Tumor metastasis is a hallmark of tumor development
and progression. Tumor metastasis involves the detachment of tumor
cells from the primary site through a series of biological
processes, including local infiltration, entry into the vascular
system, circulation, colonization of secondary tissues or organs,
and the formation of secondary tumors (1-4).
This complex process of tumor invasion and metastasis involves
multiple factors, including changes in the tumor microenvironment,
epithelial-mesenchymal transformation (EMT), tumor hypoxia,
neovascularization, decreased intercellular adhesion, cytoskeleton
remodeling, extracellular matrix degradation, and the formation of
cellular pseudopods and protuberances (5,6).
Although advances in existing treatments have improved survival
rates for some patients with cancer, distant metastasis remains a
major challenge (7,8). Once metastasis occurs, long-term
survival markedly decreases, and numerous patients succumb to the
disease shortly thereafter (9).
Tumor metastasis is one of the most important contributors to
cancer-related mortality and is a major obstacle in effective
clinical management (10).
Therefore, there is an urgent need to identify metastasis-specific
biomarkers and to explore the underlying molecular mechanisms.
Among numerous pathways, the ras homolog family
(Rho) and Rho-associated coiled-coil containing protein kinase
(ROCK) signaling pathway serves a key role in tumor metastasis.
This signaling pathway regulates cytoskeletal remodeling by
modulating myosin and actin activity, establishing cell polarity,
and organizing intermediate filaments. This pathway also
contributes to tumor cell proliferation, contraction, migration,
adhesion and cell-matrix interactions (11). The most widely studied Rho GTP
enzymes include Rho, Ras-related C3 botulinum toxin substrate (Rac)
and cell division cycle 42 (Cdc42) (12,13). These receptor proteins initiate
cytoskeleton reorganization and regulate downstream effectors that
affect tumor cell migration (14). Furthermore, the Rho/ROCK pathway
can regulate HIPPO/yes-associated protein (YAP) signaling by
modulating the actinomycin skeleton, with YAP acting as a
mechanical sensor that alters gene expression in response to
cytoskeleton dynamics (15). The
cytoskeleton, a dynamic network of protein fibers in eukaryotic
cells, serves an important role in cell division, intracellular
transport, motility and shape maintenance (16,17). The cytoskeleton comprises three
main components: Microfilaments, microtubules and intermediate
filaments (18). Microfilaments
are primarily involved in cytokinesis, cell motility and the
formation of intracellular stress fibers. Microtubules maintain
cell morphology, resist compression and facilitate intracellular
trafficking. Intermediate filaments provide mechanical stability
and coordinate cell migration. Together, these three cytoskeletal
components coordinate cellular integrity and ensure the smooth
progression of several biological functions, such as maintaining
cell morphology (16), material
transport (19) and cell
movement (20). Microtubules
serve a crucial role in cancer progression and metastasis (21,22). Microtubule-targeting agents have
emerged as promising tools to disrupt cancer cell activity and
inhibit metastasis (23,24).
Some antibody-drug conjugates, especially those
combined with microtubule-targeting compounds, have yielded
encouraging effectiveness in reducing tumor burden at metastatic
sites (25,26). Nestin, an intermediate filament
protein, is upregulated in several tumors, including pancreatic and
prostate cancer, malignant melanoma, and glioma, and is associated
with tumor aggressiveness, metastasis and poor prognosis (27,28).
Long non-coding RNAs (lncRNAs) were first identified
in the nucleus and cytoplasm of eukaryotic cells (29). Most lncRNAs lack a fully
functional open reading frame; however, previous studies have shown
that a few lncRNAs can encode a small number of functional short
peptides (30-32). Based on their location in the
genome, lncRNAs have been classified into five categories: Sense,
antisense, bidirectional, intergenic and intronic lncRNAs (33,34). Functionally, lncRNAs can be
classified into four types: Signaling, decoy, guide and scaffold
lncRNAs (35). Signaling lncRNAs
can be used as marker signals in different temporal spaces,
developmental stages and gene expression regulation (36). For example, there are three
common signaling lncRNAs: Air, Kcnq1ot1 and Xist. They mediate the
transcriptional silencing of multiple genes by interacting with
chromatin and recruiting chromatin modification mechanisms
(37). Guide lncRNAs can guide
the localization of ribonucleoprotein complexes to specific
targets. For example, the lncRNA COLDAIR can guide the polycomb
repressive complex 2 (PRC2) complex to serve a key biological role
in the chromatin of the floral repressor, flowering locus c
(38). Traditionally, proteins
have been important players in various types of scaffold complexes
(39). However, it has been
shown that lncRNAs can also serve as central platforms for the
assembly of relevant molecular components in several biological
signaling processes, and this precisely controlled property is
essential for precise control of the specificity and dynamics of
intermolecular interactions and signaling events (40). lncRNAs have been confirmed to be
associated with various cancer types, including colorectal,
gastric, hepatocellular and esophageal cancer (41-44). For example, lncRNA CCAT1 and
HOTAIR in the plasma of patients with colorectal cancer (CRC) have
been shown to serve as biological markers in CRC screening
(41). Aberrant expression of
three lncRNAs (POU3F3, SPRY4-IT1 and HNF1A-AS1) has been detected
in the sera of patients with oesophageal cancer. Notably, the
combined use of the three could effectively detect the early
development of esophageal cancer (44). lncRNAs also serve key roles in
tumor metastasis. lncRNA HIF1A-AS2 can promote the proliferation
and invasion of triple-negative breast cancer (TNBC) cells
(45). Compared with those in
normal lung tissue, lncRNA MALAT1 expression levels are elevated in
lung cancer, and abnormal MALAT1 expression can induce EMT to
further promote brain metastasis in lung cancer (46). With the continuous development of
high-throughput sequencing, numerous lncRNAs have been identified
due to their specific expression in tumor metastasis, and as
molecular biomarkers for tumor metastasis and potential therapeutic
targets for several tumors (47-49). For instance, Zhang et al
(50) developed a novel
computational framework employing six machine learning algorithms
to comprehensively analyze transcriptomic data from purified immune
cells, glioblastoma cell lines and bulk glioblastoma tissue. This
framework was utilized to screen for tumor-infiltrating immune
cell-associated lncRNAs predictive of prognosis and response to
immunotherapy in patients with glioblastoma.
In summary, among the multiple complex mechanisms of
tumor metastasis, lncRNAs are emerging as important modulators of
cytoskeleton dynamics, and likely affect tumor cell metastasis by
directly binding to cytoskeleton-related proteins or by indirectly
regulating key molecules in the Rho/ROCK signaling pathway. The
present review aims to explore the molecular mechanisms through
which lncRNAs mediate the Rho/ROCK signaling pathway during tumor
metastasis. An improved understanding of these interactions between
lncRNAs and the Rho/ROCK signaling pathway may provide novel
strategies for the early diagnosis and targeted treatment of
metastatic tumors.
The majority of the human genome (76-97%) is
transcribed into RNAs that are not translated into proteins, and
are referred to as non-coding RNAs (ncRNAs) (51). Taking 200 nt as the threshold,
ncRNAs are classified into two categories: Short ncRNAs (<200
nt) and lncRNAs (>200 nt) (52). According to their genomic
proximity to neighboring transcripts, lncRNAs can be classified
into four categories: i) Intergenic lncRNAs, which are located
between two protein-coding transcripts; ii) intronic lncRNAs, which
are present in the introns of coding transcripts; iii)
sense/antisense strand lncRNAs, which have overlapping parts with
introns and exons of different coding transcripts; and iv)
bidirectional lncRNAs, which share a promoter and are transcribed
from both sense and antisense directions of transcription start
areas (53). lncRNAs are
transcribed by polymerase II (54). Similar to mRNA, most lncRNAs
undergo 5' end capping, polyadenylation and splicing. They are
different in that the number of lncRNAs in exons is lower than that
in mRNA; therefore, lncRNAs are evolutionarily conserved to a
certain extent. Secondary and advanced structures are the most
important features of lncRNAs. lncRNAs form secondary structures,
including double helices, hairpin loops, protrusions and
pseudoknots, as well as tertiary structures with non-Watson-Crick
base pairs and more advanced structures. These structures form the
basis for the biological functions of lncRNAs. lncRNAs are unevenly
distributed in the nucleus and cytoplasm (55); however, the specific localization
of lncRNAs in the cytoplasm or nucleus, and the factors determining
such localization have not been fully confirmed (56).
In the whole genome, 50-70% of genomic DNA can be
transcribed; however, <2% of this is ultimately translated into
proteins (57), and the vast
majority of the remaining transcripts are transcribed into
non-coding ncRNAs. Although not translated, previous studies have
shown that ncRNAs are widely involved in various biological
processes of organisms (34,58-60). Compared with mRNAs, lncRNAs have
higher spatial and temporal specificity and lower interspecies
conservation, and they serve an essential role in regulating
chromatin dynamics (61,62), gene expression (63), cell proliferation (64), cell differentiation (65) and development of organisms
(63). One of the most important
functions of lncRNAs is the regulator of transcription (66,67). Among the identified action modes,
signaling, decoy, scaffolding and guidance are the most frequent
modes of action of lncRNAs in the cell (68). The main function of signaling
type lncRNAs is to regulate transcription as a molecular signal in
response to various stimuli, while the chromatin state of
regulatory elements can be further inferred from the expression of
related lncRNAs (36). In
addition, cellular stimulation can affect the transcriptional
expression of lncRNAs, which can perform regulatory functions to
affect the biological processes of organisms (69). Therefore, their production and
presence can be used as an indicator of transcriptional activity.
For example, DNA damage can induce the transcription of lncRNA
PANDA in a p53-dependent manner, which acts to limit the expression
of pro-apoptotic genes and arrest the cell cycle after interacting
with transcription factors (70). In an experiment involving the
induction of somatic cell reprogramming in pluripotent stem cells,
Loewer et al (71)
demonstrated that a large number of lncRNAs were aberrantly
expressed during this process. Among them, lncRNA RoR could
directly regulate the pluripotency factors OCT4, SOX2 and Nanog,
and served a role as a signaling molecule in the pluripotency and
reprogramming of stem cells. Decoy lncRNAs can directly bind
regulatory molecules such as transcription factors or RNA-binding
proteins, thereby sequestering them and blocking their activity and
downstream signaling pathways (72,73). For example, lncRNA PANDA inhibits
the expression of apoptotic genes by directly binding to nuclear
transcription factor Y subunit α, thus arresting the cell cycle
(74). Scaffold lncRNAs can
serve as molecular scaffolds that guide the assembly of protein
complexes onto target genes (75). For example, HOTAIR serves as a
molecular scaffold that links and targets the PRC2 and the lysine
specific demethylase 1 histone-modifying complexes, thereby
coordinating H3K27 methylation and H3K4 demethylation to reprogram
chromatin states (76). Guide
lncRNAs perform a 'guide-like' function, interacting with
ribonucleoprotein particles and taking them to specific target
genes, and serve the roles of cis-guides and trans-guides (77). In addition, some studies have
shown that a small number of lncRNAs can encode short peptides that
further function in the organism (78-80). With the widespread application of
next-generation sequencing technologies in different tumors, the
mystery of lncRNAs has been gradually unraveled. Thousands of
lncRNAs are aberrantly expressed in various tumors, suggesting that
they may serve an important role in the disease process of tumors
(81). The validated mechanisms
of lncRNAs in tumors are acting as competing endogenous RNAs
(ceRNAs), participating in chromatin remodeling, regulating
chromatin interactions and acting as natural antisense transcripts
(82). lncRNA HOTAIR is
dysregulated in a variety of cancer types (76,83,84), such as oral squamous cell
carcinoma (OSCC), hepatocellular carcinoma (HCC) and breast cancer.
Wu et al (76) found that
HOTAIR binds with EZH2 and H3K27me3 to form a complex, which can
bind to the promoter region of E-cadherin to regulate its
expression, thereby affecting OSCC metastasis. Recent studies have
revealed that lncRNAs can be encapsulated in exosomes, and
transferred from donor cells to recipient cells in the tumor
microenvironment by the fluid circulation, thereby regulating
several phenotypes of tumor processes (85,86), including malignant proliferation
and invasive metastasis of tumor cells, radiotherapy resistance of
tumor cells, formation of the tumor microenvironment and internal
angiogenesis. For example, Zhang et al (87) reported that exosomal lncRNA
MALAT1 was differentially expressed in non-small cell lung cancer
(NSCLC), high exosomal MALAT1 expression could promote metastasis
in NSCLC and the expression levels of MALAT1 were closely
associated with lymph node metastasis in patients with NSCLC. In
addition, exosomal lncRNAs released into the tumor microenvironment
have the potential to become tumor markers due to their specificity
and sensitivity (88-90).
