Introduction
Hepatocellular carcinoma (HCC) is a prevalent
malignant tumor globally, characterized by intricate survival
mechanisms, elevated morbidity, unfavorable prognosis and
significant cancer-related mortality (1,2).
HCC commonly originates from conditions such as chronic hepatitis B
and C infections, non-alcoholic fatty liver disease, heavy alcohol
use and other causes that drive ongoing liver inflammation and the
development of cirrhosis (3).
Early diagnosis is essential to improving treatment options and
reducing disease-related mortality (4). Of these patients ~30% are diagnosed
with early-stage disease and are receiving potentially curative
treatments, such as surgical resection, liver transplantation or
local ablation, which result in a median overall survival of >60
months (5). However, the
treatment of HCC, particularly advanced HCC, has been a serious
challenge (6). Hence, it is
crucial to delve into the pathogenesis of HCC and uncover effective
treatment approaches.
Cell division cycle 5-like (CDC5L) is a key
constituent of E3 ubiquitin ligase complex, which includes pre-mRNA
processing factor 19 (PRP19), pleiotropic regulator 1 and BCAS2
pre-mRNA processing factor (7).
This complex acts as a regulator of cell cycle, particularly during
G2/M phase transition and is involved in the catalytic
processes of pre-mRNA splicing and DNA damage repair (8). CDC5L has been shown to be markedly
upregulated in HCC (9) and acts
as an independent prognostic marker for unfavorable survival
outcomes in patients with HCC (10). In addition, the phosphorylation
of the CDC5L protein is involved in the metastasis of HCC (11). This indicates that CDC5L may play
a significant role in HCC tumorigenesis and could serve as a
promising therapeutic target to impede HCC progression. Predictive
analysis based on the BioGRID 4.4 database (https://thebiogrid.org/107424/summary/homo-sapiens/cdc5l.html)
showed that CDC5L interacts with the embryonic lethal abnormal
visual-like protein (ELAVL1). ELAVL1 is a widely expressed in
vivo protein that affects the target mRNA stability and
participates in post-transcriptional regulation as an RNA-binding
protein (12). The importance of
ELAVL1-mediated cell signaling, inflammation, fibrosis and cancer
development in the liver has attracted much attention (13). ELAVL1 has been shown to be
upregulated in HCC and implicated in post-transcriptional
regulation of multiple genes linked to the malignant transformation
of the liver (14). However, the
mechanism regarding the role of CDC5L and ELAVL1 in HCC is
unclear.
Pyroptosis is an innate immune response
characterized by its pro-inflammatory nature, driven by the
accumulation of large plasma membrane pores composed of subunits of
Gasdermin (GSDM) family proteins (15,16). When pathogens or other danger
signals are detected, cytokine activity and the formation of
inflammatory vesicles (cytoplasmic multiprotein complexes) trigger
pyroptosis (17). Ultimately,
this inflammatory cell death pathway results in cell lysis and
inflammatory factors IL-1β and IL-18 release (18). There is growing evidence that,
depending on the type of tumor, pyroptosis could either prevent or
promote tumor progression (19).
Pyroptosis has been shown to be crucial in various tissues and
organs, including the liver (20) and it has potential as an
effective target for treating HCC (21). Caspase 3 is a key protein that is
shared between apoptosis and pyroptosis pathways (22). An elevated Caspase 3 activity in
the liver has been observed in patients with various liver diseases
(23). Research has shown that
Caspase 3 deficiency markedly increased diethylnitrosamine-induced
HCC (24). The effect of natural
killer cells on pyroptosis in HepG2 cells treated with Schisandrin
B is primarily due to their activation of the Caspase 3-gasdermin E
(GSDME) pathway (25).
Furthermore, both the small interfering (si)RNA-mediated knockdown
of Caspase 3 and application of Caspase 3 inhibitor
Z-Asp(OMe)-Glu(OMe)-Val-Asp(OMe)-ForwardMK reduced the induction of
GSDME-dependent pyroptosis by Miltirone (26). An Encyclopedia of RNA
Interactomes 3.0 database (https://rnasysu.com/encori/rbpClipRNA.php?source=mRNA&flag=target&clade=mammal&genome=human&assembly=hg38&RBP=ELAVL1&clipNum=1®ionType=None&pval=0.05&clipType=None&panNum=0&target=CASP3)-based
analysis showed that ELAVL1 binds with Caspase 3. Therefore, the
aim of the present study was to investigate whether the mechanisms
underlying the effect of CDC5L and ELAVL1 in HCC pyroptosis
involves Caspase 3. The core hypothesis was that CDC5L and ELAVL1
interact in the pyroptosis of HCC and this interaction may
influence the expression of pyroptosis-related genes by regulating
Caspase 3, thereby affecting the progression of HCC. Previous
studies on CDC5L have mainly focused on its overexpression in HCC
and its role as a prognostic factor for patient survival (9,10), as well as the role of its protein
phosphorylation in HCC metastasis (11), while the mechanism of action of
CDC5L in HCC pyroptosis remains unclear. Similarly, although it has
been reported that ELAVL1 was overexpressed in HCC and was involved
in the post-transcriptional regulation of genes related to hepatic
malignant transformation (14),
the specific role of ELAVL1 in HCC pyroptosis and its interaction
mechanism with CDC5L are also not well understood. The present
study focused on exploring the mechanism of action of CDC5L and
ELAVL1 in HCC pyroptosis and whether they affect the pyroptosis
process by regulating Caspase 3, which is a significant departure
from previous studies that have only focused on the individual
roles of CDC5L or ELAVL1.
Building upon the aforementioned, the high
expression of CDC5L in HCC predicts poor prognosis (9,10). Therefore, the present study was
designed to explore the function of CDC5L in HCC pyroptosis via
both in vivo and in vitro experimental approaches.
CDC5L may be a therapeutic target for HCC in the future. These
results may offer novel perspectives and guidance for the treatment
of HCC.
Materials and methods
Clinical samples
HCC tissues and paracancerous tissues were collected
from 10 participants diagnosed with HCC at Xiangya Hospital,
Central South University (Hunan, China), through imaging,
serological, or histopathological examinations. The sex ratio was 5
males to 5 females, with an age range of 43-76 years. Patients were
recruited between January 1, 2024, and December 31, 2024. HCC and
paracancerous tissues were collected and divided into the Tumor and
Normal groups. Exclusion criteria were pathologically confirmed
diagnosis of non-HCC and patients with incomplete clinical
features. Informed consent was obtained from all participants and
approval was obtained from the medical ethics committee of Xiangya
Hospital, Central South University (Hunan, China; approval no.
2024052119).
Cell culture and treatment
The transformed human liver epithelial-2 (THLE-2)
cells (CL-0833) were purchased from Procell Life Science &
Technology Co., Ltd. SK-HEP-1 (AW-CCH036) and MHCC-97H (AW-CCH088)
human liver cancer cells Huh7 (AW-CCH237), were purchased from
Changsha Abiowell Biotechnology Co., Ltd. THLE-2 cells were
cultured in THLE-2 Cell Complete Medium (CM-0833; Procell Life
Science & Technology Co., Ltd.). Huh7, SK-HEP-1 or MHCC-97H
cells were cultured in DMEM (containing NaHCO3 1.5 g/l),
minimum essential medium or DMEM containing 10% fetal bovine serum
and 1% penicillin/streptomycin. Logarithmically grown SK-HEP-1 and
MHCC-97H cells were collected and spread inside a six-well plate
and cells were processed following wall attachment. First,
CDC5L was knocked down and divided into the Control, si-NC
(negative control siRNA), si-CDC5L-1 (CDC5L-specific
siRNA-1) and si-CDC5L-2 (CDC5L-specific siRNA-2)
groups. CDC5L was overexpressed and divided into the si-NC,
si-CDC5L, oe-NC (Negative control overexpression vector) and
oe-CDC5L (CDC5L overexpression vector) groups. Caspase
3 was knocked down and divided into the si-NC, si-CDC5L,
si-CDC5L+si-NC and si-CDC5L+si-Caspase 3
(Caspase 3-specific siRNA) groups. ELAVL1 was knocked
down and divided into the Control, si-NC, si-ELAVL1#1
(ELAVL1-specific siRNA#1) and si-ELAVL1#2
(ELAVL1-specific siRNA#2) groups. ELAVL1 was
overexpressed and divided into the si-NC, si-ELAVL1, oe-NC
and oe-ELAVL1 (ELAVL1 overexpression vector) groups.
Furthermore, groups were divided as follows: si-NC,
si-CDC5L, si-CDC5L+oe-NC and
si-CDC5L+oe-ELAVL1. Finally, GSDME was knocked
down and divided into the Control, si-NC, si-GSDME#1
(GSDME-specific siRNA#1) and si-GSDME#2
(GSDME-specific siRNA#2) groups. SK-HEP-1 and MHCC-97H cells
were treated with 1 μM CDC5L inhibitor CVT-313 (HY-15339;
MedChemExpress) for 48 h (27)
and divided into the following groups: Control, CVT-313, CVT-313 +
si-NC and CVT-313 + si-GSDME (GSDME-specific siRNA).
The transfection of all siRNA and overexpression vectors was
carried out using Lipofectamine® 2000 (cat. no.
11668019; Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. The siRNA was used at a
concentration of 50 nM. Transfection was performed at 37°C for 48
h. The time interval between transfection and subsequent
experimentation was 48 h. si-CDC5L (HG-SH001253),
oe-CDC5L (HG-HO001253), si-ELAVL1-1 (HG-SH001419),
oe-ELAVL1 (HG-HO001419), si-Caspase 3 (HG-SH032991),
si-GSDME and (HG-SH004403) were obtained from HonorGene. The
interference sequences are as follows: si-NC:
TTAACGGCGAGAGTTTAGGCC, si-CDC5L-1: GGACAGAATTCTGCAGGAAGC,
si-CDC5L-2: GCATAAAGCTGTCCAGAAAGA, si-ELAVL1-1:
GGTTTGGGCGGATCATCAACT, si-ELAVL1-2: GAGGCAATTACCAGTTTCAAT,
si-Caspase 3: GACAACAGTTATAAAATGGAT, si-GSDME:
GAGAGGAATTTCCATCCATTT.
Tumor formation in nude mice
Male nude mice aged 4 weeks were supplied by Hunan
SJA Laboratory Animal Co., Ltd. A total of 80 mice were used in
this study. The mice were housed in a controlled environment with a
temperature of 22±2°C, a 12-h light/dark cycle and humidity of
50±10%. The average weight of the mice was approximately 18-20 g at
the start of the experiment. The tumor formation model of
subcutaneous MHCC-97H cells in nude mice was established. Following
a week of acclimatization and rearing, MHCC-97H cells were injected
subcutaneously, with different treatment groups receiving
corresponding cell types: si-NC, si-CDC5L, oe-NC and
oe-CDC5L; si-NC, si-CDC5L, si-CDC5L+si-NC,
si-CDC5L+si-Caspase 3; si-NC, si-CDC5L,
si-CDC5L+oe-NC and si-CDC5L+oe-ELAVL1. Each
nude mouse received an injection of 1×107 cells in a
volume of 100 μl, administered into the left axillary region
(28). In the si-NC group, mice
were subcutaneously injected with si-NC-treated MHCC-97H cells. In
the si-CDC5L group, mice were subcutaneously injected with
si-CDC5L-treated MHCC-97H cells. In the oe-NC group, mice
were subcutaneously injected with oe-NC-treated MHCC-97H cells. In
the oe-CDC5L group, mice were subcutaneously injected with
oe-CDC5L-treated MHCC-97H cells. In the
si-CDC5L+si-NC group, mice were subcutaneously injected with
si-CDC5L and si-NC-treated MHCC-97H cells. In the
si-CDC5L+si-Caspase 3 group, mice were subcutaneously
injected with si-CDC5L and si-Caspase 3-treated
MHCC-97H cells. In the si-CDC5L+oe-NC group, mice were
subcutaneously injected with si-CDC5L and oe-NC-treated
MHCC-97H cells. In the si-CDC5L+oe-ELAVL1 group, mice
were subcutaneously injected with si-CDC5L and
oe-ELAVL1-treated MHCC-97H cells. Tumor dimensions were
assessed twice weekly following implantation. When tumors grew to a
diameter of 150-200 mm3, tumor volume and tumor weight
were weighted and measured. Tumor volume was tracked and calculated
using the formula: V (mm3)=LxW2/2, with L and
W denoting the longest and shortest diameters, respectively. The
study was terminated 28 days' post-implantation and the tumors were
prepared for image capture. To perform sacrifice of mice using
cervical dislocation, one hand used a rigid rod to firmly press
down on the head and neck area, while the other hand grasped the
tail or hind limbs. Then, quickly and forcefully the hindquarters
were pulled backwards and upwards to dislocate the cervical
vertebrae. Following cervical dislocation, the mice were observed
for a ≤5 min to ensure that life was extinct. To determine whether
life was extinct, its breathing and heartbeat were observed, while
simultaneously checking the pupils and limb reflexes. If the mouse
had stopped breathing and its heart had stopped beating following
cervical dislocation, the pupils did not react to light and the
limbs showed no reflex actions, it was more accurately confirmed
that life was extinct.
In addition, following a week of acclimatization,
4-week-old male nude mice were injected subcutaneously with
MHCC-97H cells and differently treated cells were injected into
corresponding groups: Control, CVT-313, CVT-313 + si-NC and CVT-313
+ si-GSDME. The amount of cells injected per nude mouse was
1×107 and a 100-μl volume was injected into the
left axilla. Following tumor implantation, tumor measurements were
conducted twice a week for observation. When the tumor grew to a
suitable size (10 days after planting), the drug was administered
in groups according to the tumor volume. Among them, mice in the
Control group were subcutaneously injected with normal MHCC-97H
cells. Nude mice in the CVT-313 group were subjected to CVT-313
drug intervention. Nude mice in the CVT-313 + si-NC group were
injected subcutaneously with transfected si-NC-treated MHCC-97H
cells and to CVT-313 drug intervention. Nude mice in the CVT-313 +
si-GSDME group were subcutaneously injected with
si-GSDME-treated MHCC-97H cells and underwent CVT-313 drug
intervention. CVT-313 drug intervention was performed by
intraperitoneal injection of CVT-313, administered every other day
at a dose of 0.625 mg/Kg in an injection volume of 10 ml/100 g
(29). The experiments were
terminated after 28 days of tumor seeding and tumors were arranged
to be sampled and images captured. All animal experiments were
carried out with the approval of the Animal Ethics Committee of
Xiangya Medical School, Central South University (approval no.
2024051707) and in compliance with the Declaration of Helsinki
(30).
Bioinformatics analysis
Clinical specimens were categorized into HCC tissues
and adjacent non-tumor tissues. Data were obtained from The Cancer
Genome Atlas (TCGA, https://xenabrowser.net/datapages/?cohort=TCGA%20Pan-Cancer%20(PANCAN)&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443)
data (369 tumor/50 normal). Box plots of differentially expressed
CDC5L were drawn. Kaplan-Meier survival analysis was applied to
assess the survival outcomes of patients with HCC in the high- and
low-CDC5L groups. Gene Set Enrichment Analysis (GSEA; https://www.gsea-msigdb.org/gsea/index.jsp) was
applied to explore Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway regulation. Bioinformatics analysis was conducted to the
expression of evaluate Caspases 1, 3, 6, 7, 8, 9 and 10 in HCC.
Venn diagrams displayed the intersection of CDC5L binding proteins
and Caspase 3 interacting RNA-binding proteins (RBPs).
Bioinformatics analysis was conducted to measure 22 RBPs levels in
HCC. The binding of ELAVL1 with Caspase 3 was shown based on the
University of California, Santa Cruz (UCSC) Genome Browser
(http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr4%3A184627598%2D184627850&hgsid=2947477028_2O6buwujau0adLCDAGdFsuwkAUi0).
Cell function assays
Based on previous studies, the viability of SK-HEP-1
and MHCC-97H cells was evaluated using the Cell Counting Kit-8
(CCK-8) assay. Briefly, cells were seeded in 96-well plates at a
density of 5,000 cells per well and incubated. The CCK-8 reagent
(10 μl) was added to each well, and the plates were
incubated for 2 h at 37°C. Absorbance was measured at 450 nm using
a microplate reader. A clone formation assay was performed to
assess the proliferative capacity of SK-HEP-1 and MHCC-97H cells
(31). The migration and
invasion of SK-HEP-1 and MHCC-97H cells was assessed through
Transwell assays (32). For
migration and invasion assays, 2×104 cells were seeded
in the upper chamber of Transwell inserts with or without Matrigel
coating. Transwell inserts were precoated with Matrigel and
incubated at 37°C for 30 min. After 24 h, cells that migrated or
invaded through the membrane were fixed with methanol and stained
with 0.1% crystal violet at room temperature for 5 min. The number
of cells was counted in five random fields under a light
microscope.
Reverse transcription-quantitative (RT-q)
PCR
CDC5L, Caspase 7, Caspase 9,
Caspase 6, Caspase 8, Caspase 1, Caspase
3, Caspase 10, GSDME and CDC5L mRNA levels were
quantified with RT-qPCR. Total RNA was isolated from cells at a
density of 80-90% confluence using the TRIzol® Total RNA
Extraction Kit (cat. no. 15596026; Thermo Fisher Scientific, Inc.)
and its concentration and purity were evaluated. Subsequently, mRNA
was reverse-transcribed into cDNA using mRNA Reverse Transcription
Kit (cat. no. CW2569; CWBIO) according to the manufacturer's
protocol. Target genes levels were determined on ABI 7900 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using Ultra
SYBR Mixture (CW2601; CWBIO) and calculated by 2−ΔΔCq
method (33), with β-actin
serving as internal reference gene. The PCR cycling conditions were
as follows: Initial denaturation at 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec.
