Allergic rhinitis (AR) is a chronic inflammatory
disease of the nasal mucosa mediated by immunoglobulin E (IgE)
(1). Dendritic cells (DCs),
which are a major subtype of antigen-presenting cells (APCs), serve
a key role in the immunopathogenesis of AR (1,2).
When allergens enter the body, they are presented by DCs to CD4 T
cells to trigger allergic inflammatory responses, leading to the
activation and maturation of DCs (2,3).
Exosomes are secreted during the maturation and differentiation of
DCs in AR (4,5). However, almost all cells and not
only DCs can produce exosomes (5-8).
Exosomes are cell-derived, nm-sized extracellular vesicles (EVs)
that are formed through the endocytosis and inward budding of the
endosomal membrane mediated by extracellular components and cell
surface proteins. They are distributed in almost all bodily fluids
and have been previously associated with the occurrence and
progression of several diseases (9,10). Exosomes can carry important
signaling molecules for intercellular communication and material
transfer (11). DC-derived
exosomes (Dex) are nano-scale lipid-membrane vesicles formed within
DCs by the inward budding of the endosomal membrane after DCs
receive immune signals (12,13). The composition and function of
DCs and Dex are strikingly similar. Dex, which mimics the biology
of donor DCs, can transfer functional major histocompatibility
complexes (MHC) to DCs, leading to the activation of CD8 and CD4 T
cells (14-16). In addition, Dex carry MHC and
T-cell costimulatory molecules to present allergens to induce the
production of Th2 cytokines in allergic donors, which are important
immunostimulatory factors of anaphylactic immune responses
(17,18). Therefore, allergen-carrying Dex
may be important targets for AR immunotherapy (17,18). Since the diverse and complex mode
of information transfer between Dex and various cells may serve an
integral role in the occurrence and progression of AR, Dex
engineered to carry anti-allergic drugs may have the potential to
interrupt the allergic and immune processes underlying AR on a
novel level (5,19).
DCs are a class of bone marrow-derived cells that
are typically found in blood, tissues, and lymphoid organs. They
primarily initiate immune responses by presenting antigens to naive
T cells in lymphoid tissues (20,21). Once activated, DCs increase the
expression levels of the MHC peptide complex and costimulatory
molecules, allowing them to efficiently activate T cells (22). As the most efficient type of
APCs, DCs serve a central role in the immune system. They are
typically classified according to their location, function, and
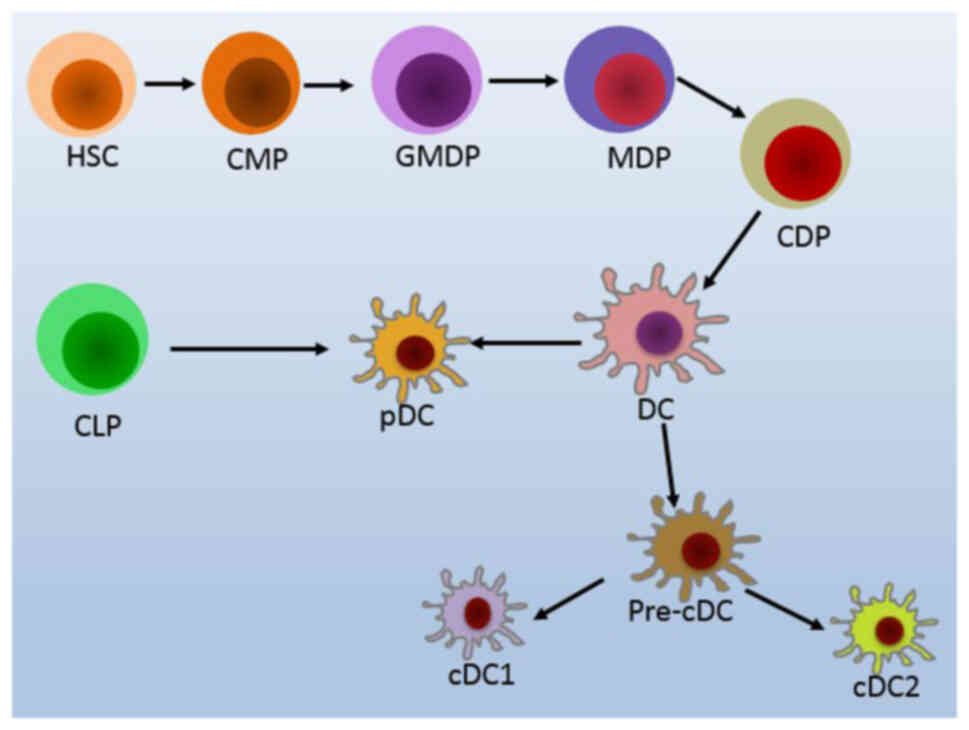
cell surface marker profile, namely plasmacytoid DCs (pDCs) and
conventional DCs (cDCs; Fig. 1)
(20,23-25). DCs can develop from different
hematopoietic or myelopoietic progenitors, where to the best of our
knowledge, no interconversion from one type to another has been
found to date (26).
pDCs are also known as 'lymphoid DCs' and form a
subset of DCs with antigen-presenting potential, accounting for
<0.3% of all blood mononuclear cells (27,28). Although they share a similar
origin with cDCs, pDCs have a different life cycle, since they
primarily accumulate in the blood and lymphoid tissues, entering
lymph nodes through the blood circulation (29). pDCs can develop in situ in
the bone marrow or from common lymphoid progenitors (CLPs; Fig. 1) (24,30). They acquire the functions of APCs
after activation, where their expression combination of
costimulatory molecules CD40, CD80, and/or CD86 can dictate which
specific T cell function is activated (24,31). After recognizing foreign nucleic
acids, pDCs will produce large quantities of IFN-I and acquire the
ability to present foreign antigens, which serve an important role
in antiviral immunity (29,30). In addition, pDCs can directly
inhibit allergic immune responses in the airway in addition to
indirectly promoting the induction of regulatory T (Treg) cells in
mice (32,33).
The majority of cDCs are short-lived hematopoietic
cells that are constantly replaced by blood-derived precursors
(29). cDCs account for a much
larger proportion of DCs compared with pDCs and are typically
distributed in most lymphoid tissues and non-lymphoid tissues. With
highly efficient antigen-presenting ability, they can capture
relevant antigens and present them to T lymphocytes after
intracellular processing (29).
Serving the role of 'sentinels', cDCs can respond to environmental
stimuli and alert the immune system to the presence of foreign
antigens, including allergens. cDCs have been reported to be
required for the initiation of Th2 immune responses (24,32). Allergens can either signal
directly through specific receptors on cDCs or indirectly by
inducing cytokine production in surrounding tissues or inflammatory
cells, which can then compel cDCs into promoting Th2 responses
(32).
DCs in different types of tissues appear to serve
different cellular functions, where various stimuli can induce the
maturation of specific and distinct DC phenotypes that mediate
different functions (34).
However, during the resting state, when the DCs are immature
(imDCs), they can acquire self-antigens but do not activate T
cells. After being stimulated by injury, pathogens, or inflammatory
cytokines, imDCs are then transformed into mature DCs (mDCs), which
then migrate to secondary lymphoid tissues, where they prime naive
T cells into initiating adaptive immune responses (35,36). In particular, only viable,
mature, and fully functional DCs migrating into lymph nodes can
stimulate T-cell responses (35). The maturation of DCs is
accompanied by the enhanced expression of MHC II, costimulatory
molecules, and chemokine receptors (37). It is mainly during the maturation
of DCs that exosomes are produced.
