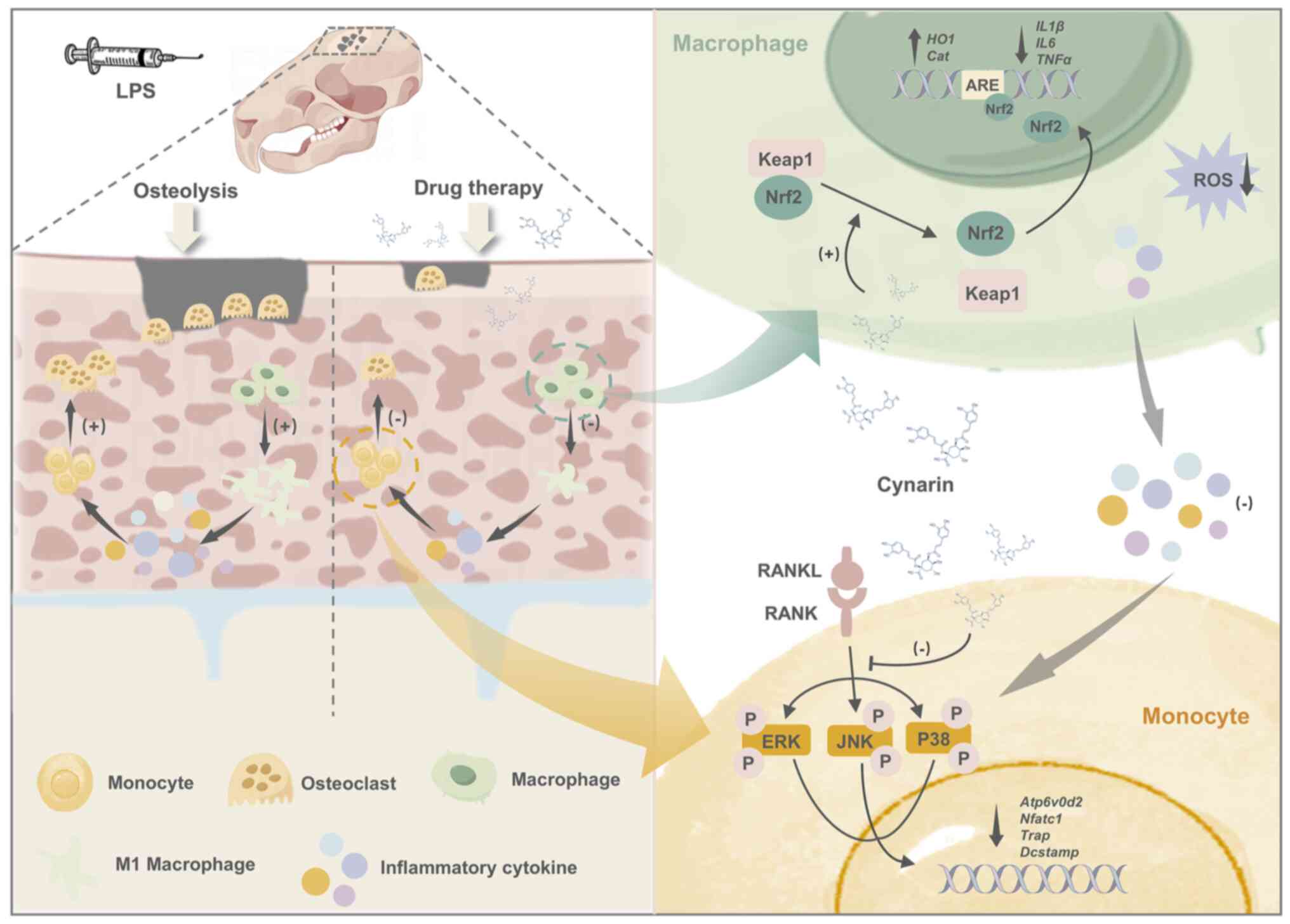
Introduction
Bone metabolism is essential for maintaining the
integrity and function of the skeleton and ensuring a balanced
process of bone formation and resorption. Under inflammatory
conditions, bone metabolism is skewed with OC-mediated bone
resorption outpacing bone formation. This imbalance accelerates
bone loss and weakens the structural integrity of the skeleton
(1). Therefore, the development
of novel therapeutic strategies that simultaneously inhibit
osteoclastogenesis and regulate inflammatory responses is crucial
for alleviating inflammatory bone resorption and improving skeletal
health.
OCs are essential effector cells in bone metabolism
that are primarily responsible for local bone resorption and play a
crucial role in inflammatory bone degradation. Multiple signalling
pathways regulate OC differentiation and activity. Receptor
activator of nuclear factor κB ligand (RANKL) activates the
mitogen-activated protein kinase (MAPK) signalling pathway, which
subsequently stimulates the expression of NFATc1, driving OC
differentiation (2,3). The nuclear factor kappa-B (NF-κB)
pathway (4,5) and the phosphoinositide 3-kinase
(PI3K) pathway (6) also play
significant roles in osteoclastogenesis and the maintenance of OC
function. Inflammation is a crucial driver of bone degradation
(7). Chronic inflammation
induces oxidative stress, leading to increased production of
reactive oxygen species (ROS), activating OCs and exacerbating bone
resorption (8,9).
Current antiresorptive therapies, including
bisphosphonates (10,11), effectively target OC formation
but lack anti-inflammatory and antioxidant effects and have limited
clinical utility (12,13). Therefore, an urgent need exists
to develop novel agents with anti-inflammatory properties that
inhibit abnormal OC production. Cynarin is a phenolic compound
isolated from artichokes (Cynara genus) with potent
antioxidant activity (14,15). In a mouse model of gouty
arthritis, cynarin has been shown to inhibit the NF-κB and JNK
pathways in macrophages, exerting anti-inflammatory and
anti-swelling effects (16). In
a mouse colitis model, it downregulated STAT3 and NF-κB (p65),
thereby suppressing M1 macrophage polarisation (17). However, the mechanism of action
of cynarin in inflammatory bone resorption remains unclear, and
further research is required to address these questions.
The present study has evaluated the potential of
cynarin as a dual-functional agent with anti-osteolytic and
anti-inflammatory properties. Specifically, its effects were
examined on RANKL-induced osteoclastogenesis, as well as on the
MAPK signalling pathway, ROS production and Nrf2 activation in
vitro. In addition, its efficacy in preventing inflammatory
bone loss was evaluated in vivo. Notably, cynarin was more
effective than alendronate in mitigating calvarial bone resorption.
These findings highlight the ability of cynarin to inhibit OC
formation and attenuate inflammation via a dual mechanism,
supporting its potential as a novel therapeutic agent for treating
inflammatory osteolysis.
Materials and methods
Reagents and antibodies
Cynarin (Selleck Chemicals) was used at a purity of
99.97% (high-performance liquid chromatography peak purity test and
13C NMR test data are available upon request from the corresponding
author). Macrophage colony-stimulating factor (M-CSF) and RANKL
were obtained from R&D Systems, Inc. Anisomycin and alendronate
were purchased from MedChemExpress, and lipopolysaccharide (LPS)
was sourced from MilliporeSigma. The tartrate-resistant acid
phosphatase (TRAP) staining kit was acquired from Solarbio Science
& Technology Co., Ltd. TRIzol reagent, PrimeScript™ RT reagent
kit and TB Green® Premix Ex Taq™ II Fast qPCR kit were
provided by Takara Biotechnology Co., Ltd. The bicinchoninic acid
(BCA) protein assay kit, commercial nuclear and cytoplasmic protein
extraction kit and phenyl-methyl-sulfonyl fluoride (PMSF) were
obtained from Beyotime Institute of Biotechnology. Primary
antibodies against histone H3 (cat. no. 9715), GAPDH (cat. no.
2118), phospho-(p)-JNK (cat. no. 4668), JNK (cat. no. 9252), p-ERK
(cat. no. 4370), ERK (cat. no. 4695), p-p38 (cat. no. 4511), p38
(cat. no. 9212) and RANK (cat. no. 4845), as well as secondary
antibodies (cat. nos. 7074 and 7076), were purchased from Cell
Signalling Technology, Inc. Primary antibodies specific to heme
oxygenase-1 (HO-1; cat. no. A1346) and RUNX2 (cat. no. A2851) were
acquired from ABclonal Biotech Co., Ltd., while the DC-STAMP (cat.
no. MABF39-I) antibody was obtained from MilliporeSigma. Antibodies
targeting cathepsin K (CTSK; cat. no. ab19027), collagen I (COL1;
cat. no. ab270993) and osteopontin (OPN; cat. no. ab218237) were
purchased from Abcam. Antibodies against Nrf2 (cat. no.
16396-1-AP), Keap1 (cat. no. 10503-2-AP), osteocalcin (OCN; cat.
no. 16157-1-AP), tumor necrosis factor-alpha (TNFα; cat. no.
17590-1-AP) and inducible nitric oxide synthase (iNOS; cat. no.
22226-1-AP) were sourced from Proteintech Group, Inc.
Cell culture and treatment
RAW264.7 cells (cat. no. TIB-71; American Type
Culture Collection) were maintained in Dulbecco's Modified Eagle
Medium (DMEM) supplemented with 5% fetal bovine serum (FBS) and 100
U/ml penicillin-streptomycin (Thermo Fisher Scientific, Inc.). Bone
marrow-derived macrophages (BMMs) were isolated and cultured
according to the protocol established by Rucci et al
(18). Primary BMMs were
incubated in α-Minimum Essential Medium containing 10% FBS, 100
U/ml penicillin-streptomycin and 30 ng/ml M-CSF. Bone
marrow-derived mesenchymal stem cells (BMSCs) were isolated from
the femurs and tibias of 4-week-old mice, as previously described
(19). BMSCs were cultured in
the same medium as BMMs, excluding M-CSF supplementation. All cells
were incubated under standard conditions in a humidified atmosphere
at 37°C with 5% CO2 to ensure optimal proliferation and
viability.
Cell viability assay
To evaluate the cytotoxic effects of cynarin,
RAW264.7 cells and BMMs were seeded in triplicate into 96-well
plates at a density of 8,000 cells per well. Cells were treated
with varying concentrations of cynarin (1, 10, 50, 100, 200 and 400
μM) and incubated for 24, 48 and 96 h to assess short-,
intermediate-, and long-term exposure effects. At each time point,
10 μl of Cell Counting Kit-8 (CCK-8; Proteintech Group,
Inc.) solution was added to each well, and the plates were
incubated for an additional 2 h. Cell viability was subsequently
quantified by measuring the absorbance at 450 nm using a microplate
reader.
