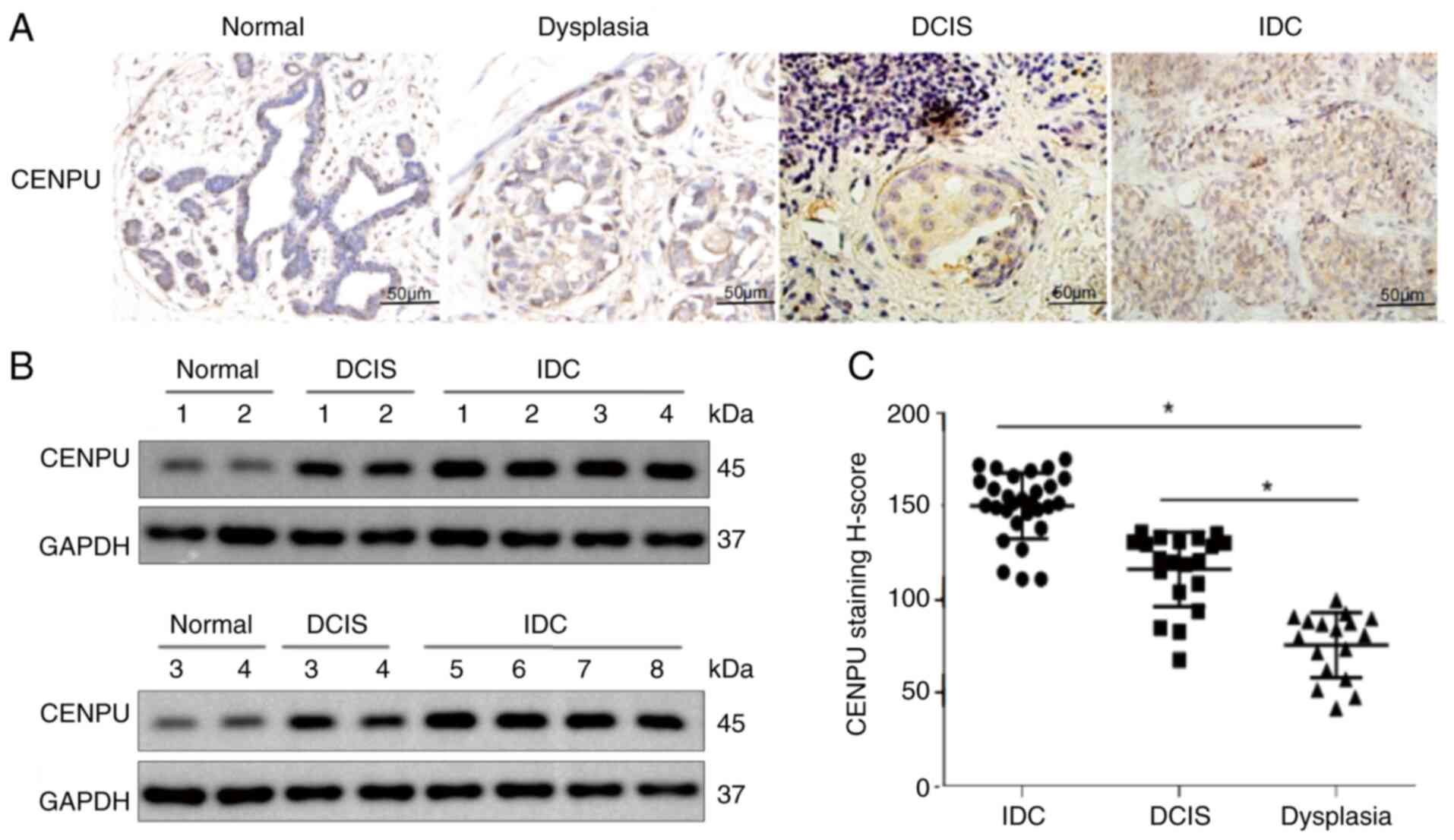
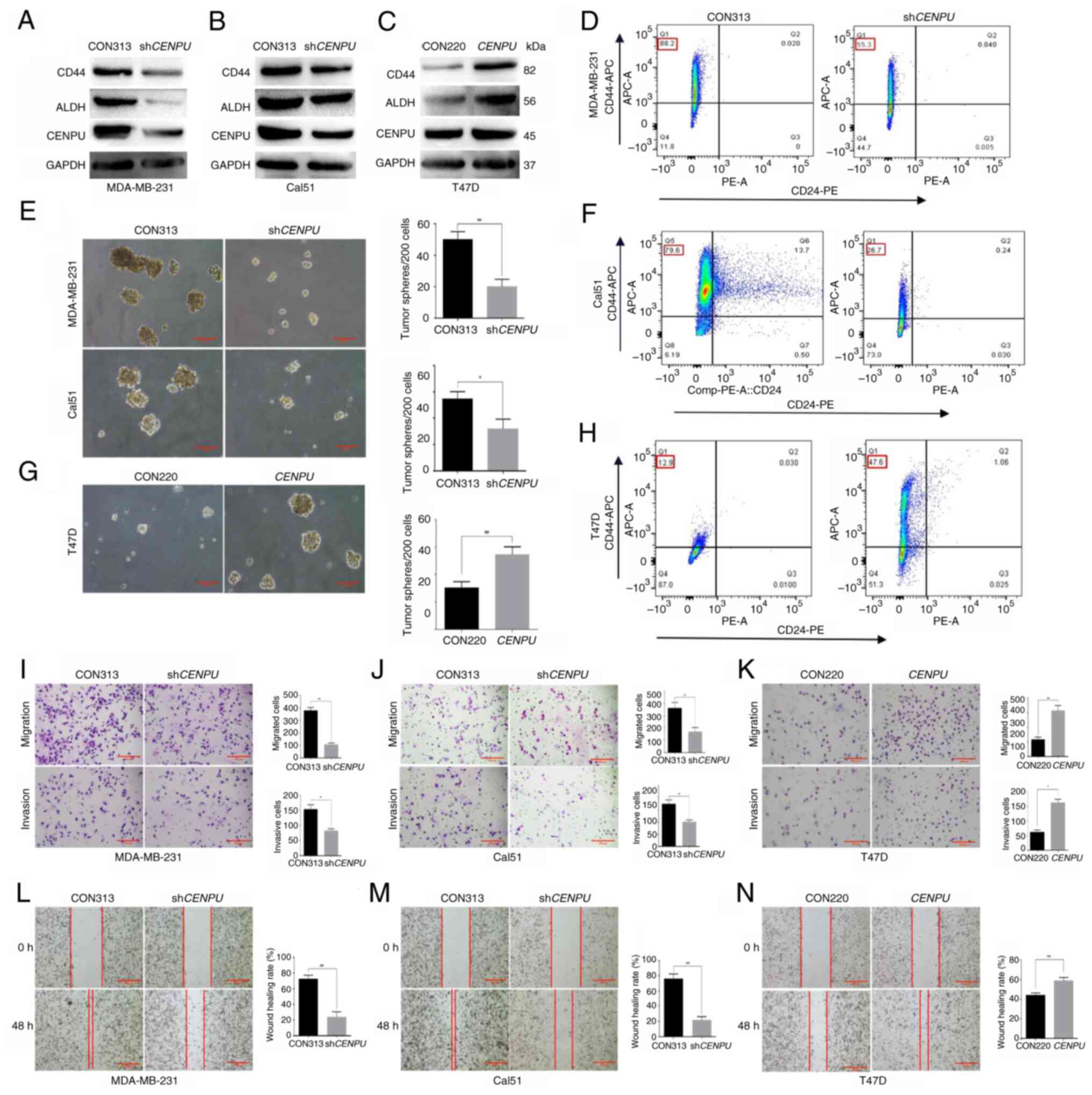
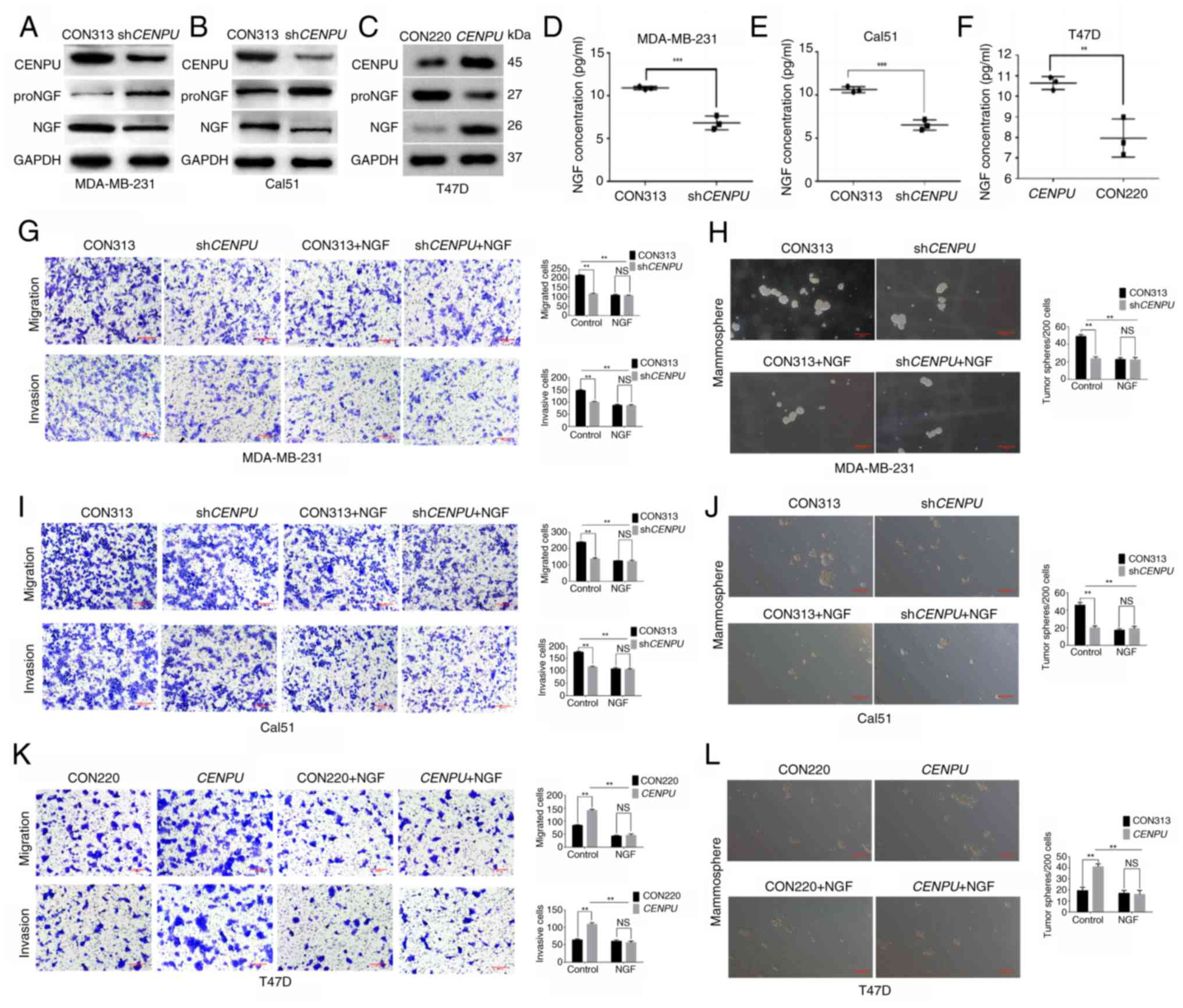
