MicroRNAs (miRNAs/miRs) are short non-coding RNAs,
which serve a role in cell differentiation, homeostasis and
organism development. miRNAs consist of ~22 nucleotides, and can
induce silencing of some genes by directing argonaute (AGO)
proteins to target mRNA at the 3′ untranslated region (UTR)
(1,2). As single-stranded short nucleic
acids, miRNAs act as guides to RNA or DNA complementary sequences
that are intended to be silenced (3). The translational repression and
further elimination of target mRNAs occur through the miRNA-induced
silencing complex (miRISC), which is formed of the miRNA, AGO
proteins and other associated proteins such as Dicer (2).
At present, it has been reported that most of the
human protein-coding genes (>60%) contain miRNA target sites
(4), and the miRNA repository
miRBase (https://www.mirbase.org) records 2,654
mature miRNAs and 1,917 precursor miRNAs (pre-miRNAs) in humans
(5).
As a hallmark of cancer and other common diseases,
the upregulated and downregulated expression of miRNAs serves a
crucial role in disease development and progression. The
upregulation of miRNAs associated with cancer promotes cancer by
targeting tumor suppressor genes (6). For example, miR-21 is often
upregulated in various cancer types, including breast, lung and
colorectal cancer (CRC). miR-21 upregulation leads to the
suppression of tumor suppressor genes such as PTEN and programmed
cell death protein 4, promoting cell proliferation and survival
(7). In a similar manner,
miR-155 is frequently upregulated in hematological malignancies,
including lymphoma and leukemias. This miRNA targets multiple genes
involved in apoptosis and cell differentiation, contributing to
tumorigenesis (8).
The upregulation of miRNAs such as tumor suppressor
miRNAs inhibits cancer via targeting of oncogenes. For instance,
miR-34a is downregulated in numerous cancer types, including
pancreatic and prostate cancer. miR-34a downregulation leads to the
upregulation of oncogenes such as MYC and BCL2, facilitating tumor
growth and resistance to apoptosis (8). Similarly, the miR-200 family of
miRNAs is often downregulated in cancer types such as ovarian and
breast cancer. The loss of miR-200 expression is associated with
epithelial-mesenchymal transition (EMT), a process that enhances
cancer cell invasion and metastasis (9).
Cancer mortality is a threat to health worldwide.
Approaches for early detection of cancer using different diagnostic
procedures and cancer treatments are needed globally. A
cancer-specific screening technique that is sensitive enough to
identify early malignancy would be ideal. This should be specific
to the cancer type and location, appropriate for the size of the
tumor, affordable, user-friendly, and safe. Currently, there are
numerous methods for identifying and measuring certain miRNAs,
including miRNA arrays, in situ hybridization and
immuneprecipitation to examine miRNA localization and specific
miRNA activity, and total miRNA measurement (10). The accuracy, cost, effectiveness
and ease of tracking miRNA dynamics vary among these methods.
Furthermore, all of these approaches have some advantages and
disadvantages for miRNA quantification in biochemical and medical
research (11,12).
The CRISPR/CRISPR-associated sequence (Cas) system
is a programmable platform, a modern world genome editing system,
which has garnered trust for the management of cancer detection and
treatment (13,14). The innovative approach of cancer
detection using the CRISPR/Cas system is more cost effective
compared with other currently known procedures (15). CRISPR/Cas systems are now a
frequently used genome editing method in molecular biology labs
worldwide because of their quick cycle, low cost, high efficiency,
good repeatability and simple design (16,17). This system consists of a set of
CRISPR-associated (Cas) genes that encode Cas proteins with
endonuclease activity as well as CRISPR repeat-spacer arrays, which
can be further translated into CRISPR RNA (crRNA) and
trans-activating crRNA (18,19).
CRISPR/CRISPR-associated sequence 13 (Cas13) is a
powerful and versatile genome-editing tool, targeting and cleaving
single-stranded RNA (ssRNA) rather than DNA, and is a class 2 type
VI CRISPR/Cas system (20).
Cas13 belongs to the class 2 type VI CRISPR/Cas system and consists
of different subtypes, including Cas13a, Cas13b, Cas13c and Cas13d.
Among all these subtypes, Cas13a (also referred to as C2c2) is the
most studied in terms of its structure and RNA editing approach
(21). The CRISPR/Cas13 system
can be effectively used for viral detection, splicing regulation,
transcript tagging and RNA knockdown (22-24).
The advancements in miRNA detection with the help of
the CRISPR/Cas13 system have shown some promising results. Some of
the key updates include direct and accurate miRNA detection,
development of amplification-free biosensors and lateral flow
assays (25-27). However, several knowledge gaps
remain, which need to be discussed further. The current methods of
miRNA detection show high sensitivity; however, the achievement of
absolute specificity in complex biological samples remains a
challenging task (11). Further
research is required to minimize false positives and improve the
robustness of these assays. The development of cost-effective and
scalable miRNA detection procedures, which are suitable for
widespread clinical use, is still a hurdle. Thus, innovations that
reduce the costs and simplify the detection processes are crucial.
Furthermore, there is still much to learn regarding the diverse
roles of miRNAs in various diseases, particularly cancer.
Therefore, comprehensive studies are needed to fully understand
their mechanisms and potential as therapeutic targets.
In the present review, current updates regarding the
biogenesis, structural and functional aspects, and dysregulation in
different cancer types of miRNAs are discussed. In addition, the
significance of altered miRNA expression in tumors is discussed.
Furthermore, the functional aspects of CRISPR/Cas13 as a
next-generation tool in different forms for the detection of miRNAs
in different cancer types are elaborated. An overview of current
methods using CRISPR/Cas13-based biosensor systems is provided,
emphasizing how these methods have made it possible to miniaturize
electrochemical transducers and enhance their sensitivity,
specificity and suitability for miRNA diagnosis. Furthermore,
challenges and prospects of the use of CRISPR/Cas13 as a miRNA
detection tool are discussed.
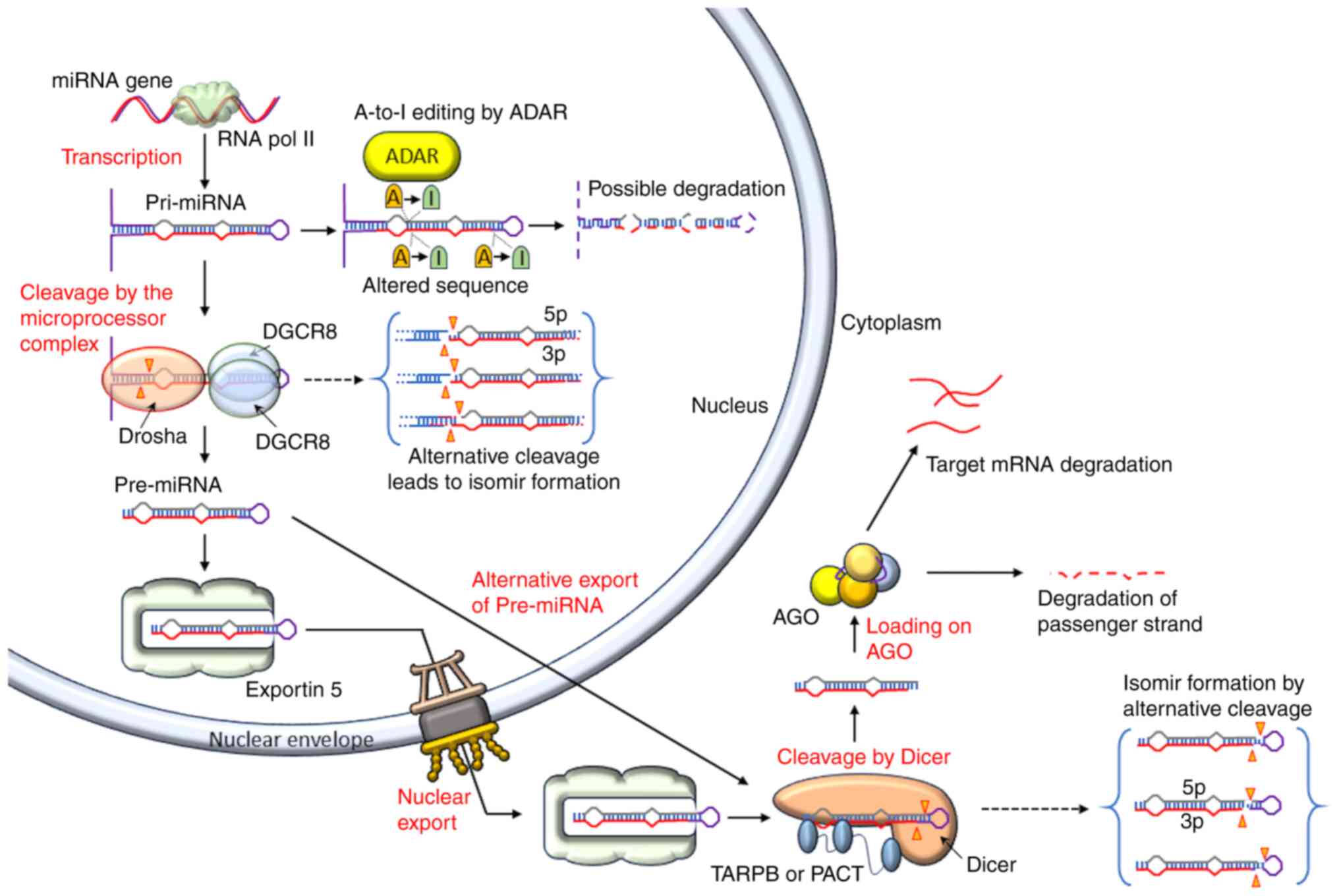
miRNA biogenesis is a multi-step process that occurs
under tight temporal and spatial control by different proteins. The
RNA polymerase II primarily transcribes them as primary miRNAs
(pri-miRNAs), which are then converted into pre-miRNAs, which are
later converted into mature miRNA duplexes (28,29) (Fig. 1). The 5p and 3p strands of the
mature miRNA are derived from the 5′ arm of the pre-miRNA. The
A-to-I editing of pri-miRNAs is performed by adenosine deaminase
proteins, which may impact subsequent biogenesis and the sequence
of the mature miRNA, or mediates pri-miRNA destruction (30).
The microprocessor complex, which includes the
DiGeorge syndrome critical region 8 (DGCR8) protein (31,32) and the RNase III enzyme Drosha
(33), excises the pri-miRNA
hairpin in the nucleus (Fig. 1).
For its action, two DGCR8 proteins bind the stem and make a proper
cleavage (34,35), while Drosha detects the junction
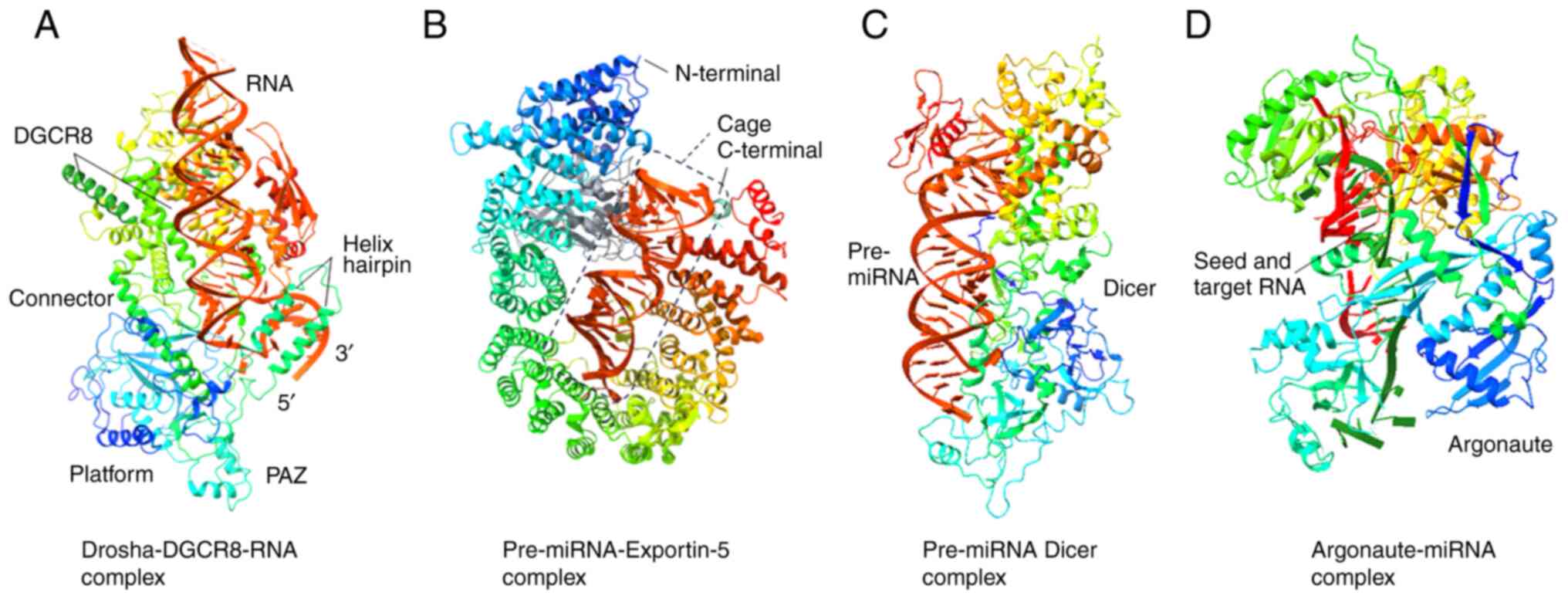
between double-stranded RNA and ssRNA at the pri-miRNA hairpin base
[Fig. 2A; obtained from Protein
Data Bank (PDB; https://www.rcsb.org) with PDB ID
6LXD and modified with Chimera X (https://www.cgl.ucsf.edu/chimerax/; version 1.8)]. The
alternative cleavage by Drosha results in the creation of isomirs
(36,37). Pre-miRNAs are hairpin-shaped RNAs
composed of ~70 nucleotides (38,39). The hairpin end has a 3′ hydroxyl,
a 5′ phosphate and a 2-nucleotide overhang at the 3′ end (40).
After identifying the overhang, exportin-5 (XPO-5; a
transport protein) transports the pre-miRNA into the cytoplasm
(41) (Figs. 1 and 2B). In a human cell line, exportin-5
(XPO-5) deletion led to decreased transportation of pre-miRNA but
did not completely stop its nuclear export, indicating the
existence of some other pre-miRNA nuclear export pathways (42). In the cytoplasm, Dicer (RNase III
enzyme) (43,44) recognizes the 5′ phosphate, 3′
overhang and loop structure (45,46), and binds the pre-miRNA (Fig. 1). Dicer acts as a 'molecular
ruler' that produces a mature miRNA duplex with a 2-nucleotide 3′
overhang (40) after cleaving
pre-miRNAs at a species-specific length (Fig. 2C). It is also possible for Dicer
to produce isomirs through alternative cleavage (37).
In vertebrates, Dicer cleavage is regulated by
protein activator of the interferon-induced protein kinase and
transactivating response-RNA-binding protein. The cleavage of
pre-miRNA by Dicer leads to the formation of two RNA strands (guide
and passenger strands). The mature miRNA 'guide' strand is loaded
into AGO protein, while the 'passenger' strand is eliminated
(47,48) (Fig. 1). The strand with the less
securely coupled 5′ end (49) is
preferentially loaded on the AGO protein (Fig. 2D).
After being transcribed into pri-miRNA transcripts,
miRNAs go through a multi-step biogenesis process that transforms
them into pre-miRNAs and mature miRNAs. The expression patterns of
miRNAs are tissue-specific (50)
and transcriptionally regulated (51). miRNAs can originate from long
non-coding RNAs or introns, and are mostly transcribed by RNA
polymerase II (52,53). Pri-miRNAs can be clusters of
frequently linked miRNAs or a single mature miRNA (54). Based on how similar their seed
sequences are, miRNAs are categorized into different families
(55). The seed region of
miRNAs, which consists of nucleotides 2-8 (counting from the 5′
end), is mostly responsible for targeting mRNAs (56).