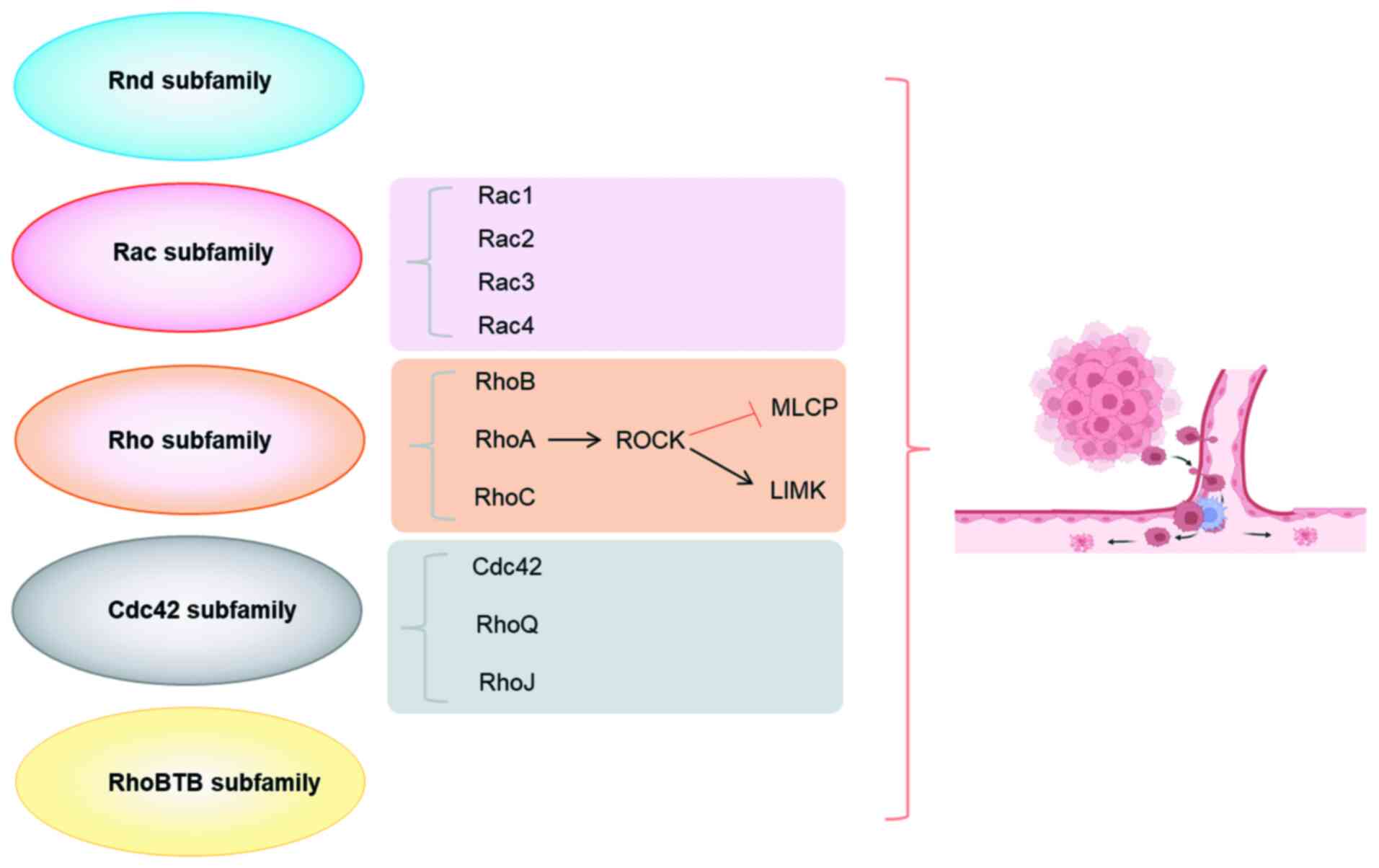
The Rho/ROCK signaling pathway consists of the
Rho-GTPase family and its downstream effector ROCK (91-93) (Fig. 1). Rho GTPases are members of the
Ras superfamily of small GTP-binding proteins and contain >20
Rho proteins that can be broadly classified into the following five
groups based on their primary sequence and functional type: Rho,
Rac, Cdc42, Rho Family GTPase (Rnd) and Rho-related BTB
domain-containing subfamilies (92). Among these, the Rho, Rac and
Cdc42 subfamilies are the most intensively investigated and best
characterized functionally. The Rho subfamily includes RhoA, RhoB
and RhoC, which are very similar in sequence, and because they
share the same set of effectors, the mode of action of the three is
presumed to be similar (94).
RhoA, RhoB and RhoC have two different states in organisms
(inactive GDP-bound and active GTP-bound forms); the molecules
cycle between the two states. The transition from the inactive
GDP-bound form to the active GTP-bound form is catalyzed by the
diffuse B-cell lymphoma family of RhoGTP guanine nucleotide
exchange factors (95). By
contrast, the transition from the active to the inactive state is
mediated by intrinsic GTPase hydrolysis stimulated by Rho
GTPase-activating proteins (96). In the active state, Rho proteins
act on >60 downstream targets, including ROCK, mammalian homolog
of Drosophila (mDia), Par6B, p21 (RAC1) activated kinase 4
and Wiskott-Aldrich syndrome proteins (97). Activation of RhoGTPase ultimately
leads to the remodeling of the cytoskeleton and changes in other
fundamental processes, such as cell division, adhesion and
migration (98,99). The ROCK family consists of two
structurally similar isoforms, ROCK1 and ROCK2. Both kinases share
>30 direct downstream substrates, including myosin
phosphatase-targeting subunit 1, myosin light chain (MLC) and LIM
domain kinase, in addition to containing an N-terminal catalytic
kinase structural domain, a central coiled-coil structural domain
and a C-terminal PH structural domain (100). In Rho/ROCK signaling, Rho
(RhoA/RhoC) activates Rho-related kinases (ROCK1/ROCK2) by binding
to their C-terminal. ROCK regulates key proteins in the
cytoskeleton by mediating the phosphorylation of MLC, and
activating Lin-11, Isl1 and MEC-3 (LIM) structural domain kinases,
which are widely involved in basic cellular functions, including
adhesion, migration, contraction, proliferation and apoptosis
(101).
The vast majority of cellular activities are
directly or indirectly influenced by Rho family proteins. Rho
family proteins regulate the actin cytoskeleton to control cell
morphology, and serve an important role in cell polarity,
endocytosis, vesicle trafficking, adhesion and migration (13). RhoA interacts with effectors at
the cell membrane (102). RhoA
mediates the formation of myosin bundles and stress fibers
(103), and serves an important
role in membrane folding and lamellipodia formation (104). RhoB is present on the outer
membranes of multivesicular bodies and in the plasma membrane and
serves a role in vesicle transport (105). RhoC is mainly located in the
plasma membrane or cytoplasmic matrix, and mainly regulates the
activity of actin and myosin and limits lamellipodial broadening
(106). The Rac subfamily
primarily stimulates the formation of lamellar pseudopods, membrane
folds and invasive foot types (107). Rac1 is commonly distributed in
tissues, and mainly regulates the formation of scaffolding
proteins, membrane ruffles and lamellipodia (108). Rac2 is confined to
haematopoietic tissues, and regulates cell adhesion and
participates in cellular immune synapse formation (109). Rac3 is a neuron-specific
protein that is important for regulating autophagy, inducing the
formation of invadopodia and degrading extracellular matrix
(110,111) The Cdc42 subfamily mediates the
formation of a third actin-based structure, the filopodia, by
binding to Wiskott-Aldrich syndrome protein or neural
Wiskott-Aldrich syndrome protein (112). Additionally, Cdc42 serves an
important role in cell polarization (113). Rho downstream effectors include
ROCK and p140mDia (114). The
ROCK family belongs to the AGC family of serine/threonine protein
kinases. ROCK1 serves a key role in the formation of actin bundles,
actin contractility and stress fibers by mediating the
cross-linking of myosin, while ROCK2 is required for phagocytosis
and cell contraction and is important in stabilizing the
cytoskeleton (115). The
Rho/ROCK signaling pathway serves a key role in central nervous
system disorders, pulmonary hypertension and other diseases
(116,117).
Metastasis, the spread of malignant cells to distant
organs, represents the ultimate stage of cancer progression and
remains a leading cause of cancer-related deaths (107). The majority of cancer deaths
result not from the primary tumor but from the secondary lesions in
vital organs. This complex process includes a series of sequential
events: Primary tumor cells gradually acquire invasive abilities,
spread through blood or lymphatic vessels (or directly invade
adjacent tissues), and finally colonize and proliferate in distant
organs (118). Although
different cancer types and molecular subtypes exhibit marked
differences in metastasis-driving genes, microenvironmental cues
and anatomical dissemination routes, several core cellular
processes are shared (119).
Among these, cell migration is a core process that is universally
present across various tumor types (120). Therefore, targeting the
signaling networks that regulate cell motility could offer an
effective strategy for the management of metastasis in multiple
cancer types (121-123).
The Rho GTPase family functions as a core molecular
switch regulating cytoskeletal dynamics and cell movement (124). Growing evidence has highlighted
the critical role of this family in tumor invasion and metastasis
(123,125). For example, Zhang et al
(126) demonstrated that
inhibition of RhoA suppressed proliferation in gastric cancer
cells. In breast cancer, altered Rho GTPase signaling disrupts
cytoskeleton architecture in cancer cells, and serves a critical
role in cell motility, migration and invasion (127). In cholangiocarcinoma, tumor
cells recruit cancer-associated fibroblasts by secreting PDGF-D,
which stimulates fibroblast migration by upregulating
platelet-derived growth factor receptor β and Rho GTPase, and
activating the JNK pathway (128). With the emergence of
pharmacological agents targeting the Rho/ROCK signaling axis,
particularly selective ROCK inhibitors, this pathway has become a
promising target for the development of antimetastatic treatments
(123).
The subcellular localization of lncRNAs is closely
related to their biological functions (129). Unlike the relatively discrete
cytoplasmic distribution of mRNAs, most lncRNAs are found in both
the nucleus and cytoplasm, with the cytoplasm often being their
primary site of localization (130,131). Structurally, lncRNAs contain
intronic and other non-coding elements that provide potential
binding sites for miRNAs. Through sequence complementarity, lncRNAs
can bind intracellular miRNAs and weaken or block their ability to
suppress target genes (24).
This competitive binding between lncRNAs and miRNAs forms ceRNA
networks, indirectly affecting components of the Rho/ROCK signaling
pathway (132).
Several studies have demonstrated that lncRNAs
influence components of the Rho/ROCK signaling pathway via ceRNA
networks, ultimately modulating tumor cell proliferation, invasion,
migration and apoptosis (38,133-181) (Table I). For example, the expression of
lncRNA DAPK1 is reduced in pancreatic cancer tissues, where it
modulates invasion and migration by sponging miR-182 to regulate
ROCK1 expression (133).
Similarly, Liu et al (134) demonstrated that lncRNA ZFAS1
promoted pancreatic cancer metastasis by acting as a sponge for
miR-3924 and subsequently regulated the RhoA/ROCK2 pathway to serve
a pro-metastatic role in pancreatic cancer. Furthermore, lncRNA
NORAD facilitates EMT in pancreatic cancer by binding miR-125a-3p,
which regulates the expression of the downstream effector molecule
RhoA (135). Liu et al
(136) reported that lncRNA
NEAT1 acted as a ceRNA by sponging miR-382-3p and upregulating
ROCK1 in ovarian cancer (OC), and thus, enhanced the metastatic
potential. NEAT1 also inhibits the progression of lung
adenocarcinoma by sponging miR-490-3p, leading to suppression of
the RhoA/ROCK pathway (137).
Furthermore, Zhang et al (138) found that lncRNA LINC01087
promoted lung adenocarcinoma progression by regulating the
miR-514a-3p/CEP55/RhoA/ROCK1 axis.
In osteosarcoma, the lncRNA DANCR sponges miR-335-5p
and miR-1972, leading to an increase in ROCK1 expression, which
promotes metastasis (139).
Similarly, Shao et al (140) demonstrated that, in
osteosarcoma, lncRNA ZNF281 suppressed invasion by upregulating
miR-144 expression, thus suppressing ROCK1 expression. Furthermore,
in osteosarcoma, lncRNA SNHG1 is overexpressed and promotes cell
metastasis by altering apoptosis and the cell cycle; functional
experiments have shown that SNHG1 regulated ROCK1 expression by
sponging miRNA-101-3p (141).
Additionally, Wang et al (142) reported that SNHG5 functioned as
a ceRNA for miR-26a and activated the ROCK signaling pathway
through the miR-26a/ROCK1 axis to promote osteosarcoma cell
malignancy. In HCC, lncRNA CDKN2BAS is upregulated in metastatic
tissues and sponges miR-153-5p to increase the expression of Rho
GTPase activating protein 18 (ARHGAP18), thus enhancing cell
migration (143). Furthermore,
lncRNA LINC00607 promotes HCC cell proliferation, migration and
invasion through the miR-584-3p/ROCK1 axis (144). Thus, cytoplasmic lncRNAs
regulate tumor metastasis by forming ceRNA networks with miRNAs
that target Rho/ROCK signaling components.
