Introduction
Aging is a fundamental biological process that
underlies the progression of various degenerative diseases,
including intervertebral disc degeneration (IVDD) (1). IVDD is a leading cause of chronic
lower back pain, particularly in older adults, and is characterized
by the breakdown of the intervertebral discs (IVD), leading to
significant musculoskeletal decline (2). Despite its prevalence, the
molecular mechanisms driving IVDD, especially those related to
immune system dysregulation, remain poorly understood (3,4).
Prevention and treatment of IVDD are challenging because of the
high incidence and recurrence rates, and associated expenses
(5,6). Current treatment options for IVDD
include conservative management with medications and surgical
intervention to alleviate compression, although both fail to
prevent further degeneration (7,8).
Hence, additional research on the pathogenesis of IVDD is
imperative to elucidate the underlying mechanisms and develop
specific treatment approaches.
It has been recently shown that microRNAs (miRNAs or
miRs) play important roles in cell proliferation, apoptosis,
pyroptosis, and regulation of cytokine release. In addition, miRNAs
have been linked to the onset and progression of IVDD (9). Therefore, the specific mechanisms
employed by miRNAs associated with the onset and progression of
IVDD must be clarified to develop miRNA-based therapies to prevent
disc degeneration. Various miRNAs play therapeutic roles in
numerous diseases by controlling inflammatory responses, thus an
in-depth understanding of the complex relationship between miRNAs
and immunity is crucial for the prevention and treatment of IVDD
(10-12). Immune cells, including
lymphocytes and various phagocytes, recognize antigens and produce
specific immune responses via complex interactions with
musculoskeletal cells. T lymphocytes and natural killer cells
promote osteoclast formation and a subsequent anabolic response
during inflammation resolution (13,14). The IVD has been identified as an
immune privileged organ because of the unique avascular structure
(15). Impairment causes changes
to the internal environment of the IVD, resulting in
pro-inflammatory and catabolic responses. Changes to the IVD
microenvironment can cause recruitment of immune cells and the
release of inflammatory factors, which can induce apoptosis,
senescence, pyroptosis, and degradation of the extracellular matrix
(ECM) in the late stage of IVDD (16,17). In addition, infiltration of the
IVD by immune cells can induce changes to the miRNA content and
subsequent immune responses in the nucleus pulposus (NP) (18-20).
IVDD is increasingly recognized as a key contributor
to age-related musculoskeletal decline. As the population ages,
understanding the molecular mechanisms underlying IVDD is critical
for developing interventions that can mitigate the debilitating
effects of this condition. Recent studies have highlighted the role
of miRNAs in modulating cellular aging processes, including
apoptosis, senescence and immune responses (10-12). However, the specific involvement
of miR-27b-3p in these aging-related pathways remains
underexplored. The present study aims to fill this gap by
investigating the role of miR-27b-3p in IVDD, particularly in the
context of its interaction with CD4+CD39+
Treg cells, which are known to play a role in immune aging.
Mendelian randomization (MR) leverages genetic
information to investigate causal relationship between exposure
variables and outcome factors (21). Genome-wide analysis is
increasingly used in association studies (22). In the present study, the MR
approach was used to examine the causal relationship between 76
miRNAs and the risk of IVDD. Then, 731 immune cell features were
selected as intermediaries to investigate potential relationships
with the complex immune response mechanism of IVDD. In addition,
in vivo and in vitro analyses were conducted to
validate the findings. The present study represents a pioneering
effort in utilizing MR to elucidate the causal relationship between
miR-27b-3p and IVDD. Furthermore, by focusing on the modulation of
CD4+CD39+ Treg cells, the present study
offers a unique perspective on the immune mechanisms driving IVDD,
positioning miR-27b-3p as a promising therapeutic target.
Materials and methods
Ethics statement
The animal experimental protocols were authorized by
the Laboratory Animal Care and Use Committee of the Institute of
Health and Medicine, Hefei Comprehensive National Science Center
(approval no. IHM-AP-2024-015; Hefei, China).
Study design
The causal relationship between 76 miRNAs in whole
blood and low back pain was assessed using a two-sample MR
approach, with single nucleotide polymorphisms (SNPs) as
instrumental variables (IVs). MR analysis relies on three key
assumptions: i) IV independence from confounders, ii) a significant
relationship between IV and the exposure, and iii) IV affecting
outcomes solely through the exposure (23). To enhance result reliability,
heterogeneity sensitivity and reverse MR analyses were performed.
In total, 731 immune cells were also selected as mediators to
improve understanding of miRNA functions in IVDD. A two-step MR
analysis was used to identify the mediating pathway (24). Furthermore, in vitro and
in vivo experiments validated the role of miR-27b-3p in
IVDD. Using the GSE205535 dataset from Gene Expression Omnibus
database (25), single-cell
suspensions were analyzed to identify immune subpopulations,
focusing on CD39+CD4+ regulatory T cells
(Tregs). UMAP dimensional reduction and clustering methods revealed
cellular heterogeneity, while differential gene expression analysis
highlighted key signaling pathways in immune regulation and disc
degeneration. This approach provided new insights into immune cell
involvement in IVDD pathogenesis at a single-cell level (Fig. S1). The present study aimed to
explore the role of miR-27b-3p in IVDD development and identify
potential treatment targets and pathways.
Data sources
Experimental datasets from individuals of European
descent were utilized to guarantee the precision of the
experiments. miRNA data was collected from a study by Huan et
al (26) that found miRNA
expression quantitative trait loci on a genome-wide scale. The
study involved analyzing miR-eQTLs in whole blood samples from
5,239 individuals, leading to the identification of 5,269
cis-miR-eQTLs in 76 mature adults (26). The 731 immune cell types utilized
for Mendelian analysis were obtained from the dataset of Orrù et
al (27), which studied over
3,000 individuals. These data were compiled to facilitate MR
analysis. Detailed information on the 731 immune cell types is
provided in Table SI. The IDD
dataset was obtained from a Finnish database of 218,792
individuals, including 19,509 cases and 199,283 controls. The human
data used in the MR analysis were sourced from a well-characterized
cohort, ensuring the representativeness of the sample for clinical
translation. This cohort provides a robust foundation for exploring
the potential of miR-27b-3p as a therapeutic target for IVDD.
Instruments selection
SNPs selected for MR analysis must have a
substantial genome-wide correlation and strong association with the
exposure of interest. Therefore, a probability (p) value
<1×10−5 and linkage disequilibrium (r2=0.001,
kb=10,000) were used to identify appropriate SNPs while preserving
data independence. The PhenoScanner database (https://github.com/phenoscanner/) was manually
screened to identify and exclude SNPs that might be influenced by
confounding factors associated with disc degeneration. The F-value
of each SNP was calculated to verify a robust relationship between
IVs and exposure consistent with the premise of MR analysis
(28,29). The F value was calculated as
[(N-K-1)/k] x [R2/(1-R2)], whereN is the sample size in a
genome-wide association study, K denotes the quantity of SNPs, and
R2 indicates the percentage of variations present in the
unauthorized database and was calculated as 2x (1-MAF) x MAF x
(β/SD), where MAF is the minor allele frequency, β is the effect
size of the allele, and SD (standard deviation) is calculated by
multiplying the standard error by the square root of the sample
size (N). SNPs with a minor allele frequency <10% were
considered inefficient and removed to ensure that all SNPs
substantially contributed to the variance of the associated
metabolite.
The false discovery rate (FDR), which was employed
to improve the stability and reliability of the MR data, was
calculated as (P-value x Rankmax)/prank). As previously described
(30), an FDR of 0.20 was
considered to have outstanding dependability.
Inverse variance weighting (IVW) was utilized as the
primary analytic approach. P<0.05 indicated a statistically
significant causal link between exposures and outcomes. The
weighted median, MR-Egger, simple mode, and weighted mode were
employed to enhance the reliability of the findings (31). Directional pleiotropy was
assessed using intercept values from MR-Egger regression. The
Cochran's Q test was utilized to assess the considerable
heterogeneity among the selected IVs. P<0.05 indicated
significant heterogeneity, thus a random effects model was utilized
to evaluate the causal relationship. Otherwise, a fixed effects
model was employed (32). The
influence of additional SNPs on IVW estimations was assessed by
'leave-one-out' sensitivity analysis. The data are presented as the
OR and 95% confidence interval (CI). P<0.05 was considered
statistically significant. The MR Pleiotropy RESidual Sum and
Outlier (MR-PRESSO) method was used to re-evaluate the existence of
several SNPs. Data analyses were conducted using R software
(version 4.3.2; https://www.r-project.org/) with the 'Two-Sample-MR'
and 'MR-PRESSO' packages.
Cell culture and treatment
NP cells were obtained from Procell Life Science
& Technology Co., Ltd. and cultivated in Dulbecco's modified
Eagle's medium supplemented with nutrient mixture F-12, 15% fetal
bovine serum, and 1% penicillin-streptomycin. The medium was
replaced every 3 days.
Western blot analysis
Cells were divided into six groups: Normal control,
lipopolysaccharide (LPS), miR-27b-3p, miR-27b-3p inhibition, LPS +
miR-27b-3p, and LPS + miR-27b-3p. After culturing, the cells were
rinsed three times with phosphate-buffered saline (PBS), detached
using a cell scraper, centrifuged at 224 × g at 5°C for 5 min, and
subsequently transferred into new Eppendorf tubes. The cells were
lysed using radio immunoprecipitation assay buffer and
phenyl-methyl-sulfonyl fluoride (Beyotime Institute of
Biotechnology). The protein content was quantified using a
bicinchoninic acid assay kit (Beyotime Institute of Biotechnology).
The proteins (10 μg per lane) were combined with loading
buffer (4:1 v/v), heated for 10 min, separated by electrophoresis
(10%), and then electroblotted on to a polyvinylidene difluoride
membrane, which was blocked with 5% non-fat dry milk in
Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at
room temperature, and then incubated overnight at 4°C with primary
antibodies (1:1,000) against aggrecan (cat. no. ab313636; Abcam),
Bax (cat. no. ET1603-34; HUABIO), Bcl-2 (cat. no. ET1702-53;
HUABIO), COL2A (cat. no. HA722809; HUABIO) and actin (cat. no.
AF7018; Affinity Biosciences), followed by incubation with a
horseradish-conjugated anti-rabbit secondary antibody (1:10,000;
cat. no. S0001; Affinity Biosciences) against immunoglobulin G at
room temperature for 90 min. The protein bands were visualized
using an enhanced chemiluminescence kit (Beyotime Institute of
Biotechnology) and imaged with a multifunctional gel imaging system
(Bio-Rad Laboratories, Inc.).
Immunofluorescence (IF)
Apoptosis was quantified using TUNEL following the
instructions provided by the reagent provider. The cells were
initially submerged in xylene for two 5-min intervals. The cells
were sequentially exposed to gradient ethanol concentrations (100,
95, 90, 80 and 70%) for 3 min each and then washed twice with PBS.
The tissues were treated with Proteinase K working solution for
15-30 min at a temperature range of 21-37°C or with a
cell-permeable solution for 8 min, followed by rinsing twice with
PBS. The TUNEL reaction mixture was produced, and the treatment
group was combined with 50 μl of TdT and 450 μl of
fluorescein-labeled dUTP solution. The negative control (NC) group
received 50 μl of fluorescein-labeled dUTP solution, whereas
the positive control group was initially treated with 100 μl
of DNase 1. The reaction took place at room temperature, ~37°C, for
10-30 min, and the subsequent stages mirrored those of the
treatment group. After drying the slides, 50 μl of TUNEL
reaction mixture (or simply 50 μl of fluorescein-labeled
dUTP solution for the NC group) was applied to the specimen. The
reaction took place in a dark, humid chamber at 37°C for 60 min,
and then the slides were ready for further use. The cells were
washed three times with PBS. A single droplet of PBS was introduced
to quantify apoptotic cells using a fluorescence microscope with an
excitation wavelength of 450-500 nm and a detection wavelength of
515-565 nm. After drying the slide, 50 μl of converter-POD
was applied to the specimen. The coverslips or sealing membranes
were then attached and the reaction took place in a dark moist box
at 37°C for 30 min. Finally, the specimen was washed with PBS three
times. Between 50 and 100 microliters of DAB substrate were applied
to the tissue, and the reaction was carried out at a temperature
between 15-25°C for 10 min. The cells were washed three times with
PBS. The slides were counterstained with hematoxylin or methyl
green and promptly washed with tap water after a brief period.