The experiments were replicated three times. The primer sequences
used are listed in Table I.
 | Table IPrimers used in this study. |
Table I
Primers used in this study.
| Name | Sequences |
|---|
| H-Caspase
1-Forward |
AGGACAACCCAGCTATGCC |
| H-Caspase
1-Reverse |
TGGATAAATCTCTGCCGACT |
| H-Caspase
3-Forward |
TGGCAACAGAATTTGAGTCCT |
| H-Caspase
3-Reverse |
ACCATCTTCTCACTTGGCAT |
| H-Caspase
6-Forward |
AGGTTCTTTTGGCACTTAACAC |
| H-Caspase
6-Reverse |
AATGTGATTGCCTTCGCCATG |
| H-Caspase
7-Forward |
ACTTCCTCTTCGCCTATTCCAC |
|
H-Caspase-7-Reverse |
TTCCAGGTCTTTTCCGTGCTC |
| H-Caspase
8-Forward C |
TGGTCTGAAGGCTGGTTGT |
| H-Caspase
8-Reverse |
GTGACCAACTCAAGGGCTCA |
| H-Caspase
9-Forward |
AAGCCAACCCTAGAAAACCTTACCC |
| H-Caspase
9-Reverse |
AGCACCGACATCACCAAATCCTC |
| H-Caspase
10-Forward |
TGCAGGTAGTAATGAGATCCTGAG |
| H-Caspase
10-Reverse |
ATGGGCTGGATTGCACTTCT |
|
H-GSDME-Forward C |
GGGCGCGCGGATAAT |
|
H-GSDME-Reverse |
GTCTCTGCCAGCACCAGAAT |
|
H-CDC5L-Forward |
GTGGCCCAATCGCTGTTACT |
|
H-CDC5L-Reverse |
CGGTATTCCTCCATACGCCC |
| M-Caspase
3-Forward |
TCTGACTGGAAAGCCGAAACTCT |
| M-Caspase
3-Reverse |
AGCCATCTCCTCATCAGTCCCA |
|
M-β-actin-Forward |
ACATCCGTAAAGACCTCTATGCC |
|
M-β-actin-Reverse |
TACTCCTGCTTGCTGATCCAC |
|
H-β-actin-Forward |
ACCCTGAAGTACCCCATCGAG |
|
H-β-actin-Reverse |
AGCACAGCCTGGATAGCAAC |
Western blotting
Western blotting was applied to assess CDC5L, GSDME,
Gasdermin E N-terminal fragment (GSDME-N), Caspase 1, 3, 6, 7, 8, 9
and 10, KH RNA binding domain containing, signal transduction
associated 1 (KHDRBS1), nudix hydrolase 21 (NUDT21), heterogeneous
nuclear ribonucleoprotein A2/B1 (HNRNPA2B1), RNA binding motif
protein X-linked (RBMX), heterogeneous nuclear ribonucleoprotein C
(HNRNPC) and ELAVL1 expressions. Total proteins were extracted
using RIPA buffer (cat. no. AWB0136; Changsha Abiowell
Biotechnology Co., Ltd.). The protein concentration was determined
using the BCA Protein Assay Kit (cat. no. 23225; Thermo Fisher
Scientific, Inc.). A total of 30 μg of protein was loaded
per lane. The proteins were separated via SDS-PAGE using a 10% gel
and transferred onto nitrocellulose membranes. Membranes were
blocked with 5% skimmed milk at room temperature for 1.5 h,
followed by incubation with primary antibodies overnight at 4°C.
The primary antibodies used were: CDC5L (cat. no. 12974-1-AP;
1:1,000; Proteintech Group, Inc.), GSDME (cat. no. ab215191;
1:1,000; Abcam), GSDME-N (cat. no. ab222408; 1:1,000; Abcam),
Caspase-7 (cat. no. ab25900; 1:1,000; Abcam), Caspase-9 (cat. no.
ab2013; 1:2,000; Abcam), Caspase-6 (cat. no. ab52951; 1:20,000;
Abcam Plc), Caspase-8 (cat. no. ab227430; 1:1,000; Abcam),
Caspase-1 (cat. no. ab286125; 1:1,000; Abcam), Caspase 3 (cat. no.
19677-1-AP; 1:1,000; Proteintech Group, Inc.), Caspase-10 (cat. no.
ab2012; 1:2,000; Abcam), KHDRBS1 (cat. no. 10222-1-AP; 1:3,000;
Proteintech Group, Inc.), NUDT21 (cat. no. 10322-1-AP; 1:5,000;
Proteintech Group, Inc.), HNRNPA2B1 (cat. no. 67445-1-Ig; 1:8,000;
Proteintech Group, Inc.), RBMX (cat. no. ab250272; 1:1,000; Abcam),
HNRNPC (cat. no. ab133607; 1:20,000; Abcam), ELAVL1 (cat. no.
ab238528; dilution, 1:1,000; Abcam) and β-actin (cat. no. AWA80002;
1:5,000; Changsha Abiowell Biotechnology Co., Ltd.). Membranes were
incubated with HRP goat anti-mouse/rabbit IgG (cat. no.
SA00001-1/SA00001-2; 1:5,000/1:6,000; Proteintech Group, Inc.) at
room temperature for 1.5 h. Next, membranes were treated with an
ECL reagent (cat. no. AWB0005, Changsha Abiowell Biotechnology Co.,
Ltd.) for 1 min and then subjected to analysis using an imaging
system (Bio-Rad ChemiDoc MP, Bio-Rad Laboratories, Inc.). The
protein levels were quantified by Quantity One 4.6.6 software
(Bio-Rad Laboratories, Inc.), with β-actin serving as internal
control.
Biochemical detection
Lactate Dehydrogenase (LDH), IL-1β and IL-18 levels
were assessed using LDH Assay Kit (cat. no. A020-2; Nanjing
Jiancheng Bioengineering Institute), IL-1β Assay Kit (human; cat.
no. KE00021; Proteintech Group, Inc.; mouse; cat. no. KE10003;
Proteintech Group, Inc.) and IL-18 Assay Kit (human: cat. no.
KE00193; Proteintech Group, Inc.; mouse; cat. no. CSB-E04609m;
Wuhan Huamei Biotech Co., Ltd.) according to the instructions.
Half-life of Caspase 3 mRNA
In cells with overexpression/knockdown of ELAVL1,
the half-life of Caspase 3 mRNA was assessed using RT-qPCR
following treatment with 5 μg/ml Act D for 0, 2, 4, 6, 8 and
10 h. In addition, in cells where CDC5L was knocked down and ELAVL1
was overexpressed, RT-qPCR was employed to measure Caspase 3
mRNA expression following treatment with 5 μg/ml Act D for
0, 2, 4, 6, 8 and 10 h, in order to determine the half-life of
Caspase 3 mRNA.
Hoechst 33342 and propidium iodide (PI)
staining
Hoechst 33342 and PI staining was performed to
observe pyroptosis. Following the corresponding time of the
aforementioned treatments, cells in each group were washed once
with PBS, followed by the addition of 1 ml of cell staining buffer.
Subsequently, 5 μl of Hoechst 33342 staining solution and 5
μl of PI staining solution were added to the cells. Cells
were then gently mixed and incubated for 30 min at 4°C. The
staining status of each group of cells was viewed under a
microscope and then the fluorescence status of each group was
viewed under a microscope and images captured.
Co-immunoprecipitation (Co-IP)
In order to investigate interaction between CDC5L
and ELAVL1, Co-IP experiments were conducted. SK-HEP-1 and MHCC-97H
cells were collected for protein extraction. The cells were lysed
in a lysis buffer containing protease inhibitors at 4°C for 30 min.
The lysate was cleared by centrifugation at 14,000 × g for 10 min
at 4°C. The protein concentration was determined using the BCA
protein assay kit (cat. no. 23225; Thermo Fisher Scientific, Inc.).
For each immunoprecipitation reaction, 500 μg of protein
lysate was incubated with 2 μg of CDC5L antibody (cat. no.
12974-1-AP; Proteintech Group, Inc.) in a total volume of 500
μl of lysis buffer at 4°C overnight. Subsequently, 30
μl of Protein A/G agarose beads (cat. no. sc-2003; Santa
Cruz Biotechnology, Inc.) added to IP mixture and allowed to
incubate for 2 h at 4°C. The beads were washed three times with 1
ml of lysis buffer to remove nonspecific binding. The entire
immunoprecipitate was eluted by boiling in 50 μl of 2X
SDS-PAGE loading buffer for 10 min at 95°C. The eluted proteins
were separated by SDS-PAGE using a 10% polyacrylamide gel and
transferred onto nitrocellulose membrane. Membranes were blocked
with 5% skimmed milk at room temperature for 1.5 h. The membrane
was probed with primary antibodies against CDC5L (cat. no.
12974-1-AP; dilution, 1:1,000; Proteintech Group, Inc.) and ELAVL1
(cat. no. AWA48828; dilution, 1:1,000; Changsha Abiowell
Biotechnology Co., Ltd.) overnight at 4°C. Membranes were incubated
with HRP goat anti-rabbit IgG (cat. no. SA00001-2; 1:6,000;
Proteintech Group, Inc.) for 1.5 h. Next, membranes were treated
with an ECL reagent (cat. no. AWB0005, Changsha Abiowell
Biotechnology Co., Ltd.) for 1 min and then subjected to analysis
using an imaging system (Bio-Rad ChemiDoc MP, Bio-Rad Laboratories,
Inc.). The protein levels were quantified by Quantity One 4.6.6
software (Bio-Rad Laboratories, Inc.).
RNA immunoprecipitation (RIP) assay
Cell RNA-binding proteins were immunoprecipitated
and fluorescence quantification was performed to detect whether
CDC5L (cat. no. 12974-1-AP; Proteintech Group, Inc.) or ELAVL1
(cat. no. 11910-1-AP; Proteintech Group, Inc.) binds to Caspase
3. Briefly, cells were lysed using RIP lysis buffer at 4°C for
30 min. The lysate was cleared by centrifugation at 14,000 × g for
10 min at 4°C. For each immunoprecipitation reaction, 100 μl
of cell lysate was incubated with 50 μl of magnetic beads
coated with Protein A (according to the manufacturer's protocol) in
RIP buffer for 2 h at 4°C. The beads were washed three times with
RIP buffer. The isolated RNA was subjected to RT-qPCR analysis,
with total RNA used as input controls. Primer sequences were
H-Caspase 3, TGG CAACAGAATTTGAGTCCT forward and H-Caspase
3, ACCATCTTCTCACTTGGCAT reverse.
Statistical analysis
Data analysis was conducted using GraphPad Prism 8.0
software (Dotmatics). Results are presented as the mean ± standard
deviation. For comparisons between two groups, Student's unpaired
t-test was used, whereas for multiple group comparisons, one-way
ANOVA followed by Tukey's post-hoc test was applied. P<0.05 was
considered to indicate a statistically significant difference.
Results
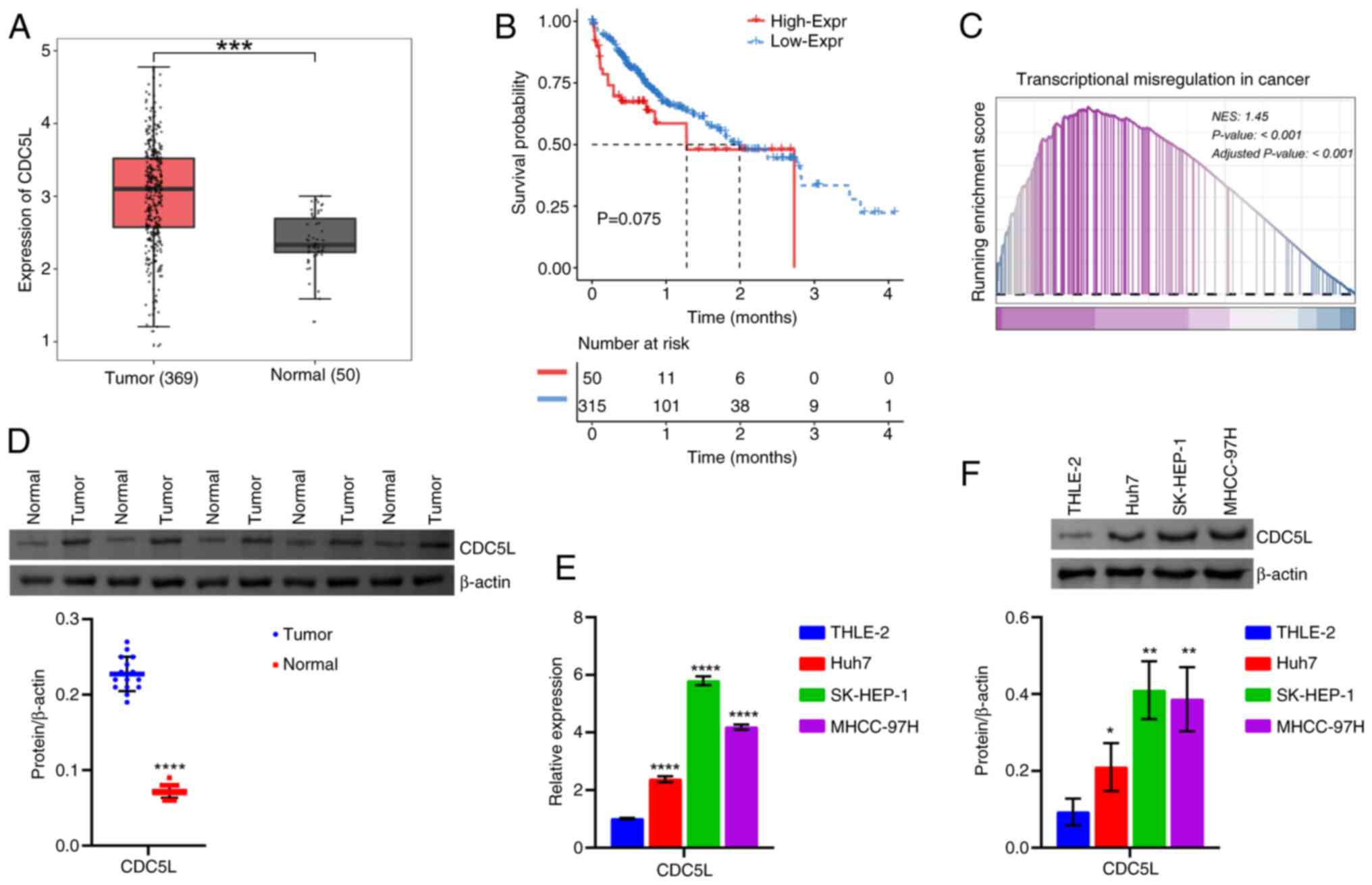
CDC5L expression is elevated in HCC
First, CDC5L expression was analyzed in HCC
based on the TCGA database. CDC5L exhibited high expression
levels in the Tumor group compared with the Normal group (Fig. 1A). In addition, Kaplan-Meier
survival analysis demonstrated that the overall survival time of
patients with HCC in the high-CDC5L group was shorter, as compared
with the low-CDC5L group (Fig.
1B). GSEA analysis showed that CDC5L was mainly enriched in
Transcriptional Misregulation In Cancer [Normalized Enrichment
Score (NES)=1.45; P<0.001; Fig.
1C]. Moreover, the functional pathways of highly expressed
CDC5L involved the MAPK signaling pathway, PI3K-Akt
signaling pathway, Wnt signaling pathway and others (Table SI). Next, western blot analysis
further revealed CDC5L expression in clinical tissues. CDC5L
expression was elevated in the HCC group compared with the
paracancerous tissue group (Fig.
1D). In addition, CDC5L expression was further validated at the
cellular level. CDC5L expression was elevated in human liver cancer
cells (Huh7, SK-HEP-1 and MHCC-97H cells) compared with THLE-2
cells. Among them, CDC5L expression was most markedly elevated in
SK-HEP-1 and MHCC-97H cells (Fig. 1E
and F) and was therefore selected for subsequent experimental
studies. The present results indicated that CDC5L expression was
upregulated in HCC.
CDC5L regulates HCC pyroptosis
Next, CDC5L was knocked down. Compared with
the si-NC group, CDC5L levels were reduced in the si-CDC5L-1
and si-CDC5L-2 groups. Among them, the si-CDC5L-2
group exhibited the most obvious reduction in CDC5L expression
(Fig. 2A) and was therefore
selected for subsequent experimental studies. Furthermore,
CDC5L was overexpressed. For cells transfected with
oe-CDC5L alone, RT-qPCR results showed a significant
increase in CDC5L mRNA expression, indicating that
oe-CDC5L transfection successfully achieved overexpression
of CDC5L (Fig. 2B).
CDC5L knockdown repressed cell viability, proliferation,
migration and invasion, but CDC5L overexpression facilitated
cell viability, proliferation, migration and invasion (Fig. 2C-E). In addition, CDC5L
knockdown promoted LDH levels, PI positive rate, GSDME and GSDME-N
levels and IL-18 and IL-1β levels. CDC5L overexpression
suppressed LDH levels, PI positive rate, GSDME and GSDME-N levels
and IL-18 and IL-1β levels (Fig.
3A-D). Moreover, the effects of low and high CDC5L
expression was verified in HCC in vivo. It was found that,
following CDC5L knockdown, the tumor volume was gradually
reduced, along with reductions in tumor size and weight.
Conversely, following CDC5L overexpression, the tumor volume
gradually increased and both tumor size and weight grew larger
(Fig. 4A-C). In addition, IL-18
and IL-1β levels were elevated following CDC5L knockdown. By
contrast, these cytokine levels were diminished following
CDC5L overexpression (Fig.
4D). CDC5L knockdown promoted GSDME and GSDME-N levels
but repressed CDC5L levels. CDC5L overexpression suppressed
GSDME and GSDME-N levels but elevated CDC5L levels (Fig. 4E). These results indicated that
CDC5L regulated HCC pyroptosis.