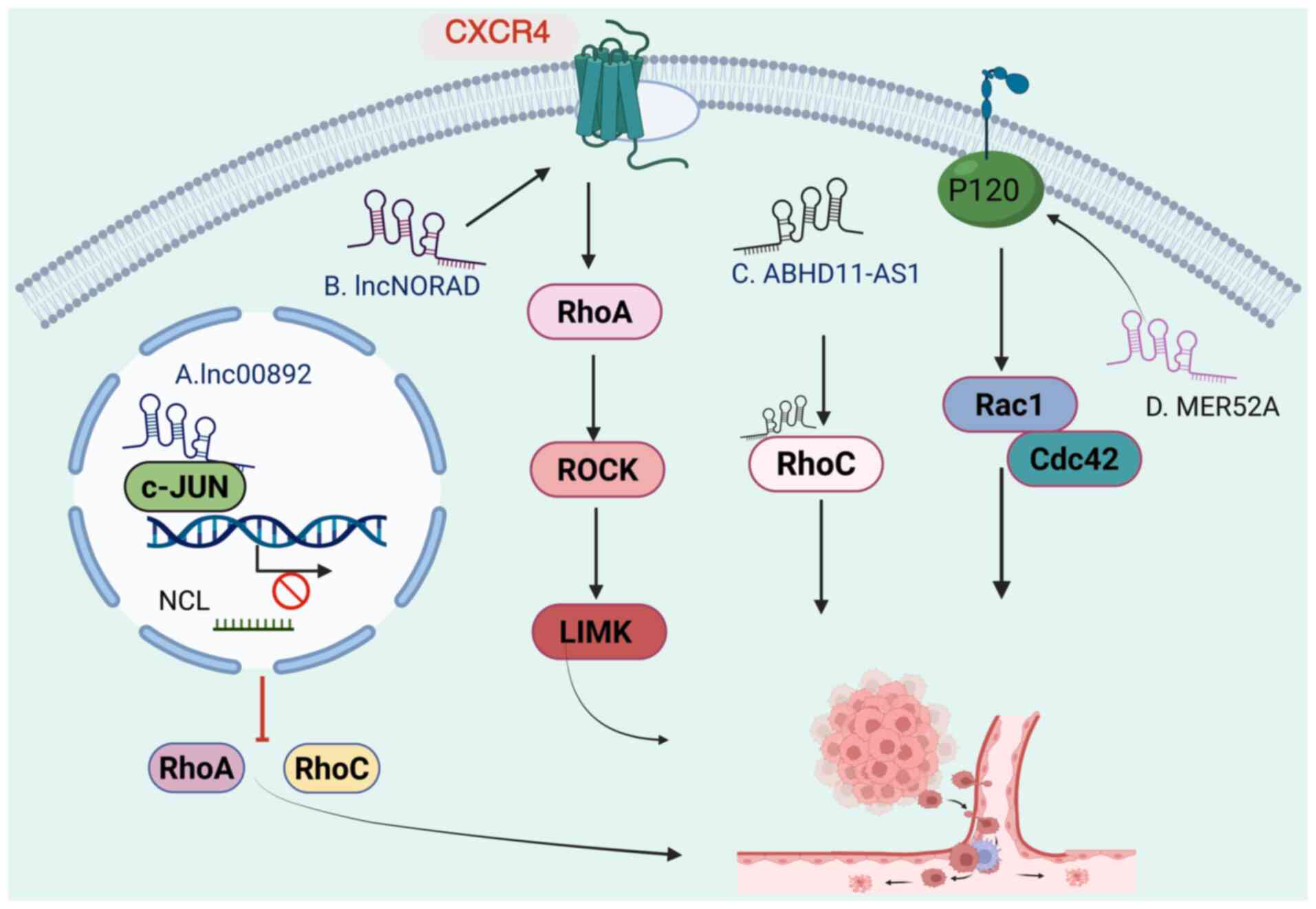
In addition to sponging miRNAs by acting as ceRNAs,
lncRNAs can also directly bind to specific proteins, including
those involved in the Rho/ROCK signaling pathway, and regulate
tumor metastasis (182)
(Fig. 2). In bladder cancer,
lncRNA lnc00892 is downregulated and is positively associated with
the prognosis of patients with bladder cancer. Mechanistically,
lnc00892 binds to c-JUN, a component of the activating protein-1
transcriptional complex, and reduces nucleolin transcription and
affects the stability of RhoA/RhoC mRNAs, which suppresses bladder
tumor metastasis (183). In
NSCLC, lncRNA NORAD inhibits metastasis by binding to and
interfering with CXC chemokine receptor 4, which in turn suppresses
the RhoA/ROCK signaling pathway (184). Similarly, in bladder cancer,
overexpressed lncRNA KTN1-AS1 recruits EP300, leading to histone H3
lysine 27 acetylation at the KTN1 promoter region. This epigenetic
regulation ultimately affects bladder cancer growth and metastasis
by mediating the Rho GTPase signaling pathway through Rac1, RhoA
and Cdc42 (185). In HCC,
overexpression of lncRNA SchLAH (also referred to as BC035072)
inhibits distant metastasis. Functional experiments have shown that
SchLAH interacts with DNA/RNA-binding protein-fusion sarcoma,
regulating the mRNA expression of downstream effector molecules
RhoA and Rac1 (186). Dai et
al (187) used mass
spectrometry and reported that a total of 93 upregulated and 352
downregulated lncRNAs were identified in patients with bladder
cancer; downregulated lncRNA MUC20-9 bound directly to ROCK1 and
inhibited tumor growth, migration and invasion. MUC20-9 was
considered to be a potential therapeutic target for bladder cancer.
In OC, lncRNA ABHD11-AS1 is upregulated and facilitates metastasis
by directly binding to RhoC (188). Similarly, Wu et al
(189) identified a novel
lncRNA, MER52A, which was highly expressed in HCC tissues.
Mechanistically, MER52A promoted HCC cell invasion and metastasis
by stabilizing P120-catenin, thus activating Rac1 and Cdc42, key
downstream Rho-GTPases. Furthermore, high MER52A expression was
strongly associated with advanced TNM stage, poor differentiation
and reduced overall survival of patients with HCC. In osteosarcoma,
AFAP1-AS1 is also highly expressed and directly interacts with RhoC
and activates the RhoC/ROCK1/p38MAPK signaling pathway, which in
turn exerts oncogenic effects (190).
Tumor metastasis remains a major contributor to
treatment failure and cancer-related mortality (191). Numerous patients with cancer
are diagnosed at intermediate or advanced stages, when clinical
symptoms appear; consequently, their prognosis is poorer than that
of patients diagnosed earlier (192). Epidemiological data indicate
that ~90% of cancer-related deaths are due to tumor metastasis
(193). Therefore, identifying
novel and reliable biomarkers and therapeutic targets associated
with the metastatic potential is critical to improve early
detection and intervention strategies.
Rho/ROCK signaling pathway-associated lncRNAs have
increasingly been explored and defined. These lncRNAs not only
serve an important role in tumor metastasis but also have a close
relationship with the clinical characteristics of patients with
tumors (133,135,136,140,145,151,153,159,165, 166, 171,173,179,180,188,189,194-204) (Table II). A previous study has
demonstrated that lncRNA AFAP1-AS1 was upregulated in patients with
HCC, and was associated with pathological stage and lymphovascular
interstitial infiltration (194). Multifactorial analysis revealed
that AFAP1-AS1 was an independent predictor for overall survival,
supporting the hypothesis that AFAP1-AS1 is a potential therapeutic
target for HCC. Similarly, lncRNA LOC284454, upregulated in
nasopharyngeal carcinoma (NPC), promotes migration and invasion by
regulating the cytoskeleton-related Rho/Rac signaling pathway, and
is strongly associated with poor prognosis in patients with NPC
(195). Hu et al
(196) demonstrated that
ARHGAP42, another pro-metastatic lncRNA in NPC, was associated with
shorter metastasis-free survival. In vitro experiments
demonstrated that ARHGAP42 promoted the metastasis and invasion of
NPC cells, suggesting that it may serve as a tumor migration
marker, a prognostic factor or a therapeutic target for patients
with NPC. In gliomas, lncRNA LINC00346 is upregulated and is
strongly associated with both disease-free and overall survival,
suggesting its usage as a potential therapeutic target and
therapeutic candidate (159).
lncRNA MAGI2-AS3 expression is downregulated in serum samples from
patients with HCC with distant metastases compared with in those
from non-metastatic patients. Overexpression of MAGI2-AS3
suppresses cell invasion and migration by modulating ROCK2, while
its downregulation is associated with distant recurrence following
surgical resection (197). In
papillary thyroid carcinoma (PTC), lncRNA DLEU1 expression is
elevated and associated with lymph node metastasis and clinical
stage (145). Follow-up
bioinformatics and dual luciferase reporter gene analysis revealed
that DLEU1 affects PTC proliferation, survival and metastasis by
regulating miR-421 binding to the 3' untranslated region of ROCK1
in TPC-1 cells. Furthermore, lncRNA CDKN2B-AS1 expression is
elevated in laryngeal squamous cell carcinoma (LSCC) tissues, and
its high expression is positively associated with overall survival,
lymph node metastasis and the clinical stage of patients with LSCC
(198).
Rho/ROCK signaling pathway-associated lncRNAs may
also serve as liquid biopsy biomarkers. Liquid biopsy, an
innovative technology, has opened novel pathways for the early
diagnosis and prognostic evaluation of cancer. Compared with
traditional tissue biopsies, liquid biopsy enables real-time
monitoring of disease progression and therapeutic responses
(205). Liquid biopsy platforms
can simultaneously detect multiple circulating biomarkers,
including circulating tumor cells (CTCs), circulating tumor DNA,
exosomes, tumor-educated platelets and circulating free RNA. Among
these, exosomes, nanosized extracellular vesicles (30-150 nm),
serve crucial roles in intercellular communication and carry active
molecules such as proteins and nucleic acids (DNA and RNA,
including lncRNAs) (206,207). Previous studies have shown that
several lncRNAs can be encapsulated in exosomes and transferred
from donor cells to recipient cells, where they regulate components
of the Rho/ROCK signaling pathway, thereby regulating cancer
progression and metastasis (154,208,209). For example, You et al
(154) demonstrated that
exosome-derived lncRNA LINC00161 sponged miR-590-3p to promote HCC
metastasis by activating the ROCK signaling pathway, suggesting
that LINC00161 may serve as a novel prognostic marker for HCC.
Furthermore, Ding et al (209) reported that CRC cell-derived
exosomal lncRNA BANCR promoted M2 macrophage polarization and
enhanced CRC cell proliferation and invasion via the RhoA/ROCK
pathway, with insulin like growth factor 2 mRNA binding protein 2
acting as a mediator. Together, these findings suggest that
Rho/ROCK pathway-associated lncRNAs, particularly those in
exosomes, are emerging as valuable molecular biomarkers with
potential applications in liquid biopsy-based diagnostics.
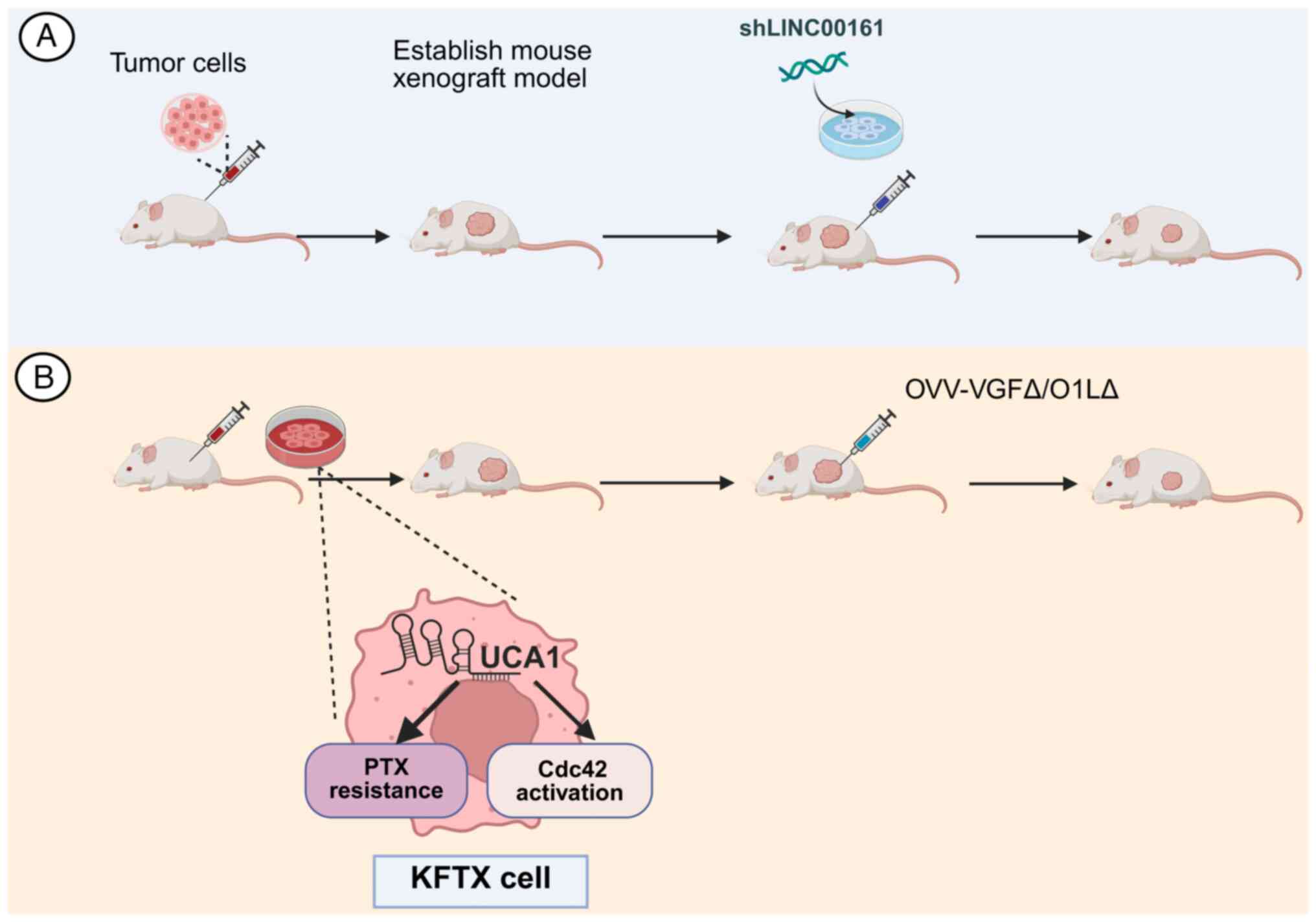
In addition to their diagnostic utility, Rho/ROCK
signaling pathway-associated lncRNAs represent potential
therapeutic targets for metastatic cancer (154,210) (Fig. 3). As shown in Fig. 3, You et al (154) established a xenograft model in
which tail-vein inoculation of HCC cells produced widespread
metastases; notably, tail-vein injection of HCC cells with stable
LINC00161 knockdown markedly attenuated both tumor initiation and
metastatic dissemination in vivo, indicating that LINC00161
silencing suppressed HCC tumorigenesis and metastasis. Horita et
al (210) reported that
lncRNA UCA1 is upregulated in OC and enhances oncolytic poxvirus
dissemination by modulating the Cdc42 signaling pathway, a key
regulator of cytoskeletal reorganization (210). Similarly, UCA1 sponges
miR-18a/miR-182 to regulate Cdc42/filopodia, thereby increasing
sensitivity to poxvirus-based virotherapy in CRC (169). Clinically, glucocorticoids,
such as dexamethasone and prednisolone, can be used in combination
with other chemotherapeutic agents for the treatment of hematologic
malignancies. In acute myeloid leukemia, lncRNA HOTAIRM1 promotes
glucocorticoid resistance by binding to the transcriptional
repressor region of ARHGAP18, which in turn activates the
RhoA/ROCK1 signaling pathway. This finding has implications for the
optimization of glucocorticoid-based leukemia treatment strategies
(211). In oral squamous cell
cancer, low expression levels of lncRNA LOC441178 are associated
with longer postoperative survival, suggesting a potential role as
a tumor suppressor and prognostic marker (212). In osteosarcoma, lncRNA CCHE1
expression is higher in patients with distant recurrence after
surgery and associated with ROCK1 expression, suggesting that CCHE1
may help guide subsequent chemotherapy or radiotherapy (213). Rho-GTPases and their downstream
signaling molecules have been shown to serve a key role in
regulating tumor angiogenesis, invasion, metastasis and EMT
(214-216). Targeting EMT is considered a
relatively promising strategy to inhibit metastasis and improve the
survival rate of patients with cancer (191). lncRNA AFAP1-AS1 expression is
upregulated in osteosarcoma, and is involved in the growth and
metastasis of osteosarcoma by interacting with RhoC and regulating
EMT. Therefore, AFAP1-AS1 inhibitors may serve as therapeutic
agents in osteosarcoma (190).