OC formation and TRAP staining
analysis
BMMs were seeded in triplicate in 96-well plates to
ensure statistical robustness. After 24 h, osteoclastogenesis was
induced by adding RANKL (50 ng/ml) in the presence of cynarin at 0,
10, 50, 100, or 200 μM. Culture medium was replenished every
other day to maintain nutrient levels and compound stability. To
preserve cellular morphology during the differentiation period,
cells were fixed in 4% paraformaldehyde for 20 min at room
temperature. Following fixation, TRAP staining was performed
according to the manufacturer's protocol. TRAP-positive OCs were
identified, counted, and images were captured using an Olympus BX53
microscope equipped for brightfield microscopy at appropriate
magnification. Quantification of OC number and size was carried out
with ImageJ software, ensuring objective assessment of
differentiation across treatment groups.
Bone resorption assay
BMMs were cultured under the same conditions as
aforementioned. To initiate OC-mediated bone resorption, cells were
treated with RANKL (50 ng/ml) and cynarin (10, 50, 100, or 200
μM) until mature OC-like cells formed on bone slices. After
removing the cells, the bone slices were fixed in a 4%
paraformaldehyde + 2.5% glutaraldehyde mixed fixative for 24 h at
4°C. The samples after decalcification were dehydrated through a
graded ethanol series, subjected to critical point drying, and
sputter-coated with a 15 nm layer of gold/palladium. Images of
resorption pits were captured by scanning electron microscopy (SEM;
FEI Quanta 250).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
For RT-qPCR, total RNA was extracted using TRIzol
reagent, and its purity and concentration were assessed by
measuring the A260/A280 ratios on a NanoDrop
2000/2000C spectrophotometer (Thermo Fisher Scientific, Inc.); only
samples with a ratio between 1.9-2.1 were used. A total of 1
μg of RNA was reverse transcribed to cDNA using the
PrimeScript™ RT reagent kit according to the manufacturer's
instructions. qPCR was performed on a Light Cycler® 480
Instrument II (Roche) with TB Green® Premix Ex Taq™ II
FAST Kit using the following protocol: Initial denaturation at 95°C
for 30 sec; 40 cycles of 95°C for 5 sec and 60°C for 30 sec. Gene
expression was calculated using the 2−ΔΔCq method
(20). Primer sequences are
listed in Table SI.
Western blotting (WB)
The treated cells were lysed on ice in RIPA buffer
(cat. no. P0013C; Beyotime Institute of Biotechnology), and the
lysates were clarified by centrifugation at 12,000 × g for 15 min
at 4°C. A BCA kit was used to quantify protein concentrations.
Equal amounts of protein (30 μg) were separated on 4-10%
SDS-polyacrylamide gels (cat. no. M00657; GenScript) and
transferred to PVDF membranes (Thermo Fisher Scientific, Inc.).
Membranes were blocked in 5% skim milk at 37°C for 1 h, then
incubated overnight at 4°C with primary antibodies (all at a
dilution of 1:1,000). After washing, secondary antibodies
(HRP-linked; 1:1,000) were administered and incubated at 37°C for 1
h to enable the detection of primary antibody-protein complexes.
Following the removal of unbound secondary antibodies, bands were
visualised using enhanced chemiluminescence (ECL; Thermo Fisher
Scientific, Inc.) and images were captured on an Odyssey 9120
system (LI-COR Biosciences). The grey value of each band was
analysed by ImageJ 24.0 software (National Institutes of
Health).
Immunofluorescence (IF) analysis of
podosome actin belt
OC differentiation was induced as aforementioned.
Upon the appearance of multinucleated OCs in RANKL-stimulated
control wells, cells were fixed in 4% paraformaldehyde for 20 min
at room temperature to preserve cellular morphology. For
intracellular antibody staining, cells were permeabilised with 0.5%
Triton X-100 (MilliporeSigma) for 5 min, followed by washing to
remove any residual detergent. F-actin structures were stained with
FITC-conjugated phalloidin (MedChemExpress) for 30 min at room
temperature, while nuclei were counterstained with DAPI (cat. no.
C1006; Beyotime Institute of Biotechnology). All staining
procedures were conducted in the dark to prevent photobleaching.
After washing with phosphate-buffered saline (PBS), images of actin
ring borders were captured using a Nikon Eclipse Ti inverted
fluorescence microscope (Nikon Corporation). The distribution of
podosome actin belts was quantified using ImageJ software.
Assessment of the generation of ROS
Intracellular ROS levels were assessed using the ROS
detection kit (cat. no. S0033; Beyotime Institute of
Biotechnology). Following thorough washing with PBS, cells were
incubated with 2 μM/l 2',7'-dichlorodihydrofluorescein
diacetate (DCFH-DA) for 20 min. The non-fluorescent DCFH-DA was
internalised by cells and hydrolysed by intracellular esterases to
form DCFH, which was subsequently oxidised by ROS to produce the
highly fluorescent compound DCF. Fluorescence intensity was
measured using a microplate reader, with excitation at 488 nm and
emission detection at 525 nm.
Preparation of nuclear extracts and
cytoplasmic extracts
RAW264.7 cells were washed with ice-cold PBS and
then centrifuged at 500 × g for 5 min at 4°C. The cell pellet was
resuspended in cytoplasmic protein extraction reagent A (containing
PMSF) and vortexed vigorously for 5 sec. After incubating on ice
for 15 min, Reagent B was added, and the mixture was vortexed for 1
min before ultracentrifugation at 4°C. The cytoplasmic fraction was
extracted from the resulting supernatant. For nuclear protein
extraction, nuclear protein extraction reagent (containing PMSF)
was added to the pellet. The sample was vortexed at high speed for
30 min, followed by ultracentrifugation. The nuclear fraction was
collected from the supernatant. Protein concentrations were
quantified as aforementioned.
RNA sequencing (RNA-seq) followed by
bioinformatics evaluation
RAW264.7 cells were cultured under OC
differentiation conditions for 5 days, with or without cynarin.
Total RNA was extracted using TRIzol reagent and stored at -80°C.
RNA quality and integrity were verified using a NanoDrop
spectrophotometer (Thermo Fisher Scientific, Inc.) to ensure
A260/A280 ratios between 1.9-2.1, and an
Agilent 2100 Bioanalyzer with the RNA Nano 6000 Assay Kit (Agilent
Technologie, Inc.s) to confirm RNA Integrity Number values >9.0.
RNA sequencing was performed by Beijing Biomarker Technologies Co.,
Ltd. (http://www.biomarker.com.cn/)
following their standard Illumina sequencing protocols.
Differential expression analysis was performed using DESeq2
(version 1.20.0). Raw counts were normalised using the
median-of-ratios approach implemented in DESeq2, and differential
expression was defined as |Log2FC|≥0.58 (FC ≥1.5) with
unadjusted P<0.05. Volcano plots were generated to visualise
distribution of differentially expressed genes (DEGs), highlighting
the magnitude and statistical significance of expression changes.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway analyses were conducted using the BioMarker BioCloud
Platform (www.biocloud.org) (21).
LPS-induced skull osteolysis in vivo
mouse model
Animal experiments were conducted in accordance with
the laboratory animal management guidelines. They were approved by
the Animal Ethics Committee of the Ninth People's Hospital,
affiliated with Shanghai Jiao Tong University School of Medicine
(approval no. SH9H-2020-A1-1; Shanghai, China). Mice were housed
with a standard temperature (20-26°C) and humidity (50-60%), under
a 12-h light/dark cycle and fed standard rodent diet and water
ad libitum. Mice were monitored daily for weight loss and
general health and euthanized when humane endpoints (signs of
severe distress, unresponsiveness to treatment, and/or significant
weight loss) were reached. A calvarial osteolysis model was
established based on previously reported methodologies (22,23). A total of 48 male C57/BL6 mice
(6-8 weeks, 18-22 g) were randomly assigned to six groups: i) Sham
group (subcutaneous injection of 50 μl PBS); ii) LPS group
(subcutaneous injection of 10 mg/kg LPS in 50 μl); iii)
alendronate group (subcutaneous injection of 10 mg/kg LPS in 50
μl; intraperitoneal injection of 10 μg/kg alendronate
in 100 μl) (24,25); iv) cynarin low-dose group
(subcutaneous injection of 10 mg/kg LPS and 100 μM cynarin
in 50 μl); v) cynarin medium-dose group (subcutaneous
injection of 10 mg/kg LPS and 250 μM cynarin in 50
μl); and vi) cynarin high-dose group (subcutaneous injection
of 10 mg/kg LPS and 500 μM cynarin in 50 μl). During
the modelling process, anaesthesia was induced via intraperitoneal
injection of a 3% sodium pentobarbital solution at a dose of 50
mg/kg. Collagen sponges (4×4×2 mm), soaked in either PBS or LPS,
were implanted at the sagittal midline suture of the calvaria to
induce bone loss. All mice received subcutaneous or intraperitoneal
injections every other day for 10 days. No mice reached humane
endpoints. The mice were euthanised by filling with carbon dioxide
at the rate of 30% CO2 tank volume/min. Tissues were
collected after death confirmation: Absent pulse and breathing,
lost corneal reflexes, no response to deep toe stimulation and
mucosal greying. Blood was collected from anesthetised mice's
eyeballs and transferred into heparinised anticoagulant tubes.
After centrifugation, the plasma was separated and used to measure
biochemical indicators, including alanine aminotransferase (ALT),
aspartate aminotransferase (AST) and blood urea nitrogen (BUN), for
assessing liver and kidney function. The major organs (heart,
liver, spleen, lungs and kidneys) were dissected and fixed for
haematoxylin and eosin (H&E) staining. The calvarial bones were
isolated and preserved for further analysis.
Micro-computed tomography (CT)
scanning
Micro-CT scans were performed using a SkyScan 1076
system (Bruker Corporation) with a resolution of 9 μm. The
scan parameters were set to 29 kV, 175 μA, and a 300 msec
exposure time. Structural features of the bone were quantified
using SCANCO software (version 6.5-3; Scanco Medical AG).
Histological staining and analysis
After decalcifying in EDTA solution for 2 weeks, the
skull was paraffin-embedded, and 4-μm sections were
prepared. Serial sections were deparaffinized in xylene and
rehydrated through a graded ethanol series. H&E and TRAP
staining were performed to evaluate tissue morphology and OC
activity. For immunohistochemical (IHC) staining, endogenous
peroxidase activity was quenched by incubation in 3%
H2O2 in methanol for 15 min at room
temperature. Non-specific binding was blocked with 5% normal goat
serum (Beijing Solarbio Science & Technology Co., Ltd.) in PBS
for 1 h at room temperature. Sections were then incubated with
primary antibodies (RANK, CTSK, OCN, RUNX2, TNFα, iNOS, p-P38,
p-ERK1/2 and p-JNK1/2, all at a dilution of 1:100) overnight at
4°C. After washing, sections were incubated with HRP-conjugated
secondary antibodies (1:500; Thermo Fisher Scientific, Inc.) for 1
h at room temperature. Signal was developed using a DAB substrate
kit (cat. no. DA1016; Beijing Solarbio Science & Technology
Co., Ltd.) and sections were counterstained with haematoxylin. For
IF staining, a similar protocol was followed. After blocking,
sections were incubated with primary antibodies against Nrf2
(1:100) and Keap1 (1:100) overnight at 4°C, followed by incubation
with Alexa Fluor® 647-conjugated secondary antibodies
(1:500; Cell Signalling Technology, Inc.) for 1 h at room
temperature in the dark. Nuclei were counterstained with DAPI.
