Introduction
Membrane proteins (MPs) are proteins integrated into
biological membranes, where they interact with lipid domains to
execute a range of essential functions within living organisms
(1). These proteins are key for
maintaining the structural integrity and functional operation of
cells, engaging in numerous processes such as ion transport, signal
transduction and regulation of cellular homeostasis (2). MPs are central to the regulation of
ion gradients across membranes, modulation of cellular
communication and responsiveness to extracellular signals (3). In addition to their physiological
roles, MPs also carry out a pivotal role in the development of
multidrug resistance (MDR), rendering them central to mechanisms of
therapeutic resistance (2).
Research has expanded the understanding of MPs, revealing that
their functions extend beyond basic cellular roles to include key
involvement in complex physiological processes and disease states,
particularly in different types of cancer such as colorectal,
ovarian, lung breast (1-5).
In the majority of mammalian cells, MPs assist in
maintaining membrane structure, facilitate the transport of ions,
nutrients and waste products or facilitate intercellular
communication (3). They are
integral to the formation of cellular microenvironments that enable
efficient communication between cells, regulating processes such as
immune responses, tissue remodeling and apoptosis (6). MPs are also involved in the
establishment of the blood-brain barrier, the regulation of
neurotransmitter transport and cellular metabolic processes
(7). This array of functions
underscores their key roles in maintaining cellular homeostasis and
regulating the essential biological processes necessary for
life.
Emerging research has emphasized the key role of MPs
in the pathogenesis of cancer, including breast cancer, colorectal
cancer, lung cancer, ovarian cancer and brain tumors (1,3-7).
For example, subfamilies of tyrosine kinase receptors, including
epidermal growth factor receptors (EGFRs) and fibroblast growth
factor receptors (FGFRs), are key in signaling pathways that
regulate cell proliferation, survival and migration, all important
processes that drive tumorigenesis (8,9).
These receptors become activated upon binding of ligands to their
respective receptors, inducing conformational changes that initiate
signal transduction to the cytoplasm via processes such as
dimerization (10). Upon
activation, receptors trigger downstream signaling cascades that
regulate gene expression, cell cycle progression and metabolic
reprogramming (11). In cancer
cells, dysregulation of receptor signaling contributes to tumor
growth, metastasis and resistance to chemotherapy, highlighting MPs
as potential therapeutic targets for the management of cancer,
including breast cancer and pancreatic cancer (12,13). MPs carry out a key role not only
in maintaining normal cellular activities but also in cancer
development and chemotherapy resistance (3-5).
The tyrosine kinase receptor family, including EGFR and FGFR, has
garnered increasing attention for its role in regulating cancer
cell proliferation, survival and migration, particularly in the
context of chemotherapy resistance (3-5,8,9).
Cancer cells often exploit dysregulated membrane protein signaling
pathways to drive tumor growth and metastasis, with resistance
mechanisms frequently associated with alterations in membrane
protein function (13-15). For example, discoidin domain
receptor 2 interactions with collagen type I promote breast cancer
cell proliferation and enhance resistance to chemotherapy,
highlighting its potential role in chemoresistance in breast cancer
(13-15). Moreover, transmembrane proteins
(TPs) such as transmembrane (TMEM) 45A, TMEM158, TMEM88, TMEM205
and TMEM16A have been identified as key mediators of
chemoresistance in several types of cancer (4,5,13,14). These proteins regulate cell
proliferation, inhibit apoptosis or modulate the tumor
microenvironment (TME) to increase resistance of cancer cells to
chemotherapeutic agents (1,4-6,13,14). Therefore, MPs not only serve as
key targets in cancer therapy but can also help in understanding
and addressing chemotherapy resistance mechanisms.
The complexity of the nexus of signal transduction
mediated by MPs presents substantial challenges in elucidating
their precise roles in cellular processes and disease mechanisms
(16). One of the primary
challenges in research on MPs lies in establishing the
structure-function relationship that governs ligand activation and
enzyme activity specificity (17). Structural studies of MPs have
been hindered by their amphipathic nature, which complicates their
isolation and analysis in vitro (1,3-5,18). However, recent advances in
cryo-electron microscopy (cryo-EM) and other high-resolution
imaging techniques have enabled the structural elucidation of key
MPs, offering insights into their functional mechanisms at the
atomic level (19). These
technological advances have markedly improved the ability to study
MPs in their natural environments, thus facilitating the
development of targeted drugs with high specificity and efficacy
(19,20).
MPs are not only essential for cellular function but
also act as key therapeutic targets, making them central to drug
discovery efforts (21). They
are involved in a range of physiological processes, many of which
can be modulated by therapeutic agents targeting MPs to achieve
clinical effects. Drug discovery efforts have concentrated on
identifying small molecules, large molecules, peptides,
polysaccharides and antibodies, particularly those that can
specifically interact with MPs to regulate their activity (22). For example, monoclonal antibodies
targeting EGFR and human epidermal growth factor receptor 2 (HER2)
have been developed successfully and are now used in the treatment
of several types of cancer, including glioma and breast cancer
(22,23). Large molecule drugs, including
polysaccharides and peptides, are seen as promising alternatives to
traditional small molecule drugs, offering various therapeutic
benefits such as reduced toxicity and enhanced efficacy when used
in combination with conventional chemotherapy. For example,
polysaccharides derived from medicinal plants such as
Anemarrhena asphodeloides and Houttuynia cordata have
demonstrated notable bioactivities, including immune modulation and
anti-inflammatory effects, which can enhance the effectiveness of
cancer treatments, including lung cancer and liver cancer (24,25). Furthermore, recent studies have
highlighted the ability of peptides to target specific cancer cell
pathways, enhancing treatment outcomes in several types of cancer,
including pancreatic cancer (PC) (14,26). These findings underscore the
potential of utilizing large-molecule drugs in cancer therapies,
particularly in addressing the challenge of chemoresistance.
Furthermore, several PMPs, including ion channels, transporters and
G protein-coupled receptors (GPCRs), are well-established
therapeutic targets for diseases ranging from cardiovascular
disorders to neurological conditions (27-29). Given the key role of MPs in the
pathophysiology of numerous diseases, understanding their
structural and functional properties is essential for the
development of novel drugs and therapies.
Biological processes, particularly those involving
MPs, constitute a complex, interconnected network within the cell.
Certain MPs can modulate interactions between various signaling
pathways that govern fundamental cellular processes, such as
metabolism, apoptosis and immune responses (30,31). The cyclic nature of these
networks is particularly key for understanding how drugs influence
cellular processes. The majority of therapeutic agents exert their
effects by binding to MPs, thereby modulating signaling pathways
essential for disease progression (32). Therefore, MPs serve as promising
targets for novel therapeutic strategies targeting specific disease
mechanisms at the molecular level. The expanding body of research
on MPs and their interactions with drugs continues to reveal novel
opportunities for drug development and personalized therapies
(21,33).
MPs are essential to cellular function and disease
pathogenesis (34). The study of
MPs is essential for advancing the understanding of both normal
physiology and disease mechanisms. Given their roles in signaling,
cellular maintenance and disease progression, MPs serve as key
targets for therapeutic intervention (7,12). Ongoing advancements in structural
biology, coupled with novel drug discovery technologies, highlight
novel opportunities for exploiting MPs as drug targets. Research on
MPs holds potential for developing innovative treatments for a
range of diseases, including cancer, cardiovascular disorders and
neurological conditions (35-37).
Structure and function of the TMEM
proteins
MPs are classified according to their interactions
with membranes, which are categorized into peripheral MPs, integral
MPs and lipid-anchored MPs (38). Integral MPs contain ≥1
transmembrane segment, commonly referred to as a TP (4). TMEM proteins represent a subgroup
of integral MPs, defined by their ability to traverse the lipid
bilayer, thus establishing stable anchors within the membrane.
These proteins are essential in the transport of molecules across
the cellular membrane (5,39).
A protein is classified as a member of the TMEM family if it
contains at least one predicted transmembrane domain that spans the
biological membrane, either partially (in monomeric form) or fully
(in multimeric form) (4).
Due to their amphipathic nature, members of the TMEM
protein family adopt specific secondary structures when
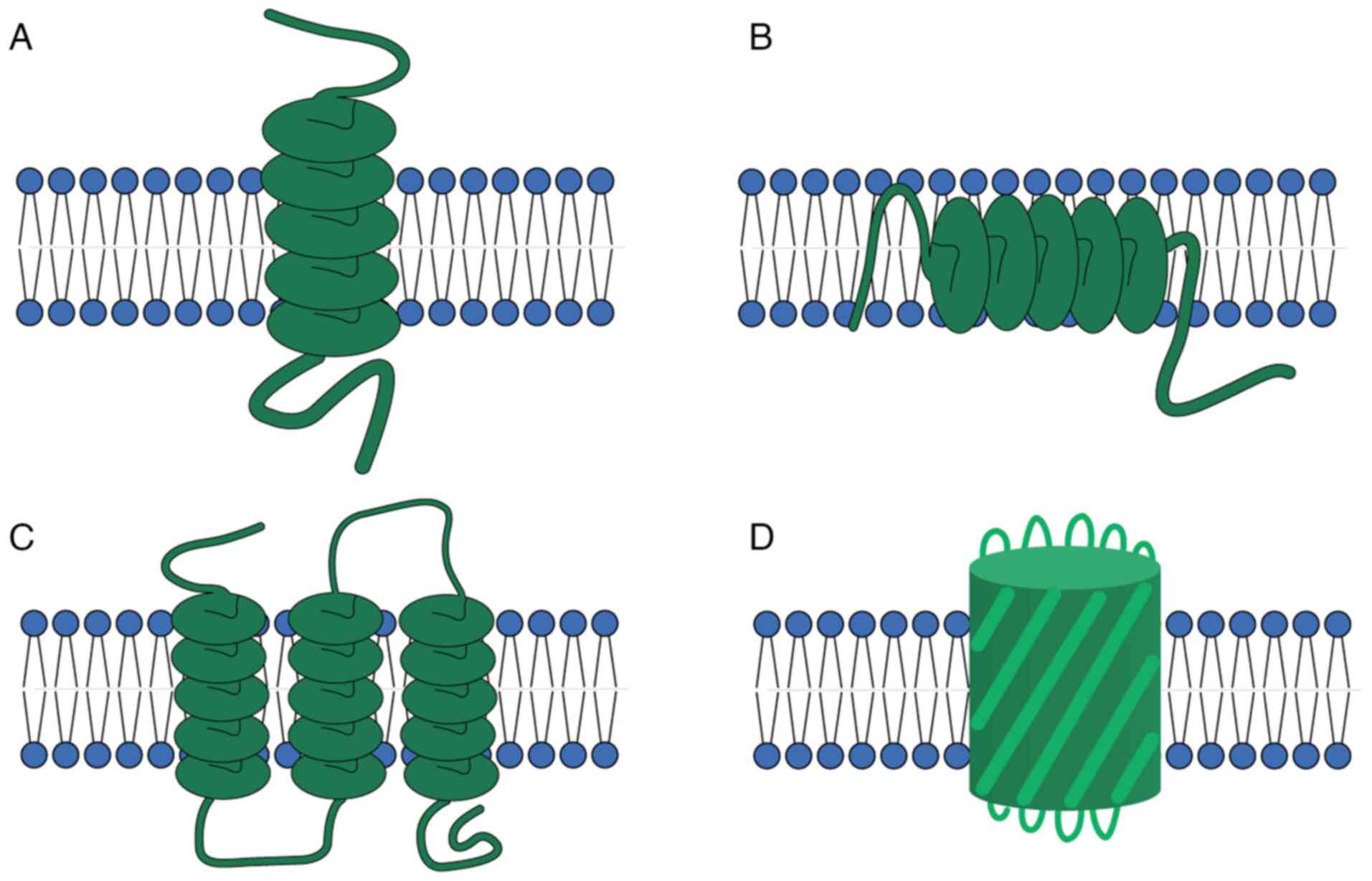
incorporation into the membrane (40,41). Based on their different
structures, TMEMs can be divided into two categories: α-helical
proteins and β-barrel proteins (Fig.
1). These two structures are the dominant structures of all
transmembrane proteins, with α-helical proteins being mostly found
in the cytoplasm and subcellular septum (Fig. 1A-C), while β-barrel proteins are
mostly found in chloroplasts, bacteria and mitochondrial membranes
(Fig. 1D) (40,41). These structural features allow
TMEM proteins to modulate essential processes, including nutrient
transport, waste removal and signal transduction (40,41).
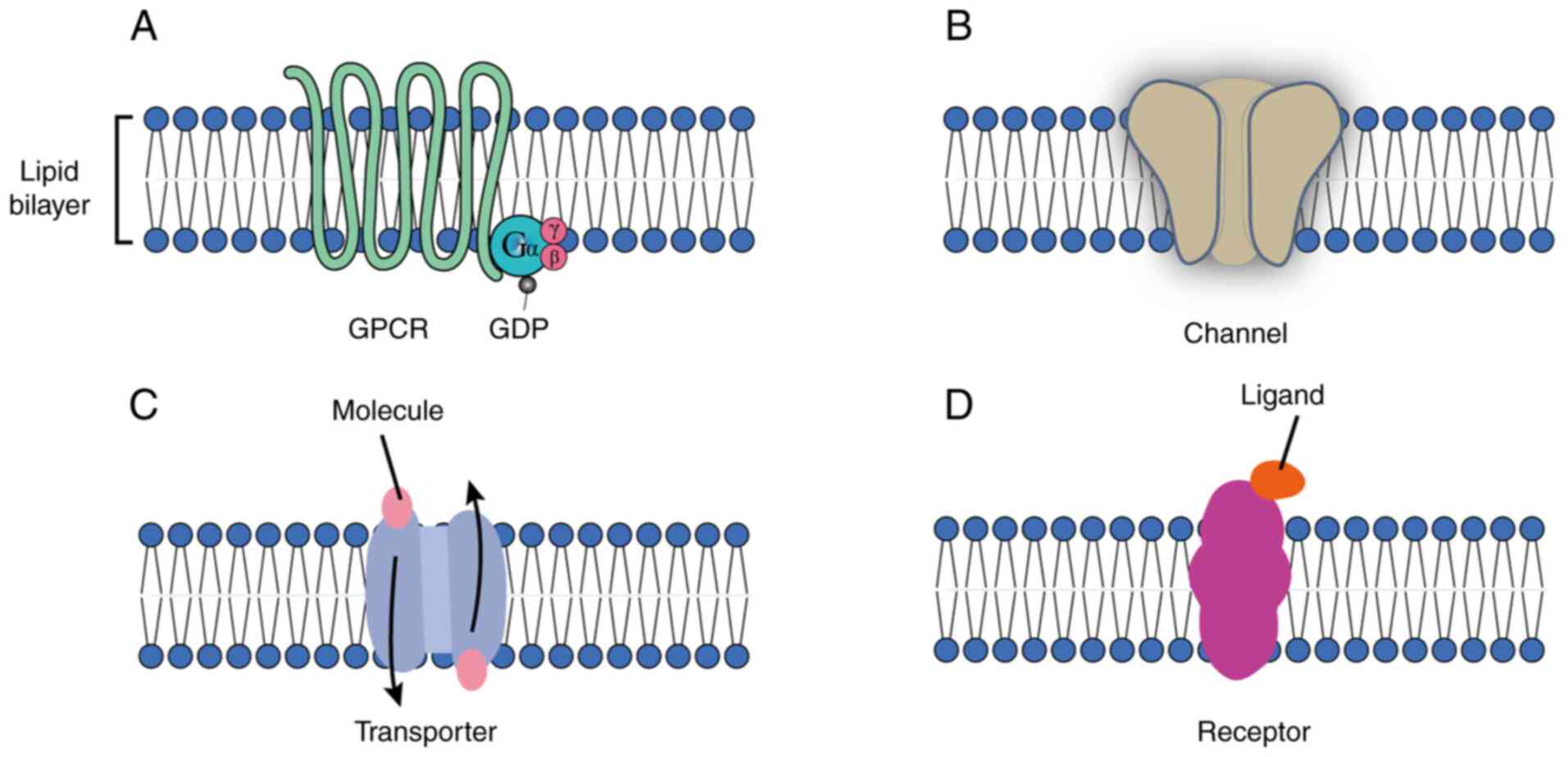
TMEM proteins exhibit considerable functional
diversity. These proteins are classified into distinct functional
groups, including GPCRs, ion channels, transporters, carriers and
other receptors (Fig. 2). GPCRs,
one of the largest and most versatile classes of TMEM proteins, are
involved in a range of signaling pathways that regulate essential
physiological processes, including sensory perception, immune
responses and cellular communication (42). Notably, TMEM proteins, including
GPCRs (Fig. 2A), carry out key
roles in cancer cell signaling, influencing tumor progression and
metastasis (43). Ion channels
(Fig. 2B) are TPs that form
pores, allowing ions (such as Na+, K+,
Ca2+ and Cl−) to passively flow across the
membrane. This movement is vital for processes such as nerve signal
transmission, muscle contraction and maintaining cellular ion
balance (44). Transporter
proteins (Fig. 2C) facilitate
the movement of molecules across the lipid bilayer, typically
against their concentration gradients. This process requires energy
and carries out a key role in nutrient uptake, waste removal and
the maintenance of cellular homeostasis (43,44). Dysfunction of these proteins is
implicated in a variety of diseases, including neurological and
cardiovascular disorders (43,44). TMEM proteins also function as
transmembrane receptors (Fig.
2D), such as the well-known EGFR, which is involved in
regulating cell growth, differentiation and survival. Dysregulation
of these transmembrane receptors is frequently associated with the
development and progression of various types of cancer, including
breast, lung, ovarian, cervical, bladder, esophageal, brain, and
head and neck cancer (45,46).
Previous studies have underscored the role of TMEM
proteins in various disease mechanisms, which highlights their
potential as therapeutic targets for drug development (47,48). For example, mutations in TMEM
proteins have been associated with several genetic disorders,
including cystic fibrosis, leading to defects in ion channels and
disruption of chloride transport (47,48). Moreover, TMEM proteins, such as
glucose transporters (GLUTs), carry out a pivotal role in cancer
metabolism by facilitating glucose uptake, therefore promoting
tumor growth, especially under nutrient-limited conditions
(49). This highlights the
potential of targeting TMEM proteins for therapeutic interventions
in a range of diseases.
The increasing understanding of the structural and
functional properties of TMEM proteins offers potential for drug
development. Their involvement in cellular signaling and molecular
transport makes them strong candidates for targeted therapies
(50). Advances in structural
biology, particularly the application of cryo-EM, have facilitated
the determination of the atomic structures of key TMEM proteins,
offering valuable insights into their functional mechanisms
(51-53). The TMEM family proteins are
essential components of the cellular membrane system, carrying out
key roles in cellular signaling and the transport of ions and small
molecules (39). Specific TMEM
proteins interact with hormone or neurotransmitter receptors,
inducing structural changes that initiate signaling cascades
important for cellular function. These proteins mediate the
selective transfer of ions or molecules across biological
membranes, thereby establishing concentration gradients essential
for cellular homeostasis and function (54). TMEM proteins are essential not
only to the plasma membrane but also to the membranes of various
intracellular organelles, including mitochondria, the endoplasmic
reticulum (ER), lysosomes and the Golgi apparatus, where they
participate in key biological processes (55). These processes carry out a key
role in an array of physiological functions and diseases, including
immune response regulation and tumorigenesis. Examples of key TMEM
proteins include TMEM45A in epidermal keratinization (56), TMEM16 in smooth muscle
contraction (57), TMEM165 in
protein glycosylation (58) and
TMEM9B in immune response modulation (59).
TMEM proteins are key contributors to cancer
biology, carrying out key roles in tumor initiation, progression
and metastasis. Their differential expression in various types of
cancer not only serves as biomarkers but also offers new targets
for therapeutic intervention (4,60). Furthermore, alterations in TMEM
protein expression levels or function are closely associated with
tumor progression, metastasis and drug resistance (60). Recent studies have emphasized the
vital role of TMEM proteins in cancer cell survival and
proliferation under harsh conditions, such as hypoxia or nutrient
deprivation, which are commonly associated with the TME (4,5,12-16,60,61). For example, TMEM16, an ion
channel, has been shown to regulate chloride and calcium ion flux,
thereby influencing cell proliferation and migration in cancer
cells (62). Likewise, TMEM65
has been implicated in regulating autophagy, an essential process
for maintaining cellular homeostasis and promoting cancer cell
survival (63). Given their
upregulated expression in various types of cancer, targeting
specific TMEM proteins presents a viable strategy for therapeutic
intervention. Therapeutic strategies may include the development of
small-molecule inhibitors or monoclonal antibodies that selectively
target these proteins, thereby inhibiting their function and
disrupting the signaling pathways they regulate (64). These approaches may address
certain limitations of conventional cancer therapies, such as drug
resistance and off-target effects, offering a more personalized and
effective treatment regimen (65).
The TMEM protein family is important for maintaining
cellular homeostasis and regulating various physiological
processes. Their involvement in the pathogenesis of cancer and
other diseases makes them potential candidates for targeted
therapies. Ongoing research into the molecular mechanisms of TMEM
proteins and their roles in disease progression is likely to lead
to novel therapeutic strategies, particularly for several types of
cancer and other disorders associated with abnormal TMEM protein
activity.
Research on the correlation between TMEM
proteins and cancer
TMEM family members as tumor suppressor
genes
TMEM7
Multiple regions on chromosome 3p are commonly
affected by loss of heterozygosity in several types of cancer
(66). TMEM7 is a candidate
tumor suppressor gene located at the 3p21.3 region of the human
genome. It is a 232-amino acid, single-pass membrane protein,
primarily expressed in the liver (67) and is integral to liver
carcinogenesis (Fig. 3). It
shares considerable sequence homology with the 28 kDa interferon-α
(IFN-α)-responsive protein, suggesting a potential role in immune
response modulation (67). TMEM7
has been implicated as a candidate tumor suppressor gene due to its
frequent downregulation or inactivation in several types of cancer.
Zhou et al (67) examined
primary hepatocellular carcinoma (HCC) tissues and HCC cell lines,
and found that TMEM7 mRNA expression was downregulated or silenced
in 85% of primary HCC samples and 33% of HCC cell lines. This
recurrent loss of function suggests that TMEM7 carries out a key
role in suppressing hepatocarcinogenesis (67). Mechanistically, the
downregulation of TMEM7 is primarily driven by epigenetic
modifications such as DNA methylation and histone deacetylation,
rather than by genomic deletions or mutations (67-69). To the best of our knowledge, no
homozygous deletions or mutations of TMEM7 have been identified in
HCC tissues or cell lines.