Exosomes are EVs with lipid bilayer structures
formed by extracellular components together with proteins, lipids,
metabolites, small molecules, ions, and other liquids through the
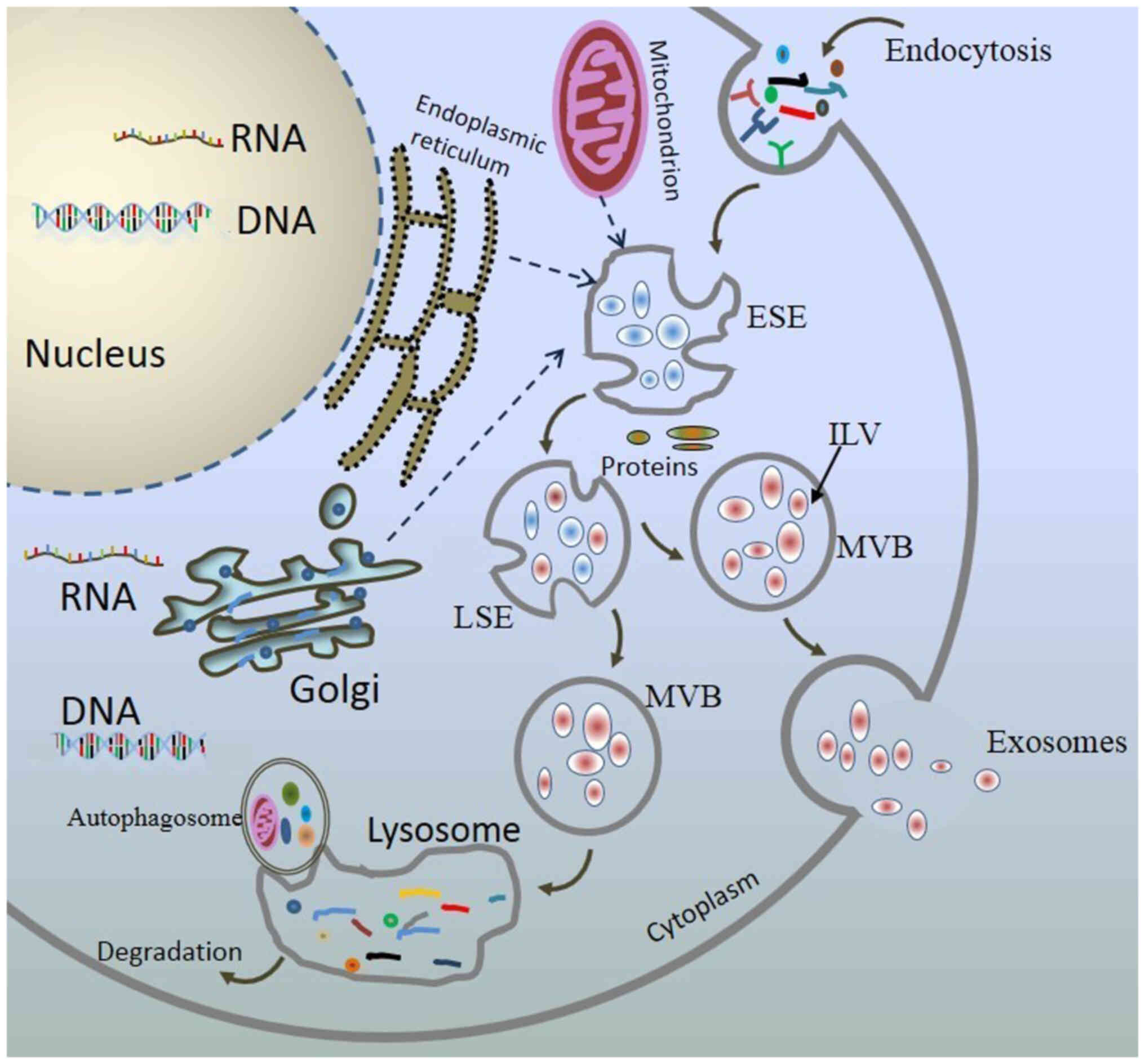
endocytosis and inward budding of the plasma membrane (9,38). The inward budding of the plasma
membrane then forms early-sorting endosomes (ESEs) in association
with the trans-Golgi network, mitochondria, and endoplasmic
reticulum. These mature ESEs subsequently form late-sorting
endosomes (LSEs) under the control of the endocytosis-sorting
complex and other related proteins. After the specific sorting and
encapsulation of proteins, lipids, and nucleic acids, LSEs then
form multiple intraluminal vesicles (ILVs) through a second
indentation. This process allows for the entry of cytoplasmic
components into the newly formed ILVs, which are the precursors of
exosomes (6,9,39,40). This sequential invagination of
the plasma membrane eventually leads to the development of multiple
ILVs into multivesicular bodies. They can either fuse with
lysosomes or autophagosomes for degradation or fuse with the plasma
membrane to release the vesicles out of the cell through
extravasation (Fig. 2) (41-44).
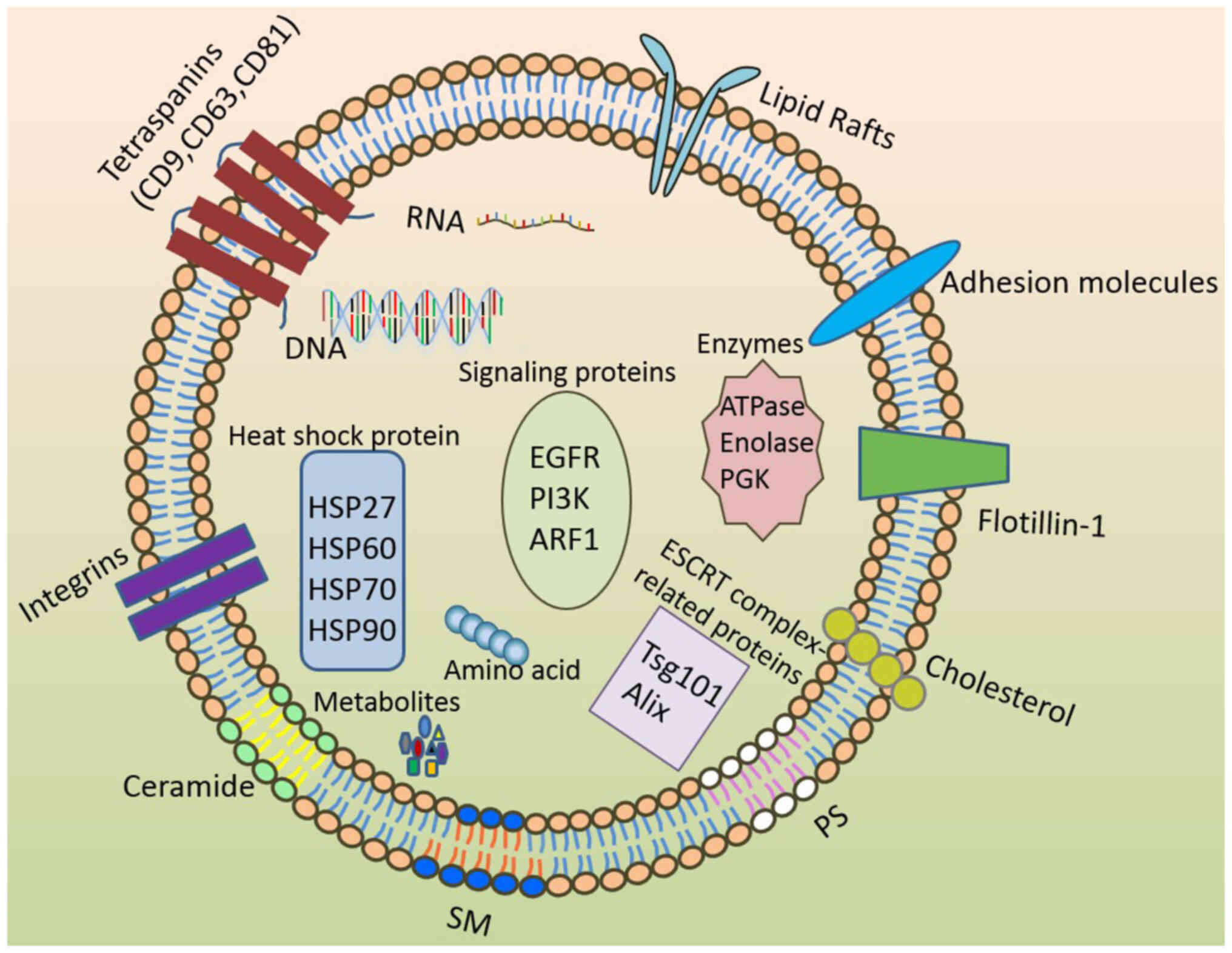
Exosomes contain a variety of components, including
lipids, proteins, amino acids, metabolites, RNA, and DNA. The
majority of these components can exert biological functions and
define the transport capacity of the exosome (45,46). Proteins that are commonly found
in exosomes include transmembrane proteins CD9, CD63, CD81, CD82,
CD151, Ras-related proteins, immunomodulatory proteins, heat shock
proteins (HSP), cell type-specific molecules, proteases, MHC
molecules, tumor susceptibility gene 101 protein, apoptosis-linked
gene 2-interacting protein X, integrins, and flotillin. They can be
found on the surfaces, in between lipid bilayers, or within
exosomes, with the yield of protein content from exosomes dependent
on the type of cells that secreted them (9,47,48). Lipids form another important
component that makes up the exosomes. They not only contribute to
their support structures but are also important participants in
their formation and release into the extracellular environment
(49,50). Major lipid components of exosomes
include sphingomyelin (SM), phosphatidylserine (PS),
phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol
(PI), phosphatidic acid, and cholesterol (Table I). The distribution of lipids in
the exosome bilayer is typically asymmetric, where SM is primarily
located in the outer layer, whilst PS is largely distributed in the
inner layer (Fig. 3) (49-51).
