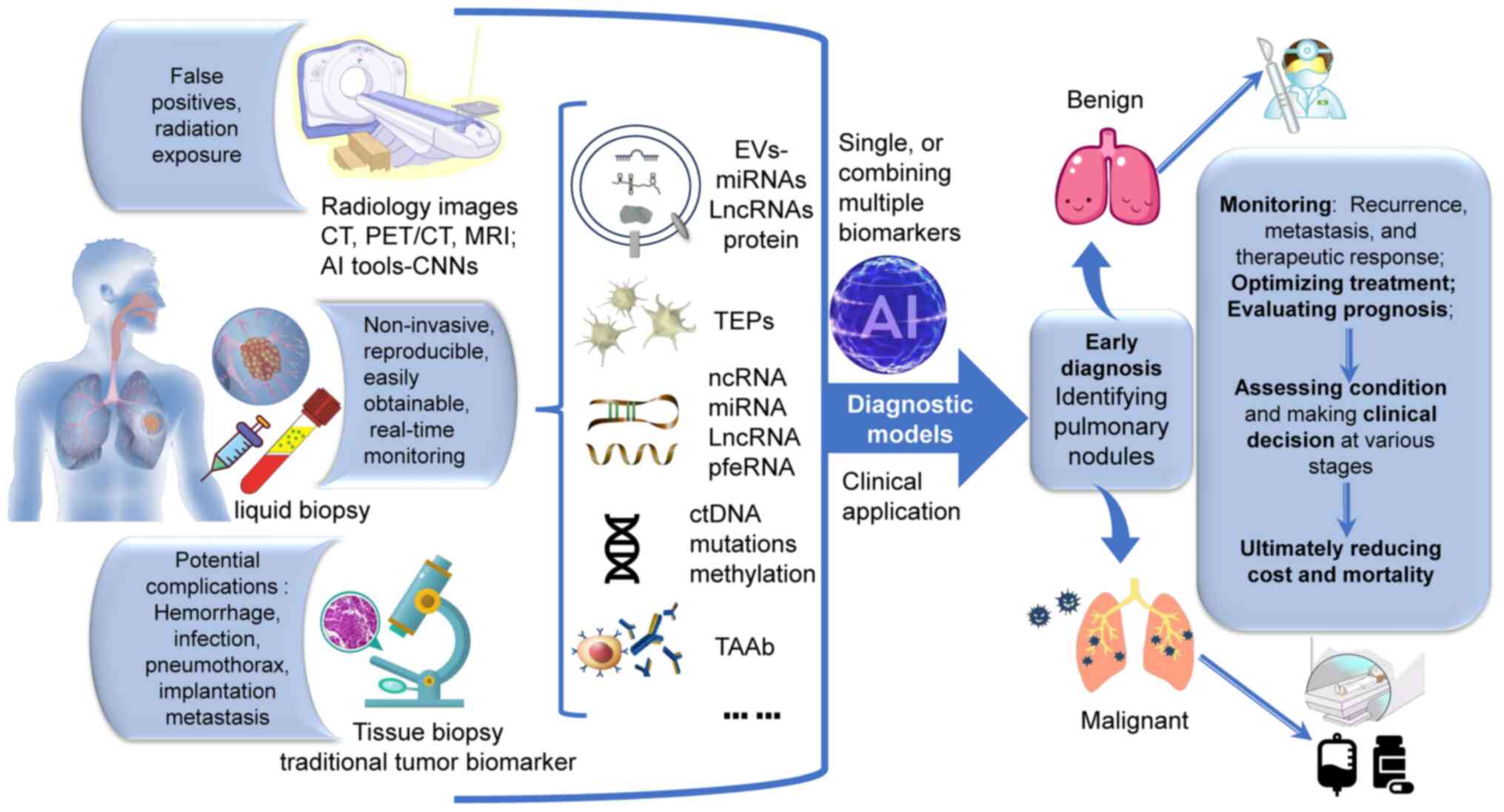
According to the Global Cancer Statistics 2020, lung
cancer is the second most common cancer and the leading cause of
cancer-related death (1). The
distinction of pulmonary nodules and early diagnosis of lung cancer
are crucial for improving prognosis and enabling surgical
intervention. However, current screening methods, such as imaging
and tissue biopsy, have limitations, such as false positives,
radiation exposure and potential complications like hemorrhage,
infection, pneumothorax and implantation metastasis. Therefore,
there is an urgent need for more reliable, testable and safer
biomarkers. Liquid biopsy has emerged as a non-invasive examination
that can reveal important tumor features, including gene mutations
and metabolic changes. It is increasingly recognized as a valuable
alternative for diagnosing and differentiating pulmonary nodules.
The present review aimed to discuss the current status of pulmonary
nodule diagnosis and summarize the latest applications of liquid
biopsy in identifying malignant and benign pulmonary nodules (BPN)
(Table I; Fig. 1).
Pulmonary nodules, defined as rounded or abnormally
cloudy lesions smaller than 30 mm in diameter, can be well or
poorly demarcated and surrounded by an inflated lung on
radiological imaging (2). They
are classified as benign or malignant, with significant prognostic
differences (3). Thus, improving
existing methods or exploring new approaches for early diagnosis of
pulmonary nodules is crucial. Tissue biopsy and imaging techniques,
such as computerized tomography (CT), positron emission tomography
(PET)/CT and magnetic resonance imaging (MRI), are currently the
main diagnostic methods. The application of artificial intelligence
(AI) tools on radiology images and digital pathology (4) can effectively enhance diagnostic
efficiency, optimize treatment, evaluate prognosis and ultimately
reduce mortality (5).
Various CT characteristics (nodule size, growth
rate, location, morphology, enhancement) are associated with the
nature of pulmonary nodules (6).
In cases where the probability of malignancy is low, such as with
constant, perifissural, well-circumscribed nodules, satellite
nodules with benign imaging features (stippled, laminated, dense
central or popcorn patterns of calcification), or individual
nodules without any risk factors (a mixed ground glass opacity
<6 mm in diameter), further imaging follow-up may not be
necessary if these features remain unchanged for two years or more.
However, with nodules larger than 10-20 mm, the risk of malignancy
increases significantly and medical follow-up is important
(7). It should be noted that CT
imaging for screening pulmonary nodules has limitations, as it can
detect both invasive and inert tumors (8). Due to the high sensitivity of
low-dose computed tomography (LDCT), numerous non-neoplastic
solitary pulmonary nodules (SPN) are also detected, resulting in an
increase in false positives and leading to follow-up repeat CT
scans and potential issues with radiation exposure (9).
MRI holds promise for longitudinally assessing lung
disease and function, offering an alternative to LDCT in patients
with lung cancer. With excellent soft tissue contrast and high
spatial resolution, MRI provides both morphologic and functional
information through diffusion-weighted and perfusion sequences
(11). However, a critical
challenge in lung MRI is its susceptibility to motion artifacts
caused by cardiac and respiratory movements. To mitigate these
effects, implementing breath-gated or breath-holding techniques
during image acquisition is recommended (12).
New techniques such as computational radiomics and
deep learning-based AI show promise in differentiating between
malignant and benign nodules (6). Computer-aided diagnosis systems
traditionally rely on imaging characteristics, including image
segmentation, feature value extraction and classification, and are
now being enhanced with convolutional neural networks (CNNs)-based
models. These models use a multi-view strategy to improve
sensitivity for pulmonary nodules. CNN models are widely employed
not only in CT images but also in cytopathological (13) and histological imaging (14). Certain studies utilize CNN models
to identify key gene mutations or investigate the development
mechanisms of cancer (15,16). However, nodule features may
influence the predictive performance of CNN models. In addition,
training complex CNN models with limited training sets can lead to
overfitting (14).
Liquid biopsy is a non-invasive and reproducible
method for real-time monitoring of tumors, allowing for the rapid
retrieval of pathological information from patient body fluids. It
offers valuable insights into the molecular mechanisms involved in
tumorigenesis and progression, making it a promising alternative to
tumor tissue samples in clinical settings (17). Various biomarkers, including
extracellular vesicles (EVs), circulating tumor DNA (ctDNA),
platelets, plasma RNA and circulating antibodies, have been
identified as non-invasive markers for diagnosing pulmonary nodules
(18). The integration of
advanced technologies has further enhanced the efficient capture of
biomarkers for liquid biopsy, revolutionizing clinical
decision-making at various stages of lung cancer management
(18-21).