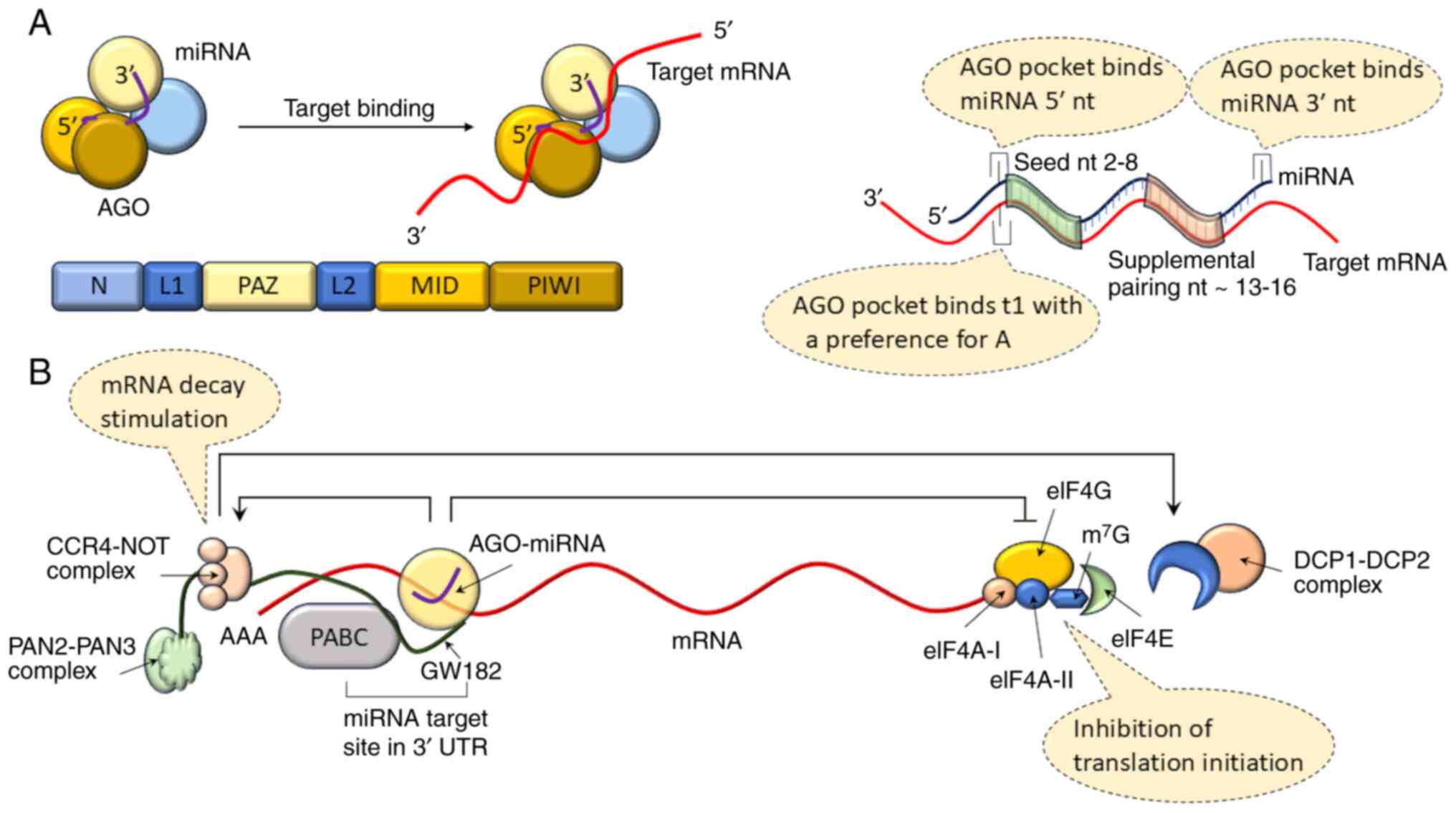
Mature miRNAs act inside a single polypeptide chain
AGO protein, with four distinctive domains (Fig. 3A). The four domains are the
N-terminal (N), P-element induced wimpy testes (PIWI)-AGO-Zwille
(PAZ), PIWI and MID domains. A linker domain (L1) is present
between the N and PAZ domains, while the L2 domain is present
between the PAZ and MID domains. The MID and PIWI domains make up
the second lobe of AGO, while the N and PAZ domains make up the
first (57,58). The PAZ domain binds the 3′
nucleotides of the miRNA (59,60), whereas the MID and PIWI domains
retain the 5′ end of the miRNA (Fig.
3A). It has been reported that four AGO proteins (AGO1-4) are
encoded by the mammalian genome. AGO2 is the most abundant and
unique among the AGO proteins, with target cleaving activity when
completely complementary to the guide strand of the miRNA (61,62). Numerous human miRNAs bind to all
AGO proteins, although some are selectively packaged into AGO
proteins (63,64). Typically found in the 3′ UTR of
mRNAs, miRNA target sites exhibit high complementarity to the seed
region, the primary requirement for the prediction of target sites
(65,66).
The strongest canonical (seed matching) target sites
are those that complement miRNA nucleotides 2-8 and have an adenine
opposite miRNA nucleotide 1 (referred to as 't1A'), followed by
those complementing nucleotides 2-8 without a t1A and those
complementing nucleotides 2-7 with t1A (67). A binding site in AGO (68,69) recognizes t1A instead of the miRNA
guide strand (Fig. 3A). Target
sites that complement miRNA nucleotides 2-7 or 3-8 are still
considered canonical, although they have a weaker affinity
(65). According to structural
and single-molecule investigations, the MID and PIWI domains
pre-organize the initial nucleotides 2-6 of the seed in helical
conformation (59,70). This two-step mechanism is
considered to be responsible for target identification (71).
The biological role of non-canonical sites has been
contested because small RNA and miRNA transfection data showed no
discernible suppression from these locations (65). Through translational suppression
and mRNA decay, the 3′ UTR binding with AGO-miRNA results in gene
silencing (72) (Fig. 3B). Furthermore,
glycine-tryptophan 182 (GW182; 182 kDa), a member of the
glycine-tryptophan protein family, is recruited by AGO for RNA
silencing (73).
By attracting the poly(A)-nuclease deadenylation
complex subunit 2 (PAN2)-PAN3 and carbon catabolite repressor
protein 4 (CCR4)-NOT complexes (74,75), the interaction between
polyadenylate binding protein and GW182 promotes the deadenylation
of mRNA. Because deadenylation encourages decapping by the
mRNA-decapping enzyme subunit 1 (DCP1)-DCP2 complex (76), 5′-3′ exoribonuclease 1 can
quickly degrade the mRNA (77)
(Fig. 3B). Through the
recruitment of the likely ATP-dependent RNA helicase DEAD-box
helicase 6, GW182-mediated recruitment of CCR4-NOT also results in
translation suppression (78).
Eukaryotic initiation factor 4A-I (eIF4A-I) and
eIF4A-II interference also inhibits translation initiation
(79,80). Although the exact method of
interference in initiation factors is not fully understood yet,
most studies suggest that miRISC causes them to separate from
target mRNAs (81,82), which prevents scanning by the
ribosome and the formation of the eIF4F translation initiation
unit. Translation inhibition requires the Trp-binding pockets in
AGO that mediate binding with GW182, according to a study conducted
in human and Drosophila melanogaster cells (83,84).
Complete understanding of miRISC-mediated
translation inhibition remains lacking, and it has been
hypothesized that the two methods of miRISC-mediated gene silencing
are related (2). Ribosome
profiling tests have shown that mRNA degradation typically accounts
for 66-90% of silencing (72,85). The observation that translation
inhibition can be restored while mRNA degradation is permanent
raises the possibility that regulated pauses, or blocks, in the
metabolic cascade leading to mRNA degradation could allow
translation suppression without mRNA decay (86). Furthermore, multiple miRNAs can
regulate the same gene (87),
and it has also been reported that hundreds of genes can be muted
by a single miRNA (88).
Furthermore, individual miRNAs or miRNA clusters can
control whole cellular pathways (89). Cooperative suppression can be
caused by nearby target site binding with miRNA on a target mRNA
(90,91), which may help justify the
functioning of non-canonical sites relying on the occupancy of
nearby canonical sites. The development of multivalent protein
interactions between GW182 and AGO proteins help to explain
cooperativity (84).
Furthermore, miRNAs can either inhibit or regulate protein
expression (92), protecting
against variations (or 'noise') in gene expression levels (93).
Human cancers exhibit dysregulated miRNA expression.
Cancer development through chromosome abnormalities,
transcriptional regulation alterations, epigenetic modifications
and flaws in the miRNA biogenesis machinery are some of the
underlying mechanisms (94,95). miRNA expression is regulated at
different stages of cellular activities as subsequently
described.
Changes in genomic miRNA copy numbers and gene
positions (amplification, deletion or translocation) are frequently
related to the cause of abnormal miRNA expression in malignant
cells (96). The loss of the
miR-15a/16-1 cluster gene at chromosome 13q14, which is commonly
seen in individuals with B-cell chronic lymphocytic leukemia, is
the earliest example of a change in miRNA gene placement (97). In addition, the 5q33 region that
contains miR-143 and miR-145 is frequently deleted in lung cancer,
which lowers the expression levels of both miRNAs (96). On the other hand, B-cell lymphoma
(98) and lung cancer (99) exhibit amplification of the
miR-17-92 cluster gene, and T-cell acute lymphoblastic leukemia
(100) exhibits translocation
of this cluster gene, resulting in upregulation of these miRNAs in
these cancer types.
High-resolution array-based comparative genomic
hybridization has been used to confirm the high frequency of
genomic changes in miRNA loci in 227 human cancer samples of
ovarian cancer, breast cancer and melanoma (101). Numerous miRNA genes are found
in genomic areas linked to cancer, according to additional
genome-wide studies (102,103). These areas on chromosomes may
be fragile sites, common breakpoint regions or minimal regions of
amplification, which may include oncogenes, or minimal regions of
loss of heterozygosity, which may carry tumor suppressor genes
(102). Considering all the
observations, the findings suggest that the amplification or
deletion of specific genomic regions containing miRNA genes may be
the cause of abnormal miRNA expression in malignant cells.
Since a number of transcription factors tightly
regulate miRNA production, dysregulation of some important
transcription factors, including p53 and c-Myc, may be linked to
aberrant miRNA expression in cancer (104,105). By binding to E-box regions in
the miR-17-92 promoter, c-Myc is commonly increased through
transcriptional activation, in a number of malignancies to control
cell proliferation and death, and promotes the transcription of the
oncogenic miR-17-92 cluster (106). In keeping with its carcinogenic
function, c-Myc also inhibits the transcriptional activity of
tumor-suppressive miRNAs, including the let-7, miR-26, miR-29 and
mir-15a families (107).
In hepatocellular carcinoma (HCC), c-Myc and the
tumor suppressor miR-122 are reciprocally regulated. By attaching
itself to its promoter, c-Myc inhibits the synthesis of miR-122
and, by targeting the transcription factor Dp2 and the
transcription factor E2F transcription factor 1 (E2f1), indirectly
stops c-Myc transcription. Therefore, the disruption of this
feedback loop between miR-122 and c-Myc is necessary for the
development of HCC (108). HCC
is brought on by direct binding of c-Myc to the promoters of the
miR-148a-5p and miR-363-3p genes, which inhibits their synthesis
and encourages the G1 to S phase transition.
miR-148a-5p, on the other hand, directly targets and suppresses the
production of c-Myc, while miR-363-3p destabilizes c-Myc by
targeting ubiquitin-specific protease (106,109). One of the most frequently
altered genes in human cancer is TP53, which encodes the tumor
suppressor p53. Another manner in which transcriptional factors
control miRNA expression to exert a tumor suppressive function is
the p53-miR-34 regulatory axis (110). A complex p53 network that
controls cell-cycle progression and apoptosis is formed by the
p53-regulated expression of several genes, including miRNA genes.
Similar to p53-mediated phenotypes, the miR-34 family, comprising
miR-34a, miR-34b and miR-34c, causes cell-cycle arrest, cell
senescence and apoptosis in cancer (111), suggesting that p53 and miR-34
are involved in the same regulatory mechanism.
Additional research has revealed that p53 regulates
the expression of several miRNAs, including miR-605 (112), miR-1246 (113) and miR-107 (114), to carry out its role.
Additional transcriptional factors have been identified to control
miRNA production in addition to the two most well researched
transcriptional factors, c-Myc and p53. For instance, miR-223 is
inhibited in a variety of cancer types, including HCC and acute
myeloid leukemia (AML), and it is predominantly expressed in the
hematopoietic system, which serves important roles in the formation
of myeloid lineages (115).
As aforementioned, several enzymes and regulatory
proteins, including Drosha, Dicer, DGCR8, AGO and XPO-5,
intricately regulate miRNA biogenesis, enabling proper maturation
of miRNA from pri-miRNA precursors. Therefore, aberrant expression
of miRNAs may result from mutations or aberrant expression of any
protein in the miRNA biogenesis pathway. Two important RNase III
endonucleases in miRNA maturation, Drosha and Dicer, oversee the
creation of pre-miRNA and the miRNA duplex (116,117). According to previous studies,
both enzymes are dysregulated in some cancer types such as breast
and bladder cancer (118,119).
A portion of miRNAs are controlled during the
Drosha-processing stage, and this control affects miRNA expression
in cancer and during embryonic development (120). It has been reported that 15% of
534 Wilms tumors had single-nucleotide substitution/deletion
mutations in DGCR8 and Drosha, which resulted in reduced expression
of mature Let-7a and the miR-200 family (121). In terms of Dicer dysregulation,
it has been noted that CRC cells with impaired Dicer acquire a
higher propensity for tumor initiation and metastasis (122). Furthermore, patients with
ovarian cancer with higher Dicer and Drosha mRNA levels have a
higher median survival rate (123). On the other hand, patients with
lower Dicer expression have a considerably lower survival rate
(124,125).
Reduced let-7 expression and poor postoperative
survival have been linked to lower Dicer mRNA levels in patients
with lung cancer (126). As
with Dicer and Drosha, cancer also leads to deregulation of AGO.
For instance, kidney tumors caused by Wilms disease frequently
exhibit loss of the human eukaryotic initiation factor 2C1/human
argonaute gene (127). Human
AGO proteins are crucial in regulating cell-dependent gene
expression. AGO2 expression levels, for example, are considerably
higher in basic gastric cancer and the lymph node metastases
(128), but AGO2 expression is
lower in melanoma, which corresponds to a lower RNA interference
efficiency, compared with in primary melanocytes (129).
A fraction of human cancers with microsatellite
instability have inactivating mutations in the XPO-5 gene. The
insertion of an 'A' in exon 32 in CRC cells (HCT-15 and DLD-1
cells), results in an early termination codon, which causes a
frameshift mutation and the creation of a shortened protein. This
shortened XPO-5 is no longer able to export pre-miRNAs. As a
result, there is less miRNA processing since pre-miRNAs are
confined to the nucleus. The restoration of XPO-5 activities
possesses tumor-suppressor properties and restores the defective
export of pre-miRNAs (130). It
has also been noted that ERK phosphorylates XPO-5, preventing XPO-5
from moving pre-miRNAs from the nucleus to the cytoplasm in HCC
(131).
A well-known characteristic of cancer is epigenetic
modification, which includes the disruption of histone
modification, aberrant DNA hypermethylation of tumor suppressor
genes and widespread genomic DNA hypomethylation. Similar to
protein coding genes, miRNAs are considered to be subjected to
epigenetic modifications (132,133). For example, it has been
reported that acute myeloid leukemia 1/eight twenty one, the most
prevalent AML-associated fusion protein, epigenetically suppresses
miR-223 expression via CpG methylation (134). A study reported that the
expression levels of 17 out of 313 human miRNAs were more than
three times higher in T24 bladder cancer cells after concomitant
treatment with DNA methylation and histone acetylation inhibitors
(134).
The downregulation of the proto-oncogene BCL6
coincides with markedly increased expression of miR-127, which is
embedded in a CpG island and not expressed in cancer cells
(135). These findings suggest
that miRNAs may function as tumor suppressors and can be activated
through DNA demethylation and histone deacetylase inhibition. In
addition, it has been reported that miR-148a and the miR-34b/c
cluster are susceptible to hypermethylation-associated silencing in
cancer cells (136).
Furthermore, in vivo metastasis formation was suppressed,
tumor growth was decreased and cancer cell motility was inhibited
when these miRNAs were restored (137). Similarly, DNA hypermethylation
has been linked to lower expression levels of miR-9-1, miR-124a and
miR-145-5p in colon, lung and breast cancer, respectively (138,139). This suggests that the abnormal
histone acetylation and DNA methylation of miRNA genes could be
useful indicators for the diagnosis and prognosis of cancer. These
findings emphasize the significance of epigenetic control in miRNA
production during carcinogenesis.
