Cataracts, characterized by progressive
opacification of the ocular lens, are the leading cause of visual
impairment and blindness worldwide (1). Cataracts can be classified into
age-related cataracts (ARCs), congenital cataracts and secondary
cataracts caused by other reasons (2). ARCs, also known as senile
cataracts, constitute the most prevalent cataract subtype, with
typical onset after 50-60 years of age (3). Currently, no clinically proven
interventions exist to prevent ARC progression. Surgery remains the
gold-standard therapeutic approach, achieving rapid visual
rehabilitation in most cases (4). Despite surgical efficacy, ARCs
persist as a critical global health challenge due to high patient
volume, substantial surgical costs and potential postoperative
complications (5,6). Therefore, continuous and dedicated
efforts are required to develop improved therapeutic strategies for
ARC prevention and treatment.
ARCs develop through the long-term interplay of
multiple pathogenic factors, with complex and heterogeneous
underlying mechanisms. To date, multiple risk factors, including
aging, diabetes, genetic predisposition, oxidative stress and
ultraviolet (UV) radiation exposure, have been implicated in ARC
pathogenesis (3). The current
understanding of ARC pathogenesis strongly implicates reactive
oxygen species (ROS) accumulation as a key driver, with numerous
studies demonstrating the pathogenic roles of superoxide anion
(O2−), hydrogen peroxide
(H2O2) and hydroxyl radicals (·OH) generated
during aerobic metabolism (7-9).
Excessive ROS induce oxidative stress by chemically modifying and
damaging critical cellular components (10). Oxidative stress, in turn, causes
modifications in lens proteins (including enzymes, crystallins and
other chaperones), thereby reducing their solubility and stability,
and ultimately promoting crystallin aggregation (11). These alterations induce light
scattering within the lens, progressively leading to lens opacity
and subsequent visual acuity impairment (12). Oxidative stress also directly
damages lens epithelial cells (LECs), which are critical for
maintaining lens transparency and metabolic homeostasis, through
the peroxidation of proteins, lipids and DNA, while concurrently
activating signalling pathways and transcription factors (11,13-17). As the most metabolically active
components of the lens, LECs possess a sophisticated antioxidant
defence system to mitigate oxidative stress (18,19). However, when ROS levels exceed
the antioxidant capacity of LECs, oxidative damage to the LECs
triggers apoptosis, an early event in ARC pathogenesis (20,21).
Epigenetics refers to alterations in gene expression
without changes to the DNA sequence in response to environmental,
developmental and nutritional factors (22,23). These alterations are reversible
and subject to dynamic regulation, leading to heritable changes in
gene expression and cellular phenotypes (24). Extensive research has
demonstrated that epigenetic mechanisms function as pivotal
regulators of disease pathogenesis, influencing initiation,
progression, prevention and treatment (25,26). The emergence of aberrant
epigenetic changes is associated with the pathogenesis of various
diseases, including cancer, diabetes, Alzheimer's disease and other
age-related disorders (27-30). Epigenetic modifications not only
underlie the molecular mechanisms driving disease development but
also serve as promising biomarkers for clinical diagnosis and
enable precision-targeted therapeutic interventions (31,32). Epigenetic modifications primarily
encompass DNA methylation, histone modifications, RNA
modifications, non-coding RNAs (ncRNAs) and chromatin remodelling.
A substantial body of research has established the critical
involvement of epigenetic processes in gene expression regulation,
DNA damage repair, and cell cycle control and aging, highlighting
their fundamental importance in molecular biology and genetics
(33-35).
Breakthroughs in epigenetic research related to
ocular diseases have only emerged within the past decade.
Epigenetic modifications have grown into a leading frontier in
biomedical research, with accumulating studies revealing their
regulatory impact on several ocular diseases such as cataracts,
glaucoma, diabetic retinopathy and age-related macular degeneration
(36-39). Epigenetic research can deepen the
understanding of the molecular mechanisms of ocular diseases and
open novel avenues for the development of epigenome-targeted
treatments. The present review summarizes current advances in
epigenetic research on ARCs, discussing the implications of these
research advances for future investigations and the therapeutic
potential of epigenetic interventions in ARCs.
Epigenetic modifications represent heritable changes
in gene expression that occur independently of alterations in the
underlying DNA sequence (40).
Epigenetic modifications, including ncRNAs, DNA methylation,
histone modifications and N6-methyladenosine
(m6A) modification, are now widely acknowledged as key
molecular drivers in the development of ARCs (41-43). A deeper understanding of these
epigenetic mechanisms will elucidate disease pathogenesis and
enable targeted epigenetic therapies for ARCs. The subsequent
sections will first introduce key epigenetic alterations, including
ncRNAs, DNA methylation, histone modifications and m6A
modification, and then systematically examine their roles in ARC
pathogenesis.
The central dogma of molecular biology describes the
flow of genetic information from DNA to RNA to protein (44,45). For decades, research on cataract
pathogenesis has predominantly focused on protein-coding genes.
However, advances in sequencing technologies have enabled the
identification of numerous unique ncRNAs and confirmed their
cellular presence. ncRNAs represent a class of RNA molecules that
lack protein-coding sequences and are consequently not translated
into polypeptides or proteins. This category includes microRNAs
(miRNAs/miRs), long non-coding RNAs (lncRNAs), circular RNAs
(circRNAs), transfer RNA-derived small RNAs (tsRNAs) and small
nuclear RNAs (46). Through
specific interactions with DNA, RNA and proteins, ncRNAs precisely
regulate nearly all biological pathways, including signal
transduction (47),
transcriptional and post-transcriptional regulation (48,49), translational repression (50), and translation initiation
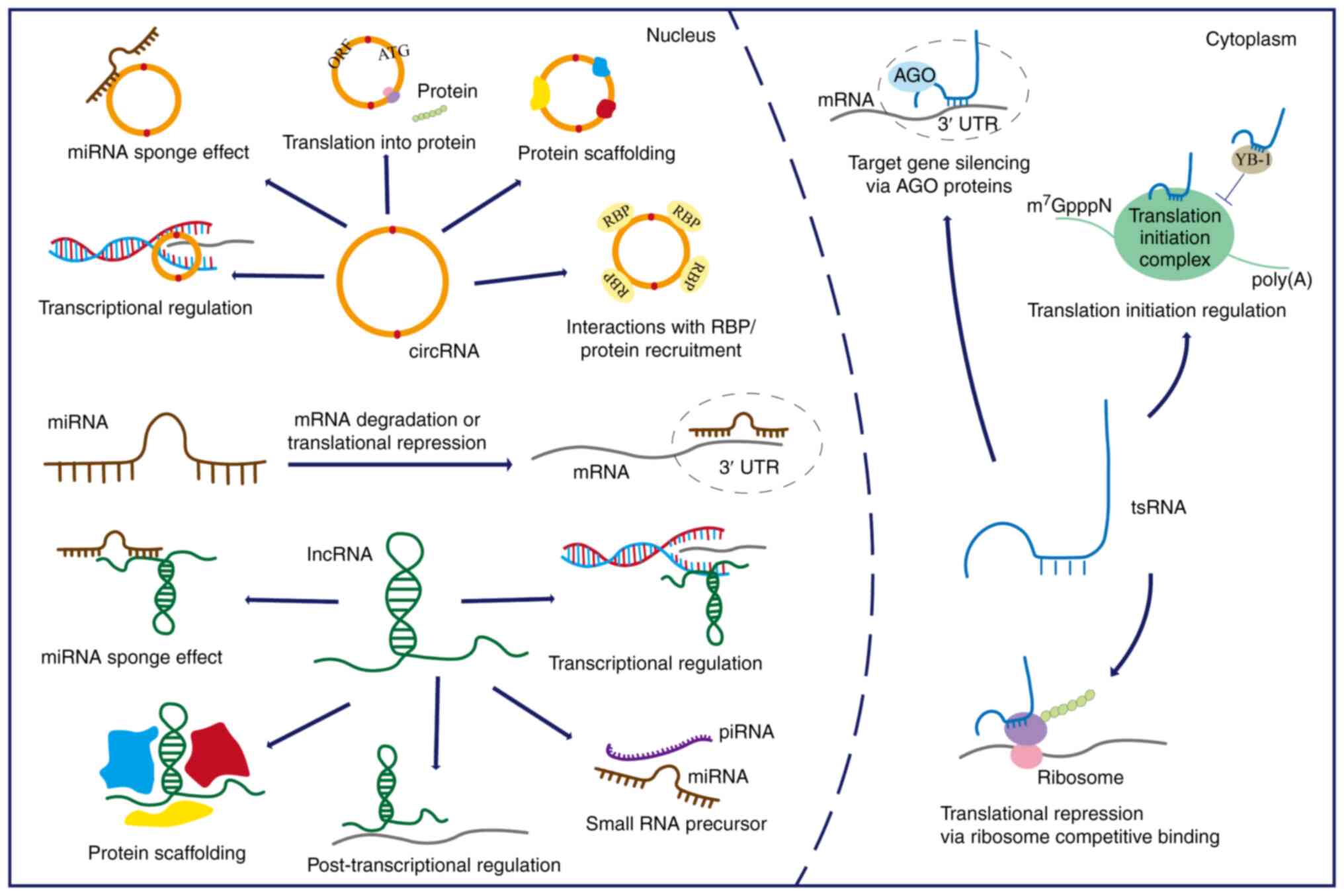
regulation (51) (Fig. 1). ncRNAs operate in both
physiological and pathological contexts, exhibiting strong
potential as novel molecular biomarkers and therapeutic targets
(52-54). Consequently, ncRNAs serve a
pivotal role in the pathogenesis and progression of ARCs, as
extensively documented in several studies (55-57). The present review primarily
focuses on reviewing advances in research on the four major ncRNAs
(miRNAs, lncRNAs, circRNAs and tsRNAs) and their regulatory
networks in ARCs.
miRNAs are the most extensively studied and best
characterized small ncRNAs to date. miRNAs are small ncRNA
molecules, typically 20-30 nucleotides in length, that serve
critical roles in structural, enzymatic and regulatory processes
across biological systems (58).
Precursor miRNAs undergo a tightly regulated series of nuclear and
cytoplasmic processing steps mediated by the endoribonucleases
DROSHA and DICER, respectively, ultimately yielding mature miRNAs.
The mature miRNAs are then incorporated into the RNA-induced
silencing complex and regulate gene expression by binding to the 3'
untranslated region (UTR) of target mRNAs, leading to either mRNA
degradation or translational repression (58,59). miRNAs have also been reported to
interact with other regions, such as the 5' UTR, coding sequence
and gene promoters (60,61). Furthermore, accumulating evidence
has demonstrated that miRNAs can post-transcriptionally activate
gene expression in specific contexts, challenging the conventional
paradigm of miRNA-mediated silencing and highlighting their
functional versatility (62-64). Notably, miRNAs form intricate,
highly interconnected regulatory networks, wherein individual
miRNAs typically regulate multiple mRNA targets, and conversely,
most mRNAs are subject to combinatorial regulation by multiple
miRNAs (65). Diverse
physiological and pathological processes, including apoptosis
(66), autophagy (67), oxidative stress (68), senescence (69) and DNA repair (70), have been shown to influence the
biological characteristics of LECs in ARC, as evidenced by both
in vitro experiments and animal studies. miRNAs have been
demonstrated to serve important roles in these processes, making it
plausible that miRNAs are closely associated with cataract
formation (Table SI).
Studies have revealed alterations in miRNA
expression profiles between transparent and cataractous lenses
(71-73). For example, miRNA let-7b
expression exhibits a positive association with patient age, and
elevated miRNA let-7b expression is associated with higher nuclear
(N), cortical (C) and posterior subcapsular (P) cataract scores
according to the Lens Opacities Classification System III (74) in patients with ARCs (73). Similarly, miR-34a expression
levels vary among patients of different ages, with moderate
associations observed between high N, C and P cataract scores and
high miR-34a levels (75).
Elevated levels of miR-210-3p have been identified in the aqueous
humour of patients with ARCs, and the elevation demonstrated strong
diagnostic potential for ARCs while being associated with oxidative
stress marker levels (76).
miR-15a and miR16-1 are expressed at a higher level in the LECs of
patients with cataracts compared with normal controls (77). Notably, ARCs can be classified
into distinct subtypes based on the location of opacification,
including cortical cataracts, nuclear cataracts (NCs), and
posterior and anterior subcapsular cataracts (ASCs), while the
remainder is classified as mixed cataracts, and differential
expression of miRNAs has been observed among these subtypes
(72). The expression levels of
let-7a-5p, let-7d-5p, let-7g-5p and miR-23b-3p differ between
patients with NC and ASC, as measured in lens epithelium samples
(72). Thus, aberrant miRNA
expression levels may not only act as risk factors in the formation
and progression of ARC, but also serve as potential biomarkers for
ARCs based on their differential expression patterns in different
severity stages and subtypes of ARCs. However, no miRNAs are used
for clinical diagnosis currently, requiring further validation in
clinical practice.
miRNAs typically function by binding to target mRNAs
and inhibiting translation (58,59). Thus, specific miRNAs indirectly
contribute to cataract formation by regulating key factors involved
in cellular processes such as apoptosis, proliferation and
oxidative stress. For instance, miR-23b-3p promotes apoptosis and
inhibits autophagy in LECs under oxidative stress by directly
targeting and downregulating silent information regulator 1
(SIRT1), a critical mediator of oxidative stress resistance,
thereby contributing to ARC pathogenesis (78). Similarly, miR-211 induces
apoptosis and decreases cell viability by suppressing SIRT1
expression (79). E2F
transcription factor 3 (E2F3), a member of the E2F transcription
factor family, serves essential roles in regulating cell
proliferation, apoptosis and differentiation (80,81). As a key cell cycle regulator,
E2F3 promotes cell cycle re-entry in quiescent lens cells. However,
aberrant cell cycle re-entry may trigger programmed cell death
(82-84), and has been observed in LECs,
which is closely linked to ARC pathogenesis (85). Experimental evidence has
identified miR-34a (86),
miR-15a (87), miR-221 (88), miR-630 and miR-378a-5p (89), as direct post-transcriptional
suppressors of E2F3, collectively contributing to lens opacity
progression. miR-34a, the expression of which is increased in the
cataractous lens, triggers mitochondria-mediated apoptosis and
oxidative stress by suppressing Notch2 (90). In addition, the upregulation of
miR-34a together with the downregulation of hexokinase 1 (HK1)
inhibits the proliferation and induces the apoptosis of LECs, thus
accelerating the opacification of mouse lenses via the HK1/caspase
3 signalling pathway (91).
Furthermore, Cao et al (67) demonstrated that miRNA let-7c-5p
directly targeted excision repair cross-complementing rodent repair
deficiency, complementation group 6 (ERCC6), and thus, inhibited
its role in autophagy, a process necessary for maintaining lens
transparency through degradation of misfolded proteins and damaged
organelles. miR-378a is upregulated in human cataract tissues and
inhibits LEC proliferation while promoting apoptosis by modulating
the ROS/PI3K/AKT signalling pathway (92).
Emerging evidence has indicated that certain miRNAs
function as cataract suppressors by preventing lens opacification
(68,93,94). Given that ARC pathogenesis is
associated with apoptotic cell death in the lens, extensive
research has highlighted dysregulated apoptosis of LECs as a key
pathogenic mechanism in cataract development (95-97). Bcl-2 family proteins are
essential for the intrinsic apoptotic pathway (98). B-cell lymphoma-2-like-2 (BCL2L2),
a Bcl-2 family member, is an important regulator of apoptosis
(99). Zhang et al
(93) identified BCL2L2 as a
direct target of miR-133b, demonstrating that miR-133b-mediated
downregulation of BCL2L2 expression led to apoptosis inhibition.
While BCL2L2 promoted apoptosis, miR-133b functioned as an
anti-apoptotic regulator by suppressing BCL2L2 expression (93). Likewise, miR-182, which is
implicated in various ophthalmic disorders, including pterygium
(100), glaucoma (101) and age-related macular
degeneration (102), protects
LECs against oxidative stress-induced apoptosis by regulating the
nicotinamide adenine dinucleotide phosphate oxidase subunit 4
(NOX4) and p38 MAPK signalling pathway (68). Furthermore, miR-125b is
downregulated in ARC lens tissue, where it suppresses p53 mRNA
expression and exerts anti-apoptotic effects on LECs (94). Although numerous miRNAs have been
demonstrated to attenuate lens opacity progression, experimental
validation by in vivo studies conducted in animals, such as
rats or rabbits, remains limited. To the best of our knowledge,
only three in vivo studies have been reported at present.
Administration of antagomirs targeting miR-326 (103), miR-29a-3p (104) and miR-187 (105) independently inhibited cataract
formation in animal models of ARC, suggesting their potential as
non-surgical therapeutic candidates.
Single nucleotide polymorphisms (SNPs) within miRNA
binding sites can affect the base pairing between miRNAs and their
target mRNAs, thereby altering miRNA-mediated gene regulation
(106). Oxidative DNA damage
and impaired DNA repair capacity in LECs are strongly involved in
ARC pathogenesis (107,108). Accumulating data have
demonstrated that SNPs within miRNA binding sites of oxidative DNA
damage repair genes may modulate ARC susceptibility (70,109,110). Zou et al (70) systematically analysed 10
miRNA-binding SNPs located in the 3' UTRs of seven oxidative
damage-related genes. Their findings demonstrated that the C allele
of XPC-rs2229090 increased susceptibility to the nuclear type of
ARC (ARNC). Mechanistically, the C allele exhibited stronger
binding affinity for hsa-miR-589-5p compared with the G allele,
resulting in reduced XPC expression and impaired DNA repair
capacity in LECs (70).
Furthermore, Kang et al (109) demonstrated that the protective
effect of Nei-like DNA glycosylase 2 (NEIL2)-rs4639T in ARCs may be
mediated through maintenance of normal NEIL2 expression levels,
potentially via disruption of the binding of rs4639T with
hsa-miR-3912-5p. Additionally, the rs78378222 polymorphism in the
p53 3' UTR contributes to ARC pathogenesis by regulating
miR-125b-mediated apoptosis in LECs (110). Taken together, these findings
suggest that SNP-mediated alterations in miRNA-dependent
post-transcriptional regulation constitute a novel pathogenic
mechanism in ARCs and reveal novel therapeutic targets.
