Introduction
Osteoblastic cells (OBCs) in bone marrow (BM)
produce the bone matrix, participate in bone mineralization and
regulate the balance of calcium and phosphate ions in developing
bone (1,2). Furthermore, radiosensitive
hematopoietic stem/progenitor cells (HSPCs) form a niche with OBCs
to maintain the immature state and self-renewal of the BM
microenvironment. This tissue is susceptible to the formation of
cancerous bone metastases that originate from circulating cells of
a primary tumor, represent the terminal stage of cancer and are
associated with a reduction in quality of life (3,4).
One of the applied treatment modalities is radiation therapy, which
is noninvasive. In patients with castration-resistant prostate
cancer, bone metastases are curatively treated with radium-223
dichloride (223RaCl2). This therapeutic drug
went through a clinical trial called ALSYMPCA and is the world's
first internal therapy drug that uses α-radiation. The mechanism of
action of this agent is that it accumulates in bone, concentrates
on the tumor and has a very large biological effect (5,6).
However, a major problem is that the efficacy and
side effects of this therapeutic technique have a high degree of
individual variability and patient response is difficult to
predict. Hematological adverse events, such as anemia,
thrombocytopenia and neutropenia, occur in at least 5% of patients
receiving radium-223 therapy (5). The mechanisms underlying the
development of these adverse events are unknown; however,
α-radiation-induced modulation of OBC activity may be a
contributing factor. The self-renewal and differentiation of HSPCs
in BM are maintained by cytokines secreted from surrounding cells,
including OBCs (7,8). However, unpredictable BM toxicity
remains a challenge and it is currently difficult to predict which
patients will experience hematologic adverse effects. As OBCs play
a central role in maintaining the BM microenvironment through
cytokine and metabolite secretion, The present study hypothesized
that α-radiation induced specific molecular alterations in the OBC
secretome and regulatory RNAs, which could affect hematopoietic
cell behavior or serve as biomarkers of radiation-induced OBC
damage. Therefore, the aim of the present study was to characterize
the proteomic, lipidomic and miRNA expression profiles of OBCs
exposed to α-radiation and to explore the potential of these
molecules as indicators of radiosensitivity or predictors of BM
toxicity in the context of α-emitting radionuclide therapy.
The present study analyzed the role of OBCs in the
regulation of HSPC radiosensitivity. It created an in vitro
osteoblast metabolic model from mouse primary BM and analyzed the
proteins and lipids metabolized by OBCs exposed to α-radiation.
Furthermore, The present study focused on the proteomics,
lipidomics and transcriptomics of these cells and investigated
whether there is a radioresistant component of the hematopoietic
system in OBC response to α-radiation.
Materials and methods
Mouse BM cells
The mice study was approved by the Animal Welfare
Body in Stockholm University (Djurskyddsorgan; approval number
64-2019) and the Institutional Animal Care and Use Committee in
Hirosaki University (approval number AE05-2024-004). All animal
experiments were performed in accordance with national animal
welfare guidelines and the ARRIVE (Animal Research: Reporting of
In Vivo Experiments) guidelines (9). Mice were
anesthetized using the inhalation anesthetic isoflurane (Pfizer,
Inc.). Anesthesia was induced with 4-5% isoflurane and maintained
at 2-3% using a small animal anesthesia system. Euthanasia was
performed by cervical dislocation under deep anesthesia and death
was confirmed by the absence of respiration and heartbeat.
Immediately after euthanasia, fresh BM cells were collected from
the femurs. C57BL/6N male mice (20-25 g) were delivered at seven
weeks of age from a breeding facility, Charles River Laboratories.
Upon arrival, the mice were housed under specific pathogen-free
conditions with a controlled environment: Room temperature
(20°C), 12-h light/dark cycle and 40-60% relative humidity.
At eight weeks of age, mice were anesthetized and euthanized to
collect the fresh BM cells. The femurs were excised and cut at both
ends using sterile scissors and BM cells were isolated by flushing
the marrow cavity with phosphate-buffered saline containing
ethylenediaminetetraacetic acid. In total, 30 male C57BL/6N mice
were used in the present study. Three groups were based on the
α-particle doses: 0 Gy (non-irradiated), 0.5 and 1.0 Gy (n=10 per
group). For each mouse, a portion of the collected BM cells was
used for the clonogenic cell-forming capacity (CFC) assay and the
remaining cells were cultured under osteoblastic differentiation
conditions. After differentiation, the cultured cells were used for
transcriptomic analysis, while the culture supernatants were
analyzed for proteomic and lipidomic profiles.
Differentiation of osteoblastic cells and
exposure to α-radiation
The isolated BM cells were seeded in 100-mm round
dishes filled with 5 ml of RPMI1640 medium supplemented with 20%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and
incubated for 7 days at 37°C in a 5% CO2
atmosphere (1×107 cells/dish). On day 7, the cells were
checked for viability and the differentiation of OBCs was initiated
by adding a differentiation medium (20-mM β-glycerol phosphate
disodium salt pentahydrate, 100-nM dexamethasone and 50-μM
L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate) for 7
days (10). On day 14, cells
were exposed to 0.5- or 1-Gy α-radiation (0.22 Gy/min). The
radiation exposure was performed on a custom-made dish covered with
2.5 μm thick Mylar foil that allowed the exposure of the
cell monolayer to α-radiation from a 241Am source (cat.
no. AP1 s/n 101; Eckert and Ziegler) (11). The cells exposed to radiation
were incubated for 15 days and the cells were harvested and stored
at −80°C as cell pellets until analysis. For proteomics and
lipidomics analyses, a cell pellet was suspended in 7-M urea and 2%
sodium dodecyl sulfate and the protein fraction was precipitated by
adding acetone. For transcriptomics analyses, total RNAs were
extracted using an RNA extraction kit (Qiagen GmbH).
Flow cytometry
The number of BM cells and OBCs expressing the CD45
cell surface antigen was analyzed by flow cytometry system (Moxi GO
II; Orflo Technologies) and the results were analyzed by Kaluza
software (v.2.2.1; Beckman Coulter Inc.). For this, samples
containing 2×105 cells were incubated with the relevant
phycoerythrin-cyanin-5-forochrome tandem conjugated anti-mouse CD45
monoclonal antibody (mAb) for 30 min at 4°C (cat. no.
103109; Biolegend Inc.). Then, erythrocytes including sample cells,
were washed using RBC lysis buffer (Thermo Fisher Scientific,
Inc.). The intracellular proteins RUNX2 (cat. no. NBP1-77462AF488)
and BAP (cat. no. NB110-3638AF488) polyclonal antibody (pAb)
(Bio-Techne) were also analyzed using a cell membrane permeation
reagent kit (BD Biosciences). Isotype-matched mAb (PE/Cyanine5 Rat
IgG2b,κ (cat. no. 400609; Biolegend Inc.) for CD45) or pAb (IgG2a
Kappa-FITC (cat. no. 400207; Biolegend Inc.) for BAP and Mouse
IgG-FITC (#NBP1-96789, bio-techne Inc.) for RUNX2) was used as
negative controls for flow cytometry.
Clonogenic potency assay of hematopoietic
BM cells
The clonogenic potency of hematopoietic cells was
analyzed using a colony-forming cell (CFC) assay. CFCs, including
colony-forming unit-granulocyte and macrophage (CFU-G/GM),
burst-forming unit-erythroid (BFU-E) and colony-forming
unit-granulocyte, erythroid, macrophage and megakaryocyte
(CFU-GEMM), were assayed using the methylcellulose culture kit
(MethoCult; Stemcell Technologies, Inc.). The extracted cells from
BM or cultured cells were seeded into each well of a 24-well cell
culture plate with 300 μl of methylcellulose medium
containing recombinant IL-3 (100 ng/ml), recombinant SCF (100
ng/ml), IL-6 (100 ng/ml), G-CSF (10 ng/ml), EPO (4 U/ml),
penicillin (100 U/ml; Stemcell Technologies, Inc.) and streptomycin
(100 μg/ml) (FUJIFILM Wako Pure Chemical Corporation). The
cell culture plate was incubated at 37°C in a humidified
atmosphere containing 5% CO2/95% air for 7 days.