Stained sections (H&E, TRAP, IHC) were examined using a Leica
DM4000B light microscope. IF-stained sections were examined using
the same microscope equipped for epifluorescence. Positive staining
was quantified using ImageJ software.
Statistical analysis
Group data are presented as the mean ± standard
deviation (SD) to indicate average values and variability. All
statistical analyses were conducted using GraphPad Prism software
(version 10.3.1; Dotmatics). One-way analysis of variance (ANOVA)
followed by Tukey's multiple comparisons post hoc test was used for
comparisons of multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Cynarin attenuates LPS-induced calvarial
osteolysis in vivo
An LPS-induced osteolytic murine model was used to
assess the therapeutic effects of cynarin on inflammatory bone
loss. Throughout the experimental period, all the subjects
maintained stable conditions until a specified terminal time point.
Examination of histological sections from the vital organs of the
mice demonstrated no discernible damage, suggesting favourable
biocompatibility of the drug (Fig.
S1A). Serum biochemistry showed mild elevations in hepatic
indices (ALT, AST and total bilirubin) in the LPS group, which were
found to be significantly improved by alendronate and a medium-dose
of cynarin (P<0.05 vs. LPS for all). The renal indices (BUN,
creatinine and uric acid) remained unchanged among the groups
(Fig. S1B). Micro-CT
demonstrated that the medium-dose cynarin (Cy-M) group had
preserved bone microarchitecture, as evidenced by increased bone
volume, trabecular number, trabecular thickness and decreased
trabecular separation relative to LPS alone. These changes were
comparable to those observed with alendronate (Fig. 1A and B). H&E staining
confirmed reduced bone porosity in the Cy-M group (Fig. 1C and D), and TRAP staining
revealed a significant reduction in the number of TRAP-positive OCs
(Fig. 1C and E).
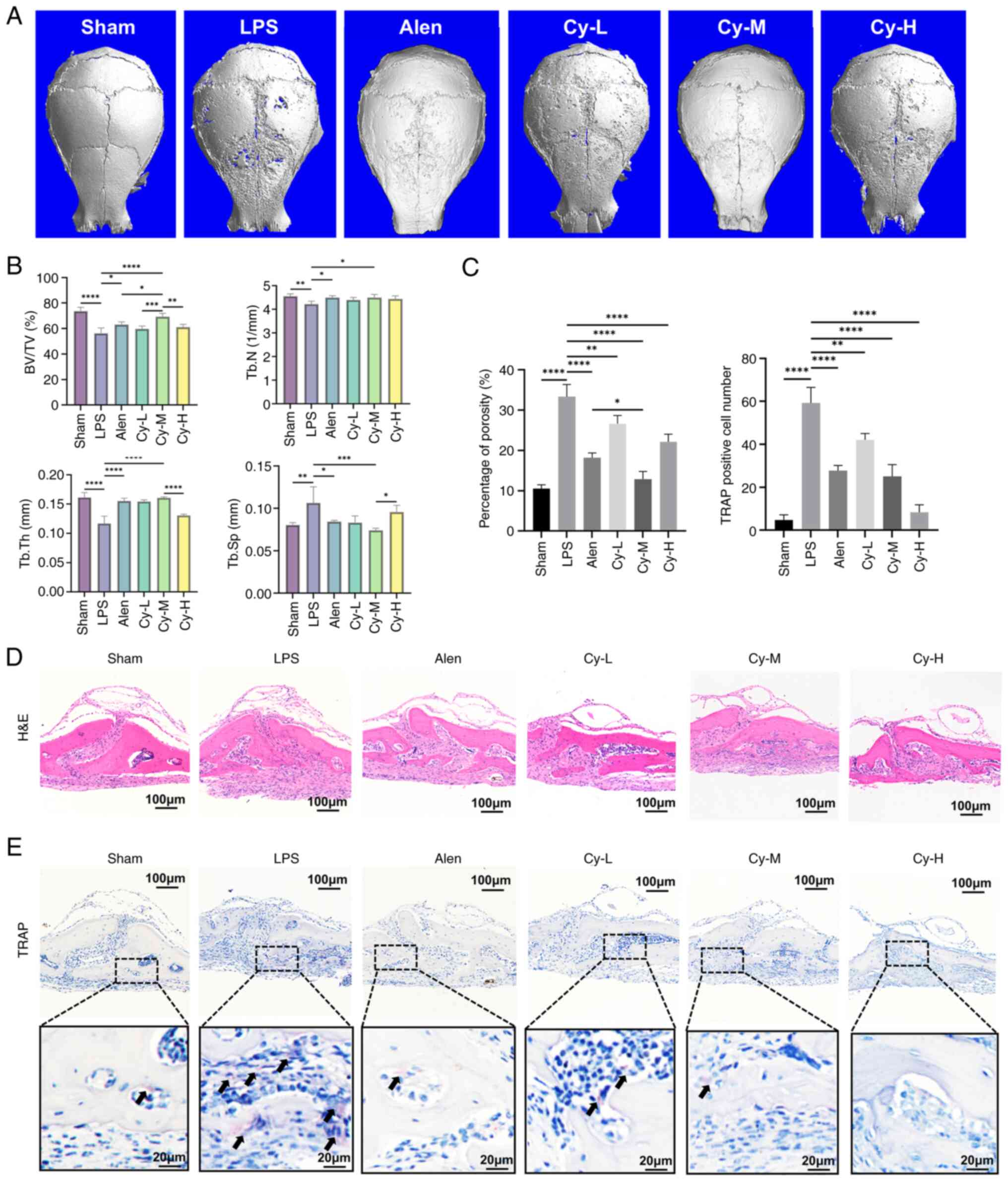 | Figure 1Cynarin shows therapeutic efficacy
comparable to alendronate in murine model of LPS-induced
osteolysis. (A) The micro-computed tomography reconstructions of
each group's murine calvaria. 6 weeks old C57/B6 mice were used to
establish model and were administrated by cynarin in indicated dose
for 10 days. (B) BV/TV, Tb.N, Tb.Th and Tb.Sp were evaluated by
quantitative analysis. (C) Quantitative analysis of porosity
percentage in H&E staining and TRAP positive cells number in
TRAP staining. (D) Representative images of calvarial histology
stained with H&E (magnification, ×100). (E) Representative
images of calvarial histology stained with TRAP (magnification,
×100). TRAP-positive cells are shown by black arrows. Animals were
randomly assigned to each group using a computer-generated
sequence. Histological scoring was conducted by an investigator who
was blinded to the treatment groups. All experiments were performed
with three independent biological replicates (n=3). The data are
presented as the mean ± SD. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001 vs. Sham and LPS group separately. LPS,
lipopolysaccharide; BV/TV, bone volume to total volume ratio; Tb.N,
trabecular number; Tb.Th, trabecular thickness; Tb.Sp, trabecular
separation; TRAP, tartrate-resistant acid phosphatase; Alen,
alendronate; Cy-M, medium-dose cynarin. |
IHC further showed that cynarin reduced RANK and
CTSK expression while also increasing OCN and RUNX2 expression in
calvarial sections (Fig. 2A and
B), suggesting the inhibition of OC differentiation and the
support of osteoblast activity. Although cynarin modestly enhanced
osteoblast differentiation in vitro, the mechanism was not
investigated in the present study (Fig. S2). Moreover, cynarin
significantly decreased TNF-α and iNOS expression in calvarial
tissues (Fig. 2C and D),
indicating suppression of inflammatory signalling. Inflammatory
signals play a crucial role in the pathogenesis of osteolysis, and
inhibition of inflammation helps alleviate osteolysis. These
findings suggest that cynarin effectively preserves the bone
structure and mitigates bone resorption by inhibiting OC
differentiation and inflammation.
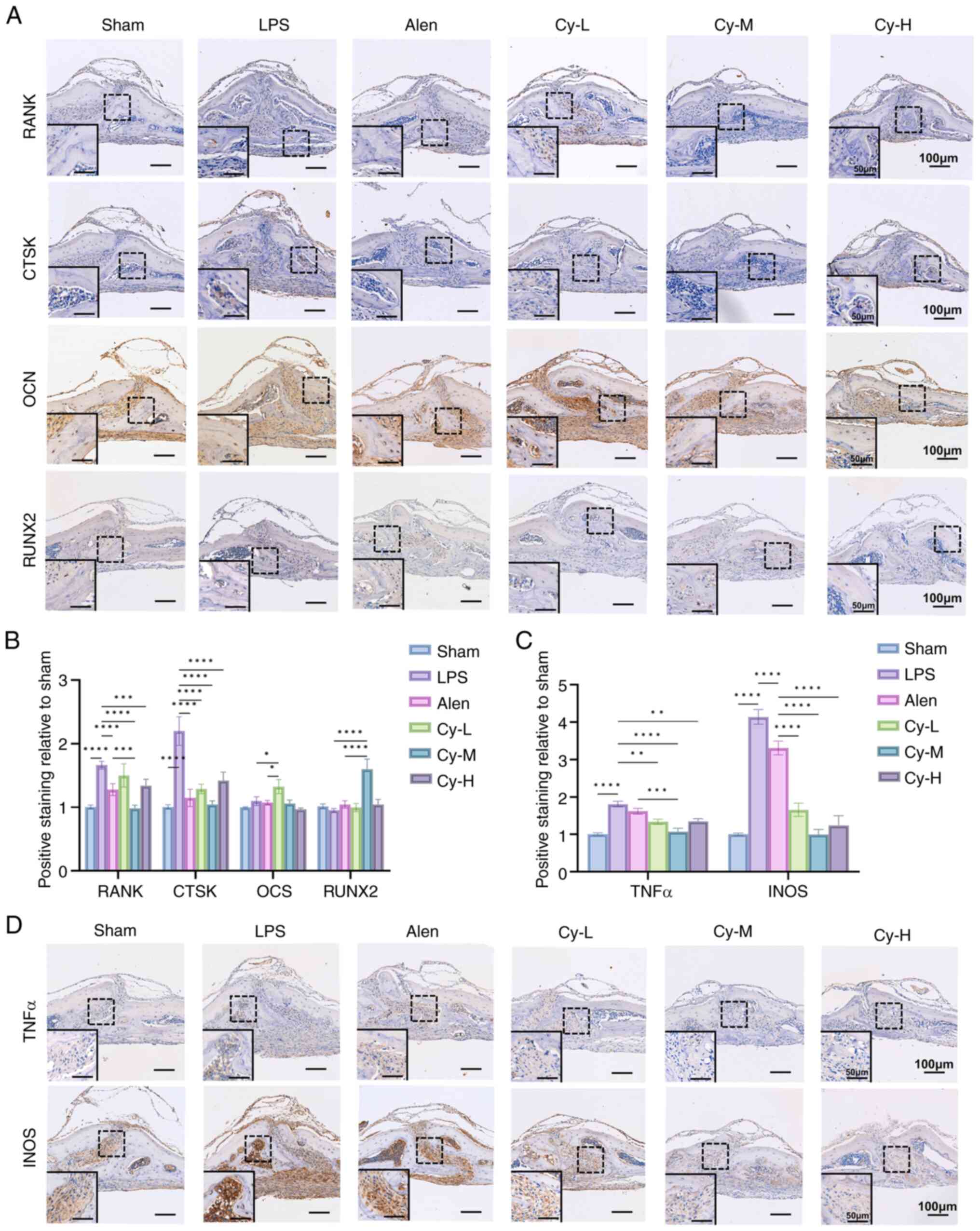 | Figure 2Cynarin exerts a more potent effect
in inhibiting osteoclast differentiation and reducing inflammation
in vivo. (A) Representative images of calvarial histology
stained with RANK, CTSK, OCN and RUNX2 (magnification, ×100). (B)
Quantitative analysis of the expression of RANK, CTSK, OCN and
RUNX2 in the IHC staining. (C) Quantitative analysis of the
expression of TNFα and iNOS in the IHC staining. (D) Representative
images of calvarial histology stained with TNFα and iNOS
(magnification, ×100). All experiments were performed with three
independent biological replicates (n=3). The data are presented as
the mean ± SD. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. Sham,
LPS and Alen group separately. RANK, receptor activator of nuclear
factor κB; CTSK, cathepsin K; OCN, osteocalcin; IHC,
immunohistochemical; iNOS; inducible nitric oxide synthase; LPS,
lipopolysaccharide; Alen, alendronate; Cy-M, medium-dose
cynarin. |
Cynarin inhibits OC differentiation and
bone resorption in vitro
The chemical formula of cynarin is illustrated in
Fig. 3A. The performed cell
viability test revealed no cytotoxic effects of cynarin at
concentrations <200 μM (Fig. 3B). To explore the direct effects
of cynarin on OC differentiation, BMMs and RAW264.7 cells treated
with RANKL were used to induce osteoclastogenesis. TRAP staining
revealed a significant dose-dependent reduction in the number of
TRAP-positive OCs after cynarin treatment (Fig. 3C-E). The performed bone
resorption assay revealed that cynarin markedly reduced resorption
pits area at concentrations exceeding 50 μM after 9 days of
differentiation (Fig. 3F).