However, it is important to note that exosomes from
different sources can contain different ingredients, even if they
originated from the same cell. As such, exosomal contents mostly
likely depend on the status of the cell from which they were
produced (8,45,48). They may vary under different
physiological or pathological conditions, where changes in the
external environment (such as various modes of stress, hypoxia, and
inflammation) will influence the molecular profile of exosomes
(46,57). The different molecular
compositions of exosomes will likely have an impact on their
transport capacity and messaging function.
Exosomes were initially considered to be carriers of
waste products from intracellular metabolism (66). However, subsequent studies have
revealed that exosomes can serve to not only remove waste products
of metabolism from the cell but also perform a variety of
functions, such as intercellular communication, transport of
intracellular, extracellular substances, and genetic material, as
well as maintenance of cellular stability and removing cellular
debris (67-70). In addition, exosomes can regulate
innate and adaptive immune responses, specifically in antigen
presentation and intercellular signaling. Akin to 'communicators',
exosomes can serve as intercellular immune mediators regulating
cell proliferation, differentiation, and migration, allowing them
to mature and adapt rapidly to environmental changes (71,72). Although the biological functions
of exosomes can vary depending on their origin, they have important
reported roles in cell differentiation, maturation, and apoptosis
(8,73). The core functions of exosomes are
mainly determined by the proteins, lipids, and nucleic acids
contained within their parental cells (73).
Exosomes with multiple biological functions can be
used for the diagnosis, treatment and prognostic evaluation of
numerous diseases such as cardiovascular disease, neurodegenerative
diseases, and HIV (74,75). Efficient and high purity but
simple methods for exosome isolation and purification form the
first step for optimizing the field of exosome research (74). Over the past decade, rapid
progress has been made in the study of exosomes. However, several
outstanding obstacles must be overcome, such as cumbersome
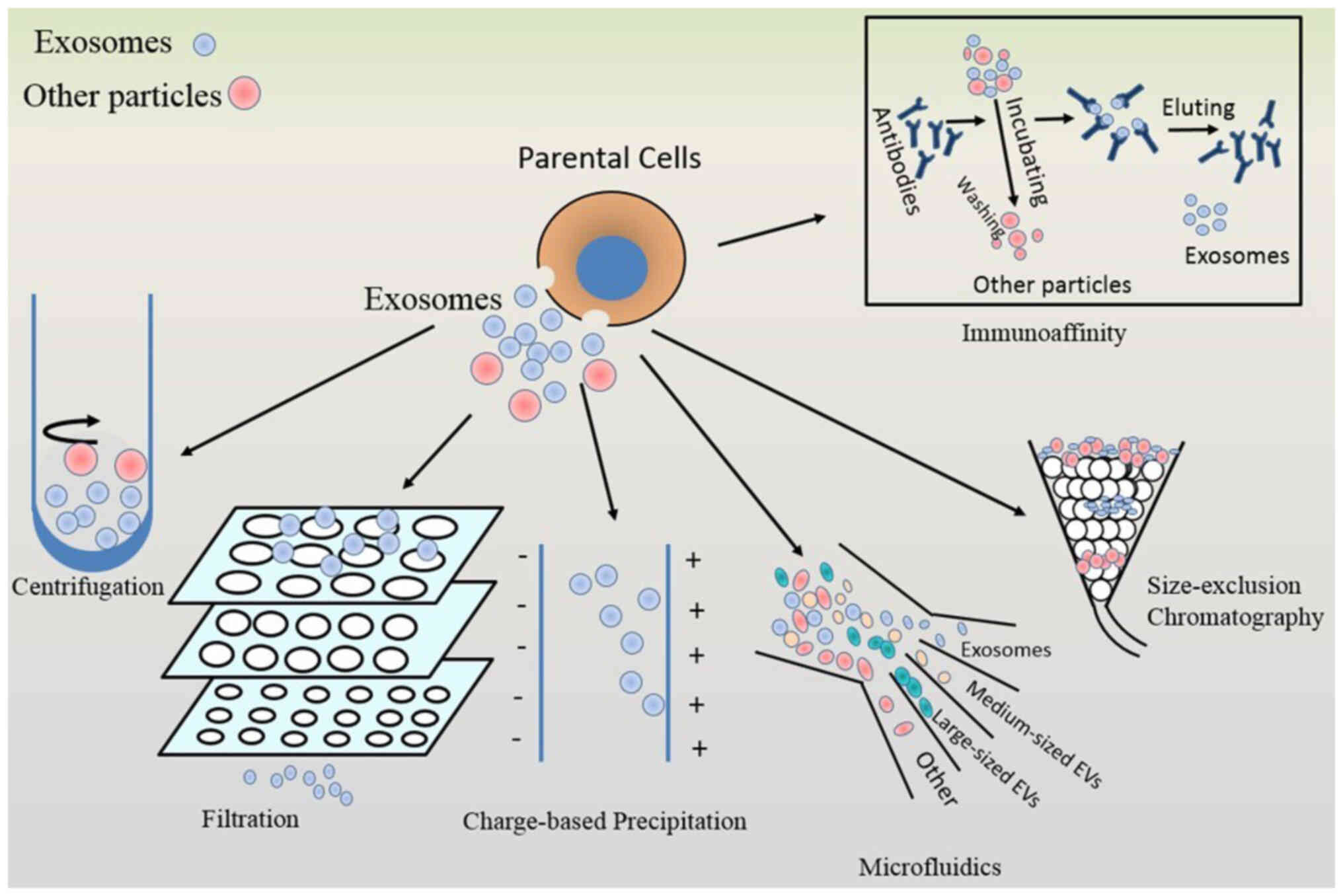
separation methods, low speed, low yield, and purity (76). Common exosome isolation and
purification methods include ultracentrifugation, ultrafiltration,
precipitation, immunoaffinity capture, and volume exclusion
chromatography (75). All these
aforementioned methods share similar disadvantages (Fig. 4; Table II). As this field develops,
emerging methods for exosome isolation and purification are
currently being found, such as microfluidics, electricity,
centrifugal force, and acoustic force, which can be exploited to
isolate exosomes of high purity in a high-throughput manner
(Fig. 4; Table II) (74).
Dex are nm-sized vesicles formed within the cell by
the inward budding of the endosomal membrane (12). There are a variety of proteins in
Dex, such as integrin α and β chains (αMβ2), immunoglobulin family
member intercellular adhesion molecule (ICAM), and milk fat globule
epidermal growth factor 8 (MFG-E8), cytoskeleton proteins and
anti-apoptosis-related proteins, which dock their membranes onto
those of host cells. CD9, CD63, and CD81 are also components that
are frequently found on the Dex surface membrane (83-85). The composition of Dex membranes
differs from those of DCs in that they are richer in sphingolipid
content but poorer in phosphatidylcholine content, in addition to
being deprived of cholesterol (13). The lipid composition of Dex
membranes can have an impact on their function (86). HSP70, HSP90, and heat shock
cognate protein 73 have also been found in Dex, which can increase
the immunogenicity of Dex (87).
There is also a variety of different types of RNAs in Dex, which
can transfer onto other cells. In particular, Dex has been
documented to contain several immunomodulatory molecules with
different structures and biochemical properties, depending on the
intracellular origin of Dex (88). The composition and features of
Dex are shown in Table
III.