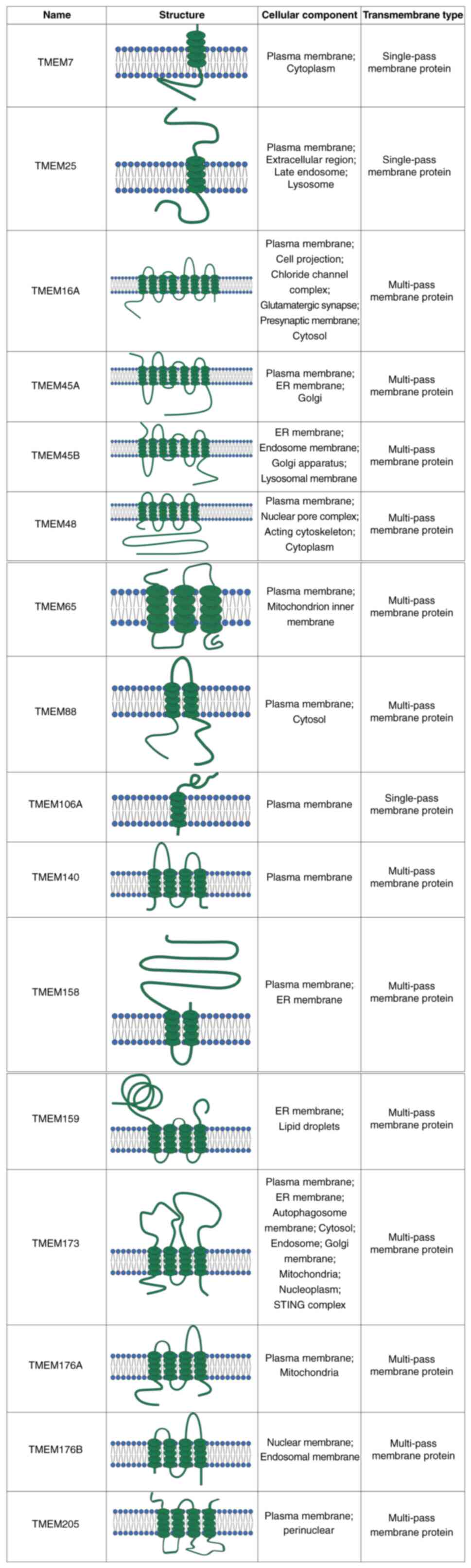 | Figure 3List of TMEM protein structures.
Representative schematic diagrams of 16 TMEM proteins, TMEM7,
TMEM25, TMEM16A, TMEM45A, TMEM45B, TMEM48, TMEM65, TMEM88,
TMEM106A, TMEM140, TMEM158, TMEM159, TMEM173, TMEM176A, TMEM176B
and TMEM205, illustrating their predicted transmembrane structures,
subcellular localization and classification as single-pass or
multi-pass membrane proteins. TMEM, transmembrane. |
Zhou et al (67) demonstrated that treatment with
DNA methylation inhibitors (such as 5-aza-2'-deoxycytidine) and
histone deacetylase inhibitors (such as trichostatin A) can restore
TMEM7 expression in knockdown cell lines, confirming the role of
epigenetic alterations in its silencing. Through the ectopic
expression of TMEM7 in two HCC cell lines with TMEM7 knockdown,
cell proliferation, colony formation and cell migration were
inhibited, and tumor formation in nude mice was reduced. After 7
days of IFN-α treatment in two highly invasive HCC cell lines,
TMEM7 expression was markedly increased and cell migration was
inhibited, further establishing the role of TMEM7 in liver cancer
metastasis (67). These findings
also suggest that modulating TMEM7 expression via IFN-α may have
therapeutic potential for certain patients with HCC. While the role
of TMEM7 in HCC is well-documented, its interaction with other
genes on chromosome 3p further expands its roles. In a study by
Kholodnyuk et al (68)
involving human chromosome 3-mouse fibrosarcoma hybrids, the
transcription of TMEM7 and its neighboring genes (such as LTF and
SLC38A3) was found to be impaired in renal cell carcinoma (RCC)
cell lines, suggesting a broader tumor-suppressive role across
multiple cancer types (68).
The epigenetic silencing of TMEM7 during
hepatocarcinogenesis highlights the functional loss of TMEM7 as a
key factor in liver cancer development, while its restoration or
upregulation (by IFN-α, for example) offers potential therapeutic
benefits (67-69). Understanding the precise role of
TMEM7 in liver cancer progression may pave the way for targeted
therapies aimed at improving outcomes for patients with this
aggressive malignancy.
TMEM25
TMEM25, a member of the immunoglobulin superfamily,
carries out a role in various cellular processes, including immune
responses, growth factor signaling and cell adhesion (70,71). Its expression is markedly altered
in several types of cancer, particularly in triple-negative breast
cancer (TNBC), colorectal cancer (CRC) and clear cell RCC (ccRCC),
making it a promising biomarker for diagnosis, prognosis and
therapeutic targeting. The functional importance of TMEM25 in
cancer is underscored by a study demonstrating its epigenetic
regulation, particularly through DNA methylation and histone
modifications, which affect its expression and role in
tumorigenesis (71).
In colon adenocarcinoma, Hrašovec et al
(70) observed a marked
downregulation of TMEM25 mRNA in 68% of tumor tissues from patients
with primary colon cancer. This downregulation was linked to
hypermethylation of a specific CpG site within the 5' untranslated
region of the TMEM25 gene, suggesting that epigenetic silencing
through DNA methylation is a key mechanism responsible for the
decreased expression of TMEM25 in colon adenocarcinoma. The inverse
relationship between TMEM25 hypermethylation and its expression
levels underscores the key role of epigenetic regulation in
modulating TMEM25 expression during colon carcinogenesis (70). In breast cancer, the expression
of TMEM25 is reduced in 50% of tumor samples compared with normal
tissues, as reported by Doolan et al (72). Increased TMEM25 expression levels
were associated with improved clinical outcomes, particularly in
patients undergoing adjuvant chemotherapy, where it was associated
with prolonged survival (72).
By contrast, TMEM25 expression was often undetectable in TNBC, a
subtype of breast cancer that is difficult to treat and associated
with poor prognosis (70-72).
In TNBC, TMEM25 acts as a suppressor of tumor progression by
inhibiting the activation of the STAT3 signaling pathway.
Specifically, TMEM25 physically interacts with the EGFR monomer,
preventing EGFR-mediated STAT3 phosphorylation. This interaction
sequesters unphosphorylated STAT3 in the cytoplasm, thereby
inhibiting its nuclear translocation and subsequent tumor-promoting
gene transcription (12,71,72). However, in TMEM25-deficient TNBC
cells, EGFR monomers can phosphorylate STAT3 independently of
ligand binding, leading to basal STAT3 activation and tumor
progression. Restoring TMEM25 expression through adeno-associated
virus delivery has been revealed to suppress STAT3 activation and
TNBC progression in preclinical models, highlighting its potential
as a therapeutic target (12,71,72). These findings indicate that
TMEM25 may act as a valuable prognostic marker in breast cancer,
particularly in predicting patient response to chemotherapy and
providing insights into the molecular mechanisms underlying
chemotherapy resistance. Growing evidence also indicates that
TMEM25 carries out a tumor-suppressive role in ccRCC, a subtype of
kidney cancer with limited therapeutic options (73). In ccRCC tissues, elevated DNA
methylation of TMEM25 has been observed, suggesting epigenetic
silencing in this type of cancer. Moreover, mutations in TMEM25
have been revealed to notably impact patient prognosis, with
reduced TMEM25 expression associating with a more aggressive
disease phenotype. Notably, the expression of TMEM25 in ccRCC is
strongly associated with several immune-related factors, including
immune checkpoint molecules, immune suppressive markers and major
histocompatibility complex molecules, implying that TMEM25 may be
involved in modulating the immune landscape of ccRCC (73). This interaction between TMEM25
and immune-related factors further underscores its role in immune
evasion within the TME.
TMEM25 has been identified as a promising prognostic
biomarker in colon cancer, breast cancer and ccRCC (70-72). Its expression is regulated
epigenetically, primarily through DNA methylation and histone
modifications, which suggests that reversing its epigenetic
silencing could offer a promising therapeutic approach.
Furthermore, the involvement of TMEM25 in immune modulation,
especially through its interactions with immune checkpoint
molecules, presents considerable promise for immunotherapeutic
interventions. This establishes TMEM25 as a key therapeutic target
in several types of cancer where its expression is reduced or
absent. Given its pivotal role in cancer progression and immune
regulation, TMEM25 may serve as a promising therapeutic target.
Future research should focus on elucidating the molecular pathways
through which TMEM25 influences tumor progression and immune
responses. Restoring or enhancing TMEM25 expression could
considerably improve treatment outcomes in colon cancer, breast
cancer, and ccRCC, thereby positioning it as a promising target in
future cancer therapies (70-72).
TMEM88
TMEM88 is a dual-transmembrane protein that
participates in several key biological processes, particularly the
regulation of human stem cell differentiation and embryonic
development (74,75). Previous studies have indicated
that TMEM88 carries out a key role in tumorigenesis, with its
expression being altered in several types of cancer (74,75). Notably, TMEM88 binds to
Dishevelled (DVL), preventing its interaction with LRP5/6 and
inhibiting the canonical Wnt pathway. This leads to decreased
β-catenin activity and downregulation of Wnt target genes,
including c-Myc and cyclin D1 (75). While TMEM88 is widely expressed
in various types of tumors, it is typically downregulated in the
majority of tumor cells, suggesting its role as a potential tumor
suppressor (76).
The expression of TMEM88 is also markedly decreased
in thyroid cancer (TC) tissues and cell lines (75-77). Studies have revealed that TMEM88
inhibits the proliferation, colony formation and invasion of TC
cancer cells, primarily through binding to DVL (75-77). In both in vitro and in
vivo models, overexpression of TMEM88 has been shown to
suppress tumorigenic properties, further supporting the role of
TMEM88 as a tumor suppressor in TC. These findings suggest that
restoring TMEM88 expression or inhibiting its methylation may offer
a promising therapeutic strategy for treating TC. The involvement
of TMEM88 in bladder cancer has been further emphasized instudies
(78,79). Upregulation of TMEM88 in bladder
cancer leads to a marked reduction in cellular proliferation and
invasion (78,79). Bioinformatic analysis revealed
that TMEM88 expression was considerably reduced in bladder cancer
tissues compared with normal tissues (79). Zhao et al (79) provided evidence that loss of
TMEM88 expression in bladder cancer cells enhances invasive and
proliferative capacities, while re-expression of TMEM88 effectively
reverses these phenotypes (79).
Additionally, overexpression of TMEM88 in bladder cancer xenograft
models inhibited tumor growth (79), indicating that TMEM88 acts as a
tumor suppressor gene in bladder cancer as well.
The biological role of TMEM88 depends on its
subcellular localization. Membrane-localized TMEM88 inhibits Wnt
signaling, while cytosolic TMEM88 promotes tumor progression
through alternative pathways. Zhang et al (80) reported that cytosolic
localization of TMEM88 in non-small cell lung cancer (NSCLC) is
associated with poor differentiation, a high TNM stage, lymph node
metastasis and worse overall survival (OS). TMEM88 was shown to
promote invasion and metastasis by activating the p38-GSK3 β-Snail
signaling pathway. Overexpression of TMEM88 in NSCLC cells
inhibited excessive cell proliferation, invasion and migration,
leading to the prevention of tumor growth in xenograft models
(80). Furthermore,
hypermethylation of the TMEM88 promoter has been observed in NSCLC
tissues, which is associated with a worse prognosis. Demethylation
of TMEM88 resulted in the inhibition of cell proliferation,
migration and invasion, suggesting that TMEM88 expression and its
epigenetic regulation carry out a key role in NSCLC progression
(81). In TNBC, TMEM88
overexpression in the cytosol stimulated invasion and metastasis by
interacting with DVL proteins and promoting Snail expression while
inhibiting tight junction proteins, such as zonula occludens-1 and
occluding (82). Thus, increased
TMEM88 expression was associated with improved prognosis in HCC and
bladder cancer, while its cytosolic localization was predictive of
a worse outcome in NSCLC and TNBC.
TMEM88 carries out a key role in regulating cell
proliferation, migration and invasion, and has demonstrated notable
potential as both a diagnostic biomarker and therapeutic target
across various types of cancer (79-82). Its expression is frequently
downregulated in tumors, and promoter hypermethylation of TMEM88 is
associated with a worse prognosis in NSCLC (80,81). These findings highlight TMEM88 as
a potential marker for disease progression and survival prediction.
The epigenetic regulation of TMEM88, particularly through DNA
methylation, offers a novel therapeutic avenue. Demethylating
agents can restore TMEM88 expression (81), thereby suppressing tumor cell
growth and metastasis. As a negative regulator of the Wnt/β-catenin
signaling pathway, TMEM88 exhibits tumor-suppressive activity in
NSCLC, TC and bladder cancer. Therapeutic strategies aimed at
restoring or enhancing TMEM88 expression could thus represent a
viable approach for cancer treatment. Ongoing research into the
molecular mechanisms of TMEM88 is expected to further clarify its
value as both a therapeutic target and a prognostic biomarker in
clinical oncology.
TMEM106A
TMEM106A is located on chromosome 17 and consists of
262 amino acids. TMEM106A is a conserved type II TP that has been
implicated in suppressing tumorigenesis across multiple types of
cancer (83,84). TMEM106A is typically expressed in
normal gastric tissue and is often downregulated or silenced in
gastric cancer (GC), due to methylation of the promoter region, and
its methylation is associated with smoking and tumor metastasis
(83,84). Frequent methylation of TMEM106A
in GC tissues highlights its potential role as a tumor suppressor.
Xu et al (84)
demonstrated that overexpression of TMEM106A markedly inhibited GC
cell proliferation and induced apoptosis, while also delaying tumor
growth in xenograft models (84). Mechanistic studies suggested that
these effects were closely associated with the activation of
caspase-2, caspase-9 and caspase-3, as well as the inactivation of
poly(ADP-ribose) polymerase (PARP) (84). Thus, TMEM106A has emerged as a
functional tumor suppressor in the pathogenesis of GC.
Methylation of TMEM106A is generally associated with
its reduced expression and the promoter region methylation serves
as a key regulatory mechanism in GC. The downregulation of TMEM106A
expression in patients with GC is associated with a worse
prognosis, emphasizing its clinical relevance as a prognostic
biomarker (85). In renal
cancer, TMEM106A expression is markedly reduced, and its
restoration in cancer cells leads to attenuated proliferation,
reduced migration and enhanced caspase-3-dependent apoptosis
(86). Similarly, the expression
of TMEM106A in HCC tissues is notably reduced compared with normal
liver tissue, accompanied by frequent promoter methylation in HCC
tissues, further reinforcing its potential as a tumor suppressor
(87). Treatment with
5-Aza-2'-deoxycytidine in HCC cells with high methylation restored
TMEM106A expression and markedly suppressed the malignant behaviors
of these cells, including reduced cell migration, invasion and lung
metastasis (87). In vivo
experiments demonstrated that overexpression of TMEM106A in HCC
cells resulted in considerable tumor suppression, and this was
associated with the suppression of epithelial-mesenchymal
transition (EMT) and the inactivation of the ERK1/2/Slug signaling
pathway (87). Moreover, in
NSCLC, TMEM106A expression was revealed to be markedly decreased in
tumor tissues. Overexpression of TMEM106A in NSCLC cells reduced
proliferation, migration and invasion, while inducing apoptosis. It
also repressed EMT by modulating E-cadherin, N-cadherin and
vimentin expression, and inhibited the PI3K/Akt/NF-κB signaling
pathway (88). In urothelial
carcinoma, TMEM106A was identified as a novel biomarker, with its
methylation levels in urine samples revealing high diagnostic
accuracy for early detection (89).
TMEM106A also exhibits antiviral activity,
specifically in the IFN-mediated antiviral immune response.
TMEM106A was revealed to interact with the receptor SCARB2 of
EV-A71 and CV-A16, thus disrupting viral binding to host cells and
preventing viral infection (90). This finding not only highlights a
novel mechanism of TMEM106A in antiviral immunity but also proposes
a potential therapeutic target for antiviral strategies. In
addition to its antiviral function, TMEM106A regulates macrophage
activation and immune responses, especially in bacterial
infection-induced inflammation. Its downregulation enhanced M1
macrophage polarization and exacerbated the LPS-induced
inflammatory response through the MAPK and NF-κB signaling
pathways, underscoring the key role of TMEM106A in immune
homeostasis (91).
TMEM106A carries out a key tumor-suppressive role in
various types of cancer, such as gastrointestinal tumors, liver
cancer and NSCLC, with its dysregulated expression, including
methylation, potentially serving as a valuable clinical prognostic
biomarker (84,86-91). Furthermore, the diverse functions
of TMEM106A in immune responses and antiviral defense establish it
as a potential target for future cancer treatments and immune
modulation strategies. Further research should prioritize
elucidating the specific mechanisms by which TMEM106A operates in
various types of cancer and exploring its clinical
applications.
TMEM173
TMEM173, also known as stimulator of interferon
genes (STING), is a key TP located on the outer membrane of the ER
and carries out a key role in cancer immunology and immunotherapy.
It is essential in innate immunity and is primarily expressed in
hematopoietic cells and immune organs such as the thymus and spleen
(92,93). Composed of four transmembrane
domains, TMEM173 forms dimers that allow ligand binding and
activation of immune responses, in addition to mediating reactions
to cytosolic DNA, which can result from infections, cellular stress
or genomic instability (94).
TMEM173 carries out a key role in host defense against pathogenic
infections by regulating type I IFN (IFN-I) signaling,
pro-inflammatory cytokines and innate immunity (95). In addition to its role in innate
immunity, TMEM173 also contributes to autoimmune inflammation,
oxidative stress and autophagy (96).
While TMEM173 has demonstrated tumor-suppressive
functions in various cancer types, including CRC, prostate cancer
and melanoma (92); it is
important to note that TMEM173 activation can act as a double-edged
sword in cancer. It promotes antitumor immunity by enhancing
dendritic cell (DC) maturation, T cell activation and macrophage
polarization toward an M1 phenotype, which supports tumor
elimination (97-99). T cell infiltration into tumors is
largely dependent on IFN-I signaling, and activation of TMEM173
enhances immune surveillance against tumors. For example, TMEM173
agonists have been revealed to amplify the cGAS-STING pathway,
leading to increased IFN-β secretion and T cell-mediated immune
responses in colorectal and breast cancer models (97). Additionally, TMEM173 activation
can reprogram tumor-associated macrophages (TAMs) from an
immunosuppressive M2 phenotype to an immunostimulatory M1
phenotype, further enhancing antitumor immunity (97,100). TMEM173 expression is
downregulated in various types of cancer, including GC and HCC, and
this downregulation associates with tumor size, invasiveness and
lymph node metastasis. In vitro experiments have revealed
that knockout of TMEM173 promotes clonal formation, migration and
invasion of GC cells, whereas its activation induces apoptosis and
autophagy in malignant cells, inhibiting tumor growth (101). However, STING signaling can
also promote immune evasion and tumor progression. For example,
TMEM173 activation in tumor monocytes induces programmed death
(PD)-ligand 1 expression, which suppresses T cell activity and
contributes to resistance against TMEM173 agonist therapy (102). Similarly, TMEM173 signaling can
expand regulatory B cells that produce IL-35, which attenuates
natural killer (NK) cell function and compromises antitumor
immunity (103). TMEM173 may
carry out a key role in metastasis formation. Activation of TMEM173
promotes metastatic tumor formation and spread, partly through the
production of TNF-α and other cytokines (104). For example, Bu et al
(105) reported that, in
clinical samples from patients with hepatitis B virus (HBV)-induced
liver cancer, TMEM173 downregulation may contribute to immune
evasion and subsequently facilitate tumor progression.
TMEM173 is not only a key regulator of tumor
immunity but also holds substantial therapeutic potential within
cancer immunotherapy. Targeting TMEM173, either by modulating its
upstream or downstream signaling pathways, may provide novel
therapeutic strategies to enhance cancer treatment outcomes,
particularly when combined with immune checkpoint blockade
therapies (106). Therefore,
TMEM173 agonists are emerging as promising candidates for cancer
immunotherapy. Nanoparticle-based delivery systems have been
developed to enhance the spatiotemporal activation of TMEM173,
improving its therapeutic efficacy while minimizing off-target
effects (107). For example,
TME-responsive nanoparticles have been designed to simultaneously
activate TMEM173 and TLR4, leading to enhanced IFN-β production and
T cell responses in colorectal and metastatic breast cancer models
(97). Additionally, combining
TMEM173 agonists with immune checkpoint inhibitors, such as
anti-PD-1 antibodies, has shown synergistic effects in preclinical
models (97,100). However, challenges remain in
translating TMEM173 agonists to clinical practice. Issues such as
poor drug stability, limited tumor targeting and immune-related
adverse events need to be addressed (103,108). Nanomedicine approaches,
including the use of radiosensitizers and targeted delivery
systems, are being explored to overcome these limitations.
TMEM173 is a central player in cancer immunology,
with its activation driving both antitumor immunity and immune
evasion. While TMEM173 agonists hold potential for cancer
immunotherapy, their clinical translation requires careful
consideration of the TME and potential off-target effects. Future
research should focus on developing targeted delivery systems and
combination therapies to maximize the therapeutic potential of
TMEM173 activation in cancer treatment.
TMEM176A/TMEM176B
TMEM176A and TMEM176B, both belonging to the
membrane-spanning 4-domains family, have emerged as key regulators
of immune responses and cancer progression. TMEM176A is localized
on chromosome 7q36.1 and has been identified as a potential tumor
suppressor in various types of cancer, including esophageal
squamous cell carcinoma (ESCC) and CRC (109). In addition, TMEM176B, also
known as tolerance-related and induced, carries out a key role in
immune modulation and cancer progression (110).
TMEM176A has been revealed to suppress the growth
of ESCC cells both in vitro and in vivo (110-112), suggesting its potential as a
diagnostic and prognostic biomarker for ESCC. In ESCC, the loss of
TMEM176A expression is associated with promoter hypermethylation
(111). TMEM176A promoter
methylation occurs in ~66% of primary ESCC tumors, which associates
with its reduced expression, poor tumor differentiation, reduced
survival rates and aggressive cancer phenotypes (111). The restoration of TMEM176A
expression using demethylation agents, such as
5'-aza-2'-deoxycytidine, inhibits cell migration and invasion,
thereby promoting apoptosis in ESCC cells and suppressing cancer
progression. Additionally, in CRC, TMEM176A promoter methylation is
observed in nearly half of the primary tumors, with TMEM176A
overexpression resulting in suppressed cell proliferation,
migration, invasion and induced apoptosis (112). Notably, TMEM176A is also
involved in regulating glioma cell cycles through pathways such as
ERK1/2, further validating its role as a tumor suppressor (112). Collectively, these findings
establish TMEM176A as a potential tumor suppressor gene, with its
expression levels serving as valuable prognostic indicators across
multiple cancer types (113).