The biological characteristics of tumor development
include continued proliferative signaling, evasion of growth
suppressors, replicative immortality, initiation of invasion and
metastasis, induction of angiogenesis, and prevention of cell death
(140). Dysregulated miRNAs may
impact one or more of these cancer hallmarks for tumor initiation
and development. In different situations, miRNAs act as tumor
suppressors or oncogenes, depending on the genes they target.
Dysregulated miRNAs affect tumorigenesis in different manners as
described subsequently.
The primary cause of tumorigenesis is aberrant cell
proliferation, which is the most significant characteristic of
cancer. Certain miRNAs functionally incorporate into several cell
proliferation pathways, and the dysregulation of these miRNAs
directs the cancer cells to continue proliferative signaling to
avoid growth suppressors. miRNAs serve a role in controlling the
expression of E2F proteins (cell proliferation regulators).
E2F1-deficient animals develop a wide range of malignancies, and
this member of the E2F family is characterized as a tumor
suppressor and stimulates target gene transcription during the
G1 to S transition (141). miR-17-92, once activated by
c-Myc, suppresses the translation of E2F1 (106). E2F1 protein levels may not
increase markedly in response to c-Myc activation if the miR-17-92
cluster acts as a brake on this putative positive feedback loop,
given that c-Myc also directly stimulates E2F1 production (142). Furthermore, the miR-17-92
cluster has been demonstrated to regulate E2F2 and E2F3 translation
(143). Additionally, the
expression of the miR-17-92 cluster might be triggered by the E2F
transcription factors (144).
Therefore, under normal conditions, the feedback loop between E2F
and the miR-17-92 cluster provides a means of preserving regular
cell-cycle progression. However, miR-17-92 upregulation, which is
common in several cancer types such as colorectal cancer and
gastric cancer, disrupts the feedback loop that encourages cell
proliferation (145).
Different cyclins, Cdks and their inhibitors,
extensively regulated by miRNAs, are essential for cell-cycle
progression. The G1/S transition inhibition of
Drosophila germline stem cells with Dicer-1 deletion suggests that
miRNAs are required for germline stem cells to pass through the
normal G1/S checkpoint (146). Additionally, Dacapo, a member
of the p21/p27 family of Cdk inhibitors, is upregulated in
Dicer-deficient germline stem cells, suggesting that miRNAs
negatively control this protein to encourage cell-cycle progression
(147). In glioblastoma cells,
miR-221/222 has been found to directly target the Cdk inhibitor
p27Kip1 (148), a finding that
has been subsequently validated in original tumor samples and other
cancer cell lines (149,150).
While its inhibition causes G1 cell-cycle arrest in
malignant cells, ectopic expression of miR-221/222 enhances cell
proliferation. Furthermore, several human malignancies have been
reported to be associated with increased expression levels of
miR-221/222, suggesting that regulation of p27Kip1 by miR-221/222
is a valid carcinogenic pathway (151).
To meet the demands of food and oxygen during tumor
growth and metastasis, the highly coordinated process of
angiogenesis produces new blood vessels from pre-existing ones
(152). Because the oxygen
concentration of tumor tissues is lower than that of the
surrounding normal tissues, hypoxia serves a critical role in the
tumor microenvironment by facilitating the proliferation and
maintenance of cancer cells. In response to hypoxia, a crucial
transcription factor, hypoxia-inducible factor (HIF), affects the
expression of several genes, including miRNAs (153). As a key angiogenic factor, VEGF
instructs endothelial cells to form new vessels when it binds to
its receptors (154).
Therefore, angiogenesis is likely to be impacted by miRNAs that
target the VEGF or HIF signaling pathways.
Hypoxia causes endothelial cells to produce miR-424,
which targets cullin 2, a ubiquitin ligase scaffold protein, to
stimulate angiogenesis. By stabilizing HIF-1α, this step enables
stabilized HIF-1α to transcriptionally trigger the expression of
VEGF (160). miR-21 is another
miRNA that promotes angiogenesis. To activate the downstream
Akt/ERK signaling pathways and increase HIF-1α and VEGF expression,
it targets PTEN (161). By
inhibiting VEGF and/or HIF-1α, on the other hand, miR-20b and
miR-519c negatively control angiogenesis (162,163). The downregulation of miR-107
increases tumor angiogenesis under hypoxic conditions because it
not only regulates HIF-1α but also inhibits the production of
HIF-1β (114).
The biology of metastasis is an intricate,
multi-step and dynamic process. The loss of cell adhesion due to
E-cadherin suppression and the activation of genes linked to
invasion and motility are the hallmarks of the EMT, which is
regarded as an initial and crucial stage of metastasis (164). Numerous miRNAs alter the
expression of genes, including MMPs and their inhibitors, that
control extracellular matrix (ECM) turnover and breakdown (165). Proliferation, migration,
differentiation and apoptosis are among the cell phenotypic
alterations that are influenced by ECM turnover or remodeling
(166). Zinc-dependent
endopeptidases, known as important MMPs such as MMP-1, -2, -3, -7
and -9, are crucial for cell migration, adhesion, dispersion,
differentiation and ECM modeling (167). They can cause cell surface
receptors to cleave ECM proteins such as type IV collagen,
interestitial collagen, elastin and casein for degradation
(168). Exogenous miR-143
expression in osteosarcoma cells leads to downregulated MMP-13
protein levels and reduced cell invasion (169). In clinical samples, MMP-13 was
found in cases with lung metastases and low miR-143 expression,
while it was not found in patients without metastases and with high
miR-143 expression (170).
It has been revealed that miR-206 decreased the
amount of the cell division cycle 42, MMP-2 and MMP-9 proteins in
human breast cancer (171). Due
to the control of actin cytoskeleton remodeling, including
filopodia formation, this protein level regulation inhibited
MDA-MB-231 cell invasion and migration (171). Furthermore, downregulation of
miR-340 has been associated with aggressive activity in several
breast cancer cell lines (172). Because it directly targets
c-Met, the stimulation of miR-340 expression suppresses the
motility and invasion of breast tumor cells, thereby modulating the
production of MMP-2 and MMP-9 (172).
Numerous signaling pathways, including the TGF-β
pathway, are considered to govern EMT. These pathways converge on
important transcription factors, including Zinc finger
E-box-binding homeobox (ZEB), SNAIL and TWIST (173). TGF-β-regulated miRNAs
participate in TGF-β signaling to trigger EMT and promote
metastasis in advanced cancer. miR-155 is implicated in this
regulation, is transcriptionally activated by TGF-β/SMAD4 signaling
and is upregulated in several malignancies such as colorectal and
breast cancer (174).
Furthermore, miR-155 stimulates EMT by targeting RhoA GTPase, an
essential modulator of cellular polarity and the formation and
maintenance of tight junctions. Additionally, miR-155 suppression
prevents TGF-β-induced EMT, tight junction disruption, cell
migration and invasion (174).
In contrast to miR-155, TGF-β inhibits miR-200 and miR-203, and it
has been demonstrated that the miR-200 family influences EMT by
suppressing the expression of the transcriptional repressors (ZEB1
and ZEB2) of E-cadherin (175).
ZEB1 and ZEB2, in turn, also suppress the miR-200 transcript,
creating a double-negative feedback loop between the miR-200 family
and ZEB1/ZEB2 (176).
An important characteristic of tumor growth is
evasion of apoptosis, which is considered to be controlled by
miRNAs (177,178). Different strategies are
developed by tumor cells to prevent or delay apoptosis. The most
prevalent of these is the loss of p53 tumor suppressor activity
(179). Other strategies to
avoid apoptosis include the suppression of proapoptotic proteins,
upregulation of anti-apoptotic regulators and blocking the death
pathways (180). miRNAs either
inhibit or activate the components involved in anti-apoptosis
mechanisms. It has been demonstrated that numerous p53-regulated
miRNAs contribute to p53 functions; some of these miRNAs can
feedback-modify p53 activity (181). For example, in multiple
myeloma, p53 transcriptionally activates three miRNAs (miR-192,
miR-194 and miR-215) to bind directly with the mRNA of mouse double
minute 2 homolog (Mdm2) and prevent p53 from being destroyed, thus
inhibiting Mdm2 expression (182). The development of multiple
myeloma is influenced by the downregulation of these miRNAs, which
are positive regulators of p53 (183).
Another negative feedback regulation takes place
between p53 and miR-122 by targeting cyclin G1 (184) and cytoplasmic polyadenylation
element-binding protein (185).
The creation of a chemotherapy and miRNA-based therapeutic
combination for HCC is made possible by the stimulation of p53
activity by miR-122, which also increases cell sensitivity to
doxorubicin (186). In
addition, cancer cells are resistant to death due to other
dysregulated p53-regulated miRNAs. For example, the miR-17-92
cluster represents a novel target for p53-mediated transcriptional
repression during hypoxia. While its upregulation prevents
apoptosis, its downregulation makes cells more vulnerable to
hypoxia-induced apoptosis. Consequently, tumor cells that express
more miR-17-92 may be able to evade the apoptosis caused by hypoxia
(187). All these
aforementioned findings demonstrate that, in healthy circumstances,
p53 and miRNAs regulate a network that intricately determines cell
destiny. However, dysregulation of p53 or its target miRNAs may
allow cancer cells to evade cell death.
Some miRNAs that serve a role in cell death may
target proapoptotic factors (Bax, Bim and Puma) and anti-apoptotic
regulators (Bcl-2 and Bcl-xL). In chronic lymphocytic leukemia,
miR-15a and miR-16-1 are downregulated, and their expression is
inversely associated with that of Bcl-2 (97). Additional research revealed that
these two miRNAs caused apoptosis and suppressed Bcl-2 expression
(188). Other miRNAs, including
miR-204 (189), miR-148a
(113) and miR-365 (190), also control Bcl-2 expression.
miR-491-5p effectively causes ovarian cancer cells to undergo
apoptosis by causing Bim accumulation and directly suppressing
Bcl-xL expression (191).
The role of some miRNAs and their relationship with
tumor occurrence, development, metastasis and survival is shown in
Table I.
Different approaches are currently available for
the identification and quantification of miRNAs, which have the
capacity to measure either total miRNAs or specific miRNAs, and
their localization within cells. For the quantification of total
miRNA, these methods have been devised based on hybridization,
amplification, sequencing and enzyme-based approaches (10). The hybridization-based methods
include northern blotting, microarrays and bead array-based
profiling. The amplification-based methods of miRNA quantification
include reverse transcription-quantitative PCR (RT-qPCR), rolling
circle amplification (RCA) and next-generation sequencing. The
enzyme-based methods of miRNA detection include invader assays,
luciferase-based assays, assay using miRNA in vivo activity
reporters and the molecular beacon imaging method. All these
detection and quantification assays vary in efficiency, cost,
accuracy and monitoring convenience (10). The currently used miRNA detection
approaches using different detections tools, and their strengths
and limitations are summarized in Table II.
CRISPR/Cas systems have been widely used for
biotechnological and clinical research following the identification
of RNA-programmable nucleases from the prokaryotic adaptive immune
system (192,193). Among different types of
CRISPR/Cas systems, the type VI system exclusively targets ssRNA
(194,195). In this system, different
subtypes have been identified as type VIA (having Cas13a)-VID
(having Cas13d), Cas13X and Cas13Y (196,197), among which Cas13a is the best
known so far. Innovative diagnostic methods for the identification
of certain miRNAs have been developed using this platform.
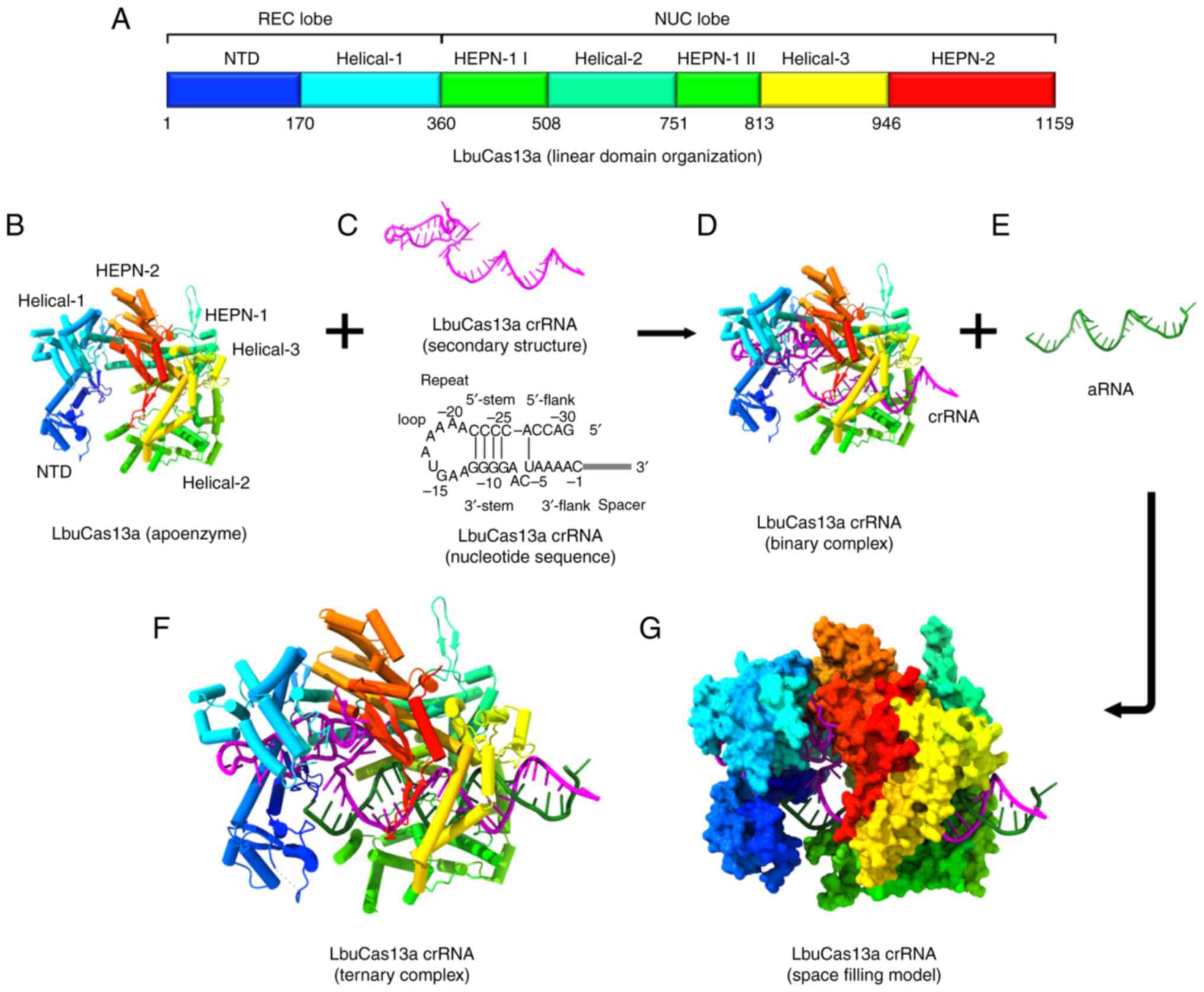
The V-shaped structure of the helix-1 domain is
formed by seven α-helices. The crRNA binding channel is formed by
the positively charged surface of the helix-1 domain facing the NTD
domain (204). The two
conserved higher eukaryotic and prokaryotic nucleotide-binding
(HEPN) domains (HEPN-1 and HEPN-2), a linker that joins the two
HEPN domains, and a helix-2 domain are all found in the NUC lobe
(202). The helix-1 and HEPN
domains are responsible for the two enzymatic functions of Cas13a.
The helix-2 domain further sub-divides the HEPN-1 domain into two
subdomains. Three α-helices make up the HEPN 1-II subdomain,
whereas four α-helices and a brief β-hairpin make up the HEPN-1 I
subdomain (Fig. 4B). Seven
α-helices plus a double-stranded β-sheet directly make up the HEPN2
domain structure (202).
Between the two HEPN-1 subdomains lies the helix-2 domain, which is
made up of eight bean-shaped α-helices (204). The Cas13a/crRNA complex targets
the matching RNA under the direction of crRNA, triggering the
protection of prokaryotes against RNA viruses (205,206) (Fig. 4F and G).
