1. Introduction
Long non-coding RNAs (lncRNAs) represent a distinct
class of noncoding RNAs (ncRNAs) that exceed 200 nucleotides in
length and have been found to modulate cell proliferation,
apoptosis, invasiveness, drug resistance, and other pivotal
biological functions. These functions can be influenced at the
transcriptional, posttranscriptional, and epigenetic levels
(1,2). Numerous investigations in recent
years have underscored the critical role lncRNAs play in multiple
stages of disease pathogenesis (3). Among the early identified lncRNAs,
lncRNA H19 has been garnering significant interest across a variety
of disciplines. This review provides a comprehensive overview of
the characteristics and recent advancements in research pertaining
to lncRNA H19 in the field of gynecology.
2. Overview of lncRNA
Definition and function of lncRNA
The most recent update from the human GENCODE
project (accessible online: https://www.gencodegenes.org/) reveals that a mere
34.09% of the total human genes, numbering 19,901, encode proteins
(4). The broad category of RNA is
bifurcated into messenger RNA (mRNA), capable of protein encoding
and ncRNA, which lacks this capability. lncRNA, defined by a length
exceeding 200 nucleotides and absence of an open reading frame for
protein translation, forms the majority within the ncRNA category
(5). lncRNAs share several
defining characteristics (6): i)
The chromatin states of their coding genes, particularly in
relation to promoter H3K4me3 and transcription region H3K36me3,
mirror those of protein-encoding genes; ii) Their expression is
governed by a diverse array of shared transcription factors; iii)
As with protein-encoding genes, lncRNAs are transcribed by RNA
polymerase II, typically undergo spliceosomal splicing and possess
polyadenylated tails.
Historically, lncRNAs were considered functionally
irrelevant, mere 'noise' in the process of gene transcription.
However, advances in genome splicing array analysis and
high-throughput transcriptomic sequencing have revealed that
lncRNAs are not genomic byproducts, but are involved in numerous
physiological processes, affecting vital biological functions such
as cell proliferation, differentiation, chromosomal remodeling,
epigenetic regulation, transcription and posttranscriptional
modification (7-11). Hence, the potential involvement of
lncRNAs in human diseases has attracted considerable research
interest. Despite the identification of over 8,000 different types
of lncRNAs through the human genomic project, only a limited subset
of ~200 lncRNAs has been extensively studied (12). The majority of lncRNAs have been
found to reside in the nucleus, cytoplasm and organelles (primarily
the nucleus) and their expression displays marked tissue- and
time-specificity, along with significant variability across
different tissues, within different regions of the same organ and
across different disease stages (13-15).
Classification and biological mechanisms
of lncRNAs
lncRNAs can be categorized based on several
criteria. Functionally, they are segregated into cis-lncRNAs and
trans-lncRNAs (16). Genomically,
lncRNAs can be classified according to the location of the encoding
DNA fragment as intragenic lncRNAs, intergenic lncRNAs, divergent
lncRNAs, antisense lncRNAs, or enhancer lncRNAs (17). In terms of temporal and functional
characteristics, lncRNAs can be divided into
transcription-regulating lncRNAs and posttranscription-regulating
lncRNAs (18).
Presently, lncRNA studies predominantly focus on
data mining and functional verification. Data mining studies employ
methods such as lncRNA microarray detection, library construction
based on ribosomal RNA (rRNA) removal, lncRNA-seq and
high-throughput sequencing. Functional verification studies, on the
other hand, involve in vitro cell-based assays and in
vivo animal model experiments (19). lncRNAs exert their biological
effects via several mechanisms, which include: i) Gene
transcription regulation: lncRNAs can repress gene transcription by
forming complementary sequences with gene promoters, or they can
stimulate gene transcription by binding to the enhancers of
proximal protein-coding genes; ii) Posttranscriptional processing:
lncRNAs can influence mRNA splicing patterns by forming
double-stranded RNA through pairing with pre-mRNA. Additionally,
lncRNAs can modulate mRNA splicing by regulating the activity of
splicing factors (20); iii)
Translation regulation: lncRNA can function as a competitive
endogenous RNA (ceRNA) of microRNA (miRNA/miR) to modulate mRNA
expression levels, thereby effecting the translation of downstream
proteins (21,22); and iv) Epigenetic modification
regulation: lncRNAs can recruit chromatin modification complexes to
facilitate epigenetic modifications, leading to altered target gene
expression (23-25).
3. Multifaceted role of lncRNA H19
Structure and biological function of
lncRNA H19
The lncRNA H19 gene, found at the locus 11p15.5 on
human chromosome and at the terminus of chromosome 7 in mice, lies
~90 kilobases (kb) distant from the insulin-like growth factor 2
(IGF-2) gene. This gene extends over 2.5 kb, encompassing 5 exons
and 4 introns and forms a secondary structure of 16 helices and
several hairpin configurations (26). The lncRNA H19 gene transcribes an
RNA molecule of 2.3 kb length with 35 open reading frames. Notably,
this RNA molecule does not engage in RNA transcription and
translation processes, thus being incapable of protein encoding.
Therefore, this molecule is classified as a lncRNA and referred to
as lncRNA H19 (25,26).
lncRNA H19, one of the earliest discovered and most
extensively studied lncRNAs, exhibits significant conservation
across evolutionary species. It expresses maternal allelic
dominance and paternal imprinting, with the imprinting regulatory
region or differentially methylated region located 4 kb upstream,
modulating its expression and biological processes (26).
Predominantly, lncRNA H19 expression is pronounced
in embryonic and placental tissues, particularly in endodermal and
mesodermal derivatives during embryonic development. This pattern
implies the pivotal role of lncRNA H19 in embryogenesis and fetal
growth (27,28). Postnatal expression of lncRNA H19
diminishes significantly but maintains a residual level in select
tissues, including breast, uterus, myocardium, skeletal muscle and
the adrenal gland (29-34). During tissue regeneration or
tumorigenesis, lncRNA H19 function reactivates, participating in
damage regulation and repair, or acting as either a tumor
suppressor gene or an oncogene (35,36). Furthermore, the expression of
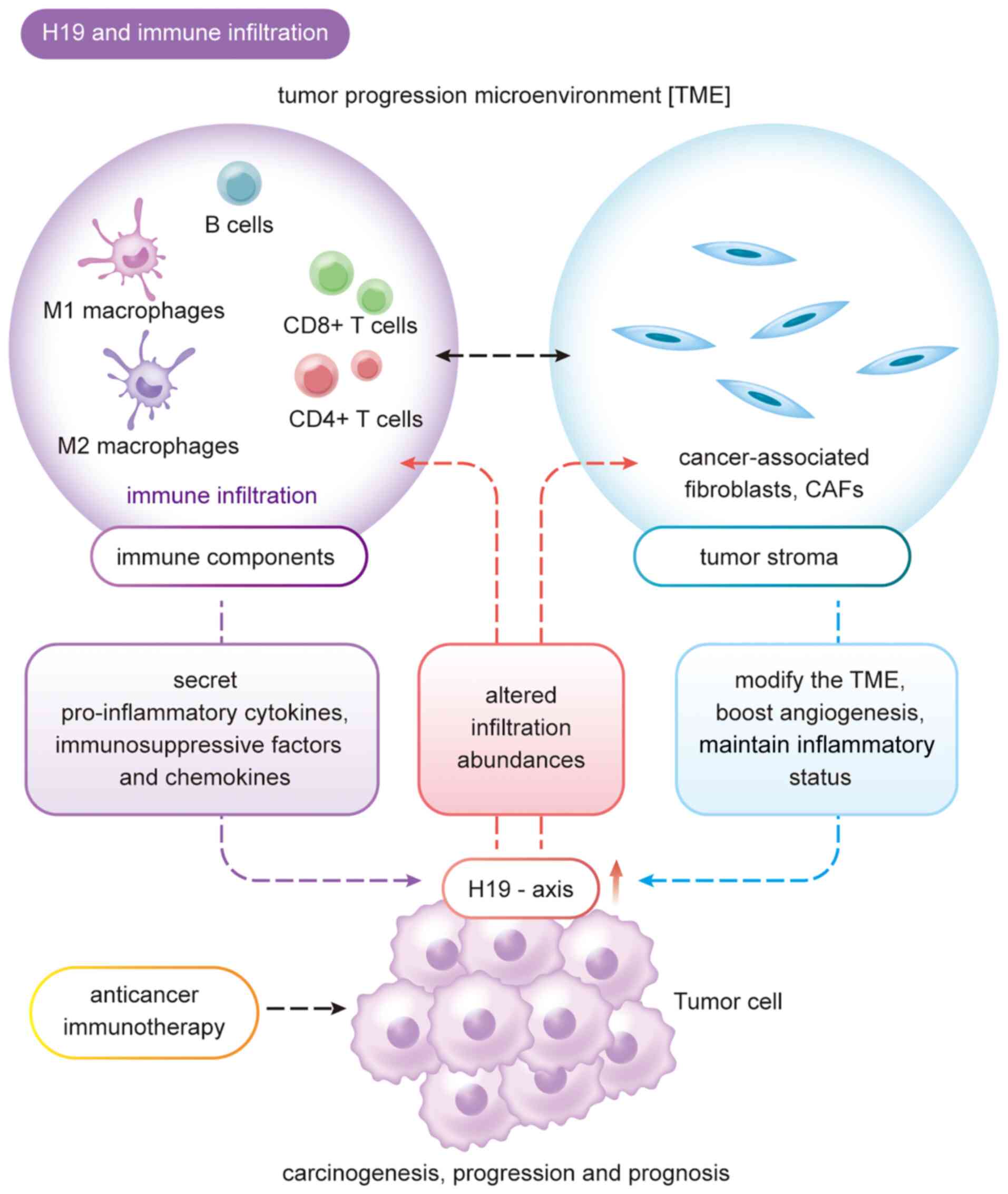
lncRNA H19 positively correlates with the infiltration of various
immune cells such as CD4+ T cells, CD8+ T cells, B cells, dendritic
cells, neutrophils and macrophages, as well as with diverse immune
markers in malignant tumors (37). Thus, lncRNA H19 probably serves a
critical role in malignant tumor genesis and progression, offering
insights for the identification of novel diagnostic biomarkers and
the development of targeted therapeutics (Fig. 1).