Dehydrated with a gradient of alcohol, made transparent using
xylene, then sealed with neutral gum. A drop of PBS or glycerol was
placed in the field of vision 200 to 500 cells were counted using a
light microscope, and images were captured.
Detection of
CD4+CD39+ Treg cells
The purpose of this experiment was to detect and
quantify CD4+CD39+ Treg cells in rat models,
focusing on their potential role in IVDD. Specifically, it was
aimed to identify the presence of CD4+CD39+
Treg cells in the NP tissue and their interaction with miR-27b-3p.
To detect CD4+CD39+ Treg cells in rat models,
CD4+CD39+ Tregs were obtained from Yubo
Biotech (cat. no. YB-72865; https://www.ubiotech.com). For in vitro
analysis, CD4+CD39+ Tregs were cultured with
miR-27b-3p in triplicate wells of 6-well plates for 24 h. The cells
were then transferred to flow cytometry tubes (grouped by
experimental conditions) and incubated with fluorescein
isothiocyanate-labeled monoclonal antibodies against CD4 (1:200;
cat. no. YB70948; https://www.yubiotech.com) and CD39 (1:100;
LM-6526R-FITC; LMAI Biotech) at 4°C for 30 min in the dark. After
incubation, the cells were washed twice with PBS, fixed with 4%
paraformaldehyde (20-25°C), and analyzed by flow cytometry
(FACSAria II, BD Biosciences) to quantify
CD4+CD39+ Tregs. For detecting
CD4+CD39+ Treg cells in the NP tissue, the
tissue samples were harvested from rat IVDD models. These tissue
samples were processed and stained (4°C, 12-16 h) with monoclonal
antibodies against CD4 and CD39 as aforementioned (4°C). After
fixation with 4% paraformaldehyde for 15 min at room temperature
(20-25°C), flow cytometry was used to analyze the expression of CD4
and CD39, identifying and quantifying
CD4+CD39+ Tregs in the NP tissue.
Animal research
Male Sprague-Dawley rats (body weight 22-250 g;
obtained from Qinglongshan Zoo) were randomly assigned to one of
six groups (n=6 per group; total 36 animals). The animals were ~10
weeks of age at the start of the experiment. Rats were housed in a
specific pathogen-free (SPF) facility under controlled
environmental conditions: temperature 22±2°C, relative humidity
50-60%, and a 12 h light/dark cycle (lights on at 07:00). Animals
had free access to standard laboratory chow and water ad
libitum throughout the study: the NC (Normal Control) group,
PBS group, LPS (lipopolysaccharide) group, and LPS + miR-27b-3p
group. The rats were anesthetized with 1% pentobarbital sodium (40
mg/kg) for the procedure. A 20 G needle was inserted into the
lamina, and the injection site was held for 30 sec. The NP tissues
of the rats in the experimental groups were injected with PBS,
miR-27b-3p, or LPS at the Co5/6 and Co6/7 discs using a 22 G
needle. The injections were repeated 4 weeks later to assess disc
degeneration and treatment effects.
Numerous studies have used acupuncture of the
coccygeal vertebra and LPS to generate rat models of disc
degeneration. As compared with the NC group, LPS increased
expression of MMP-13 in the disc degeneration model, thus creating
a metabolic imbalance of the ECM to induce disc degeneration. The
PBS treatment in this experiment did not exacerbate the
degeneration of NP tissue. Instead, the PBS injection was used as a
control in the context of the disc puncture model. The puncture
itself induces disc degeneration, and the injection of PBS was
intended to simulate the physical injury and maintain consistency
in the experimental design. It was not expected to worsen the
degeneration beyond the effect of the puncture. In addition, LPS
also increased expression of caspase-3 and BMP-2 in IVD tissues,
which accelerated inflammation-induced chondrocyte degeneration
(33-35). The Co5/6 and Co6/7 discs were
specifically selected for observation due to their anatomical
relevance and accessibility in the rat cervical spine. These discs
are frequently involved in IVDD in animal models and are critical
for understanding the progression of cervical disc degeneration,
which has translational relevance to human disc diseases. Moreover,
the Co5/6 and Co6/7 discs are easily accessible in rats without
excessive tissue damage, making them ideal for injection
treatments. Previous studies have demonstrated that these discs
provide consistent results in modeling IVDD, ensuring reliable and
reproducible outcomes for studying the effects of miR-27b-3p on
disc degeneration (36,37). The present study was conducted in
accordance with the ARRIVE guidelines (38).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
To evaluate the differential expression of
miR-27b-3p in the IVDD model, total RNA was isolated from blood
samples collected from rats with disc degeneration and NCs using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's instructions. The concentration
and purity of the total RNA was assessed with a spectrophotometer
(ND-2000; NanoDrop Technologies LLC), and samples with an A260/280
ratio >1.8 were considered to have acceptable purity. Quality
RNA was reverse transcribed into complementary DNA using a reverse
transcription kit (Biosharp Life Sciences) with the primers listed
in Table I, in accordance with
the manufacturer's instructions. and qPCR was performed using a
universal fluorescent quantitative PCR kit (Biosharp Life Sciences)
on a [qPCR platform, for example, Applied Biosystems 7500 system].
Amplification was monitored with SYBR Green fluorophore. GAPDH was
used as the internal reference gene. Thermocycling conditions were
as follows: initial denaturation at 95°C for 30 sec, followed by 40
cycles of denaturation at 95°C for 5 sec, annealing at 60°C for 30
sec, and extension at 72°C for 30 sec. A melting curve analysis was
performed to confirm amplification specificity. Relative expression
levels were calculated using the 2−ΔΔCq method (39), where Cq values were normalized to
GAPDH. Due to the small size of the miRNA sequence, conventional
primers are not effective for its amplification. To overcome this
limitation, a stem-loop structure was introduced to elongate the
miRNA sequence, which allowed us to design primers for efficient
amplification. The reverse primer was designed based on this
stem-loop structure, which is why the reverse primers for different
miRNAs are consistent. The Rno-miR-27b-3p RT was specifically used
to add the stem-loop sequence during reverse transcription,
enabling accurate quantification of Rno-miR-27b-3p.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Primer name | Sequence
(5′-3′) |
|---|
| Rno-miR-27b-3p | F:
CCGGATTCACAGTGGCTAAG
R: AGTGCAGGGTCCGAGGTATT |
| Rno-miR-27b-3p
reverse transcription |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAGAA |
| Rat-U6 | F:
CTCGCTTCGGCAGCACA |
| Rat-U6 | R:
AACGCTTCACGAATTTGCGT |
Magnetic resonance imaging (MRI)
The rats were sacrificed at 8 weeks after spinal
puncture by injection with sodium pentobarbital (200 mg/kg). The
structure of the Co5/6 and Co6/7 discs was examined using a 3.0T
MRI scanner (Bruker BioSpin GmbH) and analyzed with RadiAnt DICOM
Viewer software (https://www.radiantviewer.com/). The extent of IVDD
was assessed with the Pfirrmann grading scale.
Immunohistochemical analysis
Caudal vertebrae specimens from rats were fixed in
10% neutral-buffered formalin at room temperature (20-25°C) for
24-48 h, and subsequently decalcified, dried, embedded in paraffin,
and then sectioned at 4-5 μm. The paraffin-embedded sections
were deparaffinized in xylene and rehydrated through a graded
ethanol series (100, 95, 70%) to distilled water, stained with
hematoxylin and eosin (H&E) and saffron O-fast green, dried,
mounted on glass slides, and examined under a fluorescent
microscope. Two of the authors evaluated the degree of IVDD in each
group.
Single-cell transcriptomic analysis
The publicly available single-cell RNA sequencing
dataset GSE205535 from normal (Normal_NP) and degenerated
(Degenerate_NP) IVD samples (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205535)
was analyzed. First, data quality control, normalization, and
principal component analysis were performed using the Seurat
package, followed by UMAP for dimensionality reduction to identify
different cell populations within the samples.
CD4+CD39+ Tregs were identified and clustered
based on the CD4 and ENTPD1 (CD39) markers. Differential expression
analysis (using the FindMarkers function) was conducted to compare
CD4+CD39+ Tregs between normal and
degenerated disc samples, identifying significantly differentially
expressed genes. Gene Ontology (GO) enrichment analysis was
performed to explore the functions of these genes. To further
investigate the regulatory role of miR-27b-3p in
CD4+CD39+ Tregs, GO enrichment analysis was
performed using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/). To further investigate
the regulatory role of miR-27b-3p in
CD4+CD39+ Tregs, target genes were predicted
using multiple databases, including TargetScanHuman 8.0 (https://www.targetscan.org/vert_80/), miRDB
(http://mirdb.org/), and miRTarBase (https://mirtarbase.cuhk.edu.cn/). The expression
of these predicted target genes was then compared between normal
and degenerated samples. Finally, GO functional enrichment analysis
and a volcano plot were used to visualize the differential
expression of miR-27b-3p target genes, revealing the potential role
of miR-27b-3p in regulating CD4+CD39+
Tregs.
Statistical analysis
All tests were conducted in triplicate or with a
minimum of three distinct samples. The mean ± standard deviation
was used to report continuous data in SPSS 22.0 program (IBM
Corp.). An unpaired Student's t-test was used to compare the mean
values of two independent groups. A one-way ANOVA followed by
Tukey's post hoc test was conducted to compare the mean values
across multiple independent groups, with statistically significant
difference determined by P<0.05. Statistical maps were generated
using GraphPad Prism 8 software (Dotmatics).
Statistics and reproducibility
All experiments were repeated at least three times,
and results are expressed as the mean ± standard error of the mean
(SEM). Data analysis was performed using GraphPad Prism 9 and R
language. Various statistical packages in R were utilized,
including ggplot2 for data visualization, lme4 for linear
mixed-effects model analysis, survival for survival analysis, and
DESeq2 for differential gene expression analysis. Key experiments
were independently repeated under different conditions,
demonstrating consistent results to ensure reproducibility. Sample
sizes were determined by power analysis to ensure adequate
statistical power.