The structure of the crRNA is composed of a spacer
sequence [guide RNA (gRNA)] that mediates target recognition by
RNA-RNA hybridization and a repeat stem-loop region (referred to as
the 5′-handle) that prevents this region from being cleaved during
the cleavage of the target RNA by Cas13a (207) (Fig. 4C). The stem-loop is composed of
two single-stranded bases at the base, a nine-base loop,
neighboring motifs at both ends and a stem made up of five base
pairings. The repeat region of crRNA can twist slightly to generate
a helical shape in addition to a stem-loop secondary structure
(202). This structure is
primarily sustained by the creation of strong hydrogen bonds
between the HEPN2 domain and the backbone of the crRNA (204). It is possible to cause the
Cas13a protein to recognize the stem-loop structure of the crRNA in
a sequence-specific way by altering stem nucleotides, which is
necessary for its nuclease cleavage activity (204) (Fig. 4D).
Within the crRNA guide region, one to four
nucleotides oversee the 5′-end being attached to the void created
by the HEPN-1 and helix-2 domains, making a U-turn. The linker and
the groove created by the HEPN-2 domain bind the final three or
four nucleotides (16). To
identify the target RNA, the Cas13 protein buries a spacer of eight
or nine nucleotides at the 5′-end, while the middle region and
3′-end are exposed to the peripheral solvent (204). To determine the ideal gRNA,
researchers have created a computational model, and identified that
the properties of crRNA and the environment of the target RNA are
important constraints on the cleavage effectiveness of Cas protein
by assessing the activities of 24,460 gRNAs and looking for
mismatches between gRNAs and the target sequence (208).
Cas13 action does not require crRNA maturation; in
fact, pre-crRNA can detect target RNAs (also referred to as
activator RNA; Fig. 4E)
(209). The ribonucleoprotein
complex (RNP) undergoes a conformational change when crRNA binds to
a target RNA (Fig. 4F). A
catalytic site is formed in part by the close interaction of two
HEPN domains (201). The
binding of crRNA is anticipated to result in the cleavage of the
target RNA as well as other ssRNAs surrounding the RNP complex,
possibly including host RNA, because of the distance between the
catalytic site and the crRNA-RNA duplex. This activity of the
CRISPR/Cas13 system, known as collateral damage or collateral
cleavage, refers to the off-target cleavage of endogenous RNA
(205,210).
According to one study, sequence-specific
activation of non-specific RNA cleavage may help to protect the
nearby cells by causing cell dormancy or death, or it may improve
the prevention of phage replication by removing all RNA from a cell
in bulk (195). This ability
has been widely used in RNA knockout strategies, disease treatment,
interference with viral infection, screening of loss of function
mutants, molecular detection of biological agents and CRISPR-based
antimicrobials (211-213). Therefore, the CRISPR/Cas13
system can simultaneously cleave target RNA and non-target RNA
(206,214). Furthermore, in Cas13, the
mutation of arginine in the HEPN domain, which is responsible for
RNA cleavage, results in the formation of catalytically inactive
Cas13, known as dead Cas13 (dCas13). This form of Cas13 has been
used to increase the application of Cas13 in RNA research. Through
the binding of fluorescent proteins or enzymes, mutant dCas13
persistently attaches to target RNA and offers a platform for
detecting transcripts in living cells (20,215).
miRNA expression profiling is technically difficult
as mature miRNAs are small in size, highly similar and not too
abundant in bodily fluids (216). Different platforms have been
engineered that link CRISPR-based methods for miRNA detection with
the advantages of enzyme-assisted signal amplification and
enzyme-free amplification biosensing technologies (217). Electrochemical detection using
biosensors is a novel low-cost and sensitive diagnostic strategy
for the identification of nucleic acids. This approach offers
excellent sensitivity, accuracy and fewer diagnostic constraints as
this strategy is inexpensive, relying on effective sensors and a
straightforward, miniature readout (218).
RCA is a common isothermal enzymatic DNA
replication technique for creating DNA, RNA and protein sensors
(219,220). In an RCA reaction, a short DNA
or RNA strand is amplified to produce a long, single-stranded DNA
(ssDNA) or RNA using a circular DNA template by a specialized
isothermal strand displacement DNA or RNA polymerases (219,220). The RCA process results in a
concatemer that has >109 tandem DNA repeats that are
complementary to the circular DNA or RNA template (221). This visual detection platform
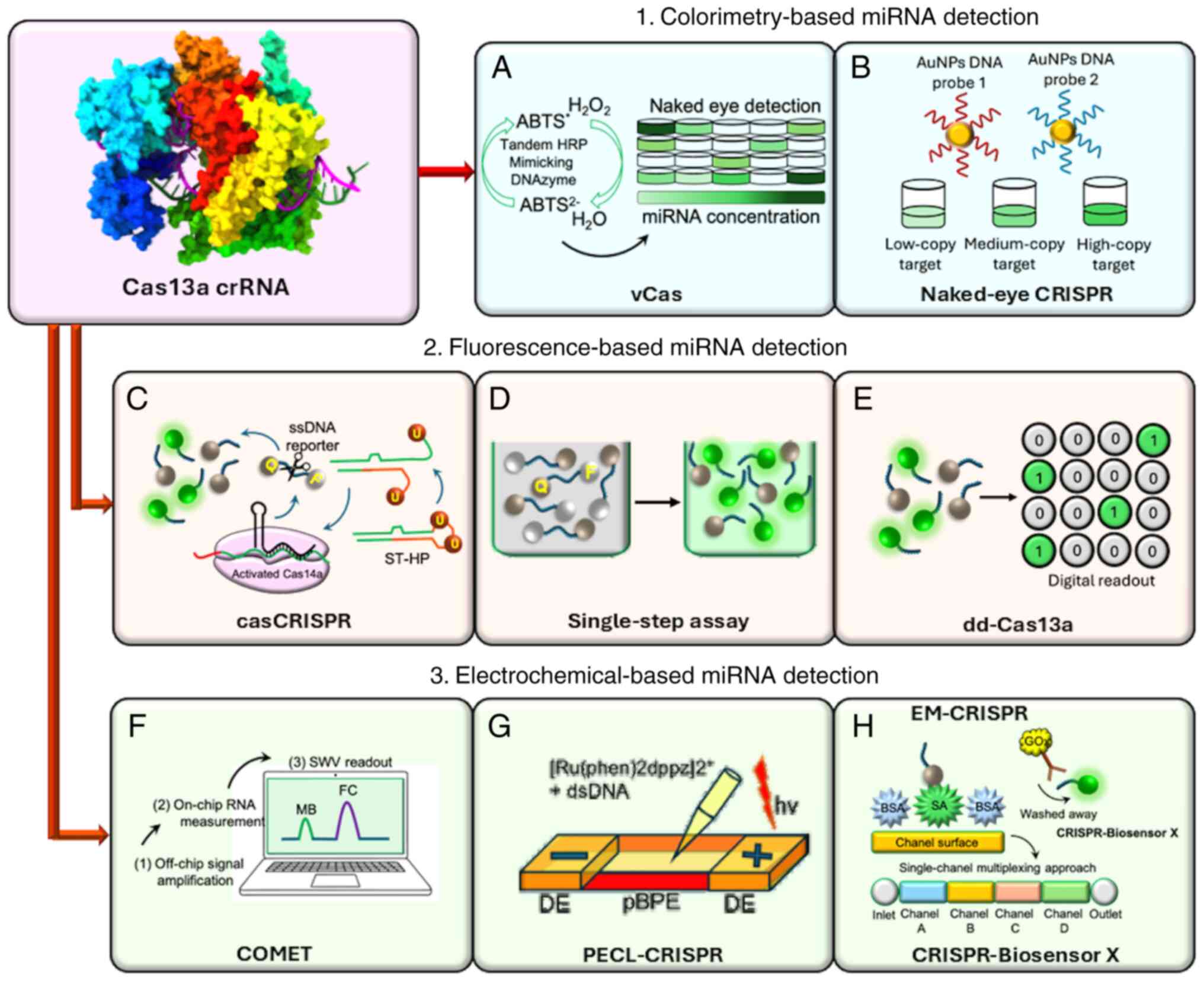
utilizing a CRISPR/Cas13 system, known as the visual Cas (vCas)
system, has been used for the specific and sensitive detection of
miRNA (222) (Fig. 5A).
Theoretically, when Cas13a/crRNA recognizes the
target miRNA, it collaterally cleaves the pre-primer that contains
uracil ribonucleotide (rU). DNA polymerase-mediated RCA can be
started using the leftover 5′-DNA fragment of the pre-primer.
Following its 3′-end, a T4 polynucleotide kinase restores the
aforementioned process to produce a lengthy G-rich repeat sequence
that can form a tandem G-quadruplex to function as a DNAzyme that
mimics HRP and T4 polynucleotide kinase catalyzes the oxidation of
the 2,2′-azino-bis (3-ethylbenzthiazoline-6-sulfonic acid)
diammonium salt (223)
(Fig. 5A). Consequently, within
10 min, the colorless mixture turns green, allowing the target
miRNA to be observed (223).
The results using this approach show that vCas can be used to
evaluate miRNAs in serum and cell extracts (222), and can offer a limit of
detection (LOD) of 1 fM for miR-10b, a type of miRNA highly
expressed in breast cancer cells (224,225). However, compared with one-step
Cas13a detection platforms, the LOD of this approach is four orders
of magnitude lower (222).
Colorimetric assays typically examine the color
shift within the test solution. Without the need for complex
equipment, miRNA detection can be performed quickly and easily with
the use of a colorimeter or the human eye (226). Once Cas12a/crRNA, Cas13a/crRNA
or Cas14/crRNA identify their target DNA or RNA, they exhibit a
transition to the active state, where their collateral cleavage
activity cleaves the ssRNA or ssDNA substrates (227,228). With the aid of CRISPR/Cas
recognition for miRNA sensing, researchers have created a DNA
probes and gold nanoparticles (AuNPs)-based platform.
Cas12a/crRNA's programmable recognition of DNA and Cas13a/crRNA's
programmable recognition of RNA initiates ssRNA or trans-ssDNA
cleavage of their corresponding complementary target (229). For the intended AuNPs-DNA probe
pair, target-induced trans-ssDNA or -ssRNA cleavage results in a
change in aggregation behavior, which allows direct visual
observation in an hour of various samples, including miRNAs
(229). For naked-eye gene
detection platforms, a universal linker ssRNA or ssDNA acts as a
trans-cleavage substrate for the CRISPR/Cas13a or CRISPR/Cas12a
systems. Furthermore, two universal AuNPs DNA probes have been
created to hybridize with either ssRNA or linker ssDNA (229).
Since trans-cleavage is not triggered in the
absence of a target (such as miRNA sequences), the linker ssDNA or
ssRNA stays intact during the reaction. An aggregated state is
created by hybridization-induced cross-linking of the AuNPs-DNA
probe pair (229). After the
trigger of trans-cleavage activity, the linker nucleotides are
broken down once the CRISPR/Cas systems identify their target
nucleotides. By identifying cross-linked and scattered AuNPs-DNA
probes (229), visual detection
can be approximated (Fig. 5B).
The sensitivity of miRNA detection is as low as 500 fM, which is on
par with or better than that of techniques based on dual-labeled
fluorescent ssRNA reporters (229). When comparing 1.0 nM target
miRNA-17 with miRNA-10b, miRNA-21 and miRNA-155 by colorimetry at
the same concentration, this approach demonstrated a strong level
of specificity (229).
Furthermore, family miRNA members with highly related sequences,
such as miRNA-17, miRNA-20a, miRNA-20b and miRNA-106a, can be
distinguished by single- or double-nucleotide variations at a
concentration of 1.0 nM using CRISPR/Cas-based colorimetric assays
(229).
A specific and sensitive biosensor, known as
casCRISPR has been developed for quick and precise miRNA detection,
even in cell extracts and serum samples, without the need for a
target amplification procedure such as PCR or recombinase
polymerase amplification (RPA) (230). By using this method, the
trans-cleavage activity of Cas13a/crRNA is triggered following
miRNA recognition. Such trans-ribonuclease activity has been
investigated using a hairpin-structured DNA oligonucleotide (stem
loop-hairpin or 'locked trigger') with an unpaired rU in the loop.
Accordingly, the phosphodiester bond adjacent to the rU of the
Cas14a/single guide RNA (sgRNA) 'locked-trigger' is broken
(Fig. 5C). Through strand
displacement, the latter causes the 'locked-trigger' to change from
a stable structure to a 5′-toehold duplex structure that can start
the interaction with Cas14a/sgRNA (230). The quencher, black hole
quencher 1 (BHQ1) and fluorophore [6-carboxyfluorescein (6-FAM)]
present on the ssDNA reporter are efficiently separated once the
'locked-trigger' is cleaved, resulting in Cas14a-based
trans-cleavage and the production of fluorescence signals (230) (Fig. 5C). In casCRISPR, the
trans-cleavage activity of Cas13a and Cas14a enables the detection
of miR-17 with a LOD of 1.33 fM, even by a difference of one
nucleotide (single base specificity), which is ~1,000 times lower
than that of direct Cas13a-based miRNA detection (199). As a promising tool for miRNA
diagnostics, casCRISPR may offer greater miRNA detection
specificity without amplification of the target as required for
RT-qPCR because of the Cas13a/crRNA high-fidelity recognition
capacity (230). Researchers
have engineered casCRISPR v2, a second version, by adding Cas13a
and Cas12a coupled with casCRISPR, and have compared this with
ordinary casCRISPR (version 1). This platform exhibited an advanced
performance, because Cas12a also functions as a ssDNA-activated
DNase (230,231). The fluorescence intensity
curves showed that casCRISPR v1 could detect miR-17 at as low as
6.25 fM, whereas casCRISPR v2 could detect miR-17 at as low as 100
fM. However, Cas13a-Cas14a (casCRISPR v1) could reach a lower
detection limit and a greater signal to background ratio than
Cas13a-Cas12a, which may be explained by the fact that Cas14a has
an improved cleavage efficiency compared with Cas12a when ssDNA is
utilized as the activator (230).
A novel approach known as dd-Cas13a increases the
concentration of local molecules for an effective reaction or
detection (234). Researchers
have adopted a droplet microfluidic technology to increase the
local concentration of the RNA reporter and target sequence for the
Cas13a system in cell-sized volumes (235,236). Thousands of picolitre-sized
droplet reactors are filled with an oil-emulsified combination of
target RNAs and Cas13a. Such droplets can be illuminated by the
cumulative fluorescence signal from the collateral cleavage of a
single RNA target-activated Cas13a (235,236) (Fig. 5E). Under these circumstances, a
single-target RNA can cause the cleavage of >100 quenched
fluorescent RNA reporters following recognition by a crRNA,
producing a fluorescence-positive droplet. Therefore, dd-Cas13a
eliminates the necessity for reverse transcription and
amplification, and enables absolute digital quantification of
single unlabeled RNA molecules (235,236). The utility of the dd-Cas13
assay was assessed by measuring miRNA-17 expression in several cell
lines, such as human glioma cells, normal human breast cells and
adenocarcinoma cells, and 100% results that corresponded with
RT-qPCR data were obtained (235).
The CRISPR/Cas13a platform and a CHDC can be
integrated on an electrochemical biosensor, known as COMET. This
approach consists of a two-stage signal amplification system for
high-sensitivity estimation of six RNAs related to non-small cell
lung cancer (NSCLC), including miR-17, miR-19b, thyroid
transcription factor-1 RNA, miR-155, EGFR mRNA and miR-210. All
components are carefully combined to enable dual signal
amplification for the CRISPR/Cas13a platform and catalyzed hairpin
DNA circuit (Cas-CHDC)-powered RNA detection (237).
To achieve quick RNA detection on a small
point-of-care (POC) testing device, scientists have incorporated
chip-based Cas-CHDC amplification into an electrochemical
biosensing technology (237)
(Fig. 5F). The COMET chip could
differentiate between miR-20a and miR-20b (two-base mismatches) and
between miR-17 and miR-106a (1-base mismatch) (237). Furthermore, a 1:1,000 M mixture
of miR-17 with miR-155 and thyroid transcription factor-1 mRNA has
been used to assess the capacity of the COMET chip to identify
lower expression target RNAs in special samples such as benign lung
disease and NSCLC samples (237).