DCs can secrete different types of exosomes to
regulate the adaptive immune response. In addition, exosomes from
different sources can modulate the differentiation, maturation, and
function of DCs (77). Dex, in
addition to the known immunostimulatory capabilities of DCs, has
been reported to regulate a variety of immune processes, including
antigen presentation, immunomodulation, and signal transduction
(12,92). With the ability to activate naive
T-cells and facilitate the transfer of MHC complexes between DCs,
Dex can be produced in large quantities and efficiently diffuse
into tissues, rendering them potentially more potent compared with
DCs in activating T lymphocytes and natural killer cells (93-95). The immunomodulatory effect of Dex
is closely associated with the maturation status of DCs (96). imDCs and mDCs secrete exosomes
with similar morphology, where the potency of exosomes secreted by
mDCs is substantially higher compared with that of exosomes from
imDCs (97). In general,
exosomes from mDCs (mDex) have higher levels of immune-related
molecules and superior antigen presentation compared with those
from imDCs, which are prone to exosome production but do not
effectively stimulate T-cell responses (84,93,94,97).
DCs have been extensively reported to be involved in
the pathogenesis of AR. Therefore, it is highly likely that Dex
will play a potentially important role in AR. Dex can be recaptured
by DCs and remain on the cell surface, where they can present
allergens and induce Th2 cytokine production in allergic donors to
elicit allergic immune responses (18,101). Dex share similarities with DCs
in promoting allergic immune responses (102). Dex can present antigens
directly to T-cells or transport MHC complexes back to the surfaces
of DCs for presentation to T-cells after docking onto APCs
(103). CD40 on Dex has been
found to induce T-cell responses to promote IgE production
(104-106). In addition, CD63 and CD81 in
Dex have been shown to inhibit FcεRI-induced degranulation by mast
cells (MCs), which further affects signaling that normally mediates
allergic inflammation (107).
Costimulatory molecules, such as CD80 and CD86, on the Dex surface
can also contribute to the maturation of DCs and promote Th2-type
inflammation, leading to an imbalance in the differentiation of
naive T-cells towards to Th2 subtype (108). In addition, CD80 and CD86 can
activate allergen-specific Th2 cells to potentiate antigen-specific
immune responses (109,110). By contrast, miRNAs in Dex can
regulate serum IgE levels and the severity of allergy symptoms
through blood, nasal mucosa, and nasal secretions in AR (111). Therefore, miRNA cargoes in Dex
can be used to determine the extent of allergic inflammation and
immune response (106). Changes
in the expression levels of ICAM and MFG-E8 can also regulate the
immune response (112). SM can
drive allergic inflammation and promote airway hyperresponsiveness
by serving as important signaling molecules for mediating
inflammatory and immune responses (113-115). This suggests that a wide
variety of cargoes carried by Dex can mediate an impact on AR,
though different cargoes are likely to exert different effects on
AR.
The immunostimulatory or suppressive function of Dex
is dependent on the type or maturity stage of DCs that secrete them
(116). Therefore, Dex in
different states is also likely to have different effects on AR.
Exosomes from imDCs (imDex), which primarily reduce
T-cell-dependent immune activation, contribute to inhibiting the
Th17 response whilst enhancing the population of Treg cells
(15,117). By contrast, mDex can directly
act on T-cells to exert specific immune responses (118). By functioning as an
antigen-presenting molecule, Dex can modulate immunity and
inflammation by perpetuating the response of Th2 cells to DCs
(119). Dex carries leukotriene
synthase, which stimulates granulocyte translocation to promote the
recruitment and migration of immune cells to sites of inflammation
(77,110,120). Choi et al (121) previously showed that DCs can
excrete allergen-bound Dex, which can trigger the degranulation of
adjacent MCs, leading to anaphylaxis. Furthermore, DCs have been
reported to secrete TNF-α and other proinflammatory cytokines in
response to Dex stimulation, leading to increased inflammation
(122). In another study, Huang
et al (123) activated
DCs using thymic stromal lymphopoietin (TSLP), which induced Dex
release and in turn promoted the proliferation and differentiation
of CD4+ T-cells into the Th2 subtype through the OX40
ligand. These studies suggest that Dex can serve a significant role
in the pathogenesis of AR through different types of cargo.
It is noteworthy that Dex can not only aggravate
allergic reactions but also prevent them. Exosome-mediated transfer
of allergens can promote allergic inflammation whereas regulatory
and/or tolerogenic exosomes can suppress allergic and
hypersensitivity reactions (19). Immunotherapy using Dex typically
involves loading antigens directly into Dex or by modifying them
(122). DCs can be modified to
produce immunosuppressive Dex for the treatment of allergic
inflammation (19). Dex can also
be modified to carry anti-allergic drugs that can reduce allergic
airway inflammation. In addition, lipids and proteins carried by
Dex can enhance the permeability of biological membranes, which
facilitates the efficiency of the delivery of the anti-allergic
drugs they carry (19,124,125). In particular, mDex can promote
the activation of T and B cells, leading to Th1-type immune
responses and increased IgG titers (101). Since high IgG titers inhibit
IgE-mediated effector function, it also suppresses the allergic
inflammation that causes AR (101). In addition, imDCs treated with
IL-10 and IL-4 have been reported to produce tolerogenic Dex to
attenuate Th2 cell responses, thereby inhibiting inflammatory
responses, in a mice model of delayed-type hypersensitivity (DTH)
(84,126). The inhibitory capacity of
IL-10-treated imDCs depends on the presence of CD80 and CD86
(127). Bianco et al
(128) previously revealed that
Dex overexpressing indoleamine 2,3-dioxygenase has
anti-inflammatory effects in a mouse model of DTH, which are
dependent on the costimulatory molecule B7. Furthermore, Jia et
al (129) showed that
DCs-derived forkhead box p3-exosomes inhibited the proliferation of
CD4+T cells, which in turn reduced the population of Th1
and Th17 cells whilst increasing that of Treg cells without
affecting the level of Th2 cells. In another previous study, Yu
et al (130) found that
Dex modified with IL-2 can upregulate Treg differentiation to
suppress allergic inflammation. Kim et al (131) also showed that genetically
modified Dex derived from FasL-expressing DCs can exert
anti-inflammatory and immunosuppressive effects by suppressing DTH
in an antigen-specific and MHC-II-dependent manner, though this
process was independent of MHC I. These aforementioned findings
suggest that Dex can either be processed or modified to inhibit
allergic inflammation and thereby treat AR. Since Dex as a
candidate for the treatment of AR is less susceptible to the
effects of the surrounding environment (132), these findings could inspire
further exploration into novel methods for treating AR with
Dex.
Dex, with superior biocompatibility,
biodegradability, and safety, can activate various immune cells and
hold significant advantages in terms of delivery efficiency
(85). Dex is more stable, can
be stored for longer, and more immunogenic than DCs, which are
highly susceptible to external factors that induce their maturation
under pro-inflammatory conditions and promote immune responses, in
addition to inducing their tolerance and moderate immune responses
in response to IL-10 and TGF-β stimulation (85,133). These advantages of Dex suggest
their viability for the treatment of AR. Treating AR with Dex not
only eliminates the need for direct contact with natural allergens
but is also less prone to triggering IgE/MC reactions. Therefore,
they tend to be safer and more effective for the treatment of AR
(19). The feasibility and
safety of Dex therapy has the potential to be one of the
alternatives to conventional treatments of AR (110). It is encouraging that Dex-based
therapies are already in clinical trials (101). However, the scope of clinical
trials for Dex is limited compared to DC vaccines, where their
potential for application has not been fully evaluated (122). The role of exosomal vaccines is
dependent on the environment, antigen, and cell type (122), which will require more in-depth
research in the future.
From the aforementioned studies, it is likely that
Dex is involved in the pathogenesis and can be exploited for the
diagnosis and treatment of AR. However, their roles can vary during
the different stages of AR. The role of Dex in AR opens another
door to understanding the pathogenesis of AR, furthering the
potential to design interventions and/or sensitizations to the
immunotherapy of AR on a novel level. Although rapid progress has
been made in the understanding of Dex (12), the clinical exploitation of Dex
remains hindered by a series of problems, such as low efficiency,
poor yields, difficulty associated with expression, and low purity
(10,122). In addition, research on Dex
biomarkers or targeted therapies for AR remains in the early stages
and requires further development (77). The complexity of Dex requires
thorough understanding. In addition, it remains necessary to
carefully monitor the potential adverse events associated with Dex
in future trials (12).