Furthermore, targeting lncRNAs involved in drug resistance has also
shown promise. In cholangiocarcinoma, lncRNA CCAT1 modulates EMT
via ROCK2. CCAT1 knockdown increased the susceptibility of an
erlotinib-resistant cholangiocarcinoma cell line xenograft to
erlotinib in vivo, suggesting that targeting the
miR-181a-5p/ROCK2 signaling axis could overcome resistance
(217). Using a small-molecule
microarray, Abulwerdi et al (218) identified several chemotypes
that bind the MALAT1 element for nuclear expression triplex-binding
chemotypes (referred to as MALAT1-IN-1). Notably, compounds 5 and
16 lowered MALAT1 RNA levels and suppressed branching morphogenesis
in mammary tumor organoids, establishing triplex-targeting
scaffolds as preclinical leads for anticancer therapeutics and
molecular probes.
Inhibitors targeting the Rho/ROCK signaling pathway
have gained attention in drug research for tumor-targeted therapy
(91). These inhibitors of the
Rho/ROCK pathway are divided into three categories: Inhibitors of
ROCK, geranylgeranyltransferase-1 and
3-hydroxy-3-methyl-glutaryl-CoA reductase (219-222). MBQ-167, a newly developed Rac
and Cdc42 inhibitor, has been shown to inhibit p21 activated
protein kinase signaling, metastatic cell migration and mammosphere
growth in TNBC. Its short half-life and low toxicity make this
inhibitor a promising candidate for future TNBC therapy (219). NecroX-5 has demonstrated
anti-metastatic effects in lung cancer, breast cancer and melanoma
models by suppressing the expression of Cdc42, Rac1 and RhoA
(220). NSC23766, the first
Rac1-specific inhibitor, blocks Rac1 activation by targeting the
guanine nucleotide exchange factors. NSC23766 inhibits the invasion
and migration of human HCC by interfering with the Rac1/JNK or LIM
and cysteine-rich domains 1-Rac1 pathways (221). Inhibition of Rac1 by NSC23766
affects the proliferation and migration of NSCLC (222). Overall, Rho/ROCK pathway
inhibitors represent a promising strategy to overcome drug
resistance and prevent tumor metastasis. Therefore, the development
of Rho/ROCK pathway inhibitors remains a key clinical strategy in
cancer therapy.
Tumorigenesis is a complex process that involves
multiple malignant phenotypes, among which metastasis is a leading
contributor to cancer-related mortality. Tumor metastasis involves
a series of complex pathophysiological changes, with cytoskeletal
reorganization serving a critical role in cancer cell invasion and
migration. Among the classical signaling pathways implicated in
cytoskeletal reorganization, the Rho/ROCK signaling pathway has
attracted considerable attention. Accumulating evidence has also
demonstrated that lncRNAs regulate tumorigenic processes, including
proliferation, migration, metastasis and resistance to radiotherapy
(198,223). With the development of
high-throughput sequencing and related technologies, an increasing
number of lncRNAs have been identified that interact, either
directly or indirectly, with key cytoskeletal regulators such as
ROCK1 and ROCK2, mediating tumor metastasis by altering the
three-dimensional structure of cancer cells and regulating the
reconstruction of the cytoskeleton. As research continues to
uncover the specific mechanisms and clinical implications of
Rho/ROCK signaling pathway-associated lncRNAs, several promising
applications have emerged. First, Rho/ROCK signaling
pathway-associated lncRNAs may serve as pathological and liquid
biopsy markers for patients with tumors. Unlike protein-based
pathological tissue biomarkers, which often reflect downstream
changes, the aberrant expression of lncRNAs often occurs earlier
than changes at the protein level, providing greater tissue
specificity and expression sensitivity for early cancer diagnosis
(224,225). Their tissue specificity,
dynamic expression and distinct subcellular localization make them
particularly suitable for precise classification and prognosis
(226). While tissue biopsy
remains the gold standard for diagnosing tumor subtypes and grades,
its invasiveness limits its applications for dynamic monitoring of
disease progression. Liquid biopsy, on the other hand, offers a
minimally invasive monitoring strategy by analyzing circulating
tumor components in bodily fluids such as blood and urine (227). In addition, circulating lncRNAs
exhibit high stability and resistance to nuclease degradation,
offering advantages over other biomarkers such as cell-free DNA
(including circulating tumor DNA), CTCs and exosomal RNA (228). Several lncRNAs, including those
detectable in liver, colorectal, gastric and prostate cancer, have
already shown promise as reliable diagnostic and prognostic markers
(229-231). Second, next-generation
sequencing, combined with artificial intelligence (AI) and machine
learning, has accelerated the collaborative development of basic
cancer research and clinical oncology (232-234). Tumor genomic databases
constructed from high-throughput sequencing systematically analyze
disease-specific expression profiles and tumor microenvironment
characteristics. Researchers can leverage advanced machine learning
algorithms to deeply mine sequencing data, successfully identifying
functional lncRNAs that can serve as biomarkers for personalized
treatment decisions or prognostic predictions. Deep learning
algorithms enable the precise identification of Rho/ROCK signaling
pathway-associated lncRNAs, as well as the prediction of their
interacting proteins and binding miRNAs, providing a novel strategy
for the development of cancer-specific diagnostic and prognostic
biomarkers. Third, despite the potential of RNA-based therapeutics,
clinical translation is challenged by issues such as target
specificity, low delivery efficiency and in vivo stability
(235). Notably, while a few
miRNA drugs have entered phase II/III clinical trials, the
development of inhibitors targeting lncRNAs remains in the
preclinical stage (236-238).
However, nanotechnology provides a promising strategy to overcome
these limitations. Nanomaterials can enable precise targeting at
the tissue and even subcellular levels, delivering complex
therapeutic molecules to key metastatic sites (such as lymph nodes,
liver and lungs) and specific subcellular regions (239). Advances in translational
medicine have focused on co-delivering lncRNA inhibitors with small
molecule drugs or immune checkpoint inhibitors via nanoparticle
systems, synergistically enhancing antitumor efficacy (240-243). Based on this research, the
future development of nanoparticle carriers encapsulating Rho/ROCK
signaling pathway-associated lncRNA inhibitors may improve the
prognosis of patients with metastatic cancer.
Nevertheless, existing research on Rho/ROCK
signaling pathway-associated lncRNAs still has several limitations.
First, although high-throughput studies have identified numerous
dysregulated lncRNAs in tumor tissues (244,245), the specific molecular
mechanisms by which Rho/ROCK signaling pathway-associated lncRNAs
influence metastasis require further elucidation. Second, Rho/ROCK
signaling pathway-associated lncRNAs may regulate gene expression
through multiple miRNA sponging interactions or binding proteins,
but the specific target axis may not be limited to one or two axes.
Third, some lncRNAs implicated in the Rho/ROCK signaling pathway
may also regulate other signaling cascades. In addition, the
Rho/ROCK signaling pathway mainly promotes tumor metastasis;
however, a study has reported that the Rho/ROCK signaling pathway
may induce apoptosis (246).
Street et al (247)
demonstrated that the ROCK inhibitor Y27632 could enhance the
survival of neuroblastoma cells. Therefore, future studies must
validate pathway specificity when proposing drug targets. Fourth,
whether Rho/ROCK signaling pathway-associated lncRNAs can be used
as therapeutic targets still requires further research at present.
Existing studies are only at the stage of animal experiments
(135,146,150,153,154), and there is still a long way to
go from basic research to clinical application. Therefore, the
relationship between Rho/ROCK signaling pathway-associated lncRNAs
and tumor metastasis needs to be further clarified. Fifth, although
the drug delivery systems of nanoparticle-based lncRNA inhibitors
show potential (239-241), further clinical validation is
needed to ensure their efficacy, safety and long-term effects of
the delivery of nucleic acids in humans.
In summary, lncRNAs associated with the Rho/ROCK
signaling pathway are emerging as key regulators of cytoskeletal
remodeling and tumor metastasis. Through interactions with miRNAs,
proteins and exosomal transport mechanisms, these lncRNAs regulate
key processes involved in cancer cell migration, invasion and
metastasis. They hold promise not only as novel biomarkers for
early cancer diagnosis but also as therapeutic targets to prevent
metastasis. Advances in high-throughput sequencing, AI-driven
bioinformatics and nanomedicine provide novel opportunities to
leverage lncRNAs in personalized treatment strategies and improve
prognostic predictions. However, further research is needed to
fully elucidate the mechanisms by which lncRNAs regulate the
Rho/ROCK pathway and to confirm their clinical applicability. The
present review highlights the growing potential of Rho/ROCK
signaling pathway-associated lncRNAs as promising diagnostic and
therapeutic targets, with the potential to improve the management
and prognosis of metastatic cancer.
Not applicable.
HN and XY performed the literature search. HN and
XY prepared the first draft of the manuscript. QC, XH and YH wrote
and edited the manuscript. HN and YH drew the figures. QH and CO
prepared the tables and were responsible for revising the
manuscript. CO obtained funding support. Data authentication is not
applicable. All authors reviewed the manuscript, and have read and
approved of the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 82373062) and the Central South
University Innovation-Driven Research Programme (grant no.
2023CXQD075).
|
1
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Han Y, Wang D, Peng L, Huang T, He X, Wang
J and Ou C: Single-cell sequencing: A promising approach for
uncovering the mechanisms of tumor metastasis. J Hematol Oncol.
15:592022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou H, He X, He Y, Ou C and Cao P:
Exosomal circRNAs: Emerging players in tumor metastasis. Front Cell
Dev Biol. 9:7862242021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liao Z, Nie H, Wang Y, Luo J, Zhou J and
Ou C: The emerging landscape of long non-coding RNAs in colorectal
cancer metastasis. Front Oncol. 11:6413432021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Liu F, Cai Q, Deng L, Ouyang Q,
Zhang XH and Zheng J: Invasion and metastasis in cancer: Molecular
insights and therapeutic targets. Signal Transduct Target Ther.
10:572025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Francoeur AA, Monk BJ and Tewari KS:
Treatment advances across the cervical cancer spectrum. Nat Rev
Clin Oncol. 22:182–199. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kaur R, Bhardwaj A and Gupta S: Cancer
treatment therapies: Traditional to modern approaches to combat
cancers. Mol Biol Rep. 50:9663–9676. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Klein CA: Cancer progression and the
invisible phase of metastatic colonization. Nat Rev Cancer.
20:681–694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gerstberger S, Jiang Q and Ganesh K:
Metastasis. Cell. 186:1564–1579. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Johan MZ and Samuel MS: Rho-ROCK signaling
regulates tumor-microenvironment interactions. Biochem Soc Trans.
47:101–108. 2019. View Article : Google Scholar
|
|
12
|
Hall A: Rho family GTPases. Biochem Soc
Trans. 40:1378–1382. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu C, Chen S, Zhang Y, Zhou X, Wang H,
Wang Q and Lan X: Mechanisms of Rho GTPases in regulating tumor
proliferation, migration and invasion. Cytokine Growth Factor Rev.
