Introduction
Colorectal cancer (CRC) is an aggressive malignancy,
ranking third and second globally in terms of morbidity and
mortality rates, respectively (1). The incidence of CRC in China is
also rising steadily (2). The
primary treatment approach for CRC combines surgery with
chemotherapy and radiotherapy; however, not all patients respond to
these standard therapies (3).
Despite ongoing advancements in combined treatment strategies,
including surgery, radiotherapy, chemotherapy, targeted therapy and
immunotherapy, the survival rate of CRC remains suboptimal, with a
high recurrence rate (3,4).
Oxaliplatin, a third-generation cisplatin analog, is
widely used in CRC treatment (5). While the 5-fluorouracil, folinic
acid and oxaliplatin regimen is initially effective, ~50% of
patients with stage II and III CRC develop resistance to
oxaliplatin-based adjuvant therapy (6). Previous studies have identified
several mechanisms of oxaliplatin resistance, including
anti-apoptotic signaling, ferroptosis, DNA repair, autophagy and
altered drug transport (7,8).
However, the precise mechanisms underlying oxaliplatin resistance
remain unclear.
KPT-330, a selective exportin 1 (XPO1) inhibitor,
has shown marked efficacy in tumor treatment (9,10). Ferreiro-Neira et al
(11) demonstrated that KPT-330
enhances the sensitivity of CRC to radiotherapy, suggesting a
potential novel therapeutic model, the combination therapy with
KPT-330 and radiotherapy. A separate report has indicated that
sequential administration of XPO1 and an ataxia telangiectasia and
Rad3-related protein inhibitor (AZD-6738) improved therapeutic
outcomes in TP53-mutated CRC (12). Additionally, KPT-330 has been
found to enhance the efficacy of platinum-based chemotherapy in
lung and ovarian cancer (13,14). However, to the best of our
knowledge, no studies have explored the relationship between
KPT-330 and oxaliplatin resistance in CRC. Thus, the combination of
oxaliplatin and KPT-330 warrants investigation as a potential
strategy for overcoming resistance in CRC, with further exploration
of its underlying mechanisms.
We hypothesized that KPT-330 could restore
oxaliplatin sensitivity in resistant CRC cells. To test this
hypothesis, the present study aimed to systematically evaluate the
therapeutic potential of KPT-330 in combination with oxaliplatin in
oxaliplatin-resistant CRC cells. The present investigation focused
on the underlying mechanisms of this combination regimen. Changes
in the mitochondrial membrane potential (MMP) were monitored to
assess mitochondrial function, and the effects on cell death
pathways, including apoptosis and ferroptosis, were investigated.
Furthermore, the present study examined the role of key molecular
players by analyzing p53 nuclear translocation, p21 expression and
the expression of the ferroptosis-related gene solute carrier
family 7 member 11 (SLC7A11). Finally, a subcutaneous xenograft
model was established to evaluate the in vivo efficacy of
this combination regimen.
Materials and methods
Reagents and antibodies
HCT116 (cat. no. CCL-247), HCT8 (cat. no. CCL-244)
and FHC (cat. no. CRL-1831) cell lines were obtained from American
Type Culture Collection. The β-actin (cat. no. 4967S), GAPDH (cat.
no. 2118), p21 (cat. no. 2947), cleaved poly (ADP-ribose)
polymerase (PARP) (cat. no. 5625T), cleaved caspase-3 (cat. no.
9664L), Bax (cat. no. 5023T), PARP (cat. no. 9532S), caspase-3
(cat. no. 9668S) and Bcl-2 (cat. no. 15071T) antibodies were
sourced from Cell Signaling Technology, Inc. CDK1 (cat. no.
A11420), cyclin B1 (CCNB1; cat. no. A19037), XPO1 (cat. no. A0299),
glutathione peroxidase 4 (GPX4; cat. no. A11243) and SLC7A11 (cat.
no. A2413) antibodies were obtained from ABclonal Biotech Co., Ltd.
The Lamin B1 (cat. no. AF5161) antibody was obtained from Affinity
Biosciences, Ltd. The p53 (cat. no. sc-126) and Ki67 (cat. no.
sc-23900) antibodies were purchased from Santa Cruz Biotechnology,
Inc. HRP-conjugated secondary antibodies (cat. nos. ZB-2306 and
ZB-2305) were obtained from Beijing Zhongshan Jinqiao Biotechnology
Co., Ltd.
Cell culture
All cells were cultured in a 37°C incubator with 5%
CO2. The oxaliplatin-resistant HCT116 and HCT8 cell
lines (HCT8/L-OHP and HCT116/L-OHP) were developed by progressively
increasing oxaliplatin concentrations, starting at 2 μM and
reaching a final concentration of 14 μM, over a period of ≥9
months. HCT8, HCT8/L-OHP and HCT116/L-OHP cells were cultured in
RPMI-1640 medium (cat. no. CR-31800; Cienry Co., Ltd.) supplemented
with 10% fetal bovine serum (cat. no. BC-SE-FBS01C; Bio-Channel
Biotechnology Co., Ltd.) and 10,000 U/ml penicillin-streptomycin
(cat. no. ant-pm-05; InvivoGen). The HCT116 cell line was cultured
in McCoy's 5A medium (cat. no. CR-16600; Cienry Co., Ltd.) with 10%
fetal bovine serum and 10,000 U/ml penicillin-streptomycin, while
FHC cells were cultured in DMEM (cat. no. CR-12800; Cienry Co.,
Ltd.) with 10% fetal bovine serum and 10,000 U/ml
penicillin-streptomycin.
Cell viability test
For cell viability assays, 5×103 cells
per well were plated on 96-well plates and treated with different
drugs at 37°C for 48 h. For single-agent dose-response curves,
cells were treated with oxaliplatin (0.33, 1, 3.3, 10, 33 and 100
μM; cat. no. HY-17371; MedChemExpress) or KPT-330 (0.001,
0.003, 0.01, 0.03, 0.1, 0.3, 1 and 3 μM; cat. no. S7252;
Selleck Chemicals). For two-agent dose-response curves, cells were
treated with a combination of oxaliplatin (0.33, 1, 3.3, 10, 33 and
100 μM) and KPT-330 (10 and 33 nM). After silencing of XPO1,
cells were treated with oxaliplatin (0.5, 1, 2, 4, 8, 16 and 32
μM). For experiments involving combination therapy and
specific inhibitors, the following treatments were performed on
HCT116/L-OHP and HCT8/L-OHP cells: Cells were treated with single
agents, including oxaliplatin (10 μM) and KPT-330 (33 nM),
as well as their combination. For inhibitor assays, a combination
of oxaliplatin (10 μM) and KPT-330 (33 nM) was used along
with either Z-VAD-FMK (10 μM; cat. no. HY-16658B;
MedChemExpress) or Fer-1 (1 μM; cat. no. HY-100579;
MedChemExpress). Separately, oxaliplatin (2, 10 and 40 μM)
was used to treat CRC (HCT8, HCT116, HCT116/L-OHP and HCT8/L-OHP
cells) and normal cells (FHC cells). Cell viability was then
assessed using the Cell Counting Kit-8 (cat. no. MA0218; Dalian
Meilun Biology Technology Co., Ltd.). Cells were incubated with
Cell Counting Kit-8 solution at 37°C for 2 h according to the
manufacturer's instructions.
Annexin V and PI staining
Apoptosis was detected using the Annexin V-FITC/PI
kit [cat. no. AT101C; MultiSciences (Lianke) Biotech Co., Ltd.].
Following treatment with different drugs (oxaliplatin, 10
μM; KPT-330, 33 nM; Z-VAD-FMK, 10 μM) at 37°C for 48
h, cells were harvested and stained with annexin V and PI for 15
min, and analyzed by flow cytometry (Fortessa; BD Biosciences).
Apoptotic cells were quantified as the sum of early apoptotic cells
(annexin V+/PI-) in the lower-right quadrant
and late apoptotic cells (annexin V+/PI+) in
the upper-right quadrant. Data were analyzed using FlowJo software
(version 10.8; BD Biosciences).
Cell cycle assay
For cell cycle analysis, 2×105 cells per
well were seeded on 6-well plates and cultured for 24 h in
serum-free medium until they reached 30-40% confluence. Cells were
then treated with drugs (oxaliplatin, 10 μM; KPT-330, 33 nM;
or the combination) for 48 h at 37°C in a CO2 incubator.
In a separate experiment, HCT116 and HCT116/L-OHP cells were
treated with oxaliplatin (2 μM) for 48 h at 37°C. After
treatment, cells were harvested, stained using the Cell Cycle
Staining Kit [cat. no. CCS012; MultiSciences (Lianke) Biotech Co.,
Ltd.] for 15 min at room temperature in the dark, and analyzed by
flow cytometry (Fortessa; BD Biosciences). Data were analyzed using
FlowJo software (version 10.8; BD Biosciences).
Reactive oxygen species (ROS) assay
Cells (2×105 per well) were seeded on
6-well plates and cultured until they reached 30-40% confluence.
The cells were then treated with drugs (oxaliplatin, 10 μM;
KPT-330, 33 nM; or the combination) for 48 h at 37°C in a
CO2 incubator. In a separate experiment, HCT116 and
HCT116/L-OHP cells were treated with oxaliplatin (2 μM) for
48 h at 37°C. After treatment, the cells were incubated in
serum-free medium containing 2',7'-dichlorodihydrofluorescein
diacetate (cat. no. S0033; Beyotime Institute of Biotechnology),
MitoSOX Red (cat. no. HY-D1055; MedChemExpress), a fluorescent
probe specifically designed for the detection of mitochondrial
superoxide, or the C11-BODIPY probe (cat. no. D3861; Invitrogen;
Thermo Fisher Scientific, Inc.), a fluorescent probe for the
detection of lipid peroxidation in cells, at 37°C for 20 min.
Following incubation, the fluorescence intensity was measured by
flow cytometry (CytoFLEX; Beckman Coulter, Inc.). Data were
analyzed using FlowJo software (version 10.8; BD Biosciences).
Measurement of malondialdehyde (MDA)
levels
Cells (2×105 per well) were seeded into
6-cm dishes and cultured overnight. After 48 h of drug treatment
(oxaliplatin, 10 μM; KPT-330, 33 nM; or the combination) at
37°C, cells were harvested and lysed. The BCA Protein Assay Kit
(cat. no. P0012; Beyotime Institute of Biotechnology) was used to
quantify the protein concentration. Following protein
quantification, MDA (cat. no. S0131S; Beyotime Institute of
Biotechnology) working solution was added to the lysate, and the
mixture was heated at 100°C for 15 min. After centrifugation at
1,000 × g for 10 min at room temperature, the supernatant was
collected and its absorbance was measured at 532 nm using a
microplate reader.
Measurement of glutathione (GSH)
levels
Cells (2×105 per well) were seeded into
6-cm dishes and cultured overnight. After 48 h of drug treatment
(oxaliplatin, 10 μM; KPT-330, 33 nM; or the combination) at
37°C, cells were harvested, processed according to the GSH and GSSG
Assay Kit instructions (cat. no. S0053; Beyotime Institute of
Biotechnology), and the absorbance was finally measured
spectrophotometrically at a wavelength of 412 nm.