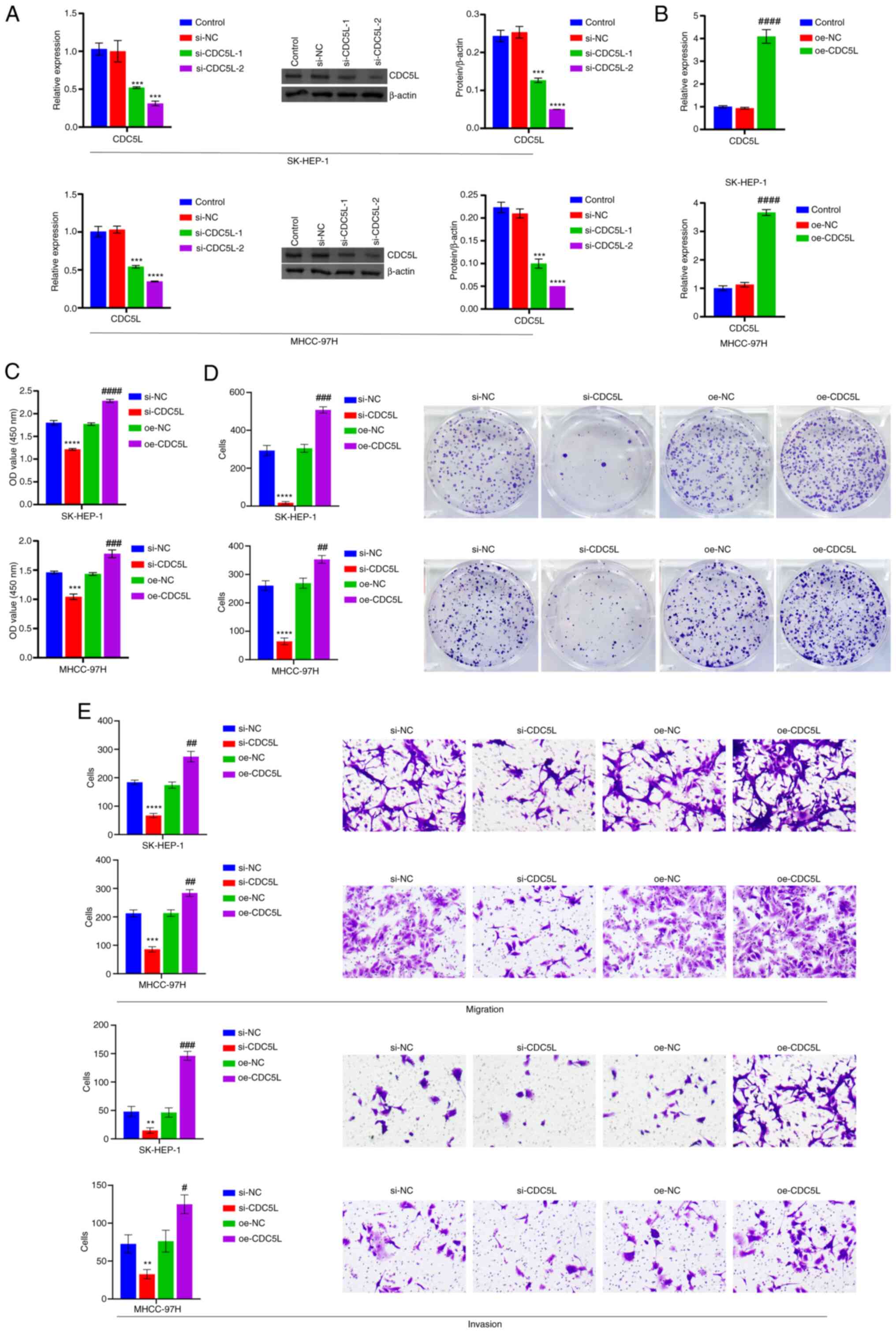 | Figure 2CDC5L regulates HCC pyroptosis. (A)
RT-qPCR and western blot analysis of CDC5L expression.
***P<0.001 and ****P<0.0001 vs. si-NC.
(B) Detection of CDC5L mRNA expression after oe-CDC5L
transfection by RT-qPCR. (C) Measurement of cell viability by CCK-8
assay. (D) Evaluation of cell proliferation via clone formation
assays. (E) Analysis of cell migration and invasion through
Transwell assays. Scale bar, 100 μm, magnification, ×100.
**P<0.01, ***P<0.001 and
****P<0.0001 vs. si-NC; #P<0.05,
##P<0.01, ###P<0.001 and
####P<0.0001 vs. oe-NC. CDC5L, cell division cycle 5
like; HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription
quantitative PCR; CCK-8, Cell Counting Kit-8; si, short
interfering; si-NC, negative control siRNA; si-CDC5L,
CDC5L-specific siRNA; oe-NC, negative control overexpression
vector; oe-CDC5L, CDC5L overexpression vector. |
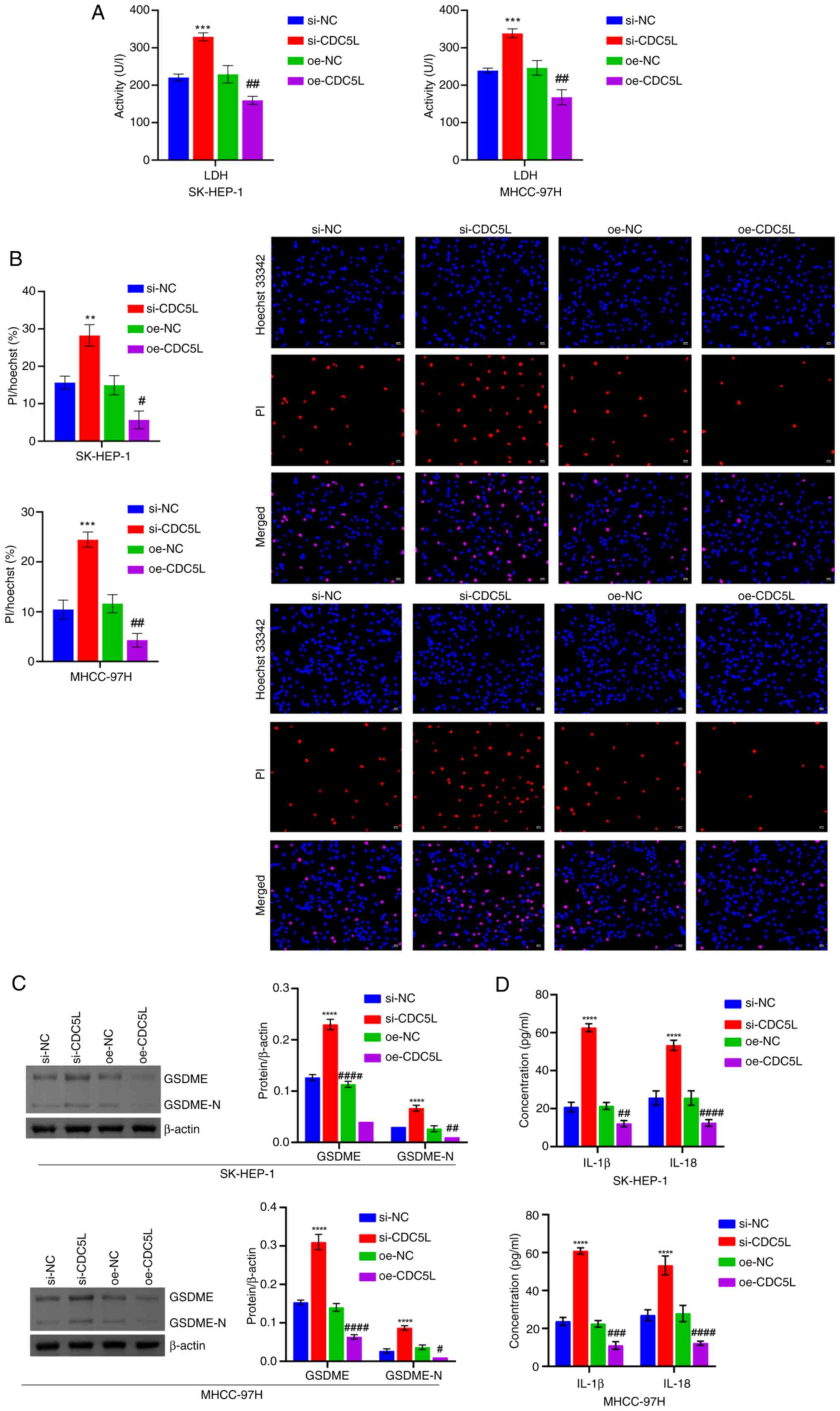 | Figure 3Detection of pyroptosis-related
levels. (A) LDH level in cell supernatant. (B) Hoechst 33342 and PI
staining of pyroptosis. Scale bar, 50 μm, magnification,
×200. (C) Western blot analysis of GSDME and GSDME-N levels. (D)
IL-18 and IL-1β levels were assessed. **P<0.01,
***P<0.001 and ****P<0.0001 vs. si-NC;
#P<0.05, ##P<0.01,
###P<0.001 and ####P<0.0001 vs. oe-NC.
LDH, lactate dehydrogenase; PI, propidium iodide; GSDME, Caspase
3/Caspase 3-gasdermin E; GSDME-N, Gasdermin E N-terminal fragment;
si, short interfering; si-NC, negative control siRNA;
si-CDC5L, CDC5L-specific siRNA; oe-NC, negative
control overexpression vector; oe-CDC5L, CDC5L
overexpression vector. |
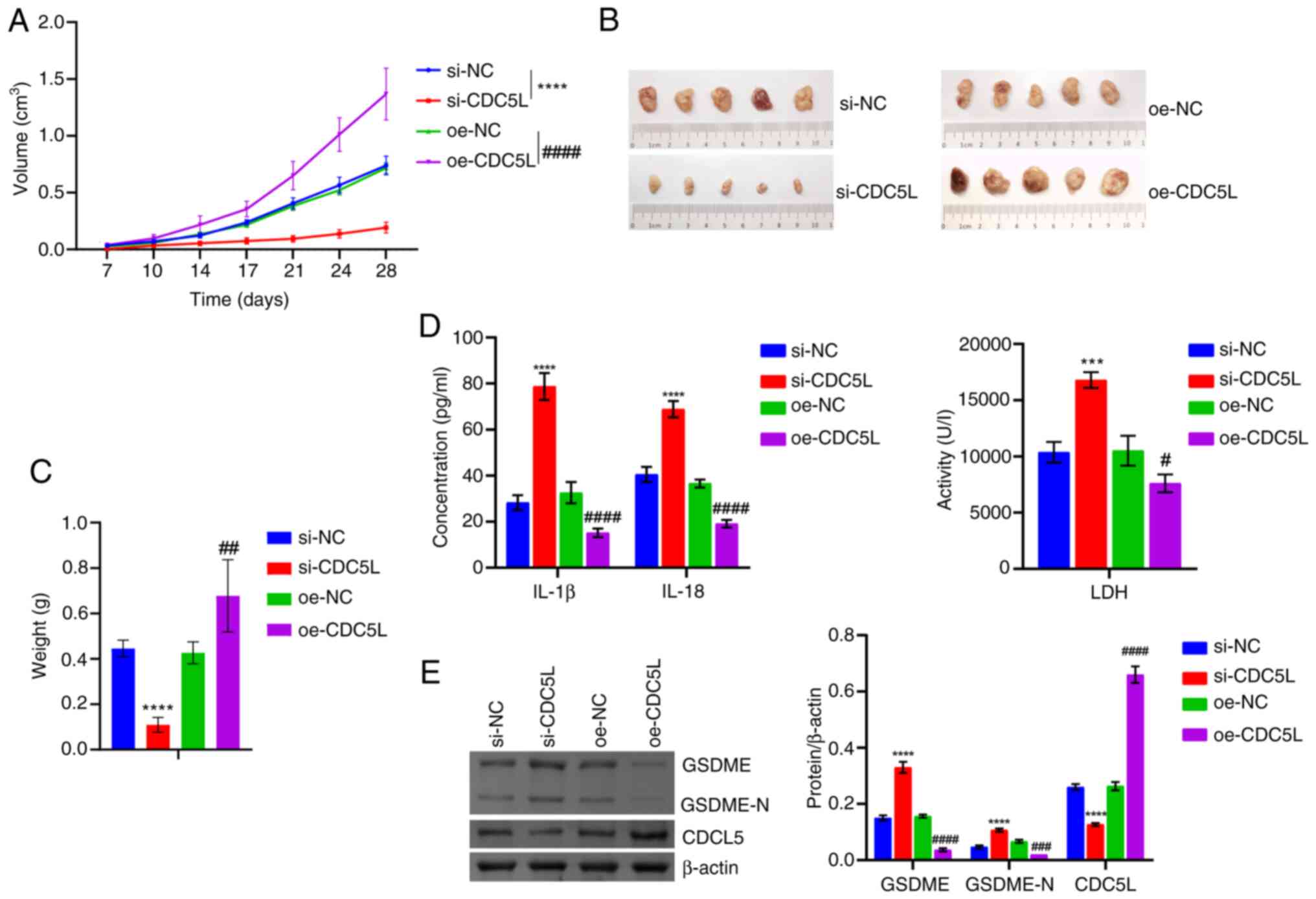 | Figure 4Validation at the animal level that
CDC5L regulates pyroptosis in HCC. (A) Tumor volume. (B) Tumor
images. (C) Tumor weight. (D) IL-18, IL-1β and LDH levels in tissue
supernatants. (E) Western blot analysis of CDC5L, GSDME and GSDME-N
levels in the tumor tissue. ***P<0.001 and
****P<0.0001 vs. si-NC; #P<0.05,
##P<0.01, ###P<0.001 and
####P<0.0001 vs. oe-NC. LDH, lactate dehydrogenase;
CDC5L, cell division cycle 5 like; GSDME, Caspase 3/Caspase
3-gasdermin E; GSDME-N, Gasdermin E N-terminal fragment; si, short
interfering; si-NC, negative control siRNA; si-CDC5L,
CDC5L-specific siRNA; oe-NC, negative control overexpression
vector; oe-CDC5L, CDC5L overexpression vector. |
The regulation of CDC5L on pyroptosis of
HCC depends on the Caspase family
Based on GeneCards search results, Caspases 1-10
were seen to be involved in the progression of HCC. The Caspase
family genes common to both the high and low expression groups of
CDC5L were Caspases 1, 2, 3, 6, 7, 8, 9 and 10. According to
GeneCards search results, among them, Caspases 1, 3, 6, 7, 8, 9 and
10 mediated the process of pyroptosis (Fig. 5A). The expression of Caspases 1,
3, 6, 7, 8, 9 and 10 was validated using RT-qPCR and western
blotting. Caspase 3, a key protein involved in both pyroptosis and
apoptosis, exerted tumor cytotoxicity via GSDME upon activation
(34). Combining the results of
mRNA and protein, it was observed CDC5L knockdown substantially
elevated Caspase 3 expression, but CDC5L overexpression result in a
markedly reduction in Caspase 3 expression (Fig. 5B). Therefore, Caspase 3 was
selected for subsequent experiments. RIP was further used to detect
the binding of CDC5L to Caspase 3 mRNA. The results showed
that CDC5L did not bind to Caspase 3 mRNA (Fig. 5C). Next, Caspase 3 was
used to interfere. For cells transfected with si-Caspase 3
alone, RT-qPCR results showed that compared with the si-NC group,
the expression of Caspase 3 mRNA was markedly reduced in
both the si-Caspase 3#1 and si-Caspase 3#2 groups.
Among them, the si-Caspase 3#2 group exhibited the most
pronounced reduction in Caspase 3 mRNA expression and was
used for subsequent experiments. This indicated that si-Caspase
3 transfection successfully achieved knockdown of Caspase
3 (Fig. 5D). Compared with
the si-NC group, si-CDC5L group exhibited a reduced the expression
of CDC5L and increased the expression of Caspase 3. Upon the
additional knockdown of Caspase 3, its expression decreased, while
CDC5L expression remained largely unchanged (Fig. 5E). Cellular function experiments
showed that, compared with si-NC group, cell viability,
proliferation, migration and invasion capabilities in the si-CDC5L
group were diminished. Following the further knockdown of Caspase
3, cell viability, proliferation, migration and invasion
capabilities increased (Figs. 5F
and 6A-B). In addition, compared
with the si-NC group, LDH, IL-18 and IL-1β levels, PI positive rate
and GSDME and GSDME-N protein levels in the si-CDC5L group
increased. Following the further knockdown of Caspase 3, LDH, IL-18
and IL-1β levels decreased, PI positive rate decreased and GSDME
and GSDME-N protein levels decreased (Figs. 6C-E and S1). Furthermore, the effects of
interfering with CDC5L and Caspase 3 were verified on HCC in
vivo. It was found that, following the knockdown of histone
deacetylase 6 (HDAC6), tumor volume gradually decreased and tumor
size and tumor weight also decreased. Following the further
knockdown of Caspase 3, tumor volume gradually increased and tumor
size weight also increased (Fig.
7A-C). In addition, following the knockdown of HDAC6, LDH,
IL-18 and IL-1β levels in the tissue supernatant increased.
Following the further knockdown of Caspase 3, LDH, IL-18 and IL-1β
levels decreased (Fig. 7D).
Following the knockdown of HDAC6, CDC5L expressions in the tumor
tissue decreased and Caspase 3, GSDME and GSDME-N expression
increased. Following the further knockdown of Caspase 3, there was
no significant change in CDC5L expression and Caspase 3, GSDME and
GSDME-N expression decreased (Fig.
7E). These results indicated that CDC5L regulated pyroptosis in
HCC in a Caspase 3-dependent manner.