EVs, which are endosome-derived nano vesicles
(30-1,000 nm) involved in intercellular communication, play a role
in the immune escape, metastasis, metabolic reprogramming and drug
resistance of lung cancer (22).
For instance, a study found that cancer cell-derived EVs containing
circUSP7 inhibited the function of CD8+ T cells, promoting the
progression of non-small cell lung cancer (NSCLC) and resistance to
anti-programmed death 1 therapy (23). EVs carrying snail-1 released by
cancer-associated fibroblasts induce epithelial-mesenchymal
transition (EMT) (24). Cancer
cells can utilize glycolysis and glutaminolysis to meet their
metabolic needs (25). EVs
carrying circSHKBP1 enhance glycolysis by sponging microRNA
(miR)-1294, leading to increased expression of the glycolytic
enzyme pyruvate kinase isozyme type M2 (PKM2) and ultimately
affecting the function of NSCLC cells and macrophages (26). EVs derived from cancer-associated
fibroblasts contained long intergenic non-coding RNA (LINC)01614,
which enhanced glutamine uptake in lung cells by upregulating the
expression of glutamine transporters (27). In the study of therapeutic
resistance, hypoxia-induced EVs were found to transmit cisplatin
resistance to sensitive NSCLC cells by delivering PKM2 (28). In addition, EV transfer of
wild-type EGFR was also shown to promote resistance to the drug
Osimertinib (29). Therefore,
targeting the secretion and transfer of specific cargo in EVs, such
as PKM2 and EGFR, may be a promising approach to overcome treatment
resistance.
i) EV-associated miRNAs: Numerous studies on EV
miRNA have primarily focused on the early diagnosis of lung cancer.
EV miR-29c-3p and miR-1290 have been identified as superior
diagnostic biomarkers for distinguishing between lung cancer and
benign BPN. Their expression levels show significant differences in
lung cancer, with miR-29c-3p decreased and miR-1290 elevated. These
miRNAs exhibit high sensitivity (89.47 and 97.37%), specificity
(84.21 and 89.47%) and area under the curve (AUC) values (0.868 and
0.934) in discriminating lung cancer from BPNs. Furthermore, they
demonstrate a strong discriminative ability between NSCLC and SCLC
with AUC values of 0.842 and 0.810 (30). Utilizing multiple EV miRNAs
enhances diagnostic accuracy. A diagnostic signature consisting of
four EV-derived miRNAs (miR-106b-3p, miR-125a-5p, miR-3615 and
miR-450b-5p) has been developed for the early detection of LUAD. In
training cohorts, the signature exhibited an AUC value of 0.917, a
sensitivity of 83.8% and a specificity of 87.1%. These diagnostic
capabilities were further validated in test cohorts (31). Mechanistic studies have
elucidated the roles of these miRNAs. EV miR-106b targets
phosphatase and tensin homolog, promoting migration and invasion of
lung cancer cells (32). In
NSCLC, miR-125a-5p inhibits the expression of histone
methyltransferase Suv39H1, leading to cancer suppression both in
vitro and in vivo (33). Furthermore, miR-125a-5p exerts a
tumor-inhibiting effect by targeting STAT3 (34). As the target RNA of two competing
endogenous long non-coding (lnc) RNAs, miR-450b-5p demonstrates a
tumor-suppressive function in NSCLC (35). Not only the quantity, but also
the ratio of EV miRNAs, plays a significant role in distinguishing
between benign and malignant pulmonary nodules. For instance, the
miR-21/Let-7a ratio is markedly elevated in NSCLC compared to BPNs,
with an AUC of 0.754 and a specificity of 82.61% (36).
The main challenge in using EV-associated miRNAs for
diagnosing lung nodules is the difficulty in effectively capturing
and enriching tumor-derived EVs from complex blood systems. A
recent study introduced a Glyexo-capture strategy using lentil
lectin-magnetic beads to target exosome membranes. This dual-target
method enhances the detection of valuable exosomal biomarkers with
increased sensitivity and specificity. For instance, a miRNA panel
consisting of miR-4732-5p, miR-451a, miR-486-5p and miR-139-3p
showed promise in screening for early LUAD from BPNs, achieving an
AUC of 0.855 with 91.07% sensitivity and 66.36% specificity
(43). These miRNAs play crucial
roles in inhibiting lung cancer through various pathways:
miR-4732-5p targets xenotropic and polytropic retrovirus receptor 1
to suppress LUAD migration and metastasis (44), miR-451a inhibits invasion by
targeting activating transcription factor-2 (45), miR-486-5p hinders the mTOR
pathway by targeting ribosomal proteins (46) and miR-139-3p reduces lung
squamous carcinoma viability by targeting checkpoint kinase 1
(47). In addition, exosomal
miR-4497 is also a tumor suppressor marker, showing diagnostic
efficacy in distinguishing NSCLC from BPNs with 73.3% sensitivity,
72.6% specificity and an AUC of 0.748. Importantly, miR-4497
demonstrates potential for monitoring tumor malignancy [size, tumor
node metastasis (TNM) stage and metastasis] and overall survival
(48).
ii) EV-associated long RNAs (exLRs): While previous
studies have primarily focused on miRNAs, their limited presence in
EVs hinders their effectiveness as biomarkers for lung cancer
diagnosis (49). By contrast,
exLRs, such as mRNAs, circRNAs and lncRNAs, are abundant in
peripheral blood EVs and have shown promise as diagnostic
biomarkers of lung cancer (50).
Specifically, a panel of 23 exLRs, including 6 upregulated and 17
downregulated genes, identified in EVs can differentiate LUAD from
BPNs with high sensitivity (93.75%), specificity (85.71%), and
accuracy (88.24%). Furthermore, a signature of 17 exLRs, comprising
2 upregulated and 15 downregulated genes, can effectively
distinguish between adenocarcinoma in situ and minimally
invasive or invasive adenocarcinoma with exceptional sensitivity
(93.33%), specificity (98.00%) and accuracy (96.25%) (51). In a recent study, a diagnostic
model incorporating three exLRs (PGM5-AS1, SFTA1P and CTA-384D8.35)
showed a strong predictive ability for NSCLC, with an AUC of 0.97
(52). Additionally, researchers
found that EV lncRNA RP5-977B1 expression was elevated in NSCLC
compared to healthy controls and patients with pulmonary
tuberculosis, showing superior discriminatory power (AUC 0.890)
over traditional markers carcino-embryonic antigen (CEA) (0.761)
and cytokeratin 19 fragment (CYFRA21-1) (0.670). This comparative
advantage was also observed in distinguishing early-stage NSCLC
from controls (53). To improve
diagnostic accuracy, a novel index combining gasdermin 5 and CEA
was developed, achieving an AUC of 0.929, sensitivity of 89.06% and
specificity of 90.00% for NSCLC diagnosis (54).
iii) EV-associated protein: EV-associated proteins,
such as fibrinogen beta chain (FGB) and fibrinogen gamma chain
(FGG), have been implicated in EMT progression of lung cancer
(55). Researchers have
demonstrated the utility of FGB and FGG levels in plasma EVs as
diagnostic biomarkers for distinguishing benign and malignant
pulmonary nodules. Compared to benign nodules, patients with lung
cancer exhibited elevated FGB and FGG expression levels. When used
individually, FGB showed a sensitivity of 0.628, specificity of
0.800 and AUC of 0.741, while FGG had a sensitivity of 0.535,
specificity of 0.850 and AUC of 0.659. Combining FGB and FGG
detection improved the diagnostic accuracy, with a sensitivity of
0.700 and AUC of 0.794, suggesting that these proteins could serve
as sensitive biomarkers for distinguishing benign from malignant
pulmonary nodules (56).
Plasma EV versican, a chondroitin sulfate
glycoprotein was found to be significantly elevated in patients
with NSCLC, with expression levels correlating with TNM stages and
clinical parameters. Combining plasma versican and plasma EV
versican showed superior diagnostic performance in identifying
patients with NSCLC and those with metastasis compared to
traditional biomarkers [neuron-specific enolase (NSE), CYFRA21-1
and squamous cell carcinoma antigen] (57). Yang et al (58) identified differential expression
of immunoglobulin heavy variable 4-4 and immunoglobulin lambda
variable 1-40 in serum EVs of patients with NSCLC, with the
combination showing a high diagnostic capacity with a sensitivity
of 88.73%, a specificity of 85.00% and AUC of 0.93. Recent research
identified 150 altered EV proteins in patients with NSCLC,
primarily involved in cell adhesion, differentiation, motility and
osmoregulation, suggesting their potential as biomarkers for early
NSCLC diagnosis [panel of FGB, FGG and von Willebrand factor] and
metastasis prediction (panel of complement factor H related protein
5, complement component 9 and mannose-binding lectin 2) (59).
