Introduction
Cancer cells cleverly evade T cell-mediated immune
responses by manipulating immune cells and leveraging immune
checkpoints to communicate with the host's defense mechanisms
within the tumor microenvironment (TME) (1). This process disrupts immune
surveillance, allowing tumors to escape detection and avoid
destruction. A critical component in this mechanism is programmed
death-ligand 1 (PD-L1; also known as B7-H1 or CD274), a membrane
glycoprotein frequently overexpressed across various cancer types.
PD-L1 acts as a critical immune checkpoint molecule through its
interaction with programmed cell death protein-1 (PD-1; also called
CD279), which is expressed on the surface of activated T
lymphocytes (2). This
interaction inhibits the activation of cytotoxic T cells, impairing
the immune system's ability to identify and eliminate cancer cells.
By hijacking this pathway, cancer cells can effectively evade
immune eradication, thereby promoting their survival and enabling
further progression (3).
Immunotherapies targeting PD-1 or PD-L1 with
antibodies have shown significant and durable clinical responses in
treating various cancers, including non-small cell lung cancer
(NSCLC), melanoma and renal cell carcinoma (4,5).
These therapies function by inhibiting the binding of PD-1 on
immune cells to PD-L1 on tumor cells, thereby enhancing the immune
system's ability to recognize and eradicate cancer cells. The
success of immune checkpoint blockade highlights its potential to
transform cancer treatment paradigms, providing new options for
patients who previously had limited alternatives (6). However, despite their notable
efficacy, these therapies do not benefit all patients and face
limitations such as poor tumor tissue penetration, adverse immune
reactions and lack of oral bioavailability (7). Additionally, targeting PD-L1 for
degradation, rather than merely blocking its interaction with PD-1,
might offer a more effective strategy by thoroughly disrupting the
PD-1/PD-L1 signaling axis, potentially enhancing therapeutic
efficacy (8,9).
PD-L1 expression is regulated through multiple
mechanisms, including the activation of oncogenic transcription
factors such as c-Myc and NF-κB, which promote PD-L1 upregulation
at the gene transcription level. Additionally, post-translational
modifications, such as glycosylation and ubiquitination, are
essential for stabilizing PD-L1 and regulating its functional
protein levels (10). For
example, activating β-catenin during epithelial-mesenchymal
transition (EMT) induces the transcription and synthesis of the
STT3 gene (a catalytic subunit of the oligosaccharyltransferase
(OST) complex). This, in turn, promotes PD-L1 glycosylation,
thereby inhibiting its degradation in cancer stem cells (11). Furthermore, cyclin D-CDK4 and
glycogen synthase kinase 3 beta (GSK3β) facilitate PD-L1
ubiquitination, marking it for proteasomal degradation (12,13). Notably, the oncogenic epidermal
growth factor receptor (EGFR) plays a key role in regulating PD-L1
expression. Inhibition of EGFR signaling destabilizes PD-L1 through
GSK3β activation, ultimately enhancing antitumor immunity (14). These findings suggested that
agents targeting EGFR may represent a promising approach to enhance
immune responses by promoting PD-L1 degradation.
GMI, a fungal immunomodulatory protein (FIP) from
Ganoderma microsporum, is a dietary supplement with both
nutritional and therapeutic benefits (15). Beyond its origin as an edible
mushroom, GMI has drawn significant attention for its diverse
biological properties, especially its potent anticancer activity.
GMI downregulates heat shock protein expression, which is
associated with apoptosis in lung cancer cells (16). In addition, it has shown
synergistic anticancer effects when combined with therapeutic
agents such as temozolomide and sotorasib in glioblastoma
multiforme (GBM) and KRASG12C mutant lung
cancers, respectively (17,18). GMI has been identified as an EGFR
degrader capable of disrupting EGFR-driven oncogenic signaling and
suppressing tumor growth in EGFR-positive cancers (19). However, whether GMI regulates
PD-L1 expression and enhances antitumor immune responses remains
unclear.
In the present study, the effects of GMI on PD-L1
expression were investigated and it was found that it promotes
GSK3β-mediated proteasomal degradation of PD-L1, leading to reduced
membrane PD-L1 levels. Furthermore, GMI enhances T cell-mediated
suppression of tumor cells. These findings collectively suggested
that GMI may serve as a functional food with potential therapeutic
benefits for enhancing antitumor immunity through modulating PD-L1
expression.
Materials and methods
Reagents
GMI was produced by MycoMagic Biotechnology. The
purification procedures and the detailed structural
characterization (Protein Data Bank ID: 3KCW) have been previously
documented (20). Bafilomycin A1
(Baf A1) and cycloheximide (CHX) were purchased from
MilliporeSigma. MG132 and lithium chloride (LiCl) were obtained
from ITW Reagents (PanReac AppliChem) and Enzo Life Sciences,
respectively. LY294002 and Osimertinib were purchased from
MedChemExpress and TargetMol, respectively. Anti-mouse PD-1
antibody (CD279; clone RMP1-14) was purchased from Bio X Cell.
Experimental cell lines and culture
conditions
Lewis lung carcinoma (LLC1), GBM8401 and Jurkat T
cells were obtained from the Bioresource Collection and Research
Center. A549 cell line was purchased from the American Type Culture
Collection. H1975 and T98G cells were obtained from Elabscience
Biotechnology, Inc. and the Japanese Cancer Research Resources
Bank, respectively. PC9 and CL1-5 cells were generously provided by
Dr Tsai of the National Yang Ming University and Dr P.C. Yang of
the National Taiwan University, respectively. U87, a human
glioblastoma cell line of unknown origin (21), was kindly provided by Dr T.H. Tu
of Taipei Veterans General Hospital. The LLC1-hPD-L1 cell line was
a gift from Dr K.L. Lan of the National Yang Ming Chiao Tung
University.
LLC1-hPD-L1 cells were cultured in Dulbecco's
Modified Eagle's Medium supplemented with 10% fetal bovine serum
(VWR Life Science Seradigm), 1% penicillin/streptomycin, 1% sodium
pyruvate, 1% L-glutamine (all from GIBCO; Thermo Fisher Scientific,
Inc.) and 200 μg/ml hygromycin B (MedChemExpress). Short
tandem repeat (STR) analysis was used for authentication in H1975,
A549 and GBM8401 cell lines. Detailed culture conditions for the
other cell lines have been previously described (18,19). All cell cultures were incubated
at 37°C in a humidified environment containing 5%
CO2.
Kyoto Encyclopedia of Genes and Genomes
(KEGG) analysis
The extraction of membrane proteins used for
LC-MS/MS analysis has been previously described (22). Briefly, membrane proteins were
extracted from GMI-treated cells and subjected to proteomic
analysis. Differentially expressed proteins (DEPs) regulated by GMI
were identified based on a fold change (GMI/CTL) of <0.75 or
>1.33. These DEPs were then analyzed for pathway enrichment
using ShinyGO V0.80 (23), and
the selected enriched pathway was visualized using KEGG diagrams,
with DEPs highlighted in red (24,25).
Western blotting and immunoprecipitation
(IP) assay
Western blot analysis was performed as previously
described (26). In brief,
treated cells were harvested and lysed in a cell lysis buffer
containing protease and phosphatase inhibitors (MilliporeSigma).
The lysates were cleared by centrifugation at 16,200 × g for 10 min
at 4°C, and protein concentrations in the supernatants were
quantified prior to analysis using the Bradford protein assay
(Bio-Rad Laboratories, Inc.), with bovine serum albumin (BSA;
Thermo Fisher Scientific, Inc.) as the calibration standard.
Subsequently, 30 μg of total protein from each sample was
separated by 10% SDS-polyacrylamide gel electrophoresis and
transferred onto PVDF membranes (0.45 μm; MilliporeSigma).
The membranes were incubated with primary antibodies at 4°C
overnight, followed by incubation with species-specific secondary
antibodies at room temperature for 1 h prior to chemiluminescence
detection. For IP assay, cells were pretreated with a proteasome
inhibitor, followed by GMI treatment for the indicated time. Cell
lysates were prepared and centrifuged in IP lysis buffer containing
EDTA at 16,200 × g for 10 min at 4°C. Then, 400 μg of
supernatant was incubated overnight at 4°C with a PD-L1 antibody
(1:50) for immunoprecipitation. Ubiquitinated PD-L1 was
subsequently detected by western blotting. Antibodies for detecting
PD-L1 (rabbit; 1:1,000; cat. no. 13684), phosphorylated (p-)GSK3β
(Ser 9; rabbit; 1:1,000; cat. no. 5558), p-AKT (Ser473; rabbit;
1:1,000; cat. no. 4060), p-EGFR (Tyr1068; rabbit; 1:1,000; cat. no.
3777) and ubiquitin (mouse; 1:1,000; cat. no. 3936) were acquired
through Cell Signaling Technology, Inc. Antibodies specific to
GSK3β (rabbit; 1:1,000; cat. no. GTX111192), caveolin (rabbit;
1:1,000; cat. no. GTX100205), EGFR (rabbit; 1:1,000; cat. no.
GTX121919), tubulin (rabbit; 1:10,000; cat. no. GTX112141), goat
anti-mouse IgG antibody (HRP; 1:10,000; cat. no. GTX213111-01) and
goat anti-rabbit IgG antibody (HRP; 1:10,000; cat. no.
GTX213110-01) were obtained via GeneTex International Corporation.
AKT (rabbit; 1:1,000; cat. IR171-666) antibody was purchased from
iReal Biotechnology. Protein band intensities were quantified by
densitometric analysis using ImageJ software (version 1.53a;
National Institutes of Health), with values normalized to the
respective loading controls.
Extraction of plasma membrane PD-L1
The experimental procedures have been previously
documented (19). Briefly,
plasma membrane proteins were isolated using the Trident Membrane
Protein Extraction Kit (GeneTex International Corporation),
following the manufacturer's instructions. The extracted membrane
proteins were subsequently evaluated using western blot
analysis.
Immunofluorescence assay
H1975 and CL1-5 cells (2×104) were seeded
on coverslips and treated with GMI and/or LiCl for 3 h. Cells were
then subjected to fixation at room temperature for 15 min with 4%
paraformaldehyde (Bioman Scientific Company, Ltd.) and
permeabilized in PBS containing 0.1% Triton X-100 (MilliporeSigma)
for another 10 min. Blocking was performed with PBS containing 1%
BSA for 30 min. Cells were then incubated overnight at 4°C with
anti-PD-L1 primary antibodies (1:400; Abcam; cat. no. ab205921).
After washing with PBS supplemented with 0.05% Tween 20, the cells
were incubated at room temperature for 1 h with goat anti-rabbit
IgG antibody (DyLight488; 1:1,000; GeneTex International
Corporation; cat. no. GTX213110-04). Nuclei were visualized by
mounting cells with Fluoroshield containing DAPI (Millipore; cat.
no. F6057). Observations and imaging were performed using an
ImageXpress Pico digital microscope (Molecular Devices, LLC).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using the Quick-RNA Miniprep
Kit (Zymo Research Corp.), following the manufacturer's
instructions. Complementary DNA (cDNA) synthesis was performed from
500 ng of RNA using the PrimeScript™ RT reagent Kit (Takara Bio,
Inc.) in accordance with the manufacturer's protocol. Amplification
of the cDNA was performed with TB Green® Premix Ex Taq
II (Takara Bio, Inc.) on a StepOnePlus™ Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were set as follows: initial denaturation
at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C
for 5 sec and annealing/extension at 60°C for 30 sec. The specific
primers for the genes of interest were as follows: PD-L1 forward,
5'-CAATGTGACCAGCACACTGAGAA-3' and reverse,
5'-GGCATAATAAGATGGCTCCCAGAA-3'; and GAPDH forward,
5'-TGGTATCGTGGAAGGACTCA-3' and reverse, 5'-AGTGGGTGTCGCTGTTGAAG-3'.
