Introduction
Triple-negative breast cancer (TNBC) is a distinct
BC subtype characterized by the absence of estrogen receptor,
progesterone receptor and human epidermal growth factor receptor 2
expression, along with unique molecular expression characteristics
(such as frequent mutations in TP53 and PIK3CA, and
dysregulation of MYC, EGFR and Wnt/β-catenin pathways), biological
behavior (such as early visceral metastasis and 'metastatic burst'
within 3 years post-diagnosis) and clinicopathological features
(such as higher histological grade, stromal tumor-infiltrating
lymphocytes and poorer 5-year survival) (1). TNBC exhibits a high rate of distant
metastasis (liver: 34.5%; brain: 9.8%; lung: 32.4%), high odds of
visceral and brain metastases, rapid progression (3-year distant
metastasis-free rate: 57.9% vs. 85.2% in luminal A; 1-year
recurrence rate after surgery: 18.4%), and limited treatment
options (2-6). Understanding the molecular basis of
TNBC occurrence and progression and identifying efficient
therapeutic targets are key issues in TNBC treatment that warrant
urgent attention.
BC stem cells (BCSCs) are tumor-initiating cells
that can self-renew and are highly tumorigenic. An increasing
amount of studies suggest that cancer stem cells promote
tumorigenesis, metastasis, recurrence and drug resistance as well
as are the major source for heterogeneity of cancer cells (7-9).
In 2003, Al-Hajj et al (10) purified BCSCs from BC patient
samples for the first time and showed that tumor stem cells, even
in limited quantities, induced tumorigenesis in immunodeficient
mice. At present, strategies for identifying BCSCs are primarily
based on two key biological characteristics of BCSCs, namely their
self-renewal ability and their differentiation ability, which
enable them to evade the effects of most cytotoxic chemotherapeutic
drugs (11). BCSCs also
frequently exhibit high levels of drug efflux and a superior
metabolic capacity. This makes them more likely to survive
chemotherapy than non-BCSCs, which leads to tumor recurrence and
metastasis (12,13). Therefore, combined treatment with
cancer stem cell antagonists and conventional cancer treatment
tools is a promising therapeutic approach for reducing BC
recurrence and metastasis.
The centromere protein U (CENPU) gene,
located on chromosome 4q35.1, participates in mitosis as an
important part of the centromere complex (14). In our previous study, it was
demonstrated that CENPU promoted angiogenesis in TNBC by
suppressing the ubiquitin-related degradation of cyclooxygenase-2
(15). In non-small cell lung
cancer, CENPU enhances the proliferative, migratory and invasive
potential of cells by activating the Wnt/β-catenin pathway
(16). However, the mechanisms
underlying the effects of CENPU on BC formation are not
well-documented.
Nerve growth factor (NGF) is an important
neurotrophic factor involved in the regulation of cell
differentiation and target neuron survival. Both NGF and NGF
precursor protein (proNGF) are biologically active, but the two
proteins may have contrasting biological functions (17-20). The maturation of proNGF to NGF
requires cleavage by furin precursor protein-processing enzymes
(21,22).
Furin is the earliest discovered mammalian precursor
protein-processing enzyme and is primarily responsible for the
cleavage of functional proteins in an organism (23). Excessive or aberrant cleavage by
furin is associated with diseases, such as cancer (24). The shearing action of furin on
proNGF contributes to the paradoxical biological function of its
downstream receptors that activate different pathways depending on
the form of its ligand. Unprocessed proNGF exerts its
apoptosis-promoting effects by binding to low-affinity p75
neurotrophin receptor. By contrast, furin-processed NGF mediates
survival in tumor cells through high-affinity binding to the
tropomyosin receptor kinase A proto-oncogene receptor, which
interacts with and phosphorylates STAT3 to enhance gene
transcription and promote BCSC properties in TNBC and HER2-enriched
BC (20,25).
The study of BCSCs and their role in tumor
progression is crucial for developing novel therapeutic strategies.
The objective of the present study was to investigate the
expression of CENPU in different types of breast lesions and its
potential role in BCSC self-renewal, tumorigenesis and the invasive
behavior of BC cells. Understanding the mechanisms underlying CENPU
function could provide insights into the molecular pathways that
drive BC aggression and metastasis. To achieve these aims, a
combination of immunohistochemistry, functional assays and
mechanistic studies was employed to explore the expression patterns
and functional significance of CENPU in BC cell lines.
Materials and methods
Cell culture and treatment
Human MDA-MB-231 (TNBC) and T47D (BC) cells were
acquired from the American Type Culture Collection. Cal51 (TNBC)
cells were acquired from the German Collection of Microorganisms
and Cell Cultures GmbH. Cell culture was performed as previously
described (15). Specifically,
Cal51 was cultured in 15% DMEM/F12 medium, MDA-MB-231 in 10% DMEM
high-glucose medium, and T47D (luminal) in 10% RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.). All the media were
supplemented with 10% FBS (cat. no. 04-001-1ACS; Biological
Industries; Sartorius AG) and 1% penicillin-streptomycin (cat. no.
P1400; Beijing Solarbio Science & Technology Co., Ltd.). The
cells were maintained in a humidified incubator at 37°C with 5%
CO2 and sub-cultured at 75-85% confluence. Cycloheximide
(CHX; cat. no. HY-12320) (20 μM) was purchased from
MedChemExpress. MG132 (cat. no. Y210207) (10 μM) and
chloroquine (CQ; cat. no. Y210184) (100 μM) were purchased
from Beyotime Institute of Biotechnology. Cell treatments were
performed at 37°C for 12 h. NGF-neutralizing antibody (cat. no.
ALM-006-250UG; Thermo Fisher Scientific, Inc.) was used at a final
concentration of 1 μg/ml in complete culture medium at 37°C
for 24 h before subsequent assays.
Immunoblotting
MDA-MB-231, T47D and Cal51 cells were lysed with
RIPA lysis buffer (Beyotime Institute of Biotechnology)
supplemented with 1% PMSF. Cellular proteins were quantified using
a BCA Assay kit (MilliporeSigma). Equal volumes of total protein
(30 μg) were resolved using 10% SDS-PAGE and
electro-transferred onto PVDF membranes (MilliporeSigma), and the
membranes were blocked with 5% non-fat milk for 1 h at room
temperature. Next, the membranes were incubated with primary
antibodies overnight at 4°C. The primary antibodies used were as
follows: CENPU (1:1,000; cat. no. YN1585; Immunoway Biotechnology
Company), aldehyde dehydrogenase (ALDH; 0.5 μg/ml; cat. no.
ab131068; Abcam), CD44 (1:1,000; cat. no. ab243894; Abcam), NGF (1
μg/ml; cat. no. ab6199; Abcam), proNGF (1:500; cat. no.
ab221609; Abcam), furin (1:1,000; cat. no. 18413-1-AP; Proteintech
Group, Inc.) and GAPDH (1:10,000; cat. no. 60004-1-Ig; Proteintech
Group, Inc.). Subsequently, incubation with HRP-conjugated
secondary antibodies for 1 h at room temperature was performed. The
secondary antibodies used were anti-mouse HRP-conjugated IgG
(1:4,500; cat. no. AS003; ABclonal Biotech Co., Ltd.) and
anti-rabbit HRP-conjugated IgG (1:4,500; cat. no. AS014; ABclonal
Biotech Co., Ltd.). Detection was performed using an ECL system
(PerkinElmer, Inc.). The target protein levels were normalized to
those of GAPDH. The band densities were analyzed using ImageJ
software (version 1.50i; National Institutes of Health).
Clinical specimen collection
BC and adjacent (>2 cm from the cancer tissue)
non-cancerous breast tissue specimens from 67 female patients
(median age, 45 years; range, 32-65 years) enrolled between January
2023 and June 2024 in Shandong Cancer Hospital and Institute,
Shandong First Medical University and Shandong Academy of Medical
Sciences (Jinan, China) were used in immunoblotting and
immunohistochemistry (IHC) experiments. The study protocol was
approved by the Institutional Review Board of Shandong Cancer
Hospital and Institute, Shandong First Medical University and
Shandong Academy of Medical Sciences (approval no.
SDTHEC2022009017; Jinan, China). The patient inclusion criteria
were as follows: i) First diagnosis of breast cancer; ii) patients
had not received any chemotherapy or hormone treatment; iii)
without breast surgery history; and iv) without motion artifact.
Breast cancer diagnosis was validated using ultrasound and MRI.
IHC
After being fixed in 10% neutral buffered formalin
at room temperature for 24 h, the tissue was embedded in paraffin
and cut into 5-μm sections. Tissue sections were dewaxed by
treatment with xylene, followed by rehydration in an ethanol
gradient (5 min each). Subsequently, the sections were treated with
an antigen retrieval solution in a microwave oven for 10 min,
incubated in 3% H2O2 at 37°C for 15 min,
washed with PBS and blocked with 3% BSA (cat. no. A8020; Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
for 30 min. The sections were then incubated with anti-CENPU
(1:100; cat. no. 13186-1-AP; Proteintech Group, Inc.), anti-proNGF
(1:200; cat. no. ANT-005; Alomone Labs), anti-NGF (1:300; cat. no.
ab216419; Abcam) and anti-furin (10 μg/ml; cat. no.
ab231573; Abcam) antibodies overnight at 4°C. Next, the sections
were incubated with secondary antibodies (1:1,000; cat. no. ab6721;
Abcam) at room temperature for 1 h. The signal was developed using
a 3,3'-diaminobenzidine color reagent, and the sections were
stained with hematoxylin at 37°C for 3 min, sealed and observed
under a light microscope (Nikon Corporation). Stained slides
scanned by Panoramic SCAN II (3DHISTECH, Ltd.) and images were
captured by CaseViewer software (version 2.9.0; 3dhistech.
com/news/caseviewer-becomes-slideviewer/). The H-score is
determined by the formula H-SCORE=∑(PI × I), where PI represents
the percentage of cells with a specific staining intensity and I
represents the corresponding staining intensity level.
Specifically, it is computed as (percentage of weakly-stained cells
x1) + (percentage of moderately-stained cells x2) + (percentage of
strongly-stained cells x3). The resulting H-score ranges from 0 to
300. The staining intensity and extent of positivity were evaluated
using the aforementioned scoring system to provide a
semi-quantitative estimation of overall staining patterns. The
composite scores were used to compare expression trends among
groups.
Transfection
A short hairpin RNA (sh/shRNA) against CENPU
(shCENPU) and a scrambled control shRNA (CON313) from
Syngentech Co., Ltd. were used. The target sequences of the shRNAs
were as follows: CON313, 5'-TTCTCCGAACGTGTCACGT-3'; and
shCENPU, 5'-CAGGTATGAGCTATAATAA-3'. Cal51 and MDA-MB-231
cells were infected with shRNA-encoding lentiviruses to generate
stable cell lines. The lentiviral transfer vector pLKO.1-puro was
utilized within a third-generation system comprising packaging
plasmids pMD2.G and psPAX2. Transient transfection was performed in
293T cells (American Type Culture Collection) using Lipofectamine
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) with 4 μg
total DNA at a 4:3:1 ratio (transfer vector:psPAX2:pMD2.G).
