Epigenetics is the study of heritable phenotypes
that do not involve changes in the actual nuclear DNA sequence
(1-4). Examples of known epigenetic
modulations include DNA methylation, expression interference by
non-coding RNAs (ncRNA) and histone modifications (5-8).
Genetic information is encoded by DNA, which is transcribed into
coding mRNA or non-coding RNA. mRNA is then translated into
functional proteins (9).
Epigenetic inheritance not only serves a role in transcription and
translation, but is also involved in post-transcriptional mRNA
regulation (10).
Some of the most commonly studied RNA methylation
modification processes include N6-methyladenosine (m6A),
5-methylcytosine (m5C) and 7-methylguanosine (m7G) (11). m6A is the most common epigenetic
modification mechanism of RNA molecules, which was first identified
in eukaryotic mRNAs in the early 1970s (12). In 2011, it was then discovered
that the m6A process is reversible, when fat mass and
obesity-associated (FTO) protein was shown to function as an m6A
mRNA demethylase (13).
m6A methylation was subsequently found to occur in
all species of eukaryotes, prokaryotes and viruses (14,15). m6A modifications can occur on
mRNAs, ribosomal (rRNAs), small nuclear (snRNAs) and microRNAs
(miRNAs) (16). To date, m6A
sites have been identified in >7,000 coding RNAs and 300
non-coding RNAs (17,18). Functionally, m6A can regulate
almost all processes during the mRNA life cycle, including
stability, splicing, export and translation (19,20). m6A modifications generally occur
on 'RRACH' RNA sequences, where R can be A or G bases and H can be
A, C or U bases (21). Qualcomm
test sequencing previously revealed that the m6A signals typically
clustered near the 3'-untranslated region (UTR) and stop codons
(22). This likely aids in
maintaining the structural stability of the mRNA (22). By contrast, clustering near the
5'-UTR or long internal exons is considered to be involved in mRNA
splicing, translation and degradation (23-25). Specifically, YTH domain-family
proteins (YTHD) 2 can bind m6A in the 5'-UTR of mRNAs, promoting
their cap-independent translation. mRNAs containing the m6A
modification in the 5'-UTR can be identified to promote
cap-independent translation initiation by direct binding to
eukaryotic initiation factor (eIF)3 (23).
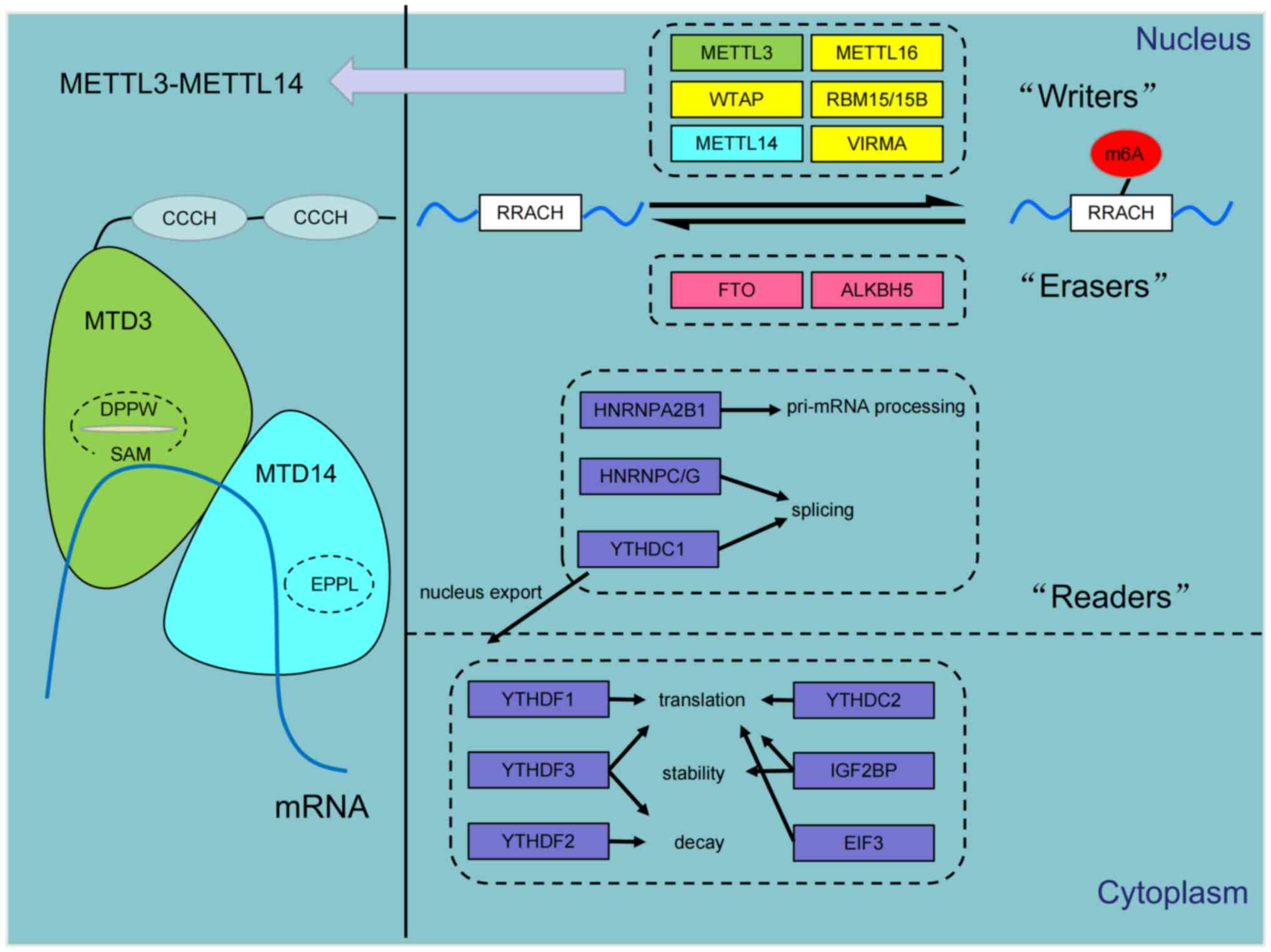
At present, m6A regulators can be classified into
methyltransferases ('writers'), demethylases ('erasers) and
RNA-binding proteins ('readers') (26). Specifically, 'Writers' catalyze
m6A methylation and include methyltransferase-like (METTL)
proteins, such as METTL3 (27-29), METTL14 (30,31), METTL16 (32,33), Wilms tumor 1-associated protein
(WTAP) (26,34,35), RNA-binding motif protein 15
(26) and Vir-like m6A
methyltransferase (36-38). Alternatively, 'erasers', such as
FTO, AlkB homolog (ALKBH)3 and ALKBH5, generally catalyze the
demethylation of m6A residues in an iron (II) and
α-ketoglutarate-dependent manner (39-42). The renders m6A methylation to be a
dynamically reversible process (10). The role of 'readers' are to
identify the methylation modifications and transmit regulatory
signals to downstream functional proteins to facilitate various
biological processes, such as RNA splicing, stabilization, export
and mRNA translation (21,43).