Therefore, the role of Dex in AR will need to be enhanced further
with a specific focus on the problems currently obstructing
progress to adequately refine and improve the application of Dex in
AR therapy. It is hoped that Dex can be used as an important marker
for the diagnosis, treatment, and prognosis of patients with AR in
the future, which requires more in-depth research on the isolation
and purification, sensitivity, and cost-effectiveness of Dex. In
addition, a more thorough exploration into the composition and
mechanism of action mediated by the various cargoes contained
within Dex needs to be performed.
Is it possible to adjust or change the composition
and function of Dex as needed to diagnose or treat AR? It is
possible to design Dex to stimulate or inhibit immune responses as
needed? Can Dex be a candidate for the treatment of AR? How can Dex
be produced for the personalized clinical diagnosis and treatment
of AR according to the situation? These questions need to be
addressed in future studies. A more thorough understanding of the
role of Dex in AR could help prevent AR or develop more effective
treatment strategies. This provides a novel insight for exploration
into the pathogenesis of AR and offers a new direction for the
efficient treatment of AR.
Overall, the application of Dex in the diagnosis and
treatment of AR is a promising approach, where novel insights in
this field will drive the development of new therapeutic or
preventive measures. However, the role of Dex in AR warrants
further investigation in the future.
Not applicable.
CK and HH conceived and drafted the manuscript. CK,
HH, JL, SQ, PL, YL, XL, JZ, HR, XZ, and HZ reviewed and edited the
manuscript. Data authentication is not applicable. All authors have
read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by grants Natural Science Foundation of
China (grant no. 81700888), Guangdong Basic and Applied Basic
Research Foundation (grant no. 2021A1515010971), Shenzhen Science
and Technology Program for Basic Research (grant no.
JCYJ20220531091417040), Shenzhen Science and Technology Program
(grant no. JCYJ20210324142207019), Shenzhen Key Medical Discipline
Construction Fund (grant no. SZXK039), Science and Technology
Development Special Fund of Shenzhen Longgang District (grant nos.
LGKCYLWS2019000864 and LGKCZSYS2019000046), Science and Technology
Innovation Special-Technology Tackling Project of Shenzhen Longgang
District (grant no. LGKCYLWS2022032).
|
1
|
Bousquet J, Anto JM, Bachert C, Baiardini
I, Bosnic-Anticevich S, Walter Canonica G, Melén E, Palomares O,
Scadding GK, Togias A and Toppila-Salmi S: Allergic rhinitis. Nat
Rev Dis Primers. 6:952020.
|
|
2
|
Zhang Y, Lan F and Zhang L: Update on
pathomechanisms and treatments in allergic rhinitis. Allergy.
77:3309–3319. 2022.
|
|
3
|
Liu P, Kang C, Zhang J, Liu Y, Liu J, Hu
T, Zeng X and Qiu S: The role of dendritic cells in allergic
diseases. Int Immunopharmacol. 113:1094492022.
|
|
4
|
Hodge AL, Baxter AA and Poon IKH: Gift
bags from the sentinel cells of the immune system: The diverse role
of dendritic cell-derived extracellular vesicles. J Leukoc Biol.
111:903–920. 2022.
|
|
5
|
Zheng Z and Yu Y: A review of recent
advances in exosomes and allergic rhinitis. Front Pharmacol.
13:10969842022.
|
|
6
|
Zhang Y, Bi J, Huang J, Tang Y, Du S and
Li P: Exosome: A review of its classification, isolation
techniques, storage, diagnostic and targeted therapy applications.
Int J Nanomedicine. 15:6917–6934. 2020.
|
|
7
|
Elashiry M, Elsayed R, Elashiry MM, Rashid
MH, Ara R, Arbab AS, Elawady AR, Hamrick M, Liu Y, Zhi W, et al:
Proteomic characterization, biodistribution, and functional studies
of immune-therapeutic exosomes: Implications for inflammatory lung
diseases. Front Immunol. 12:6362222021.
|
|
8
|
Ibrahim A and Marbán E: Exosomes:
Fundamental biology and roles in cardiovascular physiology. Annu
Rev Physiol. 78:67–83. 2016.
|
|
9
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020.
|
|
10
|
Liu J, Ren L, Li S, Li W, Zheng X, Yang Y,
Fu W, Yi J, Wang J and Du G: The biology, function, and
applications of exosomes in cancer. Acta Pharm Sin B. 11:2783–2797.
2021.
|
|
11
|
Liu Q, Li S, Dupuy A, Mai HL, Sailliet N,
Logé C, Robert JH and Brouard S: Exosomes as new biomarkers and
drug delivery tools for the prevention and treatment of various
diseases: Current perspectives. Int J Mol Sci. 22:77632021.
|
|
12
|
Pitt JM, André F, Amigorena S, Soria JC,
Eggermont A, Kroemer G and Zitvogel L: Dendritic cell-derived
exosomes for cancer therapy. J Clin Invest. 126:1224–1232.
2016.
|
|
13
|
Nikfarjam S, Rezaie J, Kashanchi F and
Jafari R: Dexosomes as a cell-free vaccine for cancer
immunotherapy. J Exp Clin Cancer Res. 39:2582020.
|
|
14
|
Wu R, Gao W, Yao K and Ge J: Roles of
exosomes derived from immune cells in cardiovascular diseases.
Front Immunol. 10:6482019.
|
|
15
|
Elashiry M, Elsayed R and Cutler CW:
Exogenous and endogenous dendritic cell-derived exosomes: Lessons
learned for immunotherapy and disease pathogenesis. Cells.
11:1152021.
|
|
16
|
Viaud S, Terme M, Flament C, Taieb J,
André F, Novault S, Escudier B, Robert C, Caillat-Zucman S, Tursz
T, et al: Dendritic cell-derived exosomes promote natural killer
cell activation and proliferation: A role for NKG2D ligands and
IL-15Ralpha. PLoS One. 4:e49422009.
|
|
17
|
Wang X, He L, Huang X, Zhang S, Cao W, Che
F, Zhu Y and Dai J: Recent progress of exosomes in multiple
myeloma: Pathogenesis, diagnosis, prognosis and therapeutic
strategies. Cancers (Basel). 13:16352021.
|
|
18
|
Vallhov H, Gutzeit C, Hultenby K, Valenta
R, Grönlund H and Scheynius A: Dendritic cell-derived exosomes
carry the major cat allergen Fel d 1 and induce an allergic immune
response. Allergy. 70:1651–1655. 2015.
|
|
19
|
Nazimek K, Bryniarski K and Askenase PW:
Functions of exosomes and microbial extracellular vesicles in
allergy and contact and delayed-type hypersensitivity. Int Arch
Allergy Immunol. 171:1–26. 2016.
|
|
20
|
Collin M and Bigley V: Human dendritic
cell subsets: An update. Immunology. 154:3–20. 2018.
|
|
21
|
Palucka K and Banchereau J: Cancer
immunotherapy via dendritic cells. Nat Rev Cancer. 12:265–277.
2012.
|
|
22
|
Pearce EJ and Everts B: Dendritic cell
metabolism. Nat Rev Immunol. 15:18–29. 2015.
|
|
23
|
Onai N and Manz MG: The STATs on dendritic
cell development. Immunity. 28:490–492. 2008.
|
|
24
|
Macri C, Pang ES, Patton T and O'Keeffe M:
Dendritic cell subsets. Semin Cell Dev Biol. 84:11–21. 2018.
|
|
25
|
Kumar S, Jeong Y, Ashraf MU and Bae YS:
Dendritic cell-mediated Th2 immunity and immune disorders. Int J
Mol Sci. 20:21592019.
|
|
26
|
Austyn JM: Dendritic cells in the immune
system-history, lineages, tissues, tolerance, and immunity.
Microbiol Spectr. 4:2016.
|
|
27
|
Manz MG: Plasmacytoid dendritic cells:
Origin matters. Nat Immunol. 19:652–654. 2018.
|
|
28
|
Upham JW: The role of dendritic cells in
immune regulation and allergic airway inflammation. Respirology.
8:140–148. 2003.
|
|
29
|
Merad M, Sathe P, Helft J, Miller J and
Mortha A: The dendritic cell lineage: Ontogeny and function of
dendritic cells and their subsets in the steady state and the
inflamed setting. Annu Rev Immunol. 31:563–604. 2013.
|
|
30
|
Reizis B: Plasmacytoid dendritic cells:
Development, regulation, and function. Immunity. 50:37–50.
