Google Scholar
|
|
19
|
Huang CC, Narayanan R, Alapati S and
Ravindran S: Exosomes as biomimetic tools for stem cell
differentiation: Applications in dental pulp tissue regeneration.
Biomaterials. 111:103–115. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jarmalavičiūtė A, Tunaitis V, Pivoraitė U,
Venalis A and Pivoriūnas A: Exosomes from dental pulp stem cells
rescue human dopaminergic neurons from 6-hydroxy-dopamine-induced
apoptosis. Cytotherapy. 17:932–939. 2015. View Article : Google Scholar
|
|
21
|
Swanson WB, Zhang Z, Xiu K, Gong T, Eberle
M, Wang Z and Ma PX: Scaffolds with controlled release of
pro-mineralization exosomes to promote craniofacial bone healing
without cell transplantation. Acta Biomater. 118:215–232. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu X, Zhong Y, Kong Y, Chen Y, Feng J and
Zheng J: Lineage-specific exosomes promote the odontogenic
differentiation of human dental pulp stem cells (DPSCs) through
TGFβ1/smads signaling pathway via transfer of microRNAs. Stem Cell
Res Ther. 10:1702019. View Article : Google Scholar
|
|
23
|
Izar B, Tirosh I, Stover EH, Wakiro I,
Cuoco MS, Alter I, Rodman C, Leeson R, Su MJ, Shah P, et al: A
single-cell landscape of high-grade serous ovarian cancer. Nat Med.
26:1271–1279. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rochereau N, Roblin X, Michaud E, Gayet R,
Chanut B, Jospin F, Corthésy B and Paul S: NOD2 deficiency
increases retrograde transport of secretory IgA complexes in
Crohn's disease. Nat Commun. 12:2612021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fornabaio G, Barnhill RL, Lugassy C,
Bentolila LA, Cassoux N, Roman-Roman S, Alsafadi S and Bene FD:
Angiotropism and extravascular migratory metastasis in cutaneous
and uveal melanoma progression in a zebrafish model. Sci Rep.
8:104482018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goerge T, Ho-Tin-Noe B, Carbo C, Benarafa
C, Remold-O'Donnell E, Zhao BQ, Cifuni SM and Wagner DD:
Inflammation induces hemorrhage in thrombocytopenia. Blood.
111:4958–4964. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marcinkiewicz AL, Lieknina I, Kotelovica
S, Yang X, Kraiczy P, Pal U, Lin YP and Tars K: Eliminating factor
H-Binding activity of borrelia burgdorferi CspZ combined with
virus-like particle conjugation enhances its efficacy as a lyme
disease vaccine. Front Immunol. 9:1812018. View Article : Google Scholar :
|
|
28
|
Albelda SM, Muller WA, Buck CA and Newman
PJ: Molecular and cellular properties of PECAM-1 (endoCAM/CD31): A
novel vascular cell-cell adhesion molecule. J Cell Biol.
114:1059–1068. 2018. View Article : Google Scholar
|
|
29
|
Ju Lee H, Bartsch D, Xiao C, Guerrero S,
Ahuja G, Schindler C, Moresco JJ, Yates JR III, Gebauer F, Bazzi H,
et al: A post-transcriptional program coordinated by CSDE1 prevents
intrinsic neural differentiation of human embryonic stem cells. Nat
Commun. 8:14562017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zeng H, Castillo-Cabrera J, Manser M, Lu
B, Yang Z, Strande V, Begue D, Zamponi R, Qiu S, Sigoillot F, et
al: Genome-wide CRISPR screening reveals genetic modifiers of
mutant EGFR dependence in human NSCLC. Elife. 19:e502232019.
View Article : Google Scholar
|
|
31
|
Kumar S, Jiang MS, Adams JL and Lee JC:
Pyridinylimidazole compound SB 203580 inhibits the activity but not
the activation of p38 mitogen-activated protein kinase. Biochem
Biophys Res Commun. 263:825–831. 2017. View Article : Google Scholar
|
|
32
|
Tu GW, Ju MJ, Zheng YJ, Hao GW, Ma GG, Hou
JY, Zhang XP, Luo Z and Lu LM: CXCL16/CXCR6 is involved in
LPS-induced acute lung injury via P38 signalling. J Cell Mol Med.
