Introduction
Glioma, the most common malignant tumor of the
central nervous system, poses a significant challenge in the field
of neuro-oncology due to its high invasiveness and poor response to
conventional treatments (1).
Despite advancements in surgical interventions, radiotherapy and
pharmacotherapy, the prognosis of glioma remains unfavorable, with
a short survival period (2). In
recent years, researchers have explored targeted therapies and
immunotherapies in hopes of improving treatment outcomes and
survival rates for these patients (3). However, despite progress in
existing treatment modalities, the high recurrence rate and
therapeutic resistance of gliomas significantly complicate their
clinical management (4). Hence,
investigating these molecular mechanisms not only aids in
elucidating the biological characteristics of gliomas but also
provides a theoretical basis for the development of new targeted
therapeutic strategies.
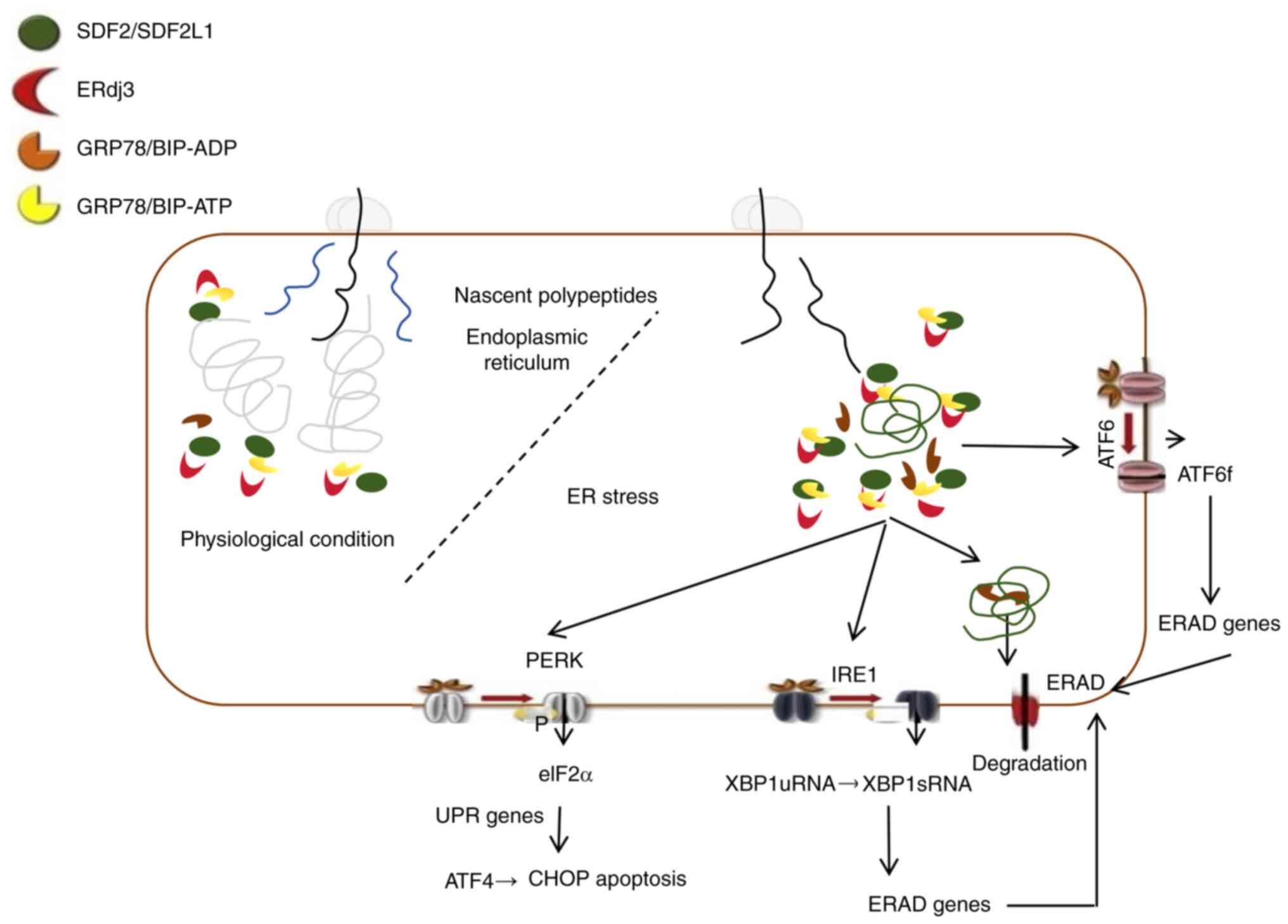
The endoplasmic reticulum (ER) is a continuous
network of membrane-bound tubular structures within the cytoplasm
that is primarily responsible for the folding, modification,
transport and degradation of proteins (5). ER-associated degradation (ERAD) is
a highly conserved quality control mechanism that plays a crucial
role in identifying and disposing of misfolded proteins by
retro-translocating them into the cytoplasm for proteasomal
degradation (6). This process
involves several key steps, including substrate recognition,
extraction, polyubiquitination, and subsequent degradation via
ERAD, in which molecular chaperones and lectins recognize unfolded
or misfolded nascent polypeptides (7). These misfolded proteins bind to
chaperones and are then delivered to ERAD ligands (7,8).
Ultimately, the polyubiquitinated substrates are translocated
through the retro-translocation channel into the cytoplasm, where
they are degraded by the 26S proteasome (8,9).
Glucose-regulated protein 78 (GRP78, Bip), a prominent ER
chaperone, not only assists in protein folding but also plays a
pivotal role in the ERAD pathway by recognizing misfolded proteins
and facilitating their retro-translocation, highlighting its
importance in maintaining ER homeostasis and cellular health
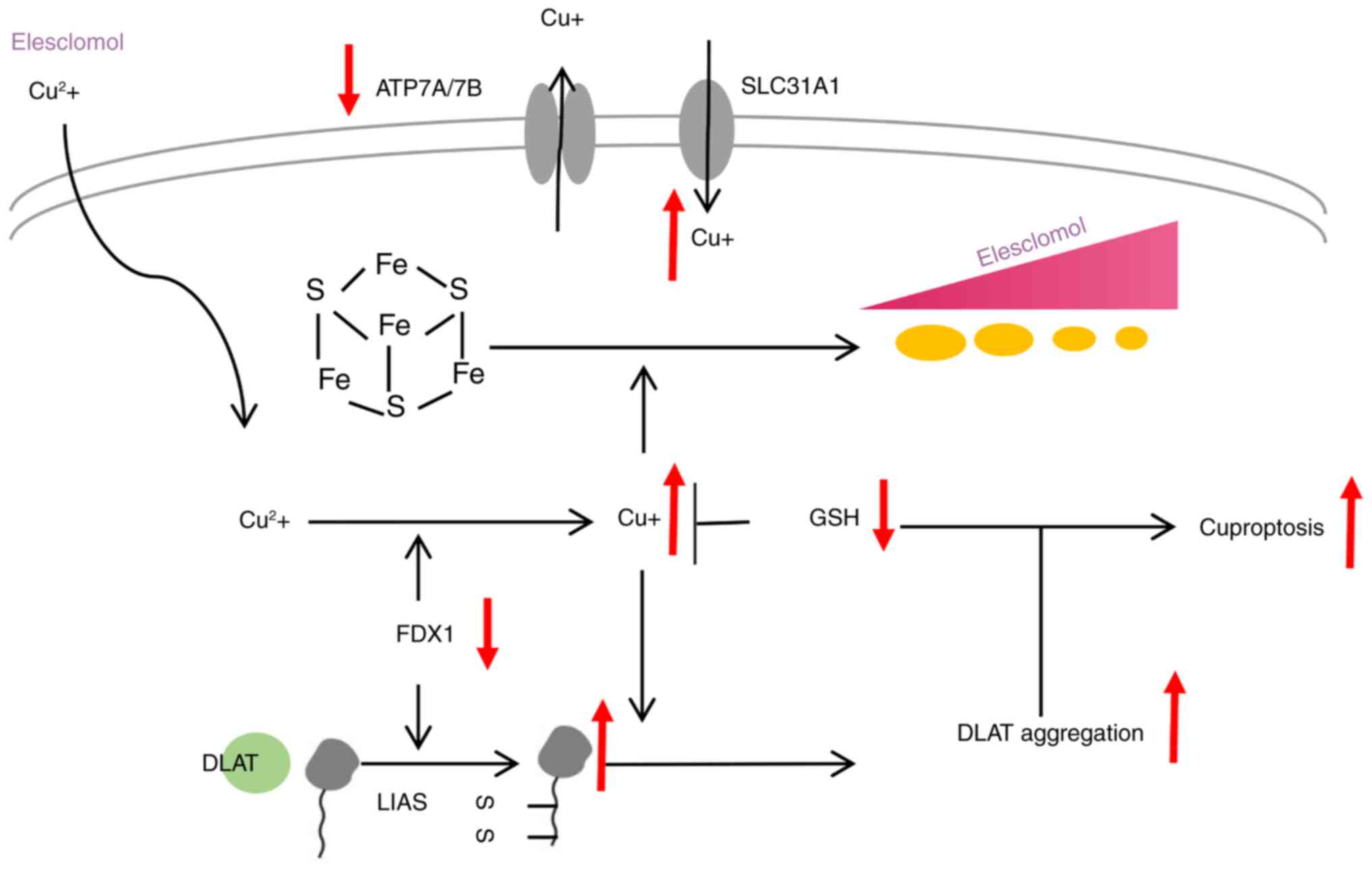
(Fig. 1) (10). Cuproptosis is a recently
identified form of programmed cell death characterized by the
accumulation of copper ions, which bind to lipoylated proteins in
the mitochondrial tricarboxylic acid cycle (Fig. 2). This interaction leads to
protein aggregation, loss of iron-sulfur cluster proteins,
proteotoxic stress, and ultimately, cell death. Emerging evidence
indicates a significant interplay between cuproptosis and ER stress
pathways (11,12). In cervical cancer, the
curcuminoid analog PBPD
(1-propyl-3,5-bis(2-bromobenzylidene)-4-piperidinone) has been
shown to induce cuproptosis by modulating the
Notch1/RBP-J/NRF2/FDX1 signaling axis (11). Specifically, PBPD suppresses
Notch1 and RBP-J expression, leading to decreased NRF2 activity and
subsequent upregulation of FDX1. This cascade results in increased
oxidative stress, activation of ER stress responses, and induction
of cuproptosis in cervical cancer cells. In lung adenocarcinoma, it
has been demonstrated that copper accumulation activates the
unfolded protein response (UPR), particularly through the spliced
form of X-box binding protein 1 (XBP1s) (12). Hence, targeting the molecular
crosstalk between ER stress and copper-induced cell death pathways
may offer novel therapeutic strategies for cancers characterized by
disrupted copper homeostasis and ER stress responses.