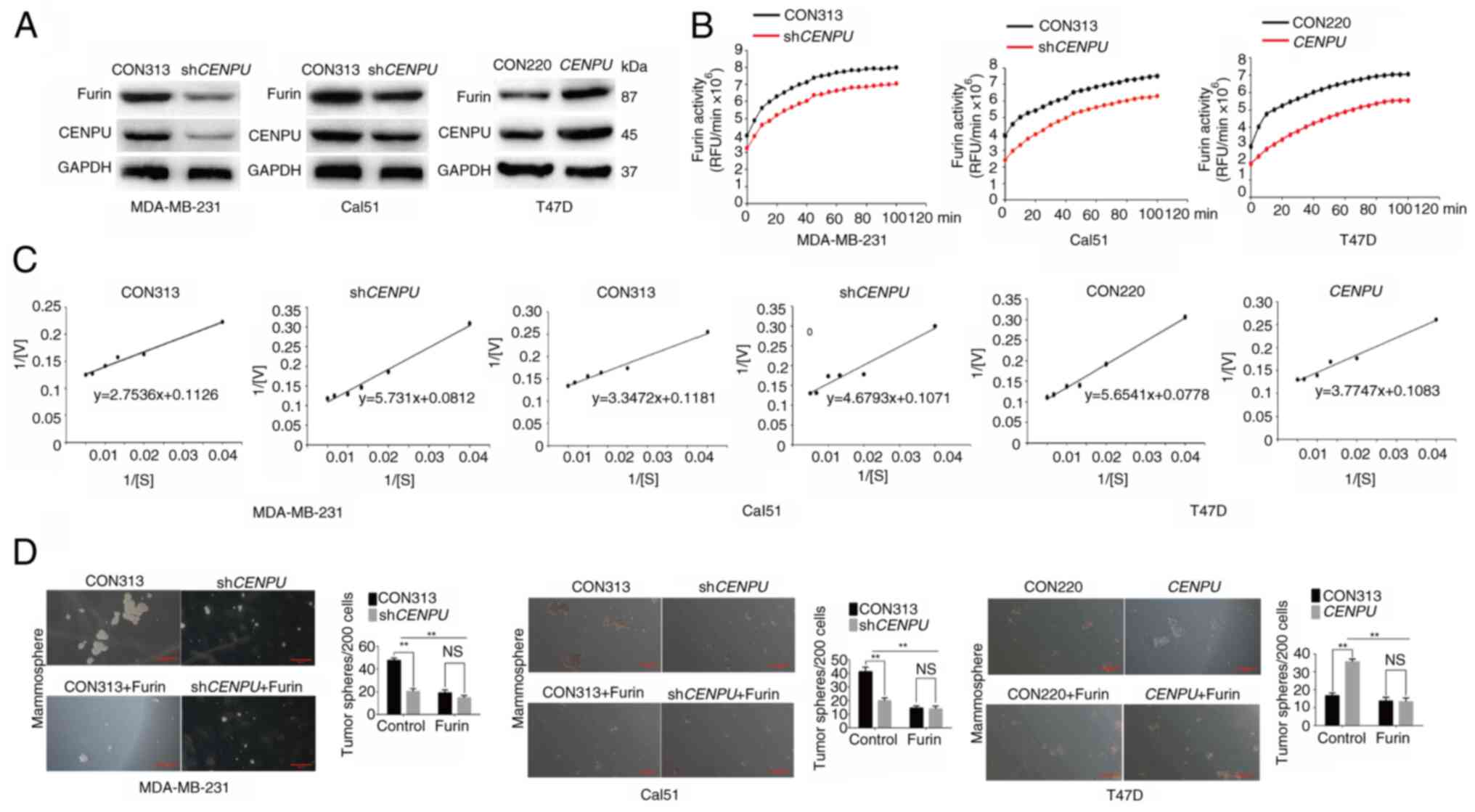
|
1
|
Derakhshan F and Reis-Filho JS:
Pathogenesis of triple-negative breast cancer. Annu Rev Pathol.
17:181–204. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahtani R, Kittaneh M, Kalinsky K,
Mamounas E, Badve S, Vogel C, Lower E, Schwartzberg L and Pegram M;
Breast Cancer Therapy Expert Group (BCTEG): Advances in therapeutic
approaches for triple-negative breast cancer. Clin Breast Cancer.