Research progress of lncRNA H19
In recent years, investigations into lncRNA H19 have
witnessed significant advancements across various diseases. The
majority of these studies concentrated on the mechanistic role of
lncRNA H19 in tumorigenesis, metastasis and drug resistance,
particularly focusing on the lncRNA H19-miRNA regulatory network
(Table I).
 | Table ILong non-coding RNA H19 and its
targeted microRNAs acting as competitive endogenous RNAs. |
Table I
Long non-coding RNA H19 and its
targeted microRNAs acting as competitive endogenous RNAs.
| First author,
year | Disease | MicroRNA | Target gene | Regulation
(microRNA/target gene) | (Refs.) |
|---|
| Wei, 2019 | Hepatocellular
carcinoma | miR-326 | Twist1 | Down/Up | (38) |
| Li, 2018 | Colorectal
adenocarcinoma | miR-194-5p | FoxM1 | Down/Up | (40) |
| Ding, 2018 | Colorectal
cancer | miR-29b-3p | PGRN | Down/Up | (41) |
| Lv, 2017 | Bladder cancer | miR-29b-3p | DNMT3B | Down/Up | (43) |
| Xiong, 2020 | Breast cancer | let-7 | LIN28 | Down/Up | (44) |
| Luan, 2018 | Malignant
melanoma | miR-106a-5p | E2F3 | Down/Up | (46) |
| Zheng, 2019 | Lung cancer | miR-675-5p | p53 | Down/Up | (47) |
| Sun, 2021 | Gastric cancer | let-7 | LDHA | Down/Up | (48) |
| He, 2017 | Renal
carcinoma | miR-29a-3p | E2F1 | Down/Up | (49) |
| Zhao, 2017 | Acute myelocytic
leukemia | miR-19a,b | ID2 | Down/Up | (50) |
| Sun, 2019 | Pancreatic
cancer | miR-194 | PFTK1 | Down/Up | (51) |
| Zhu, 2014 | Prostate
cancer | miR-675 | TGFBI | Up/Down | (52) |
| Fang, 2018 | Sepsis | miR-874 | AQP1 | Up/Down | (54) |
| Lu, 2018 | Pulmonary
fibrosis | miR-196a | COL1A1 | Down/Up | (55) |
One notable study reported elevated lncRNA H19 and
decreased miR-326 expression in a liver cancer cell line.
Bioinformatics analysis revealed that lncRNA H19 regulates cell
growth, migration and invasion by targeting Twist-related protein 1
(TWIST1), a downstream target of miR-326, acting as a ceRNA for
miR-326. Consequently, the lncRNA H19/miR-326/Twist1 axis was
confirmed as instrumental in the pathogenesis of liver cancer
(38). A separate study by Ye
et al (39) proposed that
lncRNA H19 fosters hepatocellular carcinoma (HCC) invasiveness by
triggering and activating the miR-193b/MAPK1 axis, thereby
mediating interplay between HCC and its immune
microenvironment.
Epithelial-mesenchymal transition (EMT) serves an
indispensable role in malignant tumor pathogenesis, mainly
disrupting cell adhesion and facilitating tumor metastasis. Li
et al (40) demonstrated
that the lncRNA H19/miR-194-5p/Forkhead box protein M1 axis
influences EMT, contributing to colorectal cancer progression via a
series of gene knockdown and overexpression experiments.
Furthermore, Ding et al (41) discovered that the lncRNA
H19/miR-29b-3p/progranulin axis encourages EMT in colorectal cancer
cell lines by modulating the Wnt signaling pathway. However, the
potential interaction between these two pathways remains uncertain,
necessitating further research.
In a groundbreaking study, Chen et al
(42) elucidated that lncRNA H19
inhibits vitamin D receptor expression by modulating miR-675-5p,
potentially explaining resistance to 1, 25 (OH) 2D3 in advanced
colon cancer. Concurrently, numerous studies on other malignancies,
including bladder, breast, glioma, melanoma, lung, gastric, renal
cancers and leukemia, demonstrated that lncRNA H19 regulates the
biological functions of downstream target genes by competitively
binding with miRNA, thereby affecting the immune infiltration level
and contributing to the poor prognosis of malignant tumor patients.
Thus, lncRNA H19 has been identified as a promising new target for
immunotherapy (43-51).
By contrast, lncRNA H19 can also serve as a tumor
suppressor, impeding metastasis in certain malignancies. One
seminal study showed reduced expression of lncRNA H19 and miR-675
in prostate cancer, particularly in M12 cell lines with high
tumorigenicity and metastatic potential. lncRNA H19 was found to
affect cell migration by modulating miR-675, significantly
inhibiting cell metastatic ability following lncRNA H19
overexpression. Subsequent double luciferase analysis demonstrated
that miR-675 could bind with the 3′ end of transforming growth
factor β inducing protein (TGFβI) mRNA, inhibiting its translation.
Hence, lncRNA H19 regulates the translation process of TGFBI by
modulating miR-675, thereby suppressing the metastasis of prostate
cancer (52). Another study
revealed a higher lncRNA H19 level in liver cancer tissues compared
with adjacent normal tissues. In this context, lncRNA H19 was shown
to bind to the hnRNPU/PCAF/RNAPol II protein complex, thereby
activating the miRNA-200 family, affecting EMT and consequently,
inhibiting metastasis (53).
Research pertaining to lncRNA H19's role in benign
diseases remains limited. In patients with sepsis, serum levels of
lncRNA H19 and aquaporin 1 (AQP1) were significantly diminished,
concomitant with increased miR-874 levels (54). The expression of lncRNA H19 was
negatively correlated with miR-874 and positively correlated with
AQP1. Moreover, the upregulation of lncRNA H19 significantly
reversed the abnormal expression of miR-874 and AQP1, as well as
lipopolysaccharide-induced anti-inflammatory cytokine secretion and
myocardial dysfunction in cardiomyocytes. Idiopathic pulmonary
fibrosis, characterized by pulmonary fibroblast aggregation and
extracellular matrix deposition, was associated with increased
lncRNA H19 expression in fibroblast proliferation induced by
transforming growth factor-1 (TGF-1) and bleomycin-induced
pulmonary fibrosis (55). lncRNA
H19 was found to target collagen, type I, α 1 by acting as a ceRNA
for miR-196a. The downregulation of lncRNA H19 was found to
ameliorate fibroblast activation and pulmonary fibrosis, affirming
the critical regulatory role of lncRNA H19 and its potential as a
prognostic indicator in idiopathic pulmonary fibrosis (55). Similarly, liver-specific lncRNA
H19 was discovered to enhance the enrichment of CD3+γδ+,
interleukin-4 and interleukin-17 producing CD4+ and CD8+ immune
cell populations in the livers and spleens of lncRNA H19-BDL mice.
This mechanism is facilitated through the zinc finger E-box-binding
homeobox 1 (ZEB1)/epithelial cell adhesion molecule signaling
pathway, leading to cholestatic liver fibrosis (56).
In conclusion, the biological significance of lncRNA
H19 extends across a multitude of physiological and pathological
processes. It is involved in embryonic development, tissue
regeneration, tumorigenesis, immune cell infiltration and serves a
role in both malignant and benign diseases. Furthermore, lncRNA H19
demonstrates both oncogenic and tumor-suppressive properties,
depending on the specific cellular context. It is clear that a more
profound understanding of the regulatory networks involving lncRNA
H19 could potentially unlock novel therapeutic strategies for a
wide array of diseases, ranging from benign conditions to malignant
tumors. However, given the current state of research, further
studies are necessary to conclusively elucidate the versatile and
context-dependent roles of lncRNA H19 in gynecological
pathologies.
4. lncRNA H19 and gynecologic
malignancies
lncRNA H19 and cervical cancer
Cervical cancer, a common gynecological malignancy,
exhibits the highest incidence in China, contributing to
approximately 10-15% of global female malignancy-related
mortalities, with an annual incidence of around 100,000 new cases
(57). A persistent infection
with high-risk strains of human papillomavirus (HPV) is
acknowledged as the primary predisposing factor for cervical
cancer. In a comprehensive analysis of oncogenic human viruses,
Hoppe-Seyler and Hoppe-Seyler (58) illuminated the potential role of
lncRNAs in modulating the HPV-induced carcinogenic process,
influencing the transcriptional regulation of target genes and
proteins either positively or negatively and thereby determining
the progression and eventual development of cervical cancer.
A seminal study conducted in 2012 by Gibb et
al (59) employed serial
analysis of gene expression to scrutinize 16 tissues exhibiting
varying grades of cervical intraepithelial neoplasia (CIN). They
discovered 1,056 lncRNAs with differential expression in CIN
compared with healthy cervical tissues, making this the first time
a differential lncRNA profile for CIN had been reported. Subsequent
research unearthed 617 lncRNAs implicated primarily in metabolic
and immunologic pathways associated with cervical cancer, including
the p53 and cAMP signaling pathways. The researchers developed a
prognostic signature based on eight lncRNAs (RUSC1-AS1, LINC01990,
LINC01411, LINC02099, H19, LINC00452, ADPGK-AS1 and C1QTNF1-AS1)
and revealed that the interaction among these lncRNAs correlated
significantly with adverse prognosis in cervical cancer patients
(60).
The role of lncRNA H19 in cervical cancer has been
under investigation for approximately two decades. An early study
in 2002 by Kim et al (61)
and associates examined the relationship between imprinting
regulatory deficiency and tumorigenesis at the epigenetic level in
32 cases of cervical squamous cell carcinoma across different
clinical stages. Their findings suggested that alterations in the
expression levels of lncRNA H19 and IGF-2 were implicated in
cervical cancer progression. In another study, Iempridee (62) demonstrated that lncRNA H19
expression was markedly elevated in cervical cancer cell lines,
with both overexpression and knockdown experiments indicating that
lncRNA H19 spurred cell proliferation and multicellular tumor
sphere formation. However, this did not significantly effect cell
apoptosis or migration. Subsequent research further elucidated the
role of lncRNA H19 in cervical cancer, indicating a higher
expression in cancerous compared with adjacent healthy tissues and
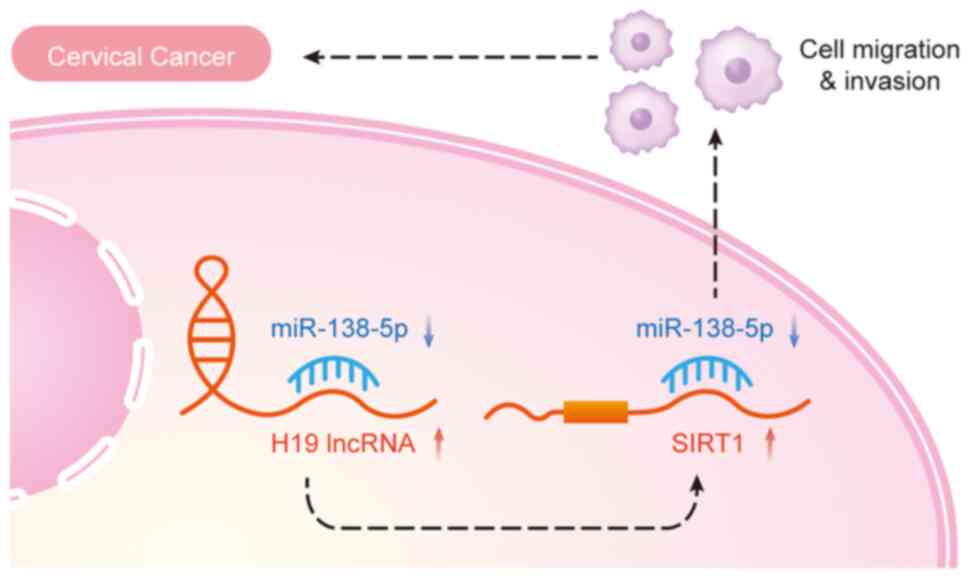
a negative correlation with miR-138-5p expression. Furthermore,
upregulation of miR-138-5p following lncRNA H19 knockdown with
small interfering RNA (siRNA) in HeLa and SIHA cells was observed.