Stromal cell-derived factor 2 (SDF2) acts as an ER
chaperone protein, primarily facilitating protein folding,
enhancing chaperone activity, maintaining ER functionality, and
regulating stress responses (13). SDF2 forms a complex with ER DnaJ
heat shock protein 3 (ERdj3), helping to maintain the folded state
of newly synthesized proteins and preventing misfolding and
aggregation (14). Its presence
enhances the chaperone activity of ERdj3, making it more effective
at inhibiting protein aggregation under normal physiological
conditions. By associating with ERdj3 and SDF2-like protein 1, SDF2
contributes to the maintenance of ER quality control mechanisms,
ensuring the proper function of intracellular proteins (14). Under ER stress conditions, the
role of SDF2 may be linked to the secretion of ERdj3 and its
extracellular functions, aiding cells in addressing the challenges
of protein folding (15).
Therefore, the primary function of SDF2 in the ER is to serve as a
molecular chaperone that promotes and maintains the correct folding
of proteins, thereby ensuring normal physiological activities and
stress responses in cells (Fig.
1) (15). However, its role
in glioma remains unexplored. The objective of the present study
was to investigate the specific functions and mechanisms of SDF2 in
glioma development and progression.
Materials and methods
Cell culture
The glial cell lines U87 (cat. no. CL-0238), U251
(cat. no. CL-0237) and HMC3 (cat. no. CL-0620; all from Procell
Life Science & Technology Co., Ltd.) were cultured in complete
MEM (HyClone; Cytiva) supplemented with 10% fetal bovine serum
(FBS), 1% non-essential amino acids, sodium pyruvate and
penicillin-streptomycin. The cells were maintained in a humidified
incubator at 37°C with 5% CO2. When the cells reached
~85% confluence, they were passaged using trypsinization. All cell
lines were authenticated using STR profiling.
MG132 treatment
To investigate the involvement of the proteasome
degradation pathway in the downregulation of ATP7A and ATP7B, U87
and U251 glioma cells were treated with the proteasome inhibitor
MG132. Specifically, cells were incubated with MG132 (cat. no.
HY-13259; MedChemExpress) at a final concentration of 10 μM
for 24 h.
Western blotting
Protein extraction was performed from cells or
tissue samples using RIPA lysis buffer (Beijing Solarbio Science
& Technology Co., Ltd.). The lysates were centrifuged at 10,000
× g for 15 min to remove cellular debris, and the supernatant
containing the total protein was collected. Protein concentrations
were quantified using a BCA assay. Equal amounts of protein samples
(15 μg/lane) were then subjected to 10% SDS-PAGE. Following
electrophoresis, the proteins were transferred onto a PVDF membrane
(MilliporeSigma). The membrane was then blocked with 5% non-fat
milk or BSA (cat. no. A8010; Beijing Solarbio Science &
Technology Co., Ltd.) in TBST (TBS with 0.1% Tween-20) for 1 h at
room temperature. The membrane was incubated overnight at 4°C with
primary antibodies specific to the target proteins SDF2 (cat. no.
PK47764), SDF2L1 (cat. no. PK17377), ERdj3 (cat. no. PA2683), GRP78
(cat. no. T55166), ATP7A (cat. no. PA7106), ATP7B (cat. no.
TA0410), dihydrolipoamide S-acetyltransferase (DLAT; cat. no.
T58125) and GAPDH (cat. no. M20006; 1:1,000; all purchased from
Abmart Pharmaceutical Technology Co., Ltd.). Lipoic acid (cat. no.
437695) was purchased from MilliporeSigma. After washing with TBST
to remove unbound primary antibodies, the membrane was incubated
with HRP-conjugated secondary antibodies (cat. no. ZB-2301 for
rabbit; cat. no. ZB-2305 for mouse) Beijing Solarbio Science &
Technology Co., Ltd.) for 1 h at room temperature. Following
several washes with TBST, the bound antibodies were visualized
using an enhanced chemiluminescence (ECL) detection system (Beijing
Solarbio Science & Technology Co., Ltd.). The intensity of each
protein band was quantified using ImageJ analysis software (version
10; Media Cybernetics, Inc.). GAPDH was used as an internal
control.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using TRIzol
reagent, and cDNA was synthesized following the instructions
provided with the PrimeScript™ RT Master Mix (Perfect Real Time)
(Takara Bio, Inc.). The reverse transcription conditions were set
at 37°C for 15 min, followed by 85°C for 5 sec and then 4°C
indefinitely. For qPCR, a 20-μl reaction mixture consisting
of 10 μl of Applied Biosystems™ SYBR Green Master Mix
(Thermo Fisher Scientific, Inc.), 1 μl each of forward and
reverse primers, 2 μl of cDNA, and 6 μl of
ddH2O was prepared. The PCR protocol included initial
denaturation at 95°C for 30 sec, followed by 40 cycles of
amplification with denaturation at 95°C for 5 sec, annealing at
60°C for 30 sec, and extension at 72°C for 30 sec. qPCR was
conducted on a LightCycler 480 system (Roche Diagnostics) to
quantify gene expression. The primers used in the present study
were listed as follows: SDF2 forward, 5′-GGAGCTTGGCATCATGGACT-3′
and reverse, 5′-GGAGCTTCATAGCGTCTCCA-3′; CHOP forward,
5′-ATGAACGGCTCAAGCAGGAA-3′ and reverse, 5′-GGGAAAGGTGGGTAGTGTGG;
GRP78 forward, 5′-TCAGGCCAAGCCCAATACAG-3′ and reverse,
5′-TCCACGGTAGTGAGAGCCTT-3′; XBP1s forward,
5′-TGCTGAGTCCGCAGCAGGTG-3′ and reverse, 5′-GCTGGCAGGCTCTGGGGAAG-3′;
XBP1u forward, 5′-ACGGGACCCCTAAAGTTCTG-3′ and reverse,
5′-TGCACGTAGTCTGAGTGCTG-3′; and GAPDH forward,
5′-CCAGCAAGAGCACAAGAGGA-3′ and reverse, 5′-GGGGAGATTCAGTGTGGTGG-3′.
The 2−ΔΔCq method was used for quantification assay
(16).
Human samples
Human tumor and adjacent non-tumor tissue samples
were obtained from patients with glioma during surgical procedures,
with informed consent obtained from all participants. A total of 10
patients were enrolled in the present study, including 4 females
and 6 males. The median age was 66 years (range, 53-67 years).
Ethical approval for sample collection and study was granted by the
institutional review board of the First Affiliated Hospital of
Xi'an Jiaotong University (approval no. 2024SF-FAHX-212; Xi'an,
China).
Additionally, the GEPIA database (http://gepia.cancer-pku.cn/) was used to analyze SDF2
mRNA expression levels across various tissue types and pathological
conditions. Statistical significance was assessed using GEPIA's
integrated analytical tools, providing insights into the potential
role of SDF2 in disease pathology.
Detection of malondialdehyde (MDA),
reduced glutathione (GSH) and adenosine triphosphate (ATP)
Cells from each group were collected, and the levels
of MDA, reduced GSH and ATP were measured using the appropriate
assay kits in strict accordance with the manufacturers' protocols.
The lipid peroxidation level was assessed using an MDA Assay kit
(cat. no. S0131S), ATP levels were quantified using an ATP Assay
kit (cat. no. S0026), and GSH and oxidized glutathione (GSSG)
levels were measured using a GSH and GSSG Assay kit (cat. no.
S0053) (all from Beyotime Institute of Biotechnology).
EdU (5-Ethynyl-2′-deoxyuridine)
detection
The procedure was conducted using a BeyoClick EdU
Cell Proliferation Kit (DAB method) (cat. no. C0085S; Beyotime
Institute of Biotechnology). Briefly, cells were incubated in
medium containing 10 μM EdU for 2 h. After EdU labeling, the
medium was removed, and 1 ml of 4% paraformaldehyde (PFA; cat. no.
P0099; Beyotime Institute of Biotechnology) was added to each well
before incubation at room temperature for 15 min. The fixative was
then removed, and the cells were washed three times with 1 ml of
wash solution per well, with each wash lasting 5 min. Following the
wash steps, 1 ml of PBS containing 0.3% Triton X-100 was added to
each well, and the cells were incubated at room temperature for 15
min to permeabilize the cells. Next, 0.5 ml of Click reaction
solution was added to each well, and the cells were incubated in
the dark at room temperature for 30 min. The Click reaction
solution was then aspirated, and the cells were washed three times
with wash solution, with each wash lasting 5 min. Finally, the
cells were mounted with antifade mounting medium containing DAPI (a
final concentration of 1 μg/ml) and observed under a
fluorescence microscope.
Transwell assay
The bottom membrane of the Transwell insert
(Corning, Inc.) was coated with Matrigel matrix (Corning, Inc.) at
37°C for 1 h. U87 and U251 glioma cells, which were cultured to
~85% confluence, were harvested using trypsinization and
centrifuged at 1,000 × g for 15 min. The cell density was adjusted
to 5×103 cells/100 μl, and the cells were
resuspended in serum-free medium to create a single-cell
suspension. Transwell inserts (8 μm pore size) were placed
in a 24-well plate, 600 μl of serum-containing medium was
added to the lower chamber to act as a chemoattractant, and 200
μl of the treated cell suspension was added to the upper
chamber. After 24 h of incubation, the inserts were removed, and
the remaining medium was aspirated. The inserts were washed with
PBS and fixed with 4% PFA for 30 min at room temperature, followed
by staining with 0.1% crystal violet for 15 min. The cells that had
migrated through the membrane were visualized and images were
captured under an inverted light microscope (Keyence Corporation).
The number of invasive cells was quantified using ImageJ
software.
Flow cytometric analysis of apoptosis
using Annexin V-PE/7-AAD staining
Cells in the logarithmic growth phase were harvested
by trypsinization, collected in centrifuge tubes, and resuspended
in PBS. After the cells were counted, 5×105 cells were
collected and washed with PBS. The cells were then resuspended in
binding buffer from an Annexin V-PE/7-AAD Apoptosis Detection Kit
(cat. no. E-CK-A216; Elabscience Biotechnology, Inc.) and incubated
in the dark at room temperature for 20 min. Following incubation,
the cells were washed, resuspended and analyzed for apoptosis by
flow cytometry (CytoFLEX; Beckman Coulter, Inc.). The data were
analyzed using FlowJo 10 (FlowJo LLC). The experiment was performed
in triplicate for statistical reliability.
Quantification of intracellular reactive
oxygen species (ROS) levels by flow cytometry
Intracellular ROS levels were measured by flow
cytometry using a fluorescent probe-based ROS detection kit (Wuhan
Servicebio Technology Co., Ltd.). Briefly, the cells were washed
twice with PBS, centrifuged at 800 × g for 5 min to remove the
supernatant at room temperature, and collected. DCFH-DA working
solution was then added to each sample at a density of
1×105 cells/ml, and the cells were incubated at 37°C in
a CO2 incubator protected from light for 30 min. After
incubation, the cells were centrifuged at 800 × g for 5 min to
remove the DCFH-DA solution, followed by washing with PBS 3 times
to ensure the removal of excess probe. The cells were finally
resuspended in PBS and immediately analyzed by flow cytometry for
ROS detection.
siRNA transfection
For transfection, cells were seeded in 6-well plates
at a density of 4×105 cells per well 24 h prior to
transfection to achieve optimal confluency. Small interfering RNA
(siRNA, 5′-AAAUAGAACAUUUUGAAGGUG-3′) targeting the gene of interest
and a non-targeting control siRNA (NC, 5′-UUCUCCGAACGUGUCACGUTT-3′)
were obtained from Shanghai GenePharma Co., Ltd. Transfection was
performed using HiPerFect Transfection Reagent (cat. no. 301705;
Qiagen, Inc.) following the manufacturer's protocol. Briefly, 150
ng of siRNA (final concentration of 10 nM) was diluted in 100
μl of serum-free DMEM. Subsequently, 6 μl of
HiPerFect Transfection Reagent was added to the diluted siRNA
solution, mixed gently, and incubated at room temperature for 10
min to allow complex formation. The siRNA-lipid complexes were then
added dropwise to each well containing cells in 2 ml of complete
growth medium. Cells were assessed 48 h post-transfection.