Transfected cells were maintained at 37°C for 72 h, and viral
supernatants were collected at 48/72 h, filtered (0.45 μm)
and concentrated by centrifugation (16 h at 6,800 rpm) at 4°C.
Target cells (MDA-MB-231/Cal51) were infected at MOI=10 with 8
μg/ml polybrene for 24 h. Post-transduction, cells underwent
puromycin selection (2 μg/ml for MDA-MB-231; 1 μg/ml
for Cal51) for 7 days, followed by maintenance at half-selection
dose. Functional assays were conducted ≥14 days post-selection to
ensure stable expression.
A CENPU overexpression plasmid and control
plasmid (CON220) from Syngentech Co., Ltd. were also used.
Transient transfection of MDA-MB-231 and Cal51 cells used
pcDNA3.1(+) with 2 μg plasmid DNA (A260/A280=1.85±0.05) per
35-mm well dish. Transfections employed Lipofectamine 3000 at a 1:2
DNA (μg):reagent (μl) ratio. Cells were incubated
with DNA-lipid complexes at 37°C for 6 h before medium replacement.
Further analyses were performed 24-48 h post-transfection.
ELISA
ELISA was performed using MDA-MB-231, Cal51 and T47D
cell lysates. Cells were resuspended in PBS at a density of
1×106 cells/ml and subjected to ultrasonication on ice
using a Covaris E220 system (Covaris, LLC) under the following
parameters: 20 kHz frequency, 10-sec pulses per cycle (3 cycles in
total), with 10-sec intervals between pulses. After centrifugation
at 2,000-3,000 rpm at 2-8°C for 20 min, the supernatant was
collected and ELISA was performed using an NGF ELISA kit (cat. no.
LP-H017695; Shanghai Lanpai Biotechnology Co., Ltd.).
Cell migration and invasion assays
A wound healing assay was performed to assess the
migratory potential of cells. MDA-MB-231, Cal51 and T47D cells were
seeded in 6-well plates and cultured until the confluency was
>90%. After serum starvation, the cells were scratched with a
10-μl pipette tip to artificially create wounds on the cell
surface, and were imaged under a light microscope at 0 and 48 h.
ImageJ software (version 1.50i) was used for quantification of the
wound.
A Matrigel-precoated Transwell chamber (with
8-μm pores) was used for the invasion assay, while the
migration assay was performed using Transwell inserts without
Matrigel precoating, as previously described (15). A total of 150 μl Matrigel
diluent (Matrigel:DMEM=1:8; cat. no. 356234; Corning, Inc.) was
spread evenly on the cell inserts, which were placed in a 37°C and
5% CO2 incubator until the Matrigel solidified. A total
of 5×104 cells/ml were diluted in DMEM without FBS. A
total of 150 μl of the diluted cells were placed in the
upper layer of the Transwell chamber and 600 μl of DMEM
supplemented with 20% FBS was added to the lower chamber. Following
incubation (10-12 h for MDA-MB-231 and Cal51 cells and 24-36 h for
T47D cells), the cells in the upper chamber were gently wiped with
a cotton swab, fixed with 4% paraformaldehyde at room temperature
for 15 min, stained with 0.1% crystal violet at room temperature
for 15 min and photographed under a light microscope. ImageJ
software (version 1.50i) was used for quantification.
Mammosphere formation assay
MDA-MB-231, Cal51 and T47D cells in the logarithmic
growth phase were collected, washed twice with serum-free DMEM/F12,
counted, and resuspended in DMEM/F12 without serum, supplemented
with B27, 20 ng/ml epidermal growth factor and 20 ng/ml basic
fibroblast growth factor (all from Invitrogen; Thermo Fisher
Scientific, Inc.). A total of 2×103 cells/well were
seeded in an ultra-low adhesion culture plate (Corning, Inc.).
After 7 days of culture at 37°C, images were acquired under an
inverted light microscope and the mammospheres formed (defined as
cell clusters with a diameter ≥30 μm, with clear boundaries
and intact morphology) were counted. ImageJ software (version
1.50i) was used for quantification.
Co-immunoprecipitation
Lysates from MDA-MB-231 and Cal51 cells prepared in
RIPA buffer (Thermo Fisher Scientific, Inc.) were centrifuged at
12,000 × g, 4°C for 10 min to remove the debris. Per reaction, 500
μg total protein was incubated overnight with: Anti-CENPU
(1:2,000, cat. no. YN1585; ImmunoWay Biotechnology Company),
anti-furin (1:1,000, cat. no. ab3467; Abcam), anti-proNGF (1:1,000;
cat. no. ab52918; Abcam) or rabbit IgG control (2
μg/reaction; cat. no. 2729; Cell Signaling Technology,
Inc.). Immune complexes were captured using the Pierce Co-IP kit
(cat. no. 88804; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions, employing Protein A/G magnetic beads
(cat. no. 88802; Thermo Fisher Scientific, Inc.). The precipitated
proteins were analyzed by western blotting.
4D-data-independent acquisition (DIA)
quantitative proteomics
MDA-MB-231 cell lines transfected with the control
vector CON313, with knockdown of the CENPU gene (shCENPU),
and overexpression of CENPU were used, and the protein precipitates
obtained after acetone precipitation of the cell lysates were
collected as experimental samples. The samples were treated with
liquid nitrogen and grinded into powder, and were subsequently
incubated in lysis buffer (7 M urea, 2 M thiourea, 4% SDS, 40 mM
Tris-HCl; pH 8.5) containing 1 mM PMSF and 2 mM EDTA (final
concentration) at 4°C for 5 min. The samples were mixed thoroughly
and sonicated using an ultrasonic disruptor at 25% power for 10
min. Subsequently, they were centrifuged at 12,000 × g at 4°C for
10 min, and the supernatant was collected. Four times the volume of
cold acetone was added to the supernatant, and it was allowed to
precipitate overnight at −20°C, followed by centrifugation at
12,000 × g at 4°C for 5 min. The precipitate was retained, washed
with cold acetone, and centrifuged again at 12,000 × g at 4°C for 5
min. The precipitate was again retained, and the washing step was
repeated three times. The protein pellets were air-dried and
resuspended in 8 M urea/100 mM triethylammonium bicarbonate (pH
8.0). The BCA protein quantitation assay was used to measure the
total protein concentration. Subsequently, protein hydrolysis and
desalination are carried out. Liquid chromatography (LC) was
performed on a nanoElute UHPLC (Bruker Corporation). In total, ~200
ng peptides were separated within 40 min at a flow rate of 0.3
μl/min on a commercially available reverse-phase C18 column
with an integrated CaptiveSpray Emitter (25 cm × 75 μm; 1.6
μm particle size; Aurora Series with CSI; IonOpticks). The
separation temperature was kept by an integrated Toaster column
oven at 50°C. Mobile phases A and B were produced with 0.1% formic
acid in water and 0.1% formic acid in acetonitrile. Mobile phase B
was increased from 2 to 22% over the first 25 min, increased to 35%
over the next 5 min, further increased to 80% over the next 5 min,
and then held at 80% for 5 min. The LC process was coupled online
to a hybrid timsTOF Pro2 mass spectrometer via a CaptiveSpray
nano-electrospray ion source (Bruker Corporation). To establish the
applicable acquisition windows, timsTOF Pro2 was operated in
Data-Independent Parallel Accumulation-Serial Fragmentation (PASEF)
mode with four PASEF MS/MS frames in one complete frame. The
capillary voltage was set to 1,500 V, and the MS and MS/MS spectra
were acquired from 100 to 1,700 m/z. As for ion mobility range
(1/), 0.85 to 1.3 Vs/was used. The 'target value' of 10,000 was
applied to a repeated schedule, and the intensity threshold was set
at 2,500. The range of charge state was set from 0 to 5. The
collision energy was ramped linearly as a function of mobility from
45eV at 1/=1.3 Vs/to 27 eV at 1/=0.85 Vs/. The quadrupole isolation
width was set to 2 Thomson for m/z <700 and 3 Th for m/z
>800. Raw mass spectrometry data were analyzed with DIA-NN
software (v1.8.1) (https://github.com/vdemichev/DiaNN) using the
library-free method. The Homo sapiens SwissProt database
(https://www.uniprot.org/; 20,425 entries) was
used to create a spectral library with deep learning algorithms of
neural networks. Match Between Runs was applied to align precursor
retention times and m/z values across DIA runs, enabling cross-run
matching of peptide identifications; a spectral library from DIA
data was created and the data were reanalyzed using this library.
To comprehensively elucidate the functional characteristics of
various proteins, R software (clusterProfiler) version 3.10.1
(https://bioconductor.org/packages/clusterProfiler) was
employed to conduct functional annotation and enrichment analysis
on both the identified proteins and differentially expressed
proteins in each comparison group. This analysis encompassed Kyoto
Encyclopedia of Genes and Genomes (KEGG) and Disease Ontology (DO)
(Fig. S1A and B), wherein all
differentially expressed proteins were mapped to the corresponding
terms in the KEGG (https://www.kegg.jp/) and DO (http://www.disease-ontology.org/) databases. The
number of proteins associated with each term was calculated,
followed by a hypergeometric test (equivalent to Fisher's exact
test; implemented in the R stats package version 4.3.2; https://www.r-project.org/) to identify significantly
enriched KEGG and DO terms among the differentially expressed
proteins. KEGG/DO terms were considered significantly enriched if
they met FDR-adjusted P<0.05 and fold change >2 (log2FC
>1).
Flow cytometry
MDA-MB-231, Cal51 and T47D cells were used for the
experiment. After resuspending the cells with PBS, the proportion
of CD24−CD44+ cells was detected using a flow
cytometer. The cells were incubated with CD44 APC (1:40; cat. no.
559942; BD Biosciences) and CD24 PE (1:50; cat. no. 555428; BD
Biosciences) at 4°C for 30 min in the dark. Isotype controls, APC
IgG (1:100; cat. no. 555751; BD Biosciences) and PE IgG (1:100;
cat. no. 555749; BD Biosciences), were incubated under the same
conditions. A BD FACSCanto II flow cytometer (BD Biosciences) was
used for data acquisition, and the data were analyzed using FlowJo
v10.8.1 (Becton, Dickinson and Company).
Furin activity measurement
Furin activity in MDA-MB-231, Cal51 and T47D cells
was measured by assessing the ability of the cells to digest
pERTKR-MCA. Briefly, each cell extract was incubated with
pERTKR-MCA (100 M) in a solution composed of 25 mM Tris, 25 mM
methyl-ethane-sulfonic acid and 2.5 mM CaCl2 (pH 7.4) at
37°C. Measurements were taken every 5 min for a total duration of
100 min. At a pH of 6.0 and a Ca2+ concentration of 1 mM, the
catalytic activity of furin was dynamically detected using a
fluorometer. The Km value was derived from the
Michaelis-Menten curve fitted to the enzymatic activity data
(velocity vs. substrate concentration). In the present study,
Km is indirectly reflected by the substrate
concentration at which the reaction velocity reaches half of
Vmax. A spectrofluorometer (FLUOstar OPTIMA; BMG
Labtech GmbH) was used for analysis. A furin inhibitor,
decanoyl-RVKR-chloromethyl ketone (DECRVKR-CMK) purchased from
Calbiochem (Merck KGaA), was used at a concentration of 10
μM. Cells were treated with this inhibitor at 37°C for 7
days.
