Various types of DNA damage can occur when cells are
exposed to endogenous or exogenous factors, including alterations
in base pairs, errors during DNA replication, and twisting and
breaking of the DNA double helix strand (1,2).
Exogenous factors, such as toxic heavy metals and ionizing
radiation are known to cause severe DNA damage (3-7).
Endogenous factors are often released during the metabolism of
exogenous substances in the body or as a result of cell damage and
the loss of cell membrane integrity (8). It is estimated that cells experience
~70,000 DNA lesions per day (9).
While the majority of these lesions are single-strand breaks, there
are also a few instances of DNA double-strand breaks (DSBs). To
cope with this continuously occurring damage, eukaryotic cells have
developed a complex and efficient DNA damage response (DDR) system
that consists of numerous DNA damage repair pathways (10-12). The primary molecular pathways for
DSB repair are homologous recombination (HR) and non-homologous end
joining (NHEJ). DSBs are particularly harmful and pose a serious
threat to cells. If DSBs are not effectively repaired or undergo
error-prone repair, they can lead to carcinogenesis or cell death
(13). DNA damage also serves as
the foundation of cancer therapy. Chemotherapy and radiotherapy,
which are based on inducing severe DNA damage to and the apoptosis
of cancer cells, are the preferred treatment regimens for the
majority of malignancies (14).
However, the activation of DNA damage repair pathways can promote
resistance to genotoxic drugs, which remains a significant obstacle
in the successful treatment of cancer (15,16). Therefore, it is crucial to
elucidate the molecular mechanisms underlying DNA damage repair in
order to improve the effectiveness of DNA damage-based anticancer
therapies.
Long non-coding RNAs (lncRNAs) are a class of RNAs
that exceed 200 nucleotides in length and lack protein-coding
potential (17). They have
garnered significant attention in recent years. There is mounting
evidence to suggest that numerous lncRNAs are dysregulated in
various types of cancer and play crucial roles in cancer
development and progression (18). The involvement of lncRNAs in drug
resistance has also been extensively reported (19,20). Through their interactions with
RNA, DNA, or proteins, lncRNAs have emerged as potent regulators of
numerous cellular processes (21). RNA-binding proteins (RBPs), a
group of proteins, are known to directly bind to single- or
double-stranded lncRNAs, participating in lncRNA-mediated
regulatory activities (22).
Furthermore, the function of certain lncRNAs is dependent on their
interactions with specific proteins (23). The interplay between lncRNAs and
RBPs plays a critical role in regulating cancer development,
progression and drug resistance by influencing DNA damage repair.
However, the underlying mechanisms involved in this interplay
remain poorly understood, thus necessitating further
investigations.
The present review provides a comprehensive summary
of the mechanisms through which lncRNAs regulate DDR, DNA DSB
repair and influence the sensitivity and resistance to chemotherapy
and radiotherapy in DNA damage/repair processes in cancer cells by
binding to RBPs. The aim of the present review was to elucidate the
regulatory mechanisms of lncRNAs and RBPs associated with cancer
development, progression and treatment, thereby aiding the
development of novel strategies for cancer therapy. A systematic
literature search was conducted using PubMed, employing keywords
such as 'long non-coding RNA', 'RNA-binding protein', 'DNA damage',
'DNA repair', 'DNA damage repair', 'DNA double-strand break' and
'cancer'. Articles discussing the regulation of DNA damage repair
in cancer by lncRNAs through interactions with RBPs were screened
and analyzed.
Genomic DNA in organisms is highly susceptible to
both exogenous and endogenous damage. To maintain genomic integrity
and prevent genetic instability, cells and organisms rely on
mechanisms to preserve the integrity of their genomic DNA (14,15,24). One crucial mechanism is the DDR, a
series of rapid cellular processes that are activated upon the
detection of DNA damage (25).
The DDR pathway comprises sensors, receptors and effectors that
sense DNA damage, propagate signals and initiate appropriate
responses, including cell cycle arrest, DNA repair or apoptosis
(26-28). In addition to its role in precise
cell replication and genome maintenance, there is increasing
evidence to indicate that he DDR is involved in resistance to DNA
damage-based chemotherapy and radiotherapy (29). It has been observed that molecules
expressed as proteins in the DDR pathway can modulate the effects
of chemotherapy and radiotherapy (30). Following DNA damage, the ataxia
telangiectasia mutated (ATM) kinase is activated through
autophosphorylation at the site of damage. ATM, in turn,
phosphorylates downstream substrates, including the tumor
suppressor p53, breast cancer type 1 susceptibility protein (BRCA1)
and checkpoint kinase (CHK)2. These effector molecules transmit DNA
damage signals and activate cell cycle checkpoints, DNA repair and
apoptosis (24,31,32). Research has highlighted the
significant regulatory role of lncRNAs in the DDR, with a number of
proteins binding to lncRNAs and participating in their regulatory
activities (22). Therefore, the
present review provides a comprehensive summary of the role of
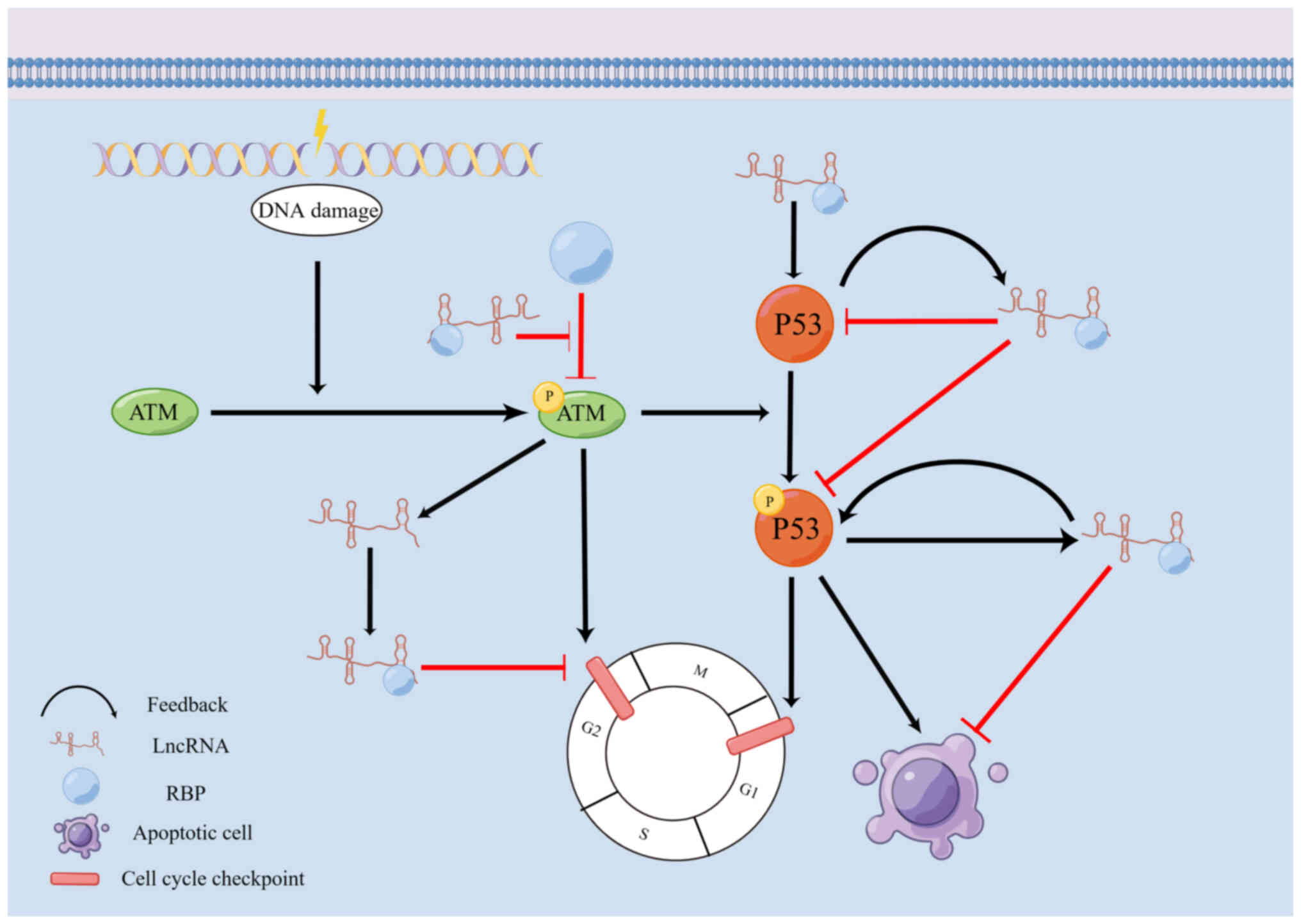
lncRNA binding to RBPs in these DDR processes (Fig. 1 and Table I).