Colonies containing >50 cells were counted under 4×
magnification using an inverted microscope (Olympus Corporation).
After benzidine staining, the blue and colorless colonies were
scored as BFU-E and CFU-G/GM, respectively. Statistical analyses
were performed using the Origin software package (OriginLab Pro
ver. 9.1; OriginLab Corporation). Radiation dose-survival curves
were fitted using the algorithm of Levenberg-Marquardt, which
combines Gauss-Newton and steepest-descent methods, nonlinear
models based on the equation
y=1-[1-exp(-x/D0)]n, where x indicates the
dose in Gy. The values for D0 (the mean lethal radiation
dose) and n (the number of targets) were determined by a single-hit
multitarget equation.
Proteomics and lipidomics
The proteins in the harvested cells were
precipitated by acetone and lysed with 50% TFE. Reduction and
alkylation of proteins were conducted using 10 mM DTT and 20 mM
iodoacetamide, respectively. The protein samples were then
incubated with trypsin (AB Sciex) overnight at 37°C. The
tryptic peptides were desalted and purified using MonoSpin C18 (GL
Sciences). The resulting samples were analyzed using liquid
chromatography (LC)-MS/MS with a nanoLC Eksigent 400 system (AB
Sciex) coupled with a TripleTOF6600 mass spectrometer (AB Sciex).
Peptides were separated using a nano C18 reverse-phase capillary
tip column (75 μm × 125 mm, 3 μm; Nikkyo Technos,
Co., Ltd.). Peptide separation was performed at 300 nl/min with a
90 min linear gradient of 8-30% acetonitrile in 0.1% formic acid,
and then, with a 10 min linear gradient of 30-40% acetonitrile in
0.1% formic acid. Spectra acquired with data-dependent acquisition
in positive ion mode were searched using ProteinPilot software
(5.0.1; AB Sciex) with the UniProt-reviewed database (https://www.uniprot.org/). The peak areas of
individual peptides were extracted using Peakview software (v2.2.0;
AB Sciex) with the resulting group files and data-independent
acquisition sequential window acquisition of all theoretical
fragment-ion spectra (SWATH) spectral data. The peak area values
for individual proteins, consisting of the sum of the peak areas of
peptides, were normalized to the sum of all proteins detected.
Comparisons between sample groups were analyzed using SIMCA
software (version 15.0.2; InfoCom Corporation) with multivariate
analysis by orthogonal partial least squares discriminant
analysis.
Secreted lipids in the culture medium were measured
using QTRAP6500+ (AB Sciex) with multiple reaction monitoring
methods in the positive ion mode. The transition channels of
lysophosphatidylcholine (lysoPC) with acyl residues, acyl-acyl
phosphatidylcholine (PC aa), phosphatidylcholine with acyl-alkyl
residue sum (PC ae) and sphingomyelin are shown in Table I (10). Lipids were extracted at room
temperature using liquid-liquid extraction. The cell-cultured
medium was added to 50% methanol and 25% dichloromethane in a glass
screw-cap tube. The samples were mixed with internal standards of
15:0-18:1-d7-PC and 18:1-d7 Lyso PC (Merck KGaA). After inverting
the tubes 10 times, they were centrifuged at 1,500 × g for 10 min
at room temperature. The lower layer was collected and evaporated.
The residues were resuspended in methanol containing 0.1% formic
acid. The samples were analyzed in the flow injection analysis
using a high-performance liquid chromatography system (ExionLC AD;
AB SCIEX). The flow injection analysis (FIA) was done by injecting
20 μl of the sample into the flow of the FIA program,
lasting for 5 min and pumping the FIA mobile phase (methanol
containing 0.1% formic acid). QTRAP6500+ settings for FIA mode are
as follows: Curtain gas, 30; ion spray voltage, 4,500 V;
temperature, 300°C; ion source gas 1, 50 psi; ion source gas
2, 60 psi; CAD gas, 8 psi; entrance potential, 10 V; collision cell
exit potential, 15 V. Flow rate settings for FIA are as follows:
0.03 ml/min in 1.6 min; 0.03 to 0.2 ml/min in 0.8 min; 0.2 ml/min
in 0.4 min; 0.2 to 0.03 ml/min in 0.2 min. Multiple reaction
monitoring (MRM) settings are shown in Table SI. PCs and lysoPCs were
normalized to the internal standards. For semi-quantitation of all
lipids, the peak areas of individual lipids were normalized to the
sum of the peak areas of all lipids detected.
 | Table ITargeted lipids. |
Table I
Targeted lipids.
| Lipid class | Number of
analyses | Analyte
abbreviation |
|---|
| Sphingomyeline,
Hydroxysphingomyelins | 15 | SM (OH) C14:1, SM
(OH) C16:1, SM (OH) C22:1, SM (OH) C22:2, SM (OH) C24:1, SM C16:0,
SM C16:1, SM C18:0, SM C18:1, SM C20:2, SM C22:3, SM C24:0, SM
C24:1, SM C26:0, SM C26:1 |
| Diacyl
phosphatidylcholine | 38 | PC aa C24:0, PC aa
C26:0, PC aa C28:1, PC aa C30:0, PC aa C30:2, PC aa C32:0, PC aa
C32:1, PC aa C32:2, PC aa C32:3, PC aa C34:1, PC aa C34:2, PC aa
C34:3, PC aa C34:4, PC aa C36:0, PC aa C36:1, PC aa C36:2, PC aa
C36:3, PC aa C36:4, PC aa C36:5, PC aa C36:6, PC aa C38:0, PC aa
C38:1, PC aa C38:3, PC aa C38:4, PC aa C38:5, PC aa C38:6, PC aa
C40:1, PC aa C40:2, PC aa C40:3, PC aa C40:4, PC aa C40:5, PC aa
C40:6, PC aa C42:0, PC aa C42:1, PC aa C42:2, PC aa C42:4, PC aa
C42:5, PC aa C42:6 |
| Acyl-alkyl
phosphatidylcholine | 38 | PC ae C30:0, PC ae
C30:1, PC ae C30:2, PC ae C32:1, PC ae C32:2, PC ae C34:0, PC ae
C34:1, PC ae C34:2, PC ae C34:3, PC ae C36:0, PC ae C36:1, PC ae
C36:2, PC ae C36:3, PC ae C36:4, PC ae C36:5, PC ae C38:0, PC ae
C38:1, PC ae C38:2, PC ae C38:3, PC ae C38:4, PC ae C38:5, PC ae
C38:6, PC ae C40:1, PC ae C40:2, PC ae C40:3, PC ae C40:4, PC ae
C40:5, PC ae C40:6, PC ae C42:0, PC ae C42:1, PC ae C42:2, PC ae
C42:3, PC ae C42:4, PC ae C42:5, PC ae C44:3, PC ae C44:4, PC ae
C44:5, PC ae C44:6 |
|
Lysophosphatidylcholine | 14 | lysoPC a C14:0,
lysoPC a C16:0, lysoPC a C16:1, lysoPC a C17:0, lysoPC a C18:0,
lysoPC a C18:1, lysoPC a C18:2, lysoPC a C20:3, lysoPC a C20:4,
lysoPC a C24:0, lysoPC a C26:0, lysoPC a C26:1, lysoPC a C28:0,
lysoPC a C28:1 |
| Total targeted
lipids | 105 | |
For pathway enrichment analysis, markedly altered
proteins and lipid species were subjected to Kyoto Encyclopedia of
Genes and Genomes (KEGG)-based annotation and analyzed using
OmicsNet 2.0 (https://www.omicsnet.ca/) (12). Functional enrichment analyses
were performed to identify affected biological pathways and
metabolic processes. The input data consisted of proteins and lipid
metabolites that showed statistically significant differences
following α-radiation exposure. The six proteins [ITIH3
(Q61704), ATPB (P56480), PDIA3 (P27773), ANT3 (P32261), PDIA6
(Q922R8) and RPL4 (Q9D8E6)] and seven lipid species (PC aa C24:0,
PC aa C26:0, PC aa C34:4, PC ae C42:3, PC ae C42:2, PC ae C44:4 and
lysoPC a C26:0) selected for KEGG enrichment were identified based
on statistically significant fold changes (adjusted P<0.05)
observed in α-irradiated OBCs compared to controls.