In general, research on miRNA alterations in ARCs
has reached a relatively mature stage, with comprehensive studies
demonstrating the crucial roles of miRNAs in mediating LEC lesions
in vitro and cataract progression in vivo (103-105). Exosomal miRNAs have become a
research focus; however, their relationship with ARCs remains
poorly understood. At present, only two studies have demonstrated
the roles of exosomal miR-222-3p in ARC formation and miR-125a-3p
in disease progression (66,111). Exosomes are 50-200-nm
extracellular nanovesicles derived from multivesicular bodies in
the cytoplasm and serve as an important medium of intercellular
communication (112,113). Exosomes regulate receptor cells
and tissues by transmitting effectors, including proteins, mRNAs or
miRNAs (114). Various types of
cells can secret miRNAs via exosomes, a process that protects
miRNAs from degradation and ensures their function in target cells
(115). Exosomes transport
miRNAs to target cells without being degraded by RNAses, making
exosomal miRNAs more stable and reliable biomarkers than their
non-exosomal counterparts (116). Exosomal miRNAs are emerging as
promising biomarkers for aging and age-related diseases (117-119). Furthermore, exosomes represent
promising drug delivery vehicles due to their inherent advantages,
including high biocompatibility, high biological barrier
penetration and high circulatory stability (120). Therefore, investigating
exosomal miRNAs could help to elucidate the pathogenesis of ARCs,
facilitate biomarker identification and enable the development of
innovative intervention strategies.
Alongside short miRNAs, a wide array of longer
transcripts, referred to as lncRNAs, has been increasingly
characterized. lncRNAs are >200 bp long, and exhibit relatively
low expression levels, poor stability and limited evolutionary
conservation (121). Therefore,
lncRNA research presents considerable challenges due to these
inherent molecular characteristics, and is only recently undergoing
a surge. Notably, similar to miRNAs, most lncRNAs are associated
with human diseases; however, in contrast to their smaller
counterparts, lncRNAs exhibit cell type-specific expression
patterns and distinct subcellular localization (122). lncRNAs perform diverse
regulatory functions, including acting as miRNA sponges, signal
mediators, molecular decoys, chromatin modifiers, and regulators of
gene expression and pre-mRNA splicing (123,124). Among these functions, their
role as competing endogenous RNAs (ceRNAs) or miRNA sponges is
particularly significant. The ceRNA hypothesis, first proposed by
Salmena et al (125) in
2011, suggests that lncRNAs regulate gene expression
post-transcriptionally by competitively binding to miRNAs via
shared miRNA response elements, thereby preventing miRNA-mRNA
interactions. Through diverse molecular mechanisms, lncRNAs
regulate critical biological processes such as stem cell
maintenance, cellular differentiation and chromatin remodelling, as
well as transcription, splicing, translation, degradation and
transport of mRNA (126,127).
Previous studies have indicated that lncRNAs are highly active in
ARCs, where they may either promote or suppress disease initiation
and progression (Table SII)
(128-130).
Emerging evidence has established the pivotal role
of numerous lncRNAs as pathogenic drivers in ARC pathogenesis. As
aforementioned, lncRNAs can act as ceRNAs or endogenous miRNA
sponges, effectively sequestering miRNAs and attenuating their
post-transcriptional repression of target mRNAs (125). Building upon this theoretical
framework, Jin et al (133) experimentally validated miR-214
as a direct target of lncKCNQ1OT1. Their findings demonstrated that
lncKCNQ1OT1 knockdown upregulated miR-214 expression and
subsequently downregulated the expression of its downstream
effector caspase-1, indicating the pathogenic role of the
lncKCNQ1OT1/miR-214/caspase-1 axis in cataract formation (133). Furthermore, lncKCNQ1OT1
functions as a molecular sponge for miR-223-3p, which directly
targets BCL2L2. Through the lncKCNQ1OT1/miR-223-3p/BCL2L2 axis,
lncKCNQ1OT1 promotes apoptosis and pyroptosis, while exacerbating
oxidative damage in H2O2-treated LECs
(134). Another study
demonstrated that lncKCNQ1OT1 downregulation protected LECs from
oxidative stress-induced damage via the miR-124-3p/BCL-2-like 11
pathway (135). In addition to
lncKCNQ1OT1, TUG1 is another well-characterized lncRNA that
exacerbates the pathogenesis of ARC, and has been shown to promote
apoptosis in H2O2 or UV irradiation-treated
LECs through three distinct molecular pathways: TUG1/miR-196a-5p
(136), TUG1/miR-421/caspase-3
(20) and TUG1/miR-29b/second
mitochondria-derived activator of caspases (137). Furthermore, research has
identified multiple novel pathogenic lncRNAs, including NEAT1
(129,138), MEG3 (139,140) and OIP5-AS1 (141), with additional candidates
expected to emerge as research progresses.
By contrast, other studies have indicated that
certain lncRNAs exert protective effects to prevent ARC
progression. For example, lncRNA phospholipase C δ3 (PLCD3)-OT1 may
act as a ceRNA to promote PLCD3 expression by sponging miR-224-5p,
thus promoting cell proliferation and viability, and inhibiting
apoptosis, under oxidative stress conditions (142). Similarly, lncRNA
NONHSAT143692.2 modulates 8-oxoguanine DNA glycosylase (OGG1)
expression in LECs by competitively binding to miR-4728-5p, thereby
protecting LECs from cell apoptosis and oxidative damage (130). Furthermore, Cheng et al
(143) revealed that lncRNA H19
suppressed apoptosis, mitigated oxidative damage, and promoted
proliferation and viability in UV irradiation-treated LECs through
the functional interplay of lncRNA H19, miR-29a and thymine DNA
glycosylase.
Although acting as miRNA sponges represents a
well-established function of lncRNAs (144-146), their other regulatory functions
are also implicated in cataract pathogenesis. lncRNAs are typically
classified into five major classes according to their relative
genomic locations: Antisense (AS), intergenic, overlapping,
intronic and full lapping (147). Within these lncRNA classes, AS
lncRNAs, which are reverse-strand complements of their endogenous
protein-coding counterparts, are generally non-protein-coding and
account for a substantial proportion of the entire long non-coding
transcriptome (148-150). Previous studies have
demonstrated that natural AS transcripts participate in diverse
biological processes by regulating sense gene expression at
multiple levels, including transcriptional regulation via gene
promoter activation, and post-transcriptional regulation through
mRNA stabilization and translational modulation (151,152). Glutathione peroxidase 3
(GPX3)-AS has been found to upregulate GPX3 expression at both the
mRNA and protein levels, enhancing the activity of this ROS
scavenger to inhibit LEC apoptosis, thereby revealing a potential
therapeutic target for ARCs (153). Furthermore, lncRNAs can also
serve as direct precursors for miRNA biogenesis (154). A representative example is
lncRNA H19, which serves as the precursor for miR-675, where H19
downregulation leads to reduced miR-675 expression. lncRNA H19
modulates LEC function via miR-675-mediated regulation of
crystallin αA (CRYAA) expression, offering novel insights into ARNC
pathogenesis (155).
Overall, lncRNAs participate in ARC pathogenesis
through multifaceted regulation of LEC biology, including oxidative
stress responses and apoptotic pathways. Given their functional
versatility and multi-target nature, the precise roles and
regulatory networks of several lncRNAs in ARC pathogenesis have yet
to be comprehensively characterized. Furthermore, although in
vitro experiments offer valuable mechanistic insights, their
translational relevance to whole organisms remains limited. To the
best of our knowledge, no in vivo models investigating
lncRNAs in ARCs have been reported at present. Animal models, which
better recapitulate the complexity of physiological systems, are
crucial for validating the observed in vitro effects.
circRNAs are single-stranded, covalently closed RNA
molecules without 5' caps and 3' poly-A tails, and are usually
produced through a back-splicing process (156,157). The absence of both 5' caps and
3' polyadenylated tails renders circRNAs resistant to
exonuclease-mediated degradation, thus ensuring their stability and
longevity in cellular processes (158). circRNAs possess distinctive
properties and diverse functions that continue to be elucidated.
Similar to lncRNAs, circRNAs can function as efficient miRNA
sponges, representing one of their most well-characterized
regulatory modes (159-161). Beyond their well-known role as
miRNA sponges, studies have revealed that circRNAs can also
interact with RNA-binding proteins (162), regulate RNA splicing and
transcription (163,164), and undergo translation into
functional peptides or proteins (165,166). circRNAs also participate in a
wide range of biological processes, including fibrosis, cell
apoptosis, proliferation, differentiation and angiogenesis
(167). In recent years, their
roles as vital regulators in multiple ocular diseases have
attracted increasing attention (168-170). Similar to linear ncRNAs,
circRNAs are implicated in cataract formation (Table SIII) (171-173). The circRNA/miRNA/mRNA
regulatory network has advanced the understanding of the molecular
basis of cataractogenesis.
Studies have characterized specific circRNAs that
contribute to ARC pathogenesis. For example, hsa_circ_0105558
promotes apoptosis and aggravates the oxidative damage of LECs
under H2O2 exposure through the
miR-182-5p/activating transcription factor 6 axis (174). hsa_circ_0007905 upregulates
eukaryotic translation initiation factor 4E binding protein 1
(EIF4EBP1) expression by competitively binding to miR-6749-3p,
consequently enhancing LEC apoptosis while inhibiting LEC
proliferation, resulting in ARC progression (175). Notably, hsa_circ_0007905
expression in LECs is upregulated through methyltransferase like 3
(METTL3)-mediated m6A modification (175). A comprehensive discussion of
m6A modifications of circRNAs is presented in the
m6A modification section. Notably, circMRE11A_013
(circMRE11A) acts primarily as a molecular scaffold that interacts
with UBX domain-containing protein 1 rather than functioning as a
miRNA sponge, and promotes excessive activation of
ataxia-telangiectasia mutated kinase (ATM), ultimately inducing LEC
cell cycle arrest and senescence through the ATM/p53/p21 signalling
pathway (176). Furthermore,
recombinant adeno-associated virus vector virions of circMRE11A
have been injected into the mouse vitreous cavity, providing in
vivo evidence that circMRE11A drives lens aging and
opacification in mice (176).
In addition to circMRE11A, circSTRBP (hsa_circ_0088,427) exerts its
biological effects independently of miRNA sponging activity by
interacting with insulin-like growth factor 2 mRNA-binding protein
1 (IGF2BP1), and enhances the stability and expression of NOX4 mRNA
in LECs (171). circSTRBP
exacerbates H2O2-induced oxidative stress and
apoptosis in LECs by enhancing NOX4 mRNA stability through IGF2BP1
recruitment, providing novel perspectives for ARC intervention
(171).
Emerging evidence has identified multiple protective
circRNAs in ARC pathogenesis, with several candidates exhibiting
potent antioxidant and anti-apoptotic effects in LECs. circRNA
homeodomain interacting protein kinase 3 (circHIPK3) promotes
viability and proliferation, inhibits apoptosis, and alleviates
oxidative damage in H2O2-treated LECs via
three distinct molecular pathways: circHIPK3/miR-221-3p/PI3K/AKT
(177),
circHIPK3/miR-193a/CRYAA (178)
and circHIPK3/miR-495-3p/histone deacetylase 4 (HDAC4) (179). circ_0060,144 regulates HIPK3
expression by sponging miR-23b-3p, thereby influencing LEC
homeostasis through regulation of proliferation, apoptosis and the
oxidative stress response (180). UV radiation downregulates
circRNA erythrocyte membrane protein band 4.1 expression, which
subsequently reduces 3'(2'), 5'-bisphosphate nucleotidase 1 levels
via miR-24-3p binding, thus inhibiting proliferation and promoting
apoptosis in LECs (181).
Furthermore, circRNA 06209 acts as a ceRNA by sequestering
miR-6848-5p to neutralize its suppressive effect on arachidonate
15-lipoxygenase, ultimately promoting proliferation and inhibiting
apoptosis in H2O2-treated LECs (182). Notably, injection of a
circRNA_06209-overexpressing plasmid into a
Na2SeO3-induced cataract model attenuated
lens opacity while reducing insoluble protein aggregation and
increasing soluble protein levels in the lens, demonstrating the
anti-cataract efficacy of circRNA_06209 in both in vitro and
in vivo settings (182).
Given that circRNAs exhibit high stability,
evolutionary conservation across species and tissue specificity
(183), investigating
ARC-related circRNAs as potential biomarkers or therapeutic targets
could open novel avenues for the diagnosis, prevention and
treatment of ARCs. However, only a limited number of circRNAs in
ARCs have well-characterized biological functions and mechanistic
pathways. While circRNAs employ diverse molecular mechanisms and
functions, current research on their roles in ARCs has largely
focused on their function as miRNA sponges and their interactions
with proteins (171,174,184-186), suggesting that novel functions
or mechanisms remain to be identified.
tsRNAs constitute a family of small ncRNAs produced
by the cleavage of precursor or mature transfer RNAs (tRNAs) via
specific endonucleases (187).
tsRNAs are mainly classified into two types: tRNA halves and
tRNA-derived fragments, based on the cleavage position of the
precursor or mature tRNAs (188). Advances in high-throughput
sequencing technologies have revealed that tsRNAs serve crucial
roles in the regulation of gene expression, post-transcriptional
modifications, apoptotic inhibition and protein translation
(189-191), while their dysregulation
contributes to various pathological processes in diseases such as
cancer, cardiovascular diseases and Alzheimer's disease (192-194). Investigations have begun to
reveal the crucial involvement of tsRNAs in ophthalmic diseases
(195-197). Through integrated
next-generation sequencing analysis, a study conducted in 2022
systematically established the first evidence of the changes in
tsRNA profiles in Emory mice, a well-characterized model for human
ARCs (198). A total of 422
differentially expressed tsRNAs were identified in 8-month-old mice
(156 upregulated and 266 downregulated), with Gene Ontology (GO)
analysis revealing that these target genes were predominantly
enriched in processes including but not limited to camera-type eye
development and sensory organ development (198). As tsRNA research remains in its
infancy, further investigations focusing on the association between
tsRNAs and ARCs, as well as their underlying molecular mechanisms,
are needed to provide novel insights into cataractogenic processes
and ARC disease management.
Understanding ncRNA functions in isolation becomes
increasingly difficult due to intricate regulatory networks among
ncRNAs. These networks involve multiple layers of interaction:
ncRNAs frequently share common protein-coding mRNA targets; miRNAs
functionally interact with other ncRNA species rather than
operating in isolation; and lncRNAs and circRNAs, in turn, regulate
the abundance of available miRNAs (199). ncRNA networks serve a pivotal
role in ARC pathogenesis by modulating gene expression, oxidative
stress responses and pathological processes in LECs, indicating
potential targets for early diagnosis and therapeutic intervention
(134,200).
In addition to the networks among different types
of ncRNAs, such as the aforementioned ceRNAs, interactions also
exist between the same type of ncRNAs. Emerging research across
various diseases, including cancers and cardiovascular diseases,
has revealed that certain miRNAs can be involved in the same
biological pathways either as effectors or regulators (201). Krek et al (202) demonstrated that myotrophin
(Mtpn) was a direct target of miR-124, let-7b and miR-375,
as evidenced by small interfering RNA (siRNA)-mediated
downregulation of Mtpn protein levels and suppression of
luciferase activity in dual-luciferase reporter assay.
Cotransfection of the luciferase reporter and a pool of siRNA
duplexes designed to mimic the function of miR-124, let-7b and
miR-375, resulted in normalized luciferase activity that was
substantially less than the activity in any of the groups
transfected with a single siRNA, revealing cooperative regulation
of Mtpn by these miRNAs (202). Similarly, lncRNAs could
interact with each other by co-regulating genes participating in
the same or similar biological functions (203). Multiple lncRNAs, such as
OIP5-AS1, TUG1 and NEAT1, have been implicated in the synergistic
regulation of genes and pathways in cancer (204). Current research on ncRNAs in
ARCs primarily focuses on their dysregulation in ARCs and the ceRNA
mechanism (104,140,174,205). The cooperative interactions
among ncRNAs remain poorly characterized and require further
analyses and validation to achieve a comprehensive understanding of
the cooperative behaviours of ncRNAs in ARC pathogenesis.
DNA methylation, a fundamental epigenetic
modification, stably modulates gene expression through heritable
regulatory mechanisms, influencing diverse physiological processes
and disease pathogenesis (206). In mammals and humans, DNA
methylation critically participates in numerous biological
processes, including genomic imprinting (207), X-chromosome inactivation
(208), transcriptional
regulation (209) and
tumorigenesis (210). DNA
methylation is a covalent chemical modification involving the
selective addition of methyl groups to DNA molecules, catalysed by
DNA methyltransferases (211).
Although DNA methylation occurs at various sites, including the C5
position of cytosine (212), N6
position of adenine (213) and
N7 position of guanine (214),
5-methylcytosine (m5C) remains the most prevalent and
extensively studied modification in humans, representing the
principal focus of DNA methylation research (215-217). The term 'DNA methylation' in
general primarily refers to the methylation of the 5th carbon atom
of cytosine within cytosine-phosphate-guanine (CpG) sites, yielding
m5C (215,218).