Because non-target RNAs are highly expressed in the
circulation of patients, scientists have identified that the COMET
chip could detect miR-17 at zeptomolar (10−21 M)
concentrations and as low as 0.1% of background RNA mixture. This
is crucial for future clinical use, and the chip and reagents can
be purchased for as little as USD 0.27 per test (237).
Based on the ECL approach and employing the
trans-cleavage activity of CRISPR/Cas13a, scientists have
engineered a sophisticated platform known as PECL-CRISPR, which
mediates the exponential amplification of the ultrasensitive
detection of miRNAs that differ even by a single-base (232).
Quantitative miRNA detection is made possible by
the ECL signal on a bipolar electrode (BPE), which directly
reflects the oxidation reaction of the ruthenium (II) polypyridyl
complex [Ru(phen)2dppz]2+ and DNA at the BPE anode, and
is directly related to the miRNA concentration (232) (Fig. 5G). By identifying miR-17 from
various human cancer cells, the application of PECL-CRISPR has been
examined, proving that the platform has great potential for miRNA
diagnosis (232).
By logically designing the crRNA, the platform
exhibits the ability to differentiate between highly homologous
members of the miR-17 and let-7 families by introducing a mismatch
at the precise location of the crRNA, independent of the single
distinct base locations at the 5′ or 3′ end of the target (233). The LOD of this platform for
miR-17 was 1.0×10−15 M, which is approximately three
orders of magnitude lower than that of the direct detection of
miR-17 (LOD, 1.0×10−12 M) using LbuCas13a (233).
For on-site detection of miRNAs, a
CRISPR/Cas13-based biosensor has been created as a user-friendly
and cheaper electrochemical microfluidic platform
(EM-CRISPR/Cas13a), which substitutes a Cas13a-driven signal
amplification for synthetic nucleic acid amplification steps
(238). In this assay, a
streptavidin is applied to the chip inlet to functionalize the
immobilized surface area of the biosensor for the
CRISPR/Cas13a-powered miRNA detection. All unbound biomolecules are
eliminated by vacuuming the channel inlet (238). To activate Cas13a and perform
the collateral cleavage of the reporter RNA (reRNA), the samples
(biotin and 6-FAM-tagged reRNA), and the other sample (containing
the target miRNA and the Cas13a effector with its target-specific
crRNA) are mixed separately (238) (Fig. 5H).
An enzymatic readout of the assay is made possible
by the addition of anti-fluorescein antibodies bonded with glucose
oxidase (GOx), which bind only to the uncleaved reRNAs (Fig. 5H). Since GOx catalyzes its
substrate to produce H2O2, which is
amperometrically measured in the electrochemical cell, a glucose
solution is used in this assay for the readout (238). Without performing nucleic acid
amplification, researchers have used this new combination to detect
the miRNA levels of the putative brain tumor markers miR-19b and
miR-20a in serum samples. With a setup time of <4 h and a
readout time of 9 min, the EM-CRISPR/Cas13a biosensor achieved a
detection limit of 10 pM with a measuring volume of <0.6 liters.
Additionally, this biosensor platform was able to identify miR-19b
in serum samples of children with brain cancer, proving that this
electrochemical CRISPR-powered system is a low-cost, readily
scalable and target amplification-free molecular approach for
miRNA-based diagnostics (238).
The development of diagnostic tools and techniques
for multiplexing approaches is based on analyzing the abundance of
a number of specific components in individual samples from a
patient. However, the variety of Cas protein types and the
potential for cross-reactions between distinct Cas13a proteins may
restrict the use of Cas13a (239).
This might be resolved using a multichannel
microfluidic chip technique.
Researchers have developed various multiform
versions of electrochemical microfluidic biosensors by segmenting
the channels into subsections, using the same principle as the
EM-CRISPR/Cas13a system (Fig.
5H). This resulted in four unique chip designs for the
simultaneous quantification of a maximum of eight miRNAs using the
innovative system called CRISPR-Biosensor X. Without altering the
sensor or measurement configuration, this system can work on a
single clinical sample with a single effector protein (239).
Because of their high sensitivity, selectivity and
low instrument costs, electrochemical biosensors are emerging as a
potent detection technique platform (240). Using the CRISPR/Cas13a platform
in conjunction with the catalytic hairpin assembly (CHA) process,
an ultrasensitive electrochemical biosensing platform has been
designed for miRNA-21 detection (241). CHA is a high-efficiency,
isothermal, enzyme-free amplification technique that may be used in
a variety of analytical formats, such as electrophoretic (242), colorimetric (243), fluorescence (231), surface plasmon resonance
(244) and electrochemical
approaches (245).
This electrochemical biosensor exhibited a strong
biosensing activity from 10 fM to 1.0 nM with an LOD of 2.6 fM
using the CRISPR/Cas13a-mediated cascade signal amplification
approach (241). The use of the
biosensing platform for miRNA-21 detection was assessed. A clinical
sample (human serum; diluted ten times), was used to test the
analytical reliability before being tested for various
concentrations of miRNA-21 (10 fM to 1.0 nM). The electrochemical
recoveries were ~98.2%, demonstrating that the CRISPR/CHDC assay
can be used to identify miRNA-21 in biological materials (241).
The off-target consequences of the CRISPR/Cas13a
system are minimal. One to two mismatches in the core of a gRNA
diminish or eliminate its activity, enabling a mismatch control for
each targeting gRNA (20,246).
In this context, the specific high-sensitivity enzymatic reporter
unlocking (SHERLOCK) detection system, employs the CRISPR/Cas13a
system as a quick DNA or RNA detection technique with aM
(10−18 M) sensitivity and single-base mismatch
specificity (247). This
approach uses isothermal RPA, transcription and detection by Cas13
platforms (247). The signal
can be recorded on a colorimetric lateral-flow strip or can be
observed by fluorescence signal readout to facilitate the rapid
identification of most types of suitable Cas13 RNA target. SHERLOCK
enables single base-pair mismatch specificity, quick (setup time,
<15 min) and accurate nucleic acid detection at concentrations
as low as ~2 aM (248).
The use of innovative methods for miRNA detection
is revealing novel information regarding the processing and
function of miRNAs, as well as providing a basic understanding of
how important miRNA factors and substrates function. There are next
steps to follow for most of the experimental procedures that have
been described. For instance, structural investigations of miRNA
biogenesis factors have only evaluated a small number of miRNAs.
Since the biogenesis efficiency of endogenous miRNAs varies
greatly, it will be instructive for both microprocessor and Dicer
substrates to include a variety of miRNAs in future investigations.
This could provide information regarding microprocessor cofactors
that serve a role in the synthesis of clustered and/or inefficient
miRNAs.
Direct viewing of dynamic miRNA processing and
regulatory complexes is made possible only by single-molecule
imaging. In vivo experiments have largely focused on AGO and
not enough research has been performed on other miRNA biogenesis
factors. In vitro investigations of microprocessors have
been lacking. The subcellular environments of microprocessors and
Dicers are functionally significant and require additional
investigation in the future.
Furthermore, the implications of miRNA biology,
such as whether organismal phenotypes are actually mediated by the
regulation of specific miRNA targets, must not be overlooked. Most
of the research on miRNA-target biology is still correlative, and
just because numerous derepressed targets are found when a miRNA is
lost, this does not always suggest that they all serve a functional
role in miRNA phenotypes.
CRISPR mutagenesis has demonstrated the potent
phenotypic influence of target mRNAs and validated the growing
number of target-directed miRNA degradation target sites that
regulate miRNA abundance. To completely comprehend the role of
miRNA in development and disease, it is necessary to establish
clear links between target site-mediated gene regulation and in
vivo phenotypes.
Prior to the full clinical use of CRISPR/Cas13
technologies, a few issues need to be resolved, including: i) As
the fidelity of the CRISPR/Cas13 platform is directly linked to the
tolerance of RNA mismatches, a wider range of mismatch tolerance by
Cas13 effectors may encourage off-target activity (22); ii) it may be challenging to meet
the specific protospacer flanking sequence requirement for Cas13
effectors; iii) RNA recognition of small RNA target sequences
(<22 nt) because of the requirement that crRNAs be long enough
for binding (205); and iv) RNA
segments derived from effective ssRNA cleavage mediated by Cas13
may be toxic in eukaryotic cells (206,249).
miRNAs serve a role in the regulation of gene
expression. It is now well documented that dysregulated miRNA
expression is a fundamental cause of different diseases. including
cancer. The dysregulation of miRNA expression can occur via the
amplification or deletion of their genes, aberrant transcriptional
control, dysregulated epigenetic modifications and flaws in their
biogenesis machinery. Aberrant miRNA expression leads a cell to
achieve the ability to maintain the proliferative signals, avoid
growth suppressors, initiate invasion and metastasis, trigger
angiogenesis, and withstand cell death, and all these changes lead
to the transformation to cancerous cells. The exploration of
programmable RNA-targeting regulators makes it easier to detect
endogenous RNA and track various mRNA species in real time within
cells. It is important to develop methods that could identify and
target miRNA sequences in a broad manner, especially those with low
expression levels. CRISPR/Cas13-based miRNA detection strategies
have proven to be a versatile platform, as this approach of
miRNA-detection is beneficial in all aspects compared with other
currently used methods, which are expensive, and require special
expertise and instruments. CRISPR/Cas13-based miRNA detection
systems hold promise for the future with the potential to
revolutionize diagnostic and personalized medicine.
CRISPR/Cas13-based miRNA detection with high sensitivity and
specificity can enable early and accurate detection of diseases
such as cancer, even when miRNA levels are low. Future developments
may allow for precise differentiation between miRNA isoforms. This
can provide insights into disease mechanisms and guide targeted
therapies. CRISPR/Cas13-based assays can be designed for rapid, POC
diagnostics, eliminating the need for complex laboratory setups.
Overall, the future of CRISPR/Cas13-based miRNA detection is
bright. With continued research and development, this technology
has the potential to transform the field of diagnostics, enabling
early disease detection, personalized medicine and potentially even
miRNA-based therapies.
Not applicable.
AAA and AAK conceived the study. AAA, AMA and AAK
wrote the original draft. AAA, AMA, AA, BFA, AHR and AAK reviewed
and edited the manuscript. Data authentication is not applicable.
All authors have read and approved the final version of the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
The researchers would like to thank the Deanship of
Graduate Studies and Scientific Research at Qassim University for
financial support (QU-APC-2025).
The present study was supported by the Deanship of Graduate
Studies and Scientific Research at Qassim University
(QU-APC-2025).
|
1
|
Shenoy A and Blelloch RH: Regulation of
microRNA function in somatic stem cell proliferation and
differentiation. Nat Rev Mol Cell Biol. 15:565–576. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jonas S and Izaurralde E: Towards a
molecular understanding of microRNA-mediated gene silencing. Nat
Rev Genet. 16:421–433. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Swarts DC, Makarova K, Wang Y, Nakanishi
K, Ketting RF, Koonin EV, Patel DJ and Van Der Oost J: The
evolutionary journey of argonaute proteins. Nat Struct Mol Biol.
21:743–753. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
5
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar :
|
|
6
|
Otmani K, Rouas R and Lewalle P: OncomiRs
as noncoding RNAs having functions in cancer: Their role in immune
suppression and clinical implications. Front Immunol.
13:9139512022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ali Syeda Z, Langden SSS, Munkhzul C, Lee
M and Song SJ: Regulatory mechanism of MicroRNA expression in
cancer. Int J Mol Sci. 21:17232020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Otmani K and Lewalle P: Tumor suppressor
miRNA in cancer cells and the tumor microenvironment: Mechanism of
deregulation and clinical implications. Front Oncol. 11:7087652021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yousefnia S and Negahdary M: Role of
miRNAs in cancer: Oncogenic and tumor suppressor miRNAs, Their
regulation and therapeutic applications. Interdisciplinary Cancer
Research. Springer; Cham: pp. 1–27. 2024
|
|
10
|
Siddika T and Heinemann IU: Bringing
MicroRNAs to light: Methods for MicroRNA quantification and
visualization in live cells. Front Bioeng Biotechnol. 8:6195832021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mockly S and Seitz H: Inconsistencies and
limitations of current MicroRNA target identification methods.
Methods Mol Biol. 1970:291–314. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koshiol J, Wang E, Zhao Y, Marincola F and
Landi MT: Strengths and limitations of laboratory procedures for
microRNA detection. Cancer Epidemiol Biomarkers Prev. 19:907–911.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Allemailem KS, Alsahli MA, Almatroudi A,
Alrumaihi F, Alkhaleefah FK, Rahmani AH and Khan AA: Current
updates of CRISPR/Cas9-mediated genome editing and targeting within
tumor cells: An innovative strategy of cancer management. Cancer
Commun (Lond). 42:1257–1287. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Allemailem KS, Almatroudi A, Alrumaihi F,
Alradhi AE, Theyab A, Algahtani M, Alhawas MO, Dobie G, Moawad AA,
Rahmani AH and Khan AA: Current updates of CRISPR/Cas system and
anti-CRISPR proteins: Innovative applications to improve the genome
editing strategies. Int J Nanomedicine. 19:10185–10212. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Chen Y, Dang L, Liu Y, Huang S, Wu
S, Ma P, Jiang H, Li Y, Pan Y, et al: EasyCatch, a convenient,
sensitive and specific CRISPR detection system for cancer gene
mutations. Mol Cancer. 20:1572021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang Z, Fang J, Zhou M, Gong Z and Xiang
T: CRISPR-Cas13: A new technology for the rapid detection of
pathogenic microorganisms. Front Microbiol. 13:10113992022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Allemailem KS, Alsahli MA, Almatroudi A,
Alrumaihi F, Al Abdulmonem W, Moawad AA, Alwanian WM, Almansour NM,
Rahmani AH and Khan AA: Innovative strategies of reprogramming
immune system cells by targeting CRISPR/Cas9-based genome-editing
tools: A new era of cancer management. Int J Nanomedicine.
18:5531–5559. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koonin EV and Makarova KS: CRISPR-Cas: An
adaptive immunity system in prokaryotes. F1000 Biol Rep. 1:952009.
View Article : Google Scholar
|
|
19
|
Allemailem KS, Almatroudi A, Rahmani AH,
Alrumaihi F, Alradhi AE, Alsubaiyel AM, Algahtani M, Almousa RM,
Mahzari A, Sindi AA, et al: Recent updates of the CRISPR/Cas9
genome editing system: Novel approaches to regulate its
spatiotemporal control by genetic and physicochemical strategies.
Int J Nanomedicine. 19:5335–5363. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abudayyeh OO, Gootenberg JS,
Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DB,
Kellner MJ, Regev A, et al: RNA targeting with CRISPR-Cas13.
Nature. 550:280–284. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
O'Connell MR: Molecular mechanisms of RNA
targeting by Cas13-containing type VI CRISPR-Cas systems. J Mol
Biol. 431:66–87. 2019. View Article : Google Scholar
|
|
22
|
Ali Z, Mahas A and Mahfouz M: CRISPR/Cas13
as a tool for RNA interference. Trends Plant Sci. 23:374–378. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang LZ, Wang Y, Li SQ, Yao RW, Luan PF,
Wu H, Carmichael GG and Chen LL: Dynamic imaging of RNA in living
cells by CRISPR-Cas13 systems. Mol Cell. 76:981–997.e7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Granados-Riveron JT and Aquino-Jarquin G:
CRISPR-Cas13 precision transcriptome engineering in cancer. Cancer
Res. 78:4107–4113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Wang Q and Wang Y: Direct and
accurate miRNA detection based on CRISPR/Cas13a-triggered
exonuclease-iii-assisted colorimetric assay. J Anal Sci Technol.
15:212024. View Article : Google Scholar
|
|
26
|
Kim JJ, Hong JS, Kim H, Choi M, Winter U,
Lee H and Im H: CRISPR/Cas13a-assisted amplification-free miRNA
biosensor via dark-field imaging and magnetic gold nanoparticles.
Sens Diagn. 3:1310–1318. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang M, Cai S, Wu Y, Li Q, Wang X, Zhang Y
and Zhou N: A lateral flow assay for miRNA-21 based on
CRISPR/Cas13a and MnO2 nanosheets-mediated recognition
and signal amplification. Anal Bioanal Chem. 416:3401–3413. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leitão AL and Enguita FJ: A structural
view of miRNA biogenesis and function. Noncoding RNA.
8:102022.PubMed/NCBI
|
|
29
|
Shang R, Lee S, Senavirathne G and Lai EC:
microRNAs in action: Biogenesis, function and regulation. Nat Rev
Genet. 24:816–833. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nishikura K: A-to-I editing of coding and
non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 17:83–96. 2016.