SIRT1 was confirmed as a target of miR-138-5p in cervical cancer,
with the latter inhibiting tumor formation via SIRT1. This
suggested a possible mechanism by which lncRNA H19 might contribute
to cervical cancer, potentially by acting as a sponge for
miR-138-5p, thereby targeting SIRT1 (63) (Fig.
2).
The prognostic value of lncRNA H19 in cervical
cancer has been previously appraised. The analysis of patient data
with cervical cancer, featuring lncRNA H19 overexpression from The
Cancer Genome Atlas database, was carried out using Kaplan-Meier
survival curves and the Cox proportional hazard model. This
analysis evaluated parameters such as the hazard ratio (HR),
overall survival, disease-free survival, relapse-free survival,
metastasis-free survival and progression-free survival. Results
denoted lncRNA H19 as an independent prognostic indicator for
cervical cancer (HR=4.099; P<0.05) (64).
lncRNA H19 and endometrial cancer
Endometrial cancer, representing the most prevalent
gynecological malignancy in industrialized nations, has exhibited
an alarming upward trajectory in incidence over recent years. This
trend, particularly noticeable in North America and Eastern Europe,
aligns with demographic shifts such as aging and increasing obesity
rates. In 2018, the United States documented 63,230 new cases,
alongside 11,350 mortalities, underscoring the gravity of this
public health concern (65,66).
Endometrial cancer manifests predominantly in two
subtypes. Type I, constituting 80-90% of incidences, typically
afflicts premenopausal or postmenopausal women, frequently
evidencing positivity for estrogen and progesterone receptors
within tumor tissues and generally offering patients a favorable
prognosis (67). By contrast,
Type II emerges from postmenopausal atrophic endometrium and is
often associated with a dismal prognosis (68). Utilizing resources like the Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
databases, researchers have identified 172 differentially expressed
lncRNAs in endometrial cancer tissues relative to the normal
endometrium (69). However,
rigorous investigations into the implications of these lncRNAs in
endometrial cancer remain scarce. Available studies predominantly
center on MALAT1, HOTAIR, lncRNA H19 and SRA (70). Intriguingly, the expression
patterns of lncRNAs in endometrial cancer are subtype-specific; for
instance, the ovarian carcinoma magnifying lncRNA is downregulated
in type II and upregulated in type I endometrial cancer (71).
In an earlier study conducted in 2004, Tanos et
al (72) and colleagues
examined lncRNA H19 and IGF-2 gene expressions in normal,
hyperplastic and cancerous endometrial samples. They found an
elevated frequency and level of lncRNA H19 in endometrial cancer,
which correlated with the degree of differentiation, while IGF-2
expression remained consistent. In line with the mechanisms
observed in other cancers, lncRNA H19 primarily exerts its
biological influence through the lnc-miRNA axis and affects the EMT
process. Zhang et al (73)
revealed a significant upregulation of lncRNA H19 and Homeobox
protein Hox-A10 (HOXA10), coupled with miR-612 downregulation, in
endometrial cancer. Survival analysis suggested an inverse
correlation between lncRNA H19 and patient survival rates. In
vitro experiments confirmed that lncRNA H19 regulates cellular
proliferation by competitively targeting HOXA10 via miR-612 in
endometrial cancer, contributing to its pathogenesis (73).
Further research employing RT-PCR to determine the
expression profile of lncRNA H19 in endometrial cancer revealed
markedly elevated levels in tumor tissues compared with adjacent
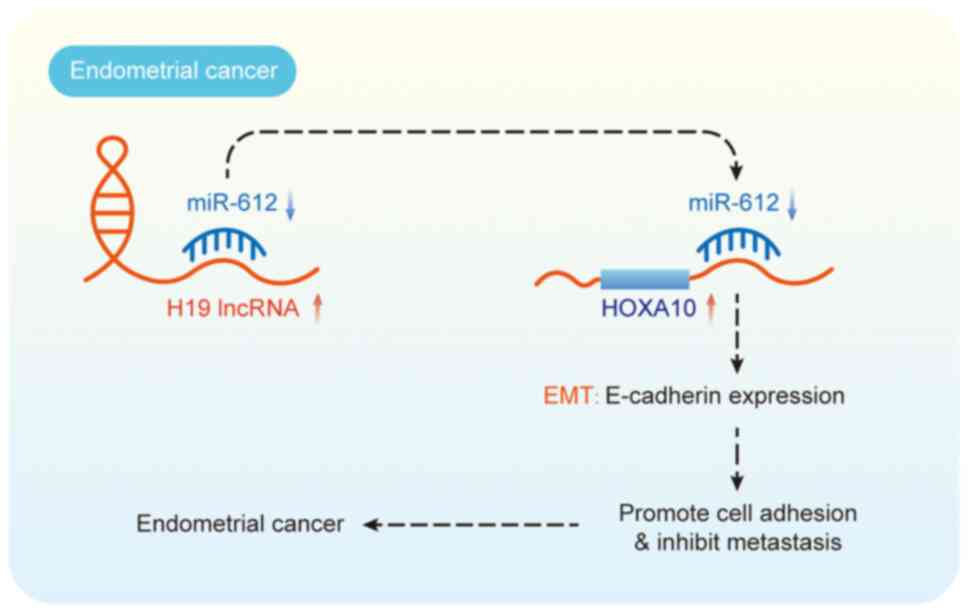
tissues. Knockdown experiments demonstrated that lncRNA H19
suppression curtailed the migratory and invasive potential of
endometrial cancer cells without influencing cell growth.
Concurrently, increased E-cadherin expression and unchanged
vimentin levels indicated a partial EMT reversal, suggesting that
lncRNA H19 modulates endometrial cancer cell invasion by regulating
EMT (74) (Fig. 3).
Research conducted by Peng et al (64) and colleagues underlined the
prognostic significance of lncRNA H19 in female malignancies,
noting its overexpression correlates with poorer endometrial cancer
prognosis. Given that diabetes and obesity represent risk factors
for endometrial cancer, metformin (a common therapeutic
intervention for type 2 diabetes) has been used in preventative and
therapeutic strategies against malignancies, including endometrial
cancer (75). The molecular
underpinnings of its anticancer effects, however, remain elusive.
Notably, Yan et al (76)
demonstrated that metformin inhibited the migration and invasion of
endometrial cancer cells by downregulating lncRNA H19 expression
via DNA methylation. This compelling finding underlines a potential
mechanistic rationale for the use of metformin in the prevention
and treatment of endometrial cancer.
lncRNA H19 and ovarian cancer
Ovarian cancer represents a principal source of
mortality among gynecological malignancies, with diagnosis often
occurring at advanced stages due to the paucity of efficacious
detection methodologies. According to 2018 data, ovarian cancer
engendered 22,240 new cases and led to 14, 070 mortalities in the
United States alone (65). The
application of GO and KEGG pathway analyses in discerning the
lncRNA profile of 30 human ovarian carcinoma tissues in relation to
20 normal ovarian tissues has yielded salient results: 795
upregulated and 2,075 downregulated lncRNAs were detected in
ovarian cancer tissues compared with their normal counterparts. The
identified differentially expressed lncRNAs were categorized into
four classes, namely, enhancer lncRNAs adjacent to coding genes,
HOX cluster, long intergenic non-coding RNAs (lincRNAs) adjacent to
coding genes and Rinn lincRNAs. This classification laid the
groundwork for subsequent in-depth functional studies (77).
In high-grade serous ovarian cancer, a different
study documented 1,511 upregulated and 2,778 downregulated lncRNAs
associated with normal fallopian tube tissues, corroborating the
upregulation of FAS-AS1, AK130076, RP11-199F11.2 and AC093818.1 and
the downregulation of GTSE1-AS1 in ovarian cancer tissues (78). Noteworthy is the observed
variation in the biological roles of lncRNAs across the various
stages of ovarian cancer (79).
Correlations were found between the expression level of lncRNAs and
diagnostic time, prognosis and chemotherapy resistance (80). Furthermore, the involvement of
estriol-regulated lncRNA and its polymorphic methylation in ovarian
cancer was elucidated (81).
Additionally, a total of 26 lncRNAs, comprising 5 upregulated and
21 downregulated, were flagged as potential multidrug-resistant
lncRNAs based on data integration of published research and
cisplatin-resistant lncRNAs in ovarian cancer cell lines or
patients. Importantly, lncRNA CTD-2589M5.4 demonstrated
coexpression with the multidrug resistance genes ABCB1, ABCB4,
ABCC3 and ABCG2, thereby providing key insights into the roles of
lncRNAs in the multidrug resistance process (81).
Scientific inquiry into the role of lncRNA H19 in
ovarian malignancies began with explorations of the H19 and IGF-2
genes. An early study in 2000 by Chen et al (82) evaluated the imprinting status of
the H19 gene and IGF-2 in a selection of ovarian cancer samples,
ovarian tumors of low malignant potential and normal ovarian
tissues. The results unveiled an imprinting deletion in all cases
of advanced stage, hinting at a potential association between the
H19 gene and IGF-2 imprinting deletion and advanced ovarian cancer.
Subsequent research by Zhu et al (83) confirmed elevated expression of
lncRNA H19 in ovarian cancer samples compared with normal ovarian
tissue. Upon knockdown of lncRNA H19, cell cycle arrest was
observed alongside aberrant expression of apoptosis-related
proteins and inhibited proliferation in OV90 and SKOV3 ovarian
cancer cell lines. Li et al (84) probed the mechanistic underpinnings
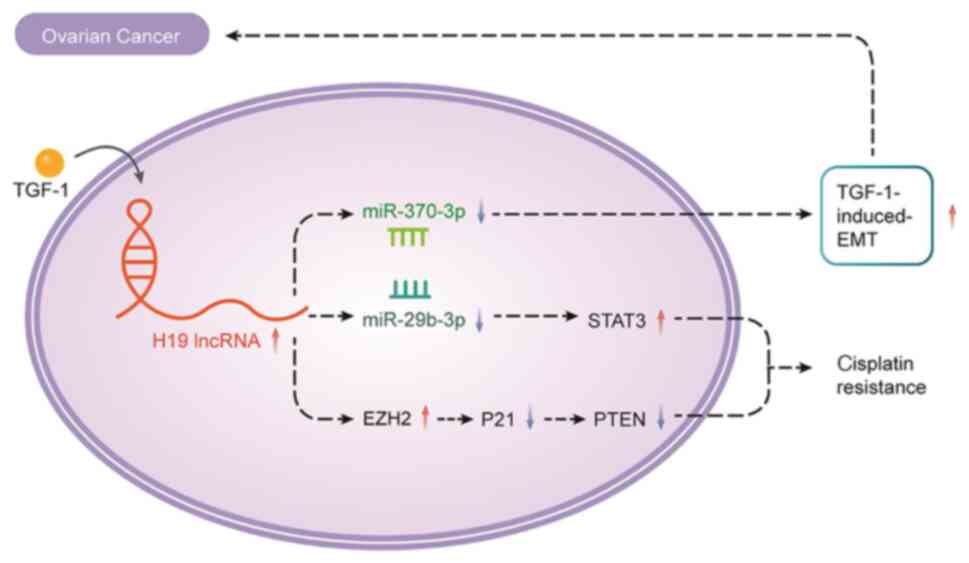
of the involvement of lncRNA H19 in ovarian cancer and found that
TGF-1 treatment led to an upregulation of lncRNA H19 and a
downregulation of miR-370-3p. Moreover, H19 gene knockdown or
miR-370-3p overexpression attenuated the TGF-1-induced EMT, while
H19 overexpression or miR-370-3p gene knockdown facilitated this
process. Consequently, this study affirmed the role of lncRNA H19
as a ceRNA sponging miR-370-3p, thereby influencing the promotion
of TGF-1-induced EMT (Fig.