80:168–174. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alarcon VB and Marikawa Y: Trophectoderm
formation: Regulation of morphogenesis and gene expressions by RHO,
ROCK, cell polarity, and HIPPO signaling. Reproduction.
164:R75–R86. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Agrawal A, Scott ZC and Koslover EF:
Morphology and transport in eukaryotic cells. Annu Rev Biophys.
51:247–266. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ando D, Korabel N, Huang KC and Gopinathan
A: Cytoskeletal network morphology regulates intracellular
transport dynamics. Biophys J. 109:1574–1582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fletcher DA and Mullins RD: Cell mechanics
and the cytoskeleton. Nature. 463:485–492. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park J, Wu Y, Kim JS, Byun J, Lee J and Oh
YK: Cytoskeleton-modulating nanomaterials and their therapeutic
potentials. Adv Drug Deliv Rev. 211:1153622024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Leduc C and Etienne-Manneville S:
Intermediate filaments in cell migration and invasion: The unusual
suspects. Curr Opin Cell Biol. 32:102–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li M, Peng L, Wang Z, Liu L, Cao M, Cui J,
Wu F and Yang J: Roles of the cytoskeleton in human diseases. Mol
Biol Rep. 50:2847–2856. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thapa N, Wen T, Cryns VL and Anderson RA:
Regulation of cell adhesion and migration via microtubule
cytoskeleton organization, cell polarity, and phosphoinositide
signaling. Biomolecules. 13:14302023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mangaonkar S, Nath S and Chatterji BP:
Microtubule dynamics in cancer metastasis: Harnessing the
underappreciated potential for therapeutic interventions. Pharmacol
Ther. 263:1087262024. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fanale D, Bronte G, Passiglia F, Calò V,
Castiglia M, Di Piazza F, Barraco N, Cangemi A, Catarella MT,
Insalaco L, et al: Stabilizing versus destabilizing the
microtubules: A double-edge sword for an effective cancer treatment
option? Anal Cell Pathol (Amst). 2015:6909162015.PubMed/NCBI
|
|
25
|
Dumontet C, Reichert JM, Senter PD,
Lambert JM and Beck A: Antibody-drug conjugates come of age in
oncology. Nat Rev Drug Discov. 22:641–661. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cai J, He M, Wang Y, Zhang H, Xu Y, Wang
Y, You C and Gao H: Discovery of a novel microtubule destabilizing
agent targeting the colchicine site based on molecular docking.
Biochem Pharmacol. 234:1168042025. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Castaldo V, Minopoli M, Di Modugno F,
Sacconi A, Liguoro D, Frigerio R, Ortolano A, Di Martile M,
Gesualdi L, Madonna G, et al: Upregulated expression of miR-4443
and miR-4488 in drug resistant melanomas promotes migratory and
invasive phenotypes through downregulation of intermediate filament
nestin. J Exp Clin Cancer Res. 42:3172023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ishiwata T, Matsuda Y and Naito Z: Nestin
in gastrointestinal and other cancers: Effects on cells and tumor
angiogenesis. World J Gastroenterol. 17:409–418. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Atianand MK, Caffrey DR and Fitzgerald KA:
Immunobiology of long noncoding RNAs. Annu Rev Immunol. 35:177–198.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang D, Han Y, Peng L, Huang T, He X, Wang
J and Ou C: Crosstalk between N6-methyladenosine (m6A) modification
and noncoding RNA in tumor microenvironment. Int J Biol Sci.
19:2198–2219. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ou C, Sun Z, He X, Li X, Fan S, Zheng X,
Peng Q, Li G, Li X and Ma J: Targeting YAP1/LINC00152/FSCN1
signaling axis prevents the progression of colorectal cancer. Adv
Sci (Weinh). 7:19013802020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He X, Yu B, Kuang G, Wu Y, Zhang M, Cao P
and Ou C: Long noncoding RNA DLEU2 affects the proliferative and
invasive ability of colorectal cancer cells. J Cancer. 12:428–437.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan H and Bu P: Non-coding RNA in cancer.
Essays Biochem. 65:625–639. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen LL and Kim VN: Small and long
non-coding RNAs: Past, present, and future. Cell. 187:6451–6485.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nadhan R, Isidoro C, Song YS and
Dhanasekaran DN: Signaling by LncRNAs: Structure, cellular
homeostasis, and disease pathology. Cells. 11:25172022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mohammad F, Mondal T and Kanduri C:
Epigenetics of imprinted long noncoding RNAs. Epigenetics.
4:277–286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heo JB and Sung S: Vernalization-mediated
epigenetic silencing by a long intronic noncoding RNA. Science.
331:76–79. 2011. View Article : Google Scholar
|
|
39
|
Good MC, Zalatan JG and Lim WA: Scaffold
proteins: Hubs for controlling the flow of cellular information.
Science. 332:680–686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Spitale RC, Tsai MC and Chang HY: RNA
templating the epigenome: Long noncoding RNAs as molecular
scaffolds. Epigenetics. 6:539–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao W, Song M, Zhang J, Kuerban M and
Wang H: Combined identification of long non-coding RNA CCAT1 and
HOTAIR in serum as an effective screening for colorectal carcinoma.
Int J Clin Exp Pathol. 8:14131–14140. 2015.
|
|
42
|
Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu
Z, Ye G, Zhang X, Xiao B and Guo J: Gastric juice long noncoding
RNA used as a tumor marker for screening gastric cancer. Cancer.
120:3320–3328. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji
J and Sun B: HULC and Linc00152 act as novel biomarkers in
predicting diagnosis of hepatocellular carcinoma. Cell Physiol
Biochem. 37:687–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tong YS, Wang XW, Zhou XL, Liu ZH, Yang
TX, Shi WH, Xie HW, Lv J, Wu QQ and Cao XF: Identification of the
long non-coding RNA POU3F3 in plasma as a novel biomarker for
diagnosis of esophageal squamous cell carcinoma. Mol Cancer.
14:32015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang YZ, Liu YR, Xu XE, Jin X, Hu X, Yu
KD and Shao ZM: Transcriptome analysis of triple-negative breast
cancer reveals an integrated mRNA-lncRNA signature with predictive
and prognostic value. Cancer Res. 76:2105–2114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen L, Chen L, Wang Y, Jiang X, Xia H and
Zhuang Z: Long noncoding RNA MALAT1 promotes brain metastasis by
inducing epithelial-mesenchymal transition in lung cancer. J
Neurooncol. 121:101–108. 2015. View Article : Google Scholar
|
|
47
|
Yu W, Ma Y, Hou W, Wang F, Cheng W, Qiu F,
Wu P and Zhang G: Identification of immune-related lncRNA
prognostic signature and molecular subtypes for glioblastoma. Front
Immunol. 12:7069362021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kong X, Qi J, Yan Y, Chen L, Zhao Y, Fang
Z, Fan J, Liu M and Liu Y: Comprehensive analysis of differentially
expressed profiles of lncRNAs, mRNAs, and miRNAs in laryngeal
squamous cell carcinoma in order to construct a ceRNA network and
identify potential biomarkers. J Cell Biochem. 120:17963–17974.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jing Z, Guo S, Zhang P and Liang Z:
LncRNA-Associated ceRNA network reveals novel potential biomarkers
of laryngeal squamous cell carcinoma. Technol Cancer Res Treat.
19:15330338209857872020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang H, Zhang N, Wu W, Zhou R, Li S, Wang
Z, Dai Z, Zhang L, Liu Z, Zhang J, et al: Machine learning-based
tumor-infiltrating immune cell-associated lncRNAs for predicting
prognosis and immunotherapy response in patients with glioblastoma.
Brief Bioinform. 23:bbac3862022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nemeth K, Bayraktar R, Ferracin M and
Calin GA: Non-coding RNAs in disease: From mechanisms to
therapeutics. Nat Rev Genet. 25:211–232. 2024. View Article : Google Scholar
|
|
52
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View Article : Google Scholar
|
|
53
|
Katayama S, Tomaru Y, Kasukawa T, Waki K,
Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et
al: Antisense transcription in the mammalian transcriptome.
Science. 309:1564–1566. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bunch H, Lawney BP, Burkholder A, Ma D,
Zheng X, Motola S, Fargo DC, Levine SS, Wang YE and Hu G: RNA
polymerase II promoter-proximal pausing in mammalian long
non-coding genes. Genomics. 108:64–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nojima T and Proudfoot NJ: Mechanisms of
lncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev
Mol Cell Biol. 23:389–406. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Collins FS, Morgan M and Patrinos A: The
human genome project: Lessons from large-scale biology. Science.
300:286–290. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Green ED, Watson JD and Collins FS: Human
genome project: Twenty-five years of big biology. Nature.
526:29–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Koffler-Brill T, Noy Y and Avraham KB: The
long and short: Non-coding RNAs in the mammalian inner ear. Hear
Res. 428:1086662023. View Article : Google Scholar :
|
|
61
|
Zhao J, Sun BK, Erwin JA, Song JJ and Lee
JT: Polycomb proteins targeted by a short repeat RNA to the mouse X
chromosome. Science. 322:750–756. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang F, Deng X, Ma W, Berletch JB, Rabaia
N, Wei G, Moore JM, Filippova GN, Xu J, Liu Y, et al: The lncRNA
Firre anchors the inactive X chromosome to the nucleolus by binding
CTCF and maintains H3K27me3 methylation. Genome Biol. 16:522015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Martianov I, Ramadass A, Barros AS, Chow N
and Akoulitchev A: Repression of the human dihydrofolate reductase
gene by a non-coding interfering transcript. Nature. 445:666–670.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bailey C, Pich O, Thol K, Watkins TBK,
Luebeck J, Rowan A, Stavrou G, Weiser NE, Dameracharla B, Bentham
R, et al: Origins and impact of extrachromosomal DNA. Nature.
635:193–200. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lopez-Pajares V, Qu K, Zhang J, Webster
DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao
S, et al: A LncRNA-MAF:MAFB transcription factor network regulates
epidermal differentiation. Dev Cell. 32:693–706. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao
L, Wu M, Xiong J, Guo X and Liu H: Endogenous miRNA sponge
lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem
cell self-renewal. Dev Cell. 25:69–80. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rapicavoli NA, Qu K, Zhang J, Mikhail M,
Laberge RM and Chang HY: A mammalian pseudogene lncRNA at the
interface of inflammation and anti-inflammatory therapeutics.
Elife. 2:e007622013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Loewer S, Cabili MN, Guttman M, Loh YH,
Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al:
Large intergenic non-coding RNA-RoR modulates reprogramming of
human induced pluripotent stem cells. Nat Genet. 42:1113–1117.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jiang M, Zhang S, Yang Z, Lin H, Zhu J,
Liu L, Wang W, Liu S, Liu W, Ma Y, et al: Self-Recognition of an
inducible host lncRNA by RIG-I feedback restricts innate immune
response. Cell. 173:906–919. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ferrer J and Dimitrova N: Transcription
regulation by long non-coding RNAs: Mechanisms and disease
relevance. Nat Rev Mol Cell Biol. 25:396–415. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yang L, Froberg JE and Lee JT: Long
noncoding RNAs: Fresh perspectives into the RNA world. Trends
Biochem Sci. 39:35–43. 2014. View Article : Google Scholar :
|
|
76
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bonasio R, Tu S and Reinberg D: Molecular
signals of epigenetic states. Science. 330:612–616. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ingolia NT, Lareau LF and Weissman JS:
Ribosome profiling of mouse embryonic stem cells reveals the
complexity and dynamics of mammalian proteomes. Cell. 147:789–802.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhao S, Meng J and Luan Y: LncRNA-Encoded
short peptides identification using feature subset recombination
and ensemble learning. Interdiscip Sci. 14:101–112. 2022.
View Article : Google Scholar
|
|
80
|
Zhao S, Meng J, Kang Q and Luan Y:
Identifying LncRNA-encoded short peptides using optimized hybrid
features and ensemble learning. IEEE/ACM Trans Comput Biol
Bioinform. 19:2873–2881. 2022. View Article : Google Scholar
|
|
81
|
Jiang W, Zhang X, Xu Z, Cheng Q, Li X, Zhu
Y, Lu F, Dong L, Zeng L, Zhong W, et al: High-throughput
single-nucleus RNA profiling of minimal puncture FFPE samples
reveals spatiotemporal heterogeneity of cancer. Adv Sci (Weinh).
12:e24107132025. View Article : Google Scholar
|
|
82
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang H: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang BR, Chu DX, Cheng MY, Jin Y, Luo HG
and Li N: Progress of HOTAIR-microRNA in hepatocellular carcinoma.
Hered Cancer Clin Pract. 20:42022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang J, Luo Q, Li X, Guo J, Zhu Q, Lu X,
Wei L, Xiang Z, Peng M, Ou C and Zou Y: Novel role of
immune-related non-coding RNAs as potential biomarkers regulating
tumour immunoresponse via MICA/NKG2D pathway. Biomark Res.
11:862023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhou M, He X, Mei C and Ou C: Exosome
derived from tumor-associated macrophages: Biogenesis, functions,
and therapeutic implications in human cancers. Biomark Res.