2019.
|
|
31
|
Barrat FJ and Su L: A pathogenic role of
plasmacytoid dendritic cells in autoimmunity and chronic viral
infection. J Exp Med. 216:1974–1985. 2019.
|
|
32
|
Lamiable O, Mayer JU, Munoz-Erazo L and
Ronchese F: Dendritic cells in Th2 immune responses and allergic
sensitization. Immunol Cell Biol. 98:807–818. 2020.
|
|
33
|
Kool M, van Nimwegen M, Willart MAM,
Muskens F, Boon L, Smit JJ, Coyle A, Clausen BE, Hoogsteden HC,
Lambrecht BN and Hammad H: An anti-inflammatory role for
plasmacytoid dendritic cells in allergic airway inflammation. J
Immunol. 183:1074–1082. 2009.
|
|
34
|
Ghislat G and Lawrence T: Autophagy in
dendritic cells. Cell Mol Immunol. 15:944–952. 2018.
|
|
35
|
Sabado RL, Balan S and Bhardwaj N:
Dendritic cell-based immunotherapy. Cell Res. 27:74–95. 2017.
|
|
36
|
Gardner A, de Mingo Pulido Á and Ruffell
B: Dendritic cells and their role in immunotherapy. Front Immunol.
11:9242020.
|
|
37
|
Koltsova EK and Ley K: How dendritic cells
shape atherosclerosis. Trends Immunol. 32:540–547. 2011.
|
|
38
|
Zhu C, Li L, Wang Z, Irfan M and Qu F:
Recent advances of aptasensors for exosomes detection. Biosens
Bioelectron. 160:1122132020.
|
|
39
|
Zhang L and Yu D: Exosomes in cancer
development, metastasis, and immunity. Biochim Biophys Acta Rev
Cancer. 1871:455–468. 2019.
|
|
40
|
Liang Y, Duan L, Lu J and Xia J:
Engineering exosomes for targeted drug delivery. Theranostics.
11:3183–3195. 2021.
|
|
41
|
Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang
JH and Zhang SJ: Exosome biogenesis: Machinery, regulation, and
therapeutic implications in cancer. Mol Cancer. 21:2072022.
|
|
42
|
Hessvik NP and Llorente A: Current
knowledge on exosome biogenesis and release. Cell Mol Life Sci.
75:193–208. 2018.
|
|
43
|
Shao H, Im H, Castro CM, Breakefield X,
Weissleder R and Lee H: New technologies for analysis of
extracellular vesicles. Chem Rev. 118:1917–1950. 2018.
|
|
44
|
Zhang Y, Liu Y, Liu H and Tang WH:
Exosomes: Biogenesis, biologic function and clinical potential.
Cell Biosci. 9:192019.
|
|
45
|
He C, Zheng S, Luo Y and Wang B: Exosome
theranostics: Biology and translational medicine. Theranostics.
8:237–255. 2018.
|
|
46
|
Gurunathan S, Kang MH and Kim JH: A
comprehensive review on factors influences biogenesis, functions,
therapeutic and clinical implications of exosomes. Int J
Nanomedicine. 16:1281–1312. 2021.
|
|
47
|
Li W, Li C, Zhou T, Liu X, Liu X, Li X and
Chen D: Role of exosomal proteins in cancer diagnosis. Mol Cancer.
16:1452017.
|
|
48
|
Gurunathan S, Kang MH, Jeyaraj M, Qasim M
and Kim JH: Review of the isolation, characterization, biological
function, and multifarious therapeutic approaches of exosomes.
Cells. 8:3072019.
|
|
49
|
Skotland T, Hessvik NP, Sandvig K and
Llorente A: Exosomal lipid composition and the role of ether lipids
and phosphoinositides in exosome biology. J Lipid Res. 60:9–18.
2019.
|
|
50
|
Skotland T, Sandvig K and Llorente A:
Lipids in exosomes: Current knowledge and the way forward. Prog
Lipid Res. 66:30–41. 2017.
|
|
51
|
Donoso-Quezada J, Ayala-Mar S and
González-Valdez J: The role of lipids in exosome biology and
intercellular communication: Function, analytics and applications.
Traffic. 22:204–220. 2021.
|
|
52
|
Kugeratski FG, Hodge K, Lilla S, McAndrews
KM, Zhou X, Hwang RF, Zanivan S and Kalluri R: Quantitative
proteomics identifies the core proteome of exosomes with syntenin-1
as the highest abundant protein and a putative universal biomarker.
Nat Cell Biol. 23:631–641. 2021.
|
|
53
|
Zhang H, Freitas D, Kim HS, Fabijanic K,
Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, et al:
Identification of distinct nanoparticles and subsets of
extracellular vesicles by asymmetric flow field-flow fractionation.
Nat Cell Biol. 20:332–343. 2018.
|
|
54
|
Zhu L, Sun HT, Wang S, Huang SL, Zheng Y,
Wang CQ, Hu BY, Qin W, Zou TT, Fu Y, et al: Isolation and
characterization of exosomes for cancer research. J Hematol Oncol.
13:1522020.
|
|
55
|
Gebeyehu A, Kommineni N, Meckes DG Jr and
Sachdeva MS: Role of exosomes for delivery of chemotherapeutic
drugs. Crit Rev Ther Drug Carrier Syst. 38:53–97. 2021.
|
|
56
|
Yu W, Hurley J, Roberts D, Chakrabortty
SK, Enderle D, Noerholm M, Breakefield XO and Skog JK:
Exosome-based liquid biopsies in cancer: Opportunities and
challenges. Ann Oncol. 32:466–477. 2021.
|
|
57
|
Meng W, Hao Y, He C, Li L and Zhu G:
Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer.
18:572019.
|
|
58
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18:1162019.
|
|
59
|
Sun Z, Yang S, Zhou Q, Wang G, Song J, Li
Z, Zhang Z, Xu J, Xia K, Chang Y, et al: Emerging role of
exosome-derived long non-coding RNAs in tumor microenvironment. Mol
Cancer. 17:822018.
|
|
60
|
Huang S, Nishiumi S, Asaduzzaman M, Pan Y,
Liu G, Yoshitake K, Maeyama K, Kinoshita S, Nagai K, Watabe S, et
al: Exosome-derived small non-coding RNAs reveal immune response
upon grafting transplantation in Pinctada fucata (Mollusca). Open
Biol. 12:2103172022.
|
|
61
|
Marzec ME, Rząca C, Moskal P and Stępień
E: Study of the influence of hyperglycemia on the abundance of
amino acids, fatty acids, and selected lipids in extracellular
vesicles using TOF-SIMS. Biochem Biophys Res Commun. 622:30–36.
2022.
|
|
62
|
Onozato M, Kobata K, Sakamoto T, Ichiba H
and Fukushima T: LC-MS/MS analysis of Thiol-containing amino acids
in exosomal fraction of serum. J Chromatogr Sci. 58:636–640.
2020.
|
|
63
|
Zhu Q, Huang L, Yang Q, Ao Z, Yang R,
Krzesniak J, Lou D, Hu L, Dai X, Guo F and Liu F: Metabolomic
analysis of exosomal-markers in esophageal squamous cell carcinoma.
Nanoscale. 13:16457–16464. 2021.
|
|
64
|
Zebrowska A, Skowronek A, Wojakowska A,
Widlak P and Pietrowska M: Metabolome of exosomes: Focus on
vesicles released by cancer cells and present in human body fluids.
Int J Mol Sci. 20:34612019.
|
|
65
|
Williams C, Royo F, Aizpurua-Olaizola O,
Pazos R, Boons GJ, Reichardt NC and Falcon-Perez JM: Glycosylation
of extracellular vesicles: Current knowledge, tools and clinical
perspectives. J Extracell Vesicles. 7:14429852018.
|
|
66
|
Gudbergsson JM and Johnsen KB: Exosomes
and autophagy: Rekindling the vesicular waste hypothesis. J Cell
Commun Signal. 13:443–450. 2019.
|
|
67
|
Gonzalez MJ, Kweh MF, Biava PM, Olalde J,
Toro AP, Goldschmidt-Clermont PJ and White IA: Evaluation of
exosome derivatives as bio-informational reprogramming therapy for
cancer. J Transl Med. 19:1032021.
|
|
68
|
Kumar V, Kiran S, Kumar S and Singh UP:
Extracellular vesicles in obesity and its associated inflammation.