Such 'readers' include YTHDC1/2 and YTHDF1/2/3 (44,45), heterogenous nuclear
ribonucleoproteins (hnRNP)C, hnRNPG and hnRNPA2B1 (46), eIF3 (38) and insulin-like growth factor-2
mRNA-binding proteins (IGF2BP; Fig.
1) (44,47).
The m6A methylation 'writer' METTL14 has been
characterized to be a weak methyltransferase that cannot catalyze
methylation independently (6).
Complete catalysis is achieved by the METTL3/METTL14 complex, a
stoichiometrically 1:1 heterodimer (48). These two methylase components were
initially discovered by isolating nuclear extracts from HeLa cells,
such that the key 70-kDa METTL3 subunit was extracted from the
200-kDa mRNA adenosine methylase complex (49). Through its open methyltransferase
domain (MTD), the internal DPPW motif on METTL3 binds to the free
methyl donor S-adenosylmethionine methionine (SAM) to generate the
metabolite S-adenosine homocysteine (SAH). SAH then competitively
regulates methylation homeostasis, by promoting the association of
the tyrosine residue Y406 on the METTL3 with the mRNA (50,51). By contrast, METTL14 also contains
an MTD, but its EPPL motif that confers catalytic cavity adopts a
closed conformation, obstructing its access to SAM (52,53). Additionally, METTL3 has a unique
Cys-Cys-Cys-His (CCCH)-type zinc-binding motif, which has been
reported to bind nucleic acids and be required for RNA
methyltransferase activity (50,54,55). The current working model of this
METTL3/METTL14 complex is that the main function of METTL14 is to
recognize the RNA substrate whilst enhancing the activity and
catalytic efficiency of METTL3 (52). However, METTL3 and METTL14 have
also been found to serve opposing regulatory roles in cancer. One
previous study found the opposite expression and prognostic value
of METTL3 and METTL14 in hepatocellular carcinoma (HCC) (56). In addition, separately
downregulating the expression of METTL3 and METTL14 resulted in the
alteration of distinct mRNA signaling pathways and biological
processes in HCC (56). Apart
from METTL3/METTL14 dimerization, WTAP can contribute to
METTL3/METTL14 complex stabilization and localization to the
nucleus (57,58).
During neoplastic transformation, m6A modification
can regulate tumor development and progression by modulating the
structures of oncogene and suppressor gene mRNAs (4,61-63). Recently, various studies have
reported that METTL14 can serve a diverse array of roles in
digestive system malignancies, including regulation of
proliferation, invasion, metastasis, angiogenesis and drug
resistance. Therefore, current research progress (until December
2022) on the functions of METTL14 in digestive system tumors was
summarized in this section and Table
I.
To date, great progress has been made in the
development of treatment methods for gastric cancer (GC). Immuno-
and targeted therapies have contributed favorably to the prognosis
of patients with GC. However, the mortality rate from GC remains to
rank highly in China, ranking second after lung cancer in 2022.
This is due in part to its high propensity for metastasis and
invasion (64,65). m6A RNA methylation has been
previously associated with the occurrence and aggressiveness of GC
(66).
HCC is the fourth leading cause of cancer-related
mortality worldwide, accounting for >80% of all cases of primary
liver cancer (74,75). In total, 6% of China's population
is affected by hepatitis B infection and/or HCC (76). The low 5-year survival and high
mortality rates for HCC are largely due to metastasis, poor
response to chemotherapy and frequent postoperative recurrence
(77). Epigenetic studies are
therefore important for the discovery of novel therapeutic targets
for HCC.
However, at present there is no unanimous consensus
on the role of METTL14 in liver cancer. A number of studies have
suggested that METTL14 can serve as an oncogene in liver
cancer. Yang et al (86)
found that the levels of METTL14 and METTL3 expression are higher
in HCC cell lines and human HCC tissue samples (86). Overexpression of METTL14 directly
increased m6A levels on the ATP citrate lyase and stearyl-CoA
desaturase 1 mRNA transcripts in HCC cells. This enhanced the
production of triglycerides and cholesterol, in addition to the
accumulation of lipid droplets in HCC, leading to HCC
proliferation, angiogenesis and the HCC progression (86). The pathogenesis of HCC is
associated with pathogen-associated molecular patterns, where
lipopolysaccharides (LPS) can enter the liver through the
entero-liver axis to promote the progression of HCC. LPS may be one
of the regulators upregulated by METTL14 in HCC (87). LPS can upregulate METTL14 to
enhance the m6A methylation level of miR-155 host gene (MIR155HG),
which then stabilizes the MIR155HG mRNA through the 'reader'
protein abnormal vision-like 1. In addition, LPS can increase the
expression of programmed death-ligand 1 (PD-L1) by upregulating the
expression of miR-155HG (87). In
addition, overexpression of MIR155HG can inhibit the expression of
miR-223-3p, which then increases the expression of STAT1 and the
expression of PD-L1 in HCC. Based on the m6A-sequencing data from
HCC cells, Liu et al (88)
previously observed that rho GTPase-activating protein 5
(ARHGAP5)-antisense (AS)1 is the long non-coding RNA
(lncRNA) showing the highest level of m6A modification. Silencing
METTL14 expression was found to reduce m6A and ARHGAP5-AS1
expression (88). Furthermore,
inhibition of cold shock domain-containing E1 degradation by
ARHGAP5-AS1 was found to promote the translation of vimentin
and Ras-related C3 botulinum toxin substrate 1 (RAC1) to activate
the ERK pathway in HCC. ERK activation then in turn promoted HCC
cell proliferation, migration and invasion (88). Another previous study revealed
that miR-628-5p in the exosomes secreted by M1
macrophages can inhibit expression of METTL14 in HCCs following
transportation (89). The m6A
modification of Circ-fucosyltransferase 8 (circFUT8)
is positively regulated by METTL14, where CircFUT8 is upregulated
in HCC cells. CircFUT8 overexpression can inhibit HCC apoptosis
whilst promoting cell proliferation through the miR-552-3p/charged
multivesicular body protein 4B pathway (89).
CRC is one of the most common malignancies in
adults, ranking third in the cases of cancer-related mortality
worldwide (90) in 2020. The
mortality rate in patients with CRC remains high largely due to
high rates of metastasis and recurrence (65,91). Studying epigenetics can provide
insight into the mechanisms regulating the progression of
colorectal cancer.