CpG sites, consisting of cytosine-guanine
dinucleotides, are distributed throughout the genome (219). These sites frequently cluster
in CpG islands, genomic regions with a high density of CpG
dinucleotides, which are predominantly located near gene promoters
(220). CpG islands are
critical regulatory elements for gene expression, as promoter DNA
methylation can inhibit transcription factor binding or recruit
methyl-CpG binding domain (MBD) proteins and HDACs, ultimately
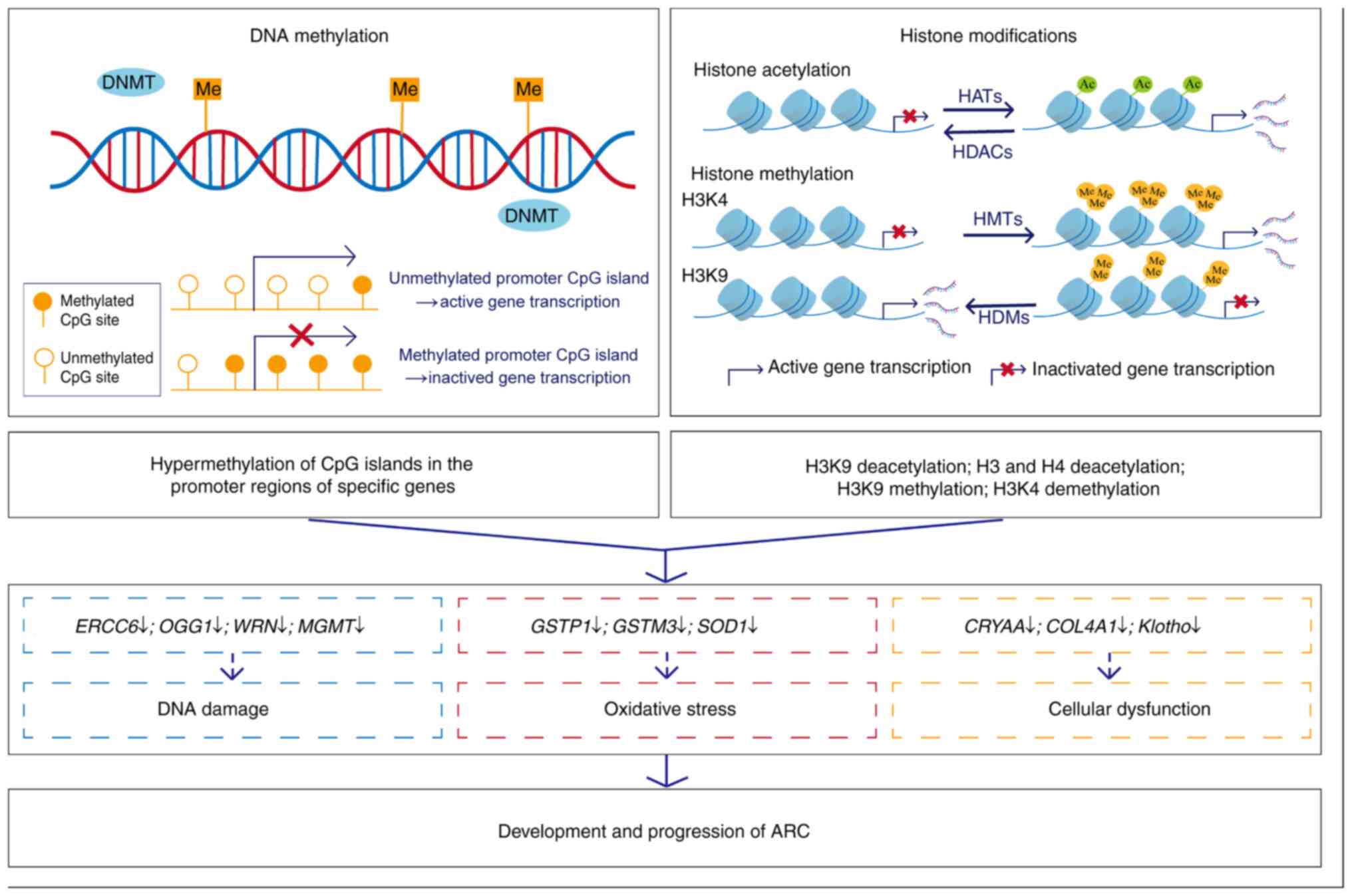
inducing gene silencing (221,222). Hypermethylation of CpG islands
in promoter regions is strongly associated with transcriptional
silencing, whereas unmethylated states are typically associated
with active gene expression (108,223).
Previous studies in ophthalmology have indicated
that aberrant DNA methylation disrupts the expression of key genes,
such as RHO, OPN1LW and TUBA1A (227,228), contributing to the pathogenesis
of various ocular diseases, such as corneal and conjunctival
disorders, glaucoma (229),
cataracts (230), retinal
diseases (231), and ocular
tumours (232). As the leading
global cause of blindness, cataracts, particularly ARCs, serve as a
paradigm of ocular aging, wherein epigenetic dysregulation
synergizes with cumulative environmental stressors, including UV
radiation and chemical exposures (233,234). While congenital and traumatic
cataracts result from genetic mutations or physical injury, ARCs
stem from progressive lens opacification associated with multiple
factors, including oxidative stress, protein aggregation, and
critically, DNA methylation-mediated silencing of protective genes
in LECs (Table SIV) (108,235,236). For example, age-dependent
hypermethylation of the Klotho gene, a key anti-aging factor,
progressively silences its expression, leading to marked reduction
or even complete loss of Klotho expression at both mRNA and protein
levels, ultimately participating in ARC pathogenesis (237). Furthermore, both mRNA and
protein expression of DNA methylation-related genes (DNMT3B and
MBD3) are elevated in LECs of ARCs, further substantiating the
involvement of DNA methylation in ARC development (238).
To characterize the epigenetic landscape of ARCs,
several studies have examined genome-wide methylation patterns in
ARC samples. Chen et al (239) employed the MethylationEPIC
BeadChip (850K) to profile DNA methylation in anterior lens capsule
membranes from patients with ARCs and control subjects. By
integrating GO and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analyses with their previously published
transcriptome sequencing data, the authors identified
differentially methylated genes. Their analysis revealed 52,705
differentially methylated sites in the ARC group (13,858
hypermethylated and 38,847 hypomethylated) compared with the
control group, which consisted of non-ARC patients, with GO and
KEGG analyses highlighting functional associations with cell
membrane components, calcium signalling pathways and their
potential molecular mechanisms (239). Similarly, a study conducted in
2018 used DNA methylation microarrays, a high-throughput sequencing
approach, to identify methylated genes, and revealed that aberrant
methylation of collagen type IV α1 chain (COL4A1), gap junction
protein α3 and signal induced proliferation associated 1 like 3 was
associated with ARCs (240).
Further investigation is warranted to elucidate the role of these
DNA methylation changes in ARC pathogenesis.
Oxidative stress is a well-established driver of
LEC pathophysiology and a key contributor to ARC pathogenesis
(7-9). Emerging evidence has further linked
oxidative stress-induced DNA damage in LECs to cataract formation
(241,242). Therefore, DNA
methylation-mediated dysregulation of oxidative stress-related
genes and DNA damage repair genes has emerged as a primary research
focus, given its pivotal role in ARC development.
Methylation changes in oxidative stress-related
genes, as epigenetic modifications of the antioxidant defence
system, likely contribute to oxidative stress-induced lens damage
in ARC (223,235,243).
One of the main antioxidant defence mechanisms is
nuclear factor erythroid 2-related factor 2 (Nrf2) activation. Nrf2
is a central nuclear transcriptional factor, which controls the
transcription of >200 protective genes, including 20 genes
encoding antioxidant enzymes (244). The activity of the Nrf2 pathway
is regulated through multiple mechanisms, both upstream and
downstream, as well as by other signalling pathways (245). Kelch-like ECH-associated
protein 1 (Keap1) is an oxidative stress sensor and a negative
regulator of Nrf2 (246). Under
basal conditions, Keap1 binds to Nrf2 in the cytoplasm, promoting
its ubiquitin-mediated proteasomal degradation, and thus,
maintaining Nrf2 at basal levels (247). During oxidative stress,
elevated ROS disrupt the Keap1-Nrf2 interaction, enabling Nrf2
nuclear translocation. Within the nucleus, Nrf2 binds to
antioxidant response elements in target gene promoters, inducing
the expression of antioxidant enzymes that mediate ROS clearance
(248). The Nrf2/Keap1 pathway
serves as a master regulator of redox homeostasis, orchestrating
the expression of key antioxidant enzymes, including heme
oxygenase-1, GPX and superoxide dismutase (SOD), to mitigate
oxidative damage (249,250).
The Nrf2/Keap1 pathway is a critical regulator of
antioxidant defence in the lens, and its dysregulation may
contribute to ARC pathogenesis (246). Age-dependent demethylation of
the Keap1 promoter (223)
upregulates Keap1 expression, enhancing Nrf2 proteasomal
degradation and impairing Nrf2-mediated antioxidant defences. This
redox imbalance, driven by Keap1 promoter demethylation, gives rise
to ROS accumulation in LECs, ultimately promoting ARC development
(223). The methylation status
of the Keap1 promoter can be influenced by multiple exogenous and
endogenous compounds. Valproic acid (VPA), an antiepileptic drug,
alters the expression profiles of passive DNA demethylation pathway
enzymes such as Dnmt1, Dnmt3a and Dnmt3b, and the active DNA
demethylation pathway enzyme ten-eleven translocation 1 (TET1),
leading to DNA demethylation in the Keap1 promoter of LECs
(251). Similarly, sodium
selenite, a widely used experimental cataract-inducing agent,
suppresses Nrf2-dependent antioxidant protection while facilitating
lenticular protein oxidation and cataract formation (246). Furthermore, methylglyoxal
treatment in LECs results in loss of Keap1 promoter methylation and
impairment of the Nrf2-dependent antioxidant system (252). By contrast, ferulic acid, a
compound with well-documented antioxidant activity (253-255), protects LECs against
ultraviolet A-triggered oxidative stress via inhibition of Keap1
promoter demethylation (245).
Furthermore, co-treatment with exogenous acetyl-L-carnitine
effectively counteracts homocysteine (Hcy)-induced Keap1 gene
demethylation in LECs, while regulating Nrf2/Keap1 mRNA expression,
restoring antioxidant protein levels and mitigating Hcy-driven ROS
overproduction (256).
Under physiological conditions, the lens maintains
high endogenous glutathione (GSH) levels, which serve an essential
role in antioxidant defence by scavenging ROS and preserving
proteins in their reduced state (257). GSH-S-transferases (GSTs) are a
diverse group of phase II detoxification enzymes that catalyse the
conjugation of GSH to both endogenous oxidative stress products and
exogenous electrophilic compounds (258,259). All eukaryotic species possess
multiple cytosolic and membrane-bound GST isoenzymes, and the
cytosolic enzymes are encoded by at least five distantly related
gene families (α, μ, π, σ and θ classes) (260). Furthermore, several GST
isozymes modulate the MAPK signalling cascade, which mediates
cellular responses to oxidative stress (261,262).
NER is a highly conserved DNA repair pathway
responsible for processing helix-destabilizing and/or -distorting
DNA lesions, such as UV-induced photoproducts (269). During NER, Cockayne syndrome
complementation group B (CSB), encoded by the ERCC6 gene, recruits
NER repair factors to DNA damage sites and facilitates subsequent
repair (67). CSB protein
deficiency has been implicated in Cockayne syndrome (270). The expression of ERCC6 at both
mRNA and protein levels is reduced in LECs of ARNCs, and is
associated with methylation of a CpG site in the ERCC6 promoter,
suggesting epigenetic regulation of ERCC6 in LECs of ARNCs
(108). Furthermore,
ultraviolet-B (UVB) exposure contributes to ARNC development by
inducing DNA damage, which is typically repaired via the NER
mechanism (108). UVB-treated
human lens epithelium B3 (HLE-B3) and 239T cells exhibit
hypermethylation at the-441 CpG site (relative to the transcription
start site) within the Sp1 transcription factor binding region of
the ERCC6 promoter, along with increased recruitment of DNMT3B to
this site (108).
OGG1 is a key enzyme in the BER pathway that
specifically recognizes and excises the oxidatively damaged base
8-oxo-7,8-dihydroguanine, a mutagenic lesion that induces G:C→T:A
transversions (271). Loss of
functional OGG1 is associated with the pathogenesis of multiple
age-related diseases, including Alzheimer's disease, age-related
macular degeneration and ARCs (272-274). Hypermethylation of the
first-exon CpG island of the OGG1 gene has been observed in the
lens cortex of ARCs, and was associated with low OGG1 expression
and impaired DNA repair function, and eventually contributed to
lens opacity (275).
In addition, two other DNA repair genes exhibit
epigenetic silencing in ARC: The Werner syndrome gene (WRN) and
O6-methylguanine-methyltransferase (MGMT). WRN, which maintains
genomic stability by repairing DNA DSBs (276), is hypermethylated in the
promoter CpG island of ARC anterior lens capsules and its
expression is decreased compared with those in normal controls
(43). Similarly, MGMT, an
enzyme that removes mutagenic adducts from the O-6 position of
guanine, thereby protecting the genome against guanine-to-adenine
transitions (277), has been
observed to be hypermethylated and downregulated in ARCs (278). These deficiencies lead to the
accumulation of unrepaired DNA lesions, driving cell dysfunction
and genomic instability, which further promotes ARC progression
(108).
Numerous studies have established the indispensable
role of lens crystallins in maintaining lens clarity, optical
transparency and refractive function (279,280). CRYAA and αB-crystallin, the
subunits of oligomeric α-crystallin, constitute ~35% of total lens
proteins (281). In addition to
serving as a major structural protein component of the lens, CRYAA
exhibits molecular chaperone-like activity that protects other
crystallins from thermally induced inactivation or aggregation,
thereby being essential for maintaining lens transparency (280-282). Furthermore, CRYAA mediates
cytoprotective effects against both thermal and oxidative stress in
LECs (283), and a study has
demonstrated that CRYAA can delay cataract formation by trapping
denatured proteins prone to aggregation (279). CRYAA expression levels have
been reported to be reduced in ARC lenses, with hypermethylation of
the CRYAA gene promoter CpG islands representing a key mechanism
for CRYAA downregulation (230). Evidence indicates that
methylation of CpG sites in the CRYAA promoter directly reduces the
DNA-binding capacity of the transcription factor Sp1 (236). Treatment with the DNA
demethylating agent zebularine increases CRYAA expression in LECs,
suggesting a potential novel therapeutic strategy for ARCs
(230,236).
COL4A1 encodes the α1 chain of type IV collagen,
the major collagen component of basement membranes (284). Genome-wide methylation analysis
has revealed hypermethylation of the COL4A1 promoter region in
anterior lens capsule samples from patients with ARCs (240). In both HLE-B3 cells and
anterior lens capsules of rats, UVB irradiation induces DNA
hypermethylation of COL4A1 promoter CpG islands, and thus,
suppresses COL4A1 expression, indicating that epigenetic
alterations of the COL4A1 promoter might be involved in UVB-induced
lens damage in ARCs (285).
In summary, emerging evidence has highlighted the
role of DNA methylation in the pathogenesis of ARCs, linking
epigenetic dysregulation to oxidative stress, DNA damage and
cellular dysfunction of LECs. Aberrant DNA methylation patterns in
key genes may serve as potential biomarkers or therapeutic targets.
Further studies based on large-scale clinical research and model
systems are needed to identify and validate additional methylation
signatures, elucidate underlying mechanisms and explore targeted
epigenetic interventions.
As essential structural components, histone
proteins form the fundamental building blocks of chromatin.
Histones undergo posttranslational modifications (PTMs), which
modulate their affinity for DNA, and thus, influence chromatin
compaction states (286). These
modifications are highly dynamic and continuously changing in
response to histone-modifying enzyme activity, ultimately
generating specific PTM patterns linked to epigenetic diseases
(287). Posttranslational
histone modifications include acetylation, methylation,
phosphorylation, citrullination, ubiquitination and adenosine
diphosphate ribosylation (288,289). Of these, histone acetylation
and methylation have been most extensively studied, with
substantial evidence supporting their associations with ARCs
(Table SV) (43,108,235,243,290).
Histone acetylation is a reversible modification of
multiple lysine residues on histone tails, and dynamically
regulated by histone acetyltransferases (HATs) and HDACs (291). HATs are a class of enzymes that
catalyse the transfer of acetyl groups to lysine residues on
histone tails, promoting transcriptional activation by disrupting
the interaction between histone tails and nucleosomal DNA, thus
facilitating chromatin opening (292). By contrast, HDACs catalyse the
removal of acetyl groups from histone tails and suppress
transcriptional activity by producing a more condensed chromatin
state (293). To date, HDACs
have been classified into four major groups (classes I-IV)
(294). Among these, SIRT1, a
class III NAD+-dependent deacetylase and a member of the
sirtuin family, may be critical in protecting against oxidative
stress-induced ocular damage, including ARCs (295). Notably, SIRT1 expression is
upregulated in human ARCs, suggesting a compensatory mechanism in
response to stressors such as UV radiation and oxidative damage.
This upregulation may confer protection by modulating oxidative
stress responses and inhibiting apoptosis of LECs (296,297).
Histone methylation refers to the transfer of
methyl groups from SAM to specific lysine or arginine residues on
histone proteins (298). This
epigenetic modification is catalysed by histone methyltransferases,
while its removal is mediated by histone demethylases (299). Lysine residues can undergo
mono-, di- or tri-methylation, whereas arginine methylation is
limited to mono- or di-methylated states (299). The functional impact of histone
methylation on transcriptional regulation depends on both the
modified residue and the degree of methylation (298). For instance, methylation at
H3K4, H3K36 and H3K79 is associated with transcriptional
activation, whereas methylation at H3K9, H3K27 and H4K20 promotes
transcriptional repression (300). As histone methylation can
either activate or repress gene expression, it serves as a critical
regulator of transcriptional activity in LECs (235).
Histone modifications, along with DNA methylation,
influence the expression of certain genes in LECs. Dysregulation of
these genes exacerbates oxidative stress damage and DNA damage
while impairing normal cellular functions, thereby promoting the
development and progression of ARC (43,108,235-237,243,275,278,285,290) (Fig. 2). Notably, numerous genes
implicated in ARCs are dually regulated by both DNA methylation and
histone modifications (43,108,235,243). These epigenetic mechanisms do
not operate in isolation but instead engage in dynamic crosstalk,
wherein each modification reciprocally enhances or inhibits the
other to modulate gene expression (301). For instance, one study has
demonstrated that endogenous DNMT3A and DNMT3B are associated with
HDAC activity, where DNMT3A facilitates transcriptional repression
via specific molecular interactions between its ATRX-like PHD
domain and both HDAC1 and the co-repressor RP58 (302). Consequently, future research
examining the effects of DNA methylation and histone modifications
on target genes should systematically consider the crosstalk
between these two epigenetic mechanisms.