Measurement of the MMP
Cells (2×105 per well) were seeded on
6-well plates and cultured overnight. After 48 h of drug treatment
(oxaliplatin, 10 μM; KPT-330, 33 nM; or their combination)
at 37°C in a CO2 incubator, the treated cells were
incubated with 1 ml JC-1 staining buffer (cat. no. C2006; Beyotime
Institute of Biotechnology) for 20 min at 37°C in the dark. In a
separate experiment, HCT116 and HCT116/L-OHP cells were also
treated with oxaliplatin (2 μM) for 48 h at 37°C and then
stained with 1 ml JC-1 staining buffer for 20 min at 37°C in the
dark. After incubation, the supernatant was removed, and cells were
centrifuged at 600 × g for 4 min at 4°C. The cells were then washed
twice with JC-1 washing buffer (1X) and resuspended in the washing
buffer. JC-1 fluorescence was assessed by flow cytometry (Fortessa;
BD Biosciences). Data were analyzed using FlowJo software (version
10.8; BD Biosciences).
Xenograft tumor model
All animal procedures were approved by the Ethical
Review Committee and Laboratory Animal Welfare Committee of the Sir
Run Run Shaw Hospital, Zhejiang University School of Medicine
(approval no. SRRSH2025-0023; Hangzhou, China). The experimental
mice were purchased from Hangzhou Ziyuan Laboratory Animal
Technology Co., Ltd. A total of 20 female BALB/c nude mice (16-18
g; 6 weeks old) were used in the present study. The mice were
housed in a standard specific-pathogen-free environment for 1 week
before the experiment for acclimation. Housing conditions were
maintained at 22±2°C and 55±10% humidity, with a 12/12 h light/dark
cycle (7:00-19:00), with free access to food and sterile water.
For tumor implantation, 2×106
HCT116/L-OHP cells suspended in 100 μl PBS were
subcutaneously injected into the right flank of each mouse. After 1
week, the mice were randomly assigned to four treatment groups (5
mice per group): Vehicle control (5% DMSO, 40% PEG300, 5% Tween 80
and 50% ddH2O; once per week), oxaliplatin (cat. no.
HY-17371; MedChemExpress) alone (10 mg/kg; once per week), KPT-330
(cat. no. S7252; Selleck Chemicals) alone (10 mg/kg; once per
week), or a combination of oxaliplatin and KPT-330.
Tumor size was measured with Vernier calipers on the
indicated days following the start of treatment: Days 1, 3, 5, 8,
10, 12, 15, 17, 20, 22, 24, 27 and 29. The total time interval from
injection to final tumor measurement was 35 days. Euthanasia was
performed at the end of the pre-defined 4-week treatment period. At
that time, the maximum tumor diameter was 16.8 mm, and the maximum
tumor volume was 1,209.6 mm3. Euthanasia was performed
by gradually filling a chamber with CO2 at a
displacement rate of 35% of the chamber volume per min, with
exposure for ≥5 min after visible cessation of respiration. Death
was confirmed by the loss of respiration, loss of muscle tone and
subsequent cervical dislocation after CO2 inhalation.
Tumors were dissected and embedded in paraffin on day 29 after the
start of treatment. The tumor volume was calculated using the
following formula: Tumor volume (mm3)=tumor length ×
tumor width2/2.
Transmission electron microscopy
(TEM)
Before being fixed for TEM, HCT116/L-OHP cells were
treated with oxaliplatin (10 μM), KPT-330 (33 nM) or a
combination of both. In a separate experiment, HCT116 and
HCT116/L-OHP cells were treated with oxaliplatin (2 μM). All
treatments were performed for 48 h at 37°C. Experimental cells were
fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer
(pH 7.4) for 2 h at 25°C, and then stored at 4°C in the same
fixative overnight. After three washes with PBS, the samples were
incubated in 1% osmium tetroxide for 1 h at room temperature,
followed by three washes with water. The samples were then
incubated in 2% uranyl acetate for 30 min at room temperature.
After dehydration through a series of ethanol steps (15 min each in
50, 70, 90 and 100% ethanol) and two 20-min washes in 100% acetone,
the samples were embedded in epoxy resin for 24 h at 60°C and
sectioned into ultrathin slices (90 nm). The sections were imaged
using a Tecnai G2 Spirit 120kV transmission electron microscope
(FEI; Thermo Fisher Scientific, Inc.).
Immunohistochemistry (IHC) assay
The tissues were fixed in 4% paraformaldehyde
overnight at 4°C, followed by dehydration, clearing and paraffin
embedding. The paraffin sections (5 μm) were then baked at
60°C for 2 h, deparaffinized in xylene and rehydrated through a
descending series of graded alcohols. Antigen retrieval was carried
out using sodium citrate buffer for 10 min at 100°C. Endogenous
peroxidase activity was blocked with 3% H2O2
for 10 min at room temperature. Non-specific binding was blocked
with 10% goat serum (cat. no. SL038; Beijing Solarbio Science &
Technology Co., Ltd.) for 30 min at room temperature. The sections
were then incubated with primary antibodies against Ki67 (1:1,000;
cat. no. sc-23900; Santa Cruz Biotechnology, Inc.) or p53 (1:1,000;
cat. no. sc-126; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
After washing, the sections were incubated with an HRP-conjugated
goat anti-mouse IgG (1:5,000; ZB-2305; Beijing Zhongshan Jinqiao
Biotechnology Co., Ltd.) secondary antibody for 1 h at 37°C. Color
was developed with a DAB kit (cat. no. ZLI-9017; Beijing Zhongshan
Jinqiao Biotechnology Co., Ltd.) for 30 sec at room temperature,
and the sections were counterstained with hematoxylin for 1 min at
room temperature before being mounted and observed under a light
microscope (Whole Slide Scanning System; Olympus VS200; Olympus
Corporation). To evaluate the percentage of Ki67-positive nuclei,
five fields per group were randomly selected and a total of 1,000
cells were counted. The Ki67 proliferation index was calculated as
the percentage of Ki67-positive nuclei out of the total number of
cells counted.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis of mRNA expression
Total RNA from treated cells was extracted using the
AG RNAex Pro Reagent (cat. no. AG21101; Hunan Accurate Bio-Medical
Technology Co., Ltd.) and the SteadyPure RNA Extraction Kit (cat.
no. AG21024; Hunan Accurate Bio-Medical Technology Co., Ltd.).
Prior to RNA extraction, cells were treated with oxaliplatin (10
μM), KPT-330 (33 nM) or a combination of both for 48 h at
37°C. RNA (~1.0 μg) was reverse-transcribed to generate the
first strand of cDNA using the Evo M-MLV RT Mix Kit with gDNA Clean
for qPCR Ver.2 (cat. no. AG11728; Hunan Accurate Bio-Medical
Technology Co., Ltd.). The reverse transcription reaction was
performed at 37°C for 15 min, followed by heating at 85°C for 5
sec. qPCR was performed using ChamQ Universal SYBR qPCR Master Mix
(cat. no. Q711-02; Vazyme Biotech Co., Ltd.). The thermocycling
conditions were as follows: Initial denaturation at 95°C for 30
sec, followed by 40 cycles of 95°C for 10 sec (denaturation) and
60°C for 30 sec (annealing/extension). GAPDH was used as the
internal reference gene. The relative gene expression levels were
quantified using the 2-ΔΔCq method (15). The primer pairs used for qPCR
were as follows: GAPDH forward, 5'-GGAGCGAGATCCCTCCAAAAT-3' and
reverse, 5'-GGCTGTTGTCATACTTCTCATGG-3'; CDK1 forward,
5'-AAACTACAGGTCAAGTGGTAGCC-3' and reverse,
5'-TCCTGCATAAGCACATCCTGA-3'; CCNB1 forward,
5'-AATAAGGCGAAGATCAACATGGC-3' and reverse,
5'-TTTGTTACCAATGTCCCCAAGAG-3'; SLC7A11 forward,
5'-TCTCCAAAGGAGGTTACCTGC-3' and reverse,
5'-AGACTCCCCTCAGTAAAGTGAC-3'; and GPX4 forward,
5'-ACAAGAACGGCTGCGTGGTGAA-3' and reverse,
5'-GCCACACACTTGTGGAGCTAGA-3'.
Western blot analysis
Treated cells and tissues were washed 2-3 times with
PBS, then lysed using RIPA buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology). Prior to protein analysis, HCT8,
HCT116, HCT116/L-OHP and HCT8/L-OHP cells were treated with a range
of oxaliplatin concentrations (1, 2, 4, 8 and 16 μM).
HCT116/L-OHP and HCT8/L-OHP cells were also treated with a series
of KPT-330 concentrations (10, 33, 100, 333 and 1,000 nM).
HCT116/L-OHP and HCT8/L-OHP cells were treated with oxaliplatin (10
μM), KPT-330 (33 nM) or a combination of both. Additionally,
after silencing of p53, HCT116/L-OHP cells were treated with
oxaliplatin (10 μM), KPT-330 (33 nM) or a combination of
both. All cell treatments were conducted for 48 h at 37°C. The
lysates were centrifuged at 12,000 × g for 15 min at 4°C to collect
the protein. Protein concentrations were quantified using a BCA
assay (cat. no. MA0082-2; Dalian Meilun Biology Technology Co.,
Ltd.). Proteins (20 μg/lane) were separated by 10% SDS-PAGE
and transferred onto PVDF membranes (cat. no. ISEQ00010;
Sigma-Aldrich; Merck KGaA). The membranes were blocked with 5% BSA
(cat. no. 9048-46-8; Dalian Meilun Biology Technology Co., Ltd.)
for 1 h at room temperature, and incubated with the specified
primary antibodies specific to the target proteins β-ACTIN (cat.
no. 4967S), GAPDH (cat. no. 2118), p21 (cat. no. 2947), cleaved
PARP (cat. no. 5625T), cleaved caspase-3 (cat. no. 9664L), Bax
(cat. no. 5023T), PARP (cat. no. 9532S), caspase-3 (cat. no.
9668S), Bcl-2 (cat. no. 15071T), CDK1 (cat. no. A11420), CCNB1
(cat. no. A19037), XPO1 (cat. no. A0299), GPX4 (cat. no. A11243),
SLC7A11 (cat. no. A2413), Lamin B1 (cat. no. AF5161) and p53 (cat.
no. sc-126), all at a dilution of 1:1,000, overnight at 4°C.
Afterwards, the membranes were washed three times with PBS and
incubated with the appropriate HRP-conjugated secondary antibodies
(cat. nos. ZB-2306 and ZB-2305; 1:10,000) for 1 h at room
temperature. Protein bands were visualized using an FDbio-Dura ECL
kit (cat. no. FD8020; Hangzhou Fude Biological Technology Co.,
Ltd.), and semi-quantified using ImageJ analysis software (version
8; National Institutes of Health).
A fractionation assay [HCT8/L-OHP and HCT116/L-OHP
cells were treated with oxaliplatin (10 μM), KPT-330 (33 nM)
or a combination of both for 48 h at 37°C] was performed to obtain
nuclear and cytoplasmic protein fractions using the Nuclear and
Cytoplasmic Protein Extraction Kit (cat. no. P0028B; Beyotime
Institute of Biotechnology) according to the manufacturer's
instructions. The separated fractions were then subjected to
western blotting as aforementioned.
