1. Introduction
All living organisms must cope with a number of
adversities during their lifetime and have to maintain a complex
dynamic equilibrium called homeostasis (1). The state of threatened (or perceived
as such) homeostasis is known as stress and the intrinsic or
extrinsic forces that lead to such a state are called stressors
(2). Severe or prolonged stress
has been associated with several pathological conditions, such as
the deregulation of the immune system, cardiovascular disease,
neuropsychiatric disorders, metabolic disorders, endocrine
disorders, and growth, as well as development impairments (3,4).
Thus, in response to stressors, an organism activates multiple
complex and dynamic processes in an effort to restore homeostasis,
while the involved processes form the stress response system
(5). The stress response system
leads to several behavioral and physiological changes, such as
increased awareness and enhanced analgesia, as well as an increased
respiratory rate and the inhibition of general vegetative functions
(6). The stress response system
is comprised mainly of two components, the
hypothalamic-pituitary-adrenal cortex (HPA) axis and the locus
coeruleus/norepinephrine autonomic nervous system (7).
The HPA axis is the neuroendocrine link between
stress and an organism's physiological response to such a state
(8). A stressor induces a chain
of events in the brain that activates the HPA axis. Specifically,
neurons whose bodies are located in the paraventricular nucleus of
the hypothalamus secrete corticotropin-releasing hormone (CRH) and
arginine-vasopressin (AVP) into the hypophysial portal system.
Then, they target the anterior lobe of the pituitary gland. There,
CRH and AVP stimulate proopiomelanocortin cells, which in turn
release adrenocorticotropic hormone (ACTH), also known as
corticotropin. Finally, ACTH is released into the bloodstream and
acts on the cortex of the adrenal glands, thus triggering
glucocorticoid (GC) production (cortisol in humans and
corticosterone in rats) (9,10).
GCs subsequently self-regulate the HPA axis through a negative
feedback loop (11). Although
elevations in cortisol levels are the physiological response of the
body to fear or threat and are beneficial to promoting survival,
chronic exposure to stress results in long-term cortisol exposure.
This prolonged exposition can lead to a broad range of issues,
including the emergence of metabolic syndrome, obesity, cancer,
mental health disorders, cardiovascular disease and increased
susceptibility to infections (5).
GC signaling is related to the function of
non-coding RNAs (ncRNAs) which have been extensively studied for
their post-transcriptional involvement in the regulation of gene
expression. It has been shown that microRNAs (miRNAs/miRs) can
function as direct regulators of GC receptors (GRs) via the
hybridization of various miRNAs with the 3′untranslated region
(3′UTR) of the GR transcript, interrupting protein synthesis
(12). Furthermore, miRNAs are
directly or indirectly influenced by GCs, while it has been shown
that the expression level of miRNAs is dependent on endogenous GC
levels (13). In addition, long
ncRNAs (lncRNAs), such as growth arrest-specific 5 (GAS5) (14) and EDN1-AS (15), regulate the expression levels of
GRs, interfering with the GC signaling pathways. It is therefore
paramount to increase our understanding of how nc-RNAs, mainly via
the action of miRNAs and lncRNAs, regulate GR and are regulated by
GCs, building an intricate regulatory network to fine-tune the
body's response to various stimuli.
2. Glucocorticoid signaling
Endogenous GCs, as the final products of the HPA
axis, are the main regulators of the stress response system
(16). GCs are steroid hormones
and as main regulators of the stress response system, are involved
in numerous biological processes, including immune response,
metabolism, as well as developmental, cognitive and behavioral
functions (17). Endogenous GC
secretion displays both ultradian and circadian rhythms. The
circadian rhythm refers to physical, mental and behavioral changes
that follow a 24-h cycle. Specifically, the circadian peak in GC
release occurs in the early morning in diurnal animals and in the
early night in nocturnal animals. On the other hand, the ultradian
pulses, which are cycles having a duration shorter than a day, but
longer than an hour, display a frequency of approximately one or
two per hour (18,19), with nocturnal animals exhibiting
an increase in amplitude towards the end of the day (20). GCs can bind either the GR or the
mineralocorticoid receptor (MR) (21). The MR has a 10-fold higher
affinity for naturally occurring GCs than the GR, leading to large
receptor occupancy, whereas GR is only activated during circadian
peak and stress response (22).
Therefore, the actions of GCs are mainly exerted through the GR.
GRs are expressed in almost every cell of the body and there are
several GR isoforms generated from one gene, due to alternative
splicing and various post-translational modifications (14). In particular, GCs function as
ligands that bind GR transcripts and enable their action, with the
full GRα transcript being the predominant isoform (23,24). It should also be mentioned that
due to this wide range of functions GCs display, primarily via
their capacity to influence immune response, synthetic GCs, such as
dexamethasone and fludrocortisone have been developed and are
widely prescribed for the treatment of several diseases, including
rheumatic, pulmonary, gastroenterological, and cutaneous diseases
(25).
Both GR and MR are members of a structurally-related
protein superfamily known as nuclear receptors (NRs) (26). NRs function as transcription
factors, regulating gene expression (27). NRs exhibit a characteristic
structure, and so does GR. In particular, the GR is encoded by the
NR subfamily 3 group C member 1 (NR3C1) gene and its
functional and structural domains include an N-terminal domain
(NTD), a DNA-binding domain (DBD), a hinge region (HR) and a
C-terminal ligand-binding domain (LBD) (17). More specifically, the NTD contains
an activation function (AF)-1 region that interacts with
coregulators (28). Additionally,
the highly conserved DNA-binding domain contains two zinc finger
motifs and binds specific DNA sequences called GC response elements
(GREs) (29). The highly flexible
HR contains an amino terminus that is an essential part of the DBD
and is involved in receptor dimerization, while the flexibility
provided by HR to the GR allows a receptor dimmer to interact with
multiple GREs (30). The somewhat
conserved C-terminal LBD binds ligands, which in turn lead to
conformational changes within the LBD that modulate a second AF
region (AF-2) and enable interaction with specific coregulators
(28).
In the absence of GCs, GR is located in the
cytoplasm, where it is bound to a number of chaperone proteins that
render it inactive (29). GR is
first bound by heat shock protein (Hsp) 40 kDa (Hsp40), heat shock
cognate 71 kDa protein and the Hsp70-Hsp90 organizing protein
(Hop), while at later stages, it is bound by Hsp90, FK506-binding
proteins (FKBPs) and prostaglandin E synthase 3 (PTGES3/p23)
(31). Specifically, following
receptor translation, Hsp70 binds the receptor in the cytosol,
which leads to the unfolding of the GR and LBD inactivation. This
process is accelerated by Hsp40 binding. The Hsp40/Hsp70-GR complex
is then recruited to interact with Hsp90 via Hop. Hop, Hsp40 and
Hsp70 are then dislodged from the Hsp90-GR complex upon Hsp90
binding, ATP and the subsequent association of FKBPs cochaperones,
such as FKBP51 and FKBP52. Hsp90 interaction with GR folds the
receptor and leads to a functional LBD. The heterocomplex is
stabilized via p23 binding through the N-terminus and middle
domains, promoting a conformation with a high affinity for
corticosteroids (17,32). The binding of a ligand to the
receptor's LBD leads to conformational alterations that change the
proteins which comprise the heterocomplex, while also promoting GR
dimerization and the subsequent translocation to the nucleus, where
it can act as a transcriptional regulator (33,34). GR import is a rapid and active
process that relies on GR association with Hsp90, FKBP52 and
importin-a. The GR complex is transported into the nucleus by
dynein along the cytoskeleton and through the nuclear pore complex
(34). Once in the nucleus, the
activated GR can either activate or repress gene transcription. As
regards the GR signaling pathways, there are three primary ways the
receptors interact with molecules and intervene in gene expression.
For instance, GR can either bind directly to DNA in specific
sequences, or can tether itself to other DNA-bound transcription
factors. The third is via direct binding to DNA and interaction
with neighboring DNA-bound transcription factors. In particular,
transactivation can be achieved directly through GR homodimer
binding to a GRE found in gene promoter regions, or indirectly,
where GR functions as a monomer and co-operates with other
transcription factors to induce transcription (35,36). Transrepression can also be either
direct, via GR homodimer or, preferably, monomer binding to a
negative GRE, or indirect via GR monomer binding to a
pro-inflammatory transcription factor, such as NF-κB (35-37). GR remains bound to DNA for a
specific time period which could be influenced by the bound ligand.
This influence may be due to differences in ligand-induced
conformational changes (38).
Following ligand disengagement, GR dissociates from DNA and is
either degraded by the proteasome or exported from the nucleus,
which is an inactive process, most likely occurring through passive
diffusion (34). This system
enables the cell to rapidly respond to environmental changes and
exercise its effects via the intricate networks established around
GR activity.
3. GR pathways
The anti-inflammatory abilities of GCs highlight
their critical role in the regulation of the immune system.
Specifically, GR inhibits major regulators of pro-inflammatory
pathways, such as transcription factors activator protein-1 (AP-1)
and the aforementioned NF-κB (39). AP-1 is a protein complex composed
of a Jun protein family member dimerized with another Jun protein
or with a Fos protein, and enhances the expression of a number of
cytokines, such as IL-1 and IL-2. In particular, GR binds the
ubiquitously expressed c-Fos/c-Jun dimers via a sequence-specific
to c-Fos and inhibits the DNA binding and transactivation abilities
of this AP-1 heterocomplex (40).
In mammals, the NF-κB protein family of transcription factors
includes several proteins, such as p65 and p105/p50, which interact
and form distinct homodimers or heterodimers with transcriptional
regulation abilities (41). NF-κB
binds to specific response elements and regulates the expression of
genes encoding proteins, such as pro-inflammatory cytokines,
chemokines, receptors and adhesion molecules (42). GR can inhibit NF-κB directly by
interacting with the p65 and p50 subunits, or indirectly by
inducing the expression of the TSC22D3 gene, which encodes
GC-induced leucine zipper, a protein that binds NF-κB and
suppresses its function (39).