Relative PD-L1 mRNA levels were quantified by the comparative
2−ΔΔCq method (27),
using GAPDH as the endogenous control and normalized to the control
group.
Short hairpin RNA (shRNA)
transfection
Lentiviral shRNA transfection was conducted as
previously described (18).
Briefly, one CTL-shRNA (shCTL; TRCN0000208001; pLKO.1 empty-vector
negative control lacking an shRNA hairpin insert) and two
GSK3β-shRNA lentivirus constructs were obtained from the National
RNAi Core Facility (Taipei, Taiwan) and generated in 293T cells.
These lentiviruses were subsequently employed to transduce H1975
and CL1-5 cells, followed by selection with 5 μg/ml
puromycin (MilliporeSigma). The target sequences for GSK3β were
5'-AGCAAATCAGAGAAATGAAC-3' (shGSK3β#1; TRCN0000039565) and
5'-CCCAAACTACACAGAATTTAA-3' (shGSK3β#2; TRCN0000039999).
T cell-mediated tumor cell killing
assay
T cell cytotoxicity assay was conducted as
previously described (28,29). Primary T cells were isolated from
the spleens of LLC1-bearing mice using a magnetic cell separation
system with the mouse CD3ε MicroBead Kit (Miltenyi Biotec GmbH;
cat. no. 130-094-973), following the manufacturer's protocols.
Jurkat and isolated T cells were activated for 48 h by incubation
with Dynabeads™ Human T-Activator CD3/CD28 (Invitrogen; Thermo
Fisher Scientific, Inc.; cat. no. 11161D) and Dynabeads™ Mouse
T-Activator CD3/CD28 (Invitrogen; Thermo Fisher Scientific, Inc.;
cat. no. 11452D), respectively. Next, GMI-treated green fluorescent
protein (GFP)-expressing tumor cells were co-cultured with the
activated T cells for 72 h at the following effector-target ratios:
1:2 for LLC1-GFP cells, 1:1 for H1975-GFP cells, and 1:10 for
PC9-GFP cells. After removing the supernatant, the cells were
washed, trypsinized, centrifuged at 120 × g for 5 min at room
temperature, and resuspended in PBS. GFP fluorescence was measured
using a Varioskan LUX multimode microplate reader (Thermo Fisher
Scientific, Inc.), with excitation set at 485 nm and emission at
520 nm.
Measurement of interleukin-2 (IL-2) and
granzyme B secreted by activated Jurkat T cells using a co-culture
assay and enzyme-linked immunosorbent assay (ELISA)
Jurkat T cells were activated for 48 h using
Dynabeads™ Human T-Activator CD3/CD28 (Invitrogen; Thermo Fisher
Scientific, Inc.; cat. no. 11161D). H1975-GFP and PC9-GFP lung
cancer cells were pretreated with GMI for 24 h, detached with
trypsin, and then co-cultured with activated T cells at a 1:1 ratio
for both H1975-GFP and PC9-GFP cells. Supernatants were collected,
and levels of secreted IL-2 and granzyme B were measured using
Human IL-2 and Granzyme B ELISA Kit (BioLegend, Inc.; cat. no.
431804; and R&D Systems, Inc.; cat. no. DY2906-05,
respectively). The assay was conducted according to the
manufacturer's protocols, as previously described (20). In addition, to directly examine
the effect of GMI on T cell function, Jurkat T cells were treated
with GMI for 24 h. Culture supernatants were collected, and IL-2
secretion was measured by ELISA.
Syngeneic tumor model
Animal studies were approved by the Institutional
Animal Care and Use Committee (IACUC) at National Yang Ming Chiao
Tung University (IACUC approval no. 1070805) and were conducted in
accordance with the established guidelines, following procedures as
described previously (17). A
total of 42 male C57BL/6 (National Laboratory Animal Center,
Taipei, Taiwan) mice were used in the present study. The animals
were 6-8 weeks of age and weighed 20-22 g at the start of the
experiments. They were housed under controlled conditions
(temperature 20-22°C; relative humidity 50±10%) with a 12/12-h
light/dark cycle and had free access to food and water. In
addition, group sizes were chosen in line with published LLC1
syngeneic practice and institutional guidance, where similar cohort
sizes have been sufficient to detect significant differences
(30-33). Briefly, mice were subcutaneously
injected with 1×105 LLC1 or LLC1-hPD-L1 cells, and then
treated intraperitoneally with GMI (5 mg/kg) or vehicle control
(PBS) every 3 days. In the tumor model involving GMI with or
without anti-PD-1 antibody treatment, mice were randomly divided
into four distinct treatment groups: control (CTL), anti-PD-1 mAb
(200 μg/mice), GMI (5 mg/kg), and GMI + anti-PD-1 mAb.
Treatments were administered every 2 days. Tumor volume was
calculated using the formula: length × width2 × 1/2. At
the study endpoint, the mice were humanely euthanized by carbon
dioxide (CO2) inhalation using a gradual fill rate of
30-70% of the chamber volume per min without pre-filling, and
tumors were excised and weighed.
Tumor-infiltrating lymphocytes (TILs) were isolated
from GMI- and PBS-treated LLC1 tumors. Non-specific binding was
blocked using CD16/CD32 antibodies (Thermo Fisher Scientific, Inc.;
cat. no. 14-0161-82), followed by staining on ice for 30 min with
anti-mouse CD3 (BioLegend, Inc.; cat. no. 100235), CD8 (BioLegend,
Inc.; cat. no. 100707) and CD4 (BioLegend, Inc.; cat. no. 100405)
antibodies, targeting immune markers. Samples were analyzed using a
CytoFLEX Flow Cytometer (Beckman Coulter, Inc.) and the
CD8+/CD4+ T cell ratio was determined. In a
separate assay, Jurkat T cells were treated with GMI for 24 h,
stained with PE anti-human CD69 antibody (BioLegend, Inc.; cat. no.
310905), and subsequently analyzed for surface CD69 expression
using flow cytometry.
Statistical analysis
All data were represented as the mean ± standard
deviation (SD) from three independent experiments. Comparisons
between two groups were performed using two-tailed unpaired
Student's t-tests. For comparisons among more than two groups,
one-way analysis of variance (ANOVA) was used, followed by
appropriate post hoc tests. Specifically, Dunnett's test was
applied when all groups were compared with the control group, while
Tukey's test was used for comparisons among all groups. Analyses
were performed using GraphPad Prism 10 software (Dotmatics).
P<0.05 was considered to indicate a statistically significant
difference.
Results
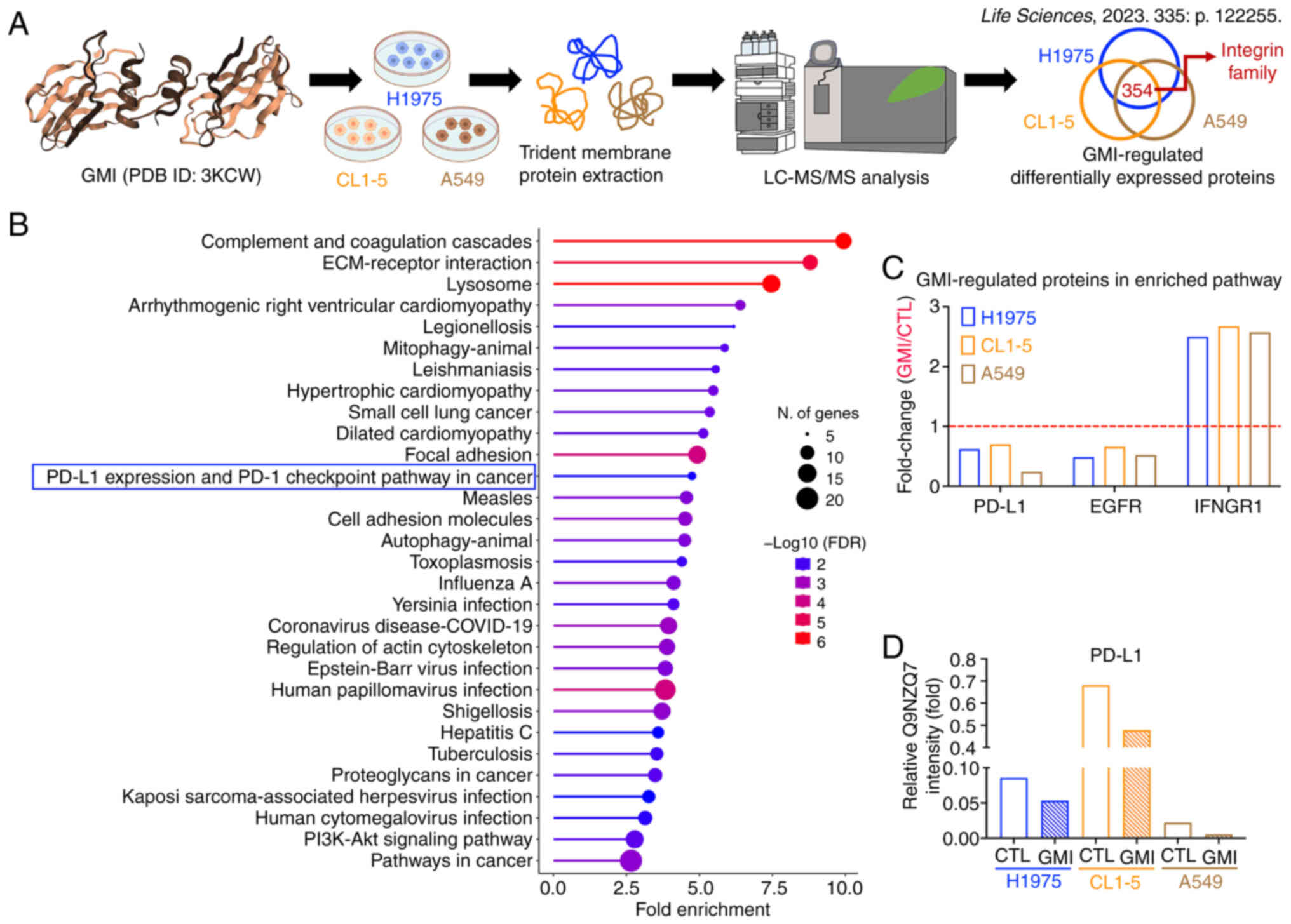
GMI reduces relative membrane PD-L1
intensity in three lung cancer cell lines
A previous proteomic analysis of membrane protein
extractions by the authors revealed that GMI downregulated members
of the integrin family in H1975, CL1-5 and A549 lung cancer cells
(Fig. 1A) (22). To further characterize the
functions of GMI-regulated DEPs, the KEGG pathway enrichment
analysis was performed. Among the enriched pathways, the 'PD-L1
expression and PD-1 checkpoint pathway in cancer' was identified
(Fig. 1B and Table SI). Within this pathway, PD-L1
and EGFR were downregulated by GMI, while IFNGR1 was upregulated
(Figs. 1C and S1, and Table SII). Given that GMI is a
known EGFR degrader and that EGFR is closely associated with the
regulation of PD-L1 protein levels, the mechanism underlying
GMI-induced reduction of membrane PD-L1 intensity was investigated
in these three lung cancer cell lines (Fig. 1D).