In addition to demonstrating its function in CRC
proliferation, invasion and migration, a number of studies have
also reported METTL14 to regulate other biological processes. Wang
et al (97) found that
decreased expression of METTL3 and METTL14 can increase
IFN-γ/STAT1/IFN regulatory factor 1 (IRF1) signaling by stabilizing
IFN-γ, C-X-C motif chemokine ligand (CXCL)9 and CXCL10 secreted by
tumor-infiltrating CD8+ cytotoxic T cells in the tumor
microenvironment. This then increases the IFN content in the tumor
microenvironment and increase the recruitment of immune cells
(97). IFN-γ/STAT1/IRF1 signaling
may provide insight into the mechanism of poor response to immune
checkpoint inhibitor (ICIs) therapy in mismatch-repair-proficient
or microsatellite low instability CRC (97). Another recent study found that the
downregulation of METTL14 m6A methylation in C1q+ tumor-associated
macrophages can increase the levels of the cytokine IL-35 subunit
Epstein-Barr virus-induced protein 3 (EBI3) mRNA transcript. This
EBI3 elevation in turn promoted CD8+ T cell differentiation into a
dysfunctional state, which have weaker proliferative capabilities
and reduced response potency to tumors, leading to CRC progression
(98). Consistent with the
findings in liver cancer, METTL14 was also found to regulate miRNA
expression by modifying pri-miRNA splicing in CRC (99). Enterotoxigenic bacteroides
fragilis in the gut flora was shown to positively regulate the
expression of pri-miR-149 through METTL14-mediated m6A methylation
in CRC (99). Reducing miR-149-3p
expression in the plasma exosomes of patients with CRC was found to
increase proliferation and induce an inflammatory response in CRC
cells (99). Functionally,
miR-149-3p increases the expression of the PHD finger protein
5A gene downstream to stabilize the splicing factor 3b complex
and transactivate superoxide dismutase 2 (SOD2), by regulating
lysine acetyltransferase 2A mRNA alternative splicing and increase
the expression of SOD2 mRNA, to induce colorectal carcinogenesis
(99). Another previous study
documented that oxidative stress induced by chemotherapeutic drugs
can upregulate the methylation of pleckstrin homology-like
domain family B member 2 (PHLDB2) by METTL14 in CRC,
which increases protein expression (100). Nuclear EGFR is a transcriptional
cofactor that can function to regulate gene expression and promote
DNA repair (100). Increased
PHLDB2 expression can stabilize EGFR and promotes its nuclear
translocation, which promotes cetuximab resistance (100). Rab11A is a common EGFR recycling
protein that facilitate EGFR translocation back to the cell
membrane, the mechanism by which PHLDB2 can promote the nuclear
translocation of EGFR may be mediated by reducing the binding
affinity between EGFR and Rab11A (100).
PC is one of the most aggressive malignancies, with
a 5-year mortality rate of ~95% after diagnosis (65). Additionally, majority of patients
with PC do not survive for >7 years following surgical treatment
(101,102). For patients with advanced
pancreatic cancer, chemotherapy or chemo-radiotherapy is typically
applied, but chemotherapy resistance frequently leads to worse
prognosis (103). Unlike other
digestive system malignancies, existing evidence actually suggest
METTL14 to be an oncogene in PC.
A previous study showed that the overexpression of
METTL14 in PC cells can increase methylation of p53 effector
related to peripheral myelin protein 22 (PERP)
downstream. PERP mRNA methylation reduces PERP protein
expression, which promotes PC cell proliferation and migration
(104). Decreased m6A
modification of METTL3 and METTL14 inhibits the expression of
miR-380-3p in PC cells, which was demonstrated by Jiang et
al (105). By contrast, the
expression level of miR-380-3p was previously found to be
significantly higher in PC tissues and cells. Further experiments
found that miR-380-3p can promote PC cell proliferation and
tumorigenesis through Akt signaling, EMT by suppressing PTEN
expression (105). In addition,
drug-resistant PC tissues tend to express higher levels of METTL14
according to Kong et al (106). Accordingly, knocking down
METTL14 expression was found to enhance both apoptosis and
autophagy in PC cells through activating 5'AMP-activated protein
kinase (AMPKα) and ERK1/2 signaling to restrain mTOR signaling,
allowing for increased sensitivity to cisplatin. Cisplatin is
essential therapeutic agent for the treatment of patients with
breast cancer gene (BRCA)1/2 or partner and localizer of
BRCA2 mutations (107).
Therefore, these findings may provide guidance for chemotherapy
regimen selection in patients with PC. Furthermore, another study
found that METTL14 can promote resistance to gemcitabine in PC
cells, which can be reversed with METTL14 knockdown
(108). METTL14 expression was
observed to be upregulated in gemcitabine-resistant human PC cells,
where increased METTL14 expression was also found after gemcitabine
treatment. Further studies then revealed that the NF-κB
transcription factor p65 can target the promoter region of
METTL14 to positively its mRNA and protein expression. This
in turn upregulated the expression of cytidine deaminase to
inactivate gemcitabine.
Contrary to the aforementioned findings, certain
studies have also reported that METTL14 can function as a
tumor suppressor in PC. PI3K catalytic subunit (PIK3CB) expression
is frequently increased in PTEN-deficient PC cells (109). PIK3CB overexpression was found
to reduce the expression of components in the METTL3/METTL14/WTAP
complex and activate AKT signaling, which enhanced the
proliferative and migratory capacity of PC cells. Cell division
cycle 2-like kinase 1 (CLK1) was also found to be upregulated in PC
tissues, which enhances the phosphorylation of serine/arginine-rich
splicing factor 5 (SRSF5) to suppress METTL14 exon 10 skipping.
METTL14 exon 10 skipping enhances the degree of m6A modification on
its gene. CLK1 can also enhance PC cell proliferation, migration
and invasion by decreasing the m6A level of METTL14, in addition to
promoting cyclin L2 exon 6.3 skipping (110). However, the downstream
regulatory mechanism of METTL14 remains unclear.
In cholangiocarcinoma (CCA), lower expression levels
of stromal interaction molecule 2 (STIM2) is associated with poor
prognosis (111). m6A
modification mediated by METTL14 and YTHDC2 was found to decrease
the expression of STIM2 mRNA (111). Consequently, knockdown of
STIM2 can increase keratin 8 (KRT8) expression to facilitate
extrahepatic CCA metastasis (111).
The Cancer Genome Atlas (TCGA) is a useful tool for
determining DNA, RNA and protein expression levels, in addition to
exploring gene function, in a variety of cancers (112). Preliminary understanding can
first be obtained, which can then be verified using experimental
data (112). This section
summarizes the results of m6A-related studies obtained by
bioinformatics analysis alone.
The majority of the bioinformatics studies performed
focused on the expression levels of METTL14 and their prognostic
value. Zhou et al (78)
found that METTL14 expression in HCC tissues is significantly lower
compared with those in adjacent tissues. Furthermore, METTL14
expression in HCC tissues is associated with tumor size and
tumor-node-metastasis (TNM) stage in patients with HCC. Lower
expression levels of METTL14 in HCC tissues were significantly
associated with poorer prognosis, with the overall survival and
tumor-free survival rates being significantly shortened. Liu et
al (56) also showed that
METTL14 expression is lower in HCC tissues and associated with
poorer prognosis. By contrast, the opposite trend was observed in
terms of the prognostic values of METTL3 in HCC (56). Kong et al (113) found that the expression of
METTL14 is upregulated in HCC tissues, but the downregulation of
METTL14 is associated with poor prognosis (113), possibly due to lower levels of
immune cell infiltration (113).
Another study revealed that METTL14 may participate in the
malignant progression of HCC by regulating the expression levels of
cysteine sulfinic acid decarboxylase, glutamate-oxaloacetate
transaminase 2 and suppressors of cytokine signaling 2 (114). The author used LASSO analysis to
analyze a total of 124 prognostic genes in 307 TCGA samples to find
real hub genes that are associated with patient prognosis, but the
specific mechanism needs to be verified by further experiments
(114). The results of the
aforementioned analyses suggest that METTL14 is upregulated in HCC
and inversely associated with clinical prognosis. Xu et al
(115) previously suggested that
METTL14 expression is upregulated in PC, which displayed a U-shaped
association with clinical stage. Specifically, expression in stages
I and IV was higher compared with that at stages II and III.