Similar to DNA modifications, RNA modifications are
dynamically regulated by specific enzymes that catalyse the
addition or removal of chemical groups on RNA nucleotides. These
modifications serve critical roles in molecular processes,
including pre-mRNA splicing, nuclear export, transcript stability
maintenance and translation initiation (303). Prevalent RNA modifications
include m6A, N1-methyladenosine, m5C and
7-methylguanosine (m7G) (304). Among these modifications,
m6A is the best-characterized modification, and is the
primary focus in this section.
Epigenetic regulation is implicated in oxidative
stress-related processes crucial in the initiation and development
of ARCs (223). ncRNA networks
regulate the expression of oxidative stress-related genes such as
NOX4, SIRT1 and GPX4, and aberrant levels of ncRNAs alter the
levels of pro-oxidant markers and antioxidant enzymes in LECs
(68,79,140). Clinically, associations have
been observed between abnormal ncRNA expression and changes in
oxidative stress markers in patients with ARCs (76). Wu et al (318) explored the relationship between
miRNAs and oxidative stress-related genes, and found that
cataract-regulated miRNAs could promote cataract formation not only
by targeting the 3' UTR, which is a classic regulatory pathway, but
also by binding to the TATA-box region of oxidative stress-related
genes, leading to the subsequent elevation of pro-oxidative genes
and inhibition of anti-oxidative genes. Furthermore, numerous
critical genes that serve protective roles in oxidative stress
processes, such as GSTP1, GSTM3 and SOD1, are regulated by DNA
methylation and histone modifications (235,243,290). Additional oxidative
stress-related genes implicated in ARC pathogenesis remain to be
fully characterized, particularly regarding their functional
contributions to disease progression and regulation through
epigenetic mechanisms.
Oxidative stress induced by exogenous stimuli can
also induce epigenetic alterations, since ROS can regulate the
activity of epigenetic enzymes involved in DNA, RNA and histone
modifications (319-321). For example, UVB exposure
induces the recruitment of DNMT3b and HDAC1 at the ERCC6 promoter,
thereby resulting in epigenetic alterations, including specific
hypermethylation of the CpG site and H3K9 deacetylation of ERCC6 in
LECs (108). At both cellular
and organismal levels, UVB exposure enhances the expression of
DNMTs, including DNMT1/2/3, and reduces the expression of TETs,
including TET1/2/3, leading to hypermethylation of COL4A1 promoter
CpG islands and subsequent inhibition of COL4A1 expression
(285). Furthermore, ROS can
directly induce the oxidation of m5C to
5-hydroxymethylcytosine (5hmC) (322). 5hmC may activate DNA
demethylation processes and serve as an intermediate of DNA
demethylation, thus causing DNA hypomethylation (322). Numerous studies have indicated
that dysregulation of 5hmC could be involved in multiple diseases,
including cancer and neurodegenerative diseases (323,324). However, the association between
5hmC and ARCs remains to be fully elucidated.
Epigenetic modulators, including DNA
methyltransferase inhibitors (DNMTis) and histone deacetylase
inhibitors (HDACis), represent a novel class of therapeutic agents
capable of correcting aberrant epigenetic modifications, providing
innovative intervention strategies against molecular defects
underlying cancer and other diseases. These compounds offer unique
therapeutic advantages due to their reversible and targetable
nature, enabling precise gene regulation without modifying DNA
sequences (325-327). The DNMTis azacitidine and
decitabine are approved by the US Food and Drug Administration
(FDA) for the treatment of myelodysplastic syndromes and acute
myeloid leukaemia, and exert their effects by inhibiting DNMTs and
reversing tumour-suppressor gene silencing through DNA
demethylation (328,329). Several other epigenetic drugs,
such as vorinostat, belinostat and romidepsin, are FDA-approved for
the treatment of cutaneous T-cell lymphoma and peripheral T-cell
lymphomas (330-332). Although DNMTis and HDACis have
been extensively tested in clinical and preclinical trials for
human diseases (333-335), their effects in ARCs remain
poorly understood. The DNMTi 5-aza-2'-deoxycytidine (decitabine)
restores the transcriptional activities of GSTP1 in LECs (235), and trichostatin A (TSA; HDACi)
corrects the anacardic acid (HAT inhibitor)-induced imbalance
between HATs and HDACs, causing increased SOD1 expression by
reversing histone acetylation (290). These findings provide evidence
that DNMTis and HDACis could rescue LECs from dysregulated
expression of key genes induced by aberrant epigenetic alterations
during ARC development. Notably, HDACis also exhibit antioxidant
properties, as evidenced by multiple studies (336,337). Qiu et al (338) compared the protective effects
of four HDACis, β-hydroxybutyrate, TSA, suberoylanilide hydroxamic
acid (SAHA) and VPA, on HLECs following UVB exposure, and found
that low concentrations of HDACis (1 μmol/l SAHA) mildly
attenuated oxidative stress. Although these promising in
vitro findings support these drugs as possible candidates for
ARC intervention, further validation through well-controlled in
vivo studies is required for their clinical application.
Alterations in the expression patterns of various
ncRNAs have been reported in ARCs (41,72,131,198), suggesting that targeted
modulation of aberrant ncRNA activity may hold therapeutic
potential. Over the past decade, miRNA mimics and anti-miRNAs have
demonstrated therapeutic promise in clinical trials for various
diseases, such as advanced solid tumours, ulcerative colitis and
hepatitis C virus infection (339-341), whereas no lncRNA drug has
reached the clinical development stage (342). miRNA mimics replicate
endogenous miRNA functions to compensate for miRNA deficiency in
pathological conditions, while anti-miRNAs (antagomirs) are
engineered to suppress endogenous miRNAs that contribute to disease
progression (342). As
aforementioned, administration of anti-miRNAs targeting miR-326,
miR-29a-3p and miR-187 independently enhanced lens transparency and
ameliorated lens damage in ARC animal models, indicating their
potential as viable non-surgical alternatives for ARC intervention
(103-105). Furthermore, with increasing
recognition of the regulatory functions of circRNAs in disease
pathogenesis, circRNAs are increasingly being considered as targets
for therapeutic intervention (56,176,182). For instance, circRNA 06209
overexpression through plasmid injection in a rat model of ARCs
reduced lens opacity (182).
However, before ncRNA-based therapies advance to the preclinical
stage, several challenges must be addressed, including RNA
instability, immune activation and off-target effects (342).
Epigenetic modifiers hold potential for the
prevention and treatment of ARCs, but there is still a long way
ahead before these agents can be routinely implemented in clinical
practice. Future studies should focus on: i) Identifying novel
therapeutic targets and developing corresponding treatments; ii)
investigating and optimizing intraocular drug delivery systems with
high efficiency and biocompatibility; and iii) evaluating long-term
safety and efficacy in both animal models and clinical trials.
Technological breakthroughs and cutting-edge
research have sparked growing interest in the pivotal role of
epigenetics in ocular disease. Advances in omics and
high-throughput sequencing technologies have facilitated the
elucidation of the role of epigenetics in disease pathogenesis and
the identification of potential targets. This section highlights
two emerging technologies, single-cell multi-omics and spatial
transcriptomics, and discusses their transformative potential in
deciphering the epigenetic landscape of ARCs.
Single-cell multi-omics sequencing enables
simultaneous single-cell profiling of chromatin accessibility,
histone modifications, transcription factor binding, DNA
methylation and transcriptome analysis (343). This approach has revealed cell
type-specific epigenetic regulatory mechanisms in brain development
(344), cancer evolution
(345) and HIV-associated
immune exhaustion (346).
Tangeman et al (347)
described the first single-cell multiomic atlas of lens
development, depicting a comprehensive portrait of lens fibre cell
differentiation and exploring congenital cataract-linked regulatory
networks. However, to the best of our knowledge, single-cell
multi-omics has not yet been applied in research on ARCs. Current
research considers different omics levels individually, drawing
conclusions from each level separately, without data integration
for a cohesive understanding of the overall mechanisms. Single-cell
multi-omics could revolutionize ARC research by offering a
comprehensive understanding of the complex biological processes
involved in ARC pathogenesis.
Spatial transcriptomics allows high-resolution
profiling of gene expression in intact cell and tissue samples
(348), and has made
significant advances in recent years, exerting notable influence on
diverse fields, such as tissue architecture, developmental biology
and disease research, particularly in cancer and neurodegenerative
diseases (349). Given the
differential miRNA expression profiles among ARC subtypes defined
by the location of lens opacity (72), there may be differences in gene
expression profiles across different regions of the lens. To the
best of our knowledge, spatial transcriptomics remains unexplored
in epigenetic studies of ARCs. Spatial transcriptomics allows the
quantification and illustration of gene expression profiles within
the native spatial context (350), which could facilitate the
identification of epigenetic biomarkers and microenvironmental
interactions critical for understanding cataract pathogenesis.
The epigenetic clock, also known as DNA methylation
age (DNAmAge), is a biomarker for biological age prediction based
on DNA methylation patterns at specific CpG sites (351). By modelling linear correlations
between methylation levels and chronological aging, it serves as a
molecular 'timer' that can estimate cellular aging rates more
accurately than chronological age, with implications for disease
risk and lifespan prediction (352). Epigenetic age acceleration
(EAA) exhibits strong associations with age-related diseases by
reflecting discrepancies between DNAmAge and chronological age,
acting as a predictive biomarker for conditions such as
cardiovascular disorders, neurodegenerative diseases and type 2
diabetes (353).
To investigate the causal relationship between
epigenetic clocks and common age-related eye diseases or glaucoma
endophenotypes, Chen et al (354) conducted bidirectional
two-sample Mendelian randomization and found that ARCs were linked
to decreased HannumAge, a measure of extrinsic aging based on 71
age-related CpGs from whole blood samples of adults. Further
research is warranted to determine whether quantifying EAA can
effectively identify ARC susceptibility at early stages and whether
it may serve as a basis for developing targeted interventions to
delay lens opacification.
Breakthroughs in omics, bioinformatics and
high-throughput sequencing technologies have accelerated
discoveries regarding the role of epigenetics in ARCs, enabling a
deeper understanding of the initiation and progression mechanisms
of ARCs while potentially revolutionizing disease management
approaches. The present review comprehensively summarizes advances
in epigenetic research on ARCs, examining how ncRNAs, DNA
methylation, histone modifications and m6A modification
contribute to ARC pathogenesis, and exploring their clinical
potential for innovative therapeutic strategies. Epigenetics in
ARCs is an emerging field with immense potential for diagnosis,
treatment and prevention of lens opacities, as innovations over the
past decade have identified numerous epigenetic alterations as
potential biomarkers and intervention targets. Further studies are
required to identify therapeutic targets and biomarkers, elucidate
the precise mechanisms through which epigenetics influences disease
pathogenesis, evaluate the efficacy and safety of novel epigenetic
therapies, and translate these therapies into clinical
practice.
Not applicable.
WY collected the literature and drafted the
manuscript. YZ, SC, JG and ZP reviewed the manuscript and made
revisions to the manuscript. YY directed the work and revised the
manuscript. Data authentication is not applicable. All authors have
read and approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 82371036), and the Key Research and
Development Program of Zhejiang Province (grant no.
2025C02156).
|
1
|
Cicinelli MV, Buchan JC, Nicholson M,
Varadaraj V and Khanna RC: Cataracts. Lancet. 401:377–389. 2023.
View Article : Google Scholar
|
|
2
|
Shiels A and Hejtmancik JF: Mutations and
mechanisms in congenital and age-related cataracts. Exp Eye Res.
156:95–102. 2017. View Article : Google Scholar :
|
|
3
|
Asbell PA, Dualan I, Mindel J, Brocks D,
Ahmad M and Epstein S: Age-related cataract. Lancet. 365:599–609.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abdelkader H, Alany RG and Pierscionek B:
Age-related cataract and drug therapy: Opportunities and challenges
for topical antioxidant delivery to the lens. J Pharm Pharmacol.
67:537–550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meacock WR, Spalton DJ, Boyce J and
Marshall J: The effect of posterior capsule opacification on visual
function. Invest Ophthalmol Vis Sci. 44:4665–4669. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang C, An Q, Zhou H and Ge H: Research
progress on the impact of cataract surgery on corneal endothelial
cells. Adv Ophthalmol Pract Res. 4:194–201. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Beebe DC, Holekamp NM and Shui YB:
Oxidative damage and the prevention of age-related cataracts.
Ophthalmic Res. 44:155–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kulbay M, Wu KY, Nirwal GK, Bélanger P and
Tran SD: Oxidative stress and cataract formation: Evaluating the
efficacy of antioxidant therapies. Biomolecules. 14:10552024.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lou MF: Redox regulation in the lens. Prog
Retin Eye Res. 22:657–682. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rahman K: Studies on free radicals,
antioxidants, and co-factors. Clin Interv Aging. 2:219–236.
2007.PubMed/NCBI
|
|
11
|
Berthoud VM and Beyer EC: Oxidative
stress, lens gap junctions, and cataracts. Antioxid Redox Signal.
11:339–353. 2009. View Article : Google Scholar :
|
|
12
|
Petrash JM: Aging and age-related diseases
of the ocular lens and vitreous body. Invest Ophthalmol Vis Sci.
54:ORSF54–ORSF59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Babizhayev MA and Costa EB: Lipid peroxide
and reactive oxygen species generating systems of the crystalline
lens. Biochim Biophys Acta. 1225:326–337. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dische Z and Zil H: Studies on the
oxidation of cysteine to cystine in lens proteins during cataract
formation. Am J Ophthalmol. 34:104–113. 1951. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kleiman NJ and Spector A: DNA single
strand breaks in human lens epithelial cells from patients with
cataract. Curr Eye Res. 12:423–431. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kruk J, Kubasik-Kladna K and Aboul-Enein
HY: The role oxidative stress in the pathogenesis of eye diseases:
Current status and a dual role of physical activity. Mini Rev Med
Chem. 16:241–257. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kubota M, Shui YB, Liu M, Bai F, Huang AJ,
Ma N, Beebe DC and Siegfried CJ: Mitochondrial oxygen metabolism in
primary human lens epithelial cells: Association with age, diabetes
and glaucoma. Free Radic Biol Med. 97:513–519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brennan LA, McGreal RS and Kantorow M:
Oxidative stress defense and repair systems of the ocular lens.
Front Biosci (Elite Ed). 4:141–155. 2012. View Article : Google Scholar
|
|
19
|
Cekić S, Zlatanović G, Cvetković T and
Petrović B: Oxidative stress in cataractogenesis. Bosn J Basic Med
Sci. 10:265–269. 2010. View Article : Google Scholar
|
|
20
|
Li G, Song H, Chen L, Yang W, Nan K and Lu
P: TUG1 promotes lens epithelial cell apoptosis by regulating
miR-421/caspase-3 axis in age-related cataract. Exp Cell Res.
356:20–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li WC, Kuszak JR, Dunn K, Wang RR, Ma W,
Wang GM, Spector A, Leib M, Cotliar AM, Weiss M, et al: Lens
epithelial cell apoptosis appears to be a common cellular basis for
non-congenital cataract development in humans and animals. J Cell
Biol. 130:169–181. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Xu H, Shu X and Song CX: Mapping
epigenetic modifications by sequencing technologies. Cell Death
Differ. 32:56–65. 2025. View Article : Google Scholar :
|
|
23
|
Chen Y, Hong T, Wang S, Mo J, Tian T and
Zhou X: Epigenetic modification of nucleic acids: From basic
studies to medical applications. Chem Soc Rev. 46:2844–2872. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mazzio EA and Soliman KF: Basic concepts
of epigenetics: Impact of environmental signals on gene expression.
Epigenetics. 7:119–130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peixoto P, Cartron PF, Serandour AA and
Hervouet E: From 1957 to nowadays: A brief history of epigenetics.
Int J Mol Sci. 21:75712020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gangisetty O, Cabrera MA and Murugan S:
Impact of epigenetics in aging and age related neurodegenerative
diseases. Front Biosci (Landmark Ed). 23:1445–1464. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nikolac Perkovic M, Videtic Paska A,
Konjevod M, Kouter K, Svob Strac D, Nedic Erjavec G and Pivac N:
Epigenetics of Alzheimer's disease. Biomolecules. 11:1952021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suárez R, Chapela SP, Álvarez-Córdova L,
Bautista-Valarezo E, Sarmiento-Andrade Y, Verde L, Frias-Toral E
and Sarno G: Epigenetics in obesity and diabetes mellitus: New
insights. Nutrients. 15:8112023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun L, Zhang H and Gao P: Metabolic
reprogramming and epigenetic modifications on the path to cancer.
Protein Cell. 13:877–919. 2022. View Article : Google Scholar :
|
|
31
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225.e12. 2015. View Article : Google Scholar
|
|
32
|
Trnkova L, Buocikova V, Mego M, Cumova A,
Burikova M, Bohac M, Miklikova S, Cihova M and Smolkova B:
Epigenetic deregulation in breast cancer microenvironment:
Implications for tumor progression and therapeutic strategies.
Biomed Pharmacother. 174:1165592024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Henikoff S and Greally JM: Epigenetics,
cellular memory and gene regulation. Curr Biol. 26:R644–R648. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi
Y and Cao L: DNA methylation, its mediators and genome integrity.