While EV biomarkers offer numerous advantages,
challenges persist in their clinical application. EVs can be
secreted by various cells and those from cells with pathological
changes may be overshadowed by those from normal cells. Multi-level
screening is an important strategy for enhancing tumor relevance
and tissue specificity of EV markers. In a study analyzing EV
proteomes from paired tumor and adjacent tissues, exclusive
proteins HIV-1 Tat interactive protein 2 and methyltransferase like
1 were identified as specific markers for LUAD. Furthermore,
comparing plasma and tissue-derived EV proteins revealed plasma EV
signatures that could distinguish between lung cancers even at
early stages (60). In another
study using lung cancer serum and cell culture supernatant, EV
fibronectin emerged as a promising biomarker. It was able to
effectively differentiate patients with NSCLC from healthy controls
(AUC=0.844, P<0.001) and showed a significant increasing trend
correlating with cancer progression (advanced NSCLC > early
NSCLC > healthy controls, P<0.001) (61).
Platelets are rich in RNA species and functional
spliceosomes, undergoing specific splicing in response to the tumor
microenvironment, leading to changes in RNA content. There has been
an increasing focus on the role of platelets in tumorigenesis,
metastasis, immune evasion and chemotherapy resistance. As tumors
progress, cancer cells can educate platelets, altering their
transcriptome and molecular makeup (62,63). For instance, in patients with
lung cancer, TEPs exhibit significant alterations in specific RNA
species, including downregulation of linc-GTF2H2-1 and
RP3-466P17.2, and upregulation of lnc-ST8SIA4-12, even at early
stages of the disease. Integrating TEP lincRNA, CEA, NSE and
CYFRA21-1 can effectively distinguish patients with advanced-stage
lung cancer from early-stage ones with an AUC of 0.899 (64).
TEPs content can be transferred via MVs. Platelets
can take up tumor-derived MVs and release their own protumoral MVs,
thereby establishing a blood-based network for distributing
tumor-derived RNA or protein. By analyzing changes in the platelet
transcriptome through sequencing and proteomics, diagnostic models
based on platelet activity can be developed and used in
differentiating pulmonary nodules. TEP integrin alpha 2b (TEP
ITGA2B) is a promising marker for improving the identification
accuracy for patients with stage I NSCLC, distinguishing malignant
tumours from BPNs. TEP ITGA2B levels were significantly elevated in
patients with NSCLC, with an AUC of 0.940, sensitivity of 96.4% and
specificity of 81.7% for identifying stage I NSCLC from BPNs.
Compared to serum CEA, TEP ITGA2B demonstrated superior performance
in distinguishing benign from malignant lung nodules, particularly
in the case of SPN. Further research indicated that a nomogram
incorporating ITGA2B and CEA may enhance diagnostic accuracy and
predict overall survival (65).
Several studies have explored the association
between pulmonary nodule with platelet characteristics, such as
platelet-to-lymphocyte ratio (PLR). A higher PLR has been
associated with an increased risk of positive nodules and lung
cancer [odds ratio=1.29 (95% CI, 0.99-1.68)] (66). To enhance diagnostic accuracy and
reduce bias, a multi-index diagnostic model called Sichuan Hospital
of Cancer (SCHC), incorporating platelet features (platelet counts
in platelet-rich plasma samples, plateletcrit in platelet-rich
plasma samples and plateletcrit in whole-blood samples), age and
nodule size, has shown promising results in distinguishing benign
from malignant nodules. The SCHC model outperformed other clinical
models (Veterans Affairs, Mayo Clinic, Brock University) by
minimizing misclassification of malignant tumors and significantly
improving reclassification metrics such as net reclassification
improvement and integrated discrimination improvement. This
platelet-based model could aid in accurately diagnosing early-stage
malignancies and guiding optimal patient management in clinical
settings (67).
ncRNA refers to a group of RNA molecules that do not
encode proteins but play important regulatory roles in cellular
processes. Epigenetic-related ncRNAs include miRNAs, small
interfering (si)RNAs, PiWi-interacting RNAs and lncRNAs.
miRNAs are small endogenous RNAs that target mRNAs,
leading to post-transcriptional silencing and potentially
influencing tumor development and metastasis (68). Dysregulation of specific miRNAs
or miRNA groups is closely associated with cancer progression
(69). Research has explored the
use of circulating miRNAs for diagnosing pulmonary nodules.
miR-499a (70) and a group of
other miRNAs, including miRNA-17, -146a, -200b, -182, -221, -205,
-7, -21, -145 and -210 (71),
were significantly elevated in NSCLC compared to BPN, and they both
have potential in differentiating between benign and malignant
pulmonary nodules. In addition to single miRNAs, combining multiple
miRNAs can enhance the accuracy of diagnosis. A panel of miRNAs
(miR-199a-3p, -148a-3p, -210-3p, -378d and -138-5p) in blood has
been validated for early diagnosis of LUAD from pulmonary nodules.
The use of this miRNA panel alongside CT scans significantly
reduces false positives. For instance, the false-positive rate of
CT imaging for nodules and ground glass nodules was reduced from
33.1 to 3.2% when positive miRNA panel results were combined with
nodule size (72). In addition,
miR-21 and miR-210 levels were higher, while miR-486-5p levels were
lower in patients with malignant lung nodules compared to benign
ones. A combination of three miRNAs achieved an AUC of 0.86 in
distinguishing lung cancer from BPN with 75.00% sensitivity and
84.95% specificity (73).
Furthermore, paired miRNA ratios were used in the differential
diagnosis of NSCLC and BPN. Five miRNA ratios
(miR-92a-3p/miR-146b-3p, miR-20a-5p/miR-146b-3p,
miR-19a-3p/miR-146b-3p, miR-15b-5p/miR-146b-3p and
miR-16-5p/miR-146b-3p) showed higher expression levels in NSCLC
compared to BPN, with a sensitivity of 0.70 and specificity of 0.90
(74).
PfeRNAs are a unique type of small ncRNA that
directly binds and regulates phosphorylated proteins involved in
lung cancer tumorigenesis (79).
Unlike miRNAs and siRNAs, pfeRNAs enhance the function of their
target proteins instead of degrading them (80). Recent research has demonstrated
that pfeRNAs can serve as diagnostic markers for pulmonary nodules.
An 8-pfeRNA classifier (pfeRNAa to pfeRNAh) identified through deep
sequencing can effectively differentiate between malignant and
BPNs, with a sensitivity of 77.1% and specificity of 74.25%
(81).
ctDNA is fragmented DNA from tumors found in the
bloodstream, typically released by necrotic or apoptotic tumor
cells in the tumor microenvironment (82). Mutated or methylated ctDNA can be
identified using PCR or DNA sequencing to track lung cancer
development. ctDNA exhibits strong tissue specificity and
correlates well with tumor tissue DNA, making it a valuable
biomarker for monitoring lung cancer progression.
Gene mutations play a crucial role in tumor
development by activating oncogenes and deactivating tumor
suppressor genes. Mutations in ctDNA directly reflect tumor
mutations, providing a valuable tool for distinguishing between
malignant and BPNs. However, the sensitivity of ctDNA may be
limited by interference from wild-type sequences (83). Recent studies utilizing targeted
next-generation sequencing have identified specific gene mutations
in ctDNA, such as RNF213, with high specificity of 100% in
distinguishing between benign and malignant pulmonary nodules
(84). Plasma ctDNA shows
promise in early cancer detection, particularly when combined with
clinical information and traditional biomarkers. Analysis of 65
lung cancer-related genes in plasma ctDNA revealed increasing
mutation levels with tumor progression, particularly in driving
genes. The ctDNA assay had a sensitivity of 69% and specificity of
96% for distinguishing lung nodule nature. Combining ctDNA, serum
biomarkers and patient age boosted sensitivity to 80% and
specificity to 99% (85).