Cell Counting Kit-8 (CCK-8) assay
U87 and U251 cells were adjusted to a density of
1,000 cells/μl and seeded into a 96-well plate. The cells
were incubated at 37°C in a 5% CO2 incubator for 24 h.
After incubation, the medium was removed, and the wells were washed
with PBS. Fresh medium (100 μl) was added to each well,
followed by 10 μl of CCK-8 solution (Beijing Solarbio
Science & Technology Co., Ltd.). The plate was then incubated
for an additional 1 h. Absorbance was measured at 450 nm using a
microplate reader to assess cell viability. The experiments
included the following groups: Ad-NC and Ad-shSDF2. To investigate
the mechanisms underlying the reduction in cell viability induced
by Ad-shSDF2, several inhibitors were added to the Ad-shSDF2 group:
z-VAD (20 μM), Nec-1 (10 μM), VX-765 (10 μM),
Fer-1 (1 μM), and TTM (10 μM). Cell viability was
calculated based on absorbance (A450) values, allowing for the
assessment of each inhibitor's effect on Ad-shSDF2-induced cell
viability reduction and identification of the specific cell death
pathways involved.
Detection of copper ion content
The intracellular copper ion content was quantified
using a Copper (Cu2+) Colorimetric Assay kit
(Elabscience Biotechnology, Inc.). For the assay, 15 μl of
each of the eight copper standards at different concentrations, as
well as 15 μl of each test sample, were added to individual
wells. Then, 230 μl of chromogenic working solution was
added to each well. The plates were covered with sealing film and
incubated at 37°C for 5 min. The optical density (OD) of each well
was measured at 580 nm using a microplate reader.
Detection of intracellular copper
ions
The intracellular copper ion distribution was
visualized using the fluorescent probe Coppersensor 1
(MedChemExpress). Cells were incubated with the probe at a final
concentration of 5 μM in each well, protected from light, at
37°C for 30 min. After incubation, the cells were observed under a
fluorescence microscope to assess the localization and intensity of
copper ion fluorescence.
Detection of mitochondrial ROS
Mitochondrial ROS levels were detected using the
specific fluorescent probe MitoTracker Red CMXRos (Beyotime
Institute of Biotechnology). Once the cells reached the desired
density, the culture medium was removed and replaced with
MitoTracker Red CMXRos working solution at a final concentration of
1 μM. The cells were incubated at 37°C for 30 min. After
incubation, the working solution was removed, and prewarmed fresh
culture medium was added. Fluorescence was observed using a
fluorescence microscope to assess the mitochondrial ROS levels.
Immunofluorescence (IF)
U87 and U251 cells were seeded in six-well plates at
a density of 5×105 cells per well and incubated at 37°C
with 5% CO2 until adherence. After 24 h, the cells were
fixed with 500 μl of 4% PFA for 30 min, followed by
permeabilization with 500 μl of 0.1% Triton X-100 for 30
min. The cells were then blocked with IF blocking buffer for 1 h.
After the blocking solution was removed, a mixture of primary
antibodies against TOM20 and DLAT (Abcam) was added (50 μl
per well), and the cells were incubated overnight at 4°C. The next
day, the primary antibodies were removed, and the cells were
incubated with the appropriate secondary antibodies (500 μl
per well) at room temperature, protected from light, for 1 h. DAPI
staining solution (a final concentration of 1 μg/ml) was
then applied for 5 min to stain the nuclei. Images were captured
and stored using an inverted fluorescence microscope.
Co-immunoprecipitation (co-IP)
For these experiments, a co-IP kit (cat. no. P0401;
GENESEED; https://www.geneseed.com.cn/web/lxwm.html) was used.
Briefly, protein extracts were prepared using RIPA lysis buffer
(Beijing Solarbio Science & Technology Co., Ltd.). A specific
antibody against GRP78 was added to the protein samples to allow
binding with the target protein, and the samples were incubated at
4°C for several hours or overnight. 30 μl of protein A or G
magnetic beads (50% concentration) were then added to bind the
antibody-protein complexes, which were collected using magnetic
separation. The beads were washed multiple times with lysis buffer
to remove non-specifically bound proteins. The immunoprecipitated
proteins were then eluted from the beads by heating or by adding an
elution buffer. Finally, the eluted proteins were separated by
SDS-PAGE and analyzed by western blotting to detect
co-immunoprecipitated proteins. By normalizing the Co-IP results to
GAPDH levels, any potential discrepancies in protein loading were
accounted for and it was ensured that the observed decrease in
ATP7A and ATP7B co-precipitation was specifically attributable to
the reduced availability of GRP78 for interaction, rather than
other experimental variations.
Construction of adenoviral vectors
Adenoviral vectors were constructed by Vigene
Biosciences for the overexpression and knockdown of the SDF2 or
GRP78 proteins. Adenoviral vectors carrying negative control genes
(Ad-NC, UUCUCCGAACGUGUCACGUTT) or shRNA sequences (Ad-sh-SDF2,
UCUUUUUGCCCACUGAUAGGU) targeting SDF2 were designed to infect
cells. Adenoviral vectors were added at a concentration of
109 PFU/ml, with 20 μl viral suspension per well
in six-well plates. After incubation at 37°C for 24 h, the
viral-containing medium was removed, cells were washed with PBS,
and fresh medium was added.
Xenograft tumor model in mice
A total of 15 SPF-grade BALB/c nude male mice (6-8
weeks old, 18-22 g, Cyagen Biosciences; https://www.lascn.net/SupplyDemand/Site/Index.aspx?id=30)
were randomly assigned into three experimental groups (Ad-NC,
Ad-SDF2, and Ad-SDF2 + Ad-GRP78), with five mice per group, using a
computer-generated randomization schedule. This sample size was
selected based on precedent studies and accepted practices in
glioma xenograft models, where comparable group sizes have been
sufficient to detect statistically significant effects (17-19). Investigators responsible for data
collection and outcome assessment were blinded to group allocation
to minimize potential bias. All animals were acclimatized to the
laboratory environment for one week prior to experimentation and
housed under standard specific pathogen-free (SPF) conditions
[22±2°C, 55±10% humidity, 12/12-h light/dark cycle (7:00-19:00),
with ad libitum access to diet and sterile water].
Logarithmically growing U251 cells were collected from each
treatment group. Cell pellets were resuspended in 200 μl of
ice-cold PBS to maintain viability and kept on ice until injection.
Each mouse was subcutaneously injected with 1×106 cells
into the inguinal region. Tumor implantation was designated as day
0. Tumor growth and general health were monitored every two days.
Tumor volume (mm3) was measured using calipers and
calculated according to the formula: volume=1/2 × longest diameter
(mm) × [shortest diameter (mm)]2. At the experimental
endpoint, mice were euthanized by cervical dislocation.
Subcutaneous tumors were excised, weighed, and processed for
further analysis. The maximum tumor diameter observed was 8.2 mm,
corresponding to a volume of 106.64 mm3. All animal
procedures were approved by the Institutional Animal Care and Use
Committee of the Animal Experimentation Center, Xi'an Jiaotong
University (approval no. XJTUAE2023-1944; Xi'an, China).
Immunohistochemistry (IHC)
Tumor tissue sections were subjected to Ki-67 IHC.
In brief, the tissues (5 μm) were deparaffinized in xylene,
rehydrated in a graded ethanol series, and treated with 3% hydrogen
peroxide to inactivate endogenous peroxidase activity. Antigen
retrieval was performed, followed by blocking with goat serum (cat.
no. C0625; Beyotime Institute of Biotechnology). Ki-67 primary
antibody (1:200; cat. no. AB2008; Beyotime Institute of
Biotechnology) was applied, and the samples were incubated
overnight at 4°C. The sections were then incubated with
HRP-conjugated goat anti-rabbit IgG (cat. no. A0208; Beyotime
Institute of Biotechnology) at 37°C for 30 min, followed by the
application of DAB substrate for color development (30 sec).
Hematoxylin counterstaining was performed for 1 min, and
differentiation was achieved in 1% hydrochloric acid ethanol. The
sections were then dehydrated through a graded ethanol series,
cleared in xylene, and mounted with neutral resin. Images were
captured using a light microscope. In each field, the number of
Ki-67-positive tumor cell nuclei and the total number of tumor cell
nuclei were manually counted. The proliferation index was then
calculated as the percentage of Ki-67-positive cells among the
total tumor cells in each field, and the average of these five
fields was taken as the final proliferation index for the
sample.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism software (version 10.0; Dotmatics). The data are presented as
the mean ± standard deviations (SDs). For relative expression
graphs, the control group was normalized to a fixed value (for
example, 1 or 100%). For comparisons between two groups, an
unpaired two-tailed Student's t-test was employed. For comparisons
among multiple groups, one-way analysis of variance was conducted,
followed by Tukey's post hoc test to assess pairwise differences
between group means. P<0.05 was considered to indicate a
statistically significant difference.