Under pathological conditions, stress-induced GCs may suppress
cell-mediated immunity and may thus lead to viral infection
susceptibility and tumor development (43). To illustrate the multifaceted
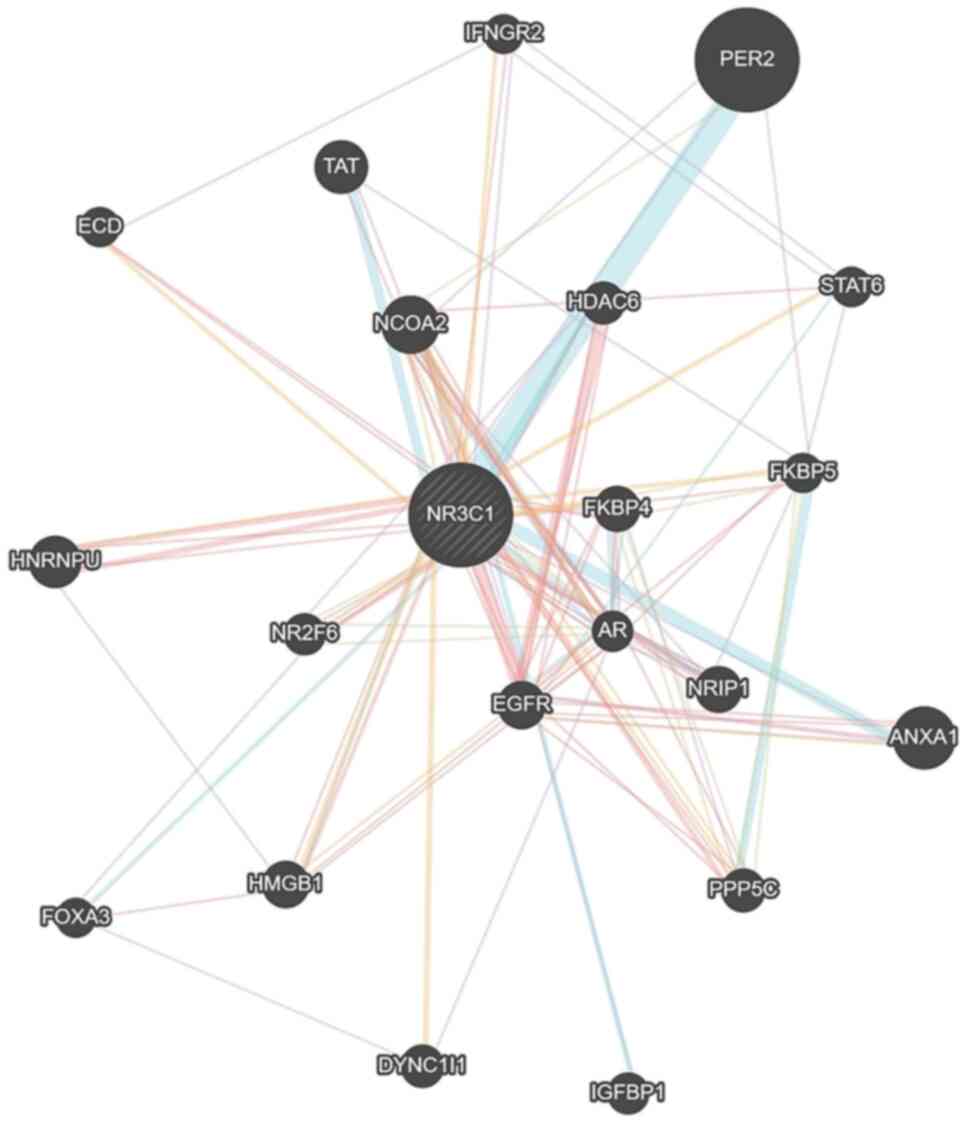
interaction of NR3C1 with other proteins, a gene association
network of genes related to the NR3C1 gene was developed
using the Genemania algorithm (44) (Fig.
1). A total of 21 genes were detected in this network,
including FKBPs, which interact with, or are influenced by GRα.
GCs also play a pivotal role in development. In
utero GC levels influence embryonic development and affect
adult physiology and pathophysiology, with a prime example being
increased susceptibility to cardiovascular disease (45). In particular, GR in cardiomyocytes
may induce the expression of genes, such as peroxisome
proliferator-activated receptor γ coactivator 1α, which regulates
cardiac mitochondrial capacity and genes encoding other regulators
of cardiac metabolism, such as peroxisome proliferator-activated
receptor α, Krüppel-like factor 15 and lipin 1 (lpn1). These
regulators of cardiac metabolism play critical roles in the
transition from the fetal to the neonatal heart (46). Additionally, GR expression in
mesenchymal cells has been proven to be necessary for lung
maturation. Mesenchymal GR regulates the development, morphological
differentiation and remodeling of the lungs in neonates.
Mesenchymal and GR-null mice have been shown to exhibit lung cell
hyperplasia, suggesting a crucial function of GC signaling during
lung development, via a decrease in proliferation and the thinning
of interstitial cells for effective gas exchange (47). Furthermore, GC excess may lead to
numerous developmental issues, culminating in congenital
disabilities. Specifically, GR can induce the expression of
Dickkopf-related protein 1, one of the main inhibitors of the
Wnt/β-Catenin pathway which among others, participates in stem cell
renewal, cell differentiation and cell proliferation during
development (48).
Cardiovascular diseases are directly connected to
stress (49). Exposure to a high
concentration of GCs, either endogenous or exogenous, is associated
with an increased risk of heart failure, ischemic heart disease and
hypertension. The cardiovascular and pulmonary system needs to
maintain a specific balance of GC levels, while either too high or
too low concentrations have adverse effects on the system
functions. It has been shown that mouse models with either GR
deletion or an altered GR expression in cardiomyocytes exhibit
cardiovascular pathologies, irregularly shaped and disorganized
myofibrils at embryonic stages, larger hearts, as well as cardiac
fibrosis. Males also develop cardiomyocyte hypertrophy, suggesting
that the sexually dimorphic actions of GCs can occur in the heart
as well (50). Furthermore,
research on GR functions in rapid GC-induced hypertension has
revealed that following vascular smooth muscle-specific GR
deletion, there is a decrease in hypertension in mice (51). The mechanism used by GCs to
regulate blood pressure is through the vascular GRs. Additionally,
the endothelial GR appears to protect against lipopolysaccharide
(LPS)-induced septic shock, via the repression of the release of
NF-κB and IL-6, two inflammatory cytokines. On the other hand, the
overexpression of GR is associated with atrio-ventricular block and
bradycardia, with the first being easily reversed, once the levels
of GR are restored (52).
GCs also participate in several metabolic processes.
In the liver, GCs can stimulate gluconeogenesis by inducing
phosphoenolpyruvate carboxykinase and glucose-6-phosphatase action,
while in skeletal muscle and white adipose tissue, they lower
glucose uptake and utilization by interfering with the insulin
signaling pathway (53,54). In skeletal muscle specifically,
GCs decrease insulin receptor substrate 1 transcription, a
downstream signaling molecule of the insulin signaling pathway, and
increase the transcription of protein tyrosine phosphatase type 1B
and p38 mitogen-activated protein kinases (p38 MAPK), which counter
insulin action (55). Of note,
excessive GC action has been found to be associated with several
metabolic diseases, such as type II diabetes and obesity (56).
GCs influence several central nervous system (CNS)
processes. GCs' effect on the CNS is both cell-type and stress-type
specific (57). GCs have been
shown to regulate stress responses, apoptosis and long-term
potentiation (58). These
hormones can cross the blood-brain-barrier and alter synaptic
physiology, stress responsiveness and behavior (59). Specifically, in the brain,
following a GC peak, areas involved in emotional responses and
simple behavioral strategies exhibit an enhanced activity, while in
the aftermath of stress, areas involved in higher cognitive
functions are activated and allow individuals to associate
stressful events with a specific context and thus store information
for future use (60). Maladaptive
behavioral responses to stress have been shown to be associated
with mood disorders (61).
According to in vivo studies, transgenic mice where the GR
was deleted in the CNS cells (brain-specific GR knockout),
exhibited increased basal corticosterone levels and reduced
anxiety. Considering that corticosterone is the rodent analog of
human cortisol, the results can be attributed to the loss of
central feedback inhibition in the HPA axis stress system.
Moreover, mice which did not produce GR in the forebrain exhibited
elevated MR expression in the hippocampus. Several variations of GR
knockout mice in the hypothalamus resulted in sex specific
differences in the HPA axis function, where females exhibited
elevated corticosterone levels at the lowest point of the circadian
cycle, while males did the same after intense stress. Notably, the
depletion of GR in dopamine-receptive neurons resulted in mice with
phenotypes of social aversion due to chronic stress (47).
The involvement of stress system dysregulation and
more specifically, the dysregulation of the HPA axis in
neuropsychiatric disorders is equally unsurprising, given the
robust and dynamic nature of stress biology. Affective disorders,
including major depressive disorder, bipolar disorder, anxiety and
panic disorders, schizophrenia and post-traumatic stress disorder
are considered anxiety disorders in which the key neural pathways
that regulate the stress response do not function optimally
(62). The increased reactivity
to threatening stimuli, the reduced ability to finish the stress
response, and/or the suboptimal coupling between internal emotional
states and the external environment are part of stress system
dysregulation. Of these, the latter dysfunction may contribute to
mood shifting in an extreme and seemingly random manner in bipolar
illness or to 'stick' in a negative manner in major depression.
Although both disorders are primarily inherited (63,64), vulnerability to these is directly
linked to how the individual responds to the environment (65). Indeed, according to the study by
Wray et al (66), who
examined ~460,000 individuals for the genetics of depression, it
appeared that 'all people carry a greater or lesser number of
genetic risk factors for major depression', where these are due to
the dysregulation of the HPA axis in depression. Major depression
is the result of the dysregulation of various genes related to
cellular development, cell repair and growth factors. An example of
these genes is the fibroblast growth factor (FGF) gene family,
which is dysregulated in major depression (67). FGF2, which is a member of
the FGF family, has been detected to function as an 'endogenous
antidepressant'. Its levels in the hippocampus and frontal cortex
are low in depression in humans, and they are reduced in rodents as
a result of repeated social stress, and have been shown to modulate
the HPA axis via GR (68). On the
other hand, the levels of FGF9, another gene member of the
FGF family, are increased in the depressed brain, as this gene
functions as a vulnerability factor and is increased by chronic
stress in animal models (67),
and its selective inhibition in the hippocampus reduces anxiety
behavior. Of note, treatment with FGF2 during early life leads to
epigenetic changes to GR in the hippocampus, increasing its
association with a repressive histone, H3K9Me3 (69), which leads to reduced levels of
NR3C1 expression and with a lower number of GRs in the hippocampus
(70). In addition to major
depression, schizophrenia and bipolar disorder are also associated
with the dysregulation of HPA axis activity, under basal conditions
and during stress. The decreased mRNA expression of the GR has been
observed in both illnesses using multiple post-mortem tissue
cohorts and brain regions (71).
Moreover, the increased expression of a truncated GR protein
isoform has been reproducibly demonstrated in the prefrontal cortex
of patients with schizophrenia and bipolar disorder (72,73). Notably, in conjunction with the
decreased expression of multiple GR mRNA transcripts and the
increased expression of the functional, truncated GRα-D1 protein
isoform 35, the altered expression, and the dysregulation of FKBP5,
PTGES3 and BAG1 mRNAs have been also detected. Taken together,
widespread stress-signaling alterations are detected in both
schizophrenia and bipolar disorder (71).
4. miRNAs in glucocorticoid signaling
miRNAs are a class of small ncRNAs ~22 nt in length,
which exert their effects by binding to the 3′UTR region of target
mRNAs. Compared to other ncRNA classes, they have been the focus of
extensive research for the past two decades, due to both their
post-transcriptional regulatory capacity, as well as their
therapeutic potential (74). To
date, >1,900 miRNAs have been discovered in the human genome and
more specifically, according to miRbase (release 22.1), there are
1,917 precursors and 2,654 mature miRNAs (GRCh38) (75-81), with regulatory activity spanning
across a multitude of biological processes, such as development,
metabolism, inflammation, as well as GR protein expression. miRNAs
have long been established as dynamic regulators during
development, but most importantly during environmental adaptation.
As such, their physiological function in the regulation of GR
expression is also predominantly in CNS development, as well as the
brain's response to both pre- and post-natal challenges (82). Nevertheless, prolonged exposure to
environmental stressors can lead to extensive neuronal
reprogramming accompanied by altered GC response (83,84). Additionally, due to the extensive
use of GCs for cancer treatment (85), accompanied by the central roles of
miRNAs in numerous types of cancer (86), investigating the association
between miRNAs and GC signaling is pivotal for developing
successful therapeutic approaches.