Reverse transcription-quantitative
PCR
Total RNA from cells in each group was extracted
using the TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). First-strand cDNA synthesis was carried out
according to the manufacturer's instructions with a first-strand
cDNA synthesis kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Primer Express software (version 3.0; Applied Biosystems;
Thermo Fisher Scientific, Inc.) was utilized to design
gene-specific primers for each target gene. The primer sequences
were as follows: CENPU: 5'-GAAAAGAAAAGGCAGCGTATGA-3' (forward) and
5'-AATATGCTGCATTCCTAAGGGA-3' (reverse); GAPDH:
5'-GGAGCGAGATCCCTCCAAAAT-3' (forward) and
5'-GGCTGTTGTCATACTTCTCATGG-3' (reverse). The reaction conditions
included pre-denaturation at 95°C for 2 min, denaturation at 95°C
for 30 sec, annealing at 60°C for 30 sec, and extension at 72°C for
30 sec for a total of 40 cycles. CENPU mRNA expression was
calculated using the 2−ΔΔCq method (26).
Xenograft model
The experimental protocol was approved by the Animal
Care Committee of Shandong Cancer Hospital and Institute, Shandong
First Medical University and Shandong Academy of Medical Sciences
(approval no. SDTHEC2022009018; Jinan, China). To investigate the
tumorigenic potential of Cal51-shCENPU and Cal51-CON313
cells at varying concentrations, 1×106,
1×105, 1×104 and 1×103 cells
(resuspended in PBS) were injected into the mammary fat pads of NSG
mice (27,28). NSG mice were obtained from
Nanjing Biomedical Research Institute of Nanjing University. A
total of 32 mice (4-6 weeks old; female; 18-20 g) were randomly
divided into 8 groups (n=4/group). Mice were maintained under
specific-pathogen-free (SPF) conditions (temperature, 25±1°C;
humidity, 55±5%; 12/12-h light/dark cycle) with ad libitum
access to irradiated food and autoclaved water. The tumor sizes
formed by the Cal51-shCENPU and Cal51-CON313 cell groups at
these different concentrations were measured and subjected to
statistical analysis to determine significant differences.
MDA-MB-231-shCENPU and MDA-MB-231-CON313 cells
(1×106 cells/specimen; resuspended in PBS) were injected
into the fat pads of 4-week-old female nude mice (Nanjing
Biomedical Research Institute of Nanjing University). A total of 15
female nude mice (age, 4 weeks; weight, 16-18 g) were randomly
divided into 3 groups (n=5/group). Mice were maintained under SPF
conditions (temperature: 25±2°C; humidity: 50±10%; 12/12-h
light/dark cycle) with free access to autoclaved food and acidified
water. The tumor dimensions were measured at 3-day intervals for 21
days. When the size of each tumor was ~100 mm3,
MDA-MB-231-CON313 mice were intraperitoneally injected with a furin
inhibitor (5 mg/kg DECRVKR-CMK) three times per week for 3 weeks.
The following humane endpoints were predefined in the present
study: i) Clear signs of pain or unremitting suffering; ii) rapid
weight loss; iii) persistent lack of appetite or abnormal behavior;
and iv) inability to stand or extreme weakness after excluding
anesthesia-related reasons (however, no anesthesia was administered
to the mice in the present study). However, during the experiment
no such signs were observed, indicating that the tumors did not
severely affect the health of the mice until the end of the study.
The tumor volumes were calculated using the following formula:
Volume=0.5 × (width2 × length). The maximum tumor volume
observed in the first experiment (Cal51 cells) was 936
mm3, and the maximum diameter was 1.3 cm. The maximum
tumor volume observed in the second experiment (MDA-MB-231 cells)
was 1,764 mm3, and the maximum diameter was 1.8 cm. At
study completion, the mice were euthanized using CO2
inhalation in accordance with the American Veterinary Medical
Association 2020 Guidelines (29). The procedure involved an initial
concentration of 30% CO2, a flow rate controlled at 50%
of the chamber volume per minute, and a gradual increase to 70% to
ensure rapid and humane loss of consciousness. Death was confirmed
by observing pupillary dilation and cessation of cardiac activity.
The tumors were then extracted, measured and used for subsequent
evaluation.
Statistical analysis
Statistical processing was implemented with SPSS
software v18.0 (SPSS, Inc.). Intergroup comparisons were performed
using unpaired Student's t-test (two-group comparisons) or one-way
ANOVA (multi-group comparisons) followed by the Least Significant
Difference pos hoc test (for equal variances between groups). When
two types of treatment were used, a two-way ANOVA followed by Sidak
post hoc test was performed. All statistical differences were
established using two-tailed probability measures. Experimental
procedures were replicated in triplicate, and the data are
presented as the mean ± SD. P<0.05 was considered to indicate a
statistically significant difference.
Results
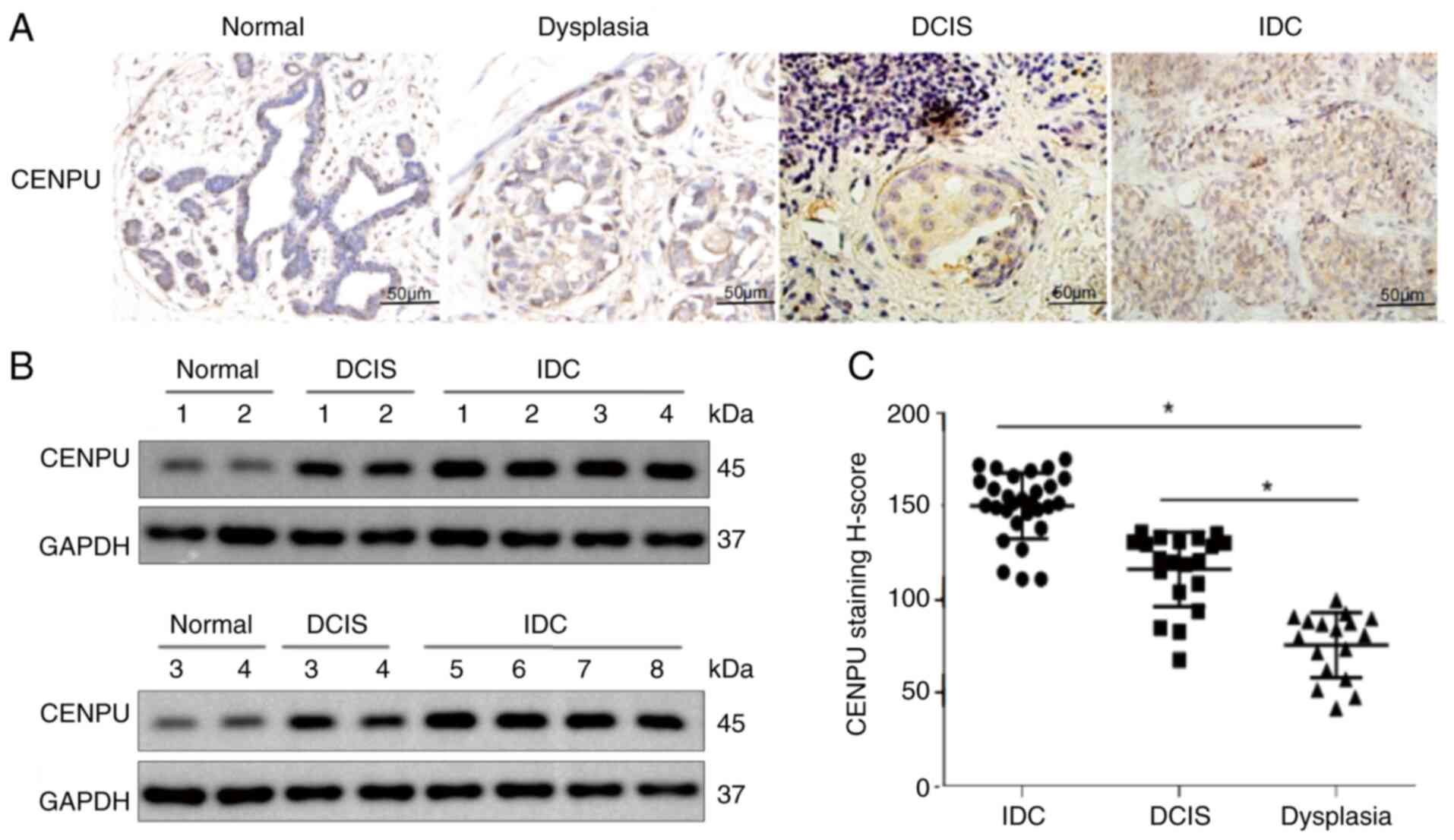
CENPU is expressed at high levels in
breast invasive ductal carcinoma
Our previous study showed that CENPU was markedly
upregulated in TNBC tissues compared with luminal breast cancer
tissues, and this upregulation was associated with a poor prognosis
(15). The present study
examined whether CENPU expression is associated with the degree of
invasiveness in BC. A total of 67 tissue samples were collected,
including 17 cases of breast dysplasia, 20 cases of ductal
carcinoma in situ and 30 cases of invasive ductal carcinoma.
IHC results confirmed that CENPU expression was positively
associated with the degree of invasiveness in BC (Fig. 1A and C). Next, four normal breast
tissue samples (adjacent tissues of the same patients with BC),
four ductal carcinoma in situ tissue samples and eight
breast invasive ductal carcinoma tissue samples were used to verify
the findings by western blotting. The results demonstrated that
compared with the normal breast and ductal carcinoma in situ
tissue samples, the invasive ductal carcinoma tissues exhibited
higher CENPU levels (Fig.
1B).
CENPU promotes the stem cell-like
properties of BCSCs
Cancer stem cells serve an important role in
tumorigenesis and recurrence. Given that our previous study showed
that high CENPU expression was associated with poor overall
survival (15), it was assessed
whether CENPU expression promotes the stem cell-like properties of
BCSCs. Firstly, the overexpression efficiency of CENPU in cells was
verified through PCR or western blotting (Fig. S2). Subsequently, a
CD24− CD44+ cell identification experiment
and a mammosphere formation assay were performed to determine the
association between CENPU expression and the stem cell-like
properties of BCSCs. Western blotting revealed that CD44 and ALDH
(30,31) were downregulated in
CENPU-knockdown MDA-MB-231 and Cal51 cells, where it had
been confirmed that CENPU was highly expressed in our previous
study (15) (Fig. 2A and B), and upregulated in
CENPU-overexpressing T47D cells, where it had been confirmed
that CENPU was expressed at a low level (15) (Fig. 2C).