TMEM176B has been extensively studied in various
types of cancer, where it often promotes tumor progression. In CRC,
TMEM176B inhibits the NOD-, LRR-, and pyrin domain-containing
protein 3 inflammasome, thereby suppressing NK cell activity and
promoting EMT, a key process in metastasis. Silencing TMEM176B
reduces tumor growth, metastasis and EMT while enhancing apoptosis
and immune activation (114).
In GC, TMEM176B overexpression drives tumor progression by
regulating the PI3K-Akt-mTOR signaling pathway and amino acid
metabolism. Its knockdown inhibits cell proliferation, migration
and invasion, while promoting apoptosis. Clinically, TMEM176B
expression is associated with a worse prognosis, making it a
potential prognostic marker (115). TMEM176B also carries out a role
in ovarian cancer (OC), where it acts as a tumor suppressor. Its
downregulation is associated with increased EMT, proliferation and
metastasis, mediated through the Wnt/β-catenin signaling pathway.
Restoring TMEM176B expression inhibits these processes,
highlighting its therapeutic potential (116).
TMEM176B also carries out a key role in regulating
immune responses, especially in DCs (117). TMEM176B is recognized as a key
modulator of allograft tolerance and immune system function
(117). A study involving
autologous tolerogenic DCs (ATDCs) demonstrated that TMEM176B
expression in ATDCs was essential for inducing donor-specific
immune tolerance and prolonging graft survival (118). TMEM176B-deficient ATDCs failed
to cross-present antigens effectively, impairing their ability to
induce regulatory CD8+ T cells and hindering allograft
tolerance (118). TMEM176B
inhibits inflammasome activation, which dampens anti-tumor
immunity. Targeting TMEM176B unleashes inflammasome activity,
increasing CD8+ T cell-mediated tumor control and
enhancing the effects of anti-CTLA-4 and anti-PD-1 therapies
(119). These findings
underscore the importance of TMEM176B in modulating immune
responses, particularly through its influence on antigen
cross-presentation and regulation of phagosomal pH, both of which
are important for immune tolerance (120). Additionally, TMEM176B has been
implicated in regulating immune checkpoint responses, particularly
in the context of PD-1 blockade therapy. Elevated TMEM176B
expression in tumor stromal cells is associated with a worse
prognosis, suggesting that it may act as a negative modulator of
anti-tumor immunity (121). Its
role in immune evasion has prompted investigations into its
potential as a biomarker for predicting patient response to immune
checkpoint inhibitors, including anti-PD-1 therapies. TMEM176B
negatively regulated the antitumor activity of CD146+
TAMs. Inhibition of TMEM176B enhanced TAM-mediated anti-tumor
immunity, highlighting a potential immunotherapeutic strategy
(122). Thus, as an
immune-regulating cation channel, TMEM176B modulates immune cell
trafficking and function, further highlighting its potential as a
therapeutic target (123).
TMEM176A and TMEM176B are known to physically
interact and co-localize at both the plasma membrane and vesicular
compartments within immune cells (124). In lymphoma, overexpression of
both proteins has been associated with immune evasion, enabling
cancer cells to evade immune surveillance and develop resistance to
chemotherapy (124,125). This interaction suggested that
the two proteins may function synergistically to regulate immune
responses and contribute to tumor progression. Both TMEM176A and
TMEM176B are downregulated during DC maturation, with their
overexpression inhibiting this process and suggesting a role in
maintaining an immature state that facilitates immune tolerance
(123).
TMEM176A and TMEM176B have emerged as key factors
in cancer biology, particularly in immune regulation and tumor
suppression. Their roles in immune modulation, especially through
the regulation of DC function and antigen cross-presentation,
position them as promising targets for novel therapeutic strategies
aimed at enhancing immune checkpoint blockade efficacy and
improving cancer immunotherapy outcomes. Future research should
focus on more comprehensively elucidating the precise molecular
mechanisms by which TMEM176A and TMEM176B regulate immune responses
and tumor progression. Clinical trials targeting these proteins,
either alone or in combination with existing immune checkpoint
inhibitors, may provide key insights into their therapeutic
potential.
TMEM family members as oncogenes
TMEM16A
TMEM16 proteins, also known as anoctamins, are a
family of ten MPs with various tissue expression and subcellular
localization. TMEM16A, also referred to as anoctamin 1 (ANO1), is a
calcium-activated chloride channel (CaCC) mapped to chromosome
11q13. It contains eight transmembrane domains and a highly
conserved, functionally undefined domain (DUF590), with its
expression predominantly observed in secretory epithelial cells,
smooth muscle tissue and sensory neurons (126). TMEM16A is acknowledged as a key
modulator of tumor progression. It is frequently amplified in a
variety of malignancies, including breast cancer, esophageal cancer
and head and neck squamous cell carcinomas (HNSCCs), where its
expression is associated with a poor prognosis, increased tumor
size and advanced clinical stages (127).
TMEM16A carries out a key role in tumorigenesis by
activating key signaling pathways, including the EGFR and
calcium/calmodulin-dependent protein kinase pathways (128). Silencing of TMEM16A has been
revealed to inhibit the proliferation of esophageal and HNSCC cells
and induce apoptosis through the MAPK and AKT signaling pathways.
TMEM16A is involved in EMT, a key process in cancer metastasis, by
regulating the transition from an epithelial to a mesenchymal
state, thus enhancing cell migration and invasion (129). In CRC, TMEM16A carries out a
key role in disease progression by promoting cell migration and
invasion. It activates the Wnt/β-catenin signaling pathway by
upregulating key components such as Frizzled protein 1 and
β-catenin, while downregulating GSK3β (129,130). The overexpression of TMEM16A is
notably elevated in HNSCC, esophageal and prostate cancer, where it
is associated with distant metastasis and a poor prognosis
(131).
The molecular mechanisms underlying these effects
include a role independent of its channel function in promoting
cell proliferation. Research suggests that TMEM16A regulates cancer
cell growth by interacting with other MPs rather than exclusively
through its chloride conductance (131). For example, TMEM16A has been
revealed to interact with EGFR, thereby potentiating EGFR signaling
in HNSCC cells (128). This
interaction not only stabilizes EGFR but also increases TMEM16A
protein expression, thereby establishing a feedback loop between
TMEM16A and EGFR signaling pathways, which likely contributes to
enhanced cancer cell proliferation and survival. Knockdown of
TMEM16A has been revealed to reduce myeloid cell leukemia 1
expression and promote the perinuclear redistribution of
p27Kip1, thereby inducing cell cycle arrest, suppressing
tumor proliferation and promoting apoptosis (132). However, Shiwarski et al
(133) reported that stable
reduction of TMEM16A expression enhanced the motility of HNSCC
cells and facilitated metastasis, independent of 11q13
amplification. Additionally, TMEM16A overexpression is associated
with increased Erk1/2 activity, reduced Bim expression and
decreased apoptotic activity, contributing to tumor progression and
cisplatin resistance (134).
These conflicting findings underscore the multifaceted role of
TMEM16A in HNSCC progression. Further studies revealed that TMEM16A
expression is associated with tumor subtype: In general, in HNSCC,
elevated TMEM16A levels associate with shorter survival and
increased risk of distant metastasis (135,136), whereas in human papilloma virus
(HPV)-positive HNSCC, its expression was not associated with
prognostic significance (136).
This suggests that TMEM16A-targeted therapies may be inappropriate
for HPV-positive patients. Collectively, these findings indicate
that the function of TMEM16A is context-dependent, shaped by the
TME and specific cellular subtypes. Continued clinical and
molecular investigations are required to clarify its precise role
in cancer progression.
TMEM16A has emerged as a potential biomarker for
several types of cancer, including esophageal and gastrointestinal
squamous cell carcinoma (127).
Its expression is associated with clinical staging and disease
progression, making it a valuable prognostic indicator. Moreover,
TMEM16A functions as a predictor of therapeutic response to
EGFR-targeted therapies, particularly in HNSCC, where TMEM16A
overexpression may mediate resistance to EGFR inhibitors (137). Given its key role in tumor
progression and interaction with EGFR signaling, TMEM16A represents
a compelling therapeutic target. Targeting TMEM16A in conjunction
with EGFR may offer a strategy to bypass resistance mechanisms and
enhance the efficacy of existing EGFR-targeted therapies in cancer
treatment (128).
Amplification of chromosome 11q13, where TMEM16A
(ANO1) is located, is associated with a poor prognosis in patients
with breast cancer. TMEM16A amplification and overexpression are
associated with a higher tumor grade and worse outcomes, primarily
through activation of EGFR and CaMKII signaling pathways to promote
cell proliferation (138).
TMEM16A expression also associates with immunohistochemical markers
such as β-catenin, cyclin D1 and E-cadherin, and has been
identified as an independent prognostic marker (139). Functional studies revealed that
TMEM16A inhibition induced G0/G1 cell cycle arrest and reduced the
invasiveness of breast cancer cells, underscoring its potential as
a therapeutic target (139,140). However, conflicting data
regarding its association with hormone receptors (HER2, ER and PR)
suggested that the role of TMEM16A may vary across molecular
subtypes and cellular contexts (140). Upregulation of TMEM16A is
notably associated with CRC progression and higher TNM staging
(141). Sui et al
(142) confirmed that TMEM16A
knockout inhibited the growth, migration and invasion of CRC cells
by suppressing the MAPK/ERK signaling pathway. It has been reported
that endogenous TMEM16A knockout suppressed CRC cell proliferation
and induced apoptosis by inactivating the downstream Wnt/β-catenin
signaling pathway (142). Li
et al (143) proposed
that TMEM16A mRNA expression was associated with lymph node
metastasis in CRC and may thus serve as an independent predictive
marker for lymphatic involvement. These findings establish the key
role of TMEM16A in CRC progression and metastasis. Overexpression
of TMEM16A is associated with TNM staging and Gleason scoring in
prostate cancer, carrying out a key role in tumor growth,
progression and metastasis (144).
Silencing or inhibiting endogenous TMEM16A has been
revealed to suppress prostate cancer growth, induce apoptosis and
increase TNF-α expression levels (145). Notably, natural products such
as resveratrol and luteolin serve both as TMEM16A inhibitors and
antitumor agents, effectively reducing prostate cancer cell
proliferation and migration (146,147). These findings highlight the
therapeutic potential of targeting TMEM16A in prostate cancer
treatment. TMEM16A is considerably upregulated in epithelial OC
cells, and its upregulation is associated with increased staging
and lower differentiation (148). Downregulation of TMEM16A
suppressed OC cell growth by reducing PI3K/Akt phosphorylation and
disrupting the PI3K/Akt signaling pathway (148). These findings highlight the key
role of TMEM16A in OC progression, with its expression associated
with advanced disease and a poor prognosis. Given its role in
cancer progression, TMEM16A has emerged as a promising therapeutic
target. Inhibition of TMEM16A suppresses tumor growth, induces
apoptosis and enhances the efficacy of existing therapies (144-149). For example, allicin, a natural
compound from garlic, was shown to inhibit the chloride ion current
through the TMEM16A channel and exhibit synergistic anticancer
effects with cisplatin in lung cancer (149). Similarly, idebenone, a novel
TMEM16A inhibitor, can block chloride channel activity and was
shown to induce apoptosis in normal prostate and PC cells (150). Furthermore, TMEM16A inhibitors
such as T16Ainh-A01 and CaCCinh-A01 have demonstrated efficacy in
reducing tumor growth and invasiveness in prostate cancer (144).
TMEM16A, as a CaCC, carries out a key role in
calcium-activated chloride secretion, which is involved in the
pathogenesis of secretory diarrhea. Yin et al (151) revealed in rotavirus-induced
diarrhea that viral enterotoxin NSP4 enhanced intracellular calcium
levels, activating TMEM16A-mediated chloride secretion, which,
in-turn, contributed to fluid loss and diarrhea. Given that
chemotherapy-induced diarrhea in patients with cancer often
involves dysregulated epithelial ion transport, TMEM16A may
similarly contribute to this adverse effect. Thus, TMEM16A may
serve as a mechanistic link between increased chloride secretion
and diarrhea in both infectious and iatrogenic contexts.
TMEM16A, through its dual function as both a
chloride channel and a signaling modulator, carries out a pivotal
role in tumor biology. The regulation of cell proliferation,
migration and metastasis by TMEM16A, in addition to its association
with EGFR signaling, highlights its potential as both a prognostic
biomarker and a therapeutic target for various malignancies.
TMEM45A
TMEM45A consists of 275 amino acids, composed of
5-7 transmembrane domains and is primarily localized in the Golgi
apparatus (4). It is highly
expressed in the skin, where it carries out a key role in epidermal
differentiation and keratinization, processes essential for
preserving the structural integrity of the skin and barrier
function (152). Beyond its
role in normal cellular functions, TMEM45A is involved in the
pathogenesis of various types of cancer, including gliomas, breast
cancer, HCC, RCC and OC, where it is commonly upregulated (153).
TGF-β is a key factor regulating various cellular
processes such as proliferation, migration and invasion. In OC, the
expression of TMEM45A is closely associated with the activation of
the TGF-β signaling pathway. Silencing TMEM45A not only reduced
cell proliferation, adhesion and invasion, but also downregulated
cancer-promoting factors such as TGF-β1, TGF-β2, RhoA and ROCK2
(154). This association has
been revealed to contribute to tumor progression and metastasis and
a similar mechanism has been identified in glioma cell lines,
providing further evidence for the oncogenic potential of TMEM45A
in these cancer types (152,153). Notably, increased TMEM45A
expression in breast cancer and cervical cancer (CC) has been
associated with poor OS, indicating that TMEM45A could function as
an important prognostic biomarker for these malignancies (155).
TMEM45A short hairpin RNA inhibited the
proliferation of CC cells, arrested the cell cycle at the S phase
and promoted apoptosis. It also suppressed EMT by downregulating
Vimentin and N-cadherin, and upregulating E-cadherin, thereby
reducing the invasive and migratory ability of the cells. The
expression of TMEM45A in cisplatin-resistant cell lines SiHa/DDP
and HeLa/DDP was revealed to be increased compared with their
parental cells, and inhibiting TMEM45A markedly reduced the
IC50 of these resistant cells to cisplatin. Silencing
TMEM45A inhibited the proliferation, invasion, migration and EMT of
HPV-positive CC cells, regulated cell cycle distribution and
promoted apoptosis (156). Sun
et al (153) revealed
that TMEM45A was overexpressed in glioma tissues compared with
non-tumorous brain tissues. TMEM45A mRNA levels progressively
increased with higher histological grades of glioma and were
associated with shorter survival times of patients. Knockout of
TMEM45A in glioma cell lines markedly inhibited cell proliferation
and induced G1 phase arrest, and reduced the migratory and invasive
abilities of glioma cells. Notably, treatment with TMEM45A small
interfering (si)RNA downregulated key proteins involved in cell
cycle progression (Cyclin D1, CDK4 and proliferating cell nuclear
antigen) as well as invasion-related proteins (MMP-2 and MMP-9),
providing a potential mechanistic explanation for its role in
glioma.
TMEM45A is closely associated with intratumoral
heterogeneity, a major challenge in cancer treatment. In lung
adenocarcinoma, TMEM45A was revealed to be overexpressed in tumor
tissues compared with healthy tissues, and its expression was
associated with markers such as GLUT1, MCT4 and CA9, which are
associated with a hypoxic microenvironment and altered metabolism
(157). Similarly, in ccRCC,
TMEM45A upregulation associated with poor OS, disease-free survival
and advanced clinicopathological features such as higher
histological grade and TNM stage (158).
TMEM45A is a multifunctional protein associated
with tumorigenesis, chemotherapy resistance and fibrosis. Its
ability to modulate key signaling pathways, including TGF-β and
Notch, positions it as a promising candidate for therapeutic
targeting. In GC, increased TMEM45A expression was associated with
poor prognosis and immune cell infiltration, making it a potential
independent prognostic marker (159). In breast cancer, TMEM45A
expression is associated with poor survival and resistance to
CDK4/6 inhibitors such as palbociclib, further underscoring its
prognostic value (160).
Furthermore, the association of TMEM45A with a poor prognosis in
various types of cancer and its involvement in MDR highlights its
potential as a biomarker for cancer progression and therapeutic
resistance (155,161,162). In breast cancer, engineered
exosomes loaded with siRNA targeting TMEM45A restored sensitivity
to palbociclib and suppressed tumor growth (160). Similarly, in CC, TMEM45A
knockdown reversed cisplatin resistance and inhibited
proliferation, migration and EMT in HPV-positive cell lines
(156). Future research
focusing on the molecular mechanisms underlying the functions of
TMEM45A in these contexts are essential for the development of
targeted therapies aimed at overcoming resistance and improving
patient outcomes.
TMEM45B
TMEM45B has attracted considerable attention due to
its involvement in various types of cancer, including lung cancer,
PC and GC (163). Okada et
al (163) have highlighted
that TMEM45B as an oncogene that plays a pivotal role in cancer
pathogenesis by regulating essential cellular processes, including
proliferation, invasion, migration, and apoptosis.
In lung cancer, TMEM45B overexpression is
associated with poor patient survival. Knockdown of TMEM45B reduced
cell proliferation, induced cell cycle arrest and apoptosis, and
inhibited cell migration and invasion. These effects were mediated
through the regulation of cell cycle-related proteins (CDK2 and
CDC25A), apoptosis-related proteins (Bcl2 and Bax) and
metastasis-related proteins (MMP-9, Twist and Snail). TMEM45B may
thus serve as a potential prognostic marker and therapeutic target
in lung cancer (164). TMEM45B
is highly expressed in PC tissues and cell lines. Its knockdown
inhibited cell proliferation, invasion and migration while inducing
cell cycle arrest and apoptosis. Conversely, TMEM45B overexpression
promoted these processes and inhibited apoptosis. Gene set
enrichment analysis revealed that TMEM45B regulated genes that are
involved in the cell cycle and metastasis pathways, further
supporting its oncogenic role in PC (165). Shen et al (166) also demonstrated that in GC,
TMEM45B was overexpressed in both tissues and cell lines. Silencing
TMEM45B inhibited cell proliferation, migration, invasion and EMT.
This suppression was mediated through the inhibition of the
JAK2/STAT3 signaling pathway, as evidenced by reduced levels of
phosphorylated JAK2 and STAT3 upon TMEM45B knockdown (166).
Moreover, in osteosarcoma, Li et al
(167) verified the
upregulation of TMEM45B expression and revealed that its knockdown
in U2OS cells effectively suppressed proliferation, migration and
invasion in vitro, and inhibited tumor growth in xenograft
models. This effect was associated with the downregulation of key
proteins such as β-catenin, cyclin D1 and c-Myc, which are
important for cancer cell proliferation and metastasis (167). These findings collectively
indicate that TMEM45B is a potent regulator of tumorigenesis across
various cancer types. Its dysregulated expression may act as a
potential prognostic biomarker for cancer progression and it holds
promise as a therapeutic target. Furthermore, previous studies have
demonstrated that the regulatory effects of TMEM45B extend to
modulating the TME, influencing immune responses and potentially
modulating the response to chemotherapeutic agents (167,168). Similarly, TMEM45B has been
identified as a novel predictive biomarker for prostate cancer
progression and metastasis. Its expression is notably increased in
tumor cell lines with high metastatic potential and is associated
with biochemical recurrence, distant metastasis and a poor OS.
Multivariate analysis confirmed that TMEM45B was an independent
risk factor for metastasis, making it a promising prognostic marker
for patients with prostate cancer (169).
TMEM45B exerts its oncogenic effects through
multiple signaling pathways, including Wnt/β-catenin, JAK2/STAT3
and TGF-β. It regulates key cellular processes such as
proliferation, apoptosis, migration and invasion by modulating the
expression of downstream target genes and proteins. Additionally,
the involvement of TMEM45B in EMT and chemotherapy resistance
further underscores its role in cancer progression. Given its
central role in regulating key pathways involved in cancer
progression, TMEM45B may be developed as a targeted therapy,
thereby improving patient outcomes.
TMEM48
TMEM48 is a member of the nuclear pore complex
(NPC) family of proteins and is essential for maintaining nuclear
membrane integrity and enabling nuclear transport (170). The protein consists of six
transmembrane domains and is vital for NPC assembly, which is
important for regulating several cellular processes, including
chromosome segregation, gene transcription, DNA replication, DNA
damage repair and apoptosis (170,171). Alterations in the expression of
NPC proteins, including TMEM48, have been associated with the onset
and progression of various malignancies, such as breast, melanoma,
pancreatic, colon, gastric, prostate, esophageal, lung cancer and
lymphoma (170-172).
Previous studies have highlighted the involvement
of TMEM48 in NSCLC (170-173). Qiao et al (173) demonstrated that TMEM48
expression was markedly increased in NSCLC tissues compared with
adjacent normal tissues. This overexpression was associated with a
poor prognosis, including reduced survival times, increased tumor
size and lymph node metastasis. TMEM48 regulates essential cellular
processes, such as proliferation, migration and invasion, by
regulating genes involved in the cell cycle and DNA replication.
Knockdown of TMEM48 in NSCLC cell lines led to reduced cell
proliferation, induction of G1 phase cell cycle arrest and
inhibition of migration and invasion. Furthermore, silencing of
TMEM48 triggered apoptosis in NSCLC cells. In an in vivo
study, silencing TMEM48 led to a notable reduction in tumor weight
in xenograft models (173).
Moreover, the study by Akkafa et al
(6) suggests that microRNA
(miR)-421, an miR targeting TMEM48, potentiates apoptotic signaling
pathways in NSCLC. Suppression of TMEM48 by miR-421 upregulated the
expression of apoptosis-related proteins, such as caspase-3, PTEN
and p53, which collectively promote cell death. This indicated that
targeting TMEM48 may serve as a therapeutic strategy for improving
the efficacy of treatments in NSCLC. TMEM48 has been identified in
other types of cancer, such as CC (174). In CC, TMEM48 is overexpressed
and its knockdown inhibited cell proliferation, migration and
invasion both in vitro and in vivo (174). Jiang et al (174) indicated that TMEM48 promotes CC
progression by activating the Wnt/β-catenin signaling pathway, as
evidenced by the reduced expression of β-catenin, TCF1 and AXIN2
following TMEM48 knockdown. This suggested that TMEM48 is important
in tumorigenesis by modulating key signaling pathways involved in
cancer progression.