Int J Biol Sci. 11:604–617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu Z, Zhang W, Qu J and Liu GH: Emerging
epigenetic insights into aging mechanisms and interventions. Trends
Pharmacol Sci. 45:157–172. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Desmettre TJ: Epigenetics in age-related
macular degeneration (AMD). J Fr Ophtalmol. 41:e407–e415. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kapuganti RS and Alone DP: Current
understanding of genetics and epigenetics in pseudoexfoliation
syndrome and glaucoma. Mol Aspects Med. 94:1012142023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kowluru RA, Kowluru A, Mishra M and Kumar
B: Oxidative stress and epigenetic modifications in the
pathogenesis of diabetic retinopathy. Prog Retin Eye Res. 48:40–61.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y and Guan H: The role of DNA
methylation in lens development and cataract formation. Cell Mol
Neurobiol. 37:979–984. 2017. View Article : Google Scholar
|
|
40
|
Ling C and Rönn T: Epigenetics in human
obesity and type 2 diabetes. Cell Metab. 29:1028–1044. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li P, Yu H, Zhang G, Kang L, Qin B, Cao Y,
Luo J, Chen X, Wang Y, Qin M, et al: Identification and
characterization of N6-methyladenosine CircRNAs and
methyltransferases in the lens epithelium cells from age-related
cataract. Invest Ophthalmol Vis Sci. 61:132020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wei YL and Sun H: Identification of
hsa-mir-34a, hsa-mir-124, and hsa-mir-204 as signatures for
cataract. J Cell Physiol. 234:10709–10717. 2019. View Article : Google Scholar
|
|
43
|
Zhu X, Zhang G, Kang L and Guan H:
Epigenetic regulation of Werner syndrome gene in age-related
cataract. J Ophthalmol. 2015:5796952015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Crick F: Central dogma of molecular
biology. Nature. 227:561–563. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Crick FH: On protein synthesis. Symp Soc
Exp Biol. 12:138–163. 1958.PubMed/NCBI
|
|
46
|
Kim SS and Lee SJV: Non-coding RNAs in
caenorhabditis elegans aging. Mol Cells. 42:379–385.
2019.PubMed/NCBI
|
|
47
|
Yao ZT, Yang YM, Sun MM, He Y, Liao L,
Chen KS and Li B: New insights into the interplay between long
non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun
(Lond). 42:117–140. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Smillie CL, Sirey T and Ponting CP:
Complexities of post-transcriptional regulation and the modeling of
ceRNA crosstalk. Crit Rev Biochem Mol Biol. 53:231–245. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Luo S, He F, Luo J, Dou S, Wang Y, Guo A
and Lu J: Drosophila tsRNAs preferentially suppress general
translation machinery via antisense pairing and participate in
cellular starvation response. Nucleic Acids Res. 46:5250–5268.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kim HK, Xu J, Chu K, Park H, Jang H, Li P,
Valdmanis PN, Zhang QC and Kay MA: A tRNA-derived small RNA
regulates ribosomal protein S28 protein levels after translation
initiation in humans and mice. Cell Rep. 29:3816–3824.e4. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ardekani AM and Naeini MM: The role of
MicroRNAs in human diseases. Avicenna J Med Biotechnol. 2:161–179.
2010.PubMed/NCBI
|
|
53
|
Condrat CE, Thompson DC, Barbu MG, Bugnar
OL, Boboc A, Cretoiu D, Suciu N, Cretoiu SM and Voinea SC: miRNAs
as biomarkers in disease: Latest findings regarding their role in
diagnosis and prognosis. Cells. 9:2762020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wawrzyniak O, Zarębska Ż, Rolle K and
Gotz-Więckowska A: Circular and long non-coding RNAs and their role
in ophthalmologic diseases. Acta Biochim Pol. 65:497–508.
2018.PubMed/NCBI
|
|
55
|
Chen S, Zhang C, Shen L, Hu J, Chen X and
Yu Y: Noncoding RNAs in cataract formation: Star molecules emerge
in an endless stream. Pharmacol Res. 184:1064172022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang C, Hu J and Yu Y: CircRNA is a
rising star in researches of ocular diseases. Front Cell Dev Biol.
8:8502020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zheng JL, Sun J, Zhang H and Zhang Y: Role
of microRNA and lncRNA in lens development and cataract formation.
Zhonghua Yan Ke Za Zhi. 54:390–395. 2018.In Chinese. PubMed/NCBI
|
|
58
|
Michlewski G and Cáceres JF:
Post-transcriptional control of miRNA biogenesis. RNA. 25:1–16.
2019. View Article : Google Scholar :
|
|
59
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Broughton JP, Lovci MT, Huang JL, Yeo GW
and Pasquinelli AE: Pairing beyond the seed supports MicroRNA
targeting specificity. Mol Cell. 64:320–333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Majid S, Dar AA, Saini S, Yamamura S,
Hirata H, Tanaka Y, Deng G and Dahiya R: MicroRNA-205-directed
transcriptional activation of tumor suppressor genes in prostate
cancer. Cancer. 116:5637–5649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Place RF, Li LC, Pookot D, Noonan EJ and
Dahiya R: MicroRNA-373 induces expression of genes with
complementary promoter sequences. Proc Natl Acad Sci USA.
105:1608–1613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Vasudevan S: Posttranscriptional
upregulation by microRNAs. Wiley Interdiscip Rev RNA. 3:311–330.
2012. View Article : Google Scholar
|
|
65
|
Selbach M, Schwanhäusser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kang L, Luo J, Li P, Zhang G, Wei M, Ji M
and Guan H: miR-125a-3p regulates apoptosis by suppressing TMBIM4
in lens epithelial cells. Int Ophthalmol. 43:1261–1274. 2023.
View Article : Google Scholar
|
|
67
|
Cao Y, Li P, Zhang G, Kang L, Zhou T, Wu
J, Wang Y, Wang Y, Chen X and Guan H: MicroRNA Let-7c-5p-mediated
regulation of ERCC6 disrupts autophagic flux in age-related
cataract via the binding to VCP. Curr Eye Res. 46:1353–1362. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li ZN, Ge MX and Yuan ZF: MicroRNA-182-5p
protects human lens epithelial cells against oxidative
stress-induced apoptosis by inhibiting NOX4 and p38 MAPK
signalling. BMC Ophthalmol. 20:2332020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang C, Chen M, Zhou N and Qi Y:
Metformin prevents H2O2-induced senescence in
human lens epithelial B3 cells. Med Sci Monit Basic Res.
26:e9233912020.
|
|
70
|
Zou X, Kang L, Yang M, Wu J and Guan H:
MicroRNA binding mediated Functional sequence variant in 3'-UTR of
DNA repair gene XPC in age-related cataract. Sci Rep. 8:151982018.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wu C, Lin H, Wang Q, Chen W, Luo H, Chen W
and Zhang H: Discrepant expression of microRNAs in transparent and
cataractous human lenses. Invest Ophthalmol Vis Sci. 53:3906–3912.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kim YJ, Lee WJ, Ko BW, Lim HW, Yeon Y, Ahn
SJ and Lee BR: Investigation of MicroRNA expression in anterior
lens capsules of senile cataract patients and MicroRNA differences
according to the cataract type. Transl Vis Sci Technol. 10:142021.
View Article : Google Scholar
|
|
73
|
Peng CH, Liu JH, Woung LC, Lin TJ, Chiou
SH, Tseng PC, Du WY, Cheng CK, Hu CC, Chien KH and Chen SJ:
MicroRNAs and cataracts: Correlation among let-7 expression, age
and the severity of lens opacity. Br J Ophthalmol. 96:747–751.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chylack LT Jr, Wolfe JK, Singer DM, Leske
MC, Bullimore MA, Bailey IL, Friend J, McCarthy D and Wu SY: The
lens opacities classification system III. The longitudinal study of
cataract study group. Arch Ophthalmol. 111:831–836. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chien KH, Chen SJ, Liu JH, Chang HM, Woung
LC, Liang CM, Chen JT, Lin TJ, Chiou SH and Peng CH: Correlation
between microRNA-34a levels and lens opacity severity in
age-related cataracts. Eye (Lond). 27:883–888. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Xu C, Xu J, Zhang W, Zheng F and Lou X:
Expression of miR-210-3p in the aqueous humor of patients with
age-related cataracts and its effect on human lens epithelial cell
injury induced by hydrogen peroxide. Arq Bras Oftalmol.
87:e202202742023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Y, Liu S, Zhang F, Jiang P, Wu X and
Liang Y: Expression of the microRNAs hsa-miR-15a and hsa-miR-16-1
in lens epithelial cells of patients with age-related cataract. Int
J Clin Exp Med. 8:2405–2410. 2015.PubMed/NCBI
|
|
78
|
Zhou W, Xu J, Wang C, Shi D and Yan Q:
miR-23b-3p regulates apoptosis and autophagy via suppressing SIRT1
in lens epithelial cells. J Cell Biochem. 120:19635–19646. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lu B, Christensen IT, Ma LW, Wang XL,
Jiang LF, Wang CX, Feng L, Zhang JS and Yan QC: miR-211 promotes
lens epithelial cells apoptosis by targeting silent mating-type
information regulation 2 homolog 1 in age-related cataracts. Int J
Ophthalmol. 11:201–207. 2018.PubMed/NCBI
|
|
80
|
Maiti B, Li J, de Bruin A, Gordon F,
Timmers C, Opavsky R, Patil K, Tuttle J, Cleghorn W and Leone G:
Cloning and characterization of mouse E2F8, a novel mammalian E2F
family member capable of blocking cellular proliferation. J Biol
Chem. 280:18211–18220. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Shen ZG, Liu XZ, Chen CX and Lu JM:
Knockdown of E2F3 inhibits proliferation, migration, and invasion
and increases apoptosis in glioma cells. Oncol Res. 25:1555–1566.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chen Q, Liang D, Yang T, Leone G and
Overbeek PA: Distinct capacities of individual E2Fs to induce cell
cycle re-entry in postmitotic lens fiber cells of transgenic mice.
Dev Neurosci. 26:435–445. 2004. View Article : Google Scholar
|
|
83
|
Chong JL, Tsai SY, Sharma N, Opavsky R,
Price R, Wu L, Fernandez SA and Leone G: E2f3a and E2f3b contribute
to the control of cell proliferation and mouse development. Mol
Cell Biol. 29:414–424. 2009. View Article : Google Scholar :
|
|
84
|
Wenzel PL, Chong JL, Sáenz-Robles MT,
Ferrey A, Hagan JP, Gomez YM, Rajmohan R, Sharma N, Chen HZ, Pipas
JM, et al: Cell proliferation in the absence of E2F1-3. Dev Biol.
351:35–45. 2011. View Article : Google Scholar
|
|
85
|
Yan Q, Liu JP and Li DWC: Apoptosis in
lens development and pathology. Differentiation. 74:195–211. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Xiang W, Lin H, Wang Q and Chen W, Liu Z,
Chen H, Zhang H and Chen W: miR-34a suppresses proliferation and
induces apoptosis of human lens epithelial cells by targeting E2F3.
Mol Med Rep. 14:5049–5056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li Q, Pan H and Liu Q: MicroRNA-15a
modulates lens epithelial cells apoptosis and proliferation through
targeting B-cell lymphoma-2 and E2F transcription factor 3 in
age-related cataracts. Biosci Rep. 39:BSR201917732019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gong W, Li J, Wang Y, Meng J and Zheng G:
miR-221 promotes lens epithelial cells apoptosis through
interacting with SIRT1 and E2F3. Chem Biol Interact. 306:39–46.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gao W, Zhou X and Lin R: miR-378a-5p and
miR-630 induce lens epithelial cell apoptosis in cataract via
suppression of E2F3. Braz J Med Biol Res. 53:e96082020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Fan F, Zhuang J, Zhou P, Liu X and Luo Y:
MicroRNA-34a promotes mitochondrial dysfunction-induced apoptosis
in human lens epithelial cells by targeting Notch2. Oncotarget.
8:110209–110220. 2017. View Article : Google Scholar
|
|
91
|
Feng L, Wei Y, Sun Y, Zhou L, Bi S, Chen W
and Xiang W: MIR34A modulates lens epithelial cell apoptosis and
cataract development via the HK1/caspase 3 signaling pathway. Aging
(Albany NY). 15:6331–6345. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Liu Y, Li H and Liu Y: microRNA-378a
regulates the reactive oxygen species (ROS)/phosphatidylinositol
3-kinases (PI3K)/AKT signaling pathway in human lens epithelial
cells and cataract. Med Sci Monit. 25:4314–4321. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Zhang F, Meng W and Tong B:
Down-regulation of MicroRNA-133b suppresses apoptosis of lens
epithelial cell by up-regulating BCL2L2 in age-related cataracts.
Med Sci Monit. 22:4139–4145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Qin Y, Zhao J, Min X, Wang M, Luo W, Wu D,
Yan Q, Li J, Wu X and Zhang J: MicroRNA-125b inhibits lens
epithelial cell apoptosis by targeting p53 in age-related cataract.
Biochim Biophys Acta. 1842:2439–2447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gao M, Huang Y, Wang L, Huang M, Liu F,
Liao S, Yu S, Lu Z, Han S, Hu X, et al: HSF4 regulates lens fiber
cell differentiation by activating p53 and its downstream
regulators. Cell Death Dis. 8:e30822017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li L, Fan DB, Zhao YT, Li Y, Kong DQ, Cai
FF and Zheng GY: Two novel mutations identified in ADCC families
impair crystallin protein distribution and induce apoptosis in
human lens epithelial cells. Sci Rep. 7:178482017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Peng J, Zheng TT, Liang Y, Duan LF, Zhang
YD, Wang LJ, He GM and Xiao HT: p-Coumaric acid protects human lens
epithelial cells against oxidative stress-induced apoptosis by MAPK
signaling. Oxid Med Cell Longev. 2018:85490522018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Youle RJ and Strasser A: The BCL-2 protein
family: Opposing activities that mediate cell death. Nat Rev Mol
Cell Biol. 9:47–59. 2008. View Article : Google Scholar
|
|
99
|
Zhang DJ, Du FF, Jing XY, Wang L, Liu D
and Yang XQ: Sequence and expression regulation of the BCL2L2 gene
in pigs. Gene. 851:1469922023. View Article : Google Scholar
|
|
100
|
İçme G, Yilmaz A, Dinç E, Görür A, Fidanci
ŞB and Tamer L: Assessment of miR-182, miR-183, miR-184, and
miR-221 expressions in primary pterygium and comparison with the
normal conjunctiva. Eye Contact Lens. 45:208–211. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu Y, Bailey JC, Helwa I, Dismuke WM, Cai
J, Drewry M, Brilliant MH, Budenz DL, Christen WG, Chasman DI, et
al: A common variant in MIR182 is associated with primary
open-angle glaucoma in the NEIGHBORHOOD consortium. Invest
Ophthalmol Vis Sci. 57:4528–4535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Szemraj M, Oszajca K, Szemraj J and
Jurowski P: MicroRNA expression analysis in serum of patients with
congenital hemochromatosis and age-related macular degeneration
(AMD). Med Sci Monit. 23:4050–4060. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ren H, Tao H, Gao Q, Shen W, Niu Z, Zhang
J, Mao H, Du A and Li W: MiR-326 antagomir delays the progression
of age-related cataract by upregulating FGF1-mediated expression of
betaB2-crystallin. Biochem Biophys Res Commun. 505:505–510. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Fang R, Li HL, Lv NX, Yue PL, Jia YX, Liu
ZC, Zhou HG and Song XD: Inhibition of miR-29a-3p alleviates
apoptosis of lens epithelial cells via upregulation of CAND1. Curr
Eye Res. 49:391–400. 2024. View Article : Google Scholar
|
|
105
|
Wang F and Ren Y: Nanofluorescence probe
in detection of miR-187 and its correlation with oxidative stress
response in cataracts. Altern Ther Health Med. 29:73–79. 2023.
|
|
106
|
Vymetalkova V, Pardini B, Rosa F,
Jiraskova K, Di Gaetano C, Bendova P, Levy M, Veskrnova V, Buchler
T, Vodickova L, et al: Polymorphisms in microRNA binding sites of
mucin genes as predictors of clinical outcome in colorectal cancer
patients. Carcinogenesis. 38:28–39. 2017. View Article : Google Scholar
|
|
107
|
Liu XC, Liu XF, Hu ZD and Li ZH:
Polymorphisms of DNA repair genes XPD (Lys751Gln) and XRCC1
(Arg399Gln), and the risk of age-related cataract: A meta-analysis.
Curr Eye Res. 40:676–682. 2015. View Article : Google Scholar
|
|
108
|
Wang Y, Li F, Zhang G, Kang L and Guan H:
Ultraviolet-B induces ERCC6 repression in lens epithelium cells of
age-related nuclear cataract through coordinated DNA
hypermethylation and histone deacetylation. Clin Epigenetics.
8:622016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Kang L, Zou X, Zhang G, Xiang J, Wang Y,
Yang M, Chen X, Wu J and Guan H: A variant in a microRNA binding
site in NEIL2 3'UTR confers susceptibility to age-related
cataracts. FASEB J. 33:10469–10476. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhao Y, Li X and Zhu S: rs78378222
polymorphism in the 3'-untranslated region of TP53 contributes to
development of age-associated cataracts by modifying
microRNA-125b-induced apoptosis of lens epithelial cells. Mol Med
Rep. 14:2305–2310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Luo J, Li P, Kang L, Ji M, Zhou T, Qin B,
Zhang J and Guan H: Exosomal microRNA-222-3p increases UVB
sensitivity of lens epithelium cells by suppressing MGMT. Int
Ophthalmol. 43:1611–1628. 2023. View Article : Google Scholar
|
|
112
|
Denzer K, Kleijmeer MJ, Heijnen HF,
Stoorvogel W and Geuze HJ: Exosome: From internal vesicle of the
multivesicular body to intercellular signaling device. J Cell Sci.
113:3365–3374. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Meldolesi J: Exosomes and ectosomes in
intercellular communication. Curr Biol. 28:R435–R444. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Perez-Hernandez J, Olivares D, Forner MJ,
Ortega A, Solaz E, Martinez F, Chaves FJ, Redon J and Cortes R:
Urinary exosome miR-146a is a potential marker of albuminuria in
essential hypertension. J Transl Med. 16:2282018. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Creemers EE, Tijsen AJ and Pinto YM:
Circulating microRNAs: Novel biomarkers and extracellular
communicators in cardiovascular disease? Circ Res. 110:483–495.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Turchinovich A, Samatov TR, Tonevitsky AG
and Burwinkel B: Circulating miRNAs: Cell-cell communication
function? Front Genet. 4:1192013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Fan Q, Yang L, Zhang X, Peng X, Wei S, Su
D, Zhai Z, Hua X and Li H: The emerging role of exosome-derived
non-coding RNAs in cancer biology. Cancer Lett. 414:107–115. 2018.