DDR involves a series of networks linking tumor
suppressor genes to DNA repair pathways, damage tolerance
processes, cell cycle checkpoints and apoptosis (24,33). The ATM kinase plays a crucial role
as a sensor in the DDR pathway, particularly in detecting DNA DSBs.
The ATM-mediated phosphorylation of downstream target proteins
initiates signaling cascades that activate cell cycle checkpoints
and DNA repair mechanisms (34).
lncRNAs have the ability to directly or indirectly
regulate the activation or repression of cell cycle checkpoints
through their interactions with RBPs, thereby influencing the DDR.
Wan et al (35) discovered
that lncRNA ANRIL was induced by the E2F1 transcription factor in
an ATM-dependent manner following DNA damage. ANRIL interacted with
polycomb repressor complex (PRC)1 and PRC2 to suppress the
expression of INK4B-ARF-INK4A motifs, specifically p15(INK4b),
p16(INK4a) and p14(ARF). This inhibitory effect on gene expression
led to the suppression of cell cycle checkpoint activation,
promoting cell proliferation and maintaining the DDR (35). In another study, Wan et al
(36) found that lncRNA JADE
inhibited the DNA damage checkpoint and enhanced cell
proliferation. Similarly, lncRNA JADE expression was induced in an
ATM-dependent manner following DNA damage. JADE acted in
collaboration with BRCA1 to mediate the transcriptional induction
of JADE1 following DNA damage, resulting in the upregulation of
JADE1 expression and increased histone H4 acetylation. These
molecular events disrupted the DNA damage checkpoint regulation,
impaired the DDR and promoted cancer progression (36). Telomeric repeat
sequence-containing RNA (TERRA) is a large non-coding RNA localized
in mammalian cells and is a component of telomeric heterochromatin
(37,38). The inhibition of telomeric-repeat
binding factor 2 (TRF2), a protein involved in telomere
maintenance, triggers an ATM-dependent DDR pathway and activates
cell cycle checkpoints (39,40). Zhang et al (41) demonstrated that TERRA can form a
complex with the G-tetraspanin quinoline derivative, CK1-14, which
binds to the TERRA G-quadruplex. This complex disrupts the binding
of TRF2 to telomeric double-stranded DNA, leading to the induction
of a DDR in U2OS cells. Consequently, the cell cycle checkpoint is
activated, resulting in cell cycle arrest, the inhibition of cell
proliferation and apoptosis. CK1-14 exhibits potential as a lead
compound for further development as a novel target for cancer
therapy (41).
p53 is a crucial transcription factor involved in
stress and the DDR. It plays a pivotal role in activating cell
cycle arrest, DNA repair and apoptosis (42). In non-stressed cells, p53 levels
are maintained at low levels, while p53 levels are significantly
increased during stress (43). In
response to stress signals such as DNA damage, p53 is stabilized
and activated to perform its function as a sequence-specific
transcription factor, inducing genes involved in cell cycle arrest,
apoptosis and the expression of its negative regulators (44,45). Apart from protein-coding genes, an
increasing number of lncRNAs are recognized as targets of p53 and
contribute to p53 regulation and its effector functions (46). lncRNAs can also be involved in the
regulation of p53 function through their interactions with RBPs,
thereby influencing the DDR.
The transcriptional response of p53 involves the
activation and repression of numerous genes. It has been discovered
that lncRNAs play crucial regulatory roles in the p53
transcriptional response (50).
Huarte et al (51)
reported that lincRNA p21, located upstream of the CDKN1A gene, was
activated by p53 following DNA damage. lincRNA P21 interacted with
hnRNPK to participate in a p53-dependent transcriptional repression
response. It inhibited the expression of downregulated genes that
are typically part of the p53 transcriptional response, thereby
promoting apoptosis and contributing to the regulation of the DDR
(51). By contrast, Hung et
al (52) found that an lncRNA
termed PANDA, induced in a p53-dependent manner, restricted
apoptosis in the DDR. PANDA was found to bind to NF-YA, preventing
or repelling NF-YA from binding to chromatin. This suppression of
NF-YA binding led to the downregulation of pro-apoptotic genes,
cell cycle arrest and the subsequent regulation of the DDR
(52).
DNA repair is a critical biological process that
ensures the integrity of genomic DNA and enables normal
physiological functions, such as cell division (10,53). Under normal conditions, cells
possess six major DNA repair pathways that precisely repair DNA
damage, thus maintaining genomic stability (54). Among the various types of DNA
damage, DSBs are particularly harmful and challenging to repair
(55). Fortunately, cells have
two primary pathways for repairing DSBs: HR and NHEJ (56). These pathways are typically
mediated by proteins belonging to the phosphatidylinositol
3-kinase-like protein kinase family, such as ATM, ATM- and
Rad3-related (ATR), and DNA-dependent protein kinase catalytic
subunits (DNA-PKcs) (10). The
selection of the repair pathway is influenced by the cell cycle
phase (57). In the G1 phase,
DSBs are primarily repaired through error-prone NHEJ, involving the
direct rejoining of DNA ends (58). By contrast, during the S/G2 phase,
HR becomes the predominant pathway and utilizes homologous DNA
template sequences for error-free repair (59). HR is considered more conserved and
error-free due to its reliance on sister chromatids (60,61). However, this property restricts
the ability of the HR pathway to repair DSBs to the S/G2 phase,
while the NHEJ pathway can repair DSBs throughout the cell cycle
(62-64).
The regulatory mechanisms of DNA damage repair play
a critical role in the identification of tumor markers and the
development of more effective targeted therapies. While the
functions of lncRNAs have been extensively studied (65), only a limited number of lncRNAs
have been implicated in DNA repair processes (66-68). Furthermore, lncRNAs can bind to
RBPs to regulate DNA damage repair. Therefore, it is essential to
investigate the regulatory mechanisms involving lncRNAs and RBPs in
the two repair pathways, HR and NHEJ, specifically in DSB repair
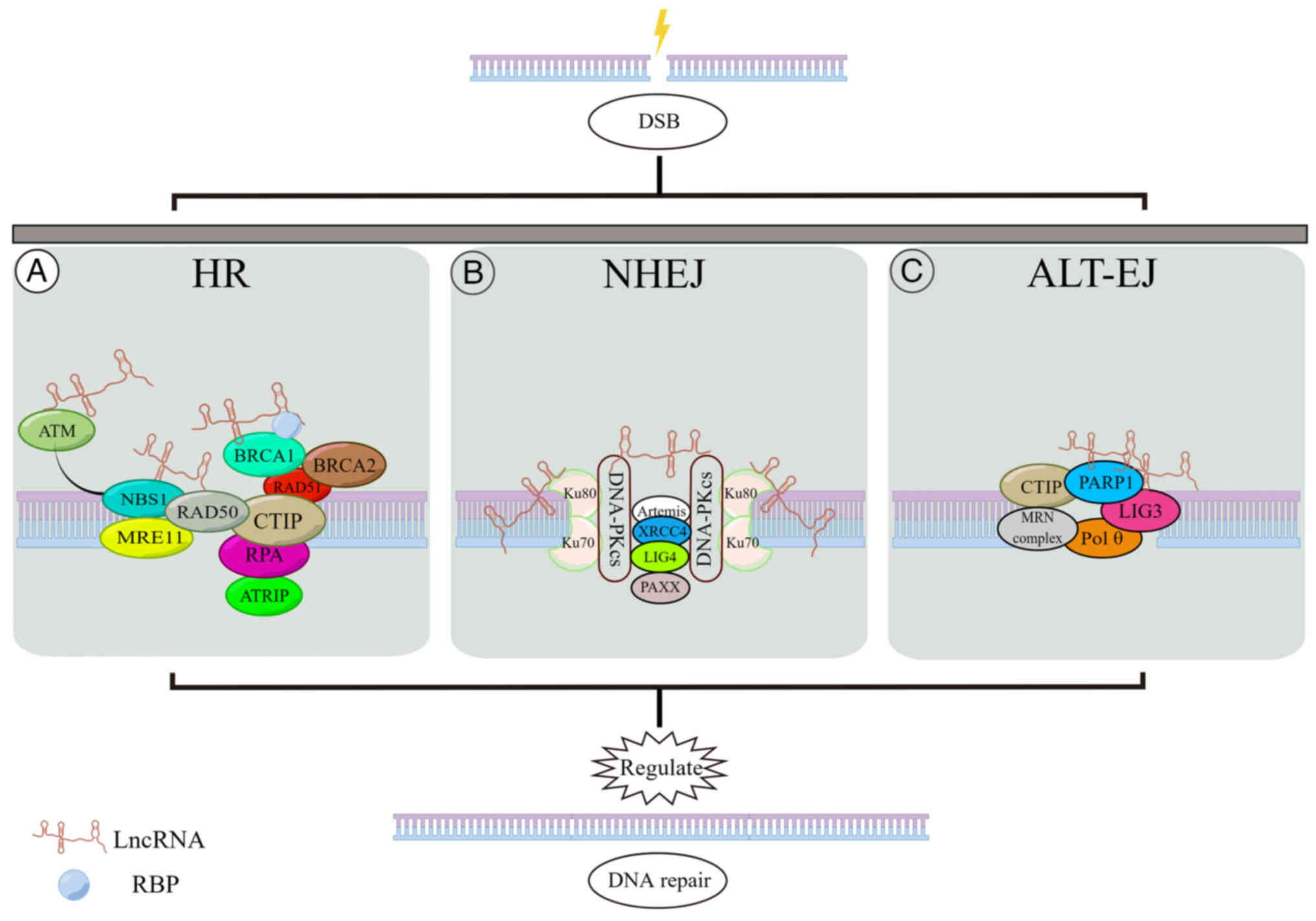
(Fig. 2 and Table II).