TMEM48 is a promising prognostic marker and
therapeutic target. Given its central role in regulating cellular
functions key for tumor progression, TMEM48 represents a valuable
target for future therapeutic strategies focused on inhibiting
cancer growth and metastasis. Further research is needed to explore
the therapeutic potential of TMEM48 in various malignancies,
including its modulation by miRNAs.
TMEM65
TMEM65 is a mitochondrial inner membrane protein
whose deficiency has been implicated in a mitochondrial disorder
characterized by a complex encephalomyopathic phenotype (175,176). Clinical manifestations include
microcephaly, craniofacial dysmorphism, psychomotor regression,
generalized hypotonia, growth retardation, lactic acidosis,
intractable seizures and dyskinetic movements, notably in the
absence of cardiomyopathy (175). Previously, TMEM65 has emerged
as a pivotal factor in cancer biology, with its involvement
reported across various malignancies, including CRC, TNBC, GC and
HCC (175-180). TMEM65 carries out a
multifaceted role in tumorigenesis by modulating mitochondrial
dynamics, driving metabolic reprogramming and regulating signaling
pathways associated with cancer progression and therapeutic
resistance. This contrast between its role in congenital
mitochondrial disease and acquired malignancies underscores its
biological value and potential as a therapeutic target.
In CRC, TMEM65 is transcriptionally regulated by
the CHD6-TCF4 axis, which is activated by both EGF and Wnt
signaling pathways. CHD6, a chromatin remodeler, binds to Wnt
signaling transcription factor TCF4 to facilitate TMEM65
expression. This axis promotes mitochondrial dynamics and ATP
production, essential for cancer cell proliferation, migration and
invasion. Targeting the CHD6-TMEM65 axis with Wnt inhibitors and
anti-EGFR therapies showed promise in restricting CRC growth
(176). TMEM65 acts as an
oncogene in TNBC, where it was revealed to be upregulated by the
transcription factor MYC and DNA demethylase TET3. TMEM65 enhanced
mitochondrial oxidative phosphorylation and reactive oxygen species
production, which, in turn, activates hypoxia inducible factor
(HIF)-1α and SERPINB3 and promotes cancer stemness, progression,
and cisplatin resistance. Inhibition of MYC and TET3 attenuated
TMEM65-driven TNBC progression, highlighting its potential as a
therapeutic target (177).
TMEM65 amplification and overexpression are common in GC and are
associated with a poor prognosis. TMEM65 promoted tumorigenesis by
activating the PI3K-Akt-mTOR signaling pathway through its
interaction with YWHAZ, a protein that inhibits ubiquitin-mediated
degradation. Silencing TMEM65 suppresses tumor growth and
metastasis, making it a promising therapeutic target in GC
(163). In HBV-related HCC,
TMEM65 amplification was driven by HBV integration-induced genomic
rearrangements. TMEM65 contributes to tumorigenesis by promoting
mitochondrial function and metabolic reprogramming, which are key
for cancer cell survival and proliferation (178).
TMEM65 expression is associated with tumor immune
infiltration, particularly CD8+ T effector cells and
immune checkpoint markers. Its role in modulating the TME and
immune response makes it a potential biomarker for predicting
prognosis and the efficacy of immunotherapy (179). TMEM65 may serve as a target for
chimeric antigen receptor T-cell therapies and antibody-drug
conjugates. Its overexpression in cancer tissues and low expression
in normal tissues make it an attractive candidate for immune-based
interventions (180). TMEM65 is
a pivotal oncogene that drives cancer progression through its roles
in mitochondrial dynamics, metabolic reprogramming and signaling
pathway activation. Its overexpression is associated with poor
prognosis across multiple cancer types, making it a promising
biomarker and therapeutic target. Future research should focus on
developing targeted therapies that exploit the oncogenic function
of TMEM65 while minimizing off-target effects.
TMEM140
TMEM140, also known as FLJ11000, is a TP encoded by
a gene located on chromosome 7q33, consisting of 185 amino acids
(181). Although initially
identified for its role in supporting hematopoietic stem cells
in vitro (181-183), TMEM140 has since been
associated with various biological processes, including
tumorigenesis and viral infection (182). Guan et al (183) have highlighted the dual
functions of TMEM140 as both a prognostic biomarker and a potential
therapeutic target in cancer and its role in inhibiting herpes
simplex virus 1 (HSV-1) replication. TMEM140 has emerged as a key
player in various types of cancer, including gliomas and GC.
Gliomas are among the most common and aggressive primary brain
tumors, characterized by a poor prognosis and limited treatment
options. TMEM140 is overexpressed in gliomas compared with normal
brain tissues and its high expression is associated with a larger
tumor size, higher histologic grade and shorter OS (181). An In vitro study using
glioma cell lines have revealed that silencing TMEM140 inhibited
cell proliferation, migration and invasion, with concurrent arrest
in the G1 phase of the cell cycle and activation of apoptotic
pathways (181). It regulates
cell cycle progression by shortening the cell cycle and reducing
apoptosis, thereby promoting tumor cell proliferation.
Additionally, TMEM140 enhanced tumor cell adhesion and survival by
modulating the expression of adhesion and anti-apoptotic proteins
(181-183). In vivo silencing of
TMEM140 led to a marked reduction in tumor volume and weight
(183), highlighting its
pivotal role in tumor growth. These findings identify TMEM140 as a
novel prognostic biomarker and therapeutic target for gliomas,
underscoring its potential for clinical application in glioma
treatment. Beyond gliomas, TMEM140 has been implicated in other
cancer types through its co-expression with long-chain
acyl-coenzyme A synthetase 5 and this was predictive of improved
prognosis in breast, ovarian and lung cancer (184). This suggests that TMEM140 may
carry out a context-dependent role in cancer progression,
potentially acting as a tumor suppressor in certain malignancies
(184). Furthermore, TMEM140
has been identified as a candidate gene in HTLV-1-infected cancer
cells, indicating its broader relevance in cancer biology (185).
TMEM140 is a multifunctional protein that is
involved in several processes in cancer biology, autoimmune
diseases and viral infections. In systemic sclerosis, the
involvement of TMEM140 in DNA methylation changes indicates a
potential key role in disease pathogenesis, particularly in
fibroblast function (186).
Additionally, its role in inhibiting HSV-1 replication highlights
its promise as an antiviral target. Given its diverse functions,
TMEM140 may serve as a compelling candidate for further
investigation, with implications for therapeutic strategies across
a variety of diseases.
TMEM158
TMEM158, located on chromosome 3p21.3, also known
as BBP, Ras-induced senescence 1 protein, p40BBP and HBB, was
initially recognized as an upregulated potential tumor suppressor
gene in the context of Ras V12 lentiviral infection-induced
senescence in fibroblasts (187). Li et al (188) have highlighted its key role in
the development of various types of cancer. Importantly, TMEM158 is
overexpressed in Wilms tumor (nephroblastoma), with somatic
mutations in the β-catenin gene, indicating a connection between
Ras and Wnt signaling pathways (189). In lung cancer, TMEM158 is
highly expressed in advanced-stage tumors and is associated with
poor OS (61). It promotes EMT
and enhanced cell migration, contributing to aggressive tumor
behavior (61). Additionally,
TMEM158 expression is induced under hypoxic conditions in a
HIF-1α-dependent manner, associating it with tumor hypoxia, a
hallmark of cancer progression (61). Moreover, TMEM158 has emerged as a
potential biomarker for predicting the efficacy of cisplatin-based
therapy in NSCLC, with its expression correlating with improved
therapeutic outcomes (61,190).
In PC, TMEM158 overexpression promotes cell
proliferation, migration and invasion through the activation of the
TGFβ1 and PI3K/AKT signaling pathways, highlighting its oncogenic
potential (191). TMEM158 is
amplified in GC tissues and is associated with poor prognosis
(192). In GC, it was revealed
to accelerate GC cell proliferation by activating the PI3K/AKT
signaling pathway and its knockdown inhibited tumor growth,
suggesting its potential as a biomarker and therapeutic target
(192). In CRC, downregulation
of TMEM158 expression notably impairs tumor cell proliferation,
migration and MDR, highlighting its value in both tumor progression
and chemoresistance (193).
Furthermore, TMEM158 is involved in regulating EMT in various types
of cancer, enhancing metastasis and aggressiveness. In OC, TMEM158
expression was shown to be considerably upregulated, thereby
promoting tumorigenic properties, including cell proliferation,
invasion, adhesion and migration (194). Silencing TMEM158 resulted in
downregulation of intercellular adhesion molecules (ICAM1 and
VCAM1) and disruption of the TGF-β signaling pathway, further
emphasizing its role as an oncogene (194). It also contributed to
doxorubicin resistance by upregulating ABCG2, a drug efflux
transporter (195). TMEM158 is
highly expressed in TNBC and was shown to be associated with poor
clinical outcomes and to drive EMT and tumor metastasis via
activation of the TGF-β signaling pathway, highlighting it as a
potential therapeutic target for this aggressive breast cancer
subtype (196).
In glioblastoma (GBM), overexpression of TMEM158
was associated with a poor prognosis, whereas silencing it
suppressed cell migration and invasion by interfering with STAT3
signaling pathways (188).
Unlike its oncogenic role in other types of cancer, TMEM158 is
downregulated in prostate cancer and is associated with advanced
disease features and poor survival. Its expression is negatively
regulated by androgen receptor signaling, and it may carry out a
tumor-suppressive role in this context (197,198). Additionally, TMEM158 was shown
to be involved in immune modulation, correlating with tumor
mutational burden, microsatellite instability and immune cell
infiltration in the TME (190,197,198).
In NSCLC, TMEM158 is considered to modulate immune
checkpoints, thereby enhancing the therapeutic efficacy of immune
checkpoint inhibitors such as anti-PD-1 and anti-CTLA-4 (190,197,198). This emerging role in
immuno-oncology highlights a promising avenue for therapeutic
interventions targeting TMEM158. Additionally, pan-cancer analyses
demonstrated that TMEM158 is upregulated in a range of cancer
types, including melanoma and HCC, where its elevated expression is
associated with tumor aggressiveness and poor prognosis (197). In these contexts, TMEM158 has
been implicated in hypoxia-induced pathways, thus facilitating
tumor growth in hypoxic conditions. These findings collectively
highlight the key role of TMEM158 in regulating tumor progression,
metastasis, immune evasion and chemoresistance across various
cancer types. Its involvement in signaling pathways, including
TGFβ, PI3K/AKT, STAT3 and Wnt, as well as its potential in immune
modulation, establishes TMEM158 as both a prognostic biomarker and
a promising therapeutic target in oncology.
TMEM159
TMEM159, also known as promethin, is a small TP
consisting of 161 amino acids with four adjacent transmembrane
helices forming two helical hairpins (199). It carries out a key role in
lipid droplet (LD) biogenesis through its interaction with Seipin,
a key regulator of lipid storage in the ER (200). The TMEM159-Seipin complex
co-purified with triacylglycerol and facilitated the initiation of
LD formation. Upon maturation of LD, TMEM159 dissociated and
relocated to the LD surface, thereby participating in the
maintenance of lipid homeostasis (200).
Although extensively studied in the context of
lipid metabolism, TMEM159 has recently gained attention for its
potential involvement in cancer progression (199-202). Notably, in GBM, enhanced LD
formation, which is associated with a poor prognosis under
metabolic stress, has been associated with altered TMEM159
expression (201). TMEM159 may
contribute to glioma malignancy by modulating EGFR signaling
pathways, which are important for cancer cell proliferation,
migration and survival (201).
These findings suggest that TMEM159 could serve as both a molecular
biomarker and therapeutic target in lipid-associated malignancies
such as GBM (201,202).
Beyond cancer, TMEM159 has also been associated
with neuropsychiatric disorders through genome-wide association
studies (199-205), but its mechanistic involvement
in tumorigenesis, particularly through pathways associated with
lipid regulation and oncogenic signaling, may represent promising
directions for future cancer-focused research. Continued
investigation is warranted to fully elucidate the multifaceted role
of TMEM159 in lipid metabolism and cancer biology (203-205).
TMEM205
TMEM205 is a TP encoded by a gene located on
chromosome 19p13.2, and is formed of 189 amino acids. TMEM205 has
emerged as a notable player in cancer biology, particularly in the
context of drug resistance, tumor progression and immune
modulation. TMEM205 has shown promise as a biomarker for
chemoresistance and prognosis. In high-grade serous OC, TMEM205 was
found to be differentially expressed in extracellular vesicles
derived from platinum-resistant patients with OC, demonstrating
high diagnostic accuracy for platinum resistance (206). In HCC, low TMEM205 expression
was independently associated with poor OS and disease-specific
survival, highlighting it as a potential prognostic marker
(207). TMEM205 is associated
with chemoresistance, particularly to platinum-based therapies such
as cisplatin (208). Moreover,
TMEM205 was also shown to carry out a role in tumor progression and
immune evasion (208).
Targeting TMEM205 may offer a novel strategy to overcome
chemoresistance and improve treatment outcomes. In ovarian clear
cell carcinoma, the combination of oncolytic viruses and cisplatin
reduced TMEM205 expression and sensitized cells to chemotherapy
(208). Similarly, small
molecule inhibitors such as L-2663 have shown efficacy in enhancing
platinum sensitivity in OC (209). These findings underscore the
potential of TMEM205 as a therapeutic target in combination
therapies.
TMEM205 is a multifaceted protein involved in
chemoresistance, tumor progression and immune modulation across
various types of cancer. Its upregulation is associated with poor
treatment outcomes, while its inhibition enhances chemotherapy
efficacy. As a biomarker, TMEM205 may serve as a diagnostic and
prognostic value, particularly in various types of
platinum-resistant cancer (206-209). Future research should focus on
developing targeted therapies to modulate TMEM205 activity,
potentially improving patient outcomes in various types of
resistant cancer.
TMEM proteins involved in TME
The TME, composed of immune cells, stromal
components, extracellular matrix and soluble factors, carries out a
pivotal role in cancer progression, immune evasion and therapy
response. Growing evidence shows that TPs not only regulate tumor
cell behavior but also engage in bidirectional crosstalk with the
TME, contributing to its remodeling and tumor development (4,5).
Several TMEM proteins have been shown to regulate
tumor-immune interactions. For example, TMEM173 carries out a key
role in the activation of innate immune responses and can modulate
T-cell infiltration and antitumor immunity within the TME (210-212). Evidence from several types of
cancer, including SCLC, TNBC, CC and GBM, demonstrates that
activation of the cGAS-STING pathway can reshape the TME by
promoting chemokine production (such as CXCL10 and CCL5),
increasing CD8+ T cell infiltration and potentiating
antitumor immunity (210,212-214). Microbiota-derived STING
agonists can reprogram monocytes and DC in the TME, enhancing IFN-I
production and improving the efficacy of immune checkpoint blockade
(215). Moreover, STING
signaling can be triggered not only by endogenous cytosolic DNA but
also by tumor-derived exosomes, indicating a dynamic
extracellular-intracellular feedback loop that facilitates immune
regulation (213). STING was
also shown to contribute to immunosuppression in certain contexts,
such as by promoting regulatory T cell (Treg) differentiation or
inducing ER stress and T cell apoptosis through non-canonical
pathways (213,216). Senescent tumor cells release
mitochondrial DNA that activates the cGAS-STING pathway in
myeloid-derived suppressor cells, promoting immunosuppression and
tumor progression (107). These
multifaceted roles highlight TMEM173 as a key modulator of immune
surveillance and tumor progression, reflecting the complex
bidirectional communication between TMEM proteins and the TME.
TMEM205 carries out a key role in modulating TME
and influencing cancer progression. In HCC, TMEM205 expression was
shown to be associated with the immune landscape of the tumor,
where low TMEM205 expression was associated with a poor prognosis
and survival outcomes (207,209). TMEM205 interacted with various
immune cell populations in the TME, particularly macrophages and
Tregs (207,209). The expression of TMEM205 was
positively associated with the proportion of macrophages, including
M1 macrophages, while it was negatively associated with
immunosuppressive M2 macrophage markers and Treg markers. This
suggested that TMEM205 may exert its antitumor effects by reducing
the levels of immunosuppressive cells such as M2 macrophages and
Tregs, while promoting the infiltration of cytotoxic
CD8+ T cells into the TME. Therefore, extracellular
feedback through TMEM205 may regulate immune cell infiltration,
enhancing antitumor immunity and potentially improving therapeutic
responses, especially in combination therapies targeting immune
checkpoints (207,209).
TMEM16A also carries out a key role in shaping the
TME. In PC, high TMEM16A expression is associated with an
immunosuppressive TME, characterized by increased cancer-associated
fibroblasts (CAFs) and reduced CD8+ T cell infiltration
(217). Similarly, in
gastrointestinal cancer, TMEM16A contributed to immunotherapy
resistance by inhibiting cancer ferroptosis and recruiting CAFs,
thereby impairing CD8+ T cell-mediated anti-tumor
immunity (218). TMEM101 also
carried out a key role in modulating the HCC TME and influencing
HCC progression. In HCC, TMEM101 expression was markedly
upregulated, and was associated with a poor prognosis and advanced
tumor grade and stage (219).
TMEM101 was shown to interact with various immune cell populations
within the TME, particularly influencing macrophage infiltration
and T cell activity. High TMEM101 expression is associated with
increased M0 macrophage infiltration, which are pro-tumorigenic and
decreased infiltration of anti-tumor immune cells such as M1
macrophages and resting memory CD4+ T cells. This shift
in immune cell populations suggests that TMEM101 promotes tumor
progression by modifying the immune landscape in favor of tumor
growth. Furthermore, intracellular feedback regulation by TMEM101
was shown to impact pathways related to cell cycle, DNA replication
and repair, facilitating tumor proliferation and survival (219). Thus, TMEM101 not only serves as
a diagnostic and prognostic biomarker for HCC but also acts as an
oncogene that influences the TME, contributing to HCC initiation
and progression (219).
TMEM proteins also mediate extracellular and
intracellular feedback regulation. Extracellularly, they can
interact with cytokines, growth factors and adhesion molecules,
modulating upstream signaling events. TMEM52B is a novel regulator
that modulates the activity of both E-cadherin and EGFR, exhibiting
tumor suppressor-like functions. Lee et al (220) revealed that peptides derived
from the extracellular domain (ECD) of TMEM52B considerably inhibit
cancer cell survival, invasion and anchorage-independent growth,
while reducing the phosphorylation of ERK1/2 and AKT. These
peptides stabilize E-cadherin at cell-cell junctions, leading to
decreased β-catenin activity, and directly bind to the E-cadherin
ECD to block the interaction between soluble E-cadherin and EGFR,
thereby suppressing EGFR activation. These findings suggest that
TMEM52B ECD-derived peptides not only possess strong anti-cancer
potential but also serve as valuable tools for investigating the
regulatory role of TMEM52B in the crosstalk between E-cadherin and
EGFR (220). Intracellularly,
TMEM proteins often participate in key oncogenic pathways such as
MAPK, PI3K/AKT and Wnt, forming regulatory loops that either
amplify or attenuate cellular responses to environmental cues.
These feedback mechanisms may affect cell proliferation, apoptosis,
migration and ultimately, cancer progression (4,5,220).
Collectively, the bidirectional interaction between
TMEM proteins and the TME contributes to tumor plasticity, immune
evasion and chemoresistance. Although this area remains
underexplored, it represents a promising direction for future
research. An improved understanding of how TMEM proteins influence
the TME may lead to the identification of novel therapeutic targets
and strategies to modulate tumor-host interactions.
TMEM proteins in tumor chemoresistance
TMEM45A
Hypoxia is a defining characteristic of solid
tumors, markedly contributing to resistance to both radiotherapy
and chemotherapy, which increases the likelihood of tumor
recurrence (221,222). This phenomenon is particularly
prominent in liver and breast cancer, where the hypoxic TME
provides a protective effect against chemotherapy-induced cell
death. In these conditions, TMEM45A has been shown to serve as a
key mediator of tumor progression and resistance to
chemotherapeutic agents under hypoxic conditions (156-161,221,222). Previous studies have
demonstrated an upregulation of TMEM45A in hypoxic tumor cells,
such as HepG2 and MDA-MB-231 cells, exposed to chemotherapeutic
agents such as Taxol or etoposide. Under these conditions, TMEM45A
protects against apoptosis, a key mechanism that drives
chemotherapy resistance (156-161,221,222). Specifically, silencing TMEM45A
using siRNA alleviated the protective effect of hypoxia and
sensitized the cells to chemotherapy-induced cell death. This
indicates that TMEM45A carries out a key role in sustaining
chemoresistance under hypoxic stress (156-161,221,222). The role of TMEM45A in
modulating chemosensitivity depends on the cancer type, with its
activation potentially acting as either a pro-apoptotic or
anti-apoptotic factor.
In HNSCC and RCC, TMEM45A is upregulated, and its
upregulation is associated with a poorer prognosis. In these types
of cancer, silencing TMEM45A enhanced sensitivity to cisplatin, a
commonly used chemotherapeutic agent, by modulating DNA damage
repair mechanisms and the unfolded protein response pathway
(156-161,221,222). Conversely, in breast and liver
cancer cells, TMEM45A contributed to chemoresistance by inhibiting
apoptotic pathways, thus preventing cell death induced by Taxol and
etoposide (156-161,221,222). Further investigation into the
mechanisms by which TMEM45A regulates chemoresistance uncovered its
involvement in regulating the EMT, a process associated with
enhanced tumor invasion and metastasis (156-161,221,222). In CC, silencing TMEM45A
downregulated EMT markers such as Vimentin and N-cadherin and
increased the expression of the epithelial marker E-cadherin. This
indicated that TMEM45A not only influenced chemoresistance but also
promoted tumor progression through EMT, thereby enhancing the
invasive potential of cancer cells (156-161,221,222). The association between TMEM45A
expression and chemotherapy resistance has been demonstrated in
HPV-positive CC cells. In this context, TMEM45A was highly
expressed in SiHa and HeLa cells and silencing TMEM45A notably
reduced cisplatin resistance, as evidenced by a decrease in the
IC50 values of cisplatin-treated SiHa/DDP and HeLa/DDP
cells. This finding highlights the potential of TMEM45A as a
therapeutic target for overcoming chemoresistance in HPV-positive
CC (156-161,221,222).
TMEM45A expression is upregulated in
Palbociclib-resistant HR+ breast cancer cells and is
associated with accelerated tumor progression and increased
glycolysis (156-161,221,222). Its overexpression drove
Palbociclib resistance, while silencing TMEM45A enhanced
sensitivity to the drug, induced cell cycle arrest and apoptosis,
and inhibited cancer cell proliferation. Mechanistic analysis
revealed that TMEM45A downregulated EMT markers and
glycolysis-related proteins, and this reduced tumor invasiveness.