View Article : Google Scholar
|
|
118
|
Machida T, Tomofuji T, Ekuni D, Maruyama
T, Yoneda T, Kawabata Y, Mizuno H, Miyai H, Kunitomo M and Morita
M: MicroRNAs in salivary exosome as potential biomarkers of aging.
Int J Mol Sci. 16:21294–21309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zamani P, Fereydouni N, Butler AE,
Navashenaq JG and Sahebkar A: The therapeutic and diagnostic role
of exosomes in cardiovascular diseases. Trends Cardiovasc Med.
29:313–323. 2019. View Article : Google Scholar
|
|
120
|
Li J, Wang J and Chen Z: Emerging role of
exosomes in cancer therapy: Progress and challenges. Mol Cancer.
24:132025. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Moreno JA, Hamza E, Guerrero-Hue M,
Rayego-Mateos S, García-Caballero C, Vallejo-Mudarra M, Metzinger L
and Metzinger-Le Meuth V: Non-coding RNAs in kidney diseases: The
long and short of them. Int J Mol Sci. 22:60772021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar
|
|
124
|
Javed Z, Khan K, Sadia H, Raza S, Salehi
B, Sharifi-Rad J and Cho WC: LncRNA & Wnt signaling in
colorectal cancer. Cancer Cell Int. 20:3262020. View Article : Google Scholar
|
|
125
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Gong C and Maquat LE: lncRNAs
transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs
via Alu elements. Nature. 470:284–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hong Y, Wu J, Sun Y, Zhang S, Lu Y and Ji
Y: ceRNA network construction and identification of hub genes as
novel therapeutic targets for age-related cataracts using
bioinformatics. PeerJ. 11:e150542023. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Zhou T, Yang M, Zhang G, Kang L, Yang L
and Guan H: Long non-coding RNA nuclear paraspeckle assembly
transcript 1 protects human lens epithelial cells against
H2O2 stimuli through the nuclear factor kappa
b/p65 and p38/mitogen-activated protein kinase axis. Ann Transl
Med. 8:16532020. View Article : Google Scholar
|
|
130
|
Zhou T, Zhang J, Qin B, Xu H, Zhang S and
Guan H: Long non-coding RNA NONHSAT143692.2 is involved in
oxidative DNA damage repair in the lens by regulating the
miR-4728-5p/OGG1 axis. Int J Mol Med. 46:1838–1848. 2020.PubMed/NCBI
|
|
131
|
Zhang N, Zhang C, Wang X and Qi Y:
High-throughput sequencing reveals novel lincRNA in age-related
cataract. Int J Mol Med. 40:1829–1839. 2017.PubMed/NCBI
|
|
132
|
Li H, Gao L, Du J, Ma T, Ye Z and Li Z:
Differentially expressed gene profiles and associated ceRNA network
in ATG7-deficient lens epithelial cells under oxidative stress.
Front Genet. 13:10889432022. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Jin X, Jin H, Shi Y, Guo Y and Zhang H:
Long non-coding RNA KCNQ1OT1 promotes cataractogenesis via miR-214
and activation of the caspase-1 pathway. Cell Physiol Biochem.
42:295–305. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Zhang M and Cheng K: Long non-coding RNA
KCNQ1OT1 promotes hydrogen peroxide-induced lens epithelial cell
apoptosis and oxidative stress by regulating miR-223-3p/BCL2L2
axis. Exp Eye Res. 206:1085432021. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Xu Y, Zheng Y, Shen P and Zhou L: Role of
long noncoding RNA KCNQ1 overlapping transcript
1/microRNA-124-3p/BCL-2-like 11 axis in hydrogen peroxide
(H2O2)-stimulated human lens epithelial
cells. Bioengineered. 13:5035–5045. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Shen Q and Zhou T: Knockdown of lncRNA
TUG1 protects lens epithelial cells from oxidative stress-induced
injury by regulating miR-196a-5p expression in age-related
cataracts. Exp Ther Med. 22:12862021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Sun M, Li K, Li X, Wang H, Li L and Zheng
G: lncRNA TUG1 regulates Smac/DIABLO expression by competitively
inhibiting miR-29b and modulates the apoptosis of lens epithelial
cells in age-related cataracts. Chin Med J (Engl). 136:2340–2350.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Guo X, Li C, Wang Y, Jiang C and Yang L:
Long non-coding RNA nuclear paraspeckle assembly transcript 1
downregulation protects lens epithelial cells from oxidative
stress-induced apoptosis by regulating the
microRNA-124-3p/death-associated protein kinase 1 axis in
age-related cataract. Int Ophthalmol. 43:3413–3424. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Tu Y, Xie L, Chen L, Yuan Y, Qin B, Wang
K, Zhu Q, Ji N, Zhu M and Guan H: Long non-coding RNA MEG3 promotes
cataractogenesis by upregulating TP53INP1 expression in age-related
cataract. Exp Eye Res. 199:1081852020. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhang X, Zheng C, Zhao J, Xu X and Yao J:
LncRNA MEG3 regulates ferroptosis of lens epithelial cells via
PTBP1/GPX4 axis to participate in age-related cataract. J Cell
Physiol. 239:e313302024. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Jing R, Ma B, Qi T, Hu C, Liao C, Wen C,
Shao Y and Pei C: Long noncoding RNA OIP5-AS1 promotes cell
apoptosis and cataract formation by blocking POLG expression under
oxidative stress. Invest Ophthalmol Vis Sci. 61:32020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Xiang J, Chen Q, Kang L, Zhang G, Wang Y,
Qin B, Wu J, Zhou T, Han Y and Guan H: LncRNA PLCD3-OT1 functions
as a CeRNA to prevent age-related cataract by sponging miR-224-5p
and regulating PLCD3 expression. Invest Ophthalmol Vis Sci.
60:4670–4680. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Cheng T, Xu M, Qin B, Wu J, Tu Y, Kang L,
Wang Y and Guan H: lncRNA H19 contributes to oxidative damage
repair in the early age-related cataract by regulating miR-29a/TDG
axis. J Cell Mol Med. 23:6131–6139. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Alkan AH and Akgül B: Endogenous miRNA
sponges. Methods Mol Biol. 2257:91–104. 2022. View Article : Google Scholar
|
|
145
|
Alshahrani SH, Al-Hadeithi ZSM, Almalki
SG, Malviya J, Hjazi A, Mustafa YF, Alawady AHR, Alsaalamy AH,
Joshi SK and Alkhafaji AT: LncRNA-miRNA interaction is involved in
colorectal cancer pathogenesis by modulating diverse signaling
pathways. Pathol Res Pract. 251:1548982023. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Karagkouni D, Karavangeli A,
Paraskevopoulou MD and Hatzigeorgiou AG: Characterizing
miRNA-lncRNA Interplay. Methods Mol Biol. 2372:243–262. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhang Y, Cai S, Jia Y, Qi C, Sun J, Zhang
H, Wang F, Cao Y and Li X: Decoding noncoding RNAs: Role of
MicroRNAs and long noncoding RNAs in ocular neovascularization.
Theranostics. 7:3155–3167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Magistri M, Faghihi MA, St Laurent G III
and Wahlestedt C: Regulation of chromatin structure by long
noncoding RNAs: Focus on natural antisense transcripts. Trends
Genet. 28:389–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Xue Y, Ma G, Zhang Z, Hua Q, Chu H, Tong
N, Yuan L, Qin C, Yin C, Zhang Z and Wang M: A novel antisense long
noncoding RNA regulates the expression of MDC1 in bladder cancer.
Oncotarget. 6:484–493. 2015. View Article : Google Scholar :
|
|
150
|
Zhang CL, Zhu KP and Ma XL: Antisense
lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by
increasing the expression of FOXC2. Cancer Lett. 396:66–75. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
He A, He S, Li X and Zhou L: ZFAS1: A
novel vital oncogenic lncRNA in multiple human cancers. Cell
Prolif. 52:e125132019. View Article : Google Scholar
|
|
152
|
Zong X, Nakagawa S, Freier SM, Fei J, Ha
T, Prasanth SG and Prasanth KV: Natural antisense RNA promotes 3'
end processing and maturation of MALAT1 lncRNA. Nucleic Acids Res.
44:2898–2908. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Tu Y, Li L, Qin B, Wu J, Cheng T, Kang L
and Guan H: Long noncoding RNA glutathione peroxidase 3-antisense
inhibits lens epithelial cell apoptosis by upregulating glutathione
peroxidase 3 expression in age-related cataract. Mol Vis.
25:734–744. 2019.PubMed/NCBI
|
|
154
|
Zhu M, Dong Q, Bing J, Songbuerbatu, Zheng
L, Dorjee T, Liu Q, Zhou Y and Gao F: Combined lncRNA and mRNA
expression profiles identified the lncRNA-miRNA-mRNA modules
regulating the cold stress response in Ammopiptanthus nanus. Int J
Mol Sci. 24:65022023. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Liu X, Liu C, Shan K, Zhang S, Lu Y, Yan B
and Luo Y: Long non-coding RNA H19 regulates human lens epithelial
cells function. Cell Physiol Biochem. 50:246–260. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Harland R and Misher L: Stability of RNA
in developing Xenopus embryos and identification of a destabilizing
sequence in TFIIIA messenger RNA. Development. 102:837–852. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
159
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Panda AC: Circular RNAs Act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Tong KL, Tan KE, Lim YY, Tien XY and Wong
PF: CircRNA-miRNA interactions in atherogenesis. Mol Cell Biochem.
477:2703–2733. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16:582017. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Qiu JN, Shan K, Xiang J, Gu JY, Zhou RM,
Zhang XL, Zhang CR and Xu JJ: Comprehensive analysis of
circRNA-associated-ceRNA networks in human corneal endothelial
dysfunction. Cornea. 41:1545–1552. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Qu B, Wang J, Li Y, Wu X and Zhang M:
Hsa_circ_0023826 protects against glaucoma by regulating
miR-188-3p/MDM4 axis. Acta Biochim Pol. 70:253–260. 2023.PubMed/NCBI
|
|
170
|
Tian H, Zhao L, Li H, Huang Y and Wang Y:
Circular RNA in retina: A potential biomarker and therapeutic
target. Ophthalmic Res. 66:516–528. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Li D, Che X, Gao N and Li J: CircSTRBP
contributes to H2O2-induced lens epithelium
cell dysfunction through increasing NOX4 mRNA stability by
recruiting IGF2BP1. Exp Eye Res. 241:1098172024. View Article : Google Scholar
|
|
172
|
Wang Y, Zhang G, Li P, Kang L, Qin B, Cao
Y, Luo J, Chen X, Qin M and Guan H: Profiling and integrated
analysis of the ERCC6-regulated circRNA-miRNA-mRNA network in lens
epithelial cells. Curr Eye Res. 46:1341–1352. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Xu X, Gao R, Li S, Jiang K, Sun X and
Zhang J: Circular RNA circZNF292 regulates H2
O2-induced injury in human lens epithelial HLE-B3 cells
depending on the regulation of the miR-222-3p/E2F3 axis. Cell Biol
Int. 45:1757–1767. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Mao W and Zhang Z: The
Hsa_circ_0105558/miR-182-5p/ATF6 cascade affects
H2O2-triggered oxidative damage and apoptosis
of human lens epithelial cells. Biochem Genet. 63:1241–1257. 2025.
View Article : Google Scholar
|
|
175
|
Li R, Zhu H, Li Q, Tang J, Jin Y and Cui
H: METTL3-mediated m6A modification of has_circ_0007905 promotes
age-related cataract progression through miR-6749-3p/EIF4EBP1.
PeerJ. 11:e148632023. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Liu J, Zhang J, Zhang G, Zhou T, Zou X,
Guan H and Wang Y: CircMRE11A_013 binds to UBXN1 and integrates ATM
activation enhancing lens epithelial cells senescence in
age-related cataract. Aging (Albany NY). 13:5383–5402. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Cui G, Wang L and Huang W: Circular RNA
HIPK3 regulates human lens epithelial cell dysfunction by targeting
the miR-221-3p/PI3K/AKT pathway in age-related cataract. Exp Eye
Res. 198:1081282020. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Liu X, Liu B, Zhou M, Fan F, Yu M, Gao C,
Lu Y and Luo Y: Circular RNA HIPK3 regulates human lens epithelial
cells proliferation and apoptosis by targeting the miR-193a/CRYAA
axis. Biochem Biophys Res Commun. 503:2277–2285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Chen S, Wang M, Jian R, Li H, Liu G, Zhou
C, Xiong Y and Wang W: Circ_HIPK3 Inhibits
H2O2-induced lens epithelial cell injury in
age-related cataract depending on the regulation of
miR-495-3p/HDAC4 pathway. Biochem Genet. 61:565–577. 2023.
View Article : Google Scholar
|
|
180
|
Liu T, Zhao L, Yan D and Wang N:
Circ_0060,144 inhibits the occurrence and development of
age-related cataract via the miR-23b-3p/HIPK3 axis. Exp Eye Res.
222:1091792022. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Zhou C, Huang X, Li X and Xiong Y:
Circular RNA erythrocyte membrane protein band 4.1 assuages
ultraviolet irradiation-induced apoptosis of lens epithelial cells
by stimulating 5'-bisphosphate nucleotidase 1 in a
miR-24-3p-dependent manner. Bioengineered. 12:8953–8964. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Fang R, Li JH, Li HL, Yue PL, Ding XF, Jia
YX, Liu ZC, Zhou HG, Yang C and Song XD: CircRNA 06209 inhibits
cataract development by sponging miR-6848-5p and regulating ALOX15
expression. Exp Eye Res. 235:1096402023. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020. View Article : Google Scholar
|
|
184
|
Cui F, Sun Z, Zhang X and Liu C:
CircMAP3K4 suppresses H2O2-induced human lens
epithelial cell injury by miR-630/ERCC6 axis in age-related
cataract. Curr Eye Res. 49:487–495. 2024. View Article : Google Scholar
|
|
185
|
Sun L, Li F, Bai S and Bi C: CircRNA HLCS
regulates lens epithelial cell apoptosis via miR-338-3p/BPNT1 axis.
Int Ophthalmol. 44:1422024. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Wu Q, Liu H, Ma B and Wang C: Circular RNA
Circ_0122396 regulates human lens epithelial cell progression by
regulating miR-23a-3p and MMP16 in age-related cataract. Curr Eye
Res. 49:1161–1170. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Liu B, Cao J, Wang X, Guo C, Liu Y and
Wang T: Deciphering the tRNA-derived small RNAs: Origin,
development, and future. Cell Death Dis. 13:242021. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Xie Y, Yao L, Yu X, Ruan Y, Li Z and Guo
J: Action mechanisms and research methods of tRNA-derived small
RNAs. Signal Transduct Target Ther. 5:1092020. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Pan J, Liu Z, Shen B, Xu J, Dai G, Xu W,
Wang J, Li L and Cheng L: tsRNA-04002 alleviates intervertebral
disk degeneration by targeting PRKCA to inhibit apoptosis of
nucleus pulposus cells. J Orthop Surg Res. 18:4132023. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Zeng T, Hua Y, Sun C, Zhang Y, Yang F,
Yang M, Yang Y, Li J, Huang X, Wu H, et al: Relationship between
tRNA-derived fragments and human cancers. Int J Cancer.
147:3007–3018. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Zhang L, Liu J and Hou Y: Classification,
function, and advances in tsRNA in non-neoplastic diseases. Cell
Death Dis. 14:7482023. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Wang S, Luo Z, Yuan L, Lin X, Tang Y, Yin
L, Liang P and Jiang B: tRNA-derived small RNAs: Novel insights
into the pathogenesis and treatment of cardiovascular diseases. J
Cardiovasc Transl Res. 16:300–309. 2023. View Article : Google Scholar
|
|
193
|
Zhang X, Trebak F, Souza LAC, Shi J, Zhou
T, Kehoe PG, Chen Q and Feng Earley Y: Small RNA modifications in
Alzheimer's disease. Neurobiol Dis. 145:1050582020. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Zhou M, He X, Zhang J, Mei C, Zhong B and
Ou C: tRNA-derived small RNAs in human cancers: Roles, mechanisms,
and clinical application. Mol Cancer. 23:762024. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Han X, Cai L, Lu Y, Li D and Yang J:
Identification of tRNA-derived fragments and their potential roles
in diabetic cataract rats. Epigenomics. 12:1405–1418. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Ma Y, Zhang Y, Zhang HY, Zhao Y, Li XM,
Jiang YF, Yao MD, Jiang Q and Yan B: Dual anti-angiogenic and
anti-inflammatory action of tRNA-Cys-5-0007 in ocular vascular
disease. J Transl Med. 22:5622024. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Peng Y, Zou J, Wang JH, Zeng H, Tan W,
Yoshida S, Zhang L, Li Y and Zhou Y: Small RNA sequencing reveals
transfer RNA-derived small RNA expression profiles in retinal
neovascularization. Int J Med Sci. 17:1713–1722. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Zhang G, Kang L, Li P, Ran Q, Chen X, Ji M
and Guan H: Genome-wide repertoire of transfer RNA-derived
fragments in a mouse model of age-related cataract. Curr Eye Res.
47:1397–1404. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar
|
|
200
|
Ma Y, Liu Y, Shu B, Yang J, Lv L, Zhou L,
Wang L and Shi Z: CircMAP3K4 protects human lens epithelial cells
from H2O2-induced dysfunction by targeting
miR-193a-3p/PLCD3 axis in age-related cataract. Cell Cycle.
22:303–315. 2023. View Article : Google Scholar
|
|
201
|
Xu J, Shao T, Ding N, Li Y and Li X:
miRNA-miRNA crosstalk: From genomics to phenomics. Brief Bioinform.