Additionally, METTL14 expression is negatively associated with the
T phase (115). Similarly,
another previous analysis by Zhang et al (116) revealed that METTL14 is highly
expressed in PC tissues. Additionally, two studies on rectal cancer
found that METTL14 expression is associated with worse prognosis
(117,118). In addition, METTL14 expression
was positively correlated with the degree of immune cell
infiltration, as found by Cai et al (117). In GC, Xu et al (119) found that lower METTL14
expression significantly correlated with the expression levels of
PD-L1 and PD-1 (119). Using
LASSO regression analysis, a study on esophageal cancer (ESCA)
found that METTL14 expression can be used to predict overall
survival in patients with ESCA (120).
As a methyltransferase, METTL14 can mediate m6A
methylation to regulate various physiological processes in
malignancies, such as proliferation, invasion, metastasis and drug
resistance (10). In addition,
METTL14 expression can be used as a marker for the diagnosis and
prognosis of digestive system tumors. Using bioinformatics, several
studies have analyzed the expression profile of METTL14 in
digestive system cancers and developed various prognostic risk
models (114,115). Further validation is required to
explore the specificity and sensitivity of METTL14 as a marker.
METTL14 expression may also confer benefits for predicting
efficacy. The role of METTL14 in cisplatin and gemcitabine
resistance in PC suggests that it can be used for predicting
chemotherapy outcome (108).
Wang et al (97)
previously found that METTL14 can regulate the response of CRC
cells to immunotherapy, suggesting that METTL14 can enhance the
effectiveness of immunotherapy, either when combined with other
drugs or as a potential independent therapeutic target (97).
METTL14 mediate different regulatory roles in
different tumors. In GC and CRC, the majority of the evidence
suggest that METTL14 serves as a tumor suppressor, whereas in PC
they suggest METTL14 to be an oncogene. In HCC, the role of METTL14
remains controversial. Whether METTL14 itself is an oncogene
or tumor suppressor remains a matter of debate. In addition, the
mechanism underlying the regulation of METTL14 remain elusive. A
study did previously find that the SUMOylation of METTL3
significantly inhibited its m6A methyltransferase activity in
NSCLC, resulting in reduced m6A levels. However, this did not
affect METTL14 stability, localization or its interaction with WTAP
(121).
In terms of the possible upstream mechanisms that
can regulate METTL14 expression, binding to its promoters and
subsequent inhibition of METT14 transcription or direct regulation
of METTL14 mRNA expression has been considered. Upstream regulators
include RBPs, transcription factors, exosomes from M1
macrophages and endotoxins produced by the gut flora (Table II). In liver cancer, FA can
reduce the m6A of METTL14 in HepG2 cells to subsequently inhibit
p53 gene expression (81). The
transcription factor HIF-1α can also inhibit METTL14 expression and
promote the development of HCC under hypoxic conditions (85). In addition, LPS treatment was
found to increase the expression of METTL14 in Huh7 cells whilst
increasing the m6A methylation level of MIR155HG (87). Exosomes of M1
macrophages inhibited METTL14 expression by transferring miR-628-5p
to HCC cells, which inhibited the m6A modification of circFUT8
(89). In CRC, KDM5C-mediated
demethylation of histone H3K4me3 in the promoter region inhibited
METTL14 transcription, which may account for the reduction of
METTL14 expression found in CRC (94). MeCP2 was also found to inhibit the
m6A methylation of METTL14 in CRC cells, mechanistically due to
MeCP2 competitively disrupting the interaction between METTL3 and
METTL14 (95). In addition, the
RBP HuR can inhibit METTL14 mRNA expression by binding to its
promoter (96), whereas TCF4
deficiency significantly increased the ubiquitination level of
METTL14 and promoted its protein degradation (96). In PC, CLK1 enhanced the
phosphorylation of SRSF5 on Ser-250, which inhibited METTL14 exon
10 skipping and increased the level of m6A modification (110). Therefore, the regulatory
mechanism of METTL14 function in the individual tumor types must be
strictly defined. In addition, the mechanism of action of the
METTL3/METTL14 complex remains unclear. In particularly, whether
the two function allosterically or synergistically remain unknown.
A previous study in HCC reported that the expression and function
profiles of the two are distinct and do not share common pathways
downstream (56).
The PI3K/AKT signaling pathway is frequently found
to be associated with cancer development, metastasis and EMT.
Previous studies in LC, GC and CRC have confirmed that the PI3K/AKT
signaling pathway is a downstream regulatory pathway of METTL14
(67,68,82,94). HIF-1α can inhibit the expression
of METTL14 in HCC, but HIF-1α can be regulated by METTL14 in GC as
one of its downstream targets (71,85). The EGFR signaling pathway was
found to serve downstream of METTL14 in LC and CRC (82,100), whereas the ERK signaling pathway
was found to serve downstream of METTL14 in HCC and PC (88,107). In conclusion, the mechanism of
regulation by METTL14 in digestive system malignancies is one of
the most important directions for future research.
Anti-angiogenic therapy is an important modality
for digestive system tumor treatment (122). Due to the low mutation rate of
anti-angiogenic genes in CRC (122), epigenetic modifications may
provide a more appropriate target for blocking tumor angiogenesis.
The role of m6A regulation in tumor angiogenesis is currently a
topic of intense research. Chen et al (123) previously found that
METTL14-mediated m6A methylation can increase TNF
receptor-associated factor RNA stability and inhibit
sunitinib-mediated anti-angiogenic effects in renal cancer cells
(123). In addition, Wen et
al (124) revealed that
basic leucine zipper activating transcription factors-like
transcription factor 2 (BATF2) expression is downregulated in
tongue squamous cell carcinoma (TSCC). Downregulated BATF2 serves a
role in promoting tumor growth, metastasis and angiogenesis
(124). METTL14-mediated m6A
modification was found to inhibit BATF2 mRNA expression and
increase the expression of vascular endothelial growth factor A
(124). The role of METTL14 in
angiogenesis in digestive system tumors remains uncharacterized,
exploring it would be of great clinical significance for patients
with digestive system tumors in addition to providing a potential
research direction.
In conclusion, the expression and role of METTL14
in different digestive system cancers remain controversial. In
particular, the regulatory mechanism of METTL14 require further
study. Although METTL14 has been demonstrated to serve an important
role in digestive system tumors, its implications for clinical
application also requires further investigation.
In summary, METTL14 serves diverse roles in
digestive system cancers and has diagnostic and therapeutic
potential. Further exploration of the regulatory mechanisms of
METTL14 is anticipated, which is expected to accelerate their
incorporation into clinical treatment.
Not applicable.
JXH and HSL contributed to conception and design.
JXH and CW contributed to literature search. JXH contributed to
manuscript writing. QS and BWC approved the final version of the
manuscript, and agree to be accountable for all aspects of the work
in ensuring that questions related to the accuracy or integrity of
any part of the work are appropriately investigated and
resolved.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
Supported by Beijing Natural Science Foundation Program (grant
no. 7222032), the National Natural Science Foundation of China
(grant no. 82173056) and the Beijing Natural Science Foundation
(grant no. 7214220 to HL), the Friendship Seed Project (grant no.