DNA methylation is a well-researched epigenetic
modification, particularly in gene promoter regions, often leading
to tumor suppressor gene inactivation and cancer development
(86). Analyzing ctDNA
methylation has emerged as a novel, sensitive and non-invasive
method for early lung cancer detection and distinguishing lung
cancers from BPN. DNA bisulfite sequencing was conducted on 309
pulmonary nodule tissue specimens to identify cancer-specific
patterns. From 3,886 hypermethylated regions in the tissue, 71
regions found in plasma were refined to select 9 markers for a
diagnostic model. In a validation set, the model had 79.5%
sensitivity and 85.2% specificity in distinguishing lung cancer
from BPNs (87). In addition to
hypermethylation changes, ctDNA hypomethylation can also aid in
diagnosing pulmonary nodules. Research indicates that methylation
levels of seven sites of fucosyltransferase 7 in lung cancer were
significantly lower than in normal controls. Furthermore, the
levels of methylation at CpG-4 and CpG-7 were lower in lung cancer
compared to BPN (88).
ctDNA is typically scarce and can be surrounded by a
background of DNA from healthy tissues (89). To improve sensitivity and
specificity, a combination of diagnostic markers and technological
innovations was employed. A study involving 246 CT-detected
patients with small pulmonary nodules (diameter, ≤3.0 cm)
discovered elevated methylation levels of tachykinin precursor 1
(TAC1), cysteine dioxygenase type 1 (CDO1), homeobox A7 (HOXA7) and
SRY-box transcription factor 17 (SOX17) in peripheral blood in
cases of NSCLC compared to benign cases. The combination of SOX17,
CDO1 and HOXA7 achieved a sensitivity of 90%, specificity of 71%
and an AUC of 0.88 for diagnosis (90). To further promote the clinical
application of these biomarkers, a new methylation analysis
technique, multiplex digital methylation-specific PCR (MSP), was
developed by Zhao et al (91), which increased the sensitivity
from 88 to 90% and specificity from 60 to 82% for the combination
of TAC1, CDO1, HOXA7 and SOX17. Another novel testing technology
(92) utilized multi-locus qPCR
to screen methylation markers, selecting the highest AUC marker as
the anchor. This approach, combined with 10×4-fold
cross-validations for each addition, resulted in the creation of
two models: LunaCAM-D with 6 methylation markers to distinguish
lung cancer from benign diseases and LunaCAM-S with 5 markers to
classify lung cancer from healthy controls. In a recent study,
methylated DNA (prostaglandin E receptor 4, ras association domain
family 1A and short stature homeobox gene 2) was combined with a
radiologic feature (pulmonary nodule diameter) to create prediction
models. These models achieved an AUC value of 0.948 with
sensitivity and specificity of 89.5 and 95.4%, respectively, for
identifying malignant from BPN (93). Another study integrated ctDNA
methylation, clinical features and CT imaging features to develop a
composite model named PULMOSEEK Plus. This model demonstrated a
sensitivity of ≥95% across all stages of lung cancer, an AUC value
of 0.98 in early-stage lung cancer and an AUC value of 0.99 in
indeterminate nodules (5-10 mm). By reclassifying pulmonary nodules
with two cutoffs (0.65 and 0.89), unnecessary invasive procedures
could have been reduced in 85% of BPN and delayed treatment avoided
in 72% of malignant nodules (94).
Autoantibodies targeting tumor-associated antigens
(TAAb) are commonly found in the preclinical phase of lung cancer
(95), indicating their
potential as promising noninvasive biomarkers with satisfactory
sensitivity and specificity (96). Studies have highlighted the
effectiveness of multi-TAAb panels, such as the Fc gamma receptor
IIa, erythrocyte B membrane protein band 4.1 like 3, Nogo
receptor-interacting protein-1 and S100 calcium binding protein A7
like 2 combination, in accurately distinguishing indeterminate
pulmonary nodules (8-20 mm) with an AUC value of 0.78, 91.7%
sensitivity and 57.1% specificity (97). In a seven-TAAb panel (tumor
protein 53, protein gene product 9.5, SOX2, G antigen 7, GBU4-5,
melanoma-associated antigen A1 and cancer-associated gene),
sensitivity and specificity were 59.7 and 81.1% for early lung
nodule differentiation. In addition, integrating these seven
autoantibodies and imaging features into a machine learning model
markedly increased the AUC from 0.75 to 0.96 in distinguishing
patients with pulmonary nodules (98). A recent study utilized
high-throughput protein microarrays to identify TAAb and developed
a random forest model with six autoantibodies (annexin 2,
dermcidin, MID1 interacting protein 1, paraneoplastic antigen MA1,
TATA-box binding protein associated factor 10, zinc finger protein
696) to detect high-risk pulmonary nodules suitable for LDCT scans
(99). In addition, another
study investigated antibody responses against bacterial and viral
proteins in patients with lung cancer, finding more prevalent
antibodies among BPNs than among LUAD. Then a panel of 20
antibodies was created to distinguish LUAD from BPNs with an AUC of
0.80 (100).
Each liquid biopsy method has distinct advantages,
limitations, diagnostic performance and cost-effectiveness
profiles, necessitating biomarker selection based on clinical
context (Table II). However,
all of these detection approaches face technical challenges in
clinical translation. For instance, ultracentrifugation remains the
gold standard for EVs isolation, its limitations-including being
time-consuming, low-throughput and potentially damaging to
vesicles-along with protein aggregate contamination, necessitate
more efficient techniques such as nanosensors (101) and microfluidic chips (102). Similarly, TEPs face challenges
in specificity due to blood cell contamination during isolation
(103) and an incomplete
understanding of spliceosome regulation (104), requiring further multicenter
validation (105). ncRNAs,
despite their detectability, have shortcomings of low abundance and
interference in bodily fluids, demanding improved quantification
methods (106). Although ctDNA
enables real-time tumor monitoring, its low concentration
necessitates costly high-sensitivity assays (e.g.,
nanoparticle-based DNA extraction/qMSP), complicating analysis
(107). Circulating antibodies,
while promising, risk false positives (cross-reactivity with
infections/autoimmunity) and false negatives (immunosuppression),
underscoring the need for multi-analyte diagnostic panels.
Collectively, advancing isolation technologies, standardizing
detection and integrating multi-omics approaches are critical for
robust clinical translation of liquid biopsy.
Liquid biopsy is a transformative tool in lung
cancer management, offering significant clinical benefits in
several clinical applications, such as early detection and
screening (108), providing
comprehensive molecular profiling, guiding therapy (109), identifying minimal residual
disease (110), and monitoring
disease progression and treatment response (111). As an important complement or
alternative to traditional diagnostic methods, liquid biopsy has
significantly enhanced the precision and efficiency of lung cancer
diagnosis and management. In particular, the integration of liquid
biopsy with current diagnostic methods, such as imaging techniques,
tissue biopsy and molecular profiling, can leverage the strengths
of each method to provide a comprehensive understanding of the
disease (108,112-117) (Table III). The cost-effectiveness of
liquid biopsy is demonstrated through multiple key aspects:
Population-based screening feasibility (108), reduction of unnecessary
invasive procedures (118,119), accelerated diagnostic
turnaround time (120) and
facilitation of personalized treatment strategies (111). These advantages collectively
contribute to significant cost reduction in the clinical management
pathway for pulmonary nodules and lung cancer. Although the upfront
costs of liquid biopsy can be high, its potential to reduce
unnecessary treatments, complications and hospitalizations makes it
a cost-effective option in numerous scenarios (119) (Table III). As technology advances and
costs decrease, liquid biopsy is likely to become an integral part
of lung cancer care, improving outcomes for patients and optimizing
healthcare resource utilization.
Liquid biopsy, a non-invasive and convenient
diagnostic tool, is increasingly utilized in clinical practice.
Circulating biomarkers provide insights into the mechanisms of lung
cancer, aiding in early detection, screening, diagnosis, monitoring
and treatment. However, the sensitivity and specificity of EVs,
TEPs, ncRNA, ctDNA and TAAb as lung cancer biomarkers may be
limited by low blood concentrations and potential interference from
molecules secreted by normal cells. The heterogeneous and atypical
nature of tumors necessitates a refined temporal and spatial
variation map of these biomarkers, with research focusing on
combining multiple biomarkers (the construction of clinical models,
including imaging features and patient characteristics) or levels
(the establishment of biomarkers from cells, tissue, peripheral
circulation) for accurate differentiation of malignant tumour and
BPN (Fig. 1). Furthermore,
AI-driven analysis has the potential to efficiently analyze vast
and complex datasets, thereby enabling the development of diverse
and efficient diagnostic and predictive models (121). Standardization of liquid biopsy
methods and critical level determination is essential for clinical
application. Ultimately, liquid biopsy holds promise for enhancing
early pulmonary nodule diagnosis.
