Introduction
Sepsis is defined as life-threatening organ
dysfunction resulting from a dysregulated host response to
infection (1). Each year, an
estimated 48.9 million people worldwide develop sepsis, leading to
approximately 11 million deaths and accounting for nearly one-fifth
of all global mortality (2,3).
Sepsis is a challenging health problem worldwide with lingering
sequelae (4-6). Sepsis is considered a pathway to
death initiated by the host immune defense system failing to
restore homeostasis in response to an invading pathogen (7-12). Understanding the potential
molecular and cellular features involved in complex immune
pathological mechanisms is key for sepsis management. Neutrophils,
as the most abundant type of white blood cell, are essential
frontline responders to infection (13). In different stages of sepsis,
neutrophils exhibit different transcriptomic profiles and
biological functions (14),
providing phenotypical diversity. Neutrophil subpopulations have
been classified with different subsets causing various
pathophysiological changes in the complex environment of sepsis,
such as immune dysregulation, coagulation dysfunction and organ
damage (15).
Neutrophil heterogeneity in sepsis has been
extensively investigated (16-21). In disagreement with the previous
consensus of neutrophil homogeneity, studies have indicated that
neutrophils may remain in the circulation long enough to interpret
environmental signals and execute specific molecular programs,
providing a rationale for neutrophil diversity in vivo and
promoting neutrophil heterogeneity research (16-21). Researchers have identified
different subsets of neutrophils via surface proteins, such as CD
molecules (22-25). However, there are no uniform
standards for different neutrophil subpopulations distinguished by
phenotypes (26), resulting in
confusing neutrophil nomenclature and disagreements regarding
neutrophil function. It remains unclear whether neutrophil subsets
present in sepsis result from bone marrow mobilization or are
specific subsets formed under the influence of sepsis. Moreover,
whether the neutrophil response to sepsis represents a transient
and reversible activation, or profound gene reprogramming with
long-lasting consequences for host immunity remains unknown. Ng
et al (21) proposed that
the integration of protein and transcript composition, functional
properties, tissue distribution, and genomic organization provides
a more definitive framework for classifying immune cells, thereby
offering a more precise approach to defining true neutrophil
heterogeneity (21). To define
these features, multi-omics analyses provide a more comprehensive
understanding of cellular heterogeneity.
Omics analyses evaluate the complete set of
molecular data, comprising genome, transcriptome, proteome and
metabolome (27). Single omics
studies, predominantly single-cell RNA sequencing (scRNA-seq), have
revealed notable heterogeneity of neutrophils (28-30). High-throughput analysis based on
scRNA-seq enables broad screening of neutrophil heterogeneity,
allowing precise identification of neutrophil subsets combined with
more practical methods, such as flow cytometry (28). As a combination of single omics
approaches, multi-omics unveils the information of a single
regulatory layer and the association between different layers
(31,32). Sepsis is heterogeneous and has a
complicated immunological mechanism, resulting in modulation of
neutrophil diversity (7,33). Multi-omics analyses can help to
elucidate underlying differences of neutrophils in sepsis that have
not been identified by previous classification methods.
The present review summarizes the phenotypical and
functional heterogeneity of neutrophils, as well as advances in the
discovery of neutrophil subsets in sepsis using single omics
approaches and the complex biological mechanisms of neutrophils in
sepsis using multi-omics analysis. The holistic perspective of
multi-omics clarifies the dynamic interplay between neutrophils and
the host immune response, and provides a foundation for the
identification of novel biomarkers and therapeutic targets.
Neutrophil heterogeneity in sepsis
In sepsis, exposure to local or systemic extrinsic
factors may modify neutrophil properties, resulting in the
diversity of neutrophils (21).
Neutrophil phenotypes and functions reflect key features of
neutrophil heterogeneity (Fig.
1).
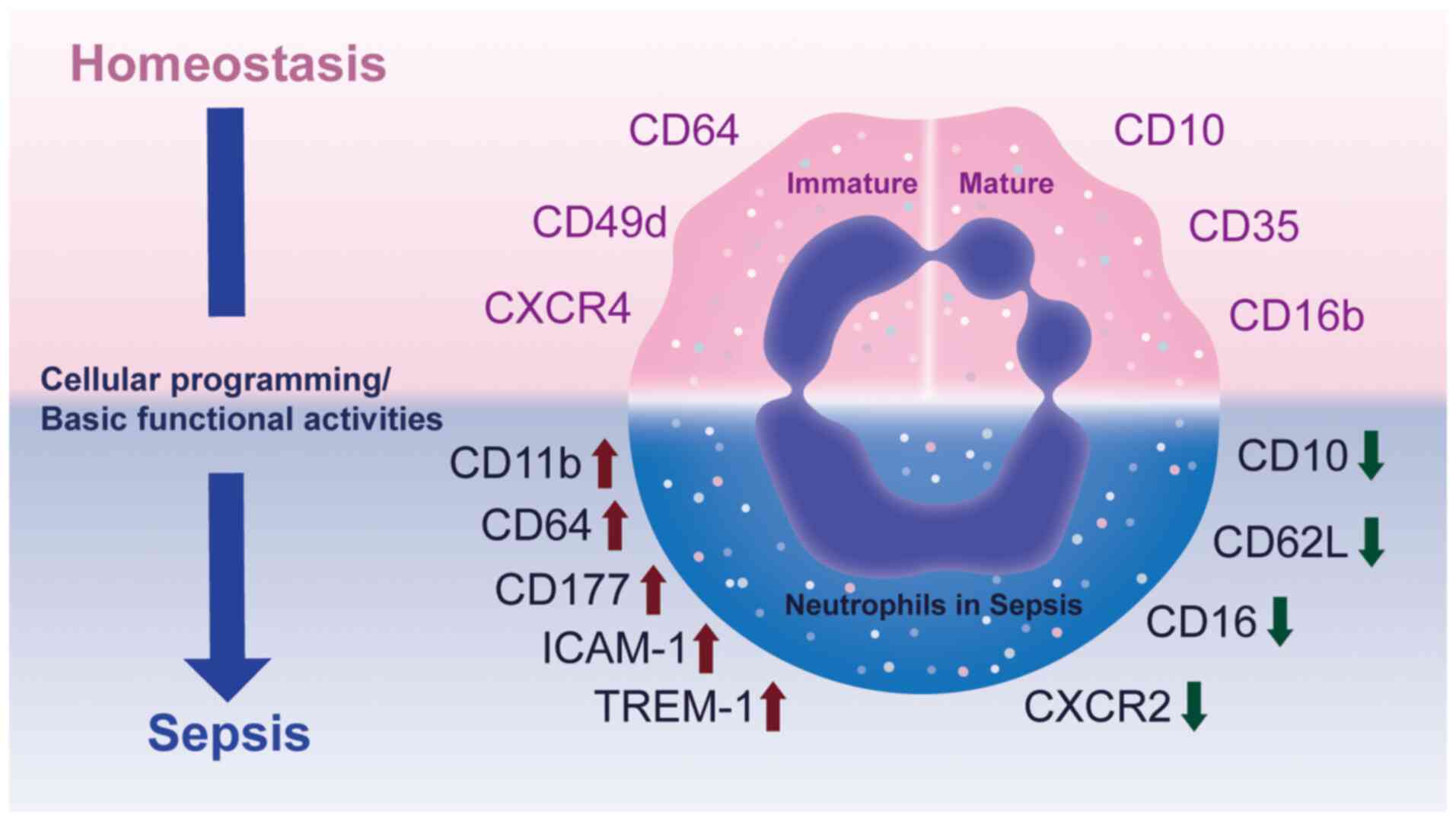 | Figure 1Phenotypical heterogeneity of
neutrophils in homeostasis and sepsis. Under homeostatic
conditions, immature neutrophils are characterized by expression of
CD64, CD49d and CXCR4, whereas mature neutrophils typically express
CD10, CD16b and CD35. During sepsis, both immature and mature
neutrophils are exposed to inflammatory cues that reprogram
cellular activities and functional states, leading to marked
phenotypical shifts. Specifically, surface molecules such as CD11b,
CD64, CD177, ICAM-1 and TREM-1 are upregulated, while CD10, CD62L,
CD16 and CXCR2 are downregulated. These alterations reflect changes
in neutrophil activation, adhesion and migration, however, the
precise functional consequences and regulatory mechanisms
underlying these transitions remain incompletely understood. CXCR,
C-X-C chemokine receptor; ICAM-1, intercellular adhesion
molecule-1; TREM-1, triggering receptor expressed on myeloid
cells-1. |
Phenotypical diversity of neutrophils in
sepsis
Morphologically, the stages of neutrophil
development include promyelocytes, myelocytes, metamyelocytes, band
cells and segmented neutrophils, among which the segmented
neutrophil is considered the mature form (34). Because of their characteristic
multilobed nuclei at the mature stage, neutrophils are often
referred to as polymorphonuclear leukocytes (PMNs).
CD11b+ CD66b+ CD15+
CD14- are commonly used phenotypical markers for human
neutrophils (22). In
homeostasis, as neutrophils mature, neutrophil surface markers
changed to facilitate altered function. Immature neutrophils
express more C-X-C chemokine receptor type 4 (CXCR4) than mature
neutrophils, which may promote its retention in bone marrow
(23). CD16b, CD35 and CD10
appear after neutrophil maturation, whereas CD49d and CD64
disappear (24).
During sepsis, neutrophils present with phenotypical
alterations. Seree-Aphinan et al (25) found that a decrease in CXCR2
surface levels is associated with sepsis due to internalization of
the CXCR2 receptor induced by circulating chemokines (35,36). Other studies also support
decreased expression of the chemokine receptor CXCR2 on the surface
of neutrophils in patients with septic shock (37,38). In addition, CD11b is decreased,
but CXCR1 expression does not change significantly (37,38). Demaret et al demonstrated
that CD10dim CD16dim neutrophils have an
increased frequency in patients with septic shock (39). Neutrophils exhibit increased
CD11b expression and decreased CD62L expression following
stimulation with IL-8 or N-formyl-methionyl-leucyl-pheny lalanine
(39). Similarly, suppressor
cells mobilized during acute inflammation are characterized by
normal expression of CD16, low expression of CD62L and high
expression of CD11b and CD11c (40). Geng et al identified a
third immune regulatory neutrophil in different inflammation
conditions (41). This hybrid
neutrophil subset extravasates at sites of inflammation or
infection and expresses dendritic cell markers CD11c, major
histocompatibility complex class II and costimulatory molecules.
Ode et al (42), using a
mouse model of sepsis, found that the frequency and number of
Intercellular adhesion molecule-1 positive neutrophils are
increased. CD64, a high-affinity immunoglobulin Fcγ receptor I,
mediates phagocytosis of bacteria. Under homeostatic conditions,
the expression of CD64 on neutrophils is comparatively low
(42). By contrast, during
infection, proinflammatory cytokines induce a 10-fold increase in
CD64 expression (43,44). In addition, CD64 expression on
neutrophils is specific for bacterial infection (45,46). Triggering receptor expressed on
myeloid cells-1, a member of the immunoglobulin superfamily, is
upregulated when PMNs are exposed to bacteria (47). Demaret et al (48) revealed that the expression of
CD177 mRNA and protein is increased in circulating neutrophils in
sepsis.
When differentiating solely by markers, however, it
remains unclear whether certain neutrophil subsets are true
lineages or simply represent differentiation and maturation states
induced by the tissue environment.