View Article : Google Scholar :
|
|
31
|
Denli AM, Tops BBJ, Plasterk RHA, Ketting
RF and Hannon GJ: Processing of primary microRNAs by the
Microprocessor complex. Nature. 432:231–235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gregory RI, Yan KP, Amuthan G, Chendrimada
T, Doratotaj B, Cooch N and Shiekhattar R: The microprocessor
complex mediates the genesis of microRNAs. Nature. 432:235–240.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nguyen TA, Jo MH, Choi YG, Park J, Kwon
SC, Hohng S, Kim VN and Woo JS: Functional anatomy of the human
microprocessor. Cell. 161:1374–1387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kwon SC, Nguyen TA, Choi YG, Jo MH, Hohng
S, Kim VN and Woo JS: Structure of human DROSHA. Cell. 164:81–90.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim B, Jeong K and Kim VN: Genome-wide
mapping of DROSHA cleavage sites on primary microRNAs and
noncanonical substrates. Mol Cell. 66:258–269.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Neilsen CT, Goodall GJ and Bracken CP:
IsomiRs-the overlooked repertoire in the dynamic microRNAome.
Trends Genet. 28:544–549. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nicholson AW: Ribonuclease III mechanisms
of double-stranded RNA cleavage. Wiley Interdiscip Rev RNA.
5:31–48. 2014. View Article : Google Scholar
|
|
41
|
Okada C, Yamashita E, Lee SJ, Shibata S,
Katahira J, Nakagawa A, Yoneda Y and Tsukihara T: A high-resolution
structure of the pre-microRNA nuclear export machinery. Science.
326:1275–1279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim YK, Kim B and Kim VN: Re-evaluation of
the roles of DROSHA, export in 5, and DICER in microRNA biogenesis.
Proc Natl Acad Sci USA. 113:E1881–E1889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bernstein E, Caudy AA, Hammond SM and
Hannon GJ: Role for a bidentate ribonuclease in the initiation step
of RNA interference. Nature. 409:363–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Grishok A, Pasquinelli AE, Conte D, Li N,
Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G and Mello CC: Genes
and mechanisms related to RNA interference regulate expression of
the small temporal RNAs that control C. elegans developmental
timing. Cell. 106:23–34. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tsutsumi A, Kawamata T, Izumi N, Seitz H
and Tomari Y: Recognition of the pre-miRNA structure by Drosophila
Dicer-1. Nat Struct Mol Biol. 18:1153–1158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tian Y, Simanshu DK, Ma JB, Park JE, Heo
I, Kim VN and Patel DJ: A phosphate-binding pocket within the
platform-PAZ-connector helix cassette of human Dicer. Mol Cell.
53:606–616. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Matranga C, Tomari Y, Shin C, Bartel DP
and Zamore PD: Passenger-strand cleavage facilitates assembly of
siRNA into Ago2-containing RNAi enzyme complexes. Cell.
123:607–620. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zamore PD, Tuschl T, Sharp PA and Bartel
DP: RNAi: Double-stranded RNA directs the ATP-dependent cleavage of
mRNA at 21 to 23 nucleotide intervals. Cell. 101:25–33. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Suzuki HI, Katsura A, Yasuda T, Ueno T,
Mano H, Sugimoto K and Miyazono K: Small-RNA asymmetry is directly
driven by mammalian argonautes. Nat Struct Mol Biol. 22:512–521.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cai X, Hagedorn CH and Cullen BR: Human
microRNAs are processed from capped, polyadenylated transcripts
that can also function as mRNAs. RNA. 10:1957–1966. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Berezikov E: Evolution of microRNA
diversity and regulation in animals. Nat Rev Genet. 12:846–860.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lim LP, Lau NC, Weinstein EG, Abdelhakim
A, Yekta S, Rhoades MW, Burge CB and Bartel DP: The microRNAs of
Caenorhabditis elegans. Genes Dev. 17:991–1008. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hall TMT: Structure and function of
argonaute proteins. Structure. 13:1403–1408. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang J, Cho WC and Zheng Y: Argonaute
proteins: Structural features, functions and emerging roles. J Adv
Res. 24:317–324. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Schirle NT and MacRae IJ: The crystal
structure of human argonaute2. Science. 336:1037–1040. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sheu-Gruttadauria J and MacRae IJ:
Structural foundations of RNA silencing by argonaute. J Mol Biol.
429:2619–2639. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Diederichs S and Haber DA: Dual role for
argonautes in microRNA processing and posttranscriptional
regulation of microRNA expression. Cell. 131:1097–1108. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu J, Carmell MA, Rivas FV, Marsden CG,
Thomson JM, Song JJ, Hammond SM, Joshua-Tor L and Hannon GJ:
Argonaute2 is the catalytic engine of mammalian RNAi. Science.
305:1437–1441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Burroughs AM, Ando Y, de Hoon MJL, Tomaru
Y, Suzuki H, Hayashizaki Y and Daub CO: Deep-sequencing of human
argonaute-associated small RNAs provides insight into miRNA sorting
and reveals Argonaute association with RNA fragments of diverse
origin. RNA Boil. 8:158–177. 2011. View Article : Google Scholar
|
|
64
|
Dueck A, Ziegler C, Eichner A, Berezikov E
and Meister G: microRNAs associated with the different human
argonaute proteins. Nucleic Acids Res. 40:9850–9862. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar :
|
|
67
|
Gebert LF and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar :
|
|
68
|
Schirle NT, Sheu-Gruttadauria J and MacRae
IJ: Structural basis for microRNA targeting. Science. 346:608–613.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Schirle NT, Sheu-Gruttadauria J,
Chandradoss SD, Joo C and MacRae IJ: Water-mediated recognition of
t1-adenosine anchors argonaute2 to microRNA targets. Elife.
4:e076462015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina
M, Meister G, Chen HY, Dauter Z, Tuschl T and Patel DJ: Crystal
structure of A. aeolicus argonaute, a site-specific DNA-guided
endoribonuclease, provides insights into RISC-mediated mRNA
cleavage. Mol Cell. 19:405–419. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li Z, Li Z, Zhang Y, Zhou L, Xu Q, Li L,
Zeng L, Xue J, Niu H, Zhong J, et al: Mammalian PIWI-piRNA-target
complexes reveal features for broad and efficient target silencing.
Nat Struct Mol Biol. 31:1222–1231. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Braun JE, Huntzinger E and Izaurralde E:
The role of GW182 proteins in miRNA-mediated gene silencing. Adv
Exp Med Biol. 768:147–163. 2013. View Article : Google Scholar
|
|
74
|
Chekulaeva M, Mathys H, Zipprich JT, Attig
J, Colic M, Parker R and Filipowicz W: miRNA repression involves
GW182-mediated recruitment of CCR4-NOT through conserved
W-containing motifs. Nat Struct Mol Biol. 18:1218–1226. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fabian MR, Cieplak MK, Frank F, Morita M,
Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF and
Sonenberg N: miRNA-mediated deadenylation is orchestrated by GW182
through two conserved motifs that interact with CCR4-NOT. Nat
Struct Mol Biol. 18:1211–1217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Chen CYA, Zheng D, Xia Z and Shyu AB:
Ago-TNRC6 triggers microRNA-mediated decay by promoting two
deadenylation steps. Nat Struct Mol Biol. 16:1160–1166. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Braun JE, Truffault V, Boland A,
Huntzinger E, Chang CT, Haas G, Weichenrieder O, Coles M and
Izaurralde E: A direct interaction between DCP1 and XRN1 couples
mRNA decapping to 5′ exonucleolytic degradation. Nat Struct Mol
Biol. 19:1324–1331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Mathys H, Basquin J, Ozgur S,
Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny
M, Conti E and Filipowicz W: Structural and biochemical insights to
the role of the CCR4-NOT complex and DDX6 ATPase in microRNA
repression. Mol Cell. 54:751–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Fukaya T, Iwakawa HO and Tomari Y:
MicroRNAs block assembly of eIF4F translation initiation complex in
Drosophila. Mol Cell. 56:67–78. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Fukao A, Mishima Y, Takizawa N, Oka S,
Imataka H, Pelletier J, Sonenberg N, Thoma C and Fujiwara T:
MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target
mRNAs in humans. Mol Cell. 56:79–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nishihara T, Zekri L, Braun JE and
Izaurralde E: miRISC recruits decapping factors to miRNA targets to
enhance their degradation. Nucleic Acids Res. 41:8692–8705. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Jungers CF and Djuranovic S: Modulation of
miRISC-mediated gene silencing in eukaryotes. Front Mol Biosci.
9:8329162022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kuzuoğlu-Öztürk D, Bhandari D, Huntzinger
E, Fauser M, Helms S and Izaurralde E: miRISC and the CCR4-NOT
complex silence mRNA targets independently of 43S ribosomal
scanning. EMBO J. 35:1186–1203. 2016. View Article : Google Scholar
|
|
84
|
Elkayam E, Faehnle CR, Morales M, Sun J,
Li H and Joshua-Tor L: Multivalent recruitment of human argonaute
by GW182. Mol Cell. 67:646–658.e3. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Eichhorn SW, Guo H, McGeary SE,
Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J and
Bartel DP: mRNA destabilization is the dominant effect of mammalian
microRNAs by the time substantial repression ensues. Mol Cell.
56:104–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Bhattacharyya SN, Habermacher R, Martine
U, Closs EI and Filipowicz W: Relief of microRNA-mediated
translational repression in human cells subjected to stress. Cell.
125:1111–1124. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Uhlmann S, Mannsperger H, Zhang JD, Horvat
EÁ, Schmidt C, Küblbeck M, Henjes F, Ward A, Tschulena U, Zweig K,
et al: Global microRNA level regulation of EGFR-driven cell-cycle
protein network in breast cancer. Mol Syst Biol. 8:5702012.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Mestdagh P, Boström AK, Impens F, Fredlund
E, Van Peer G, De Antonellis P, Von Stedingk K, Ghesquière B,
Schulte S, Dews M, et al: The miR-17-92 microRNA cluster regulates
multiple components of the TGF-β pathway in neuroblastoma. Mol
Cell. 40:762–773. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Saetrom P, Heale BS, Snøve O Jr, Aagaard
L, Alluin J and Rossi JJ: Distance constraints between microRNA
target sites dictate efficacy and cooperativity. Nucleic Acids Res.
35:2333–2342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Broderick JA, Salomon WE, Ryder SP, Aronin
N and Zamore PD: Argonaute protein identity and pairing geometry
determine cooperativity in mammalian RNA silencing. RNA.
17:1858–1869. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Tsang J, Zhu J and van Oudenaarden A:
MicroRNA-mediated feedback and feedforward loops are recurrent
network motifs in mammals. Mol Cell. 26:753–767. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Suzuki H, Maruyama R, Yamamoto E and Kai
M: Epigenetic alteration and microRNA dysregulation in cancer.
Front Genet. 4:2582013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Calin GA and Croce CM: MicroRNAs and
chromosomal abnormalities in cancer cells. Oncogene. 25:6202–6210.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Tagawa H and Seto M: A microRNA cluster as
a target of genomic amplification in malignant lymphoma. Leukemia.
19:2013–2016. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Mavrakis KJ, Wolfe AL, Oricchio E,
Palomero T, De Keersmaecker K, McJunkin K, Zuber J, James T, Khan
AA, Leslie CS, et al: Genome-wide RNA-mediated interference screen
identifies miR-19 targets in Notch-induced T-cell acute
lymphoblastic leukaemia. Nat Cell Boil. 12:372–379. 2010.
View Article : Google Scholar
|
|
101
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
et al: microRNAs exhibit high frequency genomic alterations in
human cancer. Proc Natl Acad Sci USA. 103:9136–9141. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wijnhoven BP, Michael MZ and Watson DI:
MicroRNAs and cancer. Br J Surg. 94:23–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Qin S, Xu J, Yi Y, Jiang S, Jin P, Xia X
and Ma F: Transcription factors and methylation drive prognostic
miRNA dysregulation in hepatocellular carcinoma. Front Oncol.
11:6911152021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. PProc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar
|
|
106
|
O'Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Chang TC, Yu D, Lee YS, Wentzel EA, Arking
DE, West KM, Dang CV, Thomas-Tikhonenko A and Mendell JT:
Widespread microRNA repression by Myc contributes to tumorigenesis.
Nat Genet. 40:43–50. 2008. View Article : Google Scholar
|
|
108
|
Wang B, Hsu SH, Wang X, Kutay H, Bid HK,
Yu J, Ganju RK, Jacob ST, Yuneva M and Ghoshal K: Reciprocal
regulation of microRNA-122 and c-Myc in hepatocellular cancer: Role
of E2F1 and transcription factor dimerization partner 2.
Hepatology. 59:555–566. 2014. View Article : Google Scholar
|
|
109
|
Han H, Sun D, Li W, Shen H, Zhu Y, Li C,
Chen Y, Lu L, Li W, Zhang J, et al: A c-Myc-MicroRNA functional
feedback loop affects hepatocarcinogenesis. Hepatology.
57:2378–2389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
He L, He X, Lim LP, De Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Hermeking HJ: The miR-34 family in cancer
and apoptosis. Cell Death Differ. 17:193–199. 2010. View Article : Google Scholar
|
|
112
|
Xiao J, Lin H, Luo X, Luo X and Wang Z:
miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive
feedback loop in response to stress. EMBO J. 30:524–532. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhang Y, Liao JM, Zeng SX and Lu H: p53
downregulates Down syndrome-associated DYRK1A through miR-1246.
EMBO Rep. 12:811–817. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yamakuchi M, Lotterman CD, Bao C, Hruban
RH, Karim B, Mendell JT, Huso D and Lowenstein CJ: P53-induced
microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad
Sci USA. 107:6334–6339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Eyholzer M, Schmid S, Schardt JA,
Haefliger S, Mueller BU and Pabst T: Complexity of miR-223
regulation by CEBPA in human AML. Leuk Res. 34:672–676. 2010.
View Article : Google Scholar
|
|
116
|
Hessam S, Sand M, Skrygan M, Gambichler T
and Bechara FG: Inflammation induced changes in the expression
levels of components of the microRNA maturation machinery Drosha,
Dicer, Drosha co-factor DGRC8 and Exportin-5 in inflammatory
lesions of hidradenitis suppurativa patients. J Dermatol Sci.
82:166–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Kim J, Park WJ, Jeong KJ, Kang SH, Kwon
SY, Kim S and Park JW: Racial differences in expression levels of
miRNA machinery-related genes, dicer, drosha, DGCR8, and AGO2, in
Asian Korean papillary thyroid carcinoma and comparative validation
using the cancer genome atlas. Int J Genomics. 2017:57897692017.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yan M, Huang HY, Wang T, Wan Y, Cui SD,
Liu ZZ and Fan QX: Dysregulated expression of dicer and drosha in
breast cancer. Pathol Oncol Res. 18:343–348. 2012. View Article : Google Scholar
|
|
119
|
Zhang Z, Zhang G, Kong C, Bi J, Gong D, Yu
X, Shi D, Zhan B and Ye P: EIF2C, Dicer, and Drosha are
up-regulated along tumor progression and associated with poor
prognosis in bladder carcinoma. Tumour Biol. 36:5071–5079. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Thomson JM, Newman M, Parker JS,
Morin-Kensicki EM, Wright T and Hammond SM: Extensive
post-transcriptional regulation of microRNAs and its implications
for cancer. Genes Dev. 20:2202–2207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Walz AL, Ooms A, Gadd S, Gerhard DS, Smith
MA, Auvil JMG, Meerzaman D, Chen QR, Hsu CH, Yan C, et al:
Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable
histology Wilms tumors. Cancer Cell. 27:286–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Iliou MS, da Silva-Diz V, Carmona FJ,
Ramalho-Carvalho J, Heyn H, Villanueva A, Muñoz P and Esteller M:
Impaired DICER1 function promotes stemness and metastasis in colon
cancer. Oncogene. 33:4003–4015. 2014. View Article : Google Scholar :
|
|
123
|
Merritt WM, Lin YG, Han LY, Kamat AA,
Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick
AM, et al: Dicer, Drosha, and outcomes in patients with ovarian
cancer. N Engl J Med. 359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Pampalakis G, Diamandis EP, Katsaros D and
Sotiropoulou G: Down-regulation of dicer expression in ovarian
cancer tissues. Clin Biochem. 43:324–327. 2010. View Article : Google Scholar
|
|
125
|
Faggad A, Budczies J, Tchernitsa O,
Darb-Esfahani S, Sehouli J, Müller BM, Wirtz R, Chekerov R,
Weichert W, Sinn B, et al: Prognostic significance of Dicer
expression in ovarian cancer-link to global microRNA changes and
oestrogen receptor expression. J Pathol. 220:382–391. 2010.