Immunofluorescence analysis
HCT116/L-OHP and HCT8/L-OHP cells (5×104
per well) were seeded in confocal culture dishes (cat. no. 801001;
Wuxi NEST Biotechnology Co., Ltd.) and cultured overnight. The
cells were then treated with oxaliplatin (10 μM), KPT-330
(33 nM) or a combination of both for 48 h at 37°C. Following
treatment, the cells were fixed with 4% paraformaldehyde overnight
at 4°C, permeabilized with 0.1% Triton X-100 at room temperature
for 15 min and blocked with 10% goat serum (cat. no. MB4508-1;
Dalian Meilun Biology Technology Co., Ltd.) at room temperature for
1 h. After blocking, cells were incubated with a mouse anti-p53
antibody (cat. no. sc-126; 1:500; Santa Cruz Biotechnology, Inc.)
at 4°C overnight. The cells were then incubated with a goat
anti-mouse secondary antibody (cat. no. A-11001; Alexa Fluor 488;
Thermo Fisher Scientific, Inc.) for 1 h at room temperature.
Finally, the cells were stained with DAPI (1 mg/ml; cat. no. D3571;
Thermo Fisher Scientific, Inc.) for 5 min at room temperature to
visualize the nuclei, and images were captured using a laser
scanning confocal fluorescence microscope. The fractional area of
p53 in the nucleus was quantified by analyzing 10 cells from each
treatment group, and was analyzed using ImageJ analysis software
(version 8; National Institutes of Health).
Cell transfection
The target sequences of small interfering RNAs
(siRNAs; Table SI) were
synthesized by Suzhou GenePharma Co., Ltd. For transient
transfection, Lipofectamine RNAiMAX (cat. no. 13778030; Thermo
Fisher Scientific, Inc.) was used according to the manufacturer's
protocol. A total of 5 μl of si-RNA (20 μM), 100
μl Opti-MEM (cat. no. CR22600; Cienry Co., Ltd.) and 6
μl Lipofectamine RNAiMAX were mixed at room temperature for
20 min. This complex was then added to 0.9 ml RPMI-1640 medium
(resulting in a final siRNA concentration of 100 nM), and the cells
were transfected for 8 h at 37°C. After transfection, cells were
allowed to recover for 24 h at 37°C before being used in subsequent
experiments. The transfection efficiency was determined by western
blot analysis.
Ethynyldeoxyuridine (EdU) assay
Cell proliferation was assessed using an EdU assay
kit (cat. no. C10310; Guangzhou RiboBio Co., Ltd.). Cells
(1×105 per well) were seeded on 12-well plates and
cultured overnight. After 48 h of drug treatment (oxaliplatin, 10
μM; KPT-330, 33 nM; or the combination) at 37°C, cells were
incubated with EdU at a final concentration of 50 μM for 2 h
at 37°C. The cells were then fixed, permeabilized and stained
according to the manufacturer's instructions of the EdU assay kit.
Finally, the images were captured using a laser scanning confocal
fluorescence microscope.
Crystal violet assay
Cells (1×105 per well) were seeded on
12-well plates. When the cells reached 30-40% confluence, they were
treated with oxaliplatin (5 or 10 μM), KPT-330 (10 or 33 nM)
or a combination of the two (KPT-330 at 10 or 33 nM with
oxaliplatin at 5 or 10 μM) for 48 h at 37°C in a
CO2 incubator. The cells were then washed three times
with PBS and fixed with 4% paraformaldehyde at room temperature for
15 min. Afterwards, the cells were washed twice with PBS and
stained with crystal violet at room temperature for 20 min. The
cells were washed three times with water, and images were
captured.
Statistical analysis
Data are presented as the mean ± SD of at least
three independent biological replicates (n≥3). All statistical
analyses were performed using GraphPad Prism 8.0 (Dotmatics).
Differences among multiple groups were compared using one-way
ANOVA. If the data met the assumption of homogeneity of variances,
Tukey's Honestly Significant Difference post hoc test was used. If
the assumption was violated, Welch's ANOVA with Dunnett's T3 post
hoc test was performed. P<0.05 was considered to indicate a
statistically significant difference.
Results
KPT-330 and oxaliplatin combined inhibit
the proliferation of oxaliplatin-resistant CRC cells
Oxaliplatin-resistant cell lines were established
using oxaliplatin-sensitive CRC cell lines (HCT116 and HCT8)
(Fig. 1A). Notably, HCT116/L-OHP
and HCT8/L-OHP cells exhibited higher IC50 values (45.94
μM for HCT116/L-OHP cells and 50.91 μM for HCT8/L-OHP
cells) compared with their parental counterparts HCT116 (2.979
μM) and HCT8 (4.847 μM) cells (Fig. 1B). Previous research has
suggested that XPO1 expression is negatively associated with the
prognosis of patients with CRC, positioning XPO1 as a potential
therapeutic target for CRC (16). Furthermore, KPT-185, a selective
XPO1 inhibitor, synergistically induces apoptosis in human
pancreatic and colon cancer cell lines when combined with
oxaliplatin (17). In line with
these findings, XPO1 expression was higher in the newly established
oxaliplatin-resistant cell lines compared with their parental
counterparts (Fig. S1A).
Oxaliplatin treatment induced a concentration-dependent increase in
XPO1 expression in both HCT116/L-OHP and HCT8/L-OHP cells (Fig. 1C). Additionally, a similar
induction of XPO1 expression was observed in both HCT116 and HCT8
cells following oxaliplatin treatment (Fig. S1B).
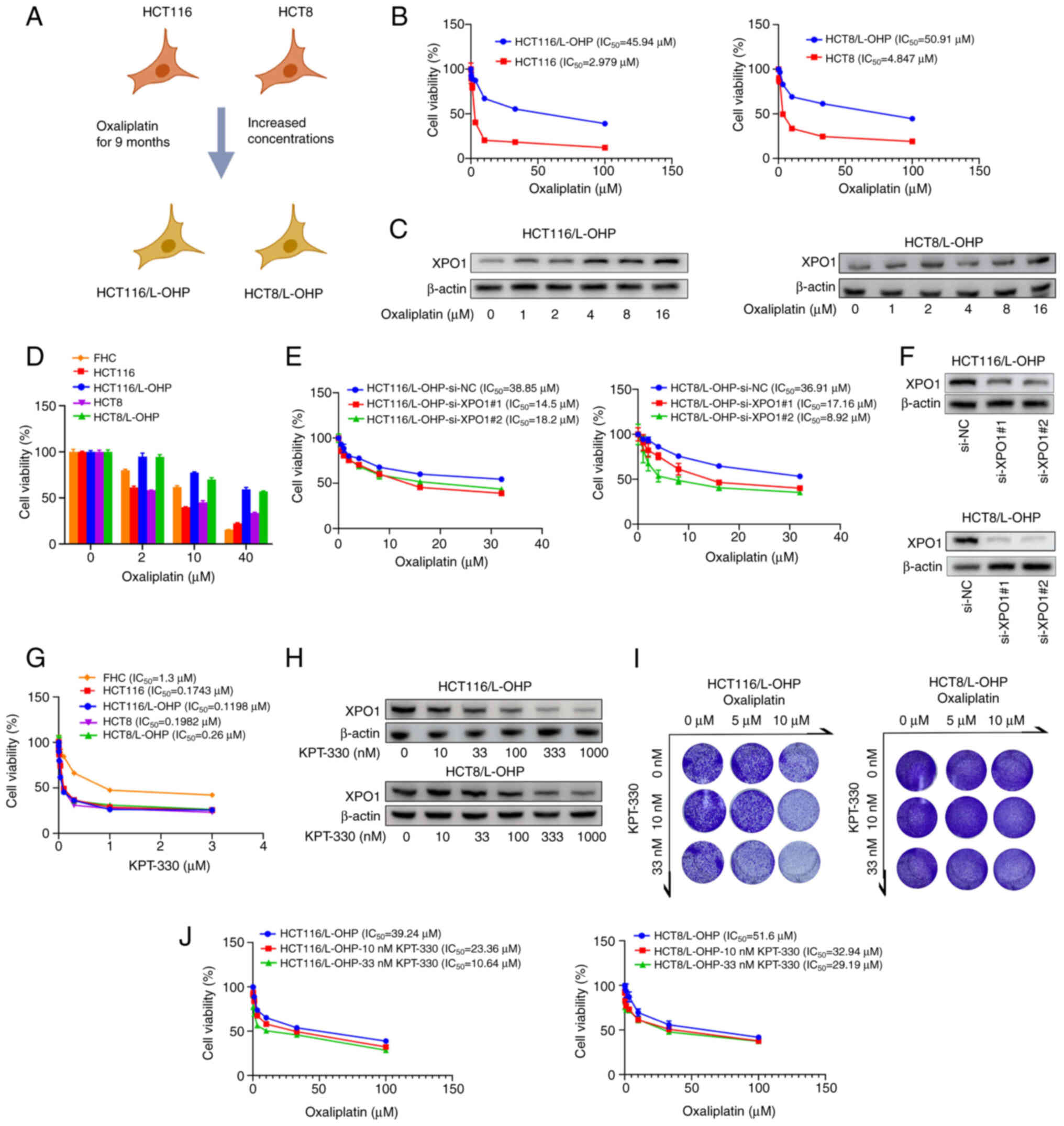 | Figure 1Combination of KPT-330 and
oxaliplatin synergistically inhibits the viability of
oxaliplatin-resistant CRC cells. (A) Schematic illustrating the
process of constructing oxaliplatin-resistant HCT116/L-OHP and
HCT8/L-OHP cell lines. (B) Dose-response curves for oxaliplatin in
the four CRC cell lines, with cell viability measured using the
CCK-8 assay after 48 h of treatment. (C) Immunoblot analysis of
HCT116/L-OHP and HCT8/L-OHP cells treated with oxaliplatin for 48
h. (D) Viability of four CRC cell lines and FHC cells treated with
oxaliplatin for 48 h, measured using a CCK-8 assay. (E) Viability
of indicated cancer cell lines after silencing of XPO1 or
transfection with si-NC, followed by 48 h of oxaliplatin treatment.
(F) Immunoblot analysis of HCT116/L-OHP and HCT8/L-OHP cells after
silencing of XPO1 or transfection with si-NC. (G) Dose-response
curves for KPT-330 in the four CRC cell lines and FHC cells, with
cell viability assessed using a CCK-8 assay after 48 h of
treatment. (H) Immunoblot analysis of HCT116/L-OHP and HCT8/L-OHP
cells treated with KPT-330 for 48 h. (I) Crystal violet assay for
indicated cancer cell lines treated with different dosages of
oxaliplatin and KPT-330. (J) Dose-response curves for oxaliplatin
in the two CRC cell lines with a fixed dose of KPT-330, with cell
viability measured using a CCK-8 assay after 48 h of treatment.
CCK-8, Cell Counting Kit-8; CRC, colorectal cancer; NC, negative
control; si, small interfering RNA; XPO1, exportin 1. |
The cytotoxic effects of oxaliplatin and KPT-330
were subsequently assessed in vitro using four CRC cell
lines (HCT8, HCT8/L-OHP, HCT116 and HCT116/L-OHP) and the
non-cancerous FHC cells. Low concentrations of oxaliplatin
decreased the viability of sensitive CRC cells, whereas high
concentrations of oxaliplatin decreased the viability of FHC cells.
The inhibitory effect on resistant CRC cells was less pronounced
than that on sensitive CRC cells and FHC cells (Fig. 1D). The IC50 values for
KPT-330 ranged between 0.1 and 0.3 μM in CRC cells, while
1.3 μM KPT-330 suppressed the viability of non-cancerous FHC
cells (Fig. 1G). Silencing of
XPO1 using two distinct siRNAs in HCT116/L-OHP and HCT8/L-OHP
cells, which are resistant to oxaliplatin, enhanced their
sensitivity to oxaliplatin (Fig. 1E
and F). The inhibition of XPO1 expression by KPT-330 was found
to be dose-dependent, and KPT-330 could enhance the sensitivity of
oxaliplatin-resistant CRC cells to oxaliplatin by downregulating
XPO1 expression (Fig. 1H-J).