By activating the AKT/mTOR pathway, TMEM45A regulated the cell
cycle and glycolysis (4,5,156-161,221,222). In mouse models, TMEM45A
knockdown restored Palbociclib sensitivity and inhibited tumor
growth (156-161,221,222). Exosome-mediated delivery of
TMEM45A-targeting siRNA in patient-derived xenografts enhanced
sensitivity to CDK4/6 inhibitors without toxicity (160).
Furthermore, TMEM45A carries out a key role in MDR,
especially in CRC. Studies have shown that TMEM45A expression is
upregulated in 5-fluorouracil (5-FU)-resistant CRC cells (155-162). Knockdown of TMEM45A increased
the sensitivity of these cells to 5-FU, induced apoptosis and
increased the intracellular accumulation of drugs. Additionally,
inhibition of TMEM45A suppressed cell migration and invasion and
reduced the activation of the TGF-β signaling pathway, which is
known to promote EMT and contribute to drug resistance in cancer
(155-162). These findings underscore the
potential of TMEM45A as a target for overcoming MDR in CRC.
In conclusion, TMEM45A is a key regulator of
chemoresistance in various types of cancer, particularly when a
hypoxic environment is present. Its dual role in promoting either
pro-apoptotic or anti-apoptotic pathways, which varies depending on
the cancer type, highlights its complexity as a therapeutic target.
Targeting TMEM45A may, therefore, serve as a promising strategy to
enhance the efficacy of chemotherapy, especially in hypoxic tumors
prone to resistance. Further studies are needed to fully elucidate
the molecular pathways through which TMEM45A exerts its effects on
tumor progression and drug resistance, offering key insights into
the development of novel therapeutic strategies (156-162,221,222).
TMEM158
TMEM158 carries out a key role in mediating
chemoresistance in several types of cancer, including CRC.
Overexpression of TMEM158 increases resistance to cisplatin and
5-FU by upregulating MDR-related proteins such as MDR1, MRP1 and
Bcl-2, thereby enhancing drug efflux and inhibiting apoptosis
(188,190-194). Silencing TMEM158 decreased the
levels of these MDR proteins, restoring drug sensitivity.
Conversely, its overexpression in drug-resistant CRC cells
increased the IC50 values for cisplatin and 5-FU,
highlighting its role in chemoresistance (188,190-194). Modulation of the intrinsic
apoptotic pathway by TMEM158 further contributed to its impact on
tumor survival and drug resistance (193). In cisplatin-resistant NSCLC
PC-9/CDDP cell lines, knockdown of TMEM158 enhanced the sensitivity
of the cells to cisplatin, demonstrating that TMEM158 carries out a
key role in the development of chemoresistance (194). Moreover, TMEM158 expression is
not only associated with resistance to cisplatin in NSCLC but also
acts as a predictive biomarker for treatment response. Given its
role in regulating tumor progression and drug resistance, TMEM158
may serve as a potential biomarker for predicting treatment
response and as a potential therapeutic target for overcoming
chemoresistance.
TMEM88
TMEM88 is a dual-transmembrane protein implicated
in several types of cancer, including breast, ovarian, lung and
testicular cancer. Research suggests that TMEM88 carries out a key
role in mediating tumor progression and is notably associated with
chemoresistance. TMEM88 modulates essential cellular processes,
including cell proliferation, differentiation and apoptosis, by
regulating the Wnt signaling pathway (74-77,223,224).
In OC, TMEM88 expression is associated with
cisplatin resistance, a commonly used chemotherapeutic agent.
Studies revealed that in cisplatin-resistant OC cells, the promoter
methylation of TMEM88 was markedly reduced, leading to its
overexpression (223). TMEM88
inhibited the canonical Wnt signaling pathway and its
overexpression or knockdown affected tumor cell sensitivity to
chemotherapy. Silencing TMEM88 in resistant cells enhanced
sensitivity to cisplatin and increased the expression of Wnt target
genes, such as cyclin D1 and c-Myc, further confirming its role in
modulating Wnt signaling (75,223). Treatment with DNA
methyltransferase inhibitors, such as SGI-110, restored TMEM88
expression, reversing chemoresistance and increasing cellular
sensitivity to cisplatin (223,224). TMEM88 inhibited canonical Wnt
signaling by interacting with DVL, a key mediator in the pathway.
This inhibition carries out a dual role, suppressing tumor
progression and influencing chemoresistance by modulating the
TME.
TMEM88 has emerged as a promising therapeutic
target for overcoming chemoresistance across several types of
cancer. Targeting TMEM88 through epigenetic modulation or gene
therapy may reverse chemoresistance and enhance chemotherapy
efficacy. The role of TMEM88 in the Wnt signaling pathway offers a
molecular basis for its involvement in tumor progression and drug
resistance, highlighting novel avenues for therapeutic strategies
in the management of cancer. Further studies are required to fully
elucidate the mechanisms underlying the role of TMEM88 in tumor
progression and chemoresistance, which may lead to the development
of novel therapeutic interventions to restore chemotherapy
sensitivity (74-82,223,224).
TMEM205
TMEM205 has been identified as a key player in
cisplatin resistance across several types of cancer, including OC
and GC (207-209). It mediates resistance through
various mechanisms, particularly by enhancing drug efflux via the
exosomal pathway. TMEM205 has been associated with increased
exosome release, which can expel cisplatin from cancer cells,
reducing its intracellular accumulation and thus contributing to
resistance (209,225,226).
In OC, TMEM205 was revealed to be upregulated in
cisplatin-resistant cell lines, and its inhibition using specific
small molecules or genetic knockdown notably increased cisplatin
accumulation in these cells, enhancing their sensitivity to the
drug (208,227). Additionally, TMEM205 was shown
to co-localize with RAB8, a protein involved in vesicular
trafficking, further suggesting its role in the regulation of
exosome-mediated drug efflux (208,227). Combining TMEM205 inhibition
with cisplatin or oncolytic virus therapy was found to restore
chemosensitivity and inhibit tumor growth (209,226).
In GC, TMEM205 also carries out a key role in
promoting cisplatin resistance by inducing M2 polarization of TAMs,
which enhanced cancer cell stemness, migration and EMT. These
processes contribute to the malignant progression of the disease
(208,225). The modulation of TMEM205
expression can alter the immune environment of the tumor, further
influencing chemoresistance (227).
Overall, TMEM205 represents a potential therapeutic
target to overcome cisplatin resistance, and strategies such as its
inhibition, combined with other treatments, may improve the
efficacy of chemotherapy in different types of resistant
cancer.
TMEM16A
Recent research has highlighted the key role of
TMEM16A in mediating chemoresistance, particularly to
platinum-based therapies in HNSCC (228). TMEM16A overexpression has been
revealed to increase oxidative stress, which upregulates ATP7B, a
copper-transporting ATPase that facilitates lysosomal sequestration
of platinum compounds, such as cisplatin, thereby diminishing their
cytoplasmic efficacy. This mechanism underpins a novel resistance
pathway, as demonstrated by a positive association between TMEM16A
and ATP7B mRNA levels in squamous cell carcinoma of the head and
neck tumor specimens (133).
Notably, interventions with copper chelators (such as cuprizone and
bathocuproine disulfonic acid) and antioxidants (for example,
N-acetylcysteine) effectively reversed ATP7B induction and restored
cisplatin sensitivity, especially in TMEM16A-overexpressing cells
(229). These findings suggest
that TMEM16A contributes to a copper-dependent resistance
mechanism, offering potential therapeutic implications.
Additionally, TMEM16A expression may serve as a biomarker to guide
the selection of chemotherapy. For example, patients with
cisplatin-resistant oral squamous cell carcinoma in high-risk
groups showed enhanced sensitivity to docetaxel and shikonin,
indicating the potential of TMEM16A in risk stratification and
treatment optimization (229).
Discussion
The present review discusses the roles of various
TMEM proteins in tumorigenesis and their potential contributions to
chemoresistance, providing insights into their mechanisms and
potential therapeutic implications (Table I). TMEM proteins have been
implicated in multiple types of cancer, in which they regulate
fundamental cellular processes such as proliferation, invasion,
migration and apoptosis. The ability of TMEM proteins to influence
tumor progression and resistance to therapy highlights their
potential as targets for cancer treatment (Fig. 4). TMEM proteins carry out a key
role in regulating cancer cell proliferation, migration and
invasion through various molecular mechanisms and signaling
pathways, which can be categorized into four key aspects (Table I and Fig. 4).
 | Table IList of TMEM proteins involved in
cancer. |
Table I
List of TMEM proteins involved in
cancer.
| Name | Amino acids | Location | Involvement in
cancer | Main function | Mechanism | (Refs.) |
|---|
| TMEM7 | 232 | Chromosome
3p21.3 | Tumor suppressor;
involved in liver cancer. | Inhibits the
proliferation, migration, and carcinogenesis of HCC cells. | Mediates epigenetic
silencing through DNA methylation and histone deacetylation. | (66-69) |
| TMEM16A | 986 | Chromosome
11q13.3 | Oncogene; involved
in breast, esophageal, head and neck cancers. | Chloride channel;
regulates cell proliferation, migration and EMT. | Interacts with EGFR
signaling pathways; involved in the activation of EGFR and CAMK
pathways, promoting metastasis through EMT. | (126-151, 217,218, 228,229) |
| TMEM25 | 366 | Chromosome
11q23.3 | Tumor suppressor,
involved in colon, breast, and renal cell carcinoma. | Modulates immune
response; regulates growth factor signaling; affects cell adhesion;
suppresses tumor progression. | Mediates epigenetic
silencing through DNA methylation and histone deacetylation;
inhibits EGFR-mediated STAT3 activation; facilitates immune
evasion. | (12,70-73) |
| TMEM45A | 275 | Chromosome
3q12.1 | Oncogene; involved
in gliomas, breast, liver, renal, ovarian cancer. | Mediator of
chemoresistance under hypoxic and drug-stressed conditions;
involved in epidermal differentiation and keratinization. | Activates the TGF-β
pathway; enhances chemoresistance in hypoxic conditions and
metabolic adaptation; promotes EMT; regulates the cell cycle and
apoptosis. | (4,152-162,221, 222) |
| TMEM45B | 275 | Chromosome
11q24.3 | Oncogene; involved
in lung, pancreatic, gastric cancer. | Promotes cell
proliferation, migration, invasion; inhibits apoptosis. | Regulates
STAT3/JAK2 pathways; modulates tumor microenvironment by
influencing immune responses and hypoxic adaptation. | (163-169) |
| TMEM48 | 674 | Chromosome
1p32.3 | Oncogene; involved
in NSCLC, cervical cancer. | Regulates nuclear
transport and membrane integrity; promotes tumor cell
proliferation, migration, and invasion. | Activates the
Wnt/β-catenin signaling pathway; regulates the cell cycle and
apoptosis. | (170-174) |
| TMEM65 | 240 | Chromosome
8q24.1 | Oncogene; involved
in TNBC, HCC, CRC and GC | Regulates
mitochondrial dynamics and OXPHOS; promotes cell proliferation and
stemness; modulates immune response and tumor
microenvironment. | Transcriptionally
activated by CHD6-TCF4 in CRC, by MYC and TET3 in TNBC; Activates
PI3K-Akt-mTOR pathway via interaction with YWHAZ in GC; enhances
ROS-HIF-1α-SERPINB3 axis to promote stemness and cisplatin
resistance in TNBC. | (175-180) |
| TMEM88 | 159 | Chromosome
17p13.1 | Tumor suppressor;
involved in breast, ovarian, lung, thyroid cancer. | Inhibits Wnt
signaling and regulates chemoresistance; involved in cardiomyocyte
differentiation. | Epigenetic
modulation; inhibits the Wnt pathway by directly interacting with
DVL; subcellular localization-dependent functional switch. | (74-82, 223,224) |
| TMEM106A | 262 | Chromosome
17q21.3 | Tumor suppressor;
involved in gastric and liver cancer. | Immune modulation;
induces apoptosis, delays tumor growth, activates the caspase
pathway. | Promoter
methylation; caspase activation; inhibition of EMT; inactivation of
the ERK1/2/Slug pathway; regulation of macrophage activation and
inflammatory responses. | (83-91) |
| TMEM140 | 185 | Chromosome
7q33 | Oncogene; involved
in gliomas, gastric cancer. | Supports glioma
cell proliferation, migration and invasion; enhances tumor cell
adhesion and survival. | Promotes tumor
aggressiveness and metastasis by regulating the cell cycle (G1
arrest) and inhibiting apoptosis, enhances expression of adhesion
and anti-apoptotic proteins. | (181-186) |
| TMEM158 | 300 | Chromosome
3p21.3 | Oncogene; involved
in NSCLC, pancreatic, colorectal, ovarian cancer. | Promotes cell
proliferation and survival; regulates EMT. | Activation of
TGF-β, PI3K/AKT and STAT3 pathways; implicated in Ras signaling and
involved in tumor progression; regulates immune responses. | (61,187-198) |
| TMEM159 | 161 | Chromosome
16p12.3 | Oncogene; involved
in glioblastoma | Regulates lipid
droplet biogenesis, associated wth lipid metabolism; promotes cell
proliferation and survival. | Interacts with
Seipin; modulates lipid metabolism; implicated in psychiatric
disorders; affects EGFR pathways. | (199-205) |
| TMEM173 | 379 | Chromosome
5q31.2 | Tumor suppressor;
involved in various types of cancer including gastric and liver
cancer. | Regulates type I
interferon signaling, innate immunity, antitumor immunity. | Modulates autophagy
and oxidative stress response; involved in T cell activation,
immune checkpoint modulation, and macrophage polarization. | (92-108, 210-216) |
| TMEM176A | 235 | Chromosome
7q36.1 | Tumor suppressor;
involved in different types of cancer such as ESCC and CRC, linked
with promoter methylation. | Suppresses tumor
growth, regulates immune responses. | Acts through
promoter methylation; inhibits ERK1/2 pathways; modulates immune
responses. | (111-113) |
| TMEM176B | 270 | Chromosome
7q36.1 | Tumor suppressor;
involved in lung adenocarcinoma and GC progression. | Modulates immune
tolerance and antigen cross-presentation; regulates immune
checkpoint responses. | Regulates
phagosomal pH, induces CD8+ T cell activation, interacts
with immune checkpoints, and may serve as a predictor of immune
checkpoint inhibitor response; facilitates immune evasion; involves
tumor-associated macrophages. | (114-123) |
| TMEM205 | 189 | Chromosome
19p13.2 | Oncogene; involved
in GC, OC and HCC. | Mediates
chemoresistance, tumor growth, and immune modulation. | Involves the
exosomal pathway and drug efflux; modulates macrophage polarization
and immune evasion; promotes immune cell infiltration and cytotoxic
T cell activity. | (206-209, 225-227) |
Epigenetic modifications and DNA
methylation
TMEM proteins are frequently regulated at the gene
level by epigenetic modifications, such as DNA methylation and
histone modifications (67-69). For example, TMEM7 and TMEM106A
expression in HCC and GC is silenced by promoter methylation, which
is associated with a poor prognosis (67-69,83,84). Studies have revealed that the
downregulation of these TMEM proteins can be reversed through DNA
demethylation, leading to the restoration of their
tumor-suppressive functions (67-69,83,84). Similarly, TMEM25 in colon cancer
and TMEM176A in esophageal cancer exhibit hypermethylation at their
promoters, which suppresses their expression and promotes tumor
progression (70,71,111,112). A common theme across these
studies is that epigenetic changes, especially promoter
methylation, not only regulate the expression and function of TMEM
proteins but also influence treatment responses (112). For example, TMEM88 in OC has
been associated with the differential sensitivity to chemotherapy
based on its methylation status (223). Moreover, TMEM proteins such as
TMEM173 and TMEM176B are involved in immune regulation, suggesting
that reversing epigenetic silencing of TMEMs could offer dual
therapeutic benefits: Restoring tumor suppressor activity and
enhancing anti-tumor immune responses (97,100,101,119-121). Epigenetic silencing of TMEM
proteins thus represents a notable mechanism of tumorigenesis and a
promising target for therapeutic intervention.
Pathway level, EGFR and other signaling
pathways
TMEM proteins are involved in the modulation of key
signaling pathways that regulate cancer cell behaviors. For
example, TMEM16A interacts with the EGFR in HNSCC and colon cancer,
enhancing EGFR signaling and contributing to cancer cell
proliferation, migration and survival (128). TMEM16A stabilizes EGFR and
forms a positive feedback loop that potentiates cancer progression
(128). Additionally, TMEM45A
is associated with the TGF-β signaling pathway in OC, where it
regulates proliferation and migration (154). TMEM158 has been implicated in
modulating the Wnt/β-catenin signaling pathway in various types of
cancer, facilitating tumor progression and metastasis (189). These interactions with key
oncogenic pathways highlight the importance of TMEM proteins in
driving cancer development and their potential as therapeutic
targets.
Functional mechanisms, regulation of the
cell cycle and apoptosis
TMEM proteins also directly influence cancer cell
function, particularly in terms of regulating the cell cycle and
apoptosis. For example, TMEM106A and TMEM158 have been shown to
induce apoptosis in cancer cells by activating caspases and
inactivating PARPs, which are involved in DNA damage repair
(83,84,193,194). Additionally, TMEM45A regulates
cell cycle progression and promotes cell proliferation by
modulating the TGF-β and Notch pathways, which are central to tumor
growth and invasion (157-160).
TME, hypoxia and immune regulation
TMEM proteins also interact with the TME,
influencing cancer cell behavior and immune responses. TMEM173
(STING) carries out a key role in modulating immune responses
within the TME by regulating innate immunity and immune
surveillance (210-212). The activation of STING
signaling enhances T cell infiltration into tumors, thereby
potentiating antitumor immunity (210,212-214). However, in certain contexts,
TMEM173 can contribute to immune suppression by promoting the
differentiation of Tregs or inducing ER stress (213,216). TMEM158, another key protein, is
involved in immune evasion by regulating immune checkpoints such as
PD-1 and CTLA-4, thereby enhancing the ability of tumors to resist
immune system attacks (190,197,198). Furthermore, TMEM proteins, such
as TMEM205, might improve the prognosis of HCC patients by reducing
the levels of immunosuppressive cells (M2 macrophages and Tregs)
and facilitating the infiltration of cytotoxic T cells into the TME
(207). These interactions with
the TME contribute to tumor plasticity, immune evasion and
resistance to chemotherapy. TMEM proteins influence cancer cell
proliferation, migration and invasion through a combination of
epigenetic regulation, modulation of key signaling pathways (such
as EGFR, TGF-β and Wnt), functional mechanisms related to cell
cycle regulation and apoptosis, and interactions with the TME.
Their complex roles in cancer progression make them promising
therapeutic targets, and further research into their molecular
mechanisms could pave the way for novel cancer therapies.
The TMEM family of proteins carries out pivotal
roles in mediating tumor chemoresistance through a range of
mechanisms across multiple cancer types. TMEM45A is upregulated
under hypoxic conditions and contributes to chemoresistance by
inhibiting apoptosis and promoting EMT, with its effects varying by
cancer type (156-161,221,222). TMEM158 enhances resistance by
upregulating MDR proteins and suppressing apoptosis, particularly
in colorectal and lung cancer (193,194). TMEM88 regulates the Wnt
signaling pathway and is epigenetically modulated, with its
overexpression associated with cisplatin resistance in OC (75,223). TMEM205 promotes cisplatin
resistance via exosome-mediated drug efflux and by shaping the
tumor immune microenvironment, particularly through macrophage
polarization in GC (209,225-227). Collectively, these TMEM
proteins are key regulators of drug resistance, making them
promising targets for overcoming chemoresistance and improving the
efficacy of chemotherapy. TMEM16A mediates platinum-based
chemotherapy resistance in HNSCC by increasing oxidative stress and
upregulating ATP7B, which facilitates lysosomal sequestration of
platinum compounds and may serve as a biomarker to guide
chemotherapy selection (229).
However, there are limitations to the current body
of knowledge regarding the roles of TMEM proteins in cancer.
Variations in study design, population demographics and
methodological approaches introduce inconsistencies, leading to
challenges in generalizing findings across different cancer types.
Further research should address these limitations by employing
standardized methods and exploring longitudinal analyses to improve
the definition of the causal relationships between TMEM protein
expression with tumor progression and chemoresistance. Future
directions for research should focus on mechanistic studies to
elucidate the molecular pathways through which TMEM proteins exert
their effects on tumor biology and therapy resistance. With
advancements in cryo-EM technology, researchers have overcome
challenges such as low expression levels, hydrophobicity and
conformational instability, enabling the high-resolution
determination of several MP structures. Additionally, cryo-EM
allows for the visualization of TPs at near-atomic resolution,
providing detailed insights into their conformational states,
ligand-binding sites and interactions with other proteins. This
capability has greatly enhanced the understanding of TMEM protein
structures and their roles in cellular signaling and chemotherapy
resistance (20,230-232).
Chang et al (233) used cryo-EM to resolve the
structure of a previously unsolved amyloid fibril composed of a
135-amino acid C-terminal fragment of TMEM106B, which is a common
finding in various human neurodegenerative diseases. Cryo-EM
allowed the determination of the TMEM106B fibril structure at a
resolution of 2.7 Å from postmortem human brain tissue. The
prevalence of amyloid fibrils made of TMEM106B across a range of
debilitating human disorders suggests a shared fibrillization
pathway that could initiate or accelerate neurodegeneration. Yuan
et al (234) employed
cryo-EM to analyze the structures of human GLUT4 bound to the small
molecule inhibitor cytochalasin B, revealing an inward-open
conformation at a resolution of 3.3 Å. Despite the nearly identical
transmembrane domain conformation to GLUT1, the cryo-EM structure
of GLUT4 reveals an extracellular glycosylation site and an
intracellular helix that is invisible in the GLUT1 crystal
structure. This structural study using cryo-EM lays the foundation
for understanding the regulatory mechanisms of GLUT4 transport, and
the methods developed for analyzing TMEMs will facilitate the
structural determination of other small solute carrier. Shang et
al (235) used cryo-EM to
reveal the structural transitions of full-length STING/TMEM173 from
both chicken and human. The structure was solved in its inactive
dimeric state and in the dimeric and tetrameric states upon binding
with cGAMP. These structural studies demonstrate how the
transmembrane and cytoplasmic regions interact to form a
domain-swapped dimeric assembly. Additionally, the closure of the
ligand-binding domain, induced by cGAMP, triggers a conformational
change in STING, ultimately facilitating the formation and
activation of the STING oligomer. Zhang et al (236) reported four distinct cryo-EM
structures revealing the apo states of IGF1R, a single
transmembrane protein with a flexible structure. IGF1R carries out
a key role in regulating cellular metabolism and growth. These
conformations were classified as 'Resting states' and 'Active
states', based on the orientation of α-CT helices and structural
symmetry. Moreover, a 'Ligand-pocket' formed in the active
conformations, providing a new perspective on the conformational
changes of apo-IGF1R.