The present review mainly focuses on two aspects of
the miRNA regulation of GC signaling. In the first part, focus is
placed on miRNAs that have been shown to be direct regulators of
GRs, investigating their effects in both brain disorders, as well
as in cancer development and treatment. In the second part, miRNAs
directly or indirectly influenced by GCs are reviewed (Table I).
 | Table IMicroRNAs and glucocorticoid
activity. |
Table I
MicroRNAs and glucocorticoid
activity.
| GR regulators | Brain-related | Cancer-related |
Immune/inflammatory-related | Other |
|---|
| miR-124 | (90-92) | (99) | | (91) |
| miR-18a | (94) | | | |
| miR-137 | (96) | | | |
| miR-142-3p | (100) | (99) | | |
| miR-101a, -96,
-433 | (101) | | | |
| miR-130b, -181,
-636 | | (103) | | |
| miR-1-3p, -128,
-370, -28 | | | | (122) |
| miR-29a | | | | (122,123) |
| miR-340 | (128) | | | |
|
| Regulated by
GCs | Brain-related | Cancer-related |
Immune/inflammatory-related | Other |
|
| miR-155 | | (104) | (105-107) | |
| miR-511 | | | (108-110) | |
| miR-98 | | | (112) | |
| miR-101 | | | (113) | |
| miR-17-92
cluster | | (114-116) | | |
| miR-221/222 | | (118) | | |
| miR-125b | | (120) | | |
miRNAs and GRs
NR3C1, the mRNA transcript of GRα protein,
has been shown to harbor a number of conserved miRNA target sites
in its 3′UTR (12). Using the
well-established miRNA target prediction algorithm TargetScan
(release 7.2; March, 2018) (87),
135 conserved putative miRNA 8-mer binding sites (with a <-0.1
context score) were predicted for the human NR3C1 transcript
(Table SI). Extending this
search to the gene-network level with the use of the Genemania
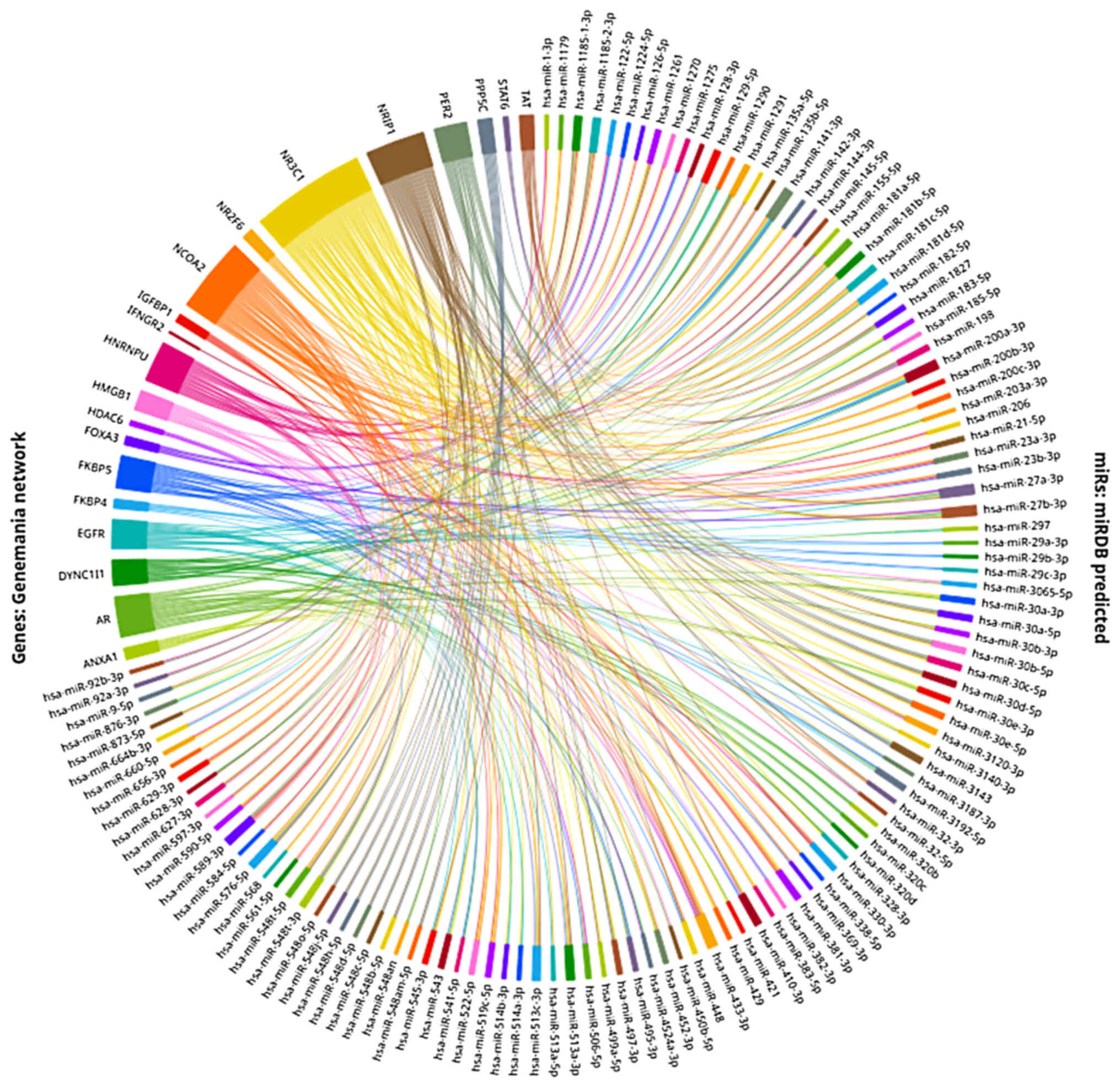
NR3C1 network, as shown in Fig. 1, which includes 21 proteins
predicted to interact and form a network with GRα and miRDB
(88,89), a miRNA target prediction on the
3′UTR region algorithm which allows for the concurrent
investigation of multiple genes. Thus, this made it possible to
identify 120 miRNAs predicted to regulate more than one of the
proteins of the Genemania network, via targeting their respective
mRNA 3′UTRs (Fig. 2). This simple
analysis serves to show the extensive regulatory effects multiple
miRNAs can have on different levels of the GC network, as well as
the great fine-tuning potential they can apply on GRα levels. Thus
far, a number of these miRNAs have already been shown to interact
with GRα and thus exert their regulatory effects.
First and foremost is miR-124, a miRNA induced by GC
treatment, which has been the subject of extensive studies in
relation to GR expression. Using in vitro experiments in
Jurkat T-cells and T-cells from healthy volunteers and patients
with sepsis, miR-124 was previously shown to decrease the
expression of GRα by direct binding of the 3′UTR of the
NR3C1 transcript (90).
This direct binding was also further validated in experiments using
293/293T cells (91). Notably,
miR-124 levels have been found to be increased in patients with
major depressive disorder (92,93), while depression-like behaviors in
mice were able to be effectively treated by miR-124 antagomir
administration, effectively blocking the inhibitory activity of
miR-124 and establishing it as both a biomarker and an interesting
therapeutic target for depression treatment (91). miR-124a, which is exclusively
expressed in the brain and is the most abundant miRNA in the
vertebrate central nervous system, was shown to regulate GR
expression by direct binding of the GR mRNA in P19 cells, a
well-established neuronal differentiation cell line (94). A similar inhibitory activity was
also observed for miR-18, although the binding potential for the GR
transcript was not observed in vitro (94). Nevertheless, additional studies
have implicated miR-18a as a direct post-transcriptional regulator
of GR. Direct binding was observed in cultured neuronal cells,
while miR-18a overexpression was concomitant with suppressed GR
protein levels in a model of stress-susceptible rats, further
establishing the direct GR regulation potential of the miR-18
family (95). miR-137, a miRNA
identified as a potential regulator in schizophrenia, as well as
other brain disorders, has been implicated in neuronal plasticity
through GR-dependent signaling, while in silico target prediction
algorithms identified NR3C1 transcript as its putative
target in both humans and mice (96). In a model of depression, following
chronic unpredictable mild stress, miR-382-5p levels were found to
be elevated with the concomitant suppression of GR levels in the
hippocampus of rats. si-miR-382-5p treatment restored GR levels, as
well as its downstream target levels of BDNF and p-TrkB62 (97).
A number of miRNAs that regulate GR expression have
been implicated in various types of cancer. In the case of miR-124,
it has also been shown that it can exert indirect regulatory
effects on the GC response and sensitivity. The upregulation of GRα
expression, accompanied by increased cAMP levels and the decreased
expression of phosphodiesterase 4B, has been observed under stable
miR-124 expression, in diffuse large B-cells lymphoma cell lines
(98). Additionally, in patients
with acute lymphoblastic leukemia (ALL), miR-124 levels have been
shown to be increased, while this miRNA contributes to GC
resistance, decreased apoptosis and decreased GR expression
(99). Another miRNA that has
been shown to be able to directly bind the NR3C1 transcript
and regulate its expression in various instances is miR-142-3p.
This miRNA, which is upregulated in patients with leukemia, has
been shown to be able to regulate GC response in T-leukemic cells
by directly binding with the 3′UTR of the GRα mRNA (100). miR-142-3p was also identified in
the study by Riester et al (101), where following ACTH stimulation,
miR-101a, miR-142-3p, miR-96 and miR-433 were identified as
putative direct regulators of GRα. Following modification of the
3′UTR of the NR3C1 transcript in an in vitro
experiment, it was shown that these four miRNAs could directly bind
the 3′UTR and inhibit GRα expression by up to 40% (101). By investigating GC sensitivity
in multiple myeloma cell lines, Tessel et al (102) identified miR-130b, miR-181a and
miR-636 as miRNAs with direct binding potential to the 3′UTR of the
NR3C1 transcript, while also exhibiting differential
expression between GC-sensitive and resistant cell lines. miR-130b
overexpression in vitro caused decreased GR expression,
concomitant with GC resistance, and decreased GC-induced apoptotic
effects. Due to the common usage of GCs as a treatment for various
forms of cancer, including leukemia and multiple myeloma, miRNA
expression patterns can be used to identify GC responsiveness and
resistance development (12,103).
GC regulation of miRNAs
Several miRNAs are involved in the mechanisms of the
immune and inflammatory response, which are modulated and regulated
by GCs. One of the first oncogenic miRNAs with elevated levels in
several types of cancer (104),
is miR-155. Increased levels of miR-155 have been reported in
fibroblasts and macrophages in rheumatoid arthritis, where this
miRNA appears to contribute to increased expression of chemokines
and pro-inflammatory cytokines (105,106). It was previously demonstrated
that the administration of GC dexamethasone to primary macrophages
and macrophage cell lines, spleen and liver cells of LPS-injected
mice, inhibited the expression of miR-155, suggesting that a
decrease in miR-155 expression is an anti-inflammatory result of
GCs (107,108).
miR-511 is a miRNA produced by the fifth intron of
mRNA encoding the C-type mannose receptor CD206. More specifically,
miR-511-5p contributes to impaired sensitivity to LPS and reduced
expression of the classically activated (M1) macrophage marker
IL-12, targeting the Toll-like receptor 4 and IL-12p40 subunit
(109). In addition, miR-511-5p
targets the p55 TNF receptor mRNA, thus providing TNF resistance,
in cases where there is increased expression of this miRNA due to
high levels of endogenous GCs (110). In conclusion, changes in the
expression level of miR-511 are dependent on levels of endogenous
GCs and have an impact on the differentiation and activation of
myeloid cells, as a byproduct of altered MRC1 gene
expression (111).