These results collectively demonstrated a synergistic effect of
KPT-330 and oxaliplatin in overcoming resistance in CRC cells.
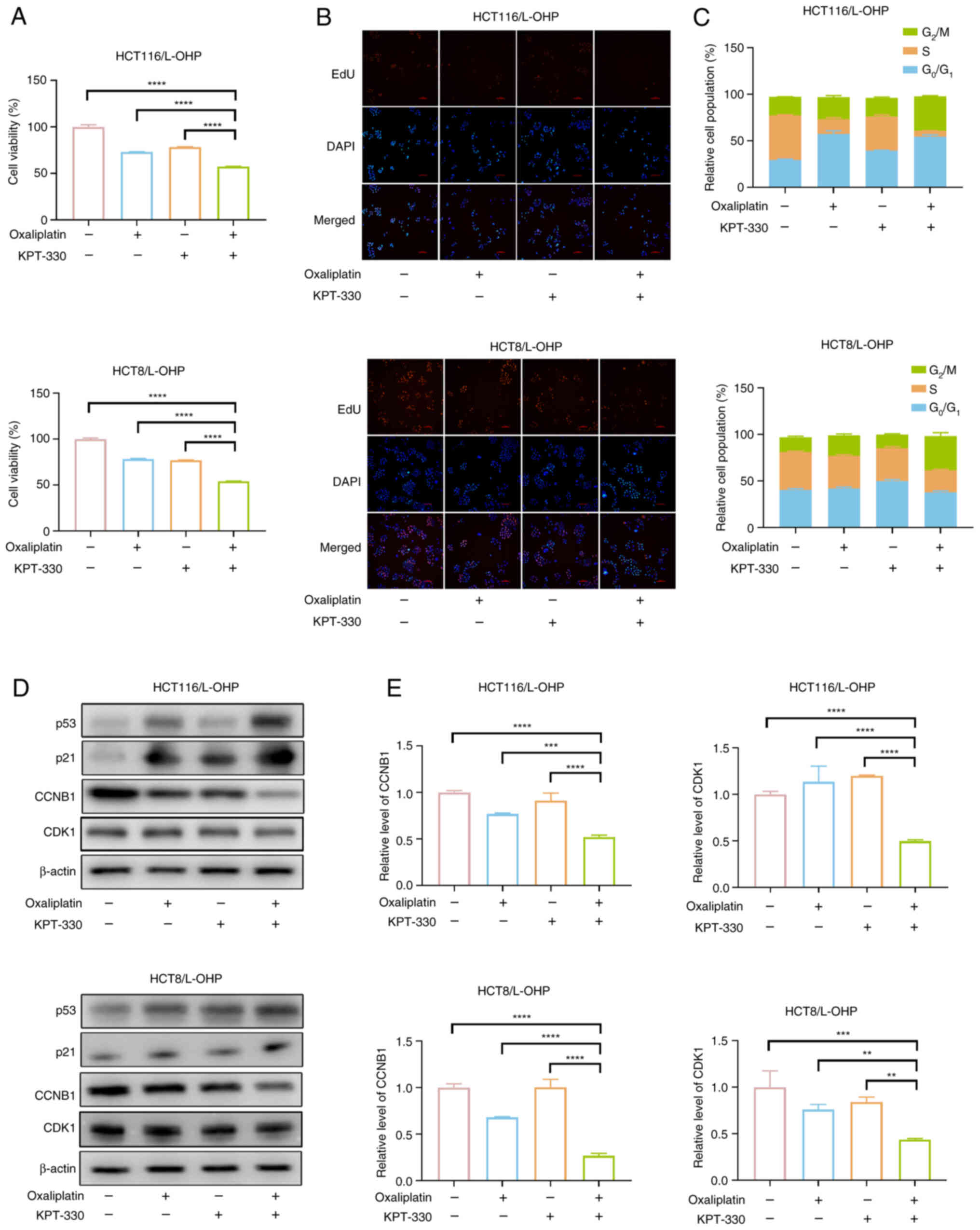
Combination of KPT-330 and oxaliplatin
induces G2/M cell cycle arrest in oxaliplatin-resistant
CRC cells
In subsequent experiments, the combination treatment
for 48 h resulted in a more significant reduction in the viability
of HCT116/L-OHP and HCT8/L-OHP cells compared with oxaliplatin
alone (Fig. 2A). EdU staining
was employed to assess the impact of the KPT-330 and oxaliplatin
combination on cell proliferation, revealing a pronounced
inhibition of proliferation in both HCT116/L-OHP and HCT8/L-OHP
cells following combined treatment (Fig. 2B). Oxaliplatin primarily induces
G2/M cell cycle arrest and a transient delay in the S
phase (18). When comparing the
cell cycle distribution of HCT116 and HCT116/L-OHP cells after 48 h
of oxaliplatin (2 μM) treatment, a clear accumulation in the
G2/M phase was observed in HCT116 cells, accompanied by
a reduction in cells in the S and G0/G1
phases (Fig. S2A). In
HCT116/L-OHP cells, the combination treatment further reduced the
proportion of cells in the S phase and notably increased the
proportion of cells in the G2/M phase compared with
oxaliplatin (10 μM) alone (Fig. S2B and C). In HCT8/L-OHP cells,
where oxaliplatin as a single agent had minimal effects, the
combination therapy increased the proportion of cells in the
G2/M phase compared with oxaliplatin (10 μM)
alone (Fig. S2B and C). These
results suggested that KPT-330 and oxaliplatin influenced proteins
associated with the G2/M phase, as evidenced by the
increase in the G2/M cell population following
combination treatment. To explore the underlying mechanism of the
cell cycle arrest induced by the combination, the expression of key
G2/M arrest markers was analyzed by western blotting.
The protein and mRNA levels of CCNB1 and CDK1, which are critical
regulators of the G2/M phase, were reduced following
combined treatment (Fig. 2D and
E). Given the known role of p53 and p21 in modulating the
CDK1-CCNB1 complex (19), the
present results demonstrated that the combination treatment
upregulated p53 and p21 protein expression, as evidenced by a
subsequent downregulation of CDK1 and CCNB1 protein levels
(Fig. 2D). These results
collectively indicated that the KPT-330 and oxaliplatin combination
induced G2/M cell cycle arrest in oxaliplatin-resistant
CRC cells.
Combination of KPT-330 and oxaliplatin
induces apoptosis in oxaliplatin-resistant CRC cells
Compared with HCT116/L-OHP cells, oxaliplatin
induced a significant increase in apoptosis in HCT116 cells
(Fig. S3A), indicating that
resistance to oxaliplatin in CRC is associated with anti-apoptotic
mechanisms (20). To further
explore whether the combination of oxaliplatin and KPT-330
synergistically induces apoptosis in oxaliplatin-resistant CRC
cells, apoptosis levels were quantitatively assessed. While
treatment with either oxaliplatin or KPT-330 alone did not
significantly induce apoptosis in HCT116/L-OHP or HCT8/L-OHP cells,
the combination therapy markedly enhanced apoptosis (Fig. 3A). To investigate whether the
observed apoptosis was caspase-dependent, Z-VAD-FMK, a pan-caspase
inhibitor, was employed (21,22). The results revealed that
Z-VAD-FMK effectively blocked apoptosis induced by the combination
therapy (Fig. 3B and C). Western
blotting further demonstrated that oxaliplatin combined with
KPT-330 increased cleaved PARP levels (Fig. 3D). Collectively, these results
suggested that KPT-330 potentiated oxaliplatin-induced apoptosis in
CRC cells.
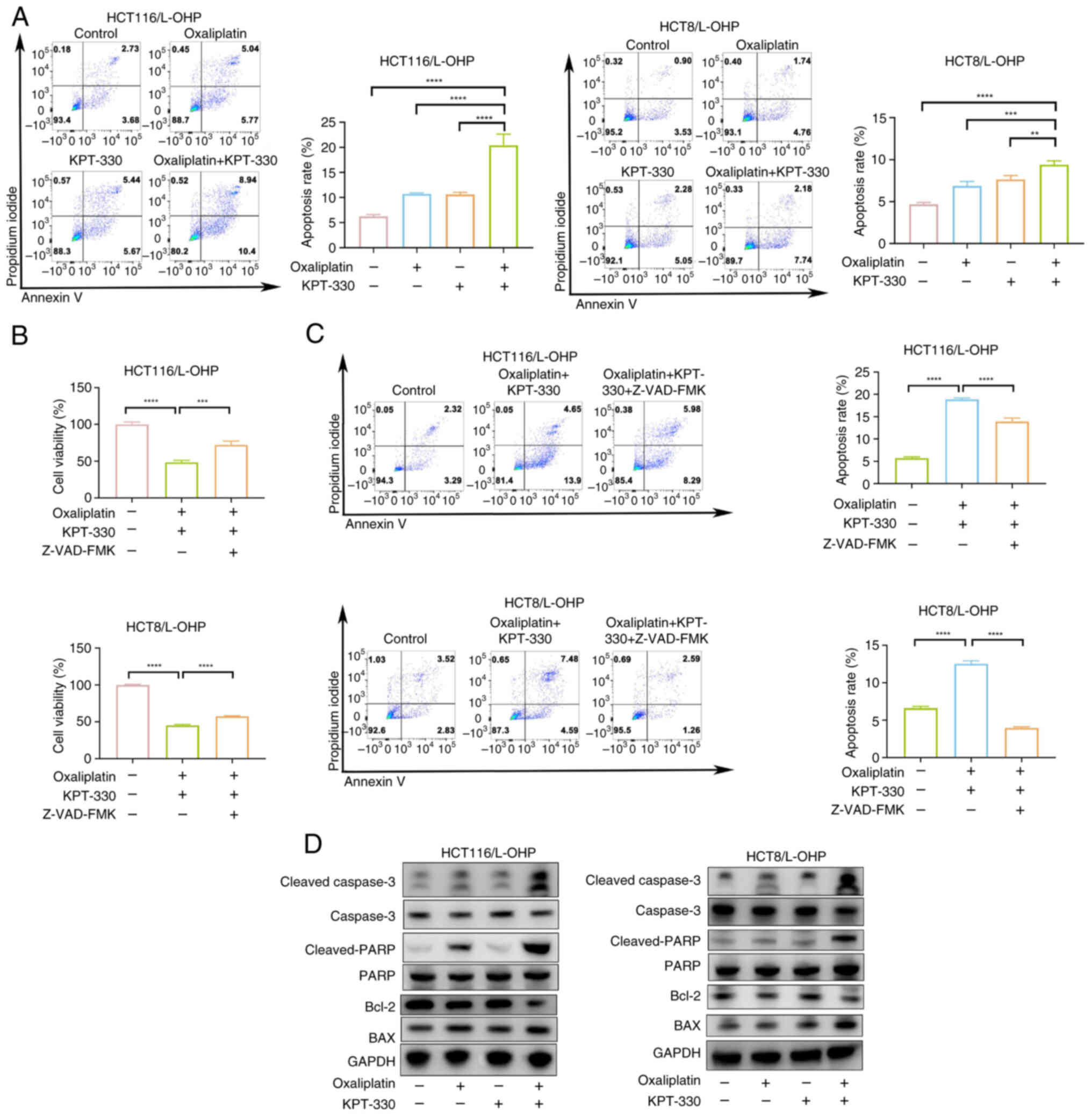 | Figure 3Combination of KPT-330 and
oxaliplatin induces apoptosis in oxaliplatin-resistant colorectal
cancer cells. (A) Apoptosis rate of HCT116/L-OHP and HCT8/L-OHP
cells treated with oxaliplatin, KPT-330 or the combination for 48
h, assessed by flow cytometry. The sum of early apoptotic cells
(annexin V+/PI-) in the lower-right quadrant
and late apoptotic cells (annexin V+/PI+) in
the upper-right quadrant indicates apoptotic cells, and was used
for quantification. (B) Viability of HCT116/L-OHP and HCT8/L-OHP
cells treated with oxaliplatin and KPT-330 or the combination with
Z-VAD-FMK for 48 h, analyzed using a Cell Counting Kit-8 assay. (C)
Apoptosis rate of HCT116/L-OHP and HCT8/L-OHP cells treated with
oxaliplatin and KPT-330 or the combination with Z-VAD-FMK for 48 h,
analyzed by flow cytometry. The sum of early apoptotic cells
(annexin V+/PI-) in the lower-right quadrant
and late apoptotic cells (annexin V+/PI+) in
the upper-right quadrant indicates apoptotic cells, and was used
for quantification. (D) Immunoblot analysis of HCT116/L-OHP and
HCT8/L-OHP cells treated with oxaliplatin, KPT-330 or the
combination for 48 h. **P<0.01,
***P<0.001, ****P<0.0001. PARP, poly
(ADP-ribose) polymerase. |
Combination of KPT-330 and oxaliplatin
increases the levels of ROS and induces mitochondrial
dysfunction
Apoptosis is a multifaceted process involving
caspase activation, MMP changes, the balance between pro-apoptotic
and anti-apoptotic proteins, and other signaling pathways (23). Compared with HCT116/L-OHP cells,
HCT116 cells exhibited greater sensitivity to oxaliplatin-induced
changes in the MMP (Fig. S3B).
