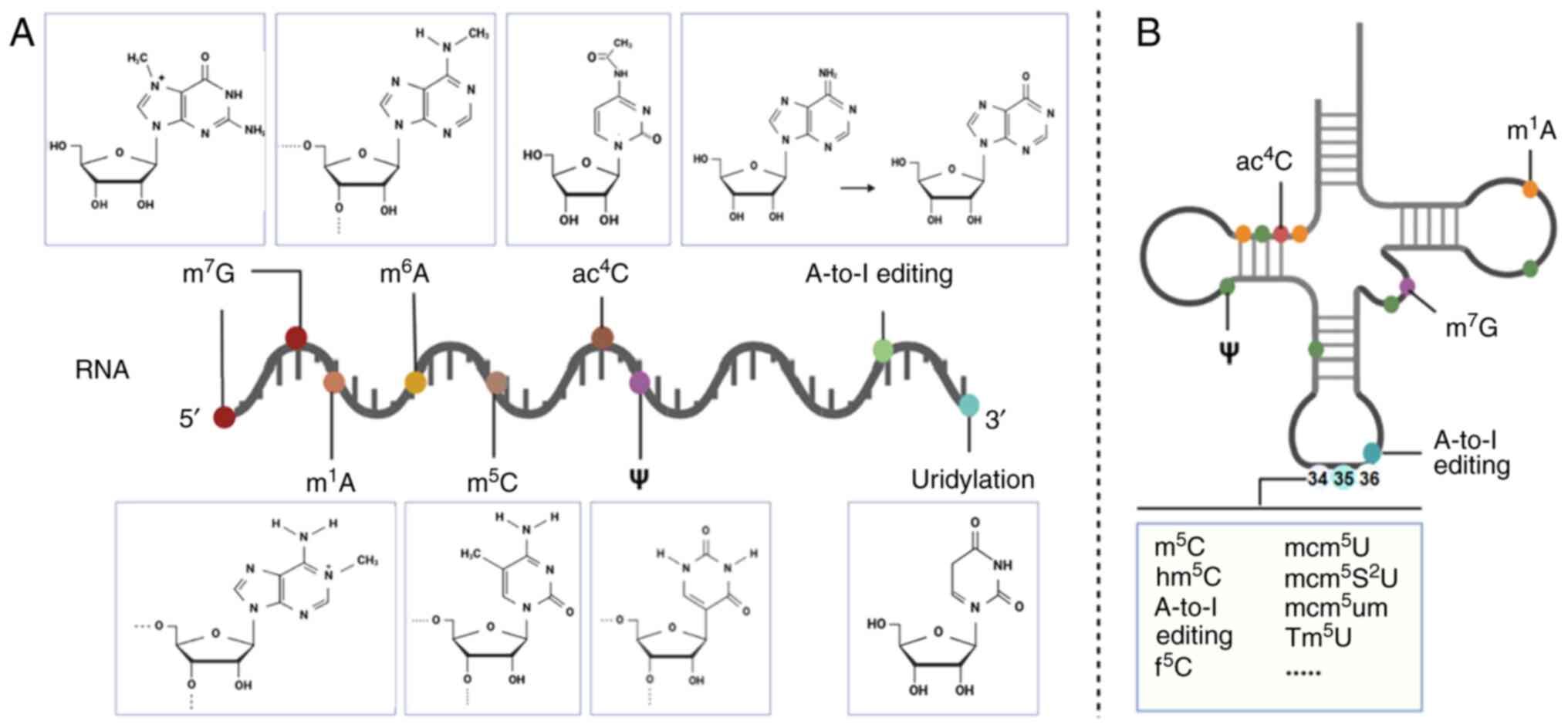
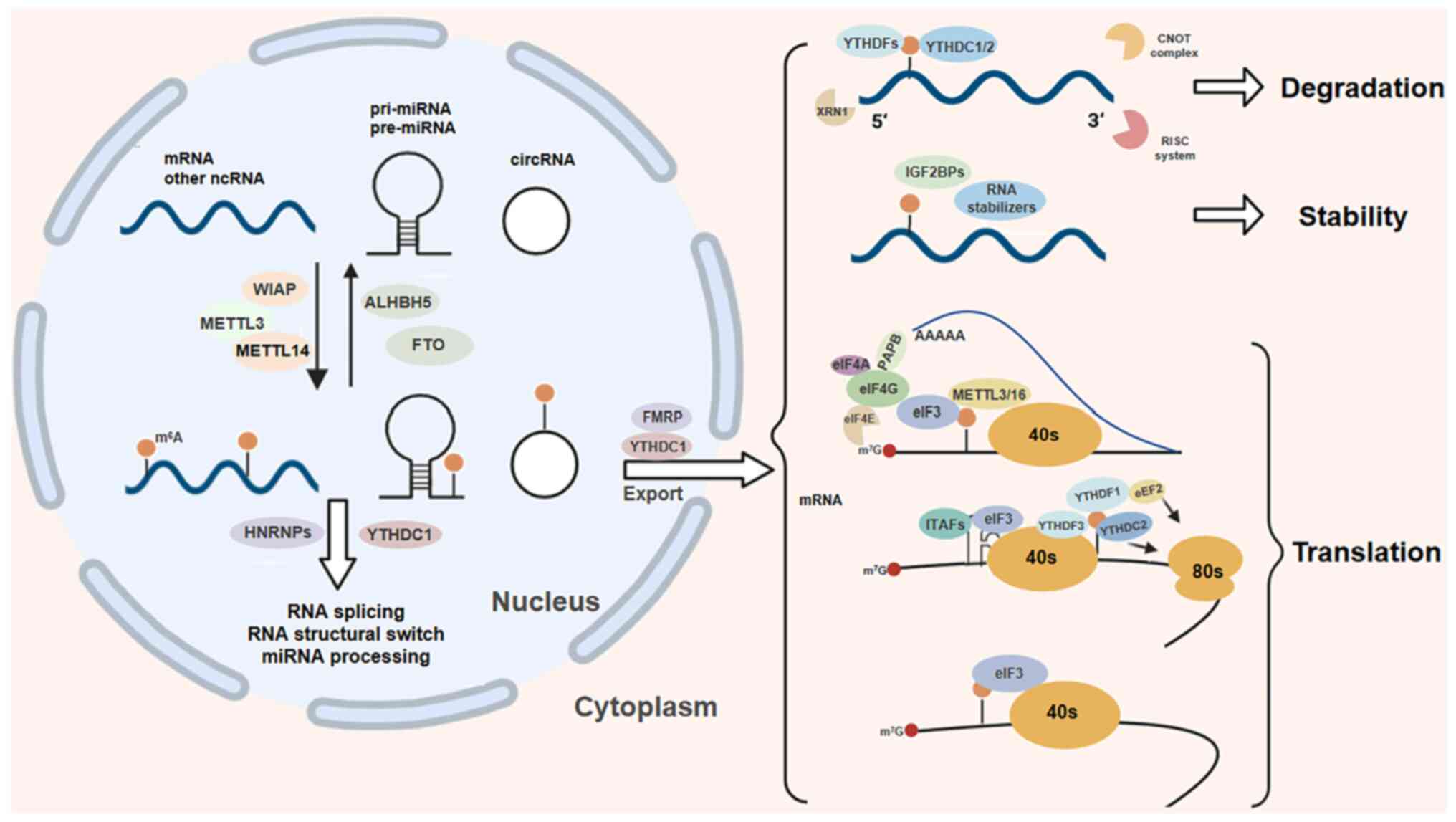
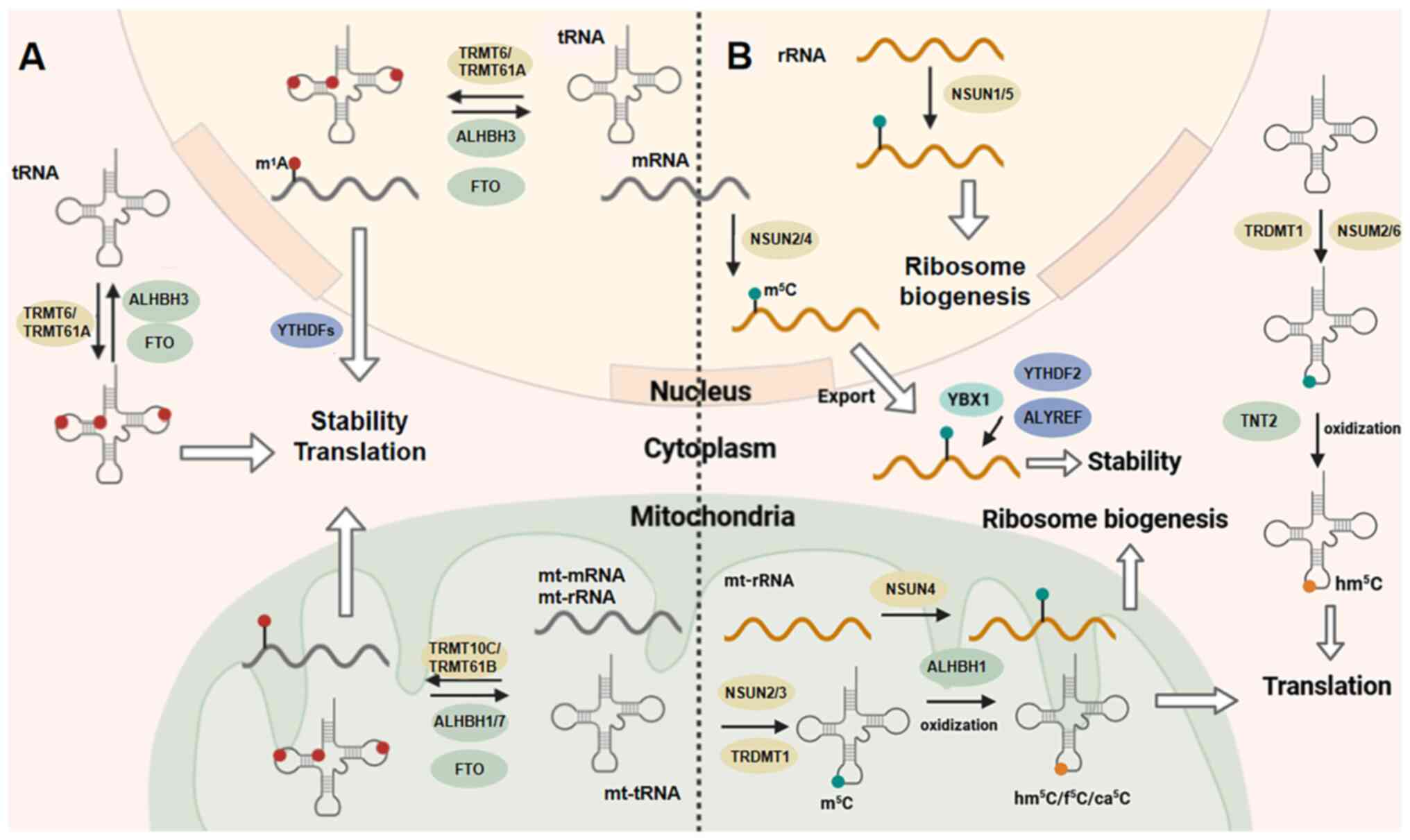
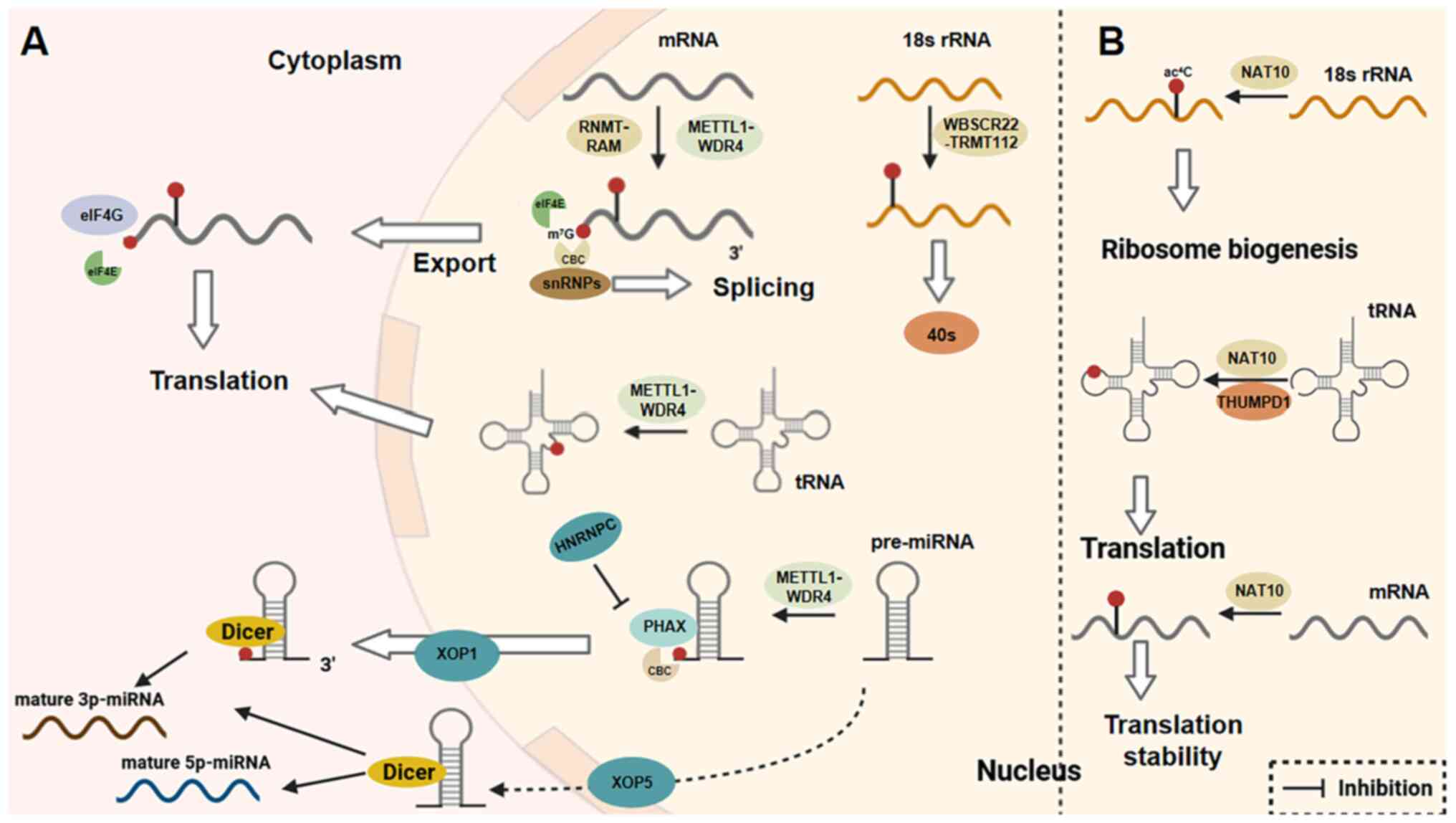
|
1
|
Liu M, López de Juan Abad B and Cheng K:
Cardiac fibrosis: Myofibroblast-mediated pathological regulation
and drug delivery strategies. Adv Drug Deliv Rev. 173:504–519.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Talman V and Ruskoaho H: Cardiac fibrosis
in myocardial infarction-from repair and remodeling to
regeneration. Cell Tissue Res. 365:563–581. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ivey MJ and Tallquist MD: Defining the
cardiac fibroblast. Circ J. 80:2269–2276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kendall RT and Feghali-Bostwick CA:
Fibroblasts in fibrosis: Novel roles and mediators. Front
Pharmacol. 5:1232014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tallquist MD and Molkentin JD: Redefining
the identity of cardiac fibroblasts. Nat Rev Cardiol. 14:484–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pellman J, Zhang J and Sheikh F:
Myocyte-fibroblast communication in cardiac fibrosis and
arrhythmias: Mechanisms and model systems. J Mol Cell Cardiol.
94:22–31. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hinz B: Myofibroblasts. Exp Eye Res.
142:56–70. 2016. View Article : Google Scholar
|
|
8
|
Thomas TP and Grisanti LA: The dynamic
interplay between cardiac inflammation and fibrosis. Front Physiol.
11:5290752020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hinderer S and Schenke-Layland K: Cardiac
fibrosis-A short review of causes and therapeutic strategies. Adv
Drug Deliv Rev. 146:77–82. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li T, Zhuang Y, Yang W, Xie Y, Shang W, Su
S, Dong X, Wu J, Jiang W, Zhou Y, et al: Silencing of METTL3
attenuates cardiac fibrosis induced by myocardial infarction via
inhibiting the activation of cardiac fibroblasts. FASEB J.