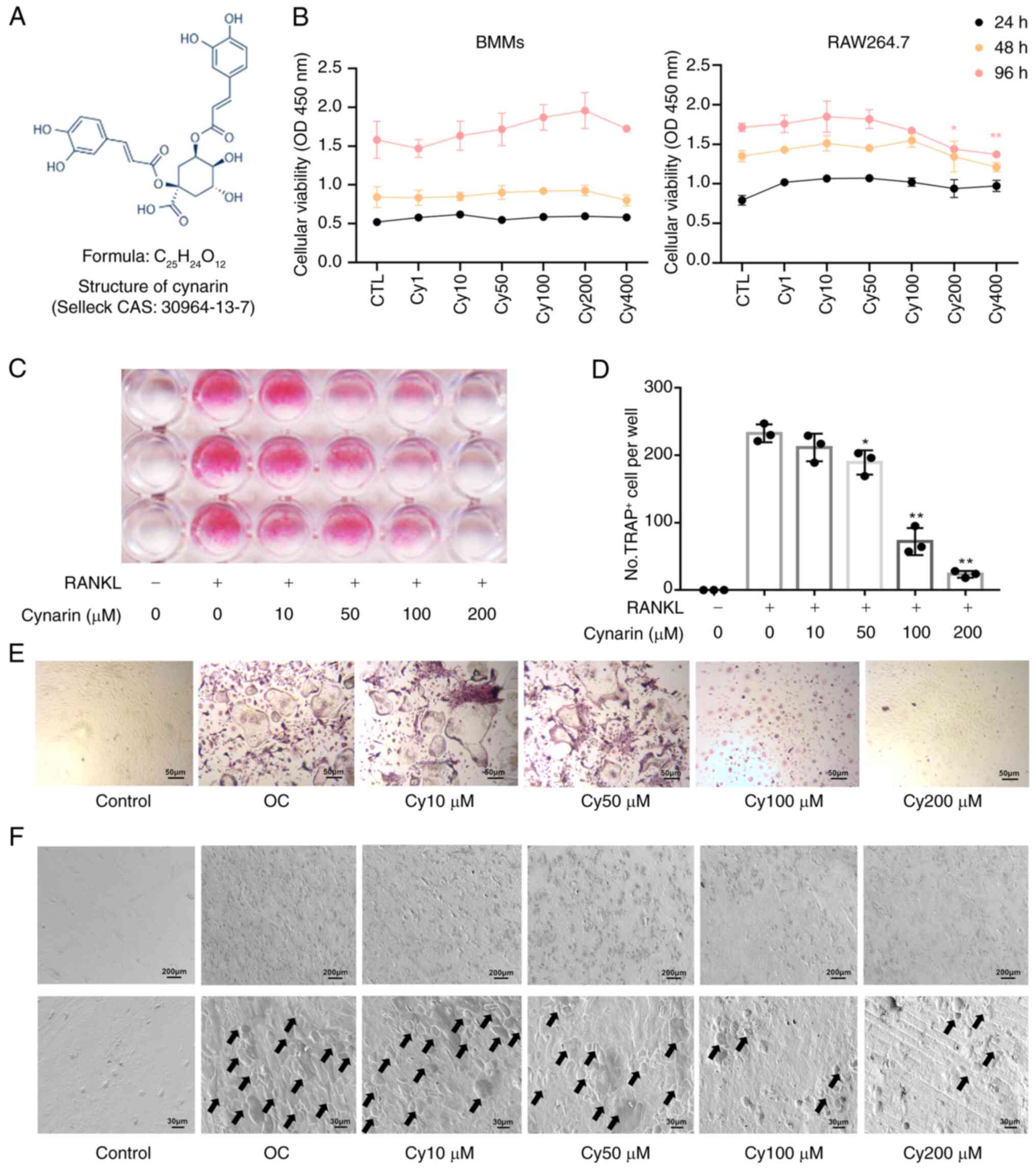 | Figure 3Cynarin restrains RANKL-induced OC
differentiation in vitro. (A) Chemical formula for cynarin.
(B) BMMs and RAW264.7 cells were treated with gradient
concentrations of cynarin for 24, 48 and 96 h, after which cells
proliferation and viability were detected by Cell Counting Kit-8
assay (n=3). *P<0.05 and **P<0.01 vs.
CTL group. (C) BMMs were incubated with gradient concentrations of
cynarin for 5-7 days. Cells were stained for TRAP staining. Scan of
a 96-well plate after TRAP staining (n=3). (D) The count of
TRAP+ OCs per well was determined (n=3).
*P<0.05 and **P<0.01 vs. RANKL (+) and
Cynarin (-) group. (E) Representative images of TRAP staining
(magnification, ×200; (n=3). (F) BMMs were incubated with gradient
concentrations of cynarin for 9 days. The bone slides were
visualised under the scanning electron microscope (magnification,
×50, first row; ×300, second row). Black arrows indicate
osteoclastic absorption pits (n=3). The data are presented as the
mean ± SD. RANKL, receptor activator of nuclear factor κB ligand;
OC, osteoclast; BMMs, bone marrow-derived macrophages; TRAP,
tartrate-resistant acid phosphatase. |
Further molecular analysis using RT-qPCR showed that
cynarin lowered the expression of Nfatc1, Dcstamp,
Trap, Atp6v0d2, Ctsk and Ctr (Fig. 4A and D). Notably, the suppressive
effect of cynarin on OC-related gene expression was most prominent
on day four (Fig. 4D). WB
revealed that cynarin reduced DC-STAMP and CTSK protein levels
during RANKL-induced differentiation (Fig. 4B). Furthermore, Actin staining
revealed that cynarin blocked the fusion of BMM precursor cells and
development of podosomal actin structures (Fig. 4C). Collectively, these results
suggested that cynarin inhibited OC differentiation and bone
resorption.
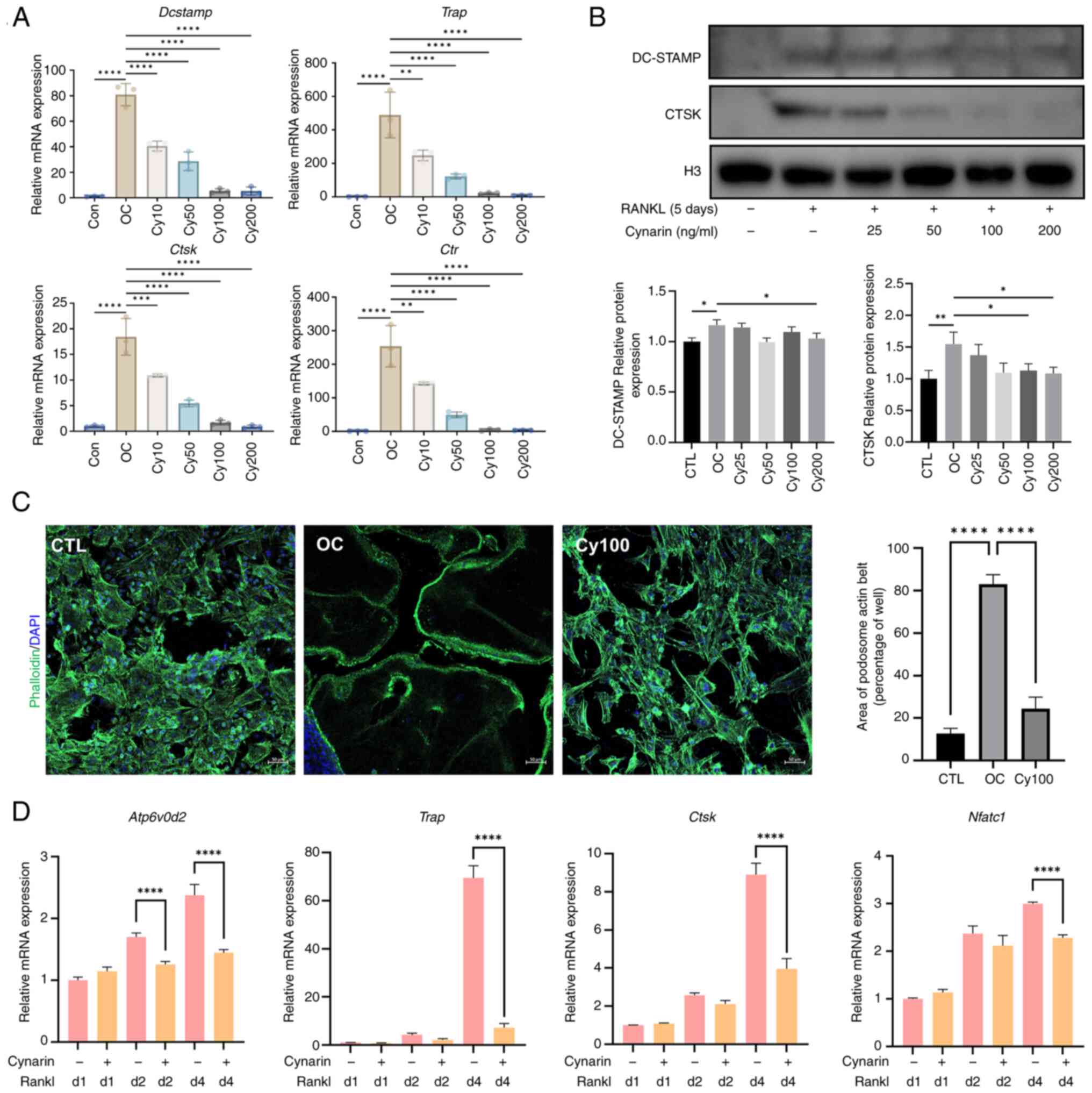 | Figure 4Cynarin downregulates
osteoclast-specific genes and proteins during osteoclastogenesis.