YYZZ202034 to HL).
|
1
|
Li S, Kuo HC, Yin R, Wu R, Liu X, Wang L,
Hudlikar R, Peter RM and Kong AN: Epigenetics/epigenomics of
triterpenoids in cancer prevention and in health. Biochem
Pharmacol. 175:1138902020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goel A and Boland CR: Epigenetics of
colorectal cancer. Gastroenterology. 143:1442–1460.e1. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou S, Treloar AE and Lupien M: Emergence
of the noncoding cancer genome: A target of genetic and epigenetic
alterationsthe noncoding cancer genome. Cancer Discov. 6:1215–1229.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Shi Y, Shen H and Xie W:
m6A-binding proteins: The emerging crucial performers in
epigenetics. J Hematol Oncol. 13:352020. View Article : Google Scholar
|
|
5
|
Ghavami S, Zamani M, Ahmadi M, Erfani M,
Dastghaib S, Darbandi M, Darbandi S, Vakili O, Siri M, Grabarek BO,
et al: Epigenetic regulation of autophagy in gastrointestinal
cancers. Biochim Biophys Acta Mol Basis Dis. 1868:1665122022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jones PA, Issa JP and Baylin S: Targeting
the cancer epigenome for therapy. Nat Rev Genet. 17:630–641. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu X, Wang P, Teng X, Zhang Z and Song S:
Comprehensive analysis of expression regulation for RNA m6A
regulators with clinical significance in human cancers. Front
Oncol. 11:6243952021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song N, Cui K, Zhang K, Yang J, Liu J,
Miao Z, Zhao F, Meng H, Chen L, Chen C, et al: The role of m6A RNA
methylation in cancer: Implication for nature products anti-cancer
research. Front Pharmacol. 13:9333322022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen DH, Zhang JG, Wu CX and Li Q:
Non-Coding RNA m6A modification in cancer: Mechanisms and
therapeutic targets. Front Cell Dev Biol. 9:7785822021. View Article : Google Scholar
|
|
12
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yi YC, Chen XY, Zhang J and Zhu JS: Novel
insights into the interplay between m6A modification and
noncoding RNAs in cancer. Mol Cancer. 19:1212020. View Article : Google Scholar
|
|
15
|
Li H, Wu H, Wang Q, Ning S, Xu S and Pang
D: Dual effects of N6-methyladenosine on cancer
progression and immunotherapy. Mol Ther Nucleic Acids. 24:25–39.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng Y, Xie W, Pickering BF, Chu KL,
Savino AM, Yang X, Luo H, Nguyen DT, Mo S, Barin E, et al:
N6-Methyladenosine on mRNA facilitates a phase-separated
nuclear body that suppresses myeloid leukemic differentiation.
Cancer Cell. 39:958–972.e8. 2021. View Article : Google Scholar
|
|
17
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3' UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Peng C, Chen J, Chen D, Yang B, He
B, Hu W, Zhang Y, Liu H, Dai L, et al: WTAP facilitates progression
of hepatocellular carcinoma via m6A-HuR-dependent epigenetic
silencing of ETS1. Mol Cancer. 18:1272019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Y, Zhao Y, Chen J, Peng C, Zhang Y,
Tong R, Cheng Q, Yang B, Feng X, Lu Y, et al: ALKBH5 suppresses
malignancy of hepatocellular carcinoma via m6A-guided epigenetic
inhibition of LYPD1. Mol Cancer. 19:1232020. View Article : Google Scholar :
|
|
21
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible m6A RNA
methylation. Nat Rev Genet. 15:293–306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ke S, Alemu EA, Mertens C, Gantman EC, Fak
JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al:
A majority of m6A residues are in the last exons, allowing the
potential for 3' UTR regulation. Genes Dev. 29:2037–2053. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He PC and He C: m6A RNA
methylation: From mechanisms to therapeutic potential. EMBO J.
40:e1059772021. View Article : Google Scholar
|
|
24
|
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang
Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential
m6A mRNA methylation in 3' UTR and near stop codon and
associates with alternative polyadenylation. Cell Discov. 4:102018.
View Article : Google Scholar
|
|
25
|
Wei CM and Moss B: Nucleotide sequences at
the N6-methyladenosine sites of HeLa cell messenger ribonucleic
acid. Biochemistry. 16:1672–1676. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen XY, Zhang J and Zhu JS: The role of
m6A RNA methylation in human cancer. Mol Cancer.
18:1032019. View Article : Google Scholar
|
|
27
|
Wang N, Huo X, Zhang B, Chen X, Zhao S,
Shi X, Xu H and Wei X: METTL3-Mediated ADAMTS9 suppression
facilitates angiogenesis and carcinogenesis in gastric cancer.
Front Oncol. 12:8618072022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu T, Liu J, Wang Y, Chen W, Liu Z, Zhu L
and Zhu W: METTL3 promotes colorectal cancer metastasis by
stabilizing PLAU mRNA in an m6A-dependent manner. Biochem Biophys
Res Commun. 614:9–16. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z,
Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, et al: METTL3-mediated
m6A modification of HDGF mRNA promotes gastric cancer
progression and has prognostic significance. Gut. 69:1193–1205.
2020. View Article : Google Scholar
|
|
30
|
Zhou H, Yin K, Zhang Y, Tian J and Wang S:
The RNA m6A writer METTL14 in cancers: Roles, structures, and
applications. Biochim Biophys Acta Rev Cancer. 1876:1886092021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W,
Lu S, Xu D, Wu Y, Chen Q, et al: LNC942 promoting METTL14-mediated
m6A methylation in breast cancer cell proliferation and
progression. Oncogene. 39:5358–5372. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ruszkowska A: METTL16,
methyltransferase-like protein 16: Current insights into structure
and function. Int J Mol Sci. 22:21762021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Su R, Dong L, Li Y, Gao M, He PC, Liu W,
Wei J, Zhao Z, Gao L, Han L, et al: METTL16 exerts an
m6A-independent function to facilitate translation and
tumorigenesis. Nat Cell Biol. 24:205–216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Trindade F, Tellechea Ó, Torrelo A,
Requena L and Colmenero I: Wilms tumor 1 expression in vascular
neoplasms and vascular malformations. Am J Dermatopathol.
33:569–572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang LJ, Xue Y, Li H, Huo R, Yan Z, Wang
J, Xu H, Wang J, Cao Y and Zhao JZ: Wilms' tumour 1-associating
protein inhibits endothelial cell angiogenesis by m6A-dependent
epigenetic silencing of desmoplakin in brain arteriovenous
malformation. J Cell Mol Med. 24:4981–4991. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu W, Wang JZ, Wei JF and Lu C: Role of
m6A methyltransferase component VIRMA in multiple human cancers.