4).
Platinum-based chemotherapy represents a cornerstone
of advanced ovarian cancer treatment, but the emergence of platinum
resistance presents a significant impediment to successful
therapeutic outcomes. The mechanism underlying the platinum
resistance of ovarian cancer is complex, governed by a myriad of
molecular entities. One study by Tian et al (85) discovered that lncRNA-H19 and STAT3
levels were significantly elevated, while miR-29b-3p levels were
diminished, in carboplatin-resistant epithelial ovarian cancer.
Furthermore, lncRNA-H19 functioned as a ceRNA of miR-29b-3p,
instigating the derepression of the downstream miR-29b-3p target,
STAT3 and consequently conferring chemoresistance. Importantly,
silencing of lncRNA-H19 augmented carboplatin efficacy, suggesting
a potential pathway for enhancing chemotherapy.
Ongoing efforts are underway to discover novel
therapeutic strategies for patients exhibiting platinum resistance.
Sajadpoor et al discerned that valproic acid (VPA), a
prevalent antiepileptic drug, could mitigate ovarian cancer via
lncRNA regulation (86). Post-VPA
treatment, the cisplatin-resistant ovarian cancer cell line
A2780-DR exhibited downregulated levels of lncRNA H19 and EZH2 and
upregulated levels of p21 and PTEN. Moreover, lncRNA H19 knockdown
instigated apoptosis and revived the sensitivity of A2780/CP cells
to cisplatin, intimating that VPA may attenuate lncRNA H19 in
ovarian cancer and inhibit cisplatin resistance via the lncRNA
H19/EZH2/p21/PTEN signaling pathway (86).
Ginsenoside, an active constituent of traditional
Chinese medicine Panax ginseng, exerts antitumor effects via a
multitude of intricate mechanisms. Zheng et al (87) revealed that the Warburg effect, a
fundamental metabolic characteristic of malignant tumors, is
modulated in part by lncRNAs. Ginsenoside 20 (S)-RG3 mitigated the
competitive inhibitory effect of lncRNA H19 on miR-324-5p in
ovarian cancer, enhancing the inhibitory effect of miR-324-5p on
PKM2, thereby suppressing the Warburg effect and ultimately
inhibiting tumorigenesis. This discovery provides a promising
foundation for the therapeutic application of ginsenosides in
ovarian cancer (87).
5. lncRNA H19 and benign gynecological
diseases
lncRNA H19 and endometriosis
Endometriosis is pathologically characterized by the
ectopic growth of active endometrial tissue beyond the confines of
the uterine cavity, a condition that precipitates associated
clinical symptoms. This disease, predominantly affecting 10-15% of
women within their reproductive years, manifests as a benign
disorder that paradoxically displays malignant biological
tendencies, thereby detrimentally affecting women's physical and
psychological wellbeing (88,89). The etiological underpinnings of
endometriosis remain elusive, with an unmet need for robust
biological markers capable of early-stage diagnosis and predicting
disease recurrence. An examination of reports pertaining to ncRNAs
and endometriosis, extracted from PubMed, MEDLINE and Google
Scholar databases between 2000 and 2016, suggested that ncRNA
regulatory dysregulation significantly contributes to
endometriosis. Importantly, microRNAs, lncRNAs and siRNAs serve
pivotal roles in endometriosis by modulating inflammatory
processes, cellular proliferation, angiogenesis and tissue
remodeling (90). The
identification of ncRNA biomarkers relevant to endometriosis offers
novel avenues for diagnostic methods and therapeutic targets,
potentially revolutionizing clinical management for patients
suffering from endometriosis (90).
Over recent years, the lncRNA expression profile in
endometriosis has been elucidated (91-94). Notably, Cui et al (91) explored the differences in lncRNA
expression between eutopic and normal endometrium during the
proliferative phase, predicting potential lncRNA targets based on
cis- and trans-regulatory actions. Additionally, they annotated the
functions of coexpressed mRNAs, identifying a total of 9,924 novel
ncRNA transcripts. Of these, 86 lncRNAs and 1,228 mRNAs
demonstrated differential expression between endometriosis patients
and controls. GO and KEGG analyses indicated these lncRNAs were
significantly involved in the biological processes and signaling
pathways characteristic of endometriosis. Wang et al
(93) identified a total of 1,277
differentially expressed lncRNAs (comprising 488 upregulated and
789 downregulated lncRNAs) between the eutopic and normal
endometrium in the late secretory phase. These lncRNAs interacted
with 1,216 differentially expressed mRNAs in a coexpression network
encompassing both coding and noncoding genes. Notably, upregulated
lncRNAs were predominantly associated with cell cycle regulation,
including DNA replication and cell cycle progression, while
downregulated lncRNAs were linked to immune-related pathways, such
as tumor necrosis factor (TNF), Wnt and mitogen-activated protein
kinase (MAPK) signaling pathways. According to these findings,
lncRNA expression profiles in endometriosis are subject to
fluctuations across menstrual cycle phases. A study by Wang et
al (94) further explored
serum lncRNA levels in endometriosis patients, identifying 1,682
differentially expressed lncRNAs, with 125 lncRNAs concurrently
detected in serum and tissue samples. Multiple reports have
detailed the mechanisms through which lncRNAs contribute to
endometriosis (95-98). For instance, actin filament
associated protein 1 antisense RNA1 (AFAP1-AS1) has been
demonstrated to promote EMT by modulating the transcription factor
ZEB1 (95). Additionally, the
lncRNA MALAT1 has been found to mediate the autophagy of
endometrial stromal cells under hypoxic conditions (96). The long intergenic non-protein
coding RNA01279 (lincRNA01279) has been implicated in endometriosis
progression via regulation of the cell cycle (97).
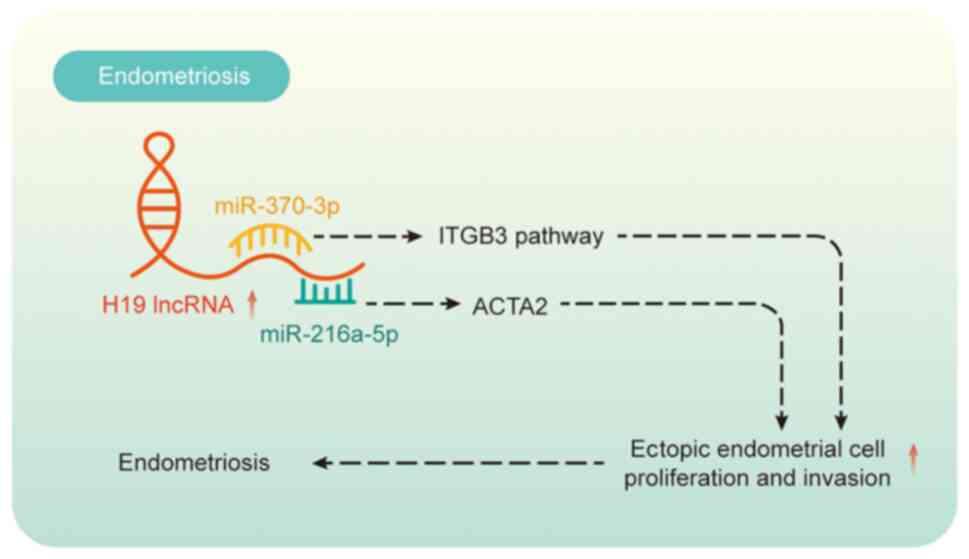
lncRNA H19 represents the first lncRNA subjected to
functional research within the context of endometriosis. In a prior
investigation, the current authors identified that perturbations in
the lncRNA H19/miR-124-3p/integrin β3 (ITGB3) pathway modulated
ectopic endometrial cell proliferation and invasion in
endometriosis (99). Another
study found that lncRNA H19 regulated the invasion and migration of
endometriosis eutopic endometrial stromal cells by modulating
miR-216a-5p and actin α2 and that estrogen promoted eutopic
endometrial stromal cell invasion and migration via lncRNA H19
(100) (Fig. 5). The current authors also
examined the overexpression of lncRNA H19 in endometriosis and its
potential as a novel biomarker for predicting disease recurrence.
Their findings indicated that lncRNA H19 expression in ectopic
endometrium correlated with infertility, disease recurrence,
bilateral ovarian lesions, elevated CA125 levels and revised
American Fertility Society stages. Moreover, the receiver operating
characteristic curve analysis revealed that, when lncRNA H19
expression in ectopic endometrium exceeded 0.0277, the sensitivity
and specificity for predicting recurrence were 90.9 and 61.0%
respectively, implying the potential involvement of lncRNA H19 in
the pathogenesis of endometriosis and its potential utility as a
novel predictor of disease recurrence (101).
The immune system serves a vital role in the
pathophysiology of endometriosis, particularly Th17 cells, which
are a subset of pro-inflammatory T-helper cells known to exacerbate
the disease. According to a study of immune cells, lncRNA H19 and
IER3 expressions were found to be downregulated in mononuclear
cells from peritoneal fluid (PFMCs) of patients with endometriosis
or under Th17 differentiation conditions (102). By contrast, miR-342-3p
expression was up-regulated and the percentage of Th17 cells was
increased in these PFMCs. The overexpression of lncRNA H19
decreased IL-17 level and the percentage of Th17 cells/CD4+ T
cells, indicating that lncRNA H19 overexpression can suppress Th17
cell differentiation and endometrial stromal cell (ESC)
proliferation through the miR-342-3p/IER3 pathway (102). A possible therapeutic approach
could involve strategies to overexpress lncRNA H19 in PFMCs of
patients with endometriosis. Overexpression of lncRNA H19 could be
achieved using gene therapy techniques, such as the use of viral
vectors to deliver the lncRNA H19 gene into target cells. However,
these techniques would need to be carefully optimized to ensure
targeted delivery and avoid off-target effects. Monitoring the
efficacy of the treatment would be crucial. This could be done by
tracking the expression levels of lncRNA H19, miR-342-3p and IER3,
as well as the percentage of Th17 cells in the PFMCs of patients.