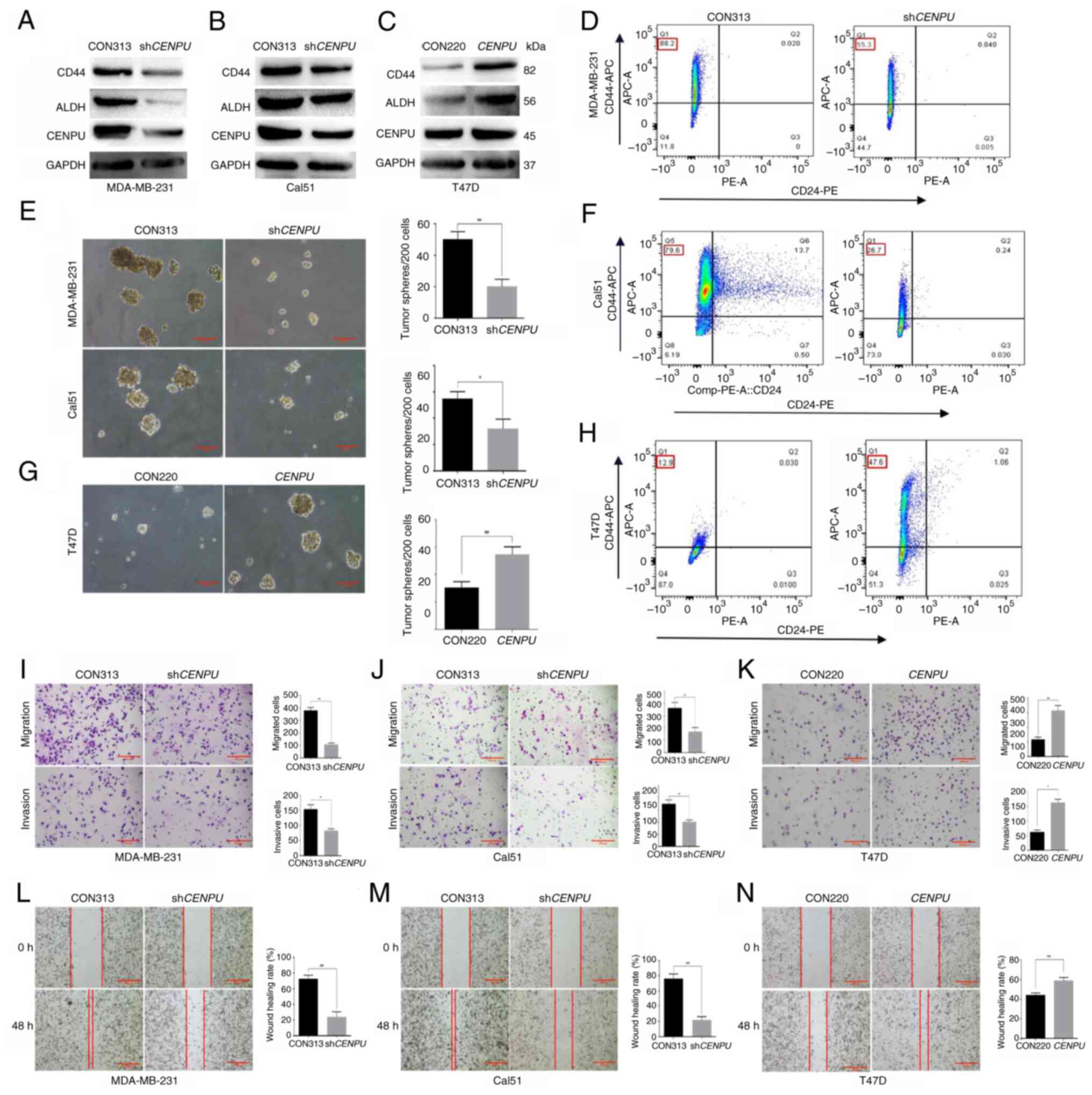 | Figure 2CENPU promotes tumorigenesis, stem
cell-like properties, migration and invasion in breast cancer
cells. Western blotting was performed to detect the expression
levels of CD44 and ALDH in (A) MDA-MB-231, (B) Cal51 and (C) T47D
cells with different CENPU expression levels. Association between
CENPU expression and the number of mammospheres per 200 cells in
(E) MDA-MB-231 and Cal51 cells and (G) T47D cells (scale bar, 100
μm). CD24− CD44+ flow cytometry
results showed that CENPU expression levels affected the percentage
of the CD24− CD44+ population among (D)
MDA-MB-231, (F) Cal51 and (H) T47D cells. Transwell assays were
performed to determine the effect of CENPU levels on the migration
and invasiveness of (I) MDA-MB-231, (J) Cal51 and (K) T47D cells
(scale bar, 200 μm). A wound healing assay was performed to
determine the effect of CENPU on the migration potential of (L)
MDA-MB-231, (M) Cal51 and (N) T47D cells (scale bar, 200
μm). *P<0.05; **P<0.01. CENPU,
centromere protein U; sh, short hairpin RNA; APC, allophycocyanin;
ALDH, aldehyde dehydrogenase; PE, phycoerythrin. |
The mammosphere generation assay showed that
CENPU knockdown decreased the number and volume of
mammospheres (Fig. 2E).
Conversely, CENPU overexpression markedly enhanced the
number and volume of mammospheres (Fig. 2G). CENPU knockdown reduced
the rate of CD24− CD44+ cells (30,32,33) (Fig. 2D and F), whereas CENPU
overexpression exerted a contrasting effect (Fig. 2H). The aforementioned data
indicated that CENPU overexpression promoted the stem
cell-like properties of BCSCs.
The role of CENPU in BC progression was examined
using wound healing and Transwell assays, to determine the effect
of CENPU on cell migration and invasion. CENPU knockdown
inhibited the migratory and invasive potential of MDA-MB-231 and
Cal51 cells (Fig. 2I, J, L and
M), whereas CENPU overexpression enhanced the migratory
and invasive potential of T47D cells (Fig. 2K and N).
CENPU promotes the maturation conversion
of proNGF to NGF
Comparative DO and KEGG enrichment analyses of
MDA-MB-231 cell lines with CENPU knockdown (shCENPU) and
overexpression revealed that CENPU is significantly associated with
BC and related cancer signaling pathways. (Fig. S1A and B), while 4D-DIA
quantitative differential protein enrichment analysis showed that
the shCENPU group exhibited notably lower NGF expression
compared with the CON313 group (Fig. S1C). NGF is a critical
neurotrophic factor that controls differentiation and survival in
target neurons. NGF matures upon undergoing furin-mediated protease
cleavage of its proNGF form (34). Therefore, western blotting and an
ELISA were performed to examine whether CENPU knockdown and
overexpression affect the expression of NGF and proNGF. Western
blotting and ELISA results revealed that NGF expression was
markedly suppressed in MDA-MB-231 and Cal51 cells with CENPU
knockdown (Fig. 3A, B, D and E).
Conversely, NGF expression was markedly enhanced upon CENPU
upregulation in CENPU-overexpressing T47D cells (Fig. 3C and F). Furthermore, the changes
in CENPU expression affected proNGF and NGF expression in
contrasting manners. MDA-MB-231-shCENPU,
Cal51-shCENPU and T47D-CENPU cells, and their
corresponding controls, were treated with NGF-neutralizing
antibodies. The ability of CENPU to promote migration, invasion and
mammosphere formation was significantly suppressed when the cells
were treated with NGF-neutralizing antibodies (Fig. 3G-L). No significant differences
were observed between each overexpression or knockdown group and
the corresponding control treated with NGF. This indicated that NGF
may serve a critical role in the CENPU-induced promotion of BC
induction and progression.
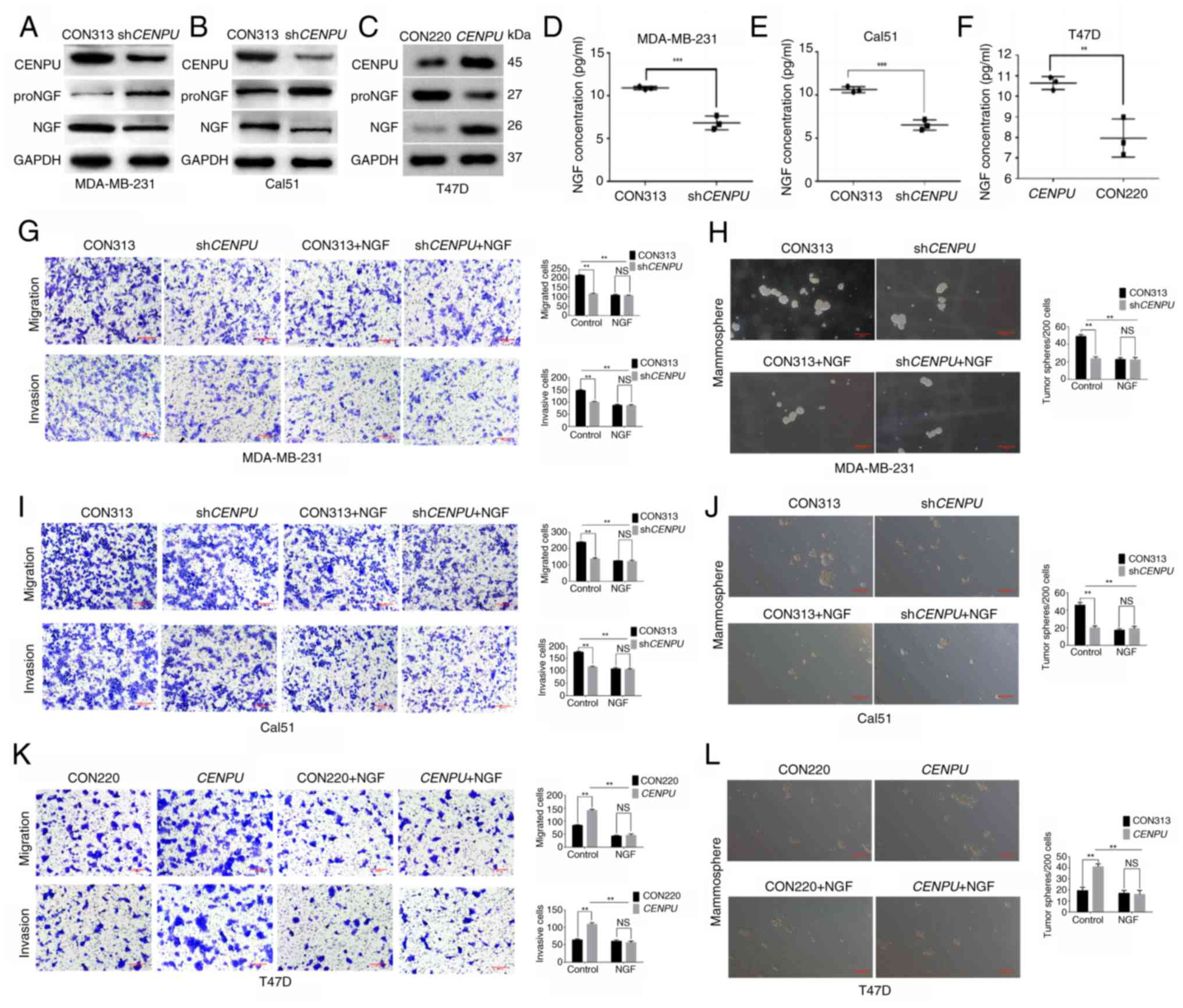 | Figure 3CENPU promotes the conversion of
proNGF to NGF. Western blotting experiments were performed to
detect the expression of NGF and proNGF in (A)
MDA-MB-231-shCENPU, (B) Cal51-shCENPU and (C)
T47D-CENPU cells and their corresponding control groups. The
intracellular NGF levels in (D) MDA-MB-231, (E) Cal51 and (F) T47D
cells after 48 h of treatment were analyzed by ELISA using
ultrasonicated cell supernatants. After NGF-neutralizing antibodies
were added to (G) MDA-MB-231-shCENPU, (I)
Cal51-shCENPU and (K) T47D-CENPU cells, and their
corresponding control groups, Transwell migration and invasion
assays were conducted (scale bar, 200 μm). After
NGF-neutralizing antibodies were added to the (H)
MDA-MB-231-shCENPU, (J) Cal51-shCENPU and (L)
T47D-CENPU cells, and their corresponding control groups, a
mammosphere formation assay was performed to measure the number of
mammospheres per 200 cells that formed (scale bar, 100 μm).