View Article : Google Scholar
|
|
126
|
Karube Y, Tanaka H, Osada H, Tomida S,
Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S,
Mitsudomi T and Takahashi T: Reduced expression of Dicer associated
with poor prognosis in lung cancer patients. Cancer Sci.
96:111–115. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Dome JS and Coppes MJ: Recent advances in
Wilms tumor genetics. Curr Opin Pediatr. 14:5–11. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhang J, Fan XS, Wang CX, Liu B, Li Q and
Zhou XJ: Up-regulation of Ago2 expression in gastric carcinoma. Med
Oncol. 30:6282013. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Völler D, Reinders J, Meister G and
Bosserhoff AK: Strong reduction of AGO2 expression in melanoma and
cellular consequences. Br J Cancer. 109:3116–3124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Melo SA, Moutinho C, Ropero S, Calin GA,
Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya
G, et al: A genetic defect in exportin-5 traps precursor microRNAs
in the nucleus of cancer cells. Cancer Cell. 18:303–315. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Chu R, Mo G, Duan Z, Huang M, Chang J, Li
X and Liu P: miRNAs affect the development of hepatocellular
carcinoma via dysregulation of their biogenesis and expression.
CCell Commun Signal. 12:452014. View Article : Google Scholar
|
|
132
|
Han L, Witmer PD, Casey E, Valle D and
Sukumar S: DNA methylation regulates MicroRNA expression. Cancer
Biol Ther. 6:1290–1294. 2007. View Article : Google Scholar
|
|
133
|
Saito Y and Jones PM: Epigenetic
activation of tumor suppressor microRNAs in human cancer cells.
Cell Cycle. 5:2220–2222. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Fazi F, Racanicchi S, Zardo G, Starnes LM,
Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco
F, et al: Epigenetic silencing of the myelopoiesis regulator
microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 12:457–466.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Saito Y, Liang G, Egger G, Friedman JM,
Chuang JC, Coetzee GA and Jones PA: Specific activation of
microRNA-127 with downregulation of the proto-oncogene BCL6 by
chromatin-modifying drugs in human cancer cells. Cancer Cell.
9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Lujambio A, Calin GA, Villanueva A, Ropero
S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso
MS, Faller WJ, et al: A microRNA DNA methylation signature for
human cancer metastasis. Proc Natl Acad Sci USA. 105:13556–13561.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Baharudin R, Rus Bakarurraini NQ, Ismail
I, Lee LH and Ab Mutalib NS: MicroRNA methylome signature and their
functional roles in colorectal cancer diagnosis, prognosis, and
chemoresistance. Int J Mol Sci. 23:72812022. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Lehmann U, Hasemeier B, Christgen M,
Müller M, Römermann D, Länger F and Kreipe H: Epigenetic
inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J
Pathol. 214:17–24. 2008. View Article : Google Scholar
|
|
139
|
Donzelli S, Mori F, Bellissimo T, Sacconi
A, Casini B, Frixa T, Roscilli G, Aurisicchio L, Facciolo F,
Pompili A, et al: Epigenetic silencing of miR-145-5p contributes to
brain metastasis. Oncotarget. 6:35183–35201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Trimarchi JM and Lees JA: Sibling rivalry
in the E2F family. Nat Rev Mol Cell Biol. 3:11–20. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Coller HA, Forman JJ and Legesse-Miller A:
'Myc'ed messages': myc induces transcription of E2F1 while
inhibiting its translation via a microRNA polycistron. PLoS Genet.
3:e1462007. View Article : Google Scholar
|
|
143
|
Sylvestre Y, De Guire V, Querido E,
Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G and Chartrand P:
An E2F/miR-20a autoregulatory feedback loop. J Biol Chem.
282:2135–2143. 2007. View Article : Google Scholar
|
|
144
|
Woods K, Thomson JM and Hammond SM: Direct
regulation of an oncogenic micro-RNA cluster by E2F transcription
factors. J Biol Chem. 282:2130–2134. 2007. View Article : Google Scholar
|
|
145
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Hatfield SD, Shcherbata HR, Fischer KA,
Nakahara K, Carthew RW and Ruohola-Baker H: Stem cell division is
regulated by the microRNA pathway. Nature. 435:974–978. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Yu JY, Reynolds SH, Hatfield SD,
Shcherbata HR, Fischer KA, Ward EJ, Long D, Ding Y and
Ruohola-Baker H: Dicer-1-dependent Dacapo suppression acts
downstream of Insulin receptor in regulating cell division of
Drosophila germline stem cells. Development. 136:1497–1507. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Gillies JK and Lorimer IAJ: Regulation of
p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 6:2005–2009.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem.
282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
le Sage C, Nagel R, Egan DA, Schrier M,
Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et
al: Regulation of the p27(Kip1) tumor suppressor by miR-221 and
miR-222 promotes cancer cell proliferation. EMBO J. 26:3699–3708.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Zhang C, Kang C, You Y, Pu P, Yang W, Zhao
P, Wang G, Zhang A, Jia Z, Han L and Jiang H: Co-suppression of
miR-221/222 cluster suppresses human glioma cell growth by
targeting p27kip1 in vitro and in vivo. Int J Oncol. 34:1653–1660.
2009.PubMed/NCBI
|
|
152
|
Carmeliet P: Mechanisms of angiogenesis
and arteriogenesis. Nat Med. 6:389–395. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Choudhry H and Harris AL: Advances in
hypoxia-inducible factor biology. Cell Metab. 27:281–298. 2018.
View Article : Google Scholar
|
|
154
|
Ferrara N: VEGF and the quest for tumour
angiogenesis factors. Nat Rev Cancer. 2:795–803. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Tiwari A, Mukherjee B and Dixit M:
MicroRNA key to angiogenesis regulation: MiRNA biology and therapy.
Curr Cancer Drug Targets. 18:266–277. 2018. View Article : Google Scholar
|
|
156
|
Landskroner-Eiger S, Moneke I and Sessa
WC: miRNAs as modulators of angiogenesis. Cold Spring Harb Perspect
Med. 3:a0066432013. View Article : Google Scholar
|
|
157
|
Camps C, Buffa FM, Colella S, Moore J,
Sotiriou C, Sheldon H, Harris AL, Gleadle JM and Ragoussis J:
hsa-miR-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Fasanaro P, D'Alessandra Y, Di Stefano V,
Melchionna R, Romani S, Pompilio G, Capogrossi MC and Martelli F:
MicroRNA-210 modulates endothelial cell response to hypoxia and
inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol
Chem. 283:15878–15883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Lou YL, Guo F, Liu F, Gao FL, Zhang PQ,
Niu X, Guo SC, Yin JH, Wang Y and Deng ZF: miR-210 activates notch
signaling pathway in angiogenesis induced by cerebral ischemia. Mol
Cell Biochem. 370:45–51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Ghosh G, Subramanian IV, Adhikari N, Zhang
X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y and
Ramakrishnan S: Hypoxia-induced microRNA-424 expression in human
endothelial cells regulates HIF-α isoforms and promotes
angiogenesis. J Clin Invest. 120:4141–4154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Liu LZ, Li C, Chen Q, Jing Y, Carpenter R,
Jiang Y, Kung HF, Lai L and Jiang BH: MiR-21 induced angiogenesis
through AKT and ERK activation and HIF-1α expression. PLoS One.
6:e191392011. View Article : Google Scholar
|
|
162
|
Lei Z, Li BO, Yang Z, Fang H, Zhang GM,
Feng ZH and Huang B: Regulation of HIF-1alpha and VEGF by miR-20b
tunes tumor cells to adapt to the alteration of oxygen
concentration. PLoS One. 4:e76292009. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Cha ST, Chen PS, Johansson G, Chu CY, Wang
MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH, et al: MicroRNA-519c
suppresses hypoxia-inducible factor-1alpha expression and tumor
angiogenesis. Cancer Res. 70:2675–2685. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Loh CY, Chai JY, Tang TF, Wong WF, Sethi
G, Shanmugam MK, Chong PP and Looi CY: The E-cadherin and
N-cadherin switch in epithelial-to-mesenchymal transition:
Signaling, therapeutic implications, and challenges. Cells.
8:11182019. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Rutnam ZJ, Wight TN and Yang BB: miRNAs
regulate expression and function of extracellular matrix molecules.
Matrix Biol. 32:74–85. 2013. View Article : Google Scholar
|
|
166
|
Raines EW: The extracellular matrix can
regulate vascular cell migration, proliferation, and survival:
Relationships to vascular disease. Int J Exp Pathol. 81:173–182.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Zitka O, Kukacka J, Krizkova S, Huska D,
Adam V, Masarik M, Prusa R and Kizek R: Matrix metalloproteinases.
Curr Med Chem. 17:3751–3768. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Chambers AF and Matrisian LM: Changing
views of the role of matrix metalloproteinases in metastasis. J
Natl Cancer Inst. 89:1260–1270. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Wu D, Huang P, Wang L, Zhou Y, Pan H and
Qu P: MicroRNA-143 inhibits cell migration and invasion by
targeting matrix metalloproteinase 13 in prostate cancer. Mol Med
Rep. 8:626–630. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Liu H, Cao YD, Ye WX and Sun YY: Effect of
microRNA-206 on cytoskeleton remodelling by downregulating Cdc42 in
MDA-MB-231 cells. Tumori. 96:751–755. 2010. View Article : Google Scholar
|
|
172
|
Wu ZS, Wu Q, Wang CQ, Wang XN, Huang J,
Zhao JJ, Mao SS, Zhang GH, Xu XC and Zhang N: miR-340 inhibition of
breast cancer cell migration and invasion through targeting of
oncoprotein c-Met. Cancer. 117:2842–2852. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Kong W, Yang H, He L, Zhao JJ, Coppola D,
Dalton WS and Cheng JQ: MicroRNA-155 is regulated by the
transforming growth factor beta/Smad pathway and contributes to
epithelial cell plasticity by targeting RhoA. Mol Cell Biol.
28:6773–6784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Bracken CP, Gregory PA, Kolesnikoff N,
Bert AG, Wang J, Shannon MF and Goodall GJ: A double-negative
feedback loop between ZEB1-SIP1 and the microRNA-200 family
regulates epithelial-mesenchymal transition. Cancer Res.
68:7846–7854. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Lima RT, Busacca S, Almeida GM, Gaudino G,
Fennell DA and Vasconcelos MH: MicroRNA regulation of core
apoptosis pathways in cancer. Eur J Cancer. 47:163–174. 2011.
View Article : Google Scholar
|
|
178
|
Li C, Hashimi SM, Good DA, Cao S, Duan W,
Plummer PN, Mellick AS and Wei MQ: Apoptosis and microRNA
aberrations in cancer. Clin Exp Pharmacol Physiol. 39:739–146.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Cadwell C and Zambetti GP: The effects of
wild-type p53 tumor suppressor activity and mutant p53
gain-of-function on cell growth. Gene. 277:15–30. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
O'Brien MA and Kirby R: Apoptosis: A
review of pro-apoptotic and anti-apoptotic pathways and
dysregulation in disease. J Vet Emerg Crit Care (San Antonio).
18:572–585. 2008. View Article : Google Scholar
|
|
181
|
Hermeking H: MicroRNAs in the p53 network:
Micromanagement of tumour suppression. Nat Rev Cancer. 12:613–626.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Pichiorri F, Suh SS, Rocci A, De Luca L,
Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder
H, et al: RETRACTED: Downregulation of p53-inducible microRNAs 192,
194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple
myeloma development. Cancer Cell. 18:367–381. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Bi C and Chng WJ: MicroRNA: Important
player in the pathobiology of multiple myeloma. Biomed Res Int.
2014:5215862014. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Fornari F, Gramantieri L, Giovannini C,
Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM,
Tavolari S, et al: MiR-122/cyclin G1 interaction modulates p53
activity and affects doxorubicin sensitivity of human
hepatocarcinoma cells. Cancer Res. 69:5761–5767. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Burns DM, D'Ambrogio A, Nottrott S and
Richter JD: CPEB and two poly(A) polymerases control miR-122
stability and p53 mRNA translation. Nature. 473:105–108. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Hajizadeh M, Hajizadeh F, Ghaffarei S,
Doustvandi MA, Hajizadeh K, Yaghoubi SM, Mohammadnejad F, Khiabani
NA, Mousavi P and Baradaran B: MicroRNAs and their vital role in
apoptosis in hepatocellular carcinoma: miRNA-based diagnostic and
treatment methods. Gene. 888:1478032023. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Yan HL, Xue G, Mei Q, Wang YZ, Ding FX,
Liu MF, Lu MH, Tang Y, Yu HY and Sun SH: Repression of the
miR-17-92 cluster by p53 has an important function in
hypoxia-induced apoptosis. EMBO J. 28:2719–2732. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Jiang X, Liu Y, Zhang G, Lin S, Wu J, Yan
X, Ma Y and Ma M: Aloe-emodin induces breast tumor cell apoptosis
through upregulation of miR-15a/miR-16-1 that suppresses BCL2. Evid
Based Complement Alternat Med. 2020:51082982020. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Sacconi A, Biagioni F, Canu V, Mori F, Di
Benedetto A, Lorenzon L, Ercolani C, Di Agostino S, Cambria AM,
Germoni S, et al: miR-204 targets Bcl-2 expression and enhances
responsiveness of gastric cancer. Cell Death Dis. 3:e4232012.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Li M, Yang Y, Kuang Y, Gan X, Zeng W, Liu
Y and Guan H: miR-365 induces hepatocellular carcinoma cell
apoptosis through targeting Bcl-2. Exp Ther Med. 13:2279–2285.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Denoyelle C, Lambert B, Meryet-Figuière M,
Vigneron N, Brotin E, Lecerf C, Abeilard E, Giffard F, Louis MH,
Gauduchon P, et al: miR-491-5p-induced apoptosis in ovarian
carcinoma depends on the direct inhibition of both BCL-XL and EGFR
leading to BIM activation. Cell Death Dis. 5:e14452014. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Burmistrz M, Krakowski K and
Krawczyk-Balska A: RNA-targeting CRISPR-Cas systems and their
applications. Int J Mol Sci. 21:11222020. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Donohoue PD, Barrangou R and May AP:
Advances in industrial biotechnology using CRISPR-Cas systems.
Trends Biotechnol. 36:134–146. 2018. View Article : Google Scholar
|
|
194
|
Konermann S, Lotfy P, Brideau NJ, Oki J,
Shokhirev MN and Hsu PD: Transcriptome engineering with
RNA-targeting type VI-D CRISPR effectors. Cell. 173:665–676 e14.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Yan WX, Chong S, Zhang H, Makarova KS,
Koonin EV, Cheng DR and Scott DA: Cas13d is a compact RNA-targeting
type VI CRISPR effector positively modulated by a
WYL-domain-containing accessory protein. Mol Cell. 70:327–339.e5.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Makarova KS, Wolf YI, Iranzo J, Shmakov
SA, Alkhnbashi OS, Brouns SJ, Charpentier E, Cheng D, Haft DH,
Horvath P, et al: Evolutionary classification of CRISPR-Cas
systems: A burst of class 2 and derived variants. Nat Rev
Microbiol. 18:67–83. 2020. View Article : Google Scholar
|
|
197
|
Xu C, Zhou Y, Xiao Q, He B, Geng G, Wang
Z, Cao B, Dong X, Bai W, Wang Y, et al: Programmable RNA editing
with compact CRISPR-Cas13 systems from uncultivated microbes. Nat
Methods. 18:499–506. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Kannan S, Altae-Tran H, Jin X, Madigan VJ,
Oshiro R, Makarova KS, Koonin EV and Zhang F: Compact RNA editors
with small Cas13 proteins. Nat Biotechnol. 40:194–197. 2022.