23:5380–5389. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Z, Wu H, Jiang K, Wang Y, Zhang W, Chu
Q, Li J, Huang H, Cai T, Ji H, et al: MAPK-Mediated YAP activation
controls mechanical-tension-induced pulmonary alveolar
regeneration. Cell Rep. 16:1810–1819. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koch S and Claesson-Welsh L: Signal
transduction by vascular endothelial growth factor receptors. Cold
Spring Harb Perspect Med. 2:a0065022012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kurogane Y, Miyata M, Kubo Y, Nagamatsu Y,
Kundu RK, Uemura A, Ishida T, Quertermous T, Hirata KI and Rikitake
Y: FGD5 mediates proangiogenic action of vascular endothelial
growth factor in human vascular endothelial cells. Arterioscler
Thromb Vasc Biol. 32:988–996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rousseau S, Houle F, Landry J and Huot J:
p38 MAP kinase activation by vascular endothelial growth factor
mediates actin reorganization and cell migration in human
endothelial cells. Oncogene. 15:2169–2177. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Armstrong SC, Delacey M and Ganote CE:
Phosphorylation state of hsp27 and p38 MAPK during preconditioning
and protein phosphatase inhibitor protection of rabbit
cardiomyocytes. J Mol Cell Cardiol. 31:555–567. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Evans IM, Britton G and Zachary IC:
Vascular endothelial growth factor induces heat shock protein (HSP)
27 serine 82 phosphorylation and endothelial tubulogenesis via
protein kinase D and independent of p38 kinase. Cell Signal.
20:1375–1384. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou T, Yang Z, Chen Y, Chen Y, Huang Z,
You B, Peng Y and Chen J: Estrogen accelerates cutaneous wound
healing by promoting proliferation of epidermal keratinocytes via
Erk/Akt signaling pathway. Cell Physiol Biochem. 38:959–968. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xie L, Guan Z, Zhang M, Lyu S, Thuaksuban
N, Kamolmattayakul S and Nuntanaranont T: Exosomal circLPAR1
promoted osteogenic differentiation of homotypic dental pulp stem
cells by competitively binding to hsa-miR-31. Biomed Res Int.
2020:63193952020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Venugopal CKS, Rai KS, Pinnelli VB, Kutty
BM and Dhanushkodi A: Neuroprotection by human dental pulp
mesenchymal stem cells: From billions to nano. Curr Gene Ther.
18:307–323. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pivoraitė U, Jarmalavičiūtė A, Tunaitis V,
Ramanauskaitė G, Vaitkuvienė A, Kašėta V, Biziulevičienė G, Venalis
A and Pivoriūnas A: Exosomes from human dental pulp stem cells
suppress carrageenan-induced acute inflammation in mice.
Inflammation. 38:1933–1941. 2015. View Article : Google Scholar
|
|
43
|
Han Y, Ren J, Bai Y, Pei X and Han Y:
Exosomes from hypoxia-treated human adipose-derived mesenchymal
stem cells enhance angiogenesis through VEGF/VEGF-R. Int J Biochem
Cell Biol. 109:59–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bakhtyar N, Jeschke MG, Herer E,
Sheikholeslam M and Amini-Nik S: Exosomes from acellular Wharton's
jelly of the human umbilical cord promotes skin wound healing. Stem
Cell Res Ther. 9:1932018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tonnesen MG, Feng X and Clark RA:
Angiogenesis in wound healing. J Investig Dermatol Symp Proc.
5:40–46. 2000. View Article : Google Scholar
|
|
46
|
Ribeiro MF, Zhu H, Millard RW and Fan GC:
Exosomes function in pro- and anti-angiogenesis. Curr Angiogenes.
2:54–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu P, Zhang B, Shi H, Qian H and Xu W:
MSC-exosome: A novel cell-free therapy for cutaneous regeneration.