35:e211622021. View Article : Google Scholar
|
|
11
|
Huang W, Chen TQ, Fang K, Zeng ZC, Ye H
and Chen YQ: N6-methyladenosine methyltransferases: Functions,
regulation, and clinical potential. J Hematol Oncol. 14:1172021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
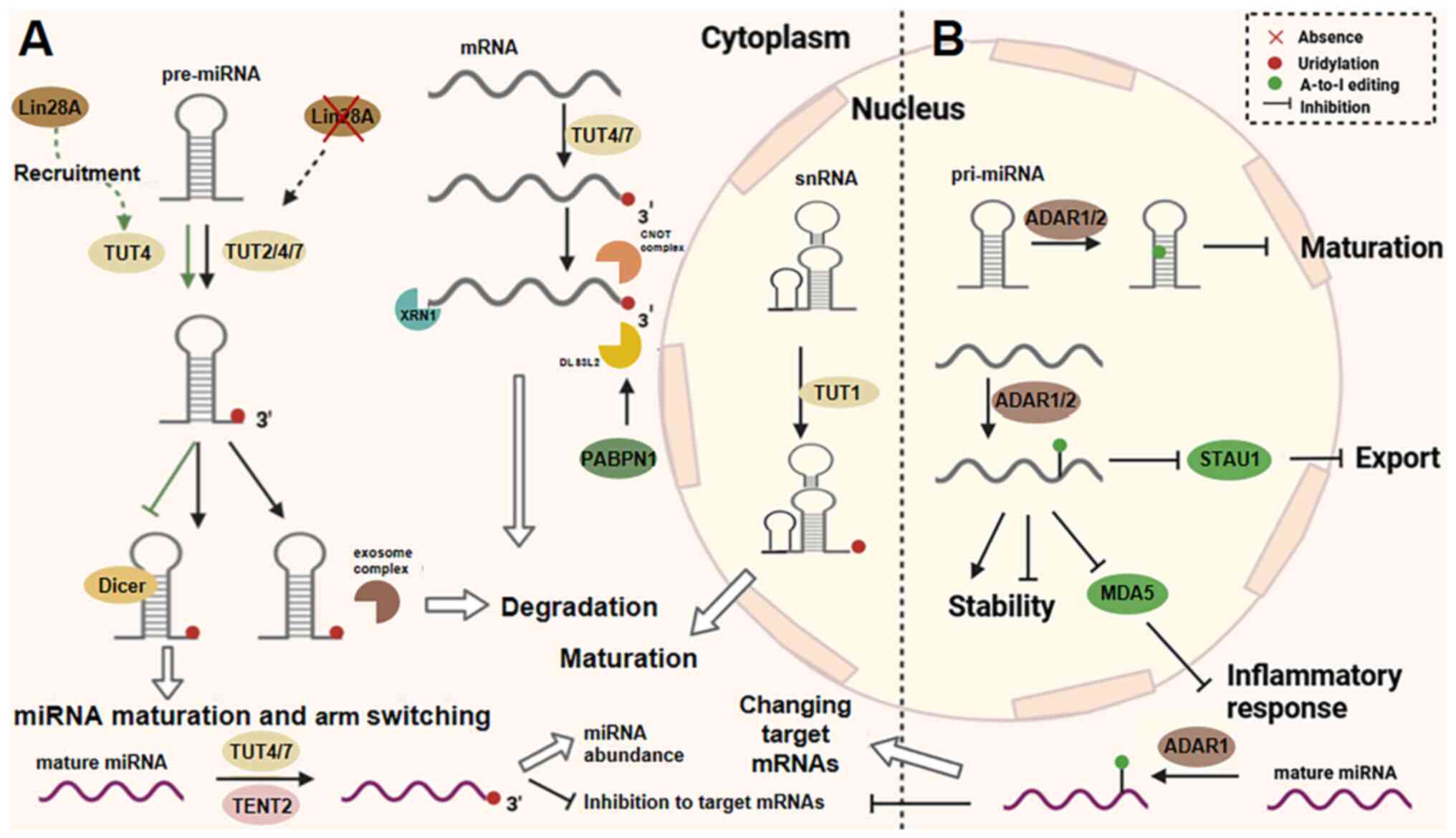
|
Wei CM, Gershowitz A and Moss B:
Methylated nucleotides block 5' terminus of HeLa cell messenger
RNA. Cell. 4:379–386. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Furuichi Y, Morgan M, Shatkin AJ, Jelinek
W, Salditt-Georgieff M and Darnell JE: Methylated, blocked 5
termini in HeLa cell mRNA. Proc Natl Acad Sci USA. 72:1904–1908.
1975. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3' UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zaccara S, Ries RJ and Jaffrey SR:
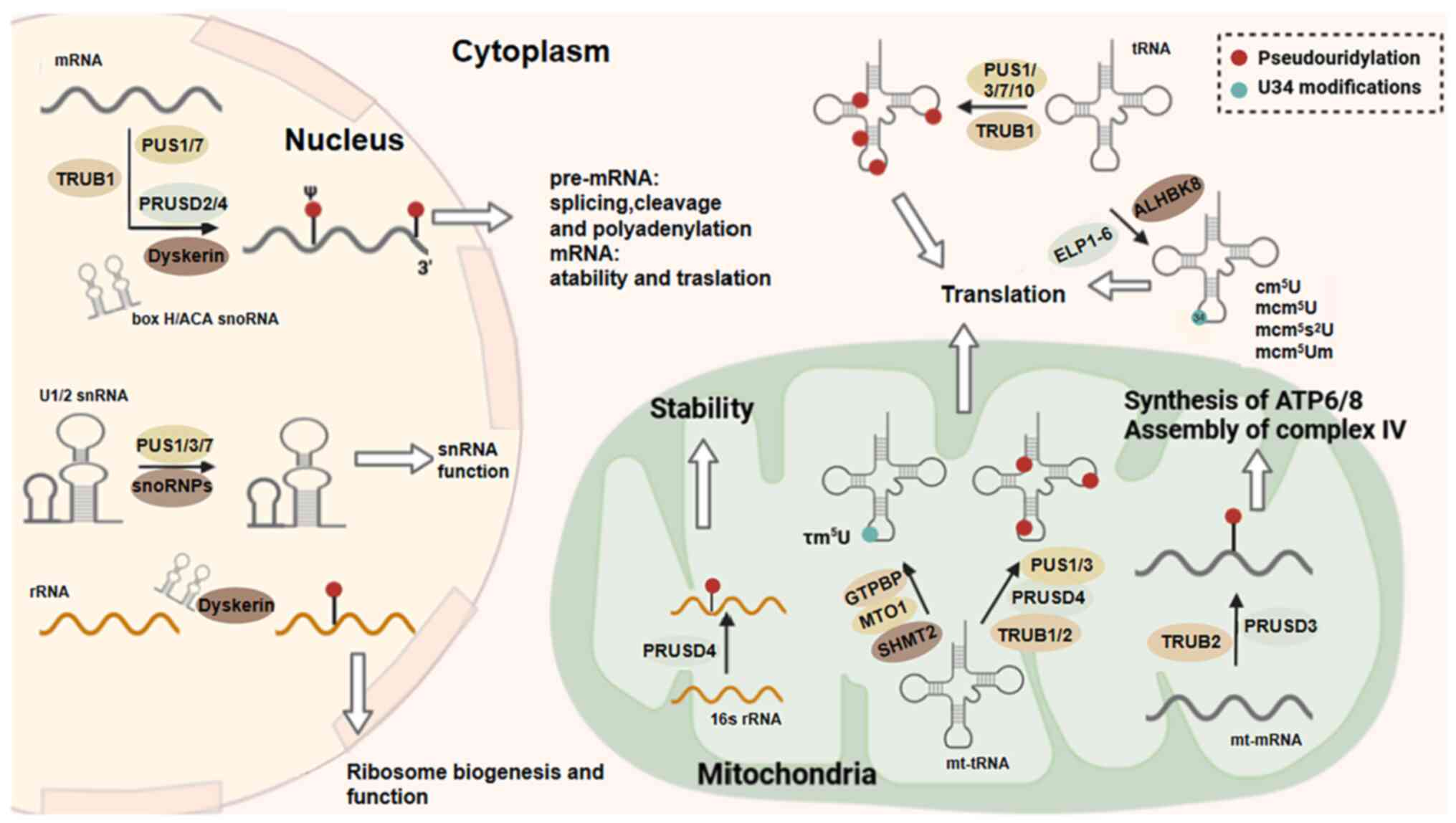
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bokar JA, Shambaugh ME, Polayes D, Matera
AG and Rottman FM: Purification and cDNA cloning of the
AdoMet-binding subunit of the human mRNA
(N-6-adenosine)-methyltransferase. RNA. 3:1233–1247.