A member of the let-7 miRNA family miR-98 is another
GC-induced miRNA that reduces the expression of the p75 TNF
receptor in T-lymphocytes by targeting Tnfrsf1b mRNA.
Moreover, this miRNA is considered to target 3′UTR of IL-13, which
plays a pathogenic role in asthma. In this way, miR-98 may
contribute to the therapeutic effects of GCs on asthma (112). miR-101 is involved in
inflammatory responses in myeloid and other cells and targets the
3′UTR of dual specificity phosphatase 1 (DUSP1) to regulate the
activation of p38 MAPK in macrophages. Numerous studies have
demonstrated that GCs in combination with pro-inflammatory stimuli,
enhance DUSP1 expression, leading to the inactivation of p38 MAPK.
In the study by Zhu et al (113), it was shown that GCs inhibit the
expression of miR-101, thus leading to prolonged expression of
DUSP1, which in turn led to reduced activation of p38 MARK and
consequently to a decreased expression of p38 MARK-dependent
inflammatory mediators.
The miR-17-92 cluster consists of six miRNAs,
miR-17, -18a, -19a, -19b-1, -20a and 92a-1, which are derived from
a precursor RNA transcribed from chromosome 13 (114). This complex targets the PTEN
tumor suppressor, which regulates inositol phosphate signaling, and
a pro-apoptotic member of the Bcl-2 protein family, Bim which
regulates cytochrome c-induced apoptosis (115,116). According to several studies, GCs
function as inhibitors of this cluster, thus exerting several
therapeutic effects on lymphoma, while the failure of GCs to
inhibit the miR-17-92 cluster resulted in resistance to apoptosis.
In conclusion, the miR-17-92 cluster plays an important role in
regulating responses to pro- and anti-apoptotic signals (116,117).
Finally, two miRNAs that have been linked to
GC-induced apoptosis resistance in multiple myeloma (MM) are
miR-221/222 (118) and miR-125b
(119). The increased expression
of these miRNAs has been shown to be associated with the
attenuation of cell death pathways, such as apoptosis led by tumor
protein 53. miR-125b expression levels are regulated by GCs as part
of a possible self-limiting mechanism of GCs pro-apoptotic effects
(119). On the other hand, a
detailed analysis of miR-221 expression demonstrates the
specificity of its expression in different types of cancer. More
specifically, the increased expression of miR-221 in MM has been
shown to induce resistance to GC-induced apoptosis (118), contrary to certain types of ALL,
where resistance to GC-induced apoptosis is due to the reduced
expression of miR-221 (120).
Performing RNA-sequencing and microarray expression
analysis of male and female mouse hearts with a
cardiomyocyte-specific knockout of the GR, Cruz-Topete et al
(121) were able to identify 130
miRNAs whose expression was sex- and GR-dependent. Of these miRNAs,
25 were responsible for the vast majority of the differences
observed between male and female hearts, including prominent heart
failure biomarkers, such as miR-1-3p, miR-128, miR-370 and miR-28.
miR-29a and miR-340 overexpression in L929 cells was previously
found to be associated with a significant reduction in GR
expression, suggesting a direct binding role for both miRNAs and
the GR receptor transcript (122). Of note, in another study, the
overexpression of miR-29a in vivo ameliorated GC-induced
bone tissue destruction (123),
while miR-340 overexpression decreased GR protein levels in the
mouse placenta and affected sensitivity against activity-based
anorexia (124).
Last but not least, a few recent studies have also
identified GR as a potential miRNA regulator. In an in vitro
study, miR-22 expression was shown to be elevated in AR42J cells
following the induction of apoptosis. A GR binding site was
identified in the promoter region of miR-22, while GR was shown to
be able to repress miR-22 expression (125). In addition, a recent profiling
study in triple-negative breast cancer suggested the ability of GR
to influence multiple miRNA expression profiles (126), while in the study by Tejos-Bravo
et al (127),
neuron-specific GR knockout mice exhibited altered miRNA expression
profiles in a sex-dependent manner. Such effects exhibit the great
complexity inherent in miRNA regulation of GC activity, but also
suggest an additional fine-tuning potential via the possible
integration of feedback loops.
miRNA regulation of GR-chaperone
proteins
As previously mentioned, until the moment of GC
activation GR remains located in the cytoplasm, forming a complex
with various chaperone proteins. As such, a number of
secondary-degree regulatory potential exists via miRNA-mediated
regulation of these chaperone proteins. While not thoroughly
investigated, such effects have already been identified. miR-511,
whose effects were previously discussed, was found to be able to
directly bind the 3′UTR of FKBP5, suppressing GC-induced
FKBP51 expression, while increasing neuronal development (128). In a similar manner, miR-124 was
also shown to be able to target FKBP5 (129). Of note miR-142 and miR-340,
miRNAs with the established binding of GRα transcript, were
predicted also to target the 3′UTR of FKBP5 (128). Taking into account the numerous
chaperone proteins that GRα interacts with, as well as the
indicative gene regulatory network already established (Fig. 1), it is clear that the
miRNA-mediated regulation of GC response is a very complex and
multilayered process, which has only begun to be explored.
5. lncRNAs in glucocorticoid signaling
As already mentioned, GC signaling involves various
signal transduction cascades in the cell. In recent years, studies
focusing on the roles of ncRNAs have increased, including their
roles in regulating the transcriptional activity of GR and other
NRs (Table II). lncRNAs are a
class of ncRNAs, which consist of >200 nucleotides and are
derived from various regions in the genome, such as promoters,
enhancers, introns, UTRs, overlapping or non-coding isoforms of
coding genes, antisense to other transcripts and pseudogenes
(130,131). According to previous studies,
lncRNAs have been observed in the majority of organisms, such as
animals (132), plants (133), fungi (134) and even viruses (135), without however evolving
conservation among species. Following technological advancements
and novel laboratory techniques, numerous data related to the roles
of lncRNAs in a variety of vital biological processes have been
gathered (136) and more
specifically in transcription (137), alternative splicing (138), translation, cell cycle (139), apoptosis (140) and heat shock response (141). Several lncRNAs create
RNA-protein, RNA-DNA and RNA-RNA complexes, while they have also
been associated with chromatin modification and guiding
transcription factors to specific genomic DNA targets. Last but not
least, a number of lncRNAs have been found to be associated with
various diseases, including cancer, myocardial infractions and
Alzheimer's disease (142,143).
 | Table IIlncRNAs and glucocorticoid
activity. |
Table II
lncRNAs and glucocorticoid
activity.
| lncRNAs | Type of
interaction |
Metabolism-related |
Immune/inflammatory-related | Cancer-related | Other |
|---|
| GR-related | | | | | |
| GAS5 | GR regulator | (14) | (144-145,147) | (148) | (146) |
| EDN1-AS | Regulated by
GR | | | | (15,149-150) |
| PSORS1C3 | Regulated by
GR | | | | (152-154) |
| Related to other
receptors | | | | | |
| SRA | AR, ER, GR and PR
regulator | | | (156) | (155) |
| PRNCR1,
PCGEM1 | AR regulators | | | (157) | |
GAS5 is a lncRNA that has been of immense interest
to researchers in recent years and is involved in GR activity. As
its name suggests, GAS5 inhibits cell growth caused by a lack of
nutrients or growth factors. In the study by Kino et al
(14), it was shown that GAS5
functions as an inhibitor of the transcriptional activity of GR and
other steroid receptors (SRs). More specifically, the sequence of
GAS5 contains two GC response elements-mimetic sequences at
nucleotides 539-544 (GRE-1) and 553-559 (GRE-2). Thus, GAS5 acts as
a competitor for GR binding through the GRE regions, decreasing
GR-mediated gene activation and ultimately affecting cell survival
and metabolic activity during nutrient deficiency. GAS5
overexpression significantly inhibits the transcription of GR
target genes, including genes encoding the cellular inhibitor of
apoptosis 2 and serum/GC-responsive kinase 1 (14).
However, an interesting observation was the fact
that during growth inhibition or lack of nutrients in cells, the
accumulation of GAS5 was reported in the organs of mice which are
involved in metabolism, i.e., in the liver and adipose tissue, by
modifying the mTOR signaling pathway, contrary to organs involved
in the immune system, such as the thymus gland, spleen and the
brain. Recent studies have demonstrated that the levels of GAS5 in
cells can vary. In particular, it has been suggested that GAS5
exerts regulatory activity on GR in the immune system,
independently of nutrient availability. This is evidenced by the
different expression of GAS5 in leukocytes of patients with
inflammatory or autoimmune diseases (144), as well as its role in the GC
response of children with diseases, such as inflammatory bowel
disease (145), multiple
sclerosis (146), human beta
cell dysfunction (147) and
acute myeloid leukemia (148).
EDN1-AS is a lncRNA, which appears to interact with
GR. This lncRNA is located antisense of the endothelin 1 gene,
which is a peptide hormone that acts on the vascular system as a
vasoconstrictor, while in the kidneys it affects blood pressure
through diuretic and natriuretic effects (15). Its aberrant quantity has been
shown to be associated with pathological conditions, such as
hypertension (149) and chronic
kidney disease (150). EDN1-AS
is expressed in multiple human cell types, including the kidneys.
In the study by Douma et al (15) in a human kidney proximal tubule
cell line (HK-2), the promoter of lncRNA EDN1-AS appeared to have a
GRE sequence, which could be recognized and bound by GR, as well as
the MR, representing a new mechanism for regulating ET-1
expression. Using CRISPR/Cas9 for the deletion of the GRE element
from the EDN1-AS promoter, abolished the binding of GR to EDN1-AS
and resulted in increased EDN1-AS expression with a concomitant
increase in endothelin 1 expression and cell proliferation. The
inhibitory effect of GR binding to the EDN1-AS promoter is
therefore inferred (15).
Psoriasis susceptibility 1 candidate 3 (PSORS1C3) is
a lncRNA whose sequence is adjacent to the octamer-binding
transcription factor 4 (OCT4) gene. As is well known, the
transcription factor OCT4 plays regulatory roles in oncogenesis,
stemness and in response to stress. PSORS1C3 has shown to be
associated with diseases, such as psoriasis (151) and other immune-mediated
diseases, such as acute anterior uveitis (152), as well as major depressive
disorder (153). According to
the study by Mirzadeh Azad et al (154), this lncRNA has two endogenously
active promoters, promoters 0 and 1 and two sets of transcripts,
small and large variants. A GRE sequence was identified upstream of
promoter 0 of PSORS1C3, where GR binds and acts as either an
enhancer or a repressor of the expression of target genes. More
specifically, that study demonstrated the positive effect of GR on
the expression level of OCT4 and small variants of PSORS1C3,
which may reflect a new regulatory pathway of cell proliferation
and the stress response through the regulation of expression levels
of OCT4. On the other hand, the binding of GR to promoter 0
of large variants of lncRNA PSORS1C3 exerted an inhibitory effect,
suggesting its function as a pro-inflammatory factor. Thus, GR
functions as a regulator of the expression of the lncRNA PSORS1C3,
which in turn regulates and moderates the expression of the OCT4
factor in non-multipotent cells, as the PSORS1C3 promoter 0 region
acts as an enhancer for the neighboring OCT4 gene (154).
In general, several lncRNAs have been recorded to
regulate the transcriptional activity of several SRs, including GR.