18:1002–1011. 2017.
|
|
202
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
203
|
Shao T, Wang G, Chen H, Xie Y, Jin X, Bai
J, Xu J, Li X, Huang J, Jin Y and Li Y: Survey of miRNA-miRNA
cooperative regulation principles across cancer types. Brief
Bioinform. 20:1621–1638. 2019. View Article : Google Scholar
|
|
204
|
Chiu HS, Somvanshi S, Patel E, Chen TW,
Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS; Cancer Genome
Atlas Research Network; et al: Pan-cancer analysis of lncRNA
regulation supports their targeting of cancer genes in each tumor
context. Cell Rep. 23:297–312.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Zhang K, Chen L, Qu L and Yan H: A
comprehensive investigation of identifying miRNA biomarkers and
their potential role in age-related cataract by meta-analysis and
bioinformatics analysis. Graefes Arch Clin Exp Ophthalmol.
263:1307–1325. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Michalak EM, Burr ML, Bannister AJ and
Dawson MA: The roles of DNA, RNA and histone methylation in ageing
and cancer. Nat Rev Mol Cell Biol. 20:573–589. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Dossin F, Pinheiro I, Żylicz JJ, Roensch
J, Collombet S, Le Saux A, Chelmicki T, Attia M, Kapoor V, Zhan Y,
et al: SPEN integrates transcriptional and epigenetic control of
X-inactivation. Nature. 578:455–460. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Grigoryan A, Pospiech J, Krämer S, Lipka
D, Liehr T, Geiger H, Kimura H, Mulaw MA and Florian MC: Attrition
of X chromosome inactivation in aged hematopoietic stem cells. Stem
Cell Reports. 16:708–716. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Kulis M, Queirós AC, Beekman R and
Martín-Subero JI: Intragenic DNA methylation in transcriptional
regulation, normal differentiation and cancer. Biochim Biophys
Acta. 1829:1161–1174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Nishiyama A and Nakanishi M: Navigating
the DNA methylation landscape of cancer. Trends Genet.
37:1012–1027. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
May MS and Hattaman S: Deoxyribonucleic
acid-cytosine methylation by host- and plasmid-controlled enzymes.
J Bacteriol. 122:129–138. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Kohli RM and Zhang Y: TET enzymes, TDG and
the dynamics of DNA demethylation. Nature. 502:472–479. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Harris AJ and Goldman AD: The complex
phylogenetic relationships of a 4mC/6mA DNA methyltransferase in
prokaryotes. Mol Phylogenet Evol. 149:1068372020. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Thomas B, Matson S, Chopra V, Sun L,
Sharma S, Hersch S, Rosas HD, Scherzer C, Ferrante R and Matson W:
A novel method for detecting 7-methyl guanine reveals aberrant
methylation levels in Huntington disease. Anal Biochem.
436:112–120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Hashimoto H, Pais JE, Zhang X, Saleh L, Fu
ZQ, Dai N, Corrêa IR Jr, Zheng Y and Cheng X: Structure of a
Naegleria Tet-like dioxygenase in complex with 5-methylcytosine
DNA. Nature. 506:391–395. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Angeloni A and Bogdanovic O: Enhancer DNA
methylation: Implications for gene regulation. Essays Biochem.
63:707–715. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Zeng Y and Chen T: DNA methylation
reprogramming during mammalian development. Genes (Basel).
10:2572019. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Zong FF, Jia DD, Huang GK, Pan M, Hu H,
Song SY, Xiao L, Wang RW and Liang L: New perspectives on DNA
methylation modifications in ocular diseases. Int J Ophthalmol.
18:340–350. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Li Y, Chen X and Lu C: The interplay
between DNA and histone methylation: Molecular mechanisms and
disease implications. EMBO Rep. 22:e518032021. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Boyes J and Bird A: DNA methylation
inhibits transcription indirectly via a methyl-CpG binding protein.
Cell. 64:1123–1134. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Cvekl A and Mitton KP: Epigenetic
regulatory mechanisms in vertebrate eye development and disease.
Heredity (Edinb). 105:135–151. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Gao Y, Yan Y and Huang T: Human
age-related cataracts: epigenetic suppression of the nuclear factor
erythroid 2-related factor 2-mediated antioxidant system. Mol Med
Rep. 11:1442–1447. 2015. View Article : Google Scholar
|
|
224
|
Jin B and Robertson KD: DNA
methyltransferases, DNA damage repair, and cancer. Adv Exp Med
Biol. 754:3–29. 2013. View Article : Google Scholar
|
|
225
|
Kim DJ: The role of the DNA
methyltransferase family and the therapeutic potential of DNMT
inhibitors in tumor treatment. Curr Oncol. 32:882025. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Asada M, Hayashi H and Takagi N: Possible
involvement of DNA methylation and protective effect of zebularine
on neuronal cell death after glutamate excitotoxity. Biol Pharm
Bull. 45:770–779. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Liu S, Hu C, Luo Y and Yao K: Genome-wide
DNA methylation profiles may reveal new possible epigenetic
pathogenesis of sporadic congenital cataract. Epigenomics.
12:771–788. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Song J, VanBuskirk JA and Merbs SL:
Regulation of opsin gene expression by DNA methylation and histone
acetylation. Int J Mol Sci. 23:14082022. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Johnson WM, Finnegan LK, Hauser MA and
Stamer WD: lncRNAs, DNA methylation, and the pathobiology of
exfoliation glaucoma. J Glaucoma. 27:202–209. 2018. View Article : Google Scholar
|
|
230
|
Zhou P, Luo Y, Liu X, Fan L and Lu Y:
Down-regulation and CpG island hypermethylation of CRYAA in
age-related nuclear cataract. FASEB J. 26:4897–4902. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Baird PN and Wei L: Age-related macular
degeneration and DNA methylation. Epigenomics. 5:239–241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Chai P, Jia R, Li Y, Zhou C, Gu X, Yang L,
Shi H, Tian H, Lin H, Yu J, et al: Regulation of epigenetic
homeostasis in uveal melanoma and retinoblastoma. Prog Retin Eye
Res. 89:1010302022. View Article : Google Scholar
|
|
233
|
Wolff SP: Cataract and UV radiation. Doc
Ophthalmol. 88:201–204. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Li X, Xie J, Xu J, Deng L, Cao G, Huang S,
Zeng C, Liu C, Zhu S, He G, et al: Long-term exposure to ambient
PM2.5 and age-related cataracts among chinese
middle-aged and older adults: Evidence from two national cohort
studies. Environ Sci Technol. 57:11792–11802. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Chen J, Zhou J, Wu J, Zhang G, Kang L, Ben
J, Wang Y, Qin B and Guan H: Aberrant epigenetic alterations of
glutathione-S-transferase P1 in age-related nuclear cataract. Curr
Eye Res. 42:402–410. 2017. View Article : Google Scholar
|
|
236
|
Liu X, Zhou P, Fan F, Li D, Wu J, Lu Y and
Luo Y: CpG site methylation in CRYAA promoter affect transcription
factor Sp1 binding in human lens epithelial cells. BMC Ophthalmol.
16:1412016. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Jin SL, Zhang Y, Chen ZH, Qian DW, Qine
YJ, Yongjie Q, He SK and Guo HK: Epigenetic changes of the Klotho
gene in age-related cataracts. Eur Rev Med Pharmacol Sci.
19:2544–2553. 2015.PubMed/NCBI
|
|
238
|
Wang Y, Zhang G, Kang L and Guan H:
Expression profiling of DNA methylation and transcriptional
repression associated genes in lens epithelium cells of age-related
cataract. Cell Mol Neurobiol. 37:537–543. 2017. View Article : Google Scholar
|
|
239
|
Chen X, Su D, Sun Z, Fu Y, Hu Y, Zhang Y,
Zhang X, Wei Q, Zhu W, Ma X and Hu S: Preliminary study on whole
genome methylation and transcriptomics in age-related cataracts.
Gene. 898:1480962024. View Article : Google Scholar
|
|
240
|
Wang L, Li P and Guo X: Screening of
methylation genes in age-related cataract. Int J Ophthalmol.
11:1102–1107. 2018.PubMed/NCBI
|
|
241
|
Pendergrass W, Penn P, Possin D and Wolf
N: Accumulation of DNA, nuclear and mitochondrial debris, and ROS
at sites of age-related cortical cataract in mice. Invest
Ophthalmol Vis Sci. 46:4661–4670. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Zhang Y, Zhang L, Zhang L, Bai J, Ge H and
Liu P: Expression changes in DNA repair enzymes and mitochondrial
DNA damage in aging rat lens. Mol Vis. 16:1754–1763.
2010.PubMed/NCBI
|
|
243
|
Li B, Zhou J, Zhang G, Wang Y, Kang L, Wu
J, Chen J and Guan H: Relationship between the altered expression
and epigenetics of GSTM3 and age-related cataract. Invest
Ophthalmol Vis Sci. 57:4721–4732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Rushmore TH, Morton MR and Pickett CB: The
antioxidant responsive element. Activation by oxidative stress and
identification of the DNA consensus sequence required for
functional activity. J Biol Chem. 266:11632–11639. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Ling X, Zhu L, Yan Y, Qian H, Kang Z, Ye
W, Xie Z and Xue C: Ferulic acid protects human lens epithelial
cells against UVA-induced oxidative damage by downregulating the
DNA demethylation of the keap1 promoter. J Biochem Mol Toxicol.
38:e700312024. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Palsamy P, Bidasee KR and Shinohara T:
Selenite cataracts: Activation of endoplasmic reticulum stress and
loss of Nrf2/Keap1-dependent stress protection. Biochim Biophys
Acta. 1842:1794–1805. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Rabbani PS, Soares MA, Hameedi SG, Qian H,
Kang Z, Ye W, Xie Z and Xue C: Dysregulation of Nrf2/Keap1 redox
pathway in diabetes affects multipotency of stromal cells.
Diabetes. 68:141–155. 2019. View Article : Google Scholar :
|
|
248
|
Raghunath A, Sundarraj K, Nagarajan R,
Arfuso F, Bian J, Kumar AP, Sethi G and Perumal E: Antioxidant
response elements: Discovery, classes, regulation and potential
applications. Redox Biol. 17:297–314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Li J, Wang T, Liu P, Yang F, Wang X, Zheng
W and Sun W: Hesperetin ameliorates hepatic oxidative stress and
inflammation via the PI3K/AKT-Nrf2-ARE pathway in oleic
acid-induced HepG2 cells and a rat model of high-fat diet-induced
NAFLD. Food Funct. 12:3898–3918. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Osama A, Zhang J, Yao J, Yao X and Fang J:
Nrf2: A dark horse in Alzheimer's disease treatment. Ageing Res
Rev. 64:1012062020. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Palsamy P, Bidasee KR and Shinohara T:
Valproic acid suppresses Nrf2/Keap1 dependent antioxidant
protection through induction of endoplasmic reticulum stress and
Keap1 promoter DNA demethylation in human lens epithelial cells.
Exp Eye Res. 121:26–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Palsamy P, Bidasee KR, Ayaki M, Augusteyn
RC, Chan JY and Shinohara T: Methylglyoxal induces endoplasmic
reticulum stress and DNA demethylation in the Keap1 promoter of
human lens epithelial cells and age-related cataracts. Free Radic
Biol Med. 72:134–148. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Zduńska K, Dana A, Kolodziejczak A and
Rotsztejn H: Antioxidant properties of ferulic acid and its
possible application. Skin Pharmacol Physiol. 31:332–336. 2018.
View Article : Google Scholar
|
|
254
|
Rampelotto CR, Pereira VG, da Silva
Silveira L, Rossato A, Machado AK, Sagrillo MR, Gündel A, Burger
ME, Schaffazick SR and de Bona da Silva C: Ferulic acid-loaded
nanocapsules: Evaluation of mucosal interaction, safety and
antioxidant activity in human mononucleated cells. Toxicol In
Vitro. 78:1052592022. View Article : Google Scholar
|
|
255
|
Mathew S and Abraham TE: Ferulic acid: an
antioxidant found naturally in plant cell walls and feruloyl
esterases involved in its release and their applications. Crit Rev
Biotechnol. 24:59–83. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Yang SP, Yang XZ and Cao GP:
Acetyl-l-carnitine prevents homocysteine-induced suppression of
Nrf2/Keap1 mediated antioxidation in human lens epithelial cells.
Mol Med Rep. 12:1145–1150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Giblin FJ: Glutathione: A vital lens
antioxidant. J Ocul Pharmacol Ther. 16:121–135. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Sheehan D, Meade G, Foley VM and Dowd CA:
Structure, function and evolution of glutathione transferases:
Implications for classification of non-mammalian members of an
ancient enzyme superfamily. Biochem J. 360:1–16. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
259
|
Whalen R and Boyer TD: Human glutathione
S-transferases. Semin Liver Dis. 18:345–358. 1998. View Article : Google Scholar
|
|
260
|
Hayes JD and Pulford DJ: The glutathione
S-transferase super-gene family: Regulation of GST and the
contribution of the isoenzymes to cancer chemoprotection and drug
resistance. Crit Rev Biochem Mol Biol. 30:445–600. 1995. View Article : Google Scholar
|
|
261
|
Choi YJ, Yeo HJ, Shin MJ, Youn GS, Park
JH, Yeo EJ, Kwon HJ, Lee LR, Kim NY, Kwon SY, et al: Tat-GSTpi
inhibits dopaminergic cells against MPP+-induced
cellular damage via the reduction of oxidative stress and MAPK
activation. Biomedicines. 11:8362023. View Article : Google Scholar
|
|
262
|
Laborde E: Glutathione transferases as
mediators of signaling pathways involved in cell proliferation and
cell death. Cell Death Differ. 17:1373–1380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Hayes JD and McLellan LI: Glutathione and
glutathione-dependent enzymes represent a co-ordinately regulated
defence against oxidative stress. Free Radic Res. 31:273–300. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Babizhayev MA: Generation of reactive
oxygen species in the anterior eye segment. Synergistic codrugs of
N-acetylcarnosine lubricant eye drops and mitochondria-targeted
antioxidant act as a powerful therapeutic platform for the
treatment of cataracts and primary open-angle glaucoma. BBA Clin.
6:49–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Sorte K, Sune P, Bhake A, Shivkumar VB,
Gangane N and Basak A: Quantitative assessment of DNA damage
directly in lens epithelial cells from senile cataract patients.
Mol Vis. 17:1–6. 2011.PubMed/NCBI
|
|
266
|
Xu B, Kang L, Zhang G, Wu J, Zhu R, Yang M
and Guan H: The changes of 8-OHdG, hOGG1, APE1 and Pol β in lenses
of patients with age-related cataract. Curr Eye Res. 40:378–385.
2015. View Article : Google Scholar
|
|
267
|
Chatterjee N and Walker GC: Mechanisms of
DNA damage, repair, and mutagenesis. Environ Mol Mutagen.
58:235–263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Maugeri-Saccà M, Bartucci M and De Maria
R: DNA damage repair pathways in cancer stem cells. Mol Cancer
Ther. 11:1627–1636. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Kong M, Liu L, Chen X, Driscoll KI, Mao P,
Böhm S, Kad NM, Watkins SC, Bernstein KA, Wyrick JJ, et al:
Single-molecule imaging reveals that Rad4 employs a dynamic DNA
damage recognition process. Mol Cell. 64:376–387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
270
|
Licht CL, Stevnsner T and Bohr VA:
Cockayne syndrome group B cellular and biochemical functions. Am J
Hum Genet. 73:1217–1239. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
271
|
Fleming AM and Burrows CJ:
8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator
versus initiator of mutagenesis. DNA Repair (Amst). 56:75–83. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
272
|
Dinçer Y, Akkaya Ç, Mutlu T, Yavuzer S,
Erkol G, Bozluolcay M and Guven M: DNA repair gene OGG1
polymorphism and its relation with oxidative DNA damage in patients
with Alzheimer's disease. Neurosci Lett. 709:1343622019. View Article : Google Scholar : PubMed/NCBI
|
|
273
|
Synowiec E, Blasiak J, Zaras M, Szaflik J
and Szaflik JP: Association between polymorphisms of the DNA base
excision repair genes MUTYH and hOGG1 and age-related macular
degeneration. Exp Eye Res. 98:58–66. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
274
|
Wu X, Lai W, Lin H and Liu Y: Association
of OGG1 and MTHFR polymorphisms with age-related cataract: A
systematic review and meta-analysis. PLoS One. 12:e01720922017.
View Article : Google Scholar : PubMed/NCBI
|
|
275
|
Wang Y, Li F, Zhang G, Kang L, Qin B and
Guan H: Altered DNA methylation and expression profiles of
8-oxoguanine DNA glycosylase 1 in lens tissue from age-related
cataract patients. Curr Eye Res. 40:815–821. 2015. View Article : Google Scholar
|
|
276
|
Yan H, McCane J, Toczylowski T and Chen C:
Analysis of the Xenopus Werner syndrome protein in DNA
double-strand break repair. J Cell Biol. 171:217–227. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Tranah GJ, Bugni J, Giovannucci E, Ma J,
Fuchs C, Hines L, Samson L and Hunter DJ: O6-methylguanine-DNA
methyltransferase Leu84Phe and Ile143Val polymorphisms and risk of
colorectal cancer in the nurses' health study and physicians'
health study (United States). Cancer Causes Control. 17:721–731.
2006.PubMed/NCBI
|
|
278
|
Li F, Wang Y, Zhang G, Zhou J, Yang L and
Guan H: Expression and methylation of DNA repair genes in lens
epithelium cells of age-related cataract. Mutat Res. 766-767:31–36.
2014. View Article : Google Scholar
|
|
279
|
Andley UP: Effects of alpha-crystallin on
lens cell function and cataract pathology. Curr Mol Med. 9:887–892.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Horwitz J: Alpha-crystallin can function
as a molecular chaperone. Proc Natl Acad Sci USA. 89:10449–10453.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Thampi P, Hassan A, Smith JB and Abraham
EC: Enhanced C-terminal truncation of alphaA- and
alphaB-crystallins in diabetic lenses. Invest Ophthalmol Vis Sci.