(A) RAW264.7 cells were treated with RANKL and gradient
concentrations of cynarin for 5 days. Dcstamp, Trap,
Ctsk and Ctr expression levels were normalised to
Gapdh expression by RT-qPCR (n=3). (B) RAW264.7 cells were
treated with RANKL and gradient concentrations of cynarin for 5
days. Expression levels of DC-STAMP and CTSK were detected by
western blotting. Quantitative densitometric analysis was performed
to normalize DC-STAMP and CTSK (n=3). (C) After 5 days of
macrophage colony-stimulating factor and RANKL induction, bone
marrow-derived macrophages were stained for immunofluorescence
analysis of podosome actin belt (magnification, ×200; (n=3). (D)
RAW264.7 cells were incubated with or without 100 μM cynarin
for 1, 2 and 4 days. Atp6v0d2, Trap, Ctsk, and
Nfatc1 expression levels were normalised to GAPDH
expression by RT-qPCR (n=3). The data are presented as the mean ±
SD. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. RANKL,
receptor activator of nuclear factor κB ligand; RT-qPCR, reverse
transcription-quantitative PCR; CTSK, cathepsin K; TRAP,
tartrate-resistant acid phosphatase; OC, osteoclast. |
Cynarin activates Nrf2 to suppress
inflammation and oxidative stress in vitro
Since inflammation plays a critical role in
promoting osteoclastogenesis, the anti-inflammatory effects of
cynarin on LPS-stimulated RAW264.7 macrophages were investigated.
Cynarin treatment significantly reduced the expression of
proinflammatory cytokines, including Tnf, IL1β,
IL6 and Nos2 at the mRNA level (Fig. 5A). Additionally, ROS levels were
significantly reduced in the cynarin-treated macrophages (Fig. 5B), indicating that cynarin
attenuated oxidative stress, which is a key contributor to
inflammation.
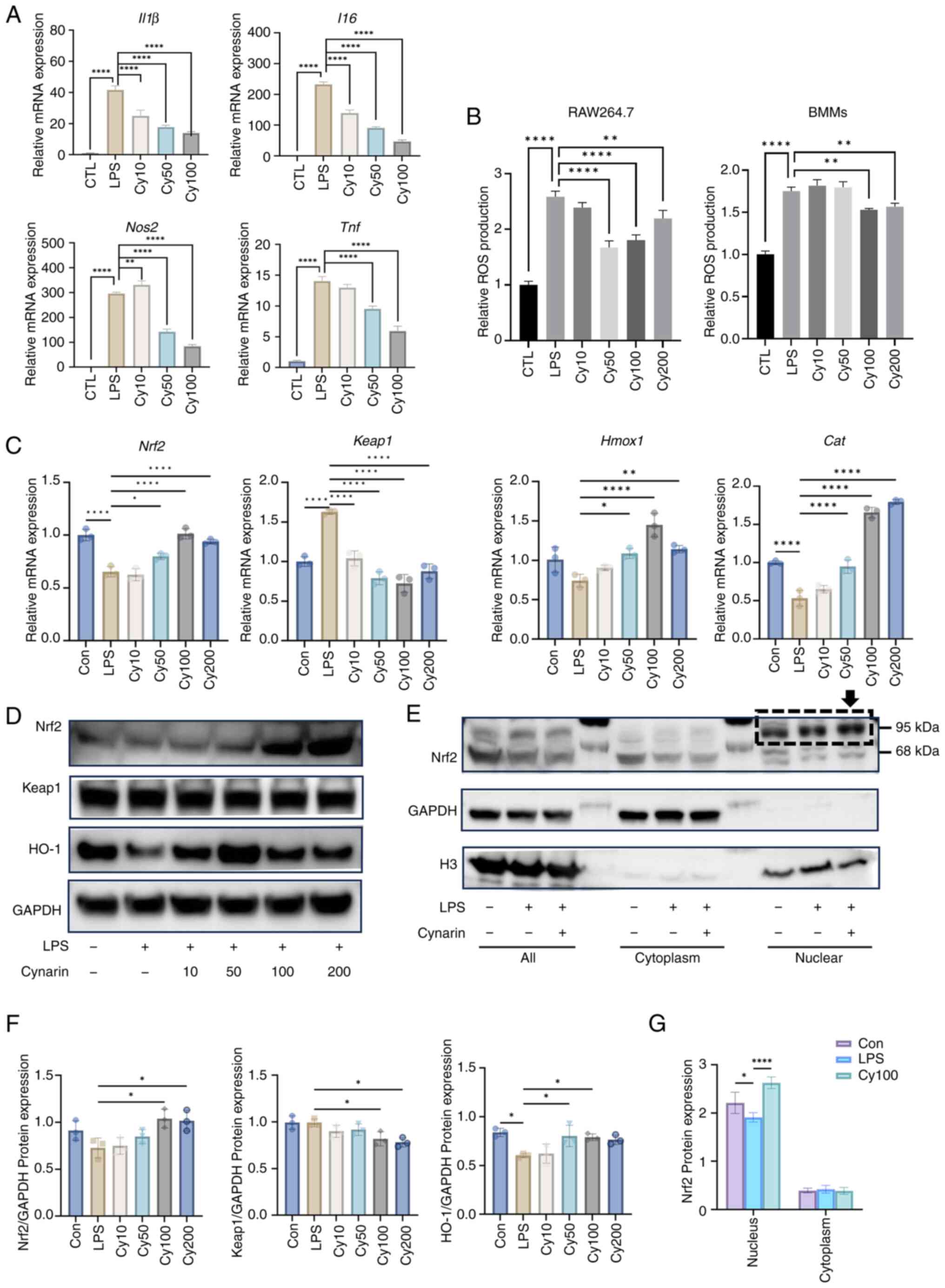 | Figure 5Cynarin suppresses
macrophage-mediated inflammation through activation of the
Nrf2-Keap1 signalling pathway. (A) RAW264.7 cells were exposed to
cynarin (10, 50 or 100 μM) for 24 h and then treated with
LPS (1 μg/ml) for 12 h. Expression levels of IL1β,
IL6, Nos2 and Tnf were detected by RT-qPCR
(n=3). (B) RAW264.7 cells and bone marrow-derived macrophages were
exposed to cynarin (10, 50, 100 or 200 μM) for 24 h and then
treated with LPS (1 μg/ml) for 12 h. Intracellular ROS
levels were measured with a ROS assay kit (n=3). (C) RAW264.7 cells
were exposed to cynarin (10, 50, 100 or 200 μM) for 6 h and
then treated with LPS (1 μg/ml) for 12 h. Expression levels
of Nrf2, Keap1, Hmox1 and Cat were
detected by RT-qPCR (n=3). (D) RAW264.7 cells were exposed to
cynarin (10, 50, 100 or 200 μM) for 24 h and then treated
with LPS (1 μg/ml) for 24 h. Expression levels of Nrf2,
Keap1 and HO-1 were detected by WB. (E) RAW264.7 cells were exposed
to 100 μM cynarin for 24 h and then treated with LPS (1
μg/ml) for 24 h. Nuclear extracts and cytoplasmic extracts
were used to test expression of Nrf2 by WB. (F) Quantitative
densitometric analysis was performed to normalize Nrf2, Keap1 and
HO-1 expression levels from D (n=3). (G) Quantitative densitometric
analysis was performed to normalize Nrf2 expression levels from E
(n=3). The data are presented as the mean ± SD.
*P<0.05, **P<0.01 and
****P<0.0001 vs. CTL and LPS group separately. LPS,
lipopolysaccharide; RT-qPCR, reverse transcription-quantitative
PCR; ROS, reactive oxygen species; HO-1, heme oxygenase-1; WB,
western blotting. |
Mechanistically, cynarin treatment led to the
upregulation of Nrf2 expression and the downregulation of Keap1,
thus resulting in the induction of antioxidant enzymes, such as
HO-1 and catalase (Fig. 5C, D and
F). Moreover, nuclear fractionation assays confirmed that
cynarin promoted Nrf2 nuclear translocation, validating its role in
reducing oxidative stress and inflammation via Nrf2 activation
(Fig. 5E and G). These results
support the hypothesis that cynarin exerts its anti-inflammatory
effects by activating the Nrf2-Keap1 pathway activation.
Cynarin suppresses MAPK signalling and
engages the Nrf2-Keap1 axis
To further elucidate the mechanism of cynarin
suppressing OC differentiation, cynarin was used for RNA-seq in
RANKL-induced RAW264.7 cells. A volcano plot illustrates that
cynarin treatment upregulated 499 genes and downregulated 726 genes
(Fig. 6A). To assess the
possible impact of cynarin on biological processes, GO enrichment
analysis was conducted. As depicted in Fig. 6B, the regulation of OC
differentiation and inflammatory response were prominently
associated with the therapeutic mechanisms of cynarin. KEGG pathway
enrichment analysis demonstrated profound modulation of the MAPK
signalling pathway following cynarin treatment (Fig. 6C).