The structural insights gained from cryo-EM have
considerable implications for drug screening and discovery,
particularly in the identification of novel drug targets. For
instance, cryo-EM structures of GPCRs and ion channels have
unveiled detailed molecular interactions with drugs, enabling the
rational design of therapeutics with improved specificity and
efficacy (237-239). Cryo-EM has also facilitated the
study of drug-target complexes, such as the interaction between
statins and the liver transporter OATP1B1, offering a mechanistic
understanding of drug transport and inhibition (240). High-resolution cryo-EM
structures provide a solid foundation for structure-based drug
design, enabling the rational development of small molecules,
peptides and biologics that modulate membrane protein function. For
example, the identification of ligand-binding sites and allosteric
pockets in cryo-EM structures has guided the design of inhibitors
and activators with enhanced specificity and potency (241,242). Moreover, cryo-EM studies of
drug-resistant MPs, such as efflux transporters, have revealed the
structural basis of resistance mechanisms. These insights can
inform the development of next-generation therapeutics that
overcome resistance by targeting alternative binding sites or
modulating protein dynamics (243). Additionally, interventional
studies focused on TMEM-targeted therapies, including epigenetic
modulation and immune checkpoint inhibition, are essential to
validate their therapeutic potential. Expanding on these findings
with multi-omics approaches, such as genomics and proteomics, may
uncover new interactions between TMEM proteins and the TME, thereby
enhancing therapeutic efficacy.
Conclusions
The present review provides a timely and
comprehensive synthesis of the roles that TMEM proteins carry out
in cancer pathogenesis and therapy resistance, emphasizing their
emerging potential as targets for diagnosis, prognosis and
therapeutic intervention. Although individual TMEM family members
have been studied, an integrative analysis of their structural
features, physiological functions and regulatory mechanisms,
particularly their dual roles in tumor progression and
chemoresistance, has been lacking. The present review fills that
gap through a systematic and multi-dimensional analysis across
various types of cancer.
The involvement of TMEM proteins in tumorigenesis,
the TME and drug resistance was discussed, with Table I and Fig. 4 offering summaries of their
functional roles. Furthermore, the regulation of TMEM proteins via
epigenetic modifications, their participation in key signaling
pathways and their influence on cell cycle control, apoptosis, and
immune regulation provide further insight into their multifaceted
contributions to cancer biology.
Importantly, the present review highlights emerging
TMEM proteins with clinical relevance and discuss how targeting
these molecules could offer promising strategies to overcome
therapeutic resistance, particularly in hypoxic tumors. These
findings support the continued investigation of TMEM proteins as
diagnostic biomarkers and therapeutic targets. Personalized
treatment strategies based on TMEM expression profiles may improve
therapeutic efficacy while minimizing adverse effects. Ultimately,
advancing research on TMEM proteins could lead to innovative
diagnostic tools and targeted therapies, improving clinical
outcomes for patients with different types of cancer.
Availability of data and materials
Not applicable.
Authors' contributions
JS, DZ, BY, QL, HX and HP were responsible for the
overall conceptualization and writing of the present review. JI,
DZ, BY, QL, HX and HP contributed to drafting key content. HX and
HP oversaw the critical review of key content and coordinated the
review. All authors read and approved the final manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
CC
|
cervical cancer
|
|
CRC
|
colorectal cancer
|
|
ESCC
|
esophageal squamous cell
carcinoma
|
|
GBM
|
glioblastoma
|
|
GC
|
gastric cancer
|
|
HCC
|
hepatocellular carcinoma
|
|
MDR
|
multidrug resistance
|
|
MPs
|
membrane proteins
|
|
NSCLC
|
non-small cell lung cancer
|
|
OC
|
ovarian cancer
|
|
OS
|
overall survival
|
|
PC
|
pancreatic cancer
|
|
RCC
|
renal cell carcinoma
|
|
TC
|
thyroid cancer
|
|
TME
|
tumor microenvironment
|
|
TMEM
|
transmembrane
|
|
TNBC
|
triple-negative breast cancer
|
|
TPs
|
transmembrane proteins
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science
Foundation of China (grant no. 82173032,82203682), Liaoning
Provincial Science and Technology Plan Project (grant no.
2023JH2/101700156), the Science and Technology Planning Project of
Shenyang (grant no. 22-321-33-50), the Fundamental Research Funds
for the Central Universities (LD202503).
References
|
1
|
Scalise M and Jaakola VP: Membrane
proteins: New approaches to probes, technologies, and drug design.
SLAS Discov. 24:865–866. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Curnow P: Designing minimalist membrane
proteins. Biochem Soc Trans. 47:1233–1245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pandey A, Shin K, Patterson RE, Liu XQ and
Rainey JK: Current strategies for protein production and
purification enabling membrane protein structural biology. Biochem
Cell Biol. 94:507–527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmit K and Michiels C: TMEM proteins in
cancer: A review. Front Pharmacol. 9:13452018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marx S, Dal Maso T, Chen JW, Bury M,
Wouters J, Michiels C and Le Calvé B: Transmembrane (TMEM) protein
family members: Poorly characterized even if essential for the
metastatic process. Semin Cancer Biol. 60:96–106. 2020. View Article : Google Scholar
|
|
6
|
Akkafa F, Koyuncu İ, Temiz E, Dagli H,
Dïlmec F and Akbas H: miRNA-mediated apoptosis activation through
TMEM 48 inhibition in A549 cell line. Biochem Biophys Res Commun.
503:323–329. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun Z, Zhao H, Yang S, Liu R, Yi L, Gao J,
Liu S, Chen Y and Zhang Z: Edaravone Dexborneol protects against
blood-brain barrier disruption following cerebral
ischemia/reperfusion by upregulating pericyte coverage via
vitronectin-integrin and PDGFB/PDGFR-β signaling. Free Radic Biol
Med. 225:758–766. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sabbah DA, Hajjo R and Sweidan K: Review
on epidermal growth factor receptor (EGFR) structure, signaling
pathways, interactions, and recent updates of EGFR inhibitors. Curr
Top Med Chem. 20:815–834. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krook MA, Reeser JW, Ernst G, Barker H,
Wilberding M, Li G, Chen HZ and Roychowdhury S: Fibroblast growth
factor receptors in cancer: Genetic alterations, diagnostics,
therapeutic targets and mechanisms of resistance. Br J Cancer.
124:880–892. 2021. View Article : Google Scholar :
|
|
10
|
Rodrigues Toledo C, Tantawy AA, Lima
Fuscaldi L, Malavolta L and de Aguiar Ferreira C: EGFR- and
integrin α(V)β(3)-targeting peptides as potential
radiometal-labeled radiopharmaceuticals for cancer theranostics.
Int J Mol Sci. 25:85532024. View Article : Google Scholar
|
|
11
|
Chiaranunt P, Ferrara JL and Byersdorfer
CA: Rethinking the paradigm: How comparative studies on fatty acid
oxidation inform our understanding of T cell metabolism. Mol
Immunol. 68C:564–574. 2015. View Article : Google Scholar
|
|
12
|
Mohan CD, Rangappa KS and Sethi G:
Transmembrane protein 25 abrogates monomeric EGFR-driven STAT3
activation in triple-negative breast cancer. MedComm (2020).
5:e4922024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li S, Yue M, Xu H, Zhang X, Mao T, Quan M,
Ma J, Wang Y, Ge W, Wang Y, et al: Chemotherapeutic drugs-induced
pyroptosis mediated by gasdermin E promotes the progression and
chemoresistance of pancreatic cancer. Cancer Lett. 564:2162062023.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee W, Song G and Bae H: Review of
synergistic anticancer effects of natural compounds combined with
conventional therapeutics against chemoresistance and progression
of pancreatic cancer. Mol Cell Toxicol. 21:455–471. 2025.
View Article : Google Scholar
|
|
15
|
Sato A, Takagi K, Yoshida M,
Yamaguchi-Tanaka M, Sagehashi M, Miki Y, Miyashita M and Suzuki T:
Discoidin domain receptor 2 contributes to breast cancer
progression and chemoresistance by interacting with collagen type
I. Cancers. 16:42852024. View Article : Google Scholar :
|
|
16
|
Hegde RS and Keenan RJ: The mechanisms of
integral membrane protein biogenesis. Nat Rev Mol Cell Biol.
23:107–124. 2022. View Article : Google Scholar
|
|
17
|
Li F, Egea PF, Vecchio AJ, Asial I, Gupta
M, Paulino J, Bajaj R, Dickinson MS, Ferguson-Miller S, Monk BC and
Stroud RM: Highlighting membrane protein structure and function: A
celebration of the Protein Data Bank. J Biol Chem. 296:1005572021.
View Article : Google Scholar
|
|
18
|
Tao X, Zhao C and MacKinnon R: Membrane
protein isolation and structure determination in cell-derived
membrane vesicles. Proc Natl Acad Sci USA. 120:e23023251202023.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vénien-Bryan C and Fernandes CAH: Overview
of membrane protein sample preparation for single-particle
cryo-electron microscopy analysis. Int J Mol Sci. 24:147852023.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thonghin N, Kargas V, Clews J and Ford RC:
Cryo-electron microscopy of membrane proteins. Methods.
147:176–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gong J, Chen Y, Pu F, Sun P, He F, Zhang
L, Li Y, Ma Z and Wang H: Understanding membrane protein drug
targets in computational perspective. Curr Drug Targets.
20:551–564. 2019. View Article : Google Scholar
|
|
22
|
Gulezian E, Crivello C, Bednenko J, Zafra
C, Zhang Y, Colussi P and Hussain S: Membrane protein production
and formulation for drug discovery. Trends Pharmacol Sci.
42:657–674. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dodd RB, Wilkinson T and Schofield DJ:
Therapeutic monoclonal antibodies to complex membrane protein
targets: Antigen generation and antibody discovery strategies.
BioDrugs. 32:339–355. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao S, Xu T, Wang W, Li J, Shan Y, Wang Y
and Tan H: Polysaccharides from Anemarrhena asphodeloides Bge, the
extraction, purification, structure characterization, biological
activities and application of a traditional herbal medicine. Int J
Biol Macromol. 311:1434972025. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao S, Wang W, Li J, Wang Y, Shan Y and
Tan H: Unveiling polysaccharides of Houttuynia cordata thunb:
Extraction, purification, structure, bioactivities, and
structure-activity relationships. Phytomedicine. 138:1564362025.
View Article : Google Scholar
|
|
26
|
Gao S, Li J, Wang W, Wang Y, Shan Y and
Tan H: Rabdosia rubescens (Hemsl.) H. Hara: A potent anti-tumor
herbal remedy - Botany, phytochemistry, and clinical applications
and insights. J Ethnopharmacol. 340:1192002025. View Article : Google Scholar
|
|
27
|
Chu XP and Xiong ZG: Acid-sensing ion
channels in pathological conditions. Adv Exp Med Biol. 961:419–431.
2013. View Article : Google Scholar :
|
|
28
|
Nandi PR: Pain in neurological conditions.
Curr Opin Support Palliat Care. 6:194–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jones F, Gamper N and Gao H: Kv7 channels
and excitability disorders. Handb Exp Pharmacol. 267:185–230. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Liu Y, Xue C, Hu Y, Zhao Y, Cai K,
Li M and Luo Z: A protein-based cGAS-STING nanoagonist enhances T
cell-mediated anti-tumor immune responses. Nat Commun. 13:56852022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang SP, Chen FY, Dong LX, Zhang YQ, Chen
HY, Qiao K and Wang KJ: A novel innexin2 forming membrane
hemichannel exhibits immune responses and cell apoptosis in Scylla
paramamosain. Fish Shellfish Immunol. 47:485–499. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hauser AS, Attwood MM, Rask-Andersen M,
Schiöth HB and Gloriam DE: Trends in GPCR drug discovery: New
agents, targets and indications. Nat Rev Drug Discov. 16:829–842.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Snijder HJ and Hakulinen J: Membrane
protein production in E. coli for applications in drug discovery.
Adv Exp Med Biol. 896:59–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kandasamy P, Gyimesi G, Kanai Y and
Hediger MA: Amino acid transporters revisited: New views in health
and disease. Trends Biochem Sci. 43:752–789. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kampen KR: Membrane proteins: the key
players of a cancer cell. J Membr Biol. 242:69–74. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Singh V, Kaur R, Kumari P, Pasricha C and
Singh R: ICAM-1 and VCAM-1: Gatekeepers in various inflammatory and
cardiovascular disorders. Clin Chim Acta. 548:1174872023.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pliego-Arreaga R, Cervantes-Montelongo JA,
Silva-Martínez GA, Tristán-Flores FE, Pantoja-Hernández MA and
Maldonado-Coronado JR: Joint hypermobility syndrome and membrane
proteins: A comprehensive review. Biomolecules. 14:4722024.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guan Q, Bhowmick B, Upadhyay A and Han Q:
Structure and functions of bacterial outer membrane protein A, a
potential therapeutic target for bacterial infection. Curr Top Med
Chem. 21:1129–1138. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kang H and Lee CJ: Transmembrane proteins
with unknown function (TMEMs) as ion channels: Electrophysiological
properties, structure, and pathophysiological roles. Exp Mol Med.
56:850–860. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li L and Li J: Dimerization of
transmembrane proteins in cancer immunotherapy. Membranes.
13:3932023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wesoly J, Pstrąg N, Derylo K,
Michalec-Wawiórka B, Derebecka N, Nowicka H, Kajdasz A, Kluzek K,
Srebniak M, Tchórzewski M, et al: Structural, topological, and
functional characterization of transmembrane proteins TMEM213, 207,
116, 72 and 30B provides a potential link to ccRCC etiology. Am J
Cancer Res. 13:1863–1883. 2023.PubMed/NCBI
|
|
42
|
Nieto Gutierrez A and McDonald PH: GPCRs:
Emerging anti-cancer drug targets. Cell Signal. 41:65–74. 2018.
View Article : Google Scholar
|
|
43
|
Dorsam RT and Gutkind JS:
G-protein-coupled receptors and cancer. Nat Rev Cancer. 7:79–94.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lu X, Luo C, Wu J, Deng Y, Mu X, Zhang T,
Yang X, Liu Q, Li Z, Tang S, et al: Ion channels and transporters
regulate nutrient absorption in health and disease. J Cell Mol Med.
27:2631–2642. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Talukdar S, Emdad L, Das SK and Fisher PB:
EGFR: An essential receptor tyrosine kinase-regulator of cancer
stem cells. Adv Cancer Res. 147:161–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu Q, Huang J, Yan W, Liu Z, Liu S and
Fang W: FGFR families: Biological functions and therapeutic
interventions in tumors. MedComm (2020). 4:e3672023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Riordan R, Saxton A, McMillan PJ, Kow RL,
Liachko NF and Kraemer BC: TMEM106B C-terminal fragments aggregate
and drive neurodegenerative proteinopathy. bioRxiv [Preprint]:
2024.06.11.598478. 2024.
|
|
48
|
Eber E, Trawinska-Bartnicka M, Sands D,
Bellon G, Mellies U, Bolbás K, Quattrucci S, Mazurek H, Widmann R,
Schoergenhofer C, et al: Aerosolized lancovutide in adolescents
(≥12 years) and adults with cystic fibrosis-a randomized trial. J
Cyst Fibros. 20:61–67. 2021. View Article : Google Scholar
|
|
49
|
Vovdenko S, Morozov A, Ali S, Kogan E and
Bezrukov E: Role of monocarboxylate transporters and glucose
transporters in prostate cancer. Urologia. 90:491–498. 2023.
View Article : Google Scholar
|
|
50
|
Pérez-Herrero E and Fernández-Medarde A:
Advanced targeted therapies in cancer: Drug nanocarriers, the
future of chemotherapy. Eur J Pharm Biopharm. 93:52–79. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Perneel J, Neumann M, Heeman B, Cheung S,
Van den Broeck M, Wynants S, Baker M, Vicente CT, Faura J,
Rademakers R and Mackenzie IRA: Accumulation of TMEM106B C-terminal
fragments in neurodegenerative disease and aging. Acta Neuropathol.
145:285–302. 2023. View Article : Google Scholar
|
|
52
|
Granholm AE, Englund E, Gilmore A, Head E,
Yong WH, Perez SE, Guzman SJ, Hamlett ED and Mufson EJ:
Neuropathological findings in Down syndrome, Alzheimer's disease
and control patients with and without SARS-COV-2: Preliminary
findings. Acta Neuropathol. 147:922024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tran Q, Park J, Lee H, Hong Y, Hong S,
Park S, Park J and Kim SH: TMEM39A and human diseases: A brief
review. Toxicol Res. 33:205–209. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jankauskas SS, Varzideh F, Kansakar U, Al
Tibi G, Densu Agyapong E, Gambardella J and Santulli G: Insights
into molecular and cellular functions of the Golgi
calcium/manganese-proton antiporter TMEM165. J Biol Chem.
300:1075672024. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dutta D, Kanca O, Shridharan RV,
Marcogliese PC, Steger B, Morimoto M, Frost FG, Macnamara E;
Undiagnosed Diseases Network; Wangler MF, et al: Loss of the
endoplasmic reticulum protein Tmem208 affects cell polarity,
development, and viability. Proc Natl Acad Sci USA.
121:e23225821212024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hayez A, Roegiers E, Malaisse J, Balau B,
Sterpin C, Achouri Y, De Rouvroit CL, Poumay Y, Michiels C and De
Backer O: TMEM45A is dispensable for epidermal morphogenesis,
keratinization and barrier formation. PLoS One. 11:e01470692016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gallos G, Remy KE, Danielsson J, Funayama
H, Fu XW, Chang HY, Yim P, Xu D and Emala CW Sr: Functional
expression of the TMEM16 family of calcium-activated chloride
channels in airway smooth muscle. Am J Physiol Lung Cell Mol
Physiol. 305:L625–L634. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dulary E, Potelle S, Legrand D and
Foulquier F: TMEM165 deficiencies in congenital disorders of
glycosylation type II (CDG-II): Clues and evidences for roles of
the protein in Golgi functions and ion homeostasis. Tissue Cell.
49A:150–156. 2017. View Article : Google Scholar
|
|
59
|
Dodeller F, Gottar M, Huesken D, Iourgenko
V and Cenni B: The lysosomal transmembrane protein 9B regulates the
activity of inflammatory signaling pathways. J Biol Chem.
283:21487–21494. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Herrera-Quiterio GA and
Encarnación-Guevara S: The transmembrane proteins (TMEM) and their
role in cell proliferation, migration, invasion, and
epithelial-mesenchymal transition in cancer. Front Oncol.
13:12447402023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Jeon M, Yoo S, Park S, Choi Y, An J, Noh
YR and Kim I: Over-expression of transmembrane protein 158 predicts
aggressive tumor behavior and poor prognosis in lung cancer.
Anticancer Res. 44:4885–4893. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pedemonte N and Galietta LJ: Structure and
function of TMEM16 proteins (anoctamins). Physiol Rev. 94:419–459.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shi L, Wang X, Guo S, Gou H, Shang H,
Jiang X, Wei C, Wang J, Li C, Wang L, et al: TMEM65 promotes
gastric tumorigenesis by targeting YWHAZ to activate PI3K-Akt-mTOR
pathway and is a therapeutic target. Oncogene. 43:931–943. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shepard HM, Phillips GL, D Thanos C and
Feldmann M: Developments in therapy with monoclonal antibodies and
related proteins. Clin Med. 17:220–232. 2017. View Article : Google Scholar
|
|
65
|
Posner J, Barrington P, Brier T and
Datta-Mannan A: Monoclonal antibodies: Past, present and future.
Handb Exp Pharmacol. 260:81–141. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shaikh MH, Barrett JW, Khan MI, Kim HAJ,
Zeng PYF, Mymryk JS and Nichols AC: Chromosome 3p loss in the
progression and prognosis of head and neck cancer. Oral Oncol.
109:1049442020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhou X, Popescu NC, Klein G and Imreh S:
The interferon-alpha responsive gene TMEM7 suppresses cell
proliferation and is downregulated in human hepatocellular
carcinoma. Cancer Genet Cytogenet. 177:6–15. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kholodnyuk ID, Kozireva S, Kost-Alimova M,
Kashuba V, Klein G and Imreh S: Down regulation of 3p genes, LTF,
SLC38A3 and DRR1, upon growth of human chromosome 3-mouse
fibrosarcoma hybrids in severe combined immunodeficiency mice. Int
J Cancer. 119:99–107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kiss H, Darai E, Kiss C, Kost-Alimova M,
Klein G, Dumanski JP and Imreh S: Comparative human/murine sequence
analysis of the common eliminated region 1 from human 3p21.3. Mamm
Genome. 13:646–655. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hrašovec S, Hauptman N, Glavač D, Jelenc F
and Ravnik-Glavač M: TMEM25 is a candidate biomarker methylated and
down-regulated in colorectal cancer. Dis Markers. 34:93–104. 2013.
View Article : Google Scholar
|
|
71
|
Bi J, Wu Z, Zhang X, Zeng T, Dai W, Qiu N,
Xu M, Qiao Y, Ke L, Zhao J, et al: TMEM25 inhibits monomeric
EGFR-mediated STAT3 activation in basal state to suppress
triple-negative breast cancer progression. Nat Commun. 14:23422023.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Doolan P, Clynes M, Kennedy S, Mehta JP,
Germano S, Ehrhardt C, Crown J and O'Driscoll L: TMEM25, REPS2 and
Meis 1: Favourable prognostic and predictive biomarkers for breast
cancer. Tumour Biol. 30:200–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xi P, Zhang Z, Liu Y, Nie Y, Gong B, Liu
J, Huang H, Liu Z, Sun T and Xie W: Multidimensional comprehensive
and integrated analysis of the potential function of TMEM25 in
renal clear cell carcinoma with low expression status. Aging
(Albany NY). 16:367–388. 2024.PubMed/NCBI
|
|
74
|
Cai M, Ni WJ, Wang YH, Wang JJ and Zhou H:
Targeting TMEM88 as an attractive therapeutic strategy in malignant
tumors. Front Oncol. 12:9063722022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ge YX, Wang CH, Hu FY, Pan LX, Min J, Niu
KY, Zhang L, Li J and Xu T: New advances of TMEM88 in cancer
initiation and progression, with special emphasis on Wnt signaling
pathway. J Cell Physiol. 233:79–87. 2018. View Article : Google Scholar
|
|
76
|
Li LY, Yang CC, Li SW, Liu YM, Li HD, Hu
S, Zhou H, Wang JL, Shen H, Meng XM, et al: TMEM88 modulates the
secretion of inflammatory factors by regulating YAP signaling
pathway in alcoholic liver disease. Inflamm Res. 69:789–800. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Geng Q, Chen X and Chen N: Transmembrane
protein 88 exerts a tumor-inhibitory role in thyroid cancer through
restriction of Wnt/β-catenin signaling. Exp Cell Res.