Functional heterogeneity of neutrophils
in sepsis
As first-line immune cells, neutrophils are required
to adapt to diverse environments and respond promptly. To address
this challenge, neutrophils have multiple capabilities. Typical
functions of neutrophils encompass granule generation and
degranulation, secretion of antimicrobial proteins, production of
reactive oxygen species (ROS), phagocytosis, and formation of
neutrophil extracellular traps (NETs) (49). NETs are DNA scaffolds containing
granule-derived proteins, such as enzymatically active proteases
and anti-microbial peptides, formed to immobilize invading
microorganisms or in response to sterile stimuli (50). Table I summarizes the functional
diversity of neutrophils in sepsis (39,40,51-65).
 | Table IHeterogeneity of neutrophil functions
in sepsis. |
Table I
Heterogeneity of neutrophil functions
in sepsis.
| Functional
aspect | Observations in
sepsis | (Refs.) |
|---|
| NET formation
(NETosis) | Decreased NETosis
observed in septic patients; associated antimicrobial defense | (51) |
| Apoptosis
regulation | Increased
neutrophil production and decreased apoptosis in bone marrow;
delayed apoptosis in circulating neutrophils | (39,51,64) |
| Phagocytosis | Enhanced phagocytic
function in septic shock; immature neutrophils show decreased
phagocytosis | (39,52,64-66) |
| Oxidative burst/ROS
production | Upregulated ROS
generation early in sepsis; decreased oxidative burst in late
sepsis; excessive ROS may drive tissue injury | (39,40,56,58-61) |
|
Chemotaxis/migration | Strongly inhibited
chemotaxis; defective migration in septic patients; mediated by GRK
activation via LPS and cytokines | (39,52-57,64,65) |
| Proinflammatory
cytokine release | High production of
TNF-α, IL-1β, chemokines, leukotrienes, adhesion molecules, ROS and
nitric oxide; immature neutrophils show high TNF-α/IL-10 ratio | (62,64) |
| Immunosuppressive
features | Neutrophil
subpopulation suppresses T cell proliferation via hydrogen peroxide
release | (40) |
| Mature vs. immature
neutrophils | Immature
neutrophils exhibit prolonged lifespan, decreased
phagocytosis/migration and proinflammatory profile; mature
neutrophils exhibit stronger innate immune functions | (64,65) |
| Aged
neutrophils | Decreased return to
bone marrow; preferential migration to inflammation sites; enhanced
phagocytic activity compared with non-aged neutrophils | (66) |
A UK cohort study identified neutrophil dysfunction
in sepsis, including a notable and sustained reduction in NETosis
(active NET release accompanied by cell death), along with
defective neutrophil migration and delayed apoptosis (51). In addition, Demaret et al
(39) found increased neutrophil
production and decreased apoptosis in the bone marrow in sepsis.
The oxidative burst and phagocytic function of neutrophils in
patients with septic shock are increased, but their chemotactic
function is strongly inhibited (39,52). Several studies have shown that
the migration ability of neutrophils in sepsis patients is reduced
(53-56). The underlying mechanism may be
that lipopolysaccharide (LPS) and cytokines in sepsis activate G
protein-coupled receptor kinase on circulating neutrophils, thereby
inducing neutrophils to become desensitized to chemoattractant
(57). Martins et al
(58) also demonstrated that ROS
production is upregulated in neutrophils during sepsis. Notably,
neutrophil oxidative burst is decreased in the late stages of
sepsis (39). These opposing
findings suggested that septic shock may involve a transition from
immune activation to immune suppression, during which neutrophils
may exhibit immunosuppressive features that compromise the host
antimicrobial defense (40,59). For example, Pillay et al
(40) identified a neutrophil
subpopulation that mediates the suppression of T cell proliferation
by releasing neutrophils via local hydrogen peroxide.
The recruitment of bone marrow neutrophils increases
and their apoptosis decreases during sepsis. Functional
characteristics of neutrophils in patients with septic shock
included enhanced respiratory burst and phagocytosis, as well as
inhibited chemotaxis. High production of ROS and proinflammatory
cytokines by neutrophils at sites far from the initial infection
may be part of the pathophysiology of sepsis (56,60,61). Proinflammatory factors, including
tumor necrosis factor-α (TNF-α), IL-1β, chemokines, leukotrienes,
adhesion molecules, ROS, and nitric oxide, serve an important role
in amplifying inflammation, recruiting immune cells and inducing
tissue injury during sepsis (62). Decreased motility associated with
acute neutrophil activation is hypothesized to play a role in the
development of multiorgan failure following sepsis (63).
The functions of mature and immature neutrophils in
sepsis are heterogenous. Drifte et al reported that immature
circulating neutrophils of patients with septic shock support
innate immune defenses to a lesser extent than mature neutrophils
(64). Moreover, immature
neutrophils possess a longer lifespan and a stronger capacity to
resist spontaneous apoptosis (64). Compared with mature granulocytes,
the phagocytosis and migration abilities of immature neutrophils
are lower (64,65). The high TNF-α/IL-10 ratio in
immature neutrophils suggests these cells adopt a proinflammatory
profile, characterized by predominant production of inflammatory
cytokines (64). Aged
neutrophils also have different characteristics. Using a mouse
model, Uhl et al (66)
reported that the number of aged neutrophils returning to the bone
marrow is decreased during an acute inflammatory response to
endotoxemia, owing to rapid migration to sites of inflammation.
Upon reaching inflamed tissues, aged neutrophils exhibit higher
phagocytic activity than subsequently recruited non-aged
neutrophils (66).
The abundant phenotypical and functional
heterogeneity of neutrophils has been extensively studied, but the
underlying mechanisms of diversity remain unclear (21,67). Neutrophils exhibit plasticity,
allowing them to respond and adapt to various stimuli. Such
context-dependent and reversible changes may resemble stable
subsets, thereby confounding the interpretation of true
heterogeneity. Because the methodologies used to study neutrophil
heterogeneity need improvement and investigations of neutrophil
heterogeneity are in an exploratory stage, additional studies are
needed to understand whether neutrophil heterogeneity is the result
of cell programming or the basic functional activity of cells.
Neutrophil heterogeneity in sepsis by
omics
The intricate nature of sepsis and its impact on
neutrophil function has prompted researchers to investigate the
molecular underpinnings of neutrophil heterogeneity using omics
technologies (68-76). Each omics layer sheds light on
neutrophil behavior in sepsis, providing insight into their roles
in both host defense and immune dysregulation. Table II provides a brief summary of
omics techniques (68-76).
 | Table IIOmics levels, research scope and
technology. |
Table II
Omics levels, research scope and
technology.
| Omics level | Definition | Research scope |
Technology/platform | Explanation | (Refs.) |
|---|
| Genomics | Comprehensive
analysis of the genome, including DNA sequence, structural variants
and mutations | WGS; WES | Short-read seq
(Illumina, Inc.); long-read seq (PacBio, ONT) | WGS covers the
entire genome to detect variants; WES focuses on protein-coding
regions; long-read seq resolves structural variants and repetitive
elements | (68-70) |
| Epigenomics | Study of
genome-wide epigenetic modifications regulating gene expression
without altering DNA sequence | DNA methylation
profiling; chromatin accessibility mapping; protein-DNA interaction
profiling | Bisulfite seq;
ATAC-seq; ChIP-seq | DNA methylation
profiling reveals CpG methylation; ATAC-seq measures chromatin
accessibility; ChIP-seq identifies histone modifications or TF
binding | (71) |
|
Transcriptomics | Profiling of RNA
expression levels to capture gene activity | Bulk RNA-seq;
scRNA-seq; spatial transcriptomics | Short-read
sequencing (Illumina, Inc.); in situ capture platforms (10x
Genomics, Slide-seq) | Bulk RNA-seq
quantifies average transcript abundance; scRNA-seq resolves
cell-to-cell heterogeneity; spatial transcriptomics retains tissue
architecture while measuring gene expression | (72) |
| Proteomics | Global profiling of
protein abundance, modification and interactions | Protein
identification and quantification | MS-based proteomics
(LC-MS/MS, DIA, TMT); antibody-based proteomics (RPPA, CyTOF) | MS-based methods
enable high-throughput protein identification and quantification;
antibody-based methods provide sensitive detection of specific
proteins | (73,74) |
| Metabolomics | Systematic
identification and quantification of metabolites reflecting cell
metabolic state | Targeted and
untargeted metabolomics | LC-MS, GC-MS, NMR
platforms | Targeted
metabolomics measures predefined metabolites with high sensitivity;
untargeted metabolomics broadly surveys metabolites to capture
global metabolic changes | (75,76) |
Genome
Genomic studies have revealed polymorphisms and
mutations within genes associated with neutrophil functions that
may predispose individuals to sepsis or influence disease severity
(77-80). Andiappan et al (77) investigated the expression
quantitative trait loci (eQTL) of neutrophils and identified 21,210
eQTLs on 832 unique genes. The aforementioned study used Ingenuity
Pathway Analysis to reveal an enrichment of neutrophil eQTLs in
inflammatory disease, consistent with the established role of
neutrophils in host defense against pathogens (77). Utilizing a genome-wide
association study, Wang et al (78) investigated NET biomarkers to
explore the causal association between NET and sepsis.
Myeloperoxidase (MPO)-DNA complex is a biomarker of NET (79). With every standard deviation
increase in the levels of the MPO-DNA complex, there is an ~18%
increase in the risk of sepsis, a 51% increase in the risk of
28-day death from sepsis, ~38% rise in the risk of requiring
intensive care due to sepsis and ~125% higher risk of 28-day death
from sepsis requiring intensive care (80). Elevated NET levels may elevate
the risk of sepsis onset, progression and mortality (80).
Beyond the genome, the epigenome provides an
additional layer of regulation that shapes neutrophil
heterogeneity. Epigenetic mechanisms such as histone modification,
DNA methylation and chromatin accessibility dynamically modulate
transcriptional programs, enabling neutrophils to adopt diverse
functional states in response to microenvironmental cues. Using
chromatin immunoprecipitation sequencing of histone H3K4me3, Piatek
et al (80) characterized
the epigenetic regulation of human neutrophil plasticity and
heterogeneity following stimulation with LPS, TNF-α or IL-10. The
aforementioned study revealed that changes in H3K4me3-marked
transcriptional start sites are associated with diverse functional
programs, including neutrophil activation, cytokine production,
apoptosis, histone remodeling and NF-κB signaling pathways
(80). IL-10 induces a distinct
subset of apoptotic yet transcriptionally active neutrophils, which
display a non-canonical NF-κB driven cytokine profile while
simultaneously suppressing the canonical NF-κB pathway (80). These findings highlight that
epigenomic profiling uncovers previously unrecognized heterogeneity
in neutrophil functional states, and that H3K4me3-associated DNA
binding sites may serve as potential therapeutic targets for
immunomodulation.
Transcriptome
Building on the genomic foundation, transcriptomic
studies have advanced understanding of neutrophil heterogeneity
(81-88).