Results
Increased expression of SDF2 in glioma
tissues
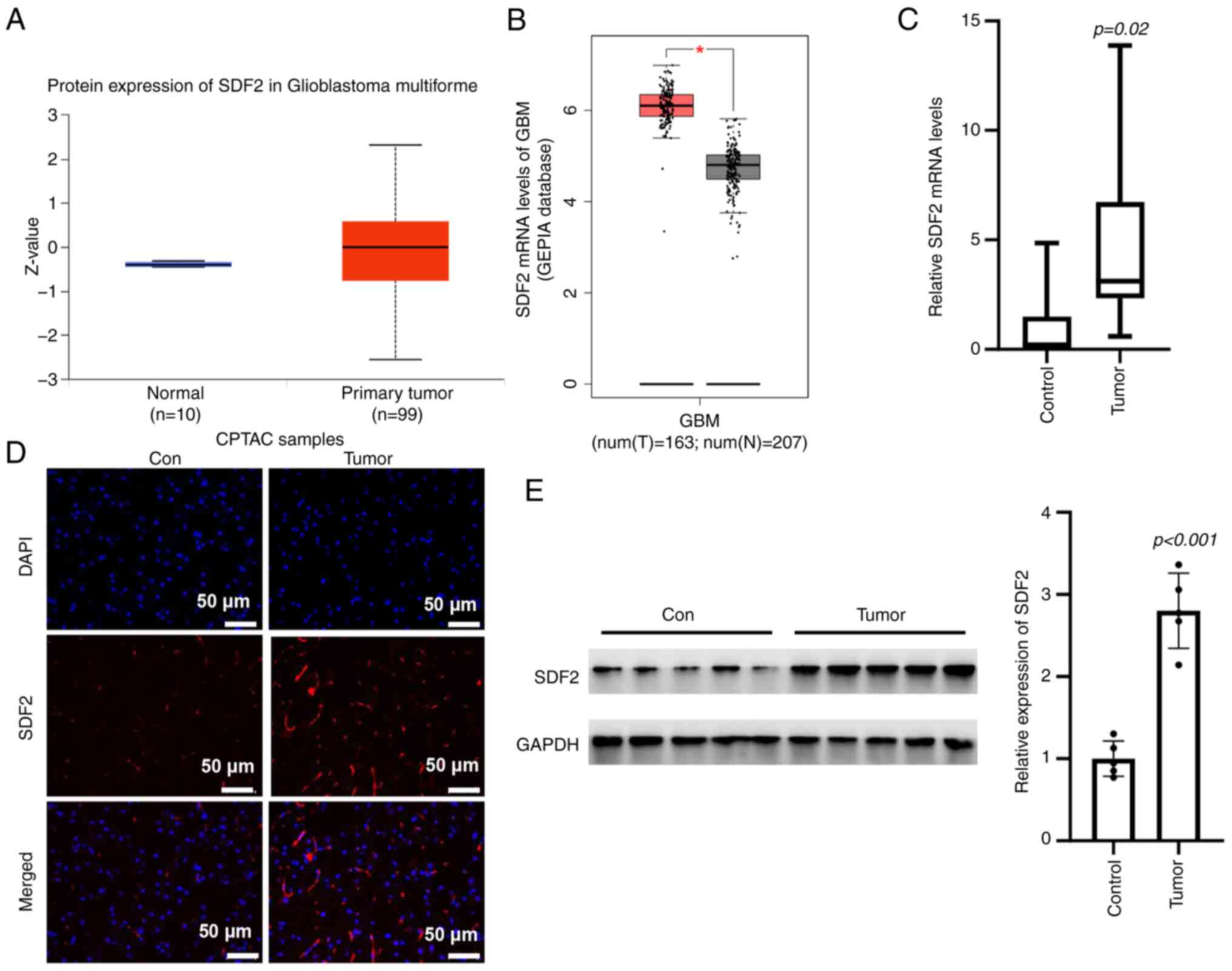
Bioinformatic predictions revealed an increase in
SDF2 mRNA expression in glioma tissues compared with control
tissues (Fig. 3A and B).
Consistently, RT-qPCR analysis revealed significantly elevated SDF2
mRNA levels in tumor tissues relative to adjacent normal tissues
(Fig. 3C). IF staining further
demonstrated an increase in SDF2 fluorescence intensity in glioma
tissues compared with control tissues (Fig. 3D). Western blot analysis
confirmed higher SDF2 protein expression in glioma samples than in
adjacent non-tumor tissues (Fig.
3E). These findings collectively suggest that SDF2 expression
is upregulated in glioma, potentially implicating it in glioma
pathogenesis.
SDF2 promotes malignant proliferation in
glioma cells
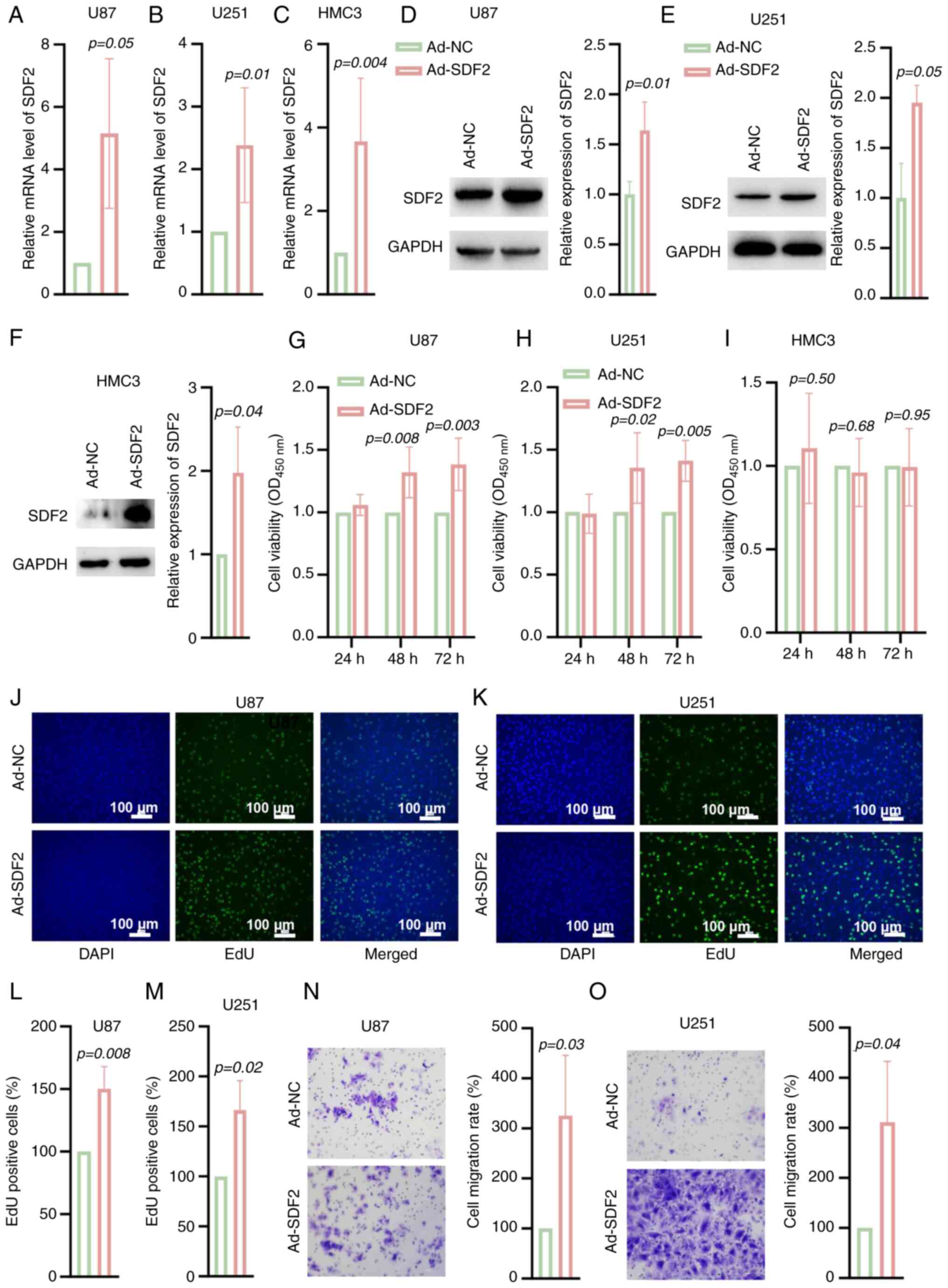
U87 and U251 cells were transfected with Ad-SDF2 to
overexpress SDF2. Following SDF2 overexpression, both the mRNA and
protein levels of SDF2 were significantly elevated in U87, U251 and
HMC3 cells (Fig. 4A-F).
Additionally, cell viability assays demonstrated that, compared
with the control, SDF2 overexpression enhanced the viability of U87
and U251 cells in a time-dependent manner (Fig. 4G and H), but the viability of
HMC3 cells did not change (Fig.
4I). The EdU staining results further confirmed that SDF2
overexpression increased the proliferative capacity of U87 and U251
cells relative to that of the controls (Fig. 4J-M). Moreover, Transwell assays
indicated that SDF2 overexpression promoted the migratory ability
of U87 and U251 cells (Fig. 4N and
O). These findings suggest that SDF2 plays a role in promoting
the malignant proliferation and migration of glioma cells.
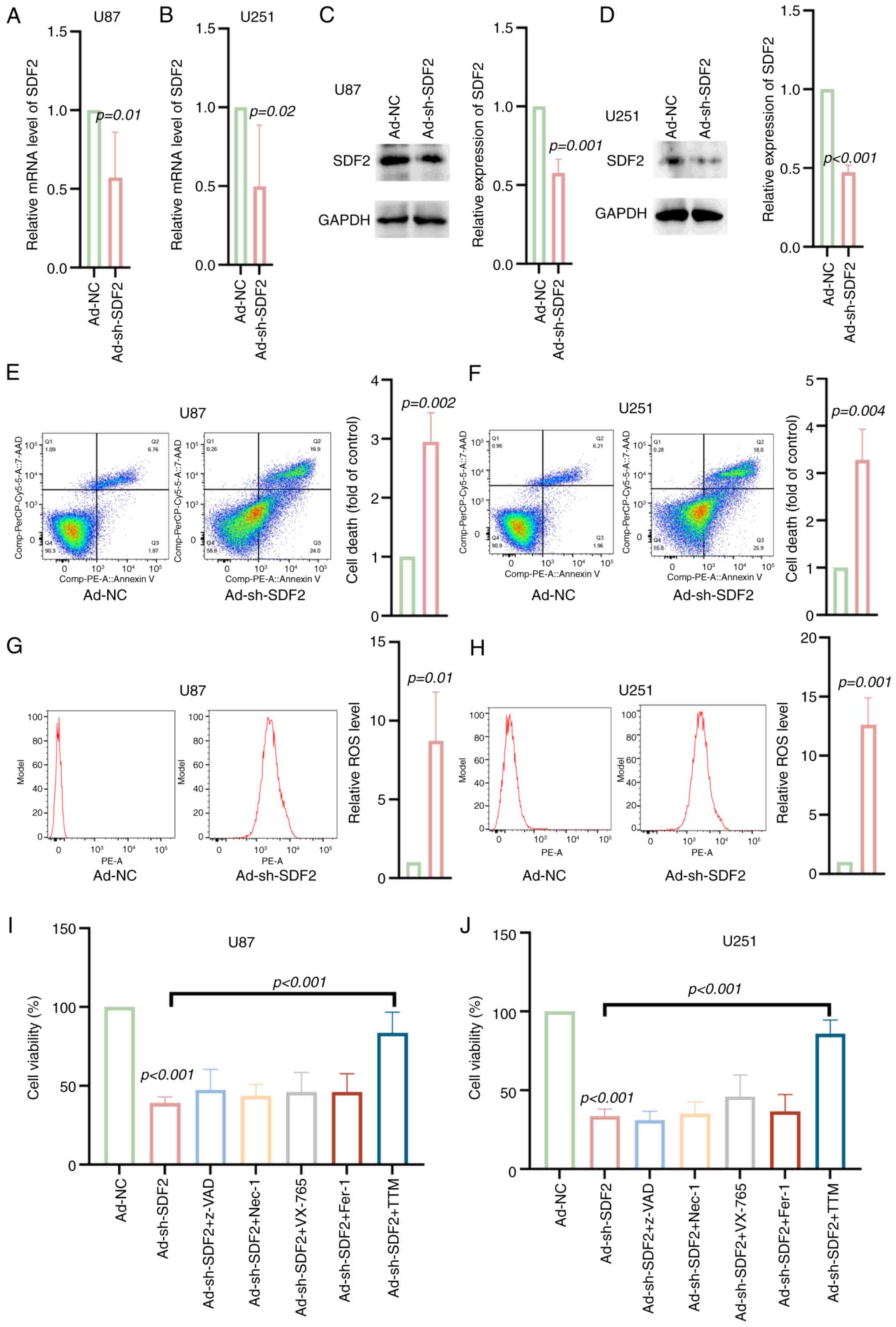
Knockdown of SDF2 increases ROS levels
and cuproptosis in glioma cells
U87 and U251 cells were transfected with Ad-shSDF2
to achieve SDF2 knockdown. The mRNA and protein levels were
decreased in U87 and U251 cells transfected with Ad-shSDF2
(Fig. 5A-D). Compared with the
control, Ad-shSDF2 transfection resulted in a significant increase
in the proportion of late apoptotic and necrotic cells in both the
U87 and U251 lines (Fig. 5E and
F). Additionally, ROS levels were significantly elevated
following SDF2 knockdown in U87 and U251 cells, indicating
increased oxidative stress (Fig. 5G
and H). Interestingly, only the cuproptosis inhibitor TTM
reversed the decrease in viability induced by Ad-shSDF2 in U87 and
U251 cells (Fig. 5I and J).