Results
Effects of miRNAs on IVDD
MR analyses were used to assess the causal
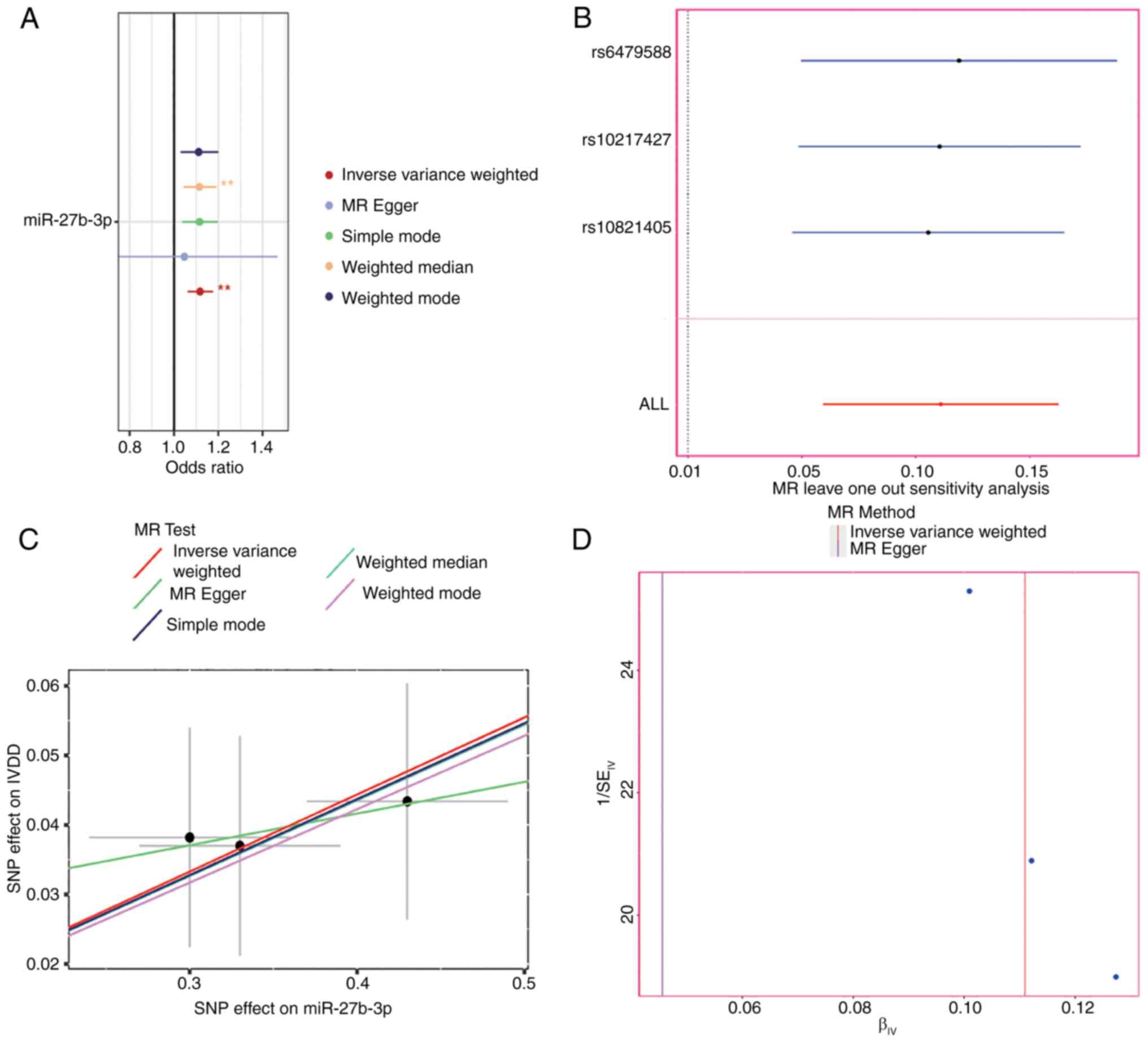
relationship between 76 miRNAs and IVDD. As shown in Fig. 1A, only miR-27a-3p had a
significant causal relationship with IVDD [odds ratio (OR)=1.12;
95% confidence interval (CI)=1.06-1.18; P<0.001] (Table SII). Then, the leave-one-out
method was used to separately remove the exposure of SNPs and the
MR analysis was repeated. The overall results were not affected by
a single SNP (Fig. 1B). A
scatter plot generated to summarize the MR analysis results between
miR-27a-3p and IVDD showed that the lines between different
algorithms were generally upward, indicating that the risk of IVDD
increased with the expression level of miR-27a-3p (Fig. 1C). The funnel plot results
eliminated possible outliers and the opportunity for miR-27a-3p
levels to be multidirectional (Fig.
1D).
Effects of immune cell on IVDD
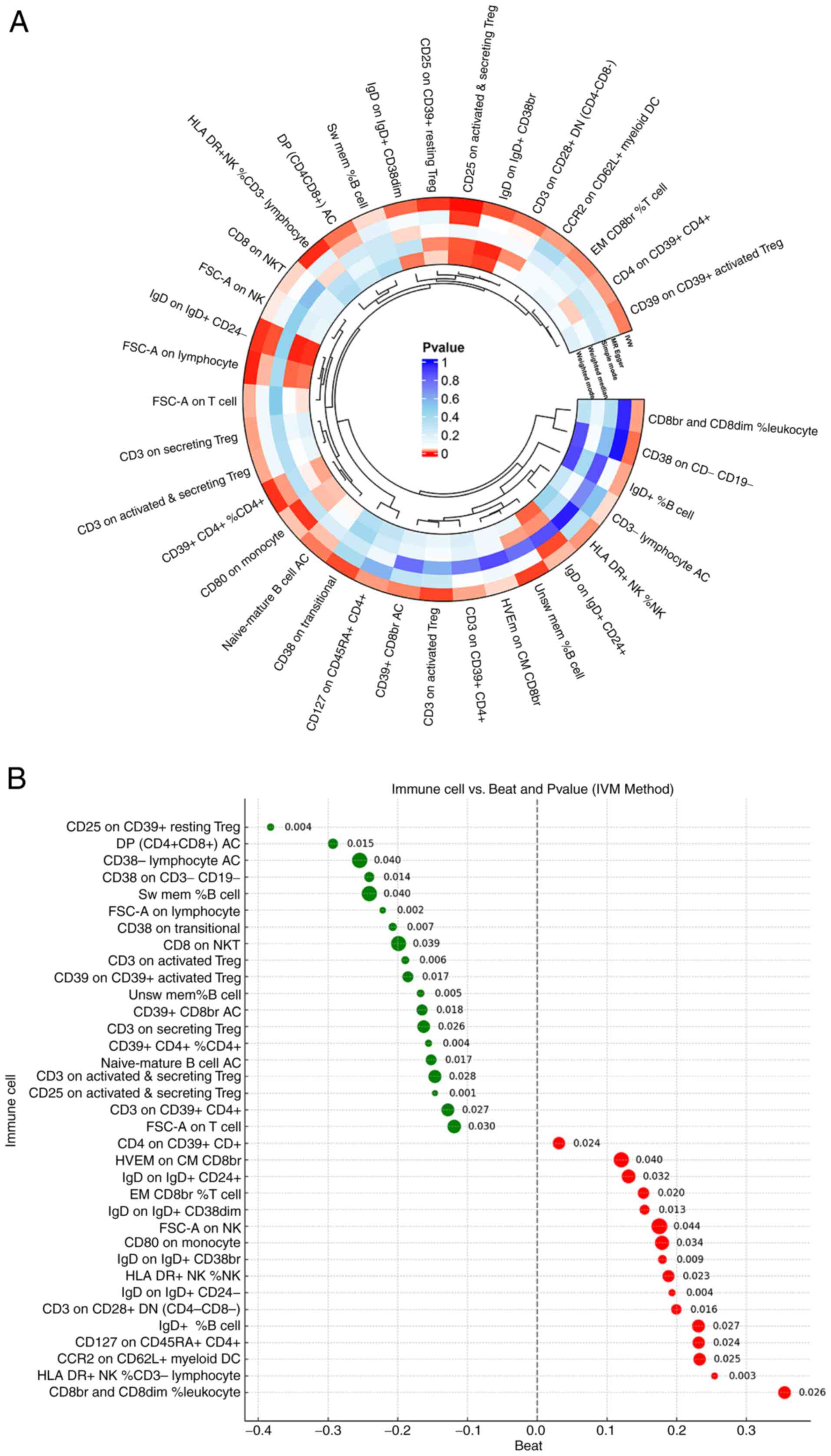
A total of five MR analysis modalities were employed
to identify potential relationships between immune cells and IVDD.
The findings indicated that 35 types of immune cells were
significantly associated with IVDD (P<0.05 with the IVW
technique) (Fig. 2A; Table SIII). Moreover, 19 types of
immune cells were positively correlated to IVDD when the value was
≤0, whereas 16 types of immune cells were positively correlated to
IVDD when the value was >1 (Fig.
2B). Then, in order to exclude false positive results, FDR
correction was performed on the MR results obtained from the batch
analysis. The results identified that 28 types of immune cells were
causally related to IVDD (Table
SIV).
CD4 on CD4+CD39+
Treg cells is positively correlated with miR-27a-3p
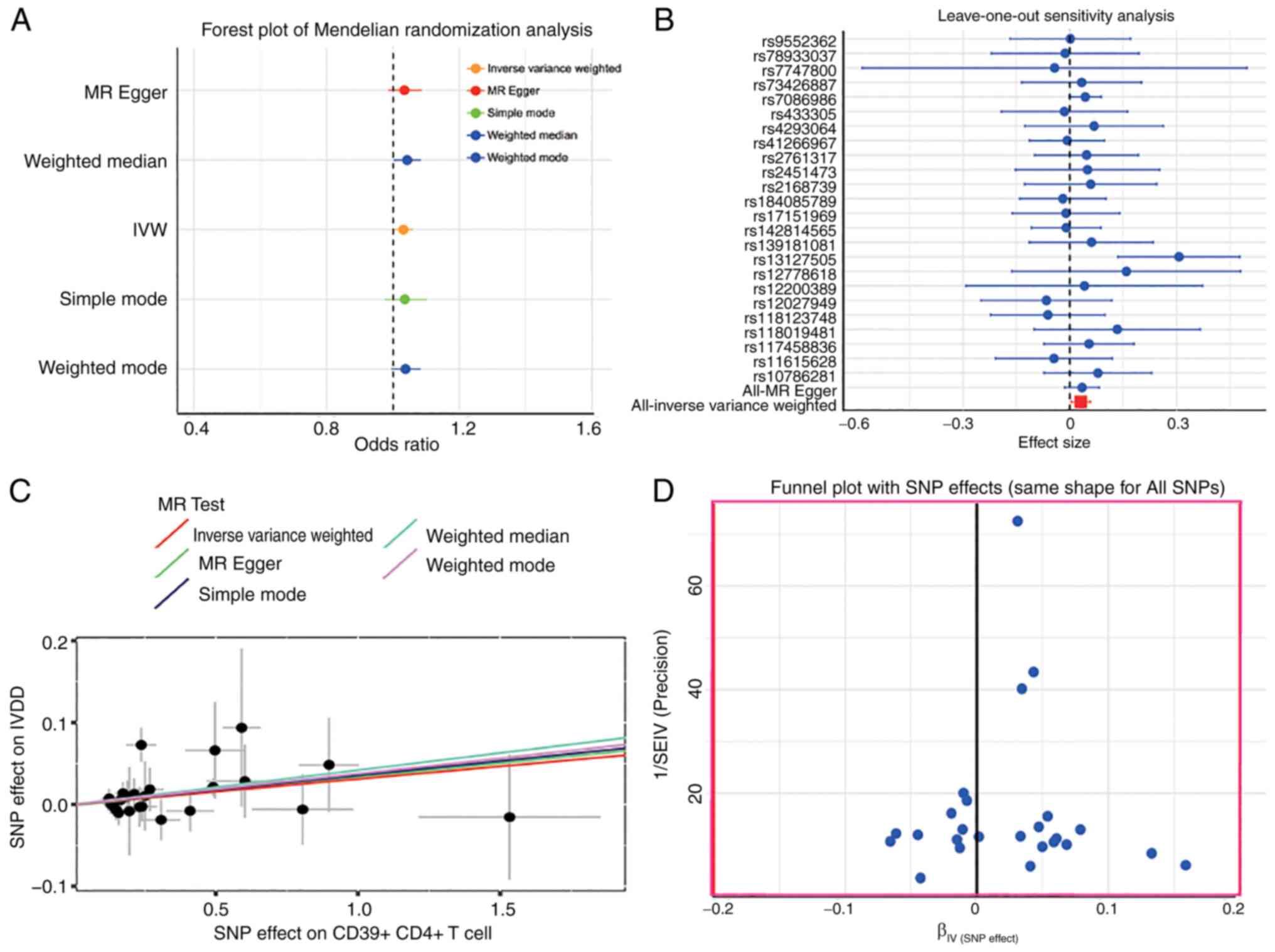
Two-sample MR analysis was conducted with 35 types
of immune cells as outcome factors and miR-27a-3p as the exposure
factor to assess the relationship between miR-27a-3p and IVDD. The
results revealed a significant causal relationship between
miR-27a-3p and CD4 on CD4+CD39+ Tregs
(OR=1.17, 95% CI=1.03-1.34, P=0.02) (Fig. 3A) (Table SV). The leave-one-out results
revealed no bias effect of individual SNPs on the overall results
(Fig. 3B). The scatter plot
results showed an overall negative correlation of miR-27a-3P and
CD4 on CD4+CD39+ Tregs (Fig. 3C). The funnel plot (Fig. 3D) excluded any potential outliers
and the possibility of multi-directivity of miR-27a-3P levels.