Bulk transcriptomic profiling in patients with
sepsis has identified 37 differentially expressed genes, with
functional enrichment and protein-protein interaction analyses
highlighting immune and inflammatory signaling pathways, including
the PI3K/AKT axis, as key regulators of neutrophil specialization
(81). Validation in whole-blood
neutrophil samples from patients with sepsis patients further
confirmed these transcriptional alterations (81). These findings demonstrate
sepsis-associated transcriptional alterations, but the limited
resolution of bulk transcriptomics obscures cellular heterogeneity
and prevents the identification of distinct neutrophil subsets.
scRNA-seq provides a deeper resolution. Xie et al (28) identified eight transcriptional
neutrophil clusters in mice, with G0-G4 representing bone marrow
developmental stages and G5a-c representing three transcriptionally
distinct mature neutrophil subsets in peripheral blood. Bacterial
infection accelerates the transition from immature to mature states
without altering overall heterogeneity but induces stage-specific
transcriptional reprogramming: Early progenitors upregulate genes
regulating immune effector functions and ROS metabolism, while
mature subsets exhibit increased cytokine production (28). Infection also reshapes
transcription factor networks, with defense-associated factors such
as interferon regulatory factor 7 activated in immature cells and
metabolic regulators such as forkhead box protein P1 (Foxp1) and
CCCTC-binding factor (Ctcf) downregulated in mature subsets,
suggesting a redistribution of cell resources toward host defense
(28). However, a murine model
raises questions about their direct applicability to human sepsis,
where immune and inflammatory contexts may differ (28). In human sepsis, Xu et al
(82) identified novel
neutrophil subtypes enriched in late differentiation stages,
characterized by upregulation of alkaline phosphatase,
liver/bone/kidney (ALPL), CD177 molecule (CD177), S100
calcium-binding protein A8 (S100A8), S100A9), and syntaxin
binding protein 2 (STXBP2), providing potential biomarkers for
therapy. Hong et al (83)
further categorized neutrophils into four subsets (Neu1-Neu4) by
single-cell transcriptomic profiling, with Neu1 expansion
associated with septic shock and higher Sequential Organ Failure
Assessment (SOFA) scores. A Neu1-specific gene module, including
NFKBIA (NFKB inhibitor alpha, an inhibitor of NF-κB signaling),
CXCL8 (C-X-C motif chemokine ligand 8, encoding interleukin-8),
G0S2 (G0/G1 switch gene 2, a regulator of cell cycle and
apoptosis), and FTH1 (ferritin heavy chain 1, involved in iron
storage and oxidative stress response), demonstrates strong
predictive value for shock. Consistently, Neu1 expresses cell
surface markers such as CD123, CD38 and CD69, which have been
linked to poor prognosis (84-87). However, whether these subsets
represent stable cell states or transient activation phenotypes
remains unresolved.
Beyond proinflammatory subtypes, immunosuppressive
phenotypes have also been reported: Qi et al (88) identified a PD-L1high
neutrophil subset induced via the p38α pathway, involving mitogen-
and stress-activated kinase 1 (MSK1) and MAPK-activated protein
kinase 2 (MK2),, capable of suppressing T cell activation and
promoting apoptosis or trans-differentiation (88).
Collectively, transcriptomics reveals that
neutrophil heterogeneity in sepsis arises from dynamic
transcriptional reprogramming, encompassing both proinflammatory
and immunosuppressive subsets with distinct clinical relevance.
Proteome
Transcriptomic profiling has advanced understanding
of neutrophil gene expression patterns in sepsis, but does not
directly link transcriptional changes to protein function.
Proteomics fills this gap by quantifying protein abundance and
activity, thereby capturing more immediate functional adaptations
of neutrophils.
Tak et al (89) identified CD62Ldim
neutrophils as a distinct subset that typically resides outside
circulation but is recruited during acute inflammation. Their
proteomic signature, enriched in proteins associated with adhesion,
activation and immune regulation, underscores functional
specialization beyond conventional morphological classification
(89). However, whether these
proteomic differences translate into stable functional phenotypes
or reflect transient activation states remains uncertain,
especially since CD62L shedding can occur in multiple inflammatory
contexts (89). This raises the
question of whether CD62Ldim neutrophils represent a
subset or activation-driven state detectable by proteomics but less
evident in physiological conditions. In parallel, hypoxia, a
hallmark of sepsis pathophysiology, triggers notable proteome
remodeling (90). Watts et
al demonstrated that hypoxic neutrophils upregulate
inflammatory receptors, including formyl peptide receptor (FPR) and
granulocyte-macrophage colony-stimulating factor receptor beta
chain (GM-CSF receptor β), enhance lysosomal protein scavenging and
sustain biosynthesis of granule and cytoskeletal proteins through
de novo synthesis (91).
These findings highlight the metabolic adaptability of neutrophils
under stress (91). The reliance
on murine models and controlled hypoxic exposure limits direct
extrapolation to human sepsis, where hypoxia is heterogeneous,
dynamic and often accompanied by additional insults such as
acidosis or oxidative stress (90,91). Furthermore, whether such
proteomic adaptations enhance host defense or contribute to
maladaptive inflammation remains unresolved.
Taken together, proteomic profiling demonstrates
that neutrophils are not passive effectors but metabolically
flexible cells capable of remodeling the proteome in response to
inflammatory and metabolic stressors. Nevertheless, studies are
largely descriptive and rely on either ex vivo stimulation
or animal models, which may not capture the temporal and spatial
complexity of sepsis in patients (89-91).
Metabolome
Proteomics reveals alterations in signaling
molecules, surface receptors and effector proteins that directly
regulate neutrophil behavior but does not capture the intricate
biochemical pathways that govern these changes in cell function and
energy metabolism. Metabolic reprogramming refers to the dynamic
reshaping of cell metabolic pathways in response to environmental
and functional demands (92). In
neutrophils, metabolomics has revealed that this process extends
beyond glycolysis to include oxidative phosphorylation and the
pentose phosphate pathway, enabling distinct effector functions
such as chemotaxis, ROS generation and NET formation (93). This metabolic adaptability
constitutes a fundamental basis of neutrophil heterogeneity,
particularly in sepsis.
In a recent study, Li et al (94) observed significant alterations in
neutrophil metabolism as severe corona virus disease 2019
(COVID-19) progresses, particularly in amino acid, redox and
central carbon metabolism. Metabolic changes in neutrophils are
associated with decreased activity of the glycolytic enzyme GAPDH
(94). When GAPDH is inhibited,
glycolysis is suppressed, leading to an increase in pentose
phosphate pathway activity, but this also diminishes the neutrophil
respiratory burst (94).
Furthermore, inhibiting GAPDH triggers the formation of NETs, which
depends on the activity of neutrophil elastase (94). This inhibition results in an
increase in neutrophil pH and blocking this pH rise prevents both
cell death and NET formation (94). These findings indicated that
neutrophils in severe COVID-19 exhibit a heterogeneous and
dysfunctional metabolic profile, which contributes to their
impaired function.
Multi-omics to explore neutrophil
heterogeneity in sepsis
Each layer of omics offers insight into genomic
variants, transcriptional changes, protein modification or
metabolic shift. However, they often present a limited view of the
complex interplay between these biological layers. For example,
genomics identifies genetic predispositions that shape neutrophil
responses, but does not capture the dynamic processes that occur
during gene expression and protein translation. Similarly,
transcriptomics reveals altered gene expression profiles associated
with sepsis but may overlook the functional implications of changes
at the protein level. Transitioning from single to multi-omics
allows for a more holistic perspective by integrating data across
different layers of biological information. Multi-omics approaches
not only facilitate the identification of distinct neutrophil
subsets and their functional states in real time but also enhance
understanding of how metabolic changes influence neutrophil
activity during sepsis (Fig.
2).
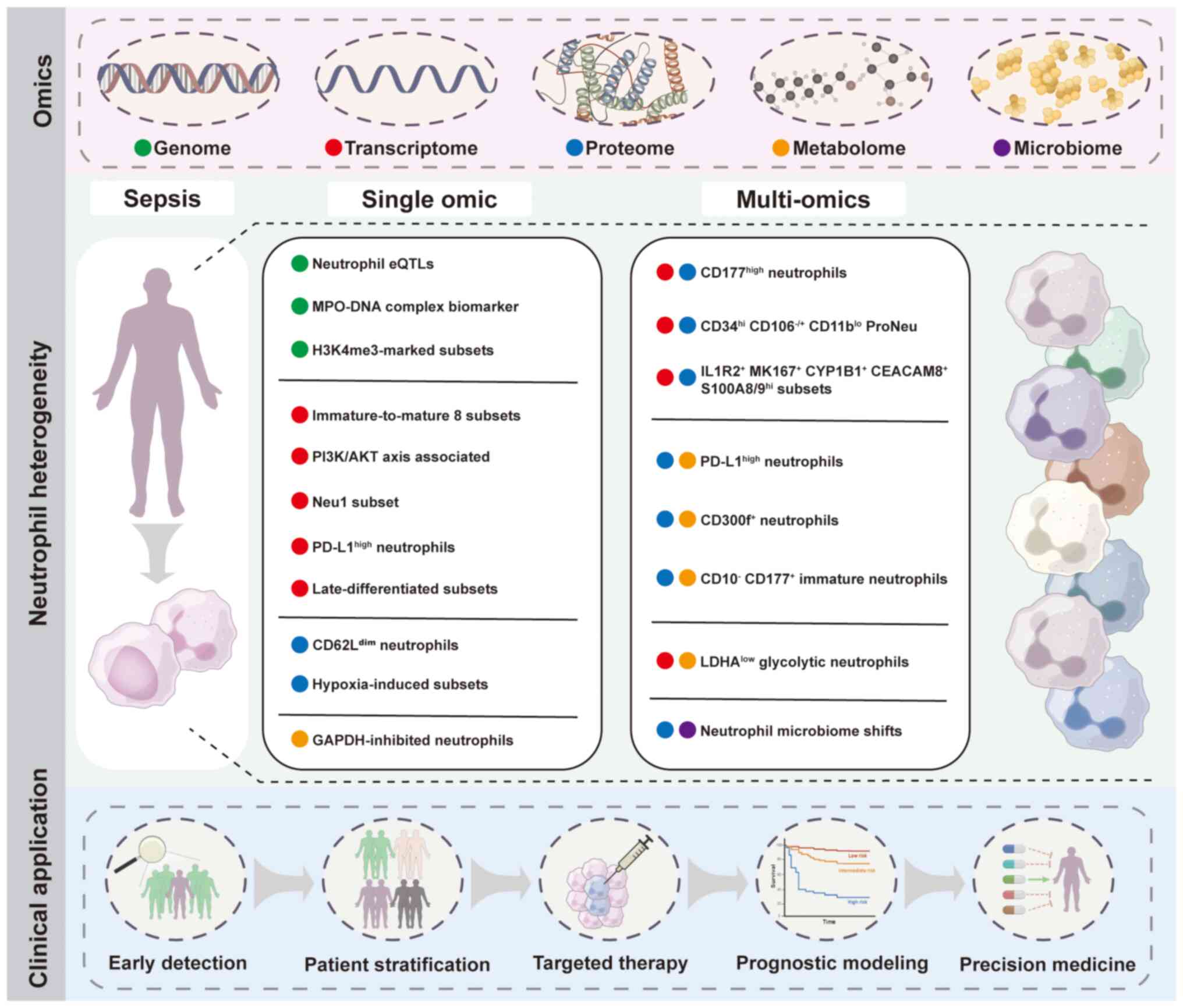 | Figure 2Multi-omics approaches reveal
neutrophil heterogeneity and clinical implications in sepsis. Omics
layers include genome, transcriptome, proteome, metabolome and
microbiome. Neutrophil subsets and characteristics are identified
through single-omics studies and multi-omics integration in sepsis.
Potential clinical applications of neutrophil heterogeneity
profiling include early detection, patient stratification, targeted
therapy, prognostic modeling and precision medicine. eQTL,
expression quantitative trait loci; MPO, myeloperoxidase; H3K4me3,
histone H3 lysine 4 trimethylation; PI3K/AKT, phosphatidylinositol
3-kinase/protein kinase B; PD-L1, programmed death-ligand 1; MKI67,
marker of proliferation Ki-67; CYP1B1, cytochrome P450 family 1
subfamily B member 1; CEACAM8, carcinoembryonic antigen-related
cell adhesion molecule 8; S100A8/9, S100 calcium-binding protein
A8/9; LDHA, lactate dehydrogenase A; Neu1, neutrophil subtype
1. |
Transcriptome and proteome
Integration of transcriptomic and proteomic data has
advanced understanding of neutrophil heterogeneity and its
functional consequences during sepsis. By linking gene expression
with protein abundance and post-translational modification, these
approaches not only delineate the molecular programs of neutrophils
but also capture how these programs are dynamically reconfigured
under septic conditions.