These results suggest that SDF2 knockdown induces oxidative stress,
increases apoptosis, and may involve copper-dependent cell death
pathways in glioma cells.
Knockdown of SDF2 induces copper ion
accumulation in glioma cells
In both the U87 and U251 cell lines, SDF2 knockdown
led to a significant increase in the intracellular copper ion
concentration compared with that in the controls. This finding was
confirmed through fluorescence staining (Fig. 6A) and further quantified using
colorimetric assays (Fig. 6B and
C). Additionally, the expression of the copper-exporting
proteins ATP7A and ATP7B, as well as FDX and lipid-DLAT, was
reduced in U87 and U251 cells following SDF2 knockdown, whereas
DLAT protein levels remained unchanged (Fig. 6D and E). Furthermore, SDF2
knockdown resulted in decreased intracellular GSH levels (Fig. 6F) and increased ROS and MDA
levels, indicating elevated oxidative stress and lipid peroxidation
within the cells (Fig. 6G and
H). These results suggest that SDF2 plays a role in maintaining
copper homeostasis, antioxidant capacity, and oxidative stress
regulation in glioma cells.
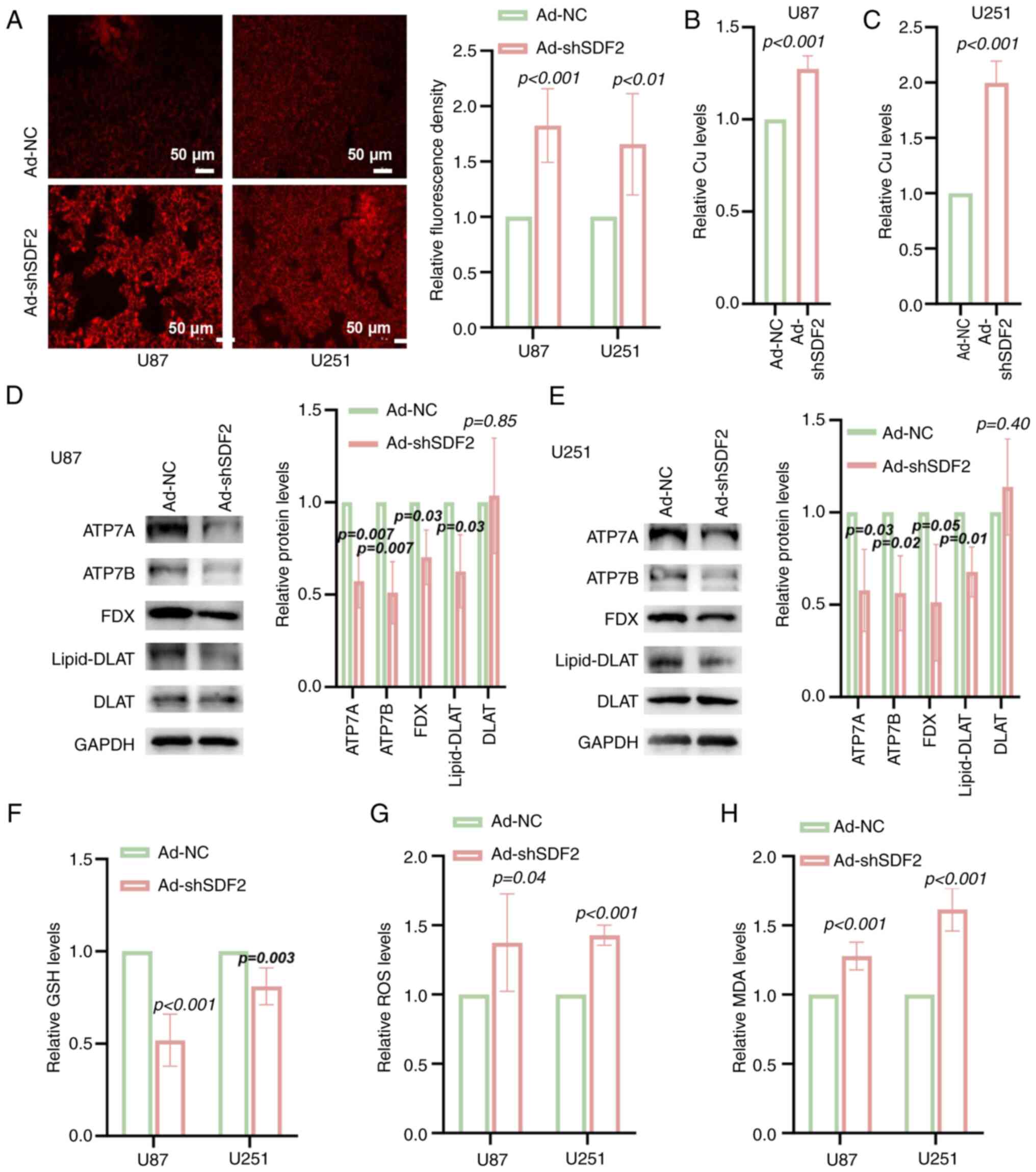 | Figure 6Knockdown of SDF2 increases
intracellular copper ion accumulation and oxidative stress in
glioma cells. (A) Fluorescence staining suggested that
intracellular copper ion levels were elevated in U87 and U251 cells
following SDF2 knockdown compared with those in control cells (n=3,
scale bar, 50 μm). (B and C) Quantitative assessment of
intracellular copper ion levels revealed significant copper
accumulation in U87 and U251 cells upon SDF2 knockdown (n=5). (D
and E) Western blot analysis revealed that ATP7A, ATP7B, FDX and
lipid-DLAT expression was reduced, whereas DLAT protein expression
did not significantly change (n=3). (F) A decrease in GSH was
identified following SDF2 knockdown in U87 and U251 cells (n=5). (G
and H) Increased ROS and MDA levels were detected in U87 and U251
cells upon SDF2 knockdown (n=3 for each group). SDF2, stromal
cell-derived factor 2; DLAT, dihydrolipoamide S-acetyltransferase;
GSH, reduced glutathione; ROS, reactive oxygen species; MDA,
malondialdehyde; sh-, short hairpin; NC, negative control. |
SDF2 mediates the downregulation of ATP7A
and ATP7B through the ERAD pathway
The effects of SDF2 on key proteins in the ER stress
pathway were investigated. Compared with Ad-NC, Ad-shSDF2 increased
GRP78 mRNA levels and the ratio of XBP1s/XBP1u in U87 and U251
cells, whereas CHOP mRNA levels remained unchanged (Fig. 7A and C). Additionally, Ad-shSDF2
increased GRP78 protein expression in both cell lines (Fig. 7B and D). To confirm the
involvement of the proteasome degradation pathway in ATP7A and
ATP7B downregulation, U87 and U251 cells were co-incubated with the
proteasome inhibitor MG132. Compared with the control treatment,
SDF2 knockdown reduced ATP7A and ATP7B expression, whereas MG132
treatment increased ATP7A and ATP7B expression (Fig. 7E-H). Notably, pretreatment with
MG132 significantly reversed the Ad-shSDF2-induced reduction in
ATP7A and ATP7B levels (Fig. 7E and
F). Furthermore, compared with Ad-NC, Ad-shSDF2 increased DLAT
aggregation within mitochondria; however, MG132 pretreatment
reduced DLAT aggregation (Fig. 7G
and H). These findings indicate that SDF2 knockdown promotes
ATP7A and ATP7B degradation through a GRP78-mediated ER-associated
degradation pathway, with potential downstream effects on
mitochondrial DLAT accumulation.