To investigate whether the combination treatment induced apoptosis
by affecting the MMP, JC-1, a fluorescent indicator of the MMP, was
used. In both HCT116/L-OHP and HCT8/L-OHP cells, combination
treatment with oxaliplatin and KPT-330 induced greater
mitochondrial depolarization than monotherapy (Fig. 4A). However, in HCT8/L-OHP cells,
low-dose KPT-330 treatment did not affect mitochondrial
depolarization (Fig. 4A).
Notably, the MMP loss in both KPT-330 and oxaliplatin monotherapy
groups was lower than that of the control group. This finding
suggests that while the single agents induced apoptosis, they might
be triggering a different or less direct mitochondrial pathway in
these resistant cells. Alternatively, the observed decrease in MMP
loss could be a result of the adaptive mechanisms of the cells,
which enables them to maintain mitochondrial function and viability
under single-agent stress, thereby contributing to drug resistance.
These results suggested that combination therapy may induce
mitochondrial dysfunction. To further determine if this dysfunction
is linked to increased ROS production, ROS levels were measured in
the cells. Oxaliplatin treatment had a negligible effect on ROS
induction in HCT116/L-OHP cells, in contrast to its marked effect
on sensitive HCT116 cells (Fig.
S3C), combination therapy increased ROS levels in both
HCT116/L-OHP and HCT8/L-OHP cells (Fig. 4B). Given that most intracellular
ROS are generated within mitochondria, the MitoSOX Red reagent, a
mitochondria-targeted form of dihydroethidium, was used to examine
mitochondrial ROS levels. The combination of oxaliplatin and
KPT-330 further elevated mitochondrial ROS levels in both
HCT116/L-OHP and HCT8/L-OHP cells compared with either monotherapy
treatment (Fig. 4C). TEM images
revealed morphological changes in the mitochondria following
treatment with oxaliplatin, KPT-330 or the combination. Notably,
the combination therapy resulted in increased membrane density and
a shrunken mitochondrial morphology (Fig. 4D). Additionally, compared with
those in HCT116/L-OHP cells, mitochondria in HCT116 cells displayed
impaired morphology following oxaliplatin treatment (Fig. S3D). These results suggested that
the combination of oxaliplatin and KPT-330 induced mitochondrial
dysfunction through the elevation of ROS levels.
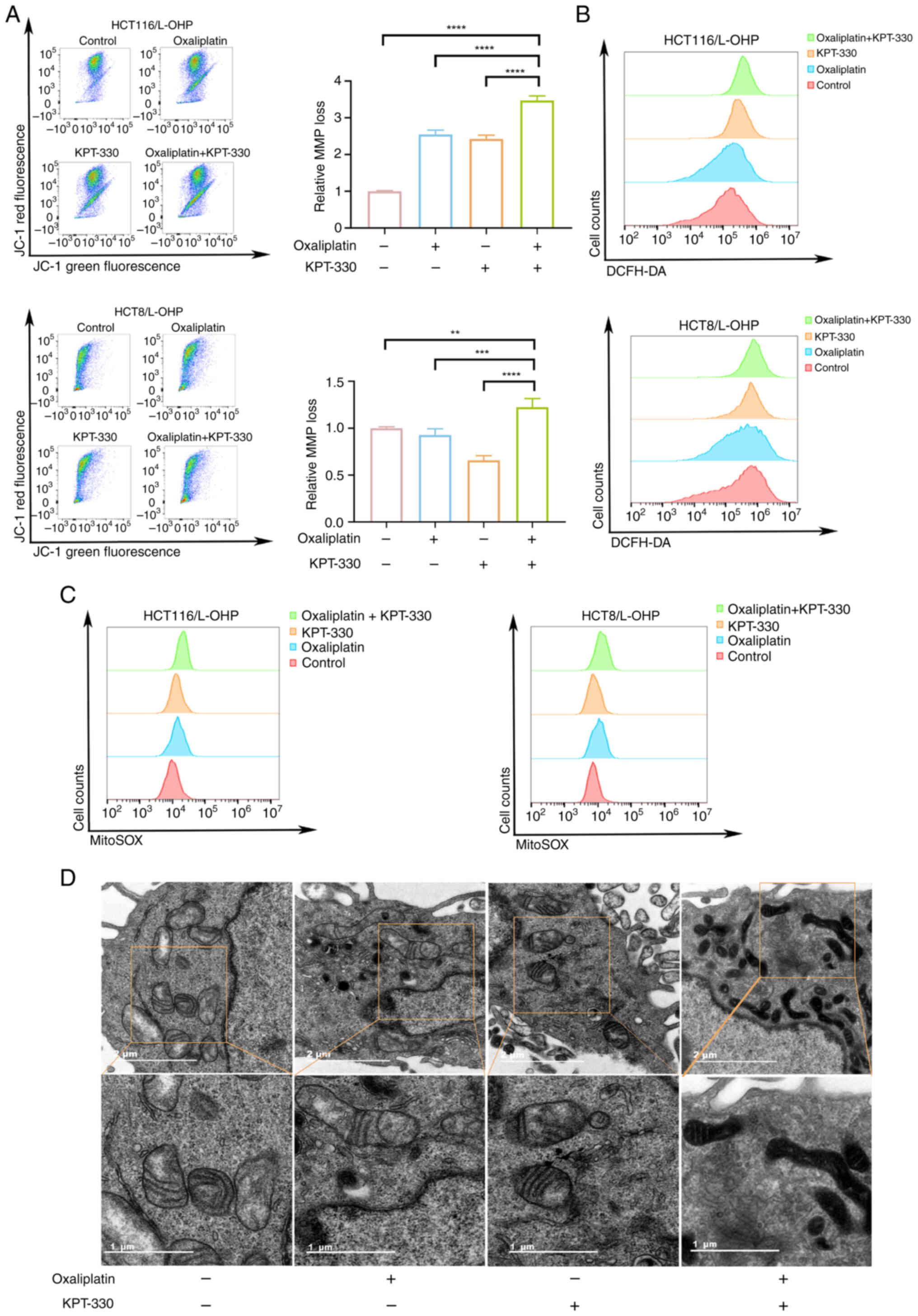 | Figure 4Combination of KPT-330 and
oxaliplatin increases the levels of reactive oxygen species and
induces mitochondrial dysfunction. (A) HCT116/L-OHP and HCT8/L-OHP
cells were treated with oxaliplatin, KPT-330 or the combination for
48 h, stained with JC-1, and analyzed by flow cytometry. The bar
chart shows the relative MMP loss in the four treatment groups. (B)
HCT116/L-OHP and HCT8/L-OHP cells were treated with oxaliplatin,
KPT-330 or the combination for 48 h, stained with DCFH-DA, and
analyzed by flow cytometry. (C) HCT116/L-OHP and HCT8/L-OHP cells
were treated with oxaliplatin, KPT-330 or the combination for 48 h,
stained with MitoSOX, and analyzed by flow cytometry. (D)
Representative transmission electron microscopy images of
mitochondrial morphology in HCT116/L-OHP cells treated with
oxaliplatin, KPT-330 or the combination for 48 h. Scale bars, 2
μm (upper panel) or 1 μm (lower panel).
**P<0.01, ***P<0.001,
****P<0.0001. DCFH-DA,
2',7'-dichlorodihydrofluorescein diacetate; MMP, mitochondrial
membrane potential. |
Combination of KPT-330 and oxaliplatin
inhibits SLC7A11 and GPX4 expression to induce ferroptosis
The present TEM results (Fig. 4D) revealed that HCT116/L-OHP
cells treated with combination therapy exhibited distinct
morphological features characteristic of ferroptosis, an
iron-dependent, non-apoptotic form of cell death marked by the
accumulation of cytotoxic lipid ROS, leading to lipid membrane
damage and perforation (24).
Inducing ferroptosis can overcome oxaliplatin resistance in CRC
cells (25,26). To further explore whether
combination therapy sensitizes oxaliplatin-resistant cells to
oxaliplatin by triggering ferroptosis, several experiments were
conducted. Both lipid peroxidation and MDA levels were increased
following combination treatment with oxaliplatin and KPT-330
(Fig. 5A and B). Inhibition of
cell viability by the combination therapy was partially reversed
using the ferroptosis inhibitor ferrostatin-1 (Fig. 5C). The present results
demonstrated that combination therapy enhanced p53 expression. As a
key regulator in tumorigenesis and development, p53 serves a
critical role in both oxaliplatin resistance and ferroptosis
(27). Based on this, it was
hypothesized that combination therapy promotes p53 expression to
activate ferroptosis-related signaling. Combination treatment
reduced the mRNA and protein levels of SLC7A11 and GPX4 (Fig. 5D and E), and similarly decreased
GSH levels (Fig. 5F). Knockdown
of p53 partially counteracted the reduction in SLC7A11 and GPX4
expression induced by the combination therapy (Figs. S4 and 5G). These results indicated that the
combination of KPT-330 and oxaliplatin induced ferroptosis by
promoting p53 expression, which in turn suppressed SLC7A11 and GPX4
expression.