The source of neutrophil heterogeneity remains
debated. On one hand, studies have suggested that circulating
neutrophils follow a predefined differentiation trajectory derived
from hematopoietic progenitor cells (95,96). On the other hand, in vitro
experiments have shown that stimulation of mature neutrophils with
inflammation-associated molecules can also reshape their
transcriptome, indicating that microenvironmental cues serve a
crucial role in driving functional diversity (88,97). Thus, both developmental origin
and peripheral reprogramming may contribute to heterogeneity.
Kaiser et al (98) used
transcriptomics and proteomics to reveal mechanisms associated with
functional reprogramming of peripheral neutrophils during acute
infection. Transcriptome analysis revealed increased expression of
classical neutrophil markers such as CXCL8, SOD2, S100A8/A9, CSF3
receptor and myeloid cell nuclear differentiation antigen (MNDA)
(98). Activation markers and
antimicrobial genes are upregulated, including cystatin F (CST7),
S100 calcium binding protein A12 (S100A12), interleukin 1 receptor
type 2 (IL1R2) and annexin A1 (ANXA1) (98). The aforementioned study used mass
spectrometry to assess whether post-infection transcriptomic
alterations are reflected in the proteome of human neutrophils
(98). Corresponding
upregulation at both transcriptional and protein levels included
interferon-induced transmembrane protein 3 and alkaline phosphatase
(98). In addition, patients
with acute bacterial infections show enrichment of
CD177high neutrophils (98). Both transcription and protein
expression of CD177 are positively associated with disease severity
(98). These results suggest
that the transcriptome response of neutrophils is effectively
translated into the proteome during bacterial inflammation.
However, the aforementioned study did not resolve how
transcriptomic shifts intersect with upstream regulatory mechanisms
such as chromatin accessibility and transcription factor activity
(95), leaving the drivers of
neutrophil reprogramming incompletely understood. Moreover,
although CD177 expression is associated with disease severity, its
biological role remains unclear, especially since up to 10% of
individuals lack CD177 without apparent immune defects (99-101). This raises the possibility that
CD177 serves as a context-dependent marker rather than a direct
effector. Finally, the cohort size and baseline heterogeneity may
limit generalizability, and larger longitudinal studies are
required to clarify the stability and functional relevance of
CD177+ neutrophils in bacterial infection (99-101).
In parallel, Kwok et al (102) identified a programmed
neutrophil lineage within granulocyte-monocyte progenitors (GMPs)
and analyzed the heterogeneous neutrophil subsets at different
stages of this lineage in sepsis. During the differentiation of
GMPs into preNeus, the aforementioned study described two
phenotypically distinct types of neutrophil progenitors, termed
proNeus (102). These include
the CD34hi CD106- CD11blo proNeu1
and the CD34lo CD106+ CD11bhi
proNeu2 subset (102).
Proteomics shows progressive upregulation of CD11b and
downregulation of CD34 expression during the differentiation of GMP
into preNeus (102). Other
differentially expressed cell surface markers include CD81, CD49a,
CD106 and CD63, which may serve as positive or exclusive markers
(102). Transcriptomics
confirms the downregulated expression of lineage-associated genes
and upregulated expression of granule protein genes in the GMP
differentiation trajectory during sepsis, suggesting that sepsis
promotes ProNeu1 differentiation (102). ProNeu1 and proNeu2 are both
early neutrophil progenitor cell populations, but their functions
are not identical (102).
ProNeu1 exhibits a stronger capacity for proliferation than proNeu2
(102). During the early stages
of sepsis inflammation, proNeu1 expands specifically and
extensively, which decreases monocyte differentiation (102). However, proNeu2 remains largely
unchanged during infection (102).
A recent multi-omics study by Kwok et al
demonstrated that septic neutrophils acquire immunosuppressive
properties and are enriched in patients with the sepsis response
signature group 1 (SRS1) (103). Single-cell transcriptomics and
cell surface protein profiling reveal the expansion of immature and
functionally distinct neutrophil subsets, including
CEACAM8+ degranulating cells, S100A8/9hi
cells, IL1R2+, peptidyl arginine deiminase type
4+, MPO+ and proliferative MK167+
CYP1B1+ neutrophils, many of which are specific to
sepsis rather than sterile inflammation (103). Coculture assays further
demonstrate that these septic subsets inhibit CD4+ T
cell activation (103).
Epigenomic profiling of hematopoietic stem and progenitor cells
revealed CCAAT/enhancer-binding protein α (CEBPA)- and β
(CEBPB)-driven regulatory programs consistent with the activation
of both steady-state and emergency granulopoiesis, linking
neutrophil reprogramming to systemic alteration in hematopoiesis
(103). Importantly, within the
SRS1 subtype, neutrophil subsets such as IL1R2+ and
MK167+ CYP1B1+ cells are significantly
enriched and exhibit STAT3- and CEBPB-dependent gene expression
programs, indicating that neutrophil heterogeneity is not only a
reflection of sepsis pathology but also a driver of
immunosuppressive disease endotypes (103). However, the single-cell cohorts
analyzed may not represent the spectrum of sepsis, and causal
involvement of transcription factors such as CEBPB and STAT3
requires direct experimental validation.
Transcriptome and metabolome
Integration of transcriptomic and metabolomic
approaches provides insights into how metabolic reprogramming
shapes neutrophil heterogeneity during sepsis.
Neutrophils are reliant on glycolysis to fuel key
antimicrobial functions, including chemotaxis, phagocytosis,
oxidative burst and NET formation (104-106), yet their metabolic flexibility
is altered in the septic milieu. Pan et al (107) demonstrated that sepsis-tolerant
neutrophils exhibit reduced glycolytic activity associated with
downregulation of LDHA via the PI3K/Akt/hypoxia-inducible factor
(HIF)-1α pathway, leading to impaired chemotactic and phagocytic
capacity. Metabolomic profiling further revealed that lactate
levels are diminished in septic neutrophils compared with
non-septic infected controls, highlighting a distinct metabolic
state of glycolytic suppression (107).
Lactate, long regarded as a metabolic byproduct, is
a immunomodulatory metabolite capable of exerting feedback control
over immune cell metabolism (108,109). While evidence in monocytes and
macrophages has demonstrated lactate-driven immunosuppression
(110,111), its role in neutrophil biology
remains less well understood. Pan et al (107) suggested that reduced lactate
production in sepsis may represent a unique metabolic signature of
neutrophil dysfunction, warranting further exploration of
lactate-mediated feedback in neutrophil plasticity. At the
molecular level, transcriptomic changes in key glycolytic enzymes,
including LDHA, pyruvate dehydrogenase kinase 1, glucose
transporter 1 (GLUT1) and pyruvate kinase M2 (PKM2) converge with
metabolomic alterations, reinforcing the key role of glycolysis in
neutrophil effector responses (107). Stabilization of HIF-1α restores
LDHA expression and glycolytic activity, thereby rescuing
neutrophil chemotaxis and phagocytosis, underscoring the
PI3K/Akt/HIF-1α axis as a central regulator of neutrophil function
in sepsis (107).
Collectively, combined transcriptomic and
metabolomic analyses uncover a metabolically defined neutrophil
subpopulation in sepsis characterized by glycolytic suppression and
functional impairment (107).
This multi-omics perspective not only advances understanding of
neutrophil heterogeneity but also highlights metabolic checkpoints
such as glycolysis and HIF-1α signaling as potential therapeutic
targets to restore neutrophil immunity in sepsis (107).
Proteome and metabolome
Exploring the association between protein
expression profiles and metabolic signatures reveals key regulatory
hubs and pathways that influence neutrophil activation, survival
and effector functions.
Using a proteomic approach, Parthasarathy et
al (112) examined the
differential expression of neutrophil subsets in patients with
sepsis. The expression of CD10, CD16 and CD86 is downregulated,
while human leukocyte antigen- DR isotype (HLA-DR), CD11b, CD80,
CD184, CD63 and CD66b are upregulated in sepsis (39,112,113). The aforementioned study
identified a specific mature neutrophil subset with high expression
of CD274 (PD-L1) and CD300f. Previous studies have shown that
blocking of PD-L1 improves survival in patients with sepsis
(114-116), and CD300f deletion stimulates
neutrophil recruitment to the site of infection and decreases
septic death in mice (117).
According to the expression of CD177, immature neutrophil subsets
are divided into two subgroups: CD10- CD177+
and CD10- CD177- (113). CD177 rapidly mobilizes specific
particles to the cell surface following cell activation (112). Differential expression of CD184
(CXCR4) and HLA-DR in CD10- CD177+ immature
neutrophil subsets is observed in patients with sepsis (118). Aged neutrophils expressing
CD184 exhibit a higher migratory activity and phagocytic capacity
than the subsequently recruited non-aged neutrophils (65). HLA-DR expression was increased in
CD10- CD177+ septic neutrophils (112). The expression and role of
HLA-DR on neutrophils remains unclear (119,120). Soluble factors pentraxin 3
(PTX3), angiopoietin-2 (Ang-2), endothelial cell-specific
molecule-1 (Endocan), growth arrest-specific 6 (Gas6) and the
inflammatory marker procalcitonin are upregulated in sepsis
(112). These factors are
elevated in patients with sepsis and contribute to vascular leak
and endothelial dysfunction (121,122). Correlation analysis shows that
CD10 is inversely correlated with these factors (112). Notably, immature neutrophils
store and release PTX3 during inflammation, and this factor
predicts disease severity and mortality in sepsis (123-126). Immature neutrophils may be a
driver of vascular inflammation or leak in sepsis.
While these observations provide insights into the
functional diversity of neutrophil subsets and their potential
links to soluble mediators and vascular pathology, limitations
remain. Distinguishing sepsis from other infections or organ
failure remains clinically challenging. Integrated multi-omics
approaches have revealed associations between neutrophil
phenotypes, soluble mediators and metabolic alterations. However,
these findings are largely observational and derived from limited
patient cohorts (112).
Therefore, further mechanistic studies are needed to validate
whether specific neutrophil subsets and their products directly
drive vascular leakage and immune dysregulation in sepsis.
Proteome and microbiome
The combined investigation of proteome and
microbiome has revealed the functional dynamics of neutrophils in
the context of sepsis.
Wang et al (127) isolated neutrophils from
patients with sepsis after surgery and characterized intracellular
bacterial communities, also termed as the neutrophil-specific
microbiome. The aforementioned study showed that the proportion of
actinobacteria decreased, while the levels of proteobacteria
increased (127). Compared with
healthy controls, the abundance of Escherichia/Shigella,
Klebsiella and Bradyrhizobium is higher in patients with
sepsis (127). The
neutrophil-specific microbiome of patients with sepsis exhibits
heterogeneity (127).
Dysregulation of the circulating microbiota may increase the risk
of postoperative infectious events (128). In addition, quantitative
proteomic analysis of neutrophils derived from patients with
demonstrates proteins involved in bactericidal activities of
neutrophils are downregulated, especially in patients with septic
shock (127). Significant
downregulation of some immunomodulatory-associated proteins is also
observed in sepsis patients, including integrin α-M, IgA Fc
receptor and lactotransferrin (127). MMP9 is significantly
downregulated in patients with septic shock, indicating that the
migratory activity of neutrophils is impaired (128). Proteomic analysis reveals a
decrease in neutrophil function in sepsis (127).