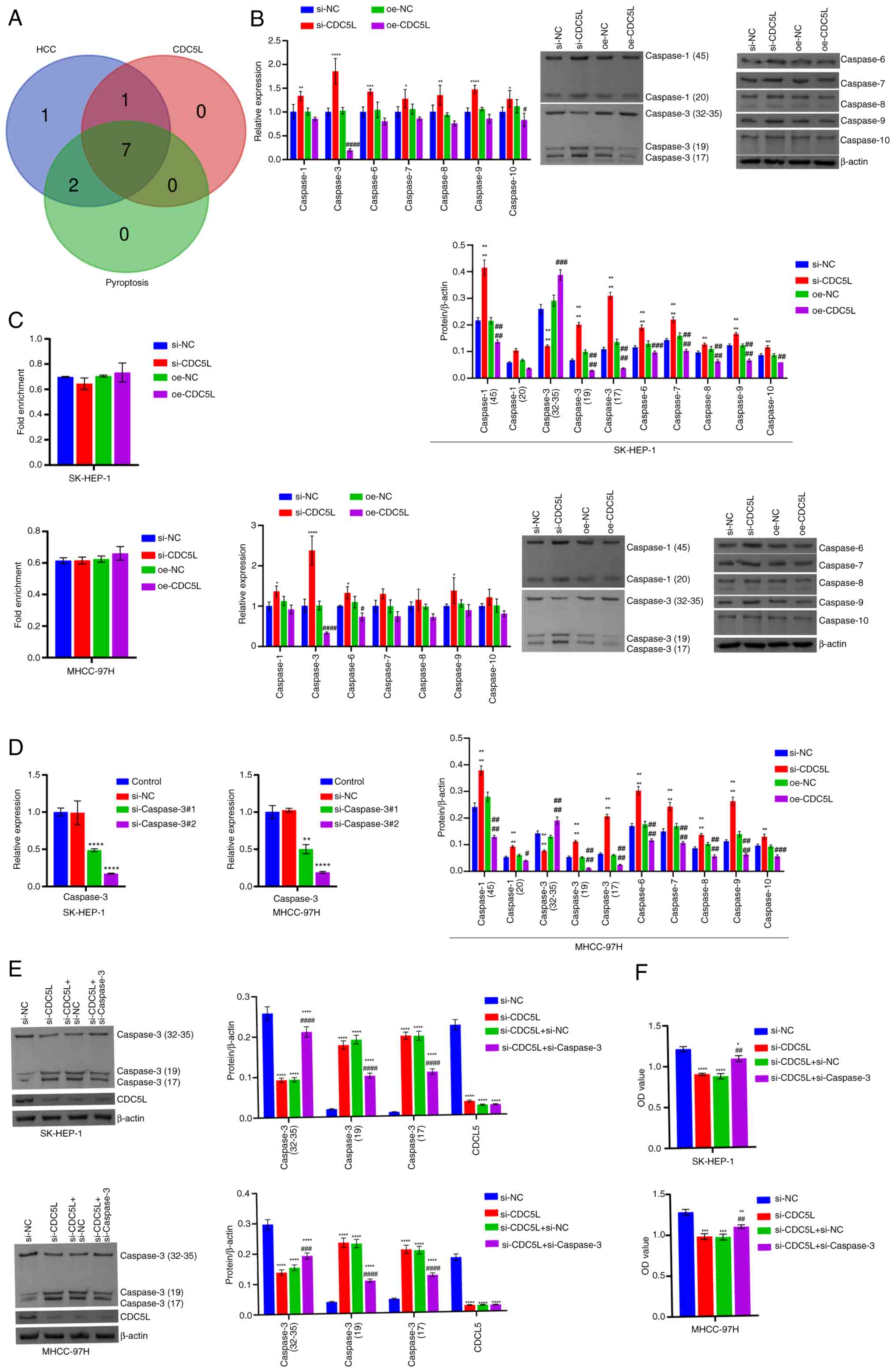 | Figure 5The regulation of CDC5L on pyroptosis
of HCC depends on the Caspase family. (A) Bioinformatics analysis
of the expression of Caspases 1, 3, 6, 7, 8, 9 and 10 levels in
HCC. (B) RT-qPCR and western blot analysis of Caspases 1, 3, 6, 7,
8, 9 and 10 levels. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. si-NC;
#P<0.05, ##P<0.01,
###P<0.001 and ####P<0.0001 vs. oe-NC.
(C) Detection of the binding of CDC5L to Caspase 3 mRNA by
RIP. (D) Detection of Caspase 3 mRNA expression after si-Caspase
3 transfection by RT-qPCR. (E) Western blot analysis of CDC5L
and Caspase 3 levels. (F) Assessment of cell viability using the
CCK-8 assay. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. si-NC;
##P<0.01, ###P<0.001 and
####P<0.0001 vs. si-CDC5L+si-NC. CDC5L, cell
division cycle 5 like; HCC, hepatocellular carcinoma; RIP, RNA
immunoprecipitation; RT-qPCR, reverse transcription
quantitative PCR; CCK-8, Cell Counting Kit-8; si, short
interfering; si-NC, negative control siRNA; si-CDC5L,
CDC5L-specific siRNA; oe-NC, negative control overexpression
vector; oe-CDC5L, CDC5L overexpression vector. |
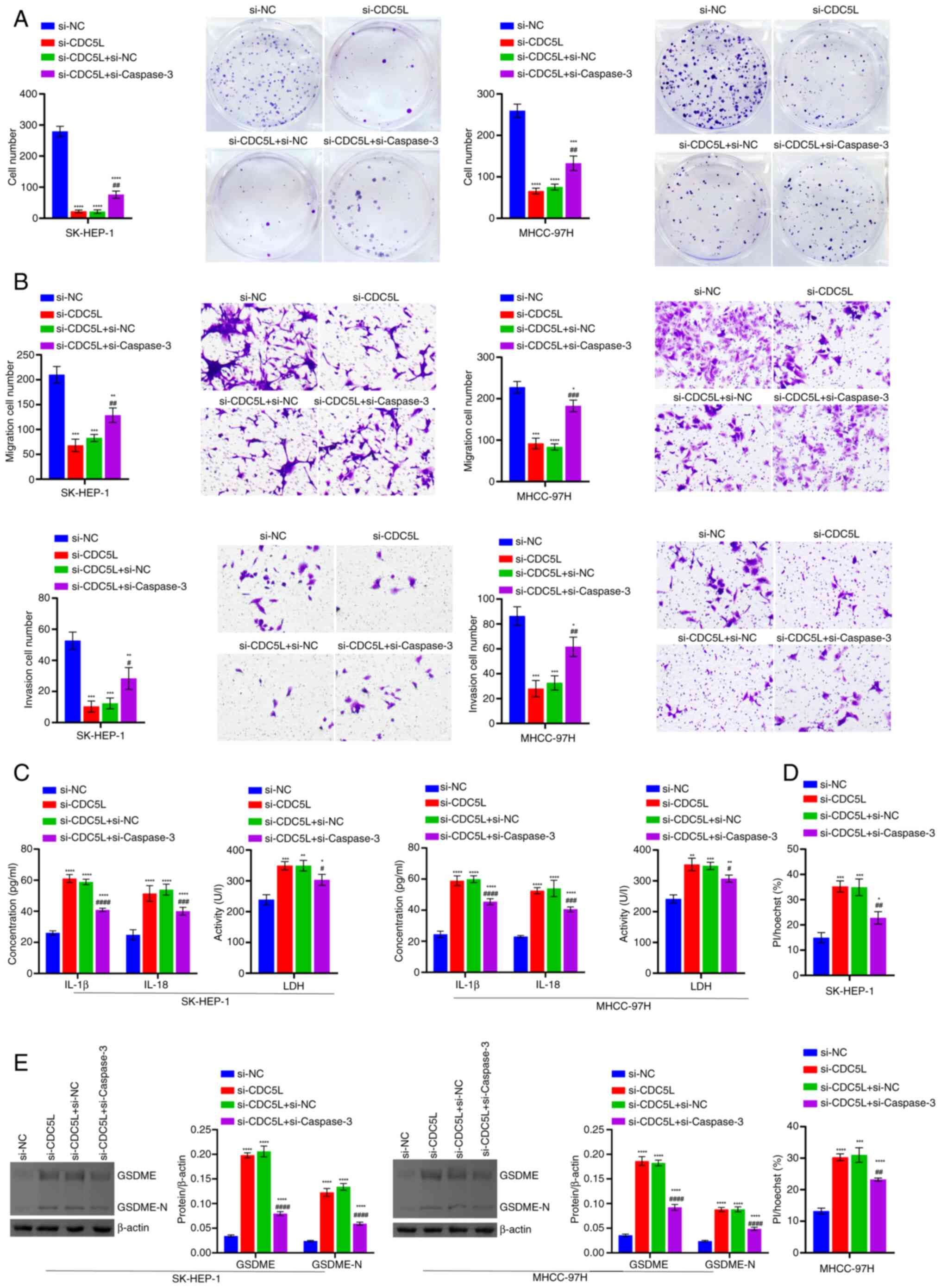 | Figure 6Detection of cell function and
pyroptosis-related levels. (A) Evaluation of cell proliferation
through clone formation assays. (B) Analysis of cell migration and
invasion through Transwell assays. Scale bar, 100 μm,
magnification, ×100. (C) LDH, IL-18 and IL-1β levels in the cell
supernatant. (D) Results of Hoechst 33342 and PI staining of
pyroptosis. Scale bar, 50 μm, ×200. (E) Western blot
analysis of GSDME and GSDME-N levels. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001 vs. si-NC; #P<0.05,
##P<0.01, ###P<0.001 and
####P<0.0001 vs. si-CDC5L+si-NC. LDH, lactate
dehydrogenase; PI, propidium iodide; GSDME, Caspase 3/Caspase
3-gasdermin E; GSDME-N, Gasdermin E N-terminal fragment; si, short
interfering; si-NC, negative control siRNA; si-CDC5L,
CDC5L-specific siRNA; si-Caspase 3, Caspase
3-specific siRNA. |
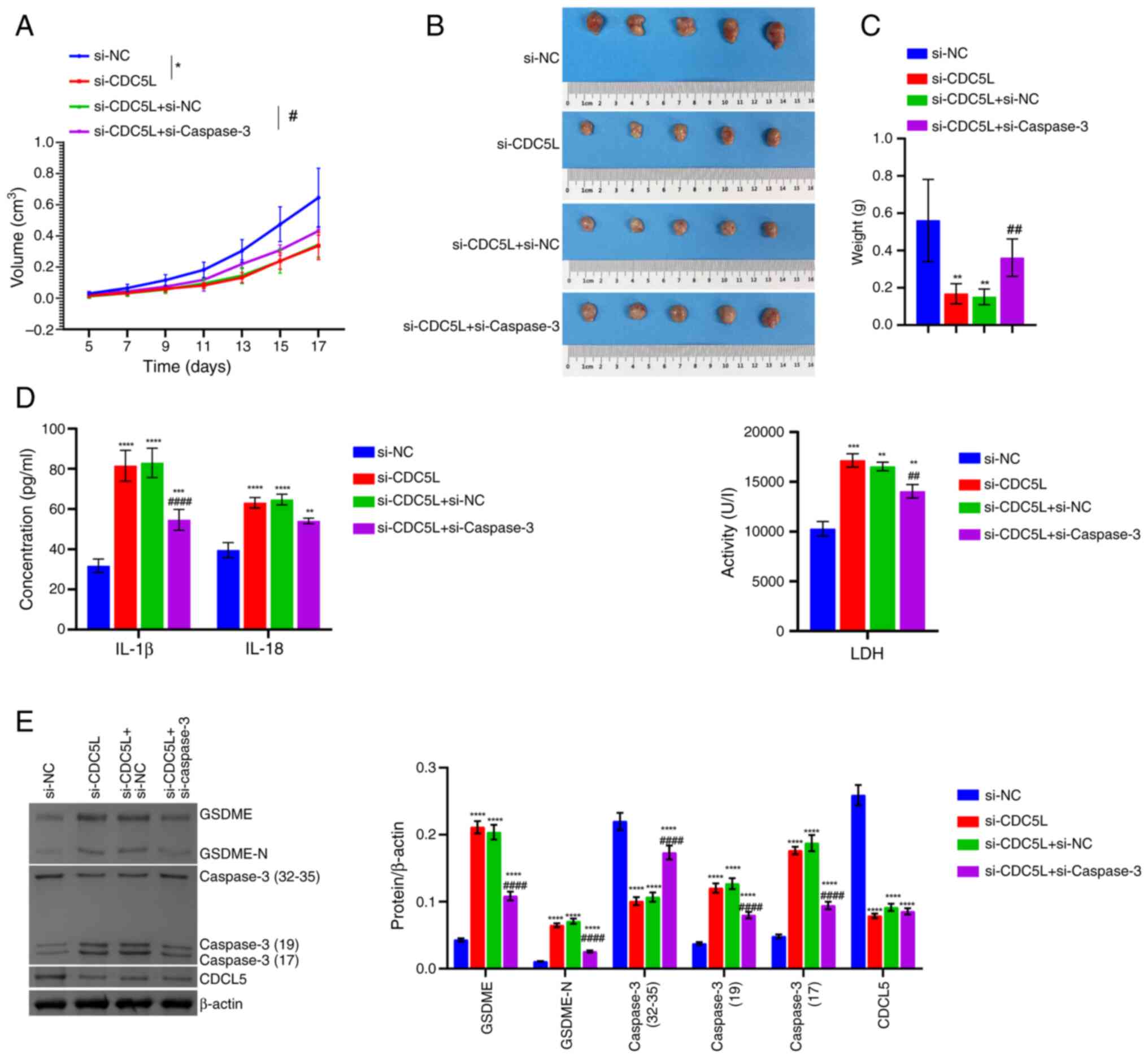 | Figure 7Validation at the animal level that
the regulation of pyroptosis in HCC by CDC5L depends on the Caspase
family. (A) Tumor volume. (B) Tumor images. (C) Tumor weight. (D)
IL-18, IL-1β and LDH levels in the tissue supernatants. (E) Western
blot analysis of CDC5L, GSDME, Caspase 3 and GSDME-N levels in the
tumor tissue. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. si-NC;
#P<0.05, ##P<0.01 and
####P<0.0001 vs. si-CDC5L+si-NC. LDH, lactate
dehydrogenase; CDC5L, cell division cycle 5 like; GSDME, Caspase
3/Caspase 3-gasdermin E; GSDME-N, Gasdermin E N-terminal fragment;
si, short interfering; si-NC, negative control siRNA;
si-CDC5L, CDC5L-specific siRNA; si-Caspase 3,
Caspase 3-specific siRNA. |
CDC5L competitively binds to ELAVL1 to
inhibit the binding of ELAVL1 and Caspase 3 mRNA to regulate the
pyroptosis of HCC
The earlier results in the present study indicated
that CDC5L downregulated Caspase 3 and reduced the stability of
Caspase 3 mRNA. It was proposed in the present study that
CDC5L might influence the stability of Caspase 3 mRNA
through specific RBPs. Therefore, starting from CDC5L, a search for
interacting proteins was performed and starting from Caspase
3 mRNA a search for interacting RBPs was performed; the RBPs
were determined by finding the intersection of these two ways.
First, the Venn diagram revealed the intersection between CDC5L
binding proteins and Caspase 3 interacting RBPs, with a total of 22
(Fig. 8A). Bioinformatics
analysis of the expression of these 22 RBPs in HCC was conducted to
screen significant RBPs (Fig.
8B). Next, the expression of markedly altered RBP proteins
KHDRBS1, NUDT21, HNRNPA2B1, RBMX, HNRNPC and ELAVL1 was validated
in clinical tissues. As compared with the Normal group, KHDRBS1,
NUDT21, HNRNPA2B1, RBMX, HNRNPC and ELAVL1 levels were elevated in
the Tumor group. Among them, the expression of ELAVL1 was the most
prominently increased, so it was selected for further analysis
(Fig. 8C). Subsequently,
ELAVL1 was knocked down and overexpressed. Compared with the
si-NC group, ELAVL1 levels in the si-ELAVL1#1 and
si-ELAVL1#2 groups were repressed, with the most significant
decrease observed in the si-ELAVL1#2 group. As compared with
the oe-NC group, ELAVL1 expression in the oe-ELAVL1 group
was markedly elevated (Fig. 8D).
The present study confirmed that si-ELAVL1 and
oe-ELAVL1 were successfully transfected. Based on the UCSC
Genome Browser, it was shown that ELAVL1 bound with Caspase
3 (Fig. 8E). RIP further
confirmed the binding of ELAVL1 to Caspase 3 mRNA. Following
the knockdown of ELAVL1, the binding of ELAVL1 to Caspase 3
mRNA was weakened. Following the overexpression of ELAVL1,
the binding of ELAVL1 to Caspase 3 mRNA was enhanced
(Fig. 8F). Furthermore,
ELAVL1 knockdown inhibited the half-life of Caspase 3
mRNA following treatment with 5 μg/ml Act D for 0, 2, 4, 6,
8 and 10 h. ELAVL1 overexpression extended the half-life of
Caspase 3 mRNA following treatment with 5 μg/ml Act D
for 0, 2, 4, 6, 8 and 10 h (Fig.
8G). In addition, ELAVL1 knockdown promoted Caspase 3,
LDH, IL-1β, IL-18, GSDME and GSDME-N levels. ELAVL1
overexpression repressed Caspase 3, LDH, IL-1β, IL-18, GSDME and
GSDME-N levels (Fig. 9A-C). In
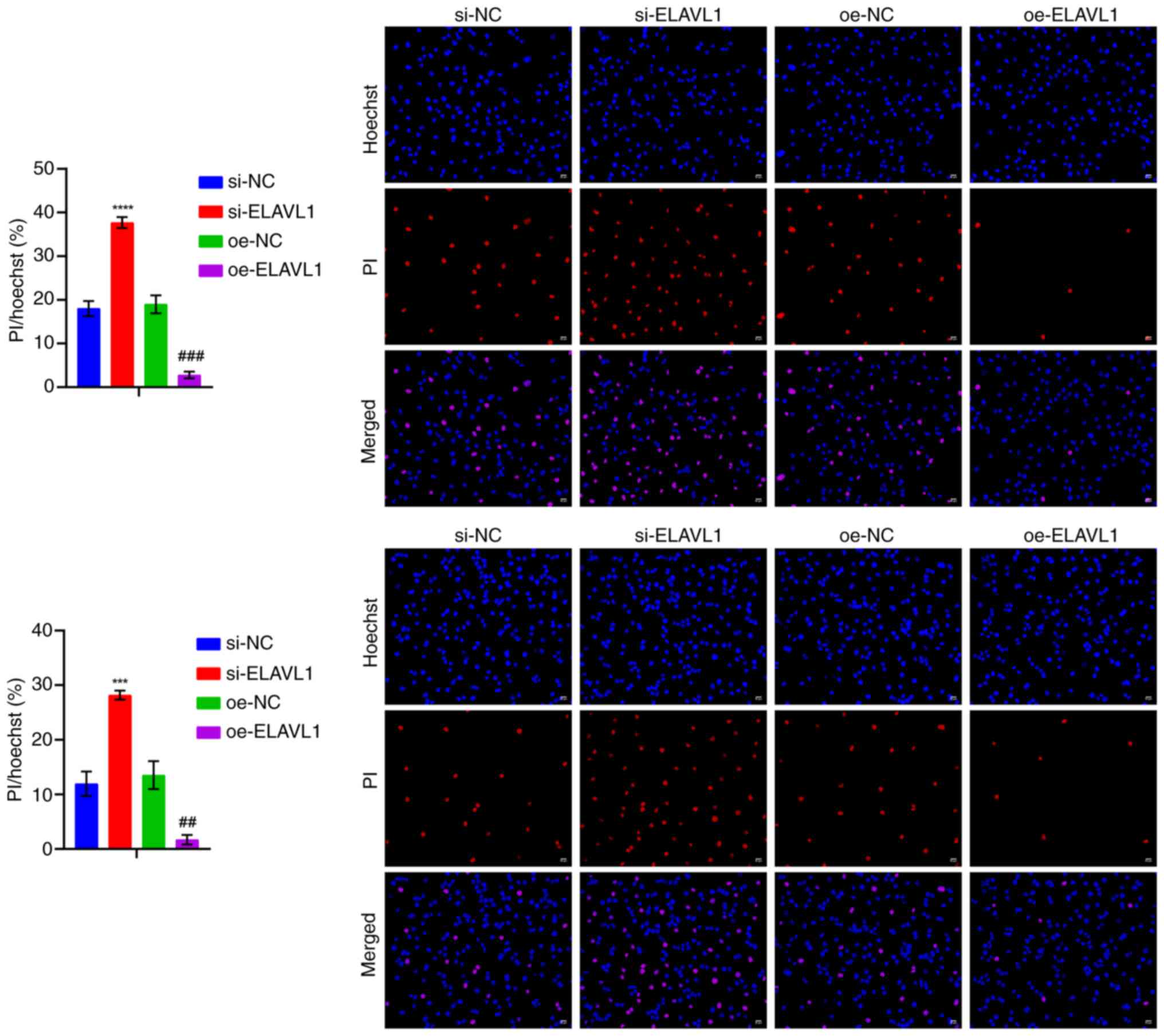
addition, ELAVL1 knockdown promoted cell pyroptosis, while
ELAVL1 overexpression inhibited cell pyroptosis (Fig. 10).