Cancer Cell Int. 21:1722021. View Article : Google Scholar
|
|
37
|
Panneerdoss S, Eedunuri VK, Yadav P,
Timilsina S, Rajamanickam S, Viswanadhapalli S, Abdelfattah N,
Onyeagucha BC, Cui X, Lai Z, et al: Cross-talk among writers,
readers, and erasers of m6A regulates cancer growth and
progression. Sci Adv. 4:eaar82632018. View Article : Google Scholar
|
|
38
|
Choe J, Lin S, Zhang W, Liu Q, Wang L,
Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al: mRNA
circularization by METTL3-eIF3h enhances translation and promotes
oncogenesis. Nature. 561:556–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shan K, Zhou RM, Xiang J, Sun YN, Liu C,
Lv MW and Xu JJ: FTO regulates ocular angiogenesis via
m6A-YTHDF2-dependent mechanism. Exp Eye Res. 197:1081072020.
View Article : Google Scholar
|
|
40
|
Mathiyalagan P, Adamiak M, Mayourian J,
Sassi Y, Liang Y, Agarwal N, Jha D, Zhang S, Kohlbrenner E,
Chepurko E, et al: FTO-dependent N6-methyladenosine
regulates cardiac function during remodeling and repair.
Circulation. 139:518–532. 2019. View Article : Google Scholar :
|
|
41
|
Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E,
He J and Cai Z: RNA demethylase ALKBH5 in cancer: From mechanisms
to therapeutic potential. J Hematol Oncol. 15:82022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao Y, Hu J, Sun X, Yang K, Yang L, Kong
L, Zhang B, Li F, Li C, Shi B, et al: Loss of m6A Demethylase
ALKBH5 Promotes post-ischemic Angiogenesis via post-transcriptional
Stabilization of WNT5A. Clin Transl Med. 11:e4022021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pendleton KE, Chen B, Liu K, Hunter OV,
Xie Y, Tu BP and Conrad NK: The U6 snRNA m6A methyltransferase
METTL16 regulates SAM synthetase intron retention. Cell.
169:824–835.e14. 2017. View Article : Google Scholar :
|
|
44
|
Yan H, Zhang L, Cui X, Zheng S and Li R:
Roles and mechanisms of the m6A reader YTHDC1 in
biological processes and diseases. Cell Death Discov. 8:2372022.
View Article : Google Scholar
|
|
45
|
Dai XY, Shi L, Li Z, Yang HY, Wei JF and
Ding Q: Main N6-methyladenosine readers: YTH family proteins in
cancers. Front Oncol. 11:6353292021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao LB, Zhu XL, Shi JX, Yang L, Xu ZQ and
Shi SL: HnRNPA2B1 promotes the proliferation of breast cancer MCF-7
cells via the STAT3 pathway. J Cell Biochem. 122:472–484. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li J, Xie H, Ying Y, Chen H, Yan H, He L,
Xu M, Xu X, Liang Z Liu B, et al: YTHDF2 mediates the mRNA
degradation of the tumor suppressors to induce AKT phosphorylation
in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer.
19:1522020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schöller E, Weichmann F, Treiber T, Ringle
S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G:
Interactions, localization, and phosphorylation of the
m6A generating METTL3-METTL14-WTAP complex. RNA.
24:499–512. 2018. View Article : Google Scholar
|
|
49
|
Śledź P and Jinek M: Structural insights
into the molecular mechanism of the m(6)A writer complex. Elife.
5:e184342016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3–METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar
|
|
53
|
Bujnicki JM, Feder M, Radlinska M and
Blumenthal RM: Structure prediction and phylogenetic analysis of a
functionally diverse family of proteins homologous to the MT-A70
subunit of the human mRNA: m(6)A methyltransferase. J Mol Evol.
55:431–444. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Han SH and Choe J: Diverse molecular
functions of m6A mRNA modification in cancer. Exp Mol
Med. 52:738–749. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wen J, Lv R, Ma H, Shen H, He C, Wang J,
Jiao F, Liu H, Yang P, Tan L, et al: Zc3h13 regulates nuclear RNA
m6A methylation and mouse embryonic stem cell
self-renewal. Mol Cell. 69:1028–1038.e6. 2018. View Article : Google Scholar
|
|
56
|
Liu X, Qin J, Gao T, Li C, Chen X, Zeng K,
Zeng K, Xu M, He B, Pan B, et al: Analysis of METTL3 and METTL14 in
hepatocellular carcinoma. Aging (albany NY). 12:21638–21659. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou
Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, et al:
Mettl3-/Mettl14-mediated mRNA N6-methyladenosine
modulates murine spermatogenesis. Cell Res. 27:1216–1230. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
59
|
Lin R, Zhan M, Yang L, Wang H, Shen H,
Huang S, Huang X, Xu S, Zhang Z, Li W, et al: Deoxycholic acid
modulates the progression of gallbladder cancer through
N6-methyladenosine-dependent microRNA maturation.
Oncogene. 39:4983–5000. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang Y, Qian Cai Q, Sheng Fu L, Wei Dong
Y, Fan F and Zhong Wu X: Reduced N6-Methyladenosine Mediated by
METTL3 Acetylation Promotes MTF1 expression and hepatocellular
carcinoma cell growth. Chem Biodivers. 19:e2022003332022.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang N, Zuo Y, Peng Y and Zuo L: Function
of N6-methyladenosine modification in tumors. J Oncol.
2021:64615522021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lan Q, Liu PY, Haase J, Bell JL,
Hüttelmaier S and Liu T: The critical role of RNA m6A
methylation in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N6-methyladenosine modification in cancers:
Current status and perspectives. Cell Res. 28:507–517. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hu BE, Wang XY, Gu XY, Zou C, Gao ZJ,
Zhang H and Fan Y: N6-methyladenosine (m6A) RNA
modification in gastrointestinal tract cancers: Roles, mechanisms,
and applications. Mol Cancer. 18:1782019. View Article : Google Scholar
|
|
67
|
Zhang C, Zhang M, Ge S, Huang W, Lin X,
Gao J, Gong J and Shen L: Reduced m6A modification predicts
malignant phenotypes and augmented Wnt/PI3K-Akt signaling in
gastric cancer. Cancer Med. 8:4766–4781. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu X, Xiao M, Zhang L, Li L, Zhu G, Shen
E, Lv M, Lu X and Sun Z: The m6A methyltransferase METTL14 inhibits
the proliferation, migration, and invasion of gastric cancer by
regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal.
35:e236552021.
|
|
69
|
Yao Q, He L, Gao X, Tang N, Lin L, Yu X
and Wang D: The m6A methyltransferase METTL14-mediated
N6-methyladenosine modification of PTEN mRNA inhibits tumor growth
and metastasis in stomach adenocarcinoma. Front Oncol.
11:6997492021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fan HN, Chen ZY, Chen XY, Chen M, Yi YC,
Zhu JS and Zhang J: METTL14-mediated m6A modification of circORC5
suppresses gastric cancer progression by regulating
miR-30c-2-3p/AKT1S1 axis. Mol Cancer. 21:512022. View Article : Google Scholar :
|
|
71
|
Lin JX, Lian NZ, Gao YX, Zheng QL, Yang
YH, Ma YB, Xiu ZS, Qiu QZ, Wang HG, Zheng CH, et al: m6A
methylation mediates LHPP acetylation as a tumour aerobic
glycolysis suppressor to improve the prognosis of gastric cancer.