The NHEJ pathway is an error-prone mechanism
initiated by the binding of DNA break ends to DNA-PK complexes
(69). Upon encountering DNA
DSBs, Ku80-Ku70 heterodimers bind to the broken ends, forming a
clamp complex that recruits DNA-PKcs to the injury site. Two
DNA-PKcs molecules interact with the DSB site, forming a synaptic
complex that immobilizes the DSB end and protects it from nuclease
digestion. Following DNA end processing by Artemis, DNA ligase
(LIG)4 and XRCC4 mediate DNA ligation to facilitate the repair of
the broken ends (70).
Ku is an RBP that stabilizes the initial synaptic
complex in classical NHEJ DSB repair (71). lncRNAs can regulate this repair
pathway by binding to Ku. Zhang et al (72) discovered that in triple-negative
breast cancer, lncRNA LINP1 interacted with Ku80 and DNA-PKcs,
acting as a molecular scaffold. This interaction enhanced the
molecular interactions between Ku80 and DNA-PKcs, stabilized the
Ku80-DNA-PKcs complex and promoted NHEJ-mediated DNA repair
(72). Similarly, in patients
with cervical cancer, Wang et al (73) observed the elevated expression of
lncRNA LINP1, which promoted NHEJ-mediated DNA repair through the
same mechanism described above. Thapar et al (71) further revealed that the
interaction between LINP1 and Ku effectively substituted the
auxiliary NHEJ protein PAXX in the NHEJ complex. LINP1 enhanced
NHEJ-mediated DNA repair by increasing the net concentration of
NHEJ factors at DSBs and facilitating the joining of two Ku
heterodimers via DSBs, thereby effectively replacing PAXX and
achieving efficient NHEJ (71).
The early and long-term binding of repair factors has been shown to
play a crucial role in the initiation and signal transduction of
DNA damage and repair (74).
Repair factors, including DNA-PKcs, XRCC4, LIG4 and XLF, bind to
DSBs following the perception of damage by the Ku70-Ku80
heterodimer (75). In various
cancer cells, lncRNA LRIK is upregulated upon the induction of DNA
damage. LRIK interacts with Ku70-Ku80 heterodimers, prolonging
their binding to DSB sites and promoting the recruitment of XRCC4
and DNA-PKcs, thereby enhancing the formation of repair complexes
at DSB sites in chromatin and facilitating effective DNA damage
repair through the NHEJ pathway (76). By contrast, the study by Guo et
al (77) reported that
linc00312 expression was downregulated in nasopharyngeal carcinoma,
leading to a significant decrease in patient survival. Further
investigations revealed that linc00312 directly bound to DNA-PKcs
and inhibited its recruitment to Ku80, thereby impairing NHEJ
repair (77). This, in turn,
reduced the viability of nasopharyngeal carcinoma cells and
promoted apoptosis (77).
Upon DNA damage, the meiotic recombination 11
homolog 1 (MRE11)-RAD50-Nijmegen breakage syndrome 1 protein (NBS1)
(MRN) complex acts as a sensor for DNA DSBs and binds to the
damaged site. BRCA1 and CTIP are subsequently recruited to the site
of damage. The MRN complex facilitates the activation of ATM
through autophosphorylation (78,79). Activated ATM phosphorylates
various DNA repair factors, including core histone variants H2AX,
CTIP, BRCA1, and the exonuclease EXO1 (59). BRCA1 interacts with CTIP, leading
to the activation of MRE11 and stimulating the exonuclease and
endonuclease activities necessary for excising the 5'-3'DNA strand
and generating a 3'single-stranded DNA (ssDNA) overhang. The ssDNA
is then coated by replication protein A (RPA), which prevents the
formation of DNA secondary structures. The RPA-coated ssDNA
activates CHK1 and CHK2 through ATRIP, resulting in cell cycle
arrest to allow time for repair (79-82). BRCA2 binds to BRCA1 and promotes
the recruitment of RAD51 to the RPA-coated ssDNA, displacing RPA
and forming a stable RAD51-ssDNA complex. BRCA2 also inhibits the
ATPase activity of RAD51 and stabilizes the RAD51-ssDNA complex
(10,79,83,84). Subsequently, a homology search and
DNA repair through strand invasion take place (85).
The MRN complex plays a central role in DNA damage
repair by sensing damaged DNA, processing broken DNA ends, and
activating DNA damage repair pathways (93,94). In osteosarcoma, lncRNA H19
interacts with RBBP8 (also known as CTIP) and participates in the
MRN complex in DNA end resection, promoting HR-mediated DSB repair
(95). Under endoplasmic
reticulum stress, HITTERS interacts with both RAD50 and MRE11,
promoting the formation of MRN complexes. This interaction
increases the expression of proteins involved in DNA damage repair,
facilitates the repair of the HR pathway, protects oral squamous
cell carcinoma from endoplasmic reticulum stress-induced apoptosis,
and promotes cancer development (96). MRN complexes also contribute to
ATM phosphorylation, subsequently triggering the phosphorylation of
various ATM effector proteins (97). Zhao et al (98) reported that lncRNA hypoxia
inducible factor-1α (HIF-1α) inhibitor at translation level (HITT)
was induced and maintained at high levels following DSB in HCT116
cells. HITT bound to the NBS1 binding site in ATM, masking the site
on ATM that binds to NBS1 (98).
This binding inhibited the association between ATM and NBS1,
preventing the NBS1-mediated recruitment of ATM to the DSB and
inhibiting HR repair. This highlights the potential role of HITT in
sensitizing cancer to genotoxic treatment (98).
During the G1 phase of the cell cycle, the 53BP1
protein blocks the accumulation of BRCA1 at the DSB site and
promotes NHEJ (99-101). The E3 ubiquitin ligase RNF169
has been found to replace 53BP1 at the DSB site, facilitating the
initiation of HR repair (102,103). Deng et al (56) discovered that lncRNA PRLH1
specifically bound to RNF169 via two GCUUCA boxes in its 5'terminal
region, forming a stable repair complex. This complex stabilized
RNF169 and controled the recruitment and retention of RNF169 at the
DSB site, replacing 53BP1 and facilitating HR repair (56).
In addition to the NHEJ and HR pathways, an
alternative end-joining repair pathway, known as ALT-EJ or
microhomologous gene-mediated end joining, is responsible for the
repair of residual DSBs that cannot be resolved by NHEJ or HR.
ALT-EJ is associated with frequent chromosomal abnormalities, such
as deletions, translocations, inversions and complex rearrangements
(104). It is Ku-independent and
is dependent on the microhomologous regions on either side of the
break site (70). Several
proteins have been identified to be involved in the ALT-EJ repair
pathway in mammals, including CTIP in complex with MRN, PARP1, LIG3
and DNA polymerase Pol θ (105).