Changes in these markers would provide valuable information about
the biological effect of the therapy and its effect on the disease
progression.
lncRNA H19 and infertility
Animal studies have revealed differential lncRNA
profiles pre- and post-pregnancy, as well as across distinct stages
of embryo implantation, pointing towards a critical role of lncRNAs
in the regulation of embryo implantation (103,104). Inadequate endometrial
receptivity constitutes the primary reason for failed pregnancies.
In patients undergoing in vitro fertilization-embryo
transfer with repeated implantation failure (RIF), the endometrial
expression of lncRNAs and mRNAs revealed 1,202 differentially
expressed genes compared with healthy endometrium, including 742
lncRNAs and 460 mRNAs. The implicated targets were primarily
associated with tumor necrosis factor, Toll-like receptor and
nuclear factor-kappa B (NF-κB) signaling pathways (105).
Huang et al (106) revealed a distinct lncRNA profile
in the endometrium of patients with RIF compared with healthy
counterparts, as determined by weighted gene coexpression network
analysis, suggesting the potential involvement of lncRNAs in
modulating endometrial receptivity. Building on this, Feng et
al (107) constructed a
lncRNA-mRNA network associated with implantation failure and
identified six crucial lncRNAs and their ceRNA subnetwork through
unsupervised clustering, GO, KEGG and coexpression module analyses.
This revealed key biological processes such as immune activity,
growth factor binding, vascular proliferation, apoptosis and
steroid synthesis, suggesting that lncRNAs may work in concert to
ensure endometrial receptivity for embryo implantation (107).
The H19 gene expression in the endometrium was found
to be cyclical, fluctuating across the menstrual cycle (108). Early research demonstrated a
decline in H19 expression with ovulation, which remained low until
the early secretory phase, gradually increased on the 21st day of
the cycle, reaching a peak and persisting until the end of the
menstrual cycle (108).
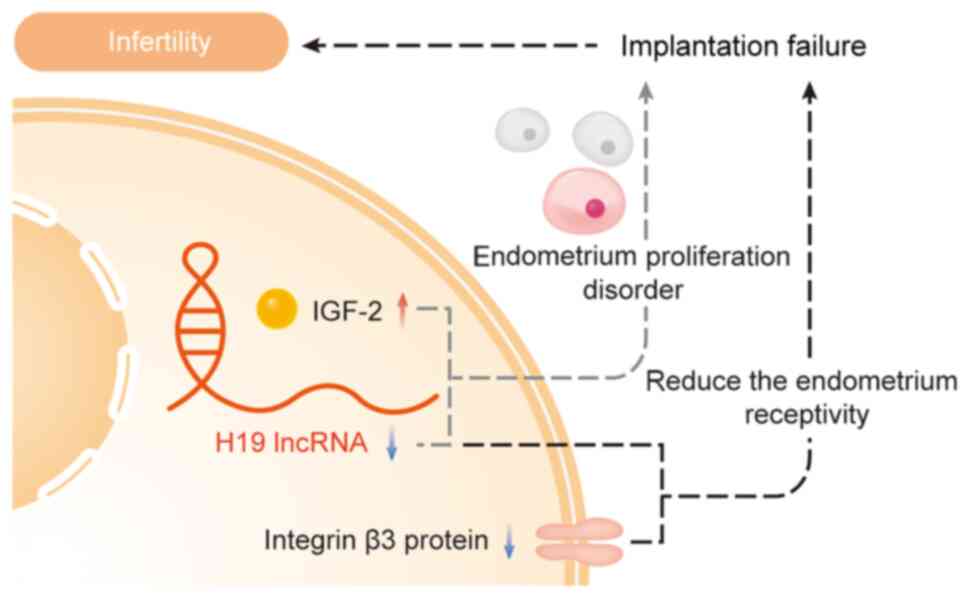
Furthermore, Korucuoglu et al (109) discovered that decreased H19 and
increased IGF-2 expressions in the endometrium of unexplained
infertility cases negatively affected endometrial proliferation,
leading to implantation failure, thus proposing H19 as a potential
biomarker or gene therapy target in assisted reproductive
technologies (109). Zeng et
al (110) reported decreased
levels of lncRNA H19 and integrin β3 protein in the endometrium
during the middle luteal phase in infertile patients with RIF
compared with controls. They proposed that the downregulation of
lncRNA H19 diminished integrin β3 protein expression, consequently
reducing endometrial receptivity and causing implantation failure
(Fig. 6).
Post-implantation, a proportion of pregnancies
suffer loss during embryonic development, with the etiology largely
undetermined. Existing literature posits an essential role for
lncRNAs in embryogenesis and fetal growth (27,28), prompting investigations into the
link between lncRNAs and spontaneous abortions. Wang et al
(111) performed lncRNA profile
analyses of blastocysts and decidua from 16 spontaneous abortion
cases and 16 induced abortion cases, identifying differentially
expressed lncRNAs implicated in six biological pathways, including
infection and inflammation, metabolism, signal and transcription
regulation, smooth muscle contraction, cell process and coagulation
and infection and inflammation. The lncRNAs associated with these
pathways were proposed as the key etiological factors for
spontaneous abortions. In the embryonic chorion tissue from
spontaneous abortion cases, lncRNA H19 was observed to regulate
trophoblastic spheroid adhesion to endometrial stromal cells by
targeting ITGB3 via let-7 modulation (112).
In a further study, Hu et al (113) evaluated the expression of lncRNA
H19, miR-200 and ZEB1 in the chorionic tissues of 20 patients with
spontaneous abortions and 20 women who had undergone induced
abortions. The findings showed diminished lncRNA H19 and ZEB1
expressions in the spontaneous abortion group compared with the
induced abortion group, with moderate positive correlation between
their levels. By contrast, there was no discernible difference in
miR-200 expression across the two groups, suggesting the possible
involvement of lncRNA H19 in spontaneous abortions via ZEB1
regulation.
Moreover, it was reported that the lncRNA H19
expression was notably lower in the spontaneous abortion group
compared with the control group, with positive associations
observed with Bcl-2 and GPX4 expressions and negative correlation
with Bax expression. Furthermore, the silencing of lncRNA H19 was
found to downregulate Bcl-2 and GPX4 expressions, but upregulated
Bax expression in HTR-8/SVneo trophoblast cells. These observations
suggest that lncRNA H19 may have significant roles in spontaneous
abortions by facilitating apoptosis and ferroptosis (114).
6. Effects of lncRNA H19 on immunity
lncRNAs have gained significant attention for their
wide-ranging roles in gene regulation, cellular functions and
disease etiology. Among the multitude of lncRNAs under rigorous
examination, lncRNA H19 stands out due to its multifaceted
involvement in immune response modulation across various
pathological contexts. In gastric cancer (GC), a highly prevalent
malignancy, lncRNA H19 has been found to regulate immune cell
infiltration via miR-378a-5p/SERPINH1 signaling (115). This mechanism is believed to
contribute significantly to GC progression, highlighting the
dynamic interaction between H19 and immune cells. Moreover, lncRNA
H19 has been implicated in the regulation of aerobic glycolysis and
cell proliferation, serving an instrumental role in immune evasion
in GC cells via the miR-519d-3p/lactate dehydrogenase A axis
(48). Markedly, research has
indicated that knocking down H19 can reduce the immunosuppressive
effect of GC cells, suggesting a promising therapeutic avenue
(48). Further emphasizing the
immune-related effects of lncRNA H19 is its role in thyroid
carcinoma (THCA). lncRNA H19 has been found to be differentially
expressed in THCA, with its expression associated with immune cell
infiltration in the disease. Specifically, it was found to be
positively correlated with the infiltration level of various immune
cells such as CD4+ T cells, CD8+ T cells, B cells, dendritic cells,
neutrophils and macrophages. Moreover, lncRNA H19 was associated
with multiple immune markers, underscoring its potential role in
shaping the immune landscape of THCA (116).
In the context of systemic lupus erythematosus
(SLE), an autoimmune disease, lncRNA H19 exhibits a significant
upregulation and is associated with immune dysregulation in bone
marrow-derived mesenchymal stem cells (BMMSCs) (117). The mechanism, notably, involves
H19 inhibiting the production of the interleukin-2 cytokine, an
important modulator of immune responses. This finding not only
broadens the known landscape of lncRNA H19-mediated
immunomodulation but also positions H19 as a potential therapeutic
target for SLE. Beyond its association with cancer and autoimmune
disease, lncRNA H19 also serves a critical role in mammary
epithelial cells. Overexpression of lncRNA H19 promotes cell
proliferation and enhances the expression of proteins related to
cell structure and function, such as β-casein and tight
junction-related proteins (118). Furthermore, it influences immune
responses by increasing the expression of inflammatory factors
[TNF-α, interleukin 6, chemokine (C-X-C motif) ligand 2 and
chemokine (C-C motif) ligand 5] and activating the NF-κB signal
pathway, thus potentially linking H19 to inflammatory disorders of
the breast, including mastitis and possibly even breast cancer
(118,119).
Overall, the effect of lncRNA H19 on immune
regulation is diverse and context-dependent, with its involvement
in a wide range of diseases and cellular functions. A recurring
theme in the literature is the relationship between lncRNA H19 and
inflammatory responses. lncRNA H19 has been associated with
heightened inflammation, frequently observed in diseases such as
endometriosis and ovarian cancer, probably by modulating immune
response genes and regulating cytokine production. The upregulation
of lncRNA H19 perpetuates an inflammatory microenvironment,
favoring disease progression through several mechanisms including
promoting cytokine production, fostering immune cell recruitment
and proliferation and enhancing expression of inflammatory genes
(120). The potential value of
lncRNA H19 as a therapeutic target to dampen excessive
inflammation, offering potential treatment avenues for inflammatory
gynecological conditions.
The effect of H19 on the immune response and
gynecological diseases does not exist in isolation. The intricate
network of microRNAs, epigenetic modifications and signal
transduction pathways form a convoluted regulatory network with H19
at the center (121,122). The functionality of this network
depends on a delicate balance, the disturbance of which can lead to
pathological states (37,120). Consequently, a comprehensive
understanding of these interactions is key for the development of
effective therapeutic strategies targeting lncRNA H19 (37). However, despite the wealth of
evidence linking lncRNA H19 with immune regulation, the precise
mechanisms through which lncRNA H19 exerts its effects remain to be
elucidated. The inherent complexities of lncRNAs, combined with the
multifaceted nature of immune responses, present challenges to
fully elucidating the roles of H19 in gynecological disease
pathogenesis (123). Future
studies, perhaps employing advanced techniques such as single-cell
RNA sequencing and high-throughput chromatin conformation capture,
could aid in painting a more comprehensive picture of H19's
functional role in these diseases.