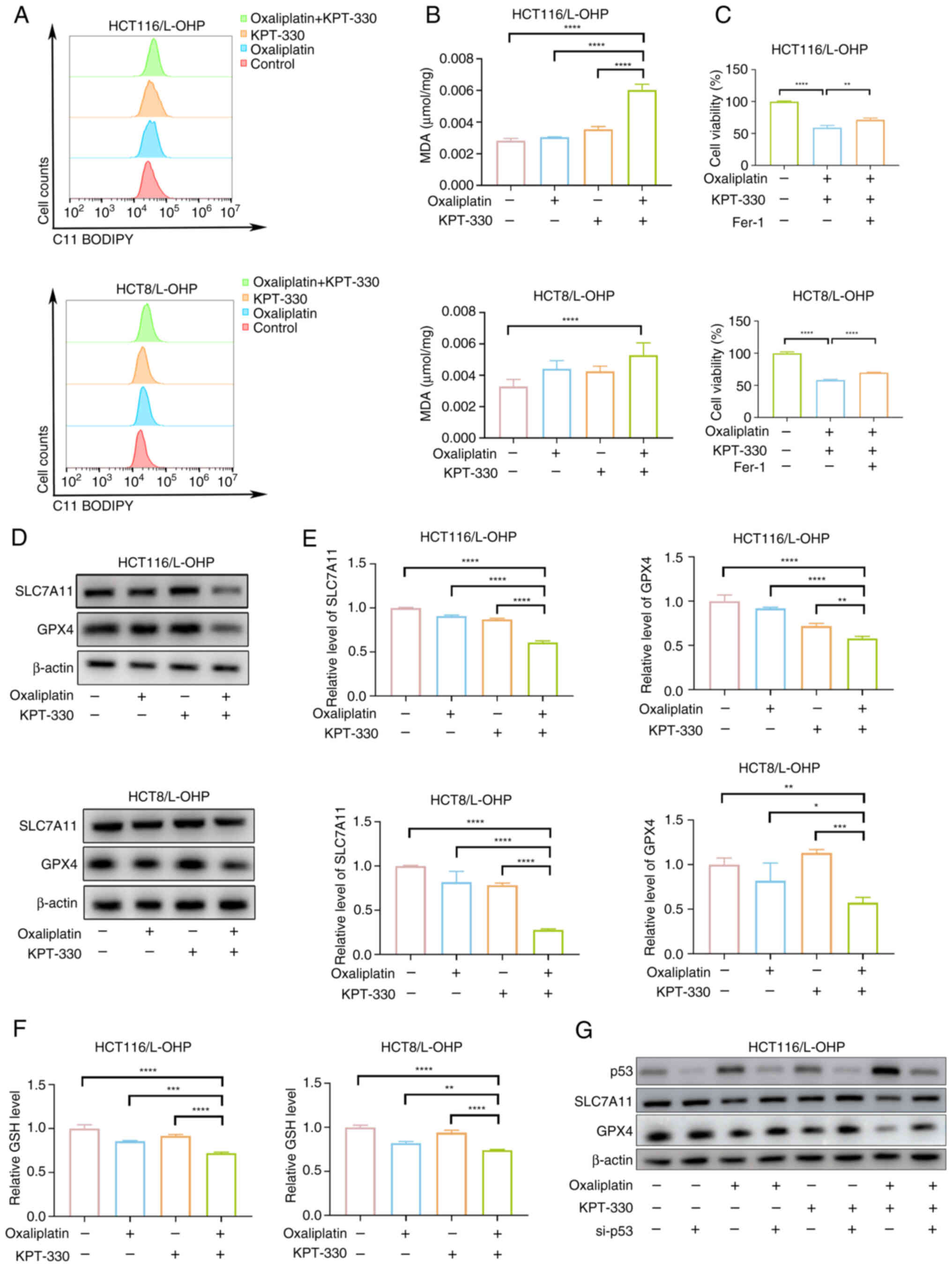 | Figure 5Combination of KPT-330 and
oxaliplatin inhibits SLC7A11 and GPX4 expression to induce
ferroptosis. (A) HCT116/L-OHP and HCT8/L-OHP cells were treated
with oxaliplatin, KPT-330 or the combination for 48 h, stained with
C11-BODIPY, and analyzed by flow cytometry. (B) MDA levels after
treatment of HCT116/L-OHP and HCT8/L-OHP cells with oxaliplatin,
KPT-330 or the combination for 48 h. (C) Viability of HCT116/L-OHP
and HCT8/L-OHP cells treated with KPT-330 and oxaliplatin or the
combination with Fer-1 for 48 h, analyzed using a Cell Counting
Kit-8 assay. (D) Immunoblot analysis of HCT116/L-OHP and HCT8/L-OHP
cells treated with oxaliplatin, KPT-330 or the combination for 48
h. (E) mRNA expression levels of SLC7A11 and GPX4 in HCT116/L-OHP
and HCT8/L-OHP cells treated with oxaliplatin, KPT-330 or the
combination for 48 h. (F) Relative GSH levels assessed after
treating HCT116/L-OHP and HCT8/L-OHP cells with oxaliplatin,
KPT-330 or the combination for 48 h. (G) Immunoblot analysis of
HCT116/L-OHP cells after knockdown of p53, and treatment with
oxaliplatin, KPT-330 or the combination for 48 h.
*P<0.05, **P<0.01,
***P<0.001, ****P<0.0001. Fer-1,
ferrostatin-1; GPX4, glutathione peroxidase 4; GSH, glutathione;
MDA, malondialdehyde; si, small interfering RNA; SLC7A11, solute
carrier family 7 member 11. |
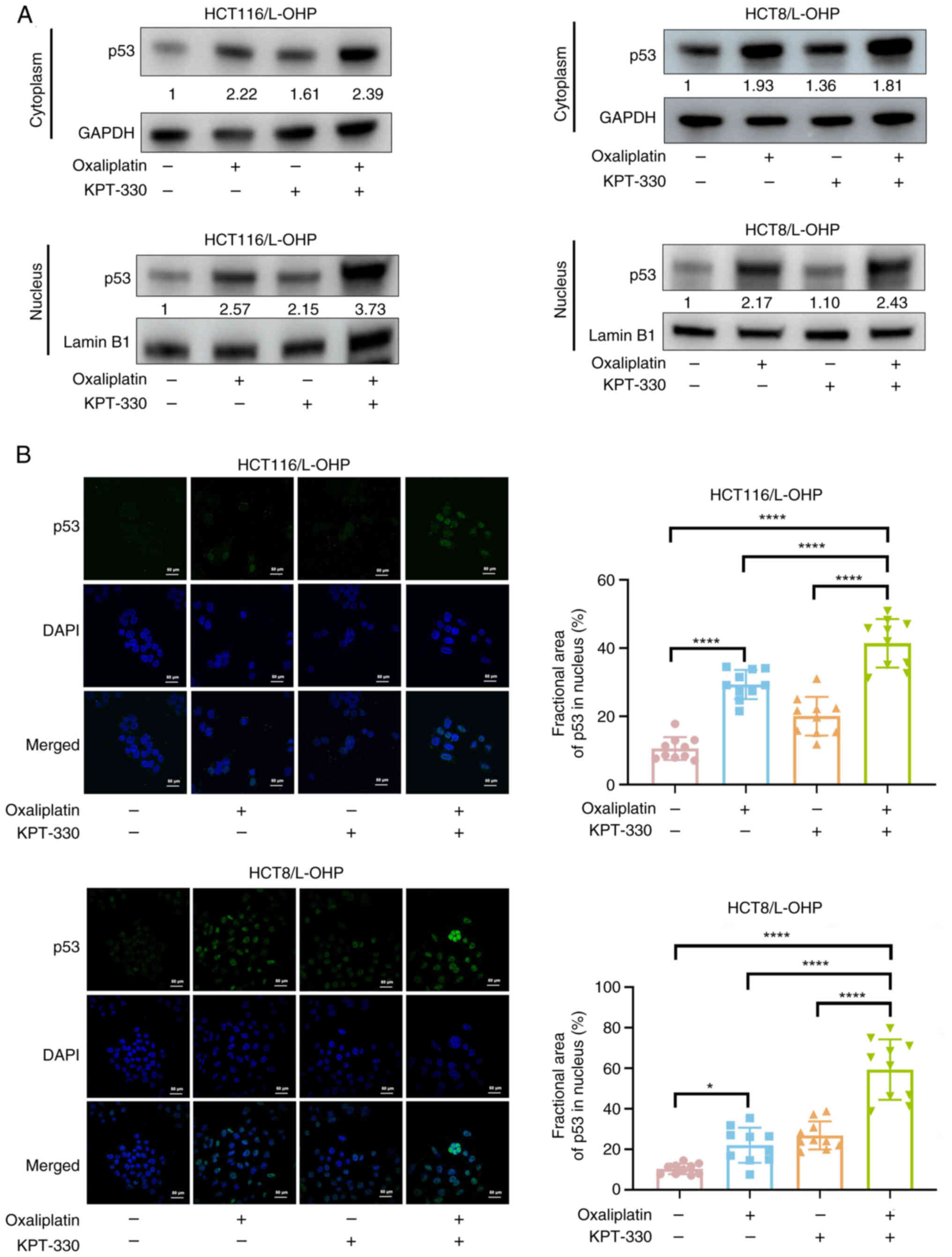
Combination of KPT-330 and oxaliplatin
induces p53 nuclear retention
Combination treatment enhanced p53 expression. Given
that XPO1 mediates the nuclear export of p53, the balance between
its nuclear and cytoplasmic localization is a key mechanism that
influences oxaliplatin sensitivity (28). Additionally, increased
cytoplasmic p53 levels diminish cellular sensitivity to oxaliplatin
(28,29). Cell fractionation experiments
revealed that oxaliplatin monotherapy resulted in elevated p53
levels in both the cytoplasmic and nuclear fractions (Fig. 6A). Immunofluorescence analysis
further demonstrated that oxaliplatin treatment alone notably
promoted nuclear accumulation of p53 (Fig. 6A and B). In HCT116/L-OHP cells,
combination therapy further enhanced nuclear p53 accumulation
(Fig. 6A and B). In HCT8/L-OHP
cells, the combination therapy not only reduced p53 nuclear export
(Fig. 6A) but also increased
nuclear p53 expression (Fig. 6A and
B). In conclusion, these results indicated that the combination
of KPT-330 and oxaliplatin induced p53 nuclear retention.
Combination of KPT-330 and oxaliplatin
effectively suppresses tumor growth in a mouse xenograft model
Assessing the effectiveness of synergistic
inhibition in animal models is crucial. A subcutaneous xenograft
model was established by injecting HCT116/L-OHP cells
subcutaneously. At 1 week after tumor implantation, four treatment
groups were formed: Vehicle (once a week), oxaliplatin (10 mg/kg;
once a week), KPT-330 (10 mg/kg; once a week) and combination
treatment (Fig. 7A). After 4
weeks of treatment, the body weight of mice in the combination
therapy group was not significantly different from those of the
single-drug (oxaliplatin or KPT-330) and vehicle control groups
(Fig. 7B). Additionally, after 4
weeks of treatment, the combination therapy group showed a
significant reduction in both tumor volume and mass compared with
the monotherapy (oxaliplatin or KPT-330) and control groups
(Fig. 7C-E), supporting the
present in vitro findings. Further IHC staining revealed
that the combination treatment reduced the proliferation index, as
indicated by decreased Ki67 expression (Fig. 7F and G). Combination therapy with
KPT-330 significantly enhanced the nuclear accumulation of p53, an
effect that was only partially achieved with oxaliplatin treatment
alone (Fig. 6A and B). IHC
analysis of xenograft tumors revealed a similar trend, with the
combination treatment promoting p53 accumulation, particularly in
the nucleus (Fig. 7F).
Additionally, western blot analysis revealed that oxaliplatin
treatment increased XPO1 expression (Fig. 7H). These data demonstrated the
effectiveness of the combination of oxaliplatin and KPT-330 in
overcoming oxaliplatin resistance in CRC.