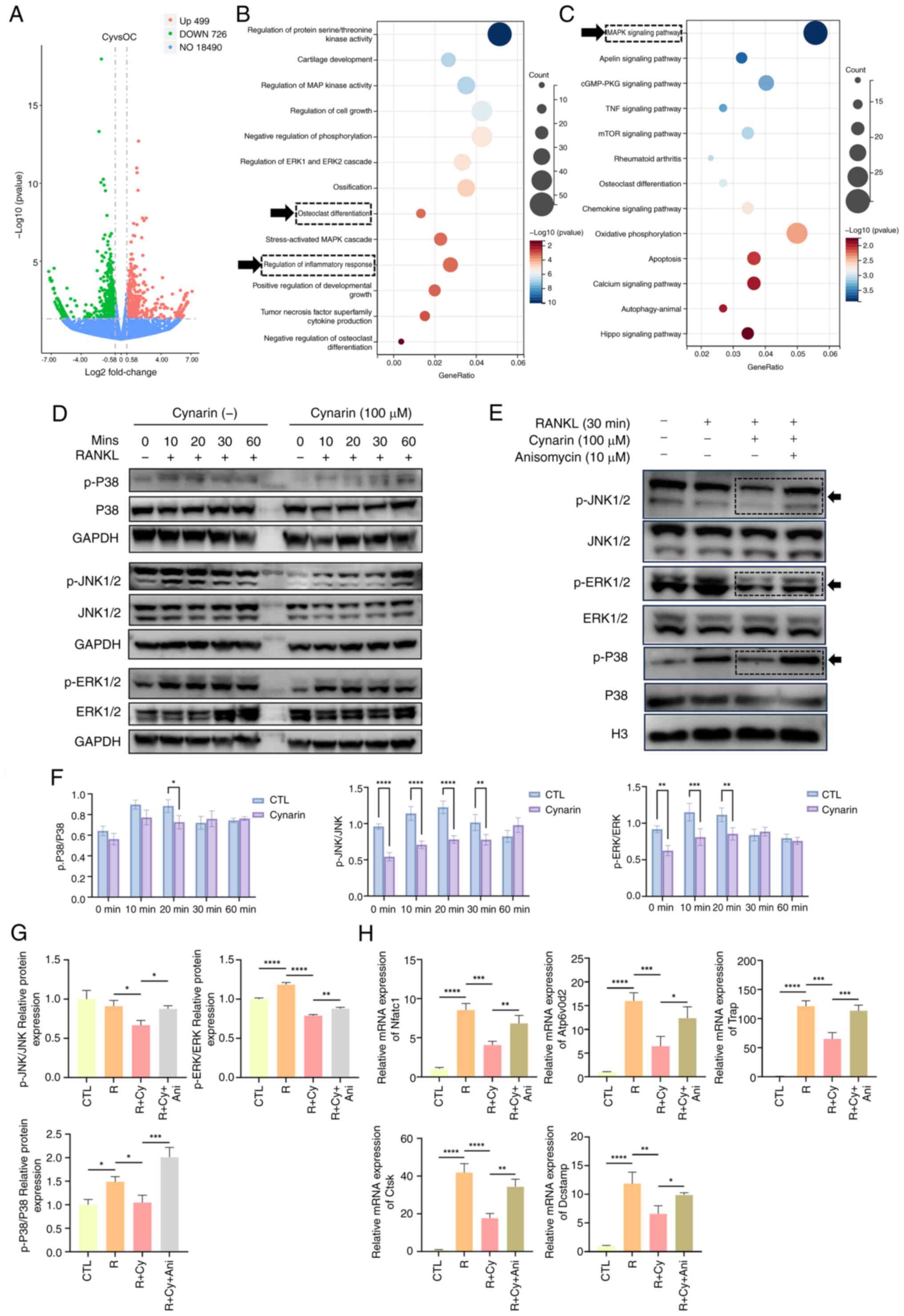 | Figure 6Cynarin suppresses the MAPK
signalling pathway in RANKL-induced osteoclastogenesis. (A) RANKL
induced RAW264.7 cells for 5 days with or without 100 μM
cynarin. Total RNA was extracted and sent for RNA sequencing. Genes
exhibiting |Log2FC|≥0.58 (FC ≥1.5) and unadjusted P≤0.05
were considered to be differentially expressed. Volcano plot of the
distinct upregulated and downregulated genes of Cy vs. OC (n=3).
(B) Gene Ontology enrichment analysis of differential genes of Cy
vs. OC (n=3). (C) Kyoto Encyclopedia of Genes and Genomes pathway
analysis of differential genes of Cy vs. OC (n=3). (D) RAW264,7
cells were pretreated with 100 μM cynarin for 6 h and
stimulated with 50 ng/ml RANKL for 10 to 60 min. Expression levels
of total and phosphorylated forms of JNK, ERK, and P38 were
detected by WB. (E) RAW264.7 cells were treated with 100 μM
cynarin, 10 μM anisomycin for 30 min and stimulated with 50
ng/ml RANKL for 30 min. The ratio of phosphorylated JNK to JNK,
p-ERK to ERK and p-P38 to P38 were quantitatively determined by WB.
(F) Quantitative densitometric analysis was performed to normalize
p-JNK/JNK, p-ERK/ERK and p-P38/P38 expression levels from D (n=3).
(G) Quantitative densitometric analysis was performed to normalize
p-JNK/JNK, p-ERK/ERK and p-P38/P38 expression from E (n=3). (H)
RAW264,7 cells were treated with 100 μM cynarin, 10
μM anisomycin for 4 days in RANKL-induced
osteoclastogenesis. Expression levels of OC-specific genes were
detected by reverse transcription-quantitative PCR (n=3). The data
are presented as the mean ± SD. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001. RANKL, receptor activator of nuclear
factor κB ligand; OC, osteoclast; p-, phosphorylated; WB, western
blotting; Ani, anisomycin. |
WB demonstrated that cynarin attenuated
RANKL-induced phosphorylation of JNK (10-30 min), ERK (10-20 min),
and p38 (20 min) (Fig. 6D and
F), indicating that cynarin suppressed osteoclastogenesis by
inhibiting MAPK signalling. To confirm this mechanism, anisomycin
(MAPK activator) was used. WB revealed that anisomycin counteracted
the inhibitory effects of cynarin on p-JNK, p-ERK and p-P38
(Fig. 6E and G). RT-qPCR
confirmed that anisomycin reversed the inhibitory effects of
cynarin on OC differentiation (Fig.
6H). These findings provide strong evidence that cynarin
inhibited osteoclastogenesis by suppressing MAPK signalling.
In vivo validation of the pathway regulation
showed that cynarin significantly increased Nrf2 expression,
decreased Keap1 expression, and promoted Nrf2 nuclear translocation
in calvarial tissues (Fig. 7A and
C). Furthermore, IHC staining showed that cynarin
administration significantly decreased p-P38, p-ERK1/2 and p-JNK1/2
levels compared with those in the LPS group (Fig. 7B and D). Collectively, cynarin
exerts dual regulatory effects in vivo by inhibiting OC
differentiation by suppressing the MAPK pathway and mitigating
inflammation and oxidative stress through activation of the
Nrf2-Keap1 axis. These combined actions therefore underscore the
potential of cynarin as a therapeutic candidate for the treatment
of inflammatory bone diseases.
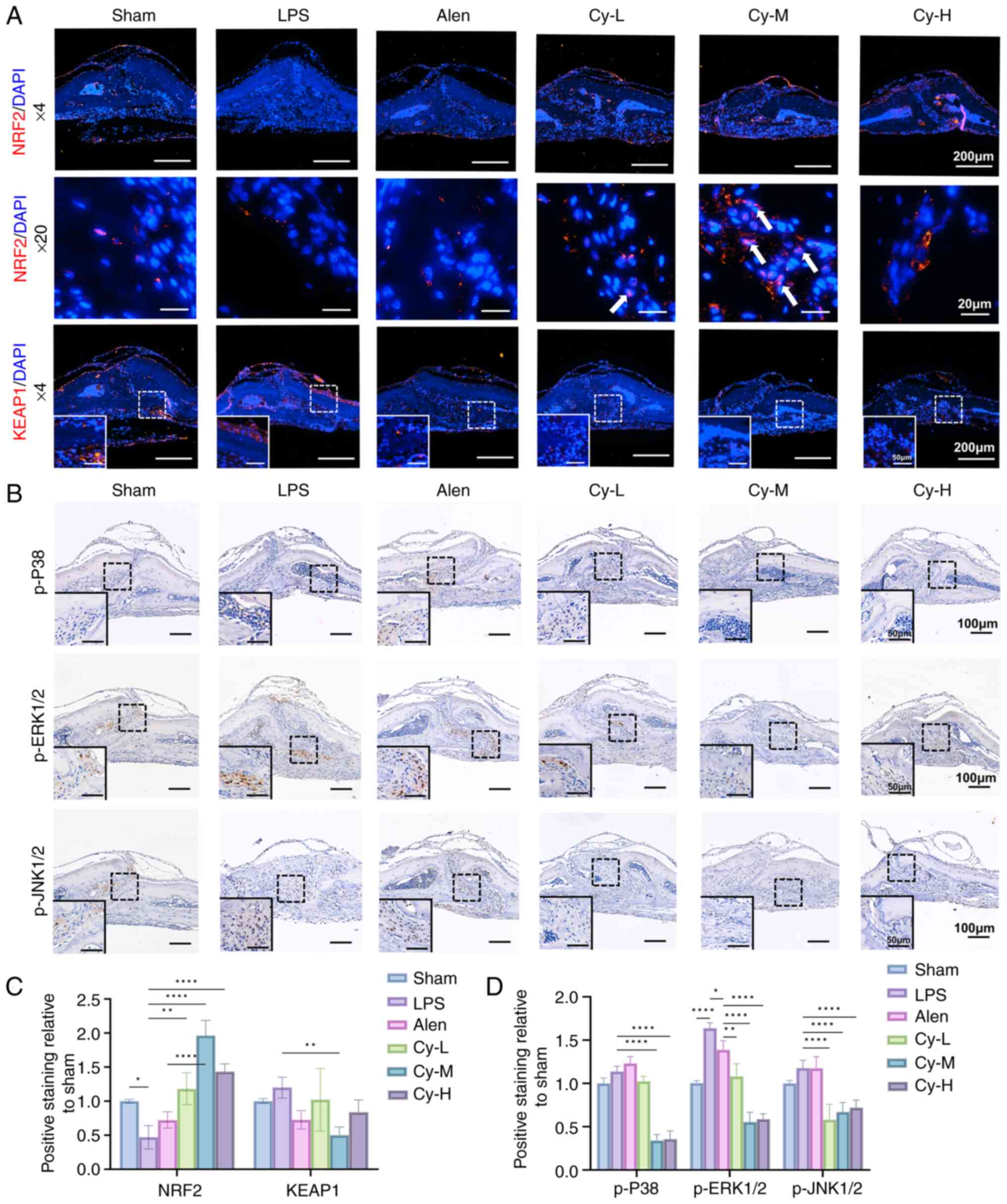 | Figure 7Cynarin suppresses osteoclastogenesis
and inflammation via the MAPK and Nrf2-Keap1 pathways in
inflammatory bone resorption. (A) Representative images of
calvarial histology stained with Nrf2 and Keap1 (magnification,
×40, first row; ×200, second row; ×40, third row). (B)
Representative images of calvarial histology stained with p-ERK1/2,
p-JNK1/2 and p-P38 (magnification, ×100). (C) Quantitative analysis
of the expression of Nrf2 and Keap1 in the immunofluorescence
staining. (D) Quantitative analysis of the expression of p-ERK1/2,
p-JNK1/2 and p-P38 in the immunohistochemical staining. All
experiments were performed with three independent biological
replicates (n=3). The data are presented as the mean ± SD.