A prototype lncRNA is the steroid RNA coactivator (SRA) which
increases the transcriptional activity of the androgen receptor
(AR), estrogen receptor, GR and progesterone receptor. The SRA
regulates the transcriptional activity by binding to the SRA
stem-loop-interacting RNA binding-protein and the RNA-induced
silencing complex complex (155,156). In addition, according to Yang
et al (157), lncRNAs
PRNCR1 and PCGEM1, which are expressed primarily in the prostate
gland, bind AR to its DNA binding domain, and suppress receptor
activity, playing a significant role in the development of prostate
cancer. In summary, lncRNAs appear to regulate the expression
levels of various target genes of NRs, including GR. For this
reason, future research is required to elucidate the function of
lncRNAs in regulating gene expression through their interaction
with transcription factors, such as GR in GC signaling
pathways.
6. Discussion
The stress response system is related to the
production of endogenous GCs. This system, particularly the
interaction between cortisol and GR, which is described by the GR
pathways, has attracted increasing attention in recent years due to
its medical interest in developing therapeutic approaches. Although
GR is derived from a single gene, its function differs based on the
different isomers which are produced due to alternative splicing
and alternative translation initiation mechanisms. It has been
shown that GCs bind in a similar manner to all the isomers
(158); however, GRs differ in
their subcellular distribution and gene regulatory profiles,
affecting the human organism via multiple mechanisms. It should be
noted that some GR polymorphisms have been linked to GC resistance
and a healthier metabolic profile, whereas others seem to be
associated with GC hypersensitivity increasing cardiovascular risk
(159). It has been proposed
that the regulation of GC signaling is related to pathological
conditions, such as cancer, heart diseases, diabetes and other
metabolic disorders.
Recently, numerous studies have implicated GC
signaling in cancer progression or prevention, depending on the
cell type. In one case, animal models of human breast cancer
revealed that GCs inhibit tumor cell apoptosis, while in other
cases, synthetic GCs are used to induce apoptotic cell death in
malignant lymphoid cells (e.g., lymphoma). Even though
pharmacologic GC therapy is frequently administered to cancer
patients to reduce the associated side effects of chemotherapy,
further investigations on the results of the treatment on patients
need to be conducted as GC application may contribute to tumor
growth (160). Other studies
have suggested that GR signaling in cardiomyocytes is critical for
the normal development and function of the heart (161,162). In a previous study (161), cardiomyocyte-specific GR
overexpression led to bradycardia, while GR inhibition resulted in
cardiac hypertrophy, systolic dysfunction, and impaired maturation.
Further research is required in order to determine the precise
molecular pathways and genes through which cardiomyocyte GC
signaling can either promote or protect against heart pathology
(161). It should also be
mentioned that, apart from the endogenous GCs, studies have
suggested the use of dexamethasone for the treatment of
post-operative nausea and vomiting, as it is a synthetic GC with
anti-inflammatory and immunosuppressant properties, with 20- to
30-fold the binding affinity for GR of endogenous cortisol
(163).
Due to the ability of miRNAs to regulate multiple
targets, while every 3′UTR can harbor multiple binding sites for
different microRNAs, it is easily apparent that
post-transcriptional regulation via miRNAs forms a highly complex
and sensitive network. Under physiological conditions, the
miRNA-mediated modulation of GR expression is mainly involved in
fine-tuning GC responses during CNS development. However, a number
of pathological responses have implicated the interaction between
the mRNA transcript of GR protein and different miRNAs, as observed
in brain disorders, as well as in cancer development and treatment.
Taking into account the negative role of miRNAs in steroidogenesis,
the association between miRNAs and GR expression warrants further
in-depth investigations. Currently, numerous miRNAs have already
been identified that have the capacity to directly bind
NR3C1 3′UTR and exercise their regulatory effects, such as
miR-124-3p and miR-142-3p, among several others. Nevertheless, the
concept of a single cause-single target, while beneficial for past
drug development, is gradually being pushed aside, as the
multi-factorial nature of regulation becomes more and more
apparent. These types of interactions offer increased sensitivity
and fine-tuning potential to environmental changes. More systematic
approaches are therefore necessary that will offer a holistic
regulatory view. Combining existing target prediction tools with
network generation packages, their complexity instantly emerges
(Fig. 2). Additional regulatory
levels are also exercised through alternative splicing and the
production of other GR isoforms. This is the case for miR-124-3p,
where differential splicing produces the GRb isoform, no longer
harboring its binding site, which in turn acts as a negative
inhibitor of GRα (90,162). Tying this to recent studies,
demonstrating that the GR can itself influence miRNA expression
profiles (125-127), the intricacies of miRNA-GR
regulation and response to GCs are becoming exponentially
complex.
Similar to miRNAs, lncRNAs have a variety of
regulatory roles in gene expression. Their ability to form
RNA-protein, RNA-DNA and RNA-RNA complexes enable them to be
involved in cellular processes, such as apoptosis, translation,
cell cycle and heat shock response. They play key roles in various
disease cases, such as numerous types of cancer, myocardial
infraction and neurodegenerative diseases, such as Alzheimer's
disease (142,143). lncRNAs are further involved in
various biological processes, including GC signaling, which
connects to extensive pathways related to the immune, nervous and
metabolic systems. An example is lncRNA GAS5, which regulates the
GR response via direct binding through its GRE sequences, thus
having an impact on the gene expression of the GR target-genes
(14). EDN1-AS and PSORS1C3 are
two additional lncRNAs whose expression is regulated by the GR. In
both cases, these lncRNAs are bound with the GR via GREs, which in
turn acts as their inhibitor, affecting the expression of genes
that interact with them, the EDN1 (15) and OCT4 (154) genes, respectively.
It thus becomes clear that GC signaling has
extensive and important functions in the immune, nervous system and
related metabolic responses in the context of homeostasis. Its
activity has been associated with numerous pathological conditions,
such as cancer, metabolic and neurodegenerative diseases. It is
easily apparent that GC signaling is part of an intricate
regulatory network, heavily involving post-transcriptional
regulation via ncRNAs, in an effort to maintain the fine-tuning
potential the body needs to respond to ever-shifting environmental
conditions. Such a network is comprised of numerous miRNAs, lncRNAs
and potentially yet unverified actors. Their complex associations
are just beginning to be unraveled, but are already demanding new
analysis paradigms to be adopted. To this aim, combinatory
bioinformatic approaches need to be employed and new tools
developed that can investigate such effects in a more systematic
manner. Only then can a better understanding of GC activity be
obtained and the utilization of its therapeutic potential can
effectively be achieved.
7. Conclusions
ncRNAs are an intriguing field of study. Their
unique properties, as well as their ability to be involved in vital
cellular processes, render them suitable pharmaceutical targets and
biomarkers. GC signaling participates in a regulatory network that
includes post-transcriptional regulation via ncRNAs, including
various miRNAs and lncRNAs. These molecules can either act as
regulators of GR activity or be regulated by endogenous GCs,
affecting the expression of GC-mediated genes. Generally, GC
activity is associated with several pathological conditions,
including cancer, neurodegenerative and metabolic diseases.
Therefore, the development of more effective therapies for these
diseases requires a better understanding of GC signaling that
includes interacting regulatory ncRNAs.
Supplementary Data
Availability of data and materials
Not applicable.
Authors' contributions
All authors (KP, LP, TM, EP, ID, SL, MS, KD, DAS,
FB, GPC, GNG, EE and DV) contributed to the conceptualization and
design of the study, as well as in the writing, drafting, revising,
editing and reviewing of the manuscript. Data authentication is not
applicable. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The authors would like to acknowledge funding from the following
organizations: i) AdjustEBOVGP-Dx (RIA2018EF-2081): Biochemical
Adjustments of native EBOV Glycoprotein in Patient Sample to Unmask
target Epitopes for Rapid Diagnostic Testing. A European and
Developing Countries Clinical Trials Partnership (EDCTP2) under the
Horizon 2020 'Research and Innovation Actions' DESCA; and ii)
'MilkSafe: A novel pipeline to enrich formula milk using omics
technologies', a research co-financed by the European Regional
Development Fund of the European Union and Greek national funds
through the Operational Program Competitiveness, Entrepreneurship
and Innovation, under the call RESEARCH-CREATE-INNOVATE (project
code: T2EDK-02222).
Abbreviations:
|
ACTH
|
adrenocorticotropic hormone
|
|
AF
|
activation function
|
|
ALL
|
acute lymphoblastic leukemia
|
|
AR
|
androgen receptor
|
|
AVP
|
arginine-vasopressin
|
|
CNS
|
central nervous system
|
|
CRH
|
corticotropic-releasing hormone
|
|
DBD
|
DNA-binding domain
|
|
DUSP1
|
dual specificity phosphatase 1
|
|
FKBPs
|
FK506-binding proteins
|
|
GAS5
|
growth arrest-specific 5
|
|
GC
|
glucocorticoid
|
|
GR
|
glucocorticoid receptor
|
|
GRE
|
glucocorticoid response element
|
|
Hop
|
Hsp70-Hsp90 organizing protein
|
|
HPA
|
hypothalamic-pituitary-adrenal
cortex
|
|
HR
|
hinge region
|
|
Hsp
|
heat shock protein
|
|
LBD
|
ligand-binding domain
|
|
lncRNA
|
long non-coding RNA
|
|
LPS
|
lipopolysaccharide
|
|
miRNAs/miRs
|
microRNAs
|
|
MM
|
multiple myeloma
|
|
MR
|
mineralocorticoid receptor
|
|
ncRNA
|
non-coding RNA
|
|
NTD
|
N-terminal domain
|
|
PTGES3/p23
|
prostaglandin E synthase 3
|
|
SRs
|
steroid receptors
|
References
|
1
|
Chrousos GP: Stress and disorders of the
stress system. Nat Rev Endocrinol. 5:374–381. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tsigos C, Kyrou I, Kassi E and Chrousos
GP: Stress: Endocrine Physiology and Pathophysiology. Endotext.
Feingold KR, Anawalt B, Boyce A, Chrousos G, de Herder WW, Dungan
K, Grossman A, Hershman JM, Hofland J, Kaltsas G, et al: MDText.com, Inc. Copyright© 20002021, MDText.com, Inc. South Dartmouth, MA: 2000
|
|
3
|
Charmandari E, Kino T, Souvatzoglou E and
Chrousos GP: Pediatric stress: Hormonal mediators and human
development. Horm Res. 59:161–179. 2003.PubMed/NCBI
|
|
4
|
Yaribeygi H, Panahi Y, Sahraei H, Johnston
TP and Sahebkar A: The impact of stress on body function: A review.
EXCLI J. 16:1057–1072. 2017.PubMed/NCBI
|
|
5
|
Russell G and Lightman S: The human stress
response. Nat Re Endocrinol. 15:525–534. 2019. View Article : Google Scholar
|
|
6
|
Smith SM and Vale WW: The role of the
hypothalamicpituitary-adrenal axis in neuroendocrine responses to
stress. Dialogues Clin Neurosci. 8:383–395. 2006. View Article : Google Scholar
|
|
7
|
Nicolaides NC, Charmandari E, Kino T and
Chrousos GP: Stress-related and circadian secretion and target
tissue actions of glucocorticoids: Impact on Health. Front
Endocrinol (Lausanne). 8:70. 2017. View Article : Google Scholar
|
|
8
|
Dunlavey CJ: Introduction to the
Hypothalamic-pituitary-adrenal axis: Healthy and dysregulated
stress responses, developmental stress and neurodegeneration. J
Undergrad Neurosci Educ. 16:R59–R60. 2018.PubMed/NCBI
|
|
9
|
DeMorrow S: Role of the
Hypothalamic-pituitary-adrenal axis in health and disease. Int J
Mol Sci. 19:9862018. View Article : Google Scholar
|
|
10
|
Chrousos GP: Stressors, stress, and
neuroendocrine integration of the adaptive response. The 1997 hans
selye memorial lecture. Ann N Y Acad Sci. 851:311–335. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Evanson NK, Tasker JG, Hill MN, Hillard CJ
and Herman JP: Fast feedback inhibition of the HPA axis by
glucocorticoids is mediated by endocannabinoid signaling.