Prior to pathway enrichment and integrated network
analyses, all raw proteomics and lipidomics data were deposited in
the public database MetaboBank under the accession number MTBKS262
(https://ddbj.nig.ac.jp/public/metabobank/study/MTBKS262/).
Integrated network analysis
To investigate the potential associations among the
identified microRNAs, proteomics and lipidomics datasets, The
present study performed an integrated network analysis using
OmicsNet 2.0. The analysis incorporated markedly downregulated
miRNAs (adjusted P<0.05) obtained from the microarray results,
alongside radiation-responsive proteins and lipids identified in
the omics datasets. Protein-protein interaction data were sourced
from the STRING database ver. 12 (https://string-db.org/), and miRNA-target
relationships were collected from TargetScan ver.8.0 (https://www.targetscan.org/vert_80/) and
miRTarBase ver. 9.0 (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2025/php/index.php).
Special attention was given to two microRNA target genes, ELF4 and
LARS2, which were selected based on their expression patterns and
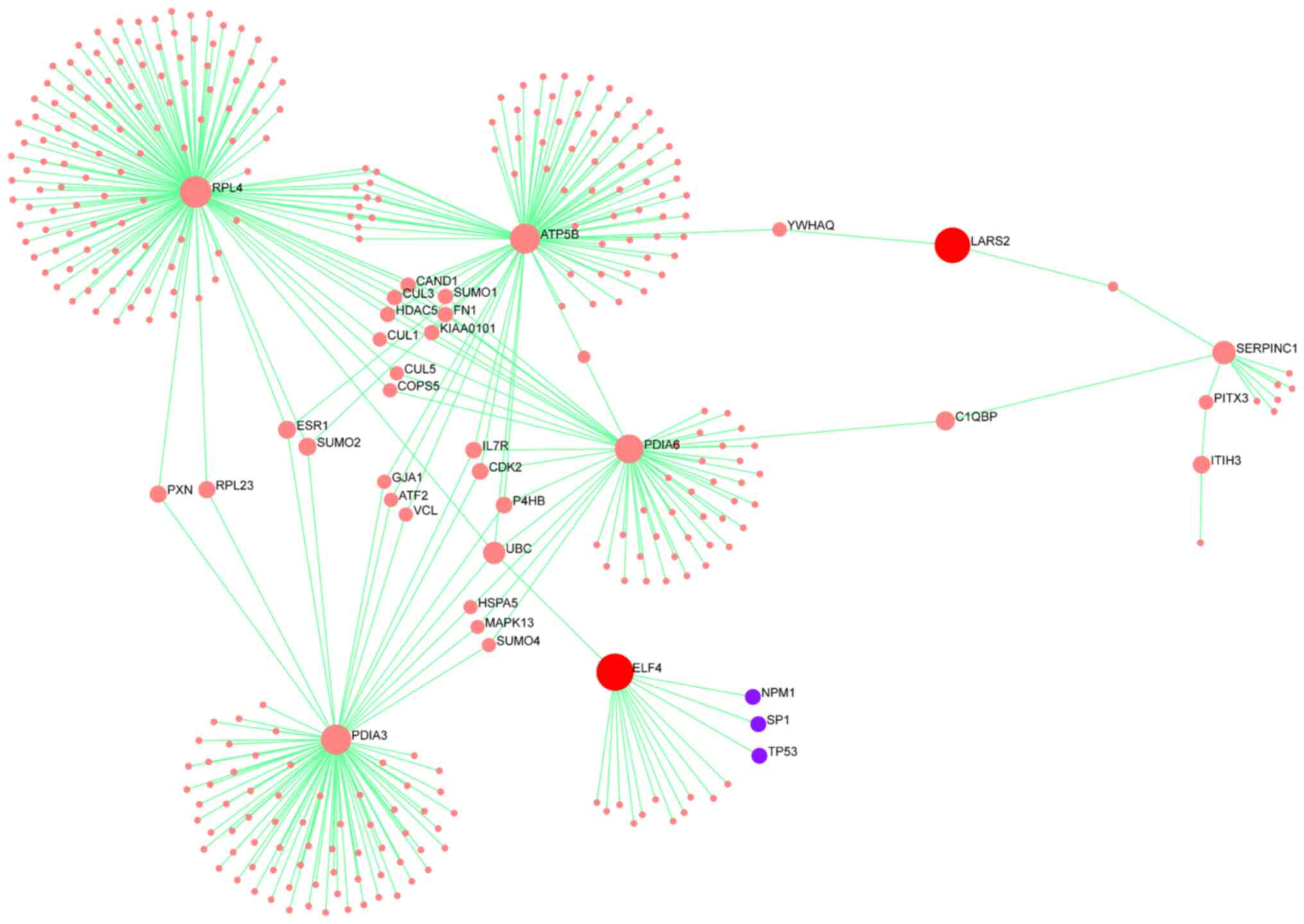
biological relevance in the transcriptome data. The interaction
networks were constructed to identify potential indirect
connections between these genes and the proteins detected in the
dataset of the present study. Network visualizations were used to
highlight subnetworks suggesting functional interactions related to
stress response, mitochondrial activity and transcriptional
regulation.
MicroRNA microarray analysis
Total RNAs from cultured BM cells were extracted
using the RNeasy kit (Qiagen GmbH) according to the manufacturer's
instructions. Furthermore, the concentration of total RNAs
extracted from cells or tissues was assessed using a NanoDrop
spectrophotometer (Thermo Fisher Scientific Inc.). All RNA samples
had 260/280-nm absorbance ratios of 1.8-2.0. The quality of small
RNAs included in the total RNAs was confirmed by an RNA 6000 Pico
kit (Agilent Technologies, Inc.) and a system of Agilent 2100
Bioanalyzer according to the manufacturer's instructions.
Cy3-labeled miRNA was synthesized from 30 ng total RNAs using a
miRNA Complete Labeling Reagent and Hyb kit (Agilent Technologies,
Inc.). A SurePrint G3 miRNA microarray slide (8×60 K, ver. 21.0)
for the mouse was hybridized with the Cy3-labeled miRNA in a
hybridization solution prepared with a Gene Expression
Hybridization Kit (Agilent Technologies, Inc.). Fluorescence signal
images of Cy3 on the slide were obtained by a microarray scanner
(SureScan; Agilent Technologies, Inc.) and processed by
Feature Extraction (version 10.7; Agilent Technologies, Inc.). The
raw data of microRNA microarray that was analyzed in the present
study was uploaded onto the Gene Expression Omnibus database
(GSE286553; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE286553).