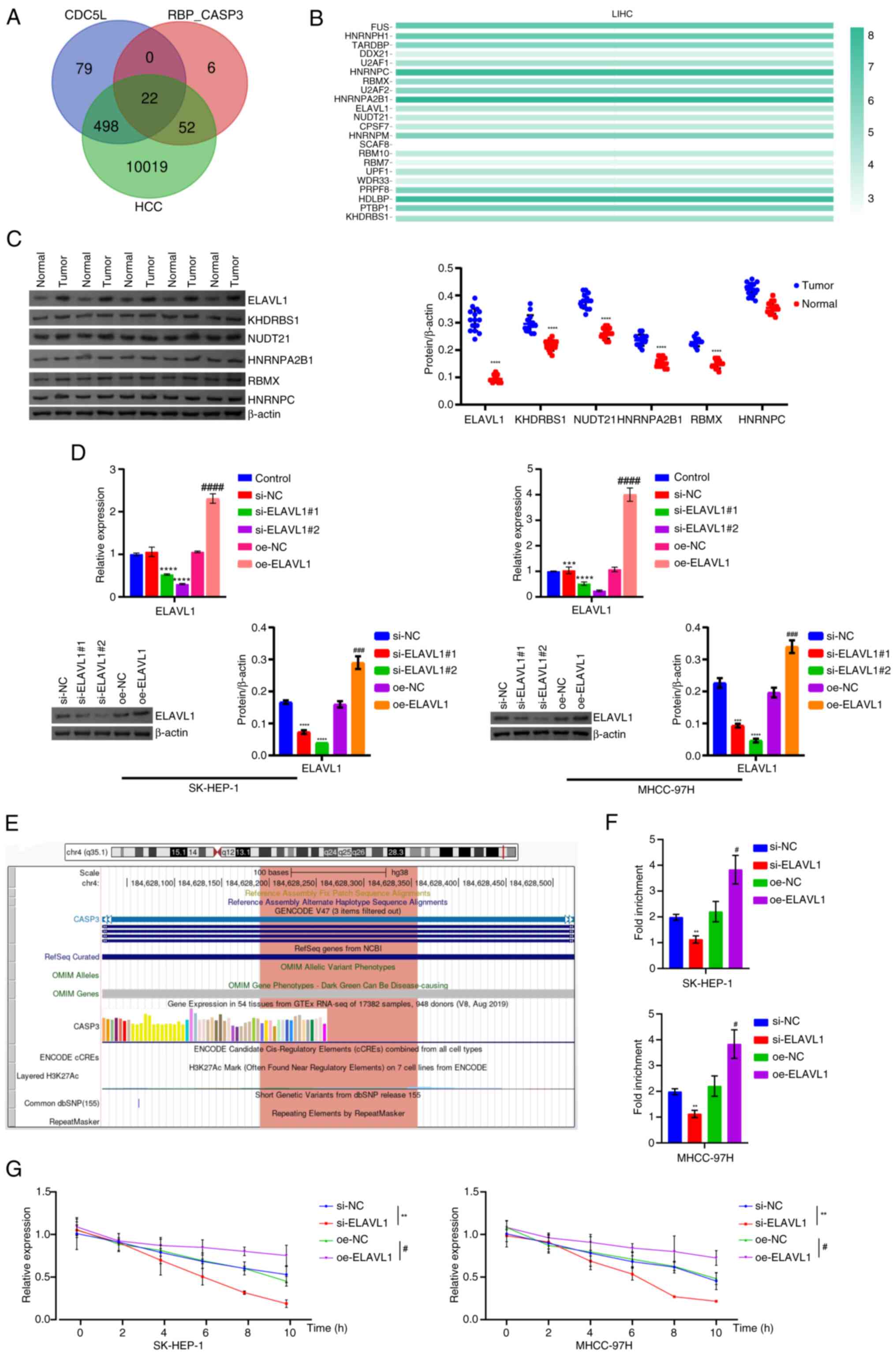 | Figure 8Mechanism of knockdown and
overexpression of ELAVL1 in pyroptosis in HCC. (A) Venn diagram
showing the intersection of CDC5L binding proteins and Caspase 3
interacting RBPs, with a total of 22. (B) Bioinformatics analysis
of the expression of these 22 RBPs in HCC was conducted to screen
significant RBPs. (C) Western blot analysis of RBP protein levels.
(D) Western blot analysis of ELAVL1 levels. (E) UCSC Genome Browser
showed that ELAVL1 binds with Caspase 3. (F) RIP detection
of the binding of ELAVL1 to Caspase 3 mRNA in cells. (G) RT-qPCR
detection of the half-life of Caspase 3 mRNA in cells
overexpressing/silencing ELAVL1 following treatment with 5
μg/ml Act D for 0, 2, 4, 6, 8 and 10 h.
**P<0.01, ***P<0.001 and
****P<0.0001 vs. si-NC; #P<0.05,
###P<0.001 and ####P<0.0001 vs. oe-NC.
ELAVL1, ELAV like RNA binding protein 1; HCC, hepatocellular
carcinoma; CDC5L, cell division cycle 5 like; RBPs, RNA-binding
proteins; UCSC, University of California, Santa Cruz; RIP, RNA
immunoprecipitation; si, short interfering; si-NC, negative control
siRNA; si-ELAVL1, ELAVL1-specific siRNA; oe-NC,
negative control overexpression vector; oe-ELAVL1,
ELAVL1 overexpression vector. |
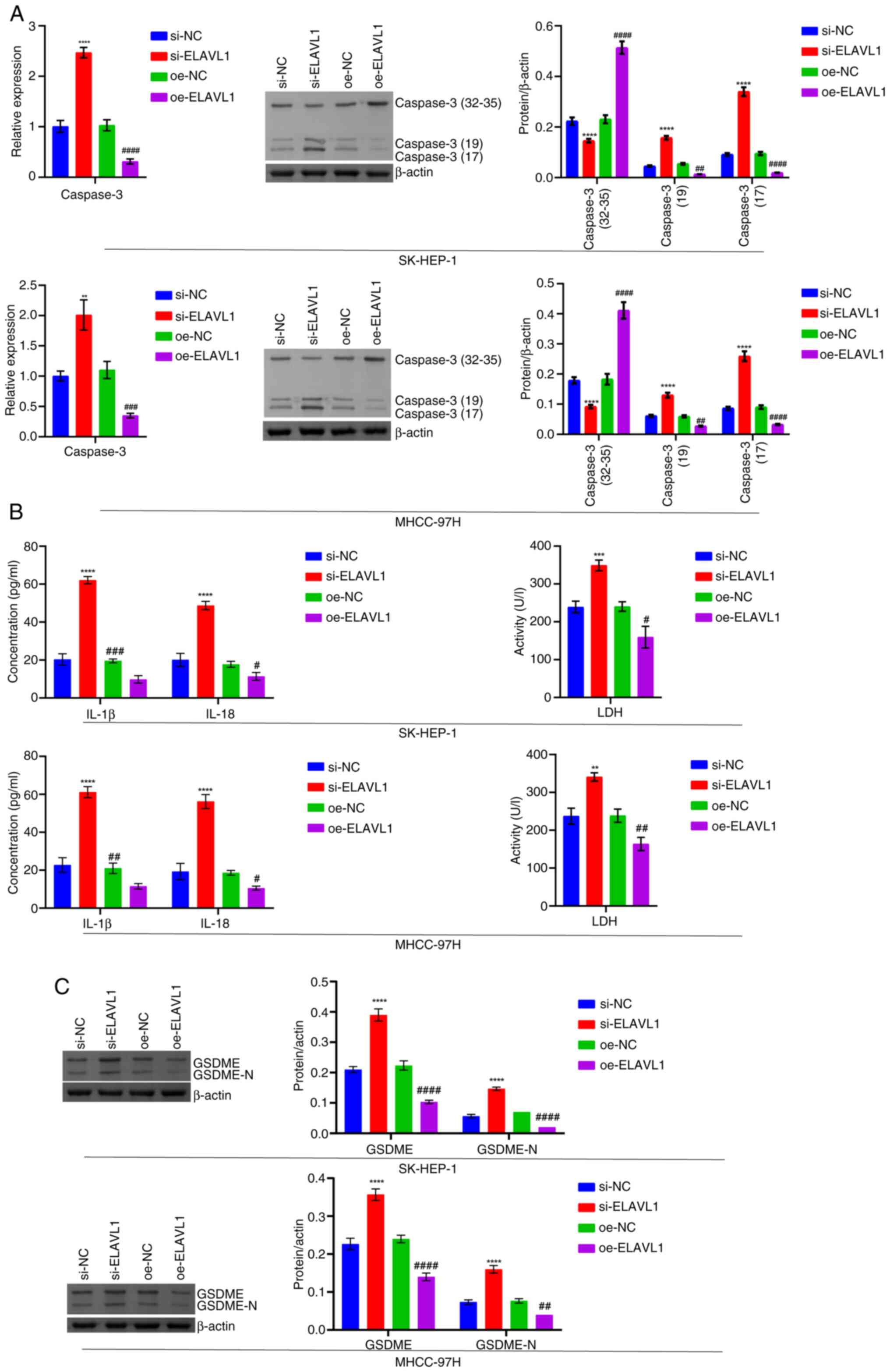 | Figure 9Detection of pyroptosis-related
levels following knockdown and overexpression of ELAVL1. (A)
RT-qPCR and western blot analysis of Caspase 3 levels. (B) LDH,
IL-1β and IL-18 levels in the cell supernatant. (C) Western blot
analysis of GSDME and GSDME-N levels. **P<0.01,
***P<0.001 and ****P<0.0001 vs. si-NC;
#P<0.05, ##P<0.01,
###P<0.001 and ####P<0.0001 vs. oe-NC.
RT-qPCR, reverse transcription-quantitative PCR; LDH, lactate
dehydrogenase; GSDME, Caspase 3/Caspase 3-gasdermin E; GSDME-N,
Gasdermin E N-terminal fragment; si, short interfering; si-NC,
negative control siRNA; si-ELAVL1, ELAVL1-specific
siRNA; oe-NC, negative control overexpression vector;
oe-ELAVL1, ELAVL1 overexpression vector. |
Furthermore, compared with the si-NC group, ELAVL1
expression in the si-CDC5L group was decreased. Compared
with the oe-NC group, ELAVL1 expression in the oe-CDC5L
group was markedly elevated (Fig.
11A). Co-IP verified the binding of CDC5L to ELAVL1 protein
(Fig. 11B). In addition,
following CDC5L knockdown, Caspase 3 expression increased.
Following CDC5L overexpression, Caspase 3 expression
decreased (Fig. 11C). RIP
further verified the binding of ELAVL1 to Caspase 3 mRNA.
Following CDC5L knockdown, the binding of ELAVL1 to
Caspase 3 mRNA was enhanced. Following CDC5L overexpression,
the binding of ELAVL1 to Caspase 3 mRNA was weakened
(Fig. 11D). Next, the effects
of interfering with CDC5L and overexpressing ELAVL1
on HCC pyroptosis was explored in vitro and in vivo.
In vitro, CDC5L knockdown suppressed ELAVL1
expression. Upon further overexpression of ELAVL1, its
expression increased (Fig.
11E). CDC5L knockdown promoted Caspase 3 expression.
Upon further overexpression of ELAVL1, Caspase 3 expression
decreased (Fig. 11F). RIP
verified the binding stability of ELAVL1 protein with Caspase
3 mRNA. CDC5L knockdown enhanced the binding stability
of ELAVL1 protein with Caspase 3 mRNA. Upon further
overexpression of ELAVL1, it inhibited the binding stability
of ELAVL1 protein with Caspase 3 mRNA (Fig. 12A). In addition, CDC5L
knockdown promoted an increase in LDH, IL-1β, IL-18, GSDME and
GSDME-N levels. Upon further overexpression of ELAVL1, it
suppressed LDH, IL-1β, IL-18, GSDME and GSDME-N levels (Fig. 12B-C). Furthermore, CDC5L
knockdown promoted cell pyroptosis. Upon further overexpression of
ELAVL1, it inhibited cell pyroptosis (Fig. 12D). In addition, CDC5L
knockdown extended the half-life of Caspase 3 mRNA following
treatment with 5 μg/ml Act D for 0, 2, 4, 6, 8, 10 h.
Overexpression of ELAVL1 inhibited the half-life of
Caspase 3 mRNA following treatment with 5 μg/ml Act D
for 0, 2, 4, 6, 8, 10 h (Fig.
12E). In vivo, it was found that following CDC5L
knockdown, tumor volume gradually decreased and tumor size and
weight were also reduced. Following further overexpression of
ELAVL1, the tumor volume gradually increased and the tumor
size and weight also increased (Fig. 13A-C). In addition, CDC5L
knockdown promoted an increase in LDH, IL-1β, IL-18, CAPS3, GSDME
and GSDME-N levels. Following further overexpression of
ELAVL1, it suppressed CAPS3, LDH, IL-1β, IL-18, GSDME and
GSDME-N levels (Fig. 13D -
Forward). These results indicated that CDC5L competitively bound to
ELAVL1 to inhibit the binding of ELAVL1 with Caspase 3 mRNA,
thereby regulating pyroptosis in HCC.
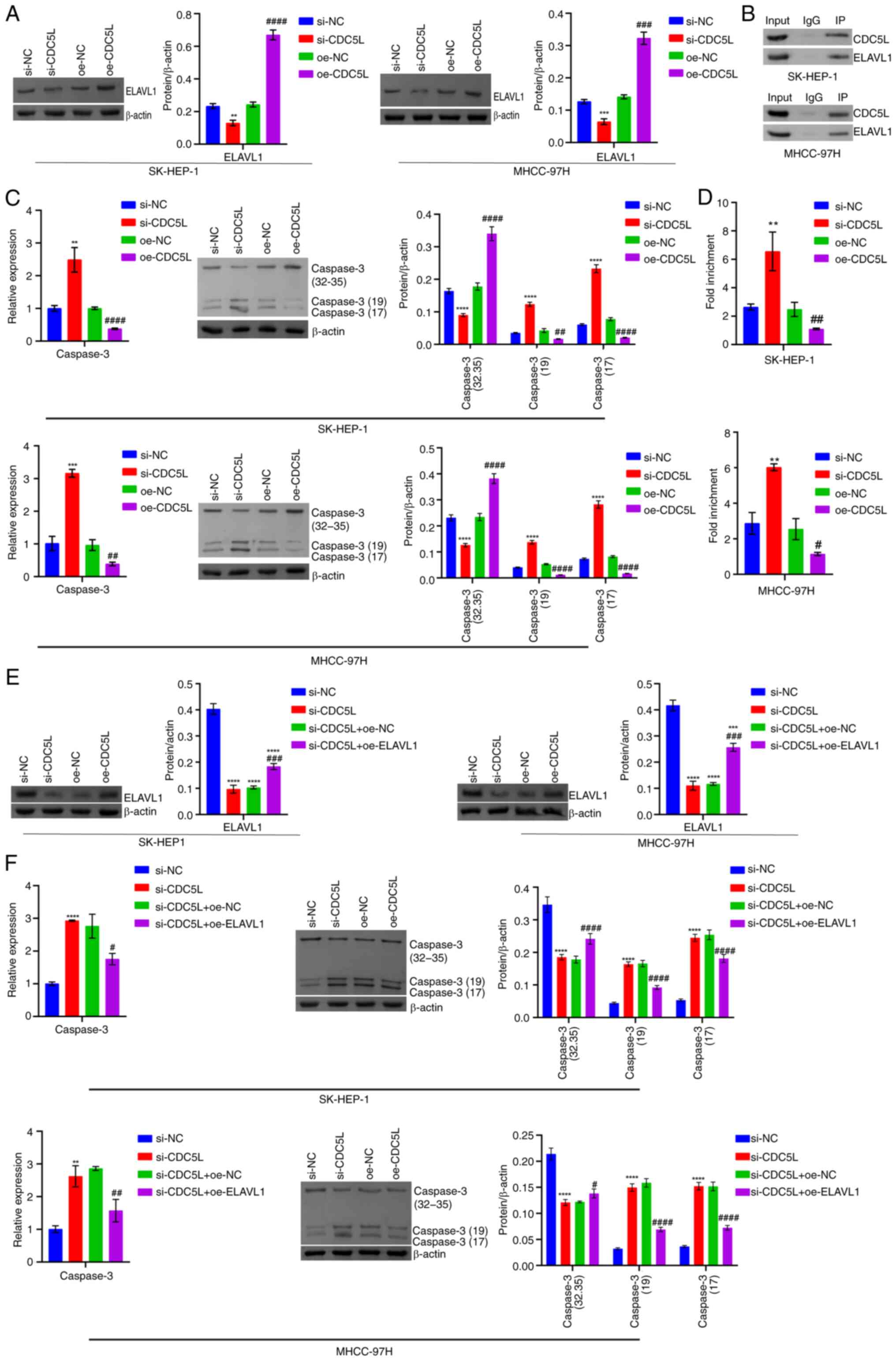 | Figure 11Mechanism of CDC5L knockdown and
overexpression of ELAVL1 in pyroptosis in HCC. (A) Western blot
analysis of ELAVL1 levels. (B) Co-IP validation of interaction
between CDC5L and ELAVL1 protein. (C) RT-qPCR and western blot
analysis of Caspase 3 levels. (D) RIP verification of the binding
of ELAVL1 to Caspase 3 mRNA in cells. **P<0.01,
***P<0.001 and ****P<0.0001 vs. si-NC;
#P<0.05, ##P<0.01,
###P<0.001 and ####P<0.0001 vs. oe-NC.