Endocrinology. 151:4811–4819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang H, Gou X, Jiang T and Ouyang J: The
effects of microRNAs on glucocorticoid responsiveness. J Cancer Res
Clin Oncoly. 143:1005–1011. 2017. View Article : Google Scholar
|
|
13
|
Kawa MP, Sobuś A, Litwińska Z,
Osowicz-Korolonek L, Cymbaluk-Płoska A, Stecewicz I, Zagrodnik E,
Romanowska H, Walczak M, Syrenicz A and Machaliński B: Expression
of selected angiogenesis-related small microRNAs in patients with
abnormally increased secretion of glucocorticoids. Endokryno Pol.
70:489–495. 2019. View Article : Google Scholar
|
|
14
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Douma LG, Solocinski K, Masten SH, Barral
DH, Barilovits SJ, Jeffers LA, Alder KD, Patel R, Wingo CS, Brown
KD, et al: EDN1-AS, a novel long non-coding rna regulating
endothelin-1 in human proximal tubule cells. Front Physiol.
11:2092020. View Article : Google Scholar :
|
|
16
|
Silverman MN and Sternberg EM:
Glucocorticoid regulation of inflammation and its functional
correlates: From HPA axis to glucocorticoid receptor dysfunction.
Ann N Y Acad Sci. 1261:55–63. 2012. View Article : Google Scholar
|
|
17
|
Timmermans S, Souffriau J and Libert C: A
general introduction to glucocorticoid biology. Front Immunol.
10:1545. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Flynn BP: Glucocorticoid ultradian
rhythms. Curr Opin Endocrine Metabolic Res. 25:1003622022.
View Article : Google Scholar
|
|
19
|
Kalafatakis K, Russell GM, Ferguson SG,
Grabski M, Harmer CJ, Munafò MR, Marchant N, Wilson A, Brooks JC,
Thakrar J, et al: Glucocorticoid ultradian rhythmicity
differentially regulates mood and resting state networks in the
human brain: A randomised controlled clinical trial.
Psychoneuroendocrinology. 124:1050962021. View Article : Google Scholar :
|
|
20
|
Dickmeis T: Glucocorticoids and the
circadian clock. J Ndocrinol. 200:3–22. 2009. View Article : Google Scholar
|
|
21
|
Sevilla LM and Pérez P: Roles of the
Glucocorticoid and mineralocorticoid receptors in skin
pathophysiology. Int J Mol Sci. 19:19062018. View Article : Google Scholar :
|
|
22
|
Sarabdjitsingh RA, Meijer OC and de Kloet
ER: Specificity of glucocorticoid receptor primary antibodies for
analysis of receptor localization patterns in cultured cells and
rat hippocampus. Brain Res. 1331:1–11. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Desmet SJ and De Bosscher K:
Glucocorticoid receptors: Finding the middle ground. J Clin Invest.
127:1136–1145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nicolaides NC, Skyrla E, Vlachakis D,
Psarra AM, Moutsatsou P, Sertedaki A, Kossida S and Charmandari E:
Functional characterization of the hGRαT556I causing Chrousos
syndrome. Eur J Clin Invest. 46:42–49. 2016. View Article : Google Scholar
|
|
25
|
Paragliola RM, Papi G, Pontecorvi A and
Corsello SM: Treatment with synthetic glucocorticoids and the
hypothalamus-pituitary-Adrenal Axis. Int J Mol Sci. 18:22012017.
View Article : Google Scholar :
|
|
26
|
Mazaira GI, Zgajnar NR, Lotufo CM,
Daneri-Becerra C, Sivils JC, Soto OB, Cox MB and Galigniana MD: The
nuclear receptor field: A historical overview and future
challenges. Nucl Receptor Res. 5:1013202018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Porter BA, Ortiz MA, Bratslavsky G and
Kotula L: Structure and function of the nuclear receptor
superfamily and current targeted therapies of prostate cancer.
Cancers (Basel). 11:18522019. View Article : Google Scholar
|
|
28
|
Weikum ER, Okafor CD, D'Agostino EH,
Colucci JK and Ortlund EA: Structural analysis of the
glucocorticoid receptor ligand-binding domain in complex with
triamcinolone acetonide and a fragment of the atypical coregulator,
small heterodimer partner. Mol Pharmacol. 92:12–21. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan CK and Wahli W: A trilogy of
glucocorticoid receptor actions. Proc Natl Acad Sci USA.
113:1115–1117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nicolaides NC, Galata Z, Kino T, Chrousos
GP and Charmandari E: The human glucocorticoid receptor: Molecular
basis of biologic function. Steroids. 75:1–12. 2010. View Article : Google Scholar :
|
|
31
|
Kaziales A, Barkovits K, Marcus K and
Richter K: Glucocorticoid receptor complexes form cooperatively
with the Hsp90 co-chaperones Pp5 and FKBPs. Sci Rep. 10:10733.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Baker JD, Ozsan I, Rodriguez Ospina S,
Gulick D and Blair LJ: Hsp90 heterocomplexes regulate steroid
hormone receptors: From stress response to psychiatric disease. Int
J Mol Sci. 20:792018. View Article : Google Scholar
|
|
33
|
Louw A: GR Dimerization and the Impact of
GR Dimerization on GR protein stability and half-life. Front
Immunol. 10:1693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Robertson S, Hapgood JP and Louw A:
Glucocorticoid receptor concentration and the ability to dimerize
influence nuclear translocation and distribution. Steroids.
78:182–194. 2013. View Article : Google Scholar
|
|
35
|
Frego L and Davidson W: Conformational
changes of the glucocorticoid receptor ligand binding domain
induced by ligand and cofactor binding, and the location of
cofactor binding sites determined by hydrogen/deuterium exchange
mass spectrometry. Protein Sci. 15:722–730. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vandevyver S, Dejager L and Libert C: On
the trail of the glucocorticoid receptor: Into the nucleus and
back. Traffic. 13:364–374. 2012. View Article : Google Scholar
|
|
37
|
Hudson WH, Youn C and Ortlund EA: The
structural basis of direct glucocorticoid-mediated transrepression.
Nat Struct Mol Biol. 20:53–58. 2013. View Article : Google Scholar :
|
|
38
|
Groeneweg FL, van Royen ME, Fenz S, Keizer
VI, Geverts B, Prins J, de Kloet ER, Houtsmuller AB, Schmidt TS and
Schaaf MJ: Quantitation of glucocorticoid receptor DNA-binding
dynamics by single-molecule microscopy and FRAP. PLoS One.
9:e905322014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Quatrini L and Ugolini S: New insights
into the cell- and tissue-specificity of glucocorticoid actions.
Cell Mol Immunol. 18:269–278. 2021. View Article : Google Scholar
|
|
40
|
Petta I, Dejager L, Ballegeer M, Lievens
S, Tavernier J, De Bosscher K and Libert C: The Interactome of the
glucocorticoid receptor and its influence on the actions of
glucocorticoids in combatting inflammatory and infectious diseases.
Microbiol Mol Biol Rev. 80:495–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oeckinghaus A and Ghosh S: The NF-kappaB
family of transcription factors and its regulation. Cold Spring
Harb Perspect Biol. 1:a0000342009. View Article : Google Scholar
|
|
42
|
Rao NA, McCalman MT, Moulos P, Francoijs
KJ, Chatziioannou A, Kolisis FN, Alexis MN, Mitsiou DJ and
Stunnenberg HG: Coactivation of GR and NFKB alters the repertoire
of their binding sites and target genes. Genome Res. 21:1404–1416.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shimba A and Ikuta K: Control of immunity
by glucocorticoids in health and disease. Semin Immunopathol.
42:669–680. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wilson KS, Tucker CS, Al-Dujaili EA,
Holmes MC, Hadoke PW, Kenyon CJ and Denvir MA: Early-life
glucocorticoids programme behaviour and metabolism in adulthood in
zebrafish. J Endocrinol. 230:125–142. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rog-Zielinska EA, Craig MA, Manning JR,
Richardson RV, Gowans GJ, Dunbar DR, Gharbi K, Kenyon CJ, Holmes
MC, Hardie DG, et al: Glucocorticoids promote structural and
functional maturation of foetal cardiomyocytes: A role for PGC-1α.
Cell Death Differ. 22:1106–1116. 2015. View Article : Google Scholar
|
|
47
|
Whirledge S and DeFranco DB:
Glucocorticoid signaling in health and disease: Insights from
tissue-specific GR knockout mice. Endocrinology. 159:46–64. 2018.
View Article : Google Scholar :
|
|
48
|
Meszaros K and Patocs A: Glucocorticoids
influencing Wnt/β-catenin pathway; multiple sites, heterogeneous
effects. Molecules. 25:14892020. View Article : Google Scholar
|
|
49
|
Steptoe A and Kivimäki M: Stress and
cardiovascular disease. Nat Rev Cardiol. 9:360–370. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Duma D, Collins JB, Chou JW and Cidlowski
JA: Sexually dimorphic actions of glucocorticoids provide a link to
inflammatory diseases with gender differences in prevalence. Sci
Signal. 3:ra742010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Goodwin JE, Zhang J and Geller DS: A
critical role for vascular smooth muscle in acute
glucocorticoid-induced hypertension. J Am Soc Nephrol.
19:1291–1299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Goodwin JE, Feng Y, Velazquez H and Sessa
WC: Endothelial glucocorticoid receptor is required for protection
against sepsis. Proc Natl Acad Sci USA. 110:306–311. 2013.
View Article : Google Scholar :
|
|
53
|
Akalestou E, Genser L and Rutter GA:
Glucocorticoid metabolism in obesity and following weight loss.
Front Endocrinol (Lausanne). 11:592020. View Article : Google Scholar
|
|
54
|
Kuo T, McQueen A, Chen TC and Wang JC:
Regulation of glucose homeostasis by glucocorticoids. Adv Exp Med
Biol. 872:99–126. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ferris HA and Kahn CR: New mechanisms of
glucocorticoid-induced insulin resistance: Make no bones about it.
J Clin Invest. 122:3854–3857. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Vegiopoulos A and Herzig S:
Glucocorticoids, metabolism and metabolic diseases. Mol Cell
Endocrinol. 275:43–61. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Madalena KM and Lerch JK: The effect of
glucocorticoid and glucocorticoid receptor interactions on brain,
spinal cord, and glial cell plasticity. Neural Plast.
2017:86409702017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen H, Lombès M and Le Menuet D:
Glucocorticoid receptor represses brain-derived neurotrophic factor
expression in neuron-like cells. Mol Brain. 10:122017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Myers B, McKlveen JM and Herman JP:
Glucocorticoid actions on synapses, circuits, and behavior:
Implications for the energetics of stress. Front Neuroendocrinol.