*P<0.05, **P<0.01 and
****P<0.0001. p-, phosphorylated; Cy-M, medium-dose
cynarin; LPS, lipopolysaccharide; Alen, alendronate. |
Discussion
The development of novel therapeutic strategies that
simultaneously suppress osteoclastogenesis and modulate
inflammation is critical for alleviating inflammatory bone
resorption and improving bone health. Naturally derived plant
compounds have garnered considerable attention because of their
favourable biological activities and safety profiles. Cynarin, a
bioactive compound extracted from artichoke (Cynara
scolymus), shows promise in this regard. The present study
aimed to evaluate the therapeutic potential of cynarin in the
prevention of bone resorption and inflammation-induced bone loss.
These findings demonstrate that cynarin significantly inhibits OC
differentiation and activity, suppresses inflammatory responses,
and modulates the key signalling pathways involved in
osteoclastogenesis and inflammation.
Our in vivo findings have shown that cynarin
effectively reduced LPS-induced osteolysis and preserved the bone
structure, with effects comparable to those of the widely used drug
alendronate sodium. Improvements in the liver and kidney function
indices following cynarin treatment provide a critical basis for
assessing its safety and therapeutic potential. However, further
clinical translation will require comprehensive pharmacokinetic
studies and the optimisation of the dosage forms to enhance
bioavailability and determine the appropriate dosage. Histological
evaluation showed decreased bone porosity and fewer TRAP-positive
OCs in the cynarin-treated mice. Additionally, cynarin
significantly reduced the expression of key OC differentiation
markers, suggesting that it inhibited OC differentiation and
activity. These findings align with those of previous studies that
emphasised the bone protective effects of various natural compounds
in inflammatory bone diseases. Well-known natural anti-inflammatory
agents such as resveratrol and curcumin show similar protective
effects by reducing OC activity (26). One limitation of the study is
that only male mice were used. Although this reduces confounding
variables such as the periodic fluctuations of female hormones
during the initial efficacy screening, it is necessary to be
cautious when applying our conclusions directly to female models or
patients. Future work will include both sexes to rigorously assess
the potential gender-specific effects of cynarin on cranial bone
repair. However, the present study offers a new perspective by
demonstrating that cynarin treatment significantly inhibits
inflammation and downregulates the levels of inflammatory factors
such as TNF-α and iNOS. Osteolysis is closely associated with
proinflammatory cytokine release and OC activation (27,28). In conclusion, cynarin may prevent
osteolysis and preserve the bone microstructure by regulating OC
production and inflammation, providing a dual mechanism of
action.
The inhibitory effect of cynarin on
osteoclastogenesis observed in our in vitro study was
consistent with the findings of our in vivo study. It was
next discovered that cynarin concentrations >50 μM
significantly suppressed OC formation as evidenced by TRAP staining
and SEM. To the best of our knowledge, this finding has not been
previously reported. The performed cell viability assays confirmed
that cynarin concentrations <200 μM exhibited no
significant cytotoxicity. It was demonstrated that varying
concentrations of cynarin suppressed Nfatc1, Dcstamp,
Trap, Ctsk and Atp6v0d2, and 100 μM was
identified as the optimal concentration. FITC-labelled phalloidin
staining indicated that 100 μM cynarin significantly
inhibited OC development. This effect was most evident after four
days of RANKL exposure.
Accumulating evidence has suggested that Nrf2 is a
key regulator of bone oxidative homeostasis. Transcriptome analysis
of Nrf2 or Keap1 knockout models revealed that Nrf2 deficiency
upregulates genes associated with mitochondrial oxidative
phosphorylation, including Car2, Calcr and
Mmp12, and increases OC production under oxidative stress
(29). Dong et al
(30) found that the Nrf2
activator bitopertin blocks Keap1-Nrf2 binding, reduces
intracellular iron levels, and inhibits OC formation. Nrf2
regulates the activation of genes that enhance antioxidant defence,
thereby reducing ROS accumulation (31). Under normal conditions, Nrf2 is
maintained at low levels because of its binding to Keap1. When
Keap1 is inhibited, Nrf2 translocates to the nucleus where it
activates genes involved in detoxification and antioxidant
responses. It was demonstrated that cynarin inhibits Keap1 during
inflammation, promotes Nrf2 nuclear translocation, and enhances the
expression of HO1 and Cat. In vivo experiments confirmed
that, compared with LPS and alendronate, medium-dose cynarin
treatment significantly increased Nrf2 expression, decreased Keap1
expression, and activated nuclear translocation. Inflammatory
conditions are commonly associated with elevated ROS, which are key
regulators of bone metabolism and function (32,33). In addition, it was demonstrated
that cynarin significantly reduces macrophages' inflammatory
response, inhibiting the production of IL-1β, IL-6, iNOS and TNF-α
while also reducing ROS production. The present results are
consistent with those of previous studies showing that Nrf2
activation is critical for controlling inflammation and preventing
cellular oxidative damage (34,35). The present study, to the best of
our knowledge, is the first to demonstrate that cynarin regulates
both oxidative stress and inflammation through the Nrf2-Keap1
signalling pathway. These results suggest that cynarin enhances the
antioxidant response by activating the Nrf2 pathway, thereby
inhibiting oxidative stress-driven OC differentiation. Compounds
such as curcumin, resveratrol and epigallocatechin-3-gallate (EGCG)
have been demonstrated to inhibit OC differentiation and activity
(26,36). However, curcumin, resveratrol and
EGCG exerted their effects mainly through inhibition of NF-κB and
MAPK pathways, while the additional activation of Nrf2-Keap1 by
cynarin provided anti-inflammatory effects. These findings,
therefore, highlight the therapeutic potential of cynarin and
provide valuable insights into its use as a dual-target therapeutic
agent for inflammatory bone diseases.
The RNA-seq results indicated that cynarin inhibited
RANKL-induced OC differentiation via the MAPK pathway. MAPK are
well-known regulators of cell proliferation and differentiation
(37,38). The current findings indicated
that cynarin significantly inhibited P38, JNK and ERK
phosphorylation. This inhibition was reversed by treatment with
anisomycin (a MAPK agonist). Our in vivo experiments have
also confirmed that moderate-dose cynarin treatment significantly
reduced the expression of p-P38, p-ERK1/2 and p-JNK1/2 compared
with LPS treatment and alendronate sodium. In conclusion, cynarin
suppressed RANKL-induced OC differentiation by targeting the MAPK
pathway.
Increasing evidence suggests a crosstalk between
MAPK and Nrf2 signalling during osteoclastogenesis. Nrf2 activation
elevates the expression of detoxifying enzymes (for example, HO-1
and NQO1) that reduce intracellular ROS, thereby attenuating
ROS-dependent MAPK activation and downstream osteoclastogenic
transcription factors such as NFATc1. In osseous echinococcosis,
Echinococcus granulosus-driven inhibition of Nrf2 shifts the
balance toward unchecked MAPK activation and OC differentiation
(39). By contrast, carnosic
acid-induced Nrf2 restores antioxidant defences and suppresses MAPK
phosphorylation, forming a negative feedback loop that limits
RANKL-driven osteoclastogenesis (40). However, the interaction between
the Nrf2 pathway and MAPK signalling in the present study remains
to be elucidated. In future studies, the authors plan to use Nrf2
inhibitors such as ML385 and Nrf2-specific small interfering RNA to
transfect OC precursor cells. It was further elucidated whether
Nrf2 knockdown acts upstream of MAPK inhibition. The effects of
cynarin on osteoblast function were also investigated. The results
showed that cynarin promoted the expression of key osteogenic
markers, such as OCN and RUNX2, in both in vivo and in
vitro models. A potential role of cynarin in supporting bone
formation has also been demonstrated. However, further studies are
required to fully understand how cynarin affects osteoblast
activity at the molecular level.
Beyond transcriptional regulation, epigenetic
alterations such as DNA methylation, histone
acetylation/deacetylation and non-coding RNA expression are
increasingly being recognised as diagnostic biomarkers and
therapeutic entry points for OC-driven bone loss. Clinically
applicable assays, such as methylation-specific PCR or targeted
bisulphite sequencing panels, can quantify the promoter methylation
of osteoclastogenic genes or inflammatory mediators. Circulating
microRNAs (for example miR-21, miR-155 and miR-223) have been
proposed as minimally invasive biomarkers that mirror OC activity
and inflammatory status. Notably, the concept of leveraging
nucleotide-level variation to stratify patients has already been
demonstrated in other inflammatory conditions. For example,
Antonino et al (41)
systematically reviewed single-nucleotide polymorphisms associated
with chronic rhinosinusitis, underscoring the translational value
of molecular variation in diagnosis and management. Given that both
the MAPK and Nrf2-Keap1 axes are subject to epigenetic regulation,
integrating epigenetic testing could help identify patients who are
most likely to benefit from cynarin or combination regimens.
In conclusion, the present study has identified
cynarin as a potent anti-inflammatory and bone-preserving agent and
demonstrated its ability to inhibit OC differentiation and modulate
inflammatory processes via the MAPK and Nrf2-Keap1 pathways for the
first time (Fig. 8). This
dual-target therapeutic strategy effectively addresses OC-driven
bone resorption while mitigating the inflammatory processes
associated with diseases, such as rheumatoid arthritis and
osteoporosis. Given the ability of cynarin to modulate bone
resorption and inflammation, future clinical applications should
explore its potential to enhance bone healing in implant therapies
or in combination treatments for chronic inflammatory bone
diseases.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The data generated in the
present study may be found in the NCBI under accession number
PRJNA1301579 or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1301579.
Authors' contributions
RC conceptualized the study, developed methodology
and wrote the original draft. YXW curated data and developed
methodology. ZL developed methodology and validated data. THW and
YM curated data. XRX and LS validated data. WFX and XZC
conceptualized the study. SYZ conceptualized and supervised the
study, and acquired funding. All authors contributed to writing,
reviewing and editing the manuscript. RC and SYZ confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Animal experiments were conducted in accordance with
the laboratory animal management guidelines. They were approved
(approval no. SH9H-2020-A1-1) by the Animal Ethics Committee of the
Ninth People's Hospital of Shanghai Jiao Tong University School of
Medicine (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 82370979 and 82301108), Shanghai's
Top Priority Research Center (grant no. 2022ZZ01017), CAMS
Innovation Fund for Medical Sciences (grant no. 2019-I2M-5-037),
Cross-disciplinary Research Fund of Shanghai Ninth People's
Hospital, Shanghai JiaoTong University School of Medicine (grant
no. JYJC202218) and the Program of Shanghai Academic/Technology
Research Leader (grant no. 21XD1431500).
References
|
1
|
Nakano S, Inoue K, Xu C, Deng Z,
Syrovatkina V, Vitone G, Zhao L, Huang XY and Zhao B: G-protein
Gα13 functions as a cytoskeletal and mitochondrial
regulator to restrain osteoclast function. Sci Rep. 9:42362019.