Int Rev Immunol. 41:30–44. 2022.
|
|
69
|
Mora EM, Álvarez-Cubela S and Oltra E:
Biobanking of exosomes in the era of precision medicine: Are we
there yet? Int J Mol Sci. 17:132015.
|
|
70
|
Krylova SV and Feng D: The machinery of
exosomes: Biogenesis, release, and uptake. Int J Mol Sci.
24:13372023.
|
|
71
|
Li N, Zhao L, Wei Y, Ea VL, Nian H and Wei
R: Recent advances of exosomes in immune-mediated eye diseases.
Stem Cell Res Ther. 10:2782019.
|
|
72
|
Buzas EI: The roles of extracellular
vesicles in the immune system. Nat Rev Immunol. 23:236–250.
2023.
|
|
73
|
Kim SB: Function and therapeutic
development of exosomes for cancer therapy. Arch Pharm Res.
45:295–308. 2022.
|
|
74
|
Chen J, Li P, Zhang T, Xu Z, Huang X, Wang
R and Du L: Review on strategies and technologies for exosome
isolation and purification. Front Bioeng Biotechnol.
9:8119712021.
|
|
75
|
Lai JJ, Chau ZL, Chen SY, Hill JJ, Korpany
KV, Liang NW, Lin LH, Lin YH, Liu JK, Liu YC, et al: Exosome
processing and characterization approaches for research and
technology development. Adv Sci (Weinh). 9:e21032222022.
|
|
76
|
Chen Y, Zhu Q, Cheng L, Wang Y, Li M, Yang
Q, Hu L, Lou D, Li J, Dong X, et al: Exosome detection via the
ultrafast-isolation system: EXODUS. Nat Methods. 18:212–218.
2021.
|
|
77
|
Zhang X, Xu D, Song Y, He R and Wang T:
Research progress in the application of exosomes in immunotherapy.
Front Immunol. 13:7315162022.
|
|
78
|
Pathan M, Fonseka P, Chitti SV, Kang T,
Sanwlani R, Van Deun J, Hendrix A and Mathivanan S: Vesiclepedia
2019: A compendium of RNA, proteins, lipids and metabolites in
extracellular vesicles. Nucleic Acids Res. 47(D1): D516–D519.
2019.
|
|
79
|
Martínez-Greene JA, Hernández-Ortega K,
Quiroz-Baez R, Resendis-Antonio O, Pichardo-Casas I, Sinclair DA,
Budnik B, Hidalgo-Miranda A, Uribe-Querol E, Ramos-Godínez MDP, et
al: Quantitative proteomic analysis of extracellular vesicle
subgroups isolated by an optimized method combining polymer-based
precipitation and size exclusion chromatography. J Extracell
Vesicles. 10:e120872021.
|
|
80
|
Gandham S, Su X, Wood J, Nocera AL, Alli
SC, Milane L, Zimmerman A, Amiji M and Ivanov AR: Technologies and
standardization in research on extracellular vesicles. Trends
Biotechnol. 38:1066–1098. 2020.
|
|
81
|
Lee K, Shao H, Weissleder R and Lee H:
Acoustic purification of extracellular microvesicles. ACS Nano.
9:2321–2327. 2015.
|
|
82
|
Lin S, Yu Z, Chen D, Wang Z, Miao J, Li Q,
Zhang D, Song J and Cui D: Progress in microfluidics-based exosome
separation and detection technologies for diagnostic applications.
Small. 16:e19039162020.
|
|
83
|
Yao Y, Fu C, Zhou L, Mi QS and Jiang A:
DC-derived exosomes for cancer immunotherapy. Cancers (Basel).
13:36672021.
|
|
84
|
Viaud S, Théry C, Ploix S, Tursz T,
Lapierre V, Lantz O, Zitvogel L and Chaput N: Dendritic
cell-derived exosomes for cancer immunotherapy: What's next? Cancer
Res. 70:1281–1285. 2010.
|
|
85
|
Xia J, Miao Y, Wang X, Huang X and Dai J:
Recent progress of dendritic cell-derived exosomes (Dex) as an
anti-cancer nanovaccine. Biomed Pharmacother. 152:1132502022.
|
|
86
|
Pelissier Vatter FA, Cioffi M, Hanna SJ,
Castarede I, Caielli S, Pascual V, Matei I and Lyden D:
Extracellular vesicle- and particle-mediated communication shapes
innate and adaptive immune responses. J Exp Med.
218:e202025792021.
|
|
87
|
Pitt JM, Charrier M, Viaud S, André F,
Besse B, Chaput N and Zitvogel L: Dendritic cell-derived exosomes
as immunotherapies in the fight against cancer. J Immunol.
193:1006–1011. 2014.
|
|
88
|
Kowal J and Tkach M: Dendritic cell
extracellular vesicles. Int Rev Cell Mol Biol. 349:213–249.
2019.
|
|
89
|
Robbins PD and Morelli AE: Regulation of
immune responses by extracellular vesicles. Nat Rev Immunol.
14:195–208. 2014.
|
|
90
|
Zeng F and Morelli AE: Extracellular
vesicle-mediated MHC cross-dressing in immune homeostasis,
transplantation, infectious diseases, and cancer. Semin
Immunopathol. 40:477–490. 2018.
|
|
91
|
Deb A, Gupta S and Mazumder PB: Exosomes:
A new horizon in modern medicine. Life Sci. 264:1186232021.
|
|
92
|
Wu J, Zhao R, Dai J, Lai G, Khan AU, Yu X,
Wu S, Ouyang J and Sang H: Analysis of differential expression of
long non-coding RNAs in exosomes derived from mature and immature
dendritic cells. Mol Med Rep. 23:1322021.
|
|
93
|
Du Z, Huang Z, Chen X, Jiang G, Peng Y,
Feng W and Huang N: Modified dendritic cell-derived exosomes
activate both NK cells and T cells through the NKG2D/NKG2D-L
pathway to kill CML cells with or without T315I mutation. Exp
Hematol Oncol. 11:362022.
|
|
94
|
Harvey BT, Fu X, Li L, Neupane KR, Anand
N, Kolesar JM and Richards CI: Dendritic cell membrane-derived
nanovesicles for targeted T cell activation. ACS Omega.
7:46222–46233. 2022.
|
|
95
|
Huang F, Jia H, Zou Y, Yao Y and Deng Z:
Exosomes: An important messenger in the asthma inflammatory
microenvironment. J Int Med Res. 48:3000605209032202020.
|
|
96
|
Wiklander OPB, Brennan MÁ, Lötvall J,
Breakefield XO and El Andaloussi S: Advances in therapeutic
applications of extracellular vesicles. Sci Transl Med.
11:eaav85212019.
|
|
97
|
Segura E, Amigorena S and Théry C: Mature
dendritic cells secrete exosomes with strong ability to induce
antigen-specific effector immune responses. Blood Cells Mol Dis.
35:89–93. 2005.
|
|
98
|
Delcayre A, Shu H and Le Pecq JB:
Dendritic cell-derived exosomes in cancer immunotherapy: Exploiting
nature's antigen delivery pathway. Expert Rev Anticancer Ther.
5:537–547. 2005.
|
|
99
|
Rao Q, Ma G, Li M, Wu H, Zhang Y, Zhang C,
Ma Z and Huang L: Targeted delivery of triptolide by dendritic
cell-derived exosomes for colitis and rheumatoid arthritis therapy
in murine models. Br J Pharmacol. 180:330–346. 2023.
|
|
100
|
Zhang Y, Cai Z, Shen Y, Lu Q, Gao W, Zhong
X, Yao K, Yuan J and Liu H: Hydrogel-load exosomes derived from
dendritic cells improve cardiac function via Treg cells and the
polarization of macrophages following myocardial infarction. J
Nanobiotechnology. 19:2712021.
|
|
101
|
Engeroff P and Vogel M: The potential of
exosomes in allergy immunotherapy. Vaccines (Basel).
10:1332022.
|
|
102
|
Sangaphunchai P, Todd I and Fairclough LC:
Extracellular vesicles and asthma: A review of the literature. Clin
Exp Allergy. 50:291–307. 2020.
|
|
103
|
Wang Y, Xiang Y, Xin VW, Wang XW, Peng XC,
Liu XQ, Wang D, Li N, Cheng JT, Lyv YN, et al: Dendritic cell
biology and its role in tumor immunotherapy. J Hematol Oncol.