35:180–196. 2014. View Article : Google Scholar
|
|
60
|
Joëls M: Corticosteroids and the brain. J
Endocrinol. 238:R121–R130. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fietta P and Fietta P: Glucocorticoids and
brain functions. Riv Biol. 100:403–418. 2007.
|
|
62
|
McEwen BS and Akil H: Revisiting the
stress concept: Implications for affective disorders. J Neurosci.
40:12–21. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Smoller JW and Finn CT: Family, twin, and
adoption studies of bipolar disorder. Am J Med Genet C Semin Med
Genet. 123C:48–58. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Geschwind DH and Flint J: Genetics and
genomics of psychiatric disease. Science. 349:1489–1494. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Akil H, Gordon J, Hen R, Javitch J,
Mayberg H, McEwen B, Meaney MJ and Nestler EJ: Treatment resistant
depression: A multi-scale, systems biology approach. Neurosci
Biobehav Rev. 84:272–288. 2018. View Article : Google Scholar
|
|
66
|
Wray NR, Ripke S, Mattheisen M,
Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM,
Andlauer TMF, et al: Genome-wide association analyses identify 44
risk variants and refine the genetic architecture of major
depression. Nat Genet. 50:668–681. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Aurbach EL, Inui EG, Turner CA, Hagenauer
MH, Prater KE, Li JZ, Absher D, Shah N, Blandino P Jr, Bunney WE,
et al: Fibroblast growth factor 9 is a novel modulator of negative
affect. Proc Natl Acad Sci USA. 112:11953–11958. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Salmaso N, Stevens HE, McNeill J, ElSayed
M, Ren Q, Maragnoli ME, Schwartz ML, Tomasi S, Sapolsky RM, Duman R
and Vaccarino FM: Fibroblast growth factor 2 modulates hypothalamic
pituitary axis activity and anxiety behavior through glucocorticoid
receptors. Biol Psychiatry. 80:479–489. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chaudhury S, Aurbach EL, Sharma V,
Blandino P Jr, Turner CA, Watson SJ and Akil H: FGF2 is a target
and a trigger of epigenetic mechanisms associated with differences
in emotionality: Partnership with H3K9me3. Proc Natl Acad Sci USA.
111:11834–11839. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tyrka AR, Parade SH, Eslinger NM, Marsit
CJ, Lesseur C, Armstrong DA, Philip NS, Josefson B and Seifer R:
Methylation of exons 1D, 1F, and 1H of the glucocorticoid receptor
gene promoter and exposure to adversity in preschool-aged children.
Dev Psychopathol. 27:577–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sinclair D, Fillman SG, Webster MJ and
Weickert CS: Dysregulation of glucocorticoid receptor co-factors
FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness.
Sci Rep. 3:35392013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sinclair D, Tsai SY, Woon HG and Weickert
CS: Abnormal glucocorticoid receptor mRNA and protein isoform
expression in the prefrontal cortex in psychiatric illness.
Neuropsychopharmacology. 36:2698–2709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sinclair D, Webster MJ, Fullerton JM and
Weickert CS: Glucocorticoid receptor mRNA and protein isoform
alterations in the orbitofrontal cortex in schizophrenia and
bipolar disorder. BMC Psychiatry. 12:842012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar :
|
|
76
|
Griffiths-Jones S: miRBase: MicroRNA
Sequences and Annotation. Curr Protoc Bioinformatics. Chapter 12:
Unit 12.9.1-10. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar :
|
|
78
|
Griffiths-Jones S: miRBase: The microRNA
sequence database. Methods Mol Biol. 342:129–138. 2006.PubMed/NCBI
|
|
79
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: MicroRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar :
|
|
80
|
Griffiths-Jones S: The microRNA Registry.
Nucleic Acids Res. 32:D109–D111. 2004. View Article : Google Scholar :
|
|
81
|
Kozomara A, Birgaoanu M and
Griffiths-Jones S: miRBase: From microRNA sequences to function.
Nucleic Acids Res. 47:D155–D162. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hollins SL and Cairns MJ: MicroRNA: Small
RNA mediators of the brains genomic response to environmental
stress. Prog Neurobiol. 143:61–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
de Kloet ER, Fitzsimons CP, Datson NA,
Meijer OC and Vreugdenhil E: Glucocorticoid signaling and
stress-related limbic susceptibility pathway: About receptors,
transcription machinery and microRNA. Brain Res. 1293:129–141.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Moisiadis VG and Matthews SG:
Glucocorticoids and fetal programming part 2: Mechanisms. Nat Rev
Endocrinol. 10:403–411. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Pufall MA: Glucocorticoids and cancer. Adv
Exp Med Biol. 872:315–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar :
|
|
88
|
Chen Y and Wang X: miRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48:D127–D131. 2020. View Article : Google Scholar :
|
|
89
|
Liu W and Wang X: Prediction of functional
microRNA targets by integrative modeling of microRNA binding and
target expression data. Genome Biol. 20:182019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ledderose C, Möhnle P, Limbeck E, Schütz
S, Weis F, Rink J, Briegel J and Kreth S: Corticosteroid resistance
in sepsis is influenced by microRNA-124-induced downregulation of
glucocorticoid receptor-α. Crit Care Med. 40:2745–2753. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang SS, Mu RH, Li CF, Dong SQ, Geng D,
Liu Q and Yi LT: microRNA-124 targets glucocorticoid receptor and
is involved in depression-like behaviors. Prog Neuropsychopharmacol
Biol Psychiatry. 79:417–425. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Roy B, Dunbar M, Shelton RC and Dwivedi Y:
Identification of MicroRNA-124-3p as a putative epigenetic
signature of major depressive disorder. Neuropsychopharmacology.
42:864–875. 2017. View Article : Google Scholar :
|
|
93
|
Dwivedi Y: microRNA-124: A putative
therapeutic target and biomarker for major depression. Expert Opin
Ther Targets. 21:653–656. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Vreugdenhil E, Verissimo CS, Mariman R,
Kamphorst JT, Barbosa JS, Zweers T, Champagne DL, Schouten T,
Meijer OC, de Kloet ER and Fitzsimons CP: MicroRNA 18 and 124a
down-regulate the glucocorticoid receptor: Implications for
glucocorticoid responsiveness in the brain. Endocrinology.
150:2220–2228. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Uchida S, Nishida A, Hara K, Kamemoto T,
Suetsugi M, Fujimoto M, Watanuki T, Wakabayashi Y, Otsuki K, McEwen
BS and Watanabe Y: Characterization of the vulnerability to
repeated stress in Fischer 344 rats: Possible involvement of
microRNA-mediated down-regulation of the glucocorticoid receptor.
Eur J Neurosci. 27:2250–2261. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Vallès A, Martens GJ, De Weerd P, Poelmans
G and Aschrafi A: MicroRNA-137 regulates a glucocorticoid
receptor-dependent signalling network: Implications for the
etiology of schizophrenia. J Psychiatry Neurosc. 39:312–320. 2014.
View Article : Google Scholar
|
|
97
|
Li S, Ma H, Yuan X, Zhou X, Wan Y and Chen
S: MicroRNA-382-5p targets nuclear receptor subfamily 3 group C
member 1 to regulate depressive-like behaviors induced by chronic
unpredictable mild stress in rats. Neuropsychiatr Dis Treat.
16:2053–2061. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kim J, Jeong D, Nam J, Aung TN, Gim JA,
Park KU and Kim SW: MicroRNA-124 regulates glucocorticoid
sensitivity by targeting phosphodiesterase 4B in diffuse large B
cell lymphoma. Gene. 558:173–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Liang YN, Tang YL, Ke ZY, Chen YQ, Luo XQ,
Zhang H and Huang LB: MiR-124 contributes to glucocorticoid
resistance in acute lymphoblastic leukemia by promoting
proliferation, inhibiting apoptosis and targeting the
glucocorticoid receptor. J Steroid Biochem Mol Biol. 172:62–68.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang
H, Hong M, Jiang T, Jiang Q, Lu J, et al: An oncogenic role of
miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by
targeting glucocorticoid receptor-alpha and cAMP/PKA pathways.
Leukemia. 26:769–777. 2012. View Article : Google Scholar
|
|
101
|
Riester A, Issler O, Spyroglou A, Rodrig
SH, Chen A and Beuschlein F: ACTH-dependent regulation of microRNA
as endogenous modulators of glucocorticoid receptor expression in
the adrenal gland. Endocrinology. 153:212–222. 2012. View Article : Google Scholar
|
|
102
|
Tessel MA, Benham AL, Krett NL, Rosen ST
and Gunaratne PH: Role for microRNAs in regulating glucocorticoid
response and resistance in multiple myeloma. Horm Cancer.
2:182–189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Sionov RV: MicroRNAs and
glucocorticoid-induced apoptosis in lymphoid malignancies. ISRN
Hematol. 2013:3482122013. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Vigorito E, Kohlhaas S, Lu D and Leyland
R: miR-155: An ancient regulator of the immune system. Immunol Rev.
253:146–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Elmesmari A, Fraser AR, Wood C, Gilchrist
D, Vaughan D, Stewart L, McSharry C, McInnes IB and
Kurowska-Stolarska M: MicroRNA-155 regulates monocyte chemokine and
chemokine receptor expression in Rheumatoid Arthritis. Rheumatology
(Oxford). 55:2056–2065. 2016. View Article : Google Scholar
|
|
106
|
Kurowska-Stolarska M, Alivernini S,
Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna
M, Fraser AR, Stolarski B, et al: MicroRNA-155 as a proinflammatory
regulator in clinical and experimental arthritis. Proc Natl Acad
Sci USA. 108:11193–11198. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang ZH, Liang YB, Tang H, Chen ZB, Li ZY,
Hu XC and Ma ZF: Dexamethasone down-regulates the expression of
microRNA-155 in the livers of septic mice. PLoS One. 8:e805472013.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zheng Y, Xiong S, Jiang P, Liu R, Liu X,
Qian J, Zheng X and Chu Y: Glucocorticoids inhibit
lipopolysaccharide-mediated inflammatory response by downregulating
microRNA-155: A novel anti-inflammation mechanism. Free Radic Biol
Med. 52:1307–1317. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Curtale G, Renzi TA, Drufuca L, Rubino M
and Locati M: Glucocorticoids downregulate TLR4 signaling activity
via its direct targeting by miR-511-5p. Eur J Immunol.
47:2080–2089. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Puimège L, Van Hauwermeiren F, Steeland S,
Van Ryckeghem S, Vandewalle J, Lodens S, Dejager L, Vandevyver S,
Staelens J, Timmermans S, et al: Glucocorticoid-induced
microRNA-511 protects against TNF by down-regulating TNFR1. EMBO
Mol Med. 7:1004–1017. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Clayton SA, Jones SW, Kurowska-Stolarska M
and Clark AR: The role of microRNAs in glucocorticoid action. J
Biol Chemistry. 293:1865–1874. 2018. View Article : Google Scholar
|
|
112
|
Davis TE, Kis-Toth K, Szanto A and Tsokos
GC: Glucocorticoids suppress T cell function by up-regulating
microRNA-98. Arthritis Rheum. 65:1882–1890. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhu QY, Liu Q, Chen JX, Lan K and Ge BX:
MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation
of MAPKs in macrophages. J Immunol. 185:7435–7442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: A comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Molitor is JK, McColl KS and Distelhorst
CW: Glucocorticoid-mediated repression of the oncogenic microRNA
cluster miR-17~92 contributes to the induction of Bim and
initiation of apoptosis. Mol Endocrinol. 25:409–420. 2011.