*P<0.05; **P<0.01. CENPU, centromere
protein U; sh, short hairpin RNA; NGF, nerve growth factor; proNGF,
NGF precursor; NS, not significant. |
CENPU promotes NGF maturation by
enhancing the enzymatic activity of furin
The previous experiment demonstrated that CENPU
could affect the expression levels of proNGF and NGF. Furin is the
sole enzyme capable of cleaving proNGF, thereby facilitating its
conversion into NGF. Therefore, it was examined whether CENPU
regulates the expression of proNGF and NGF by affecting furin
activity. Western blotting revealed lower furin expression levels
in shCENPU-expressing MDA-MB-231 and Cal51 cells, and higher
furin expression levels in CENPU-overexpressing T47D cells
(Fig. 4A). Next, furin activity
was assayed by measuring the fluorescence intensity of the
furin-specific fluorescence substrate pERTKR-MCA. Furin activity
was suppressed in shCENPU-expressing MDA-MB-231 and Cal51
cells, and elevated in CENPU-overexpressing T47D cells
(Fig. 4B).
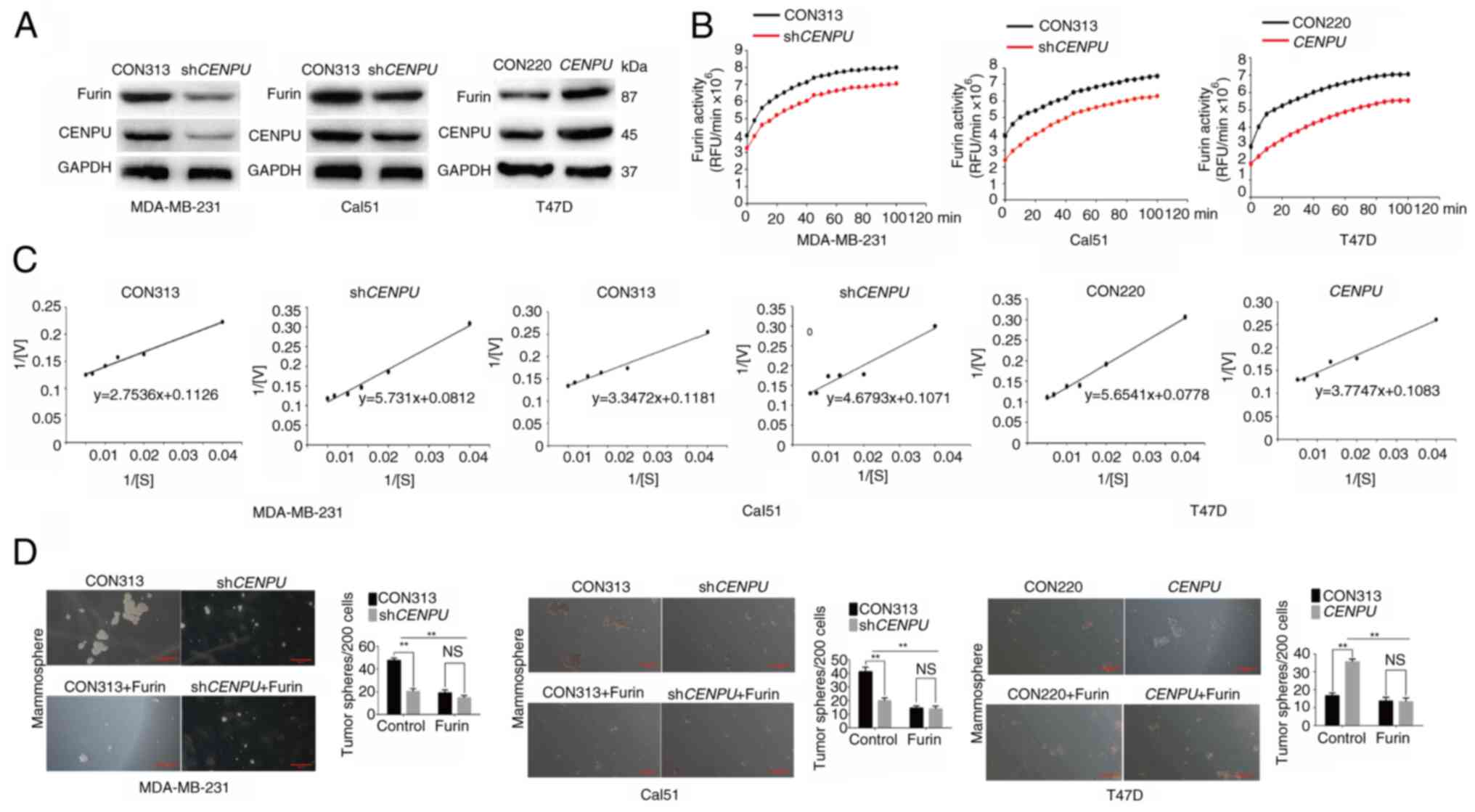 | Figure 4CENPU enhances the activity of furin.
(A) Western blotting was performed to determine the expression
levels of furin in MDA-MB-231, Cal51 and T47D cells. (B) Furin
activity in MDA-MB-231, Cal51 and T47D cells was measured based on
fluorescence intensity. (C) Michaelis constant values in the
experimental group and control group of MDA-MB-231, Cal51 and T47D
cells were obtained using double reciprocal plotting. (D)
Mammosphere formation assays were performed after treatment with a
furin inhibitor, and the number of tumor spheres per 200 cells was
quantified (scale bar, 100 μm). **P<0.01.
CENPU, centromere protein U; sh, short hairpin RNA; NS, not
significant; RFU, relative fluorescence units; V, reaction
velocity; S, substrate concentration. |
There are two methods to increase enzymatic
activity: i) Increasing the enzyme concentration; and ii)
increasing the affinity of the enzyme to the substrate, as
indicated by the Michaelis constant (Km). The
Km value is known as the meter constant, which
refers to the substrate concentration of the reaction that reaches
1/2Vmax, where Vmax is the
reaction velocity. Km is generally expressed in
mol/l, and this index is only determined by the intrinsic
characteristics of the enzyme but is independent of its
concentration. The larger the Km value, the
smaller the affinity of the enzyme to the substrate. Using the
double reciprocal plotting method, 1/V is plotted against 1/[S]
(where S is the substrate concentration) to generate a straight
line. The y-intercept of the line is 1/Vmax, and
the x-intercept is the absolute value of 1/Km
(35). At a pH of 6.0 and
Ca2+ concentration of 1 mM, the Km
value of pERTKR-MCA for furin in shCENPU-expressing
MDA-MB-231 cells was 68.63 μmol/l. The Km
in the control group was 24.45 μmol/l. The
Km values of pERTKR-MCA for furin were 43.70
μmol/l in shCENPU-expressing Cal51 cells and 24.45
μmol/l in the control group. The Km values
of pERTKR-MCA for furin were 34.86 and 72.67 μmol/l in the
CENPU-overexpressing T47D cells and the control group,
respectively (Fig. 4C). These
results showed that CENPU knockdown could reduce the
affinity of furin for its substrates, whereas CENPU
overexpression could increase this affinity.
MDA-MB-231-shCENPU, Cal51-shCENPU and
T47D-CENPU cells, and their corresponding controls, were
treated with a furin inhibitor. The results of the mammosphere
formation experiment indicated that the ability of CENPU to promote
mammosphere formation was inhibited when the cells were treated
with a furin inhibitor (Fig.
4D). No significant differences were observed between each
overexpression or knockdown group and the corresponding control
treated with furin.
CENPU enhances furin activity by
inhibiting its degradation in the lysosomal pathway
The previous experiment showed that CENPU could
influence both furin expression and activity. The presence of two
conserved LXXLL sequences and two superhelical zinc finger
structures at the C-terminal of CENPU suggest that CENPU may form a
dimer with other proteins and act as a receptor-binding protein in
signal transduction (36). CENPU
and furin co-immunoprecipitated with each other in MD-MB-231 and
Cal51 cells (Fig. 5A). Using
co-immunoprecipitation, it was also demonstrated that furin formed
protein complexes with proNGF (Fig.
5B).