13:1072020.
|
|
104
|
Metcalfe DD, Pawankar R, Ackerman SJ, Akin
C, Clayton F, Falcone FH, Gleich GJ, Irani AM, Johansson MW, Klion
AD, et al: Biomarkers of the involvement of mast cells, basophils
and eosinophils in asthma and allergic diseases. World Allergy
Organ J. 9:72016.
|
|
105
|
Sastre B, Cañas JA, Rodrigo-Muñoz JM and
Del Pozo V: Novel modulators of asthma and allergy: Exosomes and
MicroRNAs. Front Immunol. 8:8262017.
|
|
106
|
Srinivasan A and Sundar IK: Recent updates
on the role of extracellular vesicles in the pathogenesis of
allergic asthma. Extracell Vesicles Circ Nucl Acids. 2:127–147.
2021.
|
|
107
|
Kraft S, Jouvin MH, Kulkarni N, Kissing S,
Morgan ES, Dvorak AM, Schröder B, Saftig P and Kinet JP: The
tetraspanin CD63 is required for efficient IgE-mediated mast cell
degranulation and anaphylaxis. J Immunol. 191:2871–2878. 2013.
|
|
108
|
Teng ZX, Zhou XC, Xu RT, Zhu FY, Bing X,
Guo N, Shi L, Qi WW, Liu CC and Xia M: Tfh exosomes derived from
allergic rhinitis promote DC maturation through
miR-142-5p/CDK5/STAT3 pathway. J Inflamm Res. 15:3187–3205.
2022.
|
|
109
|
Wahlund CJE, Güclüler G, Hiltbrunner S,
Veerman RE, Näslund TI and Gabrielsson S: Exosomes from
antigen-pulsed dendritic cells induce stronger antigen-specific
immune responses than microvesicles in vivo. Sci Rep.
7:170952017.
|
|
110
|
Mortaz E, Alipoor SD, Varahram M, Jamaati
H, Garssen J, Mumby SE and Adcock IM: exosomes in severe asthma:
Update in their roles and potential in therapy. Biomed Res Int.
2018:28621872018.
|
|
111
|
Liu Y, Sha J, Meng C and Zhu D: The role
of small extracellular vesicles and MicroRNAs in the diagnosis and
treatment of allergic rhinitis and nasal polyps. Mediators Inflamm.
2022:44286172022.
|
|
112
|
Hazrati A, Soudi S, Malekpour K, Mahmoudi
M, Rahimi A, Hashemi SM and Varma RS: Immune cells-derived exosomes
function as a double-edged sword: Role in disease progression and
their therapeutic applications. Biomark Res. 10:302022.
|
|
113
|
Cañas JA, Sastre B, Rodrigo-Muñoz JM and
Del Pozo V: Exosomes: A new approach to asthma pathology. Clin Chim
Acta. 495:139–147. 2019.
|
|
114
|
Hovhannisyan L, Czechowska E and
Gutowska-Owsiak D: The role of non-immune cell-derived
extracellular vesicles in allergy. Front Immunol.
12:7023812021.
|
|
115
|
Hough KP, Wilson LS, Trevor JL,
Strenkowski JG, Maina N, Kim YI, Spell ML, Wang Y, Chanda D, Dager
JR, et al: Unique lipid signatures of extracellular vesicles from
the airways of asthmatics. Sci Rep. 8:103402018.
|
|
116
|
Kim SH, Bianco NR, Shufesky WJ, Morelli AE
and Robbins PD: Effective treatment of inflammatory disease models
with exosomes derived from dendritic cells genetically modified to
express IL-4. J Immunol. 179:2242–2249. 2007.
|
|
117
|
Yin W, Ouyang S, Li Y, Xiao B and Yang H:
Immature dendritic cell-derived exosomes: A promise subcellular
vaccine for autoimmunity. Inflammation. 36:232–240. 2013.
|
|
118
|
Lin J, Huang N, Li M, Zheng M, Wang Z,
Zhang X, Gao H, Lao Y, Zhang J and Ding B: Dendritic cell-derived
exosomes driven drug co-delivery biomimetic nanosystem for
effective combination of malignant melanoma immunotherapy and gene
therapy. Drug Des Devel Ther. 17:2087–2106. 2023.
|
|
119
|
Huda MN, Nafiujjaman M, Deaguero IG,
Okonkwo J, Hill ML, Kim T and Nurunnabi M: Potential use of
exosomes as diagnostic biomarkers and in targeted drug delivery:
Progress in clinical and preclinical applications. ACS Biomater Sci
Eng. 7:2106–2149. 2021.
|
|
120
|
Esser J, Gehrmann U, D'Alexandri FL,
Hidalgo-Estévez AM, Wheelock CE, Scheynius A, Gabrielsson S and
Rådmark O: Exosomes from human macrophages and dendritic cells
contain enzymes for leukotriene biosynthesis and promote
granulocyte migration. J Allergy Clin Immunol. 126:1032–1040.
1040.e1–e4. 2010.
|
|
121
|
Choi HW, Suwanpradid J, Kim IH, Staats HF,
Haniffa M, MacLeod AS and Abraham SN: Perivascular dendritic cells
elicit anaphylaxis by relaying allergens to mast cells via
microvesicles. Science. 362:eaao06662018.
|
|
122
|
Li Q, Wang H, Peng H, Huyan T and Cacalano
NA: Exosomes: Versatile nano mediators of immune regulation.
Cancers (Basel). 11:15572019.
|
|
123
|
Huang L, Zhang X, Wang M, Chen Z, Yan Y,
Gu W, Tan J, Jiang W and Ji W: Exosomes from thymic stromal
lymphopoietin-activated dendritic cells promote Th2 differentiation
through the OX40 ligand. Pathobiology. 86:111–117. 2019.
|
|
124
|
Zhu H, Wang K, Wang Z, Wang D, Yin X, Liu
Y, Yu F and Zhao W: An efficient and safe MUC1-dendritic
cell-derived exosome conjugate vaccine elicits potent cellular and
humoral immunity and tumor inhibition in vivo. Acta Biomater.
138:491–504. 2022.
|
|
125
|
Johnsen KB, Gudbergsson JM, Skov MN,
Pilgaard L, Moos T and Duroux M: A comprehensive overview of
exosomes as drug delivery vehicles-endogenous nanocarriers for
targeted cancer therapy. Biochim Biophys Acta. 1846:75–87.
2014.
|
|
126
|
Bianco NR, Kim SH, Morelli AE and Robbins
PD: Modulation of the immune response using dendritic cell-derived
exosomes. Methods Mol Biol. 380:443–455. 2007.
|
|
127
|
Schülke S: Induction of interleukin-10
producing dendritic cells as a tool to suppress allergen-specific T
helper 2 responses. Front Immunol. 9:4552018.
|
|
128
|
Bianco NR, Kim SH, Ruffner MA and Robbins
PD: Therapeutic effect of exosomes from indoleamine
2,3-dioxygenase-positive dendritic cells in collagen-induced
arthritis and delayed-type hypersensitivity disease models.
Arthritis Rheum. 60:380–389. 2009.
|
|
129
|
Jia Z, Liu J, Li B, Yi L, Wu Y, Xing J,
Wang L, Wang J and Guo L: Exosomes with FOXP3 from gene-modified
dendritic cells ameliorate the development of EAE by regulating the
balance of Th/Treg. Int J Med Sci. 19:1265–1274. 2022.
|
|
130
|
Yu D, Liu JQ, Mo LH, Luo XQ, Liu ZQ, Wu
GH, Yang LT, Liu DB, Wang S, Liu ZG and Yang PC: Specific
antigen-guiding exosomes inhibit food allergies by inducing
regulatory T cells. Immunol Cell Biol. 98:639–649. 2020.
|
|
131
|
Kim SH, Bianco N, Menon R, Lechman ER,
Shufesky WJ, Morelli AE and Robbins PD: Exosomes derived from
genetically modified DC expressing FasL are anti-inflammatory and
immunosuppressive. Mol Ther. 13:289–300. 2006.
|
|
132
|
Näslund TI, Gehrmann U, Qazi KR, Karlsson
MC and Gabrielsson S: Dendritic cell-derived exosomes need to
activate both T and B cells to induce antitumor immunity. J
Immunol. 190:2712–2719. 2013.
|
|
133
|
Iberg CA and Hawiger D: Natural and
induced tolerogenic dendritic cells. J Immunol. 204:733–744.
2020.
|