Not applicable.
YT designed this study and revised the manuscript.
MP, JG and YT wrote the manuscript. TA and HC generated the table
and figure. LC, MC and JL participated in the literature search and
collation. All authors have read and approved the final manuscript.
Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This study was funded by the Youth Interdisciplinary Special
Fund of Zhongnan Hospital of Wuhan University (grant no.
ZNQNJC2022008), the Natural Science Foundation of Hubei Province
(grant no. 2021CFB415) and the National Natural Science Foundation
of China (grant no. 81702273).
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Larici AR, Farchione A, Franchi P,
Ciliberto M, Cicchetti G, Calandriello L, Del Ciello A and Bonomo
L: Lung nodules: Size still matters. Eur Respir Rev. 26:1700252017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Groome PA, Bolejack V, Crowley JJ, Kennedy
C, Krasnik M, Sobin LH and Goldstraw P; IASLC International Staging
Committee; Cancer Research and Biostatistics; Observers to the
Committee; Participating Institutions: The IASLC lung cancer
staging project: Validation of the proposals for revision of the T,
N, and M descriptors and consequent stage groupings in the
forthcoming (seventh) edition of the TNM classification of
malignant tumours. J Thorac Oncol. 2:694–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Viswanathan VS, Toro P, Corredor G,
Mukhopadhyay S and Madabhushi A: The state of the art for
artificial intelligence in lung digital pathology. J Pathol.
257:413–429. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pei Q, Luo Y, Chen Y, Li J, Xie D and Ye
T: Artificial intelligence in clinical applications for lung
cancer: Diagnosis, treatment and prognosis. Clin Chem Lab Med.
60:1974–1983. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim TJ, Kim CH, Lee HY, Chung MJ, Shin SH,
Lee KJ and Lee KS: Management of incidental pulmonary nodules:
Current strategies and future perspectives. Expert Rev Respir Med.
14:173–194. 2020. View Article : Google Scholar
|
|
7
|
Ali K and Bal S: Management of Solitary
Pulmonary Nodule. Recent concepts in minimal access surgery. Sharma
D and Hazrah P: 1. Springer Singapore; Singapore: pp. 401–418.
2022, View Article : Google Scholar
|
|
8
|
Hasan N, Kumar R and Kavuru MS: Lung
cancer screening beyond low-dose computed tomography: The role of
novel biomarkers. Lung. 192:639–648. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nooreldeen R and Bach H: Current and
future development in lung cancer diagnosis. Int J Mol Sci.
22:86612021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dalli A, Selimoglu Sen H, Coskunsel M,
Komek H, Abakay O, Sergi C and Cetin Tanrikulu A: Diagnostic value
of PET/CT in differentiating benign from malignant solitary
pulmonary nodules. J BUON. 18:935–941. 2013.
|
|
11
|
Khalil A, Majlath M, Gounant V, Hess A,
Laissy JP and Debray MP: Contribution of magnetic resonance imaging
in lung cancer imaging. Diagn Interv Imaging. 97:991–1002. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Periaswamy G, Arunachalam VK,
Varatharajaperumal R, Kalyan G, Selvaraj R, Mehta P and Cherian M:
Comparison of ultrashort TE lung MRI and HRCT lungs for detection
of pulmonary nodules in oncology patients. Indian J Radiol Imaging.
32:497–504. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Y, Guan S, Ou Z, Li W, Yan L and Situ
B: Advances in AI-based cancer cytopathology. Interdiscip Med.
1:e202300132023. View Article : Google Scholar
|
|
14
|
Li Y, Wu X, Yang P, Jiang G and Luo Y:
Machine learning for lung cancer diagnosis, treatment, and
prognosis. Genomics Proteomics Bioinformatics. 20:850–866. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
AbdulJabbar K, Raza SEA, Rosenthal R,
Jamal-Hanjani M, Veeriah S, Akarca A, Lund T, Moore DA, Salgado R,
Al Bakir M, et al: Geospatial immune variability illuminates
differential evolution of lung adenocarcinoma. Nat Med.
26:1054–1062. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Coudray N, Ocampo PS, Sakellaropoulos T,
Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N and Tsirigos
A: Classification and mutation prediction from non-small cell lung
cancer histopathology images using deep learning. Nat Med.
24:1559–1567. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W,
Tang XM, Sun F, Lu HM, Deng J, et al: Liquid biopsy in lung cancer:
Significance in diagnostics, prediction, and treatment monitoring.
Mol Cancer. 21:252022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Levy B, Hu ZI, Cordova KN, Close S, Lee K
and Becker D: Clinical utility of liquid diagnostic platforms in
non-small cell lung cancer. Oncologist. 21:1121–1130. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng H, Wu X, Yin J, Wang S, Li Z and You
C: Clinical applications of liquid biopsies for early lung cancer
detection. Am J Cancer Res. 9:2567–2579. 2019.
|
|
20
|
Bao H, Min L, Bu F, Wang S and Meng J:
Recent advances of liquid biopsy: Interdisciplinary strategies
toward clinical decision-making. Interdiscip Med. 1:e202300212023.
View Article : Google Scholar
|
|
21
|
Zhu Y, Li W, Lan F, Chen S, Chen X, Zhang
X, Yan X and Zhang Y: DNA nanotechnology in tumor liquid biopsy:
Enrichment and determination of circulating biomarkers. Interdiscip
Med. 2:e202300432024. View Article : Google Scholar
|
|
22
|
El Andaloussi S, Mäger I, Breakefield XO
and Wood MJA: Extracellular vesicles: Biology and emerging
therapeutic opportunities. Nat Rev Drug Discov. 12:347–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D,
Long X, Lin K, Lu F, Xu JJ and Wu YB: Cancer cell-derived exosomal
circUSP7 induces CD8+ T cell dysfunction and anti-PD1
resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol
Cancer. 20:1442021. View Article : Google Scholar
|
|
24
|
You J, Li M, Cao LM, Gu QH, Deng PB, Tan Y
and Hu CP: Snail1-dependent cancer-associated fibroblasts induce
epithelial-mesenchymal transition in lung cancer cells via
exosomes. QJM. 112:581–590. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen W, Tang D, Lin J, Huang X, Lin S,
Shen G and Dai Y: Exosomal circSHKBP1 participates in non-small
cell lung cancer progression through PKM2-mediated glycolysis. Mol
Ther Oncolytics. 24:470–485. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu T, Han C, Fang P, Ma Z, Wang X, Chen
H, Wang S, Meng F, Wang C, Zhang E, et al: Cancer-associated
fibroblast-specific lncRNA LINC01614 enhances glutamine uptake in
lung adenocarcinoma. J Hematol Oncol. 15:1412022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang D, Zhao C, Xu F, Zhang A, Jin M,
Zhang K, Liu L, Hua Q, Zhao J, Liu J, et al: Cisplatin-resistant
NSCLC cells induced by hypoxia transmit resistance to sensitive
cells through exosomal PKM2. Theranostics. 11:2860–2875. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu S, Luo M, To KKW, Zhang J, Su C, Zhang
H, An S, Wang F, Chen D and Fu L: Intercellular transfer of
exosomal wild type EGFR triggers osimertinib resistance in
non-small cell lung cancer. Mol Cancer. 20:172021. View Article : Google Scholar :
|
|
30
|
Zhang Q, Zheng K, Gao Y, Zhao S, Zhao Y,
Li W, Nan Y, Li Z, Liu W, Wang X, et al: Plasma exosomal miR-1290
and miR-29c-3p as diagnostic biomarkers for lung cancer. Heliyon.
9:e210592023. View Article : Google Scholar :
|
|
31
|
Gao S, Guo W, Liu T, Liang N, Ma Q, Gao Y,
Tan F, Xue Q and He J: Plasma extracellular vesicle microRNA
profiling and the identification of a diagnostic signature for
stage I lung adenocarcinoma. Cancer Sci. 113:648–659. 2022.
View Article : Google Scholar
|
|
32
|
Sun S, Chen H, Xu C, Zhang Y, Zhang Q,
Chen L, Ding Q and Deng Z: Exosomal miR-106b serves as a novel
marker for lung cancer and promotes cancer metastasis via targeting
PTEN. Life Sci. 244:1172972020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Open Biology Editorial Team: Retraction
'Reduced miR-125a-5p level in non-small-cell lung cancer is
associated with tumour progression'. Open Biol. 10:2002052020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhong L, Sun S, Shi J, Cao F, Han X and
Chen Z: MicroRNA-125a-5p plays a role as a tumor suppressor in lung
carcinoma cells by directly targeting STAT3. Tumour Biol.