Statistical analysis
Data are presented as mean ± SD. Statistical
analyses were performed using Origin software package (OriginLab
Pro ver. 9.1; OriginLab). For comparisons involving more than two
groups, one-way analysis of variance was conducted followed by
Tukey-Kramer's multiple comparisons test to control for familywise
error. All tests were two-tailed unless otherwise specified.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clonal growth of BM cells exposed to
α-radiation
Freshly isolated BM cells were exposed to 0.5- or
1-Gy α-radiation and plated in methylcellulose-based
semisolid culture supplemented with an optimal cytokine combination
(Fig. 1). Fig. 2A presents the radiation
dose-response curves of hematopoietic progenitors (CFCs). The
surviving fractions of CFCs were markedly lower after α-radiation
compared with X-irradiation, indicating a higher radiosensitivity.
The D0 and n values are summarized in
Table II, confirming this
greater sensitivity (D0: 0.98±0.42 Gy for
α-radiation vs. 0.84±0.00 Gy for X-irradiation; n: 1±0.44 vs.
23±0.01, respectively). The relative biological effectiveness (RBE)
of α-radiation was calculated to be 2.96 at 30% survival and 2.0 at
10%. Representative images of CFU-GEMM, BFU-E and CFU-G/GM colonies
are shown in Fig. 2B-D, with
their quantified colony numbers displayed in Fig. 2E. To assess the effects of
radiation on hematopoietic and osteoblastic markers, The present
study performed flow cytometry analysis. The corresponding
quantitative data are shown in Fig.
2F-H. α-radiation markedly modulated both hematopoietic and
osteoblastic differentiation markers in BM-derived cells. Fig. 2I-K shows representative
histograms of CD45, RUNX2 and BAP expression, respectively.
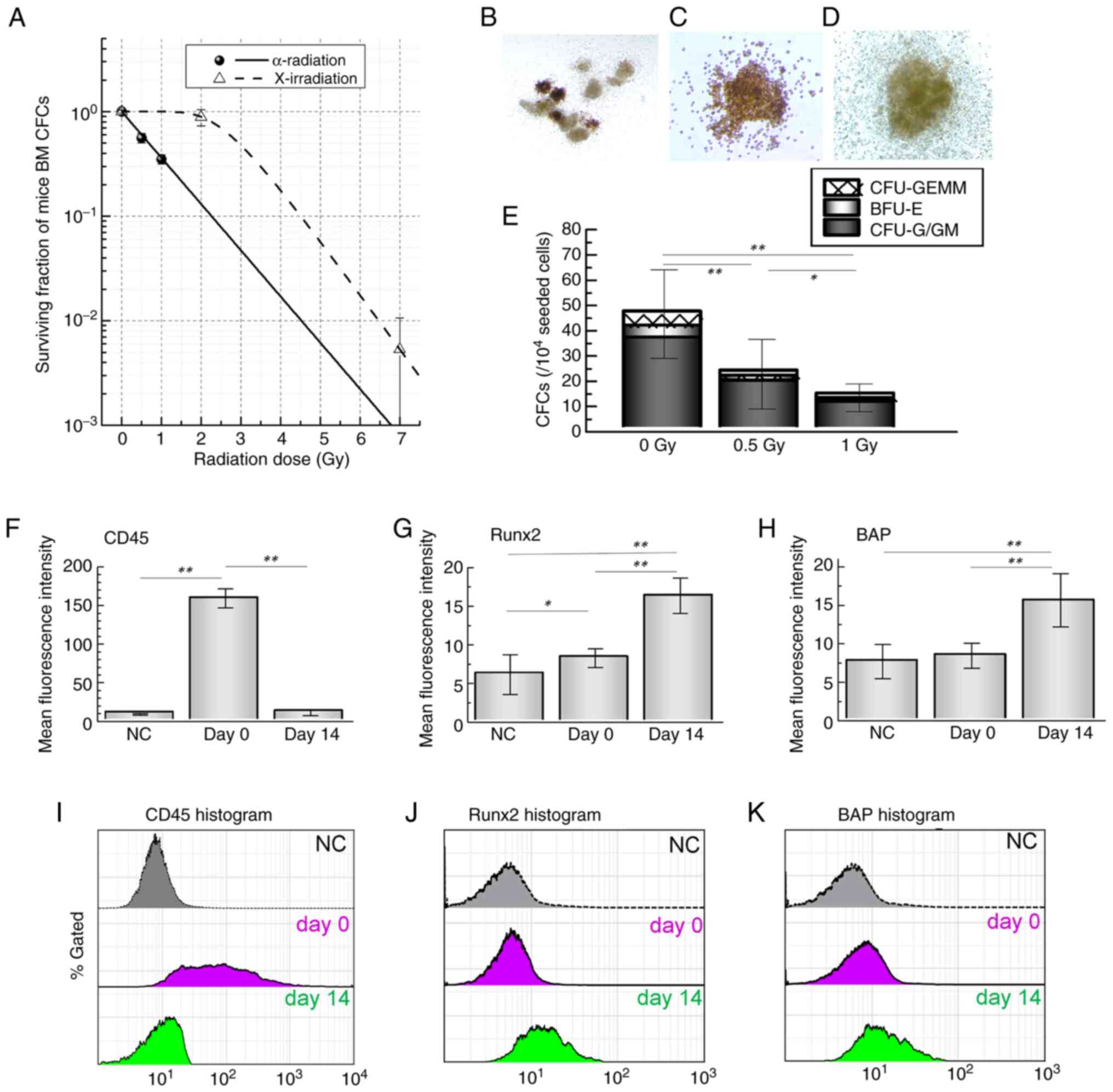 | Figure 2Radiation dose-response of HSPCs and
osteoblastic differentiation in mouse BM cells. (A) BM cells were
irradiated with α-radiation or X-irradiation and dose-response
curves of total CFCs were plotted. Representative images of
colony-forming cells (B) CFU-GEMM, (C) BFU-E and (D) CFU-G/GM from
BM cultures are shown. (E) Quantification of each colony type is
presented. (F-J) Flow cytometric analysis of BM cultures was
performed. Quantitative values (F-H) and representative histograms
(I-K) are shown for (F and I) CD45 expression and osteoblastic
differentiation markers (G and J) RUNX2 and (H and K) BAP. Each
value represents the mean ± SD of 8-10 mice per group. Statistical
analysis was performed using one-way ANOVA followed by
Tukey-Kramer's multiple comparisons test. *P<0.05,
**P<0.01 vs. undifferentiated BM-MNCs. HSPCs,
hematopoietic stem/progenitor cells; BM, bone marrow; CFCs,
colony-forming cells; CFU-GEMM, colony-forming unit-granulocyte,
erythroid, macrophage and megakaryocyte; BFU-E, burst-forming
unit-erythroid; CFU-G/GM, colony-forming unit-granulocyte and
macrophage; MNCs, mononuclear cells ; NC, negative control. |
 | Table IIRadiosensitivity of hematopoietic
stem/progenitor cells in murine bone marrow. |
Table II
Radiosensitivity of hematopoietic
stem/progenitor cells in murine bone marrow.
| Radiation type | Radiosensitive
parameter | Total CFCs |
|---|
| α-radiation | D0 | 0.98±0.42 |
| n | 1.00±0.44 |
| X-radiation | D0 | 0.84±0.00 |
| n | 23.01±0.00 |
| RBE | 30% survival | 2.96±0.23 |
| 10% survival | 2.00±0.09 |
Proteomics and lipidomics analysis
The differentiation of BM stromal cells into OBCs
was confirmed by the osteoblastic differentiation markers CD45,
RUNX2 and BAP (Fig. 2F-K).