Reverse MR analysis, which was conducted to improve the reliability
of the results, found no reverse causality of miR-27a-3P and CD4 on
CD4+CD39+ Tregs.
Effect of miR-27a-3p on IVDD was achieved
by regulating CD4 on CD4+CD39+ Tregs
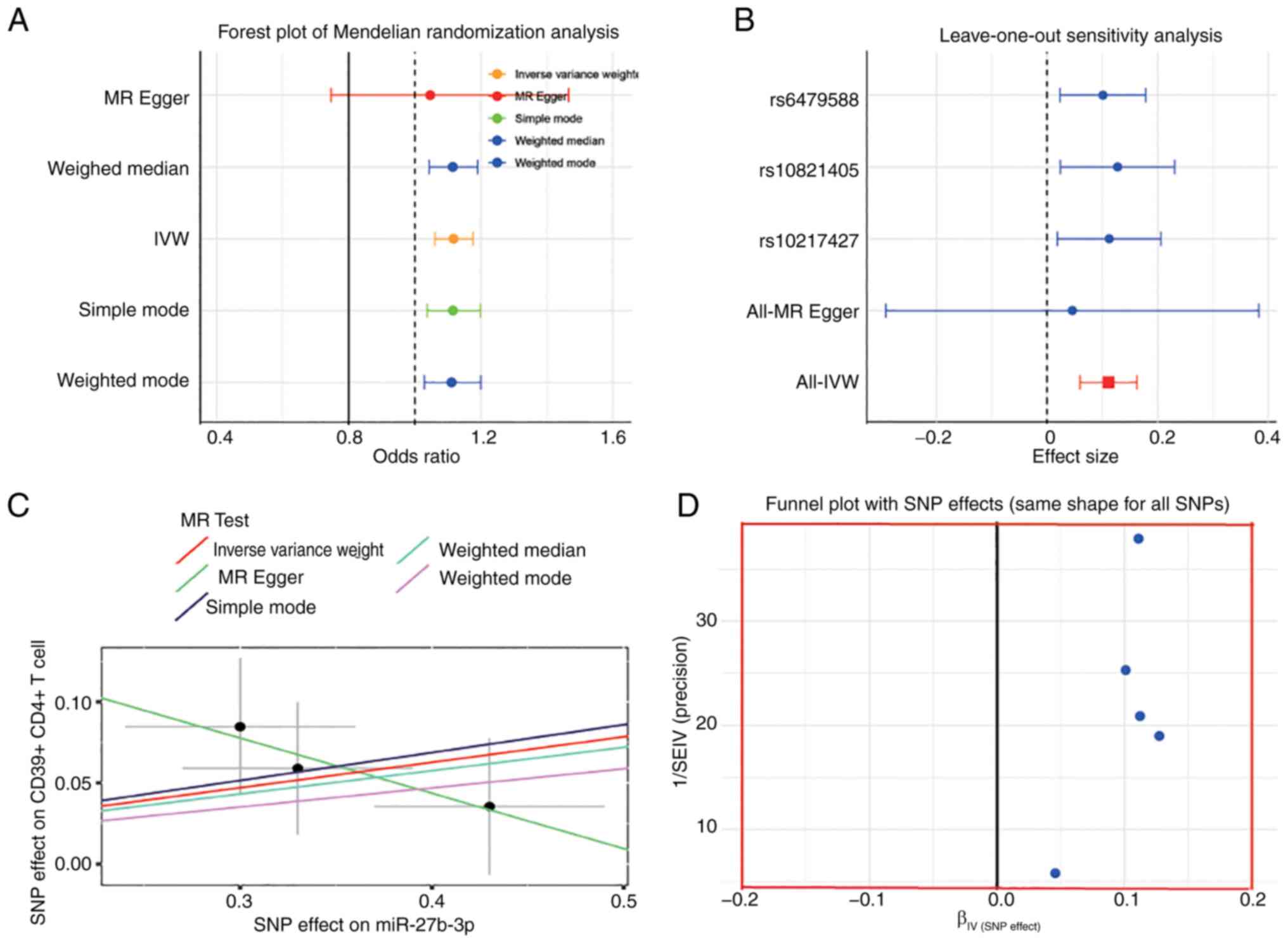
Multiple MR analyses confirmed a causal relationship
between IVDD and CD4 on CD4+CD39+ Tregs
(OR=1.03; 95% CI=1.004-1.060; P=0.024) (Fig. 4A). Analysis with the
leave-one-out method showed no deviation from the overall result of
each SNP (Fig. 4B). The scatter
plot results demonstrated that the lines of the five algorithms
generally tilted upward, indicating a positive correlation between
CD4+CD39+ Treg cells and IVDD (Fig. 4C). The funnel plot (Fig. 4D) excluded possible outliers and
opportunities for multi-directionality of
CD4+CD39+ Treg cell levels. In addition,
reverse MR analysis to improve the reliability of the results found
no reverse causality between IVDD and
CD4+CD39+ Tregs.
MiR-27a-3p enhances LPS-induced apoptosis
and modulates ECM markers in NP cells
Additional experiments were conducted using LPS,
miR-27a-3p, miR-27a-3p inhibition, LPS + miR-27a-3p and LPS +
miR-27a-3p inhibition to investigate the function of miR-27a-3p in
IVDD. The results of the TUNEL assay indicated that both LPS and
miR-27a-3p promoted apoptosis of NP cells as compared with the NC
group. In addition, miR-27a-3p enhanced LPS-induced apoptosis of NP
cells. The inclusion of the miR-27a-3p inhibitor and LPS +
miR-27a-3p inhibitor groups further confirmed the involvement of
miR-27a-3p in mediating apoptosis in NP cells (Fig. 5A). The results of western blot
analysis indicated that LPS and miR-27a-3p lowered the expression
levels of aggrecan and COL2A in NP cells. Expression of the
apoptosis-related protein Bax was increased. Furthermore,
miR-27a-3p enhanced the effect of LPS on the expression levels of
these proteins (Fig. 5B-E).
Suppression of miR-27a-3p activity prevented the decrease in
aggrecan and COL2A expression in LPS-treated NP cells, while
decreasing expression of the apoptosis-related protein Bax
(Fig. 5F-I). Although BAX is
directly related to the regulation of NP cell apoptosis, aggrecan
and COL2A are usually used as markers for assessing IVDD, mainly
for evaluating the degradation of ECM and the integrity of the IVD.
Aggrecan and COL2A were selected to assess the degenerative changes
of NP cells, which is a key feature of IVDD, while BAX and TUNEL
focus on apoptosis.
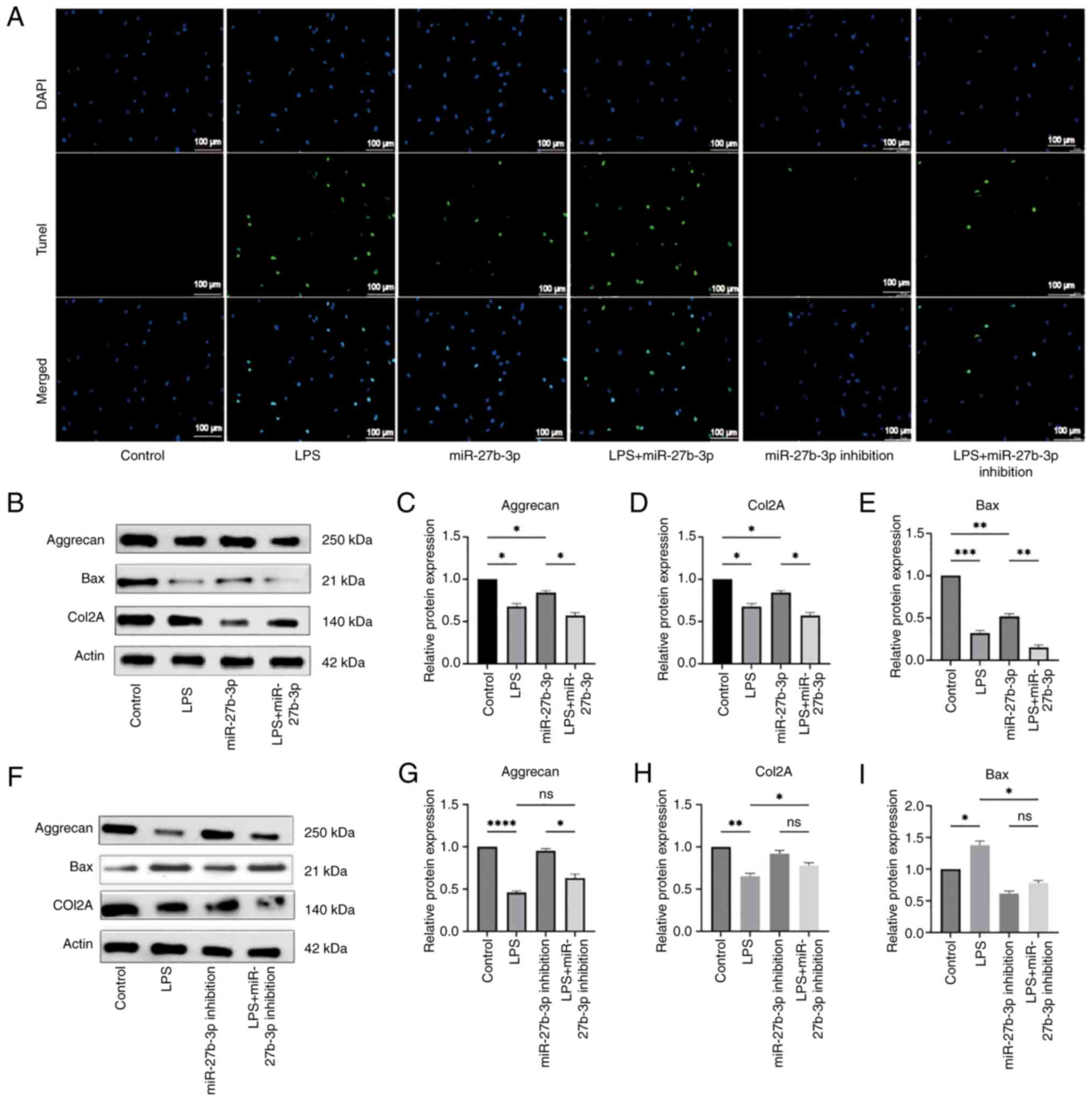 | Figure 5Apoptosis of NP cells is induced by
miR-27a-3p. (A) TUNEL analysis of cells in the NC, LPS, miR-27a-3p,
LPS + miR-27a-3p, miR-27a-3p, inhibition, and LPS + miR-27a-3p
inhibition groups. (B-E) Expression levels of apoptosis-related
proteins and IVDD-related proteins of cells in the NC, LPS,
miR-27a-3p, and LPS + miR-27a-3p groups. (F-I) Expression levels of
apoptosis-related proteins and IVDD-related proteins of cells in
the NC, LPS, miR-27a-3p inhibition and LPS + miR-27a-3p inhibition
groups. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. NP,
nucleus pulposus; miR, microRNA; NC, negative control; LPS,
lipopolysaccharide; IVDD, intervertebral disc degeneration; ns, not
significant. |
Increasing the proportion of
CD4+CD39+ Tregs by miR-27a-3p promotes