GMI decreases PD-L1 protein levels in
H1975, CL1-5 and A549 cells
A previous study by the authors showed that 0.6
μM GMI markedly modulated EGFR (19); therefore, this dose was selected
for subsequent assays. Membrane protein fractionation assays were
performed to evaluate the effects of GMI on plasma membrane PD-L1
levels. Consistent with our proteomic findings, a significant
decrease in membrane PD-L1 following GMI treatment was observed
(Fig. 2A). Immunofluorescence
analysis further confirmed that membrane PD-L1 accumulation was
significantly reduced in GMI-treated H1975 and CL1-5 cells
(Fig. 2B). Furthermore, PD-L1
protein expression was significantly reduced in all examined cell
lines after both short-term and long-term exposure to GMI (Fig. 2C and D). Basal PD-L1 levels were
assessed across a panel of lung cancer cell lines, revealing
relatively lower PD-L1 expression in A549 cells and higher
expression in H1975 cells (Fig.
S2A), echoing previous studies (34,35). Given that CL1-5 cells exhibited
intermediate PD-L1 levels, H1975 and CL1-5 cell lines were selected
for further investigation. In GBM cell lines, PD-L1 was highly
expressed in GBM8401 cells, minimally expressed in T98G cells, and
intermediate in U87 cells (Fig.
S2A). Notably, GMI reduced PD-L1 levels in both GBM8401 and U87
cells in a dose-dependent manner (Fig. S2B and C). Collectively, these
findings suggested that GMI may play a role in modulating PD-L1
expression by effectively reducing membrane PD-L1 levels.
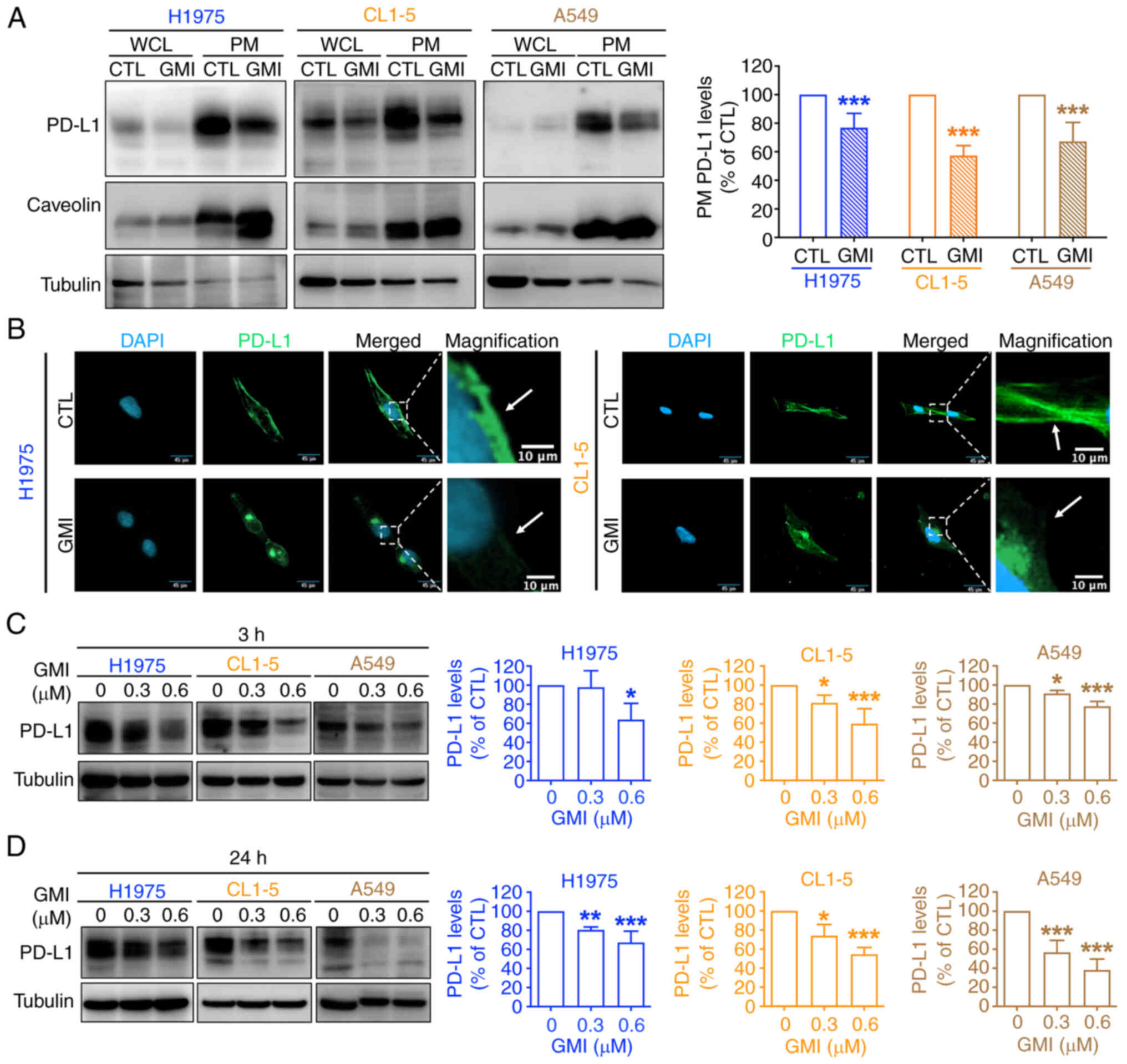 | Figure 2GMI reduces membrane PD-L1 protein
levels in lung cancer cells. (A) Left: Immunoblot analysis of PD-L1
in WCL and PM fractions isolated from H1975, CL1-5 and A549 cells
after 24 h treatment with 0.6 μM GMI. Caveolin and tubulin
were used as markers for PM and WCL fractions, respectively. Right:
quantification of PD-L1 levels in PM fractions normalized to
caveolin. (B) Immunofluorescence staining of PD-L1 (green) and
nuclei (DAPI, blue) in H1975 and CL1-5 cells treated with 0.6
μM GMI for 3 h. Scale bars, 45 μm (main images) and
10 μm (magnified insets). (C and D) Western blot analysis of
PD-L1 protein levels in H1975, CL1-5 and A549 cells treated with
indicated concentrations of GMI for 3 h (C) and 24 h (D). Tubulin
served as the loading control. Data are presented as the mean ±
standard deviation from three independent experiments.
*P<0.05, **P<0.01 and
***P<0.001. GMI, Ganoderma microsporum
immunomodulatory protein; PD-L1, programmed death-ligand 1; WCL,
whole-cell lysates; PM, plasma membrane. |
GMI suppresses PD-L1 mRNA levels and
promotes proteasomal PD-L1 degradation
It was next examined whether GMI influences
PD-L1 transcription. RT-qPCR assay revealed an initial
increase in PD-L1 mRNA levels at 1 h, followed by a
significant decrease at 24 h post-treatment (Fig. 3A). To assess the impact of GMI on
PD-L1 protein stability, CHX, the protein synthesis inhibitor, was
used. The findings of the present study revealed that PD-L1 protein
degradation was accelerated in cells exposed to GMI (Fig. 3B), indicating that GMI promotes
the post-translational regulation of PD-L1, leading to enhanced
protein degradation.
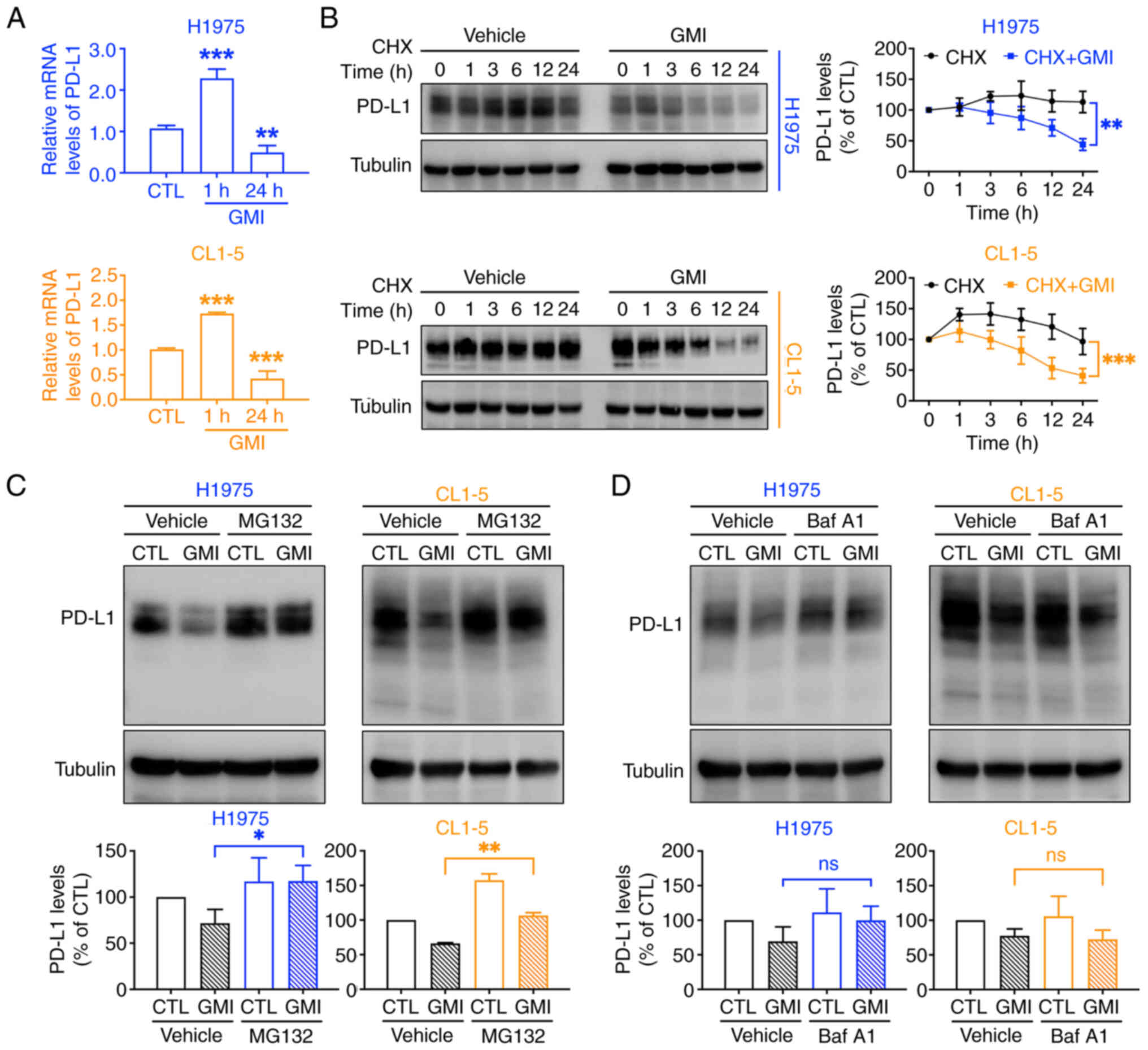 | Figure 3GMI induces proteasome-dependent
degradation of PD-L1. (A) Reverse transcription-quantitative PCR
analysis of PD-L1 mRNA expression in H1975 and CL1-5 cells
following treatment with 0.6 μM GMI for 1 and 24 h. (B) CHX
chase assay of H1975 and CL1-5 cells exposed to 200 μg/ml
CHX for 30 min, followed by incubation with or without 0.6
μM GMI for the indicated time points to assess PD-L1 protein
stability. (C and D) H1975 and CL1-5 cells were pretreated with 10
μM MG132 (C), a proteasome inhibitor, or 5 nM Baf A1 (D), a
lysosome inhibitor, and were then exposed to 0.6 μM GMI for
24 h. Data are presented as the mean ± standard deviation from
three independent experiments. *P<0.05,
**P<0.01 and ***P<0.001. GMI,
Ganoderma microsporum immunomodulatory protein; PD-L1,
programmed death-ligand 1; CHX, cycloheximide; Baf A1, bafilomycin
A1; ns, not significant. |
PD-L1 can be degraded through either the proteasomal
or lysosomal pathway, both leading to reduced PD-L1 presence on the
cell membrane (36). To identify
the specific signaling pathway implicated in GMI-mediated PD-L1
reduction, cells were exposed to GMI combined with specific
inhibitors: MG132 for proteasomes and Baf A1 for lysosomes. The
findings revealed that GMI-triggered acceleration of PD-L1
breakdown was strongly mitigated in the presence of MG132 (Fig. 3C), implicating the proteasomal
pathway in this process. By contrast, Baf A1 did not inhibit the
degradation (Fig. 3D),
indicating that the lysosomal pathway is not significantly
involved. Furthermore, under endogenous (non-overexpression)
conditions, an in-cell ubiquitination assay showed a slight
increase in PD-L1 ubiquitination following GMI treatment, although
the effect was not pronounced (Fig.