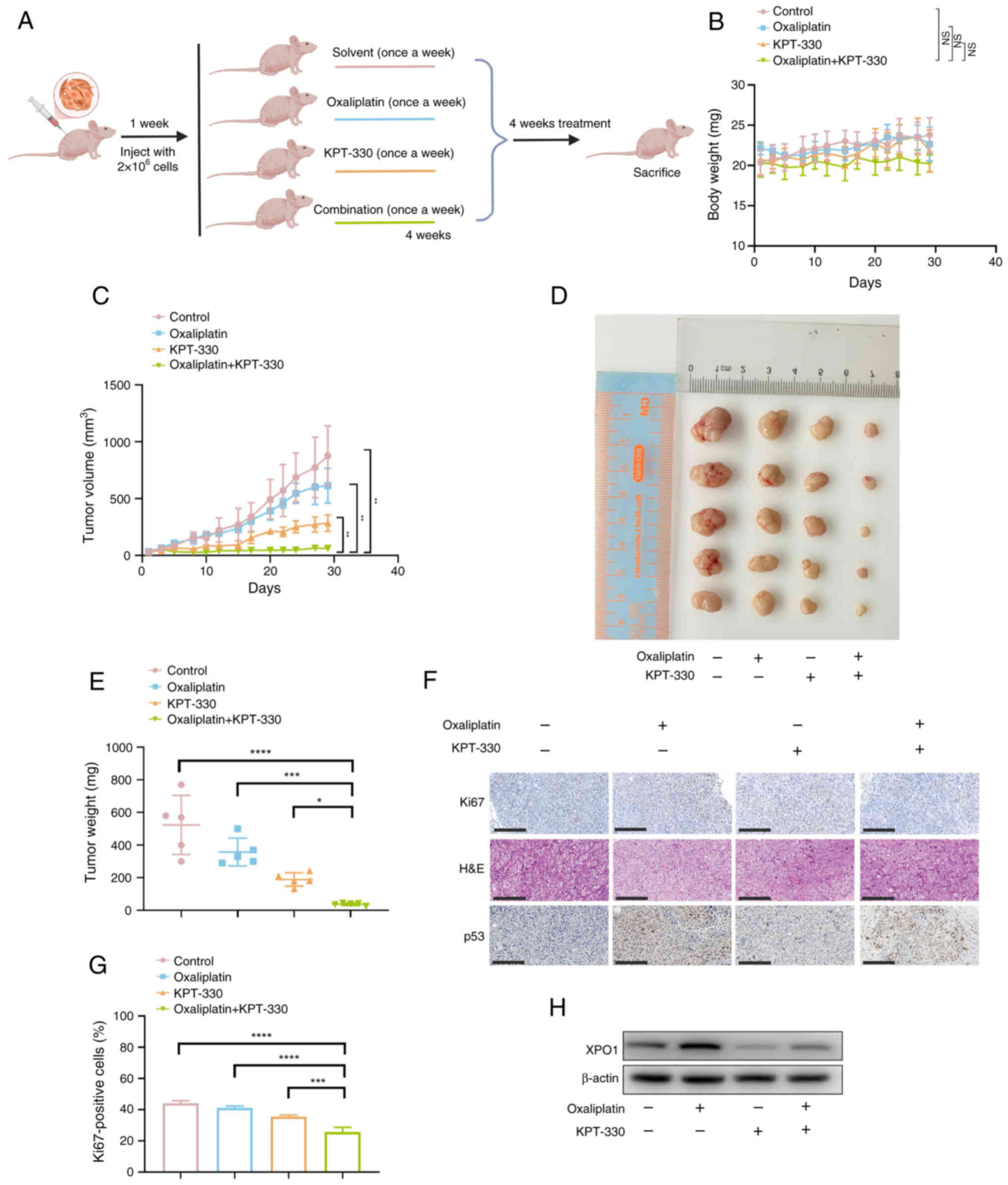 | Figure 7Combination of KPT-330 and
oxaliplatin effectively suppresses tumor growth in a mouse
xenograft model. (A) A subcutaneous tumor xenograft model was
established by injecting HCT116/L-OHP cells into the subcutaneous
tissue of nude mice. At 1 week after tumor establishment, four
treatment groups were formed: Vehicle control, oxaliplatin, KPT-330
and combination. (B) Body weight curves of mice during the in
vivo efficacy assessment of oxaliplatin and KPT-330 in
HCT116/L-OHP xenografts. The NS symbol indicates that the weight in
the combination therapy group was not significantly different from
those in the single-drug (oxaliplatin or KPT-330) and vehicle
control groups at the final timepoint. (C) Growth curve of tumor
volume in the in vivo efficacy assessment of oxaliplatin and
KPT-330 in HCT116/L-OHP xenografts. (D) Representative images of
tumors from all groups (n=5). (E) Tumor weights in each group
(n=5). (F) Representative staining images of Ki67, H&E and p53
in HCT116/L-OHP xenograft tumors. Scale bar, 50 μm. (G)
Quantification of Ki67 in HCT116/L-OHP xenograft tumors treated
with oxaliplatin, KPT-330 or the combination. (H) Immunoblot
analysis of HCT116/L-OHP xenograft tumors treated with oxaliplatin,
KPT-330 or the combination. *P<0.05,
**P<0.01, ***P<0.001,
****P<0.0001. NS, not significant; XPO1, exportin
1. |
Discussion
Oxaliplatin is a widely used chemotherapeutic agent
in the treatment of CRC (7).
However, the development of resistance to oxaliplatin limits its
clinical efficacy (30). This
resistance is often associated with the adaptive tolerance of tumor
cells to cell death mechanisms (31). Therefore, strategies aimed at
enhancing tumor cell sensitivity to oxaliplatin are critical for
developing effective approaches to overcome chemoresistance. In the
present study, a mechanism was proposed to explain how KPT-330
overcomes oxaliplatin resistance. First, it was observed that
oxaliplatin treatment alone increased the expression levels of
XPO1, which in turn, enhanced the nuclear and cytoplasmic levels of
p53. This mislocalization of p53 prevented it from performing its
tumor-suppressing function in the nucleus, thereby reducing the
sensitivity of the CRC cells to oxaliplatin. KPT-330 combined with
oxaliplatin restored oxaliplatin sensitivity by promoting the
nuclear localization of p53. As shown in our model, KPT-330 blocked
the XPO1-mediated nuclear export of p53 by inhibiting XPO1, leading
to its accumulation in the nucleus. The nuclear p53 then acts as a
transcription factor, leading to the downregulation of SLC7A11 and
the upregulation of p21. This, along with a decrease in MMP,
collectively promotes both apoptosis and ferroptosis in the CRC
cells, effectively overcoming oxaliplatin resistance (Fig. S5).
Several tumor suppressor proteins (TSPs), such as
p53 and p21, are crucial for maintaining cellular integrity by
regulating cell proliferation, apoptosis and DNA damage repair
(32). Consequently,
mislocalization of TSPs to the cytoplasm can contribute to tumor
progression (33). One approach
to prevent such mislocalization is to inhibit the nuclear export of
these proteins (34). XPO1
serves as the sole nuclear export receptor for multiple TSPs
(35). Upregulation of XPO1
expression has been linked to poor prognosis and chemoresistance in
various cancer types, including breast cancer, CRC and leukemia
(34-36). The XPO1 inhibitor KPT-330 induces
nuclear accumulation of TSPs and restores their tumor-suppressive
activity (14). In the present
study, oxaliplatin not only increased XPO1 expression but also
upregulated p53 expression in both the cytoplasm and nucleus in
oxaliplatin-resistant cells. This suggests that targeting XPO1
could be a promising therapeutic strategy for cancer treatment.
In the cytoplasm, p53 is rapidly degraded via the
ubiquitin-proteasome pathway, making it barely detectable (37). Maintaining p53 activity is a
hallmark of sensitivity to oxaliplatin (38). p53 remains in the cytoplasm of
platinum-resistant cells in lung and ovarian cancer (39,40). The mislocalization of p53 results
in its inactivation, which is a critical factor in chemoresistance
(41,42). In the present study, the
combination of KPT-330 and oxaliplatin treatment increased p53
nuclear localization, downregulated SLC7A11 and GPX4 expression,
and promoted ferroptosis compared with either oxaliplatin or
KPT-330 monotherapy. Conversely, inhibition of p53 accumulation by
transfection with si-p53 partially restored SLC7A11 and GPX4
expression. These results suggested that the detection of p53
nucleocytoplasmic localization could serve as a potential biomarker
for predicting CRC response to oxaliplatin.
While the present study demonstrated the
synergistic efficacy of KPT-330 and oxaliplatin in overcoming
oxaliplatin resistance in CRC, several limitations remain. Direct
validation of these findings in clinical samples from patients with
CRC is essential. Additionally, the present study did not predict
responsiveness based on p53 nucleocytoplasmic localization in
relevant clinical samples. Future research will focus on analyzing
XPO1 expression and p53 localization in patient tissues,
particularly those from patients with CRC exhibiting oxaliplatin
resistance, to confirm these results and further explore the
translational potential of this combination therapy.
While the present in vivo experiments
indicated no significant changes in body weight, suggesting a lack
of acute severe toxicity during the treatment period, a thorough
evaluation of the long-term safety and potential systemic side
effects of combination therapy is crucial. Future research will
focus on rigorous toxicology studies and comprehensive assessments
of organ-specific toxicities, which are essential for its eventual
clinical translation.
Furthermore, while the present study
comprehensively outlined the role of the p53-XPO1 axis in the
ability of KPT-330 to restore oxaliplatin sensitivity in CRC, the
full range of the mechanistic actions of KPT-330 may extend beyond
p53. Other factors contributing to increased cytoplasmic p53
levels, such as the ubiquitin-proteasome pathway and nuclear
transport mechanisms, were not fully explored. For instance,
impaired p53 nuclear import could contribute to its cytoplasmic
retention (42). As a selective
XPO1 inhibitor, KPT-330 may also affect the nuclear-cytoplasmic
transport of other critical proteins, including additional tumor
suppressors or factors (such as ERK5, DEAD-box helicase 17 and
FOXO1) involved in drug resistance (33,43). Future investigations should
explore these potential targets and alternative signaling pathways
to gain a more comprehensive understanding of the multifaceted role
of KPT-330 in overcoming oxaliplatin resistance.
In conclusion, KPT-330 promoted p53 nuclear
accumulation and enhanced the sensitivity of oxaliplatin-resistant
CRC to oxaliplatin. These findings suggest a promising strategy for
overcoming oxaliplatin resistance in CRC and highlight the
potential of KPT-330 as a novel sensitizing agent for this
chemotherapy.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CL, XJ, YC, KC, FW, QZ, XX, ZSC, LX and SD
contributed to the conception and design of the study. CL, XJ, YC,
KC, FW, QZ and XX contributed to data acquisition. CL, LX and SD
contributed to data analysis and interpretation. CL and LX
contributed to the writing of the original draft. SD and ZSC
contributed to mentoring, writing, review and editing. CL, XJ and
LX confirm the authenticity of all the raw data. All authors have
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Animal welfare and experimental procedures were
carried out in accordance with the Guide for the Care and Use of
Laboratory Animals and were approved by the Ethical Review
Committee and Laboratory Animal Welfare Committee of the Sir Run
Run Shaw Hospital, Zhejiang University School of Medicine (approval
no. SRRSH2025-0023, Hangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 82072628 and 32200593), the Key
Research and Development Program of Zhejiang (grant no.