1997.PubMed/NCBI
|
|
18
|
Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou
Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, et al:
Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine
spermatogenesis. Cell Res. 27:1216–1230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
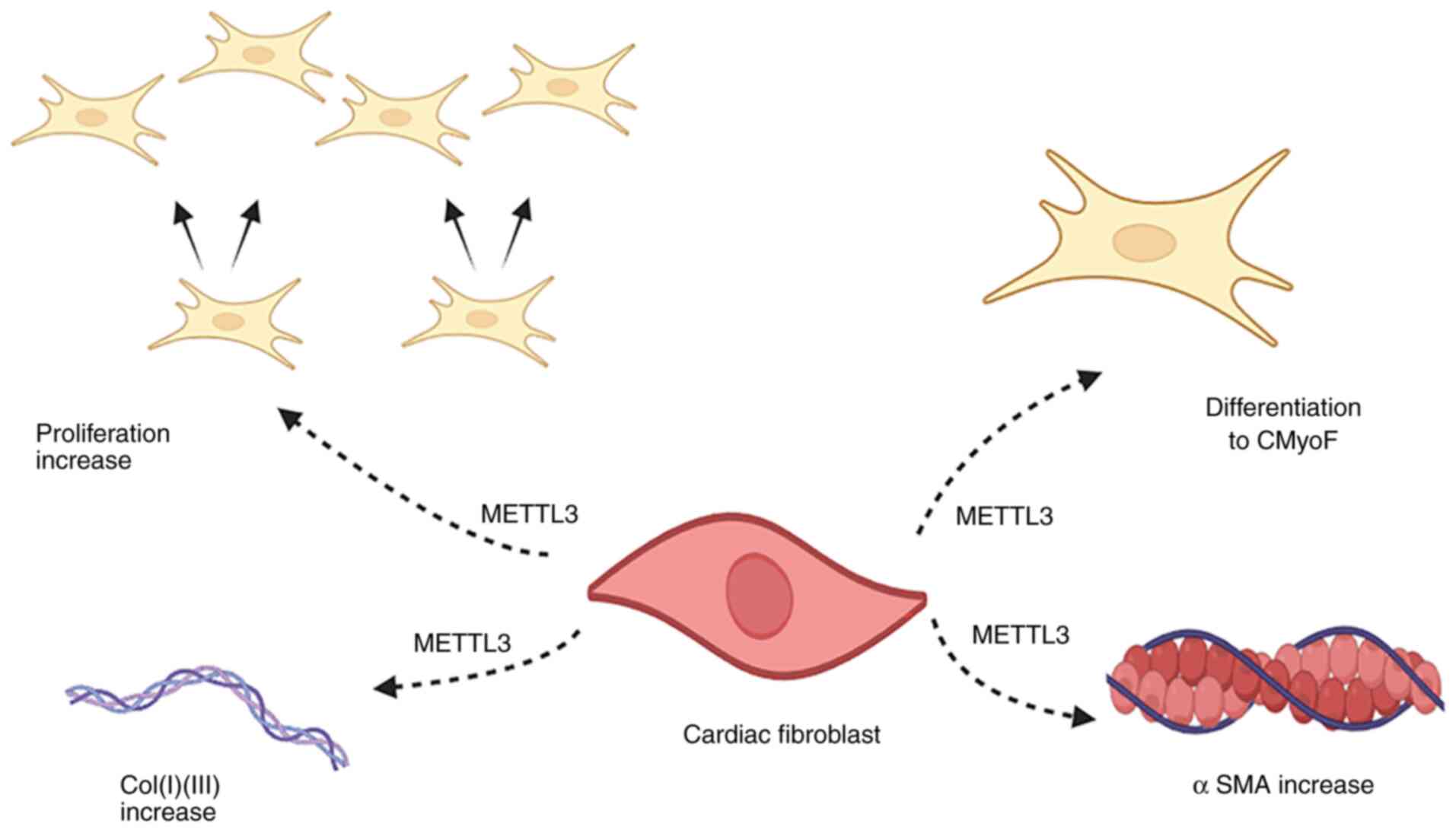
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
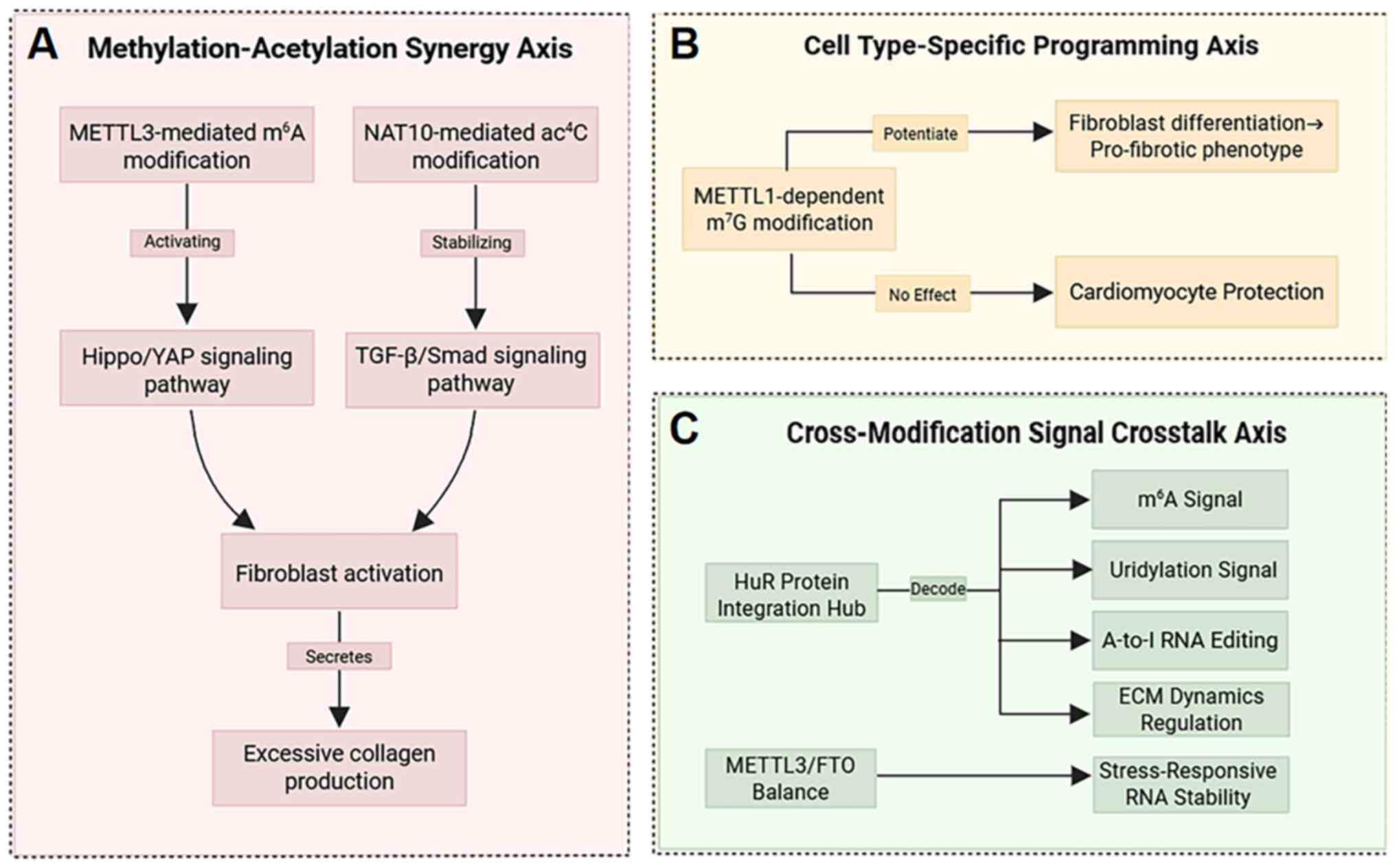
|
20
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D,
Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Structural basis of
N6-adenosine methylation by the METTL3-METTL14 complex. Nature.
534:575–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sledz P and Jinek M: Structural insights
into the molecular mechanism of the m6A writer complex. Elife.
5:e184342016. View Article : Google Scholar :
|
|
23
|
Liu JZ, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
24
|
Huang Q, Mo J, Liao Z, Chen X and Zhang B:
The RNA m6A writer WTAP in diseases: Structure, roles, and
mechanisms. Cell Death Dis. 13:8522022. View Article : Google Scholar :
|
|
25
|
Yue YA, Liu J, Cui X, Cao J, Luo G, Zhang
Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential
m6A mRNA methylation in 3'UTR and near stop codon and associates
with alternative polyadenylation. Cell Discov. 4:102018. View Article : Google Scholar
|
|
26
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m6A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Zhang L, Ren H, Ma L, Guo J, Mao
D, Lu Z, Lu L and Yan D: Role of Hakai in m6A modification pathway
in Drosophila. Nat Commun. 12:21592021. View Article : Google Scholar :
|
|
28
|
Knuckles P, Lence T, Haussmann IU, Jacob
D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, et
al: Zc3h13/Flacc is required for adenosine methylation by bridging
the mRNA-binding factor Rbm15/Spenito to the m6A machinery
component Wtap/Fl(2)d. Genes Dev. 32:415–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang XL, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du Y, Hou G, Zhang H, Dou J, He J, Guo Y,
Li L, Chen R, Wang Y, Deng R, et al: SUMOylation of the m6A-RNA
methyltransferase METTL3 modulates its function. Nucleic Acids Res.
46:5195–5208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu Z, Lv B, Qin Y and Zhang B: Emerging
roles and mechanism of m6A methylation in cardiometabolic diseases.
Cells. 11:11012022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao S, Sun H and Xu C: YTH Domain: A
family of N6-methyladenosine (m6A) Readers. Genomics Proteomics
Bioinformatics. 16:99–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schöller E, Weichmann F, Treiber T, Ringle
S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G:
Interactions, localization, and phosphorylation of the m6A
generating METTL3-METTL14-WTAP complex. RNA. 24:499–512. 2018.
View Article : Google Scholar
|
|
34
|
Zou S, Toh JD, Wong KH, Gao YG, Hong W and
Woon EC: N6-Methyladenosine: A conformational marker that regulates
the substrate specificity of human demethylases FTO and ALKBH5. Sci
Rep. 6:256772016. View Article : Google Scholar :
|
|
35
|
Grozhik AV, Olarerin-George AO, Sindelar
M, Li X, Gross SS and Jaffrey SR: Antibody cross-reactivity
accounts for widespread appearance of m1A in 5'UTRs. Nat Commun.
10:51262019. View Article : Google Scholar
|
|
36
|
Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo
T, Ishigami Y, Chemla Y, Blumberg A, Schlesinger O, Bieri P, Greber
B, et al: Mitochondrial 16S rRNA is methylated by tRNA
methyltransferase TRMT61B in all vertebrates. PLoS Biol.
14:e10025572016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chujo T and Suzuki T: Trmt61B is a
methyltransferase responsible for 1-methyladenosine at position 58
of human mitochondrial tRNAs. RNA. 18:2269–2276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vilardo E, Nachbagauer C, Buzet A,
Taschner A, Holzmann J and Rossmanith W: A subcomplex of human
mitochondrial RNase P is a bifunctional methyltransferase-extensive
moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res.