39:10104283176975792017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ye P, Lv X, Aizemaiti R, Cheng J, Xia P
and Di M: H3K27acactivated LINC00519 promotes lung squamous cell
carcinoma progression by targeting miR-450b-5p/miR-515-5p/YAP1
axis. Cell Prolif. 53:e127972020. View Article : Google Scholar
|
|
36
|
Yang G, Wang T, Qu X, Chen S, Han Z, Chen
S, Chen M, Lin J, Yu S, Gao L, et al: Exosomal miR-21/Let-7a ratio
distinguishes non-small cell lung cancer from benign pulmonary
diseases. Asia Pac J Clin Oncol. 16:280–286. 2020. View Article : Google Scholar
|
|
37
|
Zhong Y, Ding X, Bian Y, Wang J, Zhou W,
Wang X, Li P, Shen Y, Wang JJ, Li J, et al: Discovery and
validation of extracellular vesicle-associated miRNAs as
noninvasive detection biomarkers for early-stage non-small-cell
lung cancer. Mol Oncol. 15:2439–2452. 2021. View Article : Google Scholar :
|
|
38
|
Tang CP, Zhou HJ, Qin J, Luo Y and Zhang
T: MicroRNA-520c-3p negatively regulates EMT by targeting IL-8 to
suppress the invasion and migration of breast cancer. Oncol Rep.
38:3144–3152. 2017. View Article : Google Scholar
|
|
39
|
Gulhane P and Singh S: MicroRNA-520c-3p
impacts sphingolipid metabolism mediating PI3K/AKT signaling in
NSCLC: Systems perspective. J Cell Biochem. 123:1827–1840. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu X, Lu X, Sun J and Shu Y: microRNA
expression profiling of side population cells in human lung cancer
and preliminary analysis. Zhongguo Fei Ai Za Zhi. 13:665–669.
2010.In Chinese.
|
|
41
|
Squadrito ML, Baer C, Burdet F, Maderna C,
Gilfillan GD, Lyle R, Ibberson M and De Palma M: Endogenous RNAs
modulate microRNA sorting to exosomes and transfer to acceptor
cells. Cell Rep. 8:1432–1446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qi Y, Jin C, Qiu W, Zhao R, Wang S, Li B,
Zhang Z, Guo Q, Zhang S, Gao Z, et al: The dual role of glioma
exosomal microRNAs: Glioma eliminates tumor suppressor miR-1298-5p
via exosomes to promote immunosuppressive effects of MDSCs. Cell
Death Dis. 13:4262022. View Article : Google Scholar :
|
|
43
|
Chen X, Yu L, Hao K, Yin X, Tu M, Cai L,
Zhang L, Pan X, Gao Q and Huang Y: Fucosylated exosomal miRNAs as
promising biomarkers for the diagnosis of early lung
adenocarcinoma. Front Oncol. 12:9351842022. View Article : Google Scholar :
|
|
44
|
Hu Y, Bai J, Zhou D, Zhang L, Chen X, Chen
L, Liu Y, Zhang B, Li H and Yin C: The miR-4732-5p/XPR1 axis
suppresses the invasion, metastasis, and epithelial-mesenchymal
transition of lung adenocarcinoma via the PI3K/Akt/GSK3β/Snail
pathway. Mol Omics. 18:417–429. 2022. View Article : Google Scholar
|
|
45
|
Shen YY, Cui JY, Yuan J and Wang X:
MiR-451a suppressed cell migration and invasion in non-small cell
lung cancer through targeting ATF2. Eur Rev Med Pharmacol Sci.
22:5554–5561. 2018.PubMed/NCBI
|
|
46
|
Ding L, Tian W, Zhang H, Li W, Ji C, Wang
Y and Li Y: MicroRNA-486-5p suppresses lung cancer via
downregulating mTOR signaling in vitro and in vivo. Front Oncol.
11:6552362021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng X, Zhang Y, Wu S, Jiang B and Liu Y:
MiR-139-3p targets CHEK1 modulating DNA repair and cell viability
in lung squamous carcinoma cells. Mol Biotechnol. 64:832–840. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zheng B, Peng M, Gong J, Li C, Cheng H, Li
Y and Tang Y: Circulating exosomal microRNA-4497 as a potential
biomarker for metastasis and prognosis in non-small-cell lung
cancer. Exp Biol Med (Maywood). 248:1403–1413. 2023. View Article : Google Scholar
|
|
49
|
Jin X, Chen Y, Chen H, Fei S, Chen D, Cai
X, Liu L, Lin B, Su H, Zhao L, et al: Evaluation of tumor-derived
exosomal miRNA as potential diagnostic biomarkers for early-stage
non-small cell lung cancer using next-generation sequencing. Clin
Cancer Res. 23:5311–5319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li Y, Zhao J, Yu S, Wang Z, He X, Su Y,
Guo T, Sheng H, Chen J, Zheng Q, et al: Extracellular vesicles long
RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human
blood as potential biomarkers for cancer diagnosis. Clin Chem.
65:798–808. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Liu W, Zhang H, Sun B, Chen T, Hu
M, Zhou H, Cao Y, Han B and Wu L: Extracellular vesicle long RNA
markers of early-stage lung adenocarcinoma. Int J Cancer.
152:1490–1500. 2023. View Article : Google Scholar
|
|
52
|
Wang N, Yao C, Luo C, Liu S, Wu L, Hu W,
Zhang Q, Rong Y, Yuan C and Wang F: Integrated plasma and exosome
long noncoding RNA profiling is promising for diagnosing non-small
cell lung cancer. Clin Chem Lab Med. 61:2216–2228. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Min L, Zhu T, Lv B, An T, Zhang Q, Shang
Y, Yu Z, Zheng L and Wang Q: Exosomal LncRNA RP5-977B1 as a novel
minimally invasive biomarker for diagnosis and prognosis in
non-small cell lung cancer. Int J Clin Oncol. 27:1013–1024. 2022.
View Article : Google Scholar
|
|
54
|
Li C, Lv Y, Shao C, Chen C, Zhang T, Wei
Y, Fan H, Lv T, Liu H and Song Y: Tumor-derived exosomal lncRNA
GAS5 as a biomarker for early-stage non-small-cell lung cancer
diagnosis. J Cell Physiol. 234:20721–20727. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang H, Meyer CA, Fei T, Wang G, Zhang F
and Liu XS: A systematic approach identifies FOXA1 as a key factor
in the loss of epithelial traits during the
epithelial-to-mesenchymal transition in lung cancer. BMC Genomics.
14:6802013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kuang M, Peng Y, Tao X, Zhou Z, Mao H,
Zhuge L, Sun Y and Zhang H: FGB and FGG derived from plasma
exosomes as potential biomarkers to distinguish benign from
malignant pulmonary nodules. Clin Exp Med. 19:557–564. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chang W, Zhu J, Yang D, Shang A, Sun Z,
Quan W and Li D: Plasma versican and plasma exosomal versican as
potential diagnostic markers for non-small cell lung cancer. Respir
Res. 24:1402023. View Article : Google Scholar
|
|
58
|
Yang P, Zhang Y, Zhang R, Wang Y, Zhu S,
Peng X, Zeng Y, Yang B, Pan M, Gong J and Ba H: Plasma-derived
exosomal immunoglobulins IGHV4-4 and IGLV1-40 as new non-small cell
lung cancer biomarkers. Am J Cancer Res. 13:1923–1937.
2023.PubMed/NCBI
|
|
59
|
Luo B, Que Z, Lu X, Qi D, Qiao Z, Yang Y,
Qian F, Jiang Y, Li Y, Ke R, et al: Identification of exosome
protein panels as predictive biomarkers for non-small cell lung
cancer. Biol Proced Online. 25:292023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hoshino A, Kim HS, Bojmar L, Gyan KE,
Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H,
Heissel S, et al: Extracellular vesicle and particle biomarkers
define multiple human cancers. Cell. 182:1044–1061.e18. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
An T, Qin S, Sun D, Huang Y, Hu Y, Li S,
Zhang H, Li B, Situ B, Lie L, et al: Unique protein profiles of
extracellular vesicles as diagnostic biomarkers for early and
advanced non-small cell lung cancer. Proteomics. 19:e18001602019.
View Article : Google Scholar
|
|
62
|
Schlesinger M: Role of platelets and
platelet receptors in cancer metastasis. J Hematol Oncol.