apoptosis of NP cells
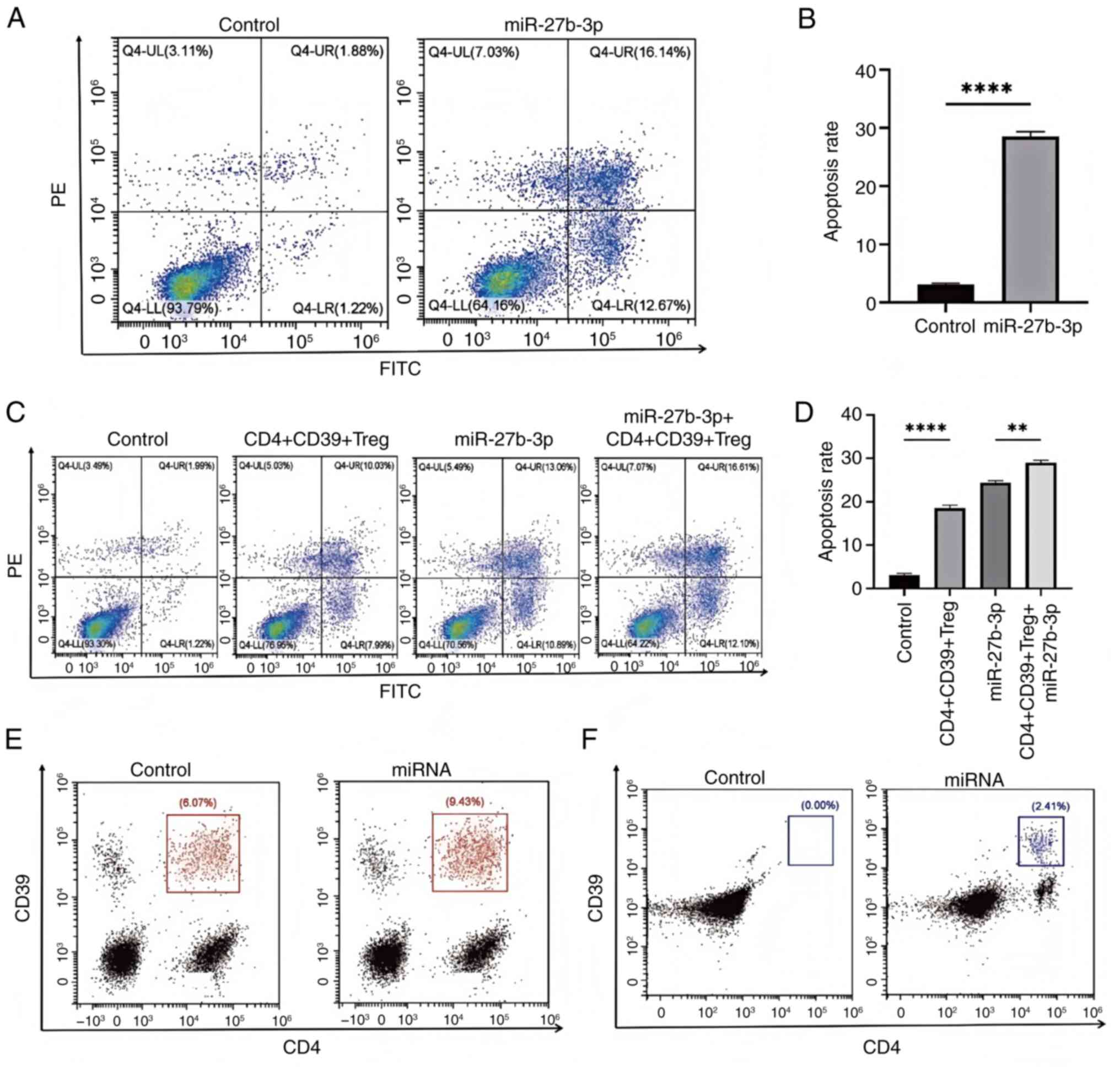
The addition of miR-27b-3p significantly increased
the apoptotic rate of CD4+CD39+ Tregs as
compared with the NC group (Fig. 6A
and B). CD4+CD39+ Tregs and miR-27b-3p
combined increased the proportion of apoptotic NP cells (Fig. 6C and D). The expression of
CD4+CD39+ Tregs in blood was increased in
rats with more severe disc degeneration (Fig. 6E). In rats with more severe disc
degeneration, the expression of CD4+CD39+
Tregs in the NP of the IVD was increased (Fig. 6F).
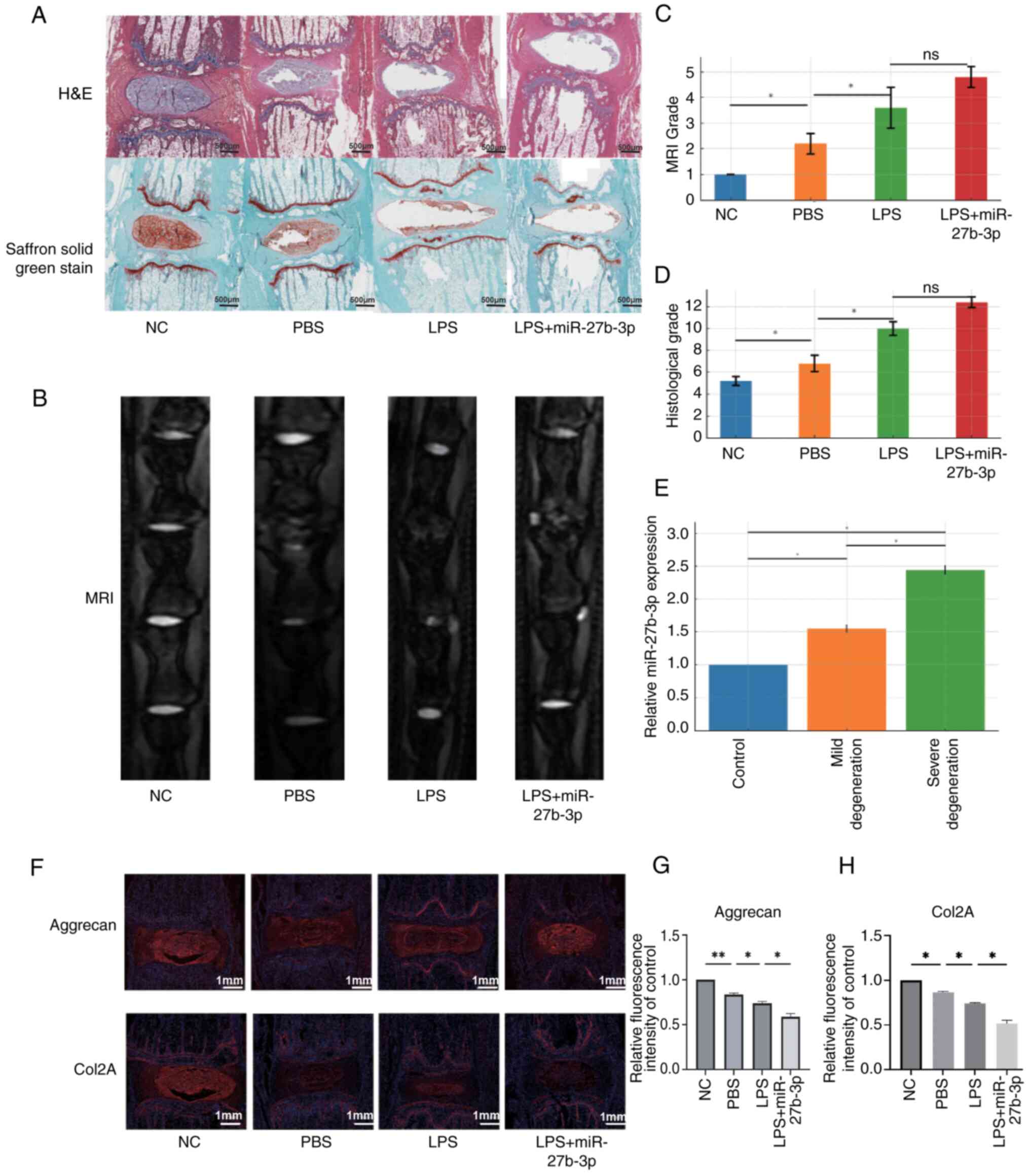
Progression of IVDD is accelerated by
miR-27a-3p
Histological analysis was conducted of the caudal
vertebra of rats following injection of PBS, LPS, and LPS +
miR-27a-3p. Staining with H&E and carthamin fast green (0.5%;
MilliporeSigma) revealed that the disc shape of the NC group was
complete and the NP and annulus fibrosus (AF) tissues were neatly
arranged. Injection of PBS and LPS altered the integrity of the
discs, resulting in a reduction in NP tissue. As compared with the
LPS group, LPS + miR-27a-3p reduced IVD integrity and the amount of
NP tissue (Fig. 7A and C). MRI
of the degenerated discs showed structural variability, changes in
signal strength, and blurred distinction between NP and AF tissues.
MRI also showed that injection of PBS and LPS promoted disc
degeneration as compared with the NC group. Moreover, disc
degeneration was more severe in the LPS + miR-27a-3p group as
compared with the LPS group (Fig. 7B
and D). Comparisons of the blood parameters of normal rats and
rats with IVDD by RT-qPCR analysis demonstrated that serum levels
of miR-27a-3p were higher in IVDD rats than the control rats
(Fig. 7E). In the NC group,
Aggrecan and Col2A are highly expressed with intact disc structure.
The PBS group (Control) showed a slight reduction in fluorescence,
indicating mild degeneration. The LPS group (Degeneration Model)
had significantly lower expression, with weakened signals and clear
signs of degeneration. In the LPS + miR-27b-3p group (Degeneration
with Treatment), further reduced fluorescence suggests the
treatment worsens disc degeneration (Fig. 7F-H). To further verify the role
of CD4+CD39+ Tregs in IVDD, the IF staining
data of the PBS group and the LPS + miR-27b-3p group were
supplemented. The results identified a significant increase in
CD4+CD39+ Tregs in the LPS + miR-27b-3p
group, further supporting that miR-27b-3p promotes the
immune-mediated process of IVDD by regulating immune cells,
especially CD4+CD39+ Tregs (Fig. S2).
Single-cell transcriptomic analysis
reveals the regulatory role of miR-27b-3p in
CD4+CD39+ Tregs during IVDD
In the present study,
CD4+CD39+ Tregs from normal and degenerated
IVD samples were analyzed using the single-cell transcriptomic
dataset GSE205535. The UMAP dimensionality reduction (Fig. 8A and B) revealed notable
differences in the distribution and clustering of
CD4+CD39+ Tregs between the Normal_NP and
Degenerate_NP disc samples. Specifically, the Normal_NP samples
exhibited a more homogeneous distribution of
CD4+CD39+ Tregs, while the Degenerate_NP
samples showed a more scattered distribution, suggesting a shift in
the composition of these immune cells in the context of disc
degeneration (Fig. 8C and D).