Beyond elucidating the molecular heterogeneity of
neutrophils, multi-omics suggests potential biomarkers and
therapeutic targets with translational relevance. Table III summarizes representative
neutrophil-associated biomarkers identified from multi-omics, along
with their potential diagnostic or prognostic value in sepsis
(28,77,78,80-83,88,89,91, 94,98,102,103,107,112,115,117,127).
 | Table IIIClinical translation and biomarker
potential of neutrophil signatures identified by multi-omics in
sepsis. |
Table III
Clinical translation and biomarker
potential of neutrophil signatures identified by multi-omics in
sepsis.
|
Biomarker/subtype | Omics source | Clinical
relevance | (Refs.) |
|---|
| Neutrophil
eQTL | Genomics | Identification of
21,210 neutrophil eQTLs (832 genes) underscores the role of
neutrophils in host defense and inflammation | (77) |
| MPO-DNA
complex | Genomics | Elevated MPO-DNA
associated with increased risk of sepsis (18%), 28-day mortality
(51%) and ICU requirement (38%) and mortality (125%) | (78) |
| H3K4me3-marked
subsets | Epigenome | IL-10 induces
transcriptionally active neutrophils, contributing to inflammation
resolution; H3K4me3 positioning suggests potential
immunotherapeutic targets | (80) |
| Transcriptional
neutrophil clusters | Transcriptome
(scRNA-seq) | Infection
accelerates immature-to-mature transition; developmental
stage-dependent neutrophil responses to infection | (28) |
| DEGs | Transcriptome (bulk
RNA-seq) | DEGs enriched in
immune/inflammatory signaling and PI3K/AKT pathway; AKT1 validated
as a potential therapeutic target in sepsis | (81) |
| Neutrophil subtypes
enriched at late differentiation stage | Transcriptome
(scRNA-seq) | A total of two
novel subsets enriched in human sepsis; five hub genes validated in
mice; candidate biomarkers and therapeutic targets (ALPL, CD177,
S100A8/A9, STXBP2) | (82) |
| Neu1 | Transcriptome
(scRNA-seq) | Neu1 associated
with septic shock and SOFA score; Neu1_C module predictive for
shock (AUC=0.81); expresses poor prognosis markers (CD123, CD38,
CD69) | (83) |
|
PD-L1high neutrophils | Transcriptome
(scRNA-seq) | Immunosuppressive
subset in sepsis (p38α/MSK1/MK2 dependent); directly inhibits T
cell response; associated with immune suppression and poor host
defense; potential therapeutic target via PD-L1 blockade | (88) |
| CD62Ldim
neutrophils | Proteome | Distinct subset
recruited during acute inflammation; proteomic profile enriched in
adhesion/activation pathways; potential biomarker of systemic
inflammation | (89) |
| Hypoxia-induced
neutrophil proteomic remodeling | Proteome | Inflammatory
receptor upregulation (FPR, GM-CSFRβ) and lysosomal protein
scavenging sustain neutrophil metabolism under stress; metabolic
pathways as potential therapeutic targets in hypoxic inflamed
tissue | (91) |
| GAPDH-inhibited
neutrophils in severe COVID-19 | Metabolome | Dysregulated amino
acid, redox and carbon metabolism; suppressed glycolysis with PPP
shift; GAPDH inhibition triggers pathogenic NET formation; links
neutrophil metabolic dysfunction to tissue damage and highlights
GAPDH as a potential therapeutic target | (94) |
|
CD177high neutrophils | Transcriptome and
proteome | Expanded during
acute bacterial infection; TLR4/NF-κB driven transcriptional
plasticity enhances antibacterial programs; CD177 upregulation is
associated with inflammation severity and may serve as a marker of
acute infectious states | (98) |
CD34hi
CD106-
CD11blo ProNeu1 | Transcriptome and
proteome | Early committed
progenitor; expands in early sepsis with high proliferative
capacity, suppressing monocyte output; potential biomarker of
emergency granulopoiesis | (102) |
CD34lo
CD106+
CD11bhi ProNeu2 | Transcriptome and
proteome | Intermediate
progenitor; stable during sepsis; reference subset for maturation
trajectory | (102) |
IL1R2+,
MK167+
CYP1B1+, CEACAM8+, S100A8/9hi | Transcriptome
(scRNA-seq) and proteome | Sepsis-specific;
inhibit CD4+ T cells; enriched in SRS1 endotype;
associated with STAT3/CEBPB-driven immunosuppression | (103) |
| LDHAlow
glycolytic neutrophils | Transcriptome and
metabolome | Exhibit suppressed
glycolysis with impaired chemotaxis and phagocytosis; dysfunction
mediated by PI3K-Akt-HIF-1α-LDHA axis; reversible by HIF-1α
activation, suggesting therapeutic potential for restoring immune
competence in sepsis | (107) |
| CD10-
CD177+ immature neutrophils | Proteome and
metabolome | Associated with
SOFA score and endothelial dysfunction (PTX3, Ang-2, endocan); PTX3
release associated with severity and mortality; associated with
metabolic dysregulation and identified as a feature for patient
stratification | (112) |
|
PD-L1high neutrophils | Proteome and
metabolome | PD-L1 blockade
improves survival in murine CLP sepsis model | (115) |
| CD300f+
neutrophils | Proteome and
metabolome | Disruption or
deletion of CD300f decreases septic mortality in murine CLP model
of sepsis | (117) |
| Neutrophil
microbiome shifts | Proteome and
microbiome | Associated with
impaired bactericidal activity and migration; downregulation of
ITGAM, LTF and MMP9; associated with septic shock | (127) |
Challenges and limitations
Despite the advances in understanding neutrophil
heterogeneity in sepsis through multi-omics, key challenges hinder
the full potential of these approaches.
Patient heterogeneity is a key obstacle to
characterizing neutrophil heterogeneity in sepsis through
multi-omics approaches. Variations in demographic and physiological
backgrounds (age, sex, genetic factors) shape neutrophil
development and lifespan, potentially masking sepsis-specific
changes (129-132). Underlying comorbid conditions
may introduce baseline alterations in neutrophil function, thereby
contributing to heterogeneity and complicating the attribution of
omics signatures solely to sepsis (133). In addition, pathogen type,
primary infection site and therapeutic intervention may induce
distinct neutrophil activation programs, while clinical
trajectories range from hyperinflammation to immunosuppression,
generating variable molecular patterns (33,134-136). These sources of variability
make datasets harder to compare, decrease reproducibility and blur
the true sepsis-associated neutrophil signatures. To overcome this
challenge, future studies should incorporate strategies such as
stratified analyses, matched cohort designs and standardized
metadata collection to distinguish patient-level variability from
the intrinsic biological diversity of neutrophils in sepsis.
Sample preparation is a key step in multi-omics
studies, but introduces inherent biases, particularly during cell
isolation and processing. For example, methods based on density
gradient centrifugation may inadvertently exclude low-density
neutrophils (LDNs), which are typically found in the peripheral
blood mononuclear cell fraction (137). By contrast, commonly used
methods obtain samples from peripheral blood or tissue, followed by
red blood cell lysis or enzymatic digestion (138). The cells are centrifuged,
resuspended, washed and filtered to ensure high-quality viable
cells. Neutrophils are purified by either positive or negative
selection using magnetic beads or fluorescence-activated cell
sorting and subjected to scRNA-seq for downstream analysis
(28). Other improvements
include initial selection of total neutrophils using magnetic beads
followed by density gradient separation of LDNs (139,140). In addition, density
gradient-based approaches to neutrophil isolation may result in
preparations containing small numbers of contaminating leukocytes,
primarily eosinophils, a potential source of bias despite the small
contribution of these leukocytes to the overall gene expression
profile (141). These technical
variations can lead to discrepancies between in vitro or
ex vivo findings and the actual in vivo behavior of
neutrophils, particularly within the complex microenvironment of
sepsis.
The temporal dynamics of neutrophil changes during
sepsis are not well understood. The heterogeneity of neutrophils in
sepsis results from a complex interaction between intrinsic factors
and disease-associated changes over time. Sepsis is a dynamic
condition, marked by rapid shifts in immune response, from
excessive inflammation in the early stage to immunosuppression in
the later stages (33). However,
most current multi-omics studies focus on single time-point
analyses, providing a static snapshot of neutrophil phenotypes,
which may overlook the ongoing changes in neutrophils during the
immune transition in sepsis (83,112). Therefore, longitudinal
multi-omics studies are necessary to capture the immune switching
of neutrophils over time and improve understanding of this dynamic
process.
Each omics layer provides a unique resolution and
focus for capturing biological heterogeneity, yet these differences
also impose analytical limitations. The integration of multi-omics
data from high-throughput platforms inherently faces challenges due
to their diverse characteristics (142). Picard et al (143) highlighted that the
heterogeneity of data complicates the integration process. For
example, transcriptomic data is often subjected to RNA-seq
normalization, while proteomic data typically rely on mass
spectrometry-specific scaling methods, leading to differing data
ranges and distribution patterns. These disparities necessitate
alignment procedures before integration. Data quality also poses a
concern. Issues such as noise, missing values and batch effects
notably impact the outcomes of the analysis (144). In addition, there are kinetic
differences between RNA and protein expression. For example,
Hoogendijk et al (145)
reported that nearly 30% of the transcriptome -proteome pairs
showed inconsistent dynamics during neutrophil differentiation.
This discrepancy may contribute to the inconsistency between the
transcriptome data from scRNA-seq and the protein-based surface
marker data from mass cytometry. These mismatches complicate the
classification of cell populations and the annotation of functional
roles.
Conclusion
Sepsis is a life-threatening syndrome characterized
by a systemic inflammatory response to infection, leading to
multiorgan dysfunction and potential mortality. The pathophysiology
of sepsis involves complex interactions between pathogens, the
immune system and various host factors. Advancements in research
have demonstrated the key role of immune cells in sepsis,
particularly neutrophils, which are frontline responders in the
immune system. The functional and phenotypical heterogeneity of
these immune cells during sepsis can influence outcomes.
In terms of phenotypical diversity, neutrophils in
sepsis exhibit various surface markers and morphologies that
reflect their activation state, origin and roles in the
inflammatory response. Functionally, neutrophils exhibit
heterogeneity in their ability to degranulate, phagocytose, release
ROS, form NETs, migrate, undergo apoptosis and mediate
immunosuppression. Understanding these functional differences is
key for developing targeted therapeutic strategies aimed at
modulating neutrophil activity during sepsis.
The study of neutrophil heterogeneity in sepsis
through multi-omics approaches has provided insights into the
complex and adaptive nature of these immune cells. Multi-omics
studies have revealed that the differentially expressed genes,
proteins and metabolites underlying neutrophil heterogeneity are
not independent events but reflect interconnected layers of
regulation. Transcriptomic analyses have identified altered
expression of transcription factors and signaling molecules that
reprogram neutrophil activation, survival and differentiation,
while proteomic profiling has identified changes in effector
functions, including degranulation, phagocytosis and cytokine
release (81,88). Metabolomic data have further
indicated shifts in glycolysis, oxidative phosphorylation and amino
acid metabolism that provide energetic and biosynthetic support for
these functions. In addition, epigenomic profiling has elucidated
associations between chromatin accessibility and histone
modifications with NF-κB signaling, apoptosis and cytokine
regulation, thereby uncovering previously unrecognized neutrophil
subsets (80,107). Multi-omics integration has
identified transcription factor-driven programs, such as
CEBPA/CEBPB- and STAT3-dependent networks, which connect emergency
granulopoiesis with the emergence of sepsis-specific neutrophil
subsets (103). Collectively,
these interconnected regulatory networks provide a mechanistic
basis by which neutrophils acquire distinct functional states,
thereby contributing to the phenotypical and functional
heterogeneity observed in sepsis.
Availability of data and materials
Not applicable.
Authors' contributions
ZT designed the study. DC, PZ, JL, SC, SG, YC, YS,
TT, LD and TC performed the literature review. ZL wrote the
manuscript. CZ edited the manuscript. All authors have read and
approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
scRNA-seq
|
single-cell RNA sequencing
|
|
PMN
|
polymorphonuclear leukocyte
|
|
ROS
|
reactive oxygen species
|
|
NET
|
neutrophil extracellular trap
|
|
LPS
|
lipopolysaccharide
|
|
TNF-α
|
tumor necrosis factor-α
|
|
eQTL
|
expression quantitative trait
loci
|
|
IPA
|
ingenuity pathway analysis
|
|
MPO
|
myeloperoxidase
|
|
COVID-19
|
coronavirus disease 2019
|
|
GMP
|
granulocyte-monocyte progenitor
|
|
SRS1
|
sepsis response signature group 1
|
|
CEBPB
|
CCAAT/enhancer binding protein β
|
|
LDN
|
low-density neutrophil
|
|
AI
|
artificial intelligence
|
Acknowledgements
Not applicable.