Proteins extracted from OBCs were measured in the data-dependent
acquisition mode and 171 proteins were identified with 95%
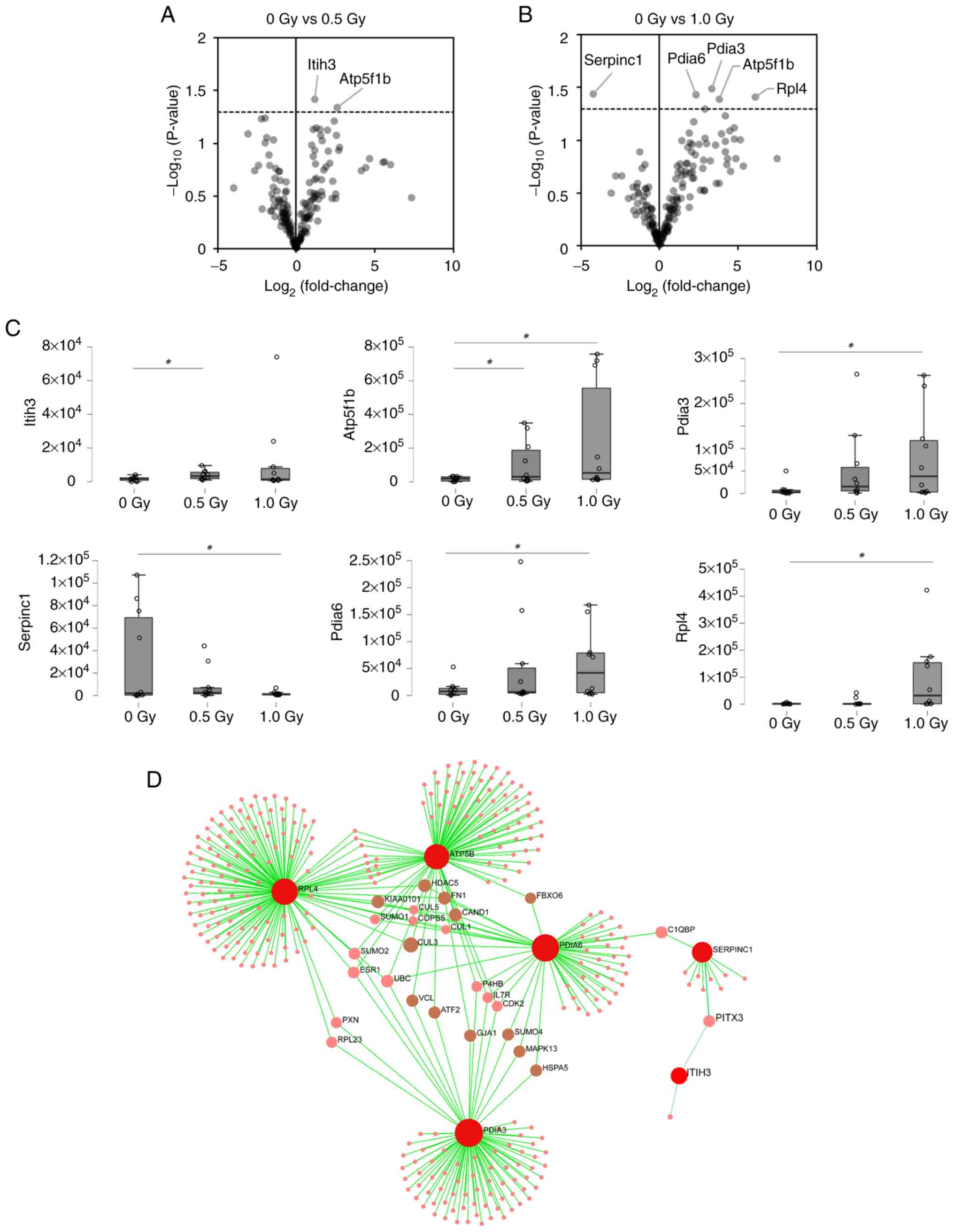
confidence, satisfying the criteria for SWATH measurement. Of
these, two and five proteins were markedly changed in OBCs exposed
to 0.5 and 1 Gy of α-radiation, respectively, compared to
nonirradiated controls (Fig. 3).
The proteins 'protein disulfide-isomerase A3', 'antithrombin-III',
'protein disulfide-isomerase A6', '60S ribosomal protein L4', 'ATP
synthase subunit β, mitochondrial', and 'inter-α-trypsin inhibitor
heavy chain H3' were detected as specific responses to α-radiation.
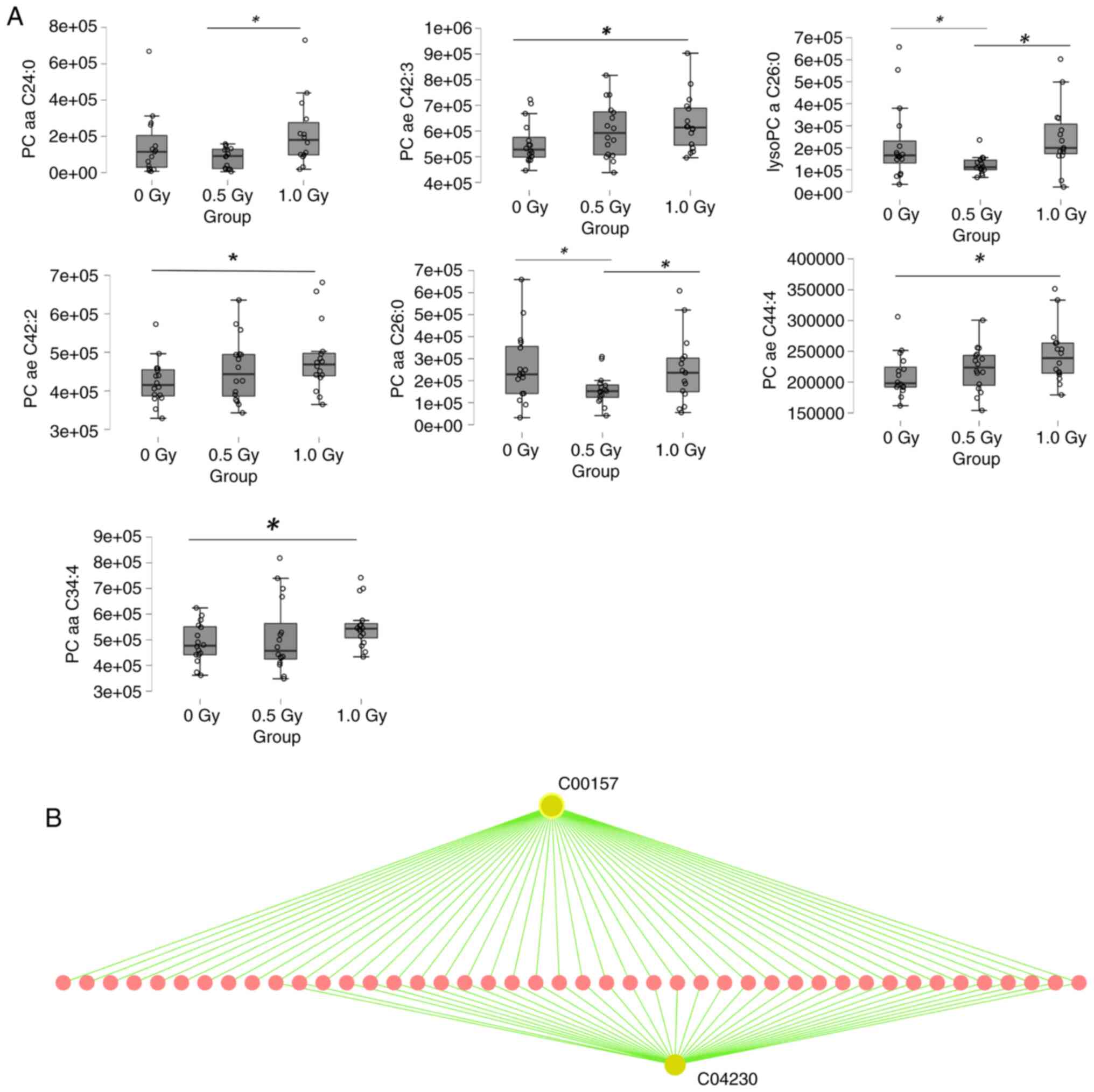
In the lipidomic analysis, several phosphatidylcholine (PC) and
lysophpsphatidylcholine (lysoPC) species were markedly changed
following α-radiation (Fig. 4A).