(E) Western blot analysis of ELAVL1 levels. (F) RT-qPCR and western
blot analysis of Caspase 3 levels. **P<0.01,
***P<0.001 and ****P<0.0001 vs. si-NC;
#P<0.05, ##P<0.01,
###P<0.001 and ####P<0.0001 vs.
si-CDC5L+oe-NC. CDC5L, cell division cycle 5 like; ELAVL1,
ELAV like RNA binding protein 1; HCC, hepatocellular carcinoma;
Co-IP, Co-immunoprecipitation; RT-qPCR, reverse
transcription-quantitative PCR; RIP, RNA immunoprecipitation; si,
short interfering; si-NC, negative control siRNA; si-CDC5L,
CDC5L-specific siRNA; oe-NC, negative control overexpression
vector; oe-CDC5L, CDC5L overexpression vector;
oe-ELAVL1, ELAVL1 overexpression vector. |
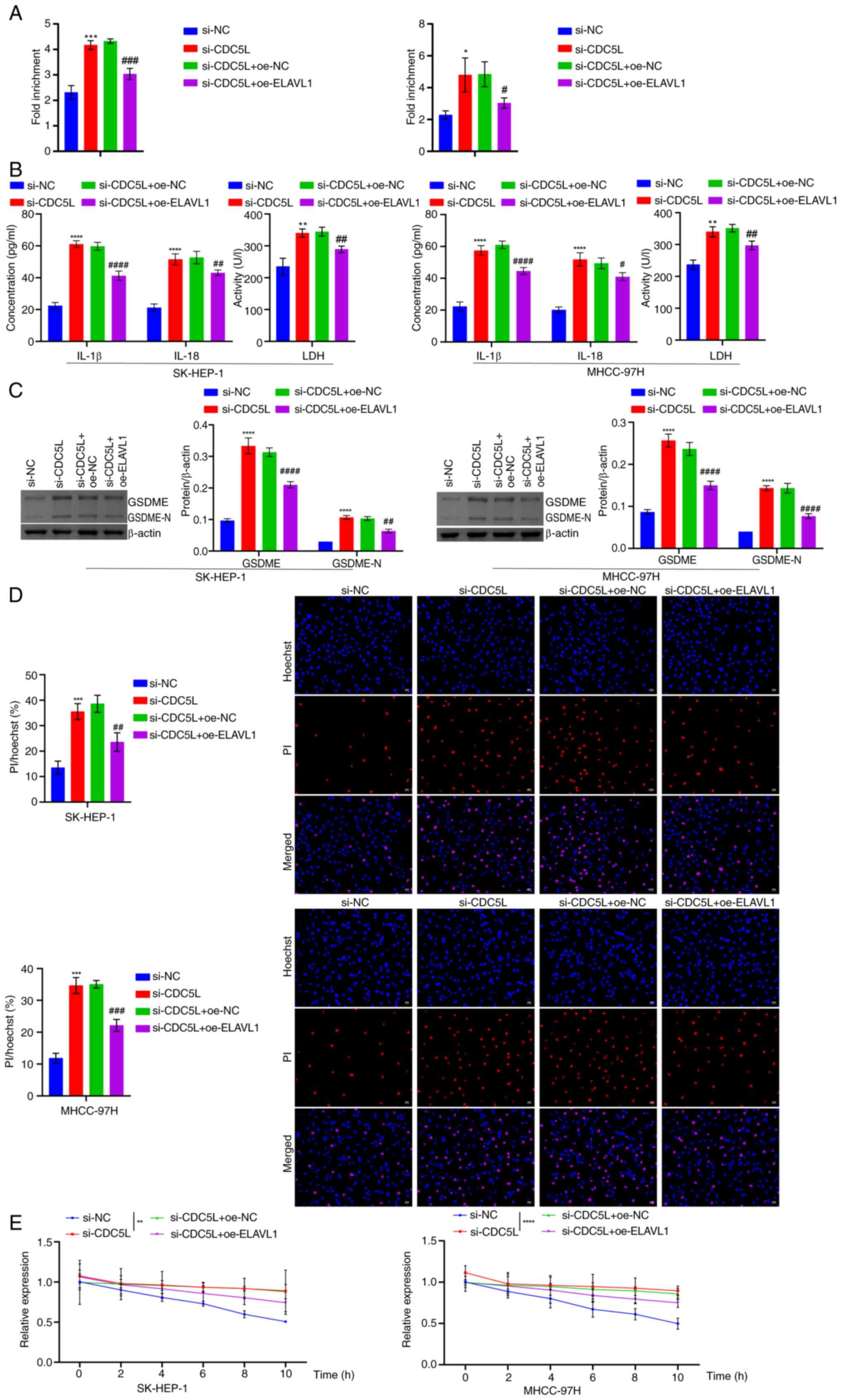 | Figure 12Detection of pyroptosis-related
levels following CDC5L knockdown and overexpression of ELAVL1. (A)
RIP detection of the binding stability of ELAVL1 protein to Caspase
3 mRNA. (B) LDH, IL-1β and IL-18 levels in the cell supernatant.
(C) Western blot analysis of GSDME and GSDME-N levels. (D) Hoechst
33342 and PI staining of pyroptosis. Scale bar, 50 μm,
magnification, ×200. (E) Following treatment with 5 μg/ml
Act D for 0, 2, 4, 6, 8 and 10 h, the expression of Caspase
3 mRNA was measured using RT-qPCR and the half-life of
Caspase 3 mRNA was assessed. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001 vs. si-NC; #P<0.05,
##P<0.01, ###P<0.001 and
####P<0.0001 vs. si-CDC5L+oe-NC. CDC5L, cell
division cycle 5 like; ELAVL1, ELAV like RNA binding protein 1;
RIP, RNA immunoprecipitation; LDH, lactate dehydrogenase; GSDME,
Caspase 3/Caspase 3-gasdermin E; GSDME-N, Gasdermin E N-terminal
fragment; PI, propidium iodide; RT-qPCR, reverse
transcription-quantitative PCR; si, short interfering; si-NC,
negative control siRNA; si-CDC5L, CDC5L-specific
siRNA; oe-NC, negative control overexpression vector;
oe-CDC5L, CDC5L overexpression vector;
oe-ELAVL1, ELAVL1 overexpression vector. |
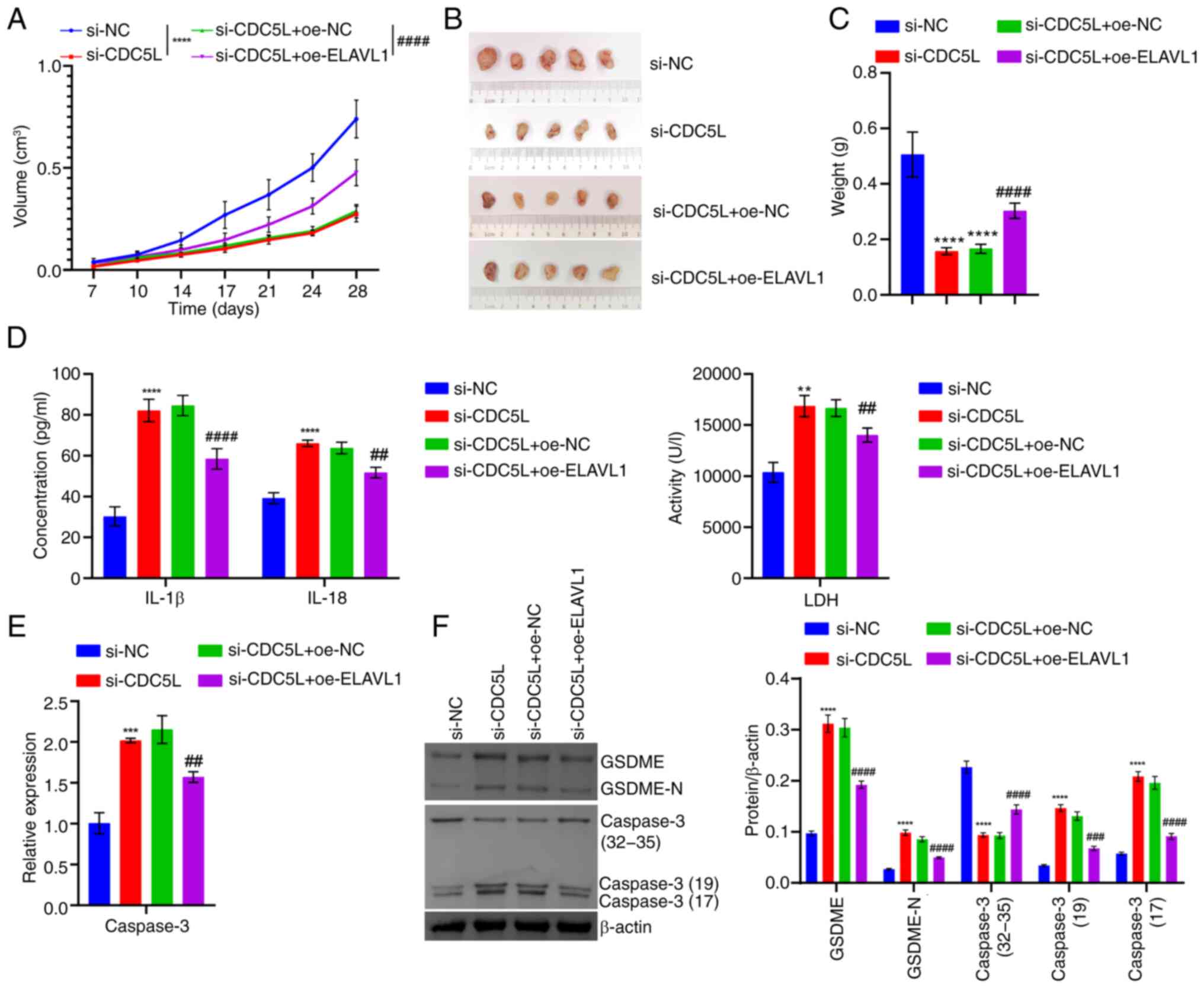 | Figure 13Validation of the mechanism of CDC5L
knockdown and overexpression of ELAVL1 in pyroptosis in HCC at the
animal level. (A) Tumor volume. (B) Tumor images. (C) Tumor weight.
(D) IL-18, IL-1β and LDH levels in tissue supernatants. (E) RT-qPCR
determination of Caspase 3 levels in tumor tissue. (F)
Western blot analysis of Caspase 3, GSDME and GSDME-N levels in the
tumor tissue. **P<0.01, ***P<0.001 and
****P<0.0001 vs. si-NC; ##P<0.01,
###P<0.001 and ####P<0.0001 vs.
si-CDC5L+oe-NC. CDC5L, cell division cycle 5 like; ELAVL1,
ELAV like RNA binding protein 1; LDH, lactate dehydrogenase;
RT-qPCR, reverse transcription-quantitative PCR; GSDME, Caspase
3/Caspase 3-gasdermin E; GSDME-N, Gasdermin E N-terminal fragment;
si, short interfering; si-NC, negative control siRNA;
si-CDC5L, CDC5L-specific siRNA; oe-NC, negative
control overexpression vector; oe-CDC5L, CDC5L
overexpression vector; oe-ELAVL1, ELAVL1
overexpression vector. |
CDC5L/ELAVL1 silencing regulates Caspase
3/GSDME to promote pyroptosis and inhibit tumor progression in
HCC
Finally, the effect of CDC5L inhibitor CVT-313 and
GSDME interference on HCC pyroptosis and tumor progression
was investigated the in vitro and in vivo. RT-qPCR
was performed on cells transfected with si-GSDME alone and
the results showed that compared with the si-NC group, the
expression of GSDME mRNA was markedly reduced in both the
si-GSDME#1 and si-GSDME#2 groups. Among them, the
si-GSDME#1 group exhibited the most pronounced reduction in
GSDME mRNA expression and was used for subsequent
experiments. This indicated that si-GSDME transfection
successfully achieved knockdown of GSDME (Fig. 14A). At the cellular level,
compared with the Control group, GSDME, GSDME-N and Caspase 3
levels were elevated, while those of CDC5L and ELAVL1 were reduced
in the CVT-313 group. Following the additional knockdown of
GSDME, GSDME and GSDME-N levels decreased, whereas Caspase
3, CDC5L and ELAVL1 levels remained largely unchanged (Fig. 14B-C). Cellular function
experiments revealed that, as compared with the Control group, cell
viability decreased and proliferation, migration and invasion
capabilities were reduced in the CVT-313 group. Upon further
knockdown of GSDME, cell viability increased and
proliferation, migration and invasion capabilities increased
(Figs. 14D-E and 15A). In addition, as compared with the
Control group, LDH levels in the cell supernatant and pyroptosis
increased in the CVT-313 group. Upon further knockdown of GSDME,
LDH levels in the cell supernatant decreased and pyroptosis reduced
(Fig. 15B-C). At the animal
level, relative to the Control group, tumor volume decreased, tumor
size reduced and tumor weight was alleviated in the CVT-313 group.
Upon further knockdown of GSDME, tumor volume gradually increased
and tumor size and weight also increased (Fig. 16A-C). In addition, as compared
with the Control group, IL-1β, IL-18, LDH, Caspase 3, GSDME and
GSDME-N levels increased, while CDC5L and ELAVL1 levels decreased
in the CVT-313 group. upon further knockdown of GSDME,
IL-1β, IL-18, LDH, GSDME and GSDME-N levels decreased, with no
significant changes in CDC5L, ELAVL1 and Caspase 3 levels (Fig. 16D-E). These results indicated
that CDC5L/ELAVL1 silencing regulated Caspase 3/GSDME to promote
HCC pyroptosis and repress tumor progression.