Although LIG3 lacks an RNA-binding structural domain, it can
interact with PARP1 through the presence of the PARP and DNA-ligase
Zn-finger (zf-PARP) region (106). PARP1 and LIG3 are key molecules
in the ALT-EJ DNA repair pathway (107). Hu et al (108) reported that in multiple myeloma,
lncRNA MALAT1 bound directly to PARP1 and indirectly to LIG3,
facilitating the recognition of DSBs γH2AX sites on DNA by the
PARP1/LIG3 complex and promoting DNA repair via A-NHEJ (108). It is worth noting that PARP1 has
three zinc finger structural domains, with only the Zn3 structural
domain capable of binding to RNA (109). Huang et al (110) further demonstrated in NSCLC
cells that PARP1 bound to MALAT1 through the Zn3 structural domain,
thereby regulating the ALT-EJ repair pathway. Additionally, MALAT1
was found to promote the HR pathway by regulating the expression of
BRCA1 for DNA repair (110).
In summary, lncRNAs play a significant role in
various DSB repair pathways through their interactions with RBPs,
influencing cancer progression. Therefore, further investigations
into the regulation of DSB repair in cancer by lncRNAs binding to
RBPs are warranted.
Resistance to chemotherapy and radiation therapy
remains a significant challenge in clinical cancer treatment. DNA
damage serves as a fundamental mechanism of action for these
treatments. DSBs represent the most harmful form of DNA damage that
can arise from radiotherapy or DNA-based chemotherapy (15). While radiation and chemotherapy
are designed to induce substantial DNA damage in cancer cells, the
activation of DNA damage repair systems in the body can limit their
effectiveness (111). Therefore,
it is crucial to investigate the effects of lncRNA binding to RBPs
through DNA damage repair on chemotherapy and radiotherapy for
cancer (Table III).
lncRNAs play a significant role in the regulation of
various physiological and pathological cellular processes at three
distinct levels: Transcriptional, post-transcriptional and
epigenetic. Moreover, they are closely associated with the
development, progression and prognosis of cancer (112). There is increasing evidence to
support the association between lncRNAs and resistance to
chemotherapy and radiotherapy in cancer treatment, thereby
highlighting the potential of lncRNAs as biomarkers (113-115). One mechanism by which lncRNAs
exert their functions is through their interaction with specific
binding proteins (77). Taking
into account the existing literature, lncRNAs combined with RBPs
are mainly discussed herein to regulate DNA damage repair through
transcriptional and post-transcriptional levels, which in turn
affects cancer cell chemotherapy and radiotherapy. The epigenetic
regulation is not further discussed.
The CDKN1A (p21) gene plays a critical role in cell
cycle checkpoint control and facilitates cell cycle arrest
(116). Liu et al
(117) discovered that in
gastric cancer, lncRNA PANDAR was overexpressed and competitively
bound to p53 protein, leading to the suppression of CDKN1A gene
transcription. This response to DNA damage inhibited apoptosis,
promoted gastric cancer cell proliferation and contributes to
chemoresistance. The depletion of PANDAR combined with a p53
activator demonstrated notable efficacy in cancer therapy in
vivo (117). PANDAR emerged
not only as a potent diagnostic biomarker for patients with gastric
cancer, but also as a promising target for cancer therapy (117). Additionally, TROY has been
identified as a contributor to DNA damage repair (118). lncRNA SNHG8 exhibits an
upregulated expression in multiple types of cancer (119-123). Zhu et al (124) revealed that in gastric cancer,
lncRNA SNHG8 bound to hnRNPA1, leading to the stabilization of TROY
expression. This interaction promoted DNA damage repair, inhibited
apoptosis and ultimately promoted chemotherapeutic resistance in
gastric cancer (124). The
inhibition of SNHG8 impeded DNA damage repair and reduced the
resistance of gastric cancer cells to chemotherapy, providing
insight into a novel molecular mechanism underlying drug resistance
in gastric cancer (124).
Furthermore, in hepatocellular carcinoma, linc01134 has been shown
to interact with the IGF2BP2 protein, enhancing MAPK1 mRNA
stability and promoting MAPK1 expression, regulating DDR (125). This interaction inhibits
apoptosis, accelerates cancer cell proliferation, and augments
radiotherapy resistance. Consequently, linc01134 may represent a
potential therapeutic target for enhancing the effectiveness of
radiotherapy in hepatocellular carcinoma (125). Similarly, Sun et al
(126) reported that the DNA
damage-activated non-coding RNA NORAD competitively bound to PUM1
of pri-miR-199a1, impeding the processing of pri-miR-199a1.
Consequently, the expression of miR-199a-5p was suppressed,
resulting in the upregulation of EEPD1 expression (126). This process enhanced the HR
repair pathway in DNA DSBs and inhibited cell apoptosis, thereby
conferring resistance to radiotherapy in ESCC cells (126). Previous research by Yao et
al (127) demonstrated that
ANKHD1 was highly expressed in colorectal cancer (CRC) and promoted
CRC cell proliferation, invasion and migration through the
activation of YAP1. Subsequent investigations revealed that ANKHD1
interacted with both lncRNA MALAT1 and YAP1 in CRC. Both ANKHD1 and
MALAT1 positively regulated the transcriptional activity of YAP1,
which in turn promoted ATM-CHK2 phosphorylation by activating AKT.
Consequently, this cascade upregulated MRE11 expression,
facilitating DNA DSB repair and ultimately promoting radiotherapy
resistance in CRC. This ANKHD1/MALAT1/YAP1 interaction loop, along
with the downstream YAP1/AKT axis, may represent a potential
therapeutic target for comprehensive CRC treatment (128).
lncRNAs can exert their influence on resistance to
chemo- and radiotherapy in cancer cells through
post-transcriptional regulation. PTBP1 is an RBP known for its
involvement in premature RNA splicing events and its association
with cancer progression (129).
Huan et al (130)
discovered that in CRC, lncRNA LUCAT1 was induced by HIF-1α
transcription under hypoxic stress. Elevated levels of LUCAT1
interacted with PTBP1 protein, regulating the selective splicing of
downstream target genes (APP, CD44, CLSTN1, MBNL1 and ZNF207)
(130). This interaction
inhibited DNA damage and apoptosis, leading to chemoresistance and
promoting CRC cell survival. These findings suggest that LUCAT1 may
serve as a predictive indicator and therapeutic target for patients
with CRC undergoing chemotherapy (130).
Resistance to chemotherapy and radiotherapy
primarily arises from the induction of DNA DSBs. In response, three
important DNA damage sensors, ATM, ATR and DNA-PKcs, are
immediately activated to assist cancer cells in evading the damage
caused by chemo- and radiotherapy. This evasion is accomplished
through enhanced DNA repair mechanisms (131-133). Zhang et al (72) reported that LINP1 was highly
expressed in triple-negative breast cancer and that the inhibition
of LINP1 expression impaired DNA repair activity, thereby
sensitizing the cancer cells to radiation therapy. Their study also
revealed a positive correlation between LINP1 and epidermal growth
factor receptor (EGFR) expression (72). Further investigations demonstrated
that EGFR pathway activation, followed by MAPK (RAS-MEK-ERK)
pathway activation and AP1 transcription factor induction, led to
an increased LINP1 transcription (72). Elevated LINP1 levels stabilized
the interaction between Ku80 and DNA-PKcs, enhancing NHEJ-mediated
DNA repair activity. This, in turn, increased cancer cell survival
and contributed to radiotherapy resistance (72). Similar mechanisms have been
observed in cervical cancer, where LINP1 played a role in radiation
resistance and served as a prognostic marker and potential
therapeutic target (73).
Conversely, another study demonstrated that linc00312 expression
was downregulated in nasopharyngeal carcinoma and this was
associated with a reduced patient survival (77). Subsequent analyses demonstrated
that linc00312 directly bound to DNA-PKcs, inhibiting its
recruitment to Ku80 and impairing NHEJ repair. This resulted in the
decreased viability and increased apoptosis of nasopharyngeal
carcinoma cells (77). Moreover,
linc00312 inhibited radiation-induced AKT-DNA-PKcs, MRN-ATM-CHK2
and ATR-CHK1 signaling, leading to impaired DNA damage sensing,
processing and repair. Consequently, the sensitivity to radiation
therapy was increased. These findings provide new insight into the
regulation of radiosensitivity by linc00312 in nasopharyngeal
carcinoma (77). Additionally,
Zhao et al (98) reported
that lncRNA HITT was downregulated in multiple types of cancer.
However, under DSB induction, HITT transcription was upregulated
and maintained at high levels. HITT bound to the NBS1 binding site
in ATM, preventing the association between ATM and NBS1 (98). This inhibition hindered the
recruitment of ATM to the DSB site, impairing HR pathway repair.