Cytotherapy. 20:291–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yu M, Liu W, Li J, Lu J, Lu H, Jia W and
Liu F: Exosomes derived from atorvastatin-pretreated MSC accelerate
diabetic wound repair by enhancing angiogenesis via AKT/eNOS
pathway. Stem Cell Res Ther. 11:3502020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu S, Uppal H, Demaria M, Desprez PY,
Campisi J and Kapahi P: Simvastatin suppresses breast cancer cell
proliferation induced by senescent cells. Sci Rep. 5:178952015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu J, He X, Corbett SA, Lowry SF, Graham
AM, Fässler R and Li S: Integrins are required for the
differentiation of visceral endoderm. J Cell Sci. 122:233–242.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lamalice L, Houle F, Jourdan G and Huot J:
Phosphorylation of tyrosine 1214 on VEGFR2 is required for
VEGF-induced activation of Cdc42 upstream of SAPK2/p38. Oncogene.
23:434–445. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lamalice L, Houle F and Huot J:
Phosphorylation of Tyr1214 within VEGFR-2 triggers the recruitment
of Nck and activation of Fyn leading to SAPK2/p38 activation and
endothelial cell migration in response to VEGF. J Biol Chem.
281:34009–34020. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Feng PC, Ke XF, Kuang HL, Pan LL, Ye Q and
Wu JB: BMP2 secretion from hepatocellular carcinoma cell HepG2
enhances angiogenesis and tumor growth in endothelial cells via
activation of the MAPK/p38 signaling pathway. Stem Cell Res Ther.
10:2372019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Correa AR, Berbel AC, Papa MP, Morais AT,
Peçanha LM and Arruda LB: Dengue virus directly stimulates
polyclonal B cell activation. PLoS One. 10:e01433912015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Morrison VL, James MJ, Grzes K, Cook P,
Glass DG, Savinko T, Lek HS, Gawden-Bone C, Watts C, Millington OR,
et al: Loss of beta2-integrin-mediated cytoskeletal linkage
reprogrammes dendritic cells to a mature migratory phenotype. Nat
Commun. 5:53592014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Maji S, Chaudhary P, Akopova I, Nguyen PM,
Hare RJ, Gryczynski I and Vishwanatha JK: Exosomal annexin II
promotes angiogenesis and breast cancer metastasis. Mol Cancer Res.
15:93–105. 2017. View Article : Google Scholar
|
|
57
|
Liang B, Liang JM, Ding JN, Xu J, Xu JG
and Chai YM: Dimethyloxaloylglycine-stimulated human bone marrow
mesenchymal stem cell-derived exosomes enhance bone regeneration
through angiogenesis by targeting the AKT/mTOR pathway. Stem Cell
Res Ther. 10:3352019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou X, Yan T, Huang C, Xu Z, Wang L,
Jiang E, Wang H, Chen Y, Liu K, Shao Z and Shang Z: Melanoma
cell-secreted exosomal miR-155-5p induce proangiogenic switch of
cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling
pathway. J Exp Clin Cancer Res. 37:2422018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xue C, Shen Y, Li X, Li B, Zhao S, Gu J,
Chen Y, Ma B, Wei J, Han Q and Zhao RC: Exosomes derived from
hypoxia-treated human adipose mesenchymal stem cells enhance
angiogenesis through the PKA signaling pathway. Stem Cells Dev.
27:456–465. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Park S, Guo Y, Negre J, Preto J, Smithers
CC, Azad AK, Overduin M, Murray AG and Eitzen G: Fgd5 is a
rac1-specific Rho GEF that is selectively inhibited by
aurintricarboxylic acid. Small GTPases. 12:147–160. 2021.
View Article : Google Scholar :
|
|
61
|
Heldin J, O'Callagha n P, Vera RH, Fuchs
PF, Gerwins P and Kreuger J: FGD5 sustains vascular endothelial
growth factor A (VEGFA) signaling through inhibition of
proteasome-mediated VEGF receptor 2 degradation. Cell Signal.
40:125–132. 2017. View Article : Google Scholar : PubMed/NCBI
|