395:1121932020. View Article : Google Scholar
|
|
78
|
Nusse R and Clevers H: Wnt/β-Catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhao X, Li G, Chong T, Xue L, Luo Q, Tang
X, Zhai X, Chen J and Zhang X: TMEM88 exhibits an antiproliferative
and anti-invasive effect in bladder cancer by downregulating
Wnt/β-catenin signaling. J Biochem Mol Toxicol. 35:e228352021.
View Article : Google Scholar
|
|
80
|
Zhang X, Yu X, Jiang G, Miao Y, Wang L,
Zhang Y, Liu Y, Fan C, Lin X, Dong Q, et al: Cytosolic TMEM88
promotes invasion and metastasis in lung cancer cells by binding
DVLS. Cancer Res. 75:4527–4537. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ma R, Feng N, Yu X, Lin H, Zhang X, Shi O,
Zhang H, Zhang S, Li L, Zheng M, et al: Promoter methylation of
Wnt/β-Catenin signal inhibitor TMEM88 is associated with
unfavorable prognosis of non-small cell lung cancer. Cancer Biol
Med. 14:377–386. 2017. View Article : Google Scholar
|
|
82
|
Yu X, Zhang X, Zhang Y, Jiang G, Mao X and
Jin F: Cytosolic TMEM88 promotes triple-negative breast cancer by
interacting with Dvl. Oncotarget. 6:25034–25045. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Mao D, Yan F, Zhang X and Gao G: TMEM106A
inhibits enveloped virus release from cell surface. iScience.
25:1038432022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Xu D, Qu L, Hu J, Li G, Lv P, Ma D, Guo M
and Chen Y: Transmembrane protein 106A is silenced by promoter
region hypermethylation and suppresses gastric cancer growth by
inducing apoptosis. J Cell Mol Med. 18:1655–1666. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Choi B, Han TS, Min J, Hur K, Lee SM, Lee
HJ, Kim YJ and Yang HK: MAL and TMEM220 are novel DNA methylation
markers in human gastric cancer. Biomarkers. 22:35–44. 2017.
View Article : Google Scholar
|
|
86
|
Wu C, Xu J, Wang H, Zhang J, Zhong J, Zou
X, Chen Y, Yang G, Zhong Y, Lai D, et al: TMEM106a is a novel tumor
suppressor in human renal cancer. Kidney Blood Press Res.
42:853–864. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Shi S, Wang B, Wan J, Song L, Zhu G, Du J,
Ye L, Zhao Q, Cai J, Chen Q, et al: TMEM106A transcriptionally
regulated by promoter methylation is involved in invasion and
metastasis of hepatocellular carcinoma. Acta Biochim Biophys Sin.
54:1008–1020. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu J and Zhu H: TMEM106A inhibits cell
proliferation, migration, and induces apoptosis of lung cancer
cells. J Cell Biochem. 120:7825–7833. 2019. View Article : Google Scholar
|
|
89
|
Hou J, Niu Y, Yan J, Tian J, Yu W, Zhang
G, Li T and Wang Z: Non-invasive diagnosis for urothelial carcinoma
using a dual-target DNA methylation biomarker panel. Clin Chim
Acta. 569:1201642025. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Guo X, Zeng S, Ji X, Meng X, Lei N, Yang H
and Mu X: Type I Interferon-induced TMEM106A blocks attachment of
EV-A71 virus by interacting with the membrane protein SCARB2. Front
Immunol. 13:8178352022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang X, Feng T, Zhou X, Sullivan PM, Hu
F, Lou Y, Yu J, Feng J, Liu H and Chen Y: Inactivation of TMEM106A
promotes lipopolysaccharide-induced inflammation via the MAPK and
NF-κB signaling pathways in macrophages. Clin Exp Immunol.
203:125–136. 2021. View Article : Google Scholar
|
|
92
|
Sun Z and Hornung V: cGAS-STING signaling.
Curr Biol. 32:R730–R734. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen C and Xu P: Cellular functions of
cGAS-STING signaling. Trends Cell Biol. 33:630–648. 2023.
View Article : Google Scholar
|
|
94
|
Luo X, Li H, Ma L, Zhou J, Guo X, Woo SL,
Pei Y, Knight LR, Deveau M, Chen Y, et al: Expression of STING is
increased in liver tissues from patients with NAFLD and promotes
macrophage-mediated hepatic inflammation and fibrosis in mice.
Gastroenterology. 155:1971–1984.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yu Y, Liu Y, An W, Song J, Zhang Y and
Zhao X: STING-mediated inflammation in Kupffer cells contributes to
progression of nonalcoholic steatohepatitis. J Clin Invest.
129:546–555. 2019. View Article : Google Scholar :
|
|
96
|
Chin KH, Tu ZL, Su YC, Yu YJ, Chen HC, Lo
YC, Chen CP, Barber GN, Chuah ML, Liang ZX and Chou SH: Novel
c-di-GMP recognition modes of the mouse innate immune adaptor
protein STING. Acta Crystallogr D Biol Crystallogr. 69:352–366.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Liu D, Liang S, Ma K, Meng QF, Li X, Wei
J, Zhou M, Yun K, Pan Y, Rao L, et al: Tumor
microenvironment-responsive nanoparticles amplifying STING
signaling pathway for cancer immunotherapy. Adv Mater.
36:e23048452024. View Article : Google Scholar
|
|
98
|
Garland KM, Sheehy TL and Wilson JT:
Chemical and biomolecular strategies for STING pathway activation
in cancer immunotherapy. Chem Rev. 122:5977–6039. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Liang S, Ma H, Liu Y, Hai L, Tian Y, Sun Y
and Wang Z: Nano-immunomodulator amplifies STING activation in
tumor-associated macrophages for cancer immunotherapy. J Control
Release. 383:1138462025. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Deng A, Fan R, Hai Y, Zhuang J, Zhang B,
Lu X, Wang W, Luo L, Bai G, Liang L, et al: A STING agonist prodrug
reprograms tumor-associated macrophage to boost colorectal cancer
immunotherapy. Theranostics. 15:277–299. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ishikawa T, Tamura E, Kasahara M, Uchida
H, Higuchi M, Kobayashi H, Shimizu H, Ogawa E, Yotani N, Irie R, et
al: Severe liver disorder following liver transplantation in
STING-Associated vasculopathy with onset in infancy. J Clin
Immunol. 41:967–974. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Song H, Chen L, Pan X, Shen Y, Ye M, Wang
G, Cui C, Zhou Q, Tseng Y, Gong Z, et al: Targeting tumor
monocyte-intrinsic PD-L1 by rewiring STING signaling and enhancing
STING agonist therapy. Cancer Cell. 43:503–518.e0. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li S, Mirlekar B, Johnson BM, Brickey WJ,
Wrobel JA, Yang N, Song D, Entwistle S, Tan X, Deng M, et al:
STING-induced regulatory B cells compromise NK function in cancer
immunity. Nature. 610:373–380. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Larabi A, Barnich N and Nguyen HTT: New
insights into the interplay between autophagy, gut microbiota and
inflammatory responses in IBD. Autophagy. 16:38–51. 2020.
View Article : Google Scholar :
|
|
105
|
Bu Y, Liu F, Jia QA and Yu SN: Decreased
expression of TMEM173 predicts poor prognosis in patients with
hepatocellular carcinoma. PLoS One. 11:e01656812016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang J, Li S, Wang M, Wang X, Chen S, Sun
Z, Ren X, Huang G, Sumer BD, Yan N, et al: STING licensing of type
I dendritic cells potentiates antitumor immunity. Sci Immunol.
9:eadj39452024. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Lai P, Liu L, Bancaro N, Troiani M, Calì
B, Li Y, Chen J, Singh PK, Arzola RA, Attanasio G, et al:
Mitochondrial DNA released by senescent tumor cells enhances
PMN-MDSC-driven immunosuppression through the cGAS-STING pathway.
Immunity. 58:811–825.e7. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zeng Q, Liu M, Wang Z, Zhou R and Ai K:
Enhancing radiotherapy-induced anti-tumor immunity via
nanoparticle-mediated STING agonist synergy. Mol Cancer.
24:1762025. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Riegman PH, Burgart LJ, Wang KK,
Wink-Godschalk JC, Dinjens WN, Siersema PD, Tilanus HW and van
Dekken H: Allelic imbalance of 7q32.3-q36.1 during tumorigenesis in
Barrett's esophagus. Cancer Res. 62:1531–1533. 2002.PubMed/NCBI
|
|
110
|
Hill M, Russo S, Olivera D, Malcuori M,
Galliussi G and Segovia M: The intracellular cation channel
TMEM176B as a dual immunoregulator. Front Cell Dev Biol.
10:10384292022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wang Y, Zhang Y, Herman JG, Linghu E and
Guo M: Epigenetic silencing of TMEM176A promotes esophageal
squamous cell cancer development. Oncotarget. 8:70035–70048. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Gao D, Han Y, Yang Y, Herman JG, Linghu E,
Zhan Q, Fuks F, Lu ZJ and Guo M: Methylation of TMEM176A is an
independent prognostic marker and is involved in human colorectal
cancer development. Epigenetics. 12:575–583. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Liu Z, An H, Song P, Wang D, Li S, Chen K
and Pang Q: Potential targets of TMEM176A in the growth of
glioblastoma cells. Onco Targets Ther. 11:7763–7775. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Qian W, Xu CY, Hong W, Li ZM and Xu DG:
Transmembrane protein 176B promotes epithelial-mesenchymal
transition in colorectal cancer through inflammasome inhibition.
World J Gastrointest Oncol. 17:976732025. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li J, Fang Z, Dal E, Zhang H, Yu K, Ma M,
Wang M, Sun R, Lu M, Wang H and Li Y: Transmembrane protein 176B
regulates amino acid metabolism through the PI3K-Akt-mTOR signaling
pathway and promotes gastric cancer progression. Cancer Cell Int.
24:952024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yan L, Song Z, Yi L, Tian C, Zhang R, Qin
X, Wang X, Ren S, Ma X, Wang X, et al: TMEM176B inhibits ovarian
cancer progression by regulating EMT via the Wnt/β-catenin
signaling pathway. J Transl Med. 23:3502025. View Article : Google Scholar
|
|
117
|
Condamine T, Le Texier L, Howie D, Lavault
A, Hill M, Halary F, Cobbold S, Waldmann H, Cuturi MC and
Chiffoleau E: Tmem176B and Tmem176A are associated with the
immature state of dendritic cells. J Leukoc Biol. 88:507–515. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Segovia M, Louvet C, Charnet P, Savina A,
Tilly G, Gautreau L, Carretero-Iglesia L, Beriou G, Cebrian I, Cens
T, et al: Autologous dendritic cells prolong allograft survival
through Tmem176b-dependent antigen cross-presentation. Am J
Transplant. 14:1021–1031. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Segovia M, Russo S, Jeldres M, Mahmoud YD,
Perez V, Duhalde M, Charnet P, Rousset M, Victoria S, Veigas F, et
al: Targeting TMEM176B enhances antitumor immunity and augments the
efficacy of immune checkpoint blockers by unleashing inflammasome
activation. Cancer Cell. 35:767–781.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Victoria S, Castro A, Pittini A, Olivera
D, Russo S, Cebrian I, Mombru AW, Osinaga E, Pardo H, Segovia M and
Hill M: Formulating a TMEM176B blocker in chitosan nanoparticles
uncouples its paradoxical roles in innate and adaptive antitumoral
immunity. Int J Biol Macromol. 279:1353272024. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Jiang L, Yang Y, Liu F, Ma M, Gao J, Sun
L, Chen Y, Shen Z and Wu D: A potential diagnostic and prognostic
biomarker TMEM176B and its relationship with immune infiltration in
skin cutaneous melanoma. Front Cell Dev Biol. 10:8599582022.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Jing L, An Y, Cai T, Xiang J, Li B, Guo J,
Ma X, Wei L, Tian Y, Cheng X, et al: A subpopulation of CD146(+)
macrophages enhances antitumor immunity by activating the NLRP3
inflammasome. Cell Mol Immunol. 20:908–923. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Picotto G, Morse LR, Nguyen N, Saltzman J
and Battaglino R: TMEM176A and TMEM176B are candidate regulators of
inhibition of dendritic cell maturation and function after chronic
spinal cord injury. J Neurotrauma. 37:528–533. 2020. View Article : Google Scholar :
|
|
124
|
Cuajungco MP, Podevin W, Valluri VK, Bui
Q, Nguyen VH and Taylor K: Abnormal accumulation of human
transmembrane (TMEM)-176A and 176B proteins is associated with
cancer pathology. Acta Histochem. 114:705–712. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Li H, Zhang M, Linghu E, Zhou F, Herman
JG, Hu L and Guo M: Epigenetic silencing of TMEM176A activates ERK
signaling in human hepatocellular carcinoma. Clin Epigenetics.
10:1372018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Al-Hosni R, Ilkan Z, Agostinelli E and
Tammaro P: The pharmacology of the TMEM16A channel: therapeutic
opportunities. Trends Pharmacol Sci. 43:712–725. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Okuyama K and Yanamoto S: TMEM16A as a
potential treatment target for head and neck cancer. J Exp Clin
Cancer Res. 41:1962022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Bill A, Gutierrez A, Kulkarni S, Kemp C,
Bonenfant D, Voshol H, Duvvuri U and Gaither LA: ANO1/TMEM16A
interacts with EGFR and correlates with sensitivity to
EGFR-targeting therapy in head and neck cancer. Oncotarget.
6:9173–9188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang F, Zhang Y, Gao M and Zeng X: TMEM16A
inhibits renal tubulointerstitial fibrosis via Wnt/β-catenin
signaling during hypertension nephropathy. Cell Signal.
117:1110882024. View Article : Google Scholar
|
|
130
|
Yan Y, Ding X, Han C, Gao J, Liu Z, Liu Y
and Wang K: Involvement of TMEM16A/ANO1 upregulation in the
oncogenesis of colorectal cancer. Biochim Biophys Acta Mol Basis
Dis. 1868:1663702022. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Li H, Yu Z, Wang H, Wang N, Sun X, Yang S,
Hua X and Liu Z: Role of ANO1 in tumors and tumor immunity. J
Cancer Res Clin Oncol. 148:2045–2068. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Filippou A, Pehkonen H, Karhemo PR,
Väänänen J, Nieminen AI, Klefström J, Grénman R, Mäkitie AA,
Joensuu H and Monni O: ANO1 expression orchestrates
p27Kip1/MCL1-mediated signaling in head and neck squamous cell
carcinoma. Cancers. 13:11702021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Shiwarski DJ, Shao C, Bill A, Kim J, Xiao
D, Bertrand CA, Seethala RS, Sano D, Myers JN, Ha P, et al: To
'grow' or 'go': TMEM16A expression as a switch between tumor growth
and metastasis in SCCHN. Clin Cancer Res. 20:4673–4688. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Godse NR, Khan N, Yochum ZA, Gomez-Casal
R, Kemp C, Shiwarski DJ, Seethala RS, Kulich S, Seshadri M, Burns
TF and Duvvuri U: TMEM16A/ANO1 inhibits apoptosis via
downregulation of bim expression. Clin Cancer Res. 23:7324–7332.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Ayoub C, Wasylyk C, Li Y, Thomas E, Marisa
L, Robé A, Roux M, Abecassis J, de Reyniès A and Wasylyk B: ANO1
amplification and expression in HNSCC with a high propensity for
future distant metastasis and its functions in HNSCC cell lines. Br
J Cancer. 103:715–726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Dixit R, Kemp C, Kulich S, Seethala R,
Chiosea S, Ling S, Ha PK and Duvvuri U: TMEM16A/ANO1 is
differentially expressed in HPV-negative versus HPV-positive head
and neck squamous cell carcinoma through promoter methylation. Sci
Rep. 5:166572015. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Liu F, Cao QH, Lu DJ, Luo B, Lu XF, Luo RC
and Wang XG: TMEM16A overexpression contributes to tumor invasion
and poor prognosis of human gastric cancer through TGF-β signaling.
Oncotarget. 6:11585–11599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Britschgi A, Bill A, Brinkhaus H, Rothwell
C, Clay I, Duss S, Rebhan M, Raman P, Guy CT, Wetzel K, et al:
Calcium-activated chloride channel ANO1 promotes breast cancer
progression by activating EGFR and CAMK signaling. Proc Natl Acad
Sci USA. 110:E1026–E1034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Bae JS, Park JY, Park SH, Ha SH, An AR,
Noh SJ, Kwon KS, Jung SH, Park HS, Kang MJ and Jang KY: Expression
of ANO1/DOG1 is associated with shorter survival and progression of
breast carcinomas. Oncotarget. 9:607–621. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Rhodes A, Jasani B, Balaton AJ, Barnes DM
and Miller KD: Frequency of oestrogen and progesterone receptor
positivity by immunohistochemical analysis in 7016 breast
carcinomas: correlation with patient age, assay sensitivity,
threshold value, and mammographic screening. J Clin Pathol.
53:688–696. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Meng Y, Liu Z, Zhai C, Di T, Zhang L,
Zhang L, Xie X, Lin Y, Wang N, Zhao J, et al: Paeonol inhibits the
development of 1-chloro-2,4-dinitrobenzene-induced atopic
dermatitis via mast and T cells in BALB/c mice. Mol Med Rep.
19:3217–3229. 2019.PubMed/NCBI
|
|
142
|
Sui Y, Sun M, Wu F, Yang L, Di W, Zhang G,
Zhong L, Ma Z, Zheng J, Fang X and Ma T: Inhibition of TMEM16A
expression suppresses growth and invasion in human colorectal
cancer cells. PLoS One. 9:e1154432014. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Li H, Yang Q, Huo S, Du Z, Wu F, Zhao H,
Chen S, Yang L, Ma Z and Sui Y: Expression of TMEM16A in colorectal
cancer and its correlation with clinical and pathological
parameters. Front Oncol. 11:6522622021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Liu W, Lu M, Liu B, Huang Y and Wang K:
Inhibition of Ca(2+)-activated Cl(-) channel ANO1/TMEM16A
expression suppresses tumor growth and invasiveness in human
prostate carcinoma. Cancer Lett. 326:41–51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Song Y, Gao J, Guan L, Chen X, Gao J and
Wang K: Inhibition of ANO1/TMEM16A induces apoptosis in human
prostate carcinoma cells by activating TNF-α signaling. Cell Death
Dis. 9:7032018. View Article : Google Scholar
|
|
146
|
Jeon D, Jo M, Lee Y, Park SH, Phan HTL,
Nam JH and Namkung W: Inhibition of ANO1 by cis- and
trans-resveratrol and their anticancer activity in human prostate
cancer PC-3 cells. Int J Mol Sci. 24:11862023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Seo Y, Ryu K, Park J, Jeon DK, Jo S, Lee
HK and Namkung W: Inhibition of ANO1 by luteolin and its
cytotoxicity in human prostate cancer PC-3 cells. PLoS One.
12:e01749352017. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Liu Z, Zhang S, Hou F, Zhang C, Gao J and
Wang K: Inhibition of Ca(2+) -activated chloride channel ANO1
suppresses ovarian cancer through inactivating PI3K/Akt signaling.
Int J Cancer. 144:2215–2226. 2019. View Article : Google Scholar
|
|
149
|
Bai X, Cheng Y, Wan H, Li S, Kang X and
Guo S: Natural compound allicin containing thiosulfinate moieties
as transmembrane protein 16A (TMEM16A) Ion channel inhibitor for
food adjuvant therapy of lung cancer. J Agric Food Chem.
71:535–545. 2023. View Article : Google Scholar
|
|
150
|
Seo Y, Park J, Kim M, Lee HK, Kim JH,
Jeong JH and Namkung W: Inhibition of ANO1/TMEM16A chloride channel
by idebenone and its cytotoxicity to cancer cell lines. PLoS One.
10:e01336562015. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Yin L, Menon R, Gupta R, Vaught L,
Okunieff P and Vidyasagar S: Glucose enhances rotavirus
enterotoxin-induced intestinal chloride secretion. Pflugers Arch.
469:1093–1105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Hayez A, Malaisse J, Roegiers E, Reynier
M, Renard C, Haftek M, Geenen V, Serre G, Simon M, de Rouvroit CL,
et al: High TMEM45A expression is correlated to epidermal
keratinization. Exp Dermatol. 23:339–344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Sun W, Qiu G, Zou Y, Cai Z, Wang P, Lin X,
Huang J, Jiang L, Ding X and Hu G: Knockdown of TMEM45A inhibits
the proliferation, migration and invasion of glioma cells. Int J
Clin Exp Pathol. 8:12657–12667. 2015.
|
|
154
|
Guo J, Chen L, Luo N, Yang W, Qu X and
Cheng Z: Inhibition of TMEM45A suppresses proliferation, induces
cell cycle arrest and reduces cell invasion in human ovarian cancer
cells. Oncol Rep. 33:3124–3130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Manawapat-Klopfer A, Thomsen LT, Martus P,
Munk C, Russ R, Gmuender H, Frederiksen K, Haedicke-Jarboui J,
Stubenrauch F, Kjaer SK and Iftner T: TMEM45A, SERPINB5 and
p16INK4A transcript levels are predictive for development of
high-grade cervical lesions. Am J Cancer Res. 6:1524–1536.
2016.PubMed/NCBI
|
|
156
|
Liu Y, Liu L and Mou ZX: TMEM45A affects
proliferation, apoptosis, epithelial-mesenchymal transition,
migration, invasion and cisplatin resistance of HPV-Positive
cervical cancer cell lines. Biochem Genet. 60:173–190. 2022.