S3A and B). These findings suggested that GMI reduces PD-L1
protein levels not only by suppressing its transcription but also
by enhancing the proteasome-dependent system and subsequent
degradation.
GMI destabilizes PD-L1 by promoting
GSK3β-mediated proteasomal degradation
Activated EGFR induces AKT-mediated phosphorylation
of GSK3β at serine 9 (Ser9), rendering it inactive and thereby
stabilizing PD-L1 by preventing its proteasomal degradation
(37). It was hypothesized that
GSK3β signaling contributes to GMI-mediated PD-L1 degradation. The
present results demonstrated that GMI, as an EGFR degrader,
effectively suppressed EGFR expression and downstream AKT
activation (Fig. 4A and B;
Fig. S4A and B). Moreover,
treatment with GMI (0.6 μM) significantly reduced GSK3β Ser9
phosphorylation after both short-and long-term treatments,
indicating GSK3β activation concomitant with PD-L1 suppression
(Fig. 4A and B). Similar
reductions in PD-L1 expression were observed upon treatment with
osimertinib, an EGFR inhibitor, and LY294002, a PI3K/AKT inhibitor
(Fig. S4C). Conversely,
co-treatment with LiCl, a GSK3β inhibitor, significantly rescued
PD-L1 levels reduced by GMI (Fig.
4C), as confirmed by immunofluorescence analysis (Fig. S5). To further validate GSK3β's
role, shRNA-mediated knockdown of GSK3β in both H1975 and CL1-5
cells attenuated GMI-induced PD-L1 degradation (Fig. 4D), resulting in elevated PD-L1
compared with GMI-treated parental cells (Fig. 4E and F). Collectively, these
findings demonstrated that GSK3β activation is crucial for
GMI-induced proteasomal degradation of PD-L1.
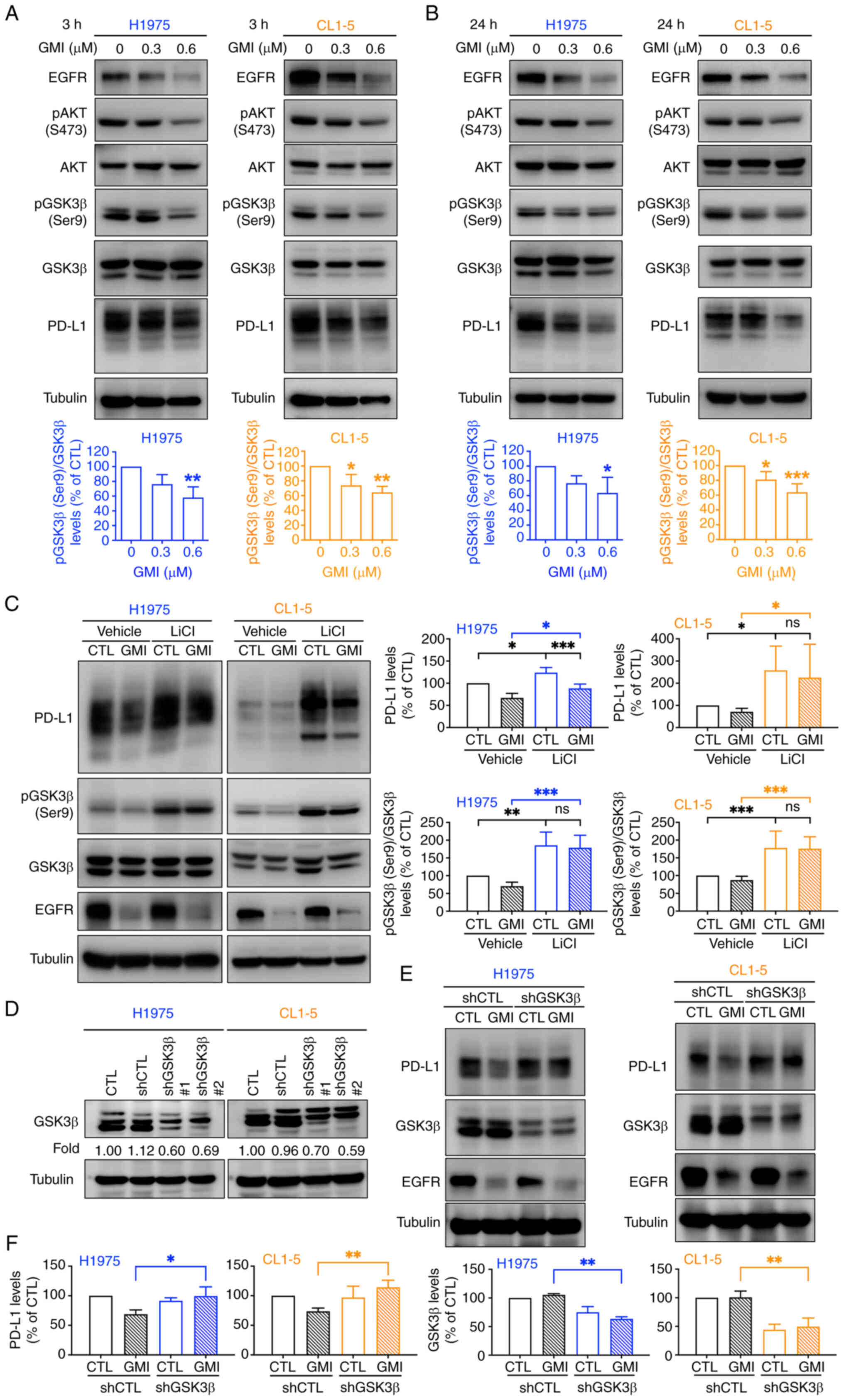 | Figure 4GMI facilitates PD-L1 degradation
through activation of GSK3β. (A and B) H1975 and CL1-5 cells were
treated with indicated concentrations of GMI for 3 h (A) and 24 h
(B). Protein expressions of EGFR, p-AKT (Ser473), p-GSK3β (Ser9)
and PD-L1 were analyzed by immunoblotting, with AKT, GSK3β and
tubulin as loading controls, and quantification of p-AKT and
p-GSK3β was performed relative to their corresponding total
proteins (AKT and GSK3β). (C) H1975 and CL1-5 cells were pretreated
with 25 mM LiCl, a GSK3β inhibitor, and were exposed to 0.6
μM GMI for 24 h. (D) Immunoblotting of GSK3β in H1975 and
CL1-5 cells expressing different GSK3β-targeting shRNAs. (E)
Immunoblotting analysis of PD-L1 and GSK3β in cancer cells
transduced with either GSK3β shRNA (shGSK3β#1) or control shRNA
(shCTL) lentiviruses and treated with or without 0.6 μM GMI
for 24 h. (F) Quantification of PD-L1 and GSK3β expression levels
shown in (E). Data are presented as the mean ± standard deviation
from three independent experiments. *P<0.05,
**P<0.01 and ***P<0.001. GMI,
Ganoderma microsporum immunomodulatory protein; PD-L1,
programmed death-ligand 1; p-, phosphorylated; LiCl, lithium
chloride; shRNA, short hairpin RNA; p-, phosphorylated; ns, not
significant. |
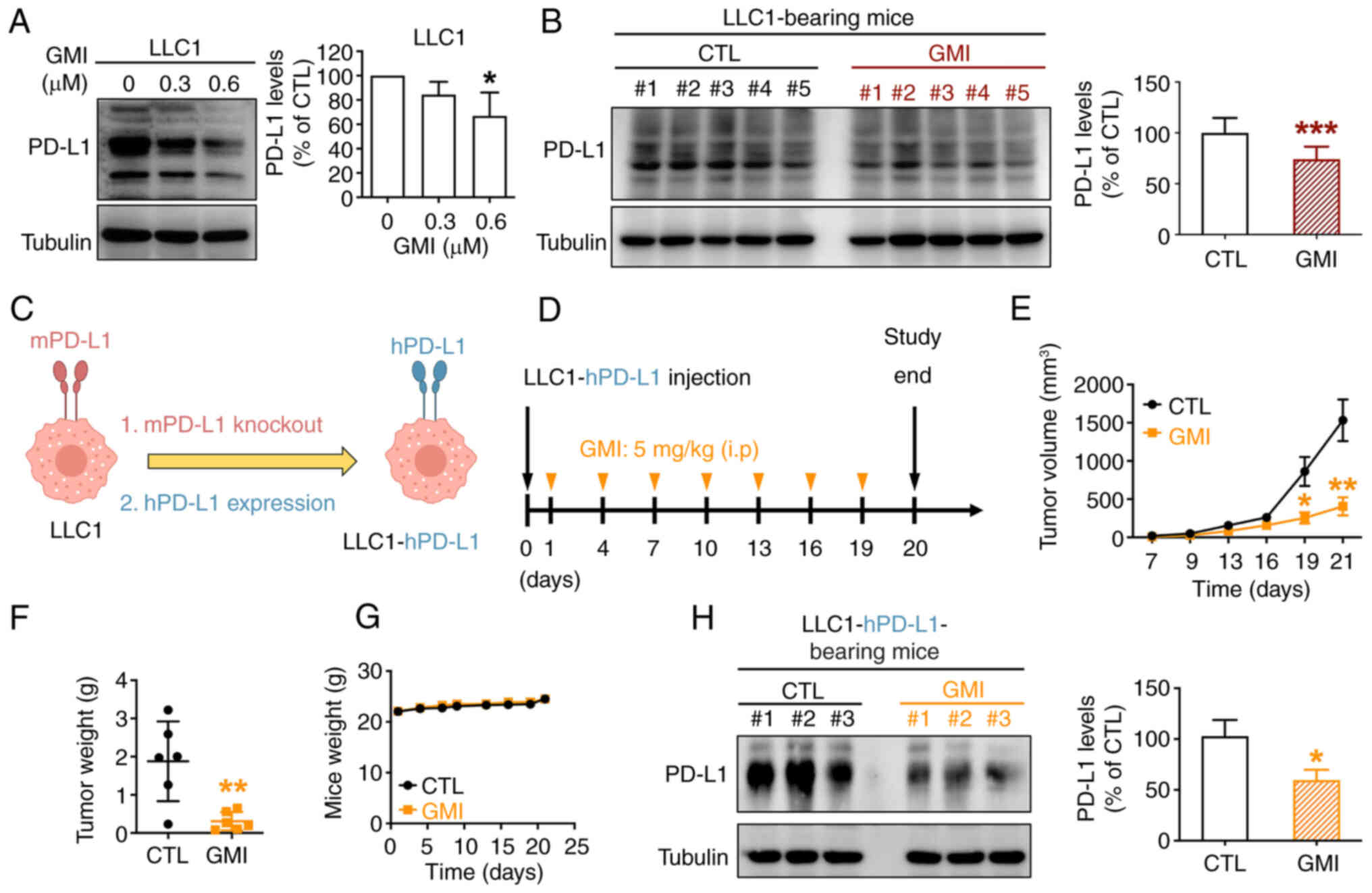
GMI suppresses tumor growth and reduces
PD-L1 expression in vivo
A previous study by the authors showed that GMI
effectively suppressed tumor growth in a LLC1-bearing mouse model
(19). In the present study, it
was further explored whether GMI can downregulate PD-L1 expression
in LLC1 cells both in vitro and in vivo. First, it
was observed that GMI treatment reduced PD-L1 levels in LLC1 cells
in vitro (Fig. 5A).