21:383–390. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang M, Deng H, Hu R, Chen F, Dong S,
Zhang S, Guo W, Yang W and Chen W: Patterns and prognostic
implications of distant metastasis in breast cancer based on SEER
population data. Sci Rep. 15:267172025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao S, Zuo WJ, Shao ZM and Jiang YZ:
Molecular subtypes and precision treatment of triple-negative
breast cancer. Ann Transl Med. 8:4992020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y, Zhan Z, Yin X, Fu S and Deng X:
Targeted therapeutic strategies for triple-negative breast cancer.
Front Oncol. 11:7315352021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bou Zerdan M, Ghorayeb T, Saliba F, Allam
S, Bou Zerdan M, Yaghi M, Bilani N, Jaafar R and Nahleh Z: Triple
negative breast cancer: Updates on classification and treatment in
2021. Cancers (Basel). 14:12532022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang R, Tu J and Liu S: Novel molecular
regulators of breast cancer stem cell plasticity and heterogeneity.
Semin Cancer Biol. 82:11–25. 2022. View Article : Google Scholar
|
|
8
|
Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y,
Jiang G, Lu M, Zhang Z, Yin J, et al: Downregulation of FOXO3a by
DNMT1 promotes breast cancer stem cell properties and
tumorigenesis. Cell Death Differ. 27:966–983. 2020. View Article : Google Scholar :
|
|
9
|
Zhu K, Xie V and Huang S: Epigenetic
regulation of cancer stem cell and tumorigenesis. Adv Cancer Res.
148:1–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo F, Zhang M, Sun B, Xu C, Yang Y, Zhang
Y, Li S, Chen G, Chen C, Li Y and Feng H: LINC00115 promotes
chemoresistant breast cancer stem-like cell stemness and metastasis
through SETDB1/PLK3/HIF1α signaling. Mol Cancer. 23:602024.
View Article : Google Scholar
|
|
12
|
Lu H, Chen I, Shimoda LA, Park Y, Zhang C,
Tran L, Zhang H and Semenza GL: Chemotherapy-induced
Ca2+ release stimulates breast cancer stem cell
enrichment. Cell Rep. 18:1946–1957. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mannello F: Understanding breast cancer
stem cell heterogeneity: Time to move on to a new research
paradigm. BMC Med. 11:1692013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh P, Pesenti ME, Maffini S, Carmignani
S, Hedtfeld M, Petrovic A, Srinivasamani A, Bange T and Musacchio
A: BUB1 and CENP-U, primed by CDK1, are the main PLK1 kinetochore
receptors in mitosis. Mol Cell. 81:67–87.e9. 2021. View Article : Google Scholar :
|
|
15
|
Pan T, Zhou D, Shi Z, Qiu Y, Zhou G, Liu
J, Yang Q, Cao L and Zhang J: Centromere protein U (CENPU) enhances
angiogenesis in triple-negative breast cancer by inhibiting
ubiquitin-proteasomal degradation of COX-2. Cancer Lett.