11:1002023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y,
Li X, Wang Y and Ming H: Serum long non coding RNA MALAT-1
protected by exosomes is up-regulated and promotes cell
proliferation and migration in non-small cell lung cancer. Biochem
Biophys Res Commun. 490:406–414. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang W, Wang Q, Yang Y, Zhou S, Zhang P
and Feng T: The role of exosomal lncRNAs in cancer biology and
clinical management. Exp Mol Med. 53:1669–1673. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tao Y, Tang Y, Yang Z, Wu F, Wang L, Yang
L, Lei L, Jing Y, Jiang X, Jin H, et al: Exploration of serum
exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for
diagnosis of non-small cell lung cancer. Int J Biol Sci.
16:471–482. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Sun W, Jiang C, Liu Q, Wang N, Huang R,
Jiang G and Yang Y: Exosomal noncoding RNAs: Decoding their role in
thyroid cancer progression. Front Endocrinol (Lausanne).
15:13372262024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Hanifa M, Singh M, Randhawa PK, Jaggi AS
and Bali A: A focus on Rho/ROCK signaling pathway: An emerging
therapeutic target in depression. Eur J Pharmacol. 946:1756482023.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Narumiya S, Tanji M and Ishizaki T: Rho
signaling, ROCK and mDia1, in transformation, metastasis and
invasion. Cancer Metastasis Rev. 28:65–76. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chin VT, Nagrial AM, Chou A, Biankin AV,
Gill AJ, Timpson P and Pajic M: Rho-associated kinase signalling
and the cancer microenvironment: novel biological implications and
therapeutic opportunities. Expert Rev Mol Med. 17:e172015.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Burridge K and Wennerberg K: Rho and Rac
take center stage. Cell. 116:167–179. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zheng Y: Dbl family guanine nucleotide
exchange factors. Trends Biochem Sci. 26:724–732. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Moon SY and Zheng Y: Rho GTPase-activating
proteins in cell regulation. Trends Cell Biol. 13:13–22. 2003.
View Article : Google Scholar
|
|
97
|
Jin D, Durgan J and Hall A: Functional
cross-talk between Cdc42 and two downstream targets, Par6B and
PAK4. Biochem J. 467:293–302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Seetharaman S and Etienne-Manneville S:
Cytoskeletal crosstalk in cell migration. Trends Cell Biol.
30:720–735. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Navarro-Lérida I, Sánchez-Álvarez M and
Del Pozo M: Post-Translational modification and subcellular
compartmentalization: Emerging concepts on the regulation and
physiopathological relevance of RhoGTPases. Cells. 10:19902021.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fukata Y, Amano M and Kaibuchi K:
Rho-Rho-kinase pathway in smooth muscle contraction and
cytoskeletal reorganization of non-muscle cells. Trends Pharmacol
Sci. 22:32–39. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Hall A: Rho GTPases and the control of
cell behaviour. Biochem Soc Trans. 33:891–895. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Bishop AL and Hall A: Rho GTPases and
their effector proteins. Biochem J. 348:241–255. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ridley AJ, Paterson HF, Johnston CL,
Diekmann D and Hall A: The small GTP-binding protein rac regulates
growth factor-induced membrane ruffling. Cell. 70:401–410. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Patel S, McKeon D, Sao K, Yang C, Naranjo
NM, Svitkina TM and Petrie RJ: Myosin II and Arp2/3 cross-talk
governs intracellular hydraulic pressure and lamellipodia
formation. Mol Biol Cell. 32:579–589. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Eckenstaler R, Hauke M and Benndorf RA: A
current overview of RhoA, RhoB, and RhoC functions in vascular
biology and pathology. Biochem Pharmacol. 206:1153212022.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Lou Y, Jiang Y, Liang Z, Liu B, Li T and
Zhang D: Role of RhoC in cancer cell migration. Cancer Cell Int.
21:5272021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Steffen A, Koestler SA and Rottner K:
Requirements for and consequences of Rac-dependent protrusion. Eur
J Cell Biol. 93:184–193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Acevedo A and González-Billault C:
Crosstalk between Rac1-mediated actin regulation and ROS
production. Free Radic Biol Med. 116:101–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Donkó A, Sharapova SO, Kabat J, Ganesan S,
Hauck FH, Bergerson JRE, Marois L, Abbott J, Moshous D, Williams
KW, et al: Clinical and functional spectrum of RAC2-related
immunodeficiency. Blood. 143:1476–1487. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Donnelly SK, Cabrera R, Mao SPH, Christin
JR, Wu B, Guo W, Bravo-Cordero JJ, Condeelis JS, Segall JE and
Hodgson L: Rac3 regulates breast cancer invasion and metastasis by
controlling adhesion and matrix degradation. J Cell Biol.
216:4331–4349. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
He D, Xu L, Wu Y, Yuan Y, Wang Y, Liu Z,
Zhang C, Xie W, Zhang L, Geng Z, et al: Rac3, but not Rac1,
promotes ox-LDL induced endothelial dysfunction by downregulating
autophagy. J Cell Physiol. 235:1531–1542. 2020. View Article : Google Scholar
|
|
112
|
Nobes CD and Hall A: Rho, rac, and cdc42
GTPases regulate the assembly of multimolecular focal complexes
associated with actin stress fibers, lamellipodia, and filopodia.
Cell. 81:53–62. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Nobes CD and Hall A: Rho GTPases control
polarity, protrusion, and adhesion during cell movement. J Cell
Biol. 144:1235–1244. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Watanabe N, Madaule P, Reid T, Ishizaki T,
Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM and Narumiya
S: p140mDia, a mammalian homolog of Drosophila diaphanous, is a
target protein for Rho small GTPase and is a ligand for profilin.
EMBO J. 16:3044–3056. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Jerrell RJ and Parekh A: Matrix rigidity
differentially regulates invadopodia activity through ROCK1 and
ROCK2. Biomaterials. 84:119–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Kubo T, Yamaguchi A, Iwata N and Yamashita
T: The therapeutic effects of Rho-ROCK inhibitors on CNS disorders.
Ther Clin Risk Manag. 4:605–615. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Oka M, Fagan KA, Jones PL and McMurtry IF:
Therapeutic potential of RhoA/Rho kinase inhibitors in pulmonary
hypertension. Br J Pharmacol. 155:444–454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Naxerova K: Evolutionary paths towards
metastasis. Nat Rev Cancer. 25:545–560. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Majidpoor J and Mortezaee K: Steps in
metastasis: An updated review. Med Oncol. 38:32021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Bakir B, Chiarella AM, Pitarresi JR and
Rustgi AK: EMT, MET, plasticity, and tumor metastasis. Trends Cell
Biol. 30:764–776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Guo S, Huang J, Li G, Chen W, Li Z and Lei
J: The role of extracellular vesicles in circulating tumor
cell-mediated distant metastasis. Mol Cancer. 22:1932023.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Matsuoka T and Yashiro M: Rho/ROCK
signaling in motility and metastasis of gastric cancer. World J
Gastroenterol. 20:13756–13766. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Crosas-Molist E, Samain R, Kohlhammer L,
Orgaz JL, George SL, Maiques O, Barcelo J and Sanz-Moreno V: Rho
GTPase signaling in cancer progression and dissemination. Physiol
Rev. 102:455–510. 2022. View Article : Google Scholar
|
|
125
|
Itoh K, Yoshioka K, Akedo H, Uehata M,
Ishizaki T and Narumiya S: An essential part for Rho-associated
kinase in the transcellular invasion of tumor cells. Nat Med.
5:221–225. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhang S, Tang Q, Xu F, Xue Y, Zhen Z, Deng
Y, Liu M, Chen J, Liu S, Qiu M, et al: RhoA regulates G1-S
progression of gastric cancer cells by modulation of multiple INK4
family tumor suppressors. Mol Cancer Res. 7:570–580. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Kazmi N, Robinson T, Zheng J, Kar S,
Martin RM and Ridley AJ: Rho GTPase gene expression and breast
cancer risk: A mendelian randomization analysis. Sci Rep.
12:14632022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Cadamuro M, Nardo G, Indraccolo S,
Dall'olmo L, Sambado L, Moserle L, Franceschet I, Colledan M,
Massani M, Stecca T, et al: Platelet-derived growth factor-D and
Rho GTPases regulate recruitment of cancer-associated fibroblasts
in cholangiocarcinoma. Hepatology. 58:1042–1053. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Bridges MC, Daulagala AC and Kourtidis A:
LNCcation: lncRNA localization and function. J Cell Biol.
220:e2020090452021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Nie H, Liao Z, Wang Y, Zhou J, He X and Ou
C: Exosomal long non-coding RNAs: Emerging players in cancer
metastasis and potential diagnostic biomarkers for personalized
oncology. Genes Dis. 8:769–780. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang Y, Nie H, He X, Liao Z, Zhou Y, Zhou
J and Ou C: The emerging role of super enhancer-derived noncoding
RNAs in human cancer. Theranostics. 10:11049–11062. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Kimura T, Horikoshi Y, Kuriyagawa C and
Niiyama Y: Rho/ROCK pathway and noncoding RNAs: Implications in
ischemic stroke and spinal cord injury. Int J Mol Sci.
22:115732021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Xu X, Wang X, Geng C, Nie X and Bai C:
Long-chain non-coding RNA DAPK1 targeting miR-182 regulates
pancreatic cancer invasion and metastasis through ROCK-1/rhoa
signaling pathway. Int J Clin Exp Pathol. 10:9273–9283.
2017.PubMed/NCBI
|
|
134
|
Liu J, Zhu Y and Ge C: LncRNA ZFAS1
promotes pancreatic adenocarcinoma metastasis via the RHOA/ROCK2
pathway by sponging miR-3924. Cancer Cell Int. 20:2492020.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C and Shen B: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Liu Y, Wang Y, Fu X and Lu Z: Long
non-coding RNA NEAT1 promoted ovarian cancer cells' metastasis
through regulation of miR-382-3p/ROCK1 axial. Cancer Sci.
109:2188–2198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhao L, Ren C, Fang Y, Li P, Zhang H, Zuo
L, Zhang J and Zhao J: MiR-490-3p sponged by lncRNA NEAT1 can
attenuate lung adenocarcinoma progression by suppressing the
RhoA/ROCK signaling pathway. Ann Clin Lab Sci. 53:42–51.
2023.PubMed/NCBI
|
|
138
|
Zhang X, Wang DJ, Jia L and Zhang W:
N6-methyladenosine-mediated LINC01087 promotes lung adenocarcinoma
progression by regulating miR-514a-3p to upregulate centrosome
protein 55. Kaohsiung J Med Sci. 40:801–818. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Wang Y, Zeng X, Wang N, Zhao W, Zhang X,
Teng S, Zhang Y and Lu Z: Long noncoding RNA DANCR, working as a
competitive endogenous RNA, promotes ROCK1-mediated proliferation
and metastasis via decoying of miR-335-5p and miR-1972 in
osteosarcoma. Mol Cancer. 17:892018. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Shao Y, Tong Z, Wei J and Yang T:
LncRNA-zinc finger protein 281 downregulates rho-associated
coiled-coil containing protein kinase 1 by upregulating miR-144 in
osteosarcoma. Oncol Lett. 20:792020. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Deng R, Zhang J and Chen J: lncRNA SNHG1
negatively regulates miRNA-101-3p to enhance the expression of
ROCK1 and promote cell proliferation, migration and invasion in
osteosarcoma. Int J Mol Med. 43:1157–1166. 2019.
|
|
142
|
Wang Z, Wang Z, Liu J and Yang H: Long
non-coding RNA SNHG5 sponges miR-26a to promote the tumorigenesis
of osteosarcoma by targeting ROCK1. Biomed Pharmacother.
107:598–605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Chen J, Huang X, Wang W, Xie H, Li J, Hu
Z, Zheng Z, Li H and Teng L: LncRNA CDKN2BAS predicts poor
prognosis in patients with hepatocellular carcinoma and promotes
metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging
(Albany NY). 10:3371–3381. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Dong S, Wang W, Liao Z, Fan Y, Wang Q and
Zhang L: MYC-activated LINC00607 promotes hepatocellular carcinoma
progression by regulating the miR-584-3p/ROCK1 axis. J Gene Med.
25:e34772023. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Li R, Wan T, Qu J, Yu Y and Zheng R: Long
non-coding RNA DLEUI promotes papillary thyroid carcinoma
progression by sponging miR-421 and increasing ROCK1 expression.
Aging (Albany NY). 12:20127–20138. 2020.PubMed/NCBI
|
|
146
|
Lian Y, Xiong F, Yang L, Bo H, Gong Z,
Wang Y, Wei F, Tang Y, Li X, Liao Q, et al: Long noncoding RNA
AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to
facilitate nasopharyngeal carcinoma metastasis through regulating
the Rho/Rac pathway. J Exp Clin Cancer Res. 37:2532018. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Feng Z, Li X, Qiu M, Luo R, Lin J and Liu
B: LncRNA EGFR-AS1 upregulates ROCK1 by sponging miR-145 to promote
esophageal squamous cell carcinoma cell invasion and migration.
Cancer Biother Radiopharm. 35:66–71. 2020.
|
|
148
|
Zhou Y, Si L, Liu Z, Shi Y and Agula B:
Long noncoding RNA zfas1 promotes progression of NSCLC via
regulating of miR-590-3p. Cell Transplant. 29:9636897209194352020.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Song S, Bian WG, Qin Z, Zeng D, Xu JJ and
Tang HC: LncRNA BCYRN1 promotes cell migration and invasion of
non-small cell lung cancer via the miR-30b-3p/ROCK1 axis.