40:11583–11593. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li X, Xiong X, Wang K, Wang L, Shu X, Ma S
and Yi C: Transcriptome-wide mapping reveals reversible and dynamic
N(1)-methyladenosine methylome. Nat Chem Biol. 12:311–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J,
Lu Z, Zheng Z, Dai Q and Wang H: Transfer RNA demethylase ALKBH3
promotes cancer progression via induction of tRNA-derived small
RNAs. Nucleic Acids Res. 47:2533–2545. 2019. View Article : Google Scholar :
|
|
41
|
Liu F, Clark W, Luo G, Wang X, Fu Y, Wei
J, Wang X, Hao Z, Dai Q, Zheng G, et al: ALKBH1-Mediated tRNA
demethylation regulates translation. Cell. 167:816–828.e16. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wei JB, Liu F, Lu Z, Fei Q, Ai Y, He PC,
Shi H, Cui X, Su R, Klungland A, et al: Differential m6A, m6Am, and
m1A Demethylation Mediated by FTO in the cell nucleus and
cytoplasm. Mol Cell. 71:973–985.e5. 2018. View Article : Google Scholar
|
|
43
|
Qin G, Sun Y-W, Guo Y-D, Li J-W, Liang C,
Feng H and Yong S: SND1 recognize the methylation sites of TINCR to
promote growth of keloid fibroblasts. Med J Chin PLA. 46:1068–1076.
2021.In Chinese.
|
|
44
|
Guo GQ, Pan K, Fang S, Ye L, Tong X, Wang
Z, Xue X and Zhang H: Advances in mRNA 5-methylcytosine
modifications: Detection, effectors, biological functions, and
clinical relevance. Mol Ther Nucleic Acids. 26:575–593. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Squires JE, Patel HR, Nousch M, Sibbritt
T, Humphreys DT, Parker BJ, Suter CM and Preiss T: Widespread
occurrence of 5-methylcytosine in human coding and non-coding RNA.
Nucleic Acids Res. 40:5023–5033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang T, Chen W, Liu J, Gu N and Zhang R:
Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat
Struct Mol Biol. 26:380–388. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Van Haute L, Dietmann S, Kremer L, Hussain
S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S,
et al: Deficient methylation and formylation of mt-tRNA (Met)
wobble cytosine in a patient carrying mutations in NSUN3. Nat
Commun. 7:120392016. View Article : Google Scholar
|
|
48
|
Liu RJ, Long T, Li J, Li H and Wang ED:
Structural basis for substrate binding and catalytic mechanism of a
human RNA:m5C methyltransferase NSun6. Nucleic Acids Res.
45:6684–6697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang ZX, Li J, Xiong QP, Li H, Wang ED
and Liu RJ: Position 34 of tRNA is a discriminative element for
m5C38 modification by human DNMT2. Nucleic Acids Res.
49:13045–13061. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Janin M, Ortiz-Barahona V, de Moura MC,
Martínez-Cardús A, Llinàs-Arias P, Soler M, Nachmani D, Pelletier
J, Schumann U, Calleja-Cervantes ME, et al: Epigenetic loss of
RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a
stress adaptive translational program. Acta Neuropathol.
138:1053–1074. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liao H, Gaur A, McConie H, Shekar A, Wang
K, Chang JT, Breton G and Denicourt C: Human NOP2/NSUN1 regulates
ribosome biogenesis through non-catalytic complex formation with
box C/D snoRNPs. Nucleic Acids Res. 50:10695–10716. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang L, Ren Z, Yan S, Zhao L, Liu J, Zhao
L, Li Z, Ye S, Liu A, Li X, et al: Nsun4 and Mettl3 mediated
translational reprogramming of Sox9 promotes BMSC chondrogenic
differentiation. Commun Biol. 5:4952022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Spåhr H, Habermann B, Gustafsson CM,
Larsson NG and Hallberg BM: Structure of the human MTERF4-NSUN4
protein complex that regulates mitochondrial ribosome biogenesis.
Proc Natl Acad Sci USA. 109:15253–15258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dai X, Gonzalez G, Li L, Li J, You C, Miao
W, Hu J, Fu L, Zhao Y, Li R, et al: YTHDF2 Binds to
5-Methylcytosine in RNA and modulates the maturation of ribosomal
RNA. Anal Chem. 92:1346–1354. 2020. View Article : Google Scholar :
|
|
55
|
Xue C, Gu X, Zheng Q, Shi Q, Yuan X, Su Y,
Jia J, Jiang J, Lu J and Li L: ALYREF mediates RNA m5C modification
to promote hepatocellular carcinoma progression. Signal Transduct
Target Ther. 8:1302023. View Article : Google Scholar :
|
|
56
|
Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu
Y, Li Z, Li X, Zhao K, Wang C, et al: Tet2 promotes pathogen
infection-induced myelopoiesis through mRNA oxidation. Nature.
554:123–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang WL, Qiu W, Zhang T, Xu K, Gu ZJ, Zhou
Y, Xu HJ, Yang ZZ, Shen B, Zhao YL, et al: Nsun2 coupling with
RoRγt shapes the fate of Th17 cells and promotes colitis. Nat
Commun. 14:8632023. View Article : Google Scholar
|
|
58
|
Muthukrishnan S, Both GW, Furuichi Y and
Shatkin AJ: 5'-Terminal 7-methylguanosine in eukaryotic mRNA is
required for translation. Nature. 255:33–37. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Adams JM and Cory S: Modified nucleosides
and bizarre 5'-termini in mouse myeloma mRNA. Nature. 255:28–33.
1975. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Létoquarta J, Huvelle E, Wacheul L,
Bourgeois G, Zorbas C, Graille M, Heurgué-Hamard V and Lafontaine
DL: Structural and functional studies of Bud23-Trm112 reveal 18S
rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes.
Proc Natl Acad Sci USA. 111:E5518–E5526. 2014.
|
|
61
|
Aregger M, Kaskar A, Varshney D,
Fernandez-Sanchez ME, Inesta-Vaquera FA, Weidlich S and Cowling VH:
CDK1-Cyclin B1 activates RNMT, coordinating mRNA Cap methylation
with G1 phase transcription. Mol Cell. 61:734–746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pandolfini L, Barbieri I, Bannister AJ,
Hendrick A, Andrews B, Webster N, Murat P, Mach P, Brandi R, Robson
SC, et al: METTL1 Promotes let-7 MicroRNA processing via m7G
methylation. Mol Cell. 74:1278–1290.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Dai ZH, Liu H, Liao J, Huang C, Ren X, Zhu
W, Zhu S, Peng B, Li S, Lai J, et al: N7-Methylguanosine tRNA
modification enhances oncogenic mRNA translation and promotes
intrahepatic cholangiocarcinoma progression. Mol Cell.
81:3339–3355.e8. 2021. View Article : Google Scholar
|
|
64
|
Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo
G, Zhang Z, Zhang L, Hu L, Dong X and He C: Transcriptome-wide
mapping of internal N7-methylguanosine methylome in mammalian mRNA.
Mol Cell. 74:1304–1316.e8. 2019. View Article : Google Scholar
|
|
65
|
Furuichi Y, LaFiandra A and Shatkin AJ:
5'-Terminal structure and mRNA stability. Nature. 266:235–239.
1977. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Konarska MM, Padgett RA and Sharp PA:
Recognition of cap structure in splicing in vitro of mRNA
precursors. Cell. 38:731–736. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hsu CL and Stevens A: Yeast-cells lacking
5'-3' exoribonuclease-1 contain mRNA species that are poly(A)
deficient and partially lack the 5' cap structure. Mol Cell Biol.
13:4826–4835. 1993.PubMed/NCBI
|
|
68
|
Ma JY, Han H, Huang Y, Yang C, Zheng S,
Cai T, Bi J, Huang X, Liu R, Huang L, et al: METTL1/WDR4-mediated
m7G tRNA modifications and m7G codon usage promote mRNA translation
and lung cancer progression. Mol Ther. 29:3422–3435. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lin S, Liu Q, Lelyveld VS, Choe J, Szostak
JW and Gregory RI: Mettl1/Wdr4-Mediated m7G tRNA methylome is
required for normal mRNA translation and embryonic stem cell
self-renewal and differentiation. Mol Cell. 71:244–255.e5. 2018.