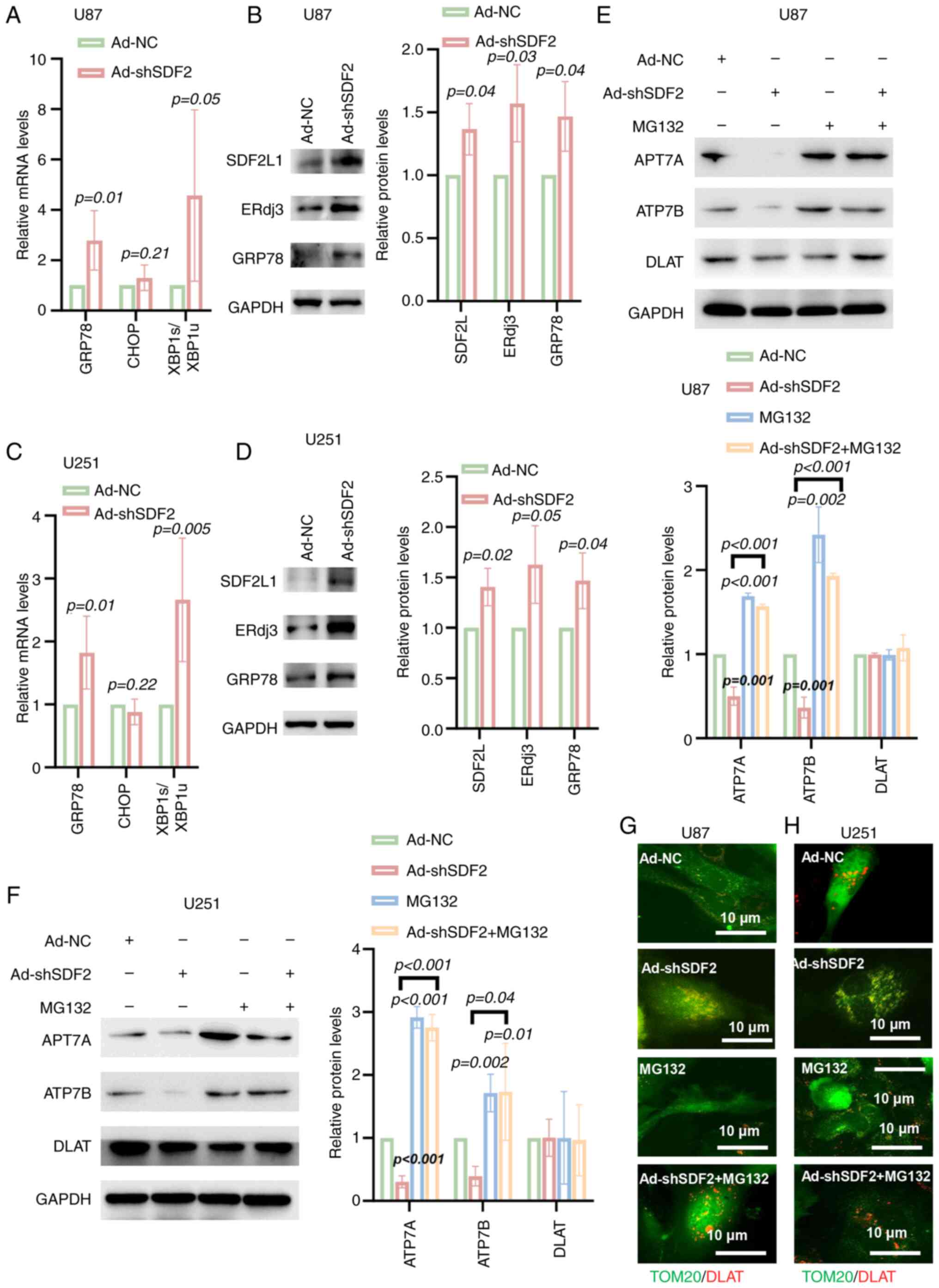 | Figure 7SDF2 knockdown mediates the
downregulation of ATP7A and ATP7B via the endoplasmic
reticulum-associated degradation pathway. (A and C) Reverse
transcription-quantitative PCR analysis revealed that GRP78 mRNA
levels and the XBP1s/XBP1u ratio were increased in U87 and U251
cells following Ad-shSDF2 transfection (n=5). (B and D) Western
blot analysis demonstrated that the protein expression of GRP78,
SDF2L and ERdj3 was increased in U87 and U251 cells after SDF2
knockdown (n=3). (E and F) Western blot analysis revealed that in
U87 and U251 cells, SDF2 knockdown reduced ATP7A and ATP7B
expression compared with that in control cells, whereas
pretreatment with MG132 significantly reversed the
Ad-shSDF2-induced reduction in ATP7A and ATP7B levels (n=3). (G and
H) Immunofluorescent analysis revealed that DLAT aggregation was
increased following Ad-shSDF2 transfection in U87 and U251 cells
(n=3; scale bar, 10 μm). SDF2, stromal cell-derived factor
2; GRP78, glucose-related protein 78; XBP1, X-box binding protein
1; sh-, short hairpin; DLAT, dihydrolipoamide S-acetyltransferase;
ERdj3, endoplasmic reticulum DnaJ heat shock protein 3; NC,
negative control. |
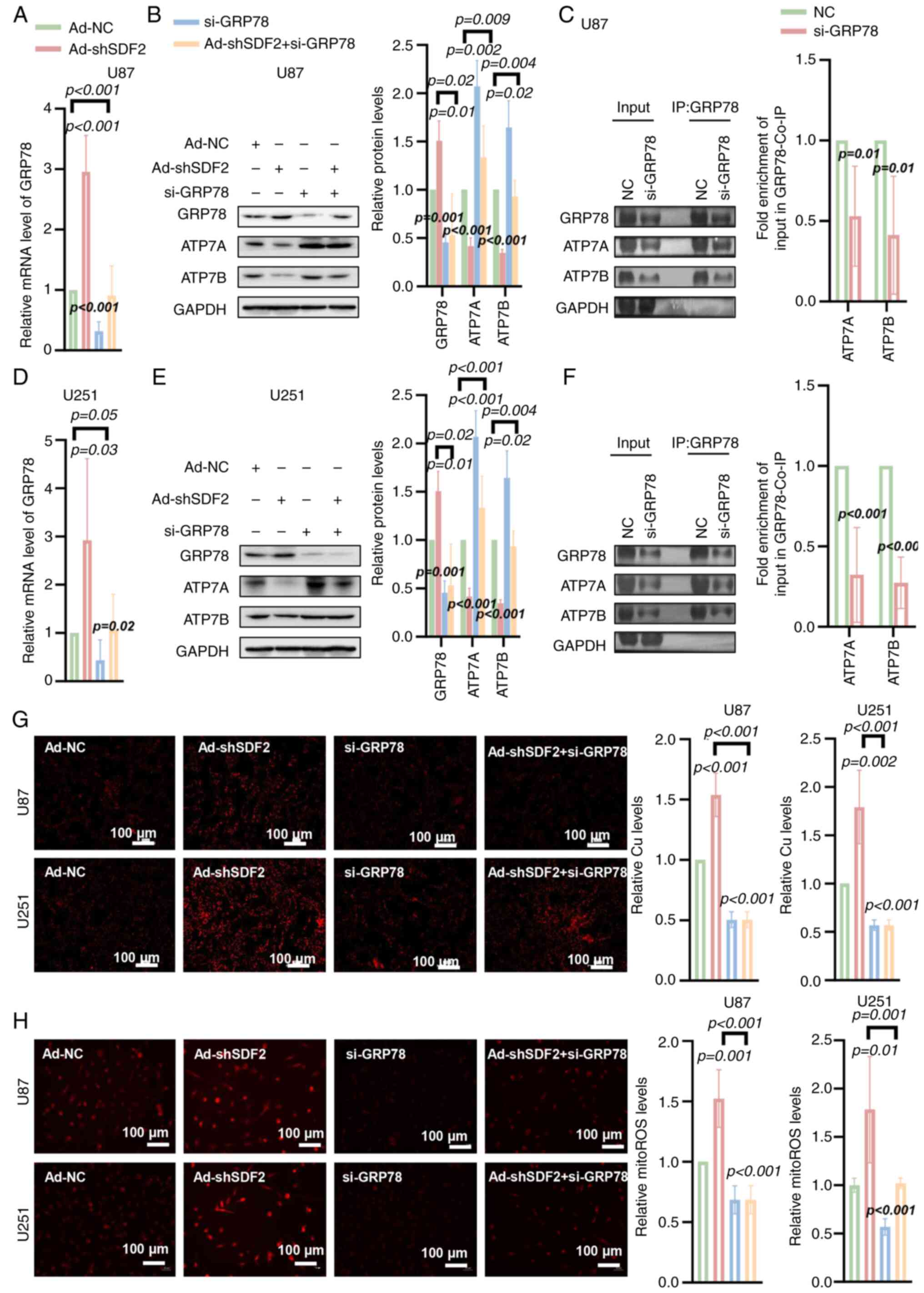
SDF2 suppresses ATP7A and ATP7B
expression via a GRP78-mediated mechanism
To investigate the role of GRP78 in this process,
si-GRP78 was utilized to specifically knock down GRP78 expression.
RT-qPCR analysis confirmed a significant reduction in GRP78 mRNA
levels in U87 and U251 cells following si-GRP78 transfection
(Fig. 8A and D). Western blot
analysis further verified that si-GRP78 effectively suppressed
GRP78 protein expression in both glioma cell lines (Fig. 8B and E). Importantly, GRP78
silencing also blocked the Ad-shSDF2-induced downregulation of
ATP7A and ATP7B in U87 and U251 cells (Fig. 8B and E). Co-IP results
demonstrated that knockdown of GRP78 led to a reduction in the
amount of ATP7A and ATP7B co-precipitated with GRP78. This decrease
was observed after normalizing the Co-IP signals to GAPDH levels in
the corresponding Input samples (Fig. 8C and F). These findings suggest
that GRP78 may play a role in regulating ATP7A and ATP7B stability
in glioma cells. Additionally, GRP78 silencing partially reversed
the Ad-shSDF2-induced increase in the intracellular Cu2+
and mito-ROS levels in U87 and U251 cells (Fig. 8G and H). These findings indicate
that SDF2 suppresses ATP7A and ATP7B expression through a
GRP78-dependent mechanism, impacting copper ion homeostasis and
mitochondrial oxidative stress in glioma cells.
Ad-GRP78 reverses Ad-SDF2-induced tumor
growth in vivo
Compared with Ad-NC, Ad-SDF2 significantly increased
tumor volume, and weight in a nude mouse model (Fig. 9A-C). IHC analysis revealed that
Ad-SDF2 increased the Ki-67 positivity rate in tumors, whereas the
co-expression of Ad-GRP78 reversed this increase, suggesting an
impact on cell proliferation (Fig.
9D). Further analysis revealed that Ad-SDF2 increased the GSH
level in tumor tissues, which was effectively reversed by Ad-GRP78
(Fig. 9E). Similarly, Ad-SDF2
overexpression led to decreased MDA levels, whereas co-expression
of Ad-GRP78 partially restored MDA levels (Fig. 9F). Moreover, Ad-SDF2 increased
ATP levels and reduced Cu2+ concentrations in tumor
tissues, while co-expression with Ad-GRP78 restored both parameters
(Fig. 9G and H). Additionally,
Ad-SDF2 overexpression suppressed GRP78 expression while increasing
the expression of copper transporters ATP7A and ATP7B.
Co-expression of Ad-GRP78 significantly reversed these changes
(Fig. 9I). These findings
indicate that GRP78 overexpression can counteract the effects of
SDF2-induced changes in tumor growth and metabolism, highlighting
the regulatory role of GRP78 in modulating SDF2-mediated changes in
glioma growth and oxidative stress.