11:1252018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Denis MM, Tolley ND, Bunting M, Schwertz
H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda
KJ, et al: Escaping the nuclear confines: Signal-dependent pre-mRNA
splicing in anucleate platelets. Cell. 122:379–391. 2005.
View Article : Google Scholar
|
|
64
|
Li X, Liu L and Song X, Wang K, Niu L, Xie
L and Song X: TEP linc-GTF2H2-1, RP3-466P17.2, and lnc-ST8SIA4-12
as novel biomarkers for lung cancer diagnosis and progression
prediction. J Cancer Res Clin Oncol. 147:1609–1622. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xing S, Zeng T, Xue N, He Y, Lai YZ, Li
HL, Huang Q, Chen SL and Liu WL: Development and validation of
tumor-educated blood platelets integrin alpha 2b (ITGA2B) RNA for
diagnosis and prognosis of non-small-cell lung cancer through
RNA-seq. Int J Biol Sci. 15:1977–1992. 2019. View Article : Google Scholar
|
|
66
|
Tian T, Lu J, Zhao W, Wang Z, Xu H, Ding
Y, Guo W, Qin P, Zhu W, Song C, et al: Associations of systemic
inflammation markers with identification of pulmonary nodule and
incident lung cancer in Chinese population. Cancer Med.
11:2482–2491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zu R, Wu L, Zhou R, Wen X, Cao B, Liu S,
Yang G, Leng P, Li Y, Zhang L, et al: A new classifier constructed
with platelet features for malignant and benign pulmonary nodules
based on prospective real-world data. J Cancer. 13:2515–2527. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar :
|
|
69
|
He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi
S, Xie H, Peng X, Yin W, Tao Y and Wang X: miRNA-based biomarkers,
therapies, and resistance in Cancer. Int J Biol Sci. 16:2628–2647.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ge N, Mao C, Yang Q, Han B, Wang Y, Xu L,
Yang X, Jiao W and Li C: Single nucleotide polymorphism rs3746444
in miR-499a affects susceptibility to non-small cell lung carcinoma
by regulating the expression of CD200. Int J Mol Med. 43:2221–2229.
2019.PubMed/NCBI
|
|
71
|
Xi KX, Zhang XW, Yu XY, Wang WD, Xi KX,
Chen YQ, Wen YS and Zhang LJ: The role of plasma miRNAs in the
diagnosis of pulmonary nodules. J Thorac Dis. 10:4032–4041. 2018.
View Article : Google Scholar
|
|
72
|
He Y, Ren S, Wang Y, Li X, Zhou C and
Hirsch FR: Serum microRNAs improving the diagnostic accuracy in
lung cancer presenting with pulmonary nodules. J Thorac Dis.
10:5080–5085. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Shen J, Liu Z, Todd NW, Zhang H, Liao J,
Yu L, Guarnera MA, Li R, Cai L, Zhan M and Jiang F: Diagnosis of
lung cancer in individuals with solitary pulmonary nodules by
plasma microRNA biomarkers. BMC Cancer. 11:3742011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fan L, Sha J, Teng J, Li D, Wang C, Xia Q,
Chen H, Su B and Qi H: Evaluation of serum paired MicroRNA ratios
for differential diagnosis of non-small cell lung cancer and benign
pulmonary diseases. Mol Diagn Ther. 22:493–502. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Huang Z, Wang Z, Xia H, Ge Z, Yu L, Li J,
Bao H, Liang Z, Cui Y and Xu Y: Long noncoding RNA HAND2-AS1: A
crucial regulator of malignancy. Clin Chim Acta. 539:162–169. 2023.
View Article : Google Scholar
|
|
76
|
Karger A, Mansouri S, Leisegang MS,
Weigert A, Günther S, Kuenne C, Wittig I, Zukunft S, Klatt S,
Aliraj B, et al: ADPGK-AS1 long noncoding RNA switches macrophage
metabolic and phenotypic state to promote lung cancer growth. EMBO
J. 42:e1116202023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen X, Zhu X, Yan W, Wang L, Xue D, Zhu
S, Pan J, Li Y, Zhao Q and Han D: Serum lncRNA THRIL predicts
benign and malignant pulmonary nodules and promotes the progression
of pulmonary malignancies. BMC Cancer. 23:7552023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jiang N, Meng X, Mi H, Chi Y, Li S, Jin Z,
Tian H, He J, Shen W, Tian H, et al: Circulating lncRNA XLOC_009167
serves as a diagnostic biomarker to predict lung cancer. Clin Chim
Acta. 486:26–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Fackche NT, Mei Y, Ito T, Garner M and
Brock M: Abstract 1822: A mitochondrial pfeRNA associates with far
upstream element binding protein 1 (FUBP1) to promote lung
adenocarcinoma tumorigenesis. Cancer Res. 79(Suppl 13): S18222019.
View Article : Google Scholar
|
|
80
|
Brock M and Mei Y: Protein functional
effector sncRNAs (pfeRNAs) in lung cancer. Cancer Lett.
403:138–143. 2017. View Article : Google Scholar
|
|
81
|
Liu W, Wang Y, Huang H, Fackche N, Rodgers
K, Lee B, Nizam W, Khan H, Lu Z, Kong X, et al: A cost-effective
and non-invasive pfeRNA-based test differentiates benign and
suspicious pulmonary nodules from malignant ones. Noncoding RNA.
7:802021.
|
|
82
|
Ponomaryova AA, Rykova EY, Solovyova AI,
Tarasova AS, Kostromitsky DN, Dobrodeev AY, Afanasiev SA and
Cherdyntseva NV: Genomic and transcriptomic research in the
discovery and application of colorectal cancer circulating markers.
Int J Mol Sci. 24:124072023. View Article : Google Scholar :
|
|
83
|
Schwarzenbach H, Hoon DSB and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar
|
|
84
|
Jiang N, Zhou J, Zhang W, Li P, Liu Y, Shi
H, Zhang C, Wang Y, Zhou C, Peng C, et al: RNF213 gene mutation in
circulating tumor DNA detected by targeted next-generation
sequencing in the assisted discrimination of early-stage lung
cancer from pulmonary nodules. Thorac Cancer. 12:181–193. 2021.
View Article : Google Scholar
|
|
85
|
Peng M, Xie Y, Li X, Qian Y, Tu X, Yao X,
Cheng F, Xu F, Kong D, He B, et al: Resectable lung lesions
malignancy assessment and cancer detection by ultra-deep sequencing
of targeted gene mutations in plasma cell-free DNA. J Med Genet.
56:647–653. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Hung CS, Wang SC, Yen YT, Lee TH, Wen WC
and Lin RK: Hypermethylation of CCND2 in lung and breast cancer is
a potential biomarker and drug target. Int J Mol Sci. 19:30962018.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liang W, Zhao Y, Huang W, Gao Y, Xu W, Tao
J, Yang M, Li L, Ping W, Shen H, et al: Non-invasive diagnosis of
early-stage lung cancer using high-throughput targeted DNA
methylation sequencing of circulating tumor DNA (ctDNA).
Theranostics. 9:2056–2070. 2019. View Article : Google Scholar :
|
|
88
|
Fang Y, Qu Y, Ji L, Sun H, Li J, Zhao Y,
Liang F, Wang Z, Su J, Liu J, et al: Novel blood-based FUT7 DNA
methylation is associated with lung cancer: Especially for lung
squamous cell carcinoma. Clin Epigenetics. 14:1672022. View Article : Google Scholar
|
|
89
|
Crowley E, Di Nicolantonio F, Loupakis F
and Bardelli A: Liquid biopsy: Monitoring cancer-genetics in the
blood. Nat Rev Clin Oncol. 10:472–484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chen C, Huang X, Yin W, Peng M, Wu F, Wu
X, Tang J, Chen M, Wang X, Hulbert A, et al: Ultrasensitive DNA
hypermethylation detection using plasma for early detection of
NSCLC: A study in Chinese patients with very small nodules. Clin
Epigenetics. 12:392020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhao Y, O'Keefe CM, Hsieh K, Cope L, Joyce
SC, Pisanic TR, Herman JG and Wang TH: Multiplex digital
methylation-specific PCR for noninvasive screening of lung cancer.
Adv Sci (Weinh). 10:e22065182023. View Article : Google Scholar
|
|
92
|
Wang Z, Xie K, Zhu G, Ma C, Cheng C, Li Y,
Xiao X, Li C, Tang J, Wang H, et al: Early detection and
stratification of lung cancer aided by a cost-effective assay
targeting circulating tumor DNA (ctDNA) methylation. Respir Res.