These findings highlight the altered immune landscape in
degenerated discs, with potential implications for immune-mediated
processes in IVDD. Differential expression analysis identified
significantly altered genes in CD4+CD39+
Tregs in the degenerated samples compared with normal samples,
particularly genes related to apoptosis and immune regulation
(Fig. 8E and F). GO enrichment
analysis further showed that these differentially expressed genes
are involved in key immune-related biological processes such as
monocyte activation and zinc ion response (Fig. 8F). Trajectory analysis using
pseudotime ordering (Fig. 8G)
revealed a dynamic progression of CD4+CD39+
Tregs from normal to degenerated samples, suggesting state
transitions associated with disc pathology. Furthermore,
pseudotime-associated expression patterns (Fig. 8H) showed that key genes,
including ACAN, BAX, BCL2, and COL2A1, exhibited distinct trends
across the trajectory, highlighting altered regulation of
extracellular matrix homeostasis and apoptosis-related pathways
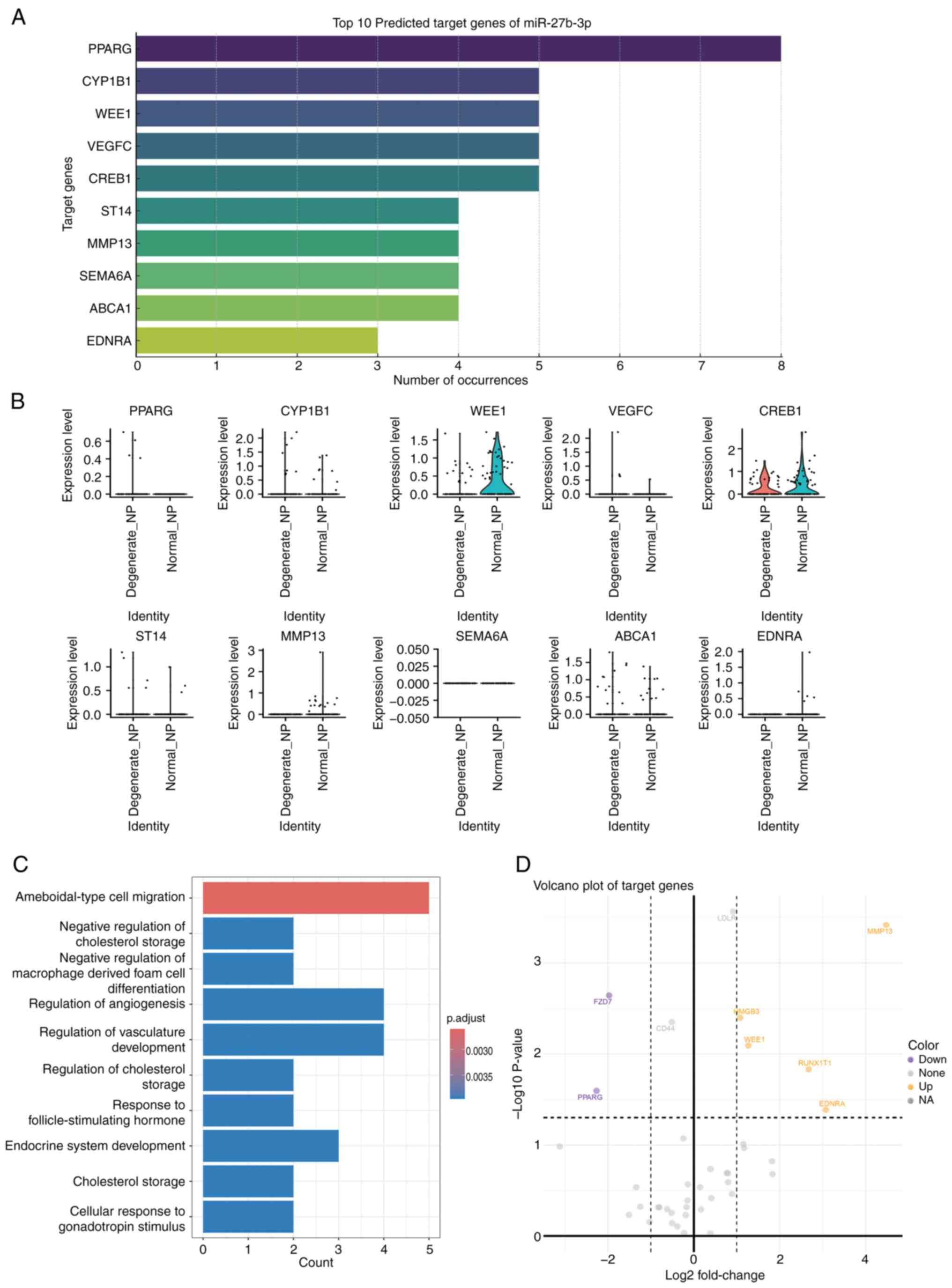
during degeneration. Additionally, bioinformatics tools were used
to predict the target genes of miR-27b-3p, and distinct expression
patterns of these target genes were found in
CD4+CD39+ Tregs (Fig. 9A). Notably, genes such as PPARG,
WEE1 and MMP13 were significantly upregulated or downregulated in
degenerated discs (Fig. 9B). GO
functional enrichment analysis indicated that these target genes
are closely associated with processes such as immune cell
migration, regulation of immune responses and cholesterol
metabolism (Fig. 9C). The
volcano plot further revealed the differential expression patterns
of these target genes (Fig. 9D),
suggesting that miR-27b-3p may exert its role in IVDD by regulating
CD4+CD39+ Tregs and their associated immune
functions.
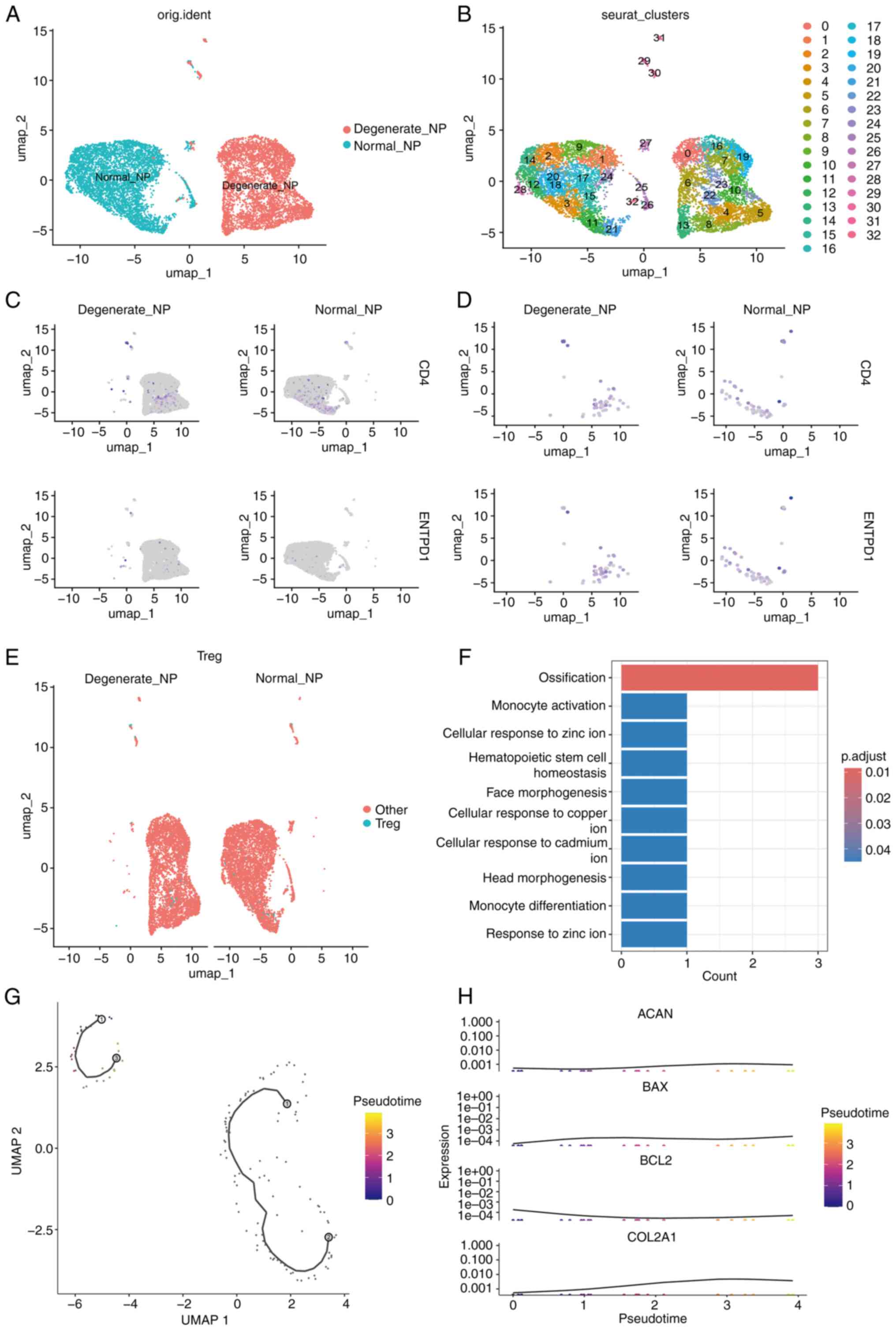 | Figure 8UMAP and functional analysis of
CD4+CD39+ Tregs in normal and degenerated
intervertebral discs. (A and B) UMAP plots showing the distribution
of CD4+CD39+ Tregs in Normal_NP) and
Degenerate_NP intervertebral disc samples. The cells are clustered
according to their gene expression profiles, revealing distinct
populations in each condition. Normal_NP samples exhibit a more
compact and uniform distribution of Tregs, while Degenerate_NP
samples show a more dispersed distribution, suggesting a shift in
Treg cell composition during disc degeneration. (C and D) Feature
plots highlight the expression of CD4 and ENTPD1 (CD39), used to
identify Tregs. (E) Distribution of CD4+CD39+
Tregs across different conditions. (F) Results of Gene Ontology
enrichment analysis, showing significant involvement of these cells
in biological processes such as monocyte activation and zinc ion
response. (G) Pseudotime trajectory analysis demonstrating dynamic
state transitions of CD4+CD39+ Tregs. (H)
Pseudotime-associated gene expression trends of ACAN,
BAX, BCL2, and COL2A1, illustrating altered
regulation of extracellular matrix and apoptosis pathways during
degeneration. Tregs, regulatory T cells; NP, nucleus pulposus. |
Discussion
Our study breaks new ground by demonstrating the
causal role of miR-27b-3p in IVDD through MR, a method rarely
applied in this context. This approach minimizes confounding
factors and strengthens the evidence for miR-27b-3p as a potential
therapeutic target. Additionally, the discovery of its interaction
with CD4+CD39+ Treg cells adds a novel
dimension to our understanding of immune-related degeneration
processes, making this research highly relevant to both the fields
of immunology and aging.MR analysis and experimental validation
both in vitro and in vivo yielded compelling evidence
on the association between miR-27a-3p and IVDD. Two-sample MR
analysis of 76 miRNAs and IVDD revealed a significant causal
association between miR-27a-3p and IVDD. A total of 731 immune cell
types were incorporated as mediators to gain a deeper understanding
of potential causal links. MR analysis of 731 immune cell types and
IVDD identified 35 immune cell types causally linked to IVDD.
Two-sample MR analysis was conducted using immune cells as the
dependent variable and miR-27a-3p as the instrumental variable. The
MR analysis using human samples revealed a significant causal
relationship between miR-27b-3p and IVDD. These results suggest
that miR-27b-3p could serve as a viable biomarker for early
diagnosis or as a target for therapeutic intervention in clinical
settings. The findings indicate that miR-27b-3p plays a crucial
role in the progression of IVDD by inducing NP cell apoptosis, a
key feature of disc degeneration. This effect was demonstrated both
in vitro and in vivo. Additionally, miR-27b-3p
modulates the immune response by influencing
CD4+CD39+ Treg cells, which are important
regulators of inflammation and immune responses. It was observed
that miR-27b-3p affects apoptosis-related proteins, such as Bax,
promoting NP cell apoptosis, while simultaneously impacting the
expression of ECM components such as COL2A and aggrecan, which are
critical for maintaining disc structure and function. The use of
LPS in IVDD models was crucial for inducing inflammation, providing
a platform to study the inflammatory component of IVDD. miR-27b-3p
was found to enhance the LPS-induced inflammatory response, further
exacerbating disc degeneration by promoting apoptosis and matrix
degradation. In addition, the current findings revealed that
miR-27b-3p regulates several key genes involved in inflammation and
matrix remodeling. PPARG, a nuclear receptor involved in immune
regulation, was found to be downregulated by miR-27b-3p, suggesting
that miR-27b-3p may contribute to inflammatory responses by
interfering with PPARG signaling (40). Similarly, miR-27b-3p also
modulates WEE1, a cell cycle regulator, which could impact the
survival of NP cells by regulating cell cycle progression and
response to DNA damage. Finally, miR-27b-3p was shown to regulate
MMP13, a matrix metalloproteinase responsible for collagen
degradation, contributing to the breakdown of the ECM in
degenerated discs. The interaction of miR-27b-3p with these key
regulatory pathways further supports its role as a central
modulator in the pathogenesis of IVDD (41). This novel connection between
miR-27b-3p and CD4+CD39+ Tregs highlights a
previously unexplored pathway, where miR-27b-3p not only
contributes to the apoptosis of NP cells but also regulates
immune-mediated degeneration processes. These findings open new
avenues for targeting miR-27b-3p in the treatment of IVDD, offering
potential strategies for both immune modulation and preservation of
disc integrity. It was chosen to focus on
CD4+CD39+ Tregs in relation to IVDD primarily
due to their role as intermediaries in the miR-27b-3p-mediated
regulation of IVDD. Our MR analysis examined 731 immune cell types
and their potential causal relationships with IVDD. Among these,
CD4+CD39+ Tregs were identified as
significantly associated with the progression of IVDD. However, it
is important to note that the focus on these cells was not due to
their direct association with IVDD, but rather because they serve
as key regulators of the immune response in the disc, specifically
through their interaction with miR-27b-3p. It was hypothesized that
miR-27b-3p influences the function of
CD4+CD39+ Tregs, and in doing so, regulates
NP cell apoptosis and ECM degradation during IVDD progression. Our
MR analysis highlighted these cells as critical intermediators in
the pathway through which miR-27b-3p affects disc degeneration
(42).