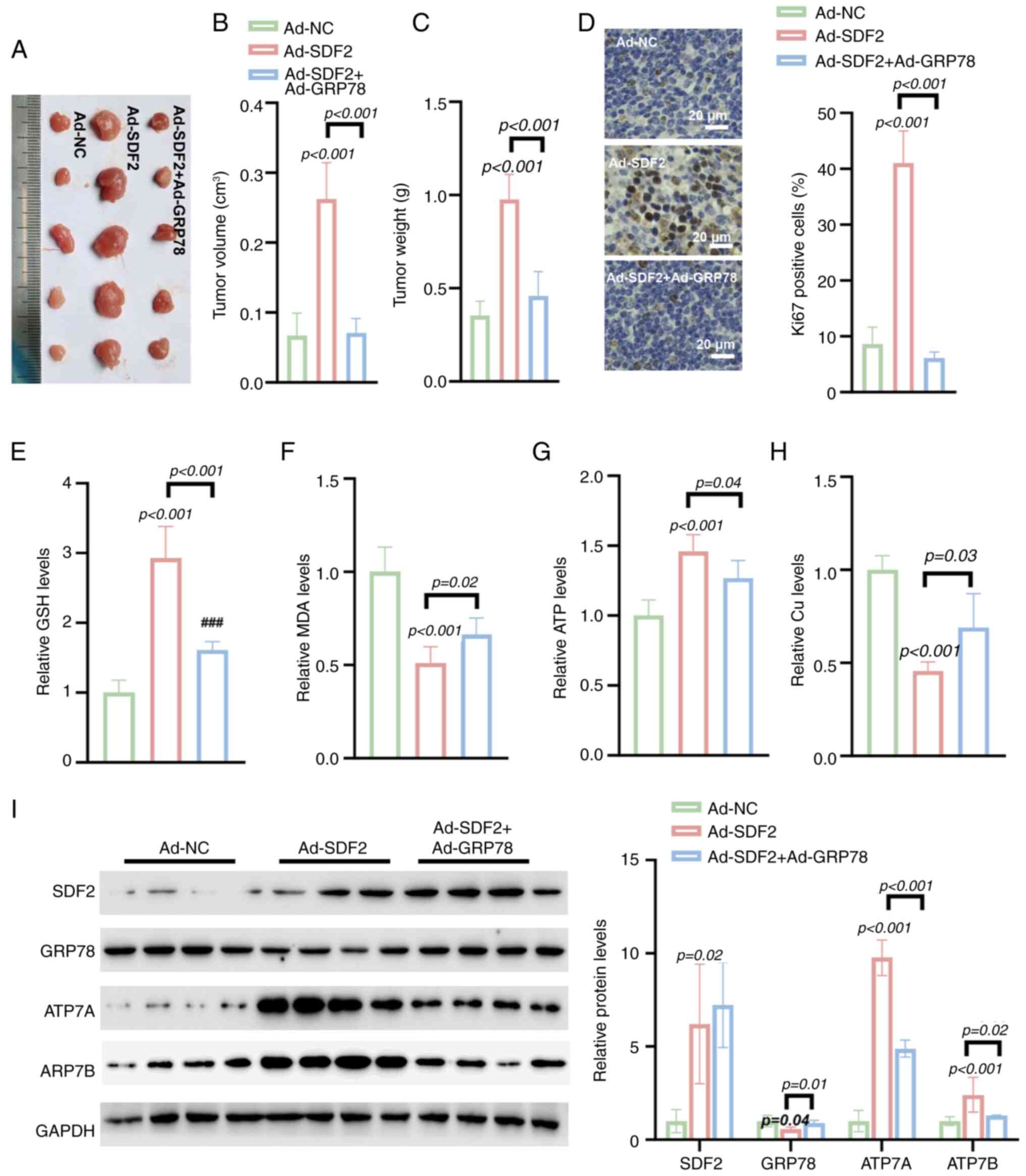 | Figure 9Ad-GRP78 reverses Ad-SDF2-induced
tumor growth and metabolic changes in a nude mouse model. (A-C)
Tumor size, volume and weight in nude mice injected with Ad-SDF2 or
Ad-NC (n=5). (D) Immunohistochemical staining demonstrated that
Ad-GRP78 co-expression reversed the Ad-SDF2-induced increase in
Ki-67 positivity (n=5; scale bar, 20 μm). (E) Ad-SDF2
increased GSH levels, whereas Ad-GRP78 restored GSH levels in tumor
tissues (n=5). (F) Ad-SDF2 reduced MDA levels, while Ad-GRP78
partially restored them (n=5). (G and H) Ad-SDF2 increased ATP and
decreased Cu2+ levels, which were restored by Ad-GRP78
co-expression (n=5). (I) Western blot analysis showing that
co-expressing Ad-GRP78 counteracted the Ad-SDF2-induced increases
in ATP7A and ATP7B levels in tumor tissues (n=5). GRP78,
glucose-related protein 78; SDF2, stromal cell-derived factor 2;
GSH, reduced glutathione; MDA, malondialdehyde; NC, negative
control. |
Discussion
Previous studies have shown that the expression of
SDF2 is closely related to the development of breast and gastric
cancers, highlighting its importance in tumor biology (20,21). Additionally, changes in SDF2
expression are associated with the occurrence of gestational
diabetes and preeclampsia, further enhancing its potential as a
biomarker (13). However,
research on the role of SDF2 in glioma remains limited. The present
study is the first to confirm the elevated expression of SDF2 in
glioma samples. In vitro experiments demonstrated that SDF2
promotes the malignant proliferation and migration of glioma cells.
These findings suggest that SDF2 may play a crucial role in the
pathogenesis and progression of glioma. Further investigations are
warranted to elucidate the underlying molecular mechanisms and
explore the potential clinical applications of targeting SDF2 in
glioma therapy.
In the present study, it was observed that the
knockdown of SDF2 significantly increased the levels of ROS and
enhanced the death of glioma cells. Transfection of U87 and U251
cells with Ad-shSDF2 resulted in a notable increase in the
proportion of late apoptotic and necrotic cells, indicating that
SDF2 depletion leads to increased cellular stress and compromised
cell viability. Further analysis revealed that ROS levels were
markedly elevated following SDF2 knockdown, suggesting a state of
heightened oxidative stress. This finding is consistent with
previous findings that link ROS accumulation to tumor progression
and cell death in various cancer types, where oxidative stress has
been shown to promote apoptotic pathways and influence tumor
behavior (22,23). Interestingly, when the mechanisms
underlying the observed decrease in cell viability were assessed,
only the cuproptosis inhibitor TTM effectively reversed the effects
of Ad-shSDF2. This finding highlights the specific involvement of
copper-dependent cell death pathways in glioma cells, aligning with
recent studies that highlight cuproptosis as a novel form of
regulated cell death distinct from apoptosis and other forms of
cell death, such as autophagy and ferroptosis (24,25). The selective reversal of cell
viability impairment by the cuproptosis inhibitor suggested that
SDF2 may play a role in modulating copper metabolism and its
downstream effects on glioma cell survival.
SDF2-L1 is a close homologue of SDF2, thus, the
expression of SDF2-L1 was detected in SDF2 knockdown cells. The
present data showed that knockdown of SDF2 elevated the expression
of SDF2-L1. These data indicated that SDF2-L1 may compensate the
deficiency of SDF2. ERdj3, an abundant and soluble ER co-chaperone
within the regulatory network of the Hsp70 family member BiP,
stimulates BiP's ATPase activity to enhance its affinity for
substrate proteins (26). Our
findings confirmed that knockdown of SDF2 upregulated ERdj3
expression. It was hypothesized that SDF2 knockdown leads to
dissociation of the SDF2-ERdj3 complex, resulting in accumulation
of unfolded proteins, which triggers ER stress and subsequently
induces the upregulation of both ERdj3 and GRP78 expression. Under
homeostatic conditions, the SDF2-ERdj3 complex maintains ER
homeostasis by suppressing the aggregation of unfolded proteins,
thereby restraining the activation of UPR signaling pathways such
as PERK and IRE1 (15). It is
proposed that knockdown of SDF2 disrupts this complex, leading to
the accumulation of unfolded proteins, which triggers ER stress and
subsequently activates the UPR. This activation drives the
transcriptional upregulation of GRP78 (27,28). In the present study, it was found
that knockdown of SDF2 elevated XBPs/XBPu, indicating the
activation of unfolded protein response (URP). Following activation
of URP in glioma cells, the expression of GRP78 was increased. As a
multifunctional chaperone, GRP78 recruits core ERAD components (for
example, Sec61, Derlin-1 and ubiquitin ligases) under stress
conditions to retro-translocate misfolded protein to the cytoplasm
for proteasomal degradation (29). Notably, GRP78 may facilitate the
maturation of ATP7A/7B through its chaperone activity under
non-stressed conditions but shifts toward a pro-degradative role
during persistent ER stress. In the present study, it was observed
that SDF2 knockdown in glioma cells led to increased levels of
spliced XBP1, indicating activation of the UPR, which in turn
upregulated GRP78 expression. This upregulation may shift GRP78's
role from promoting the maturation of ATP7A and ATP7B to
facilitating their degradation via ERAD, thereby impacting their
expression levels.
The expression of FDX and Lipid-DLAT was also
detected. The current data showed that the expression of FDX and
Lipid-DLAT was reduced in U251 and U87 cells transfected with
Ad-sh-SDF2 compared with that of Ad-NC. These data strengthened the
cuproptosis link. In the process of cuproptosis, the aggregation of
DLAT is a key feature. Studies have shown that during cuproptosis,
DLAT aggregation results from the binding of copper ions to its
lipoylated sites, forming insoluble aggregates, while the levels of
DLAT are not changed (30-32). In the present study, it was
observed that although DLAT aggregation increased in the Ad-SDF2
group, the total protein expression level of DLAT did not change
significantly. This phenomenon may be due to the binding of copper
ions to lipoylated DLAT, inducing the formation of
high-molecular-weight aggregates, leading to loss of protein
function, but these aggregates are not necessarily rapidly
degraded, thus maintaining the total protein level unchanged. This
suggests that DLAT aggregation is not due to protein degradation
pathways reducing its expression level.
The in vivo experiments demonstrated that
SDF2 overexpression led to increased tumor size, volume and weight
in a nude mouse model, suggesting a significant role in promoting
tumor proliferation, as indicated by increased Ki-67 positivity.
This effect, however, was counteracted by co-expressing Ad-GRP78,
which reversed the SDF2-induced increase in tumor growth and
proliferation. These findings suggest that GRP78 plays a key
regulatory role in modulating SDF2-mediated effects on glioma
progression. GRP78, a well-established ER chaperone protein,
facilitates protein folding and guides misfolded proteins to the
ERAD pathway for subsequent degradation (33). In the context of cellular stress,
GRP78 binds to misfolded proteins and initiates their transport to
the ERAD system, where they are ubiquitinated and targeted for
proteasomal degradation (34).
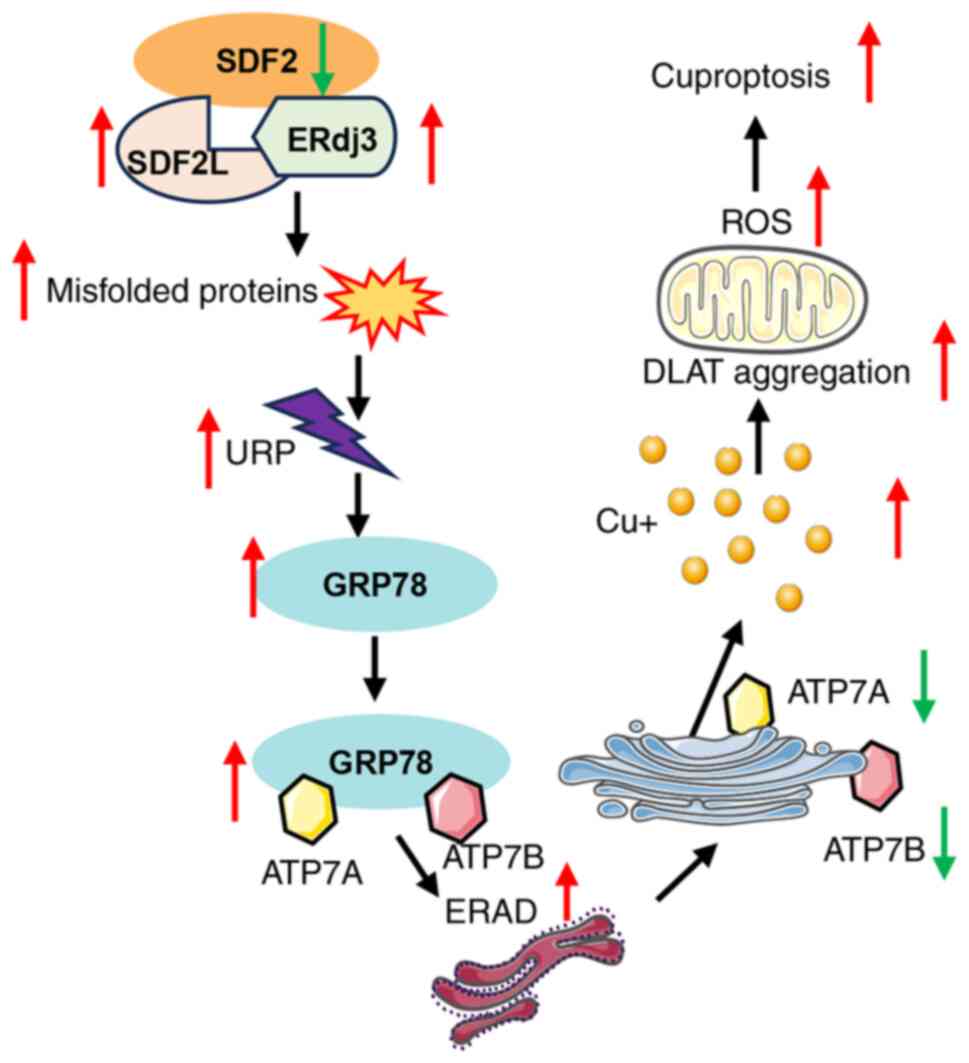
In the present study, SDF2 overexpression downregulated GRP78,
suggesting that reduced GRP78 expression impairs its ability to
guide ATP7A and ATP7B to the ERAD pathway. Consequently, this leads
to decreased degradation of these copper transport proteins,
resulting in their increased expression (Fig. 10).