7. Conclusion
In summary, the lncRNA H19 has been discerned to
serve a critical role in the etiology of diverse benign and
malignant pathologies in gynecological health, employing a
multiplicity of mechanisms. This molecular entity holds promise as
an efficacious biomarker for early-stage detection and prognostic
assessment of gynecological conditions, while also potentially
offering a novel avenue for therapeutic intervention. However, the
existing body of research concerning lncRNA H19 in the realm of
gynecology is still in its nascent stage and the intricate role
that H19 serves in disease initiation, advancement and modulation
of the immune milieu in gynecology remains predominantly
elusive.
Therefore, it is imperative that the labyrinthine
regulatory network of lncRNA H19 be further elucidated,
particularly in the context of its influence on various
gynecological diseases. Advancements in cellular and molecular
biology, coupled with cutting-edge gene technology, pave the way
for this endeavor. Moreover, the execution of additional clinical
and fundamental experiments, utilizing larger sample sizes, will
augment our understanding of the functionality and mechanistic
underpinnings of lncRNA H19 in gynecological diseases. In turn,
this acquired knowledge can expedite the evolution of diagnostic
procedures and therapeutic strategies in gynecology, thereby
elevating patient care standards.
Availability of data and materials
Not applicable.
Authors' contributions
WX conceived the idea for the present study and
drafted and revised the manuscript. YW and SL contributed to the
revision, the provision of some of the data and databases, and also
contributed to the proofreading and language correction of the
manuscript. WX and KH performed the literature search and prepared
the figures. YW and SL critically discussed and revised the
manuscript, and provided some updates on the literature and
clinical trials. All authors read and approved the final
manuscript. Data sharing is not applicable to this article.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Research Foundation for
Clinical Project of Shanghai Municipal Health Commission (grant no.
202140366), the Talents of Jinshan Hospital of Fudan University
(grant no. JYRC-2020-1), the Key Subject Development Project of
Jinshan Hospital of Fudan University-Obstetrics and Gynecology
(grant no. ZDXK-2023-6) and the National Natural Science Foundation
of China (grant no. 82001500).
References
|
1
|
Li D, Zhang J, Li X, Chen Y, Yu F and Liu
Q: Insights into lncRNAs in Alzheimer's disease mechanisms. RNA
Biol. 18:1037–1047. 2021. View Article : Google Scholar
|
|
2
|
Zhu L, Li N, Sun L, Zheng D and Shao G:
Non-coding RNAs: The key detectors and regulators in cardiovascular
disease. Genomics. 113:1233–1246. 2021. View Article : Google Scholar
|
|
3
|
Lu Q, Guo P, Liu A, Ares I,
Martínez-Larrañaga MR, Wang X, Anadón A and Martínez MA: The role
of long noncoding RNA in lipid, cholesterol, and glucose metabolism
and treatment of obesity syndrome. Med Res Rev. 41:1751–1774. 2021.
View Article : Google Scholar
|
|
4
|
Ferlita A, Battaglia R, Andronico F,
Caruso S, Cianci A, Purrello M and Pietro CD: Non-coding RNAs in
endometrial physiopathology. Int J Mol Sci. 19:21202018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar
|
|
6
|
Bin X, Hongjian Y, Xiping Z, Bo C, Shifeng
Y and Binbin T: Research progresses in roles of LncRNA and its
relationships with breast cancer. Cancer Cell Int. 18:1792018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liau WS, Samaddar S, Banerjee S and Bredy
TW: On the functional relevance of spatiotemporally-specific
patterns of experience-dependent long noncoding RNA expression in
the brain. RNA Biol. 18:1025–1036. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yarani R, Mirza AH, Kaur S and Pociot F:
The emerging role of lncRNAs in inflammatory bowel disease. Exp Mol
Med. 50:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peng H, Wan LY, Liang JJ, Zhang YQ, Ai WB
and Wu JF: The roles of lncRNA in hepatic fibrosis. Cell Biosci.
8:632018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jantrapirom S, Koonrungsesomboon N,
Yoshida H, M Candeias M, Pruksakorn D and Lo Piccolo L: Long
noncoding RNA-dependent methylation of nonhistone proteins. Wiley
Interdiscip Rev RNA. 12:e16612021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khalili-Tanha G and Moghbeli M: Long
non-coding RNAs as the critical regulators of doxorubicin
resistance in tumor cells. Cell Mol Biol Lett. 26:392021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li N, Zhang H, Hu K and Chu J: A novel
long non-coding RNA-based prognostic signature for renal cell
carcinoma patients with stage IV and histological grade G4.
Bioengineered. 12:6275–6285. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bo C, Li N, He L, Zhang S and An Y: Long
non-coding RNA ILF3-AS1 facilitates hepatocellular carcinoma
progression by stabilizing ILF3 mRNA in an m6A-dependent
manner. Hum Cell. 34:1843–1854. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo Y, Ge P, Wang M and Chen H, Liu J, Wei
T, Jiang Y, Qu J and Chen H: Research progress of DLX6-AS1 in human
cancers. Hum Cell. 34:1642–1652. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bhatti GK, Khullar N, Sidhu IS, Navik US,
Reddy AP, Reddy PH and Bhatti JS: Emerging role of non-coding RNA
in health and disease. Metab Brain Dis. 36:1119–1134. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nojima T and Proudfoot NJ: Mechanisms of
lncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev
Mol Cell Biol. 23:389–406. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yan W, Hu H and Tang B: Progress in
understanding the relationship between long noncoding RNA and
endometriosis. Eur J Obstet Gynecol Reprod Biol X. 5:1000672019.
View Article : Google Scholar
|
|
20
|
Constanty F and Shkumatava A: lncRNAs in
development and differentiation: From sequence motifs to functional
characterization. Development. 148:dev1827412021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang H, Bian C, Tu S, Yin F, Guo P, Zhang
J, Song X, Liu Q, Chen C and Han Y: Integrated analysis of
lncRNA-miRNA-mRNA ceRNA network in human aortic dissection. BMC
Genomics. 22:7242021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Qian Y, Zhang C, Wang W, Qiao Y,
Song H, Li L, Guo J, Lu D and Deng X: LncRNA LINC00473 is involved
in the progression of invasive pituitary adenoma by upregulating
KMT5A via ceRNA-mediated miR-502-3p evasion. Cell Death Dis.
12:5802021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Irwin AB, Bahabry R and Lubin FD: A
putative role for lncRNAs in epigenetic regulation of memory.
Neurochem Int. 150:1051842021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Xie R, Gu P, Huang M, Han J, Dong
W, Xie W, Wang B, He W, Zhong G, et al: Long noncoding RNA LBCS
inhibits self-renewal and chemoresistance of bladder cancer stem
cells through epigenetic silencing of SOX2. Clin Cancer Res.
25:1389–1403. 2019. View Article : Google Scholar
|
|
25
|
Wang D, Sun Y, Lin L, Sang Y, Yang F,
Zhang J, Jia L, Xu Z and Zhang W: Long non-coding RNA H19 and the
underlying epigenetic function in response to DNA damage of lung
cancer cells. Am J Transl Res. 13:5835–5850. 2021.PubMed/NCBI
|
|
26
|
Hernández-Aguilar AI, Luciano-Villa CA,
Tello-Flores VA, Beltrán-Anaya FO, Zubillaga-Guerrero MI and
Flores-Alfaro E: Dysregulation of lncRNA-H19 in cardiometabolic
diseases and the molecular mechanism involved: A systematic review.
Expert Rev Mol Diagn. 21:809–821. 2021. View Article : Google Scholar
|
|
27
|
Matsuzaki H, Okamura E, Takahashi T,
Ushiki A, Nakamura T, Nakano T, Hata K, Fukamizu A and Tanimoto K:
De novo DNA methylation through the 5′-segment of the H19 ICR
maintains its imprint during early embryogenesis. Development.
142:3833–3844. 2015.PubMed/NCBI
|
|
28
|
Gabory A, Jammes H and Dandolo L: The H19
locus: role of an imprinted non-coding RNA in growth and
development. Bioessays. 32:473–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Sun J and Yang F: The role of long
non-coding RNA H19 in breast cancer. Oncol Lett. 19:7–16.
2020.PubMed/NCBI
|
|
30
|
Wang Y and Gao WJ: Long non-coding RNA-H19
promotes ovarian cancer cell proliferation and migration via the
microRNA-140/Wnt1 axis. Kaohsiung J Med Sci. 37:768–775. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Zhang Y, Hu Q, Egranov SD, Xing Z,
Zhang Z, Liang K, Ye Y, Pan Y, Chatterjee SS, et al: Functional
significance of gain-of-function H19 lncRNA in skeletal muscle
differentiation and anti-obesity effects. Genome Med. 13:1372021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang L and Qi L: The role and mechanism of
long non-coding RNA H19 in stem cell osteogenic differentiation.
Mol Med. 27:862021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Su W, Huo Q, Wu H, Wang L, Ding X, Liang
L, Zhou L, Zhao Y, Dan J and Zhang H: The function of LncRNA-H19 in
cardiac hypertrophy. Cell Biosci. 11:1532021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tian X, Wang Y, Lu Y, Wang W, Du J, Chen
S, Zhou H, Cai W and Xiao Y: Conditional depletion of macrophages
ameliorates cholestatic liver injury and fibrosis via lncRNA-H19.
Cell Death Dis. 12:6462021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ghafouri-Fard S, Esmaeili M and Taheri M:
H19 lncRNA: Roles in tumorigenesis. Biomed Pharmacother.
123:1097742020. View Article : Google Scholar
|
|
36
|
Yang J, Qi M, Fei X, Wang X and Wang K:
LncRNA H19: A novel oncogene in multiple cancers. Int J Biol Sci.
17:3188–3208. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shermane Lim YW, Xiang X, Garg M, Le MT,
Li-Ann Wong A, Wang L and Goh BC: The double-edged sword of H19
lncRNA: Insights into cancer therapy. Cancer Lett. 500:253–262.
2021. View Article : Google Scholar
|
|
38
|
Wei LQ, Li L, Lu C, Liu J, Chen Y and Wu
H: Involvement of H19/miR-326 axis in hepatocellular carcinoma
development through modulating TWIST1. J Cell Physiol.
234:5153–5162. 2019. View Article : Google Scholar
|
|
39
|
Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu
P, Yu W, Xu L, Zhao Y and Yu J: Macrophages-induced long noncoding
RNA H19 up-regulation triggers and activates the miR-193b/MAPK1
axis and promotes cell aggressiveness in hepatocellular carcinoma.
Cancer Lett. 469:310–322. 2020. View Article : Google Scholar
|
|
40
|
Li CF, Li YC, Wang Y and Sun LB: The
effect of LncRNA H19/miR-194-5p axis on the epithelial-mesenchymal
transition of colorectal adenocarcinoma. Cell Physiol Biochem.