Recent studies have implicated miRNAs in several
signaling pathways as significant contributors to the development
of various illnesses (43,44). These tiny non-coding RNAs have
the ability to inhibit translation, leading to reduced expression
levels of certain genes (45).
IVDD is a significant contributor to low back pain (46,47). Current treatments help to
alleviate symptoms but are not curative. It is increasingly
recognized that the dysregulation of miRNAs plays a crucial role in
the progression of IVDD (48).
Several miRNAs have been shown to regulate key processes involved
in IVDD, such as inflammation, matrix degradation and apoptosis,
making them promising therapeutic targets. For example, previous
studies have identified miR-21 and miR-133a as key miRNAs involved
in the degeneration of IVDs. miR-21 has been reported to contribute
to IVDD by regulating the inflammatory response and promoting
matrix degradation (49).
Similarly, miR-133a has been shown to regulate ECM remodeling, and
its downregulation is associated with increased disc degeneration
(50). These findings highlight
the importance of miRNAs in IVDD, yet the exact mechanisms by which
they mediate disc degeneration remain under explored. In contrast
to these studies, the present study focuses on miR-27b-3p, a member
of the miR-27 family that has not been extensively studied in the
context of IVDD (51).
miR-27b-3p has been implicated in regulating immune responses and
apoptosis, processes known to contribute significantly to IVDD
progression. The current findings suggest that miR-27b-3p directly
modulates CD4+CD39+ Tregs, which are key
players in immune regulation and inflammation. It was observed that
miR-27b-3p promotes NP cell apoptosis and matrix degradation by
influencing the immune microenvironment in degenerated discs. This
provides a novel insight into the immune-modulatory role of
miR-27b-3p in IVDD, distinguishing our study from previous research
that has mainly focused on other miRNAs like miR-21 and miR-133a.
While previous studies have demonstrated the dysregulation of
miR-21 and miR-133a in IVDD, the present study adds a new dimension
by identifying miR-27b-3p as a key regulator of immune cells,
specifically CD4+CD39+ Tregs, in the context
of IVDD. Several studies have indicated that miR-27a plays a
crucial role in modulation of polymorphisms, carcinogenesis,
proliferation, apoptosis, invasion, migration, and angiogenesis
(52). A recent study revealed
that expression of miR-27a-5p was increased in the exosomes of mice
as models of Alzheimer's disease (53).
CD4 T cells are crucial in facilitating adaptive
immunity against several infections and have also been implicated
in autoimmunity, asthma, allergic responses and tumor immunity
(54,55). Naive CD4 T cells can develop into
diverse T helper (Th) cell lineages, such as Th1, Th2, Th17 and
induced Tregs, via TCR activation in particular cytokine
environments. These lineages are characterized by cytokine
production and specific functions (56,57). Treg and Th17 cells are two
subgroups of CD4+ T cells with opposing effects. Th17
cells contribute to inflammation, whereas Tregs play a vital role
in preserving immunological balance (58,59). Altered activity of Tregs plays a
crucial role in the onset and progression of immuno-senescence,
leading to greater vulnerability to age-related immune-mediated
illnesses (60). The IVD is
considered an immune privileged organ due to the lack of blood
vessels. During IVDD, immune cells and inflammatory factors can
enter the IVD by penetrating the cartilage endplate and AF tissues,
leading to the development of IVDD. In recent decades, there has
been an increasing number of publications on the inflammatory
response in the IVD, which has garnered increasing interest in a
causal link between CD4 Tregs and IVDD, highlighting prospective
treatment approaches (61,62). CD4+ Tregs exert their
regulatory effects by producing anti-inflammatory cytokines such as
IL-10 and TGF-β, which inhibit the activation of pro-inflammatory T
cells and macrophages. In the context of IVDD, the presence of
Tregs in the IVD may help control the inflammatory cascade, reduce
matrix degradation, and potentially promote tissue repair.
Importantly, miR-27b-3p, as identified in the present study, may
enhance Treg cell activity by regulating immune signaling pathways,
further supporting its potential therapeutic value in modulating
immune responses in IVDD. MR analysis strengthened the reliability
of the findings of the first part of the present study. The
consistency of five MR analysis modalities (weighted median,
MR-Egger, simple mode, weighted mode and IVW) enhanced the validity
of the results (63). The
MR-PRESSO approach was utilized to re-evaluate the presence of
diverse SNPs, while Cochran's Q test was employed to evaluate the
notable heterogeneity among the chosen instrumental factors
(64,65). The miRNA identified by MR
analysis was utilized in the IVDD model in conjunction with in
vitro and in vivo investigations. The advantage of
utilizing MR analysis is the use of existing data from large-scale
genome-wide association studies, which can reveal potential causal
relationships between modifiable factors and disease, while
allowing the inclusion of risk factors with suitable genetic
variants. Genetic variants typically have relatively small effects
on most risk factors and explain only a small portion of the
variation, while decreasing the statistical power of MR analyses
and the risk of false-negative results. To avoid the risk of
false-positive results, in vivo and in vitro
experiments were designed based on the MR findings, thereby
completing an evidence triangle and providing new research ideas
for the prevention and treatment of IVDD.
There were certain limitations to the present study
that should be addressed. First, the data were derived exclusively
from Europeans. Second, although both in vitro and in
vivo investigations demonstrated that miR-27a-3p can enhance
the development of IVDD by promoting apoptosis of NP cells, the
role CD4+CD39+ Treg cells in this process
remains unclear. Finally, a miR-27b-3p alone treatment group was
not included because our primary objective was to assess the role
of miR-27b-3p in the context of LPS-induced inflammation, which is
a key aspect of IVDD. LPS was used to model the inflammatory
environment commonly associated with IVDD, and the aim was to
observe the interaction between miR-27b-3p and inflammation.
Although a miR-27b-3p alone group was not included, the influence
of miR-27b-3p on NP cell apoptosis in the LPS + miR-27b-3p group
was determined based on the significant changes observed in
apoptosis-related markers (Bax) and ECM proteins (Aggrecan and
COL2A). The fact that miR-27b-3p enhanced LPS-induced apoptosis
suggests that miR-27b-3p plays an important role in regulating
apoptosis in an inflammatory environment. However, it is
acknowledged that a miR-27b-3p alone group would have provided
additional clarity regarding its independent effect. Future studies
could include such a group to improve understanding of miR-27b-3p's
standalone effects.
The findings of the present study not only advance
our understanding of miR-27b-3p's role in IVDD but also contribute
to the broader understanding of aging-related degeneration. The
modulation of CD4+CD39+ Tregs by miR-27b-3p
suggests a potential mechanism through which immune aging
accelerates musculoskeletal decline. These results align with
emerging theories of 'inflammaging', where chronic low-grade
inflammation contributes to age-related tissue degeneration.
Targeting miR-27b-3p may, therefore, represent a novel therapeutic
strategy to slow the progression of IVDD and possibly other
aging-associated disorders. Future research could further explore
the role of miR-27a-3p in other degenerative diseases, especially
those involving immune system regulation. Given the high prevalence
of IVDD and its significant impact on quality of life, the results
of the present study are expected to drive the development of new
diagnostic biomarkers and offer novel therapeutic targets for
personalized treatment strategies. By targeting miR-27a-3p or the
immune cells it regulates, more effective therapies could be
developed to slow down or even reverse the progression of IVDD,
which holds broad potential for clinical applications.
Additionally, future studies could investigate the generalizability
of miR-27a-3p-related pathways in other inflammation-related
degenerative diseases, paving the way for innovative cross-disease
therapeutic strategies. Future studies could explore the broader
applicability of these findings by investigating whether miR-27b-3p
plays a similar role in other age-related degenerative diseases.
Additionally, clinical trials targeting miR-27b-3p or its
downstream pathways in patients with IVDD could provide critical
insights into the therapeutic potential of this approach.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
found in the Gene Expression Omnibus under accession number
GSE205535 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205535.
Authors' contributions
QWL designed the study, performed the Mendelian
randomization analysis, and interpreted the data. CHZ conducted the
experimental work, including in vivo and in vitro
analyses, and contributed to data interpretation. PLJ made
substantial contributions to the acquisition of data, assisted with
data analysis and interpretation, and revised the manuscript
critically for important intellectual content. CLS contributed to
the conception and design of the study, supervised the research,
provided critical revisions, and finalized the manuscript. All
authors read and approved the final version of the manuscript,
agreed to be accountable for all aspects of the work, and confirm
their contributions are consistent with the ICMJE criteria. QWL and
CHZ confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The animal experimental protocols were approved
(approval no. IHM-AP-2024-015) by the Laboratory Animal Care and
Use Committee of the Institute of Health and Medicine of Hefei
Comprehensive National Science Center (Hefei, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 82272551 and 81772408).
References
|
1
|
Saberi M, Zhang X and Mobasheri A:
Targeting mitochondrial dysfunction with small molecules in
intervertebral disc aging and degeneration. Geroscience.
43:517–537. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang F, Cai F, Shi R, Wang XH and Wu XT:
Aging and age related stresses: A senescence mechanism of
intervertebral disc degeneration. Osteoarthritis Cartilage.
24:398–408. 2016. View Article : Google Scholar
|
|
3
|
Wang Y, Cheng H, Wang T, Zhang K, Zhang Y
and Kang X: Oxidative stress in intervertebral disc degeneration:
Molecular mechanisms, pathogenesis and treatment. Cell Prolif.
56:e134482023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiang Q, Zhao Y and Li W: Identification
and validation of ferroptosis-related gene signature in
intervertebral disc degeneration. Front Endocrinol (Lausanne).
14:10897962023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yi J, Zhou Q, Huang J, Niu S, Ji G and
Zheng T: Lipid metabolism disorder promotes the development of
intervertebral disc degeneration. Biomed Pharmacother.
166:1154012023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koroth J, Buko EO, Abbott R, Johnson CP,
Ogle BM, Stone LS, Ellingson AM and Bradley EW: Macrophages and
intervertebral disc degeneration. Int J Mol Sci. 24:13672023.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Samanta A, Lufkin T and Kraus P:
Intervertebral disc degeneration: Current therapeutic options and
challenges. Front Public Health. 11:11567492023. View Article : Google Scholar
|
|
8
|
Silwal P, Nguyen-Thai AM, Mohammad HA,
Wang Y, Robbins PD, Lee JY and Vo NV: Cellular senescence in
intervertebral disc aging and degeneration: Molecular mechanisms
and potential therapeutic opportunities. Biomolecules. 13:6862023.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang HS, Lin S and Yu HM: Exosome-mediated
repair of intervertebral disc degeneration: The potential role of
miRNAs. Curr Stem Cell Res Ther. 19:798–808. 2024. View Article : Google Scholar
|
|
10
|
Sharma Y, Saini AK, Kashyap S, Chandan G,
Kaur N, Gupta VK, Thakur VK, Saini V and Saini RV: Host miRNA and
immune cell interactions: Relevance in nano-therapeutics for human
health. Immunol Res. 70:1–18. 2022. View Article : Google Scholar
|
|
11
|
Schell SL and Rahman ZSM: miRNA-mediated
control of B cell responses in immunity and SLE. Front Immunol.
12:6837102021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bronevetsky Y and Ansel KM: Regulation of
miRNA biogenesis and turnover in the immune system. Immunol Rev.