482:102–111. 2020. View Article : Google Scholar
|
|
16
|
Lou Y, Lu J, Zhang Y, Gu P, Wang H, Qian
F, Zhou W, Zhang W, Zhong H and Han B: The centromere-associated
protein CENPU promotes cell proliferation, migration, and
invasiveness in lung adenocarcinoma. Cancer Lett. 532:2155992022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bruno F, Arcuri D, Vozzo F, Malvaso A,
Montesanto A and Maletta R: Expression and signaling pathways of
nerve growth factor (NGF) and pro-NGF in breast cancer: A
systematic review. Curr Oncol. 29:8103–8120. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marsland M, Dowdell A, Jiang CC, Wilmott
JS, Scolyer RA, Zhang XD, Hondermarck H and Faulkner S: Expression
of NGF/proNGF and their receptors TrkA, p75NTR and
sortilin in melanoma. Int J Mol Sci. 23:42602022. View Article : Google Scholar
|
|
19
|
Morisse M, Bourhis T, Lévêque R, Guilbert
M, Cicero J, Palma M, Chevalier D, le Bourhis X, Toillon RA and
Mouawad F: Influence of EGF and pro-NGF on EGFR/SORTILIN
interaction and clinical impact in head and neck squamous cell
carcinoma. Front Oncol. 13:6617752023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bradshaw RA, Pundavela J, Biarc J,
Chalkley RJ, Burlingame AL and Hondermarck H: NGF and ProNGF:
Regulation of neuronal and neoplastic responses through receptor
signaling. Adv Biol Regul. 58:16–27. 2015. View Article : Google Scholar :
|
|
21
|
Marcinkiewicz M, Marcinkiewicz J, Chen A,
Leclaire F, Chrétien M and Richardson P: Nerve growth factor and
proprotein convertases furin and PC7 in transected sciatic nerves
and in nerve segments cultured in conditioned media: Their presence
in Schwann cells, macrophages, and smooth muscle cells. J Comp
Neurol. 403:471–485. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan R, Yalinca H, Paoletti F, Gobbo F,
Marchetti L, Kuzmanic A, Lamba D, Gervasio FL, Konarev PV, Cattaneo
A and Pastore A: The structure of the pro-domain of mouse proNGF in
contact with the NGF domain. Structure. 27:78–89.e3. 2019.
View Article : Google Scholar
|
|
23
|
Thomas G: Furin at the cutting edge: From
protein traffic to embryogenesis and disease. Nat Rev Mol Cell
Biol. 3:753–766. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
He Z, Khatib AM and Creemers JWM: The
proprotein convertase furin in cancer: More than an oncogene.
Oncogene. 41:1252–1262. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Regua AT, Aguayo NR, Jalboush SA, Doheny
DL, Manore SG, Zhu D, Wong GL, Arrigo A, Wagner CJ, Yu Y, et al:
TrkA interacts with and phosphorylates STAT3 to enhance gene
transcription and promote breast cancer stem cells in
triple-negative and HER2-enriched breast cancers. Cancers (Basel).
13:23402021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Raninga PV, Zeng B, Moi D, Trethowan E,
Saletta F, Venkat P, Mayoh C, D'Souza RCJ, Day BW, Shai-Hee T, et
al: CBL0137 and NKG2A blockade: A novel immuno-oncology combination
therapy for Myc-overexpressing triple-negative breast cancers.
Oncogene. 44:893–908. 2025. View Article : Google Scholar :
|
|
28
|
Tang X, Cromwell CR, Liu R, Godbout R,
Hubbard BP, McMullen TPW and Brindley DN: Lipid phosphate
phosphatase-2 promotes tumor growth through increased c-Myc
expression. Theranostics. 12:5675–5690. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
American Veterinary Medical Association:
AVMA guidelines for the euthanasia of animals. 2020 edition. AVMA;
Schaumburg, IL: 2020
|
|
30
|
Yan Y, Zuo X and Wei D: Concise review:
Emerging role of CD44 in cancer stem cells: A promising biomarker
and therapeutic target. Stem Cells Transl Med. 4:1033–1043. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Namekawa T, Ikeda K, Horie-Inoue K, Suzuki
T, Okamoto K, Ichikawa T, Yano A, Kawakami S and Inoue S: ALDH1A1
in patient-derived bladder cancer spheroids activates retinoic acid
signaling leading to TUBB3 overexpression and tumor progression.
Int J Cancer. 146:1099–1113. 2020. View Article : Google Scholar
|
|
32
|
Ghebeh H, Sleiman GM, Manogaran PS,
Al-Mazrou A, Barhoush E, Al-Mohanna FH, Tulbah A, Al-Faqeeh K and
Adra CN: Profiling of normal and malignant breast tissue show
CD44high/CD24low phenotype as a predominant stem/progenitor marker
when used in combination with Ep-CAM/CD49f markers. BMC Cancer.
13:2892013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
DA Cruz Paula A and Lopes C: Implications
of different cancer stem cell phenotypes in breast cancer.