In vitro and in vivo analyses demonstrated that the
HITT-mediated inhibition of ATM increased the death of cancer cells
treated with doxorubicin, suggesting its significant role in
enhancing chemosensitivity. Blocking the NBS1/ATM interaction may
thus be a potential target for anticancer therapy (98).
In recent years, the role of RBPs and their partners
in cancer progression and treatment has garnered increasing
attention (136,137). RBPs were once considered
'non-druggable'; however, the identification of small molecules or
chemically modified antisense oligonucleotides targeting RBPs has
opened up new possibilities for the treatment of certain diseases
(138,139). Zhu et al (140) discovered that a highly expressed
RBP, zinc finger CCHC domain-containing protein 4 (ZCCHC4), was
associated with a poor prognosis in several types of cancer. ZCCHC4
and the previously unidentified lncRNA AL133467.2 formed nuclear
complexes with the DNA damage indicator γH2AX in
oxaliplatin-induced DDR. ZCCHC4 attenuated AL133467.2 and γH2AX,
resulting in a downregulation of DNA damage intensity in cancer
cells (140). This interaction
inhibited DNA damage-induced apoptosis in hepatocellular carcinoma
cells and promoted chemoresistance. These findings provide a novel
understanding of the mechanisms through which RBPs and their
interacting molecules regulate cancer progression and
chemoresistance. The epigenetic role of RBPs and their partners in
solid cancer chemoresistance remains poorly understood and thus
requires further investigation (140).
DNA damage, DDR, and repair are crucial factors in
cancer development, progression and therapy. Despite previous
perceptions of lncRNAs as 'junk RNA' due to their lack of
protein-coding capacity, it is now evident that they play
significant roles in various aspects of cancer biology. lncRNAs
interact with RBPs and contribute to numerous cellular processes,
including the regulation of DNA damage repair in cancer cells. The
present review provides insight into the molecular mechanisms
underlying the interaction between lncRNAs and RBPs, specifically
in the context of DNA damage repair in cancer cells. This knowledge
may open up new avenues for cancer treatment strategies aimed at
enhancing the effectiveness of DNA damage-repair-based therapies.
Although substantial research has been conducted to elucidate the
functions and mechanisms of lncRNAs and their impact on cancer
therapy, the precise underlying mechanisms remain largely unknown.
Therefore, further investigations are warranted to enhance the
current understanding of this intricate interplay.
Not applicable.
SZ and KW conceived the study. SZ drafted the
manuscript, and prepared the figures and tables. XG participated in
the literature search and in the analysis of the data to be
included in the review. KW edited and revised the manuscript. Data
authentication is not applicable. All authors have read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present review was supported by the National Natural Science
Foundation of China (grant no. 82160575) and the Outstanding Young
Technological and Innovative Talent Cultivation Project of Zunyi
Municipal Science and Technology Bureau, 2021 (no. 10).
|
1
|
Ragunathan K, Upfold NLE and Oksenych V:
Interaction between fibroblasts and immune cells following DNA
Damage induced by ionizing radiation. Int J Mol Sci. 21:86352020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marshall CJ and Santangelo TJ: Archaeal
DNA repair mechanisms. Biomolecules. 10:14722020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maremonti E, Brede DA, Olsen AK, Eide DM
and Berg ES: Ionizing radiation, genotoxic stress, and
mitochondrial DNA copy-number variation in Caenorhabditis elegans:
Droplet digital PCR analysis. Mutat Res Genet Toxicol Environ
Mutagen. 858-860:5032772020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pariset E, Malkani S, Cekanaviciute E and
Costes SV: Ionizing radiation-induced risks to the central nervous
system and countermeasures in cellular and rodent models. Int J
Radiat Biol. 97(Suppl): S132–S150. 2021. View Article : Google Scholar
|
|
5
|
Wu R, Hogberg J, Adner M, Ramos-Ramirez P,
Stenius U and Zheng H: Crystalline silica particles cause rapid
NLRP3-dependent mitochondrial depolarization and DNA damage in
airway epithelial cells. Part Fibre Toxicol. 17:392020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dussert F, Arthaud PA, Arnal ME, Dalzon B,
Torres A, Douki T, Herlin N, Rabilloud T and Carriere M: Toxicity
to RAW264.7 macrophages of silica nanoparticles and the E551 food
additive, in combination with genotoxic agents. Nanomaterials
(Basel). 10:14182020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang R, Yu T, Li Y and Hu J: Upregulated
has-miR-4516 as a potential biomarker for early diagnosis of
dust-induced pulmonary fibrosis in patients with pneumoconiosis.
Toxicol Res (Camb). 7:415–422. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gupta N, Khetan D, Chaudhary R and Shukla
JS: Prospective cohort study to assess the effect of storage
duration, Leuko-filtration, and gamma irradiation on cell-free DNA
in red cell components. Transfus Med Hemother. 47:409–419. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lindahl T and Barnes DE: Repair of
endogenous DNA damage. Cold Spring Harb Symp Quant Biol.
65:127–133. 2000. View Article : Google Scholar
|
|
10
|
Jackson SP and Bartek J: The DNA-damage
response in human biology and disease. Nature. 461:1071–1078. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aguilera A and Garcia-Muse T: Causes of
genome instability. Annu Rev Genet. 47:1–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aguilera A and Gomez-Gonzalez B: Genome
instability: A mechanistic view of its causes and consequences. Nat
Rev Genet. 9:204–217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li J, Sun H, Huang Y, Wang Y, Liu Y and
Chen X: Pathways and assays for DNA double-strand break repair by
homologous recombination. Acta Biochim Biophys Sin (Shanghai).
51:879–889. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Connor MJ: Targeting the DNA damage
response in cancer. Mol Cell. 60:547–560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lord CJ and Ashworth A: The DNA damage
response and cancer therapy. Nature. 481:287–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pilie PG, Tang C, Mills GB and Yap TA:
State-of-the-art strategies for targeting the DNA damage response
in cancer. Nat Rev Clin Oncol. 16:81–104. 2019. View Article : Google Scholar
|
|
17
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fanale D, Castiglia M, Bazan V and Russo
A: Involvement of Non-coding RNAs in Chemo- and Radioresistance of
colorectal Cancer. Adv Exp Med Biol. 937:207–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou XL, Wang WW, Zhu WG, Yu CH, Tao GZ,
Wu QQ, Song YQ, Pan P and Tong YS: High expression of long
non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor
prognosis in patients with esophageal squamous cell carcinoma
treated with definitive chemoradiotherapy. Mol Carcinog.
55:2095–2105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haemmig S, Yang D, Sun X, Das D, Ghaffari
S, Molinaro R, Chen L, Deng Y, Freeman D, Moullan N, et al: Long
noncoding RNA SNHG12 integrates a DNA-PK-mediated DNA damage
response and vascular senescence. Sci Transl Med. 12:eaaw18682020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Tao Y, Li Y, Zhao J, Zhang L,
Zhang X, Dong C, Xie Y, Dai X, Zhang X and Liao Q: The regulatory
network analysis of long noncoding RNAs in human colorectal cancer.
Funct Integr Genomics. 18:261–275. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y and Wang Y, Luo W, Song X, Huang L,
Xiao J, Jin F, Ren Z and Wang Y: Roles of long non-coding RNAs and
emerging RNA-binding proteins in innate antiviral responses.
Theranostics. 10:9407–9424. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ciccia A and Elledge SJ: The DNA damage
response: Making it safe to play with knives. Mol Cell. 40:179–204.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Michelini F, Pitchiaya S, Vitelli V,
Sharma S, Gioia U, Pessina F, Cabrini M, Wang Y, Capozzo I,
Iannelli F, et al: Damage-induced lncRNAs control the DNA damage
response through interaction with DDRNAs at individual
double-strand breaks. Nat Cell Biol. 19:1400–1411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Surova O and Zhivotovsky B: Various modes
of cell death induced by DNA damage. Oncogene. 32:3789–3797. 2013.
View Article : Google Scholar
|
|
27
|
Roos WP, Thomas AD and Kaina B: DNA damage
and the balance between survival and death in cancer biology. Nat
Rev Cancer. 16:20–33. 2016. View Article : Google Scholar
|
|
28
|
Sun X, Wang Y, Ji K, Liu Y, Kong Y, Nie S,
Li N, Hao J, Xie Y, Xu C, et al: NRF2 preserves genomic integrity
by facilitating ATR activation and G2 cell cycle arrest. Nucleic
Acids Res. 48:9109–9123. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu R, Hu Y, Zhang S, Li X, Tang M, Yang M,
Wu X, Li Z, Liao X, Xu Y, et al: LncRNA CTBP1-DT-encoded
microprotein DDUP sustains DNA damage response signalling to
trigger dual DNA repair mechanisms. Nucleic Acids Res.