253:304–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wythe SE, Nicolaidou V and Horwood NJ:
Cells of the immune system orchestrate changes in bone cell
function. Calcif Tissue Int. 94:98–111. 2014. View Article : Google Scholar
|
|
14
|
Sun K, Jiang J, Wang Y, Sun X, Zhu J, Xu
X, Sun J and Shi J: The role of nerve fibers and their
neurotransmitters in regulating intervertebral disc degeneration.
Ageing Res Rev. 81:1017332022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu H, Li J, Fei Q and Jiang L:
Contribution of immune cells to intervertebral disc degeneration
and the potential of immunotherapy. Connect Tissue Res. 64:413–427.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Song C, Zhou Y, Cheng K, Liu F, Cai W,
Zhou D, Chen R, Shi H, Fu Z, Chen J and Liu Z: Cellular
senescence-molecular mechanisms of intervertebral disc degeneration
from an immune perspective. Biomed Pharmacother. 162:1147112023.
View Article : Google Scholar
|
|
17
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar
|
|
18
|
Sun Z, Liu ZH, Chen YF, Zhang YZ, Wan ZY,
Zhang WL, Che L, Liu X, Wang HQ and Luo ZJ: Molecular immunotherapy
might shed a light on the treatment strategies for disc
degeneration and herniation. Med Hypotheses. 81:477–480. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang T, He Z, Zhu Z, Wang F, Chen H, Zhang
F, Zhou J, Wang J, Li B, Liu X, et al: Identification of novel gene
signatures and immune cell infiltration in intervertebral disc
degeneration using bioinformatics analysis. Front Mol Biosci.
10:11697182023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Zhang J, Sun Z, Wang H, Ning R,
Xu L, Zhao Y, Yang K, Xi X and Tian J: MAPK8 and CAPN1 as potential
biomarkers of intervertebral disc degeneration overlapping immune
infiltration, autophagy, and ceRNA. Front Immunol. 14:11887742023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen LG, Tubbs JD, Liu Z, Thach TQ and
Sham PC: Mendelian randomization: Causal inference leveraging
genetic data. Psychol Med. 54:1461–1474. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang A, Zhao Y, Yang P, Zhang X and
Giovannucci EL: Vitamin D and human health: Evidence from Mendelian
randomization studies. Eur J Epidemiol. 39:467–490. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Swanson SA, Labrecque J and Hernán MA:
Causal null hypotheses of sustained treatment strategies: What can
be tested with an instrumental variable? Eur J Epidemiol.
33:723–728. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Evans DM and Davey Smith G: Mendelian
randomization: New applications in the coming age of
hypothesis-free causality. Annu Rev Genomics Hum Genet. 16:327–350.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang P, Li Z and Ye D: Single-cell RNA-seq
analysis reveals the Wnt/Ca2+ signaling pathway with
inflammation, apoptosis in nucleus pulposus degeneration. BMC
Musculoskelet Disord. 25:3212024. View Article : Google Scholar
|
|
26
|
Huan T, Rong J, Liu C, Zhang X, Tanriverdi
K, Joehanes R, Chen BH, Murabito JM, Yao C, Courchesne P, et al:
Genome-wide identification of microRNA expression quantitative
trait loci. Nat Commun. 6:66012015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Orrù V, Steri M, Sidore C, Marongiu M,
Serra V, Olla S, Sole G, Lai S, Dei M, Mulas A, et al: Complex
genetic signatures in immune cells underlie autoimmunity and inform
therapy. Nat Genet. 52:1036–1045. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burgess S and Thompson SG; CRP CHD
Genetics Collaboration: Avoiding bias from weak instruments in
Mendelian randomization studies. Int J Epidemiol. 40:755–764. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bowden J, Del Greco MF, Minelli C, Zhao Q,
Lawlor DA, Sheehan NA, Thompson J and Davey Smith G: Improving the
accuracy of two-sample summary-data Mendelian randomization: Moving
beyond the NOME assumption. Int J Epidemiol. 48:728–742. 2019.
View Article : Google Scholar :
|
|
30
|
Glickman ME, Rao SR and Schultz MR: False
discovery rate control is a recommended alternative to
Bonferroni-type adjustments in health studies. J Clin Epidemiol.
67:850–857. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dudbridge F: Polygenic Mendelian
randomization. Cold Spring Harb Perspect Med. 11:a0395862021.
View Article : Google Scholar
|
|
32
|
Greco MFD, Minelli C, Sheehan NA and
Thompson JR: Detecting pleiotropy in Mendelian randomisation
studies with summary data and a continuous outcome. Stat Med.
34:2926–2940. 2015. View Article : Google Scholar
|
|
33
|
Fan C, Wang W, Yu Z, Wang J, Xu W, Ji Z,
He W, Hua D, Wang W, Yao L, et al: M1 macrophage-derived exosomes
promote intervertebral disc degeneration by enhancing nucleus
pulposus cell senescence through LCN2/NF-κB signaling axis. J
Nanobiotechnology. 22:3012024. View Article : Google Scholar
|
|
34
|
Lin Z, Xu G, Lu X, Liu S, Zou F, Ma X,
Jiang J, Wang H and Song J: Chondrocyte-targeted exosome-mediated
delivery of Nrf2 alleviates cartilaginous endplate degeneration by
modulating mitochondrial fission. J Nanobiotechnology. 22:2812024.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dai J, Liu J, Shen Y, Zhang B, Li C and
Liu Z: Regulation of endoplasmic reticulum stress on autophagy and
apoptosis of nucleus pulposus cells in intervertebral disc
degeneration and its related mechanisms. PeerJ. 12:e172122024.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao W, Bao J, Zhang Y, He D, Zhang L,
Zhang J, Pan H and Wang D: Injectable kaempferol-loaded fibrin glue
regulates the metabolic balance and inhibits inflammation in
intervertebral disc degeneration. Sci Rep. 13:200012023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen X, Zhang A, Zhao K, Gao H, Shi P,
Chen Y, Cheng Z, Zhou W and Zhang Y: The role of oxidative stress
in intervertebral disc degeneration: Mechanisms and therapeutic
implications. Ageing Res Rev. 98:1023232024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo W, Mu K, Geng JC, Xing HY, Dong Y, Liu
WD, Wang SC, Shi JX, Xing BR, Zhao JY and Li XM: ATF1 and
miR-27b-3p drive intervertebral disc degeneration through the
PPARG/NF-κB signaling axis. Commun Biol. 8:7512025. View Article : Google Scholar
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
40
|
Rouas R, Merimi M, Najar M, El Zein N,
Fayyad-Kazan M, Berehab M, Agha D, Bron D, Burny A, Rachidi W, et
al: Human CD8+ CD25+ CD127low
regulatory T cells: microRNA signature and impact on TGF-β and
IL-10 expression. J Cell Physiol. 234:17459–17472. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ferragut Cardoso AP, Banerjee M, Nail AN,
Lykoudi A and States JC: miRNA dysregulation is an emerging
modulator of genomic instability. Semin Cancer Biol. 76:120–131.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vafadar A, Shirazi-Tehrani E, Vosough P,
Khalili Alashti S, Kargar Jahromi H, Bagheri Lankarani K,
Savardashtaki A and Ehtiati S: Non-coding RNAs in celiac disease.
Clin Chim Acta. 576:1204172025. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Y, Kang J, Guo X, Zhu D, Liu M, Yang
L, Zhang G and Kang X: Intervertebral disc degeneration models for
pathophysiology and regenerative therapy-benefits and limitations.
J Invest Surg. 35:935–952. 2022. View Article : Google Scholar
|
|
45
|
Hu A, Xing R, Jiang L, Li Z, Liu P, Wang
H, Li X and Dong J: Thermosensitive hydrogels loaded with
human-induced pluripotent stem cells overexpressing growth
differentiation factor-5 ameliorate intervertebral disc
degeneration in rats. J Biomed Mater Res B Appl Biomater.
108:2005–2016. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kilkenny C, Browne WJ, Cuthill IC, Emerson
M and Altman DG: Improving bioscience research reporting: The
ARRIVE guidelines for reporting animal research. PLoS Biol.
8:e10004122010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Griffith JF, Wang YXJ, Antonio GE, Choi
KC, Yu A, Ahuja AT and Leung PC: Modified Pfirrmann grading system
for lumbar intervertebral disc degeneration. Spine (Phila Pa 1976).
32:E708–E712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Genedy HH, Humbert P, Laoulaou B, Le Moal
B, Fusellier M, Passirani C, Le Visage C, Guicheux J, Lepeltier É
and Clouet J: MicroRNA-targeting nanomedicines for the treatment of
intervertebral disc degeneration. Adv Drug Deliv Rev.
207:1152142024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Y, Deng M, Wu Y, Zheng C, Zhang F,
Guo C, Zhang B, Hu C, Kong Q and Wang Y: A multifunctional
mitochondria-protective gene delivery platform promote
intervertebral disc regeneration. Biomaterials. 317:1230672025.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xu YQ, Zhang ZH, Zheng YF and Feng SQ:
Dysregulated miR-133a mediates loss of type II collagen by directly
targeting matrix metalloproteinase 9 (MMP9) in human intervertebral
disc degeneration. Spine (Phila Pa 1976). 41:E717–E724. 2016.
View Article : Google Scholar
|
|
51
|
Li X, Xu M, Ding L and Tang J: MiR-27a: A
novel biomarker and potential therapeutic target in tumors. J
Cancer. 10:2836–2848. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang J, Cao Z, Yang G, You L, Zhang T and
Zhao Y: MicroRNA-27a (miR-27a) in solid tumors: A review based on
mechanisms and clinical observations. Front Oncol. 9:8932019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Su L, Li R, Zhang Z, Liu J, Du J and Wei
H: Identification of altered exosomal microRNAs and mRNAs in
Alzheimer's disease. Ageing Res Rev. 73:1014972022. View Article : Google Scholar
|
|
54
|
Speiser DE, Chijioke O, Schaeuble K and
Münz C: CD4+ T cells in cancer. Nat Cancer. 4:317–329.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sun L, Su Y, Jiao A, Wang X and Zhang B: T
cells in health and disease. Signal Transduct Target Ther.
8:2352023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhu J, Yamane H and Paul WE:
Differentiation of effector CD4 T cell populations (*). Annu Rev
Immunol. 28:445–489. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Borst J, Ahrends T, Bąbała N, Melief CJM
and Kastenmüller W: CD4+ T cell help in cancer
immunology and immunotherapy. Nat Rev Immunol. 18:635–647. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Thomas R, Qiao S and Yang X: Th17/Treg
imbalance: Implications in lung inflammatory diseases. Int J Mol
Sci. 24:48652023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sakaguchi S, Mikami N, Wing JB, Tanaka A,
Ichiyama K and Ohkura N: Regulatory T cells and human disease. Annu
Rev Immunol. 38:541–566. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Churov AV, Mamashov KY and Novitskaia AV:
Homeostasis and the functional roles of CD4+ Treg cells
in aging. Immunol Lett. 226:83–89. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wei B, Zhao Y, Li W, Zhang S, Yan M, Hu Z
and Gao B: Innovative immune mechanisms and antioxidative therapies
of intervertebral disc degeneration. Front Bioeng Biotechnol.
10:10238772022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dou Y, Zhang Y, Liu Y, Sun X, Liu X, Li B
and Yang Q: Role of macrophage in intervertebral disc degeneration.
Bone Res. 13:152025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lee S and Lee W: A review of Mendelian
randomization: Assumptions, methods, and application to
obesity-related diseases. J Obes Metab Syndr. 34:14–26. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen B, Yan Y, Wang H and Xu J:
Association between genetically determined telomere length and
health-related outcomes: A systematic review and meta-analysis of
Mendelian randomization studies. Aging Cell. 22:e138742023.
View Article : Google Scholar
|
|
65
|
Ni F, Liu X and Wang S: Impact of negative
emotions and insomnia on sepsis: A mediation Mendelian
randomization study. Comput Biol Med. 180:1088582024. View Article : Google Scholar : PubMed/NCBI
|