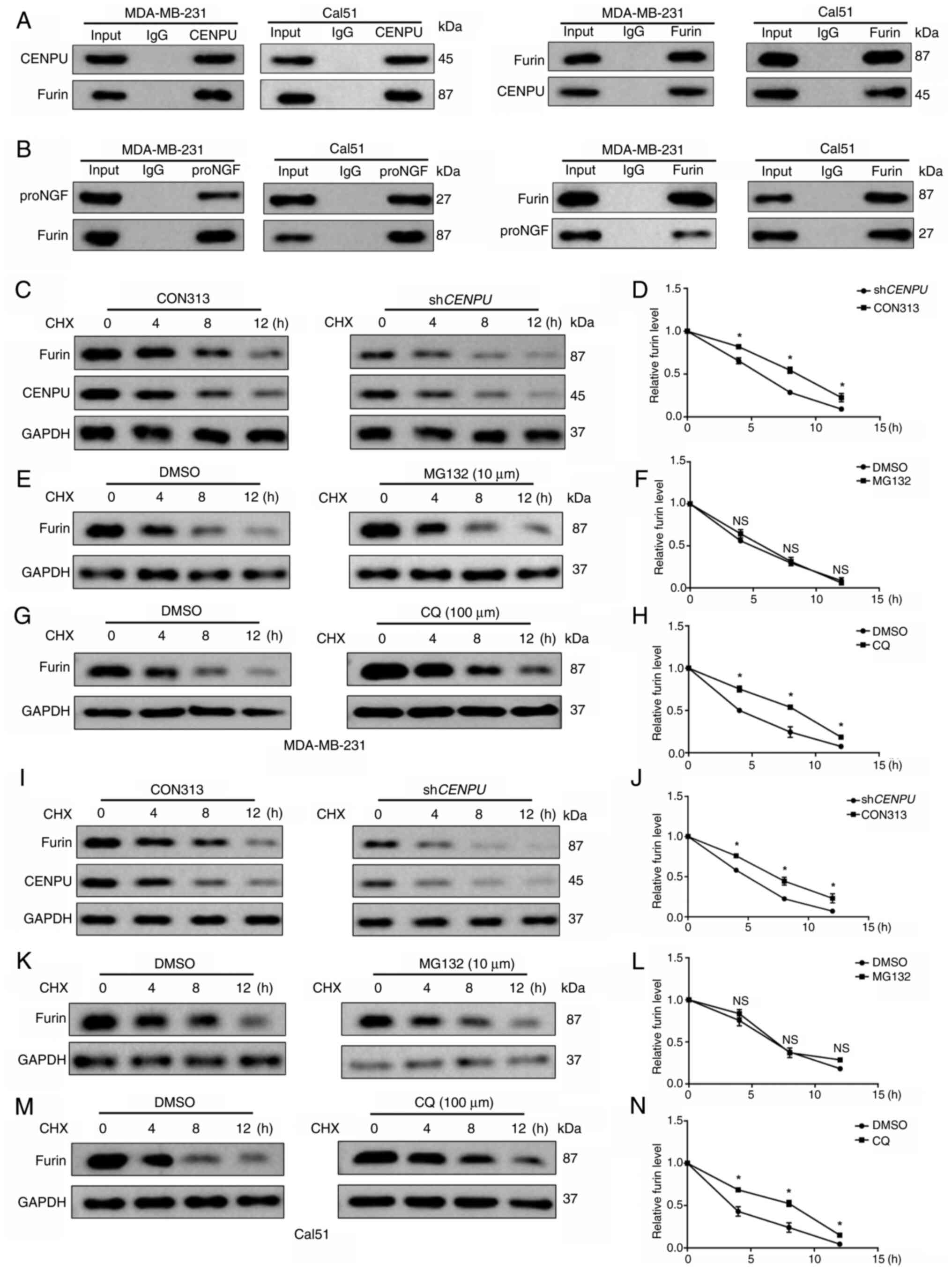 | Figure 5CENPU enhances furin activity by
inhibiting its degradation via the lysosomal pathway. (A) CENPU and
furin were subjected to co-immunoprecipitation. (B) Furin and
proNGF were subjected to co-immunoprecipitation. (C)
MDA-MB-231-shCENPU cells and their corresponding controls
were treated with CHX (20 μM) for the specified duration,
and protein levels were measured by western blotting. (D) Relative
expression of furin. (E) MDA-MB-231 cells were treated with MG132
(10 μM) for 12 h, and then also treated with CHX (20
μM) for the indicated times. Protein levels were detected by
western blotting. (F) Relative expression of furin. (G) MDA-MB-231
cells were treated with CQ (100 μM) for 12 h, and then
treated with CHX (20 μM) for the indicated times. Protein
levels were detected by western blotting. (H) Relative expression
of furin. (I) Cal51-shCENPU cells and their corresponding
controls were treated with CHX (20 μM) for the specified
duration, and protein levels were measured by western blotting. (J)
Relative expression of furin. (K) Cal51 cells were treated with
MG132 (10 μM) for 12 h, and then treated with CHX (20
μM) for the indicated times. Protein levels were detected by
western blotting. (L) Relative expression of furin. (M) Cal51 cells
were treated with CQ (100 μM) for 12 h, and then treated
with CHX (20 μM) for the indicated times. Protein levels
were detected by western blotting. (N) Relative expression of
furin. *P<0.05 vs. DMSO or CON313 group. The
experimental and control groups in panels C-M were run on the same
gel/membrane and incubated with the corresponding primary
antibodies simultaneously. CENPU, centromere protein U; sh, short
hairpin RNA; NS, not significant; proNGF, nerve growth factor
precursor; CHX, cycloheximide; CQ, chloroquine. |
The precise mechanism by which CENPU upregulated
furin expression in BC cells was then explored. We hypothesized
that CENPU interacts with furin to inhibit its degradation.
CENPU knockdown promoted furin degradation in MDA-MB-231 and
Cal51 cells treated with cycloheximide, a protein biosynthesis
inhibitor, at indicated at 4, 8 and 12 h after treatment (Fig. 5C, D, I and J). The proteasomal
and lysosomal pathways are key pathways regulating protein
degradation in cells (37).
Therefore, the potential pathways involved in CENPU-regulated furin
degradation were investigated. Proteasome suppression by MG132 did
not affect furin degradation in cells with CENPU knockdown
compared with the control group (Fig. 5E, F, K and L). However, the
suppression of lysosomal activity by chloroquine further inhibited
furin degradation in MDA-MB-231-shCENPU and
Cal51-shCENPU cells (Fig. 5G,
H, M and N). The aforementioned findings indicated that CENPU
suppressed furin degradation via the lysosomal pathway.
CENPU promotes tumorigenesis in TNBC in
vivo
Since an important function of CENPU in
tumorigenesis in the cultured cells was identified, the role of
CENPU in a mouse xenograft model was then examined. Female NSG mice
were injected with Cal51-shCENPU and Cal51-CON313 cells to
determine whether CENPU promotes tumorigenesis in vivo. The
mice injected with 1×104, 1×105 and
1×106 cells with stable CENPU knockdown exhibited
smaller xenograft tumors compared with that injected with CON313.
None of the mice injected with 1×103 cells with stable
CENPU knockdown exhibited formation of xenograft tumors.
This suggested that CENPU knockdown inhibited tumorigenesis
(Fig. 6A and B). IHC and western
blotting experiments showed that the furin and NGF protein levels
were considerably lower, and the proNGF protein level was higher in
shCENPU-Cal51 tumor samples compared with the control
samples (Fig. 6C and D).
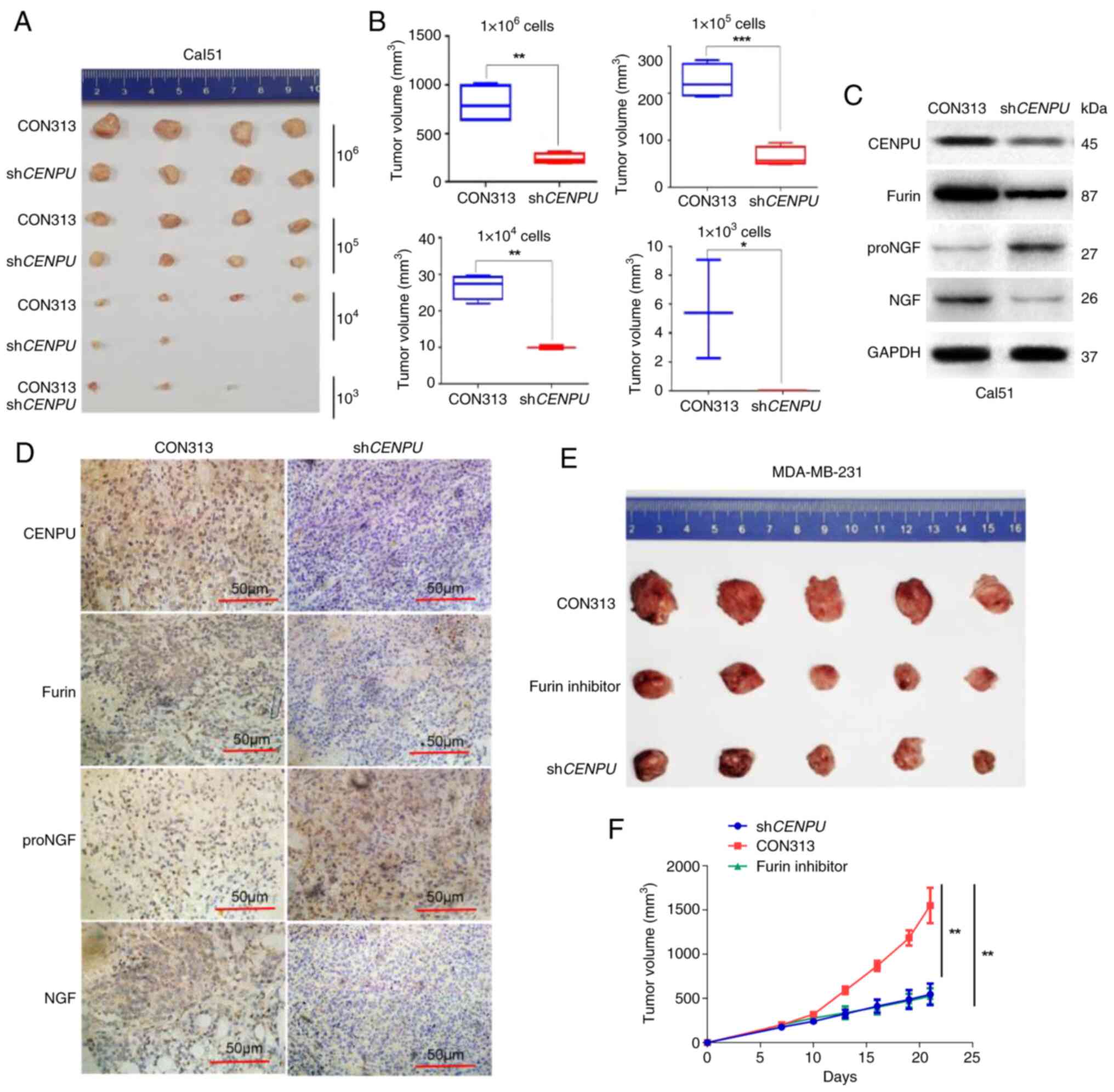 | Figure 6CENPU knockdown can inhibit
tumorigenesis. (A) A total of 1×106, 1×105,
1×104 and 1×103 Cal51-shCENPU and
Cal51-CON313 cells were injected into the mammary fat pad of NSG
mice. (B) Sizes of the tumors formed in the Cal51-shCENPU
and Cal51-CON313 groups using different cell concentrations were
measured and statistically analyzed. The tumor tissues collected
from the Cal51-shCENPU and Cal51-CON313 groups were (C)
analyzed by western blotting and (D) subjected to
immunohistochemical staining (scale bar, 50 μm). (E) A total
of 1×106 MDA-MB-231-shCENPU and MDA-MB-231-CON313
cells were injected into the mammary fat pad of nude mice. When the
tumor size was ~100 mm3, the MDA-MB-231-CON313 mice were
administered a furin inhibitor (DECRVKR-CMK) by intraperitoneal
injection. (F) The volume of the final tumor was measured,
indicating that the furin inhibitor significantly inhibited tumor
growth in the MDA-MB-231-CON313 mice. Scale bar, 50 μm.
*P<0.05; **P<0.01;
***P<0.001. CENPU, centromere protein U; sh, short
hairpin RNA; NS, not significant; NGF, nerve growth factor; proNGF,
NGF precursor. |
Furin inhibitor treatment can inhibit
CENPU-induced tumor growth
MDA-MB-231-shCENPU and MDA-MB-231-CON313
cells were injected into the mammary fat pads of nude mice. When
the tumor size was ~100 mm3, MDA-MB-231-CON313 mice were
intraperitoneally injected with the furin inhibitor DECRVKR-CMK.