43:3265–3272. 2002.PubMed/NCBI
|
|
282
|
Horwitz J: Alpha-crystallin. Exp Eye Res.
76:145–153. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Christopher KL, Pedler MG, Shieh B, Ammar
DA, Petrash JM and Mueller NH: Alpha-crystallin-mediated protection
of lens cells against heat and oxidative stress-induced cell death.
Biochim Biophys Acta. 1843:309–315. 2014. View Article : Google Scholar :
|
|
284
|
Wang Z, Sun Y, Zhang Y, Zhang Y, Zhang R,
Li C, Liu X, Pan F, Qiao D, Shi X, et al: Identification of seven
variants in the col4a1 gene that alter RNA splicing by minigene
assay. Clin Genet. 106:336–341. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
285
|
Wang L, Zhu D, Yang Y, He Y, Sun J, Li YM,
Wang ZJ and Li P: DNA hypermethylation of COL4A1 in
ultraviolet-B-induced age-related cataract models in vitro and in
vivo. Int J Ophthalmol. 17:1791–1799. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
286
|
Fujisawa T and Filippakopoulos P:
Functions of bromodomain-containing proteins and their roles in
homeostasis and cancer. Nat Rev Mol Cell Biol. 18:246–262. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
287
|
Bhaumik SR, Smith E and Shilatifard A:
Covalent modifications of histones during development and disease
pathogenesis. Nat Struct Mol Biol. 14:1008–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
288
|
Zhang Z, Fu J, Rack JGM, Li C, Voorneveld
J, Filippov DV, Ahel I, Luo ZQ and Das C: Legionella metaeffector
MavL reverses ubiquitin ADP-ribosylation via a conserved
arginine-specific macrodomain. Nat Commun. 15:24522024. View Article : Google Scholar : PubMed/NCBI
|
|
289
|
Zhu D, Zhang Y and Wang S: Histone
citrullination: A new target for tumors. Mol Cancer. 20:902021.
View Article : Google Scholar : PubMed/NCBI
|
|
290
|
Rong X, Qiu X, Jiang Y, Li D, Xu J, Zhang
Y and Lu Y: Effects of histone acetylation on superoxide dismutase
1 gene expression in the pathogenesis of senile cataract. Sci Rep.
6:347042016. View Article : Google Scholar : PubMed/NCBI
|
|
291
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
292
|
de Lera AR and Ganesan A: Epigenetic
polypharmacology: From combination therapy to multitargeted drugs.
Clin Epigenetics. 8:1052016. View Article : Google Scholar : PubMed/NCBI
|
|
293
|
Scarano N, Brullo C, Musumeci F, Millo E,
Bruzzone S, Schenone S and Cichero E: Recent advances in the
discovery of SIRT1/2 inhibitors via computational methods: A
perspective. Pharmaceuticals (Basel). 17:6012024. View Article : Google Scholar : PubMed/NCBI
|
|
294
|
Li G, Jiang H, Chang M, Xie H and Hu L:
HDAC6 α-tubulin deacetylase: A potential therapeutic target in
neurodegenerative diseases. J Neurol Sci. 304:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
295
|
Mimura T, Kaji Y, Noma H, Funatsu H and
Okamoto S: The role of SIRT1 in ocular aging. Exp Eye Res.
116:17–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
296
|
Ağaoğlu NB, Varol N, Yıldız SH,
Karaosmanoğlu C, Duman R, Özdemir Erdoğan M and Solak M:
Relationship between SIRT1 gene expression level and disease in
age-related cataract cases. Turk J Med Sci. 49:1068–1072. 2019.
View Article : Google Scholar
|
|
297
|
Zheng T and Lu Y: Changes in SIRT1
expression and its downstream pathways in age-related cataract in
humans. Curr Eye Res. 36:449–455. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
298
|
Zentner GE and Henikoff S: Regulation of
nucleosome dynamics by histone modifications. Nat Struct Mol Biol.
20:259–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
299
|
Bannister AJ and Kouzarides T: Regulation
of chromatin by histone modifications. Cell Res. 21:381–395. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
300
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
301
|
Cheng X: Structural and functional
coordination of DNA and histone methylation. Cold Spring Harb
Perspect Biol. 6:a0187472014. View Article : Google Scholar : PubMed/NCBI
|
|
302
|
Bachman KE, Rountree MR and Baylin SB:
Dnmt3a and Dnmt3b are transcriptional repressors that exhibit
unique localization properties to heterochromatin. J Biol Chem.
276:32282–32287. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
303
|
Shi H, Chai P, Jia R and Fan X: Novel
insight into the regulatory roles of diverse RNA modifications:
Re-defining the bridge between transcription and translation. Mol
Cancer. 19:782020. View Article : Google Scholar : PubMed/NCBI
|
|
304
|
Tang J, Zhou C, Ye F, Zuo S, Zhou M, Lu L,
Chai P and Fan X: RNA methylation homeostasis in ocular diseases:
All eyes on Me. Prog Retin Eye Res. 105:1013352025. View Article : Google Scholar : PubMed/NCBI
|
|
305
|
Jia G, Fu Y and He C: Reversible RNA
adenosine methylation in biological regulation. Trends Genet.
29:108–115. 2013. View Article : Google Scholar :
|
|
306
|
Kumari R, Ranjan P, Suleiman ZG, Goswami
SK, Li J, Prasad R and Verma SK: mRNA modifications in
cardiovascular biology and disease: With a focus on m6A
modification. Cardiovasc Res. 118:1680–1692. 2022. View Article : Google Scholar :
|
|
307
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
308
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3' UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
309
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
310
|
Chen X, Wang Y, Wang JN, Cao QC, Sun RX,
Zhu HJ, Zhang YR, Ji JD and Liu QH: m6A modification of
circSPECC1 suppresses RPE oxidative damage and maintains retinal
homeostasis. Cell Rep. 41:1116712022. View Article : Google Scholar
|
|
311
|
Sheng Z, Pan Y, Shao L and Bao Y: METTL3
mediates CPB1 expression by regulating transcription factor BACH2
to promote apoptosis and oxidative stress of lens epithelial cells.
J Bioenerg Biomembr. 57:161–171. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
312
|
Wang T, Kong S, Tao M and Ju S: The
potential role of RNA N6-methyladenosine in cancer progression. Mol
Cancer. 19:882020. View Article : Google Scholar : PubMed/NCBI
|
|
313
|
Kang L, Bao S, Li P, Zhang G, Zhu X, Ji M
and Guan H: METTL14-mediated depression of NEIL1 aggravates
oxidative damage and mitochondrial dysfunction of lens epithelial
cells through regulating KEAP1/NRF2 pathways. Cell Signal.
127:1116232025. View Article : Google Scholar : PubMed/NCBI
|
|
314
|
Ye HF, Zhang X, Zhao ZN, Zheng C, Fei P,
Xu Y, Lyu J, Chen JL, Guo XX, Zhu H and Zhao PQ: Characterization
of N6-methyladenosine long non-coding RNAs in sporadic
congenital cataract and age-related cataract. Int J Ophthalmol.
17:1973–1986. 2024. View Article : Google Scholar :
|
|
315
|
Wang Y, Li P, Wang C, Bao S, Wang S, Zhang
G, Zou X, Wu J, Guan Y, Ji M and Guan H: Lens epithelium cell
ferroptosis mediated by m6A-lncRNA and GPX4 expression
in lens tissue of age-related cataract. BMC Ophthalmol. 23:5142023.
View Article : Google Scholar
|
|
316
|
Luo G, Xu W, Chen X, Wang S, Wang J, Dong
F, Hu DN, Reinach PS and Yan D: NSUN2-mediated RNA m5C
modification modulates uveal melanoma cell proliferation and
migration. Epigenetics. 17:922–933. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
317
|
Zhao Y, Kong L, Pei Z, Li F, Li C, Sun X,
Shi B and Ge J: m7G methyltransferase METTL1 promotes
post-ischemic angiogenesis via promoting VEGFA mRNA translation.
Front Cell Dev Biol. 9:6420802021. View Article : Google Scholar
|
|
318
|
Wu C, Liu Z, Ma L, Pei C, Qin L, Gao N, Li
J and Yin Y: MiRNAs regulate oxidative stress related genes via
binding to the 3' UTR and TATA-box regions: A new hypothesis for
cataract pathogenesis. BMC Ophthalmol. 17:1422017. View Article : Google Scholar : PubMed/NCBI
|
|
319
|
Gu X, Sun J, Li S, Wu X and Li L:
Oxidative stress induces DNA demethylation and histone acetylation
in SH-SY5Y cells: Potential epigenetic mechanisms in gene
transcription in Aβ production. Neurobiol Aging. 34:1069–1079.
2013. View Article : Google Scholar
|
|
320
|
Niu Y, DesMarais TL, Tong Z, Yao Y and
Costa M: Oxidative stress alters global histone modification and
DNA methylation. Free Radic Biol Med. 82:22–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
321
|
Li Q, Li X, Tang H, Jiang B, Dou Y,
Gorospe M and Wang W: NSUN2-mediated m5C methylation and
METTL3/METTL14-mediated m6A methylation cooperatively enhance p21
translation. J Cell Biochem. 118:2587–2598. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
322
|
Wu Q and Ni X: ROS-mediated DNA
methylation pattern alterations in carcinogenesis. Curr Drug
Targets. 16:13–19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
323
|
Chouliaras L, Mastroeni D, Delvaux E,
Grover A, Kenis G, Hof PR, Steinbusch HW, Coleman PD, Rutten BP and
van den Hove DL: Consistent decrease in global DNA methylation and
hydroxymethylation in the hippocampus of Alzheimer's disease
patients. Neurobiol Aging. 34:2091–2099. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
324
|
Sjöström M, Zhao SG, Levy S, Zhang M, Ning
Y, Shrestha R, Lundberg A, Herberts C, Foye A, Aggarwal R, et al:
The 5-hydroxymethylcytosine landscape of prostate cancer. Cancer
Res. 82:3888–3902. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
325
|
Vatapalli R, Rossi AP, Chan HM and Zhang
J: Cancer epigenetic therapy: Recent advances, challenges, and
emerging opportunities. Epigenomics. 17:59–74. 2025. View Article : Google Scholar :
|
|
326
|
Gomez S, Tabernacki T, Kobyra J, Roberts P
and Chiappinelli KB: Combining epigenetic and immune therapy to
overcome cancer resistance. Semin Cancer Biol. 65:99–113. 2020.
View Article : Google Scholar :
|
|
327
|
Zhang P and Zhang M: Epigenetic
alterations and advancement of treatment in peripheral T-cell
lymphoma. Clin Epigenetics. 12:1692020. View Article : Google Scholar : PubMed/NCBI
|
|
328
|
Raj K and Mufti GJ: Azacytidine
(Vidaza(R)) in the treatment of myelodysplastic syndromes. Ther
Clin Risk Manag. 2:377–388. 2006. View Article : Google Scholar
|
|
329
|
Welch JS, Petti AA, Miller CA, Fronick CC,
O'Laughlin M, Fulton RS, Wilson RK, Baty JD, Duncavage EJ, Tandon
B, et al: TP53 and decitabine in acute myeloid leukemia and
myelodysplastic syndromes. N Engl J Med. 375:2023–2036. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
330
|
Duvic M and Vu J: Vorinostat in cutaneous
T-cell lymphoma. Drugs Today (Barc). 43:585–599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
331
|
McDermott J and Jimeno A: Belinostat for
the treatment of peripheral T-cell lymphomas. Drugs Today (Barc).
50:337–345. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
332
|
Smolewski P and Robak T: The discovery and
development of romidepsin for the treatment of T-cell lymphoma.
Expert Opin Drug Discov. 12:859–873. 2017.PubMed/NCBI
|
|
333
|
Kim YH, Bagot M, Pinter-Brown L, Rook AH,
Porcu P, Horwitz SM, Whittaker S, Tokura Y, Vermeer M, Zinzani PL,
et al: Mogamulizumab versus vorinostat in previously treated
cutaneous T-cell lymphoma (MAVORIC): An international, open-label,
randomised, controlled phase 3 trial. Lancet Oncol. 19:1192–1204.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
334
|
Geissler K, Koristek Z, Del Castillo TB,
Novák J, Rodríguez-Macías G, Metzelder SK, Illes A, Mayer J, Arnan
M, Keating MM, et al: Oral decitabine/cedazuridine versus
intravenous decitabine for acute myeloid leukaemia: A randomised,
crossover, registration, pharmacokinetics study. Br J Haematol.
205:1734–1745. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
335
|
Fu S, Hu W, Iyer R, Kavanagh JJ, Coleman
RL, Levenback CF, Sood AK, Wolf JK, Gershenson DM, Markman M, et
al: Phase 1b-2a study to reverse platinum resistance through use of
a hypomethylating agent, azacitidine, in patients with
platinum-resistant or platinum-refractory epithelial ovarian
cancer. Cancer. 117:1661–1669. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
336
|
Mathew OP, Ranganna K and Milton SG:
Involvement of the antioxidant effect and anti-inflammatory
response in butyrate-inhibited vascular smooth muscle cell
proliferation. Pharmaceuticals (Basel). 7:1008–1027. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
337
|
Ziemka-Nalecz M and Zalewska T:
Neuroprotective effects of histone deacetylase inhibitors in brain
ischemia. Acta Neurobiol Exp (Wars). 74:383–395. 2014. View Article : Google Scholar
|
|
338
|
Qiu X, Rong X, Yang J and Lu Y: Evaluation
of the antioxidant effects of different histone deacetylase
inhibitors (HDACis) on human lens epithelial cells (HLECs) after
UVB exposure. BMC Ophthalmol. 19:422019. View Article : Google Scholar : PubMed/NCBI
|
|
339
|
Hong DS, Kang YK, Borad M, Sachdev J,
Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, et al: Phase 1
study of MRX34, a liposomal miR-34a mimic, in patients with
advanced solid tumours. Br J Cancer. 122:1630–1637. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
340
|
Ottosen S, Parsley TB, Yang L, Zeh K, van
Doorn LJ, van der Veer E, Raney AK, Hodges MR and Patick AK: In
vitro antiviral activity and preclinical and clinical resistance
profile of miravirsen, a novel anti-hepatitis C virus therapeutic
targeting the human factor miR-122. Antimicrob Agents Chemother.
59:599–608. 2015. View Article : Google Scholar :
|
|
341
|
Vermeire S, Sands BE, Tilg H, Tulassay Z,
Kempinski R, Danese S, Bunganič I, Nitcheu J, Santo J, Scherrer D,
et al: ABX464 (obefazimod) for moderate-to-severe, active
ulcerative colitis: a phase 2b, double-blind, randomised,
placebo-controlled induction trial and 48 week, open-label
extension. Lancet Gastroenterol Hepatol. 7:1024–1035. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
342
|
Winkle M, El-Daly SM, Fabbri M and Calin
GA: Noncoding RNA therapeutics-challenges and potential solutions.
Nat Rev Drug Discov. 20:629–651. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
343
|
Zhang X, Qiu H, Zhang F and Ding S:
Advances in single-cell multi-omics and application in
cardiovascular research. Front Cell Dev Biol. 10:8838612022.
View Article : Google Scholar : PubMed/NCBI
|
|
344
|
Armand EJ, Li J, Xie F, Luo C and Mukamel
EA: Single-cell sequencing of brain cell transcriptomes and
epigenomes. Neuron. 109:11–26. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
345
|
Nam AS, Chaligne R and Landau DA:
Integrating genetic and non-genetic determinants of cancer
evolution by single-cell multi-omics. Nat Rev Genet. 22:3–18. 2021.
View Article : Google Scholar :
|
|
346
|
Liu X, Zhang L, Li X, Chen L, Lu L, Yang
Y, Wu Y, Zheng L, Tang J, Wang F, et al: Single-cell multi-omics
profiling uncovers the immune heterogeneity in HIV-infected
immunological non-responders. EBioMedicine. 115:1056672025.
View Article : Google Scholar : PubMed/NCBI
|
|
347
|
Tangeman JA, Rebull SM, Grajales-Esquivel
E, Bendezu-Sayas S, Robinson ML, Lachke SA and Del Rio-Tsonis K:
Integrated single-cell multiomics uncovers foundational regulatory
mechanisms of lens development and pathology. Development.
151:dev2022492024. View Article : Google Scholar : PubMed/NCBI
|
|
348
|
Vickovic S, Eraslan G, Salmén F,
Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R,
Bergenstråhle L, Navarro JF, et al: High-definition spatial
transcriptomics for in situ tissue profiling. Nat Methods.
16:987–990. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
349
|
Robles-Remacho A, Sanchez-Martin RM and
Diaz-Mochon JJ: Spatial transcriptomics: emerging technologies in
tissue gene expression profiling. Anal Chem. 95:15450–15460. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
350
|
Cilento MA, Sweeney CJ and Butler LM:
Spatial transcriptomics in cancer research and potential clinical
impact: A narrative review. J Cancer Res Clin Oncol. 150:2962024.
View Article : Google Scholar : PubMed/NCBI
|
|
351
|
Duan R, Fu Q, Sun Y and Li Q: Epigenetic
clock: A promising biomarker and practical tool in aging. Ageing
Res Rev. 81:1017432022. View Article : Google Scholar : PubMed/NCBI
|
|
352
|
Horvath S, Lu AT, Haghani A, Zoller JA, Li
CZ, Lim AR, Brooke RT, Raj K, Serres-Armero A, Dreger DL, et al:
DNA methylation clocks for dogs and humans. Proc Natl Acad Sci USA.
119:e21208871192022. View Article : Google Scholar : PubMed/NCBI
|
|
353
|
Margiotti K, Monaco F, Fabiani M, Mesoraca
A and Giorlandino C: Epigenetic clocks: In aging-related and
complex diseases. Cytogenet Genome Res. 163:247–256. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
354
|
Chen J, Yuan XL, Zhou X, Xu J, Zhang X and
Duan X: Mendelian randomization implicates causal association
between epigenetic age acceleration and age-related eye diseases or
glaucoma endophenotypes. Clin Epigenetics. 16:1062024. View Article : Google Scholar : PubMed/NCBI
|