50:196–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding D, Li C, Zhao T, Li D, Yang L and
Zhang B: LncRNA H19/miR-29b-3p/PGRN axis promoted
epithelial-mesenchymal transition of colorectal cancer cells by
acting on Wnt signaling. Mol Cells. 41:423–435. 2018.PubMed/NCBI
|
|
42
|
Chen S, Bu D, Ma Y, Zhu J, Chen G, Sun L,
Zuo S, Li T, Pan Y, Wang X, et al: H19 overexpression induces
resistance to 1,25(OH)2D3 by targeting VDR through miR-675-5p in
colon cancer cells. Neoplasia. 19:226–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lv M, Zhong Z, Huang M, Tian Q, Jiang R
and Chen J: lncRNA H19 regulates epithelial-mesenchymal transition
and metastasis of bladder cancer by miR-29b-3p as competing
endogenous RNA. Biochim Biophys Acta Mol Cell Res. 1864:1887–1899.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xiong H, Shen J, Chen Z, Yang J, Xie B,
Jia Y, Jayasinghe U, Wang J, Zhao W, Xie S, et al: H19/let-7/Lin28
ceRNA network mediates autophagy inhibiting epithelial-mesenchymal
transition in breast cancer. Int J Oncol. 56:794–806.
2020.PubMed/NCBI
|
|
45
|
Zhao W, Lin X, Han H, Zhang H, Li X, Jiang
C and Feng M: Long noncoding RNA H19 contributes to the
proliferation and autophagy of glioma cells through mTOR/ULK1
pathway. Neuroreport. 32:352–358. 2021.PubMed/NCBI
|
|
46
|
Luan W, Zhou Z, Ni X, Xia Y, Wang J, Yan Y
and Xu B: Long non-coding RNA H19 promotes glucose metabolism and
cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J
Cancer Res Clin Oncol. 144:531–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng ZH, Wu DM, Fan SH, Zhang ZF, Chen GQ
and Lu J: Upregulation of miR-675-5p induced by lncRNA H19 was
associated with tumor progression and development by targeting
tumor suppressor p53 in non-small cell lung cancer. J Cell Biochem.
120:18724–18735. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun L, Li J, Yan W, Yao Z, Wang R, Zhou X,
Wu H, Zhang G, Shi T and Chen W: H19 promotes aerobic glycolysis,
proliferation, and immune escape of gastric cancer cells through
the microRNA-519d-3p/lactate dehydrogenase A axis. Cancer Sci.
112:2245–2259. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
He H, Wang N, Yi X, Tang C and Wang D:
Long non-coding RNA H19 regulates E2F1 expression by competitively
sponging endogenous miR-29a-3p in clear cell renal cell carcinoma.
Cell Biosci. 7:652017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao TF, Jia HZ, Zhang ZZ, Zhao XS, Zou
YF, Zhang W, Wan J and Chen XF: LncRNA H19 regulates ID2 expression
through competitive binding to hsa-miR-19a/b in acute myelocytic
leukemia. Mol Med Rep. 16:3687–3693. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun Y, Zhu Q, Yang W, Shan Y, Yu Z, Zhang
Q and Wu H: LncRNA H19/miR-194/PFTK1 axis modulates the cell
proliferation and migration of pancreatic cancer. J Cell Biochem.
120:3874–3886. 2019. View Article : Google Scholar
|
|
52
|
Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng
R, Wang Y, Huang J, Xu M, Yan J and Yu J: lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting TGFBI. FEBS J.
281:3766–3775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou
WP, Huo XS, Xu D, Bi HS, Wang F and Sun SH: Epigenetic activation
of the MiR-200 family contributes to H19-mediated metastasis
suppression in hepatocellular carcinoma. Carcinogenesis.
34:577–586. 2013. View Article : Google Scholar
|
|
54
|
Fang Y, Hu J, Wang Z, Zong H, Zhang L,
Zhang R and Sun L: LncRNA H19 functions as an aquaporin 1
competitive endogenous RNA to regulate microRNA-874 expression in
LPS sepsis. Biomed Pharmacother. 105:1183–1191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lu Q, Guo Z, Xie W, Jin W, Zhu D, Chen S
and Ren T: The lncRNA H19 mediates pulmonary fibrosis by regulating
the miR-196a/COL1A1 axis. Inflammation. 41:896–903. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Song Y, Liu C, Liu X, Trottier J, Beaudoin
M, Zhang L, Pope C, Peng G, Barbier O, Zhong X, et al: H19 promotes
cholestatic liver fibrosis by preventing ZEB1-mediated inhibition
of epithelial cell adhesion molecule. Hepatology. 66:1183–1196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
You L, Lv Z, Li C, Ye W, Zhou Y, Jin J and
Han Q: Worldwide cancer statistics of adolescents and young adults
in 2019: A systematic analysis of the global burden of disease
study 2019. ESMO Open. 6:1002552021. View Article : Google Scholar
|
|
58
|
Hoppe-Seyler F and Hoppe-Seyler K:
Emerging topics in human tumor virology. Int J Cancer.
129:1289–1299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gibb EA, Becker-Santos DD, Enfield KS,
Guillaud M, Niekerk Dv, Matisic JP, Macaulay CE and Lam WL:
Aberrant expression of long noncoding RNAs in cervical
intraepithelial neoplasia. Int J Gynecol Cancer. 22:1557–1563.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhong Q, Lu M, Yuan W, Cui Y, Ouyang H,
Fan Y, Wang Z, Wu C, Qiao J and Hang J: Eight-lncRNA signature of
cervical cancer were identified by integrating DNA methylation,
copy number variation and transcriptome data. J Transl Med.
19:582021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kim SJ, Park SE, Lee C, Lee SY, Jo JH, Kim
JM and Oh YK: Alterations in promoter usage and expression levels
of insulin-like growth factor-II and H19 genes in cervical
carcinoma exhibiting biallelic expression of IGF-II. Biochim
Biophys Acta. 1586:307–315. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Iempridee T: Long non-coding RNA H19
enhances cell proliferation and anchorage-independent growth of
cervical cancer cell lines. Exp Biol Med (Maywood). 242:184–193.
2017. View Article : Google Scholar
|
|
63
|
Ou L, Wang D, Zhang H, Yu Q and Hua F:
Decreased expression of miR-138-5p by lncRNA H19 in cervical cancer
promotes tumor proliferation. Oncol Res. 26:401–410. 2018.
View Article : Google Scholar
|
|
64
|
Peng L, Yuan XQ, Liu ZY, Li WL, Zhang CY,
Zhang YQ, Pan X, Chen J, Li YH and Li GC: High lncRNA H19
expression as prognostic indicator: Data mining in female cancers
and polling analysis in non-female cancers. Oncotarget.
8:1655–1667. 2017. View Article : Google Scholar :
|
|
65
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Takenaka K, Chen BJ, Modesitt SC, Byrne
FL, Hoehn KL and Janitz M: The emerging role of long non-coding
RNAs in endometrial cancer. Cancer Genet. 209:445–455. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Urick ME and Bell DW: Clinical
actionability of molecular targets in endometrial cancer. Nat Rev
Cancer. 19:510–521. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Brooks RA, Fleming GF, Lastra RR, Lee NK,
Moroney JW, Son CH, Tatebe K and Veneris JL: Current
recommendations and recent progress in endometrial cancer. CA
Cancer J Clin. 69:258–279. 2019.PubMed/NCBI
|
|
69
|
Xu J, Qian Y, Ye M, Fu Z, Jia X, Li W, Xu
P, Lv M, Huang L, Wang L, et al: Distinct expression profile of
lncRNA in endometrial carcinoma. Oncol Rep. 36:3405–3412. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Smolle MA, Bullock MD, Ling H, Pichler M
and Haybaeck J: Long non-coding RNAs in endometrial carcinoma. Int
J Mol Sci. 16:26463–26472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Vallone C, Rigon G, Gulia C, Baffa A,
Votino R, Morosetti G, Zaami S, Briganti V, Catania F, Gaffi M, et
al: Non-coding RNAs and endometrial cancer. Genes (Basel).
9:1872018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tanos V, Ariel I, Prus D, De-Groot N and
Hochberg A: H19 and IGF2 gene expression in human normal,
hyperplastic, and malignant endometrium. Int J Gynecol Cancer.
14:521–525. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang L, Wang DL and Yu P: LncRNA H19
regulates the expression of its target gene HOXA10 in endometrial
carcinoma through competing with miR-612. Eur Rev Med Pharmacol
Sci. 22:4820–4827. 2018.PubMed/NCBI
|
|
74
|
Zhao L, Li Z, Chen W, Zhai W, Pan J, Pang
H and Li X: H19 promotes endometrial cancer progression by
modulating epithelial-mesenchymal transition. Oncol Lett.
13:363–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ahmed ZSO, Golovoy M, Abdullah Y, Ahmed
RSI and Dou QP: Repurposing of metformin for cancer therapy:
Updated patent and literature review. Recent Pat Anticancer Drug
Discov. 16:161–186. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yan L, Zhou J, Gao Y, Ghazal S, Lu L,
Bellone S, Yang Y, Liu N, Zhao X, Santin AD, et al: Regulation of
tumor cell migration and invasion by the H19/let-7 axis is
antagonized by metformin-induced DNA methylation. Oncogene.
34:3076–3084. 2015. View Article : Google Scholar
|
|
77
|
Lu YM, Wang Y, Liu SQ, Zhou MY and Guo YR:
Profile and validation of dysregulated long non-coding RNAs and
mRNAs in ovarian cancer. Oncol Rep. 40:2964–2976. 2018.PubMed/NCBI
|
|
78
|
Lou Y, Jiang H, Cui Z, Wang X, Wang L and
Han Y: Gene microarray analysis of lncRNA and mRNA expression
profiles in patients with high-grade ovarian serous cancer. Int J
Mol Med. 42:91–104. 2018.PubMed/NCBI
|
|
79
|
Zhong Y, Gao D, He S, Shuai C and Peng S:
Dysregulated expression of long noncoding RNAs in ovarian cancer.
Int J Gynecol Cancer. 26:1564–1570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ren C, Li X, Wang T, Wang G, Zhao C, Liang
T, Zhu Y, Li M, Yang C, Zhao Y and Zhang GM: Functions and
mechanisms of long noncoding RNAs in ovarian cancer. Int J Gynecol
Cancer. 25:566–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Xu J, Wu J, Fu C, Teng F, Liu S, Dai C,
Shen R and Jia X: Multidrug resistant lncRNA profile in
chemotherapeutic sensitive and resistant ovarian cancer cells. J
Cell Physiol. 233:5034–5043. 2018. View Article : Google Scholar
|
|
82
|
Chen CL, Ip SM, Cheng D, Wong LC and Ngan
HY: Loss of imprinting of the IGF-II and H19 genes in epithelial
ovarian cancer. Clin Cancer Res. 6:474–479. 2000.PubMed/NCBI
|
|
83
|
Zhu Z, Song L, He J, Sun Y, Liu X and Zou
X: Ectopic expressed long non-coding RNA H19 contributes to
malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol.