50:8060–8079. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu CH, Chen CY, Yeh CT and Lin KH:
Radiosensitization of hepatocellular carcinoma through targeting
radio-associated MicroRNA. Int J Mol Sci. 21:18592020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kitagawa R and Kastan MB: The
ATM-dependent DNA damage signaling pathway. Cold Spring Harb Symp
Quant Biol. 70:99–109. 2005. View Article : Google Scholar
|
|
32
|
Matsuoka S, Ballif BA, Smogorzewska A,
McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini
N, Lerenthal Y, et al: ATM and ATR substrate analysis reveals
extensive protein networks responsive to DNA damage. Science.
316:1160–1166. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bartek J and Lukas J: DNA damage
checkpoints: From initiation to recovery or adaptation. Curr Opin
Cell Biol. 19:238–245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shiloh Y: ATM and related protein kinases:
Safeguarding genome integrity. Nat Rev Cancer. 3:155–168. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wan G, Mathur R, Hu X, Liu Y, Zhang X,
Peng G and Lu X: Long non-coding RNA ANRIL (CDKN2B-AS) is induced
by the ATM-E2F1 signaling pathway. Cell Signal. 25:1086–1095. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wan G, Hu X, Liu Y, Han C, Sood AK, Calin
GA, Zhang X and Lu X: A novel non-coding RNA lncRNA-JADE connects
DNA damage signalling to histone H4 acetylation. EMBO J.
32:2833–2847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schoeftner S and Blasco MA:
Developmentally regulated transcription of mammalian telomeres by
DNA-dependent RNA polymerase II. Nat Cell Biol. 10:228–236. 2008.
View Article : Google Scholar
|
|
38
|
Xu Y and Komiyama M: Structure, function
and targeting of human telomere RNA. Methods. 57:100–105. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Karlseder J, Broccoli D, Dai Y, Hardy S
and de Lange T: p53- and ATM-dependent apoptosis induced by
telomeres lacking TRF2. Science. 283:1321–1325. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Okamoto K, Bartocci C, Ouzounov I,
Diedrich JK, Yates JR III and Denchi EL: A two-step mechanism for
TRF2-mediated chromosome-end protection. Nature. 494:502–505. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Zeng D, Cao J, Wang M, Shu B,
Kuang G, Ou TM, Tan JH, Gu LQ, Huang ZS and Li D: Interaction of
Quindoline derivative with telomeric repeat-containing RNA induces
telomeric DNA-damage response in cancer cells through inhibition of
telomeric repeat factor 2. Biochim Biophys Acta Gen Subj.
1861:3246–3256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang A, Zhou N, Huang J, Liu Q, Fukuda K,
Ma D, Lu Z, Bai C, Watabe K and Mo YY: The human long non-coding
RNA-RoR is a p53 repressor in response to DNA damage. Cell Res.
23:340–350. 2013. View Article : Google Scholar :
|
|
43
|
Meek DW and Anderson CW: Posttranslational
modification of p53: Cooperative integrators of function. Cold
Spring Harb Perspect Biol. 1:a0009502009. View Article : Google Scholar
|
|
44
|
Zilfou JT and Lowe SW: Tumor suppressive
functions of p53. Cold Spring Harb Perspect Biol. 1:a0018832009.
View Article : Google Scholar :
|
|
45
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang A, Xu M and Mo YY: Role of the
lncRNA-p53 regulatory network in cancer. J Mol Cell Biol.
6:181–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shihabudeen Haider Ali MS, Cheng X, Moran
M, Haemmig S, Naldrett MJ, Alvarez S, Feinberg MW and Sun X: LncRNA
Meg3 protects endothelial function by regulating the DNA damage
response. Nucleic Acids Res. 47:1505–1522. 2019. View Article : Google Scholar :
|
|
48
|
Wen D, Huang Z, Li Z, Tang X, Wen X, Liu J
and Li M: LINC02535 co-functions with PCBP2 to regulate DNA damage
repair in cervical cancer by stabilizing RRM1 mRNA. J Cell Physiol.
235:7592–7603. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li N and Richard S: Sam68 functions as a
transcriptional coactivator of the p53 tumor suppressor. Nucleic
Acids Res. 44:8726–8741. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
van Gent DC, Hoeijmakers JH and Kanaar R:
Chromosomal stability and the DNA double-stranded break connection.
Nat Rev Genet. 2:196–206. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K
and Linn S: Molecular mechanisms of mammalian DNA repair and the
DNA damage checkpoints. Annu Rev Biochem. 73:39–85. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sharma V, Khurana S, Kubben N, Abdelmohsen
K, Oberdoerffer P, Gorospe M and Misteli T: A BRCA1-interacting
lncRNA regulates homologous recombination. EMBO Rep. 16:1520–1534.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Deng B, Xu W, Wang Z, Liu C, Lin P, Li B,
Huang Q, Yang J, Zhou H and Qu L: An LTR retrotransposon-derived
lncRNA interacts with RNF169 to promote homologous recombination.
EMBO Rep. 20:e476502019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Branzei D and Foiani M: Regulation of DNA
repair throughout the cell cycle. Nat Rev Mol Cell Biol. 9:297–308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lieber MR: The mechanism of human
nonhomologous DNA end joining. J Biol Chem. 283:1–5. 2008.
View Article : Google Scholar
|
|
59
|
San Filippo J, Sung P and Klein H:
Mechanism of eukaryotic homologous recombination. Annu Rev Biochem.
77:229–257. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kumar A, Purohit S and Sharma NK: Aberrant
DNA Double-strand break repair threads in breast carcinoma:
Orchestrating genomic insult survival. J Cancer Prev. 21:227–234.
2016. View Article : Google Scholar
|
|
61
|
Yao Y, Li X, Chen W, Liu H, Mi L, Ren D,
Mo A and Lu P: ATM promotes RAD51-mediated meiotic DSB repair by
inter-sister-chromatid recombination in Arabidopsis. Front Plant
Sci. 11:8392020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Trenner A and Sartori AA: Harnessing DNA
Double-strand break repair for cancer treatment. Front Oncol.
9:13882019. View Article : Google Scholar
|
|
63
|
Gomez-Mejiba SE and Ramirez DC: Trapping
of DNA radicals with the nitrone spin trap 5,5-dimethyl-1-pyrroline
N-oxide and genotoxic damage: Recent advances using the immuno-spin
trapping technology. Mutat Res Rev Mutat Res. 782:1082832019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Dasika GK, Lin SC, Zhao S, Sung P,
Tomkinson A and Lee EY: DNA damage-induced cell cycle checkpoints
and DNA strand break repair in development and tumorigenesis.
Oncogene. 18:7883–7899. 1999. View Article : Google Scholar
|
|
65
|
Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y,
Li Z, Bu D, Sun N, Zhang MQ and Chen R: NONCODE 2016: An
informative and valuable data source of long non-coding RNAs.
Nucleic Acids Res. 44:D203–D208. 2016. View Article : Google Scholar :
|
|
66
|
Dimitrova N, Zamudio JR, Jong RM, Soukup
D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA and Jacks
T: LincRNA-p21 activates p21 in cis to promote Polycomb target gene
expression and to enforce the G1/S checkpoint. Mol Cell.
54:777–790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Schmitt AM, Garcia JT, Hung T, Flynn RA,
Shen Y, Qu K, Payumo AY, Peres-da-Silva A, Broz DK, Baum R, et al:
An inducible long noncoding RNA amplifies DNA damage signaling. Nat
Genet. 48:1370–1376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu X, Li D, Zhang W, Guo M and Zhan Q:
Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6
mRNA decay. EMBO J. 31:4415–4427. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shen L, Wang Q, Liu R, Chen Z, Zhang X,
Zhou P and Wang Z: LncRNA lnc-RI regulates homologous recombination
repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a
competitive endogenous RNA. Nucleic Acids Res. 46:717–729. 2018.
View Article : Google Scholar :
|
|
70
|
Huang R and Zhou PK: DNA damage repair:
Historical perspectives, mechanistic pathways and clinical
translation for targeted cancer therapy. Signal Transduct Target
Ther. 6:2542021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Thapar R, Wang JL, Hammel M, Ye R, Liang
K, Sun C, Hnizda A, Liang S, Maw SS, Lee L, et al: Mechanism of
efficient double-strand break repair by a long non-coding RNA.