2022C03032), and the Natural Science Foundation of Zhejiang
Province, China (grant no. LQ23H160040).
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bullock AJ, Schlechter BL, Fakih MG,
Tsimberidou AM, Grossman JE, Gordon MS, Wilky BA, Pimentel A,
Mahadevan D, Balmanoukian AS, et al: Botensilimab plus balstilimab
in relapsed/refractory microsatellite stable metastatic colorectal
cancer: A phase 1 trial. Nat Med. 30:2558–2567. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tripathi PK, Mittal KR, Jain N, Sharma N
and Jain CK: KRAS pathways: A potential gateway for cancer
therapeutics and diagnostics. Recent Pat Anticancer Drug Discov.
19:268–279. 2024. View Article : Google Scholar
|
|
5
|
Capdevila J, Elez E, Peralta S, Macarulla
T, Ramos FJ and Tabernero J: Oxaliplatin-based chemotherapy in the
management of colorectal cancer. Expert Rev Anticancer Ther.
8:1223–1236. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sargent D, Sobrero A, Grothey A, O'Connell
MJ, Buyse M, Andre T, Zheng Y, Green E, Labianca R, O'Callaghan C,
et al: Evidence for cure by adjuvant therapy in colon cancer:
Observations based on individual patient data from 20,898 patients
on 18 randomized trials. J Clin Oncol. 27:872–877. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng K, Li W, Wang Y, Zhang Z, Zhang L,
Zhang W, Xing Y and Zhou C: Inhibition of CDK1 overcomes
oxaliplatin resistance by regulating ACSL4-mediated ferroptosis in
colorectal cancer. Adv Sci (Weinh). 10:e23010882023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Gan Y, Liu J, Li J, Zhou Z, Tian R,
Sun R, Liu J, Xiao Q, Li Y, et al: Downregulation of MEIS1 mediated
by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and
oxaliplatin resistance in colorectal cancer. Signal Transduct
Target Ther. 7:872022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abdul Razak AR, Mau-Soerensen M, Gabrail
NY, Gerecitano JF, Shields AF, Unger TJ, Saint-Martin JR, Carlson
R, Landesman Y, McCauley D, et al: First-in-class, first-in-human
phase I study of selinexor, a selective inhibitor of nuclear
export, in patients with advanced solid tumors. J Clin Oncol.
34:4142–4150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ben-Barouch S and Kuruvilla J: Selinexor
(KTP-330)-a selective inhibitor of nuclear export (SINE):
Anti-tumor activity in diffuse large B-cell lymphoma (DLBCL).
Expert Opin Investig Drugs. 29:15–21. 2020. View Article : Google Scholar
|
|
11
|
Ferreiro-Neira I, Torres NE, Liesenfeld
LF, Chan CH, Penson T, Landesman Y, Senapedis W, Shacham S, Hong TS
and Cusack JC: XPO1 inhibition enhances radiation response in
preclinical models of rectal cancer. Clin Cancer Res. 22:1663–1673.
2016. View Article : Google Scholar
|
|
12
|
Inoue A, Robinson FS, Minelli R, Tomihara
H, Rizi BS, Rose JL, Kodama T, Srinivasan S, Harris AL, Zuniga AM,
et al: Sequential administration of XPO1 and ATR inhibitors
enhances therapeutic response in TP53-mutated colorectal cancer.
Gastroenterology. 161:196–210. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Quintanal-Villalonga A, Taniguchi H, Hao
Y, Chow A, Zhan YA, Chavan SS, Uddin F, Allaj V, Manoj P, Shah NS,
et al: Inhibition of XPO1 sensitizes small cell lung cancer to
first- and second-line chemotherapy. Cancer Res. 82:472–483. 2022.
View Article : Google Scholar :
|
|
14
|
Chen Y, Camacho SC, Silvers TR, Razak AR,
Gabrail NY, Gerecitano JF, Kalir E, Pereira E, Evans BR, Ramus SJ,
et al: Inhibition of the nuclear export receptor XPO1 as a
therapeutic target for platinum-resistant ovarian cancer. Clinical
Cancer Research. 23:1552–1563. 2017. View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Zhou J, Lei Z, Chen J, Liao S, Chen Y, Liu
C, Huang S, Li L, Zhang Y, Wang P, et al: Nuclear export of BATF2
enhances colorectal cancer proliferation through binding to CRM1.
Clin Transl Med. 13:e12602023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Azmi AS, Kauffman M, McCauley D, Shacham S
and Mohammad RM: Novel small-molecule CRM-1 inhibitor for GI cancer
therapy. J Clin Oncol. 30(suppl 4): abstr 245. 2012. View Article : Google Scholar
|
|
18
|
Chiu SJ, Lee YJ, Hsu TS and Chen WS:
Oxaliplatin-induced gamma-H2AX activation via both p53-dependent
and -independent pathways but is not associated with cell cycle
arrest in human colorectal cancer cells. Chem Biol Interact.
182:173–182. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schmidt AK, Pudelko K, Boekenkamp JE,
Berger K, Kschischo M and Bastians H: The p53/p73-p21(CIP1) tumor
suppressor axis guards against chromosomal instability by
restraining CDK1 in human cancer cells. Oncogene. 40:436–451. 2021.
View Article : Google Scholar
|
|
20
|
Martinez-Balibrea E, Martinez-Cardus A,
Gines A, Ruiz de Porras V, Moutinho C, Layos L, Manzano JL, Bugés
C, Bystrup S, Esteller M and Abad A: Tumor-related molecular
mechanisms of oxaliplatin resistance. Mol Cancer Ther.
14:1767–1776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yapasert R, Khaw-On P and Banjerdpongchai
R: Coronavirus Infection-associated cell death signaling and
potential therapeutic targets. Molecules. 26:74592021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arango D, Wilson AJ, Shi Q, Corner GA,
Arañes MJ, Nicholas C, Lesser M, Mariadason JM and Augenlicht LH:
Molecular mechanisms of action and prediction of response to
oxaliplatin in colorectal cancer cells. Br J Cancer. 91:1931–1946.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu X, Lai Y and Hua ZC: Apoptosis and
apoptotic body: Disease message and therapeutic target potentials.
Biosci Rep. 39:BSR201809922019. View Article : Google Scholar :
|
|
24
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang C, Zhang Y, Lin S, Liu Y and Li W:
Suppressing the KIF20A/NUAK1/Nrf2/GPX4 signaling pathway induces
ferroptosis and enhances the sensitivity of colorectal cancer to
oxaliplatin. Aging (Albany NY). 13:13515–13534. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin JF, Hu PS, Wang YY, Tan YT, Yu K, Liao
K, Wu QN, Li T, Meng Q, Lin JZ, et al: Phosphorylated NFS1 weakens
oxaliplatin-based chemosensitivity of colorectal cancer by
preventing PANoptosis. Signal Transduct Target Ther. 7:542022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Di Y, Zhang X, Wen X, Qin J, Ye L, Wang Y,
Song M, Wang Z and He W: MAPK Signaling-mediated RFNG
phosphorylation and nuclear translocation restrain
oxaliplatin-induced apoptosis and ferroptosis. Adv Sci (Weinh).
11:e24027952024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Z, Zhan Y, Xu J, Wang Y, Sun M, Chen
J, Liang T, Wu L and Xu K: β-Sitosterol reverses multidrug
resistance via BCRP suppression by inhibiting the p53-MDM2
interaction in colorectal cancer. J Agric Food Chem. 68:3850–3858.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Brate A and Giannakakou P: The
importance of p53 location: Nuclear or cytoplasmic zip code? Drug
Resist Updat. 6:313–322. 2003. View Article : Google Scholar
|
|
30
|
Kanemitsu Y, Shimizu Y, Mizusawa J, Inaba
Y, Hamaguchi T, Shida D, Ohue M, Komori K, Shiomi A, Shiozawa M, et
al: Hepatectomy followed by mFOLFOX6 versus hepatectomy alone for
liver-only metastatic colorectal cancer (JCOG0603): A phase II or
III randomized controlled trial. J Clin Oncol. 39:3789–3799. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo S, Yue M, Wang D, Lu Y, Wu Q and Jiang
J: Breaking the barrier: Epigenetic strategies to combat platinum
resistance in colorectal cancer. Drug Resist Updat. 77:1011522024.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kau TR, Way JC and Silver PA: Nuclear
transport and cancer: From mechanism to intervention. Nat Rev
Cancer. 4:106–117. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lai C, Xu L and Dai S: The nuclear export
protein exportin-1 in solid malignant tumours: From biology to
clinical trials. Clin Transl Med. 14:e16842024. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Turner JG, Dawson JL, Grant S, Shain KH,
Dalton WS, Dai Y, Meads M, Baz R, Kauffman M, Shacham S and
Sullivan DM: Treatment of acquired drug resistance in multiple
myeloma by combination therapy with XPO1 and topoisomerase II
inhibitors. J Hematol Oncol. 9:732016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Saenz-Ponce N, Pillay R, de Long LM,
Kashyap T, Argueta C, Landesman Y, Hazar-Rethinam M, Boros S,
Panizza B, Jacquemyn M, et al: Targeting the XPO1-dependent nuclear
export of E2F7 reverses anthracycline resistance in head and neck
squamous cell carcinomas. Sci Transl Med. 10:eaar72232018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim J, McMillan E, Kim HS, Venkateswaran
N, Makkar G, Rodriguez-Canales J, Villalobos P, Neggers JE,
Mendiratta S, Wei S, et al: XPO1-dependent nuclear export is a
druggable vulnerability in-mutant lung cancer. Nature. 538:114–117.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oren M and Rotter V: Mutant p53
gain-of-function in cancer. Cold Spring Harb Perspect Biol.
2:a0011072010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Toscano F, Parmentier B, Fajoui ZE,
Estornes Y, Chayvialle JA, Saurin JC and Abello J: p53 dependent
and independent sensitivity to oxaliplatin of colon cancer cells.
Biochem Pharmacol. 74:392–406. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nishitsuji K, Mito R, Ikezaki M, Yano H,
Fujiwara Y, Matsubara E, Nishikawa T, Ihara Y, Uchimura K, Iwahashi
N, et al: Impacts of cytoplasmic p53 aggregates on the prognosis
and the transcriptome in lung squamous cell carcinoma. Cancer Sci.
115:2947–2960. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Komlodi-Pasztor E, Trostel S, Sackett D,
Poruchynsky M and Fojo T: Impaired p53 binding to importin: A novel
mechanism of cytoplasmic sequestration identified in
oxaliplatin-resistant cells. Oncogene. 28:3111–3120. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Prokocimer M and Peller S: Cytoplasmic
sequestration of wild-type p53 in a patient with therapy-related
resistant AML: First report. Med Oncol. 29:1148–1150. 2012.
View Article : Google Scholar
|
|
42
|
Fang J, Zou M, Yang M, Cui Y, Pu R and
Yang Y: TAF15 inhibits p53 nucleus translocation and promotes HCC
cell 5-FU resistance via post-transcriptional regulation of UBE2N.
J Physiol Biochem. 80:919–933. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Z, Pan B, Yao Y, Qiu J, Zhang X, Wu X
and Tang N: XPO1 intensifies sorafenib resistance by stabilizing
acetylation of NPM1 and enhancing epithelial-mesenchymal transition
in hepatocellular carcinoma. Biomed Pharmacother. 160:1144022023.
View Article : Google Scholar : PubMed/NCBI
|