Funding
The present study was supported by Hubei Province health and
family planning scientific research project (grant no.
WJ2023M015).
References
|
1
|
Singer M, Deutschman CS, Seymour CW,
Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche
JD, Coopersmith CM, et al: The third international consensus
definitions for sepsis and septic shock (Sepsis-3). JAMA.
315:801–810. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rudd KE, Johnson SC, Agesa KM, Shackelford
KA, Tsoi D, Kievlan DR, Colombara DV, Ikuta KS, Kissoon N, Finfer
S, et al: Global, regional, and national sepsis incidence and
mortality, 1990-2017: Analysis for the Global Burden of Disease
Study. Lancet. 395:200–211. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fleischmann C, Scherag A, Adhikari NK,
Hartog CS, Tsaganos T, Schlattmann P, Angus DC and Reinhart K;
International Forum of Acute Care Trialists: Assessment of global
incidence and mortality of hospital-treated sepsis. Current
estimates and limitations. Am J Respir Crit Care Med. 193:259–272.
2016. View Article : Google Scholar
|
|
4
|
Prescott HC and Angus DC: Enhancing
recovery from sepsis: A review. JAMA. 319:62–75. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Prescott HC, Langa KM and Iwashyna TJ:
Readmission diagnoses after hospitalization for severe sepsis and
other acute medical conditions. JAMA. 313:1055–1057. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shankar-Hari M, Saha R, Wilson J, Prescott
HC, Harrison D, Rowan K, Rubenfeld GD and Adhikari NKJ: Rate and
risk factors for rehospitalisation in sepsis survivors: Systematic
review and meta-analysis. Intensive Care Med. 46:619–636. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Poll T, van de Veerdonk FL,
Scicluna BP and Netea MG: The immunopathology of sepsis and
potential therapeutic targets. Nat Rev Immunol. 17:407–420. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen S, Zhang C, Luo J, Lin Z, Chang T,
Dong L, Chen D and Tang ZH: Macrophage activation syndrome in
Sepsis: from pathogenesis to clinical management. Inflamm Res.
73:2179–2197. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang TD, Chen D, Luo JL, Wang YM, Zhang
C, Chen SY, Lin ZQ, Zhang PD, Tang TX, Li H, et al: The different
paradigms of NK cell death in patients with severe trauma. Cell
Death Dis. 15:6062024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen S, Zhang C, Chen D, Dong L, Chang T
and Tang ZH: Advances in attractive therapeutic approach for
macrophage activation syndrome in COVID-19. Front Immunol.
14:12002892023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen D, Zhang C, Luo J, Deng H, Yang J,
Chen S, Zhang P, Dong L, Chang T and Tang ZH: Activated autophagy
of innate immune cells during the early stages of major trauma.
Front Immunol. 13:10903582023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Chang T, Tang L, Deng H, Chen D,
Luo J, Wu H, Tang T, Zhang C, Li Z, et al: Increased expression of
Tim-3 is associated with depletion of NKT Cells In SARS-CoV-2
infection. Front Immunol. 13:7966822022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fine N, Tasevski N, McCulloch CA,
Tenenbaum HC and Glogauer M: The Neutrophil: Constant defender and
first responder. Front Immunol. 11:5710852020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kangelaris KN, Clemens R, Fang X, Jauregui
A, Liu T, Vessel K, Deiss T, Sinha P, Leligdowicz A, Liu KD, et al:
A neutrophil subset defined by intracellular olfactomedin 4 is
associated with mortality in sepsis. Am J Physiol Lung Cell Mol
Physiol. 320:L892–L902. 2021. View Article : Google Scholar :
|
|
15
|
Shen XF, Cao K, Jiang JP, Guan WX and Du
JF: Neutrophil dysregulation during sepsis: an overview and update.
J Cell Mol Med. 21:1687–1697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Silvestre-Roig C, Hidalgo A and Soehnlein
O: Neutrophil heterogeneity: Implications for homeostasis and
pathogenesis. Blood. 127:2173–2181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Naranbhai V, Fairfax BP, Makino S, Humburg
P, Wong D, Ng E, Hill AV and Knight JC: Genomic modulators of gene
expression in human neutrophils. Nat Commun. 6:75452015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Coit P, Yalavarthi S, Ognenovski M, Zhao
W, Hasni S, Wren JD, Kaplan MJ and Sawalha AH: Epigenome profiling
reveals significant DNA demethylation of interferon signature genes
in lupus neutrophils. J Autoimmun. 58:59–66. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pillay J, den Braber I, Vrisekoop N, Kwast
LM, de Boer RJ, Borghans JA, Tesselaar K and Koenderman L: In vivo
labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days.
Blood. 116:625–627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ericson JA, Duffau P, Yasuda K,
Ortiz-Lopez A, Rothamel K, Rifkin IR and Monach PA; ImmGen
Consortium: Gene expression during the generation and activation of
mouse neutrophils: Implication of novel functional and regulatory
pathways. PLoS One. 9:e1085532014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ng LG, Ostuni R and Hidalgo A:
Heterogeneity of neutrophils. Nat Rev Immunol. 19:255–265. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grecian R, Whyte MKB and Walmsley SR: The
role of neutrophils in cancer. Br Med Bull. 128:5–14. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Day RB and Link DC: Regulation of
neutrophil trafficking from the bone marrow. Cell Mol Life Sci.
69:1415–1423. 2012. View Article : Google Scholar
|
|
24
|
Elghetany MT: Surface antigen changes
during normal neutrophilic development: A critical review. Blood
Cells Mol Dis. 28:260–274. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Seree-Aphinan C, Vichitkunakorn P,
Navakanitworakul R and Khwannimit B: Distinguishing sepsis from
infection by neutrophil dysfunction: A promising role of CXCR2
surface level. Front Immunol. 11:6086962020. View Article : Google Scholar
|
|
26
|
Silvestre-Roig C, Fridlender ZG, Glogauer
M and Scapini P: Neutrophil diversity in health and disease. Trends
Immunol. 40:565–583. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Fan D, Yang Y, Gimple RC and Zhou
S: Integrative multi-omics approaches to explore immune cell
functions: Challenges and opportunities. iScience. 26:1063592023.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su
J, Yu H, Park SY, Guo R, Ren Q, et al: Single-cell transcriptome
profiling reveals neutrophil heterogeneity in homeostasis and
infection. Nat Immunol. 21:1119–1133. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shaath H, Vishnubalaji R, Elkord E and
Alajez NM: Single-cell transcriptome analysis highlights a role for
neutrophils and inflammatory macrophages in the pathogenesis of
severe COVID-19. Cells. 9:23742020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deerhake ME, Reyes EY, Xu-Vanpala S and
Shinohara ML: Single-Cell transcriptional heterogeneity of
neutrophils during acute pulmonary cryptococcus neoformans
infection. Front Immunol. 12:6705742021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Civelek M and Lusis AJ: Systems genetics
approaches to understand complex traits. Nat Rev Genet. 15:34–48.
2014. View Article : Google Scholar :
|
|
32
|
Johnson Chavarria EC: A primer of human
genetics. Yale J Biol Med. 89:6032016.
|
|
33
|
van der Poll T, Shankar-Hari M and
Wiersinga WJ: The immunology of sepsis. Immunity. 54:2450–2464.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Borregaard N: Neutrophils, from marrow to
microbes. Immunity. 33:657–670. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stadtmann A and Zarbock A: CXCR2: From
bench to bedside. Front Immunol. 3:2632012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Phillipson M and Kubes P: The neutrophil
in vascular inflammation. Nat Med. 17:1381–1390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chishti AD, Shenton BK, Kirby JA and
Baudouin SV: Neutrophil chemotaxis and receptor expression in
clinical septic shock. Intensive Care Med. 30:605–611. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rios-Santos F, Alves-Filho JC, Souto FO,
Spiller F, Freitas A, Lotufo CM, Soares MB, Dos Santos RR, Teixeira
MM and Cunha FQ: Down-regulation of CXCR2 on neutrophils in severe
sepsis is mediated by inducible nitric oxide synthase-derived
nitric oxide. Am J Respir Crit Care Med. 175:490–497. 2007.
View Article : Google Scholar
|
|
39
|
Demaret J, Venet F, Friggeri A, Cazalis
MA, Plassais J, Jallades L, Malcus C, Poitevin-Later F, Textoris J,
Lepape A and Monneret G: Marked alterations of neutrophil functions
during sepsis-induced immunosuppression. J Leukoc Biol.
98:1081–1090. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pillay J, Kamp VM, van Hoffen E, Visser T,
Tak T, Lammers JW, Ulfman LH, Leenen LP, Pickkers P and Koenderman
L: A subset of neutrophils in human systemic inflammation inhibits
T cell responses through Mac-1. J Clin Invest. 122:327–336. 2012.
View Article : Google Scholar :
|
|
41
|
Geng S, Matsushima H, Okamoto T, Yao Y, Lu
R, Page K, Blumenthal RM, Ward NL, Miyazaki T and Takashima A:
Emergence, origin, and function of neutrophil-dendritic cell
hybrids in experimentally induced inflammatory lesions in mice.
Blood. 121:1690–1700. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ode Y, Aziz M and Wang P: CIRP increases
ICAM-1(+) phenotype of neutrophils exhibiting elevated iNOS and
NETs in sepsis. J Leukoc Biol. 103:693–707. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hoffmann JJ: Neutrophil CD64: A diagnostic
marker for infection and sepsis. Clin Chem Lab Med. 47:903–916.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hoffmann JJ: Neutrophil CD64 as a sepsis
biomarker. Biochem Med (Zagreb). 21:282–290. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cid J, Aguinaco R, Sánchez R, García-Pardo
G and Llorente A: Neutrophil CD64 expression as marker of bacterial
infection: A systematic review and meta-analysis. J Infect.
60:313–319. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li S, Huang X, Chen Z, Zhong H, Peng Q,
Deng Y, Qin X and Zhao J: Neutrophil CD64 expression as a biomarker
in the early diagnosis of bacterial infection: A meta-analysis. Int
J Infect Dis. 17:e12–e23. 2013. View Article : Google Scholar
|
|
47
|
Bouchon A, Facchetti F, Weigand MA and
Colonna M: TREM-1 amplifies inflammation and is a crucial mediator
of septic shock. Nature. 410:1103–1107. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Demaret J, Venet F, Plassais J, Cazalis
MA, Vallin H, Friggeri A, Lepape A, Rimmelé T, Textoris J and
Monneret G: Identification of CD177 as the most dysregulated
parameter in a microarray study of purified neutrophils from septic
shock patients. Immunol Lett. 178:122–130. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Amulic B, Cazalet C, Hayes GL, Metzler KD
and Zychlinsky A: Neutrophil function: From mechanisms to disease.
Annu Rev Immunol. 30:459–489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Boeltz S, Amini P, Anders HJ, Andrade F,
Bilyy R, Chatfield S, Cichon I, Clancy DM, Desai J, Dumych T, et
al: To NET or not to NET: Current opinions and state of the science
regarding the formation of neutrophil extracellular traps. Cell
Death Differ. 26:395–408. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Patel JM, Sapey E, Parekh D, Scott A,
Dosanjh D, Gao F and Thickett DR: Sepsis Induces a Dysregulated
Neutrophil Phenotype That Is Associated with Increased Mortality.