Nucleic Acids Res. 48:10953–10972. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang
Z, Yuan J, Shan W, Li C, Hu X, et al: Long noncoding RNA LINP1
regulates repair of DNA double-strand breaks in triple-negative
breast cancer. Nat Struct Mol Biol. 23:522–530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang X, Liu H, Shi L, Yu X, Gu Y and Sun
X: LINP1 facilitates DNA damage repair through non-homologous end
joining (NHEJ) pathway and subsequently decreases the sensitivity
of cervical cancer cells to ionizing radiation. Cell Cycle.
17:439–447. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Soutoglou E and Misteli T: Activation of
the cellular DNA damage response in the absence of DNA lesions.
Science. 320:1507–1510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Downs JA and Jackson SP: A means to a DNA
end: The many roles of Ku. Nat Rev Mol Cell Biol. 5:367–378. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang D, Zhou Z, Wu E, Ouyang C, Wei G,
Wang Y, He D, Cui Y, Zhang D, Chen X, et al: LRIK interacts with
the Ku70-Ku80 heterodimer enhancing the efficiency of NHEJ repair.
Cell Death Differ. 27:3337–3353. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Guo Z, Wang YH, Xu H, Yuan CS, Zhou HH,
Huang WH, Wang H and Zhang W: LncRNA linc00312 suppresses
radiotherapy resistance by targeting DNA-PKcs and impairing DNA
damage repair in nasopharyngeal carcinoma. Cell Death Dis.
12:692021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Uziel T, Lerenthal Y, Moyal L, Andegeko Y,
Mittelman L and Shiloh Y: Requirement of the MRN complex for ATM
activation by DNA damage. EMBO J. 22:5612–5621. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Prakash R, Zhang Y, Feng W and Jasin M:
Homologous recombination and human health: The roles of BRCA1,
BRCA2, and associated proteins. Cold Spring Harb Perspect Biol.
7:a0166002015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gorgoulis VG, Pefani DE, Pateras IS and
Trougakos IP: Integrating the DNA damage and protein stress
responses during cancer development and treatment. J Pathol.
246:12–40. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Heyer WD, Ehmsen KT and Liu J: Regulation
of homologous recombination in eukaryotes. Annu Rev Genet.
44:113–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Maréchal A and Zou L: DNA damage sensing
by the ATM and ATR kinases. Cold Spring Harb Perspect Biol.
5:a0127162013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Renkawitz J, Lademann CA and Jentsch S:
Mechanisms and principles of homology search during recombination.
Nat Rev Mol Cell Biol. 15:369–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ranjha L, Howard SM and Cejka P: Main
steps in DNA double-strand break repair: An introduction to
homologous recombination and related processes. Chromosoma.
127:187–214. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yu N, Qin H, Zhang F, Liu T, Cao K, Yang
Y, Chen Y and Cai J: The role and mechanism of long non-coding RNAs
in homologous recombination repair of radiation-induced DNA damage.
J Gene Med. 25:e34702023. View Article : Google Scholar
|
|
86
|
Ohta T, Sato K and Wu W: The BRCA1
ubiquitin ligase and homologous recombination repair. FEBS Lett.
585:2836–2844. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kim H, Chen J and Yu X: Ubiquitin-binding
protein RAP80 mediates BRCA1-dependent DNA damage response.
Science. 316:1202–1205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hu Y, Scully R, Sobhian B, Xie A,
Shestakova E and Livingston DM: RAP80-directed tuning of BRCA1
homologous recombination function at ionizing radiation-induced
nuclear foci. Genes Dev. 25:685–700. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Coleman KA and Greenberg RA: The
BRCA1-RAP80 complex regulates DNA repair mechanism utilization by
restricting end resection. J Biol Chem. 286:13669–13680. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hu Y, Petit SA, Ficarro SB, Toomire KJ,
Xie A, Lim E, Cao SA, Park E, Eck MJ, Scully R, et al: PARP1-driven
poly-ADP-ribosylation regulates BRCA1 function in homologous
recombination-mediated DNA repair. Cancer Discov. 4:1430–1447.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Hu Z, Mi S, Zhao T, Peng C, Peng Y, Chen
L, Zhu W, Yao Y, Song Q, Li X, et al: BGL3 lncRNA mediates
retention of the BRCA1/BARD1 complex at DNA damage sites. EMBO J.
39:e1041332020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang ZW, Pan JJ, Hu JF, Zhang JQ, Huang L,
Huang Y, Liao CY, Yang C, Chen ZW, Wang YD, et al: SRSF3-mediated
regulation of N6-methyladenosine modification-related lncRNA ANRIL
splicing promotes resistance of pancreatic cancer to gemcitabine.
Cell Rep. 39:1108132022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Syed A and Tainer JA: The MRE11-RAD50-NBS1
complex conducts the orchestration of damage signaling and outcomes
to stress in DNA replication and repair. Annu Rev Biochem.
87:263–294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Stracker TH and Petrini JH: The MRE11
complex: Starting from the ends. Nat Rev Mol Cell Biol. 12:90–103.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Xu A, Huang MF, Zhu D, Gingold JA, Bazer
DA, Chang B, Wang D, Lai CC, Lemischka IR, Zhao R and Lee DF:
LncRNA H19 suppresses Osteosarcomagenesis by regulating snoRNAs and
DNA repair protein complexes. Front Genet. 11:6118232020.
View Article : Google Scholar
|
|
96
|
Wu C, Chen W, Yu F, Yuan Y, Chen Y, Hurst
DR, Li Y, Li L and Liu Z: Long noncoding RNA HITTERS protects oral
squamous cell carcinoma cells from endoplasmic reticulum
stress-induced apoptosis via promoting MRE11-RAD50-NBS1 complex
formation. Adv Sci (Weinh). 7:20027472020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Paull TT: Mechanisms of ATM Activation.
Annu Rev Biochem. 84:711–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhao K, Wang X, Xue X, Li L and Hu Y: A
long noncoding RNA sensitizes genotoxic treatment by attenuating
ATM activation and homologous recombination repair in cancers. PLoS
Biol. 18:e30006662020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Bunting SF, Callén E, Wong N, Chen HT,
Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao
L, et al: 53BP1 inhibits homologous recombination in
Brca1-deficient cells by blocking resection of DNA breaks. Cell.
141:243–254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Escribano-Díaz C, Orthwein A,
Fradet-Turcotte A, Xing M, Young JT, Tkáč J, Cook MA, Rosebrock AP,
Munro M, Canny MD, et al: A cell cycle-dependent regulatory circuit
composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway
choice. Mol Cell. 49:872–883. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zimmermann M, Lottersberger F, Buonomo SB,
Sfeir A and de Lange T: 53BP1 regulates DSB repair using Rif1 to
control 5' end resection. Science. 339:700–704. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Poulsen M, Lukas C, Lukas J, Bekker-Jensen
S and Mailand N: Human RNF169 is a negative regulator of the
ubiquitin-dependent response to DNA double-strand breaks. J Cell
Biol. 197:189–199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Hu Q, Botuyan MV, Cui G, Zhao D and Mer G:
Mechanisms of Ubiquitin-nucleosome recognition and regulation of
53BP1 chromatin recruitment by RNF168/169 and RAD18. Mol Cell.
66:473–487.e479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Muvarak N, Kelley S, Robert C, Baer MR,
Perrotti D, Gambacorti-Passerini C, Civin C, Scheibner K and
Rassool FV: c-MYC generates repair errors via increased
transcription of Alternative-NHEJ Factors, LIG3 and PARP1, in
tyrosine kinase-activated leukemias. Mol Cancer Res. 13:699–712.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ahrabi S, Sarkar S, Pfister SX, Pirovano
G, Higgins GS, Porter AC and Humphrey TC: A role for human
homologous recombination factors in suppressing
microhomology-mediated end joining. Nucleic Acids Res.
44:5743–5757. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Leppard JB, Dong Z, Mackey ZB and
Tomkinson AE: Physical and functional interaction between DNA
ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA
single-strand break repair. Mol Cell Biol. 23:5919–5927. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Chiruvella KK, Liang Z and Wilson TE:
Repair of double-strand breaks by end joining. Cold Spring Harb
Perspect Biol. 5:a0127572013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Hu Y, Lin J, Fang H, Fang J, Li C, Chen W,
Liu S, Ondrejka S, Gong Z, Reu F, et al: Targeting the
MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in
multiple myeloma. Leukemia. 32:2250–2262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Langelier MF, Ruhl DD, Planck JL, Kraus WL
and Pascal JM: The Zn3 domain of human poly(ADP-ribose)
polymerase-1 (PARP-1) functions in both DNA-dependent
poly(ADP-ribose) synthesis activity and chromatin compaction. J
Biol Chem. 285:18877–18887. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Huang J, Lin C, Dong H, Piao Z, Jin C, Han
H and Jin D: Targeting MALAT1 induces DNA damage and sensitize
non-small cell lung cancer cells to cisplatin by repressing BRCA1.