The furin inhibitor could significantly inhibit CENPU-induced tumor
growth. This observation corroborated the aforementioned in
vitro findings (Fig. 6E and
F).
Discussion
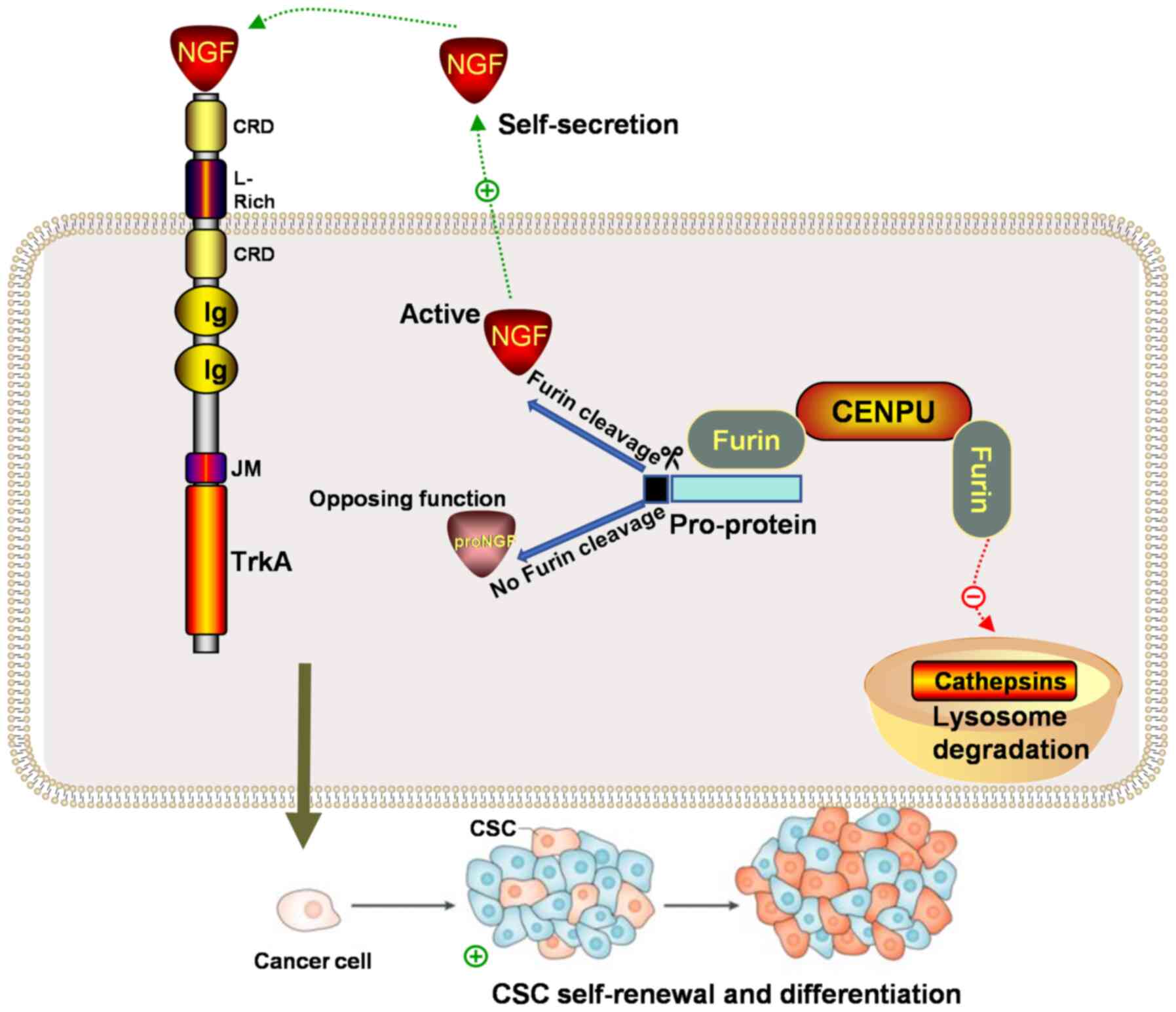
The present study identified a potential role of
CENPU in TNBC tumorigenesis. Furthermore, the molecular mechanism
by which CENPU potentially promotes TNBC tumorigenesis and the stem
cell-like properties of BCSCs by enhancing NGF maturation was also
established. Specifically, the current study showed that CENPU
inhibited the lysosomal degradation of furin, and could bind to the
furin protein, increase its expression and enhance its enzymatic
activity. The enhancement of furin activity promoted the maturation
and transformation of proNGF into NGF. NGF promoted the BCSC-like
properties of TNBC cells (Fig.
7). Collectively, these data provided novel insights into how
CENPU may promote TNBC tumorigenesis and the stem cell-like
properties of TNBC cells.
Our previous study reported that CENPU was notably
upregulated in BC tissues compared with adjacent normal breast
tissues and expressed at higher levels in TNBC than in luminal BC
(15). The present showed that
CENPU expression was significantly associated with the degree of
invasiveness in BC. The effects of CENPU on TNBC tumorigenesis,
BCSC properties, migration and invasion were further investigated,
and the results demonstrated that CENPU promoted these
features.
CD44 is a multifunctional class I transmembrane
glycoprotein. In BCSCs, its expression is linked to self-renewal,
tumorigenesis, adhesion, invasion and metastasis. The
CD44+CD24−-low phenotype is widely used as a
marker for BCSC identification and isolation. ALDH1A1, as a stem
cell marker, has a high expression level in BCSCs and can promote
proliferation and spheroid formation (30-32). The present study demonstrated
that knockdown of CENPU reduced the proportion of CD44+
CD24− cells, while western blotting analysis revealed
downregulation of CD44 and ALDH expression. Conversely,
overexpression of CENPU exhibited the opposite effect. Therefore,
we hypothesize that overexpression of CENPU can promote the
stem-like characteristics of BCSCs.
NGF is a critical neurotrophic factor that
regulates the differentiation and survival of target neurons
(25). NGF is biosynthesized as
proNGF, and NGF maturation involves furin-associated protease
cleavage (38). Hayakawa et
al (39) reported that NGF
promoted gastric cancer development by altering cholinergic
signaling. Tomellini et al (40) demonstrated that NGF promoted
symmetric self-renewal, quiescence and epithelial-to-mesenchymal
transition, thereby enlarging the BCSC compartment. In the present
study, NGF expression decreased with a decrease in the CENPU level,
whereas proNGF expression increased. Furthermore, after
NGF-neutralizing antibodies were added to the culture medium of BC
cells, the ability of CENPU to promote BC migration, invasion and
mammosphere formation was inhibited. Therefore, we hypothesize that
CENPU may promote NGF maturation.
Furin is a precursor protein-processing enzyme that
helps process multiple bioactive polypeptides or protein precursors
in the brain, glands and other peripheral tissues (41). There are large volumes of furin
present in tumor tissues, and studies have shown that furin
expression increases with the tumor proliferation rate (24,42,43). Furthermore, furin inhibition
suppresses the proliferation and infiltration of tumor cells
(44-46). The present results showed that
CENPU inhibited the lysosomal degradation of furin, bound to furin
and increased its expression, thereby enhancing its enzymatic
effect on proNGF. A furin inhibitor (DECRVKR-CMK), which was
administered in both in vivo and in vitro settings,
inhibited the ability of CENPU to promote tumorigenesis in
TNBC.
Collectively, the results of the present study
indicated that CENPU promoted TNBC invasion, tumorigenesis and BCSC
properties through the furin-proNGF-NGF signaling pathway. CENPU
enhanced the conversion of proNGF to NGF by inhibiting furin
degradation via the lysosomal pathway. However, there are certain
limitations to the present study that warrant discussion. Two
common strategies for enhancing enzyme activity were identified:
Increasing the enzymatic affinity for its substrates and raising
the enzymatic concentration. The present study demonstrated that
the overexpression of CENPU could elevate furin activity. Both the
substrate affinity of furin and its expression levels were
examined, and it was observed that CENPU enhanced the activity and
increased the expression of furin. However, while an increase in
furin activity and expression levels was observed, the underlying
molecular mechanisms that mediate these effects were not fully
elucidated. Furthermore, a previous study has suggested that the
activity of furin is regulated by its propeptide occupying the
active site in its proenzyme form, thus preventing substrate
binding (47). The present
findings, however, indicate that CENPU may modulate furin activity
through protein-protein interactions, which is not well-documented
in the existing literature. Therefore, further research should be
conducted to verify this hypothesis and explore the potential
mechanisms.
In summary, the current findings revealed a novel
mechanism underlying the tumor-promoting effect of CENPU,
specifically in TNBC. The inhibition of CENPU expression and its
downstream activated signaling pathway may be a novel strategy for
TNBC treatment.
Supplementary Data
Availability of data and materials
The proteomics data generated in the present study
may be found in the ProteomeXchange (iProX) repository under
accession number PXD061628 or at the following URLs: https://www.iprox.cn/page/project.html?id=IPX0011300000
and http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD061628.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
SS performed data collection, analysis and
interpretation. ZH and LQ conducted data collection and analysis.
DZ designed the study and supervised the data interpretation. SS
and DZ confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The human and animal experiments in the present
study were approved by the Ethics Committee of Shandong Cancer
Hospital and Institute, Shandong First Medical University and
Shandong Academy of Medical Sciences (approval nos.
SDTHEC2022009017 and SDTHEC2022009018, respectively; Jinan, China).
The human samples used in the present study were obtained with
written informed consent from patients or their families (due to
patient's physical reasons or low educational level) for use in
research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Youth Natural Science
Foundation of Shandong Province (grant nos. ZR2023QH128,
ZR2023QH069 and ZR202211090247).
References
|
1
|
Derakhshan F and Reis-Filho JS:
Pathogenesis of triple-negative breast cancer. Annu Rev Pathol.
17:181–204. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahtani R, Kittaneh M, Kalinsky K,
Mamounas E, Badve S, Vogel C, Lower E, Schwartzberg L and Pegram M;
Breast Cancer Therapy Expert Group (BCTEG): Advances in therapeutic
approaches for triple-negative breast cancer. Clin Breast Cancer.
21:383–390. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang M, Deng H, Hu R, Chen F, Dong S,
Zhang S, Guo W, Yang W and Chen W: Patterns and prognostic
implications of distant metastasis in breast cancer based on SEER
population data. Sci Rep. 15:267172025. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao S, Zuo WJ, Shao ZM and Jiang YZ:
Molecular subtypes and precision treatment of triple-negative
breast cancer. Ann Transl Med. 8:4992020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li Y, Zhan Z, Yin X, Fu S and Deng X:
Targeted therapeutic strategies for triple-negative breast cancer.
Front Oncol. 11:7315352021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bou Zerdan M, Ghorayeb T, Saliba F, Allam
S, Bou Zerdan M, Yaghi M, Bilani N, Jaafar R and Nahleh Z: Triple
negative breast cancer: Updates on classification and treatment in
2021. Cancers (Basel). 14:12532022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang R, Tu J and Liu S: Novel molecular
regulators of breast cancer stem cell plasticity and heterogeneity.