Mediators Inflamm. 2018:40653622018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Martins PS, Kallas EG, Neto MC, Dalboni
MA, Blecher S and Salomão R: Upregulation of reactive oxygen
species generation and phagocytosis, and increased apoptosis in
human neutrophils during severe sepsis and septic shock. Shock.
20:208–212. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alves-Filho JC, Spiller F and Cunha FQ:
Neutrophil paralysis in sepsis. Shock. 34(Suppl 1): S15–S21. 2010.
View Article : Google Scholar
|
|
54
|
Reddy RC and Standiford TJ: Effects of
sepsis on neutrophil chemotaxis. Curr Opin Hematol. 17:18–24. 2010.
View Article : Google Scholar
|
|
55
|
Tavares-Murta BM, Zaparoli M, Ferreira RB,
Silva-Vergara ML, Oliveira CH, Murta EF, Ferreira SH and Cunha FQ:
Failure of neutrophil chemotactic function in septic patients. Crit
Care Med. 30:1056–1061. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Brown KA, Brain SD, Pearson JD, Edgeworth
JD, Lewis SM and Treacher DF: Neutrophils in development of
multiple organ failure in sepsis. Lancet. 368:157–169. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Arraes SM, Freitas MS, da Silva SV, de
Paula Neto HA, Alves-Filho JC, Auxiliadora Martins M, Basile-Filho
A, Tavares-Murta BM, Barja-Fidalgo C and Cunha FQ: Impaired
neutrophil chemotaxis in sepsis associates with GRK expression and
inhibition of actin assembly and tyrosine phosphorylation. Blood.
108:2906–2913. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Martins PS, Brunialti MK, Martos LS,
Machado FR, Assunçao MS, Blecher S and Salomao R: Expression of
cell surface receptors and oxidative metabolism modulation in the
clinical continuum of sepsis. Crit Care. 12:R252008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kalyan S and Kabelitz D: When neutrophils
meet T cells: Beginnings of a tumultuous relationship with
underappreciated potential. Eur J Immunol. 44:627–633. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kovach MA and Standiford TJ: The function
of neutrophils in sepsis. Curr Opin Infect Dis. 25:321–327. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen D, Tang TX, Deng H, Yang XP and Tang
ZH: Interleukin-7 biology and its effects on immune cells: Mediator
of generation, differentiation, survival, and homeostasis. Front
Immunol. 12:7473242021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bone RC, Grodzin CJ and Balk RA: Sepsis: A
new hypothesis for pathogenesis of the disease process. Chest.
112:235–243. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Alves-Filho JC, de Freitas A, Spiller F,
Souto FO and Cunha FQ: The role of neutrophils in severe sepsis.
Shock. 30(Suppl 1): S3–S9. 2008. View Article : Google Scholar
|
|
64
|
Drifte G, Dunn-Siegrist I, Tissières P and
Pugin J: Innate immune functions of immature neutrophils in
patients with sepsis and severe systemic inflammatory response
syndrome. Crit Care Med. 41:820–832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Taneja R, Sharma AP, Hallett MB, Findlay
GP and Morris MR: Immature circulating neutrophils in sepsis have
impaired phagocytosis and calcium signaling. Shock. 30:618–622.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Uhl B, Vadlau Y, Zuchtriegel G, Nekolla K,
Sharaf K, Gaertner F, Massberg S, Krombach F and Reichel CA: Aged
neutrophils contribute to the first line of defense in the acute
inflammatory response. Blood. 128:2327–2337. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yang P, Li Y, Xie Y and Liu Y: Different
faces for different places: Heterogeneity of neutrophil phenotype
and function. J Immunol Res. 2019:80162542019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
van Dijk EL, Jaszczyszyn Y, Naquin D and
Thermes C: The third revolution in sequencing technology. Trends
Genet. 34:666–681. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Warr A, Robert C, Hume D, Archibald A,
Deeb N and Watson M: Exome sequencing: Current and future
perspectives. G3 (Bethesda). 5:1543–1550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Goldman AD and Landweber LF: What is a
genome? PLoS Genet. 12:e10061812016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang KC and Chang HY: Epigenomics:
Technologies and applications. Circ Res. 122:1191–1199. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View Article : Google Scholar
|
|
73
|
Timp W and Timp G: Beyond mass
spectrometry, the next step in proteomics. Sci Adv. 6:eaax89782020.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fu Q, Vegesna M, Sundararaman N, Damoc E,
Arrey TN, Pashkova A, Mengesha E, Debbas P, Joung S, Li D, et al: A
proteomics pipeline for generating clinical grade biomarker
candidates from data-independent acquisition mass spectrometry
(DIA-MS) discovery. Angew Chem Int Ed Engl. 63:e2024094462024.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Moco S, Vervoort J, Moco S, Bino RJ, De
Vos RC and Bino R: Metabolomics technologies and metabolite
identification. TrAC Trends in Analytical Chemistry.
2007.26:855–866. 2007. View Article : Google Scholar
|
|
76
|
Bedair M and Sumner LW: Current and
emerging mass-spectrometry technologies for metabolomics. TrAC
Trends in Analytical Chemistry. 27:238–250. 2008. View Article : Google Scholar
|
|
77
|
Andiappan AK, Melchiotti R, Poh TY, Nah M,
Puan KJ, Vigano E, Haase D, Yusof N, San Luis B, Lum J, et al:
Genome-wide analysis of the genetic regulation of gene expression
in human neutrophils. Nat Commun. 6:79712015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang J, Zhang Y, Cheng L, Geng Y, Lu J and
Zhou J: Neutrophil extracellular trap increase the risk of sepsis:
A two-sample, one-way Mendelian randomization study. Zhonghua Wei
Zhong Bing Ji Jiu Yi Xue. 35:1045–1052. 2023.In Chinese. PubMed/NCBI
|
|
79
|
Zhang H, Wang Y, Qu M, Li W, Wu D, Cata JP
and Miao C: Neutrophil, neutrophil extracellular traps and
endothelial cell dysfunction in sepsis. Clin Transl Med.
13:e11702023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Piatek P, Namiecinska M, Lewkowicz N,
Kulińska-Michalska M, Jabłonowski Z, Matysiak M, Dulska J,
Michlewska S, Wieczorek M and Lewkowicz P: Changes WIthin
H3K4me3-marked histone reveal molecular background of neutrophil
functional plasticity. Front Immunol. 13:9063112022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liu Z, Chen Y, Pan T, Liu J, Tian R, Sun
S, Qu H and Chen E: Comprehensive analysis of common different gene
expression signatures in the neutrophils of sepsis. Biomed Res Int.
2021:66554252021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Xu P, Tao Z and Zhang C: Integrated
multi-omics and artificial intelligence to explore new neutrophils
clusters and potential biomarkers in sepsis with experimental
validation. Front Immunol. 15:13778172024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Hong Y, Chen L, Sun J, Xing L, Yang Y, Jin
X, Cai H, Dong L, Zhou L and Zhang Z: Single-cell transcriptome
profiling reveals heterogeneous neutrophils with prognostic values
in sepsis. iScience. 25:1053012022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Goswami DG, Garcia LF, Dodoo C, Dwivedi
AK, Zhou Y, Pappas D and Walker WE: Evaluating the Timeliness and
Specificity of CD69, CD64, and CD25 as Biomarkers of Sepsis in
Mice. Shock. 55:507–518. 2021. View Article : Google Scholar
|
|
85
|
Zhou Y, Zhang Y, Johnson A, Venable A,
Griswold J and Pappas D: Combined CD25, CD64, and CD69 biomarker
panel for flow cytometry diagnosis of sepsis. Talanta. 191:216–221.
2019. View Article : Google Scholar
|
|
86
|
Meghraoui-Kheddar A, Chousterman BG,
Guillou N, Barone SM, Granjeaud S, Vallet H, Corneau A, Guessous K,
de Roquetaillade C, Boissonnas A, et al: Two new neutrophil subsets
define a discriminating sepsis signature. Am J Respir Crit Care
Med. 205:46–59. 2022. View Article : Google Scholar
|
|
87
|
Wang P, Wang J, Li YH, Wang L, Shang HC
and Wang JX: Phenotypical changes of hematopoietic stem and
progenitor cells in sepsis patients: Correlation with immune
status? Front Pharmacol. 11:6402032020. View Article : Google Scholar
|
|
88
|
Qi X, Yu Y, Sun R, Huang J, Liu L, Yang Y,
Rui T and Sun B: Identification and characterization of neutrophil
heterogeneity in sepsis. Crit Care. 25:502021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tak T, Wijten P, Heeres M, Pickkers P,
Scholten A, Heck AJR, Vrisekoop N, Leenen LP, Borghans JAM,
Tesselaar K and Koenderman L: Human CD62L(dim) neutrophils
identified as a separate subset by proteome profiling and in vivo
pulse-chase labeling. Blood. 129:3476–3485. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mallat J, Rahman N, Hamed F, Hernandez G
and Fischer MO: Pathophysiology, mechanisms, and managements of
tissue hypoxia. Anaesth Crit Care Pain Med.
41:1010872022.PubMed/NCBI
|
|
91
|
Watts ER, Howden AJ, Morrison T, Sadiku P,
Hukelmann J, von Kriegsheim A, Ghesquiere B, Murphy F, Mirchandani
AS, Humphries DC, et al: Hypoxia drives murine neutrophil protein
scavenging to maintain central carbon metabolism. J Clin Invest.
131:e1340732021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Sun L, Yang X, Yuan Z and Wang H:
Metabolic reprogramming in immune response and tissue inflammation.
Arterioscler Thromb Vasc Biol. 40:1990–2001. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kumar S and Dikshit M: Metabolic insight
of neutrophils in health and disease. Front Immunol. 10:20992019.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Li Y, Hook JS, Ding Q, Xiao X, Chung SS,
Mettlen M, Xu L, Moreland JG and Agathocleous M: Neutrophil
metabolomics in severe COVID-19 reveal GAPDH as a suppressor of
neutrophil extracellular trap formation. Nat Commun. 14:26102023.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Khoyratty TE, Ai Z, Ballesteros I, Eames
HL, Mathie S, Martín-Salamanca S, Wang L, Hemmings A, Willemsen N,
von Werz V, et al: Distinct transcription factor networks control
neutrophil-driven inflammation. Nat Immunol. 22:1093–1106. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Grieshaber-Bouyer R, Radtke FA, Cunin P,
Stifano G, Levescot A, Vijaykumar B, Nelson-Maney N, Blaustein RB,
Monach PA and Nigrovic PA; ImmGen Consortium: The neutrotime
transcriptional signature defines a single continuum of neutrophils
across biological compartments. Nat Commun. 12:28562021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Tsukahara Y, Lian Z, Zhang X, Whitney C,
Kluger Y, Tuck D, Yamaga S, Nakayama Y, Weissman SM and Newburger
PE: Gene expression in human neutrophils during activation and
priming by bacterial lipopolysaccharide. J Cell Biochem.
89:848–861. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kaiser R, Gold C, Joppich M, Loew Q,
Akhalkatsi A, Mueller TT, Offensperger F, Droste Zu, Senden A, Popp
O, di Fina L, et al: Peripheral priming induces plastic
transcriptomic and proteomic responses in circulating neutrophils
required for pathogen containment. Sci Adv. 10:eadl17102024.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Grieshaber-Bouyer R and Nigrovic PA:
Neutrophil heterogeneity as therapeutic opportunity in
immune-mediated disease. Front Immunol. 10:3462019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Eulenberg-Gustavus C, Bähring S, Maass PG,
Luft FC and Kettritz R: Gene silencing and a novel monoallelic
expression pattern in distinct CD177 neutrophil subsets. J Exp Med.