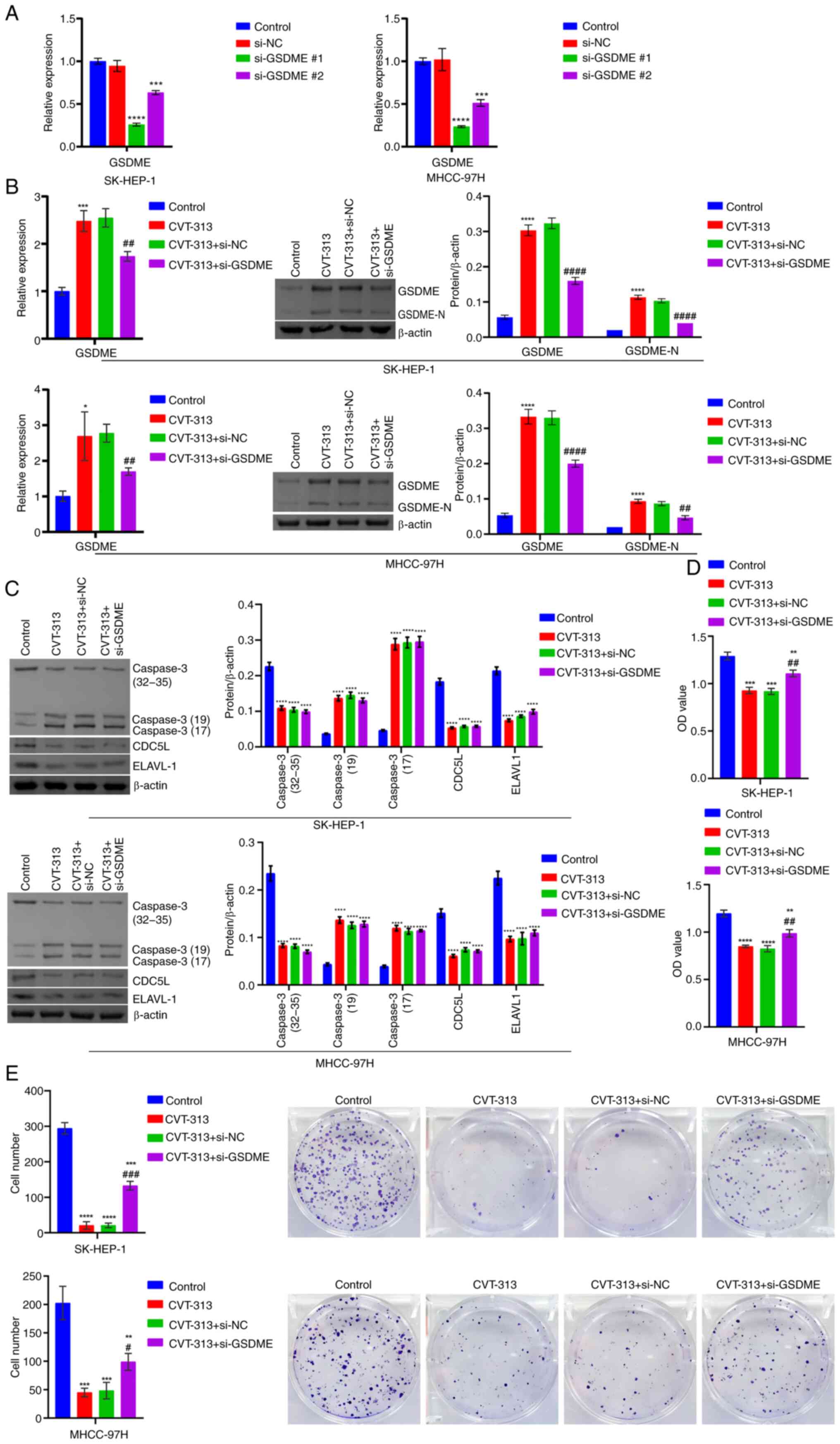 | Figure 14The silencing of CDC5L/ELAVL1
regulated Caspase 3/GSDME. (A) Detection of GSDME mRNA
expression after si-GSDME transfection by RT-qPCR. (B)
RT-qPCR and western blot analysis of GSDME levels. (C) Western blot
analysis of CDC5L, ELAVL1 and Caspase 3 levels. (D) Evaluation of
cell viability by CCK-8 assay. (E) Assessment of cell proliferation
through clone formation experiments. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001 vs. Control; #P<0.05,
##P<0.01, ###P<0.001 and
####P<0.0001 vs. CVT-313 + si-NC. CDC5L, cell
division cycle 5 like; ELAVL1, ELAV like RNA binding protein 1;
GSDME, Caspase 3/Caspase 3-gasdermin E; si, short interfering; HCC,
hepatocellular carcinoma; RT-qPCR, reverse
transcription-quantitative PCR; CCK-8, Cell Counting Kit-8; si,
short interfering; si-NC, Negative control siRNA; si-GSDME,
GSDME-specific siRNA. |
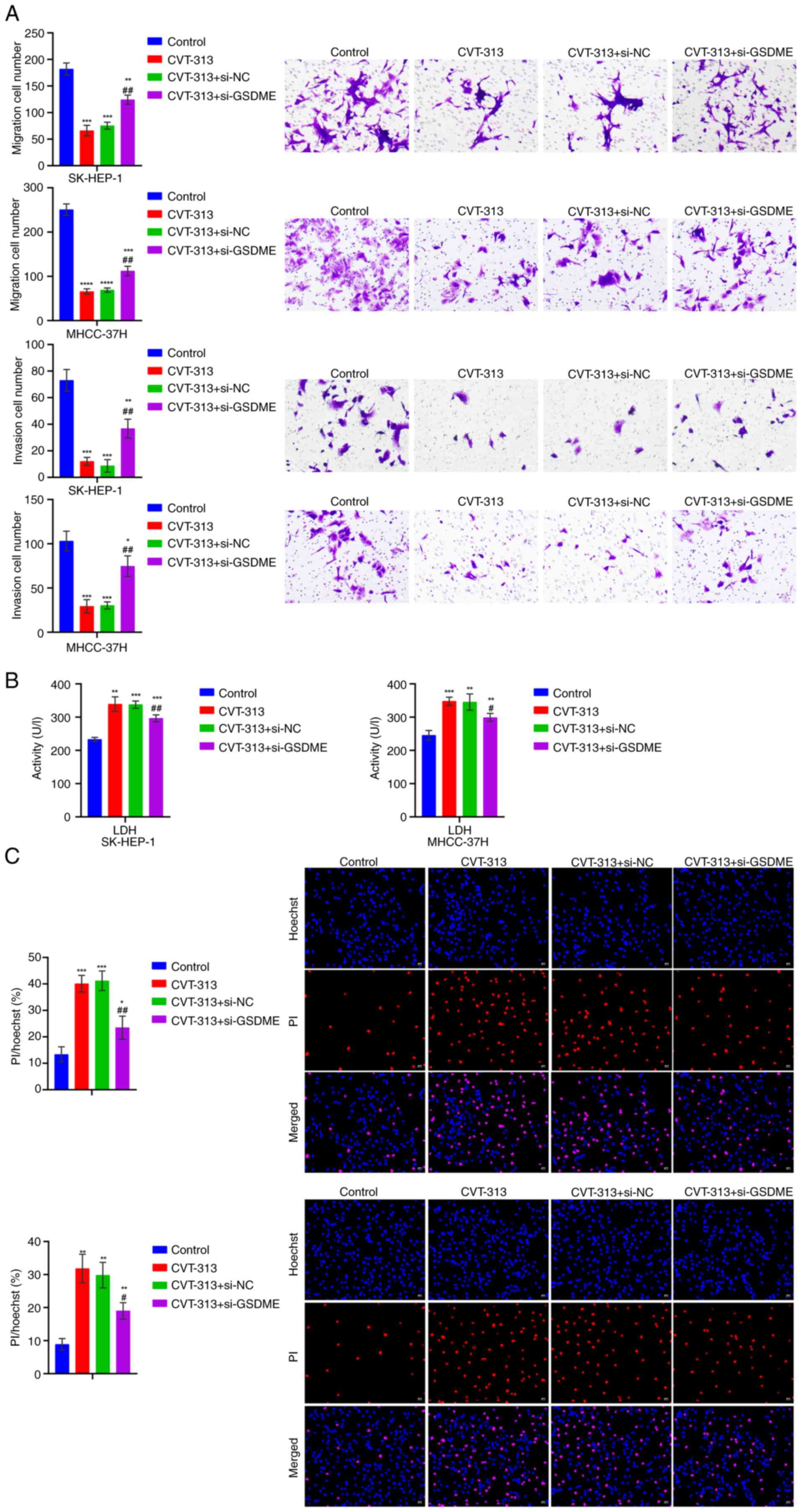 | Figure 15The silencing of CDC5L/ELAVL1
regulated Caspase 3/GSDME to promote pyroptosis in HCC. (A)
Examination of cell migration and invasion using Transwell assays.
Scale bar, 100 μm, magnification, ×100. (B) LDH level in
cell supernatant. (C) Hoechst 33342 and PI staining for the
detection of pyroptosis. Scale bar, 50 μm, magnification,
×200. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs.
Control; #P<0.05 and ##P<0.01 vs.
CVT-313 + si-NC. LDH, lactate dehydrogenase; PI, propidium iodide;
si, short interfering; si-NC, negative control siRNA;
si-GSDME, GSDME-specific siRNA. |
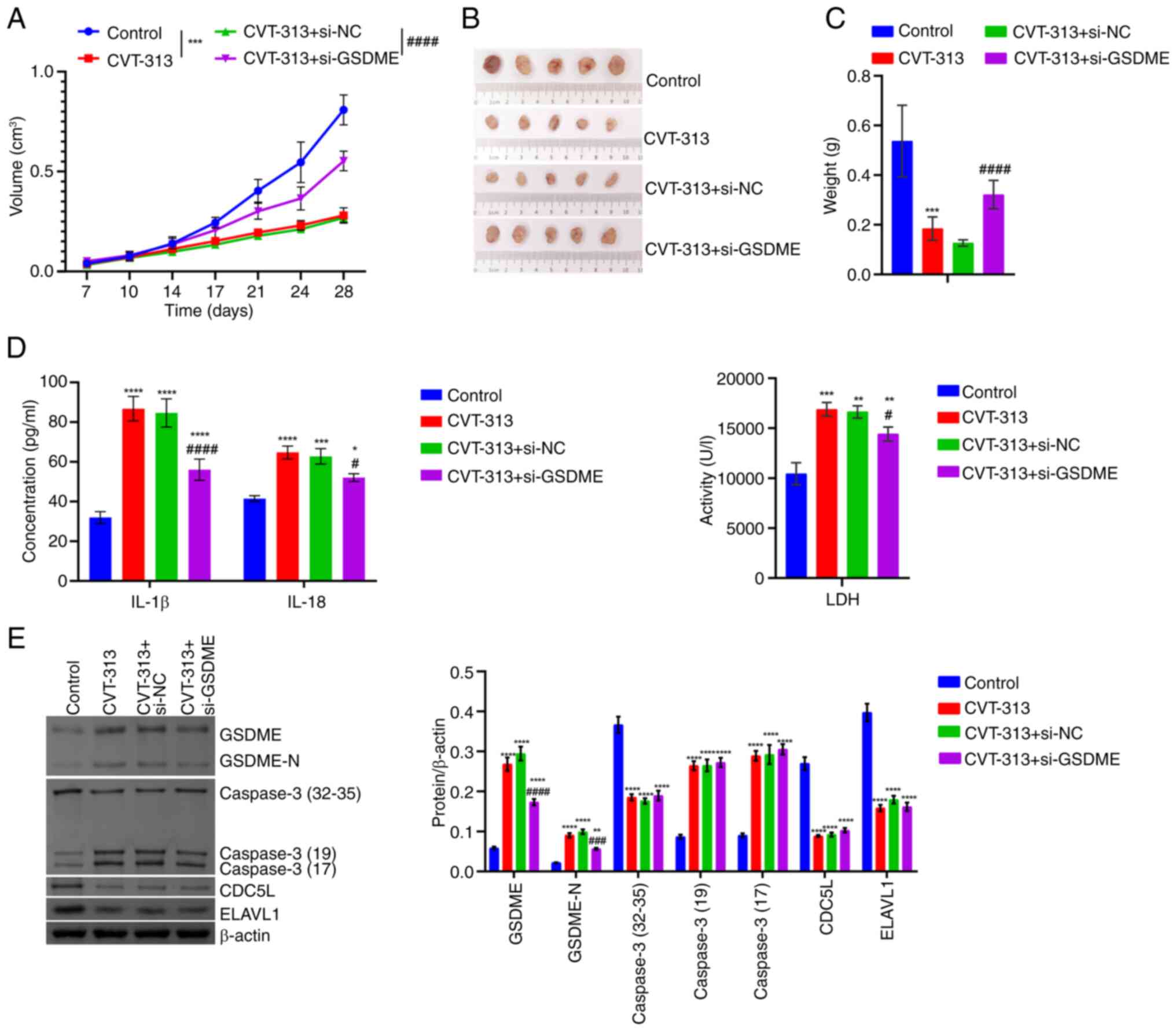 | Figure 16The silencing of CDC5L/ELAVL1
regulated Caspase 3/GSDME to inhibit tumor progression in HCC. (A)
Tumor volume. (B) Tumor images. (C) Tumor weight. (D) IL-1β, IL-18
and LDH levels in tissue supernatants. (E) Western blot analysis of
CDC5L, ELAVL1, Caspase 3 and GSDME levels in tumor tissue.
*P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs.
Control; #P<0.05, ###P<0.001 and
####P<0.0001 vs. CVT-313 + si-NC. CDC5L, cell
division cycle 5 like; ELAVL1, ELAV like RNA binding protein 1;
LDH, lactate dehydrogenase; GSDME, Caspase 3/Caspase 3-gasdermin E;
si, short interfering; si-NC, negative control siRNA;
si-GSDME, GSDME-specific siRNA. |
Discussion
HCC is highly heterogeneous disease with distinct
etiologies, which leads to different driver mutations that enhance
a unique tumor immune microenvironment (35). Despite substantial advancements
in cancer treatment, HCC, the most common form of liver cancer,
remains a significant global public health challenge (36). In the present study, the role of
CDC5L on HCC tumorigenesis and pyroptosis was explored both in
vivo and in vitro. It was found that CDC5L bound to
ELAVL1 to inhibit pyroptosis in HCC through the Caspase 3/GSDME
signaling pathway. To the best of the authors' knowledge, this is
the first study to report a CDC5L-induced regulation of ELAVL1 in
HCC pyroptosis. It was also the first to demonstrate that CDC5L
mediated the expression of Caspase 3 mRNA by binding to
ELAVL1.
HCC is highly malignant and prone to metastasis,
due to the heterogeneity and immunosuppressive nature of the tumor
microenvironment (37). HCC
development is a complex process involving the aberrant activation
or inactivation of multiple signaling pathways (38). CDC5L is a novel human telomerase
reverse transcriptase promoter binding protein (39). Mechanistic studies have shown
that silencing PRP19 suppresses CDC5L expression in HCC cells by
inhibiting CDC5L mRNA translation and promoting lysosome-mediated
CDC5L degradation (9). CDC5L
phosphorylation might contribute to HCC metastasis through the
regulation of mRNA processing and RNA splicing (11). Furthermore, the high expression
of methylated CDC5L-cg05671347 has been linked to improved overall
survival in patients with HCC (40). In the present study, high CDC5L
levels were found to be associated with poor HCC prognosis.
Therefore, the mechanism of CDC5L in HCC was further explored.
Evidence has indicated that tumor cells undergoing
regulated death might modulate immunogenicity in the tumor
microenvironment to a certain degree, potentially rendering it more
conducive to inhibiting cancer progression and metastasis (41). The induction of pyroptosis is
considered an anticancer therapy, due to its ability to unleash an
anticancer immune response (42). Upon detecting exogenous or
endogenous signals, cells initiate processes such as the assembly
of inflammatory vesicles, cleavage of GSDMs and release of
pro-inflammatory cytokines, ultimately resulting in inflammatory
cell death (43). Recent studies
have highlighted that pyroptosis is a crucial factor in HCC
development (44,45). In the present study, CDC5L
knockdown repressed cell viability, proliferation, migration and
invasion, at the cellular level but promoted pyroptosis. Also, at
the animal level, CDC5L knockdown led to a gradual decrease in
tumor volume and a reduction in tumor size and weight. CDC5L
overexpression exerted the opposite effect. The results of the
present study revealed that CDC5L acting in HCC could affect
pyroptosis.
Pyroptosis and apoptosis are both forms of
Caspase-mediated programmed cell death. Caspase 3 is an important
protein in pyroptosis and apoptosis, controlling tumor cell
toxicity when activated, while the expression of GSDME modulates
this process (34). The Caspase
3/GSDME signaling pathway functions as a regulatory 'switch' that
determines the balance between apoptosis and pyroptosis in cancer
cells. When GSDME expression is high, Caspase 3 cleaves GSDME to
induce pyroptosis; by contrast, when GSDME expression is low,
Caspase 3 triggers apoptosis (22). Research has shown that cisplatin
induced acute liver injury through triggering Caspase
3/GSDME-mediated cellular pyroptosis (46). Furthermore, Germacrone induced
GSDME-dependent pyroptosis by activating the Caspase 3/GSDME
pathway, at least in part by damaging mitochondria and enhancing
ROS production, thus supporting the possibility of Germacrone as a
candidate drug for the prevention and treatment of liver cancer
(47). These studies suggest
that the Caspase 3/GSDME pathway is highly significant in liver
cancer, not only participating in the processes of apoptosis and
pyroptosis but also potentially serving as a target for liver
cancer treatment. In the present study, it was further discovered
the regulation of HCC pyroptosis by CDC5L depends on the Caspase
family. Therefore, the mechanisms of CDC5L and Caspase 3 in HCC
pyroptosis will be further explored.
As an RNA-binding protein, the distribution and
function of ELAVL1 within cells may be regulated by multiple
signaling pathways (48). ELAVL1
functions as a crucial post-transcriptional regulator of specific
RNAs in both normal and disease states, including cancer (49). ELAVL1 stabilizes target mRNAs
involved in cellular de-differentiation, proliferation and survival
(50). ELAVL1 is a gatekeeper of
liver homeostasis and can prevent HCC (51). ELAVL1 has been reported to
regulate hepatitis B virus replication and growth in HCC cells
(52). In the nucleus, CCAT2
binds to ELAVL1 to promote HCC progression (53). In addition, cisplatin triggered
acute kidney injury and pyroptosis in mice through exosomal
miR-122/ELAVL1 regulatory pathway (54). However, the mechanism regarding
the role of ELAVL1 and CDC5L in HCC pyroptosis is unclear. Research
has shown that ELAVL1 regulates post-transcriptional gene
expression in cancer cells and is subject to cleavage by
stress-activated Caspase 3 (55). Qiu et al (56) confirmed that pre-mRNA processing
factor 19 upregulated CDC5L expression, which bound to MAPK1,
thereby promoting gastric cancer progression via the MAPK
pathway-mediated homologous recombination. Additionally, IGF2BP1
acted as a post-transcriptional enhancer of CDC5L in an
m6A-dependent manner to promote the proliferation of multiple
myeloma cells with chromosome 1q gain (57). Jokoji et al (58) revealed that CDC5L promoted early
chondrocyte differentiation and proliferation by modulating
pre-mRNA splicing of SOX9, COL2A1 and WEE1. The KEGG analysis in
the present study revealed that the functional pathways of highly
expressed CDC5L involved the MAPK signaling pathway, PI3K-Akt
signaling pathway, Wnt signaling pathway and others. In gastric
cancer, high expression of CDC5L promoted homologous recombination
by activating the MAPK signaling pathway, thereby driving the
progression of gastric cancer (56). Research has shown that CDC5L mRNA
was upregulated in the exosomes of osteosarcoma patients with poor
chemotherapeutic responses. Moreover, bioinformatics analysis has
found that miRNA patterns associated with poor chemotherapeutic
responses were enriched in the PI3K-Akt signaling pathway (59). Therefore, high expression of
CDC5L may affect the chemotherapeutic response in osteosarcoma by
influencing the activity of the PI3K/Akt signaling pathway. Lan
et al (60) reported that
TRIP13 was primarily enriched in the PI3K-Akt-mTOR signaling
pathway, suggesting that TRIP13 may promote the progression of
breast cancer by influencing the activity of the PI3K/Akt signaling
pathway. Since CDC5L is one of the hub genes highly associated with
TRIP13, it can be inferred that CDC5L may indirectly influence the
activity of the PI3K/Akt signaling pathway through its interaction
with TRIP13. A study on lung adenocarcinoma demonstrated that CDC5L
promoted the progression of lung adenocarcinoma by
transcriptionally regulating WNT7B to activate the Wnt/β-catenin
signaling pathway (61).
Additionally, the ELAVL1/PI3K/NF-κB pathway regulated by
lactoferrin was verified to participate in the protection of
infantile intestinal immune barrier damage in the study by Li et
al (62). These studies
indicated that CDC5L and ELAVL1 play important roles in various
cancers and related pathological processes through interactions
with multiple key signaling pathways. CDC5L promotes cancer
progression by activating the MAPK, PI3K-Akt and Wnt signaling
pathways, while ELAVL1, as an RNA-binding protein, regulates the
stability of specific mRNAs, affecting cell dedifferentiation,
proliferation and survival. Although their roles in different
cancers have been revealed in existing studies, the specific
mechanisms of action of CDC5L and ELAVL1 in HCC pyroptosis still
need further exploration. In the present study, at the cellular
level, CDC5L was found to competitively bind to ELAVL1 to inhibit
the binding of ELAVL1 with Caspase 3 mRNA, thereby regulating HCC
pyroptosis. This discovery is significant for understanding the
role of pyroptosis in tumor suppression. Finally, it was confirmed
through cellular and animal experiments that silencing CDC5L/ELAVL1
regulated Caspase 3/GSDME to promote HCC pyroptosis and inhibit
tumor progression. Therefore, targeting the CDC5L/ELAVL1 axis could
pave the way for new therapeutic methods that increase the
chemosensitivity of live cancer cells, thereby inhibiting tumor
progression. Notably, the present study is the first to reveal the
mechanism by which CDC5L interacts with ELAVL1 and regulates
Caspase 3/GSDME signaling pathway in the pyroptosis of HCC. This
finding not only provides a new potential therapeutic target for
HCC, but also offers a new perspective for understanding the tumor
microenvironment and immune regulation in HCC.