View Article : Google Scholar :
|
|
70
|
Ito S, Horikawa S and Suzuki T, Kawauchi
H, Tanaka Y and Suzuki T and Suzuki T: Human NAT10 Is an
ATP-dependent RNA acetyltransferase responsible for
N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA). J Biol
Chem. 289:35724–35730. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Arango D, Sturgill D, Alhusaini N, Dillman
AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler
MD, et al: Acetylation of cytidine in mRNA promotes translation
efficiency. Cell. 175:1872–1886.e24. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Arango D, Sturgill D, Yang R, Kanai T,
Bauer P, Roy J, Wang Z, Hosogane M, Schiffers S and Oberdoerffer S:
Direct epitranscriptomic regulation of mammalian translation
initiation through N4-acetylcytidine. Mol Cell. 82:2797–2814.e11.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yu XM, Li SJ, Yao ZT, Xu JJ, Zheng CC, Liu
ZC, Ding PB, Jiang ZL, Wei X, Zhao LP, et al: N4-acetylcytidine
modification of lncRNA CTC-490G23.2 promotes cancer metastasis
through interacting with PTBP1 to increase CD44 alternative
splicing. Oncogene. 42:1101–1116. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yang W, Li HY, Wu YF, Mi RJ, Liu WZ, Shen
X, Lu YX, Jiang YH, Ma MJ and Shen HY: ac4C acetylation of RUNX2
catalyzed by NAT10 spurs osteogenesis of BMSCs and prevents
ovariectomy-induced bone loss. Mol Ther Nucleic Acids. 26:135–147.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang GP, Zhang M, Zhang Y, Xie Y, Zou J,
Zhong J, Zheng Z, Zhou X, Zheng Y, Chen B and Liu C: NAT10-mediated
mRNA N4-acetylcytidine modification promotes bladder cancer
progression. Clin Transl Med. 12:e7382022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Feng ZY, Li K, Qin K, Liang J, Shi M, Ma
Y, Zhao S, Liang H, Han D, Shen B, et al: Retraction Note: The
LINC00623/NAT10 signaling axis promotes pancreatic cancer
progression by remodeling ac4C modification of mRNA. J Hematol
Oncol. 16:1092023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zheng X, Wang Q, Zhou Y, Zhang D, Geng Y,
Hu W, Wu C, Shi Y and Jiang J: N-acetyltransferase 10 promotes
colon cancer progression by inhibiting ferroptosis through
N4-acetylation and stabilization of ferroptosis suppressor protein
1 (FSP1) mRNA. Cancer Commun (Lond). 42:1347–1366. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jin C, Wang T, Zhang D, Yang P, Zhang C,
Peng W, Jin K, Wang L, Zhou J, Peng C, et al: Acetyltransferase
NAT10 regulates the Wnt/β-catenin signaling pathway to promote
colorectal cancer progression via ac4C acetylation of KIF23 mRNA. J
Exp Clin Cancer Res. 41:3452022. View Article : Google Scholar
|
|
79
|
Liao L, He Y, Li SJ, Yu XM, Liu ZC, Liang
YY, Yang H, Yang J, Zhang GG, Deng CM, et al: Lysine
2-hydroxyisobutyrylation of NAT10 promotes cancer metastasis in an
ac4C-dependent manner. Cell Res. 33:355–371. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nance KD, Gamage ST, Alam MM, Yang A, Levy
MJ, Link CN, Florens L, Washburn MP, Gu S, Oppenheim JJ and Meier
JL: Cytidine acetylation yields a hypoinflammatory synthetic
messenger RNA. Cell Chem Biol. 29:312–320.e7. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chang H, Lim J, Ha M and Kim VN: TAIL-seq:
Genome-wide determination of poly(A) tail length and 3' end
modifications. Mol Cell. 53:1044–1052. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lim J, Ha M, Chang H, Kwon SC, Simanshu
DK, Patel DJ and Kim VN: Uridylation by TUT4 and TUT7 marks mRNA
for degradation. Cell. 159:1365–1376. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yang A, Bofill-De Ros X, Stanton R, Shao
TJ, Villanueva P and Gu S: TENT2, TUT4, and TUT7 selectively
regulate miRNA sequence and abundance. Nat Commun. 13:52602022.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho
J, Yeom KH, Han J and Kim VN: TUT4 in Concert with Lin28 suppresses
MicroRNA biogenesis through Pre-MicroRNA Uridylation. Cell.
138:696–708. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yi H, Park J, Ha M, Lim J, Chang H and Kim
VN: PABP Cooperates with the CCR4-NOT complex to promote mRNA
deadenylation and block precocious decay. Mol Cell.
70:1081–1088.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Mullen TE and Marzluff WF: Degradation of
histone mRNA requires oligouridylation followed by decapping and
simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'.
Genes Dev. 22:50–65. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Schmidt MJ, West S and Norbury CJ: The
human cytoplasmic RNA terminal U-transferase ZCCHC11 targets
histone mRNAs for degradation. RNA. 17:39–44. 2011. View Article : Google Scholar :
|
|
88
|
Malecki M, Viegas SC, Carneiro T, Golik P,
Dressaire C, Ferreira MG and Arraiano CM: The exoribonuclease
Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J.
32:1842–1854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ustianenko D, Pasulka J, Feketova Z,
Bednarik L, Zigackova D, Fortova A, Zavolan M and Vanacova S:
TUT-DIS3L2 is a mammalian surveillance pathway for aberrant
structured non-coding RNAs. EMBO J. 35:2179–2191. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Koppers-Lalic D, Hackenberg M, Bijnsdorp
IV, van Eijndhoven MAJ, Sadek P, Sie D, Zini N, Middeldorp JM,
Ylstra B, de Menezes RX, et al: Nontemplated nucleotide additions
distinguish the small RNA composition in cells from exosomes. Cell
Rep. 8:1649–1658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Liu X, Zheng Q, Vrettos N, Maragkakis M,
Alexiou P, Gregory BD and Mourelatos Z: A MicroRNA precursor
surveillance system in quality control of MicroRNA synthesis. Mol
Cell. 55:868–879. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ripin N, Boudet J, Duszczyk MM, Hinniger
A, Faller M, Krepl M, Gadi A, Schneider RJ, Šponer J, Meisner-Kober
NC and Allain FH: Molecular basis for AU-rich element recognition
and dimerization by the HuR C-terminal RRM. Proc Natl Acad Sci USA.
116:2935–2944. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Loh XY, Sun QY, Ding LW, Mayakonda A,
Venkatachalam N, Yeo MS, Silva TC, Xiao JF, Doan NB, Said JW, et
al: RNA-binding protein ZFP36L1 suppresses hypoxia and cell-cycle
signaling. Cancer Res. 80:219–233. 2020. View Article : Google Scholar
|
|
94
|
Rataj F, Planel S, Denis J, Roelants C,
Filhol O, Guyon L, Feige JJ and Cherradi N: Targeting AU-rich
element-mediated mRNA decay with a truncated active form of the
zinc-finger protein TIS11b/BRF1 impairs major hallmarks of mammary
tumorigenesis. Oncogene. 38:5174–5190. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gu L, Wang H, Wang J, Guo Y, Tang Y, Mao
Y, Chen L, Lou H and Ji G: Reconstitution of HuR-Inhibited CUGBP1
expression protects cardiomyocytes from acute myocardial
infarction-induced injury. Antioxid Redox Signal. 27:1013–1026.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Rau F, Freyermuth F, Fugier C, Villemin
JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, et
al: Misregulation of miR-1 processing is associated with heart
defects in myotonic dystrophy. Nat Struct Mol Biol. 18:840–845.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon
SC, Chang H and Kim VN: Mono-Uridylation of Pre-MicroRNA as a key
step in the biogenesis of group II let-7 MicroRNAs. Cell.
151:521–532. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ansari MY, Khan NM, Ahmad N, Green J,
Novak K and Haqqi TM: Genetic Inactivation of ZCCHC6 suppresses
interleukin-6 expression and reduces the severity of experimental
osteoarthritis in mice. Arthritis Rheumatol. 71:583–593. 2019.
View Article : Google Scholar :
|
|
99
|
Nishikura K: A-to-I editing of coding and
non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 17:83–96. 2016.
View Article : Google Scholar :
|
|
100
|
Chung HC, Calis JJA, Wu X, Sun T, Yu Y,
Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR and
Rice CM: Human ADAR1 prevents endogenous RNA from triggering
translational shutdown. Cell. 172:811–824.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yang CC, Chen YT, Chang YF, Liu H, Kuo YP,
Shih CT, Liao WC, Chen HW, Tsai WS and Tan BC: ADAR1-mediated 3'
UTR editing and expression control of antiapoptosis genes
fine-tunes cellular apoptosis response. Cell Death Dis.
8:e28332017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Chalk AM, Taylor S, Heraud-Farlow JE and
Walkley CR: The majority of A-to-I RNA editing is not required for
mammalian homeostasis. Genome Biol. 20:2682019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wu X, Wang L, Wang K, Li J, Chen R, Wu X,
Ni G, Liu C, Das S, Sluijter JPG, et al: ADAR2 increases in
exercised heart and protects against myocardial infarction and
doxorubicin-induced cardiotoxicity. Mol Ther. 30:400–414. 2022.
View Article : Google Scholar :
|
|
104
|
Chen J, Liu HF, Qiao LB, Wang FB, Wang L,
Lin Y and Liu J: Global RNA editing identification and
characterization during human pluripotent-to-cardiomyocyte
differentiation. Mol Ther Nucleic Acids. 26:879–891. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang FJ, He J, Liu S, Gao A, Yang L, Sun
G, Ding W, Li CY, Gou F, He M, et al: A comprehensive RNA editome
reveals that edited Azin1 partners with DDX1 to enable
hematopoietic stem cell differentiation. Blood. 138:1939–1952.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Jiang L, Hao Y, Shao C, Wu Q, Prager BC,
Gimple RC, Sulli G, Kim LJ, Zhang G, Qiu Z, et al: ADAR1-mediated
RNA editing links ganglioside catabolism to glioblastoma stem cell
maintenance. J Clin Invest. 132:e1433972022. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Solomon O, Di Segni A, Cesarkas K, Porath
HT, Marcu-Malina V, Mizrahi O, Stern-Ginossar N, Kol N,
Farage-Barhom S, Glick-Saar E, et al: RNA editing by ADAR1 leads to
context-dependent transcriptome-wide changes in RNA secondary
structure. Nat Commun. 8:14402017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kokot KE, Kneuer JM, John D, Rebs S,
Möbius-Winkler MN, Erbe S, Müller M, Andritschke M, Gaul S, Sheikh
BN, et al: Reduction of A-to-I RNA editing in the failing human
heart regulates formation of circular RNAs. Basic Res Cardiol.