24:1632023. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xing W, Sun H, Yan C, Zhao C, Wang D, Li M
and Ma J: A prediction model based on DNA methylation biomarkers
and radiological characteristics for identifying malignant from
benign pulmonary nodules. BMC Cancer. 21:2632021. View Article : Google Scholar
|
|
94
|
He J, Wang B, Tao J, Liu Q, Peng M, Xiong
S, Li J, Cheng B, Li C, Jiang S, et al: Accurate classification of
pulmonary nodules by a combined model of clinical, imaging, and
cell-free DNA methylation biomarkers: A model development and
external validation study. Lancet Digit Health. 5:e647–e656. 2023.
View Article : Google Scholar
|
|
95
|
Seijo LM, Peled N, Ajona D, Boeri M, Field
JK, Sozzi G, Pio R, Zulueta JJ, Spira A, Massion PP, et al:
Biomarkers in lung cancer screening: achievements, promises, and
challenges. J Thorac Oncol. 14:343–357. 2019. View Article : Google Scholar
|
|
96
|
Zhang X, Liu M, Zhang X, Wang Y and Dai L:
Autoantibodies to tumor-associated antigens in lung cancer
diagnosis. Adv Clin Chem. 103:1–45. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lastwika KJ, Kargl J, Zhang Y, Zhu X, Lo
E, Shelley D, Ladd JJ, Wu W, Kinahan P, Pipavath SNJ, et al:
Tumor-derived autoantibodies identify malignant pulmonary nodules.
Am J Respir Crit Care Med. 199:1257–1266. 2019. View Article : Google Scholar :
|
|
98
|
Xu L, Chang N, Yang T, Lang Y, Zhang Y,
Che Y, Xi H, Zhang W, Song Q, Zhou Y, et al: Development of
diagnosis model for early lung nodules based on a seven
autoantibodies panel and imaging features. Front Oncol.
12:8835432022. View Article : Google Scholar
|
|
99
|
Auger C, Moudgalya H, Neely MR, Stephan
JT, Tarhoni I, Gerard D, Basu S, Fhied CL, Abdelkader A, Vargas M,
et al: Development of a novel circulating autoantibody biomarker
panel for the identification of patients with 'actionable'
pulmonary nodules. Cancers (Basel). 15:22592023. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Shome M, Gao W, Engelbrektson A, Song L,
Williams S, Murugan V, Park JG, Chung Y, LaBaer J and Qiu J:
Comparative microbiomics analysis of antimicrobial antibody
response between patients with lung cancer and control subjects
with benign pulmonary nodules. Cancer Epidemiol Biomarkers Prev.
32:496–504. 2023. View Article : Google Scholar
|
|
101
|
Sina AAI, Vaidyanathan R, Dey S,
Carrascosa LG, Shiddiky MJA and Trau M: Real time and label free
profiling of clinically relevant exosomes. Sci Rep. 6:304602016.
View Article : Google Scholar
|
|
102
|
Fang S, Tian H, Li X, Jin D, Li X, Kong J,
Yang C, Yang X, Lu Y, Luo Y, et al: Clinical application of a
microfluidic chip for immunocapture and quantification of
circulating exosomes to assist breast cancer diagnosis and
molecular classification. PLoS One. 12:e01750502017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Morales-Pacheco M, Valenzuela-Mayen M,
Gonzalez-Alatriste AM, Mendoza-Almanza G, Cortés-Ramírez SA,
Losada-García A, Rodríguez-Martínez G, González-Ramírez I,
Maldonado-Lagunas V, Vazquez-Santillan K, et al: The role of
platelets in cancer: From their influence on tumor progression to
their potential use in liquid biopsy. Biomark Res. 13:272025.
View Article : Google Scholar
|
|
104
|
Didychuk AL, Butcher SE and Brow DA: The
life of U6 small nuclear RNA, from cradle to grave. RNA.
24:437–460. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Najafi S, Asemani Y, Majidpoor J, Mahmoudi
R, Aghaei-Zarch SM and Mortezaee K: Tumor-educated platelets. Clin
Chim Acta. 552:1176902024. View Article : Google Scholar
|
|
106
|
Khan J, Lieberman JA and Lockwood CM:
Variability in, variability out: best practice recommendations to
standardize pre-analytical variables in the detection of
circulating and tissue microRNAs. Clin Chem Lab Med. 55:608–621.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Sánchez-Herrero E, Provencio M and Romero
A: Clinical utility of liquid biopsy for the diagnosis and
monitoring of EML4-ALK NSCLC patients. Adv Lab Med.
1:201900192020.
|
|
108
|
Lennon AM, Buchanan AH, Kinde I, Warren A,
Honushefsky A, Cohain AT, Ledbetter DH, Sanfilippo F, Sheridan K,
Rosica D, et al: Feasibility of blood testing combined with PET-CT
to screen for cancer and guide intervention. Science.
369:eabb96012020. View Article : Google Scholar :
|
|
109
|
Cescon DW, Bratman SV, Chan SM and Siu LL:
Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer.
1:276–290. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Chaudhuri AA, Chabon JJ, Lovejoy AF,
Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL,
Zhou L, et al: Early detection of molecular residual disease in
localized lung cancer by circulating tumor DNA profiling. Cancer
Discov. 7:1394–1403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Fagery M, Khorshidi HA, Wong SQ, Vu M and
Ijzerman M: Health economic evidence and modeling challenges for
liquid biopsy assays in cancer management: A systematic literature
review. PharmacoEconomics. 41:1229–1248. 2023. View Article : Google Scholar :
|
|
112
|
Kammer MN, Lakhani DA, Balar AB, Antic SL,
Kussrow AK, Webster RL, Mahapatra S, Barad U, Shah C, Atwater T, et
al: Integrated biomarkers for the management of indeterminate
pulmonary nodules. Am J Respir Crit Care Med. 204:1306–1316. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar
|
|
115
|
Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim
HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WSME, et
al: Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung
cancer. N Engl J Med. 376:629–640. 2017. View Article : Google Scholar
|
|
116
|
Gandara DR, Paul SM, Kowanetz M,
Schleifman E, Zou W, Li Y, Rittmeyer A, Fehrenbacher L, Otto G,
Malboeuf C, et al: Blood-based tumor mutational burden as a
predictor of clinical benefit in non-small-cell lung cancer
patients treated with atezolizumab. Nat Med. 24:1441–1448. 2018.
View Article : Google Scholar
|
|
117
|
Oxnard GR, Paweletz CP, Kuang Y, Mach SL,
O'Connell A, Messineo MM, Luke JJ, Butaney M, Kirschmeier P,
Jackman DM and Jänne PA: Noninvasive detection of response and
resistance in EGFR-mutant lung cancer using quantitative
next-generation genotyping of cell-free plasma DNA. Clin Cancer
Res. 20:1698–1705. 2014. View Article : Google Scholar
|
|
118
|
Rolfo C, Mack P, Scagliotti GV, Aggarwal
C, Arcila ME, Barlesi F, Bivona T, Diehn M, Dive C, Dziadziuszko R,
et al: Liquid biopsy for advanced NSCLC: A consensus statement from
the international association for the study of lung cancer. J
Thorac Oncol. 16:1647–1662. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Pennell NA, Mutebi A, Zhou ZY, Ricculli
ML, Tang W, Wang H, Guerin A, Arnhart T, Dalal A, Sasane M, et al:
Economic impact of next-generation sequencing versus single-gene
testing to detect genomic alterations in metastatic non-small-cell
lung cancer using a decision analytic model. JCO Precis Oncol.
3:1–9. 2019. View Article : Google Scholar
|
|
120
|
Leighl NB, Page RD, Raymond VM, Daniel DB,
Divers SG, Reckamp KL, Villalona-Calero MA, Dix D, Odegaard JI,
Lanman RB and Papadimitrakopoulou VA: Clinical utility of
comprehensive cell-free DNA analysis to identify genomic biomarkers
in patients with newly diagnosed metastatic non-small cell lung
cancer. Clin Cancer Res. 25:4691–4700. 2019. View Article : Google Scholar
|
|
121
|
Xie H, Jia Y and Liu S: Integration of
artificial intelligence in clinical laboratory medicine:
Advancements and challenges. Interdiscip Med. 2:e202300562024.
View Article : Google Scholar
|