Semin Cancer Biol. 82:11–25. 2022. View Article : Google Scholar
|
|
8
|
Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y,
Jiang G, Lu M, Zhang Z, Yin J, et al: Downregulation of FOXO3a by
DNMT1 promotes breast cancer stem cell properties and
tumorigenesis. Cell Death Differ. 27:966–983. 2020. View Article : Google Scholar :
|
|
9
|
Zhu K, Xie V and Huang S: Epigenetic
regulation of cancer stem cell and tumorigenesis. Adv Cancer Res.
148:1–26. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo F, Zhang M, Sun B, Xu C, Yang Y, Zhang
Y, Li S, Chen G, Chen C, Li Y and Feng H: LINC00115 promotes
chemoresistant breast cancer stem-like cell stemness and metastasis
through SETDB1/PLK3/HIF1α signaling. Mol Cancer. 23:602024.
View Article : Google Scholar
|
|
12
|
Lu H, Chen I, Shimoda LA, Park Y, Zhang C,
Tran L, Zhang H and Semenza GL: Chemotherapy-induced
Ca2+ release stimulates breast cancer stem cell
enrichment. Cell Rep. 18:1946–1957. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mannello F: Understanding breast cancer
stem cell heterogeneity: Time to move on to a new research
paradigm. BMC Med. 11:1692013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh P, Pesenti ME, Maffini S, Carmignani
S, Hedtfeld M, Petrovic A, Srinivasamani A, Bange T and Musacchio
A: BUB1 and CENP-U, primed by CDK1, are the main PLK1 kinetochore
receptors in mitosis. Mol Cell. 81:67–87.e9. 2021. View Article : Google Scholar :
|
|
15
|
Pan T, Zhou D, Shi Z, Qiu Y, Zhou G, Liu
J, Yang Q, Cao L and Zhang J: Centromere protein U (CENPU) enhances
angiogenesis in triple-negative breast cancer by inhibiting
ubiquitin-proteasomal degradation of COX-2. Cancer Lett.
482:102–111. 2020. View Article : Google Scholar
|
|
16
|
Lou Y, Lu J, Zhang Y, Gu P, Wang H, Qian
F, Zhou W, Zhang W, Zhong H and Han B: The centromere-associated
protein CENPU promotes cell proliferation, migration, and
invasiveness in lung adenocarcinoma. Cancer Lett. 532:2155992022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bruno F, Arcuri D, Vozzo F, Malvaso A,
Montesanto A and Maletta R: Expression and signaling pathways of
nerve growth factor (NGF) and pro-NGF in breast cancer: A
systematic review. Curr Oncol. 29:8103–8120. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marsland M, Dowdell A, Jiang CC, Wilmott
JS, Scolyer RA, Zhang XD, Hondermarck H and Faulkner S: Expression
of NGF/proNGF and their receptors TrkA, p75NTR and
sortilin in melanoma. Int J Mol Sci. 23:42602022. View Article : Google Scholar
|
|
19
|
Morisse M, Bourhis T, Lévêque R, Guilbert
M, Cicero J, Palma M, Chevalier D, le Bourhis X, Toillon RA and
Mouawad F: Influence of EGF and pro-NGF on EGFR/SORTILIN
interaction and clinical impact in head and neck squamous cell
carcinoma. Front Oncol. 13:6617752023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bradshaw RA, Pundavela J, Biarc J,
Chalkley RJ, Burlingame AL and Hondermarck H: NGF and ProNGF:
Regulation of neuronal and neoplastic responses through receptor
signaling. Adv Biol Regul. 58:16–27. 2015. View Article : Google Scholar :
|
|
21
|
Marcinkiewicz M, Marcinkiewicz J, Chen A,
Leclaire F, Chrétien M and Richardson P: Nerve growth factor and
proprotein convertases furin and PC7 in transected sciatic nerves
and in nerve segments cultured in conditioned media: Their presence
in Schwann cells, macrophages, and smooth muscle cells. J Comp
Neurol. 403:471–485. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan R, Yalinca H, Paoletti F, Gobbo F,
Marchetti L, Kuzmanic A, Lamba D, Gervasio FL, Konarev PV, Cattaneo
A and Pastore A: The structure of the pro-domain of mouse proNGF in
contact with the NGF domain. Structure. 27:78–89.e3. 2019.
View Article : Google Scholar
|
|
23
|
Thomas G: Furin at the cutting edge: From
protein traffic to embryogenesis and disease. Nat Rev Mol Cell
Biol. 3:753–766. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
He Z, Khatib AM and Creemers JWM: The
proprotein convertase furin in cancer: More than an oncogene.
Oncogene. 41:1252–1262. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Regua AT, Aguayo NR, Jalboush SA, Doheny
DL, Manore SG, Zhu D, Wong GL, Arrigo A, Wagner CJ, Yu Y, et al:
TrkA interacts with and phosphorylates STAT3 to enhance gene
transcription and promote breast cancer stem cells in
triple-negative and HER2-enriched breast cancers. Cancers (Basel).
13:23402021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Raninga PV, Zeng B, Moi D, Trethowan E,
Saletta F, Venkat P, Mayoh C, D'Souza RCJ, Day BW, Shai-Hee T, et
al: CBL0137 and NKG2A blockade: A novel immuno-oncology combination
therapy for Myc-overexpressing triple-negative breast cancers.
Oncogene. 44:893–908. 2025. View Article : Google Scholar :
|
|
28
|
Tang X, Cromwell CR, Liu R, Godbout R,
Hubbard BP, McMullen TPW and Brindley DN: Lipid phosphate
phosphatase-2 promotes tumor growth through increased c-Myc
expression. Theranostics. 12:5675–5690. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
American Veterinary Medical Association:
AVMA guidelines for the euthanasia of animals. 2020 edition. AVMA;
Schaumburg, IL: 2020
|
|
30
|
Yan Y, Zuo X and Wei D: Concise review:
Emerging role of CD44 in cancer stem cells: A promising biomarker
and therapeutic target. Stem Cells Transl Med. 4:1033–1043. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Namekawa T, Ikeda K, Horie-Inoue K, Suzuki
T, Okamoto K, Ichikawa T, Yano A, Kawakami S and Inoue S: ALDH1A1
in patient-derived bladder cancer spheroids activates retinoic acid
signaling leading to TUBB3 overexpression and tumor progression.
Int J Cancer. 146:1099–1113. 2020. View Article : Google Scholar
|
|
32
|
Ghebeh H, Sleiman GM, Manogaran PS,
Al-Mazrou A, Barhoush E, Al-Mohanna FH, Tulbah A, Al-Faqeeh K and
Adra CN: Profiling of normal and malignant breast tissue show
CD44high/CD24low phenotype as a predominant stem/progenitor marker
when used in combination with Ep-CAM/CD49f markers. BMC Cancer.
13:2892013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
DA Cruz Paula A and Lopes C: Implications
of different cancer stem cell phenotypes in breast cancer.
Anticancer Res. 37:2173–2183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seidah NG, Benjannet S, Pareek S, Savaria
D, Hamelin J, Goulet B, Laliberte J, Lazure C, Chrétien M and
Murphy RA: Cellular processing of the nerve growth factor precursor
by the mammalian pro-protein convertases. Biochem J. 314:951–960.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deichmann U, Schuster S, Mazat JP and
Cornish-Bowden A: Commemorating the 1913 michaelis-menten paper die
kinetik der invertinwirkung: Three perspectives. FEBS J.
281:435–463. 2014. View Article : Google Scholar
|
|
36
|
Hanissian SH, Akbar U, Teng B, Janjetovic
Z, Hoffmann A, Hitzler JK, Iscove N, Hamre K, Du X, Tong Y, et al:
cDNA cloning and characterization of a novel gene encoding the
MLF1-interacting protein MLF1IP. Oncogene. 23:3700–3707. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Goldberg AL: Protein degradation and
protection against misfolded or damaged proteins. Nature.
426:895–899. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lim KC, Tyler CM, Lim ST, Giuliano R and
Federoff HJ: Proteolytic processing of proNGF is necessary for
mature NGF regulated secretion from neurons. Biochem Biophys Res
Commun. 361:599–604. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hayakawa Y, Sakitani K, Konishi M, Asfaha
S, Niikura R, Tomita H, Renz BW, Tailor Y, Macchini M, Middelhoff
M, et al: Nerve growth factor promotes gastric tumorigenesis
through aberrant cholinergic signaling. Cancer Cell. 31:21–34.
2017. View Article : Google Scholar :
|
|
40
|
Tomellini E, Touil Y, Lagadec C, Julien S,
Ostyn P, Ziental-Gelus N, Meignan S, Lengrand J, Adriaenssens E,
Polakowska R and Le Bourhis X: Nerve growth factor and proNGF
simultaneously promote symmetric self-renewal, quiescence, and
epithelial to mesenchymal transition to enlarge the breast cancer
stem cell compartment. Stem Cells. 33:342–353. 2015. View Article : Google Scholar
|
|
41
|
Rockwell NC, Krysan DJ, Komiyama T and
Fuller RS: Precursor processing by kex2/furin proteases. Chem Rev.
102:4525–4548. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen C, Gupta P, Parashar D, Nair GG,
George J, Geethadevi A, Wang W, Tsaih SW, Bradley W, Ramchandran R,
et al: ERBB3-induced furin promotes the progression and metastasis
of ovarian cancer via the IGF1R/STAT3 signaling axis. Oncogene.
39:2921–2933. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou B and Gao S: Pan-cancer analysis of
FURIN as a potential prognostic and immunological biomarker. Front
Mol Biosci. 8:6484022021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ji S, Li S, Gao H, Wang J, Wang K, Nan W,
Chen H and Hao Y: An AIEgen-based 'turn-on' probe for sensing
cancer cells and tiny tumors with high furin expression. Biomater
Sci. 11:2221–2229. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen M, Pan Y, Liu H, Ning F, Lu Q, Duan
Y, Gan X, Lu S, Hou H, Zhang M, et al: Ezrin accelerates breast
cancer liver metastasis through promoting furin-like
convertase-mediated cleavage of Notch1. Cell Oncol (Dordr).
46:571–587. 2023. View Article : Google Scholar
|
|
46
|
Liu Z, Gu X, Li Z, Shan S, Wu F and Ren T:
Heterogeneous expression of ACE2, TMPRSS2, and FURIN at single-cell
resolution in advanced non-small cell lung cancer. J Cancer Res
Clin Oncol. 149:3563–3573. 2023. View Article : Google Scholar
|
|
47
|
Schaale D, Laspa Z, Balmes A, Sigle M,
Dicenta-Baunach V, Hochuli R, Fu X, Serafimov K, Castor T, Harm T,
et al: Hemin promotes platelet activation and plasma membrane
disintegration regulated by the subtilisin-like proprotein
convertase furin. FASEB J. 38:e701552024. View Article : Google Scholar : PubMed/NCBI
|