Subsequently, LLC1-bearing immune-competent C57BL/6 mice were
administered GMI or PBS (CTL) via intraperitoneal injection every 3
days (Fig. S6A). The results
demonstrated that GMI significantly suppressed tumor growth
(Fig. S6B), echoing our
previous findings, without affecting overall body weight, thus
revealing favorable tolerability of GMI (Fig. S6C). Western blot analysis of
harvested tumors revealed that GMI treatment significantly
decreased PD-L1 expression compared with the CTL group, indicating
that GMI induces PD-L1 degradation in tumor-bearing mice (Fig. 5B).
To further investigate whether GMI-induced
degradation of human PD-L1 (hPD-L1) in vivo, LLC1-hPD-L1
cells were utilized, where murine PD-L1 (mPD-L1) was replaced with
hPD-L1 (Figs. 5C and S2A). GMI suppressed PD-L1 levels in
LLC1-hPD-L1 cells in vitro (Fig. S7A) and promoted PD-L1
degradation through the ubiquitin-mediated proteasomal pathway,
consistent with observations in H1975 and CL1-5 cells (Fig. S7B and C). LLC1-hPD-L1 cells were
subcutaneously inoculated into C57BL/6 mice to evaluate the in
vivo efficacy of GMI (Fig.
5D). Treatment with GMI significantly inhibited tumor growth
and reduced tumor weight without causing body weight loss (Figs. 5E-G and S7D). Tumor tissue analysis confirmed
GMI-mediated downregulation of PD-L1 expression (Fig. 5H). These findings indicated that
GMI suppresses PD-L1 expression and may enhance antitumor immunity
by facilitating PD-L1 degradation in tumor-bearing mice.
T cell suppresses the viability of
GMI-pretreated cancer cells
The binding of PD-L1 on cancer cell membranes to
PD-1 on T cells suppresses T cell activation, facilitating immune
evasion and tumor progression (38). Given the previous finding that
GMI promotes PD-L1 degradation on cancer cell surfaces, it was
hypothesized that this would enhance secretion of T cell cytokines,
such as IL-2 and granzyme B, thereby boosting the cytotoxic
response against tumor cells (29). To test this, co-culture
experiments were performed using T cells isolated from the spleens
of LLC1 tumor-bearing mice (Fig.
6A). Activated spleen-derived T cells significantly suppressed
the viability of GMI-treated LLC1 cells compared with untreated
controls (Fig. 6B). Next, lung
cancer cell lines H1975 and PC9, both harboring EGFR
mutations and stably expressing GFP, were pre-treated with GMI and
co-cultured with CD3/CD28-activated Jurkat T cells (29). This co-culture led to a
significant increase in IL-2 and granzyme B secretion compared with
co-cultures without GMI pretreatment (Fig. S8A and B). Furthermore,
tumor-eliminating activity assessed by relative fluorescence
intensity showed a significant decrease in viability of
GMI-pretreated lung cancer cells co-cultured with
CD3/CD28-stimulated Jurkat T cells (Figs. S8C-E). These results suggested
that GMI-mediated reduction of membrane PD-L1 may enhance
CD3/CD28-stimulated T cell suppression of cancer cell viability.
Interestingly, without tumor cells, GMI elevated CD69 expression
and enhanced IL-2 secretion in Jurkat T cells (Fig. S8F and G), indicating a direct
stimulatory effect on T cells. In addition, the presence of TILs,
including CD8+ and CD4+ T cells, in LLC1
tumor tissues was analyzed (Fig.
5B). Flow cytometry revealed a higher proportion of
CD8+ TILs in GMI-treated tumors (Figs. 6C and S9), indicating that GMI may promote
the infiltration of activated CD8+ cytotoxic T cells
into the TME through PD-L1 degradation. Collectively, the
aforementioned findings suggested that GMI-facilitated PD-L1
degradation may promote antitumor immunity.
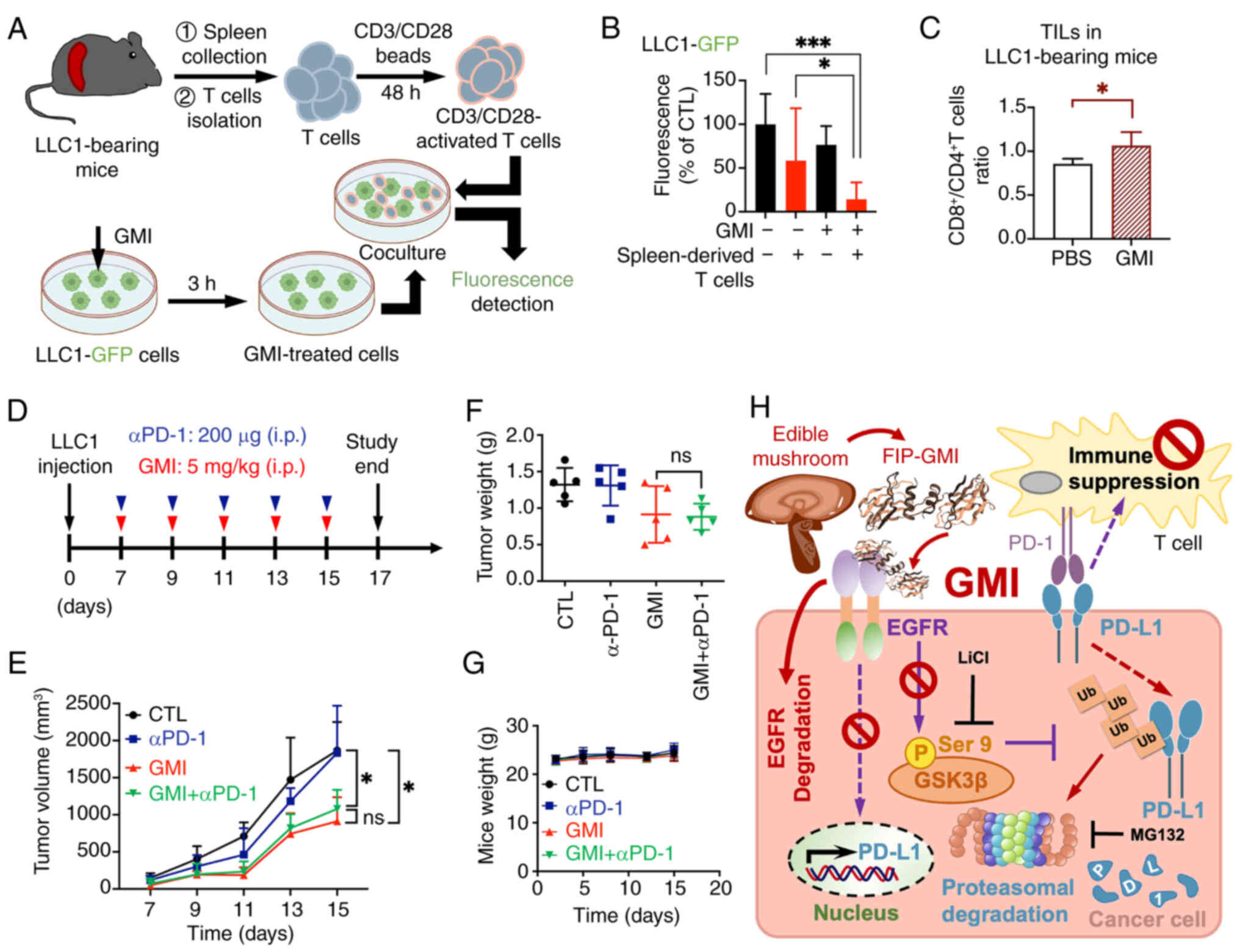 | Figure 6GMI regulates T cell-mediated
suppression of tumor cells ex vivo and promotes antitumor
immunity in vivo. (A) Schematic diagram illustrating the
co-culture protocol. Activated spleen-derived T cells isolated from
LLC1-bearing mice were co-cultured with LLC1-GFP cells pretreated
with 0.6 μM GMI for 3 h. After 72 h of co-culture, GFP
fluorescence in LLC1-GFP cells was analyzed. (B) Quantification of
GFP fluorescence intensity in LLC1-GFP cells following co-culture
with activated T cells. (C) Flow cytometric analysis of
tumor-infiltrating lymphocytes from LLC1-bearing mice treated with
control (PBS) or GMI (Fig. 5B).
The ratio of CD8+ to CD4+ T cells is shown.
(D) Treatment schedule for LLC1 syngeneic C57BL/6 mice receiving
intraperitoneal injections of GMI (5.0 mg/kg), anti-PD-1 antibody
(200 μg/mice), or their combination every 2 days. (E-G)
Tumor volume (E), tumor weight (F) and body weight (G) were
monitored at specified intervals throughout the experiment. Data
are presented as the mean ± SEM (n=5 mice per group). (H) Schematic
model illustrating how GMI enhances antitumor immunity by driving
PD-L1 breakdown. GMI, a FIP derived from an edible mushroom and
known as an EGFR degrader, reduces PD-L1 levels by downregulating
its mRNA expression and promoting GSK3β-mediated ubiquitination and
proteasomal degradation. This process further diminishes T cell
immune suppression and enhances antitumor immunity.
*P<0.05 and ***P<0.001. Ganoderma
microsporum immunomodulatory protein; GFP, green fluorescent
protein; PD-1, programmed cell death protein-1; FIPs, fungal
immunomodulatory proteins; LiCl, lithium chloride; ns, not
significant. |
Finally, analysis on whether combining GMI with PD-1
blockade (Fig. 6D) can further
enhance antitumor efficacy revealed that combination treatment (GMI
+ anti-PD-1 mAb) significantly suppressed tumor growth (Fig. 6E) and reduced tumor weight
(Figs. 6F and S10) compared with the control (PBS)
group. However, tumor suppression in the combined treatment group
was not significantly different from the GMI-only group.
Furthermore, in line with our earlier findings from the syngeneic
mouse model, body weights remained stable across all groups
(Fig. 6G), indicating that GMI
is safe for promoting PD-L1 degradation as a therapeutic
strategy.