Cancer Chemother Pharmacol. 86:663–672. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Goldstein M and Kastan MB: The DNA damage
response: Implications for tumor responses to radiation and
chemotherapy. Annu Rev Med. 66:129–143. 2015. View Article : Google Scholar
|
|
112
|
Yao RW, Wang Y and Chen LL: Cellular
functions of long noncoding RNAs. Nat Cell Biol. 21:542–551. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Kang M, Ren M, Li Y, Fu Y, Deng M and Li
C: Exosome-mediated transfer of lncRNA PART1 induces gefitinib
resistance in esophageal squamous cell carcinoma via functioning as
a competing endogenous RNA. J Exp Clin Cancer Res. 37:1712018.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Xiong XD, Ren X, Cai MY, Yang JW, Liu X
and Yang JM: Long non-coding RNAs: An emerging powerhouse in the
battle between life and death of tumor cells. Drug Resist Updat.
26:28–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li Z, Zhou Y, Tu B, Bu Y, Liu A and Kong
J: Long noncoding RNA MALAT1 affects the efficacy of radiotherapy
for esophageal squamous cell carcinoma by regulating Cks1
expression. J Oral Pathol Med. 46:583–590. 2017. View Article : Google Scholar
|
|
116
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Liu J, Ben Q, Lu E, He X, Yang X, Ma J,
Zhang W, Wang Z, Liu T, Zhang J and Wang H: Long noncoding RNA
PANDAR blocks CDKN1A gene transcription by competitive interaction
with p53 protein in gastric cancer. Cell Death Dis. 9:1682018.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Shao L, Zuo X, Yang Y, Zhang Y, Yang N,
Shen B, Wang J, Wang X, Li R, Jin G, et al: The inherited
variations of a p53-responsive enhancer in 13q12.12 confer lung
cancer risk by attenuating TNFRSF19 expression. Genome Biol.
20:1032019. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zhen Y, Ye Y, Wang H, Xia Z, Wang B, Yi W
and Deng X: Knockdown of SNHG8 repressed the growth, migration, and
invasion of colorectal cancer cells by directly sponging with
miR-663. Biomed Pharmacother. 116:1090002019. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Liu J, Yang C, Gu Y, Li C, Zhang H, Zhang
W, Wang X, Wu N and Zheng C: Knockdown of the lncRNA SNHG8 inhibits
cell growth in Epstein-Barr virus-associated gastric carcinoma.
Cell Mol Biol Lett. 23:172018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Tian X, Liu Y, Wang Z and Wu S: lncRNA
SNHG8 promotes aggressive behaviors of nasopharyngeal carcinoma via
regulating miR-656-3p/SATB1 axis. Biomed Pharmacother.
131:1105642020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Miao W, Lu T, Liu X, Yin W and Zhang H:
LncRNA SNHG8 induces ovarian carcinoma cells cellular process and
stemness through Wnt/β-catenin pathway. Cancer Biomark. 28:459–471.
2020. View Article : Google Scholar
|
|
123
|
Fan D, Qiu B, Yang XJ, Tang HL, Peng SJ,
Yang P, Dong YM, Yang L, Bao GQ and Zhao HD: LncRNA SNHG8 promotes
cell migration and invasion in breast cancer cell through
miR-634/ZBTB20 axis. Eur Rev Med Pharmacol Sci. 24:11639–11649.
2020.PubMed/NCBI
|
|
124
|
Zhu W, Tan L, Ma T, Yin Z and Gao J: Long
noncoding RNA SNHG8 promotes chemoresistance in gastric cancer via
binding with hnRNPA1 and stabilizing TROY expression. Dig Liver
Dis. 54:1573–1582. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Wang Z, Wang X, Rong Z, Dai L, Qin C, Wang
S and Geng W: LncRNA LINC01134 contributes to radioresistance in
hepatocellular carcinoma by regulating DNA damage response via MAPK
signaling pathway. Front Pharmacol. 12:7918892021. View Article : Google Scholar
|
|
126
|
Sun Y, Wang J, Ma Y, Li J, Sun X, Zhao X,
Shi X, Hu Y, Qu F and Zhang X: Radiation induces NORAD expression
to promote ESCC radiotherapy resistance via EEPD1/ATR/Chk1
signalling and by inhibiting pri-miR-199a1 processing and the
exosomal transfer of miR-199a-5p. J Exp Clin Cancer Res.
40:3062021. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Yao P, Li Y, Shen W, Xu X, Zhu W, Yang X,
Cao J and Xing C: ANKHD1 silencing suppresses the proliferation,
migration and invasion of CRC cells by inhibiting YAP1-induced
activation of EMT. Am J Cancer Res. 8:2311–2324. 2018.PubMed/NCBI
|
|
128
|
Yao PA, Wu Y, Zhao K, Li Y, Cao J and Xing
C: The feedback loop of ANKHD1/lncRNA MALAT1/YAP1 strengthens the
radioresistance of CRC by activating YAP1/AKT signaling. Cell Death
Dis. 13:1032022. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Takahashi H, Nishimura J, Kagawa Y, Kano
Y, Takahashi Y, Wu X, Hiraki M, Hamabe A, Konno M, Haraguchi N, et
al: Significance of Polypyrimidine Tract-binding Protein 1
expression in colorectal cancer. Mol Cancer Ther. 14:1705–1716.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Huan L, Guo T, Wu Y, Xu L, Huang S, Xu Y,
Liang L and He X: Hypoxia induced LUCAT1/PTBP1 axis modulates
cancer cell viability and chemotherapy response. Mol Cancer.
19:112020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Jin MH and Oh DY: ATM in DNA repair in
cancer. Pharmacol Ther. 203:1073912019. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Cimprich KA and Cortez D: ATR: An
essential regulator of genome integrity. Nat Rev Mol Cell Biol.
9:616–627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Panzarino NJ, Krais JJ, Cong K, Peng M,
Mosqueda M, Nayak SU, Bond SM, Calvo JA, Doshi MB, Bere M, et al:
Replication gaps underlie BRCA deficiency and therapy response.
Cancer Res. 81:1388–1397. 2021. View Article : Google Scholar
|
|
134
|
Zhang B, Bao W, Zhang S, Chen B, Zhou X,
Zhao J, Shi Z, Zhang T, Chen Z, Wang L, et al: LncRNA HEPFAL
accelerates ferroptosis in hepatocellular carcinoma by regulating
SLC7A11 ubiquitination. Cell Death Dis. 13:7342022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Jiang Y, Guo H, Tong T, Xie F, Qin X, Wang
X, Chen W and Zhang J: lncRNA lnc-POP1-1 upregulated by VN1R5
promotes cisplatin resistance in head and neck squamous cell
carcinoma through interaction with MCM5. Mol Ther. 30:448–467.
2022. View Article : Google Scholar :
|
|
136
|
Choi PS and Thomas-Tikhonenko A:
RNA-binding proteins of COSMIC importance in cancer. J Clin Invest.
131:e1516272021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Fabbri L, Chakraborty A, Robert C and
Vagner S: The plasticity of mRNA translation during cancer
progression and therapy resistance. Nat Rev Cancer. 21:558–577.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Duffy AG, Makarova-Rusher OV, Ulahannan
SV, Rahma OE, Fioravanti S, Walker M, Abdullah S, Raffeld M,
Anderson V, Abi-Jaoudeh N, et al: Modulation of tumor eIF4E by
antisense inhibition: A phase I/II translational clinical trial of
ISIS 183750-an antisense oligonucleotide against eIF4E-in
combination with irinotecan in solid tumors and
irinotecan-refractory colorectal cancer. Int J Cancer.
139:1648–1657. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Shen L and Pelletier J: Selective
targeting of the DEAD-box RNA helicase eukaryotic initiation factor
(eIF) 4A by natural products. Nat Prod Rep. 37:609–616. 2020.
View Article : Google Scholar
|
|
140
|
Zhu H, Chen K, Chen Y, Liu J, Zhang X,
Zhou Y, Liu Q, Wang B, Chen T and Cao X: RNA-binding protein ZCCHC4
promotes human cancer chemoresistance by disrupting
DNA-damage-induced apoptosis. Signal Transduct Target Ther.
7:2402022. View Article : Google Scholar : PubMed/NCBI
|