214:2089–2101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wu Z, Liang R, Ohnesorg T, Cho V, Lam W,
Abhayaratna WP, Gatenby PA, Perera C, Zhang Y, Whittle B, et al:
Heterogeneity of human neutrophil CD177 expression results from
CD177P1 pseudogene conversion. PLoS Genet. 12:e10060672016.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kwok I, Becht E, Xia Y, Ng M, The YC, Tan
L, Evrard M, Li JLY, Tran HTN, Tan Y, et al: Combinatorial
single-cell analyses of granulocyte-monocyte progenitor
heterogeneity reveals an early uni-potent neutrophil progenitor.
Immunity. 53:303–318.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Kwok AJ, Allcock A, Ferreira RC,
Cano-Gamez E, Smee M, Burnham KL, Zurke YX; Emergency Medicine
Research Oxford (EMROx); McKechnie S, Mentzer AJ, et al:
Neutrophils and emergency granulopoiesis drive immune suppression
and an extreme response endotype during sepsis. Nat Immunol.
24:767–779. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Borregaard N and Herlin T: Energy
metabolism of human neutrophils during phagocytosis. J Clin Invest.
70:550–557. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bao Y, Ledderose C, Graf AF, Brix B,
Birsak T, Lee A, Zhang J and Junger WG: mTOR and differential
activation of mitochondria orchestrate neutrophil chemotaxis. J
Cell Biol. 210:1153–1164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Rodríguez-Espinosa O, Rojas-Espinosa O,
Moreno-Altamirano MM, López-Villegas EO and Sánchez-García FJ:
Metabolic requirements for neutrophil extracellular traps
formation. Immunology. 145:213–224. 2015. View Article : Google Scholar :
|
|
107
|
Pan T, Sun S, Chen Y, Tian R, Chen E, Tan
R, Wang X, Liu Z, Liu J and Qu H: Immune effects of
PI3K/Akt/HIF-1α-regulated glycolysis in polymorphonuclear
neutrophils during sepsis. Crit Care. 26:292022. View Article : Google Scholar
|
|
108
|
Ratter JM, Rooijackers HMM, Hooiveld GJ,
Hijmans AGM, de Galan BE, Tack CJ and Stienstra R: In vitro and in
vivo effects of lactate on metabolism and cytokine production of
human primary PBMCs and monocytes. Front Immunol. 9:25642018.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Dietl K, Renner K, Dettmer K, Timischl B,
Eberhart K, Dorn C, Hellerbrand C, Kastenberger M, Kunz-Schughart
LA, Oefner PJ, et al: Lactic acid and acidification inhibit TNF
secretion and glycolysis of human monocytes. J Immunol.
184:1200–1209. 2010. View Article : Google Scholar
|
|
110
|
Nolt B, Tu F, Wang X, Ha T, Winter R,
Williams DL and Li C: Lactate and immunosuppression in sepsis.
Shock. 49:120–125. 2018. View Article : Google Scholar :
|
|
111
|
Colegio OR, Chu NQ, Szabo AL, Chu T,
Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC,
Phillips GM, et al: Functional polarization of tumour-associated
macrophages by tumour-derived lactic acid. Nature. 513:559–563.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Parthasarathy U, Kuang Y, Thakur G, Hogan
JD, Wyche TP, Norton JE Jr, Killough JR, Sana TR, Beakes C, Shyong
B, et al: Distinct subsets of neutrophils crosstalk with cytokines
and metabolites in patients with sepsis. iScience. 26:1059482023.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Marini O, Costa S, Bevilacqua D, Calzetti
F, Tamassia N, Spina C, De Sabata D, Tinazzi E, Lunardi C, Scupoli
MT, et al: Mature CD10(+) and immature CD10(-) neutrophils present
in G-CSF-treated donors display opposite effects on T cells. Blood.
129:1343–1356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang JF, Li JB, Zhao YJ, Yi WJ, Bian JJ,
Wan XJ, Zhu KM and Deng XM: Up-regulation of programmed cell death
1 ligand 1 on neutrophils may be involved in sepsis-induced
immunosuppression: An animal study and a prospective case-control
study. Anesthesiology. 122:852–863. 2015. View Article : Google Scholar
|
|
115
|
Zhang Y, Zhou Y, Lou J, Li J, Bo L, Zhu K,
Wan X, Deng X and Cai Z: PD-L1 blockade improves survival in
experimental sepsis by inhibiting lymphocyte apoptosis and
reversing monocyte dysfunction. Crit Care. 14:R2202010. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Brahmamdam P, Inoue S, Unsinger J, Chang
KC, McDunn JE and Hotchkiss RS: Delayed administration of anti-PD-1
antibody reverses immune dysfunction and improves survival during
sepsis. J Leukoc Biol. 88:233–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Izawa K, Maehara A, Isobe M, Yasuda Y,
Urai M, Hoshino Y, Ueno K, Matsukawa T, Takahashi M, Kaitani A, et
al: Disrupting ceramide-CD300f interaction prevents septic
peritonitis by stimulating neutrophil recruitment. Sci Rep.
7:42982017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Hu N, Mora-Jensen H, Theilgaard-Mönch K,
Doornbosvan der Meer B, Huitema MG, Stegeman CA, Heeringa P,
Kallenberg CG and Westra J: Differential expression of
granulopoiesis related genes in neutrophil subsets distinguished by
membrane expression of CD177. PLoS One. 9:e996712014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Davis RE, Sharma S, Conceição J, Carneiro
P, Novais F, Scott P, Sundar S, Bacellar O, Carvalho EM and Wilson
ME: Phenotypic and functional characteristics of HLA-DR(+)
neutrophils in Brazilians with cutaneous leishmaniasis. J Leukoc
Biol. 101:739–749. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Chakravarti A, Rusu D, Flamand N, Borgeat
P and Poubelle PE: Reprogramming of a subpopulation of human blood
neutrophils by prolonged exposure to cytokines. Lab Invest.
89:1084–1099. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Vincent JL and Beumier M: Diagnostic and
prognostic markers in sepsis. Expert Rev Anti Infect Ther.
11:265–275. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Parlato M and Cavaillon JM: Host response
biomarkers in the diagnosis of sepsis: A general overview. Methods
Mol Biol. 1237:149–211. 2015. View Article : Google Scholar
|
|
123
|
Daigo K and Hamakubo T: Host-protective
effect of circulating pentraxin 3 (PTX3) and complex formation with
neutrophil extracellular traps. Front Immunol. 3:3782012.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Jaillon S, Peri G, Delneste Y, Frémaux I,
Doni A, Moalli F, Garlanda C, Romani L, Gascan H, Bellocchio S, et
al: The humoral pattern recognition receptor PTX3 is stored in
neutrophil granules and localizes in extracellular traps. J Exp
Med. 204:793–804. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Caironi P, Masson S, Mauri T, Bottazzi B,
Leone R, Magnoli M, Barlera S, Mamprin F, Fedele A, Mantovani A, et
al: Pentraxin 3 in patients with severe sepsis or shock: The ALBIOS
trial. Eur J Clin Invest. 47:73–83. 2017. View Article : Google Scholar
|
|
126
|
Lee YT, Gong M, Chau A, Wong WT, Bazoukis
G, Wong SH, Lampropoulos K, Xia Y, Li G, Wong MCS, et al:
Pentraxin-3 as a marker of sepsis severity and predictor of
mortality outcomes: A systematic review and meta-analysis. J
Infect. 76:1–10. 2018. View Article : Google Scholar
|
|
127
|
Wang C, Li Q, Tang C, Zhao X, He Q, Tang X
and Ren J: Characterization of the blood and neutrophil-specific
microbiomes and exploration of potential bacterial biomarkers for
sepsis in surgical patients. Immun Inflamm Dis. 9:1343–1357. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kalkoff M, Cursons RT, Sleigh JW and
Jacobson GM: The use of real time rtPCR to quantify inflammatory
mediator expression in leukocytes from patients with severe sepsis.
Anaesth Intensive Care. 32:746–755. 2004. View Article : Google Scholar
|
|
129
|
Lu RJ, Taylor S, Contrepois K, Kim M,
Bravo JI, Ellenberger M, Sampathkumar NK and Benayoun BA:
Multi-omic profiling of primary mouse neutrophils predicts a
pattern of sex-and age-related functional regulation. Nat Aging.
1:715–733. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Bongers SH, Chen N, van Grinsven E, van
Staveren S, Hassani M, Spijkerman R, Hesselink L, Lo Tam Loi AT,
van Aalst C, Leijte GP, et al: Kinetics of neutrophil subsets in
acute, subacute, and chronic inflammation. Front Immunol.
12:6740792021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Kingren MS, Starr ME and Saito H:
Divergent sepsis pathophysiology in older adults. Antioxid Redox
Signal. 35:1358–1375. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Arcaroli J, Fessler MB and Abraham E:
Genetic polymorphisms and sepsis. Shock. 24:300–312. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Shurtz-Swirski R, Sela S, Herskovits AT,
Shasha SM, Shapiro G, Nasser L and Kristal B: Involvement of
peripheral polymorphonuclear leukocytes in oxidative stress and
inflammation in type 2 diabetic patients. Diabetes Care.
24:104–110. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
van der Poll T and Opal SM: Host-pathogen
interactions in sepsis. Lancet Infect Dis. 8:32–43. 2008.
View Article : Google Scholar
|
|
135
|
Ma Y, Zhao Y and Zhang X: Factors
affecting neutrophil functions during sepsis: Human microbiome and
epigenetics. J Leukoc Biol. 116:672–688. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Shukla P, Rao GM, Pandey G, Sharma S,
Mittapelly N, Shegokar R and Mishra PR: Therapeutic interventions
in sepsis: Current and anticipated pharmacological agents. Br J
Pharmacol. 171:5011–5031. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Sun R, Huang J, Yang Y, Liu L, Shao Y, Li
L and Sun B: Dysfunction of low-density neutrophils in peripheral
circulation in patients with sepsis. Sci Rep. 12:6852022.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Dagur PK and McCoy JP Jr: Collection,
storage, and preparation of human blood cells. Curr Protoc Cytom.
73:5.1.–5.1.16. 2015.
|
|
139
|
Hardisty GR, Llanwarne F, Minns D, Gillan
JL, Davidson DJ, Gwyer Findlay E and Gray RD: High purity isolation
of low density neutrophils casts doubt on their exceptionality in
health and disease. Front Immunol. 12:6259222021. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Yennemadi AS, Keane J and Leisching G: The
isolation and characterization of low-and normal-density
neutrophils from whole blood. J Vis Exp. 7(216)2025.
|
|
141
|
Thomas HB, Moots RJ, Edwards SW and Wright
HL: Whose gene is it anyway? The effect of preparation purity on
neutrophil transcriptome studies. PLoS One. 10:e01389822015.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Bersanelli M, Mosca E, Remondini D,
Giampieri E, Sala C, Castellani G and Milanesi L: Methods for the
integration of multi-omics data: mathematical aspects. BMC
Bioinformatics. 17(Suppl 2): S152016. View Article : Google Scholar
|
|
143
|
Picard M, Scott-Boyer MP, Bodein A, Périn
O and Droit A: Integration strategies of multi-omics data for
machine learning analysis. Comput Struct Biotechnol J.
19:3735–3746. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Flores JE, Claborne DM, Weller ZD,
Webb-Robertson BM, Waters KM and Bramer LM: Missing data in
multi-omics integration: Recent advances through artificial
intelligence. Front Artif Intell. 6:10983082023. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Hoogendijk AJ, Pourfarzad F, Aarts CEM,
Tool ATJ, Hiemstra IH, Grassi L, Frontini M, Meijer AB, van den
Biggelaar M and Kuijpers TW: Dynamic transcriptome-proteome
correlation networks reveal human myeloid differentiation and
neutrophil-specific programming. Cell Rep. 29:2505–2519.e4. 2019.
View Article : Google Scholar : PubMed/NCBI
|
