Discussion
PD-L1 on tumor cells interacts with the PD-1
receptor on T lymphocytes, a key mechanism of immune evasion in
multiple cancer types. This interaction effectively suppresses the
immune system's ability to detect and eliminate malignant cells,
contributing to tumor progression. The development and approval of
multiple antibodies targeting the PD-1/PD-L1 pathway represent a
major advancement in cancer immunotherapy by disrupting this immune
checkpoint and restoring immune function in patients (39). Beyond inhibiting the PD-1/PD-L1
interaction between cancer cells and immune T cells, a more direct
strategy targeting PD-L degradation has gained attention. By
reducing PD-L1 expression on tumor cell surface, this approach may
diminish immune evasion and strengthen antitumor immunity,
positioning PD-L1 degradation as a promising therapeutic avenue
(40). Since EGFR activation
stabilizes PD-L1 by inhibiting its degradation, agents targeting
the EGFR pathway can modulate PD-L1 levels (12). Bioactive compounds derived from
medicinal mushrooms, including FIPs and polysaccharides, have
attracted considerable interest for their therapeutic potential,
particularly in cancer treatment (41). To the best of our knowledge, the
present study is the first to demonstrate that FIP-GMI, an approved
dietary ingredient and EGFR degrader, regulates antitumor immunity
by promoting PD-L1 degradation (Fig.
6H). Notably, previous studies by the authors revealed that GMI
can induce proteasomal degradation of key proteins, including EGFR
and Slug in lung and brain cancer cells (18,19), and ACE2 in SARS-CoV-2 infection
models (20). In the present
study, it was further demonstrated that GMI facilitates
GSK3β-mediated proteasomal degradation of PD-L1, suggesting that
GMI targets multiple substrates through this pathway across various
cell types. Moreover, proteomic analysis revealed that GMI
regulates a broad spectrum of membrane proteins (Fig. 1A), though the precise mechanisms
remain to be elucidated.
PD-L1 transcription is regulated by various
transcription factors. Notably, c-Myc binds directly to the PD-L1
gene promoter, with its inhibition reducing both PD-L1 mRNA and
protein levels, thereby boosting antitumor immunity (42). Similarly, Hippo pathway
effectors, such as the transcriptional activator yes-associated
protein and signal transducer and activator of transcription 3
(STAT3), have been shown to regulate PD-L1 transcription in various
cancer types (43,44). EGFR inhibitors significantly
suppress PD-L1 mRNA in lung cancer cells with activating mutations
(14,45) and block EGF-induced PD-L1
upregulation, indicating that inhibiting EGFR activity regulates
PD-L1 mRNA expression. Previous research has shown that GMI reduces
STAT3 phosphorylation, hinting at transcriptional regulation of
PD-L1 (31). The current
findings confirmed that GMI significantly downregulates PD-L1 mRNA
expression in lung cancer cells within 24 h, likely by targeting
these critical transcriptional regulators. Additional
investigations could be carried out to examine how GMI influences
these transcription factors involved in regulating PD-L1 mRNA.
Moreover, other signaling pathways, including those involving
receptor tyrosine kinases (RTKs) such as anaplastic lymphoma kinase
(46) and neurotrophic RTK 1
(47), as well as the
PI3K-AKT-mTOR axis (48), also
upregulate both PD-L1 mRNA and protein expression.
Beyond transcriptional regulation, GMI promotes
PD-L1 degradation via GSK3β activity in lung cancer cells. Future
investigations could explore whether GMI exerts effects on broader
immune checkpoint regulatory pathways, extending beyond GSK3β, and
further evaluate its potential to disrupt both transcriptional and
post-translational processes governing PD-L1 expression.
GSK3β facilitates PD-L1 identification by the E3
ubiquitin ligase complex, which encompasses beta-transducing
repeat-containing protein (β-TrCP), leading to ubiquitination and
proteasomal degradation. By contrast, EGFR activation inhibits
GSK3β via AKT-mediated phosphorylation, stabilizing PD-L1 and
promoting immune resistance (12). EGFR inhibitors, such as gefitinib
and osimertinib, counteract this stabilization effect by blocking
EGFR signaling, which can activate GSK3β (45). Consistently, the present study
demonstrated that GMI, the EGFR degrader, activates GSK3β, and
GSK3β inhibition or knockdown reverses GMI-induced PD-L1 reduction,
confirming its crucial role. Of note, short-term EGF exposure
partially rescued GMI-induced GSK3β activation (data not shown);
however, an EGFR overexpression rescue experiment was not
performed, suggesting that other RTKs may also regulate GSK3β
activity. Contrary to the previous study (12), the present findings revealed that
β-TrCP knockdown did not fully prevent PD-L1 degradation by GMI
(data not shown), suggesting involvement of alternative E3
ubiquitin ligases. Candidates include membrane-associated RING-CH 8
(MARCH8) and ariadne-1 homolog (ARIH1), which mediate PD-L1
degradation in EGFR-mutant NSCLC cells exposed to EGFR inhibitors
(14,49). Thus, GMI appears to regulate
PD-L1 via a β-TrCP-independent yet GSK3β-dependent mechanism,
warranting further exploration of novel E3 ubiquitin ligases
involved in GMI-mediated PD-L1 degradation.
Numerous PD-L1-targeting strategies, such as
proteolysis-targeting chimeras, lysosomal-targeted chimeras and
GlueBody chimeras, have shown considerable promise in promoting
PD-L1 degradation (50,51). However, these approaches often
face limitations associated with suboptimal drug delivery
efficiency, large molecular size and restricted tumor tissue
penetration (52). By contrast,
GMI is a naturally derived FIP with a favorable safety profile
(53). The findings of the
present study demonstrated that GMI effectively downregulates PD-L1
expression by repressing its transcription and enhancing
GSK3β-mediated proteasomal degradation. Unlike synthetic degraders
that frequently require intricate molecular design and chemical
modification, GMI offers potential advantages in terms of
biocompatibility, low toxicity and feasibility as a functional
dietary supplement. Overall, these characteristics underscore the
therapeutic significance of GMI and support its promise as a novel
and practical alternative to existing PD-L1-targeting
approaches.
Immune resistance and the immunosuppressive TME
remain major challenges in treating solid tumors (54). Elevated PD-L1 correlates with
tumor progression, reduced T-cell activity, and the development of
immune evasion. A promising therapeutic strategy that disrupts
PD-L1/PD-1 binding can re-engage T cell-mediated immunity.
Promoting PD-L1 degradation enhances immune responses and T cell
function, thereby improving therapeutic outcomes (55). The aqueous extract derived from
the medicinal herb Centipeda minima alleviates tumor burden
in mice harboring NSCLC cells by regulating PD-L1 expression and
enhancing CD8+ T cell cytotoxicity (56). Moreover, the natural marine
compound benzosceptrin C strongly promotes PD-L1 degradation and
exerts immune antitumor effects, effectively inhibiting tumor
growth in MC38 tumor-bearing mice (57). Collectively, these studies
indicate that using active compounds from natural products or
traditional Chinese medicine herbs may offer promising therapeutic
strategies for combating tumors and enhancing antitumor immunity.
Furthermore, FIPs, including FIP-vvo derived from Volvariella
volvacea, modulate immune responses and may support health by
boosting immune function (58).
The present study introduces fungal-derived GMI as a novel PD-L1
degrader. In vitro co-cultures showed that GMI effectively
reduces PD-L1 in cancer cells, significantly enhancing
CD3/CD28-stimulated T cell-mediated suppression of cancer cell
viability. Consistently, in vivo experiments demonstrated
significant tumor growth inhibition accompanied by increased
infiltration of CD8+ TILs, supporting enhanced antitumor
immunity within the TME. Notably, in the absence of tumor cells,
GMI upregulated Jurkat T-cell CD69 expression and increased IL-2
secretion (Fig. S8F and G),
indicating that, beyond downregulating PD-L1 on tumor cells, GMI
directly stimulates T cells and may thereby potentiate antitumor
immune responses. Future studies will further explore the molecular
mechanisms underlying this direct activation and its implications
for cancer immunotherapy.
No synergistic effect was observed when GMI was
administered in combination with anti-PD-1 in LLC1-bearing mice,
consistent with LLC tumors being relatively 'cold' and resistant to
anti-PD-1/PD-L1 therapy (30).
This may explain the limited efficacy of anti-PD-1 antibodies in
our in vivo model. Prior studies indicate that checkpoint
inhibitors beyond PD-1/PD-L1, such as CTLA-4 blockade, which
modulates the CTLA-4/CD80 (CD86) axis or TIGIT blockade (targeting
the TIGIT/PVR axis), can elicit distinct immune responses that vary
with tumor type and microenvironment (59). Although CD80 and CD86 were not
detected in our proteomic analysis, PVR, an immune checkpoint
ligand involved in immune evasion (60), was unexpectedly identified,
suggesting that the TIGIT pathway may warrant further
investigation. Accordingly, investigating the combination of GMI
with blocking antibodies against other immune checkpoints, such as
CTLA-4 or TIGIT, presents an intriguing direction for future
research to better elucidate potential synergistic effects. It is
essential to highlight that no significant weight loss or visible
adverse effects were observed in our tumor-bearing model,
indicating that GMI is well-tolerated. In addition, recent research
by the authors demonstrated that GMI ameliorates SARS-CoV-2
envelope protein-induced inflammation and exerts anti-inflammatory
effects in mouse models of excessive immune activation (61). These results suggested that,
beyond its role in PD-L1 regulation, GMI may contribute to immune
homeostasis, potentially mitigating the risk of immune-related
adverse effects. Furthermore, a favorable safety profile for GMI
has been previously demonstrated, further supporting its potential
as a safe option in therapeutic applications (53). Collectively, the aforementioned
findings support GMI as a valuable supplement in cancer therapy by
targeting both tumor growth and immune evasion.
While PD-L1 has traditionally been studied for its
role as an immune checkpoint ligand, evidence highlights its tumor
cell-intrinsic functions, which may complicate its use as a
biomarker for immunotherapy. These intrinsic roles extend beyond
immune evasion and include promoting cancer cell survival,
invasion, EMT, stemness, metabolic reprogramming and therapy
resistance. Emerging studies have shown that PD-L1 can activate
intracellular signaling pathways, such as PI3K/AKT and MAPK, which
are crucial for cancer cell proliferation and survival (62,63). Moreover, intrinsic PD-L1 activity
in tumor cells drives NSCLC progression by facilitating EMT, and
elevated EMT levels in PD-L1-high NSCLC may serve as a predictor of
poor outcomes following immune checkpoint inhibitor therapy
(64). Additionally, PD-L1 has
been implicated in enhancing glycolysis, thereby supporting the
metabolic demands of rapidly growing tumors (65). Other intrinsic functions include
its involvement in the anti-apoptotic processes and chemotherapy
resistance, where PD-L1 helps protect cancer cells from these
therapeutic agents (66,67). Importantly, previous studies by
the authors showed that GMI suppresses lung and brain cancer cell
migration and metastasis by inhibiting Slug, a transcription factor
associated with EMT (18,22).
This mechanism may intersect with PD-L1's roles in promoting EMT
and metastasis, highlighting a potential link between GMI's effects
and the intrinsic functions of PD-L1. Future studies could clarify
whether GMI's suppression of EMT and metastasis is directly
mediated through its impact on PD-L1 signaling pathways. Such
investigations could provide deeper insights into the dual role of
GMI in targeting both the immune and non-immune functions of PD-L1,
further advancing its therapeutic potential.