These specific proteins and lipids, showing consistent and
statistically significant alterations across biological replicates
in response to 0.5 or 1.0 Gy α-radiation, were selected to ensure
robust input for pathway enrichment. KEGG pathway enrichment
analysis using OmicsNet revealed that the differentially expressed
proteins were involved in pathways such as Basal transcription
factors, Apoptosis-multiple species and MicroRNAs in cancer
(Fig. 3D; Table SII). Similarly, enrichment
analysis of the altered lipid species predicted associations with
pathways including Glycerophospholipid metabolism, EGFR tyrosine
kinase inhibitor resistance, Ether lipid metabolism and
Cytokine-cytokine receptor interaction (Fig. 4B, Table SIII).
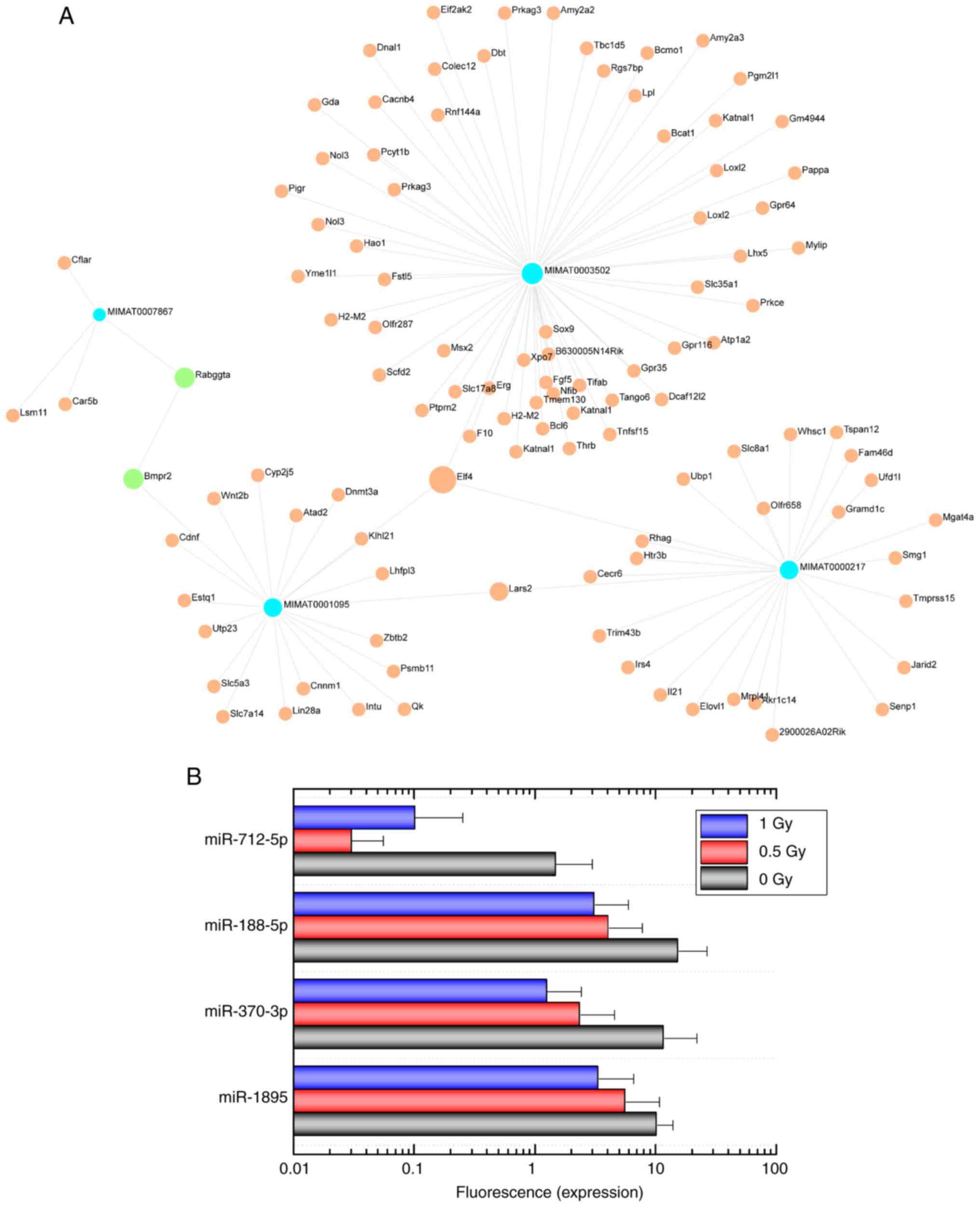
miRNA expression
RNA microarray analysis of OBCs exposed to
α-radiation revealed that 24 miRNAs were markedly downregulated.
The data has been deposited in the Gene Expression Omnibus (GEO:
GSE286553). The following miRNAs were identified: miR-1894-3p,
miR-1895, miR-3072-5p, miR-370-3p, miR-5107-5p, miR-652-5p,
miR-6984-5p, miR-7011-5p, miR-7040-5p, miR-7226-5p, miR-8094,
miR-8102, miR-188-5p, miR-3081-5p, miR-3098-5p, miR-6538,
miR-6969-5p, miR-7020-5p, miR-7044-5p, miR-7048-5p, miR-7082-5p,
miR-712-5p, miR-721 and miR-8101. The functions of the genes
targeted by these miRNAs were identified using the database
OmicsNet 2.0 and included 'negative regulation of metabolic
process, endothelial cell proliferation' and 'regulation of protein
metabolic process' (Table
III). Furthermore, the cellular components 'organelle membrane'
and 'Golgi membrane' were identified as targets of these miRNAs
(Table SIV). Most target genes
were regulated by miR-1895, miR-370-3p, miR-188-5p and miR-712-5p
(Fig. 5).
 | Table IIIFunctional of predictive targeted
genes from 24 miRNAs by Gene Ontology (Biological process). |
Table III
Functional of predictive targeted
genes from 24 miRNAs by Gene Ontology (Biological process).
| Function name | Number of
predictive genes | P-value |
|---|
| Negative regulation
of metabolic process | 12 | 0.0455 |
| Endothelial cell
proliferation | 10 | 0.038 |
| Macromolecule
catabolic process | 6 | 0.0337 |
| Cytokinesis | 6 | 0.0337 |
| Regulation of
protein metabolic process | 5 | 0.0369 |
| Sexual
reproduction | 5 | 0.042 |
| RNA catabolic
process | 5 | 0.0425 |
| Regulation of
translation | 4 | 0.00728 |
| Negative regulation
of growth | 3 | 0.0314 |
| Translation | 3 | 0.0418 |
| Positive regulation
of translation | 2 | 0.0111 |
| RNA 3′-end
processing | 2 | 0.0153 |
| Regulation of
endothelial cell proliferation | 2 | 0.0309 |
| Regulation of Rho
GTPase activity | 2 | 0.0336 |
| Pattern
specification process | 2 | 0.0364 |
| Cell projection
assembly | 2 | 0.0385 |
| Regulation of
signal transduction | 2 | 0.0445 |
Integrated network analysis of
multi-omics data
To investigate potential mechanistic links between
the miRNA, proteomics and lipidomics datasets, an integrated
network analysis was performed using OmicsNet 2.0. Two
representative miRNA target genes, ELF4 and LARS2, were selected
based on their biological relevance and expression changes
following α-radiation. Although these genes were not directly
identified in the proteomic dataset, the analysis revealed that
ELF4 was indirectly connected to radiation-responsive proteins such
as RPL4, PDIA3, ATP5B and PDIA6 through interaction networks.