Neoplasma. 69:583–593. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M,
Li X, Li J, Mi C, Zhang J, et al: Long noncoding RNA LCAT1
functions as a ceRNA to regulate RAC1 function by sponging
miR-4715-5p in lung cancer. Mol Cancer. 18:1712019. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Xiao H, Zhu Q and Zhou J: Long non-coding
RNA MALAT1 interaction with miR-429 regulates the proliferation and
EMT of lung adenocarcinoma cells through RhoA. Int J Clin Exp
Pathol. 12:419–430. 2019.
|
|
152
|
Chen K and Zhang L: LINC00339 regulates
ROCK1 by miR-152 to promote cell proliferation and migration in
hepatocellular carcinoma. J Cell Biochem. 120:14431–14443. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Wang W, Yang T, Li D, Huang Y, Bai G and
Li Q: LINC00491 promotes cell growth and metastasis through
miR-324-5p/ROCK1 in liver cancer. J Transl Med. 19:5042021.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
You LN, Tai QW, Xu L, Hao Y, Guo WJ, Zhang
Q, Tong Q, Zhang H and Huang WK: Exosomal LINC00161 promotes
angiogenesis and metastasis via regulating miR-590-3p/ROCK axis in
hepatocellular carcinoma. Cancer Gene Ther. 28:719–736. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Zhou Y, Fan RG, Qin CL, Jia J, Wu XD and
Zha WZ: LncRNA-H19 activates CDC42/PAK1 pathway to promote cell
proliferation, migration and invasion by targeting miR-15b in
hepatocellular carcinoma. Genomics. 111:1862–1872. 2019. View Article : Google Scholar
|
|
156
|
Guo D, Li Y, Chen Y, Zhang D, Wang X, Lu
G, Ren M, Lu X and He S: DANCR promotes HCC progression and
regulates EMT by sponging miR-27a-3p via ROCK1/LIMK1/COFILIN1
pathway. Cell Prolif. 52:e126282019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Ma Y, Xue Y, Liu X, Qu C, Cai H, Wang P,
Li Z, Li Z and Liu Y: SNHG15 affects the growth of glioma
microvascular endothelial cells by negatively regulating miR-153.
Oncol Rep. 38:3265–3277. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Li HL, Han PH, Pan DQ, Chen G, Lu XH and
Li J: LncRNA XIST regulates cell proliferation, migration and
invasion of glioblastoma via regulating miR-448 and ROCK1. J Biol
Regul Homeost Agents. 34:2049–2058. 2020.PubMed/NCBI
|
|
159
|
Chen X, Li D, Chen L, Hao B, Gao Y, Li L,
Zhou C, He X and Cao Y: Long noncoding RNA LINC00346 promotes
glioma cell migration, invasion and proliferation by up-regulating
ROCK1. J Cell Mol Med. 24:13010–13019. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Wang Z, Han Y, Li Q, Wang B and Ma J:
LncRNA DLGAP1-AS1 accelerates glioblastoma cell proliferation
through targeting miR-515-5p/ROCK1/NFE2L1 axis and activating Wnt
signaling pathway. Brain Behav. 11:e23212021. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Liu Y, Gao X and Tian X: High expression
of long intergenic non-coding RNA LINC00662 contributes to
malignant growth of acute myeloid leukemia cells by upregulating
ROCK1 via sponging microRNA-340-5p. Eur J Pharmacol.
859:1725352019. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Bai H, Li X and Wu S: Up-regulation of
long non-coding RNA LOXL1-AS1 functions as an oncogene in cervical
squamous cell carcinoma by sponging miR-21. Arch Physiol Biochem.
129:143–147. 2023. View Article : Google Scholar
|
|
163
|
Chen X, Zhang Z, Ma Y, Su H, Xie P and Ran
J: LINC02381 promoted cell viability and migration via targeting
miR-133b in cervical cancer cells. Cancer Manag Res. 12:3971–3979.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Yao X, Li X, Luo Y, Xu X, Liu J and Bu J:
LncRNA GAS5 regulates osteosarcoma cell proliferation, migration,
and invasion by regulating RHOB via sponging miR-663a. Cancer Manag
Res. 12:8253–8261. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Wang Y, Yang T, Zhang Z, Lu M, Zhao W,
Zeng X and Zhang W: Long non-coding RNA TUG1 promotes migration and
invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells.
Cancer Sci. 108:859–867. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Cui M, Wang J, Li Q, Zhang J, Jia J and
Zhan X: Long non-coding RNA HOXA11-AS functions as a competing
endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p
in osteosarcoma. Biomed Pharmacother. 92:437–444. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Yu X, Wang D, Wang X, Sun S, Zhang Y, Wang
S, Miao R, Xu X and Qu X: CXCL12/CXCR4 promotes inflammation-driven
colorectal cancer progression through activation of RhoA signaling
by sponging miR-133a-3p. J Exp Clin Cancer Res. 38:322019.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Kong W, Li H, Xie L, Cui G, Gu W, Zhang H,
Ma W and Zhou Y: LncRNA MCF2L-AS1 aggravates the malignant
development of colorectal cancer via targeting miR-105-5p/RAB22A
axis. BMC Cancer. 21:10692021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Horita K, Kurosaki H, Nakatake M, Ito M,
Kono H and Nakamura T: Long noncoding RNA UCA1 enhances sensitivity
to oncolytic vaccinia virus by sponging miR-18a/miR-182 and
modulating the Cdc42/filopodia axis in colorectal cancer. Biochem
Biophys Res Commun. 516:831–838. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Tian Y, Zhong L, Gao S, Yu Y, Sun D, Liu
X, Ji J, Yao Y, Liu Y and Jiang Z: LncRNA LINC00974 downregulates
miR-122 to upregulate RhoA in oral squamous cell carcinoma. Cancer
Biother Radiopharm. 36:18–22. 2021.
|
|
171
|
Yang J, Wang WG and Zhang KQ: LINC00452
promotes ovarian carcinogenesis through increasing ROCK1 by
sponging miR-501-3p and suppressing ubiquitin-mediated degradation.
Aging (Albany NY). 12:21129–21146. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Yang Q and Dong YJ: LncRNA SNHG20 promotes
migration and invasion of ovarian cancer via modulating the
microRNA-148a/ROCK1 axis. J Ovarian Res. 14:1682021. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Chen S, Wang LL, Sun KX, Xiu YL, Zong ZH,
Chen X and Zhao Y: The role of the long non-coding RNA TDRG1 in
epithelial ovarian carcinoma tumorigenesis and progression through
miR-93/RhoC pathway. Mol Carcinog. 57:225–234. 2018. View Article : Google Scholar
|
|
174
|
Liu Y, Zong ZH, Guan X, Wang LL and Zhao
Y: The role of long non-coding RNA PCA3 in epithelial ovarian
carcinoma tumorigenesis and progression. Gene. 633:42–47. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Li HL, Wang CP, Zhang Y, Du JX, Han LH and
Yang HY: Long non-coding RNA PVT1 facilitates cell migration and
invasion by regulating miR-148a-3p and ROCK1 in breast cancer. Clin
Transl Oncol. 24:882–891. 2022. View Article : Google Scholar
|
|
176
|
Chou J, Wang B, Zheng T, Li X, Zheng L, Hu
J, Zhang Y, Xing Y and Xi T: MALAT1 induced migration and invasion
of human breast cancer cells by competitively binding miR-1 with
cdc42. Biochem Biophys Res Commun. 472:262–269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Chen Y, Sheng HG, Deng FM and Cai LL:
Downregulation of the long noncoding RNA SNHG1 inhibits tumor cell
migration and invasion by sponging miR-195 through targeting Cdc42
in oesophageal cancer. Kaohsiung J Med Sci. 37:181–191. 2021.
View Article : Google Scholar
|
|
178
|
Wang Z, Liu J, Wang R, Wang Q, Liang R and
Tang J: Long non-coding RNA taurine upregulated gene 1 (TUG1)
downregulation constrains cell proliferation and invasion through
regulating cell division cycle 42 (CDC42) expression Via MiR-498 in
esophageal squamous cell carcinoma cells. Med Sci Monit.
26:e9197142020.PubMed/NCBI
|
|
179
|
Zong W, Feng W, Jiang Y, Cao Y, Ke Y, Shi
X, Ju S, Cong H, Wang X, Cui M and Jing R: LncRNA CTC-497E21.4
promotes the progression of gastric cancer via modulating
miR-22/NET1 axis through RhoA signaling pathway. Gastric Cancer.
23:228–240. 2020. View Article : Google Scholar
|
|
180
|
Chen Z, Xu C, Pan X, Cheng G, Liu M, Li J
and Mei Y: lncRNA DSCR8 mediates miR-137/Cdc42 to regulate gastric
cancer cell proliferation, invasion, and cell cycle as a
competitive endogenous RNA. Mol Ther Oncolytics. 22:468–482. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Wang J, He Z, Xu J, Chen P and Jiang J:
Long noncoding RNA LINC00941 promotes pancreatic cancer progression
by competitively binding miR-335-5p to regulate ROCK1-mediated
LIMK1/Cofilin-1 signaling. Cell Death Dis. 12:362021. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Ferrè F, Colantoni A and Helmer-Citterich
M: Revealing protein-lncRNA interaction. Brief Bioinform.
17:106–116. 2016. View Article : Google Scholar
|
|
183
|
Ren S, Zhang N, Shen L, Lu Y, Chang Y, Lin
Z, Sun N, Zhang Y, Xu J, Huang H and Jin H: Lnc00892 competes with
c-Jun to block NCL transcription, reducing the stability of
RhoA/RhoC mRNA and impairing bladder cancer invasion. Oncogene.
40:6579–6589. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Wu Y, Shen QW, Niu YX, Chen XY, Liu HW and
Shen XY: LncNORAD interference inhibits tumor growth and lung
cancer cell proliferation, invasion and migration by
down-regulating CXCR4 to suppress RhoA/ROCK signaling pathway. Eur
Rev Med Pharmacol Sci. 24:5446–5455. 2020.PubMed/NCBI
|
|
185
|
Hu X, Xiang L, He D, Zhu R, Fang J, Wang Z
and Cao K: The long noncoding RNA KTN1-AS1 promotes bladder cancer
tumorigenesis via KTN1 cis-activation and the consequent initiation
of Rho GTPase-mediated signaling. Clin Sci (Lond). 135:555–574.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Ge Z, Cheng Z, Yang X, Huo X, Wang N, Wang
H, Wang C, Gu D, Zhao F, Yao M, et al: Long noncoding RNA SchLAH
suppresses metastasis of hepatocellular carcinoma through
interacting with fused in sarcoma. Cancer Sci. 108:653–662. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Dai R, Zhou Y, Chen Z, Zou Z, Pan Z, Liu P
and Gao X: Lnc-MUC20-9 binds to ROCK1 and functions as a tumor
suppressor in bladder cancer. J Cell Biochem. 121:4214–4225. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Wu DD, Chen X, Sun KX, Wang LL, Chen S and
Zhao Y: Role of the lncRNA ABHD11-AS(1) in the tumorigenesis and
progression of epithelial ovarian cancer through targeted
regulation of RhoC. Mol Cancer. 16:1382017. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Wu Y, Zhao Y, Huan L, Zhao J, Zhou Y, Xu
L, Hu Z, Liu Y, Chen Z, Wang L, et al: An LTR
retrotransposon-derived long noncoding RNA lncMER52A promotes
hepatocellular carcinoma progression by binding p120-catenin.
Cancer Res. 80:976–987. 2020. View Article : Google Scholar
|
|
190
|
Shi D, Wu F, Mu S, Hu B, Zhong B, Gao F,
Qing X, Liu J, Zhang Z and Shao Z: LncRNA AFAP1-AS1 promotes
tumorigenesis and epithelial-mesenchymal transition of osteosarcoma
through RhoC/ROCK1/p38MAPK/Twist1 signaling pathway. J Exp Clin
Cancer Res. 38:3752019. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Huang Y, Hong W and Wei X: The molecular
mechanisms and therapeutic strategies of EMT in tumor progression
and metastasis. J Hematol Oncol. 15:1292022. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Kunc M, Skrzypkowska P, Pęksa R and
Biernat W: Tumor-to-tumor metastases: Systematic review and
meta-analysis of 685 reported cases. Clin Exp Metastasis.
42:142025. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Peng L, Wang D, Han Y, Huang T, He X, Wang
J and Ou C: Emerging role of cancer-associated fibroblasts-derived
exosomes in tumorigenesis. Front Immunol. 12:7953722021. View Article : Google Scholar
|
|
194
|
Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q,
Wu JY, Qin J, Jin T and Xu J: Long noncoding RNA AFAP1-AS1
indicates a poor prognosis of hepatocellular carcinoma and promotes
cell proliferation and invasion via upregulation of the RhoA/Rac2
signaling. Int J Oncol. 48:1590–1598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Fan C, Tang Y, Wang J, Wang Y, Xiong F,
Zhang S, Li X, Xiang B, Wu X, Guo C, et al: Long non-coding RNA
LOC284454 promotes migration and invasion of nasopharyngeal
carcinoma via modulating the Rho/Rac signaling pathway.