Cell Death Dis. 13:4632022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jin H, Wu Z, Tan B, Liu Z, Zu Z, Wu X, Bi
Y and Hu X: Ibuprofen promotes p75 neurotrophin receptor expression
through modifying promoter methylation and
N6-methyladenosine-RNA-methylation in human gastric cancer cells.
Bioengineered. 13:14595–14604. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hu N and Ji H: N6-methyladenosine
(m6A)-mediated up-regulation of long noncoding RNA LINC01320
promotes the proliferation, migration, and invasion of gastric
cancer via miR495-5p/RAB19 axis. Bioengineered. 12:4081–4091. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Balogh J, Victor D III, Asham EH,
Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM and Monsour
HP Jr: Hepatocellular carcinoma: A review. J Hepatocell Carcinoma.
3:41–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Raoul JL and Edeline J: Systemic treatment
of hepatocellular carcinoma: Standard of care in China and
elsewhere. Lancet Oncol. 21:479–481. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li ZL, Yan WT, Zhang J, Zhao YJ, Lau WY,
Mao XH, Zeng YY, Zhou YH, Gu WM, Wang H, et al: Identification of
actual 10-year survival after hepatectomy of HBV-related
hepatocellular carcinoma: A multicenter study. J Gastrointest Surg.
23:288–296. 2019. View Article : Google Scholar
|
|
77
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar
|
|
78
|
Zhou T, Ren Z and Chen C: METTL14 as a
predictor of postoperative survival outcomes of patients with
hepatocellular carcinoma. Nan Fang Yi Ke Da Xue Xue Bao.
40:567–572. 2020.In Chinese. PubMed/NCBI
|
|
79
|
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH,
Wang F, Wang TT, Xu QG, Zhou WP and Sun SH: METTL14 suppresses the
metastatic potential of hepatocellular carcinoma by modulating
N6-methyladenosine-dependent primary MicroRNA
processing. Hepatology. 65:529–543. 2017. View Article : Google Scholar
|
|
80
|
Laptenko O and Prives C: Transcriptional
regulation by p53: One protein, many possibilities. Cell Death
Differ. 13:951–961. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ghazi T, Nagiah S and Chuturgoon AA:
Fusaric acid decreases p53 expression by altering promoter
methylation and m6A RNA methylation in human hepatocellular
carcinoma (HepG2) cells. Epigenetics. 16:79–91. 2021. View Article : Google Scholar :
|
|
82
|
Shi Y, Zhuang Y, Zhang J, Chen M and Wu S:
METTL14 inhibits hepatocellular carcinoma metastasis through
regulating EGFR/PI3K/AKT signaling pathway in an m6A-dependent
manner. Cancer Manag Res. 12:13173–13184. 2020. View Article : Google Scholar :
|
|
83
|
Zhou T, Li S, Xiang D, Liu J, Sun W, Cui
X, Ning B, Li X, Cheng Z, Jiang W, et al: m6A RNA
methylation-mediated HNF3γ reduction renders hepatocellular
carcinoma dedifferentiation and sorafenib resistance. Signal
Transduct Target Ther. 5:2962020. View Article : Google Scholar
|
|
84
|
Du L, Li Y, Kang M, Feng M, Ren Y, Dai H,
Wang Y, Wang Y and Tang B: USP48 is upregulated by Mettl14 to
attenuate hepatocellular carcinoma via regulating SIRT6
stabilization. Cancer Res. 81:3822–3834. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Fan Z, Yang G, Zhang W, Liu Q, Liu G, Liu
P, Xu L, Wang J, Yan Z, Han H, et al: Hypoxia blocks ferroptosis of
hepatocellular carcinoma via suppression of METTL14 triggered
YTHDF2-dependent silencing of SLC7A11. J Cell Mol Med.
25:10197–10212. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yang Y, Cai J, Yang X, Wang K, Sun K, Yang
Z, Zhang L, Yang L, Gu C, Huang X, et al: Dysregulated m6A
modification promotes lipogenesis and development of non-alcoholic
fatty liver disease and hepatocellular carcinoma. Mol Ther.
30:2342–2353. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Peng L, Pan B, Zhang X, Wang Z, Qiu J,
Wang X and Tang N: Lipopolysaccharide facilitates immune escape of
hepatocellular carcinoma cells via m6A modification of lncRNA
MIR155HG to upregulate PD-L1 expression. Cell Biol Toxicol.
38:1159–1173. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu J, Zhang N, Zeng J, Wang T, Shen Y, Ma
C and Yang M: N6-methyladenosine-modified lncRNA
ARHGAP5-AS1 stabilises CSDE1 and coordinates oncogenic RNA regulons
in hepatocellular carcinoma. Clin Transl Med. 12:e11072022.
View Article : Google Scholar
|
|
89
|
Wang L, Yi X, Xiao X, Zheng Q, Ma L and Li
B: Exosomal miR-628-5p from M1 polarized macrophages hinders m6A
modification of circFUT8 to suppress hepatocellular carcinoma
progression. Cell Mol Biol Lett. 27:1062022. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Teng S, Li YE, Yang M, Qi R, Huang Y, Wang
Q, Zhang Y, Chen S, Li S, Lin K, et al: Tissue-specific
transcription reprogramming promotes liver metastasis of colorectal
cancer. Cell Res. 30:34–49. 2020. View Article : Google Scholar :
|
|
92
|
Tian J, Ying P, Ke J, Zhu Y, Yang Y, Gong
Y, Zou D, Peng X, Yang N, Wang X, et al: ANKLE1
N6-Methyladenosine-related variant is associated with
colorectal cancer risk by maintaining the genomic stability. Int J
Cancer. 146:3281–3293. 2020. View Article : Google Scholar
|
|
93
|
Yang X, Zhang S, He C, Xue P, Zhang L, He
Z, Zang L, Feng B, Sun J and Zheng M: METTL14 suppresses
proliferation and metastasis of colorectal cancer by
down-regulating oncogenic long non-coding RNA XIST. Mol Cancer.
19:462020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B,
Li C, Sun L, Qin J, Xu T, et al: METTL14-mediated
N6-methyladenosine modification of SOX4 mRNA inhibits tumor
metastasis in colorectal cancer. Mol Cancer. 19:1062020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wang S, Gan M, Chen C, Zhang Y, Kong J,
Zhang H and Lai M: Methyl CpG binding protein 2 promotes colorectal
cancer metastasis by regulating N6-methyladenosine
methylation through methyltransferase-like 14. Cancer Sci.
112:3243–3254. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wang H, Wei W, Zhang ZY, Liu Y, Shi B,
Zhong W, Zhang HS, Fang X, Sun CL, Wang JB and Liu LX: TCF4 and HuR
mediated-METTL14 suppresses dissemination of colorectal cancer via
N6-methyladenosine-dependent silencing of ARRDC4. Cell Death Dis.
13:32021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang L, Hui H, Agrawal K, Kang Y, Li N,
Tang R, Yuan J and Rana TM: m6A RNA methyltransferases
METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J.
39:e1045142020. View Article : Google Scholar
|
|
98
|
Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li
J, Liu Y, Liu J, Chang R, Li Y, et al: The loss of RNA
N6-adenosine methyltransferase Mettl14 in
tumor-associated macrophages promotes CD8+ T cell
dysfunction and tumor growth. Cancer Cell. 39:945–957.e10. 2021.