A previous study (19) by the authors demonstrated that
GMI is a novel EGFR degrader capable of disrupting EGFR-driven
oncogenic signaling and inhibiting tumor progression in
EGFR-positive cancers. In xenograft models, GMI effectively
suppresses tumor growth in nude mice harboring tumors with both
EGFR wild-type and activating mutations, indicating a direct
cytotoxic effect (17,19). Notably, in immune-competent mouse
models, GMI inhibits tumor metastasis in a tail-vein injection
model and suppresses the growth of subcutaneous LLC1 tumors in
C57BL/6 mice (22). These
findings indicated that, beyond its direct cytotoxic effects, GMI
may also exert antitumor activity by enhancing endogenous immune
defense mechanisms. Building on this, the current study further
demonstrated that GMI suppresses tumor growth and induces PD-L1
downregulation in vivo, suggesting a dual mechanism of
action. One potential anticancer mechanism of GMI is its ability to
enhance antitumor immunity through PD-L1 degradation, thereby
alleviating immune suppression. While PD-L1 reduction contributes
to enhanced antitumor immunity, tumor suppression in nude mouse
models with defective T cell function suggests an additional direct
cytotoxic effect. This is consistent with the mechanisms of other
FDA-approved EGFR-tyrosine kinase inhibitors, such as osimertinib
and erlotinib, which not only disrupt EGFR signaling and induce
tumor cell apoptosis but also regulate immune responses by reducing
PD-L1 expression and modulating the TME (12,14). The observed reduction in PD-L1
levels and the concurrent increase in CD8+ T cell
infiltration in the TME indicated that GMI may exert its effects by
modulating the T cell landscape to combat tumors. This suggests
that GMI's effects extend beyond simply eliminating cancer cells,
potentially encompassing tumor suppression through PD-L1
downregulation on malignant cells.
The present study has several limitations. Firstly,
while the co-culture system employing Jurkat T cells and cancer
cells represents a well-established and widely adopted approach for
assessing the functional activation of Jurkat T cells (68), it fails to fully replicate
antigen-specific T cell-mediated antitumor responses or to provide
a reliable evaluation of T cell-mediated tumor cell cytotoxicity.
Alternative models, such as the co-culture of human peripheral
blood mononuclear cells with GMI-treated human cancer cells or the
co-culture of OVA-specific T cells with GMI-treated OVA-expressing
cancer cells (56), could better
elucidate the mechanisms underlying T cell-mediated antitumor
immunity in the context of GMI treatment. Secondly, while GMI
enhances antitumor immunity by promoting GSK3β-mediated PD-L1
degradation, without a T cell depletion assay, in vivo
antitumor effects in immune-competent mice remain unclear. In
addition, p-GSK3β (Ser9) was not quantified in the tumor samples;
therefore, while in vitro data support GSK3β-dependent PD-L1
degradation, GSK3β activation in vivo could not be directly
confirmed in this cohort. Lastly, proteomic analysis revealed that
GMI does not significantly alter surface MHC class I
(HLA-A/B/C/E/G/F) expression on several lung cancer cell lines
following GMI treatment (data not shown), suggesting no direct
effect on antigen presentation pathways. However, broader indirect
effects on immune modulation require further investigation.
In conclusion, the present study provides evidence
for the first time that the naturally derived FIP GMI, beyond its
cytotoxic effects as an EGFR degrader, promotes PD-L1 degradation
by decreasing its mRNA expression and inducing GSK3β-mediated
proteasomal degradation. This reduction of PD-L1 enhances
CD3/CD28-stimulated T cell-mediated suppression of cancer cell
viability and inhibits tumor growth with increased CD8+
T cell infiltration in vivo. Collectively, these results
highlight the promising potential of GMI as a therapeutic agent or
functional dietary supplement for cancer immunotherapy.
Supplementary Data
Acknowledgements
The authors are sincerely grateful to Ms Y-C Chen
of National Yang Ming Chiao Tung University for providing technical
assistance.
Funding
The present study was supported by grants from the Ministry of
Science and Technology of Taiwan (grant no. Young Scholar
Fellowship Einstein Program; MOST 111-2636-B-A49-009) and the
National Science and Technology Council of Taiwan (grant no. NSTC
112-2320-B-A49-010-MY3, NSTC 113-2320-B-A49-028-MY3, NSTC
113-2811-B-A49A-038 and NSTC 114-2811-B-A49A-015).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The data generated in the
present study may be found in the NYCU Dataverse under accession
number 10.57770/QNCRKI or at the following URL: https://doi.org/10.57770/QNCRKI.
Authors' contributions
LLL, WHH and TYL conceptualized the study. WJH
performed experiments, collected and analyzed data, interpreted the
results, and wrote the manuscript. WJH and LCH performed
bioinformatic analysis. LCH, ZHL, YRC and KFL performed in
vitro and in vivo experiments. TYL reviewed and edited
the manuscript. WJH and TYL confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
All procedures involving animals were conducted in
accordance with the guidelines established by the Institutional
Animal Care and Use Committee (IACUC; approval no. 1070805) at
National Yang Ming Chiao Tung University (Taipei, Taiwan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
GMI
|
Ganoderma microsporum
immunomodulatory protein
|
|
PD-L1
|
programmed death-ligand 1
|
|
EGFR
|
epidermal growth factor receptor
|
|
FIPs
|
fungal immunomodulatory proteins
|
|
PD-1
|
programmed cell death protein-1
|
|
GSK3β
|
glycogen synthase kinase 3 beta
|
|
LiCl
|
lithium chloride
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
NSCLC
|
non-small cell lung cancer
|
|
DEPs
|
differentially expressed proteins
|
|
TILs
|
tumor-infiltrating lymphocytes
|
References
|
1
|
Zou W: Immunosuppressive networks in the
tumour environment and their therapeutic relevance. Nat Rev Cancer.
5:263–274. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Han Y, Liu D and Li L: PD-1/PD-L1 pathway:
Current researches in cancer. Am J Cancer Res. 10:727–742.
2020.PubMed/NCBI
|
|
3
|
Sun C, Mezzadra R and Schumacher TN:
Regulation and function of the PD-L1 checkpoint. Immunity.
48:434–452. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iwai Y, Hamanishi J, Chamoto K and Honjo
T: Cancer immunotherapies targeting the PD-1 signaling pathway. J
Biomed Sci. 24:262017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sharma P and Allison JP: Immune checkpoint
targeting in cancer therapy: Toward combination strategies with
curative potential. Cell. 161:205–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naidoo J, Page DB, Li BT, Connell LC,
Schindler K, Lacouture ME, Postow MA and Wolchok JD: Toxicities of
the anti-PD-1 and anti-PD-L1 immune checkpoint antibodies. Ann
Oncol. 26:2375–2391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang N, Dou Y, Liu L, Zhang X, Liu X,
Zeng Q, Liu Y, Yin M, Liu X, Deng H and Song D: SA-49, a novel
aloperine derivative, induces MITF-dependent lysosomal degradation
of PD-L1. EBioMedicine. 40:151–162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang T, Cai S, Cheng Y, Zhang W, Wang M,
Sun H, Guo B, Li Z, Xiao Y and Jiang S: Discovery of small-molecule
inhibitors of the PD-1/PD-L1 axis that promote PD-L1
internalization and degradation. J Med Chem. 65:3879–3893. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cha JH, Chan LC, Li CW, Hsu JL and Hung
MC: Mechanisms controlling PD-L1 expression in cancer. Mol Cell.
76:359–370. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsu JM, Xia W, Hsu YH, Chan LC, Yu WH, Cha
JH, Chen CT, Liao HW, Kuo CW, Khoo KH, et al: STT3-dependent PD-L1
accumulation on cancer stem cells promotes immune evasion. Nat
Commun. 9:19082018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo
CW, Khoo KH, Chang SS, Cha JH, Kim T, et al: Glycosylation and
stabilization of programmed death ligand-1 suppresses T-cell
activity. Nat Commun. 7:126322016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Bu X, Wang H, Zhu Y, Geng Y,
Nihira NT, Tan Y, Ci Y, Wu F, Dai X, et al: Cyclin D-CDK4 kinase
destabilizes PD-L1 via cullin 3-SPOP to control cancer immune
surveillance. Nature. 553:91–95. 2018. View Article : Google Scholar
|
|
14
|
Qian G, Guo J, Vallega KA, Hu C, Chen Z,
Deng Y, Wang Q, Fan S, Ramalingam SS, Owonikoko TK, et al:
Membrane-associated RING-CH 8 functions as a novel PD-L1 E3 ligase
to mediate PD-L1 degradation induced by EGFR inhibitors. Mol Cancer
Res. 19:1622–1634. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su YW, Huang WY, Lin SH and Yang PS:
Effects of reishimmune-S, a fungal immunomodulatory peptide
supplement, on the quality of life and circulating natural killer
cell profiles of patients with early breast cancer receiving
adjuvant endocrine therapy. Integr Cancer Ther.
23:153473542412421202024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin TY, Hua WJ, Yeh H and Tseng AJ:
Functional proteomic analysis reveals that fungal immunomodulatory
protein reduced expressions of heat shock proteins correlates to
apoptosis in lung cancer cells. Phytomedicine. 80:1533842021.
View Article : Google Scholar
|
|
17
|
Hua WJ, Hwang WL, Yeh H, Lin ZH, Hsu WH
and Lin TY: Ganoderma microsporum immunomodulatory protein combined
with KRAS(G12C) inhibitor impedes intracellular AKT/ERK network to
suppress lung cancer cells with KRAS mutation. Int J Biol Macromol.
259:1292912024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tseng AJ, Tu TH, Hua WJ, Yeh H, Chen CJ,
Lin ZH, Hsu WH, Chen YL, Hsu CC and Lin TY: GMI, Ganoderma
microsporum protein, suppresses cell mobility and increases
temozolomide sensitivity through induction of Slug degradation in
glioblastoma multiforme cells. Int J Biol Macromol. 219:940–948.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hua WJ, Yeh H, Lin ZH, Tseng AJ, Huang LC,
Qiu WL, Tu TH, Wang DH, Hsu WH, Hwang WL and Lin TY: Ganoderma
microsporum immunomodulatory protein as an extracellular epidermal
growth factor receptor (EGFR) degrader for suppressing
EGFR-positive lung cancer cells. Cancer Lett. 578:2164582023.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yeh H, Vo DNK, Lin ZH, Ho HPT, Hua WJ, Qiu
WL, Tsai MH and Lin TY: GMI, a protein from Ganoderma microsporum,
induces ACE2 degradation to alleviate infection of SARS-CoV-2
Spike-pseudotyped virus. Phytomedicine. 103:1542152022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re3532016. View Article : Google Scholar
|
|
22
|
Lo HC, Hua WJ, Yeh H, Lin ZH, Huang LC,
Ciou YR, Ruan R, Lin KF, Tseng AJ, Wu ATH, et al: GMI, a Ganoderma
microsporum protein, abolishes focal adhesion network to reduce
cell migration and metastasis of lung cancer. Life Sci.
335:1222552023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ge SX, Jung D and Yao R: ShinyGO: A
graphical gene-set enrichment tool for animals and plants.
Bioinformatics. 36:2628–2629. 2020. View Article : Google Scholar :
|
|
24
|
Luo W and Brouwer C: Pathview: An
R/Bioconductor package for pathway-based data integration and
visualization. Bioinformatics. 29:1830–1831. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M, Furumichi M, Sato Y,
Ishiguro-Watanabe M and Tanabe M: KEGG: Integrating viruses and
cellular organisms. Nucleic Acids Res. 49:D545–D551. 2021.