Carcinogenesis. 40:380–391. 2019. View Article : Google Scholar
|
|
196
|
Hu Q, Lin X, Ding L, Zeng Y, Pang D,
Ouyang N, Xiang Y and Yao H: ARHGAP42 promotes cell migration and
invasion involving PI3K/Akt signaling pathway in nasopharyngeal
carcinoma. Cancer Med. 7:3862–3874. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Fang G, Wang J, Sun X, Xu R, Zhao X, Shao
L, Sun C and Wang Y: LncRNA MAGI2-AS3 is downregulated in the
distant recurrence of hepatocellular carcinoma after surgical
resection and affects migration and invasion via ROCK2. Ann
Hepatol. 19:535–540. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Liu F, Xiao Y, Ma L and Wang J: Regulating
of cell cycle progression by the lncRNA CDKN2B-AS1/miR-324-5p/ROCK1
axis in laryngeal squamous cell cancer. Int J Biol Markers.
35:47–56. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Ma T, Ma H, Zou Z, He X, Liu Y, Shuai Y,
Xie M and Zhang Z: The long intergenic noncoding RNA 00707 promotes
lung adenocarcinoma cell proliferation and migration by regulating
Cdc42. Cell Physiol Biochem. 45:1566–1580. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Song L, Wang L, Pan X and Yang C: lncRNA
OIP5-AS1 targets ROCK1 to promote cell proliferation and inhibit
cell apoptosis through a mechanism involving miR-143-3p in cervical
cancer. Braz J Med Biol Res. 53:e88832020. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Pan L, Meng Q, Li H, Liang K and Li B:
LINC00339 promotes cell proliferation, migration, and invasion of
ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomed
Pharmacother. 120:1094232019. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Chen S, Wang LL, Sun KX, Liu Y, Guan X,
Zong ZH and Zhao Y: LncRNA PCGEM1 induces ovarian carcinoma
tumorigenesis and progression through RhoA pathway. Cell Physiol
Biochem. 47:1578–1588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Yuan S, Luan X, Chen H, Shi X and Zhang X:
Long non-coding RNA EGFR-AS1 sponges micorRNA-381 to upregulate
ROCK2 in bladder cancer. Oncol Lett. 19:1899–1905. 2020.PubMed/NCBI
|
|
204
|
Shang W, Yang Y, Zhang J and Wu Q: Long
noncoding RNA BDNF-AS is a potential biomarker and regulates cancer
development in human retinoblastoma. Biochem Biophys Res Commun.
497:1142–1148. 2018. View Article : Google Scholar
|
|
205
|
Nikanjam M, Kato S and Kurzrock R: Liquid
biopsy: Current technology and clinical applications. J Hematol
Oncol. 15:1312022. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
He X, Kuang G, Wu Y and Ou C: Emerging
roles of exosomal miRNAs in diabetes mellitus. Clin Transl Med.
11:e4682021. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Zhuang T, Wang S, Yu X, He X, Guo H and Ou
C: Current status and future perspectives of platelet-derived
extracellular vesicles in cancer diagnosis and treatment. Biomark
Res. 12:882024. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Wu S, Cui Y, Zhao H, Xiao X, Gong L, Xu H,
Zhou Q, Ma D and Li X: Trophoblast exosomal UCA1 induces
endothelial Injury through the PFN1-RhoA/ROCK pathway in
preeclampsia: A human-specific adaptive pathogenic mechanism. Oxid
Med Cell Longev. 2022:21989232022. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Ding AX, Wang H, Zhang JM, Yang W and
Kuang YT: lncRNA BANCR promotes the colorectal cancer metastasis
through accelerating exosomes-mediated M2 macrophage polarization
via regulating RhoA/ROCK signaling. Mol Cell Biochem. 479:13–27.
2024. View Article : Google Scholar
|
|
210
|
Horita K, Kurosaki H, Nakatake M, Kuwano
N, Oishi T, Itamochi H, Sato S, Kono H, Ito M, Hasegawa K, et al:
lncRNA UCA1-mediated Cdc42 signaling promotes oncolytic vaccinia
virus cell-to-cell spread in ovarian cancer. Mol Ther Oncolytics.
13:35–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Liang L, Gu W, Li M, Gao R, Zhang X, Guo C
and Mi S: The long noncoding RNA HOTAIRM1 controlled by AML1
enhances glucocorticoid resistance by activating RHOA/ROCK1 pathway
through suppressing ARHGAP18. Cell Death Dis. 12:7022021.
View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Xu K, Tian H, Zhao S, Yuan D, Jiang L, Liu
X, Zou B and Zhang J: Long noncoding RNA LOC441178 reduces the
invasion and migration of squamous carcinoma cells by targeting
ROCK1. Biomed Res Int. 2018:43576472018. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Zhang Z, Yu T and Geng W: Long non-coding
RNA CCHE1 participates in postoperative distant recurrence but not
local recurrence of osteosarcoma possibly by interacting with
ROCK1. BMC Musculoskelet Disord. 21:4622020. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Maldonado MDM and Dharmawardhane S:
Targeting Rac and Cdc42 GTPases in cancer. Cancer Res.
78:3101–3111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Guo X, Stafford LJ, Bryan B, Xia C, Ma W,
Wu X, Liu D, Songyang Z and Liu M: A Rac/Cdc42-specific exchange
factor, GEFT, induces cell proliferation, transformation, and
migration. J Biol Chem. 278:13207–13215. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Hosseini K, Frenzel A and
Fischer-Friedrich E: EMT induces characteristic changes of Rho
GTPases and downstream effectors with a mitosis-specific twist.
Phys Biol. 20:2023. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Zhou W, Li X, Zhang B, Peng H, Quan C,
Xiao X, Luo M, Huang Y, Xu D, Huang K, et al: The long non-coding
RNA CCAT1 promotes erlotinib resistance in cholangiocarcinoma by
inducing epithelial-mesenchymal transition via the
miR-181a-5p/ROCK2 axis. Am J Cancer Res. 14:2852–2867. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Abulwerdi FA, Xu W, Ageeli AA, Yonkunas
MJ, Arun G, Nam H, Schneekloth JS Jr, Dayie TK, Spector D, Baird N
and Le Grice SFJ: Selective small-molecule targeting of a triple
helix encoded by the long noncoding RNA, MALAT1. ACS Chem Biol.
14:223–235. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Cruz-Collazo A, Ruiz-Calderon JF, Picon H,
Borrero-Garcia LD, Lopez I, Castillo-Pichardo L, Del Mar Maldonado
M, Duconge J, Medina JI, Bayro MJ, et al: Efficacy of Rac and Cdc42
inhibitor MBQ-167 in triple-negative breast cancer. Mol Cancer
Ther. 20:2420–2432. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Moon GT, Lee JH, Jeong SH, Jin SW and Park
YM: NecroX-5 can suppress melanoma metastasis by reducing the
expression of Rho-family GTPases. J Clin Med. 10:27902021.
View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Li D, Ding X, Xie M, Huang Z, Han P, Tian
D and Xia L: CAMSAP2-mediated noncentrosomal microtubule
acetylation drives hepatocellular carcinoma metastasis.
Theranostics. 10:3749–3766. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Gastonguay A, Berg T, Hauser AD, Schuld N,
Lorimer E and Williams CL: The role of Rac1 in the regulation of
NF-κB activity, cell proliferation, and cell migration in non-small
cell lung carcinoma. Cancer Biol Ther. 13:647–656. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Zhou M, He X, Zhang J, Mei C, Zhong B and
Ou C: tRNA-derived small RNAs in human cancers: Roles, mechanisms,
and clinical application. Mol Cancer. 23:762024. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Sarfi M, Abbastabar M and Khalili E: Long
noncoding RNAs biomarker-based cancer assessment. J Cell Physiol.
234:16971–16986. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Mouraviev V, Lee B, Patel V, Albala D,
Johansen TE, Partin A, Ross A and Perera RJ: Clinical prospects of
long noncoding RNAs as novel biomarkers and therapeutic targets in
prostate cancer. Prostate Cancer Prostatic Dis. 19:14–20. 2016.
View Article : Google Scholar
|
|
226
|
Goyal B, Yadav SRM, Awasthee N, Gupta S,
Kunnumakkara AB and Gupta SC: Diagnostic, prognostic, and
therapeutic significance of long non-coding RNA MALAT1 in cancer.
Biochim Biophys Acta Rev Cancer. 1875:1885022021. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Alix-Panabières C and Pantel K: Liquid
biopsy: From discovery to clinical application. Cancer Discov.
11:858–873. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Diez-Fraile A, Ceulaer J, Derpoorter C,
Spaas C, Backer T, Lamoral P, Abeloos J and Lammens T: Circulating
non-coding RNAs in head and neck cancer: Roles in diagnosis,
prognosis, and therapy monitoring. Cells. 10:482020. View Article : Google Scholar
|
|
229
|
Guo X, Peng Y, Song Q, Wei J, Wang X, Ru
Y, Xu S, Cheng X, Li X, Wu D, et al: A liquid biopsy signature for
the early detection of gastric cancer in patients.
Gastroenterology. 165:402–413. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Raza A, Khan AQ, Inchakalody VP, Mestiri
S, Yoosuf ZSKM, Bedhiafi T, El-Ella DMA, Taib N, Hydrose S, Akbar
S, et al: Dynamic liquid biopsy components as predictive and
prognostic biomarkers in colorectal cancer. J Exp Clin Cancer Res.
41:992022. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Kabzinski J, Kucharska-Lusina A and
Majsterek I: RNA-based liquid biopsy in head and neck cancer.
Cells. 12:19162023. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Zhang N, Zhang H, Wu W, Zhou R, Li S, Wang
Z, Dai Z, Zhang L, Liu F, Liu Z, et al: Machine learning-based
identification of tumor-infiltrating immune cell-associated lncRNAs
for improving outcomes and immunotherapy responses in patients with
low-grade glioma. Theranostics. 12:5931–5948. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Lu Q, Li J, Chen W, Wang Z, Wang D, Liu C,
Sun Y, Jiang H, Zhang C, Chang Y, et al: NetLnc: A network-based
computational framework to identify immune checkpoint-related
lncRNAs for Immunotherapy response in melanoma. Int J Mol Sci.
26:45572025. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Liu Y, Chen J, Yang D, Liu C, Tang C, Cai
S and Huang Y: Machine learning combined with multi-omics to
identify immune-related LncRNA signature as biomarkers for
predicting breast cancer prognosis. Sci Rep. 15:238632025.
View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Zhu Y, Zhu L, Wang X and Jin H: RNA-based
therapeutics: An overview and prospectus. Cell Death Dis.
13:6442022. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Puri B, Majumder S and Gaikwad AB: LncRNA
MALAT1 as a potential diagnostic and therapeutic target in kidney
diseases. Pathol Res Pract. 266:1557832025. View Article : Google Scholar
|
|
237
|
Chen Y, Li Z, Chen X and Zhang S: Long
non-coding RNAs: From disease code to drug role. Acta Pharm Sin B.
11:340–354. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Botti G, Marra L, Malzone MG, Anniciello
A, Botti C, Franco R and Cantile M: LncRNA HOTAIR as prognostic
circulating Marker and potential therapeutic target in patients
with tumor diseases. Curr Drug Targets. 18:27–34. 2017. View Article : Google Scholar
|
|
239
|
Prabhakaran R, Thamarai R, Sivasamy S,
Dhandayuthapani S, Batra J, Kamaraj C, Karthik K, Shah MA and
Mallik S: Epigenetic frontiers: miRNAs, long non-coding RNAs and
nanomaterials are pioneering to cancer therapy. Epigenetics
Chromatin. 17:312024. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Yang L, Li Z, Huang X, Huang L, Fu Y, Zhao
B, Zhang Y, Ma L, Jing S, Fu L, et al: VPS9D1-AS1 antisense therapy
via lipid nanoparticles reprograms cold tumors and enhances
immunotherapy in colorectal cancer. J Control Release.
384:1138652025. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
He X, Lin F, Jia R, Xia Y, Liang Z, Xiao
X, Hu Q, Deng X, Li Q and Sheng W: Coordinated modulation of long
non-coding RNA ASBEL and curcumin co-delivery through
multicomponent nanocomplexes for synchronous triple-negative breast
cancer theranostics. J Nanobiotechnology. 21:3972023. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Jia F, Li Y, Deng X, Wang X, Cui X, Lu J,
Pan Z and Wu Y: Self-assembled fluorescent hybrid
nanoparticles-mediated collaborative lncRNA CCAT1 silencing and
curcumin delivery for synchronous colorectal cancer theranostics. J
Nanobiotechnology. 19:2382021. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Zhang Y, Liu F, Tan L, Li X, Dai Z, Cheng
Q, Liu J, Wang Y, Huang L, Wang L and Wang Z: LncRNA-edited
biomimetic nanovaccines combined with anti-TIM-3 for augmented
immune checkpoint blockade immunotherapy. J Control Release.
361:671–680. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Herman AB, Tsitsipatis D and Gorospe M:
Integrated lncRNA function upon genomic and epigenomic regulation.
Mol Cell. 82:2252–2266. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Yip CW, Sivaraman DM, Prabhu AV and Shin
JW: Functional annotation of lncRNA in high-throughput screening.
Essays Biochem. 65:761–773. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Street CA and Bryan BA: Rho kinase
proteins-pleiotropic modulators of cell survival and apoptosis.
Anticancer Res. 31:3645–3657. 2011.PubMed/NCBI
|
|
247
|
Street CA, Routhier AA, Spencer C, Perkins
AL, Masterjohn K, Hackathorn A, Montalvo J, Dennstedt EA and Bryan
BA: Pharmacological inhibition of Rho-kinase (ROCK) signaling
enhances cisplatin resistance in neuroblastoma cells. Int J Oncol.
37:1297–1305. 2010.PubMed/NCBI
|