8:10082–10091. 2015.PubMed/NCBI
|
|
84
|
Li J, Huang Y, Deng X, Luo M, Wang X, Hu
H, Liu C and Zhong M: Long noncoding RNA H19 promotes transforming
growth factor-β-induced epithelial-mesenchymal transition by acting
as a competing endogenous RNA of miR-370-3p in ovarian cancer
cells. Onco Targets Ther. 11:427–440. 2018. View Article : Google Scholar :
|
|
85
|
Tian X, Zuo X, Hou M, Li C and Teng Y:
LncRNA-H19 regulates chemoresistance to carboplatin in epithelial
ovarian cancer through microRNA-29b-3p and STAT3. J Cancer.
12:5712–5722. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sajadpoor Z, Amini-Farsani Z, Teimori H,
Shamsara M, Sangtarash MH, Ghasemi-Dehkordi P and Yadollahi F:
Valproic acid promotes apoptosis and cisplatin sensitivity through
downregulation of H19 noncoding RNA in ovarian A2780 cells. Appl
Biochem Biotechnol. 185:1132–1144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zheng X, Zhou Y, Chen W, Chen L, Lu J, He
F, Li X and Zhao L: Ginsenoside 20(S)-Rg3 prevents PKM2-targeting
miR-324-5p from H19 sponging to antagonize the warburg effect in
ovarian cancer cells. Cell Physiol Biochem. 51:1340–1353. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hansen S, Sverrisdóttir UÁ and Rudnicki M:
Impact of exercise on pain perception in women with endometriosis:
A systematic review. Acta Obstet Gynecol Scand. 100:1595–1601.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Liu S, Xin W, Lu Q, Tang X, Wang F, Shao
W, Zhang Y, Qiu J and Hua K: Knockdown of lncRNA H19 suppresses
endometriosis in vivo. Braz J Med Biol Res. 54:e101172021.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Panir K, Schjenken JE, Robertson SA and
Hull ML: Non-coding RNAs in endometriosis: A narrative review. Hum
Reprod Update. 24:497–515. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Cui D, Ma J, Liu Y, Lin K, Jiang X, Qu Y,
Lin J and Xu K: Analysis of long non-coding RNA expression profiles
using RNA sequencing in ovarian endometriosis. Gene. 673:140–148.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wu J, Huang H, Huang W, Wang L, Xia X and
Fang X: Analysis of exosomal lncRNA, miRNA and mRNA expression
profiles and ceRNA network construction in endometriosis.
Epigenomics. 12:1193–1213. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wang Y, Li Y, Yang Z, Liu K and Wang D:
Genome-wide microarray analysis of long non-coding RNAs in eutopic
secretory endometrium with endometriosis. Cell Physiol Biochem.
37:2231–2245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wang WT, Sun YM, Huang W, He B, Zhao YN
and Chen YQ: Genome-wide long non-coding RNA analysis identified
circulating LncRNAs as novel non-invasive diagnostic biomarkers for
gynecological disease. Sci Rep. 6:233432016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lin D, Huang Q, Wu R, Dai S, Huang Z, Ren
L, Huang S and Chen Q: Long non-coding RNA AFAP1-AS1 promoting
epithelial-mesenchymal transition of endometriosis is correlated
with transcription factor ZEB1. Am J Reprod Immunol. 81:e130742019.
View Article : Google Scholar
|
|
96
|
Liu H, Zhang Z, Xiong W, Zhang L, Du Y,
Liu Y and Xiong X: Long non-coding RNA MALAT1 mediates
hypoxia-induced pro-survival autophagy of endometrial stromal cells
in endometriosis. J Cell Mol Med. 23:439–452. 2019. View Article : Google Scholar
|
|
97
|
Liu J, Wang Q, Zhang R, Zhang C, Lin J and
Huang X: Identification of LINC01279 as a cell cycle-associated
long non-coding RNA in endometriosis with GBA analysis. Mol Med
Rep. 18:3850–3858. 2018.PubMed/NCBI
|
|
98
|
Huan Q, Cheng SC, Du ZH, Ma HF and Li C:
LncRNA AFAP1-AS1 regulates proliferation and apoptosis of
endometriosis through activating STAT3/TGF-β/Smad signaling via
miR-424-5p. J Obstet Gynaecol Res. 47:2394–2405. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Liu S, Qiu J, Tang X, Cui H, Zhang Q and
Yang Q: LncRNA-H19 regulates cell proliferation and invasion of
ectopic endometrium by targeting ITGB3 via modulating miR-124-3p.
Exp Cell Res. 381:215–222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xu Z, Zhang L, Yu Q, Zhang Y, Yan L and
Chen ZJ: The estrogen-regulated lncRNA H19/miR-216a-5p axis alters
stromal cell invasion and migration via ACTA2 in endometriosis. Mol
Hum Reprod. 25:550–561. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu S, Xin W, Tang X, Qiu J, Zhang Y and
Hua K: LncRNA H19 overexpression in endometriosis and its utility
as a novel biomarker for predicting recurrence. Reprod Sci.
27:1687–1697. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Liu Z, Liu L, Zhong Y, Cai M, Gao J, Tan
C, Han X, Guo R and Han L: LncRNA H19 over-expression inhibited
Th17 cell differentiation to relieve endometriosis through
miR-342-3p/IER3 pathway. Cell Biosci. 9:842019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wang Y, Hu T, Wu L, Liu X, Xue S and Lei
M: Identification of non-coding and coding RNAs in porcine
endometrium. Genomics. 109:43–50. 2017. View Article : Google Scholar
|
|
104
|
Wang Y, Hua R, Xue S, Li W, Wu L, Kang T
and Lei M: mRNA/lncRNA expression patterns and the function of
fibrinogen-like protein 2 in Meishan pig endometrium during the
preimplantation phases. Mol Reprod Dev. 86:354–369. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Chen MY, Liao GD, Zhou B, Kang LN, He YM
and Li SW: Genome-wide profiling of long noncoding RNA expression
patterns in women with repeated implantation failure by RNA
sequencing. Reprod Sci. 26:18–25. 2019. View Article : Google Scholar
|
|
106
|
Huang J, Song N, Xia L, Tian L, Tan J,
Chen Q, Zhu J and Wu Q: Construction of lncRNA-related competing
endogenous RNA network and identification of hub genes in recurrent
implantation failure. Reprod Biol Endocrinol. 19:1082021.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Feng C, Shen JM, Lv PP, Jin M, Wang LQ,
Rao JP and Feng L: Construction of implantation failure related
lncRNA-mRNA network and identification of lncRNA biomarkers for
predicting endometrial receptivity. Int J Biol Sci. 14:1361–1377.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ariel I, Weinstein D, Voutilainen R,
Schneider T, Lustig-Yariv O, de Groot N and Hochberg A: Genomic
imprinting and the endometrial cycle. The expression of the
imprinted gene H19 in the human female reproductive organs. Diagn
Mol Pathol. 6:17–25. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Korucuoglu U, Biri AA, Konac E, Alp E,
Onen IH, Ilhan MN, Turkyilmaz E, Erdem A, Erdem M and Menevse S:
Expression of the imprinted IGF2 and H19 genes in the endometrium
of cases with unexplained infertility. Eur J Obstet Gynecol Reprod
Biol. 149:77–81. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zeng H, Fan X and Liu N: Expression of H19
imprinted gene in patients with repeated implantation failure
during the window of implantation. Arch Gynecol Obstet.
296:835–839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wang H, Cao Q, Ge J, Liu C, Ma Y, Meng Y,
Wang Y, Zhao X, Liu R, Li C, et al: LncRNA-regulated infection and
inflammation pathways associated with pregnancy loss: Genome wide
differential expression of lncRNAs in early spontaneous abortion.
Am J Reprod Immunol. 72:359–375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
He D, Zeng H, Chen J, Xiao L, Zhao Y and
Liu N: H19 regulates trophoblastic spheroid adhesion by
competitively binding to let-7. Reproduction. 157:423–430. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hu L, Zeng H and Liu N: Expression of H19
long non-coding RNA and ZEB1 in the trophoblast of women with
spontaneous abortion. Zhong Nan Da Xue Xue Bao Yi Xue Ban.
43:179–183. 2018.In Chinese. PubMed/NCBI
|
|
114
|
Bai RX and Tang ZY: Long non-coding RNA
H19 regulates Bcl-2, Bax and phospholipid hydroperoxide glutathione
peroxidase expression in spontaneous abortion. Exp Ther Med.
21:412021. View Article : Google Scholar
|
|
115
|
Li J, Han T, Wang X, Wang Y, Chen X, Chen
W and Yang Q: H19 may regulate the immune cell infiltration in
carcinogenesis of gastric cancer through miR-378a-5p/SERPINH1
signaling. World J Surg Oncol. 20:2952022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sahin Y: LncRNA H19 is a potential
biomarker and correlated with immune infiltration in thyroid
carcinoma. Clin Exp Med. 23:841–851. 2023. View Article : Google Scholar
|
|
117
|
Chen X, Luo X, Wei Y, Sun H, Dai L,
Tangzhou Y, Jin H and Yin Z: LncRNA H19 induces immune
dysregulation of BMMSCs, at least partly, by inhibiting IL-2
production. Mol Med. 27:612021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Li X, Wang H, Zhang Y, Zhang J, Qi S,
Zhang Y and Gao MQ: Overexpression of lncRNA H19 changes basic
characteristics and affects immune response of bovine mammary
epithelial cells. PeerJ. 7:e67152019. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Yang S, Fang F, Yu X, Yang C, Zhang X,
Wang L, Zhu L, Shao K and Zhu T: Knockdown of H19 inhibits the
pathogenesis of acne vulgaris by targeting the miR-196a/TLR2/NF-κB
axis. Inflammation. 43:1936–1947. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wang B, Suen CW, Ma H, Wang Y, Kong L, Qin
D, Lee YWW and Li G: The roles of H19 in regulating inflammation
and aging. Front Immunol. 11:5796872020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Hashemi M, Moosavi MS, Abed HM, Dehghani
M, Aalipour M, Heydari EA, Behroozaghdam M, Entezari M,
Salimimoghadam S, Gunduz ES, et al: Long non-coding RNA (lncRNA)
H19 in human cancer: From proliferation and metastasis to therapy.
Pharmacol Res. 184:1064182022. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ye Y, Shen A and Liu A: Long non-coding
RNA H19 and cancer: A competing endogenous RNA. Bull Cancer.
106:1152–1159. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Alipoor B, Parvar SN, Sabati Z, Ghaedi H
and Ghasemi H: An updated review of the H19 lncRNA in human cancer:
Molecular mechanism and diagnostic and therapeutic importance. Mol
Biol Rep. 47:6357–6374. 2020. View Article : Google Scholar : PubMed/NCBI
|