View Article : Google Scholar
|
|
99
|
Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B,
Shen C, Ma Y, Jiang S, Ma D, et al: Enterotoxigenic Bacteroides
fragilis promotes intestinal inflammation and malignancy by
inhibiting exosome-packaged miR-149-3p. Gastroenterology.
161:1552–1566.e12. 2021. View Article : Google Scholar
|
|
100
|
Luo M, Huang Z, Yang X, Chen Y, Jiang J,
Zhang L, Zhou L, Qin S, Jin P, Fu S, et al: PHLDB2 mediates
cetuximab resistance via interacting with EGFR in latent metastasis
of colorectal cancer. Cell Mol Gastroenterol Hepatol. 13:1223–1242.
2022. View Article : Google Scholar :
|
|
101
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Wolfgang CL, Herman JM, Laheru DA, Klein
AP, Erdek MA, Fishman EK and Hruban RH: Recent progress in
pancreatic cancer. CA Cancer J Clin. 63:318–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Hidalgo M, Cascinu S, Kleeff J, Labianca
R, Löhr JM, Neoptolemos J, Real FX, Van Laethem JL and Heinemann V:
Addressing the challenges of pancreatic cancer: Future directions
for improving outcomes. Pancreatology. 15:8–18. 2015. View Article : Google Scholar
|
|
104
|
Wang M, Liu J, Zhao Y, He R, Xu X, Guo X,
Li X, Xu S, Miao J, Guo J, et al: Upregulation of METTL14 mediates
the elevation of PERP mRNA N6 adenosine methylation
promoting the growth and metastasis of pancreatic cancer. Mol
Cancer. 19:1302020. View Article : Google Scholar
|
|
105
|
Jiang Z, Song X, Wei Y, Li Y, Kong D and
Sun J: N(6)-methyladenosine-mediated miR-380-3p maturation and
upregulation promotes cancer aggressiveness in pancreatic cancer.
Bioengineered. 13:14460–14471. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Kong F, Liu X, Zhou Y, Hou X, He J, Li Q,
Miao X and Yang L: Downregulation of METTL14 increases apoptosis
and autophagy induced by cisplatin in pancreatic cancer cells. Int
J Biochem Cell Biol. 122:1057312020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Nabors LB, Portnow J, Ahluwalia M,
Baehring J, Brem H, Brem S, Butowski N, Campian JL, Clark SW,
Fabiano AJ, et al: Central nervous system cancers, version 3.2020,
NCCN clinical practice guidelines in oncology. J Natl Compr Canc
Netw. 18:1537–1570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhang C, Ou S, Zhou Y, Liu P, Zhang P, Li
Z, Xu R and Li Y: m6A Methyltransferase METTL14-Mediated
upregulation of cytidine deaminase promoting gemcitabine resistance
in pancreatic cancer. Front Oncol. 11:6963712021. View Article : Google Scholar
|
|
109
|
Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D,
Peng X, Ying P, Zhang M, Niu S, et al:
N6-methyladenosine mRNA methylation of PIK3CB regulates
AKT signalling to promote PTEN-deficient pancreatic cancer
progression. Gut. 69:2180–2192. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Chen S, Yang C, Wang ZW, Hu JF, Pan JJ,
Liao CY, Zhang JQ, Chen JZ, Huang Y, Huang L, et al: CLK1/SRSF5
pathway induces aberrant exon skipping of METTL14 and Cyclin L2 and
promotes growth and metastasis of pancreatic cancer. J Hematol
Oncol. 14:602021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen FQ, Zheng H, Gu T, Hu YH, Yang L,
Huang ZP, Qiao GL and Li HJ: Modification of STIM2 by
m6A RNA methylation inhibits metastasis of
cholangiocarcinoma. Ann Transl Med. 10:402022. View Article : Google Scholar
|
|
112
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19(1A): A68–A77. 2015.PubMed/NCBI
|
|
113
|
Kong F, Wang K and Wang L: Systematic
analysis of the expression profile and prognostic significance of
m6A regulators and PD-L1 in hepatocellular carcinoma. Discov Oncol.
13:1312022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Li Z, Li F, Peng Y, Fang J and Zhou J:
Identification of three m6A-related mRNAs signature and risk score
for the prognostication of hepatocellular carcinoma. Cancer Med.
9:1877–1889. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Xu F, Zhang Z, Yuan M, Zhao Y, Zhou Y, Pei
H and Bai L: M6A regulatory genes play an important role in the
prognosis, progression and immune microenvironment of pancreatic
adenocarcinoma. Cancer Invest. 39:39–54. 2021. View Article : Google Scholar
|
|
116
|
Zhang T, Sheng P and Jiang Y: m6A
regulators are differently expressed and correlated with immune
response of pancreatic adenocarcinoma. J Cancer Res Clin Oncol.
149:2805–2822. 2023. View Article : Google Scholar
|
|
117
|
Cai C, Long J, Huang Q, Han Y, Peng Y, Guo
C, Liu S, Chen Y, Shen E, Long K, et al: M6A 'Writer' gene METTL14:
A favorable prognostic biomarker and correlated with immune
infiltrates in rectal cancer. Front Oncol. 11:6152962021.
View Article : Google Scholar
|
|
118
|
Chen Y, Wang S, Cho WC, Zhou X and Zhang
Z: Prognostic Implication of the m6A RNA Methylation
Regulators in Rectal Cancer. Front Genet. 12:6042292021. View Article : Google Scholar
|
|
119
|
Xu Z, Chen Q, Shu L, Zhang C, Liu W and
Wang P: Expression profiles of m6A RNA methylation regulators,
PD-L1 and immune infiltrates in gastric cancer. Front Oncol.
12:9703672022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wang H, Zhang Y, Chen L, Liu Y, Xu C,
Jiang D, Song Q, Wang H, Wang L, Lin Y, et al: Identification of
clinical prognostic features of esophageal cancer based on m6A
regulators. Front Immunol. 13:9503652022. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Du Y, Hou G, Zhang H, Dou J, He J, Guo Y,
Li L, Chen R, Wang Y, Deng R, et al: SUMOylation of the m6A-RNA
methyltransferase METTL3 modulates its function. Nucleic Acids Res.
46:5195–5208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Chen HM, Li H, Lin MX, Fan WJ, Zhang Y,
Lin YT and Wu SX: Research progress for RNA modifications in
physiological and pathological angiogenesis. Front Genet.
13:9526672022. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Chen Y, Lu Z, Qi C, Yu C, Li Y, Huan W,
Wang R, Luo W, Shen D, Ding L, et al:
N6-methyladenosine-modified TRAF1 promotes sunitinib
resistance by regulating apoptosis and angiogenesis in a
METTL14-dependent manner in renal cell carcinoma. Mol Cancer.
21:1112022. View Article : Google Scholar
|
|
124
|
Wen H, Tang J, Cui Y, Hou M and Zhou J:
m6A modification-mediated BATF2 suppresses metastasis and
angiogenesis of tongue squamous cell carcinoma through inhibiting
VEGFA. Cell Cycle. 22:100–116. 2023. View Article : Google Scholar
|