117:322022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Liddicoat BJ, Piskol R, Chalk AM,
Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH and Walkley
CR: RNA EDITING RNA editing by ADAR1 prevents MDA5 sensing of
endogenous dsRNA as nonself. Science. 349:1115–1120. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Garcia-Gonzalez C, Dieterich C, Maroli G,
Wiesnet M, Wietelmann A, Li X, Yuan X, Graumann J, Stellos K, Kubin
T, et al: ADAR1 prevents autoinflammatory processes in the heart
mediated by IRF7. Circ Res. 131:580–597. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
de Reuver R, Verdonck S, Dierick E,
Nemegeer J, Hessmann E, Ahmad S, Jans M, Blancke G, Van
Nieuwerburgh F, Botzki A, et al: ADAR1 prevents autoinflammation by
suppressing spontaneous ZBP1 activation. Nature. 607:784–789. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Chan TH, Lin CH, Qi L, Fei J, Li Y, Yong
KJ, Liu M, Song Y, Chow RK, Ng VH, et al: A disrupted RNA editing
balance mediated by ADARs (Adenosine DeAminases that act on RNA) in
human hepatocellular carcinoma. Gut. 63:832–843. 2014. View Article : Google Scholar
|
|
113
|
Chan TH, Qamra A, Tan KT, Guo J, Yang H,
Qi L, Lin JS, Ng VH, Song Y, Hong H, et al: ADAR-Mediated RNA
editing predicts progression and prognosis of gastric cancer.
Gastroenterology. 151:637–650.e10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Shen H, An O, Ren X, Song Y, Tang SJ, Ke
XY, Han J, Tay DJT, Ng VHE, Molias FB, et al: ADARs act as potent
regulators of circular transcriptome in cancer. Nat Commun.
13:15082022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Tan MH, Li Q, Shanmugam R, Piskol R,
Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, et
al: Dynamic landscape and regulation of RNA editing in mammals.
Nature. 550:249–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Karijolich J, Yi CQ and Yu YT:
Transcriptome-wide dynamics of RNA pseudouridylation. Nat Rev Mol
Cell Biol. 16:581–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Davis DR: Stabilization of RNA stacking by
pseudouridine. Nucleic Acids Res. 23:5020–5026. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Zhao BS and He C: Pseudouridine in a new
era of RNA modifications. Cell Res. 25:153–154. 2015. View Article : Google Scholar :
|
|
119
|
Li X, Zhu P, Ma S, Song J, Bai J, Sun F
and Yi C: Chemical pulldown reveals dynamic pseudouridylation of
the mammalian transcriptome. Nat Chem Biol. 11:592–597. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Guegueniat J, Halabelian L, Zeng H, Dong
A, Li Y, Wu H, Arrowsmith CH and Kothe U: The human pseudouridine
synthase PUS7 recognizes RNA with an extended multi-domain binding
surface. Nucleic Acids Res. 49:11810–11822. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ni J, Tien AL and Fournier MJ: Small
nucleolar RNAs direct site-specific synthesis of pseudouridine in
ribosomal RNA. Cell. 89:565–573. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ruggero D, Grisendi S, Piazza F, Rego E,
Mari F, Rao PH, Cordon-Cardo C and Pandolfi PP: Dyskeratosis
congenita and cancer in mice deficient in ribosomal RNA
modification. Science. 299:259–262. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Li L and Ye K: Crystal structure of an
H/ACA box ribonucleoprotein particle. Nature. 443:302–307. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
McCleverty CJ, Hornsby M, Spraggon G and
Kreusch A: Crystal structure of human Pus10, a novel pseudouridine
synthase. J Mol Biol. 373:1243–1254. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Song J, Zhuang Y, Zhu C, Meng H, Lu B, Xie
B, Peng J, Li M and Yi C: Differential roles of human PUS10 in
miRNA processing and tRNA pseudouridylation. Nat Chem Biol.
16:160–169. 2020. View Article : Google Scholar
|
|
126
|
Bykhovskaya Y, Casas K, Mengesha E, Inbal
A and Fischel-Ghodsian N: Missense mutation in pseudouridine
synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic
anemia (MLASA). Am J Hum Genet. 74:1303–1308. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Jia Z, Meng F, Chen H, Zhu G, Li X, He Y,
Zhang L, He X, Zhan H, Chen M, et al: Human TRUB1 is a highly
conserved pseudouridine synthase responsible for the formation of
ψ55 in mitochondrial tRNAAsn, tRNAGln, tRNAGlu and tRNAPro. Nucleic
Acids Res. 50:9368–9381. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Shaheen R, Han L, Faqeih E, Ewida N,
Alobeid E, Phizicky EM and Alkuraya FS: A homozygous truncating
mutation in PUS3 expands the role of tRNA modification in normal
cognition. Hum Genet. 135:707–713. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lin TY, Smigiel R, Kuzniewska B,
Chmielewska JJ, Kosińska J, Biela M, Biela A, Kościelniak A, Dobosz
D, Laczmanska I, et al: Destabilization of mutated human PUS3
protein causes intellectual disability. Hum Mutat. 43:2063–2078.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Cui Q, Yin K, Zhang X, Ye P, Chen X, Chao
J, Meng H, Wei J, Roeth D, Li L, et al: Targeting PUS7 suppresses
tRNA pseudouridylation and glioblastoma tumorigenesis. Nat Cancer.
2:932–949. 2021. View Article : Google Scholar
|
|
131
|
Eyler DE, Franco MK, Batool Z, Wu MZ,
Dubuke ML, Dobosz-Bartoszek M, Jones JD, Polikanov YS, Roy B and
Koutmou KS: Pseudouridinylation of mRNA coding sequences alters
translation. Proc Natl Acad Sci USA. 116:23068–23074. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Dai Q, Zhang LS, Sun HL, Pajdzik K, Yang
L, Ye C, Ju CW, Liu S, Wang Y, Zheng Z, et al: Quantitative
sequencing using BID-seq uncovers abundant pseudouridines in
mammalian mRNA at base resolution. Nat Biotechnol. 41:344–354.
2023. View Article : Google Scholar :
|
|
133
|
Karijolich J and Yu YT: Converting
nonsense codons into sense codons by targeted pseudouridylation.
Nature. 474:395–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Anderson BR, Muramatsu H, Nallagatla SR,
Bevilacqua PC, Sansing LH, Weissman D and Karikó K: Incorporation
of pseudouridine into mRNA enhances translation by diminishing PKR
activation. Nucleic Acids Res. 38:5884–5892. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Martinez NM, Su A, Burns MC, Nussbacher
JK, Schaening C, Sathe S, Yeo GW and Gilbert WV: Pseudouridine
synthases modify human pre-mRNA co-transcriptionally and affect
pre-mRNA processing. Mol Cell. 82:645–659.e9. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Rapino F, Zhou Z, Roncero Sanchez AM,
Joiret M, Seca C, El Hachem N, Valenti G, Latini S, Shostak K,
Geris L, et al: Wobble tRNA modification and hydrophilic amino acid
patterns dictate protein fate. Nat Commun. 12:21702021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Rosu A, El Hachem N, Rapino F,
Rouault-Pierre K, Jorssen J, Somja J, Ramery E, Thiry M, Nguyen L,
Jacquemyn M, et al: Loss of tRNA-modifying enzyme Elp3 activates a
p53-dependent antitumor checkpoint in hematopoiesis. J Exp Med.
218:e202006622021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Cheng H, Li L, Xue J, Ma J and Ge J: TNC
accelerates hypoxia-induced cardiac injury in a METTL3-dependent
manner. Genes (Basel). 14:5912023. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zhuang Y, Li T, Hu X, Xie Y, Pei X, Wang
C, Li Y, Liu J, Tian Z, Zhang X, et al: MetBil as a novel molecular
regulator in ischemia-induced cardiac fibrosis via METTL3-mediated
m6A modification. FASEB J. 37:e227972023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Cai X, Zou P, Hong L, Chen Y, Zhan Y, Liu
Y and Shao L: RNA methylation reading protein YTHDF2 relieves
myocardial ischemia-reperfusion injury by downregulating BNIP3 via
m6A modification. Hum Cell. 36:1948–1964. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Ding JF, Sun H, Song K, Zhou Y, Tu B, Shi
KH, Lu D, Xu SS and Tao H: IGFBP3 epigenetic promotion induced by
METTL3 boosts cardiac fibroblast activation and fibrosis. Eur J
Pharmacol. 942:1754942023. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Liu ZY, You QY, Liu ZY, Lin LC, Yang JJ
and Tao H: m6A control programmed cell death in cardiac fibrosis.
Life Sci. 353:1229222024. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wang L, Zhou J, Kong L, Ying G, Sha J, Yi
D, Zeng J, Xiong W and Wen T: Fibroblast-specific knockout of
METTL1 attenuates myocardial infarction-induced cardiac fibrosis.
Life Sci. 329:1219262023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Kurian L and Brandes RP: RNA modification
that breaks the heart: RNA Acetylase Nat10 promotes fibrosis. Circ
Res. 133:1003–1005. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Wang XX, Zhao YM, Zhang QY, Zhao JX, Yin
DH, Zhang ZZ, Jin XY, Li SN, Ji HY, Chen HY, et al: Acetylcytidine
modification of Amotl1 by N-acetyltransferase 10 contributes to
cardiac fibrotic expansion in mice after myocardial infarction.