While the present study has made significant
progress in exploring the mechanisms of CDC5L and ELAVL1 in HCC,
there are still some limitations that need to be further improved
in future research. First, the relatively small cohort size of the
present study, which was limited by available resources and initial
research design, may affect the universality and reliability of the
results despite the significant statistical findings. Future
research should expand the sample size to further validate the
findings and reduce potential bias. Second, potential selection
bias in TCGA analysis is also a concern. Although strict screening
criteria were applied and data were validated multidimensionally,
public database analyses are generally subject to factors such as
sample sources and data quality and potential bias cannot be
completely ruled out. Therefore, future research needs to combine
more independent datasets for validation to improve the reliability
and universality of the results. In addition, the present study
relied on only two HCC cell lines, which may limit the generality
of the results. Although these two cell lines are widely used and
representative in HCC research, significant biological differences
may exist between different cell lines. To more comprehensively
evaluate the mechanisms of CDC5L and ELAVL1, future research plans
to introduce more HCC cell lines and explore the differences and
commonalities between them. Finally, the lack of in situ or
immunologically active in vivo models in the present study
somewhat limits our comprehensive understanding of the HCC
pathological process. In vitro experiments and animal models
have limitations in simulating human diseases. For an improved
simulation of the HCC pathological process, future research plans
to develop in situ HCC models and explore immunologically
active models to more comprehensively assess the mechanisms of
CDC5L and ELAVL1.
In conclusion, in the present study, a preliminary
investigation into the mechanism of CDC5L in HCC was conducted and
its potential mode was found to be associated with its interaction
with ELAVL1 and pyroptosis. These findings suggested a new way of
considering the treatment of HCC. Furthermore, gaining a deeper
understanding of the involvement of pyroptosis in HCC may provide a
basis for exploring and developing new therapeutic approaches to
combat HCC.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SL and KY conceived the design, ZZ, SL, SZ, YT, MX
and XG carried out the experiments, KY carried out data
visualization, SL and KY confirm the authenticity of all the raw
data, SL wrote the first draft and ZZ, SZ, YT, MX, XG and KY
directed the revision. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The clinical study was approved by Medical Ethics
Committee of Xiangya Hospital, Central South University (Hunan,
China; approval no. 2024052119) and all patients afforded informed
consent. The animal experiment was approved by the Animal Ethics
Committee of Xiangya Medical School, Central South University
(approval no. 2024051707) and all animal experiments were conducted
in accordance with the guidelines of the Ethics Committee.
Patient consent for publication
All patients gave informed consent.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
the present study was funded by the Enterprise Joint Fund of
Natural Science Foundation of Hunan Province (grant no.
2025JJ90293) and the Natural Science Foundation of Hunan Province
(grant no. 2023JJ30910).
References
|
1
|
Wang W, Zhou Y, Li W, Quan C and Li Y:
Claudins and hepatocellular carcinoma. Biomed Pharmacother.
171:1161092024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mo Y, Zou Z and Chen E: Targeting
ferroptosis in hepatocellular carcinoma. Hepatol Int. 18:32–49.
2024. View Article : Google Scholar
|
|
3
|
Sankar K, Gong J, Osipov A, Miles SA,
Kosari K, Nissen NN, Hendifar AE, Koltsova EK and Yang JD: Recent
advances in the management of hepatocellular carcinoma. Clin Mol
Hepatol. 30:1–15. 2024. View Article : Google Scholar :
|
|
4
|
Butaye E, Somers N, Grossar L, Pauwels N,
Lefere S, Devisscher L, Raevens S, Geerts A, Meuris L, Callewaert
N, et al: Systematic review: Glycomics as diagnostic markers for
hepatocellular carcinoma. Aliment Pharmacol Ther. 59:23–38. 2024.
View Article : Google Scholar
|
|
5
|
Llovet JM, Pinyol R, Yarchoan M, Singal
AG, Marron TU, Schwartz M, Pikarsky E, Kudo M and Finn RS: Adjuvant
and neoadjuvant immunotherapies in hepatocellular carcinoma. Nat
Rev Clin Oncol. 21:294–311. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen X, Kou L, Xie X, Su S, Li J and Li Y:
Prognostic biomarkers associated with immune checkpoint inhibitors
in hepatocellular carcinoma. Immunology. 172:21–45. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang N, Kaur R, Akhter S and Legerski RJ:
Cdc5L interacts with ATR and is required for the S-phase cell-cycle
checkpoint. EMBO Rep. 10:1029–1035. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Z, Mao W, Wang L, Liu M, Zhang W, Wu
Y, Zhang J, Mao S, Geng J and Yao X: Depletion of CDC5L inhibits
bladder cancer tumorigenesis. J Cancer. 11:353–363. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang R, Xue R, Qu D, Yin J and Shen XZ:
Prp19 arrests cell cycle via Cdc5L in hepatocellular carcinoma
cells. Int J Mol Sci. 18:7782017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qiu H, Zhang X, Ni W, Shi W, Fan H, Xu J,
Chen Y, Ni R and Tao T: Expression and clinical role of Cdc5L as a
novel cell cycle protein in hepatocellular carcinoma. Dig Dis Sci.
61:795–805. 2016. View Article : Google Scholar
|
|
11
|
Tian M, Cheng H, Wang Z, Su N, Liu Z, Sun
C, Zhen B, Hong X, Xue Y and Xu P: Phosphoproteomic analysis of the
highly-metastatic hepatocellular carcinoma cell line, MHCC97-H. Int
J Mol Sci. 16:4209–4225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma Q, Lu Q, Lei X, Zhao J, Sun W, Huang D,
Zhu Q and Xu Q: Relationship between HuR and tumor drug resistance.
Clin Transl Oncol. 25:1999–2014. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu R, Wu K, Li Y, Sun R and Li X: Human
antigen R: A potential therapeutic target for liver diseases.
Pharmacol Res. 155:1046842020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Papatheofani V, Levidou G, Sarantis P,
Koustas E, Karamouzis MV, Pergaris A, Kouraklis G and Theocharis S:
HuR protein in hepatocellular carcinoma: Implications in
development, prognosis and treatment. Biomedicines. 9:1192021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Imre G: Pyroptosis in health and disease.
Am J Physiol Cell Physiol. 326:C784–C794. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chao L, Zhang W, Feng Y, Gao P and Ma J:
Pyroptosis: A new insight into intestinal inflammation and cancer.
Front Immunol. 15:13649112024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wen J, Xuan B, Liu Y, Wang L, He L, Meng
X, Zhou T and Wang Y: NLRP3 inflammasome-induced pyroptosis in
digestive system tumors. Front Immunol. 14:10746062023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wallace HL and Russell RS: Inflammatory
consequences: Hepatitis C virus-induced inflammasome activation and
pyroptosis. Viral Immunol. 37:126–138. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zou Z, Zhao M, Yang Y, Xie Y, Li Z, Zhou
L, Shang R and Zhou P: The role of pyroptosis in hepatocellular
carcinoma. Cell Oncol (Dordr). 46:811–823. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao H, Liu H, Yang Y and Wang H: The role
of autophagy and pyroptosis in liver disorders. Int J Mol Sci.
23:62082022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng C, Hsu SK, Chen YC, Liu W, Shu ED,
Chien CM, Chiu CC and Chang WT: Burning down the house: Pyroptosis
in the tumor microenvironment of hepatocellular carcinoma. Life
Sci. 347:1226272024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang M, Qi L, Li L and Li Y: The
caspase-3/GSDME signal pathway as a switch between apoptosis and
pyroptosis in cancer. Cell Death Discov. 6:1122020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lorente L, Rodriguez ST, Sanz P,
González-Rivero AF, Pérez-Cejas A, Padilla J, Díaz D, González A,
Martín MM, Jiménez A, et al: High serum caspase-3 levels in
hepatocellular carcinoma prior to liver transplantation and high
mortality risk during the first year after liver transplantation.
Expert Rev Mol Diagn. 19:635–640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shang N, Bank T, Ding X, Breslin P, Li J,
Shi B and Qiu W: Caspase-3 suppresses diethylnitrosamine-induced
hepatocyte death, compensatory proliferation and
hepatocarcinogenesis through inhibiting p38 activation. Cell Death
Dis. 9:5582018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song A, Ding T, Wei N, Yang J, Ma M, Zheng
S and Jin H: Schisandrin B induces HepG2 cells pyroptosis by
activating NK cells mediated anti-tumor immunity. Toxicol Appl
Pharmacol. 472:1165742023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang X, Zhang P, An L, Sun N, Peng L,
Tang W, Ma D and Chen J: Miltirone induces cell death in
hepatocellular carcinoma cell through GSDME-dependent pyroptosis.
Acta Pharm Sin B. 10:1397–1413. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gräub R, Lancero H, Pedersen A, Chu M,
Padmanabhan K, Xu XQ, Spitz P, Chalkley R, Burlingame AL, Stokoe D
and Bernstein HS: Cell cycle-dependent phosphorylation of human
CDC5 regulates RNA processing. Cell Cycle. 7:1795–1803. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du F, Zhao X and Fan D: Tumorigenicity
assay in nude mice. Bio Protoc. 7:e23642017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Somarelli JA, Roghani RS, Moghaddam AS,
Thomas BC, Rupprecht G, Ware KE, Altunel E, Mantyh JB, Kim SY,
McCall SJ, et al: A precision medicine drug discovery pipeline
identifies combined CDK2 and 9 inhibition as a novel therapeutic
strategy in colorectal cancer. Mol Cancer Ther. 19:2516–2527. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ashall VD, Morton D and Clutton E: A
declaration of helsinki for animals. Vet Anaesth Analg. 50:309–314.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He GN, Bao NR, Wang S, Xi M, Zhang TH and
Chen FS: Ketamine induces ferroptosis of liver cancer cells by
targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des Devel Ther.
15:3965–3978. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng
B, Chen C, Chang W, Ping Y, Ji P, et al: Exosomal circPACRGL
promotes progression of colorectal cancer via the
miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 19:1172020.
View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Bhat AA, Thapa R, Afzal O, Agrawal N,
Almalki WH, Kazmi I, Alzarea SI, Altamimi ASA, Prasher P, Singh SK,
et al: The pyroptotic role of Caspase-3/GSDME signalling pathway
among various cancer: A review. Int J Biol Macromol.
242:1248322023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lehrich BM and Delgado ER: Lipid
nanovesicle platforms for hepatocellular carcinoma precision
medicine therapeutics: Progress and perspectives. Organogenesis.
20:23136962024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Akbulut Z, Aru B, Aydın F and Demirel GY:
Immune checkpoint inhibitors in the treatment of hepatocellular
carcinoma. Front Immunol. 15:13796222024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu Y, Chen D, Hong M, Liu J, Li Y, Hao J,
Lu L, Yin Z and Wu Y: Apoptosis, pyroptosis, and ferroptosis
conspiringly induce immunosuppressive hepatocellular carcinoma
microenvironment and γδ T-Cell imbalance. Front Immunol.
13:8459742022. View Article : Google Scholar
|
|
38
|
Xin X, Cheng X, Zeng F, Xu Q and Hou L:
The role of TGF-β/SMAD signaling in hepatocellular carcinoma: From
mechanism to therapy and prognosis. Int J Biol Sci. 20:1436–1451.
2024. View Article : Google Scholar :
|
|
39
|
Li J, Zhang N, Zhang R, Sun L, Yu W, Guo
W, Gao Y, Li M, Liu W, Liang P, et al: CDC5L promotes hTERT
expression and colorectal tumor growth. Cell Physiol Biochem.
41:2475–2488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhen L, Ning G, Wu L, Zheng Y, Yang F,
Chen T, Xu W, Liu Y, Xie C and Peng L: Prognostic value of
aberrantly expressed methylation genes in human hepatocellular
carcinoma. Biosci Rep. 40:BSR201925932020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tong X, Tang R, Xiao M, Xu J, Wang W,
Zhang B, Liu J, Yu X and Shi S: Targeting cell death pathways for
cancer therapy: Recent developments in necroptosis, pyroptosis,
ferroptosis, and cuproptosis research. J Hematol Oncol. 15:1742022.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Khan M, Ai M, Du K, Song J, Wang B, Lin J,
Ren A, Chen C, Huang Z, Qiu W, et al: Pyroptosis relates to tumor
microenvironment remodeling and prognosis: A pan-cancer
perspective. Front Immunol. 13:10622252022. View Article : Google Scholar
|
|
43
|
Yang F, Bettadapura SN, Smeltzer MS, Zhu H
and Wang S: Pyroptosis and pyroptosis-inducing cancer drugs. Acta
Pharmacol Sin. 43:2462–2473. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng Y, Yang S, Dai W, Wang J, Bi S,
Zhang X, Zheng Z, Sun Y, Wu S and Kong J: CHMP3 promotes the
progression of hepatocellular carcinoma by inhibiting
caspase-1-dependent pyroptosis. Int J Oncol. 64:82024. View Article : Google Scholar
|
|
45
|
Xiao C, Gong J, Jie Y, Liang W, Tai Y, Qin
W, Lu T, Chong Y, Hei Z, Hu B and Zhang Q: E2F1-mediated
up-regulation of NCAPG promotes hepatocellular carcinoma
development by inhibiting pyroptosis. J Clin Transl Hepatol.
12:25–35. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu PP, Shen XJ and Zheng SS: Cisplatin
induces acute liver injury by triggering caspase-3/GSDME-mediated
cell pyroptosis. Hepatobiliary Pancreatic Dis Int. 24:177–187.
2024. View Article : Google Scholar
|
|
47
|
Sun X, Zhong X, Ma W, Feng W, Huang Q, Ma
M, Lv M, Hu R, Han Z, Li J and Zhou X: Germacrone induces
caspase-3/GSDME activation and enhances ROS production, causing
HepG2 pyroptosis. Exp Ther Med. 24:4562022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rothamel K, Arcos S, Kim B, Reasoner C,
Lisy S, Mukherjee N and Ascano M: ELAVL1 primarily couples mRNA
stability with the 3' UTRs of interferon-stimulated genes. Cell
Rep. 35:1091782021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lachiondo-Ortega S, Delgado TC,
Baños-Jaime B, Velázquez-Cruz A, Díaz-Moreno I and Martínez-Chantar
ML: Hu Antigen R (HuR) protein structure, function and regulation
in hepatobiliary tumors. Cancers (Basel). 14:26662022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fernández-Ramos D and Martínez-Chantar ML:
NEDDylation in liver cancer: The regulation of the RNA binding
protein Hu antigen R. Pancreatology. 15(4 Suppl): S49–S54. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Subramanian P, Gargani S, Palladini A,
Chatzimike M, Grzybek M, Peitzsch M, Papanastasiou AD, Pyrina I,
Ntafis V, Gercken B, et al: The RNA binding protein human antigen R
is a gatekeeper of liver homeostasis. Hepatology. 75:881–897. 2022.
View Article : Google Scholar
|
|
52
|
Kanzaki H, Chiba T, Kaneko T, Ao J, Kan M,
Muroyama R, Nakamoto S, Kanda T, Maruyama H, Kato J, et al: The
RNA-Binding protein ELAVL1 regulates hepatitis B Virus replication
and growth of hepatocellular carcinoma cells. Int J Mol Sci.
23:78782022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shi J, Guo C and Ma J: CCAT2 enhances
autophagy-related invasion and metastasis via regulating miR-4496
and ELAVL1 in hepatocellular carcinoma. J Cell Mol Med.
25:8985–8996. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu B, He J, Ye X, Pei X, Bai Y, Gao F,
Guo L, Yong H and Zhao W: Role of cisplatin in inducing acute
kidney injury and pyroptosis in mice via the exosome miR-122/ELAVL1
regulatory axis. Physiol Res. 72:753–765. 2023. View Article : Google Scholar
|
|
55
|
Janakiraman H, House RP, Talwar S,
Courtney SM, Hazard ES, Hardiman G, Mehrotra S, Howe PH, Gangaraju
V and Palanisamy V: Repression of caspase-3 and RNA-binding protein
HuR cleavage by cyclooxygenase-2 promotes drug resistance in oral
squamous cell carcinoma. Oncogene. 36:3137–3148. 2017. View Article : Google Scholar :
|
|
56
|
Qiu S, Wang F, Gao X, Guan W, Dai T, Yin
L, Wang F, Sun J, Guo P, Wu H, et al: Prp19/CDC5L promotes gastric
cancer via activation of the MAPK pathway-mediated homologous
recombination. Int J Biol Sci. 21:1603–1618. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xu J, Wang Y, Ren L, Li P and Liu P:
IGF2BP1 promotes multiple myeloma with chromosome 1q gain via
increasing CDC5L expression in an m6A-dependent manner.
Genes Dis. 12:1012142025. View Article : Google Scholar
|
|
58
|
Jokoji G, Maeda S, Oishi K, Ijuin T,
Nakajima M, Tawaratsumida H, Kawamura I, Tominaga H, Taketomi E,
Ikegawa S and Taniguchi N: CDC5L promotes early chondrocyte
differentiation and proliferation by modulating pre-mRNA splicing
of SOX9, COL2A1, and WEE1. J Biol Chem. 297:1009942021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xu JF, Wang YP, Zhang SJ, Chen Y, Gu HF,
Dou XF, Xia B, Bi Q and Fan SW: Exosomes containing differential
expression of microRNA and mRNA in osteosarcoma that can predict
response to chemotherapy. Oncotarget. 8:75968–75978. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lan J, Huang J, Tao X, Gao Y, Zhang L,
Huang W, Luo J, Liu C, Deng Y, Liu L and Liu X: Evaluation of the
TRIP13 level in breast cancer and insights into potential molecular
pathways. J Cell Mol Med. 26:2673–2685. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yu N, Wu Y, Wei Q, Li X, Li M and Wu W:
m6A modification of CDC5L promotes lung adenocarcinoma
progression through transcriptionally regulating WNT7B expression.
Am J Cancer Res. 14:3565–3583. 2024. View Article : Google Scholar :
|
|
62
|
Li C, Liu X, Huang Z, Zhai Y, Li H and Wu
J: Lactoferrin alleviates lipopolysaccharide-induced infantile
intestinal immune barrier damage by regulating an ELAVL1-related
signaling pathway. Int J Mol Sci. 23:137192022. View Article : Google Scholar : PubMed/NCBI
|