Similarly, LARS2 was associated with a subnetwork including ATP5B
and SERPINC1 (ANT3). These findings suggested indirect regulatory
relationships linking α-radiation-induced protein expression
changes to downstream gene regulation via miRNA targets. The
resulting interaction networks are visualized in Fig. 6, showing the predicted
interconnectivity between altered proteins and target genes of
radiation-responsive miRNAs.
Discussion
The present study investigated how α-radiation
alters the molecular phenotype of BM-derived OBCs, focusing on
secreted metabolites and regulatory RNAs. As OBCs play a key role
in maintaining the hematopoietic niche through cytokine and
metabolite secretion, their radiation-induced dysfunction may
influence HSPC radiosensitivity and contribute to BM toxicity
observed during α-emitting radionuclide therapies such as
223Ra. To test this hypothesis, the present study
applied an integrated omics approach combining proteomic, lipidomic
and transcriptomic analyses in murine OBCs irradiated in
vitro. This multi-layered dataset enabled the examination not
only of the direct biochemical effects of α-radiation on OBCs, but
also potential signaling pathways involved in hematopoietic
dysregulation. It is well established that HSPCs are highly
radiosensitive and that cytokine-mediated signaling can modulate
this sensitivity (13-15). A-radiation, characterized as high
linear energy transfer (LET) radiation, exerts greater biological
damage per unit dose than low-LET radiation such as X-rays
(16). The findings of the
present study were consistent with this notion.
Internal radioactive nuclide therapy using
223Ra, an α-emitting nuclide, markedly enhanced the
level of BM toxicity, particularly neutropenia and
thrombocytopenia. It is not possible to predict which patients are
at increased risk of toxicities (17,18). The self-renewal and
differentiation of HSPCs in BM are maintained by various cytokines
secreted from surrounding cells, including OBCs (7,8).
As OBCs secrete numerous growth factors, differentiation factors
and other regulatory cytokines, their secretome may affect the BM
response to α-radiation.
The present study accurately quantified the
metabolites secreted by OBCs exposed to α-radiation. The
concentrations of five proteins [protein disulfide-isomerase A3
(P27773), protein disulfide-isomerase A6 (Q922R8), 60S ribosomal
protein L4 (Q9D8E6), inter-α-trypsin inhibitor heavy chain H3
(Q61704) and ATP synthase subunit beta, mitochondrial (P56480)]
were markedly upregulated following exposure to α-radiation
compared with the nonirradiated control. By contrast,
antithrombin-III (P32261) levels were markedly decreased. In the
lipidomics analysis, the concentrations of four lipids (PC ae
C42:3, PC ae C42:2, PC ae C44:4 and PC aa C34:4) exhibited a
significant upregulation following exposure to 1-Gy α-radiation
compared with the nonirradiated controls.
The reason why the secretion of the aforementioned
metabolites is modulated by α-radiation is unclear; however, they
may act directly or indirectly on cell surface receptors. Another
possibility is that they are released due to cell death caused by
radiation stress. The cell membrane is a lipid bilayer structure
containing phospholipids rich in PC and lysoPC. When apoptosis is
induced, cellular tissues are degraded and membrane lipids are
released (19).
Although these metabolites are secreted, the
expression of small RNAs also changes. In particular, the
expression of miR-1895, miR-370-3p, miR-188-5p and miR-712-5p was
markedly downregulated by exposure to α-radiation. miRNAs regulate
gene expression by binding to the 3′-untranslated regions of target
mRNAs, which modulate protein synthesis by increasing mRNA
degradation or inhibiting translation (20).
Based on the obtained results, The present study
predicted the expression of related mRNAs using OmicsNet and their
functions, inferred from Gene Ontology analysis, included 'negative
regulation of the metabolic process', 'endothelial cell
proliferation', 'macromolecule catabolic process', 'cytokinesis',
and 'regulation of protein metabolic process'. Endothelial cells
and BM microvascular cells support BM hematopoiesis (21). Furthermore, changes in gene
expression related to cell membranes are expected to be related to
the disruption of membrane formation induced by apoptosis.
To explore the interplay among different omics
layers, the present study conducted an integrated network analysis
using OmicsNet 2.0. Although ELF4 and LARS2 were not directly
identified in the proteomic dataset, the network analysis revealed
biologically plausible associations between these miRNA target
genes and radiation-responsive proteins. Specifically, ELF4 was
indirectly linked to RPL4, PDIA3, ATP5B and PDIA6; proteins
involved in ribosome biogenesis, endoplasmic reticulum stress
response and mitochondrial energy production. LARS2 was associated
with a subnetwork involving ATP5B and SERPINC1 (ANT3), suggesting
its potential involvement in mitochondrial translation and
coagulation-related stress pathways. These indirect associations
suggest that the downregulation of miRNAs regulating ELF4 and LARS2
may contribute to radiation-induced cellular responses through
downstream proteomic alterations. The integrated network, presented
in Fig. 6, underscores the
potential cross-talk between transcriptional and
post-transcriptional regulation in the α-radiation response. The
present systems-level view not only strengthened the mechanistic
understanding of OBC damage, but also offered potential molecular
targets for evaluating or mitigating radiation-induced bone marrow
toxicity.
The enrichment analysis was based on a subset of
proteins and lipids that were markedly altered following radiation
exposure, chosen for their reproducibility and magnitude of change.
This targeted approach enhanced the reliability of functional
pathway predictions. Furthermore, KEGG-based enrichment analysis
was conducted to explore the functional relevance of
radiation-induced changes at the proteomic and lipidomic levels.
The six proteins showing altered expression following α-radiation
were associated with key pathways such as 'Basal transcription
factors', 'Apoptosis-multiple species', and 'MicroRNAs in cancer',
implying involvement in transcriptional regulation and cell death
processes. Similarly, the altered lipid species were predicted to
affect 'Glycerophospholipid metabolism', 'EGFR tyrosine kinase
inhibitor resistance', and 'Cytokine-cytokine receptor
interaction', all of which are functionally linked to membrane
remodeling, inflammatory signaling and cell survival under
radiation-induced stress. These enrichment results provided
mechanistic insight into how OBCs respond to α-radiation at the
molecular level and support their potential utility as
biodosimetric markers. Currently, no publicly available omics
datasets exist for α-radiation-exposed osteoblastic or stromal
cells. Unlike prior studies focusing on hematopoietic cells, the
present study uniquely applied integrated multi-omics to bone
marrow OBCs exposed to α-radiation, revealing novel molecular
signatures that may underlie radiation-induced bone marrow
toxicity. Thus, the present study served as a foundational in
vitro model to identify candidate radiation-responsive markers.