Acta Pharmacol Sin. 45:1425–1437. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Hao Y, Li B, Yin F and Liu W: tRNA-derived
small RNA (tsr007330) regulates myocardial fibrosis after
myocardial infarction through NAT10-mediated ac4C acetylation of
EGR3 mRNA. Biochim Biophys Acta Mol Basis Dis. 1870:1672672024.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhou B, Perel P, Mensah GA and Ezzati M:
Global epidemiology, health burden and effective interventions for
elevated blood pressure and hypertension. Nat Rev Cardiol.
18:785–802. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Touyz RM, Alves-Lopes R, Rios FJ, Camargo
LL, Anagnostopoulou A, Arner A and Montezano AC: Vascular smooth
muscle contraction in hypertension. Cardiovasc Res. 114:529–539.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Jain M, Mann TD, Stulić M, Rao SP, Kirsch
A, Pullirsch D, Strobl X, Rath C, Reissig L, Moreth K, et al: RNA
editing of Filamin A pre-mRNA regulates vascular contraction and
diastolic blood pressure. EMBO J. 37:e948132018. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Marcadenti A, Fuchs FD, Matte U, Sperb F,
Moreira LB and Fuchs SC: Effects of RS9939906 and MC4R RS17782313
on obesity, type 2 diabetes mellitus and blood pressure in patients
with hypertension. Cardiovasc Diabetol. 12:1032013. View Article : Google Scholar
|
|
151
|
Liu S, Jiang X, Lu H, Xing M, Qiao Y,
Zhang C and Zhang W: HuR (Human Antigen R) regulates the
contraction of vascular smooth muscle and maintains blood pressure.
Arterioscler Thromb Vasc Biol. 40:943–957. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Klöss S, Rodenbach D, Bordel R and Mülsch
A: Human-antigen R (HuR) expression in hypertension-:
Downregulation of the mRNA stabilizing protein HuR in genetic
hypertension. Hypertension. 45:1200–1206. 2005. View Article : Google Scholar
|
|
153
|
Botros M, Fadah K and Mukherjee D: The
role of inflammatory response in the development of
atherosclerosis, myocardial infarction, and remodeling. Vessel
Plus. 8:312024.
|
|
154
|
Chien CS, Li JY, Chien Y, Wang ML,
Yarmishyn AA, Tsai PH, Juan CC, Nguyen P, Cheng HM, Huo TI, et al:
METTL3-dependent N6-methyladenosine RNA modification mediates the
atherogenic inflammatory cascades in vascular endothelium. Proc
Natl Acad Sci USA. 118:e20250701182021. View Article : Google Scholar :
|
|
155
|
Jian D, Wang Y, Jian L, Tang H, Rao L,
Chen K, Jia Z, Zhang W, Liu Y, Chen X, et al: METTL14 aggravates
endothelial inflammation and atherosclerosis by increasing FOXO1
N6-methyladeosine modifications. Theranostics. 10:8939–8956. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Zheng Y, Li Y, Ran X, Wang D, Zheng X,
Zhang M, Yu B, Sun Y and Wu J: Mettl14 mediates the inflammatory
response of macrophages in atherosclerosis through the NF-κB/IL-6
signaling pathway. Cell Mol Life Sci. 79:3112022. View Article : Google Scholar
|
|
157
|
Sun ZW, Chen W, Wang Z, Wang S, Zan J,
Zheng L and Zhao W: Matr3 reshapes m6A modification complex to
alleviate macrophage inflammation during atherosclerosis. Clin
Immunol. 245:1091762022. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Chen J, Ning Y, Zhang H, Song N, Gu Y, Shi
Y, Cai J, Ding X and Zhang X: METTL14-dependent m6A regulates
vascular calcification induced by indoxyl sulfate. Life Sci.
239:1170342019. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Zhou T, Han D, Liu J, Shi J, Zhu P, Wang Y
and Dong N: Factors influencing osteogenic differentiation of human
aortic valve interstitial cells. J Thorac Cardiovasc Surg.
161:e163–e185. 2021. View Article : Google Scholar
|
|
160
|
Wang L, Wang J, Yu P, Feng J, Xu GE, Zhao
X, Wang T, Lehmann HI, Li G, Sluijter JPG and Xiao J: METTL14 is
required for exercise-induced cardiac hypertrophy and protects
against myocardial ischemia-reperfusion injury. Nat Commun.
13:67622022. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Wang C, Hou X, Guan Q, Zhou H, Zhou L, Liu
L, Liu J, Li F, Li W and Liu H: RNA modification in cardiovascular
disease: Implications for therapeutic interventions. Signal
Transduct Target Ther. 8:4122023. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Yu S, Sun Z, Ju T, Liu Y, Mei Z, Wang C,
Qu Z, Li N, Wu F, Liu K, et al: The m7G Methyltransferase Mettl1
drives cardiac hypertrophy by regulating SRSF9-mediated splicing of
NFATc4. Adv Sci (Weinh). 11:e23087692024. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Maron MS and Maron BJ: Hypertrophic
cardiomyopathy-Authors reply. Lancet. 381:1457–1458. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Schultheiss HP, Fairweather D, Caforio
ALP, Escher F, Hershberger RE, Lipshultz SE, Liu PP, Matsumori A,
Mazzanti A, McMurray J and Priori SG: Dilated cardiomyopathy. Nat
Rev Dis Primers. 5:322019. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Muchtar E, Blauwet LA and Gertz MA:
Restrictive cardiomyopathy: Genetics, pathogenesis, clinical
manifestations, diagnosis, and therapy. Circ Res. 121:819–837.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Ghezzi D, Baruffini E, Haack TB,
Invernizzi F, Melchionda L, Dallabona C, Strom TM, Parini R,
Burlina AB, Meitinger T, et al: Mutations of the mitochondrial-tRNA
Modifier MTO1 cause hypertrophic cardiomyopathy and lactic
acidosis. Am J Hum Genet. 90:1079–1087. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Perks KL, Rossetti G, Kuznetsova I, Hughes
LA, Ermer JA, Ferreira N, Busch JD, Rudler DL, Spahr H, Schöndorf
T, et al: PTCD1 Is Required for 16S rRNA maturation complex
stability and mitochondrial ribosome assembly. Cell Rep.
23:127–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Gao S, Sun H, Chen K, Gu X, Chen H, Jiang
L, Chen L, Zhang S, Liu Y, Shi D, et al: Depletion of m6A reader
protein YTHDC1 induces dilated cardiomyopathy by abnormal splicing
of Titin. J Cell Mol Med. 25:10879–10891. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Meng L, Lin H, Huang X, Weng J, Peng F and
Wu S: METTL14 suppresses pyroptosis and diabetic cardiomyopathy by
downregulating TINCR lncRNA. Cell Death Dis. 13:382022. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Peng T, Liu M, Hu L, Guo D, Wang D, Qi B,
Ren G, Hu C, Zhang F, Chun HJ, et al: LncRNA Airn alleviates
diabetic cardiac fibrosis by inhibiting activation of cardiac
fibroblasts via a m6A-IMP2-p53 axis. Biol Direct. 17:322022.
View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Zhuang S, Ma Y, Zeng Y, Lu C, Yang F,
Jiang N, Ge J, Ju H, Zhong C, Wang J, et al: METTL14 promotes
doxorubicin-induced cardiomyocyte ferroptosis by regulating the
KCNQ1OT1-miR-7-5p-TFRC axis. Cell Biol Toxicol. 39:1015–1035. 2023.
View Article : Google Scholar
|
|
172
|
Nakamura M and Sadoshima J: Mechanisms of
physiological and pathological cardiac hypertrophy. Nat Rev
Cardiol. 15:387–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Berulava T, Buchholz E, Elerdashvili V,
Pena T, Islam MR, Lbik D, Mohamed BA, Renner A, von Lewinski D,
Sacherer M, et al: Changes in m6A RNA methylation contribute to
heart failure progression by modulating translation. Eur J Heart
Fail. 22:54–66. 2020. View Article : Google Scholar
|
|
174
|
Zhang B, Jiang H, Wu J, Cai Y, Dong Z,
Zhao Y, Hu Q, Hu K, Sun A and Ge J: m6A demethylase FTO attenuates
cardiac dysfunction by regulating glucose uptake and glycolysis in
mice with pressure overload-induced heart failure. Signal Transduct
Target Ther. 6:3772021. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Dorn LE, Lasman L, Chen J, Xu X, Hund TJ,
Medvedovic M, Hanna JH, van Berlo JH and Accornero F: The
N6-Methyladenosine mRNA Methylase METTL3 controls cardiac
homeostasis and hypertrophy. Circulation. 139:533–545. 2019.
View Article : Google Scholar :
|
|
176
|
Gao XQ, Zhang YH, Liu F, Ponnusamy M, Zhao
XM, Zhou LY, Zhai M, Liu CY, Li XM, Wang M, et al: The piRNA CHAPIR
regulates cardiac hypertrophy by controlling METTL3-dependent
N6-methyladenosine methylation of Parp10 mRNA. Nat Cell Biol.
22:1319–1331. 2020. View Article : Google Scholar : PubMed/NCBI
|