View Article : Google Scholar
|
|
116
|
Harada M, Pokrovskaja-Tamm K, Söderhäll S,
Heyman M, Grander D and Corcoran M: Involvement of miR17 pathway in
glucocorticoid-induced cell death in pediatric acute lymphoblastic
leukemia. Leuk Lymphoma. 53:2041–2050. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Palagani A, Op de Beeck K, Naulaerts S,
Diddens J, Sekhar Chirumamilla C, Van Camp G, Laukens K, Heyninck
K, Gerlo S, Mestdagh P, et al: Ectopic microRNA-150-5p
transcription sensitizes glucocorticoid therapy response in MM1S
multiple myeloma cells but fails to overcome hormone therapy
resistance in MM1R cells. PLoS One. 9:e1138422014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z,
Jiang M, Chen M, Wang X, Kang Y, Zhou Y, et al: Targeting the
miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance
in multiple myeloma. Cancer Res. 75:4384–4397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Murray MY, Rushworth SA, Zaitseva L,
Bowles KM and Macewan DJ: Attenuation of dexamethasone-induced cell
death in multiple myeloma is mediated by miR-125b expression. Cell
Cycle. 12:2144–2153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Kotani A, Ha D, Hsieh J, Rao PK, Schotte
D, den Boer ML, Armstrong SA and Lodish HF: miR-128b is a potent
glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia
cells and exerts cooperative effects with miR-221. Blood.
114:4169–4178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Cruz-Topete D, Oakley RH, Xu X and
Cidlowski JA: Glucocorticoid receptor signaling is critical for
microRNA Gender-specific regulation of gene expression in the adult
mouse heart. FASEB J. 31:687.4. 2017.
|
|
122
|
Jung SH, Wang Y, Kim T, Tarr A, Reader B,
Powell N and Sheridan JF: Molecular mechanisms of repeated social
defeat-induced glucocorticoid resistance: Role of microRNA. Brain
Behav Immun. 44:195–206. 2015. View Article : Google Scholar
|
|
123
|
Ko JY, Chuang PC, Ke HJ, Chen YS, Sun YC
and Wang FS: MicroRNA-29a mitigates glucocorticoid induction of
bone loss and fatty marrow by rescuing Runx2 acetylation. Bone.
81:80–88. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Schroeder M, Jakovcevski M, Polacheck T,
Drori Y, Luoni A, Röh S, Zaugg J, Ben-Dor S, Albrecht C and Chen A:
Placental miR-340 mediates vulnerability to activity based anorexia
in mice. Nat Commun. 9:15962018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Fu Q, Liu CJ, Zhang X, Zhai ZS, Wang YZ,
Hu MX, Xu XL, Zhang HW and Qin T: Glucocorticoid receptor regulates
expression of microRNA-22 and downstream signaling pathway in
apoptosis of pancreatic acinar cells. World J Gastroenterol.
24:5120–5130. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Buschmann D, González R, Kirchner B,
Mazzone C, Pfaffl MW, Schelling G, Steinlein O and Reithmair M:
Glucocorticoid receptor overexpression slightly shifts microRNA
expression patterns in triple-negative breast cancer. Int J Oncol.
52:1765–1776. 2018.PubMed/NCBI
|
|
127
|
Tejos-Bravo M, Oakley RH, Whirledge SD,
Corrales WA, Silva JP, García-Rojo G, Toledo J, Sanchez W,
Román-Albasini L, Aliaga E, et al: Deletion of hippocampal
Glucocorticoid receptors unveils sex-biased microRNA expression and
neuronal morphology alterations in mice. Neurobiol Stress.
14:1003062021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zheng D, Sabbagh JJ, Blair LJ, Darling AL,
Wen X and Dickey CA: MicroRNA-511 binds to FKBP5 mRNA, which
encodes a chaperone protein, and regulates neuronal
differentiation. J Biol Chemistry. 291:17897–17906. 2016.
View Article : Google Scholar
|
|
129
|
Pelleymounter LL, Moon I, Johnson JA,
Laederach A, Halvorsen M, Eckloff B, Abo R and Rossetti S: A novel
application of pattern recognition for accurate SNP and indel
discovery from high-throughput data: Targeted resequencing of the
glucocorticoid receptor co-chaperone FKBP5 in a Caucasian
population. Mol Genet Metab. 104:457–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Clemson CM, McNeil JA, Willard HF and
Lawrence JB: XIST RNA paints the inactive X chromosome at
interphase: Evidence for a novel RNA involved in nuclear/chromosome
structure. J Cell Biol. 132:259–275. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Swiezewski S, Liu F, Magusin A and Dean C:
Cold-induced silencing by long antisense transcripts of an
arabidopsis polycomb target. Nature. 462:799–802. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Houseley J, Rubbi L, Grunstein M,
Tollervey D and Vogelauer M: A ncRNA modulates histone modification
and mRNA induction in the yeast GAL gene cluster. Mol Cell.
32:685–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Reeves MB, Davies AA, McSharry BP,
Wilkinson GW and Sinclair JH: Complex I binding by a virally
encoded RNA regulates mitochondria-induced cell death. Science.
316:1345–1348. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Clark MB and Mattick JS: Long noncoding
RNAs in cell biology. Semin Cell Dev Biol. 22:366–376. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Martianov I, Ramadass A, Serra Barros A,
Chow N and Akoulitchev A: Repression of the human dihydrofolate
reductase gene by a non-coding interfering transcript. Nature.
445:666–670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Place RF and Noonan EJ: Non-coding RNAs
turn up the heat: An emerging layer of novel regulators in the
mammalian heat shock response. Cell Stress Chaperones. 19:159–172.
2014. View Article : Google Scholar :
|
|
142
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Mayama T, Marr AK and Kino T: Differential
expression of glucocorticoid receptor noncoding RNA repressor Gas5
in autoimmune and inflammatory diseases. Horm Metab Res.
48:550–557. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Lucafò M, Di Silvestre A, Romano M, Avian
A, Antonelli R, Martelossi S, Naviglio S, Tommasini A, Stocco G,
Ventura A, Decorti G and De Iudicibus S: Role of the long
non-coding RNA growth Arrest-Specific 5 in glucocorticoid response
in children with inflammatory bowel disease. Basic Clin Pharmacol
Toxicol. 122:87–93. 2018. View Article : Google Scholar
|
|
146
|
Gharesouran J, Taheri M, Sayad A,
Ghafouri-Fard S, Mazdeh M and Omrani MD: The growth arrest-specific
Transcript 5 (GAS5) and nuclear receptor Subfamily 3 Group C Member
1 (NR3C1): Novel Markers involved in multiple sclerosis. Int J Mol
Cell Med. 7:102–110. 2018.PubMed/NCBI
|
|
147
|
Esguerra JLS, Ofori JK, Nagao M, Shuto Y,
Karagiannopoulos A, Fadista J, Sugihara H, Groop L and Eliasson L:
Glucocorticoid induces human beta cell dysfunction by involving
riborepressor GAS5. LincRNA Mol Metab. 32:160–167. 2020. View Article : Google Scholar
|
|
148
|
Ketab FNG, Gharesouran J, Ghafouri-Fard S,
Dastar S, Mazraeh SA, Hosseinzadeh H, Moradi M, Javadlar M,
Hiradfar A, Rezamand A, et al: Dual biomarkers long non-coding RNA
GAS5 and its target, NR3C1, contribute to acute myeloid leukemia.
Exp Mol Pathol. 114:1043992020. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Pulido T, Adzerikho I, Channick RN,
Delcroix M, Galiè N, Ghofrani HA, Jansa P, Jing ZC, Le Brun FO,
Mehta S, et al: Macitentan and morbidity and mortality in pulmonary
arterial hypertension. N Engl J Med. 369:809–818. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Speed JS and Pollock DM: Endothelin,
kidney disease, and hypertension. Hypertension. 61:1142–1145. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Holm SJ, Sánchez F, Carlén LM, Mallbris L,
Ståhle M and O'Brien KP: HLA-Cw*0602 associates more strongly to
psoriasis in the Swedish population than variants of the novel
6p213 gene PSORS1C3. Acta Derm Venereol. 85:2–8. 2005. View Article : Google Scholar
|
|
152
|
Robinson PC, Leo PJ, Pointon JJ, Harris J,
Cremin K, Bradbury LA; Wellcome Trust Case Control Consortium;
Australasian Osteoporosis Genetics Consortium (AOGC); Stebbings S,
Harrison AA, et al: The genetic associations of acute anterior
uveitis and their overlap with the genetics of ankylosing
spondylitis. Genes Immun. 17:46–51. 2016. View Article : Google Scholar
|
|
153
|
Murphy TM, Crawford B, Dempster EL, Hannon
E, Burrage J, Turecki G, Kaminsky Z and Mill J: Methylomic
profiling of cortex samples from completed suicide cases implicates
a role for PSORS1C3 in major depression and suicide. Transl
Psychiatry. 7:e9892017. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Mirzadeh Azad F, Malakootian M and Mowla
SJ: lncRNA PSORS1C3 is regulated by glucocorticoids and fine-tunes
OCT4 expression in non-pluripotent cells. Sci Rep. 9:83702019.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Redfern AD, Colley SM, Beveridge DJ, Ikeda
N, Epis MR, Li X, Foulds CE, Stuart LM, Barker A, Russell VJ, et
al: RNA-induced silencing complex (RISC) Proteins PACT, TRBP, and
Dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad
Sci USA. 110:6536–6541. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Hatchell EC, Colley SM, Beveridge DJ, Epis
MR, Stuart LM, Giles KM, Redfern AD, Miles LE, Barker A, MacDonald
LM, et al: SLIRP, a small SRA binding protein, is a nuclear
receptor corepressor. Mol Cell. 22:657–668. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Vitellius G, Trabado S, Bouligand J,
Delemer B and Lombès M: Pathophysiology of Glucocorticoid
Signaling. Ann Endocrinol (Paris). 79:98–106. 2018. View Article : Google Scholar
|
|
159
|
Nicolaides NC, Geer EB, Vlachakis D,
Roberts ML, Psarra AM, Moutsatsou P, Sertedaki A, Kossida S and
Charmandari E: A novel mutation of the hGR gene causing Chrousos
syndrome. Eur J Clin Invest. 45:782–791. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Volden PA and Conzen SD: The influence of
glucocorticoid signaling on tumor progression. Brain Behav Immun.
30(Suppl): S26–S31. 2013. View Article : Google Scholar
|
|
161
|
Oakley RH and Cidlowski JA: Glucocorticoid
signaling in the heart: A cardiomyocyte perspective. J Steroid
Biochem Mol Biol. 153:27–34. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Oakley RH and Cidlowski JA: The biology of
the glucocorticoid receptor: New signaling mechanisms in health and
disease. J Allergy Clin Immunol. 132:1033–1044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Zabirowicz ES and Gan TJ: 34-Pharmacology
of Postoperative Nausea and Vomiting. Pharmacology and Physiology
for Anesthesia (Second Edition). Hemmings HC and Egan TD: Elsevier;
Philadelphia: pp. 671–692. 2019, View Article : Google Scholar
|
|
164
|
Flor M: R interface to D3 chord diagrams.
Chorddiag. https://github.com/mattflor/chorddiag. Accessed
February 18, 2022.
|