Future studies involving clinical samples from patients treated
with α-emitting radionuclides (for example, 223Ra) are
warranted to validate the translational potential of these
findings. Although the present study provided novel insights into
the molecular responses of OBCs to α-radiation in vitro,
further validation using in vivo models or clinical BM
samples from patients treated with α-emitting radionuclide
therapies is necessary to confirm the relevance of these candidate
biomarkers in physiological settings. The present study used a
limited number of biological replicates (8-10 mice per dose group)
due to the technical challenges and resource demands of primary OBC
culture and comprehensive multi-omics analyses. While consistent
and statistically significant trends were observed, the present
study acknowledged that a larger sample size would enhance the
robustness and generalizability of these findings. Future studies
with increased biological replicates and independent validations
are warranted to confirm and extend these results. In addition,
although the present study identified radiation-responsive
metabolites and miRNAs in OBCs, The present study did not perform
functional assays (such as gene knockdown or overexpression) to
directly establish causal relationships. Such mechanistic
validations were beyond the scope of this initial exploratory study
but remain essential future directions to clarify the biological
roles of these candidate molecules.
In summary, the results indicate that exposure of
OBCs to α-radiation leads to altered secretion of metabolites and
changes in the expression of related miRNAs. The metabolites may be
OBC damage markers in α-radiation therapy to predict individual
therapy responses.
In conclusion, the present study presents a
comprehensive multi-omics characterization of osteoblastic bone
marrow cells exposed to α-radiation, providing new insights into
their molecular responses. While the findings highlight key
biological alterations associated with high-LET exposure, several
limitations remain. Notably, the present study did not include
comparisons with other types of radiation, such as γ or β rays.
Although the present study focused on α-radiation due to its
clinical relevance in 223Ra-targeted therapy, future
work should include comparative analyses with other radiation types
(such as X-rays, γ rays, or β radiation) to delineate the
specificity of the observed omics responses to high-LET radiation.
Such comparisons would provide a more comprehensive understanding
of radiation-induced molecular changes across different LET
contexts.
Supplementary Data
Availability of data and materials
The datasets generated and analyzed during the
current study are available as follows. The raw microRNA microarray
data have been deposited in the NCBI GEO under accession number
GSE286553 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE286553).
The raw proteomic and lipidomic data have been deposited in
MetaboBank under accession number MTBKS262 (https://ddbj.nig.ac.jp/public/metabobank/study/MTBKS262/).
All datasets are publicly available and can be accessed through the
respective databases.
Authors' contributions
SM, YM and AW designed the study, drafted the
manuscript and actively participated in its revision. SM, MC and YT
examined and analyzed the experimental data. SM and AW oversaw the
manuscript and provided the final approval of the version submitted
and published. SM and AW confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
The authors are grateful to Mrs. Miyu Miyazaki of
the Scientific Research Facility Center of Hirosaki University
Graduate School of Medicine for the mass spectrometry
assistance.
Funding
The present study was supported by JSPS KAKENHI, Grants-in-Aid
for Scientific Research (B) (Project No. 21H028 61/23K21419, Satoru
Monzen) and JSPS KAKENHI, Fund for the Promotion of Joint
International Research (Fostering Joint International Research;
project no. 17KK0181, Satoru Monzen).
References
|
1
|
Amarasekara DS, Kim S and Rho J:
Regulation of osteoblast differentiation by cytokine networks. Int
J Mol Sci. 22:28512021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mizoguchi T and Ono N: The diverse origin
of bone-forming osteoblasts. J Bone Miner Res. 36:1432–1447. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Clézardin P, Coleman R, Puppo M, Ottewell
P, Bonnelye E, Paycha F, Confavreux CB and Holen I: Bone
metastasis: Mechanisms, therapies and biomarkers. Physiol Rev.
101:797–855. 2021. View Article : Google Scholar
|
|
4
|
Hofbauer LC, Bozec A, Rauner M, Jakob F,
Perner S and Pantel K: Novel approaches to target the
microenvironment of bone metastasis. Nat Rev Clin Oncol.
18:488–505. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parker C, Nilsson S, Heinrich D, Helle SI,
O'Sullivan JM, Fosså SD, Chodacki A, Wiechno P, Logue J, Seke M, et
al: Alpha emitter radium-223 and survival in metastatic prostate
cancer. N Engl J Med. 369:213–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hoskin P, Sartor O, O'Sullivan JM,
Johannessen DC, Helle SI, Logue J, Bottomley D, Nilsson S,
Vogelzang NJ, Fang F, et al: Efficacy and safety of radium-223
dichloride in patients with castration-resistant prostate cancer
and symptomatic bone metastases, with or without previous docetaxel
use: A prespecified subgroup analysis from the randomised,
double-blind, phase 3 ALSYMPCA trial. Lancet Oncol. 15:1397–1406.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilson A and Trumpp A: Bone-marrow
haematopoietic-stem-cell niches. Nat Rev Immunol. 6:93–106. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Morrison SJ and Scadden DT: The bone
marrow niche for haematopoietic stem cells. Nature. 505:327–334.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kilkenny C, Browne WJ, Cuthill IC, Emerson
M and Altman DG: Improving bioscience research reporting: The
ARRIVE guidelines for reporting animal research. J Pharmacol
Pharmacother. 1:94–99. 2010. View Article : Google Scholar
|
|
10
|
Tatara Y and Monzen S: Proteomics and
secreted lipidomics of mouse-derived bone marrow cells exposed to a
lethal level of ionizing radiation. Sci Rep. 13:88022023.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Staaf E, Brehwens K, Haghdoost S,
Pachnerová-Brabcová K, Czub J, Braziewicz J, Nievaart S and Wojcik
A: Characterisation of a setup for mixed beam exposures of cells to
241Am alpha particles and X-rays. Radiat Prot Dosimetry.
151:570–579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou G, Pang Z, Lu Y, Ewald J and Xia J:
OmicsNet 2.0: A web-based platform for multi-omics integration and
network visual analytics. Nucleic Acids Res. 50:W527–W533. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hérodin F and Drouet M: Cytokine-based
treatment of accidentally irradiated victims and new approaches.
Exp Hematol. 33:1071–1080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Takahashi K, Monzen S, Yoshino H, Abe Y,
Eguchi-Kasai K and Kashiwakura I: Effects of a 2-step culture with
cytokine combinations on megakaryocytopoiesis and thrombopoiesis
from carbon-ion beam-irradiated human hematopoietic stem/progenitor
cells. J Radiat Res. 49:417–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patterson AM, Wu T, Chua HL, Sampson CH,
Fisher A, Singh P, Guise TA, Feng H, Muldoon J, Wright L, et al:
Optimizing and profiling prostaglandin E2 as a medical
countermeasure for the hematopoietic acute radiation syndrome.
Radiat Res. 195:115–127. 2021.
|
|
16
|
Goodhead DT: Mechanisms for the biological
effectiveness of high-LET radiations. J Radiat Res. 40(Suppl):
S1–S13. 1999. View Article : Google Scholar
|
|
17
|
Parlani M, Boccalatte F, Yeaton A, Wang F,
Zhang J, Aifantis I and Dondossola E: 223Ra induces
transient functional bone marrow toxicity. J Nucl Med.
63:1544–1550. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heidegger I, Pichler R, Heidenreich A,
Horninger W and Pircher A: Radium-223 for metastatic
castration-resistant prostate cancer: Results and remaining open
issues after the ALSYMPCA trial. Transl Androl Urol. 7:S132–S134.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagata S: Apoptosis and clearance of
apoptotic cells. Annu Rev Immunol. 36:489–517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rafii S, Shapiro F, Pettengell R, Ferris
B, Nachman RL, Moore MA and Asch AS: Human bone marrow
microvascular endothelial cells support long-term proliferation and
differentiation of myeloid and megakaryocytic progenitors. Blood.
86:3353–3363. 1995. View Article : Google Scholar : PubMed/NCBI
|