View Article : Google Scholar
|
|
157
|
Neuperger P, Balog J, Tiszlavicz L, Furák
J, Gémes N, Kotogány E, Szalontai K, Puskás LG and Szebeni GJ:
Analysis of the single-cell heterogeneity of adenocarcinoma cell
lines and the investigation of intratumor heterogeneity reveals the
expression of transmembrane protein 45A (TMEM45A) in lung
adenocarcinoma cancer patients. Cancers. 14:1442021. View Article : Google Scholar
|
|
158
|
Jiang H, Chen H, Wan P, Liang M and Chen
N: Upregulation of TMEM45A promoted the progression of clear cell
renal cell carcinoma in vitro. J Inflamm Res. 14:6421–6430. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Xie Q, Guo T, Deng H, Yu C and Fang C:
Pan-cancer analysis of TMEM45A and exploration of its prognostic
value and mechanism in gastric cancer. Cell Mol Biol. 70:218–229.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Chen C, Chen Z, Zhao J, Wen X, Yao H, Weng
Z, Xiong H, Zheng Z and Wu J: TMEM45A enhances palbociclib
resistance and cellular glycolysis by activating AKT/mTOR signaling
pathway in HR+ breast cancer. Cell Death Discov.
11:472025. View Article : Google Scholar
|
|
161
|
Flamant L, Roegiers E, Pierre M, Hayez A,
Sterpin C, De Backer O, Arnould T, Poumay Y and Michiels C: TMEM45A
is essential for hypoxia-induced chemoresistance in breast and
liver cancer cells. BMC Cancer. 12:3912012. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Zhu M, Jiang B, Yan D, Wang X, Ge H and
Sun Y: Knockdown of TMEM45A overcomes multidrug resistance and
epithelial-mesenchymal transition in human colorectal cancer cells
through inhibition of TGF-β signalling pathway. Clin Exp Pharmacol
Physiol. 47:503–516. 2020. View Article : Google Scholar
|
|
163
|
Okada N, Yamamoto T, Watanabe M, Yoshimura
Y, Obana E, Yamazaki N, Kawazoe K, Shinohara Y and Minakuchi K:
Identification of TMEM45B as a protein clearly showing thermal
aggregation in SDS-PAGE gels and dissection of its amino acid
sequence responsible for this aggregation. Protein Expr Purif.
77:118–123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Hu R, Hu F, Xie X, Wang L, Li G, Qiao T,
Wang M and Xiao H: TMEM45B, up-regulated in human lung cancer,
enhances tumorigenicity of lung cancer cells. Tumour Biol.
37:12181–12191. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Zhao LC, Shen BY, Deng XX, Chen H, Zhu ZG
and Peng CH: TMEM45B promotes proliferation, invasion and migration
and inhibits apoptosis in pancreatic cancer cells. Mol Biosyst.
12:1860–1870. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Shen K, Yu W, Yu Y, Liu X and Cui X:
Knockdown of TMEM45B inhibits cell proliferation and invasion in
gastric cancer. Biomed Pharmacother. 104:576–581. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Li Y, Guo W, Liu S, Zhang B, Yu BB, Yang
B, Kan SL and Feng SQ: Silencing transmembrane protein 45B
(TNEM45B) inhibits proliferation, invasion, and tumorigenesis in
osteosarcoma cells. Oncol Res. 25:1021–1026. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Tanioku T, Nishibata M, Tokinaga Y, Konno
K, Watanabe M, Hemmi H, Fukuda-Ohta Y, Kaisho T, Furue H and
Kawamata T: Tmem45b is essential for inflammation- and tissue
injury-induced mechanical pain hypersensitivity. Proc Natl Acad Sci
USA. 119:e21219891192022. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Luo F, Yang K, Wang YZ and Lin D: TMEM45B
is a novel predictive biomarker for prostate cancer progression and
metastasis. Neoplasma. 65:815–821. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Stavru F, Hülsmann BB, Spang A, Hartmann
E, Cordes VC and Görlich D: NDC1: A crucial membrane-integral
nucleoporin of metazoan nuclear pore complexes. J Cell Biol.
173:509–519. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
D'Angelo MA and Hetzer MW: Structure,
dynamics and function of nuclear pore complexes. Trends Cell Biol.
18:456–466. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Mahipal A and Malafa M: Importins and
exportins as therapeutic targets in cancer. Pharmacol Ther.
164:135–143. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Qiao W, Han Y, Jin W, Tian M, Chen P, Min
J, Hu H, Xu B, Zhu W, Xiong L and Lin Q: Overexpression and
biological function of TMEM48 in non-small cell lung carcinoma.
Tumour Biol. 37:2575–2586. 2016. View Article : Google Scholar
|
|
174
|
Jiang XY, Wang L, Liu ZY, Song WX, Zhou M
and Xi L: TMEM48 promotes cell proliferation and invasion in
cervical cancer via activation of the Wnt/β-catenin pathway. J
Recept Signal Transduct Res. 41:371–377. 2021. View Article : Google Scholar
|
|
175
|
Nazli A, Safdar A, Saleem A, Akhtar M,
Brady LI, Schwartzentruber J and Tarnopolsky MA: A mutation in the
TMEM65 gene results in mitochondrial myopathy with severe
neurological manifestations. Eur J Hum Genet. 25:744–751. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Zhang B, Liu Q, Wen W, Gao H, Wei W, Tang
A, Qin B, Lyu H, Meng X, Li K, et al: The chromatin remodeler CHD6
promotes colorectal cancer development by regulating
TMEM65-mediated mitochondrial dynamics via EGF and Wnt signaling.
Cell Discov. 8:1302022. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Zhang YL, Huang MY, Yang SY, Cai JY, Zhao
Q, Zhang FL, Hu X, Shao ZM, Liao L, Cao AY and Li DQ:
MYC/TET3-regulated TMEM65 activates OXPHOS-SERPINB3 pathway to
promote progression and cisplatin resistance in triple-negative
breast cancer. Adv Sci. 12:e004212025. View Article : Google Scholar
|
|
178
|
Qian Z, Liang J, Huang R, Song W, Ying J,
Bi X, Zhao J, Shi Z, Liu W, Liu J, et al: HBV integrations
reshaping genomic structures promote hepatocellular carcinoma. Gut.
73:1169–1182. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Song X, Wang P, Feng R, Chetry M, Li E, Wu
X, Liu Z, Liao S and Lin J: Pan-cancer analysis of prognostic and
immune infiltrates for the TMEM65, especially for the breast
cancer. Evid Based Complement Alternat Med. 2023:93494942023.
View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Li X, Zhou J, Zhang W, You W, Wang J, Zhou
L, Liu L, Chen WW and Li H: Pan-cancer analysis identifies tumor
cell surface targets for CAR-T cell therapies and antibody drug
conjugates. Cancers. 14:56742022. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Li B, Huang MZ, Wang XQ, Tao BB, Zhong J,
Wang XH, Zhang WC and Li ST: TMEM140 is associated with the
prognosis of glioma by promoting cell viability and invasion. J
Hematol Oncol. 8:892015. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Shimizu N, Noda S, Katayama K, Ichikawa H,
Kodama H and Miyoshi H: Identification of genes potentially
involved in supporting hematopoietic stem cell activity of stromal
cell line MC3T3-G2/PA6. Int J Hematol. 87:239–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Guan Y, Guo L, Yang E, Liao Y, Liu L, Che
Y, Zhang Y, Wang L, Wang J and Li Q: HSV-1 nucleocapsid egress
mediated by UL31 in association with UL34 is impeded by cellular
transmembrane protein 140. Virology. 464-465:1–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Chen WC, Wang CY, Hung YH, Weng TY, Yen MC
and Lai MD: Systematic analysis of gene expression alterations and
clinical outcomes for long-chain acyl-coenzyme a synthetase family
in cancer. PLoS One. 11:e01556602016. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Refaat A, Owis M, Abdelhamed S, Saiki I
and Sakurai H: Retrospective screening of microarray data to
identify candidate IFN-inducible genes in a HTLV-1 transformed
model. Oncol Lett. 15:4753–4758. 2018.PubMed/NCBI
|
|
186
|
Baker Frost D, da Silveira W, Hazard ES,
Atanelishvili I, Wilson RC, Flume J, Day KL, Oates JC, Bogatkevich
GS, Feghali-Bostwick C, et al: Differential DNA methylation
landscape in skin fibroblasts from African Americans with systemic
sclerosis. Genes. 12:1292021. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Barradas M, Gonos ES, Zebedee Z, Kolettas
E, Petropoulou C, Delgado MD, León J, Hara E and Serrano M:
Identification of a candidate tumor-suppressor gene specifically
activated during Ras-induced senescence. Exp Cell Res. 273:127–137.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Li J, Wang X, Chen L, Zhang J, Zhang Y,
Ren X, Sun J, Fan X, Fan J, Li T, et al: TMEM158 promotes the
proliferation and migration of glioma cells via STAT3 signaling in
glioblastomas. Cancer Gene Ther. 29:1117–1129. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Zirn B, Samans B, Wittmann S, Pietsch T,
Leuschner I, Graf N and Gessler M: Target genes of the
WNT/beta-catenin pathway in Wilms tumors. Genes Chromosomes Cancer.
45:565–574. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Mohammed Ael S, Eguchi H, Wada S, Koyama
N, Shimizu M, Otani K, Ohtaki M, Tanimoto K, Hiyama K, Gaber MS and
Nishiyama M: TMEM158 and FBLP1 as novel marker genes of cisplatin
sensitivity in non-small cell lung cancer cells. Exp Lung Res.
38:463–474. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Fu Y, Yao N, Ding D, Zhang X, Liu H, Ma L,
Shi W, Zhu C and Tang L: TMEM158 promotes pancreatic cancer
aggressiveness by activation of TGFβ1 and PI3K/AKT signaling
pathway. J Cell Physiol. 235:2761–2775. 2020. View Article : Google Scholar
|
|
192
|
Cui X, Lu J, Zhao C and Duan Y: Oncogenic
transmembrane protein 158 drives the PI3K/Akt signaling pathway to
accelerate gastric cancer cell growth. Braz J Med Biol Res.
56:e129432023. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Liu L, Zhang J, Li S, Yin L and Tai J:
Silencing of TMEM158 inhibits tumorigenesis and multidrug
resistance in colorectal cancer. Nutr Cancer. 72:662–671. 2020.
View Article : Google Scholar
|
|
194
|
Cheng Z, Guo J, Chen L, Luo N, Yang W and
Qu X: Overexpression of TMEM158 contributes to ovarian
carcinogenesis. J Exp Clin Cancer Res. 34:752015. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Zhu X, Liu T and Yin X: TMEM158, as plasma
cfRNA marker, promotes proliferation and doxorubicin resistance in
ovarian cancer. Pharmacogenomics J. 24:342024. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Tong J, Li H, Hu Y, Zhao Z and Li M:
TMEM158 regulates the canonical and non-canonical pathways of TGF-β
to mediate EMT in triple-negative breast cancer. J Cancer.
13:2694–2704. 2022. View Article : Google Scholar :
|
|
197
|
Li J, Hou H, Sun J, Ding Z, Xu Y and Li G:
Systematic pan-cancer analysis identifies transmembrane protein 158
as a potential therapeutic, prognostic and immunological biomarker.
Funct Integr Genomics. 23:1052023. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Huang J, Liu W, Zhang D, Lin B and Li B:
TMEM158 expression is negatively regulated by AR signaling and
associated with favorite survival outcomes in prostate cancers.
Front Oncol. 12:10234552022. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Jawinski P, Kirsten H, Sander C, Spada J,
Ulke C, Huang J, Burkhardt R, Scholz M, Hensch T and Hegerl U:
Human brain arousal in the resting state: A genome-wide association
study. Mol Psychiatry. 24:1599–1609. 2019. View Article : Google Scholar
|
|
200
|
Goodman JM: LDAF1 holds the key to seipin
function. Dev Cell. 51:544–545. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Shi J, Zhang Y, Chen Y, Guo T and Piao H:
Identification of TMEM159 as a biomarker of glioblastoma
progression based on immune characteristics. Biocell. 48:1241–1263.
2024. View Article : Google Scholar
|
|
202
|
Geng F, Cheng X, Wu X, Yoo JY, Cheng C,
Guo JY, Mo X, Ru P, Hurwitz B, Kim SH, et al: Inhibition of SOAT1
suppresses glioblastoma growth via blocking SREBP-1-mediated
lipogenesis. Clin Cancer Res. 22:5337–5348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Castro IG, Eisenberg-Bord M, Persiani E,
Rochford JJ, Schuldiner M and Bohnert M: Promethin is a conserved
seipin partner protein. Cells. 8:2682019. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Bohnert M: New friends for seipin -
Implications of seipin partner proteins in the life cycle of lipid
droplets. Semin Cell Dev Biol. 108:24–32. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Chartschenko E, Hugenroth M, Akhtar I,
Droste A, Kolkhof P, Bohnert M and Beller M: CG32803 is the fly
homolog of LDAF1 and influences lipid storage in vivo. Insect
Biochem Mol Biol. 133:1035122021. View Article : Google Scholar
|
|
206
|
Wagner V, Morton M, Dorayappan KDP,
Gonzalez A, Yu L, Sakaue T, Conrads T, Maxwell GL, Cosgrove C,
Backes F, et al: Circulating extracellular vesicles protein
expression for early prediction of platinum-resistance in
high-grade serous ovarian cancer. Oncogene. 44:1197–1203. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Rao J, Wu X, Zhou X, Deng R and Ma Y:
TMEM205 is an independent prognostic factor and is associated with
immune cell infiltrates in hepatocellular carcinoma. Front Genet.
11:5757762020. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Saini U, Smith BQ, Dorayappan KDP, Yoo JY,
Maxwell GL, Kaur B, Konishi I, O'Malley D, Cohn DE and Selvendiran
K: Targeting TMEM205 mediated drug resistance in ovarian clear cell
carcinoma using oncolytic virus. J Ovarian Res. 15:1302022.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Calo CA, Smith BQ, Dorayappan KDP, Saini
U, Lightfoot M, Wagner V, Kalaiyarasan D, Cosgrove C, Wang QE,
Maxwell GL, et al: Aberrant expression of TMEM205 signaling
promotes platinum resistance in ovarian cancer: An implication for
the antitumor potential of DAP compound. Gynecol Oncol.
164:136–145. 2022. View Article : Google Scholar
|
|
210
|
Sen T, Rodriguez BL, Chen L, Corte CMD,
Morikawa N, Fujimoto J, Cristea S, Nguyen T, Diao L, Li L, et al:
Targeting DNA damage response promotes antitumor immunity through
STING-Mediated T-cell activation in small cell lung cancer. Cancer
Discov. 9:646–661. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Jiang M, Chen P, Wang L, Li W, Chen B, Liu
Y, Wang H, Zhao S, Ye L, He Y and Zhou C: cGAS-STING, an important
pathway in cancer immunotherapy. J Hematol Oncol. 13:812020.
View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Pantelidou C, Sonzogni O, De Oliveria
Taveira M, Mehta AK, Kothari A, Wang D, Visal T, Li MK, Pinto J,
Castrillon JA, et al: PARP inhibitor efficacy depends on CD8(+)
T-cell recruitment via intratumoral STING pathway activation in
BRCA-Deficient models of triple-negative breast cancer. Cancer
Discov. 9:722–737. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Ni H, Zhang H, Li L, Huang H, Guo H, Zhang
L, Li C, Xu JX, Nie CP, Li K, et al: T cell-intrinsic STING
signaling promotes regulatory T cell induction and
immunosuppression by upregulating FOXP3 transcription in cervical
cancer. J Immunother Cancer. 10:e0051512022. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Zhang P, Rashidi A, Zhao J, Silvers C,
Wang H, Castro B, Ellingwood A, Han Y, Lopez-Rosas A, Zannikou M,
et al: STING agonist-loaded, CD47/PD-L1-targeting nanoparticles
potentiate antitumor immunity and radiotherapy for glioblastoma.
Nat Commun. 14:16102023. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Lam KC, Araya RE, Huang A, Chen Q, Di
Modica M, Rodrigues RR, Lopès A, Johnson SB, Schwarz B, Bohrnsen E,
et al: Microbiota triggers STING-type I IFN-dependent monocyte
reprogramming of the tumor microenvironment. Cell.
184:5338–5356.e21. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Wu J, Chen YJ, Dobbs N, Sakai T, Liou J,
Miner JJ and Yan N: STING-mediated disruption of calcium
homeostasis chronically activates ER stress and primes T cell
death. J Exp Med. 216:867–883. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Zhang G, Shu Z, Yu J, Li J, Yi P, Wu B,
Deng D, Yan S, Li Y, Ren D, et al: High ANO1 expression is a
prognostic factor and correlated with an immunosuppressive tumor
microenvironment in pancreatic cancer. Front Immunol.
15:13412092024. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Jiang F, Jia K, Chen Y, Ji C, Chong X, Li
Z, Zhao F, Bai Y, Ge S, Gao J, et al: ANO1-mediated inhibition of
cancer ferroptosis confers immunotherapeutic resistance through
recruiting cancer-associated fibroblasts. Adv Sci. 10:e23008812023.
View Article : Google Scholar
|
|
219
|
Kuang L, Pang Y and Fang Q: TMEM101
expression and its impact on immune cell infiltration and prognosis
in hepatocellular carcinoma. Sci Rep. 14:318472024. View Article : Google Scholar
|
|
220
|
Lee Y, Ko D, Yoon J and Kim S:
TMEM52B-derived peptides inhibit generation of soluble E-cadherin
and EGFR activity to suppress colon cancer growth and early
metastasis. J Transl Med. 23:1462025. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Sermeus A, Cosse JP, Crespin M, Mainfroid
V, de Longueville F, Ninane N, Raes M, Remacle J and Michiels C:
Hypoxia induces protection against etoposide-induced apoptosis:
Molecular profiling of changes in gene expression and transcription
factor activity. Mol Cancer. 7:272008. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Berezin IV, Kershengol'ts BM and Ugarova
NN: Microenvironment of enzymes as 1 of the factors determining
enzyme stability. Stabilization of soluble and immobilized
horseradish peroxidase. Dokl Akad Nauk SSSR. 223:1256–1259. 1975.In
Russian. PubMed/NCBI
|
|
223
|
Lee PS, Teaberry VS, Bland AE, Huang Z,
Whitaker RS, Baba T, Fujii S, Secord AA, Berchuck A and Murphy SK:
Elevated MAL expression is accompanied by promoter hypomethylation
and platinum resistance in epithelial ovarian cancer. Int J Cancer.
126:1378–1389. 2010. View Article : Google Scholar
|
|
224
|
Lee H and Evans T: TMEM88 Inhibits Wnt
signaling by promoting Wnt signalosome localization to
multivesicular bodies. iScience. 19:267–280. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Shen DW and Gottesman MM: RAB8 enhances
TMEM205-mediated cisplatin resistance. Pharm Res. 29:643–650. 2012.
View Article : Google Scholar :
|
|
226
|
Shen DW, Ma J, Okabe M, Zhang G, Xia D and
Gottesman MM: Elevated expression of TMEM205, a hypothetical
membrane protein, is associated with cisplatin resistance. J Cell
Physiol. 225:822–828. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Fu Q, Wu X, Lu Z, Chang Y, Jin Q, Jin T
and Zhang M: TMEM205 induces TAM/M2 polarization to promote
cisplatin resistance in gastric cancer. Gastric Cancer.
27:998–1015. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Hu Y, Zhang Y, He J, Rao H, Zhang D, Shen
Z and Zhou C: ANO1: central role and clinical significance in
non-neoplastic and neoplastic diseases. Front Immunol.
16:15703332025. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Vyas A, Duvvuri U and Kiselyov K:
Copper-dependent ATP7B up-regulation drives the resistance of
TMEM16A-overexpressing head-and-neck cancer models to platinum
toxicity. Biochem J. 476:3705–3719. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Carvalho V, Pronk JW and Engel AH:
Characterization of membrane proteins using cryo-electron
microscopy. Curr Protoc Protein Sci. 94:e722018. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Duan J, He XH, Li SJ and Xu HE:
Cryo-electron microscopy for GPCR research and drug discovery in
endocrinology and metabolism. Nat Rev Endocrinol. 20:349–365. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Robertson MJ, Meyerowitz JG and Skiniotis
G: Drug discovery in the era of cryo-electron microscopy. Trends
Biochem Sci. 47:124–135. 2022. View Article : Google Scholar
|
|
233
|
Chang A, Xiang X, Wang J, Lee C, Arakhamia
T, Simjanoska M, Wang C, Carlomagno Y, Zhang G, Dhingra S, et al:
Homotypic fibrillization of TMEM106B across diverse
neurodegenerative diseases. Cell. 185:1346–1355.e15. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Yuan Y, Kong F, Xu H, Zhu A, Yan N and Yan
C: Cryo-EM structure of human glucose transporter GLUT4. Nat
Commun. 13:26712022. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Shang G, Zhang C, Chen ZJ, Bai XC and
Zhang X: Cryo-EM structures of STING reveal its mechanism of
activation by cyclic GMP-AMP. Nature. 567:389–393. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Zhang X, Wu C, Wei T, Lu Y, Liu C and
Zhang J: Cryo-EM studies of the apo states of human IGF1R. Biochem
Biophys Res Commun. 618:148–152. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
García-Nafría J and Tate CG: Cryo-electron
microscopy: Moving beyond X-ray crystal structures for drug
receptors and drug development. Annu Rev Pharmacol Toxicol.
60:51–71. 2020. View Article : Google Scholar
|
|
238
|
Khorn PA, Luginina AP, Pospelov VA,
Dashevsky DE, Khnykin AN, Moiseeva OV, Safronova NA, Belousov AS,
Mishin AV and Borshchevsky VI: Rational design of drugs targeting
G-Protein-coupled receptors: A structural biology perspective.
Biochemistry. 89:747–764. 2024.PubMed/NCBI
|
|
239
|
Madej MG and Ziegler CM: Dawning of a new
era in TRP channel structural biology by cryo-electron microscopy.
Pflugers Arch. 470:213–225. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Sung MW, Hu K, Hurlimann LM, Lees JA,
Fennell KF, West MA, Costales C, Rodrigues AD, Zimmermann I, Dawson
RJP, et al: Cyclosporine A sterically inhibits statin transport by
solute carrier OATP1B1. J Biol Chem. 301:1084842025. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Ansell TB, Song W, Coupland CE, Carrique
L, Corey RA, Duncan AL, Cassidy CK, Geurts MMG, Rasmussen T, Ward
AB, et al: LipIDens: Simulation assisted interpretation of lipid
densities in cryo-EM structures of membrane proteins. Nat Commun.
14:77742023. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Ackle F, Thavarasah S, Earp JC and Seeger
MA: Rigid enlargement of sybodies with antibody fragments for
cryo-EM analyses of small membrane proteins. Sci Rep. 15:94602025.
View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Sun C and Gennis RB: Single-particle
cryo-EM studies of transmembrane proteins in SMA copolymer
nanodiscs. Chem Phys Lipids. 221:114–119. 2019. View Article : Google Scholar : PubMed/NCBI
|