View Article : Google Scholar :
|
|
199
|
Granados-Riveron JT and Aquino-Jarquin G:
CRISPR/Cas13-based approaches for ultrasensitive and specific
detection of microRNAs. Cells. 10:16552021. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Wang H, La Russa M and Qi LS: CRISPR/Cas9
in genome editing and beyond. Annu Rev Biochem. 85:227–264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Liu L, Li X, Ma J, Li Z, You L, Wang J,
Wang M, Zhang X and Wang Y: The molecular architecture for
RNA-guided RNA cleavage by Cas13a. Cell. 170:714–726.e10. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Watanabe S, Cui B, Kiga K, Aiba Y, Tan XE,
Sato'o Y, Kawauchi M, Boonsiri T, Thitiananpakorn K, Taki Y, et al:
Composition and diversity of CRISPR-Cas13a systems in the genus
Leptotrichia. Front Microbiol. 10:28382019. View Article : Google Scholar
|
|
203
|
Jain I: CRISPR Cas adaptive immunity in
Leptotrichia shahii type VI-A system. Rutgers University Community
Repository. 1132022.
|
|
204
|
Liu L, Li X, Wang J, Wang M, Chen P, Yin
M, Li J, Sheng G and Wang Y: Two distant catalytic sites are
responsible for C2c2 RNase activities. Cell. 168:121–134.e12. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Abudayyeh OO, Gootenberg JS, Konermann S,
Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E,
Minakhin L, et al: C2c2 is a single-component programmable
RNA-guided RNA-targeting CRISPR effector. Science. 353:aaf55732016.
View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Mahas A, Aman R and Mahfouz M:
CRISPR-Cas13d mediates robust RNA virus interference in plants.
Genome Boil. 20:2632019. View Article : Google Scholar
|
|
207
|
Yang L and Chen LL: Enhancing the RNA
engineering toolkit. Science. 358:996–997. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Wessels HH, Méndez-Mancilla A, Guo X,
Legut M, Daniloski Z and Sanjana NE: Massively parallel Cas13
screens reveal principles for guide RNA design. Nat Biotechnol.
38:722–727. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
East-Seletsky A, O'Connell MR, Burstein D,
Knott GJ and Doudna JA: RNA targeting by functionally orthogonal
type VI-A CRISPR-Cas enzymes. Mol Cell. 66:373–383.e3. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Smargon AA, Cox DBT, Pyzocha NK, Zheng K,
Slaymaker IM, Gootenberg JS, Abudayyeh OA, Essletzbichler P,
Shmakov S, Makarova KS, et al: Cas13b is a type VI-B
CRISPR-associated RNA-guided RNase differentially regulated by
accessory proteins Csx27 and Csx28. Mol Cell. 65:618–630. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Cox DBT, Gootenberg JS, Abudayyeh OO,
Franklin B, Kellner MJ, Joung J and Zhang F: RNA editing with
CRISPR-Cas13. Science. 358:1019–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Dong Y, Zhang B, Wei Y, Murashev A, Wang
S, Wu Y, Ma W and Liu T: Development of Cas13a-based therapy for
cancer treatment. Mol Biol Rep. 51:942024. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Bot JF, van der Oost J and Geijsen N: The
double life of CRISPR-Cas13. Curr Opin Biotechnol. 78:1027892022.
View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Fonfara I, Richter H, Bratovič M, Le Rhun
A and Charpentier E: The CRISPR-associated DNA-cleaving enzyme Cpf1
also processes precursor CRISPR RNA. Nature. 532:517–521. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Palaz F, Kalkan AK, Can Ö, Demir AN,
Tozluyurt A, Özcan A and Ozsoz M: CRISPR-Cas13 system as a
promising and versatile tool for cancer diagnosis, therapy, and
research. ACS Synth Biol. 10:1245–1267. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Chen H, Guo R, Li G, Zhang W and Zhang Z:
Comparative analysis of similarity measurements in miRNAs with
applications to miRNA-disease association predictions. BMC
Bioinformatics. 21:1762020. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Liu S, Jiang X, Wan F, Jia S and Si S: A
novel detection of MicroRNA based on homogeneous electrochemical
sensor with enzyme-assisted signal amplification. Talanta.
256:1242632023. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Zhang H, Fan M, Jiang J, Shen Q, Cai C and
Shen J: Sensitive electrochemical biosensor for MicroRNAs based on
duplex-specific nuclease-assisted target recycling followed with
gold nanoparticles and enzymatic signal amplification. Anal Chim
Acta. 1064:33–39. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Lizardi PM, Huang X, Zhu Z, Bray-Ward P,
Thomas DC and Ward DC: Mutation detection and single-molecule
counting using isothermal rolling-circle amplification. Nat Genet.
19:225–232. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
220
|
de la Torre TZG, Mezger A, Herthnek D,
Johansson C, Svedlindh P, Nilsson M and Strømme M: Detection of
rolling circle amplified DNA molecules using probe-tagged magnetic
nanobeads in a portable AC susceptometer. Biosens Bioelectron.
29:195–199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Liu K, Tong H, Li T, Wang X and Chen Y:
Research progress in molecular biology related quantitative methods
of MicroRNA. Am J Transl Res. 12:3198–3211. 2020.PubMed/NCBI
|
|
222
|
Zhou T, Huang M, Lin J, Huang R and Xing
D: High-fidelity CRISPR/Cas13a trans-cleavage-triggered rolling
circle amplified DNAzyme for visual profiling of microRNA. Anal
Chem. 93:2038–2044. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Albada HB, Golub E and Willner I: Rational
design of supramolecular hemin/G-quadruplex-dopamine aptamer
nucleoapzyme systems with superior catalytic performance. Chem Sci.
7:3092–3101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Singh R, Pochampally R, Watabe K, Lu Z and
Mo YY: Exosome-mediated transfer of miR-10b promotes cell invasion
in breast cancer. Mol Cancer. 13:2562014. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Yoo B, Kavishwar A, Ross A, Wang P,
Tabassum DP, Polyak K, Barteneva N, Petkova V, Pantazopoulos P,
Tena A, et al: Combining miR-10b-targeted nanotherapy with low-dose
doxorubicin elicits durable regressions of metastatic breast
cancer. Cancer Res. 75:4407–4415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Ye J, Xu M, Tian X, Cai S and Zeng S:
Research advances in the detection of miRNA. J Pharm Anal.
9:217–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Harrington LB, Burstein D, Chen JS,
Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF
and Doudna JA: Programmed DNA destruction by miniature CRISPR-Cas14
enzymes. Science. 362:839–842. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Li SY, Cheng QX, Liu JK, Nie XQ, Zhao GP
and Wang J: CRISPR-Cas12a has both cis- and trans-cleavage
activities on single-stranded DNA. Cell Res. 28:491–493. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Yuan C, Tian T, Sun J, Hu M, Wang X, Xiong
E, Cheng M, Bao Y, Lin W, Jiang J, et al: Universal and naked-eye
gene detection platform based on the clustered regularly
interspaced short palindromic repeats/Cas12a/13a system. Anal Chem.
92:4029–4037. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Sha Y, Huang R, Huang M, Yue H, Shan Y, Hu
J and Xing D: Cascade CRISPR/cas enables amplification-free
microRNA sensing with fM-sensitivity and single-base-specificity.
Chem Commun (Camb). 57:247–250. 2021. View Article : Google Scholar
|
|
231
|
Chen JS, Ma E, Harrington LB, Da Costa M,
Tian X, Palefsky JM and Doudna JA: CRISPR-Cas12a target binding
unleashes indiscriminate single-stranded DNase activity. Science.
360:436–439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Zhou T, Huang R, Huang M, Shen J, Shan Y
and Xing D: CRISPR/Cas13a powered portable electrochemiluminescence
chip for ultrasensitive and specific MiRNA detection. Adv Sci
(Weinh). 7:19036612020. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Shan Y, Zhou X, Huang R and Xing D:
High-fidelity and rapid quantification of miRNA combining crRNA
programmability and CRISPR/Cas13a trans-cleavage activity. Anal
Chem. 91:5278–5285. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Russo G, Zegar C and Giordano A:
Advantages and limitations of microarray technology in human
cancer. Oncogene. 22:6497–6507. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Tian T, Shu B, Jiang Y, Ye M, Liu L, Guo
Z, Han Z, Wang Z and Zhou X: An ultralocalized Cas13a assay enables
universal and nucleic acid amplification-free single-molecule RNA
diagnostics. ACS Nano. 15:1167–1178. 2020. View Article : Google Scholar
|
|
236
|
Aquino-Jarquin G: Recent progress on rapid
SARS-CoV-2/COVID-19 detection by CRISPR-Cas13-based platforms. Drug
Discov Today. 26:2025–2035. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Sheng Y, Zhang T, Zhang S, Johnston M,
Zheng X, Shan Y, Liu T, Huang Z, Qian F, Xie Z, et al: A
CRISPR/Cas13a-powered catalytic electrochemical biosensor for
successive and highly sensitive RNA diagnostics. Biosens
Bioelectron. 178:1130272021. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Bruch R, Baaske J, Chatelle C, Meirich M,
Madlener S, Weber W, Dincer C and Urban GA: CRISPR/Cas13a-powered
electrochemical microfluidic biosensor for nucleic acid
amplification-free miRNA diagnostics. Adv Mater. 31:19053112019.
View Article : Google Scholar
|
|
239
|
Bruch R, Johnston M, Kling A, Mattmüller
T, Baaske J, Partel S, Madlener S, Weber W, Urban GA and Dincer C:
CRISPR-powered electrochemical microfluidic multiplexed biosensor
for target amplification-free miRNA diagnostics. Biosens
Bioelectron. 177:1128872021. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Kimmel DW, LeBlanc G, Meschievitz ME and
Cliffel DE: Electrochemical sensors and biosensors. Anal Chem.
84:685–707. 2012. View Article : Google Scholar :
|
|
241
|
Cui Y, Fan S, Yuan Z, Song M, Hu J, Qian
D, Zhen D, Li J and Zhu B: Ultrasensitive electrochemical assay for
microRNA-21 based on CRISPR/Cas13a-assisted catalytic hairpin
assembly. Talanta. 224:1218782021. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Zou L, Wu Q, Zhou Y, Gong X, Liu X and
Wang F: A DNAzyme-powered cross-catalytic circuit for amplified
intracellular imaging. Chem Commun (Camb). 55:6519–6522. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Park C, Park H, Lee HJ, Lee HS, Park KH,
Choi CH and Na S: Double amplified colorimetric detection of DNA
using gold nanoparticles, enzymes and a catalytic hairpin assembly.
Mikrochim Acta. 186:342018. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Li J, Lei P, Ding S, Zhang Y, Yang J,
Cheng Q and Yan Y: An enzyme-free surface plasmon resonance
biosensor for real-time detecting microRNA based on allosteric
effect of mismatched catalytic hairpin assembly. Biosens
Bioelectron. 77:435–441. 2016. View Article : Google Scholar
|
|
245
|
Zhao RN, Feng Z, Zhao YN, Jia LP, Ma RN,
Zhang W, Shang L, Xue QW and Wang HS: A sensitive electrochemical
aptasensor for Mucin 1 detection based on catalytic hairpin
assembly coupled with PtPdNPs peroxidase-like activity. Talanta.
200:503–510. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Xu D, Cai Y, Tang L, Han X, Gao F, Cao H,
Qi F and Kapranov P: A CRISPR/Cas13-based approach demonstrates
biological relevance of vlinc class of long non-coding RNAs in
anticancer drug response. Sci Rep. 10:17942020. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Gootenberg JS, Abudayyeh OO, Lee JW,
Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer
NM, Freije CA, et al: Nucleic acid detection with
CRISPR-Cas13a/C2c2. Science. 356:438–442. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Kellner MJ, Koob JG, Gootenberg JS,
Abudayyeh OO and Zhang F: SHERLOCK: Nucleic acid detection with
CRISPR nucleases. Nat Protoc. 14:2986–3012. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Makarova KS, Zhang F and Koonin EV:
SnapShot: Class 2 CRISPR-Cas systems. Cell. 168:328–328.e1. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Sheedy P and Medarova Z: The fundamental
role of miR-10b in metastatic cancer. Am J Cancer Res. 8:1674–1688.
2018.PubMed/NCBI
|
|
251
|
Kolenda T, Guglas K, Kopczyńska M,
Sobocińska J, Teresiak A, Bliźniak R and Lamperska K: Good or not
good: Role of miR-18a in cancer biology. Rep Pract Oncol Radiother.
25:808–819. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Feng YH and Tsao CJ: Emerging role of
microRNA-21 in cancer. Biomed Rep. 5:395–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Zhang L, Liao Y and Tang L: MicroRNA-34
family: A potential tumor suppressor and therapeutic candidate in
cancer. J Exp Clin Cancer Res. 38:532019. View Article : Google Scholar : PubMed/NCBI
|
|
254
|
Mahesh G and Biswas R: MicroRNA-155: A
master regulator of inflammation. J Interferon Cytokine Res.
39:321–330. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Lam J, van den Bosch M, Wegrzyn J, Parker
J, Ibrahim R, Slowski K, Chang L, Martinez-Høyer S, Condorelli G,
Boldin M, et al: miR-143/145 differentially regulate hematopoietic
stem and progenitor activity through suppression of canonical TGFβ
signaling. Nat Commun. 9:24182018. View Article : Google Scholar
|
|
256
|
Cavallari I, Ciccarese F, Sharova E, Urso
L, Raimondi V, Silic-Benussi M, D'agostino DM and Ciminale V: The
miR-200 family of microRNAs: Fine tuners of epithelial-mesenchymal
transition and circulating cancer biomarkers. Cancers (Basel).
13:58742021. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Zaccagnini G, Maimone B, Fuschi P, Maselli
D, Spinetti G, Gaetano C and Martelli F: Overexpression of miR-210
and its significance in ischemic tissue damage. Sci Rep.
7:95632017. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Torres-Berrío A, Nouel D, Cuesta S, Parise
EM, Restrepo-Lozano JM, Larochelle P, Nestler EJ and Flores C:
MiR-218: A molecular switch and potential biomarker of
susceptibility to stress. Mol Psychiatry. 25:951–964. 2020.
View Article : Google Scholar
|
|
259
|
Abak A, Amini S, Sakhinia E and Abhari A:
MicroRNA-221: Biogenesis, function and signatures in human cancers.
Eur Rev Med Pharmacol Sci. 22:3094–3117. 2018.PubMed/NCBI
|
|
260
|
Ahmad W, Gull B, Baby J and Mustafa F: A
comprehensive analysis of Northern versus liquid hybridization
assays for mRNAs, small RNAs, and miRNAs using a non-radiolabeled
approach. Curr Issues Mol Biol. 43:457–484. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
261
|
Gaarz A, Debey-Pascher S, Classen S, Eggle
D, Gathof B, Chen J, Fan JB, Voss T, Schultze JL and
Staratschek-Jox A: Bead array-based microrna expression profiling
of peripheral blood and the impact of different RNA isolation
approaches. J Mol Diagn. 12:335–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Mehta N: RT-qPCR made simple: A
comprehensive guide on the methods, advantages, disadvantages, and
everything in between. Undergrad Res Nat Clin Sci Technol J. 6:1–6.
2022.
|
|
263
|
Tan M, Liao C, Liang L, Yi X, Zhou Z and
Wei G: Recent advances in recombinase polymerase amplification:
Principle, advantages, disadvantages and applications. Front Cell
Infect Microbiol. 12:10190712022. View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Pervez MT, Hasnain MJU, Abbas SH, Moustafa
MF, Aslam N and Shah SSM: A comprehensive review of performance of
next-generation sequencing platforms. Biomed Res Int.
2022:34578062022. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Li X, Liao L, Jiang B, Yuan R and Xiang Y:
Invader assay-induced catalytic assembly of multi-DNAzyme junctions
for sensitive detection of single nucleotide polymorphisms. Anal
Chim Acta. 1224:3402252022. View Article : Google Scholar : PubMed/NCBI
|
|
266
|
Jin Y, Chen Z, Liu X and Zhou X:
Evaluating the microRNA targeting sites by luciferase reporter gene
assay. Methods Mol Biol. 936:117–127. 2013. View Article : Google Scholar :
|
|
267
|
Oh SW, Hwang DW and Lee DS: In vivo
monitoring of microRNA biogenesis using reporter gene imaging.
Theranostics. 3:1004–1011. 2013. View Article : Google Scholar
|
|
268
|
Hwang JY, Kim ST, Kwon J, Lee J, Chun YO,
Han JS and Han HS: Ultrasensitive fluorescence monitoring and in
vivo live imaging of circulating tumor cell-derived miRNAs using
molecular beacon system. ACS Sens. 3:2651–2659. 2018. View Article : Google Scholar : PubMed/NCBI
|