View Article : Google Scholar
|
|
2
|
Udagawa N, Koide M, Nakamura M, Nakamichi
Y, Yamashita T, Uehara S, Kobayashi Y, Furuya Y, Yasuda H, Fukuda C
and Tsuda E: Osteoclast differentiation by RANKL and OPG signaling
pathways. J Bone Miner Metab. 39:19–26. 2021. View Article : Google Scholar
|
|
3
|
Park-Min KH, Lim E, Lee MJ, Park SH,
Giannopoulou E, Yarilina A, van der Meulen M, Zhao B, Smithers N,
Witherington J, et al: Inhibition of osteoclastogenesis and
inflammatory bone resorption by targeting BET proteins and
epigenetic regulation. Nat Commun. 5:54182014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boyce BF, Li J, Yao Z and Xing L: Nuclear
factor-kappa B regulation of osteoclastogenesis and
osteoblastogenesis. Endocrinol Metab (Seoul). 38:504–521. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo Q, Jin Y, Chen X, Ye X, Shen X, Lin M,
Zeng C, Zhou T and Zhang J: NF-κB in biology and targeted therapy:
New insights and translational implications. Signal Transduct
Target Ther. 9:532024. View Article : Google Scholar
|
|
6
|
Liu L, Geng H, Mei C and Chen L:
Zoledronic acid enhanced the antitumor effect of cisplatin on
orthotopic osteosarcoma by ROS-PI3K/AKT signaling and attenuated
osteolysis. Oxid Med Cell Longev. 2021:66615342021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Place DE, Malireddi RKS, Kim J, Vogel P,
Yamamoto M and Kanneganti TD: Osteoclast fusion and bone loss are
restricted by interferon inducible guanylate binding proteins. Nat
Commun. 12:4962021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cauley JA, Barbour KE, Harrison SL,
Cloonan YK, Danielson ME, Ensrud KE, Fink HA, Orwoll ES and
Boudreau R: Inflammatory markers and the risk of hip and vertebral
fractures in men: The osteoporotic fractures in men (MrOS). J Bone
Miner Res. 31:2129–2138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Domazetovic V, Marcucci G, Iantomasi T,
Brandi ML and Vincenzini MT: Oxidative stress in bone remodeling:
Role of antioxidants. Clin Cases Miner Bone Metab. 14:209–216.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moreau MF, Guillet C, Massin P, Chevalier
S, Gascan H, Baslé MF and Chappard D: Comparative effects of five
bisphosphonates on apoptosis of macrophage cells in vitro. Biochem
Pharmacol. 73:718–723. 2007. View Article : Google Scholar
|
|
11
|
Kimachi K, Kajiya H, Nakayama S, Ikebe T
and Okabe K: Zoledronic acid inhibits RANK expression and migration
of osteoclast precursors during osteoclastogenesis. Naunyn
Schmiedebergs Arch Pharmacol. 383:297–308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sakaguchi O, Kokuryo S, Tsurushima H,
Tanaka J, Habu M, Uehara M, Nishihara T and Tominaga K:
Lipopolysaccharide aggravates bisphosphonate-induced osteonecrosis
in rats. Int J Oral Maxillofac Surg. 44:528–534. 2015. View Article : Google Scholar
|
|
13
|
Cohen SB, Dore RK, Lane NE, Ory PA,
Peterfy CG, Sharp JT, van der Heijde D, Zhou L, Tsuji W and Newmark
R; Denosumab Rheumatoid Arthritis Study Group: Denosumab treatment
effects on structural damage, bone mineral density, and bone
turnover in rheumatoid arthritis: A twelve-month, multicenter,
randomized, double-blind, placebo-controlled, phase II clinical
trial. Arthritis Rheum. 58:1299–1309. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adzet T, Camarasa J and Laguna JC:
Hepatoprotective activity of polyphenolic compounds from Cynara
scolymus against CCl4 toxicity in isolated rat hepatocytes. J Nat
Prod. 50:612–617. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Topal M, Gocer H, Topal F, Kalin P, Köse
LP, Gülçin İ, Çakmak KC, Küçük M, Durmaz L, Gören AC and Alwasel
SH: Antioxidant, antiradical, and anticholinergic properties of
cynarin purified from the Illyrian thistle (Onopordum illyricum
L.). J Enzyme Inhib Med Chem. 31:266–275. 2016. View Article : Google Scholar
|
|
16
|
Wu C, Chen S, Liu Y, Kong B, Yan W, Jiang
T, Tian H, Liu Z, Shi Q, Wang Y, et al: Cynarin suppresses gouty
arthritis induced by monosodium urate crystals. Bioengineered.
13:11782–11793. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen S, Tang S, Zhang C and Li Y: Cynarin
ameliorates dextran sulfate sodium-induced acute colitis in mice
through the STAT3/NF-κB pathway. Immunopharmacol Immunotoxicol.
46:107–116. 2024. View Article : Google Scholar
|
|
18
|
Rucci N, Zallone A and Teti A: Isolation
and generation of osteoclasts. Methods Mol Biol. 1914:3–19. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mao Y, Xie X, Sun G, Yu S, Ma M, Chao R,
Wan T, Xu W, Chen X, Sun L and Zhang S: Multifunctional prosthesis
surface: modification of titanium with cinnamaldehyde-loaded
hierarchical titanium dioxide nanotubes. Adv Healthc Mater.
13:e23033742024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Chen X, Li C, Cao X, Jia X, Chen X, Wang
Z, Xu W, Dai F and Zhang S: Mitochondria-targeted supramolecular
coordination container encapsulated with exogenous itaconate for
synergistic therapy of joint inflammation. Theranostics.
12:3251–3272. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Chen X, Zhou Z, Mao Y, Wang Y, Ma
Z, Xu W, Qin A and Zhang S: Nirogacestat suppresses RANKL-Induced
osteoclast formation in vitro and attenuates LPS-Induced bone
resorption in vivo. Exp Cell Res. 382:1114702019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Chen X, Zhou Z, Qin A, Wang Y, Fan
B, Xu W and Zhang S: LY411575, a potent γ-secretase inhibitor,
suppresses osteoclastogenesis in vitro and LPS-induced calvarial
osteolysis in vivo. J Cell Physiol. 234:20944–20956. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwarz EM, Benz EB, Lu AP, Goater JJ,
Mollano AV, Rosier RN, Puzas JE and Okeefe RJ: Quantitative
small-animal surrogate to evaluate drug efficacy in preventing wear
debris-induced osteolysis. J Orthop Res. 18:849–855. 2000.
View Article : Google Scholar
|
|
25
|
Tsutsumi R, Hock C, Bechtold CD, Proulx
ST, Bukata SV, Ito H, Awad HA, Nakamura T, O'Keefe RJ and Schwarz
EM: Differential effects of biologic versus bisphosphonate
inhibition of wear debris-induced osteolysis assessed by
longitudinal micro-CT. J Orthop Res. 26:1340–1346. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Inchingolo AD, Inchingolo AM, Malcangi G,
Avantario P, Azzollini D, Buongiorno S, Viapiano F, Campanelli M,
Ciocia AM, De Leonardis N, et al: Effects of resveratrol, curcumin
and quercetin supplementation on bone metabolism-a systematic
review. Nutrients. 14:35192022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu L, Guo Q, Yang J and Ni B: Tumor
necrosis factor alpha promotes osteoclast formation via PI3K/Akt
pathway-mediated blimp1 expression upregulation. J Cell Biochem.
118:1308–1315. 2017. View Article : Google Scholar
|
|
28
|
Lin TH, Tamaki Y, Pajarinen J, Waters HA,
Woo DK, Yao Z and Goodman SB: Chronic inflammation in
biomaterial-induced periprosthetic osteolysis: NF-κB as a
therapeutic target. Acta Biomater. 10:1–10. 2014. View Article : Google Scholar
|
|
29
|
Sakai E and Tsukuba T: Transcriptomic
characterization reveals mitochondrial involvement in
Nrf2/Keap1-mediated osteoclastogenesis. Antioxidants (Basel).
13:15752024. View Article : Google Scholar
|
|
30
|
Dong Y, Kang H, Peng R, Liu Z, Liao F, Hu
SA, Ding W, Wang P, Yang P, Zhu M, et al: A clinical-stage Nrf2
activator suppresses osteoclast differentiation via the
iron-ornithine axis. Cell Metab. 36:1679–1695.e6. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baird L and Dinkova-Kostova AT: The
cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol.
85:241–272. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tao H, Ge G, Liang X, Zhang W, Sun H, Li M
and Geng D: ROS signaling cascades: Dual regulations for osteoclast
and osteoblast. Acta Biochim Biophys Sin (Shanghai). 52:1055–1062.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Joo JH, Huh JE, Lee JH, Park DR, Lee Y,
Lee SG, Choi S, Lee HJ, Song SW, Jeong Y, et al: A novel pyrazole
derivative protects from ovariectomy-induced osteoporosis through
the inhibition of NADPH oxidase. Sci Rep. 6:223892016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li X, Chen Y, Mao Y, Dai P, Sun X, Zhang
X, Cheng H, Wang Y, Banda I, Wu G, et al: Curcumin protects
osteoblasts from oxidative stress-induced dysfunction via
GSK3β-Nrf2 signaling pathway. Front Bioeng Biotechnol. 8:6252020.
View Article : Google Scholar
|
|
35
|
Saha S, Buttari B, Panieri E, Profumo E
and Saso L: An overview of Nrf2 signaling pathway and its role in
inflammation. Molecules. 25:54742020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu H, Liu T, Jia Y, Li J, Jiang L, Hu C,
Wang X and Sheng J: (-)-Epigallocatechin-3-gallate inhibits
osteoclastogenesis by blocking RANKL-RANK interaction and
suppressing NF-κB and MAPK signaling pathways. Int Immunopharmacol.
95:1074642021. View Article : Google Scholar
|
|
37
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Morrison DK: MAP kinase pathways. Cold
Spring Harb Perspect Biol. 4:a0112542012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Y, Li J, Zhang Z, Li Q, Tian Y, Wang
S, Shi C and Sun H: Echinococcus granulosus promotes MAPK
pathway-mediated osteoclast differentiation by inhibiting Nrf2 in
osseous echinococcosis. Vet Res. 56:812025. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Thummuri D, Naidu VGM and Chaudhari P:
Carnosic acid attenuates RANKL-induced oxidative stress and
osteoclastogenesis via induction of Nrf2 and suppression of NF-κB
and MAPK signalling. J Mol Med (Berl). 95:1065–1076. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Antonino M, Nicolò M, Jerome Renee L,
Federico M, Chiara V, Stefano S, Maria S, Salvatore C, Antonio B,
Calvo-Henriquez C, et al: Single-nucleotide polymorphism in chronic
rhinosinusitis: A systematic review. Clin Otolaryngol. 47:14–23.
2022. View Article : Google Scholar
|