View Article : Google Scholar :
|
|
26
|
Hsu WH, Hua WJ, Qiu WL, Tseng AJ, Cheng HC
and Lin TY: WSG, a glucose-enriched polysaccharide from Ganoderma
lucidum, suppresses tongue cancer cells via inhibition of
EGFR-mediated signaling and potentiates cisplatin-induced
apoptosis. Int J Biol Macromol. 193:1201–1208. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Zhang Y, Huang Y, Yu D, Xu M, Hu H, Zhang
Q, Cai M, Geng X, Zhang H, Xia J, et al: Demethylzeylasteral
induces PD-L1 ubiquitin-proteasome degradation and promotes
antitumor immunity via targeting USP22. Acta Pharm Sin B.
14:4312–4328. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu CS, Lin CW, Chang YH, Chen HY, Chung
WC, Lai WY, Ho CC, Wang TH, Chen CY, Yeh CL, et al: Antimetabolite
pemetrexed primes a favorable tumor microenvironment for immune
checkpoint blockade therapy. J Immunother Cancer. 8:e0013922020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He K, Barsoumian HB, Puebla-Osorio N, Hu
Y, Sezen D, Wasley MD, Bertolet G, Zhang J, Leuschner C, Yang L, et
al: Inhibition of STAT6 with antisense oligonucleotides enhances
the systemic antitumor effects of radiotherapy and Anti-PD-1 in
metastatic non-small cell lung cancer. Cancer Immunol Res.
11:486–500. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang TY, Yu CC, Hsieh PL, Liao YW, Yu CH
and Chou MY: GMI ablates cancer stemness and cisplatin resistance
in oral carcinomas stem cells through IL-6/Stat3 signaling
inhibition. Oncotarget. 8:70422–70430. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin TY, Hsu HY, Sun WH, Wu TH and Tsao SM:
Induction of Cbl-dependent epidermal growth factor receptor
degradation in Ling Zhi-8 suppressed lung cancer. Int J Cancer.
140:2596–2607. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hsin IL, Chiu LY, Ou CC, Wu WJ, Sheu GT
and Ko JL: CD133 inhibition via autophagic degradation in
pemetrexed-resistant lung cancer cells by GMI, a fungal
immunomodulatory protein from Ganoderma microsporum. Br J Cancer.
123:449–458. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin PL, Wu TC, Wu DW, Wang L, Chen CY and
Lee H: An increase in BAG-1 by PD-L1 confers resistance to tyrosine
kinase inhibitor in non-small cell lung cancer via persistent
activation of ERK signalling. Eur J Cancer. 85:95–105. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu M, Wang X, Li W, Yu X,
Flores-Villanueva P, Xu-Monette ZY, Li L, Zhang M, Young KH, Ma X
and Li Y: Targeting PD-L1 in non-small cell lung cancer using CAR T
cells. Oncogenesis. 9:722020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lemma EY, Letian A, Altorki NK and McGraw
TE: Regulation of PD-L1 trafficking from synthesis to degradation.
Cancer Immunol Res. 11:866–874. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Feng C, Zhang L, Chang X, Qin D and Zhang
T: Regulation of post-translational modification of PD-L1 and
advances in tumor immunotherapy. Front Immunol. 14:12301352023.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zuazo M, Gato-Cañas M, Llorente N,
Ibañez-Vea M, Arasanz H, Kochan G and Escors D: Molecular
mechanisms of programmed cell death-1 dependent T cell suppression:
Relevance for immunotherapy. Ann Transl Med. 5:3852017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ghosh C, Luong G and Sun Y: A snapshot of
the PD-1/PD-L1 pathway. J Cancer. 12:2735–2746. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gou Q, Dong C, Xu H, Khan B, Jin J, Liu Q,
Shi J and Hou Y: PD-L1 degradation pathway and immunotherapy for
cancer. Cell Death Dis. 11:9552020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Venturella G, Ferraro V, Cirlincione F and
Gargano ML: Medicinal Mushrooms: Bioactive Compounds, Use, and
Clinical Trials. Int J Mol Sci. 22:2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Casey SC, Tong L, Li Y, Do R, Walz S,
Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M and Felsher
DW: MYC regulates the antitumor immune response through CD47 and
PD-L1. Science. 352:227–231. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Miao J, Hsu PC, Yang YL, Xu Z, Dai Y, Wang
Y, Chan G, Huang Z, Hu B, Li H, et al: YAP regulates PD-L1
expression in human NSCLC cells. Oncotarget. 8:114576–114587. 2017.
View Article : Google Scholar
|
|
44
|
Zhang N, Zeng Y, Du W, Zhu J, Shen D, Liu
Z and Huang JA: The EGFR pathway is involved in the regulation of
PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in
EGFR-mutated non-small cell lung cancer. Int J Oncol. 49:1360–1368.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang XM, Xu YL, Huang MY, Zhang LL, Su
MX, Chen X and Lu JJ: Osimertinib (AZD9291) decreases programmed
death ligand-1 in EGFR-mutated non-small cell lung cancer cells.
Acta Pharmacol Sin. 38:1512–1520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ota K, Azuma K, Kawahara A, Hattori S,
Iwama E, Tanizaki J, Harada T, Matsumoto K, Takayama K, Takamori S,
et al: Induction of PD-L1 expression by the EML4-ALK oncoprotein
and downstream signaling pathways in non-small cell lung cancer.
Clin Cancer Res. 21:4014–4021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Konen JM, Rodriguez BL, Fradette JJ,
Gibson L, Davis D, Minelli R, Peoples MD, Kovacs J, Carugo A,
Bristow C, et al: Ntrk1 promotes resistance to PD-1 checkpoint
blockade in mesenchymal Kras/p53 mutant lung cancer. Cancers
(Basel). 11:4622019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lastwika KJ, Wilson W III, Li QK, Norris
J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, et
al: Control of PD-L1 expression by oncogenic activation of the
AKT-mTOR pathway in non-small cell lung cancer. Cancer Res.
76:227–238. 2016. View Article : Google Scholar
|
|
49
|
Wu Y, Zhang C, Liu X, He Z, Shan B, Zeng
Q, Zhao Q, Zhu H, Liao H, Cen X, et al: ARIH1 signaling promotes
antitumor immunity by targeting PD-L1 for proteasomal degradation.
Nat Commun. 12:23462021. View Article : Google Scholar
|
|
50
|
Ren X, Wang L, Liu L and Liu J: PTMs of
PD-1/PD-L1 and PROTACs application for improving cancer
immunotherapy. Front Immunol. 15:13925462024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang F, Jiang R, Sun S, Wu C, Yu Q,
Awadasseid A, Wang J and Zhang W: Recent advances and mechanisms of
action of PD-L1 degraders as potential therapeutic agents. Eur J
Med Chem. 268:1162672024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gao H, Sun X and Rao Y: PROTAC technology:
Opportunities and challenges. ACS Med Chem Lett. 11:237–240. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fu HY and Hseu RS: Safety assessment of
the fungal immunomodulatory protein from Ganoderma microsporum
(GMI) derived from engineered Pichia pastoris: Genetic toxicology,
a 13-week oral gavage toxicity study, and an embryo-fetal
developmental toxicity study in Sprague-Dawley rats. Toxicol Rep.
9:1240–1254. 2022. View Article : Google Scholar :
|
|
54
|
Spitzer MH, Carmi Y, Reticker-Flynn NE,
Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR,
Chabon J, Bendall SC, et al: systemic immunity is required for
effective cancer immunotherapy. Cell. 168:487–502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yi M, Niu M, Xu L, Luo S and Wu K:
Regulation of PD-L1 expression in the tumor microenvironment. J
Hematol Oncol. 14:102021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang M, Guo H, Sun BB, Jie XL, Shi XY, Liu
YQ, Shi XL, Ding LQ, Xue PH, Qiu F, et al: Centipeda minima and
6-O-angeloylplenolin enhance the efficacy of immune checkpoint
inhibitors in non-small cell lung cancer. Phytomedicine.
132:1558252024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang Q, Wang J, Yu D, Zhang Q, Hu H, Xu M,
Zhang H, Tian S, Zheng G, Lu D, et al: Benzosceptrin C induces
lysosomal degradation of PD-L1 and promotes antitumor immunity by
targeting DHHC3. Cell Rep Med. 5:1013572024. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li JP, Lee YP, Ma JC, Liu BR, Hsieh NT,
Chen DC, Chu CL and You RI: The enhancing effect of fungal
immunomodulatory protein-volvariella volvacea (FIP-vvo) on
maturation and function of mouse dendritic cells. Life (Basel).
11:4712021.PubMed/NCBI
|
|
59
|
Sharma P, Goswami S, Raychaudhuri D,
Siddiqui BA, Singh P, Nagarajan A, Liu J, Subudhi SK, Poon C, Gant
KL, et al: Immune checkpoint therapy-current perspectives and
future directions. Cell. 186:1652–1669. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Freed-Pastor WA, Lambert LJ, Ely ZA,
Pattada NB, Bhutkar A, Eng G, Mercer KL, Garcia AP, Lin L, Rideout
WM III, et al: The CD155/TIGIT axis promotes and maintains immune
evasion in neoantigen-expressing pancreatic cancer. Cancer Cell.
39:1342–1360. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lin ZH, Yeh H, Lo HC, Hua WJ, Ni MY, Wang
LK, Chang TT, Yang MH and Lin TY: GMI, a fungal immunomodulatory
protein, ameliorates SARS-CoV-2 envelope protein-induced
inflammation in macrophages via inhibition of MAPK pathway. Int J
Biol Macromol. 241:1246482023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Almozyan S, Colak D, Mansour F, Alaiya A,
Al-Harazi O, Qattan A, Al-Mohanna F, Al-Alwan M and Ghebeh H: PD-L1
promotes OCT4 and Nanog expression in breast cancer stem cells by
sustaining PI3K/AKT pathway activation. Int J Cancer.
141:1402–1412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gao L, Guo Q, Li X, Yang X, Ni H, Wang T,
Zhao Q, Liu H, Xing Y, Xi T and Zheng L: MiR-873/PD-L1 axis
regulates the stemness of breast cancer cells. EBioMedicine.
41:395–407. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Jeong H, Koh J, Kim S, Song SG, Lee SH,
Jeon Y, Lee CH, Keam B, Lee SH, Chung DH and Jeon YK:
Epithelial-mesenchymal transition induced by tumor cell-intrinsic
PD-L1 signaling predicts a poor response to immune checkpoint
inhibitors in PD-L1-high lung cancer. Br J Cancer. 131:23–36. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chang CH, Qiu J, O'Sullivan D, Buck MD,
Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ,
et al: Metabolic competition in the tumor microenvironment is a
driver of cancer progression. Cell. 162:1229–1241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ghebeh H, Lehe C, Barhoush E, Al-Romaih K,
Tulbah A, Al-Alwan M, Hendrayani SF, Manogaran P, Alaiya A,
Al-Tweigeri T, et al: Doxorubicin downregulates cell surface B7-H1
expression and upregulates its nuclear expression in breast cancer
cells: Role of B7-H1 as an anti-apoptotic molecule. Breast Cancer
Res. 12:R482010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu S, Chen S, Yuan W, Wang H, Chen K and
Li D and Li D: PD-1/PD-L1 interaction up-regulates MDR1/P-gp
expression in breast cancer cells via PI3K/AKT and MAPK/ERK
pathways. Oncotarget. 8:99901–99912. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zheng Y, Fang YC and Li J: PD-L1
expression levels on tumor cells affect their immunosuppressive
activity. Oncol Lett. 18:5399–5407. 2019.PubMed/NCBI
|