However, it is accepted that ATP7A and ATP7B are key
copper-exporting transporters, and their downregulation may lead to
intracellular copper accumulation, thereby triggering cuproptosis.
The importance of rescue experiments was also acknowledged to
validate this mechanism. While such experiments were not included
in the current study, the authors plan to perform overexpression of
ATP7A/B in future studies to further investigate their role in
copper accumulation and cell death.
In summary, these findings collectively indicate
that GRP78 overexpression counters SDF2-driven tumor growth and
oxidative stress by enhancing the degradation of ATP7A and ATP7B
through the ERAD pathway. These findings highlight the potential
therapeutic value of modulating GRP78 to control SDF2-mediated
effects in glioma, suggesting a novel approach to mitigate tumor
growth and associated metabolic disruptions.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
TW and JS designed the experiments and revised the
manuscript. AL and XL performed the experiments. AL analyzed the
data and wrote the manuscript. TW and JS confirmed the authenticity
of all the raw data. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Human tumor and adjacent nontumor tissue samples
were obtained from patients during surgical procedures, with
informed consent obtained from all participants. Ethical approval
for the collection and use of human samples was granted by the
Institutional Review Board of the First Affiliated Hospital of
Xi'an Jiaotong University (approval no. 2024SF-FAHX-212; Xi'an,
China), in accordance with the ethical principles outlined in the
Declaration of Helsinki. All animal experiments were conducted in
compliance with institutional, national, and international
guidelines, specifically adhering to the revised Animals
(Scientific Procedures) Act 1986 (UK) and Directive 2010/63/EU
(Europe). Approval for all animal experiments was granted by the
Institutional Ethics Committee of the Animal Experimentation
Center, Xi'an Jiaotong University (approval no. XJTUAE2023-1944;
Xi'an, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Shaanxi Key Research and
Development Plan Project (grant no. 2024SF-YBXM-215).
References
|
1
|
Wang LM, Englander ZK, Miller ML and Bruce
JN: Malignant glioma. Adv Exp Med Biol. 1405:1–30. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weller M, Wen PY, Chang SM, Dirven L, Lim
M, Monje M and Reifenberger G: Glioma. Nat Rev Dis Primers.
10:332024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hakar MH and Wood MD: Updates in pediatric
glioma pathology. Surg Pathol Clin. 13:801–816. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei R, Zhou J, Bui B and Liu X: Glioma
actively orchestrate a self-advantageous extracellular matrix to
promote recurrence and progression. BMC Cancer. 24:9742024.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Krshnan L, van de Weijer ML and Carvalho
P: Endoplasmic reticulum-associated protein degradation. Cold
Spring Harb Perspect Biol. 14:a0412472022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou Z, Torres M, Sha H, Halbrook CJ, Van
den Bergh F, Reinert RB, Yamada T, Wang S, Luo Y, Hunter AH, et al:
Endoplasmic reticulum-associated degradation regulates
mitochondrial dynamics in brown adipocytes. Science. 368:54–60.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Turk SM, Indovina CJ, Miller JM, Overton
DL, Runnebohm AM, Orchard CJ, Tragesser-Tiña ME, Gosser SK, Doss
EM, Richards KA, et al: Lipid biosynthesis perturbation impairs
endoplasmic reticulum-associated degradation. J Biol Chem.
299:1049392023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stevenson J, Huang EY and Olzmann JA:
Endoplasmic reticulum-associated degradation and lipid homeostasis.
Annu Rev Nutr. 36:511–542. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu Y and Fang D: Endoplasmic
reticulum-associated degradation and beyond: The multitasking roles
for HRD1 in immune regulation and autoimmunity. J Autoimmun.
109:1024232020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun S, Shi G, Sha H, Ji Y, Han X, Shu X,
Ma H, Inoue T, Gao B, Kim H, et al: IRE1α is an endogenous
substrate of endoplasmic-reticulum-associated degradation. Nat Cell
Biol. 17:1546–1555. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang MJ, Shi M, Yu Y, Ou R, Ge RS and
Duan P: Curcuminoid PBPD induces cuproptosis and endoplasmic
reticulum stress in cervical cancer via the Notch1/RBP-J/NRF2/FDX1
pathway. Mol Carcinog. 63:1449–1466. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gu Y, Wang H, Xue W, Zhu L, Fu C, Zhang W,
Mu G, Xia Y, Wei K and Wang J: Endoplasmic reticulum stress related
super-enhancers suppress cuproptosis via glycolysis reprogramming
in lung adenocarcinoma. Cell Death Dis. 16:3162025. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lorenzon-Ojea AR, Yung HW, Burton GJ and
Bevilacqua E: The potential contribution of stromal cell-derived
factor 2 (SDF2) in endoplasmic reticulum stress response in severe
preeclampsia and labor-onset. Biochim Biophys Acta Mol Basis Dis.
1866:1653862020. View Article : Google Scholar
|
|
14
|
Hanafusa K, Wada I and Hosokawa N:
SDF2-like protein 1 (SDF2L1) regulates the endoplasmic reticulum
localization and chaperone activity of ERdj3 protein. J Biol Chem.
294:19335–19348. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fujimori T, Suno R, Iemura SI, Natsume T,
Wada I and Hosokawa N: Endoplasmic reticulum proteins SDF2 and
SDF2L1 act as components of the BiP chaperone cycle to prevent
protein aggregation. Genes Cells. 22:684–698. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
DiVita Dean B, Wildes T, Dean J, Yegorov
O, Yang C, Shin D, Francis C, Figg JW, Sebastian M, Font LF, et al:
Immunotherapy reverses glioma-driven dysfunction of immune system
homeostasis. J Immunother Cancer. 11:e0048052023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Xiang Z, Chen L, Deng X, Liu H
and Peng X: PSMA2 promotes glioma proliferation and migration via
EMT. Pathol Res Pract. 256:1552782024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zamler DB, Shingu T, Kahn LM, Huntoon K,
Kassab C, Ott M, Tomczak K, Liu J, Li Y, Lai I, et al: Immune
landscape of a genetically engineered murine model of glioma
compared with human glioma. JCI Insight. 7:e1489902022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gong W, Martin TA, Sanders AJ, Jiang A,
Sun P and Jiang WG: Location, function and role of stromal
cell-derived factors and possible implications in cancer (Review).
Int J Mol Med. 47:435–443. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Zheng M, Du S, Wang P, Zhang T,
Zhang X and Zu G: Clinicopathological and prognostic significance
of stromal cell derived factor 2 in the patients with gastric
cancer. BMC Gastroenterol. 24:3252024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Slika H, Mansour H, Wehbe N, Nasser SA,
Iratni R, Nasrallah G, Shaito A, Ghaddar T, Kobeissy F and Eid AH:
Therapeutic potential of flavonoids in cancer: ROS-mediated
mechanisms. Biomed Pharmacother. 146:1124422022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jelic MD, Mandic AD, Maricic SM and
Srdjenovic BU: Oxidative stress and its role in cancer. J Cancer
Res Ther. 17:22–28. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Li S, Guo Y, Zhao C, Chen Y, Ning
W, Yang J and Zhang H: Cuproptosis-related gene SLC31A1 expression
correlates with the prognosis and tumor immune microenvironment in
glioma. Funct Integr Genomics. 23:2792023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang B, Xie L, Liu J, Liu A and He M:
Construction and validation of a cuproptosis-related prognostic
model for glioblastoma. Front Immunol. 14:10829742023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo F and Snapp EL: ERdj3 regulates BiP
occupancy in living cells. J Cell Sci. 126:1429–1439.
2013.PubMed/NCBI
|
|
27
|
Lorenzon-Ojea AR, Guzzo CR, Kapidzic M,
Fisher SJ and Bevilacqua E: Stromal cell-derived factor 2: A novel
protein that interferes in endoplasmic reticulum stress pathway in
human placental cells. Biol Reprod. 95:412016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ibrahim IM, Abdelmalek DH and Elfiky AA:
GRP78: A cell's response to stress. Life Sci. 226:156–163. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cesaratto F, Sasset L, Myers MP, Re A,
Petris G and Burrone OR: BiP/GRP78 mediates ERAD targeting of
proteins produced by membrane-bound ribosomes stalled at the
STOP-Codon. J Mol Biol. 431:123–141. 2019. View Article : Google Scholar
|
|
30
|
Hu F, Huang J, Bing T, Mou W, Li D, Zhang
H, Chen Y, Jin Q, Yu Y and Yang Z: Stimulus-responsive copper
complex nanoparticles induce cuproptosis for augmented cancer
immunotherapy. Adv Sci (Weinh). 11:e23093882024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qi RQ, Chen YF, Cheng J, Song JW, Chen YH,
Wang SY, Liu Y, Yan KX, Liu XY, Li J and Zhong JC: Elabela
alleviates cuproptosis and vascular calcification in
vitaminD3-overloaded mice via regulation of the PPAR-γ/FDX1
signaling. Mol Med. 30:2232024. View Article : Google Scholar
|
|
32
|
Xiao Y, Yin J, Liu P, Zhang X, Lin Y and
Guo J: Triptolide-induced cuproptosis is a novel antitumor strategy
for the treatment of cervical cancer. Cell Mol Biol Lett.
29:1132024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cai X, Ito S, Noi K, Inoue M, Ushioda R,
Kato Y, Nagata K and Inaba K: Mechanistic characterization of
disulfide bond reduction of an ERAD substrate mediated by
cooperation between ERdj5 and BiP. J Biol Chem. 299:1052742023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akinyemi AO, Simpson KE, Oyelere SF, Nur
M, Ngule CM, Owoyemi BCD, Ayarick VA, Oyelami FF, Obaleye O, Esoe
DP, et al: Unveiling the dark side of glucose-regulated protein 78
(GRP78) in cancers and other human pathology: A systematic review.
Mol Med. 29:1122023. View Article : Google Scholar : PubMed/NCBI
|