Anticancer Res. 37:2173–2183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seidah NG, Benjannet S, Pareek S, Savaria
D, Hamelin J, Goulet B, Laliberte J, Lazure C, Chrétien M and
Murphy RA: Cellular processing of the nerve growth factor precursor
by the mammalian pro-protein convertases. Biochem J. 314:951–960.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deichmann U, Schuster S, Mazat JP and
Cornish-Bowden A: Commemorating the 1913 michaelis-menten paper die
kinetik der invertinwirkung: Three perspectives. FEBS J.
281:435–463. 2014. View Article : Google Scholar
|
|
36
|
Hanissian SH, Akbar U, Teng B, Janjetovic
Z, Hoffmann A, Hitzler JK, Iscove N, Hamre K, Du X, Tong Y, et al:
cDNA cloning and characterization of a novel gene encoding the
MLF1-interacting protein MLF1IP. Oncogene. 23:3700–3707. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Goldberg AL: Protein degradation and
protection against misfolded or damaged proteins. Nature.
426:895–899. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lim KC, Tyler CM, Lim ST, Giuliano R and
Federoff HJ: Proteolytic processing of proNGF is necessary for
mature NGF regulated secretion from neurons. Biochem Biophys Res
Commun. 361:599–604. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hayakawa Y, Sakitani K, Konishi M, Asfaha
S, Niikura R, Tomita H, Renz BW, Tailor Y, Macchini M, Middelhoff
M, et al: Nerve growth factor promotes gastric tumorigenesis
through aberrant cholinergic signaling. Cancer Cell. 31:21–34.
2017. View Article : Google Scholar :
|
|
40
|
Tomellini E, Touil Y, Lagadec C, Julien S,
Ostyn P, Ziental-Gelus N, Meignan S, Lengrand J, Adriaenssens E,
Polakowska R and Le Bourhis X: Nerve growth factor and proNGF
simultaneously promote symmetric self-renewal, quiescence, and
epithelial to mesenchymal transition to enlarge the breast cancer
stem cell compartment. Stem Cells. 33:342–353. 2015. View Article : Google Scholar
|
|
41
|
Rockwell NC, Krysan DJ, Komiyama T and
Fuller RS: Precursor processing by kex2/furin proteases. Chem Rev.
102:4525–4548. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen C, Gupta P, Parashar D, Nair GG,
George J, Geethadevi A, Wang W, Tsaih SW, Bradley W, Ramchandran R,
et al: ERBB3-induced furin promotes the progression and metastasis
of ovarian cancer via the IGF1R/STAT3 signaling axis. Oncogene.
39:2921–2933. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou B and Gao S: Pan-cancer analysis of
FURIN as a potential prognostic and immunological biomarker. Front
Mol Biosci. 8:6484022021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ji S, Li S, Gao H, Wang J, Wang K, Nan W,
Chen H and Hao Y: An AIEgen-based 'turn-on' probe for sensing
cancer cells and tiny tumors with high furin expression. Biomater
Sci. 11:2221–2229. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen M, Pan Y, Liu H, Ning F, Lu Q, Duan
Y, Gan X, Lu S, Hou H, Zhang M, et al: Ezrin accelerates breast
cancer liver metastasis through promoting furin-like
convertase-mediated cleavage of Notch1. Cell Oncol (Dordr).
46:571–587. 2023. View Article : Google Scholar
|
|
46
|
Liu Z, Gu X, Li Z, Shan S, Wu F and Ren T:
Heterogeneous expression of ACE2, TMPRSS2, and FURIN at single-cell
resolution in advanced non-small cell lung cancer. J Cancer Res
Clin Oncol. 149:3563–3573. 2023. View Article : Google Scholar
|
|
47
|
Schaale D, Laspa Z, Balmes A, Sigle M,
Dicenta-Baunach V, Hochuli R, Fu X, Serafimov K, Castor T, Harm T,
et al: Hemin promotes platelet activation and plasma membrane
disintegration regulated by the subtilisin-like proprotein
convertase furin. FASEB J. 38:e701552024. View Article : Google Scholar : PubMed/NCBI
|